- 1Centre for Crop Disease Management, Department of Environment and Agriculture, Curtin University, Bentley, WA, Australia
- 2Plant Sciences Division, Research School of Biology, Australian National University, Canberra, ACT, Australia
- 3School of Chemistry and Biochemistry, University of Western Australia, Perth, WA, Australia
Necrotrophic diseases of wheat cause major losses in most wheat growing areas of world. Tan spot (caused by Pyrenophora tritici-repentis) and septoria nodorum blotch (SNB; Parastagonospora nodorum) have been shown to reduce yields by 10–20% across entire agri-ecological zones despite the application of fungicides and a heavy focus over the last 30 years on resistance breeding. Efforts by breeders to improve the resistance of cultivars has been compromised by the universal finding that resistance was quantitative and governed by multiple quantitative trait loci (QTL). Most QTL had a limited effect that was hard to measure precisely and varied significantly from site to site and season to season. The discovery of necrotrophic effectors has given breeding for disease resistance new methods and tools. In the case of tan spot in West Australia, a single effector, PtrToxA and its recogniser gene Tsn1, has a dominating impact in disease resistance. The delivery of ToxA to breeders has had a major impact on cultivar choice and breeding strategies. For P. nodorum, three effectors – SnToxA, SnTox1, and SnTox3 – have been well characterized. Unlike tan spot, no one effector has a dominating role. Genetic analysis of various mapping populations and pathogen isolates has shown that different effectors have varying impact and that epistatic interactions also occur. As a result of these factors the deployment of these effectors for SNB resistance breeding is more complex. We have deleted the three effectors in a strain of P. nodorum and measured effector activity and disease potential of the triple knockout mutant. The culture filtrate causes necrosis in several cultivars and the strain causes disease, albeit the overall levels are less than in the wild type. Modeling of the field disease resistance scores of cultivars from their reactions to the microbially expressed effectors SnToxA, SnTox1, and SnTox3 is significantly improved by including the response to the triple knockout mutant culture filtrate. This indicates that one or more further effectors are secreted into the culture filtrate. We conclude that the in vitro-secreted necrotrophic effectors explain a very large part of the disease response of wheat germplasm and that this method of resistance breeding promises to further reduce the impact of these globally significant diseases.
Introduction
Effectors are defined as molecules that are produced by microbial pathogens that interact with specific “recognition” gene products in the plant host so as to affect the outcome of the contact – disease susceptibility or resistance (Tyler and Rouxel, 2012; Rovenich et al., 2014; Vleeshouwers and Oliver, 2014). Pathogen species are believed to harbor up to a few hundred such effectors and plant host species contain a much larger number of recognition genes to cope with the plethora of pathogens with which they will come into contact.
Plant pathogens are conventionally described as either biotrophic, in which the infected host tissue remains alive or necrotrophic, where host tissues are killed. In typical biotrophic interactions, recognition of an effector by a specific plant recognition gene (normally termed an R-gene) leads to a defense response and elimination of the pathogen. Recognition of just one such effector is sufficient to render the others redundant (Stotz et al., 2014). Such functional dominance is characteristic of interactions of biotrophic pathogens and accounts for the marked differential between resistance and susceptibility in these diseases.
Necrotrophic pathogens also produce effectors that induce a defense like response upon recognition by an R-gene-like recogniser protein (Lorang et al., 2007; Faris et al., 2010) but are distinguished by their ability to survive in the affected plant tissue and go on to proliferate and sporulate. Indeed, necrotrophic disease is clearly promoted by the recognition of effectors (Oliver and Solomon, 2010).
Like biotrophic pathogens, necrotrophs also produce many effectors. The question we address here is how do the multiple effector/recogniser interactions cooperate to cause disease? Necrotrophic diseases are typically quantitative in nature; the null-hypothesis is that each effector/recogniser interaction operating in a given situation acts additively to produce the degree of necrosis and that this directly translates into a corresponding level of disease. Depending on the context, the level of disease can be defined in terms of either as loss of yield or quantity of pathogen sporulation.
This question has practical importance as effector recognition has been adopted by breeders as a partial proxy for field resistance testing (Vleeshouwers and Oliver, 2014). Can the disease resistance of new cultivars be accurately predicted from the response to effectors of the input germplasm?
Parastagonospora (syn. ana, Stagonospora; teleo, Phaeosphaeria) nodorum (Berk.; Quaedvlieg, Verkley, and Crous) is the causal agent of Septoria nodorum blotch (SNB) on wheat (Solomon et al., 2006a; Quaedvlieg et al., 2013) and is responsible for significant yield losses in some areas of the world (Murray and Brennan, 2009; Oliver et al., 2012). Losses in the West Australian wheat belt amount to greater than AUD$100 m pa. Breeding for disease resistance has been a priority but has been hampered by the quantitative nature of the interaction. Wheat genetic analysis using infection assays as the phenotype has revealed a multitude of quantitative trait loci (QTL) and efforts to define molecular markers acceptable to breeders has proved frustrating (Oliver et al., 2012).
The discovery of multiple necrotrophic effectors has provided a clear framework to dissect the disease and has provided breeders with much needed tools (Friesen et al., 2006, 2008, 2009, 2012; Chu et al., 2010; Faris et al., 2011; Waters et al., 2011; Abeysekara et al., 2012; Crook et al., 2012; Liu et al., 2012; Oliver et al., 2012; Tan et al., 2012; McDonald et al., 2013). Our working hypothesis is that the disease can be explained by the interaction of effectors (which all appear to be small proteins secreted into culture media) and their corresponding recognition genes. Genetic analysis of the response to purified effectors has identified several wheat genetic loci that correspond to regions conferring susceptibility to the disease.
Thus far, three necrotrophic effector genes have been cloned from Parastagonospora nodorum. These are SnToxA (for which the recognition gene Tsn1 has been cloned; Liu et al., 2006; Faris et al., 2010), SnTox3 (Liu et al., 2009), and SnTox1 (Liu et al., 2012). The corresponding recognition genes Snn3 and Snn1 have been mapped but not yet cloned. Furthermore, it is clear that several other effectors operate in this pathosystem (Friesen et al., 2012; Tan et al., 2014; Gao et al., 2015).
We have previously examined the degree of correlation between the effector sensitivities of current cultivars of West Australian cultivars and their reported field resistance (Tan et al., 2014). Unlike the tan spot system for which there is a clear dominance of one effector/recogniser interaction (ToxA/Tsn1; Moffat et al., 2014), no single effector had a similarly dominating role in SNB. One consideration was that all the current cultivars were sensitive to at least one of these three effectors SnToxA, 1 or 3.
Our overall goal is to model the disease susceptibility of wheat cultivars from knowledge of their effector sensitivities. We seek to understand the relative importance of each known effectors, formulate strategies to identify novel effectors/their corresponding host recogniser genes/QTLs and determine how they interact in the SNB interaction.
In this study, we have constructed a P. nodorum strain deleted in SnToxA, SnTox1, and SnTox3. This approach allows us to detect further secreted effectors and determine how recognition corresponds to disease expression without the interference of SnToxA, SnTox1, and SnTox3. We demonstrate that the removal of the three effectors reduced but did not entirely eliminate pathogenicity. We conclude that the secreted necrotrophic effector-recogniser model remains sufficient to explain the disease and provides a useful framework for cultivar resistance breeding.
Materials and Methods
Wheat Cultivars
All wheat cultivars used in this study were obtained from the Australian Winter Cereal Collection (Tamworth, NSW, Australia) and grown in vermiculite in a growth chamber at 21°C with a 12 h light/dark cycle for 2 weeks prior to infection or infiltration. Current SNB disease resistance ratings (DRR) of commercial cultivars were obtained from the Department of Agriculture and Food Western Australia (DAFWA; Shackley et al., 2013). For statistical purposes, a numerical scoring system was assigned to all DRR categories: (1) very susceptible; (2) susceptible–very susceptible; (3) susceptible; (4) moderately susceptible–susceptible; (5) moderately susceptible; (6) moderately resistant–moderately susceptible. Note that no cultivars are scored in categories 7 to 10.
SnToxA, SnTox1, and SnTox3 Triple Gene Deletions in P. nodorum
Parastagonospora nodorum SN15 strains deleted in ToxA, SnTox1, and SnTox3 (toxa13) were created through sequential transformations using homologous-gene knockout vectors that were generated from fusion PCR (Solomon et al., 2006b) and Gibson assembly (Gibson et al., 2009; Table 1). All PCR amplifications were performed with Phusion Taq DNA polymerase (New England Biolabs, Ipswich, MA, USA). The SnTox3 deletion construct harboring a phleomycin resistance cassette described in Tan et al. (2014) was transformed into SN15 tox18 carrying a SnToxA deletion to facilitate gene knockout. PCR was used to identify the appropriate mutants deleted in SnTox3. A robust quantitative PCR was used to determine the integration copy number of SnTox3 deletion constructs in all transformants (Solomon et al., 2008). The mutant toxa3-10 carrying a single copy SnTox3 deletion vector insertion was subsequently selected for SnTox1 deletion (Supplementary Figure S1). The SnTox1 deletion vector was constructed using the Gibson assembly mix (New England Biolabs, Ipswich, MA, USA). The 5′ and 3′ UTR regions of SnTox1 were PCR amplified from genomic DNA using 5_Tox1Fbar, 5_Tox1Rbar, 3_Tox1Fbar, and 3_Tox1Rbar. Both flanking regions were simultaneously fused to the Bar gene (phosphinothricin acetyl transferase) derived from pBARKS1 (obtained from Fungal Genetics Stock Center) and pGEMT-Easy for propagation (Table 1; Figure 1). The resulting gene knockout vector was PCR-amplified for transformation to facilitate gene knockout according to Solomon et al. (2004) with modifications. Glufosinate was extracted from commercial herbicide Basta containing 200 g.l-1 glufosinate ammonium (Bayer Cropscience, Monheim, Germany) using chloroform as described previously (Nayak et al., 2006). An equal volume of chloroform was mixed vigorously with the Basta herbicide and centrifuged at 6,000g for 30 min. The upper aqueous layer containing glufosinate was retained. Bar transformants were selected on minimal medium containing 13 mM NH4Cl as the sole nitrogen source and 8 μl.ml-1 of extracted glufosinate. PCR was used to identify the appropriate mutants deleted in SnTox1. Quantitative PCR was used to determine the integration copy number of SnTox1 deletion constructs in all transformants according to Solomon et al. (2008) with modifications. 5_Tox1qPCRF (5′-CGTAAAGAGCCGAAGATATGCC-3′) and 5_Tox1qPCRR (5′- ATAGCCCAACAGATAGGCCC-3′) were used to amplify 123 bp of the 5′ UTR homologous region immediately adjacent to the Bar marker of the KO cassette. Wild-type SnTox1 was as a standard control for copy number determination of the SnTox1 knockout cassette in the mutants. All fungal strains used in this study were described in Table 2.
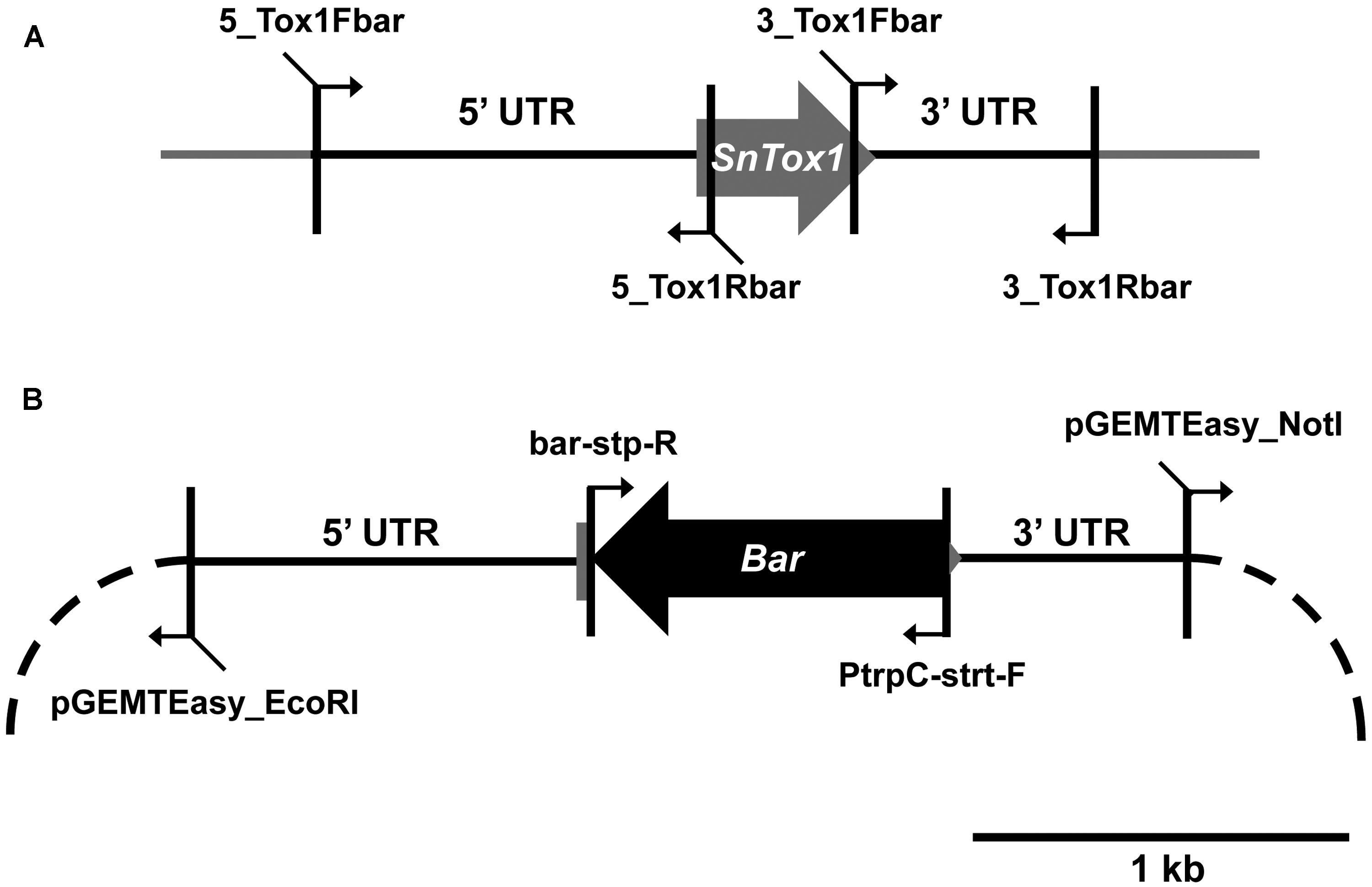
FIGURE 1. Construction of the SnTox1 deletion vector. (A) 5′ and 3′ UTR of SnTox1 were amplified with primers that contain flanking sequences for Bar. (B) All four fragments were simultaneously recombined using the Gibson assembly reaction into pGEMT-Easy (dash). The resulting vector containing the SnTox1-Bar knockout construct was PCR-amplified with 5_Tox1Fbar and 3_Tox1Fbar for transformation in toxa3-10 to result in SnToxA, SnTox1, and SnTox3 deletant mutants.
Production of Necrotrophic Effectors and Infiltration
Necrotrophic effectors were produced from growth in Fries 3 medium broth as described in Liu et al. (2004). Culture filtrate containing effectors were sequentially filtered gauze, miracloth, Whatman paper, and 0.22 μm sterilizers prior to infiltration with a needleless 1 cc syringe. The necrosis reaction was scored at 10 days according to visual score of 0 to 3 as previously described (Waters et al., 2011). A score of 0 indicates insensitivity (no reaction); 1, slight chlorosis; 2, extensive chlorosis; and 3, necrosis. Varieties that scored 1 were considered weakly sensitive, whereas those that scored 2 or 3 were considered highly sensitive to the effector preparation. Infiltration assays were performed on the first leaf of 2-week old seedlings.
Whole Plant Infection Assay
Whole plant infection assay was performed on 2 week old wheat seedlings as described (Solomon et al., 2005). Briefly, 2 week old wheat seedlings were sprayed with 1 × 106 pycnidiospores suspended with 0.5% w/v gelatin using an air brush system. To facilitate the infection process, all seedlings were covered for 2 days to increase humidity. After this, plants were uncovered and the infection process was allowed to continue for an additional 5 days prior the assessment of the disease symptom. A score of 1 indicates no disease symptoms were observed and a score of 9 indicates a fully necrotised plant.
Statistical Analyses
Statistical analysis was performed using JMP 10.0.0 (SAS Institute, Cary, NC, USA) using a 2 × 2 Pearson’s chi-square test was used to test effector sensitivity datasets and SNB DRR for evidence of correlation (Tan et al., 2014). As Chi-square analyses on individual effector sensitivity scores vesus SNB DRR classes resulted in expected values less than one. As such, combining classes was used to overcome this problem on a 2 × 2 Pearson’s chi-square test (Tan et al., 2014). This approach enables a significant association to be demonstrated between SNB DRR and effector insensitivity. Crude culture filtrate sensitivity scores of 2 and 3 were pooled and scores of 0 and 1 were similarly pooled. As previously described in Tan et al. (2014), wheat cultivars that carry SNB DRR of 5 and 6 were pooled separately from scores 1 to 4. Cultivars with mixed effector sensitivity were treated as missing values by the statistical software.
Results
Deletion of SnToxA, SnTox1, and SnTox3 in P. nodorum SN15
SN15 is an aggressive P. nodorum wildtype isolate that carries SnToxA, SnTox3, and SnTox1 (Hane et al., 2007; Syme et al., 2013). To develop a strain deleted for all three genes, we sequentially removed SnTox3 and SnTox1 from P. nodorum tox18, a SN15 transformant deleted in SnToxA (Friesen et al., 2006). Using the previously described SnTox3-knockout vector (Tan et al., 2014), we were able to generate 14 phleomycin resistant transformants, of which two carried the desired SnTox3 deletion as determined by PCR. This represents 14.3% homologous recombination efficiency. We then selected toxa3-10 for SnTox1 deletion. The transformation yielded 27 glufosinate-resistant transformants, of which five contained the desired SnTox1 deletion. This represents 18.5% homologous recombination efficiency. We then selected four SnTox1 knockout strains from the toxa3-10 background to analyse for insert copy number (Supplementary Figure S2). All strains possess a single integration of the SnTox1 knockout-Bar cassette at the SnTox1 locus. From here, toxa13-6 was selected for subsequent analyses.
Characterization of P. nodorum toxa13-6
Parastagonospora nodorum toxa13-6 was tested for its ability to produce SnToxA, SnTox1, and SnTox3 in vitro. Culture filtrate of the mutant was infiltrated into wheat cultivars BG261 (Tsn1, snn1, and snn3), Chinese Spring (tsn1, Snn1, and snn3), and BG220 (tsn1, snn1, and Snn3). Necrotic/chlorotic symptoms that are associated with compatible effector responses were not observed confirming the deletion of the genes (data not shown).
The activity of the toxa13-6 culture filtrate was assessed on 46 Australian commercial wheat cultivars (Figure 2). Ten cultivars are highly sensitive to the culture filtrate, resulting in significant chlorosis and necrosis whereas nine were mildly sensitive. It was observed that Cv. Magenta segregated in sensitivity to the culture filtrate.
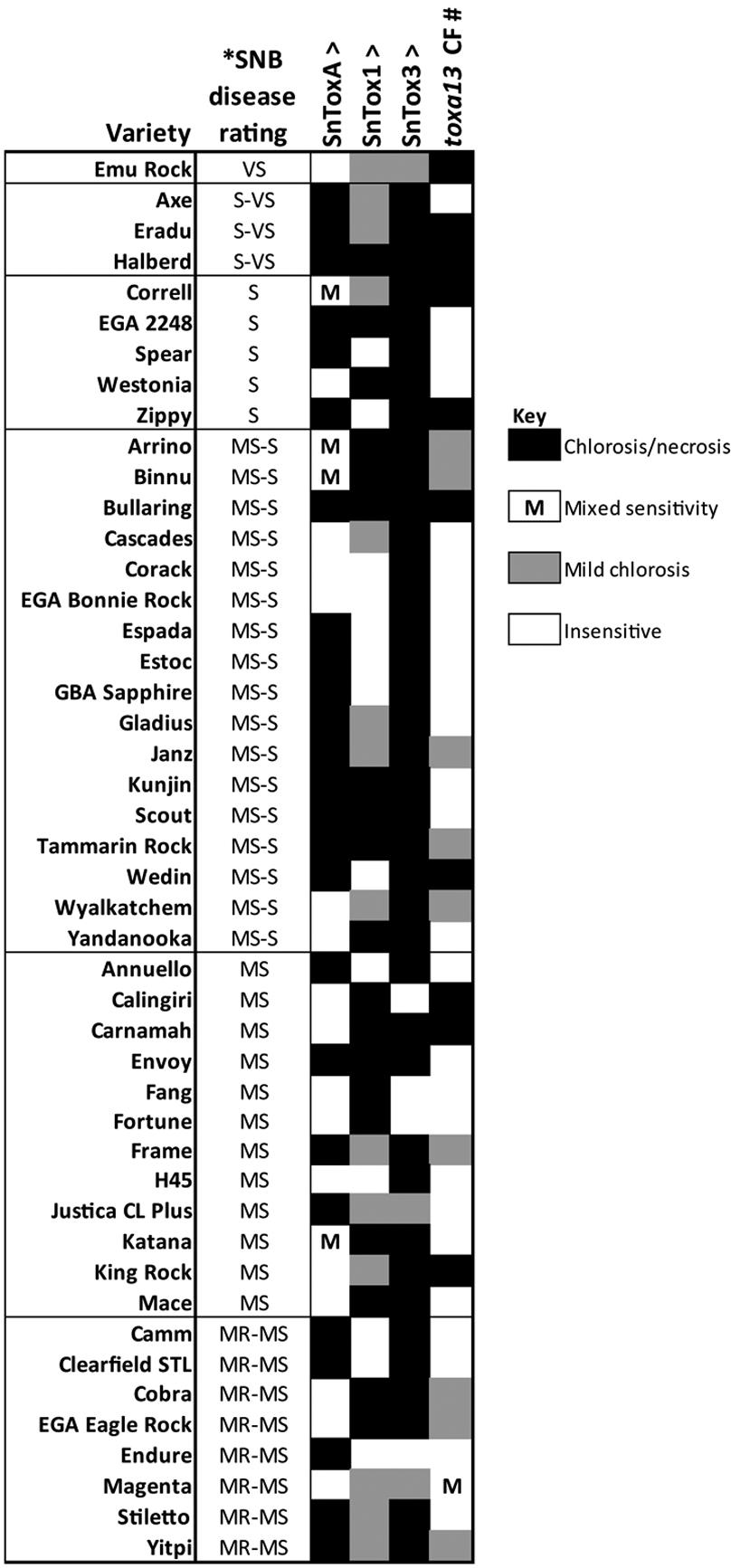
FIGURE 2. Reactions of 46 Australian wheat cultivars to effectors [∧, from Tan et al. (2014)] and the Parastagonospora nodorum toxa13-6 culture filtrate [#, this study]. ∗SNB disease rating was obtained from Shackley et al. (2013). VS, very susceptible; S-VS, susceptible-very susceptible; S, susceptible; MS-S, moderately susceptible-susceptible; MS, moderately susceptible; MR-MS, moderately resistant-moderately susceptible. Effector sensitivity scores are described in Supplementary Table S1.
The triple deletion of SnToxA, SnTox1, and SnTox3 evidently produces further n necrosis-inducing factors. We then compared toxa13-6 culture filtrate sensitivity and SNB DRR using frequency counts in a 2 × 2 mosaic plot (Figure 3A). No significant correlation was observed between toxa13-6 culture filtrate sensitivity and SNB DRR (p = 0.6508). The combined SnToxA, SnTox1, and SnTox3 sensitivity scores correlated with the variety DRRs correlated marginally above the p = 0.05 significance threshold (Figure 3B). However, when toxa13-6 culture filtrate scores were combined with the SnToxA, SnTox1, and SnTox3 sensitivity scores of each wheat variety (Figure 3C), a strongly significant correlation was observed with the SNB DRR (p = 0.0239). This indicates that novel necrosis inducing factors in the toxa13-6 culture filtrate positively contribute to the severity of SNB.
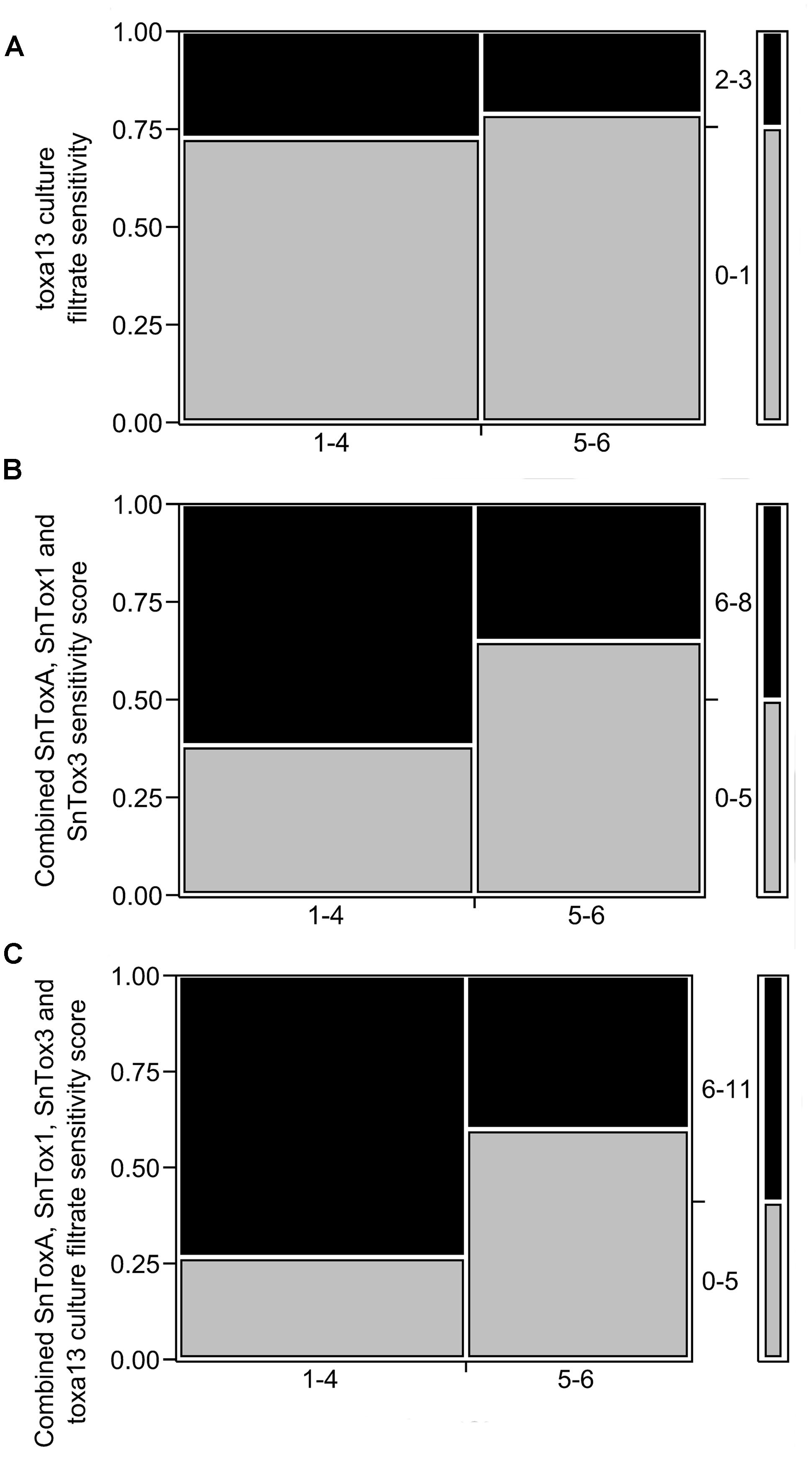
FIGURE 3. Relationship between the distribution of 2013 septoria nodorum blotch (SNB) disease resistance rating (DRR) and reactions to (A) toxa13-6 culture filtrate (p = 0.6553); (B) combined SnToxA, SnTox1, and SnTox3 sensitivity scores (p = 0.0743); and (C) combined SnToxA, SnTox1, SnTox3, and toxa13-6 culture filtrate sensitivity scores (p = 0.0239). The y-axis represents the proportion of effector sensitivity for each DRR score designated on the x-axis. The right column demonstrates the distribution of effector sensitivity scores. Effector sensitivity scores are described in Supplementary Table S1. Statistical analysis was performed using frequency counts in 2 × 2 mosaic plots. The Pearson’s chi-square test was set at a significance threshold of p ≤ 0.05 as previously described (Tan et al., 2014).
The ability of toxa13-6 to infect wheat was assessed using a whole plant spray on selected wheat cultivars that are highly sensitive to the culture filtrate. It was observed P. nodorum toxa13-6 can infect Calingiri, Emu Rock, and Halberd similarly to the wildtype SN15 (Figure 4). The other four P. nodorum toxa13 mutants produced chlorosis/necrosis-inducing culture filtrates and remained infective on Calingiri, Emu Rock, and Halberd (data not shown).

FIGURE 4. Septoria nodorum blotch and culture filtrate symptoms of P. nodorum toxa13-6 on Calingiri, Emu Rock, and Halberd. (A) P. nodorum toxa13 remained pathogenic on Calingiri, Emu Rock, and Halberd. (B) Distinct chlorotic and necrotic symptoms were observed after 10 days post infiltration with P. nodorum toxa13-6 culture filtrate.
Discussion
Classical genetic studies indicate that SNB resistance in wheat is a polygenic trait (Wicki et al., 1999; Czembor et al., 2003). Research since 2001 (reviewed in Oliver et al., 2012) has shown that the SNB interaction involves a complex interplay of fungal effector and host dominant susceptibility genes that are necessary and sufficient to explain the polygenic and quantitative nature of the interaction. Most wheat varieties are sensitive to more than one effector and most pathogen isolates produce more than one effector.
All wheat cultivars used in this study are sensitive to one or more known effectors. This hinders the discovery of novel effector discovery through the use of P. nodorum culture filtrate infiltration. Furthermore, the presence of multiple QTLs which could due to the presence of multiple effector/receptor interactions can make the study of a targeted single interaction difficult as other interactions may introduce bias that mask its effect. Therefore, positional gene cloning may proof impossible under these circumstance. To overcome these difficulties, we developed pathogenic P. nodorum strains that are deleted in SnToxA, SnTox1, and SnTox3 as a tool to discover novel effectors and SNB/sensitivity QTLs in wheat that were previously masked or unassigned. We have achieved this through the use of selectable marker genes that confer resistance to hygromycin (Solomon et al., 2004; Oliver et al., 2012) and phleomycin (Tan et al., 2008). In this study, we have implemented Bar as a third selectable marker for P. nodorum transformation. Bar has been adapted for use as a selectable marker in other fungal system (Avalos et al., 1989; Nayak et al., 2006; Chooi et al., 2010). Nourseothricin and G418 were tested on P. nodorum SN15 as potential antibiotics for fungal transformation using their respective selectable marker genes. However, P. nodorum showed a high level of natural tolerance to these antibiotics and cannot be used for the development of transgene resistance.
The acquisition of the triple effector knockout strain is a tool that can be used to assess the presence of further effectors relevant to commercially important wheat cultivars. These novel effectors can then be identified using biochemical separation methods. In addition, their role in virulence will be assessed through the generation of gene deletion mutants. This approach will require the deletion of additional genes in the P. nodorum toxa13 background. To overcome limitations in marker-based selection, a selectable marker recycling system using Cre-loxP recombination is being developed and adapted for functional gene analysis in P. nodorum (Mizutani et al., 2012). Novel effectors that are verified for their role in the establishment of SNB can be implemented as a tool in resistance breeding (Vleeshouwers and Oliver, 2014).
A broad correlation between disease severity and the additive effect of effectors was observed (Figure 4). Ten wheat varieties showed strong sensitivity reactions to the toxa13-6 culture filtrate. We also demonstrated that three of these wheat cultivars are highly susceptible to SNB caused by the mutant. This clearly indicates that the major effectors are secreted and function as disease determinants (Figure 3). This approach will enable the selection of wheat cultivars that show differential sensitivity to the P. nodorum toxa13 culture filtrate and the construction of wheat mapping populations to genetically identify novel QTLs that confer effector sensitivity and disease susceptibility/resistance. From here, reliable genetic markers that are closely linked with QTLs of interest will be identified and used as a tool in resistance breeding in parallel with effectors to facilitated the ultimate removal of dominant sensitivity traits in wheat (Oliver et al., 2013; Vleeshouwers and Oliver, 2014).
The response of cultivars to necrosis and chlorosis-inducing effectors in the culture filtrate secretome can be compared to the field responses. The correlation between effector sensitivity and DRR is complex. We showed previously that there a significant correlation to SnTox3 (Tan et al., 2014) using DRR data available at that time. Here we show that the best correlation with the current DRR is when reactions to the three cloned effectors plus the culture filtrate from the triple mutants are combined. The correlation is significant (Figure 3B) and so this indicates that breeding by selecting germplasm that is insensitive to the three effectors and the culture remains a viable strategy but that functional redundancy exists between effectors. Purification of the effectors and their individual use should improve the correlation still further.
Nonetheless, whilst the correlation is significant, it is not a simple additive reaction. Epistatic effects have been observed, whereby SnToxA or SnTox1 sensitivity has been found to eliminate the reaction to SnTox3 (Oliver et al., 2012). Different effectors have different effector activity, different variants of effectors have different effector activity and different recogniser genes have different responses (Tan et al., 2012). This indicates that whilst elimination of effector sensitivities from breeding programs will lead ultimately to improved resistance, individual steps may have an impact that is too small to be noticeable or indeed may be zero. Conversely, the impact of elimination of effector sensitivities is predicted never to be negative. Trade-offs in disease resistance between resistance and susceptibility have been found in the case of some genes conferring resistance to biotrophic pathogens, such effects have not yet been found for necrotrophic effectors sensitivities (Oliver et al., 2013). Whilst we need to be vigilant for cases where the elimination of effector sensitivities has a negative pleotropic effect, the current necrotrophic effector model is both necessary and sufficient to explain all that is known about these disease interactions and to inform strategies for disease resistance.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This study was funded by the Grains Research and Development Corporation research grant CUR00023 (Programme 3).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00501
FIGURE S1 | Copy number of the SnTox3-phleomycin resistance gene knockout cassette normalised to a single copy of actin (Act1). The experiment was performed in biological triplicates. Standard error bars are shown.
FIGURE S2 | Copy number of the SnTox1-glufosinate resistance gene knockout cassette normalised to a single copy of actin (Act1). The experiment was performed in biological triplicates. The background strain toxa3-10 was included as a single copy control. The toxa13-2 ectopic mutant was included as a 5’ UTR SnTox1 multicopy control. Standard error bars are shown.
References
Abeysekara, N. S., Faris, J. D., Chao, S., McClean, P. E., and Friesen, T. L. (2012). Whole-genome QTL analysis of Stagonospora nodorum blotch resistance and validation of the SnTox4-Snn4 interaction in hexaploid wheat. Phytopathology 102, 94–104. doi: 10.1094/PHYTO-02-11-0040
Avalos, J., Geever, R. F., and Case, M. E. (1989). Bialaphos resistance as a dominant selectable marker in Neurospora crassa. Curr. Genet. 16, 369–372. doi: 10.1007/BF00340716
Chooi, Y. H., Cacho, R., and Tang, Y. (2010). Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem. Biol. 17, 483–494. doi: 10.1016/j.chembiol.2010.03.015
Chu, C. G., Faris, J. D., Xu, S. S., and Friesen, T. L. (2010). Genetic analysis of disease susceptibility contributed by the compatible Tsn1-SnToxA and Snn1-SnTox1 interactions in the wheat-Stagonospora nodorum pathosystem. Theor. Appl. Genet. 120, 1451–1459. doi: 10.1007/s00122-010-1267-z
Crook, A. D., Friesen, T. L., Liu, Z. H., Ojiambo, P. S., and Cowger, C. (2012). Novel necrotrophic effectors from Stagonospora nodorum and corresponding host sensitivities in winter wheat germplasm in the southeastern United States. Phytopathology 102, 498–505. doi: 10.1094/PHYTO-08-11-0238
Czembor, P. C., Arseniuk, E., Czaplicki, A., Song, Q., Cregan, P. B., and Ueng, P. P. (2003). QTL mapping of partial resistance in winter wheat to Stagonospora nodorum blotch. Genome 46, 546–554. doi: 10.1139/g03-036
Faris, J. D., Zhang, Z., Lu, H., Lu, S., Reddy, L., Cloutier, S., et al. (2010). A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. U.S.A. 107, 13544–13549. doi: 10.1073/pnas.1004090107
Faris, J. D., Zhang, Z., Rasmussen, J. B., and Friesen, T. L. (2011). Variable expression of the Stagonospora nodorum effector SnToxA among isolates is correlated with levels of disease in wheat. Mol. Plant Microbe Interact. 24, 1419–1426. doi: 10.1094/MPMI-04-11-0094
Friesen, T. L., Chu, C. G., Liu, Z. H., Xu, S. S., Halley, S., and Faris, J. D. (2009). Host-selective toxins produced by Stagonospora nodorum confer disease susceptibility in adult wheat plants under field conditions. Theor. Appl. Genet. 118, 1489–1497. doi: 10.1007/s00122-009-0997-2
Friesen, T. L., Chu, C., Xu, S. S., and Faris, J. D. (2012). SnTox5-Snn5: a novel Stagonospora nodorum effector-wheat gene interaction and its relationship with the SnToxA-Tsn1 and SnTox3-Snn3-B1 interactions. Mol. Plant Pathol. 13, 1101–1109. doi: 10.1111/j.1364-3703.2012.00819.x
Friesen, T. L., Faris, J. D., Solomon, P. S., and Oliver, R. P. (2008). Host-specific toxins: effectors of necrotrophic pathogenicity. Cell Microbiol. 10, 1421–1428. doi: 10.1111/j.1462-5822.2008.01153.x
Friesen, T. L., Stukenbrock, E. H., Liu, Z. H., Meinhardt, S., Ling, H., Faris, J. D., et al. (2006). Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956. doi: 10.1038/ng1839
Gao, Y., Faris, J. D., Liu, Z., Kim, Y., Syme, R. A., Oliver, R. P., et al. (2015). Identification and characterization of the SnTox6-Snn6 interaction in the Parastagonospora nodorum - wheat pathosystem. Mol. Plant Microbe Interact. 28, 615–625. doi: 10.1094/MPMI-12-14-0396-R
Gibson, D. G., Young, L., Chuang, R. Y., Venter, J. C., Hutchison, C. A. III, and Smith, H. O. (2009). Enzymatic assembly of, D. NA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. doi: 10.1038/nmeth.1318
Hane, J. K., Lowe, R. G., Solomon, P. S., Tan, K. C., Schoch, C. L., Spatafora, J. W., et al. (2007). Dothideomycete plant interactions illuminated by genome sequencing and EST analysis of the wheat pathogen Stagonospora nodorum. Plant Cell 19, 3347–3368. doi: 10.1105/tpc.107.052829
Liu, Z., Faris, J. D., Oliver, R. P., Tan, K. C., Solomon, P. S., McDonald, M. C., et al. (2009). SnTox3 acts in effector triggered susceptibility to induce disease on wheat carrying the Snn3 gene. PLoS Pathog 5:e1000581. doi: 10.1371/journal.ppat.1000581
Liu, Z. H., Faris, J. D., Meinhardt, S. W., Ali, S., Rasmussen, J. B., and Friesen, T. L. (2004). Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host-selective toxin produced by Stagonospora nodorum. Phytopathology 94, 1056–1060. doi: 10.1094/PHYTO.2004.94.10.1056
Liu, Z., Friesen, T. L., Ling, H., Meinhardt, S. W., Oliver, R. P., Rasmussen, J. B., et al. (2006). The Tsn1-ToxA interaction in the wheat-Stagonospora nodorum pathosystem parallels that of the wheat-tan spot system. Genome 49, 1265–1273. doi: 10.1139/g06-088
Liu, Z., Zhang, Z., Faris, J. D., Oliver, R. P., Syme, R., McDonald, M. C., et al. (2012). The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog 8:e1002467. doi: 10.1371/journal.ppat.1002467
Lorang, J. M., Sweat, T. A., and Wolpert, T. J. (2007). Plant disease susceptibility conferred by a “resistance” gene. Proc. Natl. Acad. Sci. U.S.A. 104, 14861–14866. doi: 10.1073/pnas.0702572104
McDonald, M. C., Oliver, R. P., Friesen, T. L., Brunner, P. C., and McDonald, B. A. (2013). Global diversity and distribution of three necrotrophic effectors in Phaeosphaeria nodorum and related species. New Phytol. 199, 241–251. doi: 10.1111/nph.12257
Mizutani, O., Masaki, K., Gomi, K., and Iefuji, H. (2012). Modified Cre-loxP recombination in Aspergillus oryzae by direct introduction of Cre recombinase for marker gene rescue. Appl. Environ. Microbiol. 78, 4126–4133. doi: 10.1128/AEM.00080-12
Moffat, C. S., See, P. T., and Oliver, R. P. (2014). Generation of a ToxA knockout strain of the wheat tan spot pathogen Pyrenophora tritici-repentis. Mol. Plant Pathol. 15, 918–926.
Murray, G. M., and Brennan, J. P. (2009). Estimating disease losses to the Australian wheat industry. Australas. Plant Pathol. 38, 558–570. doi: 10.1071/AP09053
Nayak, T., Szewczyk, E., Oakley, C. E., Osmani, A., Ukil, L., Murray, S. L., et al. (2006). A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172, 1557–1566. doi: 10.1534/genetics.105.052563
Oliver, R. P., Friesen, T. L., Faris, J. D., and Solomon, P. S. (2012). Stagonospora nodorum: from pathology to genomics and host resistance. Annu. Rev. Phytopathol. 50, 23–43. doi: 10.1146/annurev-phyto-081211-173019
Oliver, R., Lichtenzveig, J., Tan, K. C., Waters, O., Rybak, K., Lawrence, J., et al. (2013) Absence of detectable yield penalty associated with insensitivity to Pleosporales necrotrophic effectors in wheat grown in the West Australian wheat belt. Plant Pathol. 63, 1027–1032. doi: 10.1111/ppa.12191
Oliver, R. P., and Solomon, P. S. (2010). New developments in pathogenicity and virulence of necrotrophs. Curr. Opin. Plant Biol. 13, 415–419. doi: 10.1016/j.pbi.2010.05.003
Quaedvlieg, W., Verkley, G. J., Shin, H. D., Barreto, R. W., Alfenas, A. C., Swart, W. J., et al. (2013). Sizing up septoria. Stud. Mycol. 75, 307–390. doi: 10.3114/sim0017
Rovenich, H., Boshoven, J. C., and Thomma, B. P. (2014). Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol. 20, 96–103. doi: 10.1016/j.pbi.2014.05.001
Shackley, B., Zaicou-Kunusch, C., Dhammu, H., Shankar, M., Amjad, M., and Young, K. (2013). Wheat Variety Guide for WA 2013. Perth, WA: Department of Agriculture and Food.
Solomon, P. S., IpCho, S. V. S., Hane, J. K., Tan, K. C., and Oliver, R. P. (2008). A quantitative PCR approach to determine gene copy number. Fungal Genet. Rep. 55, 5–8.
Solomon, P. S., Lowe, R. G. T., Tan K-C, Waters, O. D. C., and Oliver, R. P. (2006a). Stagonospora nodorum: cause of stagonospora nodorum blotch of wheat. Mol. Plant Pathol. 7, 147–156. doi: 10.1111/j.1364-3703.2006.00326.x
Solomon, P. S., Rybak, K., Trengove, R. D., and Oliver, R. P. (2006b). Investigating the role of calcium/calmodulin-dependent protein kinases in Stagonospora nodorum. Mol. Microbiol. 62, 367–381. doi: 10.1111/j.1365-2958.2006.05380.x
Solomon, P. S., Tan, K. C., and Oliver, R. P. (2005). Mannitol 1-phosphate metabolism is required for sporulation in planta of the wheat pathogen Stagonospora nodorum. Mol. Plant Microbe Interact. 18, 110–115. doi: 10.1094/MPMI-18-0110
Solomon, P. S., Tan, K. C., Sanchez, P., Cooper, R. M., and Oliver, R. P. (2004). The disruption of a Galpha subunit sheds new light on the pathogenicity of Stagonospora nodorum on wheat. Mol. Plant Microbe Interact. 17, 456–466. doi: 10.1094/MPMI.2004.17.5.456
Stotz, H. U., Mitrousia, G. K., de Wit, P. J., and Fitt, B. D. (2014). Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 19, 491–500. doi: 10.1016/j.tplants.2014.04.009
Syme, R. A., Hane, J. K., Friesen, T. L., and Oliver, R. P. (2013). Resequencing and comparative genomics of Stagonospora nodorum; sectional gene absence and effector discovery. G3(Bethesda) 3, 959–969. doi: 10.1534/g1533.1112.004994
Tan, K. C., Ferguson-Hunt, M., Rybak, K., Waters, O. D., Stanley, W. A., Bond, C. S., et al. (2012). Quantitative variation in effector activity of ToxA isoforms from Stagonospora nodorum and Pyrenophora tritici-repentis. Mol. Plant Microbe Interact. 25, 515–522. doi: 10.1094/MPMI-10-11-0273
Tan, K. C., Heazlewood, J. L., Millar, A. H., Thomson, G., Oliver, R. P., and Solomon, P. S. (2008). A signaling-regulated, short-chain dehydrogenase of Stagonospora nodorum regulates asexual development. Eukaryot. Cell 7, 1916–1929. doi: 10.1128/EC.00237-08
Tan, K. C., Waters, O. D. C., Rybak, K., Antoni, E., Furuki, E., and Oliver, R. P. (2014). Sensitivity to three Parastagonospora nodorum necrotrophic effectors in current Australian wheat cultivars and the presence of further fungal effectors. Crop Pasture Sci. 65, 150–158. doi: 10.1071/CP13443
Tyler, B. M., and Rouxel, T. (2012). “Effectors of fungi and oomycetes: their virulence and avirulence functions and translocation from pathogen to host cells,” in Molecular Plant Immunity, ed. G. Sessa (Oxford: Wiley-Blackwell). doi: 10.1002/9781118481431.ch7
Vleeshouwers, V. G., and Oliver, R. P. (2014). Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 27, 196–206. doi: 10.1094/MPMI-10-13-0313-IA
Waters, O. D. C., Lichtenzveig, J., Rybak, K., Friesen, T. L., and Oliver, R. P. (2011). Prevalence and importance of sensitivity to the Stagonospora nodorum necrotrophic effector SnTox3 in current Western Australian wheat cultivars. Crop Pasture Sci. 62, 556–562. doi: 10.1071/CP11004
Keywords: septoria, nodorum, stagonospora, phaeosphaeria, necrotrophic fungi, effectors
Citation: Tan K-C, Phan HTT, Rybak K, John E, Chooi YH, Solomon PS and Oliver RP (2015) Functional redundancy of necrotrophic effectors – consequences for exploitation for breeding. Front. Plant Sci. 6:501. doi: 10.3389/fpls.2015.00501
Received: 22 April 2015; Accepted: 22 June 2015;
Published: 08 July 2015.
Edited by:
Pietro Daniele Spanu, Imperial College London, UKReviewed by:
Guus Bakkeren, Agriculture and Agri-Food Canada, CanadaMahmut Tör, University of Worcester, UK
Copyright © 2015 Tan, Phan, Rybak, John, Chooi, Solomon and Oliver. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard P. Oliver and Kar-Chun Tan, Centre for Crop Disease Management, Department of Environment and Agriculture, Curtin University, Bentley, WA 6102, Australia,cmljaGFyZC5vbGl2ZXJAY3VydGluLmVkdS5hdQ==;a2FyLWNodW4udGFuQGN1cnRpbi5lZHUuYXU=
†These authors have contributed equally to this work.
 Kar-Chun Tan
Kar-Chun Tan Huyen T. T. Phan
Huyen T. T. Phan Kasia Rybak
Kasia Rybak Evan John1
Evan John1 Yit H. Chooi
Yit H. Chooi Peter S. Solomon
Peter S. Solomon Richard P. Oliver
Richard P. Oliver
