- 1Department of Biology, Faculty of Science, Khon Kaen University, Khon Kaen, Thailand
- 2Department of Biological Sciences and Temasek Life Sciences Laboratory, National University of Singapore, Singapore, Singapore
APETALA1 (AP1) encodes a key MADS-box transcription factor that specifies the floral meristem identity on the flank of the inflorescence meristem, and determines the identity of perianth floral organs in the model plant Arabidopsis thaliana. Orchids are members of the Orchidaceae, one of the largest families of angiosperms. Although the expression patterns of a few AP1-like genes in orchids have been reported, their actual functions in orchid reproductive development are so far largely unknown. In this study, we isolated and characterized an AP1 ortholog, DOAP1, from Dendrobium Chao Praya Smile. DOAP1 was highly expressed in reproductive tissues, including inflorescence apices and flowers at various developmental stages. Overexpression of DOAP1 resulted in early flowering in Arabidopsis, and was able to rescue the floral organ defects of Arabidopsis ap1 mutants. Moreover, we successfully created transgenic Dendrobium Chao Praya Smile orchids overexpressing DOAP1, which displayed earlier flowering and earlier termination of inflorescence meristems into floral meristems than wild-type orchids. Our results demonstrate that DOAP1 plays an evolutionarily conserved role in promoting flowering and floral meristem specification in the Orchidaceae family.
Introduction
During the floral transition in flowering plants, the vegetative shoot apical meristem that produces leaves is transformed into the inflorescence meristem that produces flowers. This process is controlled by flowering regulatory networks in response to various environmental and endogenous signals, and the underlying molecular mechanisms have been intensively studied in the model plant Arabidopsis thaliana. In Arabidopsis, environmental and endogenous flowering signals perceived by various genetic pathways mediate transcriptional regulation of two major floral pathway integrators, FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), which in turn activate two floral meristem identity genes, APETALA1 (AP1) and LEAFY (LFY), to regulate the specification and formation of floral meristems on the flank of the shoot apical meristem (Kardailsky et al., 1999; Kobayashi et al., 1999; Blazquez and Weigel, 2000; Lee et al., 2000; Samach et al., 2000; Liu et al., 2009).
AP1 encodes a MADS-box transcription factor that not only specifies the floral meristem identity, but also contributes to determination of the identity of perianth floral organs in Arabidopsis (Irish and Sussex, 1990; Mandel et al., 1992; Bowman et al., 1993). When the activity of AP1 is lost, floral meristems that would normally develop into flowers are partially converted into inflorescence meristems. In ap1 mutants, flowers exhibited transformation of sepals into leaf-like organs and failure of petal development. In contrast, overexpression of AP1 results in early flowering and transformation of inflorescence meristems into floral meristems (Mandel and Yanofsky, 1995). During the floral transition, AP1 expression is directly activated by LFY and a protein complex consisting of FT and FD (Wagner et al., 1999; Abe et al., 2005; Wigge et al., 2005). In emerging floral meristems, AP1 plays dual functions as an activator and a repressor. It activates B class homeotic genes to mediate the specification of petals (Hill et al., 1998; Ng and Yanofsky, 2001), while it also suppresses a group of flowering time genes to prevent the reversion of floral meristems into inflorescence meristems (Yu et al., 2004; Liu et al., 2007; Kaufmann et al., 2010). Moreover, AP1 directly regulates the homeostasis of cytokinin by suppressing its biosynthesis and activating its degradation to establish determinate floral meristems (Han et al., 2014).
So far AP1 orthologs have been isolated in a wide range of plant species, such as pea (Pisum sativum) (Berbel et al., 2001), apple (Malus domestica) (Kotoda et al., 2002), common wheat (Triticum aestivum) (Adam et al., 2007), moth orchid (Phalaenopsis ‘Hatsuyuki’) (Song et al., 2011), longan (Dimocarpus longan) (Winterhagen et al., 2013), trifoliate orange (Poncirus trifoliata L. Raf.) (Sun et al., 2014), Birch (Betula platyphylla) (Huang et al., 2014) and poplar (Populus tomentosa) (Chen et al., 2015). Through mainly examining their expression patterns and their effects on heterologous systems, most of these genes have been suggested to affect either the flowering process or floral organ formation.
Dendrobium is one of the largest genera in the family Orchidaceae, which is one of the largest families of angiosperms (Teixeira da Silva et al., 2014). The Dendrobium genus comprises more than 1200 species, and its members have attractive ornamental and medicinal values (Yu and Goh, 2001; Teixeira da Silva et al., 2014). The high demand of Dendrobium orchids has enabled Dendrobium growers to create many varieties and hybrids with various floral traits. However, the long vegetative phase and low survival rate of orchid seedlings under natural environmental conditions make it difficult to propagate and breed Dendrobium orchids (Yu and Goh, 2001; Chai and Yu, 2007; Wang et al., 2009). Therefore, it is important to elucidate the molecular mechanisms underlying the floral transition in orchids so as to identify appropriate regulators for targeted manipulation or screening of orchid traits. Recent studies have isolated and characterized several orthologs of Arabidopsis floral pathway integrators in orchids. Overexpression of three orthologs of FT, OnFT, DnFT and CgFT from Oncidium, Dendrobium, and Cymbidium orchids, respectively, causes early flowering in transgenic Arabidopsis or tobacco plants (Hou and Yang, 2009; Li et al., 2012; Xiang et al., 2012). In addition, overexpression of DOSOC1, an ortholog of the Arabidopsis SOC1, promotes flowering in both transgenic Arabidopsis and Dendrobium orchids (Ding et al., 2013).
In this study, we isolated an AP1 ortholog, DOAP1, from Dendrobium Chao Praya Smile, and characterized its function through examining its expression patterns and investigating its biological effects in both transgenic Arabidopsis and orchid plants. Our results suggest that DOAP1 may play a conserved role in promoting flowering and floral meristem development in the Orchidaceae family.
Materials and Methods
Plant Material and Growth Conditions
Dendrobium Chao Praya Smile (a hybrid of Dendrobium Pinky and Dendrobium Kiyomi Beauty) plants were grown in pots in the greenhouse at 28 ± 4°C under natural lighting conditions. In our in vitro culture system for Dendrobium Chao Praya Smile, protocorms developing from seeds were used as the starting materials. They were cultured in modified liquid Knudson C (KC) medium supplemented with 2% (w/v) sucrose, 15% (v/v) coconut water and 4.4 μM benzyladenine (BA) at 24°C under a 16-h photoperiod on rotary shakers at 120 rpm (Yu and Goh, 2000a; Ding et al., 2013). Plantlets at the floral transitional stage were transferred to the two-layer modified KC medium as previously described (Hee et al., 2007; Sim et al., 2007). Wild-type Arabidopsis thaliana ecotype Columbia (Col-0) and 35S:DOAP1 transgenic plants in both wild-type and ap1-10 Col-0 backgrounds were grown under long-day conditions (16 h light/8 h dark) at 23 ± 2°C.
Isolation of DOAP1
Total RNA was extracted from inflorescences of Dendrobium Chao Praya Smile using the RNeasy® Plant Mini Kit (QIAGEN). To isolate the putative AP1 orthologs, two degenerate primers, 5′-CAGCTGARGCGRATMGAGAAC-3′ and 5′-GCKMAGCATCCAWGGYGG-3′, were manually designed based on the conserved regions among AP1 orthologs in various plant species. Based on the partial cDNA sequence obtained, the full-length cDNA of DOAP1 was further amplified using the SMARTERTM RACE cDNA Amplification Kit (BD Biosciences Clontech).
Sequence Analysis
Protein sequences of AP1-like genes were retrieved from the National Center for Biotechnology Information (NCBI) database. Alignment of amino acid sequences was performed using the Clustal Omega multiple sequence alignment program1 and BOXSHADE 3.212. The phylogenetic tree was generated by the neighbor-joining algorithm using the MEGA6 software3.
Expression Analysis
Total RNA was extracted from orchids or Arabidopsis plants using the RNeasy® Plant Mini Kit (QIAGEN), and reverse-transcribed using the SuperScriptTM II Reverse Transcriptase Kit (Invitrogen) according to the manufacturer’s instructions. Real-time PCR reaction was performed in triplicates on the CFX384 Real-Time PCR Detection System (Bio-Rad) with the SYBR Green Master Mix (Toyobo). UBIQUITIN (DOUbi) in Dendrobium Chao Praya Smile was used as a reference gene (Ding et al., 2013). Calculation of relative gene expression levels was performed as previously reported (Liu et al., 2007). Semi-quantitative reverse transcription PCR (RT-PCR) was carried out as previously reported (Xu et al., 2006) using either Arabidopsis β-TUBLIN2 (TUB2) or orchid ACTIN (DOActin) as a reference gene.
Arabidopsis Transformation
The full-length DOAP1 cDNA fragment was cloned into pGreen0229-35S under the control of two CaMV 35S promoters (Yu et al., 2004). The resulting construct was introduced into Agrobacterium tumefaciens GV3101 competent cells by electroporation. 35S:DOAP1 transgenic Arabidopsis lines were created by Agrobacterium tumefaciens-mediated transformation and selected by 0.3 g/L Basta on soil.
Orchid Transformation by Agrobacterium-Mediated Transformation
Genetic transformation of Dendrobium Chao Praya Smile was performed according to the reported L-methionine sulfoximine (MSO) selection system with minor modifications (Yu et al., 2001; Chai et al., 2007; Ding et al., 2013). Dendrobium Chao Praya Smile calli were cut into small pieces 3–5 mm in diameter and then cultured in modified KC liquid medium supplemented with 2% (w/v) sucrose, 15% (v/v) coconut water and 4.4 μM BA. Agrobacterium pellet was re-suspended in KC liquid medium to be co-cultivated with prepared orchid calli. Acetosyringone (100 μM) was added to induce transformation. After co-cultivation for 2 h, calli were placed on solid KC medium in the dark for three nights. Agrobacterium-infected calli were rinsed by water containing 200 mg/L cefotaxime to remove the bacteria, and grown on solid KC medium containing 0.5 μM MSO as a selection agent. Surviving calli were subcultured onto fresh solid medium every 20 days. After four rounds of MSO selection, green calli were transferred to modified KC solid medium containing 2 μM MSO for lethal selection. After three rounds of selection, putative surviving transgenic lines were cultured on modified KC solid medium for further investigation.
Southern Blot
Genomic DNA isolated from wild-type and 35S:DOAP1 transgenic orchids was digested by EcoRI for 16 h, resolved on 1.2% (w/v) agarose gels, and then transferred onto nylon membranes. Blots were hybridized overnight with the specific digoxigenin-labeled DNA, washed and detected in accordance with the manufacturer’s instructions (DIG Application Manual for Filter Hybridization, Roche) as previously reported (Yu and Goh, 2000b).
Results
Isolation of DOAP1 from Dendrobium Chao Praya Smile
To isolate AP1-like genes from Dendrobium Chao Praya Smile, we designed a pair of degenerate primers based on the conserved amino acid sequences of AP1 orthologs in various plant species. A partial cDNA fragment was obtained by reverse transcription PCR using RNA extracted from inflorescence apices of Dendrobium Chao Praya Smile. Since this fragment showed high sequence similarity with other AP1-like genes, we further designed primers based on this fragment and obtained the corresponding full-length cDNA sequence, designated DOAP1 (GenBank accession No. KY471451), using the rapid amplification of cDNA ends (RACE) method.
DOAP1 cDNA is 897 bp in length with a 639 bp coding region. The deduced DOAP1 amino acid sequence contains a highly conserved MADS domain and a less conserved K domain, and a diverse C terminal region as found in other AP1-like genes from various plant species (Figure 1). Multiple sequence alignment showed that DOAP1 shared the highest sequence identity with other orchid AP1 orthologs, such as CeAP1 (Cymbidium ensifolium; 69.95% identity) and PjAP1 (Phalaenopsis japonica; 62.56% identity). DOAP1 also had higher sequence identity with monocot AP1 orthologs, such as TaAP1 (Triticum aestivum; 58% identity), OsAP1 (Oryza sativa; 56.72% identity) and ZmAP1 (Zea mays; 54.59% identity), than Arabidopsis AP1 (53.3% identity).
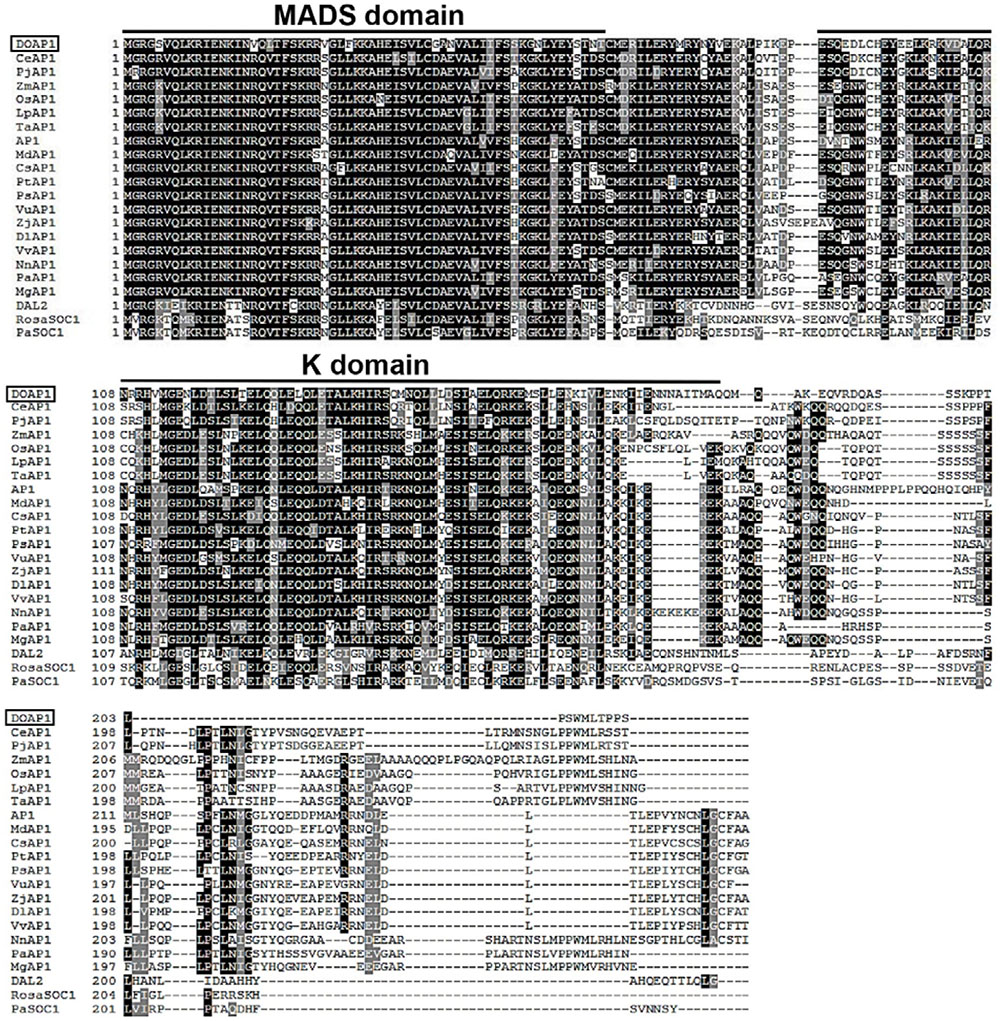
FIGURE 1. Alignment of the amino acid sequences of DOAP1 and its orthologs from other plant species. Conserved residues are shown in black, while similar residues are shown in gray. Dashes indicate gaps to maximize alignment. The conserved MADS and K domains are indicated. The species names are shown behind their corresponding protein names as follows: DOAP1, Dendrobium Chao Praya Smile; CeAP1, Cymbidium ensifolium; PjAP1: Phalaenopsis japonica; ZmAP1, Zea mays; OsAP1, Oryza sativa; LpAP1, Lolium perenne; TaAP1, Triticum aestivum; AP1, Arabidopsis thaliana; MdAP1, Malus domestica; CsAP1, Citrus sinensis; PtAP1, Populus trichocarpa; PsAP1, Paeonia suffruticosa; VuAP1, Vigna unguiculata; ZjAP1, Ziziphus jujube; DlAP1, Dimocarpus longan; VvAP1, Vitis vinifera; NnAP1, Nelumbo nucifera; PaAP1, Persea Americana; MgAP1, Magnolia grandiflora. Non-AP1-like proteins that are encoded by other MADS-box subfamily genes were used as outlier in this alignment as follows: DEFICIENS-AGAMOUS-LIKE2 (DAL2) of Picea abies, and two SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1)-like proteins from Rosa hybrid cultivar (RosaSOC1) and Picea abies (PaSOC1).
To determine the evolutionary relationship between DOAP1 and other AP1-like proteins from other plant species, we constructed a phylogenetic tree based on the analysis of amino acid sequences of MIK regions (Figure 2). The tree showed that DOAP1 was clustered together with other orchid AP1-like proteins, such as OMADS10 (Chang et al., 2009) and DOMADS2 (Yu and Goh, 2000b), in the monocotyledonous subgroup of SQUA that includes AP1-like proteins isolated from monocots (Purugganan et al., 1995; Theissen et al., 1996).
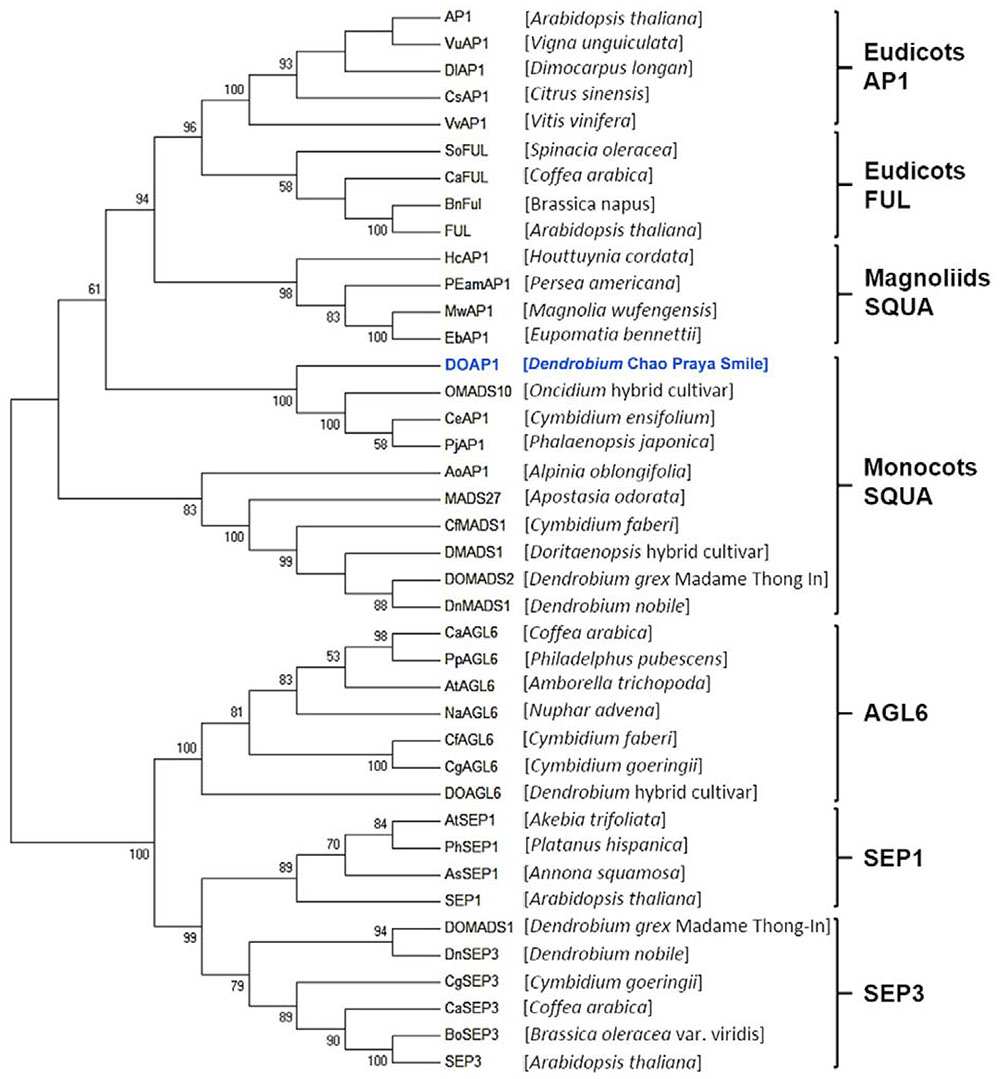
FIGURE 2. Phylogenetic tree of AP1/AGL9 -like MADS-box proteins. Amino acid sequences of AP1/AGL9 –like MADS box genes were obtained from the NCBI database. DOAP1 investigated in this study is highlighted in blue. The multiple sequence alignment was performed via Clustal Omega. The tree was constructed with MEGA6 using the neighbor-joining method. Genus names of respective species are given in the parentheses behind the corresponding protein names. Bootstrap values (>50%) in 1,000 replicates are shown below the nodes.
DOAP1 Expression Pattern in Dendrobium Chao Praya Smile
To study the expression pattern of DOAP1 in Dendrobium Chao Praya Smile, cDNA was synthesized from RNA extracted from various orchid tissues, including leaves, roots, inflorescence apices, floral buds at different stages (stages 1–3, at which buds were 0.5–1 cm, 1–2 cm, and over 2 cm in length, respectively), half-bloomed flowers and fully bloomed flowers (Figure 3A). Quantitative real-time PCR analysis showed that DOAP1 transcripts were detected at the highest level in inflorescence apices and at the lowest level in roots (Figure 3B). Notably, DOAP1 expression was significantly higher in inflorescence apices than in vegetative shoot apices. Moreover, DOAP1 expression gradually increased in developing floral buds at various stages and half-bloomed flowers, but decreased afterward in fully bloomed flowers (Figure 3B). These expression patterns indicate that DOAP1 function may be closely associated with flowering time control and flower development in Dendrobium Chao Praya Smile.
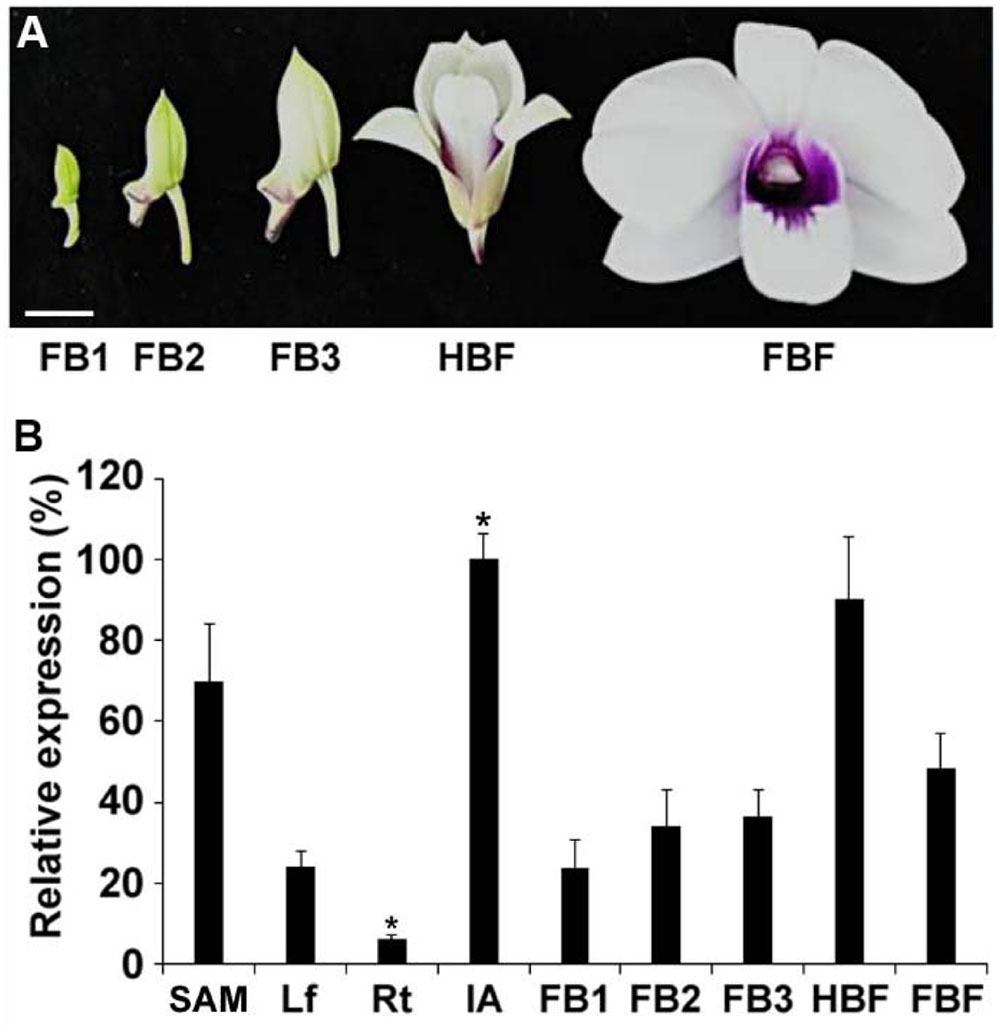
FIGURE 3. Expression of DOAP1 in various tissues of Dendrobium Chao Praya Smile. (A) Five developmental stages of flowers, including floral bud 1 (FB1), FB2, FB3, half-bloomed flower (HBF), and fully-bloomed flower (FBF), are shown. Scale bar, 1 cm. (B) Quantitative analysis of DOAP1 expression in various tissues and different floral developmental stages of Dendrobium Chao Praya Smile. Transcript levels were determined by quantitative real-time PCR analyses of three independently harvested samples. The levels of gene expression normalized to the orchid polyubiquitin gene (DOUbi) expression are shown relative to the highest level set at 100%. SAM, vegetative shoot apical meristem; Lf, leaf; Rt, root; IA, inflorescence apex; FB, floral bud; HBF, half-bloomed flower; FBF, fully bloomed flower. Error bars, mean ± SD. Asterisks indicate statistically significant differences in the expression level of DOAP1 in IA or Rt as compared to its expression levels in any other tissues examined (two-tailed paired Student’s t-test, p < 0.05).
DOAP1 Promotes Flowering and Partially Complements ap1 Mutants in Arabidopsis
In order to investigate the biological function of DOAP1, we firstly generated transgenic Arabidopsis plants harboring the DOAP1 coding sequence driven by the CaMV 35S promoter. Among 50 independent 35S:DOAP1 transgenic lines created at the T1 generation, 46 lines showed an early flowering phenotype typically producing only 5–7 rosette leaves as compared to wild-type plants, which developed 9–12 rosette leaves under long day conditions (Figures 4A,B). Semi-quantitative RT-PCR using DOAP1-specific primers confirmed that DOAP1 was overexpressed in a representative 35S:DOAP1 transgenic Arabidopsis line with a single T-DNA insertion (Figure 4C), implying that the early flowering phenotype of 35S:DOAP1 is associated with overexpression of DOAP1. In addition to the defect in flowering time, the inflorescence meristems of 35S:DOAP1 transgenic lines usually terminated as flowers after producing only a few flowers, while these flowers produced floral organs indistinguishable from those of wild-type flowers (Figures 4D,E), suggesting that overexpression of DOAP1 promotes the formation of floral meristems, but does not affect floral organ identity in Arabidopsis.
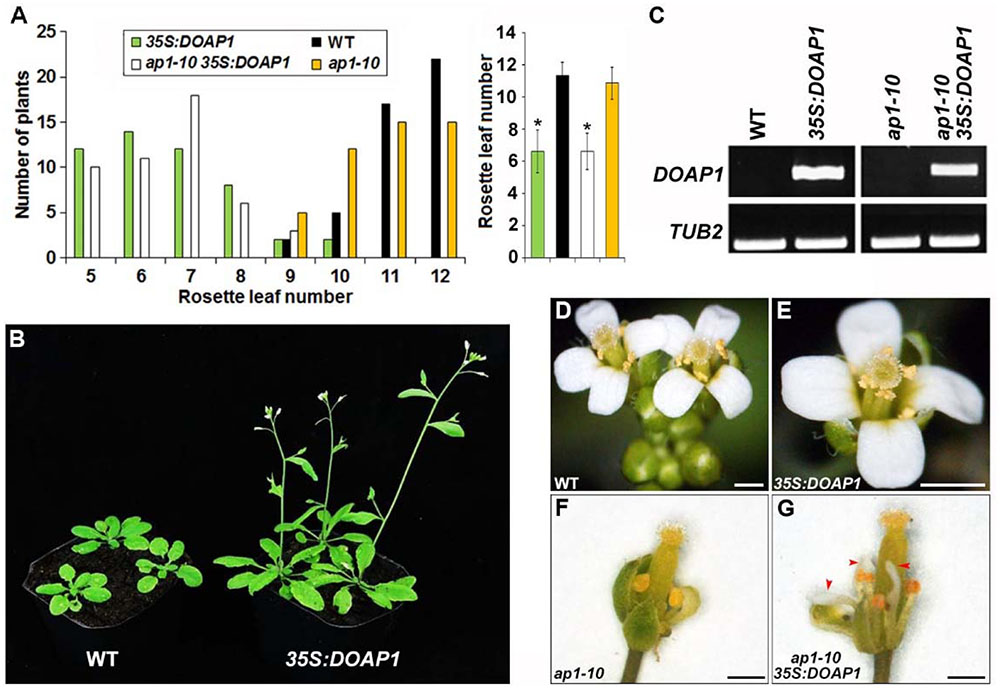
FIGURE 4. Overexpression of DOAP1 in Arabidopsis results in early flowering. (A) Flowering time of 35S:DOAP1 in wild-type and ap1-10 backgrounds. The left panel shows the distribution of flowering time in T1 transgenic lines harboring 35S:DOAP1 in wild-type and ap1-10 backgrounds grown under long day conditions, while the right panel shows the average flowering time of various transgenic lines. Error bars, mean ± SD. Asterisks indicate statistically significantly differences in flowering time of 35S:DOAP1 and ap1-10 35S:DOAP1 compared to wild-type and ap1-10, respectively (two-tailed paired Student’s t-test, p < 0.05). (B) Progenies of a representative 35S:DOAP1 line (right) flower earlier than wild-type plants (left) under long day conditions. (C) Examination of DOAP1 expression in representative 35S:DOAP1 and ap1-10 35S:DOAP1 lines by semi-quantitative RT-PCR. TUB2 was amplified as an internal control. (D–G) Floral phenotypes of 35S:DOAP1 and ap1-10 35S:DOAP1. There is no visible difference in floral morphology of wild-type (D) and 35S:DOAP1 (E) plants, whereas deficiency in petal formation in ap1-10 (F) was partially rescued in ap1 35S:DOAP1 (G). Red arrowheads indicate petals in ap1 35S:DOAP1. Bars represent 5 mm.
We further performed Agrobacterium-mediated transformation to introduce 35S:DOAP1 into ap1-10 mutants, which were generated from ethyl methanesulfonate-mutagenized populations of Arabidopsis ecotype Columbia (Schultz and Haughn, 1993), to examine whether DOAP1 could complement the loss function of AP1. While ap1-10 exhibited comparable flowering time to wild-type plants, most of the ap1-10 35S:DOAP1 lines flowered much earlier than wild-type and ap1-10 plants, a flowering pattern resembling 35S:DOAP1 in the wild-type background (Figure 4A). We further identified several ap1-10 35S:DOAP1 transgenic lines with single T-DNA insertion, which consistently displayed the similar early flowering phenotype at various generations, and selected one representative line for further investigations. Like other ap1 mutants, ap1-10 showed typical defects in the identity of perianth organs, such as homeotic transformation of petals into stamens or stamen-petal chimeric structures (Figure 4F). In contrast, petal formation was restored in ap1-10 35S:DOAP1 (Figure 4G), demonstrating that DOAP1 plays the same role as AP1 in regulating petal formation in Arabidopsis. Similarly, semi-quantitative RT-PCR detected high expression of DOAP1 in the representative ap1-10 35S:DOAP1 line (Figure 4C), substantiating a causal link between the observable phenotypes and overexpression of DOAP1 in ap1-10 35S:DOAP1.
DOAP1 Promotes Early Formation of Inflorescences in Dendrobium Chao Praya Smile
To understand the endogenous function of DOAP1 in Dendrobium Chao Praya Smile, we created transgenic orchids bearing 35S:DOAP1 in a pGreen vector (Figure 5A) using an integrated orchid gene transformation and in vitro orchid culture system (Yu et al., 2001; Chai et al., 2007; Ding et al., 2013).
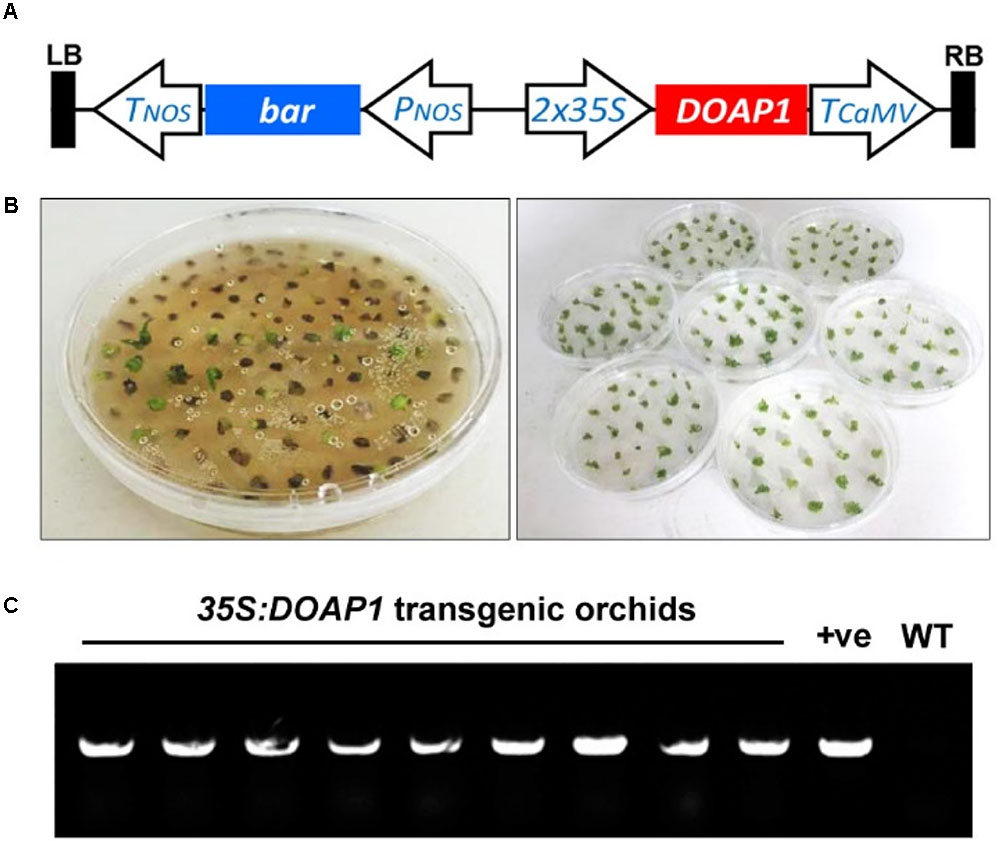
FIGURE 5. Generation of 35S:DOAP1 transgenic orchids. (A) Schematic diagram of the T-DNA region of the pGreen0229-35S:DOAP1 construct. LB, left border; Tnos, nopaline synthase terminator sequence; bar, bialaphos resistance gene; Pnos, nopaline synthase promoter sequence; 35S, Cauliflower mosaic virus 35S promoter sequence; TCaMV, Cauliflower mosaic virus terminator sequence; RB, right border. (B) Genetic transformation of Dendrobium Chao Praya Smile using the MSO selection system. After lethal section on the solid medium containing 2 μM MSO (left panel), green surviving calli were further subcultured onto fresh medium every 20 days (right panel). (C) PCR genotyping of DOAP1 transgenic orchids shows the presence of 35S:DOAP1 transgene in nine independent lines of Dendrobium Chao Praya Smile. The specific primers from the 35S promoter and DOAP1, respectively, were designed for genotyping. The 35S:DOAP1 plasmid and the genomic DNA of wild-type orchids were used as a positive control (+ve) and a negative control (WT), respectively.
After Agrobacterium-mediated transformation of Dendrobium Chao Praya Smile calli, the transformed materials were selected on the medium containing 0.5 and 2 μM MSO as a selection agent for initial and lethal selection (Figure 5B), respectively. After a total of seven rounds of selection, most of the calli turned necrotic and eventually died, while some putative transformants survived and proliferated into protocorm-like bodies, which further developed into young plantlets. We screened out nine independent 35S:DOAP1 transgenic Dendrobium Chao Praya Smile lines, and confirmed the presence of the 35S:DOAP1 transgene in these lines by PCR genotyping using the specific primers from 35S and DOAP1 (Figure 5C).
We then compared the growth status of wild-type and 35S:DOAP1 Dendrobium Chao Praya Smile plants using our established in vitro culture system (Ding et al., 2013), which allows rapid in vitro development of orchid plants from the vegetative to reproductive phase. Under our growth conditions, it took wild-type orchid plants 25–30 weeks to grow from protocorm-like bodies to plantlets with the first visible inflorescence stalks (Figure 6A), whereas 8 out of 9 35S:DOAP1 transgenic lines displayed the first visible inflorescences at 10–17 weeks of culture (Figures 6A,B). These phenotypes indicate a role of DOAP1 in promoting the transition of vegetative shoot apical meristems into inflorescence meristems. As compared to wild-type inflorescences (Ding et al., 2013), the inflorescence apices of these 35S:DOAP1 transgenic plants were usually terminated as floral buds immediately after producing only 1–2 floral buds or even without generating any other floral structure (Figure 6C), suggesting a quick transformation of inflorescence meristems into floral meristems in 35S:DOAP1. In addition, Southern blot hybridization revealed the presence of the T-DNA region containing the bialaphos resistance (bar) gene in the genome of 35S:DOAP1 #1 and #2 lines (Figure 6D), which substantiated the results showing the presence of the 35S:DOAP1 transgene in the 35S:DOAP1 genome (Figure 5C). As expected, semi-quantitative RT PCR showed that DOAP1 was overexpressed in inflorescence apices of 35S:DOAP1 transgenic orchids compared to wild-type orchids (Figure 6E). These results imply that the phenotypes of 35S:DOAP1 transgenic orchids are associated with overexpression of DOAP1 in these plants.
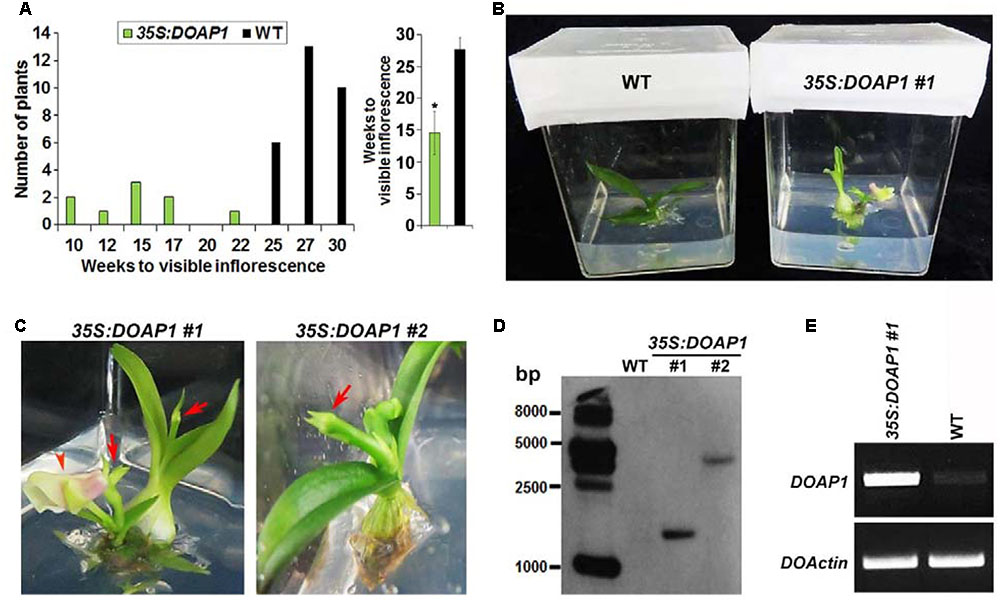
FIGURE 6. Overexpression DOAP1 in Dendrobium Chao Praya Smile causes early flowering. (A) Comparison of flowering time of wild-type and 35S:DOAP1 transgenic orchids. The left panel shows that all 9 transgenic lines produce inflorescences (indicated by the visible inflorescence stalk) earlier than 29 wild-type plants, while the right panel shows the comparison of the average time to produce visible inflorescences of 35S:DOAP1 and wild-type orchids. Error bars, mean ± SD. Asterisk indicates a statistically significant difference in flowering time between 35S:DOAP1 and wild-type (two-tailed paired Student’s t-test, p < 0.05). (B) One representative 35S:DOAP1 transgenic orchid (#1) shows earlier flowering than a wild-type (WT) orchid at 16 weeks after in vitro culture under our growth conditions. (C) Close views of two independent 35S:DOAP1 transgenic lines (#1 and #2). Arrows indicate inflorescence apices, while arrowhead indicates a flower. (D) Southern blot analysis of the presence of the 35S:DOAP1 transgene in transgenic orchids. The genomic DNA extracted from wild-type and two 35S:DOAP1 transgenic lines (#1 and #2) was digested with EcoRI, and the DNA gel blot was hybridized with the digoxigenin-labeled probe that was synthesized from the bar gene. The sizes of the DNA markers are given on the left in bases. (E) Examination of DOAP1 expression in wild-type and 35S:DOAP1 (#1) orchids by semi-quantitative RT-PCR. Total RNA was extracted from inflorescence apices, and DOActin was amplified as an internal control.
Discussion
Although the Orchidaceae is one of the largest and most widespread families of flowering plants, the molecular mechanism underlying the flowering process of the members in this family remain largely unknown. The difficulty in performing molecular genetic studies on orchids is partially due to their long vegetative phase and recalcitrant nature for gene transformation. In this study, we have taken advantage of an established Agrobacterium-mediated gene transformation system based on MSO selection coupled with in vitro orchid culture system (Yu et al., 2001; Hee et al., 2007; Sim et al., 2007; Ding et al., 2013) to study the biological function of a newly isolated AP1 ortholog, DOAP1, from Dendrobium Chao Praya Smile.
Our findings have provided several pieces of evidence to support that DOAP1 plays a conserved role in promoting the floral transition and specifying the identity of perianth floral organs. First, in addition to the conserved MADS and K domains found among AP1-like proteins, phylogenetic analysis has revealed that DOAP1 and other orchid AP1-like proteins are assigned to the same clade in the monocotyledonous subgroup of SQUA, which contains AP1/SQUA-like proteins from monocots (Purugganan et al., 1995; Theissen et al., 1996). Second, DOAP1 is highly expressed in inflorescence apices and flowers at various developmental stages, which is similar to the patterns exhibited by most of AP1-like genes studied (Becker and Theissen, 2003; Litt and Irish, 2003). Notably, DOAP1 is also expressed in vegetative shoot apices and upregulated in inflorescence apices, suggesting that it could be one of the earliest regulatory genes that promote the floral transition in orchids. Third, overexpression of DOAP1 in both Arabidopsis and orchids causes early flowering and early termination of inflorescence meristems as floral meristems, which are similar to the effects of overexpression of AP1 in Arabidopsis (Mandel and Yanofsky, 1995), indicating that like AP1, DOAP1 may serve as a floral meristem identity gene to promote the specification of floral meristems in both Arabidopsis and orchids. Similarly, overexpression of AP1 orthologs from other plant species also causes early flowering in transgenic plants (Berbel et al., 2001; Kotoda et al., 2002; Adam et al., 2007; Huang et al., 2014; Sun et al., 2014), suggesting a conserved role of AP1-like genes in accelerating flowering probably through promoting the early formation of floral meristems. Last, overexpression of DOAP1 in ap1-10 Arabidopsis mutants partially rescues the defect in petal formation, suggesting that DOAP1 and AP1 also share a similar role as class A organ identity genes (Irish and Sussex, 1990; Mandel et al., 1992; Bowman et al., 1993). Taken together, our findings suggest that DOAP1 may play evolutionarily conserved roles in promoting floral meristem formation and specification of perianth organs in the Orchidaceae family. As AP1 has been shown to prevent the formation of flowers in the axils of sepals by directly regulating the cytokinin homeostasis (Han et al., 2014), it would be interesting to investigate whether DOAP1 also has a similar role in orchids in future studies.
Despite the functional conservation of DOAP1 as discussed above, some of our results also indicate other potential functions of DOAP1 in orchid development. For example, DOAP1 is expressed in both leaves and floral buds at stage 1 almost at comparable levels (Figure 3B), implying its potential involvement in vegetative growth. In addition, there is a dramatic decrease in DOAP1 expression in fully bloomed flowers as compared to its expression in half-bloomed flowers (Figure 3B). This change could be relevant to a possible role of DOAP1 in late flower development. Consistently, several other orchid AP1-like genes also exhibit potential functions in various aspects of flower development other than their conserved roles in floral meristem development and perianth organ specification. For example, another orchid AP1-like gene, DOMADS2, isolated from Dendrobium Madame Thong-In is not only highly expressed in the shoot apical meristem during the floral transition, but also expressed in orchid reproductive organs, such as column (gynostemium, a fused structure of stigmas, styles and stamens) and ovary (Yu and Goh, 2000b), indicating that DOMADS2 may also be involved in reproductive organ development. Similarly, another AP1-like gene, EpMADS12, in Erycina pusilla is also detectable in multiple floral organs, such as lips, column, and pollinarium (Lin et al., 2016). These diverse expression patterns of AP1-like genes indicate that AP1-like genes may evolve with multiple functions in regulating flowering and flower development in orchids. Further investigation of these orchid AP1-like genes through knocking down or knocking out their expression in orchids will shed light on their endogenous functions in orchid reproductive development.
Author Contributions
NS, YW, LS, and HY conceived and designed the study. NS and SB performed the experiments. NS, SB, YW, LS, and HY analyzed data. NS, YW, LS, and HY wrote the paper. All authors read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Academic Research Fund (MOE2015-T2-1-002) from the Ministry of Education-Singapore and the Singapore National Research Foundation Investigatorship Programme (NRF-NRFI2016-02) and the intramural research support from National University of Singapore and Temasek Life Sciences Laboratory. NS was financially supported by Development and Promotion of Science and Technology Talents Project (Royal Government of Thailand Scholarship).
Footnotes
- ^http://www.ebi.ac.uk/Tools/msa/clustalo/
- ^http://www.ch.embnet.org/software/BOX_form.html
- ^http://www.megasoftware.net/
References
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Adam, H., Ouellet, F., Kane, N. A., Agharbaoui, Z., Major, G., Tominaga, Y., et al. (2007). Overexpression of TaVRN1 in Arabidopsis promotes early flowering and alters development. Plant Cell Physiol. 48, 1192–1206. doi: 10.1093/pcp/pcm089
Becker, A., and Theissen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489. doi: 10.1016/S1055-7903(03)00207-0
Berbel, A., Navarro, C., Ferrandiz, C., Canas, L. A., Madueno, F., and Beltran, J. P. (2001). Analysis of PEAM4, the pea AP1 functional homologue, supports a model for AP1-like genes controlling both floral meristem and floral organ identity in different plant species. Plant J. 25, 441–451. doi: 10.1046/j.1365-313x.2001.00974.x
Blazquez, M. A., and Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404, 889–892. doi: 10.1038/35009125
Bowman, J. L., Alvarez, J., Weigel, D., Meyerowitz, E., and Smyth, D. R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–743.
Chai, D., Lee, S. M., Ng, J. H., and Yu, H. (2007). L-methionine sulfoximine as a novel selection agent for genetic transformation of orchids. J. Biotechnol. 131, 466–472. doi: 10.1016/j.jbiotec.2007.07.951
Chai, D., and Yu, H. (2007). Recent advances in transgenic orchid production. Orchid Sci. Biotechnol. 1, 34–39.
Chang, Y.-Y., Chiu, Y.-F., Wu, J.-W., and Yang, C.-H. (2009). Four orchid (Oncidium Gower Ramsey) AP1/AGL9-like MADS box genes show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol. 50, 1425–1438. doi: 10.1093/pcp/pcp087
Chen, Z., Yang, X., Su, X., Rao, P., Gao, K., Lei, B., et al. (2015). Identification and expression analysis of APETALA1 homologues in poplar. Acta Physiol. Plant. 37, 1–10. doi: 10.1007/s11738-015-1805-z
Ding, L., Wang, Y., and Yu, H. (2013). Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the orchid Dendrobium Chao Parya Smile. Plant Cell Physiol. 54, 595–608. doi: 10.1093/pcp/pct026
Han, Y., Zhang, C., Yang, H., and Jiao, Y. (2014). Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 6840–6845. doi: 10.1073/pnas.1318532111
Hee, K. H., Loh, C. S., and Yeoh, H. H. (2007). Early in vitro flowering and seed production in culture in Dendrobium Chao Praya Smile (Orchidaceae). Plant Cell Rep. 26, 2055–2062. doi: 10.1007/s00299-007-0421-9
Hill, T. A., Day, C. D., Zondlo, S. C., Thackeray, A. G., and Irish, V. F. (1998). Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 125, 1711–1721.
Hou, C. J., and Yang, C. H. (2009). Functional analysis of FT and TFL1 orthologs from orchid (Oncidium Gower Ramsey) that regulate the vegetative to reproductive transition. Plant Cell Physiol. 50, 1544–1557. doi: 10.1093/pcp/pcp099
Huang, H., Wang, S., Jiang, J., Liu, G., Li, H., Chen, S., et al. (2014). Overexpression of BpAP1 induces early flowering and produces dwarfism in Betula platyphylla × Betula pendula. Physiol. Plant. 151, 495–506. doi: 10.1111/ppl.12123
Irish, V. F., and Sussex, I. M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2, 741–753. doi: 10.1105/tpc.2.8.741
Kardailsky, I., Shukla, V. K., Ahn, J. H., Dagenais, N., Christensen, S. K., Nguyen, J. T., et al. (1999). Activation tagging of the floral inducer FT. Science 286, 1962–1965. doi: 10.1126/science.286.5446.1962
Kaufmann, K., Wellmer, F., Muino, J. M., Ferrier, T., Wuest, S. E., Kumar, V., et al. (2010). Orchestration of floral initiation by APETALA1. Science 328, 85–89. doi: 10.1126/science.1185244
Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. doi: 10.1126/science.286.5446.1960
Kotoda, N., Wada, M., Kusaba, S., Kano-Murakami, Y., Masuda, T., and Soejima, J. (2002). Overexpression of MdMADS5, an APETALA1-like gene of apple, causes early flowering in transgenic Arabidopsis. Plant Sci. 162, 679–687. doi: 10.1016/S0168-9452(02)00024-9
Lee, H., Suh, S. S., Park, E., Cho, E., Ahn, J. H., Kim, S. G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. doi: 10.1101/gad.813600
Li, R., Wang, A., Sun, S., Liang, S., Wang, X., Ye, Q., et al. (2012). Functional characterization of FT and MFT ortholog genes in orchid (Dendrobium nobile Lindl) that regulate the vegetative to reproductive transition in Arabidopsis. Plant Cell Tiss. Org. 111, 143–151. doi: 10.1007/s11240-012-0178-x
Lin, C. S., Hsu, C. T., Liao, D. C., Chang, W. J., Chou, M. L., Huang, Y. T., et al. (2016). Transcriptome-wide analysis of the MADS-box gene family in the orchid Erycina pusilla. Plant Biotechnol. J. 14, 284–298. doi: 10.1111/pbi.12383
Litt, A., and Irish, V. F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165, 821–833.
Liu, C., Xi, W. Y., Shen, L. S., Tan, C. P., and Yu, H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711–722. doi: 10.1016/j.devcel.2009.03.011
Liu, C., Zhou, J., Bracha-Drori, K., Yalovsky, S., Ito, T., and Yu, H. (2007). Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134, 1901–1910. doi: 10.1242/dev.003103
Mandel, M. A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. doi: 10.1038/360273a0
Mandel, M. A., and Yanofsky, M. F. (1995). A gene triggering flower formation in Arabidopsis. Nature 377, 522–524. doi: 10.1038/377522a0
Ng, M., and Yanofsky, M. F. (2001). Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell 13, 739–753. doi: 10.1105/tpc.13.4.739
Purugganan, M. D., Rounsley, S. D., Schmidt, R. J., and Yanofsky, M. F. (1995). Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics 140, 345–356.
Samach, A., Onouchi, H., Gold, S. E., Ditta, G. S., Schwarz-Sommer, Z., Yanofsky, M. F., et al. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. doi: 10.1126/science.288.5471.1613
Schultz, E. A., and Haughn, G. W. (1993). Genetic analysis of the floral initiation process (FLIP) in Arabidopsis. Development 119, 745–765.
Sim, G. E., Loh, C. S., and Goh, C. J. (2007). High frequency early in vitro flowering of Dendrobium Madame Thong-In (Orchidaceae). Plant Cell Rep. 26, 383–393. doi: 10.1007/s00299-006-0242-2
Song, I.-J., Fukuda, T., Ko, S.-M., Ito, T., Yokoyama, J., Ichikawa, H., et al. (2011). Expression analysis of an APETALA1/FRUITFULL-like gene in Phalaenopsis sp. ‘Hatsuyuki’ (Orchidaceae). Hortic. Environ. Biotechnol. 52, 183–195. doi: 10.1007/s13580-011-0199-0
Sun, L.-M., Zhang, J.-Z., Mei, L., and Hu, C.-G. (2014). Molecular cloning, promoter analysis and functional characterization of APETALA 1-like gene from precocious trifoliate orange (Poncirus trifoliata L. Raf.). Sci. Hortic. 178, 95–105. doi: 10.1016/j.scienta.2014.08.015
Teixeira da Silva, J. A., Aceto, S., Liu, W., Yu, H., and Kanno, A. (2014). Genetic control of flower development, color and senescence of Dendrobium orchids. Sci. Hortic. 75, 74–86. doi: 10.1016/j.scienta.2014.05.008
Theissen, G., Kim, J. T., and Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. doi: 10.1007/pl00006110
Wagner, D., Sablowski, R. W., and Meyerowitz, E. M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285, 582–584. doi: 10.1126/science.285.5427.582
Wang, Z., Wang, L., and Ye, Q. (2009). High frequency early flowering from in vitro seedlings of Dendrobium nobile. Sci. Hortic. 122, 328–331. doi: 10.1016/j.scienta.2009.05.018
Wigge, P. A., Kim, M. C., Jaeger, K. E., Busch, W., Schmid, M., Lohmann, J. U., et al. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. doi: 10.1126/science.1114358
Winterhagen, P., Tiyayon, P., Samach, A., Hegele, M., and Wünsche, J. N. (2013). Isolation and characterization of FLOWERING LOCUS T subforms and APETALA1 of the subtropical fruit tree Dimocarpus longan. Plant Physiol. Biochem. 71, 184–190. doi: 10.1016/j.plaphy.2013.07.013
Xiang, L., Li, X., Qin, D., Guo, F., Wu, C., Miao, L., et al. (2012). Functional analysis of FLOWERING LOCUS T orthologs from spring orchid (Cymbidium goeringii Rchb. f.) that regulates the vegetative to reproductive transition. Plant Physiol. Biochem. 58, 98–105. doi: 10.1016/j.plaphy.2012.06.011
Xu, Y., Teo, L. L., Zhou, J., Kumar, P. P., and Yu, H. (2006). Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J. 46, 54–68. doi: 10.1111/j.1365-313X.2006.02669.x
Yu, H., and Goh, C. J. (2000a). Differential gene expression during floral transition in an orchid hybrid Dendrobium Madame Thong In. Plant Cell Rep. 19, 926–931. doi: 10.1007/s002990000227
Yu, H., and Goh, C. J. (2000b). Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol. 123, 1325–1336. doi: 10.1104/pp.123.4.1325
Yu, H., and Goh, C. J. (2001). Molecular genetics of reproductive biology in orchids. Plant Physiol. 127, 1390–1393. doi: 10.1104/pp.010676
Yu, H., Ito, T., Wellmer, F., and Meyerowitz, E. M. (2004). Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat. Genet. 36, 157–161. doi: 10.1038/ng1286
Keywords: DOAP1, MADS-box gene, flowering time, flower development, orchid, Arabidopsis
Citation: Sawettalake N, Bunnag S, Wang Y, Shen L and Yu H (2017) DOAP1 Promotes Flowering in the Orchid Dendrobium Chao Praya Smile. Front. Plant Sci. 8:400. doi: 10.3389/fpls.2017.00400
Received: 21 January 2017; Accepted: 08 March 2017;
Published: 23 March 2017.
Edited by:
Prem Bhalla, University of Melbourne, AustraliaReviewed by:
Fernando Andres Lalaguna, Genetic Improvement and Adaptation of Mediterranean and Tropical Plants (INRA), FranceMaurizio Trovato, Sapienza University of Rome, Italy
Copyright © 2017 Sawettalake, Bunnag, Wang, Shen and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisha Shen, bGlzaGFAdGxsLm9yZy5zZw== Hao Yu, ZGJzeXVoYW9AbnVzLmVkdS5zZw==
 Nunchanoke Sawettalake
Nunchanoke Sawettalake Sumontip Bunnag
Sumontip Bunnag Yanwen Wang
Yanwen Wang Lisha Shen
Lisha Shen Hao Yu
Hao Yu