- Cotton Fiber Bioscience and Utilization Research Unit, United States Department of Agriculture (USDA), Agricultural Research Service (ARS), Southern Regional Research Center (SRRC), New Orleans, LA, United States
Naturally colored cotton (NCC) offers an environmentally friendly fiber for textile applications. Processing white cotton fiber into textiles requires extensive energy, water, and chemicals, whereas processing of NCC skips the most polluting activity, scouring-bleaching and dyeing; therefore, NCC provides an avenue to minimize the harmful impacts of textile production. NCC varieties are suitable for organic agriculture since they are naturally insect and disease-resistant, salt and drought-tolerant. Various fiber shades, ranging from light green to tan and brown, are available in the cultivated NCC (Gossypium hirsutum L.) species. The pigments responsible for the color of brown cotton fiber are proanthocyanidins or their derivatives synthesized by the flavonoid pathway. Due to pigments, the NCC has excellent ultraviolet protection properties. Some brown cotton varieties exhibited superior thermal resistance of fiber that can be used to make fabrics with enhanced flame retardancy. Here, we review molecular mechanisms involved in the pigment production of brown cotton and challenges in breeding NCC varieties with a wide range of colors but without penalty in fiber quality. Also, we discuss opportunities for NCC with flame-retarding properties in textile applications.
Introduction
Naturally colored cotton (NCC) fibers exist in different hues of brown, red, rust, and green and can be used as eco-friendly alternatives to white cotton (Figure 1). The conventional process of dyeing white cotton requires extensive water, energy, and chemicals and contributes about 15% to the cost of the finished garment (Nimon and Beghin, 1999). NCC fabrics resist fading since colors become stronger after laundering (Günaydin et al., 2019). Due to pigments, the clothes made from NCC fibers protect the skin from ultraviolet radiation. Fibers of some brown cotton varieties exhibited flame retardant (FR) properties, making them suitable for specific end-use applications, such as automotive interiors (Parmar and Chakraborty, 2001; Hinchliffe et al., 2015, 2016; Nam et al., 2016, 2017).
Despite these advantages, typical NCC fibers are weaker and shorter than cultivated white cotton fibers, deeming them unsuitable for high-speed woven textile machinery. The breeding efforts in the 1990s resulted in the release of improved brown fiber germplasm lines (May et al., 1994). However, the major obstacle to expanding NCC production is not marketing but regulations to protect white cotton from contamination during ginning and the cost associated with cleaning equipment (Funk and Gamble, 2009). Limited color diversity is another hindrance to expanding the NCC to commercial textile production (Dutt et al., 2004; Yuan et al., 2013). Currently, NCC occupies a textile market niche promoting environmentally friendly clothing production.
Nevertheless, the use of NCC has great merit and should be expanded since global textile production generates toxic chemical waste with a negative impact on the environment. A recent forum article reviewed molecular mechanisms underlying pigmentation in brown and green cotton and suggested “omics-driven breeding” for better fiber quality NCC (Sun et al., 2021). Here, we review the regulation of pigment development in brown cotton and possible biotechnological strategies to increase hue diversity and improve fiber yield and quality. Also, we discuss the source of FR in brown cotton.
Pigments in brown cotton fibers
Brown cotton varieties are most commonly used for NCC fabrics. Proanthocyanidins (PAs) are the primary pigments responsible for brown fibers. PAs, also called condensed tannins, are polymeric flavan-3-ols with the diphenylpropane typical chemical structure (C6-C3-C6), which includes a benzopyran (A and C rings) linked with another aromatic ring (B ring) at C2 position (Figure 2) (Yu et al., 2023). The adjacent subunits of PA are linked by C4-C8 or C4-C6 carbon-carbon bonds (Xie et al., 2004). PAs were initially detected in brown fibers with DMACA (p-Dimethylaminocinnamaldehyde) staining. A comparative study of the treatment of mature white, green, and brown fibers with DMACA determined that only brown fibers turned blue, while white and green fibers did not change color (Xiao et al., 2007). However, PAs in minimal quantities were detected in developing white fibers from 5 to 15 days post-anthesis (DPA) and, after that, gradually disappeared (Li et al., 2012). In contrast, PAs were detected as early as 3 DPA in developing brown fibers and were synthesized in high quantities up to maturation (Li et al., 2012).
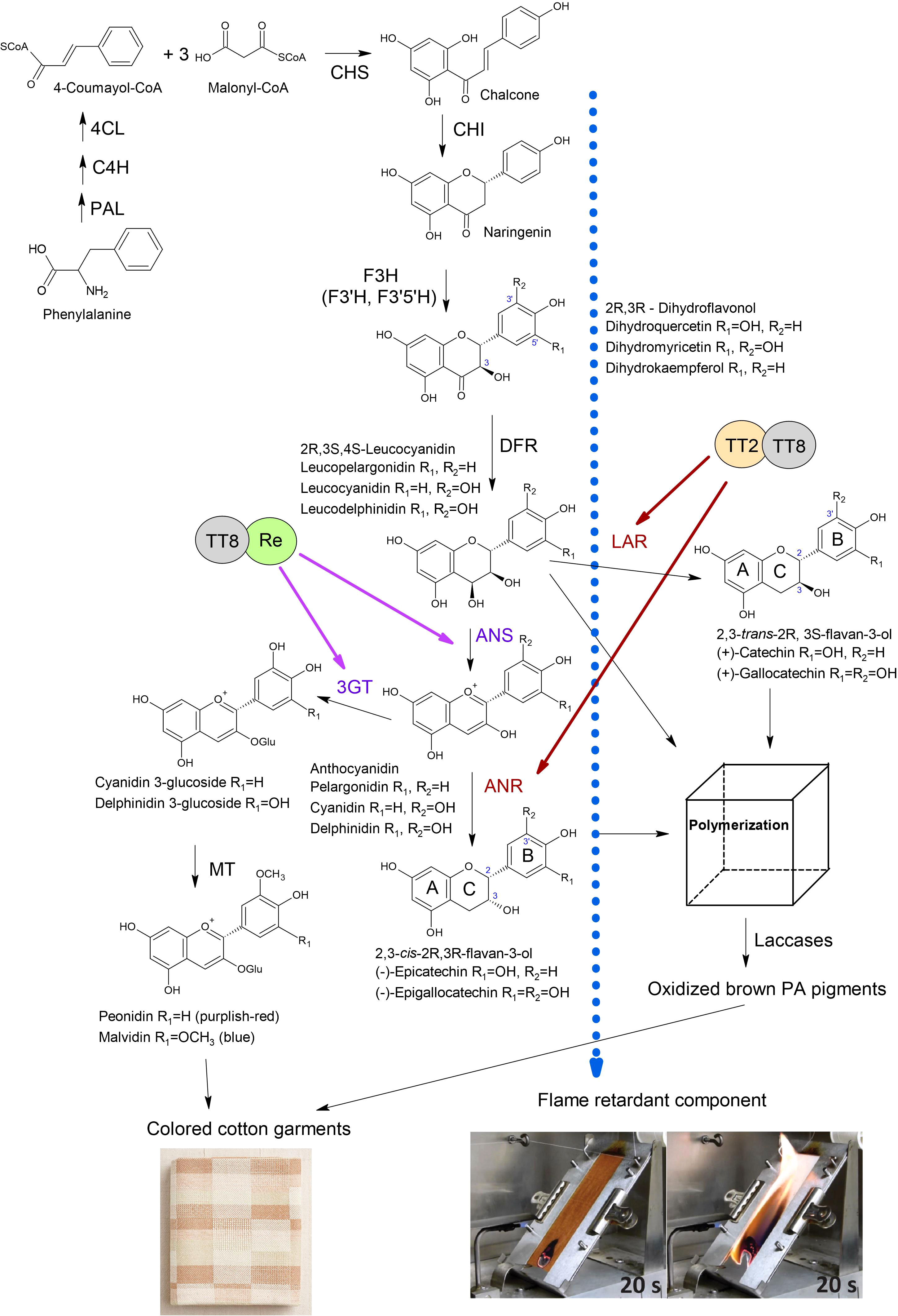
Figure 2 Anthocyanidin and proanthocyanidin biosynthesis in brown cotton. PAL, phenylalanine ammonia-lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate CoA ligase; CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3’H, flavonoid 3’-hydroxylase; F3’5’H, flavonoid 3’,5’-hydroxylase; DFR, dihydroflavonol 4-reductase; LAR, leucoanthocyanidin reductase; ANS, anthocyanidin synthase; 3GT, 3-O-glucosyltransferase; ANR, anthocyanidin reductase; MT, methyltransferase. The Blue dotted arrow indicates the transport of an unknown FR compound. Images of the flammability test of brown and white cotton fabrics were adopted from the report (Hinchliffe et al., 2016). Brown arrows indicate the activation of LAR and ANR by the TT2-TT8 complex reported by (Yan et al., 2018). Purple arrows indicate activation of ANS and 3GT by Re-TT8 complex reported by (Wang et al., 2022).
The common flavan-3-ols are (+)-catechin (2,3-trans) and (-)-epicatechin (2,3-cis) with the gallate modification of the hydroxyl group at the C3’ position on the B-ring (Figure 2) (Dixon et al., 2005; Xie and Dixon, 2005). The structure variability of PAs depends on the 2,3- stereochemistry of the starter and extension units and additional derivatizations such as O-methylation, O-acylation, C- and O-glycosylation (Dixon and Sarnala, 2020). The structure of PA units in brown fibers was studied with nuclear magnetic resonance (NMR), a matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS), and liquid-chromatography mass spectrometry (LC-MS) (Feng et al., 2014; Xiao et al., 2014). In one study, LC-MS analyses determined that the most abundant flavan-3-ols in brown fiber were in 2,3-trans form (catechin and gallocatechin), and leucoanthocyanidin reductase (LAR) was highly expressed (Xiao et al., 2014). Another work showed by NMR and MALDI-TOF MS analyses that the most predominant PA units were 2,3-cis form (epicatechin and epigallocatechin), and anthocyanidin reductase (ANR) was a key gene in brown fiber pigment biosynthesis (Feng et al., 2014). Therefore, both flavan-3-ols forms, 2,3-trans and 2,3-cis units, can be produced in brown cotton. The dimeric PAs, procyanidin (PC) and prodelphidin (PD) were detected in white and brown fibers; however, acylation modified some PAs in white fibers (Feng et al., 2014). The ratio of PC and PD was equal in white fibers, whereas brown cotton fibers contained mainly PD units with a relative ratio of 9:1. Feng et al. (2014) suggested that quinones, the oxidation products of proanthocyanidins, directly contribute to color development in brown fiber because developing fibers do not show distinct coloration until maturation.
PA biosynthesis in brown cotton
PAs are synthesized through the flavonoid pathway, a branch of the phenylpropanoid pathway (Yu et al., 2023). Phenylalanine is converted into 4-Coumaroyl-CoA through a few enzymatic steps by phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate CoA ligase (4CL) (Figure 2). Chalcone synthase (CHS) then catalyzes chalcone from one molecule of 4-coumaroyl-CoA and three molecules of malonyl-CoA. Chalcone isomerase (CHI) catalyzes the cyclization of chalcone into flavanone naringenin. Flavanone 3/3’/3’5’hydroxylases (F3H/F3’H/F3’5’H) add hydroxyl groups to the C-3, C-3’, and C-5’ of the naringenin, generating dihydrokaempferol, dihydroquercetin, or dihydromyricetin. Dihydroflavonol 4-reductase (DFR) reduces the carbonyl group at the C4 position to a hydroxyl group, producing leucoanthocyanidins (Figure 2). Leucoanthocyanidins are converted into flavan-3-ols in two ways: 1) LAR catalases the reductive removal of the hydroxyl at the C4 position producing 2,3-trans isoforms; 2) anthocyanidin synthase (ANS) catalases production of anthocyanidins, which are converted by ANR into 2,3-cis flavan-3-ol monomers (Xie et al., 2003). It has been demonstrated by in vitro enzyme assays and virus-induced gene silencing (VIGS) that the ANR pathway contributes to the biosynthesis of 2,3-cis flavan-3-ol in cotton (Zhu et al., 2015).
Transcriptomic and proteomic studies in cotton identified that a set of structural genes of the flavonoid pathway, including PAL, C4H, 4CL, CHS, CHI, F3H, F3’H, DFR, LAR, ANS, ANR, were up-regulated in brown fibers (Li et al., 2013; Feng et al., 2014; Xiao et al., 2014; Hinchliffe et al., 2016; Gao et al., 2019; Peng et al., 2020; Canavar and Rausher, 2021). Several structural genes of the flavonoid pathway were characterized in brown cotton. VIGS of GhCHS2, GhLAR1, and GhANR significantly reduced intensity of the color of brown fibers and anthocyanidin content (Gao et al., 2019). RNAi silencing of GhCHI-1 in brown cotton resulted in three fiber phenotypes of transgenic plants: brown, almost white, and green (Liu et al., 2018). In transgenic lines with almost white fibers, the GhCHI-1 and GhCHI-2 were significantly suppressed, whereas in transgenic lines with brown and green fibers were not. Interestingly, an anthocyanidin 3-O-glucosyltransferase (Gh3GT) that glucosylates the 3-position of the flavonoid C-ring was significantly up-regulated in green fiber transgenic lines. Furthermore, it was confirmed that the overexpression of the Gh3GT or At3GT gene in brown cotton lines produced green fiber. The green color of fibers could be due to the increased amount of anthocyanidins caused by the upregulation of Gh3GT that redirected the flux toward anthocyanidins (Liu et al., 2018; Canavar and Rausher, 2021). RNAi suppression of GhF3H in white cotton significantly increased naringenin content and reduced the length and micronaire of mature fibers (Tan et al., 2013). The even greater negative effect of naringenin on fiber properties was observed in brown cotton with introduced RNAi GhF3H copy by a cross. However, the overexpression of the GhF3H caused no noticeable effects on fiber phenotype (Tan et al., 2013).
Three classes of proteins, including TT2 (MYB123), TT8 (bHLH42), and TTG1 (WD40 family), often referred to as MBW complex, are primary regulators of PA biosynthesis. Experimental studies showed that TT2 and TT8 activate PA pathway genes while TTG1 stabilizes the MBW complex and is essential in maintaining MBW activity genes (Baudry et al., 2004; Yu et al., 2023). Two TT2-type MYB transcription factors (GhMYB36 and GhMYB10) isolated from cotton were able to activate promoters of LAR and ANR as well as recover the seed phenotype of Arabidopsis tt2 mutant, confirming their role in PA biosynthesis (Lu et al., 2017). In cotton, genetic studies identified at least six incompletely dominant genetic loci (Lc1 – Lc6) linked to brown fiber (Kohel, 1985). The Lc1 was linked to a 1.4 Mb inversion on chromosome A07 upstream of a TT2 homologous gene (Gh_A07G2341), which this inversion could activate (Hinchliffe et al., 2016). A fine-mapping study determined that two QTLs in the Lc1 locus regulate the shades of brown color in fibers. The qBF-A07-1 (Gh_A07G2341 - TT2) is responsible for the brown color, whereas interaction between the qBF-A07-1 and qBF-A07-2 (Gh_A07G0100 - TTG1) regulates the shades of brown fiber (Wen et al., 2018). A genome-wide association study (GWAS) revealed that epistatic interaction between qBF-A07-1 and qBF-A07-2 negatively impacts fiber yield and quality traits (Wen et al., 2018).
Progress on research to enhance color diversity in NCC
The first report that inadvertently shifted the carbon flow from colorless PAs into colorful anthocyanidins in brown fibers was trying to suppress GhCHI-1 (Liu et al., 2018). The transgenic line that failed to suppress the GhCHI produced green-colored cotton due to the activation of Gh3GT, further confirmed by transgenic overexpression of Gh3GT and At3GT in cotton. Whatever activated 3GT in brown cotton during CHI suppression is unclear; nonetheless, Liu et al. (2018) showed an experimental enhancement of the anthocyanidin biosynthesis in NCC.
Another study has exploited color diversity in NCC by modulating the expression of transcription factors controlling anthocyanin biosynthesis genes. The transcription factor GhMYB113 (Re), which regulates anthocyanin biosynthesis, has been overexpressed in cotton under fiber-specific promoter (Wang et al., 2022). A clear purple-red color was produced in developing fibers (5-10 DPA) of transgenic lines; however, the mature fiber turned brown. The anthocyanins content was high at 5-10 DPA and then gradually decreased, whereas PA content was low at 20 DPA and gradually increased at 25 DPA (Wang et al., 2022). The expression levels of UFGT (3GT) and ANR were elevated in transgenic lines. Authors suggested that anthocyanins accumulated during the early fiber developmental stage converted into PAs during the maturation and dehydration process (Wang et al., 2022).
How do we prevent the conversion of anthocyanins into PAs during the late stage of fiber development? One possible approach is activating anthocyanin biosynthesis during the later stage of fiber development, and a recent study demonstrated that it is feasible. The GhTT2-3A (Gh_A07G2341) was overexpressed under the secondary cell wall (SCW) specific promoter (FbL2A) in cotton; PA structural genes and PA biosynthesis were activated during the SCW stage in transgenic plants that resulted in brown mature fiber with lint percentage and fiber quality comparable to white fiber control (Yan et al., 2018). This study tried to reduce the adverse effects of pigment production on fiber cell development due to the known PA subunit’s cellular toxicity and the inhibitory role of naringenin on fiber elongation (Tan et al., 2013; Dixon and Sarnala, 2020). Yan et al. (2018) achieved the goal of producing brown fiber with improved quality. They demonstrated that the SCW stage can be used for biotechnological manipulations of PAs and other flavonoids in cotton fiber.
Enhanced flame retardancy of NCC
What is causing the flame retardancy (FR) of NCC still remains unclear. Few studies have been published on NCC fabrics’ thermal and burning behavior. The earlier work detected a higher value of limited oxygen index for brown than for white cotton fabric (Parmar and Chakraborty, 2001). The same study suggested that heavy metal ions and color compounds could be responsible for higher FR in brown cotton (Parmar and Chakraborty, 2001). Another study tested FR properties of needle-punched (NP) and hydroentangled (H-E) nonwoven fabrics produced from fibers of brown and white cotton (Hinchliffe et al., 2015). NP is a process for manufacturing nonwoven fabrics using barbed needles, whereas H-E is high-pressure water jets for interlocking fibers together. The NP fabrics showed significantly higher FR than H-E fabrics of brown cotton. The study suggested that the unknown FR component in the brown cotton fibers is water-soluble and was removed by water during the H-E process; the higher levels of phosphorous and boron in brown cotton corresponded with enhanced FR (Hinchliffe et al., 2015).
More recent studies suggested that PA biosynthesis plays a vital role in the natural FR of brown cotton (Figure 2). Nam et al. (2016) demonstrated by washing fibers in water and a low concentration of sodium hydroxide solution that the condensed tannins and higher sodium content in brown cotton are responsible for the FR property. Authors suggested that condensed tannins significantly enhanced the adsorption of sodium ions to the fiber, contributing to FR (Nam et al., 2016). Another study argued that condensed tannin is not the source of FR in brown cotton since enhanced FR and anthocyanin precursors appear in developing fibers well before the brown color is detectable (Hinchliffe et al., 2016). Authors suggested that enhanced FR can be developed in white cotton by activating biosynthesis of flavonoids or PA precursors but preventing PA polymerization (source of brown color); the unknown FR component is probably sequestered by PAs or PA precursors via metal-flavonoid complexes (Figure 2) (Hinchliffe et al., 2016). As proof of concept, the follow-up study found four recombinant inbred lines (RILs) with enhanced FR in the multi-parent advanced generation intercross (MAGIC) population (Thyssen et al., 2023). All four RILs have white cotton fibers and enhanced FR. Authors suggested that the accumulation of colorless flavonoids might contribute to FR in these RILs; however, metabolite analysis of developing fibers has yet to confirm this hypothesis (Thyssen et al., 2023). Identifying the natural FR compound from cotton could help minimize the use of synthetic FR additives in textiles.
Utilization of NCC
NCC has been cultivated and used for thousands of years; however, with the Industrial Revolution and the development of synthetic dyes, NCC varieties became less common as white cotton became the standard. Recently, interest in NCC has resurged due to its sustainability and environmental benefits since NCC cultivation is adaptable to dry land and organic farming (Günaydin et al., 2019; Rathinamoorthy and Parthiban, 2019). Organic NCC is softer, more breathable, and less likely to cause allergies than conventional cotton.
NCC is primarily used in high-end fashion, home textiles, upholstery fabrics, etc (Günaydin et al., 2019; Rathinamoorthy and Parthiban, 2019). The unique colors of NCC, ranging from light beige to red, rust, and brown, are highly sought by designers and consumers looking for sustainable and environmentally friendly products. These cotton varieties create a wide range of products, including clothing, bedding, and towels. Using NCC in these products adds a unique and natural element that is impossible with conventionally dyed cotton. One of the challenges of using NCC in textiles is its limited availability compared to conventional cotton. While there are efforts to increase the production of NCC, it remains a niche product in the textile industry. However, as consumer demand for sustainable and environmentally friendly products grows, the use of NCC will likely continue to increase.
Some NCC varieties possess flame-retardant properties which can be used in various applications. For example, flame-retardant textiles can be used as protective clothing for firefighters, gear for soldiers, automobile interiors, matrasses, house upholsteries, and drapes. Currently, cotton-based textiles are chemically modified to introduce multifunctional groups, making them flame-retardant. The most common method is to merge N, S, P, and Si-based polymeric, non-polymeric, polymeric/non-polymeric hybrids, inorganic, and organic/inorganic hybrids with cellulose to fabricate flame-retardant cotton textiles (Islam and van de Ven, 2021). As an eco-friendlier approach, biomass tannin and phytic acid are applied to the cotton fabric surface as a flame-retardant modification (Nam et al., 2017; Yang et al., 2023). Researchers at the U.S. Department of Agriculture’s Agricultural Research Service have recently reported studies on the FR properties of brown and four white cotton lines (Hinchliffe et al., 2015, 2016; Thyssen et al., 2023). These lines reduce the need for flame-retardant chemicals embedded in consumer products. These advancements demonstrate the potential of naturally colored cotton in creating safer and more sustainable textiles.
Concluding remarks
The biosynthesis of PAs in brown cotton has been well characterized through transgenic analysis of structural flavonoid genes. However, little is known about how PA units are modified and transported into the vacuole and how PAs are polymerized. It has been experimentally demonstrated that the SCW biosynthesis stage of fiber development is the most suitable time for biotechnological modification of the flavonoid pathway. The anthocyanin pathway can be induced in cotton fibers by overexpressing Re (GhMYB113). In future studies, it would be interesting to overexpress Re and anthocyanidin modifiers during the SCW stage of fiber development to see the effects on the color of fibers. The FR has been detected in brown and white cotton fibers. The FR compound, which is yet to be identified, could be an eco-friendly alternative to synthetic FR additives in textiles. The utilization of NCC remains a niche in the textile industry. However, usage of NCC will likely continue to increase as consumer demand for sustainable and environmentally friendly products grows.
Author contributions
MN: Writing – original draft; Writing – review & editing. DH: Writing – review & editing. GT: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The United States Department of Agriculture, Agricultural Research Service financially supported the research (CRIS project 6054-21000-019-00D). The mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by USDA. USDA is an equal opportunity provider and employer.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Canavar, Ö., Rausher, M. D. (2021). Molecular analysis of structural genes involved in flavonoids biosynthesis in naturally colored cotton. Crop Sci. 61, 1117–1126. doi: 10.1002/csc2.20410
Dixon, R. A., Sarnala, S. (2020). Proanthocyanidin biosynthesis-a matter of protection. Plant Physiol. 184, 579–591. doi: 10.1104/pp.20.00973
Dixon, R. A., Xie, D.-Y., Sharma, S. B. (2005). Proanthocyanidins: A final frontier in flavonoid research? New Phytol. 165, 9–28. doi: 10.1111/j.1469-8137.2004.012
Dutt, Y., Wang, X. D., Zhu, Y. G., Li, Y. Y. (2004). Breeding for high yield and fiber quality in colored cotton. Plant Breed. 123, 145–151. doi: 10.1046/j.1439-0523.2003.00938.x
Feng, H., Li, Y., Wang, S., Zhang, L., Liu, Y., Xue, F., et al. (2014). Molecular analysis of proanthocyanidins related to pigmentation in brown cotton fiber (Gossypium hirsutum L.). J. Exp. Bot. 65, 5759–5769. doi: 10.1093/jxb/eru286
Funk, P., Gamble, G. R. (2009). Fiber properties of saw and roller ginned naturally colored cottons. J. Cotton. Sci. 13, 166–173.
Gao, J., Shen, L., Yuan, J., Zheng, H., Su, Q., Yang, W., et al. (2019). Functional analysis of GhCHS, GhANR and GhLAR in colored fiber formation of Gossypium hirsutum L. BMC Plant Biol. 19, 1–18. doi: 10.1186/s12870-019-2065-7
Günaydin, G. K., Avinc, O., Palamutcu, S., Yavas, A., Soydan, A. S. (2019). “Naturally colored organic Cotton and Naturally Colored Cotton Fiber Production,” in Organic Cotton: Is it a Sustainable Solution? Eds. Gardetti, M. A., Muthu, S. S. (Springer Singapore, Singapore), 81–99.
Hinchliffe, D., Condon, B., Delhom, C. D., Chang, S., Montalvo, J., Madison, C., et al. (2015). Physical and combustion properties of nonwoven fabrics produced from conventional and naturally colored cottons. Textile. Res. J. 85, 1666–1680. doi: 10.1177/0040517515573410
Hinchliffe, D. J., Condon, B. D., Thyssen, G., Naoumkina, M., Madison, C. A., Reynolds, M., et al. (2016). The GhTT2_A07 gene is linked to the brown color and natural flame retardancy phenotypes of Lc1 cotton (Gossypium hirsutum L.) fibers. J. Exp. Bot. 67, 5461–5471. doi: 10.1093/jxb/erw312
Islam, M. S., van de Ven, T. G. M. (2021). Cotton-based flame-retardant textiles: A review. BioResources 16, 4354–4381.
Kohel, R. J. (1985). Genetic analysis of fiber color variants in cotton1. Crop Sci. 25, cropsci1985.0011183X0025000500017x. doi: 10.2135/cropsci1985.0011183X0025000500017x
Li, T., Fan, H., Li, Z., Wei, J., Lin, Y., Cai, Y. (2012). The accumulation of pigment in fiber related to proanthocyanidins synthesis for brown cotton. Acta Physiol. Plant. 34, 813–818. doi: 10.1007/s11738-011-0858-x
Li, Y.-J., Zhang, X.-Y., Wang, F.-X., Yang, C.-L., Liu, F., Xia, G.-X., et al. (2013). A comparative proteomic analysis provides insights into pigment biosynthesis in brown color fiber. J. Proteomics 78, 374–388. doi: 10.1016/j.jprot.2012.10.005
Liu, H.-F., Luo, C., Song, W., Shen, H., Li, G., He, Z.-G., et al. (2018). Flavonoid biosynthesis controls fiber color in naturally colored cotton. PeerJ 6, e4537. doi: 10.7717/peerj.4537
Lu, N., Roldan, M., Dixon, R. A. (2017). Characterization of two TT2-type MYB transcription factors regulating proanthocyanidin biosynthesis in tetraploid cotton, Gossypium hirsutum. Planta 246, 323–335. doi: 10.1007/s00425-017-2682-z
May, O. L., Green, C. C., Roach, S. H., Kittrell, B. U. (1994). Registration of PD 93001, PD 93002, PD 93003, and PD 93004 germplasm lines of upland cotton with brown lint and high fiber quality. Crop Sci. 34, 542–542.
Nam, S., Condon, B. D., Xia, Z., Nagarajan, R., Hinchliffe, D. J., Madison, C. A. (2017). Intumescent flame-retardant cotton produced by tannic acid and sodium hydroxide. J. Analytical. Appl. Pyrolysis. 126, 239–246. doi: 10.1016/j.jaap.2017.06.003
Nam, S., Kim, H. J., Condon, B. D., Hinchliffe, D. J., Chang, S., McCarty, J. C., et al. (2016). High resistance to thermal decomposition in brown cotton is linked to tannins and sodium content. Cellulose 23, 1137–1152. doi: 10.1007/s10570-016-0871-8
Nimon, W., Beghin, J. (1999). Are eco-labels valuable? Evidence from the apparel industry. Am. J. Agric. Econ. 81, 801–811. doi: 10.2307/1244325
Parmar, M. S., Chakraborty, M. (2001). Thermal and burning behavior of naturally colored cotton. Textile. Res. J. 71, 1099–1102. doi: 10.1177/004051750107101211
Peng, Z., Gao, Q., Luo, C., Gong, W., Tang, S., Zhang, X., et al. (2020). Flavonoid biosynthetic and starch and sucrose metabolic pathways are involved in the pigmentation of naturally brown-colored cotton fibers. Ind. Crops Products. 158, 113045. doi: 10.1016/j.indcrop.2020.113045
Rathinamoorthy, R., Parthiban, M. (2019). “Colored Cotton: Novel Eco-friendly Textile Material for the Future,” in Handbook of Ecomaterials. Eds. Martínez, L. M. T., Kharissova, O. V., Kharisov, B. I. (Springer International Publishing, Cham), 1499–1519.
Sun, J., Sun, Y., Zhu, Q.-H. (2021). Breeding next-generation naturally colored cotton. Trends Plant Sci. 26, 539–542. doi: 10.1016/j.tplants.2021.03.007
Tan, J., Tu, L., Deng, F., Hu, H., Nie, Y., Zhang, X. (2013). A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant Physiol. 162, 86–95. doi: 10.1104/pp.112.212142
Thyssen, G. N., Condon, B. D., Hinchliffe, D. J., Zeng, L., Naoumkina, M., Jenkins, J. N., et al. (2023). Flame resistant cotton lines generated by synergistic epistasis in a MAGIC population. PloS One 18, e0278696. doi: 10.1371/journal.pone.0278696
Wang, N., Zhang, B., Yao, T., Shen, C., Wen, T., Zhang, R., et al. (2022). Re enhances anthocyanin and proanthocyanidin accumulation to produce red foliated cotton and brown fiber. Plant Physiol. 189, 1466–1481. doi: 10.1093/plphys/kiac118
Wen, T., Wu, M., Shen, C., Gao, B., Zhu, D., Zhang, X., et al. (2018). Linkage and association mapping reveals the genetic basis of brown fiber (Gossypium hirsutum). Plant Biotechnol. J. 16, 1654–1666. doi: 10.1111/pbi.12902
Xiao, Y.-H., Yan, Q., Ding, H., Luo, M., Hou, L., Zhang, M., et al. (2014). Transcriptome and biochemical analyses revealed a detailed proanthocyanidin biosynthesis pathway in brown cotton fiber. PloS One 9, e86344. doi: 10.1371/journal.pone.0086344
Xiao, Y.-H., Zhang, Z.-S., Yin, M.-H., Luo, M., Li, X.-B., Hou, L., et al. (2007). Cotton flavonoid structural genes related to the pigmentation in brown fibers. Biochem. Biophys. Res. Commun. 358, 73–78. doi: 10.1016/j.bbrc.2007.04.084
Xie, D.-Y., Dixon, R. A. (2005). Proanthocyanidin biosynthesis – still more questions than answers? Phytochemistry 66, 2127–2144. doi: 10.1016/j.phytochem.2005.01.008
Xie, D. Y., Sharma, S. B., Dixon, R. A. (2004). Anthocyanidin reductases from Medicago truncatula and Arabidopsis thaliana. Arch. Biochem. Biophys. 422, 91–102. doi: 10.1016/j.abb.2003.12.011
Xie, D.-Y., Sharma, S. B., Paiva, N. L., Ferreira, D., Dixon, R. A. (2003). Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299, 396–399. doi: 10.1126/science.1078540
Yan, Q., Wang, Y., Li, Q., Zhang, Z., Ding, H., Zhang, Y., et al. (2018). Up-regulation of Gh TT 2-3A in cotton fibers during secondary wall thickening results in brown fibers with improved quality. Plant Biotechnol. J. 16, 1735–1747. doi: 10.1111/pbi.12910
Yang, M., Yang, Y., Shi, J., Rao, W. (2023). Fabrication of eco-friendly flame-retardant and hydrophobic coating for cotton fabric. Cellulose 30, 3267–3280. doi: 10.1007/s10570-023-05051-9
Yu, K., Song, Y., Lin, J., Dixon, R. A. (2023). The complexities of proanthocyanidin biosynthesis and its regulation in plants. Plant Commun. 4, 100498. doi: 10.1016/j.xplc.2022.100498
Yuan, S. N., Malik, W., Bibi, N., Wen, G. J., Ni, M., Wang, X. D. (2013). Modulation of morphological and biochemical traits using heterosis breeding in colored cotton. J. Agric. Sci. 151, 57–71. doi: 10.1017/S0021859612000172
Keywords: naturally colored cotton, proanthocyanidin pigments, flame retardance, condensed tannin, flavan-3-ols
Citation: Naoumkina M, Hinchliffe DJ and Thyssen GN (2024) Naturally colored cotton for wearable applications. Front. Plant Sci. 15:1350405. doi: 10.3389/fpls.2024.1350405
Received: 05 December 2023; Accepted: 11 March 2024;
Published: 21 March 2024.
Edited by:
Li Tian, University of California, Davis, United StatesReviewed by:
Guoli Song, Chinese Academy of Agricultural Sciences, ChinaYue Zhu, North Carolina State University, United States
Copyright © 2024 Naoumkina, Hinchliffe and Thyssen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marina Naoumkina, bWFyaW5hLm5hb3Vta2luYUB1c2RhLmdvdg==
 Marina Naoumkina
Marina Naoumkina Doug J. Hinchliffe
Doug J. Hinchliffe Gregory N. Thyssen
Gregory N. Thyssen