- Institute of Life Sciences, Jiangsu University, Zhenjiang, China
Sclerotinia sclerotiorum (Lib.) de Bary is an unusual pathogen which has the broad host range, diverse infection modes, and potential double feeding lifestyles of both biotroph and necrotroph. It is capable of infecting over 400 plant species found worldwide and more than 60 names have agriculturally been used to refer to diseases caused by this pathogen. Plant defense to S. sclerotiorum is a complex biological process and exhibits a typical quantitative disease resistance (QDR) response. Recent studies using Arabidopsis thaliana and crop plants have obtained new advances in mechanisms used by plants to cope with S. sclerotiorum infection. In this review, we focused on our current understanding on plant defense mechanisms against this pathogen, and set up a model for the defense process including three stages: recognition of this pathogen, signal transduction and defense response. We also have a particular interest in defense signaling mediated by diverse signaling molecules. We highlight the current challenges and unanswered questions in both the defense process and defense signaling. Essentially, we discussed candidate resistance genes newly mapped by using high-throughput experiments in important crops, and classified these potential gene targets into different stages of the defense process, which will broaden our understanding of the genetic architecture underlying quantitative resistance to S. sclerotiorum. We proposed that more powerful mapping population(s) will be required for accurate and reliable QDR gene identification.
Introduction
Sclerotinia sclerotiorum (Lib.) de Bary is a devastating fungal plant pathogen with a broad host range including at least 408 described species of plant from 278 genera in 75 families (Boland and Hall, 1994). It can infect many economically important dicotyledonous crops such as oilseed rape (Brassica napus), edible dry bean (Phaseolus vulgaris), soybean (Glycine max), dry pea (Pisium sativum), chickpea (Cicer arietinum), peanut (Arachis hypogaea), sunflower (Helianthus annuus), lentils (Lens culinaris), and various vegetables, and some monocotyledonous species such as tulip (Tulipa gesneriana) and onion (Allium cepa) (Boland and Hall, 1994; Bolton et al., 2006). More than 60 names have been used to refer to the resulting diseases in agriculture (Purdy, 1979) including cottony rot, watery soft rot, drop, crown rot, blossom blight, and perhaps most common, Sclerotinia stem rot (SSR) or white mold. Diseases caused by S. sclerotiorum have a worldwide distribution and cause serious crop losses around the world. For example, in China, oilseed rape yield losses caused by SSR usually range from 10 to 20% and may be up to 80% for severe SSR outbreaks seasons (Yang, 1959; Li et al., 2006; Mei et al., 2011). In United States, annual losses caused by this pathogen have exceeded $200 million (Bolton et al., 2006). Diseases caused by S. sclerotiorum have traditionally been difficult to control (Bolton et al., 2006) due to lacking high level resistance in major crops, which makes it difficult to improve resistance using classical breeding methods. Disease management depends heavily on the application of fungicides, but this may cause environmental contamination, increase farming costs and may be ineffective because of the difficulties associated with the application of sprays to thick canopies and the lack of suitable forecasting methods to enable the timely application of fungicides. Currently, molecular breeding is pursued as an important strategy to control diseases caused by this pathogen. Thus, it is important for breeders to understand the molecular basis of host genetic resistance against S. sclerotiorum.
The molecular basis of plant — S. sclerotiorum interaction is complicated. S. sclerotiorum is an ascomycete with a reported necrotrophic lifestyle, secreting cell wall degrading enzymes and toxins, such as oxalic acid, Ss-Rhs1 and so on (Yu et al., 2017), to kill host cells and derive energy. Recent research revealed that there is a potential short biotrophic phase in the lifestyle of S. sclerotiorum, and a new model depicting the lifestyle transition of the pathogen from biotrophic to necrotrophic growth was proposed (Kabbage et al., 2015), which suggests a multifaceted pathogenic strategies for this pathogen. In return, plants use a range of multifaceted defense mechanisms to accurately detect and appropriately respond to the infection of this pathogen. Genetic resistance to S. sclerotiorum shows quantitative inheritance (Bert et al., 2002; Liu et al., 2005; Zhao et al., 2006; Perchepied et al., 2010; Fusari et al., 2012). Early studies looking for resistance loci in B. napus were conducted through quantitative trait loci (QTL) mapping (Zhao and Meng, 2003; Zhao et al., 2006; Yin et al., 2010; Behla, 2017; Wu et al., 2013; Wei et al., 2014). Alternatives to QTL mapping based on a biparental population with fewer recombination events, have emerged recently, including genome-wide association study (GWAS) that uses natural plant populations as mapping populations. Further, high-throughput sequencing technology provides access to a large resource of omic data and increases options of host species, GWAS mapping population(s) and effective data analysis strategies for the identification of quantitative genes/loci.
In the past decade, new advances in mechanisms of plant defense to S. sclerotiorum infection have been obtained. Further, the recent application of omic methodologies and instrumentation enhances our understanding of defense strategies in important crops. Here, we focused on past studies on mechanisms and strategies employed by plants in coping with this pathogen infection. We also have a particular interest on defense signaling mediated by diverse signaling molecules. Finally, we discussed candidate resistance genes mapped by using High-throughput experiments. This review highlights challenges in defense mechanisms identification, which will broaden our understanding of the genetic architecture underlying this quantitative resistance.
Plant Defense Mechanisms Against S. Sclerotiorum
Facing the attack of pathogens, such as S. sclerotiorum, plants need to accurately detect and send timely signal and then appropriately respond to each of the different pathogenic strategies by deploying a range of multifaceted defense response mechanisms (Figure 1). Defense responses often begin with either recognition of pathogen (or microbe)-associated molecular patterns (PAMPs or MAMPs) by plant-cell-surface pattern recognition receptors (PRRs) or recognition of the pathogen’s virulence molecules, termed effectors, by a set of plant intracellular resistance (R) gene products, which results in MAMP (or PAMP)-triggered immunity (MTI or PTI) or effector-triggered immunity (ETI), respectively (Jones and Dangl, 2006). In 1971, ETI was also named as gene-for-gene interaction by Flor, (1971). In the plant–Sclerotinia pathosystem, ETI (or R gene-mediated resistance) has not been observed (Yang et al., 2018). In contrast, the presence of SCFE1 (Sclerotinia culture filtrate elicitor1), an elicitor in S. sclerotiorum evoking MAMP-triggered immune responses and sensed by RLP30 (Receptor-like protein30), demonstrates the relevance of MTI in resistance to S. sclerotiorum (Zhang et al., 2013). RLP30, belonging to a specific class of plasma membrane (PM)-localised receptors, presents all the hallmarks of a cell surface-located RLP with an N-terminal signal peptide, an extracellular domain containing 21 LRRs (which possibly act as the SCFE1 binding site), a single transmembrane domain, and a short cytoplasmic tail of 25 amino acid residues (Zhang et al., 2013). Other elicitors, such as HRE (A heat-released elicitor) (Bertinetti and Ugalde, 1996), SsCut (cutinase) (Zhang et al., 2014) and SsSm1 (a Cerato-platanin family protein) (Pan et al., 2018), have also been identified from S. sclerotiorum. Upon infection with S. sclerotiorum, these elicitors can inspire the host plant to produce an immune response against this invading pathogen. Interestingly, a fungus-secreted protein, the cerato-platanin protein (CP), acts as a MAMP to elicit plant defense responses (Djonovic et al., 2006; Seidl et al., 2006; Yang et al., 2009; Frias et al., 2011), but S. sclerotiorum CP1 (SsCP1) interacts with PR1, overexpression of which shows more resistance to S. sclerotiorum, in the apoplast to facilitate infection by S. sclerotiorum (Yang et al., 2018). CP also induces synthesis of ROS and triggers local cell death (Pazzagli et al., 1999; Yang et al., 2009; Frias et al., 2011), suggesting that the MTI triggered by CP may be involved in the production of HR, which may also facilitate the infection by S. sclerotiorum with necrotrophic lifestyle. In addition, a recent study showed that treatment of N. benthamiana with VmE02 (a small cysteine-rich protein), a novel PAMP from the necrotrophic fungus Valsa mali, enhances plant resistance to S. sclerotiorum and Phytophthora capsici. (Nie et al., 2019).
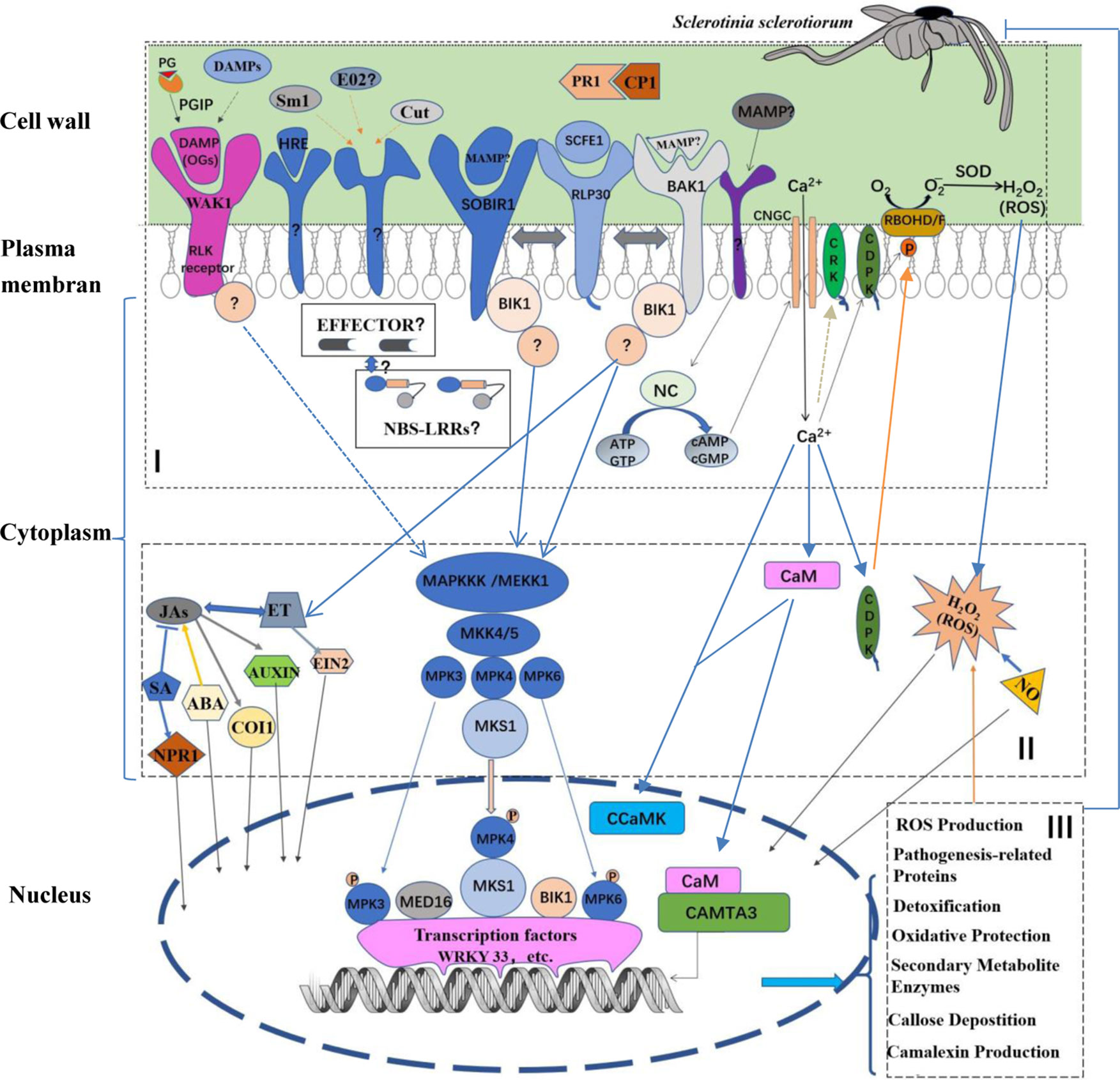
Figure 1 Schematic model of plant innate immune defense process against Sclerotinia sclerotiorum infection. In the model, plant defense process can be split into (I) recognition, (II) signal-transduction, and (III) defense response. (I) Plant recognition of MAMPs (or PAMPs) and DAMPs are detected by PRRs in apoplast. The SCFE1–RLP30/SOBIR1/BAK1 recognition, a classical PAMP–PRR recognition mode, is shown. PRRs of other MAMPs and the recognition of OGs, typical DAMPs, by WAK1 remain to be identified. Additionally, potential effectors secreted by S. sclerotiorum may recognized by potential NBS-LRRs R protein in cytoplasm. (II) The signal-transduction is performed by the MAP kinase cascade or MAPK-independent pathways. In response to S. sclerotiorum infection, the MAPK-independent pathways, such as calcium (Ca) signaling pathway, are also involved. In the Ca signaling pathway, firstly the NC may be activated by unknown PAMP(s) to produce cAMP and cGMP. Subsequently, the two cyclic nucleotides activate Ca2+ channels such as CNGCs, resulting in a Ca2+ elevation in the cytoplasm. Then the signal of the cytosolic Ca2+ elevation is transduced by Ca2+ sensor proteins, such as CaMs, CDPKs as well as CRKs. Further, CDPKs phosphorylate and activate RBOHD/F, resulting in ROS accumulation. However, substrates of CRKs are not yet identified under the pathogen infection. Importantly, many signal molecules, including SA, JA, ET, auxin, ABA, NO as well as ROS, play key roles in the signal-transduction. (III) These signals direct various nuclear proteins including transcription factors, transcription activators as well as protein kinases, such as WRKY33, CAMTA3 and CCaMK, to activate specific defense responses, including pathogenesis-related proteins, ROS production, detoxification, oxidative protection, callose deposition camalexin production and other specialized secondary metabolites. Partition of the defense process is generally according to the ref. (Corwin and Kliebenstein, 2017), and the model is expounded in the following text. PR1, pathogenesis-related protein1; MAMPs, microbial associated molecular patterns; DAMPs, damage-associated molecular patterns; PG, polygalacturonases; RLPs, receptor-like proteins; BAK1, BR insensitive1-associated receptor kinase1; SOBIR1, suppressor of BIR1 BIR1-1; WAK1, wall-associated kinase 1; RLK, receptor-like kinase; BIK1, Botrytis-induced kinase1; NBS-LRR, nucleotide-binding site-leucinerich repeat; MAPK, mitogen activated protein kinase; MAPKKKs, MAPK kinase kinases; MAPKKs, MAPK kinases; MKS1, MPK4 substrate 1; SA, salicylate; JAs, jasmonates; ET, ethylene; EIN2, ethylene insensitive 2; ABA, abscisic acid; NO, nitric oxide; NPR1, nonexpressor of PR1; COI1, the coronatine- insensitive protein 1, and MED16, mediator complex subunit; NC, nucleotidyl cyclase; CNGC, cyclic nucleotide gated channel; ATP, adenosine triphosphate; GTP, guanosine triphosphate; cAMP, cyclic adenosine monophosphate; cGMP, cyclic guanosine monophosphate; NC, nucleotidyl cyclase; CaM, calmodulin; CDPK, calcium-dependent protein kinase; CRK, CDPK-related kinase; RBOH, respiratory burst oxidase homologue; SOD, superoxide dismutase; CCaMK, Ca and CaM-dependent protein kinase; CAMTA3, CaM-binding transcription activator3.
Host damage-associated molecular patterns (DAMPs) also activate protective immune signaling in plants. Oligogalaturonides (OGs) are representative DAMPs. They are released from plant cell wall after infection of pathogens like Botrytis cinerea and S. sclerotiorum, function as DAMPs, induce plant defense response (Ferrari et al., 2013). OGs are recognized by WAK1 (WALL-ASSOCIATED KINASE 1) who is capable of activating the kinase domain of the elongation factor Tu (EF-TU) receptor (EFR). So OGs seem to be involved in MTI responses to necrotrophic pathogens (Mengiste, 2012). Interestingly, polygalacturonase inhibitor proteins (PGIPs) seem to shift a breakdown process toward generating OGs. In A. thaliana, the overexpression of B. napus PGIP2 inhibited necrotic lesions, but had no long-term effects on S. sclerotiorum disease progression (Bashi et al., 2013). Recently, a study showed that the ectopic expression of OsPGIP2 in rapeseed conferred the resistance to S. sclerotiorum at both the seedling and adult stages (Wang et al., 2018).
RLP30 belongs to a specific class of plasma membrane (PM)-localized receptors that carry an extracellular ligand-binding domain, but lack any obvious cytoplasmic signaling-competent moiety (Fritz-Laylin et al., 2005; Wang et al., 2010; Zhang et al., 2013; Liebrand et al., 2014), so following recognition of MAMPs by RLP30, the transduction of the extracellular signals to intracellular targets is dependent on additional signaling partners. Two transmembrane RLKs (receptor-like kinases), BAK1 (Brassinosteroid insensitive1-associated receptor kinase1) and SOBIR1/EVR (SUPPRESSOR OF BIR1-1/EVERSHED) are identified as RLP30 interacting and signaling partners, and are responsible for subsequent signal-transduction to cytoplasmic targets (Zhang et al., 2013). For example, activation of mitogen activated protein kinases (MAPKs), including MAPK 3, 4, and 6, by the SCFE1-containing fraction was impaired in bak and sobir1 mutant plants, indicating that BAK1 and SOBIR1 are required for the signal transduction from RLP30 to the intracellular MAPK kinase kinase(s) (MAPKKK). MAPKKK activates the MAP kinase kinase(s) (MAPKK), by which the MAP kinase(s) (MAPK or MPK) is sequential activated by MAPKK. Additionally, SCFE1-dependent production of ethylene (ET), an important signaling molecule, was abolished in bak1-5 mutant plants, and mutants of RLP30, BAK1, and SOBIR1 are more susceptible to S. sclerotiorum and the related fungus Botrytis cinerea (Zhang et al., 2013). Further, the signaling from RLP30/SOBIR1/BAK1 to MAPKKK might require the RLCK (receptor-like cytoplasmic kinase), because Botrytis-induced kinase1 (BIK1), a RLCK, can cooperate with BAK1 to regulate constitutive immunity and cell death in A. thaliana (Eckardt, 2011), which need to be further identified in the pathosystem between S. sclerotiorum and its hosts. BIK1 was initially identified as a plasma membrane localized protein (Veronese et al., 2006). A recent study showed that BIK1 can also localize to the nucleus and interact directly with WRKY transcription factors, such as WRKY33, to be involved in the jasmonic acid (JA) and salicylic acid (SA) regulation (Lal et al., 2018).
MAPKs play an important role in signal transduction from cytoplasmic to nucleus, leading to the rapid changes of transcription depending on a series of phosphorylation. It has been reported that the MAP kinase 4 (MPK4) and the WRKY33 transcription factor play an important role in resistance to S. sclerotiorum as well as Botrytis cinerea (Petersen et al., 2000; Brodersen et al., 2006; Wang et al., 2009; Wang et al., 2014; Wang et al., 2015a). MPK4 is localized in cytoplasm and nuclei, and the MPK4 substrate MKS1 is primarily localized in nuclei (Andreasson et al., 2005). In the absence of pathogens, depending on MKS1, MPK4 can exist in nuclear composited with WRKY33 (Qiu et al., 2008; Wang et al., 2014). Challenge with pathogens or MAMPs leads to the activation of MPK4 and phosphorylation of MKS1. Subsequently, the MKS1–WRKY33 complex is released from MPK4, and WRKY33 activates the expression of PHYTOALEXIN DEFICIENT 3 (PAD3) and Cytochrome P450 71A13 (CYP71A13) (Qiu et al., 2008), both of which are involved in antimicrobial camalexin synthesis (Birkenbihl et al., 2012; Liu et al., 2015). WRKY33 also positively regulates the expression of JA/ET defense pathway marker gene PLANT DEFENSIN1.2 (PDF1.2) and OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF59 (ORA59) (Birkenbihl et al., 2012; Wang et al., 2014; Wang et al., 2015a). However, how WRKY33 is released from the MKS1-WRKY33 complex remains to be determined. The MKS1–WRKY33 complex can exist both before and after phosphatase treatments of MKS1 in A. thaliana, and phospho-mimics, non-phosphorylatable, and wild-type forms of MKS1 bind WRKY33 equally well in yeast, suggesting that the complex exists independently of MKS1 phosphorylation (Qiu et al., 2008). It has been shown that WRKY33 can be phosphorylated by MPK3/MPK6 in vivo in response to the pathogen infection in A. thaliana, which may be a hint for the release of WRKY33. A recent study has shown that B. napus MPK3 is a key regulator in defense responses to S. sclerotiorum (Wang et al., 2019).
The A. thaliana mediator complex subunit MED16 can also physically interact with WRKY33 in yeast and in planta (Wang et al., 2015a). The mediator complex, a multiprotein co-activator scaffold acting as a bridge between RNA polymerase II (RNAPII) and transcription factors, is involved in host transcriptional reprogramming after pathogen challenge, being paramount in the establishment of plant defense (Samanta and Thakur, 2015). Mutations in MED16 subunit abolished induction of JA/ET cross-talk genes and reduced resistance to S. sclerotiorum and B. cinerea (Kidd et al., 2009; Zhang et al., 2012; Wang et al., 2015a). Thus, these data suggest that WRKY33 seem to be a recruitment center in defense to S. sclerotiorum, as well as B. cinerea.
In addition to the MAPK pathway, calcium (Ca) signaling pathway has been reported to play important roles in plant defense against S. sclerotiorum. A Ca2+ elevation in the cytoplasm is known to be an important early event in plant cell perception of pathogen invasion (Dangl et al., 1996; Ma and Berkowitz, 2011). Plant cyclic nucleotide gated ion channels (CNGCs) provide a pathway for Ca2+ conductance across the plasma membrane (PM) and facilitate cytosolic Ca2+ elevation in response to pathogen signals. In the case of the interaction of plant host with S. sclerotiorum, pharmacological assays showed that the putative CNGC activators cGMP (cyclic adenosine monophosphate) and cAMP (cyclic guanosine monophosphate) enhanced resistance of tomato (Solanum lycopersicum) against this pathogen (Saand et al., 2015a). Further, two tomato CNGC genes, SlCNGC1 and SlCNGC6, were reported to play a positive role in tomato resistance to S. sclerotiorum (Saand et al., 2015b). Interestingly, other CNGC genes such as SlCNGC17 and SlCNGC18, exhibited negative roles in this resistance (Saand et al., 2015a). Transient elevations of the Ca2+ concentration in the cytoplasm can be sensed by various Ca2+ sensor proteins including calmodulins (CaMs) and calcium-dependent protein kinases (CDPKs) (Ranty et al., 2006; Hamel et al., 2014). It has been reported that a CaM-binding transcription activator CAMTA3 negatively regulated PTI probably by directly targeting BAK1 under the S. sclerotiorum infection (Rahman et al., 2016). In contrast, Ca and CaM-dependent protein kinase (CCaMK), a nuclear-localized protein, positively regulates resistance to S. sclerotiorum via promoting H2O2 accumulation (Wang et al., 2015b). The plasma membrane- and the cytosol-localized CDPKs can also positively regulate H2O2 accumulation via phosphorylating and activating RBOHD/F (Liu and He, 2016). However, knock-down of a set of CDPKs did not affect resistance to S. sclerotiorum in tomato (Wang et al., 2016). In contrast, SlCRK6, a CDPK-related kinase (CRK) that probably localizes at the plasma membrane (Leclercq et al., 2005; Rigo et al., 2013), plays a positive role in this resistance (Wang et al., 2016). The CRK is another type of protein kinase closely related to CDPKs in structure. Unlike CDPKs, however, substrates of CRKs are not yet identified under the pathogen infection. Together, the increasing evidence indicated roles for Ca2+ signaling in resistance to S. sclerotiorum. Activation of Ca2+ channels, such as CNGC, requires the involvement of nucleotidyl cyclase (NC) which generates Ca2+ channel activators: cAMP and cGMP, together referred to as the cyclic nucleotide signaling system (Swiezawska et al., 2018). However, how plant NC is activated in response to S. sclerotiorum infection remains to be identified.
In fact, a large body of data has implied that plant defense to S. sclerotiorum is complex and involves multiple signaling pathways. This is supported by a recent study on the protein profile of B. napus in response to S. sclerotiorum. The study indicated that plant defense to this pathogen involves various biological processes including redox homeostasis, lipid signaling, calcium signaling, histone, and DNA methylation-mediated transcription regulation and defense-related proteins such as defensin and defensin-like proteins as well as cyanate lyase (Cao et al., 2016a). Moreover, it was reported plant defense to S. sclerotiorum is also regulated at miRNA level and probably involves PTGS (Post-Transcriptional Gene Silencing) (Cao et al., 2016b). More recently, glycolate oxidase genes, encoding crucial enzymes in photorespiration, were reported to be involved in resistance to S. sclerotiorum (Xu et al., 2018b), suggesting a potential role for photorespiration in this resistance. In addition, defense against S. sclerotiorum was found to be stage/phase-associated, and the phytohormones SA, ET, JA and abscisic acid (ABA) likely play an essential, but pathosystem-dependent, role in leaf stage associated resistance (Xu et al., 2018a).
Signaling Molecules and Their Crosstalk in Plant Response to S. Sclerotiorum
Initiation of MTI, as well as ETI, is correlated with a complex network of defense signaling pathways, resulting in defensive cellular responses and changes in expression of thousands of host genes (Zipfel et al., 2006; Zipfel, 2014; Lewis et al., 2015). The hormones, such as JA, SA, ET, ABA, and auxin, function as signaling molecules to transduce the perception signal through a huge and complex signaling network, and then complex defense responses are activated.
Early studies on global profiles of host gene expression in response to infection with S. sclerotiorum indicated that genes associated with JA and ET signal are induced, but few of SA responsive genes are identified (Zhao et al., 2007; Zhao et al., 2009). In plant immune responses, SA can antagonize the JA signaling pathway, which has already been identified in plants during S. sclerotiorum infection (Wang et al., 2012), and JA and ET are well known to synergistically induce defense responses and cooperate in resistance against pathogens (Thomma et al., 2001; Kunkel and Brooks, 2002; Glazebrook, 2005). However, a study on Arabidopsis mutants, including npr1 (nonexpressor of PR genes1) being defective in SA perception, coi1-2 impaired in JA perception, and ein2 (ethylene insensitive), revealed that defense against S. sclerotiorum in A. thaliana is dependent on all of SA-, JA- and ET-mediated signaling (Guo and Stotz, 2007). The conclusion is supported by results from B. napus (Wang et al., 2012; Nováková et al., 2014), and it is suggested that defense against S. sclerotiorum in B. napus is associated with a sequential activation of SA and JA signaling (Wang et al., 2012). This can be explained by the new model depicting the lifestyle transition of the pathogen from biotrophic to necrotrophic growth (Kabbage et al., 2015), because it has been suggested that the SA signaling protects against biotrophs, while JA/ET signaling activates defense responses against necrotrophs (Penninckx et al., 1996; Thomma et al., 1998; Glazebrook, 2005), as well as herbivorous insects (Howe and Jander, 2008; War et al., 2012). In favor of a role of ET signaling in this defense, is the recent discovery that B. napus MPK3, a positive regulator of ET signaling, positive regulates resistance to S. sclerotiorum (Wang et al., 2019).
Roles of SA and JA signaling in defense against S. sclerotiorum are challenging. By using three different mutants or transgenic lines impaired in SA production (sid1/eds5, sid2/eds16, and nahG) and one impaired in SA signaling (npr1-1), Perchepied et al. (2010) suggested that resistance to S. sclerotiorum is not dependent on SA. And they thought that the availability of bioactive jasmonates is not essential for resistance to S. sclerotiorum, because jar1-1, impaired in biosynthesis of jasmonoyl-L-amino acid, showed a completely wild-type phenotype in response to S. sclerotiorum (Perchepied et al., 2010), although coi1-1 was found to be highly susceptible to S. sclerotiorum (Guo and Stotz, 2007). However, studies have shown that SA, BTH (benzothiadiazole, a SA functional analogue) and MeJA application to B. napus leaves significantly results in increased resistance to S. sclerotiorum (Wang et al., 2012; Nováková et al., 2014). These results suggest that the role of SA or JA signaling in defense against S. sclerotiorum may vary depending on the plant species or that possible crosstalk between these signaling pathways and other regulatory pathways is likely to play a role in this defense (Perchepied et al., 2010). For example, an analysis on both jasmonate-dependent and COI1-independent defense responses against S. sclerotiorum showed that auxin signaling regulates the COI1-independent defense response pathway and plays an important role in resistance to this pathogen in A. thaliana (Stotz et al., 2011).
It has been suggested that ABA signaling would interlink with SA, JA, and ET signaling to affect pathogen resistance via a complex interplay of mechanisms (Mauch-Mani and Mauch, 2005; Adie et al., 2007; Kaliff et al., 2007). For example, ABA is required for biosynthesis of JA and expression of JA signaling defense gene in response to infection with Pythium spp., a necrotrophic pathogen (Adie et al., 2007). In the case of resistance to S. sclerotiorum, A. thaliana mutants (aba3-2, abi1-1, and abi2-1), being defective in ABA biosynthesis or perception, showed a complete loss of resistance to S. sclerotiorum (Guimaraes and Stotz, 2004; Perchepied et al., 2010). ABA-mediated regulation of guard cells is a major mechanism of this defense in these interactions, which is compatible with two views. One is that OA, the essential pathogenicity factor for S. sclerotiorum, was concluded to favor infection by both inducing stomatal opening and inhibiting ABA-mediated stomatal closure (Guimaraes and Stotz, 2004; Billon-Grand et al., 2012). Another is that the decreased ambient pH caused by S. sclerotiorum infection results in increased synthesis of photoprotective compounds of the xanthophyll cycle that serve as precursors for ABA synthesis, and thus the decrease in de novo ABA biosynthesis through decreasing xanthophyll precursors was suggested to account for enhanced plant susceptibility to S. sclerotiorum (Zhou et al., 2015). These observations also suggest that ABA signaling may be bi-directionally regulated by both S. sclerotiorum and its host plants, which needs to be fully understood by further studies. However, in other two A. thaliana mutants (aba2-3 or abi5), they didn’t show any significant changes in their response to S. sclerotiorum compared to control plants (Perchepied et al., 2010), suggesting that it remains unclear whether ABA signaling is required for this resistance.
In addition to these hormones, NO (nitric oxide) and ROSs (reactive oxygen species) are important signaling molecules (Kunkel and Brooks, 2002). Nitric Oxide (NO) is rapidly generated after recognition of pathogens (Wendehenne et al., 2004). The NO-impaired mutants showed an extremely susceptible phenotype to S. sclerotiorum, revealing the role of NO in resistance to this pathogen (Perchepied et al., 2010). It is suggested that the major role of NO in this resistance is multifaceted, such as regulating defense gene expression, interfering with the ROS signaling pathway, or modulating cell death (Perchepied et al., 2010). It has been shown that inhibition of host cell death by expressing negative regulators of mammalian apoptosis in transgenic tobacco plants leads to markedly enhanced resistance to S. sclerotiorum (Dickman et al., 2001); when ROS induction is inhibited, apoptotic-like cell death induced by oxalic acid does not occur and the PCD (programmed cell death) response is required for disease development (Kim et al., 2008). Further, the control of cell death governs the outcome of the S. sclerotiorum–plant interaction (Kabbage et al., 2013) and, once infection is established, the necrotrophic S. sclerotiorum induces the generation of plant ROS, leading to PCD of host tissue, the result of which is of direct benefit to the pathogen (Williams et al., 2011). Further experiments should be carried out to fully investigate how NO interference with the ROS signaling pathway to inhibit PCD in the context of S. sclerotiorum–plant interactions.
Polygenic Architecture of Quantitative Resistance to S. sclerotiorum
Plant immune response to necrotrophs is governed by a complex interplay of minor-effect genes, which results in a full continuum of resistance phenotypes in natural plant populations, designated as quantitative disease resistance (QDR) (Roux et al., 2014). The genetic determinants of QDR are complex, and the underlying genetic components can be common with, but are generally not limited to, PTI and ETI response genes (Iakovidis et al., 2016).
In the case of S. sclerotiorum, its host plants, such as the model plant A. thaliana, B. napus and soybean, show symptoms ranging from high susceptibility to relative tolerance to the pathogen, corresponding to a typical QDR response (Kim et al., 1999; Chen and Wang, 2005; Liu et al., 2005; Perchepied et al., 2010), which involves allelic variation at different quantitative trait loci (QTLs) since the continuous distribution of heritable phenotypes must result from combinations of genetic loci (Corwin and Kliebenstein, 2017). However, although a large body of mapping information on QTLs is available for the QDR to the pathogen (Bert et al., 2002; Zhao and Meng, 2003; Bert et al., 2004; Micic et al., 2004; Micic et al., 2005a; Micic et al., 2005b; Ronicke et al., 2005; Zhao et al., 2006; Yin et al., 2010; Behla, 2017; Wu et al., 2013; Wei et al., 2014), relatively little is known about the molecular basis of QTLs.
Recent research technologies have developed efficient omic tools to better understand the genetic and molecular mechanisms regarding plant QDR to S. sclerotiorum. Chalhoub et al. (2014) predicted a total of 181 and 245 putative NBS-LRR resistance genes on the A and C subgenomes of the B. napus genome through examining large-scale genomic data. Interestingly, from these genes, Li et al. (2015) found a total of 26 candidate NBS-LRR genes associated with resistance to S. sclerotiorum through integrating and comparing QTLs for resistance to this pathogen from previous mapping efforts. Correspondingly, a bioinformatic study revealed that the S. sclerotiorum genome encodes a large set of candidate effector proteins (Guyon et al., 2014). R gene-mediated resistance commonly results in rapid cellular desiccation and death at the site of attempted infection that constitutes a hypersensitive response (HR) (Wright and Beattie, 2004; Dodds and Rathjen, 2010). Consistently, some researchers claimed that they observed HR-like lesions on S. sclerotiorum-inoculated stems (Uloth et al., 2013; Uloth et al., 2015; Ming et al., 2016) or cotyledons (Garg et al., 2008; Garg et al., 2010; Uloth et al., 2014; Ge et al., 2015). These data suggested that R-mediated resistance seems to exist in the interaction of B. napus with S. sclerotiorum. Considering the continuous distribution of disease resistance phenotype in host populations, a typical characterization of QDR, roles of these candidate R genes in this resistance need to be confirmed further, because R-mediated resistance can be seen as an extreme of the phenotypic spectrum, in which the switch from susceptibility to resistance in plant populations is reduced to a minimum of detectable transition states (Roux et al., 2014).
With the development of high-throughput sequencing technology, genome-wide association study (GWAS) based on linkage disequilibrium (LD), has emerged as an important tool for identifying small-to-moderate effect loci associated with resistance to S. sclerotiorum. For example, based on GWAS, two recent studies identified two QDR genes, coding for the POQR prolyl oligo peptidase and the actin-related protein complex isoform 4, respectively, for S. sclerotiorum in the model plant A. thaliana (Badet et al., 2017; Badet et al., 2019). However, in most cases of crops, GWAS cannot lead directly to the gene(s) at a given locus because of insufficient marker density and linkage disequilibrium. Thus, GWAS data are usually combined with other omic experiments, such as microarray study, RNA sequencing (RNAseq), to interpret the results, which can increase the confidence in identifying candidate defense-associated (CDA) genes (Corwin and Kliebenstein, 2017). To date, this approach has been employed to interpret GWAS results associated with resistance to S. sclerotiorum in two crops, B. napus and soyabean. On the basis of data from these reports from 2015–2018 in the two crops, we focus on these candidate genes related with defense mechanisms, to our knowledge (Table 1), and find out the following features in resistance to S. sclerotiorum. Additionally, bean CDA genes identified by using a QTL meta-analysis also are considered (Table 1).
i. Resistance to S. sclerotiorum is determined by minor QTLs. These phenotypic contributions of the GWAS-identified loci was low, with each locus explaining less than 10% of the observed phenotypic variance in resistance to S. sclerotiorum. This is supported by QTL mapping in which resistance to S. sclerotiorum is a trait with very complex genetic underpinnings determined by multiple minor QTLs.
ii. The predominant group of genes linked to GWAS-identified loci as potential causal genes were those involved in downstream defense responses including pathogenesis-related proteins, ROS production, detoxification, oxidative protection and secondary metabolite enzymes. This implies that QDR to S. sclerotiorum is a function of a variety of cellular processes and not simply pathogen-detection and signal-transduction.
iii. Interestingly, many potential resistance (R) genes are identified as potential causal genes by GWAS. It has been suggested that upstream signaling components, such as R protein, in the plant pathogen response are typically encoded by medium-to-large-effect loci, which is supported by the large number of studies that investigated the quantitative genetics of wheat resistance to wheat stripe rust (Fu et al., 2009), Arabidopsis resistance to Xanthomonas campestris (Huard-Chauveau et al., 2013) and Fusarium oxysporum (Diener and Ausubel, 2005; Shen and Diener, 2013) and rice resistance to Magnaporthe oryzae (Ballini et al., 2008; Miah et al., 2013; Kang et al., 2016; Raboin et al., 2016). In the case of resistance to S. sclerotiorum, all of these potential candidate R genes are located in small-effect loci. Is ETI response to S. sclerotiorum, if it exists, a quantitative trait? A recent report has shown that an ETI response in Arabidopsis resistance to Pseudomonas syringae, a hemi-biotroph, is a quantitative trait, in which a single effector, HopAM1, was used to identify quantitative natural variation in the response to this effector (Iakovidis et al., 2016). Considering the identified large set of candidate effector protein from S. sclerotiorum genome (Guyon et al., 2014) and a potential double feeding lifestyle of S. sclerotiorum, it is possible to use these candidate effectors to test if there is quantitative variation in ETI response to S. sclerotiorum signals.
iv. In the case of B. napus, an amphidiploid formed by interspecific hybridization of the two diploid species B. rapa (AA,n = 10) and B. oleracea (CC, n = 9), the GWAS data showed that the majority of potential causal genes for the pathogen were identified in the C genome (C9 and C6), but not in A genome (Table 1), although it has been known that putative resistance-related genes in C genome also are observed in the syntenic region on A genome (Mei et al., 2013). These observations suggested that B. oleracea, not B. rapa, may be a good source of QDR genes for S. sclerotiorum. Further, It has been reported that a few Chinese B. oleracea var. capitata genotypes exhibit high level stem and leaf resistances to S. sclerotiorum (Mei et al., 2011; Ming et al., 2016). Contrastly, there is no genome specificity of QDR genes for this pathogen in soybean.
v. Comparing with existing biparental populations, the GWAS populations use more lines and also utilize the increased number of meiotic generations to provide increased recombination and potentially increased mapping resolution (Nordborg et al., 2002; Nordborg et al., 2005; Nordborg and Weigel, 2008; Atwell et al., 2010; Alonso-Blanco et al., 2016). However, so far, the GWAS-identified loci/genes for resistance to S sclerotiorum in each analysis collectively explained a small portion of the phenotypic variation, and few loci/genes could be detected repeatedly in different populations for the same species (Table 1). This suggests that these GWASs is still largely underpowered given the number of accessions and the effect of residual population structure or adaptive genetic variation unaccounted for in these studies (Platt et al., 2010; Brachi et al., 2015). Thus, future studies with even more powerful populations are required.
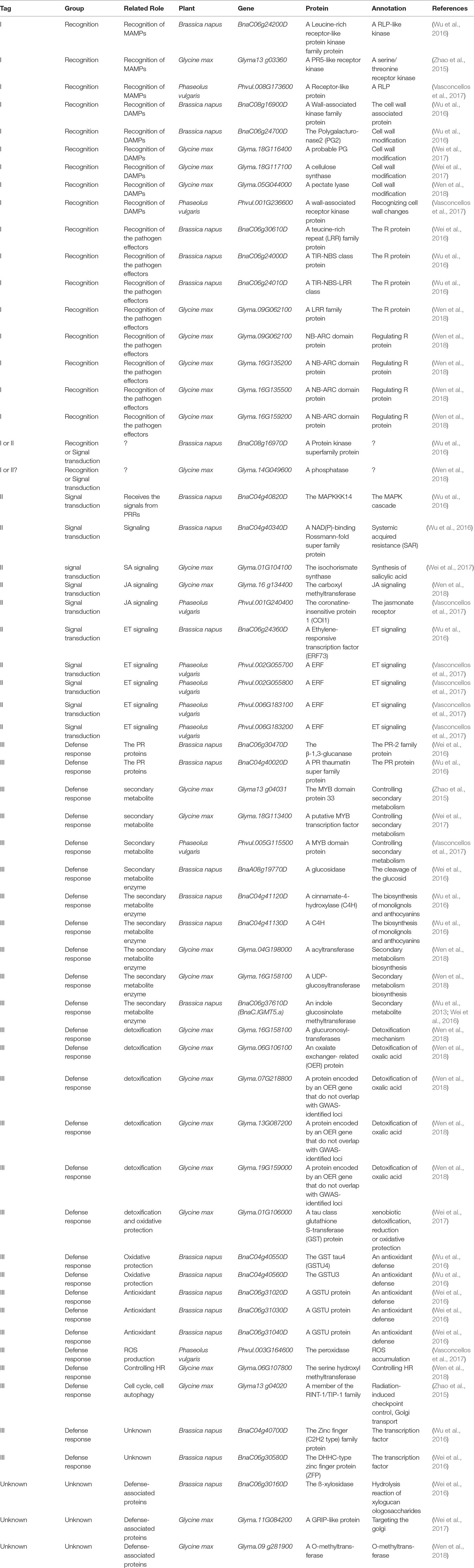
Table 1 Candidate defense-related genes mapped by genome-wide association study (GWAS) combined with other omic experiments.
Conclusions
As S. sclerotiorum has the broad host range, diverse infection modes and potential double lifestyle of nutrient acquisition, scientists have been trying to dissect the S. sclerotiorum–host interaction and to understand the mechanisms of QDR to the pathogen. Current knowledge on plant defense to S. sclerotiorum has set up basic framework including recognition of the pathogen, signaling and defense response, but a lot of effort will be needed to enrich the contents. Pyramiding the QDR genes appears to be a promising strategy for durable resistance, but considering the pyramiding power in the gene number, key question is which mechanisms play an important role in this QDR. Thus, the assessment of the phenotypic contribution of QDR genes requires robust and accurate methods. The choice of a truly quantitative readout, which can be precisely and reliably measured, may be essential for both the accuracy of QTL mapping and the validation of QDR gene function. High-throughput omic technologies will broaden the choice of more powerful mapping population(s) and data analysis strategies available for accurate and reliable QDR gene identification.
Author Contributions
ZW conceptualized and drafted the manuscript, with additions from X-LT, L-YM, JC, Y-LL, L-ND, K-MZ, and Y-HY. The figure was made by L-YM and ZW, and the table was prepared by ZW. All authors read and approved the final manuscript.
Funding
The authors acknowledge funding support from National Natural Science Foundation of China (Grant Nos. 31771836).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adie, B. A., Perez-Perez, J., Perez-Perez, M. M., Godoy, M., Sanchez-Serrano, J. J., Schmelz, E. A., et al. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19 (5), 1665–1681. doi: 10.1105/tpc.106.048041
Alonso-Blanco, C., Andrade, J., Becker, C., Bemm, F., Bergelson, J., Borgwardt, K., et al. (2016). 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166 (2), 481–491. doi: 10.1016/j.cell.2016.05.063
Andreasson, E., Jenkins, T., Brodersen, P., Thorgrimsen, S., Petersen, N. H., Zhu, S., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24 (14), 2579–2589. doi: 10.1038/sj.emboj.7600737
Atwell, S., Huang, Y. S., Vilhjalmsson, B. J., Willems, G., Horton, M., Li, Y., et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465 (7298), 627–631. doi: 10.1038/nature08800
Badet, T., Leger, O., Barascud, M., Voisin, D., Sadon, P., Vincent, R., et al. (2019). Expression polymorphism at the ARPC4 locus links the actin cytoskeleton with quantitative disease resistance to Sclerotinia sclerotiorum in Arabidopsis thaliana. New Phytol. 222 (1), 480–496. doi: 10.1111/nph.15580
Badet, T., Voisin, D., Mbengue, M., Barascud, M., Sucher, J., Sadon, P., et al. (2017). Parallel evolution of the POQR prolyl oligo peptidase gene conferring plant quantitative disease resistance. PLoS Genet. 13 (12), e1007143. doi: 10.1371/journal.pgen.1007143
Ballini, E., Morel, J. B., Droc, G., Price, A., Courtois, B., Notteghem, J. L., et al. (2008). A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant Microbe Interact. 21 (7), 859–868. doi: 10.1094/MPMI-21-7-0859
Bashi, Z. D., Rimmer, S. R., Khachatourians, G. G., Hegedus, D. D. (2013). Brassica napus polygalacturonase inhibitor proteins inhibit Sclerotinia sclerotiorum polygalacturonase enzymatic and necrotizing activities and delay symptoms in transgenic plants. Can. J. Microbiol. 59 (2), 79–86. doi: 10.1139/cjm-2012-0352
Behla, R., Hirani, A. H., Zelmer, C. D., Yu, F., Fernando, W. G. D., McVetty, P., Li, G. (2017). Identification of common QTL for resistance to Sclerotinia sclerotiorum in three doubled haploid populations of Brassica napus (L.). Euphytica, 213(11):260. doi: 10.1007/s10681-017-2047-5
Bert, P. F., Dechamp-Guillaume, G., Serre, F., Jouan, I., de Labrouhe, D. T., Nicolas, P., et al. (2004). Comparative genetic analysis of quantitative traits in sunflower (Helianthus annuus L.) 3. Characterisation of QTL involved in resistance to Sclerotinia sclerotiorum and Phoma macdonaldi. Theor. Appl. Genet. 109 (4), 865–874. doi: 10.1007/s00122-004-1701-1
Bert, P. F., Jouan, I., De Labrouhe, D. T., Serre, F., Nicolas, P., Vear, F. (2002). Comparative genetic analysis of quantitative traits in sunflower (Helianthus annuus L.) 1. QTL involved in resistance to Sclerotinia sclerotiorum and Diaporthe helianthi. Theor. Appl. Genet. 105 (6-7), 985–993. doi: 10.1007/s00122-002-1004-3
Bertinetti, C., Ugalde, R. A. (1996). Studies on the response of carrot cells to a Sclerotinia sclerotiorum elicitor: induction of the expression of an extracellular glycoprotein mRNA. Mol. Plant Microbe Interact. 9 (7), 658–663. doi: 10.1094/MPMI-9-0658
Billon-Grand, G., Rascle, C., Droux, M., Rollins, J. A., Poussereau, N. (2012). pH modulation differs during sunflower cotyledon colonization by the two closely related necrotrophic fungi Botrytis cinerea and Sclerotinia sclerotiorum. Mol. Plant Pathol. 13 (6), 568–578. doi: 10.1111/j.1364-3703.2011.00772.x
Birkenbihl, R. P., Diezel, C., Somssich, I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159 (1), 266–285. doi: 10.1104/pp.111.192641
Boland, G. J., Hall, R. (1994). Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 16, 93–108. doi: 10.1080/07060669409500766
Bolton, M. D., Thomma, B. P., Nelson, B. D. (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7 (1), 1. doi: 10.1111/j.1364-3703.2005.00316.x
Brachi, B., Meyer, C. G., Villoutreix, R., Platt, A., Morton, T. C., Roux, F., et al. (2015). Coselected genes determine adaptive variation in herbivore resistance throughout the native range of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 112 (13), 4032–4037. doi: 10.1073/pnas.1421416112
Brodersen, P., Petersen, M., Bjorn Nielsen, H., Zhu, S., Newman, M. A., Shokat, K. M., et al. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47 (4), 532–546. doi: 10.1111/j.1365-313X.2006.02806.x
Cao, J. Y., Xu, Y. P., Cai, X. Z. (2016a). TMT-based quantitative proteomics analyses reveal novel defense mechanisms of Brassica napus against the devastating necrotrophic pathogen Sclerotinia sclerotiorum. J. Proteomics 143, 265–277. doi: 10.1016/j.jprot.2016.03.006
Cao, J. Y., Xu, Y. P., Zhao, L., Li, S. S., Cai, X. Z. (2016b). Tight regulation of the interaction between Brassica napus and Sclerotinia sclerotiorum at the microRNA level. Plant Mol. Biol. 92 (1-2), 39–55. doi: 10.1007/s11103-016-0494-3
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345 (6199), 950–953. doi: 10.1126/science.1253435
Chen, Y., Wang, D. (2005). Two convenient methods to evaluate soybean for resistance to Sclerotinia sclerotiorum. Plant Dis 89 (12), 1268–1272. doi: 10.1094/PD-89-1268
Corwin, J. A., Kliebenstein, D. J. (2017). Quantitative Resistance: more than just perception of a pathogen. Plant Cell 29 (4), 655–665. doi: 10.1105/tpc.16.00915
Dangl, J. L., Dietrich, R. A., Richberg, M. H. (1996). Death don’t have no mercy: cell death programs in plant-microbe interactions. Plant Cell 8 (10), 1793–1807. doi: 10.1105/tpc.8.10.1793
Dickman, M. B., Park, Y. K., Oltersdorf, T., Li, W., Clemente, T., French, R. (2001). Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc. Natl. Acad. Sci. U.S.A. 98 (12), 6957–6962. doi: 10.1073/pnas.091108998
Diener, A. C., Ausubel, F. M. (2005). RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171 (1), 305–321. doi: 10.1534/genetics.105.042218
Djonovic, S., Pozo, M. J., Dangott, L. J., Howell, C. R., Kenerley, C. M. (2006). Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant Microbe Interact. 19 (8), 838–853. doi: 10.1094/MPMI-19-0838
Dodds, P. N., Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11 (8), 539–548. doi: 10.1038/nrg2812
Eckardt, N. A. (2011). BIK1 function in plant growth and defense signaling. Plant Cell 23 (8), 2806. doi: 10.1105/tpc.111.230811
Ferrari, S., Savatin, D. V., Sicilia, F., Gramegna, G., Cervone, F., Lorenzo, G. D. (2013). Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 4, 49. doi: 10.3389/fpls.2013.00049
Flor, H. H. (1971). Current Status of the gene-for-gene concept. Ann. Rev. Phytopathol. 9 (1), 275–296. doi: 10.1146/annurev.py.09.090171.001423
Frias, M., Gonzalez, C., Brito, N. (2011). BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192 (2), 483–495. doi: 10.1111/j.1469-8137.2011.03802.x
Fritz-Laylin, L. K., Krishnamurthy, N., Tor, M., Sjolander, K. V., Jones, J. D. G. (2005). Phylogenomic analysis of the receptor-like proteins of rice and arabidopsis. Plant Physiol. 138 (2), 611–623. doi: 10.1104/pp.104.054452
Fu, D., Uauy, C., Distelfeld, A., Blechl, A., Epstein, L., Chen, X., et al. (2009). A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323 (5919), 1357–1360. doi: 10.1126/science.1166289
Fusari, C. M., Di Rienzo, J. A., Troglia, C., Nishinakamasu, V., Moreno, M. V., Maringolo, C., et al. (2012). Association mapping in sunflower for Sclerotinia head rot resistance. BMC Plant Biol. 12, 93. doi: 10.1186/1471-2229-12-93
Garg, H., Kohn, L. M., Andrew, M., Li, H., Sivasithamparam, K., Barbetti, M. J. (2010). Pathogenicity of morphologically different isolates of Sclerotinia sclerotiorum with Brassica napus and B. juncea genotypes. Eur. J. Plant Pathol. 126 (3), 305–315. doi: 10.1007/s10658-009-9547-7
Garg, H., Sivasithamparam, K., Banga, S. S., Barbetti, M. J. (2008). Cotyledon assay as a rapid and reliable method of screening for resistance against Sclerotinia sclerotiorum in Brassica napus genotypes. Australas. Plant Pathol. 37 (2), 106–111. doi: 10.1071/AP08002
Ge, X. T., You, M. P., Barbetti, M. J. (2015). Virulence differences among Sclerotinia sclerotiorum isolates determines host cotyledon resistance responses in Brassicaceae genotypes. Eur. J. Plant Pathol. 143 (3), 527–541. doi: 10.1007/s10658-015-0696-6
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Guimaraes, R. L., Stotz, H. U. (2004). Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol. 136 (3), 3703–3711. doi: 10.1104/pp.104.049650
Guo, X., Stotz, H. U. (2007). Defense against Sclerotinia sclerotiorum in Arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Mol. Plant Microbe Interact. 20 (11), 1384–1395. doi: 10.1094/MPMI-20-11-1384
Guyon, K., Balague, C., Roby, D., Raffaele, S. (2014). Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15, 336. doi: 10.1186/1471-2164-15-336
Hamel, L. P., Sheen, J., Seguin, A. (2014). Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci. 19 (2), 79–89. doi: 10.1016/j.tplants.2013.10.009
Howe, G. A., Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Huard-Chauveau, C., Perchepied, L., Debieu, M., Rivas, S., Kroj, T., Kars, I., et al. (2013). An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PLoS Genet. 9 (9), e1003766. doi: 10.1371/journal.pgen.1003766
Iakovidis, M., Teixeira, P. J., Exposito-Alonso, M., Cowper, M. G., Law, T. F., Liu, Q., et al. (2016). Effector-triggered immune response in Arabidopsis thaliana is a quantitative trait. Genetics 204 (1), 337–353. doi: 10.1534/genetics.116.190678
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444 (7117), 323–329. doi: 10.1038/nature05286
Kabbage, M., Williams, B., Dickman, M. B. (2013). Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9 (4), e1003287. doi: 10.1371/journal.ppat.1003287
Kabbage, M., Yarden, O., Dickman, M. B. (2015). Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233, 53–60. doi: 10.1016/j.plantsci.2014.12.018
Kaliff, M., Staal, J., Myrenas, M., Dixelius, C. (2007). ABA is required for Leptosphaeria maculans resistance via ABI1- and ABI4-dependent signaling. Mol. Plant Microbe Interact. 20 (4), 335–345. doi: 10.1094/MPMI-20-4-0335
Kang, H., Wang, Y., Peng, S., Zhang, Y., Xiao, Y., Wang, D., et al. (2016). Dissection of the genetic architecture of rice resistance to the blast fungus Magnaporthe oryzae. Mol. Plant Pathol. 17 (6), 959–972. doi: 10.1111/mpp.12340
Kidd, B. N., Edgar, C. I., Kumar, K. K., Aitken, E. A., Schenk, P. M., Manners, J. M., et al. (2009). The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21 (8), 2237–2252. doi: 10.1105/tpc.109.066910
Kim, H. S., Sneller, C. H., Diers, B. W. (1999). Evaluation of soybean cultivars for resistance to Sclerotinia stem rot in field environments. Crop Sci. 39, 64–68. doi: 10.2135/cropsci1999.0011183X003900010010x
Kim, K. S., Min, J. Y., Dickman, M. B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe Interact. 21 (5), 605–612. doi: 10.1094/MPMI-21-5-0605
Kunkel, B. N., Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5 (4), 325–331. doi: 10.1016/S1369-5266(02)00275-3
Lal, N. K., Nagalakshmi, U., Hurlburt, N. K., Flores, R., Bak, A., Sone, P., et al. (2018). The receptor-like cytoplasmic kinase bik1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe 23485-497 (4), e485. doi: 10.1016/j.chom.2018.03.010
Leclercq, J., Ranty, B., Sanchez-Ballesta, M. T., Li, Z., Jones, B., Jauneau, A., et al. (2005). Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. J. Exp. Bot. 56 (409), 25–35. doi: 10.1093/jxb/eri003
Lewis, L. A., Polanski, K., de Torres-Zabala, M., Jayaraman, S., Bowden, L., Moore, J., et al. (2015). Transcriptional dynamics driving mamp-triggered immunity and pathogen effector-mediated immunosuppression in Arabidopsis Leaves following infection with pseudomonas syringae pv tomato DC3000. Plant Cell 27 (11), 3038–3064. doi: 10.1105/tpc.15.00471
Li, G. Q., Huang, H. C., Miao, H. J., Erickson, R. S., Jiang, D. H., Xiao, Y. N. (2006). Biological control of sclerotinia diseases of rapeseed by aerial applications of the mycoparasite Coniothyrium minitans. Eur. J. Plant Pathol. 114, 345–355. doi: 10.1007/s10658-005-2232-6
Li, J., Zhao, Z., Hayward, A., Cheng, H., Fu, D. (2015). Integration analysis of quantitative trait loci for resistance to Sclerotinia sclerotiorumin Brassica napus. Euphytica 205, 483–489. doi: 10.1007/s10681-015-1417-0
Liebrand, T. W. H., van den Burg, H. A., Joosten, M. H. A. J. (2014). Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 19 (2), 123–132. doi: 10.1016/j.tplants.2013.10.003
Liu, S., Kracher, B., Ziegler, J., Birkenbihl, R. P., Somssich, I. E. (2015). Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife 4, e07295. doi: 10.7554/eLife.07295
Liu, S., Wang, H., Zhang, J., Fitt, B. D., Xu, Z., Evans, N., et al. (2005). In vitro mutation and selection of doubled-haploid Brassica napus lines with improved resistance to Sclerotinia sclerotiorum. Plant Cell Rep. 24 (3), 133–144. doi: 10.1007/s00299-005-0925-0
Liu, Y., He, C. (2016). Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Rep. 35 (5), 995–1007. doi: 10.1007/s00299-016-1950-x
Ma, W., Berkowitz, G. A. (2011). Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol. 190 (3), 566–572. doi: 10.1111/j.1469-8137.2010.03577.x
Mauch-Mani, B., Mauch, F. (2005). The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 8 (4), 409–414. doi: 10.1016/j.pbi.2005.05.015
Mei, J., Ding, Y., Lu, K., Wei, D., Liu, Y., Disi, J. O., et al. (2013). Identification of genomic regions involved in resistance against Sclerotinia sclerotiorum from wild Brassica oleracea. Theor. Appl. Genet. 126 (2), 549–556. doi: 10.1007/s00122-012-2000-x
Mei, J., Qian, L., Disi, J. O., Yang, X., Li, Q., Li, J., et al. (2011). Identification of resistant sources against Sclerotinia sclerotiorum in Brassica species with emphasis on B. oleracea. Euphytica 177, 393–399. doi: 10.1007/s10681-010-0274-0
Mengiste, T. (2012). Plant immunity to necrotrophs. Annu. Rev Phytopathol. 50, 267–294. doi: 10.1146/annurev-phyto-081211-172955
Miah, G., Rafii, M. Y., Ismail, M. R., Puteh, A. B., Rahim, H. A., Asfaliza, R., et al. (2013). Blast resistance in rice: a review of conventional breeding to molecular approaches. Mol. Biol. Rep. 40 (3), 2369–2388. doi: 10.1007/s11033-012-2318-0
Micic, Z., Hahn, V., Bauer, E., Melchinger, A. E., Knapp, S. J., Tang, S., et al. (2005a). Identification and validation of QTL for Sclerotinia midstalk rot resistance in sunflower by selective genotyping. Theor. Appl. Genet. 111 (2), 233–242. doi: 10.1007/s00122-005-2004-x
Micic, Z., Hahn, V., Bauer, E., Schon, C. C., Knapp, S. J., Tang, S., et al. (2004). QTL mapping of Sclerotinia midstalk-rot resistance in sunflower. Theor. Appl. Genet. 109 (7), 1474–1484. doi: 10.1007/s00122-004-1764-z
Micic, Z., Hahn, V., Bauer, E., Schon, C. C., Melchinger, A. E. (2005b). QTL mapping of resistance to Sclerotinia midstalk rot in RIL of sunflower population NDBLOSsel x CM625. Theor. Appl. Genet. 110 (8), 1490–1498. doi: 10.1007/s00122-005-1984-x
Ming, P. Y., Uloth, M. B., Xi, X. L., Banga, S. S., Banga, S. K., Barbetti, M. J. (2016). Valuable new resistances ensure improved management of Sclerotinia stem rot (Sclerotinia sclerotiorum) in Horticultural and Oilseed Brassica Species. J. Phytopathol. 164 (5), 291–299. doi: 10.1111/jph.12456
Nie, J., Yin, Z., Li, Z., Wu, Y., Huang, L. (2019). A small cysteine-rich protein from two kingdoms of microbes is recognized as a novel pathogen-associated molecular pattern. New Phytol. 222 (2), 995–1011. doi: 10.1111/nph.15631
Nordborg, M., Borevitz, J. O., Bergelson, J., Berry, C. C., Chory, J., Hagenblad, J., et al. (2002). The extent of linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 30 (2), 190–193. doi: 10.1038/ng813
Nordborg, M., Hu, T. T., Ishino, Y., Jhaveri, J., Toomajian, C., Zheng, H., et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3 (7), e196. doi: 10.1371/journal.pbio.0030196
Nordborg, M., Weigel, D. (2008). Next-generation genetics in plants. Nature 456 (7223), 720–723. doi: 10.1038/nature07629
Nováková, M., Sašek, V., Dobrev, P. I., Valentová, O., Burketová, L. (2014). Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum - reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiol. Biochem. 80, 308–317. doi: 10.1016/j.plaphy.2014.04.019
Pan, Y., Wei, J., Yao, C., Reng, H., Gao, Z. (2018). SsSm1, a Cerato-platanin family protein, is involved in the hyphal development and pathogenic process of Sclerotinia sclerotiorum. Plant Sci. 270, 37–46. doi: 10.1016/j.plantsci.2018.02.001
Pazzagli, L., Cappugi, G., Manao, G., Camici, G., Santini, A., Scala, A. (1999). Purification, characterization, and amino acid sequence of cerato-platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani. J. Biol. Chem. 274 (35), 24959–24964. doi: 10.1074/jbc.274.35.24959
Penninckx, I. A., Eggermont, K., Terras, F. R., Thomma, B. P., De Samblanx, G. W., Buchala, A., et al. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8 (12), 2309–2323. doi: 10.1105/tpc.8.12.2309
Perchepied, L., Balague, C., Riou, C., Claudel-Renard, C., Riviere, N., Grezes-Besset, B., et al. (2010). Nitric oxide participates in the complex interplay of defense-related signaling pathways controlling disease resistance to Sclerotinia sclerotiorum in Arabidopsis thaliana. Mol. Plant Microbe Interact. 23 (7), 846–860. doi: 10.1094/MPMI-23-7-0846
Petersen, M., Brodersen, P., Naested, H., Andreasson, E., Lindhart, U., Johansen, B., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103 (7), 1111–1120. doi: 10.1016/S0092-8674(00)00213-0
Platt, A., Vilhjalmsson, B. J., Nordborg, M. (2010). Conditions under which genome-wide association studies will be positively misleading. Genetics 186 (3), 1045–1052. doi: 10.1534/genetics.110.121665
Purdy, L. H. (1979). Sclerotinia sclerotiorum: history, diseases and symptomatology, host range, geographic distribution, and impact. Phytopathology 69 (8), 875–880. doi: 10.1094/Phyto-69-875
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27 (16), 2214–2221. doi: 10.1038/emboj.2008.147
Raboin, L. M., Ballini, E., Tharreau, D., Ramanantsoanirina, A., Frouin, J., Courtois, B., et al. (2016). Association mapping of resistance to rice blast in upland field conditions. Rice (N Y) 9 (1), 59. doi: 10.1186/s12284-016-0131-4
Rahman, H., Xu, Y. P., Zhang, X. R., Cai, X. Z. (2016). Brassica napus genome possesses extraordinary high number of camta genes and camta3 contributes to pamp triggered immunity and resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 7, 581. doi: 10.3389/fpls.2016.00581
Ranty, B., Aldon, D., Galaud, J. P. (2006). Plant calmodulins and calmodulin-related proteins: multifaceted relays to decode calcium signals. Plant Signal. Behav. 1 (3), 96–104. doi: 10.4161/psb.1.3.2998
Rigo, G., Ayaydin, F., Tietz, O., Zsigmond, L., Kovacs, H., Pay, A., et al. (2013). Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 25 (5), 1592–1608. doi: 10.1105/tpc.113.110452
Ronicke, S., Hahn, V., Vogler, A., Friedt, W. (2005). Quantitative Trait Loci analysis of resistance to Sclerotinia sclerotiorum in sunflower. Phytopathology 95 (7), 834–839. doi: 10.1094/PHYTO-95-0834
Roux, F., Voisin, D., Badet, T., Balague, C., Barlet, X., Huard-Chauveau, C., et al. (2014). Resistance to phytopathogens e tutti quanti: placing plant quantitative disease resistance on the map. Mol. Plant Pathol. 15 (5), 427–432. doi: 10.1111/mpp.12138
Saand, M. A., Xu, Y. P., Li, W., Wang, J. P., Cai, X. Z. (2015a). Cyclic nucleotide gated channel gene family in tomato: genome-wide identification and functional analyses in disease resistance. Front. Plant Sci. 6, 303. doi: 10.3389/fpls.2015.00303
Saand, M. A., Xu, Y. P., Munyampundu, J. P., Li, W., Zhang, X. R., Cai, X. Z. (2015b). Phylogeny and evolution of plant cyclic nucleotide-gated ion channel (CNGC) gene family and functional analyses of tomato CNGCs. DNA Res. 22 (6), 471–483. doi: 10.1093/dnares/dsv029
Samanta, S., Thakur, J. K. (2015). Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front. Plant Sci. 6, 757. doi: 10.3389/fpls.2015.00757
Seidl, V., Marchetti, M., Schandl, R., Allmaier, G., Kubicek, C. P. (2006). Epl1, the major secreted protein of Hypocrea atroviridis on glucose, is a member of a strongly conserved protein family comprising plant defense response elicitors. FEBS J 273 (18), 4346–4359. doi: 10.1111/j.1742-4658.2006.05435.x
Shen, Y., Diener, A. C. (2013). Arabidopsis thaliana resistance to fusarium oxysporum 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genet. 9 (5), e1003525. doi: 10.1371/journal.pgen.1003525
Stotz, H. U., Jikumaru, Y., Shimada, Y., Sasaki, E., Stingl, N., Mueller, M. J., et al. (2011). Jasmonate-dependent and COI1-independent defense responses against Sclerotinia sclerotiorum in Arabidopsis thaliana: auxin is part of COI1-independent defense signaling. Plant Cell Physiol. 52 (11), 1941–1956. doi: 10.1093/pcp/pcr127
Swiezawska, B., Duszyn, M., Jaworski, K., Szmidt-Jaworska, A. (2018). Downstream targets of cyclic nucleotides in plants. Front. Plant Sci. 9, 1428. doi: 10.3389/fpls.2018.01428
Thomma, B. P., Eggermont, K., Penninckx, I. A., Mauch-Mani, B., Vogelsang, R., Cammue, B. P., et al. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 95 (25), 15107–15111. doi: 10.1073/pnas.95.25.15107
Thomma, B. P., Penninckx, I. A., Broekaert, W. F., Cammue, B. P. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13 (1), 63–68. doi: 10.1016/S0952-7915(00)00183-7
Uloth, M., You, M. P., Finnegan, P. M., Banga, S. S., Yi, H., Barbetti, M. J. (2014). Seedling resistance to Sclerotinia sclerotiorum as expressed across diverse cruciferous species. Plant Dis. 98 (2), 184–190. doi: 10.1094/PDIS-06-13-0612-RE
Uloth, M. B., Ming, P. Y., Finnegan, P. M., Banga, S. S., Banga, S. K., Sandhu, P. S., et al. (2013). New sources of resistance to Sclerotinia sclerotiorum for crucifer crops. Field Crops Res. 154 (154), 40–52. doi: 10.1016/j.fcr.2013.07.013
Uloth, M. B., You, M. P., Barbetti, M. J. (2015). Host resistance to Sclerotinia stem rot in historic and current Brassica napus and B. juncea varieties: critical management implications. Crop Pasture Sci. 66 (8), 841–848. doi: 10.1071/CP15064
Vasconcellos, R. C., Oraguzie, O. B., Soler, A., Arkwazee, H., Myers, J. R., Ferreira, J. J., et al. (2017). Meta-QTL for resistance to white mold in common bean. PLoS One 12 (2), e0171685. doi: 10.1371/journal.pone.0171685
Veronese, P., Nakagami, H., Bluhm, B., Abuqamar, S., Chen, X., Salmeron, J., et al. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18 (1), 257–273. doi: 10.1105/tpc.105.035576
Wang, C., Yao, J., Du, X., Zhang, Y., Sun, Y., Rollins, J. A., et al. (2015a). The Arabidopsis mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Physiol. 169 (1), 856–872. doi: 10.1104/pp.15.00351
Wang, G. D., Fiers, M., Ellendorff, U., Wang, Z. Z., de Wit, P. J. G. M., Angenent, G. C., et al. (2010). The diverse roles of extracellular leucine-rich repeat-containing receptor-like proteins in plants. Crit. Rev. Plant Sci. 29 (5), 285–299. doi: 10.1080/07352689.2010.502082
Wang, J. P., Munyampundu, J. P., Xu, Y. P., Cai, X. Z. (2015b). Phylogeny of plant calcium and calmodulin-dependent protein kinases (ccamks) and functional analyses of tomato ccamk in disease resistance. Front. Plant Sci. 6, 1075. doi: 10.3389/fpls.2015.01075
Wang, J. P., Xu, Y. P., Munyampundu, J. P., Liu, T. Y., Cai, X. Z. (2016). Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: genome-wide identification and functional analyses in disease resistance. Mol. Genet. Genomics 291 (2), 661–676. doi: 10.1007/s00438-015-1137-0
Wang, Z., Bao, L. L., Zhao, F. Y., Tang, M. Q., Chen, T., Li, Y., et al. (2019). BnaMPK3 is a key regulator of defense responses to the devastating plant pathogen Sclerotinia Sclerotiorum in oilseed rape. Front. Plant Sci. 10, 91. doi: 10.3389/fpls.2019.00091
Wang, Z., Fang, H., Chen, Y., Chen, K., Li, G., Gu, S., et al. (2014). Overexpression of BnWRKY33 in oilseed rape enhances resistance to Sclerotinia sclerotiorum. Mol. Plant Pathol. 15 (7), 677–689. doi: 10.1111/mpp.12123
Wang, Z., Mao, H., Dong, C., Ji, R., Cai, L., Fu, H., et al. (2009). Overexpression of Brassica napus MPK4 enhances resistance to Sclerotinia sclerotiorum in oilseed rape. Mol. Plant Microbe Interact. 22 (3), 235–244. doi: 10.1094/MPMI-22-3-0235
Wang, Z., Tan, X., Zhang, Z., Gu, S., Li, G., Shi, H. (2012). Defense to Sclerotinia sclerotiorum in oilseed rape is associated with the sequential activations of salicylic acid signaling and jasmonic acid signaling. Plant Sci. 184, 75–82. doi: 10.1016/j.plantsci.2011.12.013
Wang, Z., Wan, L., Xin, Q., Chen, Y., Zhang, X., Dong, F., et al. (2018). Overexpression of OsPGIP2 confers Sclerotinia sclerotiorum resistance in Brassica napus through increased activation of defense mechanisms. J. Exp. Bot. 69 (12), 3141–3155. doi: 10.1093/jxb/ery138
War, A. R., Paulraj, M. G., Ahmad, T., Buhroo, A. A., Hussain, B., Ignacimuthu, S., et al. (2012). Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 7 (10), 1306–1320. doi: 10.4161/psb.21663
Wei, D., Mei, J., Fu, Y., Disi, J. O., Li, J., Qian, W. (2014). Quantitative trait loci analyses for resistance to Sclerotinia sclerotiorum and flowering time in Brassica napus. Mol. Breed. 34 (4), 1797–1804. doi: 10.1007/s11032-014-0139-7
Wei, L., Jian, H., Lu, K., Filardo, F., Yin, N., Liu, L., et al. (2016). Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol. J. 14 (6), 1368. doi: 10.1111/pbi.12501
Wei, W., Mesquita, A. C. O., Figueiró, A. D. A., Xing, W., Manjunatha, S., Wickland, D. P., et al. (2017). Genome-wide association mapping of resistance to a Brazilian isolate of Sclerotinia sclerotiorum in soybean genotypes mostly from Brazil. Bmc Genomics 18 (1), 849. doi: 10.1186/s12864-017-4160-1
Wen, Z., Tan, R., Zhang, S., Collins, P. J., Yuan, J., Du, W., et al. (2018). Integrating GWAS and gene expression data for functional characterization of resistance to white mold in soybean. Plant Biotechnol. J. 16(11), 1825–1835 doi: 10.1111/pbi.12918
Wendehenne, D., Durner, J., Klessig, D. F. (2004). Nitric oxide: a new player in plant signalling and defence responses. Curr. Opin. Plant Biol. 7 (4), 449–455. doi: 10.1016/j.pbi.2004.04.002
Williams, B., Kabbage, M., Kim, H. J., Britt, R., Dickman, M. B. (2011). Tipping the balance: Sclerotinia Sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7 (6), e1002107. doi: 10.1371/journal.ppat.1002107
Wright, C. A., Beattie, G. A. (2004). Pseudomonas syringae pv. tomato cells encounter inhibitory levels of water stress during the hypersensitive response of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101 (9), 3269–3274. doi: 10.1073/pnas.0400461101
Wu, J., Cai, G., Tu, J., Li, L., Liu, S., Luo, X., et al. (2013). Identification of QTLs for resistance to Sclerotinia stem rot and bnac.igmt5.aas a candidate gene of the major resistant QTLSRC6inBrassica napus. Plos One 8 (7), e67740. doi: 10.1371/journal.pone.0067740
Wu, J., Zhao, Q., Liu, S., Shahid, M., Lan, L., Cai, G., et al. (2016). Genome-wide association study identifies new loci for resistance to Sclerotinia stem rot in Brassica napus. Front. Plant Sci. 7, 1418. doi: 10.3389/fpls.2016.01418
Xu, Y. P., Lv, L. H., Xu, Y. J., Yang, J., Cao, J. Y., Cai, X. Z. (2018a). Leaf stage-associated resistance is correlated with phytohormones in a pathosystem-dependent manner. J. Integr. Plant Biol. 60 (8), 703–722. doi: 10.1111/jipb.12661
Xu, Y. P., Yang, J., Cai, X. Z. (2018b). Glycolate oxidase gene family in Nicotiana benthamiana: genome-wide identification and functional analyses in disease resistance. Sci. Rep. 8 (1), 8615. doi: 10.1038/s41598-018-27000-4
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2018). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217 (2), 739–755. doi: 10.1111/nph.14842
Yang, S. M. (1959). An investigation on the host range and some ecological aspects of the Sclerotinia disease of rape plants. Acta Phytopathol. Sinica 5, 111–122. doi: 10.13926/j.cnki.apps.1959.02.006
Yang, Y., Zhang, H., Li, G., Li, W., Wang, X., Song, F. (2009). Ectopic expression of MgSM1, a Cerato-platanin family protein from Magnaporthe grisea, confers broad-spectrum disease resistance in Arabidopsis. Plant Biotechnol. J. 7 (8), 763–777. doi: 10.1111/j.1467-7652.2009.00442.x
Yin, X., Yi, B., Chen, W., Zhang, W., Tu, J., Wgdilantha, F., et al. (2010). Mapping of QTLs detected in a Brassica napus DH population for resistance to Sclerotinia Sclerotiorum in multiple environments. Euphytica 173 (1), 25–35. doi: 10.1007/s10681-009-0095-1
Yu, Y., Xiao, J., Zhu, W., Yang, Y., Mei, J., Bi, C., et al. (2017). Ss-Rhs1, a secretory Rhs repeat-containing protein, is required for the virulence of Sclerotinia sclerotiorum. Mol. Plant Pathol. 18 (8), 1052–1061. doi: 10.1111/mpp.12459
Zhang, H., Wu, Q., Cao, S., Zhao, T., Chen, L., Zhuang, P., et al. (2014). A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants. Plant Mol. Biol. 86 (4-5), 495–511. doi: 10.1007/s11103-014-0244-3
Zhang, W., Fraiture, M., Kolb, D., Loffelhardt, B., Desaki, Y., Boutrot, F. F., et al. (2013). Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25 (10), 4227–4241. doi: 10.1105/tpc.113.117010
Zhang, X., Wang, C., Zhang, Y., Sun, Y., Mou, Z. (2012). The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24 (10), 4294–4309. doi: 10.1105/tpc.112.103317
Zhao, J., Buchwaldt, L., Rimmer, S. R., Sharpe, A., McGregor, L., Bekkaoui, D., et al. (2009). Patterns of differential gene expression in Brassica napus cultivars infected with Sclerotinia sclerotiorum. Mol. Plant Pathol. 10 (5), 635–649. doi: 10.1111/j.1364-3703.2009.00558.x
Zhao, J., Meng, J. (2003). Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.). Theor. Appl. Genet. 106 (4), 759–764. doi: 10.1007/s00122-002-1171-2
Zhao, J., Udall, J. A., Quijada, P. A., Grau, C. R., Meng, J., Osborn, T. C. (2006). Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor. Appl. Genet. 112 (3), 509–516. doi: 10.1007/s00122-005-0154-5
Zhao, J., Wang, J., An, L., Doerge, R. W., Chen, Z. J., Grau, C. R., et al. (2007). Analysis of gene expression profiles in response to Sclerotinia sclerotiorum in Brassica napus. Planta 227 (1), 13–24. doi: 10.1007/s00425-007-0586-z
Zhao, X., Han, Y., Li, Y., Liu, D., Sun, M., Zhao, Y., et al. (2015). Loci and candidate gene identification for resistance to Sclerotinia sclerotiorum in soybean (Glycine max L. Merr.) via association and linkage maps. Plant J. 82 (2), 245–255. doi: 10.1111/tpj.12810
Zhou, J., Zeng, L., Liu, J., Xing, D. (2015). Manipulation of the Xanthophyll Cycle Increases Plant Susceptibility to Sclerotinia sclerotiorum. PLoS Pathog. 11 (5), e1004878. doi: 10.1371/journal.ppat.1004878
Zipfel, C. (2014). Plant pattern-recognition receptors. Trends Immunol. 35 (7), 345–351. doi: 10.1016/j.it.2014.05.004
Keywords: Sclerotinia sclerotiorum, plant defense, signaling molecules, quantitative resistance, genome-wide association study, candidate resistance genes
Citation: Wang Z, Ma L-Y, Cao J, Li Y-L, Ding L-N, Zhu K-M, Yang Y-H and Tan X-L (2019) Recent Advances in Mechanisms of Plant Defense to Sclerotinia sclerotiorum. Front. Plant Sci. 10:1314. doi: 10.3389/fpls.2019.01314
Received: 17 May 2019; Accepted: 20 September 2019;
Published: 18 October 2019.
Edited by:
Xin Li, University of British, Columbia, CanadaCopyright © 2019 Wang, Ma, Cao, Li, Ding, Zhu, Yang and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Li Tan, eGx0YW5AdWpzLmVkdS5jbg==
 Zheng Wang
Zheng Wang Lu-Yue Ma
Lu-Yue Ma Jun Cao
Jun Cao Li-Na Ding
Li-Na Ding Ke-Ming Zhu
Ke-Ming Zhu Xiao-Li Tan
Xiao-Li Tan