- 1Graduate School of Environmental and Life Science, Okayama University, Okayama, Japan
- 2Department of Crop Sciences, University of Illinois at Urbana-Champaign, Urbana, IL, United States
- 3Graduate School of Agriculture, Kyoto University, Sakyo-ku, Japan
Sex expression in plants is often flexible and contributes to the maintenance of genetic diversity within a species. In diploid persimmons (the genus Diospyros), the sexuality is controlled by the Y chromosome-encoded small-RNA gene, OGI, and its autosomal counterpart, MeGI. Hexaploid Oriental persimmon (Diospyros kaki) evolved more flexible sex expression, where genetically male individuals carrying OGI can produce both male and female flowers (monoecy). This is due to (semi-)inactivation of OGI by the Kali-SINE retrotransposon insertion on the promoter region and the resultant DNA methylations. Instead, flower sex determination in Oriental persimmon is also dependent on DNA methylation states of MeGI. Here, we focused on a cultivar, Kumemaru, which shows stable male flower production. Our results demonstrated that cv. Kumemaru carries OGI with Kali-SINE, which was highly methylated as well as in other monoecious cultivars; nevertheless, OGI gene could have a basal expression level. Transcriptomic analysis between cv. Kumemaru and 14 cultivars that predominantly produce female flowers showed differentially expressed genes (DEGs) specific to cv. Kumemaru, which is mainly involved in stress responses. Co-expression gene networks focusing on the DEGs also suggested the involvement of stress signals, mainly via gibberellin (GA), salicylic acid (SA), and especially jasmonic acid (JA) signal pathways. We also identified potential regulators of this co-expression module, represented by the TCP4 transcription factor. Furthermore, we attempted to identify cv. Kumemaru-specific transcript polymorphisms potentially contributing to derepressed OGI expression by cataloging subsequences (k-mers) in the transcriptomic reads from cv. Kumemaru and the other 14 female cultivars. Overall, although the direct genetic factor to activate OGI remains to be solved, our results implied the involvement of stress signals in the release of silenced OGI and the resultant continuous male production.
Introduction
Sexuality is a main biological process that maintains genetic diversity within a species. Nevertheless, most angiosperms are hermaphroditic, which is thought to be the ancestral state in flowering plants (Ainsworth, 2000). Minorities have evolved distinct sexual systems, including monoecy (i.e., the formation of male and female flowers by one plant) or dioecy (i.e., separate male and female plants) (Renner, 2014). Regarding dioecy, a few genetic determinants have been identified in some lineages, including persimmons (Diospyros spp.) (Akagi et al., 2014), garden asparagus (Asparagus officinalis L.) (Murase et al., 2017; Harkess et al., 2017; Tsugama et al., 2017), and kiwifruit (Actinidia spp.) (Akagi et al., 2018, 2019). Candidates of sex determinants have been also identified in date palm (Torres et al., 2018), wild strawberries (Tennessen et al., 2018), grape (Zhou et al., 2017), and poplar (Müller et al., 2020). These studies have revealed molecular bases of the genetic sex-determining genes and the evolutionary paths to establish dioecious sex determination systems. On the other hand, mechanisms relating to transitions out of dioecy have been little understood. Transition out of dioecy often involves domestication or polyploidizations in plants (Comai, 2005; Goldberg et al., 2017; Henry et al., 2018). The transition from dioecy into hermaphroditism in domestication would be explainable with artificial selective pressures for stable production, such as in papaya and grape (VanBuren et al., 2015; Zhou et al., 2019). Yet, a distinct mechanism to connect polyploidization and transition out of dioecy remains to be solved.
In dioecious Diospyros species, the Y chromosome-encoded small-RNA (smRNA) gene, OGI, which is believed to be a single-sex determinant, is responsible for repressing the expression of the autosomal counterpart, MeGI (Akagi et al., 2014; Yang et al., 2019). A recent polyploidization event in wild diploid species that generated the cultivated hexaploid Oriental persimmon (Diospyros kaki) promoted a transition out of dioecy and established a plastic sex-determination system (Akagi et al., 2016a; Henry et al., 2018). In hexaploid persimmon, genetically male plants carrying at least one copy of the OGI gene exhibit monoecy, while individuals carrying only X chromosomes are female (Akagi et al., 2016a,b). The basic mechanism for producing the male flower in hexaploid monoecious D. kaki individuals is identical to that in diploid dioecious male individuals, in which it depends on the repression of MeGI expression. In hexaploid D. kaki, OGI expression is, however, substantially inactivated because of the presence of a highly methylated SINE-like transposon, named Kali, in the promoter region (Akagi et al., 2016a). Hypothetically, once OGI occasionally (or accidentally) works to trigger small-RNA production in MeGI, the resultant DNA methylation in the promoter region represses MeGI expression to form initial male flowers. Then, from the male parental branches, the sexuality of the derived flowers is dependent on the maintenance or release of DNA methylation in MeGI. These situations derive monoecious individuals that are genetically male (Figure 1; Akagi et al., 2016a; Henry et al., 2018). In previous studies, all of the assessed genetically male cultivars/accessions, which carry at least one copy of OGI gene, included the Kali insertion in the OGI promoter region and exhibited monoecy or female (Akagi et al., 2016a,b). This suggested that the derepression of OGI had importance in the evolutionary path to establishing the hexaploid D. kaki population.
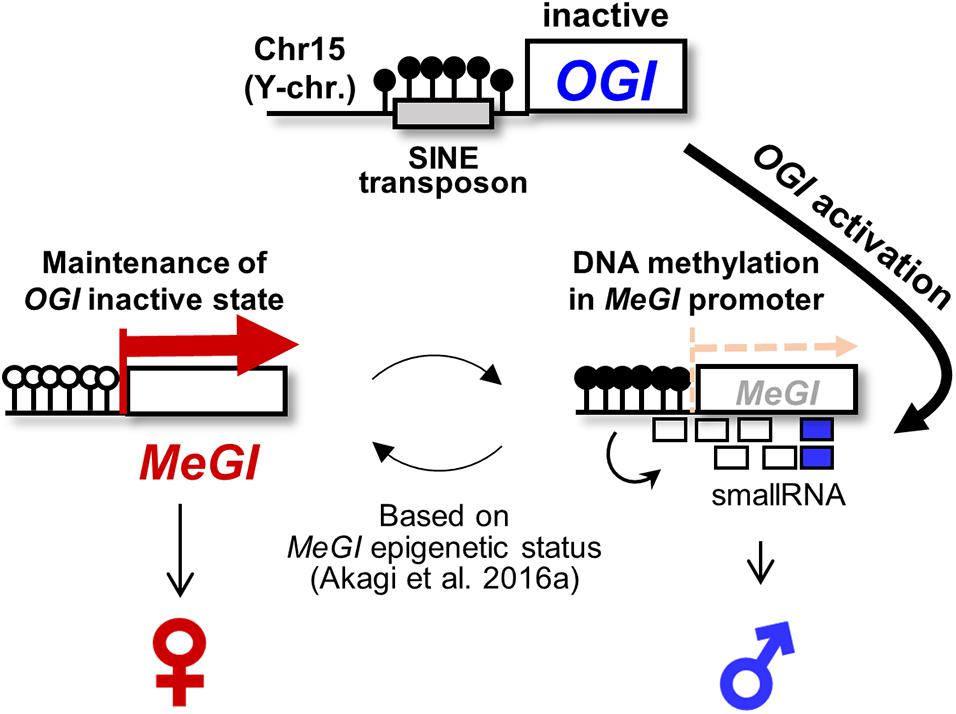
Figure 1. Epigenetic sex determination model in monoecious D. kaki. The Y chromosome-encoded small-RNA gene OGI triggers to accumulate the small-RNAs targeting its autosomal counterpart MeGI, which induces DNA methylation of the MeGI sequence, resulting in the repression of the MeGI expression. In D. kaki (Oriental persimmon), OGI is basically inactivated by the Kali-SINE insertion and the resultant DNA methylation. Hypothetically, occasional (or accidental) activation of OGI allows small-RNA production and DNA methylation in MeGI. Once the promoter region of MeGI is methylated, the fate of flower sexuality depends on the maintenance or release of DNA methylation in MeGI (Akagi et al., 2016a).
Among a wide variety of Oriental persimmon cultivars deposited to the germplasm repository in the Genebank Project, NARO, Japan,1 cv. Kumemaru2 is thought to specifically produce only male flowers. This cultivar was derived from a selection of chance seedling in the purpose of the collection of stable pollinizers at Toyama prefecture in Japan. Although the genetic factors or inheritance mode involving this derepressed male production system have not been identified, the previous study has already indicated that cv. Kumemaru carries the OGI gene with Kali-SINE insertion in its promoter region, as well as in other cultivars (Akagi et al., 2016a). To characterize the mechanism underlying the derepressed male production in cv. Kumemaru, in this study, we assessed on OGI regulation and then approached from co-expression network and comprehensive polymorphism detection. Insights into an irregularly derepressed male production system would shed light on the mechanism of the repressing in hexaploid persimmon.
Materials and Methods
Plant Materials
Developmental stages of diploid (Diospyros lotus) and Oriental persimmon (D. kaki Thunb.) buds/flowers were defined in previous studies (Akagi et al., 2016a). Flower buds in 14 predominantly female-producing cultivars, which carry at least one copy of OGI but have produced no or rare male flowers, and cv. Kumemaru were collected in early June 2017, which correspond to the male/female primordia differentiation (Akagi et al., 2014, 2016a). The 13 dominantly female-producing cultivars (cv. Atago, cv. Beniwase, cv. Chagone, cv. Kunitomi, cv. Nagara, cv. Nanshi, cv. Ogosho, cv. Okugosho, cv. Oniwa, cv. Saisho, cv. Suruga, cv. Yashima, cv. Yoshino) planted in the experimental orchard of Kyoto University (Kyoto, Japan) and cv. Kumemaru and cv. Suruga planted in the grape and persimmon research station, NIFTS, NARO, Japan (Hiroshima, Japan), were used. To construct mRNA-Seq libraries, the buds were separately collected at different positions of medial and top in branches with three to four biological replicates abbreviated as #1 and #2, respectively. The collected samples were frozen at −80°C until used for RNA extractions.
Characterization of MeGI and OGI
cDNAs of developing flower buds of cvs. Taishu and Kumemaru, and D. lotus cv. Kunsenshi-male were synthesized from total RNA with ReverTra Ace qRT-PCR Master Mix (TOYOBO, Osaka, Japan). The relative expression level of OGI and MeGI (Akagi et al., 2014, 2016b for the primer sequences) was detected by qRT-PCR using a LightCycler 480 (Roche Diagnostics, Mannheim, Germany), with four biological replicates. A constitutively expressed DkActin gene in hexaploid persimmon (Akagi et al., 2009) was used for standardizing expression levels. qPCR analyses were conducted under the following conditions: 95°C for 5 min followed by 45 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 30 s, using THUNDERBIRD SYBR qPCR Mix (TOYOBO).
Genomic DNA was extracted from young flower buds with the CTAB method (Akagi et al., 2014). The full-length and 5′ promoter regions of the OGI in cv. Kumemaru and a wide variety of Diospyros species, D. kaki, D. lotus, Diospyros glaucifolia, Diospyros digyna, Diospyros virginiana, Diospyros mespiliformis, and Diospyros rhombifolia, were amplified from gDNAs, with the OGI-prom-2F-TOPO and OGI-spR primer set (Akagi et al., 2016a). Quantitative genotyping of OGI in cv. Kumemaru was conducted according to the previous report (Akagi et al., 2016b). Alleles of Kali-SINE insertion on the OGI promoter and OGI transcript region, in cv. Kumemaru and D. kaki cultivars, were amplified from gDNAs, with primer sets OGI-441-kali-F (5′- GCTCCAGTTTAGTTATGGAAGAG-3′, Tm: 60) and OGI-441-kali-R (5′- ATTGCCAACGTTCACATCAC -3′, Tm: 63) for the Kali-SINE, and OGI-candF and OGI-spR for the OGI transcript region (Akagi et al., 2016b). For the detection of DNA methylation status in OGI, gDNA extracted from developing flower buds was treated with an EZ DNA Methylation-Gold kit (Zymo Research, United States) to deaminate and convert non-methylated cytosine residues into uracil residues. Bisulfite-treated OGI 5′ promoter sequences were amplified with Epi Taq HS (TaKaRa) and each four sense- and antisense-direction primer sets (Akagi et al., 2016a). The bisulfite amplicons were subjected to Illumina library construction described below.
Illumina Sequencing
To construct mRNA-Seq libraries, total RNA was purified with the Dynabeads mRNA Purification Kit (Invitrogen, United States). The mRNA-Seq libraries were prepared with the KAPA RNA HyperPrep Kit (Roche, Switzerland) as previously described (Yang et al., 2019), followed by a DNA cleanup step with AMPure XP beads (Beckman Coulter, United States; AMPure: reaction = 0.8:1). To construct bisulfite-PCR-Seq libraries, the promoter region of OGI was amplified from bisulfite-treated gDNA of cv. Kumemaru, according to the previous report (Akagi et al., 2016a). The bisulfite amplicons were purified with AMPure and then applied to KAPA HyperPrep Kit (Roche) as previously described (Akagi et al., 2016a). The purified libraries were quantified with a Qubit 2.0 fluorometer (Invitrogen) and then analyzed with the Illumina HiSeq 4000 system at the QB3 Genomic Sequencing Laboratory of UC Berkeley.3 The resulting SR50 sequencing reads were analyzed at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley. Raw sequencing reads were processed using custom Python scripts developed in the Comai Laboratory, which are available online,4 as previously described (Akagi et al., 2014). All Illumina sequencing data have been deposited in the DDBJ database: Short Read Archives (SRA) database (BioProject ID PRJDB9564, Run ID DRR233540-DRR233569).
Transcriptomic Data Profiling
The mRNA-Seq Illumina reads were aligned to the reference coding sequences (CDSs) of the diploid Caucasian persimmon, D. lotus5 (Akagi et al., 2020), using the default parameters of the Burrows-Wheeler Aligner (BWA) (version 0.7.12) (Li and Durbin, 2009).6 These settings allow the mapping of variable allele sequences in hexaploid D. kaki (Yang et al., 2019). The read counts per CDS were determined from the aligned SAM files using a custom R script to calculate the RPKM for each gene. To examine the gene expression dynamics in 14 predominantly female-producing cultivars and cv. Kumemaru, a PCA was conducted using the genes with RPKM > 1, with prcomp in R.
Differentially expressed genes (DEGs) between predominantly female-producing cultivars and cv. Kumemaru were detected by DESeq analysis with “fit-only” for sharing mode (FDR < 0.01). The DEGs were further filtered based on RPKM and bias values (RPKM > 1.0, bias >2). Furthermore, Kumemaru-specific DEGs were detected by comparing between cv. Kumemaru and each predominantly female-producing cultivar with edgeR (Robinson et al., 2010; McCarthy et al., 2012) using the paired-test option depending on the positions in branches (N = 2 × 14 for biological replicates). The DEGs were filtered according to RPKM and p-values (RPKM > 1.0, p < 0.1 for cvs. Nagara and Ogosho, and p < 0.05 for the rest of the 12 cultivars) (Supplementary Table 1). Putative functions of each gene were annotated with a BLASTX search of the TAIR10 database.7
Construction of the Co-expression Network
The genes detected as DEGs in the 14 comparisons of predominantly female-producing cultivars and cv. Kumemaru (N = 20,216) were selected to construct a gene co-expression network with the WGCNA package (Langfelder and Horvath, 2008). The soft-thresholding power for a signed network was set at 5, with a scale-free model fitting index R2 > 0.6. A relatively large minimum module size (30) and a medium sensitivity (deepSplit = 2) to cluster splitting were applied. In co-expression networking, genes were represented by nodes, and the correlation values (weight) between two genes were calculated by raising the Pearson’s correlation coefficient. The heatmap for expression was designed with ggplot2 packages (Csardi and Nepusz, 2006; Wickham, 2009). The genes in the same module were first visualized with the Cytospace program (Shannon et al., 2003) and only lines with a weight greater than 0.43.
Cataloging of cv. Kumemaru-Specific Subsequences From mRNA-Seq Reads
For the comprehensive caption of Kumemaru-specific polymorphisms in the expressed transcripts, we conducted “kmer cataloging” in the cDNA reads, according to the previous reports characterizing Y-linked polymorphisms in persimmon (Akagi et al., 2014) and kiwifruit (Akagi et al., 2018). We cataloged 35-bp subsequences triggered with an “A” nucleotide in the mRNA-Seq reads of cv. Kumemaru and of the predominantly female-producing cultivars pools using custom Python scripts.8 The subsequence catalogs from cv. Kumemaru and from female cultivars were compared to extract the Kumemaru-specific subsequences (“0” counts in the female cultivars pool). The Illumina reads including cv. Kumemaru-specific subsequences (k-mers) (KSK) were assembled with a CLC assembler, as described (Akagi et al., 2014). The derived contigs were filtered with the remapping of the KSK to exclude minor polymorphisms or sequencing noises (nos. of KSK < 10).
Results
Morphological Characterization of Male Flowers in cv. Kumemaru
The architectures of flowering branches and inflorescences in cv. Kumemaru were similar to those in other monoecious cultivars, where male flowers set in medial positions of the parental branches, with cyme-like trifurcated inflorescence (Figures 2A,B). In the flower developing/maturing stages (stages 1–4) previously defined (Yang et al., 2019), developmental processes of male flowers/organs in cv. Kumemaru were consistent with other monoecious cultivars (Figures 2C–N, compared with Yang et al., 2019), producing fertile anther/pollens (Figures 2F,J,N). Flower structure/components in cv. Kumemaru were identical to male flowers in monoecious cv. Taishu (Figures 2O,P). These observations suggested that the mechanism of male flower production in cv. Kumemaru is consistent with monoecious cultivars, where gene networks orchestrated by MeGI play an important role (Yang et al., 2019).
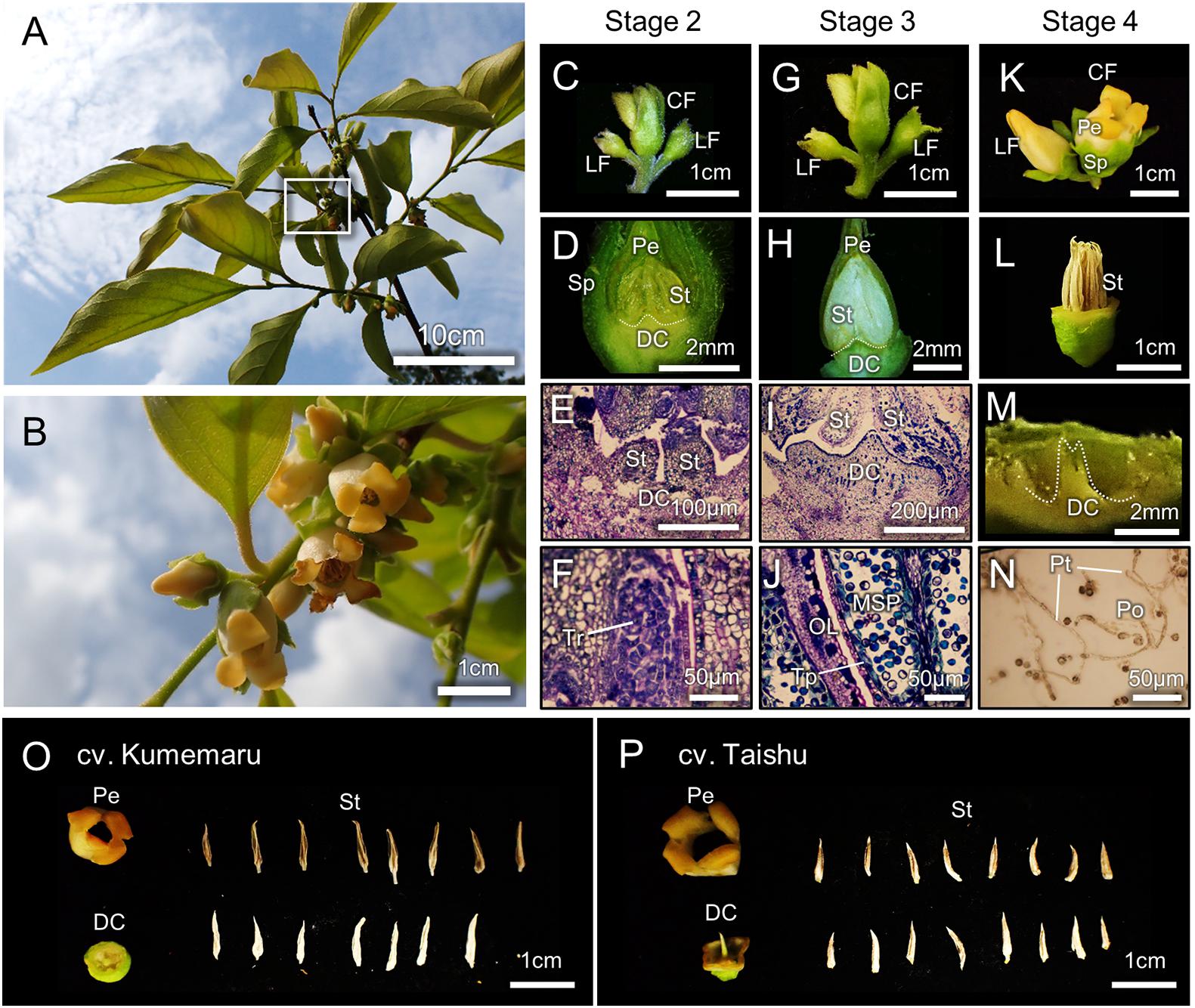
Figure 2. Morphological characterization of male flower development in cv. Kumemaru. (A,B) The appearance of flowering branches in cv. Kumemaru (A), and the closing up of the white box in panel (A), showing cyme-like trifurcated inflorescence (B). (C–N) The developmental processes of male flowers in cv. Kumemaru, in the stages defined according to Yang et al. (2019). In stage 2 (C–F), tetrads (Tr) were formed inside stamens (St) (F). In stage 3 (G–J), microspores (MSP) were developed, while tapetal layers (Tp) have not been degenerated (J). In stage 4 (K–N), flowers opened with mature stamens, producing fertile pollens (Po). Sp, sepal; Pe, petal; DC, deficient carpel; Pt, pollen tube. (O,P) Alignment of male flower components in cv. Kumemaru (O) and monoecious cv. Taishu (P), which included four gamosepalous petals, 1 deficient carpel, and ca 16 stamens.
Stable Male Production System Caused by Derepression of the OGI Silencing
To find out the basic molecular mechanism of stable male production in cv. Kumemaru, we first analyzed the regulations of OGI and MeGI in cv. Kumemaru. The expression level of MeGI in cv. Kumemaru was substantially lower than in the predominantly female-producing cultivars, at the onset of primordia development (Figure 3A), which was consistent with male flower buds in other monoecious cultivars (Akagi et al., 2016a). For the structure of OGI, cv. Kumemaru has the Kali-SINE insertion on the OGI promoter like in other cultivars (Figure 3B) and simplex OGI. The sequences of Kali-SINE insertion in the promoter and transcript region showed no specific polymorphisms in comparison to other cultivars (Supplementary Figure 1; Akagi et al., 2016b). Furthermore, bisulfite PCR-Seq analysis targeting the OGI promoter in flower bud primordia showed that the cytosine residues in the Kali-SINE insertion were highly methylated in the CG and CHG sequences (62.8 and 59.4%, respectively) (Figure 3C). The methylation pattern at each cytosine residue was also consistent with male flower buds in the monoecious cultivar (p > 0.1; Akagi et al., 2016a) (Figure 3D). These results signified “repression” of the OGI expression, as observed in other cultivars (Akagi et al., 2016a). However, in cv. Kumemaru, OGI exhibited slight but fundamental expression in the flower bud primordia (RPKM = 0.87 and 1.18 for independent buds from different positions in mRNA-Seq analysis), which was significantly higher than in other monoecious cultivars (RPKM < 0.2 in mRNA-Seq analysis) (Figures 3E,F). The expression level of OGI in cv. Kumemaru was still lower than that in diploid male Caucasian persimmon (D. lotus) (average RPKM = 2.69), which carries no Kali-SINE insertion on the promoter (Figure 3F). In the late-developing stage, consistent with the diploid male persimmon, the expression levels of OGI in buds and other tissues were substantially decreased, suggesting that derepression of OGI in cv. Kumemaru is not constitutive (Supplementary Figure 2). These results suggested that the epigenetic sex regulation mechanism, in which the expression of OGI is suppressed under the control of the Kali-SINE, is shared in common among monoecious hexaploid persimmon cultivars, but OGI is induced with an unknown mechanism in cv. Kumemaru.
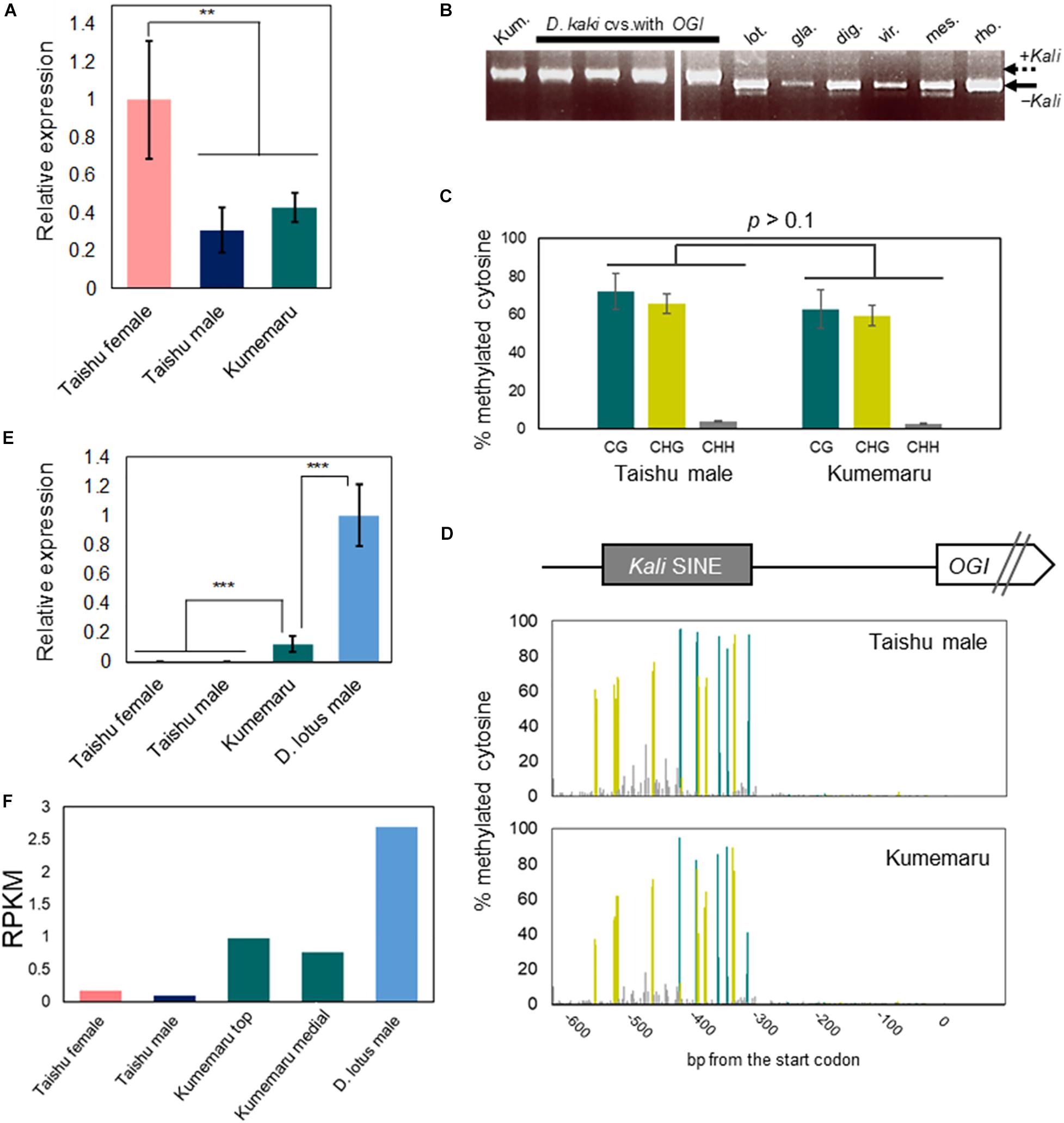
Figure 3. Regulation of OGI and MeGI in cv. Kumemaru. (A) MeGI expression level in flower buds in cv. Kumemaru, and monoecious cv. Taishu, from qRT-PCR analysis. cv. Kumemaru showed substantial reduction in the MeGI expression, similar to male flowers of cv. Taishu (**p < 0.01, N = 4 for biological replicates). (B) PCR analysis detecting the Kali SINE in D. kaki, and the relatives. cv. Kumemaru (Kum.) carries only the Kali inserted OGI, as well as other D. kaki cultivars (Akagi et al., 2016a). Other representative Diospyros species (lot. D. lotus, gla: D. glaucifolia, dig: D. digyna, vir: D. virginiana, mes: D. mespiliformis, and rho: D. rhombifolia) showed no Kali-SINE insertion in the OGI promoter region. (C,D) Cytosine methylation level (C) and methylation pattern at each cytosine residue (D) in the cv. Kumemaru and cv. Taishu OGI promoter region, in flower primordia. cv. Kumemaru showed similar methylation pattern with cv. Taishu (p > 0.1) (E,F) OGI expression level in flower buds in cvs. Kumemaru, Taishu, and diploid male D. lotus (cv. Kunsenshi-male), from qRT-PCR (E) and mRNA-Seq (F) analyses. OGI is not fundamentally expressed in monoecious cultivars including cv. Taishu (Akagi et al., 2016a), while cv. Kumemaru showed significant increase in the OGI expression (E, ***p < 0.001, N = 4 for biological replicates). The OGI expression in cv. Kumemaru was still lower than in diploid D. lotus, which carries no Kali SINE in the OGI promoter (E) (***p < 0.001, N = 4 for biological replicates). Bars indicate standard errors.
Differentially Expressed Genes in cv. Kumemaru-Specific Manner
To screen the genes associated with the mechanism for derepression of OGI, we compared mRNA-Seq data from the flower bud primordia in cv. Kumemaru and the 14 predominantly female-producing cultivars. Profiling with PCA using all the genes with RPKM > 1 (N = 32,143) indicated no specific clusters between cv. Kumemaru and the female-producing cultivars, although buds from some female-producing cultivars formed a specific cluster differentiated in the PC1 axis (Figure 4). These results indicated that cv. Kumemaru showed no dynamic expression differentiation from the female-producing cultivars.
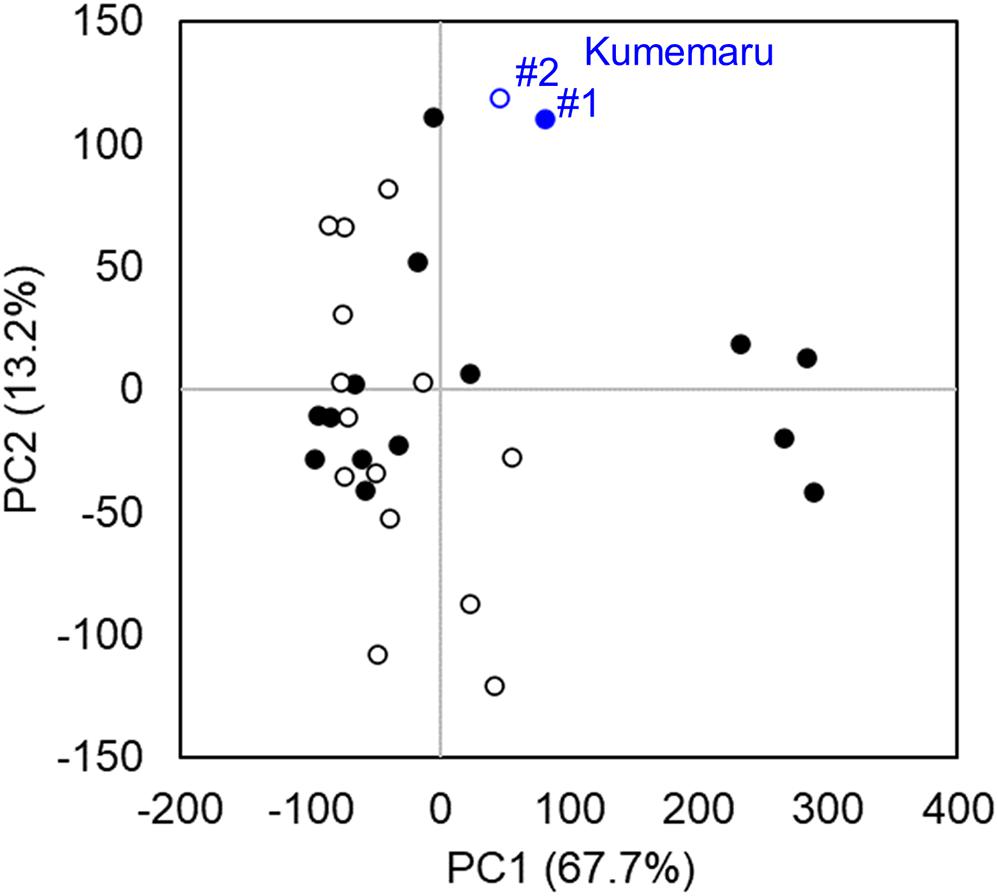
Figure 4. Transcriptomic profiles in comparison to cv. Kumemaru (Km) and 14 predominantly female-producing trees. Characterization of gene expression dynamics by PCA on transcriptomic data (RPKM > 1) of each cultivar. The buds were sampled from two positions, substantially affecting flower sexuality in monoecious cultivars (Akagi et al., 2016a). The two positions of buds in branches such as medial (#1, filled circle) and top (#2, open circle) based on the criteria in the previous report (Akagi et al., 2016a) were used as replicates in each cultivar. No significant difference was observed between the cv. Kumemaru and the other dominantly female-producing cultivars (p > 0.05).
To analyze sex-biased gene expression, we first conducted DESeq analysis between two groups, namely cv. Kumemaru vs. all 14 female-producing cultivars (N = 2 vs. 28 for biological replicates), defined as “comparison I.” We identified 3,441 upregulated and 2,931 downregulated DEGs (bias > 2, FDR < 0.01, RPKM > 1). These detected DEGs were annotated with gene ontology (GO) terms (FDR < 0.05). The 3,441 upregulated DEGs in cv. Kumemaru showed significantly enriched GO terms of protein phosphorylation and tyrosine kinase signaling (p = 1.9E-15 and 2.2E-17, respectively; Supplementary Table 2). The 2,931 downregulated DEGs in cv. Kumemaru showed enriched GO terms of post-embryonic development and response to stimulus (p = 1.4E-24 and 1.0E-18, respectively; Supplementary Table 3). Next, we conducted paired edgeR analysis between cv. Kumemaru and each female-producing cultivar, separately in 14 combinations (N = 2 × 2 for biological replicates in all 14 combinations), to detect cv. Kumemaru-specific expressions in more stringent criteria, defined as “comparison II.” We identified 21 genes that were upregulated in cv. Kumemaru in all 14 combinations and 21 genes showing downregulation (p < 0.1 for cvs. Nagara and Ogosho, p < 0.05 for the other 12 female cultivars, RPKM > 1) (Supplementary Table 4). For these DEGs, no significantly enriched GO terms were detected, presumably due to small numbers of the DEGs. Taken together, we detected the candidate genes related to the derepressed OGI silencing, which probably acts as an upstream of MeGI during early primordia differentiation.
Core Gene Networks Correlated With the Derepressed OGI Expression System in cv. Kumemaru
To understand the regulatory paths of Kumemaru-specific derepression of OGI, the co-expression patterns were visualized by applying a weighted correlation network analysis (WGCNA), which were first clustered into 27 modules (Figure 5A). Of them, 26 and 8 modules included the Kumemaru-specific DEGs detected from comparisons I and II, respectively (see Supplementary Table 5 for the details). Here we focused on module 1 (N = 993), which has the most significant enrichment of the DEGs for both comparison I (N = 840) and comparison II (N = 17) (p = 3.5e-519 and 2.5e-19, respectively, for one-sided Fisher’s exact test). Of the 612 biological process GO terms annotated to the genes in the 8 modules with the comparison II DEGs, 15 terms were specifically enriched in module 1 (p < 0.06) (Figures 5B,C). These terms included tyrosine kinase signaling pathway and response to gibberellin (GA), salicylic acid (SA), jasmonic acid (JA), and chitin (p < 0.06; Figures 5B,C), which overall implied association with stress signals (Xu et al., 1998; Devoto and Turner, 2004; Hu et al., 2017). These JA/GA/SA-associated genes were not included in the DEGs between male and female buds in diploid dioecious D. lotus, where OGI is genetically active in male (Akagi et al., 2014). Thus, these JA/SA/GA associated genes might contribute to the derepression of OGI in a cv. Kumemaru-specific manner.
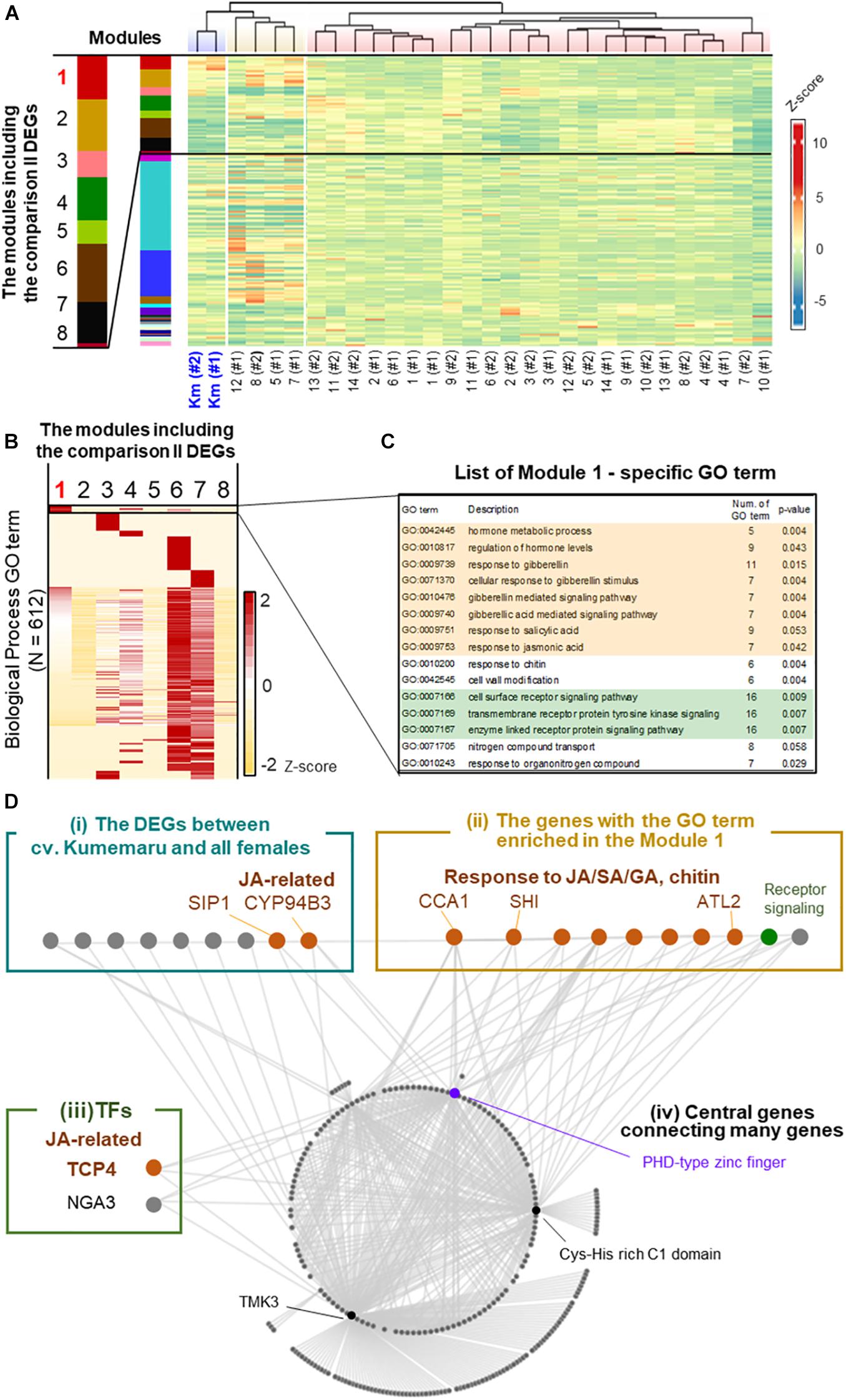
Figure 5. Co-expression network analysis in cv. Kumemaru and the predominantly female-producing cultivars. (A) Co-expression moduling of the genes based on expression patterns in cv. Kumemaru and 14 predominantly female-producing cultivars, with the WGCNA package. Modules 1–8 included the comparison II DEGs. (B) Heatmap for the enrichment of GO terms involving biological processes in these eight modules. The numbers of annotated GO terms were standardized by Z-scoring. (C) List of the GO terms specifically enriched in module 1 (p < 0.06), which included most of the cv. Kumemaru-specific DEGs. The GO terms relating to the hormonal process and receptor signaling pathway were highlighted in orange and green, respectively. (D) Visualization of the coexpressed gene network in module 1. As described in the main text, they were categorized into the following four groups: (i) the DEGs specific to cv. Kumemaru, (ii) the genes with the GO terms specifically enriched in module 1, as given in the panel (C), (iii) transcription factors, and (iv) central genes in this network.
To further dissect the key structure potentially derepressing OGI in cv. Kumemaru, we visualized the co-expression gene network in module 1 and focused on the genes highly correlated to other genes (correlation values >0.43) (Figure 5D). Here we featured the following four categories: (i) the Kumemaru-specific DEGs in comparison II, which were thought to be potential candidates to regulate Kumemaru-specific stable maleness (from 42 DEGs as described); (ii) the genes with the GO terms specifically enriched in module 1, which are mostly associated with stress signal (Supplementary Table 6) and would reflect cv. Kumemaru-specific physiological reactions; (iii) transcription factors; and (iv) central genes connecting many genes in this module. In category (i), 9 out of 17 DEGs were visualized with high correlation values with other genes (Supplementary Table 7). Among them, SIP1 mediates the abiotic stress-induced accumulation of raffinose (Egert et al., 2013), and CYP94B3 is a key enzyme in the catalytic pathway of JA biosynthesis (Heitz et al., 2011). In category (ii), focusing on JA/SA signaling, CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and ARABIDOPSIS TOXICOS EN LEVADURA 2 (ATL2) can act for the promotion of plant defense presumably via JA/SA signals (Salinas-Mondragón et al., 1999; Serrano and Guzmán, 2004; Lei et al., 2019). ATL2 is often applied as a biomarker for JA/SA signals (Serrano and Guzmán, 2004). Focusing on GA signaling, SHORT INTERNODES (SHI) is thought to involve gibberellin response to regulate internode length and structures (Fridborg et al., 2001). The other seven genes were annotated with the GO term associated with the regulation of hormone levels, receptor signaling, and nitrogen compound transport (Supplementary Table 6). For category (iii), we found two transcription factors in module 1. TCP4 can affect JA regulation by directly controlling the expression of LIPOXYGENASE2 (LOX2), which is one of the first steps of the JA biosynthesis pathway in Arabidopsis (Schommer et al., 2008). Moreover, the class II TCP family, including TCP4, was reported to regulate the expression of the other TF in category (iii), NGATHA3 (NGA3) (Ballester et al., 2015). The results so far are reminiscent of the involvement of stress signals, especially with the effect of JA signals. As the core genes [or category (iv)] predominantly connecting the module 1 network (with >50 connections), the genes annotated as the PHD-type zinc finger (Lee et al., 2009), Cys-His rich C1 domain (Bhaskar et al., 2015), and TMK3 (Dai et al., 2013) were identified. Among the three genes, the PHD-zinc finger has been reported to be associated with both gene expression and repression through histone modification (Mellor, 2006), which is possibly related to OGI derepression.
Identification of cv. Kumemaru-Specific Transcript Structure and Polymorphisms
To identify candidate genetic factors related to the derepression of OGI, kmer cataloging was conducted for the comprehensive caption of Kumemaru-specific polymorphisms. Assembly reads of Kumemaru-specific k-mers (KSK) derived 1,446 contigs (Table 1 for the representative 25 genes with >80 KSK; Supplementary Table 8 for the others). These polymorphic (or cv. Kumemaru-specific) contigs showed no overlap with the Kumemaru-specific DEGs described above. The transcript contig with the most KSK showed no homologous sequences in the persimmon genome (see text footnote 5; Akagi et al., 2020), while it showed undefined numerous virus-like sequences similar to marafiviruses. For other transcript contigs with fundamental numbers of the KSK, we could identify a structural variation of 38-bp insertion in RING-type E3 ubiquitin ligase. The other candidate contigs with the KSK showed no clear disruptions directly related to their protein functions.
Discussion
Our result of DEGs and co-expression network, focusing on the cv. Kumemaru-specific upregulated module, suggested the involvement of stress, particularly the JA-related pathway, for the activation of OGI expression in cv. Kumemaru. In addition to the genes involved in regulations of stress-induced plant hormones, other keywords (or GO terms in module 1) including chitin response and/or tyrosine kinase signaling (see Figures 5B,C) would imply the involvement of plant defenses and immunity. Tyrosine kinase has been reported to be responsible for biotic or abiotic stresses (Miyamoto et al., 2019) and is also known as a regulator of gibberellin responses (Fu et al., 2002), abscisic acid (ABA) signaling (Ghelis et al., 2008), cold stress (Sangwan et al., 2001), and sugar responses (Ritsema et al., 2009). Furthermore, one of the core genes in module 1, TMK3, as an Auxin-Binding Protein (ABP) family gene (Xu et al., 2014), is a member of the transmembrane kinase TMK subfamily, which is reported to be inducible under the stress or JA signals in Nicotiana plants (Cho and Pai, 2000). These implications might be consistent with empirical observations of flower sex bias in a tree in monoecious persimmon cultivars. Monoecious trees are supposed to produce only female flowers in vigor branches and/or at young age, while older or stressed branches tend to produce a high frequency of male flowers (Hasegawa, 1995; Hasegawa et al., 2004), although they might depend on cultivars/accessions. Furthermore, the ratio of male/female flowers in trees depends on their parental branch characters, such as sexuality, length, and bud positions (Akagi et al., 2016a). These reports suggest that not only genetic factors but also internal and external conditions of cv. Kumemaru may be involved in stresses, including specific viral infection as showed in Table 1, and may be the causal factor(s) to express derepressed male production. The viral-like sequence detected in this study was not matched to any other known viruses but most likely to be classified into the genus Marafivirus. Although the pathogenicity of this virus is little known, viral proteins encoded by another plant virus are thought to often inhibit JA signaling in Arabidopsis thaliana plants (Lozano-Duran et al., 2011) or to interact with plant histone deacetylase (Wang et al., 2018). Thus, it may be possible that virus infection can induce specific stress signaling to activate OGI, although we need further investigation to validate that in the future.
The direct effectors of derepressed OGI still remain to be solved. We can propose mainly two potential pathways to activate OGI: (i) release from the repression by the Kali-SINE insertion on the promoter region and (ii) an undefined “non-canonical” pathway that other monoecious cultivars do not have and cv. Kumemaru specifically established (Figure 6). For possibility (i), the Kali-SINE was highly methylated in cv. Kumemaru, as well as other cultivars (Figure 3D; Akagi et al., 2016a), suggesting that epigenetic conditions other than DNA methylation, such as histone methylation/acetylation, might affect the OGI expression. These histone modifications are frequently associated with the condition of DNA methylation on transposons (Qian et al., 2012; Du et al., 2015; Zhang et al., 2018). Although we have not assessed the histone methylation/acetylation in the OGI promoter, a comparison of them among a wide variety of cultivars may unveil the importance of histone modification. For possibility (ii), transcription factors that were highly expressed in cv. Kumemaru and related to JA signals such as TCP4 and NGA3 (Figure 5D) might be candidate trans-factors to regulate the non-canonical pathway to activate OGI, although k-mer cataloging has not found cv. Kumemaru-specific polymorphisms on them. On the other hand, cv. Kumemaru-specific mutations in cis-factors (or mutations in the promoter sequences) would be unlikely to be the causal factor to establish a derepressed OGI expression system. Mutations in cis-factors of OGI would affect the regulatory paths only under the control of OGI (or targeted MeGI), while the JA/SA/GA-related genes detected as DEGs specific to cv. Kumemaru are thought not to be the downstream pathways of the OGI gene (Akagi et al., 2014; Yang et al., 2019). As given in the model (Figure 6), future assessment about the factors potentially connecting stress signals and the OGI release would have importance to reveal the mechanisms not only of the cv. Kumemaru-specific derepressed male producing system but also for the evolution of monoecious system in a wide variety of hexaploid persimmon cultivars.
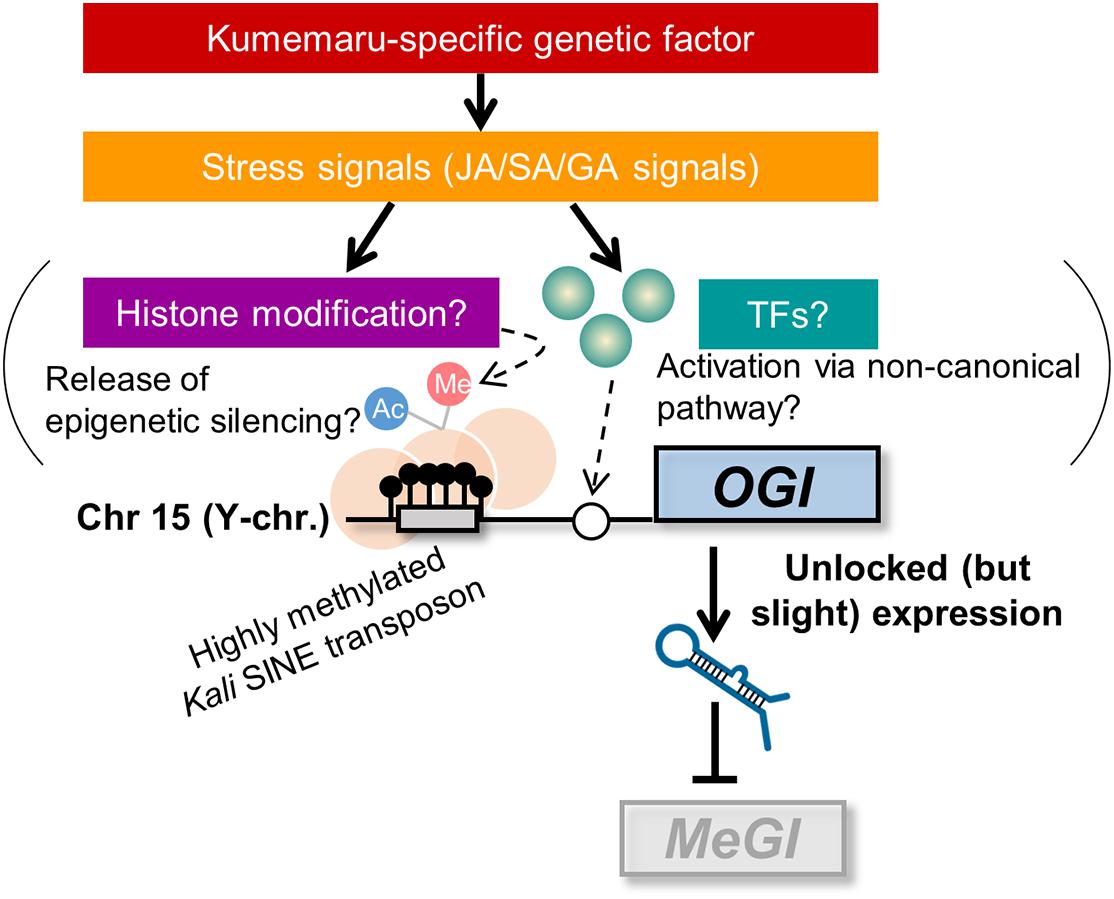
Figure 6. Model for derepressed OGI expression in cv. Kumemaru. Inactivation of OGI due to the Kali-SINE insertion and the resultant DNA methylation can be activated by two potential pathways: (i) release from the epigenetic silencing, which might result from histone modification, and (ii) an undefined “non-canonical” pathway such as activation via cv. Kumemaru-specific transcription factors. Those are likely induced by stress signals, especially JA/SA/GA signals, under the control of hypothetical Kumemaru-specific genetic (or possibly environmental) factors.
Data Availability Statement
All Illumina sequencing data have been deposited in the DDBJ database: Short Read Archives (SRA) database (BioProject ID PRJDB9564, Run ID DRR233540-DRR233569).
Author Contributions
TA designed the study. KM, HY, and TA conducted the experiments. KM, NF, and TA performed the data analyses and wrote the manuscript. KU, YK, and RT contributed to plant resources and facilities. All authors have read and approved the final manuscript.
Funding
This work was supported by PRESTO (Grant No. JPMJPR15Q1 to TA) from the Japan Science and Technology Agency (JST) by a Grant-in-Aid for Scientific Research on Innovative Areas (No. 19H04862 to TA) and for JSPS Fellows (No. 19J23361 to KM) from JSPS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Akihiko Sato, Atsushi Kono, Noriyuki Onoue, Toshihiro Saito, and Ryusuke Matsuzaki (Grape and Persimmon Research Station, NIFTS, Japan) for some plant materials including cv. Kumemaru. Some of this work was performed at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH Instrumentation (Grant No. S10 OD018174).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.567249/full#supplementary-material
Footnotes
- ^ https://www.gene.affrc.go.jp/databases_en.php?section=plant
- ^ https://www.gene.affrc.go.jp/databases-plant_search_detail_en.php?jp=117219
- ^ http://qb3.berkeley.edu/gsl/
- ^ https://github.com/Comai-Lab/allprep/blob/master/allprep-13.py
- ^ http://persimmon.kazusa.or.jp/index.html
- ^ https://github.com/Ih3/bwa
- ^ https://www.arabidopsis.org/index.jsp
- ^ https://github.com/Comai-Lab/kmer-extract-by-trigger-site
References
Ainsworth, C. (2000). Boys and girls come out to play: the molecular biology of dioecious plants. Ann. Bot. 86, 211–221. doi: 10.1006/anbo.2000.1201
Akagi, T., Henry, I. M., Ohtani, H., Beppu, K., Kataoka, I., and Tao, R. (2018). A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell 30, 780–795. doi: 10.1105/tpc.17.00787
Akagi, T., Henry, I. M., Tao, R., and Comai, L. (2014). A Y-chromosome-encoded small RNA acts as a sex determination in persimmons. Science 346, 646–650. doi: 10.1126/science.1257225
Akagi, T., Henry, M. I., Kawai, T., Comai, L., and Tao, R. (2016a). Epigenetic regulation of the sex determination gene MeGI in polyploid persimmon. Plant Cell 28, 2905–2915. doi: 10.1105/tpc.16.00532
Akagi, T., Ikegami, A., Tsujimoto, T., Kobayashi, S., Sato, A., Kono, A., et al. (2009). DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 151, 2028–2045. doi: 10.1104/pp.109.146985
Akagi, T., Kawai, T., and Tao, R. (2016b). A male determinant gene in diploid dioecious diospyros, OGI, is required for male flower production in monoecious individuals of oriental persimmon (D. kaki). Sci. Hort. 213, 243–251. doi: 10.1016/j.scienta.2016.10.046
Akagi, T., Pilkington, S. M., Varkonyi-Gasic, E., Henry, I. M., Sugano, S. S., Sonoda, M., et al. (2019). Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat. Plants 5, 801–809. doi: 10.1038/s41477-019-0489-6
Akagi, T., Shirasawa, K., Nagasaki, H., Hirakawa, H., Tao, R., Comai, L., et al. (2020). The persimmon genome reveals clues to the evolution of a lineage-specific sex determination system in plants. PLoS Genet. 16:e1008566. doi: 10.1371/journal.pgen.1008566
Ballester, P., Navarrete-Gómez, M., Carbonero, P., Oñate-Sánchez, L., and Ferrándiz, C. (2015). Leaf expansion in Arabidopsis is controlled by a TCP-NGA regulatory module likely conserved in distantly related species. Physiol. Plantarum. 155, 21–32. doi: 10.1111/ppl.12327
Bhaskar, R. V., Mohanty, B., Verma, V., Wijaya, E., and Kumar, P. P. (2015). A hormone-responsive C1-domain-containing protein At5g17960 mediates stress response in Arabidopsis thaliana. PLoS One 10:e0115418. doi: 10.1371/journal.pone.0115418
Cho, H. S., and Pai, H. S. (2000). Cloning and characterization of ntTMK1 gene encoding a TMK1-homologous receptor-like kinase in tobacco. Mol. Cells. 10, 317–324.
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. doi: 10.1038/nrg1711
Csardi, G., and Nepusz, T. (2006). The Igraph Software Package for Complex Network Research. InterJournal, Complex Systems, 1695. Available online at: https://igraph.org
Dai, N., Wang, W., Patterson, S. E., and Bleecker, A. B. (2013). The TMK subfamily of receptor-like kinases in Arabidopsis display an essential role in growth and a reduced sensitivity to auxin. PLoS One 8:e60990. doi: 10.1371/journal.pone.0060990
Devoto, A., and Turner, J. G. (2004). Jasmonate-regulated Arabidopsis stress signalling network. Physiol. Plant 123, 161–172. doi: 10.1111/j.1399-3054.2004.00418.x
Du, J., Johnson, L. M., Jacobsen, S. E., and Patel, D. (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532. doi: 10.1038/nrm4043
Egert, A., Felix, K., and Shaun, P. (2013). Abiotic stress-induced accumulation of raffinose in Arabidopsis leaves is mediated by a single raffinose synthase (RS5. At5g40390). BMC Plant Bio. 13:218. doi: 10.1186/1471-2229-13-218
Fridborg, I., Kuusk, S., Robertson, M., and Sundberg, E. (2001). The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiol. 127, 937–948. doi: 10.1104/pp.010388
Fu, X., Richards, D. E., Ait-Ali, T., Hynes, L. W., Ougham, H., Peng, J., et al. (2002). Gibberellin-mediated proteasome-dependent degradation of the barley DELLA protein SLN1 repressor. Plant Cell 14, 3191–3200. doi: 10.1105/tpc.006197
Ghelis, T., Bolbach, G., Clodic, G., Habricot, Y., Miginiac, E., Sotta, B., et al. (2008). Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol. 148, 1668–1680. doi: 10.1104/pp.108.124594
Goldberg, E. E., Otto, S. P., Vamosi, J. C., Mayrose, I., Sabath, N., Ming, R., et al. (2017). Macroevolutionary synthesis of flowering plant sexual systems. Evolution. 71, 898–912. doi: 10.1111/evo.13181
Harkess, A., Zhou, J., Xu, C., Bowers, J. E., Van Der Hulst, R., Ayyampalayam, S., et al. (2017). The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat. Comm. 8:1279. doi: 10.1038/s41467-017-01064-8
Hasegawa, K. (1995). Effects of girdling and strapping of lateral branches on male and female flowering in persimmon cv. Nishimurawase. Res. Rep. Kochi Univ. 44, 11–18.
Hasegawa, K., Fukuta, T., Kitahima, A., and Ogata, T. (2004). Effect of permanent and temporary strapping of 2-year old branches on male and female flower bud formation and return bloom in Japanese persimmon. Res. Rep. Kochi Univ. 53, 42–52.
Heitz, T., Widemann, E., Lugan, R., Miesch, L., Ullmann, P., Désaubry, L., et al. (2011). Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 287, 6296–6306. doi: 10.1074/jbc.M111.316364
Henry, I. M., Akagi, T., Tao, R., and Comai, L. (2018). One hundred ways to invent the sexes: theoretical and observed paths to dioecy in plants. Annu. Rev. Plant Biol. 69, 553–575. doi: 10.1146/annurev-arplant-042817-040615
Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J., and Yu, D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. doi: 10.1093/jxb/erx004
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. doi: 10.1186/1471-2105-9-559
Lee, W. Y., Lee, D., Chung, W., and Kwon, C. S. (2009). Arabidopsis ING and Alfin1-like protein families localize to the nucleus and bind to H3K4me3/2 via plant homeodomain fingers. Plant J. 58, 511–524. doi: 10.1111/j.1365-313X.2009.03795.x
Lei, J., Jayaprakasha, G. K., Singh, J., Uckoo, R., Borrego, E. J., Finlayson, S., et al. (2019). Circadian clock-associated1 controls resistance to aphids by altering indole glucosinolate protection. Plant Physiol. 181, 1344–1359. doi: 10.1104/pp.19.00676
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Lozano-Duran, R., Rosas-Diaz, T., Gusmaroli, G., Luna, P., Taconnat, L., Deng, X. W., et al. (2011). Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 23, 1014–1032. doi: 10.1105/tpc.110.080267
McCarthy, D. J., Chen, Y., and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
Mellor, J. (2006). It takes a PHD to read the histone code. Cell 126, 22–24. doi: 10.1016/j.cell.2006.06.028
Miyamoto, T., Uemura, T., Nemoto, K., Daito, M., Nozawa, A., Sawasaki, T., et al. (2019). Tyrosine kinase-dependent defense responses against herbivory in Arabidopsis. Front. Plant Sci. 10:1–9. doi: 10.3389/fpls.2019.00776
Müller, N. A., Kersten, B., Montalvão, A. P. L., Mähler, N., Bernhardsson, C., Bräutigam, K., et al. (2020). A single gene underlies the dynamic evolution of poplar sex determination. Nat. Plants 6, 630–637. doi: 10.1038/s41477-020-0672-9
Murase, K., Shigenobu, S., Fujii, S., Ueda, K., Murata, T., Sakamoto, A., et al. (2017). MYB transcription factor gene involved in sex determination in Asparagus officinalis. Genes Cells 22, 115–123. doi: 10.1111/gtc.12453
Qian, W., Miki, D., Zhang, H., Liu, Y., Tang, K., Kan, Y., et al. (2012). A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336, 1445–1448. doi: 10.1126/science.1219416
Renner, S. S. (2014). The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 101, 1588–1596. doi: 10.3732/ajb.1400196
Ritsema, T., Brodmann, D., Diks, S. H., Bos, C. L., Nagaraj, V., Pieterse, C. M. J., et al. (2009). Are small GTPases signal hubs in sugar-mediated induction of fructan biosynthesis? PLoS One. 4:e6605. doi: 10.1371/journal.pone.0006605
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Salinas-Mondragón, R. E., Garcidueñas-Piña, C., and Guzmán, P. (1999). Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in Arabidopsis thaliana. Plant Mol. Biol. 40, 579–590. doi: 10.1023/A:1006267201855
Sangwan, V., Foulds, I., Singh, J., and Dhindsa, R. S. (2001). Cold-activation of Brassica napus BN115 promoter is mediated by structural changes in membranes and cytoskeleton, and requires Ca2+ influx. Plant J. 27, 1–12. doi: 10.1046/j.1365-313x.2001.01052.x
Schommer, C., Palatnik, J. F., Aggarwal, P., Chételat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. doi: 10.1371/journal.pbio.0060230
Serrano, M., and Guzmán, P. (2004). Isolation and gene expression analysis of Arabidopsis thaliana mutants with constitutive expression of ATL2, an early elicitor-response RING-H2 zinc-finger gene. Genetics 167, 919–929. doi: 10.1534/genetics.104.028043
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Tennessen, J. A., Wei, N., Straub, S. C. K., Govindarajulu, R., Liston, A., and Ashman, T. L. (2018). Repeated translocation of a gene cassette drives sex-chromosome turnover in strawberries. PLoS Biol. 16:e2006062. doi: 10.1371/journal.pbio.2006062
Torres, M. F., Mathew, L. S., Ahmed, I., Al-Azwani, I. K., Krueger, R., Rivera-Nuñez, D., et al. (2018). Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nat. Commun. 9:3969. doi: 10.1038/s41467-018-06375-y
Tsugama, D., Matsuyama, K., Ide, M., Hayashi, M., Fujino, K., and Masuda, K. (2017). A putative MYB35 ortholog is a candidate for the sex- determining genes in Asparagus officinalis. Sci. Rep. 7:41497. doi: 10.1038/srep41497
VanBuren, R., Zeng, F., Chen, C., Zhang, J., Wai, C. M., Han, J., et al. (2015). Origin and domestication of papaya Yh chromosome. Genome Res. 25, 524–533. doi: 10.1101/gr.183905.114
Wang, B., Yang, X., Wang, Y., Xie, Y., and Zhou, X. (2018). Tomato yellow leaf curl virus V2 interacts with host histone deacetylase 6 to suppress methylation-mediated transcriptional gene silencing in plants. J. Virol. 92:e00036-18. doi: 10.1128/JVI.00036-18
Wickham, H. (2009). Ggplot2: Elegant Graphics for Data Analysis. Berlin: Springer Science & Business Media.
Xu, Q., Fu, H. H., Gupta, R., and Luan, S. (1998). Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell 10, 849–857. doi: 10.1105/tpc.10.5.849
Xu, T., Dai, N., Chen, J., Nagawa, S., Cao, M., Li, H., et al. (2014). Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 28, 1025–1028. doi: 10.1126/science.1245125
Yang, H., Akagi, T., Kawakatsu, T., and Tao, R. (2019). Gene networks orchestrated by MeGI: a single-factor mechanism underlying sex determination in persimmon. Plant J. 98, 97–111. doi: 10.1111/tpj.14202
Zhang, H., Lang, Z., and Zhu, J. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell. Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhou, Y., Massonnet, M., Sanjak, J. S., Cantu, D., and Gaut, S. (2017). Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. Proc. Natl. Acad. Sci. 114, 11715–11720. doi: 10.1073/pnas.1709257114
Keywords: monoecious, sex expression, polyploidy, Oriental persimmon, co-expression network
Citation: Masuda K, Fujita N, Yang H-W, Ushijima K, Kubo Y, Tao R and Akagi T (2020) Molecular Mechanism Underlying Derepressed Male Production in Hexaploid Persimmon. Front. Plant Sci. 11:567249. doi: 10.3389/fpls.2020.567249
Received: 29 May 2020; Accepted: 05 November 2020;
Published: 22 December 2020.
Edited by:
Akira Kanno, Tohoku University, JapanReviewed by:
Hideo Matsumura, Shinshu University, JapanMichael Nicolas, National Center for Biotechnology (CNB), Spain
Alex Harkess, Donald Danforth Plant Science Center, United States
Copyright © 2020 Masuda, Fujita, Yang, Ushijima, Kubo, Tao and Akagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Akagi, dGFrYXNoaWFAb2theWFtYS11LmFjLmpw
†These authors have contributed equally to this work
 Kanae Masuda
Kanae Masuda Naoko Fujita
Naoko Fujita Ho-Wen Yang2
Ho-Wen Yang2 Koichiro Ushijima
Koichiro Ushijima Yasutaka Kubo
Yasutaka Kubo Ryutaro Tao
Ryutaro Tao Takashi Akagi
Takashi Akagi