- Department of Plant Developmental Biology, Max Planck Institute for Plant Breeding Research, Cologne, Germany
The Arabidopsis thaliana circadian clock is an interconnected network highly tractable to systems approaches. Most elements in the transcriptional–translational oscillator were identified by genetic means and the expression of clock genes in various mutants led to the founding hypothesis of a positive–negative feedback loop being the core clock. The identification of additional clock genes beyond those defined in the core led to the use of systems approaches to decipher this angiosperm oscillator circuit. Kinetic modeling was first used to explain periodicity effects of various circadian mutants. This conformed in a flexible way to experimental details. Such observations allowed a recursive use of hypothesis generating from modeling, followed by experimental corroboration. More recently, the biochemical finding of new description of a DNA-binding activity for one class of clock components directed improvements in feature generation, one of which revealed that the core of the oscillator is a negative–negative feedback loop. The recursive use of modeling and experimental validation has thus revealed many essential transcriptional components that drive negative arms in the circadian oscillator. What awaits is to more fully describe the positive arms and an understanding of how additional pathways converge on the clock.
A Historical Path through the Molecular Genetics of the Plant Circadian Clock
The circadian clocks of various phyla have a rich history of modeling in molecular terms. The discovery of the first Drosophila clock gene PERIOD led to early mechanistic models of a daily oscillator (Goldbeter, 1995). As more genes were added to this animal oscillator, models became more rich and predictive (Gonze et al., 2011). Similar modeling approaches have been applied to the fungal (Tseng et al., 2012), the mammalian (Gerard and Goldbeter, 2012), and the cyanobacterial oscillators (Hatakeyama and Kaneko, 2012). Molecular differences in circadian clocks have been visible, which suggests multiple evolutionary origins (Dunlap et al., 2004).
For plants, a similarly rich use of modeling can be found (Shin and Davis, 2010). To date no less than seven distinct models of the plant clock have been published (Figure 1), and the most recent kinetic model (Pokhilko et al., 2012) and linear time invariant (LTI) model (Herrero et al., 2012) have highlighted the complexity in this system. These models, alongside other approaches, such as Boolean modeling (Akman et al., 2012), have various strengths in predicting the system. The first model that placed the plant clock in molecular terms was in 2005 (Locke et al., 2005a), where the authors presented an approach to inferring models when available molecular-genetic data are sparse and noisy. It is of interest that the recursive use of molecular genetics with mathematical modeling has served as a platform toward rational genetic understanding of the gears that make up the plant clock. Such iterative approaches should also bridge the transition of knowledge from genetic to cellular terms. In this review, we will briefly overview the important use of mathematics in unraveling the plant circadian oscillator, in an integrative sense, sometimes with “false starts,” to define where the state of the art is now. The need for new models will also be defended.
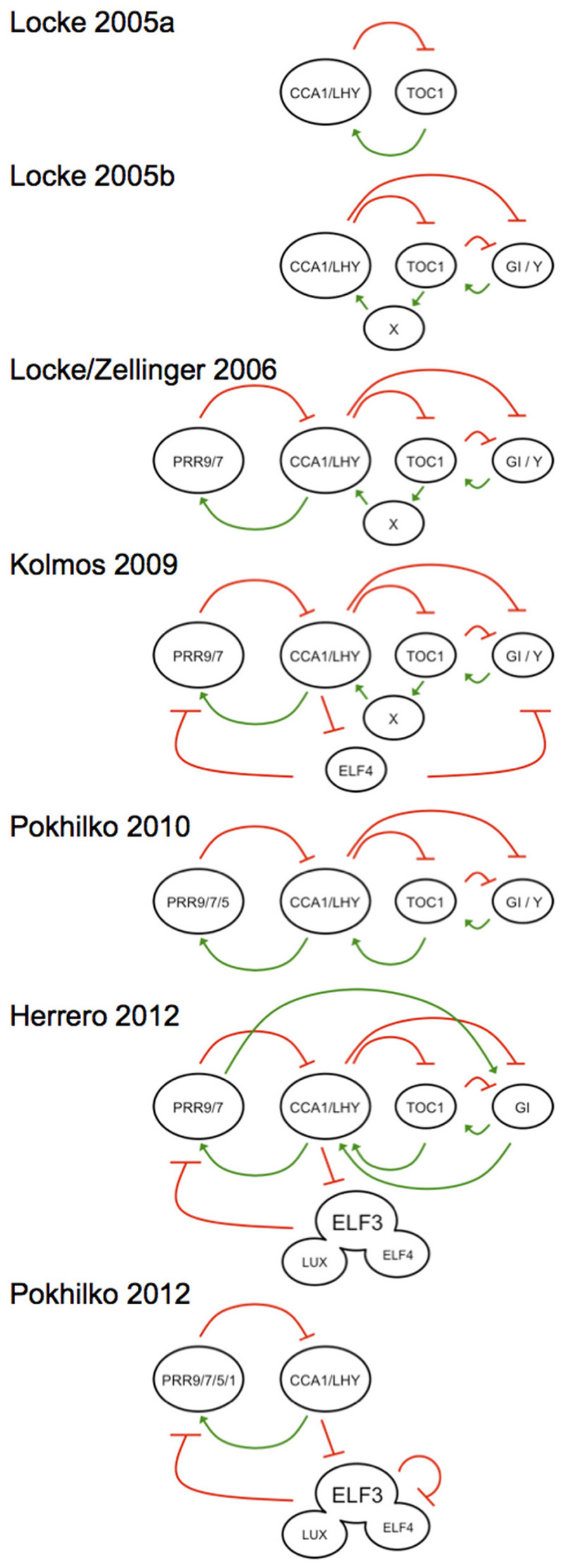
FIGURE 1. Graphical outline of the mathematical A. thaliana clock models in historical order, showing the development from a simple positive–negative feedback model (Locke et al., 2005a) toward more complicated interconnected feedback loops (Locke et al., 2005b, 2006; Zeilinger et al., 2006; Pokhilko et al., 2010). The oscillator expands with he subsequent placement of an evening complex element (Kolmos et al., 2009). Recent experiments could prove the existence of an evening complex working directly on the morning loop (Herrero et al., 2012; Pokhilko et al., 2012). Activation elements are defined by a green positive arrow and a red negative arrow indicates repression, respectively. All models are centered at the CCA1/LHY hub to illustrate the directional movement of the models over publication time. Each model is described in the text and the models are named after the primary author and publication year. Note that two models define the 2006 efforts, and are the predominant models that has driven most recent circadian work.
The Importance of Daily Rhythms in Plants
The circadian clock drives rhythms of growth and development as a pervasive force in essentially all aspects of a plant life cycle (Davis and Millar, 2001). For example, primary and secondary metabolism is timed to be coincident with available photic-energy sources and predictable heat and cold (McClung and Davis, 2010; Sanchez et al., 2011). These environmental rhythms are all a consequence of the earth’s rotation, and a plant has a fitness benefit to anticipate all three (Green et al., 2002; Dodd et al., 2005; Yerushalmi et al., 2011).
The assembly of a coherent understanding of the plant clock required suitable assay systems to, with limited intervention, measure various parameters of clock function. Oscillations can be mathematically characterized by period, phase, and amplitude. Additionally, the precision of the circadian oscillation has been described by its faithfulness to maintain constant periodicity (all parameters are graphically described in the supplement of Hanano et al., 2006). To characterize circadian mutants, two robust systems were initially employed in the model plant Arabidopsis thaliana: luciferase reporter rhythms and leaf movement rhythms (Millar et al., 1995; Swarup et al., 1999). By analyzing circadian parameters of mutants, genetic approaches have uncovered a number of genes in the clock, and these will be described below.
The Arabidopsis thaliana Clock Models
The first mutants defective in clock function provided a platform toward understanding the components of this oscillating gene network (Millar et al., 1995). One break-through in this approach led to the hypothesis that the morning acting clock genes CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY; Schaffer et al., 1998; Wang and Tobin, 1998) repress the evening acting transcriptional regulator TIME OF CAB EXPRESSION (TOC1; Strayer et al., 2000), which was then reported to induce CCA1 and LHY expression (Alabadi et al., 2001). This defined a so-called negative–positive feedback loop as the core oscillator (graphically illustrated in the Locke et al., 2005a model; Figure 1). Early genetic models of the higher plant clock realized the lack of construction features of this oscillator (McWatters et al., 2001; Staiger, 2002). Mathematical modeling of this network revealed that the clock network must be more complicated (Locke et al., 2005a). Nevertheless, this “one-loop” model provided a critical conceptual framework that guided a decade of molecular-genetics research. From this core-loop hypothesis, many other clock genes were described and placed to the clock circuitry.
Experimental data on the clock accumulated rapidly and exceeded the conceptual capacity to “understand” the network. Several landmark papers in systems biology resolved this dilemma. Firstly it was mathematically hypothesized that the oscillator is composed of interconnected loops (Locke et al., 2005b model; Figure 1). Then work from the Doyle and Millar groups separately defined similar kinetic models that incorporated most available molecular-genetic data (Locke et al., 2006; Zeilinger et al., 2006). These models took in to account the period lengthening and shortening behavior of mutations in genes defined in these models, and were often capable of recapitulating the transcript misexpression levels of genes in the clock in various reciprocal mutant combinations. Both models came to similar conclusions of how the multiple interconnected feedback loops are constructed (Locke/Zellinger 2006 model; Figure 1; Locke et al., 2006; Zeilinger et al., 2006). These working hypotheses were predictive for future experiments, and subsequent molecular-genetic tests have often conformed to mathematical predictions. One key observation predicted was that the cca1 lhy toc1 triple mutant, which lacks the core oscillator, would be arrhythmic. That was indeed experimentally observed (Ding et al., 2007; Ito et al., 2007b). Taken together, the seminal hypothesis of Doyle and Millar that the oscillator is a set of interconnected feedback loops defined for the first time a rational view of how plants tell daily time (Locke et al., 2006; Zeilinger et al., 2006). These explicit mathematical hypotheses have largely stood experimental tests (Shin and Davis, 2010), with some small additions and modifications described below, and one very large one (see below on TOC1 as a repressor).
The clock has been proposed as an interconnected feedback loop with morning, mid-day, evening, and night elements. The first practical models of the clock are illustrated in the Locke/Zellinger 2006 model (Figure 1; Locke et al., 2006; Zeilinger et al., 2006). In this, the morning expressed PSEUDORESPONSE-REGULATOR 9 and 7 (PRR9, PRR7) proteins repress CCA1 and LHY expression, whose proteins in turn activate the former. This is the so-called morning arm. TOC1 protein in turn represses its activator “Y,” an element whose activity is at the end of the day, and this can be partially ascribed with the GIGANTEA protein (GI). This has been defined as the evening arm. These models served the clock community well, although some conflicts could be noted. For instance, the Locke/Zellinger 2006 model predicted that Y/GI transcript levels would elevate in the toc1 null, but this could not be experimentally observed (Martin-Tryon et al., 2007). One explanation for this was that decreased GI expression in toc1 loss-of-function mutants is not direct (Martin-Tryon et al., 2007), and this hypothesis still awaits testing. Finally, several groups have concluded that GI has biochemically separable roles in its ability to integrate light signals, work in the clock, and control flowering time (Mizoguchi et al., 2005; Martin-Tryon et al., 2007; Oliverio et al., 2007). How GI transcriptionally fits into the clock has not been particularly well resolved, but it has been proposed to additionally work in the clock as a hub of a protein destruction complex (Kim et al., 2007).
The Locke/Zellinger 2006 models considered multiple interconnections in the oscillating circuit. One reason for this was based on the observation that none of the founding clock components in this model were arrhythmic when mutated to loss-of-function. Genetic ablation of any one loop leads to the persistence of other loops; rhythms thus persist. In an interconnected circuit, reduction of paths reduces flux. As such, cca1, lhy, and toc1 mutants were short period because there were less paths in the circuit (Locke et al., 2005a, 2006; Zeilinger et al., 2006). This could be extended in the Pokhilko et al. (2010) model (Figure 1). These described models thus did not allow for loss of function in any one gene to lead to arrhythmicity.
The notion of a single gene in the clock leading exclusively to periodicity defects needed to be reevaluated with the finding that the EARLY FLOWERING 4 (ELF4) gene (Doyle et al., 2002) was core to the oscillator, and that when mutated, the oscillator stopped (Kolmos and Davis, 2007; McWatters et al., 2007). The ELF4 gene was found to be both necessary and sufficient to promote CCA1 and LHY, and repress TOC1 (McWatters et al., 2007). This led to a preliminary hypothesis that ELF4 worked directly on these genes (Kolmos and Davis, 2007). That hypothesis could quickly be refuted. If CCA1/LHY levels were low and TOC1 levels were high in elf4, then it was a simple expectation that PRR9, PRR7, and GI levels would also be low. Experimentally the reverse was found for all cases (Kolmos et al., 2009). The Locke et al., 2006 model helped to solve this contradiction. Using parameter fitting of the observed levels of PRR9, PRR7, and GI in elf4, it was found that the oscillator would stop and that CCA1 levels would collapse and that TOC1 levels would be constantly high without rhythm. This is indeed exactly what is seen in the elf4 mutant (Kolmos et al., 2009). Partial function alleles at ELF4 conformed to this finding (Kolmos et al., 2009). Thus, in silico hypothesis testing of the Locke et al., 2006 model provided the first correct placement of an evening complex (EC) component into the oscillator (Kolmos et al., 2009 model). What this work did not address was the placement of ELF4 in mathematical terms to this oscillating circuit.
Placing the ELF3 and LUX Evening Components into the Clock Model
The multiple interconnected feedback loops were made in part to accommodate the phenotypic effects from loss-of-function data. Arrhythmic mutants could not simply be defined in the original Doyle and Millar models (Locke et al., 2006; Zeilinger et al., 2006). Arrhythmic mutants exist at three loci and these are at the EC-components ELF4, ELF3, and LUX (Hazen et al., 2005; McWatters et al., 2007; Thines and Harmon, 2010). None of these genes had been conceptualized in the core-oscillator mechanism. ELF4 association to ELF3 directs LUX action in the clock (Herrero and Davis, 2012; Herrero et al., 2012), and this complex was termed the EC (Nusinow et al., 2011).
Using LTI modeling, ELF4 and ELF3 were concluded to directly target PRR9 and PRR7 (Herrero et al., 2012 model; Figure 1). Indeed, the elf3 mutant was found to be responsive to loss-of-function as it showed increased levels of these transcripts, especially in darkness, but not to such an extent under light (Kolmos et al., 2011). The elf4 mutant had a larger effect on transcript misexpression phenotypes than elf3, and this was especially seen for the increase of PRR7 transcript levels in elf4 during the light phase, whereas in darkness, both PRR9 and PRR7 were similarly increased in elf4 (Kolmos et al., 2009). Also, epistasis experiments showed that both ELF3 and LUX act downstream of ELF4 (Herrero et al., 2012). Consistent with that, whereas the ELF4 and ELF3 proteins have both been shown capable associated to the PRR9 promoter (Dixon et al., 2011; Herrero et al., 2012), as can LUX (Helfer et al., 2011), only ELF4 has been shown to directly bind to the PRR7 promoter (Dixon et al., 2011). Notably ELF4 over-expression resulted in attenuated PRR7 accumulation to a reduced extent than that of ELF3 over-expression (Herrero et al., 2012). ELF4 thus appears to have more targets in the clock than ELF3 and LUX.
A systems analysis of the EC led to a new kinetic model that agreed with the LTI modeling of ELF4 and ELF3. Here a “repressilator” hypothesis was created with sequential waves of repression first by the transcription factors CCA1 and LHY, then by the PRRs, and finally by the EC, with LUX as the as the DNA-binding component of this complex (Pokhilko et al., 2012 model; Figure 1). Notable here was the biochemical finding that all PRRs directly associate to DNA (Gendron et al., 2012) and direct repression at their targets (Gendron et al., 2012; Huang et al., 2012). It is thought that all three EC genes are evening expressed because of direct repression by CCA1 and LHY (Kolmos and Davis, 2007; Lu et al., 2012; Pokhilko et al., 2012). The EC in turn is known to repress PRR9 directly (Dixon et al., 2011; Helfer et al., 2011; Herrero et al., 2012), and genetically, can repress PRR7 and perhaps GI (Kolmos et al., 2009, 2011; Herrero et al., 2012). It is currently unclear if the EC can directly repress GI, as mathematically predicted in one study (Kolmos et al., 2009), or if this is indirect through effects at PRR9 and PRR7, as mathematically predicted in another (Herrero et al., 2012). In the one test of this latter mathematical hypothesis, the prr9 prr7 mutant was not found to have altered mean transcript levels of GI (Salome et al., 2010). Therefore, how GI fits in the clock is not particularly well understood within a transcriptional context.
Conceptual Uses of Models: Weather Patterns as an Example
Having a firm and experimentally validated model at hand allows for future optimization to test the robustness of the complex circadian clock network hypotheses. Here, the Millar group asked which environmental cues and following downstream regulations demanded such a highly complex clock network and followed an in silico approach to test whether the proposed oscillator is plausible in contexts of environmental variation seen in nature. For instance, they selected for networks that correctly predicted particular phases of the day under a light/dark cycle (Troein et al., 2009). The general conclusion was that changes in environmental cues demand for a high complexity in the clock network in order to encompass the details of environmental perturbations typical of daily weather or annual photoperiod variation. This finding provided a validation for the benefits of modeling. Tests of mathematical models often show their limitations. Community willingness to flexibly perform molecular-genetic and biochemical tests of these models has allowed for new model generation to account for such proven discrepancies.
While the existing models reveal a rational view of how an oscillating circuit can resist weather-related environmental changes, and still be sensitive to daily entrainment cues, the models have also been insightful into placing the vernalization effect on clock speed. Prolonged cold of winter results in the derepression of MADS domain transcription factors, of which the most notable is FLOWERING LOCUS C (FLC; Michaels and Amasino, 1999). Its expression correlates with period length, as flc is described as a short-period mutant and FLC over-expression displays a long period (Salathia et al., 2006). As predicted, vernalization effects that lower FLC transcript accumulation lead to a decrease in period length. This is genetically partially dependent on the allelic state of FLC (Salathia et al., 2006). From this, modeling could predict that this FLC action on the clock is through the expression of the EC component LUX (Edwards et al., 2006). Whether FLC binds to the LUX promoter, and whether FLC regulates the other EC components, has not yet been established.
An Example of a Technical Limitation of Models: TOC1 as a Repressor
It was shown that TOC1 loss-of-function results in low CCA1 and LHY transcript levels, which implies TOC1 as a transcriptional activator. However, TOC1 over-expression also results in low CCA1/LHY expression (Makino et al., 2002; Mas et al., 2003). Recent publications resolved this discrepancy and have shown that TOC1 acts as a transcriptional repressor of CCA1 and LHY, as well as of PRR9, PRR7, and GI (Gendron et al., 2012; Huang et al., 2012; Pokhilko et al., 2012). Pokhilko et al. (2012) recently modeled this effect in kinetic terms, and included the EC elements, using the data of the cca1/lhy double mutant, in which only the evening loop sustains rhythmicity. They also included in this the post-translational modification of ELF3 by the ubiquitin E3 ligase COP1 and modulation of the EC complex by GI and ZEITLUPE (ZTL) (Kevei et al., 2006; Kim et al., 2007; Yu et al., 2008). This Pokhilko et al. (2012) model revealed consistency with the observed ztl and prr9/7 double mutant data, when TOC1 was included as an inhibitor for CCA1/LHY, which resolved the inconsistency of the previously proposed activator role of TOC1 and the available experimental data (Pokhilko et al., 2012).
Conclusively, the initial model of the oscillator with reciprocal transcriptional positive–negative feedback loops has been revised. Indeed all PRR-proteins have now been shown biochemically and molecularly to possess repressive function (Nakamichi et al., 2010; Gendron et al., 2012; Huang et al., 2012). This allows for current modeling efforts to define numerous negative–negative feedbacks. In the current Pokhilko et al. (2012) kinetic model, only CCA1 and LHY are defined as positive elements. This model also predicts that the EC controls CCA1/LHY and TOC1 expression through the multi-loop of PRRs, which is consistent with the previous experimental observation of higher transcript levels of the CCA1, LHY, TOC1, and PRR9 in the elf3, elf4, and lux mutants. Consequently, the Pokhilko et al. (2012) model shows the importance of repression of TOC1 and PRR9 by the EC for robust anticipation of dawn.
The recent work of Pokhilko et al. (2012) additionally considered the role of light as an input factor to the clock when investigating the phase change of the clock by light pulses. This model predicted that the acute activation of CCA1/LHY expression by light is required for the observed phase advance or delays at a given time during the night. The ability of the model to predict such an observation emphasizes the importance of incorporation of input signals to circadian modeling. From this, we need to start investigating effects of feedback signals from further downstream processes, such as hormonal signaling (Hanano et al., 2006) and as a consequence of metabolic changes (Dalchau et al., 2011), and cellular coordination of said processes (Wenden et al., 2012).
A general conclusion of recent kinetic and linear models could lead one to consider the clock network as “solved.” This is not the case. Two-component limit cycle oscillators can exist if at least one component is “autocatalytic” and there is also a negative feedback (Tyson, 2002; Novak and Tyson, 2008). Here, “limit cycle” means that every cycle is the same, and thus, there is no dampening or noise. If the plant circadian oscillator is not built in such a way, to make this oscillator, the circuit is anticipated to have a minimum of three components and positive and negative arms must exist within it: repressor networks need activators (Sprinzak and Elowitz, 2005). It is thus plausible that the current model of the plant clock lacks adequate activators to be rationally defined. How such activators fit into the clock system of course requires their discovery and subsequent integration to the models. As these putative activators are found, probably by molecular-genetic methods, the elements that define the core of the clock could be moved toward being considered as centrally defined. Another important consideration is to define the details of feedback signals from metabolic rhythms (Stitt and Zeeman, 2012) and their role in the redox circadian oscillator (Edgar et al., 2012). The whole clock community awaits those integrative results (van Ooijen and Millar, 2012).
Model Needs and Proposed Further Uses
There are several areas where modeling has yet to place the clock in a signal context of observed findings. This seems relevant as numerous transcription factors fine-tune clock parameters, implying massive signal interconnections of divergent and disparate signaling systems to and from the clock (Hanano et al., 2008). One example is that multiple phytohormones have distinct effects on clock parameters (Hanano et al., 2006; Yin et al., 2007), but to date, modeling has not explained how this feedback is plausible. Here current efforts to reciprocally link the stress hormone abscisic acid (ABA) to the clock seem particularly relevant (Legnaioli et al., 2009). This could relate physiological connections to drought and salinity on the physiology of clock performance. Additionally, as auxin signaling rises with increasing warmth (Gray et al., 1998), and as auxin application phenocopies the effect of warmth to create more stochastic noise in the oscillator (Hanano et al., 2006), this thermal-dampening mechanism could relate to an ability of increasing temperatures to increase auxin signaling flux as a modulator of circadian amplitude. Modeling this hypothesis could direct the plausibility of this. Other hormones have distinct effects on phase and period, and these could act on light signaling to the clock, but that is not yet described in mathematical terms. Modeling signaling cross-talk to and from the clock seems ripe for future investigation.
Light has two main modes to set the clock. Light intensity increases lead to periodicity decreases (Somers et al., 1998a). This speeding up of the clock by increased light perception leads to an eventual phase shift of the clock back to a correct resonance, and this is called parametric entrainment. In contrast, the discontinuous nearly immediate setting of the clock happens at dawn and needs extended light far beyond that which activates light-regulated gene expression (Millar and Kay, 1996), and this sudden clock setting is called non-parametric entrainment. In some manner, the phytochromes and cryptochromes have a role in these setting mechanisms (Somers et al., 1998a; Devlin and Kay, 2000). Interestingly, a photoreceptor complex (Mas et al., 2000) is genetically interactive in clock function (Devlin and Kay, 2000). A mechanistic hypothesis for photoreceptor input to the clock has not yet been generated. Although, it has been shown biochemically that light controls degradation of PRR7, PRR9, TOC1, and GI proteins (Mas et al., 2003; David et al., 2006; Farre and Kay, 2007; Ito et al., 2007a). These photic effects then act on outputs within a diurnal context that changes in duration throughout the season (Davis, 2002; Salazar et al., 2009; Troein et al., 2009; Guerriero et al., 2012; Song et al., 2012).
Low-fluence rate UV-B light has been shown to control development, promote photomorphogenesis, and drive gene expression (Heijde and Ulm, 2012). UVR8 and COP1 are crucial for physiological UV-B responses and entrainment of the clock by UV-B light (Feher et al., 2011). Although under supplemented UV-B light, COP1 induces ELONGATED HYPOPCOTYL 5 (HY5) and HY5 HOMOLOGY (HYH), HY5 and HYH are not required for clock entrainment by UV-B (Feher et al., 2011). With the identification of UVR8 as the UV-B receptor (Heijde and Ulm, 2012), this is another input signal to the oscillator that must also be mathematically defined as an input cue.
Interestingly, under far-red (FR) light the otherwise arrhythmic elf3 and elf4 mutants regain rhythmicity (Kolmos et al., 2011; Wenden et al., 2011). Phytochrome A (phyA) has been shown to be required for controlling clock-regulated gene expression under these conditions (Wenden et al., 2011), yet the effect of FR-light input to the clock is not well understood. As shade alters the red/FR ratio, this observation suggests a different clock entrainment under these environmental conditions. Here, genetics could profit from mathematical modeling.
Ambient temperature effects on the clock work in two discreet ways. In one sense, the oscillator resists changes in mean ambient temperature to run at about 24 h over a fairly wide range of temperatures. Modeling has been able to explain this as effects at transcript abundance of clock genes (Gould et al., 2006). In contrast, the daily oscillation of daytime warmth with evening coolness can set the oscillator (Somers et al., 1998b; Boikoglou et al., 2011). This form of entrainment is completely unresolved (McClung and Davis, 2010). Modeling efforts have not yet been conducted to predict inputs generated from temperature entrainment. Another point is that stress temperatures of cold can lead to oscillator arrest (Bieniawska et al., 2008). How stress temperature stops the clock is as yet non-explored in a systems sense (McClung and Davis, 2010).
Stress and metabolic signals enter the clock. Redox effects by photosynthesis, and alterations in sucrose and starch have been connected to normal oscillator function (Dalchau et al., 2011). Relations to ABA signaling appear to intersect here (Sanchez et al., 2011). How redox and carbon as photosynthesis-related processes enter the clock are not known. Modeling is likely to add useful hypotheses to this point. Other metabolites can act on oscillator parameters, including cyclic ADP ribose (cADPR), and this has been modeled (Dodd et al., 2007). Primary metabolites could act as energy intermediates, and trehalose-6-phosphate has been hypothesized to signal in homeostasis (Schluepmann et al., 2012). In contrast, secondary metabolites, such as glucosinolates that act on clock parameters (Kerwin et al., 2011), are more difficult to rationalize as just a metabolic effect. Numerous secondary compounds are perhaps probable as direct signaling molecules in clock fine-tuning. Placing all of these metabolic effects to the clock will likely be aided by informatics and systems approaches.
Clock genes in A. thaliana display extensive sequence variation manifested in quantitative variation within a population (Swarup et al., 1999; Boikoglou et al., 2011; Undurraga et al., 2012) and this is also seen in the monocot barley (Stracke et al., 2009; Faure et al., 2012). Furthermore, ploidy changes that are prevalent in plants also act on clock behavior at a physiologic (Ni et al., 2009) and genomic scale (Lou et al., 2012). Future clock models should be able to predict how subtle allelic variants lead to expressed-trait effects on clock parameters. As the analysis of clock-gene expression in barley did not exactly follow that of A. thaliana (Campoli et al., 2012), models will need to be generated that consider the evolutionary divergence between monocots and dicots. Finally, modeling is also likely to be useful in predicting how the assembly of a larger nucleus in new polyploids, and the effect of larger gene dosage, is buffered.
Moving beyond transcription, numerous clock proteins are subjected to post-transcriptional and post-translational regulation (Staiger and Koster, 2011). Phosphorylation and regulated protein degradation can be a directing force for the input of environmental signals to the oscillator (Herrero and Davis, 2012). From there, another layer within the system can be seen in regulated protein-complex assembly and the action of alteration of protein localization and DNA association capacity (Schoning and Staiger, 2005; Herrero and Davis, 2012). Together, these dynamics at the protein level need to be considered in new modeling efforts. Perhaps such an approach could lead to an improved placement of GI into the clock.
Conclusion
Mathematical models of the A. thaliana circadian oscillator have motivated hypothesis-driven experimental studies that have largely resolved this system. In this way, the plant circadian network serves as an example for how other plant-signaling systems can profit from interactive modeling-experimental efforts.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge financial funding provided from the Max-Planck-Gesellschaft and Deutsche Forschungsgemeinschaft (DA1061/4-1; DA1061/5-1; DA1061/6-1; DA1061/7-1). The authors also thank Amanda M. Davis for critical reading of the article and feedback.
References
Akman, O. E., Watterson, S., Parton, A., Binns, N., Millar, A. J., and Ghazal, P. (2012). Digital clocks: simple Boolean models can quantitatively describe circadian systems. J. R. Soc. Interface 9, 2365–2382.
Alabadi, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Mas, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Bieniawska, Z., Espinoza, C., Schlereth, A., Sulpice, R., Hincha D. K., and Hannah, M. A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147, 263–279.
Boikoglou, E., Ma, Z., von Korff, M., Davis, A. M., Nagy, F., and Davis, S. J. (2011). Environmental memory from a circadian oscillator: the Arabidopsis thaliana clock differentially integrates perception of photic vs. thermal entrainment. Genetics 189, 655–664.
Campoli, C., Shtaya, M., Davis S. J., and von Korff, M. (2012). Expression conservation within the circadian clock of a monocot: natural variation at barley Ppd-H1 affects circadian expression of flowering time genes, but not clock orthologs. BMC Plant Biol. 12:97. doi: 10.1186/1471-2229-12-97
Dalchau, N., Baek, S. J., Briggs, H. M., Robertson, F. C., Dodd, A. N., Gardner, M. J., et al. (2011). The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. U.S.A. 108, 5104–5109.
David, K. M., Armbruster, U., Tama N., and Putterill, J. (2006). Arabidopsis GIGANTEA protein is post-transcriptionally regulated by light and dark. FEBS Lett. 580, 1193–1197.
Davis, S. J. (2002). Photoperiodism: the coincidental perception of the season. Curr. Biol. 12, R841–R843.
Davis, S. J., and Millar, A. J. (2001). Watching the hands of the Arabidopsis biological clock. Genome Biol. 2, REVIEWS1008.
Devlin P. F., and Kay, S. A. (2000). Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12, 2499–2510.
Ding, Z., Doyle, M. R., Amasino, R., and Davis, S. J. (2007). A complex genetic interaction between Arabidopsis thaliana TOC1 and CCA1/LHY in driving the circadian clock and in output regulation. Genetics 176, 1501–1510.
Dixon, L. E., Knox, K., Kozma-Bognar, L., Southern, M. M., Pokhilko A., and Millar, A. J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 21, 120–125.
Dodd, A. N., Gardner, M. J., Hotta, C. T., Hubbard, K. E., Dalchau, N., Love, J., et al. (2007). The Arabidopsis circadian clock incorporates a cADPR-based feedback loop. Science 318, 1789–1792.
Dodd, A. N., Salathia, N., Hall, A., Kevei, E., Toth, R., Nagy, F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633.
Doyle, M. R., Davis, S. J., Bastow, R. M., McWatters, H. G., Kozma-Bognar, L., Nagy, F., et al. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419, 74–77.
Dunlap, J. C., Loros J. J., and DeCoursey, P. J. (2004). Chronobiology: Biological Timekeeping, Sunderland: Sinauer Associates.
Edgar, R. S., Green, E. W., Zhao, Y., van Ooijen, G., Olmedo, M., Qin, X., et al. (2012). Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464.
Edwards, K. D., Anderson, P. E., Hall, A., Salathia, N. S., Locke, J. C., Lynn, J. R., et al. (2006). FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18, 639–650.
Farre, E. M., and Kay, S. A. (2007). PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. Plant J. 52, 548–560.
Faure, S., Turner, A. S., Gruszka, D., Christodoulou, V., Davis, S. J., von Korff, M., et al. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. U.S.A. 109, 8328–8333.
Feher, B., Kozma-Bognar, L., Kevei, E., Hajdu, A., Binkert, M., Davis, S. J., et al. (2011). Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J. 67, 37–48.
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172.
Gerard, C., and Goldbeter, A. (2012). Entrainment of the mammalian cell cycle by the circadian clock: modeling two coupled cellular rhythms. PLoS Comput. Biol. 8: e1002516. doi: 10.1371/journal.pcbi.1002516
Goldbeter, A. (1995). A model for circadian oscillations in the Drosophila period protein (PER). Proc. Biol. Sci. 261, 319–324.
Gonze, D., Abou-Jaoude, W., Ouattara, D. A., and Halloy, J. (2011). How molecular should your molecular model be? On the level of molecular detail required to simulate biological networks in systems and synthetic biology. Methods Enzymol. 487, 171–215.
Gould, P. D., Locke, J. C., Larue, C., Southern, M. M., Davis, S. J., Hanano, S., et al. (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18, 1177–1187.
Gray, W. M., Östin, A., Sandberg, G., Romano C. P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 7197–7202.
Green, R. M., Tingay, S., Wang, Z. Y., and Tobin, E. M. (2002). Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 129, 576–584.
Guerriero, M. L., Pokhilko, A., Fernandez, A. P., Halliday, K. J., Millar, A. J., and Hillston, J. (2012). Stochastic properties of the plant circadian clock. J. R. Soc. Interface 9, 744–756.
Hanano, S., Domagalska, M. A., Nagy, F., and Davis, S. J. (2006). Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 11, 1381–1392.
Hanano, S., Stracke, R., Jakoby, M., Merkle, T., Domagalska, M. A., Weisshaar, B., et al. (2008). A systematic survey in Arabidopsis thaliana of transcription factors that modulate circadian parameters. BMC Genomics 9:182. doi: 10.1186/1471-2164-9-182
Hatakeyama, T. S., and Kaneko, K. (2012). Generic temperature compensation of biological clocks by autonomous regulation of catalyst concentration. Proc. Natl. Acad. Sci. U.S.A. 109, 8109–8114.
Hazen, S. P., Schultz, T. F., Pruneda-Paz, J. L., Borevitz, J. O., Ecker, J. R., and Kay, S. A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 102, 10387–10392.
Heijde, M., and Ulm, R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17, 230–237.
Helfer, A., Nusinow, D. A., Chow, B. Y,.Gehrke, A. R., Bulyk M. L., and Kay, S. A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21, 126–133.
Herrero, E., and Davis, S. J., (2012). Time for a nuclear meeting: protein trafficking and chromatin dynamics intersect in the plant circadian system. Mol. Plant 5, 554–565.
Herrero, E., Kolmos, E.,Bujdoso, N., Yuan, Y., Wang, M., Berns, M. C., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24, 428–443.
Huang, W., Perez-Garcia, P., Pokhilko, A., Millar, A. J., Antoshechkin, I., Riechmann J. L., et al. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79.
Ito, S., Nakamichi, N., Kiba, T., Yamashino T., and Mizuno, T. (2007a). Rhythmic and light-inducible appearance of clock-associated pseudo-response regulator protein PRR9 through programmed degradation in the dark in Arabidopsis thaliana. Plant Cell Physiol. 48, 1644–1651.
Ito, S., Nakamichi, N., Nakamura, Y., Niwa, Y., Kato, T., Murakami, M., et al. (2007b). Genetic linkages between circadian clock-associated components and phytochrome-dependent red light signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 48, 971–983.
Kerwin, R. E., Jimenez-Gomez, J. M., Fulop, D., Harmer, S. L., Maloof J. N., and Kliebenstein, D. J. (2011). Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 23, 471–485.
Kevei, E., Gyula, P., Hall, A., Kozma-Bognar, L., Kim, W. Y., Eriksson, M. E., et al. (2006). Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol. 140, 933–945.
Kim, W. Y., Fujiwara, S., Suh, S. S., Kim, J., Kim, Y., Han, L., et al. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360.
Kolmos, E., and Davis, S. J. (2007). ELF4 as a Central Gene in the Circadian Clock. Plant Signal. Behav. 2, 370–372.
Kolmos, E., Herrero, E., Bujdoso, N., Millar, A. J., Toth, R., Gyula, P., et al. (2011). A reduced-function allele reveals that EARLY FLOWERING3 repressive action on the circadian clock is modulated by phytochrome signals in Arabidopsis. Plant Cell 23, 3230–3246.
Kolmos, E., Nowak, M., Werner, M., Fischer, K., Schwarz, G., Mathews, S., et al. (2009). Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 3, 350–366.
Legnaioli, T., Cuevas J., and Mas, P. (2009). TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 28, 3745–3757.
Locke, J. C., Kozma-Bognar, L., Gould, P. D., Feher, B., Kevei, E., Nagy, F., et al. (2006). Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2, 59.
Locke, J. C., Millar, A. J., and Turner, M. S., (2005a). Modelling genetic networks with noisy and varied experimental data: the circadian clock in Arabidopsis thaliana. J. Theor. Biol. 234, 383–393.
Locke, J. C., Southern, M. M., Kozma-Bognar, L., Hibberd, V., Brown, P. E., Turner, M. S., et al. (2005b). Extension of a genetic network model by iterative experimentation and mathematical analysis. Mol. Syst. Biol. 1, 2005.0013.
Lou, P., Wu, J., Cheng, F., Cressman, L. G., Wang X., and McClung, C. R. (2012). Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell 24, 2415–2426.
Lu, S. X., Webb, C. J., Knowles, S. M., Kim, S. H., Wang Z., and Tobin, E. M. (2012). CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 158, 1079–1088.
Makino, S., Matsushika, A., Kojima, M., Yamashino, T., and Mizuno, T. (2002). The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 43, 58–69.
Martin-Tryon, E. L., Kreps, J. A., and Harmer, S. L., (2007). GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143, 473–486.
Mas, P., Devlin, P. F., Panda, S., and Kay, S. A. (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408, 207–211.
Mas, P., Kim, W. Y., Somers, D. E., and Kay, S. A. (2003). Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570.
McClung, C. R., and Davis, S. J. (2010). Ambient thermometers in plants: from physiological outputs towards mechanisms of thermal sensing. Curr. Biol. 20, R1086–R1092.
McWatters, H. G., Kolmos, E., Hall, A., Doyle, M. R., Amasino, R. M.,Gyula, P., et al. (2007). ELF4 is required for oscillatory properties of the circadian clock. Plant Physiol. 144, 391–401.
McWatters, H. G., Roden, L. C., and Staiger, D. (2001). Picking out parallels: plant circadian clocks in context. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1735–1743.
Michaels, S. D., and Amasino, R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956.
Millar, A. J., and Kay, S. A. (1996). Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 93, 15491–15496.
Millar, A. J., Carre, I. A., Strayer, C. A., Chua N. H., and Kay, S. A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267, 1161–1163.
Mizoguchi, T., Wright, L., Fujiwara, S., Cremer, F., Lee, K., Onouchi, H., et al. (2005). Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–2270.
Nakamichi, N., Kiba, T., Henriques, R., Mizuno, T., Chua N. H., and Sakakibara, H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605.
Ni, Z., Kim, E. D., Ha, M., Lackey, E., Liu, J., Zhang, Y., et al. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457, 327–331.
Novak, B., and Tyson, J. J. (2008). Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 9, 981–991.
Nusinow, D. A., Helfer, A., Hamilton, E. E., King, J. J., Imaizumi, T., Schultz, T. F., et al. (2011). The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402.
Oliverio, K. A., Crepy, M., Martin-Tryon, E. L., Milich, R., Harmer, S. L.,Putterill, J., et al. (2007). GIGANTEA regulates phytochrome A-mediated photomorphogenesis independently of its role in the circadian clock. Plant Physiol. 144, 495–502.
Pokhilko, A., Fernandez, A. P., Edwards, K. D., Southern, M. M., Halliday, K. J., and Millar, A. J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8, 574.
Pokhilko, A., Hodge, S. K., Stratford, K., Knox, K., Edwards, K. D.,Thomson, A. W., et al. (2010). Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol. Syst. Biol. 6, 416.
Salathia, N., Davis, S. J., Lynn, J. R., Michaels, S. D., Amasino, R. M., and Millar, A. J. (2006). FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant Biol. 6:10. doi: 10.1186/1471-2229-6-10
Salazar, J. D., Saithong, T., Brown, P. E., Foreman, J., Locke, J. C., Halliday, K. J., et al. (2009).Prediction of photoperiodic regulators from quantitative gene circuit models. Cell 139, 1170–1179.
Salome, P. A., Weigel D., and McClung, C. R. (2010). The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22, 3650–3661.
Sanchez, A., Shin, J., and Davis, S. J. (2011). Abiotic stress and the plant circadian clock. Plant Signal. Behav. 6, 223–231.
Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I. A., et al. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229.
Schluepmann, H., Berke L., and Sanchez-Perez, G. F. (2012). Metabolism control over growth: a case for trehalose-6-phosphate in plants. J. Exp. Bot. 63, 3379–3390.
Schoning, J. C., and Staiger, D. (2005). At the pulse of time: protein interactions determine the pace of circadian clocks. FEBS Lett. 579, 3246–3252.
Shin, J., and Davis, S. J. (2010). Recent advances in computational modeling as a conduit to understand the plant circadian clock. F1000 Biol. Rep. 2, 49.
Somers, D. E., Devlin P. F., and Kay, S. A. (1998a). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1490.
Somers, D. E., Webb, A. A., Pearson M., and Kay, S. A. (1998b). The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125, 485–494.
Song, Y. H., Smith, R. W., To, B. J., Millar A. J., and Imaizumi, T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049.
Staiger, D., and Koster, T. (2011). Spotlight on post-transcriptional control in the circadian system. Cell Mol. Life Sci. 68, 71–83.
Staiger, D. (2002). Circadian rhythms in Arabidopsis: time for nuclear proteins. Planta 214, 334–344.
Stitt, M., and Zeeman, S. C., (2012). Starch turnover: pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15, 282–292.
Stracke, S., Haseneyer, G., Veyrieras, J. B., Geiger, H. H., Sauer, S., Graner A., et al. (2009). Association mapping reveals gene action and interactions in the determination of flowering time in barley. Theor. Appl. Genet. 118, 259–273.
Strayer, C., Oyama, T., Schultz, T. F., Raman, R.,Somers, D. E., Mas, P. S., et al. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771.
Swarup, K., Alonso-Blanco, C., Lynn, J. R., Michaels, S. D., Amasino, R. M., Koornneef, M., et al. (1999). Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 20, 67–77.
Thines, B., and Harmon, F. G., (2010). Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. U.S.A. 107, 3257–3262.
Troein, C., Locke, J. C., Turner M. S., and Millar, A. J. (2009). Weather and seasons together demand complex biological clocks. Curr. Biol. 19, 1961–1964.
Tseng, Y. Y., Hunt, S. M., Heintzen, C., Crosthwaite, S. K., and Schwartz, J. M. (2012). Comprehensive modelling of the Neurospora circadian clock and its temperature compensation. PLoS Comput. Biol. 8:e1002437. doi: 10.1371/journal.pcbi.1002437
Tyson, J. J. (2002). “Biochemical oscillations,” in Computational Cell Biology, ed. I. A. Mathematics (New York: Springer).
Undurraga, S. F., Press, M. O., Legendre, M., Bujdoso, N., Bale, J., Wang, H., et al. (2012). Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc. Natl. Acad. Sci. U.S.A. 109, 19363–19367.
van Ooijen, G., and Millar, A. J. (2012). Non-transcriptional oscillators in circadian timekeeping. Trends Biochem. Sci. 37, 484–492.
Wang, Z. Y., and Tobin, E. M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217.
Wenden, B., Kozma-Bognar, L., Edwards, K. D., Hall, A. J., Locke J. C., and Millar, A. J. (2011). Light inputs shape the Arabidopsis circadian system. Plant J. 66, 480–491.
Wenden, B., Toner, D. L., Hodge, S. K., Grima, R., and Millar, A. J., (2012). Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proc. Natl. Acad. Sci. U.S.A. 109, 6757–6762.
Yerushalmi, S., Yakir, E., and Green, R. M. (2011). Circadian clocks and adaptation in Arabidopsis. Mol. Ecol. 20, 1155–1165.
Yin, X. J., Volk, S., Ljung, K., Mehlmer, N., Dolezal, K., Ditengou, F., et al. (2007). Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19, 1898–1911.
Yu, J. W., Rubio, V., Lee, N. Y., Bai, S., Lee, S. Y., Kim, S. S., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32, 617–630.
Keywords: Arabidopsis thaliana, circadian clock, mathematical modeling, light signal transduction, temperature acclimation, hormone signal integration, metabolic signal integration, stress signal integration
Citation: Bujdoso N and Davis SJ (2013) Mathematical modeling of an oscillating gene circuit to unravel the circadian clock network of Arabidopsis thaliana. Front. Plant Sci. 4:3. doi: 10.3389/fpls.2013.00003
Received: 01 September 2012; Accepted: 03 January 2013;
Published online: 25 January 2013.
Edited by:
Roeland Merks, Centrum Wiskunde & Informatica, NetherlandsReviewed by:
Roeland Merks, Centrum Wiskunde & Informatica, NetherlandsÖnder Kartal, Eidgenössische Technische Hochschule Zürich, Switzerland
Copyright: © 2013 Bujdoso and Davis. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Seth J. Davis, Department of Plant Developmental Biology, Max Planck Institute for Plant Breeding Research, Carl-von-Linné-Weg 10, 50829 Cologne, Germany. e-mail:ZGF2aXNAbXBpcHoubXBnLmRl

