- 1College of Life Sciences, Qufu Normal University, Qufu, China
- 2State Key Laboratory of Crop Biology, College of Agronomic Sciences, Shandong Agricultural University, Taian, China
Plant architecture is a culmination of the features necessary for capturing light energy and adapting to the environment. An ideal architecture can promote an increase in planting density, light penetration to the lower canopy, airflow as well as heat distribution to achieve an increase in crop yield. A number of plant architecture-related genes have been identified by map cloning, quantitative trait locus (QTL) and genome-wide association study (GWAS) analysis. LIGULELESS1 (LG1) belongs to the squamosa promoter-binding protein (SBP) family of transcription factors (TFs) that are key regulators for plant growth and development, especially leaf angle (LA) and flower development. The DRL1/2-LG1-RAVL pathway is involved in brassinosteroid (BR) signaling to regulate the LA in maize, which has facilitated the regulation of plant architecture. Therefore, exploring the gene regulatory functions of LG1, especially its relationship with LA genes, can help achieve the precise regulation of plant phenotypes adapted to varied environments, thereby increasing the yield. This review comprehensively summarizes the advances in LG1 research, including its effect on LA and flower development. Finally, we discuss the current challenges and future research goals associate with LG1.
1 Introduction
Plant growth and development are inextricably linked to the absorption of light energy (Ort et al., 2015). A reasonable leaf angle (LA) throughout different parts of the plant is crucial for making optimum use of solar energy and increasing biomass (Mantilla-Perez and Salas Fernandez, 2017). Specifically, plant varieties with narrow angles of the upper leaves promote an increased in planting densities and yields that is beneficial to meet the rising food demand (Cao et al., 2022b). For example, maize yields have increased more than 7-fold since the mid-twentieth century, primarily because of increased tolerance to high planting density and effectiveness of hybrids from increased heterosis (Duvick, 2005; Mansfield and Mumm, 2014; Wang et al., 2020).
Current advances in biotechnology have facilitated the study of the developmental and regulatory mechanisms of LA in plants. During the past three decades, LG1 has been identified as the most critical transcription factor (TF) regulating LA and flower development (Moreno et al., 1997; Lewis et al., 2014). LG1 is a member of the SBP family, which are special transcription factors in plants that contribute to growth and development as well as various physiological and biochemical processes (Klein et al., 1996; Cardon et al., 1999; Birkenbihl et al., 2005; Xie et al., 2006; Guo et al., 2008; Chen et al., 2010; Preston and Hileman, 2013; Zhang et al., 2016; Peng et al., 2019). SBP family proteins comprise 80 conserved amino acids, two conserved zinc finger domains (Zn1 and Zn2) and a C-terminal nuclear localization signal. The zinc finger domains specifically bind the GTAC core sequence in the downstream gene promoter, and SBP function is also conservatively regulated by miR156 throughout the growth period (Cardon et al., 1999; Birkenbihl et al., 2005; Guo et al., 2008; Chen et al., 2010; Peng et al., 2016). In Arabidopsis, overexpression of AtSPL8, a ZmLG1 homologous gene, causes non-dehiscence of anthers through the gibberellin (GA) pathway and affects fertility (Zhang et al., 2007; Xing et al., 2010; Preston and Hileman, 2013) (Figure 1). In maize, ZmLG1 affects the development of the ligular region. The homozygous Zmlg1 mutant shows a phenotype with no ligule or auricles, thereby reducing the LA. It was later observed that ZmLG1 is regulated by, for example, ZmWAB1and ZmDRL1/2, which affected the development of flowers and LA (Becraft et al., 1990; Sylvester et al., 1990; Becraft and Freeling, 1991; Moreno et al., 1997; Hay and Hake, 2004; Lewis et al., 2014; Tian et al., 2019). Recent studies have shown that ZmLG1 is associated with resistance to northern leaf blight, which has improved the understanding of ZmLG1 function (Kolkman et al., 2020). ZmLG1 is involved in leaf morphological development, flower development and disease resistance, and its transcriptional and post-transcriptional regulation has received considerable attention. Hormone activity is critical for the development of the ligular region. Studies have found that Zmlg1 mutants exhibit aberrant ZmPIN1a expression, and cannot form a continuous distribution in the preligule band (PLB) zone. However, there is no direct evidence that ZmLG1 is involved in auxin signaling in maize (Johnston et al., 2014). In addition to auxin, ZmLG1 directly regulates ZmRVAL1 to participate in the signaling involving brassinosteroids (BRs), which are the most important hormones shown to regulate LA (Tian et al., 2019). Based on the important role of LG1 and the extensive research in recent years, we summarize the mechanisms of transcriptional and molecular regulation of LG1. We also valuate the challenges and the future prospects of LG1 research.
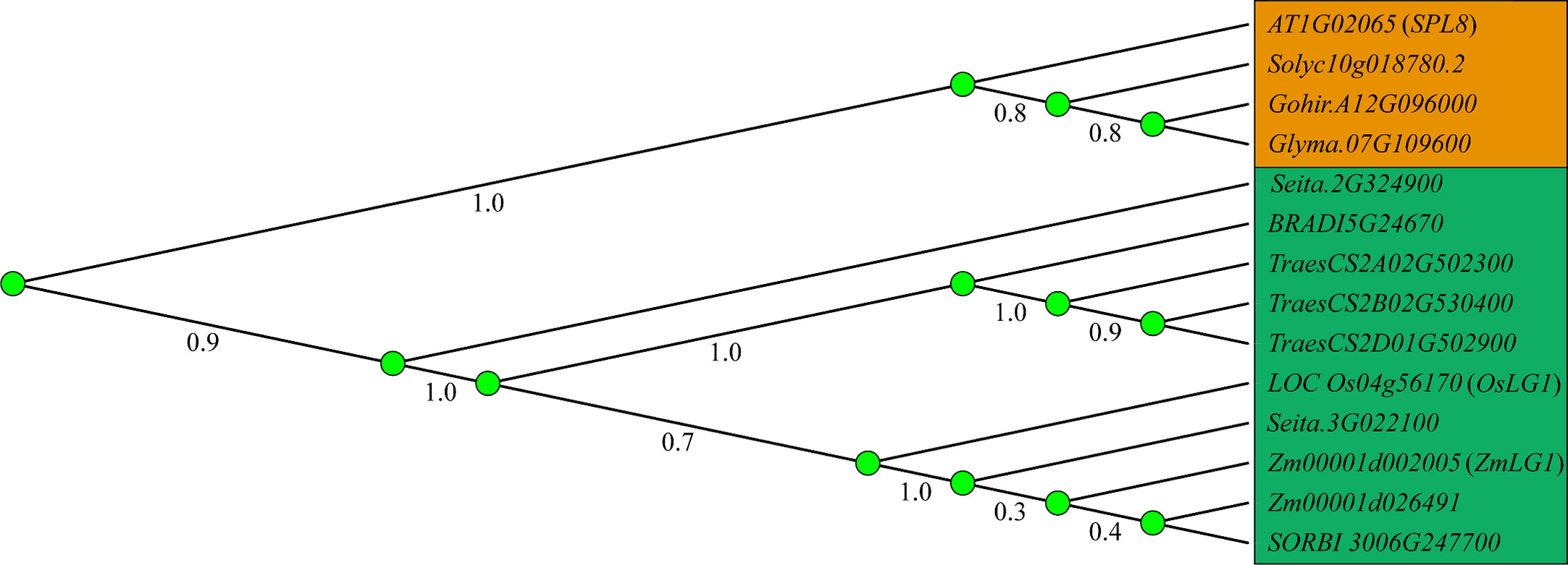
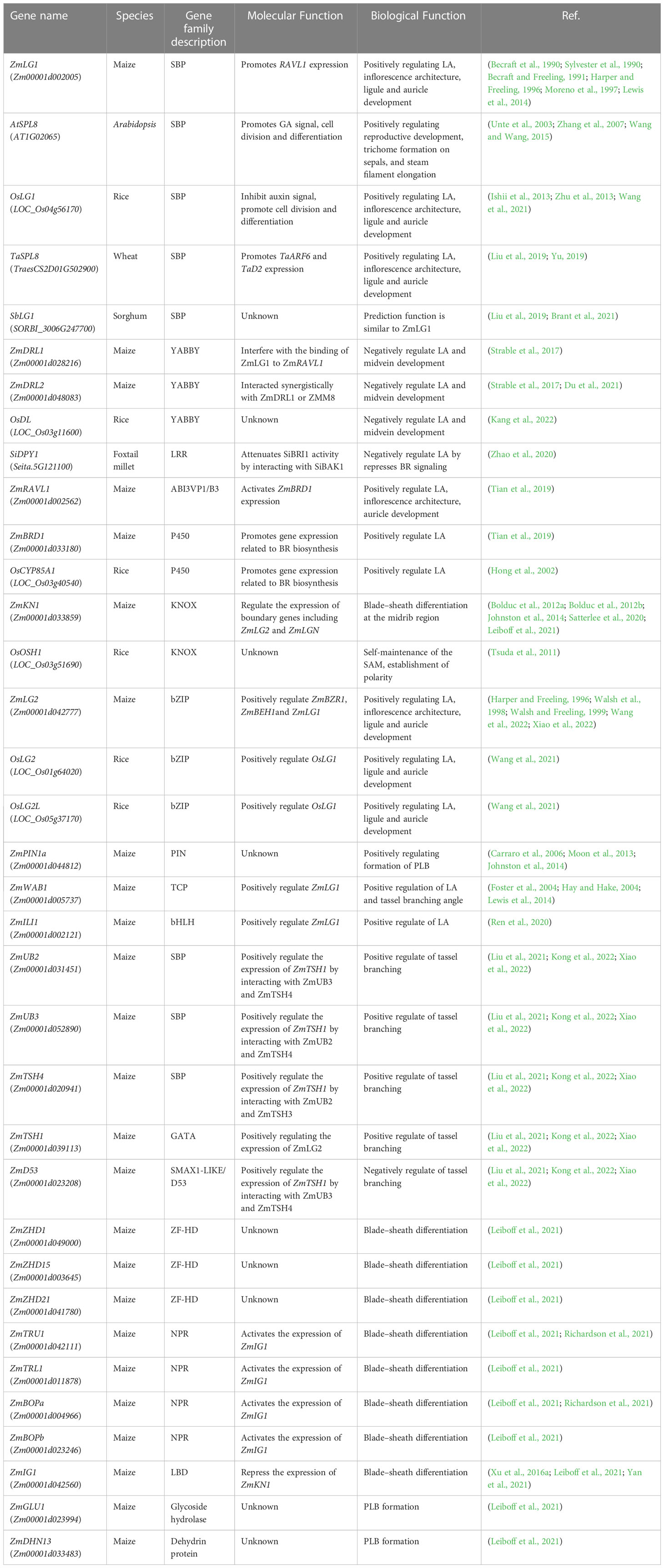
Figure 1 Phylogenetic relationship of LG1 orthologs. Phylogenetic relationship of LG1 in dicotyledon (orange) such as Arabidopsis, tomato, cotton, soybean, and monocotyledon (green) such as rice, maize, wheat, sorghum, foxtail millet, Brachypodium. The phylogenetic tree was created using MEGAX with LG1 orthologs sequences by neighbor-joining method, and 1000 bootstrap replicates.
2 Tissue-specific expression of LG1 leads to various biological functions
The ZmLG1 gene sequence was obtained in 1997 in maize; this gene was found to be expressed at a very low level in the ligular region of leaf primordia, and it may be expressed earlier than the plastochron6 (P6) phase (Becraft et al., 1990; Sylvester et al., 1990; Becraft and Freeling, 1991; Moreno et al., 1997). ZmLG1 mRNA are expressed autonomously on the adaxial and abaxial PLB at the P6 stage and play a role in signal reception (Figure 2). LIGULELESS2 (LG2) is another important gene of the bZIP family and affects the development of the ligular region. Its expression period occurs earlier than does that of ZmLG1, and it transmits signals in a noncell-independent manner. Although many studies have shown that LG1 and LG2 play a role in the same signaling pathway, there is no actual direct evidence to suggest this relationship (Harper and Freeling, 1996; Walsh et al., 1998; Johnston et al., 2014; Strable and Nelissen, 2021). Later, ZmLG1 specifically expressed PLB in the P6-P10 stage, and the expression level was found to be higher in P9-P10 than in P6-P7 but not in P1-P5 or in the blades, and OsLG1 promoter-driven ß-glucuronidase (GUS) expression also showed similar results in rice (Foster et al., 2004; Wang et al., 2021). ZmLG1 affects the positioning and function of Wavy auricle in blade (ZmWab1) through an unknown pathway, and sheath-like cells appear in the blades of the dominant mutant Zmwab1, which is more obvious in lg1/wab1 double mutants (Foster et al., 2004).
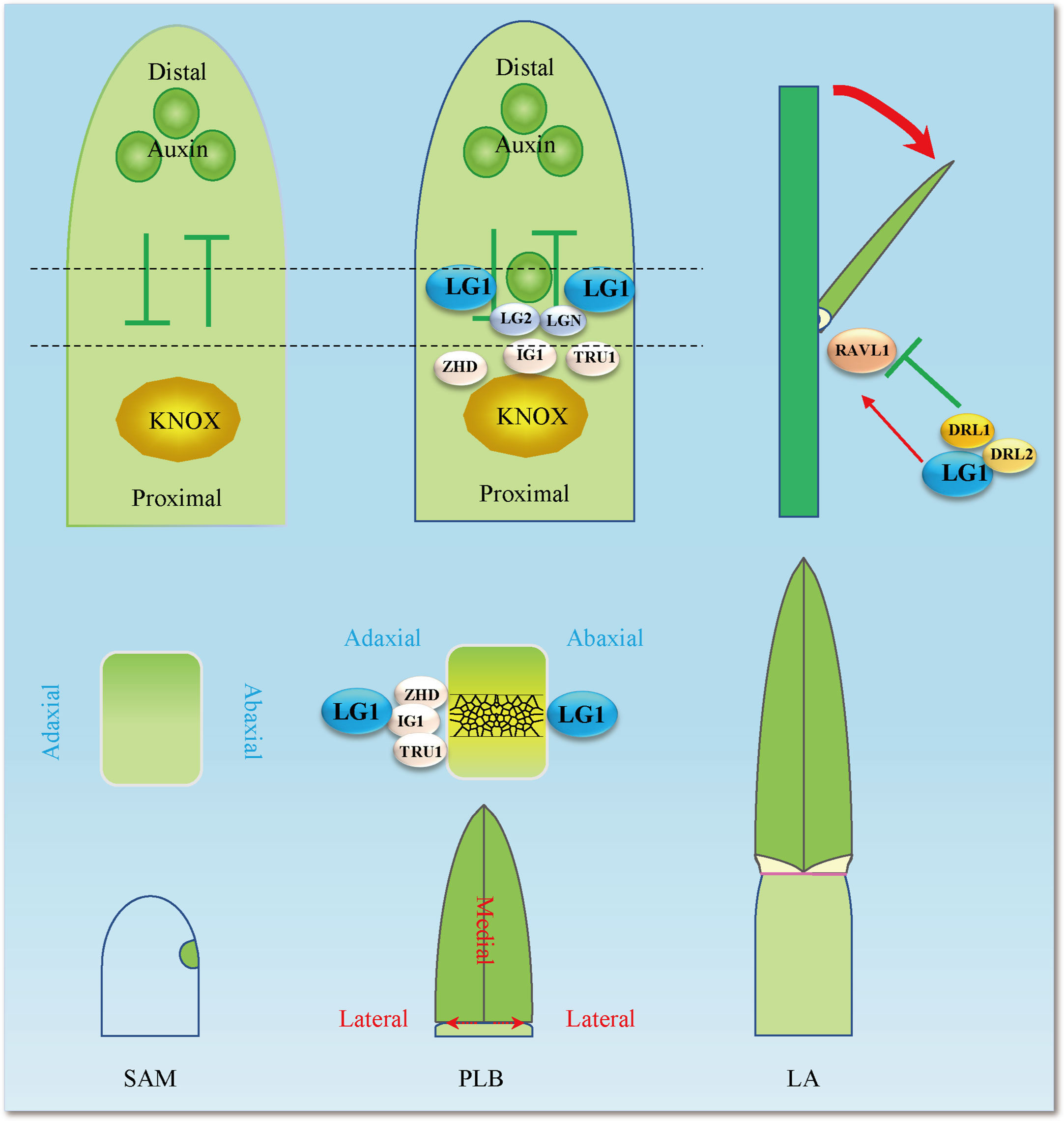
Figure 2 Establishment of LA mediated by auxin and LA-related genes. Auxin and KNOX establish polarity at the distal and proximal ends, respectively, and boundary genes including ZHD, IG1, TRU1, LG2, LGN, LG1, DRL1, DRL2 and RAVL1 participate in the formation of PLB (preligule band) and the establishment of LA (leaf angle). Black dotted lines indicate the position at the junction of blade and sheath. Red arrows indicate positive regulation, while green lines ended with perpendicular bars represent suppressive regulation.
ZmLG1 was detected using a specific antibody and found to be expressed not only in PLB, but also in the axils of developing tassel branches (Lewis et al., 2014). The ZmWab1 gene is specifically expressed in flowers, which is necessary for ZmLG1 function in flower development (Figure 2). The specific tassel branch angle decreased in the Zmwab1 recessive mutant, while the LA remained unchanged. The dominant ZmWAB1-R/+ mutant has a greater tassel branch angle due to the increased ZmWAB1 expression level (Lewis et al., 2014). At the same time, ZmWAB1-R/+ caused abnormal accumulation of ZmLG1, resulting in increased LA. ZmWAB1 is a member of the TCP family, and the TCP-miR319 module promotes the transition from cell division to cell differentiation in Arabidopsis. When TCP expression is high, the TCP-miR319 module inhibits the expansion of the margins, while the miR396-GRF/GIF module of the leaf promotes the expansion of proximal cells (Du et al., 2018; Qin et al., 2022a). The abnormal expression of ZmWAB1 disrupts the developmental gradient of leaves and changes the expression of ZmLG1, thus affecting the growth of leaves (Lewis et al., 2014; Du et al., 2018). Boundary genes such as ZmLG1 often regulate the development of flowers while regulating the LA. ZmLG2 has been reported to be involved in tassel branching and the transition from vegetative growth to reproductive growth, and Unbranched3 (UB3), Tasselsheath4 (TSH4) and LG2 jointly contribute to flower development in a dose-dependent manner (Walsh and Freeling, 1999; Xiao et al., 2022) (Figure 3). OsLG1 is highly expressed in the panicles, sheaths, ligules, leaves and stems of rice, and is related to the control of cell flower development (Zhu et al., 2013) (Figure 1). Like that in maize and rice, the expression level of SevLG1 is also higher in the inflorescences, and this gene is involved in the formation of the lateral organ position boundary and branch meristems in Setaria viridis (Zhu et al., 2018) (Figure 1). Taken together, these results indicate that LG1 plays a role in the early development of the ligular region and inflorescence in different species.
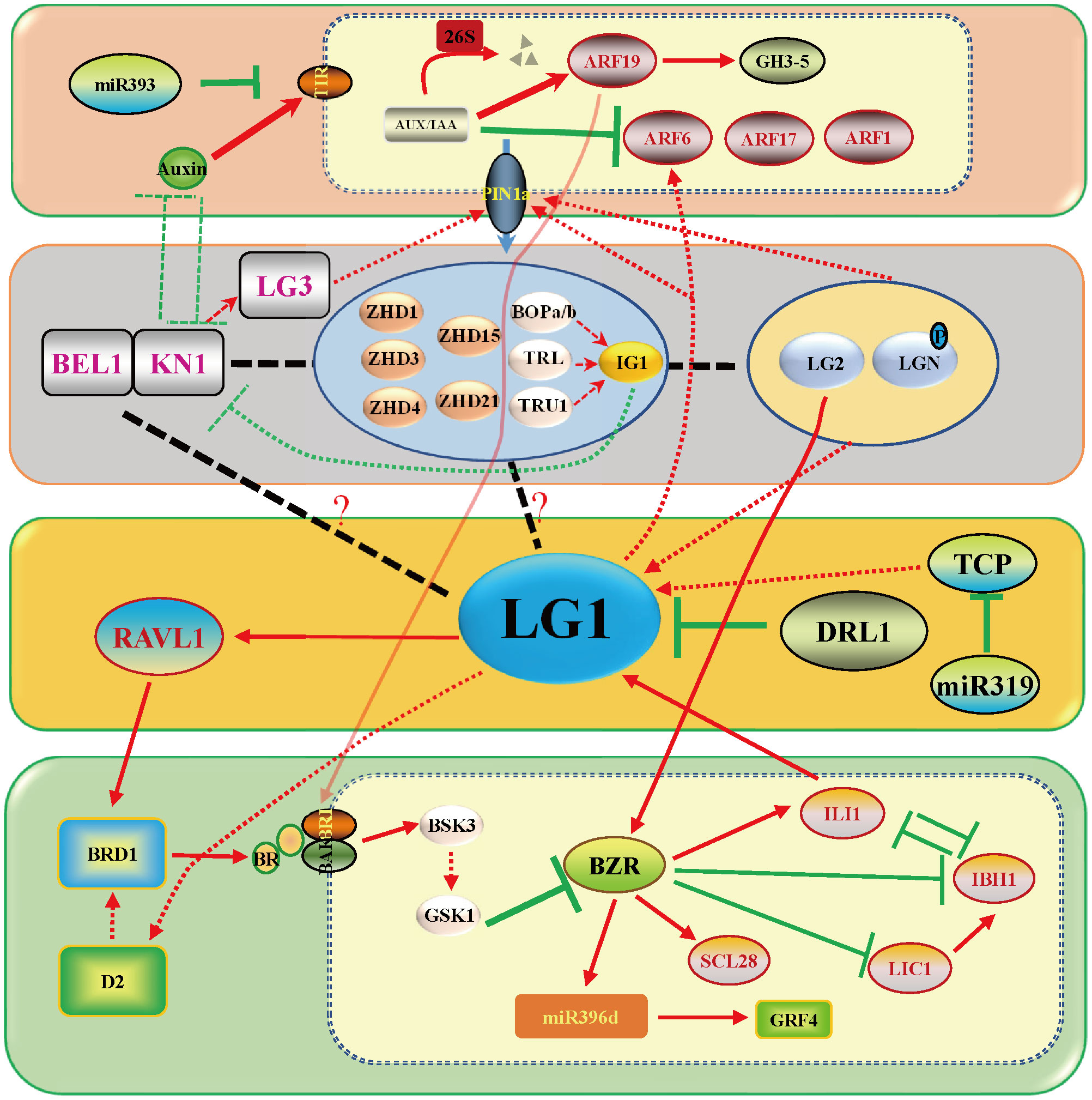
Figure 3 LG1 participates in the complex LA control network. LG1 regulates RAVL1 and then participates in the pathway of BR regulating LA by BRD1, which includes positive regulation of the LA gene RAVL1, BRD1, BRI1, BSK3, BZR1, SCL28, ILI1 and miR396, and negative regulation of the LA gene DRL1, IBH1, LIC1 and GRF4. LG1 is involved in the regulation of LA by auxin and boundary genes including PIN1a and LG2, although the involvement is not entirely clear. Red arrows indicate positive regulation, while green lines ended with perpendicular bars represent suppressive regulation. The dotted arrows and black lines indicate undetermined relationships.
3 Molecular mechanism of LA regulated by LG1
The LA is the angle formed by the leaves away from the stem to capture sunlight and which is regulated by complex hormones and the external environment (Mantilla-Perez and Salas Fernandez, 2017; Xu et al., 2021). The precise regulation of plant architecture is facilitated by the study of the molecular regulation of LA genes, such as LG1, which is extremely important for crop production (Duvick, 2005; Mantilla-Perez and Salas Fernandez, 2017; Wang et al., 2020) (Table 1). Phenotypic observations and genetic evidence indicate a role of LG1 in LA development. In particular, LG1 is an excellent gene with natural variation affecting LA at the population level, and the molecular regulatory effects of LG1 on LA, especially the related hormone pathways, have also been discovered in recent years.
3.1 Role of LG1 in LA at the population level
At the population level, the change in LA is determined by the accumulation of small variations in many genes. Many studies have cloned LA genes via QTL and GWAS (Tang et al., 2021). According to GWAS analysis, liguleless genes, especially ZmLG1, have a greater contribution in upright leaves (Tian et al., 2011). Twenty-five QTLs were identified in a population constructed with Yu82 and Shen137, and QTL qLA2 was found to harbor a key gene, ZmTAC1 (Tilleranglecontrol1), that regulates LA (Ku et al., 2010; Ku et al., 2011; Ku et al., 2012). Since that discovery, researchers have continuously conducted GWAS or QTL analyses and have identified a large number of LA genes in different populations (Lu et al., 2018; Peng et al., 2021; Tang et al., 2021; Zhang and Huang, 2021). There are 308 QTLs distributed across maize chromosomes, of which chromosome 1 contains the most 63 (QTLs), while chromosome 6 contains a minimum of 15 QTLs. These QTLs are divided into 17 important intervals, including 7 reported LA regulatory genes, such as ZmDRL1, ZmDWF and ZmLG1. A large number of QTL hot spots exist in which the target genes have not been identified (Qin et al., 2022b). Two important QTL sites, UPA1 and UPA2, have been identified from a population obtained by crossing maize and its ancestor teosinte. UPA2 is located 9.5 kb upstream of ZmRAVL1, which is a binding site of ZmDRL1/2. ZmLG1 activates the expression of ZmRAVL1, and the interaction between ZmDRL1/2 and ZmLG1 inhibits the activation of ZmRAVL1(Figures 2, 3). This study reveals the regulatory network of maize LA from the perspective of domestication and opens the door to the transformation from molecular breeding to application (Tian et al., 2019). QTL/GWAS analysis has suggested that ZmLG1 plays an important role in regulating LA in the population. How to identify more loci and complex remote regulatory mechanisms requires further research, especially involving QTLs with specific leaf positions, which deserve more attention (Tang et al., 2021).
3.2 Molecular regulatory effects of LG1 on hormone-regulated LA
Much research has been done on the regulation of LA by plant hormones. Auxin represents the basis for promoting the formation of PLB and its content is thought to have a negative role in LA. BRs have a more significant function in LA, and a large number of genes involved in BR synthesis and signaling pathways participate in regulating LA. In recent years, it has been found that LG1 participates in the auxin and BR pathways to regulate LA. Two well-defined regulatory pathways are known, namely, DRL1/2-LG1-RAVL1 and ILI1-LG1, which are involved in BR signaling, and two poorly understood pathways, KN1-LG2-LG1-ARF and TCP-LG1-ARF, are known to be involved in auxin signaling (Xu et al., 2021; Cao et al., 2022b) (Figure 3).
Hormones play an important role in the whole process of plant growth and development. At the early stage, auxin accumulates at this position of leaf primordium initiation in the shoot apical meristem (SAM) (Bolduc et al., 2012a). The growth polarity of proximal–distal patterning is established between KNOX and auxin. The boundary gene represented by ZmLG1 plays a role in the connection. Zmlg1 mutants are accompanied by changes in the expression of ZmPIN1a (Bolduc et al., 2012a; Johnston et al., 2014). Auxin is an important signal for the development of veins and promotes the development of tertiary veins in leaves (Robil and McSteen, 2022). It is still unclear whether auxin signaling to ZmLG1 participates in the PLB formation process from the midvein to the leaf margin. At present, auxin is located distal to the leaf primordium, ZmKN1 is located proximal to the leaf primordium, and boundary genes such as ZmLG1, ZmLG2, ZmBell and ZmBOP are located at the junction promote the formation of the ligular region (Carraro et al., 2006; Osmont et al., 2006; Johnston et al., 2014; Richardson and Hake, 2018; Toriba et al., 2019). Combined with the transcriptome data including that of Zmlg1 at the PLB early stage, a gene coexpression network was constructed. The network module of ZmLG1 includes 270 genes (Johnston et al., 2014; Leiboff et al., 2021). According to this coexpression network, a model of the early transcriptomic proximodistal prepattern was proposed. At the bottom of leaf primordia, ZmKN1 regulates downstream genes including ZmZHDs, ZmBOPs, ZmBOPs, activating the expression of the ASYMMETRIC LEAVES2 (AS2)-related gene (INDETERMINANT GAMETOPHYTE1, IG1). ZmLG1, ZmLG2, ZmLGN and ZmRAVL are coexpressed in PLB, and previous genetic evidence also shows that they are in the same signaling pathway. The establishment of this development gradient provided a basis for the subsequent formation of LA (Leiboff et al., 2021). According to the present data, there is a ZmKN1-ZmLG2-ZmLG1 pathway closely related to auxin, which is affected more by the gene regulation that regulates LA (Figure 3). The interaction of OsGRF7 and OsGIFs promotes an increase in auxin content and inhibits the LA size in rice (Chen et al., 2020), and miR393 inhibits the expression of OsTIR1 and OsAFB2 to promote LA size (Bian et al., 2012).
TaSPL8, a homologous gene of ZmLG1 (Figure 1), directly activates the expression of TaARF6, which is a core auxin signaling pathway gene that regulates LA. The TaSPL8-TaARF6 pathway provides the possibility for ZmLG1 to regulate LA through the auxin signaling pathway in a similar way (Liu et al., 2019) (Figure 3). The maize seedling indoleacetic acid (IAA) concentration gradient showed a significant difference between the wild type and Zmlpa1 mutant after treatment with exogenous 100 µmol•L–1 IAA (Ji et al., 2022). Nonetheless, the response of the Zmlg1 mutant to exogenous auxin treatment has not been reported and needs to be explored. As a member of the SBP family, the LG1 homolog has also been reported to be related to auxin in rice. OsSPL14 of the SBP family binds to the promoter of OsPIN1b to regulate the distribution of auxin and inhibit the growth of tillers (Li et al., 2022). Functioning upstream of ZmLG1, two ZmLG2 homologous genes, OsLG2 and OsLG2L, were identified to redundantly regulate the LA of rice. Moreover, the expression level of auxin signal-related genes was increased in OsLG1-cri, OsLG2-cri and OsLG2L-cri knockout mutant (Wang et al., 2021). Therefore, it was proposed that there was a quantitative effect at play to regulate the LA in the same signal pathway between OsLG1 and OsLG2/2L in rice. OsLG1 and OsLG2/2L negatively regulate the expression of auxin-related genes to promote ligular region division and development (Wang et al., 2021). The auxin-related genes GH3.6/DFL1, axi1-like and PIN5 and the TF-encoding genes ARF6, ARF9 and ARF16 were highly expressed in the PLB, and their expression levels decreased in the Zmlg1 mutant. These are potential ZmLG1 downstream regulatory genes (Johnston et al., 2014). The auxin signal-related genes OsARF19, OsARF6, OsARF11 and OsARF17 were also reported to participate in the development of the LA of rice (Zhang et al., 2015; Liu et al., 2018; Huang et al., 2021). On the other hand, ZmKN1 directly binds and regulates the expression of ZmLG2, and the KN1-LG2-LG1-ARF pathway regulates LA (Bolduc et al., 2012b; Liu et al., 2019; Wang et al., 2021). The regulation of ZmWAB1, a TCP family member, is redundant in LA. Zmwab1 does not exhibit a reduced-LA phenotype, but overexpression of ZmWAB1 can ultimately increase LA by increasing the expression of ZmLG1 (Lewis et al., 2014). Therefore, there is a way for the TCP-LG1-ARF pathway to alter the LA, but other TCP members involved need to continue to be identified.
BRs constitute one of the most critical hormones affecting plant architecture. A large number of genes related to LA are directly or indirectly involved in BR biosynthesis and signaling pathways (Mantilla-Perez and Salas Fernandez, 2017; Gruszka, 2020; Li et al., 2020; Nolan et al., 2020; Xu et al., 2021; Cao et al., 2022b). There are no reports about exogenous BR treatment in the Zmlg1 mutant, but it can be determined that ZmLG1 is closely related to BR signaling. Through experimental identification, it was found that ZmLG1 directly binds to the promoter of ZmRAV1, and ZmRAVL1 binds and activates the expression of the downstream gene ZmBRD1, which is the key BR biosynthesis-related gene (Tian et al., 2019). Although ZmRAV1 functions downstream of ZmLG1, the Zmravl1 mutant does not show defective ligules, but the width of the ligular band and the mature auricle margin decrease, and the number of adaxial sclerenchyma cell layers increases (Tian et al., 2019). ZmRAV1 is one of the many redundant regulatory genes that functions downstream of ZmLG1, which indicates that the regulation of LA not only is controlled by the genes in the early stage but also is important in the later stage. This small change can realize the application of genes in breeding without affecting other traits. In addition, ZmLG1 interacts with ZmDRL1 to inhibit the function of ZmLG1 activation of ZmRAVL1(Tian et al., 2019). ZmDRL1 belongs to the YABBY family and promotes the development of the midvein of the leaf blade. Zmdrl1 mutations lead to an increase in LA as leaves were noticeably droopy because the strength of leaves midribs was reduced(Strable et al., 2017). SiDPY1, an LRR receptor-like kinase in foxtail millet, interacts with the BR signaling receptor SiBAK1 and negatively regulates SiBRI1 (Zhao et al., 2020). Therefore, DRL1-LG1-RAVL is involved in the pathway by which BRs regulate LA. Functioning as a downstream gene of OsBZR1 in BR signaling, Increased Lamina Inclination1 (OsILI1) positively regulates the LA of rice. At the same time, OsILI1 interacts with ILI1 binding bHLH1 (OsIBH1) to jointly regulate LA (Zhang et al., 2009). In maize, ZmILI1 binds specifically to the promoter (-1868 bp) of ZmLG1 and activates its expression (Ren et al., 2020). ZmIBH1-1 has a negative regulatory effect on LA by mapping. In addition, through CRISPR−Cas9-mediated genome editing in Setaria, the effects of its homologous gene Sv.IBH1-1 and Sv.IBH1-2 result in a phenotype of increased LA (Cao et al., 2020). Therefore, we propose a hypothesis that there may be an interaction between ZmILI1 and ZmIBH1 to jointly regulate the expression of ZmLG1. There is additional evidence that ZmLG1 participates in the BR pathway. In wheat, TaSPL8 directly binds to GTAC the core sequence in the TaD2 promoter, and activates TaD2 expression (Liu et al., 2019). Therefore, ZmLG1 possibly participates in the BR pathway in a conservative way like wheat TaSPL-TaD2. A recent study showed that another important LA gene, ZmLG2 also participates in the BR signaling pathway. ZmLG2 directly combined with and activated the expression of the downstream genes ZmBZR1 and ZmBEH1, further activating the expression of the downstream ZmSCL28 to regulate LA (Wang et al., 2022). According to previous studies, ZmLG1 and ZmLG2 participate in the same pathway (Harper and Freeling, 1996; Moreno et al., 1997; Tian et al., 2019), so ZmLG1 participating in the complex process of BR signaling is reasonable (Figure 3).
4 Molecular regulation of LG1 in the flower development
Flowering is the most important feature for plants to adapt to the environment, and is the characteristic of transformation from vegetative growth to reproductive growth (Walsh and Freeling, 1999). Due to the different sunshine durations at different latitudes of the earth, plants require a specific time to flower. Therefore, plants have difficulty adapting to the local environment due to changing latitudes, showing delayed or even an inability to flower (Huang et al., 2018). Increasing evidence shows that members of the SBP/SPL family play an important role in the transformation from vegetative growth to reproductive growth. The expression of SPLs is inhibited by microRNA156s (miR156s) at the vegetative growth stage, the expression of miR156s decreases with age, and the increase in SPL expression is the main process of reproductive transformation (Unte et al., 2003; Preston and Hileman, 2013; Wang and Wang, 2015) (Figure 4). In Arabidopsis, the ZmLG1 homolog AtSPL8 is necessary for fertility during spore development in a gibberellic acid (GA)-dependent manner (Zhang et al., 2007; Xing et al., 2010; Wang and Wang, 2015). In rice, OsLG1, which affects the angle of panicle branches, has also been reported. An SPR3 locus including a 9.3 kb genomic region of the OsLG1 promoter regulates the branch angle of the panicle and seed shattering (Ishii et al., 2013). Subsequently, through the use of multiple inbred lines, it was found that the region controlling panicle architecture was a 3.3 kb region 10 kb upstream of the OsLG1 translation start site, and the variation of one single-nucleotide polymorphisms6 (SNP6) might affect panicle architecture by changing the methylation level (Zhu et al., 2013) (Figure 5). In maize, ZmLG1 affects panicle architecture, but it is still unclear whether there is a regulatory mechanism at play similar to that of OsLG1 upstream of the promoter (Lewis et al., 2014). However, it has been confirmed that ZmLG1 genetically regulates panicle architecture through ZmWab1 (a member of the TCP family). Considering the conservation of the SPL family in plants, we speculate that ZmWAB1 may bind to a region upstream of the ZmLG1 promoter to regulate the panicle architecture of maize (Wang and Wang, 2015; Xu et al., 2016b). ZmLG2, another key gene that regulates LA, plays an important role in the transformation from vegetative growth to reproductive growth, and the number of panicle branches is reduced in the Zmlg2 mutant (Walsh and Freeling, 1999). Together, with AtBOP and AtROXY, the ZmLG2 homologous gene AtTGA9 cooperatively regulates pollen fertility (Hepworth et al., 2005; Li et al., 2009; Murmu et al., 2010). In maize, the SPL family genes UB2, UB3, TSH4 and LG2 form a pathway to inhibit the growth of bracts and increase the number of inflorescence branches (Liu et al., 2021; Xiao et al., 2022) (Figure 4). Through simulated shade experiments, it was found that UB2, UB3 and TSH4 constituted the core of the shade response and regulated the expression of the downstream genes Barren inflorescence2 (BIF2) and Zea mays teosinte branched1/cycloidea/proliferating cell factor30 (ZmTCP30) (Kong et al., 2022). In addition, UB3 is also regulated by GIF1 and affects ear development (Zhang et al., 2018). Whether LG1 is involved in this process is still unclear, but the earliest expression of SevLG2 can be identified in inflorescence development in Setaria viridis, and SevLG1 expression regulates the development of flowers (Zhu et al., 2018).
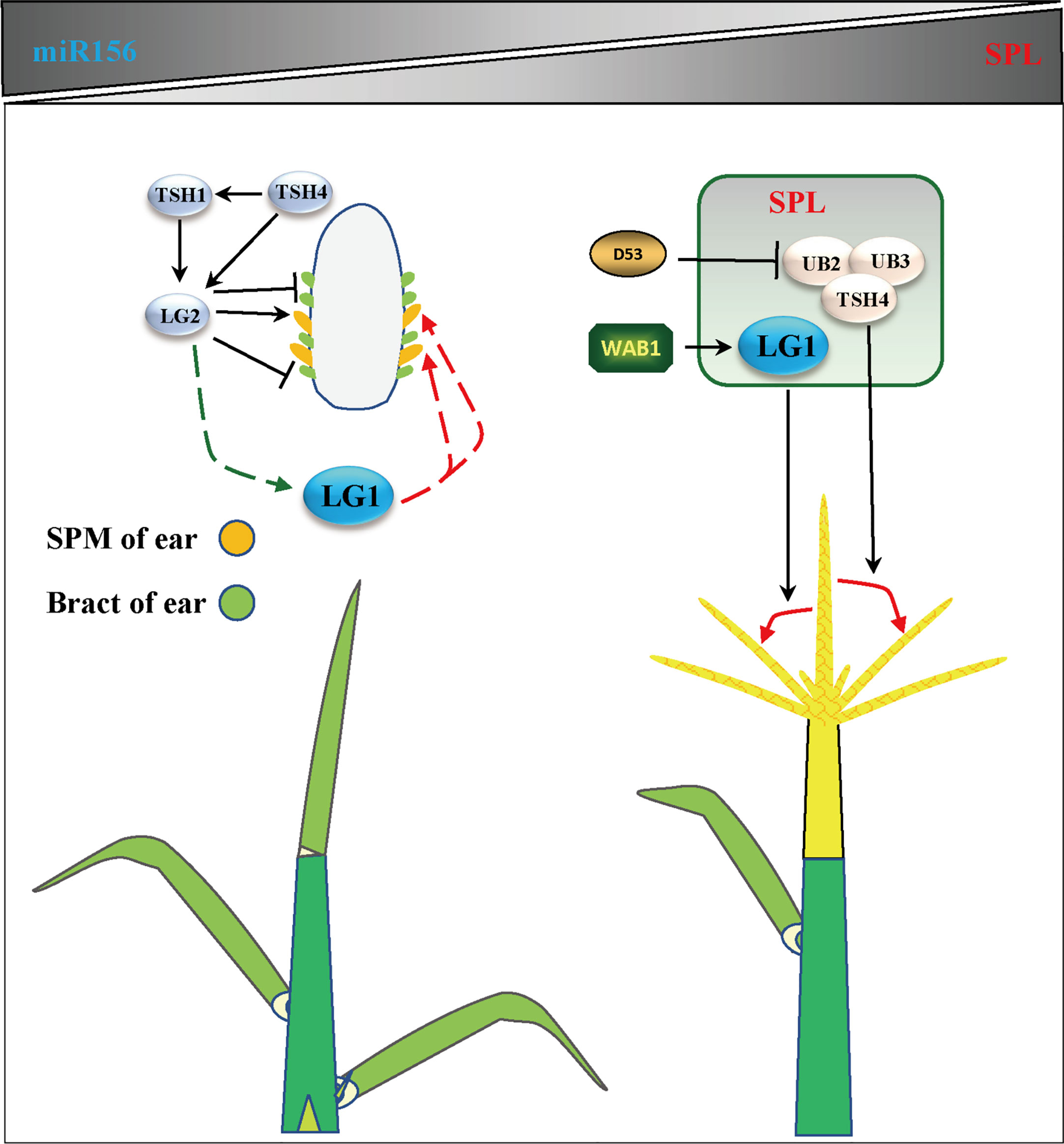
Figure 4 LG1 participates in flower architecture. Genes related to tassel branching or ear development include LG2, TSH1, TSH4, UB3, UB2, WAB1 and D53, which participates in the regulation of flower architecture of LG1 with aging. LG1 may be regulated by LG2 and participate in the development of SPM (spikelet pair meristem) or bract in ear. LG1 regulated by WAB1 and participate in the development of tassel branching. The triangle vertical gradual shadings underneath miR156, SPL genes indicate expression level changes with age, LG1 cooperates with miR156-targeted SPLs to regulate tassel branching. Black arrows indicate positive regulation, while black lines ended with perpendicular bars represent suppressive regulation. Red and green dotted lines arrows indicate undetermined relationships.
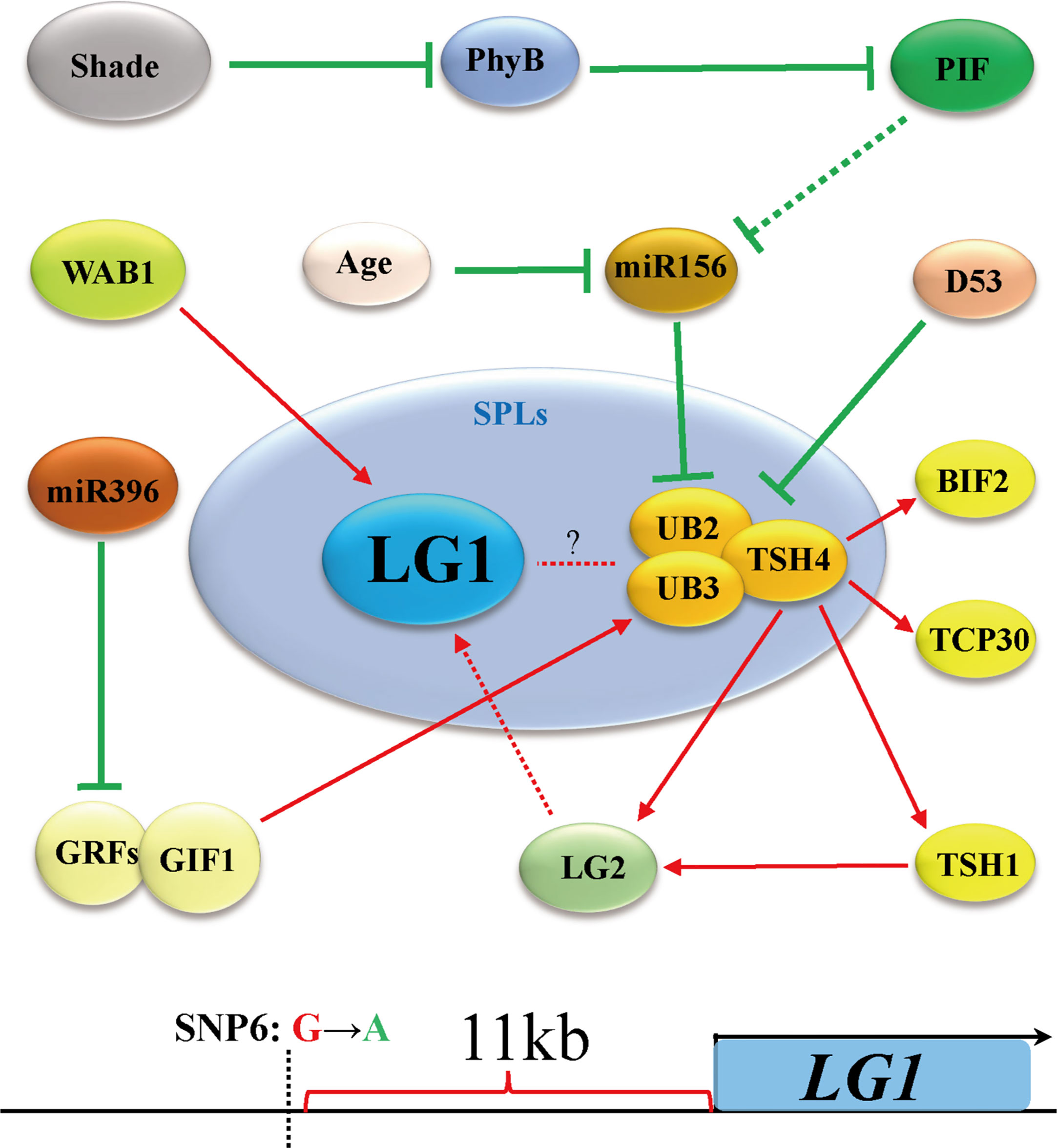
Figure 5 LG1 may participate in the regulation of flower architecture via the shade pathway. Shading negatively regulates miR156 through PHYB and PIF, thereby promoting the expression of UB2/UB3/TSH4. LG1 may participate in this way downstream of LG2. The OsLG1 promoter contains an important element for regulating flowers architecture at a long distance. Red arrows indicate positive regulation, while green lines ended with perpendicular bars represent suppressive regulation. Red and green dotted lines arrows indicate undetermined relationships.
5 Perspectives
Plant architecture is very important for crop production. Erect leaves can make full use of light energy to achieve yield increases by dense planting, increasing light penetration to the lower canopy, improving airflow as well as heat distribution (Duvick, 2005; Mansfield and Mumm, 2014; Ort et al., 2015; Wang et al., 2020). LG1, a major gene that regulates LA, has a conservative function in plants and is highly valuable for molecular breeding (Becraft et al., 1990; Sylvester et al., 1990; Moreno et al., 1997; Foster et al., 2004; Osmont et al., 2006; Lee et al., 2007; Ahn et al., 2008; Zeng et al., 2009; Liu et al., 2019; Yu, 2019). Much evidence shows that LG1 is involved in the pathway of regulating LA by hormones such as auxin and BRs and is the core regulator of LA and inflorescence development (Moon et al., 2013; Johnston et al., 2014; Okagaki et al., 2018; Liu et al., 2019; Tian et al., 2019; Ren et al., 2020; Dou et al., 2021; Guo et al., 2021). Other hormones, including GA, cytokinin (CK), ethylene (ET), directly or indirectly affect BR synergetic regulation of LA (Li et al., 2020; Cao et al., 2022b). Therefore, there is still much room to explore the LG1 regulatory network related to the development of other ligular regions in plants (Cao et al., 2022a; Liu et al., 2022). As a member of the SBP family, LG1 shows essential functions in reproductive transformation, especially in flower development (Wang and Wang, 2015; Xu et al., 2016b). Nonetheless, further exploration of the function of ZmLG1 and genetic experiments are needed.
Based on the development of the LG1 and SBP families in LA and flowers, we propose several problems need of solving. First is the source and signal of the development of PLB. Although transcriptome data are available for the early stage of PLB development and concerning the antagonistic effects of auxin on the distal and proximal KNOX families in leaf primordia, the transmission signal of PLB and the LG1-specific receiving signal pathway is still unclear (Johnston et al., 2014; Leiboff et al., 2021). Recently, according to information obtained from developmental genetics and computational models, there has been a new discussion on the origin of sheaths in plant species, and the petiole-sheath hypothesis has been proposed (Richardson et al., 2021). Similarly, exploring the precise formation process of PLB by the use of the model could help us understand the development mechanism of PLB more in depth. Another feasible method is to accurately analyze the changes in PLB development through single-cell technology, especially single-cell analysis in maize SAM development, which provides one possible direction (Satterlee et al., 2020). Second is, the LG1 regulatory network and its response to the external environment. LG1 is involved in BR signaling through RAVL1 in regulating LA, but there is still no research on whether LG1 responds to BRs and whether it participates in the regulation of the BR biosynthesis pathway (Liu et al., 2019; Tian et al., 2019) With respect to the SPL family, it is proposed that light signaling may be associated with miR156-SPL through the PHYB-PIF pathway, but whether LG1 participates in this process needs further experimental verification. It is also unclear whether DRL1/2 interacting with LG1 participates in PLB signal initiation, although BR signaling can be suppressed through SiBAK1 interaction with SiDPY in Setaria (Strable et al., 2017; Wei et al., 2018; Tian et al., 2019; Zhao et al., 2020). Third, the application of LG1 in breeding. Because of its extremely severe phenotype, ZmLG1 has not been availably applied in molecular design breeding. There are SNP near the 10 kb upstream region of OsLG1 that regulate flower development in rice, so it is reasonable to believe that they are similar in maize, and even precisely control the LA via different loci (Ishii et al., 2013; Zhu et al., 2013). Recently, a practical strategy has emerged. The promoter of OsIPA1 (Ideal Plant Architecture 1), which is a member of the SBP family, was screened in a high-yield variety that could break the linkage between favorable and unfavorable traits to obtain the ideal plant architecture through tiling-deletion-based CRISPR–Cas9 screening technology, which also provides a new idea for ZmLG1 in the future (Song et al., 2022). Therefore, we should make full use of new technologies and means to explore the functions of LG1 and its potential value.
Author contributions
HZ and LQ designed and drafted the work. All authors approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32201762), the Natural Science Foundation of Shandong Province (ZR2022QC003), and the China Postdoctoral Science Foundation (2022M723460).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
LA, leaf angle; PLB, preligule band; BSB, blade-sheath boundary; P6, plastochron 6; TF, transcription factor; LG1, LIGULELESS1; LG2, LIGULELESS2; LGN, LIGULELESS NARROW; QTL, Quantitative Trait Loci; GWAS, Genome-wide association studies; BR, brassinolide; GA, gibberellin acid; CK, cytokinins; ABA, Abscisic Acid; SAM, shoot apical meristem; KNOX, KNOTTED-like Homeobox; KN1, KNOTTED1; ZHD, ZINCFINGER HOMEODOMAIN; TRU1, TASSELS REPLACE UPPER EARS1; TRL1, RU1-LIKE1; BOP, BLADE-ONPETIOLE; IG1, INDETERMINANT GAMETOPHYTE1; AS2, ASYMMETRIC LEAVES2; RAVL1, Related to ABI3/VP1-Like 1; BEL1, BEL1-like Homeodomain; DRL1, Drooping leaf1; BRD1, Brassinosteroid-deficient dwarf1; SCL28, SCARECROW‐LIKE28; ILI1, Increased Lamina Inclination1; IBH1, ILI1 binding bHLH; LIC1, LEAF AND TILLER ANGLE INCREASED CONTROLLER; D2, DWARF1; GIF, GRF-interacting factor; GRF, The growth regulatory factor; SPL, Squamosa Promoter Binding Protein-LIKE; UB2, Unbranched2; UB3, Unbranched3; TSH4, Tasselsheath4; TSH1, Tasselsheath4; WAB1, Wavy Auricle in Blade; SPM, spikelet pair meristem; BIF2, Barren inflorescence2; TCP30, teosinte branched1/cycloidea/proliferating cell factor30; PIF, phytochrome-interacting factors.
References
Ahn, B.-O., Ji, S.-H., Yun, D.-W., Ji, H.-S., Park, Y.-H., Park, S.-H., et al. (2008). Isolation and characterization of a ds-tagged liguleless mutant in rice (Oryza sativa. l). J. Crop Sci. Biotechnol. 11, 237–242.
Becraft, P. W., Bongard-Pierce, D. K., Sylvester, A. W., Poethig, R. S., Freeling, M. (1990). The liguleless-1 gene acts tissue specifically in maize leaf development. Dev. Biol. 141, 220–232. doi: 10.1016/0012-1606(90)90117-2
Becraft, P. W., Freeling, M. (1991). Sectors of liguleless-1 tissue interrupt an inductive signal during maize leaf development. Plant Cell 3, 801–807. doi: 10.1105/tpc.3.8.801
Bian, H., Xie, Y., Guo, F., Han, N., Ma, S., Zeng, Z., et al. (2012). Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 196, 149–161. doi: 10.1111/j.1469-8137.2012.04248.x
Birkenbihl, R. P., Jach, G., Saedler, H., Huijser, P. (2005). Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352, 585–596. doi: 10.1016/j.jmb.2005.07.013
Bolduc, N., O’Connor, D., Moon, J., Lewis, M., Hake, S. (2012a). How to pattern a leaf. Cold Spring Harb. Symp. Quant. Biol. 77, 47–51. doi: 10.1101/sqb.2012.77.014613
Bolduc, N., Yilmaz, A., Mejia-Guerra, M. K., Morohashi, K., O’Connor, D., Grotewold, E., et al. (2012b). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26, 1685–1690. doi: 10.1101/gad.193433.112
Brant, E. J., Baloglu, M. C., Parikh, A., Altpeter, F. (2021). CRISPR/Cas9 mediated targeted mutagenesis of LIGULELESS-1 in sorghum provides a rapidly scorable phenotype by altering leaf inclination angle. Biotechnol. J. 16, e2100237. doi: 10.1002/biot.202100237
Cao, Y., Dou, D., Zhang, D., Zheng, Y., Ren, Z., Su, H., et al. (2022a). ZmDWF1 regulates leaf angle in maize. Plant Sci. 325, 111459. doi: 10.1016/j.plantsci.2022.111459
Cao, Y., Zeng, H., Ku, L., Ren, Z., Han, Y., Su, H., et al. (2020). ZmIBH1-1 regulates plant architecture in maize. J. Exp. Bot. 71, 2943–2955. doi: 10.1093/jxb/eraa052
Cao, Y., Zhong, Z., Wang, H., Shen, R. (2022b). Leaf angle: a target of genetic improvement in cereal crops tailored for high-density planting. Plant Biotechnol. J. 20, 426–436. doi: 10.1111/pbi.13780
Cardon, G., Höhmann, S., Klein, J., Nettesheim, K., Saedler, H., Huijser, P. (1999). Molecular characterisation of the arabidopsis SBP-box genes. Gene 237, 91–104. doi: 10.1016/S0378-1119(99)00308-X
Carraro, N., Forestan, C., Canova, S., Traas, J., Varotto, S. (2006). ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol. 142, 254–264. doi: 10.1104/pp.106.080119
Chen, Y., Dan, Z., Gao, F., Chen, P., Fan, F., Li, S. (2020). Rice GROWTH-REGULATING FACTOR7 modulates plant architecture through regulating GA and indole-3-Acetic acid metabolism. Plant Physiol. 184, 393–406. doi: 10.1104/pp.20.00302
Chen, X., Zhang, Z., Liu, D., Zhang, K., Li, A., Mao, L. (2010). SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J. Integr. Plant Biol. 52, 946–951. doi: 10.1111/j.1744-7909.2010.00987.x
Dou, D., Han, S., Cao, L., Ku, L., Liu, H., Su, H., et al. (2021). CLA4 regulates leaf angle through multiple hormone signaling pathways in maize. J. Exp. Bot. 72, 1782–1794. doi: 10.1093/jxb/eraa565
Du, F., Guan, C., Jiao, Y. (2018). Molecular mechanisms of leaf morphogenesis. Mol. Plant 11, 1117–1134. doi: 10.1016/j.molp.2018.06.006
Du, Y., Lunde, C., Li, Y., Jackson, D., Hake, S., Zhang, Z. (2021). Gene duplication at the fascicled ear1 locus controls the fate of inflorescence meristem cells in maize. Proc. Natl. Acad. Sci. U.S.A. 118. doi: 10.1073/pnas.2019218118
Duvick, D. (2005). Genetic progress in yield of united states maize (Zea mays l.). Maydica 50, 193. doi: 10.1300/J064v25n04_08
Foster, T., Hay, A., Johnston, R., Hake, S. (2004). The establishment of axial patterning in the maize leaf. Development 131, 3921–3929. doi: 10.1242/dev.01262
Gruszka, D. (2020). Exploring the brassinosteroid signaling in monocots reveals novel components of the pathway and implications for plant breeding. Int. J. Mol. Sci. 21, 354. doi: 10.3390/ijms21010354
Guo, J., Li, W., Shang, L., Wang, Y., Yan, P., Bai, Y., et al. (2021). OsbHLH98 regulates leaf angle in rice through transcriptional repression of OsBUL1. New Phytol. 230, 1953–1966. doi: 10.1111/nph.17303
Guo, A. Y., Zhu, Q. H., Gu, X., Ge, S., Yang, J., Luo, J. (2008). Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418, 1–8. doi: 10.1016/j.gene.2008.03.016
Harper, L., Freeling, M. (1996). Interactions of liguleless1 and liguleless2 function during ligule induction in maize. Genetics 144, 1871–1882. doi: 10.1093/genetics/144.4.1871
Hay, A., Hake, S. (2004). The dominant mutant wavy auricle in blade1 disrupts patterning in a lateral domain of the maize leaf. Plant Physiol. 135, 300–308. doi: 10.1104/pp.103.036707
Hepworth, S. R., Zhang, Y., McKim, S., Li, X., Haughn, G. W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in arabidopsis. Plant Cell 17, 1434–1448. doi: 10.1105/tpc.104.030536
Hong, Z., Ueguchi-Tanaka, M., Shimizu-Sato, S., Inukai, Y., Fujioka, S., Shimada, Y., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, c-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. doi: 10.1046/j.1365-313X.2002.01438.x
Huang, G., Hu, H., van de Meene, A., Zhang, J., Dong, L., Zheng, S., et al. (2021). AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints. Plant Cell 33, 3120–3133. doi: 10.1093/plcell/koab175
Huang, C., Sun, H., Xu, D., Chen, Q., Liang, Y., Wang, X., et al. (2018). ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. 115, E334–E341. doi: 10.1073/pnas.1718058115
Ishii, T., Numaguchi, K., Miura, K., Yoshida, K., Thanh, P. T., Htun, T. M., et al. (2013). OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 45 462-465, 465e461–462. doi: 10.1038/ng.2567
Ji, X., Gao, Q., Chen, F., Bai, M., Zhuang, Z., Peng, Y. (2022). Mutant lpa1 analysis of ZmLPA1 gene regulates maize leaf-angle development through the auxin pathway. Int. J. Mol. Sci. 23, 4886. doi: 10.3390/ijms23094886
Johnston, R., Wang, M., Sun, Q., Sylvester, A. W., Hake, S., Scanlon, M. J. (2014). Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell 26, 4718–4732. doi: 10.1105/tpc.114.132688
Kang, S. G., Lee, D. S., Do, G. S., Pandeya, D., Matin, M. N. (2022). Genetic analysis of a DROOPING LEAF mutant allele dl-6 associated with a twisted and folded leaf base caused by a deficiency in midrib development in rice. J. Plant Physiol. 279, 153837. doi: 10.1016/j.jplph.2022.153837
Klein, J., Saedler, H., Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. MGG 250, 7–16. doi: 10.1007/BF02191820
Kolkman, J. M., Strable, J., Harline, K., Kroon, D. E., Wiesner-Hanks, T., Bradbury, P. J., et al. (2020). Maize introgression library provides evidence for the involvement of liguleless1 in resistance to northern leaf blight. G3 (Bethesda) 10, 3611–3622. doi: 10.1534/g3.120.401500
Kong, D., Li, C., Xue, W., Wei, H., Ding, H., Hu, G., et al. (2022). UB2/UB3/TSH4-anchored transcriptional networks regulate early maize inflorescence development in response to simulated shade. Plant Cell. 35, 717–737. doi: 10.1093/plcell/koac352
Ku, L., Wei, X., Zhang, S., Zhang, J., Guo, S., Chen, Y. (2011). Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays l.). PloS One 6, e20621. doi: 10.1371/journal.pone.0020621
Ku, L. X., Zhang, J., Guo, S. L., Liu, H. Y., Zhao, R. F., Chen, Y. H. (2012). Integrated multiple population analysis of leaf architecture traits in maize (Zea mays l.). J. Exp. Bot. 63, 261–274. doi: 10.1093/jxb/err277
Ku, L., Zhao, W., Zhang, J., Wu, L., Wang, C., Wang, P., et al. (2010). Quantitative trait loci mapping of leaf angle and leaf orientation value in maize (Zea mays l.). Theor. Appl. Genet. 121, 951–959. doi: 10.1007/s00122-010-1364-z
Lee, J., Park, J. J., Kim, S. L., Yim, J., An, G. (2007). Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 65, 487–499. doi: 10.1007/s11103-007-9196-1
Leiboff, S., Strable, J., Johnston, R., Federici, S., Sylvester, A. W., Scanlon, M. J. (2021). Network analyses identify a transcriptomic proximodistal prepattern in the maize leaf primordium. New Phytol. 230, 218–227. doi: 10.1111/nph.17132
Lewis, M. W., Bolduc, N., Hake, K., Htike, Y., Hay, A., Candela, H., et al. (2014). Gene regulatory interactions at lateral organ boundaries in maize. Development 141, 4590–4597. doi: 10.1242/dev.111955
Li, Y., He, Y., Liu, Z., Qin, T., Wang, L., Chen, Z., et al. (2022). OsSPL14 acts upstream of OsPIN1b and PILS6b to modulate axillary bud outgrowth by fine-tuning auxin transport in rice. Plant J. 111, 1167–1182. doi: 10.1111/tpj.15884
Li, S., Lauri, A., Ziemann, M., Busch, A., Bhave, M., Zachgo, S. (2009). Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in arabidopsis thaliana. Plant Cell 21, 429–441. doi: 10.1105/tpc.108.064477
Li, X., Wu, P., Lu, Y., Guo, S., Zhong, Z., Shen, R., et al. (2020). Synergistic interaction of phytohormones in determining leaf angle in crops. Int. J. Mol. Sci. 21, 5052. doi: 10.3390/ijms21145052
Liu, K., Cao, J., Yu, K., Liu, X., Gao, Y., Chen, Q., et al. (2019). Wheat TaSPL8 modulates leaf angle through auxin and brassinosteroid signaling. Plant Physiol. 181, 179–194. doi: 10.1104/pp.19.00248
Liu, S., Magne, K., Daniel, S., Sibout, R., Ratet, P. (2022). Brachypodium distachyon UNICULME4 and LAXATUM-a are redundantly required for development. Plant Physiol. 188, 363–381. doi: 10.1093/plphys/kiab456
Liu, Y., Wu, G., Zhao, Y., Wang, H. H., Dai, Z., Xue, W., et al. (2021). DWARF53 interacts with transcription factors UB2/UB3/TSH4 to regulate maize tillering and tassel branching. Plant Physiol. 187, 947–962. doi: 10.1093/plphys/kiab259
Liu, X., Yang, C. Y., Miao, R., Zhou, C. L., Cao, P. H., Lan, J., et al. (2018). DS1/OsEMF1 interacts with OsARF11 to control rice architecture by regulation of brassinosteroid signaling. Rice (N Y) 11, 46. doi: 10.1186/s12284-018-0239-9
Lu, S., Zhang, M., Zhang, Z., Wang, Z., Wu, N., Song, Y., et al. (2018). Screening and verification of genes associated with leaf angle and leaf orientation value in inbred maize lines. PloS One 13, e0208386. doi: 10.1371/journal.pone.0208386
Mansfield, B. D., Mumm, R. H. (2014). Survey of plant density tolerance in U.S. Maize Germplasm. Crop Sci. 54, 157–173. doi: 10.2135/cropsci2013.04.0252
Mantilla-Perez, M. B., Salas Fernandez, M. G. (2017). Differential manipulation of leaf angle throughout the canopy: current status and prospects. J. Exp. Bot. 68, 5699–5717. doi: 10.1093/jxb/erx378
Moon, J., Candela, H., Hake, S. (2013). The liguleless narrow mutation affects proximal-distal signaling and leaf growth. Development 140, 405–412. doi: 10.1242/dev.085787
Moreno, M. A., Harper, L. C., Krueger, R. W., Dellaporta, S. L., Freeling, M. (1997). liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 11, 616–628. doi: 10.1101/gad.11.5.616
Murmu, J., Bush, M. J., DeLong, C., Li, S., Xu, M., Khan, M., et al. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 154, 1492–1504. doi: 10.1104/pp.110.159111
Nolan, T. M., Vukasinovic, N., Liu, D., Russinova, E., Yin, Y. (2020). Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32, 295–318. doi: 10.1105/tpc.19.00335
Okagaki, R. J., Haaning, A., Bilgic, H., Heinen, S., Druka, A., Bayer, M., et al. (2018). ELIGULUM-a regulates lateral branch and leaf development in barley. Plant Physiol. 176, 2750–2760. doi: 10.1104/pp.17.01459
Ort, D. R., Merchant, S. S., Alric, J., Barkan, A., Blankenship, R. E., Bock, R., et al. (2015). Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl. Acad. Sci. U.S.A. 112, 8529–8536. doi: 10.1073/pnas.1424031112
Osmont, K. S., Sadeghian, N., Freeling, M. (2006). Mosaic analysis of extended auricle1 (eta1) suggests that a two-way signaling pathway is involved in positioning the blade/sheath boundary in zea mays. Dev. Biol. 295, 1–12. doi: 10.1016/j.ydbio.2005.11.012
Peng, H., He, X., Gao, J., Luo, M., Pan, G., Zhang, Z. (2016). Genome-wide identification and function analysis of SBP gene family in maize. Acta Agronomica Sin. 42, 201–211. doi: 10.3724/SP.J.1006.2016.00201
Peng, X., Wang, Q., Zhao, Y., Li, X., Ma, Q. (2019). Comparative genome analysis of the SPL gene family reveals novel evolutionary features in maize. Genet. Mol. Biol. 42, 380–394. doi: 10.1590/1678-4685-gmb-2017-0144
Peng, B., Zhao, X., Wang, Y., Li, C., Li, Y., Zhang, D., et al. (2021). Genome-wide association studies of leaf angle in maize. Mol. Breed. 41, 50. doi: 10.1007/s11032-021-01241-0
Preston, J. C., Hileman, L. C. (2013). Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 4, 80. doi: 10.3389/fpls.2013.00080
Qin, L., Chen, H., Wu, Q., Wang, X. (2022a). Identification and exploration of the GRF and GIF families in maize and foxtail millet. Physiol. Mol. Biol. Plants 28, 1717–1735. doi: 10.1007/s12298-022-01234-z
Qin, W.-X., Liu, X., Long, Y., Dong, Z.-Y., Wan, X.-Y. (2022b). Progress on genetic analysis and molecular dissection on maize leaf angle traits. China Biotechnol. 41, 74–87. doi: 10.1007/s12298-022-01234-z
Ren, Z., Wu, L., Ku, L., Wang, H., Chen, Y. (2020). ZmILI1 regulates leaf angle by directly affecting liguleless1 expression in maize. Plant Biotechnol. J. 18, 881–883. doi: 10.1111/pbi.13255
Richardson, A., Cheng, J., Johnston, R., Kennaway, R., Conlon, B., Rebocho, A., et al. (2021). Evolution of the grass leaf by primordium extension and petiole-lamina remodeling. Science 374, 1377–1381. doi: 10.1126/science.abf9407
Richardson, A. E., Hake, S. (2018). Drawing a line: grasses and boundaries. Plants (Basel) 8, 4. doi: 10.3390/plants8010004
Robil, J. M., McSteen, P. (2022). Hormonal control of medial-lateral growth and vein formation in the maize leaf. New Phytol. 238, 125–141. doi: 10.1111/nph.18625
Satterlee, J. W., Strable, J., Scanlon, M. J. (2020). Plant stem-cell organization and differentiation at single-cell resolution. Proc. Natl. Acad. Sci. U.S.A. 117, 33689–33699. doi: 10.1073/pnas.2018788117
Song, X., Meng, X., Guo, H., Cheng, Q., Jing, Y., Chen, M., et al. (2022). Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat. Biotechnol. 40, 1403–1411. doi: 10.1038/s41587-022-01281-7
Strable, J., Nelissen, H. (2021). The dynamics of maize leaf development: patterned to grow while growing a pattern. Curr. Opin. Plant Biol. 63, 102038. doi: 10.1016/j.pbi.2021.102038
Strable, J., Wallace, J. G., Unger-Wallace, E., Briggs, S., Bradbury, P. J., Buckler, E. S., et al. (2017). Maize YABBY genes drooping leaf1 and drooping leaf2 regulate plant architecture. Plant Cell 29, 1622–1641. doi: 10.1105/tpc.16.00477
Sylvester, A. W., Cande, W. Z., Freeling, M. (1990). Division and differentiation during normal and liguleless-1 maize leaf development. Development 110, 985–1000. doi: 10.1242/dev.110.3.985
Tang, D., Chen, Z., Ni, J., Jiang, Q., Li, P., Wang, L., et al. (2021). Identification of QTL for leaf angle at canopy-wide levels in maize. Euphytica 217. doi: 10.1007/s10681-021-02781-4
Tian, F., Bradbury, P. J., Brown, P. J., Hung, H., Sun, Q., Flint-Garcia, S., et al. (2011). Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 43, 159–162. doi: 10.1038/ng.746
Tian, J., Wang, C., Xia, J., Wu, L., Xu, G., Wu, W., et al. (2019). Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365, 658–664. doi: 10.1126/science.aax5482
Toriba, T., Tokunaga, H., Shiga, T., Nie, F., Naramoto, S., Honda, E., et al. (2019). BLADE-ON-PETIOLE genes temporally and developmentally regulate the sheath to blade ratio of rice leaves. Nat. Commun. 10, 619. doi: 10.1038/s41467-019-08479-5
Tsuda, K., Ito, Y., Sato, Y., Kurata, N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23, 4368–4381. doi: 10.1105/tpc.111.090050
Unte, U. S., Sorensen, A. M., Pesaresi, P., Gandikota, M., Leister, D., Saedler, H., et al. (2003). SPL8, an SBP-box gene that affects pollen sac development in arabidopsis. Plant Cell 15, 1009–1019. doi: 10.1105/tpc.010678
Walsh, J., Freeling, M. (1999). The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot apex. Plant J. 19, 489–495. doi: 10.1046/j.1365-313X.1999.00541.x
Walsh, J., Waters, C. A., Freeling, M. (1998). The maize geneliguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade–sheath boundary. Genes Dev. 12, 208–218. doi: 10.1101/gad.12.2.208
Wang, B., Lin, Z., Li, X., Zhao, Y., Zhao, B., Wu, G., et al. (2020). Genome-wide selection and genetic improvement during modern maize breeding. Nat. Genet. 52, 565–571. doi: 10.1038/s41588-020-0616-3
Wang, R., Liu, C., Chen, Z., Sun, S., Wang, X. (2021). Oryza sativa LIGULELESS 2s determine lamina joint positioning and differentiation by inhibiting auxin signaling. New Phytol. 229, 1832–1839. doi: 10.1111/nph.16970
Wang, H., Wang, H. (2015). The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant 8, 677–688. doi: 10.1016/j.molp.2015.01.008
Wang, X., Wang, X., Sun, S., Tu, X., Lin, K., Qin, L., et al. (2022). Characterization of regulatory modules controlling leaf angle in maize. Plant Physiol. 190, 500–515. doi: 10.1093/plphys/kiac308
Wei, H., Zhao, Y., Xie, Y., Wang, H. (2018). Exploiting SPL genes to improve maize plant architecture tailored for high-density planting. J. Exp. Bot. 69, 4675–4688. doi: 10.1093/jxb/ery258
Xiao, Y., Guo, J., Dong, Z., Richardson, A., Patterson, E., Mangrum, S., et al. (2022). Boundary domain genes were recruited to suppress bract growth and promote branching in maize. Sci. Adv. 8, eabm6835. doi: 10.1126/sciadv.abm6835
Xie, K., Wu, C., Xiong, L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142, 280–293. doi: 10.1104/pp.106.084475
Xing, S., Salinas, M., Hohmann, S., Berndtgen, R., Huijser, P. (2010). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in arabidopsis. Plant Cell 22, 3935–3950. doi: 10.1105/tpc.110.079343
Xu, M., Hu, T., Zhao, J., Park, M. Y., Earley, K. W., Wu, G., et al. (2016b). Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in arabidopsis thaliana. PloS Genet. 12, e1006263. doi: 10.1371/journal.pgen.1006263
Xu, C., Luo, F., Hochholdinger, F. (2016a). LOB domain proteins: beyond lateral organ boundaries. Trends Plant Sci. 21, 159–167. doi: 10.1016/j.tplants.2015.10.010
Xu, J., Wang, J. J., Xue, H. W., Zhang, G. H. (2021). Leaf direction: lamina joint development and environmental responses. Plant Cell Environ. 44, 2441–2454. doi: 10.1111/pce.14065
Yan, Y., Zhu, J., Qi, X., Cheng, B., Liu, C., Xie, C. (2021). Establishment of an efficient seed fluorescence reporter-assisted CRISPR/Cas9 gene editing in maize. J. Integr. Plant Biol. 63, 1671–1680. doi: 10.1111/jipb.13086
Yu, Y. (2019). Liguleless1, a conserved gene regulating leaf angle and a target for yield improvement in wheat. Plant Physiol. 181, 4–5. doi: 10.1104/pp.19.00872
Zeng, D., Hu, J., Dong, G., Liu, J., Zeng, L., Zhang, G., et al. (2009). Quantitative trait loci mapping of flag-leaf ligule length in rice and alignment with ZmLG1 gene. J. Integr. Plant Biol. 51, 360–366. doi: 10.1111/j.1744-7909.2008.00803.x
Zhang, L. Y., Bai, M. Y., Wu, J., Zhu, J. Y., Wang, H., Zhang, Z., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and arabidopsis. Plant Cell 21, 3767–3780. doi: 10.1105/tpc.109.070441
Zhang, N., Huang, X. (2021). Mapping quantitative trait loci and predicting candidate genes for leaf angle in maize. PloS One 16, e0245129. doi: 10.1371/journal.pone.0245129
Zhang, W., Li, B., Yu, B. (2016). Genome-wide identification, phylogeny and expression analysis of the SBP-box gene family in maize (Zea mays). J. Integr. Agric. 15, 29–41. doi: 10.1016/S2095-3119(14)60955-2
Zhang, Y., Schwarz, S., Saedler, H., Huijser, P. (2007). SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in arabidopsis. Plant Mol. Biol. 63, 429–439. doi: 10.1007/s11103-006-9099-6
Zhang, D., Sun, W., Singh, R., Zheng, Y., Cao, Z., Li, M., et al. (2018). GRF-interacting factor1 regulates shoot architecture and meristem determinacy in maize. Plant Cell 30, 360–374. doi: 10.1105/tpc.17.00791
Zhang, S., Wang, S., Xu, Y., Yu, C., Shen, C., Qian, Q., et al. (2015). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38, 638–654. doi: 10.1111/pce.12397
Zhao, M., Tang, S., Zhang, H., He, M., Liu, J., Zhi, H., et al. (2020). DROOPY LEAF1 controls leaf architecture by orchestrating early brassinosteroid signaling. Proc. Natl. Acad. Sci. U.S.A. 117, 21766–21774. doi: 10.1073/pnas.2002278117
Zhu, Z., Tan, L., Fu, Y., Liu, F., Cai, H., Xie, D., et al. (2013). Genetic control of inflorescence architecture during rice domestication. Nat. Commun. 4, 2200. doi: 10.1038/ncomms3200
Keywords: plant architecture, LG1, leaf angle, flower development, SBP
Citation: Qin L, Wu X and Zhao H (2023) Molecular and functional dissection of LIGULELESS1 (LG1) in plants. Front. Plant Sci. 14:1190004. doi: 10.3389/fpls.2023.1190004
Received: 21 March 2023; Accepted: 24 May 2023;
Published: 12 June 2023.
Edited by:
Junfei Gu, Yangzhou University, ChinaReviewed by:
Thomas L. Slewinski, Bayer CropScience, BelgiumMaria Jazmin Abraham-Juarez, National Polytechnic Institute of Mexico (CINVESTAV), Mexico
Copyright © 2023 Qin, Wu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hang Zhao, aGFuZ3poYW9sZ2xAMTYzLmNvbQ==
 Lei Qin
Lei Qin Xintong Wu1
Xintong Wu1 Hang Zhao
Hang Zhao