- Department of Biology, Stanford University, Stanford, CA, USA
Mu killer contains a partial inverted duplication of the mudrA transposase gene and two copies of the terminal inverted repeat A (TIRA) region of the master MuDR element of maize. Mu killer can effectively silence single copy MuDR/Mu lines, and it is proposed that a ∼4 kb hairpin RNA is generated by read through transcription from a flanking gene and that this transcript serves as a substrate for siRNA production. Mu killer was sequenced, except for a recalcitrant portion in the center of the locus, and shown to be co-linear with mudrA as originally proposed. The ability of the dominant Mu killer locus to silence a standard high copy number MuDR/Mu transposon line was evaluated. After two generations of exposure, about three quarters of individuals were silenced indicating reasonable effectiveness as measured by the absence of mudrA transposase transcripts. Mu killer individuals that effectively silenced MuDR expressed two short antisense transcripts. In contrast, Mu killer individuals that failed to silence MuDR expressed multiple sense transcripts, derived from read through transcription initiating in a flanking gene, but no antisense transcripts were detected.
Introduction
MuDR/Mu comprise the mutator family of maize Class II transposable elements (reviewed in Lisch, 2002; Walbot and Rudenko, 2002). These high copy number elements are very effective mutagens, because they target RNA Polymerase II transcription units (Fernandes et al., 2004); MuDR/Mu in active mutator lines increase the forward mutation frequency 50–100-fold per locus (Walbot and Rudenko, 2002). MuDR contains two convergently transcribed genes: mudrA encodes transposase and mudrB encodes a protein of unknown function. Transcripts of both genes are subject to alternative splicing, which results in multiple protein isoforms (Walbot and Rudenko, 2002). The diverse elements share ∼215 bp terminal inverted repeats (TIRs) within which there is a highly conserved MURA transposase binding site in all mobile Mu (Benito and Walbot, 1997) and the gene promoters (Raizada et al., 2001a,b). A key feature of the family is the high copy number of ∼50–100 mobile Mu elements in active mutator plants. Utilizing replicative transposition in which parental transposon locations are preserved, MuDR/Mu elements double in copy number late in the lifecycle by inserting throughout the genome. Consequently, progeny of an outcross to a non-mutator line have about the same number of elements as the mutator parent (Alleman and Freeling, 1986; Walbot and Warren, 1988).
When MuDR/Mu copy number increases substantially, as after self-pollination, the likelihood of transposon silencing increases (Robertson, 1986; Walbot, 1986). Historically Mu element silencing was followed using visible phenotypes, either loss of somatic excision from mutable alleles, for example the switch from spotted to colorless kernels (Robertson, 1986; Walbot, 1986), or by activation of gene expression at a locus suppressed by a nearby Mu insertion, for example converting pale green to darker leaf pigmentation (Martienssen et al., 1990). Molecular assays for silencing include DNA methylation within Mu elements (Chandler and Walbot, 1986) and decreased mudrA transposase transcripts (Rudenko et al., 2003). Although some individuals switch abruptly from an active to inactive status, in many cases loss of Mu element insertion and excision occur over a span of time. Silencing is progressive, both within the sequential organs of a developing plant (Martienssen et al., 1990) and over several generations (Takumi and Walbot, 2007). The trigger for silencing other than the correlation with high MuDR/Mu copy number has been difficult to define, although there are clearly separable steps for imposition and maintenance of silencing (Woodhouse et al., 2006).
The stochastic appearance and incomplete penetrance of silencing have been barriers to establishing the specific components of the silencing machinery. All maize lines contain defective hMuDR elements (Rudenko and Walbot, 2001) that generate both sense and antisense transcripts, however, these elements are insufficient to direct silencing efficiently in crosses with active mutator individuals. Similarly introduction of antisense transgenes is not an efficient silencing method (Kim and Walbot, 2003). In an unusual single copy Mu1 plus single copy MuDR line, called minimal mutator (Lisch et al., 1995), spontaneous silencing is exceedingly rare (Lisch and Freeling, 1994). This permitted detection of a newly arisen dominant factor, Mu killer (GenBank accession: DQ011286), that could swiftly silence the minimal mutator system (Slotkin et al., 2003). The Mu killer locus has an internally deleted and partially duplicated MuDR element (Figure 1); the two TIRs are both derived from the mudrA adjacent element end and there are two partial copies of mudrA that terminate in an inverted duplication. The precise DNA sequence of Mu killer has not been reported. It is hypothesized that transcripts containing the inverted mudrA duplicated region form fold back RNA that is processed into small interfering RNAs that direct silencing of the single MuDR in the line (Slotkin et al., 2005).
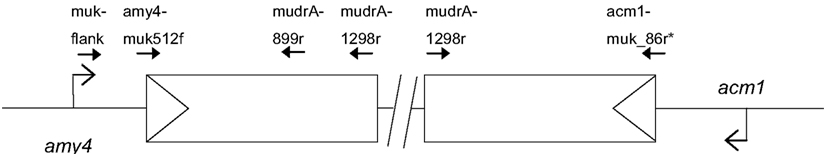
Figure 1. Schematic of the Mu killer locus and the two flanking maize genes amy4 and acm1. The triangles indicate the TIRA sequences. Primers used in the analysis of Mu killer genomic DNA and RT-PCR products are indicated.
We have reported a transcriptomic and proteomic comparison of anthers from sister lines that remained active or were recently fully silenced. In standard, high copy number active mutator lines the transcriptome is altered ∼25% in comparison to the silenced line, including activation of stress pathways (Skibbe et al., 2009), indicating that mutator activity has a major impact on the host without disrupting normal anther development. In the current study we sought to determine if Mu killer could silence a standard high element copy number mutator line, because these lines are non-responsive to ectopically expressed antisense RNA (Kim and Walbot, 2003). Second, if Mu killer can invoke silencing in such lines is the impact on the transcriptome similar to what occurs during spontaneous silencing? Third, we wished to better define the structure of Mu killer as an aid to understanding the structure of predicted RNA transcripts that might be mechanistically involved in MuDR/Mu silencing.
Materials and Methods
Genetic Materials
A Mu killer, a1-mu1//a1 single copy MuDR minimal mutator stock was obtained from Damon Lisch. This line was maintained by crossing to an a1 minimal mutator line or to an a1 tester grown at Stanford CA or on Moloka’i HI. High copy MuDR/Mu lines with bz2-mutable alleles were described previously (Skibbe et al., 2009). The crossing program for evaluating the capacity of the Mu killer line to silence the high copy number lines is presented in Figure 2.
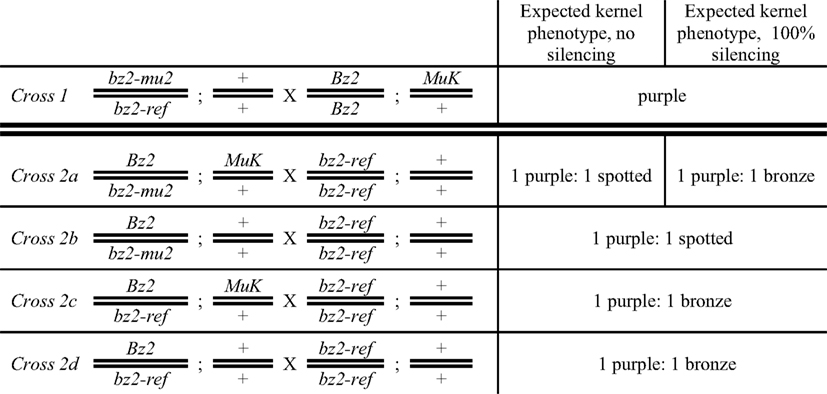
Figure 2. Crossing scheme to test for Mu killer-induced silencing of a high copy mutator line. Half of the progeny ears contain the bz2-mu2 reporter allele and are expected to segregate 1:1 purple:spotted (Crosses 2a and 2b); the other half (Crosses 2c and 2d) lack the reporter allele and are expected to segregate 1:1 purple:bronze. Mu killer is segregating 1:1 in Crosses 2a and 2c.
Sequencing Mu killer
The Mu killer locus was PCR amplified in two halves (Figure 1). A 1.8 kb amplicon was obtained for the “left” half using the muk-flank (5′ CCTTTGGTCAGTTCGGTTATCTCTG 3′) and mudrA-1298r (5′GCACCCATTTGTGGTTTCTT 3′) primers. A 1.3 kb amplicon was obtained for the “right” half using the acm1-Mu killer chimeric primer acm1-muk_86r (5′ CCCCTACTTGTTGTTGAGATAATTG 3′) and mudrA-1298r primer. Genomic DNA targets were amplified in a 25 μl reaction using 20 ng genomic DNA as template, 0.5 μM of each primer, 2 mM MgSO4, 0.2 mM dNTPs, 1× HiFi Buffer, and 1 unit HiFi Platinum Taq (Invitrogen Life Science, Carlsbad CA, USA) per reaction under the following conditions: 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 60°C for 45 s, and 72° for 1.5 min; there was a final extension at 72°C for 10 min. The left and right Mu killer amplicons were purified using the Qiagen (Valencia, CA, USA) PCR Purification Kit, TOPO subcloned, and sequenced using M13 forward and M13 reverse primers. The Mu killer sequence segments were assembled and validated by replicate sequencing and are available under GenBank numbers JX067398 (left half) and JX067399 (right half).
Genotyping
Genomic DNA was extracted from adult leaf tissue using the CTAB method of Rogers and Bendich (1985). Genotyping was performed using the PCR parameters described above. Target, amplicon size, and corresponding primer pairs are: Mu killer, 0.7 kb amplicon with the muk-flank (5′ CCTTTGGTCAGTTCGGTTATCTCTG 3′) and mudr-219r (5′ AGGAGAGACGGTGACAAGAGGAGTA 3′) primers; bz2-mu2: 0.2 kb amplicon with the bz2-637f (5′ CAGAGAGGTGCCAACAGAAGT 3′) and Mu-TIR (5′ AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC 3′) primers.
RNA Analysis
Total RNA was extracted from either whole spikelets containing 1 mm (pre-meiotic) to ∼2 mm (late prophase I meiotic stage) anthers or dissected 1–2 mm anthers using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. RNA quantity and quality were assessed with the Quant-iT RiboGreen RNA Assay Kit (Invitrogen) and agarose gel electrophoresis, respectively. Approximately 1 μg of total RNA was treated with DNase I and reverse transcribed into cDNA with an oligo(dT) primer using the SuperScript III Reverse Transcription system (Invitrogen) as per manufacturer’s instructions. Approximately 20 ng cDNA was used as template for RT-PCR. Target, expected amplicon size, and corresponding primer pairs are: Mu killer, 0.5 and 0.7 kb amplicons with the muk-flank (5′ CCTTTGGTCAGTTCGGTTATCTCTG 3′) and mudr-219r (5′ AGGAGAGACGGTGACAAGAGGAGTA 3′) primers; 0.5 kb (spliced) and 0.6 kb (alternatively spliced) mudrA with the mudrA-2345f (5′ AGGAATGGCAACACACTGGGAAAC′ 3′) and mudrA-2938r (5′ TTCAGTGACTTCCTCTGCTACGTC 3′) primers; and tubulin6 (a DNase I control transcript), 0.5 kb (cDNA) with the tub6-475f (5′ AGTATGCCACTCCCTTGGTG 3′) and tub6-979r (5′ GGCACACATCATGTTCTTGG 3′) primers. Other primers used to amplify Mu killer were: amy-muk512f (5′AGTCCAGTCCGAGATAATTGC 3′) and mudrA-899r (5′ TTGTCCGTATCCAAACTTCCCT 3′). PCR amplification parameters were identical to those described above. PCR products were resolved on a 2% agarose/TBE gel stained with ethidium bromide.
Transcriptome Profiling
Individuals from ears segregating for mutator active and siblings silenced by Mu killer were used as generated from Cross 2 (Figure 2). Spotted (mutator active) or bronze (Mu killer) kernels were selected and grown in Stanford, California in summer 2008, and the presence or absence of Mu killer was confirmed via PCR on genomic DNA. Anthers were staged from the upper florets and then dissected onto dry ice and stored at −80°C. Total RNA was extracted from four biological replicates of mutator active and Mu killer anthers (15–20 anther sample size) and quantified and qualitatively assessed as described above. DNase I-treated total RNA (1 μg) was amplified and labeled with either Cy3 or Cy5 using a Low RNA Input Fluorescent Linear Amplification Kit (Agilent Technologies, Santa Clara, CA, USA). Eight hundred twenty-five nanograms of each labeled cRNA was hybridized for 17 h at 60°C on a 4 K × 44 K Agilent in situ synthesized oligonucleotide platform along with spike-in controls for calculating target RNA concentrations. Data acquisition, image processing, spot flagging and removal, and normalization were performed as described in Skibbe et al., 2009. Probes were considered “on” if fluorescence values were greater than a cutoff of 2.6 SD above the average background for all four replicates. Probes with only three replicates above the cutoff were also considered “on” if the average intensity was above the median for the set of probes with only three replicates above the cutoff. All microarray data associated with these experiments are available at GEO1 under the series accession number GSE38314.
Results
Mu killer Transcriptional Silencing of MuDR in High Copy Lines is Relatively Efficient but Incomplete
Mu killer acts as a dominant locus that effectively silences the minimal mutator system (Slotkin et al., 2003); based on the presence of a large inverted duplication of ∼1300 bp of the mudrA gene (Figure 1), it is hypothesized that Mu killer encodes a large fold back, double-stranded RNA transcript directing MuDR silencing by targeting transposase-encoding mudrA transcripts. Mu killer effectiveness in standard, high copy number mutator lines has not been reported. To address this question, a line with a single copy of Mu killer in a Bz2 line was crossed (Cross 1) to a high copy mutator bz2-mu2//bz2-ref spotted kernel line that has been demonstrated to have both a high forward mutation frequency and a large impact on the transcriptome of developing maize anthers (Skibbe et al., 2009). When the mutator system is active, late somatic excision from the bz2-mu2 reporter allele results in fine purple spots on a beige (bronze) background; when mutator activity is silenced, kernels are uniformly bronze. After Cross 1 all progeny have purple seed, requiring genotyping to identify the Mu killer carriers. As expected, all these individuals were also carriers of the MuDR/Mu transposon system. Because Mu killer-mediated silencing occurs most effectively when Mu killer is inherited through the female parent, an additional cross was performed by a bz2 tester strain. If Mu killer is highly effective, then the expectation was that half of the progeny ears would segregate 1:1 purple:bronze and half of the progeny ears would be fully bronze. Among the fully bronze cohort, there are four classes. Two of the classes lack the bz2-mu2 reporter gene and will be bronze only, independent of Mu killer. Classes 2a and 2b (Figure 2) inherit the bz2-mu2 reporter allele; both of these classes were exposed to Mu killer in the previous generation and one class retains Mu killer (Class 2a, segregating 1:1 in the progeny).
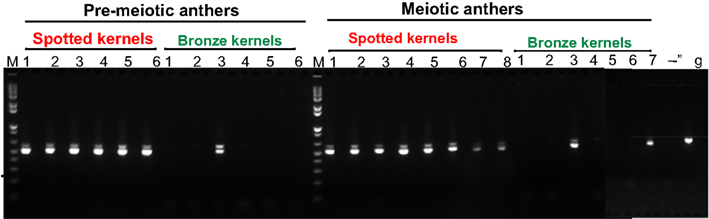
In Crosses 2a and 2b, ears contained the expected 50% purple kernels, but the remaining kernels were a mixture of spotted (active mutator) and bronze (loss of mutator activity). The mutator system can silence spontaneously, and there is a very wide variation in this frequency and in the percentage of spotted kernels during progressive spontaneous silencing (Takumi and Walbot, 2007). Therefore, interpreting loss of the somatic kernel phenotype is problematic. Instead, we conducted a molecular test independent of the spotted kernel phenotype to assess the ability of Mu killer to eliminate mudrA transcripts. Plants were grown from bronze kernels of Cross 2a (Mu killer carrier parent), using genotyping to pick plants that inherited Mu killer, to compare to plants derived from spotted kernels of the parental bz2-mu2/bz2 active mutator line. Anthers were chosen for analysis because they are the final organs differentiated and thus represent the location most likely to experience silencing.
The selected primers span the third intron of mudrA; this region of MuDR is absent from the Mu killer locus. The primers amplify a single genomic DNA band (Figure 3, lane g); however, near-complete splicing of the third intron, as expected (Hershberger et al., 1995), generated a smaller cDNA product with a faint larger band in active mutator samples. RT-PCR confirmed that all spotted kernels from the standard mutator line resulted in plants that amplified this 3′ region of mudrA transcripts indicative of maintenance of active mutator status; note that individuals 7 and 8 in the meiotic anther samples contain lowered mudrA transcript, an indication of higher likelihood of spontaneous loss of mutator activity (Rudenko et al., 2003). In contrast, only 3/13 of the Mu killer carrier bronze kernel plants retained mudrA transcripts (pre-meiotic anther sample 3 and meiotic anther samples 3 and 7). These initial results are consistent with Mu killer acting as a reasonably efficient silencing agent of high copy mutator lines when present for two generations of plant growth.
Attempts to Fully Sequence the Mu killer locus
The structure of the Mu killer transcript has been proposed to be a hairpin ∼4 kb in length, with an inversion point ∼1.3 kb into the mudrA region of the MuDR transposon. The structure of the Mu killer locus is based primarily on Southern blot evidence. To examine the proposed structure in more detail, the Mu killer locus was amplified using primers that are specific to mudrA, the TIR adjacent to mudrA, and the flanking amy4 and the acm1 genes to validate the gene order. The left and right halves of Mu killer were amplified in contiguous molecules of 1.8 and 1.3 kb, respectively, and fully sequenced and deposited into GenBank. The Mu killer locus begins with terminal inverted repeat A (TIRA) of MuDR and extends 1.298 kb into the mudrA gene on both the left and right halves. PCR amplification attempts using primers internal to 1.298 kb were unsuccessful indicating that the central region of MuDR is not present. Attempts to span the two halves using long-range PCR methods also failed; this result raises the possibility that an insertion/deletion polymorphism, a large rearrangement of unknown origin, or an exceptionally stable DNA hairpin structure between the left and right halves of the Mu killer locus prevents PCR amplification.
Unexpected Transcripts Encoded by Mu killer
In their analysis of the Mu killer locus, Slotkin et al. (2005) proposed novel transcription regulation in which the promoter of one of the flanking genes drives expression of Mu killer transcripts proposed to cause mutator silencing. This model invokes the unexpected feature that both TIRA promoters are inactive and that the TIRs fail to “insulate” the element from read through transcription.
To test these aspects of the model, progeny from Cross 1 were classified as active (gDNA “+”) or inactive (gDNA “−”) in silencing based on PCR confirmation of Mu killer using genomic DNA. In an effort to identify Mu killer transcripts associated with silencing activity, five gDNA “−” and five gDNA “+” individuals were tested via RT-PCR for accumulation of MuDR and Mukiller-derived transcripts in whole spikelets. As expected, all of the gDNA “−” individuals accumulated MuDR-derived transcript but failed to amplify detectable levels of Mukiller transcript (Figure 4, lanes 6–10). Surprisingly, only three of the five gDNA “+” individuals (Figure 4, lanes 1, 3, and 4, top row) amplified Mu killer transcript types. Furthermore, although only a 0.7 kb transcript was previously reported, an additional type was detected here. The remaining two gDNA “+” individuals (Figure 4, lanes 2 and 5, top row), failed to amplify detectable levels of Mukiller transcript. The MuDR and tubulin 6 control primers amplified as expected for each individual.
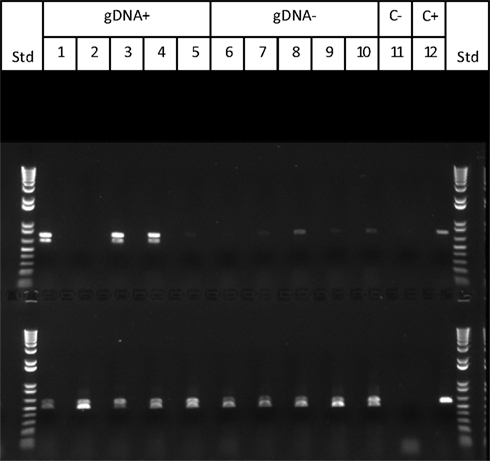
Figure 4. Mu killer transcript accumulation in whole spikelets. The upper panel primer pair amplifies Mu killer; the lower panel primer pair amplifies the 3′ end of MuDR (not present in Mu killer). Lanes 1–5 are five Mu killer gDNA “+” individuals; Lanes 6–10 are five Mu killer gDNA “−” individuals; Lane 11 is the negative PCR control; Lane 12 is the positive PCR control.
The two individuals that contained Mu killer but lacked Mu killer cDNA, along with two individuals that did accumulate mudrA cDNA were selected for further analysis along with positive and negative controls. The transcript accumulation in these four individuals was interrogated using primers positioned at four locations: (1) ∼500 bp upstream of Mukiller (amy4-flank) (2) at the junction of the amy4/Mu killer (amy4-muk_512f), (3) ∼900 bp into the Mu killer gene (mudrA-899r), and (4) ∼1300 bp into the Mu killer gene (mudrA-1298r). When the amy4-flank primer was paired with the two internal Mu killer primers, amplicons of the expected size (plus a splice variant ∼100 bp smaller) accumulated in the two Mu killer transcript “+” individuals but no transcripts were detected in either of the Mu killer transcript “−” individuals. When the amy4/Mu killer chimeric primer was paired with the two internal Mukiller primers, amplicons of the expected size (without any splice variants) accumulated in the two Mu killer transcript “+” individuals. In the Mu killer transcript “−” individuals, amplicons of an unexpected size were detected (Figure A1 in Appendix for the RT-PCR results). All amplicons were purified, sequenced, and the results are diagrammed in Figure 5.
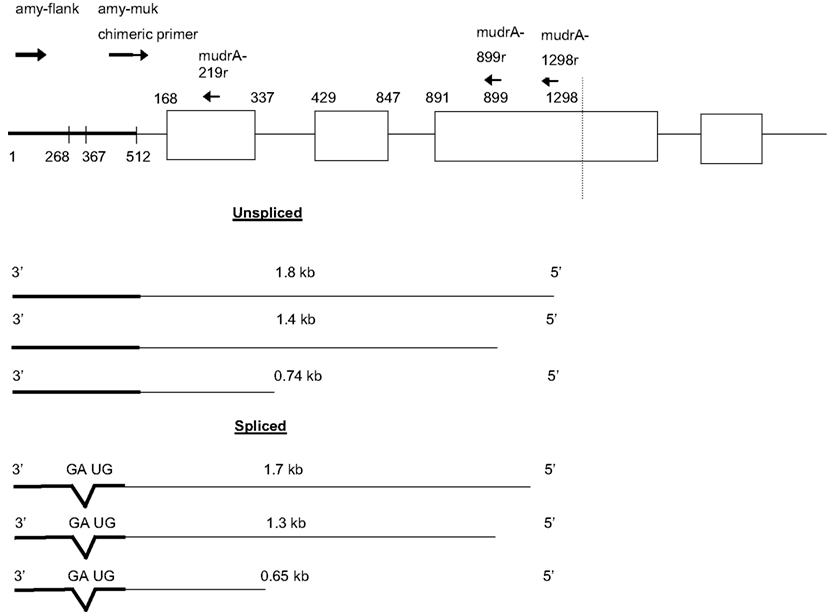
Figure 5. Antisense transcripts initiating in Mu killer and continuing into amy4 occur in plants with Mu killer-induced mutator silencing. Individuals were scored as having experienced Mu killer-induced silencing based on having genomic DNA positive for Mukiller but cDNA that lacked mudrA 3′ transcripts indicative of mutator activity (Figure 1).
Several unexpected molecular observations were made while assessing accumulation of Mu killer and mudrA transcripts in whole spikelets and anthers of these effective silencing cases. First, two families of antisense transcripts initiating within Mu killer and proceeding toward the amy4 locus accumulated in whole spikelets (Figure 5). A 0.74 kb amplicon was reported previously (Slotkin et al., 2005); however, none of the larger amplicons or the smaller 0.65 kb size class has been described. Approximately 80% of total transcript is in the 0.65 kb class, therefore, the 0.65 and 0.74 kb transcripts were cloned and sequenced to elucidate their structures. The 0.74 kb transcript type is antisense to mudrA. It is co-linear with an internal region contained in both Mu killer and MuDR (Hershberger et al., 1995) and with TIRA; the transcript terminates within amy4 (at position 1, Figure 5). Therefore, we conclude that the 0.74 kb transcripts initiate at a previously unreported antisense promoter within the mudrA sequence (within the sense intron 1) shared by Mu killer and MuDR. Additional analysis using RT-PCR amplification employing oligonucleotides further into Mu killer (mudrA-899r and mudrA-1298r) demonstrated that longer transcripts (Figure 5) can originate at least 1.298 kb into the Mu killer locus, starting within or beyond the region of Mu killer that could not be sequenced; it is possible that some transcripts initiate within the right side TIRA of Mu killer producing sense mudrA-containing RNA (5′ half of the transcripts) plus antisense mudrA (3′ half) in the region shared with the 0.74 kb transcript type.
Interestingly, the major 0.65 kb transcript results from splicing of a short intron with canonical 5′ donor and 3′ acceptor sites in the amy4 flanking region. Similarly, the minor 1.8 and 1.4 kb transcripts diagrammed in Figure 5 are inferred to be unspliced forms, and the minor 1.7 and 1.3 kb forms involve removal of the same intron documented in the 0.65 kb transcripts. Tissue-specific variation in the ratio of the 0.74 kb and alternatively spliced 0.65 kb transcripts was observed in whole spikelets (containing the vegetative glumes, lemma, and palea plus anthers), in anthers, and in adult leaves but the 0.65 kb form is always predominant.
Next, transcripts were analyzed from two individuals that failed to silence MuDR: they contained Mu killer but generated mudrA cDNA. In these individuals, no antisense transcript types were identified that extended into the amy4 flanking primer region, that is, both the 0.74 and 0.65 kb transcript types were missing. We therefore propose that production of the antisense 0.74 and 0.65 kb transcripts is required for Mu killer-mediated silencing of mudrA. In individuals that failed to silence mutator, RT-PCR amplification products were only identified for the combination of the chimeric amy4/Mu killer primer paired with the two internal mudrA oligonucleotides (Figure 6). These amplicons are in the sense orientation relative to mudrA but were shorter in length than canonical mudrA transcripts. Sequence analysis demonstrated that several alternatively spliced transcript types are present; analysis of the acceptor and donor sites confirmed that these transcripts are in the sense orientation with respect to the normal mudrA transcript. The introns identified are not spliced from normal mudrA transcripts, although alternative splicing does occur in master element transcripts (Hershberger et al., 1995).
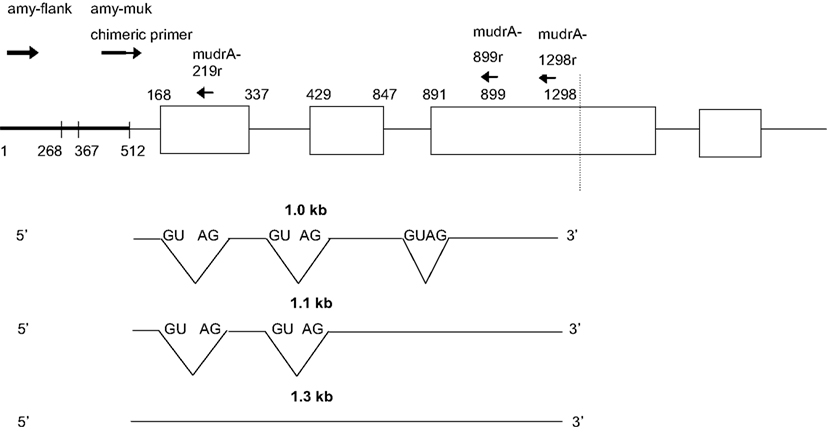
Figure 6. Sense transcripts contained within Mu killer occur in lines failing to silence MuDR. The two plants analyzed were genotyped as Mu killer carriers, however, the cDNA is negative for Mu killer antisense transcripts and positive for mudrA sense transcripts covering the 3′ end of the gene characteristic of mudrA (Figure 1).
Collectively, transcript analyses demonstrated that Mu killer individuals that silence MuDR accumulate mudrA antisense transcripts that extend into the amy4 flanking gene region; the promoter for these transcripts is not yet defined but it resides inside Mu killer. This result disproves the proposed Slotkin et al. (2005) model that antisense transcripts would originate from read through transcription from flanking maize genes. The results also demonstrate that TIRA lacks a transcription terminator in the context of the Mu killer structure at this locus. In contrast, Mu killer individuals that fail to silence MuDR accumulate sense transcript types that are predominately alternatively spliced compared to authentic mudrA transcripts. These sense transcripts appear to arise from transcriptional read through from the amy4 locus, as opposed to the Slotkin et al. (2005) model where antisense transcripts derive from flanking gene promoters. In our analysis such transcripts were determined to be in the sense orientation and not involved in mutator silencing. The existence of read through transcripts indicates that the TIRA in the context of Mu killer fails to insulate the Mu element from flanking gene transcriptional activity.
Mu killer and Spontaneous Silencing Result in Distinctive Gene Expression Changes in Anthers
A third question we addressed is how similar the consequences are of Mu killer-mediated compared to spontaneous epigenetic silencing in anther development. In a previous study, 1 and 2 mm anthers were shown to express more than 30,000 transcripts (Skibbe et al., 2009). In the current study the same criteria to be considered expressed or “on” were employed after analyzing four biological replicates of the mitotic (1 mm) and meiotic (2 mm) anther stages in Mu active and Mu killer silenced lines (Figure 7).
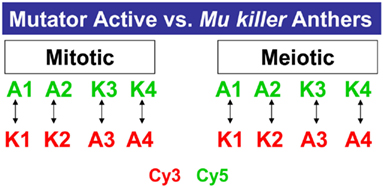
Figure 7. Microarray profiling experimental design. Four biological replicates of Mu active (A) or Mu killer (K) maize anthers were collected for two stages. For each stage, two samples were labeled with the Cy3 fluor (Green) or Cy5 fluor (Red) and hybridized as shown on the 4 K × 44 K Agilent maize oligonucleotide array platform.
We found that the pre-meiotic 1 mm anther transcriptome contains 34,338 transcripts in an active mutator background and slightly more (35,056) transcript types in Mu killer. The 2 mm meiotic anther samples expressed a similar number of transcripts with 34,508 (Mu active) and 33,700 (Mu killer). There were many examples of stage-specific transcripts in both biological sample types. Two types of differences in transcript expression comparing Mu active to Mu killer silenced lines were analyzed: (1) “Off/On” differences represent expression that was detected in one sample but not in the other and (2) “Up/Down” differences are the differential expression class and include probes where expression was at least 1.5-fold higher in one sample than the other (p-value < 0.05). Venn diagrams summarizing On/Off differences for Mu active vs. Mu killer backgrounds for both anther stages are shown in Figure 8. Both stages have distinct sets of more than 1500 transcripts not detected in the Mu active background but expressed in Mu killer (green numbers).
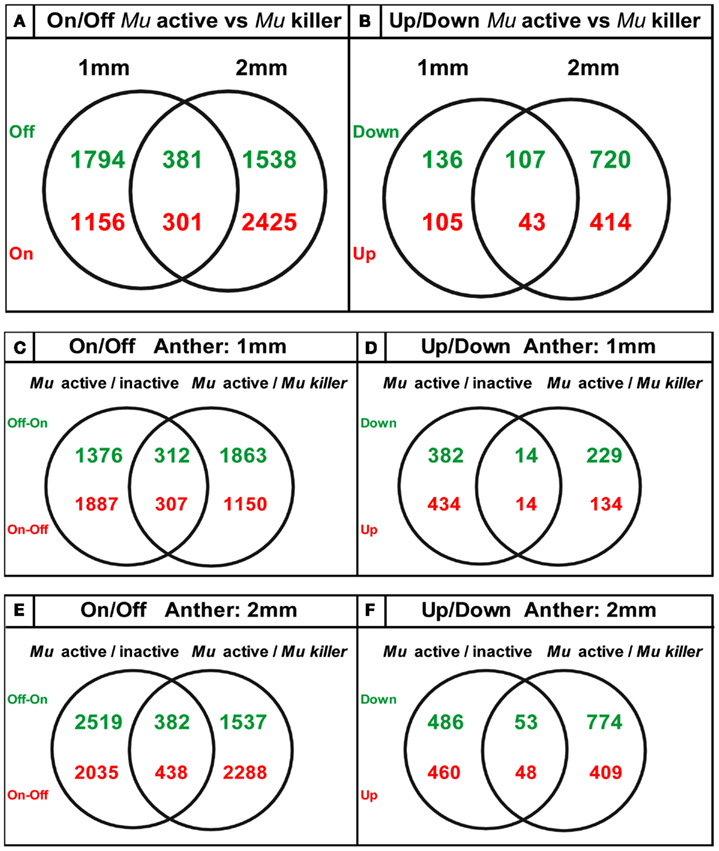
Figure 8. Venn diagrams showing unique sets of differentially expressed transcripts result from epigenetic and genetic inactivation of the mutator transposon family.
The ectopic activation of gene expression indicates that Mu killer has a major impact on the anther transcriptome compared to an active mutator line. Mu killer impact is dynamic, because only 381 transcript types were OFF in Mu active anthers at both developmental stages. In the reverse comparison (present in Mu active but absent in Mu killer) there are 1156 transcripts at 1 mm and 2425 at 2 mm with 301 transcripts absent in Mu killer for both anther lengths. Collectively, the differences between the two lines are more than 10% of the anther transcriptome. It is already established that high copy mutator lines express a diverse suite of stress response genes, compared to silenced sister lines (or to similar stage fertile anthers in a standard inbred line; Skibbe et al., 2009). Although not the largest GO category, 29 of the 1457 transcript types present in Mu active but missing from Mu killer at 1 mm and 47 of the 2726 transcript types at 2 mm are stress-associated. Surprisingly, there were far fewer differentially expressed transcript types than transcripts present in the On/Off category (Figures 8A,B). At 1 mm less than 1% of the transcriptome was impacted in the differential gene expression analysis, with only 2% altered at the 2 mm stage. It appears, therefore, that most transcriptome changes in Mu active compared to Mu killer lines involve complete suppression or de novo activation of suites of genes rather than modulation of transcript levels.
The transcriptome data was next analyzed with regard to how similar epigenetic silencing is to the silencing directed by Mu killer. We compared the differentially expressed transcripts determined from the previous study of Mu active vs. Mu inactive lines (Skibbe et al., 2009) with the differentially expressed transcripts found here in the Mu active vs. Mu killer lines. For both On/Off and Up/Down differences (Figures 8C + E,D + F) distinctive sets of transcripts are affected at each stage with a much smaller number of transcripts affected in common. These data suggest that Mu killer inactivation reprograms gene expression using a different route and perhaps involving different mechanisms that result in such distinctive consequences compared to spontaneous epigenetic silencing.
The transcripts that were “on” in Mu killer but “off” in Mu active lines at the 2 mm stage, as well as transcripts in the reverse situation, were mapped to version 2 of the MaizeCyc metabolic pathways database2 using the Omics Viewer feature of the Pathway Tools application3. The Omics Viewer displays metabolic pathways grouped into pathway classes (e.g., carbohydrate cycles or secondary metabolites biosynthesis) to acquire a general picture of the experiment effects. The On/Off status of the Mu killer line as compared to the Mu inactive line was used to color-code the affected reactions in each pathway; an opposite status is indicated by blue (“off” Mu killer, “on” Mu inactive) or red (“on” Mu killer, “off” Mu inactive) and the same status by teal (both “off”) or orange (both “on”). Affected pathways within a sample of five pathway classes were then categorized by their color scheme. As seen in Table 1, the majority of affected pathways were those with transcripts “off” in Mu killer but “on” in Mu inactive (blue). The combined “different” effects (27, blue and red) is more than the combined “same” effects (20, teal and orange), again indicating that Mu killer has distinctive aspects to its silencing mechanism. The many pathways affected in the “Carbohydrate biosynthesis” pathway class are displayed in Figure 9; five pathways have “off” reactions only with Mu killer (blue) and four pathways have “on” reactions only in Mu killer (red).
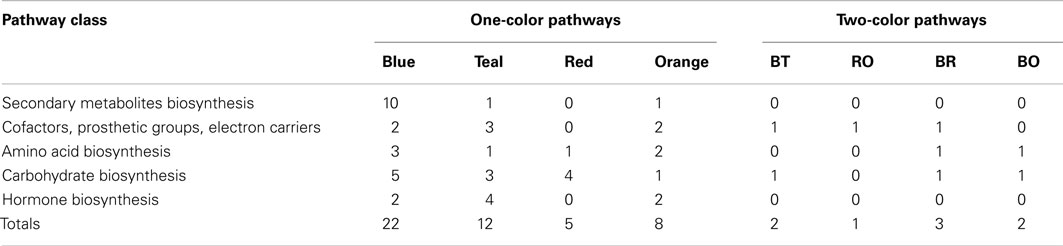
Table 1. Five metabolic pathway classes showing affects by Mu killer in common or different from Mu inactive lines.
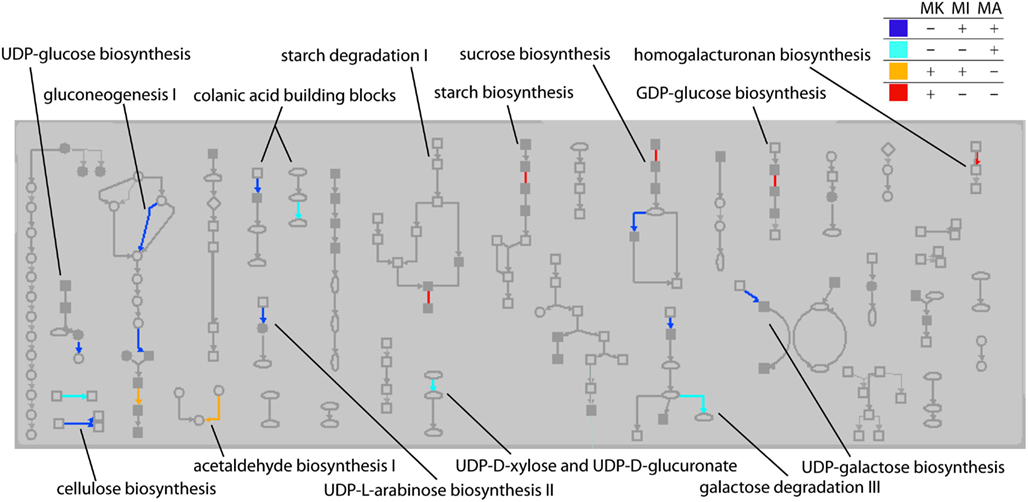
Figure 9. Carbohydrate biosynthesis pathway class showing reactions affected by Mu killer (MK) On/Off transcripts compared to Mu inactive (MI) and Mu Active (MA). Reactions in a pathway that were affected only by Mu killer are blue (“off” in Mu killer) or red (“on” in Mu killer). Reactions affected by both Mu killer and Mu inactive but not Mu active are either teal (“off” in Mu killer) or orange (“on” in Mu killer).
Interestingly, three of the pathway classes in Table 1 had pathways with multiple reactions affected in different ways; these are shown by non-zero counts in the last four columns. A web page was generated from the Pathway Tools with the full Omics Viewer display and is available at http:/www.stanford.edu/~walbot/maizecyc/mukiller_onoff.html. These insights can guide future work in metabolomics and proteomics to discover whether the transcriptomic differences reported here result in distinctive cellular properties.
Discussion
This study was formulated to examine three aspects of Mu killer: effectiveness in silencing standard high Mu copy number lines, molecular organization and expression of the locus, and impact on the anther transcriptome. After two generations of Mu killer exposure, 77% (10/13) high copy number MuDR/Mu individuals tested were silenced based on loss of transcripts characteristic of mudrA. Thus Mu killer-mediated silencing is considerably more frequent than the spontaneous ∼10–20% loss of mutator activity per generation generally observed (Robertson, 1986; Walbot, 1986) and is a useful practical tool to accelerate mutator silencing. Second, sequencing Mu killer confirmed the overall structure originally proposed without resolving the nature of the element center, which failed to amplify in any PCR strategy attempted. On the other hand, detailed analysis of transcript types documented numerous additional transcript types, clarified the strandedness of these and a previously documented transcript, discovered an association between two short antisense transcripts and mutator silencing, and found accumulation of sense transcripts in Mu killer individuals that failed to silence MuDR elements. Third, the impact of Mu killer silencing activity on the anther transcriptome is different from the impact of spontaneous silencing.
Standard MuDR elements contain promoters and transcription start sites within the TIRs (at about +165 and at sequences just internal to TIRA) that generate sense mudrA and mudrB RNA while antisense transcripts result from termination failure in the region separating these two genes (Walbot and Rudenko, 2002). Transgenic maize expressing additional antisense RNA failed to suppress MuDR sense transcript accumulation (Kim and Walbot, 2003); nonetheless, in this study a single copy Mu killer element producing 0.74 and 0.65 kb antisense mudrA transcripts was associated with effective silencing. Because these transcripts terminate in the flanking amy4 locus, it is possible that antisense RNA stability or effectiveness depends on the sequences contributed by the flanking gene. Both antisense transcripts are complementary to a short region of about 250 bases in authentic mudrA transcripts, including part of the second exon and the first exon including the 5′ untranslated region. Because the 0.65 and 0.74 kb RNAs cannot form large hairpin structures, it is presumed that small RNAs derived from them target either translation or transcription of mudrA. This is a new model to explain how Mu killer silences MuDR. It is possible that the minor longer transcript types (Figure 6) also contribute through the previously proposed mechanism of long hairpin formation (Slotkin et al., 2005).
In Mu killer individuals that failed to silence MuDR, we found sense rather than antisense transcripts derived from Mu killer. These read through transcripts initiated in the amy4 flanking locus, and also included the non-transcribed portion of TIRA, and mudrA-homologous sequences. A complex splicing pattern was observed, utilizing canonical 5′ donor and 3′ acceptor sites that are not used in authentic mudrA transcripts (Figure 7). Because MuDR is transcribed independent of its insertion sites, it is thought that transcription at MuDR loci is driven primarily by the enhancer elements located within the element TIRs; there is no evidence for read through transcripts when MuDR is inserted within the coding region of bz2-mu4 (Hershberger et al., 1995) although the TIRs function as promoters when assayed in heterologous constructs (Benito and Walbot, 1994; Raizada et al., 2001a,b). Thus the unusual structure of Mu killer and the flanking sequence context unexpectedly highlight that both MuDR promoters and the insulator functions of the TIRs can be suppressed.
If there is an “arms race” between transposon and host mechanisms to suppress them, aberrant transposon forms could be a potent weapon in host-mediated defenses as originally proposed for Mu killer. Endogenous hMuDR elements reside in every maize line tested; they are co-linear with MuDR but contain point mutations and other small defects that make them incompetent to program Mu element activities. They are also impotent in regulating MuDR (Rudenko and Walbot, 2001) suggesting that in this case MuDR persisted, or won the contest. The newly arisen Mu killer element can silence MuDR with reasonable frequency; however, we also found that when Mu killer fails to silence MuDR it switches from producing antisense to producing sense mudrA transcripts. In this case too, it appears that MuDR is victorious. The stability of the Mu killer transcription states is unknown, but it will be interesting in future studies to assess the persistence of Mu killer over a multiple generation time scale to determine if, like hMuDR elements, it ultimately fails to contain MuDR/Mu elements.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Supported by NSF grant 07-01880 from the Plant Genome Research Program.
Footnotes
References
Alleman, M., and Freeling, M. (1986). The Mu transposable elements of maize: evidence for transposition and copy number regulation during development. Genetics 112, 107–119.
Benito, M.-I., and Walbot, V. (1994). Promoter elements active in maize cells are located within the terminal inverted repeat sequences of MuDR. Maydica 39, 255–264.
Benito, M.-I., and Walbot, V. (1997). Characterization of the maize mutator transposable element MURA transposase as a DNA-binding protein. Mol. Cell. Biol. 17, 5165–5175.
Chandler, V. L., and Walbot, V. (1986). DNA modification of a transposable element of maize correlates with loss of activity. Proc. Natl. Acad. Sci. U.S.A. 83, 1767–1771.
Fernandes, J., Dong, Q. F., Schneider, B., Morrow, D. J., Nan, G. L., Brendel, V., and Walbot, V. (2004). Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol. 5, 82.
Hershberger, R. J., Benito, M.-I., Hardeman, K. J., Warren, C., Chandler, V. L., and Walbot, V. (1995). Convergent transcripts, antisense RNA, and splicing failure in the maize mutator element MuDR. Genetics 140, 1087–1098.
Kim, S.-H., and Walbot, V. (2003). Structural and functional analysis of antisense MuDR transcripts: insensitivity of maize mutator transposon activities to endogenous and transgene-encoded antisense RNA. Plant Cell 15, 2430–2447.
Lisch, D., Chomet, P., and Freeling, M. (1995). Genetic characterization of the mutator system in maize: behavior and regulation of Mu elements in a minimal line. Genetics 139, 1777–1796.
Lisch, D., and Freeling, M. (1994). Loss of mutator activity in a minimal line. Maydica 39, 289–300.
Martienssen, R., Barkan, A., and Taylor, W. C. (1990). Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize. Genes Dev. 4, 331–343.
Raizada, M. N., Benito, M.-I., and Walbot, V. (2001a). The MuDR transposon terminal inverted repeat contains a complex plant promoter directing distinct somatic and germinal programs. Plant J. 25, 1–15.
Raizada, M. N., Brewer, K. V., and Walbot, V. (2001b). A maize MuDR transposon promoter shows limited autoregulation. Mol. Genet. Genomics 265, 82–94.
Rogers, S. O., and Bendich, A. J. (1985). Extraction of DNA from milligram amounts of fresh herbarium and mummified plant tissues. Plant Mol. Biol. 5, 69–76.
Rudenko, G. N., Ono, A., and Walbot, V. (2003). Initiation of silencing of maize MuDR/Mu transposable elements. Plant J. 33, 1013–1025.
Rudenko, G. N., and Walbot, V. (2001). Expression and post-transcriptional regulation of maize transposable element MuDR and its derivatives. Plant Cell 13, 553–570.
Skibbe, D. S., Fernandes, J. F., Medzihradszky, K., Burlingame, A. L., and Walbot, V. (2009). mutator transposon activity reprograms the transcriptome and proteome of developing maize anthers. Plant J. 59, 622–633.
Slotkin, R. K., Freeling, M., and Lisch, D. (2003). Mu killer causes the heritable inactivation of the mutator family of transposable elements in Zea mays. Genetics 165, 781–797.
Slotkin, R. K., Freeling, M., and Lisch, D. (2005). Heritable silencing of a transposon family is initiated by a naturally occurring inverted repeat derivative. Nat. Genet. 137, 641–644.
Takumi, S., and Walbot, V. (2007). Epigenetic silencing and unstable inheritance of MuDR activity monitored at four bz2-mu alleles in maize (Zea mays L.). Genes Genet. Syst. 82, 387–401.
Walbot, V. (1986). Inheritance of mutator activity in Zea mays as assayed by somatic instability of the bz2-mu1 allele. Genetics 114, 1293–1312.
Walbot, V., and Rudenko, G. N. (2002). “MuDR/Mu transposons of maize,” in Mobile DNA II, eds N. L. Craig, R. Craigie, M. Gellert, and A. Lambowitz (Washington, DC: American Society for Microbiology), 533–564.
Walbot, V., and Warren, C. (1988). Regulation of Mu element copy number in maize lines with an active or inactive mutator transposable element system. Mol. Gen. Genetics 211, 27–34.
Woodhouse, M., Freeling, M., and Lisch, D. (2006). Initiation, establishment and maintenance of MuDR transposon silencing require distinct factors. PLoS Biol. 4, e339. doi:10.1371/journal.pbio.0040339/
Appendix
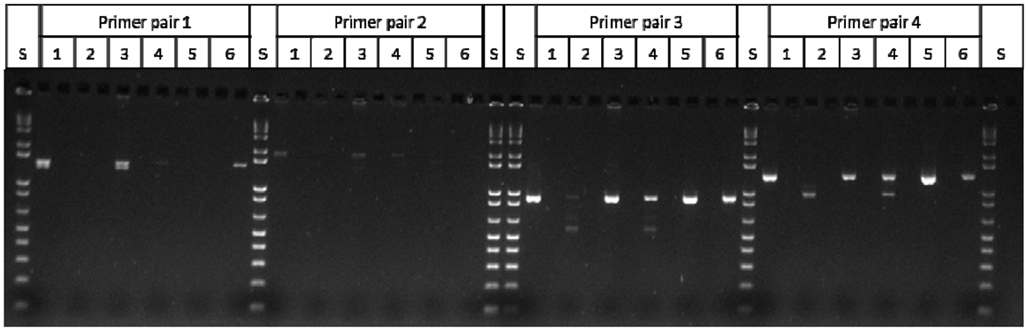
Figure A1. Mu killer individuals undergoing Mu killer silencing accumulate aberrant Mu killer transcript types. S = 1 kb plus DNA ladder; Lanes 1–4 = Mu killer gDNA “+” individuals; Lanes 1 and 3 accumulate expected transcript types; Lanes 2 and 4 accumulate aberrant transcript types; Lane 5 = mudrA control DNA; Lane 6 = Mu killer control DNA; Primer Pair 1: amy4-flank + mudrA-899r (expected size = 1400 bp); Primer Pair 2: amy4-flank + mudrA-1298r (expected size = 1819 bp); Primer Pair 3: amy4-muk_512f + mudrA-899r (expected size = 910 bp); Primer Pair 4: amy4-muk_512f + mudrA-1298r (expected size = 1308 bp). The amy4-muk_512f primer is a chimeric primer that is expected to amplify both Mu killer and MuDR loci.
Keywords: transposon, mutator, Mu killer, anther, maize, splicing
Citation: Skibbe DS, Fernandes JF and Walbot V (2012) Mu killer-mediated and spontaneous silencing of Zea mays mutator family transposable elements define distinctive paths of epigenetic inactivation. Front. Plant Sci. 3:212. doi: 10.3389/fpls.2012.00212
Received: 30 May 2012; Accepted: 23 August 2012;
Published online: 13 September 2012.
Edited by:
Bernie Carroll, The University of Queensland, AustraliaReviewed by:
Ruairidh Sawers, Laboratorio Nacional de Genomica para la Biodiversidad, MexicoPaula Casati, Centro de Estudios Fotosinteticos-CONICET, Argentina
Copyright: © 2012 Skibbe, Fernandes and Walbot. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: J. F. Fernandes and Virginia Walbot, Department of Biology, 385 Serra Mall, Stanford University, Stanford, CA 94305-5020, USA. e-mail:amZlcm5hbmRAc3RhbmZvcmQuZWR1;d2FsYm90QHN0YW5mb3JkLmVkdQ==
†Present address: David S. Skibbe, Syngenta, 317 330th Street, Stanton, MN 55018, USA.
Gene expression data from these data are available at GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE38314.