- 1Guizhou Subcenter of National Wheat Improvement Center, College of Agronomy, Guizhou University, Guiyang, China
- 2State Key Laboratory of Crop Biology, Shandong Key Laboratory of Crop Biology, College of Agronomy, Shandong Agricultural University, Taian, China
- 3College of Life Science, Zaozhuang University, Zaozhuang, China
In this study, the intergeneric hybrids F1, F2, BC1F1, BC1F2, and BC2F1 from Elytrigia elongata and Triticum aestivum crosses were produced to study their chromosome pairing behavior. The average E. elongata chromosome configuration of the two F1 hybrids agreed with the theoretical chromosome configuration of 21I+7II, indicating that the genomic constitution of this F1 hybrid was ABDStStEeEbEx. Compared with the BC1F1 generation, the BC2F1 generation showed a rapid decrease in the number of E. elongata chromosomes and the BC1F2 generation showed a more extensive distribution of E. elongata chromosomes. In addition, pairing between wheat and E. elongata chromosomes was detected in each of the wheat-E. elongata hybrid progenies, albeit rarely. Our results demonstrated that genomic in situ hybridization (GISH) using an E. elongata genomic DNA probe offers a reliable approach for characterizing chromosome pairing in wheat and E. elongata hybrid progenies.
Introduction
Modern cultivation strategies have diminished the genetic base of common wheat (Triticum aestivum). A number of wild relatives and related species were popularly used to increase the genetic diversity available to wheat breeders. Elytrigia elongata (Host) Nevisk. [Syn. Thinopyrum ponticum (Podp.) Barkworth] (2n = 10x = 70) was initially hybridized with wheat approximately 70 years ago because of its resistance to several wheat diseases, as well as its stress tolerance and high crossing ability with various Triticum species (Sepsi, 2010; Hu et al., 2011; Fu et al., 2012; Ayala-Navarrete et al., 2013; He et al., 2013; Zheng et al., 2014; Li et al., 2016). Many desirable genes, such as Sr25, Sr43, Lr19, Cmc2, and Pm51, have been characterized and transferred from this wild grass species into wheat. These translocations have supported the development of several wheat germplasms that are used in wheat improvement programs throughout the world (Li and Wang, 2009; Niu et al., 2014; Zhan et al., 2014). The genomic composition of the decaploid species E. elongata has been a subject of interest for quite some time and is designated JJJJJJJsJsJsJs (Chen et al., 1998) or StStStStEeEeEbEbExEx (Zhang et al., 1996). There is some evidence that the St chromosomes in E. elongata are closely related to those of Pseudoroegneria strigosa and that the J/Eb and Js/Ee genomes are closely related to the Thinopyrum bessarabicum and/or Thinopyrum elongatum genomes (Chen et al., 2001). However, the genomic composition of E. elongata has not yet been clarified.
Chromosome engineering is the procedure of altering ploidy, chromosome structure, and/or chromosome number of an organism intended for genetic improvement. This technology has been used to incorporate favorable genes from wild species into the wheat genome for germplasm and variety development. These favorable genes can be introduced into wheat from wild species through chromosome addition, substitution, and translocation. Alien chromosome addition and substitution, which introduce one or more entire foreign chromosomes into the wheat genome, usually include desirable genes, as well as undesirable genes. There is a general demand to quickly utilize those lines in wheat breeding. Chromosome translocation, which integrates alien chromosome segments containing the gene of interest into the wheat genome, has been the most effective approach for alien gene introgression (Guo et al., 2015; Li et al., 2016). The translocations generally result from meiotic recombination between wheat chromosomes and their homoeologous complements from wild species (Bagherikia et al., 2014; Song et al., 2016).
The corresponding chromosomes of the A, B, and D genomes are genetically closely related. However, the pairing propinquity between genetically analogous chromosomes of these genomes is suppressed, largely by the activity of the Ph1 gene in the long arm of chromosome 5B (Sears, 1976). Ph1 represses homoeologous pairing so that only homologous partners can pair. So far, allelic variation inducing different levels of homoeologous pairing in wheat or in wheat hybrids has not been found in Ph1. Such variation can best be discovered in intergeneric hybrids where homologs are not present and homoeologous pairing is normally very low so that any change in the level of pairing can be demonstrably detected. While in several intergeneric hybrids, the action of Ph1 is counterbalanced by pairing promoters of the alien species, and in most intergeneric wheat hybrids there is either little or no effect of the alien genome on homoeologous pairing (Qi et al., 2007).
Metaphase I (MI) pairing reflects cross-formation that might be associated with recombination. Metamorphic chromosomal pairing from meiosis between interspecific or intraspecific hybrids is an efficient method for estimating interphase gene transfer and revealing phylogenetic relationships among these species (Bao et al., 2014; Su et al., 2016). Cytogenetic studies on intergeneric hybrids between Elytrigia species have shown close relationships between J/Eb, Js/Ee, and St chromosomes (Chen et al., 2001; Liu et al., 2007). Although some information on chromosome pairing in Elytrigia and wheat hybrids is available (Roundy, 1985; Cai and Jones, 1997), little is known about the pairing frequency between E. elongata and wheat chromosomes because of the complexity of wheat-E. elongata chromosome pairings and the difficulty of distinguishing chromosomes in hybrids using conventional chromosome techniques.
In this study, hybrid progeny involving F1, F2, BC1F1, BC1F2, and BC2F1 were created by hybridizing T. aestivum with E. elongata to transfer desirable traits from E. elongata into wheat. The objective of this work was to characterize the meiotic behavior and genomic composition of the progeny from wheat-E. elongata hybrids using cytogenetic analysis and genomic in situ hybridization (GISH) technology.
Materials and Methods
Plant Material
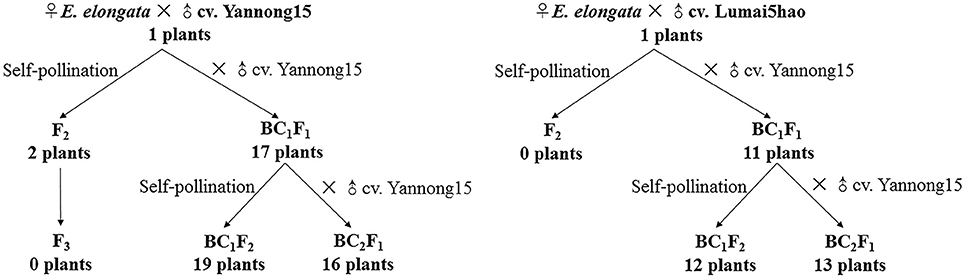
E. elongata was provided by Prof. Zhensheng Li, formerly of the Northwest Institute of Botany, Chinese Academy of Sciences, Yangling, China. The E. elongata × T. aestivum (cv. Yannong15) and E. elongata × T. aestivum (cv. Lumai5hao) were obtained from Prof. Honggang Wang (College of Agronomy, Shandong Agricultural University, Taian, China). All plant materials were maintained through selfing at the Tai'an Subcenter of the National Wheat Improvement Center, Shandong, China. The crosses and results of offspring production are described in Figure 1.
Meiotic Preparations
When the plants reached the flag leaf stage, spikes were sampled, stages of meiosis were determined in acetocarmine squashes of 1 of 3 anthers per flower. If appropriate stages were present, the remaining 2 anthers were fixed in ethanol-acetic acid (3:1) for 24 h and stored at 4°C in 70% alcohol until use. Preparations were made from pollen mother cells (PMCs) by squashing pieces of anthers in 45% acetic acid. Slide preparations were examined using phase-contrast microscopy and then placed on dry ice to remove the cover glass. The images were captured with an Olympus BX-60.
Gish Techniques
Elytrigia elongata DNA was labeled with fluorescein-12-dUTP by nick translation to be used as a probe. Sheared genomic DNA from Yannong15 (AABBDD, 2n = 42) was used as blocking DNA. Detailed procedures of the hybridization mixture were performed as previously described (Kato et al., 2004). The slides were counterstained with propidium iodide (PI, 0.25 mg/mL) in Vectashield mounting medium (Vector Laboratories, USA).
Statistical Analyses
The data concerning the number of univalents, bivalents, trivalents, quadrivalents, pentavalents, and hexavalents for all PMCs of BC1F1, BC1F2, and BC2F1 hybrids studied were considered binomial responses, with the appropriate totals, obtained in a one-way classification. They were analyzed by the generalized linear model with logit link function to estimate mean values for plants and to test the significance of differences between plants. The calculation of mean values, standard deviations and coefficient of variation were analyzed by Excel 2013 with the statistics function. ANOVA analysis was carried out using Excel 2013, and the statistical significance (P) is shown in the Tables S1–S3.
Results
Chromosome Pairing in F1 Hybrids
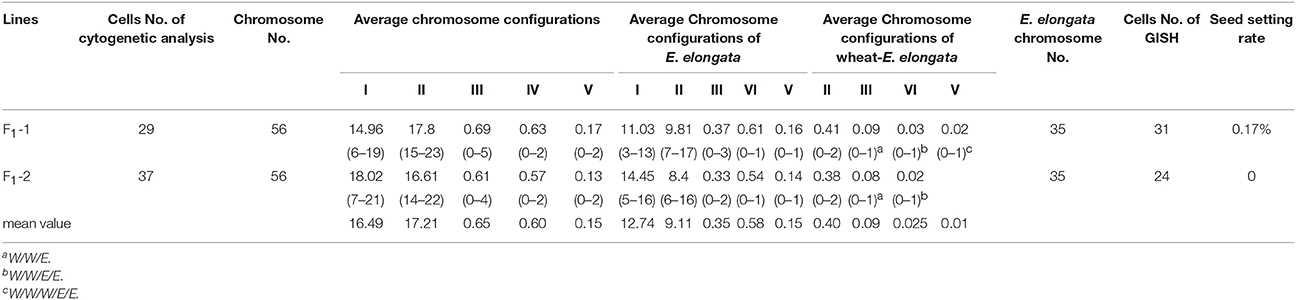
The F1 hybrids from the E. elongata × T. aestivum cross exhibited a low setting percentage and were morphologically different from the 2 parents, except for a similar perennial of E. elongata. All plants had 56 somatic chromosomes with 35 chromosomes from E. elongata. Meiotic association was determined in 29 PMCs at the MI stage from E. elongata × T. aestivum cv. Yannong15 (F1-1) and 37 PMCs at the MI stage from E. elongata × T. aestivum cv. Lumai5hao (F1-2) (Table 1), and the average chromosome configurations were 14.96I+17.8II+0.69III+0.63IV+0.17V (F1-1, Figure 2A) and 18.02I+16.61II+0.61III+0.57IV+0.13V (F1-2, Figure 2B), respectively. Chromosome pairing configurations in the hybrid PMCs were very complex, and a high frequency of univalent and a variety of trivalent and tetravalent configurations were observed.

Figure 2. Chromosome configuration of PMC MI in wheat-E. elongata F1 hybrids. (A) Chromosome configurations of F1-1: 2n = 7I+20II+3III; (B) Chromosome configurations of F1-2: 2n = 6I+20II+2III+1IV; (C) E. elongata chromosome configurations of F1-1: 2n = 3I+16II. Wheat chromosomes were detected in red and E. elongata chromosomes or chromosome segments were visualized in green. Bar = 10 μm
GISH was performed to detect E. elongata chromosomes in F1-1 and F1-2 (Figure 2C) using total genomic DNA from E. elongata as a probe and ABD-genomic DNA from Yannong15 wheat as a blocker. The mean E. elongata chromosome configurations determined after GISH analysis were 11.03I+9.81II+0.37III+0.61IV+0.16V and 14.45I+8.4II+0.33III+0.54IV+0.12V, respectively (Table 1). The chromosome configurations of wheat-E. elongata in the hybrid included bivalents, one type of trivalent (W/W/E), one chain quadrivalent (W/W/E/E), and one chain pentavalent (W/W/W/E/E) (Table 1).
Chromosome Pairing in F2 Progeny
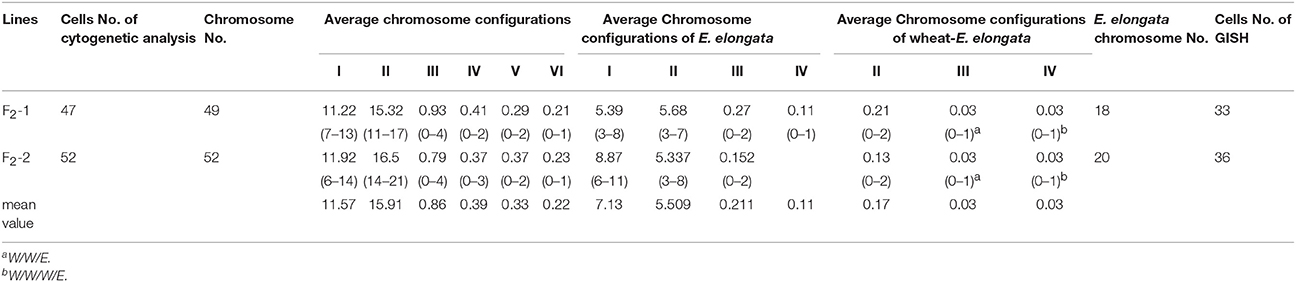
Although five of the F1-1 selfed F2 seeds were obtained, wherein two survived, the F1-2 and these two F2 plants were self-sterile. These F2 plants (F2-1 and F2-2) were identified by cytogenetic analysis and GISH (Table 2). The F2-1 plant had 49 chromosomes, 18 of which were from E. elongata, and the F2-2 plant had 52 chromosomes, 20 of which were from E. elongata. The average chromosome configurations were 11.22I+15.32II+0.93III+0.41IV+0.29V+0.21VI (F2-1, Figure 3A) and 11.92I+16.5II+0.79III+0.37IV+0.37V+0.23VI (F2-2, Figure 3B), respectively. GISH analysis showed that the average E. elongata chromosome configurations were 5.39I+5.68II+0.27III+0.11IV (F2-1, Figure 3C) and 8.87I+5.337II+0.152III (F2-2, Figure 3D), respectively. The chromosome configurations of wheat-E. elongata in the hybrid included bivalents, one kind of trivalent (W/W/E), and one chain quadrivalent (W/W/W/E) (Table 1). In addition, a translocation or interspecific chromosome pairing between wheat and E. elongata chromosomes was also detected in some of these plants (Figure 3D, arrows).
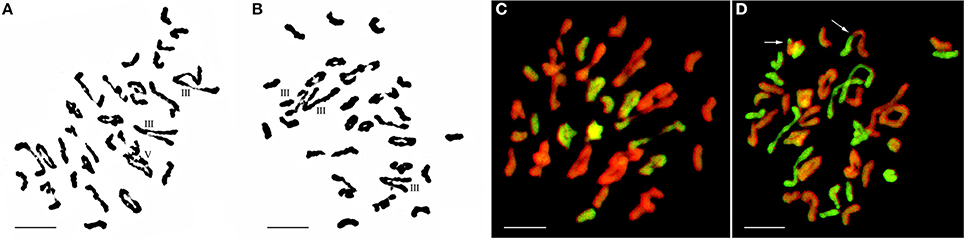
Figure 3. Chromosome configuration in PMC MI for wheat-E. elongata F2 hybrids. (A) Chromosome configuration of F2-1: 2n = 10I+14II+2III+1V; (B) Chromosome configuration of F2-2: 2n = 7I+18II+3III; (C) E. elongata chromosome configuration of F2-1: 2n = 5I+6II; (D) E. elongata chromosome configuration of F2-2: 2n = 10I+5II. Wheat chromosomes were detected in red and E. elongata chromosomes or chromosome segments were visualized in green. The arrows indicate pairing between wheat and E. elongata chromosomes. Bar = 10 μm.
Chromosome Pairing and Separation Trend in Hybrid Derivatives
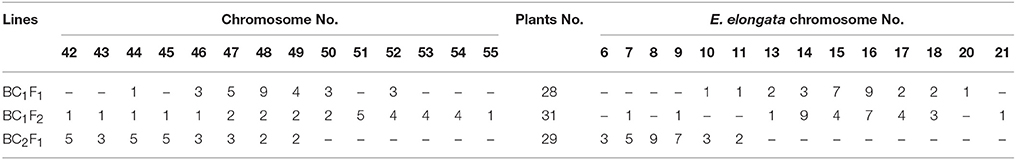
Seventeen plants were produced from F1-1 hybrids with T. aestivum cv. Yannong15, and 11 plants were produced from F1-2 hybrids with T. aestivum cv. Yannong15. The PMCs from these 28 BC1F1 hybrid plants were analyzed with cytogenetic and GISH techniques (Table S1). The mean chromosome number of the BC1F1 progeny was 2n = 48.25. Most lines (18 plants) had 2n = 47–49; the distribution range was 44–52 (Table 3). The combinations of average chromosome configurations included 6.17–11.92 univalents, 15.07–18.6 bivalents, 0.31–1.52 trivalents, 0.1–0.79 tetravalents, 0–0.41 pentavalents and 0–0.23 hexavalents (Table S1, Figures 4A–F). GISH analysis revealed that 10–20 E. elongata chromosomes were detected in BC1F1 progeny (Table 3); the distribution range of average E. elongata chromosome configurations was 1.96–8.87 univalents, 2.62–6.41 bivalents, 0.12–1.04 trivalents, and 0.12–1.04 tetravalents (Table S1, Figures 5A,B). The average pairing configuration for wheat-E. elongata chromosomes included 0.15–0.32 bivalents, 0.02–0.06 trivalents, 0–0.03 tetravalents, and 0–0.04 pentavalents (Table S1).
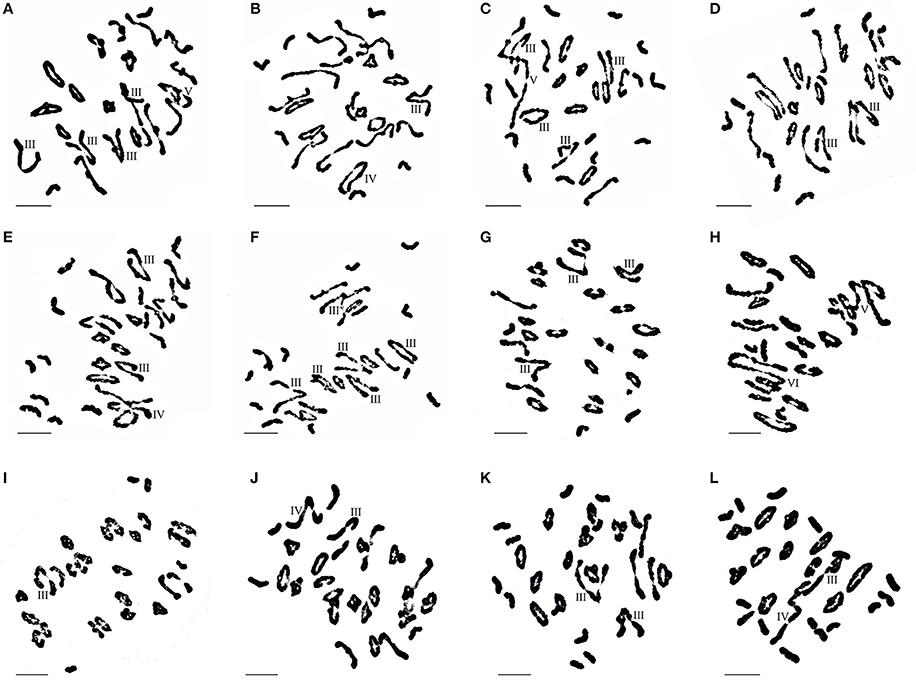
Figure 4. Chromosome configuration of wheat- E. elongata BC1F1, BC1F2 and BC2F1 hybrids. (A–F) Chromosome configurations in BC1F1 (A) 2n = 3I+15II+4III+1IV; (B) 2n = 6I+17II+1III+1IV; (C) 2n = 12I+10II+4III+1V; (D) 2n = 8I+16II+2III; (E) 2n = 11I+12II+2III+1IV; (F) 2n=10I+9II+6III; (G–J) Chromosome configurations in BC1F2 (G) 2n = 3I+19II+3III; (H) 2n = 5I+14II+1V+1VI; (I) 2n = 7I+21II+1III; (J) 2n = 3I+20II+1III+1IV; (K–L) Chromosome configurations in BC2F1 (K) 2n = 10I+13II+2III; (L) 2n = 15I+11II+1III+1IV. Bar = 10 μm.
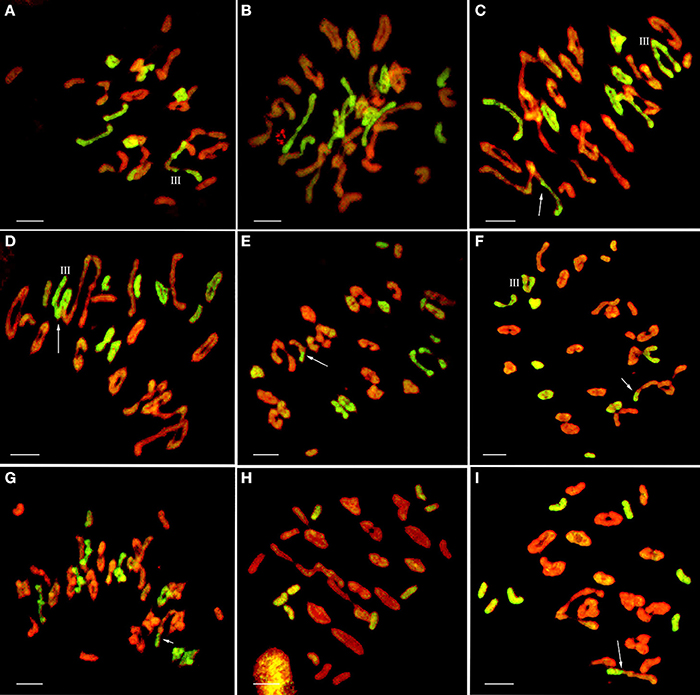
Figure 5. GISH patterns of PMC MI in wheat-E. elongata BC1F1, BC1F2, and BC2F1 hybrids. (A,B) E. elongata chromosome configurations in BC1F1 (A) 2n = 1I+6II+1III; (B) 2n = 3I+7II; (C–G) E. elongata chromosome configurations in BC1F2 (C) 2n = 1I+6II+1III; (D) 2n = 3I+4II+1III; (E) 2n = 5I+6II; (F) 2n = 2I+6II+1III; (G) 2n = 3I+7II; (H–I) E. elongata chromosome configurations in BC2F1 (H) 2n = 8I; (I) 2n = 8I. Wheat chromosomes were detected in red and E. elongata chromosomes or chromosome segments were visualized in green. The arrows indicate pairing between wheat and E. elongata chromosomes. Bar = 10 μm.
Thirty-one BC1F2 plants were randomly selected from BC1F1 self-fertilization progeny for further cytogenetic analysis. The mean chromosome number of the progenies was 2n = 50.13; the distribution range was 42–55 (Table 3). The distribution range of average chromosome configuration at meiotic metaphase I in BC1F2 PMCs included 2.51–16.01 univalents, 11.01–24.25 bivalents, 0.17–2.67 trivalents, 0–1.37 quadrivalents, 0–1.17 pentavalents and 0–0.83 hexavalents (Table S2, Figures 4G–J). GISH analysis during meiosis revealed 7-21 chromosomes with hybridization signals in these 31 plants (Table 3). The average pairing configuration of E. elongata chromosomes included 0.34–6.69 univalents, 0.5–7.94 bivalents, 0–1.14 trivalents and 0–0.54 tetravalents (Table S2, Figures 5C–G). The distribution range of average wheat-E. elongata chromosome configurations was 0.15–0.3 bivalents, 0.02–0.07 trivalents, 0–0.04 tetravalents, and 0–0.05 pentavalents (Table S2).
Twenty-nine BC2F1 plants produced from BC1F1 hybrids with T. aestivum cv. Yannong15 were analyzed by cytogenetic techniques and GISH. Overall, 42–50 total chromosomes and 6–11 E. elongata chromosomes were detected in these plants (Table 3). The distribution range of average chromosome configurations included 8.21–11.77 univalents, 12.68–17.34 bivalents, 0–1.77 trivalents, 0–1.38 tetravalents, 0–0.31 pentavalents and 0–0.1 hexavalents (Table S3, Figures 4K,L). GISH analysis revealed that the average pairing configuration for E. elongata chromosomes included 0.33–0.67 univalents, 4.21–9.64 bivalents, and 0.15–2.39 trivalents (Table S3, Figure 5H,I). The average pairing configuration of wheat-E. elongata chromosomes included 0.17–0.39 bivalents, 0.03–0.14 trivalents, 0–0.04 tetravalents, and 0–0.04 pentavalents (Table S3).
The separation trend is the chromosome variation amplitude of total chromosome number and E. elongata chromosome number of BC1F2 and BC2F1 compared with BC1F1. Obviously, the number of bivalents, trivalents and tetravalents among BC1F1, BC1F2, and BC2F1 plants were different. The numbers of chromosomes increased after selfing according to the result, and the pairing chromosome number also increased after selfing and backcrossing. Additionally, exogenous chromosomes decreased after backcrossing. These results were consistent with the theoretical hypothesis.
Discussion
E. elongata is an influential perennial Triticeae species with a considerable number of traits with the potential to improve wheat. Several studies have reported wide hybridization between E. elongata and other species of Triticeae (Fu et al., 2012; Ayala-Navarrete et al., 2013; Guo et al., 2015). A higher seed set was usually obtained when T. aestivum was used as the female parent, whereas hybrid seed development was usually less successful. In wide hybridization between wheat and E. elongata, a 15.9% (0–76.9%) average seed setting rate in the dozens of combinations showed a very low crossability (Group of Eemote and Northwestern Institute, 1977). It is difficult to obtain offspring from the wheat and E. elongata hybrid; over the years, we have only obtained two perennial F1 plants. Early studies in our laboratory found that, in distant hybridization, when T. aestivum cv. Yannong15 was a parent, the seed setting rate and seed survival rate of the offspring were the highest. Therefore, in order to obtain more seeds, we use T. aestivum cv. Yannong15 as a backcross parent. In this study, we harvested only five seeds from E. elongata × T. aestivum cv. Yannong15 offspring, and only two survived. This may be due to the genome ploidy gap between wheat and E. elongata, although the genetic relationship between them is very close, and may also be caused by the difference between common wheat varieties.
In recent decades, several wheat-E. elongata amphiploid, addition, substitution, and translocation lines have been developed in various laboratories throughout the world and are promising sources of multiple disease resistance (Fu et al., 2012; Zheng et al., 2014; Li et al., 2016). However, few studies have focused on the transmission characteristics of E. elongata chromosomes in the T. aestivum background. GISH has proved to be a useful technique to genetically differentiate closely related genomes, to distinguish alien chromosomes from wheat chromosomes, and to identify wheat-alien translocated chromosomes in a wheat background (Jiang and Gill, 2006; Scoles et al., 2010; Guo et al., 2015). In this study, GISH using E. elongata DNA as a probe was a powerful tool to differentiate chromosomes from T. aestivum and E. elongata hybrid progeny in PMCs at the MI stage. This differentiation allowed the precise analysis of the chromosome composition and the relationships between E. elongata and wheat chromosomes in a wheat genetic background. Using this approach, the genomic composition of the wheat-E. elongata BC1F1, BC1F2, and BC2F1 hybrid progenies was clearly identified in the MI stage and was shown to contain 10–20, 7–21, and 6–11 E. elongata chromosomes, respectively. In the backcross generation, the number of E. elongata chromosomes decreased rapidly; the distribution of E. elongata chromosomes was more extensive in self-progeny. This observation indicated that backcrossing will promote cytological stability and that inbreeding will increase variability.
The genomic composition of E. elongata has been reported to be decaploid, with the genomic designation JJJJJJJsJsJsJs (Chen et al., 1998) or StStStStEeEeEbEbExEx (Zhang et al., 1996). The F1 hybrids were expected to have the genomic constitution of ABDJJJJsJs or ABDStStEeEbEx (2n = 56 chromosomes), and the theoretical E. elongata chromosome configuration of these F1 should be 7II+7III (JJJJsJs) or 21I+7II (StStEeEbEx). In this study, the average E. elongata chromosome configurations of F1 hybrids after GISH analysis were 11.03I+9.81II+0.37III+0.61IV+0.16V and 14.45I+8.4II+0.33III+0.54IV+0.12V. The earlier conclusion that the St and J/Eb (including J/Eb and Js/Ee) genomes are very closely related was drawn from molecular and cytogenetic studies (Liu et al., 2007; Mahelka et al., 2013; Kantarski et al., 2017; Linc et al., 2017). In meiotic metaphase I, these closely related chromosomes may be associated with allosyndetic pairing, thereby reducing the number of univalents and increasing the number of bivalents and multivalents. Thus, in the actual statistical chromosome configuration, the univalents will be less than the theoretical value, while the bivalents and multivalents will be greater than the theoretical value. The average E. elongata chromosome configuration of these two F1 lines accorded with the theoretical chromosome configuration of 21I+7II. Therefore, the genomic composition of E. elongata should be StStStStEeEeEbEbExEx.
The strict pairing of homologous chromosomes in hexaploid wheat reflects a delicate balance between genes that inhibit homologous pairing, such as Ph1 and Ph2, and genes that promote pairing, such as those located on homologous groups 2, 3, and 5 (Naranjo and Benavente, 2015). A similar theory was suggested for Elytrigia species. Dvorák (1987) proposed that the chromosome arms 3ES, 3EL, 4ES, and 5Ep and chromosome 6E of T. elongatum had genes that induce homoeologous chromosome pairing. Charpentier et al. (1988) further demonstrated that the role of 5E in the wheat and Agropyron elongatum hybrid was similar to the deletion of the Ph1 gene. Later, Zhang et al. (1995) implied that two basic chromosomes in E. elongata encode genes that promote homoeologous chromosome pairing and might have additive effects. Although more recent studies observed similar inferences, there is no direct evidence to confirm these hypotheses. In the present study, pairing between wheat and E. elongata was detected in each of the wheat-E. elongata hybrid progenies, albeit rarely. This result suggests a close genetic relationship between wheat and E. elongata chromosomes. Similar results were detected on meiotic chromosomes at MI in trigeneric hybrids produced from a heterozygous Langdon Ph mutant (Ph1ph1b) or Langdon 5D (5B) disomic substitution line (without Ph1) hybridization with the JJEE amphidiploids using multicolor fluorescent GISH by Jauhar et al. (2004) and (Jauhar and Peterson (2006)). The pairing between wheat and E. elongata chromosomes can be used as direct evidence that genes promoting homoeologous chromosome pairing or Ph suppressor genes exist in E. elongata. Although it is worthwhile for E. elongata chromosomes to promote homoeologous pairing or inhibit Ph gene effects, the use of these genotypes might promote the homoeologous pairing of E. elongata and wheat chromosomes and facilitate alien gene transfer into the wheat genome.
Common wheat is a major, global cereal crop that accounts for approximately 20% of the calories consumed by humans (Brenchley et al., 2012). However, effective wheat breeding has been hindered by a narrow genetic base (Friebe et al., 1996). Genes from wild relatives have been exploited to confer desirable agronomic traits to wheat, as illustrated by the application of many wheat-alien translocation lines (Lukaszewski, 2001). For example, Lr26/Sr31/Yr9/Pm8 have endowed the translocation line T1RS·1BL with improved environmental adaption and enhanced kernel numbers (Friebe et al., 1996). Both T. aestivum-Thinopyrum bessarabicum T2JS-2BS·2BL and T. aestivum-Dasypyrum villosum T2VS·2DL translocation lines have been reported with elevated grain numbers per spike (Qi et al., 2010; Zhang et al., 2015). However, the formation of these translocation lines is rarely reported. GISH patterns of meiotic chromosomes at MI in these hybrids of wheat with E. elongata indicated that chromosome pairing in these hybrids mainly occurred among wheat chromosomes and among E. elongata chromosomes and that allosyndetic pairing between wheat and E. elongata chromosomes was very rare (Table 1). The much higher frequencies of autosyndetic pairing than allosyndetic pairing in these hybrids of wheat with E. elongata demonstrated that the relationships among T. aestivum genomes and among E. elongata genomes are much closer than the relationship between T. aestivum and E. elongata genomes. Meanwhile, these allosyndetic pairings promote the recombination between homologous chromosomes, enrich the genetic diversity of distant hybrid progeny, and improve the frequency of the offspring to obtain a translocation line, which will benefit from the selection of excellent genetic resources, and thus applied to wheat breeding. Our results demonstrate that GISH using E. elongata genomic DNA as a probe provided a reliable approach to discriminate the identity of chromosomes involved in pairing. This observation might significantly improve our understanding of the genomic relationships within Triticeae. Knowledge of the relationships between wheat and grass genomes also improves our understanding of characteristic inheritance to generate efficient strategies for transferring target gene(s) from E. elongata to wheat. With the advancement and development of technology, multicolor GISH (mcGISH) has been widely used in academic research to simultaneously visualize two or more genomes in a polyploid species (Zheng et al., 2014; Guo et al., 2015). Although there are few reports analyzing chromosome pairing behavior using mcGISH, our future research will focus on these types of analyses. This approach might extend the analysis of chromosomes, genomes and phylogenies, especially for the analysis of complex polyploids and their hybrids in wheat.
Author Contributions
FH, PX, and YB performed the experiments, analyzed the data and wrote the manuscript. MR, SL, YW, XL, and HW designed the study and discussed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was funded by the National Key Research and Development Plan of China (2016YFD0102004), the National Natural Science Foundation of China (Grant No. 31501298, No. 31671675 and No. 31660390), the National Key Research and Program of China (2017YFD0100900) and the Provincial Science and Technology Plan for Colleges in Shandong Province (Grant No. J17KA151).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.02161/full#supplementary-material
References
Ayala-Navarrete, L. I., Mechanicos, A. A., Gibson, J. M., Singh, D., Bariana, H. S., Fletcher, J., et al. (2013). The Pontin series of recombinant alien translocations in bread wheat: single translocations integrating combinations of Bdv2, Lr19 and Sr25 disease-resistance genes from Thinopyrum intermedium and Th. ponticum. Theor. Appl. Genet. 126, 2467–2475. doi: 10.1007/s00122-013-2147-0
Bagherikia, S., Karimzadeh, G., and Naghavi, M. R. (2014). Distribution of 1AL.1RS and 1BL.1RS wheat-rye translocations in Triticum aestivum using specific PCR. Biochem. Syst. Ecol. 55, 20–26. doi: 10.1016/j.bse.2014.02.001
Bao, Y., Wu, X., Zhang, C., Li, X., He, F., Qi, X., et al. (2014). Chromosomal constitutions and reactions to powdery mildew and stripe rust of four novel wheat-Thinopyrum intermedium partial amphiploids. J. Genet. Genomics 41, 663–666. doi: 10.1016/j.jgg.2014.11.003
Brenchley, R., Spannagl, M., Pfeifer, M., Barker, G. L. A., D'Amore, R., Allen, A. M., et al. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705–710. doi: 10.1038/nature11650
Cai, X., and Jones, S. (1997). Direct evidence for high level of autosyndetic pairing in hybrids of Thinopyrum intermedium and Th. ponticum with Triticum aestivum. Theor. Appl. Genet. 95, 568–572. doi: 10.1007/s001220050597
Charpentier, A., Cauderon, Y., and Feldman, M. (1988). “Control of chromosome pairing in Agropyron elongatum”, in Proceedings of the Seventh International Wheat Genetics Symposium, eds T. E. Miller and R. M. D. Koebner (Cambridge: Published by the Institute of Plant Science Research, Cambridge Laboratory).
Chen, Q., Conner, R., Laroche, A., and Ahmad, F. (2001). Molecular cytogenetic evidence for a high level of chromosome pairing among different genomes in Triticum aestivum-Thinopyrum intermedium hybrids. Theor. Appl. Genet. 102, 847–852. doi: 10.1007/s001220000496
Chen, Q., Conner, R., Laroche, A., and Thomas, J. (1998). Genome analysis of Thinopyrum intermedium and Thinopyrum ponticum using genomic in situ hybridization. Genome 41, 580–586. doi: 10.1139/g98-055
Dvorák, J. (1987). Chromosomal distribution of genes in diploid Elytrigia elongata that promote or suppress pairing of wheat homoeologous chromosomes. Genome 29, 34–40. doi: 10.1139/g87-006
Friebe, B., Jiang, J., Raupp, W. J., McIntosh, R. A., and Gill, B. S. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91, 59–87. doi: 10.1007/BF00035277
Fu, S., Lv, Z., Qi, B., Guo, X., Li, J., Liu, B., et al. (2012). Molecular cytogenetic characterization of wheat-Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium Head Blight. J. Genet. Genomics 39, 103–110. doi: 10.1016/j.jgg.2011.11.008
Group of Eemote and Northwestern Institute (1977). The cross breeding and its genetic analysis between Triticum aestivum and Agropyron elongatum. J. Genet. Genomics (04), 283–293.
Guo, X., Shi, Q., Wang, J., Hou, Y., Wang, Y., and Han, F. (2015). Characterization and genome changes of new amphiploids from wheat wide hybridization. J. Genet. Genomics 42, 459–461. doi: 10.1016/j.jgg.2015.06.006
He, F., Xu, J., Qi, X., Bao, Y., Li, X., Zhao, F., et al. (2013). Molecular cytogenetic characterization of two partial wheat-Elytrigia elongata amphiploids resistant to powdery mildew. Plant Breed. 132, 553–557. doi: 10.1111/pbr.12104
Hu, L. J., Li, G. R., Zeng, Z. X., Chang, Z. J., Liu, C., and Yang, Z. J. (2011). Molecular characterization of a wheat-Thinopyrum ponticum partial amphiploid and its derived substitution line for resistance to stripe rust. J. Appl. Genet. 52, 279–285. doi: 10.1007/s13353-011-0038-0
Jauhar, P. P., Dogramaci, M., and Peterson, T. (2004). Synthesis and cytological characterization of trigeneric hybrids of durum wheat with and without Ph1. Genome 47, 1173–1181. doi: 10.1139/g04-082
Jauhar, P., and Peterson, T. (2006). Cytological analyses of hybrids and derivatives of hybrids between durum wheat and Thinopyrum bessarabicum, using multicolour fluorescent GISH. Plant Breed. 125, 19–26. doi: 10.1111/j.1439-0523.2006.01176.x
Jiang, J., and Gill, B. S. (2006). Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49, 1057–1068. doi: 10.1139/g06-076
Kantarski, T., Larson, S., Zhang, X., DeHaan, L., Borevitz, J., Anderson, J., et al. (2017). Development of the first consensus genetic map of intermediate wheatgrass (Thinopyrum intermedium) using genotyping-by-sequencing. Theor. Appl. Genet. 130, 137–150. doi: 10.1007/s00122-016-2799-7
Kato, A., Lamb, J. C., and Birchler, J. A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. U.S.A. 101, 13554–13559. doi: 10.1073/pnas.0403659101
Li, H., and Wang, X. (2009). Thinopyrum ponticum and Th. intermedium: the promising source of resistance to fungal and viral diseases of wheat. J. Genet. Genomics 36, 557–565. doi: 10.1016/S1673-8527(08)60147-2
Li, H., Zheng, Q., Pretorius, Z. A., Li, B., Tang, D., and Li, Z. (2016). Establishment and characterization of new wheat-Thinopyrum ponticum addition and translocation lines with resistance to Ug99 races. J. Genet. Genomics 43, 573–575. doi: 10.1016/j.jgg.2016.07.004
Linc, G., Gaal, E., Molnar, I., Icso, D., Badaeva, E., and Molnar-Lang, M. (2017). Molecular cytogenetic (FISH) and genome analysis of diploid wheatgrasses and their phylogenetic relationship. PLoS ONE 12:e0173623. doi: 10.1371/journal.pone.0173623
Liu, Z., Li, D., and Zhang, X. (2007). Genetic relationships among Five Basic Genomes St, E., A, B and D in Triticeae Revealed by Genomic Southern and in situ hybridization. J. Integr. Plant Biol. 49, 1080–1086. doi: 10.1111/j.1672-9072.2007.00462.x
Lukaszewski, A. J. (2001). Breeding behavior of the cytogenetically engineered wheat-rye translocation chromosomes 1RS.1BL. Crop Sci. 41, 1062–1065. doi: 10.2135/cropsci2001.4141062x
Mahelka, V., Kopecky, D., and Baum, B. R. (2013). Contrasting patterns of evolution of 45S and 5S rDNA families uncover new aspects in the genome constitution of the agronomically important grass Thinopyrum intermedium (Triticeae). Mol. Biol. Evol. 30, 2065–2086. doi: 10.1093/molbev/mst106
Naranjo, T., and Benavente, E. (2015). “The mode and regulation of chromosome pairing in wheat–alien hybrids (Ph Genes, an Updated View),” in Alien Introgression in Wheat, eds M. Molnar, C. Ceoloni, and J. DoleŽel (Cham: Springer), 133–162. doi: 10.1007/978-3-319-23494-6_6
Niu, Z., Klindworth, D. L., Yu, G., T, L. F., Chao, S., Jin, Y., et al. (2014). Development and characterization of wheat lines carrying stem rust resistance gene Sr43 derived from Thinopyrum ponticum. Theor. Appl. Genet. 127, 969–980. doi: 10.1007/s00122-014-2272-4
Qi, L., Friebe, B., Zhang, P., and Gill, B. S. (2007). Homoeologous recombination, chromosome engineering and crop improvement. Chromosome Res. 15, 3–19. doi: 10.1007/s10577-006-1108-8
Qi, Z., Du, P., Qian, B., Zhuang, L., Chen, H., Chen, T., et al. (2010). Characterization of a wheat–Thinopyrum bessarabicum (T2JS-2BS∙2BL) translocation line. Theor. Appl. Genet. 121, 589–597. doi: 10.1007/s00122-010-1332-7
Roundy, B. A. (1985). Emergence and establishment of basin wildrye and tall wheatgrass in relation to moisture and salinity. J. Range Manag. 38, 126–131. doi: 10.2307/3899254
Scoles, G., Wang, Q., Xiang, J., Gao, A., Yang, X., Liu, W., et al. (2010). Analysis of chromosomal structural polymorphisms in the St, P., and Y genomes of Triticeae (Poaceae). Genome 53, 241–249. doi: 10.1139/G09-098
Sears, E. R. (1976). Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 10, 31–51. doi: 10.1146/annurev.ge.10.120176.000335
Sepsi, A. I. (2010). Molecular Cytogenetic Characterisation of a Leaf-Rust Resistant Wheat-Thinopyrum pontificum Partial Amphiploid. Doctoral Dissert, Eotvos Lorand Univesity of Science, Budapest.
Song, L., Lu, Y., Zhang, J., Pan, C., Yang, X., Li, X., et al. (2016). Cytological and molecular analysis of wheat - Agropyron cristatum translocation lines with 6P chromosome fragments conferring superior agronomic traits in common wheat. Genome 59, 840–850. doi: 10.1139/gen-2016-0065
Su, Y., Zhang, D., Li, Y., and Li, S. (2016). Nonhomologous chromosome pairing in aegilops-secale hybrids. Cytogenet. Genome Res. 147, 268–273. doi: 10.1159/000444435
Zhan, H., Li, G., Zhang, X., Li, X., Guo, H., Gong, W., et al. (2014). Chromosomal location and comparative genomics analysis of powdery mildew resistance gene Pm51 in a putative wheat-Thinopyrum ponticum introgression line. PLoS ONE 9:e113455. doi: 10.1371/journal.pone.0113455
Zhang, R., Hou, F., Feng, Y., Zhang, W., Zhang, M., and Chen, P. (2015). Characterization of a Triticum aestivum–Dasypyrum villosum T2VS•2DL translocation line expressing a longer spike and more kernels traits. Theor. Appl. Genet. 128, 2415–2425. doi: 10.1007/s00122-015-2596-8
Zhang, X., Dong, Y., and Wang, R. R.-C. (1996). Characterization of genomes and chromosomes in partial amphiploids of the hybrid Triticum aestivum× Thinopyrum ponticum by in situ hybridization, isozyme analysis, and RAPD. Genome 39, 1062–1071. doi: 10.1139/g96-133
Zhang, X., Dong, Y., and Yang, X. (1995). Cytogenitic research on hybrids of Triticum with both Thinopyrum ponticum and Th. Intermedium as well as their derivatives- III. Primary detection of genrtic base for introgression of useful genes from the two alien species to wheat. J. Genet. Genomics 22, 217–222
Keywords: E. elongata, T. aestivum, chromosome pairing, hybrid progenies, genomic in situ hybridization
Citation: He F, Xing P, Bao Y, Ren M, Liu S, Wang Y, Li X and Wang H (2017) Chromosome Pairing in Hybrid Progeny between Triticum aestivum and Elytrigia elongata. Front. Plant Sci. 8:2161. doi: 10.3389/fpls.2017.02161
Received: 24 July 2017; Accepted: 07 December 2017;
Published: 19 December 2017.
Edited by:
Mónica Pradillo, Complutense University of Madrid, SpainReviewed by:
Pilar Prieto, Consejo Superior de Investigaciones Científicas (CSIC), SpainIsabelle Colas, James Hutton Institute, United Kingdom
Copyright © 2017 He, Xing, Bao, Ren, Liu, Wang, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingfeng Li, bGl4ZkBzZGF1LmVkdS5jbg==
Honggang Wang, aGd3YW5nQHNkYXUuZWR1LmNu
 Fang He
Fang He Piyi Xing2
Piyi Xing2 Yinguang Bao
Yinguang Bao Xingfeng Li
Xingfeng Li