- 1Department of Molecular Genetics, College of Natural Resources and Life Science, Dong-A University, Busan, South Korea
- 2Department of Agricultural Biotechnology, National Institute of Agricultural Science, Rural Development Administration, Jeonju, South Korea
- 3Biosafety Division, National Institute of Agricultural Science, Rural Development Administration, Jeonju, South Korea
- 4Department of Bioindustry and Bioresource Engineering, Plant Engineering Research Institute, Sejong University, Seoul, South Korea
Soybean is a major crop that is used as a source of vegetable oil for human use. To develop transgenic soybean with high α-linolenic acid (ALA; 18:3) content, the FAD3-1 gene isolated from lesquerella (Physaria fendleri) was used to construct vectors with two different seed-specific promoters, soybean β-conglycinin (Pβ-con) and kidney bean phaseolin (Pphas), and one constitutive cauliflower mosaic virus 35S promoter (P35S). The corresponding vectors were used for Agrobacterium-mediated transformation of imbibed mature half seeds. The transformation efficiency was approximately 2%, 1%, and 3% and 21, 7, and 17 transgenic plants were produced, respectively. T-DNA insertion and expression of the transgene were confirmed from most of the transgenic plants by polymerase chain reaction (PCR), quantitative real-time PCR (qPCR), reverse transcription PCR (RT-PCR), and Southern blot analysis. The fatty acid composition of soybean seeds was analyzed by gas chromatography. The 18:3 content in the transgenic generation T1 seeds was increased 7-fold in Pβ-con:PfFAD3-1, 4-fold in Pphas : PfFAD3-1, and 1.6-fold in P35S:PfFAD3-1 compared to the 18:3 content in soybean “Kwangankong”. The increased content of 18:3 in the Pβ-con:PfFAD3-1 soybean (T1) resulted in a 52.6% increase in total fatty acids, with a larger decrease in 18:1 content than 18:2 content. The increase in 18:3 content was also maintained and reached 42% in the Pphas : PfFAD3-1 transgenic generation T2. Investigations of the agronomic traits of 12 Pβ-con:PfFAD3-1 transgenic lines (T1) revealed that plant height, number of branches, nodes, pods, total seeds, and total seed weight were significantly higher in several transgenic lines than those in non-transgenic soybean. Especially, an increase in seed size was observed upon expression of the PfFAD3-1 gene with the β-conglycinin promoter, and 6%–14% higher seed lengths were measured from the transgenic lines.
Introduction
Soybean (Glycine max (L.) Merr.) is an important crop that serves as a significant source of oil (~20%) and protein (~40%). The agricultural importance of soybean has been recognized owing to its various beneficial effects on human health (Oksman-Caldentey and Hiltunen, 1996; Zeng et al., 2004; Manavalan et al., 2009; John et al., 2016). Given its amenability to genetic transformation, soybean has been subjected to gene transfer. Soybean is the world's largest genetically modified crop because of its applications in food, industrial, and pharmaceutical products (Li et al., 2017; Chen et al., 2018). Initially, research on biotech soybean focused on agronomic traits for securing yields. There has been great interest in the development of biotech crops with value-added traits to improve nutritional value and industrial applications (Homrich et al., 2012). Increasing interest in the production of functional crops has propelled the development of soybean crops with specific new nutrients and increased functionality. Trials to produce or increase the levels of functional compounds, such as isoflavone, β-carotene, and syringin, were conducted in soybean calluses and seeds using Agrobacterium-mediated transformation (Jiang et al., 2010; Kim et al., 2012; Kwon et al., 2017).
Increasing the oil and protein content of soybean seed has been a task for breeders to meet the demand of the rapidly growing human population and the rising concern of food shortage (El-Hamidi and Zaher, 2018). Both quantity and quality are important factors to improve soybean oil production (Al Amin et al., 2019; Kanai et al., 2019). The fatty acids in soybean seed oil include palmitic acid (11%), stearic acid (4%), oleic acid (23%), linoleic acid (LA) (54%), and α-linolenic acid (ALA) (8%). At present, soybean oil is mainly used for frying; hence, molecular breeding is being utilized to reduce the production of trans fat from polyunsaturated fatty acids at high temperatures. The main goal of breeding was to increase the content of oleic acid and reduce the LA and ALA levels (Flores et al., 2008; Pham et al., 2012; Yang et al., 2018; Al Amin et al., 2019). However, some contradictory reports have mentioned the benefits of high LA and ALA to human health (Rao et al., 2008; Amjad Khan et al., 2017). With the increase in market demand for functional food materials and industrial feedstock, there is a growing need to develop a soybean variety that produces high levels of functional omega-3 fatty acids. ALA, an omega-3 fatty acid in plants, is essential for humans and is obtained only from the diet. ALA is converted to eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) in the human body. In addition, ALA is used as an environmentally friendly coating material such as linoleum used for floor covering (Jhala and Hall, 2010). Omega-3 fatty acids are critical for the human biological system and particularly helpful for the prevention of cardiovascular diseases (Psota et al., 2006; Khan et al., 2017). However, the content of omega-3 fatty acids in soybean seed is ~8%, which is relatively lower than that in other plants rich in omega-3. The recommended intake ratio of omega-6/omega-3 in healthy diets varies from 5:1 to 1:1 (Simopoulos, 2000). Therefore, it is desirable to increase the level of omega-3 ALA and lower the content of omega-6 LA in soybean. The enzyme involved in the synthesis of ALA is microsomal omega-3 fatty acid desaturase 3 (FAD3), which forms a double bond between the 15th and 16th carbon atoms of LA to synthesize ALA (Arondel et al., 1992). Soybean possesses four FAD3 genes, three of which (GmFAD3-1a, GmFAD3-1b, and GmFAD3-2a) are expressed in the seeds and control ALA content in the seed oil (Bilyeu et al., 2003; Anai et al., 2005).
We had previously cloned two genes, PfFAD3-1 and PfFAD3-2, from lesquerella (Physaria fendleri), a new oil crop that produces an industrially useful hydroxy fatty acid, lesquerolic acid (20:1-OH). Introduction of the PfFAD3-1 gene into FAD3-deficient Arabidopsis resulted in an increase in the ALA content from 1.6% to 30% (Lee et al., 2019). Although the soybean genome contains three FAD3 genes that are expressed in the seeds, ALA content in the seeds is low (8%). The reason could be that soybean FAD3s have low enzyme activity or this enzymatic activity is impacted by feedback inhibition. Thus, we decided to use heterologous PfFAD3-1. In the present study, PfFAD3-1 was transformed into soybean to develop healthy functional soybean varieties with increased omega-3 ALA content.
Materials and Methods
Construction of Three Types of PfFAD3-1 Expression Vectors for Soybean Transformation
The gene PfFAD3-1 (Accession No. MF611845) was amplified using the first-strand cDNA of the total RNA from the developing seeds of the lesquerella germplasm line WCL-LY2 (Dierig et al., 2001) as a template for polymerase chain reaction (PCR) using PfFAD3-1 forward primer (5ʹ-CACCATGGTGGTTGCTATGGACA-3ʹ) and reverse primer (5ʹ-TAATTGATTTTAGATTTGTCAG-3ʹ). The resulting PCR product was subcloned into the vector pENTR/D-TOPO (Invitrogen, USA) and, then, recombined into the three desired destination vectors, pB2GW7.0-Pβ-conglycinin (pB2GW7.0 vector carrying a soybean seed-specific β-conglycinin promoter), pB2GW7.0-P35S (pB2GW7.0 vector with the CaMV 35S promoter), and pB2GW7.0-Pphaseolin (pB2GW7.0 vector carrying a seed-specific phaseolin promoter derived from Phaseolus vulgaris) (VIB-Ghent University, Ghent, Belgium) using the Gateway cloning method. These three constructed plasmids (Pβ-con:PfFAD3-1, Pphas : PfFAD3-1, and P35S:PfFAD3-1; Figure 1A) were transformed into Agrobacterium tumefaciens EHA105 (Karimi et al., 2002) as per the protocol described by Kim et al. (2013; 2016).
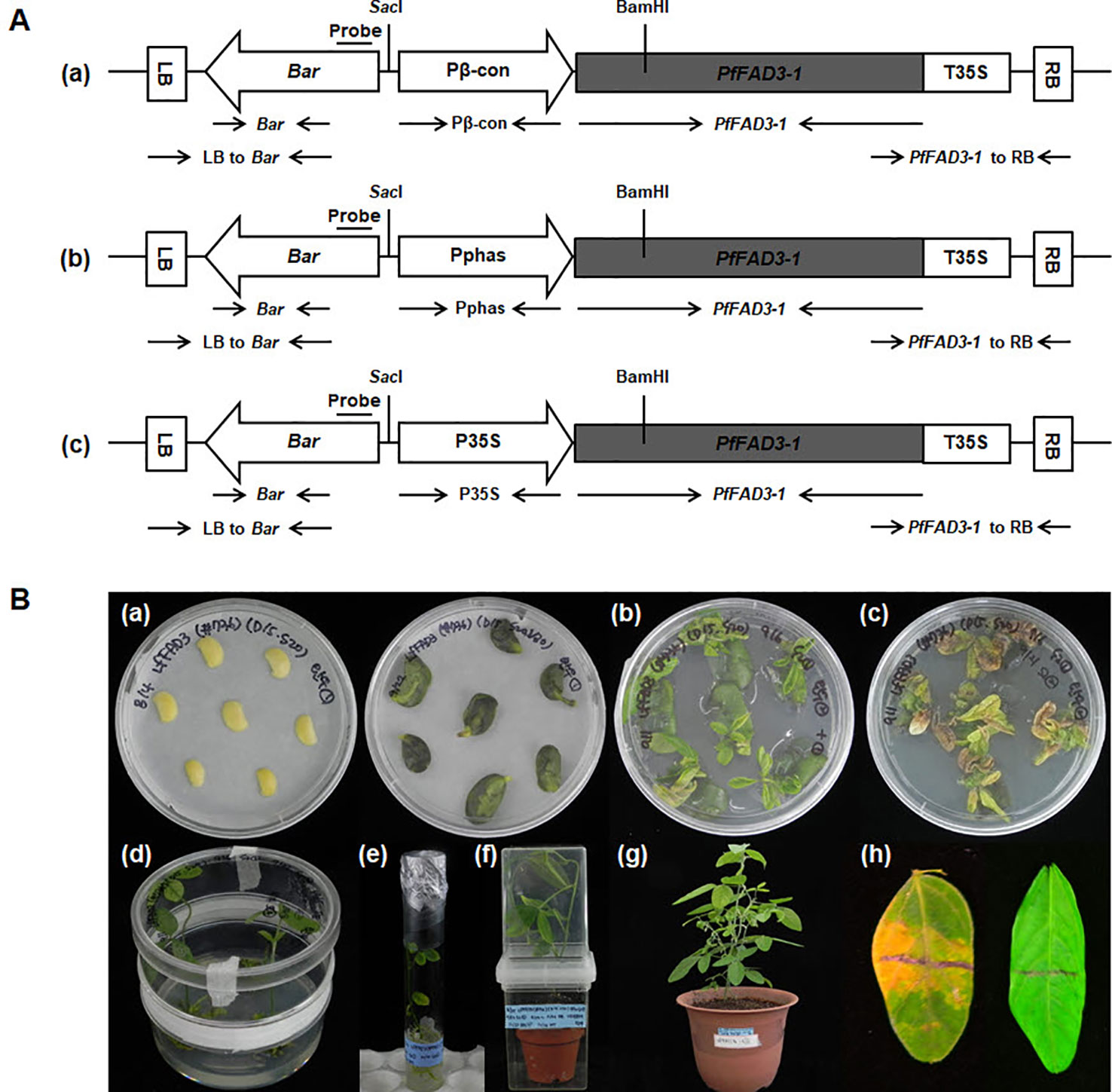
Figure 1 Production of soybean transgenic plants with the PfFAD3-1 gene via Agrobacterium-mediated transformation. (A) Vectors used for soybean transformation. The amplified PfFAD3-1 (1,164 bp size) was subcloned into the pB2GW7.0 vector carrying the β-conglycinin promoter (548 bp size) (a), phaseolin promoter (1,543 bp size) (b), and CaMV-35S promoter (953 bp size) (c). LB/RB, left/right T-DNA border; Pβ-con, soybean seed-specific β-conglycinin promoter; Pphas, seed-specific phaseolin promoter derived from Phaseolus vulgaris; P35S/T35S, cauliflower mosaic virus (CaMV) 35S promoter/terminator; Bar, coding region of the DL-phosphinothricin acetyltransferase gene that confers resistance to the herbicide glufosinate. SacI and BamHI restriction enzymes sites are marked. (B) Production of PfFAD3-1 soybean transgenic plants. (a) Half-seed explants after inoculation (left) and at 5 days after inoculation (right). (b) Shoot induction on SIM without PPT selection for 14 days. (c) Shoot induction on SIM with 10 mg/L PPT for another 14 days. (d) Shoot elongation on SEM with 5 mg/L PPT. (e) Root formation on RM. (f) Acclimation of a putative transgenic plant in a small pot. (g) Transgenic plant (T0) cultivated in a large pot in a greenhouse. (h) Non-transgenic plant leaf was sensitive (left), whereas the leaf from the transgenic plant was resistant (right) 5 days after herbicide (100 mg/L PPT) leaf painting.
Selection of PfFAD3-1 Transformed Soybeans
Mature soybean seeds of the Korean cultivar (Glycine max L. “Kwangankong”) were used, following the half-seed method for soybean transformation (Paz et al., 2006) with modifications. Six batches of transformation were carried out with 100–120 seeds per batch. Sterilized soybean seeds were soaked in distilled water for 12 h at 24°C. The Agrobacterium culture was centrifuged for 15 min at 20°C, 7,000 rpm. The cell pellet was resuspended in 15 ml liquid CCM (Co-Cultivation Medium) containing 0.32 g·L−1 Gamborg B5 salt with B5 vitamins, 4.26 g·L−1 2-(N-morpholino)ethanesulfonic acid (MES), 3% sucrose, 1.67 mg·L–1 6-benzylaminopurine, 0.25 mg·L−1 gibberellic acid, 0.2 mM acetosyringone, 3.3 mM L-cysteine, 1.0 mM sodium thiosulfate, and 1.0 mM dithiothreitol (DTT), pH 5.4. The cotyledons were separated by a longitudinal cut, and the seed coat was removed. The embryonic axis found at the junction between the hypocotyl and the cotyledon was excised to obtain the half-seed explant. The junction area was wounded eight times using a No. 11 scalpel blade (Feather, Japan) and, then, treated with the 15 ml liquid CCM containg Agrobacterium suspension as a dipping solution. Approximately 50 half-seed explants were immersed in 15 ml Agrobacterium suspension and inoculated for 30 min at room temperature. After inoculation, the explants were placed adaxial side down on CCM containing 5 g·L−1 agar overlaid with filter paper (Advantec, Japan). The co-cultivation was continued for 5 d at 24°C under an 18-h photoperiod. After 5 d of co-cultivation, the half-seed explants were briefly washed in liquid SIM (shoot induction medium) containing 3.2 g·L−1 B5 salt with B5 vitamins, 0.6 g·L−1 MES, 3% sucrose, 1.67 mg·L−1 +6-benzyladenine (BA), 250 mg·L−1 cefotaxime, 100 mg·L−1 ticarcillin, and 50 mg·L−1 vancomycin for 10 min. The explants were embedded in SIM containing 5.4 g·L−1 agar with the flat side up. Shoot induction was carried out for 14 d at 24°C under an 18-h photoperiod in a tissue culture room. Explants were also transferred to fresh SIM containing 10 mg·L−1 DL-phosphinothricin (PPT) for 14 d. After 4 weeks of shoot induction, the explants were transferred to SEM (shoot elongation medium) containing 4.4 g·L−1 MS salt with B5 vitamins, 0.6 g·L−1 MES, 3% sucrose, 5.4 g·L−1 agar, 0.5 mg·L−1 gibberellic acid, 50 mg·L−1 asparagine, 100 mg·L−1 L-pyroglutamic acid, 0.1 mg·L−1 indole-3-acetic acid, 1 mg·L−1 zeatin riboside, 250 mg·L−1 cefotaxime, 100 mg·L−1 ticarcillin, 50 mg·L−1 vancomycin, and 5 mg·L−1 phosphinothricin (PPT), pH 5.6. The explants were transferred to fresh SEM every 2 weeks. During shoot elongation, the elongated shoots were cut using a No. 11 scalpel blade, dipped in 1 mg·L−1 indole-3-butyric acid for 3 min, and, then, transferred to RM (rooting medium) containing 4.4 g·L−1 MS salt with B5 vitamins, 0.6 g·L−1 MES, 3% sucrose, 4.4 g·L−1 agar, 25 mg·L−1 asparagine, 25 mg·L−1 L-pyroglutamic acid, 50 mg·L−1 cefotaxime, 50 mg·L−1 ticarcillin, and 50 mg·L−1 vancomycin, pH 5.6. The rooted plantlets were rinsed with water to wash off the agar medium and transplanted to soil containing a mixture of Biosoil (Hongnong Horticulture, Korea) and vermiculite at a ratio of 3:1 in a magenta jar (SPL, Korea) for 1–3 weeks. T0 plants were then transplanted to large pots and grown in a greenhouse. Two trifoliate leaves of the T0 plants were screened using an herbicide assay for the identification of putative transformants expressing the Bar gene. The herbicide assay was performed on the upper surface of a leaf painted with a mixture of 100 mg·L−1 PPT and Tween-20. The herbicide-resistant T0 plants were cultivated in a greenhouse, and the seeds harvested (Figure 1B).
To identify gene insertions in the soybean transgenic plants, total genomic DNA was extracted. Specifically, DNA extraction using the CTAB (cetyltrimethylammonium bromide) method was conducted with leaf samples from non-transgenic (NT) and transgenic soybean plants (Lipp et al., 1999). Each sample (200 mg) was transferred to a 2-ml sterile reaction tube and 1 ml of CTAB extraction buffer (20 g·L−1 CTAB, 1.4 M NaCl, 0.1 M Tris/HCl, 200 mM EDTA) and 1.4 M β-mercaptoethanol were added. The mixture was vortexed and incubated at 65°C for 60 min. The solution was then centrifuged for 10 min at 12,000 × g. The supernatant was transferred to a new 2-ml sterile reaction tube. Ten microliters of RNase A (10 mg·ml−1) was added and the mixture was incubated at 37°C for 60 min. The mixture was then extracted with 800 μl of chloroform:isoamyl alcohol (24:1) and centrifuged for 10 min at 12,000 × g. The upper layer was transferred to a new reaction tube. This step was repeated. Isopropanol (0.6 volumes) was added to the upper phase; the mixture was mixed and incubated for 30 min at −20°C. After incubation, the mixture was centrifuged for 10 min at room temperature and the supernatant was discarded. The pellet was washed with 1 ml of 70% ethanol and centrifuged for 5 min at 12,000 × g. The supernatant was discarded, and the pellet was dried at 37°C for 30 min. Then, the dried pellet was dissolved in 100 μl of deionized water and stored at −20°C.
PCR analysis was conducted using KOD FX (TOYOBO, Japan), according to the manufacturer's instructions, and two primer sets designed to amplify specific regions of the PfFAD3-1 (1,146 bp) and Bar (548 bp) genes. The inserted promoters, including β-conglycinin (548 bp), phaseolin (1,543 bp), and CaMV 35S (953 bp) were also amplified. To verify T-DNA insertion into the soybean plant genome, the DNA sequence from the left border (LB) of the Bar gene and that from the PfFAD3-1 gene to the right border (RB) were amplified to represent both ends of the vector (Table 1, Figure 1). The primers for Bar, the promoters, and PfFAD3-1 were designed to amplify full sequences. The PCR reactions were conducted using a thermal cycler (Takara, Japan) under the following conditions: 95°C for 5 min, followed by 35 cycles at 95°C for 30 s, 50–65°C for 30 s, and 72°C for 30–90 s, with a final extension at 72°C for 10 min.
For Southern blot analysis, 10 µg genomic DNA from NT and transgenic leaf tissues was digested overnight with HindIII (Takara, Japan), fractionated on 0.8% agarose gels by electrophoresis, and transferred onto a Hybond N+ nylon membrane (Amersham Pharmacia, USA). Hybridization, washing, and detection were carried out with a digoxigenin (DIG)-labeled DNA probe and chemiluminescence system (Roche, Germany), according to the manufacturer's instructions. The Bar primers (5ʹ-AACTTCCGTACCGAGCCGCA-3ʹ/5ʹ-TCGTAGGCGTTGCGTGCCTT-3ʹ) were used to generate the DIG-labeled probe by PCR amplification. Real-time PCR (qPCR) was also performed with NT and transgenic leaf tissues to examine the transgene insertion events, using the CFX-96™ Real-Time System (Bio-Rad, Hercules, CA, USA), following the reaction described by Kim et al. (2016). Each reaction contained 4 μl of 3.3 ng·μl−1 DNA, 1 μl of a mixture of 5 pmol·μl−1 forward primer (5ʹ-AACTTCCGTACCGAGCCGCA-3ʹ) and 5 pmol·μl−1 reverse primer (5ʹ-TCGTAGGCGTTGCGTGCCTT-3ʹ), 5 μl of water, and 10 μl of iQ™SYBR® Green Supermix (Bio-Rad, Ca, USA), to make a total volume of 20 μl. The amplification conditions were as follows: 95°C for 3 min, 40 cycles of 10 s at 95°C, and 30 s at 60°C, and, finally, 95°C for 10 s. To verify amplification specificity, a dissociation curve was generated by increasing the temperature from 65°C–95°C. The Bar primers (5ʹ-AACTTCCGTACCGAGCCGCA-3ʹ/5ʹ-TCGTAGGCGTTGCGTGCCTT-3ʹ) were used. A homozygous transgenic plant already confirmed to have a single Bar gene introgression was used as a single copy control.
Analysis of PfFAD3-1 Gene Expression in Soybean Transformants
Total RNA was isolated from the leaves (T0) and seeds (T2) of NT and transgenic plants using Plant RNA Purification Reagent (Invitrogen, USA), according to the manufacturer's instructions. One gram of leaves and three seeds were ground using a mortar and pestle with liquid nitrogen and Tris-HCl (pH 9.0), respectively. Each sample (200 mg) was transferred to a 2-ml sterile reaction tube. Subsequently, 1 ml of Plant RNA Purification Reagent was added. The mixture was vortexed and incubated at room temperature for 5 min. The solution was then centrifuged for 10 min at 12,000 × g. The supernatant was transferred to a new 2-ml sterile reaction tube and 150 μl of 5 M NaCl and 450 μl of chloroform were added. The mixture was then vortexed and centrifuged for 10 min at 4°C and 12,000 × g. The upper layer was transferred to a new reaction tube. An equal volume of isopropanol was added and the mixture was incubated at room temperature for 10 min. After incubation, the mixture was centrifuged for 10 min at 4°C and 12,000 × g and the supernatant was discarded. The pellet was washed with 1 ml of 75% ethanol and centrifuged for 3 min at 12,000 × g. The supernatant was discarded and the pellet was dried for 20 min. Then, the dried pellet was dissolved in 20 μl of RNase-free water and stored at −70°C. Reverse transcription PCR (RT-PCR) was performed using the RT-PCR Premix Kit (Genetbio, South Korea), as per the manufacturer's instructions. Primers for the PfFAD3-1 (1,146 bp product) and Bar (548 bp product) genes were used to confirm expression levels (Table 2). The constitutively expressed TUB gene (256 bp) was used as a reference to normalize the amplified test genes. The PCR reactions were conducted using a thermal cycler (Takara, Japan) under the following conditions: 45°C for 30 min and 95°C for 5 min, followed by 35 cycles at 95°C for 30 s, 59°C–65°C for 30 s, and 72°C for 30–60 s, with a final extension at 72°C for 5 min.
Fatty Acid Analysis
Ten milligrams of seed powder obtained by crushing several seeds (n = ~5) with a metal ball in a mill was transferred to a 10-ml glass tube with a Teflon-sealed cap. In total, 0.5 ml toluene and 0.5 ml 5% sulfuric acid (H2SO4) (v/v) in methanol containing 100 μg of pentadecanoic acid (15:0) as an internal standard were added to each sample. Fatty acids were extracted and transmethylated in a water bath at 90°C for 90 min. Each sample was treated with 1.0 ml of 0.9% sodium chloride (NaCl) solution and 0.5 ml n-hexane and vigorously shaken for extraction. The upper phase containing fatty acid methyl esters (FAMEs) was transferred to a new uncapped glass tube after centrifugation at 2,000 rpm for 2 min. The FAMEs collected after three extractions with 0.5 ml n-hexane were dried with nitrogen gas and dissolved in 0.3 ml n-hexane. The FAMEs were analyzed using a GC-2010 Plus (Shimadzu, Japan) gas chromatograph coupled to a flame ionization detector (FID) and a 30 m × 0.25 mm (inner diameter) HP-FFAP column (Agilent, USA). The oven temperature was increased from 190°C to 232°C at 3°C/min. Nitrogen was used as the carrier gas. The fatty acid content was calculated as the average of three biologically independent sample measurements.
Analysis of Plant Growth and Phenotype of Soybean Transgenic Plants
NT and transgenic soybean seeds (T1) were planted in a seedling tray in June, 2018, and the early leaves were screened by herbicide painting (100 mg·L−1 PPT). Herbicide-resistant seedlings were then transplanted into a GMO field (Gunwi, South Korea). Agronomic traits, including plant height; number of branches, nodes, pods, and total seeds; and total seed weight, of the transgenic plants (T1) were determined in October, 2018, and compared with those of the NT plants (n = 10 each). To compare the seed size between NT and transgenic soybean seeds, NT and T2 soybean seeds harvested from the GMO field were randomly selected and horizontally placed in a row (n = 10 each). The size of the NT and T2 seeds was measured from 10 different sets of samples.
Statistical Analysis of Data
Statistical analysis was performed using the Excel T Test program to confirm significant differences between means. Asterisks indicate significant differences compared to NT plants (*P < 0.05; **P < 0.01).
Results
Production of Transgenic Soybean Plants With Three Types of PfFAD3-1 Expression Vectors
To produce transgenic soybean plants expressing high ALA levels, the PfFAD3-1 gene was cloned with two different types of promoters, namely, the seed-specific promoters from soybean β-conglycinin (Pβ-con) and kidney bean phaseolin (Pphas) for soybean seed-specific expression (Sengupta-Gopalan et al., 1985; Doyle et al., 1986) and CaMV 35S (P35S) for constitutive overexpression. The two seed-specific promoters were used to compare the efficiency of each promoter in soybean seed. The corresponding Pβ-con:PfFAD3-1, Pphas : PfFAD3-1 and P35S:PfFAD3-1 plasmids (Figure 1A) were used for Agrobacterium-mediated transformation of imbibed mature half seeds of the Korean soybean cultivar “Kwangankong” following the modified half-seed method described by Kim et al. (2013; 2016) (Figure 1B). According to the herbicide assay results, the transformation efficiency was approximately 2%, 1%, and 3% with Pβ-con:PfFAD3-1, Pphas : PfFAD3-1, and P35S:PfFAD3-1, respectively. In total, 21, 7, and 17 transgenic plants were produced with Pβ-con:PfFAD3-1, Pphas : PfFAD3-1, and P35S:PfFAD3-1, respectively.
Confirmation of Introduced Genes in PfFAD3-1 Transformed Soybeans
Leaf tissues from the 21 Pβ-con:PfFAD3-1, 7 Pphas : PfFAD3-1, and 17 P35S:PfFAD3-1 transgenic plants (T0) were used to confirm the integration of the transgene with PCR using PfFAD3-1 and Bar primers that amplified 1,146 and 548 bp DNA fragments, respectively. In addition, the DNA regions of the β-conglycinin, phaseolin, and CaMV 35S promoters were amplified as 548, 1,543, and 953 bp fragments, respectively (Figure 2). The results of the PCR analysis for the 21 Pβ-con:PfFAD3-1 transgenic lines confirmed the insertion of the transgene and β-conglycinin promoter sequence in all lines except line #15 (missing a region of the PfFAD3-1 gene) (Figure 2A). All seven putative Pphas : PfFAD3-1 transgenic plants successfully represented the transgene and phaseolin promoter (Figure 2B). Among the 17 P35S:PfFAD3-1 transgenic lines, only line #7 was missing regions of both the PfFAD3-1 gene and CaMV 35S promoter (Figure 2C). The selectable marker (Bar gene) was introduced in all PfFAD3-1-transformed soybean plants. Moreover, T-DNA insertion was verified by the amplification of both end regions of the vector construct (data not shown).
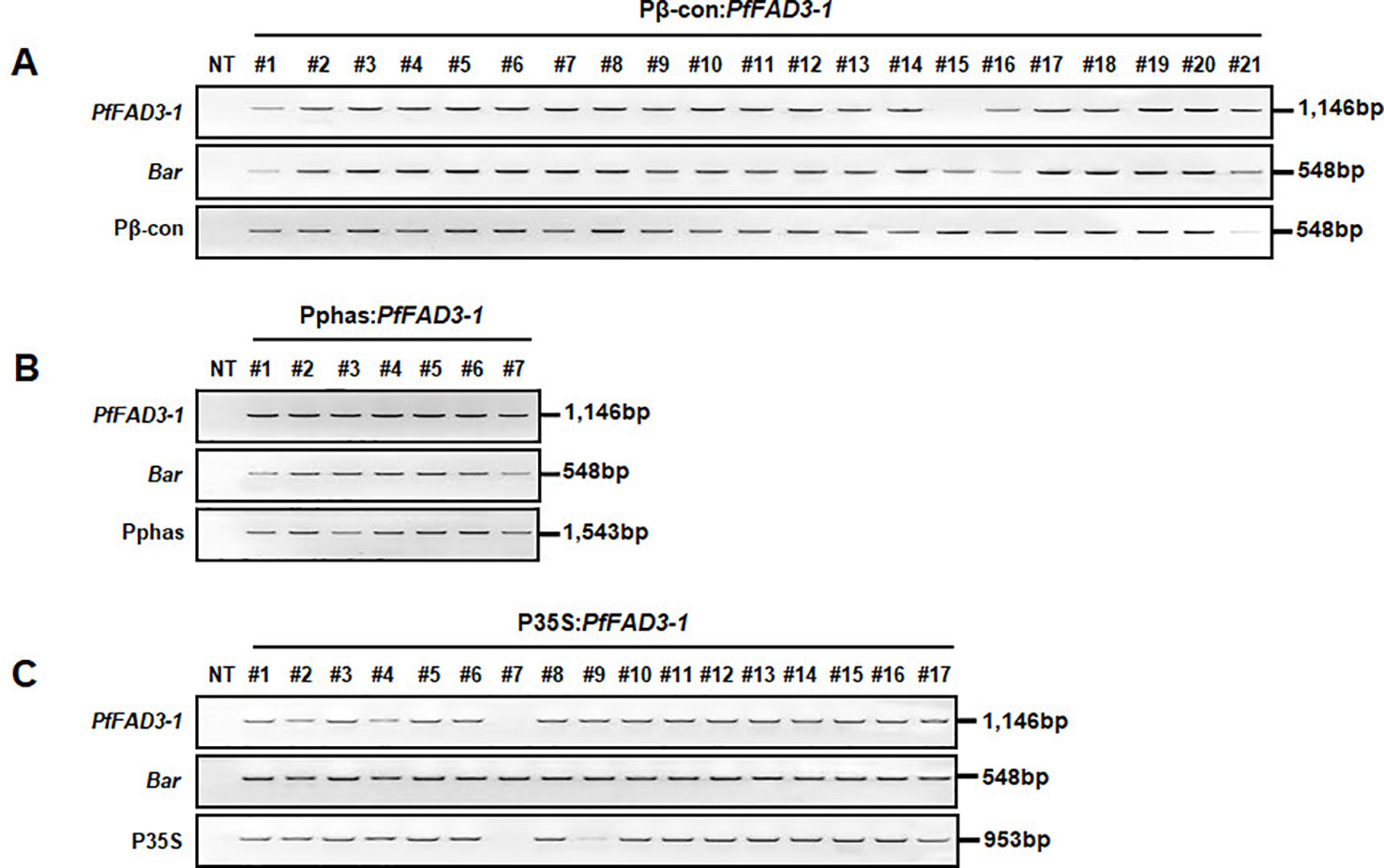
Figure 2 Confirmation of introduced genes from PfFAD3-1 transformants (T0) using PCR. Genomic DNA was extracted from Pβ-con:PfFAD3-1 (A), Pphas : PfFAD3-1 (B), and P35S:PfFAD3-1 (C) T0 transgenic leaf tissues. PfFAD3-1, PfFAD3-1 gene; Bar, Bar gene; Pβ-con, β-conglycinin promoter; Pphas, phaseolin promoter; P35S, CaMV 35S promoter; NT, non-transgenic plant; #1–#21 (part A), Pβ-con:PfFAD3-1 transgenic lines (T0); #1–#7 (part B), Pphas : PfFAD3-1 transgenic lines (T0); #1–#17 (part C), P35S:PfFAD3-1 transgenic lines (T0).
We determined the copy numbers of transgene insertions by performing genomic Southern blots of the 21 Pβ-con:PfFAD3-1 and 16 P35S:PfFAD3-1 transgenic plants (T0) (Figure 3). Hybridization with a Bar probe revealed that Pβ-con:PfFAD3-1 transgenic lines #6, #8, #11, and #12 had single insertions and lines #2, #3, #4, #5, #7, #10, #14, #17, #19, and #20 contained multiple copies (Figure 3A). Integration of the transgene in the P35S:PfFAD3-1 transgenic plants (except line #17, which completely withered) was also confirmed, indicating a low copy number of the transgene in lines #5, #7, #11, and #16 and multiple insertions in lines #3, #12, #13, and #15 (Figure 3B). Given the limited number of plants and leaf tissue samples of the Pphas : PfFAD3-1 transformants, their copy numbers were estimated by qPCR of the Bar gene, instead of Southern blot analysis (Supplementary Figure 1). Most transgenic lines of the Pphas : PfFAD3-1 transformants possessed low insert copy numbers, except line #2. Considering the homozygous single copy control of the Bar gene, most transgenic lines contained a single copy.
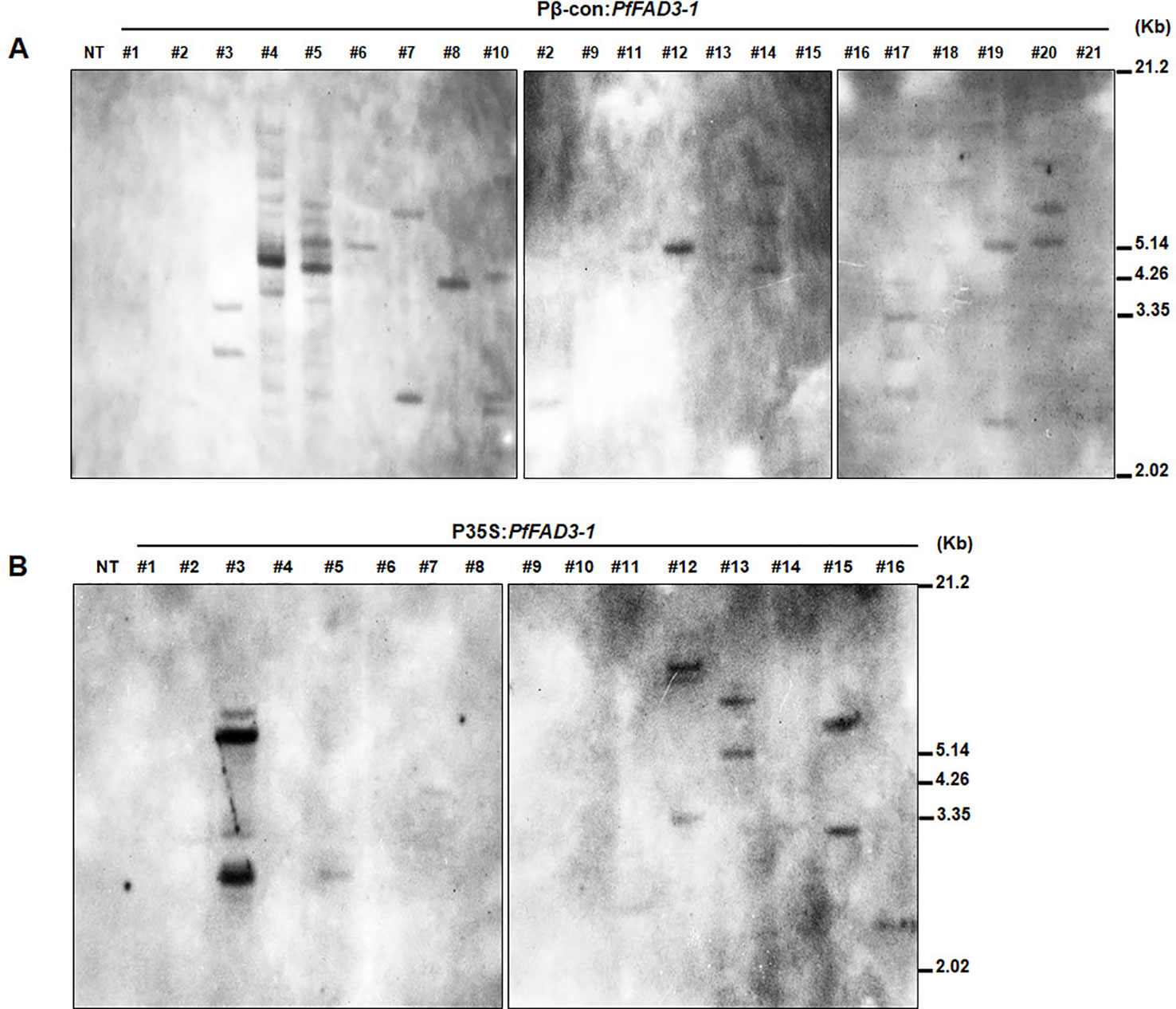
Figure 3 Genomic Southern blot analysis of PfFAD3-1 transgenic soybean (T0). Ten micrograms of genomic DNA from Pβ-con:PfFAD3-1 (A) and P35S:PfFAD3-1 (B) transgenic plants (T0) were digested with HindIII and hybridized with the Bar probe. Approximate DNA size markers are indicated on the right. NT, non-transgenic plant; #1–#21 (part A), Pβ-con:PfFAD3-1 transgenic lines (T0); #1–#16 (part B), P35S:PfFAD3-1 transgenic lines (T0).
Transgene Expression in PfFAD3-1 Transformed Soybeans
The expression levels of the PfFAD3-1 and Bar genes were analyzed in the Pβ-con:PfFAD3-1, Pphas : PfFAD3-1 and P35S:PfFAD3-1 transgenic plants (T0) using RT-PCR (Figure 4). The results from the 21 Pβ-con:PfFAD3-1 transgenic lines confirmed the expression of the PfFAD3-1 gene in all lines except line #15. The relative level of expression was remarkably higher in lines #6, #8, #10, #11, and #18 than in the other transgenic lines (Figure 4A). Expression of the PfFAD3-1 gene was confirmed in all seven Pphas : PfFAD3-1 transgenic lines. Of these, lines #1, #5, and #6 showed noticeably high expression of the PfFAD3-1 gene (Figure 4B). The integrated PfFAD3-1 gene was expressed in all 17 P35S:PfFAD3-1 transgenic lines as expected, except line #7 (Figure 4C). Expression of the selectable marker Bar was confirmed in all PfFAD3-1-transformed soybean plants except Pβ-con:PfFAD3-1 line #16. In addition, the PfFAD3-1 and Bar genes were not detected in the NT plants. Using RT-PCR, the expression level of the PfFAD3-1 gene was also analyzed in Pβ-con:PfFAD3-1 (lines #3, #6, #8, #10, #11, #12, and #18), Pphas : PfFAD3-1 (lines #1 and #5), and P35S:PfFAD3-1 (lines #15 and #17) transgenic seeds (T2) with higher 18:3 content (Figure 4D). PfFAD3-1 expression was much higher with the seed-specific β-conglycinin promoter (Pβ-con:PfFAD3-1) than with the seed-specific phaseolin promoter (Pphas : PfFAD3-1) and 35S promoter (P35S:PfFAD3-1). Expression from the seed-specific phaseolin promoter (Pphas : PfFAD3-1) was higher than or similar to the 35S promoter (P35S:PfFAD3-1).
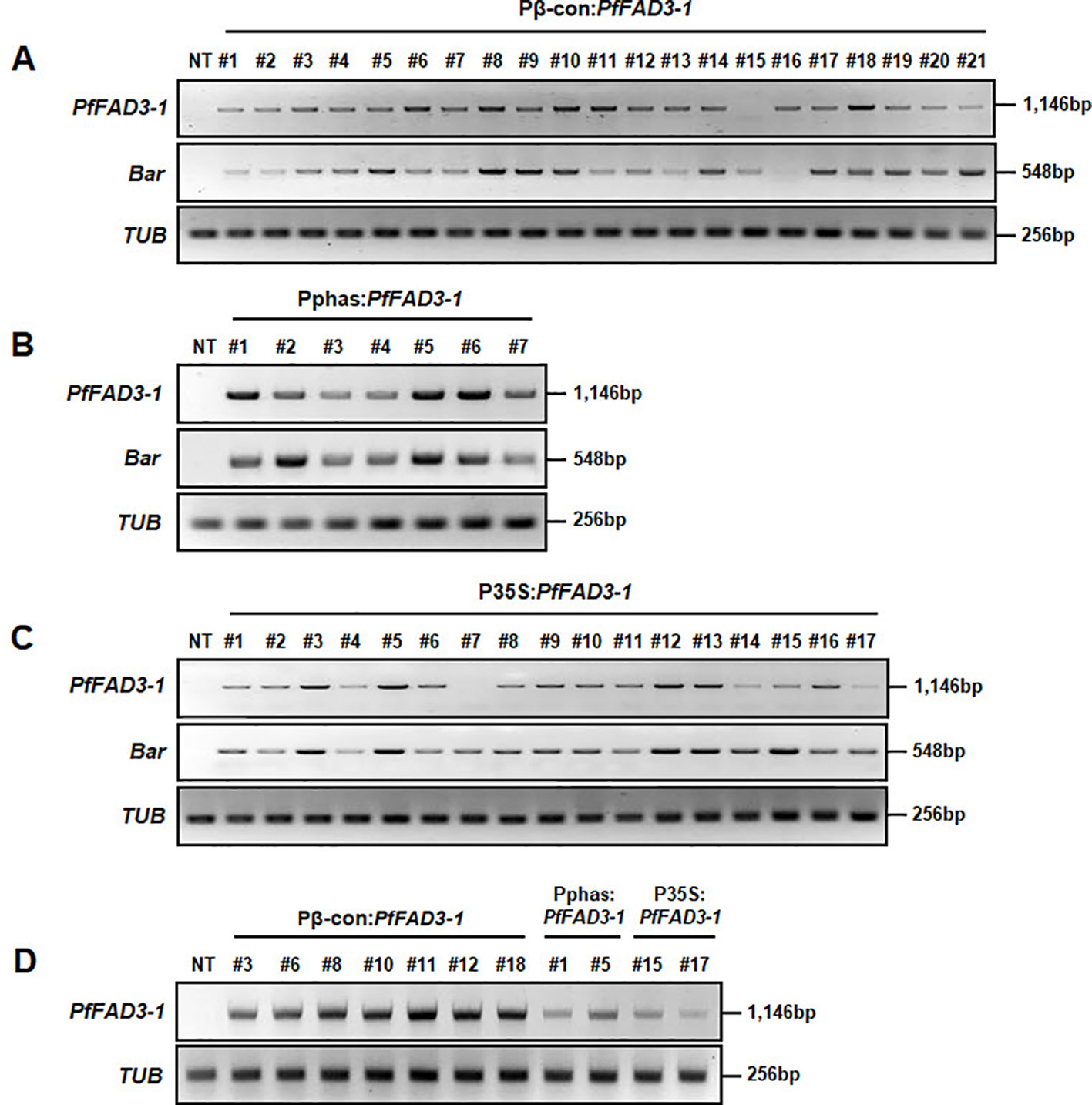
Figure 4 Transcript levels of PfFAD3-1 and Bar genes in transgenic plants (T0) and seeds (T2) using reverse transcriptase PCR (RT-PCR). Total RNA was extracted from Pβ-con:PfFAD3-1 (A), Pphas : PfFAD3-1 (B), and P35S:PfFAD3-1 (C) T0 transgenic leaf tissues and from T2 seeds (D). The TUB gene was used as a quantitative control. NT, non-transgenic plant; #1–#21 (part A), Pβ-con:PfFAD3-1 transgenic lines (T0); #1–#7 (part B), Pphas : PfFAD3-1 transgenic lines (T0); #1–#17 (part C), P35S:PfFAD3-1 transgenic lines (T0); #3, #6, #8, #11, #12, and #18, T2 seeds of Pβ-con:PfFAD3-1 transgenic lines; #1 and #5, T2 seeds of Pphas : PfFAD3-1 transgenic lines; #15 and #17, T2 seeds of P35S:PfFAD3-1 transgenic lines (part D).
Seed Fatty Acids in Soybean Transformants
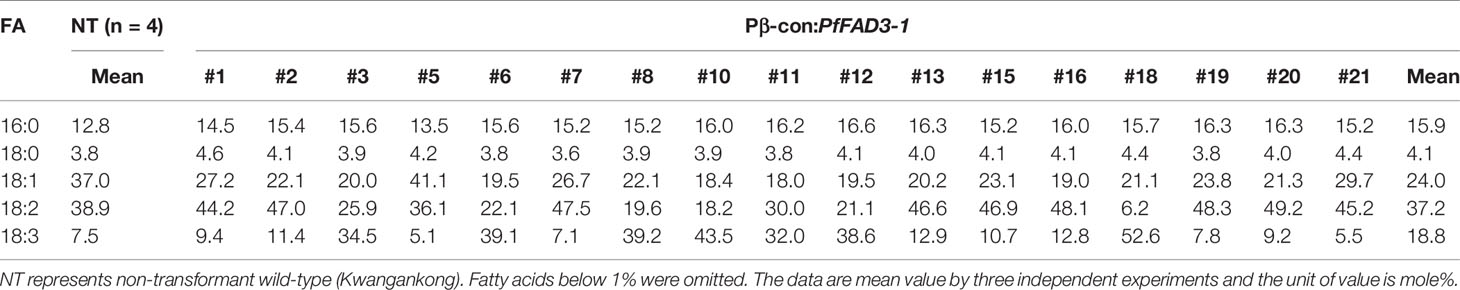
The fatty acid composition of the soybean seeds was analyzed using gas chromatography to determine the change in seed fatty acid composition upon heterologous PfFAD3-1 expression. The 18:3 content was 7.5% in the NT soybean “Kwangankong” and increased up to 52.4% in Pβ-con:PfFAD3-1, a 7-fold increase (Figure 5A, Table 3). In the case of Pphas : PfFAD3-1, the content increased up to 33.3%, which was 4.4 times the level reported for “Kwangankong” (Figure 5B, Table 4). However, the 18:3 content increased up to 12.1% in P35S:PfFAD3-1, an increase of only 1.6 fold (Figure 5C, Table 4).
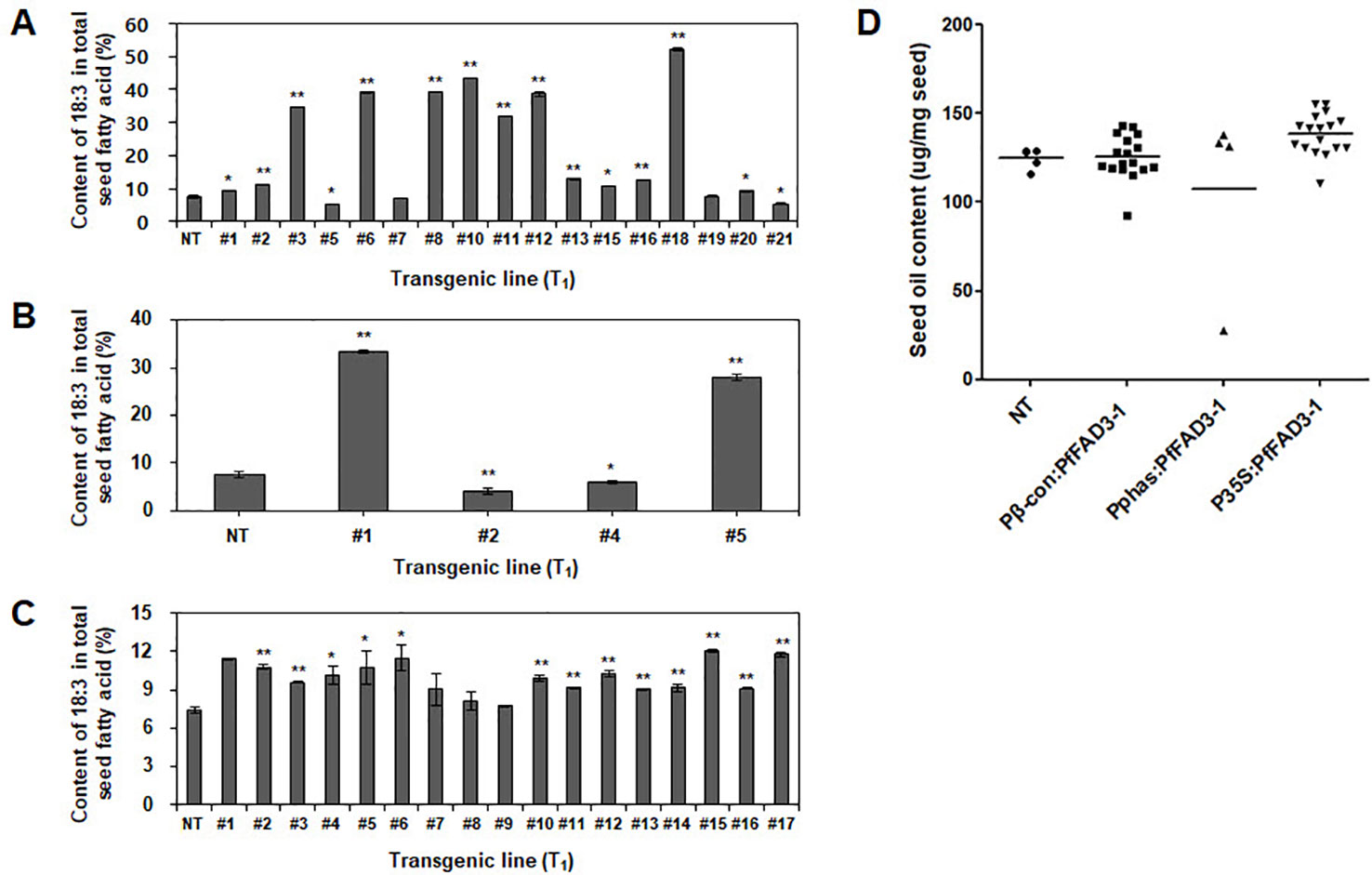
Figure 5 Content of α-linolenic acid (ALA) and seed oil in transgenic soybean seeds (T1). Content of ALA in Pβ-con:PfFAD3-1 (A), Pphas : PfFAD3-1 (B), and P35S:PfFAD3-1 (C) was analyzed using gas chromatography. (D) Seed oil content analysis of three PfFAD3-1 transgenic soybeans. NT, non-transgenic plant; #1–#21 (part A), T1 seeds of Pβ-con:PfFAD3-1 transgenic lines; #1–#5 (part B), T1 seeds of Pphas : PfFAD3-1 transgenic lines; #1–#17 (part C), T1 seeds of P35S:PfFAD3-1 transgenic lines. In parts A–C, error bars indicate the means ± standard deviations; asterisks indicate significant changes compared to NT values (* = p < 0.05; ** = p < 0.01). In part D, dots represent the seed oil content from each line (triplicate measurements); bars indicate mean values.
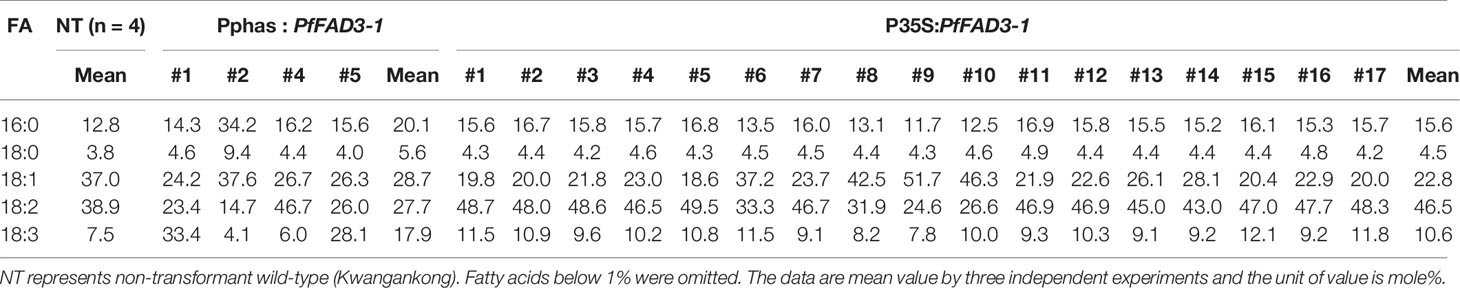
Table 4 Fatty acid composition of Pphas : PfFAD3-1 and P35S:PfFAD3-1 transgenic soybeans (T1 seeds).
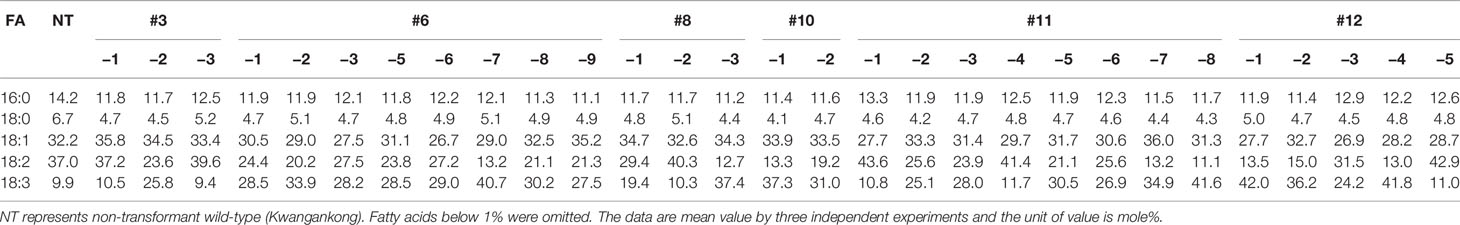
Seed oil content in the three types of transgenic soybean were analyzed and compared with that in wild-type plants. The average seed oil content for Pβ-con:PfFAD3-1, Pphas : PfFAD3-1, and P35S:PfFAD3-1 soybean was 125.4, 107.5, and 138.3 μg/mg seed, respectively (Figure 5D). For wild-type plants, the average seed oil content was 124.7 μg/mg seed. No statistically significant difference was observed in the seed oil content between transgenic and wild-type soybeans by one-way analysis of variance (ANOVA). In all PfFAD3-1 transgenic soybeans, the content of saturated fatty acid increased by 1%–5% and that of 18:1 and 18:2 decreased; the content of 18:3 increased compared with that in the wild-type soybean. In particular, 38% or less increase in the content of 18:3 in the Pβ-con:PfFAD3-1 soybean resulted in a larger decrease in 18:1 content than 18:2 content. Meanwhile, a >38% increase in the content of 18:3 induced a larger decrease in 18:2 content than 18:1 content (Table 3). For the Pphas : PfFAD3-1 soybean, the 18:2 content decreased more than the 18:1 content, with a rise in 18:3 content to ≥28%. The content of 18:3 slightly increased in the P35S:PfFAD3-1 soybeans compared with that in the wild-type plants and reached a maximum of 4.6%. Regardless of the magnitude of increase in 18:3 content, an inverse relationship was observed between 18:1 and 18:2 content (Tables 3 and 4). Furthermore, the increase in fatty acid content was maintained in the T2 generation. The 18:3 content of the Pβ-con:PfFAD3-1 T2 seeds increased up to 42%, showing a 4.2-fold rise compared with that in the wild-type “Kwangankong” (Figure 6, Table 5). Unlike the fatty acid composition of the Pβ-con:PfFAD3-1 T1 seeds, the saturated fatty acid content decreased by 3%–5% in T2 compared with that in the wild-type soybean (Table 5). A comparison of the fatty acid compositions of the T1 and T2 seeds revealed an average decrease of 4.6% in the saturated fatty acid content in T2 (Tables 3 and 5). The decrease in 18:1 content in T2 was not significant. One transgenic line (#11–7) had a 3.7% increase in 18:1 content, although the 18:3 content was greater than 30%.
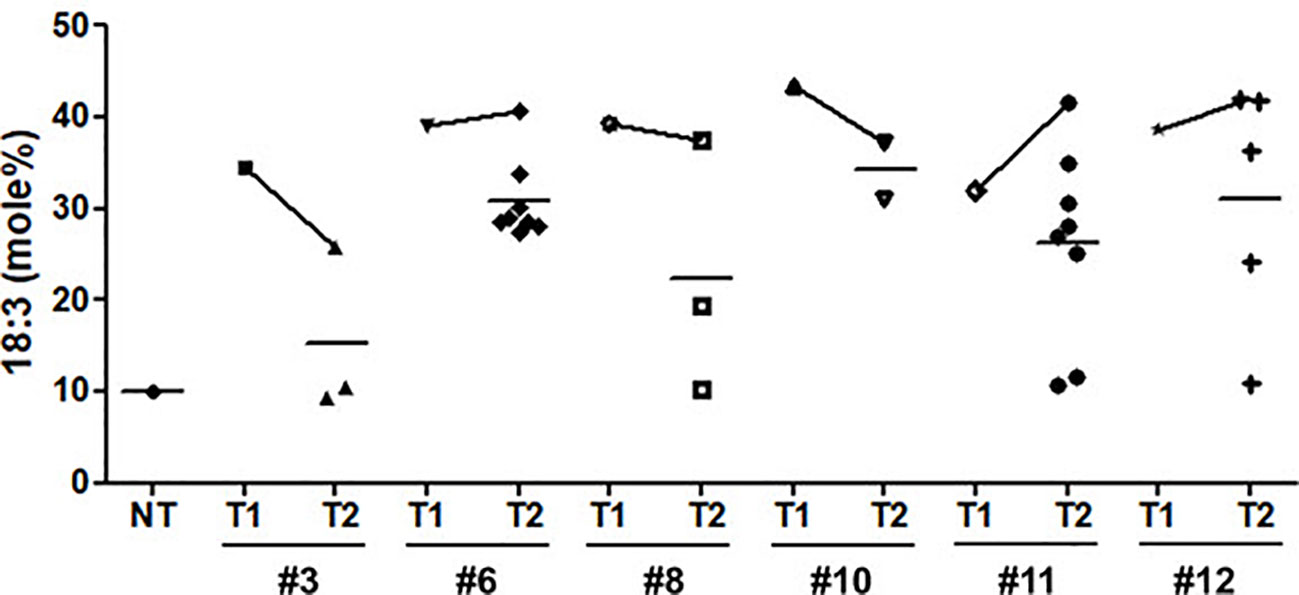
Figure 6 Comparison of α-linolenic acid (ALA) content between T1 and T2 seeds of Pβ-con:PfFAD3-1 transgenic soybeans. NT, non-transgenic. Symbols represent the ALA content of each line performed in triplicate. Horizontal bars indicate mean values.
Yield Components and Seed Phenotypes of Pβ-con:PfFAD3-1 Transformed Soybeans
Agronomic traits, including plant height, number of branches, nodes, pods, and total seeds and total seed weight, were investigated in the GMO field (Gunwi, South Korea) from the 12 Pβ-con:PfFAD3-1 transgenic lines (T1) (Figure 7). The heights of the transgenic lines #1, #6, #11, #12, #13, and #15 were significantly higher than those of the NT plants (p < 0.05 for line #6 and p < 0.01 for lines #1, #11, #12, #13, and #15) (Figure 7A). The transgenic lines had more nodes but similar or higher numbers of branches than the NT plants (Figures 7B, C). The number of pods was higher in the transgenic lines than in the NT plants, representing a significant increase of approximately 45%, 70%, 147%, 61%, 99%, 82%, and 87% in lines #1, #7, #10, #11, #12, #13, and #15, respectively (p < 0.05 for line #7 and p < 0.01 for lines #1, #10, #11, #12, #13, and #15) (Figure 7D). The total number of seeds and total seed weight, which exerts a decisive influence on yield, were investigated from the 12 Pβ-con:PfFAD3-1 transgenic lines (T1). The total number of seeds from transgenic lines #1, #7, #10, #11, #12, #13, and #15 increased by approximately 54%, 72%, 167%, 69%, 100%, 93%, and 84%, respectively (p < 0.05 for lines #7 and #13 and p < 0.01 for lines #1, #10, #11, #12, and #15) (Figure 7E). The total seed weights of transgenic lines #1, #6, #7, #8, #10, #11, #12, #13, and #15 were approximately 57%, 48%, 78%, 88%, 176%, 75%, 106%, 108%, and 86% higher than those of the NT plants (p < 0.05 for lines #6, #7, and #8 and p < 0.01 for lines #1, #10, #11, #12, #13, and #15) (Figure 7F). Phenotypic observations of the dried seeds indicated an increase in seed size upon expression of the PfFAD3-1 gene with the β-conglycinin promoter (Figure 8). Ten randomly selected Pβ-con:PfFAD3-1 T2 seeds were arranged in a row, and the average length was measured using 10 repeats. As a result, the seed length of the Pβ-PfFAD3-1-transformed soybean was 6%–14% higher than that of the wild-type soybean seeds. In particular, the highest increase (14%) among the 12 transgenic lines was observed for lines #3, #6, #8, and #10. In summary, the Pβ-con:PfFAD3-1 transgenic soybeans had higher yields than the wild-type soybeans and the increase in seed size contributed to the increased yield by affecting the total seed weight.
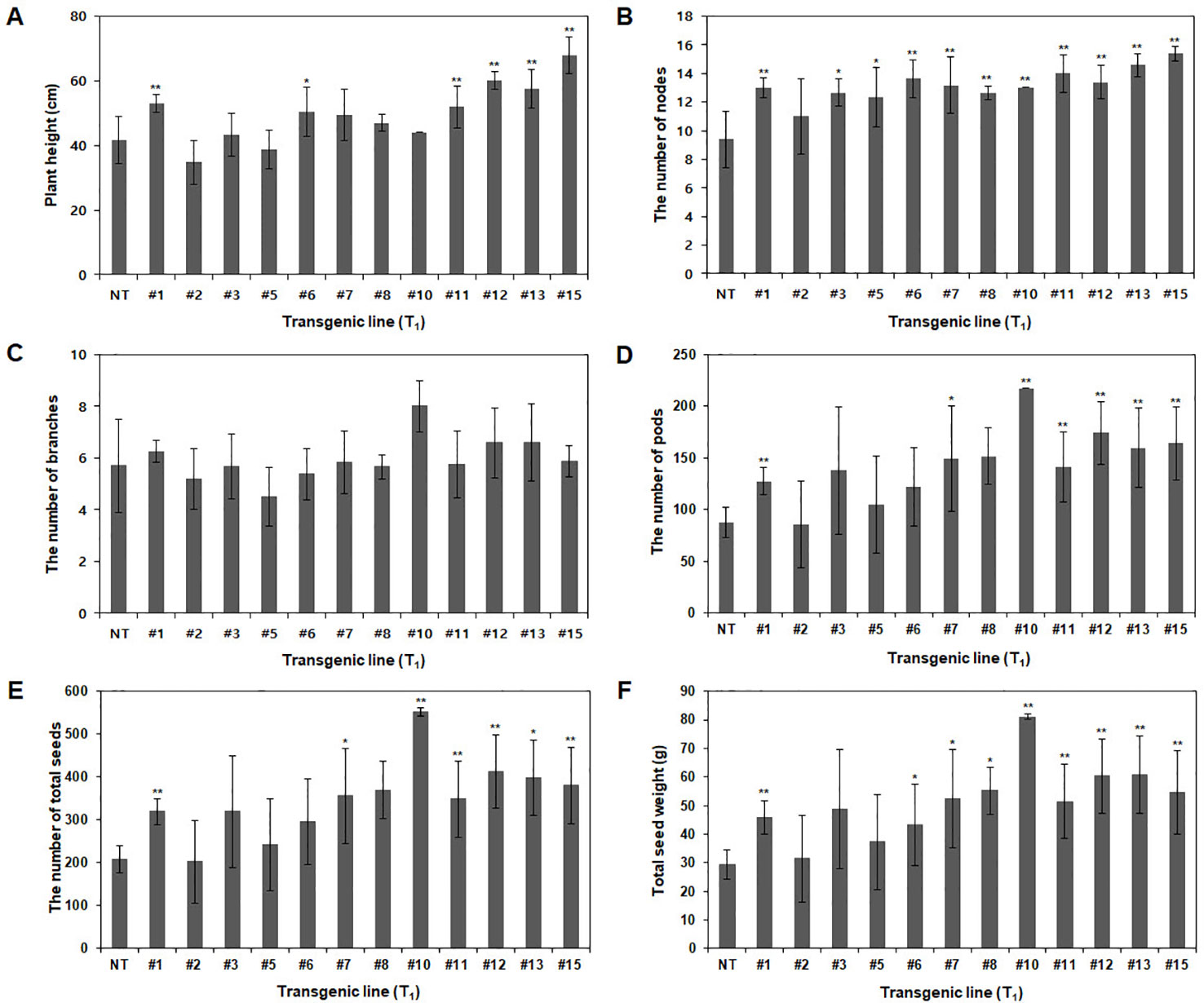
Figure 7 Agronomic characteristics of Pβ-con:PfFAD3-1 transgenic plants (T1) in a GMO field. Non-transgenic and T1 Pβ-con:PfFAD3-1 transgenic plants were cultivated in a GMO field, and agronomic traits such as plant height (A), number of nodes (B), number of branches (C), number of pods (D), total number of seeds (E) and total seed weight (F) were investigated. NT, non-transgenic plants; #1–#15, Pβ-con:PfFAD3-1 transgenic lines (T1). Data represent the means (± SD) from ten independent biological replicates for each line. Asterisks indicate significant changes compared to NT values (* = p < 0.05; ** = p < 0.01).
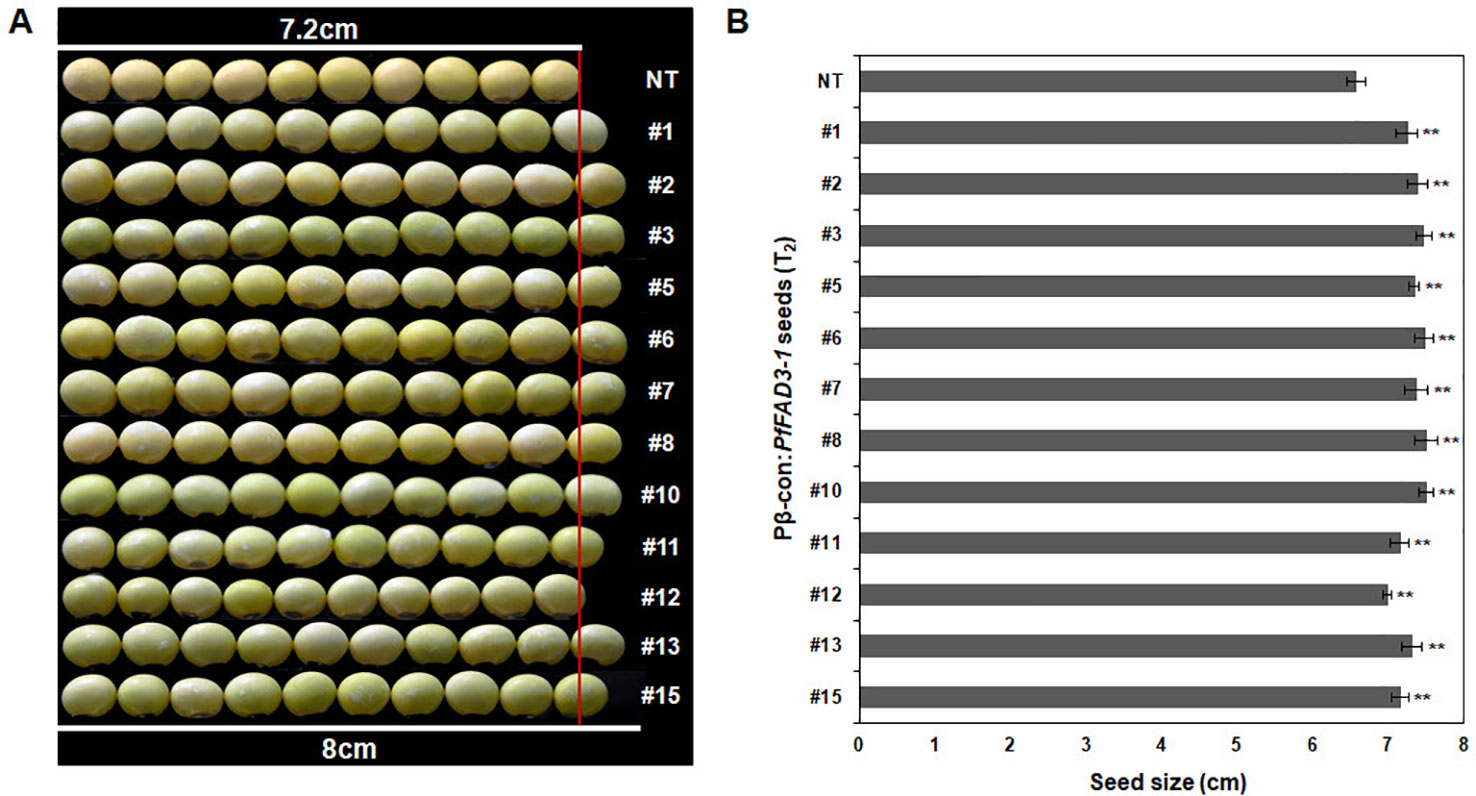
Figure 8 Comparison of sizes of T2 seeds from Pβ-con:PfFAD3-1 transgenic lines and non-transgenic soybean seeds. (A) Comparison of seed size between non-transgenic and T2 Pβ-con:PfFAD3-1 soybean seeds. Non-transgenic and T2 Pβ-con:PfFAD3-1 soybean seeds were randomly chosen and horizontally placed in a row (n = 10 each). (B) Measurement of soybean seed size. The size of non-transgenic and T2 Pβ-con:PfFAD3-1 soybean seeds was measured ten times. NT, non-transgenic soybean seeds; #1–#15, T2 Pβ-con:PfFAD3-1 soybean seeds. Bars indicate the means ± standard deviations. Asterisks indicate significant changes compared to NT values (** = p < 0.01).
Discussion
In this study, we successfully transformed soybean with PfFAD3-1 and developed a new soybean transformant that produces up to 42% ALA in seed oil. Soybean transformation is based on the cotyledonary node (CN) method (Hinchee et al., 1988) and can be improved by various methods. One of these methods involves the use of half-seed explants (Paz et al., 2006). Since the introduction of the half-seed method, we have utilized this efficient transformation method and overcome many difficulties, such as efficiency, reproducibility, and genotype dependency. With this relatively new and modified transformation protocol, various soybean transgenic plants expressing agronomically important genes, such as those imparting tolerance to drought and salt stress, affording resistance to soybean mosaic virus, expressing high content of secondary metabolites, and exhibiting better yields, have been produced (Jiang et al., 2010; Kim et al., 2012; Kim et al., 2013; Kim et al., 2016; Kim et al., 2017; Kwon et al., 2017; Kim et al., 2018; Cho et al., 2019; Park et al., 2019).
Based on our modified Agrobacterium-mediated transformation protocol, the PfFAD3-1 gene was transformed into soybean to increase the ALA content in the seeds. Soybean seed oil is mainly used for frying in the United States and Europe. Therefore, soybeans containing low amounts of polyunsaturated fatty acids, LA and ALA, which are easily oxidized, and high oleic acid content, which is stable to oxidation at high temperatures, have been developed using genetic engineering or mutant breeding methods (Buhr et al., 2002; Graef et al., 2009; Pham et al., 2010; Pham et al., 2012). The genome editing method with clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 was recently used to knockout the soybean FAD2-2 gene to achieve better production of oleic acid (Al Amin et al., 2019). However, soybean is used as a health functional food in the form of soymilk in Korea and East Asia. To improve the oil components of the seeds, transgenic plants with increased content of omega-3 fatty acids were produced in the present study. ALA is the longest carbon omega-3 fatty acid produced by plants and serves as an essential fatty acid consumed in food. ALA is converted to EPA and DHA by the human body and is involved in metabolism. The soybean genome contains four copies of FAD3, three of which are expressed in developing seeds (Bilyeu et al., 2003; Anai et al., 2005). A single copy of FAD3 is expressed in Arabidopsis, which synthesizes 20% of the ALA in the seeds. Perilla (Perilla frutescens L.) has two copies of FAD3, which synthesize 60% of the ALA in the seeds (Arondel et al., 1992; Lee et al., 2016; Lee et al., 2019). Despite the presence of a higher copy number of FAD3 in soybean than in other plants, the ALA content in soybean seeds is as low as 8%. Thus, the FAD3 genes in soybean have weak activity at the transcriptional or translational level or their expression is weak in embryos and endosperms, the sites of oil accumulation. As PfFAD3-1 isolated from lesquerella showed efficient conversion of LA to ALA in Arabidopsis (Lee et al., 2019), the transformation of PfFAD3-1 to soybean was also expected to enhance ALA content. PfFAD3 was expressed in soybean using two seed-specific promoters and the 35S promoter. As expected, PfFAD3 expression under the control of the three promoters resulted in an increase in ALA content in the soybean seeds compared with that in the NT controls. The β-conglycinin and phaseolin promoters showed higher synthesis of ALA than the 35S promoter (Figure 5). Thus, the seed-specific promoters induced more PfFAD3 expression during seed development than the 35S constitutive promoter (Figure 4D). Among the two seed-specific promoters, β-conglycinin showed a higher synthesis of ALA than phaseolin (Figure 5). Being derived from the same species, β-conglycinin may be more favorable for the transcriptional activity of PfFAD3 in soybean than phaseolin from kidney bean. Another possibility is that the regulation of PfFAD3 expression by β-conglycinin may be more consistent with soybean ALA biosynthesis than the regulation of PfFAD3 expression by phaseolin.
The activity of the PfFAD3 transgene was well maintained over generations. ALA content increased by up to 42% in the seeds of the T2 generation plants that were considered to be homozygous for the transgene (Figure 6). This ALA content is 45% that of flax (Linum usitatissimum L.) and 60% that of perilla, the two oil crops with the highest ALA content (Rao et al., 2008). Considering the cultivation area and production of seed oil compared with other oil crops, high omega-3 soybeans that produce 42% ALA may provide a great opportunity for increased omega-3 production.
During the investigation of agronomic traits, such as plant height, number of branches, nodes, pods, and total seeds, and total seed weight, several Pβ-con:PfFAD3-1 transgenic lines (T1) showed a significant increase in most traits (Figure 7A). In particular, the number of pods and total seeds and the total seed weight significantly increased. This increase in yield parameters could reflect the apparent increase in the size of seeds from the Pβ-con:PfFAD3-1 transgenic lines (Figure 8). The seed length of Pβ-PfFAD3-1-transformed soybeans (lines #3, #6, #8, and #10) increased by more than 10% compared to that of wild-type soybean plants. Further detailed analysis of the seeds is currently underway and we have confirmed an approximately 10% increase in cell number and area (data not shown). Currently, we do not know how the high ALA content of the Pβ-con:PfFAD3 transformants increases seed yield and seed size. ALA is a precursor to the jasmonic acid (JA) phytohormone. It has been reported that JA treatment increases the grain yield of amaranth in the absence of drought stress (Délano-Frier et al., 2004). ALA treatment of Arabidopsis cell suspension culture has been shown to induce the expression of methionine sulfoxide reductase and alkenal reductase genes that protect against abiotic and oxidative stresses (Mata-Pérez et al., 2015). Therefore, it is possible to increase the expression of genes that offer resistance to abiotic stress to increase adaptability to the growing environment or increase growth by inducing resistance to oxidative stress even under normal growth conditions. Investigating the various metabolic changes in plants with an increased ALA content will help to determine the mechanism of increased yields.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
WY, HJK, K-RL, HC, SJ, and J-YK performed the experiments. HJ and S-WO analyzed the data. HUK and Y-SC wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by a grant from the Next-Generation BioGreen 21 Program, (Project No. PJ01366501), Rural Development Administration, South Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ALA: α-linolenic acid (18:3), FAME: fatty acid methyl ester, NT: non-transgenic, CN: cotyledonary node, Pβ: β-conglycinin promoter, Pphas: phaseolin promoter
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01812/full#supplementary-material
Supplementary Figure 1 | Determination of transgene copy number in Pphas : PfFAD3-1 transgenic plants with real-time PCR. Genomic DNA was extracted from Pphas : PfFAD3-1 transgenic plants (T0) to determine transgene copy number. Bar, target Bar gene; Con, homozygous single copy transgenic plant; NT, non-transgenic plant; #1–#7, Pphas : PfFAD3-1 transgenic lines (T0).
References
Al Amin, N., Ahmad, N., Wu, N., Pu, X., Ma, T., Du, Y., et al. (2019). CRISPR-Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max.L). BMC Biotechnol. 19, 9. doi: 10.1186/s12896-019-0501-2
Amjad Khan, W., Chun-Mei, H., Khan, N., Iqbal, A., Lyu, S.-W., Shah, F. (2017). Bioengineered Plants Can Be a Useful Source of Omega-3 Fatty Acids. BioMed. Res. Int. 2017, 7348919. doi: 10.1155/2017/7348919
Anai, T., Yamada, T., Kinoshita, T., Rahman, S. M., Takagi, Y. (2005). Identification of corresponding genes for three low-α-linolenic acid mutants and elucidation of their contribution to fatty acid biosynthesis in soybean seed. Plant Sci. 168, 1615–1623. doi: 10.1016/j.plantsci.2005.02.016
Arondel, V., Lemieux, B., Hwang, I., Gibson, S., Goodman, H. M., Somerville, C. R. (1992). Map-based cloning of a gene controlling omega-3 fatty acid desaturation in Arabidopsis. Science 258, 1353–1355. doi: 10.1126/science.1455229
Bilyeu, K. D., Palavalli, L., Sleper, D. A., Beuselinck, P. R. (2003). Three microsomal omega-3 fatty-acid desaturase denes contribute to soybean linolenic acid levels. Crop Sci. 43, 1833–1838. doi: 10.2135/cropsci2003.1833
Buhr, T., Sato, S., Ebrahim, F., Xing, A., Zhou, Y., Mathiesen, M., et al. (2002). Ribozyme termination of RNA transcripts down-regulate seed fatty acid genes in transgenic soybean. Plant J. 30, 155–163. doi: 10.1046/j.1365-313X.2002.01283.x
Chen, L., Cai, Y., Liu, X., Yao, W., Guo, C., Sun, S., et al. (2018). Improvement of soybean Agrobacterium-mediated transformation efficiency by adding glutamine and asparagine into the culture media. Int. J. Mol. Sci. 19 (10), 3039. doi: 10.3390/ijms19103039
Cho, H. S., Lee, D. H., Jung, H. W., Oh, S. W., Kim, H. J., Chung, Y. S. (2019). Evaluation of yield components from transgenic soybean overexpressing chromatin architecture-controlling ATPG8 and ATPG10 genes. Plant Breed. Biotech. 7 (1), 34–41. doi: 10.9787/PBB.2019.7.1.34
Délano-Frier, J. P., Martínez-Gallardo, N. A., Martínez-de la Vega, O., Salas-Araiza, M. D., Barbosa-Jaramillo, E. R., Torres, A., et al. (2004). The effect of exogenous jasmonic acid on induced resistance and productivity in amaranth (Amaranthus hypochondriacus) is influenced by environmental conditions. J. Chem. Ecol. 30, 1001–1034. doi: 10.1023/B:JOEC.0000028464.36353.bb
Dierig, D. A., Tomasi, P. M., Dahlquist, G. H. (2001). Registration of WCL-LY2 high oil Lesquerella fendleri germplasm. Crop Sci. 41, 604–604. doi: 10.2135/cropsci2001.412604x
Doyle, J. J., Schuler, M. A., Godette, W. D., Zenger, V., Beachy, R. N., Slightom, J. L. (1986). The glycosylated seed storage proteins of Glycine max and Phaseolus vulgaris. Structural homologies of genes and proteins. J. Biol. Chem. 261, 9228–9238.
El-Hamidi, M., Zaher, F. A. (2018). Production of vegetable oils in the world and in Egypt: an overview. Bull. Natl. Res. Centre 42, 19. doi: 10.1186/s42269-018-0019-0
Flores, T., Karpova, O., Su, X., Zeng, P., Bilyeu, K., Sleper, D. A., et al. (2008). Silencing of GmFAD3 gene by siRNA leads to low alpha-linolenic acids (18:3) of fad3-mutant phenotype in soybean [Glycine max (Merr.)]. Transgenic Res. 17, 839–850. doi: 10.1007/s11248-008-9167-6
Graef, G., LaVallee, B. J., Tenopir, P., Tat, M. E., Schweiger, B. J., Kinney, A. J., et al. (2009). A high oleic acid and low palmitic acid soybean: agronomic performance and evaluation as a feedstock for biodiesel. Plant Biotechnol. J. 7, 411–421. doi: 10.1111/j.1467-7652.2009.00408.x
Hinchee, M. A. W., Connor-Ward, D. V., Newell, C. A., McDonnell, R. E., Sato, S. J., Gasser, C. S., et al. (1988). Production of transgenic soybean plants using Agrobacterium-mediated DNA transfer. Nat. Biotechnol. 6, 915–922. doi: 10.1038/nbt0888-915
Homrich, M. S., Wiebke-Strohm, B., Weber, R. L. M., Bodanese-Zanettini, M. H. (2012). Soybean genetic transformation: A valuable tool for the functional study of genes and the production of agronomically improved plants. Genet. Mol. Biol. 35suppl, 998–1010. doi: 10.1590/S1415-47572012000600015
Jhala, A. J., Hall, L. M. (2010). Flax (Linum usitatissimum L.): Current uses and future applications. Aust. J. Basic Appl. Sci. 4 (9), 4304–4312.
Jiang, N., Jeon, E. H., Pak, J. H., Ha, T. J., Baek, I. Y., Jung, W. S., et al. (2010). Increase of isoflavones in soybean callus by Agrobacterium-mediated transformation. Plant Biotechnol. Rep. 4, 253–260. doi: 10.1007/s11816-010-0143-2
John, K. M. M., Natarajan, S., Luthria, D. L. (2016). Metabolite changes in nine different soybean varieties grown under field and greenhouse conditions. Food Chem. 211, 347–355. doi: 10.1016/j.foodchem.2016.05.055
Kanai, M., Yamada, T., Hayashi, M., Mano, S., Nishimura, M. (2019). Soybean (Glycine max L.) triacylglycerol lipase GmSDP1 regulates the quality and quantity of seed oil. Sci. Rep. 9, 8924. doi: 10.1038/s41598-019-45331-8
Karimi, M., Inze´, ,. D., Depicker, A. (2002). GatewayTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 (5), 193–195. doi: 10.1016/S1360-1385(02)02251-3
Khan, W. A., Chun-Mei, H., Khan, N., Iqbal, A., Lyu, S. W., Shah, F. (2017). Bioengineered plants can be a useful source of omega-3 fatty acid. BioMed. Res. Int. 7348919, 9. doi: 10.1155/2017/7348919
Kim, M. J., Kim, J. K., Kim, H. J., Pak, J. H., Lee, J. H., Kim, D. H., et al. (2012). Genetic modification of the soybean to enhance the β-carotene content through seed-specific expression. PloS One 7 (10), e48287. doi: 10.1371/journal.pone.0048287
Kim, H. J., Kim, M. J., Pak, J. H., Jung, H. W., Choi, H. K., Lee, Y. H., et al. (2013). Characterization of SMV resistance of soybean produced by genetic transformation of SMV-CP gene in RNAi. Plant Biotechnol. Rep. 7, 425–433. doi: 10.1007/s11816-013-0279-y
Kim, H. J., Kim, M. J., Pak, J. H., Im, H. H., Lee, D. H., Kim, K. H., et al. (2016). RNAi-mediated soybean mosaic virus (SMV) resistance of a Korean soybean cultivar. Plant Biotechnol. Rep. 10, 257–267. doi: 10.1007/s11816-016-0402-y
Kim, H. J., Cho, H. S., Pak, J. H., Kim, K. J., Lee, D. H., Chung, Y. S. (2017). Overexpression of a chromatin architecture-controlling ATPG7 has positive effect on yield components in transgenic soybean. Plant Breed. Biotech. 5 (3), 237–242. doi: 10.9787/PBB.2017.5.3.237
Kim, H. J., Cho, H. S., Pak, H. J., Kwon, T., Lee, J. H., Kim, D. H., et al. (2018). Confirmation of drought tolerance of ectopically expressed AtABF3 gene in soybean. Mol. Cells 41 (5), 413–422. doi: 10.14348/molcells.2018.2254
Kwon, T., Kim, H. J., Yun, S. Y., Kim, J., Cho, H. S., Nam, J., et al. (2017). Enhancement of syringin contents in soybean seeds with seed-specific expression of a chimeric UGT72E3/E2 gene. Plant Biotechnol. Rep. 11, 439–447. doi: 10.1007/s11816-017-0464-5
Lee, K.-R., Lee, Y., Kim, E.-H., Lee, S.-B., Roh, K. H., Kim, J.-B., et al. (2016). Functional identification of oleate 12-desaturase and ω-3 fatty acid desaturase genes from Perilla frutescens var. frutescens. Plant Cell Rep. 35, 2523–2537. doi: 10.1007/s00299-016-2053-4
Lee, K.-R., Kim, E.-H., Jeon, I., Lee, Y., Chen, G. Q., Kim, H. U. (2019). Lesquerella FAD3-1 gene is responsible for the biosynthesis of trienoic acid and dienoic hydroxy fatty acids in seed oil. Ind. Crops Prod. 134, 257–264. doi: 10.1016/j.indcrop.2019.04.008
Li, S., Cong, Y., Liu, Y., Wang, T., Shuai, Q., Chen, N., et al. (2017). Optimization of Agrobacterium-mediated transformation in soybean. Front. Plant Sci. 8, 246. doi: 10.3389/fpls.2017.00246
Lipp, M., Brodmann, P., Pietsch, K., Pauwels, J., Anklam, E. (1999). IUPAC collaborative trial study of a method to detect genetically modified soy beans and maize in dried powder. J. AOAC Int. 82, 923–928.
Manavalan, L. P., Guttikonda, S. K., Tran, L. S. P., Nguyen, H. T. (2009). Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 50 (7), 1260–1276. doi: 10.1093/pcp/pcp082
Mata-Pérez, C., Sánchez-Calvo, B., Begara-Morales, J. C., Luque, F., Jiménez-Ruiz, J., Padilla, M. N., et al. (2015). Transcriptomic profiling of linolenic acid-responsive genes in ROS signaling from RNA-seq data in Arabidopsis. Front. Plant Sci. 17, 122. doi: 10.3389/fpls.2015.00122
Oksman-Caldentey, K.-M., Hiltunen, R. (1996). Transgenic crops for improved pharmaceutical products. Field Crops Res. 45, 57–69. doi: 10.1016/0378-4290(95)00059-3
Park, J. S., Kim, H. J., Cho, H. S., Jung, H. W., Cha, J. Y., Yun, D. J., et al. (2019). Overexpression of AtYUCCA6 in soybean crop results in reduced ROS production and increased drought tolerance. Plant Biotechnol. Rep. 13, 161–168. doi: 10.1007/s11816-019-00527-2
Paz, M. M., Martinez, J. C., Kalvig, A. B., Fonger, T. M., Wang, K. (2006). Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep. 25, 206–213. doi: 10.1007/s00299-005-0048-7
Pham, A. T., Lee, J. D., Shannon, J. G., Bilyeu, K. D. (2010). Mutant alleles of FAD2-1A and FAD2-1B combine to produce soybeans with the high oleic acid seed oil trait. BMC Plant Biol. 10, 195. doi: 10.1186/1471-2229-10-195
Pham, A. T., Shannon, J. G., Bilyeu, K. D. (2012). Combinations of mutant FAD2 and FAD3 genes to produce high oleic acid and low linolenic acid soybean oil. Theor. Appl. Genet. 125, 503–515. doi: 10.1007/s00122-012-1849-z
Psota, T. L., Gebauer, S. K., Kris-Etherton, P. (2006). Dietary omega-3 fatty acid intake and cardiovascular risk. Am. J. Cardiol. 98, 3i–18i. doi: 10.1016/j.amjcard.2005.12.022
Rao, S., Abdel-Reheem, M., Bhella, R., Mccracken, C., Hildebrand, D. (2008). Characteristics of high alpha-linolenic acid accumulation in seed oils. Lipids 43, 749–755. doi: 10.1007/s11745-008-3207-0
Sengupta-Gopalan, C., Reichert, N. A., Barker, R. F., Hall, T. C., Kemp, J. D. (1985). Developmentally regulated expression of the b-phaseolin gene in tobacco seed. Proc. Natl. Acad. Sci. U.S.A. 82, 3320–3324. doi: 10.1073/pnas.82.10.3320
Simopoulos, A. P. (2000). Human requirement for N-3 polyunsaturated fatty acids. Poult. Sci. 79, 961–970. doi: 10.1093/ps/79.7.961
Yang, J., Xing, G., Niu, L., He, H., Guo, D., Du, Q., et al. (2018). Improved oil quality in transgenic soybean seeds by RNAi-mediated knockdown of GmFAD2-1B. Transgenic. Res. 27, 155–166. doi: 10.1007/s11248-018-0063-4
Keywords: ALA: alpha-linolenic acid, FAME: fatty acid methyl ester, NT: non-transgenic, Pβ: β-conglycinin promoter, Pphas: phaseolin promoter
Citation: Yeom WW, Kim HJ, Lee K-R, Cho HS, Kim J-Y, Jung HW, Oh S-W, Jun SE, Kim HU and Chung Y-S (2020) Increased Production of α-Linolenic Acid in Soybean Seeds by Overexpression of Lesquerella FAD3-1. Front. Plant Sci. 10:1812. doi: 10.3389/fpls.2019.01812
Received: 09 September 2019; Accepted: 27 December 2019;
Published: 31 January 2020.
Edited by:
Goetz Hensel, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), GermanyReviewed by:
Joaquín J. Salas, Instituto de la Grasa (IG), SpainRaquel Iglesias Fernández, Polytechnic University of Madrid, Spain
Copyright © 2020 Yeom, Kim, Lee, Cho, Kim, Jung, Oh, Jun, Kim and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyun Uk Kim, aHVraW02NEBzZWpvbmcuYWMua3I=; Young-Soo Chung, Y2h1bmd5c0BkYXUuYWMua3I=
†These authors have contributed equally to this work
 Wan Woo Yeom1†
Wan Woo Yeom1† Kyeong-Ryeol Lee
Kyeong-Ryeol Lee Ho Won Jung
Ho Won Jung Hyun Uk Kim
Hyun Uk Kim Young-Soo Chung
Young-Soo Chung