- Section of Plant Physiology, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
Adequate water supply is of utmost importance for growth and reproduction of plants. In order to cope with water deprivation, plants have to adapt their development and metabolism to ensure survival. To maximize water use efficiency, plants use a large array of signaling mediators such as hormones, protein kinases, and phosphatases, Ca2+, reactive oxygen species, and low abundant phospholipids that together form complex signaling cascades. Phosphatidic acid (PA) is a signaling lipid that rapidly accumulates in response to a wide array of abiotic stress stimuli. PA formation provides the cell with spatial and transient information about the external environment by acting as a protein-docking site in cellular membranes. PA reportedly binds to a number of proteins that play a role during water limiting conditions, such as drought and salinity and has been shown to play an important role in maintaining root system architecture. Members of two osmotic stress-activated protein kinase families, sucrose non-fermenting 1-related protein kinase 2 and mitogen activated protein kinases were recently shown bind PA and are also involved in the maintenance of root system architecture and salinity stress tolerance. In addition, PA regulates several proteins involved in abscisic acid-signaling. PA-dependent recruitment of glyceraldehyde-3-phosphate dehydrogenase under water limiting conditions indicates a role in regulating metabolic processes. Finally, a recent study also shows the PA recruits the clathrin heavy chain and a potassium channel subunit, hinting toward additional roles in cellular trafficking and potassium homeostasis. Taken together, the rapidly increasing number of proteins reported to interact with PA implies a broad role for this versatile signaling phospholipid in mediating salt and water stress responses.
Introduction
Plants have to adapt to various changes in their environment and signals from the outside have to pass the membrane in order for the cell to respond. Environmental stress causes changes in the phospholipid composition of cellular membranes. Several lipids, which are only present in small amounts under normal conditions, are synthesized rapidly and transiently in response to stress. They act as a lipid second messenger and can form docking sites that bind different proteins and thus provide spatial and transient signals needed to adequately respond to external stimuli (Meijer and Munnik, 2003; Wang, 2004; Munnik and Testerink, 2009; Xue et al., 2009; Munnik and Vermeer, 2010). The phospholipid phosphatidic acid (PA) is one of these signaling lipids, which accumulates rapidly in response to different environmental signals (Testerink and Munnik, 2005; Li et al., 2009; Galvan-Ampudia and Testerink, 2011).
The Involvement and Generation of PA during Abiotic Stress and Developement
Induced PA formation has been described as a response of plants to abiotic stress stimuli such as dehydration (Jacob et al., 1999), salt and osmotic stress (Munnik et al., 2000) treatment with the plant hormone abscisic acid (ABA; Fan et al., 1997; Ritchie and Gilroy, 1998; Jacob et al., 1999) and also accumulates in response to biotic stress stimuli (van der Luit et al., 2000; de Torres Zabela et al., 2002; de Jong et al., 2004).
Lipidology and PA-Generating Enzymes
Different PA metabolizing pathways have been shown to contribute to PA production in response to abiotic stress (Munnik et al., 2000; Ruelland et al., 2002; Arisz et al., 2009; Bargmann et al., 2009; Li et al., 2009; Hong et al., 2010). The phospholipase D (PLD) enzyme hydrolyses primarily structural lipids such as phosphatidylcholine (PC), and phosphatidylethanolamine (PE), resulting in formation of PA and the remaining headgroup (Pappan et al., 1998). Phospholipase C (PLC) hydrolyses phosphatidylinositol lipids (PPIs) into water soluble inositol-bis or trisphosphate (IP2, IP3) and diacylglycerol (DAG), which remains in the membrane (Munnik and Vermeer, 2010). DAG can be subsequently phosphorylated to PA by DAG kinase (DGK; Arisz et al., 2009).
Twelve PLDs have been identified in the model plant species Arabidopsis, which were initially classified in two groups based on their N-terminal lipid-binding domain. These domains consist either of a pleckstrin homology (PH) and PHOX (PX) or a calcium dependent-lipid binding (C2) domain (Elias et al., 2002). Later, these classes were further subdivided into six classes based on sequence homology and in vitro enzymatic activity: three α-, two β-, three γ-, one δ-, and one ε- PLD with a C2 domain and two ζ-class PLDs that contain PH and PX domains (Qin and Wang, 2002; Bargmann and Munnik, 2006; Li et al., 2009).
In the Arabidopsis genome, nine PLCs and seven DGK genes have been identified. Initially, PLC/DGK derived PA was primarily implicated in responses to biotic stress (van der Luit et al., 2000; de Jong et al., 2004). However, abiotic stress, in particular cold stress, also induced an accumulation of PLC/DGK-mediated PA formation (Ruelland et al., 2002; Gomez-Merino et al., 2004). The DGKs involved in PA formation during cold stress have not been identified yet. T-DNA insertion lines of the seven DGKs did not alter PA formation in response to cold, which is likely due to redundancy (Arisz et al., 2013) and therefore the role of DGK-derived PA in response to abiotic stress remains largely unknown.
Genetic Evidence for PLD Requirement during Water Stress and Related Responses of Roots
Roots are the primary site of perception of salt stress, drought and low nutrient availability. To cope with these conditions, plants adapt the growth and morphology of their roots. Several PLD isoforms were found to be involved in adjusting root system architecture during abiotic stress (Galvan-Ampudia and Testerink, 2011; Figure 1A).
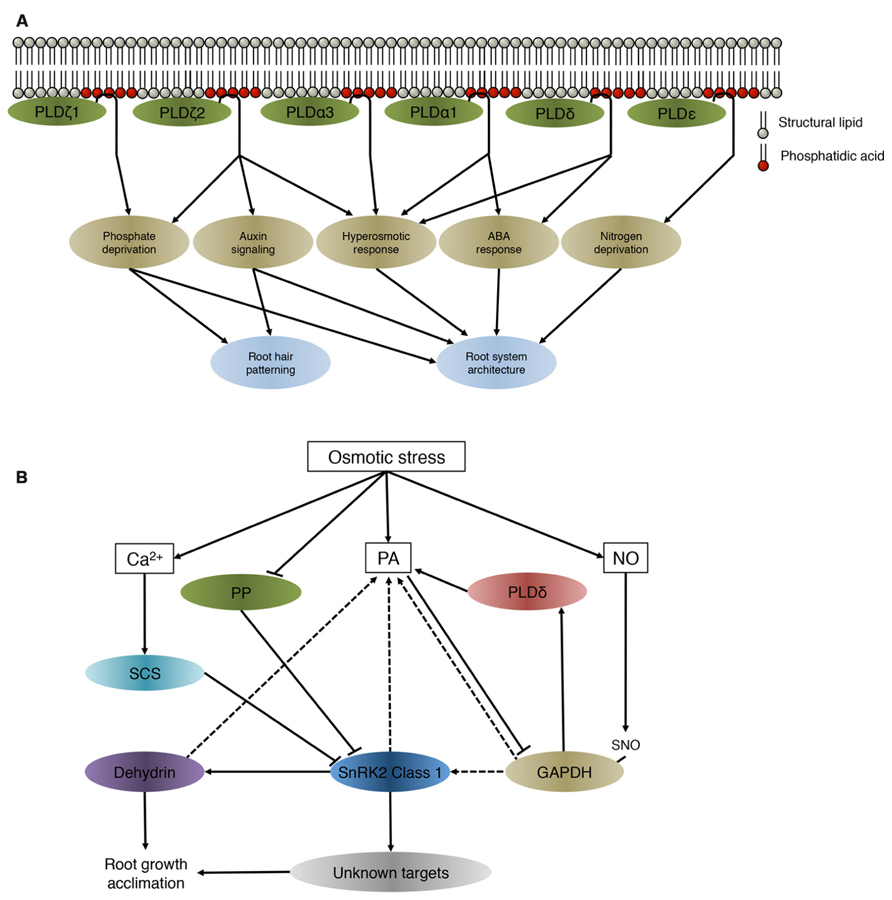
FIGURE 1. (A) PA derived from different PLDs involved in the maintenance of root system architecture during abiotic stress. PLDs regulate downstream targets through producing PA. Although all PLD isoforms hydrolyse structural lipids and generate PA in vitro, they have been identified to be involved in different processes and signaling cascades in vivo. (B) Preliminary network of osmotic stress-induced PA-SnRK2 signaling cascades in roots. This model is based on data obtained on the class 1 SnRK2 members in different plant species. SnRK2, sucrose non-fermenting 1-related protein kinase 2; SCS, SnRK2-interacting calcium sensor; PA, phosphatidic acid; NO, nitric oxide; SNO, S-nitrosylated; PP, protein phosphatase; GAPDH, glyceraldehyde-3-phosphate; PLDδ, phospholipase D δ. Solid lines indicate an activation or inhibitory effect, dashed lines show which proteins/lipids interact without a direct change in activity.
PLDζ2 is involved in directional root growth in saline conditions. Exposing one side of the root to salt increased pin-formed (PIN) 2 internalization, effectively redistributing auxin in the root tip. This redistribution resulted in bending away from saline conditions, named halotropism. A pldζ2-KO (knock-out) mutant showed reduced clathrin-dependent PIN2 internalization and reduced primary root bending (Galvan-Ampudia et al., 2013). Expression of PLDζ2 increased under low phosphate availability (Oropeza-Aburto et al., 2011) and the pldζ2-KO showed increased root hair growth when deprived of phosphate (Cruz-Ramirez et al., 2006). In accordance, less PA was formed in low phosphate conditions in the pldζ1/2 double mutant (Li et al., 2006a) and this mutant showed reduced lateral root and increased primary root growth in low phosphate conditions (Li et al., 2006b). In addition, the pldζ2-KO mutant also exhibited decreased sensitivity to auxin and a reduced root gravitropic response (Li and Xue, 2007).
Nitrogen is another important nutrient for plants and Arabidopsis PLDε-OE lines displayed an increase in lateral root and root hair elongation and primary root growth in low nitrogen conditions. This effectively increased the dry weight of the plant under these conditions and indicated an important role for PLDε in growth and nitrogen signaling (Hong et al., 2009).
PLDα1 and PLDδ are involved in different responses to abiotic stress including reactive oxygen species (ROS) signaling in response to ABA (Sang et al., 2001; Zhang et al., 2003; Zhang et al., 2009; Uraji et al., 2012) studied in stomata and leaves. The same phospholipases were also shown to play distinct roles in freezing tolerance (Welti et al., 2002; Li et al., 2004). Expression of PLDδ was elevated in response to dehydration and high salt stress (Katagiri et al., 2001). Salt stress induced formation of PA through PLDα1 and PLDδ, where both single mutants showed a reduction in primary root growth in saline conditions and during dehydration. This was even clearer in the pldα1/δ double mutant (Bargmann et al., 2009). A similar reduction in root growth was observed in pldα3, which was more susceptible to salinity and water deficiency. In hyperosmotic-stress conditions, the pldα3 mutant displays a reduction in primary root growth and a reduction in lateral roots (Hong et al., 2008).
Together, these studies show that PLDs are important for maintaining root growth in saline and hyperosmotic stress conditions amongst other functions. PA is not limited to its function as a signaling lipid; it is also an important intermediate during lipid-turnover. Therefore it is hard to separate the role of PA in lipid-turnover from its role in signaling and protein recruitment (Testerink and Munnik, 2011). To discern between the different roles of PA it is important to identify which proteins interact with PA and how these mediate the response that eventually leads to the acclimation to different stresses (Testerink and Munnik, 2005). In contrast to other signaling lipids such as phosphoinositides, no consensus PA-binding domain has been identified, which hampers the identification of new PA targets. A number of PA binding proteins have been identified involved in diverse cellular processes (Table 1).
PA Protein Targets Involved in Osmotic/Salt Stress Signaling and Root System Architecture
Metabolism
A central mediator in metabolism, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), is targeted to PA in response to salt in Arabidopsis roots (McLoughlin et al., 2013). The best-described role of GAPDH is the conversion of glyceraldehyde-3-phosphate to D-glycerate 1,3-bisphosphate in the glycolytic breakdown of glucose. PA-binding does not alter the activity of GAPDH dramatically in vitro but adding PA to seedlings did induce proteolytic cleavage of glyceraldehyde-3-phosphate dehydrogenase C2 (GAPC2; Kim et al., 2013). Adding exogenous PA also reduced primary root growth, which was more severe when GAPDH was over-expressed whilst knock-out mutants showed less reduction in growth (Kim et al., 2013) indicating that the effect of PA on root growth was partially mediated by proteolytic degradation of GAPDH. Although GAPDH has been described to be involved in different non-metabolic processes, promoting its degradation might mediate energy conservation and arrest of root growth, which are immediate and relevant responses to any osmotic stress including saline conditions (Munns and Tester, 2008).
Cytoskeleton and Membrane Cellular Trafficking
Phosphatidic acid has also emerged as an important regulator of microtubules and actin organization and re-organization. Microtubule reorganization is crucial for plants to adapt to saline conditions (Wang et al., 2007). In the pldα1 mutant background, the microtubule disorganization was more severe in response to salt and could not be recovered after the removal of salt. This effectively resulted in the plant being more salt sensitive in a microtubule-associated proteins 65-1 (MAP65-1) dependent manner. PA directly interacted with MAP65-1 and promoted the interaction of MAP65-1 with microtubules, effectively promoting the polymerization of cortical microtubules (Zhang et al., 2012).
In addition to microtubules, PA levels were also shown to be important for the behavior of actin filaments through the regulation of actin capping proteins (Li et al., 2012). PA specifically interacts with an actin capping protein (CP; Huang et al., 2006), which prevents actin filament from annealing and elongating. PA-binding reduced the activity of CP, effectively promoting actin reorganization and promoting cytoskeleton dynamics which are required for adaptation to adverse conditions.
In mammalian cells, PA and PA-generating enzymes such as PLD and DGK have been implicated in various aspects of vesicle transport (Manifava et al., 2001; Corda et al., 2002; Jang et al., 2012), but so far, little evidence is present that suggest a similar role in plants. Recently, clathrin heavy chain and clathrin assembly units were shown to recruit to the membrane in Arabidopsis roots in response to salt and to bind to PA-beads (McLoughlin et al., 2013). This likely represents an important aspect of the molecular mechanism of salt-induced PIN2 internalization which controls directional root bending in saline conditions (Galvan-Ampudia et al., 2013).
Cellular Signaling and Development
The Arabidopsis phosphoinositide-dependent kinase 1 (PDK1) binds several phosphoinositides and PA through its PH domain (Deak et al., 1999). PA activates PDK1 and indirectly, its downstream target AGC2-1 (OXI1) (Anthony et al., 2004, 2006). This signaling cascade induces a respiratory burst required for the full activation of mitogen activated protein kinase 6 (MPK6; Rentel et al., 2004), which was also shown to bind PA (Yu et al., 2010). MPK6 knock-out plants showed reduced growth in the primary root in saline conditions and the activation of MPK6 in response to salt is abolished in the pldα1 mutant background. In addition, MPK6 can phosphorylate the Na+/H+ antiporter salt overly sensitive (SOS1) in vitro and might therefore play a direct role in sodium homeostasis in roots (Yu et al., 2010). Another phosphorylation target of PDK1, PINOID (PID), was shown to bind phosphoinositides and PA. PID is involved in the asymmetric distribution of PIN auxin transporters, which are key regulators of root development (Zegzouti et al., 2006). Additionally, PA was also shown to bind the subunit of protein phosphatase 2A (PP2A); roots curl in NPA 1 (RCN1; Testerink et al., 2004). Recruitment to the membrane increased the activity of PP2A (Gao et al., 2013). PP2A in turn dephosphorylates the auxin transporter PIN1, changing its polarity and effectively altering the auxin distribution in the root, which is necessary for normal root development (Gao et al., 2013).
In a proteomics screen to identify PA targets during salt stress in Arabidopsis roots, the potassium channel β subunit KAB1 was shown to recruit to the membrane in response to salt and bind to PA (McLoughlin et al., 2013). The Arabidopsis genome encodes a single potassium channel β subunit 1 (Tang et al., 1995), which, as a tetramer, associates with the transmembrane α subunits of KAT1 channels (Tang et al., 1996). KAT1 is inactivated rapidly and internalized in response to ABA in guard cells. The inactivation occurs more rapidly than internalization, which suggests an additional mechanism. PA-recruitment could play a role in the inactivation of KAT1 by competing for KAB1, but this remains to be investigated. Since AtKAT1 expression is prevalent in leaves rather than roots, KAB1 is speculated to bind not exclusively to KAT1 but also to a different α subunit, viz. AKT1, which is selectively expressed in Arabidopsis roots (Tang et al., 1996). Recruitment of KAB1 by PA opens the possibility that PA might function as a regulator of potassium homeostasis in different parts of the plant.
The sucrose non-fermenting 1-related protein kinase 2 proteins (SnRK2s), is a family of osmotic stress-activated protein kinases (Kulik et al., 2011). SnRK2.10, a member of the class 1 subfamily, which is activated by salt and water stress but not by ABA, was identified in a PA affinity screen (Testerink et al., 2004). More recently, both SnRK2.10 and its close homolog SnRK2.4 were shown to bind PA directly. Arabidopsis snrk2.4 and 2.10-KO mutants exhibited reduced primary root length and lateral root density in saline conditions, respectively (McLoughlin et al., 2012). SnRK2.4 was shown be involved in regulating the ROS levels in roots in response to cadmium, but remarkably snrk2.4-KO mutants displayed an increase in primary root length when exposed to cadmium (Kulik et al., 2012). Overexpression of the SnRK2.4 wheat ortholog (TaSnRK2.4) in Arabidopsis caused an increase in primary root growth and resulted in more drought tolerant plants. This was explained by stronger water retention ability in these plants compared to wild-type (Mao et al., 2010). Overexpression of SAPK4 (a rice class 1 SnRK2 ortholog) increased tolerance to salinity and oxidative stress (Diedhiou et al., 2008).
Interactors of Class 1 SnRK2 Kinases and the Role of PA Herein
Arabidopsis SnRK2.4 is rapidly and transiently activated in saline conditions and is targeted to punctate structures in epidermal and cortex cells in roots (McLoughlin et al., 2012). Activity of class 1 SnRK2 kinases is not directly regulated by PA (Testerink et al., 2007), therefore PA might spatially facilitate protein–protein interactions. Several proteins that interact or are regulated by class 1 SnRK2 kinases also bind to PA (Figure 1B).
SnRK2 class 3 kinases are activated by ABA through suppression of protein phosphatases of the PP2C family, including the known PA target ABI1 (Zhang et al., 2004; Yoshida et al., 2006; Umezawa et al., 2009; Vlad et al., 2009; Soon et al., 2012). Although class 1 SnRK2s also have auto-activating capacity, their phosphorylation mechanism has been shown to be different (Burza et al., 2006; Vlad et al., 2010), and it remains to be established if they are inactivated through PP2C phosphatases or other protein phosphatases (Kulik et al., 2011). Several minutes after their activation by salt, class 1 SnRK2 kinases are inactivated again in Arabidopsis roots (McLoughlin et al., 2012), possibly through an interaction with the SnRK2-interacting Calcium Sensor (SCS) which inhibits SnRK2 kinase activity (Bucholc et al., 2011), but no evidence for a role of PA herein has been reported.
Nitric oxide (NO) activates a SnRK2.4 ortholog in tobacco; NtOSAK, but no S-nitrosylation of NtOSAK was observed after NO treatment. GAPDH was identified as a molecular partner of NtOSAK and S-nitrosylation of GAPDH in a NO dependent manner occurred several minutes after applying NO. Although S-nitrosylation of GAPDH did not influence the activity of NtOSAK it is speculated that it might be important for the recognition of cellular partners or substrates (Wawer et al., 2010). In addition to SnRK2, oxidized GAPDH binds to PLDδ, which increases the activity of PLDδ, effectively inducing PA formation and forming a feedback loop (Guo et al., 2012).
So far, no in vivo phosphorylation targets have been identified for the class 1 SnRK2 members yet, but a preferential phosphorylation affinity peptide motif was identified for SnRK2.10, which is conserved in dehydrins (Vlad et al., 2008). Dehydrins are important for cold, salt, and drought stress and function in protecting macromolecules besides several other potential protective roles (Allagulova Ch et al., 2003) and have been implicated in maintaining root growth in saline conditions (Ruibal et al., 2012). Interestingly, they have affinity for different phospholipids, including PA (Koag et al., 2003; Koag et al., 2009; Eriksson et al., 2011). Although the phosphorylation event still has to be confirmed in vivo, this is a candidate class 1 SnRK2 target to control maintenance of root growth in saline conditions.
Summarizing, many of the proteins that interact with or are targeted by class1 SnRK2s have also been described to have PA binding affinity. While PA does not directly influence the activity of class 1 SnRK2 kinases, recruitment of SnRK2.4 to the membrane is speculated be necessary to facilitate interaction with its cellular partners or to phosphorylate target proteins that function near or on the membrane (Figure 1B). Alternatively, PA-binding could play an indirect role in the rapid inactivation of the SnRK2 protein kinases through compartmentalization, which was found to occur within minutes after activation by salinity in Arabidopsis roots (McLoughlin et al., 2012).
Conclusion and Outlook
In conclusion, PA is a central phospholipid intermediate which is rapidly and transiently formed during salt and osmotic stress. PA accumulation is used as an appropriate and general sensor to monitor the extracellular environment, which could explain the versatile role of this phospholipid. The specificity in which phospholipases transduce diverse signals remains largely unknown, although differences can be expected through abundance, localization, and substrate preference of the different phospholipases. Besides evidence that PA acts as a docking site for protein recruitment to the membrane, an increasing amount of papers indicate post-translational modifications, activation/inactivation or proteolytic cleavage upon binding either directly (solely in the presence of PA) or indirectly (orchestrated with different proteins), profoundly increasing the complexity of its role. The localization of the PLDs and the effect of PLD-KO and OE lines on the functionality and localization of PA-targets have to be studied to verify their exact relation, which will ultimately result in the understanding of how PA-signaling mediates root proliferation in adverse conditions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Edgar E. Kooijman and Robert H. Vass for critically reading the manuscript. This work was supported by the Netherlands Organisation for Scientific Research (NWO), grants Vidi 700.56.429, ALW 820.02.017, ALW 846.11.002 and NGI Horizon project 93511011.
References
Allagulova, Ch. R., Gimalov, F. R., Shakirova, F. M., and Vakhitov, V. A. (2003). The plant dehydrins: structure and putative functions. Biochemistry (Mosc). 68, 945–951. doi: 10.1023/A:1026077825584
Anthony, R. G., Henriques, R., Helfer, A., Meszaros, T., Rios, G., Testerink, C., et al. (2004). A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 23, 572–581. doi: 10.1038/sj.emboj.7600068
Anthony, R. G., Khan, S., Costa, J., Pais, M. S., and Bogre, L. (2006). The Arabidopsis protein kinase PTI1-2 is activated by convergent phosphatidic acid and oxidative stress signaling pathways downstream of PDK1 and OXI1. J. Biol. Chem. 281, 37536–37546. doi: 10.1074/jbc.M607341200
Arisz, S. A., Testerink, C., and Munnik, T. (2009). Plant PA signaling via diacylglycerol kinase. Biochim. Biophys. Acta 1791, 869–875. doi: 10.1016/j.bbalip.2009.04.006
Arisz, S. A., Van Wijk, R., Roels, W., Zhu, J. K., Haring, M. A., and Munnik, T. (2013). Rapid phosphatidic acid accumulation in response to low temperature stress in Arabidopsis is generated through diacylglycerol kinase. Front. Plant Sci. 4:1. doi: 10.3389/fpls.2013.00001
Awai, K., Xu, C., Tamot, B., and Benning, C. (2006). A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc. Natl. Acad. Sci. U.S.A. 103, 10817–10822. doi: 10.1073/pnas.0602754103
Bargmann, B. O., Laxalt, A. M., Ter Riet, B., Testerink, C., Merquiol, E., Mosblech, A., et al. (2009). Reassessing the role of phospholipase D in the Arabidopsis wounding response. Plant Cell Environ. 32, 837–850. doi: 10.1111/j.1365-3040.2009.01962.x
Bargmann, B. O., and Munnik, T. (2006). The role of phospholipase D in plant stress responses. Curr. Opin. Plant Biol. 9, 515–522. doi: 10.1016/j.pbi.2006.07.011
Bucholc, M., Ciesielski, A., Goch, G., Anielska-Mazur, A., Kulik, A., Krzywinska, E., et al. (2011). SNF1-related protein kinases 2 are negatively regulated by a plant-specific calcium sensor. J. Biol. Chem. 286, 3429–3441. doi: 10.1074/jbc.M110.115535
Burza, A. M., Pekala, I., Sikora, J., Siedlecki, P., Malagocki, P., Bucholc, M., et al. (2006). Nicotiana tabacum osmotic stress-activated kinase is regulated by phosphorylation on Ser-154 and Ser-158 in the kinase activation loop. J. Biol. Chem. 281, 34299–34311. doi: 10.1074/jbc.M601977200
Camoni, L., Di Lucente, C., Pallucca, R., Visconti, S., and Aducci, P. (2012). Binding of phosphatidic acid to 14-3-3 proteins hampers their ability to activate the plant plasma membrane H+-ATPase. IUBMB Life 64, 710–716. doi: 10.1002/iub.1058
Corda, D., Hidalgo Carcedo, C., Bonazzi, M., Luini, A., and Spano, S. (2002). Molecular aspects of membrane fission in the secretory pathway. Cell. Mol. Life Sci. 59, 1819–1832. doi: 10.1007/PL00012508
Cruz-Ramirez, A., Oropeza-Aburto, A., Razo-Hernandez, F., Ramirez-Chavez, E., and Herrera-Estrella, L. (2006). Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 103, 6765–6770. doi: 10.1073/pnas. 0600863103
Deak, M., Casamayor, A., Currie, R. A., Downes, C. P., and Alessi, D. R. (1999). Characterisation of a plant 3-phosphoinositide-dependent protein kinase-1 homologue which contains a pleckstrin homology domain. FEBS Lett. 451, 220–226. doi: 10.1016/S0014-5793(99)00556-6
de Jong, C. F., Laxalt, A. M., Bargmann, B. O., De Wit, P. J., Joosten, M. H., and Munnik, T. (2004). Phosphatidic acid accumulation is an early response in the Cf-4/Avr4 interaction. Plant J. 39, 1–12. doi: 10.1111/j.1365-313X.2004.02110.x
de Torres Zabela, M., Fernandez-Delmond, I., Niittyla, T., Sanchez, P., and Grant, M. (2002). Differential expression of genes encoding Arabidopsis phospholipases after challenge with virulent or avirulent Pseudomonas isolates. Mol. Plant Microbe. Interact. 15, 808–816. doi: 10.1094/MPMI.2002.15.8.808
Diedhiou, C. J., Popova, O. V., Dietz, K. J., and Golldack, D. (2008). The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 8:49. doi: 10.1186/1471-2229-8-49
Dubots, E., Audry, M., Yamaryo, Y., Bastien, O., Ohta, H., Breton, C., et al. (2010). Activation of the chloroplast monogalactosyldiacylglycerol synthase MGD1 by phosphatidic acid and phosphatidylglycerol. J. Biol. Chem. 285, 6003–6011. doi: 10.1074/jbc.M109.071928
Elias, M., Potocky, M., Cvrckova, F., and Zarsky, V. (2002). Molecular diversity of phospholipase D in angiosperms. BMC Genomics 3:2. doi: 10.1186/1471-2164-3-2
Eriksson, S. K., Kutzer, M., Procek, J., Grobner, G., and Harryson, P. (2011). Tunable membrane binding of the intrinsically disordered dehydrin lti30, a cold-induced plant stress protein. Plant Cell 23, 2391–2404. doi: 10.1105/tpc.111.085183
Fan, L., Zheng, S., and Wang, X. (1997). Antisense suppression of phospholipase D alpha retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell 9, 2183–2196. doi: 10.1105/tpc.9.12.2183
Galvan-Ampudia, C. S., Julkowska, M. M., Darwish, E., Gandullo, J., Korver, R. A., Brunoud, G., et al. (2013). Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol. 23, 2044–2050. doi: 10.1016/j.cub.2013.08.042
Galvan-Ampudia, C. S., and Testerink, C. (2011). Salt stress signals shape the plant root. Curr. Opin. Plant Biol. 14, 296–302. doi: 10.1016/j.pbi.2011.03.019
Gao, H. B., Chu, Y. J., and Xue, H. W. (2013). Phosphatidic Acid (PA) binds PP2AA1 to regulate PP2A activity and PIN1 polar localization. Mol. Plant 6, 1692–1702. doi: 10.1093/mp/sst076
Gomez-Merino, F. C., Brearley, C. A., Ornatowska, M., Abdel-Haliem, M. E., Zanor, M. I., and Mueller-Roeber, B. (2004). AtDGK2, a novel diacylglycerol kinase from Arabidopsis thaliana, phosphorylates 1-stearoyl-2-arachidonoyl-sn-glycerol and 1,2-dioleoyl-sn-glycerol and exhibits cold-inducible gene expression. J. Biol. Chem. 279, 8230–8241. doi: 10.1074/jbc.M312187200
Guo, L., Devaiah, S. P., Narasimhan, R., Pan, X., Zhang, Y., Zhang, W., et al. (2012). Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Ddelta to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24, 2200–2212. doi: 10.1105/tpc.111.094946
Guo, L., Mishra, G., Taylor, K., and Wang, X. (2011). Phosphatidic acid binds and stimulates Arabidopsis sphingosine kinases. J. Biol. Chem. 286, 13336–13345. doi: 10.1074/jbc.M110.190892
Hong, Y., Devaiah, S. P., Bahn, S. C., Thamasandra, B. N., Li, M., Welti, R., et al. (2009). Phospholipase D epsilon and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J. 58, 376–387. doi: 10.1111/j.1365-313X.2009.03788.x
Hong, Y., Pan, X., Welti, R., and Wang, X. (2008). Phospholipase Dalpha3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20, 803–816. doi: 10.1105/tpc.107.056390
Hong, Y., Zhang, W., and Wang, X. (2010). Phospholipase D and phosphatidic acid signalling in plant response to drought and salinity. Plant Cell Environ. 33, 627–635. doi: 10.1111/j.1365-3040.2009.02087.x
Huang, S., Gao, L., Blanchoin, L., and Staiger, C. J. (2006). Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol. Biol. Cell 17, 1946–1958. doi: 10.1091/mbc.E05-09-0840
Jacob, T., Ritchie, S., Assmann, S. M., and Gilroy, S. (1999). Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. U.S.A. 96, 12192–12197. doi: 10.1073/pnas.96.21.12192
Jang, J. H., Lee, C. S., Hwang, D., and Ryu, S. H. (2012). Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners. Prog. Lipid Res. 51, 71–81. doi: 10.1016/j.plipres.2011.12.003
Jost, R., Berkowitz, O., Shaw, J., and Masle, J. (2009). Biochemical characterization of two wheat phosphoethanolamine N-methyltransferase isoforms with different sensitivities to inhibition by phosphatidic acid. J. Biol. Chem. 284, 31962–31971. doi: 10.1074/jbc.M109.022657
Katagiri, T., Takahashi, S., and Shinozaki, K. (2001). Involvement of a novel Arabidopsis phospholipase D, AtPLDdelta, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 26, 595–605. doi: 10.1046/j.1365-313x.2001.01060.x
Kieber, J. J., Rothenberg, M., Roman, G., Feldmann, K. A., and Ecker, J. R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441. doi: 10.1016/0092-8674(93)90119-B
Kim, S. C., Guo, L., and Wang, X. (2013). Phosphatidic acid binds to cytosolic glyceraldehyde-3-phosphate dehydrogenase and promotes its cleavage in Arabidopsis. J. Biol. Chem. 288, 11834–11844. doi: 10.1074/jbc.M112.427229
Klimecka, M., Szczegielniak, J., Godecka, L., Lewandowska-Gnatowska, E., Dobrowolska, G., and Muszynska, G. (2011). Regulation of wound-responsive calcium-dependent protein kinase from maize (ZmCPK11) by phosphatidic acid. Acta Biochim. Pol. 58, 589–595.
Koag, M. C., Fenton, R. D., Wilkens, S., and Close, T. J. (2003). The binding of maize DHN1 to lipid vesicles. Gain of structure and lipid specificity. Plant Physiol. 131, 309–316. doi: 10.1104/pp.011171
Koag, M. C., Wilkens, S., Fenton, R. D., Resnik, J., Vo, E., and Close, T. J. (2009). The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 150, 1503–1514. doi: 10.1104/pp.109.136697
Kulik, A., Anielska-Mazur, A., Bucholc, M., Koen, E., Szymanska, K., Zmienko, A., et al. (2012). SNF1-related protein kinases type 2 are involved in plant responses to cadmium stress. Plant Physiol. 160, 868–883. doi: 10.1104/pp.112.194472
Kulik, A., Wawer, I., Krzywinska, E., Bucholc, M., and Dobrowolska, G. (2011). SnRK2 protein kinases – key regulators of plant response to abiotic stresses. OMICS 15, 859–872. doi: 10.1089/omi.2011.0091
Li, G., and Xue, H. W. (2007). Arabidopsis PLDzeta2 regulates vesicle trafficking and is required for auxin response. Plant Cell 19, 281–295. doi: 10.1105/tpc.106.041426
Li, J., Henty-Ridilla, J. L., Huang, S., Wang, X., Blanchoin, L., and Staiger, C. J. (2012). Capping protein modulates the dynamic behavior of actin filaments in response to phosphatidic acid in Arabidopsis. Plant Cell 24, 3742–3754. doi: 10.1105/tpc.112.103945
Li, M., Hong, Y., and Wang, X. (2009). Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochim. Biophys. Acta 1791, 927–935. doi: 10.1016/j.bbalip.2009.02.017
Li, M., Qin, C., Welti, R., and Wang, X. (2006a). Double knockouts of phospholipases Dzeta1 and Dzeta2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol. 140, 761–770. doi: 10.1104/pp.105.070995
Li, M., Welti, R., and Wang, X. (2006b). Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases D zeta1 and D zeta2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 142, 750–761. doi: 10.1104/pp.106.085647
Li, W., Li, M., Zhang, W., Welti, R., and Wang, X. (2004). The plasma membrane-bound phospholipase D delta enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 22, 427–433. doi: 10.1038/nbt949
Ma, L., Zhang, H., Sun, L., Jiao, Y., Zhang, G., Miao, C., et al. (2012). NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na(+)/K(+)homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 63, 305–317. doi: 10.1093/jxb/err280
Manifava, M., Thuring, J. W., Lim, Z. Y., Packman, L., Holmes, A. B., and Ktistakis, N. T. (2001). Differential binding of traffic-related proteins to phosphatidic acid- or phosphatidylinositol (4,5)- bisphosphate-coupled affinity reagents. J. Biol. Chem. 276, 8987–8994. doi: 10.1074/jbc.M010308200
Mao, X., Zhang, H., Tian, S., Chang, X., and Jing, R. (2010). TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J. Exp. Bot. 61, 683–696. doi: 10.1093/jxb/erp331
McLoughlin, F., Galvan-Ampudia, C. S., Julkowska, M. M., Caarls, L., Van Der Does, D., Lauriere, C., et al. (2012). The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J. 72, 436–449. doi: 10.1111/j.1365-313X.2012.05089.x
McLoughlin, F., Arisz, S. A., Dekker, H. L., Kramer, G., De Koster, C. G., Haring, M. A., et al. (2013). Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem. J. 450, 573–581. doi: 10.1042/BJ20121639
Meijer, H. J., and Munnik, T. (2003). Phospholipid-based signaling in plants. Annu. Rev. Plant Biol. 54, 265–306. doi: 10.1146/annurev.arplant.54.031902.134748
Min, M. K., Kim, S. J., Miao, Y., Shin, J., Jiang, L., and Hwang, I. (2007). Overexpression of Arabidopsis AGD7 causes relocation of Golgi-localized proteins to the endoplasmic reticulum and inhibits protein trafficking in plant cells. Plant Physiol. 143, 1601–1614. doi: 10.1104/pp.106.095091
Monreal, J. A., McLoughlin, F., Echevarria, C., Garcia-Maurino, S., and Testerink, C. (2010). Phosphoenolpyruvate carboxylase from C4 leaves is selectively targeted for inhibition by anionic phospholipids. Plant Physiol. 152, 634–638. doi: 10.1104/pp.109.150326
Munnik, T., Meijer, H. J., Ter Riet, B., Hirt, H., Frank, W., Bartels, D., et al. (2000). Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 22, 147–154. doi: 10.1046/j.1365-313x.2000.00725.x
Munnik, T., and Testerink, C. (2009). Plant phospholipid signaling: “in a nutshell”. J. Lipid Res. 50(Suppl.), S260–S265. doi: 10.1194/jlr.R800098-JLR200
Munnik, T., and Vermeer, J. E. (2010). Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ. 33, 655–669. doi: 10.1111/j.1365-3040.2009.02097.x
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Oropeza-Aburto, A., Cruz-Ramirez, A., Acevedo-Hernandez, G. J., Perez-Torres, C. A., Caballero-Perez, J., and Herrera-Estrella, L. (2011). Functional analysis of the Arabidopsis PLDZ2 promoter reveals an evolutionarily conserved low-Pi-responsive transcriptional enhancer element. J. Exp. Bot. 63, 2189–2202. doi: 10.1093/jxb/err446
Pappan, K., Austin-Brown, S., Chapman, K. D., and Wang, X. (1998). Substrate selectivities and lipid modulation of plant phospholipase D alpha, -beta, and -gamma. Arch. Biochem. Biophys. 353, 131–140. doi: 10.1006/abbi.1998.0640
Petersen, J., Eriksson, S. K., Harryson, P., Pierog, S., Colby, T., Bartels, D., et al. (2012). The lysine-rich motif of intrinsically disordered stress protein CDeT11-24 from Craterostigma plantagineum is responsible for phosphatidic acid binding and protection of enzymes from damaging effects caused by desiccation. J. Exp. Bot. 63, 4919–4929. doi: 10.1093/jxb/ers173
Pleskot, R., Pejchar, P., Zarsky, V., Staiger, C. J., and Potocky, M. (2012). Structural insights into the inhibition of actin-capping protein by interactions with phosphatidic acid and phosphatidylinositol (4,5)-bisphosphate. PLoS Comput. Biol. 8:e1002765. doi: 10.1371/journal.pcbi.1002765
Pribat, A., Sormani, R., Rousseau-Gueutin, M., Julkowska, M. M., Testerink, C., Joubes, J., et al. (2012). A novel class of PTEN protein in Arabidopsis displays unusual phosphoinositide phosphatase activity and efficiently binds phosphatidic acid. Biochem. J. 441, 161–171. doi: 10.1042/BJ20110776
Qin, C., and Wang, X. (2002). The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLD zeta 1 with distinct regulatory domains. Plant Physiol. 128, 1057–1068. doi: 10.1104/pp.010928
Rentel, M. C., Lecourieux, D., Ouaked, F., Usher, S. L., Petersen, L., Okamoto, H., et al. (2004). OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature 427, 858–861. doi: 10.1038/nature02353
Ritchie, S., and Gilroy, S. (1998). Abscisic acid signal transduction in the barley aleurone is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. U.S.A. 95, 2697–2702. doi: 10.1073/pnas.95.5.2697
Ruelland, E., Cantrel, C., Gawer, M., Kader, J. C., and Zachowski, A. (2002). Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol. 130, 999–1007. doi: 10.1104/pp.006080
Ruibal, C., Salamo, I. P., Carballo, V., Castro, A., Bentancor, M., Borsani, O., et al. (2012). Differential contribution of individual dehydrin genes from Physcomitrella patens to salt and osmotic stress tolerance. Plant Sci. 190, 89–102. doi: 10.1016/j.plantsci.2012.03.009
Sang, Y., Cui, D., and Wang, X. (2001). Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol. 126, 1449–1458. doi: 10.1104/pp.126.4.1449
Soon, F. F., Ng, L. M., Zhou, X. E., West, G. M., Kovach, A., Tan, M. H., et al. (2012). Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 335, 85–88. doi: 10.1126/science.1215106
Tang, H., Vasconcelos, A. C., and Berkowitz, G. A. (1995). Evidence that plant K+ channel proteins have two different types of subunits. Plant Physiol. 109, 327–330. doi: 10.1104/pp.109.1.327
Tang, H., Vasconcelos, A. C., and Berkowitz, G. A. (1996). Physical association of KAB1 with plant K+ channel alpha subunits. Plant Cell 8, 1545–1553. doi: 10.1105/tpc.8.9.1545
Testerink, C., Dekker, H. L., Lim, Z. Y., Johns, M. K., Holmes, A. B., Koster, C. G., et al. (2004). Isolation and identification of phosphatidic acid targets from plants. Plant J. 39, 527–536. doi: 10.1111/j.1365-313X.2004.02152.x
Testerink, C., Larsen, P. B., Van Der Does, D., Van Himbergen, J. A., and Munnik, T. (2007). Phosphatidic acid binds to and inhibits the activity of Arabidopsis CTR1. J. Exp. Bot. 58, 3905–3914. doi: 10.1093/jxb/erm243
Testerink, C., and Munnik, T. (2005). Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 10, 368–375. doi: 10.1016/j.tplants.2005.06.002
Testerink, C., and Munnik, T. (2011). Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 62, 2349–2361. doi: 10.1093/jxb/err079
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
Uraji, M., Katagiri, T., Okuma, E., Ye, W., Hossain, M. A., Masuda, C., et al. (2012). Cooperative function of PLDdelta and PLDalpha1 in ABA-induced stomatal closure in Arabidopsis. Plant Physiol. 159, 450–460. doi: 10.1104/pp.112.195578
van der Luit, A. H., Piatti, T., Van Doorn, A., Musgrave, A., Felix, G., Boller, T., et al. (2000). Elicitation of suspension-cultured tomato cells triggers the formation of phosphatidic acid and diacylglycerol pyrophosphate. Plant Physiol. 123, 1507–1516. doi: 10.1104/pp.123.4.1507
Vlad, F., Droillard, M. J., Valot, B., Khafif, M., Rodrigues, A., Brault, M., et al. (2010). Phospho-site mapping, genetic and in planta activation studies reveal key aspects of the different phosphorylation mechanisms involved in activation of SnRK2s. Plant J. 63, 778–790. doi: 10.1111/j.1365-313X.2010.04281.x
Vlad, F., Rubio, S., Rodrigues, A., Sirichandra, C., Belin, C., Robert, N., et al. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21, 3170–3184. doi: 10.1105/tpc.109.069179
Vlad, F., Turk, B. E., Peynot, P., Leung, J., and Merlot, S. (2008). A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 55, 104–117. doi: 10.1111/j.1365-313X.2008.03488.x
Wang, C., Li, J., and Yuan, M. (2007). Salt tolerance requires cortical microtubule reorganization in Arabidopsis. Plant Cell Physiol. 48, 1534–1547. doi: 10.1093/pcp/pcm123
Wang, X. (2004). Lipid signaling. Curr. Opin. Plant Biol. 7, 329–336. doi: 10.1016/j.pbi.2004.03.012
Wang, Z., Anderson, N. S., and Benning, C. (2013). The phosphatidic acid binding site of the Arabidopsis trigalactosyldiacylglycerol 4 (TGD4) protein required for lipid import into chloroplasts. J. Biol. Chem. 288, 4763–4771. doi: 10.1074/jbc.M112.438986
Wang, Z., Xu, C., and Benning, C. (2012). TGD4 involved in ER-to-chloroplast lipid trafficking is a phosphatidic acid binding protein. Plant J. 70, 614–623. doi: 10.1111/j.1365-313X.2012.04900.x
Wawer, I., Bucholc, M., Astier, J., Anielska-Mazur, A., Dahan, J., Kulik, A., et al. (2010). Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem. J. 429, 73–83. doi: 10.1042/BJ20100492
Welti, R., Li, W., Li, M., Sang, Y., Biesiada, H., Zhou, H. E., et al. (2002). Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277, 31994–32002. doi: 10.1074/jbc.M205375200
Xue, H. W., Chen, X., and Mei, Y. (2009). Function and regulation of phospholipid signalling in plants. Biochem. J. 421, 145–156. doi: 10.1042/BJ20090300
Yoshida, R., Umezawa, T., Mizoguchi, T., Takahashi, S., Takahashi, F., and Shinozaki, K. (2006). The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 281, 5310–5318. doi: 10.1074/jbc.M509820200
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Zegzouti, H., Li, W., Lorenz, T. C., Xie, M., Payne, C. T., Smith, K., et al. (2006). Structural and functional insights into the regulation of Arabidopsis AGC VIIIa kinases. J. Biol. Chem. 281, 35520–35530. doi: 10.1074/jbc. M605167200
Zhang, Q., Lin, F., Mao, T., Nie, J., Yan, M., Yuan, M., et al. (2012). Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24, 4555–4576. doi: 10.1105/tpc.112. 104182
Zhang, W., Qin, C., Zhao, J., and Wang, X. (2004). Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 9508–9513. doi: 10.1073/pnas.0402112101
Zhang, W., Wang, C., Qin, C., Wood, T., Olafsdottir, G., Welti, R., et al. (2003). The oleate-stimulated phospholipase D, PLDdelta, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15, 2285–2295. doi: 10.1105/tpc.013961
Keywords: phosphatidic acid, Arabidopsis thaliana, roots, salt, drought, protein kinase
Citation: McLoughlin F and Testerink C (2013) Phosphatidic acid, a versatile water-stress signal in roots. Front. Plant Sci. 4:525. doi: 10.3389/fpls.2013.00525
Received: 07 August 2013; Paper pending published: 10 September 2013;
Accepted: 06 December 2013; Published online: 23 December 2013.
Edited by:
Yuriko Osakabe, RIKEN Plant Science Center, JapanReviewed by:
Omar Pantoja, Universidad Nacional Autonoma de Mexico, MexicoRoger Allen Leigh, University of Adelaide, Australia
Copyright © 2013 McLoughlin and Testerink. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christa Testerink, Section of Plant Physiology, Swammerdam Institute for Life Sciences, University of Amsterdam, Postbus 94215, 1090GE Amsterdam, Netherlands e-mail:Yy5zLnRlc3Rlcmlua0B1dmEubmw=
 Fionn McLoughlin
Fionn McLoughlin Christa Testerink
Christa Testerink