- 1The Key Laboratory of Biotechnology for Medicinal Plant of Jiangsu Province, School of Life Science, Jiangsu Normal University, Xuzhou, China
- 2Centre for Transformational Biotechnology of Medicinal and Food Plants, Jiangsu Normal University – The University of Edinburgh, Xuzhou, China
- 3School of Bioengineering and Biotechnology, Tianshui Normal University, Tianshui, China
- 4Institute of Molecular Plant Sciences, School of Biological Sciences, The University of Edinburgh, Edinburgh, UK
In agriculture, seed mass is one of the most important components related to seed yield. MINISEED3 (MINI3) which encodes the transcriptional activator WRKY10, is thought to be a pivotal regulator of seed mass. In Arabidopsis SHORT HYPOCOTYL UNDER BLUE1 (SHB1) associates with the promoter of MINI3, regulating embryo cell proliferation (both cell division and elongation), which, in turn, modulates seed mass. Furthermore, the recruitment of SHB1 via MINI3 to both its cognate promoter and that of IKU2 implies a two-step amplification for countering the low expression level of IKU2, which is thought to function as a molecular switch for seed cavity enlargement. However, it is largely unknown how embryo cell proliferation, which encompasses both cell division and elongation, is regulated by SHB1 and MINI3 function. Here, we show that a loss of function mutation within the transcriptional coactivator ANGUSTIFOLIA3 (AN3), increases seed mass. Further, AN3 associates with the MINI3 promoter in vivo. Genetic evidence indicates that the absence of MINI3 function suppresses the decrease of cell number observed in an3-4 mutants by regulating cell division and in turn inhibits increased cell size of the an3-4 line by controlling cell elongation. Thus, seed embryo development is modulated via an AN3-MINI3 gene cascade. This regulatory model provides a deeper understanding of seed mass regulation, which may in turn lead to increased crop yields.
Introduction
ANGUSTIFOLIA3 (AN3)/GRF-INTERACTING FACTOR1, encodes a homolog of the human transcription coactivator, synovial sarcoma translocation protein (SYT) (Horiguchi et al., 2005). AN3, a member of a small gene family in Arabidopsis (Horiguchi et al., 2005), has been implicated in modulating cell proliferation, adaxial/abaxial determination in leaf primordia and establishing cotyledon identity (Horiguchi et al., 2005, 2011; Kanei et al., 2012). AN3/GIF1 has been shown to function as a component of a complex implicated in modulating the growth and shape of leaf blades and petals (Kim and Kende, 2004). AN3 expression is restricted to within mesophyll cells and is not detected within epidermal cells (Horiguchi et al., 2011).
Seed mass is modulated through three major components – embryo, endosperm and seed coat – derived from different cells of the ovule and with distinct complements of maternal and paternal genomes. In angiosperms, seed development involves a double-fertilization process in which two polar cells fuse to form the central cell before fertilization. Thus, one sperm cell fuses with the egg cell and the other fuses with the diploid central cell to form the triploid endosperm (Lopes and Larkins, 1993). When seed maturity is completed, the seed possesses only a single layer of endosperm cells and the maternal integument forms the seed coat. The embryo is surrounded by the endosperm, which in turn is surrounded by the maternal seed coat. Therefore, the coordinated growth of maternal sporophytic and zygotic tissues determines seed mass.
The accumulating evidence suggests that seed mass is genetically regulated. The apetala2 (ap2) mutant has enhanced seed mass due to increases of both embryonic cell size and cell number. DA1 which functions as a growth-repressor, is a ubiquitin receptor with two ubiquitin interaction motifs. Further, the da1-1 mutant exhibits large seed mass due to alterations in the maternal integuments of ovules (Li et al., 2008). Mutants in an enhancer of da1-1, EOD1, encoding the E3 ubiquitin ligase, BIGBROTHER (BB) (Disch et al., 2006; Li et al., 2008), synergistically increase the seed mass phenotype of da1-1. Thus, revealing that DA1 acts synergistically with EOD1/BB to regulate seed size.
A triple mutant of the three cytokinin receptors produces twice the seed mass as the corresponding wild-type line. In this context, it has been proposed that cytokinin might regulate embryo mass through a maternal and/or endosperm based mechanism (Hutchison et al., 2006; Riefler et al., 2006). Arabidopsis auxin response factor2 (arf2) mutant plants have extra cell proliferation in the integuments of the mutant ovules and therefore have enlarged seed coats and ultimately enhanced seed mass (Schruff et al., 2006). MINISEED3 (MINI3) which encodes a WRKY transcription factor, is expressed in both endosperm and embryo (Luo et al., 2005), while HAIKU2 (IKU2), a leucine-rich repeat receptor kinase gene, is predominantly expressed in the endosperm but its expression is absent from the embryo (Luo et al., 2005). Both iku2 and mini3 plants exhibit decreased seed mass and their seed phenotypes are determined through the genotype of either embryo or endosperm (Luo et al., 2005). Therefore, IKU1, IKU2, and MINI3 function in the same pathway of seed development (Garcia et al., 2003; Luo et al., 2005; Wang et al., 2010).
The gain-of-function mutant, SHORT HYPOCOTYL UNDER BLUE 1 (SHB1), exhibits a large seed mass as a result of increased cell number and enhanced cell size (Zhou et al., 2009). In contrast, the mini3 mutant has a small seed mass as a result of a decreased cell number (Luo et al., 2005; Zhou et al., 2009). Furthermore, the binding of SHB1 to both the MINI3 and IKU2 promoters suggests a two-step amplification for countering the low expression level of IKU2, which is thought to function as a molecular switch for seed cavity enlargement (Zhou et al., 2009; Kang et al., 2013). Recently, it has been proposed that ABA negatively regulates SHB1 transcription through the action of the basic leucine zipper protein, ABI5, which directly binds to ABRE cis-elements within the SHB1 promoter, which are required for the regulation of seed development (Cheng et al., 2014). Thus, ABI5-SHB1-MINI3-IKU1 forms a gene cascade that negatively controls embryo cell proliferation and by extension also seed mass. However, it is largely unknown how embryo cell division and elongation is fine-tuned and consequently, how in detail seed mass is regulated. Uncovering this mechanism will provide a better understanding of seed mass regulation.
In the present study, we have found that the loss-of-function mutant, an3-4, exhibited increased seed mass. Further, molecular analysis indicated that MINI3 was a target gene of AN3. Genetically, AN3 acted upstream of MINI3: mutation of MINI3 inhibited the decrease of cell number (cell division) in the an3-4 mutant and suppressed the increase of cell size (cell elongation) during embryo development of an3-4 plants. Thus, an AN3-MINI3 gene cascade may regulate seed embryo development by amplification (cell division and cell elongation). Our proposed model provides a greater understanding of seed mass regulation, which may in turn help guide approaches to increase crop yields.
Materials and Methods
Plant Materials and Growth Conditions
The an3-4 and mini3-2 mutants were described previously (Horiguchi et al., 2005; Zhou et al., 2009). shb1 (SALK_128406), grf1 (SALK_001350), mini3-2(SALK_050364), iku2-4 (SALK_073260), and ap2 (SALK_071140) were in the Col background and obtained from the ABRC (Ohio State University, Columbus). The an3 mini3 double-mutant was obtained from F2 seedlings of an3-4 ×mini3-2 that had a narrow rosette leaf phenotype of plants grown in white light (the an3-4 phenotype for homozygous lines; Horiguchi et al., 2005). The an3 mini3 mutant was confirmed in F3 by PCR genotyping of MINI3 using the gene-specific primers for MINI3 described by Zhou et al. (2009). grf1 (SALK_001350) homozygous lines were obtained through herbicide selection for three or more generations and analysis of segregation ratios. Absence of gene expression in the mutant was verified by RT-PCR. pMD111-Pro MINI3:GFP (MINI3 pro:GFP) were introduced into the 35S:AN3 background by Agrobacterium mediated transformation of 35S:AN3 homozygous plants. Transformants were selected on hygromycin B for three or more generations and segregation ratios analyzed. This expression levels of transgenes in these transgenic lines was confirmed. The resulting seeds were subjected to 4°C for 3 days, and then sown onto solid Murashige and Skoog (MS) medium supplemented with 1% sucrose at pH 5.8 and 0.8% agar. The seedlings grown on agar were maintained in a growth room under 16/8 h light/dark cycles with cool white fluorescent light at 21 ± 2°C. Plants grown in soil-less media were maintained in a controlled environment growth room under 16/8 h light/dark cycles with cool white fluorescent light at 21 ± 2°C under constant light (120 μmol⋅m-2⋅s-1 light from a mixture of fluorescent and incandescent bulbs).
Semi-Quantitative RT-PCR (SQ-RT-PCR) and Quantitative PCR
Total RNA was extracted from tissues indicated in the figures by the TRIZOL reagent (Invitrogen), as described by Meng et al. (2015a,b). SYBR [Sangon Biotech (Shanghai) Co., Ltd] was used to monitor the kinetics of PCR product in real-time RT-PCR (Meng et al., 2015a,b). Briefly, first-strand cDNA samples were produced from total RNA samples via reverse transcription using an AMV reverse transcriptase first-strand cDNA synthesis kit (Life Sciences, Promega) and were used as templates for RT-PCR–based gene expression analysis. For qRT-PCR analysis, after RNA isolation, reverse transcription was performed according to the manufacturer’s protocol (M-MLV reverse transcription system; Promega), followed by qPCR analysis to determine the gene expression level (Bio-Rad iQ5). The oligonucleotide primer sequences used to amplify specific cDNAs are described in Supplementary Table S2.
For analyzing AP2, SHB1, IKU2, and MINI3 expressions in 10–12 days-after-pollination (DAP) siliques of an3-4, grf1 and wild-type (whole siliques), the primers were employed as shown in Supplementary Table S2. For analyzing AN3 expression in mini3-2, shb1 (SALK_128406), iku2-4 (SALK_073260), and ap2 (SALK_071140) 10–12 DAP siliques (whole siliques), the primers were employed as shown in Supplementary Table S2. For SQ-RT-PCR analysis, the primers were utilized as shown in Supplementary Table S2. These experiments were all repeated at least twice with similar results.
Plasmid Constructs
By promoter analysis, an AN3 (At5g28640) promoter–GUS construct was produced through inserting a ∼2.0-kb promoter fragment (pGWC vector, Gateway, Lei et al., 2007), amplified by specific primers (Supplementary Table S2). These constructs were transferred into pCB308R, as described by Lei et al. (2007). For obtaining pCB2004-35S-AN3 in the associated plasmid, primers were outlined in Supplementary Table S2. For obtaining pMD111-MINI3:GFP in the associated plasmid, the relevant primers are listed in Supplementary Table S2.
ChIP Assay
Transgenic lines over-expressing 35S:AN3:3XGFP (Kawade et al., 2010) in an an3-4 genetic background and transgenic 35S:GFP lines were used in this assay. ChIP was performed using transgenic plants 10–12 DAP whole siliques as materials. The details of the ChIP process were described by Saleh et al. (2008). Briefly, Agrobacterium with a GFP construct was introduced into an3-4 10–12 DAP siliques using a leaf disk method (Meng et al., 2016). Two days after transformation, these transgenic materials (an3-4-GFP) were used for ChIP. 1.5 g of 10–12 DAP siliques of 35S:AN3-3XGFP plants were harvested and then fixed in 1% formaldehyde for 10–15 min in vacuum and neutralized with 0.125 M Gly in vacuum for an additional 5 min. After washing twice with cold, sterile water, these tissues were ground in liquid nitrogen. Nuclei were isolated and sonicated. Sonicated chromatin supernatant (250 μL) was diluted to 3, and 20 mL of protein A-agarose bead (Upstate) was added for preclearing at 4°C for 1–2 h. Then, the chromatin was divided into two 1.5-mL aliquots. Five microliters of mouse GFP and HA tag-specific monoclonal antibody (Sigma–Aldrich) was added to one tube. After incubating at 4°C overnight, beads were washed with low-salt wash buffer, high-salt wash buffer and TE (for Tris– EDTA buffer [10 mM Tris, 1 mM EDTA, pH8.0]) buffer. Elution and reverse cross-linking were performed as previously described (Saleh et al., 2008). Eluates were treated with Proteinase K (10 mg/mL; Sigma–Aldrich) and RNase for 3.0 h at 45°C, then phenol/chloroform extracted and ethanol precipitated with the aid of 20 μg of glycogen. The purified DNA was resuspended in 50 mL of water. The enrichment of DNA fragments was measured by qPCR using primers listed in Supplementary Table S2 online.
GUS Assay
Plants were incubated at 37°C for 8–10 h in a buffer mix of 0.4 mM of K3Fe(CN)6/K4Fe(CN)6, 1 mM X-gluc, 60 mM NaPO4 buffer and 0.1% (v/v) Triton X-100. Chlorophyll was removed using sequential washes for 30 min with 30, 50, 70, 90, and 100% ethanol, as described by Meng (2015) and Meng and Yao (2015).
Protein Analysis
Total protein extracts from 15 mature seeds of wild-type, an3-4 mutants and transgenic plants harboring the 35S:AN3:3XGFP transgene in an an3-4 background were fractionated on 8% SDS-PAGE and stained with Coomassie Brilliant Blue as described by Ohto et al. (2005).
Confocal Laser Scanning for GFP Imaging
Subcellular localization of MINI3: Green Fluorescent Protein (GFP) fusion proteins was undertaken in the abaxial epidermis of 8-day-old transgenic plants harboring pMD111-MINI3:GFP. An Olympus IX-70 microscope1 was used for detecting GFP expression using λ = 488 nm and λem = 510 nm. The sections were photographed under a confocal laser scanning microscope.
Cytological Experiments
Average seed weight was tested through weighing mature dry seeds in batches of 100 using an electronic analytical balance (Mettler, Toledo). Measurements of cotyledon area were made through scanning these organs to form a digital image and then calculating area using Image J software. Mature seeds were photographed under relevant magnification using a HIROX three-dimensional video microscope.
Mature dried seeds were imbibed for 60–100 min and dissected under microscope for isolating mature embryos. The embryos were incubated overnight in buffer (30 mM sodium phosphate, pH 7.0, 10 mM EDTA, 1% Triton X-100 and 1% DMSO) at 37°C, fixed for 1 h in buffer (FAA with 10% formalin, 5% acetic acid and 45% ethanol) and 0.01% Triton X-100, and dehydrated with an ethanol series, as described by Ohto et al. (2005). The embryos were then treated for 1–2 h in Hoyer’s buffer (3:0.8:0.4 of chloral hydrate: water: glycerol). A HIROX three-dimensional video microscope, under relevant magnification, was used to observe the treated embryos. The cell size of cotyledon embryos and the cell size of globular stage embryos were measured using Image J software.
Results
an3-4 Plants Have Increased Seed Mass
While investigating drought tolerance exhibited by an3-4 plants, we observed this line had larger cotyledons than wild-type (Col-0). To confirm whether these large cotyledons were due to a lack of AN3 protein activity, we investigated the phenotype of an3-4 complemented lines (Kanei et al., 2012). These lines harboring a 35S:AN3 transgene in an an3-4 genetic background exhibited similar cotyledon size to Col-0 (Supplementary Figures S1A,B). Significantly, AN3 interacts with GRF1 to regulate leaf development (Horiguchi et al., 2005). However, a GRF1 loss-of-function mutant showed similar cotyledon size relative to wild-type (Supplementary Figures S1A,B), implying GRF1 may not be involved in the regulation of seed mass. Moreover, while Ferjani et al. (2007) reported that the an3-4 mutant presented both larger cotyledons and embryos than wild-type (Col-0), an impact on seed mass was not highlighted. Taken together our data suggests that the an3-4 plants have larger cotyledons relative to wild-type (Col-0).
Further the seeds of self-pollinated homozygous an3-4 mutant plants weighed ∼60% more than Col-0 and an3-4 complemented lines. Also, they exhibited a pale orange coloration relative to wild-type [Figures 1A(a–c),B]. The area, length and width of these seeds were all enhanced in an3-4 plants relative to wild-type [Figures 1A(a–c),C–E]. However, a GRF1 loss-of-function mutant was similar to wild-type for these traits (Figures 1A–E). Interestingly, aborted seeds can be observed in the 35S:AN3 complemented line, which may be a result of the imperfect activity of the 35S promoter in embryonic tissues [Figure 1A(c)]. AN3 is also a positive regulator of anthocyanin biosynthesis (Meng, 2015; Meng and Liu, 2015) and the 35S:AN3 an3-4 line largely restored anthocyanin accumulation in an3-4 plants [Figure 1A(c)]. Thus, AN3 is a regulator of both seed mass and anthocyanin biosynthesis, in a similar fashion to transparent testa glabra2 (ttg2) (Garcia et al., 2005). Collectively these data suggest that AN3 negatively regulates seed mass.
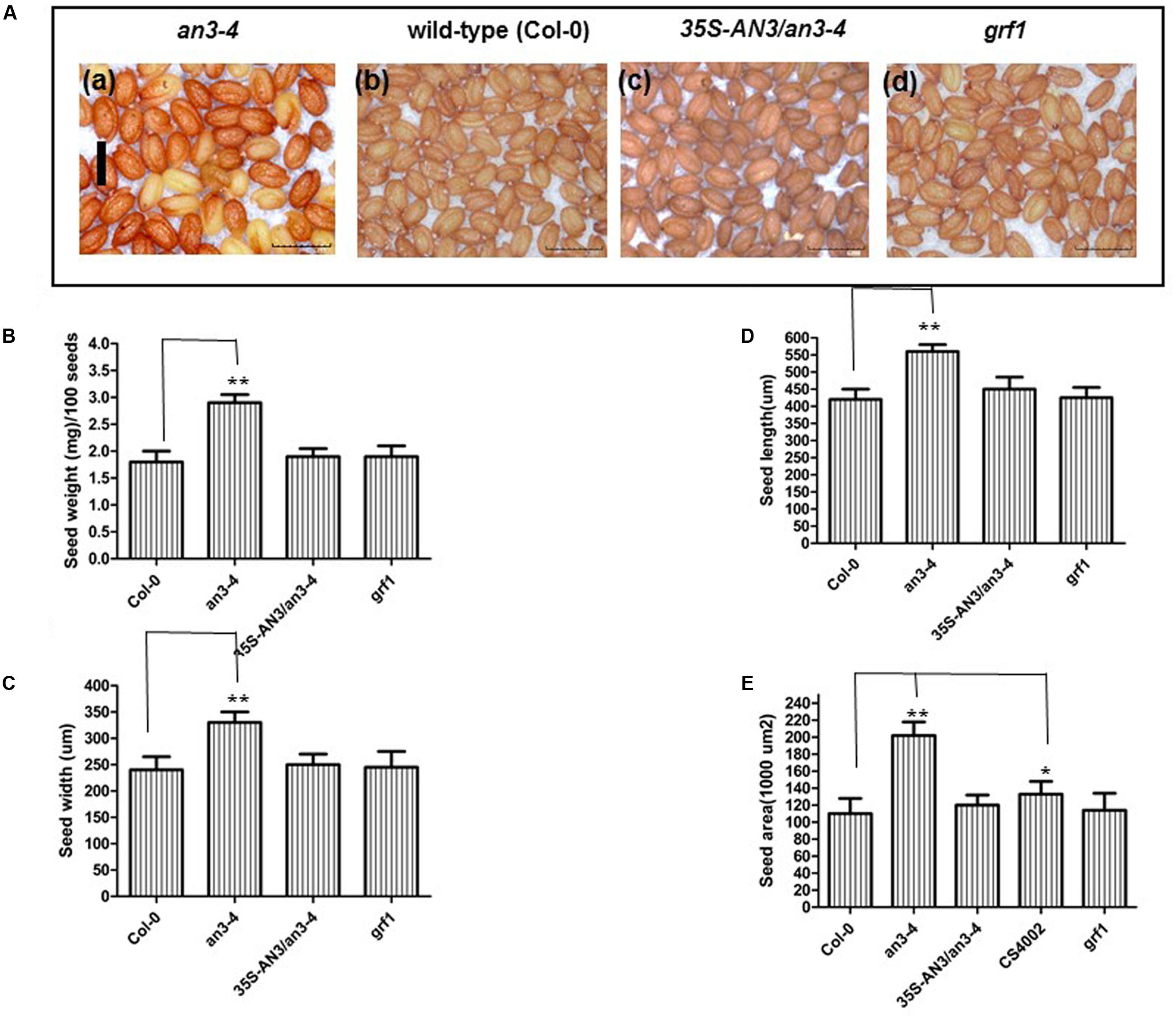
FIGURE 1. an3-4 mutant plants have increased seed mass. (A). Representative mature dry seeds of an3-4, Col-0, 35S-AN3/an3-4, and grf1 plants grown on solid MS medium with 1% sucrose, respectively. Bar = 0.1 mm for (a)–(d). (B–E) Bar graph showing the difference in average seed weight/100 seeds (B), seed width (C), seed length (D), and seed area (E) between an3-4, Col-0, an3-4+35S-AN3 and grf1 seeds. Data are means ± SD from at least 10 independently propagated lines of an3-4, WT, an3-4+35S-AN3 and grf1 (∗P < 0.05; ∗∗P < 0.01; in B, n = 4; in C–E, n = 30).
an3-4 Mutant Plants Have Lower Seed Yield
Understanding the molecular machinery underpinning the control of seed mass may help guide future improvements in crop yields. A large seed mass is associated with an altered seed yield, for example, ap2 plants exhibited increased seed mass but gave a lower seed yield (Ohto et al., 2005). However, da1 plants had a larger seed mass and conversely, a higher seed yield (Li et al., 2008). Thus, we determined the relationship between increased seed mass in an3-4 plants and seed yield. We therefore analyzed the seed/silique number, silique length, seed/silique weight, flower number, elongated silique number, and total seed weight. While the an3-4 mutant exhibited increased seed mass, those values associated with total seed yield all declined (Figure 2F). Thus, seed/silique number, silique length, and seed/silique weight were all reduced in the an3-4 mutant line relative to Col-0 plants (Figures 2A–C). However, these parameters were not significantly different in grf1 plants relative to wild-type (Figures 2A–F). Due to a deficiency in reproductive development, the decrease in elongated silique number in an3-4 mutant plants relative to flower number resulted in a reduced seed yield (Figures 2D,E). Therefore, we determined flower morphology and found that the petals and leaf blades of an3-4 plants were smaller than those of wild-type (Supplementary Figure S2; Horiguchi et al., 2005). Occasionally, flowers on the primary inflorescences of an3-4 plants did not open in a similar fashion to wild-type. Further, stamens of an3-4 plants were dramatically shorter compared with wild-type (the frequency: 28.0 ± 2.2; Supplementary Figure S2). Also, in the an3-4 line, the dehiscence of anthers was severely delayed (Supplementary Figure S2). These observations might partially explain why the elongated silique number in an3-4 mutant plants was decreased relative to flower number. In aggregate, these data imply the, lack of AN3 activity led to increased seed mass but resulted in a lower seed yield.
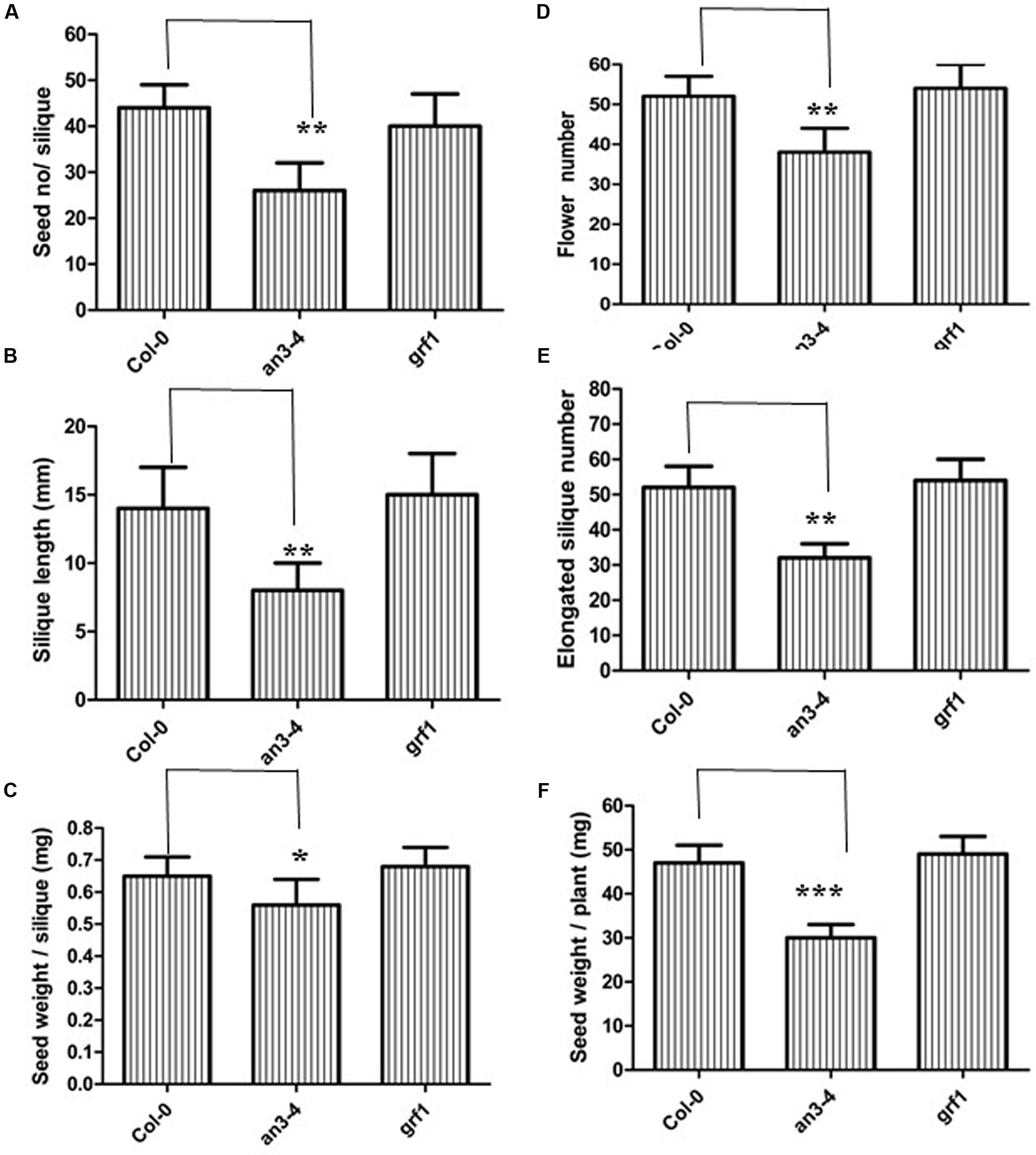
FIGURE 2. an3-4 mutant plants have low seed yield. For the given plant genotypes representative seed number/silique (A), silique length (B), seed weight/silique (C), flower number (D), elongation silique number (E), and seed weight/plant (F) are shown, respectively. Data are means ± SD from at least 10 independently propagated Col-0, mutant lines or independent transgenic lines (∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05, respectively; in A, n = 12; in B, n = 10; in C, n = 15; in D, n = 6; in E, n = 6; in F, n = 6).
The Increased Seed Mass of an3-4 Plants May Be Related with Low Fertility and the Absence of AN3
An allocation of extra resources for fewer seeds produced, due to decreased fertility, can also result in larger seed mass (Ohto et al., 2005). Thus, we determined whether increased seed mass in an3-4 plants was caused by an allocation of extra resources. We hand-pollinated five flowers from the primary inflorescences of wild-type, an3-4 plants and a male-sterile mutant (CS4002). Manual pollination ensured that all siliques had similar numbers of seeds. Thus, flowers were pollinated using pollen from the same genotype, but wild-type pollen was used as the donor in male-sterile plants. After maturity, each male-sterile mutant produced five siliques. On average, the seed weight was ∼18% higher when derived from the male-sterile plant compared to wild-type (Figure 1E), suggesting that reduced fertility can enhance seed weight. However, the average seed weight of the an3-4 mutant was ∼60% higher than that of the wild-type (Figure 1E), indicating that the increased seed mass exhibited by an3-4 plants may be partially associated with low fertility and in addition to an absence of AN3 function.
The Large Seeds of an3-4 Plants Are Due to Increased Embryo Cell Size
The embryo constitutes the major volume of a mature seed in Arabidopsis and thus changes in embryo cell size impact seed mass. Thus, we isolated and visualized mature embryos originating from the self-pollinated homozygous seeds of wild-type, complemented an3-4 lines and an3-4 plants. The an3-4 mature embryos were larger than those of wild-type and complemented an3-4 lines (Figures 3A,C). However, these parameters were similar in grf1 plants relative to wild-type (Figures 3A,C). Ferjani et al. (2007) showed that the an3-4 mutant presented larger embryos than wild-type (Col-0), but this was not mentioned by these authors. Cytological observations indicated that the average area of cotyledon embryos from an3-4 plants was ∼1.40 times that of wild-type (Figures 3B,D). On average, the epidermal cell size of an3-4 embryos was ∼1.70 times that of wild-type (Figures 3B,D). These data (i.e., 1.40/1.70 = 0.8) indicated that the increased embryo size exhibited by an3-4 plants was due to embryo cell elongation. AN3 positively regulates leaf cell proliferation (Horiguchi et al., 2005). The larger epidermal cells in an3-4 embryos are probably a result of a phenomenon termed compensation, in which a decrease in cell number triggers an increase in mature cell size (Ferjani et al., 2007; Fujikura et al., 2007). This compensation-induced cell enlargement is mostly independent from endoreduplication, however, the underpinning mechanism is unknown (Ferjani et al., 2007; Fujikura et al., 2007). Furthermore, the increased cell size phenotype (termed compensation) is a secondary effect triggered by impaired cell proliferation (Fujikura et al., 2009). Therefore, the absence of AN3 resulted in larger embryo cells which resulted in both larger embryos and seeds.
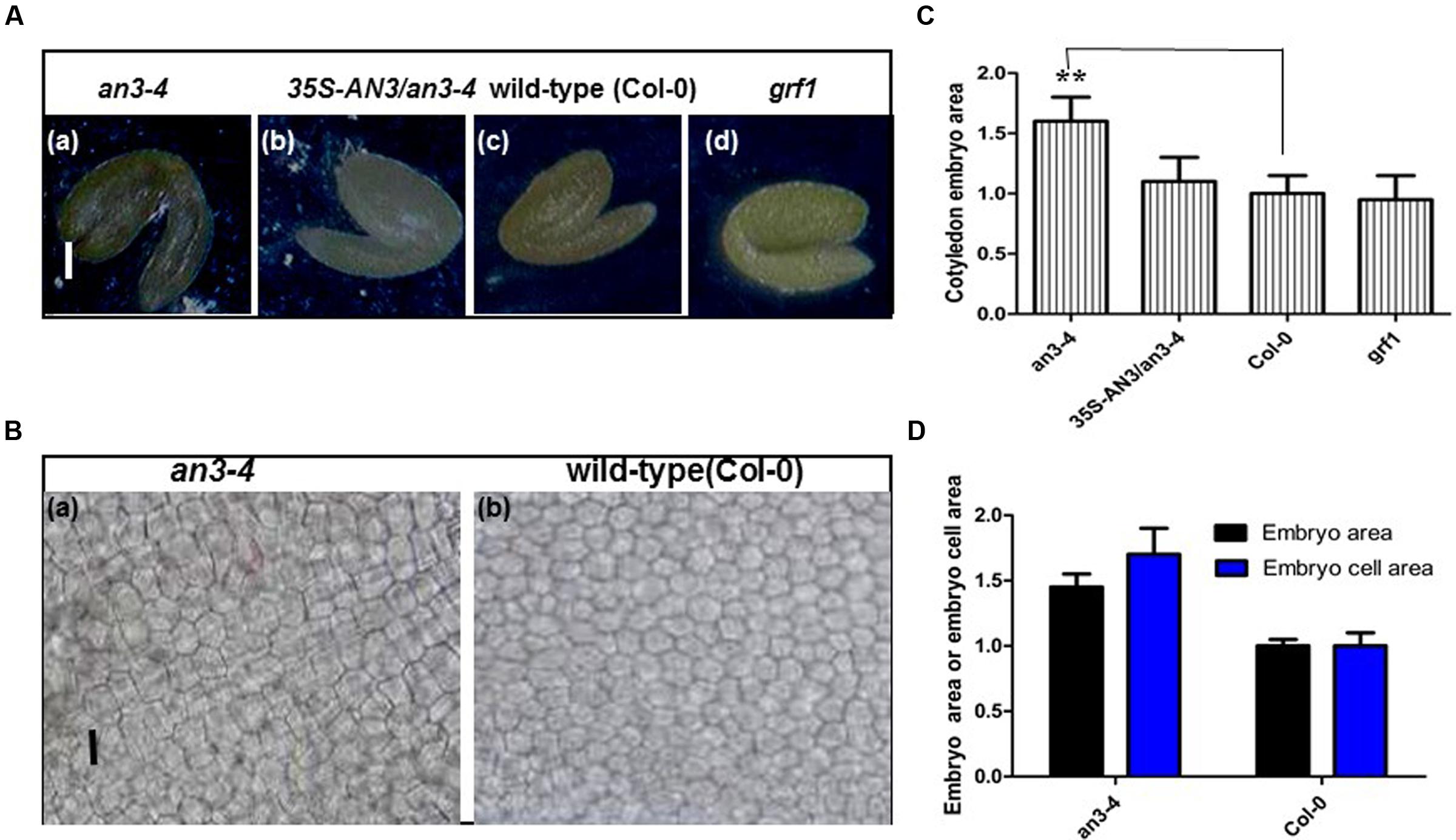
FIGURE 3. an3-4 mutant plants have large embryos caused by increased embryo cell size. (A) Representative mature embryos of an3-4, 35S-AN3/an3-4, Col-0, and grf1 plants grown on solid MS medium with 1% sucrose, respectively. Bars = 100 μm. (B) Representative epidermal cell layer derived from the central region of cotyledon embryos from an3-4 mutant and Col-0 seeds, respectively. Bars = 10 μm. (C) Bar graph showing the difference in cotyledon embryo area between an3-4, 35S-AN3/an3-4, Col-0, and grf1 plants. Data are means ± SD from at least five independently propagated Col-0, mutant lines and independent transgenic lines (∗∗P < 0.01; n = 10). Col-0 is set as 1.0. (D) Bar graph showing the difference in embryo area and embryo cell area between an3-4 and Col-0 plants. Col-0 is set as 1.0. Data are means ± SD from at least five independently propagated Col-0 and mutant lines (embryo area, n = 12; embryo cell area, n = 28).
Seeds from an3-4 Plants Exhibited an Increased Protein Content Relative to Wild-Type Seeds
Increased embryo cell size at the mature phase of seeds promotes accumulation of storage reserves (Ohto et al., 2005). Two major classes of Arabidopsis seed storage proteins are the 12S cruciferins and the 2S albumins (Heath et al., 1986; Pang et al., 1988). Therefore, we prepared protein extracts from 15 seeds each derived from an3-4, wild-type and complemented an3-4 lines. The 12S and 2S protein content form these seeds was then determined by SDS-PAGE. 12S proteins were obviously in greater abundance in seeds from the an3-4 line compared to wild-type, whereas levels of this protein did not significantly differ between complemented an3-4 lines and wild-type (Figure 4). However, the 2S albumin proteins were not retained in the 8% SDS-PAGE gel (Figure 4). These results suggest that 12S protein accumulation is increased in the absence of AN3 activity. This implied that the overall proportion of individual proteins were not affected by lack of AN3 activity.
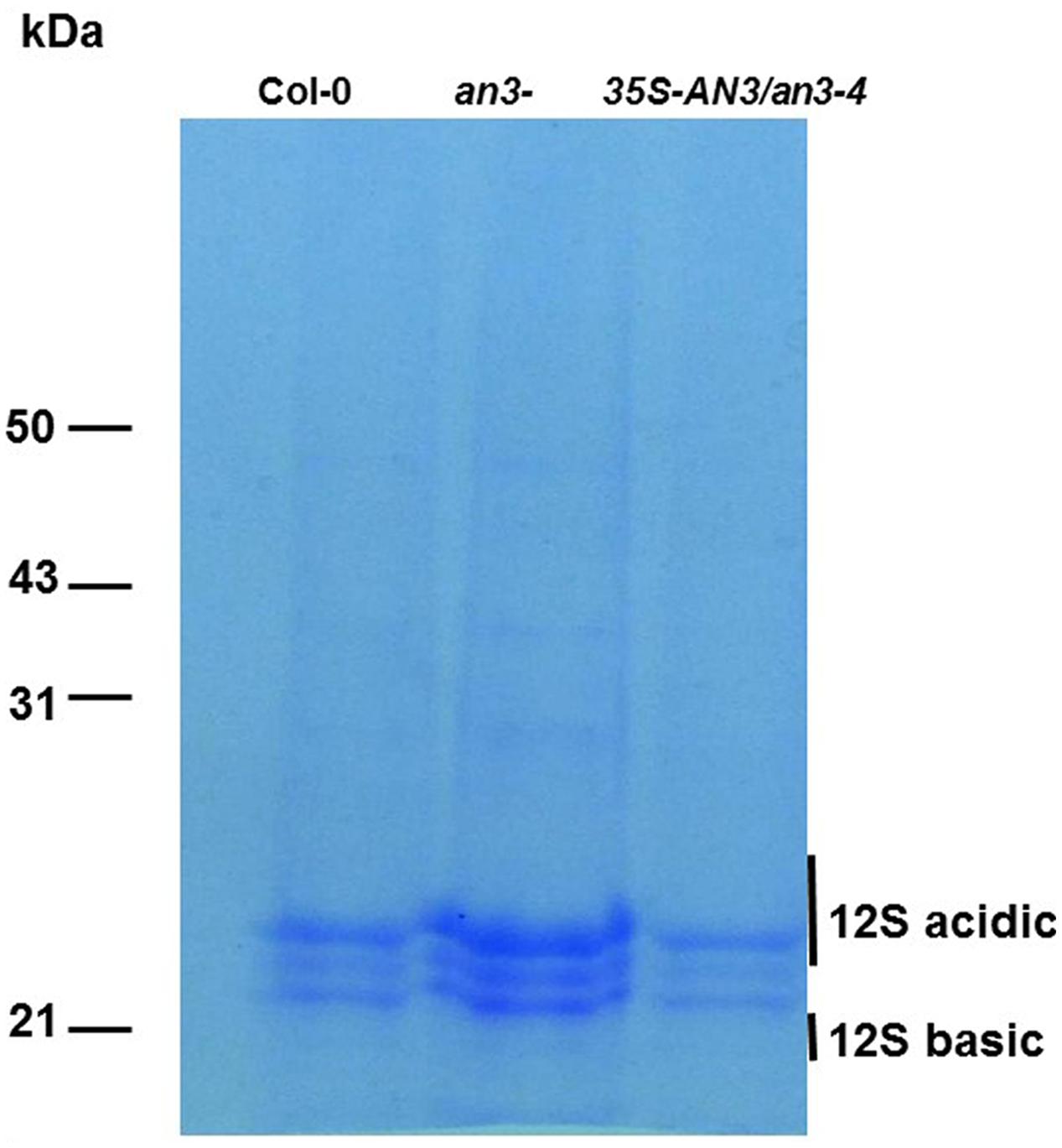
FIGURE 4. an3-4 protein analysis. an3-4 seeds contain more protein than wild-type seeds. Protein extracts from 15 wild-type and an3-4 seeds were fractionated on an 8% SDS/polyacrylamide gel and stained. Molecular mass markers are shown to the left of the gel.
AN3 Negatively Regulates MINI3 Expression during Reproductive Growth
It is well established that IKU2, MINI3, AP2, and SHB1 are involved in modulating seed mass (Luo et al., 2005; Ohto et al., 2005; Zhou et al., 2009). Therefore, we performed real-time PCR experiments to determine whether AN3 modulated expression of these genes using siliques 10–12 DAP from an3-4 and wild-type plants. MINI3 transcript levels in an3-4 plants were 4.6 times greater than those found in wild-type (P < 0.01, two-tailed Student’s t-test) (Figure 5A). Conversely, the transcript levels of complemented an3-4 lines were 0.6 times that of wild-type plants (P < 0.05). However, the expressions of IKU2, AP2 and SHB1 were not significantly different between an3-4 lines or Col-0 plants (Figure 5A). Also, the expressions of IKU2, AP2, MINI3, and SHB1 were not significantly different between the grf1 mutant and Col-0 plants (Figure 5A). Moreover, AN3 expression did not significantly differ between Col-0 plants and ap2, shb1, mini3, and iku2 mutants (Figure 5B). Thus, while MINI3 expression was increased in an an3-4 genetic background, AN3 expression was unaltered in mini3 plants relative to the wild-type line. Collectively, these data suggest that AN3 might negatively regulate MINI3 at the transcriptional level.
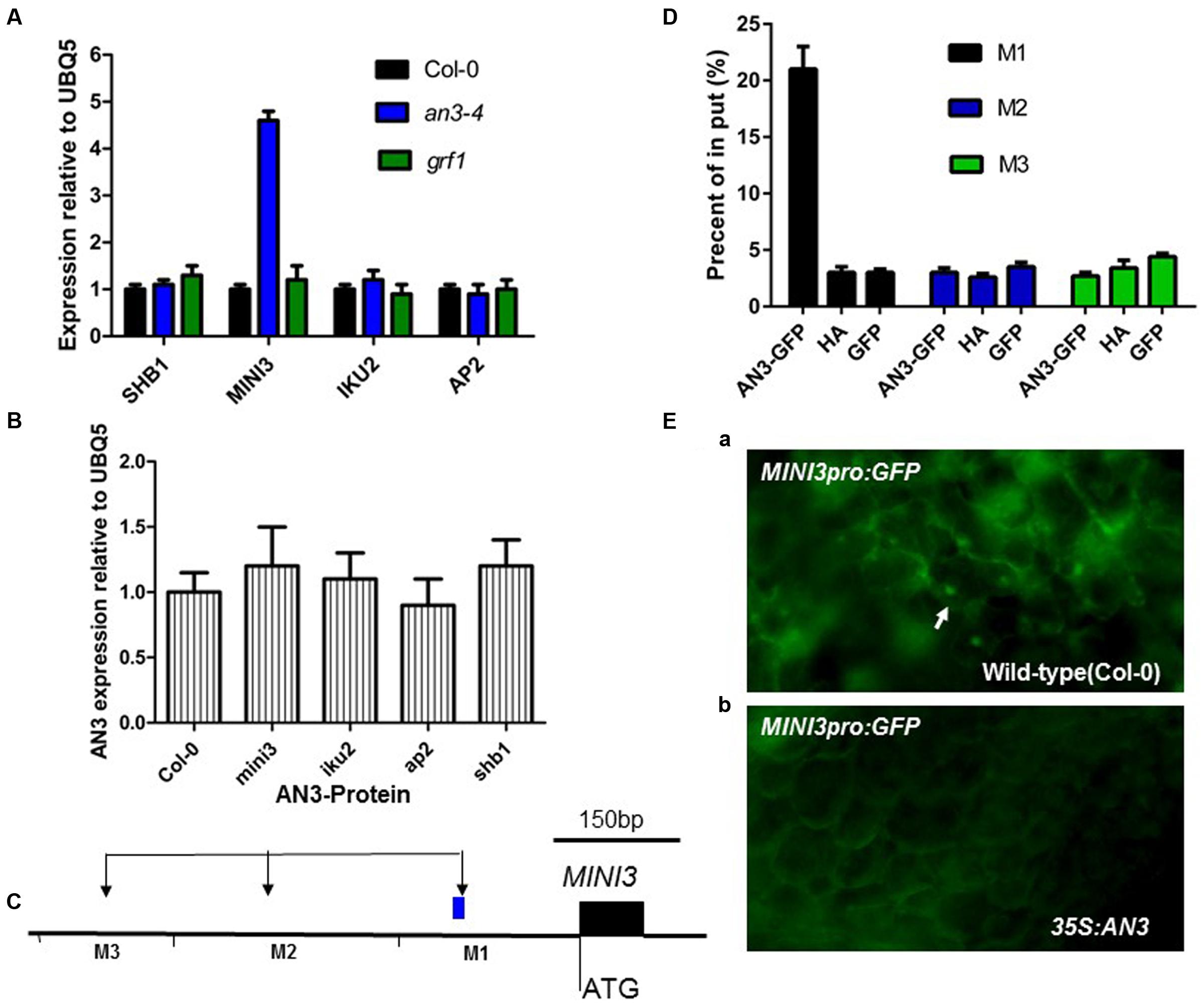
FIGURE 5. AN3 Associates with the MINI3 promoter in vivo during seed development. (A) Bar graph exhibiting the expression difference in SHB1, MINI3, IKU2, and AP2 between the WT (Col-0), an3-4, and grf1 10–12 DAP siliques. (B) Bar graph showing the difference in expression of AN3 between WT, mini3-2, iku2-4 (SALK_073260), ap2 (SALK_071140), and shb1 (SALK_128406) 10–12 DAP siliques. Three biological replicates and two technical repeats were undertaken. Error bars represent SD. (C) Schematic diagram of MINI3 and three amplicons initiating from the ATG of MINI3: M1, M2, and M3 used for ChIP analysis. (D) Bar graph showing AN3 association with the MINI3 promoter. ChIP was performed to analyze the in vivo interaction of AN3 with the MINI3 promoter. Input was chromatin before immunoprecipitation. Anti-GFP antibody was used for precipitating chromatin associated with 35S-AN3-3XGFP and 35S-GFP. Anti-HA antibody was used for precipitating chromatin associated with 35S-AN3-3XGFP. Anti-GFP antibody was used for precipitating chromatin associated with 35S-AN3-3XGFP. Anti-GFP antibody was used for precipitating chromatin associated with 35S-GFP. The MINI3 promoter region associated with AN3 was amplified by qPCR using MINI3 promoter-specific primers for distinct regions. Three biological replicates were performed. Error bars represent SD. (E) MINI3pro: GFP in WT (Col-0) (a) and 35S:AN3 (b) cotyledons. White arrows point to GFP-positive nuclei.
AN3 Represses MINI3 Promoter Activity
To confirm that AN3 regulated MINI3 expression, we performed chromatin immunoprecipitation (ChIP) analysis using 10–12 DAP siliques of transgenic plants harboring a 35S:AN3:3XGFP transgene. The 35S:GFP lines were used as a negative control. Chromatin associated with AN3–GFP and GFP was immunoprecipitated with an Anti-GFP antibody. Real-time PCR analysis was then performed with primers specific for different regions of the MINI3 promoter (Figure 5C). Regions of M1 (-210 to -370 bp) primers produced a large amount of PCR product (21% of Input), whereas less PCR product (3 or 2.7% of Input in M2 or M3, respectively) was detected in the region of M2 (-822 to -1053 bp) and M3 (-1258 to -1463 bp) primers (Figure 5D). Anti-HA used as a negative control resulted in significantly less PCR product, 3, 2.6 or 3.4% of input in the M1, M2, or M3, respectively (Figure 5D). Using the 35S:GFP lines as materials, less PCR product (3, 3.5, and 4.4% of Input in M1, M2, and M3, respectively) was detected in the region of M1, M2, and M3 primers (Figure 5D). At 8 days post-germination (dpg), wild-type plants expressed a transgene comprised of the MIN3 promoter adjacent to a gene encoding the Green Florescent Protein (MINI3pro:GFP) (13 of 15 plants). However, the MINI3pro:GFP is not expressed in a 35S:AN3 background (0 of 15 plants) [Figures 5E(a,b)], indicating that AN3 is a major factor repressing the MINI3 promoter. These results suggest that AN3 is associated with the MINI3 promoter for the control of seed mass. Moreover, AN3 was unable to bind directly to the MINI3 promoter sequences in a gel mobility shift assay (our unpublished data), which is consistent with the notion that AN3 is a transcriptional coactivator.
AN3 Expression Pattern
To further explore AN3 function, we examined the expression pattern of the β-glucuronidase (GUS) reporter gene driven by a native AN3 promoter. Analysis of Arabidopsis transformed with an approximately 2.0 kb AN3 promoter–GUS reporter gene fusion (AN3pro:GUS) showed that AN3pro:GUS was expressed in the seed coat, testa, stamens, petals, siliques, young leaves, stems and embryos, but not in endosperms, mature leaves and mature pollen grains (Figures 6A–J). Consistent with this, RT-PCR results indicated the presence of AN3 transcripts in the corresponding organs (Figure 6I). These results implied that AN3 is expressed in root, shoot, and flower organs; indicating AN3 is widely expressed. Moreover, Kanei et al. (2012) observed that AN3 can be strongly expressed in the globular, heart, torpedo, and cotyledon embryo. Thus, AN3 might function during the whole of embryo development. And Lee et al. (2014) suggested that AN3 might be expressed in the endosperm.
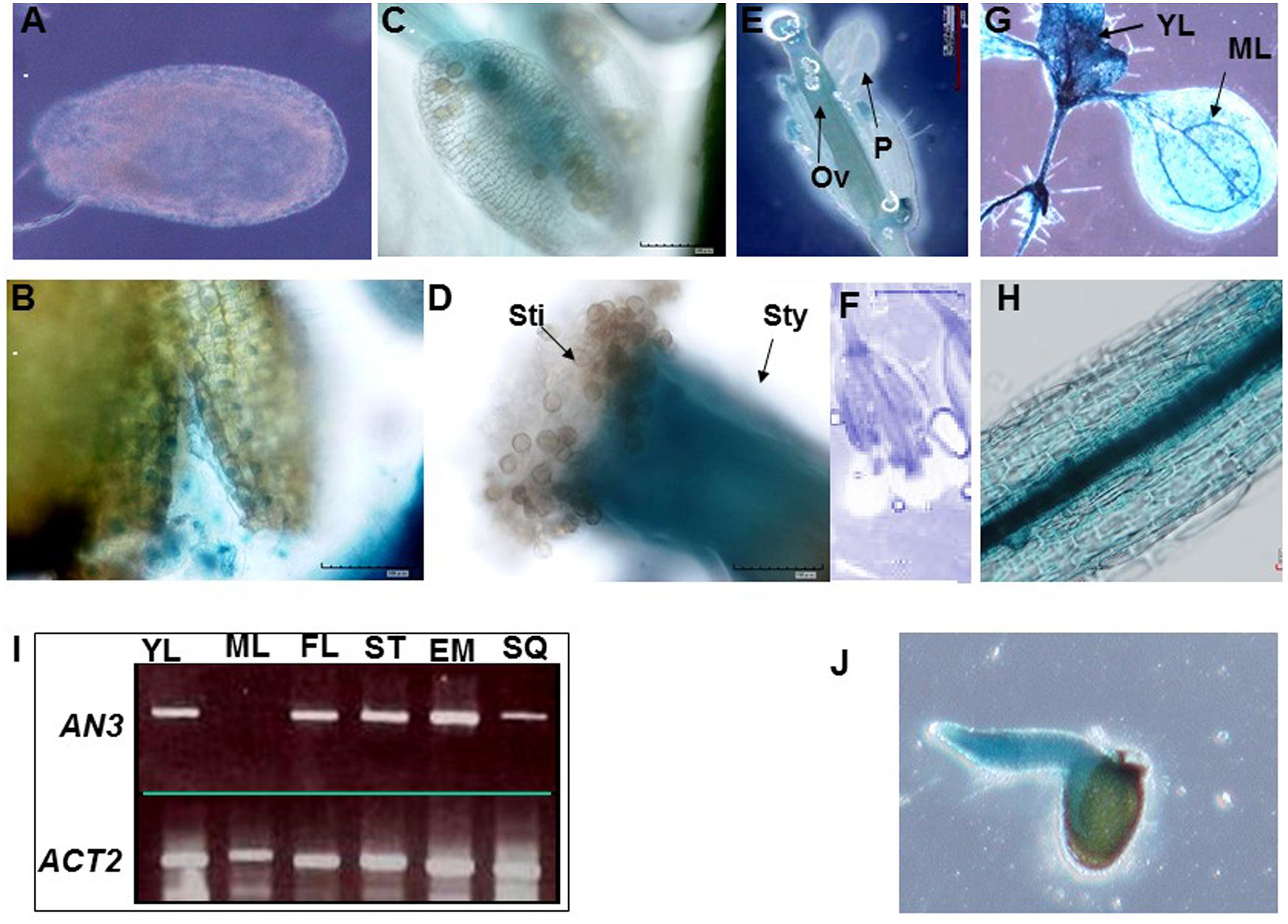
FIGURE 6. AN3 expression analysis. Expression of AN3pro:GUS in the seed coat (A), testa (B), mature pollen and stamens (C), stigma and style (D), flowers (E), and silique (F), seedlings with young leaves and mature leaves (G), stems (H), and embryos (J). Ov, ovule; P, petal; Sti, stigma; Sty, style. (I) AN3 expression analysis in several organs by SQ-RT-PCR. YL, young leaves; ML, mature leaves; FL, flower; ST, stem; EM, embryo; SQ, silique.
When an organ undergoes growth by proliferation, a fundamental event is the duration of cell proliferation, which generates a sufficient number of cells to support further growth and finally determines the overall size of the organ (Mizukami and Fischer, 2000). This process is modulated by several factors and our data suggests that AN3 contributes to this process.
AN3 Acts Maternally to Influence Seed Mass
The mass of a seed is modulated through the coordinated growth of maternal sporophytic and zygotic tissues. Reciprocal cross experiments between an3-4 and Col-0 plants were performed to determine whether AN3 functioned maternally or zygotically. When pollen of Col-0 and an3-4 was used as the donor and Col-0 plants were used as the acceptor, the seed mass resulting from the reciprocal crosses was not altered with the change of the donor (Col-0 and an3-4) (Supplementary Table S1). Similarly, when pollen of Col-0 and an3-4 was used as the donor and an3-4 plants were used as the acceptor, the seed mass of reciprocal crosses were also not altered with the change of the donor (Col-0 and an3-4) (Supplementary Table S1). On the contrary, with different acceptors (Col-0 and an3-4), the seed mass of reciprocal crosses was altered. This phenomenon indicates that in an3-4 mutant, the integument (the integument of Arabidopsis finally forms seed coat) determines the seed size (a maternal effect). Moreover, it is obvious that the genotype of the seed may be not relevant in this case of the maternal effect. These above findings suggest that an3-4 seed mass is determined by maternal patterns.
We also observed the embryo sac or seed cavity at 4 DAP in an3-4 and Col-0 plants. Cytological observations indicated that the average area of the ovule in an3-4 plants was ∼1.38 times that of wild-type (Supplementary Figures S3A–C). In a similar fashion, the average area of embryos from an3-4 plants was ∼1.40 times that of wild-type (Supplementary Figures S3A,B,D). These results indicate that the ovule and embryo in an an3-4 mutant is grown coordinately. Thus, the changes in seed mass maternally controlled by AN3 were reflected in the size of the embryo. On average, the cell area of the an3-4 maternal integument was ∼1.72 times that of wild-type (Supplementary Figures S3A,B,E). Similar with this, on average, the epidermal cell size of an3-4 embryos was ∼1.70 times that of wild-type (Figures 3B,D). These data (i.e., 1.40/1.72 = 0.8) indicated that the increased embryo (or integument) size of an3-4 plants may be due to embryo (or integument) cell elongation. Therefore, we concluded that the an3-4 mutant had a maternal influence on the regulation of seed mass, which agrees with a recent report (Lee et al., 2014). Hence, with the ovules matured, AN3/GIF1 expression is strongly detected in the chalazal portions of the ovule and in the initiating and developing integuments.
When cells in the outermost layer of the integument were counted, Lee et al. (2014) found that the gif1/an3 mutants had significantly decreased cell numbers, whereas integument mass of gif1/an3 but not gif1gif2gif3 plants was increased. This suggested that the cell size of gif1/an3 integuments was enhanced. These results further imply that AN3/GIF1 is involved in the regulation of integument development, which is consistent with the notion that the absence of AN3 activity has a maternal influence on the regulation of seed mass. Similar with AN3, KLU which encodes a cytochrome P450 and maternally influences seed mass, is expressed in the inner integument of developing ovules, where it non-cell autonomously stimulates cell proliferation, therefore determining the growth potential of both the seed coat and the seed (Adamski et al., 2009).
In a similar fashion to AN3, other reports (Garcia et al., 2005; Luo et al., 2005) have also showed the impact of maternal functions on seed mass. In the transparent testa glabra2 (ttg2) mutant, for example, endosperm growth is restricted as a consequence of prevention of cell elongation in the integument and is one of the few examples of maternal control of seed size in Arabidopsis (Garcia et al., 2005). The size of cells of the outer integument was correlated with embryo size in the mini3 mutant (Luo et al., 2005). Consequently, the mini3 mutant showed smaller seed mass. As embryo cell size is not altered in the mutant but the overall size of the embryo is smaller, the seed must contain fewer cells (Luo et al., 2005).
GRF1 Is Not Involved in Regulating MINI3 Expression during Seed Development
AN3/GIF1 and GRF1 regulate the growth and shape of leaf blades and petals through modulating cell proliferation (Kim and Kende, 2004). Therefore, we investigated whether AN3/GIF1 might act as a cofactor and interact with the GRF1 protein that is a specific transcription factor for the MINI3 promoter and mediate regulation of MINI3 transcription during seed development. grf1 plants had similar cotyledon size (Supplementary Figures S1A,B), seed mass (Figures 1A–E), seed/silique number, silique length, seed/silique weight, flower number, elongation silique number, total seed/plant weight (Figures 2A–F) and cotyledon embryo area (Figures 3A,C) to wild-type. In addition, expression levels in the developing siliques of SHB1, MINI3, IKU2, and AP2 did not significantly differ between wild-type and grf1 mutant plants (Figure 5A). These results suggest that, at least, AN3/GIF1 might act as a cofactor and interact with one or more unknown proteins that might function as specific transcription factors to regulate MINI3 transcription during seed development. However, our results suggest that this interacting protein is not GRF1.
AN3 Acts Genetically Upstream of MINI3 in Regulating Seed Mass
We performed double-mutant analysis, crossing an3-4, which has large seed mass, with mini3-2, which has small seed mass, to determine whether AN3 acted upstream of MINI3 in controlling this trait. Thus, we selected the genotypes from the segregating F3 population identified by PCR genotyping. mini3-2 obviously reduced the large seed phenotype of an3-4 (Figures 7A,C). The an3-4 mini3-2 double mutant had very similar seed mass to the mini3-2 single mutant (Figures 7A,C), indicating that mini3-2 was epistatic to an3-4. These genetic results confirmed that AN3 acted upstream of MINI3 in regulating seed mass.
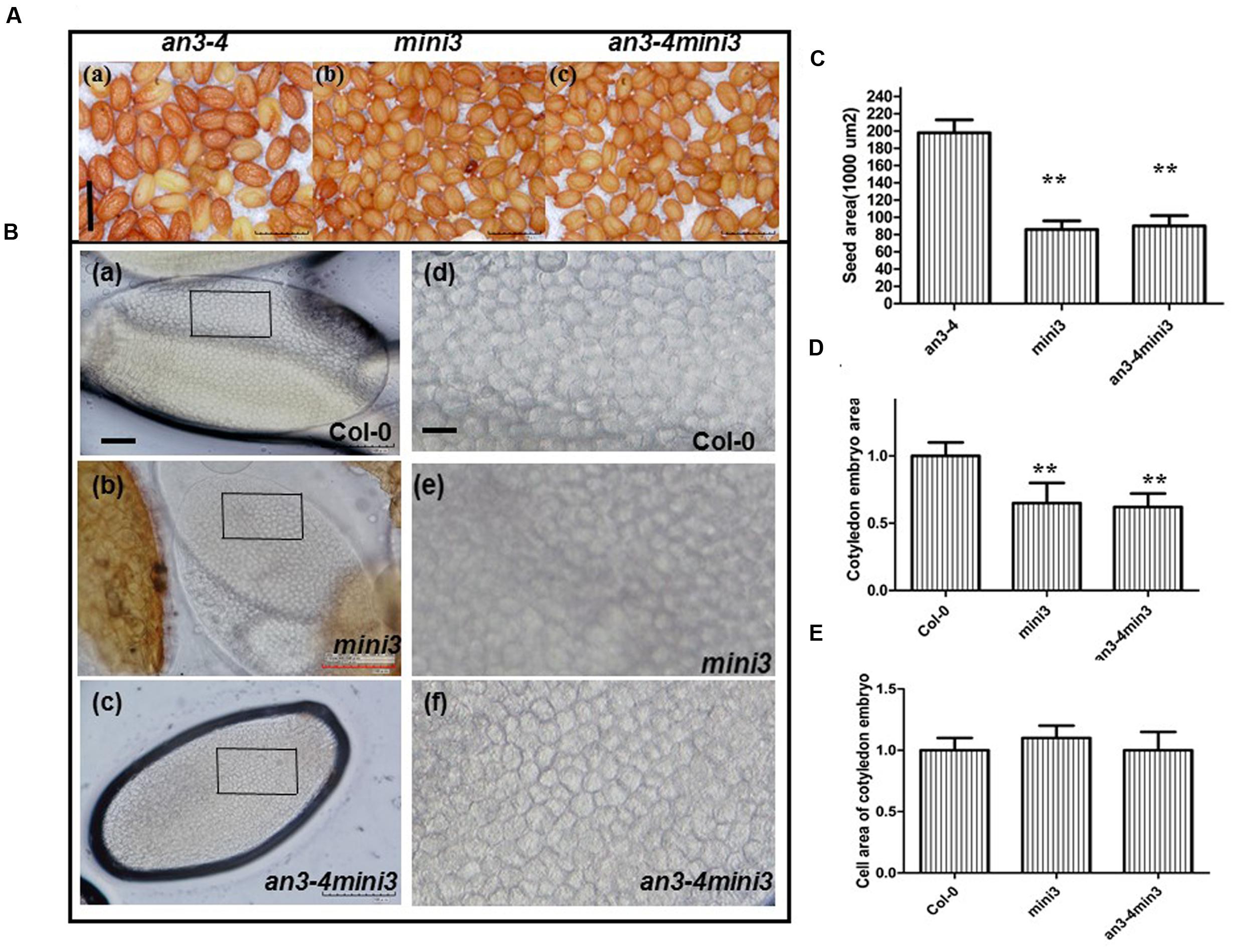
FIGURE 7. AN3 acts genetically upstream of MINI3 in regulating seed mass. (A) Representative mature seeds of an3-4, mini3, and an3-4 mini3 plants, respectively. Bar = 0.1 mm. (B) Representative mature embryos of Col-0 and mini3 plants were isolated and visualized, respectively. Pane in (a–c) was amplified to (d–f). Bars = 100 μm in (a–c); Bars = 10 μm in (d–f). (C) Bar graph exhibiting differences in seed area between an3-4, mini3, and an3-4 mini3 mutants. Data are means ± SD from at least five independently propagated lines (n = 22). (D) and (E) Bar graph exhibiting the difference in embryo area (D) and embryo cell area (E) between mini3 and Col-0 seeds. Col-0 is set as 1.0. Data are means ± SD from at least five independently propagated Col-0 and mutant lines (∗∗P < 0.01; in D, n = 15; in E, n = 32).
Reduced Seed Size Exhibited by mini3 Plants Is Due to Reduced Embryo Cell Number
We isolated and visualized mature embryos originating from wild-type and mini3 mutant seeds. The mini3 mature seeds had less mass than wild-type (Figures 7A,C). Cytological experiments showed smaller average area of mini3 cotyledon embryos than that of wild-type [Figures 7B(a,b,d,e),D]. However, the embryo cell size in mini3 cotyledons did not differ from that of wild-type [Figures 7B(a,b,d,e),E]. Thus, the data indicates that, on average, the embryo cell size exhibited by mini3 plants was not altered, rather embryo size reduction was due to the reduced number of embryo cells. Therefore, the absence of MINI3 resulted in a smaller embryo and fewer cells compared with wild-type. In addition, cytological experiments revealed a smaller average area of an3-4 mini3 cotyledon embryos relative to wild-type [Figures 7B(a,c,d,f),D]. However, the embryo cell size in cotyledons from an3-4mini3 plants was not significantly different from that of wild-type [Figures 7B(a,c,d,f),E]. Collectively, this data implies that the reduced seed size exhibited by mini3 plants is due to reduced embryo cell number.
Discussion
AN3 Negatively Regulates MINI3 Expression
AN3 is required for the expression of MINI3 (Figure 5). Double-mutant analyses revealed that the effect of an3 on seed mass was largely dependent on MINI3 function (Figure 7). MINI3 encodes WRKY10, a WRKY class transcription factor. Levels of MINI3 transcripts were higher in an3-4 than in wild-type plants. We also found that AN3 and MINI3 had different expression patterns in many organs. While AN3 was expressed in young leaves, flowers, stems, stamens, and embryos (Figure 6), expression of MINI3 was not detected in these organs (Luo et al., 2005). AN3 expression was also not detected in mature pollen grains (Figure 6) unlike MINI3 (Luo et al., 2005).
The action of AN3 in regulating seed size is maternally mediated (Supplementary Table S1). This would suggest that AN3 acts by influencing integument growth and this idea is supported by the fact the ovules of an3 plants are reported to be abnormally large, even before fertilization (Lee et al., 2014). However, the MINISEED3 protein has been described as acting zygotically to increase seed size through promotion of early endosperm expansion (Luo et al., 2005). Thus, AN3 may act maternally to repress early zygotic endosperm expansion driven by MINI3, which would explain the fact that the mini3 phenotype is epistatic to the an3 phenotype. In such a scenario epistasis would not necessitate a direct interaction of AN3 with the MINI3 promoter. On the other hand, we find that AN3 acts to repress MINI3 expression and indeed, binds the MINI3 promoter. However, direct regulation of MINI3 expression may not explain the maternal action of AN3. That is, the ChIP data indicated that AN3 is associated with MINI3 promoter, AN3 acts in the endosperm or the embryo (Figure 6; Lee et al., 2014), where MINI3 performs its function. Thus, the possible scenario to resolve the conflict is whether AN3 is a maternally imprinted gene expressed in the endosperm or the embryo. In fact, AN3 encodes a homolog of the human transcription coactivator SYT and is a putative transcription coactivator, synovial SYT (Horiguchi et al., 2005). In synovial sarcomas, chromosomal translocations of the SYT locus to the SSX locus are always observed (Horiguchi et al., 2005). Moreover, the chimeric SYT-SSX protein might possess changed regulatory functions, which can trigger tumor development (Horiguchi et al., 2005). A few reports suggested that the human transcription coactivator SYT might be an imprinted gene (Sun et al., 2006; Cironi et al., 2009). Thus, AN3 may be an imprinted gene. It will await future research to determine whether AN3 is a maternally imprinted gene expressed in the endosperm or the embryo of Arabidopsis.
Besides the above interpretation, we can provide other model, which is as followed in details. AN3 encodes a homolog of the human transcription coactivator SYT and is a putative transcription coactivator (Horiguchi et al., 2005). A similar result has been reported for many other proteins, such as FLOWERING LOCUS T (FT), GIGANTEA (GI), FLAVIN BINDING, KELCH REPEAT, F-BOX1 (FKF1), and SHB1 (Wigge et al., 2005; Sawa et al., 2007; Zhou et al., 2009). Although FT does not have DNA binding activity, it can associate with AP1 promoters by a direct interaction with FLOWERING LOCUS D (a bZIP transcription factor) that directly binds to the AP1 promoter (Wigge et al., 2005). In addition, GI and FKF1 can associate with CONSTANS promoters through a direct interaction with CYCLING DOF FACTOR1 (a Dof transcription factor) in a blue light-dependent manner (Sawa et al., 2007). SHORT HYPOCOTYL UNDER BLUE1 (SHB1) might have a direct interaction with other unknown proteins that are specific transcription factors for MINISEED3 (MINI3) and HAIKU2 (IKU2) promoters and regulate transcription of MINI3 and IKU2 during the initial phase of seed development (Zhou et al., 2009).
While AN3 was also associated with the MINI3 promoter in vivo, AN3 might not have a direct interaction with SHB1 because AN3 is a transcription coactivator not a transcription factor. Similarly, AN3 probably acts as a transcription coactivator and might interact with other unknown proteins that are specific transcription factors for the MINI3 promoter to regulate the transcription of this gene during seed mass development (Figure 8). In this context, GRF1 encodes a putative transcription factor, which interacts with AN3 to promote cell proliferation (Kim and Kende, 2004; Horiguchi et al., 2005). Therefore, AN3 might have a direct interaction with GRF1 that is a specific transcription factor and subsequently the AN3–GRF1 complex might directly bind to the MINI3 promoter regulating transcription of this target gene during modulation of seed mass. However, we found that grf1 plants had similar cotyledon size (Supplementary Figures S1A,B), seed mass (Figures 1A–E), seed number, silique length, seed weight, flower number, elongation silique number, total seed weight (Figures 2A–F), and cotyledon embryo area (Figures 3A,C) with those of wild-type. These results imply that GRF1 is not a candidate.
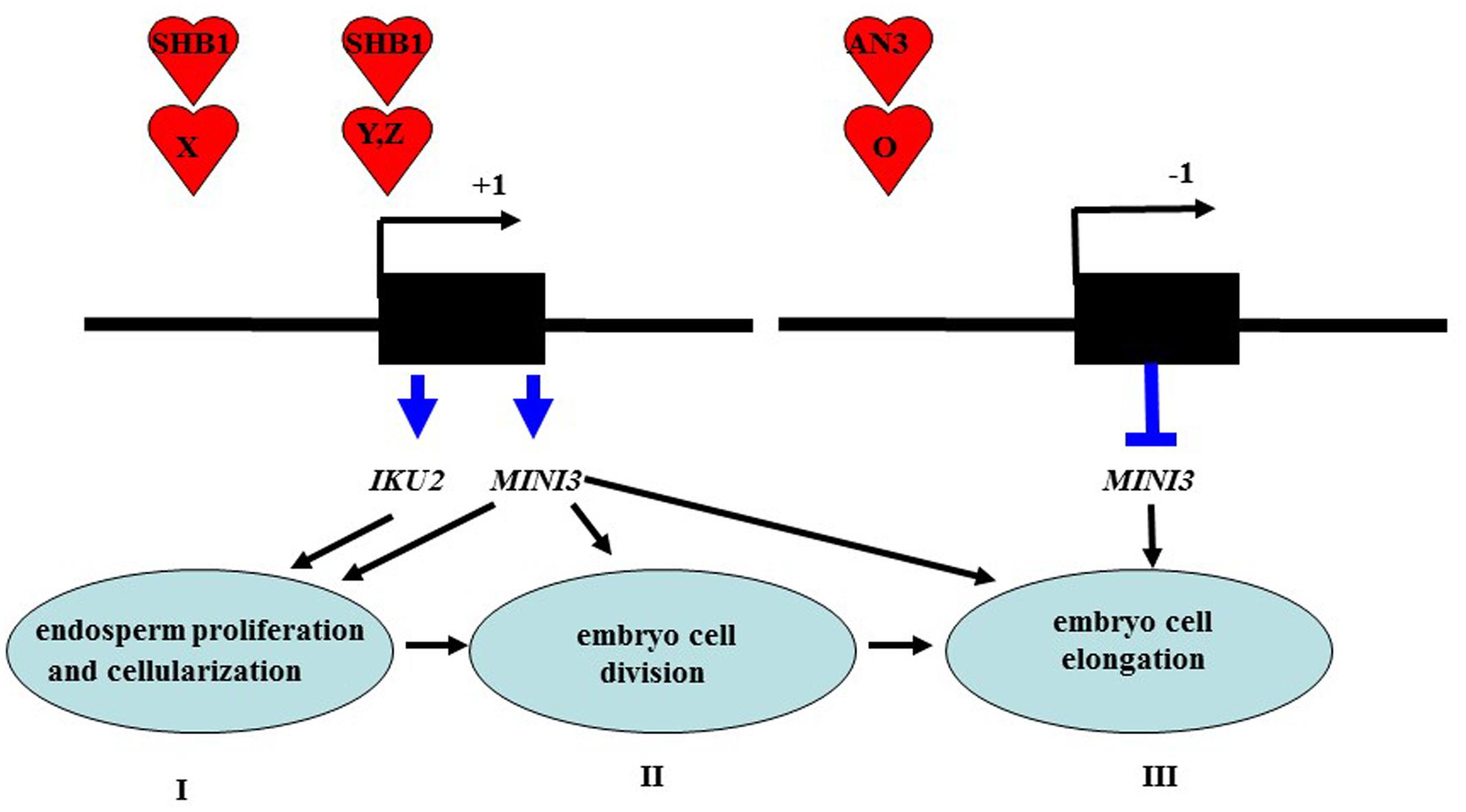
FIGURE 8. A working model is proposed for AN3 function in seed development. I stage: SHB1 is recruited to the promoters of MINI3 and IKU2 via protein X and modulates the expression of key genes including MINI3 and IKU2 that are required for endosperm proliferation and cellularization (Zhou et al., 2009). II stage: SHB1 deploys a similar mechanism to present itself by protein Y to the promoters of unknown targets including Z gene or MINI3 that are required for embryo cell proliferation and elongation (Zhou et al., 2009). II stage: AN3 is recruited to the promoters of MINI3 by protein O and negatively regulates the expression of key genes such as MINI3 that are required for embryo cell division; which in turn triggers cell elongation (terms: complementation). IKU2 is expressed uniquely in the endosperm (Luo et al., 2005), whereas MINI3 is expressed in both the endosperm, the embryo and integument (Luo et al., 2005; Zhou et al., 2009) in this work, we found that AN3 is expressed in the embryo and integument but not in the endosperm.
Regulation of Seed Embryo Development by Cell Division and Cell Elongation Mediated via an AN3-MINI3 Cascade
AN3 acted maternally to influence seed mass, suggesting AN3 regulates seed mass by controlling cell proliferation of the seed coat (integument). However, the embryo constitutes the major volume of a mature seed in Arabidopsis and the changes in seed mass were reflected in the size of the embryos. Thus, we focused our attention on the embryo. As estimated from both embryo size and embryo cell size of an3 mini3 double mutants, the cell number in an3 mini3 embryos is expected to be almost identical to parental mini3 mutants. That is, in this work, we used mini3 [for mini3-2 (SALK_050364)], and the cell number in the mini3-2 mature embryo is reduced to ∼60% of wild-type (Col-0) (Figure 7D). Consistent with this result, Zhou et al. (2009) also showed that seed size (or embryo size) in the mini3-2 mutant is reduced to ∼60% of wild-type (Col-0), whereas embryo cell size in this mutant is not altered. Embryo size is determined via both embryo cell size and cell number. Thus, the cell number in mini3-2 mature embryos is reduced to ∼60% of wild-type (Col-0). Moreover, our findings indicated that the cell number in an3-4 mini3-2 mature embryos was also reduced to ∼60% of wild-type (Col-0) (Figure 7D). However, cytological observations indicated that the average area of the an3-4 cotyledon embryos was ∼1.40 times that of wild-type (Figures 3B,D). On average, the epidermal cell size of an3-4 embryos was ∼1.70 times that of wild-type (Figures 3B,D). These data (i.e., 1.40/1.70 = 0.8) indicated that the increased embryo size of an3-4 plants was due to embryo cell elongation.
These results also indicated that the cell number in an3-4 mature embryos is reduced to ∼80% of wild-type (Col-0). This contrasts with mini3-2 and an3-4 mini3-2 plants where embryo cell number was only ∼60% of wild-type (Col-0). Therefore, the absence of MINI3 function suppresses cell division in an3-4 plants and also inhibits increased cell size during embryo development in this line. Larger cell size in an3-4 plants is thought to result from a phenomenon termed compensation where larger an3-4 cells result from a decrease in cell number which triggers an increase in mature cell size (Ferjani et al., 2007; Fujikura et al., 2007;). Thus, mutation of MINI3 leads to a decrease of an3-4 mini3-2 seed mass. These genetic findings indicate that AN3 mediated suppression of MINI3 expression appears to occur at an early stage of seed embryo development (cell division), but is also influenced at a later stage of seed embryo development (cell elongation).
Consistent with genetic data, in seed embryo development, the expression of AN3 can be detected in later embryo development (Kanei et al., 2012), whereas the expression of MINI3 cannot be observed at this stage (Luo et al., 2005). Consistent with our molecular data that suggests AN3 trans-represses MINI3 by associating with the MINI3 promoter (Figure 5). Cell division activity, directly relevant for cell number, mainly occurs during the early phase of seed development. Conversely, cell elongation, relevant for cell size, mainly occurs during the late phase of seed development (Weber et al., 1996, 1997a,b). Thus, an AN3-MINI3 gene cascade might regulate seed embryo development by controlling both cell division and cell elongation.
Compensation, such as that leading to larger cells in an3-4 embryos, is thought to be largely independent from endoreduplication, although the underpinning molecular mechanism is unknown (Ferjani et al., 2007; Fujikura et al., 2007). Here, we find that mutation of MINI3 suppresses cell division in an an3-4 mutant during early embryo development and in turn inhibits the increased cell size phenotype (cell elongation) during later embryo development. Thus, our findings reveal that the larger epidermal cells in an3-4 plants are due to compensation, which might occur as a result of the dysregulation of MINI3 expression.
Accession Numbers
Sequence data derived from this paper can be found in the Arabidopsis Genome Initiative database under the following accession numbers AN3 (At5G28640) and MINI3 (At1G55600).
Author Contributions
L-SM designed experiments. L-SM performed the experiments. L-SM, and Y-BW completed statistical analysis of data. L-SM, GL, and J-HJ wrote, edited and revised this manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. G. Horiguchi (Rikkyo University, Japan) for providing the 35S:AN3:3XGFP seeds, Prof. H. G. Nam (PoHang University of Science and Technology, Korea) for providing the an3-4 mutant seeds. We also thank Professor Hong Gil Nam (DGSIT, Korea) for his support at the initial stage of this work. This study was supported by grants from the National Science Foundation of China (31370646; 31401443; 31560164).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01645/full#supplementary-material
FIGURE S1 | an3-4 mutant plants had enlarged cotyledons. (A) Representative 10-day-old mature cotyledons of an3-4, 35S-AN3/an3-4, grf1 and wild-type (Col-0) grown on solid MS medium with 1% sucrose, respectively. Bar = 5 mm. (B) Bar graph showing the difference in the cotyledon area between an3-4, 35S-AN3/an3-4, grf1, and wild-type (Col-0). Data are means ± SD from at least 10 independently propagated lines of an3-4, 35S-AN3/an3-4, grf1 and Col-0 (∗∗P < 0.01; n = 20). Col-0 is set as 1.0.
FIGURE S2 | an3-4 mutant plants present shorter stamens than wild-type (Col-0). Arrows indicate stamen. Scale bars: 1 mm
FIGURE S3 | AN3 regulates embryo size by modulation of integument. (A) Col-0 (Col-0 embryo) × Col-0 (maternal sporophytic tissue). (B) an3-4 (an3-4 embryo) × an3-4 (maternal sporophytic tissue). Scale bars: (A), (B), 100 μm. (C) Bar graph exhibiting the difference in ovule area between an3-4 and Col-0 seeds. Data are means ± SD from at least five independently propagated lines (n = 12). (D) and (E) Bar graph exhibiting the difference in global embryo area (D) and integument cell area (E) between an3 and Col-0 seeds. Col-0 is set as 1.0. Data are means ± SD from at least five independently propagated Col-0 and mutant lines (∗∗P < 0.01; in D, n = 10; in E, n = 38).
TABLE S1 | an3-4 act maternally for regulating seed mass.
TABLE S2 | primers in this work are used.
Footnotes
References
Adamski, N. M., Anastasiou, E., Eriksson, S. O., Neill, C. M., and Lenhard, M. (2009). Local maternal control of seed size by KLUH/CYP78A5- dependent growth signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20115–20120. doi: 10.1073/pnas.0907024106
Cheng, Z. J., Zhao, X. Y., Shao, X. X., Wang, F., Zhou, C., Liu, Y. G., et al. (2014). Abscisic acid regulates early seed development in Arabidopsis by ABI5-mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26, 1053–1068. doi: 10.1105/tpc.113.121566
Cironi, L., Provero, P., Riggi, N., Janiszewska, M., Suva, D., Suva, M. L., et al. (2009). Epigenetic features of human mesenchymal stem cells determine their permissiveness for induction of relevant transcriptional changes by SYTSSX1. PLoS ONE 4:e7904. doi: 10.1371/journal.pone.0007904
Disch, S., Anastasiou, E., Sharma, V. K., Laux, T., Fletcher, J. C., and Lenhard, M. (2006). The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16, 272–279. doi: 10.1016/j.cub.2005.12.026
Ferjani, A., Horiguchi, G., Yano, S., and Tsukaya, H. (2007). Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol. 144, 988–999. doi: 10.1104/pp.107.099325
Fujikura, U., Horiguchi, G., Ponce, M. R., Micol, J. L., and Tsukaya, H. (2009). Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J. 59, 499–508. doi: 10.1111/j.1365-313X.2009.03886.x
Fujikura, U., Horiguchi, G., and Tsukaya, H. (2007). Dissection of enhanced cell expansion processes in leaves triggered by a defect in cell proliferation, with reference to roles of endoreduplication. Plant Cell Physiol. 48, 278–286. doi: 10.1093/pcp/pcm002
Garcia, D., Gerald, J. N. F., and Berger, F. (2005). Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17, 52–60. doi: 10.1105/tpc.104.027136
Garcia, D., Saingery, V., Chambrier, P., Mayer, U., Jürgens, G., and Berger, F. (2003). Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131, 1661–1670. doi: 10.1104/pp.102.018762
Heath, J. D., Weldon, R., Monnot, C., and Meinke, D. W. (1986). Analysis of storage proteins in normal and aborted seeds from embryo-lethal mutants of Arabidopsis thaliana. Planta 169, 304–312. doi: 10.1007/BF00392124
Horiguchi, G., Kim, G. T., and Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68–78. doi: 10.1111/j.1365-313X.2005.02429.x
Horiguchi, G., Nakayama, H., Ishikawa, N., Kubo, M., Demura, T., Fukuda, H., et al. (2011). ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol. 52, 112–124. doi: 10.1093/pcp/pcq178
Hutchison, C. E., Li, J., Argueso, C., Gonzalez, M., Lee, E., Lewis, M. W., et al. (2006). The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18, 3073–3087. doi: 10.1105/tpc.106.045674
Kanei, M., Horiguchi, G., and Tsukaya, H. (2012). Stable establishment of cotyledon identity during embryogenesis in Arabidopsis by ANGUSTIFOLIA3 and HANABA TARANU. Development 139, 2436–2446. doi: 10.1242/dev.081547
Kang, X., Li, W., Zhou, Y., and Ni, M. (2013). A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement. PLoS Genet. 9:e1003347. doi: 10.1371/journal.pgen.1003347
Kawade, K., Horiguchi, G., and Tsukaya, H. (2010). Non-cell-autonomously coordinated organ size regulation in leaf development. Development 137, 4221–4227. doi: 10.1242/dev.057117
Kim, J. H., and Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 13374–13379. doi: 10.1073/pnas.0405450101
Lee, B. H., Wynn, A. N., Franks, R. G., Hwang, Y. S., Lim, J., and Kim, J. H. (2014). The Arabidopsis thaliana GRF-INTERACTING FACTOR gene family plays an essential role in control of male and female reproductive development. Dev. Biol. 386, 12–24. doi: 10.1016/j.ydbio.2013.12.009
Lei, Z. Y., Zhao, P., Cao, M. J., Cui, R., Chen, X., Xiong, L. Z., et al. (2007). High-throughput binary vectors for plant gene function analysis. J. Integr. Plant Biol. 49, 556–567. doi: 10.1111/j.1744-7909.2007.00442.x
Li, Y., Zheng, L., Corke, F., Smith, C., and Bevan, M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Lopes, M. A., and Larkins, B. A. (1993). Endosperm origin, development, and function. Plant Cell 5, 1383–1399. doi: 10.1105/tpc.5.10.1383
Luo, M., Dennis, E. S., Berger, F., Peacock, W. J., and Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 102, 17531–17536. doi: 10.1073/pnas.0508418102
Meng, L. S. (2015). Transcription coactivator Arabidopsis ANGUSTIFOLIA3 modulates anthocyanin accumulation and light-induced root elongation through transrepression of constitutive photomorphogenic1. Plant Cell Environ. 38, 838–851. doi: 10.1111/pce.12456
Meng, L. S., and Liu, A. (2015). Light signaling induces anthocyanin biosynthesis via AN3 mediated COP1 expression. Plant Signal. Behav. 10:e1001223. doi: 10.1080/15592324.2014.1001223
Meng, L. S., Wang, Y. B., Yao, S. Q., and Liu, A. (2015a). Arabidopsis AINTEGUMENTA mediates salt tolerance by trans-repressing SCABP8. J. Cell Sci. 128, 2919–2927. doi: 10.1242/jcs.172072
Meng, L. S., Wang, Z. B., Cao, X. Y., Zhang, H. J., Wang, Y. B., and Jiang, J. H. (2016). ASYMMETRIC LEAVES2-LIKE15 gene, a member of AS2/LOB family, shows a dual abaxializing or adaxializing function in Arabidopsis lateral organs. Acta Physiol. Plant 38, 240. doi: 10.1007/s11738-016-2256-x
Meng, L. S., Wang, Z. B., Yao, S. Q., and Liu, A. (2015b). The ARF2–ANT– COR15A gene cascade regulates ABA signaling-mediated resistance of large seeds to drought in Arabidopsis. J. Cell Sci. 128, 3922–3932. doi: 10.1242/jcs.171207
Meng, L. S., and Yao, S. Q. (2015). Transcription co-activator Arabidopsis ANGUSTIFOLIA3 (AN3) regulates water-use efficiency and drought tolerance by modulating stomatal density and improving root architecture by the transrepression of YODA (YDA). Plant Biotechnol. J. 13, 893–902. doi: 10.1111/pbi.12324
Mizukami, Y., and Fischer, R. L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. U.S.A. 97, 942–947. doi: 10.1073/pnas.97.2.942
Ohto, M., Fischer, R. L., Goldberg, R. B., Nakamura, K., and Harada, J. J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. U.S.A. 102, 3123–3128. doi: 10.1073/pnas.0409858102
Pang, P. P., Pruitt, R. E., and Meyerowitz, E. M. (1988). Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol. Biol. 11, 805–820. doi: 10.1007/BF00019521
Riefler, M., Novak, O., Strnad, M., and Schmulling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54. doi: 10.1105/tpc.105.037796
Saleh, A., Alvarez-Venegas, R., and Avramova, Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3, 1018–1025. doi: 10.1038/nprot.2008.66
Sawa, M., Nusinow, D. A., Kay, S. A., and Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. doi: 10.1126/science.1146994
Schruff, M. C., Spielman, M., Tiwari, S., Adams, S., Fenby, N., and Scott, R. J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251–261. doi: 10.1242/dev.02194
Sun, Y., Gao, D., Liu, Y., Huang, J., Lessnick, S., and Tanaka, S. (2006). IGF2 is critical for tumorigenesis by synovial sarcoma oncoprotein SYT-SSX1. Oncogene 25, 1042–1052. doi: 10.1038/sj.onc.1209143
Wang, A., Garcia, D., Zhang, H., Feng, K., Chaudhury, A., Berger, F., et al. (2010). The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63, 670–679. doi: 10.1111/j.1365-313X.2010.04271.x
Weber, H., Borisjuk, L., Heim, U., Sauer, N., and Wobus, U. (1997a). A role for sugar transporters during seed development: molecular characterization of a hexose and a sucrose carrier in fava bean seeds. Plant Cell 9, 895–908. doi: 10.1105/tpc.9.6.895
Weber, H., Borisjuk, L., and Wobus, U. (1996). Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. Plant J. 10, 823–834. doi: 10.1046/j.1365-313X.1996.10050823.x
Weber, H., Borisjuk, L., and Wobus, U. (1997b). Sugar import and metabolism during seed development. Trends Plant Sci. 2, 169–174. doi: 10.1016/S1360-1385(97)85222-3
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Keywords: ANGUSTIFOLIA3 (AN3), MINISEED3 (MINI3), seed mass, embryo cell elongation, embryo cell division, Arabidopsis
Citation: Meng L-S, Wang Y - B, Loake GJ and Jiang J - H (2016) Seed Embryo Development Is Regulated via an AN3-MINI3 Gene Cascade. Front. Plant Sci. 7:1645. doi: 10.3389/fpls.2016.01645
Received: 10 July 2016; Accepted: 18 October 2016;
Published: 03 November 2016.
Edited by:
Jin-Gui Chen, Oak Ridge National Laboratory, USACopyright © 2016 Meng, Wang, Loake and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lai-Sheng Meng, bWVuZ2xzaEBqc251LmVkdS5jbg== Ji-Hong Jiang, amhqaWFuZ0Bqc251LmVkdS5jbg== Gary J. Loake, Z2xvYWtlQGVkLmFjLnVr
 Lai-Sheng Meng
Lai-Sheng Meng Yi-Bo Wang3
Yi-Bo Wang3 Gary J. Loake
Gary J. Loake