- 1Southern Cross Plant Science, Southern Cross University, Lismore, NSW, Australia
- 2Southern Cross GeoScience, Southern Cross University, Lismore, NSW, Australia
Glucosinolates (GSLs) represent one of the most widely studied classes of plant secondary metabolite, and have a wide range of biological activities. Their unique properties also affect livestock and human health, and have been harnessed for food and other end-uses. Since GSLs are sulfur (S)-rich there are many lines of evidence suggesting that plant S status plays a key role in determining plant GSL content. However, there is still a need to establish a detailed knowledge of the distribution and remobilization of S and GSLs throughout the development of Brassica crops, and to represent this in terms of primary and secondary sources and sinks. The increased genome complexity, gene duplication and divergence within brassicas, together with their ontogenetic plasticity during crop development, appear to have a marked effect on the regulation of S and GSLs. Here, we review the current understanding of inorganic S (sulfate) assimilation into organic S forms, including GSLs and their precursors, the intracellular and inter-organ transport of inorganic and organic S forms, and the accumulation of GSLs in specific tissues. We present this in the context of overlapping sources and sinks, transport processes, signaling molecules and their associated molecular interactions. Our analysis builds on recent insights into the molecular regulation of sulfate uptake and transport by different transporters, transcription factors and miRNAs, and the role that these may play in GSL biosynthesis. We develop a provisional model describing the key processes that could be targeted in crop breeding programs focused on modifying GSL content.
Introduction
Glucosinolates are sulfur-rich, nitrogen-containing secondary metabolites with a wide range of biological activities. Their unique properties also affect livestock and human health, and have been harnessed for food and other end-uses. Almost, 200 different GSLs have been identified to date and are predominantly reported in the order Brassicales (Capparales) (reviewed in Fahey et al., 2001; Clarke, 2010). However, some GSLs have also been identified in non-cruciferous dicotyledonous angiosperms (Rodman et al., 1996). The evolutionary role of these molecules appears to have arisen from the selective advantage conferred due to their insect anti-feedant or attraction properties (reviewed in Bones and Rossiter, 1996; Wittstock and Burow, 2010), nematicidal (Lazzeri et al., 1993; Buskov et al., 2002) and fungal disease suppression (Mari et al., 2002; Tunc et al., 2007; Bednarek et al., 2009; Hacquard et al., 2016; Hiruma et al., 2016) as well as herbicidal properties (Brown and Morra, 1995; Vaughn et al., 2006).
Until about 40 years ago oilseed crop cultivars of Brassica napus and Brassica rapa possessed high levels of GSLs, ranging from 100 to 180 μmol/g in defatted seed meal, which hindered the use of the protein-rich rapeseed meal because of its anti-nutritional effects on livestock (reviewed in Griffiths et al., 1998). Following discovery of a low GSL-containing Polish spring rape cultivar, Bronowski (Kondra and Stefansson, 1970), the production and consumption of B. napus increased rapidly and it now represents 15% of total global vegetable oil (USDA, 2016). The name ‘canola’ was first used by the Rapeseed Association of Canada, but it is now a general name for any rapeseed or oilseed rape cultivars with ‘double low’ levels of erucic acid (<2%) and GSL (<30 μmol/g in oil-free seed meal) (Canola Council, 2016).
However, GSLs and their derivatives also have a wide range of positive attributes in the context of food production, human nutrition and other end-uses such as in biofumigation. The unique S-based properties of GSLs impart the distinctive flavors and pungency of cruciferous vegetables and oils, beneficial effects on human health and anti-carcinogenic properties (Hayes et al., 2008). For example, sulforaphane, a derivative of glucoraphanin and mostly found in broccoli (B. oleracea var. italica), can act as a preventive agent for certain types of cancer (Brooks et al., 2001). Phenyl group-containing isothiocyanates of some GSLs and indole-3-carbinol derivatives of glucobrassicin are also reported to suppress the growth of mammalian carcinogenic cells (Hecht, 2000; Nakajima et al., 2001; Choi et al., 2010).
The volatile mustard oil ally-isothiocyanate (AITC), produced from defatted Brassica juncea seed meal, is used as a food flavoring agent, and as a natural preservative because it has been shown to inhibit the growth of certain fungi and bacteria (Mari et al., 2002; Rhee et al., 2003; Bednarek et al., 2009). Isothiocyanates (ITCs) can also be used as biofumigants for soil-borne pests (Brown and Morra, 2005; Bellostas et al., 2007a). Because of the desirability of many key GSLs and their hydrolyzed products – especially isothiocyanate – there has been strong interest in understanding the genetic regulation of GSLs in order to manipulate their content in crop plants through plant breeding (reviewed in Mithen, 2001).
While total GSL content in most Brassica species is typically less than 1% of the total plant dry weight (Fieldsend and Milford, 1994; Blake-Kalff et al., 1998; Bellostas et al., 2007a), a substantial amount of S can be sequestered within the GSLs – between 1.7 and 8.0% of total plant S content (reviewed in Falk et al., 2007). Many of the predominant methionine (Met) derived GSLs have three S atoms in their structure (reviewed in Falk et al., 2007; Frerigmann and Gigolashvili, 2014b), one of which derives from the amino acid precursor. Arabidopsis phytoalexinine (pad2) mutant and adenosine-5′-phosphosulfate kinase (apk1apk3apk4) mutant shown that the remaining two S atoms, in common with other GSLs, originate from glutathione (GSH) (Geu-Flores et al., 2011) and 3′-phosphoadenosine 5′-phosphosulfate (PAPS) produced in the S-assimilation pathway (Mugford et al., 2009; Figure 1). As such, GSL-containing species of the Brassicaceae such as oilseed rape have a higher S requirement than non-GSL-containing species (Klessa and Sinclair, 1989). Under S deficient conditions, synthesis of S-containing amino acids is restricted, and this can reduce the photosynthetic activity of the plant leading to reduced plant growth (Ahmad and Abdin, 2000). Moreover, there is evidence that application of relatively higher rates of S fertilizer in B. rapa can lead to preferential accumulation of aliphatic and aromatic rather than indole GSLs (Chen et al., 2006). In most field and greenhouse trials, as well as in experiments carried out in artificial media, GSL content is sensitive to increased S availability levels, with up to 10-fold increases reported in some cases (reviewed in Walker and Booth, 2003; Falk et al., 2007; Supplementary Table S1). However, few of these studies report detailed dose response curves, and ultimately any GSL response to S fertilization will depend on the existing native soil S status and will be site and season dependent.
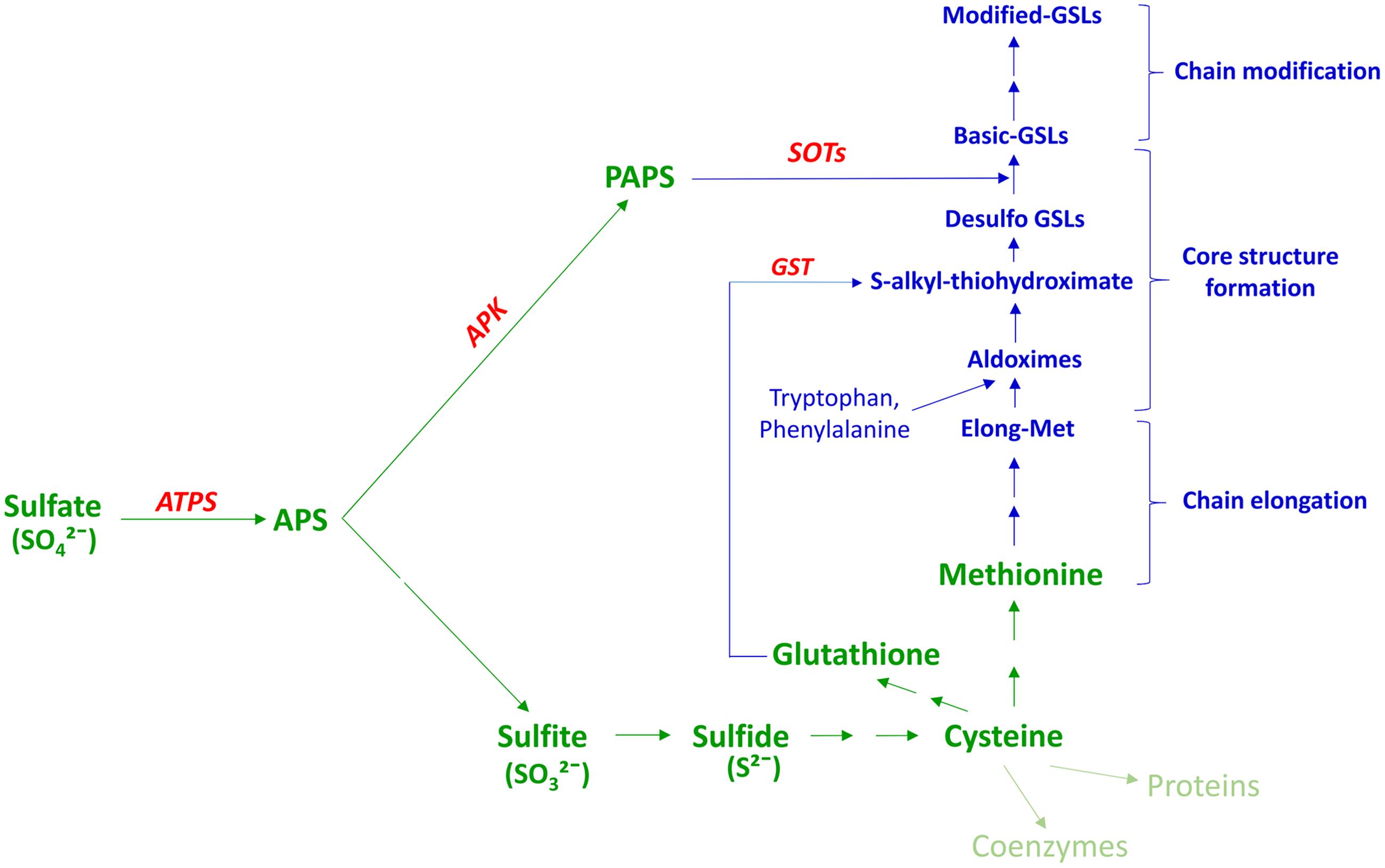
FIGURE 1. Association between plant S assimilation and GSL biosynthesis. Modified from Sonderby et al. (2010); Mugford et al. (2011) PAPS, 3′-phosphoadenosine 5′-phosphosulfate; APS, adenosine 5′-phosphosulphate; ATPS, ATP sulfurylase; APK, APS kinase; GSL, glucosinolate; GST, glutathione -S-transferase; SOT, sulfotransferase. Text in green indicates the S-assimilation process, and text in blue GSL biosynthesis. Enzymes are shown in red.
In recent years, plant S metabolism has been well-reviewed in terms of products, substrates and associated enzyme (Takahashi et al., 2011; Calderwood et al., 2014) as subsequently in terms of transport (Gigolashvili and Kopriva, 2014) and molecular regulation of the key processes of S-assimilation (Koprivova and Kopriva, 2014, 2016). However, our understanding of the molecular regulation of GSL biosynthesis in relation to S availability is less clear. Here, we review the current understanding of inorganic S (sulfate) assimilation into organic S forms, including GSLs and their precursors, the intracellular and inter-organ transport of inorganic and organic S forms, and the accumulation of GSLs in specific tissues. We present this in the context of overlapping sources and sinks, transport processes, signaling molecules and their associated molecular interactions. We refer to sources as the specific organs and tissues associated with S assimilation and GSL biosynthesis, and sinks as those associated with storage or use of assimilates. Our analysis builds on recent insights into the molecular regulation of sulfate uptake and transport by different transporters, transcription factors (TFs) and miRNAs, and the role that these may play in GSL biosynthesis. We develop a provisional model describing the key processes that may be manipulated in breeding programs focused on modifying GSL content.
Chemistry and Biosynthesis of GSLs
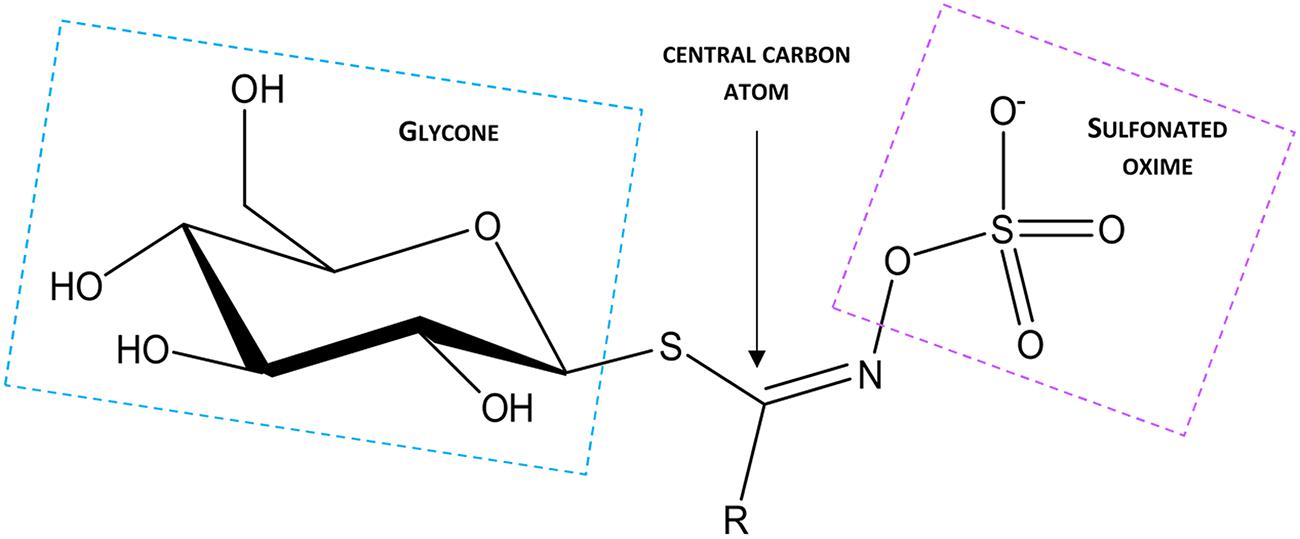
Glucosinolates have been known for 1000s of years as pungency of mustard oil and bitter flavor of cruciferous vegetables. The basic structures of singrin and sinalbin were first elucidated in Ettlinger and Lundeen (1956). Subsequently, the underlying process of GSL hydrolysis was found to be responsible for the specific flavor properties within the brassicaceae (Kjaer, 1976; Tookey et al., 1980; Fenwick et al., 1983). GSLs are anionic, with the core structure of each molecule having a centrally localized carbon atom which is linked to a glycone group via a thioglucoside bond which originates from GSH. In addition, the central carbon atom links to a sulfonated oxime via an N bond, and is also linked to an amino acid-derived variable side chain (R group) (reviewed in Fahey et al., 2001; Figure 2). In some cases, other components are also attached to the glycone moiety or oxygen (O), S, or N atoms of the R group (reviewed in Agerbirk and Olsen, 2012). The background to the chemistry and molecular regulation of glucosinolates has been addressed in many extensive reviews (Halkier and Gershenzon, 2006; Sonderby et al., 2010; Agerbirk and Olsen, 2012; Baskar et al., 2012; Ishida et al., 2014). It was the detailed characterization of GSLs and associated pathways in Arabidopsis that opened the way for a molecular understanding of the genes encoding specific enzymes associated with each synthetic and modification step.
Almost, all GSL-producing plants also possess a β-thioglucoside glycohydrolase enzyme known as myrosinase (reviewed in Rask et al., 2000; Bellostas et al., 2007b). Upon tissue damage or insect herbivory, endogenous myrosinase and vacuolar GSLs come in contact with each other, resulting in hydrolyzation of the GSL to form a variety of compounds such as isothiocyanates, epithionitriles, nitriles, thiocyanates, and oxazolidines, depending on the structure of the substrate GSL, pH and on the presence of metal ions, additional proteins and cofactors (reviewed in Bones and Rossiter, 1996, 2006; Halkier and Gershenzon, 2006; Kissen and Bones, 2009).
Glucosinolates are categorized into three major types according to their precursor amino acids. In different tissues of Brassica, aliphatic GSLs account for 70–97% of the total GSL content. This has been reported in leaves of B. oleracea (Cartea et al., 2008), leaves and stems of B. napus (Cleemput and Becker, 2012), leaves and seeds of B. juncea (Shilpa et al., 2012; Othmane, 2015) and sprouts and mature leaves of B. rapa (Wiesner et al., 2013). Aliphatic GSLs are mostly derived from Met, with a few from leucine, isoleucine, alanine, and valine. In Arabidopsis, aliphatic and indole GSLs are the most abundant in different tissues (Kliebenstein et al., 2001; Brown et al., 2003). However, aromatic GSLs are the minor components in both Brassica and Arabidopsis. The indole GSLs are derived from tryptophan (Trp), whereas aromatic GSLs are derived from phenylalanine (Phe) and tyrosine (reviewed in Fahey et al., 2001; Mithen, 2001; Halkier and Gershenzon, 2006).
There are three independent enzyme-mediated phases of GSLs biosynthesis, which have been well-characterized in Arabidopsis thaliana: amino acid side-chain elongation, core GSL structure formation, and side chain modification, the first and last of which are dependent upon substrate specificity (reviewed in Grubb and Abel, 2006; Halkier and Gershenzon, 2006; Sonderby et al., 2010; Baskar et al., 2012; Figure 1). Although, aliphatic and aromatic GSLs usually undergo side-chain elongation, this process has not been reported in the case of indole GSLs (Textor et al., 2004). The chain elongation pathway is mediated by branched chain aminotransferases (BCATs), methyl thioalkyl malate synthases (MAMs), isopropyl malate isomerase (IPMI) and isopropyl malate dehydrogenase (IPM-DH) (Schuster et al., 2006; Knill et al., 2008; Gigolashvili et al., 2009b; Sawada et al., 2009a,b; Zhang et al., 2015a). The resulting chain-elongated amino acid can either enter into the core GSL structure formation phase, or proceed through additional chain elongation cycles to add further methylene groups (reviewed in Halkier and Gershenzon, 2006). The GSL core structure formation phase involves enzymes of the CYP79 (Hull et al., 2000; Mikkelsen et al., 2000; Hansen et al., 2001; Chen et al., 2003), CYP83 (Bak and Feyereisen, 2001; Hemm et al., 2003; Naur et al., 2003), UGT74 (Grubb et al., 2004, 2014; Gachon et al., 2005) families, C-S-lyases (Mikkelsen et al., 2004) and sulfotransferases (SOTs or STs) (Piotrowski et al., 2004). These enzymes facilitate the biosynthesis of basic GSL structures from elongated and non-elongated amino acids. Notably, GSH produced in the S-assimilation process is considered as the preferred S donor for synthesis of S-alkyl-thiohydroximate via GSH-S-transferase (GST) in the core structure development phase (Schlaeppi et al., 2008; Dixon et al., 2010; Geu-Flores et al., 2011; Figure 1). On the other hand, basic GSLs are produced only after the sulfation of desulfo-GSLs with a PAPS sulfate donor, catalyzed by SOTs (Figure 1). As cysteine (Cys) is the precursor for GSH as well as Met, Cys and PAPS can be considered as the major connection between GSL biosynthesis and the S assimilation process (Figures 1, 5, and 6). After synthesis, these basic GSL structures are subjected to a range of secondary side chain modification and transformation pathways involving enzymes such as flavin mono oxygenase (FMOOXs) (Hansen et al., 2007; Li et al., 2008), GSL-AOPs (Mithen et al., 1995), GSL-OH (Hansen et al., 2008) and CYP81Fs (Pfalz et al., 2009, 2011) to generate different types of GSL structures (reviewed in Fahey et al., 2001; Clarke, 2010; Figure 1).
Candidate Genes and Signaling Molecules Involved in GSL Biosynthesis
The genes encoding enzymes associated with each of the known steps of GSL synthesis have been characterized in Arabidopsis (reviewed in Sonderby et al., 2010). Amongst the enzymes of chain elongation reaction of GSL biosynthesis, the genetic loci responsible for MAMs have been resolved within the Arabidopsis genome (Kroymann et al., 2001; Textor et al., 2007; Sawada et al., 2009b; Redovnikovic et al., 2012), and more recently in different Brassica species (Gao et al., 2006; Liu et al., 2011; Sotelo et al., 2014; Zhang et al., 2015a). An early confirmation of functional conservation within the Brassicaceae came from cloning B. oleracea homologs of AtGSL-Elong/AtMAM (BoGSL-Elong) and of AtGS-ALK (BoGSL-ALK+), with confirmation of their involvement in GSL biosynthesis by functional analysis (Li and Quiros, 2002, 2003), In particular, the function of the BoGSL-ALK gene was demonstrated by transgenic gene complementation of A. thaliana ecotype Columbia, in which the corresponding allele for GSL-ALK is non-functional. In addition, the BoGSL-PRO gene cloned from B. oleracea was found to be involved in the side chain elongation phase of aliphatic GSL synthesis (Li et al., 2001). Comparative genomic analysis has been extended, firstly via physical mapping, with B. oleracea BAC clones containing orthologs of the AOP (B21H13) and MAM (B21F5) families providing insights into gene conservation and divergence between A. thaliana and B. oleracea (Gao et al., 2004, 2006). Putative orthologs of five major genes in the GSL biosynthetic pathway (BraGSL-ELONGa, BraGSL-ALKa, BraCYP83B1, BraSUR1a, and BraST5a) were cloned from both cDNA and genomic DNA of different subspecies of B. rapa (Yang et al., 2010), with expression studies substantiating their involvement in GSL biosynthesis. Likewise, putative orthologs of Arabidopsis CYP83A1 and CYP83B1 isolated from B. rapa, including BrCYP83A1 and BrCYP83B1 were characterized by qRT-PCR analysis (Zhu et al., 2012). Twenty one ‘metabolite’-quantitative trait loci (mQTL) have been identified for total GSL in the seeds of B. napus (AC genome) (Feng et al., 2012), of which nine correspond to the QTLs reported earlier in the same species (Toroser et al., 1995; Uzunova et al., 1995; Howell et al., 2003; Quijada et al., 2006). This study (Feng et al., 2012), also identified twenty-seven compositional mQTLs for aliphatic and indole GSL accumulation in leaves, of which four correspond to those previously identified in B. rapa (A genome) for aliphatic and indole GSL accumulation in leaves (Lou et al., 2008). A further 25 mQTL for seed aliphatic GSLs were identified on the A genome (Feng et al., 2012), of which only four are thought to have been identified previously in B. juncea (AB genome) (Ramchiary et al., 2007; Bisht et al., 2009). Very recently, biparental and association mapping studies in B. juncea (Rout et al., 2015) and B. napus (Qu et al., 2015) identified additional QTLs associated with GSL compositional variation and seed GSL accumulation.
Since divergence from a common ancestor over 20 million years ago (Liu et al., 2014) the genomes of the diploid crop brassicas B. rapa (A genome), B. nigra (B) and B. oleracea (C) have undergone duplications leading to a triplicated mesopolyploid structure (Wang X. et al., 2011), which is then duplicated in the amphidiploids B. juncea (AB), B. napus (AC), and B. carinata (BC) (Figure 3). This has led to considerable diversity resulting from gene duplication and divergence, with opportunities for neo-functionalization in terms of temporal and spatial gene regulation. The divergence of the diploid Brassica genomes over the past 5–10 million years has resulted in a distinct pattern of aliphatic GSL composition associated with each of the A, B, or C genomes. Thus, GSLs with three carbon side chains derived from a single elongation reaction are found in B. nigra, whereas B. oleracea contains three or four carbon side chains, and B. rapa contains four or five carbon side chain GSLs. The three amphidiploid species each have a GSL composition corresponding to their respective diploid genome composition (reviewed in Ishida et al., 2014). The greater opportunity for gene divergence and specialization driven by the complex genome structure of brassicas also appears to provide these crop species with considerable ontogenetic plasticity during plant development, and selection of a wide range of morphotypes.
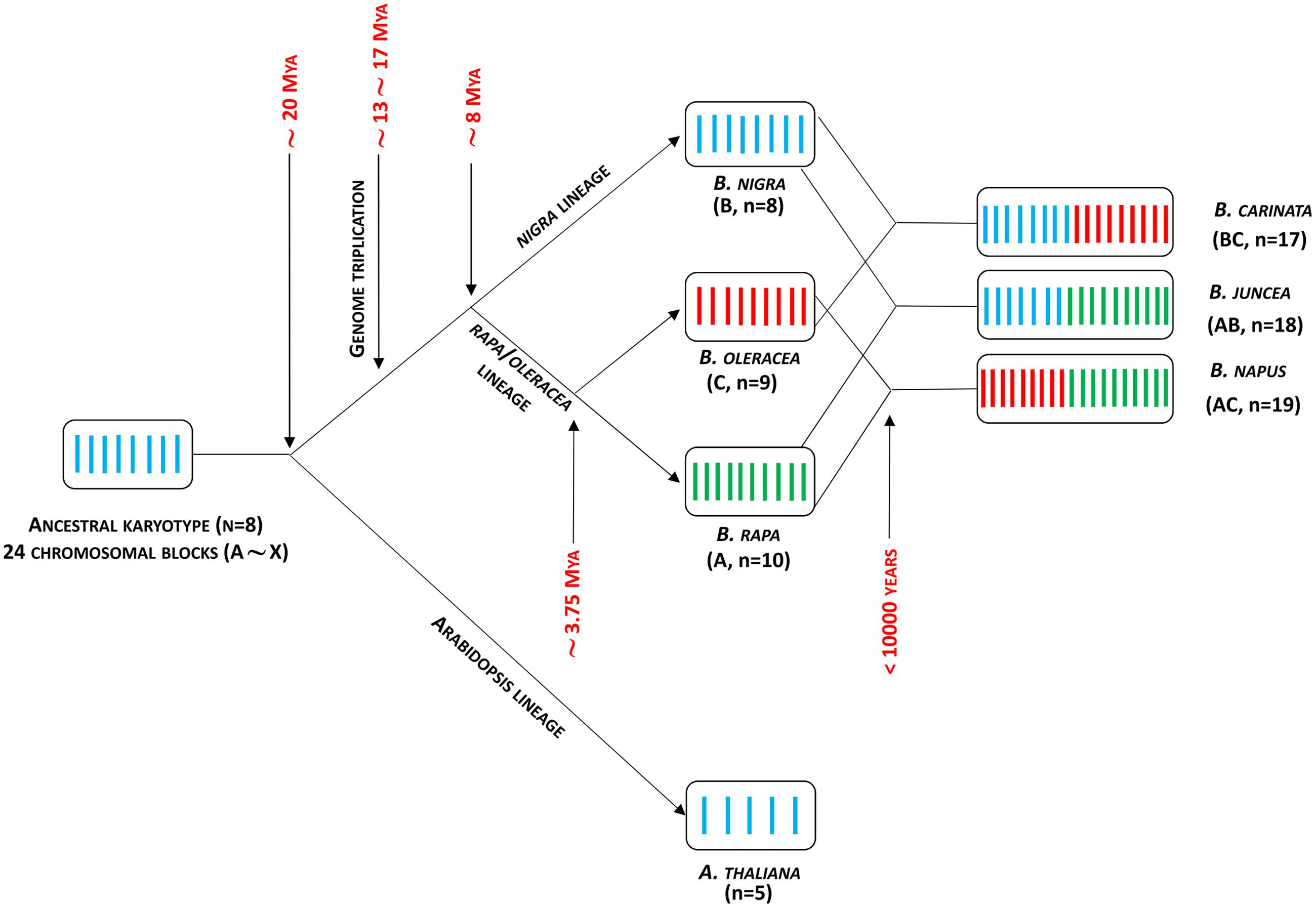
FIGURE 3. Brassica genome evolution model. Adapted from Liu et al. (2014). Mya, million years ago.
Recently, 82 mQTLs associated with GSL compositional profiles were identified in B. oleracea, which corresponded by collinearity with orthologous Arabidopsis GSL biosynthesis genes (Sotelo et al., 2014). A separate study has indicated that three paralagous genes encoding AOP2 protein involved in aliphatic GSL biosynthesis, are differentially expressed in B. rapa (Zhang et al., 2015b). Although, there is evidence of a reduction in GSL biosynthesis and related genes due to recent breeding selection in canola-type B. napus (Chalhoub et al., 2014), the ability of these polyploid genomes to carry multiple copies of biosynthetic and regulatory genes at upward of six or more paralogous loci provides an opportunity for far greater complexity. The collinearity of GSL biosynthesis genes between Arabidopsis and Brassica genomes and their proliferation in Brassica has been established through comparative whole-genome analysis in B. rapa (Wang H. et al., 2011), as well as in B. oleracea (Liu et al., 2014), B. napus (Chalhoub et al., 2014), and B. juncea (Yang et al., 2016) (Supplementary Table S2).
A complex network of TFs belonging to the R2R3-MYB family regulate the biosynthesis of GSLs in Arabidopsis. These are distinct from those involved in generic S metabolism and transport, which we discuss later. MYB28, MYB76, and MYB29 TFs influence the expression of MAMs, CYP79F1/F2, CYP83A1, SUR1, UGT74B1, and SOT17/18 genes involved in aliphatic GSL biosynthesis (Gigolashvili et al., 2007, 2008, Hirai et al., 2007; Sønderby et al., 2007). A complementary set of TFs including MYB51, MYB122, and MYB34 appear to regulate indole GSL biosynthesis via CYP79B2/B3, CYP83B1, SUR1, UGT74B1, and SOT16 genes (Yatusevich et al., 2010; Frerigmann and Gigolashvili, 2014a). Four differentially expressed AtMYB28 homologs in B. juncea can act as major transcriptional regulators of aliphatic GSLs accumulation (Augustine et al., 2013a). Moreover, targeted gene silencing of these MYBs suggests that BjMYB28 would be a potential candidate for developing low GSL lines in B. juncea (Augustine et al., 2013b). However, recent functional analysis in transgenic B. rapa of three orthologous copies of AtMYB28 revealed that BrMYB28 genes are responsible not only for aliphatic, but also for indole and aromatic GSLs biosynthesis, where they specifically act as positive regulators of the BrGSL-OH gene and negatively regulate the BrAOP2 genes (Seo et al., 2016).
Moreover, evidence from yeast-two-hybrid (Y2H) screening (Frerigmann et al., 2014) and mutant analysis in Arabidopsis (Schweizer et al., 2013) has revealed that four members of another group of TFs belonging to the bHLH subgroup IIIe, also known as MYC-bHLH TFs, regulate GSL biosynthesis in cooperation with MYB TFs (reviewed in Frerigmann, 2016). MYC-bHLHs also act as key signaling components of the jasmonic acid pathway (Fernandez-Calvo et al., 2011). Besides these six MYBs and four MYCs, two new MYBs (MYB115 and MYB118) have recently been identified which are responsible for the regulation of aliphatic GSLs in Arabidopsis (Zhang Y. et al., 2015). A further recent study based on an Arabidopsis mutant of a small heme-binding protein, known as Cytochrome b5 (CB5C), showed that it can influence GSL biosynthesis via cytochrome P450 (Vik et al., 2016). However, the mode of action of this protein is still unknown.
Sites of GSL Synthesis and Accumulation in Plants
Within intact plants, the site of GSL synthesis is physically segregated in tissues distinct from the site of GSL storage (Chen et al., 2001), most likely to protect biologically active cells from the cytotoxic effects of GSL hydrolysis products. However, this may also be the most resource efficient means by which a centrally localized biosynthetic machinery can deliver GSLs to several sink tissues (reviewed in Kissen et al., 2009; Jørgensen et al., 2015). There is additional separation of the GSL substrates and myrosinase enzymes in a stable spatial distribution system (reviewed in Winde and Wittstock, 2011) originally known as the ‘mustard oil bomb’ (Lüthy and Matile, 1984). This has been characterized by the typical localization of GSLs in specialized S-cells of high S content, with myrosinases localized in distinct myrosin cells (Koroleva and Cramer, 2011). Additional evidence from metabolite profiling in Arabidopsis has suggested a separate system wherein GSLs and myrosinase can co-exist in the same cell, with GSLs localized within vacuoles and ‘atypical myrosinase (PEN2 protein)’ in endoplasmic reticulum (ER) bodies, which are derived vesicles of the ER (Moussaieff et al., 2013). The presence of similar ER bodies in Arabidopsis, as well as in some other members of the Brassicales, suggests that this distribution is a common feature within these taxa (reviewed in Nakano et al., 2014). Moreover, a recent confocal laser scanning microscopy study of Arabidopsis pen2 mutant plants has indicated that the ‘atypical myrosinase’ has a dual-membrane targeting capability toward peroxisomes and mitochondria (Fuchs et al., 2016).
The concentration of GSLs varies between tissue types and developmental stage, with a study of 29 different plant species demonstrating that greater diversity and higher concentrations of GSLs exist in roots compared to shoots during vegetative growth (van Dam et al., 2009). In mature Arabidopsis, the highest concentrations of GSLs are found in seeds (2.5–3.3% of dry weight), followed by inflorescences and siliques (0.6–1.0%) and cauline leaves, roots, stems, and rosette leaves (0.3–1.0%) (Brown et al., 2003). Similar distributions of GSLs are observed in Brassica plants (Fahey et al., 2001; Rangkadilok et al., 2002; Bhandari et al., 2015). A GSL tissue audit in B. napus has suggested that changes within sink tissues are determined according to varying requirements at specific plant developmental stages (Clossais-Besnard and Larher, 1991; Porter et al., 1991).
It has been suggested that siliques and rosette leaves are the primary sources for the GSLs accumulated in the seeds (secondary sink) of B. napus (Magrath and Mithen, 1993; Bloem et al., 2007), B. oleracea (Bhandari et al., 2015), Sinapis alba (Du and Halkier, 1998) and Arabidopsis (Chen et al., 2001; Ellerbrock et al., 2007) (Figure 4). This is supported by recent studies with mutants of Arabidopsis GSL transporters (Nour-Eldin et al., 2012), although confirmatory evidence for primary sources of GSLs accumulated in seed (embryo) has yet to emerge (reviewed in Jørgensen et al., 2015). Roots and trichomes are also capable of aliphatic and indole-GSL biosynthesis in Arabidopsis (Frerigmann et al., 2012; Andersen et al., 2013). Based on micro-grafting experiments in Arabidopsis, distinct source-sink relationships have been found to be associated with short- and long chained aliphatic GSLs. For long-chained aliphatic GSLs, roots appear to act as a source and rosette leaves as a sink, whereas for short chained-aliphatic GSLs, rosettes can act as both source and sink (Andersen et al., 2013; Moussaieff et al., 2013; Andersen and Halkier, 2014). This evidence has been used to suggest that there must be specific regulation of GSL distribution between different tissues within the plant (reviewed in Jørgensen et al., 2015). Moreover, evidence from transcriptomic studies of sulfate transporters (SUTRs) (Buchner et al., 2004b) suggests that primary sink tissues (e.g., leaf, silique, root) associated with S assimilation at an early stage of plant development may later act as a source for primary and secondary sinks of GSLs, such as siliques and seeds (Andersen et al., 2013). However, there remains an incomplete picture of how GSL source and sink distribution changes in relation to plant and crop developmental phases.
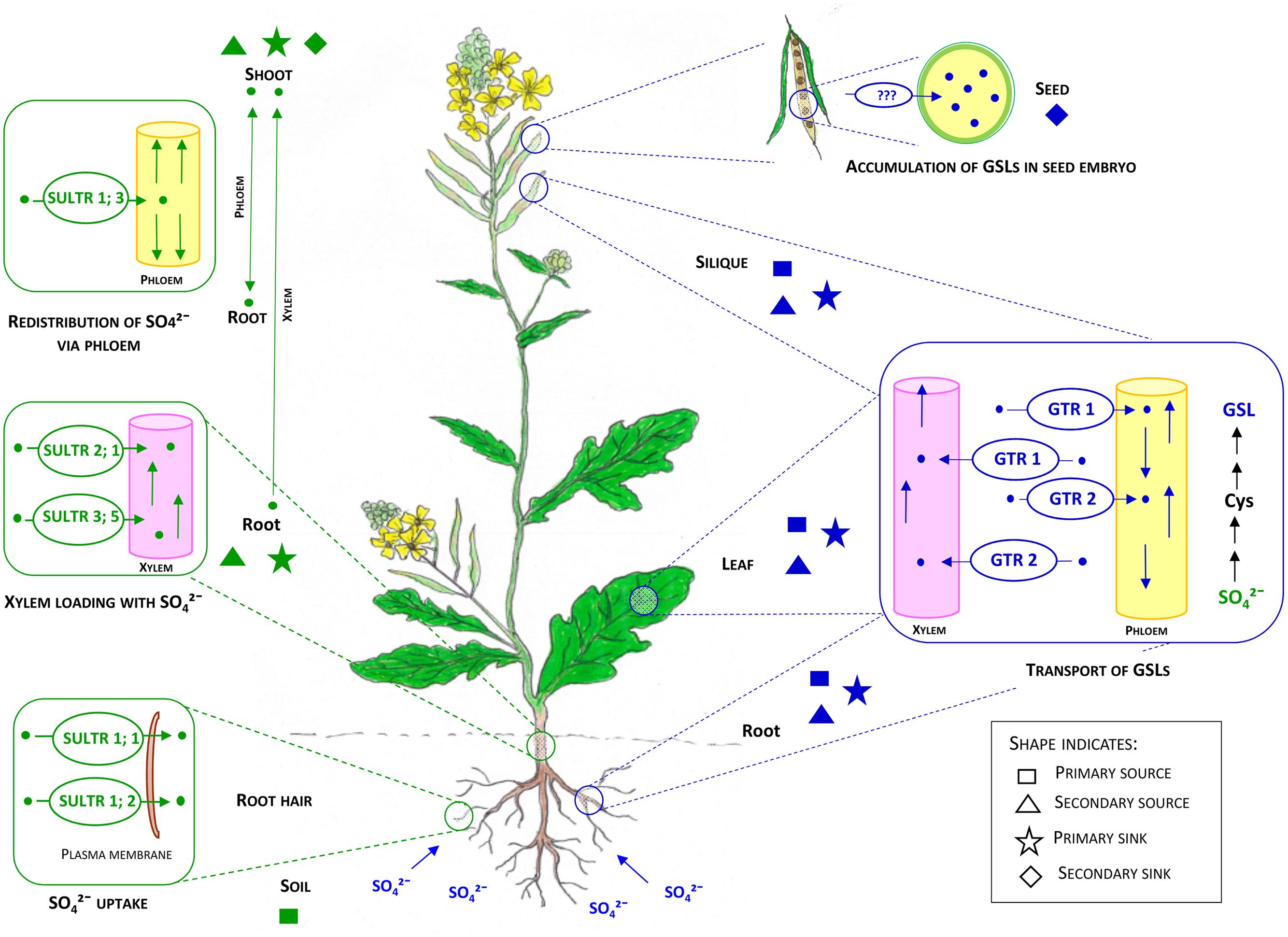
FIGURE 4. Internal transport systems of sulfate (SO42-) and GSLs in the context of primary and secondary sources and sinks in brassicas. Adapted from Frerigmann and Gigolashvili (2014b); Gigolashvili and Kopriva (2014), Jørgensen et al. (2015). Soil can be considered as the primary source for S. Roots and shoots have overlapping functions as primary and secondary sources and sinks for S. Sulfate uptake by root hairs from soil is facilitated by SULTR 1; 1 and SULTR 1; 2. Root to shoot transport of sulfate is facilitated by SULTR2; 1 and SULTR3; 5 via xylem tissues. Redistribution of sulfate is likely to occur by SULTR 1; 3, while GSLs are primarily synthesized in leaves, siliques and roots. After synthesis, GSLs can undergo bidirectional transport to the same and/or other organs or tissues by GTR 1 and GTR2. There is an overlap of primary and secondary sources and sinks for GSL (leaf, root, and silique). Seed embryos can be considered as the final sink for GSLs in mature Brassica plants. GTR1 and GTR2 facilitate the transport process by vascular and symplastic domains. The critical barrier (e.g., the funiculus) for localization of GTRs to facilitate transport of GSLs to embryo is still unknown. Text and shapes in green indicate transport, sources and sinks related to the sulfate system whereas text and shapes in blue indicate the GSL system.
Transport of Inorganic and Organic S
Sulfur is an essential macronutrient involved in numerous metabolic activities required for plant growth, especially in amino acid and protein synthesis. The primary inorganic form, sulfate, accounts for 42–94% of the total S content in different tissues of Brassica plants (Blake-Kalff et al., 1998; Castro et al., 2004), whereas other inorganic forms (sulfite and elemental sulfur) are much lower in concentration. Notably, in Brassica seeds, proteins (cruciferins and napins) and GSLs represent the major sinks of S, with each representing an approximate similar stoichiometric ratio of S relative to seed mass (Supplementary Table S3). However, this ratio is sensitive to the relative composition of cruciferins to napins, suggesting scope for manipulating GSL as an S sink.
Transport of Sulfate
Plants maintain a well-organized transport system to make nutrients available within the whole plant, and to distribute important compounds to specific organs. Sulfur assimilation primarily takes place in the shoot chloroplast despite roots containing all the necessary sulfate reductase enzymes (reviewed in Hawkesford, 2000; Hawkesford and De Kok, 2006). As a result, sulfate is the major form of S loaded in the xylem for transport from root to shoot, and is also the major form of stored S in the plant (Buchner et al., 2004b). Constant cytoplasmic sulfate concentrations are maintained throughout the plant system, whilst excess amounts are stored in the vacuole (Kaiser et al., 1989; Miller et al., 2001). Specialized sulfate transporters encoded by a multigene family play a key role in S-metabolite transport, and are divided into four distinct groups with different functions (Smith et al., 1995). In Arabidopsis, the SULTR family is well-characterized with orthologs also reported in B. rapa, B. oleracea and B. juncea (Buchner et al., 2004a; Koralewska et al., 2008; Wang X. et al., 2011; Chalhoub et al., 2014; Lancilli et al., 2014; Liu et al., 2014). In Arabidopsis the first group of SULTRs are high affinity transporters, amongst which SULTR1;1 and SULTR1;2 – found in the root hairs – facilitate the initial uptake of sulfate from the soil (Yoshimoto et al., 2002) while phloem-localized SULTR1;3 facilitates the redistribution of sulfate from secondary sources to primary and secondary sinks, i.e., root to shoot and vice versa (Yoshimoto et al., 2003; Figure 4). Group 2 SULTRs are mostly confined to vascular tissues to facilitate long distance transport of sulfate (Takahashi et al., 2000), while Group 3 transporters are the most diffuse form, localized in plastid membranes for import of sulfate (Cao et al., 2013; Figures 4 and 5). Group 4 SULTRs (SULTR4;1 and SULTR4;2) are found in the tonoplast, where they facilitate the efflux of sulfate from vacuoles into the cytoplasm (Kataoka et al., 2004; Figure 5). However, transporters responsible for sulfate influx into the vacuole are yet to be identified. It is reasonable to assume that Brassica SULTR homologs would have a similar or identical function as Arabidopsis SULTRs, due to the close phylogenic relationship of these genera (Figure 4). However, transcript analysis of SULTRs indicates a spatial contrast, with a pattern of abundance for group 3 SULTRs (SULTR3;2, SULTR3;3, and SULTR3;5) in B. oleracea root tissues, which in Arabidopsis are normally expressed in the leaves (Buchner et al., 2004a). Additionally, this study showed that under S deficient condition, groups 1, 2, and 4 SULTRs have been found to be up-regulated in the roots of B. oleracea, whereas Group 3 SULTRs are unaffected by plant S status.
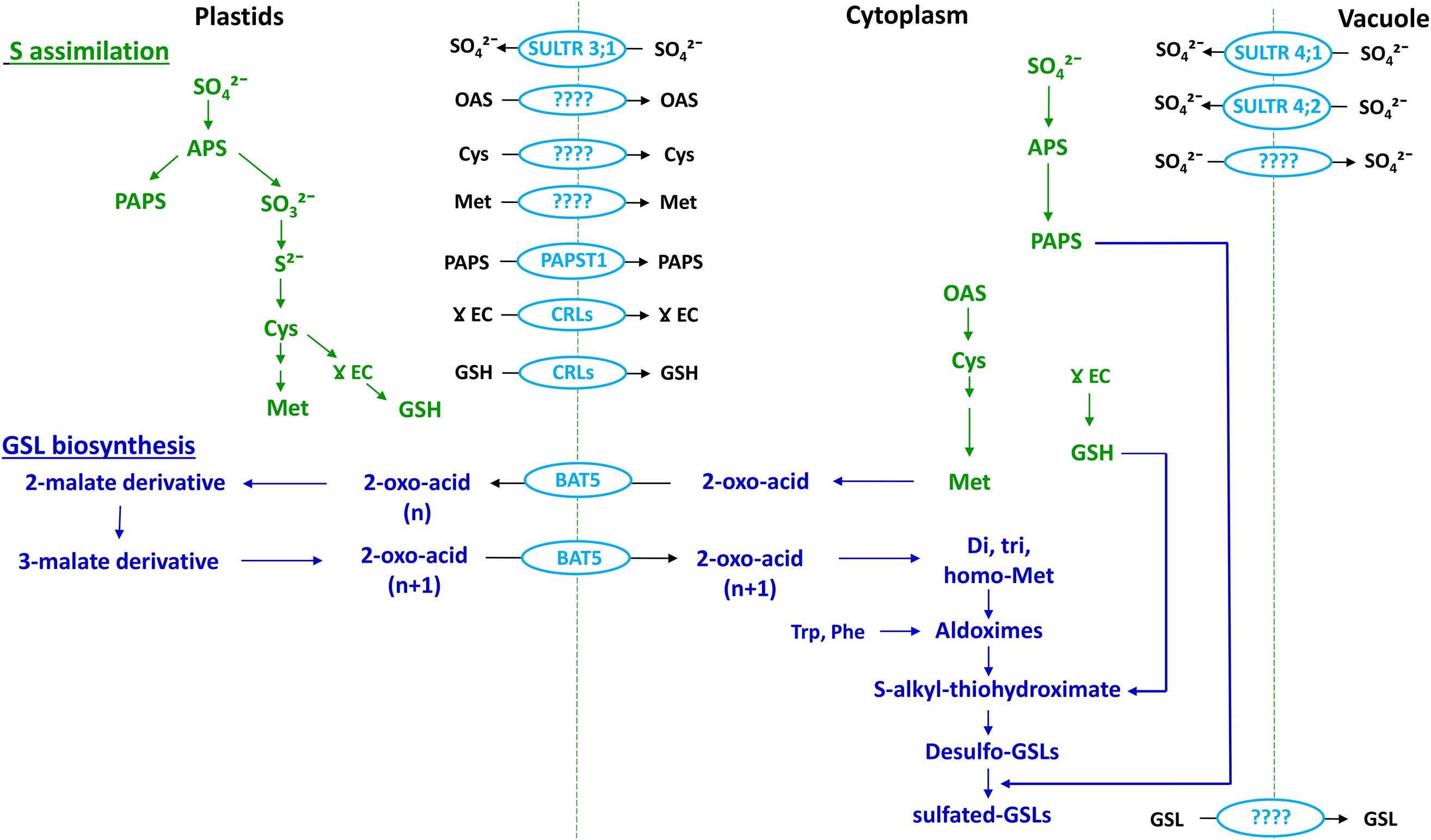
FIGURE 5. Association between the S assimilation pathway and GSL biosynthesis (emphasizing Met-derived GSLs) within a cell. Adapted from Gigolashvili and Kopriva (2014). OAS, O-acetylserine. Cellular compartments are indicated at the top of the figure. Text in green indicates S assimilation; text in dark blue indicates GSL biosynthesis and text and shapes in light blue indicate transporters.
Transport Associated with S Assimilation
Following uptake by roots, sulfate is transported to shoots and assimilated by enzymatic reduction and conversion to organic S forms. Within this process, sulfate taken up from the soil is activated to adenosine 5′-phosphosulfate (APS), which is the branching point (Figure 1) for subsequent steps of reduction of APS to form sulfide and PAPS. Sulfide is used for synthesis of Cys and thence GSH and Met, whereas PAPS is used for sulfation of desulfo-GSLs (Mugford et al., 2011; Figures 1 and 5).
Cysteine, GSH, Met, and PAPS are the major S assimilates involved in GSL synthesis, with Cys the primary form synthesized in cytoplasm, plastids, and mitochondria (Lunn et al., 1990). Although, there is evidence to support the presence of numerous transport systems for Cys, specific transporters have yet to be identified (reviewed in Gigolashvili and Kopriva, 2014; Figure 5).
Based on transcript analysis and transient transformation experiments, the gamma (γ)-glutamylcysteine (γEC) synthase and GSH synthase enzymes of GSH synthesis have been found to be located in plastids and cytoplasm, respectively (Wachter et al., 2005). This suggests the need for export of γ-EC to cytoplasm for synthesis of GSH. The chloroquine-resistance transporter like proteins (CRLs) facilitates the transport of γ-EC as well as GSH into cytoplasm for GSL biosynthesis (Pasternak et al., 2008; Figure 5). GSH is one of the assimilated forms of S and there is evidence for long distance transport of GSH within the plant (Herschbach and Rennenberg, 1995). This suggest the need for identification of cell membrane located transporters in plants (reviewed in Gigolashvili and Kopriva, 2014).
Within the S assimilation process Met is formed via the mediation of cytosolic Met synthase, and is the primary precursor for aliphatic-GSLs (Schuster et al., 2006; Figure 5). Initially, it had been thought that the synthesis of Met required homocysteine to be transported from plastids to the cytoplasm, as there was no evidence of a Met synthase enzyme in the plastids (Eichel et al., 1995). However, following the identification of plastid-localized Met synthase, it was concluded that plastids are indeed capable of de novo synthesis of Met (Ravanel et al., 2004), and so a pool of Met could be sequestered within plastids that is not available for synthesis of aliphatic GSLs (Figure 5). So far no specific amino acid transporters have been identified in plants to facilitate Met transport between organelles or cells.
The major portion of PAPS is synthesized in chloroplasts, whereas the sulfation reaction of GSLs takes place in the cytoplasm (Mugford et al., 2009, 2010; Figure 5). Notably, chloroplasts contain the SAL1 phosphatase enzyme, which can hydrolyze PAP (Estavillo et al., 2011). Therefore, there must be a plastid-localized transporter to deliver PAPS in to the cytoplasm (Figure 5). In Arabidopsis, apart from its primary function, the transporter known as thylakoid ADP/ATP carrier (TAAC) has an additional function to transport PAPS across the plastid membrane (Thuswaldner et al., 2007; Gigolashvili et al., 2012). More recently, the TAAC transporter has been renamed as PAPS transporter 1 (PAPST1) (Gigolashvili et al., 2012). However, knockout mutants of Arabidopsis PAPST1 contain 30–50% of aliphatic GSLs and have an unaltered levels of indole GSLs, which indicates the existence of an additional transporter for PAPS in the plastid membrane (Mugford et al., 2009). It should be recognized that all these studies have been carried out solely in Arabidopsis, whereas detailed investigation is required in crop brassicas in order to understand the complexity of transport system associated with S assimilation in field situations.
Transport of GSLs
Biosynthesis of GSLs is compartmentalized within plastids and the cytoplasm, where Met-derived GSLs undergo a chain elongation process in the chloroplast, prior to the synthesis of the core structure in the cytoplasm (reviewed in Halkier and Gershenzon, 2006; Sonderby et al., 2010). Transcriptome co-expression analysis of Arabidopsis knock out mutants suggested that chloroplast-localized bile acid transporter 5 (BAT5) can facilitate the import of Met-derived 2-oxo acid into the chloroplast, and can also export chain elongated 2-oxo-acid to the cytoplasm for synthesis of the GSL core structure (Gigolashvili et al., 2009a; Sawada et al., 2009a; Figure 5). A feeding experiment of the Atbat5 knock-out mutant with Met-derived 2-keto acids and different chain length amino acids confirmed that the BAT5 transporter has specific affinity toward only 2-keto acids (Gigolashvili et al., 2009a). After the sulfated-GSLs are formed in the cytoplasm, they need to be transported either to vacuoles or S -cells for storage. The transporter(s) responsible for influx of GSLs into the vacuole for storage is still unknown.
Proteomic analysis has indicated the absence of GSL biosynthesis machinery within the cytoplasm of S-cells, which is consistent with the need for transporters to account for the accumulation of GSLs in these cells (Koroleva and Cramer, 2011). It has been confirmed that only sulfated GSLs can be transported into the developing seeds, rather than desulfo-GSLs (Mugford et al., 2009). This strongly indicates the need for long distance transport of GSLs from primary and secondary source (leaf, root, and silique) to sink (leaf, root, silique, and embryo) tissue. The recent identification of nitrate/peptide group GSL transporters AtNPF2.10 (AtGTR1) and AtNPF2.11 (AtGTR2) in Arabidopsis has contributed toward an understanding of the likely transport mechanisms for GSL distribution (Nour-Eldin et al., 2012). These transporters are responsible for long distance transport of short and long-chained aliphatic GSLs from primary and secondary source tissues (root, shoot, silique) to primary and secondary sinks (leaves, embryo), by facilitating long distance transport through phloem and xylem tissues (Andersen et al., 2013; Moussaieff et al., 2013; Andersen and Halkier, 2014). However, the ability of GTRs to transport GSLs via symplastic domains suggests that they also appear to be essential for maintaining the distribution of GSLs within roots and leaves (Nour-Eldin et al., 2012). Micro-grafting experiments in Arabidopsis have suggested the existence of a specific-transporter in addition to GTR1 and 2 that is able to facilitate the transport of indole-GSLs between rosette leaves and roots (Andersen et al., 2013). It is reasonable to expect that these components within the Arabidopsis sulfate and GSL transport systems are likely to be relevant in the larger and more complex crop brassicas, but confirmation of specific roles requires further investigation (Figure 4). To date, there appear to be no reports of specific transporters from functional studies in brassicas. However, these transporters may be useful for engineering novel insect resistance, as a recent study has demonstrated that leaves of Arabidopsis mutants of gtr1gtr2 contributes to reducing the fitness of green peach aphid (Myzus persicae) due to reduced availability of GSL in phloem sap, and consequent increased GSL in the tissues surrounding the phloem (Madsen et al., 2015).
Molecular Cross-Talk Between S Availability and GSL Biosynthesis
The recent identification of feedback mechanisms between specific transporters, signaling molecules and TFs in Arabidopsis is leading to a better understanding of the molecular interactions that may mediate between S status and GSL biosynthesis. Metabolomic and transcriptomic studies have demonstrated that under S limitation, reduction of GSL levels (Hirai et al., 2003; Maruyama-Nakashita et al., 2003; Nikiforova et al., 2003) and activation of sulfate uptake and assimilation (Hirai et al., 2003) may occur simultaneously. So far, Sulfur Limitation 1 (SLIM1) has been reported to be the key TF in Arabidopsis which regulates the genes involved in sulfate uptake and S assimilation. Under S-deficient conditions, SLIM1 appears to affect both sulfate uptake and GSL biosynthesis. Transcriptome profile of slim1 Arabidopsis mutants clearly demonstrated that under S limitation, the expression of genes encoding SULTRs from group 1 (SULTR1;1 and SULTR1;2: for sulfate uptake by roots), group 3 (SULTR3;4; for import of sulfate) and group 4 (SULTR4;2: for release of sulfate from vacuole to cytoplasm in roots) are upregulated to enhance sulfate uptake from the root and S assimilation (Maruyama-Nakashita et al., 2006). In contrast, key enzymes within different phases of GSL biosynthesis, including MAM1, MAML, CYP79B2/B3, CYP83B1, GST, BCAT and MYB34, are downregulated in slim1 mutants under S limitation (Maruyama-Nakashita et al., 2006). Moreover, the expression of MYBs regulating aliphatic GSL synthesis in Arabidopsis varies under S stress. Under mild S deficient conditions, in the transgenic lines of distinct MYB genes, expression level of MYB28 can be induced, whereas MYB29 and MYB76 expression level were positively correlated with S concentrations (Li et al., 2013). It has been suggested that in Arabidopsis, S assimilation is affected by SLIM1, which can be considered as a negative regulator of the R2R3-MYB family of TF, members of which have specific interactions with different components of GSL synthesis pathways (Takahashi et al., 2011; Figure 6). qRT-PCR analysis of Arabidopsis over-expressed SLIM1 cultured cells confirmed that SLIM1 can suppress the expression of R2R3-MYBs in vitro (Frerigmann and Gigolashvili, 2014b). However, the expression level of MYB28, MYB34, MYB51, and MYB122 was neither changed nor increased upon S limitation. To justify this observation, it was suggested that a low substrate (indole or aliphatic GSL) production signal can nullify the negative regulation of MYBs by SLIM1 (Frerigmann and Gigolashvili, 2014b).
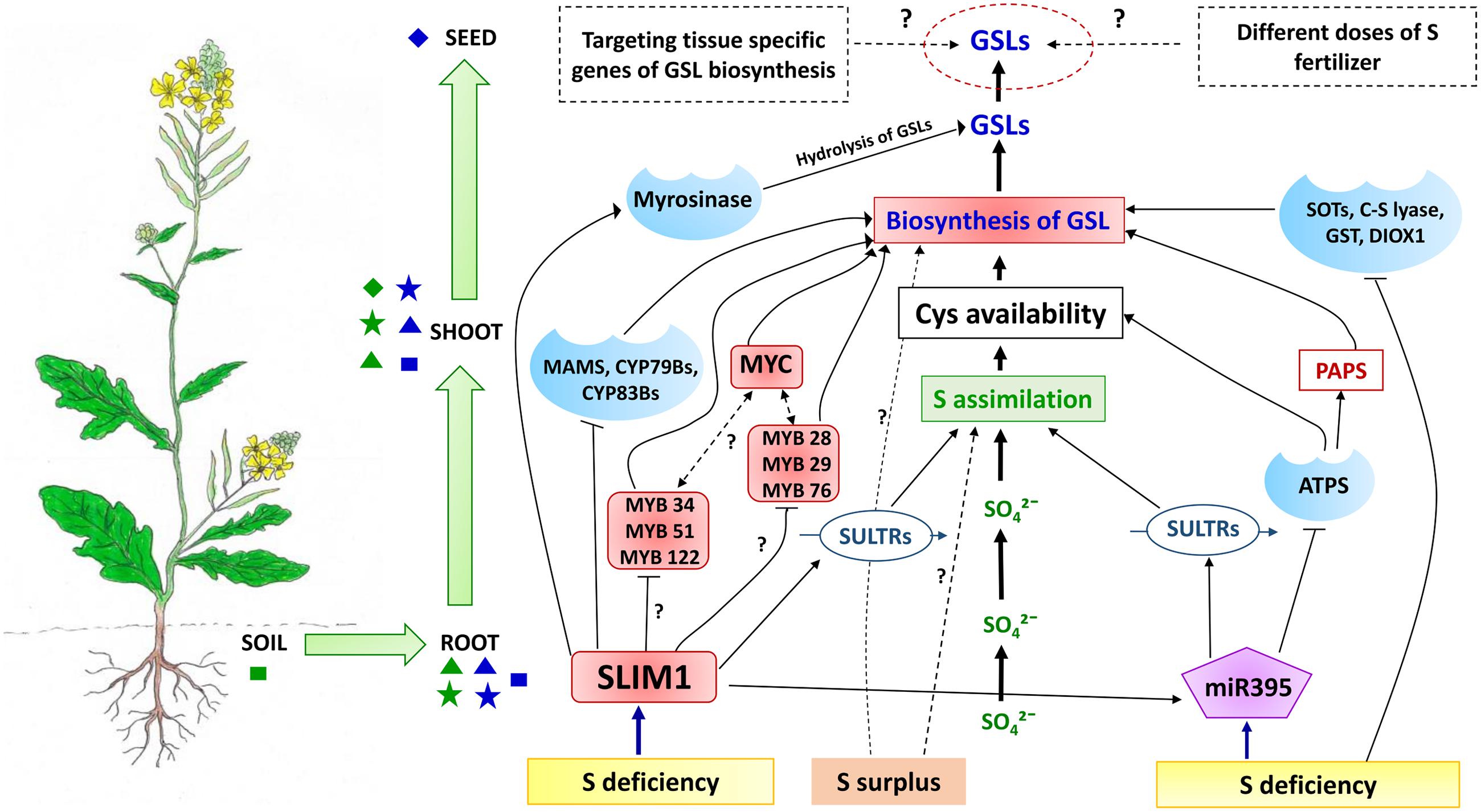
FIGURE 6. A conceptual model how soil S status affects GSL accumulation in Brassica crops. Under S deficiency, the SLIM1 TF can down regulate the expression of key GSL biosynthesis enzymes (MAMS, CYP79Bs, and CYP83Bs) and MYB TFs (Lewandowska and Sirko, 2008; Frerigmann and Gigolashvili, 2014b). Further investigation needed to understand the complete picture of regulation of MYBs targeting indole GSL biosynthesis (MYB34, MYB51, MYB 122) and aliphatic GSL synthesis (MYB28, MYB 29, MYB76) by SLIM1 under different S conditions. In contrast, under S deficient conditions SLIM1 can also upregulate the expression of SULTRs and miR395, as well as the GSL hydrolysis enzyme (myrosinase) (Takahashi et al., 2011). In addition, upregulation of miR395 via SLIM1 under S deficiency can increase the expression of SULTRs and lead to enhanced uptake of sulfate from soil. SLIM1 can also down-regulate the expression of ATPS in the first step of S assimilation (Liang et al., 2010) which can affect the availability of Cys and PAPS for GSL biosynthesis. Sulfur deficiency has been suggested as a negative regulator of SOTs, C-S lyases, GST and AOPs enzymes of the GSL biosynthesis pathway (Nikiforova et al., 2005), although additional signaling molecules may be involved in this regulation. Moreover, we would expect the presence of additional signaling molecules and TFs, which can regulate both systems (S assimilation and GSL biosynthesis) under S deficiency or varying S levels of the primary source (soil). As seeds are the ultimate sink for GSLs, optimizing the external application of S fertilizer and identifying the role of tissue-specific genes of GSL biosynthesis under different S levels would provide scope for manipulation of seed GSL levels in brassicas. Primary and secondary sources and sinks for S and GSLs are shown as indicated as in Figure 4. Shapes in green indicate sources and sinks related to the sulfate system whereas shapes in blue indicate the GSL system. Rectangular shapes indicate primary sources, triangles indicate secondary sources whereas stars indicate primary sinks and diamonds indicate secondary sinks.
Early investigation of S and GSH supply and concentrations in B. napus had suggested that under S limitation, GSLs might be degraded within intact plants by myrosinase, without tissue damage (Schnug et al., 1995). In response to S limitation, Arabidopsis plants degrade GSLs, so that the released S can be remobilized for production of primary metabolites. In this S recycling process, SLIM1 appears to play an important role, by upregulating the genes encoding enzymes for GSL degradation. This is supported by evidence from transcript analysis of a Arabidopsis slim1 mutant under S limiting conditions, where a putative thioglucosidase (i.e., myrosinase) were found to be significantly upregulated, to hydrolyze GSLs (Maruyama-Nakashita et al., 2006; Figure 6).
Several studies suggest that there is a metabolic association between indole glucosinolates and indole-3-acetic acid (IAA), with GSL being hydrolyzed in consecutive reactions into indole acetonitrile (IAN), which is then hydrolyzed by nitrilase to IAA. Moreover, under S stress hydrolysis of indole GSLs can induce the synthesis of auxin (IAA), enhancing lateral root formation to increase uptake of sulfate from the soil. Although, increased accumulation of IAA has not been reported under S limiting conditions, several investigations suggest that indole GSL hydrolysis does occur under these conditions (Frerigmann and Gigolashvili, 2014b), with associated gene induction of Trp synthesis (Nikiforova et al., 2003), activation of a GSL hydrolysis enzyme (Nikiforova et al., 2003, 2005; Hirai et al., 2004, 2005) and overexpression of nitrilases (Kutz et al., 2002).
MicroRNAs (miRNAs) have a significant regulatory function in controlling gene expression in response to nutrient deficiency, including S deficiency (Sunkar et al., 2007). In Arabidopsis, miR395 is involved in the regulation of S assimilation and sulfate transport processes (Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004), by targeting the mRNAs of three ATPS genes involved in S assimilation (Jones-Rhoades and Bartel, 2004; Matthewman et al., 2012; Figure 6). Under S starvation, expression of miR395, which can also target the transcripts of SULT2; 1, was found to be upregulated (Allen et al., 2005; Liang and Yu, 2010), which suggests that the rate of S assimilation can also be manipulated by targeting the cell-specific expression pattern of miR395 (Kawashima et al., 2011; Matthewman et al., 2012). Under S deficiency, as opposed to complete S starvation, SLIM1 TF in plants can directly or indirectly induce the expression of miR395 (Kawashima et al., 2009; Liang et al., 2010; Figure 6), as can redox signaling (Jagadeeswaran et al., 2014). It has been shown that miR395 is phloem-mobile and can play the role of a long distance signaling molecule in response to changes in plant S status (Buhtz et al., 2010). In addition, miR393, which normally targets auxin receptors, was found to be capable of redirecting the biosynthesis of secondary metabolites from camalexin toward GSLs (Robert-Seilaniantz et al., 2011). To date no such studies have unraveled the role of direct miRNA signaling within the GSL pathway.
The network of interactions outlined above (reviewed in Lewandowska and Sirko, 2008; Koprivova and Kopriva, 2014, 2016; Frerigmann, 2016) suggests that there are significant feedback mechanisms to enable cross-talk between the S and GSL systems (Figure 6). This is consistent with earlier studies which demonstrated, through combined metabolomic and transcriptomic analyses, that the major gene families of GSL biosynthesis including MAM, CYP79 and CYP83 were down-regulated under S deficiency (Maruyama-Nakashita et al., 2006). Additional genes encoding SOTs, C-S lyase, GST and AOPs of GSL biosynthesis, have also been found to be down-regulated under S deficiency (Nikiforova et al., 2003, 2005; Hirai et al., 2005; Figure 6). This appears to explain the reduction in GSL content typically observed under S starvation. Consequently, to maximize sulfate uptake, assimilation and utilization under S-deficient conditions, plants increase the expression of genes responsible for the S assimilation process (Hirai et al., 2003; Falk et al., 2007).
A Regulatory Model of GSL Response to S
Overall, there are many lines of evidence to suggest that plant S status has a considerable influence on GSL content. Here, we propose that the interactions between the S and GSL systems act to mediate the distribution of resources from the respective sources and sinks. Moreover, we propose that the specific sets of transporters, signaling molecules and TFs mediate the cross-talk between these sources and sinks in the context of the intracellular, symplastic and vascular systems. Sulfate taken up from soil (primary source) can be assimilated into either primary or secondary metabolites, including GSLs in roots or shoots which can be considered as secondary sources, as well as primary sink tissues. Notably, GSH and Met produced from Cys in the S assimilation process can enter into the GSL biosynthesis pathway within different organelles and tissues. On the other hand, PAPS associated with S assimilation acts as an S donor for sulfation of biologically active GSLs, which can then be transported to the primary (leaf, root and silique) and secondary GSL sink (seed) tissue. As mentioned, SLIM1 can affect miR395, which in turn can regulate uptake, assimilation and transport of sulfate. In order to maintain S homoeostasis, the mi395 signaling molecule provides a feedback mechanism to regulate transporters which then facilitates S assimilation within secondary sources (shoot tissues) (Figures 4 and 6) (Liang et al., 2010). As a consequence this mechanism can affect the availability of Cys, GSH, Met, and PAPS for GSL biosynthesis in primary and secondary source and sink tissues (leaf, root, and silique), and can thus limit the accumulation of GSLs in the ultimate sink (seed embryo) at maturity. Moreover, under S deficiency, the TF SLIM1 affects both systems, including S assimilation by SULTR transporters, as well as targeting the key enzymes of GSL biosynthesis (Maruyama-Nakashita et al., 2006). The GSL network is also targeted by a complex network of MYB TFs via SLIM1 as discussed in Section “Molecular Cross-Talk between S Availability and GSL Biosynthesis”. SMIL1 can negatively regulate MYBs under S deficiency, although a signal induced by low indole GSL production can nullify this effect (Li et al., 2013; Frerigmann and Gigolashvili, 2014b).
The overall model (Figure 6) provides a working framework to explore the cross-talk between S and GSL systems. However, we would expect there to be additional signaling molecules required to mediate between Cys and GSL chain elongation and core structure synthesis, PAPS and the GSL core structure synthesis, as well as between ATPS and GSL biosynthesis. In Arabidopsis there appears to be strong evidence for only six MYBs (MYB 28, 29, 76 for aliphatic and MYB 51, 122, 34 for indole GSL synthesis) being sufficient for signaling the regulation of GSL biosynthesis. However, this may not be the case in brassicas, due to the increased genome complexity, ontogenetic plasticity, and GSL diversity compared with Arabidopsis. Additionally, recent identification of four MYC-bHLHs, two new MYBs (MYB115 and MYB118 for aliphatic GSLs synthesis) and a CB5C protein suggests that there may be more distinct or subtle patterns of GSL regulation in Arabidopsis under different conditions of S starvation and sufficiency, which needs further investigation. Moreover, there may be further TFs that provide more specific interactions with signaling molecules and sulfate and GSL transporters, as well as with specific biosynthetic steps. Overall, we would expect that under different soil S levels S assimilation rates will vary, which can affect Cys, GSH, PAPS and Met availability and the transcription of genes for GSL biosynthesis.
Concluding Remarks
As GSLs represent one of the most widely studied classes of plant secondary metabolites, there is an extensive literature describing GSL biosynthesis genes available from the model plant Arabidopsis and B. oleracea (Sonderby et al., 2010; Sotelo et al., 2014). Based on this we have developed a model of crop Brassica GSL response to S (Figure 6). Seeds can be considered as the ultimate sink for GSL accumulation. In order to manipulate the content of GSLs within the seed, our model would suggest that different rates of S provided as fertilizer can make a significant contribution. Also, our model suggests that there is scope to understand the role of tissue-specific GSL biosynthesis genes under different S status.
Brassica genomes are three to five times the size of the Arabidopsis genome (Paterson et al., 2001) and are more complex, with multiple paralogous gene arising from genome duplication events. This provides considerable scope for gene neo-functionalization in terms of their spatial and temporal regulation (Kroymann, 2011; Mickelbart et al., 2015), and may lead to more sophisticated means of controls for GSL accumulation. Moreover, Brassica seeds are more than 10 times larger than those of Arabidopsis (Gorsuch et al., 1991), and contain a broader range of GSLs (reviewed in Ishida et al., 2014). Thus at present it is difficult to resolve and understand the complexity and subtlety of interactions in large crop Brassica plants based on the available literature from the Arabidopsis model, especially where loci associated with GSL response to S have yet to be characterized. Studies with the amphidiploid Brassica species are likely to yield additional valuable information to refine the model and establish a better understanding of the molecular regulation of GSL in relation to S availability.
While a number of studies have demonstrated that seed yield and GSL content of Brassica crops can increase with S fertilization, any response will ultimately depend on genotype, environment, and existing soil S status. A better understanding of cultivar-specific S requirements to achieve a given seed GSL yield is needed, with appropriate S fertilizer requirements developed according to soil S status. This will be informed by a detailed knowledge of the distribution and remobilization of S and GSLs throughout the development of Brassica crops, which will also contribute to completing the framework of our conceptual model (Figure 6).
From the evidence we have evaluated, it is clear that the underlying regulatory mechanisms affecting the interaction of S sources and GSL sinks remain to be resolved. As indicated in the model (Figure 6), only a few recent studies have identified sulfate transporters, and additionally, only one TF (SLIM1) and one microRNA (miR395) of Arabidopsis that are expressed under S deficient conditions. As yet few studies have evaluated the mechanism(s) behind the differential response of plant GSLs in the context of S availability. We are aware that a more complex or sophisticated regulatory framework may exist in the brassicas that integrates the rate of S assimilation, availability of precursors for GSL biosynthesis and seed GSL accumulation in response to different S status.
Author Contributions
PB carried out the literature review and prepared the manuscript, TR and GK each read, edited, and refined the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
PB received scholarship funding from Australian Mustard Oil Pty. Ltd, Australia, DBT-AAU Center of Assam Agricultural University, India and Southern Cross Plant Science, Southern Cross University, Australia.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01735/full#supplementary-material
Abbreviations
APK, adenosine 5′-phosphosulfate kinase; APS, adenosine 5′-phosphosulfate; ATPS, ATP sulfurylase; Cys, cysteine; GSH, glutathione; GSL, glucosinolate; GST, glutathione S-transferease; GTR, glucosinolate transporter; MAM, methylthioalkylmalate; Met, methionine; N, nitrogen; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; qRT-PCR, quantitative real time-polymerase chain reaction; QTL, quantitative trait locus; S, sulfur; SLIM1, Sulfur Limitation 1; SOTs or STs, sulfotransferases; SULTR, sulfate transporter; TF, transcription factor.
References
Agerbirk, N., and Olsen, C. E. (2012). Glucosinolate structures in evolution. Phytochemistry 77, 16–45. doi: 10.1016/j.phytochem.2012.02.005
Ahmad, A., and Abdin, M. (2000). Interactive effect of sulphur and nitrogen on the oil and protein contents and on the fatty acid profiles of oil in the seeds of rapeseed (Brassica campestris L.) and mustard (Brassica juncea L. Czern. and Coss.). J. Agron. Crop Sci. 185, 49–54. doi: 10.1046/j.1439-037X.2000.00401.x
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Andersen, T. G., and Halkier, B. A. (2014). Upon bolting the GTR1 and GTR2 transporters mediate transport of glucosinolates to the inflorescence rather than roots. Plant Signal. Behav. 9:e27740. doi: 10.4161/psb.27740
Andersen, T. G., Nour-Eldin, H. H., Fuller, V. L., Olsen, C. E., Burow, M., and Halkier, B. A. (2013). Integration of biosynthesis and long-distance transport establish organ-specific glucosinolate profiles in vegetative Arabidopsis. Plant Cell Online 25, 3133–3145. doi: 10.1105/tpc.113.110890
Augustine, R., Majee, M., Gershenzon, J., and Bisht, N. C. (2013a). Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea. J. Exp. Bot. 64, 4907–4921. doi: 10.1093/jxb/ert280
Augustine, R., Mukhopadhyay, A., and Bisht, N. C. (2013b). Targeted silencing of BjMYB28 transcription factor gene directs development of low glucosinolate lines in oilseed Brassica juncea. Plant Biotechnol. J. 11, 855–866. doi: 10.1111/pbi.12078
Bak, S., and Feyereisen, R. (2001). The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 127, 108–118.
Baskar, V., Gururani, M., Yu, J., and Park, S. (2012). Engineering glucosinolates in plants: current knowledge and potential uses. Appl. Biochem. Biotechnol. 168, 1694–1717. doi: 10.1007/s12010-012-9890-6
Bednarek, P., Piślewska-Bednarek, M., Svatoš, A., Schneider, B., Doubský, J., Mansurova, M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106. doi: 10.1126/science.1163732
Bellostas, N., Sørensen, J. C., and Sørensen, H. (2007a). Profiling glucosinolates in vegetative and reproductive tissues of four Brassica species of the U-triangle for their biofumigation potential. J. Sci. Food Agric. 87, 1586–1594. doi: 10.1002/jsfa.2896
Bellostas, N., Sørensen, J., and Sørensen, H. (2007b). Biofumigation: from the “classical” approach to the use of bio-refined glucosinolates as natural plant protection agents. GCIRC Bull. 24.
Bhandari, S. R., Jung, S. J., and Jun, G. L. (2015). Comparison of glucosinolate profiles in different tissues of nine brassica crops. Molecules 20, 15827–15841. doi: 10.3390/molecules200915827
Bisht, N. C., Gupta, V., Ramchiary, N., Sodhi, Y. S., Mukhopadhyay, A., Arumugam, N., et al. (2009). Fine mapping of loci involved with glucosinolate biosynthesis in oilseed mustard (Brassica juncea) using genomic information from allied species. Theor. Appl. Genet. 118, 413–421. doi: 10.1007/s00122-008-0907-z
Blake-Kalff, M. M. A., Harrison, K. R., Hawkesford, M. J., Zhao, F. J., and McGrath, S. P. (1998). Distribution of sulfur within Oilseed rape leaves in response to sulfur deficiency during vegetative growth. Plant Physiol. 118, 1337–1344. doi: 10.1104/pp.118.4.1337
Bloem, E., Haneklaus, S., and Schnug, E. (2007). Changes in the sulphur and glucosinolate content of developing pods and seeds of oilseed rape (Brassica napus L.) in relation to different cultivars. Landbauforschung Volkenrode 57, 297–306.
Bones, A. M., and Rossiter, J. T. (1996). The myrosinase-glucosinolate system, its organisation and biochemistry. Physiol. Plant. 97, 194–208. doi: 10.1034/j.1399-3054.1996.970128.x
Bones, A. M., and Rossiter, J. T. (2006). The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry 67, 1053–1067. doi: 10.1016/j.phytochem.2006.02.024
Bonnet, E., Wuyts, J., Rouzé, P., and Van de Peer, Y. (2004). Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20, 2911–2917. doi: 10.1093/bioinformatics/bth374
Brooks, J. D., Paton, V. G., and Vidanes, G. (2001). Potent induction of phase 2 enzymes in human prostate cells by sulforaphane. Cancer Epidemiol. Biomark. Preven. 10, 949–954.
Brown, J., and Morra, M. (2005). Glucosinolate-Containing Seed Meal as a Soil Amendment to Control Plant Pests. Moscow, ID: University of Idaho.
Brown, P. D., and Morra, M. J. (1995). Glucosinolate-containing plant tissues as bioherbicides. J. Agric. Food Chem. 43, 3070–3074. doi: 10.1021/jf00060a015
Brown, P. D., Tokuhisa, J. G., Reichelt, M., and Gershenzon, J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62, 471–481. doi: 10.1016/S0031-9422(02)00549-6
Buchner, P., Stuiver, C. E., Westerman, S., Wirtz, M., Hell, R., Hawkesford, M. J., et al. (2004a). Regulation of sulfate uptake and expression of sulfate transporter genes in Brassica oleracea as affected by atmospheric H2S and pedospheric sulfate nutrition. Plant Physiol. 136, 3396–3408. doi: 10.1104/pp.104.046441
Buchner, P., Takahashi, H., and Hawkesford, M. J. (2004b). Plant sulphate transporters: co-ordination of uptake, intracellular and long-distance transport. J. Exp. Bot. 55, 1765–1773. doi: 10.1093/jxb/erh206
Buhtz, A., Pieritz, J., Springer, F., and Kehr, J. (2010). Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol. 10:64. doi: 10.1186/1471-2229-10-64
Buskov, S., Serra, B., Rosa, E., Sørensen, H., and Sørensen, J. (2002). Effects of intact glucosinolates and products produced from glucosinolates in myrosinase-catalyzed hydrolysis on the potato cyst nematode (Globodera rostochiensis cv. Woll). J. Agric. Food Chem. 50, 690–695. doi: 10.1021/jf010470s
Calderwood, A., Kopriva, S., and Morris, R. J. (2014). Predictive sulfur metabolism - a field in flux. Front. Plant Sci. 5:646. doi: 10.3389/fpls.2014.00646
Canola Council (2016). What is Canola? Available at: http://www.canolacouncil.org/oil-and-meal/what-is-canola/ [accessed 16 June, 2016].
Cao, M. J., Wang, Z., Wirtz, M., Hell, R., Oliver, D. J., and Xiang, C. B. (2013). SULTR3; 1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J. 73, 607–616. doi: 10.1111/tpj.12059
Cartea, M. E., Velasco, P., Obregon, S., Padilla, G., and de Haro, A. (2008). Seasonal variation in glucosinolate content in Brassica oleracea crops grown in northwestern Spain. Phytochemistry 69, 403–410. doi: 10.1016/j.phytochem.2007.08.014
Castro, A., Aires, A., Rosa, E., Bloem, E., Stulen, I., and De Kok, L. J. (2004). Distribution of Glucosinolates in Brassica oleracea cultivars. Phyton-Annales Rei Bot. 44, 133–143.
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A. P., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, S., Glawischnig, E., Jorgensen, K., Naur, P., Jorgensen, B., Olsen, C. E., et al. (2003). CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J. Cell Mol. Biol. 33, 923–937. doi: 10.1046/j.1365-313X.2003.01679.x
Chen, S., Petersen, B. L., Olsen, C. E., Schulz, A., and Halkier, B. A. (2001). Long-distance phloem transport of glucosinolates in Arabidopsis. Plant Physiol. 127, 194–201. doi: 10.1104/pp.127.1.194
Chen, X.-J., Zhu, Z.-J., Ni, X.-L., and Qian, Q.-Q. (2006). Effect of nitrogen and sulfur supply on glucosinolates in Brassica campestris ssp. chinensis. Agric. Sci. China 5, 603–608. doi: 10.1016/S1671-2927(06)60099-0
Choi, H.-S., Cho, M.-C., Lee, H. G., and Yoon, D.-Y. (2010). Indole-3-carbinol induces apoptosis through p53 and activation of caspase-8 pathway in lung cancer A549 cells. Food Chem. Toxicol. 48, 883–890. doi: 10.1016/j.fct.2009.12.028
Clarke, D. B. (2010). Glucosinolates, structures and analysis in food. Anal. Methods 2, 310–325. doi: 10.1039/b9ay00280d
Cleemput, S., and Becker, H. (2012). Genetic variation in leaf and stem glucosinolates in resynthesized lines of winter rapeseed (Brassica napus L.). Genet. Resour. Crop Evol. 59, 539–546. doi: 10.1007/s10722-011-9701-x
Clossais-Besnard, N., and Larher, F. (1991). Physiological role of glucosinolates in Brassica napus. Concentration and distribution pattern of glucosinolates among plant organs during a complete life cycle. J. Sci. Food Agric. 56, 25–38. doi: 10.1002/jsfa.2740560104
Dixon, D. P., Skipsey, M., and Edwards, R. (2010). Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 71, 338–350. doi: 10.1016/j.phytochem.2009.12.012
Du, L., and Halkier, B. A. (1998). Biosynthesis of glucosinolates in the developing silique walls and seeds of Sinapis alba. Phytochemistry 48, 1145–1150. doi: 10.1016/S0031-9422(97)00877-7
Eichel, J., González, J. C., Hotze, M., Matthews, R. G., and Schröder, J. (1995). Vitamin-B12-Independent methionine synthase from a higher plant (Catharanthus roseus). Eur. J. Biochem. 230, 1053–1058. doi: 10.1111/j.1432-1033.1995.1053g.x
Ellerbrock, B. L. J., Kim, J. H., and Jander, G. (2007). Contribution of glucosinolate transport to Arabidopsis defense responses. Plant Signal. Behav. 2, 282–283. doi: 10.4161/psb.2.4.4014
Estavillo, G. M., Crisp, P. A., Pornsiriwong, W., Wirtz, M., Collinge, D., Carrie, C., et al. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012. doi: 10.1105/tpc.111.091033
Ettlinger, M. G., and Lundeen, A. J. (1956). The structures of sinigrin and sinalbin: an enzymatic rearrangement. J. Ann. Chem. Soc. 78, 4172–4173. doi: 10.1021/ja01597a090
Fahey, J. W., Zalcmann, A. T., and Talalay, P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56, 5–51. doi: 10.1016/S0031-9422(00)00316-2
Falk, K. L., Tokuhisa, J. G., and Gershenzon, J. (2007). The effect of sulfur nutrition on plant glucosinolate content: physiology and molecular mechanisms. Plant Biol. (Stuttg.) 9, 573–581. doi: 10.1055/s-2007-965431
Feng, J., Long, Y., Shi, L., Shi, J., Barker, G., and Meng, J. (2012). Characterization of metabolite quantitative trait loci and metabolic networks that control glucosinolate concentration in the seeds and leaves of Brassica napus. New Phytol. 193, 96–108. doi: 10.1111/j.1469-8137.2011.03890.x
Fenwick, R., Heaney, R. K., and Mullin, W. J. (1983). Glucosinolates and their breakdown products in food and food plants. Food Sci. Nutr. 18, 123–201.
Fernandez-Calvo, P., Chini, A., Fernandez-Barbero, G., Chico, J.-M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715.
Fieldsend, J., and Milford, G. (1994). Changes in glucosinolates during crop development in single-and double-low genotypes of winter oilseed rape (Brassica napus): I. Production and distribution in vegetative tissues and developing pods during development and potential role in the recycling of sulphur within the crop. Annal. Appl. Biol. 124, 531–542.
Frerigmann, H. (2016). Glucosinolate regulation in a complex relationship - MYC and MYB - no one can act without each other. Adv. Botan. Res. 80, 57–97. doi: 10.1016/bs.abr.2016.06.005
Frerigmann, H., Berger, B., and Gigolashvili, T. (2014). bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 166, 349–369. doi: 10.1104/pp.114.240887
Frerigmann, H., Böttcher, C., Baatout, D., and Gigolashvili, T. (2012). Glucosinolates are produced in trichomes of Arabidopsis thaliana. Front. Plant Sci. 3:242. doi: 10.3389/fpls.2012.00242
Frerigmann, H., and Gigolashvili, T. (2014a). MYB34, MYB 51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 7, 814–828. doi: 10.1093/mp/ssu004
Frerigmann, H., and Gigolashvili, T. (2014b). Update on the role of R2R3-MYBs in the regulation of glucosinolates upon sulfur deficiency. Front. Plant Sci. 5:626. doi: 10.3389/fpls.2014.00626
Fuchs, R., Kopischke, M., Klapprodt, C., Hause, G., Meyer, A. J., Schwarzlander, M., et al. (2016). Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in Arabidopsis. Plant Cell 28, 130–145.
Gachon, C. M., Langlois-Meurinne, M., Henry, Y., and Saindrenan, P. (2005). Transcriptional co-regulation of secondary metabolism enzymes in Arabidopsis: functional and evolutionary implications. Plant Mol. Biol. 58, 229–245. doi: 10.1007/s11103-005-5346-5
Gao, M., Li, G., Potter, D., McCombie, W. R., and Quiros, C. F. (2006). Comparative analysis of methylthioalkylmalate synthase (MAM) gene family and flanking DNA sequences in Brassica oleracea and Arabidopsis thaliana. Plant Cell Rep. 25, 592–598. doi: 10.1007/s00299-005-0078-1
Gao, M., Li, G., Yang, B., Mccombie, W. R., and Quiros, C. F. (2004). Comparative analysis of a Brassica BAC clone containing several major aliphatic glucosinolate genes with its corresponding Arabidopsis sequence. Genome 47, 666–679. doi: 10.1139/g04-021
Geu-Flores, F., Moldrup, M. E., Bottcher, C., Olsen, C. E., Scheel, D., and Halkier, B. A. (2011). Cytosolic gamma-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant Cell 23, 2456–2469. doi: 10.1105/tpc.111.083998
Gigolashvili, T., Engqvist, M., Yatusevich, R., Müller, C., and Flügge, U. I. (2008). HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 177, 627–642. doi: 10.1111/j.1469-8137.2007.02295.x
Gigolashvili, T., Geier, M., Ashykhmina, N., Frerigmann, H., Wulfert, S., Krueger, S., et al. (2012). The Arabidopsis thylakoid ADP/ATP carrier TAAC has an additional role in supplying plastidic phosphoadenosine 5′-phosphosulfate to the cytosol. Plant Cell 24, 4187–4204. doi: 10.1105/tpc.112.101964
Gigolashvili, T., and Kopriva, S. (2014). Transporters in plant sulfur metabolism. Front. Plant Sci. 5:442. doi: 10.3389/fpls.2014.00442
Gigolashvili, T., Yatusevich, R., Berger, B., Müller, C., and Flügge, U. I. (2007). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51, 247–261. doi: 10.1111/j.1365-313X.2007.03133.x
Gigolashvili, T., Yatusevich, R., Rollwitz, I., Humphry, M., Gershenzon, J., and Flugge, U. (2009a). The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell 21, 1813–1829. doi: 10.1105/tpc.109.066399
Gigolashvili, T., Berger, B., and Flügge, U.-I. (2009b). Specific and coordinated control of indolic and aliphatic glucosinolate biosynthesis by R2R3-MYB transcription factors in Arabidopsis thaliana. Phytochem. Rev. 8, 3–13. doi: 10.1007/s11101-008-9112-6
Gorsuch, J. W., Lower, W. R., Lewis, M. A., and Wang, W. (1991). “Plants for toxicity assessment: second volume,” in The Second Symposium on Use of Plants for Toxicity Assessment, eds J. W. Gorsuch, W. R. Lower, M. A. Lewis, and W. Wang (West Conshohocken, PA: ASTM).
Griffiths, D., Birch, A., and Hillman, J. (1998). Antinutritional compounds in the Brassicaceae: analysis, biosynthesis, chemistry and dietary effects. J. Hort. Sci. 73, 1–18.
Grubb, C. D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11, 89–100. doi: 10.1016/j.tplants.2005.12.006
Grubb, C. D., Zipp, B. J., Kopycki, J., Schubert, M., Quint, M., Lim, E.-K., et al. (2014). Comparative analysis of Arabidopsis UGT74 glucosyltransferases reveals a special role of UGT74C1 in glucosinolate biosynthesis. Plant J. 79, 92–105. doi: 10.1111/tpj.12541
Grubb, C. D., Zipp, B. J., Ludwig-Muller, J., Masuno, M. N., Molinski, T. F., and Abel, S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40, 893–908. doi: 10.1111/j.1365-313X.2004.02261.x
Hacquard, S., Kracher, B., Hiruma, K., Munch, P. C., Garrido-Oter, R., Thon, M. R., et al. (2016). Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat. Commun. 7:11362. doi: 10.1038/ncomms11362
Halkier, B. A., and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333. doi: 10.1146/annurev.arplant.57.032905.105228
Hansen, B. G., Kerwin, R. E., Ober, J. A., Lambrix, V. M., Mitchell-Olds, T., Gershenzon, J., et al. (2008). A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol. 148, 2096–2108. doi: 10.1104/pp.108.129981
Hansen, B. G., Kliebenstein, D. J., and Halkier, B. A. (2007). Identification of a flavin-monooxygenase as the S-oxygenating enzyme in aliphatic glucosinolate biosynthesis in Arabidopsis. Plant J. 50, 902–910. doi: 10.1111/j.1365-313X.2007.03101.x
Hansen, C. H., Wittstock, U., Olsen, C. E., Hick, A. J., Pickett, J. A., and Halkier, B. A. (2001). Cytochrome p450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomo-methionine and trihomo-methionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J. Biol. Chem. 276, 11078–11085. doi: 10.1074/jbc.M010123200
Hawkesford, M. J. (2000). Plant responses to sulphur deficiency and the genetic manipulation of sulphate transporters to improve S-utilization efficiency. J. Exp. Bot. 51, 131–138. doi: 10.1093/jexbot/51.342.131
Hawkesford, M. J., and De Kok, L. J. (2006). Managing sulphur metabolism in plants. Plant Cell Environ. 29, 382–395. doi: 10.1111/j.1365-3040.2005.01470.x
Hayes, J. D., Kelleher, M. O., and Eggleston, I. M. (2008). The cancer chemopreventive actions of phytochemicals derived from glucosinolates. Eur. J. Nutr. 47, 73–88. doi: 10.1007/s00394-008-2009-8
Hecht, S. S. (2000). Inhibition of carcinogenesis by isothiocyanates. Drug Metab. Rev. 32, 395–411. doi: 10.1081/dmr-100102342
Hemm, M. R., Ruegger, M. O., and Chapple, C. (2003). The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15, 179–194. doi: 10.1105/tpc.006544
Herschbach, C., and Rennenberg, H. (1995). Long-distance transport of 35S-sulphur in 3-year-old beech trees (Fagus sylvatica). Physiol. Plant. 95, 379–386. doi: 10.1034/j.1399-3054.1995.950307.x
Hirai, M. Y., Fujiwara, T., Awazuhara, M., Kimura, T., Noji, M., and Saito, K. (2003). Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 33, 651–663. doi: 10.1046/j.1365-313X.2003.01658.x
Hirai, M. Y., Klein, M., Fujikawa, Y., Yano, M., Goodenowe, D. B., and Yamazaki, Y. (2005). Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J. Biol. Chem. 280, 25590–25595. doi: 10.1074/jbc.M502332200
Hirai, M. Y., Sugiyama, K., Sawada, Y., Tohge, T., Obayashi, T., and Suzuki, A. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 6478–6483. doi: 10.1073/pnas.0611629104
Hirai, M. Y., Yano, M., Goodenowe, D. B., Kanaya, S., Kimura, T., Awazuhara, M., et al. (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 10205–10210. doi: 10.1073/pnas.0403218101
Hiruma, K., Sacristán, G. N., Nakano, S., Hacquard, R. T., Kracher, S., Neumann, B., et al. (2016). Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 165, 464–474. doi: 10.1016/j.cell.2016.02.028
Howell, P. M., Sharpe, A. G., and Lydiate, D. J. (2003). Homoeologous loci control the accumulation of seed glucosinolates in oilseed rape (Brassica napus). Genome 46, 454–460. doi: 10.1139/g03-028
Hull, A. K., Vij, R., and Celenza, J. L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 2379–2384. doi: 10.1073/pnas.040569997
Ishida, M., Hara, M., Fukino, N., Kakizaki, T., and Morimitsu, Y. (2014). Glucosinolate metabolism, functionality and breeding for the improvement of Brassicaceae vegetables. Breed. Sci. 64, 48–59. doi: 10.1270/jsbbs.64.48
Jagadeeswaran, G., Li, Y. F., and Sunkar, R. (2014). Redox signaling mediates the expression of a sulfate-deprivation-inducible microRNA395 in Arabidopsis. Plant J. 77, 85–96. doi: 10.1111/tpj.12364
Jones-Rhoades, M. W., and Bartel, D. P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 14, 787–799. doi: 10.1016/j.molcel.2004.05.027
Jørgensen, M. E., Nour-Eldin, H. H., and Halkier, B. A. (2015). Transport of defense compounds from source to sink: lessons learned from glucosinolates. Trends Plant Sci. 20, 508–514. doi: 10.1016/j.tplants.2015.04.006
Kaiser, G., Martinoia, E., Schröppel-Meier, G., and Heber, U. (1989). Active transport of sulfate into the vacuole of plant cells provides halotolerance and can detoxify SO2. J. Plant Physiol. 133, 756–763. doi: 10.1016/S0176-1617(89)80085-9
Kataoka, T., Watanabe-Takahashi, A., Hayashi, N., Ohnishi, M., Mimura, T., Buchner, P., et al. (2004). Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16, 2693–2704. doi: 10.1105/tpc.104.023960
Kawashima, C. G., Matthewman, C. A., Huang, S., Lee, B. R., Yoshimoto, N., Koprivova, A., et al. (2011). Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J. 66, 863–876. doi: 10.1111/j.1365-313X.2011.04547.x
Kawashima, C. G., Yoshimoto, N., Maruyama-Nakashita, A., Tsuchiya, Y. N., Saito, K., Takahashi, H., et al. (2009). Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 57, 313–321. doi: 10.1111/j.1365-313X.2008.03690.x
Kissen, R., and Bones, A. M. (2009). Nitrile-specifier proteins involved in glucosinolate hydrolysis in Arabidopsis thaliana. J. Biol. Chem. 284, 12057–12070. doi: 10.1074/jbc.M807500200
Kissen, R., Rossiter, J. T., and Bones, A. M. (2009). The ‘mustard oil bomb’: not so easy to assemble? Localization, expression and distribution of the components of the myrosinase enzyme system. Phytochem. Rev. 8, 69–86. doi: 10.1007/s11101-008-9109-1
Kjaer, A. (1976). “Glucosinolates in the Cruciferae,” in The Biology and Chemistry of the Cruciferae, eds J. G. Vaughan, A. J. MacLeod, and B. M. G. Jones (London: Academic Press), 207–219.
Klessa, D. A., and Sinclair, A. H. (1989). Sulphur in Soils, Fertilisers and Crops. AC Technical Note T160. Perth: Scottish Agricultural collegs.
Kliebenstein, D. J., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J., et al. (2001). Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol. 126, 811–825. doi: 10.1104/pp.126.2.811
Knill, T., Schuster, J., Reichelt, M., Gershenzon, J., and Binder, S. (2008). Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. Plant Physiol. 146, 1028–1039. doi: 10.1104/pp.107.111609
Kondra, Z., and Stefansson, B. (1970). Inheritance of the major glucosinolates of rapeseed (Brassica napus) meal. Can. J. Plant Sci. 50, 643–647. doi: 10.4141/cjps70-122
Koprivova, A., and Kopriva, S. (2014). Molecular mechanisms of regulation of sulfate assimilation: first steps on a long road. Front. Plant Sci. 5:589. doi: 10.3389/fpls.2014.00589
Koprivova, A., and Kopriva, S. (2016). Sulfation pathways in plants. Chemico-Biol. Interact. doi: 10.1016/j.cbi.2016.05.021 [Epub ahead of print].
Koralewska, A., Stuiver, C. E. E., Posthumus, F. S., Kopriva, S., Hawkesford, M. J., and De Kok, L. J. (2008). Regulation of sulfate uptake, expression of the sulfate transporters SULTR1; 1 and SULTR1; 2, and APS reductase in Chinese cabbage (Brassica pekinensis) as affected by atmospheric H2S nutrition and sulfate deprivation. Funct. Plant Biol. 35, 318–327. doi: 10.1071/FP07283
Koroleva, O. A., and Cramer, R. (2011). Single-cell proteomic analysis of glucosinolate-rich S-cells in Arabidopsis thaliana. Methods 54, 413–423. doi: 10.1016/j.ymeth.2011.06.005
Kroymann, J. (2011). Natural diversity and adaptation in plant secondary metabolism. Curr. Opin. Plant Biol. 14, 246–251. doi: 10.1016/j.pbi.2011.03.021
Kroymann, J., Textor, S., Tokuhisa, J. G., Falk, K. L., Bartram, S., Gershenzon, J., et al. (2001). A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 127, 1077–1088. doi: 10.1104/pp.010416
Kutz, A., Müller, A., Hennig, P., Kaiser, W. M., Piotrowski, M., and Weiler, E. W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 30, 95–106. doi: 10.1046/j.1365-313X.2002.01271.x
Lancilli, C., Giacomini, B., Lucchini, G., Davidian, J.-C., Cocucci, M., Sacchi, G. A., et al. (2014). Cadmium exposure and sulfate limitation reveal differences in the transcriptional control of three sulfate transporter (SULTR1;2) genes in Brassica juncea. BMC Plant Biol. 14:132. doi: 10.1186/1471-2229-14-132
Lazzeri, L., Tacconi, R., and Palmieri, S. (1993). In vitro activity of some glucosinolates and their reaction products toward a population of the nematode Heterodera schachtii. J. Agric. Food Chem. 41, 825–829. doi: 10.1021/jf00029a028
Lewandowska, M., and Sirko, A. (2008). Recent advances in understanding plant response to sulfur-deficiency stress. Acta Biochim. Pol. 55, 457–471.
Li, G., and Quiros, C. F. (2002). Genetic analysis, expression and molecular characterization of BoGSL-ELONG, a major gene involved in the aliphatic glucosinolate pathway of Brassica species. Genetics 162, 1937–1943.
Li, G., and Quiros, C. F. (2003). In planta side-chain glucosinolate modification in Arabidopsis by introduction of dioxygenase Brassica homolog BoGSL-ALK. Theor. Appl. Genet. 106, 1116–1121. doi: 10.1007/s00122-002-1161-4
Li, G., Riaz, A., Goyal, S., Abel, S., and Quiros, C. F. (2001). Inheritance of three major genes involved in the synthesis of aliphatic glucosinolates in Brassica oleracea. J. Am. Soc. Hort. Sci. 126, 427–431.
Li, J., Hansen, B. G., Ober, J. A., Kliebenstein, D. J., and Halkier, B. A. (2008). Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol. 148, 1731–1733. doi: 10.1104/pp.108.125757
Li, Y., Sawada, Y., Hirai, A., Sato, M., Kuwahara, A., Yan, X., et al. (2013). Novel insights into the function of Arabidopsis R2R3-MYB transcription factors regulating aliphatic glucosinolate biosynthesis. Plant Cell Physiol. 54, 1335–1344. doi: 10.1093/pcp/pct085
Liang, G., Yang, F., and Yu, D. (2010). MicroRNA395 mediates regulation of sulfate accumulation and allocation in Arabidopsis thaliana. Plant J. 62, 1046–1057.
Liang, G., and Yu, D. (2010). Reciprocal regulation among miR395, APS and SULTR2;1 in Arabidopsis thaliana. Plant Signal. Behav. 5, 1257–1259. doi: 10.4161/psb.5.10.12608
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5:3930. doi: 10.1038/ncomms4930
Liu, Z., Hammerlindl, J., Keller, W., McVetty, P. E., Daayf, F., Quiros, C., et al. (2011). MAM gene silencing leads to the induction of C3 and reduction of C4 and C5 side-chain aliphatic glucosinolates in Brassica napus. Mol. Breed. 27, 467–478. doi: 10.1007/s11032-010-9444-y
Lou, P., Zhao, J., He, H., Hanhart, C., Del Carpio, D. P., Verkerk, R., et al. (2008). Quantitative trait loci for glucosinolate accumulation in Brassica rapa leaves. New Phytol. 179, 1017–1032. doi: 10.1111/j.1469-8137.2008.02530.x
Lunn, J. E., Droux, M., Martin, J., and Douce, R. (1990). Localization of ATP sulfurylase and O-acetylserine (thiol) lyase in spinach leaves. Plant Physiol. 94, 1345–1352. doi: 10.1104/pp.94.3.1345
Lüthy, B., and Matile, P. (1984). The mustard oil bomb: rectified analysis of the subcellular organisation of the myrosinase system. Biochem. Physiol. Pflanzen 179, 5–12. doi: 10.1016/S0015-3796(84)80059-1
Madsen, S. R., Kunert, G., Reichelt, M., Gershenzon, J., and Halkier, B. A. (2015). Feeding on leaves of the glucosinolate transporter mutant gtr1gtr2 reduces fitness of Myzus persicae. J. Chem. Ecol. 41, 975–984. doi: 10.1007/s10886-015-0641-3
Magrath, R., and Mithen, R. (1993). Maternal effects on the expression of individual aliphatic glucosinolates in seeds and seedlings of Brassica napus. Plant Breed. 111, 249–252. doi: 10.1111/j.1439-0523.1993.tb00637.x
Mari, M., Leoni, O., Iori, R., and Cembali, T. (2002). Antifungal vapour-phase activity of allyl-isothiocyanate against Penicillium expansum on pears. Plant Pathol. 51, 231–236. doi: 10.1046/j.1365-3059.2002.00667.x
Maruyama-Nakashita, A., Inoue, E., Watanabe-Takahashi, A., Yamaya, T., and Takahashi, H. (2003). Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 132, 597–605. doi: 10.1104/pp.102.019802
Maruyama-Nakashita, A., Nakamura, Y., Tohge, T., Saito, K., and Takahashi, H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18, 3235–3251. doi: 10.1105/tpc.106.046458
Matthewman, C. A., Kawashima, C. G., Húska, D., Csorba, T., Dalmay, T., and Kopriva, S. (2012). miR395 is a general component of the sulfate assimilation regulatory network in Arabidopsis. FEBS Lett. 586, 3242–3248. doi: 10.1016/j.febslet.2012.06.044
Mickelbart, M. V., Hasegawa, P. M., and Bailey-Serres, J. (2015). Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16, 237–251. doi: 10.1038/nrg3901
Mikkelsen, M. D., Hansen, C. H., Wittstock, U., and Halkier, B. A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275, 33712–33717. doi: 10.1074/jbc.M001667200
Mikkelsen, M. D., Naur, P., and Halkier, B. A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37, 770–777. doi: 10.1111/j.1365-313X.2004.02002.x
Miller, A. J., Cookson, S. J., Smith, S. J., and Wells, D. M. (2001). The use of microelectrodes to investigate compartmentation and the transport of metabolized inorganic ions in plants. J. Exp. Bot. 52, 541–549. doi: 10.1093/jexbot/52.356.541
Mithen, R. (2001). Glucosinolates – biochemistry, genetics and biological activity. Plant Growth Regul. 34, 91–103. doi: 10.1023/A:1013330819778
Mithen, R., Clarke, J., Lister, C., and Dean, C. (1995). Genetics of aliphatic glucosinolates. III. Side chain structure of aliphatic glucosinolates in. Heredity 74, 210–215. doi: 10.1038/hdy.1995.29
Moussaieff, A., Rogachev, I., Brodsky, L., Malitsky, S., Toal, T. W., Belcher, H., et al. (2013). High-resolution metabolic mapping of cell types in plant roots. Proc. Natl. Acad. Sci. U.S.A. 110, E1232–E1241. doi: 10.1073/pnas.1302019110
Mugford, S. G., Lee, B. R., Koprivova, A., Matthewman, C., and Kopriva, S. (2011). Control of sulfur partitioning between primary and secondary metabolism. Plant J. 65, 96–105. doi: 10.1111/j.1365-313X.2010.04410.x
Mugford, S. G., Matthewman, C. A., Hill, L., and Kopriva, S. (2010). Adenosine-5’-phosphosulfate kinase is essential for Arabidopsis viability. FEBS Lett. 584, 119–123. doi: 10.1016/j.febslet.2009.11.014
Mugford, S. G., Yoshimoto, N., Reichelt, M., Wirtz, M., Hill, L., Mugford, S. T., et al. (2009). Disruption of adenosine-5’-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21, 910–927. doi: 10.1105/tpc.109.065581
Nakajima, M., Yoshida, R., Shimada, N., Yamazaki, H., and Yokoi, T. (2001). Inhibition and inactivation of human cytochrome P450 isoforms by phenethyl isothiocyanate. Drug Metab. Dispos. 29, 1110–1113.
Nakano, R. T., Yamada, K., Bednarek, P., Nishimura, M., and Hara-Nishimura, I. (2014). ER bodies in plants of the Brassicales order: biogenesis and association with innate immunity. Front. Plant Sci. 5:73. doi: 10.3389/fpls.2014.00073
Naur, P., Petersen, B. L., Mikkelsen, M. D., Bak, S., Rasmussen, H., Olsen, C. E., et al. (2003). CYP83A1 and CYP83B1, two non-redundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol. 133, 63–72. doi: 10.1104/pp.102.019240
Nikiforova, V., Freitag, J., Kempa, S., Adamik, M., Hesse, H., and Hoefgen, R. (2003). Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J. 33, 633–650. doi: 10.1046/j.1365-313X.2003.01657.x
Nikiforova, V. J., Kopka, J., Tolstikov, V., Fiehn, O., Hopkins, L., Hawkesford, M. J., et al. (2005). Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol. 138, 304–318. doi: 10.1104/pp.104.053793
Nour-Eldin, H. H., Andersen, T. G., Burow, M., Madsen, S. R., Jorgensen, M. E., Olsen, C. E., et al. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488, 531–534. doi: 10.1038/nature11285
Othmane, M. (2015). Genetic variability in glucosinolates in seed of Brassica juncea: interest in mustard condiment. J. Chem. 2015:6. doi: 10.1155/2015/606142
Pasternak, M., Lim, B., Wirtz, M., Hell, R., Cobbett, C. S., and Meyer, A. J. (2008). Restricting glutathione biosynthesis to the cytosol is sufficient for normal plant development. Plant J. 53, 999–1012. doi: 10.1111/j.1365-313X.2007.03389.x
Paterson, A. H., Lan, T.-H., Amasino, R., Osborn, T. C., and Quiros, C. (2001). Brassica genomics: a complement to, and early beneficiary of, the Arabidopsis sequence. Genome Biol, 2:REVIEWS1011. doi: 10.1186/gb-2001-2-3-reviews1011
Pfalz, M., Mikkelsen, M. D., Bednarek, P., Olsen, C. E., Halkier, B. A., and Kroymann, J. (2011). Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell 23, 716–729. doi: 10.1105/tpc.110.081711
Pfalz, M., Vogel, H., and Kroymann, J. (2009). The gene controlling the indole glucosinolate modifier quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21, 985–999. doi: 10.1105/tpc.108.063115
Piotrowski, M., Schemenewitz, A., Lopukhina, A., Muller, A., Janowitz, T., Weiler, E. W., et al. (2004). Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J. Biol. Chem. 279, 50717–50725. doi: 10.1074/jbc.M407681200
Porter, A. J. R., Morton, A. M., Kiddle, G., Doughty, K. L., and Wallsgrove, R. M. (1991). Variation in the glucosinolate content of oilseed rape (Brassica napus L.) leaves. Annal. Appl. Biol. 118, 461–467. doi: 10.1111/j.1744-7348.1991.tb05647.x
Qu, C. M., Li, S. M., Duan, X. J., Fan, J. H., Jia, L. D., Zhao, H. Y., et al. (2015). Identification of candidate genes for seed glucosinolate content using association mapping in Brassica napus L. Genes 6, 1215–1229. doi: 10.3390/genes6041215
Quijada, P. A., Udall, J. A., Lambert, B., and Osborn, T. C. (2006). Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor. Appl. Genet. 113, 549–561. doi: 10.1007/s00122-006-0323-1
Ramchiary, N., Bisht, N. C., Gupta, V., Mukhopadhyay, A., Arumugam, N., Sodhi, Y. S., et al. (2007). QTL analysis reveals context-dependent loci for seed glucosinolate trait in the oilseed Brassica juncea: importance of recurrent selection backcross scheme for the identification of ‘true’. QTL. Theor. Appl. Genet. 116, 77–85. doi: 10.1007/s00122-007-0648-4
Rangkadilok, N., Nicolas, M. E., Bennett, R. N., Premier, R. R., Eagling, D. R., and Taylor, P. W. (2002). Determination of sinigrin and glucoraphanin in Brassica species using a simple extraction method combined with ion-pair HPLC analysis. Sci. Horticul. 96, 27–41. doi: 10.1016/S0304-4238(02)00119-X
Rask, L., Andréasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B., and Meijer, J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42, 93–113. doi: 10.1023/A:1006380021658
Ravanel, S., Block, M. A., Rippert, P., Jabrin, S., Curien, G., Rébeillé, F., et al. (2004). Methionine metabolism in plants chloroplast are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J. Biol. Chem. 279, 22548–22557. doi: 10.1074/jbc.M313250200
Redovnikovic, I. R., Textor, S., Lisnic, B., and Gershenzon, J. (2012). Expression pattern of the glucosinolate side chain biosynthetic genes MAM1 and MAM3 of Arabidopsis thaliana in different organs and developmental stages. Plant Physiol. Biochem. 53, 77–83. doi: 10.1016/j.plaphy.2012.01.015
Rhee, M.-S., Lee, S.-Y., Dougherty, R. H., and Kang, D.-H. (2003). Antimicrobial effects of mustard flour and acetic acid against Escherichia coli O157: H7, Listeria monocytogenes, and Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 69, 2959–2963. doi: 10.1128/AEM.69.5.2959-2963.2003
Robert-Seilaniantz, A., MacLean, D., Jikumaru, Y., Hill, L., Yamaguchi, S., Kamiya, Y., et al. (2011). The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. Plant J. 67, 218–231. doi: 10.1111/j.1365-313X.2011.04591.x
Rodman, J. E., Karol, K., Price, R. A., and Sytsma, K. J. (1996). Molecules, morphology, and Dahlgrens expanded order Capparales. Syst. Bot. 21, 289–307. doi: 10.2307/2419660
Rout, K., Sharma, M., Gupta, V., Mukhopadhyay, A., Sodhi, Y. S., Pental, D., et al. (2015). Deciphering allelic variations for seed glucosinolate traits in oilseed mustard (Brassica juncea) using two bi-parental mapping populations. Theor. Appl. Genet. 128, 657–666. doi: 10.1007/s00122-015-2461-9
Sawada, Y., Kuwahara, A., Nagano, M., Narisawa, T., Sakata, A., Saito, K., et al. (2009b). Omics-based approaches to methionine side chain elongation in Arabidopsis: characterization of the genes encoding methylthioalkylmalate isomerase and methylthioalkylmalate dehydrogenase. Plant Cell Physiol. 50, 1181–1190. doi: 10.1093/pcp/pcp079
Sawada, Y., Toyooka, K., Kuwahara, A., Sakata, A., Nagano, M., Saito, K., et al. (2009a). Arabidopsis bile acid: sodium symporter family protein 5 is involved in methionine-derived glucosinolate biosynthesis. Plant Cell Physiol. 50, 1579–1586. doi: 10.1093/pcp/pcp110
Schlaeppi, K., Bodenhausen, N., Buchala, A., Mauch, F., and Reymond, P. (2008). The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J. 55, 774–786. doi: 10.1111/j.1365-313X.2008.03545.x
Schnug, E., Haneklaus, S., Borchers, A., and Polle, A. (1995). Relations between sulphur supply and glutathione and ascorbate concentrations in Brassica napus. J. Plant Nutr. Soil Sci. 158, 67–69. doi: 10.1002/jpln.19951580113
Schuster, J., Knill, T., Reichelt, M., Gershenzon, J., and Binder, S. (2006). Branched chain amiontransferase 4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. Plant Cell 18, 2664–2679. doi: 10.1105/tpc.105.039339
Schweizer, F., Fernandez-Calvo, P., Zander, M., Diez-Diaz, M., Fonseca, S., Glauser, G., et al. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25, 3117–3132. doi: 10.1105/tpc.113.115139
Seo, M. S., Jin, M., Chun, J., Kim, S., Park, B., Shon, S., et al. (2016). Functional analysis of three BrMYB28 transcription factors controlling the biosynthesis of glucosinolates in Brassica rapa. Plant Mol. Biol. 90, 503–516. doi: 10.1007/s11103-016-0437-z
Shilpa, G., Sangha, M. K., Kaur, G., Atwal, A. K., Banga, S., and Banga, S. S. (2012). Variability for leaf and seed glucosinolate contents and profiles in a germplasm collection of the Brassica juncea. Biochem. Anal. Biochem. 1:120. doi: 10.4172/2161-1009.1000120
Smith, F. W., Ealing, P. M., Hawkesford, M. J., and Clarkson, D. T. (1995). Plant members of a family of sulfate transporters reveal functional subtypes. Proc. Natl. Acad. Sci. U.S.A. 92, 9373–9377. doi: 10.1073/pnas.92.20.9373
Sonderby, I. E., Geu-Flores, F., and Halkier, B. A. (2010). Biosynthesis of glucosinolates – gene discovery and beyond. Trends Plant Sci. 15, 283–290. doi: 10.1016/j.tplants.2010.02.005
Sønderby, I. E., Hansen, B. G., Bjarnholt, N., Ticconi, C., Halkier, B. A., and Kliebenstein, D. J. (2007). A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE 2:e1322. doi: 10.1371/journal.pone.0001322
Sotelo, T., Soengas, P., Velasco, P., Rodriguez, V. M., and Cartea, M. E. (2014). Identification of metabolic QTLs and candidate genes for glucosinolate synthesis in Brassica oleracea leaves, seeds and flower buds. PLoS ONE 9:e91428. doi: 10.1371/journal.pone.0091428
Sunkar, R., Chinnusamy, V., Zhu, J., and Zhu, J.-K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12, 301–309. doi: 10.1016/j.tplants.2007.05.001
Takahashi, H., Kopriva, S., Giordano, M., Saito, K., and Hell, R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184. doi: 10.1146/annurev-arplant-042110-103921
Takahashi, H., Watanabe-Takahashi, A., Smith, F. W., Blake-Kalff, M., Hawkesford, M. J., and Saito, K. (2000). The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 23, 171–182. doi: 10.1046/j.1365-313x.2000.00768.x
Textor, S., Bartram, S., Kroymann, J., Falk, K. L., Hick, A., Pickett, J. A., et al. (2004). Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta 218, 1026–1035. doi: 10.1007/s00425-003-1184-3
Textor, S., de Kraker, J.-W., Hause, B., Gershenzon, J., and Tokuhisa, J. G. (2007). MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 144, 60–71. doi: 10.1104/pp.106.091579
Thuswaldner, S., Lagerstedt, J. O., Rojas-Stütz, M., Bouhidel, K., Der, C., Leborgne-Castel, N., et al. (2007). Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis. J. Biol. Chem. 282, 8848–8859. doi: 10.1074/jbc.M609130200
Tookey, H. L., VanEtten, C. H., and Daxenbichler, M. E. (1980). “Glucosinolates,” in Toxic Constitutents of Plant Foodstuffs, ed. I. E. Liner (New York, NY: Academic press), 103–142.
Toroser, D., Thormann, C. E., Osborn, T. C., and Mithen, R. (1995). RFLP mapping of quantitative trait loci controlling seed aliphatic glucosinolate content in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 91, 802–808. doi: 10.1007/BF00220963
Tunc, S., Chollet, E., Chalier, P., Preziosi-Belloy, L., and Gontard, N. (2007). Combined effect of volatile antimicrobial agents on the growth of Penicillium notatum. Int. J. Food Microbiol. 113, 263–270. doi: 10.1016/j.ijfoodmicro.2006.07.004
USDA (2016). Oil Crops Outlook. Available at: http://www.ers.usda.gov/webdocs/publications/ocs16i/ocs-16i.pdf [accessed November 10, 2016].
Uzunova, M., Ecke, W., Weissleder, K., and Robbelen, G. (1995). Mapping the genome of rapeseed (Brassica napus L.). I. Construction of an RFLP linkage map and localization of QTLs for seed glucosinolate content. Theor. Appl. Genet. 90, 194–204. doi: 10.1007/BF00222202
van Dam, N. M., Tytgat, T. O. G., and Kirkegaard, J. A. (2009). Root and shoot glucosinolates: a comparison of their diversity, function and interactions in natural and managed ecosystems. Phytochem. Rev. 8, 171–186. doi: 10.1007/s11101-008-9101-9
Vaughn, S. F., Palmquist, D. E., Duval, S. M., and Berhow, M. A. (2006). Herbicidal activity of glucosinolate-containing seedmeals. Weed Sci. 54, 743–748. doi: 10.1614/WS-06-007R.1
Vik, D., Crocoll, C., Andersen, T. G., Burow, M., and Halkier, B. A. (2016). CB5C affects the glucosinolate profile in Arabidopsis thaliana. Plant Signal. Behav. 11:e1160189. doi: 10.1080/15592324.2016.1160189
Wachter, A., Wolf, S., Steininger, H., Bogs, J., and Rausch, T. (2005). Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 41, 15–30. doi: 10.1111/j.1365-313X.2004.02269.x
Walker, K., and Booth, E. (2003). “Sulphur nutrition and oilseed quality,” in Sulphur in Plants, eds Y. P. Abrol and A. Ahmad (Dordrecht: Springer), 323–339.
Wang, H., Wu, J., Sun, S., Liu, B., Cheng, F., Sun, R., et al. (2011). Glucosinolate biosynthetic genes in Brassica rapa. Gene 487, 135–142. doi: 10.1016/j.gene.2011.07.021
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–U1157. doi: 10.1038/ng.919
Wiesner, M., Zrenner, R., Krumbein, A., Glatt, H., and Schreiner, M. (2013). Genotypic variation of the glucosinolate profile in pak choi (Brassica rapa ssp chinensis). J. Agric. Food Chem. 61, 1943–1953. doi: 10.1021/jf303970k
Winde, I., and Wittstock, U. (2011). Insect herbivore counteradaptations to the plant glucosinolate-myrosinase system. Phytochemistry 72, 1566–1575. doi: 10.1016/j.phytochem.2011.01.016
Wittstock, U., and Burow, M. (2010). Glucosinolate breakdown in Arabidopsis: mechanism, regulation and biological significance. Arabidopsis Book 8:e0134.
Yang, B., Qiu, D., and Quiros, C. F. (2010). Variation of five major glucosinolate genes in Brassica rapa in relation to Brassica oleracea and Arabidopsis thaliana. Span. J. Agric. Res. 8, 662–671. doi: 10.5424/sjar/2010083-1263
Yang, J., Liu, D., Wang, X., Ji, C., Cheng, F., Liu, B., et al. (2016). The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 48, 1225–1232. doi: 10.1038/ng.3657
Yatusevich, R., Mugford, S. G., Matthewman, C., Gigolashvili, T., Frerigmann, H., Delaney, S., et al. (2010). Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J. 62, 1–11. doi: 10.1111/j.1365-313X.2009.04118.x
Yoshimoto, N., Inoue, E., Saito, K., Yamaya, T., and Takahashi, H. (2003). Phloem-localizing sulfate transporter, SULTR1; 3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 131, 1511–1517. doi: 10.1104/pp.014712
Yoshimoto, N., Takahashi, H., Smith, F. W., Yamaya, T., and Saito, K. (2002). Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 29, 465–473. doi: 10.1046/j.0960-7412.2001.01231.x
Zhang, J., Liu, Z., Liang, J., Wu, J., Cheng, F., and Wang, X. (2015b). Three genes encoding AOP2, a protein involved in aliphatic glucosinolate biosynthesis, are differentially expressed in Brassica rapa. J. Exp. Bot. 66, 6205–6218. doi: 10.1093/jxb/erv331
Zhang, Y., Li, B., Huai, D., Zhou, Y., and Kliebenstein, D. J. (2015). The conserved transcription factors, MYB115 and MYB118, control expression of the newly evolved benzoyloxy glucosinolate pathway in Arabidopsis thaliana. Front. Plant Sci. 6:343. doi: 10.3389/fpls.2015.00343
Zhang, J., Wang, X., Cheng, F., Wu, J., Liang, J., Yang, W., et al. (2015a). Lineage-specific evolution of Methylthioalkylmalate synthases (MAMs) involved in glucosinolates biosynthesis. Front. Plant Sci. 6:18. doi: 10.3389/fpls.2015.00018
Keywords: allyl-isothiocyanate, mustard, internal signaling, sinigrin, sulfate, canola, Brassicaceae
Citation: Borpatragohain P, Rose TJ and King GJ (2016) Fire and Brimstone: Molecular Interactions between Sulfur and Glucosinolate Biosynthesis in Model and Crop Brassicaceae. Front. Plant Sci. 7:1735. doi: 10.3389/fpls.2016.01735
Received: 30 June 2016; Accepted: 03 November 2016;
Published: 21 November 2016.
Edited by:
Ralph Kissen, Norwegian University of Science and Technology, NorwayReviewed by:
Ruediger Hell, Heidelberg University, GermanyTamara Gigolashvili, University of Cologne, Germany
Copyright © 2016 Borpatragohain, Rose and King. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Graham J. King, Z3JhaGFtLmtpbmdAc2N1LmVkdS5hdQ==
 Priyakshee Borpatragohain
Priyakshee Borpatragohain Terry J. Rose
Terry J. Rose Graham J. King
Graham J. King