- 1Hainan Key Laboratory for Sustainable Utilization of Tropical Bioresources, Institute of Tropical Agriculture and Forestry, Hainan University, Haikou, China
- 2State Key Laboratory of Hybrid Rice, College of Life Sciences, Wuhan University, Wuhan, China
Rubber trees (Hevea brasiliensis) were successfully introduced to south China in the 1950s on a large-scale; however, due to the climate, are prone to cold injury during the winter season. Increased cold tolerance is therefore an important goal, yet the mechanism underlying rubber tree responses to cold stress remains unclear. This study carried out functional characterization of HbICE1 (Inducer of CBF Expression 1) from H. brasiliensis. A nucleic protein with typical features of ICEs, HbICE1 was able to bind to MYC recognition sites and had strong transactivation activity. HbICE1 was constitutively expressed in all tested tissues, with highest levels in the bark, and was up-regulated when subjected to various stresses including cold, dehydration, salinity and wounding. When overexpressed in Arabidopsis, 35S::HbICE1 plants showed enhanced cold resistance with increased proline content, reduced malondialdehyde (MDA) metabolism and electrolyte leakage, and decreased reactive oxygen species (ROS) accumulation. Expression of the cold responsive genes (COR15A, COR47, RD29A, and KIN1) was also significantly promoted in 35S::HbICE1 compared to wild-type plants under cold stress. Differentially expressed genes (DEGs) analysis showed that cold treatment changed genes expression profiles involved in many biological processes and phytohormones perception and transduction. Ethylene, JA, ABA, as well as ICE-CBF signaling pathways might work synergistically to cope with cold tolerance in rubber tree. Taken together, these findings suggest that HbICE1 is a member of the ICE gene family and a positive regulator of cold tolerance in H. brasiliensis.
Introduction
Cold stress is one of the most devastating environmental factors adversely affecting plant growth and development, significantly constraining geographic distribution and agricultural productivity. Cold stress interferes with various physiological and biochemical processes via direct inhibition of metabolic reactions and indirect induction of osmotic, oxidative and other stresses. Hevea brasiliensis, with its high rate of production and superior rubber quality, is the sole commercial source of natural rubber. A perennial tropical tree species originating from the Amazonian forests of Brazil, rubber trees were traditionally planted within a restricted region between 15° north and 15° south latitudes. Though rubber trees were successfully introduced to south China in the 1950s on a large-scale, they are prone to cold injury during the winter season. Cold stress not only affects rubber production, but also threatens the survival of rubber trees in China. Increased cold tolerance is therefore a major aim of rubber tree breeding programs.
Plants have evolved various physiological, biochemical and molecular strategies aimed at adaptation to adverse situations (Nakashima et al., 2009; Thomashow, 2010; Theocharis et al., 2012). In the past decade, significant progress has been made in deciphering the key components of the cold signaling pathway in the model plants Arabidopsis and other species (Wisniewski et al., 2014; Shi Y. et al., 2015). In Arabidopsis, three C-repeat-binding factors (CBFs), CBF1, CBF2, and CBF3, have been functionally characterized (Jaglo-Ottosen et al., 1998; Medina et al., 1999). Under low temperatures, cold stress rapidly and transiently induces CBF expression, stimulating expression of cold-responsive (COR) genes by binding to C-repeat binding factor (CRT)/dehydration-responsive element (DRE) cis-elements in the promoters of COR genes, thereby increasing accumulation of proline and total sugar and protecting membranes and proteins from damage (Stockinger et al., 1997; Liu et al., 1998; Thomashow, 1999; Chinnusamy et al., 2007; Maruyama et al., 2012; Shi Y. et al., 2015). ICE1 (inducer of CBF expression 1), a constitutively expressed MYC-like bHLH transcriptional activator, also functions during cold acclimation by inducing CBF expression via binding of MYC recognition elements in CBF promoters (Chinnusamy et al., 2003, 2010; Fursova et al., 2009). The mutant ice1 was found to exhibit reduced plant tolerance to chilling and freezing stresses due to repression of AtCBF3 expression and subsequent decreases in expression of various downstream COR genes (Chinnusamy et al., 2003; Fursova et al., 2009). In contrast, plants overexpressing either AtICE1 or AtICE2 display improved freezing tolerance via enhanced AtCBF3 and AtCBF1 expression(Chinnusamy et al., 2003; Fursova et al., 2009). Thus, the ICE-CBF-COR transcriptional regulatory cascade is a well-established plant response to cold stress (Chinnusamy et al., 2007; Shi Y. et al., 2015). Cold acclimation, which is thought to enhance freezing tolerance after exposure to low temperatures (Thomashow, 1999), is one of the major mechanisms of plant adaptation to cold stress. But whether cold acclimation can affect the cold resistance of rubber tree has not been reported so far. Besides transcriptional regulation of ICE gene, several important components have been found to regulate cold acclimation by modulating ICE-CBF pathway at posttranslational levels. The small ubiquitin-related modifier (SUMO) E3 ligase, SIZ1 (SAP and Miz 1) and the RING finger E3 ligase HOS1 (high expression of osmotically responsive genes1) have been shown to modify ICE1 posttranslationally and function in the ICE-CBF/DREB1 signaling pathway (Dong et al., 2006; Miura et al., 2007). Recent reports further suggest that ICE1 is also regulated by various other factors such as jasmonate signaling proteins JAZ 1/4 (JASMONATE ZIM-DOMAIN 1/4), which inhibit ICE1 transcriptional activity (Hu et al., 2013). Moreover, Ding et al. (2015) discovered that the protein kinase OST1 (open stomata 1), a key component in ABA signaling, interacts with and phosphorylates ICE1 protein under cold stress, stabilizing and activating ICE1 and thereby enhancing plant tolerance to freezing temperatures. ICE1 is also degraded by the E3 ligase HOS1 (high expression of osmotically responsive gene 1)-mediated 26S-proteasome pathway (Dong et al., 2006). More recently, Huang X. S. et al. (2015) reported that ICE1 from Poncirus trifoliate functions in cold tolerance by altering polyamine accumulation via interaction with arginine decarboxylase. ICE1 is therefore not only a central component in cold signaling, but also serves as a convergence point, integrating signals to regulate cold tolerance in plants.
Studies on cold stress in rubber trees were mainly focused on changes in physiological parameters and alleviating injury after cold stress, the major components of cold signaling of rubber tree were nearly unidentified except HbCBF1and the molecular mechanisms underlying how rubber tree responses to cold stress were still very poorly understood (Cheng et al., 2015). In this study, the H. brasiliensis ICE1 (HbICE1) gene was cloned and functionally characterized for its role in cold tolerance, revealing its location in the nucleus and ability to bind to MYC recognition sites. Overexpression of 35S::HbICE1 in Arabidopsis enhanced cold resistance probably due to increased proline level, reduced MDA content and electrolyte leakage, and decreased reactive oxygen species (ROS) accumulation under cold stress. Furthermore, the cold responsive genes (COR15A, COR47, RD29A, and KIN1) were significantly activated in overexpressing plants compared to the wild-type (WT) under cold conditions. Taken together, these data suggest that HbICE1 is a functional member of the ICE gene family, playing a positive role in cold resistance in H. brasiliensis.
Materials and Methods
Plant Material and Treatments
Reyan 7-33-97 rubber trees (H. brasiliensis) cultivated at the experimental plantation of Hainan University, Hainan Province, China, were used in this study. The plants were pruned annually. To examine tissue-specific expression of HbICE1, samples from six tissues (the latex, leaves, stem, bark, stamen and pistil) were collected for RNA extraction from 17-year-old mature trees tapped for the previous two years. To determine expression of HbICE1 in response to NaCl and dehydration treatment, four batches of 12 seedlings per treatment were selected. Three batches were treated with 200 mM NaCl or 10% polyethylene glycol (PEG), and one with ddH2O then the leaves collected for RNA isolation. For cold treatment, seedlings were transferred to a culture room at 4°C under a 12 h light/12 h dark cycle with 80% humidity for 1, 3, 6, 12, or 24 h then the leaves collected for RNA isolation. For wounding treatment, leaves of 5-year-old mature virgin (untapped) trees were wounded with a hemostat then the leaves collected for RNA extraction.
To generate 35S::GFP-HbICE1 transgenic Arabidopsis plants, the coding region of HbICE1 was amplified using primers 5′-gaattcATGCTTGATACCGACTGGTATGATA-3′and 5′-gaattcTCACATCATTCCATGAAAGCCAGCT-3′. The coding sequence fragment was then subcloned into the ECOR1 site of the pEGAD vector and the recombinant plasmid introduced into Arabidopsis ecotype Col-0 via Agrobacterium tumefaciens (GV3101)-mediated transformation using the floral dip method (Clough and Bent, 1998). Arabidopsis plants were sown in vermiculite in pots at 23°C under 16 h light/8 h dark conditions with 75% humidity and a light intensity of 150 mmol m−2 s−1.
To determine cold tolerance of the transgenic plants, a cold treatment assay was performed as described previously with slight modifications (Ding et al., 2015). Fourteen-day-old seedlings cultivated in 1/2 MS medium at 23°C were transferred to 4°C for 2 days then exposed to −8°C for 4 h and subsequently returned to normal conditions. Survival rates were measured after 7 days. To determine cold tolerance of the rubber tree seedlings, seedlings with or without cold acclimation (1 day at 4°C) were exposed to −16°C for 0.5 and 1 h and subsequently returned to normal conditions. The freezing treatment experiment was performed in triplicate. For Arabidopsis, T3 or T4 homozygous transgenic plants were used in this study.
Isolation and Bioinformatics Analysis of HbICE1
The CDS of HbICE1 was predicted using Bioedit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and confirmed using BLASTP on the NCBI BLAST server (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The detailed Insilico cloning procedure was carried out as previously described (Hong et al., 2015). Molecular weights (MW) and isoelectric points (pI) were predicted using ExPASy (http://www.expasy.org/tools), nuclear localization signals were predicted using the online server (http://www.predictprotein.org/), and protein secondary domains were predicted with a Motif scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan). Sequence alignment was performed using DNAMAN software and a phylogenetic tree constructed using MEGA 5.1 software.
Subcellular Localization of HbICE1
The full-length CDS of HbICE1 was fused to the C terminus of the green fluorescent protein (GFP) of the pEGAD vector, driven by the 35S promoter. GFP in the roots of homozygote transgenic Arabidopsis 35S::GFP-HbICE1 plants was examined using confocal microscopy (excitation and emission: 488 and 515 nm, respectively; 22°C).
Transcriptional Activation Assay and MYCR-Binding Assay of HbICE1
For the transcriptional activation assay, the open reading frame (ORF) of HbICE1 was amplified by PCR using primers P1 (cgGAATCCatgcttgataccgactggta) and P2 (aaCTGCAGcatcattccatgaaagccag). The coding sequence fragment was then subcloned into the EcoRI and PstI restriction sites of the pGBKT7 vector, giving pGBKT7-HbICE1. The recombinant vector pGBKT7-HbICE1, as well as pGBKT7-53+pGADT7-T (positive control) and pGBKT7 (negative control), were transformed into yeast strain AH109 according to the manufacturer's instructions (Clontech, PT4084-1). The transformed yeast was placed on plates containing SD/-Trp-Leu/X-α-Gal/Aureobasidin A, SD/-Trp-Leu, SD/-Trp, SD/-Trp-His or SD/-Trp-His-Ade medium then incubated at 28°C for 3–4 days for analysis of transformant growth.
To investigate whether HbICE1 was able to bind to the MYC recognition sequence, a yeast one–hybrid experiment (Y1H) was performed according to the manufacturer's instructions (Clontech). The ORF of HbICE1 was fused to the pGADT7 vector digested with EcoRI to create pGADT7-HbICE1. A 66-bp DNA fragment containing triple tandem repeats of the sequence containing the MYC recognition sequence (CACATG) was inserted into the pHIS2 vector, generating the recombinant vector pHIS2-MYCR. pHIS2-MYCR was then transformed into yeast strain Y187 on plates containing SD/-Trp, SD/-Trp/-His or SD/-Trp/-His/10mM 3-AT medium to verify autoactivation of pHIS2-MYCR. Both pGADT7-HbICE1 and pHIS2-MYCR were subsequently co-transformed into Y187 to verify the DNA sequence MYCR and HbICE1 protein interactions. The transformed yeast strains were placed on plates containing SD/-Trp/-Leu, SD/-Trp/-Leu/-His, or SD/-Trp/-Leu/-His/10mM 3-AT medium then incubated at 28°C for 3–4 days for analysis of transformant growth.
RNA Extraction and Quantitative Real-Time PCR
Hevea RNA was extracted from leaves of H. brasiliensis as described previously (Xia et al., 2011). Arabidopsis RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the instruction manual. All RNA samples were treated with RNase-free DNase I (Promega, Madison, WI, USA) to digest genomic DNA. The concentration and quality of DNaseI-treated total RNA was determined by spectrophotometry and agarose gel electrophoresis. As a template for first-strand cDNA synthesis, 2 μg DNase I-treated RNA was used according to the manufacturer's instructions (RevertAid First Stand cDNA Synthesis kit; Fermentas, Vilnius, Lithuania). Quantitative real-time PCR (qRT-PCR) was carried out using ABI-7500 Real-Time PCR apparatus with SYBR Green I dye (Takara, Tokyo, Japan) as follows: 95°C for 3 min followed by 40 cycles of 95°C for 20 s, 58°C for 15 s and 72°C for 20 s. The efficiency of each primer pair was evaluated prior to PCR using the primers listed in Table S1. Relative levels were calculated as 2−ΔΔCT. Each biological sample was performed with three technical repetitions, and data analysis carried out using three independent biological replicates. The whole process from plant material treatment, RNA extraction, cDNA synthesis to qRT-PCR was repeated three times per treatment as three independent biological replicates. Values were statistically analyzed by ANOVA or the Student's t-test.
Analysis of Malondialdehyde (MDA) and Proline Contents, and Electrolyte Leakage
Three-week-old seedlings of transgenic 35S::HbICE1 and WT Arabidopsis were subjected to cold stress treatment then leaves harvested to determine MDA and proline contents, and the degree of electrolyte leakage. Proline accumulation was determined as described previously (Shi H. T. et al., 2012). Briefly, 0.25 g leaf samples were harvested and extracted in 3% sulfosalicylic acid then centrifuged at 12,000 × g for 10 min. The supernatant (2 mL) was incubated with 2 mL ninhydrin reagent [2.5% (w/v) ninhydrin, 60% (v/v) glacial acetic acid, and 40% 6M phosphoric acid] and 2 mL glacial acetic acid at 100°C for 40 min, and the reaction terminated in an ice bath. MDA levels were measured using the thio-barbituric acid (TBA) method as described previously (Cai et al., 2015). Electrolyte leakage was measured as described previously (Lin et al., 2012). Eight leaves from transgenic and WT plants treated with and without cold treatment were placed in a bottle with 40 mL ddH2O2, shaken at 120 rpm for 3 h then conductivity (C1) measured using an ion leakage meter. Conductivity (C2) was measured after boiling the leaves for 30 min and shaking for 1 h. Electrolyte leakage was calculated as (C1/C2) × 100%.
Screening of Differentially Expressed Genes (Degs) Based on RNA-Seq
Total RNA was isolated from the rubber tree seedlings treated at 4°C for 3 and12 h, including three biological repeats for each condition. RNA-Seq was performed by the Beijing Genomics Institute (Shenzhen, China). Oligo (dT) magnetic beads were used to select mRNA with polyA tail, followed by DNase I reaction to remove DNA probe. The purified mRNA was used reverse transcription to double-strand cDNA (dscDNA) by N6 random primer. End of dscDNA was repaired with phosphate at 5′ end and stickiness “A” at 3′ end, then ligated and with adaptor. Two specific primers of adaptor were used to amplify the ligation product. The PCR product was denatured by heat and the single strand DNA was cyclized by splint oligo and DNA ligase. The prepared library was sequenced by (Illumina HiSeqTM 2000).
Clean reads were mapped to the hevea contigs assembly using SOAPaligner/soap2 mismatches; no more than 2 bases were allowed in the alignment. The number of clean reads for each gene was calculated and then normalized to Reads Per Kb per Million reads (RPKM), which associates the read number with the gene expression levels. Further, deep analysis were performed based on DEGs, including Gene Ontology (GO) enrichment analysis, KEGG pathway enrichment analysis, cluster analysis, protein-protein interaction network analysis and finding transcription factor.
Results
Cloning and Bioinformatics Analysis of HbICE1
Plant ICE1-like genes play a critical role in cold tolerance in a number of different plants; however, the ICE1 gene has yet to be identified in H. brasiliensis. To clone the HbICE1 gene from H. brasiliensis, the A. thaliana ICE1 protein sequence was used as a query sequence in a blast search against the Hevea EST and nucleotide database. The matching sequence with the lowest E-value was subsequently selected as a backbone for further insilico assembly of full-length cDNA. The predicted full-length HbICE1 cDNA contained an ORF of 1410 bps, as validated by PCR amplification and sequencing. The predicted ORF encoded a protein of 469 amino acid residues with an estimated MW of 51 kDa and a pI of 5.30. Multiple alignments of HbICE1 and ICE1 proteins from other plants indicated that the C terminus of all ICE1 proteins tested was highly conserved, whereas the N terminus varied (Figure 1). Furthermore, HbICE1 contained a MYC-like basic helix-loop-helix (bHLH) domain, a serine-rich region (S-rich), a zipper region (ZIP) and an ACT-UUT-ACR like domain, all of which are typical features of ICE proteins (Figure 1). A phylogenetic tree was reconstructed using the deduced amino acid sequence of HbICE1 and other plant ICEs, revealing that HbICE1 is most closely related to JcICE1 of Jatrophacurcas (Figure 2).
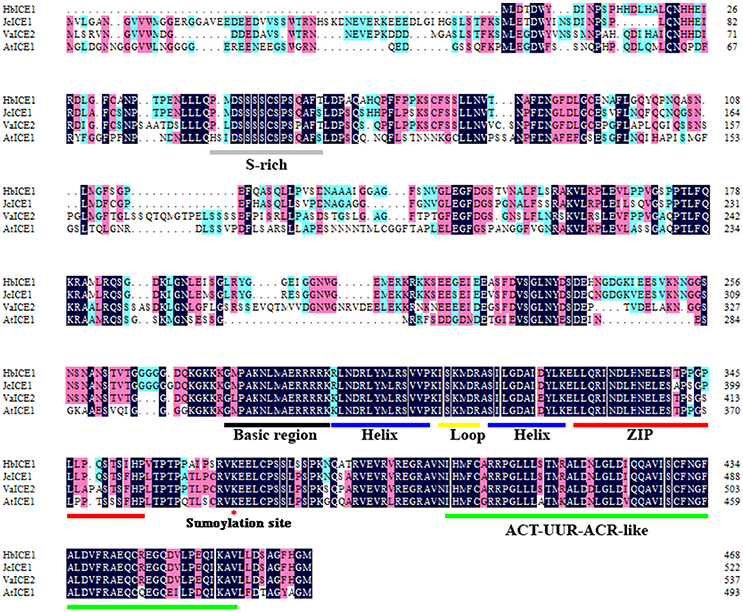
Figure 1. Amino acid sequence alignment of HbICE1 and ICEs from other plant species. Sequences and accession numbers are as follows: Jatrophacurcas (NP_001306848.1), Vitisamurensis (AGP04218.1) and Arabidopsis thaliana (NP_189309.2). Blue and pink backgrounds indicate identical and similar residues, respectively. The S-rich motif, basic-helix-loop-helix-leucine zipper (bHLH-ZIP) region, ACT-UUR-ACR-like domain, and sumoylation site are labeled.
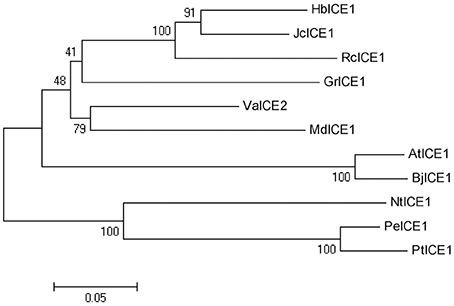
Figure 2. Phylogenetic analysis of HbICE1 and ICEs from other plant species. The neighbor-joining method was used to construct the tree. GenBank accessions of the predicted ICE protein sequences used are as follows: JcICE1 (NP_001306848.1), VaICE1 (AGP04218.1), AtICE1 (NP_189309.2), RcICE1 (XP_015570780.1), GrICE1 (XP_012489464.1), MdICE1 (XP_008379053.1), PeICE1 (XP_011040262.1), PtICE1 (XP_002318166.1), NtICE1 (XP_009625133.1) and BjICE1 (AEB97375.2).
HbICE1 Is Nuclear-Localized and Has Transactivation Activity in Yeast
As a possible transcriptional factor, HbICE1 should be located in the nucleus for transcription regulation. To verify this, we created a construct expressing the GFP-HbICE1 fusion protein by fusing GFP in-frame to the 5′ end of the HbICE1 ORF under control of the cauliflower mosaic virus 35S promoter then transforming the construct into WT Arabidopsis using the floral dip method. Following selection by Basta resistance, independent transgenic lines were selected for verification by genomic PCR and qRT-PCR analysis. QRT-PCR data revealed high expression of HbICE1 in six lines (L1, L2, L3, L6, L9, and L10), three of which (L1, L3, and L6) showing highest transcript levels were selected for further analysis (Figure 3A). Roots of homozygote 35S::GFP-HbICE1 plants were examined using confocal microscopy revealing exclusive expression of the GFP-HbICE1 protein in the nucleus (Figure 3B). This finding confirmed that HbICE1 is a nuclear protein, consistent with a previous report suggesting that Arabidopsis ICE1 is a transcription factor (Chinnusamy et al., 2003).
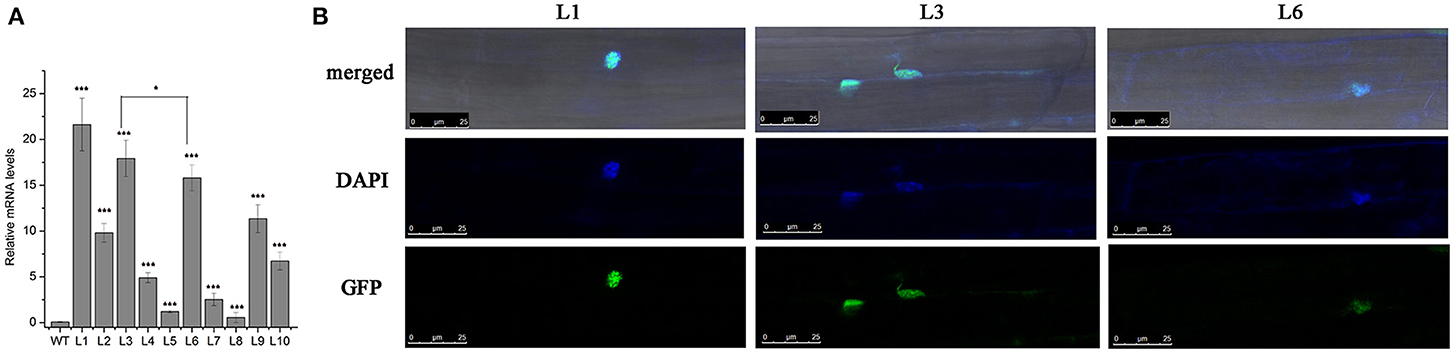
Figure 3. Subcellular localization of the HbICE1 protein. (A) Confirmation of HbICE1 expression in selected transgenic lines. mRNA levels of 35S::GFP-HbICE1 lines and the wild–type (WT) were examined by qRT–PCR analysis, using AtEIF4 as the internal standard. (B) Representative images show root cells of 5-day-old 35S::GFP-HbICE1 transgenic Arabidopsis seedlings. DAPI was used to stain nuclei (pseudo-color, blue).
Next, we assayed transactivation activity of HbICE1, another important feature of transcription factors. As shown in Figure 4A, yeast transformed with both pGBKT7-53 and pGADT7-T (CK+) grew well on SD/-Trp-Leu and SD/-Trp-Leu/X-α-Gal/Aureobasidin A medium. Moreover, while yeast cells transformed with pGBKT7-HbICE1 grew well on SD/-Trp medium, they grew normally on SD/-Trp-His-Ade medium, exhibiting fairly strong β-galactosidase activity. In contrast, yeast cells transformed with the negative control (pGBKT7) did not grow on SD/-Trp-His medium (Figure 4A). These results further confirm that HbICE1 has transcriptional activation activity.
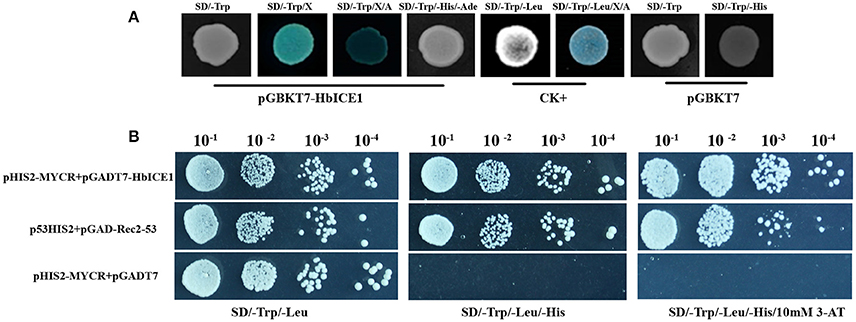
Figure 4. Transcriptional activation assay of HbICE1. (A) Growth of AH109 yeast cells transformed with vectors of the positive control (pGBKT7-53 and pGADT7-T, CK+), negative control (pGBKT7), and pGBKT7-HbICE1 on various selection media. SD/-Trp-Leu/X-α-Gal/Aureobasidin A medium is indicated by SD/-Trp-Leu/X/A. (B) Growth of Y187 yeast cells co-transformed with vectors of the positive control (p53HIS2+pGAD-Rec2-53), negative control (pHIS2-MYCR+pGADT7), and pGADT7-HbICE1 with pHIS2-MYCR on SD/-Leu-Trp and SD/-Leu-Trp-His selection medium with and without 10 mM 3-AT.
Since Arabidopsis ICE1 can bind to the MYC-recognition element (Chinnusamy et al., 2003), we also explored whether HbICE1 could bind to a sequence containing the MYC recognition element using a yeast one hybrid (Y1H) assay. As shown in Figure 4B, yeast cells co-transformed with pHIS2-MYCR and pGADT7-HbICE1 grew as well as those containing the positive control (p53HIS2+pGAD-Rec2-53) on SD/-Trp-Leu-His medium with and without 10 mM 3-AT. In contrast, cells co-transformed with the negative control (pHIS2MYCR+pGADT7) did not grow under this condition, further suggesting that HbICE1 binds to MYC recognition sites, activating the transcription of report genes in yeast.
Analysis of HbICE1 Expression Patterns
To investigate tissue-specific expression of HbICE1, HbICE1 expression in the latex, leaves, stem, bark, stamen and pistil was examined by qRT-PCR. HbICE1 was universally expressed in all tissues tested with the highest expression in bark (Figure 5A). We also examined expression profiles of HbICE1 under various abiotic stresses including cold, dehydration, wounding, and salinity. HbICE1 was significantly induced at 3 h after cold treatment, but expression decreased to a low level at 24 h (Figure 5B). Under dehydration, HbICE1 mRNA was markedly induced by nearly 4.5-fold at 6 h then quickly returned to the normal level at 12 h post treatment (Figure 5C). After wounding treatment, the HbICE1 transcripts showed strong induction at 1 and 3 h (Figure 5D), while under salt stress, expression was induced 3 h after treatment (Figure 5E). These results indicate that HbICE1 expression is regulated during plant responses to multiple abiotic stresses.
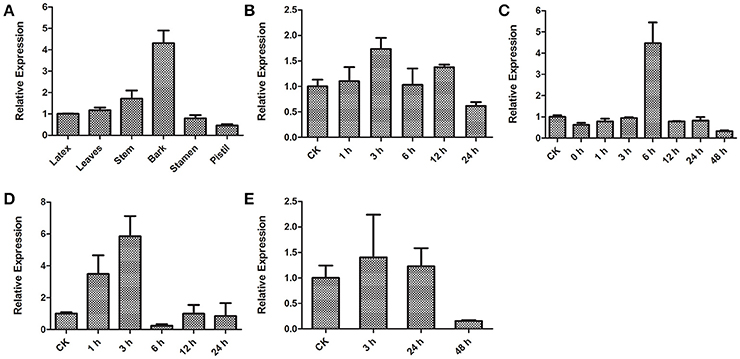
Figure 5. Transcription patterns of HbICE1 determined by qRT-PCR. (A) Differential expression of HbICE1 in various tissues (the latex, leaves, stem, bark, stamen and pistil). Time-course expression patterns of HbICE1 in response to different biotic and abiotic stresses: cold (B), dehydration (C), wounding (D), and salt stress (E).
Over-Expression of HbICE1 in Arabidopsis Increases Cold Tolerance
To further determine the role of HbICE1 in cold tolerance, HbICE1-overexpressing lines and WT plants were subjected to freezing treatment. As shown in Figure 6A, HbICE1-overexpressing lines displayed less freezing damage and increased survival ratecompared with the WT after 1-week recovery. The survival rate of WT plants was only 2.8%, significantly lower than that of the transgenic lines (77.1% for L1, 55.6% for L3, and 22.2% for L6; Figure 6B), suggesting that overexpression of HbICE1 conferred cold tolerance in transgenic Arabidopsis.
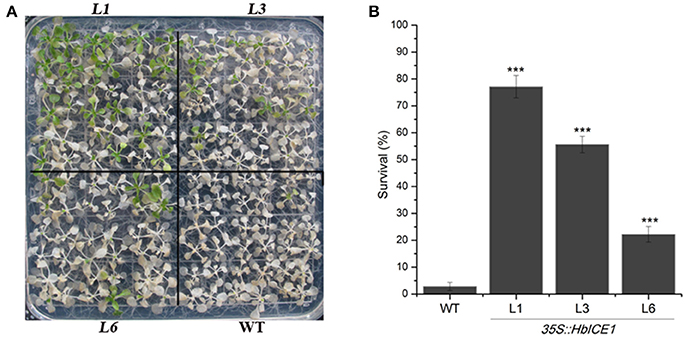
Figure 6. Overexpression of HbICE1 confers enhanced cold tolerance in Arabidopsis. Freezing phenotypes (A) and survival rates (B) of transgenic lines L1, L3, and L6 and the WT. Two-week-old Arabidopsis plants were transferred to 4°C for 2 days, exposed to −8°C for 4 h then returned to normal conditions. Photographs were taken before and after 7-d recovery. In (B), data represent the means of three replicates ± SD. Asterisks indicate significant differences compared with the WT under the same treatment condition (***P < 0.005, student's t-test).
Over-Expression of HbICE1 Affects Proline Content, Electrolyte Leakage, MDA Metabolism and H2O2 Accumulation under Cold Stress
Many physiological parameters such as proline content, electrolyte leakage, MDA metabolism and ROS accumulation are known indicators of tolerance to a wide variety of abiotic stresses (Feng et al., 2012; Xu et al., 2014; Cai et al., 2015; Huang X. S. et al., 2015). Thus, these physiological parameters were subsequently examined in HbICE1-overexpressing lines under cold stress. Proline accumulation is considered an adaptive response to various kinds of environmental stress, conferring stress tolerance by promoting osmotic adjustment, protecting membranes and proteins, and inhibiting ROS production. Under normal growth conditions, the proline content of the HbICE1-overexpressing lines was similar to that of the WT. However, after 24 and 48 h at 0°C, levels were significantly higher in the transgenic lines compared to the WT. At 48 h after treatment, proline levels were 2-fold higher in the transgenic compared to the WT plants (Figure 7A).
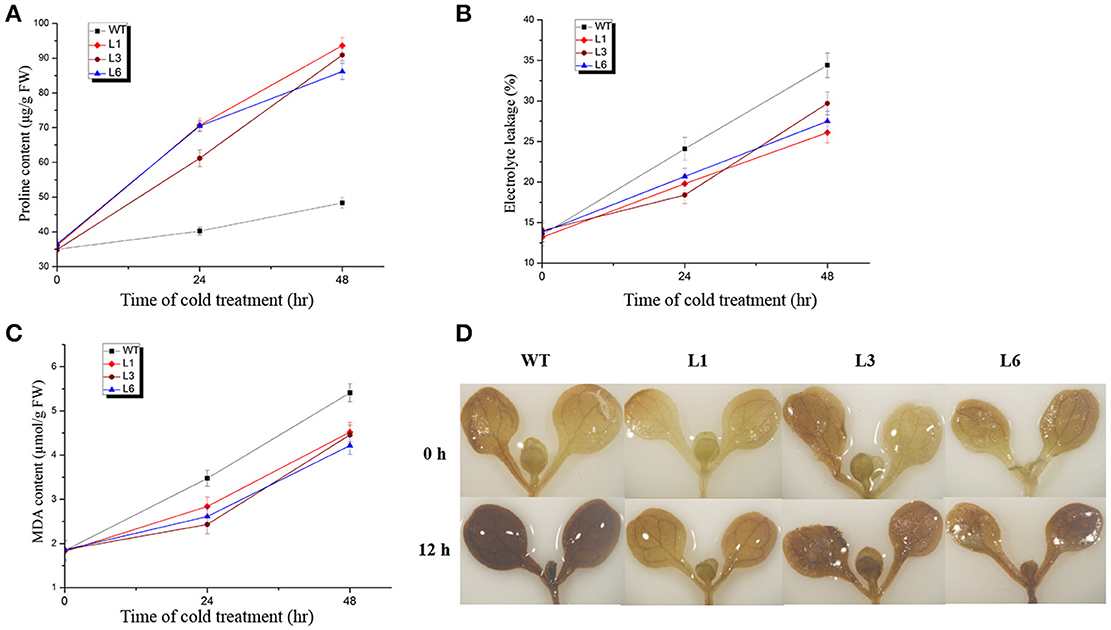
Figure 7. Changes in physiological parameters of 35S::GFP-HbICE1 lines and wild-type (WT) plants under cold stress. Three-week-old Arabidopsis plants were grown at 0°C for the time indicated then leaves collected to determine the free proline content (A), electrolyte leakage (B) and MDA content (C). In (A–C), data represent the mean ± SD (n = 3). (D) DAB staining of 6-day-old Arabidopsis plants with and without cold treatment.
Electrolyte leakage is another reliable indicator of cell membrane damage during the plant stress response (Feng et al., 2012; Huang X. et al., 2015; Cheng et al., 2016). Under normal growth conditions, the transgenic lines and WT plants showed similar levels of electrolyte leakage ranging from 13 to 15%. However, during cold treatment, electrolyte leakage was considerably less in the transgenic lines compared to the WT, suggesting a reduction in cell membrane damage in the HbICE1-overexpressing lines (Figure 7B), and therefore, improved cold tolerance. MDA is also an important index of plant oxidative stress and cell injury in response to stress conditions. While no significant differences in MDA content were observed between transgenic lines and WT plants under normal conditions, lower values were observed in the transgenic compared to WT plants after cold treatment (Figure 7C).
ROS-induced oxidative damage is associated with much of the stress-induced damage that occurs at the cellular level (Chinnusamy et al., 2007). Stress-induced ROS production results in degradation of polyunsaturated lipids, resulting in formation of MDA. In other words, the reduction in MDA content in the transgenic plants might reflect a decrease in ROS accumulation under cold stress. We therefore assayed ROS accumulation in the 35S::HbICE1 plants using DAB staining. As expected, stronger staining was observed in the WT plants compared with the transgenic plants under cold conditions (Figure 7D), suggesting higher levels of damage in the WT. Taken together, these findings suggest that the increased cold tolerance of 35S::HbICE1 plants is the result of increased proline accumulation, reduced electrolyte leakage and MDA metabolism, and a decrease in H2O2 accumulation under cold conditions.
HbICE1 Positively Regulates Cold-Responsive Gene Expression under Cold Conditions
To further elucidate the molecular mechanism of the 35S::HbICE1 plant response to cold stress, we examined transcript levels of cold-responsive genes using qRT-PCR. These cold response genes (COR15A, COR47, KIN1, and RD29A), which contain DRE or related motifs, are downstream target genes of CBF (Stockinger et al., 1997). Under normal conditions, expression levels of COR15A, COR47, KIN1, and RD29A were relatively low in both the transgenic and WT plants. In contrast, after cold exposure for 12 h, expression levels of all tested genes were significantly up-regulated in all three 35S::HbICE1 lines compared with the WT (Figures 8A–D), suggesting that overexpression of HbICE1 positively regulates cold-responsive gene expression, thereby contributing to improved cold tolerance.
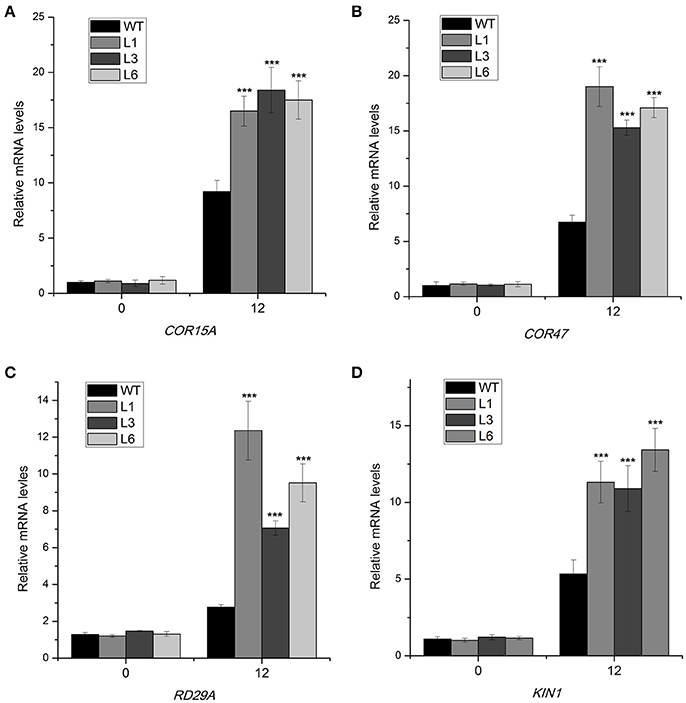
Figure 8. Expression of cold-responsive genes in HbICE1-overexpressing lines subjected to cold stress. Expression of COR15A (A), COR47 (B), RD29A (C), and KIN1(D) in wild-type (WT) and HbICE1-overexpressing lines under normal and cold conditions assayed by quantitative real-time PCR, using AtEIF4 as the internal standard. Data represent means of three replicates ± SE, and asterisks indicate significant differences at ***P < 0.005 (Student's t-test).
Analysis of Differentially Expressed Genes (DEGs) Based on RNA-Seq of Rubber Tree in Response to Cold Stress
In order to gain a global view of transcript expression in rubber tree in response to cold stress, RNA-Seq was used to analyze the differentially expressed genes, generating 24,108,424 raw sequencing reads and 24,050,817 clean reads after filtering low quality (Table S2). 96.50% reads of the control samples matched to a unique or multiple genomic locus, whereas 96.60% of the 3 h cold treatment sample and 96.16% of 12 h cold treatment matched, respectively (Table S3). A total of 8077genes showed differential expression (low false discovery rate [FDR] < 0.001 and P-value < 0.05), including 4096 genes at 3 h (1389 up-regulated, 2707 down-regulated) and 6060 genes at 12 h (3188 up-regulated, 2872 down-regulated) after cold treatment (Figure 9, Tables S4, S5). More DEGs appeared at 12 h than at 3 h after treatment. All the DEGs could be categorized into three main clusters, e.g., biological process, cellular component and molecular function according to GO classification. In the cluster of “biological process,” 17 GO terms of were significantly enriched, of which the GO terms “metabolic process,” “single-organism process,” “cellular process,” “location,” “response to stimulus” were most evidently enriched (Figure S1), suggesting that the these biological processes were responsive to cold stress in rubber tree.
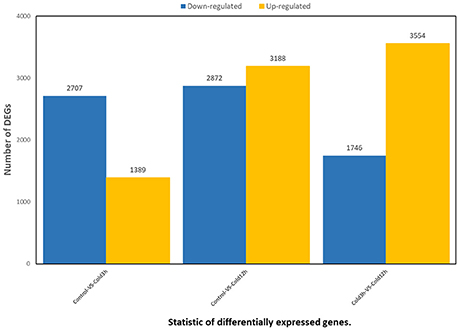
Figure 9. Statistic of differentially expressed genes. X axis represents pairwise and Y axis means number of screened DEGs. Blue bar denotes down-regulated genes and orange bar for the up-regulated.
Pathways Enrichment Analysis of DEGs
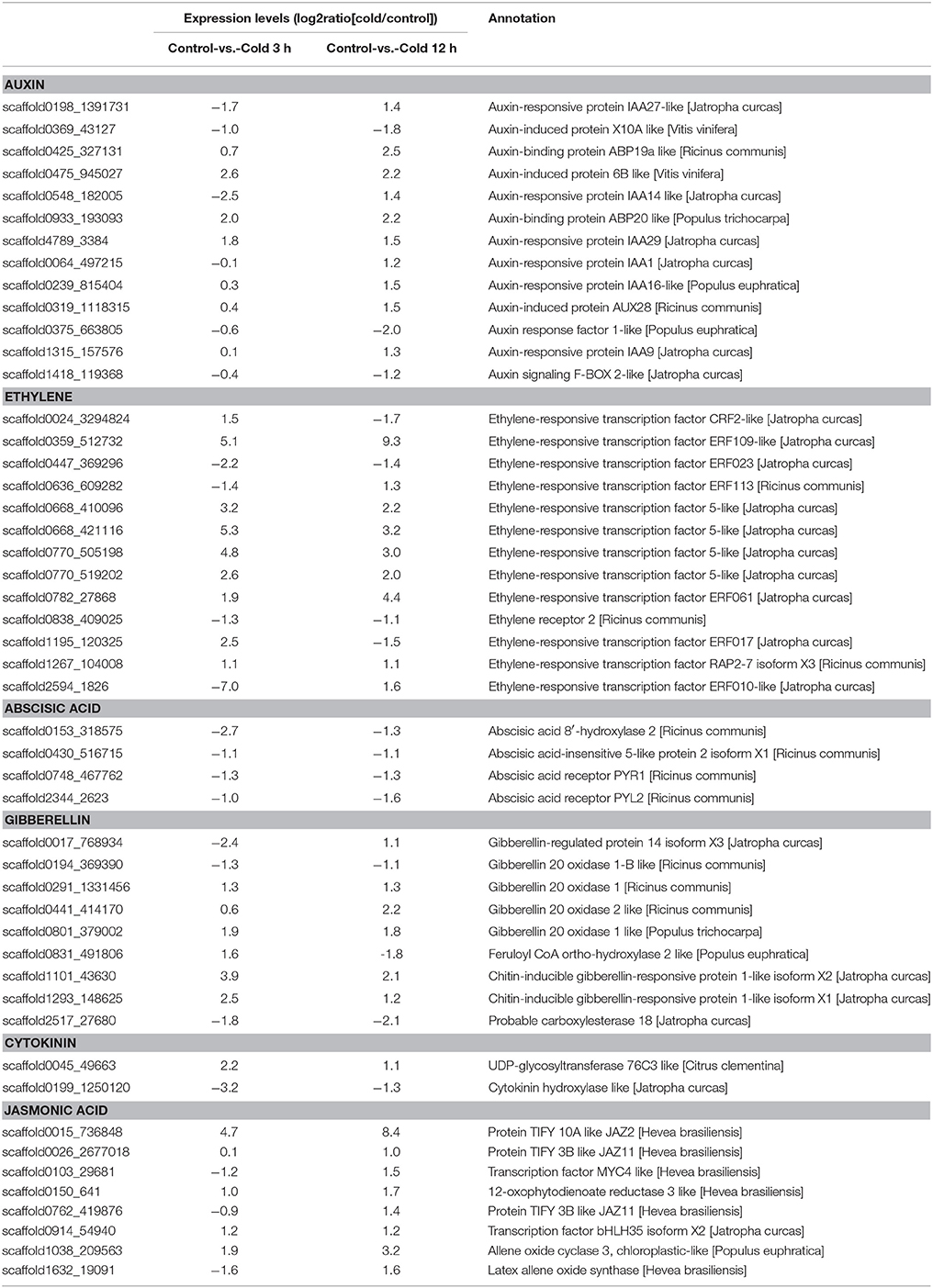
To further reveal the biological functions of cold responsive genes in rubber tree, pathway enrichment analysis of DEGs based on KEGG database were performed, generating a scatter plot of the top 20 of KEGG enrichment (Figure 10). Most DGEs were enriched in “metabolic pathway” and “biosynthesis of secondary metabolites.” Interestingly, “plant-pathogen interaction,” and “plant hormone signal transduction” pathways were also significantly enriched. Noteworthy, the DGEs enriched in “plant hormone signal transduction” pathway showed most differentially expression in both samples, suggesting that phytohormones involved in the perception and transduction of cold induced signaling in rubber tree. In total, 50 DEGs associated with biosynthesis and/or signal transduction of the phytohormones, including 13 genes related with auxin, 13 genes related with ethylene, 9 genes related with gibberellin (GA), 9 genes related with jasmonic acid (JA), 4 genes related with abscisic acid (ABA) and 2 genes related with cytokinin (CK) (Table 1). Interestingly, ABA 8′-hydroxylase 2 (scaffold0153_318575), ABSCISIC ACID-INSENSITIVE 5-like protein 2 isoform X1 (scaffold0430_516715), and two ABA receptors (scaffold2344_2623, scaffold0748_467762), were down-regulated by cold treatment at both 3 h and 12 h. Furthermore, 12 ethylene-responsive transcription factors (ERFs) showed differential expression, of which nine ERFs (scaffold0359_512732, scaffold0636_609282, scaffold0668_410096, scaffold0668_421116, scaffold0770_505198, scaffold0770_519202, scaffold0782_27868, scaffold1267_104008, scaffold2594_1826) were up-regulated and three ERFs (scaffold0024_3294824, scaffold0447_369296, scaffold1195_120325) were down-regulated at 12 h after cold treatment. Additionally, four genes related with JA biosynthesis, two encoding allene oxide cyclase (AOC, scaffold1038_209563, scaffold1142_120639), one encoding 12-oxophytodienoate reductase (OPR, scaffold0150_641) and one encoding latex allene oxide synthase (AOS, scaffold1632_19091), were up regulated at 12 h after cold treatment. Furthermore, three JAZ genes and one MYC2 were also significantly up-regulated, strongly suggesting that JA signaling pathway involved in cold response of rubber tree.
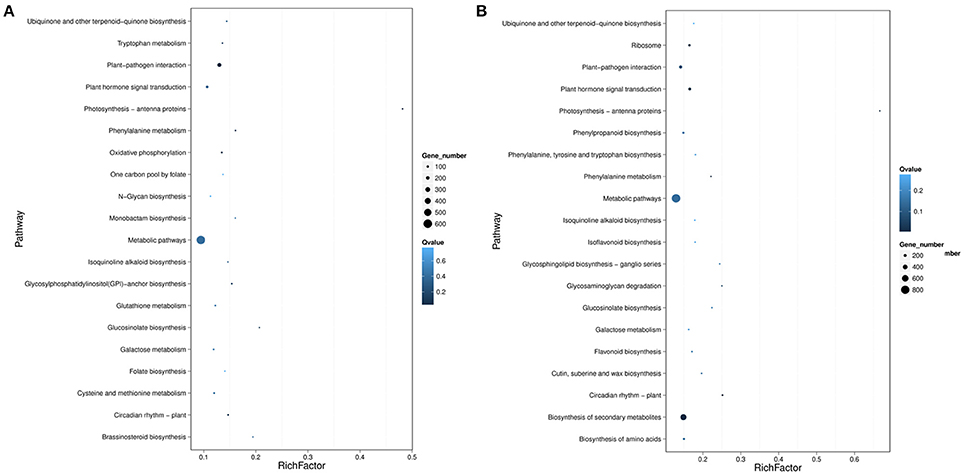
Figure 10. Statistics of pathway enrichment of DEGs in pairwise of Control-VS-Cold3 h (A) and Control-VS-Cold12 h (B). Rich Factor is the ratio of differentially expressed gene numbers annoted in this pathway term to all gene numbers annoted in this pathway term. Greater rich Fator means greater intensiveness. Q-value is corrected p-value ranging from 0 to 1, and less Q-value means greater intensiveness. We just display the top 20 of enriched pathway terms.
Identification of Transcription Factors (TFs) in Response to Cold Stress
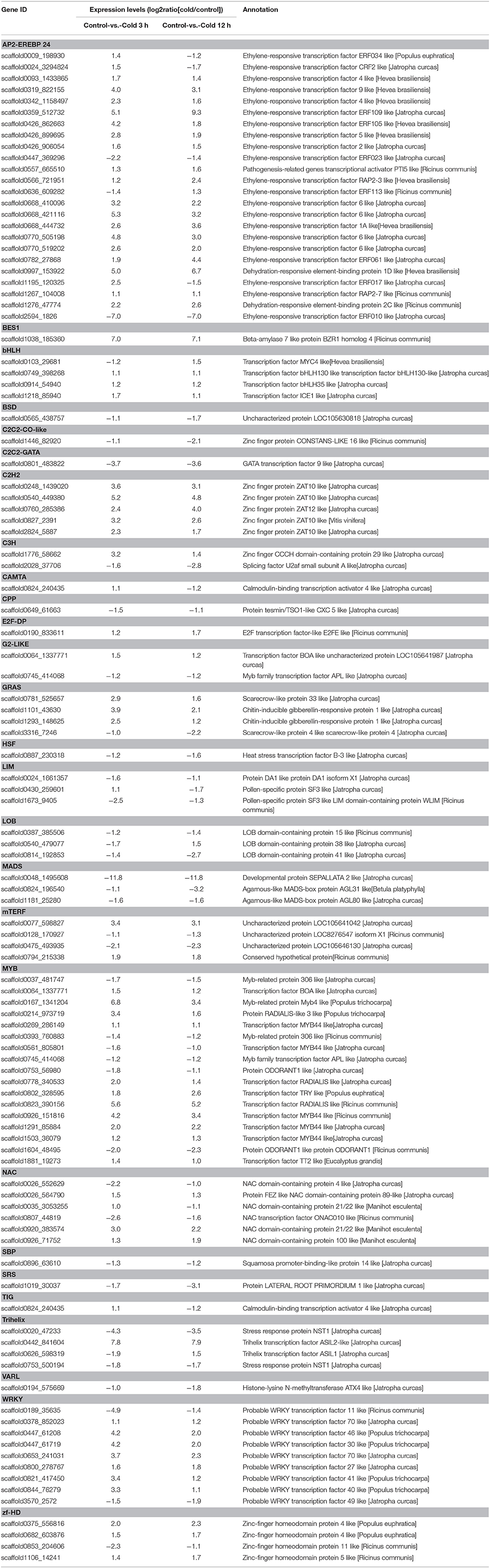
Numerous families of transcription factors (TFs) are known to play crucial roles in signal transduction and regulation when plants are subjected with various abiotic stresses, including the AP2/EREBP, MYB, MYC, WRKY, NAC, and bHLH families (Agarwal and Jha, 2010; Liu et al., 2014). In the current study, 105 putative TFs were found to be exhibited differential expression in response to the cold treatment, of which those encoding AP2/EREBP domain-containing proteins constituted the largest group (22.8%), followed by MYB proteins (16.2%), WRKY proteins (8.6%), NAC proteins (5.7%), C2H2 proteins (4.7%), bHLHs (3.8%) (Table 2). The largest group of the cold-mediated TFs belonged to the AP2/EREBP family and was composed of 24 members. Of these, two genes (scaffold0997_153922, scaffold1276_47774) were annotated as CBF/ DREB genes, which have been shown to play important roles in cold acclimation leading to freezing tolerance (Nakashima et al., 2009). In addition, four members (scaffold0103_29681, scaffold0749_398268, scaffold0914_54940, scaffold1218_8594) of bHLH family were found to be exhibited differential expression in response to the cold treatment. Of those, the expression of an ICE1-like TF (scaffold1218_8594) was observed to up-regulated by cold treatment. Our results therefore indicated that the ICE-CBF pathway is conserved in rubber tree responses to the cold stress. Among the differentially expressed C2H2 TFs, all of the four members (scaffold0248_1439020, scaffold0540_449380, scaffold0760_285386, scaffold0827_2391, scaffold2824_5887) were up-regulated by cold treatment. We also noticed that the cold stress mediated the expression of TFs in the WRKY (scaffold0378_852023, scaffold0447_61208, scaffold0447_61719, scaffold0653_241031, scaffold0800_278767, scaffold0821_417450, scaffold0844_76279) family. The roles of members of these families in cold tolerance have been well established in numerous plants (Chinnusamy et al., 2010).
Discussion
Cold is the major environmental abiotic stress adversely affecting the growth and geographical distribution of plants. Hevea brasiliensis is native to tropical rainforests in the Amazonian basin, but was expanded to sub-optimal environments worldwide in the late 1970s, prominently northeast India, southern China, highland and coastal areas of Vietnam, and southern Brazil. Cold stress has therefore become a limiting factor not only of rubber production, but also survival of rubber trees. Chilling temperatures (0–15°C) interfere with a number of metabolic and physiological processes such as chloroplast and mitochondria integrity, plastid membrane composition and photosynthetic electron transport, resulting in leaf wilt and lesions, bark breakdown and latex leakage, root system withering and frostbite, and ultimately cell death. Increased cold tolerance is therefore a major aim of rubber tree breeding programs.
As an upstream regulator, ICE1 plays a key role in cold signaling pathways in a wide range of plants species such as Arabidopsis (Shi Y. et al., 2012; Lang and Zhu, 2015), wheat, rice, Phalaenopsis aphrodite, Trifoliate orange, banana and grapevine (Badawi et al., 2008; Nakamura et al., 2011; Peng et al., 2013, 2014; Xu et al., 2014; Huang X. S. et al., 2015). It is therefore reasonable to expect that ICE1 homologs also play a key role in cold tolerance in H. brasiliensis. Characterization and functional analysis of the ICE1 gene in rubber trees is therefore a significant step in determining its cold signaling pathway.
Multiple sequence alignments indicate that HbICE1 shares typical ICE1 protein domains with other plant species such as the MYC-like basic helix-loop-helix (bHLH) domain, serine-rich region (S-rich), zipper region (ZIP) and ACT-UUT-ACR-like domain; however, it possesses a varied N terminus (Figure 1). A potential sumoylation site, which is reportedly crucial for AtICE1 activation and stability (Miura et al., 2007), was also found in HbICE1 (Figure 1), suggesting that HbICE1 activity is mediated by the SUMO E3 ligase. Furthermore, HbICE1 was confirmed as being nuclear localized, and able to bind to the MYC-recognition element. These observations imply that HbICE1 is a novel putative ICE1 homolog.
Expression profiles revealed that, like AtICE1, HbICE1 is expressed constitutively in all tissues (Chinnusamy et al., 2003). Furthermore, HbICE1 was induced by multiple abiotic stresses including cold, dehydration, wound and salinity (Figure 5). HbICE1 was only slightly induced by cold, consistent with a previous report suggesting that cold stress induces little transcriptional alteration, instead resulting in posttranslational modification of ICE1 to active the CBF pathway (Chinnusamy et al., 2003; Ding et al., 2015). Upregulation of HbICE1 following dehydration was consistent with previous reports in P. trifoliate and Pyrus ussuriensis (Huang X. et al., 2015; Huang X. S. et al., 2015), but differs from AtICE1, which is not triggered by dehydration (Chinnusamy et al., 2003). This disparity between HbICE1 and AtICE1 might be attributed to the inherent differences between plant species.
Cold acclimation is one of the major mechanisms for plant to adapt to cold stress (Thomashow, 1999). Indeed, cold acclimation was found to enhance freezing tolerance rubber tree after exposure to low temperature (Figure S2).The electrolyte leakage was less in the cold-acclimated (CA) rubber trees to the nonacclimated (NA) rubber tree, and CA rubber trees experienced less cold stress-induced H2O2 accumulation compared to the NA plants, suggesting an enhanced membrane integrity and the lower cold stress-induced H2O2 accumulation in the CA rubber tree seedlings to NA plants. Overexpression of HbICE1 in Arabidopsis enhanced cold tolerance only after cold acclimation (Figure 6), indicating that other co-factors associated with cold acclimation are essential for HbICE1-mediated cold tolerance. Similar results were reported for wheat ICE genes (Badawi et al., 2008), where overexpression of TaICE87 or TaICE41 in Arabidopsis enhanced freezing tolerance only after cold acclimation (Badawi et al., 2008). These results suggest that other co-factors induced by cold acclimation are essential for HbICE1-mediated cold tolerance. Previous reports also confirmed this hypothesis. Chinnusamy et al. (2003) showed that cold-induced modification of the AtICE1 protein or a transcriptional cofactor is necessary for AtICE1-induced activation of CBF expression. Furthermore, Miura et al. (2007) showed that SIZ1 (SAP and Miz1), mediates sumoylation of ICE1, which reduces the polyubiquitination of ICE1 to enhance its stability. A potential sumoylation site was also found in HbICE1 protein (Figure 1), suggesting that HbICE1 activity could be regulated by sumoylation via the SUMO E3 ligase.
Physiological parameters such as electrolyte leakage and contents of MDA, chlorophyll and proline are closely related to cold tolerance under the regulation of COR, which is triggered by the ICE transcription factor (Badawi et al., 2008; Peng et al., 2014; Xu et al., 2014; Liu et al., 2017). In many plant species, overexpression of ICE1 is sufficient to alter physiological parameters and enhance cold tolerance (Feng et al., 2013; Huang X. et al., 2015). In our study, the HbICE1ox transgenic lines showed improved survival rates and freezing tolerance as a result reduced electrolyte leakage and MDA metabolism, and increased proline accumulation (Figure 7), suggesting that HbICE1 plays a positive regulatory role in the response to cold stress. These findings suggest that the ICE-CBF-COR transcriptional cascade, which influences the freezing tolerance capacity of plants, exists not only in Arabidopsis but other species such as H. brasiliensis.
H2O2 is often associated with biotic and abiotic stresses (Yuan and Huang, 2016; Cao et al., 2017; Lu et al., 2017; Yuan et al., 2017). Our study further suggests that the 35S::HbICE1 plants experienced less cold stress-induced H2O2 accumulation compared to the WT, despite similar contents under normal conditions. Consistence with the decrease in MDA content and electrolyte leakage under cold stress, these data imply that enhanced membrane integrity and decreased levels of lipid peroxidation caused the lower cold stress-induced H2O2 accumulation in the transgenic compared to WT plants.
Transcription factors families such as AP2/EREBP, MYB, WRKY, C2H2, NAC, and bHLH are well known to be involved in stress tolerance in plants (Chinnusamy et al., 2010), but how they work synergistically to cope with cold tolerance requires determination. In this work, two genes (scaffold0997_153922, scaffold1276_47774) annotated as CBF/DREB and one ICE1 member of bHLH family (scaffold1218_8594) were up-regulated by cold treatment (Table 2), indicating that ICE-CBF pathway is conserved in rubber tree responses to the cold stress. The phytohormones auxin, ABA, JA, and ethylene are known to play important roles in the regulation of plant growth and abiotic stress responses (Shi Y. et al., 2015). In current study, the DGEs data suggested that many phytohormones related genes were responsive to cold treatment in rubber tree. ABA integrates various stress signals and modulates stress responses, but whether it is involved in cold responses is still debated (Tuteja, 2007). It has been suggested that plant's response to the cold stress may be ABA-independent, but increasing evidences of contrary were reported (Lang et al., 1994; Wang et al., 2015). We noticed that ABA 8′-hydroxylase, a key enzyme in ABA catabolism, was down-regulated by cold treatment. Down-regulation of the ABA 8′-hydroxylase gene implies that catabolism of ABA might be attenuated in rubber tree under cold stress, which is consistent with the previous report that ABA levels increase slightly in response to low temperature (Lang et al., 1994). In addition, we detected the decreased expression of ABA receptor PYR1 and PYL2 (Table 1), how ABA mediated signaling is involved in the cold responses of rubber tree remains further investigation. Ethylene has been well documented in the cold stress response (Shi Y. et al., 2012, 2015). In our study, the expression of ERFs was significantly regulated by cold treatment. JA was recently reported to significantly enhance plant freezing tolerance with or without cold acclimation, (Hu et al., 2013). We observed the increased expression of several key genes for JA biosynthesis, including OPR (scaffold0150_641), AOS (scaffold1038_209563) and AOC (scaffold1632_19091), and several essential genes (e.g., JAZs and MYC) in JA signaling pathways at 12 h after cold treatment, strongly suggesting that cold stress triggers JA biosynthesis and responses in rubber tree, which is consistent with the study of A. thaliana (Hu et al., 2013). JA was therefore proposed to be a positive regulator of cold responses in rubber tree.
In conclusion, a novel ICE-like transcription factor, designated HbICE1, was isolated from H. brasiliensis and functionally characterized. This nuclei protein, which has typical features of ICE proteins, was found to have transactivation activity via binding to MYC recognition sites. 35S::HbICE1 plants showed enhanced cold resistance via increased proline content, a reduction in MDA metabolism and electrolyte leakage, and a decrease in ROS accumulation, promoting expression of the cold-responsive genes. These findings suggest that HbICE1 is a member of the ICE gene family and positive regulator of cold tolerance. Differentially expressed genes (DEGs) analysis showed that cold treatment changed genes expression profiles involved in many biological processes and phytohormones perception and transduction. Ethylene, JA, ABA, as well as ICE-CBF signaling pathways might work synergistically to cope with cold tolerance in rubber tree. These findings will help elucidate the cold signaling network in H. brasiliensis, ultimately aiding breeding programs aimed at improving cold stress tolerance.
Author Contributions
HY conceived and directed this study, designed and performed the experiments, analyzed the data, wrote and revised the manuscript; YS, YL, MO, XT, and WC performed the experiments, analyzed the data; XH revised the manuscript. All authors have read and approved the final version submitted.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [grant number 31560197, 31260170], the Hainan Education Department Foundation [grant number Hnky2015-4], Hainan University Youth Foundation (hdkyxj201703) and the Startup Funding of Hainan University [grant number kyqd1437].
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01462/full#supplementary-material
Figure S1. GO functional classification on DEGs in pairwise of Control-vs.-Cold 3 h (A) and Control-vs.-Cold12 h (B). X axis means number of DEGs (the number is presented by its square root value). Y axis represents GO terms. All GO terms are grouped in to three ontologies: blue is for biological process, brown is for cellular component and orange is for molecular function.
Figure S2. Electrolyte leakage (A) and DAB staining (B) of the rubber tree seedlings with indicated freezing temperatures. Seedlings were treated at −16°C for 0.5 h or 1 h for nonacclimated (NA) and cold-acclimated (CA) plants (CA; 1 day at 4°C). Error bars show SD from three replicates. (**P < 0.01, ***P < 0.005, student's t-test).
Table S1. List of the primers used for qRT-PCR.
Table S2. Summary of sequencing data for each sample.
Table S3. Alignment statistics of reads align to reference genome.
Table S4. Differentially Expressed Genes (using Noiseq) in pairwise of Control-VS-Cold 3 h.
Table S5. Differentially Expressed Genes (using Noiseq) in pairwise of Control-VS-Cold 12 h.
Abbreviations
GFP, green fluorescent protein; ICE1, Inducer of CBF Expression 1; MDA, malondialdehyde; qRT-PCR, quantitative real-time PCR; ROS, reactive oxygen species.
References
Agarwal, P. K., and Jha, B. (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plant. 54, 201–212. doi: 10.1007/s10535-010-0038-7
Badawi, M., Reddy, Y. V., Agharbaoui, Z., Tominaga, Y., Danyluk, J., Sarhan, F., et al. (2008). Structure and functional analysis of wheat ICE (inducer of CBF expression) genes. Plant Cell Physiol. 49, 1237–1249. doi: 10.1093/pcp/pcn100
Cai, W., Liu, W., Wang, W. S., Fu, Z. W., Han, T. T., and Lu, Y. T. (2015). Overexpression of rat neurons nitric oxide synthase in rice enhances drought and salt tolerance. PLoS ONE 10:e0131599. doi: 10.1371/journal.pone.0131599
Cao, Y., Zhai, J., Wang, Q., Yuan, H., and Huang, X. (2017). Function of Hevea brasiliensis NAC1 in dehydration-induced laticifer differentiation and latex biosynthesis. Planta 245, 31–44. doi: 10.1007/s00425-016-2589-0
Cheng, H., Cai, H., Fu, H., An, Z., Fang, J., Hu, Y., et al. (2015). Functional characterization of Hevea brasiliensis CRT/DRE binding factor 1 gene revealed regulation potential in the cbf pathway of tropical perennial tree. PLoS ONE 10:e0137634. doi: 10.1371/journal.pone.0137634
Cheng, H., Chen, X., Zhu, J., and Huang, H. (2016). Overexpression of a Hevea brasiliensis ErbB-3 binding protein 1 gene increases drought tolerance and organ size in Arabidopsis. Front. Plant Sci. 7:1703. doi: 10.3389/fpls.2016.01703
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Chinnusamy, V., Zhu, J. K., and Sunkar, R. (2010). Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 639, 39–55. doi: 10.1007/978-1-60761-702-0_3
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Ding, Y., Li, H., Zhang, X., Xie, Q., Gong, Z., and Yang, S. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 32, 278–289. doi: 10.1016/j.devcel.2014.12.023
Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 103, 8281–8286. doi: 10.1073/pnas.0602874103
Feng, H. L., Ma, N. N., Meng, X., Zhang, S., Wang, J. R., Chai, S., et al. (2013). A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol. Biochem. 73, 309–320. doi: 10.1016/j.plaphy.2013.09.014
Feng, X. M., Zhao, Q., Zhao, L. L., Qiao, Y., Xie, X. B., Li, H. F., et al. (2012). The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol. 12:22. doi: 10.1186/1471-2229-12-22
Fursova, O. V., Pogorelko, G. V., and Tarasov, V. A. (2009). Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 429, 98–103. doi: 10.1016/j.gene.2008.10.016
Hong, H., Xiao, H., Yuan, H., Zhai, J., and Huang, X. (2015). Cloning and characterisation of JAZ gene family in Hevea brasiliensis. Plant Biol. 17, 618–624. doi: 10.1111/plb.12288
Hu, Y., Jiang, L., Wang, F., and Yu, D. (2013). Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25, 2907–2924. doi: 10.1105/tpc.113.112631
Huang, X., Li, K., Jin, C., and Zhang, S. (2015). ICE1 of Pyrus ussuriensis functions in cold tolerance by enhancing PuDREBa transcriptional levels through interacting with PuHHP1. Sci. Rep. 5:17620. doi: 10.1038/srep17620
Huang, X. S., Zhang, Q., Zhu, D., Fu, X., Wang, M., Moriguchi, T., et al. (2015). ICE1 of Poncirus trifoliata functions in cold tolerance by modulating polyamine levels through interacting with arginine decarboxylase. J. Exp. Bot. 66, 3259–3274. doi: 10.1093/jxb/erv138
Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O., and Thomashow, M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. doi: 10.1126/science.280.5360.104
Lang, V., Mantyla, E., Welin, B., Sundberg, B., and Palva, E. T. (1994). Alterations in water status, endogenous abscisic acid content, and expression of rab18 gene during the development of freezing tolerance in Arabidopsis thaliana. Plant Physiol. 104, 1341–1349. doi: 10.1104/pp.104.4.1341
Lang, Z., and Zhu, J. (2015). OST1 phosphorylates ICE1 to enhance plant cold tolerance. Sci. China Life Sci. 58, 317–318. doi: 10.1007/s11427-015-4822-7
Lin, A., Wang, Y., Tang, J., Xue, P., Li, C., Liu, L., et al. (2012). Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 158, 451–464. doi: 10.1104/pp.111.184531
Liu, J. H., Peng, T., and Dai, W. (2014). Critical cis-acting elements and interacting transcription factors: key players associated with abiotic stress responses in plants. Plant Mol. Biol. Rep. 32, 303–317. doi: 10.1007/s11105-013-0667-z
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Liu, Z., Jia, Y., Ding, Y., Shi, Y., Li, Z., Guo, Y., et al. (2017). Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 66, 117–128 e115. doi: 10.1016/j.molcel.2017.02.016
Lu, X., Zhou, X., Cao, Y., Zhou, M., McNeil, D., Liang, S., et al. (2017). RNA-seq analysis of cold and drought responsive transcriptomes of zea mays ssp. mexicana L. Front. Plant Sci. 8:136. doi: 10.3389/fpls.2017.00136
Maruyama, K., Todaka, D., Mizoi, J., Yoshida, T., Kidokoro, S., Matsukura, S., et al. (2012). Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49. doi: 10.1093/dnares/dsr040
Medina, J., Bargues, M., Terol, J., Perez-Alonso, M., and Salinas, J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119, 463–470. doi: 10.1104/pp.119.2.463
Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V., Miura, T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414. doi: 10.1105/tpc.106.048397
Nakamura, J., Yuasa, T., Huong, T. T., Harano, K., Tanaka, S., Iwata, T., et al. (2011). Rice homologs of inducer of CBF expression (OsICE) are involved in cold acclimation. Plant Biotechnol. 28, 303–309. doi: 10.5511/plantbiotechnology.11.0421a
Nakashima, K., Ito, Y., and Yamaguchi-Shinozaki, K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Peng, H. H., Shan, W., Kuang, J. F., Lu, W. J., and Chen, J. Y. (2013). Molecular characterization of cold-responsive basic helix-loop-helix transcription factors MabHLHs that interact with MaICE1 in banana fruit. Planta 238, 937–953. doi: 10.1007/s00425-013-1944-7
Peng, P. H., Lin, C. H., Tsai, H. W., and Lin, T. Y. (2014). Cold response in phalaenopsis aphrodite and characterization of PaCBF1 and PaICE1. Plant Cell Physiol. 55, 1623–1635. doi: 10.1093/pcp/pcu093
Shi, H. T., Li, R. J., Cai, W., Liu, W., Wang, C. L., and Lu, Y. T. (2012). Increasing nitric oxide content in Arabidopsis thaliana by expressing rat neuronal nitric oxide synthase resulted in enhanced stress tolerance. Plant Cell Physiol. 53, 344–357. doi: 10.1093/pcp/pcr181
Shi, Y., Ding, Y., and Yang, S. (2015). Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 56, 7–15. doi: 10.1093/pcp/pcu115
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24, 2578–2595. doi: 10.1105/tpc.112.098640
Stockinger, E. J., Gilmour, S. J., and Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Theocharis, A., Clement, C., and Barka, E. A. (2012). Physiological and molecular changes in plants grown at low temperatures. Planta 235, 1091–1105. doi: 10.1007/s00425-012-1641-y
Thomashow, M. F. (1999). PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Thomashow, M. F. (2010). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154, 571–577. doi: 10.1104/pp.110.161794
Tuteja, N. (2007). Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2, 135–138. doi: 10.4161/psb.2.3.4156
Wang, M., Zhang, X., and Liu, J. H. (2015). Deep sequencing-based characterization of transcriptome of trifoliate orange (Poncirus trifoliata (L.) Raf.) in response to cold stress. BMC Genomics 16:555. doi: 10.1186/s12864-015-1629-7
Wisniewski, M., Nassuth, A., Teulieres, C., Marque, C., Rowland, J., Cao, P. B., et al. (2014). Genomics of cold hardiness in woody plants. CRC. Crit. Rev. Plant Sci. 33, 92–124. doi: 10.1080/07352689.2014.870408
Xia, Z., Xu, H., Zhai, J., Li, D., Luo, H., He, C., et al. (2011). RNA-Seq analysis and de novo transcriptome assembly of Hevea brasiliensis. Plant Mol. Biol. 77, 299–308. doi: 10.1007/s11103-011-9811-z
Xu, W., Jiao, Y., Li, R., Zhang, N., Xiao, D., Ding, X., et al. (2014). Chinese wild-growing Vitis amurensis ICE1 and ICE2 encode MYC-type bHLH transcription activators that regulate cold tolerance in Arabidopsis. PLoS ONE 9:e102303. doi: 10.1371/journal.pone.0102303
Yuan, H. M., and Huang, X. (2016). Inhibition of root meristem growth by cadmium involves nitric oxide-mediated repression of auxin accumulation and signalling in Arabidopsis. Plant Cell Environ. 39, 120–135. doi: 10.1111/pce.12597
Keywords: hevea brasiliensis, ICE1, cold stress, CBF pathway, bHLH, reactive oxygen species
Citation: Yuan H-M, Sheng Y, Chen W-J, Lu Y-Q, Tang X, Ou-Yang M and Huang X (2017) Overexpression of Hevea brasiliensis HbICE1 Enhances Cold Tolerance in Arabidopsis. Front. Plant Sci. 8:1462. doi: 10.3389/fpls.2017.01462
Received: 05 April 2017; Accepted: 07 August 2017;
Published: 22 August 2017.
Edited by:
Jie Zhou, Zhejiang University, ChinaCopyright © 2017 Yuan, Sheng, Chen, Lu, Tang, Ou-Yang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Mei Yuan, eXVhbmhvbmdtZWlAaGFpbnUuZWR1LmNu
Xi Huang, eGlodWFuZ0BoYWludS5lZHUuY24=
 Hong-Mei Yuan
Hong-Mei Yuan Ying Sheng
Ying Sheng Wei-Jie Chen
Wei-Jie Chen Yu-Qing Lu
Yu-Qing Lu Xiao Tang
Xiao Tang Mo Ou-Yang
Mo Ou-Yang Xi Huang
Xi Huang