- 1Laboratory of Plant Pathology and Biotechnology, Kochi University, Kochi, Japan
- 2Research Institute of Molecular Genetics, Kochi University, Kochi, Japan
- 3Graduate School of Life and Environmental Sciences, Osaka Prefecture University, Osaka, Japan
A soil-borne bacterium Ralstonia solanacearum invading plant roots first colonizes the intercellular spaces of the root, and eventually enters xylem vessels, where it replicates at high levels leading to wilting symptoms. After invasion into intercellular spaces, R. solanacearum strain OE1-1 attaches to host cells and expression of the hrp genes encoding components of the type III secretion system (T3SS). OE1-1 then constructs T3SS and secrets effectors into host cells, inducing expression of the host gene encoding phosphatidic acid phosphatase. This leads to suppressing plant innate immunity. Then, OE1-1 grows on host cells, inducing quorum sensing (QS). The QS contributes to regulation of OE1-1 colonization of intercellular spaces including mushroom-type biofilm formation on host cells, leading to its virulence. R. solanacearum strains AW1 and K60 produce methyl 3-hydroxypalmitate (3-OH PAME) as a QS signal. The methyltransferase PhcB synthesizes 3-OH PAME. When 3-OH PAME reaches a threshold level, it increases the ability of the histidine kinase PhcS to phosphorylate the response regulator PhcR. This results in elevated levels of functional PhcA, the global virulence regulator. On the other hand, strains OE1-1 and GMI1000 produce methyl 3-hydroxymyristate (3-OH MAME) as a QS signal. Among R. solanacearum strains, the deduced PhcB and PhcS amino acid sequences are related to the production of QS signals. R. solanacearum produces aryl-furanone secondary metabolites, ralfuranones, which are extracellularly secreted and required for its virulence, dependent on the QS. Interestingly, ralfuranones affect the QS feedback loop. Taken together, integrated signaling via ralfuranones influences the QS, contributing to pathogen virulence.
Introduction
A soil-borne plant pathogenic bacterium Ralstonia solanacearum normally invades plant roots through wounds or natural openings. The pathogen first colonizes the intercellular spaces of the root, and eventually enters xylem vessels and spreads up into stems through the xylem (Figure 1A; Hikichi, 2016). Reduced sap flow caused by the presence of many bacterial cells and exopolysaccharide (EPS) slime produced by the bacteria in some xylem vessels leads to wilting symptoms (Figure 1B; Genin and Denny, 2012). Molecular traits such as EPS production in R. solanacearum infecting xylem vessels have been thus focused in its virulence mechanisms. On the contrary, colonization of intercellular spaces is required for R. solanacearum virulence (Hikichi, 2016), molecular traits regulating its colonization of intercellular spaces has remained unclear. In this mini review, we focus molecular traits regulating colonization of intercellular spaces of host plants in R. solanacearum, especially integrated intracellular/intercellular signaling with the quorum sensing (phc QS) involved in its virulence.
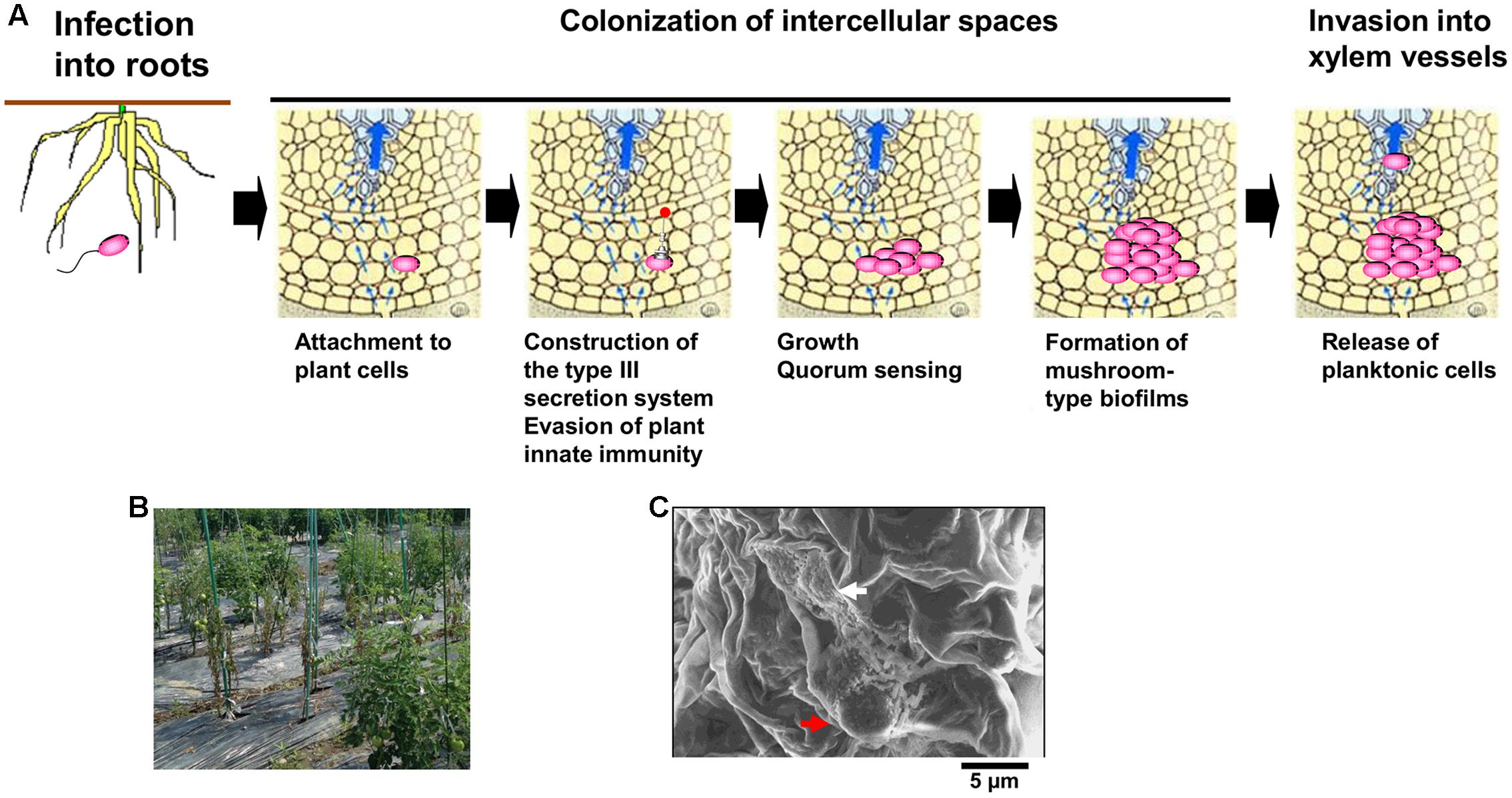
FIGURE 1. Infection route of Ralstonia solanacearum in intercellular spaces of roots (A), bacterial wilt of tomato plants caused by R. solanacearum in a field located in Kochi university, Japan (B), and microcolony (white-colored arrow) and the mushroom type-biofilm formed by cells of R. solanacearum strain OE1-1 (red-colored arrow) on tomato cells observed by the scanning electron microscope (C).
R. solanacearum Strain OE1-1 Produces Mushroom-Type Biofilms
After invading intercellular spaces, R. solanacearum strain OE1-1 first attaches to host cells and produces microcolonies following by mBFs (Figures 1A,C; Mori et al., 2016). The mBF formation is essential for colonization of intercellular spaces by OE1-1, leading to its virulence.
Regulation of hrp Genes
Ralstonia solanacearum has hrp genes encoding structural constituents of the type III secretion system (T3SS), which translocates effectors into host cells (Genin and Denny, 2012). This activation is sensed by the outer membrane receptor PrhA, which transduces signals through the PrhI and PrhR anti-sigma-sigma system and a complex regulatory cascade integrated by PrhJ, HrpG, and HrpB regulators (Figure 2A). A MarR family transcriptional regulator, PrhN, is also involved in positive regulation of T3SS (Zhang et al., 2015).
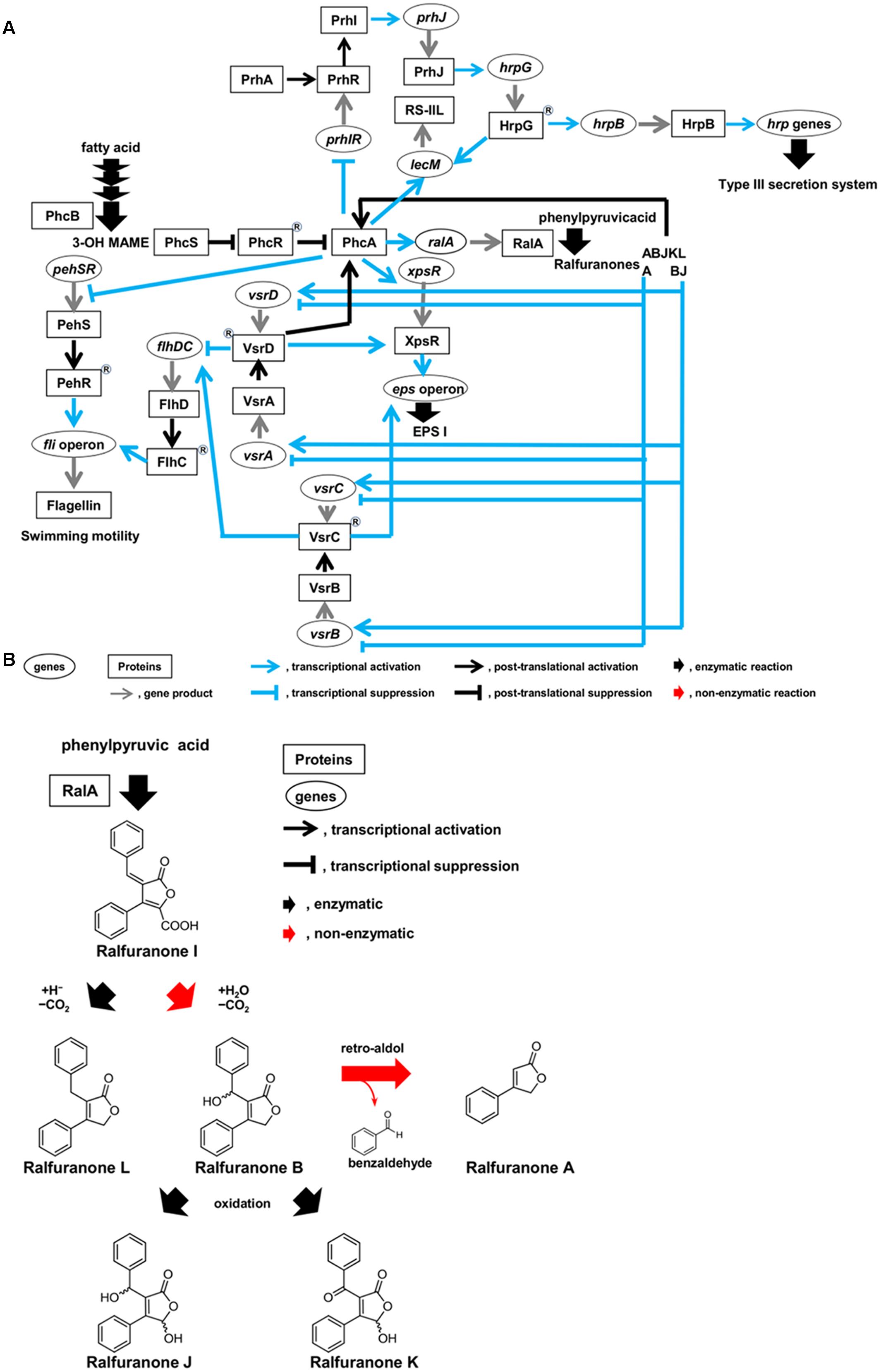
FIGURE 2. Circuits of molecular traits involved in colonization of intercellular spaces (A) and schematic diagram of synthesis route of ralfuranones (B) in R. solanacearum strain OE1-1 during activation of quorum sensing. R. solanacearum strain OE1-1 produces methyl 3-hydroxymyristate (3-OH MAME) as a quorum sensing signal by PhcB, a putative methyltransferase, and ralfuranone I as a precursor of the other ralfuranones by RalA, a tridomain non-ribosomal peptide synthetase-like furanone synthetase. Ralfuranone I is non-enzymatically converted into ralfuranone B in the supernatant. The non-enzymatic elimination of benzaldehyde from ralfuranone B produces ralfuranone A. Ralfuranones J and K are the products of enzymatic oxidation of ralfuranone B. Ralfuranone L is enzymatically synthesized from ralfuranone I.
After invading intercellular spaces, OE1-1 induces expression of lecM encoding a lectin RS-IIL by HrpG, leading to its attachment into host cells (Figure 2A; Mori et al., 2016). It is thus thought that OE1-1 synchronizes attachment into host cells with T3SS construction.
Evasion of Plant Innate Immunity by R. solanacearum Invading Intercellular Spaces of Host Plants
In tobacco plants, the invasion of intercellular spaces by R. solanacearum induces Sec14P-mediated phospholipid signaling which produces phosphatidic acid (PA) in chloroplast membranes (Kiba et al., 2012, 2014). The PA is involved in induction of the defense system mediated by jasmonic acid and reactive oxygen species. The expression of the gene encoding phosphatidic acid phosphatase (PAP) is induced in tobacco plants by invasion of the virulent R. solanacearum strain OE1-1, but not the avirulent strain 8107 (Nakano et al., 2013). The PAP dephosphorylates PA into diacylglycerol, interfering with the induction of jasmonic acid- and reactive oxygen-mediated plant innate immunity. The translocation of effectors into tobacco cells through the T3SS leads to decreased levels of PA, interfering with the induction of PA-mediated plant innate immunity and allowing OE1-1 to grow on tobacco cells (Figure 1A; Hikichi, 2016).
The Quorum Sensing of R. solanacearum
The expression of pathogenicity factors in R. solanacearum is controlled by a complex regulatory network that responds to environmental conditions, the presence of host cells, and bacterial density (Schell, 2000; Genin and Denny, 2012). At the center of this network is a LysR family transcriptional regulator, PhcA (Figure 2A; Brumbley et al., 1993), which coordinates the expression of several virulence factors including the major ESP, EPS I (Huang and Schell, 1995). The function of PhcA is regulated in response to cell density by the phc QS (Flavier et al., 1997).
After evading the innate immunity of host plants, R. solanacearum vigorously grows on host cells, inducing the phc QS (Figure 1A; Mori et al., 2016). The levels of functional PhcA via the phc QS are controlled by the phcBRS operon (Genin and Denny, 2012). The R. solanacearum strains AW1 and K60 produce methyl 3-hydroxypalmitate (3-OH PAME) as a QS signal (Flavier et al., 1997; Kai et al., 2015). Additionally, R. solanacearum strain OE1-1 produces methyl 3-hydroxymyristate (3-OH MAME) as a QS signal (Kai et al., 2015). These QS signals are synthesized by PhcB, a putative methyltransferase (Flavier et al., 1997; Kai et al., 2015). When the QS signals reach a threshold level, they induce the ability of the histidine kinase PhcS to phosphorylate the response regulator PhcR (Figure 2A; Schell, 2000; Genin and Denny, 2012; Papenfort and Bassler, 2016). It has been thought that the phosphorylation of PhcR reduced its binding activity to PhcA, resulting in elevated levels of functional PhcA. Therefore, cells of R. solanacearum at higher densities (>106 cfu/ml) have abundant functional PhcA and produce multiple virulence factors such as EPS I while suppressing the production of survival and invasion factors such as T3SS and swimming motility (Genin and Denny, 2012; Hikichi, 2016).
Phylogenetic trees constructed using amino acid sequences of PhcB and PhcS, but not PhcR, cluster 18 strains of R. solanacearum into two groups according to their QS signal types; 3-OH MAME or 3-OH PAME (Kai et al., 2015). The types of phc QS signals do not reflect the locations from which they were isolated, the phylotypes, or the host plants from which the strains were isolated, the races. Thus, the ancestors of R. solanacearum might have first coevolved the QS signal synthase (PhcB) and its receptor (PhcS), and then evolved the QS-dependent signaling for the adaptation to new and different environments.
Interestingly, functional PhcA suppresses expression of the prhIR operon, leading to the suppression of hrp gene expression (Figure 2A; Genin et al., 2005; Yoshimochi et al., 2009). The phc QS-deficient mutants lose their ability to colonize intercellular spaces and cannot invade xylem vessels and lose virulence (Mori et al., 2016), similar to hrp mutants (Hikichi, 2016). Furthermore, expression of lecM is also induced by functional PhcA through the phc QS (Meng et al., 2015; Mori et al., 2016). The lecM, of which expression is induced by functional PhcA, is involved in control development of mBFs, suggesting that the phc QS controls mBF formation by strain OE1-1 (Mori et al., 2016). Therefore, the phc QS dependent PhcA-mediated regulation allows R. solanacearum to control the elaborate and tunable regulation of its colonization of intercellular spaces, leading to its virulence.
Feedback Regulation of the phc QS by Ralfuranones
Ralstonia solanacearum synthesizes aryl-furanone secondary metabolites known as ralfuranones A, B, I, J, K, and L, which are extracellularly secreted (Figure 2B; Pauly et al., 2013; Kai et al., 2014). Ralfuranone I is a precursor of the other ralfuranones. The production of transaminase and furanone synthase, which are encoded by ralD and ralA, respectively, depends on functional PhcA via the phc QS system. Both the transaminase and furanone synthase are involved in the biosynthesis of ralfuranone I (Schneider et al., 2009; Wackler et al., 2011; Kai et al., 2014). Ralfuranone I is non-enzymatically converted into ralfuranone B in the supernatant (Kai et al., 2016). The non-enzymatic elimination of benzaldehyde from ralfuranone B produces ralfuranone A, while ralfuranones J and K are the products of enzymatic oxidation of ralfuranone B. Ralfuranone L is enzymatically synthesized from ralfuranone I. Thus, ralfuranone production is dependent on the phc QS system.
Ralfuranone productivity is involved in the full virulence of R. solanacearum strain OE1-1 (Kai et al., 2014). A ralfuranone-deficient mutant (ΔralA) exhibits significantly less EPS I production and significantly enhanced the swimming motility than strain OE1-1 (Mori et al., 2017). Quantitative real-time PCR assays reveal that ΔralA expresses phcB and phcA at levels similar to those in strain OE1-1. In contrast, R. solanacearum transcriptome data generated by RNA sequencing technology shows that ΔralA exhibits downregulated expression of more than 90% of QS-positively regulated genes including EPS I production-related genes, type VI secretion system-related genes, plant cell wall degradation enzyme genes (pme, egl, and pehC), acyl-homoserine lactones-two component system-related genes (solI and solR) and some effector genes secreted through T3SS (ripG4, ripG5, ripO1, ripTP5, ripS, and ripAU) (Mori et al., 2017). Furthermore, ΔralA exhibits upregulated expression of more than 75% of QS-negatively regulated genes including flagellar motility-related genes, T3SS-related genes, some effector genes secreted through T3SS (ripAX1, ripA2, ripAF1, ripX, ripAB, ripAC, ripS2, ripS3, ripQ, ripAZ1, and ripAD), and chemotaxis-related genes.
Ralfuranone supplementation restores EPS I production-related epsB expression, which is induced by the QS in strain OE1-1, in ΔralA, restoring its ability to aggregate dependently on EPS I production (Mori et al., 2017). Additionally, expression of flagellar motility-related fliC expression, which is negatively regulated by the phc QS, is suppressed in ΔralA by application of ralfuranones A and B, restoring its swimming motility to wild-type levels.
Together, post-translational regulation through ralfuranones may affect the QS feedback loop.
Regulation of the Two-Component Systems, VsrAd and VsrBc, By Ralfuranones
The PhcA and VsrAD two-component sensor/response regulatory systems are necessary for full activation of xpsR transcription (Figure 2A; Genin and Denny, 2012). Additionally, both the transcriptional regulator XpsR and the response regulator VsrC upregulate the expression of the eps operon (Huang et al., 1998; Garg et al., 2000). EPS I production is thus influenced by the VsrAD and VsrBC two-component systems. These two-component systems also regulate flagella biogenesis (Genin and Denny, 2012). Interestingly, ralfuranone A is involved in the negative regulation of vsrAD and vsrBC expression (Mori et al., 2017). In contrast, ralfuranones B and J positively regulate the expression of vsrA, vsrD, vsrB, and vsrC. Therefore, the integrated regulation of vsrAD and vsrBC expression by ralfuranones A, B, and J contributes to EPS I production and swimming motility. Furthermore, VsrAD is upstream of PhcA and is involved in the biosynthesis of ralfuranones (Schneider et al., 2009). Therefore, the expression of vsrAD may be feedback-regulated through ralfuranones A, B, and J, leading to the regulation of PhcA function.
Conclusion
Ralstonia solanacearum invading intercellular spaces of roots attaches to plant cells and constructs the T3SS, translocating effectors into plant cells through the T3SS and evading induction of plant innate immunity. R. solanacearum then grows on plant cells and activates the phc QS. The phc QS contributes to control of mBF formation by R. solanacearum on plant cells, leading to its colonization of intercellular spaces required for its virulence. R. solanacearum produces ralfuranones A, B, I, J, K, and L dependently on the phc QS, and the extracellular secretion of each ralfuranone by OE1-1 changes over time. During the early stages of infection, 3-OH MAME-mediated intercellular signaling activates the phc QS, leading to the production and secretion of ralfuranones. Each ralfuranone then mediates intercellular signaling between R. solanacearum cells in association with the feedback loop of the phc QS. The integrated intracellular/intercellular signaling of OE1-1 cells via each ralfuranone coupled with phc QS elaborately and tunably regulate molecular traits during colonization of intercellular spaces by R. solanacearum strain OE1-1, leading to its virulence (Figure 2A).
Author Contributions
YM, SI, and KH performed experiments and analyzed data. KO designed the research, analyzed data, and wrote the manuscript. AK designed the research. KK and YH designed the research, performed experiments, analyzed data, and wrote the manuscript.
Funding
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, the Towa Foundation for Food Science and Research, a research grant from Sumitomo Chemical Co. Ltd to YH, and a Sasakawa Scientific Research Grant from The Japan Science Society to YM.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Brumbley, S. M., Carney, B. F., and Denny, T. P. (1993). Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of PhcA, a putative LysR transcriptional regulator. J. Bacteriol. 175, 5477–5487. doi: 10.1128/jb.175.17.5477-5487.1993
Flavier, A. B., Clough, S. J., Schell, M. A., and Denny, T. P. (1997). Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26, 251–259. doi: 10.1046/j.1365-2958.1997.5661945.x
Garg, R. P., Huang, J., Yindeeyoungyeon, W., Denny, T. P., and Schell, M. A. (2000). Multicomponent transcriptional regulation at the complex promoter of the exopolysaccharide I biosynthetic operon of Ralstonia solanacearum. J. Bacteriol. 182, 6659–6666. doi: 10.1128/JB.182.23.6659-6666.2000
Genin, S., Brito, B., Denny, T. P., and Boucher, C. (2005). Control of the Ralstonia solanacearum type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 579, 2077–2081. doi: 10.1016/j.febslet.2005.02.058
Genin, S., and Denny, T. P. (2012). Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89. doi: 10.1146/annurev-phyto-081211-173000
Hikichi, Y. (2016). Interactions between plant pathogenic bacteria and host plants during the establishment of susceptibility. J. Gen. Plant Pathol. 82, 326–331. doi: 10.1007/s10327-016-0680-9
Huang, J., and Schell, M. (1995). Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol. Microbiol. 16, 977–989. doi: 10.1111/j.1365-2958.1995.tb02323.x
Huang, J., Yindeeyoungyeon, W., Garg, R. P., Denny, T. P., and Schell, M. A. (1998). Joint transcriptional control of XpsR, the unusual signal integrator of the Ralstonia solanacearum virulence gene regulatory network, by a response regulator and a LysR-type transcriptional activator. J. Bacteriol. 180, 2736–2743.
Kai, K., Ohnishi, H., Kiba, A., Ohnishi, K., and Hikichi, Y. (2016). Studies on the biosynthesis of ralfuranones in Ralstonia solanacearum. Biosci. Biotechnol. Biochem. 80, 440–444. doi: 10.1080/09168451.2015.1116931
Kai, K., Ohnishi, H., Mori, Y., Kiba, A., Ohnishi, K., and Hikichi, Y. (2014). Involvement of ralfuranone production in the virulence of Ralstonia solanacearum OE1-1. Chembiochem 15, 2590–2597. doi: 10.1002/cbic.201402404
Kai, K., Ohnishi, H., Shimatani, M., Ishikawa, S., Mori, Y., Kiba, A., et al. (2015). Methyl 3-hydroxymyristate, a diffusible signal mediating phc quorum sensing in Ralstonia solanacearum. Chembiochem 16, 2309–2318. doi: 10.1002/cbic.201500456
Kiba, A., Galis, I., Hojo, Y., Ohnishi, K., Yoshioka, H., and Hikichi, Y. (2014). SEC14 phospholipid transfer protein is involved in lipid signaling-mediated plant immune responses in Nicotiana benthamiana. PLoS ONE 9:e98150. doi: 10.1371/journal.pone.0098150
Kiba, A., Nakano, M., Vincent-Pope, P., Takahashi, H., Sawasaki, T., Endo, Y., et al. (2012). A novel Sec14 phospholipid transfer protein from Nicotiana benthamiana is upregulated in response to Ralstonia solanacearum infection, pathogen associated molecular patterns and effector molecules and involved in plant immunity. J. Plant Physiol. 169, 1017–1022. doi: 10.1016/j.jplph.2012.04.002
Meng, F., Babujee, L., Jacobs, J. M., and Allen, C. (2015). Comparative transcriptome analysis reveals cool virulence factors of Ralstonia solanacearum race 3 biovar 2. PLoS ONE 10:e0139090. doi: 10.1371/journal.pone.0139090
Mori, Y., Inoue, K., Ikeda, K., Nakayashiki, H., Higashimoto, C., Ohnishi, K., et al. (2016). The vascular plant-pathogenic bacterium Ralstonia solanacearum produces biofilms required for its virulence on the surfaces of tomato cells adjacent to intercellular spaces. Mol. Plant Pathol. 17, 890–902. doi: 10.1111/mpp.12335
Mori, Y., Ohnishi, H., Shimatani, M., Morikawa, Y., Ishikawa, S., Ohnishi, K., et al. (2017). Involvement of ralfuranones in the quorum sensing signaling pathway and virulence of Ralstonia solanacearum strain OE1-1. Mol. Plant Pathol. [Epub ahead of print]. doi: 10.1111/mpp.12537
Nakano, M., Nishihara, M., Yoshioka, H., Takahashi, H., Sawasaki, T., Ohnishi, K., et al. (2013). Suppression of DS1 phosphatidic acid phosphatase confirms resistance to Ralstonia solanacearum in Nicotiana benthamiana. PLoS ONE 8:e75124. doi: 10.1016/j.jplph.2015.06.007
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Pauly, J., Spiteller, D., Linz, J., Jacobs, J., Allen, C., Nett, M., et al. (2013). Ralfuranone thioether production by the plant pathogen Ralstonia solanacearum. Chembiochem 14, 2169–2178. doi: 10.1002/cbic.201300364
Schell, M. A. (2000). Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu. Rev. Phytopathol. 38, 263–292. doi: 10.1146/annurev.phyto.38.1.263
Schneider, P., Jacobs, J. M., Neres, J., Aldrich, C. C., Allen, C., Nett, M., et al. (2009). The global virulence regulators VsrAD and PhcA control secondary metabolism in the plant pathogen Ralstonia solanacearum. Chembiochem 10, 2730–2732. doi: 10.1002/cbic.200900510
Wackler, B., Schneider, P., Jacobs, J. M., Pauly, J., Allen, C., Nett, M., et al. (2011). Ralfuranone biosynthesis in Ralstonia solanacearum suggests functional divergence in the quinone synthetase family of enzymes. Chem. Biol. 18, 354–360. doi: 10.1016/j.chembiol.2011.01.010
Yoshimochi, T., Hikichi, Y., Kiba, A., and Ohnishi, K. (2009). The global virulence regulator PhcA negatively controls the Ralstonia solanacearum hrp regulatory cascade by repressing expression of the PrhIR signaling proteins. J. Bacteriol. 191, 3424–3428. doi: 10.1128/JB.01113-08
Zhang, Y., Luo, F., Wu, D., Hikichi, Y., Kiba, A., Igarashi, Y., et al. (2015). PrhN, a putative marR family transcriptional regulator, is involved in positive regulation of type III secretion system and full virulence of Ralstonia solanacearum. Front. Microbiol. 6:357. doi: 10.3389/fmicb.2015.00357
Keywords: Ralstonia solanacearum, colonization of intercellular spaces, quorum sensing, methyl 3-hydroxymyristate, methyl 3-hydroxypalmitate, ralfuranones, virulence
Citation: Hikichi Y, Mori Y, Ishikawa S, Hayashi K, Ohnishi K, Kiba A and Kai K (2017) Regulation Involved in Colonization of Intercellular Spaces of Host Plants in Ralstonia solanacearum. Front. Plant Sci. 8:967. doi: 10.3389/fpls.2017.00967
Received: 27 February 2017; Accepted: 23 May 2017;
Published: 08 June 2017.
Edited by:
Nemo Peeters, INRA Centre Occitanie-Toulouse, FranceReviewed by:
Hai-Lei Wei, Cornell University, United StatesMunusamy Madhaiyan, Temasek Life Sciences Laboratory, Singapore
Copyright © 2017 Hikichi, Mori, Ishikawa, Hayashi, Ohnishi, Kiba and Kai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yasufumi Hikichi, eWhpa2ljaGlAa29jaGktdS5hYy5qcA==
 Yasufumi Hikichi
Yasufumi Hikichi Yuka Mori
Yuka Mori Shiho Ishikawa1
Shiho Ishikawa1 Kouhei Ohnishi
Kouhei Ohnishi Kenji Kai
Kenji Kai