- Australian Research Council Centre of Excellence in Plant Energy Biology, School of Molecular Sciences, The University of Western Australia, Crawley, WA, Australia
RNA editing in plastids is known to be required for embryogenesis, but no single editing event had been shown to be essential. We show that the emb2261-2 mutation is lethal through a failure to express an editing factor that specifically recognizes the rps14-2 site. EMB2261 was predicted to bind the cis-element upstream of the rps14-2 site and genetic complementation with promoters of different strength followed by RNA-seq analysis was conducted to test the correlation between rps14-2 editing and EMB2261 expression. Rps14-2 is the only editing event in Arabidopsis chloroplasts that correlates with EMB2261 expression. Sequence divergence between the cis-element and the EMB2261 protein sequence in plants where rps14-2 editing is not required adds support to the association between them. We conclude that EMB2261 is the specificity factor for rps14-2 editing. This editing event converts P51 in Rps14 to L51, which is conserved among species lacking RNA editing, implying the importance of the editing event to Rps14 function. Rps14 is an essential ribosomal subunit for plastid translation, which, in turn, is essential for Arabidopsis embryogenesis.
Introduction
RNA editing is a crucial process in plant organellar gene expression. In flowering plants, it involves cytidine (C) to uridine (U) deamination (Takenaka et al., 2013b). In Arabidopsis thaliana, over 600 C-to-U editing events have been detected in mitochondria and 44 C-to-U editing events have been detected in chloroplasts (Giege and Brennicke, 1999; Chateigner-Boutin and Small, 2007; Bentolila et al., 2013; Ruwe et al., 2013). RNA editing in plant organelles is facilitated by organelle-targeted pentatricopeptide repeat (PPR) editing factors (Barkan and Small, 2014). They contain multiple tandem helix-loop-helix PPR motifs that specifically bind to the RNA sequence just 5′ to the edited nucleotide in a one-motif to one-base manner, acting as site recognition factors (Barkan and Small, 2014). Amino acids at two positions in each PPR motif specifically recognize one of the four RNA bases, denoted as the PPR-RNA recognition code (Barkan et al., 2012; Takenaka et al., 2013a; Yagi et al., 2013a). This recognition appears to involve hydrogen bonding to the aligned RNA base (Shen et al., 2016). At the C-terminus of PPR proteins, there is a deaminase-like domain that is hypothesized to be part of a larger editosome (Sun et al., 2016). Arabidopsis thaliana encodes 216 potential PPR editing factors, forming one of its largest protein families (Cheng et al., 2016). Nineteen PPR editing factors have been identified accounting for 30 out of the 34 major editing sites in Arabidopsis chloroplasts (Kotera et al., 2005; Okuda et al., 2007, 2009; Chateigner-Boutin et al., 2008; Cai et al., 2009; Hammani et al., 2009; Robbins et al., 2009; Zhou et al., 2009; Hayes et al., 2013; Yagi et al., 2013b; Wagoner et al., 2015; Yap et al., 2015). Editing factors for the following four sites remained unidentified prior to this work: ndhB-3 (96579), ndhB-1 (97016), petL (65716), and rps14-2 (37092).
Chloroplast biogenesis is essential to seed development. Mutations in genes involved in chloroplast gene expression such as those encoding ribosomal units (Tsugeki et al., 1996; Meinke et al., 2008) or splicing factors (Asakura and Barkan, 2006; Aryamanesh et al., 2017), can lead to premature arrest of embryogenesis during the globular to heart transition. RNA editing as an important post-transcriptional processing step in organelles is also known to be essential for seed development. For example, mutations in genes encoding DYW2 and NUWA required for RNA editing in both mitochondria and chloroplasts are embryo-lethal (Andres-Colas et al., 2017; Guillaumot et al., 2017). Some site-specific PPR editing factors targeted to mitochondria, such as EMP9 (Yang et al., 2017) and DEK36 (Wang et al., 2017) are also essential during seed development. However, mutants of the 19 site-specific PPR editing factors in Arabidopsis chloroplasts described prior to this work are all viable, showing a variety of phenotypes at later developmental stages, including decreased chloroplast NDH complex activity (Kotera et al., 2005; Okuda et al., 2007, 2009; Cai et al., 2009), changes in leaf pigmentation (Chateigner-Boutin et al., 2008; Cai et al., 2009; Zhou et al., 2009; Wagoner et al., 2015; Yap et al., 2015) and aberrant leaf shapes (Hayes et al., 2013). Only one embryo-lethal mutation (emb2261) that affects a potential site-specific PPR editing factor in Arabidopsis chloroplasts has been described (Cushing et al., 2005). The emb2261 mutant stalls at the heart stage during seed development. We sought to characterize the function of EMB2261 and its potential target site(s).
One approach to study embryo-lethal mutants is to perform partial complementation. The use of the seed-specific ABI3 promoter to drive the gene of interest for partial complementation has been successful in studying the embryo-lethal mutants emb506 (Despres et al., 2001), emb2394 and emb2654 (Aryamanesh et al., 2017), among which emb2654 is a chloroplast PPR splicing factor mutant. We therefore hypothesized that ABI3-promoter-driven EMB2261 constructs could partially complement the emb2261 mutant such that it could complete seed development. EMB2261 expression would then fade away as the ABI3 promoter loses its activity, leaving only the emb2261 mutant background from the seedling stage onward. Therefore, the partial complementation method would provide an opportunity to obtain enough plant tissue to examine RNA editing in the emb2261 mutant background.
During the writing up of this work, an independent manuscript reported that ECD1 (synonymous with EMB2261) is required for editing of the rps14-2 site in Arabidopsis and is required for early chloroplast development (Jiang et al., 2018). We confirm this conclusion with a different genetic approach and a different mutant allele of the EMB2261 gene, and expand these findings by considering the specificity of the EMB2261/rps14-2 interaction and the evolutionary history of the pair.
Materials and Methods
Prediction Method
The alignment of an editing factor and a site was scored by calculating the sum of the log-likelihood ratios at each position in the alignment (Yap et al., 2015). The log-likelihood ratios were derived from observed frequencies of association between amino acid combinations at the fifth and last position and the four RNA nucleotides (Supplementary Table S1). Histograms were generated using matplotlib v1.5.31. Heat maps were generated using v0.8.12.
Cloning of Plant Transformation Constructs
The EMB2261 gene fragment was amplified from Col-0 genomic DNA with the attB recombination sites introduced using PrimeSTAR polymerase (Clontech3). The EMB2261 PCR product was purified by QIAquick PCR purification kit (Qiagen4), cloned into the donor vector pDONR207 using Gateway BP Clonase (Invitrogen5). The EMB2261 gene fragment was then cloned from the entry vector pDONR207 to the plant expression vector pH7WG containing the ABI3 promoter (Aryamanesh et al., 2017) (ABI3:EMB2261), or pGWB2 (EMBL) containing the 35S promoter (35S:EMB2261), using Gateway LR Clonase (Invitrogen, see footnote 5). Cloning reactions were transformed into E. coli competent cells (DH5α). Positive clones for each construct were confirmed by Sanger sequencing. The verified plant expression constructs were transformed into Agrobacterium tumefaciens competent cells (GV3101).
Plant Growth, Transformation, and Selection
Arabidopsis seeds were surface sterilized with 70% ethanol supplemented with 0.05% Triton-X100 for 5 min and washed with 100% ethanol before being dried in the fume hood. Sterilized seeds were sowed on plates (half-strength MS medium and 0.8% agar), stratified at 4°C in the dark for 3 days, germinated and grown under long-day conditions (16 h light/8 h dark cycle, approximately 120 μmol photons m-2 s-1). Heterozygous plants of emb2261-2 (SALK_024975) were selected by genotyping using the primer pair SALK_024975_RP (CTTTCTCGAGTGCATTCAAGG) and LBb1.3 (ATTTTGCCGATTTCGGAAC) for T-DNA insertion, and with the primer pair SALK_024975_RP (CTTTCTCGAGTGCATTCAAGG) and SALK_024975_LP (TATATTTGGTGAGCATTCGGG) for genomic DNA. Plants were transformed by floral dip (Clough and Bent, 1998). Seeds harvested from the dipped plants were germinated and selected on Hygromycin B (25 μg/ml). Transformants were genotyped for homozygosity of the T-DNA insertion in the EMB2261 gene with the same set of primers listed above, except that the reverse primer SALK_024975_LP2 (GTGTATCTAAATCTCAAAGTCACC) annealing to the 3′UTR of the native EMB2261 gene was used to distinguish between the native EMB2261 gene and the EMB2261 transgenes.
RNA Analysis
Total RNA was isolated using the PureZOL reagent (Bio-Rad6) and treated with TURBO DNase (Ambion7) according to the manufacturer’s instructions. Completion of DNase treatment was verified by PCR targeting chloroplast genomic DNA. Complementary DNA (cDNA) was synthesized using random primers and SuperScript III reverse transcriptase (Invitrogen, see footnote 5) according to the manufacturer’s instructions.
The primer pair targeting the rps14-2 editing site were TCGCTAAGTGAGAAATGGAAAA (forward) and CGTCGATGAAGACGTGTAGG (reverse). The PCR cycling conditions were 40 cycles of 10 s at 98°C, 15 s at 58°C, and 4 s at 72°C, using PrimeSTAR polymerase (Clontech, see footnote 3). Poisoned primer extension (PPE) was carried out as described by Chateigner-Boutin and Small (2007) with a nucleotide mix containing dideoxythymidine (ddT). The fluorescein-labeled primer used for PPE was 6′FAM-AAATGGAAAATTCATGGAAAATTACAAT.
The quantitative PCR (qPCR) primer pair targeting the EMB2261 gene were CGTACGTTTCTTGGAGCTTGCAG (forward) and TTCCCATTTCCCTGCACAAGCG (reverse). qPCR was performed using the Quantinova mix (Qiagen, see footnote 4) according to manufacturer’s instructions on a Lightcycler 480 machine (Roche Molecular Diagnostics8). EMB2261 gene expression was normalized to expression of the reference gene CACS by the formula: (1 + EEMB2261)ˆ(35-CqEMB2261)/(1 + ECACS)ˆ(35-CqCACS).
RNA-Seq and Data Analysis
RNA-seq libraries were prepared using TruSeq Stranded Total RNA LT Kit with Ribo-zero plant (Illumina9), quantified using KAPA Library Quantification Kit for Illumina platforms (Kapa Biosystems, KK485410), and pooled in an equimolar ratio. Single-end sequencing was performed with a read length of 61 bases on an Illumina HiSeq 1500 sequencer. The sequence datasets are available in the NCBI Sequence Read Archive (SRA) repository (accession SRP141099).
The data in fastq format was trimmed using Trimmomatic v0.33 (Bolger et al., 2014) to remove the adapter sequence (ILLUMINACLIP:TruSeq_index.fasta:2:30:3, TruSeq_index =CAAGCAGAAGACGGCATACGAGAT), bases with Phred Quality score < 20 (LEADING:15 TRAILING:15 SLIDINGWINDOW:4:20) and reads shorter than 30 bases (MINLEN:30). All the reads were then reverse complemented using seqtk v1.2-r102-dirty11 before mapping with STAR v020201 (Dobin et al., 2013). The index was built upon the TAIR10 genome and annotation (Lamesch et al., 2012) with the following modifications: (1) The concatenated rps12 gene (Aryamanesh et al., 2017), namely rps12A-intron1a-intron1b-rps12B with 60 bp extra at each end, was appended to the end of the chloroplast genome (ChrC:154479–156997); (2) The coordinates of concatenated rps12 intron 1 and intron 2 are set as 154653–156142 and 156375–156911, respectively; (3) The coordinates of ycf3 intron 1 is shifted one nucleotide downstream to 43753–44466. The reads were aligned with the following parameters: –outFilterMismatchNmax 4, –outSAMprimaryFlag AllBestScore, –alignIntronMax 1 and –outSAMtype BAM SortedByCoordinate. The alignments of the highest scores were selected (view –bF 0x100) and indexed with samtools v1.3.1 (Li et al., 2009).
For the editing analysis, pileup files were generated using pysamstats v1.0.112 using the parameters –d, -D 100,000,000 and –type variation_strand and filtered as followed: (1) the nucleotide is encoded as C on the examined strand of the Arabidopsis chloroplast genome; and (2) the number of putatively edited reads (containing a T instead of a C at the site) was greater than 10, and the proportion of putatively edited reads was greater than 1%. Potential editing events induced by EMB2261 expression were looked for as follows: (1) the editing event was detected in all three samples of 35S:EMB2261; (2) the ‘edited’ reads were not simply mis-aligned, especially where ‘edited’ position is in the sequence context (T)nC(T)n on the forward strand or (A)nG(A)n on the reverse strand; and (3) the proportion of edited reads followed the pattern of ABI3:EMB2261 = < Col-0 < 35S:EMB2261.
For the splicing analysis, the splicing function of the ChloroSeq package (Castandet et al., 2016) was used with the same adjustments as described above made to the annotation files. For the gene expression analysis, the count for each gene was obtained using featureCounts v1.5.3 (Liao et al., 2014). Ribosomal RNA genes, tRNA genes, the non-concatenated rps12 exons and one copy of the inverted repeat region were excluded. The RPKM (Reads Per Kilobase of transcript per Million mapped reads) values were calculated using the formula , where C is a constant of 109, Ri is the number of reads per gene of interest, Ti is the total number of reads mapped to the gene set of interest, and L is the length of the gene of interest.
Evolution Analyses
The rps14 sequences were extracted from the chloroplast genomes deposited in NCBI Genbank (Supplementary Table S2). The orthologs of EMB2261 were identified in the plantPPR database (Cheng et al., 2016) by BLAST (v2.2.29+) search (Camacho et al., 2009). The PPR protein sequences from species where no genome sequence is available were retrieved by BLAST from NCBI Genbank13. We selected a single best sequence for each species when multiple matches to Arabidopsis EMB2261 were reported (Supplementary Table S2).
The alignment was carried out using the MAFFT v.7 online server (Katoh et al., 2017) and trimmed with trimAl using default parameters (Capella-Gutierrez et al., 2009). A phylogenetic tree was inferred by maximum likelihood with Amborella trichopoda EMB2261 as an outgroup, using IQ-Tree-omp v1.5.5 (Nguyen et al., 2015). The optimal evolutionary model (JTT + F + I + G4) was selected by ModelFinder (Kalyaanamoorthy et al., 2017). Branching support was estimated from 100 standard non-parametric bootstrap replicates.
The consensus sequence logos of the PPR binding sites and PPR motifs were generated by Weblogo14. To compare the conservation of the 12th PPR motif in different families of monocots, alignments were generated using the MAFFT v.7 online server and then submitted to the EMBOSS plot conservation tool15.
Results
PPR Editing Factor EMB2261 Is Predicted to Edit rps14-2 in Arabidopsis Chloroplasts
EMB2261 is located in chloroplasts in Arabidopsis (Tanz et al., 2013). The rps14-2 site in Arabidopsis chloroplasts is predicted to be edited by EMB2261 (encoded by AT3G49170) according to the PPR-RNA code (Barkan et al., 2012). As shown in Figure 1A, EMB2261 scores the highest among all Arabidopsis editing factors against the rps14-2 site. Moreover, as shown in Figure 1B, rps14-2 scores the highest among all major editing sites in Arabidopsis chloroplasts against EMB2261. EMB2261 motifs align with the rps14-2 editing site (Figure 1C).
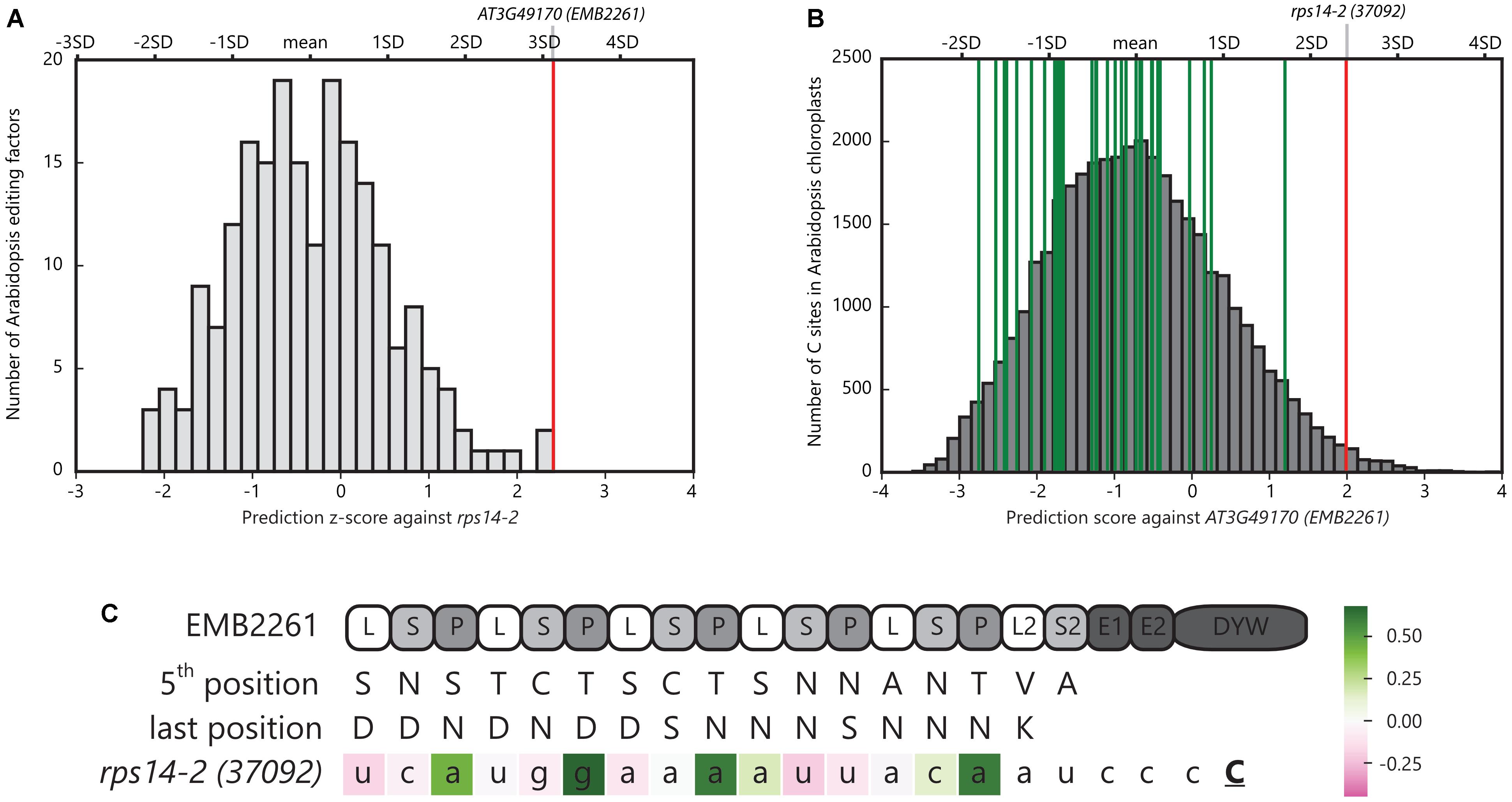
FIGURE 1. Pentatricopeptide repeat (PPR) editing factor EMB2261 is predicted to edit rps14-2 in Arabidopsis chloroplasts. (A) Histogram of z-scores of the Arabidopsis rps14-2 editing site aligned against all putative Arabidopsis PPR editing factors (n = 204). The z-scores were calculated as the number of standard deviations the rps14-2 alignment score was from the mean prediction score of all possible alignments in the chloroplast genome for that editing factor. The red line indicates the highest scoring alignment against rps14-2, representing AT3G49170 (EMB2261). (B) Distribution of alignment scores of potential editing sites (YC) across the Arabidopsis chloroplast genome (n = 37,888) against AT3G49170 (EMB2261). The red line indicates the rps14-2 alignment; green lines indicate the alignments to the other 33 major chloroplast editing sites. (C) Alignment of EMB2261 motifs with the rps14-2 editing site in Arabidopsis chloroplasts. C indicates the edited cytidine. The nucleotides are colored according to the alignment scores (Supplementary Table S1). The P1, P2, L1, and S1 motifs are labeled as P, L, and S motifs for simplicity.
Editing of rps14-2 Correlates With EMB2261 Expression and Is Essential for Arabidopsis Seed Development
Rps14-2 editing changes the 51st codon of the rps14 transcript from CCA, encoding proline (P), to CUA, encoding leucine (L). As shown in Figure 2A, the genomic rps14 sequences from E. coli, as well as examples of other species that lack RNA editing, encode L51 instead of P51, implying that L51 is important to Rps14 function. According to the structure reported for the chloroplast 70S ribosome (Bieri et al., 2017), L51 is in a loop that is in close contact with the ribosomal RNA. Proline is a poor substitute for flexible amino acids (e.g., leucine) in protein structures, and the L51P mutation is likely to change Rps14 structure and function.
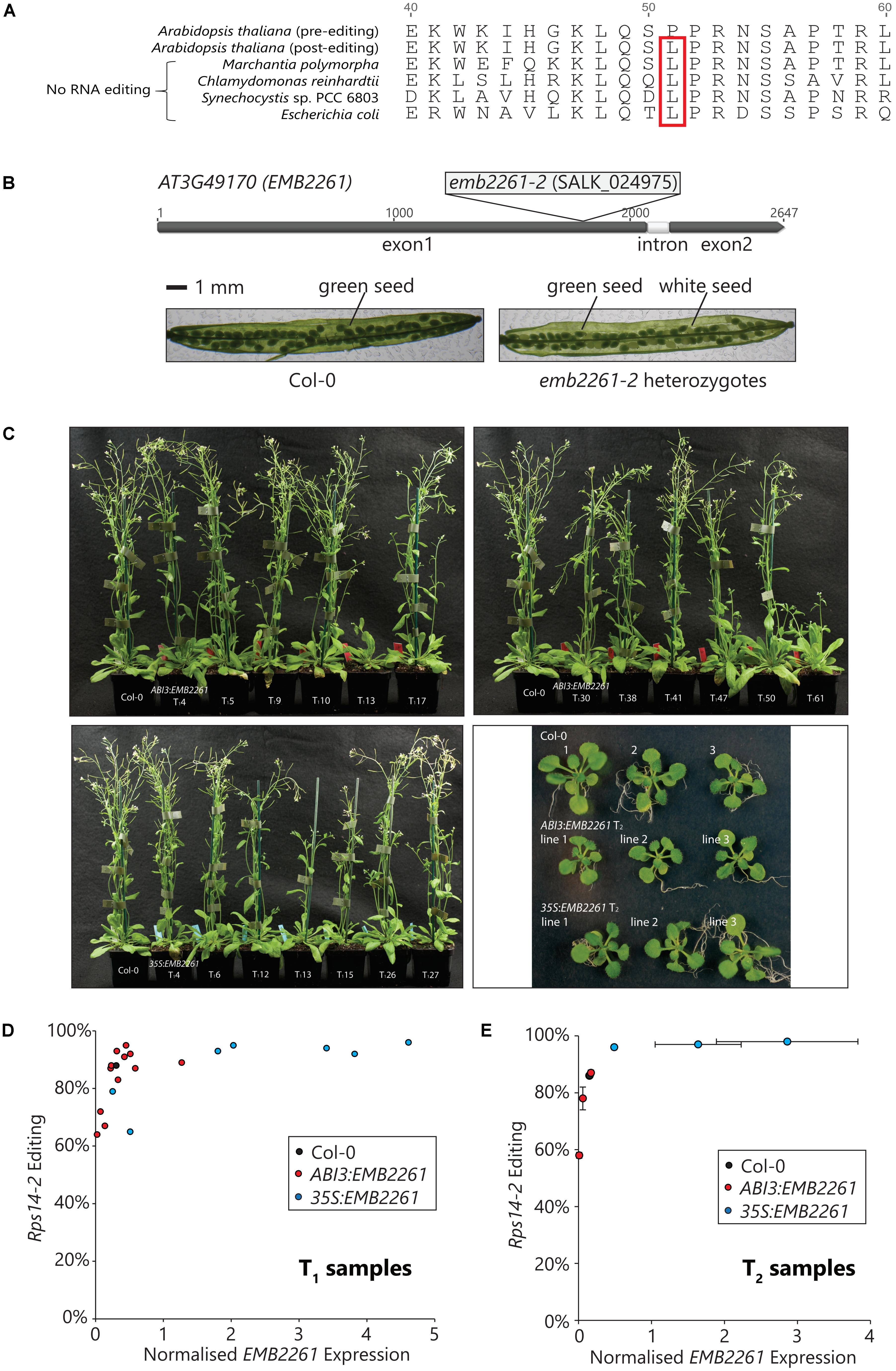
FIGURE 2. Editing of rps14-2 correlates with EMB2261 expression and is essential for Arabidopsis seed development. (A) Conservation of the edited rps14-2 codon in species lacking RNA editing. Rps14-2 editing converts the second position of the 51st codon in Rps14 from C to U, changing the amino acid P51 to L51. L51 is encoded in the rps14 genomic sequence from species lacking RNA editing, such as Marchantia polymorpha, Chlamydomonas reinhardtii, Synechocystis sp. PCC 6803, and Escherichia coli. (B) Embryo-lethality of the emb2261-2 mutation. The SALK_024975 mutant line carries a T-DNA insertion in the first exon of the EMB2261 (AT3G49170) gene. Seeds of heterozygous EMB2261-2/emb2261-2 plants segregate into two phenotypes, green and white, at a ratio of 3 to 1. No homozygous mutant plants can be recovered. (C) Complementation of the emb2261-2 mutation with ABI3:EMB2261 and 35S:EMB2261. Mature plants of primary transformants (T1) expressing ABI3:EMB2261 or 35S:EMB2261 in a homozygous emb2261-2 mutant background, in comparison with wild type Col-0; and seedlings (18-day-old) of three independent transgenic lines (T2) expressing ABI3:EMB2261 or 35S:EMB2261 in a homozygous emb2261-2 mutant background, in comparison with wild type Col-0. ABI3:EMB2261 lines 1, 2, and 3 were derived from the primary transformants T113, T141, and T161, respectively. 35S:EMB2261 lines 1, 2, and 3 were derived from the primary transformants T112, T115, and T126, respectively. (D) Correlation between rps14-2 editing and EMB2261 expression in T1 transgenic plants. From the flowering tissue of each T1 plant shown in (C), rps14-2 editing was quantified by poisoned primer extension (PPE), and EMB2261 expression was quantified by RT-qPCR normalized to the expression of the reference gene CACS (AT5G46630). Rps14-2 editing is plotted against EMB2261 expression. Each data point represents an individual transgenic line. (E) Correlation between rps14-2 editing and EMB2261 expression in T2 transgenic plants. From the whole seedling (T2) tissue shown in (c), rps14-2 editing was quantified by PPE, and EMB2261 expression was quantified by RT-qPCR normalized to the expression of the reference gene CACS (AT5G46630). Rps14-2 editing is plotted against EMB2261 expression. Each data point represents an individual transgenic line. Both horizontal and vertical error bars show SE, n = 3.
Consistent with the previous characterization of an emb2261 mutation (Cushing et al., 2005), the T-DNA insertion in the line SALK_024975 (emb2261-2) is embryo-lethal. As shown in Figure 2B, the T-DNA insertion was mapped directly after the nucleotide 1809 of the EMB2261 gene, within the region encoding the L2 motif. Siliques of the heterozygous EMB2261-2/emb2261-2 plants contain three quarter green seeds and one quarter white seeds. No homozygous plants (emb2261-2/emb2261-2) could be recovered.
To investigate the relationship between EMB2261 and rps14-2 editing, emb2261-2 heterozygotes were transformed with either the ABI3:EMB2261 or the 35S:EMB2261 construct. It was expected that the ABI3:EMB2261 primary transformants would segregate into two phenotypes. Plants that carry the transgene in wild type or heterozygous mutant background would look like wild type, and plants that carry the transgene in homozygous mutant background would show a strong chloroplast-deficient phenotype as the ABI3 promoter activity fades away. However, all plants resembled the wild type (Figure 2C). Subsequent genotyping revealed that around 1/5 (12 out of 62) seedlings carry the ABI3:EMB2261 transgene in a homozygous mutant background. These results indicate that both ABI3:EMB2261 and 35S:EMB2261 complement the embryo-lethal phenotype of emb2261-2, i.e., that the residual ABI3 promoter activity beyond the seed stage drives sufficient expression of EMB2261 for normal embryo and seedling development. Seven 35S:EMB2261 lines in the homozygous mutant background were isolated.
We then sought to check whether there were any subtle rps14-2 editing defects in the ABI3:EMB2261 lines. All the primary transformants were screened for EMB2261 gene expression and rps14-2 editing, in comparison with wild type Col-0. Flower tissues were harvested from the primary transformants and Col-0 for RNA analyses (Figure 2C). EMB2261 expression was quantified by qPCR (Supplementary Figure S1A), and Rps14-2 editing was quantified by PPE (Supplementary Figure S1B). As shown in Figure 2D, the proportion of rps14-2 editing was plotted against the normalized EMB2261 expression value. Rps14-2 editing correlates with EMB2261 expression in the primary transformants. In general, ABI3:EMB2261 lines show decreased or comparable EMB2261 expression level to wild type, correlating with lower or comparable rps14-2 editing level. 35S:EMB2261 over-expression lines show increased EMB2261 expression and almost 100% rps14-2 editing.
To confirm the correlation between rps14-2 editing and EMB2261 expression, the analysis was repeated in the second generation (T2) of the transgenic lines. Three ABI3:EMB2261 lines that showed lower than wild-type levels of EMB2261 expression and three 35S:EMB2261 lines that showed higher than wild-type levels of EMB2261 expression were selected, and whole 18-day-old seedlings were harvested for RNA analyses (Figure 2C). In Figure 2E, the proportion of rps14-2 editing (Supplementary Figure S1D) is plotted against the normalized EMB2261 expression value (Supplementary Figure S1C). The correlation between rps14-2 editing and EMB2261 gene expression is maintained in the T2 generation. Taking the above results together, we conclude that EMB2261 is an editing specificity factor for the rps14-2 site.
Rps14-2 Is the Only Chloroplast Editing Event That Positively Correlates With EMB2261 Expression
If EMB2261 edits other sites, their editing level should also correlate with EMB2261 gene expression, following a similar pattern to editing of the rps14-2 site. We quantified the editing level at all known chloroplast editing sites in ABI3:EMB2261, wild type Col-0, and 35S:EMB2261 by RNA-seq.
The transgenic line showing the lowest EMB2261 expression level was chosen for ABI3:EMB2261, and the transgenic line showing the highest EMB2261 expression level was chosen for 35S:EMB2261. A separate batch of 18-day-old T2 seedlings was obtained, including three seedlings as three biological replicates for each of ABI3:EMB2261, 35S:EMB2261, and wild type Col-0. Prior to RNA-seq library preparation, RNA quality was checked with an Agilent Screentape. There is no difference between the genotypes in terms of ribosomal RNA accumulation (Figure 3A), indicating that ribosome assembly in ABI3:EMB2261 is not greatly different from wild type, despite reduced editing of rps14-2. This observation is consistent with the lack of visible growth phenotypes in ABI3:EMB2261.
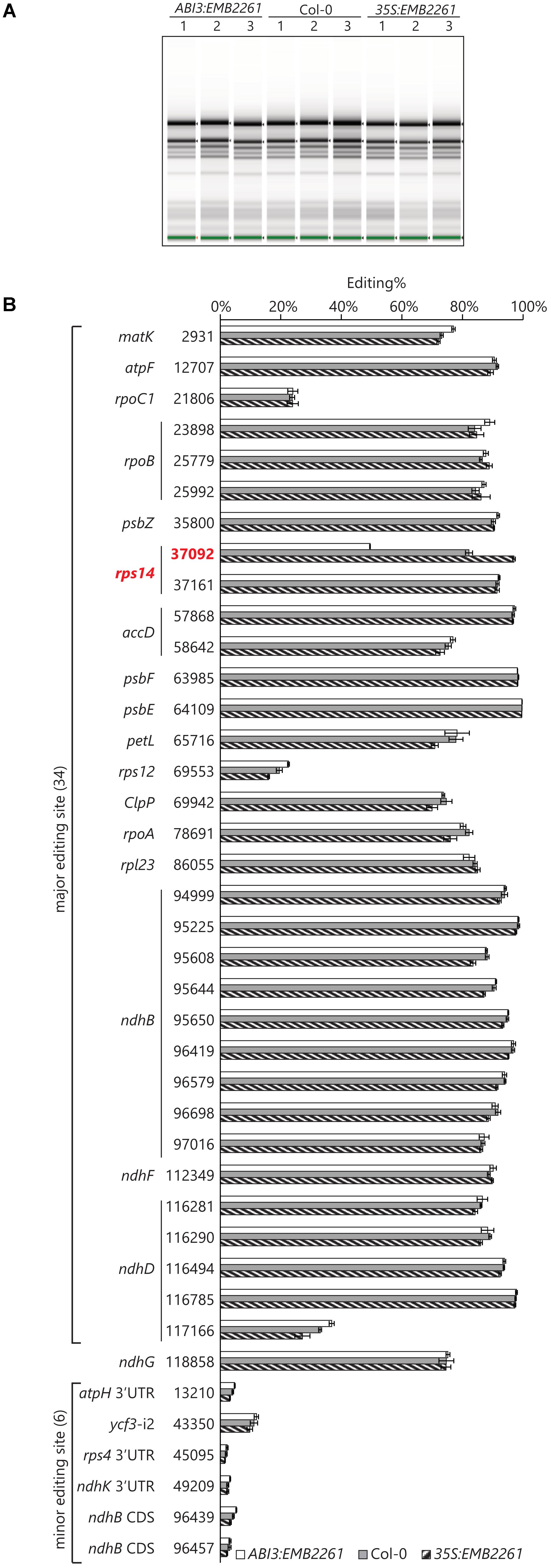
FIGURE 3. Rps14-2 is the only known chloroplast editing event that positively correlates with EMB2261 expression. (A) Screentape image of RNA samples extracted from ABI3:EMB2261 line 1 and 35S:EMB2261 line 2, in comparison with wild type Col-0. Each lane represents one RNA sample extracted from a single 18-day-old seedling (T2). (B) RNA editing quantified at known chloroplast editing sites in ABI3:EMB2261 and 35S:EMB2261, in comparison with wild type Col-0. Error bars show SE, n = 3.
About 33 million reads were obtained for each sample, roughly 60% of which aligned to the Arabidopsis chloroplast genome. Editing at known chloroplast editing sites were quantified as (number of T reads)/(number of T reads + number of C reads)%. Only rps14-2 editing positively correlates with EMB2261 expression (Figure 3B). Some major editing events [e.g., rps12(69553)] may correlate negatively with EMB2261 expression, although to a lesser extent than the positive correlation with rps14-2. In addition, six out of seven previously reported minor editing events (Bentolila et al., 2013; Ruwe et al., 2013) were detected in all nine samples, none of which correlate with EMB2261 expression (Figure 3B). After examining all possible editing events across the chloroplast transcriptome, not limited to the known chloroplast editing sites, we found no novel editing events that were induced by EMB2261 overexpression (Supplementary Table S3). Notably, potential editing sites that match EMB2261 motifs better than rps14-2 were confirmed not to be edited (Supplementary Figure S2). Lack of induced editing events upon overexpression indicates that EMB2261 is a highly specific editing factor in Arabidopsis chloroplasts. In addition, we also quantified chloroplast splicing efficiency (Supplementary Figure S3) and gene expression (Supplementary Figure S4) based on the RNA-seq data. We found no transcripts or processing events besides rps14-2 editing that strongly depended on EMB2261 expression.
Both EMB2261 and the Recognition Sequence Are Subject to Divergence in Species Where rps14-2 Editing Is no Longer Needed
The rps14-2 site has been lost three times during evolution in Solanaceae, Fabaceae and Poaceae, respectively (Figure 4A), where T instead of C is present in the chloroplast genomes. In these species, the cis-element immediately upstream of the rps14-2 editing site is less conserved than when editing is required. As shown in Figure 4B, positions -7, -15, -17, -18, and -20 show variation from the consensus once rps14-2 no longer needs to be edited. This nucleotide variation introduces variation in the corresponding amino acid sequence (Figure 4C), apparently without affecting Rps14 function.
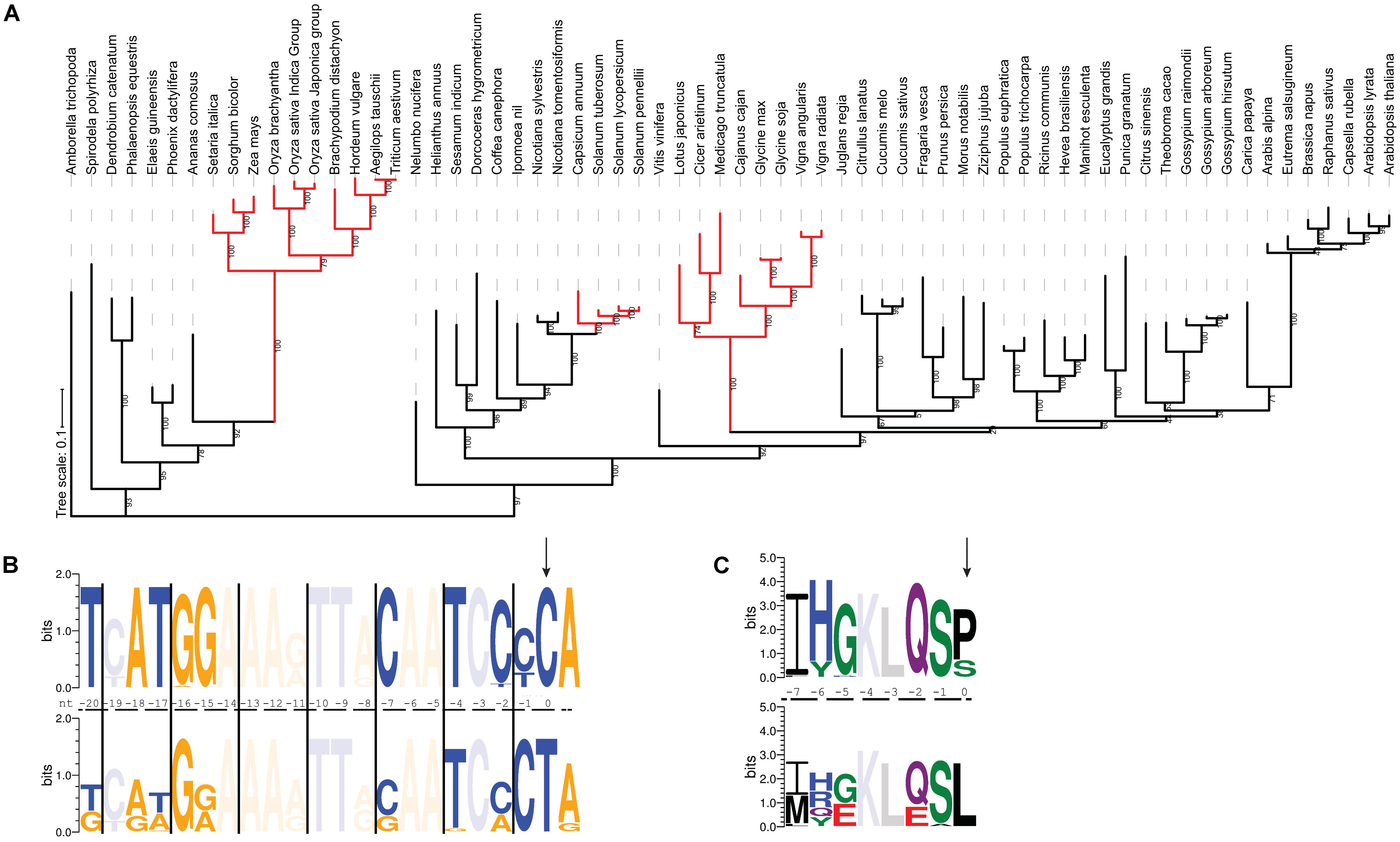
FIGURE 4. The RNA recognition sequence diverges in species where rps14-2 editing is no longer needed. (A) Maximum likelihood tree of EMB2261 orthologs from 66 species (Supplementary Table S2). Branches where rps14-2 editing is no longer needed are highlighted in red. (B) Conservation of cis-elements immediately upstream of the rps14-2 editing site. The top panel shows the sequence logo generated from rps14 sequences from 43 plant species where the rps14-2 site requires editing. The bottom panel shows the sequence logo generated from rps14 sequences from 23 plant species where the equivalent position is a T in the chloroplast genome. The rps14-2 editing site is highlighted by the arrow. Purines are indicated in orange and pyrimidines in blue. Positions where there are no new nucleotides introduced in either group are faded. (C) Conservation of the cis-elements shown in (B) translated to amino acids. The rps14-2 editing site is highlighted by the arrow. Positions where there are no new amino acids introduced in either group are faded.
Putative EMB2261 orthologs are still present in all three families. The C-terminal domains including the DYW domain are conserved (Supplementary Figure S5A). Despite the loss of rp14-2 editing site in Solanoideae (Solanum and Capsicum), a subfamily of Solanaceae, the protein sequence of putative EMB2261 orthologs remain conserved (Supplementary Table S4). EMB2261 protein sequences in Fabaceae species show slightly more variation compared to those in other dicot species (Supplementary Table S4). EMB2261 proteins in Poaceae species show more dramatic sequence variation compared to those in other monocot species, especially in one truncated P1 motif (Supplementary Figures S5B,C). Thus, the absence of rps14-2 editing can be associated with sequence divergence in EMB2261.
Discussion
With a different genetic approach based on a different mutant allele of the EMB2261 gene, we confirm the conclusion of Jiang et al. (2018), claiming that EMB2261/ECD1 is the editing specificity factor of rps14-2 in Arabidopsis chloroplasts and is essential for Arabidopsis development. Here, we discuss the implication of this finding from four aspects: (1) editing factor mutants as surrogate mutants of the corresponding genes; (2) PPR specificity factors as limiting factors in RNA editing; (3) editing specificity determinants beyond the PPR-RNA recognition code; and (4) co-evolutionary scenarios involving PPR RNA editing factors and their targets.
Lethality of Editing Defects
RNA editing events in plant organelles mainly occur in the coding region of genes and alter the corresponding protein sequences. In mutants lacking one of the site-specificity PPR editing factors, the editing site(s) targeted by the factor remain(s) completely unedited, often leading to functional defects of the gene products encoded by the affected transcripts. Therefore, editing factor mutants appear as surrogate mutants of the corresponding organellar genes. As most chloroplast genes encode subunits of the photosynthetic machinery, it is thus not surprising that most chloroplast editing mutants show photosynthetic defects. Photosynthesis is not essential for embryogenesis, but plastid translation is essential (in most plants) due to the requirement for the plastid-encoded AccD (acetyl-coA carboxylase D) gene product, the loss of which results in embryonic lethality (Bryant et al., 2011). Thus, loss of RNA editing of plastid-encoded essential components of the translation machinery could conceivably cause embryonic lethality. Rps14 is known to be an essential ribosomal subunit in tobacco chloroplasts (Ahlert et al., 2003) and in E. coli (Shoji et al., 2011). We contend that this is the most likely explanation for the emb2261 phenotype: EMB2261 is required for editing at the rps14-2 site, which in turn is required for synthesis of functional Rps14, which in turn is required for plastid translation.
EMB2261 Is a Limiting Factor for rps14-2 Editing
Partial complementation of emb2261-2 by ABI3:EMB2261 did not work as we expected. Comparing the expression profiles (Schmid et al., 2005) of ABI3, EMB2261, and the other successfully partially complemented EMB genes (Despres et al., 2001; Aryamanesh et al., 2017), we found that the expression level of the other EMB genes are 10–100 times higher than ABI3 beyond the seed stage, whereas the expression level of EMB2261 is of the same order of magnitude as ABI3 (Supplementary Figure S6). Therefore, the residual EMB2261 expression driven by ABI3 promoter beyond the seed stage is likely to have been sufficient to complement the emb2261 mutant phenotype. Since ABI3 is considered to be a seed-specific gene (Cushing et al., 2005), the required EMB2261 expression level must be very low. That rps14-2 editing is increased upon EMB2261 overexpression indicates that it is not saturated and that a limiting factor is the expression of EMB2261.
Unlike EMB2261, some PPR specificity factors lack the essential C-terminal editing domains that have to be supplied in trans (Andres-Colas et al., 2017; Diaz et al., 2017; Guillaumot et al., 2017). In these cases, the PPR specificity factor itself may not be the limiting factor for editing. For example, CRR4 lacks the DYW domain, which is complemented by the DYW1 protein in trans (Boussardon et al., 2012). The editing of ndhD-1 by CRR4 is not correlated with CRR4 expression (Kotera et al., 2005), but can be boosted by overexpression of a CRR4-DYW1 fusion (Boussardon et al., 2012).
Editing Specificity
The specificity of PPR editing factors is predominantly determined by the interaction between the fifth and last position of a PPR motif and the aligned RNA base. The one-motif to one-base recognition code forms the basis of editing site prediction methods (Barkan et al., 2012; Takenaka et al., 2013a; Yagi et al., 2013a). However, if these are the only determinants of RNA editing specificity, it is surprising that there are not more editing events in chloroplasts than are observed. For example, there are hundreds of potential editing sites in Arabidopsis chloroplasts that match the EMB2261 PPR motifs equally well or better than rps14-2 (Supplementary Figure S2), yet only one editing event was unambiguously detected in this work. The lack of off-target editing events implies that there are additional factors other than the canonical PPR-RNA code controlling the editing specificity of EMB2261.
Pentatricopeptide repeat motifs may contribute differently to RNA recognition. Although the predicted candidate sites shown in Supplementary Figure S2 score higher than rps14-2 based on the PPR-RNA code, they show different distributions of mis-matches across the PPR-RNA alignments compared to rps14-2. It may be that EMB2261 motifs matching rps14-2 play more important roles in target recognition, and the potential editing sites containing mismatches to these motifs are not likely to be recognized. There may also be non-canonical, yet sequence-specific, interactions between PPR motifs and target RNA that are not currently taken into account. In the case of EMB2261, the PPR motifs aligned to positions -16, -13, and -8 contain non-canonical combinations of amino acids, the selectivity of which is poorly understood due to lack of prior examples.
Not all predicted sites may be expressed or accessible to PPR editing factors in planta, where RNA forms secondary structures or is bound by other proteins. RNA secondary structure prevents PPR binding in vitro (Kindgren et al., 2015; Miranda et al., 2018), implying that it would inhibit RNA editing in planta. RNA structures and protein-RNA interactions can be partially modulated by RNA chaperones and helicases. For example, knock-down of chloroplast RNA helicase ISE2 leads to specific editing defects at 12 sites, including rps14-2 (Bobik et al., 2017). Therefore, rps14-2 editing by EMB2261 may also require ISE2, and lack of rps14-2 editing in ise2 null mutants may be one of the reasons that the mutation is lethal. There is also an association shown by RNA immunoprecipitation sequencing (RIP-seq) between ISE2 and rps14 transcripts as well as other edited chloroplast transcripts. Taken together, these observations imply that the chloroplast RNA helicase ISE2 may be required to remodel RNA structures and/or protein-rps14 interactions near the rps14-2 site.
Co-evolution Between Editing Factors and Editing Sites
Co-evolution between PPR editing factors and their target sites has been demonstrated in two scenarios. First, PPR editing factors and editing sites tend to be gained in parallel. For example, at the current limits of phylogenetic resolution, the mitochondrial PPR editing factor PPR_56 in Physcomitrella patens appeared simultaneously with its two editing sites on nad3 and nad4 (Beike et al., 2014). Second, PPR editing factors and editing sites tend to also be lost in parallel. For example, loss of the chloroplast editing factors CRR28 and RARE1 coincide with loss of their corresponding editing sites (Hein et al., 2016). Editing factors targeting multiple sites tend to be retained as long as a subset of their targeting sites remains (Hein et al., 2016). EMB2261 may have more than one target site in species other than Arabidopsis, which would explain the conservation of EMB2261 despite the loss of the rps14-2 editing event. The patterns of sequence variation in the cis-element immediately upstream of rps14-2 suggest that it is conserved in species that edit this site due to the requirement for RNA recognition rather than because of protein sequence conservation. When editing is no longer needed, the cis-element of the editing site is free to diverge to the extent that the corresponding amino acid changes can be functionally tolerated. Co-evolution of editing factors and their binding sites may be a powerful way of examining sequence recognition by PPR proteins, where a sufficient number of independent examples of editing loss and subsequent sequence divergence can be compared.
Author Contributions
YS, BG, AY, PK, and IS designed the research and wrote the manuscript. YS performed the experiments. YS, BG, and IS collected, analyzed, and interpreted the data.
Funding
The work was supported by grants from the Australian Research Council to IS (CE140100008 and FL140100179) and to BG (DE150101484). YS and AY were recipients of a Research Training Program Scholarship from the Australian Government.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00841/full#supplementary-material
Footnotes
- ^ www.matplotlib.org
- ^ seaborn.pydata.org
- ^ www.clontech.com
- ^ www.qiagen.com
- ^ www.thermofisher.com/au/en/home/brands/invitrogen
- ^ www.bio-rad.com
- ^ www.thermofisher.com/au/en/home/brands/invitrogen/ambion
- ^ www.molecular.roche.com
- ^ www.illumina.com
- ^ www.kapabiosystems.com
- ^ github.com/lh3/seqtk
- ^ github.com/alimanfoo/pysamstats
- ^ http://www.ncbi.nlm.nih.gov/blast/Blast.cgi
- ^ http://weblogo.berkeley.edu
- ^ http://emboss.bioinformatics.nl/cgi-bin/emboss/plotcon
References
Ahlert, D., Ruf, S., and Bock, R. (2003). Plastid protein synthesis is required for plant development in tobacco. Proc. Natl. Acad. Sci. U.S.A. 100, 15730–15735. doi: 10.1073/pnas.2533668100
Andres-Colas, N., Zhu, Q., Takenaka, M., De Rybel, B., Weijers, D., and Van Der Straeten, D. (2017). Multiple PPR protein interactions are involved in the RNA editing system in Arabidopsis mitochondria and plastids. Proc. Natl. Acad. Sci. U.S.A. 114, 8883–8888. doi: 10.1073/pnas.1705815114
Aryamanesh, N., Ruwe, H., Sanglard, L. V. P., Eshraghi, L., Bussell, J. D., Howell, K. A., et al. (2017). The pentatricopeptide repeat protein EMB2654 is essential for trans-splicing of a chloroplast small ribosomal subunit transcript. Plant Physiol. 173, 1164–1176. doi: 10.1104/pp.16.01840
Asakura, Y., and Barkan, A. (2006). Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 142, 1656–1663. doi: 10.1104/pp.106.088096
Barkan, A., Rojas, M., Fujii, S., Yap, A., Chong, Y. S., Bond, C. S., et al. (2012). A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8:e1002910. doi: 10.1371/journal.pgen.1002910
Barkan, A., and Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Beike, A. K., Von Stackelberg, M., Schallenberg-Rudinger, M., Hanke, S. T., Follo, M., Quandt, D., et al. (2014). Molecular evidence for convergent evolution and allopolyploid speciation within the Physcomitrium-Physcomitrella species complex. BMC Evol. Biol. 14:158. doi: 10.1186/1471-2148-14-158
Bentolila, S., Oh, J., Hanson, M. R., and Bukowski, R. (2013). Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 9:e1003584. doi: 10.1371/journal.pgen.1003584
Bieri, P., Leibundgut, M., Saurer, M., Boehringer, D., and Ban, N. (2017). The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 36, 475–486. doi: 10.15252/embj.201695959
Bobik, K., Mccray, T. N., Ernest, B., Fernandez, J. C., Howell, K. A., Lane, T., et al. (2017). The chloroplast RNA helicase ISE2 is required for multiple chloroplast RNA processing steps in Arabidopsis thaliana. Plant J. 91, 114–131. doi: 10.1111/tpj.13550
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boussardon, C., Salone, V., Avon, A., Berthome, R., Hammani, K., Okuda, K., et al. (2012). Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell 24, 3684–3694. doi: 10.1105/tpc.112.099507
Bryant, N., Lloyd, J., Sweeney, C., Myouga, F., and Meinke, D. (2011). Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol. 155, 1678–1689. doi: 10.1104/pp.110.168120
Cai, W., Ji, D., Peng, L., Guo, J., Ma, J., Zou, M., et al. (2009). LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiol. 150, 1260–1271. doi: 10.1104/pp.109.136812
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST + : architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Castandet, B., Hotto, A. M., Strickler, S. R., and Stern, D. B. (2016). ChloroSeq, an optimized chloroplast RNA-seq bioinformatic pipeline, reveals remodeling of the organellar transcriptome under heat stress. G3 (Bethesda) 6, 2817–2827. doi: 10.1534/g3.116.030783
Chateigner-Boutin, A. L., Ramos-Vega, M., Guevara-Garcia, A., Andres, C., De La Luz Gutierrez-Nava, M., Cantero, A., et al. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56, 590–602. doi: 10.1111/j.1365-313X.2008.03634.x
Chateigner-Boutin, A. L., and Small, I. (2007). A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 35:e114. doi: 10.1093/nar/gkm640
Cheng, S. F., Gutmann, B., Zhong, X., Ye, Y. T., Fisher, M. F., Bai, F. Q., et al. (2016). Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547. doi: 10.1111/tpj.13121
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cushing, D. A., Forsthoefel, N. R., Gestaut, D. R., and Vernon, D. M. (2005). Arabidopsis emb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221, 424–436. doi: 10.1007/s00425-004-1452-x
Despres, B., Delseny, M., and Devic, M. (2001). Partial complementation of embryo defective mutations: a general strategy to elucidate gene function. Plant J. 27, 149–159. doi: 10.1046/j.1365-313x.2001.01078.x
Diaz, M. F., Bentolila, S., Hayes, M. L., Hanson, M. R., and Mulligan, R. M. (2017). A protein with an unusually short PPR domain, MEF8, affects editing at over 60 Arabidopsis mitochondrial C targets of RNA editing. Plant J. 92, 638–649. doi: 10.1111/tpj.13709
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Giege, P., and Brennicke, A. (1999). RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. U.S.A. 96, 15324–15329. doi: 10.1073/pnas.96.26.15324
Guillaumot, D., Lopez-Obando, M., Baudry, K., Avon, A., Rigaill, G., Falcon de Longevialle, A., et al. (2017). Two interacting PPR proteins are major Arabidopsis editing factors in plastid and mitochondria. Proc. Natl. Acad. Sci. U.S.A. 114, 8877–8882. doi: 10.1073/pnas.1705780114
Hammani, K., Okuda, K., Tanz, S. K., Chateigner-Boutin, A. L., Shikanai, T., and Small, I. (2009). A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell 21, 3686–3699. doi: 10.1105/tpc.109.071472
Hayes, M. L., Giang, K., Berhane, B., and Mulligan, R. M. (2013). Identification of two pentatricopeptide repeat genes required for RNA editing and zinc binding by C-terminal cytidine deaminase-like domains. J. Biol. Chem. 288, 36519–36529. doi: 10.1074/jbc.M113.485755
Hein, A., Polsakiewicz, M., and Knoop, V. (2016). Frequent chloroplast RNA editing in early-branching flowering plants: pilot studies on angiosperm-wide coexistence of editing sites and their nuclear specificity factors. BMC Evol. Biol. 16:23. doi: 10.1186/s12862-016-0589-0
Jiang, T., Zhang, J., Rong, L., Feng, Y., Wang, Q., Song, Q., et al. (2018). ECD1 functions as an RNA editing trans-factor of rps14-149 in plastids and is required for early chloroplast development in seedlings. J. Exp. Bot. 69, 3037–3051. doi: 10.1093/jxb/ery139
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., Rozewicki, J., and Yamada, K. D. (2017). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. doi: 10.1093/bib/bbx108 [Epub ahead of print].
Kindgren, P., Yap, A., Bond, C. S., and Small, I. (2015). Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. Plant Cell 27, 403–416. doi: 10.1105/tpc.114.134189
Kotera, E., Tasaka, M., and Shikanai, T. (2005). A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330. doi: 10.1038/nature03229
Lamesch, P., Berardini, T. Z., Li, D. H., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Meinke, D., Muralla, R., Sweeney, C., and Dickerman, A. (2008). Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci. 13, 483–491. doi: 10.1016/j.tplants.2008.06.003
Miranda, R. G., Mcdermott, J. J., and Barkan, A. (2018). RNA-binding specificity landscapes of designer pentatricopeptide repeat proteins elucidate principles of PPR-RNA interactions. Nucleic Acids Res. 46, 2613–2623. doi: 10.1093/nar/gkx1288
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Okuda, K., Chateigner-Boutin, A. L., Nakamura, T., Delannoy, E., Sugita, M., Myouga, F., et al. (2009). Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21, 146–156. doi: 10.1105/tpc.108.064667
Okuda, K., Myouga, F., Motohashi, R., Shinozaki, K., and Shikanai, T. (2007). Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. U.S.A. 104, 8178–8183. doi: 10.1073/pnas.0700865104
Robbins, J. C., Heller, W. P., and Hanson, M. R. (2009). A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA 15, 1142–1153. doi: 10.1261/rna.1533909
Ruwe, H., Castandet, B., Schmitz-Linneweber, C., and Stern, D. B. (2013). Arabidopsis chloroplast quantitative editotype. FEBS Lett. 587, 1429–1433. doi: 10.1016/j.febslet.2013.03.022
Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M., et al. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506. doi: 10.1038/ng1543
Shen, C., Zhang, D., Guan, Z., Liu, Y., Yang, Z., Yang, Y., et al. (2016). Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 7:11285. doi: 10.1038/ncomms11285
Shoji, S., Dambacher, C. M., Shajani, Z., Williamson, J. R., and Schultz, P. G. (2011). Systematic chromosomal deletion of bacterial ribosomal protein genes. J. Mol. Biol. 413, 751–761. doi: 10.1016/j.jmb.2011.09.004
Sun, T., Bentolila, S., and Hanson, M. R. (2016). The unexpected diversity of plant organelle RNA editosomes. Trends Plant Sci. 21, 962–973. doi: 10.1016/j.tplants.2016.07.005
Takenaka, M., Zehrmann, A., Brennicke, A., and Graichen, K. (2013a). Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One 8:e65343. doi: 10.1371/journal.pone.0065343
Takenaka, M., Zehrmann, A., Verbitskiy, D., Hartel, B., and Brennicke, A. (2013b). RNA editing in plants and its evolution. Annu. Rev. Genet. 47, 335–352. doi: 10.1146/annurev-genet-111212-133519
Tanz, S. K., Castleden, I., Small, I. D., and Millar, A. H. (2013). Fluorescent protein tagging as a tool to define the subcellular distribution of proteins in plants. Front. Plant Sci. 4:214. doi: 10.3389/fpls.2013.00214
Tillich, M., Funk, H. T., Schmitz-Linneweber, C., Poltnigg, P., Sabater, B., Martin, M., et al. (2005). Editing of plastid RNA in Arabidopsis thaliana ecotypes. Plant J. 43, 708–715. doi: 10.1111/j.1365-313X.2005.02484.x
Tsugeki, R., Kochieva, E. Z., and Fedoroff, N. V. (1996). A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 10, 479–489. doi: 10.1046/j.1365-313X.1996.10030479.x
Wagoner, J. A., Sun, T., Lin, L., and Hanson, M. R. (2015). Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J. Biol. Chem. 290, 2957–2968. doi: 10.1074/jbc.M114.622084
Wang, G., Zhong, M., Shuai, B., Song, J., Zhang, J., Han, L., et al. (2017). E + subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol. 214, 1563–1578. doi: 10.1111/nph.14507
Yagi, Y., Hayashi, S., Kobayashi, K., Hirayama, T., and Nakamura, T. (2013a). Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One 8:e57286. doi: 10.1371/journal.pone.0057286
Yagi, Y., Tachikawa, M., Noguchi, H., Satoh, S., Obokata, J., and Nakamura, T. (2013b). Pentatricopeptide repeat proteins involved in plant organellar RNA editing. RNA Biol. 10, 1419–1425. doi: 10.4161/rna.24908
Yang, Y. Z., Ding, S., Wang, H. C., Sun, F., Huang, W. L., Song, S., et al. (2017). The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol. 214, 782–795. doi: 10.1111/nph.14424
Yap, A., Kindgren, P., Colas Des, Francs-Small, C., Kazama, T., Tanz, S. K., et al. (2015). AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J. 81, 661–669. doi: 10.1111/tpj.12756
Zhou, W., Cheng, Y., Yap, A., Chateigner-Boutin, A. L., Delannoy, E., Hammani, K., et al. (2009). The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 58, 82–96. doi: 10.1111/j.1365-313X.2008.03766.x
Keywords: PPR, RNA editing, editing specificity, evolution, chloroplast, seed development, Arabidopsis
Citation: Sun YK, Gutmann B, Yap A, Kindgren P and Small I (2018) Editing of Chloroplast rps14 by PPR Editing Factor EMB2261 Is Essential for Arabidopsis Development. Front. Plant Sci. 9:841. doi: 10.3389/fpls.2018.00841
Received: 27 April 2018; Accepted: 30 May 2018;
Published: 20 June 2018.
Edited by:
Stefan A. Rensing, Philipps University of Marburg, GermanyCopyright © 2018 Sun, Gutmann, Yap, Kindgren and Small. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ian Small, aWFuLnNtYWxsQHV3YS5lZHUuYXU=
 Yueming K. Sun
Yueming K. Sun Bernard Gutmann
Bernard Gutmann Aaron Yap
Aaron Yap Peter Kindgren
Peter Kindgren Ian Small
Ian Small