- 1Bio-Protection Research Centre, Lincoln University, Lincoln, New Zealand
- 2School of Biological Sciences, The University of Auckland, Auckland, New Zealand
- 3Department of Wine, Food and Molecular Biosciences, Lincoln University, Lincoln, New Zealand
- 4Bio-Protection Research Centre, New Zealand and Institute of Fundamental Sciences, Massey University, Wellington, New Zealand
- 5Lincoln Agritech Ltd., Lincoln, New Zealand
In Nature, almost every plant is colonized by fungi. Trichoderma virens is a biocontrol fungus which has the capacity to behave as an opportunistic plant endophyte. Even though many plants are colonized by this symbiont, the exact mechanisms by which Trichoderma masks its entrance into its plant host remain unknown, but likely involve the secretion of different families of proteins into the apoplast that may play crucial roles in the suppression of plant immune responses. In this study, we investigated T. virens colonization of maize roots under hydroponic conditions, evidencing inter- and intracellular colonization by the fungus and modifications in root morphology and coloration. Moreover, we show that upon host penetration, T. virens secretes into the apoplast an arsenal of proteins to facilitate inter- and intracellular colonization of maize root tissues. Using a gel-free shotgun proteomics approach, 95 and 43 secretory proteins were identified from maize and T. virens, respectively. A reduction in the maize secretome (36%) was induced by T. virens, including two major groups, glycosyl hydrolases and peroxidases. Furthermore, T. virens secreted proteins were mainly involved in cell wall hydrolysis, scavenging of reactive oxygen species and secondary metabolism, as well as putative effector-like proteins. Levels of peroxidase activity were reduced in the inoculated roots, suggesting a strategy used by T. virens to manipulate host immune responses. The results provide an insight into the crosstalk in the apoplast which is essential to maintain the T. virens-plant interaction.
Introduction
Trichoderma spp. are cosmopolitan soil fungi with the capacity to establish symbiotic relationships within the roots of most plant species (Harman et al., 2004). Trichoderma spp. promote plant growth, increase nutrient availability, improve crop production, and increase sensitivity to respond successfully to later pathogen invasion termed as induced systemic resistance (ISR) (Shoresh and Harman, 2008; Vinale et al., 2008; Shoresh et al., 2010). Despite of the direct benefits obtained from this mutualistic interaction, plants react to endophyte colonization via a basal immune response activation, in which, plants have evolved different strategies to recognize conserved microbial features referred to as microbe-associated molecular patterns (MAMPs) (Lorito et al., 2010; Zamioudis and Pieterse, 2012; Schmoll et al., 2016; Mendoza-Mendoza et al., 2018). Diverse MAMPs synthetized by Trichoderma have been identified (Hermosa et al., 2013), including the cerato-platanin protein Sm1 (Djonović et al., 2006) and the ethylene-inducing xylanase (EIX) (Ron and Avni, 2004). The proteinaceous elicitor Sm1 is induced during Trichoderma virens-plant interaction, which promotes the expression of pathogenesis-related genes (Djonović et al., 2007). The EIX has a dual role during plant colonization, involving both lytic enzyme activity and induction of systemic resistance in specific cultivars of tobacco and tomato (Rotblat et al., 2002; Ron and Avni, 2004).
Endophytic Trichoderma penetrates the first or second layers of plant root systems, first colonizing the root epidermis and then into the cortex area, without reaching the vascular system (Chacón et al., 2007). The initial steps of root colonization by Trichoderma starts with the attachment on the root surface followed by the formation of appressoria-like structures that may help for the penetration into the internal tissues (Yedidia et al., 1999, 2000; Viterbo and Chet, 2006). After recognition by Trichoderma MAMPs, the plant responds by depositing callose in the neighborhood cells allowing only superficial cell-colonization. Microscopic observations of early colonization of tomato roots by T. harzianum showed the capacity of the fungus to colonize inter- and intracellular spaces without disrupting cell integrity (Chacón et al., 2007). However, T. asperellum (formally called T. harzianum) induces morphological and physiological changes in cucumber plant roots, which includes necrosis of the penetration peg, high chitinase activity and formation of fluorescent products in intercellular spaces of the colonized roots (Yedidia et al., 1999). Additionally, enhanced protection against reactive oxygen species (ROS), and repression of the ethylene synthesis pathway is proposed to enable root colonization by Trichoderma as has been shown in other endophytes (Shoresh et al., 2010). Recently, was observed that the cerato-platanin elicitor Sm2 from T. virens is required for root colonization (Crutcher et al., 2015), although the mode of action is currently unknown.
Plant symbionts and pathogens have developed specific strategies to promote colonization by evading the first layer of plant defense called MAMP-triggered immunity (MTI). Plant microbes secrete molecules including effector proteins into the apoplast where they interact with their molecular targets or are translocated into the plant cell cytoplasm blocking downstream signals, thereby suppressing MTI. Examples of conventional secreted effectors, including the Cladosporium fulvum LysM effector Ecp6 which prevents elicitation of host defense by sequestering chitin oligosaccharides of the fungus (de Jonge et al., 2010), and the toxin-like ToxB which is secreted into the apoplast by the necrotrophic fungus Pyrenophora tritici-repentis and is necessary for complete disease development in wheat (Figueroa et al., 2015). Furthermore, unconventionally secreted effector proteins also play important roles in the manipulation of plant processes. The protein chorismate mutase Cmu1, secreted by Ustilago maydis, manipulates the metabolome of neighboring cells to favor pathogen infection (Djamei et al., 2011).
Recent studies have focused on the secretome of T. virens to the presence of maize roots (Lamdan et al., 2015). However, proteins from the apoplastic region, where T. virens closely interacts with host cells, were not considered in this study. Plant-associated microbes and their host plants continuously secrete an arsenal of proteins into the apoplast using a conventional secretion system, which involves the Golgi-endoplasmic reticulum pathway an approach for those proteins that carry an N-terminal signal peptide. Apoplastic proteins (APs) are also delivered into the apoplast by leaderless secretory pathways (LSPs) that constitutes, on average, 50% of the plant and fungal secretomes (Agrawal et al., 2010; Ding et al., 2012; Girard et al., 2013; Delaunois et al., 2014). APs secreted by both players have major roles in the maintenance of plant cell wall structure, stress responses, primary and secondary metabolism, defense and signaling (Alexandersson et al., 2013; Kim et al., 2013).
Proteomic tools such as mass spectrometry (MS) enables identification of key proteins of the secretome during complex physiological cell processes such as microbe-host interactions (Schmidt and Volker, 2011; Delaunois et al., 2014; Gupta et al., 2015). One clear example is the study of the apoplastic secretome of the phytopathogenic fungus Magnaporthe oryzae during interaction with rice plants, where Kim et al. (2013) identified more than 200 proteins secreted into the apoplast including putative effector proteins. Here we present the morphological changes occurring in the maize roots by their interaction with T. virens Gv29.8 in a sterile system, then we identified the fungal colonization to the plant root tissue and finally we analyzed the apoplastic secretome during the interaction between T. virens and maize roots by using two different approaches: (1) gel-based, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) couple with LC-MS/MS which allows separation and identification of a large number of proteins (Gel-LC-MS/MS), and (2) gel-free shotgun proteomics, which is more powerful technology for large-scale separation and identification of complex mixtures of proteins (González-Fernández et al., 2010; Porteus et al., 2011; Jayaraman et al., 2012). The results will provide a better understanding of how endophytic T. virens modulates host plant defensive processes and how plant responds to the presence of the fungus.
Materials and Methods
Maize Germination
Maize seeds from hybrid line 34H31 (Pioneer® Brand Products, Gisborne, New Zealand) were surface sterilized by soaking in 2% (w/v) sodium hypochlorite (NaOCl) (active ingredient) for 7 min, followed by 70% ethanol for 7 min, then washed three times with sterile nanopure water. Seeds were germinated on sterile seed germination papers (30 × 45 cm; Anchor Paper Company, MN, USA) previously soaked in sterile Hoagland's No.2 basal salt solution (Sigma-Aldrich, MO, USA) (Hoagland and Arnon, 1950), for 60 h in a humidity controlled plant growth chamber at 25°C under a 16 h light/8 h dark cycle and a relative humidity of 80%.
Inoculum Preparation
T. virens Gv 29.8 conidia was propagated on potato-dextrose agar (PDA) (Difco, Fisher Scientific, NH, USA) at 25°C under a cycle of 12 h light and 12 h dark for 7 d to induce conidiation. Conidia were collected using sterile nanopure water and filtered through a double layer of sterile of Miracloth (Millipore Merck, MA, USA).
Colonization of Maize Roots by Trichoderma virens
Sterilized maize seeds were surface inoculated with 1 × 106 conidia. After germination, seedlings were grown under hydroponic conditions without aeration in 50 mL centrifuge tubes containing 45 mL of sterile Hoagland's solution with a piece of sterile cotton to support the seedling. Seedlings were incubated for an additional 60 h as described by Lawry (2016). For sampling, fresh root tissues were washed gently with sterile nanopure water, and then 2 cm sections nearest to the seed were cut from the primary root where T. virens primarily colonize (Lawry, 2016). Un-inoculated roots samples were taken as control.
Confocal Visualization of Maize Roots Colonization by Trichoderma virens
T. virens root colonization of maize seeds was examined using confocal microscopy (Fluoview FV10i, Olympus, Tokyo, Japan). For this analysis, transverse free-hand sections of maize roots were prepared. After 5 d post inoculation (d.p.i) maize roots were collected and either washed in phosphate-buffered saline (PBS) pH 7.4 or fixed in fresh ethanol: acetic acid (3:1, v/v) solution. Two staining methods were used: for fresh tissues a wheat germ agglutinin (WGA)-Alexa Fluor™ 488 (Thermo Fisher Scientific, MA, USA) /FM4-64 dye (Thermo Fisher Scientific, USA) mixture was used, while fixed tissues were stained with WGA-Alexa Fluor™ 488/Propidium iodide (PI) (Sigma-Aldrich, USA) mixture. Fungal material was stained with WGA-Alexa Fluor™ 488 (Mochizuki et al., 2011). Plant cell walls were stained with PI, while the plasma membranes were stained with FM4-64 (Bolte et al., 2004).
For fixed tissues, roots were treated with 10% KOH for 4 h at 95°C and then transferred to PBS pH 7.4 for 1 h. Samples were infiltrated with the staining solution (20 μg/mL PI; 10 μg/mL WGA-Alexa Fluor™ 488, 0.02% Tween 20 made up in 1X PBS) for 15 min twice. Samples were distained in PBS-tween (0.02%) and stored in the dark at 4°C. For fresh tissues, samples were washed in PBS solution and infiltrated with the same staining solution mentioned above, except that PI was substituted for 5 mM FM4-64.
Isolation of Total Protein From Maize Primary Root
For total protein extraction from maize roots the methodology described by Wu et al. (2014) was used with modifications as follows. Fresh root tissue (0.25 g) was ground into a fine powder in liquid nitrogen, then homogenized in 2.5 mL of ice cold Tris/ethylenediaminetetraacetic acid (EDTA) extraction buffer, containing 1 mM EDTA, 10 mM Tris-HCL pH 8, 2% w/v polyvinylpolypyrrolidone (PVPP) and with 0.3% (v/v) Pefabloc (Sigma-Aldrich, USA). Samples were centrifuged at 5,000 × g for 30 min at 4°C, then supernatant proteins were precipitated with 10 mL of cold trichloroacetic acid (TCA)/acetone (−20°C). After centrifugation at 3,000 × g for 10 min, the pellet was washed three times with ice cold acetone containing 0.007% w/v dithiothreitol (DTT). Protein pellets were dried to evaporate any remaining acetone and stored at −80°C.
Isolation of Apoplastic Proteins
For isolation of apoplastic fluid (AF), the primary root on the proximal side nearest to the seed (2 cm section) was cut from one side and collected using preferably the infiltration-centrifugation methodology (see Supplementary Methods). Individual primary roots from 20 plants were combined for each replicate. Three replicates were used for each condition: (a) inoculated and (b) un-inoculated plants. Roots were sampled 5 d.p.i.
Primary root sections (2 cm) were placed immediately in 100 mM sodium phosphate buffer (SPB) pH 6.5 (Witzel et al., 2011) supplemented with 0.3% (v/v) Pefabloc and 10 mM EDTA. The chilled samples were vacuum infiltrated by reducing the pressure at −45 kPa for 15 min using a diaphragm vacuum pump (Rocker 400, Rocker Scientific, Taipei, Taiwan), followed by slow return to atmospheric pressure to avoid cell damage (Dragišić Maksimović et al., 2008). Roots were then placed in a 5 mL syringe without the plunger and placed inside a 15 mL centrifuge tube then centrifuged at 2,000 × g for 15 min at 4°C (Model 5810R, Eppendorf, Hamburg, Germany). The harvested AF was filtered through cellulose acetate membrane filters (0.2 μm porosity; Axiva Sichem Biotech, Delhi, India) and stored at −80°C.
APs were concentrated using the TCA-sodium deoxycholate (Na-DOC)/acetone method described by Bensadoun and Weinstein (1976) with modifications. Briefly, for every volume of AF solution, 0.1 vol. of 2% of Na-DOC and 100% TCA were added and the samples were kept at RT for 1 h. Samples were then centrifuged at 14,000 × g for 10 min at 4°C, the supernatants removed and the pellet dried. The pellet was then washed in 200 μL of ice-cold acetone, placed on ice for 15 min, then centrifuged at 14,000 × g for 10 min at 4°C. The pellet was then air dried before being re-suspended in 10 μL in PBS buffer pH 7.0.
For gel-free shotgun proteomics, modifications were carried out to improve the method described above. Root sections were placed immediately in 50 mM potassium phosphate buffer (PPB) pH 5.5 supplemented with 0.3% (v/v) Pefabloc and 10 mM EDTA. Roots were then vacuum infiltrated with the PPB solution. The root samples were centrifuged at 2,000 × g for 15 min at 4°C (Dragišić Maksimović et al., 2008) and harvested AF was immediately snap frozen in liquid nitrogen. APs samples from un-inoculated and inoculated roots were concentrated by freeze drying (Thermo Savant Micro Modulyo-115, Thermo Fisher Scientific, USA). Samples were rehydrated in 30 μL of 50 mM ammonium acetate buffer pH 5.5 for quantification and then stored at −80°C.
Malate Dehydrogenase Activity
Malate dehydrogenase (MDH) activity was performed as described by Husted and Schjoerring (1995) with modifications. MDH activity was assayed to determine cytoplasmic contamination in AF. A total of 5 μg of protein extract (total or apoplastic) were added into 3 mL reaction mixtures containing 0.094 mM β-NADH (Sigma-Aldrich, USA), 0.17 mM oxaloacetic acid (Sigma-Aldrich, USA), and 0.1 M phosphate buffer, pH 7.5. Oxidation of NADH was measured at 340 nm using an UV-Vis spectrophotometer (Genesys 10S, Thermo Fisher Scientific, USA), monitoring for 5 min at 25°C. A non-enzyme reaction mix was used as a blank. Enzymatic activity in AF was expressed as a percentage of the total root protein extract activity.
Identification of Apoplastic Proteins by Gel-LC-MS/MS
APs were separated on a 4–12% pre-cast NuPAGE bis-tris gel (Novex, Life Technologies, CA, USA) in 1X MOPS SDS running buffer (2.5 mM MOPS, 2.5 mM Tris, 0.005% SDS, 0.05 mM EDTA, pH 7.7). Prior to loading, samples were mixed with 2 μL 6X Tris-Glycine SDS sample buffer (0.378 M Tris-HCl pH 6.8, 0.6 M DTT, 12% SDS, 60% Glycerol, 0.06% Bromophenol Blue). Gels were stained with Coomassie blue solution (10% acetic acid, 50% methanol, 0.25% Coomassie blue R-250). Molecular weights were determined using the SDS-PAGE PageRuler Plus prestained protein ladder (Fermentas, MA, USA).
For identification, appropriate protein zones were excised and subjected to trypsin digestion. Briefly, excised gel plugs were washed three times with 200 μL of 1:1 acetonitrile: 50 mM ammonium bicarbonate (ABC) solution pH 8.3, and dried in a vacuum centrifuge. The plugs were then incubated with 10 mM DTT in 50 mM ABC solution pH 8.3 for 1 h at 56°C. Each plug was diced into small cubes with dimensions of 0.5–1.0 mm. For distaining, 200 μL of 50% acetonitrile (ACN)/ 50 mM ABC was added and the tubes were vortexed for 30 s. Plugs were then incubated at 45°C for 15 min in a Discoverer II System microwave (CEM Corporation, NC, USA). For gel dehydration and reduction, 200 μL of 100% ACN was added and the tubes vortexed for 30 s and placed in a heating block (Thermomixer, Eppendorf, USA) at 56°C with the lids open to allow ACN evaporation. Once the plugs were dry, 50 μL of 10 mM DTT was added and tubes were incubated at 56°C for 15 min, for protein reduction. For alkylation, the remaining liquid was removed and 50 μL of 50 mM iodoacetamide (IAM) was added and the plugs were incubated in darkness for 60 min at RT. The digestion was carried out by adding 50 μL of freshly prepared 12.5 ng/μL trypsin (Roche, Basel, Switzerland) suspended in 50 mM ABC solution supplemented with 10% ACN and incubated at 37°C for 18 h. The resulting peptides were extracted first with nanopure water and twice with water–acetonitrile–formic acid solution (45/50/5) then concentrated to a minimal volume (~10 μL), and mixed with 30 μL with 5% acetonitrile in nanopure water containing 1% formic acid.
Peptide samples were then subjected to electrospray LC-MS/MS using a Finnigan™ LTQ-FT™ tandem mass spectrometer (Thermo Fisher Scientific, USA). The peptides were separated by reversed-phase chromatography on a Zorbax SB-300 C-18 column (150 mm × 300 μm, 5 μm particle size 300 Å pore size) (Agilent Technologies, CA, USA) eluting with an acetonitrile gradient in water containing 0.1% formic acid from 5 to 60% over 40 min using a Surveyor MS pump (Thermo Fisher Scientific, USA). The eluent entered the electrospray source at a flow rate of 5–6 μL.min−1 produced by splitting the Surveyor flow of 100 μL.min−1 with an UpChurch variable flow splitter. The mass spectrometer was operated in the positive ion mode with helium as the collision gas; the mass/charge range acquired was 300–2000 m/z. The capillary temperature was set at 210°C; the source voltage set at 3.8 kV. Data were acquired using a Top 5 experiment (one full scan in both the ion trap and ICR cell (parallel mode) followed by two averaged MS/MS microscans of each of the top five ions recorded in the ion trap) in data-dependent mode with dynamic exclusion enabled. Full scan Fourier transform data were obtained at a resolution of 100,000 at m/z 400 and used to refine the database search parameters.
Identification of the peptides was undertaken using Proteome Discoverer 1.4 (Thermo Fisher Scientific, USA), which was used to search fragment ion spectra matching peptides previously digested in silico using the proteome from maize (Zea mays) (http://www.plantgdb.org) and T. virens (http://genome.jgi-psf.org/TriviGv29_8_2) databases.
Identification of Apoplastic Proteins by Gel-Free Shotgun Proteomics
For this analysis, APs (15 μg) for each biological replicate were digested separately. For each sample, 15 μL of 1 M of ABC were added, and adjusted to 100 μL with 50 mM ABC. Samples were reduced as follows: 1 M DTT was added to the samples to a final concentration of 10 mM, and then incubated at 56°C using 50 W for 15 min in a Discoverer II System microwave. The pH of the samples was adjusted to pH 8 with 1 M ABC. For alkylation, 1 M IAM was added to the samples to reach a final concentration of 50 mM. APs samples were kept in total darkness at RT for 30 min. After incubation, 1 M DTT was added to the samples to a final concentration of 20 mM. For protein digestion, 1 μg of trypsin (Promega, WI, USA) was added to 15 μg of total protein. Digestion was conducted for 60 min using microwave digestion at 45°C and 15 W power. After digestion, the reaction was quenched by addition of 2 μL of 50% formic acid. The tryptic peptides were desalted using a sensitive solid phase extraction (SPE) method with 1 mL Oasis HLB 10 mg extraction cartridges (Waters, MA, USA). Samples were diluted to 0.5 mL in 0.1% formic acid and pH was verified to a pH between 2 and 3 using pH indicator strips. A second SPE extraction was performed using 1 mL Oasis MCX 30 mg extraction cartridges (Waters, USA). APs samples were eluted with 0.3 mL of freshly prepared 50% acetonitrile in nanopure water, then concentrated in a Speedvac concentrator (Thermo Scientific, USA) to a volume between 10-15 μL, vortexed vigorously, then centrifuged at 16,000 × g for 30 s and finally diluted in 0.1% formic acid for MS analysis.
APs digests were separated on a 0.075 × 200 mm picofrit column (New Objective, Scientific Instrument Service, NJ, USA) packed with Reprosil C18 media (Dr Maisch GmbH HPLC, Entringen, Germany) using the following gradient: 0 min 5% B, 72 min 35% B, 76 min 95% B, 82 min 95% B, 83 min 5% B, 90 min 5% B, where A was 0.1% formic acid in water and B was 0.1% formic acid in acetonitrile. The gradient was formed at 250 nL/min using a NanoLC 400 UPLC system (Eksigent Technologies, CA, USA). The picofrit spray was directed into a TripleTOF 6600 Quadrupole-Time-of-Flight mass spectrometer (Sciex, MA, USA) scanning from 350 to 1600 m/z for 150 ms, followed by 40 ms. MS/MS scans on the 40 most abundant multiply-charged peptides (m/z 80–1600) for a total cycle time of 1.8 s. The mass spectrometer and HPLC system were under the control of Analyst TF 1.7 software (Sciex, USA).
The MS data were searched against a database which combined the Uniprot proteomes from Z. mays (http://www.uniprot.org/proteomes/UP000007305) and T. virens Gv29.8 (http://www.uniprot.org/proteomes/UP000007115) along with common contaminant entries (141,930 entries in total), using ProteinPilot version 5.0 (Sciex, USA) for peak picking identification selecting the following parameters: Cys-alkylation, iodoacetamide; digestion, full-trypsin digestion; and ID focus: biological modifications. Protein and peptide level false discovery rates (FDRs) were filtered to 1% and proteins with a threshold ProtScore ≤ 0.99 were discarded. The tandem MS/MS data was also analyzed using PEAKS Studio version 8 (BSI, ON, Canada). All resulting matched peptides were then confirmed by visual examination of the individual spectra.
Label-Free Quantification Analysis
The mass data was quantified by label free analysis using PEAKS software version 8 (BSI, Canada). Quantification was performed versus full tryptic digestion with a mass error tolerance of 20 ppm and 5.0 min for retention time shift tolerance. The quantification ratios were normalized using total ion current (TIC). Protein and peptide level FDRs were filtered to 1%. In addition, proteins with significance ≥ 10, fold change ≥ 1.5 and unique peptide ≥ 1 scores were selected.
Identification of Potential Functional Domains and Gene Ontology (Go) Analysis
A comprehensive pipeline was designed to identify secreted proteins and predict their characteristic features such as: (a) presence of a signal peptide (SignalP 3.0 and 4.0) (Bendtsen et al., 2004b; Petersen et al., 2011), (b) presence of a transmembrane domains (TMHMM 2.0) (Emanuelsson et al., 2007); (c) subcellular localization (WolfPsort and TargetP 1.1) (Emanuelsson et al., 2000; Horton et al., 2007); and (d) non-classical protein secretion (SecretomeP 2.0) (Bendtsen et al., 2004a) (Supplementary Figure 3). Moreover, prediction of secretory proteins was based on bioinformatics tools and parameters reported by two different fungal secretome databases: Fungal Secretome Database (FSD) (Choi et al., 2010) and FunSeckB2 (Meinken et al., 2014), and one plant secretome database: PlanSeckB (Min et al., 2014). Additionally, the prediction tool EffectorP was used to identify putative effectors-like proteins from T. virens (Sperschneider et al., 2016). Predicted functional analysis of the APs was performed using Blast2GO that combines GO (http://geneontology.org/), BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and Interpro (https://www.ebi.ac.uk/interpro) databases and searches for protein annotation (Supplementary Figure 3).
Peroxidase Activity
To determine peroxidase (POX) activity in the AF, the methodology described by Urbanek et al. (1991) was followed with modifications, using guaiacol as a hydrogen donor. The reaction mixture comprised 2 mL of a mixture containing 50 mM potassium phosphate buffer pH 6.8, 20 mM guaiacol (Sigma-Aldrich, USA) and 20 mM H2O2 (Merck Millipore, USA). The enzyme reaction was started by adding 10 μL containing 1 μg of APs and incubated for 10 min at 30°C. The reaction was stopped by adding 0.5 mL 5% (v/v) TCA and the absorbance was read at 480 nm. One unit of peroxidase activity was defined as the amount of enzyme that increased the absorbance by 0.01 expressed as units of POX/μg protein.
Statistical Analysis
Statistical analyses were performed using general analysis of variance (ANOVA) in the GenStat 18th package (VSN International, United Kingdom). Each value is the mean ± STD for 3 replicates in each group, and P ≤ 0.05 was considered as significant.
Results
Trichoderma virens-Maize Root Interaction
A hydroponic system was used to assess communication between T. virens and maize plant roots (Figure 1A). When maize seeds were inoculated with the fungal spores and then germinated in germination paper soaked with Hoagland's solution, germination was not substantially inhibited by the fungus (data not shown).
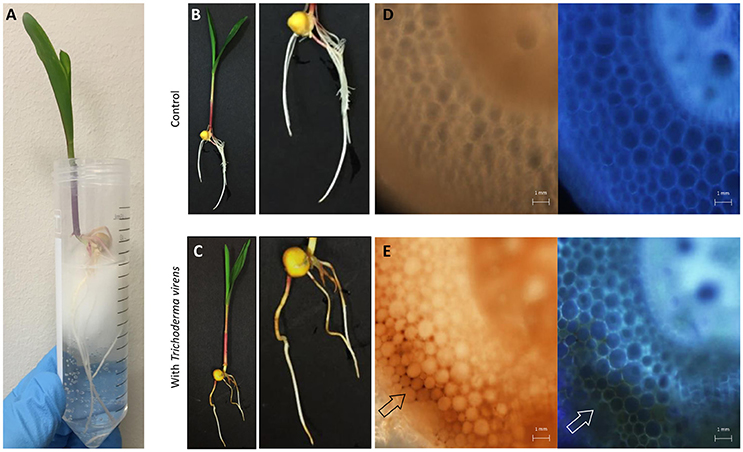
Figure 1. Overview of T. virens-maize interaction under a hydroponic system. (A) Five day old maize seedling growing aseptically under hydroponic conditions. (B) Un-inoculated and (C) inoculated plants with T. virens. Inoculated plants show phenotypical changes in their root system compared to the control. Cross section of un-inoculated primary root (D) bright-field and DAPI. Cross section of inoculated primary root (E) showing accumulation of brown pigmentation in epidermal and cortical cells, bright-field and DAPI. Images were obtained with a fluorescent microscope.
The presence of T. virens altered seedling root morphology when compared with un-inoculated seedlings (Figures 1B,C); a reduction in secondary root length and the presence of a brownish color on the surface where evident in the inoculated seedlings (Figures 1C,E). The brownish color was independent of the original T. virens spores inoculum (seedlings inoculated from 104 to 107 spores per seed, Supplementary Figure 1). This pigmentation extended from epidermal to cortical cell layers compared to un-inoculated roots (Figures 1D,E), but was not observed in the vascular system (Figure 1E).
To examine the colonization of T. virens in the root system, dual staining of the fungal cell wall (WGA- Alexa Fluor™ 488) and the plant cell wall (PI) or plant membrane (FM 4-64) was performed. Superficial fungal root colonization was predominantly found in the first two centimeters of the primary root (close to the seed) compared with other sections of the primary root (Figures 2A,C). In addition, T. virens colonized secondary roots and new root tips (Figure 2B). A transverse cut of the primary maize root enabled visualization of the internal colonization by T. virens. The fungus colonized the cortex layer adjacent to the vascular system of the primary root (Figure 2D). T. virens colonized intercellular spaces (apoplast) (Figure 2E). To visualize if T. virens colonized intracellular spaces, we used the plasma membrane dye (FM 4-64) in combination with WGA- Alexa Fluor™ 488 to follow the fungus. As observed in Figure 2F, T. virens colonized intracellular spaces and grew between the plasma membrane and the plant cell wall.
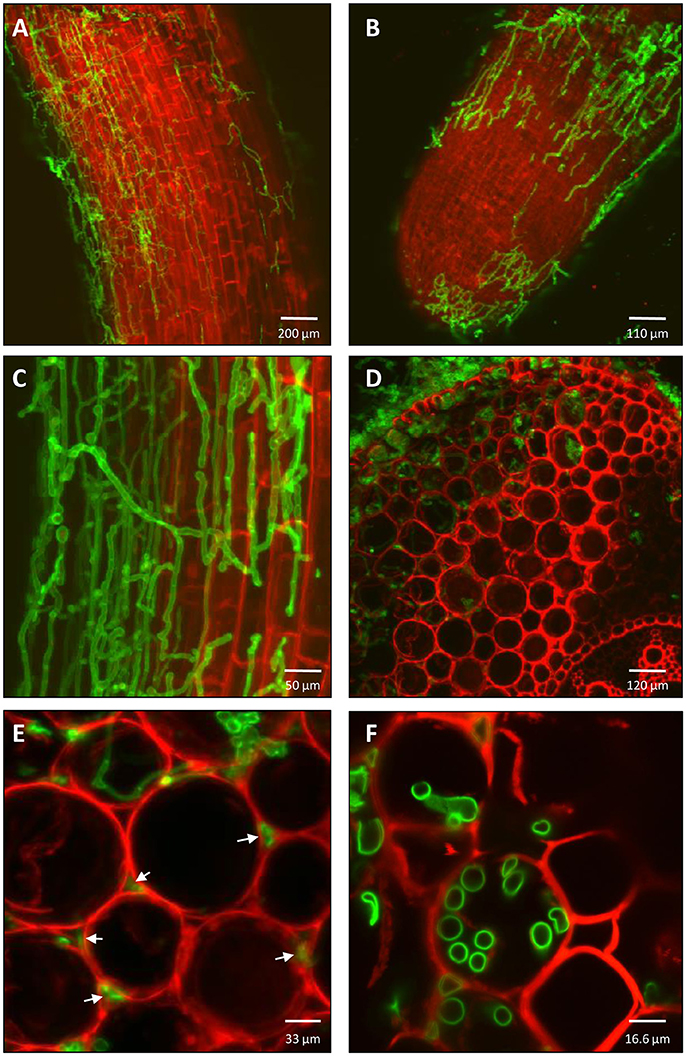
Figure 2. Colonization pattern of T. virens in maize roots. (A) After 5 days post inoculation, T. virens hyphae inhabit epidemical cells of maize primary root and (B) root tip of secondary root. (C) Close up of the hyphae occupying epidemical cells of the differentiation zone of the primary roots. (D) Cross section of primary root showing internal colonization of epidermal and cortical layers near to vascular system. (E) Intercellular and (F) intracellular colonization of cortex cells by T. virens hyphae (arrows). Fungal and plant cells were detected using WGA-Alexa Fluor 488 (green channel), propidium iodide (PI) and FM 4-64 Dye (red channel). Plant cell walls were detected with PI (A-F) and plant plasma membrane with FM 4-64 (D-F). Fungal cells were detected with WGA-Alexa Fluor 488 (A-F). Images were obtained with a confocal microscope.
Isolation and Identification of Apoplastic Proteins From Maize Root Seedlings Confronted or Not With T. virens Gv29.8
To determine the efficiency of different methodologies used for the extraction of AF from maize roots, two methods were tested. APs were successfully obtained by the infiltration-centrifugation system compared to the sorption when observed on the 1-D SDS-PAGE gel (Supplementary Figures 2A,B).
To evaluate levels of cytoplasmic contamination the activity of malate dehydrogenase (MDH) was used as a biomarker, which is commonly tested during extraction of AF (Gupta et al., 2015). The activity of MDH detected in AF extracted from un-inoculated and inoculated roots was up to 1.5% compared to the total soluble protein extract from roots (Supplementary Table 1). MDH levels below 2% are considered suitable for plant apoplast studies (Dannel et al., 1995; Dragišić Maksimović et al., 2008, 2014; Yang et al., 2015).
Multiple methods were used to obtain proteome coverage; in this study we compared Gel-LC-MS/MS and gel-free shotgun proteomics.
Identification of Apoplastic Proteins by Gel-LC-MS/MS
APs were separated by 1-D SDS-PAGE, with three biological replicates for each condition (Supplementary Figure 2A). The APs showed differences in their protein complement with distinctly different protein bands visible in inoculated plants (M+Tv) compared with un-inoculated plants (M). The patterns of protein fractions from 15 to 75 kDa and 130 to 250 kDa were largely similar in their intensity and mass separation. Differences were observed in the protein fractions between 75 and 130 kDa sections in inoculated samples compared with the un-inoculated (Supplementary Figure 2A). Specifically, two treatment-specific bands were observed; one was located above the 100 kDa fraction and the other below (Supplementary Figure 2A). Based on their mass, the protein bands were cut from the gels in five sections (Supplementary Figure 2A), and then analyzed by LC-MS/MS.
Using this gel-based proteomic approach coupled with LC-MS/MS, 13 proteins were identified, of which 12 corresponded to maize and one to T. virens. Five of these maize proteins: LRR receptor-like serine/threonine-protein kinase (A0A1D6ERY2_MAIZE), aspartic-type endopeptidase (A0A1D6F8J3_MAIZE), germin-like protein subfamily T member 1 (B4FRS8_MAIZE), barwin-like protein (Win1) (B6SH12_MAIZE), and peroxidase (Per66) (PER66_MAIZE) were common to both conditions (inoculated and un-inoculated maize samples) and were located in the 75–15 kDa fractions. Proteins that were only present in the un-inoculated samples included two peroxidases (C0PGF4_MAIZE; A0A1D6PD14_MAIZE) and a pathogenesis-related protein 1 (PR-1) (A0A1D6K5Y8_MAIZE) which were identified in the 50–15 kDa fractions. Four maize proteins identified from the inoculated samples exclusively were in the 120–30 kDa fractions: methionine synthase (Q8W529_MAIZE), heat shock protein 70 (A0A1D6MWU7_MAIZE), adenosylhomocysteinase (A0A1D6PTE3_MAIZE), and pectinesterase (B6SSX0_MAIZE). In contrast, the protein TV_29366, which encodes for a β-xylosidase, was the only protein detected from T. virens and this was present in the 120–75 kDa fractions.
Identification of Proteins by Gel-Free Shotgun Proteomics
The low number of proteins identified by Gel-LC-MS/MS base technology drove us to use a more powerful proteomic tool (gel-free shotgun proteomics) to increase the identification number of proteins present in the apoplast which has a complex protein mixture. Using the gel-free shotgun proteomics approach, 148 maize proteins were identified in the un-inoculated control roots that were present in all three biological replicates. In the inoculated roots, a total of 177 were identified, where 85 and 92 proteins corresponded to the maize and T. virens proteomes, respectively.
Interestingly, in comparison with the un-inoculated roots, the inoculated roots showed a 43% (63 proteins) reduction in the number of total maize proteins identified. These results show that an alteration in the maize proteome is triggered by the presence of T. virens. In contrast to Gel-LC-MS/MS, gel-free shotgun proteomics showed an increase of identified proteins from 13 to 272, showing that gel-free shotgun proteomics coupled with next generation LC-MS/MS instruments, is a useful technology to identify larger numbers of proteins in complex samples, such as in AF during plant-microbe interactions.
Prediction and Annotation of Secreted Apoplastic Proteins Through Bioinformatics Tools
Putative secreted proteins were classified into two classes: (a) classical secreted proteins and (b) non-classical secreted proteins. In addition, the literature was used as a point of reference for those proteins that were not predicted as being derived from classical and non-classical secretion systems by bioinformatics analysis, but have been reported as secreted proteins in other fungal and plant models. Based on the above criteria, in the un-inoculated maize roots 76 (51%) of the proteins were predicted to be secreted. Of these 56 (74%) followed the classical secretion system, while 20 (26%) were seemingly secreted by a non-classical mechanism and consequently were classified as leaderless secreted proteins (LSPs) (Supplementary Figure 4A). From the 85 maize proteins identified from the inoculated maize roots, 49 were predicted to be secreted and of these, 22 (45%) followed the classical and 27 (55%) the non-classical secretion system (Supplementary Figure 4A). In addition, un-inoculated roots showed 46 unique secreted proteins, while 18 unique secreted proteins were present in inoculated roots (Supplementary Figure 4A). Of 92 conserved T. virens proteins, 43 (46%) were predicted to be secreted, where 20 (46%) followed the Golgi-ER secretion system and 23 (54%) were secreted by non-conventional secretion systems (Supplementary Figure 4B). These results indicate that both organisms use both classical and non-classical secretion systems to deliver proteins into the apoplast.
Functional Annotation of Maize Secreted Apoplastic Proteins at 5 Days Interaction
Secreted proteins were organized into functional categories for biological processes and molecular function based on their gene ontologies (GO) (Figures 3A,B). In un-inoculated maize roots, the four major biological process groups of proteins were catabolic processes (22%), response to stress (19%), carbohydrate metabolic processes (11%) (Figure 3A), and cellular nitrogen compound metabolic process (11%). The three main molecular functions were ion binding (47%), oxidoreductase activity (35%) and glycosyl hydrolase activity (18%) (Figure 3B). By comparison in inoculated samples, the four major biological process functional groups were response to stress (27%), catabolic processes (17%), carbohydrate metabolic processes (10%), or sulfur compound biosynthetic processes (10%) (Figure 3A). The three main molecular functions were ion binding (33%), oxidoreductase activity (20%) and enzyme regulator activity (15%) (Figure 3B).
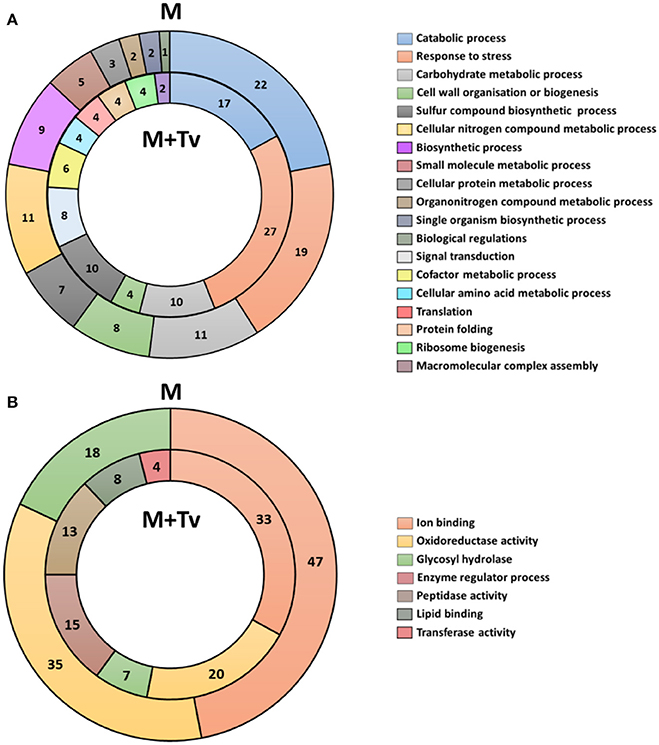
Figure 3. Functional classification of all secreted proteins from un-inoculated and inoculated maize roots at 5 days interaction. (A) Blast2GO multilevel chart for biological process of un-inoculated (M) and inoculated (M+Tv) maize roots. (B) Multilevel chart for molecular function of un-inoculated and inoculated maize roots. Score distribution represented as a percentage of each group is indicated inside the pie slices.
Multiple changes were identified in the predicted suite of functions of the proteins secreted from un-inoculated compared with inoculated roots. Response to stress was increased during the interaction with T. virens from 19 to 27%; in contrast, catabolic processes were reduced from 22 to 17%. No differences were observed in proteins belonging to carbohydrate metabolic processes; however, some differences were present in secondary metabolism, changing from nitrogen to sulfur metabolic processes (Figure 3A). Major changes in molecular functions were observed; a reduction of ion binding from 47 to 33%, oxidoreductases from 35 to 20%, and glycosyl hydrolases from 18 to 7% activity. Conversely, lipid binding, protease and transferase activities were present only in inoculated roots (Figure 3B).
Putative identification based on conserved domains and function was performed on the identified protein family members from maize roots that were secreted into the apoplast in un-inoculated and inoculated plants after 5 d interaction. The major protein groups from maize were: (a) 11 glycosyl hydrolases (GHs) that are involved in the degradation of carbohydrate complexes; (b) 9 antioxidant proteins that catalyze reactions to neutralize free radicals and ROS; (c) 15 peroxidases, that participate in the biosynthesis of the cell wall and defense responses and have multiple tissue-specific functions; (d) pathogenesis-related (PR) family proteins that are activated under biotic stresses; (e) proteases/peptidases that are responsible for the hydrolysis of peptide bonds, and (f) proteinase inhibitors (PIs) that participate in the inactivation of proteases. Additionally, other family groups that were identified belonged to DUF proteins, oxidoreductases, lipases, ribonucleases, chaperones, calmodulin, ribosomal proteins and cyclophilin (Table 1 and Supplementary Table 2).
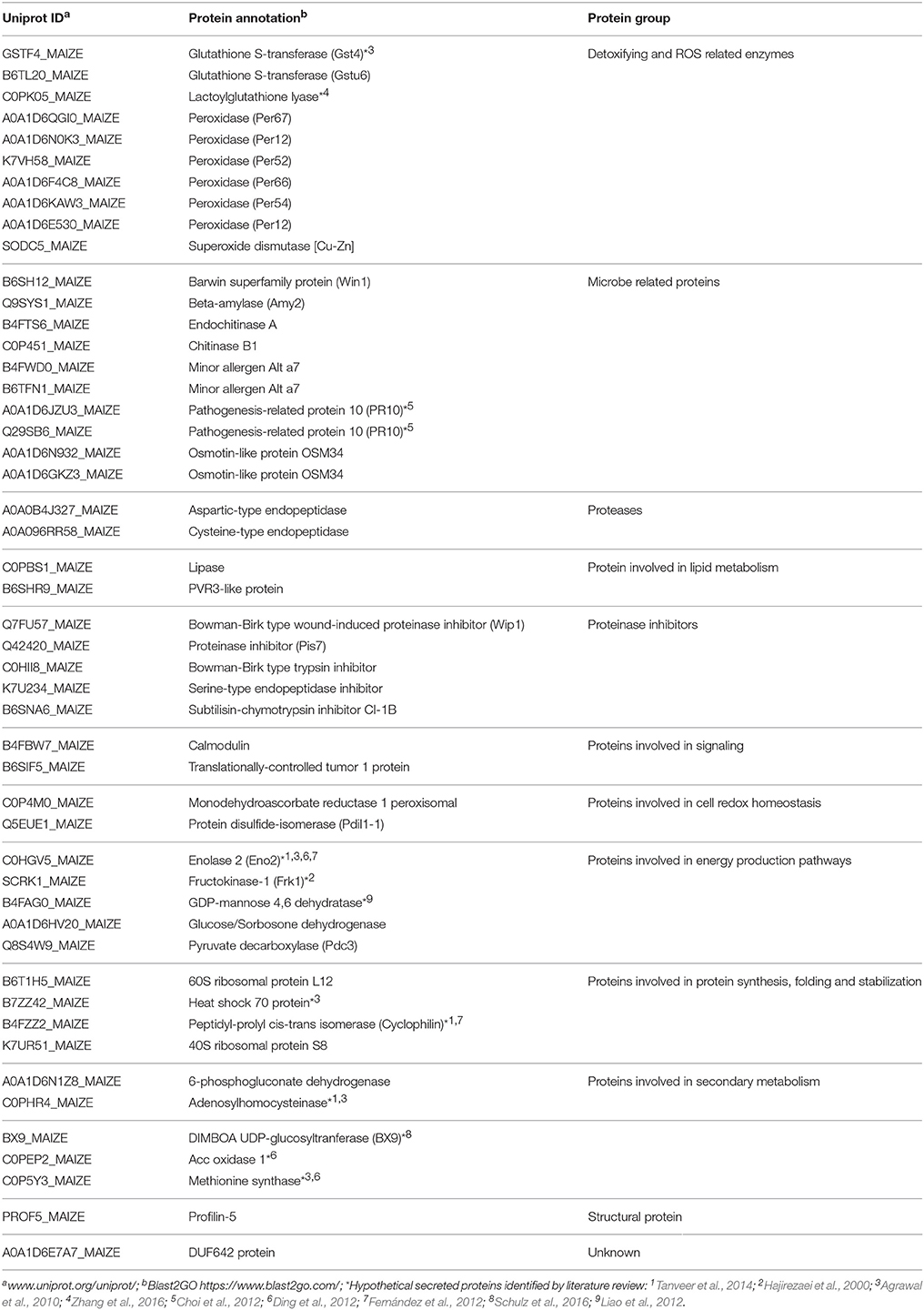
Table 1. Summary of the apoplastic proteins secreted by Zea mays after 5 days interaction (inoculated).
Interestingly, protein family groups were founded in higher abundance in un-inoculated compared to inoculated roots, for example, maize glycosyl hydrolases (GHs) were reduced from 15 to 6%, respectively. Similar results were identified in the peroxidase group where their reduction was from 19 to 13%. In contrast, pathogen-related proteins increased from 3 to 10% and protease/peptidase and PIs from 8 to 16 %. These results indicate that the maize proteome is altered by the presence of T. virens by the expression and suppression of different protein families, suggesting that the fungus is re-shaping the plant secretome.
Functional Annotation of T. virens Secreted Apoplastic Proteins at 5 Days Interaction
A total of 43 secreted proteins from T. virens were identified during the interaction with maize roots (Table 2). Secreted proteins were organized into functional categories for biological processes and molecular function based on their gene ontologies (GO) (Figures 4A,B). The four major biological process groups of proteins identified were: carbohydrate metabolic process (12%), oxidation-reduction process (12%), catabolic process (11%), and response to stress (11%) (Figure 4A). The three main molecular functions were oxidoreductase activity (41%), glycosyl hydrolase (13%) and lyase activity (10%) (Figure 4B).
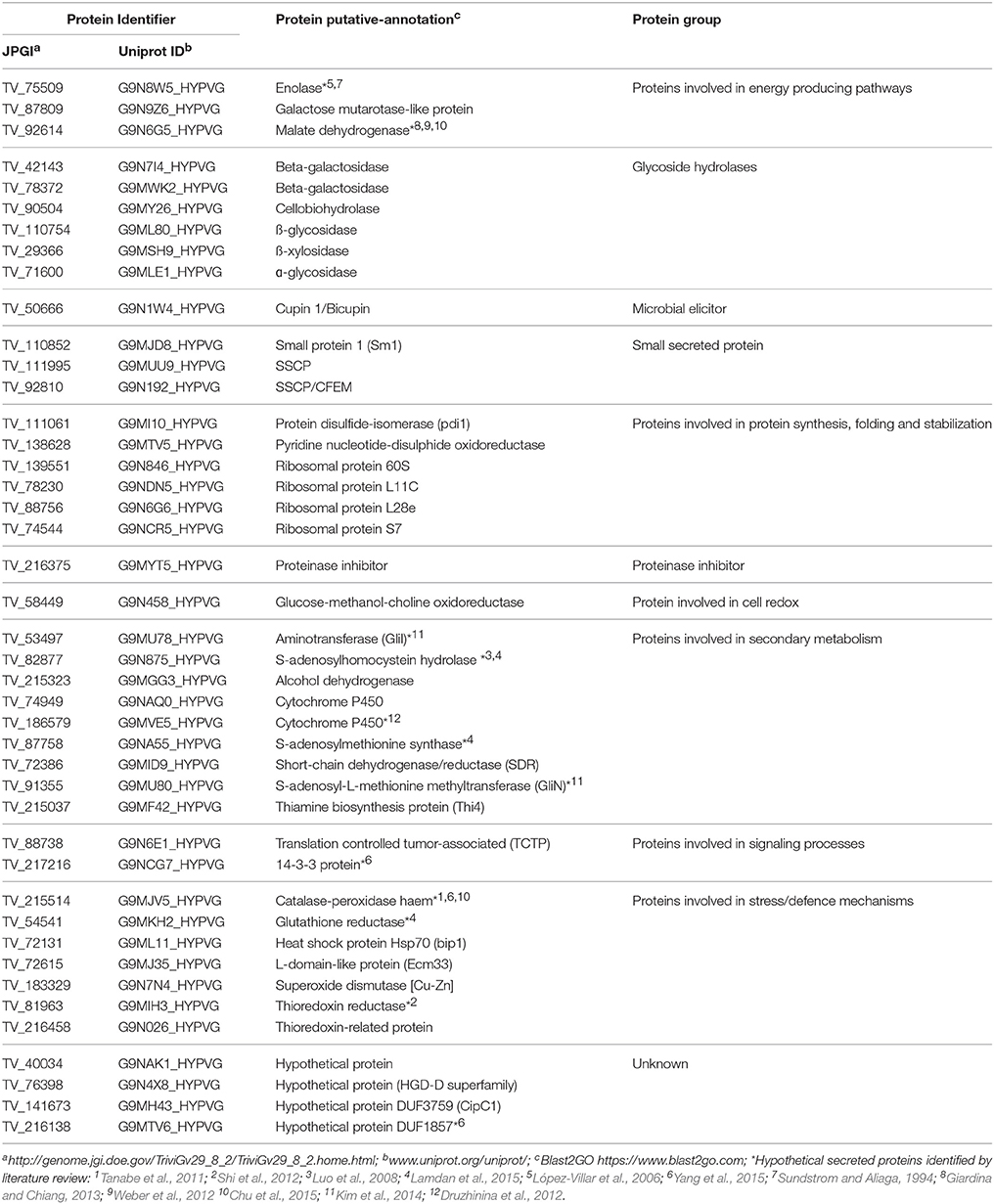
Table 2. Summary of the apoplastic proteins secreted by Trichoderma virens after 5 days interaction.
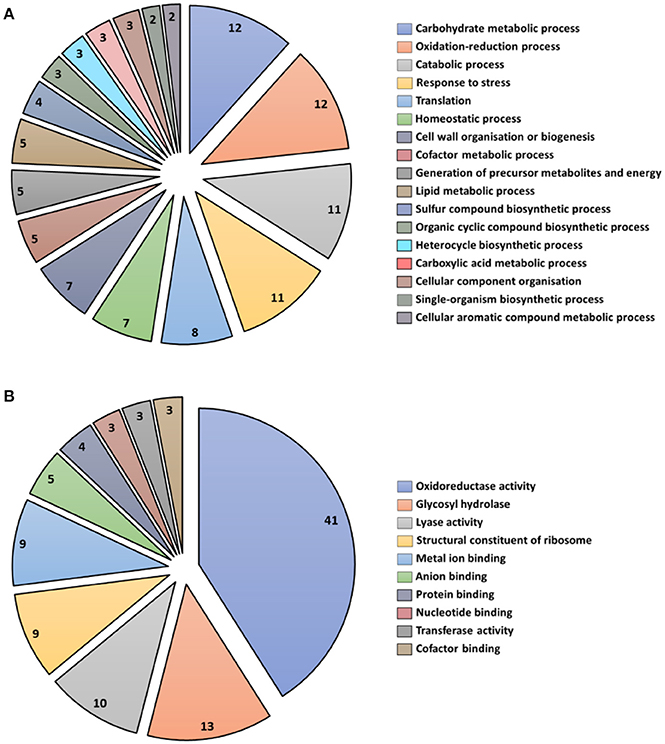
Figure 4. Functional classification of all secreted proteins from T. virens at 5 days interaction. Blast2GO multilevel chart for (A) biological process and (B) molecular function. Score distribution represented as a percentage of each group is indicated inside the pie slices.
Identification of putative proteins based on their conserved domains and function was performed on the proteins from T. virens that were secreted into the apoplast in inoculated plants after 5 d interaction. The glycosyl hydrolase (GHs) family was the highly represented in the T. virens secretome (Table 2). Proteins that participate in antioxidant processes, oxidate stress resistance and secondary metabolism were also identified (Table 2). Furthermore, in the presence of maize roots, groups of proteins corresponding to putative effector-like proteins, chaperones, 14-3-3 like proteins and ribosomal proteins were identified as part of the T. virens secretome (Table 2). Overall, these results suggest that T. virens activates different molecular mechanisms during host root colonization.
Label-Free Quantification of Apoplastic Proteins During the T. virens-Maize Interaction
Label-Free Quantification
By using the label-free quantification approach we identified 10 proteins from maize that were significantly (significance ≥10; fold change ≥1.5) different in their intensities between un-inoculated and inoculated roots (Supplementary Figure 5). Analysis of the correlation of the signal intensities showed that the biological repeats of each treatment were clustered together, with an average correlation of 0.87 for un-inoculated roots (M) and 0.90 for inoculated roots (M+Tv). Correlation between all biological repeats showed an average relationship of ≥0.70. Proteins that showed a decreased abundance during the interaction with T. virens were 40S ribosomal protein (B4FSW0_MAIZE), pathogenesis-related protein (PR-10) (Q29SB6_MAIZE), peroxidase (Per12) (B4FG39_MAIZE), cytosolic ascorbate peroxidase (Apx1) (B6TM55_MAIZE), cysteine endopeptidase (K7W288), blue copper protein (B6UHQ8_MAIZE), adenosylhomocysteinase (C0PHR4_MAIZE), protein disulphide-isomerase (Pdil1-1) (Q5EUE1_MAIZE), and ribonuclease 1 (B4FBD6_MAIZE). In contrast, the protein serine-type endopeptidase inhibitor (K7U234_MAIZE) showed an increased abundance. Overall, these results suggest that the abundance of proteins involved in different plant biological processes, including plant defense, are manipulated by the presence of T. virens.
Peroxidase Levels Influenced by T. virens
The peroxidase activity in the AF after 5 d interaction was measured in maize root tissues in un-inoculated and inoculated with T. virens. Shifts in the enzyme activity were observed, where inoculated roots with T. virens showed a significant reduction in peroxidase activity compared with un-inoculated roots (p ≤ 0.05) (Figure 5). These results indicated that the peroxidase activity was higher in un-inoculated roots compared to inoculated, showing that the peroxidase activity was directly influenced by the presence of T. virens in the root system.
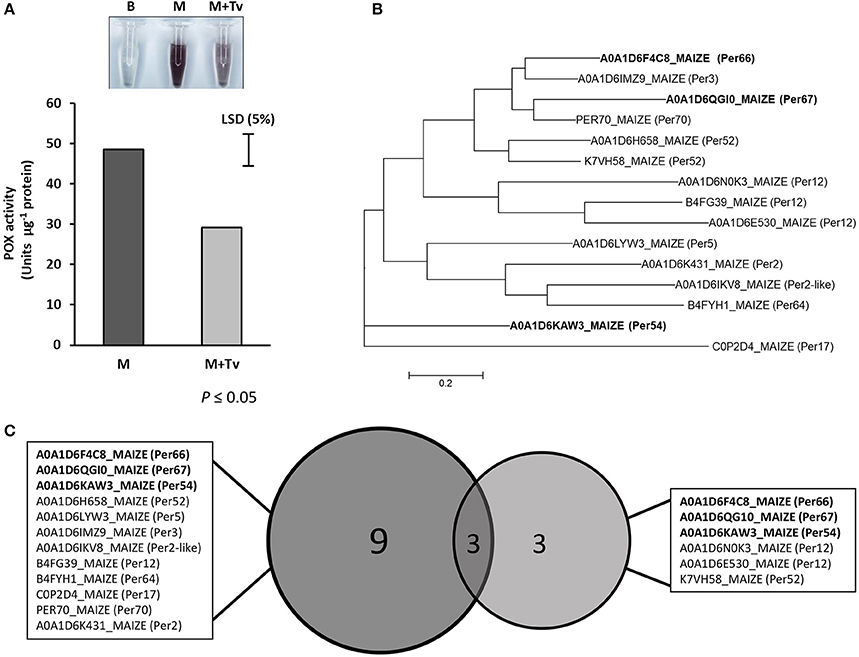
Figure 5. Peroxidase study during T. virens-maize interaction. (A) Peroxidase activity in un-inoculated (M) and inoculated (M+Tv) maize roots with T. virens after 5 days interaction (p ≤ 0.05). (B) Phylogenetic tree of peroxidases identified in the maize apoplast zone. Using Muscle, the composite proteins were aligned, and the Maximum likelihood tree, were generated in Mega 6. (C) Comparison between peroxidases expressed in maize roots with or without T. virens. Bold names represent peroxidases that were present in both conditions.
Discussion
In this study, we observed the interaction between maize roots and T. virens by confocal microscopy. In addition, we identified the secretome profile of maize roots growing alone or during the interaction with T. virens. Furthermore, the identification and function of these proteins were analyzed to understand the molecular dialogue that exists in the apoplast between T. virens and maize, and unravel the role that these proteins may play during the symbiotic interaction.
Root Interaction With T. virens
Previous reports elucidate the lifestyle of T. virens as endophytic in different host plants (Vargas et al., 2009; Moran-Diez et al., 2015; Lawry, 2016; Romão-Dumaresq et al., 2016). Our findings showed that T. virens colonized different sections of the root system, including primary and secondary roots (Figure 2). Interestingly, T. virens is able to endophytically colonize inter- and intracellular spaces of maize roots (Figure 2), suggesting that the fungus utilizes both pathways to establish itself in the host root system.
During the interaction, phenotypic responses of maize roots were detected when colonized by T. virens, for example, appearance of a brownish pigment and reduction of secondary root growth (Figure 1). Accumulation of browning of inoculated roots has been observed previously in T. virens (Moran-Diez et al., 2015) and Trichoderma harzianum (Palaniyandi et al., 2017) and in other fungal systems, including incompatible interaction of arbuscular mycorrhizal fungi with Salsola kali (Allen et al., 1989) or in detrimental interactions of pathogens such as Pythium aphanidermatum or Fusarium graminearum with their host roots (Sutton et al., 2006; Ye et al., 2013). Therefore, it could be argued that maize cells are responding to T. virens colonization by triggering the accumulation of phenolic compounds involved in initial responses to stress to reinforce plant cell walls and inhibit fungal growth (Beckman, 2000). Nevertheless, it cannot be discounted that the physiological changes on maize roots were due to the deposition of T. virens secondary metabolites such as melanin onto the root surface or into the media or that T. virens mycelia were blocking the aeration of the media creating anoxic conditions and inducing this physiological change (browning).
Endophytic colonization (both inter- and intracellular) by T. virens showed different mechanisms that the fungus undertakes to develop an interaction with its host plant and to promote fungal growth on plant tissue. A similar pattern of colonization was observed in the endophytic fungus Piriformospora indica interaction with barley plants (Deshmukh et al., 2006). This symbiotic fungus requires host cell death in differentiated barley roots, in order to proliferate and become endophytic and form a mutualistic interaction. This implies that the fungus biotrophically colonizes by digesting plant cell walls and subsequently either interferes with the host cell death program or actively kills host cells (Deshmukh et al., 2006). We suggest that under hydroponic conditions T. virens may employ similar mechanisms to colonize maize roots. Therefore, our interest was to elucidate the molecular mechanisms that occur in the apoplastic zone for developing an endophytic relationship by the manipulation of host defense responses.
Identification of Apoplastic Proteins by Gel-Based Proteomic Technology
The infiltration-centrifugation method was the most efficient approach for the extraction of AF and APs, which has been previously reported in other studies for the identification of microbe-secreted proteins in planta (Floerl et al., 2012; Shenton et al., 2012). Using the gel-based LC-MS/MS approach, maize proteins such as methionine synthase, heat shock protein 70, adenosyl homocysteine hydrolase and pectin esterase were expressed and identified in the AF during the interaction with T. virens. These proteins have been previously reported as part of the plant immune response pathways by activation of microbe elicitors (Kawalleck et al., 1992; Lionetti et al., 2007; Maimbo et al., 2007; Balmer et al., 2013); suggesting that maize roots are sensing T. virens elicitors, for example, chitin, and are responding to T. virens colonization.
The protein β-xylosidase (TV_29366) from T. virens was confirmed in this analysis, suggesting the secretion of hydrolytic enzymes into the apoplast by T. virens. The precursor of β-xylosidase has been reported as a virulence factor of Sclerotinia sclerotium (Yajima et al., 2009), and it has been observed previously in the secretome of T. virens interacting with maize roots under hydroponic conditions (Lamdan et al., 2015). β-xylosidase participates in the hydrolysis of xylan, one of the major polysaccharides present in plant cell walls.
Identification of Apoplastic Proteins by Gel-Free Proteomic Technology
Using the gel-free shotgun proteomics approach, a total of 148 and 177 proteins were identified in un-inoculated and inoculated roots, respectively.
A number of cytosolic proteins were identified in this study which may suggest levels of contamination by cytoplasm from both organisms despite a low level of cytoplasm biomarker being detected (<1.5% of total MDH activity). Similar results were observed by Dragišić Maksimović et al. (2008), using the vacuum-infiltration technique to extract AF from maize roots. Other studies have identified classical cytoplasmic proteins as part of the secretome of different organisms, for example, plants and fungi (Agrawal et al., 2010; Kim et al., 2013). Techniques used for the isolation of APs may induce mechanical damage to host cells leading to cell breakdown and possible contamination by cytoplasmic proteins, which is a constraint in plant-microbe secretomics.
A comprehensive filter-pipeline was used in this study to identify and select putative secreted proteins from both organisms using available prediction software and literature. Nevertheless, proteins that were discarded as secreted proteins in this study may have also an important function during T. virens-maize interaction.
Secretion Systems of Apoplastic Proteins During T. virens-Maize Interaction
Identification of potential secreted-APs from maize under both conditions, suggests that maize roots secrete APs by using both conventional (proteins containing a signal peptide) and unconventional secretion systems which include the leaderless secretory proteins (LSPs). These findings have been previously reviewed in plant models by Yadav et al. (2015). Interestingly, T. virens seems to induce the secretion of LSPs; the plant apoplastic LSPs increased from 26% in the absence of the fungus to 55% when the fungus was present. These changes in the population of host secreted proteins during fungal colonization has been reviewed by Agrawal et al. (2010) in different plant models, suggesting that plant LSPs are involved in the defense/stress responses against microbial invasions. This may suggest that maize roots are responding to T. virens colonization by secreting proteins related to defense pathways through conventional and unconventional secretion systems.
Remarkably, 54% of the 92 identified T. virens secreted-APs lacked a signal peptide (Supplementary Figure 4B). In fungi under biotic conditions the secretome population includes LSPs and reveals several unknown mechanisms of secretion that are active during the microbe-plant interaction (Girard et al., 2013). It was expected that different mechanisms of secretion would be found in T. virens during the interaction. This secretion pattern has previously been reported during the detrimental interaction between M. oryzea and rice (Kim et al., 2013), where 48% of the total secretome identified corresponded to LSPs. Similar results were observed by Weber et al. (2012) during the secretome analysis of the fungus Paracoccidioides; where they predicted that 52% of identified secreted proteins are released by non-classical secretion systems. The exact mechanisms of secretion for leaderless secretion pathways remain largely unknown for plant-microbe interactions.
Maize Secretome: The Influence of T. virens Colonization
Functional analysis of the maize secretome present in the apoplast during colonization by T. virens showed that T. virens is inducing several changes in plant metabolism and activating pathways of stress responses against biotic factors. Interestingly, oxidoreduction activity seems to be reduced in the presence of T. virens in the apoplast (Figure 3B). Similar results were observed in the transcriptome analysis after 5 d interaction performed by Lawry (2016), where genes involved in plant defense responses were identified, implying that maize roots are responding to fungal colonization and vice versa.
Detoxifying and ROS Related Proteins Influenced by T. virens
A larger number of proteins were observed during non-interaction conditions compared with inoculated roots, where a reduction in secreted proteins was observed in roots interacting with T. virens; for example, proteins belonging to the peroxidase family. Peroxidases are principal components in plant defense responses against pathogens either by cell wall reinforcement and/or for inducing plant oxidative bursts in the apoplast (Mehdy, 1994), creating unsuitable conditions for microbe interactions.
Differences were found in the secretion of peroxidases in this study; where 12 were secreted in the control roots; this number reduced to six during the Trichoderma-plant interaction, three of which were unique to the inoculated roots (A0A1D6N0K3_MAIZE; A0A1D6E530_MAIZE; K7VH58_MAIZE) (Table 1). This suggest that these latter peroxidases are part of the immune respond of maize to T. virens colonization. Remarkably, other peroxidases (B4FG39_MAIZE and B6TM55_MAIZE) were significantly reduced in their abundance when T. virens was present (Supplementary Figure 5). These results demonstrate that recognition of the endophyte T. virens by resistance proteins involves plant defense mechanisms and that this reduction in plant defense molecules such as peroxidases suggests that T. virens may be re-shaping plant secretome responses as a mechanism to suppress plant immunity. In the symbiotic interaction between Glomus mossea and tobacco roots oxidative reduction activity plays an important role, activity of ascorbate peroxidases was increased in mycorrhizal roots at earlier stages, but after the interaction is established the fungus is able to diminish plants immune responses (Blilou et al., 2000).
Additionally, we identified proteins in un-inoculated maize such as superoxide dismutase [Cu-Zn] (SODC5_MAIZE), peroxiredoxin-2B (B4FN24_MAIZE), thioredoxin (Trxh1) (B6SHW5_MAIZE), and glutathione S-transferase (B4FSR6_MAIZE; GSTF4_MAIZE; B6TL20_MAIZE) that are involved in detoxification of the apoplast reducing ROS, but in inoculated roots the peroxiredoxin, thioredoxin and one glutathione S-transferase weren't present. ROS play a major role in plant defense. Therefore, to maintain a balance during the plant-microbe interaction, all these enzymes are considered necessary to confer resistance to oxidative stresses. Interestingly, antioxidant activity is reduced in maize roots during the interaction with T. virens, suggesting that alterations in redox activity are influenced by fungal colonization.
Pathogenesis Related Proteins Secreted in Presence of T. virens
Plant defense-related proteins were also identified in the secretome for both conditions. Pathogenesis related proteins (PRs) are produced in plants in the event of microbial recognition, and involve antimicrobial activity. Most PRs are induced through jasmonic acid (JA), salicylic acid (SA) and ethylene (ET) defense signaling (van Loon et al., 2006). Extracellular defense-related proteins are considered the first line of defense against invading attackers before tissue penetration takes place. Several PR-like proteins were identified in the apoplast from un-inoculated and inoculated plants, for example, chitinase, endochitinase B, nine peroxidases (PR-9), two ribonucleases (PR-10), two proteinase inhibitors (PR-6), and an osmotin-like protein (PR-5) (Table 1 and Supplementary Table 2). These proteins may be considered as part of basal defense mechanisms in maize, because they were present in both conditions. Additionally, other defense-related proteins were identified that were probably induced by the mechanical wounding provoked by the manipulation of the samples, for example, the Bowman-Birk type wound-induced proteinase inhibitor. Hence we cannot discard that some of previous proteins described were activated through this process, given that several-defense proteins are induced during wounding or cold stress (van Loon et al., 2006).
In the presence of T. virens, maize roots expressed an increase of different PR-like proteins. APs such as Barwin superfamily protein (B6SH12_MAIZE), cysteine-type endopeptidase (A0A1D6ICV7_MAIZE), PR-10 (A0A1D6JZU3_MAIZE), three peroxidases (A0A1D6N0K3_MAIZE; A0A1D6E530_MAIZE; K7VH58_MAIZE), proteinase inhibitor (Pis7) (Q42420_MAIZE), serine-type endopeptidase inhibitor (K7U234_MAIZE), and osmotin-like protein (A0A096PW84_MAIZE) were activated during fungal colonization. This suggests that these proteins are specifically activated during T. virens recognition. Comparable findings during the three-way interaction between Trichoderma atroviride, a host plant and a fungal pathogen were identified, where PRs were up-regulated when T. atroviride was interacting with the plant (Marra et al., 2006). Although maize responded by constitutive expression of various PRs, they did not affect colonization by the beneficial T. virens. Similar findings were observed during the beneficial relationship between the mycorrhizal fungus G. mosseae and its host plant (Vierheilig et al., 1995). Remarkably, T. virens may directly affect the secretion of specific PRs such as ribonuclease (B4FBD6_MAIZE) and PR-10 (Q29SB6_MAIZE) as their abundance in the apoplast was reduced; however, the plant may counter attack by activating other PR proteins, for example, serine-type endopeptidase inhibitor (K7U234_MAIZE) to target proteins secreted by T. virens in the apoplast (Supplementary Figure 5). The serine-type endopeptidase inhibitor was identified as the only protein whose abundance was higher in presence of T. virens (Supplementary Figure 5).
Secreted Proteins Involved in Hormone Signaling Cell Wall Modification, Nutrient Acquisition, Protein Modification and Metabolism
Proteins involved in phytohormones signaling were identified. These were predominantly from the ethylene pathway, including proteins responsible for the biosynthesis of precursor components such as methionine synthase (COP5Y3_MAIZE), adenosylhomocysteinase (COPHR4_MAIZE) and the enzyme 1-aminocyclopropane-1-1-carboxylate (ACC) oxidase (COPEP2_MAIZE) which catalyzes the final stage of ethylene biosynthesis. Levels of adenosylhomocysteinase were influenced by the presence of T. virens and ACC oxidase was only expressed in inoculated plants suggesting that one strategy of the fungus is the manipulation of plant immunity responses via the ethylene pathway. In the pathosystem Arabidopsis-Pseudomonas syringae, Kaffarnik et al. (2009) observed that the protein abundance of methionine synthase was dramatically influenced in the apoplastic region and not intracellularly by the bacterial effector-like proteins.
Other proteins with relevance in the AF that were identified are involved in cell wall modification and nutrient acquisition such as glycosyl hydrolases (GHs) proteins, signal transduction and secondary messengers, such as, 14-3-3 protein and calmodulin, respectively (Cui et al., 2005; Lozano-Durán and Robatzek, 2015); proteins that participate in protein modification such as heat shock 70 protein (chaperone) and protein disulphide-isomerase (Pdil 1) involved in correct protein folding essential for protein functionality (Park and Seo, 2015; Porter et al., 2015); and ribosomal proteins, cyclophilin and proteins involved in lipid transport and secondary metabolism that have potential roles in defense/stress responses (Agrawal et al., 2010) were also observed in the AF. Under unstressed conditions these APs are involved in cell wall modification or maintenance (Albenne et al., 2013), whereas upon pathogenic attack they are specifically involved in cell wall remodeling and reinforcement (Hamann, 2012).
Overall, all the identified proteins above play important roles in basal and induced defense/stress responses, metabolism, signaling and protein modifications in plants (Agrawal et al., 2010) and have been previously reported as part of the apoplastic secretome in-planta (Gupta et al., 2015), where several plant response pathway were activated or inactivated by the presence of T. virens.
T. virens Secretome During the Interaction With Maize Roots
Diverse mechanisms are involved in the Trichoderma-plant interaction during the process of root colonization (Harman, 2006). In this study several protein families were identified that may have a direct influence on T. virens colonization and plant defense manipulation, which correlates with the findings previously reported by Lamdan et al. (2015), where protein families such as glycosyl hydrolases, antioxidant proteins, small secreted cysteine rich proteins and secondary metabolism proteins were secreted in the presence of maize roots.
Glycosyl Hydrolase (GH) Protein Family (Cell Wall Modification)
Secretion of both endophyte and host glycosyl hydrolases have an important role either promoting successful colonization via degradation of the host cell wall, or resisting microbe invasion through the reinforcement of host defenses via cell wall maintenance during early stages of the interaction. Seven GH proteins were identified including two -galactosidases (TV_42143 and TV_78372) that hydrolyse galactose-rich polysaccharides in plant cell walls (Ranwala et al., 1992), galactose mutarotase-like protein (TV_87809) that participates during galactose metabolism, -glycosidase (TV_71600) involved in catabolism and turnover of plant N-glycans (Minic, 2008), -glucosidase (TV_110754) involved in cellulose degradation (Tiwari et al., 2013), -xylosidase (TV_29366), and cellobiohydrolase (TV_90504) for degradation of xylan and cellulose, respectively (Nummi et al., 1983; Biely, 1985). Interestingly, the secretion of the glycosyl hydrolases and cell wall-degrading enzymes (CWDEs) identified in this study are mediated through the Golgi-ER secretion pathway. In addition, all identified enzymes were transcriptionally up-regulated after 5 d interaction with maize roots compared with axenic conditions (Lawry, 2016). CWDEs have also been proposed to act as virulence factors (effectors) in U. maydis and M. oryzea; however, the exact role they play in plant immunity is not known (Kubicek et al., 2014). The results suggest when T. virens encounters its host plant root system, to overcome the barrier of the plant cell wall and successfully penetrate and colonize internally, T. virens may secrete enzymes into the apoplast such as glycosyl hydrolases and CWDEs which focus on the degradation of cell wall polymers necessary for colonizing host tissues.
Antioxidant Secreted Proteins
The oxidative burst is one of the most critical events upon plant recognition of plant pathogens (Heller and Tudzynski, 2011). This reaction is activated by a rapid production of ROS in the apoplast such as superoxide, hydroxyl radical and hydrogen peroxide which are also involved in plant response signaling. In addition, ROS activate physiological changes in the plant such as cell wall strengthening. Although ROS induction is transient in-planta, the compounds are highly reactive and can cause detrimental oxidation of essential macromolecules causing host cell damage and triggering hypersensitive response (HR), if they are not controlled (Nanda et al., 2010). ROS are not only toxic, but are also signaling molecules involved in growth and environmental adaptation (Marschall and Tudzynski, 2016). Antioxidants can protect the cell from oxidative damage by scavenging the ROS. Therefore, cellular redox homeostasis is important to maintain plant-microbe symbiotic interactions (Marschall and Tudzynski, 2016). In this study, T. virens secreted proteins were identified that work as detoxification enzymes. The enzyme superoxide dismutase [Cu/Zn] (TV_183329) was present in the apoplast and is involved in the breakdown of ROS molecules protecting fungal integrity. Superoxide dismutase is an essential molecule during plant microbe interactions. During the beneficial relationship between the fungal endosymbiont Neotypodium lolii and ryegrass, superoxide dismutase is necessary for limited host defense and normal endophytic growth (Zhang et al., 2011). The protein catalase-peroxidase haem (TV_215514) was also identified. This enzyme exhibits both catalase and peroxidase activity, and provides protection against oxidative stress dismutating H2O2 to O2 + H2O. In the fungus M. oryzea, the secreted catalase-peroxidase (CPXB) confers resistance to H2O2 accumulated in epidermal cells of rice, but is not essential for pathogenicity (Tanabe et al., 2011). As part of the antioxidant protein arsenal, the enzyme glutathione reductase was found, which is required for protection against oxidative stress. Glutathione reductase (TV_ 54541) plays an essential role in rice blast disease, facilitating biotrophic colonization of host cells and by suppression of host ROS accumulation (Fernandez and Wilson, 2014). In the Gluconacetobacter diazotrophicus-rice system glutathione reductase was determined to be crucial for endophytic colonization (Alquéres et al., 2013). Also identified was the protein pyridine nucleotide-disulphide oxidoreductase (TV_138628) which is associated with antioxidant activity. All these antioxidant proteins secreted by T. virens have been related to inactivation of ROS during plant-microbe interactions. Moreover, the GPI-anchored protein Ecm33-like (TV_72615) from T. virens was identified in the secretome. Ecm33 protein being reported as a GPI-anchored protein that attaches into the plasma membrane in Beauveria bassiana and Metarhizium robertsii, where it contributes to multi-stress tolerance against oxidant molecules, fungicide and osmotic stress (Chen et al., 2014). In T. virens, Ecm3 was found by Lamdan et al. (2015) as part of the secretome in the presence of maize roots. This may suggest that during colonization Ecm33-like protein may play an important role due to its properties as a multi-stress tolerance molecule, protecting T. virens hyphae against the oxidative burst created by host cells. Overall, the results presented here suggest that antioxidant secreted proteins collaborate in the suppression of basal resistance through the regulation of the cellular redox state, and by modifying gene expression in the host plant to maintain symbiosis between T. virens and maize.
Secreted Proteins Involved in Secondary Metabolism Biosynthesis
Fungal secondary metabolites (SMs) are classified into four classes: polyketides, terpenoids, shikimic acid derived compounds, and non-ribosomal peptides. During plant-microbe interactions, secondary metabolites such as phytohormones and toxins play major roles in the regulation of plant metabolic and defense response processes (Pusztahelyi et al., 2015). In addition, fungal secondary metabolites can shape fungal-plant interactions in a similar way as effector proteins (Pusztahelyi et al., 2015). The protein (S-adenosylmethionine (SAM) synthase (TV_87758), involved in the biosynthesis of the precursor of ethylene (ET) was identified and, ET is one of the phytohormones involved in plant defense responses (Broekgaarden et al., 2015). Interestingly, SAM synthase is involved in the ET biosynthesis pathway and is predicted to be secreted into the apoplast through a non-classical secretion system, where has been reported to be secreted by the presence of host roots (Lamdan et al., 2015). Ethylene is a crucial phytohormone for plant defense responses during the plant-microbe interactions, where successful plant symbionts have evolved different strategies to manipulate plant responses by affecting the ethylene pathway. For example, the small secreted protein SP7 from the arbuscular mycorrhizal Glomus intraradices is translocated into the cell nucleus and interacts with host ethylene-responsive transcription factor (ERF19) interfering with the defense cascade, thereby promoting mycorrhizal colonization (Kloppholz et al., 2011). In T. harzianum, activation of the enzyme ACC deaminase promotes root elongation of canola seedlings by modulating ET levels in the host plant (Viterbo et al., 2010). Results suggest that T. virens may influence ET levels in the apoplast allowing T. virens to colonize the intercellular spaces without triggering a major immune response.
Additionally, the proteins S-adenosyl-L-methionine methyltransferase (GliN) (TV_91355) and aminotransferase (GliI) TV_53497 which are involved in gliotoxin biosynthesis and belong to the gliotoxin cluster in T. virens (Mukherjee et al., 2012) were identified. Gliotoxin is a fungistatic mycotoxin that confers protection to oxidative stress and participates in the mycoparasitism but does not have an influence in T. virens root colonization (Vargas et al., 2014). The role of mycotoxins in plant microbe interactions are mainly described as virulence or pathogenicity factors. It will be necessary to elucidate the function of Trichoderma toxins during host colonization, which may act as neutralizers of plant defense mechanisms (Pedras and Ahiahonu, 2005).
Putative Effector-Like Proteins
To establish a beneficial or detrimental interaction plant microbes deliver effector-like proteins into host tissues, where they play a central role in the suppression of plant defense responses and modulate plant physiology by reprogramming metabolic processes of colonized tissues (Doehlemann et al., 2014; Mendoza-Mendoza et al., 2018). Effector-like proteins are defined as stable small secreted proteins normally ≤ 300 amino acids as although larger proteins have been identified that play similar roles (Lo Presti et al., 2015). Effector-like proteins are secreted either into the apoplast space or translocated into the plant cells (Lo Presti et al., 2015). Putative effector-like proteins were identified in the secretome of T. virens during the interaction with maize roots. The effector-like proteins were divided into subgroups according to their predicted functions.
Thioredoxin-like proteins
As mentioned above, the oxidative burst and the progressive induction of ROS have a major impact during plant microbe interactions; hence their homoeostasis in the apoplastic region is necessary for a long-term relationship. Two thioredoxins (TV_81963 and TV_216458) and one disulphide-isomerase (TV_11061) were identified which participate in the rearrangement of -S-S- bonds in proteins. These enzymes are secreted into the apoplast by T. virens upon colonization of maize roots and may function as putative effector-like proteins, reducing ROS activity in the apoplast and, in parallel, diminishing the plant defense reactions.
Several studies have summarized the importance of thioredoxins in plant-microbe interactions. The thioredoxin GBNRx1 from V. dahlia plays a crucial role in the apoplastic immune response functioning in apoplastic ROS scavenging in cotton (Li et al., 2016). The thioredoxin system has a major influence in Botrytis cinerea pathogenicity protecting it from oxidative stress (Viefhues et al., 2014). The thioredoxin complex influences several cellular processes by affecting protein folding or their activity by thiol-disulphide redox control (Arnér and Holmgren, 2000). In Ralstonia solani, the thioredoxin TRX2 is necessary for the activation of the effector RipAy that degrades glutathione which is involved in the plant immune response (Mukaihara et al., 2016). Thioredoxins are also involved in the bacterial-plant interactions where their plays a role in melanin synthesis and contributes to signal transduction during symbiotic nitrogen fixation, giving resistance to oxidative stress (Castro-Sowinski et al., 2007). Results suggest that T. virens may use antioxidant-secreted proteins that can act as effector-like proteins to control ROS levels in the apoplast which diminishes the plant immune response thus enabling fungal colonization.
Small secreted cysteine proteins (SSCPs)
Small secreted cysteine proteins (SSCPs) are widely distributed in plant symbionts. Trichoderma spp. (T. virens, T. atroviride, and T. reesei) contain around 173 SSCPs. Of these, 129 are specific to Trichoderma species and around 25 are unique having no BLAST matches (Lawry, 2016). Three SSCPs were identified, including Sm1 (TV_110852), SSCP1 (TV_92810), and SSCP2 (TV_111995) in the secretome from T. virens in the apoplast. The small protein (Sm1) belongs to the cerato-platanin family and was reported as an elicitor that induces systemic disease resistance through activating the JA pathway in the host plant (Djonović et al., 2007). However, the specific role of Sm1 during T. virens colonization is not known. Several paralogs of Sm1 have been identified: Sm2, Sm3, and Sm4 (Crutcher et al., 2015). Sm2 is highly expressed in the presence of maize roots compared with the other homologs, is involved in colonization and is more important than Sm1 in the promotion of plant protection (Gaderer et al., 2015), however we did not identify Sm2 in the T. virens secretome. The protein SSCP1 exhibits an 8 cysteine-containing CFEM domain was reported in the secretome of T. virens in the presence of maize roots (Lamdan et al., 2015). SSCP1 was predicted to act as a negative effector, directly affecting the defense level in the plant. The SSCPs in T. virens may play essential roles additional to elicitor and effector functions such as cell surface receptors, signal transducers, or adhesion molecules during colonization. Few of the SSCPs have been characterized so far in Trichoderma (Djonović et al., 2007; Crutcher et al., 2015; Gaderer et al., 2015; Lamdan et al., 2015). Further research is required to understand the redundancy and abundance of SSCPs in T. virens during the symbiotic relationship with it host plant.
Protein inhibitors (PIs)
The proteinase inhibitor I9 (TV_216375) was identified in the T. virens secretome. This protein belongs to the protease inhibitor superfamily, which are responsible for the modulation of folding and activity of the peptidase pro-enzyme. We hypothesize that PIs may be indispensable for the inactivation of plant defense secreted proteins in the apoplast, enabling fungal colonization and inactivation of plant defenses. Several apoplastic effectors in fungal plant pathogens with protease activity have been described during successful infection, for examples, the effector Pit2 in U. maydis functions as an inhibitor of maize cysteine proteases, and is required for fungal virulence and suppression of host immunity (Mueller et al., 2013). Other examples are the protease inhibitor Avr2 secreted by the fungal plant pathogen C. fulvum that targets the tomato defense protease Rcr3 (Song et al., 2009), and recently, a candidate effector had been predicted in S. sclerotorium that has a conserved serine protease I9 domain, which may be necessary to suppress host resistance (Guyon et al., 2014). In the T. virens-maize interaction different proteases were observed such as aspartic-type endopeptidase (A0A1D6F8J3_MAIZE) and a cysteine-type proteinase (A0A1D6ICV7_MAIZE) secreted by maize that were expressed during colonization. For that reason, PIs may be essential to inactivate or block plant immune proteases.
Peroxidase Levels Influenced by T. virens
Peroxidases are defense-related enzymes that are induced in the presence of plant associated microbes triggered by elicitor-induced signal transduction pathways (Almagro et al., 2009). The fact that peroxidase activity levels were significantly reduced in the presence of T. virens at 5 d interaction (Figure 5A) may suggest that fungal colonization does not activate stronger immune responses in maize roots at this development stage of the interaction. Moreover, reduction in peroxidase activity may directly influence lignification and suberization of cell walls and crosslinking of cell wall structural proteins allowing internal colonization by T. virens as shown in Figure 2D, due to their role in different physiological process and defense reactions such as cell wall reinforcement (Hiraga et al., 2001; Almagro et al., 2009). In poplar plants, peroxidase activity was influenced by the ectomycorrhizal Paxillus involutus dependent on the compatibility of the isolate, showing that during compatible interactions changes in the level of peroxidases were not observed, but under incompatible interaction the levels were significantly higher compared to the control plants (Gafur et al., 2007).
ROS scavenging enzymes secreted by T. virens into the apoplast, such as glutathione reductase (TV_54541), catalase-peroxidase haem (TV_215514), superoxide dismutase [Cu-Zn], thioredoxin reductase (TV_81963) and thioredoxin-related protein (TV_216458) help overcome the oxidative burst, which may have an effect in the activation of robust plant defense reactions including plant peroxidases. It has been demonstrated that production of fungal ROS can result in the suppression of endophyte growth in the host plant, which is critical in the mutualistic Epichloë festuce and grass interaction (Tanaka et al., 2006). Therefore, we suggest that T. virens secretes detoxifying enzymes into the apoplastic as a strategy to maintain ROS stability to mitigate the impact of stress and to manipulate further plant immune responses diminishing the effect of peroxidases in the apoplast.
Conclusions
In closing, this study reveals possible mechanisms necessary for T. virens endophytism (Figure 6). We identified that host cell wall degradation and modification are essential during internal colonization. Several pathways are activated during the interaction. In particular, redox homeostasis is crucial to maintain a controlled environment in the apoplast. We propose that both organisms secrete proteins into the apoplast via conventional and unconventional secretory mechanisms. We identified putative effector-like proteins secreted by T. virens that may lead the symbiotic relationship by the suppression of plant immune pathways, for example, peroxidase activity. This study lays the foundation for future studies for a better understanding of plant-fungus interactions, particularly at the proteomic level.
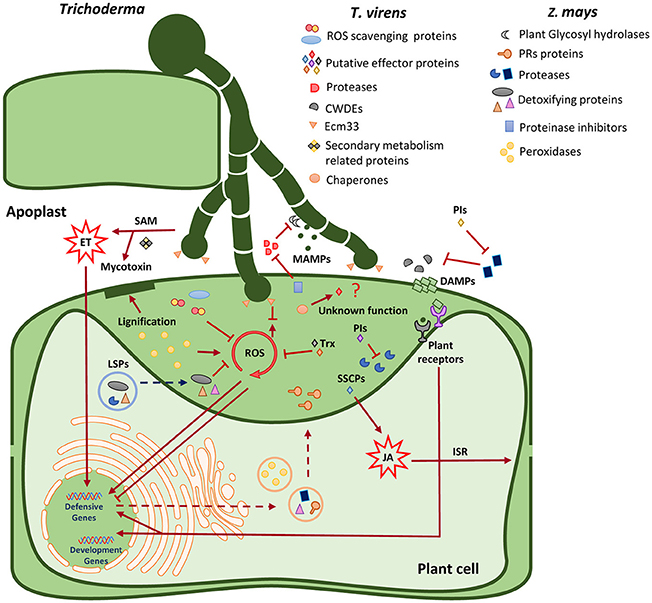
Figure 6. Model of apoplastic proteins identified in the T. virens-maize interaction. Location and putative function of apoplastic proteins secreted by T. virens and maize. ET, ethylene; JA, jasmonic acid; ISR, induced systemic resistance; ROS, reactive oxygen species; MAMPs, microbial-associated molecular patterns; DAMPS, damage-associated molecular patterns; CWDEs, Cell wall degradative enzymes; PRs, pathogenesis-related proteins; PIs, proteinase inhibitors; Trx, thioredoxin; LSPs, leaderless secretory proteins; SAM, S-adenosyl-l-methionine; SSCPs, small secreted cysteine-rich proteins.
Author Contributions
GN-L and MM performed experimental work; GN-L, AM-M, JS, CW, and DG designed the experiments; GN-L, AM-M, JS, CW, DG, CE, and MM discussed and interpreted the results; GN-L, AM-M, JS, and CW designed the research; AM-M and DG contributed to chemicals and scientific advice; GN-L, AM-M, and JS wrote the paper. All authors reviewed the final version of the paper.
Funding
This work was supported by the Tertiary Education Commission (contracts 38631 and 38651), Fast Start Marsden and Meadow Mushrooms Scholarship to AM-M and GN-L (Meadow Mushrooms Scholarship Lincoln University). GN-L was supported by Conacyt-Mexico for his PhD studies at Lincoln University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Charles Kenerley (Texas A&M, USA) for having provided us with Trichoderma virens Gv28.9 strain, Pioneer® brand products (New Zealand) are acknowledged for providing us with the maize inbred line seeds used in this work, Lee Botes from Olympus New Zealand Ltd is acknowledge for lending us the use of confocal microscope used in this work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00409/full#supplementary-material
References
Agrawal, G. K., Jwa, N. S., Lebrun, M. H., Job, D., and Rakwal, R. (2010). Plant secretome: unlocking secrets of the secreted proteins. Proteomics 10, 799–827. doi: 10.1002/pmic.200900514
Albenne, C., Canut, H., and Jamet, E. (2013). Plant cell wall proteomics: the leadership of Arabidopsis thaliana. Front. Plant Sci. 4:111. doi: 10.3389/fpls.2013.00111
Alexandersson, E., Ali, A., Resjo, S., and Andreasson, E. (2013). Plant secretome proteomics. Front. Plant Sci. 4:9. doi: 10.3389/fpls.2013.00009
Allen, M. F., Allen, E. B., and Friese, C. F. (1989). Responses of the non-mycotrophic plant Salsola kali to invasion by vesicular-arbuscular mycorrhizal fungi. New Phytol. 111, 45–49. doi: 10.1111/j.1469-8137.1989.tb04216.x
Almagro, L., Gomez Ros, L. V., Belchi-Navarro, S., Bru, R., Ros Barcelo, A., and Pedreno, M. A. (2009). Class III peroxidases in plant defence reactions. J. Exp. Bot. 60, 377–390. doi: 10.1093/jxb/ern277
Alquéres, S., Meneses, C., Rouws, L., Rothballer, M., Baldani, I., Schmid, M., et al. (2013). The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus PAL5. Mol. Plant Microbe Interact 26, 937–945. doi: 10.1094/MPMI-12-12-0286-R
Arnér, E. S., and Holmgren, A. (2000). Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267, 6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x
Balmer, D., Flors, V., Glauser, G., and Mauch-Mani, B. (2013). Metabolomics of cereals under biotic stress: current knowledge and techniques. Front. Plant Sci. 4:82. doi: 10.3389/fpls.2013.00082
Beckman, C. H. (2000). Phenolic-storing cells: keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants? Physiol. Mol. Plant Pathol. 57, 101–110. doi: 10.1006/pmpp.2000.0287
Bendtsen, J. D., Jensen, L. J., Blom, N., Von Heijne, G., and Brunak, S. (2004a). Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356. doi: 10.1093/protein/gzh037
Bendtsen, J. D., Nielsen, H., Von Heijne, G., and Brunak, S. (2004b). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. doi: 10.1016/j.jmb.2004.05.028
Bensadoun, A., and Weinstein, D. (1976). Assay of proteins in the presence of interfering materials. Anal. Biochem. 70, 241–250. doi: 10.1016/S0003-2697(76)80064-4
Biely, P. (1985). Microbial xylanolytic systems. Trends Biotechnol. 3, 286–290. doi: 10.1016/0167-7799(85)90004-6
Blilou, I., Bueno, P., Ocampo, J. A., and García-Garrido, J. M. (2000). Induction of catalase and ascorbate peroxidase activities in tobacco roots inoculated with the arbuscular mycorrhizal Glomus mosseae. Mycol. Res. 104, 722–725. doi: 10.1017/S095375629900204X
Bolte, S., Talbot, C., Boutte, Y., Catrice, O., Read, N. D., and Satiat-Jeunemaitre, B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214, 159–173. doi: 10.1111/j.0022-2720.2004.01348.x
Broekgaarden, C., Caarls, L., Vos, I. A., Pieterse, C. M. J., and Van Wees, S. C. M. (2015). Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiol. 169, 2371–2379. doi: 10.1104/pp.15.01020
Castro-Sowinski, S., Matan, O., Bonafede, P., and Okon, Y. (2007). A thioredoxin of Sinorhizobium meliloti CE52G is required for melanin production and symbiotic nitrogen fixation. Mol. Plant Microbe Interact 20, 986–993. doi: 10.1094/MPMI-20-8-0986
Chacón, M. R., Rodríguez-Galán, O., Benítez, T., Sousa, S., Rey, M., Llobell, A., et al. (2007). Microscopic and transcriptome analyses of early colonization of tomato roots by Trichoderma harzianum. Int. Microbiol. 10, 19–27. doi: 10.2436/20.1501.01.4
Chen, Y., Zhu, J., Ying, S. H., and Feng, M. G. (2014). The GPI-anchored protein Ecm33 is vital for conidiation, cell wall integrity, and multi-stress tolerance of two filamentous entomopathogens but not for virulence. Appl. Microbiol. Biotechnol. 98, 5517–5529. doi: 10.1007/s00253-014-5577-y
Choi, D. S., Hwang, I. S., and Hwang, B. K. (2012). Requirement of the cytosolic interaction between pathogenesis-related protein10 and leucine-rich repeat protein1 for cell death and defense signaling in pepper. Plant Cell 24, 1675–1690. doi: 10.1105/tpc.112.095869
Choi, J., Park, J., Kim, D., Jung, K., Kang, S., and Lee, Y. H. (2010). Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genomics 11:105. doi: 10.1186/1471-2164-11-105
Chu, J., Li, W.-F., Cheng, W., Lu, M., Zhou, K.-H., Zhu, H.-Q., et al. (2015). Comparative analyses of secreted proteins from the phytopathogenic fungus Verticillium dahliae in response to nitrogen starvation. Biochim. Biophys. Acta 1854, 437–448. doi: 10.1016/j.bbapap.2015.02.004
Crutcher, F. K., Moran-Diez, M. E., Ding, S., Liu, J., Horwitz, B. A., Mukherjee, P. K., et al. (2015). A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by Trichoderma virens. Fungal Biol. 119, 476–486. doi: 10.1016/j.funbio.2015.01.004
Cui, S., Guo, X., Chang, F., Cui, Y., Ma, L., Sun, Y., et al. (2005). Apoplastic calmodulin receptor-like binding proteins in suspension-cultured cells of Arabidopsis thaliana. J. Biol. Chem. 280, 31420–31427. doi: 10.1074/jbc.M501349200
Dannel, F., Pfeffer, H., and Marschner, H. (1995). Isolation of apoplasmic fluid from sunflower leaves and its use for studies on influence of nitrogen supply on apoplasmic pH. J. Plant Physiol. 146, 273–278. doi: 10.1016/S0176-1617(11)82053-5
de Jonge, R., Van Esse, H. P., Kombrink, A., Shinya, T., Desaki, Y., Bours, R., et al. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329, 953–955. doi: 10.1126/science.1190859
Delaunois, B., Jeandet, P., Clement, C., Baillieul, F., Dorey, S., and Cordelier, S. (2014). Uncovering plant-pathogen crosstalk through apoplastic proteomic studies. Front. Plant Sci. 5:249. doi: 10.3389/fpls.2014.00249
Deshmukh, S., Hückelhoven, R., Schäfer, P., Imani, J., Sharma, M., Weiss, M., et al. (2006). The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc. Natl. Acad. Sci. U.S.A 103, 18450–18457. doi: 10.1073/pnas.0605697103
Ding, Y., Wang, J., Wang, J., Stierhof, Y. D., Robinson, D. G., and Jiang, L. (2012). Unconventional protein secretion. Trends Plant Sci. 17, 606–615. doi: 10.1016/j.tplants.2012.06.004
Djamei, A., Schipper, K., Rabe, F., Ghosh, A., Vincon, V., Kahnt, J., et al. (2011). Metabolic priming by a secreted fungal effector. Nature 478, 395–398. doi: 10.1038/nature10454
Djonović, S., Pozo, M. J., Dangott, L. J., Howell, C. R., and Kenerley, C. M. (2006). Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant Microbe Interact 19, 838–853. doi: 10.1094/MPMI-19-0838
Djonović, S., Vargas, W. A., Kolomiets, M. V., Horndeski, M., Wiest, A., and Kenerley, C. M. (2007). A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in Maize. Plant Physiol. 145, 875–889. doi: 10.1104/pp.107.103689
Doehlemann, G., Requena, N., Schaefer, P., Brunner, F., O'connell, R., and Parker, J. E. (2014). Reprogramming of plant cells by filamentous plant-colonizing microbes. New Phytol. 204, 803–814. doi: 10.1111/nph.12938
Dragišić Maksimović, J. J., Zivanovic, B. D., Maksimovic, V. M., Mojovic, M. D., Nikolic, M. T., and Vucinic, Z. B. (2014). Filter strip as a method of choice for apoplastic fluid extraction from maize roots. Plant Sci. 223, 49–58. doi: 10.1016/j.plantsci.2014.03.009
Dragišić Maksimović, J., Maksimović, V., Živanović, B., HadŽi-Tašković Šukalović, V., and Vuletić, M. (2008). Peroxidase activity and phenolic compounds content in maize root and leaf apoplast, and their association with growth. Plant Sci. 175, 656–662. doi: 10.1016/j.plantsci.2008.06.015
Druzhinina, I. S., Shelest, E., and Kubicek, C. P. (2012). Novel traits of Trichoderma predicted through the analysis of its secretome. FEMS Microbiol. Lett. 337, 1–9. doi: 10.1111/j.1574-6968.2012.02665.x
Emanuelsson, O., Brunak, S., Von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Emanuelsson, O., Nielsen, H., Brunak, S., and Von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Fernandez, J., and Wilson, R. A. (2014). Characterizing roles for the glutathione reductase, thioredoxin reductase and thioredoxin peroxidase-encoding genes of Magnaporthe oryzae during rice blast disease. PLoS ONE 9:e87300. doi: 10.1371/journal.pone.0087300
Fernández, M. B., Pagano, M. R., Daleo, G. R., and Guevara, M. G. (2012). Hydrophobic proteins secreted into the apoplast may contribute to resistance against Phytophthora infestans in potato. Plant Physiol. Biochem. 60, 59–66. doi: 10.1016/j.plaphy.2012.07.017
Figueroa, M., Manning, V. A., Pandelova, I., and Ciuffetti, L. M. (2015). Persistence of the host-selective toxin ptr ToxB in the apoplast. Mol. Plant Microbe Interact 28, 1082–1090. doi: 10.1094/MPMI-05-15-0097-R
Floerl, S., Majcherczyk, A., Possienke, M., Feussner, K., Tappe, H., Gatz, C., et al. (2012). Verticillium longisporum infection affects the leaf apoplastic proteome, metabolome, and cell wall properties in Arabidopsis thaliana. PLoS ONE 7:e31435. doi: 10.1371/journal.pone.0031435
Gaderer, R., Lamdan, N., Frischmann, A., Sulyok, M., Krska, R., Horwitz, B. A., et al. (2015). Sm2, a paralog of the Trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of Trichoderma with maize. BMC Microbiol. 15:2. doi: 10.1186/s12866-014-0333-0
Gafur, A., Schutzendubel, A., and Polle, A. (2007). Peroxidase activity in poplar inoculated with compatible and incompetent isolates of Paxillus involutus. HAYATI J. Biosci. 14, 49–53. doi: 10.4308/hjb.14.2.49
Giardina, B. J., and Chiang, H.-L. (2013). Fructose-1,6-bisphosphatase, malate dehydrogenase, isocitrate lyase, phosphoenolpyruvate carboxykinase, glyceraldehyde-3-phosphate dehydrogenase, and cyclophilin a are secreted in Saccharomyces cerevisiae grown in low glucose. Commun. Integr. Biol. 6:e27216. doi: 10.4161/cib.27216
Girard, V., Dieryckx, C., Job, C., and Job, D. (2013). Secretomes: the fungal strike force. Proteomics 13, 597–608. doi: 10.1002/pmic.201200282
González-Fernández, R., Prats, E., and Jorrín-Novo, J. V. (2010). Proteomics of plant pathogenic fungi. J. Biomed. Biotechnol. 2010:932527. doi: 10.1155/2010/932527
Gupta, R., Lee, S. E., Agrawal, G. K., Rakwal, R., Park, S., Wang, Y., et al. (2015). Understanding the plant-pathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front. Plant Sci. 6:352. doi: 10.3389/fpls.2015.00352
Guyon, K., Balagué, C., Roby, D., and Raffaele, S. (2014). Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15:336. doi: 10.1186/1471-2164-15-336
Hajirezaei, M. R., Takahata, Y., Trethewey, R. N., Willmitzer, L., and Sonnewald, U. (2000). Impact of elevated cytosolic and apoplastic invertase activity on carbon metabolism during potato tuber development. J. Exp. Bot. 51, 439–445. doi: 10.1093/jexbot/51.suppl_1.439
Hamann, T. (2012). Plant cell wall integrity maintenance as an essential component of biotic stress response mechanisms. Front. Plant Sci. 3:77. doi: 10.3389/fpls.2012.00077
Harman, G. E. (2006). Overview of mechanisms and uses of Trichoderma spp. Phytopathology 96, 190–194. doi: 10.1094/PHYTO-96-0190
Harman, G. E., Howell, C. R., Viterbo, A., Chet, I., and Lorito, M. (2004). Trichoderma species-opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56. doi: 10.1038/nrmicro797
Heller, J., and Tudzynski, P. (2011). Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49, 369–390. doi: 10.1146/annurev-phyto-072910-095355
Hermosa, R., Rubio, M. B., Cardoza, R. E., Nicolas, C., Monte, E., and Gutierrez, S. (2013). The contribution of Trichoderma to balancing the costs of plant growth and defense. Int. Microbiol. 16, 69–80. doi: 10.2436/20.1501.01.181
Hiraga, S., Sasaki, K., Ito, H., Ohashi, Y., and Matsui, H. (2001). A large family of class III plant peroxidases. Plant Cell Physiol. 42, 462–468. doi: 10.1093/pcp/pce061
Hoagland, D. R., and Arnon, D. I. (1950). The water-culture method for growing plants without soil. Calif. Agricult. Exp. Station Circ. 347, 1–32.
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Husted, S., and Schjoerring, J. K. (1995). Apoplastic pH and ammonium concentration in leaves of Brassica napus L. Plant Physiol. 109, 1453–1460. doi: 10.1104/pp.109.4.1453
Jayaraman, D., Forshey, K. L., Grimsrud, P. A., and Ane, J. M. (2012). Leveraging proteomics to understand plant-microbe interactions. Front. Plant Sci. 3:44. doi: 10.3389/fpls.2012.00044
Kaffarnik, F. A., Jones, A. M., Rathjen, J. P., and Peck, S. C. (2009). Effector proteins of the bacterial pathogen Pseudomonas syringae alter the extracellular proteome of the host plant, Arabidopsis thaliana. Mol. Cell Proteom. 8, 145–156. doi: 10.1074/mcp.M800043-MCP200
Kawalleck, P., Plesch, G., Hahlbrock, K., and Somssich, I. E. (1992). Induction by fungal elicitor of S-adenosyl-L-methionine synthetase and S-adenosyl-L-homocysteine hydrolase mRNAs in cultured cells and leaves of Petroselinum crispum. Proc. Natl. Acad. Sci. U.S.A. 89, 4713–4717. doi: 10.1073/pnas.89.10.4713
Kim, J. Y., Wu, J., Kwon, S. J., Oh, H., Lee, S. E., Kim, S. G., et al. (2014). Proteomics of rice and Cochliobolus miyabeanus fungal interaction: insight into proteins at intracellular and extracellular spaces. Proteomics 14, 2307–2318. doi: 10.1002/pmic.201400066
Kim, S. G., Wang, Y., Lee, K. H., Park, Z. Y., Park, J., Wu, J., et al. (2013). In-depth insight into in vivo apoplastic secretome of rice-Magnaporthe oryzae interaction. J. Proteomics 78, 58–71. doi: 10.1016/j.jprot.2012.10.029
Kloppholz, S., Kuhn, H., and Requena, N. (2011). A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr. Biol. 21, 1204–1209. doi: 10.1016/j.cub.2011.06.044
Kubicek, C. P., Starr, T. L., and Glass, N. L. (2014). Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 52, 427–451. doi: 10.1146/annurev-phyto-102313-045831
Lamdan, N. L., Shalaby, S., Ziv, T., Kenerley, C. M., and Horwitz, B. A. (2015). Secretome of the biocontrol fungus Trichoderma virens co-cultured with maize roots: role in induced systemic resistance. Mol. Cell Proteomics 14, 1054–1063. doi: 10.1074/mcp.M114.046607.
Lawry, R. (2016). Cross-communication Between Trichoderma and Plants During Root Colonisation. PhD, Lincoln University.
Li, Y. B., Han, L. B., Wang, H. Y., Zhang, J., Sun, S. T., Feng, D. Q., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Liao, C., Hochholdinger, F., and Li, C. (2012). Comparative analyses of three legume species reveals conserved and unique root extracellular proteins. Proteomics 12, 3219–3228. doi: 10.1002/pmic.201100629
Lionetti, V., Raiola, A., Camardella, L., Giovane, A., Obel, N., Pauly, M., et al. (2007). Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 143, 1871–1880. doi: 10.1104/pp.106.090803
López-Villar, E., Monteoliva, L., Larsen, M. R., Sachon, E., Shabaz, M., Pardo, M., et al. (2006). Genetic and proteomic evidences support the localization of yeast enolase in the cell surface. Proteomics 6(Suppl. 1), S107–S118. doi: 10.1002/pmic.200500479
Lo Presti, L., Lanver, D., Schweizer, G., Tanaka, S., Liang, L., Tollot, M., et al. (2015). Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545. doi: 10.1146/annurev-arplant-043014-114623
Lorito, M., Woo, S. L., Harman, G. E., and Monte, E. (2010). Translational research on Trichoderma: from'omics to the field. Annu. Rev. Phytol. 48, 395–417. doi: 10.1146/annurev-phyto-073009-114314
Lozano-Durán, R., and Robatzek, S. (2015). 14-3-3 proteins in plant-pathogen interactions. Mol. Plant Microbe Interact 28, 511–518. doi: 10.1094/MPMI-10-14-0322-CR
Luo, Y., Yuan, Z., Luo, G., and Zhao, F. (2008). Expression of secreted His-tagged S-adenosylmethionine synthetase in the methylotrophic yeast Pichia pastoris and its characterization, one-step purification, and immobilization. Biotechnol. Prog. 24, 214–220. doi: 10.1021/bp0702727
Maimbo, M., Ohnishi, K., Hikichi, Y., Yoshioka, H., and Kiba, A. (2007). Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol. 145, 1588–1599. doi: 10.1104/pp.107.105353
Marra, R., Ambrosino, P., Carbone, V., Vinale, F., Woo, S. L., Ruocco, M., et al. (2006). Study of the three-way interaction between Trichoderma atroviride, plant and fungal pathogens by using a proteomic approach. Curr. Genet. 50, 307–321. doi: 10.1007/s00294-006-0091-0
Marschall, R., and Tudzynski, P. (2016). Reactive oxygen species in development and infection processes. Semin. Cell Dev. Biol. 57, 138–146. doi: 10.1016/j.semcdb.2016.03.020
Mehdy, M. C. (1994). Active oxygen species in plant defense against pathogens. Plant Physiol. 105, 467–472. doi: 10.1104/pp.105.2.467
Meinken, J., Asch, K. D., Neizer-Ashun, A. K., Guang-Hwa, C., Cooper, J. R. C., and Xiang, J. M. (2014). FunSecKB2: a fungal protein subcellular location knowledgebase. Comput. Mol. Biol. 4:7. doi: 10.5376/cmb.2014.04.0007
Mendoza-Mendoza, A., Zaid, R., Lawry, R., Hermosa, R., Monte, E., Horwitz, B. A., et al. (2018). Molecular dialogues between Trichoderma and roots: role of the fungal secretome. Fungal Biol. Rev. 32, 62–85. doi: 10.1016/j.fbr.2017.12.001
Min, X. J., Lum, G., Meinken, J., Orr, J., and Frazier, S. (2014). PlantSecKB: the plant secretome and subcellular proteome knowledgebase. Comput. Mol. Biol. 4:1. doi: 10.5376/cmb.2014.04.0001
Minic, Z. (2008). Physiological roles of plant glycoside hydrolases. Planta 227, 723–740. doi: 10.1007/s00425-007-0668-y
Mochizuki, S., Saitoh, K.-I., Minami, E., and Nishizawa, Y. (2011). Localization of probe-accessible chitin and characterization of genes encoding chitin-binding domains during rice–Magnaporthe oryzae interactions. J. Gen. Plant Pathol. 77, 163–173. doi: 10.1007/s10327-011-0310-5
Moran-Diez, M. E., Trushina, N., Lamdan, N. L., Rosenfelder, L., Mukherjee, P. K., Kenerley, C. M., et al. (2015). Host-specific transcriptomic pattern of Trichoderma virens during interaction with maize or tomato roots. BMC Genomics 16:8. doi: 10.1186/s12864-014-1208-3
Mueller, A. N., Ziemann, S., Treitschke, S., Assmann, D., and Doehlemann, G. (2013). Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. PLoS Pathog. 9:e1003177. doi: 10.1371/journal.ppat.1003177
Mukaihara, T., Hatanaka, T., Nakano, M., and Oda, K. (2016). Ralstonia solanacearum type III effector RipAY is a glutathione-degrading enzyme that is activated by plant cytosolic thioredoxins and suppresses plant immunity. MBio 7:e00359–16. doi: 10.1128/mBio.00359-16
Mukherjee, P. K., Horwitz, B. A., and Kenerley, C. M. (2012). Secondary metabolism in Trichoderma–a genomic perspective. Microbiology 158, 35–45. doi: 10.1099/mic.0.053629-0
Nanda, A. K., Andrio, E., Marino, D., Pauly, N., and Dunand, C. (2010). Reactive oxygen species during plant-microorganism early interactions. J. Integr. Plant Biol. 52, 195–204. doi: 10.1111/j.1744-7909.2010.00933.x
Nummi, M., Niku-Paavola, M. L., Lappalainen, A., Enari, T. M., and Raunio, V. (1983). Cellobiohydrolase from Trichoderma reesei. Biochem. J. 215, 677–683. doi: 10.1042/bj2150677
Palaniyandi, U., Anandaraj, M., and Benjamin, S. (2017). Endophytic interactions of Trichoderma harzianum in a tropical perennial rhizo-ecosystem. Res. J. Biotechnol. 12, 22–30.
Park, C.-J., and Seo, Y.-S. (2015). Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathol. J. 31, 323–333. doi: 10.5423/PPJ.RW.08.2015.0150
Pedras, M. S., and Ahiahonu, P. W. (2005). Metabolism and detoxification of phytoalexins and analogs by phytopathogenic fungi. Phytochemistry 66, 391–411. doi: 10.1016/j.phytochem.2004.12.032
Petersen, T. N., Brunak, S., Von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Porter, B. W., Yuen, C. Y. L., and Christopher, D. A. (2015). Dual protein trafficking to secretory and non-secretory cell compartments: clear or double vision? Plant Sci. 234, 174–179. doi: 10.1016/j.plantsci.2015.02.013
Porteus, B., Kocharunchitt, C., Nilsson, R. E., Ross, T., and Bowman, J. P. (2011). Utility of gel-free, label-free shotgun proteomics approaches to investigate microorganisms. Appl. Microbiol. Biotechnol. 90, 407–416. doi: 10.1007/s00253-011-3172-z.
Pusztahelyi, T., Holb, I. J., and Pócsi, I. (2015). Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 6:573. doi: 10.3389/fpls.2015.00573
Ranwala, A. P., Suematsu, C., and Masuda, H. (1992). The role of β-galactosidases in the modification of cell wall components during muskmelon fruit ripening. Plant Physiol. 100, 1318–1325. doi: 10.1104/pp.100.3.1318
Romão-Dumaresq, A. S., Dourado, M. N., Fávaro, L. C. D. L., Mendes, R., Ferreira, A., and Araújo, W. L. (2016). Diversity of cultivated fungi associated with conventional and transgenic sugarcane and the interaction between endophytic Trichoderma virens and the host plant. PLoS ONE 11:e0158974. doi: 10.1371/journal.pone.0158974
Ron, M., and Avni, A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16, 1604–1615. doi: 10.1105/tpc.022475
Rotblat, B., Enshell-Seijffers, D., Gershoni, J. M., Schuster, S., and Avni, A. (2002). Identification of an essential component of the elicitation active site of the EIX protein elicitor. Plant J. 32, 1049–1055. doi: 10.1046/j.1365-313X.2002.01490.x
Schmidt, F., and Volker, U. (2011). Proteome analysis of host-pathogen interactions: investigation of pathogen responses to the host cell environment. Proteomics 11, 3203–3211. doi: 10.1002/pmic.201100158
Schmoll, M., Dattenbock, C., Carreras-Villasenor, N., Mendoza-Mendoza, A., Tisch, D., Aleman, M. I., et al. (2016). The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiol. Mol. Biol. Rev. 80, 205–327. doi: 10.1128/MMBR.00040-15
Schulz, M., Filary, B., Kuhn, S., Colby, T., Harzen, A., Schmidt, J., et al. (2016). Benzoxazolinone detoxification by N-glucosylation: the multi-compartment-network of Zea mays L. Plant Signal. Behav. 11:e1119962. doi: 10.1080/15592324.2015.1119962
Shenton, M. R., Berberich, T., Kamo, M., Yamashita, T., Taira, H., and Terauchi, R. (2012). Use of intercellular washing fluid to investigate the secreted proteome of the rice-Magnaporthe interaction. J. Plant Res. 125, 311–316. doi: 10.1007/s10265-012-0473-y
Shi, L.-N., Li, F.-Q., Huang, M., Lu, J.-F., Kong, X.-X., Wang, S.-Q., et al. (2012). Immunoproteomics based identification of thioredoxin reductase GliT and novel Aspergillus fumigatus antigens for serologic diagnosis of invasive aspergillosis. BMC Microbiol. 12:11. doi: 10.1186/1471-2180-12-11
Shoresh, M., and Harman, G. E. (2008). The relationship between increased growth and resistance induced in plants by root colonizing microbes. Plant Signal. Behav. 3, 737–739. doi: 10.4161/psb.3.9.6605
Shoresh, M., Harman, G. E., and Mastouri, F. (2010). Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48, 21–43. doi: 10.1146/annurev-phyto-073009-114450
Song, J., Win, J., Tian, M., Schornack, S., Kaschani, F., Ilyas, M., et al. (2009). Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. U.S.A. 106, 1654–1659. doi: 10.1073/pnas.0809201106
Sperschneider, J., Gardiner, D. M., Dodds, P. N., Tini, F., Covarelli, L., Singh, K. B., et al. (2016). EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol. 210, 743–761. doi: 10.1111/nph.13794
Sundstrom, P., and Aliaga, G. R. (1994). A subset of proteins found in culture supernatants of Candida albicans includes the abundant, immunodominant, glycolytic enzyme enolase. J. Infect. Dis. 169, 452–456. doi: 10.1093/infdis/169.2.452
Sutton, J. C., Sopher, C. R., Owen-Going, T. N., Liu, W., Grodzinski, B., Hall, J. C., et al. (2006). Etiology and epidemiology of Pythium root rot in hydroponic crops: current knowledge and perspectives. Summa Phytopathol. 32, 307–321. doi: 10.1590/S0100-54052006000400001
Tanabe, S., Ishii-Minami, N., Saitoh, K., Otake, Y., Kaku, H., Shibuya, N., et al. (2011). The role of catalase-peroxidase secreted by Magnaporthe oryzae during early infection of rice cells. Mol. Plant Microbe Interact 24, 163–171. doi: 10.1094/MPMI-07-10-0175
Tanaka, A., Christensen, M. J., Takemoto, D., Park, P., and Scott, B. (2006). Reactive oxygen species play a role in regulating a fungus–perennial ryegrass mutualistic interaction. Plant Cell 18, 1052–1066. doi: 10.1105/tpc.105.039263
Tanveer, T., Shaheen, K., Parveen, S., Kazi, A. G., and Ahmad, P. (2014). Plant secretomics: identification, isolation, and biological significance under environmental stress. Plant Signal. Behav. 9:e29426. doi: 10.4161/psb.29426
Tiwari, P., Misra, B. N., and Sangwan, N. S. (2013). ß-glucosidases from the fungus Trichoderma: an efficient cellulase machinery in biotechnological applications. Biomed. Res. Int. 2013:203735. doi: 10.1155/2013/203735
Urbanek, H., Kuzniak-Gebarowska, E., and Herka, K. (1991). Elicitation of defence responses in bean leaves by Botrytis cinerea polygalacturonase. Acta Physiol. Plant. 13, 43–50.
van Loon, L. C., Rep, M., and Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Vargas, W. A., Mandawe, J. C., and Kenerley, C. M. (2009). Plant-derived sucrose is a key element in the symbiotic association between Trichoderma virens and maize plants. Plant Physiol. 151, 792–808. doi: 10.1104/pp.109.141291
Vargas, W. A., Mukherjee, P. K., Laughlin, D., Wiest, A., Moran-Diez, M. E., and Kenerley, C. M. (2014). Role of gliotoxin in the symbiotic and pathogenic interactions of Trichoderma virens. Microbiology 160, 2319–2330. doi: 10.1099/mic.0.079210-0
Viefhues, A., Heller, J., Temme, N., and Tudzynski, P. (2014). Redox systems in Botrytis cinerea: impact on development and virulence. Mol. Plant Microbe Interact 27, 858–874. doi: 10.1094/MPMI-01-14-0012-R
Vierheilig, H., Alt, M., Lange, J., Gut-Rella, M., Wiemken, A., and Boller, T. (1995). Colonization of transgenic tobacco constitutively expressing pathogenesis-related proteins by the vesicular-arbuscular mycorrhizal fungus Glomus mosseae. Appl. Environ. Microbiol. 61, 3031–3034.
Vinale, F., Sivasithamparam, K., Ghisalberti, E. L., Marra, R., Barbetti, M. J., Li, H., et al. (2008). A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol. Mol. Plant Pathol. 72, 80–86. doi: 10.1016/j.pmpp.2008.05.005
Viterbo, A., and Chet, I. (2006). TasHyd1, a new hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol. Plant Pathol. 7, 249–258. doi: 10.1111/j.1364-3703.2006.00335.x
Viterbo, A., Landau, U., Kim, S., Chernin, L., and Chet, I. (2010). Characterization of ACC deaminase from the biocontrol and plant growth-promoting agent Trichoderma asperellum T203. FEMS Microbiol. Lett. 305, 42–48. doi: 10.1111/j.1574-6968.2010.01910.x
Weber, S. S., Parente, A. F. A., Borges, C. L., Parente, J. A., Bailão, A. M., and De Almeida Soares, C. M. (2012). Analysis of the secretomes of paracoccidioides mycelia and yeast cells. PLoS ONE 7:e52470. doi: 10.1371/journal.pone.0052470
Witzel, K., Shahzad, M., Matros, A., Mock, H. P., and Muhling, K. H. (2011). Comparative evaluation of extraction methods for apoplastic proteins from maize leaves. Plant Methods 7:48. doi: 10.1186/1746-4811-7-48
Wu, X., Xiong, E., Wang, W., Scali, M., and Cresti, M. (2014). Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat. Protocols 9, 362–374. doi: 10.1038/nprot.2014.022
Yadav, N., Khurana, P. S. M., and Yadav, D. K. (2015). “Plant secretomics: unique initiatives,” in PlantOmics: The Omics of Plant Science, eds D. Barh, M. S. Khan, and E. Davies (New Delhi: Springer), 358–384.
Yajima, W., Liang, Y., and Kav, N. N. (2009). Gene disruption of an arabinofuranosidase/beta-xylosidase precursor decreases Sclerotinia sclerotiorum virulence on canola tissue. Mol. Plant Microbe Interact 22, 783–789. doi: 10.1094/MPMI-22-7-0783
Yang, F., Li, W., Derbyshire, M., Larsen, M. R., Rudd, J. J., and Palmisano, G. (2015). Unraveling incompatibility between wheat and the fungal pathogen Zymoseptoria tritici through apoplastic proteomics. BMC Genomics 16:362. doi: 10.1186/s12864-015-1549-6
Ye, J., Guo, Y., Zhang, D., Zhang, N., Wang, C., and Xu, M. (2013). Cytological and molecular characterization of quantitative trait locus qRfg1, which confers resistance to gibberella stalk rot in maize. Mol. Plant Microbe Interact 26, 1417–1428. doi: 10.1094/MPMI-06-13-0161-R
Yedidia, I., Benhamou, N., Kapulnik, Y., and Chet, I. (2000). Induction and accumulation of PR proteins activity during early stages of root colonization by the mycoparasite Trichoderma harzianum strain T-203. Plant Physiol. Biochem. 38, 863–873. doi: 10.1016/S0981-9428(00)01198-0
Yedidia, I. I., Benhamou, N., and Chet, I. I. (1999). Induction of defense responses in cucumber plants (Cucumis sativus L.) by the biocontrol agent Trichoderma harzianum. Appl. Environ. Microbiol. 65, 1061–1070.
Zamioudis, C., and Pieterse, C. M. J. (2012). Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 25, 139–150. doi: 10.1094/MPMI-06-11-0179
Zhang, N., Zhang, S., Borchert, S., Richardson, K., and Schmid, J. (2011). High levels of a fungal superoxide dismutase and increased concentration of a PR-10 plant protein in associations between the endophytic fungus Neotyphodium lolii and ryegrass. Mol. Plant Microbe Interact 24, 984–992. doi: 10.1094/MPMI-02-11-0028
Keywords: peroxidases, apoplast, reactive oxygen species (ROS), secretome, Trichoderma, roots, endophyte
Citation: Nogueira-Lopez G, Greenwood DR, Middleditch M, Winefield C, Eaton C, Steyaert JM and Mendoza-Mendoza A (2018) The Apoplastic Secretome of Trichoderma virens During Interaction With Maize Roots Shows an Inhibition of Plant Defence and Scavenging Oxidative Stress Secreted Proteins. Front. Plant Sci. 9:409. doi: 10.3389/fpls.2018.00409
Received: 11 December 2017; Accepted: 14 March 2018;
Published: 05 April 2018.
Edited by:
Víctor Flors, Jaume I University, SpainReviewed by:
Maria J. Pozo, Consejo Superior de Investigaciones Científicas (CSIC), SpainTanja Mimmo, Free University of Bozen-Bolzano, Italy
Copyright © 2018 Nogueira-Lopez, Greenwood, Middleditch, Winefield, Eaton, Steyaert and Mendoza-Mendoza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Artemio Mendoza-Mendoza, YXJ0ZW1pby5tZW5kb3phQGxpbmNvbG4uYWMubno=
 Guillermo Nogueira-Lopez
Guillermo Nogueira-Lopez David R. Greenwood2
David R. Greenwood2 Martin Middleditch
Martin Middleditch Artemio Mendoza-Mendoza
Artemio Mendoza-Mendoza