- 1Plant Biology, The University of Vermont, Burlington, VT, USA
- 2Ecology and Evolutionary Biology, The University of Kansas, Lawrence, KS, USA
The SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) family of transcription factors is functionally diverse, controlling a number of fundamental aspects of plant growth and development, including vegetative phase change, flowering time, branching, and leaf initiation rate. In natural plant populations, variation in flowering time and shoot architecture have major consequences for fitness. Likewise, in crop species, variation in branching and developmental rate impact biomass and yield. Thus, studies aimed at dissecting how the various functions are partitioned among different SPL genes in diverse plant lineages are key to providing insight into the genetic basis of local adaptation and have already garnered attention by crop breeders. Here we use phylogenetic reconstruction to reveal nine major SPL gene lineages, each of which is described in terms of function and diversification. To assess evidence for ancestral and derived functions within each SPL gene lineage, we use ancestral character state reconstructions. Our analyses suggest an emerging pattern of sub-functionalization, neo-functionalization, and possible convergent evolution following both ancient and recent gene duplication. Based on these analyses we suggest future avenues of research that may prove fruitful for elucidating the importance of SPL gene evolution in plant growth and development.
Introduction
SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) proteins constitute a diverse family of transcription factors that play fundamental roles in plant growth and development, and are defined by a highly conserved region of 76 amino acids called the SBP domain (Klein et al., 1996; Yang et al., 2007). The SBP domain is involved in both nuclear import and sequence-specific DNA binding to a consensus-binding site containing a GTAC core motif and gene-specific flanking regions (Birkenbihl et al., 2005; Yamasaki et al., 2006; Liang et al., 2008). SPL genes are found in all green plants, including single-celled green algae, mosses, gymnosperms, and angiosperms (Cardon et al., 1997; Arazi et al., 2005; Riese et al., 2008), and were first identified in Antirrhinum majus (snapdragon, Plantaginaceae, asterid) based on the ability of closely related AmSBP1 and AmSBP2 to bind to the promoter of the floral meristem identity gene SQUAMOSA (SQUA) (Klein et al., 1996). This review focuses on the diversification of SPL genes following both gene duplication and speciation events, and illustrates the importance of research into these genes for a better understanding of plant development and evolution.
Gene Duplication as a Mechanism for SPL Gene Diversification
Gene duplication is common in plants and plays a key role in trait evolution (Lawton-Rauh, 2003; Crow and Wagner, 2006; Kaessmann, 2010; Airoldi and Davies, 2012). The SPL gene family is an excellent system in which to determine the fate of duplicate genes due to its extensive history of gene doubling, previously identified upstream pathways and downstream targets, and wide range of developmental functions. Whereas the most common fate of gene duplication is functional loss in one copy (non-functionalization), functional evolution can occur through the partitioning of ancestral functions (sub-functionalization), or the acquisition of novel functions (neo-functionalization) in one or both descendent genes (Ohno, 1970; Zhang et al., 1998; Lynch and Conery, 2000; reviewed in Zhang, 2003). Although sub-functionalization may have little immediate impact on phenotype, increased specialization of sub-functionalized paralogs within particular developmental modules is thought to be an important pre-requisite for trait evolution by mitigating the negative effects of mutations in genes that would otherwise exhibit strong pleiotropy (Force et al., 1999; Hughes, 1999; Hittinger and Carroll, 2007). Numerous studies have revealed different fates for duplicate genes in plant development (e.g., Kramer et al., 2004; Causier et al., 2005; Yamaguchi et al., 2006). However, much still remains to be learned about the evolutionary outcome of duplicated genes – both generally and in specific gene lineages – regarding their impact on genetic pathway and trait evolution.
Genomic sequencing has revealed 16, 18, 13, and 31 SPL genes in Arabidopsis thaliana (Arabidopsis, Brassicaceae), Oryza sativa (rice, Poaceae), Physcomitrella patens (moss, Funariaceae), and Zea mays (maize, Poaceae), respectively (Cardon et al., 1999; Arazi et al., 2005; Hultquist and Dorweiler, 2008; Riese et al., 2008; Miura et al., 2010). These genes can be separated into two major groups – long and short – the latter of which are largely regulated by the microRNAs miR156 and miR157 (Cardon et al., 1997, 1999; Rhoades et al., 2002; Guo et al., 2008). Although understudied relative to other gene families, functional analyses have revealed divergent developmental roles for SPL genes in a diversity of angiosperm taxa. These include the promotion of juvenile to adult phase change (heteroblasty), reproductive transition, trichome development, apical dominance, inflorescence branching, fruit ripening, plastochron length (time between leaf initiation), pollen sac development, and copper homeostasis (Unte et al., 2003; Manning et al., 2006; Wu and Poethig, 2006; Schwarz et al., 2008; Wang et al., 2009; Yamaguchi et al., 2009; Yamasaki et al., 2009; Jiao et al., 2010; Miura et al., 2010; Preston and Hileman, 2010; Yu et al., 2010). Since variation in many of these traits accounts for both inter- and intra-specific variation in lifetime fitness (e.g., Hall and Willis, 2006; Anderson et al., 2011), functional analyses of duplicated SPL genes under different environmental conditions may foster substantial insights into the genetic basis for variation in plant life history traits including architecture and phase change evolution.
Patterns of Duplication in the SPL Gene Family
Similar to the developmentally important MADS-box transcription factor family (Becker and Theissen, 2003), phylogenetic evidence supports retention of multiple SPL paralogs following both ancient and more recent duplication events (Yang et al., 2007; Guo et al., 2008; Salinas et al., 2012) (Figure 1). Thus, it is hypothesized that duplicate SPL genes have been maintained in the genome by positive Darwinian selection following sub- or neo-functionalization. As will become clear in the following sections, gene orthology in this family does not always predict function. This suggests either common patterns of neo-functionalization or differential sub-functionalization in different SPL gene lineages.
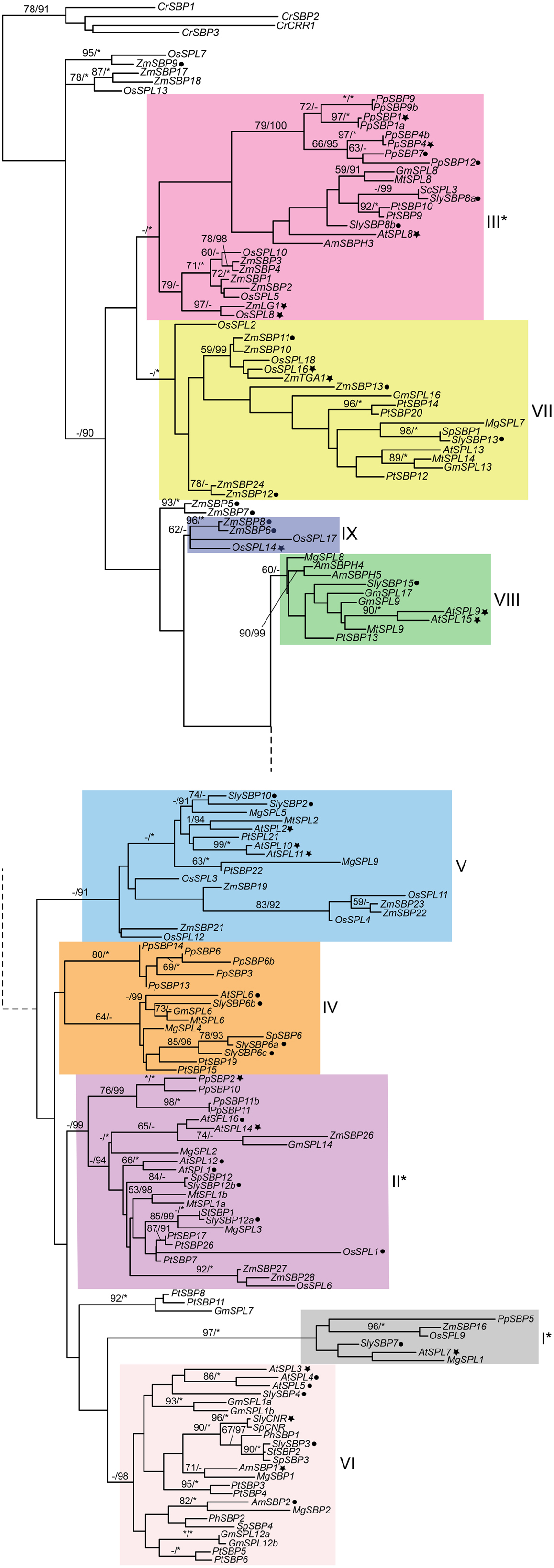
Figure 1. Phylogeny of SPL genes based on an alignment of the SBP-box domain from representative seed plants. Sequences were downloaded from Genbank/EMBL and Phytozome v8.0 (http://www.phytozome.net/) based on BLAST searches and previously published data (Table S1 in Supplementary Material). Amino acids were aligned by eye in Macclade (Maddison and Maddison, 2003) and nucleotides were subjected to maximum likelihood (ML) in GARLI (Zwickl, 2006) using the best-fitting GTR + I + Γ model of evolution based on results of ModelTest 3.7 (Posada and Crandall, 1998) with 10 random-additions. ML bootstrap values were obtained using 500 bootstrap replicates and are shown below the branches if above 50%; asterisks indicate 100%. Bayesian posterior probability values were obtained in MrBayes 3.2.1. (Ronquist and Huelsenbeck, 2003), with 12 million generations, sampling every 1000th generation, with 25% of trees discarded as burn-in. Values above 90% are indicated to the right of bootstrap values; asterisks indicate 100%. The tree was rooted on genes from the most distantly related species, the green alga Chlamydomonas reinhardtii. Genes in clades with asterisks are not regulated by miRNA156 or miRNA157. Filled star, functionally characterized; filled circle, expression data available. At, Arabidopsis thaliana; Am, Antirrhinum majus; Cr, Chlamydomonas reinhardii; Gm, Glycine max; Mg, Mimulus guttatus; Mt, Medicago truncatula; Pp, Physcomitrella patens; Pt, Populus trichocarpa; Os, Oryza sativa; Sc, Solanum chacoense; Sl, Solanum lycopersicum; Sp, Solanum phureja; St, Solanum tuberosum; Zm, Zea mays. Dotted lines connect the upper and lower parts of the tree. Clade colors match Salinas et al. (2012) where applicable.
The recent availability of several fully sequenced plant genomes has bolstered phylogenetic reconstruction of duplication in the SPL gene family. Salinas et al. (2012) recently reported eight (I-VIII) major SPL clades, at least four of which predate the diversification of embryophytes (land plants) based on a neighbor-joining algorithm, whereas Guo et al. (2008) identified six major clades. To independently determine support for these different SPL gene clades, we analyzed a slightly different dataset of SPL genes derived from sequences available in Genbank and Phytozome version 9.0 (Table S1 in Supplementary Material) under the GTR + I + γ model of evolution in a maximum likelihood (ML) framework. The model of evolution was selected according to results of MrModelTest version 3.7 (Posada and Crandall, 1998), and ML analyses were run in GARLI with 500 bootstrap replicates (Zwickl, 2006). Bayesian posterior probabilities were also obtained in MrBayes version 3.2.1. (Ronquist and Huelsenbeck, 2003) with 12 million generations, sampling every 1000th generation, and discarding 25% of trees as burn-in. Results of these analyses support the eight major clades described in Salinas et al. (2012), and suggest a possible ninth clade containing the domestication gene OsSPL14 from rice (Figure 1). Since surveys of available genomic data suggest that SPL genes are absent from fungi and metazoans, the ML tree is rooted with genes from the green alga Chlamydomonas reinhardtii, which are each other’s closest relatives in unrooted trees. Posterior probability values support the monophyly of clades I, II, III, V, VI, and VII (Figure 1). However, despite some support within clades, there is low support for relationships among major lineages (Figure 1), probably due to the limited number of informative characters within the alignable SBP-box domain (Salinas et al., 2012).
Intron numbers are highly variable in the gene family, ranging from 1 to 10 (Cardon et al., 1999; Kropat et al., 2005; Guo et al., 2008; Salinas et al., 2012). However, evidence based on available genomic sequences support conservation of exon-intron structures within clades, with the exception of clade-II and clade-IV (Figure 1) (Cardon et al., 1999; Guo et al., 2008; Salinas et al., 2012). In the following sections we summarize our knowledge on the expression and function of representative genes for each clade given available data. In order to better understand the relative timing and number of origins of SPL gene functions, the ancestral state for several traits was reconstructed using a ML framework in Mesquite version 2.01 based on pruned versions of the Figure 1 tree (Maddison and Maddison, 2003, 2008) (Figure 2). Only traits with both presence (coded 1) and absence (coded 0) data were included in the analysis. We discuss these results in the context of the importance of different SPL clades on green plant evolution and development, and suggest important directions for future research.
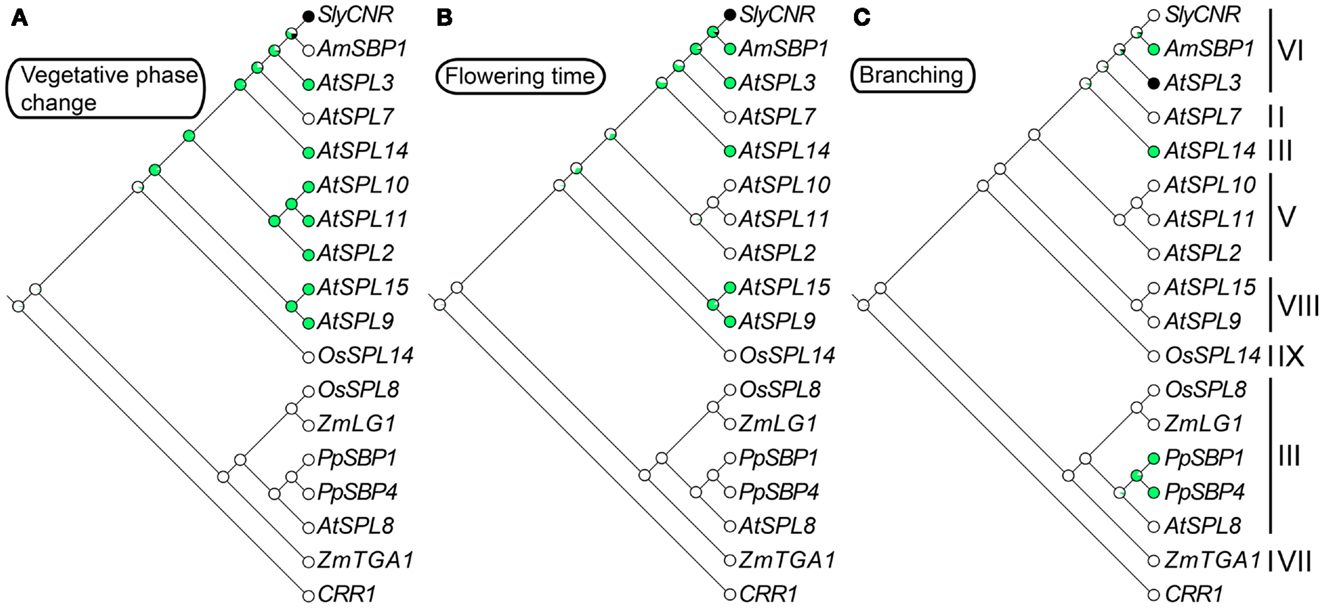
Figure 2. Ancestral character state reconstructions across the SPL gene family. The gene trees are based on Figure 1, but only include genes for which functional data are available. Character state transitions in vegetative phase change (A), flowering time (B), and branching (C) were inferred from a ML analysis in Mesquite version 2.5 (Maddison and Maddison, 2008) under the Mk1 (single rate of change between states) model of evolution. Circles denote the probability of having (green) or not having (white) a particular function. Black circles are coded as unknown.
SPL Gene Clade Evolution
Clade-I
SPL clade-I genes are characterized by their large size, lack of miRNA156 and miR157-binding sites, and near ubiquitous expression across different organs of the plant (Cardon et al., 1999; Wang et al., 2010; Salinas et al., 2012). For example, in Solanum lycopersicum (tomato) SlySBP7 is expressed in roots, leaves, stems, shoot apices, flowers, and fruits at early to late stages of development (Salinas et al., 2012). Functional analysis within clade-I is limited to AtSPL7 from Arabidopsis and supports a role for this gene in the regulation of copper homeostasis (Yamasaki et al., 2009). Copper is an essential micronutrient required for healthy plant growth and development. However, whereas low levels of copper can result in stunted growth and reduced reproductive output, high levels can be toxic. Thus, maintaining a specific copper concentration in cells has major benefits for plant fitness (Clemens et al., 2001; Puig et al., 2007).
Under low copper conditions atspl7 mutants have much lower levels of the microRNAs miR397a, miR398b, miR398c, miR408, and miR857, which in the wild type collectively and negatively regulate copper homeostasis proteins (Abdel-Ghany and Pilon, 2008; Yamasaki et al., 2009). In the case of miR398b and miR398c, decreased expression in the atspl7 mutant is due to the loss of AtSPL7 binding to the Cu-response element (CuRE), characterized by the core sequence GTAC, in the promoter region (Yamasaki et al., 2009). In turn, low levels of miR398b andmiR398c result in the loss of Cu/Zn SUPEROXIDE DISMUTASE 1 (CSD1) and CSD2 degradation, which is normally essential for the reallocation of limited copper supply to support photosynthetic functioning (Sunkar et al., 2006; Yamasaki et al., 2007, 2009; Abdel-Ghany and Pilon, 2008).
Other SPL genes that function as copper-responsive gene promoter binding proteins are COPPER RESPONSE REGULATOR 1 (CRR1) in Chlyamydomonas reinhardtii (green alga) and PpSBP2 in P. patens (clade-II) (Kropat et al., 2005; Nagae et al., 2008; Castruita et al., 2011; Strenkert et al., 2011). Similar to AtSPL7, CRR1 binds to GTAC motifs of CuREs, and in C. reinhardtii this results in the transcriptional activation of copper-deficiency target genes (e.g., CYTOCHROMEC6 and CPX1) that cause a physiological shift to copper-independent photosynthesis (Quinn and Merchant, 1995; Quinn et al., 1999). However, unlike AtSPL7, CRR1, and PpSBP2 are not members of SPL clade-I (Figure 1). This suggests either that a function in copper homeostasis has evolved multiple times independently, or has been lost multiple times following speciation and gene duplication (Figure 3). Given the lack of data for this trait across a diversity of SPL genes, future studies explicitly testing for copper homeostasis in divergent SPL protein clades will be required to distinguish between these alternative hypotheses.
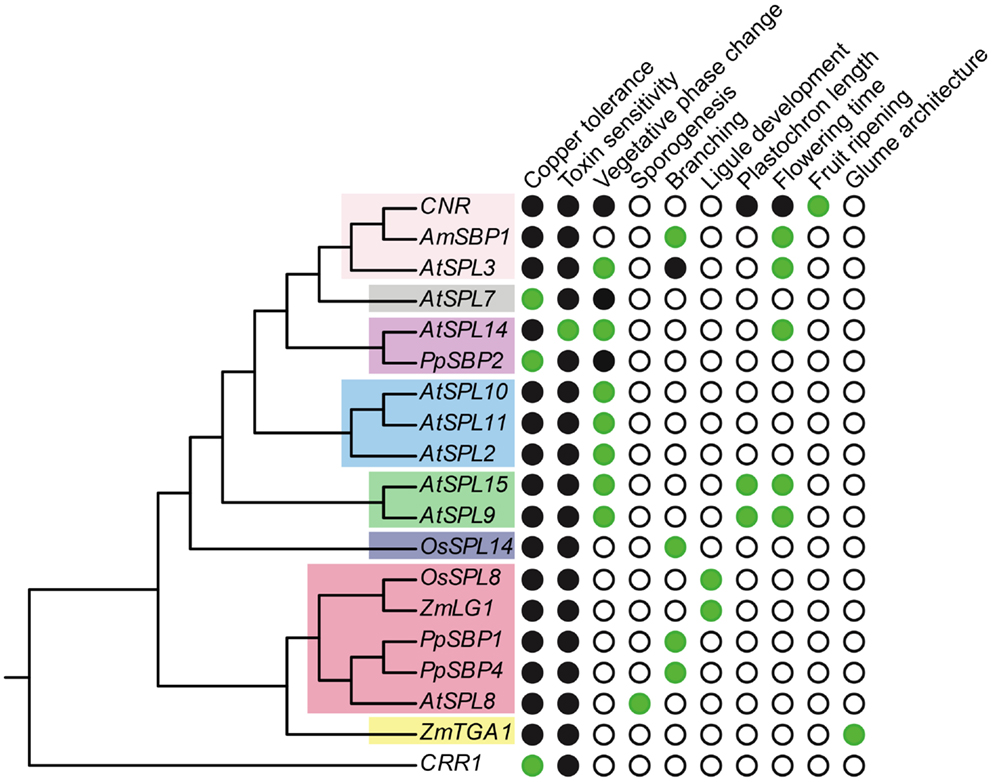
Figure 3. Functional diversity of SPL genes. The gene tree is based on Figure 1, but only includes genes for which functional data are available. Green circles, presence; white circles, absence; black circles, unknown.
Clade-II
Similar to clade-I genes, members of SPL clade-II are relatively large, are expressed widely across the plant and throughout ontogeny, and lack negative regulation by miR156 and miR157 (Cardon et al., 1999; Stone et al., 2005; Xie et al., 2006; Chuck et al., 2007; Yang et al., 2007; Wang et al., 2010; Salinas et al., 2012). Arabidopsis has four clade-II genes – AtSPL1, AtSPL12, AtSPL14, and AtSPL16 – derived from two recent and one ancient duplication event(s) (Yang et al., 2007) (Figure 1). Expressed sequence tag (EST) and microarray data (eFP Browser: http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) suggest that all four paralogs are expressed widely in seedlings, rosette, and cauline leaves, shoot apical meristems, flowers, fruits, and roots (Winter et al., 2007; Yang et al., 2007). However, whereas AtSPL16 is expressed most strongly in mid stage shoot apices and cauline leaves, AtSPL1, AtSPL12, and AtSPL14 are expressed most strongly in cauline leaves, flowers, and late stage shoot apices (Winter et al., 2007). In tomato, the clade-II genes SlySBP12a and SlySBP12b are expressed ubiquitously and constitutively from seedling to ripe fruit (Salinas et al., 2012); a similar expression pattern has been described for OsSPL1 in rice (Oryza sativa) (Yang et al., 2007).
Functional data exists for two clade-II genes, AtSPL14 and PpSBP2; these data suggest functional diversification following speciation in this clade (Stone et al., 2005; Nagae et al., 2008). Similar to its distant paralogs, CRR1 (outgroup) and AtSPL7 (clade-I), PpSBP2 is involved in the regulation of copper homeostasis in P. patens (Nagae et al., 2008) (Figure 1). Under high copper conditions abundant PpSBP2 binds to GTACT motifs in the promoter of IRON SUPEROXIDE DISMUTASE (FeSOD), resulting in its transcriptional repression (Nagae et al., 2008). However, since SODs are critical for eliminating the harmful effects of reactive oxygen species (ROS), produced as by-products of cellular metabolism, ROS accumulation is mitigated by the switch to copper/zinc SOD catalysis (Mittler, 2002). By contrast, when copper and zinc are limiting, it is hypothesized that PpSBP2 expression is low, resulting in the reinstatement of FeSOD functioning.
In the case of arabidopsis, mutations in AtSPL14 result in plants that fail to respond to the fungal toxin fumonisin B1 (FB1) (Stone et al., 2005). In wild type plants, FB1 exposure causes apoptosis, presumably as a defense mechanism against its negative effect on sphingolipid metabolism (Desai et al., 2002). By contrast, growth of atspl14 mutants is not inhibited on media containing FB1 (Stone et al., 2005). In addition to FB1 resistance, atspl14 mutants are defective in early developmental phase change (Stone et al., 2005) (Figure 3). In early development wild type arabidopsis plants undergo a physiological transition from juvenile to adult growth. This phase change is accompanied by subtle changes in leaf morphology, and results in vegetative adult plants that are competent to respond to floral inductive signals (Poethig, 1990; Telfer et al., 1997; Baürle and Dean, 2006). Reduction-of-function atspl14 mutants have a developmentally faster transition to the adult growth phase relative to wild type plants, which slightly accelerates flowering time (Stone et al., 2005). Thus, it is inferred that AtSPL14 functions to delay the juvenile to adult transition. This result is in striking contrast with other characterized arabidopsis SPL genes in clades V, VI, and VIII (see sections 8, 9, and 11) that function to accelerate vegetative phase change (Figures 2A and 3). How SPL paralogs evolved antagonistic effects on developmental phase change awaits further molecular genetic inquiry.
Clade-III
Members of clade-III SPL genes have diverse functions, and although short relative to clade-I and II genes, lack regulation by miR156 or miR157 (Cardon et al., 1999; Xie et al., 2006; Salinas et al., 2012). The clade-III gene AtSPL8 was the first SPL gene to be functionally characterized in arabidopsis (Unte et al., 2003). Mutations in AtSPL8 have no effect on phase change, but have a profound effect on seed set, petal trichome production, and root growth (Unte et al., 2003; Zhang et al., 2007). This is modulated through the positive (flowers) and negative (roots) regulation of gibberellic acid (GA) signaling (Zhang et al., 2007). At late stages of flower development stamen filaments of atspl8 mutants are shorter than wild type and anthers produce fewer pollen grains due to a reduction in sporogenous cells undergoing meiosis; a similar, but weaker, reduction in meiosis is also observed in the ovule (Unte et al., 2003). As a consequence, seed set in atspl8 mutants is strongly reduced (Unte et al., 2003; reviewed in Xing et al., 2011). Interestingly, this phenotype is amplified in quadruple spl8:spl2:spl9:spl15 mutants, although some pollen viability remains (Xing et al., 2010). Thus, since AtSPL2, AtSPL9, and AtSPL15 are members of clades V, VIII, and VIII, respectively, these data support either functional conservation or independent recruitment of SPL genes in microsporogenesis.
Despite the lack of functional data, expression of the AtSPL8 co-orthologs SlySBP8a and SlySBP8b in tomato tentatively suggests at least partial conservation of function in megasporogenesis. Both genes are expressed more highly in carpels and young versus old fruits, but have very low expression in roots, seedlings, and stamens (Salinas et al., 2012). By contrast, expression of the AtSPL8 ortholog VvSBP19 in grapevine (Vitis vinifera) has not been detected in fruits (Wang et al., 2010). The moss P. patens has between five and eight clade-III SPL genes, PpSBP1/1b, PpSBP4/4b, PpSBP7, PpSBP9/9b, and PpSBP12, two of which have been functionally characterized. Loss-of-function ppsbp1 and ppsbp4 mutants produce more branches than wild type at both early and late stages of colony development, and are defective in spore germination (Riese et al., 2008) (Figure 2C). Furthermore, PpSBP1, PpSBP4, PpSBP7, and PpSBP12 are negatively regulated by cryptochromes, blue-light absorbing photoreceptors involved in regulating phase change and the circadian clock (Guo et al., 1998; Somers et al., 1998; Devlin and Kay, 2000; Riese et al., 2008). Although functionally distinct, AtSPL8 expression is also regulated by cryptochromes. However, this regulation is positive rather than negative. It is postulated that this difference in regulation is the result of differential dominance of the gametophytic and sporophytic stages of mosses and land plants, respectively (Folta et al., 2003; Riese et al., 2008).
In maize and rice, the SPL clade-III gene liguleless1 (Zmlg1and OsSPL8) is involved in development of the ligule and auricle (Figure 3), two structures borne on the adaxial surface of grass leaves between the blade and the sheath (Moreno et al., 1997; Lee et al., 2007). In lg1 mutants, the ligule and auricle are completely missing due to the loss of longitudinal periclinal divisions, and the blade-sheath boundary is less well defined than wild type (Sylvester et al., 1990). Interestingly, grasses vary greatly in whether they possess ligules and/or auricles (Lee et al., 2007). Thus, it is hypothesized that changes in the expression or function of lg1-like genes explain variation for these leaf traits. This hypothesis waits further testing.
Clade-IV
Members of clade-IV SPL genes include AtSPL6 from arabidopsis, SlySBP6a, SlySBP6b, and SlySBP6c from tomato, and PpSBP3, PpSBP6, PpSBP6b, PpSBP13, and PpSBP14 from Physcomitrella (Figure 1). No orthologs have been found in monocots suggesting a loss of this gene lineage at least in the fully sequenced genomes of rice and other grasses. The only SPL clade-IV gene to be functionally characterized is P. patens PpSBP3 (Cho et al., 2012). Deletion of PpSBP3 accelerates and increases the number of gametophore-producing leafy buds in the moss gametophyte, suggesting that it normally represses reproductive development (Cho et al., 2012). Although not directly comparable due to the lifecycle differences of mosses and angiosperms, this function is somewhat similar to AtSPL14 (clade-II), which functions to delay the transition to adult development (Stone et al., 2005).
Sequence analyses and expression data in miR156 and miR157 mutants suggest that clade-IV SPL genes are regulated by both miR156 and miR157 (Cho et al., 2012; Salinas et al., 2012). In tomato expression of the three clade-IV SPL genes suggest divergence of function. With the exception of stems, SlySBP6a is expressed constitutively across the plant, similar to AtSPL6 in arabidopsis (Cardon et al., 1999; Salinas et al., 2012). By contrast, SlySBP6c expression is confined to shoot apical meristems and the whole inflorescence, but is not expressed in vegetative tissues, flowers, or fruits. Finally, SlySBP6b expression increases during fruit development (Salinas et al., 2012). It will be interesting to functionally test the role of SlySBP6b in fruit maturation – a key agronomic trait – and to determine whether the differential gene expression of these tomato genes reflect sub- or neo-functionalization.
Clade-V
In contrast to SPL genes that affect both the timing of and morphological features associated with phase change (see sections 9 and 11), silencing of the closely related SPL clade-V paralogs AtSPL10, AtSPL11, and AtSPL2 only affects the latter (Figure 2A) (Shikata et al., 2009). During vegetative growth, the first rosette leaves of plants with reduced AtSPL10, AtSPL11, and AtSPL2 expression are narrower, more oval, and have more serrated edges than their wild type counterparts, characteristic of later developing rosette leaves. Furthermore, following the onset of inflorescence development, the late developing cauline leaves of AtSPL10, AtSPL11, and AtSPL2 silenced plants are wider compared to late developing leaves on wild type plants, and have adaxial trichomes that are normally only found on early cauline leaves (Shikata et al., 2009). Taken together, these data show that growth and development during the vegetative and reproductive phases of the arabidopsis life cycle can be uncoupled.
In the case of AtSPL10, AtSPL11, and AtSPL2 silenced plants changes in leaf characteristics are likely the result of reduced FRUITFULL (FUL) expression, which has long been known to affect leaf development (Gu et al., 1998; Shikata et al., 2009). However, unlike ful mutants, mutations in AtSPL10, AtSPL11, and AtSPL2 have no effect on inflorescence or fruit development (Shikata et al., 2009). This suggests that FUL is differentially regulated in leaves, inflorescence meristems and fruits. It will be important to determine how this regulation is partitioned among arabidopsis SPL genes, and to see if this differential regulation is conserved in other species, including the other miR156-regulated clade-V rice genes OsSPL3, OsSPL4, OsSPL11, and OsSPL12 (Xie et al., 2006), and the constitutively expressed tomato paralogs SlySBP2 and SlySBP10 (Salinas et al., 2012) (Figure 1).
In addition to leaf morphology, AtSPL10 and AtSPL11 have been implicated in cell differentiation during early embryogenesis (Nodine and Bartel, 2010). Dicer-like 1 (dcl1) mutants have decreased miRNA expression, resulting in derepression of several hundred miRNA target genes, including AtSPL10 and AtSPL11, and precocious differentiation of early embryonic cells. However, when dcl1 mutants are crossed with atspl10:atspl11 mutants, normal embryogenesis is partially restored (Nodine and Bartel, 2010). These data demonstrate redundant roles for AtSPL10 and AtSPL11 in both early and late stage differentiation.
Clade-VI
The most widespread reported function of SPL genes is promoting the transition from juvenile to adult growth (Figure 3), which is marked by an increase in responsiveness to floral inductive signals, resulting in competence to flower (Baürle and Dean, 2006). In arabidopsis, phase change occurs primarily in response to environmental signals, such as temperature and developmental age, and is accompanied by subtle changes in leaf morphology (Poethig, 1990; Telfer et al., 1997). Overexpression analyses implicate AtSPL3, and tentatively the other two clade-VI genes AtSPL4 and AtSPL5, in the timing of and/or morphological features associated with phase change (Wu and Poethig, 2006). These functions are likely moderated through negative regulation by miR156 (Wu and Poethig, 2006; Schwarz et al., 2008; Shikata et al., 2009; Wang et al., 2009; Yamaguchi et al., 2009).
Constitutive expression of miR156, which results in a decrease in expression of 10 out of 16 arabidopsis SPL genes including AtSPL3/4/5, results in prolongation of the vegetative phase, as well as delayed flowering, and an increase in the number of juvenile leaves (Schwab et al., 2005; Wu and Poethig, 2006). In mutants that overexpress the clade-VI genes AtSPL3, AtSPL4, and AtSPL5, and the clade-VIII gene AtSPL15 (see Section 11), leaves develop adult characteristics – including abaxial trichomes, and an increase number of cells that are smaller in size – faster than their wild type counterparts (Wu and Poethig, 2006; Usami et al., 2009). This is in contrast to the overexpression of the clade-II gene AtSPL14, which results in a truncated juvenile vegetative phase (Stone et al., 2005). Based on these incomplete data, ancestral trait reconstructions suggest that a role in vegetative phase change evolved fairly early during the diversification of angiosperm SPL genes, and was followed by multiple losses of function outside arabidopsis (Figure 2A). However, with mutants available for only nine out of 16 arabidopsis SPL genes, and very few characterized SPL genes from other species, the exact timing of this evolutionary transition is equivocal (Figure 2A).
In addition to promoting vegetative phase change, overexpression data suggest that AtSPL3, AtSPL4, and AtSPL5 redundantly promote the reproductive transition by integrating signals from the autonomous, photoperiod, age, and GA pathways (Figures 2B and 4) (Cardon et al., 1997; Gandikota et al., 2007; Wang et al., 2009; Yamaguchi et al., 2009; Jung et al., 2012; Porri et al., 2012; Yu et al., 2012). Under short day conditions, all three SPL genes are negatively regulated in an age dependent manner by miR156, and are positively regulated by SUPPRESSION OF OVEREXPRESSION OF CONSTANS1 (SOC1) through the GA pathway (Jung et al., 2011, 2012). By contrast, under long day conditions, SOC1, FLOWERING LOCUS T (FT), and FLOWERING LOCUS D (FD) positively regulate AtSPL3, AtSPL4, and AtSPL5 in leaves in response to photoperiod signals (Jung et al., 2012). SPL proteins indirectly activate FT expression, probably through the direct binding of the inflorescence meristem gene FUL, and directly activate transcription of FUL, APETALA1 (AP1), and LEAFY (LFY) in the shoot apical meristem (Corbesier and Coupland, 2006; Corbesier et al., 2007; Wang et al., 2009; Yamaguchi et al., 2009) (Figure 4).
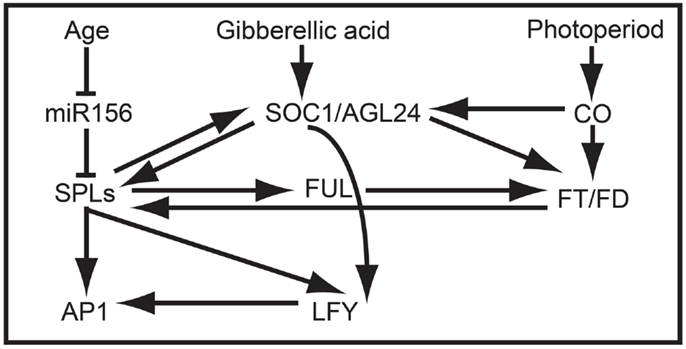
Figure 4. SPL clade-VI gene-dependent flowering network in Arabidopsis. The at least partially redundant genes AtSPL3, AtSPL4, and AtSPL5 are negatively regulated by miR156 in early development and positively regulated by SOC1 prior to flowering. In turn, AtSPL3, AtSPL4, and AtSPL5 induce the expression of FUL, AP1, LFY, and SOC1, resulting in the production of flowers.
Functional data exist for a few core eudicot clade-VI genes, allowing preliminary comparative analysis following speciation. Similar to arabidopsis, the single AtSPL3/4/5 ortholog AmSBP1 in snapdragon is involved in initiating flower development within the inflorescence. However, in contrast to arabidopsis plants constitutively expressing miR156, where flower production is delayed, silencing of AmSBP1 can eliminate flowering completely (Preston and Hileman, 2010). One hypothesis for these different phenotypes in arabidopsis and snapdragon is a tighter control of AmSBP1 versus AtSPL3/4/5 on the expression of downstream floral organ identity genes, including the AP1/FUL-like genes SQUA, DEFH28, and AmFUL, and the LFY-like gene FLORICAULA (FLO) (Klein et al., 1996; Preston and Hileman, 2010). Alternatively, the less extreme phenotype associated with simultaneously silencing AtSPL3, AtSPL4, AtSPL5, and several other arabidopsis SPL genes, versus silencing of a single snapdragon SPL gene, might be explained by antagonistic functions of arabidopsis miRNA-regulated SPL genes. To test this antagonism hypothesis, loss-of-function phenotypes must be generated and compared for all arabidopsis SPL genes, starting with the clade-VI genes AtSPL4 and AtSPL5.
In addition to the flowering phenotype, silencing of AmSBP1 also causes an increase in vegetative branching under long days (Figure 2C) (Preston and Hileman, 2010). This branched phenotype is similar to that observed under short day growth conditions, suggesting that it is a consequence of late flowering rather than a loss of apical dominance per se (Preston and Hileman, pers. obs.). By contrast in tomato, mutations in the AtSPL3/4/5 ortholog COLORLESS NON-RIPENING (CNR) result in fruits that fail to ripen (Manning et al., 2006). At present, this can be ascribed as a novel SPL gene function (Figure 3). Indeed, differential expression of the closest CNR homologs, SlySBP3, and SlySBP4, suggest that these genes are not involved in fruit ripening, but may function in earlier carpel development (Salinas et al., 2012). It will be interesting to see if tomato SPL genes other than CNR have been recruited to function in fruit ripening, as suggested by gene expression data (Salinas et al., 2012), and if paralogous SPL genes affect fruit development in related species with (e.g., Solanum melongena, eggplant, Solanaceae) or without (e.g., Petunia x hybrida, petunia, Solanaceae) fleshy fruits (Pabón-Mora and Litt, 2011).
In addition to the lineage containing AtSPL3, AtSPL4, and AtSPL5, another clade-VI gene lineage exists for which there is no arabidopsis ortholog (Figure 1). Based on data from snapdragon, this clade of genes also targets expression of AP1/FUL-, FT-, and LFY-like genes (Klein et al., 1996; Preston and Hileman, 2010). Future characterization of genes within this will potentially allow more accurate reconstruction of the ancestral functions of clade-VI genes and promises to bolster our understanding of developmental differences between rosid and asterid core eudicots.
Clade-VII
Expression and functional data in arabidopsis, maize, and rice implicate clade-VII genes in various aspects of above ground plant development. In arabidopsis, the single SPL clade-VII gene AtSPL13 has been implicated in delaying leaf outgrowth following emergence of the cotyledons during germination (Martin et al., 2010a,b). Loss of negative regulation of AtSPL13 due to mutations in the miR156 binding site significantly delays emergence of leaf primordia probably due to the concomitant upregulation of miR157, which normally represses AP2-like genes such SCHNARCHZAPFEN (SNZ) (Martin et al., 2010a,b). In addition to hypocotyls, AtSPL13 is broadly expressed in different plant organs (Martin et al., 2010b). Thus, it will be interesting to determine if AtSPL13 has other functions in plant development, such as those described for clade-VII SPL genes below.
The most extensively characterized clade-VII gene in maize is TEOSINTE GLUME ARCHITECTURE 1 (TGA1), variation in which explains the drastic difference in fruitcase morphology between cultivated maize and its ancestor teosinte (Zea mays ssp. parviglumis) (Wang et al., 2005). Whereas teosinte has hard fruitcases resulting from invagination of the inflorescence branch and hardening of the floral bract (glume), maize has a soft fruitcase that allows easy harvesting of the fruit. Evidence strongly suggests that the difference in fruitcase morphology between subspecies is due to a single amino acid substitution in TGA1 (Wang et al., 2005; Preston et al., 2012). However, amino acid and gene/protein expression analyses suggest that the parallel evolution of soft fruitcases in other grasses is under the regulation of other, as yet unknown, genes (Preston et al., 2012). Furthermore, the actual function of TGA1 in teosinte is still under investigation.
Despite the lack of correlation between TGA1 structure and expression, research on one of two TGA1 homologs in rice suggests that clade-IX SPL genes function generally in late reproductive development (Wang et al., 2012). Specifically, in a recent study it was demonstrated that increased grain size and grain number in the rice indica variety HJX74 compared to the rice indica Basmati varieties are positively correlated with OsSPL16 expression (Wang et al., 2012). These differences are mediated by changes in patterns of cell proliferation and elongation, resulting in associated changes in grain shape. Unlike SPL genes in many other clades, silencing of OsSPL16 has no affect on phase change or plant architecture. However, constitutive expression in arabidopsis and rice accelerates flowering and results in dwarf plants with fewer inflorescence branches, respectively (Wang et al., 2012). Together these data suggest that the regulatory interactions between many SPL proteins and their downstream targets (e.g., AP1/FUL-, LFY-, FT-, and SOC1-like genes) may be largely conserved (Figure 4), but that differential expression is driving broad-scale functional differences.
In addition to TGA1, maize has five other clade-VII genes, one of which has an expression profile consistent with its playing a role in feminization (Hultquist and Dorweiler, 2008) (Figure 1). Maize plants are monecious, developing a terminal male inflorescence (tassel) and lateral female inflorescences (ears). Tassels and ears differ substantially in terms of their developmental timing, branching architecture, flower and reproductive organ abortion, and floral bract (glume) morphology (Kiesselbach, 1999). In the tasselseed1 (ts1) and mediator of paramutation1 (mop1) mutants, tassels are feminized relative to wild type tassels (Dorweiler et al., 2000; Acosta et al., 2009). Interestingly, ZmSBP11 is expressed more highly in these mutant feminized tassels than in wild type tassels, suggesting a role in one or all of the ear-specific traits (Hultquist and Dorweiler, 2008). It will be of great interest to discern if and how ZmSBP11 affects ear development and to see whether similar functions can be assigned to the AtSPL13 homologs in arabidopsis, constitutively expressed SlySBP13 homolog in tomato, and the OsSPL2, and OsSPL18 homologs in rice (Salinas et al., 2012) (Figure 1).
Clade-VIII
Redundant roles in vegetative phase change and reproductive transition have been demonstrated for the closely related miR156-regulated clade-VIII genes AtSPL9 and AtSPL15 (Schwarz et al., 2008; Usami et al., 2009). Mutations in these genes cause a subtle increase in vegetative rosette leaves, but have no significant effect on inflorescence cauline leaves (Schwarz et al., 2008). However, in the double spl9 spl15 mutant, vegetative rosette leaf number is significantly increased relative to the single mutants, leaves are more rounded, and flowering time is delayed relative to wild type (Schwarz et al., 2008). Furthermore, overexpression of AtSPL9 in hyponastic leaves1 (hyl1) mutants that have lowered miR156 expression, and therefore higher SPL gene expression, results in the complete loss of the juvenile phase (Li et al., 2012). Ancestral state reconstructions suggest that vegetative phase change and flowering time function evolved in the SPL gene lineage prior to diversification of the core eudicots, but the exact timing of these transitions is ambiguous (Figures 2A,B). Thus, future work is needed to determine whether the shared function of the arabidopsis SPL clade-V, VI, and VIII genes is due to ancient neo-functionalization, followed by multiple losses, or alternatively, multiple independent evolutionary gains.
In addition to phase change, plastochron length is affected in late flowering atspl9atspl15 double mutants, suggesting dissociation between growth and development (Figure 3) (Schwarz et al., 2008). To date, this is the only clade of genes for which a role in plastochron length has been described (Figure 3). The atspl9atspl15 double mutant has a shortened plastochron relative to wild type, which is correlated with a smaller shoot apical meristem and rounded leaves (Schwarz et al., 2008). However, in the more and smaller cells 1d (msc1d) mutant that overexpresses AtSPL15 due to disruption of the miR156 binding site, the juvenile to adult phase transition is accelerated (Figure 2A). This acceleration is accompanied by an increased ratio of rosette leaves having adult characteristics – adaxial trichomes and more smaller cells – similar to overexpression lines of AtSPL3, AtSPL4, and AtSPL5 (Wu and Poethig, 2006; Usami et al., 2009). Thus, leaf number and identity is correlated with phase change, but it can be uncoupled.
Recent genetic evidence also suggests that AtSPL9 is involved in petal trichome initiation, through the activation of TRICHOMELESS (TCL1), and anthocyanin pigment accumulation in vegetative stems (Yu et al., 2010; Guo et al., 2011). In the case of pigment production, overexpression of AtSPL9 results in reduced levels of anthocyanin, suggesting a negatively regulatory interaction in wild type plants. The mechanism of this negative regulation is hypothesized to be interference of the MYB-bHLH-WD40 complex, which controls the transcription of flavonoid biosynthesis genes (Guo et al., 2011). It is as yet unknown whether a role in trichome development and pigment production is specific to AtSPL9, or whether SPL homologs have similar functions in other species.
Clade-IX
In maize, the clade-IX genes ZmSBP6 and ZmSBP8, and the currently unplaced paralogs ZmSBP5 and ZmSBP7 (Figure 1), are hypothesized to be involved in feminization (Hultquist and Dorweiler, 2008) (Figure 1). Evidence supporting this comes from gene expression analyses in ts1 and mop1 mutants, where transcript levels of both genes are higher in feminized versus non-feminized tassels (Hultquist and Dorweiler, 2008). However, since this correlation in gene expression does not imply causation, future functional studies are needed to test this hypothesis.
Convincing evidence suggesting a role for SPL clade-IX grass genes in branching, which has apparently evolved multiple times independently during SPL gene diversification (Figure 2C), comes from genetic work on the rice domestication gene OsSPL14, which is strongly associated with the WEALTHY FARMER’S PANICLE QTL for variation in rice architecture (Jiao et al., 2010; Miura et al., 2010). Increased expression of OsSPL14 in the japonica line Shaoniejing relative to the indica line Taichung Native 1, and ST-12 relative to japonica Nipponbare, results in decreased vegetative branching and increased inflorescence branching, the latter resulting in an increase in grain number (Jiao et al., 2010; Miura et al., 2010). It remains unclear how OsSPL14 is able to at once both promote and repress branching in different developmental contexts (i.e., vegetative versus inflorescence). However, expression of OsSPL14 is negatively regulated by OsmiR156 (Miura et al., 2010).
The opposite action of OsSPL14 on vegetative and inflorescence branching has major implications for breeding. In the case of rice, increased expression of this gene is favorable as it increases grain yield at the expense of biomass. However, in forage and biofuel grasses increased biomass is favorable to grain yield. Indeed, Fu et al. (2012) recently demonstrated that by differentially increasing the expression of PvmiR156 in switch grass (Panicum virgatum), some plants had good characteristics for biofuel engineering, including increased leaf number, late flowering, increased vegetative branch number, and reduced inflorescence size. This phenotype resulted from the differential targeting of eight SPL genes. Thus, research aimed at elucidating the specific function of different SPL genes in crop species and their relatives has great promise for fine-tuning plant architecture in an agricultural context.
Future Directions and Concluding Remarks
Accepting the caveat that deep branches in the most likely SPL trees are not well supported (Figure 1), phylogenetic state reconstructions based on functionally characterized SPL genes from a few distantly related taxa tentatively suggest the origin of early and late phase change function at least prior to the diversification of core eudicots (Figures 2A,B). However, if the ancestor of clades I, II, V, VI, and VIII had a dual role in regulating vegetative and reproductive transitioning, this function has subsequently been partitioned through differential sub-functionalization following both gene duplication and speciation (Figures 2A,B). Furthermore, there is evidence of both neo-functionalization (e.g., in fruit ripening and glume architecture) and parallel recruitment (e.g., in branching) in several different lineages (Figures 2C and 3).
The inferred dynamic history of SPL gene function across both gene clades and species might be explained by a combination of changes in regulation, biochemical function, and/or downstream targeting. Indeed, it is already known that most, but not all, SPL genes are regulated by microRNAs in an age dependent manner, and that there have been shifts in regulation between the cryptochrome, photoperiod, and GA pathways (Cardon et al., 1997; Guo et al., 1998; Somers et al., 1998; Devlin and Kay, 2000; Gandikota et al., 2007; Zhang et al., 2007; Riese et al., 2008; Wang et al., 2009; Yamaguchi et al., 2009; Jung et al., 2012; Porri et al., 2012). Furthermore, sequence analyses and comparisons of gene expression in SPL-silenced versus wild type plants suggest both conservation and divergence of SPL protein targets. The most intriguing case is the regulatory relationship between SPL proteins and the meristem identity AP1/FUL-like genes. Sequence and expression analyses suggest that either these regulatory relationships evolved early and have subsequently been lost multiple times following the divergence of the TGA1-like clade from other angiosperm SPL clades, or that these regulatory relationships have evolved multiple times independently (Klein et al., 1996; Shikata et al., 2009; Preston and Hileman, 2010). Testing these alterative hypotheses across multiple SPL clades and species will be important to determine the mechanistic basis for functional changes within the gene family.
In addition to the mechanistic basis of changes in function, characterization of multiple SPL genes from different taxa has generated hypotheses regarding the diversification of key ecological traits, such as the evolution of growth habit. Annual and perennial growth habits can be distinguished on the basis of quantitative differences in flowering time, meristem dormancy/identity, architecture, biomass, and/or woodiness, all of which are affected in spl and their target ap1/ful-like gene mutants (Melzer et al., 2008). Thus, SPL genes are good candidates underlying shifts between annuity and perenniality, possibly through the differential targeting of genes involved in meristem identity and phase change. A promising avenue of research will be to study related annual and perennial populations or species to see if growth habit differences can be explained by differences in expression and/or protein functional differences of microRNA-regulated SPL genes. Such studies are currently underway for different populations of Mimulus guttatus and different species of Arabidopsis that vary in several traits related to growth habit.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank María Salinas and Jane Dorweiler for access to tomato and maize SPL gene sequences, respectively, and two anonymous reviewers. This work was supported by a National Science Foundation grant (IOS-1051886) to Lena C. Hileman and Jill C. Preston.
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/Plant_Evolution_and_Development/10.3389/fpls.2013.00080/abstract
References
Abdel-Ghany, S. E., and Pilon, M. (2008). Micro-RNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283, 15932–15945.
Acosta, I. F., Laparra, H., Romero, S. P., Schmelz, E., Hamberg, M., Mottinger, J. P., et al. (2009). tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323, 262–265.
Airoldi, C. A., and Davies, B. (2012). Gene duplication and the evolution of plant MADS-box transcription factors. J. Genet. Genomics 39, 157–165.
Anderson, J. T., Lee, C.-R., and Olds, T. M. (2011). Life history QTLs and natural selection on flowering time in Boechera stricta, a perennial relative of Arabidopsis. Evolution 65, 771–787.
Arazi, T., Talmor-Neiman, M., Stav, R., Riese, M., Huijser, P., and Baulcombe, D. C. (2005). Cloning and characterization of micro-RNAs from moss. Plant J. 43, 837–848.
Baürle, I., and Dean, C. (2006). The timing of developmental transitions in plants. Cell 125, 655–664.
Becker, A., and Theissen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489.
Birkenbihl, R. P., Jach, G., Saedler, H., and Huijser, P. (2005). Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352, 585–596.
Cardon, G., Hohmann, S., Klein, J., Nettesheim, K., Saedler, H., and Huijser, P. (1999). Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237, 91–104.
Cardon, G. H., Hohmann, S., Nettesheim, K., Saedler, H., and Huijser, P. (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 12, 367–377.
Castruita, M., Casero, D., Karpowicz, S. J., Kropat, J., Vieler, A., Hsieh, S. I., et al. (2011). Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23, 1273–1292.
Causier, B., Castillo, R., Zhou, J., Ingram, R., Xue, Y., Schwarz-Sommer, Z., et al. (2005). Evolution in action: following function in duplicated floral homeotic genes. Curr. Biol 15, 1508–1512.
Cho, S. H., Coruh, C., and Axtell, M. J. (2012). miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 24, 4837–4849.
Chuck, G., Meeley, R., Irish, E., Sakai, H., and Hake, S. (2007). The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Gen 39, 1517–1521.
Clemens, S., Schroeder, J. I., and Degenkolb, T. (2001). Caenorhabditis elegans expresses a functional phytochelatin synthase. Eur. J. Biochem 268, 3640–3643.
Corbesier, L., and Coupland, G. (2006). The quest for florigen: a review of recent progress. J. Exp. Bot. 57, 3395–3403.
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 317, 1030–1033.
Crow, K. D., and Wagner, G. P. (2006). What is the role of genome duplication in the evolution of complexity and diversity? Mol. Biol. Evol. 23, 887–892.
Desai, K., Sullards, M. C., Allegood, J., Wang, E., Schmelz, E. M., Hartl, M., et al. (2002). Fumonisins and fumonisin analogs as inhibitors of ceramide synthase and inducers of apoptosis. Biochim. Biophys. Acta 1585, 188–192.
Devlin, P. F., and Kay, S. K. (2000). Cryptochromes are required for phytochrome signalling to the circadian clock but not for rhythmicity. Plant Cell 12, 2499–2509.
Dorweiler, J. E., Carey, C. C., Kubo, K. M., Hollick, J. B., Kermicle, J. L., and Chandler, V. L. (2000). Mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12, 2101–2118.
Folta, K. M., Pontin, M. A., Karlin-Neumann, G., Bottini, R., and Spalding, E. P. (2003). Genomic and physiological studies of early cryptochrome I action demonstrate role for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J. 36, 203–214.
Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y.-I., and Postlethwait, J. (1999). Preservation of duplicate genes by complementary degenerative mutations. Genetics 151, 1531–1545.
Fu, C., Sunkar, R., Zhou, C., Shen, H., Zhang, J.-Y., Matts, J., et al. (2012). Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotech. J 10, 443–452.
Gandikota, M., Birkenbihl, R. P., Hohmann, S., Cardon, G. H., Saedler, H., and Huijser, P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition of seedlings. Plant J. 49, 683–693.
Gu, Q., Ferrandiz, C., Yanofsky, M. F., and Martienssen, R. (1998). FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517.
Guo, A.-Y., Zhu, Z.-H., Gu, X., Ge, S., Yang, J., and Luo, J. (2008). Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418, 1–8.
Guo, H., Yang, H., Mockler, T. C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363.
Guo, J.-Y., Felippes, F. F., Liu, C.-J., Weigel, D., and Wang, J.-W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 23, 1512–1522.
Hall, M. C., and Willis, J. H. (2006). Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evolution 60, 2466–2477.
Hittinger, C. T., and Carroll, S. B. (2007). Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449, 677–681.
Hultquist, J. F., and Dorweiler, J. E. (2008). Feminized tassels of maize mop1 and ts1 mutants exhibit altered levels of miR156 and specific SBP-box genes. Planta 229, 99–113.
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet 42, 541–545.
Jung, J.-H., Ju, Y., Seo, P. J., Lee, J.-H., and Park, C.-M. (2012). The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69, 577–588.
Jung, J.-H., Seo, P. J., Kang, S. K., and Park, C.-M. (2011). miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant Mol. Biol. 76, 35–45.
Kaessmann, H. (2010). Origins, evolution, and phenotypic impact of new genes. Genome Res. 20, 1313–1326.
Kiesselbach, T. A. (1999). The Structure and Reproduction of Corn. 50th Anniversary Edition. New York: Cold Spring Harbor Laboratory Press.
Klein, J., Saedler, H., and Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Genet. Genomics 250, 7–16.
Kramer, E. M., Jaramillo, M. A., and Di Stilio, V. S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166, 1011–1023.
Kropat, J., Tootey, S., Birkenbihl, R. P., Depege, N., Huijser, P., and Merchant, S. (2005). A regulator of nutritional copper signalling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. U.S.A. 102, 18730–18735.
Lawton-Rauh, A. (2003). Evolutionary dynamics of duplicated genes in plants. Mol. Phylogenet. Evol. 29, 396–409.
Lee, J., Park, J.-J., Kim, S. L., Yim, J., and An, G. (2007). Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 65, 487–499.
Li, S., Yang, X., Wu, J., and He, Y. (2012). HYL1 controls the miR156-mediated juvenile phase of vegetative growth. J. Exp. Bot. 63, 2787–2798.
Liang, X., Nazarenus, T. J., and Stone, J. M. (2008). Identification of a consensus DNA-binding site for the Arabidopsis thaliana SBP domain transcription factor. Biochemistry 47, 3645–3653.
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155.
Maddison, D. R., and Maddison, W. P. (2003). MacClade: Analysis of Phylogeny and Character Evolution. Sunderland: Sinauer Associates.
Maddison, W. P., and Maddison, D. R. (2008). Mesquite: A Modular System for Evolutionary Analysis. Version 2.5. Available at: http://mesquiteproject.org
Manning, K., Tor, M., Poole, M., Hong, Y., Thompson, A. J., King, G. J., et al. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet 38, 948–952.
Martin, R. C., Asahina, M., Liu, P.-P., Kritof, J. R., Coppersmith, J. L., Pluskota, W. E., et al. (2010a). The microRNA156 and microRNA172 gene regulation cascades at post-germinative stages in Arabidopsis. Seed Sci. Res 20, 79–87.
Martin, R. C., Asahina, M., Liu, P.-P., Kritof, J. R., Coppersmith, J. L., Pluskota, W. E., et al. (2010b). The regulation of post-germination transition from the cotyledon- to vegetative-leaf stages by microRNA-targeted SQUAMOSA PROMOTER-BINDING PROTEIN LIKE13 in Arabidopsis. Seed Sci. Res. 20, 89–96.
Melzer, S., Lens, F., Gennen, J., Vanneste, S., Rohde, A., and Beeckman, T. (2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40, 1489–1492.
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410.
Miura, K., Ikeda, M., Matsubara, A., Song, X.-J., Ito, M., Asano, K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–550.
Moreno, M. A., Harper, L. C., Krueger, R. W., Dellaporta, S. L., and Freeling, M. (1997). Ligueleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 11, 616–628.
Nagae, M., Nakata, M., and Takahashi, Y. (2008). Identification of negative cis-acting elements in response to copper in the chloroplastic iron superoxide dismutase gene of the moss Barbula unguiculata. Plant Physiol. 146, 1687–1696.
Nodine, M. D., and Bartel, D. P. (2010). MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 24, 2678–2692.
Pabón-Mora, N., and Litt, A. (2011). Comparative anatomical and developmental analysis of dry and fleshy fruits of Solanaceae. Am. J. Bot. 98, 1415–1436.
Poethig, R. S. (1990). Phase change and the regulation of shoot morphogenesis in plants. Science 250, 923–930.
Porri, A., Torti, S., Romera-Branchat, M., and Coupland, G. (2012). Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long day conditions. Development 139, 2198–2209.
Posada, D., and Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818.
Preston, J. C., and Hileman, L. C. (2010). SQUAMOSA-PROMOTER BINDING PROTEIN 1 initiates flowering in Antirrhinum majus through the activation of meristem identity genes. Plant J. 62, 704–712.
Preston, J. C., Wang, H., Kursel, L., Doebley, J., and Kellogg, E. A. (2012). The role of teosinte glume architecture (tga1) in coordinated regulation and evolution of grass glumes and inflorescence axes. New Phytol. 193, 204–215.
Puig, S., Andres-Colas, N., Garcia-Molina, A., and Penarrubia, L. (2007). Copper and iron homeostasis in Arabidopsis: responses to metal deficiencies, interactions and biotechnological applications. Plant Cell Environ. 30, 271–290.
Quinn, J. M., and Merchant, S. (1995). Two copper-responsive elements associated with the Chylamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7, 623–628.
Quinn, J. M., Nakamoto, S. S., and Merchant, S. (1999). Induction of coproporphyrinogen oxidase in Chlamydomonas chloroplasts occurs via transcriptional regulation of Cpx1 mediated by copper response elements and increased translation from a copper deficiency-specific form of the transcript. J. Biol. Chem. 274, 14444–14454.
Rhoades, M. W., Reinhardt, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520.
Riese, M., Zobell, O., Saedler, H., and Huijser, P. (2008). SBP-domain transcription factors as possible effectors of cryptochrome-mediated blue light signalling in the moss Physcomitrella patens. Planta 227, 505–515.
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574.
Salinas, M., Xing, S., Hohmann, S., Berndtgen, R., and Huijser, P. (2012). Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 235, 1171–1184.
Schwab, R., Palatnik, J. F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517–527.
Schwarz, S., Grande, A. V., Bujdoso, N., Saedler, H., and Huijser, P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67, 183–195.
Shikata, M., Koyama, T., Mitsuda, N., and Ohme-Takagi, M. (2009). Arabidopsis SBP-box genes SPL10. Plant Cell Physiol. 50, 2133–2145.
Somers, D. E., Devlin, P. F., and Kay, A. K. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1490.
Stone, J. M., Liang, X., Nekl, E. R., and Stiers, J. J. (2005). Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 41, 744–754.
Strenkert, D., Schmollinger, S., Sommer, F., Schulz-Raffelt, M., and Schroda, M. (2011). Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell 23, 2285–2301.
Sunkar, R., Kapoor, A., and Zhu, J. K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065.
Sylvester, A. W., Cande, W. Z., and Freeling, M. (1990). Division and differentiation during normal and liguleless-1 maize leaf development. Development 110, 985–1000.
Telfer, A., Bollman, K. M., and Poethig, R. S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, 645–654.
Unte, U. S., Sorensen, A.-M., Pesaresi, P., Gandikota, M., Leister, D., Saedler, H., et al. (2003). SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 15, 1009–1019.
Usami, T., Horiguchi, G., Yano, S., and Tsukaya, H. (2009). The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136, 955–964.
Wang, H., Nussbaum-Wagler, T., Li, B., Zhao, Q., Vigouroux, Y., Faller, M., et al. (2005). The origin of the naked grains of maize. Nature 436, 714–719.
Wang, J.-W., Czech, B., and Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749.
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet 24, 950–954.
Wang, Y., Hu, Z., Yang, Y., Chen, X., and Chen, G. (2010). Genome-wide identification, phylogeny, and expression analysis of the SBP-box gene family in grapevine. Russ. J. Plant Physiol. 57, 273–296.
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718. doi:10.1371/journal.pone.0000718
Wu, G., and Poethig, R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133, 3539–3547.
Xie, K., Wu, C., and Xiong, L. (2006). Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 142, 280–293.
Xing, S., Salinas, M., Höhmann, S., Berndtgen, R., and Huijser, P. (2010). miR156-targed and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22, 3935–3950.
Xing, S., Salinas, M., and Huijser, P. (2011). New players unveiled in early anther development. Plant Signal. Behav. 6, 934–938.
Yamaguchi, A., Wu, M.-F., Yang, L., Wu, G., Poethig, R. S., and Wagner, D. (2009). The microRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY. Dev. Cell 17, 268–278.
Yamaguchi, T., Lee, D. Y., Miyao, A., Hirochika, H., An, G., and Hirano, H. Y. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18, 15–28.
Yamasaki, H., Abdel-Ghany, S. E., Cohu, C. M., Kobayashi, Y., Shikanai, T., and Pilon, M. (2007). Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 282, 16369–16378.
Yamasaki, H., Hayashi, M., Fukazawa, M., Kobayashi, Y., and Shikanai, T. (2009). SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21, 347–361.
Yamasaki, K., Kigawa, T., Inoue, M., Yamasaki, T., Yabuki, T., Aoki, M., et al. (2006). An Arabidopsis SBP-domain fragment with a disrupted C-terminal zinc-binding site retains its tertiary structure. FEBS Lett. 580, 2109–2116.
Yang, Z., Wang, X., Gu, S., Hu, Z., Xu, H., and Xu, C. (2007). Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 407, 1–11.
Yu, N., Cai, W.-J., Wang, S., Shan, C.-M., Wang, L.-J., and Chen, X.-Y. (2010). Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22, 2322–2335.
Yu, S., Galvao, V. C., Zhang, Y.-C., Horrer, D., Zhang, T.-Q., Hao, Y.-H., et al. (2012). Gibberellin regulates the Arabidopsis floral transition through miR156-targeted PROMOTER BINDING-LIKE transcription factors. Plant Cell 24, 3320–3332.
Zhang, J., Rosenberg, H. F., and Nei, M. (1998). Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. U.S.A. 95, 3708–3713.
Keywords: SPL genes, gene duplication, phase change, flowering time, branching architecture, developmental transitions
Citation: Preston JC and Hileman LC (2013) Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 4:80. doi: 10.3389/fpls.2013.00080
Received: 12 February 2013; Paper pending published: 25 February 2013;
Accepted: 19 March 2013; Published online: 05 April 2013.
Edited by:
Elena M. Kramer, Harvard University, USAReviewed by:
Elena M. Kramer, Harvard University, USAPablo D. Jenik, Franklin & Marshall College, USA
Copyright: © 2013 Preston and Hileman. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Jill C. Preston, Department of Plant Biology, The University of Vermont, 111 Jeffords Hall, 63 Carrigan Drive, Burlington, VT 05405, USA. e-mail:amlsbC5wcmVzdG9uQHV2bS5lZHU=

