- 1College of Life Science and Technology, Jinan University, Jinan, China
- 2State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Tai’an, China
Plant uses multiple photoreceptors and downstream components to rapidly respond to dynamic changes in environmental light. Under shade conditions, many species exhibit shade avoidance responses that promote stem and petiole elongation, thus helping plants reach the sunlight. In the last few years, the regulatory molecular mechanisms by which plants respond to shade signals have been intensively studied. This review discusses the regulatory mechanisms underlying auxin-mediated cell elongation in the shade avoidance responses. In the early response to shade signals, auxin biosynthesis, transport, and sensitivity are all rapidly activated, thus promoting cell elongation of the hypocotyls and other organs. Under prolonged shade, increased auxin sensitivity—rather than increased auxin biosynthesis—plays a major role in cell elongation. In addition, we discuss the interaction network of photoreceptors and Phytochrome-Interacting Factors, and the antagonistic regulation of Auxin/Indole Acetic Acid proteins by auxin and light. This review provides perspectives to reframe how we think about shade responses in the natural environment.
Introduction
Light is a vital resource for green plants, as it provides an energy source for photosynthesis and acts as a signal to direct plant growth and development. In leaves at the top of the canopy, chlorophylls and other pigments specifically absorb blue (400–500 nm, B) and red (600–700 nm; R) wavelengths of light. By contrast, plant tissues transmit or reflect most far-red (700–750 nm; FR) wavelengths of light, leading to strong enrichment of FR light under the canopy. Under canopy shade, the intensity of B and R light, as well as Photosynthetically Active Radiation (400–700 nm; PAR) are reduced, whereas green and FR light intensities are relatively enriched; these dynamic changes in light quality and intensity trigger specific shade responses (Sellaro et al., 2010; Casal, 2013; Pierik and de Wit, 2014; Pedmale et al., 2016; Ballare and Pierik, 2017; Fiorucci and Fankhauser, 2017). Under shade conditions, most shade-intolerant species (e.g., Arabidopsis thaliana) exhibit shade avoidance responses, including enhanced cell elongation in various organs (hypocotyl, petioles, internode, stem, and branches), increased hyponastic growth of leaves, and accelerated flowering time (Franklin, 2008; Casal, 2012, 2013). When growing taller is not an option, some plants exhibit a series of tolerance-related morphological and physiological changes, including expanded leaf size and area, decreased leaf thickness, and reduced chlorophyll a:b ratio, thus increasing plant performance and the efficiency of light capture under shade and dim-light conditions (Gommers et al., 2013). In this review, we focus on the current understanding of auxin-mediated cell elongation under various shade conditions in Arabidopsis thaliana.
Responses to Low R:FR Shade Signals
In Arabidopsis, the photoreceptor phytochrome A (phyA) mediates the response to continuous FR light and phyB mediates the response to continuous R light (Li et al., 2011; Wang and Wang, 2015). The disruption of phyB leads to constitutive shade avoidance responses, consistent with its negative role in this response (Reed et al., 1993). Arabidopsis PHYTOCHROME-INTERACTING FACTORS (PIFs, including PIF3, 4, 5, and 7), a subfamily of basic helix-loop-helix (bHLH)-type transcription factors, physically interact with phyB and positively regulate the shade avoidance response by directly inducing the transcription of growth-promoting genes (Leivar et al., 2012a,b; Li et al., 2012). Under low R:FR conditions, PIF3 has a minor role in the shade avoidance response, PIF4 and PIF5 play redundant roles, and PIF7 plays a more prominent role in mediating shade-induced cell elongation (Hornitschek et al., 2012; Leivar et al., 2012a,b; Li et al., 2012; de Wit et al., 2015). The transcription of genes encoding other bHLH, or HLH-type transcriptional regulators, including LONG HYPOCOTYL IN FAR-RED1/SLENDER IN CANOPY SHADE1 (HFR1/SICS1), PHYTOCHROME RAPIDLY REGULATED1 (PAR1), PAR2, AND PIF3-LIKE1 (PIL1), is also rapidly induced by low R:FR shade signal. These factors negatively regulate shade avoidance responses through physically interactions with PIF4 and PIF5, forming non-DNA binding heterodimers and inhibiting transcriptional activation of PIF4 and PIF5 downstream targets (Salter et al., 2003; Roig-Villanova et al., 2007; Hornitschek et al., 2009). In addition, other well-studied components of the light signaling pathway, including the basic leucine zipper (bZIP) transcription factor ELONGATED HYPOCOTYL5 (HY5), B-Box (BBX) transcription factors (including BBX21, 24, and 25), and the E3 ligase CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1), are involved in shade responses (Crocco et al., 2010; Sellaro et al., 2011; Rolauffs et al., 2012; Pacin et al., 2016; van Gelderen et al., 2018; Ortiz-Alcaide et al., 2019).
When plants perceive low R:FR, the expression levels of thousands of genes are rapidly altered, thus allowing plants to respond to shade conditions. A recent meta-analysis of public transcriptome data of shade responses identified a set of core response genes, including 98 up-regulated and 112 down-regulated genes (Sellaro et al., 2017). These core response genes include well-known markers of the shade avoidance response, such as HOMEOBOX2 (HB2), HFR1, and IAA29. Interestingly, a large proportion of core shade-upregulated genes are the direct targets of PIF3 (∼40%), PIF4 (∼80%), PIF5 (∼50%), and AUXIN RESPONSE FACTOR6 (ARF6; ∼60%), further confirming that PIFs and ARFs play critical roles in shade responses (Sellaro et al., 2017).
Auxin Biosynthesis and Transport Are Induced During the Early Shade Response
Shade Signal Perception
In Arabidopsis, rosette leaves and cotyledons are the major sites of low R:FR shade signal perception. Within 1 h of low R:FR treatment, free indole-3-acetic acid (IAA) contents in Arabidopsis shoots increased by over 50% (Tao et al., 2008; Li et al., 2012; Kohnen et al., 2016). This newly synthesized auxin is subsequently transported out to the bases of the lamina, petiole, and hypocotyl, where locally synthesized and newly transported auxin promote cell elongation in the petiole and hypocotyl (Casal, 2013; Kohnen et al., 2016; Michaud et al., 2017; Pantazopoulou et al., 2017; Iglesias et al., 2018). Subjecting the tip of the cotyledon or leaf to low R:FR significantly induces hyponastic growth. Interestingly, this only occurs in the abaxial side of the petiole of the leaf that has perceived FR, but not in other rosette leaves (Figure 1) (Muller-Moule et al., 2016; Michaud et al., 2017; Pantazopoulou et al., 2017; Kim et al., 2018). By contrast, subjecting whole plants to low R:FR conditions induces elongation of both the abaxial and adaxial sides of the petiole (Figure 1; Michaud et al., 2017; Pantazopoulou et al., 2017).
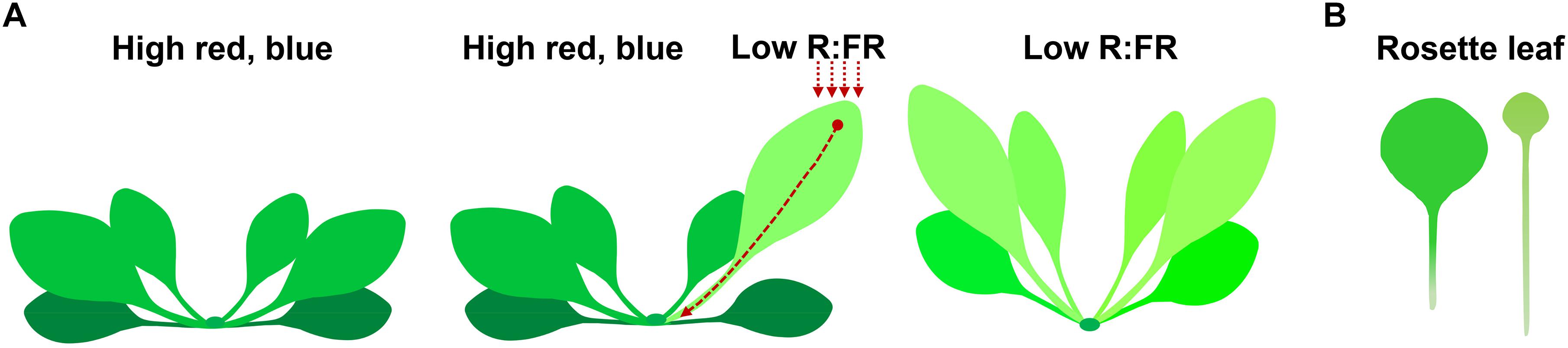
Figure 1. Morphological changes in plants under shade conditions. (A) Low R:FR shade treatment of a single leaf (middle) and whole plant (right) induces leaf hyponasty compared to the unshaded control plant (left). Arrows indicate the position of low R:FR shade treatment, which induces auxin biosynthesis at the site of shade signal perception. The newly synthesized auxin is transported out to the petiole, inducing cell elongation of petiole and hyponastic growth of leaf. (B) Prolonged shade treatment increases petiole elongation and reduces leaf size (right, compare to the unshaded control on the left).
Auxin Biosynthesis and Metabolism
In Arabidopsis, auxin biosynthesis starts with the conversion of tryptophan (Trp) to indole-3-pyruvic acid (IPA), catalyzed by TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1), which is encoded by SAV3 (SHADE AVOIDANCE3) (Tao et al., 2008). In turn, IPA is converted to IAA by the YUCCA (YUC) family of flavin monooxygenases (Won et al., 2011; Zhao, 2018). The sav3/taa1 and yuc2,5,8,9 quadruple mutants show reduced auxin accumulation and short hypocotyls under low R:FR conditions, indicating that TAA1, YUC2, 5, 8, and 9 act additively and are required for shade-induced auxin biosynthesis (Tao et al., 2008; Kohnen et al., 2016; Muller-Moule et al., 2016). Although TAA1 is required for low R:FR-induced auxin production, SAV3/TAA1 transcription is not directly induced by shade (Tao et al., 2008). In contrast to SAV3/TAA1, the transcription of YUC2, 5, 8, and 9 is rapidly and strongly induced within 1–2 h, or even 15 min, of low R:FR treatment (Tao et al., 2008; Li et al., 2012; de Wit et al., 2015; Kohnen et al., 2016; Muller-Moule et al., 2016). YUC2 and YUC5 are strongly expressed in the vascular tissues of petioles and hypocotyls, whereas YUC8 and YUC9 are strongly expressed in mesophyll and vascular cells of the leaf margin but not in hypocotyls (Challa et al., 2016; Muller-Moule et al., 2016). YUC3 is also significantly induced in hypocotyl after 45 min low R:FR treatment and is required for shade-induced hypocotyl elongation (Kohnen et al., 2016).
In addition to SAV3/TAA1 and YUCs, auxin homeostasis-related genes are also involved in shade responses. Disruption of REVERSAL OF SAV3 (VAS1) not only rescued the impaired avoidance response of the sav3/taa1 mutant under low R:FR conditions, but the vas1 sav3 plants also exhibited a mild constitutive shade response phenotype even under high R:FR conditions, indicating that VAS1 negatively regulates shade avoidance responses (Zheng et al., 2013). Biochemical analyses revealed that VAS1 and TAA1 have opposing biochemical functions; therefore, VAS1 inhibits shade response by inhibiting auxin production (Zheng et al., 2013).
VAS2/GH3.17 (GRETCHEN HAGEN3.17) catalyzes the conjugation of free IAA to inactive IAA-Glu (IAA-glutamate). Disruption of VAS2/GH3.17 resulted in accumulation of free IAA, thus enhancing shade-induced hypocotyl elongation (Zheng et al., 2016). Interestingly, low R:FR treatment suppresses the transcription of VAS2/GH3.17 and its two homologs, GH3.18 and GH3.19, suggesting that shade induces the accumulation of free IAA not only by promoting its synthesis and transport in the cotyledon, but also by reducing the conjugation of free IAA in the hypocotyl (Salter et al., 2003; Zheng et al., 2016). A recent study revealed that GH3 family protein FAR-RED INSENSITIVE 219/JASMONATE RESISTANCE1 (FIN219/JAR1/GH3.11), negatively regulates shade avoidance responses by modulating auxin homeostasis (Swain et al., 2017). These findings confirm that local auxin metabolism in the hypocotyl plays critical roles in the shade avoidance response (Figure 2A, left).
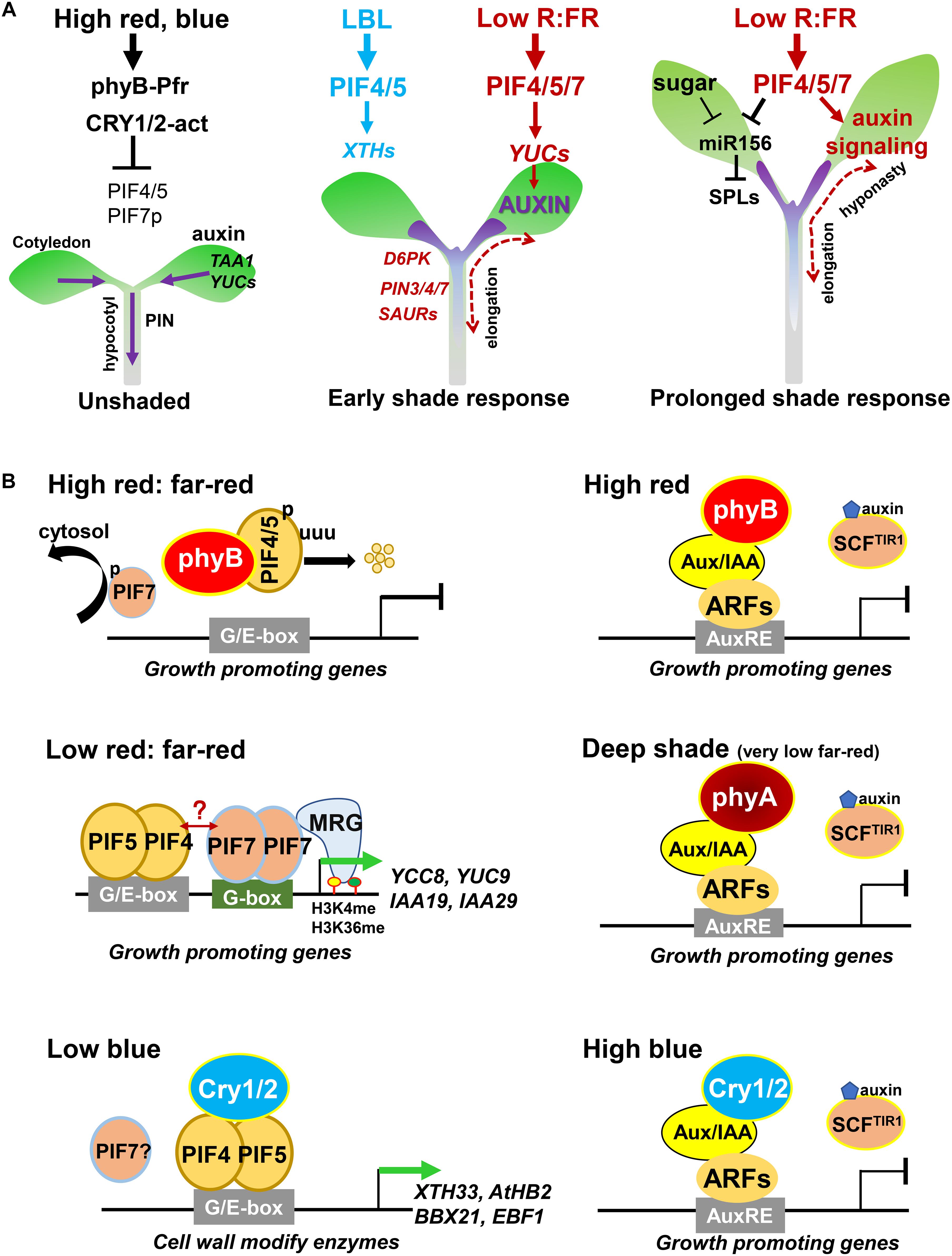
Figure 2. Regulatory mechanisms of the shade response in Arabidopsis thaliana. (A) The effects of light on hypocotyl elongation. Left, high-intensity red and blue light inhibits the functions of PIF4/5/7 by increasing the activity of phyB and CRYs under unshaded conditions. Middle, during the early shade response, LBL regulates the expression of genes encoding cell wall-modify enzymes through PIF4/5; low R:FR signal activates PIF4/5/7, thus promoting auxin biosynthesis in the cotyledon, which is then transported out to the hypocotyl where it induces cell elongation. Right, PIF4/5/7 increase auxin sensitivity and inhibit the expression of miR156 to mediate the prolonged shade avoidance response. (B) Physical interactions of different photoreceptors, PIFs, Aux/IAA, and ARF proteins under different light conditions.
Auxin Transport
In the hypocotyl, the auxin efflux carrier genes PIN3 (PIN-FORMED3) and PIN7 are rapidly induced by low R:FR treatment, and induced by low blue light (LBL) treatment (Devlin et al., 2003; Keuskamp et al., 2010, 2011; Kohnen et al., 2016). Consistent with potential functions of PIN3 and PIN7 in hypocotyl elongation, low R:FR and LBL-induced hypocotyl elongation is significantly reduced in the pin3 pin4 pin7 triple mutant. This indicates that polar auxin transport from the cotyledon to the hypocotyl is required for the shade avoidance response (Keuskamp et al., 2010; Kohnen et al., 2016).
The low induction of PIN genes compared to the extremely high induction of other auxin-related genes (such as YUC8, IAA19, and IAA29) suggests that the rapid response of PIN-type transporters to shade signals might not primarily occur through transcriptional regulation. Consistent with other levels of regulation participating in this response, the protein abundance and subcellular localization of PIN3 are rapidly regulated by low R:FR shade signal (Keuskamp et al., 2010). D6 PROTEIN KINASE (D6PK) and its homolog D6PK-LIKE1 (D6PKL1), encode two shade-induced protein kinases that function in the phosphorylation of PINs. Disruption of D6PK and D6PKL1 leads to inhibited elongation under low R:FR conditions, indicating that the phosphorylation of PINs is required for the shade response (Figure 2A, middle; Keuskamp et al., 2010; Barbosa et al., 2014; Kohnen et al., 2016).
Although PIN3, PIN4, and PIN7 are known to be involved in the shade response, how various shade signals regulate their transcription and subcellular localization, thus leading to the rapid transport of auxin from the cotyledon to the hypocotyl, remains to be explored in the future. In addition to PINs, ATP-binding cassette B (ABCB) auxin carriers also play a significant role in the shade avoidance response (Ge et al., 2017).
Auxin Signaling
SMALL AUXIN UP-REGULATED RNA (SAUR) genes are the largest family of early auxin-response genes (Ren and Gray, 2015). SAUR9, 10, 19, 20, 22, and 23 are rapidly induced by shade treatment, suggesting they might be involved in the shade avoidance response (Devlin et al., 2003; Kohnen et al., 2016). Overexpressing SAUR19 in the pif4 mutant background completely rescued the pif4 phenotype of impaired hypocotyl elongation at high temperatures and overexpressing SAUR36 also promoted hypocotyl elongation (Franklin et al., 2011; Stamm and Kumar, 2013). However, the mechanisms by which these SAURs mediate shade-induced cell elongation remain to be demonstrated. Interestingly, recent studies have shown that SAUR19-mediated elongation might involve the direct activation of plasma membrane H+-ATPase, leading to cell wall acidification and loosening, thereby facilitating cell expansion (Spartz et al., 2014, 2017; Ren and Gray, 2015; Fendrych et al., 2016). These findings suggest that SAUR proteins could mediate shade-promoted cell elongation by regulating cell wall acidification and loosening (Figure 2A, middle).
AUXIN RESPONSE FACTOR-type transcription factors and Auxin/Indole Acetic Acid (Aux/IAA)-type repressors are transcriptional regulators that form homo- and hetero-oligomers and play crucial roles in the transcriptional regulation of auxin response genes. The arf6 arf7 arf8 triple mutant fails to respond to low R:FR treatment, indicating that ARF6, ARF7, and ARF8 are required for shade-induce cell elongation (Reed et al., 2018). Although many Aux/IAA genes (e.g., IAA1, IAA5, IAA19, IAA29, and IAA30) are rapidly and strongly induced by various shade signals, how they mediate shade responses remains unclear (Kohnen et al., 2016). A recent study suggested that IAA19 and IAA29 mediate shade-induced cell elongation possibly by inhibiting the transcription of IAA17 (Pucciariello et al., 2018). Consistent with this, IAA17 is highly expressed in the hypocotyl epidermal cells and the gain-of-function mutant iaa17-1 showed impaired shade-induced hypocotyl elongation, indicating that IAA17 inhibits the shade avoidance response (Procko et al., 2016). In addition, IAA19 physically interacts with ARF7 mediating auxin- and brassinosteroid (BR)-regulated phototropic responses of hypocotyls (Harper et al., 2000; Tatematsu et al., 2004; Zhou et al., 2013). Therefore, in addition to the auxin pathway, IAA19-mediated shade responses might involve the BR signal pathway (Zhou et al., 2013).
Auxin Sensitivity Increases Under Prolonged Shade Conditions
The expression of auxin biosynthetic genes in cotyledons, the lamina, and petioles is rapidly induced within hours after plants perceive a low R:FR shade signal (de Wit et al., 2015). However, auxin contents rapidly decrease to basal levels after 24 h of low R:FR treatment, suggesting that auxin-mediated prolonged shade responses might not involve the regulation of auxin biosynthesis (de Wit et al., 2015). The response to prolonged shade conditions requires a system-wide rearrangement of auxin perception and signaling transduction (Pucciariello et al., 2018). Under these conditions, reduced phyB activity leads to decreased PIF4 levels in cotyledon mesophyll cells, and increased levels in hypocotyl vascular cells, which promotes IAA19 and IAA29 expression. IAA19 and IAA29 then suppress the expression of IAA17, thus promoting growth. Meanwhile, the transcript levels of the genes encoding the auxin receptors TRANSPORT INHIBITOR RESPONSE1 (TIR1) and AUXIN SIGNALING F-BOX (AFB) proteins are significantly increased, thus enhancing auxin perception and signal transduction under prolonged shade conditions (Pucciariello et al., 2018).
After prolonged low R:FR shade treatment, lamina width decreases but petiole length increases. This contrast in growth might be caused by differences in auxin sensitivity, because the auxin contents do not dramatically differ between the lamina and petiole (Figure 1B; de Wit et al., 2015). PIF7 is required for this contrast growth, but how PIF7 differentially modifies auxin sensitivity in the petiole vs. lamina, with contrasting effects on growth, remains poor understood (Figure 2A, right; de Wit et al., 2015, 2018).
Under prolonged shade conditions, free IAA and BR decrease to basal levels after approximately 24 h, while gibberellic acid (GA) contents continue to increase, suggesting that GA might play an important role in the response to prolonged shade (Bou-Torrent et al., 2014). In addition, PIF4 and PIF5 directly suppress the expression of MIR156; miR156 further reduces the transcript abundance of its targets SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) family genes, thus mediating the prolonged shade response (Figure 2A, right; Xie et al., 2017; Wei et al., 2018).
It is worth noting that decreased photosynthetic activity leads to reduced sucrose contents under prolonged shade conditions; this might contribute to the morphological and physiological changes in these plants. Indeed, reducing the expression of SUCROSE TRANSPORTER 4 (SUT4) significantly inhibited the shade avoidance response in potato (Kozuka et al., 2005; Chincinska et al., 2008). Consistent with a role of sugar signaling in shade responses, sugar transport in the phloem and local starch metabolism are required for shade-induced cell elongation of Arabidopsis hypocotyls (de Wit et al., 2018).
Response to Low Blue Light (LBL) Shade Signals
Arabidopsis PIF4 and PIF5 are not only involved in low R:FR responses, but also participate in LBL responses (Keller et al., 2011; de Wit et al., 2016; Goyal et al., 2016; Pedmale et al., 2016). LBL strongly induces the expression of PIF4 and PIF5 in the apical portion of the hypocotyl and increases the stability of PIF5 (Sun et al., 2013; Pedmale et al., 2016). After LBL treatment, pif4 and pif5 seedling plants show significantly reduced hypocotyl and petiole elongation, and reduced hyponastic leaf growth. These observations indicate that PIF4 and PIF5 are essential for the LBL-induced shade response, with PIF4 playing a more important role (Keller et al., 2011; de Wit et al., 2016; Pedmale et al., 2016).
Although PIF4 and PIF5 are involved in both low R:FR- and LBL-induced shade responses, the underlying mechanisms are distinct. PIF4 and PIF5 mediate the low R:FR response by regulating the expression of auxin biosynthesis, transport and signaling-related genes (Figure 2A, middle; Hornitschek et al., 2012). By contrast, they mediate the LBL response mainly by regulating the expression of genes encoding cell wall-modifying enzymes (Pedmale et al., 2016). Although free IAA levels do not rapidly increase after LBL treatment, the LBL response is significantly reduced in taa1, pin3 pin4 pin7, and tir1 afb1 afb2 afb3 seedlings, indicating that plant responses to LBL require auxin synthesis, transport, and signaling transduction (Keuskamp et al., 2011; Pedmale et al., 2016). In addition, pif7 mutant seedlings showed a partial response to LBL treatment, indicating that PIF7 is also required for LBL responses (Pedmale et al., 2016).
Interaction of Photoreceptors and PIFs During Shade Responses
High R:FR increases the ratio of the active form of phyB (Pfr, the far-red light-absorbing form) in the nucleus; Pfr physically interacts with PIFs and promotes their phosphorylation, ubiquitination, and degradation (Leivar and Quail, 2011; Leivar and Monte, 2014). Low R:FR increases the ratio of the inactive form of phyB (red light-absorbing form, Pr) in the cytoplasm, thus increasing PIF accumulation in the nucleus and promoting the expression of their downstream target genes (Figure 2B; Casal, 2013). Consequently, PIF3, PIF4, and PIF5 but not PIF7 are rapidly degraded under high R:FR conditions, but their levels rapidly increase after low R:FR shade treatment (Lorrain et al., 2008; Leivar et al., 2012b).
PIF4 and PIF5 directly bind to the promoters of auxin biosynthetic genes YUC8, TAA1, and CYP79B2 and auxin-responsive genes IAA19, IAA29, and SAUR19-24 and activate their expression in response to shade, as well as phototropic and high-temperature responses (Franklin et al., 2011; Sun et al., 2013; Ma et al., 2016). Under low PAR but high R:FR conditions, PIF4 and PIF5 also bind to the promoters of auxin biosynthesis and signaling genes, suggesting that low PAR-induced cell elongation is also dependent on PIF4- and PIF5-mediated auxin responses (Hornitschek et al., 2012).
In contrast to the effect of low R:FR on PIF4 and PIF5 stability and accumulation, low R:FR induces PIF7 de-phosphorylation and translocation from the cytoplasm to the nucleus, thus enhancing PIF7 binding to the promoters of auxin-related genes such as, YUC8, YUC9, IAA19, and GH3.3 (Hornitschek et al., 2012; Li et al., 2012; Huang et al., 2018). In turn, PIF7 interacts with H3K4me3 and H3K36me3-READER MOTIF RELATED GENE2 (MRG2) and recruits it to the coding regions of various targets of PIF7 further regulate their expression (Figure 2B; Huang et al., 2018; Peng et al., 2018). Consistent with this, disrupting both MRG1 and MRG2 resulted in a reduced shade response, indicating that they positively regulate the shade response (Peng et al., 2018).
High-intensity red and blue light promotes PIF4 and PIF5 protein phosphorylation and degradation, thus inhibiting the transcription of growth-promoting genes and suppressing hypocotyl elongation (Figure 2A, left). Under LBL, photoreceptors cryptochrome-1 (CRY1) and CRY2 physically interact with PIF4 and PIF5. In turn, CRY2 is recruited to the promoters of PIF4 and PIF5 targets and modulates their expression (Figure 2B; Pedmale et al., 2016). By contrast, under high blue light, CRY1 physically interacts with PIF4 and represses its effects on transcription, thus suppressing hypocotyl elongation under high temperature conditions (Ma et al., 2016). In addition, the UV-B photoreceptor UVR8 negatively regulates low R:FR shade and high temperature induced auxin biosynthesis and cell elongation partially by enhancing the degradation or inhibited the transcriptional activity of PIF4 on its targets (Hayes et al., 2014, 2017). Therefore, PIF proteins physically interact with multiple different kinds of photoreceptors and mediate cell elongation through different regulatory mechanisms.
Auxin and Light Antagonistically Regulate the Stability of Aux/IAA Proteins
In general, under low free IAA conditions, Aux/IAA repressors physically interact with ARF transcription factors and inhibit their function, thus negatively regulating auxin responses. Under high free IAA conditions, Aux/IAA proteins are rapidly degraded, thus releasing its repression on ARFs, which in turn promotes the expression of ARFs downstream targets and induces auxin responses (Leyser, 2018). Interestingly, the stability of Aux/IAA proteins is also controlled by light (Huq, 2018; Xu et al., 2018; Yang et al., 2018). In the dark, phyB and CRY1 are present in the cytoplasm in inactive forms that cannot physically interact with Aux/IAA proteins. Under high-intensity red- or blue-light, phyB or CRY1 interact with Aux/IAA proteins in a light-dependent manner, thus inhibiting their protein degradation and decreasing hypocotyl elongation (Xu et al., 2018; Figure 2). Under deep shade (very low R:FR) conditions, phyA protein stability significantly increases. In turn, phyA physically interacts with Aux/IAA proteins and prevents their degradation, thus negatively regulating shade avoidance responses (Yang et al., 2018). These studies revealed that Aux/IAA stability is rapidly regulated by auxin receptors and photoreceptors, which allow plants to perceive endogenous auxin content and environmental light signaling thus to fine-tune cell elongation under different light conditions.
Understanding Shade Responses: Future Perspectives
In the past decades, the mechanisms regulating the shade responses have been well studied in the model plant Arabidopsis, however, it will take much longer to transfer this knowledge to crop plants. Our recent study revealed that ectopic expression of maize PIF4 and PIF5 in Arabidopsis pifq mutant completely rescued its impaired shade avoidance response, which indicated that PIF proteins might play conserved roles in shade response (Shi et al., 2018b). Transcriptome analyses further indicated that maize and Arabidopsis might share very conserved regulatory pathways of shade avoidance response (Wang et al., 2016; Shi et al., 2018a). Besides the well-known shade avoidance response, how shade-tolerant plants increase their survival and fitness in various shade conditions remains to be further investigated. A recent study suggested that the biosynthesis and signaling of auxin, GA, and BR are not significantly affected by shade signals in the shade-tolerant Geranium robertianum, suggesting that differing patterns of hormones might be one of the reasons for the different morphogenic and physiological changes in shade tolerant vs. intolerant species (Gommers et al., 2018). Interestingly, the clonal plant Potentilla reptans uses different strategies to respond to various shade signals, including enhanced vertical growth in the presence of (simulated) short-dense neighbors, shade tolerance behavior in the presence of tall-dense neighbors, and lateral avoidance behavior in the presence of tall-sparse neighbors (Gruntman et al., 2017). Indeed, both vertical growth and lateral avoidance behaviors are phototropisms or types of directional growth, consistent with the finding that plants tend to reposition their growth towards unfiltered sunlight at the edges or gaps of natural canopies (de Wit et al., 2016; Goyal et al., 2016; Fiorucci and Fankhauser, 2017). These findings provide new perspectives about shade responses in the natural environment.
Author Contributions
Both authors conceived the study, and wrote and revised the manuscript.
Funding
This work was supported by grants from the National Science Foundation of China (31870266 and 31670249).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ballare, C. L., and Pierik, R. (2017). The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ. 40, 2530–2543. doi: 10.1111/pce.12914
Barbosa, I. C., Zourelidou, M., Willige, B. C., Weller, B., and Schwechheimer, C. (2014). D6 PROTEIN KINASE activates auxin transport-dependent growth and PIN-FORMED phosphorylation at the plasma membrane. Dev. Cell 29, 674–685. doi: 10.1016/j.devcel.2014.05.006
Bou-Torrent, J., Galstyan, A., Gallemi, M., Cifuentes-Esquivel, N., Molina-Contreras, M. J., Salla-Martret, M., et al. (2014). Plant proximity perception dynamically modulates hormone levels and sensitivity in Arabidopsis. J. Exp. Bot. 65, 2937–2947. doi: 10.1093/jxb/eru083
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427. doi: 10.1146/annurev-arplant-050312-120221
Challa, K. R., Aggarwal, P., and Nath, U. (2016). Activation of YUCCA5 by the transcription factor TCP4 integrates developmental and environmental signals to promote hypocotyl elongation in Arabidopsis. Plant Cell 28, 2117–2130. doi: 10.1105/tpc.16.00360
Chincinska, I. A., Liesche, J., Krugel, U., Michalska, J., Geigenberger, P., Grimm, B., et al. (2008). Sucrose transporter StSUT4 from potato affects flowering, tuberization, and shade avoidance response. Plant Physiol. 146, 515–528.
Crocco, C. D., Holm, M., Yanovsky, M. J., and Botto, J. F. (2010). AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 64, 551–562. doi: 10.1111/j.1365-313X.2010.04360.x
de Wit, M., George, G. M., Ince, Y. C., Dankwa-Egli, B., Hersch, M., Zeeman, S. C., et al. (2018). Changes in resource partitioning between and within organs support growth adjustment to neighbor proximity in Brassicaceae seedlings. Proc. Natl. Acad. Sci. U.S.A. 115, E9953–E9961. doi: 10.1073/pnas.1806084115
de Wit, M., Keuskamp, D. H., Bongers, F. J., Hornitschek, P., Gommers, C. M. M., Reinen, E., et al. (2016). Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr. Biol. 26, 3320–3326. doi: 10.1016/j.cub.2016.10.031
de Wit, M., Ljung, K., and Fankhauser, C. (2015). Contrasting growth responses in lamina and petiole during neighbor detection depend on differential auxin responsiveness rather than different auxin levels. New Phytol. 208, 198–209. doi: 10.1111/nph.13449
Devlin, P. F., Yanovsky, M. J., and Kay, S. A. (2003). A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 133, 1617–1629.
Fendrych, M., Leung, J., and Friml, J. (2016). TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5:e19048. doi: 10.7554/eLife.19048
Fiorucci, A. S., and Fankhauser, C. (2017). Plant strategies for enhancing access to sunlight. Curr. Biol. 27, R931–R940. doi: 10.1016/j.cub.2017.05.085
Franklin, K. A. (2008). Shade avoidance. New Phytol. 179, 930–944. doi: 10.1111/j.1469-8137.2008.02507.x
Franklin, K. A., Lee, S. H., Patel, D., Kumar, S. V., Spartz, A. K., Gu, C., et al. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. U.S.A. 108, 20231–20235. doi: 10.1073/pnas.1110682108
Ge, Y., Yan, F., Zourelidou, M., Wang, M., Ljung, K., Fastner, A., et al. (2017). SHADE AVOIDANCE 4 is required for proper auxin distribution in the hypocotyl. Plant Physiol. 173, 788–800. doi: 10.1104/pp.16.01491
Gommers, C. M., Visser, E. J., St Onge, K. R., Voesenek, L. A., and Pierik, R. (2013). Shade tolerance: when growing tall is not an option. Trends Plant Sci. 18, 65–71. doi: 10.1016/j.tplants.2012.09.008
Gommers, C. M. M., Buti, S., Tarkowská, D., Pìnèík, A., Banda, J. P., Arricastres, V., et al. (2018). Organ-specific phytohormone synthesis in two Geranium species with antithetical responses to far-red light enrichment. Plant Direct 2:e00066. doi: 10.1002/pld3.66
Goyal, A., Karayekov, E., Galvao, V. C., Ren, H., Casal, J. J., and Fankhauser, C. (2016). Shade promotes phototropism through phytochrome B-controlled auxin production. Curr. Biol. 26, 3280–3287. doi: 10.1016/j.cub.2016.10.001
Gruntman, M., Gross, D., Majekova, M., and Tielborger, K. (2017). Decision-making in plants under competition. Nat. Commun. 8:2235. doi: 10.1038/s41467-017-02147-2
Harper, R. M., Stowe-Evans, E. L., Luesse, D. R., Muto, H., Tatematsu, K., Watahiki, M. K., et al. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12, 757–770.
Hayes, S., Sharma, A., Fraser, D. P., Trevisan, M., Cragg-Barber, C. K., Tavridou, E., et al. (2017). UV-B perceived by the UVR8 photoreceptor inhibits plant thermomorphogenesis. Curr. Biol. 27, 120–127. doi: 10.1016/j.cub.2016.11.004
Hayes, S., Velanis, C. N., Jenkins, G. I., and Franklin, K. A. (2014). UV-B detected by the UVR8 photoreceptor antagonizes auxin signaling and plant shade avoidance. Proc. Natl. Acad. Sci. U.S.A. 111, 11894–11899. doi: 10.1073/pnas.1403052111
Hornitschek, P., Kohnen, M. V., Lorrain, S., Rougemont, J., Ljung, K., Lopez-Vidriero, I., et al. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71, 699–711. doi: 10.1111/j.1365-313X.2012.05033.x
Hornitschek, P., Lorrain, S., Zoete, V., Michielin, O., and Fankhauser, C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28, 3893–3902. doi: 10.1038/emboj.2009.306
Huang, X., Zhang, Q., Jiang, Y., Yang, C., Wang, Q., and Li, L. (2018). Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. eLife 7:e31636. doi: 10.7554/eLife.31636
Huq, E. (2018). Direct convergence of light and auxin signaling pathways in Arabidopsis. Mol. Plant 11, 515–517.
Iglesias, M. J., Sellaro, R., Zurbriggen, M. D., and Casal, J. J. (2018). Multiple links between shade avoidance and auxin networks. J. Exp. Bot. 69, 213–228. doi: 10.1093/jxb/erx295
Keller, M. M., Jaillais, Y., Pedmale, U. V., Moreno, J. E., Chory, J., and Ballare, C. L. (2011). Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J. 67, 195–207. doi: 10.1111/j.1365-313X.2011.04598.x
Keuskamp, D. H., Pollmann, S., Voesenek, L. A., Peeters, A. J., and Pierik, R. (2010). Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc. Natl. Acad. Sci. U.S.A. 107, 22740–22744. doi: 10.1073/pnas.1013457108
Keuskamp, D. H., Sasidharan, R., Vos, I., Peeters, A. J., Voesenek, L. A., and Pierik, R. (2011). Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J. 67, 208–217. doi: 10.1111/j.1365-313X.2011.04597.x
Kim, S., Mochizuki, N., Deguchi, A., Nagano, A. J., Suzuki, T., and Nagatani, A. (2018). Auxin contributes to the intraorgan regulation of gene expression in response to shade. Plant Physiol. 177, 847–862. doi: 10.1104/pp.17.01259
Kohnen, M. V., Schmid-Siegert, E., Trevisan, M., Petrolati, L. A., Senechal, F., Muller-Moule, P., et al. (2016). Neighbor detection induces organ-specific transcriptomes, revealing patterns underlying hypocotyl-specific growth. Plant Cell 28, 2889–2904. doi: 10.1105/tpc.16.00463
Kozuka, T., Horiguchi, G., Kim, G. T., Ohgishi, M., Sakai, T., and Tsukaya, H. (2005). The different growth responses of the Arabidopsis thaliana leaf blade and the petiole during shade avoidance are regulated by photoreceptors and sugar. Plant Cell Physiol. 46, 213–223.
Leivar, P., and Monte, E. (2014). PIFs: systems integrators in plant development. Plant Cell 26, 56–78. doi: 10.1105/tpc.113.120857
Leivar, P., Monte, E., Cohn, M. M., and Quail, P. H. (2012a). Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually negative phyB-PIF feedback loop. Mol. Plant 5, 734–749. doi: 10.1093/mp/sss031
Leivar, P., Tepperman, J. M., Cohn, M. M., Monte, E., Al-Sady, B., Erickson, E., et al. (2012b). Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24, 1398–1419. doi: 10.1105/tpc.112.095711
Leivar, P., and Quail, P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. doi: 10.1016/j.tplants.2010.08.003
Li, J., Li, G., Wang, H., and Wang Deng, X. (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9:e0148. doi: 10.1199/tab.0148
Li, L., Ljung, K., Breton, G., Schmitz, R. J., Pruneda-Paz, J., Cowing-Zitron, C., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790. doi: 10.1101/gad.187849.112
Lorrain, S., Allen, T., Duek, P. D., Whitelam, G. C., and Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323.
Ma, D., Li, X., Guo, Y., Chu, J., Fang, S., Yan, C., et al. (2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. U.S.A. 113, 224–229. doi: 10.1073/pnas.1511437113
Michaud, O., Fiorucci, A. S., Xenarios, I., and Fankhauser, C. (2017). Local auxin production underlies a spatially restricted neighbor-detection response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, 7444–7449. doi: 10.1073/pnas.1702276114
Muller-Moule, P., Nozue, K., Pytlak, M. L., Palmer, C. M., Covington, M. F., Wallace, A. D., et al. (2016). YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ 4:e2574.
Ortiz-Alcaide, M., Llamas, E., Gomez-Cadenas, A., Nagatani, A., Martinez-Garcia, J. F., and Rodriguez-Concepcion, M. (2019). Chloroplasts modulate elongation responses to canopy shade by retrograde pathways involving HY5 and abscisic acid. Plant Cell 31, 384–398. doi: 10.1105/tpc.18.00617
Pacin, M., Semmoloni, M., Legris, M., Finlayson, S. A., and Casal, J. J. (2016). Convergence of CONSTITUTIVE PHOTOMORPHOGENESIS 1 and PHYTOCHROME INTERACTING FACTOR signalling during shade avoidance. New Phytol. 211, 967–979. doi: 10.1111/nph.13965
Pantazopoulou, C. K., Bongers, F. J., Kupers, J. J., Reinen, E., Das, D., Evers, J. B., et al. (2017). Neighbor detection at the leaf tip adaptively regulates upward leaf movement through spatial auxin dynamics. Proc. Natl. Acad. Sci. U.S.A. 114, 7450–7455. doi: 10.1073/pnas.1702275114
Pedmale, U. V., Huang, S. S., Zander, M., Cole, B. J., Hetzel, J., Ljung, K., et al. (2016). Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233–245. doi: 10.1016/j.cell.2015.12.018
Peng, M., Li, Z., Zhou, N., Ma, M., Jiang, Y., Dong, A., et al. (2018). Linking PHYTOCHROME-INTERACTING FACTOR to histone modification in plant shade avoidance. Plant Physiol. 176, 1341–1351. doi: 10.1104/pp.17.01189
Pierik, R., and de Wit, M. (2014). Shade avoidance: phytochrome signalling and other aboveground neighbour detection cues. J. Exp. Bot. 65, 2815–2824. doi: 10.1093/jxb/ert389
Procko, C., Burko, Y., Jaillais, Y., Ljung, K., Long, J. A., and Chory, J. (2016). The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev. 30, 1529–1541. doi: 10.1101/gad.283234.116
Pucciariello, O., Legris, M., Costigliolo Rojas, C., Iglesias, M. J., Hernando, C. E., Dezar, C., et al. (2018). Rewiring of auxin signaling under persistent shade. Proc. Natl. Acad. Sci. U.S.A. 115, 5612–5617. doi: 10.1073/pnas.1721110115
Reed, J. W., Nagpal, P., Poole, D. S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5, 147–157.
Reed, J. W., Wu, M. F., Reeves, P. H., Hodgens, C., Yadav, V., Hayes, S., et al. (2018). Three auxin response factors promote hypocotyl elongation. Plant Physiol. 178, 864–875. doi: 10.1104/pp.18.00718
Ren, H., and Gray, W. M. (2015). SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 8, 1153–1164. doi: 10.1016/j.molp.2015.05.003
Roig-Villanova, I., Bou-Torrent, J., Galstyan, A., Carretero-Paulet, L., Portoles, S., Rodriguez-Concepcion, M., et al. (2007). Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J. 26, 4756–4767.
Rolauffs, S., Fackendahl, P., Sahm, J., Fiene, G., and Hoecker, U. (2012). Arabidopsis COP1 and SPA genes are essential for plant elongation but not for acceleration of flowering time in response to a low red light to far-red light ratio. Plant Physiol. 160, 2015–2027. doi: 10.1104/pp.112.207233
Salter, M. G., Franklin, K. A., and Whitelam, G. C. (2003). Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426, 680–683.
Sellaro, R., Crepy, M., Trupkin, S. A., Karayekov, E., Buchovsky, A. S., Rossi, C., et al. (2010). Cryptochrome as a sensor of the blue/green ratio of natural radiation in Arabidopsis. Plant Physiol. 154, 401–409. doi: 10.1104/pp.110.160820
Sellaro, R., Pacin, M., and Casal, J. J. (2017). Meta-analysis of the transcriptome reveals a core set of shade-avoidance genes in Arabidopsis. Photochem. Photobiol. 93, 692–702. doi: 10.1111/php.12729
Sellaro, R., Yanovsky, M. J., and Casal, J. J. (2011). Repression of shade-avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J. 68, 919–928. doi: 10.1111/j.1365-313X.2011.04745.x
Shi, Q., Kong, F., Zhang, H., Jiang, Y., Heng, S., Liang, R., et al. (2018a). Molecular mechanisms governing shade responses in maize. Biochem. Biophys. Res. Commun. 516, 112–119. doi: 10.1016/j.bbrc.2019.05.142
Shi, Q., Zhang, H., Song, X., Jiang, Y., Liang, R., and Li, G. (2018b). Functional characterization of the maize phytochrome-interacting factors PIF4 and PIF5. Front. Plant Sci. 8:2273. doi: 10.3389/fpls.2017.02273
Spartz, A. K., Lor, V. S., Ren, H., Olszewski, N. E., Miller, N. D., Wu, G., et al. (2017). Constitutive expression of Arabidopsis SMALL AUXIN UP RNA19 (SAUR19) in tomato confers auxin-independent hypocotyl elongation. Plant Physiol. 173, 1453–1462. doi: 10.1104/pp.16.01514
Spartz, A. K., Ren, H., Park, M. Y., Grandt, K. N., Lee, S. H., Murphy, A. S., et al. (2014). SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26, 2129–2142.
Stamm, P., and Kumar, P. P. (2013). Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep. 32, 759–769. doi: 10.1007/s00299-013-1406-5
Sun, J., Qi, L., Li, Y., Zhai, Q., and Li, C. (2013). PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25, 2102–2114. doi: 10.1105/tpc.113.112417
Swain, S., Jiang, H. W., and Hsieh, H. L. (2017). FAR-RED INSENSITIVE 219/JAR1 contributes to shade avoidance responses of Arabidopsis seedlings by modulating key shade signaling components. Front. Plant Sci. 8:1901. doi: 10.3389/fpls.2017.01901
Tao, Y., Ferrer, J. L., Ljung, K., Pojer, F., Hong, F., Long, J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049
Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M. K., Harper, R. M., et al. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379–393.
van Gelderen, K., Kang, C., Paalman, R., Keuskamp, D., Hayes, S., and Pierik, R. (2018). Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell 30, 101–116. doi: 10.1105/tpc.17.00771
Wang, H., and Wang, H. (2015). Phytochrome signaling: time to tighten up the loose ends. Mol. Plant 8, 540–551. doi: 10.1016/j.molp.2014.11.021
Wang, H., Wu, G., Zhao, B., Wang, B., Lang, Z., Zhang, C., et al. (2016). Regulatory modules controlling early shade avoidance response in maize seedlings. BMC Genomics 17:269. doi: 10.1186/s12864-016-2593-6
Wei, H., Zhao, Y., Xie, Y., and Wang, H. (2018). Exploiting SPL genes to improve maize plant architecture tailored for high density planting. J. Exp. Bot. 69, 4675–4688. doi: 10.1093/jxb/ery258
Won, C., Shen, X., Mashiguchi, K., Zheng, Z., Dai, X., Cheng, Y., et al. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523. doi: 10.1073/pnas.1108436108
Xie, Y., Liu, Y., Wang, H., Ma, X., Wang, B., Wu, G., et al. (2017). Phytochrome-interacting factors directly suppress MIR156 expression to enhance shade-avoidance syndrome in Arabidopsis. Nat. Commun. 8:348. doi: 10.1038/s41467-017-00404-y
Xu, F., He, S., Zhang, J., Mao, Z., Wang, W., Li, T., et al. (2018). Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol. Plant 11, 523–541. doi: 10.1016/j.molp.2017.12.003
Yang, C., Xie, F., Jiang, Y., Li, Z., Huang, X., and Li, L. (2018). Phytochrome a negatively regulates the shade avoidance response by increasing auxin/indole acidic acid protein stability. Dev. Cell 44, 29–41.e4. doi: 10.1016/j.devcel.2017.11.017
Zhao, Y. (2018). Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 69, 417–435. doi: 10.1146/annurev-arplant-042817-040226
Zheng, Z., Guo, Y., Novak, O., Chen, W., Ljung, K., Noel, J. P., et al. (2016). Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat. Plants 2:16025. doi: 10.1038/nplants.2016.25
Zheng, Z., Guo, Y., Novák, O., Dai, X., Zhao, Y., Ljung, K., et al. (2013). Coordination of auxin and ethylene biosynthesis by the aminotransferase VAS1. Nat. Chem. Biol. 9, 244–246. doi: 10.1038/nchembio.1178
Keywords: auxin, cell elongation, shade avoidance response, photoreceptor, PIFs
Citation: Ma L and Li G (2019) Auxin-Dependent Cell Elongation During the Shade Avoidance Response. Front. Plant Sci. 10:914. doi: 10.3389/fpls.2019.00914
Received: 09 April 2019; Accepted: 27 June 2019;
Published: 12 July 2019.
Edited by:
Rujin Chen, Lanzhou University, ChinaReviewed by:
Chuanwei Yang, Fudan University, ChinaIrma Roig-Villanova, Centre for Research in Agricultural Genomics (CRAG), Spain
Elena Monte, Centre for Research in Agricultural Genomics (CRAG), Spain
Copyright © 2019 Ma and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Ma, eXp6ZDExMTFAMTI2LmNvbQ==; Gang Li, Z2FuZ2xpQHNkYXUuZWR1LmNu
 Lin Ma
Lin Ma Gang Li
Gang Li