- 1Department of Agricultural Botany, Faculty of Agriculture, Tanta University, Tanta, Egypt
- 2Business Administration Department, Community College, King Khalid University, Abha, Saudi Arabia
- 3Plant Pathology Research Institute, Agricultural Research Center, Giza, Egypt
- 4Department of Plant Pathology, Faculty of Agriculture, Zagazig University, Zagazig, Egypt
- 5Department of Plant Pathology, Faculty of Agriculture, Minia University, Minia, Egypt
- 6Institute of Agricultural Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh, India
- 7Department of Life Sciences, Western Caspian University, Baku, Azerbaijan
- 8Department of Plant Biotechnology, Korea University, Seoul, Republic of Korea
- 9Department of Biology, College of Science, United Arab Emirates University, Al Ain, United Arab Emirates
- 10Department of Agricultural Botany, Faculty of Agriculture, Minufiya University, Shebeen El-Kom, Egypt
The necrotrophic fungal phytopathogen, Sclerotinia sclerotiorum (Lib.) de Bary, employs a multilayered strategy to infect a wide range of host plants. The current study proposed the diamine L-ornithine, a non-proteinogenic amino acid that promotes the synthesis of other essential amino acids, as an alternative management strategy to boost the molecular, physiological, and biochemical responses of common bean (Phaseolus vulgaris L.) against white mold disease caused by S. sclerotiorum. In vitro experiments showed that L-ornithine significantly inhibited the mycelial growth of S. sclerotiorum in a dose-dependent manner. Moreover, it markedly diminished the white mold severity under greenhouse conditions. Moreover, L-ornithine stimulated the growth of treated plants suggesting that the tested concentration of L-ornithine has no phytotoxicity on treated plants. Additionally, L-ornithine enhanced the non-enzymatic antioxidants (total soluble phenolics and flavonoids), the enzymatic antioxidants (CAT, POX, and PPO), and upregulated the gene expression of three antioxidant-associated genes (PvCAT1, PvSOD, and PvGR). Moreover, in silico analysis showed that the genome of S. sclerotiorum possesses a putative oxaloacetate acetylhydrolase (SsOAH) protein that is highly similar in its functional analysis, conserved domains, and topology with OAH from Aspergillus fijiensis (AfOAH) and Penicillium lagena (PlOAH). Interestingly, the addition of L-ornithine to the potato dextrose broth (PDB) medium significantly down-regulated the gene expression of SsOAH in the mycelium of S. sclerotiorum. Likewise, exogenous application of L-ornithine significantly down-regulated the gene expression of SsOAH in the fungal mycelia collected from treated plants. Finally, L-ornithine application significantly diminished the secretion of oxalic acid in the PDB medium as well as infected leaves. Collectively, L-ornithine plays a pivotal role in maintaining the redox status, in addition to boosting the defense responses of infected plants. The current study provides insights that may lead to innovative eco-friendly approaches for controlling white mold disease and mitigating its impact on common bean cultivation particularly, and other crops in general.
1 Introduction
White mold, caused by the necrotrophic fungus Sclerotinia sclerotiorum (Lib.) de Bary, is a devastating yield-limiting disease that poses a significant threat to bean (Phaseolus vulgaris L.) production worldwide (Bolton et al., 2006). S. sclerotiorum is one of the most formidable challenging soil-borne fungal phytopathogens, with a broad host range of over 600 plant species, and incites rapid host tissue maceration in a nondiscriminant manner (Liang and Rollins, 2018). Upon unfavorable conditions, it undergoes a crucial phase in its life cycle by producing black, hard, seed-like resting structures known as ‘sclerotia’ in the soil for extended periods, or it may develop in the white fuzzy growths of mycelium or the stem pith of infected plants (Schwartz et al., 2005). The ability of S. sclerotiorum to form sclerotia contributes to its long-term survival and the persistence of the disease in infested fields (Schwartz et al., 2005). Sclerotia contain nutrient assets and can persevere in the soil for extended periods, acting as a source of primary inoculum for subsequent infections (Schwartz et al., 2005). Under favorable conditions, sclerotia germinate to produce airborne spores that can infect all above-ground parts of the plant, including but not limited to flowers, stems, or pods (Schwartz et al., 2005).
Sclerotinia sclerotiorum employs a multilayered strategy to infect its host plant which involves a series of coordinated events, from sclerotia germination to symptoms development. Initially, S. sclerotiorum produces airborne spores called ascospores from mushroom-like structures called ‘apothecia’ that develop resting sclerotia on infected plant debris (Bolton et al., 2006). Subsequently, the fungus secretes oxalic acid, a pathogenicity factor, to manipulate the pH of the plant cell wall, facilitating enzymatic degradation and tissue invasion (Hegedus and Rimmer, 2005) and suppressing the oxidative burst of the host plant. This acidification process weakens the plant cell wall and offers a favorable environment for fungal cell wall degrading enzymes (CWDEs) to function properly and efficiently to enable the pathogen to breach the physical barrier and penetrate host tissues (Marciano et al., 1983). After penetration, S. sclerotiorum secretes an array of CWDEs, such as polygalacturonase and cellulases, to facilitate its spreading through the infected tissues and causing necrosis. The progression of lesions and mycelial mats contributes to the characteristic symptoms of white mold disease (Hegedus and Rimmer, 2005). Simultaneously, the host plant recognizes the pathogen-associated molecular patterns (PAMPs) through pattern recognition receptors (PRRs), triggering a series of signaling events leading to the activation of defense responses.
Despite decades of dedicated effort and due to the pathogen’s resilience, survival, and adaptability, adequate resistant germplasm is still lacking in common bean particularly, and in other economically important crops in general. Therefore, disease control is tremendously challenging and comprises integrated multifaceted strategies using a mixture of cultural practices, biological control, and chemical fungicides (O’Sullivan et al., 2021). Chemical control is the most valuable tool to manage white mold disease because fungicides can effectively control the spread of the disease, reduce the severity of infections, and minimize yield losses when applied correctly and at the right time. However, extensive use and over-reliance on fungicides can lead to the development of resistant strains of S. sclerotiorum, as well as negatively affect the non-target organisms, soil health, and water quality (Le Cointe et al., 2016; Ceresini et al., 2024). Therefore, the identification of eco-friendly alternatives has become a necessity.
Polyamines (PAs), such as putrescine, spermidine, spermine, and cadaverine, might be a promising alternative against soil-borne phytopathogens to reduce the use of hazardous chemical fungicides entirely or partially (Nehela et al., 2024; Yi et al., 2024). In higher plants, PAs are involved in numerous physiological processes, including, but not limited to, cell division, differentiation, and response to abiotic and biotic stresses (Killiny and Nehela, 2020). They can act as antioxidants and contribute to the scavenging of reactive oxygen species (ROS), maintaining redox homeostasis (Nehela and Killiny, 2023), induction of defense-related genes (Romero et al., 2018), modulation of multiple metabolic pathways (Nehela and Killiny, 2023), regulation of endogenous phytohormones (Nehela and Killiny, 2019), establishment of systemic acquired resistance (SAR), and modulation of the plant-pathogen interactions (Nehela and Killiny, 2020; Asija et al., 2022; Czerwoniec, 2022). It is worth mentioning that the exact mechanism(s) and the definite role of PAs in plant defense vary depending on the plant species, pathogen, and environmental conditions. The most common PAs found in plants are biosynthesized from the alkaline polyamine L-ornithine (Killiny and Nehela, 2020).
L-Ornithine plays several roles in plant growth and development. For instance, in rice (Oryza sativa), previous studies showed that ornithine could be associated with nitrogen reuse (Liu et al., 2018), yield formation, grain quality and aroma (Luo et al., 2020), and response to water stress (Yang et al., 2000). Moreover, exogenous application of L-ornithine significantly enhanced the drought tolerance in sugar beet (Beta vulgaris) (Hussein et al., 2019) and mitigated the salt stress in onion (Allium Cepa) (Çavuşoǧlu and Çavuşoǧlu, 2021) and cashew (Anacardium occidentale) plants (da Rocha et al., 2012). L-Ornithine’s potential role(s) in defending abiotic stress might be due to its involvement in proline accumulation in treated plants. For example, proline metabolism-related genes such as ornithine delta-aminotransferase (δOAT) and proline dehydrogenases (ProDH1 and ProDH2) were reported previously to be involved in the defense against non-host strains of Pseudomonas syringae in Nicotiana benthamiana and Arabidopsis (Senthil-Kumar and Mysore, 2012). On the other hand, fungal ornithine decarboxylase (ODC) is vital for pathogen growth (Singh et al., 2020). Targeting ODC of Fusarium oxysporum f. sp. lycopersici through host-induced gene silencing (HIGS) significantly enhanced the resistance of tomato plants against fusarium wilt disease (Singh et al., 2020). However, the potential role of the exogenous application of ornithine against biotic stress such as phytopathogens is inadequately reported. More importantly, the impacts of ornithine on disease resistance and its associated biochemical and physiological events remained largely unexplored.
Understanding the intricacies of S. sclerotiorum infection in common bean plants is pivotal for developing effective management strategies. In the current study, we aim to unveil the potential role of the diamine, L-ornithine, as a key factor in enhancing the common bean’s defense mechanisms and resilience against S. sclerotiorum. We hypothesize that the application of L-ornithine plays a pivotal role in maintaining the redox status, in addition to boosting the defense responses of infected plants. We believe that the potential role of L-ornithine is correlated with the regulation of enzymatic and non-enzymatic antioxidant defense machinery, as well as interfering with the fungal pathogenicity/virulence factors and their associated proteins. This dual function makes L-ornithine a promising candidate for sustainable strategies to mitigate the impact of white mold disease and fortify the resilience of common bean plants in the face of this formidable fungal pathogen. The current study provides insights that may lead to innovative eco-friendly approaches for controlling white mold disease and mitigating its impact on bean cultivation.
2 Materials and methods
2.1 Plant material and treatments
Throughout the current study, the sensitive commercial cultivar Giza 3 of common bean (Phaseolus vulgaris L. cv. Giza 3) was used as an experimental plant material. Healthy seeds were generously provided by the Department of Food Legumes Research, Field Crops Research Institute (FCRI), Agricultural Research Center (ARC), Egypt. Five seeds were sown in plastic pots (35 cm inner diameter and 50 cm in depth) filled with S. sclerotiorum-infested soil under greenhouse conditions (25 ± 2°C, 75 ± 1% RH, and 8 h light/16 h dark photoperiod). However, 7-10 days post-sowing (dps), seedlings were thinned and only two uniform seedlings with fully expanded trifoliate leaves were left in each pot. All pots were irrigated every other week and fertilized monthly according to the recommendations for this cultivar.
To prepare a 500 mg/L stock solution of the diamine L-ornithine (also known as (+)-(S)-2,5-Diaminovaleric acid; Sigma-Aldrich, Darmstadt, Germany), 50 mg was initially dissolved in a final volume of 100 mL sterilized distilled water. Subsequently, the stock solution has been diluted and used for subsequent experiments. Briefly, six serial concentrations of L-ornithine (12.5, 25, 50, 75, 100, and 125 mg/L) were tested in vitro individually. Besides, sterilized distilled water was used as a negative control (Mock), whereas the commercial fungicide ‘Rizolex-T’ 50% WP (Tolclofos-Methyl 20% + Thiram 30%; KZ–Kafr El Zayat Pesticides and Chemicals, Kafr El-Zayat, Gharbia, Egypt) was used as a positive control. Five concentrations of the commercial fungicide ‘Rizolex-T’ were tested in vitro (2, 4, 6, 8, and 10 mg/L).
2.2 Pathogen isolation and identification
Samples of common bean stems and pods showing typical symptoms of white mold (disease severity: 10-30%) were collected from commercial farms. While most of the infected plant materials were identified by variety/cultivar (the sensitive commercial cultivar Giza 3), others, particularly those obtained from local markets, were of unknown variety. Initially, collected infected materials were surface sterilized with 0.5% NaOCl for three min, then washed several times using sterilized distilled water, and dried between sterilized filter papers to remove the extra water. Subsequently, infected organs were chopped into small pieces from the intermediate tissues (between healthy and infected ones), cultured on potato dextrose agar (PDA) medium, and incubated at 25 ± 2°C for 5 days under 12-hour light/12-hour dark cycles to encourage sclerotia formation. The hyphal tips method was also used to purify the fungal isolates from the mixed or contaminated cultures. The purified fungal isolates were first identified based on their cultural morphology characteristics, and then the identification was confirmed using microscopic features as S. sclerotiorum. Finally, the pathogenicity of all purified isolates was tested on the sensitive commercial cultivar Giza 3 of common bean to fulfill Koch’s postulates.
Moreover, the identification of the most aggressive isolate of S. sclerotiorum (isolate #3) was further confirmed based on the sequencing of the Internal transcribed spacer (ITS) region as described by (White et al., 1990; Baturo-Ciesniewska et al., 2017). Briefly, the isolate was grown on potato dextrose broth (PDB) and incubated at 25 ± 2°C for 5-7 days. Then, the fungal mycelium was harvested, filtered through cheesecloth, washed twice with sterile water, and dried using sterile filter paper. The genomic DNA was extracted using a Quick-DNA™ Fungal/Bacterial Miniprep Kit (Kuramae-Izioka, 1997; Atallah et al., 2022, 2024). Subsequently, ITS of the rDNA was amplified using a specific primer pair ITS1/ITS4 (TCCGTAGGTGAACCTGCGG TCCTCCGCTTATTGATATGC; Expected size: 540 bp) (Baturo-Ciesniewska et al., 2017). The purified PCR product was sent for sequencing (Aoke Dingsheng Biotechnology Co., Beijing, China). Sanger sequencing was used to perform the two-directional sequencing of the ITS rDNA sequence. Subsequently, a Nucleotide-Nucleotide Basic Local Alignment Search Tool (BLASTn) was used to compare the assembled query sequence with the most recent available data in GenBank and the National Center for Biotechnology Information website (NCBI, http://www.ncbi.nlm.nih.gov/gene/). Alignment was carried out using ClustalW through the Molecular Evolutionary Genetics Analysis software package (MEGA-11; version 11) (Kumar et al., 2024) to align the query sequence with other 20 Sclerotinia strains/isolates retrieved from the recently available data in NCBI GenBank (Supplementary Table S1). Evolutionary analysis was inferred using the Maximum Likelihood method and General Time Reversible model (Nei and Kumar, 2000) of nucleotide substitutions, and the tree with the highest log likelihood was shown. The initial tree for the heuristic search was selected by choosing the tree with the superior log-likelihood between a Neighbor-Joining (NJ) tree (Kumar et al., 2024) and a Maximum Parsimony (MP) tree. The NJ tree was generated using a matrix of pairwise distances computed using the General Time Reversible model (Nei and Kumar, 2000).
2.3 In vitro antifungal activity of L-ornithine against S. sclerotiorum
The antifungal activities of L-ornithine, as well as the ‘Rizolex-T’ fungicide, were tested in vitro using the agar diffusion method. Briefly, a proper volume of L-ornithine stock solution (500 mg/L) was finely mixed with 10 mL PDA medium to obtain the final concentration of 12.5, 25, 50, 75, 100, and 125 mg/L. Moreover, five concentrations of ‘Rizolex-T’ fungicide (2, 4, 6, 8, and 10 mg/L) and sterilized distilled water were used as controls. After media solidification, a 4 mm-diameter mycelial plug of freshly prepared culture of S. sclerotiorum was transferred into the middle of the petri dish, incubated at 25 ± 2°C until the mycelial growth covered the whole control plate, then the fungal growth was recorded. The percentage of radial growth inhibition of S. sclerotiorum was calculated using Equation 1:
The experiment was repeated twice with six biological replicates per control/treatment and five pots (two plants per pot) for each biological replicate. Each biological replicate was analyzed twice (two technical replicates) to ensure the accuracy, reliability, and reproducibility of our experimental findings. Additionally, the half-maximal inhibitory concentration (IC50) and IC99 were calculated using probit regression analysis (Prentice, 1976).
2.4 Greenhouse experiments
Two successive pot experiments were carried out to evaluate the potential of L-ornithine under greenhouse conditions. Briefly, pots were filled with sterilized clay-sandy soil (3:1) and inoculated with a freshly prepared culture of S. sclerotiorum. Initially, actively growing cultures of the most aggressive isolate of S. sclerotiorum (isolate #3) were produced by bisecting a sclerotium, placing it face down on PDA, and incubating at 25°C for 4 days under continuous darkness (24 h) to encourage mycelial growth. Then, four 5-mm diameter agar plugs from the leading edge were used to inoculate 100 g of a sterile mixture of wheat and rice bran (1:1, v/v), then all flasks were incubated at 25 ± 2°C for 5 days under 12-hour light/12-hour dark cycles to encourage sclerotia formation. Prior to the soil inoculation, all flasks were thoroughly mixed to ensure homogeneity. Subsequently, 100 g of the colonized bran mixture was used to inoculate each pot to ensure consistent pathogen concentration. Inoculated pots were directly watered to activate fungal growth and left under greenhouse conditions for 7 days.
Later, five seeds of the cultivar Giza 3 were sown in each pot. For L-ornithine- and ‘Rizolex-T’ fungicide-treated pots, disinfected seeds were initially soaked in aqueous solutions of both compounds at a final concentration of the IC99 (about 250 mg/L, and 50 mg/L, respectively) for two hours, then air-dried for one-hour before sowing. On the other hand, seeds were soaked in sterilized distilled water as a negative control. At 10 dps and right before the first irrigation, seedlings were thinned and only two uniform seedlings were left in each pot. Furthermore, to warrant the infection with S. sclerotiorum stems of bean plants at the same developmental stage (10 dps) were cut in two different positions using a sterilized scalpel, and approximately 0.5 g of the colonized bran mixture was placed into each wound, then a high level of humidity was maintained to encourage infection and disease development for all inoculated plants. A similar wounding procedure was performed on the control plants and the same amount (0.5 g) of the sterile non-colonized bran mixture was placed into the wounds and maintained under high humidity conditions to simulate the environment for disease development, ensuring consistency between the treatments.
For treatments, bean seedlings were watered via soil drench with 500 mL of aqueous solutions of L-ornithine (250 mg/L) or ‘Rizolex-T’ fungicide (50 mg/L), then the treatment was repeated three times in total at 10 days intervals. Mock-treated control was irrigated with 500 mL of sterilized distilled water. All treatments were maintained under the greenhouse conditions (25 ± 2°C, 75 ± 1% RH, and 8 h light/16 h dark photoperiod). All pots were irrigated every other week and fertilized monthly using a balanced NPK fertilizer (20-20-20, in addition to 3.6% Sulfur and Microelements TE; Zain Seeds, Egypt) at a concentration of 3-4 g/L, applied via foliar spray according to the recommendations for this cultivar and the manufacturer’s instructions. Unless otherwise mentioned, the fully expanded, mature leaves (2nd and 3rd leaves from the top) were collected at 72 hours post-treatment (hpt) from each biological replicate, homogenized, mixed, and stored at −80°C for further analyses including, but not limited to, in situ histochemical localization of oxidative stress indicators, lipid peroxidation, enzymatic and non-enzymatic antioxidants, and gene expression.
2.5 Assessment of white mold disease and vegetative parameters
The severity of white mold disease was evaluated weekly till 21 days post-inoculation (dpi) using a 1-9 scale (Supplementary Table S2) according to Petzoldt and Dickson scale (Petzoldt and Dickson, 1996) as modified by Terán et al. (2006). Briefly, the stem and branches of the bean plant were examined starting from the inoculated site and observed for lesion progression along the internodes and nodes. Then, the distance of the lesion from the point of inoculation to its furthest point along the stem or branch was measured and a 1-9 score (based on the lesion position) was assigned, where a score (1) was assigned for no visible infection near the inoculant site, whereas scores (2-9) were assigned to progressively increasing lesion size and advancement across nodes/internodes (Supplementary Table S2). Subsequently, the severity of white mold disease was converted into a percentage using Equation 2:
Moreover, the area under the disease progress curve (AUDPC) was calculated according to (Shaner and Finney, 1977) which was recently adapted for the white mold disease in the common bean (Chauhan et al., 2020) using Equation 3:-
Where, Yi = Disease severity at time ti, Yi+1 = Disease severity at the next time point ti+1, ti = Time of the first measurement (in days), ti+1 = Time of the next measurement (in days), and n = Total number of time points or observations. Vegetative parameters of bean plants including plant height (cm), number of branches per plant, and number of leaves per plant were recorded weekly for 21 days for all biological replicates.
2.6 Photosynthetic pigment contents
Leaf samples (2nd and 3rd fully developed leaves from the top) were collected at 45 dps (15 days post-last treatment) from each biological replicate. Each biological replicate contains five pots (two plants per pot). Approximately 500 mg of ground tissues were used to extract the photosynthetic pigments (chlorophyll a, chlorophyll b, and carotenoids) using 80% acetone in the dark at 4°C. After 24 hours, samples were centrifuged and the supernatant was collected and the chlorophyll a, chlorophyll b, and carotenoids contents were colorimetrically assessed by measuring the absorbance at three different wavelengths (A470, A646, and A663 nm) using a UV-160A spectrophotometer (Shimadzu, Japan) as described by (Lichtenthaler, 1987). Finally, the content of photosynthetic pigments was calculated using the following Equations 4–6 as described by Lichtenthaler (1987).
2.7 Oxidative stress indicators
2.7.1 In situ histochemical localization of hydrogen peroxide (H2O2) and superoxide anion (O2•−)
Leaves (2nd and 3rd fully developed leaves from the top) were collected from each biological replicate at 72 hours post-treatment (hpt) for in situ histochemical localization of hydrogen peroxide (H2O2) and superoxide anion (O2•−). Each biological replicate contains five pots (two plants per pot). Each biological replicate was analyzed twice (two technical replicates) to ensure the accuracy, reliability, and reproducibility of the method. H2O2 and O2•− were determined using 0.1% 3,3′-Diaminobenzidine (DAB; Sigma–Aldrich, Darmstadt, Germany) or nitro blue tetrazolium (NBT; Sigma–Aldrich, Darmstadt, Germany) as described by Romero-Puertas et al. (2004) and Adám et al. (1989), respectively, with slight modifications. For in situ histochemical localization of H2O2, leaflets were vacuum-infiltrated with 0.1% DAB in 10 mM tris buffer (pH 7.8), then incubated under light at room temperature for 60 min. Leaflets were bleached in 0.15% (w/vol) TCA in ethanol: chloroform 4:1 (v/v) (Al-Gomhoria Company for medicines and medical supplies, Cairo, Egypt), then illuminated until the development of brown color. Likewise, leaflets were vacuum-infiltrated with 10 mM potassium phosphate buffer (pH 7.8) containing 0.1 w/v % NBT for in situ histochemical localization of O2•−. Leaflets were incubated under light at room temperature for 20 min before being bleached as described above, then illuminated until the appearance of dark blue/purple spots. The intensity of the developed brown color (as an indicator of H2O2) or blue/purple color (as an indicator of O2•−) was evaluated using the Fiji version of ImageJ image processing package (http://fiji.sc; accessed on 7 March 2024).
2.7.2 Assessment of lipid peroxidation
Malondialdehyde (MDA; as a lipid peroxidation marker) was assessed as described by Du and Bramlage (1992) with slight modification. Leaves (2nd and 3rd fully developed leaves from the top) were collected from each biological replicate at 72 hours post-treatment (hpt). Each biological replicate contains five pots (two plants per pot). Each biological replicate was analyzed twice (two technical replicates) to ensure the accuracy, reliability, and reproducibility of the method. Briefly, 0.5 g of ground leaf tissue was used to extract MDA using 20% trichloroacetic acid (TCA; MilliporeSigma, Burlington, MA, USA) containing 0.01% butyl hydroxyl toluene (BHT; Sigma–Aldrich, Saint Louis, MO, USA). Subsequently, MDA was colorimetrically determined in the supernatants by measuring the absorbance at 532 and 600 nm using a UV-160A spectrophotometer (Shimadzu, Japan), then expressed as nmol g−1 FW.
2.8 Non-enzymatic antioxidants
For the assessment of non-enzymatic and enzymatic antioxidants, leaves (2nd and 3rd fully developed leaves from the top) were collected from each biological replicate at 72 hours post-treatment (hpt). Each biological replicate contains five pots (two plants per pot). Each biological replicate was analyzed twice (two technical replicates). Both leaves were ground together using liquid nitrogen and subjected directly for the analysis of enzymatic, and non-enzymatic antioxidants, total amino acids, proline content, gene expression, and the quantification of oxalic acid.
2.8.1 Total soluble phenolics
Total soluble phenolics were determined using the Folin-Ciocalteu reagent (Sigma–Aldrich, Saint Louis, MO, USA) as previously described by Kähkönen et al. (1999) with minor modifications. Briefly, about 0.1 g of homogenized leaf tissues were extracted using 20 mL of 80% methanol for 24 h in the dark, centrifuged, and the supernatant was collected. For the reaction mixture, 0.1 mL of the sample extract was mixed with 0.5 mL of the Folin-Ciocalteu reagent (10%), vortexed for 30 seconds, and let stand for 5 minutes in the dark. Subsequently, 0.5 mL of 20% sodium carbonate (Na2CO3; Al-Gomhoria Company for medicines and medical supplies, Cairo, Egypt) solution was added to each tube, mixed thoroughly, then incubated in the dark for one hour at room temperature. After incubation, the absorbance of the reaction mixture was measured at 765 nm using a UV-160A spectrophotometer (Shimadzu, Japan). A calibration curve of gallic acid (Fisher Scientific, Hampton, NH, USA) was used to determine the concentration of total soluble phenolics in the sample extracts, then expressed as mg of gallic acid equivalents per gram of fresh weight (mg GAE g−1 FW).
2.8.2 Total soluble flavonoids
Total soluble flavonoids were determined using Djeridane’s method (Djeridane et al., 2006) with slight modifications. Briefly, 0.3 ml of methanolic extract as described above was mixed with 0.3 of 5% aluminum chloride (AlCl3; Fisher Scientific, Hampton, NH, USA) solution, vigorously mixed, then incubated for 5 min at room temperature, then 0.3 mL of 10% potassium acetate (Al - Gomhoria Company for medicines and medical supplies, Cairo, Egypt) solution was added, mixed thoroughly, and incubated at room temperature for 30 minutes in the dark. After incubation, the absorbance of the reaction mixture was measured at 430 nm using a UV-160A spectrophotometer (Shimadzu, Japan). A calibration curve of rutin (TCI America, Portland, OR, USA) was used to determine the concentration of total soluble flavonoids in the sample extracts, then expressed as mg of rutin equivalents per gram of fresh weight (mg RE g−1 FW).
2.9 Total amino acids and proline content
The total free amino acids in bean leaves were determined using a modified ninhydrin (Thermo Scientific Chemicals, Waltham, MA, USA) reagent, as described by Yokoyama and Hiramatsu (2003) and improved by (Sun et al., 2006). Briefly, 0.1 g of ground tissues were extracted with a pH 5.4 buffer, and then 200 μL of the supernatant was reacted with 200 μL of ninhydrin (2%) and 200 μL of pyridine (10%; Spectrum Chemical, New Brunswick, NJ, USA), incubated for 30 min in a boiling-water water bath, then cooled, and measured at 580 nm using a UV-160A spectrophotometer (Shimadzu, Japan). On the other hand, proline was determined using Bates’s method (Bates et al., 1973). Proline was extracted with 3% sulfosalicylic acid (Thermo Scientific Chemicals, Waltham, MA, USA), centrifuged, and then 0.5 mL of the supernatant was mixed with one mL of glacial acetic acid (Fisher Scientific, Hampton, NH, USA) and ninhydrin reagent, then incubated at 90°C for 45 min, then cooled, and measured at 520 nm using the same spectrophotometer described above. Calibration curves of glycine and proline (Sigma–Aldrich, Saint Louis, MO, USA) standards were used to determine the total free amino acids and proline content, respectively, in the leaves extract and expressed as mg g−1 FW.
2.10 Antioxidant enzymes activity
To determine the enzymatic activity of antioxidant-associated enzymes, about 500 mg of homogenized tissues were extracted using 3 mL of 50 mM Tris buffer (pH 7.8) containing 1 mM EDTA-Na2 (Sigma–Aldrich, Saint Louis, MO, USA) and 7.5% polyvinylpyrrolidone (PVP; Sigma–Aldrich, Saint Louis, MO, USA), centrifuged at 10000× g for 20 min under cooling (4°C), and the supernatant (crude enzyme extract) was collected (El-Nagar et al., 2023; Osman et al., 2023). Subsequently, the enzymatic activity of catalase (CAT) was assessed according to the method of Aebi (1984) with slight modifications (El-Nagar et al., 2023; Osman et al., 2023) after the reaction with 2 mL of 0.1 M sodium phosphate buffer (pH 6.5; Sigma–Aldrich, Saint Louis, MO, USA) and 100 μL of 269 mM H2O2 solution. The enzymatic activity of guaiacol-dependent peroxidases (POX) was determined using the method of Harrach et al. (2008) with slight modifications (El-Nagar et al., 2023; Osman et al., 2023) after the reaction with 2.2 mL of 100 mM sodium phosphate buffer (pH 6.0), 100 µL guaiacol (TCI chemicals, Portland, OR, USA), and 100 µL of 12 mM H2O2. Whereas the enzymatic activity of polyphenol oxidase (PPO) was determined according to the method of Malik and Singh (1980) with slight modifications (El-Nagar et al., 2023; Osman et al., 2023). After the reaction with 3 mL catechol (Thermo Scientific Chemicals, Waltham, MA, USA) solution (0.01 M), freshly prepared in 0.1 M phosphate buffer (pH 6.0). CAT activity was measured by following the decomposition of H2O2 at 240 nm (A240), POX activity was measured by following the increase in the absorption at 436 nm (A436), whereas PPO activity was measured by recording the fluctuations in the absorbance at 495 nm (A495) every 30 seconds for 3 min using a UV-160A spectrophotometer (Shimadzu, Japan).
2.11 Gene expression of antioxidant-associated genes of common bean
The transcript levels of three antioxidant-associated genes, including peroxisomal catalase (PvCAT1; GenBank accession number: KF033307.1), superoxide dismutase (PvSOD; GenBank accession number: XM_068639556.1), and glutathione reductase (PvGR; GenBank accession number: KY195009.1) were assessed from common bean leaves (2nd and 3rd fully developed leaves from the top) at 72 hpt after last treatment using real-time RT-qPCR. Briefly, RNA was extracted using a Simply P Total RNA Extraction Kit (catalog number BSC52S1; BioFlux, Bioer technology, China), according to the manufacturer’s procedure. Then, cDNA was synthesized using a TOP script™ cDNA Synthesis Kit as described in the manufacturer’s protocol. The primer sequences for the three genes mentioned above are listed in Supplementary Table S3. PvActin-3 (GenBank accession number: XM_068616709.1) was used as a housekeeping gene, and the 2−ΔΔCT method was used for the calculation of relative gene expression (Livak and Schmittgen, 2001). The stability of Actin was confirmed previously under biotic (incompatible interaction between common bean and fungus Colletotrichum lindemuthianum) as well as abiotic (drought; salinity; cold temperature) stresses (Borges et al., 2012).
2.12 In silico analysis of oxaloacetate acetylhydrolase (SsOAH) protein from S. sclerotiorum
Initially, genome-wide in silico analysis of oxaloacetate acetylhydrolase (OAH) protein in S. sclerotiorum was carried out using the protein-protein BLAST (BLASTp 2.15.0+) tool (Altschul et al., 1997, 2005). Briefly, OAH from Aspergillus fijiensis CBS 313.89 (AfOAH; taxid:1191702; GenBank Accession No. XP_040799428.1; 342 aa) and Penicillium lagena (PlOAH; taxid:94218; GenBank Accession No. XP_056833920.1; 316 aa) were used as query sequences to match their homologous proteins from S. sclerotiorum (taxid:5180). BLASTp was carried out based on recently available data about the S. sclerotiorum genome on GenBank, National Center for Biotechnology Information website (NCBI, http://www.ncbi.nlm.nih.gov/gene/).
Moreover, evolutionary analysis and phylogenetic tree of predicted OAH genes from S. sclerotiorum (SsOAH), as well as AfOAH from A. fijiensis CBS 313.89 and PlOAH from P. lagena were inferred by using the Maximum Likelihood method and JTT matrix-based model (Jones et al., 1992) in MEGA11 (Tamura et al., 2021). The phylogenetic tree was combined with multiple protein sequence alignment analysis of all predicted OAH genes from S. sclerotiorum (SsOAH), as well as the query sequences using Constraint-Based Alignment Tool (COBALT; for multiple protein sequences (https://www.ncbi.nlm.nih.gov/tools/cobalt/re_cobalt.cgi) (Papadopoulos and Agarwala, 2007). Furthermore, ClustalW (http://www.genome.jp/tools-bin/clustalw) was used to align the top-matched AA sequences of SsOAH from S. sclerotiorum with the query sequences (AfOAH and PlOAH) (Larkin et al., 2007) and ESPript tool (version 3.0; https://espript.ibcp.fr/ESPript/ESPript/index.php) was used to visualize conserved regions in the alignment.
Additionally, functionally representative domains and conserved sites of the predicted SsOAH from S. sclerotiorum were interactively classified into families using the InterPro tool (https://www.ebi.ac.uk/interpro/) (Blum et al., 2021). Finally, three-dimensional (3D) structure modeling of predicted SsOAH from S. sclerotiorum was carried out using the Protein Homology/analogy Recognition Engine (Phyre2-server version 2.0; http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index) (Kelley et al., 2015), then confirmed using the SWISS-MODEL server (https://swissmodel.expasy.org/) (Biasini et al., 2014). The predicted 3D structures (PDB format) were interactively visualized using the UCSF-Chimera package (version 1.15; https://www.cgl.ucsf.edu/chimera/) (Pettersen et al., 2004).
2.13 Gene expression of oxaloacetate acetylhydrolase (SsOAH) from S. sclerotiorum
The transcript levels of oxaloacetate acetylhydrolase (SsOAH; GenBank accession number: XM_001590428.1) were assessed from S. sclerotiorum mycelia using real-time RT-qPCR. Briefly, S. sclerotiorum was inoculated into PDB-containing flasks and incubated on an incubator shaker (Model I2400, New Brunswick Scientific Co., Edison, NJ, USA) at 150 rpm for 24 hours at 25 ± 2°C under continuous darkness (24 h) to encourage mycelial growth, then treated with L-ornithine- and ‘Rizolex-T’ fungicide at a final concentration of the IC50 (about 40, and 3.2 mg/L, respectively), then the incubation was continued at the same conditions for another 24 hours. After incubation, cultures were centrifuged at 2500 rpm for 5 min and the supernatant (fungal mycelia) was collected for gene expression analysis. Likewise, fungal mycelia were collected at 0, 24, 48, 72, 96, and 120 hpt from infected plants that produced a white mold that forms cottony mycelium on the surface of infected tissue. RNA was extracted from the fungal mycelium, and then cDNA was synthesized as described above. The primer sequences for SsOAH are listed in Supplementary Table S3. SsActin (GenBank accession number: XM_001589919.1) was used as a housekeeping gene, and the 2−ΔΔCT method was used for the calculation of relative gene expression (Livak and Schmittgen, 2001).
2.14 Quantification of oxalic acid
Oxalic acid was determined in both potato dextrose broth (PDB) containing the fungal pathogen, S. sclerotiorum, as well as the plant samples according to the method of Xu and Zhang (2000) with slight modifications. Briefly, S. sclerotiorum isolates were inoculated into PDB-containing flasks and then incubated on an incubator shaker (Model I2400, New Brunswick Scientific Co., Edison, NJ, USA) at 150 rpm for 3-5 days at 25 ± 2°C under continuous darkness (24 h) to encourage mycelial growth. After incubation, the fungal cultures were initially filtered through Whatman filter paper No. 1, then centrifuged at 2500 rpm for 5 min to remove any mycelial debris. The supernatant was collected and stored at 4°C for further quantification of oxalic acid. Approximately 0.1 g of ground tissues were extracted three times (2 ml each) using distilled water to prepare the plant sample. Subsequently, samples were centrifuged at 2500 rpm for 5 min and the supernatant was filtered dryly through Whatman No. 1 paper, then collected for further analysis.
For oxalic acid quantification, the reaction mix was prepared in a test tube fitted with a glass stopper in the order of 0.2 mL of sample (or PDB culture filtrate or standard oxalic acid solution), 0.11 ml of bromophenol blue (BPB, 1 mM; Fisher Chemical, Pittsburgh, PA, USA), 0.198 ml of 1 M sulfuric acid (H2SO4; Al - Gomhoria Company for medicines and medical supplies, Cairo, Egypt), and 0.176 ml of 100 mM potassium dichromate (K2Cr2O7; TCI chemicals, Portland, OR, USA), then the solution was made up to the mark using 4.8 ml of distilled water, vigorously mixed, and placed in the water bath immediately at 60°C. After 10 minutes, the reaction was quenched by adding 0.5 ml sodium hydroxide solution (NaOH; 0.75 M). The absorbance of the reaction mix was measured at 600 nm (A600) using a UV-160 spectrophotometer (Shimadzu, Japan). PDB and distilled water were used as a blank control for quantification in culture filtrate or plant samples, respectively. A calibration curve of oxalic acid (Thermo Scientific Chemicals, Waltham, MA, USA) was used to determine the concentration of oxalic acid in the culture filtrate and expressed as μg oxalic acid per mL PDB medium (μg.mL−1) as well as in the leaves extract which was expressed as μg oxalic acid per gram fresh weight (μg.g−1 FW).
2.15 Statistical analyses
Throughout the study, unless otherwise stated, all experiments were arranged in a completely randomized design (CRD) with six biological replicates per treatment and five pots (two plants per pot) for each biological replicate. Biological replicates were analyzed twice (two technical replicates). Technical replicates were used to test the reproducibility within the same experiment, but they were not used in the statistical analysis to avoid the possibility of pseudoreplication. Data were statistically analyzed using the analysis of variance (ANOVA), followed by the Tukey-Kramer honestly significant difference (HSD) test (p ≤ 0.05). For the in vitro experiments, probit analysis was used to calculate the IC50 and IC99 with 95% confidence intervals.
3 Results
3.1 Characterization of S. sclerotiorum isolates from common bean plants
A total of 4 isolates were collected from different bean fields in El-Gharbia Governorate, Egypt. On the PDA medium, all isolates developed creamy white mycelia that rapidly became cottony white (Figure 1A), then turned beige or brownish at the stage of sclerotia formation. Generally, the sclerotia were hard-bodied, black-colored, globose to irregular in shape, and measured 5.2 to 7.7 mm in length and 3.4 to 5.3 mm in diameter (Figure 1B). Although the four isolates showed a peripheral sclerotia pattern that developed at the edge of the culture media 10-12 days post incubation at 25 ± 2°C (Figure 1A), significant differences were observed in the number of sclerotia per plate between them (P < 0.001) with superiority for isolate #3 (32.33 ± 1.53 sclerotia per plate; Figure 1C). Likewise, isolate #3 produced more oxalic acid in PDB than other isolates (3.33 ± 0.49 μg.mL−1; Figure 1D). Isolate #3 showed typical morphological and microscopic characteristics with the phytopathogenic fungus S. sclerotiorum. For instance, on PDA, colonies of isolate #3 were fast growing with milky white (Figure 1A), and reverse beige or little salmon-buff color and took 6-7 days to fully cover the surface of a 9-cm Petri dish when incubated at 25 ± 2°C. Collectively, isolate #3 was identified as S. sclerotiorum based on the above-mentioned morphological and microscopic features.
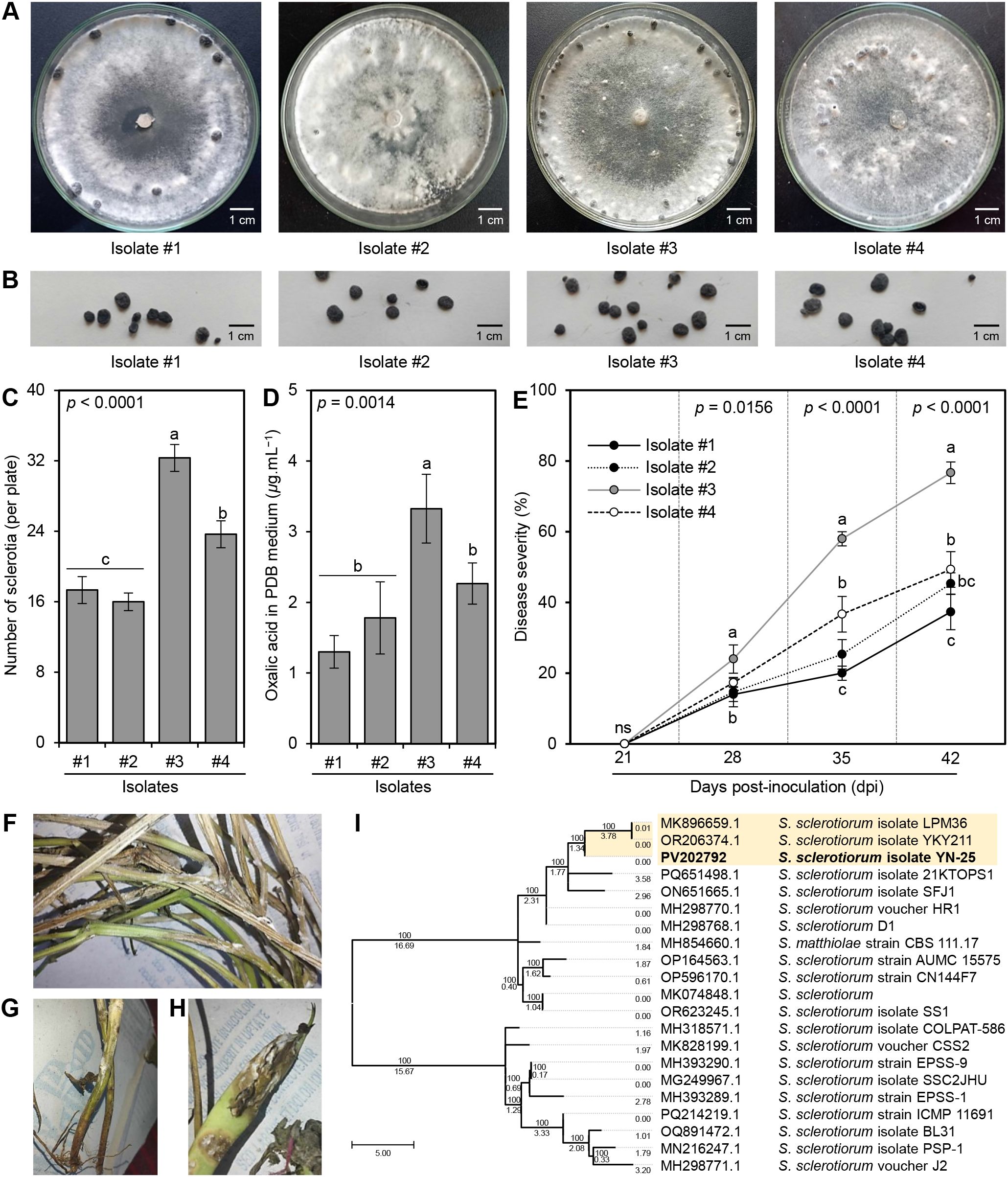
Figure 1. Characterization and Pathogenicity of S. sclerotiorum isolates from common bean plants. (A) Mycelial growth of four S. sclerotiorum isolates on PDA media at sclerotia formation stage, (B) Sclerotia of four S. sclerotiorum isolates, (C) Number of sclerotia (per plate), (D) Oxalic acid secretion in PDB media (μg.mL−1), and (E) Disease severity (%) of four S. sclerotiorum isolates on the sensitive commercial cultivar Giza 3 of common bean under greenhouse conditions. Values represent the means ± standard deviation (means ± SD) of five biological replicates (n = 5). Different letters indicate statistically significant differences between treatments (p < 0.05). (F–H) Typical white mold symptoms on above-ground parts stems, and pods, respectively, after 10 days post-inoculation (dpi) with isolate #3. (I) Evolutionary analysis by the Maximum Likelihood method of the internal transcribed spacer (ITS) region of S. sclerotiorum isolate #3 in comparison to 20 reference isolates/strains retrieved from The National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/). Numbers above the cluster lines indicate the site coverage (%), whereas, numbers below the cluster lines indicate the branch length.
Moreover, to confirm pathogenicity, Koch’s postulates were fulfilled using the four obtained isolates of S. sclerotiorum to inoculate the sensitive commercial cultivar Giza 3 of common bean under greenhouse conditions (Figure 1E). Although all obtained fungal isolates were pathogenic and were able to infect common bean (cv. Giza 3) producing typical white mold symptoms on all above-ground parts (Figure 1F), particularly stems (Figure 1G) and pods (Figure 1H) after 10 days post inoculation (dpi), isolate #3 was the most aggressive isolate in two separate experiments. Isolate #3 had the highest disease severity (%) on bean plants (24.0 ± 4.0, 58.0 ± 2.0, and 76.7 ± 3.1 at 7, 14, and 21 dpi, respectively; Figure 1F).
The identification of the most aggressive S. sclerotiorum isolate #3 was further confirmed based on the sequencing of the Internal transcribed spacer (ITS) region (Figure 1I). The phylogenetic analysis between isolate #3 and 20 reference isolates/strains showed high similarity (over 99%) between them. It is worth mentioning that S. sclerotiorum – isolate #3 (533 bp) showed high similarity with the American isolate S. sclerotiorum isolate LPM36 (GenBank accession No. MK896659.1; 540 bp) isolated from the dry pea seed and the Chinese isolate S. sclerotiorum isolate YKY211 (GenBank accession No. OR206374.1; 548 bp) the causal agent of stem rot of stock (Matthiola incana) which all were clustered separately at the top of the dendogram (Figure 1I). The new sequence was deposited in the NCBI database and named ‘S. sclerotiorum - isolate YN-25’ (GenBank Accession No. PV202792). Accordingly, it was noticeable that isolate #3 was the most aggressive isolate; consequently, it was chosen for all subsequent experiments.
3.2 In vitro antifungal activity of L-ornithine
The antifungal activity of serial concentrations of (12.5, 25, 50, 75, 100, and 125 mg/L) of the diamine, L-ornithine (Sigma-Aldrich, Darmstadt, Germany), was examined in vitro against S. sclerotiorum – isolate #3. It is worth mentioning that L-ornithine showed fungistatic action and progressively inhibited the mycelial radial growth of S. sclerotiorum in a dose-dependent manner (Figures 2A, B). At the highest tested concentration (125 mg/L), L-ornithine had the highest mycelial growth inhibition (99.62 ± 0.27%; Figure 2B) which was equivalent to the highest tested concentration (10 mg/L) of the commercial fungicide ‘Rizolex-T’ (99.45 ± 0.39% inhibition; Figure 2C), suggesting similar potency.
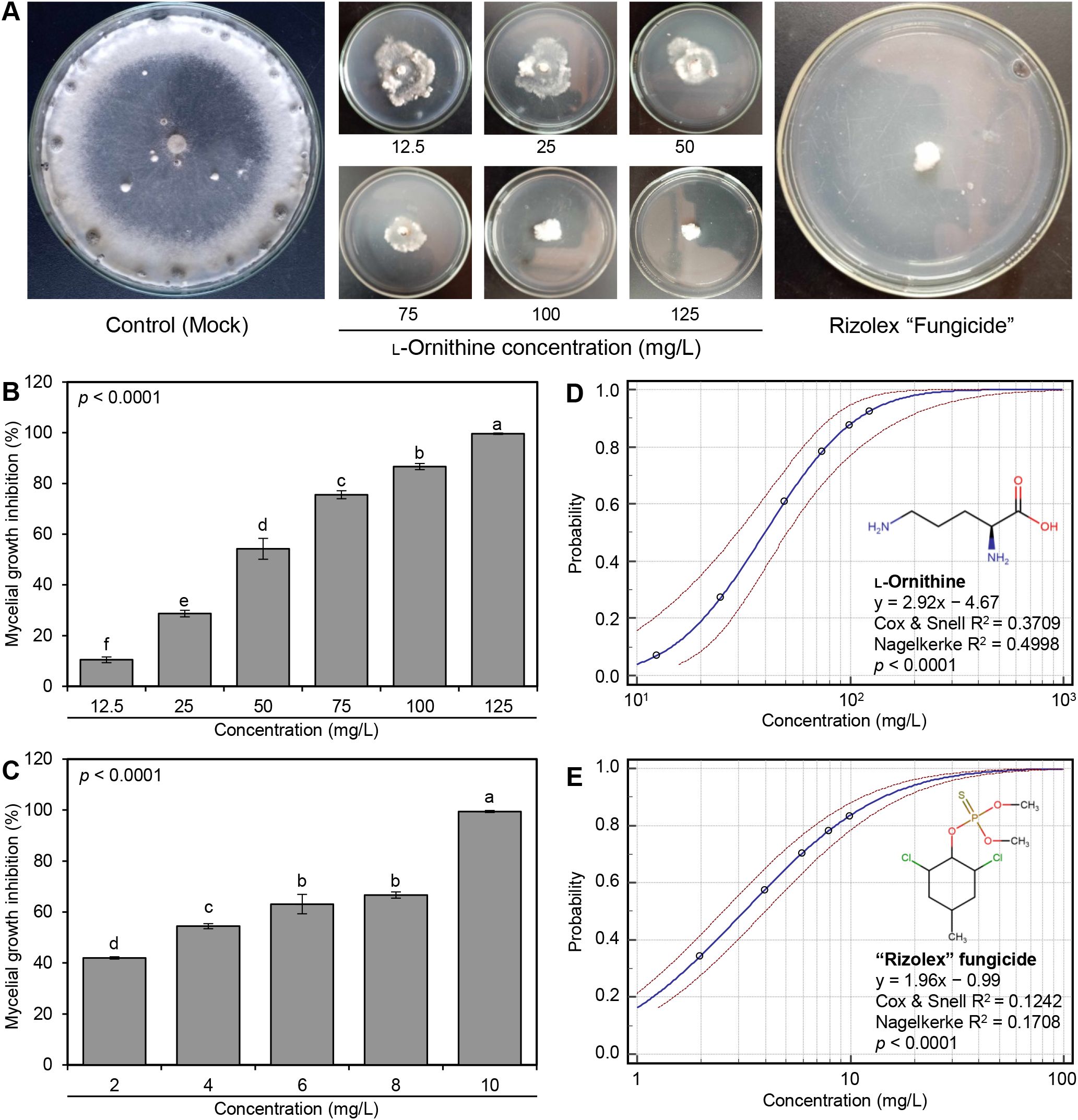
Figure 2. In vitro antifungal activity of L-ornithine against S. sclerotiorum. (A) Antifungal activity of serial concentrations of L-ornithine against S. sclerotiorum in comparison with the commercial fungicide ‘Rizolex-T’ (10 mg/L). (B, C) Mycelial growth inhibition (%) of S. sclerotiorum after the treatment with different concentrations of L-ornithine (12.5, 25, 50, 75, 100, and 125 mg/L) or ‘Rizolex-T’ fungicide (2, 4, 6, 8, and 10 mg/L), respectively. Values represent the means ± standard deviation (means ± SD) of five biological replicates (n = 5). Different letters indicate statistically significant differences between treatments (p < 0.05). (D, E) Probit regression analysis of L-ornithine and the commercial fungicide ‘Rizolex-T’, respectively. The probit regression lines are presented as blue solid lines, whereas the confidence intervals (95%) are edged with dotted red lines.
Moreover, the probit regression analysis was carried out and its associated plots are presented in Table 1 and Figures 2D, E. Briefly, the acceptable slope value of L-ornithine (y = 2.92x − 4.67) and its associated significant statistics (Cox & Snell R2 = 0.3709, Nagelkerke R2 = 0.4998, and p < 0.0001; Figures 2D) compared with the commercial fungicide ‘Rizolex-T’ (y = 1.96x − 0.99, Cox & Snell R2 = 0.1242, Nagelkerke R2 = 0.1708, and p < 0.0001) indicates a potentiating effect on antifungal activity against S. sclerotiorum (Table 1).
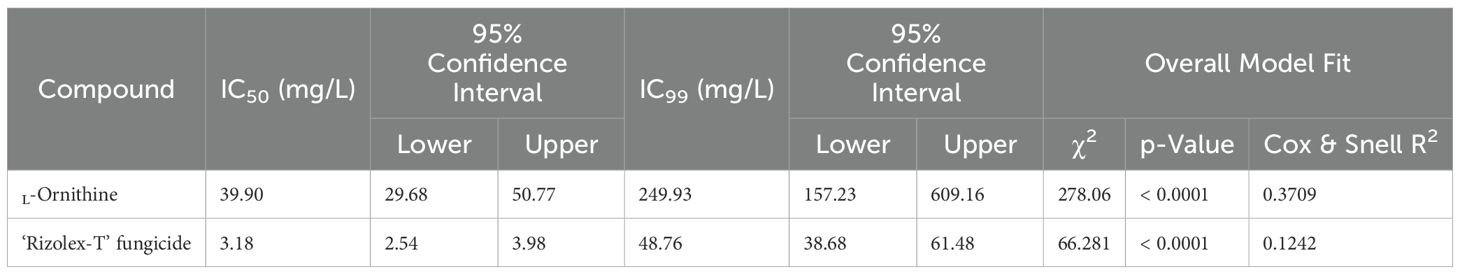
Table 1. The half-maximal inhibitory concentration (IC50) and IC99 values (mg/L) of L-ornithine and the commercial fungicide ‘Rizolex-T’ against S. sclerotiorum.
3.3 L-Ornithine reduced the development of white mold on common bean plants under greenhouse conditions
Generally, L-ornithine (250 mg/L) significantly reduced the development and severity of white mold disease on treated common bean plants compared with non-treated S. sclerotiorum-infected plants (control; Figure 3A). Briefly, although the disease severity was progressively increased in the non-treated infected control plants (52.67 ± 1.53, 83.21 ± 2.61, and 92.33 ± 3.06%), L-ornithine markedly lowered disease severity (%) throughout the experiment (8.97 ± 0.15, 18.00 ± 1.00, and 26.36 ± 3.07) at 7, 14, and, 21 days post-treatment (dpt), respectively (Figure 3A). Likewise, the area under the disease progress curve (AUDPC) declined from 1274.33 ± 33.13 (in non-treated control) to 281.03 ± 7.95 when S. sclerotiorum-infected bean plants were treated with the 250 mg/L of L-ornithine, which was just behind the positive control 50 mg/L ‘Rizolex-T’ fungicide (183.61 ± 7.71; Figure 3B). The same trend was observed in the second trial.

Figure 3. Effect of the exogenous application of L-ornithine on the development of white mold disease of common beans caused by S. sclerotiorum under greenhouse conditions. (A) Disease progress curves of white mold disease of common beans after the treatment with 250 mg/L L-ornithine. (B) The area under the disease progress curve (AUDPC) of white mold disease of common beans after the treatment with L-ornithine. Values represent the means ± standard deviation (means ± SD) of five biological replicates (n = 5). Different letters indicate statistically significant differences between treatments (p < 0.05).
3.4 L-Ornithine stimulated the growth of S. sclerotiorum-infected bean plants
Exogenous application of 250 mg/L L-ornithine progressively enhanced the plant height (Figure 4A), number of branches per plant (Figure 4B), and number of leaves per plant (Figure 4C) throughout 42 dpi. Although the commercial fungicide ‘Rizolex-T’ (50 mg/L) had the highest effect on all studied vegetative parameters, the exogenous application of L-ornithine (250 mg/L) came second highest when compared with non-treated control plants (Figures 4A–C). On the other hand, the application of L-ornithine had no significant effect on the content of both photosynthetic pigments chlorophyll a (Figure 4D) and chlorophyll b (Figure 4E), however, it slightly augmented the total carotenoids (0.56 ± 0.03 mg.g−1 FW) compared with both negative (0.44 ± 0.02 mg.g−1 FW) and positive controls (0.46 ± 0.02 mg.g−1 FW; Figure 4F). Collectively, these findings suggest that L-ornithine has no phytotoxicity on the treated bean plants, but it even stimulates their growth.
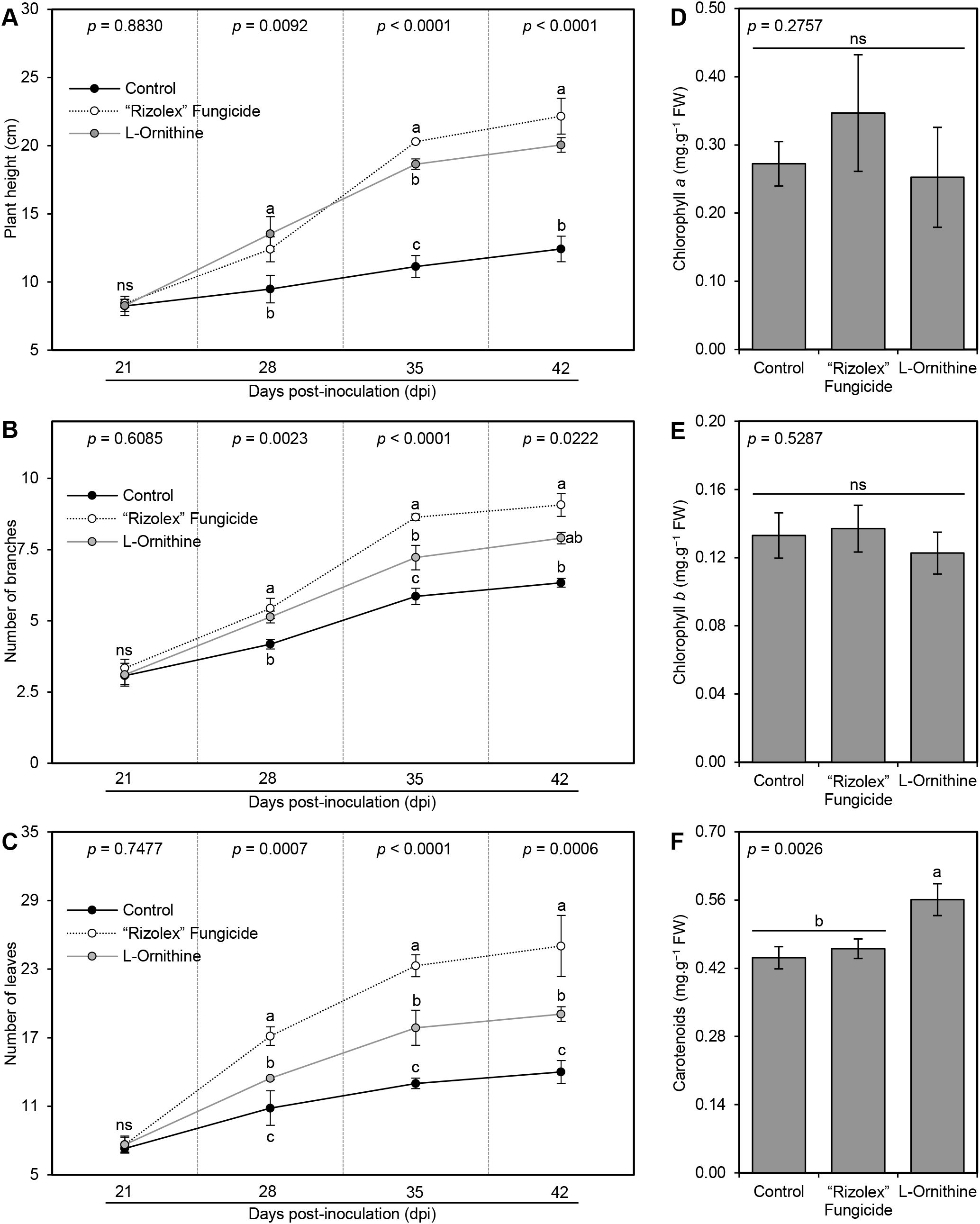
Figure 4. Effect of the exogenous application of L-ornithine on the growth features and photosynthetic pigments in the leaves of S. sclerotiorum-infected common beans under greenhouse conditions. (A) Plant height (cm), (B) number of branches per plant, (C) number of leaves per plant, (D) Chlorophyll a content (mg.g−1 FW), (E) chlorophyll b content (mg.g−1 FW), and (F) total carotenoids content (mg g−1 FW). Values represent the means ± standard deviation (means ± SD) of five biological replicates (n = 5). Different letters indicate statistically significant differences between treatments (p < 0.05).
3.5 The diamine L-ornithine mitigates the oxidative stress of S. sclerotiorum-infected bean plants
In situ histochemical localization of the reactive oxygen species (ROS; expressed as hydrogen peroxide [H2O2]) and free radicals (expressed as superoxide anion [O2•−]) showed that exogenous application of L-ornithine (250 mg/L) significantly reduced the accumulation of both H2O2 (96.05 ± 5.33 nmol.g−1 FW; Figure 5A) and O2•− (32.69 ± 8.56 nmol.g−1 FW; Figure 5B), compared to non-treated infected plants (173.31 ± 12.06 and 149.35 ± 7.94 nmol.g−1 FW, respectively) and those treated with 50 mg/L of the commercial fungicide ‘Rizolex-T’ (170.12 ± 9.50 and 157.00 ± 7.81 nmol.g−1 FW, respectively) which both accumulate higher contents of H2O2 and O2•− at 72 hpt (Figures 5A, B). Similarly, TCA -based assay of malondialdehyde (MDA) suggested that S. sclerotiorum-infected bean plants accumulate higher MDA levels (113.48 ± 10.02 nmol.g−1 FW) in their leaves (Figure 5C). Nevertheless, exogenous application of L-ornithine markedly diminished lipid peroxidation as expressed by lower MDA content within treated plants (33.08 ± 4.00 nmol.g−1 FW).
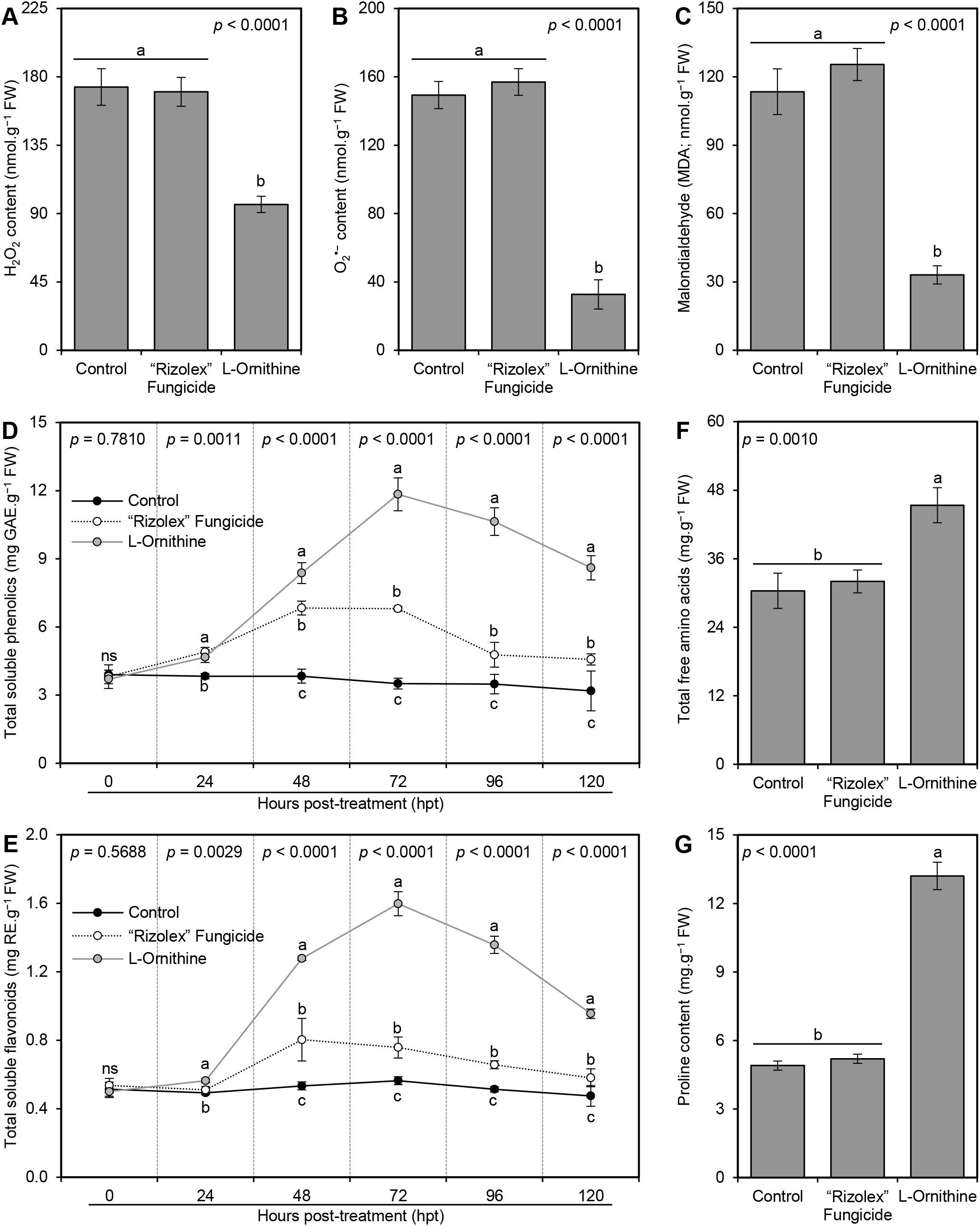
Figure 5. Effect of the exogenous application of L-ornithine on the major oxidative stress markers and non-enzymatic antioxidant defense machinery in S. sclerotiorum-infected common bean leaves at 72 hpt under greenhouse conditions. (A) hydrogen peroxide (H2O2; nmol g−1 FW) at 72 hpt, (B) superoxide anion (O2•−; nmol g−1 FW) at 72 hpt, (C) Malondialdehyde (MDA; nmol g−1 FW) at 72 hpt, (D) total soluble phenolics (mg GAE g−1 FW), (E) total soluble flavonoids (mg RE g−1 FW), (F) Total free amino acids (mg.g−1 FW) at 72 hpt, and (G) Proline content (mg.g−1 FW) at 72 hpt. Values represent the means ± standard deviation (means ± SD) of five biological replicates (n = 5). Different letters indicate statistically significant differences between treatments (p < 0.05).
3.6 L-Ornithine boosts the non-enzymatic antioxidant defense machinery of S. sclerotiorum-infected bean plants
To better understand the physicochemical mechanisms of L-ornithine within S. sclerotiorum-infected bean plants, non-enzymatic antioxidant defense machinery as expressed by total soluble phenolics (TSP; Figure 5D) and total soluble flavonoids (TSF; Figure 5E) were further investigated. Briefly, the application of L-ornithine (250 mg/L) induced the accumulation of TSP within treated S. sclerotiorum-infected bean plants to reach its highest peak at 72 hpt (11.84 ± 0.73 mg GAE.g−1 FW), compared to non-treated (3.51 ± 0.24 mg GAE.g−1 FW) and fungicide-treated controls (6.80 ± 0.10 mg GAE.g−1 FW) at the same time point (Figure 5D). Similarly, exogenous application of 250 mg/L L-ornithine dramatically increased the accumulation of TSF within treated S. sclerotiorum-infected bean plants to reach its highest peak at 72 hpt (1.60 ± 0.07 mg RE.g−1 FW), compared to non-treated (0.56 ± 0.02 mg RE.g−1 FW) and fungicide-treated controls (0.76 ± 0.06 mg RE.g−1 FW) at the same time point (Figure 5E). However, both TSP and TSF dropped thereafter when measured at 96 and 120 hpt, but still higher than both controls.
3.7 L-Ornithine enhanced the total free amino acids and proline content of S. sclerotiorum-infected bean plants
Generally, the application of L-ornithine (250 mg/L) significantly boosted the total free amino acids (45.37 ± 3.07 mg.g−1 FW; Figure 5F) and proline content (13.20 ± 0.60 mg.g−1 FW; Figure 5G) of S. sclerotiorum-infected bean plants compared with the non-treated control (30.38 ± 3.08 and 4.90 ± 0.20 mg.g−1 FW; respectively) and 50 mg/L ‘Rizolex-T’ fungicide-treated plants (32.02 ± 2.00 and 5.20 ± 0.20 mg.g−1 FW; respectively). Nevertheless, there were no significant differences between non-treated and fungicide-treated common bean plants in terms of total free amino acids and proline content (Figures 5F and G).
3.8 L-Ornithine enhances the enzymatic antioxidant defense machinery of S. sclerotiorum-infected bean plants
In agreement with its effect on non-enzymatic antioxidant defense machinery, L-ornithine (250 mg/L) gradually improved the enzymatic activity of catalase (CAT; Figure 6A), peroxidase (POX; Figure 6B), and polyphenol oxidase (PPO; Figure 6C) till they reached their highest peak at 72 hpt (129.69 ± 3.05 μM H2O2 g−1 FW min−1, 1.182 ± 0.182 μM of tetraguaiacol g−1 FW min−1, 0.676 ± 0.025 arbitrary units, respectively) compared to both controls at the same time point, then they dropped slowly at 96 and 120 hpt. Moreover, exogenous application of 250 mg/L L-ornithine unregulated three antioxidant-associated genes, including peroxisomal catalase (PvCAT1; Figure 6D), superoxide dismutase (PvSOD; Figure 6E), and glutathione reductase (PvGR; Figure 6F) compared to both controls.
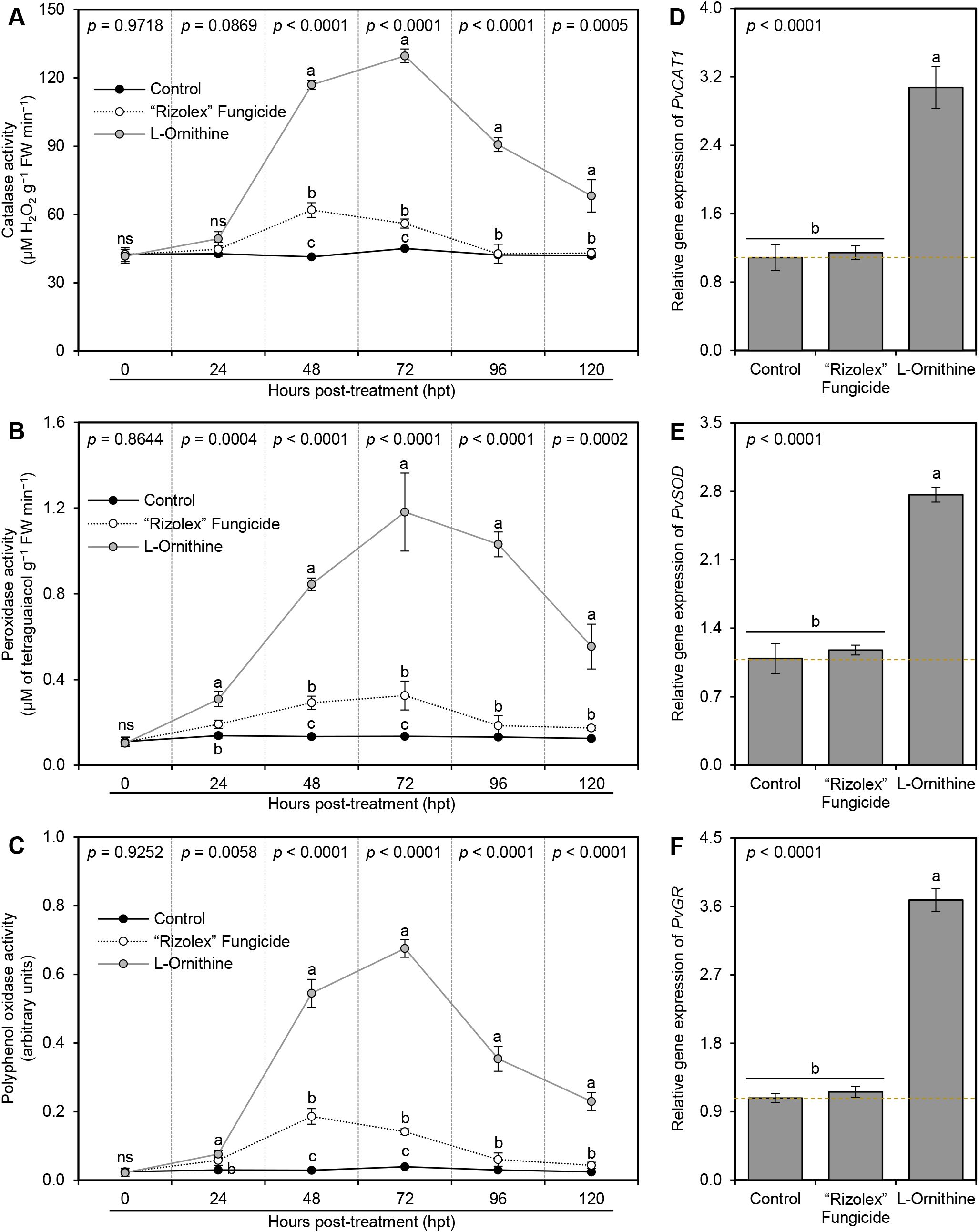
Figure 6. Effect of the exogenous application of L-ornithine on the major components of enzymatic antioxidant defense machinery of S. sclerotiorum-infected common beans at 72 hpt under greenhouse conditions. (A) catalase activity (μM H2O2 g−1 FW min−1), (B) peroxidase activity (μM Tetraguaiacol g−1 FW min−1), and (C) polyphenol oxidase activity (Arbitrary units), (D, E, and F) Relative gene expression of peroxisomal catalase (PvCAT1), superoxide dismutase (PvSOD), and glutathione reductase (PvGR), respectively, at 72 hpt. Values represent the means ± standard deviation (means ± SD) of five biological replicates (n = 5). Different letters indicate statistically significant differences between treatments (p < 0.05).
3.9 The genome of Sclerotinia sclerotiorum possesses a putative oxaloacetate acetylhydrolase (SsOAH) protein
In silico analysis showed that the S. sclerotiorum genome possesses about six putative oxaloacetate acetylhydrolase (OAH) proteins that were similar and produced significant alignment statistics with OAH protein from Aspergillus fijiensis CBS 313.89 and Penicillium lagena (Supplementary Table S4). Only three of them had an identity percentage of more than 50%, including Hypothetical protein SS1G_08218 (GenBank accession no. XP_001590478.1, 338 aa, identity = 70.28 and 61.97% with OAH from A. fijiensis and P. lagena, respectively), OAH-1 (GenBank accession no. AEY84228.1, identity = 71.08 and 69.79% with OAH from A. fijiensis and P. lagena, respectively), and OAH-2 (GenBank accession no. AEY84231.1, identity = 69.02 and 65.93% with OAH from A. fijiensis and P. lagena, respectively) (Supplementary Table S4). It is worth mentioning that the AA sequence of both OAH-1 (249 aa) and OAH-2 (185 aa) from S. sclerotiorum was a little bit shorter than the query AA sequences of OAH from Aspergillus fijiensis (342 aa) and from Penicillium lagena (316 aa), but the length of AA sequences Hypothetical protein SS1G_08218 (henceforth SsOAH; 338 aa) was approximately like them. Therefore, we focused only on the SsOAH protein for further in silico analysis.
Moreover, the multiple protein sequences alignment using COBALT analysis showed that the obtained six protein sequences were relatively highly homologous to each other, as well as OAH from A. fijiensis and P. lagena, and all had a conserved 2-methylisocitrate lyase (PrpB) domain (Figure 7A). However, the phylogenetic analysis showed that only SsOAH was phylogenetically closer to OAH from Penicillium lagena while OAH from Aspergillus fijiensis was clustered separately at the bottom of the dendrogram (Figure 7A). Likewise, multiple sequence alignment showed that the SsOAH protein from S. sclerotiorum had high similarity and conserved sequence with both query sequences (OAH from A. fijiensis and P. lagena) (Supplementary Figure S1).
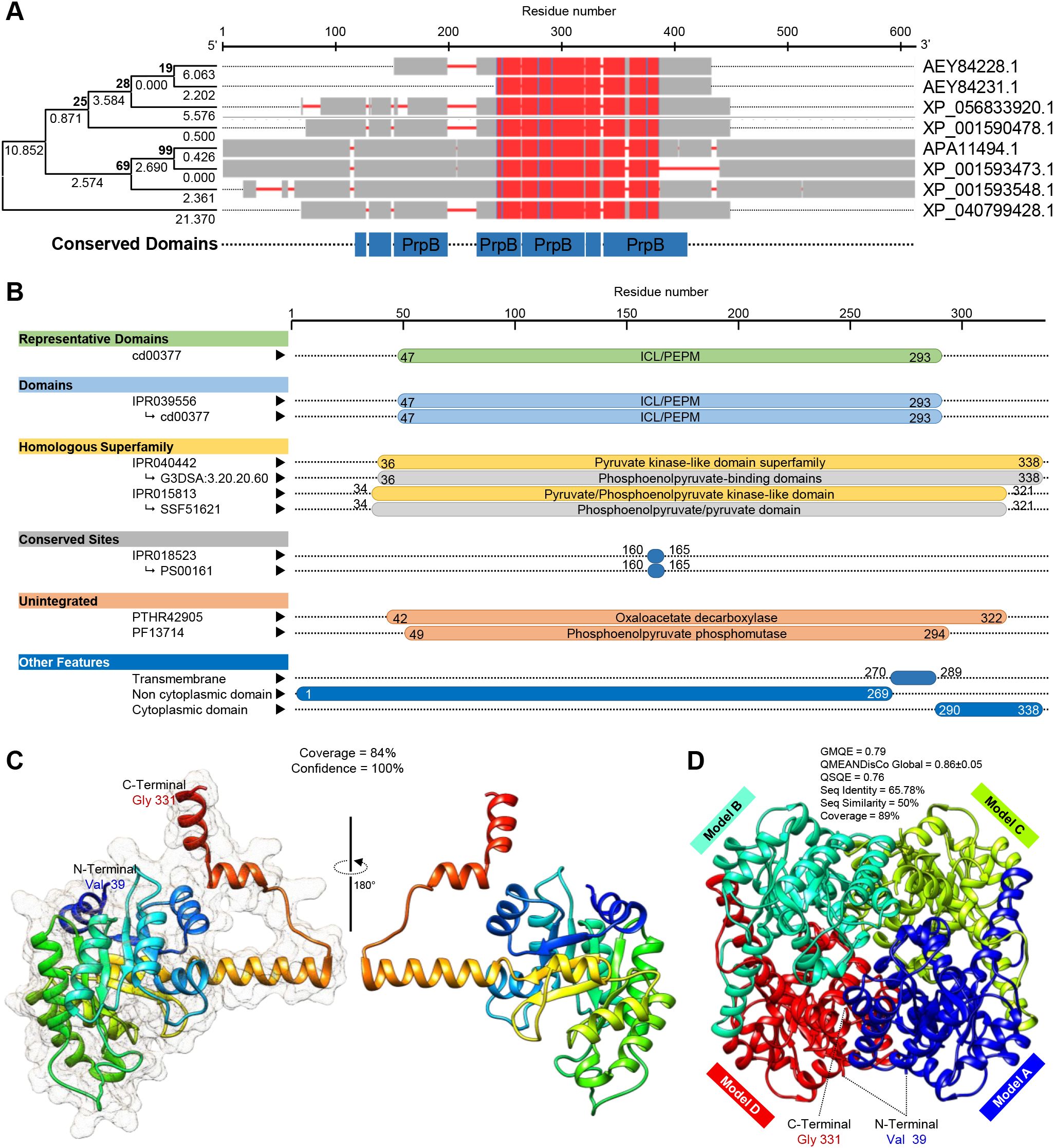
Figure 7. In silico characterization and structural modeling of oxaloacetate acetylhydrolase (SsOAH) protein from S. sclerotiorum. (A) Evolutionary analysis using the maximum likelihood method as conducted in Molecular Evolutionary Genetics Analysis software package (MEGA; version 11) (Kumar et al., 2024) and its associated multiple protein sequences alignments using Constraint-Based Alignment tool (COBALT) analysis. In the COBALT analysis, residues were colored using a column-based method according to their relative entropy threshold. The red color indicates highly conserved columns. The full list of genes, names, and accession numbers is available in Supplementary Table S4. (B) Functional and conserved domains analysis of SsOAH using the InterPro Scan tool. (C) The crystallographic tertiary structure and its associated mesh surface of putative SsOAH monomer as predicted by Phyre2 tool. The tertiary structures of SsOAH were predicted using the crystal structure of oxaloacetate acetylhydrolase in the protein data bank (PDB ID: 3lye.1.A) and refined using the X-ray method to 1.30 Å resolution with acceptable statistics and 100.0% confidence. Protein chains are colored according to the rainbow color spectrum, from blue (N-terminus) to red (C-terminus). (D) The crystallographic tertiary structure of putative SsOAH homo-tetramer as predicted by the SWISS-MODEL server. Both tertiary structures were visualized with the UCSF-Chimera package.
Additionally, the InterPro Scan tool was used for the functional analysis of proteins by classifying them into families and predicting domains and important sites of SsOAH (Figure 7B). Briefly, SsOAH protein has a phosphoenolpyruvate mutase (PEPM) isocitrate lyase (ICL) representative domain (ICL/PEPM, CDD entry: cd00377), as well as an ICL/PEPM domain (InterPro entry: IPR039556). Furthermore, SsOAH has a pyruvate kinase-like domain superfamily (InterPro entry: IPR040442) with phosphoenolpyruvate-binding domains (CATH-Gene3D entry: G3DSA:3.20.20.60), as well as a pyruvate/phosphoenolpyruvate kinase-like domain superfamily (InterPro entry: IPR015813) with phosphoenolpyruvate/pyruvate domain (SUPERFAMILY entry: SSF51621). It also has an isocitrate lyase/phosphorylmutase conserved site (InterPro entry: IPR018523) with an isocitrate lyase signature (PROSITE patterns entry: PS00161) Figure 7B). It is worth mentioning that SsOAH had an unintegrated site of isocitrate lyase/PEP mutase superfamily, oxaloacetate decarboxylase (PANTHER entry: PTHR42905) with phosphoenolpyruvate phosphomutase site (Pfam entry: PF13714). Other features of SsOAH include a region of a membrane-bound protein predicted to be embedded in the membrane (as a transmembrane site), a region of a membrane-bound protein predicted to be outside the membrane, in the extracellular region (as a non-cytoplasmic domain), and a region of a membrane-bound protein predicted to be outside the membrane, in the cytoplasm (as a cytoplasmic domain) (Figure 7B). Collectively, these findings suggest that the predicted SsOAH protein is highly similar in its functional analysis, conserved domains, and topology with known OAH proteins from other species.
The crystallographic three-dimensional (3D) structure of SsOAH (Figures 7C, D) was predicted using the crystal structure of oxaloacetate acetylhydrolase in the protein data bank (PDB ID: 3lye.1.A) and refined using the X-ray method to 1.30 Å resolution with acceptable statistics (Figures 7C, D; Supplementary Figure S2). Briefly, the Phyre2 tool showed that approximately 283 residues (84%; residues Val 39 to Gly 331) of SsOAH have been modeled with template protein with 100% confidence (Figure 7C). According to the Phyre2-based analysis, the predicted 3D structure of SsOAH is a monomer composed of 12 α-helix ribbons (approximately 40%) and eight β-sheets (about 11%; Figure 7C; Supplementary Figure S2) with considerable predicted local similarity (PLS) to the target. Interestingly, the SWISS-model protein structure homology-modeling server confirmed almost the same 3D structure. However, the SWISS-model-based predicted model of SsOAH is a homo-tetramer (sequence identity = 65.78%, sequence similarity = 50%, and coverage = 89%) with a satisfactory global model quality estimate (GMQE = 0.79), QMEANDisCo Global (0.86 ± 0.05), and quaternary structure quality estimate (QSQE = 0.76) (Figure 7D; Supplementary Figure S3).
3.10 L-Ornithine down-regulates oxaloacetate acetylhydrolase (SsOAH) of S. sclerotiorum
The addition of 40 mg/L L-ornithine to the PDB medium significantly down-regulated the gene expression of SsOAH in the mycelium of S. sclerotiorum (Figure 8A). Likewise, although the transcript levels of SsOAH were progressively increased in the fungal mycelium collected from non-treated control plants till the end of the experiment (120 hpt), exogenous application of 250 mg/L L-ornithine significantly down-regulated the gene expression of SsOAH in the fungal mycelia collected from treated plants at 48 hpt and till the end of the experiment (Figure 8A).
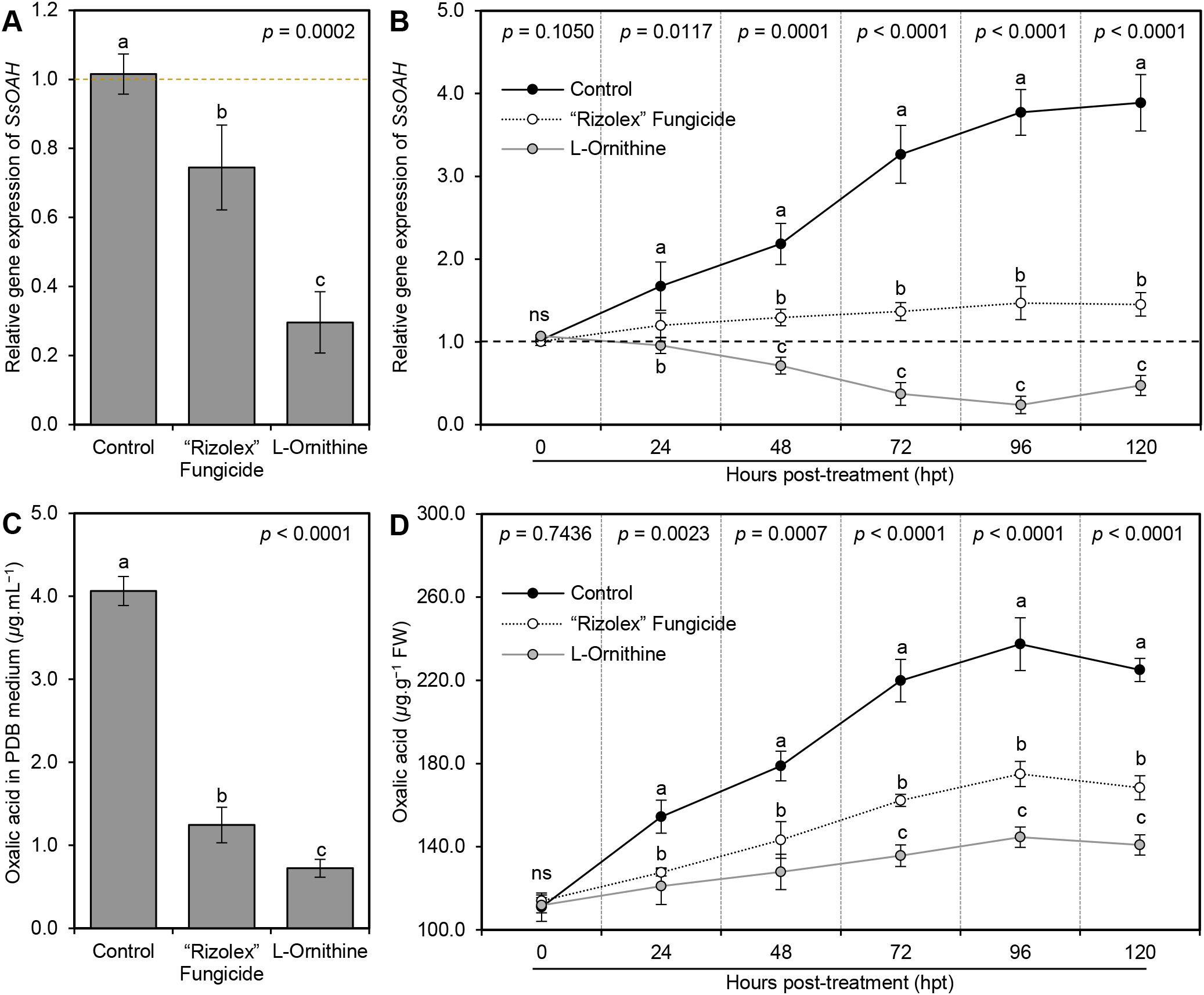
Figure 8. Effect of the exogenous application of L-ornithine on the (A) oxalic acid secretion in PDB medium (μg.mL−1) and oxalic acid content (μg.g−1 FW) of S. sclerotiorum-infected common beans under greenhouse conditions. Values represent the means ± standard deviation (means ± SD) of five biological replicates (n = 5). Different letters indicate statistically significant differences between treatments (p < 0.05).
3.11 The diamine L-ornithine application diminishes the secretion of oxalic acid
To better understand the relationship between L-ornithine and the pathogenicity factor of S. sclerotiorum, oxalic acid, the effect of L-ornithine application on the secretion of oxalic acid in liquid media (PDB), as well as infected tissues was investigated. Briefly, the addition of 40 mg L-ornithine per liter of PDB media significantly reduced the secretion of oxalic acid in the medium (1.24 ± 0.21 μg.mL−1) compared to the non-treated control (4.06 ± 0.17 μg.mL−1; Figure 8C). However, we believe that the observed reduction in oxalic acid levels could be due to the inhibited growth of S. sclerotiorum or due to direct inhibition of SsOAH activity. Therefore, the fungal mycelium was weighed to normalize oxalic acid production per gram of fungal biomass in our in vitro experiments. The results showed that L-ornithine (40 mg/L) significantly reduced oxalic acid production even after normalizing fungal biomass, suggesting a direct inhibitory effect on the enzyme’s activity or its expression. Likewise, exogenous application of L-ornithine (250 mg/L) significantly reduced the oxalic acid accumulation in S. sclerotiorum-infected bean leaves. Although the oxalic acid level in non-treated infected leaves was gradually increased over the time course until it reached its highest peak at 96 hpt (237.40 ± 12.69 μg.g−1 FW), L-ornithine application significantly lessened the oxalic acid accumulation in treated plants (Figure 8D).
4 Discussion
The phytopathogenic fungus S. sclerotiorum can infect over 600 plant species, with diverse phylogenetic backgrounds (Liang and Rollins, 2018). Therefore, it is noticeably vital to develop novel eco-friendly strategies to manage its spread among agronomically important crops, particularly because the traditional control measures have repeatedly been reported to be inadequate (Cessna et al., 2000). Recently, we proposed two non-proteinogenic amino acids (γ-aminobutyric acid (GABA) and ß-alanine) as innovative eco-friendly alternatives to mitigate oxidative stress and enhance the resistance of common bean plants against S. sclerotiorum, the causal agent of white mold disease (Nehela et al., 2024). In the current study, we proposed the diamine, L-ornithine, as a potential alternative that enhances resilience and boosts the defense mechanisms of common bean plants against S. sclerotiorum. L-Ornithine application significantly reduced the AUDPC of white mold disease by approximately 78% compared to the non-treated control. It is worth mentioning that L-Ornithine application was more efficient in reducing the AUDPC than GABA (74.12%) and ß-alanine (67.10) which both were tested in our previous study (Nehela et al., 2024). These findings suggest that non-proteinogenic amino acids (GABA, ß-alanine, and L-ornithine) might be promising eco-friendly alternatives for white mold disease management for sustainable bean production with a superiority of L-ornithine.
Initially, we isolated and characterized four S. sclerotiorum isolates. It is worth mentioning that the pathogenicity of these isolates was positively associated with oxalic acid secretion. In other words, the highly virulent isolate #3 of S. sclerotiorum was found to secrete more oxalic acid in the PDB media (El-Argawy, 2012). Although the used method in the current study is primarily designed for oxalic acid quantification (Xu and Zhang, 2000), it is possible that other organic acids with similar chemical properties could interfere with the reaction and contribute to the absorbance at 600 nm may result in minor cross-reactivity that cannot be entirely ruled out. Further studies using HPLC or GC-MS-based targeted metabolomics are required to confirm the specificity of oxalic acid and to better understand its role(s) in bean-S. sclerotiorum interaction.
The non-specific phytotoxin, oxalic acid, is an essential pathogenicity determinant of S. sclerotiorum that is associated with diverse and sometimes contradictory effects on host plants (Marciano et al., 1983; Cessna et al., 2000; Guimarães and Stotz, 2004; Hegedus and Rimmer, 2005; Williams et al., 2011; Heller and Witt-Geiges, 2013; Fagundes-Nacarath et al., 2018; Daniel et al., 2024). Initially, it was proposed to manipulate the pH of the plant cell to create a favorable environment for infection (Marciano et al., 1983; Hegedus and Rimmer, 2005). However, emerging evidence suggests that the negative effects of oxalic acid on host cells are largely independent of both its acidity and its affinity for Ca2+ (Cessna et al., 2000) and it might interfere with host defense responses in a more complex manner than previously thought (Cessna et al., 2000; Guimarães and Stotz, 2004; Williams et al., 2011). Although its role in lowering environmental pH and enhancing the activity of cell wall-degrading enzymes (CWDEs) was experimentally proven (Guimarães and Stotz, 2004; Heller and Witt-Geiges, 2013; Daniel et al., 2024), it might play a key contradictory role in reactive oxygen species (ROS) regulation (Cessna et al., 2000; Williams et al., 2011; Daniel et al., 2024). For example, oxalic acid was found to suppress the oxidative burst of the host plant in early infection (Cessna et al., 2000; Williams et al., 2011), while it induces apoptotic-like programmed cell death (PCD) that requires the generation of reactive oxygen species (ROS) in host cells (Williams et al., 2011). In other words, oxalic acid both suppresses and induces host ROS, depending on infection stage, during the compatible interaction (Cessna et al., 2000; Williams et al., 2011).
In agreement with this hypothesis, S. sclerotiorum-secreted oxalic acid did not show any phytotoxicity in early infection stages (Heller and Witt-Geiges, 2013) which might be due to the ability of the host cell to catabolize it in the early infection stage, and low concentrations, and translocate it to their vacuoles where it is stored as calcium oxalate (Heller and Witt-Geiges, 2013). However, at late infection stages and upon the accumulation of high oxalic acid levels, it deregulates guard cells and induces foliar wilting (Heller and Witt-Geiges, 2013). Additionally, recent studies proved that oxalic acid is involved in the regulation of fungal endopolygalacturonase (SSPG1) activity at low pH, reinforcing its integral role in S. sclerotiorum pathogenesis (Daniel et al., 2024). In addition to its role in virulence, oxalic acid is involved in the mediation of biochemical and physiological changes in the common bean-Sclerotinia sclerotiorum interaction, particularly reducing photosynthetic pigments and intensifying oxidative stress (Fagundes-Nacarath et al., 2018). Collectively, these findings highlight oxalic acid as a multifaceted virulence factor, influencing infection dynamics through mechanisms that are still not fully understood, warranting further research into its signaling interference and broader physiological effects.
It is worth mentioning that L-ornithine application diminishes the secretion of oxalic acid in PDB media and reduces the oxalic acid accumulation in S. sclerotiorum-infected tissues which was associated with a significant reduction in the development and severity of white mold on common bean plants under greenhouse conditions. We understand that the reduced oxalic acid levels in PDB media or within infected tissue might be due to the reduced mycelial mass of S. sclerotiorum since L-ornithine showed strong antifungal activity against the fungal pathogen and significantly reduced the mycelial growth.
Moreover, the role of polyamine, as well as non-proteinogenic amino acids, during S. sclerotiorum infection has gained increasing attention, particularly in the context of oxalic acid production and fungal development (Walker et al., 2023; Nehela et al., 2024). Recently, it was proven that silencing of Abhydrolase-3 from S. sclerotiorum via host-induced gene silencing (HIGS) in transgenic Arabidopsis thaliana (AT1703) significantly downregulated the expression of key polyamine biosynthesis genes, including Ornithine decarboxylase (ODC), Spermine synthase, and Spermidine synthase of the fungal pathogen and ultimately slowed fungal infection on AT1703 plants compared to their wild type ones (Walker et al., 2023). Our study aligns with and expands on these findings by showing that the exogenous application of L-ornithine significantly suppressed the mycelial growth of S. sclerotiorum and reduced the production of oxalic acid, which both were correlated with a significant reduction in disease severity in common bean. ODC is a rate-limiting enzyme in polyamine biosynthesis that catalyzes the decarboxylation of ornithine to produce putrescine, the first step in polyamine synthesis (Michael, 2016). We suggest that L-ornithine might interfere with this pathway in the causal agent and negatively affect fungal development. The observed in vitro antifungal activity of L-ornithine may, therefore, be linked to its interference with the polyamine biosynthesis pathway in S. sclerotiorum. Taken together, these findings suggest a novel approach for managing white mold through metabolic disruption of S. sclerotiorum via polyamine pathway interference. However, further studies are required to explore the L-ornithine- S. sclerotiorum-oxalic acid interactions as a potential disease control strategy.
It was reported previously that the oxaloacetate acetylhydrolase gene of S. sclerotiorum plays a vital role in the fungal oxalic acid accumulation and virulence (Rana et al., 2022) and its deletion mutants failed to accumulate oxalic acid in culture or during plant infection, exhibiting reduced pathogenicity, and produce limited lesions on host plants (Liang et al., 2015; Rana et al., 2022) highlighting the key role of oxalic acid in the disease development. In the current study, in silico analysis showed that the S. sclerotiorum genome possesses a putative oxaloacetate acetylhydrolase (SsOAH) protein that is highly similar in its functional analysis, conserved domains, and topology to the OAH protein from A. fijiensis and P. lagena. It is worth mentioning that the addition of L-ornithine to the PDB medium down-regulated the gene expression of SsOAH in the mycelium of S. sclerotiorum. Moreover, our findings showed that exogenous application of L-ornithine can effectively down-regulate SsOAH expression both in vitro and in infected plant tissues, leading to a reduction in the accumulation of oxalic acid and a corresponding decrease in disease severity. It was reported previously that targeting a gene encoding oxaloacetate acetylhydrolase (Ssoah1) of S. sclerotiorum via HIGS resulted in an oxalic acid deficiency and a non-pathogenic interaction on soybean (McCaghey et al., 2021). The current study builds on previous work and investigates the potential role of SsOAH in S. sclerotiorum virulence, particularly with oxalic acid production. We believe that manipulating the production of oxalic acid, either through silencing SsOAH (McCaghey et al., 2021) or inhibiting its activity with compounds like L-ornithine, can serve as an effective strategy for managing S. sclerotiorum infections. Moreover, both findings of the current investigation and previous studies highlight the importance of targeting oxalic acid biosynthesis pathway as a promising approach to control S. sclerotiorum.
Although current data does not fully differentiate the direct causes of the observed reduction in oxalic acid levels. However, our findings strongly suggest that multiple factors could contribute to this reduction. For instance, in vitro antifungal assays demonstrated that L-ornithine significantly inhibits the radial mycelial growth of S. sclerotiorum, which likely reduces the overall metabolic activity of the pathogen, including oxalic acid production. Moreover, gene expression analysis showed that L-ornithine downregulated SsOAH transcript levels in both in vitro cultures and infected plant tissues, suggesting that reduced oxalic acid production is at least partially due to transcriptional suppression of this key enzyme. Additionally, the observed reduction in oxalic acid levels, even when normalized to fungal biomass, suggests that L-ornithine may directly or indirectly inhibit the enzymatic activity of SsOAH. However, further studies are required to confirm this hypothesis.
Likewise, oxalic acid-deficient mutants of S. sclerotiorum were nonpathogenic while the oxalic acid-producing wild type was pathogenic to whole plants, stems, and leaves under growth chamber conditions (Godoy et al., 1990). Moreover, the fungal oxalic acid-deficient mutants were nonpathogenic, and apoptotic-like features were not observed following plant inoculation (Kim et al., 2008) and resulted in restricted growth, reminiscent of an HR-like response (Williams et al., 2011). Herein, our findings showed that L-ornithine application significantly down-regulated SsOAH, diminished the secretion of oxalic acid, and inhibited the growth of S. sclerotiorum to reduce its pathogenicity in treated plants in contrast to non-treated ones which had higher oxalic acid levels and showed overwhelming disease and runaway cell death. Meanwhile, L-ornithine stimulated the growth of S. sclerotiorum-infected bean plants as expressed by the plant height, number of branches, and number of leaves per plant suggesting no phytotoxicity on the treated bean plants. It was reported previously that the amine functional group of ornithine and its derivative N-acetylornithine play a key role in reducing the toxicity of fumonisins (a mycotoxin produced by numerous fungal species such as Fusarium sp. and Aspergillus sp.) (Renaud et al., 2021).
Although the specific biochemical, physiological, and molecular mechanisms responsible for the protective role of L-ornithine are not fully understood, several potential mechanisms have been proposed. An acceptable hypothesis that could explain how L-ornithine effectively enhances the resilience of common bean plants against S. sclerotiorum is due to its ability to mitigate oxidative stress and modulate the redox status within infected plants. Exogenous ornithine was reported previously to improve the plant response to abiotic stress such as drought (Hussein et al., 2019) and salinity (da Rocha et al., 2012) via contribution to proline accumulation (da Rocha et al., 2012) or modulation of oxidative stress (Hussein et al., 2019).
Our findings showed that infection with S. sclerotiorum induced oxidative stress in infected bean plants, leading to the overproduction of ROS such as H2O2 and O2•−, as well as lipid peroxidation which might contribute to disease progression (Torres et al., 2006). Nevertheless, exogenous application of L-ornithine successfully decreases ROS levels and alleviates oxidative stress within S. sclerotiorum-infected plants via the induction of multilayered antioxidant defense machinery. The complex antioxidant defense contains two key mechanisms, enzymatic and non-enzymatic antioxidant defense machinery (Sharma et al., 2022). The enzymatic antioxidants act as the front line in antioxidant defenses, however, the non-enzymatic antioxidants characterize the second line of defense against ROS (Sharma et al., 2022).
In the current study, L-ornithine boosts the non-enzymatic (total soluble phenolics and flavonoids) as well as the enzymatic (CAT, POX, PPO, PvCAT1, PvSOD, and PvGR) antioxidant defense machinery of S. sclerotiorum-infected bean plants. In agreement with these findings, D-ornithine increased the activity of major antioxidant enzymes (SOD, CAT, and POX) in tobacco cells under salinity (Ghahremani et al., 2013) and enhanced the activity of CAT and POX in sugar beet plants under drought stress (Hussein et al., 2019). Similarly, L-ornithine slightly increased the POX (at 48 hpi) and CAT activity (at 72 hpi), but not phenylalanine ammonia-lyase (PAL) in lime (Citrus aurantifolia) plants infected with the bacterial phytopathogen Xanthomonas citri subsp. citri, the causal agent of Citrus bacterial canker (Hasabi et al., 2014). Collectively, these findings demonstrate that L-ornithine substantially enhances both the enzymatic and non-enzymatic antioxidant defense machinery of bean plants to reduce ROS levels and maintain their homeostasis to ease the undesirable oxidative stress caused by S. sclerotiorum.
Another hypothesis that might explain the potential role(s) of L-ornithine against S. sclerotiorum is due to its contribution to amino acids and proline accumulation. Exogenous ornithine effectively contributes to proline accumulation in cashew leaves under salt stress (da Rocha et al., 2012). Moreover, higher levels of proline were accumulated in Arabidopsis thaliana during defense against pathogens (Monteoliva et al., 2014) and proline catabolism usually enhanced during the early stages of pathogen infection. Likewise, the proline intermediate pyrroline-5-carboxylate (P5C) might contribute to plant defense against invading pathogens.
The hypothesis that L-ornithine might be involved in the common bean defense responses against S. sclerotiorum by contributing proline and P5C accumulation is supported by several experimental evidence on proline metabolism under stress (Qamar et al., 2015; Rizzi et al., 2015). For instance, it was suggested previously that P5CDH contributes to the modulation of the proline biosynthesis pathway and its catabolism after the activation of ProDH in response to biotic and abiotic stressors (Rizzi et al., 2015). The balance between the biosynthetic and catabolic pathways is key to maintaining cellular homeostasis. Likewise, P5C was reported to play a key role in plant defense against invading pathogens (Qamar et al., 2015) where it triggers a hypersensitive response (HR) and contributes to the induction of ROS. Collectively, these findings suggest that the proline-P5C pathway, as well as its regulatory enzymes such as P5CDH and ProDH, might be involved in the activation of defense responses such as HR and ROS accumulation after pathogen attack. In the context of our study, the exogenous application of L-ornithine might boost proline levels by inducing P5C accumulation and potentially enhancing the plant’s defense against S. sclerotiorum infection through the modulation of HR-like responses and ROS production. Accordingly, the regulation of proline and P5C metabolism by L-ornithine supplementation represents a complementary mechanism by which plants combat pathogen infection, in addition to the above-discussed impact on oxalic acid production.
It is worth noting that the P5C dehydrogenase (P5CDH) mutant of A. thaliana showed higher proline dehydrogenase (ProDH) and ornithine aminotransferase (OAT) activity, as well as higher levels of ornithine and proline when challenged with exogenous proline, or infection with the bacterial pathogen Pseudomonas syringae pv. tomato (Monteoliva et al., 2014). Activation of OAT and higher ornithine and proline levels proposes that ornithine is a precursor for proline biosynthesis (Monteoliva et al., 2014) and contributes to multiple abiotic stress tolerances. Additionally, proline plays a key role in redox homeostasis and energy transfer reactions, which lead to plant resistance against pathogens or programmed cell death (PCD) (Anwar et al., 2018). Interestingly, our findings showed that exogenous L-ornithine efficiently enhanced the total amino acids and proline content of S. sclerotiorum-infected bean plants. Nevertheless, while the spectrophotometric approach provides a reliable estimate of total free amino acids, advanced techniques such as LC/MS could complement these findings by offering detailed amino acid profiles and insights into non-enzymatic antioxidant contributions.
5 Conclusion
In conclusion, our findings suggest that L-ornithine might be a promising eco-friendly alternative in management programs for white mold disease caused by S. sclerotiorum in common beans. L-Ornithine exhibited a strong antifungal in vitro and progressively reduced the development of white mold on common bean plants under greenhouse conditions. These fungistatic actions were combined with the ability of L-ornithine to mitigate the oxidative stress of S. sclerotiorum-infected bean plants via the activation of multilayered antioxidant defense machinery (enzymatic and non-enzymatic antioxidants), as well as proline accumulation (Figure 9). Moreover, L-ornithine did not show any phytotoxicity on treated plants but enhanced their growth. Collectively, all these findings establish L-ornithine as a promising eco-friendly alternative for sustainable bean production. However, further investigations are required to optimize the application strategies for L-ornithine use in the field and to explore their potential in integrated pest management programs. Moreover, further studies are required to better understand the impact on bean production, including potential effects on the number of pods, size of beans, and total yield. While our current study focuses on the antifungal activity and plant response, we acknowledge the importance of evaluating agronomic traits. Likewise, investigating the effect(s) of L-ornithine supplementation on healthy (non-infected) bean plants is another direction that requires further research, including its potential impact on growth, performance, physiological responses, and yield. This additional research might provide a more comprehensive understanding of the broader implications of L-ornithine in bean physiology particularly and other legumes in general.
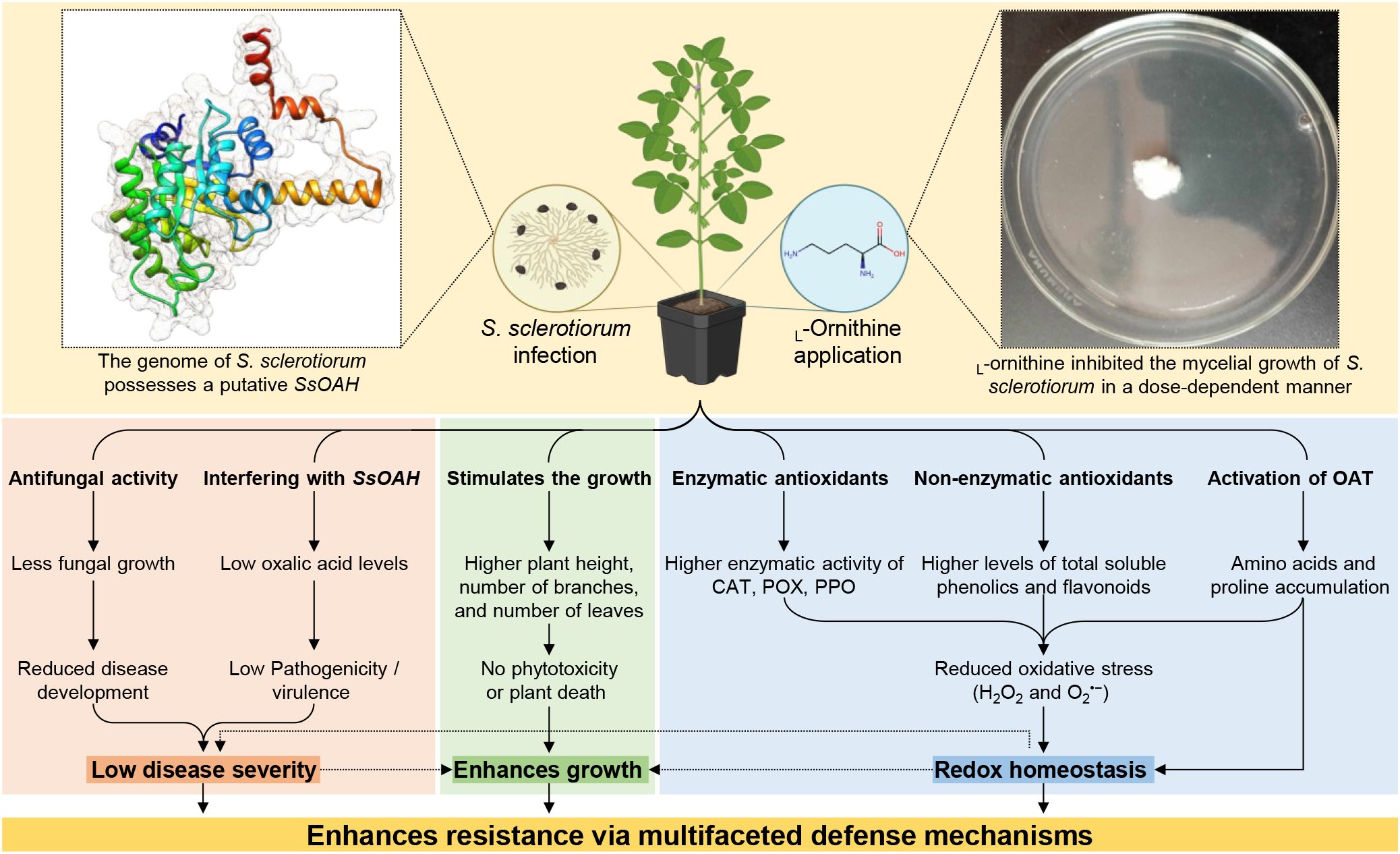
Figure 9. Hypothetical model for the potential effects of L-ornithine supplementation on the growth and response of common beans against S. sclerotiorum, the causal agent of white mold disease under greenhouse conditions. In this model, we suggest that L-ornithine supplementation via seed priming combined with root drench might enhance the response of common bean plants to the infection with the soil-borne fungal pathogen, S. sclerotiorum, via modulation of multilayered defense machinery. Briefly, (i) L-Ornithine showed antifungal activity and progressively inhibited the mycelial growth of S. sclerotiorum in a dose-dependent manner. Moreover, L-ornithine supplementation down-regulates oxaloacetate acetylhydrolase (SsOAH) of S. sclerotiorum in the PDB medium and the fungal mycelia collected from treated plants at 48 hpt. Together, the antifungal activity of L-ornithine and interfering with SsOAH diminished the development of white mold on common bean plants under greenhouse conditions. (ii) L-Ornithine supplementation progressively enhanced the plant height, number of branches per plant, and number of leaves per plant throughout 42 dpi. Collectively, these findings suggest that L-ornithine has no phytotoxicity on the treated bean plants, but it even stimulates their growth. (iii) L-Ornithine mitigated the oxidative stress of S. sclerotiorum-infected bean plants via the induction of multilayered antioxidant defense machinery including enzymatic (CAT, POX, PPO, PvCAT1, PvSOD, and PvGR) and non-enzymatic (total soluble phenolics and flavonoids) antioxidants, as well as total amino acids and proline accumulation. Collectively, these findings suggest that L-ornithine supplementation induces multilayered antioxidant defense machinery to maintain the Redox homeostasis within S. sclerotiorum-infected common bean plants.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YN: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Project administration, Resources, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing. YM: Funding acquisition, Resources, Writing – review & editing. NE: Formal Analysis, Investigation, Methodology, Validation, Writing – review & editing. OA: Formal Analysis, Investigation, Methodology, Validation, Writing – review & editing. AA: Formal Analysis, Investigation, Methodology, Validation, Writing – review & editing. SK: Formal Analysis, Investigation, Methodology, Validation, Writing – review & editing. TA: Formal Analysis, Investigation, Methodology, Validation, Writing – review & editing. QA: Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Validation, Writing – review & editing. AM: Formal Analysis, Investigation, Methodology, Validation, Writing – review & editing. WH: Conceptualization, Formal Analysis, Investigation, Methodology, Resources, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The work was funded by the Deanship of Research and Graduate Studies at King Khalid University for funding this work through a Large Research Project under grant number RGP2/249/45.
Acknowledgments
The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through a Large Research Project under grant number RGP2/249/45. YN would like to acknowledge the Graduate Student and Research Affairs Sector of Tanta University, Egypt. QA thanks the United Arab Emirates University for providing a postdoctoral grant on climate action under grant number 12S140.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1483417/full#supplementary-material
References
Adám, A., Farkas, T., Somlyai, G., Hevesi, M., Király, Z. (1989). Consequence of O2·– generation during a bacterially induced hypersensitive reaction in tobacco: deterioration of membrane lipids. Physiol. Mol. Plant Pathol. 34, 13–26. doi: 10.1016/0885-5765(89)90013-1
Aebi, H. (1984). Catalase in vitro. Methods Enzymol. 105, 121–126. doi: 10.1016/S0076-6879(84)05016-3
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Altschul, S. F., Wootton, J. C., Gertz, E. M., Agarwala, R., Morgulis, A., Schaffer, A. A., et al. (2005). Protein database searches using compositionally adjusted substitution matrices. FEBS J. 272, 5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x
Anwar, A., She, M., Wang, K., Riaz, B., Ye, X. (2018). Biological roles of ornithine aminotransferase (OAT) in plant stress tolerance: present progress and future perspectives. Int. J. Mol. Sci. 19, 3681. doi: 10.3390/IJMS19113681
Asija, S., Seth, T., Umar, S., Gupta, R. (2022). Polyamines and their crosstalk with phytohormones in the regulation of plant defense responses. J. Plant Growth Regul. 42, 5224–5246. doi: 10.1007/S00344-022-10837-5
Atallah, O., Hassanin, A., Yassin, S. M., Aloufi, A. S., Almanzalawi, E. A., Abdelkhalek, A., et al. (2024). Pathological characterization and management of Lasiodiplodia theobromae, a hemi-biotroph with an interkingdom host range. Plant Dis. 108 (11), 3243–3257. doi: 10.1094/PDIS-03-24-0713-RE
Atallah, O. O., Mazrou, Y. S. A., Atia, M. M., Nehela, Y., Abdelrhim, A. S., Nader, M. M. (2022). Polyphasic characterization of four aspergillus species as potential biocontrol agents for white mold disease of bean. J. Fungi 8, 626. doi: 10.3390/jof8060626
Bates, L. S., Waldren, R. P., Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Baturo-Ciesniewska, A., Groves, C. L., Albrecht, K. A., Smith, D. L., Grau, C. R., Willis, D. K. (2017). Molecular identification of Sclerotinia trifoliorum and Sclerotinia sclerotiorum isolates from the United States and Poland. Plant Dis. 101, 192–199. doi: 10.1094/PDIS-06-16-0896-RE/ASSET/IMAGES/LARGE/PDIS-06-16-0896-RE_F5-1481799443551.JPEG
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340
Blum, M., Chang, H. Y., Chuguransky, S., Grego, T., Kandasaamy, S., Mitchell, A., et al. (2021). The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354. doi: 10.1093/nar/gkaa977
Bolton, M. D., Thomma, B. P. H. J., Nelson, B. D. (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. doi: 10.1111/J.1364-3703.2005.00316.X
Borges, A., Tsai, S. M., Caldas, D. G. G. (2012). Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 31, 827–838. doi: 10.1007/S00299-011-1204-X/TABLES/5
Çavuşoǧlu, K., Çavuşoǧlu, D. (2021). Role of L-ornithine in mitigation of salt stress in Allium cepa L. Bangladesh J. Bot. 50, 1165–1171. doi: 10.3329/BJB.V50I4.57085
Ceresini, P. C., Silva, T. C., Vicentini, S. N. C., Júnior, R. P. L., Moreira, S. I., Castro-Ríos, K., et al. (2024). Strategies for managing fungicide resistance in the Brazilian tropical agroecosystem: Safeguarding food safety, health, and the environmental quality. Trop. Plant Pathol. 49, 36–70. doi: 10.1007/S40858-023-00632-2
Cessna, S. G., Sears, V. E., Dickman, M. B., Low, P. S. (2000). Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12, 2191–2199. doi: 10.1105/TPC.12.11.2191
Chauhan, S., Katoch, S., Sharma, S. K., Sharma, P. N., Rana, J. C., Singh, K., et al. (2020). Screening and identification of resistant sources against Sclerotinia sclerotiorum causing white mold disease in common bean. Crop Sci. 60, 1986–1996. doi: 10.1002/CSC2.20160
Czerwoniec, P. (2022). New plant resistance inducers based on polyamines. Open Chem. 20, 1591–1600. doi: 10.1515/CHEM-2022-0261/ASSET/GRAPHIC/J_CHEM-2022-0261_FIG_003.JPG
Daniel, A. I., Basson, G., Keyster, M., Klein, A., Gokul, A. (2024). Molecular mechanism of oxalic acid synthesis as virulence factor of Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 134, 102412. doi: 10.1016/J.PMPP.2024.102412
da Rocha, I. M. A., Vitorello, V. A., Silva, J. S., Ferreira-Silva, S. L., Viégas, R. A., Silva, E. N., et al. (2012). Exogenous ornithine is an effective precursor and the δ-ornithine amino transferase pathway contributes to proline accumulation under high N recycling in salt-stressed cashew leaves. J. Plant Physiol. 169, 41–49. doi: 10.1016/J.JPLPH.2011.08.001
Djeridane, A., Yousfi, M., Nadjemi, B., Boutassouna, D., Stocker, P., Vidal, N. (2006). Antioxidant activity of some Algerian medicinal plants extracts containing phenolic compounds. Food Chem. 97, 654–660. doi: 10.1016/j.foodchem.2005.04.028
Du, Z., Bramlage, W. J. (1992). Modified thiobarbituric acid assay for measuring lipid oxidation in sugar-rich plant tissue extracts. J. Agric. Food Chem. 40, 1566–1570. doi: 10.1021/JF00021A018
El-Argawy, E. (2012). Oxalic acid production by Sclerotinia sclerotiorum and its relation to pathogenicity. J. Plant Prot. Pathol. 3, 211–225. doi: 10.21608/jppp.2012.83753
El-Nagar, A., Elzaawely, A. A., El-Zahaby, H. M., Xuan, T. D., Khanh, T. D., Gaber, M., et al. (2023). Benzimidazole Derivatives Suppress Fusarium Wilt Disease via Interaction with ERG6 of Fusarium equiseti and Activation of the Antioxidant Defense System of Pepper Plants. J. Fungi 9, 244. doi: 10.3390/JOF9020244/S1
Fagundes-Nacarath, I. R. F., Debona, D., Rodrigues, F. A. (2018). Oxalic acid-mediated biochemical and physiological changes in the common bean-Sclerotinia sclerotiorum interaction. Plant Physiol. Biochem. 129, 109–121. doi: 10.1016/J.PLAPHY.2018.05.028
Ghahremani, M., Ghanati, F., Bernard, F., Gholami, M., Azad, T. (2013). Effects of exogenous ornithine enantiomers on tobacco cells under salinity conditions. Prog. Biol. Sci. 3, 100–107. doi: 10.22059/pbs.2013.32099
Godoy, G., Steadman, J. R., Dickman, M. B., Dam, R. (1990). Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 37, 179–191. doi: 10.1016/0885-5765(90)90010-U
Guimarães, R. L., Stotz, H. U. (2004). Oxalate Production by Sclerotinia sclerotiorum Deregulates Guard Cells during Infection. Plant Physiol. 136, 3703–3711. doi: 10.1104/PP.104.049650
Harrach, B. D., Fodor, J., Pogány, M., Preuss, J., Barna, B. (2008). Antioxidant, ethylene and membrane leakage responses to powdery mildew infection of near-isogenic barley lines with various types of resistance. Eur. J. Plant Pathol. 121, 21–33. doi: 10.1007/s10658-007-9236-3
Hasabi, V., Askari, H., Alavi, S. M., Zamanizadeh, H. (2014). Effect of amino acid application on induced resistance against citrus canker disease in lime plants. J. Plant Prot. Res. 54, 144–149. doi: 10.2478/jppr-2014-0023
Hegedus, D. D., Rimmer, S. R. (2005). Sclerotinia sclerotiorum: When ‘to be or not to be’ a pathogen? FEMS Microbiol. Lett. 251, 177–184. doi: 10.1016/J.FEMSLE.2005.07.040
Heller, A., Witt-Geiges, T. (2013). Oxalic acid has an additional, detoxifying function in Sclerotinia sclerotiorum pathogenesis. PloS One 8, e72292. doi: 10.1371/JOURNAL.PONE.0072292
Hussein, H. A. A., Mekki, B. B., El-Sadek, M. E. A., El Lateef, E. E. (2019). Effect of L-Ornithine application on improving drought tolerance in sugar beet plants. Heliyon 5, e02631. doi: 10.1016/J.HELIYON.2019.E02631
Jones, D. T., Taylor, W. R., Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Kähkönen, M. P., Hopia, A. I., Vuorela, H. J., Rauha, J. P., Pihlaja, K., Kujala, T. S., et al. (1999). Antioxidant activity of plant extracts containing phenolic compounds. J. Agric. Food Chem. 47, 3954–3962. doi: 10.1021/jf990146l
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Killiny, N., Nehela, Y. (2020). Citrus polyamines: structure, biosynthesis, and physiological functions. Plants 9, 426. doi: 10.3390/plants9040426
Kim, K. S., Min, J. Y., Dickman, M. B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Molecular Plant-Microbe Interactions, 21(5), 605–612.
Kumar, S., Stecher, G., Suleski, M., Sanderford, M., Sharma, S., Tamura, K., et al. (2024). MEGA12: molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 41, 1–9. doi: 10.1093/MOLBEV/MSAE263
Kuramae-Izioka, E. E. (1997). A rapid, easy and high yield protocol for total genomic DNA isolation of Colletotrichum gloeosporioides and Fusarium oxysporum. Rev. UNIMAR 19, 683–689. doi: 10.5555/19981005329
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Le Cointe, R., Simon, T. E., Delarue, P., Hervé, M., Leclerc, M., Poggi, S. (2016). Reducing the use of pesticides with site-specific application: the chemical control of rhizoctonia solani as a case of study for the management of soil-borne diseases. PloS One 11, e0163221. doi: 10.1371/JOURNAL.PONE.0163221
Liang, X., Liberti, D., Li, M., Kim, Y. T., Hutchens, A., Wilson, R., et al. (2015). Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 16, 559–571. doi: 10.1111/MPP.12211/SUPPINFO
Liang, X., Rollins, J. A. (2018). Mechanisms of broad host range necrotrophic pathogenesis in sclerotinia sclerotiorum. Phytopathology 108, 1128–1140. doi: 10.1094/PHYTO-06-18-0197-RVW/ASSET/IMAGES/LARGE/PHYTO-06-18-0197-RVW_F3.JPEG
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Liu, C., Xue, Z., Tang, D., Shen, Y., Shi, W., Ren, L., et al. (2018). Ornithine δ-aminotransferase is critical for floret development and seed setting through mediating nitrogen reutilization in rice. Plant J. 96, 842–854. doi: 10.1111/TPJ.14072
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, H. W., Xing, P. P., Liu, J. H., Lai, R. F., He, L. X., Zhang, T. T., et al. (2020). Application of ornithine-induced regulation in yield formation, grain quality and aroma of fragrant rice. Cereal Res. Commun. 48, 485–492. doi: 10.1007/S42976-020-00075-4/FIGURES/4
Malik, C. P., Singh, M. B. (1980). Plant enzymology and histo- enzymology (New Delhi, IN: Kalyani Publishers).
Marciano, P., Di Lenna, P., Magro, P. (1983). Oxalic acid, cell wall-degrading enzymes and pH in pathogenesis and their significance in the virulence of two Sclerotinia sclerotiorum isolates on sunflower. Physiol. Plant Pathol. 22, 339–345. doi: 10.1016/S0048-4059(83)81021-2
McCaghey, M., Shao, D., Kurcezewski, J., Lindstrom, A., Ranjan, A., Whitham, S. A., et al. (2021). Host-Induced Gene Silencing of a Sclerotinia sclerotiorum oxaloacetate acetylhydrolase Using Bean Pod Mottle Virus as a Vehicle Reduces Disease on Soybean. Front. Plant Sci. 12. doi: 10.3389/FPLS.2021.677631/BIBTEX
Michael, A. J. (2016). Biosynthesis of polyamines and polyamine-containing molecules. Biochem. J. 473, 2315–2329. doi: 10.1042/BCJ20160185
Monteoliva, M. I., Rizzi, Y. S., Cecchini, N. M., Hajirezaei, M. R., Alvarez, M. E. (2014). Context of action of proline dehydrogenase (ProDH) in the Hypersensitive Response of Arabidopsis. BMC Plant Biol. 14, 1–11. doi: 10.1186/1471-2229-14-21
Nehela, Y., Killiny, N. (2019). ‘Candidatus Liberibacter asiaticus’ and its vector, Diaphorina citri, augment the tricarboxylic acid cycle of their host via the γ-aminobutyric acid shunt and polyamines pathway. Mol. Plant-Microbe Interact. 32, 413–427. doi: 10.1094/MPMI-09-18-0238-R
Nehela, Y., Killiny, N. (2020). The unknown soldier in citrus plants: polyamines-based defensive mechanisms against biotic and abiotic stresses and their relationship with other stress-associated metabolites. Plant Signal. Behav. 15, 1761080. doi: 10.1080/15592324.2020.1761080
Nehela, Y., Killiny, N. (2023). Gamma-Aminobutyric Acid Accumulation Contributes to Citrus sinensis Response against ‘Candidatus Liberibacter Asiaticus’ via Modulation of Multiple Metabolic Pathways and Redox Status. Plants 12, 3753. doi: 10.3390/PLANTS12213753
Nehela, Y., Mazrou, Y. S. A., El_Gammal, N. A., Atallah, O., Xuan, T. D., Elzaawely, A. A., et al. (2024). Non-proteinogenic amino acids mitigate oxidative stress and enhance the resistance of common bean plants against Sclerotinia sclerotiorum. Front. Plant Sci. 15. doi: 10.3389/FPLS.2024.1385785/BIBTEX
O’Sullivan, C. A., Belt, K., Thatcher, L. F. (2021). Tackling control of a cosmopolitan phytopathogen: sclerotinia. Front. Plant Sci. 12. doi: 10.3389/FPLS.2021.707509/BIBTEX
Osman, H. E. M., Nehela, Y., Elzaawely, A. A., El-Morsy, M. H., El-Nagar, A. (2023). Two Bacterial Bioagents Boost Onion Response to Stromatinia cepivora and Promote Growth and Yield via Enhancing the Antioxidant Defense System and Auxin Production. Horticulturae 9, 780. doi: 10.3390/HORTICULTURAE9070780/S1
Papadopoulos, J. S., Agarwala, R. (2007). COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. doi: 10.1093/bioinformatics/btm076
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi: 10.1002/jcc.20084
Petzoldt, R., Dickson, M. H. (1996).Straw test for resistance to white mold in beans. Available online at: https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=Straw+test+for+resistance+to+white+mold+in+beans&btnG (Accessed November 7, 2023).
Prentice, R. L. (1976). A generalization of the probit and logit methods for dose response curves. Biometrics 32, 761. doi: 10.2307/2529262
Qamar, A., Mysore, K., Senthil-Kumar, M. (2015). Role of proline and pyrroline-5-carboxylate metabolism in plant defense against invading pathogens. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00503
Rana, K., Yuan, J., Liao, H., Banga, S. S., Kumar, R., Qian, W., et al. (2022). Host-induced gene silencing reveals the role of Sclerotinia sclerotiorum oxaloacetate acetylhydrolase gene in fungal oxalic acid accumulation and virulence. Microbiol. Res. 258, 126981. doi: 10.1016/J.MICRES.2022.126981
Renaud, J. B., Desrochers, N., Hoogstra, S., Garnham, C. P., Sumarah, M. W. (2021). Structure activity relationship for fumonisin phytotoxicity. Chem. Res. Toxicol. 34, 1604–1611. doi: 10.1021/ACS.CHEMRESTOX.1C00057/SUPPL_FILE/TX1C00057_SI_001.PDF
Rizzi, Y. S., Monteoliva, M. I., Fabro, G., Grosso, C. L., Laróvere, L. E., Alvarez, M. E. (2015). P5CDH affects the pathways contributing to Pro synthesis after ProDH activation by biotic and abiotic stress conditions. Front. Plant Sci. 6. doi: 10.3389/FPLS.2015.00572
Romero, F. M., Maiale, S. J., Rossi, F. R., Marina, M., Ruíz, O. A., Gárriz, A. (2018). “Polyamine metabolism responses to biotic and abiotic stress,” in Polyamines: Methods and Protocols, Methods in Molecular Biology, vol. 1694 . Eds. Alcázar, R., Tiburcio, A. F. (Humana Press, New York, NY), 37–49. doi: 10.1007/978-1-4939-7398-9_3
Romero-Puertas, M. C., Rodríguez-Serrano, M., Corpas, F. J., Gómez, M., Del Río, L. A., Sandalio, L. M. (2004). Cadmium-induced subcellular accumulation of O2.- and H2O2 in pea leaves. Plant Cell Environ. 27, 1122–1134. doi: 10.1111/j.1365-3040.2004.01217.x
Schwartz, H. F., Steadman, J. R., Hall, R., Forster, R. L. (2005). Compendium of bean diseases. 2nd ed. Eds. Schwartz, H. F., Steadman, J. R. (St. Paul, MN: APS Press, American Phytopathological Society).
Senthil-Kumar, M., Mysore, K. S. (2012). Ornithine-delta-aminotransferase and proline dehydrogenase genes play a role in non-host disease resistance by regulating pyrroline-5-carboxylate metabolism-induced hypersensitive response. Plant Cell Environ. 35, 1329–1343. doi: 10.1111/J.1365-3040.2012.02492.X
Shaner, G., Finney, R. E. (1977). The effect of nitrogen fertilization on the expression of slow-mildewing resistance in knox wheat. Phytopathology 77, 1051. doi: 10.1094/phyto-67-1051
Sharma, S. K., Singh, D., Pandey, H., Jatav, R. B., Singh, V., Pandey, D. (2022). “An overview of roles of enzymatic and nonenzymatic antioxidants in plant,” in Antioxidant Defense in Plants: Molecular Basis of Regulation. Eds. Aftab, T., Hakeem, K. R. (Springer Nature, Singapore), 1–13. doi: 10.1007/978-981-16-7981-0_1/TABLES/1
Singh, N., Mukherjee, S. K., Rajam, M. V. (2020). Silencing of the Ornithine Decarboxylase Gene of Fusarium oxysporum f. sp. lycopersici by Host-Induced RNAi Confers Resistance to Fusarium Wilt in Tomato. Plant Mol. Biol. Rep. 38, 419–429. doi: 10.1007/S11105-020-01205-2/FIGURES/5
Sun, S. W., Lin, Y. C., Weng, Y. M., Chen, M. J. (2006). Efficiency improvements on ninhydrin method for amino acid quantification. J. Food Compos. Anal. 19, 112–117. doi: 10.1016/J.JFCA.2005.04.006
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/MOLBEV/MSAB120
Terán, H., Lema, M., Schwartz, H. F., Duncan, R., Gilbertson, R., Singh, S. P. (2006).Modified Petzoldt and Dickson scale for white mold rating of common bean. Available online at: https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=Modified+Petzoldt+and+Dickson+scale+for+white+mold+rating+of+common+bean&btnG (Accessed November 7, 2023).
Torres, M. A., Jones, J. D. G., Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/PP.106.079467
Walker, P. L., Ziegler, D. J., Giesbrecht, S., McLoughlin, A., Wan, J., Khan, D., et al. (2023). Control of white mold (Sclerotinia sclerotiorum) through plant-mediated RNA interference. Sci. Rep. 13, 1–13. doi: 10.1038/s41598-023-33335-4
White, T. J., Bruns, T., Lee, S., Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications. Eds. Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. (Academic Press, Inc., New York, New York), 315–322.
Williams, B., Kabbage, M., Kim, H.-J., Britt, R., Dickman, M. B. (2011). Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PloS Pathog. 7, e1002107. doi: 10.1371/journal.ppat.1002107
Xu, X.-Q., Zhang, Z.-Q. (2000). Kinetic spectrophotometric determination of oxalic acid based on the catalytic oxidation of bromophenol blue by dichromate. Mikrochim. Acta 135, 169–172. doi: 10.1007/s006040070006
Yang, C. W., Lin, C. C., Kao, C. H. (2000). Proline, ornithine, arginine and glutamic acid contents in detached rice leaves. Biol. Plant 43, 305–307. doi: 10.1023/A:1002733117506/METRICS
Yi, Q., Park, M. J., Vo, K. T. X., Jeon, J. S. (2024). Polyamines in plant–pathogen interactions: roles in defense mechanisms and pathogenicity with applications in fungicide development. Int. J. Mol. Sci. 25, 10927. doi: 10.3390/IJMS252010927
Keywords: ornithine, white mold, bean, antioxidant, Sclerotinia, OAH, oxalic acid
Citation: Nehela Y, Mazrou YSA, EL_Gammal NA, Atallah O, Abdelrhim AS, Kumar S, Ahmed T, Ali Q, Makhlouf AH and Hussain WAM (2025) Ornithine enhances common bean growth and defense against white mold disease via interfering with SsOAH and diminishing the biosynthesis of oxalic acid in Sclerotinia sclerotiorum. Front. Plant Sci. 16:1483417. doi: 10.3389/fpls.2025.1483417
Received: 19 August 2024; Accepted: 18 March 2025;
Published: 04 April 2025.
Edited by:
Brigitte Mauch-Mani, Université de Neuchâtel, SwitzerlandReviewed by:
Nikhilesh Dhar, University of California, Davis, United StatesChaofeng Wang, University of Nebraska-Lincoln, United States
Copyright © 2025 Nehela, Mazrou, EL_Gammal, Atallah, Abdelrhim, Kumar, Ahmed, Ali, Makhlouf and Hussain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasser Nehela, eWFzc2VyLm5laGVsYUBhZ3IudGFudGEuZWR1LmVn; Qurban Ali, cmF0dGFycXVyYmFuQHVhZXUuYWMuYWU=
†Present address: Sumit Kumar, KVK, Mau, Acharya Narendra Deva University of Agriculture and Technology, Ayodhya, UP, India
 Yasser Nehela
Yasser Nehela Yasser S. A. Mazrou2
Yasser S. A. Mazrou2 Osama Atallah
Osama Atallah Abdelrazek S. Abdelrhim
Abdelrazek S. Abdelrhim Sumit Kumar
Sumit Kumar Temoor Ahmed
Temoor Ahmed Qurban Ali
Qurban Ali