- 1National Institute of Plant Genome Research, New Delhi, India
- 2Central Institute of Medicinal and Aromatic Plants, Council of Scientific and Industrial Research, Lucknow, India
- 3Department of Biological Chemistry, David Geffen School of Medicine, University of California, Los Angeles, Los Angeles, CA, USA
α-Mannosidase (α-Man), a fruit ripening-specific N-glycan processing enzyme, is involved in ripening-associated fruit softening process. However, the regulation of fruit-ripening specific expression of α-Man is not well understood. We have identified and functionally characterized the promoter of tomato (Solanum lycopersicum) α-Man to provide molecular insights into its transcriptional regulation during fruit ripening. Fruit ripening-specific activation of the α-Man promoter was revealed by analysing promoter driven expression of beta-glucuronidase (GUS) reporter in transgenic tomato. We found that RIPENING INHIBITOR (RIN), a MADS box family transcription factor acts as positive transcriptional regulator of α-Man during fruit ripening. RIN directly bound to the α-Man promoter sequence and promoter activation/α-Man expression was compromised in rin mutant fruit. Deletion analysis revealed that a promoter fragment (567 bp upstream of translational start site) that contained three CArG boxes (binding sites for RIN) was sufficient to drive GUS expression in fruits. In addition, α-Man expression was down-regulated in fruits of Nr mutant which is impaired in ethylene perception and promoter activation/α-Man expression was induced in wild type following treatment with a precursor of ethylene biosynthesis, 1-aminocyclopropane-1-carboxylic acid (ACC). Although, α-Man expression was induced in rin mutant after ACC treatment, the transcript level was less as compared to ACC-treated wild type. Taken together, these results suggest RIN-mediated direct transcriptional regulation of α-Man during fruit ripening and ethylene may acts in RIN-dependent and -independent ways to regulate α-Man expression.
Introduction
Tomato (Solanum lycopersicum) is an important component of human diet and also serves a model for biochemical and genetic analysis of fleshy fruit development and ripening process. The ripening of fleshy fruits is accompanied by a number of biochemical events, including changes in color, sugar, acidity, texture, and aroma volatiles that are crucial for the development of sensory qualities of fruits (Klee and Giovannoni, 2011; Osorio et al., 2012; Seymour et al., 2013a,b). During ripening, fruit textural changes are brought about by concerted and coordinated activities of the enzymes that act upon cell wall components such as cellulose, hemicellulose, pectin and N-glycoproteins (Giovannoni, 2001; Brummell, 2006; Meli et al., 2010; Klee and Giovannoni, 2011). An earlier report suggested that blocking of N-glycosylation can lead to delay of fruit ripening (Handa et al., 1985). Moreover, during ripening, tomato pericarp has been shown to accumulate high amount of free N-glycans as precursors of N-glycosylation or as a result of N-glycoprotein degradation (Priem et al., 1993; Nakamura et al., 2008; Hossain et al., 2009). These free N-glycans can also stimulate fruit ripening by inducing ethylene biosynthesis and signaling (Priem and Gross, 1992). A more mechanistic insight on the role of N-glycans in fruit ripening was obtained by studying two N-glycan processing enzymes, α-mannosidase (α-Man) and β-D-N-acetylhexosaminidase (β-Hex). The suppression of expression of these enzymes during tomato ripening had resulted in enhancement of fruit shelf life due to reduced softening of fruits during ripening (Meli et al., 2010; Ghosh et al., 2011; Cao et al., 2014). Although, regulation of β-Hex expression during fruit ripening was being studied in detail (Irfan et al., 2014); our knowledge of how α-Man expression is regulated during fruit ripening is still not clear.
α-Man (EC 3.2.1.24) is a member of the glycosyl hydrolase 38 (GH38) family carbohydrate acting enzymes reported in animals, plants and microorganisms. It cleaves terminal α-mannosidic linkages from both the high mannose type and complex type N-glycans present in glycoproteins (Strasser et al., 2006; Hossain et al., 2009; Liebminger et al., 2009; Hüttner et al., 2014). α-Man activity was shown to increase during ripening of several fleshy fruits such as tomato, capsicum, mango, papaya (Priya Sethu and Prabha, 1997; Suvarnalatha and Prabha, 1999; Hossain et al., 2009; Meli et al., 2010; Ghosh et al., 2011). The genes encoding fruit ripening-specific α-Man have been identified and characterized from tomato and capsicum (Hossain et al., 2010; Meli et al., 2010; Ghosh et al., 2011). Functional characterization of α-Man in climacteric fruit tomato and non-climacteric fruit capsicum revealed its involvement in ripening-associated fruit softening (Meli et al., 2010; Ghosh et al., 2011). Interestingly, the genes encoding enzymes involved in cell wall loosening, e.g., pectin methyl esterase, glucan endo1,3-β-D-glucosidase, β 1,3 glucanse, endo-xyloglucan transferase, pectin esterase, expansions, pectin acetyl esterase, α-galactosidase, pectate lyase, (1-4)-β-mannan endohydrolase and β-galactosidase were down-regulated in α-Man-RNAi tomato fruits (Meli et al., 2010). Moreover, the expression of ethylene biosynthesis genes (ACC synthase, and ACC oxidase) and transcription factors like ERFs were also down-regulated in α-Man-suppressed tomato fruits, suggesting that α-Man-mediated cleavage of terminal mannose residues attached to the cell wall N-glycoproteins may be involved in positive feed-back regulation of fruit ripening (Meli et al., 2010).
The MADS box family transcription factor, RIN (RIPENING INHIBITOR) plays a pivotal role in fruit ripening by controlling the transcription of a large number of genes involved in cellular signaling and metabolism (Giovannoni, 2007; Ito et al., 2008; Fujisawa et al., 2011, 2013; Zhong et al., 2013; Kumar et al., 2015). Tomato rin mutant fruit was characterized by enlarged sepals and completely inhibited ripening process, due to the abnormal expression of two genes encoding the RIN and MC (MACROCALYX). Although RIN regulates fruit ripening, MC is involved in sepal development (Vrebalov et al., 2002). RIN-dependent transcriptional regulation of fruit ripening has been well studied following chromatin immunoprecipitation, transcriptome and proteome analyses. This led to identification of several direct and indirect targets of RIN including those involved in biosynthesis and signaling of phytohormone ethylene, cell wall modification, and accumulation of carotenoid and aroma volatiles. (Ito et al., 2008; Fujisawa et al., 2011, 2012, 2013; Martel et al., 2011; Kumar et al., 2012, 2015; Qin et al., 2012; Zhong et al., 2013). Besides, the expression of some key transcriptional regulators namely NON-RIPENING, COLORLESS NON-RIPENING, FRUITFULL1 was also found to be under the transcriptional control of RIN (Fujisawa et al., 2011, 2012, 2013, 2014; Martel et al., 2011).
A strong correlation between α-Man transcript and protein accumulation patterns in tomato fruits (Meli et al., 2010), suggested that the fruit ripening-specific activity of α-Man is under the transcriptional control of gene expression. Therefore, to gain an insight into transcriptional regulation of α-Man during fruit ripening, the promoter of tomato α-Man was isolated and functionally characterized. The activation of α-Man promoter during fruit development and ripening was studied by analyzing promoter-driven expression of beta-glucuronidase (GUS) reporter in transgenic tomato plants. The results suggest that ethylene and RIN play important role in controlling the transcription of α-Man during fruit ripening.
Materials and Methods
Plant Materials and Growth Conditions
Seeds of Solanum lycopersicum cv. Pusa Ruby were collected from the National Seeds Corporation Ltd, New Delhi and Solanum lycopersicum cv. Ailsa Craig, rin and Nr mutants were obtained from Tomato Genetics Resource Center, University of California at Davis. Seeds were germinated in small pots and after 3 weeks, seedlings were transplanted to big pots in the greenhouse conditions; 25/25°C temperature, 70% humidity under 14/10 h light/dark regime. Flowers were tagged at anthesis and fruits from various developmental stages [3, 5, 10, 15, and 20 days after anthesis (DAA)] and ripening stages [mature green (MG), breaker (BR), pink (P), and red ripe (RR)] were harvested for various type analysis of fruits.
Isolation and In Silico Analysis of α-Man Promoter
The Universal GenomeWalkerTM Kit (Clontech, USA) was used for isolation of α-Man promoter from tomato. Genomic DNA was extracted from leaves by using cetyl trimethyl ammonium bromide (CTAB) method described by Doyle and Doyle (1987). Five genome walking libraries were prepared by digesting genomic DNA separately with PvuII, XmnI, MscI, DraI, and SspI enzymes. After that, PCR was carried out with a GenomeWalker adapter-specific primer (AP1) and a gene-specific primer (GSP1) separately for each library. This PCR product was then used as a template for nested amplification by using AP2 and GSP2 primers followed by cloning in pGEM-T Easy vector and then sequencing the clones. The promoter sequence thus obtained was further verified with sequence available on www.solgenomics.net. Tomato α-Man promoter sequence was analyzed in silico to find out putative cis-acting elements using NewPLACE (Higo et al., 1999), PlantCARE (Lescot et al., 2002), and MatInspector (Cartharius et al., 2005) servers, and the FUZZNUC program (EMBOSS package; Rice et al., 2000).
Construction of Promoter::GUS Fusion and Deletion Vectors
Tomato α-Man promoter::GUS fusion construct (MP::GUS) and deletion constructs of α-Man promoter were prepared in binary vector pBI121 by replacing CaMV 35S promoter with α-Man promoter. Tomato full length α-Man promoter (1155 bp) and deletion fragments of α-Man promoter (-779 bp, -567 bp, -373 bp, and -187 bp region from translational start site of α-Man) were PCR amplified using high fidelity pfx DNA polymerase to incorporate appropriate restriction sites. Further, these amplified regions were cloned upstream to ATG of GUS gene in pBI121 binary vector following standards restriction digestion and ligation methods. All these constructs were named as MP::GUS (with 1155 bp long promoter), MD1::GUS (with (779 bp fragment of α-Man promoter), MD2::GUS (567 bp fragment of α-Man promoter), MD3::GUS (373 bp of α-Man promoter) and MD4::GUS (187 bp of α-Man promoter) respectively. The positive clones were transformed into Agrobacterium tumefaciens (strain EHA105) following electroporation. Transformation in Agrobacterium was confirmed by colony PCR using all these plasmid DNA as template and set of primers enlisted in Supplementary Table S2.
Agrobacterium-Based Transient Assay
Agrobacterium-based transient assay was carried out as described previously (Orzaez et al., 2006) with few modifications. In the pericarp of tomato fruits (Solanum lycopersicum cv. Pusa Ruby) of mature green stage, Agrobacterium suspension was infiltrated. In brief, Agrobacterium cultures (3 mL) were grown overnight from individual colonies (transformed with appropriate construct) at 28°C in YEP medium plus selective antibiotics. Two hundred micro liter of it was transferred to 50 mL induction medium (0.5% beef extract, 0.1% yeast extract, 0.5% Peptone, 0.5% Sucrose, 2 mM MgSO4, 20 mM acetosyringone, 10 mM MES, pH 5.6) plus antibiotics, and again grown overnight until the OD600 of the culture reached 0.8–1.0. After centrifugation, cultures were recovered, resuspended in infiltration medium (10 mM MgCl2, 10 mM MES, 200 mM acetosyringone, pH 5.6) and incubated at room temperature with gentle agitation (20 rpm) for at least 2 h and then infiltrated into the fruits.
Stable Transformation of Tomato
The transgenic tomato plants were developed as described previously (Fillati et al., 1987) with few modifications. Initially, seeds were sterilized using 4% commercial bleach and kept on Murashige and Skoog (MS) medium for germination. Cotyledons from 2-week-old seedlings were cut and co-cultivated with Agrobacterium transformed with appropriate construct in MS medium containing acetosyringone (0.1 μM). Cotyledons were dried and then kept for selection on MS plates containing kanamycin (50 mgl-1), cefotaxime (250 mgl-1) and zeatine (1 ngl-1). After few days, regenerated plantlets were transferred to rooting medium [MS containing kanamycin (50 mgl-1), cefotaxime (250 mgl-1) and IAA (1 ngl-1)]. Transgenic seeds were collected and then germinated in MS medium containing kanamycin (50 mgl-1) to get the progeny plants.
RNA Isolation and qRT-PCR
RNA was extracted as protocol described previously (Menke et al., 1999) and purified using the RNeasy Mini Kit (Qiagen). Quantification of total RNA was carried out using a nanodrop (ND 1000) and five micrograms of RNA was reverse transcribed to cDNA using superscript II RT (Invitrogen). qRT-PCR analysis was performed using One Step Real Time RT PCR (Applied Biosystems) with SYBR Green as described by previously (Ghosh et al., 2013). All the qRT-PCR analysis was performed in triplicate from cDNA derived from at least two independent experiments. The data was analyzed by using the 2-ΔΔCT method (Bovy et al., 2002) and presented as fold change in gene expression or percentage of expression, normalized to the endogenous control (tomato actin gene). The primers used for qRT-PCR reaction are listed in Supplementary Table S2.
GUS Histochemical and Fluorometric Assay
Detailed histochemical analysis of the reporter gene (GUS) was carried out as method described by Jefferson et al. (1987) with few modifications. The transverse sections of fruits from different development and ripening stages, seedlings, roots, leaves, and flowers were dipped in GUS staining solution (100 mM sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% triton-X 100, 20% methanol and 1 mM X-gluc). After vacuum infiltration, the plant materials were left for overnight at 37°C in darkness. To remove chlorophyll, samples were destained in 75% ethanol and then photographs were taken by using Canon G6 powershot with 4X zoom or Canon EOS 400D DIGITAL (10.1 megapixel) and Nikon AZ100 5X microscope. Further, GUS activity was quantified by measuring the production of 4-methylumbelliferone (4-MU) as pmol 4-MU mg-1min-1. To do this, samples were homogenized in 400 μl GUS extraction buffer (50 mM sodium phosphate buffer, pH 7.0, 10 mM DTT, 10 mM EDTA, 0.1% sodium lauryl sarcosine, 0.1% triton-X 100) and then centrifuged. After that, 50 μl of supernatant was mixed to 450 μl MUG assay buffer (GUS extraction buffer containing 10 mM MUG) and incubated at 37°C for 1 h. The reaction was terminated by mixing 100 μl of aliquots with 900 μl 0.2 M Na2CO3. The fluorescence was recorded in fluorometer (Cary Eclipse, Varian) with excitation at 380 nm and emission at 454 nm. GUS activity was calculated as pmoles MU mg protein-1min-1.
Electrophoretic Mobility Shift Assay
EMSA was performed for in vitro binding of RIN (AF448522.1) protein with α-Man promoter as methods described previously (Irfan et al., 2014) with minor modifications. Briefly, 167 bp long fragment of α-Man promoter upstream of ATG of gene was PCR amplified by using primers listed in Supplementary Table S1. HindIII/XbaI digested α-Man promoter fragment was end filled with [α-P32]dCTP (3000 Ci/mmol, 50 μCi), using DNA polymerase I (Klenow) fragment (New England Biolabs) and purified using sephadex G-50 column. EMSA was performed with [α-P32]dCTP labeled α-Man promoter fragment incubated with RIN protein purified by Irfan et al. (2014) in gel shift assay binding buffer (20 mM HEPES, pH 7.5, 20% glycerol, 0.05 μg poly(dIdC):poly(dIdC), 10 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT and 25 mM NaCl) at 25°C for 30 min. For competition assay, unlabeled promoter fragment was used as specific competitive inhibitor and an unrelated DNA [200 bp region downstream of ATG of tomato actin (FJ532351.1)] was used as non-specific competitor. After incubation for 30 min, reaction was loaded onto 6% native PAGE. The gel was run at room temperature at constant current of 10 mA using 0.5X TBE running buffer. The protein-DNA complexes as well as free probes were visualized by autoradiography.
ACC Treatment
Three weeks old tomato seedlings, germinated on MS media were transferred to liquid MS media containing 1 mM ACC. The tissue was harvested at different time points and frozen immediately in liquid nitrogen. Seedlings transferred to MS liquid media without ACC were used as control.
Results
Isolation and In Silico Analysis of α-Man Promoter
Analysis of the tomato genomic sequence1 revealed that α-Man gene (Solyc06g068860.2.1) contains 30 exons interrupted by 29 introns (Figure 1A). The upstream sequence of tomato α-Man was initially isolated by directional genome walking PCR using a set of adapter and gene-specific primers based on the cDNA sequence information (EU244853) (Supplementary Figure S1). The sequence (1155 bp) upstream of the translational start site, obtained from the genome walking PCR, was verified with genomic sequence of tomato α-Man available on solgenomics network database1. In order to identify putative cis-acting regulatory elements that might control the transcription of α-Man, in silico analysis of upstream sequence, i.e., putative promoter sequence, was carried out by using NewPLACE2, PlantCARE (bioinformatics.psb.ugent.be), and MatInspector3. In addition to the ubiquitous elements including TATA, CAAT boxes, the promoter region of tomato α-Man contains sequences similar to the cis-acting regulatory elements found within the promoter of other plant genes (Supplementary Table S1). The promoter region of α-Man contains several putative functional cis-acting elements recognized by the transcription factors, which may be involved in perceiving stimulus from different plant hormones and environmental stresses (Supplementary Table S1). Interestingly, in silico analysis also revealed the presence of RIN binding sites (CArG boxes) within the α-Man promoter (Supplementary Table S1) which suggested the possible role of RIN in the regulation of α-Man transcript expression during fruit ripening.
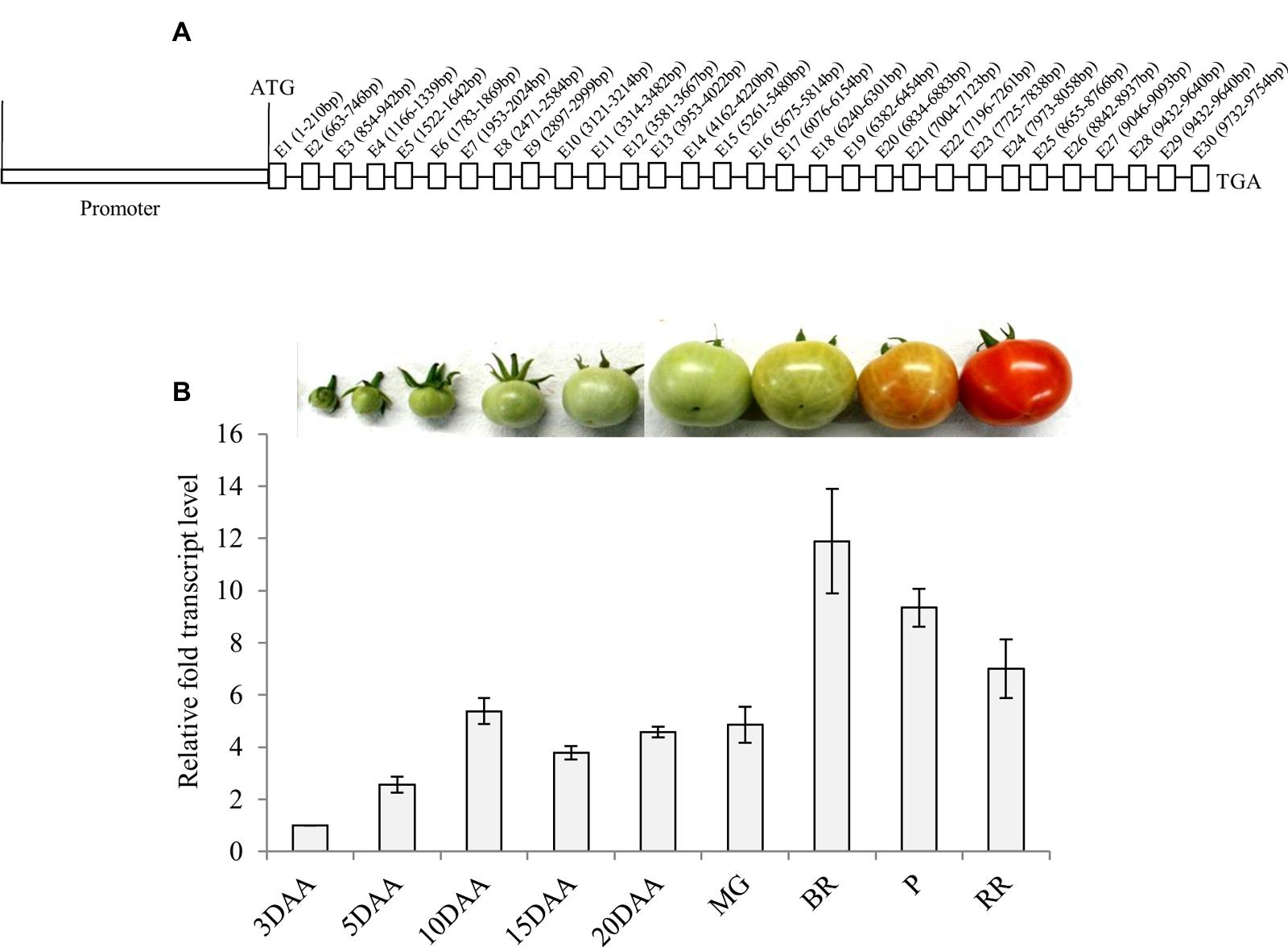
FIGURE 1. α-Man genomic organization and transcript abundance pattern during tomato fruit development and ripening. (A) Genomic organization of α-Man. Introns on α-Man gene were identified following intron finder tool (solgenomics.net). (B) Expression analysis of α-Man during fruit development and ripening. RNA was isolated from fruits of various developmental stages (3DAA–20DAA) and ripening stages (MG, BR, P and RR). The transcript level of α-Man was determined by qRT-PCR analysis. Tomato actin was used as endogenous control. Data are mean (±SE) of three biological replicates.
Histochemical, Fluorometric and Transcript Analysis of α-Man Promoter::GUS Transgenic Lines
Fruit-specific expression of α-Man during tomato ripening, with maximum transcript expression at the breaker stage, was reported earlier following northern blot analysis (Meli et al., 2010). To validate the expression pattern of α-Man during fruit ripening, transcript level was quantified by qRT-PCR during tomato fruit development and ripening stages. The results of qRT-PCR analysis corroborated earlier findings that α-Man transcript accumulates during ripening with maximum level at breaker stage (Figure 1B). However, during fruit developmental stages (3DAA, 5DAA, 10DAA, 15DAA, 20DAA) a basal level expression of α-Man was noticed (Figure 1B). In order to gain further insight into fruit ripening-specific activation, α-Man promoter driven expression of the GUS reporter was studied in transgenic tomato plants. α-Man promoter (-1 to -1155 bp upstream of ATG) was cloned into pBI121 binary vector in order to make MP::GUS fusion construct (Figure 2A). Fifteen independent transgenic events, developed with MP::GUS fusion construct were advanced for T2 generation and confirmed by PCR (Supplementary Figure S2). T2 transgenic events with single transgene copy insertion were chosen for further analysis. To examine the tissue-specific expression pattern, fruits at different development [10, 15, and 20 days after (DAA)] and ripening (mature green, breaker, pink, and red ripe) stages, leaves, roots, and flowers from transgenic plants were subjected to histochemical GUS staining as described in Material and methods (Figures 2B and 3A). Intense GUS staining of fruits during ripening with a peak at breaker stage was observed when GUS expression was driven by α-Man promoter in transgenic fruits (Figure 2B). GUS activity was apparently not detectable by visual observations of seedlings, leaves, stems and roots of transgenic plants transformed with MP::GUS fusion construct (Figure 3A). The flowers of transgenic plants harboring α-Man promoter showed very less GUS staining mainly in sepals, while there was no GUS staining in petals (Figure 3A). The results obtained from histochemical GUS staining were further validated by quantifying GUS activity through fluorometric MUG (4-methylumbelliferone glucuronide) assay. In α-Man promoter transgenic fruits, GUS activity was higher at breaker stage of fruit ripening as compared with other stages (Figure 2C). The CaMV 35S::GUS transgenic plants showed constitutive GUS activity in fruits, seedlings, leaves, stem, roots, and flowers (Figures 2B,C and 3A,B) whereas wild type (cv. Pusa Ruby) plants did not show GUS activity in any parts of the plant (Figure 2B). To further corroborate these results, GUS transcript accumulation pattern was studied following qRT-PCR. Maximum level of GUS mRNA accumulated during fruit ripening stages which was in accordance with GUS histochemical and fluorometric results (Figure 2C). Therefore, 1155 bp sequence upstream sequence from ATG can be regarded as the full-length α-Man promoter. The promoter sequence contains cis-acting elements that may be involved in fruit-ripening specific expression of α-Man (Supplementary Table S1) and the promoter was able to drive tissue-specific expression of GUS similar to that observed for the endogenous expression of α-Man (Figures 1B and 2B,C).
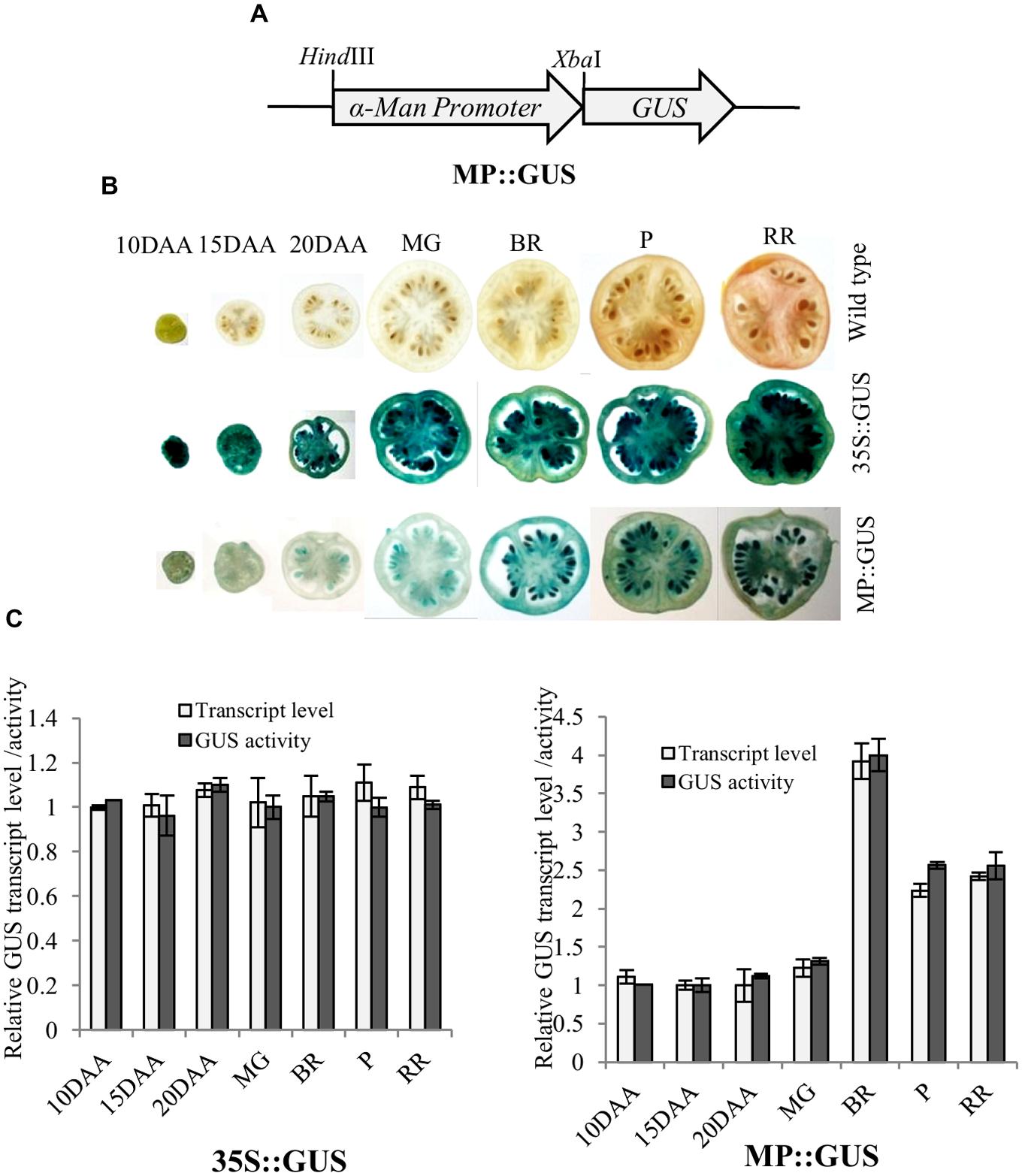
FIGURE 2. Histochemical and fluorometric assays of GUS activity in fruits. (A) Schematic representation of MP::GUS fusion construct (GUS fused with α-Man promoter). (B) Histochemical GUS staining was carried out in fruits of different developmental and ripening stages. MP::GUS and 35S::GUS denote MP and CaMV35S promoter driven expression of GUS. Wild type was used as untranformed control. (C) Accumulation of GUS transcript and activity in transgenic fruits were measured by qRT-PCR and fluorometric GUS assay, respectively. Tomato actin was used as endogenous control in qRT-PCR. Data are mean (±SE) of three biological replicates.
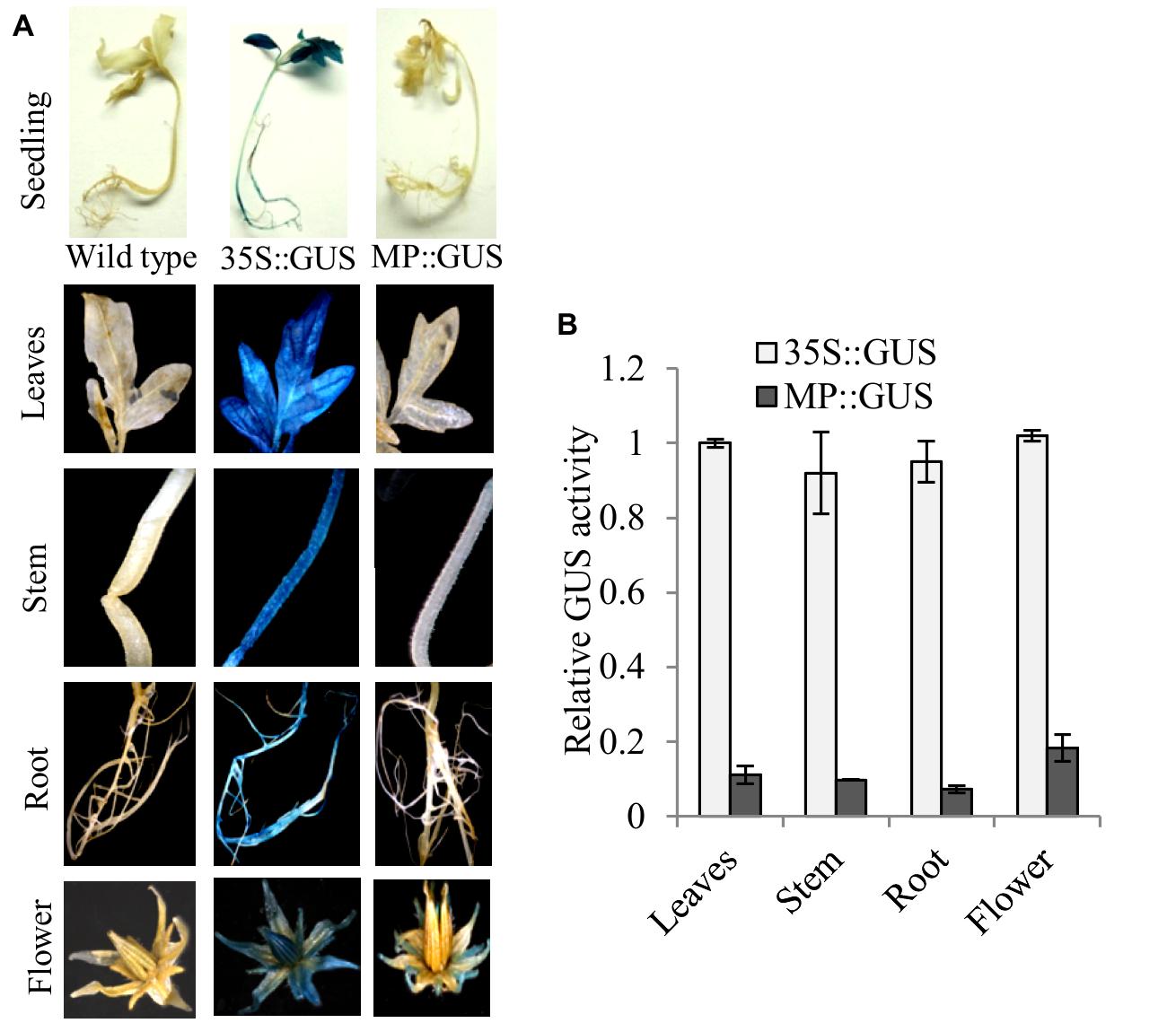
FIGURE 3. Histochemical and fluorometric assays to detect GUS activity in different plant parts. (A) Seedling, leaves, stem, root, and flower from MP::GUS, 35S::GUS and wild type (untransformed) were subjected to histochemical staining. (B) Fluorometric analysis of GUS activity. Data are presented as the mean (±SE) of three biological replicates.
Deletion Analysis of α-Man Promoter
To identify optimal promoter region required for α-Man expression in fruit, deletion analysis of the promoter was carried out. Four deletion constructs (MD1::GUS, MD2::GUS, MD3::GUS, and MD4::GUS) that included 779, 567, 373 and 187 bp upstream of ATG, respectively, were designed to study GUS reporter expression pattern, controlled by the α-Man promoter fragments (Figure 4A, Supplementary Figures S3 and S4). Functional analysis of these promoter::GUS constructs was carried out by Agrobacterium-mediated transient expression (Agroinjection) of GUS gene in tomato fruits as described in “Materials and Methods.” Fruits were harvested 3 days after agroinjection in order to assess the time period necessary for measuring reporter expression and cut into sections and analyzed for GUS activity by histochemical assay and fluorometric MUG assay. GUS expression in fruits driven by MD3 and MD4 fragment of α-Man promoter was significantly less as compare to the full length and other promoter fragments (Figure 4B). These results were further validated by fluorometric quantification of GUS activity by MUG assay. The MUG assay also demonstrated that α-Man promoter activity was affected in MD3::GUS and MD4::GUS injected tomato fruits (Figure 4B). In silico analysis of these promoter fragments revealed the presence of RIN binding sites [CArG box, C(T/A/C)(AT)6(A/T/G)G] on these region (Supplementary Figure S4; Supplementary Table S1).
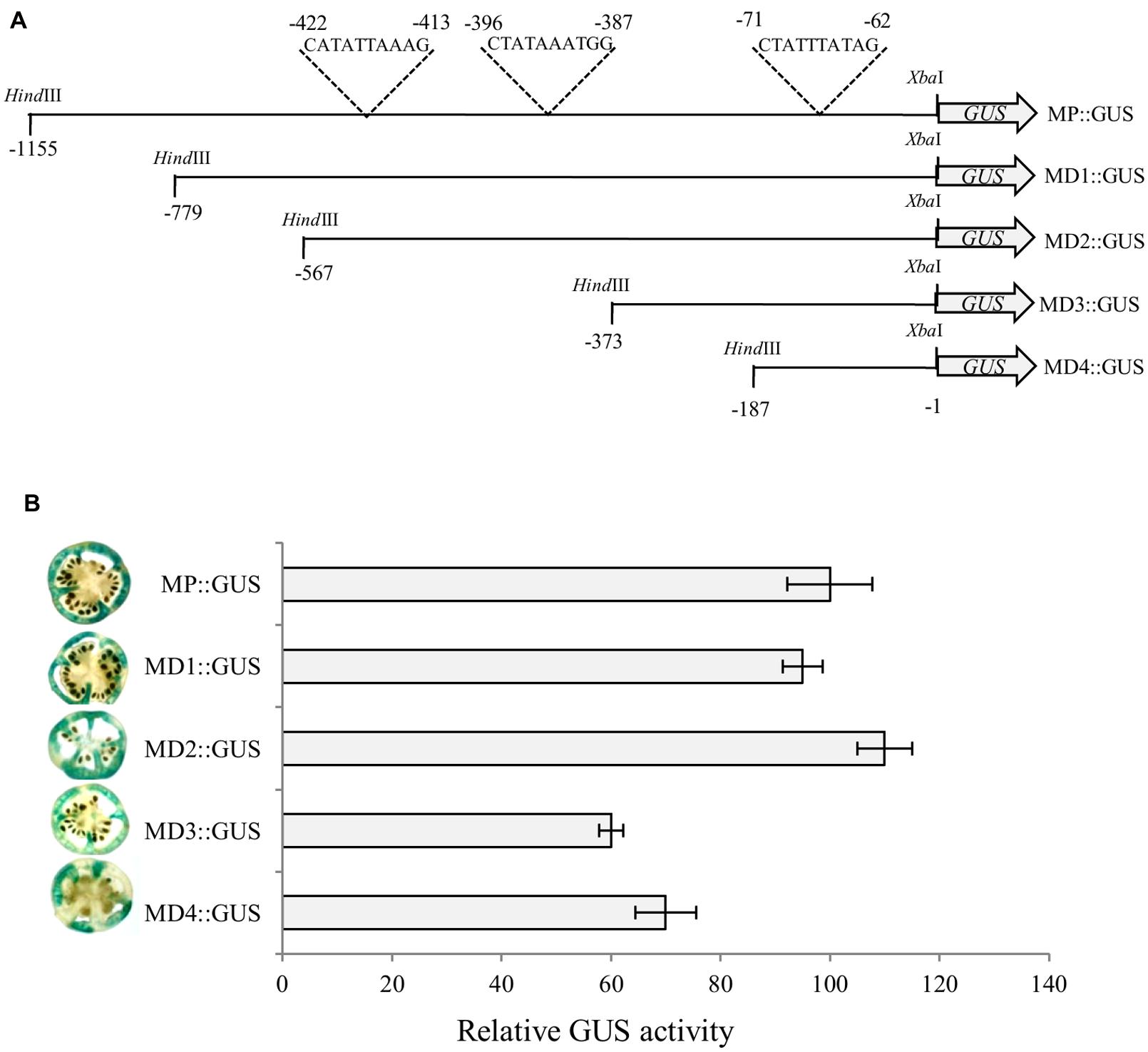
FIGURE 4. Deletion analysis of α-Man promoter. (A) Schematic representation of promoter deletion constructs. 187, 373, 567, and 779 bp regions upstream of translational start site were cloned into pBI121 binary vector in place of CaMV35S promoter to generate GUS fusion constructs (MD4::GUS, MD3::GUS, MD2::GUS and MD1::GUS respectively). (B) Analysis of promoter driven expression of GUS. Histochemical and fluorometric GUS assays were carried out in fruits after Agrobacterium-mediated transient expression. Data are presented as the mean (±SE) from three biological replicates.
Transcriptional Regulation of α-Man by RIN
The qRT-PCR analysis of α-Man transcript level in wild type and rin mutant fruit revealed about 90% suppression of transcript level in rin mutant fruits (Figure 5A; Meli et al., 2010). Moreover, in silico analysis of α-Man promoter identified three CArG boxes (Figure 4A, Supplementary Table S2). CArG boxes are required for the direct binding of RIN to the promoter of ripening-specific genes (Ito et al., 2008; Fujisawa et al., 2011, 2013). In order to understand the role of RIN in regulation of α-Man expression, direct interaction of RIN with the α-Man promoter was tested by EMSA. For this a 187 bp fragment of promoter (MD4) that contained a CArG box closest to ATG, was used (Figure 5B, Supplementary Figure S3). Recombinant RIN protein which is known to bind to promoter of fruit ripening specific gene of β-Hex was purified from Escherichia coli cells and used for the EMSA (Irfan et al., 2014). When radiolabeled probe was incubated with recombinant RIN protein a shift was detected, whereas shift was not observed when the terminal C and G of CArG box was replace to T and A, respectively (Figure 5B). Moreover, signal was not detected when we included cold competitor in binding reaction (Figure 5B). Taken together these results suggest that RIN specifically binds to the CArG box element of α-Man promoter.
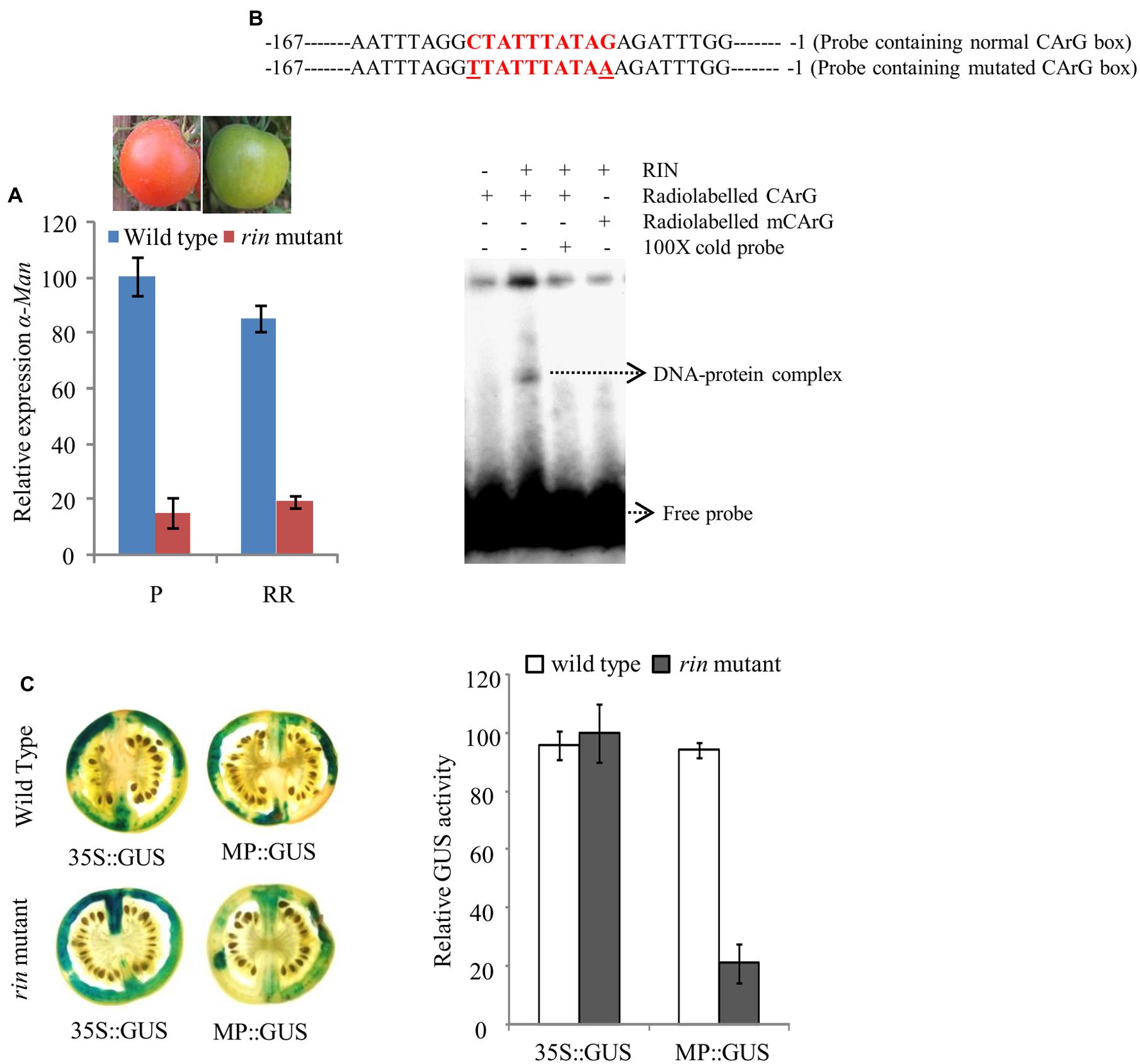
FIGURE 5. RIN regulated expression of α-Man in fruit. (A) Relative transcript abundance of α-Man in wild type (cv. Ailsa Craig) and rin mutant fruits was determined by qRT-PCR using tomato actin as endogenous control. (B) EMSA showing in vitro binding of RIN protein with the α-Man promoter. (C) CaMV35S and α-Man promoter driven expression of GUS in fruits of wild type and rin mutant. GUS activity in agroinjected fruits was analyzed by histochemical and fluorometric assays. Data are presented as the mean (±SE) of two biological replicates.
In order to further examine α-Man transcriptional regulation by RIN, in fruto transient expression assay of MP::GUS construct in rin mutant fruits was performed through Agroinjection. After histochemical staining, we observed that GUS activity in rin mutant fruit was significantly reduced as compared to wild type fruits, when GUS expression was driven by the α-Man promoter (Figure 5C). However, GUS activity was found to be almost similar in wild type and rin mutant fruits, when GUS expression was under the control of constitutive promoter. Fluorometric quantification assay also demonstrated a decrease in reporter gene activity conferred by α-Man promoter in rin mutant fruits as compared to wild type (Figure 5C). These results suggest that the transcription factor RIN positively regulates α-Man expression during tomato ripening.
Regulation of α-Man by Ethylene
Ethylene plays a vital role in ripening of climacteric fruits including tomato (Zegzouti et al., 1999; Giovannoni, 2001; Pech et al., 2012). Therefore, the role of ethylene in the regulation of α-Man expression was examined. The expression of α-Man in Nr mutant fruits was determined by qRT PCR. In Nr mutant, ripening is abolished due to mutation in NEVERRIPE (NR) receptor which percepts ethylene (Wilkinson et al., 1995). The transcript level of α-Man gene was decreased about 30% at the pink (P) and red ripe (RR) stages of Nr mutant fruit as compare to wild type fruit (Figure 6A). Through in silico analysis of α-Man promoter, ethylene responsive cis-acting elements and ethylene insensitive 3 (EIN3) like factors binding sites were also identified (Supplementary Table S1). To further understand the regulation of α-Man by ethylene, tomato seedlings were treated by ACC, the precursor of ethylene. ACC treatment to the wild type resulted in induced expression of α-Man (Figure 6B). Moreover, to check whether the promoter of α-Man was activated by ACC treatment, the expression of GUS was determined by qRT-PCR in MP::GUS transgenic seedlings after ACC treatment. The result revealed induced expression of GUS, suggesting activation of α-Man promoter after ACC treatment (Figure 6C). Although, RIN directly regulates the expression of ethylene biosynthesis and signaling genes, the expression of RIN is also under the control of ethylene (Lincoln and Fischer, 1988; Fujisawa et al., 2011, 2013; Martel et al., 2011; Osorio et al., 2011; Su et al., 2015). Therefore, we tested whether ACC treatment can affect α-Man expression in rin mutant. Interestingly, α-Man expression was induced when rin mutant was treated with ACC. However, as compared to the wild type (Ailsa craig), α-Man transcript induction level was less in rin mutant (Figures 6B,D). Therefore, ethylene may regulate α-Man expression in both RIN dependent and independent ways.
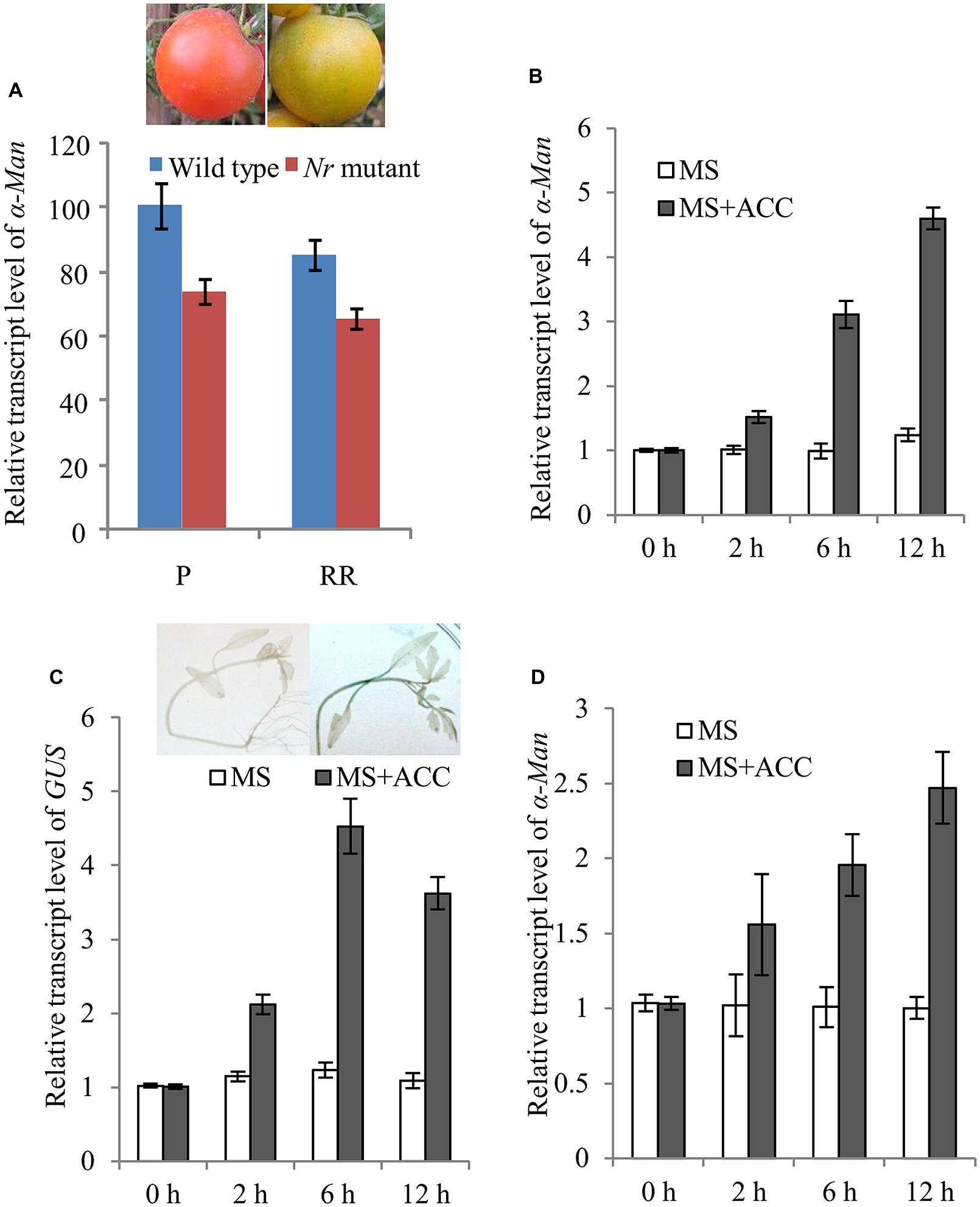
FIGURE 6. Ethylene regulated expression of α-Man. (A) qRT PCR expression analysis of α-Man in fruits of wild type (Ailsa Craig) and ethylene receptor Nr mutant. (B) Ethylene inducibility of α-Man was confirmed with qRT-PCR analysis of α-Man transcript level in wild type seedlings and (C) GUS transcript level in MP::GUS transgenic seedlings. Histochemical assay was carried out after 4 h of treatment. Data are presented as the mean (±SE) of three biological replicates. (D) Ethylene inducible expression of α-Man in rin mutant seedlings was determined by qRT-PCR analysis after ACC treatment.
Discussion
The physiological role of α-Man in ripening-associated fruit softening process has been demonstrated previously (Priya Sethu and Prabha, 1997; Suvarnalatha and Prabha, 1999; Hossain et al., 2009, 2010; Meli et al., 2010; Ghosh et al., 2011), but transcriptional regulation of α-Man during fruit ripening was not studied. To understand transcriptional regulation of α-Man during tomato fruit ripening, the promoter of α-Man was isolated from tomato and functionally characterized. In silico analysis revealed that the promoter region of α-Man contains several putative cis-acting elements, which may be involved in perceiving stimulus from different plant hormones and environmental signals during fruit ripening (Supplementary Table S1). The spatial and temporal expression patterns of the GUS reporter gene under the control of α-Man promoter have been determined through generating promoter::GUS transgenic tomato plants. In accordance to the fruit-ripening specific expression pattern of α-Man (Figure 1B; Meli et al., 2010), promoter activation during tomato ripening was also recorded (Figures 2B,C). Histochemical and fluorometric GUS assays, and transcript analysis in transgenic fruits suggested that the promoter is maximally active at the breaker stage of ripening (Figures 2B,C). As expected for fruit specific genes, α-Man promoter activity was not observed in whole seedling and in other parts (roots, stems, leaves, and flower) of transgenic plant (Figure 3A). However, sepals of flower showed very less GUS activity (Figure 3A). These results indicated that 1155 bp long promoter of α-Man used in this study was sufficient for driving fruit-ripening specific expression of GUS and contained all the cis-acting regulatory elements required for spatio-temporal regulation of the endogenous α-Man. These results substantiated the earlier findings suggesting the role of α-Man in ripening-associated fruit softening (Hossain et al., 2009, 2010; Meli et al., 2010; Ghosh et al., 2011).
In an effort to identify minimal α-Man promoter region required for expression in fruit, deletion analysis of the promoter was carried out (Figures 4A,B). Our data showed that GUS activity under the MD3 and MD4 promoter fragments was significantly less as compared to MD1 and MD2 truncated promoter and full length promoter (Figure 4B). These results suggested that the promoter region upstream to the MD3 fragment contained cis-acting element(s) required for the binding of the transcription factor involved in α-Man expression and full activity of the promoter. In silico analysis revealed that promoter region upstream to the MD3 fragment contains two CArG boxes at -413 and -387 bp (Figure 4A; Supplementary Figure S4) suggesting that α-Man expression could be regulated by the MADS box family transcription factor RIN. Although, MD3 and MD4 promoter fragments contain one CArG box at -62 bp that was not sufficient for the full activity of the α-Man promoter in fruit (Figures 4A,C). qRT-PCR analysis demonstrated that the expression of α-Man was down-regulated in rin mutant fruits (Figure 5A). Moreover, RIN also bound to the α-Man promoter in EMSA which suggested RIN-mediated direct transcriptional regulation of α-Man (Figure 5B). Moreover, α-Man promoter driven expression of GUS reporter was affected in rin mutant, further confirmed our hypothesis that RIN is involved in direct transcriptional regulation of the α-Man expression (Figure 5C). These results suggested that RIN positively regulates the transcriptional expression of α-Man during tomato ripening. RIN acts as a master regulator of fruit ripening by affecting expression of genes of various biological processes, both directly and indirectly (Ito et al., 2008; Fujisawa et al., 2011, 2012, 2013; Martel et al., 2011; Kumar et al., 2012; Qin et al., 2012). Our results demonstrate that RIN is a direct target transcriptional of α-Man during fruit ripening.
The role of ethylene in inducing tomato ripening is well established (Giovannoni, 2001). Therefore, to know the regulation of α-Man by ethylene, α-Man expression was analyzed by qRT-PCR after treatment with ACC, the precursor of ethylene. ACC was able to induce α-Man expression in wild type and promoter activation in MP::GUS transgenic seedlings (Figures 6B,C). These observations supported an essential role of ethylene in activating α-Man promoter during natural fruit ripening and early induction of α-Man at the breaker stage might be brought about by ethylene. Ethylene can regulate α-Man expression through ethylene receptors, e.g., ETR4 and NR and transcription factors such as ERF and ENI3 whose binding sites were identified in α-Man promoter. The role of ETR4 in the perception of ethylene has already been described (Kevany et al., 2008); however, its role in α-Man expression needs to be tested. The involvement of NR cannot be excluded as the expression of α-Man in Nr background was also suppressed (Figure 6A). Moreover, in silico analysis also revealed the presence of various cis-acting elements, related to the ethylene signaling, on the α-Man promoter (Supplementary Table S1). RIN is expressed prior to the onset of climacteric ethylene biosynthesis and rin mutant fruits show reduced ethylene production. Moreover, RIN regulates the expression of ethylene biosynthesis genes directly as well as indirectly (Ito et al., 2008; Fujisawa et al., 2011, 2013; Martel et al., 2011; Kumar et al., 2012; Qin et al., 2012; Zhong et al., 2013). RIN also known to regulate other ethylene signaling cascade genes including NR and ETR (Kevany et al., 2008; Martel et al., 2011; Kumar et al., 2012). Besides, the expression of RIN is also under the control of ethylene (Lincoln and Fischer, 1988; Fujisawa et al., 2013; Su et al., 2015). Interestingly, ethylene mediated induction of α-Man was less in rin mutant as compared to the wild type. Therefore, ethylene may regulate α-Man expression during fruit ripening through RIN as well as other ethylene regulated transcription factors such as EIN3 and ERFs. Taken together, these results suggest RIN-mediated direct transcriptional regulation of α-Man and ethylene may affect α-Man expression by RIN dependent and independent ways. In conclusion, the fruit ripening-specific promoter of α-Man has been identified, which could be a useful tool in fruit ripening related gene expression studies. The insights into transcriptional regulation of α-Man will also help us in understanding of molecular mechanism of fruit ripening regulation through RIN and ethylene. Further analyses of some ethylene related transcription factors and other regulators of ripening in context of α-Man and other N-glycan processing enzymes will clarify the regulation of N-glycan processing mechanism that triggers tomato fruit ripening.
Author Contributions
MI, SG, VM, AD designed research; MI, SG, VM, AK, VK performed research; MI, SG, VM, NC, SC, AD analyzed data; MI, SG, AD wrote the paper. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The present study was financially supported by a research grant (BT/01/CEIB/12/II/01) from the Department of Biotechnology and core research grant from National Institute of Plant Genome Research. MI thanks University Grant Commission for pre-doctoral fellowship.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00010
Footnotes
References
Bovy, A., de Vos, R., Kemper, M., Schijlen, E., Pertejo, M. A., Muir, S., et al. (2002). High-flavonol tomatoes resulting from the heterologous expression of the maize transcription factor gene LC and C1. Plant Cell 14, 2509–2526. doi: 10.1105/tpc.004218
Brummell, D. A. (2006). Cell wall disassembly in ripening fruit. Funct. Plant Biol. 33, 103–119. doi: 10.1071/FP05234
Cao, L., Zhao, C., Su, S., Luo, C., and Han, M. (2014). The role of β-hexosaminidase in peach (Prunus persica) fruit softening. Sci. Hortic. 169, 226–233. doi: 10.1016/j.scienta.2014.02.015
Cartharius, K., Frech, K., Grote, K., Klocke, B., Haltmeier, M., Klingenhoff, A., et al. (2005). MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942. doi: 10.1093/bioinformatics/bti473
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Fillati, J. J., Kiser, J., Rose, R., and Comai, L. (1987). Efficient transfer of a C glyphosphate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Biotechnology 5, 726–730. doi: 10.1038/nbt0787-726
Fujisawa, M., Nakano, T., and Ito, Y. (2011). Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol. 11:26. doi: 10.1186/1471-2229-11-26
Fujisawa, M., Nakano, T., Shima, Y., and Ito, Y. (2013). A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25, 371–386. doi: 10.1105/tpc.112.108118
Fujisawa, M., Shima, Y., Higuchi, N., Nakano, T., Koyama, Y., Kasumi, T., et al. (2012). Direct targets of the tomato-ripening regulator RIN identified by transcriptome and chromatin immunoprecipitation analyses. Planta 235, 1107–1122. doi: 10.1007/s00425-011-1561-2
Fujisawa, M., Shima, Y., Nakagawa, H., Kitagawa, M., Kimbara, J., Nakano, T., et al. (2014). Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26, 89–101. doi: 10.1105/tpc.113.119453
Ghosh, S., Meli, V. S., Kumar, A., Thakur, A., Chakraborty, N., Chakraborty, S., et al. (2011). The N-glycan processing enzymes α-mannosidase and β-D-N-acetylhexosaminidase are involved in ripening-associated softening in the nonclimacteric fruits of capsicum. J. Exp. Bot. 62, 571–582. doi: 10.1093/jxb/erq289
Ghosh, S., Singh, U. K., Meli, V. S., Kumar, V., Kumar, A., Irfan, M., et al. (2013). Induction of senescence and identification of differentially expressed genes in tomato in response to monoterpene. PLoS ONE 8:e76029. doi: 10.1371/journal.pone.0076029
Giovannoni, J. (2001). Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 725–749. doi: 10.1146/annurev.arplant.52.1.725
Giovannoni, J. J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10, 283–289.
Handa, A. K., Singh, N. K., and Biggs, S. M. (1985). Effect of tunicamycin on in vitro ripening of tomato pericarp tissue. Plant Physiol. 63, 417–424. doi: 10.1111/j.1399-3054.1985.tb02320.x
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hossain, M. A., Nakamura, K., and Kimura, Y. (2009). α-Mannosidase involved in turnover of plant complex type N-glycans in tomato (Lycopersicum esculentum) fruits. Biosci. Biotechnol. Biochem. 73, 140–146. doi: 10.1271/bbb.80561
Hossain, M. A., Nakano, R., Nakamura, K., Hossain, M. T., and Kimura, Y. (2010). Molecular characterization of plant acidic alpha-mannosidase, a member of glycosylhydrolase family 38, involved in the turnover of N-glycans during tomato fruit ripening. J. Biochem. 148, 603–616. doi: 10.1093/jb/mvq094
Hüttner, S., Veit, C., Vavra, U., Schoberer, J., Liebminger, E., Maresch, D., et al. (2014). Arabidopsis class I mannosidases MNS4 and MNS5 are involved in ERassociated degradation of misfolded glycoproteins. Plant Cell 26, 1712–1728. doi: 10.1105/tpc.114.123216
Irfan, M., Ghosh, S., Kumar, V., Chakraborty, N., Chakraborty, S., and Datta, A. (2014). Insights into transcriptional regulation of β-D-N-acetylhexosaminidase, an N-glycan-processing enzyme involved in ripening-associated fruit softening. J. Exp. Bot. 65, 5835–5848. doi: 10.1093/jxb/eru324
Ito, Y., Kitagawa, M., Ihashi, N., Yabe, K., Kimbara, J., Yasuda, J., et al. (2008). DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55, 212–223. doi: 10.1111/j.1365-313X.2008.03491.x
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Kevany, B., Taylor, M., and Klee, H. (2008). Fruit-specific suppression of the ethylene receptor LeETR4 results in early-ripening tomato fruit. Plant Biotechnol. J. 6, 295–300. doi: 10.1111/j.1467-7652.2007.00319.x
Klee, H. J., and Giovannoni, J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45, 41–59.
Kumar, R., Sharma, M. K., Kapoor, S., Tyagi, A. K., and Sharma, A. K. (2012). Transcriptome analysis of rin mutant fruit and in silico analysis of promoters of differentially regulated genes provides insight into LeMADS-RIN-regulated ethylene-dependent as well as ethylene-independent aspects of ripening in tomato. Mol. Genet. Genomics 287, 189–203. doi: 10.1007/s00438-011-0671-7
Kumar, V., Irfan, M., Ghosh, S., Chakraborty, N., Chakraborty, S., and Datta, A. (2015). Fruit ripening mutants reveal cell metabolism and redox state during ripening. Protoplasma doi: 10.1007/s00709-015-0836-z [Epub ahead of print].
Lescot, M., Déhais, P., Thijs, G., Moreau, Y., De Moor, B., Rouzé, P., et al. (2002). PlantCARE: a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Liebminger, E., Hüttner, S., Vavra, U., Fischl, R., Schoberer, J., Grass, J., et al. (2009). Class I alpha-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21, 3850–3867. doi: 10.1105/tpc.109.072363
Lincoln, J. E., and Fischer, R. L. (1988). Regulation of gene expression by ethylene in wild-type and rin tomato (Lycopersicon esculentum) fruit. Plant Physiol. 88, 370–374. doi: 10.1104/pp.88.2.370
Martel, C., Vrebalov, J., Tafelmeyer, P., and Giovannoni, J. J. (2011). The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157, 1568–1579. doi: 10.1104/pp.111.181107
Meli, V. S., Ghosh, S., Prabha, T. N., Chakraborty, N., Chakraborty, S., and Datta, A. (2010). Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc. Natl. Acad. Sci. U.S.A. 107, 2413–2418. doi: 10.1073/pnas.0909329107
Menke, F. L. H., Parchmann, S., Mueller, M. J., Kijne, J. W., and Memelink, J. (1999). Involvement of the octadecanoid pathway and protein phosphorylation in fungal elicitor-induced expression of terpenoid indole alkaloid biosynthesis genes in Catharanthus roseus. Plant Physiol. 119, 1289–1296. doi: 10.1104/pp.119.4.1289
Nakamura, K., Inoue, M., Yoshiie, T., Hosoi, K., and Kimura, Y. (2008). Changes in structural features of free N-glycan and endoglycosidase activity during tomato fruit ripening. Biosci. Biotechnol. Biochem. 72, 2936–2945. doi: 10.1271/bbb.80414
Orzaez, D. F., Mirabel, S., Wieland, W. H., and Granell, A. (2006). Agroinjection of tomato fruits. A tool for rapid functional analysis of transgenes directly in fruits. Plant Physiol. 140, 3–11. doi: 10.1104/pp.105.068221
Osorio, S., Alba, R., Damasceno, C. M., Lopez-Casado, G., Lohse, M., Zanor, M. I., et al. (2011). Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157, 405–425. doi: 10.1104/pp.111.175463
Osorio, S., Alba, R., Nikoloski, Z., Kochevenko, A., Fernie, A. R., and Giovannoni, J. J. (2012). Integrative comparative analyses of transcript and metabolite profiles from pepper and tomato ripening and development stages uncovers species-specific patterns of network regulatory behavior. Plant Physiol. 159, 1713–1729. doi: 10.1104/pp.112.199711
Pech, J. C., Purgatto, E., Bouzayen, M., and Latche, A. (2012). Ethylene and fruit ripening. Annu. Plant Rev. 44, 275–304.
Priem, B., Gitti, R., Bush, C. A., and Gross, K. C. (1993). Structure of ten free N-glycans in ripening tomato fruit (arabinose is a constituent of a plant N-glycan). Plant Physiol. 102, 445–458. doi: 10.1104/pp.102.2.445
Priem, B., and Gross, K. C. (1992). Mannosyl- and Xylosyl- containing glycans promote tomato (Lycopersicon esculentum Mill.) fruit ripening. Plant Physiol. 98, 399–401. doi: 10.1104/pp.98.1.399
Priya Sethu, K. M., and Prabha, T. N. (1997). α-D-manosidase from Capsicum annum. Phytochemistry 44, 383–387. doi: 10.1016/S0031-9422(96)00490-6
Qin, G., Wang, Y., Cao, B., Wang, W., and Tian, S. (2012). Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 70, 243–255. doi: 10.1111/j.1365-313X.2011.04861.x
Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: the european molecular biology open software suite. Trends Genet. 16, 276–277. doi: 10.1016/S0168-9525(00)02024-2
Seymour, G. B., Chapman, N. H., Chew, B. L., and Rose, J. K. C. (2013a). Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol. J. 11, 269–278. doi: 10.1111/j.1467-7652.2012.00738.x
Seymour, G. B., Østergaard, L., Chapman, N. H., Knapp, S., and Martin, C. (2013b). Fruit development and ripening. Annu. Rev. Plant Biol. 64, 219–241. doi: 10.1146/annurev-arplant-050312-120057
Strasser, R., Schoberer, J., Jin, C., Glössl, J., Mach, L., and Steinkellner, H. (2006). Molecular cloning and characterization of Arabidopsis thaliana Golgi alphamannosidase II, a key enzyme in the formation of complex N-glycans in plants. Plant J. 45, 789–803. doi: 10.1111/j.1365-313X.2005.02648.x
Su, L., Diretto, G., Purgatto, E., Danoun, S., Zouine, M., Li, Z., et al. (2015). Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC Plant Biol. 15:114. doi: 10.1186/s12870-015-0495-4
Suvarnalatha, G., and Prabha, T. N. (1999). α-D-Mannosidase from Lycopersicon esculentum II. Phytochemistry 50, 1111–1115. doi: 10.1016/S0031-9422(98)00693-1
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-Box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. doi: 10.1126/science.1068181
Wilkinson, J. Q., Lanahan, M. B., Yen, H. C., Giovannoni, J. J., and Klee, H. J. (1995). An ethylene-inducible component of signal transduction encoded by never-ripe. Science 270, 1807–1809. doi: 10.1126/science.270.5243.1807
Zegzouti, H., Jones, B., Frasse, P., Marty, C., Maitre, B. A., Latche, Â, et al. (1999). Ethylene-regulated gene expression in tomato fruit: characterization of novel ethylene-responsive and ripening related genes isolated by differential display. Plant J. 18, 589–600. doi: 10.1046/j.1365-313x.1999.00483.x
Keywords: fruit ripening, transcriptional regulation, fruit ripening-specific promoter, α-Man, RIN
Citation: Irfan M, Ghosh S, Meli VS, Kumar A, Kumar V, Chakraborty N, Chakraborty S and Datta A (2016) Fruit Ripening Regulation of α-Mannosidase Expression by the MADS Box Transcription Factor RIPENING INHIBITOR and Ethylene. Front. Plant Sci. 7:10. doi: 10.3389/fpls.2016.00010
Received: 19 November 2015; Accepted: 07 January 2016;
Published: 21 January 2016.
Edited by:
Thomas Debener, Leibniz University of Hanover, GermanyReviewed by:
Shogo Matsumoto, Nagoya University, JapanAntonio Ferrante, Universitá degli Studi di Milano, Italy
Copyright © 2016 Irfan, Ghosh, Meli, Kumar, Kumar, Chakraborty, Chakraborty and Datta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Asis Datta, YXNpc19kYXR0YUByZWRpZmZtYWlsLmNvbQ==
 Mohammad Irfan1
Mohammad Irfan1 Vijaykumar S. Meli
Vijaykumar S. Meli Niranjan Chakraborty
Niranjan Chakraborty Asis Datta
Asis Datta