- 1Max Planck Institute of Molecular Plant Physiology, Golm, Germany
- 2Max-Delbrück-Centrum für Molekulare Medizin, Berlin-Buch, Germany
- 3Plant Systems Biology, University of Hohenheim, Stuttgart, Germany
The plasma membrane is an important organelle providing structure, signaling and transport as major biological functions. Being composed of lipids and proteins with different physicochemical properties, the biological functions of membranes depend on specific protein–protein and protein–lipid interactions. Interactions of proteins with their specific sterol and lipid environment were shown to be important factors for protein recruitment into sub-compartmental structures of the plasma membrane. System-wide implications of altered endogenous sterol levels for membrane functions in living cells were not studied in higher plant cells. In particular, little is known how alterations in membrane sterol composition affect protein and lipid organization and interaction within membranes. Here, we conducted a comparative analysis of the plasma membrane protein and lipid composition in Arabidopsis sterol-biosynthesis mutants smt1 and ugt80A2;B1. smt1 shows general alterations in sterol composition while ugt80A2;B1 is significantly impaired in sterol glycosylation. By systematically analyzing different cellular fractions and combining proteomic with lipidomic data we were able to reveal contrasting alterations in lipid–protein interactions in both mutants, with resulting differential changes in plasma membrane signaling status.
Introduction
The plasma membrane contains numerous lipid species with different physicochemical properties. The three major classes are glycerolipids, sphingolipids and sterols. Spontaneous phase separation was shown to occur in artificial membranes involving mainly lipids of higher hydrophobicity such as sphingolipids, sterols or long-chain phospholipids (Karnovsky et al., 1982; Thompson and Tillack, 1985). These experiments extended the fluid mosaic model of Singer and Nicolson (Singer and Nicolson, 1972) of protein–lipid organization by adding sub-compartmental occurrences of specific protein and lipid compositions in the plasma membrane in so-called microdomains. Since then, biological evidence for such membrane domains came from many organisms and different biological contexts (Simons and Toomre, 2000; Lucero and Robbins, 2004; Bhat and Panstruga, 2005; Rajendran and Simons, 2005; Lingwood et al., 2009; Cacas et al., 2012). With a reported size of microdomains ranging from nanoscale up to microscale (Edidin, 2001; Cacas et al., 2012), direct visualization of such domains in living cells remains challenging. Sterol-dependent protein localization was reported in various plant systems (Borner et al., 2005; Roche et al., 2008; Kierszniowska et al., 2009; Minami et al., 2009; Carmona-Salazar et al., 2011; Navarro-Lérida et al., 2012) and selected proteins with sterol-dependent membrane location were shown to exhibit a patchy organization within the plasma membrane (Bariola et al., 2004; Parton and Hancock, 2004). The biological function of these microdomains was especially linked to signaling and transport processes in several independent studies (Simons and Toomre, 2000; Borner et al., 2005; Langhorst et al., 2005; Kierszniowska et al., 2009; Staubach and Hanisch, 2011; Stuermer, 2011). The current working model of sterol-rich microdomains involves controlled dynamic association/dissociations of particular proteins with the distinct local sterol-lipid environment. Support for this model in plants came from stimulus-dependent membrane microdomain localization of the Arabidopsis flagellin receptor and the ion channel SLAH3 using a combination of proteomic and cell biology approaches (Keinath et al., 2010; Demir et al., 2013).
Large-scale proteomic analysis of microdomain-associated proteins in plants has so far been mainly based on the treatment of purified plasma membranes with non-ionic detergents. Fractionation of the plasma membrane after detergent treatment results in a high density detergent soluble fraction (DSF) that contains membranes and proteins solubilized by detergent treatment, and a sterol-enriched detergent-resistant membrane (DRM) fraction with associated proteins (Lingwood and Simons, 2007). Proteomic analysis of such DRMs elucidated a specific set of DRM-resident proteins especially involved in signaling or transport (Shahollari and Berghöfer, 2004; Kierszniowska et al., 2009). Furthermore, the sterol-dependence of these proteins was validated by using sterol-depleting agents such as methyl-β-cyclodextrin (mβcd) (Ilangumaran and Hoessli, 1998; Zidovetzki and Levitan, 2007; Kierszniowska et al., 2009). However, these approaches were criticized as being prone to artifacts due to in vitro modification of plasma membranes after disruption of the cells (Tanner et al., 2011).
System-wide studies of membrane composition in living systems with endogenously altered sterol levels are rare and have so far only focused on single-protein examples (Lauwers and André, 2006). Endogenous alteration of sterol levels can be achieved by nutritional manipulations in sterol auxotrophic species (Entchev and Kurzchalia, 2005). In sterol autotrophic species such as plants, sterol diets obviously have no major effects. Alternatively sterol synthesis can be manipulated applying sterol-depleting drugs (He et al., 2003; Benveniste, 2004; Schrick et al., 2004a) or sterol synthesis inhibitors (He et al., 2003; Benveniste, 2004) on living cells. Nevertheless, secondary side-effects of applying drug treatments to living cells are hard to control, especially when these drugs are poisonous to the cell. In contrast, mutants inhibited in various steps of sterol biosynthesis (Schrick et al., 2000, 2002, 2004a, 2012a; Souter et al., 2002) display altered sterol profiles without the need of drug treatment. Indeed, reported sterol profiles of sterol-biosynthesis mutants showed significant shifts in total sterol composition (Schrick et al., 2002, 2004a; Boutte et al., 2008). All of these sterol-biosynthesis mutants exhibit a strong dwarf phenotype and are sterile (Schrick et al., 2000, 2002, 2004a, 2012a; Clouse, 2002). Although this pleiotropic phenotype could partially result from alterations in sterol- (He et al., 2003) and brassinosteroid-signaling (Clouse and Sasse, 1998), strong perturbations of general plasma membrane structures and microdomain functions are also likely. Therefore, these sterol-biosynthesis mutants are ideal systems for an in-depth characterization of microdomain protein composition and plasma membrane signaling status in the context of an endogenously-altered membrane sterol-composition.
We used the mutant smt1 (Schrick et al., 2002, 2004a; Willemsen et al., 2003; Fujioka, 2010) for its reported qualitative and quantitative alterations in sterol levels. smt1 encodes one of three sterol-methyl-transferases in Arabidopsis (Fujioka, 2010). Even though smt1 mutants exhibit a typical dwarf like phenotype at the whole plant level, its phenotype is less visible on non-differentiated callus systems of smt1 (Supplemental Figure 2 and Schrick et al., 2002), which makes it an ideal system for the propagation of cell material on a large scale. In contrast, the ugt80A2;B1 (DeBolt et al., 2009) double knockout mutant lacks the function of the only two sterol-UDP-glycosyltransferases in Arabidopsis. This mutant is fertile, but was shown to lack sterolglycosides (DeBolt et al., 2009; Schrick et al., 2012b) with only small effects on embryo development, root length or general sterol composition (DeBolt et al., 2009). Therefore, ugt80A2;B1 is an optimal further candidate for studying loss of sterol-related function (glycosylation) on sterol–protein interactions in the plasma membrane. We performed a systematic comparative analysis between both mutants with a particular focus on how altered sterol composition affects cellular protein–lipid interactions. Thereby, the DRM/DSF abundance-ratio of proteins and lipids (Zauber et al., 2013) was shown to be a useful proxy of sterol–protein interactions in smt1. We used a systems biology-based approach involving label-free proteomics as well as lipidomics to identify disturbed protein–sterol interactions with implications on stress signaling in these mutants.
Results
The following work aims at a thorough assessment of the role of sterols for membrane protein and lipid composition. Based on mass spectrometric analysis of proteins and lipids in the sterol-biosynthesis mutants smt1 and ugt80A2;B1 we suggest significant roles of sterols in membrane-based signaling processes.
Detection of Sterol-Dependent Protein Candidates by DRM/DSF Distribution Analysis
To characterize the role of sterol composition for the localization of proteins within membrane microdomains, we studied the distribution of proteins between sterol-rich (detergent resistant membranes, DRM) and sterol depleted (detergent sensitive fraction, DSF) membrane fractions. From 3716 identified proteins, 1435 and 1610 could be analyzed based on the abundance ratio distribution between DRM/DSF in smt1 or ugt80A2;B1, respectively (Figure 1). In smt1 most proteins showed decreased abundance in DRM while a similar number of proteins were enriched or depleted relative to DRM fractions for ugt80A2;B1. In total, 149 and 146 proteins (classified as responsive proteins) were found with a differential DRM/DSF distribution ratio in smt1 and ugt80A2;B1, respectively, as compared to the wildtype (Figure 1). Since alterations in sterol composition or glycosylation is the primary result of the mutations, we particularly expected to observe altered sterol–protein interactions. Therefore, in the following sections all distribution changes between DRM/DSF ratios will be expressed relative to the DRM fraction, which in the wildtype is the sterol-enriched membrane fraction. By comparing abundances of proteins identified in DRM and DSF against abundances in fractions of soluble proteins (SP) and intracellular membranes (IM), co-purifying proteins were defined if their highest abundance was either in SP or IM. These co-purifying proteins were found to be specifically enriched in DRM fractions of smt1 as reported previously (Zauber et al., 2013). In contrast, occurrence of co-purifying proteins was equally distributed between DRM and DSF fractions in ugt80A2;B1. Most of the co-purifying proteins were of cytosolic location, but mitochondrial, vacuolar, plastidial, nuclear and endoplasmic reticulum proteins were also present, according to SUBA3 (Tanz et al., 2013) (Supplemental Figure 2). Among the co-purifying proteins, we identified also a number of plasma membrane located proteins which showed significantly higher abundance in IM and SP. These proteins were not included in later analysis of abundance DRM/DSF ratios in order to minimize effects related to altered membrane trafficking in the mutants (Zauber et al., 2013).
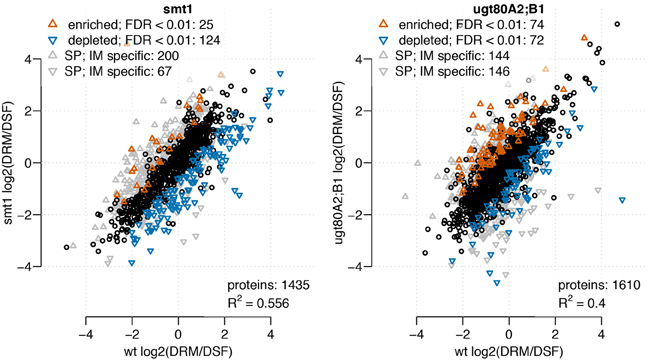
Figure 1. Scatterplot of log2 transformed DRM/DSF protein abundance ratios from smt1 (left) and ugt80A2;B1 (right), each in comparison to DRM/DSF distributions in wildtype. Proteins with Unicorn scores above the threshold defining a false discovery rate lower than 1% were considered to be significantly altered in their distribution between wildtype and the respective mutant. In smt1 more proteins exhibited a decreased DRM/DSF ratio compared to wildtype, while among enriched proteins the number of co-purifying proteins from intracellular membrane (IM) and soluble protein (SP) fractions was much higher. In ugt80A2;B1, similar numbers of proteins were found to be enriched (red) or decreased (blue) in their DRM/DSF abundance ratios.
In summary, 14 proteins showed decreased abundance in DRM in both mutants and 14 proteins were depleted in smt1 but enriched in ugt80A2;B1 (Figure 2A). Only a small fraction of proteins seems to be responsive in both mutants while the overlap based on hand-curated protein groups is much higher (Figure 2B). In this study, the largest two classes of overlapping proteins cover already 96 of all responding proteins and were represented in 9 protein groups and proteins of unknown functions. Within these overlapping protein groups, most proteins were strongly decreased in smt1 and enriched or depleted in ugt80A2;B1. Protein groups that were enriched in smt1 were only represented by one protein across all comparisons (Supplemental Table 1). However, the majority of all identified sterol-dependent proteins showed reduced abundance in DRM fractions in both smt1 and ugt80A2;B1 (Figure 2C). This included ATPases, protein kinases, leucine-rich repeat kinases (LRR kinases), GTP binding proteins, glycosyl-hydrolases, SNARE like family proteins, ABC-transporters, receptor-like kinases, SPFH protein family and “early responsive to dehydration” proteins (ERD). Cytochrome oxidases (Asard et al., 2001) with two proteins responding in both mutants were depleted and enriched in DRM of ugt80A2;B1. Additionally some co-purifying proteins were also found in this category, such as dicarboxylate transporter, glucose-6-phosphate/phosphate-translocator and an ADP-ribosylation-factor (Supplemental Table 1). However, these co-purifying groups were generally represented by one or two proteins only.
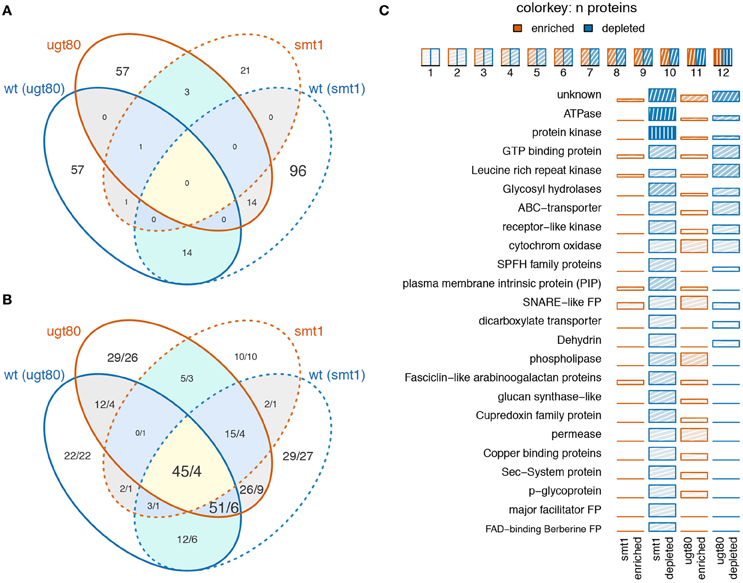
Figure 2. Venn diagram displaying the number of overlapping proteins (A) or protein groups (B) being enriched (red) or depleted (blue) in smt1 or ugt80A2;B1 (ugt80) DRM fractions. Numbers in (B) indicate the number of unique proteins represented in the overlapping protein groups, separated by a slash. Low overlap was observed between proteins (A) but analysis based on protein groups showed a higher degree of overlap (B). A quantitative analysis of significantly-altered protein abundances based on manually-curated protein groups is displayed in (C). Only protein groups represented by at least two proteins are shown. The number of proteins in protein groups is indicated by color intensity. wt, wildtype; ugt80, utg80A2;B1; FP, family protein.
The second big class of protein groups was generally depleted from DRM in smt1 but enriched in DRM in ugt80A2;B1. In total, 13 protein groups represented by 41 individual proteins showed this pattern. Examples with the highest number of unique proteins were the group of phospholipases, fasciclin like arabinogalactan proteins (FLA), plasma membrane intrinsic proteins (PIP) and cupredoxins (SKU; Figure 2C). Interestingly, the accepted plant microdomain marker remorin was depleted from the DRM fraction in smt1, but not in ugt80A2;B1 (Supplemental Table 1). In both mutants 12 protein groups showed a similar trend in their abundance distributions relative to in the DRM fraction. From these only one was enriched and 11 were strongly reduced in abundance (Table 1).
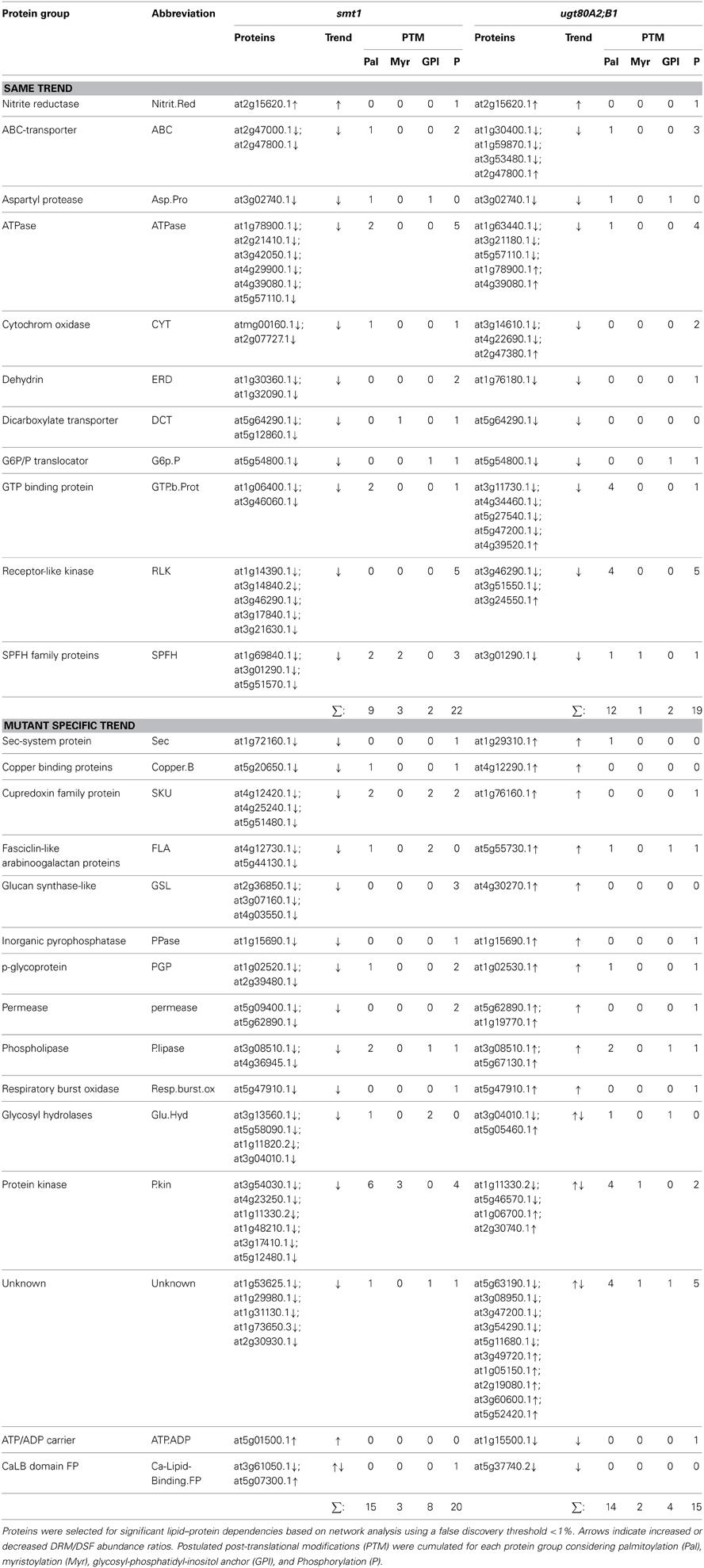
Table 1. Selected protein groups showing significant alterations in DRM/DSF protein abundance ratios in smt1 and ugt80A2;B1.
The last fraction included mainly protein groups that were previously shown to be sterol-dependent, such as ATPases, ABC-transporters, leucine-rich repeat kinases, dehydrin, receptor-like kinases, and SPFH proteins (Borner et al., 2005; Browman et al., 2007; Kierszniowska et al., 2009; Minami et al., 2009). Core protein groups being enriched in ugt80A2;B1 DRM fraction but depleted in smt1 included cupredoxins, fasciclin-like arabinogalactan proteins, plasma membrane intrinsic proteins, and lipid signaling relevant proteins with phospholipase C activity. In addition, proteins involved in membrane trafficking were differentially affected such as Sec and SNARE-like proteins.
The group “kinases” included a large number of signaling active proteins likely to alter the signaling status in both mutants. Association of the proteins with DRMs was not in general dependent on the presence of post-translational modification sites (Table 1). However, modification status could affect degree of lipid protein associations, as was suggested for SLAH3 (Demir et al., 2013).
Alteration in Lipid Profiles in Both Mutants
Changes in sterol composition are the primary effect of the mutations (Schrick et al., 2002, 2004a; DeBolt et al., 2009). Since we were interested in how this altered sterol composition and glycosylation status affected other type of lipids in the membranes we complemented the proteomic investigation by a lipidomic analysis. Both mutants exhibited an altered membrane lipid profile compared to wildtype (t-test, α = 0.05). Although there were only a few changes observed after multiple testing correction for single compounds, analysis at the class level show a contrasting lipid composition between both mutants (Figure 3B). This observation could be confirmed through a separate analysis of the sterol biosynthesis mutant cpi. cpi exhibits stronger alterations in its whole-plant sterol composition (Boutte et al., 2008) than smt1. Further, this mutant shows a similar but stronger dwarf-like phenotype as smt1 (Schrick et al., 2004a; Boutte et al., 2008). Concordantly the lipid profile of cpi shows a similar but even stronger global shift of lipid abundances as smt1 (Supplemental Figure 3). Affected plasma membrane lipid classes in smt1 and ugt80A2;B1 were ceramides (Cer), glycosylceramides (GlcCer), phosphatidylglycerol (PG; 36 acylcarbons), phosphatidylserines (PS; 40 acylcarbons), and phosphatidylcholines (PC; 36 acylcarbons) (Figure 3A; Supplemental Table 2). The largest relative change was evident for glycosylceramides (GlcCer), which showed a general increased abundance in smt1 and decreased abundance in ugt80A2;B1. Two GlcCer species with lower hydroxylation status showed interestingly a decreased abundance also in smt1.
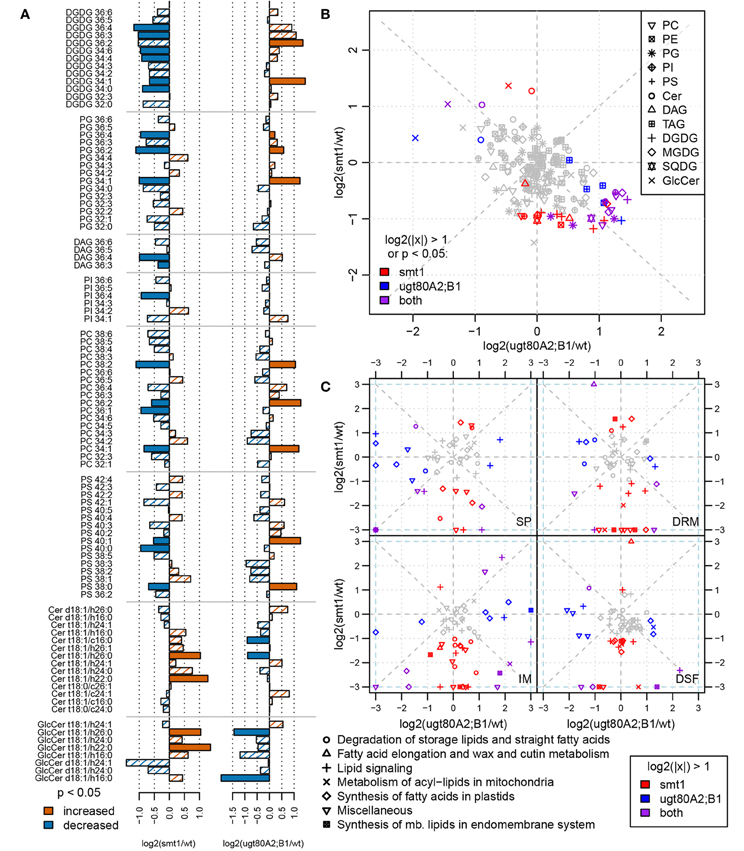
Figure 3. Comparison of log2 transformed lipid abundances from smt1 and ugt80A2;B1 related to wildtype (A). Lipids with a higher-than two-fold change or significant difference in abundance (t-test; α < 0.05) are colored as indicated in the legend. Lipid abundance changes of smt1 and ugt80A2;B1 relative to wildtype display a generally inverse pattern (B). Specific changes in proteins involved in lipid metabolism correlate with lipid distributions in respective fractions (IM, SP, DRM, DSF) (C). Isomer forms of detected ceramides and glycosylceramides are shown in Supplemental Table 2. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; Cer, ceramide; GlcCer, glycosylceramide; DAG, diacylglycerol; MGDG/DGDG, mono-/digalactosyldiacylglycerol; SQDG, sulfoquinovosyldiacylglycerol; TAG: triacylglycerol; SP, soluble proteins; IM, intracellular membranes; DRM, detergent resistant membranes; DSF, detergent soluble fractions.
Alterations in Lipid Synthesis-Related Proteins Correlate with Lipid Abundance Profiles
The observed differential lipid composition in smt1 and ugt80A2;B1 could be related to alterations in the abundance of proteins involved in lipid synthesis (Figure 3C). All four fractions SP, IM, DRM, and DSF were analyzed using two-fold changes as a significance-threshold. Large differences in the abundance of proteins involved in lipid synthesis were especially visible in the IM fraction. In this study, proteins involved in lipid degradation were strongly decreased in the smt1 IM fraction, while they showed a slight increase in ugt80A2;B1. Proteins involved in fatty acid synthesis in the plastid (Supplemental Table 3) were strongly decreased in smt1 and increased in the ugt80A2;B1 IM fraction. Interestingly, a decreased abundance was observed for acyl carrier proteins (ACP2, ACP3) in the IM fraction in both mutants. Similarly, proteins involved in the synthesis of lipids at the endoplasmic reticulum (ER) were affected, which could possibly explain the observed changes in membrane lipids. These proteins were in general decreased in the smt1 mutant, but increased in ugt80, such as putative cholinephosphate cytidylyltransferase and ethanolaminephosphate cytidylyltransferase, involved in head group synthesis; glycerol-3-phosphate dehydrogenase, involved in glycerol synthesis and involved in the synthesis of long-chain fatty acids (Beaudoin et al., 2009).
The majority of proteins involved in lipid signaling (hand curated categories, see Supplemental Table 3) shows a plasma membrane localization according to SUBA3 (Tanz et al., 2013). Therefore, alterations in abundance of these proteins should be viewed with regards to altered distributions between DRM and DSF fractions. A general decrease in the abundance of proteins involved in lipid signaling in the DRM as well as DSF could be observed for smt1. Based on DRM/DSM ratios, the phospholipases C2 and D γ 1 were depleted from smt1 DRM fractions but significantly enriched in ugt80A2;B1 DRM fractions. Interestingly, cell-wide DAG abundance in smt1 was significantly decreased indicating a reduced phospholipase C activity in smt1. Furthermore, SMT2 was found with an increased abundance in IM and DSF fractions of ugt80A2;B1 while it was not detected in smt1. SMT2 acts at the branching step of sterol synthesis which leads to sitosterol synthesis through isofucosterol (Fujioka, 2010). Isofucosterol levels were slightly but not significantly elevated in ugt80A2;B1 (DeBolt et al., 2009) which might be linked to the observed increased abundance of SMT2.
The Link Between Sterol-Dependent Proteins and Ceramides
The statistical significance of a co-occurrence between sterol-dependent proteins and specific lipid abundances was analyzed using a network based approach (Figures 4, 5). Pairwise scores between lipid abundances and sterol-dependence of proteins were calculated as a measure for lipid–protein correlation. These scores were based on log2 fold changes of lipid abundances in mutant compared to wildtype multiplied by the Unicorn scores (Zauber et al., 2013) of the identified sterol-dependent proteins. The significance of protein–lipid correlations displayed as network edges, was determined by applying an FDR threshold of 0.01%. This FDR-threshold was determined from score populations derived from randomized Unicorn-scores and randomized lipid abundance ratios. Based on this correlative relationship we propose lipid–protein interactions. Networks based on protein localizations (SUBA3; Tanz et al., 2013) showed that most protein abundances correlated significantly with abundances of glycosylceramides, which showed the highest degree among the lipid nodes in both networks (Figure 4). In smt1, proteins localized to the plasma membrane had a degree of 167 and were exclusively depleted (blue node frame) from DRM fractions. Similarly, in ugt80A2;B1 plasma membrane protein were mainly connected to glycosylceramides (GlcCer). Phospholipids and galactolipids showed a generally lower degree of connectivity and were with a few exceptions connected to plastid or plasma membrane proteins. In ugt80A2;B1, proteins showed positive as well as negative correlations to depletion of GlcCer at equal frequencies. In general, proteins assigned to vacuolar, extracellular or nuclear localizations showed an enrichment in ugt80A2;B1 while only plastidial and cytosolic proteins were enriched in smt1.
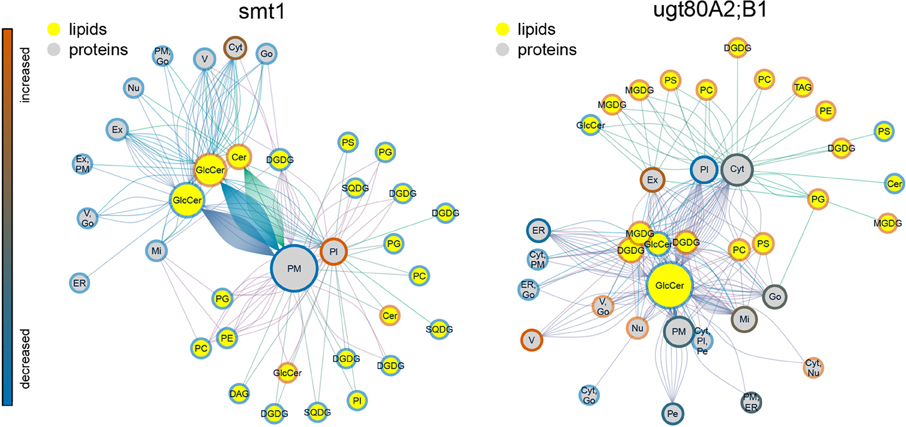
Figure 4. Lipid–protein correlation network displaying proteins according to subcellular localization in SUBA3 (Tanz et al., 2013). Edges were selected for significance using a false discovery threshold lower 1%. For both mutants glycosylceramide shows the highest degree indicated by node diameter. In particular an inverse relationship of increased (blue node frame) ceramides and plasma membrane proteins is visible for smt1, while in ugt80A2;B1 more sterol-dependent proteins correlated with decreased (red node frame) levels of glycosylceramides. In general, lipid–protein connections were grouped by their subcellular localization. Frame node color indicates decrease (blue) or increase (blue) in sterol dependency of proteins or abundance of lipids (yellow node color). Edge colors are linked to the different lipid nodes. Lipids: PC, Phosphatidylcholine; PG, Phosphatidylglycerol; PS, Phosphatidylserine; Cer, Ceramide; GlcCer, Glycosylceramide; MGDG/DGDG, Mono-/Digalactosyldiacylglycerol; SQDG, Sulfoquinovosyldiacylglycerol. Subcellular localizations: PM, plasma membrane; Pl, plastid; ER, endoplasmatic reticulum; V, vacuole; Go, Golgi apparatus; Ex, extracellular; Mi, mitochondrion; Pe, peroxysome; Nu, nucleus; Cyt, cytosol.
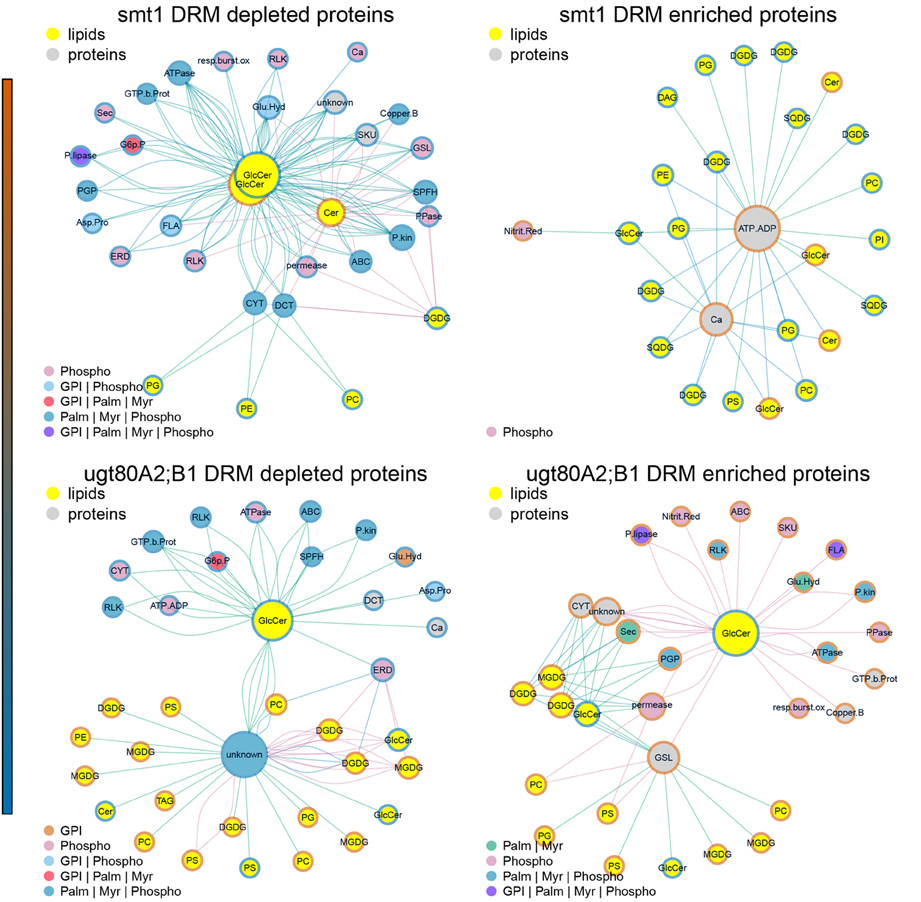
Figure 5. Subnetworks of lipid–protein interactions (Figure 4) filtered for edges between commonly identified protein groups of ugt80A2;B1 and smt1. Networks were further separated into proteins exhibiting an increased (red) or decreased (blue) DRM/DSF ratio. Abbreviations of protein groups are explained in Table 1 and Lipid species in Figure 4. Proteins could be divided into ceramide-dependent or non-dependent classes which show a high degree in edges with galactolipids or glycerolipids. From these networks a list of proteins with biologically-relevant alterations in protein abundances was obtained and is shown in Figure 1. Node color of proteins is according to postulated posttranslational modifications (PTM) relevant for protein sterol/lipid interactions. Abbreviations and color code are explained in Figure 4. GPI, Glycosyl-Phosphatidyl-Inositol; Palm, Palmitoylation; Myr, Myristoylation; Phospho, Phosphorylation.
Subnetworks were constructed from edges of commonly-identified protein groups in smt1 and ugt80A2;B1 (Figure 5). This analysis resulted in a core set of 27 protein groups represented by 63 proteins in smt1 and 59 different proteins in ugt80A2;B1. In smt1, 60 DRM-depleted proteins remained in the subnetwork and were mainly associated with the decrease of GlcCer and Cer and interestingly to one decreased GlcCer with a lower hydroxylation status. In contrast, only 3 DRM-enriched proteins remained in the smt1 network (Figure 5 left graph) including two co-purifying proteins (plastidial ATP/ADP translocator and nitrite reductase) and one calcium dependent copine like protein. Proteins in this network were mainly linked to phospho- and galactolipids in contrast to proteins depleted in smt1 DRM, which were almost exclusively linked to GlcCer and Cer. In ugt80A2;B1, proteins were almost evenly distributed between the subnetworks, with 31 depleted and 27 enriched proteins. These proteins could be divided into two classes: a larger class connected to one of the most depleted glycosylceramides and a smaller group mainly linked to enriched galacto- and phospholipids. Based on this network analysis and the observed lipid profiles, a working model for biologically relevant alterations in lipid and signaling pathways was constructed (Figure 6).
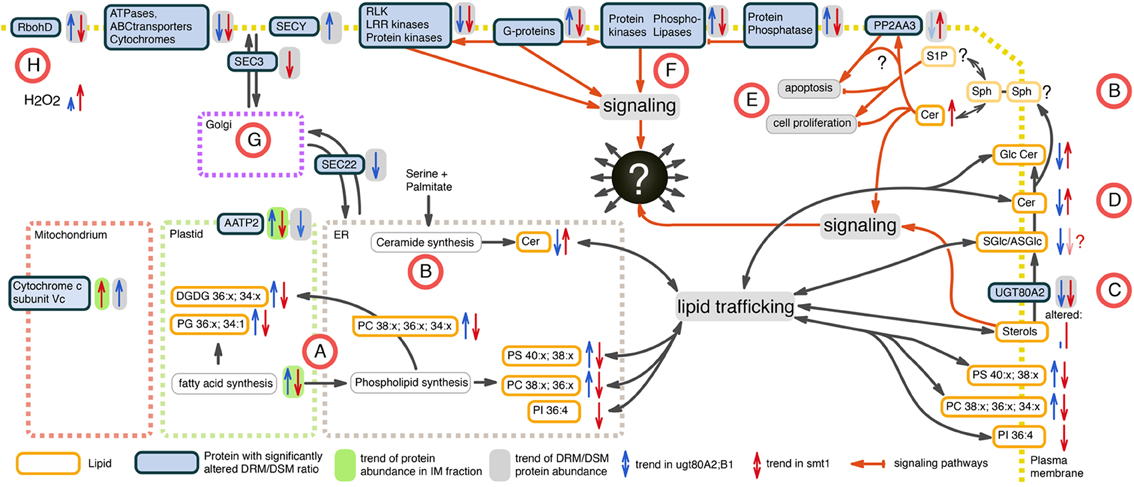
Figure 6. Working model of altered proteins and lipids in smt1 (blue) and ugt80A2;B1 (red) based on the set of identified core proteins from network analysis. (A) Fatty acid and glycerolipid synthesis is decreased in smt1 and increased in ugt80A2;B1, with significant alterations in abundances of distinct glycerolipids. (B) Alteration in ceramide (Cer) de novo synthesis was not observable. Additionally, alterations in activity of salvage synthesis pathways (Van Brocklyn and Williams, 2012) could explain changes in abundances of ceramides in the mutants. (C) Glycosylation of sterols (SGlc/ASGlc) might additionally happen in plasma membrane catalyzed by UGT80A2, which was depleted from DRM in both mutants. (D) GlcCer levels are inversely altered in both mutants implying an additional sterol-dependent pathway for ceramide glycosylation in the plasma membrane. (E) Alterations in Cer activated phosphatase PP2 (Nickels and Broach, 1996) observed in a DRM-dependent response of PP2 subunit PP2AA3 indicate an unbalance in activation of apoptosis and cell proliferation in both mutants. (F) Phospholipase signaling was sterol-dependent in a discordant manner in both mutants. (G) Vesicle trafficking is likely affected in both mutants. (H) RbohD shows discordant sterol affinity, but lead to increased ROS production in both mutants. Lipids: PC, phosphatidylcholine; PG, phosphatidylglycerol; PS, phosphatidylserine; PI, phosphatidylinositol; Cer, ceramide; GlcCer, glycosylceramide; DGDG, digalactosyldiacylglycerol; SGlc, sterolglycosides; ASGlc, acylated sterolglycosides; Sph, sphingosine.
In our study, fatty acid and glycerolipid synthesis appeared to be decreased in smt1 and increased in ugt80A2;B1 (Figure 6A). This is supported by the significant alterations in detected phospho- and galactolipids (Figure 3). We are not certain whether the de novo synthesis pathway of ceramide biosynthesis was affected as we could not identify most of the proteins involved in this process by the proteomic analysis. However, the alkaline phytoceramidase (CES1) showed a decreased DRM/DSM protein abundance ratio in ugt80A2;B1 which could indicate alterations in salvage synthesis pathways of ceramides (Van Brocklyn and Williams, 2012). In contrast, CES1 was not detectable in smt1 DSM fraction, so no conclusions regarding the CES1 DRM/DSM distribution in smt1 could be drawn (Figure 6B).
Glycosylation of sterols (SGlc/ASGlc) is likely to also happen in the plasma membrane and this pathway could be catalyzed by UGT80A2. This Glycosyl-transferase was depleted from DRM in both mutants (Figure 6C) and is listed in the PhosPhat database as a phosphorylated protein (Durek et al., 2010; Zulawski et al., 2013) and might be involved in dynamic microdomain remodeling processes. Because several GlcCer levels are inversely altered in both mutants anaylzed here, it implies an additional sterol-dependent pathway for ceramide glycosylation in the plasma membrane (Figure 6E). Therefore, a dynamic glycosylation of ceramides may also be involved in microdomain organization and remodeling. Furthermore, ceramide signaling pathways seem to be altered in the sterol-mutants and the observed elevated levels of Cer in smt1 might activate phosphatase PP2A (Nickels and Broach, 1996). A ceramide dependent activation of PP2A was previously observed in yeast triggering apoptosis (Nickels and Broach, 1996). Here we observed a sterol-dependent response of the PP2A subunit PP2AA3 (Figure 6D) that could additionally also be a result of ceramide signaling events. PP2AA3 was previously shown to be involved in various signaling processes in plants (Zhou et al., 2004; Blakeslee et al., 2008; Dai et al., 2012a,b). Therefore, misbalances in signaling events might contribute to the strong phenotype of smt1 with cells remaining in a continuous apoptotic status.
In addition, phospholipases showed opposing responses in their degree of DRM residence in both mutants (Figure 6F) and regulation of protein–protein interactions with G-proteins, protein kinases and phosphatases might involve different degrees of lipid phase separation. Vesicular transport might be affected in both mutants as indicated by differential DRM residences of vesicle trafficking proteins SEC3 and SEC22 (Figure 6G). The plant NADPH oxidase RbohD showed discordant changes in DRM/DSM ratio distributions between smt1 and ugt80 (Figure 6H). Based on the prediction of perturbed H2O2 levels, as indicated by responses of RbohD (Liu et al., 2009) (Table 1), as well as based on reported linkages of ceramide signaling to reactive oxygen signaling (summarized in Berkey et al., 2012; Van Brocklyn and Williams, 2012), we measured intrinsic H2O2 levels in both mutants studied (Supplemental Figure 4). Consistent with our prediction we observed disturbed oxidative response levels in both mutants with threefold higher H2O2 concentrations in smt1 and only one fold higher levels in ugt80A2;B1 relative to wildtype (Figure 6G).
Discussion
The proteome- and lipidome-wide alterations observed in smt1 and ugt80A2;B1 can largely be brought down to specific alterations in membrane structure and signaling properties. Membrane structures of the mutants are mainly affected by the intrinsically altered sterol composition and sterol glycosylation status (Schrick et al., 2002, 2004b; DeBolt et al., 2009). By combining information from systematic proteomics with information from lipidomics, a number of biologically-relevant processes could be identified and linked to new aspects in interpretation of the phenotypes of both mutants.
Structural Effects of Altered Membrane Lipid Environments on DRM Protein Composition
We could show that besides changed sterol composition, particularly alterations in ceramide (Cer) and glycosylceramide (GlcCer) abundances were related with changes in protein DRM/DSF distribution in the mutants. Cer abundance as well as overall glycosylation status of microdomain lipids, in addition to sterols, could potentially affect recruitment and residence times of proteins in membrane microdomains.
Recently we could show, that smt1 exhibits a protein DRM/DSF distribution that resembles methyl-β-cyclodextrin (mβcd) treated DRM compositions (Zauber et al., 2013). This suggests a structure-related impact of altered sterol–protein interactions in the plasma membrane. In contrast to smt1, the whole-plant phenotype of ugt80A2;B1 is less severe and adult plants are remarkably healthy. Consistently, the sterol profile of ugt80A2;B1 was shown not to be strongly altered, apart from a depletion of sterolglycosides. However, low abundant sterols were slightly decreased (cycloarthenol and cholesterol) or increased (isofucosterol) (DeBolt et al., 2009). Nevertheless, strong changes in the glycero- and sphingolipids and DRM/DSF protein distributions were visible for this mutant. These changes were in many aspects discordant to the changes in smt1. While the differential proteomic profile suggests plasma membrane structure can be affected in different ways by both mutant backgrounds, a general depletion of known sterol-dependent proteins was observed for both mutants. This suggests that structural perturbations in the plasma membrane could reduce sterol–protein interactions in both mutants with global effects on protein and lipid residence and recruitment to microdomains. Furthermore, the glycosylation status of microdomain lipids seems to play a crucial role for protein recruitment, as was shown in ugt80A2;B1 mutant. Glycosylresidues in microdomains might be one of the triggers for reinforcement or prevention of protein recruitment into these microdomains or, alternatively, general microdomain formation (Table 1). Glycosylhydrolases previously identified as resident to membrane microdomains (Kierszniowska et al., 2009; Gupta and Surolia, 2010) might be involved in the dynamic control of the glycosylation in microdomain lipids.
A small number of DRM-dependent microdomain proteins was identified as co-purifying in DRM and DSF fractions and could not be automatically filtered by pairwise comparisons with intracellular membrane and soluble protein fractions. The purity of plasma membrane preparations over two-phase systems was reported to reach 95% based on activity measurements of known plasma membrane proteins (Bérczi and Asard, 2003; Mika et al., 2004; Schindler and Nothwang, 2006; Lüthje, 2008). Nevertheless, major proteins of endomembrane systems can introduce biases into plasma membrane analysis (Bérczi and Asard, 2003, Griesen et al., 2004; Kjell et al., 2004; Preger et al., 2005; Lüthje, 2008). The differential distribution of these proteins in both mutants may indicate biologically relevant structural changes in membranes leading to an altered protein abundance and/or distribution. Within the core set of proteins selected from the network analysis a glucose-6-phosphate/phosphate translocator and a dicarboxilic acid transporter were significantly altered in their DRM/DSF ratios (Table 1). Analysis of copy numbers revealed, that these proteins even had higher copy numbers in DSF than in IM fraction, where they would have been expected (Supplemental Figure 5). While the DRM/DSF ratios rather reflects alterations in sterol–protein interactions particularly of the plasma membrane, sterol dependence of plastidial or mitochondrial membrane proteins is debatable due to very low amounts of sterols in these organelle membranes (Block et al., 2007). However, if protein abundances are drastically altered it is possible that these changes affect abundances across all analyzed fractions. Even though the exact structural basis for the apparent changes in sterol–protein affinities remains unclear for these co-purifying proteins, their observed alterations in smt1 may also be linked to disturbed membrane trafficking, as explained later.
Alterations in Signaling Pathways Localized at the Plasma Membrane
The strong changes in membrane lipid composition in the mutants may have direct implications on signaling pathways that are initiated at the plasma membrane. Due to the strong changes seen in cellular lipid composition, observed alterations in plasma membrane signaling can for the most part be attributed to structural alterations in the plasma membrane. Since microdomain-forming lipids, such as ceramides, are particularly affected, specific alterations in microdomain formation and dynamic protein recruitment are likely. As microdomains were shown to be related to signaling, it is likely that smt1 and ugt80A2;B1 have altered sterol-sphingolipid-protein interactions. Therefore, signaling involving the plasma membrane may not function properly in response to environmental or cellular stimuli. In the mutants, different signaling pathways seem to be affected, involving not only signaling proteins but also microdomain specific lipids. As signaling properties have been shown in particular for sterols (He et al., 2003) and ceramides (Cer) (Nickels and Broach, 1996; Chen et al., 2008; Alden et al., 2011; Van Brocklyn and Williams, 2012), a diverse set of signaling pathways may be constitutively altered in smt1 and ugt80A2;B1. The impact of these mutations on plant phenotypes is challenging to interpret as it is most likely a combination of structural effects and altered signaling properties of the membrane. Nevertheless, the systems biology-based approach applied in this study proposes two signaling pathways affecting the phenotype of both mutants.
Cer and sphingosine-1-phosphate (S1P) are both involved in signaling but regulate contrary processes (Breslow and Weissman, 2010; Van Brocklyn and Williams, 2012). In the currently accepted model, both lipids are directly related to sphingosine and the balance of the involved reactions is crucial for the activation of either apoptosis or cell proliferation processes when Cer or S1P is high, respectively. This model was shown to be ubiquitous among different species and seems to play a role in plants as well (Stunff et al., 2004). As one target of this signaling pathway, the phosphatase PP2A in yeast was shown to be activated by ceramides (Nickels and Broach, 1996). In support of this, the positive regulatory subunit PP2AA3 (Marmagne et al., 2007; Dai et al., 2012a,b) of PP2A phosphatase showed a significant increase in DRM/DSF ratios in smt1, but was slightly decreased in ugt80A2;B1, although this was not significant (Supplemental Figure 5). As PP2A is involved in cell growth limiting processes, the activity of PP2A could therefore contribute to the extreme pleiotropic phenotype of smt1. Alterations in PP2A activity can also be considered to be a direct effect of observed elevated Cer levels in smt1. If levels of S1P were raised in ugt80A2;B1 and decreased in smt1, alterations in S1P and Cer levels might explain the inverse lipid profiles observed for both mutants in comparison to wildtype. Following this hypothesis, both mutants could be oppositely unbalanced in signaling processes that regulate apoptosis and cell proliferation (Van Brocklyn and Williams, 2012) (Figure 6E). To investigate this further, sphingosine kinase, ceramide synthase, ceramidase as well as S1P-phosphatase could serve as targets of future studies.
Furthermore we could also identify an inverse abundance profile for phospholipases between both mutants. The phospholipase signaling cascade was shown to involve particularly G-proteins, protein kinases as well as phosphatases (Nickels and Broach, 1996). All these protein groups showed an inverse DRM abundance in the two mutants indicated by inverse DRM/DSF distribution ratios between ugt80A2;B1 and smt1. Phospholipase activity was shown to be linked to microdomains as recently summarized (Gardiner and Marc, 2013). Therefore, we postulate a regulation of the detected phospholipases that involves protein segregation in membrane microdomains. In this model, recruitment of phospholipases and their regulating proteins into microdomains brings them into close proximity and could trigger phospholipase activity. Thereby, phosphorylation of the identified phospholipases could be a potential mechanism for regulating the recruitment to microdomains. Accordingly, the identified phospholipase 2C was reported to be phosphorylated in sucrose and nitrate starvation/resupply experiments (PhosPhat; Durek et al., 2010) and phosphorylation-dependent recruitment of proteins to membrane microdomains has now been reported in the context of ABA signaling (Demir et al., 2013). Interestingly, sterol–protein interactions among these proteins were increased in ugt80A2;B1. This exemplifies once more that glycosylation of microdomain lipids could directly affect the recruitment of sterol-dependent proteins.
Evidence for Sterols As Donors of Glycosylresidues in a Ceramide Glycosylation Pathway
In ugt80A2;B1, the observed significant decrease of glycosylated ceramide (GlcCer) species correlates with the depletion of glycosylated sterols (DeBolt et al., 2009). GlcCer in beans were shown to be mainly present in the outer plasma membrane leaflet (Lynch et al., 1997). It was therefore suggested that glycosylresidues from sterolglycosides might serve as precursors also for ceramide glycosylation (Lynch and Phinney, 1995). Depletion of GlcCer in ugt80A2;B1 strongly supports this finding in Arabidopsis, but it does not prove location of this process at the plasma membrane. Interestingly, UGT80A2 was depleted from DRM fractions in the smt1 mutant and was shown to also be depleted in an independent mβcd treatment of plasma membranes (Zauber et al., 2013). Furthermore, according to TAIR (Poole, 2007), UGT80A2 is localized to the plasma membrane and was exclusively identified in DRM and DSF fractions in our analysis. This raises the possibility that sterol glycosylation indeed occurs at the plasma membrane in addition to sterol glycosylation events happening in the endomembrane system. Therefore, sterol glycosylation events at the plasma membrane might be particularly involved in controlling lipid remodeling processes and lipid–protein interactions induced by dynamic lipid glycosylation processes. An additional level of control could thus be imposed by phosphorylation, as UGT80A2 exhibits a treatment-specific phosphorylation status according to PhosPhat database (Durek et al., 2010; Zulawski et al., 2013). Our current working model suggests therefore, that a dynamic glycosylation status of microdomains may be specifically adjusted to environmental and cellular conditions. In case sterol glycosylation occurs at the plasma membrane, the available pools of glucose-derivates for sterol glycosylation might accumulate in ugt80A2;B1 which increases the probability for glucose derivates being incorporated into other extracellular structures, such as cell wall components. Consistent with this hypothesis, ugt80A2 and ugt80B1 mutants were shown to incorporate more glucose residues into their cell walls (DeBolt et al., 2009). This effect was also observed by an analysis of the mucilage of ugt80A2;B1 seeds (Supplemental Figure 6). Interestingly, proteins with a putative function in glucan synthesis showed altered sterol–protein interactions in smt1 as well as ugt80A2;B1 (Table 1) and smt1 was reported to have lowered cellulose levels (Schrick et al., 2004a). Thus, our data strongly supports the existence of plasma membrane-bound lipid glycosylation.
Inverse Effects on Lipid Synthesis Pathways in Both Mutants
Both mutants exhibited inverse abundance profiles in most of the altered lipids. Interestingly, the changes in lipid abundances correlated with protein abundances involved in lipid synthesis. Synthesis of acyl chains takes place in the plastid, while esterification of fatty acids occurs in both the plastid and the endoplasmic reticulum to give rise to membrane lipids (Benning, 2008). The analysis of proteins involved in the lipid synthesis pathways suggested that fatty acid synthesis in general could be decreased in smt1 and elevated in ugt80A2;B1. The overall trend of increased lipid synthesis in ugt80A2;B1 was even supported by the abundances of proteins involved in lipid degradation (Figure 3C). In general, no significant alterations could be observed in triacylglyceride abundance profiles (Supplemental Figure 3). For this reason, global starvation effects on fatty acid synthesis can presumably be excluded in the mutants. While the used callus system was suppressed in photosynthesis activity, alterations in lipid synthesis require adjustment of energy supply in form of ATP to the plastid in the mutants. Such a scenario could explain the observed altered protein abundances of the plastidial ATP/ADP transporter AATP2 in the IM fraction of both mutants (Figure 6 and Supplemental Figure 5).
General Implications of Lipid–Protein Interactions in Plasma Membrane Functions
A major function of the plasma membrane is the build-up and maintenance of a transmembrane electric potential through a number of ATPases (Flickinger et al., 2010). Further, a constitutive redox system involving cytochromes (Asard et al., 2001; Lüthje et al., 2005) and NAD(P)H oxidoreductases (Lüthje et al., 1997; Sparla et al., 1997; Bérczi and Møller, 1998a,b; Trost et al., 2000; Lüthje, 2008) was identified and linked to regulation of redox processes for inorganic nutrient uptake (Böttger and Lüthen, 1986; Döring and Lüthje, 1996; Robinson et al., 1997, 1999; Waters et al., 2002). Cytochromes and ATPases were both significantly decreased in their DRM/DSF ratio in ugt80A2;B1 and smt1, indicating the relevance of sterols and sterol glycosylation also for redox processes and transport. The detailed implications of alterations in these membrane functions are beyond the scope of our study. However, it needs to be pointed out, that phase separation in membranes was shown to also be dependent on pH gradients in giant vesicles (Staneva et al., 2012) underlining the biological significance of the observed alterations in cytochromes and ATPases identified in this study. Nevertheless, only the co-purifying cytochrome c oxidase showed an increased DRM/DSF ratio in ugt80A2;B1 and is involved in mitochondrial respiration for generating ATP. Elevated levels of cytochrome c oxidase were also observed in IM and SP fraction in both mutants, indicating a possible increase in cell respiration. Thus, elevated respiration in both mutants could be linked to perturbed cell homeostasis as a result of alterations in cell signaling status.
Membrane trafficking in cells is an obligatory process and interconnects most cellular membrane structures in the cell. In this study, we identified proteins being significantly and discordantly altered in their DRM/DSF ratio abundance between the two mutants, which indicates distinct alterations in membrane trafficking pathways. For instance, in ugt80A2;B1 membrane trafficking from ER to Golgi apparatus seems to be altered indicated by decreased DRM/DSF abundance ratio of SEC22. In contrast, vesicle movement from Golgi apparatus to the plasma membrane shows alterations particularly in smt1, with SEC3 displaying a decreased DRM/DSF ratio. Further SEC14 was shown to be a major factor for localizing recruitment of microdomain proteins to the plasma membrane (Curwin et al., 2013). Even though this protein was not significantly altered in the DRM/DSF distribution, when analyzing log2 fold-change or DRM/DSF protein abundance ratios, it showed depletion from DRM and DSF fractions in smt1 but was increased in ugt80A2;B1 (Supplemental Figure 5).
The very high amounts of co-purifying proteins in the DRM fraction of smt1 might be even linked to disturbed membrane trafficking. However, our data at this point does not allow conclusions on whether structural changes or affected signaling pathways in involved membranes are triggering these effects. Since sterols were shown to be rapidly transported by MT to the plasma membrane (Hartmann and Benveniste, 1987; Moreau et al., 1998; Grebe et al., 2003), disturbed membrane trafficking would likely cause alterations in the sterol gradient from ER over Golgi apparatus to the plasma membrane. Therefore, the high amount of co-purifying proteins in smt1 DRM fraction suggests an accumulation of sterols in non-plasma-membrane structures. This could then explain high amounts of co-purifying “non-plasma membrane” proteins by sterol-enriched patches in other membrane types in smt1. A similar effect was described for maize roots treated with fenpropimorph (Grandmougin et al., 1989). There, it was shown, that unusual sterols accumulated, but were incorporated into membranes with drastic effects on the cellular distribution of sterols leading to enrichment of free sterols in the ER (Grandmougin et al., 1989). Such an effect was also observed in smt1, in which functional categories of DRM co-purifying proteins linked to ER and related membrane structures (Supplemental Figure 7) are dominant among the co-purifying proteins with typical sterol-dependent abundance distributions. Therefore, although total sterol amounts are likely depleted in smt1 plasma membrane, subcellular sterol distributions may additionally be affected.
Conclusion
Sterol-biosynthesis mutants have strong pleiotropic phenotypes, which are obviously the result of several overlaying effects related to perturbations in membrane structure and signaling properties. In order to understand the underlying biological processes in more detail, systems biology-based approaches as exemplified in this study, are helpful in dissecting the pathways involved. In particular, the changes that were observed in lipid abundances were of a general systemic nature and have not been considered so far when studying sterol–protein relationships. Within this study we could connect alterations in the mutants' lipidomes and proteomes resulting in a consistent working model of specifically altered biological processes in ugt80A2;B1 and smt1. With respect to effects on sterol–protein interactions, we could show that (1) particularly an interplay between sterols, sphingolipids and proteins is necessary for formation and maintenance of membrane subcompartmentation in form of microdomains. We could (2) confirm disturbed H2O2 signaling in smt1 and ugt80A2;B1 connecting this signaling pathway to lipid/sterol environments. In addition, (3) modifications, such as glycosylation of lipid species (sterols, ceramides, or sphingolipids) may also serve as additional regulating factors for microdomain formation and dynamic glycosylation status of microdomains might control and alter specific recruitment of proteins to membrane subcompartments. Our work herewith provides a thorough assessment and working model of the roles of lipid–protein interactions for cellular signaling processes.
Methods
Cell Suspension Culturing
Heterozygous seeds from smt1 mutant named cphT357 (Schrick et al., 2002) with a point mutation at T357 in the smt1 (AT5G13710) locus were germinated and homozygous seedlings showing the cph typical dwarf like phenotype were selected (Supplemental Figure 8). Seedlings from homozygous ugt80A2;B1 (A2: AT3G07020; B1: AT1G43620) double mutant (DeBolt et al., 2009; Schrick et al., 2012b) (Supplemental Figure 8) were immediately used for callus initiation. Callus cultures were initialized from leafs on 6.8% Agar in full mineral Murashige-Skoog-Medium (Murashige and Skoog, 1962) with the freshly added components 3% sucrose, 200 mg/l myoinositol, 1 mg/l 2,4-dichlorophenoxyacetic acid and 0.25 mg/l kinetin for smt1, ugt80A2;B1 and corresponding wildtype Ler-0 or Col-0, respectively (A. thaliana). Established callus cultures (Supplementary Figure 1) were chopped and directed to solution culture using the same agar free medium. Arabidopsis cell suspension cultures were subcultured to fresh medium every week. For all analyses up to three or four biological replicates were collected for smt1 or ugt80A2;B1, respectively and their corresponding wildtypes.
Sample Preparation and Protein Extraction
The protocol for sample preparation was recently described (Zauber et al., 2013) and will only be briefly summarized. Frozen cell powder was mixed with 2 volumes of cold extraction buffer (100 mM Hepes-KOH pH 7.5, 250 mM sucrose, 3 mM KCL, 0.1 mM EDTA, 1 mM DTT and addition of μl/l protease inhibitor cocktail (Thermo Scientific)). Supernatant after ultra centrifugation was used for extracting soluble proteins (SP). Microsomal pellet fraction was resuspended (5 mM KH2PO4, 0.33 M sucrose, 3 mM KCL, 0.1 mM EDTA, 1 mM DTT, protein inhibitor cocktail) and plasma membrane (PM) and intracellular membranes (IM) containing organelle membranes were separated using aqueous two phase system (Schindler and Nothwang, 2006). IM pellet and PM pellet were resuspended in 25 mM Tris buffer (pH 7.5; 150 mM NaCl; 5 mM EDTA; 1 mM DTT). Samples were adjusted to equal protein amounts of 100 μg using Bradford (Bradford, 1976). All PM fractions were mixed with TritonX 100 and incubated for 30 min at 4°C using a protein detergent ratio of 1:15 at a total TritonX 100 concentration of 1%. A detergent resistant membrane fraction (DRM) and a detergent soluble fraction (DSF) was separated on a sucrose gradient (1.8 mM; 1.6 mM; 1.4 mM and 0.15 mM) by ultra centrifugation. From the four fractions DRM, DSF, SP, and IM, proteins were extracted using methanol/chloroform extraction.
Lipid Extraction
From frozen callus, aliquots of 25 mg were prepared in 1.5 mL Eppendorf tubes, maintaining the sample constantly frozen. The aliquots were suspended in 1 mL of a pre-cooled 1:3 methanol/methyl tert-butyl ether (MTBE) solution. After 10 min ultrasonication and shaking for 30 min at 4°C, 500 μl of a 3:1 water/methanol solution were added to the samples. After mixing, the tubes were spun down for 5 min in a table top centrifuge. The organic phase was dried down in a SpeedVac and stored at −20°C before being analyzed. Four to five biological replicates were analyzed per mutant or wildtype line.
LC-MS/MS Analysis and Protein Identification
Up to three replicate injections, containing 25 μg of protein, were analyzed by LC-MS/MS using nano-flow HPLC (Proxeon Biosystems) and an Orbitrap hybrid mass spectrometer (LTQ-Orbitrap XL, Thermo Scientific) as mass analyzer. Peptides were eluted from a 75 μm analytical column (Reprosil C18, Dr. Maisch GmbH) on a linear gradient, running from 5% to 80% acetonitrile in 90 min at a flow rate of 250 nL/min. Up to five data-dependent MS/MS spectra were acquired in the linear ion trap for each FTMS full-scan spectrum acquired at 60,000 full-width half-maximum resolution settings with an overall cycle time of approximately 1 s. Raw file peak extraction, protein identification and quantitation of peptides was done by MaxQuant (version 1.2.2.5) using a protein sequence database of Arabidopsis thaliana (TAIR10, 35386 entries, www.arabidopsis.org). For protein identification by Andromeda search engine implemented in MaxQuant (Cox and Mann, 2008), carbamidomethylation and N-terminal protein acetylation were used as fixed modifications and methionine oxidation as a variable modification. Trypsin was selected as a digestive enzyme and for database search, up to two missed cleavages were allowed. Precursor ion tolerance was set to 6 ppm, mass tolerance for fragment ion matching was 0.5 Da. Standard settings in MaxQuant involving peptide false-discovery rate of 0.01, minimum peptide length of 6 amino acids and enabled retention time correlation, over a time window of 2 min, were used.
Peak Identification and Quantification of Lipids
GeneData software was used to pre-process the chromatogram raw files, that is, baseline correction, chemical noise subtraction, chromatogram alignment and peak detection. Pre-processing parameters were set to the same values as described (Giavalisco et al., 2011). After pre-processing, a list of detected peaks (a retention time and m/z pair) and a matrix with their respective intensities for each sample were obtained. A targeted search for the glycerolipid species of interest was carried out using the in-house developed R package “grms,” and based on the library compiled by Giavalisco et al. (2011). The software first performs a retention time correction of the output matrix based on previously identified markers with known m/z values and retention times. Then, the compounds were searched by comparing their specific m/z, expected adduct and retention times within user-given tolerance thresholds. A mass tolerance of 5 ppm and retention time deviation of 0.5 min were used to identify the lipid species. Further confirmation was achieved by manually inspecting the chromatograms.
Statistical Analysis
Statistical analysis of all datasets was done using open source scripting language R, version 2.15.2 (Team, 2009). Analyses involved functions from the following packages: “gplots” (Warnes et al., 2013a), “igraph” (Han et al., 2010, 2011), “gdata” (Warnes et al., 2013b), “seqinr” (Charif et al., 2007), and “grImport” (Murrell et al., 2012). Lipids detected in positive and negative mode were combined before normalization. For normalization only lipids with a variation lower than the median variation of all lipids across all analyzed samples were used. Triacylglycerides (TAG) were excluded for calculating total ion-intensity sums per sample. Finally, all lipids were expressed as fraction of total ion-intensity sums. For further analysis, data was z-scaled and linearly transformed to positive values for log2 transformation. Differential abundances were tested using t-test (α = 0.05). Protein intensities were calculated from peptide ion-intensity based on the table evidence.txt derived from MaxQuant. This table was processed further with the cRacker platform (Zauber and Schulze, 2012) for automated analysis of peptide intensities. Parameters were set as described (Zauber et al., 2013). Principle steps involved peptides exclusion if measured in less than 70% of one of the fractions. Further peptide intensities within each sample were normalized to fraction of total ion-intensity sums. Normalized peptide intensities were median scaled and median averaged. cRacker exports were directly used for analyzing DRM/DSF protein abundance ratios using Unicorn. The applied algorithm was recently described (Zauber et al., 2013). Unicorn is based on bootstrapping intensities from ratios for statistical analysis of protein distribution between two fractions. False positives were controlled by calculating a data specific threshold based on randomized data. False discovery rates (FDR) were set to <1%. Significant protein candidates were further filtered out against higher abundance proteins in SP or IM fraction, applying a pairwise comparison using t-test (α = 0.01). At this step the applied statistics was used to filter out co-purifying proteins. Thats why a multiple testing correction was not applied to be more stringent in the filtering.
Network Analysis
A combined score for definition of lipid–protein correlations was calculated by multiplying lipid log2 fold changes in mutant versus wildtype with scores from comparative DRM/DSF protein abundance analysis (Unicorn scores). Thereby, only significantly altered proteins were included. Significance of lipid–protein dependence was defined by a false discovery threshold <1%. This threshold was calculated from randomized sampling of lipid ratios and protein Unicorn scores. Networks were visualized using the “igraph” (Han et al., 2010, 2011) R-package. Protein subcellular localizations were obtained from SUBA3 (Tanz et al., 2013). Phosphorylation data was obtained from PhosPhat database (Durek et al., 2010; Zulawski et al., 2013). Myristoylation data was taken from TAIR and was based on work from Thierry Meinnel's group. Protein palmitoylation was predicted using CSS-Palm software 3.0 (Ren et al., 2008). GPI-anchoring of proteins was predicted using Arabidopsis specific predictions obtained from GPI-DB (Poisson et al., 2007). Palmitoylation and GPI-anchoring predictions were FDR (FDR <0.05%) controlled, running both algorithms with randomized sequences from analyzed proteins in parallel.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Aenne Eckart for performing lipid LC-MS measurements. Further we acknowledge Aleksandar Vasilevski for the sample preparation and analysis of neutral sugar composition of seed mucilage in ugt80A2;B1, Sylwia Kierszniowska for callus initiation of the smt1 mutant and supervision during initiation time of the project, Ulrike Minzlaff and Natalie Arndt for maintenance of suspension cultures. Kathrin Schrick (UCLA) and Wolf-Rüdiger Scheible (Noble Foundation) are acknowledged for donation of seeds of the sterol biosynthesis and sterol glycosylation mutants.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00078/abstract
Supplemental Figure 1 | Photographs from callus cultures.
Supplemental Figure 2 | Subcellular localizations of responding proteins according to SUBA3 (Tanz et al., 2013) in DRM/DSF for smt1 (left) and ugt80A2;B1 mutant (right).
Supplemental Figure 3 | Barplots of log2 transformed intensity ratios of all detected lipids.
Supplemental Figure 4 | H2O2 contents in callus from smt1, ugt80A2;B1, and corresponding wildtypes.
Supplemental Figure 5 | Barplots of calculated protein abundances from all analyzed proteins across all experiments.
Supplemental Figure 6 | Barplots of amounts from mucilage side chain residues using HPLC system for detection after hydrolysis of purified mucilage from seeds (Vasilevski et al., 2012).
Supplemental Figure 7 | Mapman functional categories of co-purifiying proteins showing a significantly increased DRM/DSM protein abundance ratio in smt1.
Supplemental Figure 8 | Homozygous ugt80A2;B1 was confirmed using a PCR screen as described in DeBolt et al. (2009).
Supplemental Table 1 | List of proteins with significantly different DRM/DSF distributions in mutants versus wildtype.
Supplemental Table 2 | List of analyzed lipids.
Supplemental Table 3 | List of responding proteins involved in lipid synthesis.
References
Alden, K. P., Dhondt-Cordelier, S., McDonald, K. L., Reape, T. J., Ng, C. K.-Y., McCabe, P. F., et al. (2011). Sphingolipid long chain base phosphates can regulate apoptotic-like programmed cell death in plants. Biochem. Biophys. Res. Commun. 410, 574–580. doi: 10.1016/j.bbrc.2011.06.028
Asard, H., Kapila, J., Verelst, W., and Bérczi, A. (2001). Higher-plant plasma membrane cytochromeb 561: a protein in search of a function. Protoplasma 217, 77–93. doi: 10.1007/BF01289417
Bariola, P., Retelska, D., Stasiak, A., Kammerer, R., Fleming, A., Hijri, M., et al. (2004). Remorins form a novel family of coiled coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Mol. Biol. 55, 579–594. doi: 10.1007/s11103-004-1520-4
Beaudoin, F., Wu, X., Li, F., Haslam, R. P., Markham, J. E., Zheng, H., et al. (2009). Functional characterization of the Arabidopsis beta-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 150, 1174–1191. doi: 10.1104/pp.109.137497
Benning, C. (2008). A role for lipid trafficking in chloroplast biogenesis. Prog Lipid Res. 47, 381–389. doi: 10.1016/j.plipres.2008.04.001
Benveniste, P. (2004). Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 55, 429–457. doi: 10.1146/annurev.arplant.55.031903.141616
Bérczi, A., and Asard, H. (2003). Soluble proteins, an often overlooked contaminant in plasma membrane preparations. Trends Plant. Sci. 8, 250–251. doi: 10.1016/S1360-1385(03)00100-6
Bérczi, A., and Møller, I. M. (1998a). Characterization and solubilization of residual redox activity in salt-washed and detergent-treated plasma membrane vesicles from spinach leaves. Protoplasma 205, 59–65. doi: 10.1007/BF01279294
Bérczi, A., and Møller, I. M. (1998b). NADH-Monodehydroascorbate oxidoreductase is one of the redox enzymes in spinach leaf plasma membranes. Plant Physiol. 116, 1029–1036. doi: 10.1104/pp.116.3.1029
Berkey, R., Bendigeri, D., and Xiao, S. (2012). Sphingolipids and plant defense/disease: the “death” connection and beyond. Front Plant. Sci. 3:68. doi: 10.3389/fpls.2012.00068
Bhat, R. A., and Panstruga, R. (2005). Lipid rafts in plants. Planta 223, 5–19. doi: 10.1007/s00425-005-0096-9
Blakeslee, J. J., Zhou, H.-W., Heath, J. T., Skottke, K. R., Barrios, J. A. R., Liu, S.-Y., et al. (2008). Specificity of RCN1-mediated protein phosphatase 2A regulation in meristem organization and stress response in roots. Plant Physiol. 146, 539–553. doi: 10.1104/pp.107.112995
Block, M. A., Douce, R., Joyard, J., and Rolland, N. (2007). Chloroplast envelope membranes: a dynamic interface between plastids and the cytosol. Photosynth. Res. 92, 225–244. doi: 10.1007/s11120-007-9195-8
Borner, G. H. H., Sherrier, D. J., Weimar, T., Michaelson, L. V., Hawkins, N. D., Macaskill, A., et al. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137, 104–116. doi: 10.1104/pp.104.053041
Böttger, M., and Lüthen, H. (1986). Possible linkage between NADH-oxidation and proton secretion in Zea mays L. roots. J. Exp. Bot. 37, 666–675. doi: 10.1093/jxb/37.5.666
Boutte, Y., Palme, K., Stierhof, Y.-D., Hartmann, M.-A., Moritz, T., and Grebe, M. (2008). Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nature 10, 237–244. doi: 10.1038/ncb1686
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Breslow, D. K., and Weissman, J. S. (2010). Membranes in balance: mechanisms of sphingolipid homeostasis. Mol. Cell 40, 267–279. doi: 10.1016/j.molcel.2010.10.005
Browman, D. T., Hoegg, M. B., and Robbins, S. M. (2007). The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 17, 394–402. doi: 10.1016/j.tcb.2007.06.005
Cacas, J.-L., Furt, F., Le Guédard, M., Schmitter, J.-M., Buré, C., Gerbeau-Pissot, P., et al. (2012). Lipids of plant membrane rafts. Prog Lipid Res. 51, 272–299. doi: 10.1016/j.plipres.2012.04.001
Carmona-Salazar, L., El Hafidi, M., Enríquez-Arredondo, C., Vázquez-Vázquez, C., González de la Vara, L. E., and Gavilanes-Ruíz, M. (2011). Isolation of detergent-resistant membranes from plant photosynthetic and non-photosynthetic tissues. Anal. Biochem. 417, 220–227. doi: 10.1016/j.ab.2011.05.044
Charif, D., Lobry, J., Necsulea, A., Palmeira, L., and Penel, M. S. (2007). “SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis,” in Biological and Medical Physics, Biomedical Engineering (Berlin; Heidelberg: Springer), 207–232. doi: 10.1007/978-3-540-35306-5_10
Chen, M., Markham, J. E., Dietrich, C. R., Jaworski, J. G., and Cahoon, E. B. (2008). Sphingolipid long-chain base hydroxylation is important for growth and regulation of sphingolipid content and composition in Arabidopsis. Plant Cell Online 20, 1862–1878. doi: 10.1105/tpc.107.057851
Clouse, S. D. (2002). Arabidopsis mutants reveal multiple roles for sterols in plant development. Plant Cell 14, 1995–2000. doi: 10.1105/tpc.140930
Clouse, S. D., and Sasse, J. M. (1998). BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451. doi: 10.1146/annurev.arplant.49.1.427
Cox, J., and Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Curwin, A. J., LeBlanc, M. A., Fairn, G. D., and McMaster, C. R. (2013). Localization of lipid raft proteins to the plasma membrane is a major function of the phospholipid transfer protein sec14. PLoS ONE 8:e55388. doi: 10.1371/journal.pone.0055388
Dai, M., Terzaghi, W., and Wang, H. (2012a). Multifaceted roles of Arabidopsis PP6 phosphatase in regulating cellular signaling and plant development. Plant Signal. Behav. 8:e22508. doi: 10.4161/psb.22508.
Dai, M., Zhang, C., Kania, U., Chen, F., Xue, Q., McCray, T., et al. (2012b). A PP6-type phosphatase holoenzyme directly regulates PIN phosphorylation and auxin efflux in Arabidopsis. Plant Cell Online 24, 2497–2514. doi: 10.1105/tpc.112.098905
DeBolt, S., Scheible, W. R., Schrick, K., Auer, M., Beisson, F., Bischoff, V., et al. (2009). Mutations in UDP-glucose: sterol glucosyltransferase in Arabidopsis cause transparent testa phenotype and suberization defect in seeds. Plant Physiol. 151, 78–87. doi: 10.1104/pp.109.140582
Demir, F., Horntrich, C., Blachutzik, J. O., Scherzer, S., Reinders, Y., Kierszniowska, S., et al. (2013). Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc. Natl. Acad. Sci. U.S.A. 110, 8296–8301. doi: 10.1073/pnas.1211667110
Döring, O., and Lüthje, S. (1996). Molecular components and biochemistry of electron transport in plant plasma membranes (review). Mol. Membr. Biol. 13, 127–142. doi: 10.3109/09687689609160589
Durek, P., Schmidt, R., Heazlewood, J. L., Jones, A., MacLean, D., Nagel, A., et al. (2010). PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 38, D828–D834. doi: 10.1093/nar/gkp810
Edidin, M. (2001). Shrinking patches and slippery rafts: scales of domains in the plasma membrane. Trends Cell Biol. 11, 492–496. doi: 10.1016/S0962-8924(01)02139-0
Entchev, E. V., and Kurzchalia, T. V. (2005). Requirement of sterols in the life cycle of the nematode Caenorhabditis elegans. Semin. Cell Dev. Biol. 16, 175–182. doi: 10.1016/j.semcdb.2005.01.004
Flickinger, B., Berghöfer, T., Hohenberger, P., Eing, C., and Frey, W. (2010). Transmembrane potential measurements on plant cells using the voltage-sensitive dye ANNINE-6. Protoplasma 247, 3–12. doi: 10.1007/s00709-010-0131-y
Fujioka, S. (2010). The sterol methyltransferases SMT1, SMT2, and SMT3 influence Arabidopsis development through nonbrassinosteroid products. Plant Physiol. 153, 741–756. doi: 10.1104/pp.109.152587
Gardiner, J., and Marc, J. (2013). Phospholipases may play multiple roles in anisotropic plant cell growth. Protoplasma 250, 391–395. doi: 10.1007/s00709-012-0377-7
Giavalisco, P., Matthes, A., Eckhardt, A., Hubberten, H.-M., Hesse, H., Segu, S., et al. (2011). Elemental Formula Annotation of Polar and Lipophilic Metabolites Using (13) C, (15) N and (34) S Isotope Labelling, in Combination with High-Resolution Mass Spectrometry. Technical report, Max Planck Institute of Molecular Plant Physiology, Potsdam, Germany.
Grandmougin, A., Bouvier-Navé, P., Ullmann, P., Benveniste, P., and Hartmann, M. A. (1989). Cyclopropyl sterol and phospholipid composition of membrane fractions from maize roots treated with fenpropimorph. Plant Physiol. 90, 591–597. doi: 10.1104/pp.90.2.591
Grebe, M., Xu, J., Möbius, W., Ueda, T., Nakano, A., Geuze, H. J., et al. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13, 1378–1387. doi: 10.1016/S0960-9822(03)00538-4
Griesen, D., Su, D., Bérczi, A., and Asard, H. (2004). Localization of an ascorbate-reducible cytochrome b561 in the plant tonoplast. Plant Physiol. 134, 726–734. doi: 10.1104/pp.103.032359
Gupta, G., and Surolia, A. (2010). Glycosphingolipids in microdomain formation and their spatial organization. FEBS Lett. 584, 1634–1641. doi: 10.1016/j.febslet.2009.11.070
Han, W.-S., Lee, J., Pham, M.-D., and Yu, J. X. (2010). iGraph: a framework for comparisons of disk-based graph indexing techniques. Proc. VLDB Endow. 3, 449–459.
Han, W.-S., Pham, M.-D., Lee, J., Kasperovics, R., and Yu, J. X. (2011). “iGraph in action,” in SIGMOD '11 (New York, NY: ACM Press), 1241–1242.
Hartmann, M.-A., and Benveniste, P. (1987). “Plant membrane sterols: isolation, identification, and biosynthesis,” in Methods in Enzymology, ed R. D. Lester Packer (New York, NY: Academic Press), 632–650.
He, J.-X., Fujioka, S., Li, T.-C., Kang, S. G., Seto, H., Takatsuto, S., et al. (2003). Sterols regulate development and gene expression in Arabidopsis. Plant Physiol. 131, 1258–1269. doi: 10.1104/pp.014605
Ilangumaran, S., and Hoessli, D. C. (1998). Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem. J. 335, 433–440.
Karnovsky, M. J., Kleinfeld, A. M., Hoover, R. L., and Klausner, R. D. (1982). The concept of lipid domains in membranes. J. Cell Biol. 94, 1–6. doi: 10.1083/jcb.94.1.1
Keinath, N. F., Kierszniowska, S., Lorek, J., Bourdais, G., Kessler, S. A., Shimosato-Asano, H., et al. (2010). PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J. Biol. Chem. 285, 39140–39149. doi: 10.1074/jbc.M110.160531
Kierszniowska, S., Seiwert, B., and Schulze, W. X. (2009). Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-beta-cyclodextrin and quantitative proteomics. Mol. Cell. Proteomics 8, 612–623. doi: 10.1074/mcp.M800346-MCP200
Kjell, J., Rasmusson, A. G., Larsson, H., and Widell, S. (2004). Protein complexes of the plant plasma membrane resolved by Blue Native PAGE. Physiol. Plant 121, 546–555. doi: 10.1111/j.1399-3054.2004.00354.x
Langhorst, M. F., Reuter, A., and Stuermer, C. A. O. (2005). Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell Mol. Life Sci. 62, 2228–2240. doi: 10.1007/s00018-005-5166-4
Lauwers, E., and André, B. (2006). Association of yeast transporters with detergent-resistant membranes correlates with their cell-surface location. Traffic 7, 1045–1059. doi: 10.1111/j.1600-0854.2006.00445.x
Lingwood, D., Kaiser, H.-J., Levental, I., and Simons, K. (2009). Lipid rafts as functional heterogeneity in cell membranes. Biochem. Soc. Trans. 37, 955–960. doi: 10.1042/BST0370955
Lingwood, D., and Simons, K. (2007). Detergent resistance as a tool in membrane research. Nat. Protoc. 2, 2159. doi: 10.1038/nprot.2007.294
Liu, P., Li, R.-L., Zhang, L., Wang, Q.-L., Niehaus, K., Baluska, F., et al. (2009). Lipid microdomain polarization is required for NADPH oxidase-dependent ROS signaling in Picea meyeri pollen tube tip growth. Plant J. 60, 303–313. doi: 10.1111/j.1365-313X.2009.03955.x
Lucero, H. A., and Robbins, P. W. (2004). Lipid rafts-protein association and the regulation of protein activity. Arch. Biochem. Biophys. 426, 208–224. doi: 10.1016/j.abb.2004.03.020
Lüthje, S. (2008). Plasma membrane redox systems: lipid rafts and protein assemblies. Prog. Bot. 69, 169–200. doi: 10.1007/978-3-540-72954-9_7
Lüthje, S., Böttger, M., and Döring, O. (2005). Proton channelling b-type cytochromes in plant plasma membranes? Prog. Bot. 66, 187–217. doi: 10.1007/3-540-27043-4_9
Lüthje, S., Döring, O., Heuer, S., Lüthen, H., and Böttger, M. (1997). Oxidoreductases in plant plasma membranes. Biochim. Biophys. Acta 1331, 81–102. doi: 10.1016/S0304-4157(96)00016-0
Lynch, D. V., Criss, A. K., Lehoczky, J. L., and Bui, V. T. (1997). Ceramide glucosylation in bean hypocotyl microsomes: evidence that steryl glucoside serves as glucose donor. Arch. Biochem. Biophys. 340, 311–316. doi: 10.1006/abbi.1997.9928
Lynch, D. V., and Phinney, A. J. (1995). “The transbilayer distribution of glucosylceramide in plant plasma membrane,” in Plant Lipid Metabolism, eds J.-C. Kader and P. Mazliak (Dordrecht: Springer Netherlands), 239–241.
Marmagne, A., Ferro, M., Meinnel, T., Bruley, C., Kuhn, L., Garin, J., et al. (2007). A high content in lipid-modified peripheral proteins and integral receptor kinases features in the Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 6, 1980–1996. doi: 10.1074/mcp.M700099-MCP200
Mika, A., Minibayeva, F., Beckett, R., and Lüthje, S. (2004). Possible functions of extracellular peroxidases in stress-induced generation and detoxification of active oxygen species. Phytochem. Rev. 3, 173–193. doi: 10.1023/B:PHYT.0000047806.21626.49
Minami, A., Fujiwara, M., Furuto, A., Fukao, Y., Yamashita, T., Kamo, M., et al. (2009). Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimation. Plant Cell Physiol. 50, 341–359. doi: 10.1093/pcp/pcn202
Moreau, P., Hartmann, M., Perret, A., Sturbois-Balcerzak, B., and Cassagne, C. (1998). Transport of sterols to the plasma membrane of leek seedlings. Plant Physiol. 117, 931–937. doi: 10.1104/pp.117.3.931
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Murrell, P., Walton, R., and Murrell, M. P. (2012). Importing Vector Graphics: the grImport package for R. J. Stat. Softw. 30, 1–37.
Navarro-Lérida, I., Sánchez-Perales, S., Calvo, M., Rentero, C., Zheng, Y., Enrich, C., et al. (2012). A palmitoylation switch mechanism regulates Rac1 function and membrane organization. EMBO J. 31, 534–551. doi: 10.1038/emboj.2011.446
Nickels, J. T., and Broach, J. R. (1996). A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 10, 382–394. doi: 10.1101/gad.10.4.382
Parton, R. G., and Hancock, J. F. (2004). Lipid rafts and plasma membrane microorganization: insights from Ras. Trends Cell Biol. 14, 141–147. doi: 10.1016/j.tcb.2004.02.001
Poisson, G., Chauve, C., Chen, X., and Bergeron, A. (2007). FragAnchor: a large-scale predictor of glycosylphosphatidylinositol anchors in eukaryote protein sequences by qualitative scoring. Genomics Proteomics Bioinformatics 5, 121–130. doi: 10.1016/S1672-0229(07)60022-9
Poole, R. L. (2007). The TAIR database. Methods Mol. Biol. 406, 179–212. doi: 10.1007/978-1-59745-535-0_8
Preger, V., Scagliarini, S., Pupillo, P., and Trost, P. (2005). Identification of an ascorbate-dependent cytochrome b of the tonoplast membrane sharing biochemical features with members of the cytochrome b561 family. Planta 220, 365–375. doi: 10.1007/s00425-004-1360-0
Rajendran, L., and Simons, K. (2005). Lipid rafts and membrane dynamics. J. Cell Sci. 118, 1099. doi: 10.1242/jcs.01681
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y., and Yao, X. (2008). CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21, 639–644. doi: 10.1093/protein/gzn039
Robinson, N. J., Groom, S. J., and Groom, Q. J. (1997). The froh gene family from Arabidopsis thaliana: putative iron-chelate reductases. Plant Soil 196, 245–248. doi: 10.1023/A:1004258225806
Robinson, N. J., Procter, C. M., Connolly, E. L., and Guerinot, M. L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. doi: 10.1038/17800
Roche, Y., Gerbeau-Pissot, P., Buhot, B., Thomas, D., Bonneau, L., Gresti, J., et al. (2008). Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. FASEB J 22, 3980–3991. doi: 10.1096/fj.08-111070
Schindler, J., and Nothwang, H. (2006). Aqueous polymer two-phase systems: effective tools for plasma membrane proteomics. Proteomics 6, 5409–5417. doi: 10.1002/pmic.200600243
Schrick, K., Debolt, S., and Bulone, V. (2012a). Deciphering the molecular functions of sterols in cellulose biosynthesis. Front. Plant Sci. 3:84. doi: 10.3389/fpls.2012.00084
Schrick, K., Fujioka, S., Takatsuto, S., Stierhof, Y.-D., Stransky, H., Yoshida, S., et al. (2004a). A link between sterol biosynthesis, the cell wall, and cellulose in Arabidopsis. Plant J. 38, 227–243. doi: 10.1111/j.1365-313X.2004.02039.x
Schrick, K., Karlowski, W., and Mayer, K. (2004b). START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 5:R41. doi: 10.1186/gb-2004-5-6-r41
Schrick, K., Mayer, U., Horrichs, A., Kuhnt, C., Bellini, C., Dangl, J., et al. (2000). FACKEL is a sterol C-14 reductase required for organized cell division and expansion in Arabidopsis embryogenesis. Genes Dev. 14, 1471–1484. doi: 10.1101/gad.14.12.1471
Schrick, K., Mayer, U., Martin, G., Bellini, C., Kuhnt, C., Schmidt, J., et al. (2002). Interactions between sterol biosynthesis genes in embryonic development of Arabidopsis. Plant J. 31, 61–73. doi: 10.1046/j.1365-313X.2002.01333.x
Schrick, K., Shiva, S., Arpin, J. C., Delimont, N., Isaac, G., Tamura, P., et al. (2012b). Steryl glucoside and acyl steryl glucoside analysis of Arabidopsis seeds by electrospray ionization tandem mass spectrometry. Lipids 47, 185–193. doi: 10.1007/s11745-011-3602-9
Shahollari, B., and Berghöfer, T. P. (2004). Receptor kinases with leucine-rich repeats are enriched in Triton X-100 insoluble plasma membrane microdomains from plants. Physiol. Plantarum 122, 397–403. doi: 10.1111/j.1399-3054.2004.00414.x
Simons, K., and Toomre, D. (2000). Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1, 31–39. doi: 10.1038/35036052
Singer, S. J., and Nicolson, G. L. (1972). The fluid mosaic model of the structure of cell membranes. Science 175, 720–731. doi: 10.1126/science.175.4023.720
Souter, M., Topping, J., Pullen, M., Friml, J., Palme, K., Hackett, R., et al. (2002). Hydra mutants of Arabidopsis are defective in sterol profiles and auxin and ethylene signaling. Plant Cell 14, 1017–1031. doi: 10.1105/tpc.001248
Sparla, F., Bagnaresi, P., Scagliarini, S., and Trost, P. (1997). NADH:Fe(III)-chelate reductase of maize roots is an active cytochrome b5 reductase. FEBS Lett. 414, 571–575. doi: 10.1016/S0014-5793(97)01073-9
Staneva, G., Puff, N., Seigneuret, M., Conjeaud, H., and Angelova, M. I. (2012). Segregative clustering of LO and LD membrane microdomains induced by local pH gradients in GM1-containing giant vesicles: a lipid model for cellular polarization. Langmuir 28, 16327–16337. doi: 10.1021/la3031107
Staubach, S., and Hanisch, F.-G. (2011). Lipid rafts: signaling and sorting platforms of cells and their roles in cancer. Expert Rev. Proteomics 8, 263–277. doi: 10.1586/epr.11.2
Stuermer, C. A. O. (2011). Microdomain-forming proteins and the role of the reggies/flotillins during axon regeneration in zebrafish. Biochim. Biophys. Acta 1812, 415–422. doi: 10.1016/j.bbadis.2010.12.004
Stunff, H. L., Milstien, S., and Spiegel, S. (2004). Generation and metabolism of bioactive sphingosine-1-phosphate. J. Cell Biochem. 92, 882–899. doi: 10.1002/jcb.20097
Tanner, W., Malinsky, J., and Opekarová, M. (2011). In plant and animal cells, detergent-resistant membranes do not define functional membrane rafts. Plant Cell 23, 1191–1193. doi: 10.1105/tpc.111.086249
Tanz, S. K., Castleden, I., Hooper, C. M., Vacher, M., Small, I., and Millar, H. A. (2013). SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res. 41, D1185–D1191. doi: 10.1093/nar/gks1151
Team, R. D. C. (2009). R: A Language and Environment for Statistical Computing. Vienna Austria: R Foundation for Statistical Computing 1(09/18/2009).
Thompson, T. E., and Tillack, T. W. (1985). Organization of glycosphingolipids in bilayers and plasma membranes of mammalian cells. Annu. Rev. Biophys. Biophys. Chem. 14, 361–386. doi: 10.1146/annurev.biophys.14.1.361
Trost, P., Bérczi, A., Sparla, F., Sponza, G., Marzadori, B., Asard, H., et al. (2000). Purification of cytochrome b-561 from bean hypocotyls plasma membrane. Evidence for the presence of two heme centers. Biochim. Biophys. Acta 1468, 1–5. doi: 10.1016/S0005-2736(00)00283-2
Van Brocklyn, J. R., and Williams, J. B. (2012). The control of the balance between ceramide and sphingosine-1-phosphate by sphingosine kinase: oxidative stress and the seesaw of cell survival and death. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 163, 26–36. doi: 10.1016/j.cbpb.2012.05.006
Vasilevski, A., Giorgi, F. M., Bertinetti, L., and Usadel, B. (2012). LASSO modeling of the Arabidopsis thaliana seed/seedling transcriptome: a model case for detection of novel mucilage and pectin metabolism genes. Mol. Biosyst. 8, 2566–2574. doi: 10.1039/c2mb25096a
Warnes, G. R., Bolker, B., Bonebakker, L., Gentleman, R., Liaw, W. H. A., Lumley, T., et al. (2013a). gplots: Various R Programming Tools for Plotting Data. R Package Version 2.12.1. Available online at: http://CRAN.R-project.org/package=gplots
Warnes, G. R., Bolker, B., Gorjanc, G., Grothendieck, G., Korosec, A., Lumley, T., et al. (2013b). gdata: Various R Programming Tools for Data Manipulation.
Waters, B. M., Blevins, D. G., and Eide, D. J. (2002). Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition. Plant Physiol. 129, 85–94. doi: 10.1104/pp.010829
Willemsen, V., Friml, J., Grebe, M., van den Toorn, A., Palme, K., and Scheres, B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require sterol methyltransferase1 function. Plant Cell 15, 612–625. doi: 10.1105/tpc.008433
Zauber, H., and Schulze, W. X. (2012). Proteomics wants cracker: automated standardized data analysis of LC–MS derived proteomic data. J. Proteome Res. 11, 5548–5555. doi: 10.1021/pr300413v
Zauber, H., Szymanski, W. G., and Schulze, W. X. (2013). Unraveling sterol-dependent membrane phenotypes by analysis of protein abundance-ratio distributions in different membrane fractions under biochemical and endogenous sterol-depletion. Mol. Cell. Proteomics 12, 3732–3743. doi: 10.1074/mcp.M113.029447
Zhou, H.-W., Nussbaumer, C., Chao, Y., and DeLong, A. (2004). Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell 16, 709–722. doi: 10.1105/tpc.018994
Zidovetzki, R., and Levitan, I. (2007). Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim. Biophys. Acta 1768, 1311–1324. doi: 10.1016/j.bbamem.2007.03.026
Keywords: microdomains, sterols, sphingolipids, signaling
Citation: Zauber H, Burgos A, Garapati P and Schulze WX (2014) Plasma membrane lipid–protein interactions affect signaling processes in sterol-biosynthesis mutants in Arabidopsis thaliana. Front. Plant Sci. 5:78. doi: 10.3389/fpls.2014.00078
Received: 11 December 2013; Accepted: 18 February 2014;
Published online: 18 March 2014.
Edited by:
Sonia Osorio, Málaga University, SpainCopyright © 2014 Zauber, Burgos, Garapati and Schulze. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Waltraud X. Schulze, Plant Systems Biology, University of Hohenheim, Garbenstrasse 30, Stuttgart 70593, Germany e-mail:dy5zY2h1bHplQHVuaS1ob2hlbmhlaW0uZGU=
 Henrik Zauber
Henrik Zauber Asdrubal Burgos
Asdrubal Burgos Prashanth Garapati
Prashanth Garapati Waltraud X. Schulze
Waltraud X. Schulze