- 1State Key Laborotary of Grassland Agro-Ecosystems, College of Pastoral Agriculture Science and Technology, Lanzhou University, Lanzhou, China
- 2AgResearch Limited, Grasslands Research Centre, Palmerston North, New Zealand
Yellow sweet clover is a widely spread legume species that has potential to be used as a forage crop in Western China. However, limited information is available on the genetic variation for herbage yield, key morphological traits, and coumarin content. In this study, 40 half sib (HS) families of M. officinalis were evaluated for genotypic variation and phenotypic and genotypic correlation for the traits: LS (leaf to stem ratio), SV (spring vigor), LA (leaf area), PH (plant height), DW (herbage dry weight), SD (stem diameter), SN (stem number), Cou (coumarin content), SY (seed yield), across two locations, Yuzhong and Linze, in Western China. There was significant (P < 0.05) genotypic variation among the HS families for all traits. There was also significant (P < 0.05) genotype-by-environment interaction for the traits DW, PH, SD, SN, and SV. The estimates of HS family mean repeatability across two locations ranged from 0.32 for SN to 0.89 for LA. Pattern analysis generated four HS family groups where group 3 consisted of families with above average expression for DW and below average expression for Cou. The breeding population developed by polycrossing the selected HS families within group 3 will provide a significant breeding pool for M. officinalis cultivar development in China.
Introduction
Yellow sweet clover known as field melilot or yellow melilot, is an annual or biennial herb that belongs to the Fabaceae family. It is native to temperate and tropical Asia, and Europe (GRIN, 2000). Melilotus officinalis is one of the most common species in the Melilotus genus. This species has adaptation to environmental constraints such as drought and cold (Turkington et al., 1978) and salinity (Sherif, 2009). Melilotus is used as a ground cover in depleted soils (Allen and Allen, 1981), especially in moderately saline areas where traditional forage legumes cannot be grown (Maddaloni, 1986). Melilotus officinalis usually occurs in the northern region of China, where it is used as green manure for soil fertility improvement and also as a medicinal plant.
Species of Melilotus, including yellow sweet clover, have not been widely used in forage production due to their high coumarin content. Coumarin, a secondary plant metabolite, is associated with dicoumarol production. Dicoumarol is an anticoagulant that can cause a haemorrhagic condition known as sweet clover disease (Evans and Kearney, 2003; Nair et al., 2010). Therefore, the success of forage cultivar development based on any of the Melilotus species will depend on a combination of increasing dry matter production and decreasing coumarin content. A number of cultivars of Melilotus have been released to date; Acuma, Cumino, Denta, Polara (Smith and Gorz, 1965; Goplen, 1971) for M. albus and Norgold (Goplen, 1981), N28, N29 (Gorz et al., 1992) for M. officinalis. The Melilotus breeding program at Lanzhou University is specifically focused on the development of new cultivars with adaptation to the vast temperate grazing environments of China (Luo et al., 2014).
In any plant breeding program, the rate of genetic gain depends on the genetic diversity for a given trait in the breeding population (Hallauer and Miranda, 1981). Information on the magnitude of genetic variation for key plant attributes in breeding programs will enhance the development of appropriate breeding strategies to achieve maximum genetic gain (Moll and Stuber, 1974). Jahufer and Casler (2015) evaluated the relative merit in genetic gain using single trait selection, correlated response to selection and index selection, based on estimated genetic variation for a range of morphological and quality traits in switch grass (Panicum virgatum L.). Genetic variation for key traits have been reported for some of the important forage grasses and legumes: ryegrass (Breese and Hayward, 1972), tall fescue (Piano et al., 2007), white clover (Jahufer et al., 2002), alfalfa (Riday and Brummer, 2007).
There is a lack of quantitative genetic information for Melilotus. Few studies have been carried out on the genetic variation for agronomic traits in Melilotus species (Ivanov and Chetvertnykh, 1980; Sagalbekov, 1980). Nair et al. (2010) reported genotypic variation for coumarin content among 149 accessions of 15 Melilotus species. This study demonstrated the presence of potential genetic variation for coumarin content in Melilotus germplasm useful for breeding. However, breeding Melilotus species as a forage crop needs to focus on not only coumarin content but also biomass and associated traits. There is also a lack of information on the magnitude of genotype-by-environment interaction effects in Melilotus, which will be important for breeding for broad adaptation (Cooper et al., 1993b).
The objective of our study was to conduct a preliminary assessment of the performance of half sib (HS) families of Melilotus officinalis across two contrasting locations to: (a) estimate genotypic variation for key traits, and (b) identify families with a combination of superior agronomic performance and low coumarin expression in comparison to two commercial controls.
Materials and Methods
Plant Material
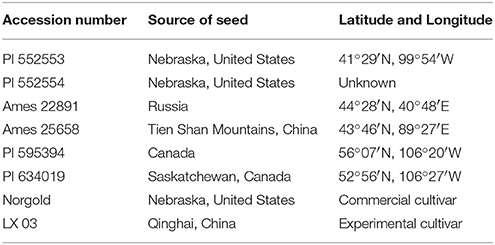
Six germplasm accessions (PI 552553 and PI 552554, PI 595394, PI 634019, Ames 22891, and Ames 25658) were selected from a set of 51 accessions that were evaluated for biomass production, agronomy and low coumarin, in Yuzhong, Gansu Province, during 2012–2013 (results not presented). Elite genotypes representing each of the germplasm accessions were polycrossed in isolation, using honey bees, to generate a breeding population to be used for cultivar development. A total of 40 HS families were generated by harvesting each of the genotypes individually. All harvested seeds from the individual genotypes were kept separately as individual HS families.
Field Trials
The M. officinalis HS families were established at two locations: Yuzhong (104°09′ E, 35°89′ N, elevation 1 653 m a.s.l.) and Linze (100°02′ E, 39°15′ N, elevation 1 390 m a.s.l.) in Gansu Province, China. There are different climate conditions between Yuzhong and linze. Yuzhong in Loess Plateau region is a medium temperate semi-arid climate, whereas Linze in the Hexi Corridor is typical desert climate and characterized by an arid climate (Su et al., 2007; Hu et al., 2012; Li et al., 2014). The average annual precipitation in Yuzhong is 295 mm and in Linze is only 117 mm. The mean monthly minimum and maximum temperatures, and total monthly rainfall during the trial period at the two locations are shown in Figure 1.
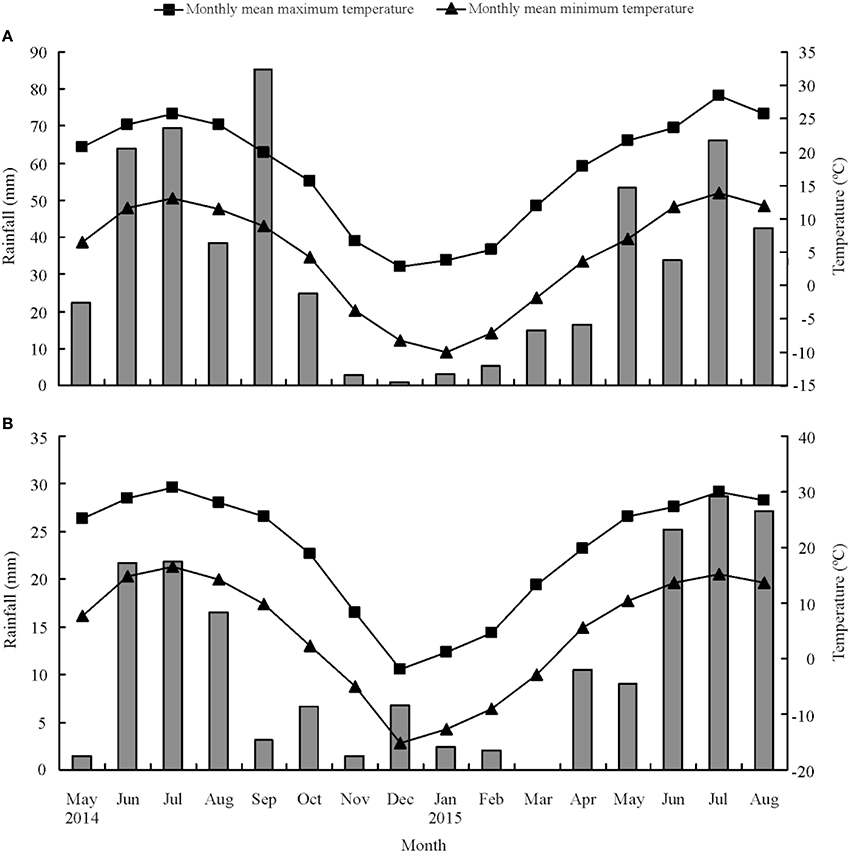
Figure 1. Mean monthly maximum and minimum temperatures (°C) and total monthly rainfall (mm) at Yuzhong (A) and Linze (B), respectively.
The soil type at each location is loessal soil at Yuzhong and meadow soil at Linze. The saline-alkali degree was much higher in Linze than in Yuzhong, the salinity is 1.8 ppt in Linze and 0.5 ppt in Yuzhong. Initial soil conditions in Yuzhong and Linze are: pH 7.0 and 7.5, total N of 0.756 g/kg and 0.803 g/kg, total P of 0.752 and 0.708 g/kg, respectively.
At each location, the experimental plots were arranged in a randomized complete block design containing three replicates. Each replicate consisted of the 40 HS families the six parental germplasm accessions and two commercial checks. The origins of these entries are provided in Table 1. The two trials were sown in 15–18 June 2014. The experimental plot size for each entry was 2.4 m2 (0.8 × 3 m). Within each plot, the seed was planted at a spacing of 30 cm within-rows and 60 cm between-rows. The plots were fertilized with 150 kg (NH4)2HPO4 ha−1 after sowing.
Measurements
The traits measured were: LS, leaf to stem ratio; SV, spring vigor; LA, leaf area (cm2); PH, plant height (cm); DW, herbage dry weight (g/plant); SD, stem diameter (cm); SN, stem number; Cou, coumarin (% of dry matter); SY, seed yield (g/plant). All the traits were measured in the second year (2015).
Visual scoring for SV was based on a scale of 1 to 5 (1 = low; 5 = high). The morphological traits (PH, SN, SD, and LA) were measured at the flowering stage (50% of the plants had open flowers), resulting in a minimum of three individuals per replicate. LA was measured from three middle leaflets per plant by using a flatbed scanner (EPSON GT-15000) and a WinSEEDLE 2011 image analysis system (Regent Instruments Inc.). Individual plant was harvested for DW measurement at the flowering stage after measuring morphological traits. At harvest, three randomly sampled plants from each replicate were cut off at 3 cm above the soil, placed in paper bags and dried at room temperature (about 20–25°C) with good ventilation until no change in weight was recorded. After measuring DW, the dried samples were hand separated into leaf blade and stem (including the inflorescence and leaf sheath) components and weighted to determine the LS ratio. Three sub samples from each field replicate at Yuzhong were combined and ground in a mill to pass through a 1 mm screen for Cou determination. SY was determined from two randomly sampled individuals taken from each replicate when 90% of the pods turned blackish brown at the Linze field. Cou was quantified using HPLC (high performance liquid chromatography, Agilent 1100 series) with a mobile phase of methanol-water (65:35) through an Agilent-XDB C18 column (Zhu and Fan, 2008).
Analysis of Variance
The data were analyzed within and across the two locations Yuzhong and Linze. The analysis across locations was conducted: (a) on only the 40 HS families to estimate genotypic variation, and (b) using all entries in the trial that consisted of the 40 HS families, the six parental germplasm accessions and two check cultivars, which enabled comparison of progeny, parents, and the commercial material. The analysis was conducted using the variance component analysis procedure, Residual Maximum Likelihood (REML) option, in GenStat 7.1 (2003). A mixed linear model was used for the analyses across the two locations using the REML algorithm.
The linear model used in the analysis was,
Where, Yijk is the value of an attribute measured from HS family i in replicate k in location j, and I = 1,…,ng, j = 1,…,nl, k = 1,…,nr; M is the overall mean; gi is the random genotypic effect of HS family i, N(0,σ2g); lj is the fixed effect of location j, N(0,σ2l); rjk is the random effect of replicate k within location j, N(0,σ2b); (gl)ij is the effect between HS family i and environment j, N(0,σ2gl); εijk is the residual effect for HS family i in replicate k in location j, N(0,σ2ε).
The mixed model analysis generated HS family means based on Best Linear Unbiased Predictors (BLUP) (White and Hodge, 1989). These BLUP values were used to construct a HS family × trait mean matrix adjusted of HS family × location interaction effects.
Genotypic Variation and Repeatability
Variation among HS families generated from a population that has gone through at least two cycles of random mating, is an estimate of ¼ additive variation of the random mating population they represent (Falconer, 1989). In our study, the 40 HS families were a result of the first random mating of selected germplasm and therefore represented only the F1 generation. Therefore, we do not refer to the variation estimated among the 40 HS families as ¼additive variation, but as genotypic variation, due to a possible combination of additive and non-additive effects. The genotypic variation for the different traits enabled calculation of repeatability, an estimation of the upper limits of their degrees of genetic determination (Falconer, 1989).
The genotypic variance components generated from the REML analysis within and across locations were used to calculate repeatability (R) (Fehr, 1987).
HS family mean repeatability at a single site:
HS family mean repeatability across locations:
Where, in both model (2) and model (3), the respective variance components and their divisors are defined in relation to linear model (1).
Phenotypic and Genotypic Correlation
Phenotypic correlation (rp) analysis was carried out using GenStat 7.1 (2003). The multivariate MANOVA procedure, within GenStat 7.1 (2003), enabled estimation sums of cross-products, using the multisite trait data from the 40 HS families. Mean cross products were then calculated and resolved to estimate genotypic covariance components. The genotypic covariance components were used together with the σ2g estimates, from REML analysis, to determine genotypic correlation coefficients (rg) according to Falconer (1989).
Pattern Analysis
Pattern analysis was conducted to: (a) provide a graphical summary of the performance of the 40 HS families, six parental germplasm accessions and the two check cultivars of M. officinalis, based on the genotype × trait BLUP adjusted mean matrix generated from variance component analysis across the two locations Yuzhong and Linze, and (b) investigate any changes in type (positive or negative) and magnitude of the association among the seven traits across Yuzhong and Linze. Pattern analysis consisted of a combination of cluster and principal component analysis (PCA) (Gabriel, 1971; Kroonenberg, 1994; Watson et al., 1995). To identify the optimum level of truncation for the resulting hierarchy from cluster analysis, the increase in the sum of squares among accession groups was monitored as the number of groups increased. The group level selected was determined by the point where the percentage of accession sum of squares among groups did not improve substantially as the number of groups increased (DeLacy, 1981).
Results
Genotypic Variance Components and HS Family Mean Repeatability of Plant Attributes of M. officinalis
The genotypic variance estimated for the different traits from the individual location, Yuzhong and Linze, analysis indicated significant (P < 0.05) variation among the 40 M. officinalis HS families (Tables 2A,B). At both these locations HS family mean repeatability estimates ranged from intermediate to very high, depending on the traits.
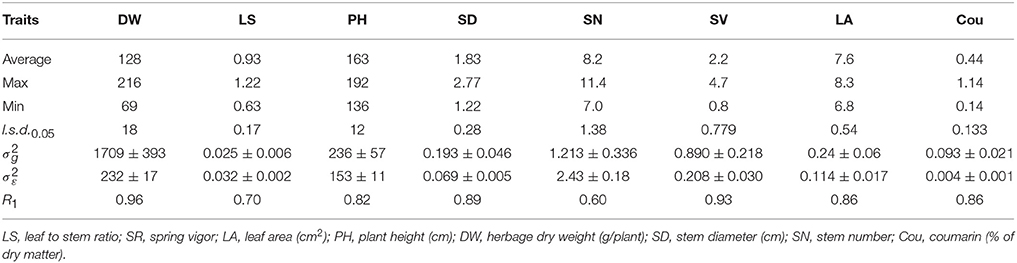
Table 2A. Average, maximum, minimum, least significant differences (l.s.d.0.05), genotypic (), and experimental error () variance components and associated standard errors (±SE), and HS family mean repeatability (R1) estimated from the 40 M. officinalis half sib families, evaluated at Yuzhong.
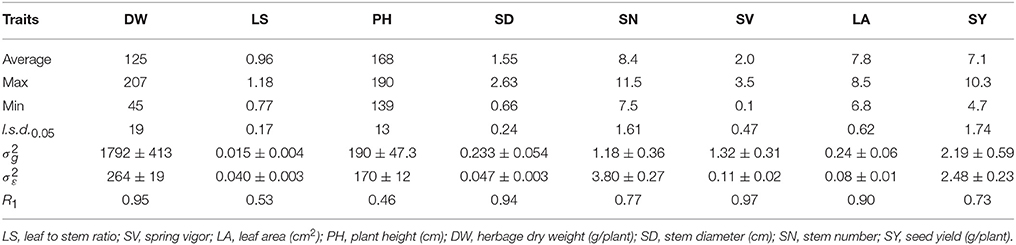
Table 2B. Average, maximum, minimum, least significant differences (l.s.d.0.05), genotypic (), and experimental error () variance components and associated standard errors (±SE), and HS family mean repeatability (R1) estimated from the 40 M. officinalis half sib families, evaluated at Linze.
At Yuzhong, the HS family mean repeatability (R1) was high for the traits DW, SD and SV, which ranged from 0.89 to 0.96 (Table 2A). For the traits PH, LA and Cou, HS family mean repeatability was high (0.82–0.86). HS family mean repeatability was intermediate (0.60 and 0.70) for SN and LS. At Linze, HS family mean repeatability was very high (0.90–0.97) for the traits LA, SD, DW, and SV (Table 2B). The traits SY and SN had high (0.73, 0.77) HS family mean repeatability. HS family mean repeatability was intermediate (0.46, 0.53) for PH and LS.
Analysis of variance for mean trait expression across the two sites Yuzhong and Linze indicated significant (P<0.05) genotypic variation among the 40 HS families. There was also significant (P < 0.05) genotype-by-location interaction, depending on the traits (Table 3). There was no significant (P > 0.05) genotype-by-location interaction for the traits LS and LA. Line mean repeatability (R2) across the two locations varied from: relatively high for the traits DW and LA; intermediate for PH, LS, SD, and SV; and low for SN.
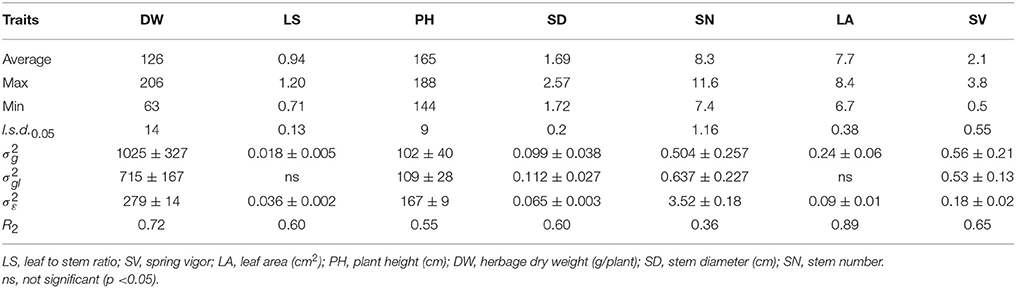
Table 3. Average, maximum, minimum, least significant differences (l.s.d.0.05), genotypic (), genotype-by-location interaction (), and experimental error () variance components and associated standard errors (±SE), and HS family mean repeatability (R2) estimated from the 40 M. officinalis half sib families, evaluated across two locations, Yuzhong and Linze.
Pattern Analysis: Principal Component Analysis (PCA)
The biplot (Figure 2) was generated from PCA of the 40 HS families, the six parental germplasm accessions and the two check cultivars of M. officinalis, based on the 9 traits LS, SV, LA, PH, DW, SD, SN, and Cou. The first principle component explained 46% of the total trait variation, and the second principle component explained 18%. The correlation structure of the traits is indicated by the directional vectors in the biplot. In this study, SD, SN, and PH showed a strong positive association with DW. The traits LS and Cou also showed a negative correlation with DW.
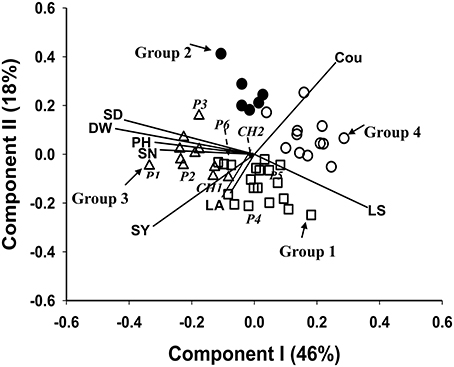
Figure 2. Biplot generated using standardized Best Linear Unbiased Predictor values for eight traits measured from: the 40 half sib families, the 6 parental germplasm accessions and the 2 check cultivars of M. officinalis, evaluated across two locations Yuzhong and Linze. Components I and II account for 46 and 18% of total variation, respectively. The different symbols indicate progeny Groups 1 to 4 generated from cluster analysis. The vectors represent the traits: LS, leaf to stem ratio; LA, leaf area; PH, plant height; DW, herbage dry weight; SD, stem diameter; SN, stem number; SY, seed yield; Cou, coumarin. The 6 parental germplasm accessions: P1 to P6. Check's: CH1, experimental cultivar; CH2, cv Norgold.
The seven plant trait responses at the locations Yuzhong and Linze are presented in the two biplots, (Figures 3A,B). In Figure 3A, based on breeding line performance at Yuzhong, the first and second principal components accounted for 43 and 19% of the total variation, respectively. Based on the line performance at Linze, the first principle component explained 51% of the total trait variation, and the second principle component explained 23% (Figure 3B).
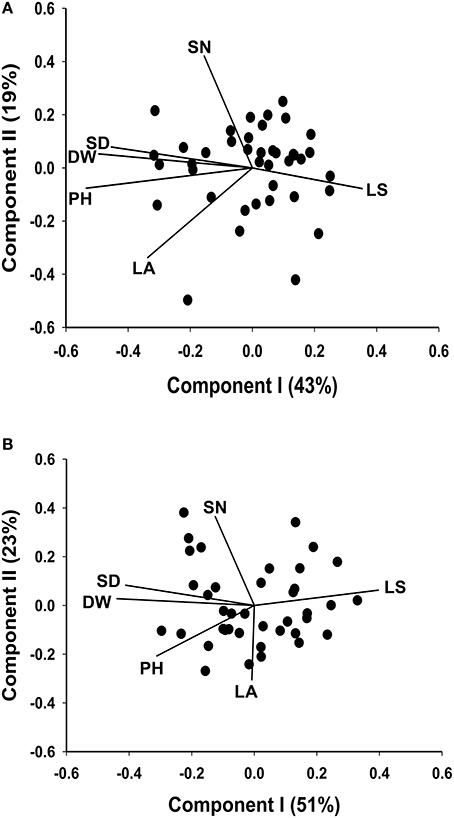
Figure 3. Biplots based on standardized Best Linear Unbiased Predictor values for seven morphological traits, measured from the 40 half sib families of M. officinalis, evaluated at Yuzhong (A) and Linze (B). In each of the biplots Components I and II account for most of the total variation. The vectors represent the traits: LS, leaf to stem ratio; LA, leaf area; PH, plant height; DW, herbage dry weight; SD, stem diameter; SN, stem number.
There were differences in trait association across the two locations Yuzhong and Linze. The traits DW, SD, PH, SN, and LA showed a strong positive correlation at Yuzhong (angles between the directional vectors are at < 45°). At Linze, DW was positively correlated with SD and PH similar to that showed in Yuzhong. However, SN and LA showed a weak positive associated with DW (Figures 3A,B).
Cluster Analysis
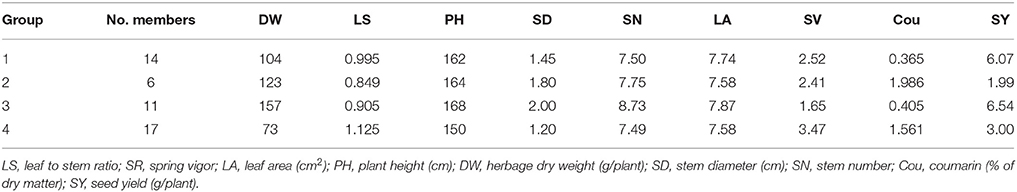
Clustering of the 40 HS families, together with the 6 parental germplasm accessions and 2 check cultivars, was truncated at the four group level. Group 4, the largest group contained 17 members, followed by group 1, group 3 and group 2, which contained 14, 11, and 6 members, respectively (Table 4). As indicated by the Figure 2, the check cultivars were both in group 1. The parental germplasm accessions P1, P2, P3 and P4, P5, P6 were in groups 3 and 1, respectively. The trait means for each group (Table 4) indicated that the members in group 3 had high DW and low coumarin content, and those in group 1 had low coumarin content and intermediate expression for traits DW, PH, SD, and SN. The members in group 4 showed characteristics of a small plant type with high coumarin content. The highest expression for coumarin was in group 2. Groups 3 and 1 had higher SY expression in comparison to groups 4 and 2.
Phenotypic and Genotypic Correlation
A range of genotypic and phenotypic correlation coefficients are presented in Table 5. These coefficients range from strong to weak positive and negative pairwise associations among the 7 traits. Of the special interest are the phenotypic and genotypic correlations between DW and the other traits. There was strong positive phenotypic correlation between DW and the traits SD, PH and SN, and strong negative phenotypic correlation with LS and SV. These results are further supported by the directional vectors in the biplots (Figures 2, 3A,B). In comparison to phenotypic correlation, the estimated genotypic correlation coefficients for all 7 traits showed similar types of pairwise association (Table 5).
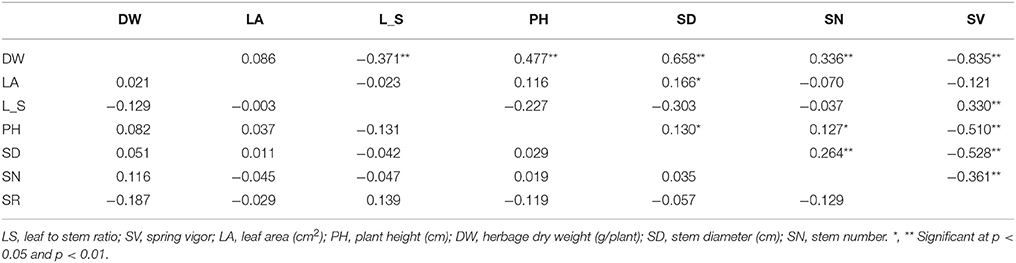
Table 5. Genotypic (rg) (lower triangle) and phenotypic (rP) (upper triangle) correlation coefficients, between traits base on the 40 M. officinalis half sib families, the six parental germplasm accessions and the two check cultivars, evaluated across two locations, Yuzhong and Linze.
Discussion
Previous studies on genotypic variation within Melilotus spp. have mainly focused on interspecific comparisons for traits such as coumarin content (Nair et al., 2010), salinity, waterlogging tolerance (Rogers et al., 2008), and also on phylogenic relationships (Di et al., 2015) and genetic diversity (Di et al., 2014; Wu et al., 2016). The significant (P < 0.05) genotypic variation and high to moderate line mean repeatability reported from our study, indicates the potential for genetic improvement of the nine traits examined. There are no reported studies in M. officinalis similar to ours that estimate the magnitude of genotypic variation for key traits such as DW, Cou, PH, and SY.
Phenotypic variation, expressed as ranges, has been reported for some morphological traits. Klebesadel (1992) reported 2 year means of PH of M. officinalis ecotypes ranging from 112 to 145 cm. Second year mean plant height (PH) measured in our study ranged from 144 to 188 cm. Martino et al. (2006) reported a range of coumarin content between 0.12 and 0.39% based on different extraction methods. Nair et al. (2010) reported coumarin content measured from 27 M. officinalis accessions ranging from 0.09 to 0.61% of dry matter. Our study indicated a coumarin content that ranged from 0.04 to 0.91% of dry matter. Herbage dry matter from single plants has been reported from experiments conducted under glasshouse conditions (Rogers et al., 2008). There is a lack of information on morphological traits measured under field conditions. Results from our study on the genotypic variation for the traits LS, SD, SN, LA, SV and SY, measured under field conditions, will be valuable to Melilotus breeders. Information on the magnitude and significance of the genotypic and environmental components of phenotypic variation for important traits will provide a basis for the development of efficient breeding methods for their improvement (Moll and Stuber, 1974). Results from the present study showed that there was significant genotypic variation among the 40 HS families at each location, Yuzhong and Linz, and also across these two locations for all the traits measured. High genotypic variation was present for DW, SV, and SD at Yuzhong and LA, SD, DW, and SV at Linze. These results, together with the relatively high HS family mean repeatabilities estimated, indicate the potential genetic variation available, within the new M. officinalis breeding population, for improvement of these traits through selection and breeding.
Forage plants are utilized across a wide range of environments, which include different climates, soil types and grazing systems (Breese, 1969). The presence of genotype-by-environment interactions complicates selection of material for broad adaptation due to unreliable performance across environments (Comstock and Moll, 1963; Cooper and Byth, 1996). Quantifying the magnitude and understanding the causes of genotype-by-environment interaction can be helpful when planning breeding strategies (Milligan et al., 1990; Basford and Cooper, 1998). Caradus (1993) reported that a range of traits in white clover, especially yield-related traits, were sensitive to genotype-by-environment interactions. A similar result in white clover was reported by Jahufer et al. (1999). In our study, the genotype-by-environment interactions were significant for most traits except for the traits LS and LA. This indicates the importance of multi-site evaluation in M. officinalis breeding programs when focusing on broad adaptation. The application of multisite testing in breeding programs to investigate the effect of genotype-by-environment interaction on line performance has been reported for forage grass and legume species such as perennial ryegrass (Easton et al., 2015), switchgrass (Jahufer and Casler, 2015), alfalfa (Hill and Baylor, 1983), and white clover (Ballizany et al., 2012).
The association among the traits measured in our study was examined using a combination of phenotypic and genotypic correlation with pattern analysis. The estimates of phenotypic and genotypic correlation coefficients supported the association among traits indicated in the biplots. The positive and significant phenotypic association of DW with traits PH, SD, and SN, predicts a positive correlated response in all these traits when any one of them is selected for individual. This relationship will be useful in a breeding program. The strong positive correlation between DW and SY shown in the biplot (Figure 2) indicates that selection for herbage yield would also result in increasing seed yield. Significant correlation of forage yield and seed yield was also demonstrated in other legumes (Iannucci and Martiniello, 1998; Guler et al., 2001; Cakmakci et al., 2006). Our study indicated negative phenotypic and genotypic correlation between DW and LS. The LS is used as an indicator of digestibility and intake in forage (Kephart et al., 1990). This result implies a trade-off between herbage yield and quality. Julier et al. (2000) also estimated significant negative correlation between DW and LS in alfalfa, which is similar to M. officinalis in vegetative form (Whitson et al., 1992).
The strong negative relationship between SV and DW suggests that measurement of spring vigor, at a very early stage of plant growth, could serve as an indirect selection criterion for increasing herbage yield for M. offcinalis grown in western China (Table 5). This will increase the efficiency of current breeding methods, especially when dealing with the biennial forage specie like M. officinalis. Similar results were reported from studies on common vetch (Cakmakci et al., 2006). The negative phenotypic correlation between the traits DW and Cou shown in our study (Figure 2) indicates the possibility of identifying HS families with a combination of high herbage dry weight and low coumarin content expression. This association will be of significant importance in our M. officinalis breeding program. Hofmann and Jahufer (2011) showed negative association between flavonoid accumulation and biomass using multivariate analysis.
Pattern analysis has been successfully used to summarize complex genotype-by-environment (Cooper et al., 1993a; Zhang et al., 2006) and genotype-by-trait (Jahufer et al., 1999; Davodi et al., 2011) data matrices. Jahufer et al. (1999) successfully identified superior white clover full-sib families based on seven morphological traits using a combination of principle component and cluster analysis. Davodi et al. (2011) used pattern analysis to summarize the performance of 200 alfalfa germplasm accessions, based on 12 traits, for use in the improvement of yield and quality. In our study, pattern analysis generated four groups (Figure 3), where group 3 consisted of HS families with above average performance for DW and below average performance for Cou. Group 3 consisted of 11 members, which included the parental germplasm accessions P1, P2, and P3. All the HS families in group 3 had a higher expression of the traits DW, SD, and SY in comparison to both commercial checks. The breeding lines in group 3 will be polycrossed to produce a breeding population that will be used in the recurrent selection program to develop new cultivars of M. officinalis with high herbage yield and low coumarin content for the Loess Plateau region in China.
Conclusion
The estimates of genotypic variation and HS family mean repeatability indicate the potential genetic variation available for all the traits examined in our study. These estimates also indicate the potential to develop cultivars with increased forage yield and low coumarin content. The significant genotype-by-environment interaction estimated for the traits DW, PH, SD, SN, and SV across the two environments, Yuzhong and Linze, indicate the importance of multi-environment evaluation trials in our M. officinalis breeding program. The breeding population developed by polycrossing the HS families within group 3, identified using pattern analysis, will provide a significant breeding pool for M. officinalis cultivar development in China.
Author Contributions
KL, MJ, JZ, and YW conceived the topic. KL, FW, HD, and XM performed the experiments. KL and MJ analyzed all statistical data. KL wrote the manuscript. All authors revised the manuscript. We thank National Plant Germplasm System (NPGS) for offering the Melilotus officinalis seeds.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by National Basic Research Program (973) of China (2014CB138704), Special Fund for Agroscientific Research in the Public Interest (20120304205), Natural Science Foundation of China (31572453), Program for Changjiang Scholars and Innovative Research Team in University (IRT13019), and Fundamental Research Funds for the Central Universities (lzujbky-2016-10).
References
Allen, O. N., and Allen, E. K. (1981). The Leguminosae: A Source Book of Characteristics, Uses and Nodulation. Madison, WI: The University of Wisconsin Press.
Ballizany, W. L., Hofmann, R. W., Jahufer, M. Z. Z., and Barrett, B. A. (2012). Genotype × environment analysis of flavonoid accumulation and morphology in white clover under contrasting field conditions. Field Crop. Res. 128, 156–166. doi: 10.1016/j.fcr.2011.12.006
Basford, K. E., and Cooper, M. (1998). Genotype × environment interactions and some considerations of their implications for wheat breeding in Australia. Aust. J. Agr. Res. 49, 153–174. doi: 10.1071/A97035
Breese, E. L. (1969). The measurement and significance of genotype – environment interactions in grasses. Heredity 24, 27–44. doi: 10.1038/hdy.1969.3
Breese, E. L., and Hayward, M. D. (1972). The genetic basis of present breeding methods in forage crops. Euphytica 21, 324–336.
Cakmakci, S., Aydinoglu, B., Karaca, M., and Bilgen, M. (2006). Heritability of yield components in common vetch (Vicia sativa L.). Acta Agric. Scand. B Soil Plant. 56, 54–59. doi: 10.1080/09064710510008531
Caradus, J. R. (1993). “White clover breeding line performance under sheep and cattle grazing,” in Proceedings of the 10th Australian Plant Breeding Conference, Vol. 2 (Gold Coast, QLD), 35–36.
Comstock, R. E., and Moll, R. H. (1963). “Genotype-environment interactions,” in Statistical Genetics and Plant Breeding, Vol. 982, eds W. D. Hanson and H. F. Robinson (Washington, DC: Nat Acad Sci Natl Res Publ), 164–196.
Cooper, M., and Byth, D. E. (1996). “Understanding plant adaptation to achieve systematic applied crop improvement - A fundamental challenge,” in Plant Adaptation and Crop Improvement, eds M. Cooper and G. L. Hammer (Wallingford, CT: CAB International), 5–23.
Cooper, M., Delacy, I. H., Byth, D. E., and Woodruff, D. R. (1993a). Predicting grain-yield in Australian environments using data from CIMMYT international wheat performance trials. 2. The application of classification to identify environmental relationships which exploit correlated response to selection. Field Crop. Res. 32, 323–342.
Cooper, M., DeLacy, I. H., and Eisemann, R. L. (1993b). “Recent advances in the study of genotype × environment interactions and their application to plant breeding,” in Proceedings of the 10th Australian Plant Breeding Conference, Vol. 1 (Gold Coast, QLD), 16–31.
Davodi, M., Jafari, A. A., Assadian, G., and Ariapour, A. (2011). Assessment of relationships among yield and quality traits in alfalfa (Medicago sativa) under dryland farming system, Hamadan, Iran. J. Rangeland Sci. 1, 1247–1254.
DeLacy, I. H. (1981). “Cluster analysis for the interpretation of genotype by environment interaction,” in Interpretation of Plant Response and Adaptation to Agricultural Environments, eds D. E. Byth and V. E. Mungomery (Brisbane, QLD: Australian Institute of Agricultural Science), 277–292.
Di, H. Y., Duan, Z., Luo, K., Zhang, D. Y., Wu, F., Zhang, J. Y., et al. (2015). Interspecific phylogenic relationships within genus Melilotus based on nuclear and chloroplast DNA. PLoS ONE 10:e0132596. doi: 10.1371/journal.pone.0132596
Di, H. Y., Luo, K., Zhang, J. Y., Duan, Z., Huo, Y. X., and Wang, Y. R. (2014). Genetic diversity analysis of Melilotus populations based on ITS and trnL-trnF Sequences. Acta Bot. Boreal. Occident. Sin. 34, 0265–0269. doi: 10.7606/j.issn.1000-4025.2014.02.0265
Easton, H. S., Jahufer, M. Z. Z., Flay, C., Schmidt, J., Rolston, M. P., Trethewey, J. A. K., et al. (2015). Environment and ryegrass genetics – multisite trial of advanced breeding populations. J. N.Z. Grasslands 77, 233–238.
Evans, P. M., and Kearney, G. A. (2003). Melilotus albus (Medik) is productive and persistent on saline soils of neutral to alkaline reaction in the high rainfall zone of south-west Victoria. Aust. J. Exp. Agric. 43, 349–355. doi: 10.1071/EA02079
Falconer, D. S. (1989). Introduction to Quantitative Genentics. New York, NY: Longman Scientific and Technical.
Fehr, W. R. (1987). Principles of Cultivar Development, Vol. 1. New York, NY: Collier Macmillan Publishers.
Gabriel, K. R. (1971). The biplot graphical display of matrices with application to principal component analysis. Biometrika 58, 453–467. doi: 10.1093/biomet/58.3.453
Goplen, B. P. (1971). Polara, a low coumarin cultivar of sweetclover. Can. J. Plant Sci. 51, 249–251. doi: 10.4141/cjps71-050
Goplen, B. P. (1981). NORGOLD - A low coumarin yellow blossom sweetclover. Can. J. Plant Sci. 61, 1019–1021. doi: 10.4141/cjps81-154
Gorz, H. J., Haskins, F. A., Manglitz, G. R., Smith, R. R., and Vogel, K. P. (1992). Registration of N28 and N29 sweetclover germplasms. Crop Sci. 32, 510.
GRIN (2000). Grin Taxonomy. United States Department of Agriculture, Agricultural, Agricultural Research Service. Baltimore, MD: The Germplasm Resources Information Network (GRIN).
Guler, M., Adak, M. S., and Ulukan, H. (2001). Determining relationships among yield and some yield components using path coefficient analysis in chickpea (Cicer arietinum L.). Eur. J. Agric. 14, 161–166. doi: 10.1016/S1161-0301(00)00086-1
Hallauer, A. R., and Miranda, J. B. (1981). Quantitative Genetics in Maize Breeding. Ames: IA State University Press.
Hill, R. R., and Baylor, J. E. (1983). Genotype × environment interaction analysis for yield in alfalfal. Crop Sci. 23, 811–815. doi: 10.2135/cropsci1983.0011183X002300050002x
Hofmann, R. W., and Jahufer, M. Z. Z. (2011). Tradeoff between biomass and flavonoid accumulation in white clover reflects contrasting plant strategies. PLoS ONE 6:e18949. doi: 10.1371/journal.pone.0018949
Hu, B., Yu, J., Zhao, Z. H., Li, F. M., and Siddique, K. H. M. (2012). Soil P availability, inorganic P fractions and yield effect in a calcareous soil with plastic-film-mulched spring wheat. Field Crop. Res. 137, 221–229. doi: 10.1016/j.fcr.2012.08.014
Iannucci, A., and Martiniello, P. (1998). Analysis of seed yield components in four mediterranean annual clovers. Field Crop. Res. 55, 235–243. doi: 10.1016/S0378-4290(97)00091-9
Ivanov, A. I., and Chetvertnykh, L. M. (1980). Evaluation of a collection of Melilotus species and varieties, and variation in their characters. Vestn. Selskokhozyaistvennoi Nauki 5, 117–122.
Jahufer, M. Z. Z., and Casler, M. (2015). Application of the smith-hazel selection index for improving biomass yield and quality of switchgrass. Crop Sci. 55, 1212–1222. doi: 10.2135/cropsci2014.08.0575
Jahufer, M. Z. Z., and Cooper, M., Ayres, J. F., and Bray, R. A. (2002). Identification of future research to improve the efficiency of conventional white clover breeding strategies in Australia – a review. Aust. J. Agric. Res. 53, 239–256. doi: 10.1071/AR01110
Jahufer, M. Z. Z., Cooper, M., Bray, R. A., and Ayres, J. F. (1999). Evaluation of white clover (Trifolium repens L.) populations for summer moisture stress adaptation in Australia. Aust. J. Agric. Res. 50, 561–574. doi: 10.1071/A98141
Julier, B., Christian, H., and Christian, E. (2000). Within- and among-cultivar genetic variation in alfalfa: forage quality, morphology, and yield. Crop Sci. 40, 365–369. doi: 10.2135/cropsci2000.402365x
Kephart, K. D., Buxton, D. R., and Hill, R. R. (1990). Digestibility and cell-wall components of alfalfa following selection for divergent herbage lignin concentration. Crop Sci. 30, 207–212. doi: 10.2135/cropsci1990.0011183X003000010045x
Klebesadel, L. J. (1992). Morphological, Physiological, and Winterhardiness Comparisons among Latitudinal Ecotypes of Biennial Sweetclover (Melilotus Species) in Subarctic Alaska. Fairbanks, AK: School of Agriculture and Land Resources Management Agricultural and Forestry Experiment Station.
Kroonenberg, P. M. K. (1994). The TUCKALS line: a suite of programs for three-way data analysis. Comput. Stat. Data Anal. 18, 73–96. doi: 10.1016/0167-9473(94)90133-3
Li, X. Y., Wang, Y. R., Wei, X., Tai, J. H., Jia, C. Z., Hu, X. W., et al. (2014). Planting density and irrigation timing affects Cleistogenes songorica seed yield sustainability. Agron. J. 106, 1690–1696. doi: 10.2134/agronj14.0111
Luo, K., Di, H.-Y., Zhang, J.-Y., Wang, Y.-R., and Li, Z.-Q. (2014). Preliminary evaluation of agronomy and quality traits of nineteen Melilotus accessions. Pratacultural Sci. 31, 2125–2134. doi: 10.11829/j.issn.1001-0629.2013-0455
Maddaloni, J. (1986). Forage production on saline and alkaline soils in the humid region of Argentina. Reclam. Reveg. Res. 5, 11–16.
Martino, E., Ramaiola, I., Urbano, M., Bracco, F., and Collina, S. (2006). Microwave-assisted extraction of coumarin and related compounds from Melilotus officinalis (L.) pallas as an alternative to soxhlet and ultrasound-assisted extraction. J. Chromatogr. A. 1125,147–151. doi: 10.1016/j.chroma.2006.05.032
Milligan, S. B., Gravois, K. A., Bischoff, K. P., and Martin, F. A. (1990). Crop effects on broad-sense heritabilities and genetic variances of sugarcane yield components. Crop Sci. 30, 344–349. doi: 10.2135/cropsci1990.0011183X003000020020x
Moll, R. H., and Stuber, C. W. (1974). Quantitative genetics-empirical results relevant to plant breeding. Advn. Agron. 26, 277–313. doi: 10.1016/S0065-2113(08)60874-3
Nair, R. M., Whittall, A., Hughes, S. J., Craig, A. D., Revell, D. K., Miller, S. M., et al. (2010). Variation in coumarin content of Melilotus species grown in South Australia. N.Z. J. Agr. Res. 53, 201–213. doi: 10.1080/00288233.2010.495743
Piano, E., Annicchiarico, P., Romani, M., and Pecetti, L. (2007). Effect of the number of parents and their combining ability on the performance of synthetic varieties in tall fescue. Aust. J. Agr. Res. 58, 1100–1105. doi: 10.1071/AR06424
Riday, H., and Brummer, C. (2007). Narrow sense heritability and additive genetic correlations in alfalfa subsp. falcata. J. Iowa Acad. Sci. 114, 28–34.
Rogers, M. E., Colmer, T. D., Frost, K., Henry, D., Cornwall, D., Hulm, E., et al. (2008). Diversity in the genus Melilotus for tolerance to salinity and waterlogging. Plant Soil 304, 89–101. doi: 10.1007/s11104-007-9523-y
Sagalbekov, U. M. (1980). Evaluation of a collection of sweet clover in the southern forest steppe of omsk province. Nauch.Tekhn. Byul. Sib. NII S. Kh. 3–6.
Sherif, E. A. (2009). Melilotus indicus (L.) all, a salt-tolerant wild leguminous herb with high potential for use as a forage crop in salt-affected soils. Flora 204, 737–746. doi: 10.1016/j.flora.2008.10.004
Smith, W. K., and Gorz, H. J. (1965). Sweetclover improvement. Adv. Agron. 17, 163–231. doi: 10.1016/S0065-2113(08)60414-9
Su, Y. Z., Zhao, W. Z., Su, P. X., Zhang, Z. H., Wang, T., and Ram, R. (2007). Ecological effects of desertification control and desertified land reclamation in an oasis-desert ecotone in an arid region: a case study in Hexi Corridor, northwest China. Ecol. Eng. 29, 117–124. doi: 10.1016/j.ecoleng.2005.10.015
Turkington, R. A., Cavers, P. B., and Rempel, E. (1978). The biology of Canadian weeds. 29. Melilotus alba Desr. and M. officinalis (L.) Lam. Can. J. Plant Sci. 58, 523–537. doi: 10.4141/cjps78-078
Watson, S. L., DeLacy, I. H., Podlich, D. W., and Basford, K. E. (1995). GEBEIL: An Analysis Package Using Agglomerative Hierarchical Classificatory and Svd Ordination Procedures for Genotype × Environment Data. Brisbane, QLA: Centre for Statistics Research Report; Department of Agriculture, University of Queensland.
White, T. L., and Hodge, G. R. (1989). “Predicting breeding values with applications in forest tree improvement,” in Forestry Sciences 33, ed P. K. R. Nair (Boston, MA: Kluwer Academic), 367.
Whitson, T. D., Burrill, L. C., Dewey, S. A., Cudney, D. W., Nelson, B. E., Lee, R. D., et al. (1992). Weeds of the West (Edited by T. D. Whitson). Laramie: The Western Society of Weed Science in cooperation with the Western United States Land Grant Universities Cooperative Extension Services and the University of Wyoming.
Wu, F., Zhang, D. Y., Ma, J. X., Luo, K., Di, H. Y., Liu, Z. P., et al. (2016). Analysis of genetic diversity and population structure in accessions of the genus Melilotus. Ind. Crop. Prod. 85, 84–92. doi: 10.1016/j.indcrop.2016.02.055
Zhang, Y., He, Z. H., Zhang, A. M., Ginkel, M. V., Pena, R. J., and Ye, G. Y. (2006). Pattern analysis on protein properties of Chinese and CIMMYT spring wheat cultivars sown in China and CIMMYT. Aust. J. Agric. Res. 57, 811–822. doi: 10.1071/AR05372
Keywords: forage breeding, genotypic variation, genotype-by-environment interactions, correlation coefficient, coumarin
Citation: Luo K, Jahufer MZZ, Wu F, Di H, Zhang D, Meng X, Zhang J and Wang Y (2016) Genotypic Variation in a Breeding Population of Yellow Sweet Clover (Melilotus officinalis). Front. Plant Sci. 7:972. doi: 10.3389/fpls.2016.00972
Received: 16 December 2015; Accepted: 20 June 2016;
Published: 12 July 2016.
Edited by:
Nicolas Rispail, Institute for Sustainable Agriculture - CSIC, SpainReviewed by:
Inger Martinussen, Norwegian Institute of Bioeconomy Research, NorwayJuan Marcelo Zabala, Universidad Nacional del Litoral, Argentina
Copyright © 2016 Luo, Jahufer, Wu, Di, Zhang, Meng, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiyu Zhang, emhhbmdqeUBsenUuZWR1LmNu;
Yanrong Wang, eXJ3YW5nQGx6dS5lZHUuY24=
†These authors have contributed equally to this work.
 Kai Luo
Kai Luo M. Z. Z. Jahufer2†
M. Z. Z. Jahufer2† Jiyu Zhang
Jiyu Zhang