- Department of Plant Agriculture, University of Guelph, Guelph, ON, Canada
Microspores of Brassica napus can be diverted from normal pollen development into embryogenesis by treating them with a mild heat shock. As microspore embryogenesis closely resembles zygotic embryogenesis, it is used as model for studying the molecular mechanisms controlling embryo formation. A previous study comparing the transcriptomes of three-day-old sorted embryogenic and pollen-like (non-embryogenic) microspores identified a gene homologous to AT1G74730 of unknown function that was upregulated 8-fold in the embryogenic cells. In the current study, the gene was isolated and sequenced from B. napus and named BnMicEmUP (B. napus microspore embryogenesis upregulated gene). Four forms of BnMicEmUP mRNA and three forms of genomic DNA were identified. BnMicEmUP2,3 was upregulated more than 7-fold by day 3 in embryogenic microspore cultures compared to non-induced cultures. BnMicEmUP1,4 was highly expressed in leaves. Transient expression studies of BnMicEmUP3::GFP fusion protein in Nicotiana benthamiana and in stable Arabidopsis transgenics showed that it accumulates in chloroplasts. The features of the BnMicEmUP protein, which include a chloroplast targeting region, a basic region, and a large region containing 11 complete leucine-rich repeats, suggest that it is similar to a bZIP PEND (plastid envelope DNA-binding protein) protein, a DNA binding protein found in the inner envelope membrane of developing chloroplasts. Here, we report that the BnMicEmUP3 overexpression in Arabidopsis increases the sensitivity of seedlings to exogenous abscisic acid (ABA). The BnMicEmUP proteins appear to be transcription factors that are localized in plastids and are involved in plant responses to biotic and abiotic environmental stresses; as well as the results obtained from this study can be used to improve crop yield.
Introduction
A striking characteristic of plants is that a wide variety of cells can be induced to develop into embryos. Microspore embryogenesis is one of the best-known examples of asexual embryo development in plants. In tissue culture, a mild stress can redirect microspores from their normal path of development, that leads to mature pollen formation, to the development of a haploid embryos, and ultimately to mature plants with a haploid number of chromosomes (Wędzony et al., 2009; Soriano et al., 2013; Humphreys and Knox, 2015). The haploid embryos or plants can be treated with colchicine to produce doubled haploid plants (Pauls, 1996; Seguí-Simarro and Nuez, 2008). Doubled haploid production from microspore (immature pollen) culture provides a faster method for developing homozygous plants than conventional selfing and selection and can improve the efficiency of plant breeding for a large number of plants, including dicot and monocot species (Pauls, 1996; Germanà, 2011; Humphreys and Knox, 2015; Shariatpanahi and Ahmadi, 2016).
Zygotic embryogenesis in plants is difficult to study because the process occurs within the ovule. In addition, it is difficult to study early stages of embryogenesis because the quantities of embryo tissue for cellular and molecular analyses are limited. In vitro haploid embryogenesis is considered an important tool for research of early embryo development, because the pattern of development during microspore embryogenesis is very similar to zygotic embryogenesis at many levels and because the liquid culture medium makes it easy to observe and manipulate the various stages of embryogenesis (Prem et al., 2012; Soriano et al., 2013). It is possible to produce thousands of embryos from microspore cultures, thus providing sufficient amounts of material for molecular and cellular studies of embryogenic processes in plants. Specifically, microspore embryogenesis in Brassica napus has been established as a model system for studies of early events in embryogenesis because of the simplicity and efficiency of this system (Schulze and Pauls, 1997; Seguí-Simarro and Nuez, 2008; Dubas et al., 2013; Soriano et al., 2013).
Several molecular and biochemical studies have demonstrated that microspore and zygotic embryos share key regulatory proteins and the expression of a number of transcription factors (Perry et al., 1999; Boutilier et al., 2002; Joosen et al., 2007; Soriano et al., 2013, 2014). B. napus microspore-derived embryo cultures were used to identify genes that are involved in the initiation of embryo development in plants. Comparative gene expression studies using embryogenic and non-embryogenic cells in B. napus, have resulted in the identification of several genes specifically expressed in embryogenic cells. Gene expression research in Brassica began with subtractive hybridization studies (Hattori et al., 1998; Tsuwamoto et al., 2007) and has continued with microarray analyses (Boutilier et al., 2002; Malik et al., 2007, 2008; Whittle et al., 2010). Hattori et al. (1998) performed a subtractive hybridization study using 4-day responsive and non-responsive microspore cultures to identify the upregulated genes in responsive cultures. This study identified and tested the function of two transcription factor genes, BNM2 and BABYBOOM (BBM). BNM2 was expressed during all stages of microspore and zygotic embryogenesis, especially during the storage accumulation period. However, BBM was expressed only during the early stages of microspore and zygotic embryogenesis in root and flower buds (non-seed tissues; Boutilier et al., 2002; Soriano et al., 2013). Genes expressed in microspore cultures have also been identified by a differential PCR technique. Custers et al. (2001) identified a gene family member of CLAVATA3/ESR (CLE) called CLE19 that is a member of a family expressed in meristem initials (Wang and Fiers, 2010; Xu et al., 2015) that also plays a role in the development of globular and heart stage microspore and zygotic embryos (Fiers et al., 2004).
Chan and Pauls (2006) found that the expression of the ROP GTPase BnRop9 was 2.5-fold higher in the embryogenic cells than in the pollen-like cells sorted from induced microspore cultures. In addition, they used Arabidopsis microarray cDNAs from the AR12K array [11,960 unique express sequence tags (EST) sequences including more than 6,000 genes] to identify genes upregulated in embryogenic microspore cultures after heat shock (Chan, 2006; Pauls et al., 2006). This study showed that 120 transcripts were upregulated, at least 2-fold, in embryogenic cells compared to non-embryogenic cells sorted from induced microspore cultures. These genes were classified into 13 groups with known functions and one group with unknown function. The genes of unknown function were upregulated more than 3–8-fold in embryogenic cells compared to pollen-like cells including a gene homologous to the Arabidopsis gene, AT1G74730, that has a motif of unknown function and was upregulated 8-fold in the embryogenic cells. The current work was initiated to confirm its upregulation in embryogenic microspore cells and gain an understanding of the function of the protein it encodes, in general.
In this work, we identify and characterize a gene that is differentially expressed during the switch from pollen development to microspore-derived embryo development, named BnMicEmUP (B. napus microspore embryogenesis upregulated gene). Our results show that BnMicEmUP encodes a unique protein, targeted to chloroplasts and containing a nucleotide-binding domain and a leucine zipper (bZIP)-like domain, similar to PEND (plastid envelope DNA-binding; Sato et al., 1998) proteins. It is preferentially expressed during microspore embryo induction and in expanded leaves and increases seedling sensitivity to exogenously applied abscisic acid (ABA).
Materials and Methods
Plant Material
Brassica napus L. cv. Topas plants were used throughout the experiments in this study. Seeds of B. napus were sown in soil in 24 cell-planting trays in a growth room at 20°C/18°C (day/night) under a 16 h photoperiod for microspore embryogenesis. Plants were fertilized with 1-1-1 ratio of nitrogen, phosphorous, and potassium and watered every 3 days. Once plants passed the three-leaf stage, they were transplanted to plastic pots (20 cm diameter, 25 cm height). Plants that were at the flower bud formation stage 45 days after sowing (DAS) of growth served as donors for flower bud collection and microspore isolation.
Unopened flower buds (2.0–3.0 mm in length) were selected from donor plants. The buds were surface sterilized in 75% ethanol for 30 s and then in 5.6% sodium hypochlorite bleach for 10 min. The sterilized buds were rinsed, three times, with sterile distilled water and homogenized in a blender with 30 ml cold 13% sucrose solution. The suspension was filtered consecutively through a 64 μm then 40 μm Nitex nylon sieves (Tetko Co., Elmsford, NY, United States). The filtrate was centrifuged for 5 min at 200 × g. The dark green supernatant was discarded and the pellet of microspores was re-suspended in wash solution (13% sucrose) and re-centrifuged. The procedure was repeated three times. The final pellet was re-suspended in full NLN medium (Nitsch and Nitsch, 1967; Lichter, 1982). The density of the microspores was adjusted to 8 × 104 microspores/ml and 10 ml of the microspore suspension was dispensed into Petri plates (150 mm wide, 10 mm deep). The microspores were incubated at constant 30°C (inductive temperature) or 25°C (non-inductive temperature) in the darkness. The cultures were monitored from 0 to 5 days of culture (initiation phase microspore embryogenesis); each day, six plates were sampled for RNA extraction from responsive (30°C) and nonresponsive cultures (25°C).
Isolation of the BnMicEmUP Gene and Southern Blotting
DNA was extracted from first leaves of 3-week old plants using CTAB method (Doyle, 1991). The genomic DNA was quantified using a Nanodrop ND1000 Spectrophotometer (Nanodrop Technologies, Inc., Wilmington, DE, United States). Approximately 50 μg of genomic DNA was digested with restriction enzymes XbaI and XhoI (New England Biolabs, Beverly, MA, United States). The digested DNA was separated on a 1.5% (w/v) agarose, 1 X TBE ethidium bromide gel at 40 V for 24 h and the results were photographed. The DNA was transferred to charged nylon membranes and left overnight (Roche Diagnostic, Laval, QC, Canada). Probes were prepared from pTopo2.1-BnMicEmUP3 templates using a PCR DIG probe synthesis kit (Roche Diagnostics, Laval, QC, Canada). Hybridization was done using the DIG Easy Hyb Kit (Roche Diagnostics, Laval, QC, Canada). The hybridized probes were immuno-detected with anti-digoxigenin-AP, Fab fragments (Roche Diagnostics, Laval, QC, Canada) and then visualized by exposing Kodak X-OMAT film (Kodak Inc., Rochester, NY, United States) to the membrane treated with CDP-star (Roche Diagnostics, Laval, QC, Canada).
RNA Extraction, Analysis, and Reverse Transcription to cDNA
Total RNA samples from 0 h, 1, 3, 5, and 7 days induced (30°C, embryogenic) and non-induced (25°C, non-induced) cultured microspores were isolated using glass bead disruption (Bio 101, Vista, CA, United States) and the modified RNeasy Midi kit (Qiagen), including on-column DNase digestion. RNA also was isolated from young leaves, old leaves, stem, buds, and root by using the RNeasy Midi kit. To remove genomic DNA from all RNA samples they were treated with Turbo DNAse, according to the manufacturer’s specifications (Ambion, Austin, TX, United States). The quality of the extracted RNA was determined with a Nanodrop-1000 (Thermo Fisher Scientific, Wilmington, DE, United States) by measuring the 260/280 ratio. The quality was tested by gel-electrophoresis through 1% agarose containing ethidium bromide (EtBr) at 80 V to visualize the 28S and 18S rRNA bands with a BIO-Rad Gel Doc System. The first strand of cDNA was synthesized from 1 μg of total RNA using the superscript first strand cDNA synthesis kit, according to manufacturer’s instructions (Invitrogen) or a SMART™ RACE cDNA Amplification Kits (Clontech, Palo Alto, CA, United States) were utilized, according to manufacturer’s instructions.
Primer Design
To amplify BnMicEmUP genes from B. napus, the primer were designed based on the AT1G74730 gene sequence available at the TAIR1 and Brassica ESTs (Genbank accession number: CN729138 and CN737378) with high sequence homology to Arabidopsis thaliana, which were found with Computational Biology and Functional Genomics Laboratory (CBFGL). The ESTs and AT1G74730 cDNA sequences were aligned with CLUSTALW version 1.812 and primers were designed for homologous regions with Primer3Plus (Untergasser et al., 2007) to amplify homologous cDNAs from Brassica cDNA (Supplementary Table 1). To isolate the upstream and downstream non-coding regions of BnMicEmUP, a SMART™ RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, United States) was utilized, according to manufacturer’s instructions. In brief, total RNA was obtained from seedlings and induced microspore cultures on day 1, 2, and 3 and primers were designed from conserved regions of different forms of BnMicEmUP (Supplementary Table 1).
Sequence Analysis and Conserved Domain Search
All sequencing results obtained were analyzed with BLAST at NCBI.3 To determine the number gene versions that exist in Brassica, the clones were aligned with CLUSTALW version 1.81 (Chenna et al., 2003).4 As a result of this alignment, a reliable set of SNPs between different forms of genes was discovered and a consensus sequence was generated.
An amino acid sequence comparison and similarities search was performed by BLAST in NCBI/N1H5 using the non-redundant database (nr) and in ExPASy.6 The search of conserved domains was carried out by NCBI BLAST and the protein sequences were compared with the Conserved Domain Database (CDD)7 as well. Although the CDD search included a search in Pfam (the database of protein domains), it does not integrate as many databases as InterProScan. Therefore, a search using InterProScan and the SMART database was performed to identify possible trans-membrane helices in the protein and signal peptides. To predict protein structure, the Phyre2 server was used (Kelley and Sternberg, 2009).
Phylogenetic Analyses
Phylogenetic and molecular evolutionary analyses of the BnMicEmUP gene family were conducted using Workbench, version 3.0.2 (CLC bio, Aarhus, Denmark) software. Translated protein sequences were aligned and 60-amino acid regions containing putative homeodomains were chosen for phylogenetic analysis. They were imported into the CLC Workbench (CLC Bio, Aarhus, Denmark) program and globally aligned by CLUSTALW. A phylogenetic tree was generated with neighbor-joining methods, and bootstrap values were calculated.
Gene Localization
Two fusion protein constructs, BnMicEmUP3-GFP and AtBnMicEmUP-GFP, were prepared to test the protein localization. To produce a fusion of BnMicEmUP3 or AtBnMicEmUP with the green fluorescent protein (GFP; Siemering et al., 1996), fragments for the coding sequences of BnMicEmUP3 or AtBnMicEmUP were amplified by PCR from clones containing the genes (BnMicEmUP3 and AT1G74730 with the forward primer (5'-GTCGTCGGCGCTCCAATATGGATCCAAGGAGATATAACAATGGCT-3') and specific reverse primers (5'-TGGATGGACTTCAAGAAGCTGACATGGGCAAGGGCGAGAAACTGTTC-3'). The reverse primers were designed to attach the 5'GFP sequence to 3' BnMicEmUP3 gene with two design features. Firstly, it contained 15 nucleotides of 5' end of the GFP sequence and 15 nucleotides of the 3' end of the BnMicEmUP3 sequence. Secondly, it was designed to remove the stop codon from the 3' end of the BnMicEmUP3 gene. This primer was also used as the forward primer for GFP amplification, which was amplified from pCAMBIA 1302 (gene fusion reverse and GFP fusion forward: GCAGAGCTCATCTTCACTTGTAGAGTTCATCCA). The two amplified gene products were fused using the gene forward specific primer containing a BamHI site and ribosome binding site and GFP reverse primer containing SacI. The BnMicEmUP3-GFP and AtBnMicEmUP-GFP fusion fragments were subcloned into BamHI-SacI-digested pBI121 (Clontech) under control the CaMV35S promoter to generate pBIN35S::BnMicEmUP3-GFP and pBIN35S::AtBnMicEmUP-GFP plasmids. These were introduced into Agrobacterium tumefaciens strain EHA105.
Chloroplast Isolation
Chloroplasts were isolated from transgenic Arabidopsis plants expressing BnMicEmUP3-GFP and AtBnMicEmUP-GFP and from wild type plants, following Takabe’s protocol (Takabe et al., 1979). The leaves (10 g) were homogenized in 0.5 M sucrose. The homogenate was filtered through eight layers of clean cheesecloth and centrifuged at 50 × g for 10 min. The pellets were resuspended in 4 ml of ice-cold 0.5 M sucrose. The pellet containing chloroplasts was examined with the confocal microscopy for GFP expression.
Microscopic Study
To observe GFP fluorescence, isolated chloroplasts were observed immediately under an upright confocal laser scanning microscopy (CLSM; Leica DM RE microscope connected to a Leica TCS SP2, Leica, Germany). Images were processed using Leica Confocal Software (LCS, version 2.61).
Gene Expression Level Assays Using Quantitative Real-Time PCR
To investigate the involvement of BnMicEmUP2,3 gene expression in microspore embryogenesis of B. napus, comparative gene expression studies were done using isolated RNA from responsive (30°C) and nonresponsive (25°C) microspore cultures at different time points. Gene expression was analyzed by real-time PCR (RT-PCR) by using gene-specific primers designed for nucleotide deletion regions in BnMicEmUP1,4 (without deletion) and BnMicEmUP2,3 (with deletion; Figures 1, 2). Primers were designed to amplify either a BnMicEmUP1 and/or a BnMicEmUP4 fragment with a smaller size. The primers were: 5 prime (5-CCGATATCGTCTCCTGCGGCTC-3) and 3 prime (5-CGGGACTATTCTTCGAAGGGA-3) to produce a 160 bp or a larger size (200 bp) band, or 5 prime (5-CCTATATCGTCTCACCCGCGAA-3) and 3 prime (5-CATCAGCCTTCTCCGTCGTCGC-3) for BnMicEmUP2 and/or BnMicEmUP3 (Supplementary Figure 6). The optimal annealing temperature (TA) for each primer set was determined using the BioRad thermocycler. Real-time PCR was performed in 25 μl reactions with 1X IQ SYBR Green Supermix (Bio-Rad Laboratories) and 0.25 μM of each primer. An optimized volume of cDNA solution (100 ng) was used for the amplification of each gene. To reduce pipetting errors, master mixes were prepared for triplicate reactions (3 × 25 μl) for each standard and gene. Twenty microliter of the reaction mixture was pipetted into wells of a 96-well plate, followed by 5 μl of a 100 ng concentration cDNA solution into each well except for a blank which served as the “no template” control. The plates were sealed with microseal film, centrifuged, and placed on the optical reaction block of the BioRad Real-Time Detection System. Quantitative real-time PCR (qRT-PCR) protocols were set up using the iCycler iQ real time detection system software (BioRad). The same qPCR protocol was used for the two genes BnMicEmUP2,3. The following PCR program was used to amplify all genes: 5 min at 95°C followed by 40 cycles of 30 s at 95°C, 30 s at 65°C, and 30 s at 72°C, followed by 4°C hold.

Figure 2. Comparison of the four forms of the BnMicEmUP cDNAs. The predicted 5'UTRs and 3'UTRs are shown in black and red sequences indicate the start and stop codons. Primers used for RT-PCR experiments are indicated with horizontal arrows. These primers were used to isolate and sequence two main classes of BnMicEmUPs namely: and BnEmMicUP1,4 (2) and BnEmMicUP2,3 (3; Supplementary Table 1).
To calculate alterations in transcription levels of target gene, standard curves with log input amount and CT values were applied for target gene and elongation factor 1-alpha (ef1alpha) for each sample using the iCycler iQ™ real time detection software (BioRad). For normalization, the amount of a target gene in each sample was divided by the amount of ef1alpha in the same sample. The normalized value was standardized by a control sample of each experiment (e.g., microspore in 0 day).
The analyses were performed on RNA isolated from three biological replications including three separate microspore cultures from 0 h, 1, 3, 5, and 7 days induced (30°C, embryogenic) and non-induced (25°C, non-induced) cultured microspores, and three preparations of vegetative tissue cDNA.
Statistical Analyses
All statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, CA, United States) and Excel. Data were analyzed with either paired Student’s t-test or one-way ANOVA to assess the changes in expression level of genes in different time during microspore culture and different tissue of B. napus. A probability level of p ≤ 0.05 was considered significant. All data are shown as mean ± standard error of mean (SEM). The confocal images were obtained as single optical sections and all images (Arabidopsis seedling leaf cells) shown in figures are representative of at least 20 cells from two independent lines.
Preparation of Binary Vector Constructs With AtBnMicEmUP2,3 Gene
The plasmids of pBI12135s::AtBnMicEmUP and pBI12135s::BnMicEmUP3 (pBI121; Jefferson et al., 1987), containing coding region of A. thaliana and B. napus for BnMicEmUP2,3, were constructed to generate overexpressed lines. The BnMicEmUP2,3 and AtBnMicEmUP genes were also cloned into the plant expression vector pFGC5941 (GenBank accession no AY310901),8 which forms a double stranded RNA (dsRNA) transcript under the control of the CaMV 35S promoter to generate constitutive gene silencing lines (RNAi lines). The RNAi construct was prepared using one highly gene-specific (unique) fragment, which was isolated from nucleotide positions 57–217. These plasmids were introduced into A. tumefaciens strain EHA105 (Hood et al., 1993). Arabidopsis plants were transformed with the constructs by the floral dip method (Clough and Bent, 1998). T0 seeds were selected on selection media as follows: Over-expressed lines produced with pBI121 were selected on kanamycin (50 mg/ml). The expression of AtBnMicEmUP genes in the Arabidopsis transformants and control lines were evaluated by RT-PCR using AtBnMicEmUP specific primers (At.25185 57-57 and At.25185 57-217; Supplementary Table 1). The orientation of the final construct was confirmed by DNA sequencing, using a pBI121 sequencing primer (35S promoter and N-terminal) and a gene specific primer. Homozygous T4 plants were used in all experiments.
SALK T-DNA insertion seeds (SALK_ 027022 and SALK_ 034644) were received from Arabidopsis Biological Resource Center (ABRC). Homozygous plants were obtained by PCR using primers designed by the SIGnALwebsite (Supplementary Table 1).9
For ABA germination assays, seeds were grown on 1/2 MS medium containing 1.5% sucrose and 0, 0.5, 1.0, and 2.0 μM ABA (Sigma, St. Louis, MO, United States) for 5 days. Three experimental repeats were carried out, each involving one independent AtBnMicEmUP mutant line (atbnmicemup-SALK_ 027022), two independent silenced lines (AtBnMicEmUP-RNAi1.1, AtBnMicEmUP-RNAi1.2), two overexpressing lines (35S::AtBnMicEmUP 1.4, 35S::AtBnMicEmUP1.6), and WT.
Results
Gene Structure
Isolation of B. napus Homologs of the Arabidopsis Gene AT1G74730
Gene sequences with homology to the gene AT1G74730 (Figure 1) were amplified from B. napus (cv. Topas) genomic DNA by PCR and from RNA by RT-PCR from induced (at 30°C) microspore cultures, buds, and young leaves using PCR primers (At.25185 atg-gta, At.25185 118-735, and At.25185 57-217). These primers were designed based on multiple sequence alignments of ESTs and genomic sequences for the AT1G74730 gene and its homolog AT5G08050 in Arabidopsis (Supplementary Table 1). Three fragments were amplified with primers At.25185 atg-gta forward and At.25185 atg-gta reverse from B. napus genomic DNA and named BnMicEmUP (B. napus Microspore Embryogenesis UPregulated) genes. The three genomic clones of BnMicEmUP had sizes of 709, 803, and 863 bp (Figure 1 and Supplementary Table 2). Genomic DNA sequences of BnMicEmUP1, BnMicEmUP2, and BnMicEmUP3 were submitted to GenBank and were given the accession numbers HQ647330, HQ647331, and HQ647332, respectively.

Figure 1. Comparison of nucleotide sequences of three forms of Brassica napus BnMicEmUP genes with the Arabidopsis AT1G74730 gene. Yellow box (intron); un-highlighted region (the 5' un-translated and 3' un-translated regions); and white background (polymorphisms).
The BnMicEmUP genes have very similar structures to AT1G74730 and consist of two exons and an intron (Figure 1). The nucleotide similarities among the BnMicEmUP genes and with AT1G74730 were highest in the exons (85–87%) but more variable for the intron (43–69%). Particularly, BnMicEmUP3, which contains the longest intron (269 bp), differed from BnMicEmUP1 intron (121 bp) by a 7 bp indel plus multiple single nucleotide polymorphisms (SNPs). The identity between the BnMicEmUP3 and BnMicEmUP2 (209 pb) introns is high (89%), but these two forms have low identities to the BnMicEmUP1 intron sequence (13%) which is the shortest intron (121 bp). The BnMicEmUP3 intron is also the most similar to the AT1G74730 intron (Figure 1 and Supplementary Table 2).
Most of the differences in the exons were SNPs, but a 3-nucleotide indel in exon 1 distinguished the B. napus genes from the Arabidopsis gene, and a 9 nucleotide insertion in BnMicEmUP2 and BnMicEmUP3 separated those forms from BnMicEmUP1 and BnMicEmUP4 (Figure 2). The cDNA sequences of BnMicEmUP1 (588 bp), BnMicEmUP2 (594 bp), BnMicEmUP3 (594 bp), and BnMicEmUP4 (588 bp) were submitted to GenBank and were given the accession numbers, HQ660218, HQ660217, HQ660216, and HQ660215, respectively. The nucleotide identities with AT1G74730cDNA were high (85–89%). The differences were mostly related to SNPs spread throughout the coding region. Most of the SNPs occurred at the first coding exon. Sequence analysis of the BnMicEmUP cDNA clones indicates that there are at least two classes of cDNAs, which differ primarily by a nine-nucleotide deletion at position 30 of the coding region of the genes (Figure 2). The cDNA types containing the nucleotide deletion include the BnMicEmUP1 and BnMicEmUP4 genes. BnMicEmUP3 and BnMicEmUP2 belong to the cDNA type with no nucleotide deletion. The nucleotide sequence identity was 83–91% between the two classes of BnMicEmUP cDNAs (Supplementary Table 2). Similarly, these two cDNA classes also contain two different upstream untranslated regions (5' UTR) of BnMicEmUP with 92% identity. BnMicEmUP1 and BnMicEmUP4 belong to one class and BnMicEmUP2 and BnMicEmUP3 belong to a second class. However, cloning and sequencing of 5' and 3' RACE products allowed isolation of the short 5' UTR region, which is one-third as long as the 5' UTR region of AT1G74730 (54–59 bp vs. to 150 bp). The 5' UTR region of BnMicEmUP has 61–72% identity with the homologous region of AT1G74730 in Arabidopsis. More extensive heterogeneity was observed among the 3' UTRs of the different BnMicEmUPs. The sequence similarity between the 3' UTRs of the two classes of BnMicEmUPs was 47% and between these classes and AT1G74730 was 41–55%. The similarities between the coding regions of different classes of BnMicEmUP cDNAs and the gDNAs allowed the matched pairs to be identified. The results indicate that because four forms of cDNA were detected, but only three forms gDNA were identified, there is a possibility that another form of gDNA exists, which corresponds with cDNA of BnMicEmUP4 (Supplementary Figure 1).
Protein Domain Characterization
The BnMicEmUP3 ORF encodes a protein of 195 amino acids with an estimated molecular mass of 20.6 kDa. The amino acid composition of BnMicEmUP3 is 9.2% acidic, 21.5–22.5% polar, 10.7% basic, and 57.4–59.0% hydrophobic amino acids (Figures 3, 4) and the calculated pI = 9.3.10 A comparison of the four forms of putative BnMicEmUP proteins and AtBnMicEmUP (Figure 3) showed that they are 83–87% identical. A comparison of the BnMicEmUP amino acid sequence against a protein sequence database identified a region of the BnMicEmUP proteins sequence that is similar to the predicted Domain of Unknown Function 1118 (DUF1118; Figure 3).
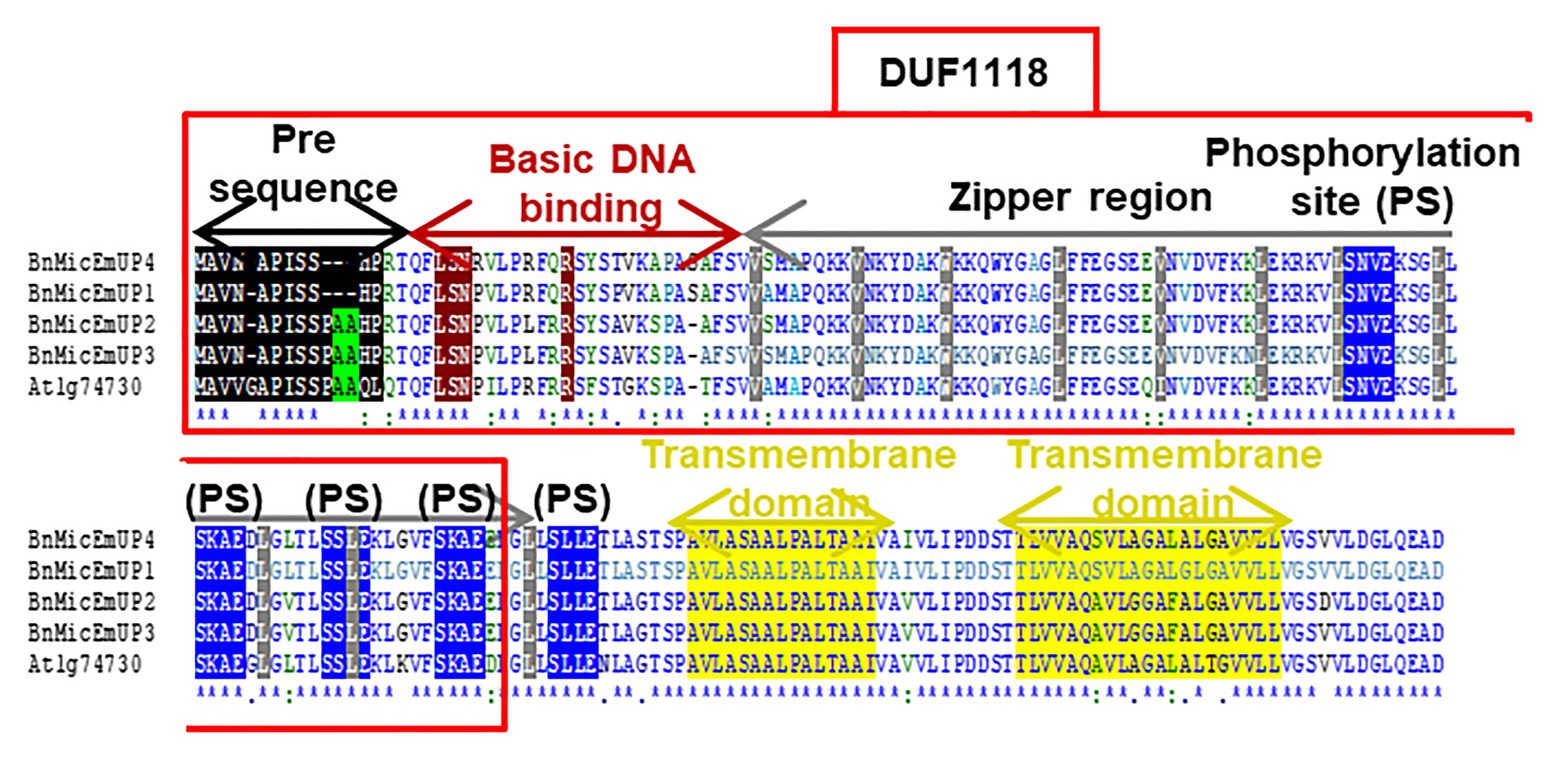
Figure 3. Comparison of four B. napus BnMicEmUP protein sequences with Arabidopsis AT1G74730. DUF1118 (domain unknown function) region is shown in the large box. Putative chloroplast transit peptides are highlighted in black and putative motifs (A/A) for chloroplast transit peptide cleavage sites are shaded in green. Basic DNA binding and leucine zipper (dimerization) domains are shaded in dark red and gray, respectively. Potential casein kinase II phosphorylation sites (S/TxxD/E) are shaded in blue. Putative transmembrane helix topologies of BnMicEmUPs are shaded in yellow.
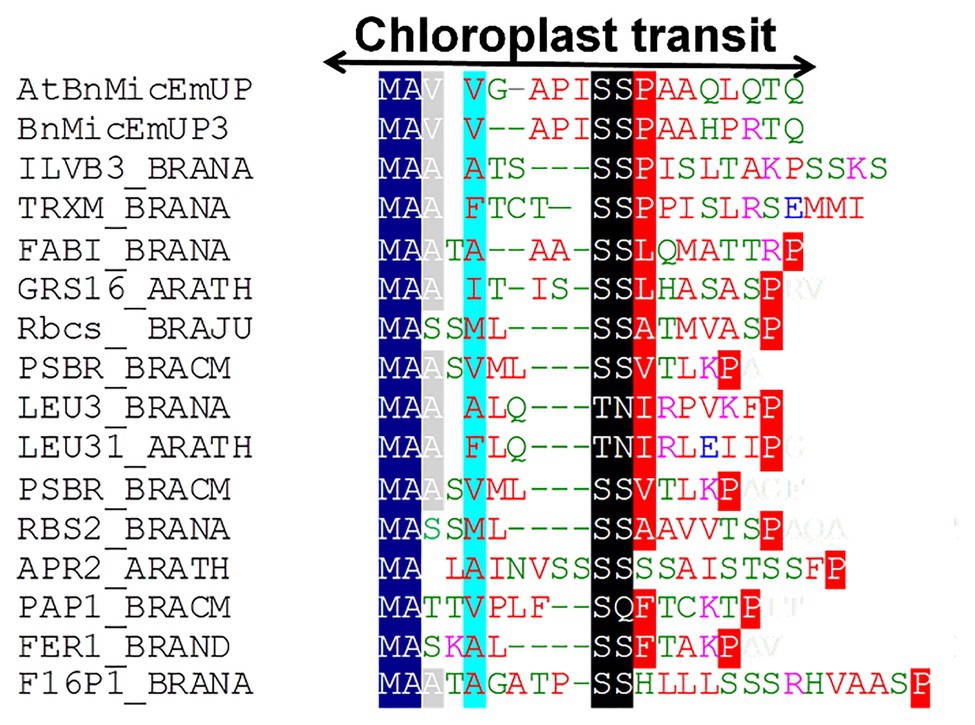
Figure 4. Alignment of predicted amino acid sequences of transit peptides of chloroplast targeted proteins with the AtBnMicEmUP and BnMicEmUP3 transit peptides. Species represented and the genes used to generate the sequence alignments are as follows: B. napus (BRANA); B. rapa subsp. campestris (BRACM); A. thaliana (ARATH); Ferritin-1 (Ferritin-1 Q96540.1); PSBR (Photosystem II 10 kDa polypeptide P49108.1); LEU3 (isopropylmalate dehydrogenase P29102.1); FABI (NADH-dependent enoyl-ACP reductase P80030.2); GSH1 (Glutamate – cysteine ligase CAD91713.1); ILVB1 (Acetolactate synthase 1 AAA62705.1); GRS16 (Monothiol glutaredoxin-S16 AEC09515.1); F16P1 (Fructose-1,6-bisphosphatase AAA82750.1); TRXM (Thioredoxin M-type Q9XGS0.1); Rubisco small subunit (Rbcs); Plastid lipid-associated protein 1 (PAP1); and 5'-adenylylsulfate reductase 2 (APR2). Conserved residues are highlighted in dark blue (block1), black (block2), and red (block 3). The consensus non-shared residues are colored according to: red, hydrophobic; purple, basic; green, hydrophilic; and blue, acidic.
Further searches for motifs in BnMicEmUP3 were performed using the domain alignments in PFAM (Bateman et al., 2002), SMART (Letunic et al., 2002), PRINTS (Attwood et al., 2002), and PROSITE databases. These analyses showed that BnMicEmUP contains a putative chloroplast transit peptide (cTP) in the N-terminus of the BnMicEmUP protein. Figure 4 shows that the putative BnMicEmUP cTP contains regions in its beginning, middle, and end sections that are well conserved in transit peptides sequences among diverse genes targeted to the chloroplast in Brassica and Arabidopsis species. In particular, all the sequences start with methionine and alanine (MA), followed by hydrophobic amino acids, like alanine or valine (A or V), a pair of serines (S), and terminate with proline P (Zybailov et al., 2008). The three-homology blocks are separated by different lengths of non-conserved sequences. A cleavage motif, consisting of two alanines (A/A), which is a unique cleavage site for cTPs (Zhang and Glaser, 2002) was also identified at the N-terminal sequences of the BnMicEmUP2 and BnMicEmUP3 putative protein sequences (Figure 3).
BnMicEmUP protein also contains DNA-binding domains that are most similar to those found in the basic leucine zipper (bZIP) family (PROSITE accession no ps00036). A common motif for bZIP proteins is a basic region at the N-terminus (between amino acids 17 and 69 or 125) that includes an invariant DNA-binding motif N-x7-R, followed by a repeat of four to ten Leu, which may be replaced by other hydrophobic residues such as isoleucine, alanine, and valine. An alignment of the BnMicEmUP DNA binding amino acid sequence with bZIP sequences from the plant transcription database11 shows the high conservation of the characteristic features of this group of transcription factors (Figures 5A, 6A). Especially, the occurrence of N, at amino acid position 19 of the BnMicEmUP protein, is highly conserved in DNA binding bZIP family proteins (Figure 3). However, BnMicEmUP has a much longer stretch of intervening amino acids, between the putative DNA binding site and the start of the zipper region, that is found in typical bZIPs (Jakoby et al., 2002; Figure 3). Moreover, the leucine zipper of BnMicEmUP protein consists (initially) an octad, rather than a conventional heptad. The consensus repeat for BnMicEmUP is V/L X4-X8 V/L.
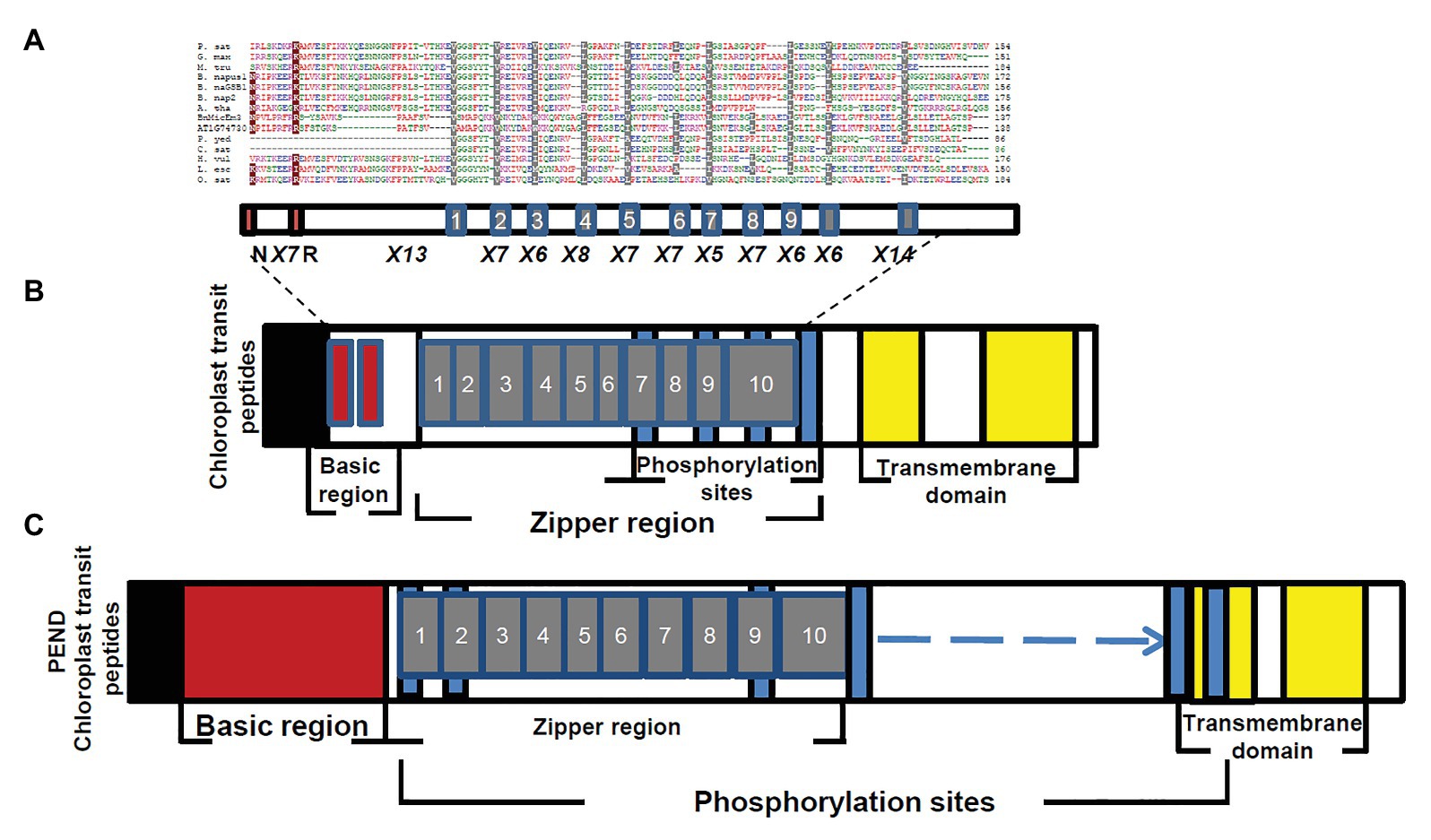
Figure 5. Comparison of BnMicEmUP3 protein to PEND proteins. (A) Alignment of PEND homologs to BnMicEmUP3 protein from various plant species. PEND sequences have been identified in various angiosperms by Terasawa and Sato, (2005). Sources of sequences: P. sat (Pisum sativa, PEND, genome: AB189736); B. nap (B. napus: GSBF1, cDNA: X91138); B. nap1 (B. napus, genome: AB189734); B. nap2 (B. napus, genome: AB189735); A. tha (A. thaliana, genome: AL094711); G. max (G. max, EST:BM308592); M. tru (Medicago truncatula, ESTs: BG644842); L. esc (Lycopersicon esculentum, ESTs: BE450861); H. vul (Hordeum vulgare, ESTs: AV833513); O. sat (Oryza sativa, genome: EST: AK106548); C. sat (Cucumis sativus, genome: AB189737); and P. yed (Prunus yedoensis, genome: AB189738) BnMicEmUP3 (B. napus, genome: HQ660216). The consensus residues are colored according to: red, hydrophobic; purple, basic; green, hydrophilic; and blue, acidic. Highlighted residues show identical (dark red) or similar (gray) amino acids between BnMicEmUP bZIP domains and bZIP domains of PEND proteins. Additional common structural domains include a short pre-sequence (black), an N-terminal DNA-binding domain (dark red), central repeat domain (gray), and C-terminal transmembrane domains (yellow). (B) Phylogenetic tree of PEND homologs. The phylogenetic tree was constructed by the neighbor-joining method with 100 bootstrap replicates using CLC software. The numbers on branches show bootstrap confidence levels based on 100 bootstrap trials.

Figure 6. Structural features of BnMicEmUP3 compared to PEND proteins. (A) Alignment of the BnMicEmUP bZIP domain with selected bZIP domains of PEND proteins. Sources of sequences: P. sat (Pisum sativa, PEND, genome: AB189736); B. nap (B. napus: GSBF1, cDNA: X91138); B. nap1 (B. napus, genome: AB189734); B. nap2 (B. napus, genome: AB189735); A. tha (A. thaliana, genome: AL094711); G. max (Glycine max, EST: BM308592); M. tru (Medicago truncatula, EST: BG644842); L. esc (Lycopersicon esculentum, EST: BE450861); H. vu l (Hordeum vulgare, EST: AV833513); O. sat (Oryza sativa, genome: EST: AK106548); C. sat (Cucumis sativus, genome: AB189737); and P. yed (Prunus yedoensis, genome: AB189738) BnMicEmUP3 (B. napus, genome: HQ660216). The consensus residues are colored according to red, hydrophobic; purple, basic; green, hydrophilic; and blue, acidic. Highlighted residues are identical (dark red) or similar (gray) amino acid residue between BnMicEmUP bZIP domains and bZIP domains of PEND protein. Similar structural domains include a short pre-sequence (black), a N-terminal DNA-binding domain (dark red), and a central repeat domain (gray). (B) Schematic diagram of the BnMicEmUP3 protein showing the locations of the chloroplast transit peptide (black), the basic DNA binding, zipper region (dark red), predicted transmembrane domains (yellow), and casein kinase II phosphorylation sites. (C) Schematic diagram of the PEND protein showing similar domains to BnMicEmUP3 including a chloroplast transit peptide, a basic DNA binding region, a zipper region, predicted transmembrane domains, and casein kinase II phosphorylation sites.
The essential features of the BnMicEmUP protein include a chloroplast-targeting region, a basic region, and a large repeat region containing 11 complete leucine-rich repeats. These features have been observed in members of the bZIP family called PEND (for plastid envelope DNA binding) proteins. This family of DNA binding proteins is found in the inner envelope membrane of developing chloroplasts and response to environmental changes (Terasawa and Sato, 2009). A multi alignment of PEND proteins from different species of angiosperms including: P. sativa, B. napus, A. thaliana, G. max, M. truncatula, L. esculentum, H. vulgare, O. sativa, C. sativus, P. yedoensis, with BnMicEmUP and AT1G74730 (Terasawa and Sato, 2005; Figures 5A,B) reveals that the basic region, zipper region, and transmembrane domains are conserved, but the central region is highly variable in these proteins. The occurrence of the essential features of PEND proteins in BnMicEmUP, including a possible chloroplast transit peptide, a DNA binding site, a leucine rich repeat region, and transmembrane domains (Figures 6A–C) supports the proposal that it can be considered to be a member of this unique protein family.
A protein analysis of the amino acid sequence for BnMicEmUP showed that this protein also contains several casein kinase II phosphorylation sites that correspond to the consensus S/TXXD/E, in the 89–129aa region (Figure 3). These four repeat sites include acidic residues aspartic acid (D) or glutamic acid (E), close to the targets of the phosphorylation, serine or threonine (S or T; Meggio and Pinna, 2003). The same analysis of the PEND protein from the Prosite database12 (Gasteiger et al., 2005; Hulo et al., 2006) showed that it also contains several casein kinase II phosphorylation sites (Figure 6C).
The protein secondary structure prediction by CLC (CLC bio, Aarhus, Denmark) showed that the BnMicEmUP protein has a characteristic α-helical structure in the region that contains the leucine-rich repeats (Supplementary Figure 2A), which is the main structural motif found in bZIP proteins (Jakoby et al., 2002). In addition, two putative trans-membrane domains were predicted by phyre2 (Kelley and Sternberg, 2009) that correspond to α-helix topologies for the same regions modeled by CLC (Figure 6). The overall structure predictions for the proteins coded by the four Brassica genes and the Arabidopsis gene were very similar. They all have extended C-terminal regions with some helical structure and end with two regions of strongly folded (alpha helix or beta strand) amino acids that are associated with transmembrane regions (Supplementary Figures 2B, 3).
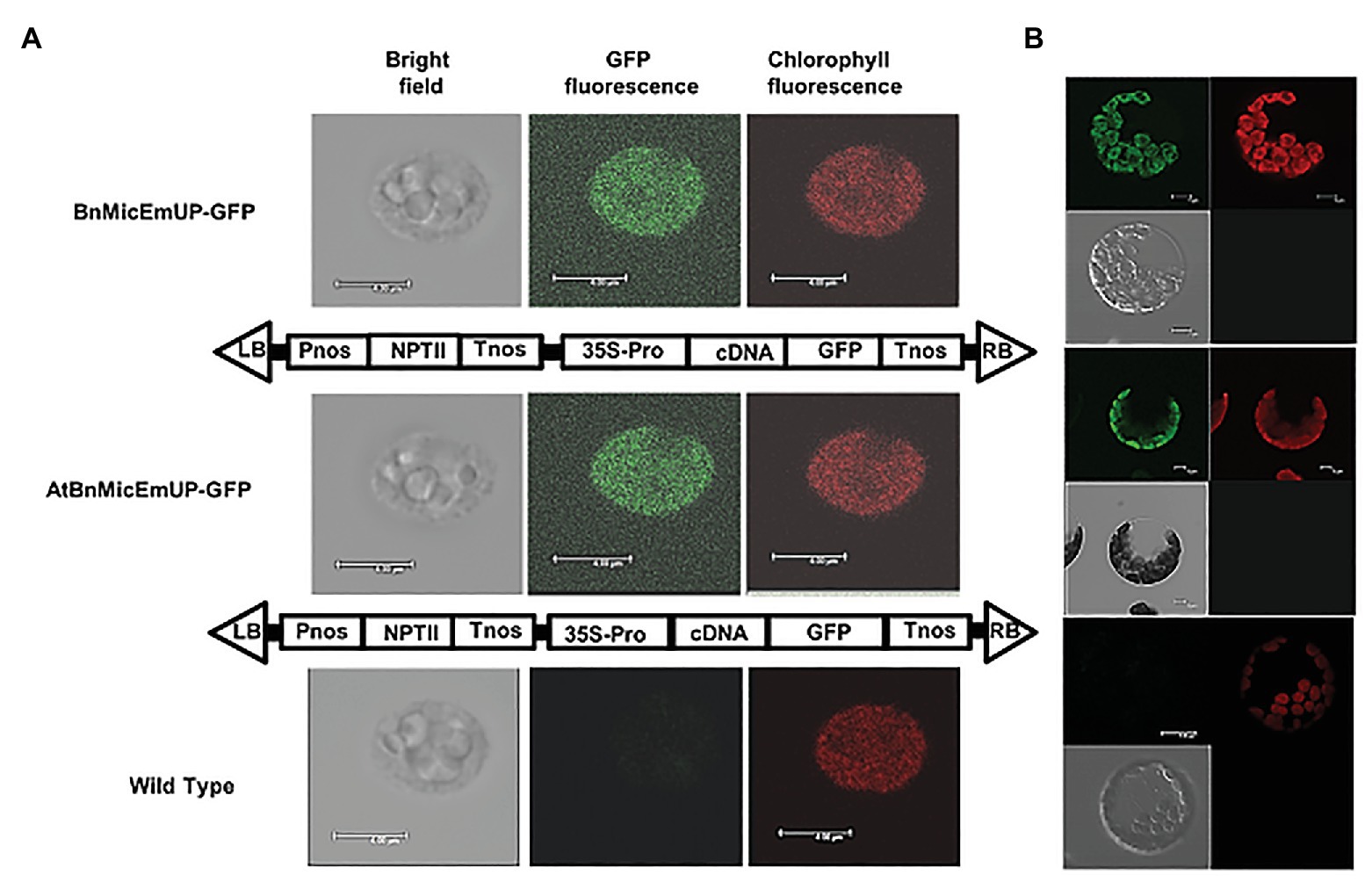
Figure 7. Targeting of AtBnMicEmUP:GFP and BnMicEmUP3::GFP to the chloroplast. (A) Chloroplasts were isolated from transgenic Arabidopsis transformed with pBI12: AtBnMicEmUP::GFP (top, diagram of the construct vector) and pBI121::BnMicEmUP3::GFP (middle, diagram of the construct vector). Right: chlorophyll fluorescence; middle: GFP fluorescence; and left: chloroplast in bright light. Scale bar = 4 μm. (B) Isolated protoplast. Right: chlorophyll fluorescence; left: GFP fluorescence; and left bottom: protoplast in bright light. Scale bar = 5 μm.
Analysis Elements in AtBnMicEmUP and BnMicEmUP3 Promoter
To characterize specific elements within the AtBnMicEmUP3 promoter, a 580 bp sequence upstream of the coding region (nucleotide 28,078,401–28,079,001) was obtained from TAIR SeqViewer.13 A 539 bp sequence upstream of the co ding region for BnMicEmUP3 (22,077,826–22,078,365) was obtained from COGE14 (Chalhoub et al., 2014) analyzed with PlantCARE15 (Lescot et al., 2002) and PLACE database (Higo et al., 1999).16 The analysis for the AtBnMicEmUP promoter identified nine potential TATA sequences at 463, 437, 429, 411, 384, 353, 230, 180, and 155 bases upstream from the start of transcription. Four CAAT sequences were found at 35, 106, 264, and 320 bases upstream from the start codon (ATG). In the BnMicEmUP3 promoter region, the analysis identified nine potential TATA sequences at 57, 135, 216, 349, 350, and 422 bases upstream from the start of transcription. Four CAAT sequences were found at 48, 76, 240, 317, 395 (−) bases upstream from the start codon (ATG). In addition to these core promoter elements, the analysis of the upstream sequence of AtBnMicEmUP and BnMicEmUP3 identified a number of cis-regulatory elements that are associated with gene regulation by light, metabolism, fungal elicitors, and growth regulators. The functions of these predicted cis-acting elements are categorized in Supplementary Table 3 and their arrangement in the upstream region of BnMicEmUP3 and AtBnMicEmUP is shown in Supplementary Figures 4, 5, respectively. The cis-acting regulatory elements involved in light responsiveness (G-box, GT1-motif, GA-motif, GAG-motif, AE-box, and BOX4) were found, and abscisic acid responsiveness (ABRE), a MeJA-responsiveness (CGTCA-motif and TGACG-motif), and auxin-responsiveness (TGA-box) elements all shared nucleotides centered at positions 461 and 410 in BnMicEmUP3 and AtBnMicEmUP, respectively. The BnMicEmUP3 and AtBnMicEmUP promoters also contain elements that are induced by physiological and environmental factors, including wounding (W box), fungal elicitors (ERE), heat shock (HSE), pathogen or stress response (TC-rich repeats), auxin (ARE), and light (G-box, GT1-motif). In addition, the promoter regions contain a MBS-MYB binding site, involved in drought-inducibility and a TGACG site, involved in the methyl jasmonate (MeJA)-responsiveness.
Gene Localization
Bioinformatics-Based Prediction of Protein Sub-Cellular Localization
Eleven different programs were used to predict the sub-cellular localization of BnMicEmUP and AtBnMicEmUP proteins. The majority of the predictions placed these proteins, in either mitochondria or plastids, especially chloroplasts (eight programs; Supplementary Table 4). Seven of the 11 programs analyzed the N-terminal sequence rather than the whole protein. Six of these programs predicted chloroplast localization for BnMicEmUP with high reliability (87–96%). PSORT predicted thylakoid membrane localization for BnMicEmUP and a mitochondrial membrane localization as a second option. Two of the programs predicted mitochondrial localization for BnMicEmUP proteins. In particular, TargetP, a neural Web-based predictor (Emanuelsson et al., 2000), predicted chloroplast and mitochondrial localization with likelihood values of 0.65 and 0.52, respectively (Guda, 2006).
Targeting of BnMicEmUP Homologs to Chloroplasts
The subcellular localization of the BnMicEmUP protein was experimentally tested by creating constructs with AtBnMicEmUP or BnMicEmUP3 tagged with GFP at their C-terminal. The majority of the chloroplasts isolated from transgenic Arabidopsis plants expressing BnMicEmUP3::GFP and AtBnMicEm P::GFP had a uniform green fluorescence (Figures 7A,B), suggesting that the BnMicEmUP3::GFP was dispersed throughout mature chloroplasts.
BnMicEmUP Copy Number
Southern blots of genomic DNA from young B. napus leaves digested with XbaI and probed with Dig-labeled BnMicEmUP3 had three bands ranging in size from 2 kb to 4.3 Kb (Figure 8A). In addition, the genes were detected in genomic DNA by PCR with specific primers (At.25185 atg-gta) that resulted approximately 800 kb in size. When the PCR product was cloned into a TOPO cloning vector, three different clones, with similar, but distinguishable inserts were obtained that were 709, 803, and 863 bp in size (Figure 8B). These results indicate that there are at least three copies of the BnMicEmUP gene in the B. napus genome (Figure 8C).
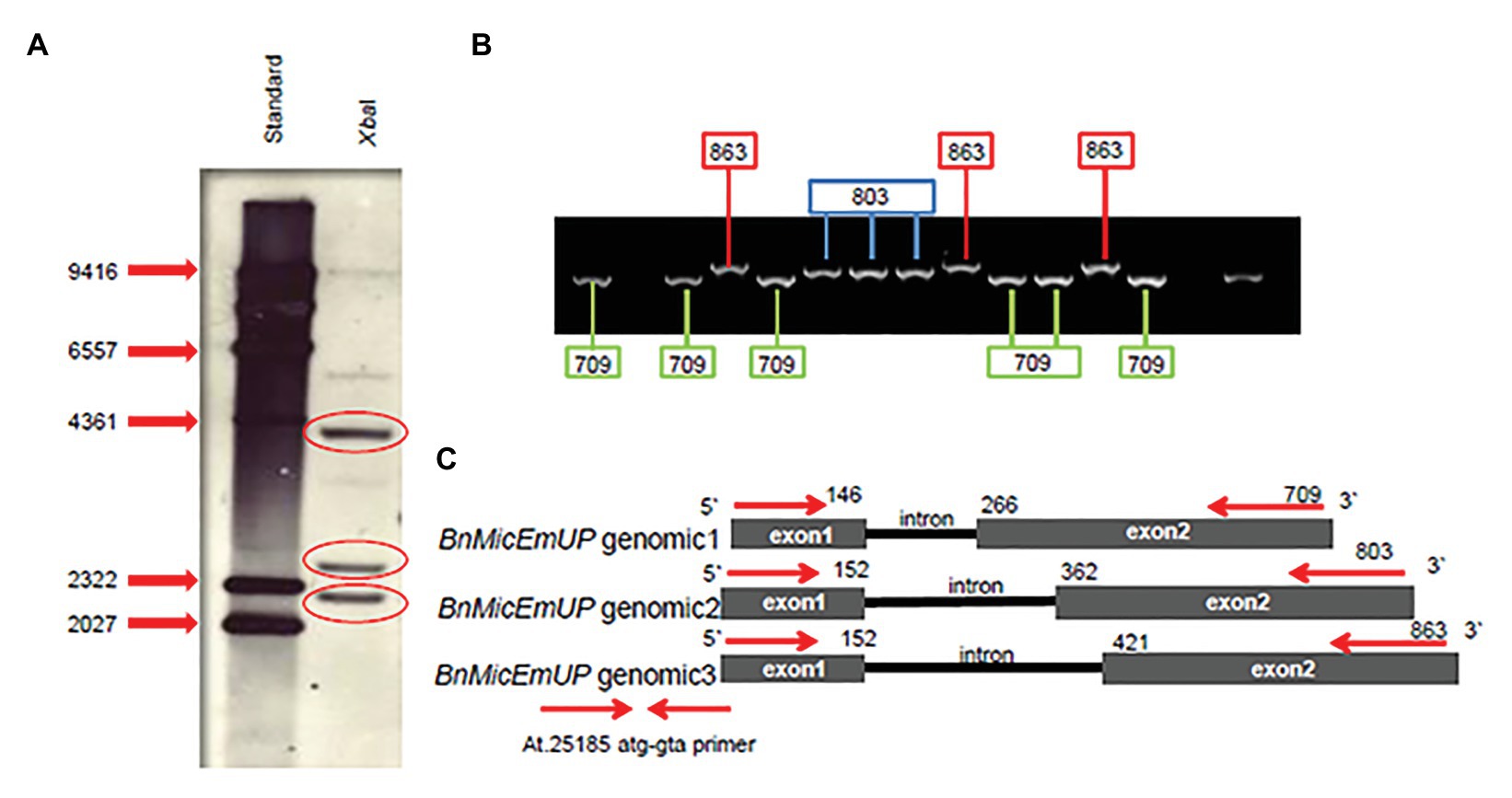
Figure 8. BnMicEmUP gene copy numbers. (A) Southern blot of B. napus DNA cut with restriction enzyme XbaI, probed with BnMicEmUP3. (B) PCR products derived from amplifications of individual colonies of PCR fragments into TOPO vectors using the At.25185 atg-gta primer. Cloning PCR fragments into TOPO vectors randomly picked from a plate and used as PCR templates contained fragments at three different discrete sizes of BnMicEmUP genomic DNA size (black line). The genomic organization of the Brassica gene is the same as the Arabidopsis, and the genomic DNA fragments are indicated with different colors. (C) The comparison of the BnMicEmUP gene structure with Arabidopsis AT1G74730. Three forms of the gene with different sizes are shown, comprised of two exons (black boxes) and one intron.
Figure 10. Comparative analysis of gene expression patterns of different forms of BnMicEmUP in induced and non-induced microspore cultures. (A,B) Expression of BnMicEmUP2,3 and BnMicEmUP1,4 from 24 to 110 h in induced (30°C, embryogenic) and non-induced (23°C, nonembryogenic) microspore cultures, values shown here relative to EF1alpha, and transcription levels of genes at day 0. (C) RNA quality evaluated by visualizing the RNA on a gel, showing two bright discrete bands that represent the 28S and 18S ribosomal species. (D) B. napus EF1alpha (housekeeping gene) as a positive control for cDNA quality. Error bars show variance (SE) among three biological replicates. Means with the same letter are not significantly different.
Genome Origins of the Three BnMicEmUP Homologs
Brassica napus is an allotetraploid (AACC) that originated by hybridization of two diploid progenitors, Brassica rapa (AA) and Brassica oleracea (CC; Nagahara, 1935). To study the genome origins of the BnMicEmUP homologs, they were BLASTed against the B. rapa and B. oleracea genomes. The BnMicEmUP1 gene exhibited highest sequence similarity (E-value < 1) with a BnMicEmUP1 homolog from B. rapa. Therefore, the B. rapa gene was named BnMicEmUP1 (Figures 9A–C). Additionally, BnMicEmUP2 and BnMicEmUP3 were predicted to be paralogous to B. olerace BnMicEmUP genes (E-value < 1). The BnMicEmUP2 and BnMicEmUP3 genes cloned from B. napus with nine nucleotide insertions in intron did not have counterparts within the B. rapa genome. Another copy of a BnMicEmUP was identified in B. rapa (which is predicted to be homologous to the unidentified genomic DNA BnMicEmUP4 (Figures 2, 9C).
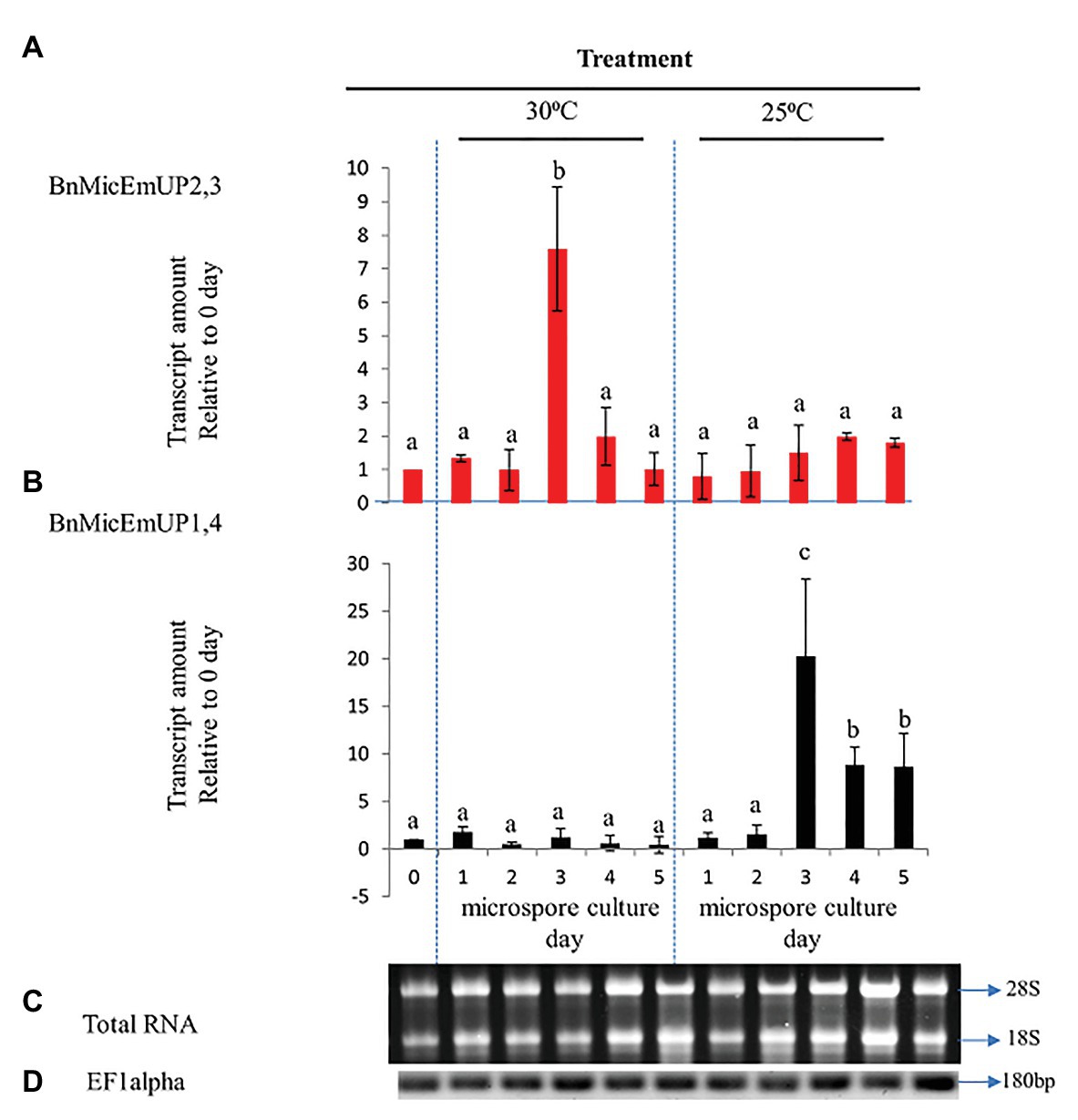
Figure 9. Sequence analysis of the BnMicEmUP genes from B. napus and its diploid progenitors, Brassica rapa, and Brassica oleracea. (A) Genomic DNA sequence alignment of the BnMicEmUP genes. (B) Phylogenetic tree of the genomic sequences of the BnMicEmUP genes. (C) Phylogenetic tree of the cDNA sequences of the BnMicEmUP genes. Sequences labeled with the blue color indicate start and stop codon and yellow color shows intron region.
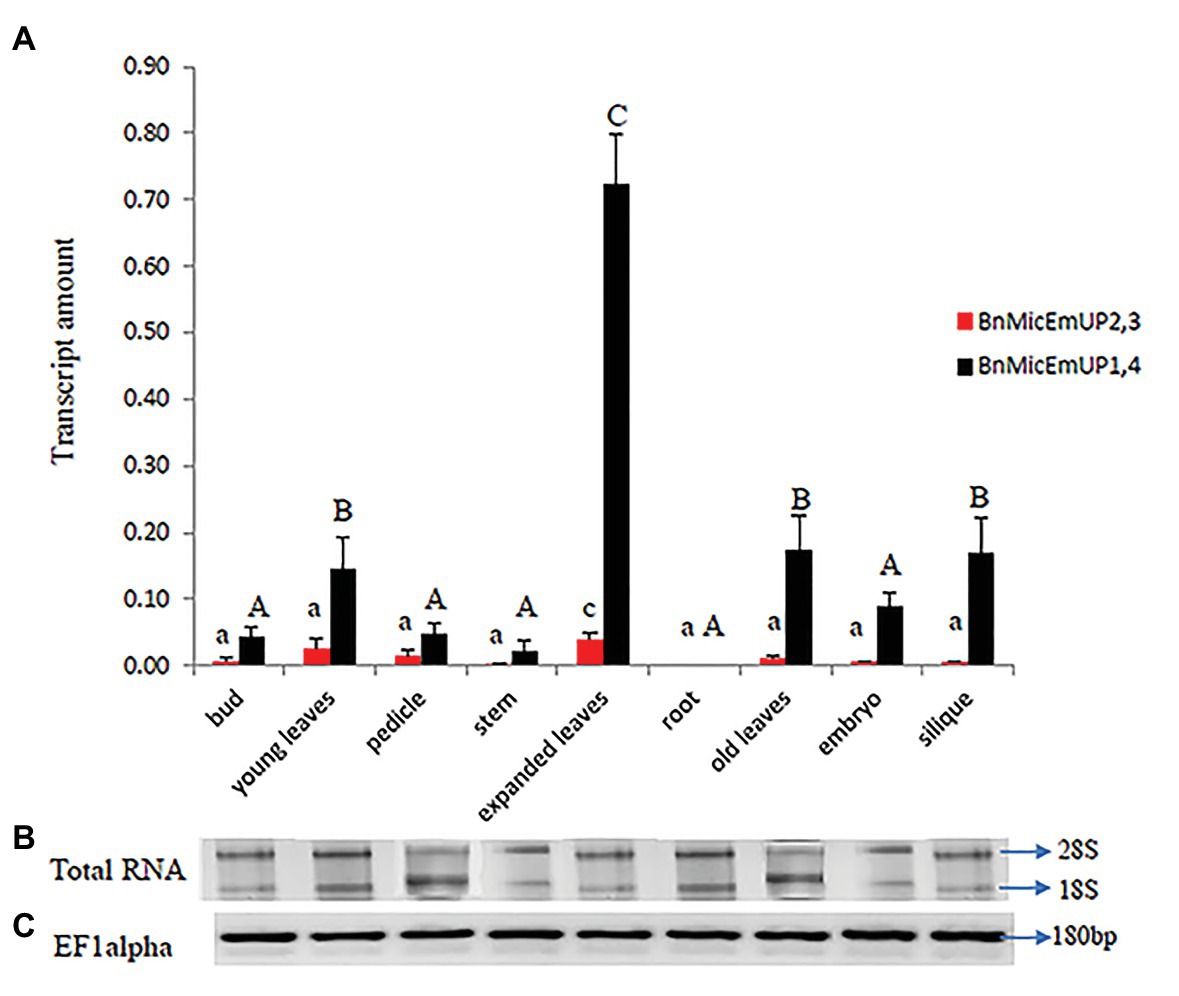
Figure 11. Real time PCR studies of BnMicEmUP2,3 and BnMicEmUP1,4 expressions in different tissues. (A) Comparison of the expression patterns of BnMicEmUP2,3 and BnMicEmUP1,4 in different tissues of B. napus. The transcription rates were expressed relative to the EF1alpha transcription rate. (B) RNA quality evaluated by visualizing the RNA on a gel, showed two bright discrete bands that represent the 28S and 18S ribosomal species. (C) B. napus Ef1alpha (housekeeping gene) expression was used as a positive control for cDNA quality. Error bars show variance (SE) among three biological replicates. Means with the same letter are not significantly different.
Gene Expression Profile of BnMicEmUP Gene in B.napus
Microspore Culture
To investigate the specificity of BnMicEmUP gene expression during microspore culture, cDNA from RT-PCR was examined by RT-PCR in reactions containing BnMicEmUP primers and RNA from freshly isolated microspores from non-responsive (25°C) or responsive (30°C) microspores cultured for 5 days. RNA isolated at different times after microspore culture initiation over 5 days in culture at 30 °C (inductive) and 25°C (non-inductive) was of good quality according to a PCR assay of the cDNA produced from it and primers for EF1 alpha. A single band of the expected size (180 bp) was observed in all of the samples. In addition, separations of the total RNA from the samples all had clearly visible 28S and 18S rRNA species, indicating that the RNA was intact (Figures 10C,D). Supplementary Figure 5 shows that the primers for these forms are highly specific. The BnMicEmUP1,4 primer amplified a band of 160 bp from the cloned BnMicEmUp1and 4-Topo2.1 but not the cloned BnMicEmUp2 and 3-Topo2.1. The BnMicEmUp2,3 primers give a larger product (200 bp) only with BnMicEmUp2,3 Topo2.1 clones (Supplementary Figure 6).
BnMicEmUP2,3 and BnMicEmUP1,4 genes showed different patterns of expression in induced microspores cultured at 30°C and non-induced microspores cultured at 25°C (Figures 10A,B). BnMicEmUP2,3 expression levels were more than 7.6-fold higher in 3 day induced cultures compared to day 0 cultures, but the other three time points did not show such clear differences in BnMicEmUP2,3 expressions (Figure 10A). One-way ANOVA showed that the difference between BnMicEmUP2,3 expressions at day 3 was significantly (p < 0.05) higher than the levels of expression at 0, 1, 2, 4, and 5 days in responsive cultures. Although BnMicEmUP2,3 expression levels were slightly higher than day 0 at day 1 and day 4, the differences were not statistically significant. No significant differences were detected between different time points for BnMicEmUp1,4 expressions in responsive (30 °C) microspore cultures. However, three patterns of expression of BnMicEmUP1,4 were observed in non-induced (25 °C) cultures. BnMicEmup1,4 expression levels in non-induced cultures were statistically significantly higher after 3, 4, or 5 days compared to day 0 as well as between day 3 and all other time points. There was no significant difference in BnMicEmUP1,4 expressions in microspore cultures between day 4 and day 5 at 25°C.
Organ-Specific Expression Profile of BnMicEmUP
BnMicEmUP1,4 was highly expressed in every organ type and developmental stage except for roots, where it was not detectable (Figure 11A). In contrast, BnMicEmUP2,3 showed very low expression levels, compared to BnMicEmUP1,4 in all of the tested tissues (Figure 11A). The expression of BnMicEmUP1,4 was 6-fold higher in expanded leaves compared to immature leaves. The ANOVA showed that the high level of BnMicEmUP1,4 expressions in expanded leaves was significantly different from its expression in other parts the plant. Expression differences for BnMicEmUP1,4 among buds, young and old leaves, embryo, and silique tissues were not significantly different and approximately one-seventh of the level seen in the expanded leaves but approximately 2-fold higher (significantly different) than the expression levels observed in the bud, pedicle, root, and embryo. BnMicEmUP1, 4 expression differences among the bud, pedicle, root, and embryo tissues were not significantly different from each other (Figure 11A). RNA integrity (18s and 26s) and RNA quality (EF1 alpha, 180 bp) are described in Figures 11B,C.
BnMicEmUP Function
Transgenic Arabidopsis over expression lines with BnMicEmUP3 under control of the cauliflower mosaic virus 35S promoter had delayed seedling development, compared to the wild type. Conversely, seedling development was accelerated in the knockdown (mutant and RNAi) Arabidopsis lines compared to wild type (Figure 12).
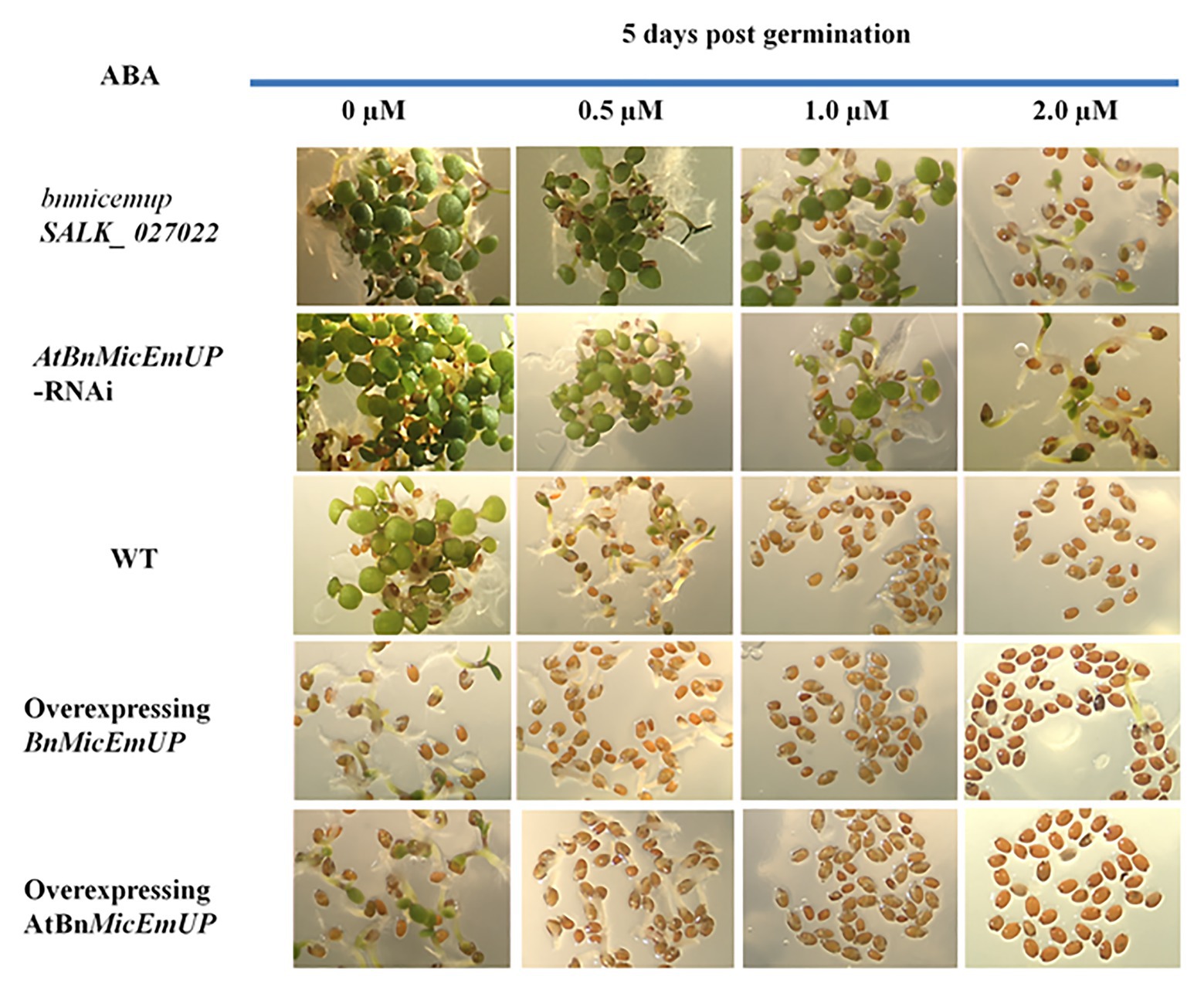
Figure 12. Effect of AtBnMicEmUP3 expressions on development of Arabidopsis seedlings in presence of different levels of abscisic acid (ABA), 5 days post-germination. Seeds from one independent AtBnMicEmUP mutant lines (atbnmicemup-SALK_ 027022), two independent silenced lines (AtBnMicEmUP-RNAi), two independent overexpressing lines (35S::AtBnMicEmUP3), and WT were germinated on plates containing 0, 0.5, 1.0, and 2.0 μM ABA for 5 days. The figures show representative responses of the experiment that was conducted three times.
Germination of wild type and overexpressing lines on various levels of ABA (between 0 and 2 μM) delayed their seedling development. In contrast, seedling sensitivity to ABA was reduced in the knockdown lines. For example, both the wild type seedlings on media without ABA (control) and BnMicEmUP3 knockdown lines on.5.0 μM ABA had similar numbers of green cotyledons, 5 days after transferring to germination medium, whereas, none of the BnMicEmUP3 overexpressing lines and wild type lines were green on 5.0 μM ABA.
Discussion
Gene Copies
The present finding that B. napus contains multiple copies of BnMicEmUP homologs was expected because B. napus is an allotetraploid (AACC) that arose from hybridization of two diploid progenitors, B. rapa (AA) and B. oleracea (CC; Nagahara, 1935). In fact, information from recent B. napus sequencing efforts,17 made it possible to identify the genomic origins of the homologs of BnMicEmUP genes identified in the present work. The BnMicEmUP2,3 homologs, with the nine nucleotide insertions, are most similar to B. oleracea sequences and, therefore, likely originated from the C genome, while the BnMicEmUP1 homolog is most similar to the sequence from B. rapa. In addition, when the recently sequenced and annotated B. napus genome18 (Chalhoub et al., 2014) was searched for BnMicEmUP sequences four BnMicEmUP genes were found (Figure 9A). Therefore, the presence of the BnMicEmUP4 genomic DNA was confirmed and the total number of BnMicEmUP genes, and their phylogenetic relationships are in agreement with the evolutionary history of Brassica species.
The greatest variability among the different copies of the BnMicEmUP gene was in the lengths of their introns. In contrast, the BnMicEmUPs exons are highly conserved. This is consistent with general findings in other organisms (Alberts et al., 2002). Although intron variations are not generally correlated with functional or developmental differences, they may affect levels of gene expression (Coulombe-Huntington and Majewski, 2007). Castillo-Davis et al. (2002) have shown that genes with shorter introns have higher levels of expression than genes with longer introns. Therefore, it is possible that variations in the intron lengths and sequences of BnMicEmUP genes are related to differential gene expression levels and expression patterns.
Gene Expression
The current finding that the BnMicEmUP2,3 homologs are highly upregulated during the first few days in embryogenic microspore cultures, confirms our previous finding that a transcript with homology to AT1G74730 was upregulated in embrogenic microspore cells (Chan, 2006; Pauls et al., 2006) suggesting that these genes play a role in the differentiation and/or commitment to embryo development-specific. In contrast, the upregulation of BnMicEmUP1,4 homologs in non-responsive microspore cultures and other tissues, such as leaves, indicates that they are markers for vegetative tissues. Differential regulation was also reported for five members of plant ROP genes (Rho-GTPases) in various organs and during androgenesis in B. napus, including upregulation of BnROP9 in embryogenic cells sorted from induced microspore cultures (Chan and Pauls, 2006).
Supporting evidence for the expression patterns for BnMicEmUP genes was obtained with the eFP browser, “a visualization tool for exploring publicly available microarray data sets” (Winter et al., 2007). The pictorial representations of gene expression that this tool (Supplementary Figure 7) confirmed that AtBnMicEmUP is highly expressed in expanded leaves, young leaves, and the globular embryo stage in Arabidopsis but, is weakly expressed in the roots and mature leaves, and is not expressed in ripened siliques. Perhaps, because the gene encodes a putative chloroplast-targeted protein, its high expression in expanded leaves is related to the relatively high numbers of chloroplasts that would be expected in growing and expanding leaf cells (Possingham and Saurer, 1969; Pyke, 1999).
No information about the expression of the AtBnMicEmUP during embryo initiation (day 0–5) is available in the eFP browser; however, high expression levels of the gene are shown for globular stage embryos, which occur 7 days after fertilization (Supplementary Figure 7B). These analyses also revealed that the level of expression of this gene was higher at day 7 than at day 15, when the embryos were at the heart and torpedo stages, respectively (Borkird et al., 1988). The high expression of AtBnMicEmUP in the globular stage of embryo development also supports the relationship between AtBnMicEmUP expression and chloroplast development because chloroplast biogenesis occurs during early globular stage embryogenesis (Tejos et al., 2010) and it has been suggested that the ability of cells to develop chloroplasts can be used as a marker for cell differentiation during embryogenesis. In addition, loss of chloroplast function in Arabidopsis leads to embryo lethality (Tzafrir et al., 2004).
Moreover, the eFP browser revealed that the expression of AtBnMicEmUP (homolog of BnMicEmUP) was downregulated by abiotic stresses, such as osmotic, heat, and UV stress. Interestingly, it was shown that heat stress (3 h at 38°C followed by recovery at 25°C) in seedlings and shoots downregulated AtBnMicEmUP after 3, 4, and 6 h of treatment (Winter et al., 2007). This contrasts with the current results that indicate that the BnMicEmUP2,3 genes in Brassica is highly upregulated by day 3 in embryogenic microspores after exposure to a mild heat stress. Perhaps, differences in heat treatments (i.e., 30°C in the current study vs. 38°C for the previous work) and tissues tested (i.e., microspores vs. seedlings) account for the different responses to heat stress. Together, the results suggest that expression of BnMicEmUP is regulated by stress.
Promoter Analysis
The central regions of the BnMicEmUP3 promoters have combinations of different light-responsive elements, stress-response elements, and hormone-response elements that allow for different levels of expression during plant growth and development and environmental conditions (Yamaguchi-Shinozaki and Shinozaki, 2005). They may also provide interesting links between specific developmental and stress responses. The multiple TATA (nine) and CAAT (four) box sequences found in BnMicEmUP3, which are defined as core promoter elements (Kozak, 1981), may function to promote high levels of transcription (Kiran et al., 2006). Multiple TATA boxes may also promote differential transcription, depending on the environmental conditions (Doyle and Han, 2001). In particular, the multiple TATA boxes in BnMicEmUP3 promoters may be related to the stress inducibility of these genes (Bae et al., 2015). The ABRE elements found in the BnMicEmUP3 gene promoters are important cis-acting elements in ABA-responsive genes that mediate gene expression responses to abiotic stress (Xu et al., 1996). The cis-acting elements that are responsive to jasmonic acid (Wasternack, 2007; Balbi and Devoto, 2008) in the BnMicEmUP3 promoters may support the response of these genes to insect pests. The W box type elements that occur in BnMicEmUP3 promoters are binding sites for WRKY transcription factors, which also have roles in gene expression responses to wounding associated with pathogenesis and herbivory (Hara et al., 2000). WRKY transcription factors are also known to modulate gene expression during a variety of developmental and physiological processes including embryogenesis (Lagacé and Matton, 2004), and stem cell maintenance in apical meristems (Rose et al., 2006).
The large number of light responsive cis-elements, such as the GT1 motif and G-Box, in the upstream sequences of BnMicEmUP3 genes may indicate that they are regulated by light. However, these elements may also be an indication of sensitivity to stress since they also occur in the promoters of genes that respond to various environmental stressors and ABA (Williams et al., 1992; Komarnytsky and Borisjuk, 2003). Some light responsive cis-elements are found in promoters of genes that are not light-regulated (Kuno and Furuya, 2000). Therefore, the presence of LREs in the promoter of BnMicEmUP3 is not enough to consider this gene as a photo-regulated gene. Nevertheless, it could be a link between the chloroplast localization of the protein and some roles of light in the stress induced microspore embryogenesis, since previous experiments with B. napus microspore cultures showed that embryo production was significantly promoted by growing the donor plants at high light (Thurling and Chay, 1984).
A very interesting finding was the extensive overlap of cis-acting regulatory elements for abscisic acid (ABRE), MeJA (CGTCA-motif and TGACG-motif), and auxin responsiveness (TGA-box) in one region of the promoter at position 461, next to circadian cis-elements. This region may have an important role in coordinating the expression of BnMicEmUP3 in response to various signaling pathways, through competition for common promoter sites. Other work has shown that ABI3, an ABA responsive transcription factor, binds to both ABA and auxin response element (Nag et al., 2005).
Characteristics of the BnMicEmUP Predicted Protein
The results presented here demonstrate that BnMicEmUP is a PEND protein with a short pre-sequence, an N-terminal DNA-binding domain (cbZIP), a central repeat domain, and a C-terminal transmembrane domain. The analysis of the polypeptide encoded by BnMicEmUP strongly indicated that it contains a putative bZIP domain, which makes it a member of one of the largest transcription factor (TF) families in plant genomes (Jakoby et al., 2002).
This assignment as a putative bZIP transcription factor was based on the identification of a basic DNA binding motif and leucine zipper motifs (Figures 3, 5A). Additionally, the bioinformatics and experimental evidence for the chloroplast localization of BnMicEmUP characterizes it as a cbZIP. A previous comparison of cbZIP domains with typical bZIP domains (Terasawa and Sato, 2005) showed that there were clear differences that related to variation in the structure of the central region that separates the DNA binding and leucine zipper motifs. For example, A PEND protein from pea, a cbZIP DNA-binding protein in the inner envelope of the chloroplast (Terasawa and Sato, 2005), contains a central region exceeding 20 amino acid residues (Sato et al., 1998). This variation in the central region was also reported in genes homologous to PEND, such as the GS-box binding factor 1 (GSBF1) of B. napus and GSBF1 homolog of A. thaliana (Sato et al., 1998). Similarly, the BnMicEmUP central region contains 13 amino acids, which is longer than the nine amino acid residues typically found in bZIP domains. Surprisingly, the common motif of basic DNA binding domains, consisting of invariant Asn and Arg residues (Miller, 2009), is not conserved in the PEND from pea (Sato et al., 1993) and its homologs (Sato et al., 1998), but it is conserved in the basic region of the BnMicEmUP protein. In accordance with a previous comparison of the amino acid sequences of bZIP proteins, which showed more similarities in their basic domains than in their leucine zippers (Vettore et al., 1998), the current study showed that leucine residues are replaced by other hydrophobic residues in many of the heptads in BnMicEmUP (Figure 5). In addition, a variation in the number of amino acids between the basic region and heptad repeat of hydrophobic amino acids has been observed in many members of the pea PEND (cbZIP) protein.
The transmembrane domains in the C-terminal end of the BnMicEmUP polypeptide were identified based on the occurrence of regions that contain ~20 hydrophobic residues. Such an arrangement of amino acids is necessary for the formation of a helix to span a biological membrane (Rost et al., 1995). A model for the BnMicEmUP peptide structure that is similar to PEND (Terasawa and Sato, 2009) has the C-terminal transmembrane domain of the processed protein inserted into the inner chloroplast membrane and the N-terminal DNA-binding domain oriented toward the stroma so that it can bind to chloroplast DNA.
The N-terminal end of the BnMicEmUP polypeptide also contains a signal peptide region as a distinct and separate region from the basic region (BR) that serves as either a nuclear localization signal (NLS; van der Krol and Chua, 1991) or chloroplast localization signal (Terasawa and Sato, 2005) in other proteins. The transit peptide in the N-terminus of AtBnMicEmUP identified by PSORT is only 15 amino acids long, which is shorter than most known chloroplast presequences (Heins et al., 1998; Keegstra and Cline, 1999). However, similar lengths of chloroplast protein presequences were reported for the PEND and E37 proteins (Terasawa and Sato, 2009). The three amino acid homology in the transit peptides of the BnMicEmUPs (Figure 5) has also been identified in the transit peptides of two nuclear-encoded chloroplast proteins, including chlorophyll a/b protein and the small subunit of ribulose-1,5-bisphosphate carboxylase (Rubisco; Karlin-Neumann and Tobin, 1986). Chloroplast import pre-sequences are cleaved during import and a putative processing site consisting of the A/A motif (Zhang and Glaser, 2002) was identified at the N-terminal end of the chloroplast presequence of the BnMicEmUP protein.
In agreement with the present results (C-terminal GFP/BnMicEmUP fusion protein), a study to identify proteins targeted to the plastid (Koo and Ohlrogge, 2002) indicated that AT1G74730 (AtBnMicEmUP) was localized in the chloroplast envelope, but a study by Friso et al. (2009) suggests that it is located in the thylakoid membrane. The absence of detectable levels of GFP fluorescence in other organelles indicated that the GFP/BnMicEmUP fusion protein was very efficiently targeted to chloroplasts. Yokoyama et al. (2016) recently suggested that two homologs of this same gene encode proteins (they called RIQ1 and RIQ2) that affect the organization of the light-harvesting complex II and grana stacking in Arabidopsis, however, did not comment on their DNA binding motifs.
The BnMicEmUP protein is not the only transcription factor located in the plastid. Other transcription factors, either PEND or non-PEND proteins, have been shown to be located in plastids. Examples of these proteins (Nakano et al., 2003): a plastid envelope DNA binding protein (PEND) from P. sativum containing a basic domain followed by leucine zipper motif (Thurling and Chay, 1984), proteins named PD1 and PD3 also from P. sativum (Sato et al., 1995). A protein designated as Plastid Transcription Factor 1 (PTF1) containing a basic helix-loop-helix (bHLH) motif from A. thaliana has also been suggested as a plastid transcription factor (Baba et al., 2001). The NtWIN4 protein, a bHLH motif containing a wound-induced protein from N. tabacum, has also been reported to be a transcription repressor with roles in adaption to biotic and abiotic stresses (Kodama and Sano, 2006). Kodama and Sano (2007) suggested that these proteins have evolved from eukaryotic nuclear transcription factors into plastid functional proteins. Furthermore, a current hypothesis is that these genes were altered evolutionarily to allow plants to cope with environmental stresses (Sato, 2001).
Growth Arrest Mediated by BnMicEmUP
The delay in development observed in the present study for the constitutively expressing 35S::BnMicEmUP3 transgenic Arabidopsis seedlings supports the hypothesis that BnMicEmUP3 expression inhibits plant growth and development. Interestingly, a similar delay in seedling growth was reported for Arabidopsis seedlings overexpressing ABSCISIC ACID INSENSITIVE 5 (ABI5), during the first 3 days after seed germination (Lopez-Molina et al., 2001; Garcia et al., 2008; Wang et al., 2013). Many studies have shown that the addition of endogenous ABA maintains seed dormancy, resulting in the inhibition of seed germination (Nambara and Marion-Poll, 2003; Leymarie et al., 2008; Rodríguez-Gacio et al., 2009). The similarity of the delay in seedling growth, therefore, suggests that ABA may be associated with BnMicEmUP3 function and the finding that BnMicEmUP overexpression results in an enhanced response to ABA supported this conclusion.
BnMicEmUP is also structurally similar to ABI5, since both have elements of basic leucine zipper (bZIP) transcription factors. Moreover, both BnMicEmUP and ABI5 contain casein kinase phosphorylation sites, which may be phosphorylated by SnRK2s (Finkelstein et al., 2005; Nakashima et al., 2009). Furthermore, the identification of conserved ABA-responsive elements (ABRE; PyACGTGGC), which control ABA-regulated gene expression, in the promoter of BnMicEmUP3 suggests that BnMicEmUP3 is stress inducible and may itself be regulated by ABA.
ABA May Be Associated With BnMicEmUP Function During the Induction Phase of Microspore Embryogenesis
Several lines of evidence suggest that an ABA signal may promote the observed upregulation of BnMicEmUP2,3 during the induction phase of microspore embryogenesis. In particular, different kinds of stress, which would increase endogenous ABA levels (Sah et al., 2016), are widely applied as pretreatments in anther and microspore cultures of tobacco (Imamura and Harada, 1980), barley (van Bergen et al., 1999). It has also been shown that androgenesis is improved by the addition of exogenous ABA in wheat and tobacco (Kyo and Harada, 1986; Hu et al., 1995). Heat-stress induction of embryogenesis in microspore cultures of B. napus is associated with calcium release and alkalization of the microspore (Pauls et al., 2006). Consequently, an ABA signaling pathway may be directly or indirectly involved in the activation of the gene expression during microspore embryogenesis (Hays et al., 1999; Tsuwamoto et al., 2007). Furthermore, inhibition of ABA synthesis reduces regeneration efficiency in androgenesis (Hoekstra et al., 1997). In addition, the ABI3 gene, which regulates ABA responsiveness in plants (Brady et al., 2003), is a necessary embryogenesis factor (McCourt, 1999) and is considered a molecular marker gene for microspore embryogenesis in B. napus (Malik et al., 2007). Interestingly, a subtracted cDNA library composed of upregulated genes during androgenic initiation shows that these genes contain ABA-inducible cis-elements, such as the ABRE motif in their promoter regions (Tsuwamoto et al., 2007).
Model for BnMicEmUP Protein Involvement in ABA-Mediated Transcriptional Regulation
A model for BnMicEmUP involvement in ABA signaling is proposed in Figure 13. In this model, the signaling system includes: an ABA receptor (PYR/PYL/RCAR; Ma et al., 2009; Park et al., 2009), a type 2C protein phosphatase (Umezawa et al., 2009; Vlad et al., 2009; PP2C), a SNF1-related protein kinase (2SnRK2; Fujii and Zhu, 2009; Umezawa et al., 2010) and leucine zipper (bZIP) transcription factors as target of SnRK2. This system can regulate ABA-mediated ABRE-dependent gene expression either negatively or positively. The PP2C inactivates SnRK2 by dephosphorylation in absence of ABA, whereas, in the presence of ABA, PYR/PYL/RCAR binds to ABA, resulting in PP2C inhibition and SnRK2 activation. Then SnRK2 phosphorylates a basic-domain leucine zipper (bZIP) transcription factor (Cutler et al., 2010; Hubbard et al., 2010; Klingler et al., 2010; Raghavendra et al., 2010). In the model, BnMicEmUP functions as a transcription factor that is stored in chloroplasts. Stress causes it to be released from the plastid envelope and translocated to the nucleus where it functions as an ABRE-binding proteins (AREBs) that is regulated through phosphorylation mediated by SnRK2s (Finkelstein et al., 2005; Nakashima et al., 2009). Interestingly, BnMicEmUP3 can function as an ABA-regulated gene because it has ABRE elements in its promoter that are targets for AREBs, such as ABI5. In this model ABI5 and BnMicEmUP3 have similar functions. Synergistic interactions between ABI5 and several other bZIP ABA response factors have been reported (Reeves et al., 2011).
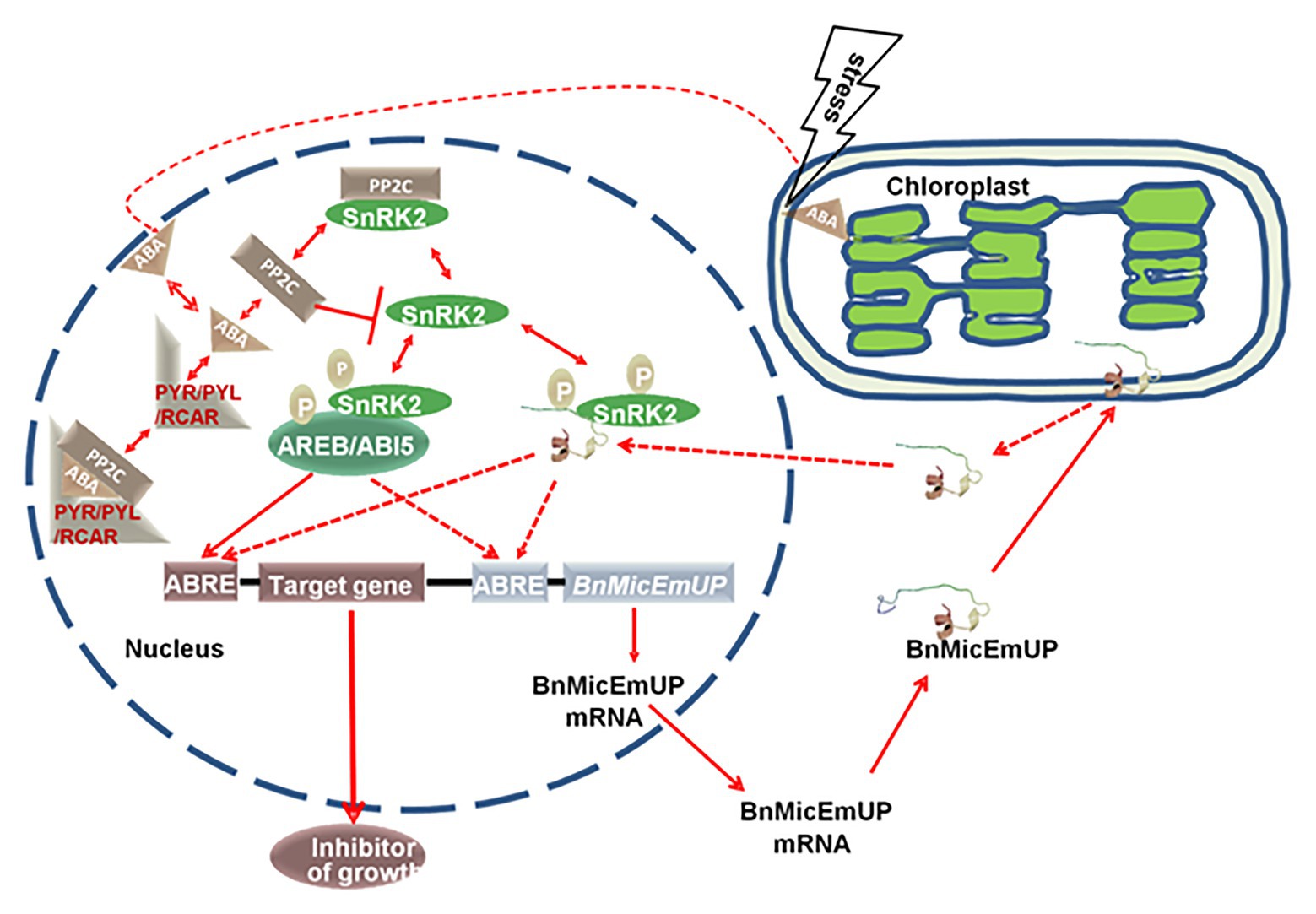
Figure 13. A proposed model for a dual targeting of BnMicEmUP protein and its involvement in ABA-mediated transcriptional regulation. BnMicEmUP protein is first sequestered to chloroplasts by transit through the outer membrane and is possibly associating with the inner membrane through interaction with the transmembrane domains. Stress leads to the release of BnMicEmUP and shuttling to the nucleus. In the nucleus, in the absence of ABA, PP2C inhibits autophosphorylation of a SnRK kinase by dephosphorylation. In the presence of ABA, PYR/PYL/RCAR binds ABA and sequesters PP2C. As a result, SnRK2 becomes auto-activated and can subsequently phosphorylate downstream factors, such as the ABREB/ABI5 or transcription factor or BnMicEmUP. AB15 or BnMicEmUP either could stimulate transcription of a downstream inhibitory protein or may stimulate its own transcription in a self-regulatory manner.
In summary, it can be proposed that the BnMicEmUP protein has two structural forms that act at different times and different locations. The complete structure of protein contains a putative cTP in the N-terminus and trans-membrane domains in C-terminus, analogous to a PEND protein, could be involved in sequestering and storage of the protein before activation in chloroplast. The second structure would be derived from the first after activation by a stress stimulus that is perceived by chloroplasts. It is proposed that the process involves movement of the sequestered protein from chloroplast to the nucleus, dimerization, and interaction with DNA, through the basic DNA binding motif, to act as a transcription factor. However, additional studies are needed to confirm the dual localization of BnMicEmUP and demonstrate its DNA binding and gene regulation capacity.
Conclusion
BnMicEmUP encodes a protein with a predicted molecular mass of 19.8 kDa that is homologous to bZIP-type transcription factors. The BnMicEmUP proteins appear to be transcription factors that are localized in plastids and are involved in plant responses to biotic and abiotic environmental stresses. Seeds harvested from transgenic Arabidopsis plants that over-express BnMicEmUP3 had lower germination rates than seed from untransformed control plants, and from transgenics or mutants with reduced levels of BnMicEmUP2,3 gene expression.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
FS conducted the experiment, analyzed the data, and wrote the manuscript. KP developed the overall strategy, designed experiments, coordinated the project, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Natural Sciences and Engineering Research Council of Canada (NSERC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Michaela Strüder-Kypke for her assistance with the confocal microscopy, Laima Kott and Kathy Perry for their advice on the B. napus microspore culture system, and Dietmar Scholz for maintaining the growth chamber facilities. Special thanks to Jan Brazolot for providing technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.576008/full#supplementary-material
Footnotes
1. https://www.arabidopsis.org/portals/masc/projects.jsp and http://www.arabidopsis.org
2. http://www.qiagenbioinformatics.com/
3. http://www.ncbi.nlm.nih.gov/BLAST/
5. http://www.ncbi.nlm.nih.gov/BLAST/
6. https://www.expasy.org/resources/blast
7. http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml
9. http://signal.salk.edu/cgi-bin/tdnaexpress
11. http://planttfdb.gao-lab.org
12. https://www.expasy.org/resources/prosite
14. https://genomevolution.org/CoGe
15. http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
16. http://www.dna.affrc.go.jp/PLACE/
References
Alberts, B., Johnson, A., Lewis, J., Raff, M., Roberts, K., and Walter, P. (2002). Molecular biology of the cell. New York: Garland Science.
Attwood, T. K., Blythe, M. J., Flower, D. R., Gaulton, A., Mabey, J. E., Maudling, N., et al. (2002). PRINTS and PRINTS-S shed light on protein ancestry. Nucleic Acids Res. 30, 239–241. doi: 10.1093/nar/30.1.239
Baba, K., Nakano, T., Yamagishi, K., and Yoshida, S. (2001). Involvement of a nuclear-encoded basic helix-loop-helix protein in transcription of the light-responsive promoter of psbD. Plant Physiol. 125, 595–603. doi: 10.1104/pp.125.2.595
Bae, S. H., Han, H. W., and Moon, J. (2015). Functional analysis of the molecular interactions of TATA box-containing genes and essential genes. PLoS One 10:e0120848. doi: 10.1371/journal.pone.0120848
Balbi, V., and Devoto, A. (2008). Jasmonate signaling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol. 177, 301–318. doi: 10.1111/j.1469-8137.2007.02292.x
Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., et al. (2002). The Pfam protein families’ database. Nucleic Acids Res. 30, 276–280. doi: 10.1093/nar/30.1.276
Borkird, C., Choi, J. H., Jin, Z., Franz, G., Hatzopoulos, P., Chorneau, R., et al. (1988). Developmental regulation of embryonic genes in plants. Proc. Natl. Acad. Sci. U. S. A. 85, 6399–6403. doi: 10.1073/pnas.85.17.6399
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749. doi: 10.1105/tpc.001941
Brady, S. M., Sarkar, S. F., Bonetta, D., and McCourt, P. (2003). The abscisic acid insensitive 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 34, 67–75. doi: 10.1046/j.1365-313X.2003.01707.x
Castillo-Davis, C., Mekhedov, S., Hart, D., Koonin, E., and Kondrashov, F. (2002). Selection for short introns in highly expressed genes. Nat. Genet. 31, 415–418. doi: 10.1038/ng940
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chan, J. (2006). A comparison of transcriptomes and the specific gene expression patterns of embryogenic and pollen-like cells in the Brassica napus microspore culture system. Dissertation, University of Guelph.
Chan, J., and Pauls, K. P. (2006). Brassica napus Rop GTPases and their expression in microspore cultures. Planta 225, 469–484. doi: 10.1007/s00425-006-0362-5
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G., et al. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. doi: 10.1093/nar/gkg500
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coulombe-Huntington, J., and Majewski, J. (2007). Characterization of intron loss events in mammals. Genet. Res. 17, 23–32. doi: 10.1101/gr.5703406
Custers, J. B. M., Coredewener, J. H. G., Fiers, M. A., Maassen, B. T. H., van Lookeren Campagne, M. M., and Lui, C. M. (2001). “Androgenesis in Brassica: a model system to study the initiation of plant embryogenesis” in Current trends in embryology of angiosperms. eds. S. S. Bhojwani and W. Y. Soh (Dordrecht: Kluwer Academic), 451–470.
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. l61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Doyle, J. J. (1991). “DNA protocols for plants” in Molecular techniques in taxonomy. eds. G. Hewitt, A. W. B. Johnson, and J. P. W. Young (Berlin: Springer-Verlag), 283–293.
Doyle, M. C., and Han, I. S. (2001). The roles of two TATA boxes and 3-flanking region of soybean ß-tubulin gene (tubb1) in light-sensitive expression. Mol. Cell 12, 197–203.
Dubas, E., Custers, J., Kieft, H., Wedzony, M., and van Lammeren, A. A. (2013). Characterization of polarity development through 2– and 3-D imaging during the initial phase of microspore embryogenesis in Brassica napus L. Protoplasma 251, 103–113. doi: 10.1007/s00709-013-0530-y
Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their n-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Fiers, M., Hause, G., Boutilier, K., Casamitjana-Martinez, E., Weijers, D., Offringa, R., et al. (2004). Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327, 37–49. doi: 10.1016/j.gene.2003.11.014
Finkelstein, R., Gampala, S. S. L., Lynch, T. J., Thomas, T. L., and Rock, C. D. (2005). Redundant and distinct functions of the aba response loci aba-insensitive (ABI5) and abre-binding factor (ABF3). Plant Mol. Biol. 59, 253–267. doi: 10.1007/s11103-005-8767-2
Friso, G., Giacomelli, L., Ytterberg, A. J., Peltier, J. B., Rudella, A., Sun, Q., et al. (2009). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database(W). Plant Cell 16, 478–499. doi: 10.1105/tpc.017814
Fujii, H., and Zhu, J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U. S. A. 106, 8380–8385. doi: 10.1073/pnas.0903144106
Garcia, M. E., Lynch, T., Peeters, J., Snowden, C., and Finkelstein, R. (2008). A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 67, 643–658. doi: 10.1007/s11103-008-9344-2
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server” in The proteomics protocols handbook. eds. J. M. Walker and N. Totowa (NJ, USA: Humana Press), 571–607.
Germanà, M. A. (2011). Anther culture for haploid and doubled haploid production. Plant Cell Tissue Organ Cult. 104, 283–300. doi: 10.1007/s11240-010-9852-z
Guda, C. (2006). pTARGET: a web server for predicting protein subcellular localization. Nucleic Acids Res. 34, 210–213. doi: 10.1093/nar/gkl093
Hara, K., Yagi, M., Kusano, T., and Sano, H. (2000). Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol. Gen. Genet. 263, 30–37. doi: 10.1007/PL00008673
Hattori, J. B., Boutilier, K. A., Campagne, M. M. L., and Miki, B. L. (1998). A conserved BURP domain defines a novel group of plant proteins with unusual primary structures. Mol. Gen. Genet. 259, 424–428. doi: 10.1007/s004380050832
Hays, D. B., Wilen, R. W., Sheng, C. X., Moloney, M. M., and Pharis, R. P. (1999). Embryo-specific gene expression in microspore-derived embryos of Brassica napus. An interaction between abscisic acid and jasmonic acid. Plant Physiol. 119, 1065–1072. doi: 10.1104/pp.119.3.1065
Heins, L., Collinson, I., and Soll, J. (1998). The protein translocation apparatus of chloroplast envelopes. Trends Plant Sci. 3, 56–61. doi: 10.1016/S1360-1385(97)01161-8
Higo, K. U., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hoekstra, S., van Bergen, S., van Brouwershaven, I. R., Schilperoort, R. A., and Wang, M. (1997). Androgenesis in Hordeum vulgare L.: effects of mannitol, calcium and abscisic acid on anther pretreatment. Plant Sci. 126, 211–218. doi: 10.1016/S0168-9452(97)00096-4
Hood, E. E., Gelvin, S. B., Melchers, L. S., and Hoekema, A. (1993). New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2, 208–218. doi: 10.1007/BF01977351
Hu, T., Ziauddin, A., Simion, E., and Kasha, K. (1995). Isolated microspore culture of wheat (Triticum aestivum L.) in a defined media I. Effects of pretreatment, isolation methods, and hormones. In Vitro Cell. Dev. Biol. Plant 31, 79–83. doi: 10.1007/BF02632241
Hubbard, K. E., Nishimura, N., Hitomi, K., Getzoff, E. D., and Schroeder, J. I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24, 1695–1708. doi: 10.1101/gad.1953910
Hulo, N., Bairoch, A., Bulliard, V., Cerutti, L., De Castro, E., Langendijk-Genevaux, P., et al. (2006). The PROSITE database. Nucleic Acids Res. 34, 227–230. doi: 10.1093/nar/gkj063
Humphreys, D. G., and Knox, R. E. (2015). “Doubled haploid breeding in cereals” in Advances in plant breeding strategies: Breeding, biotechnology and molecular tools. eds. J. Al-Khayri, H. Jain, and D. Johnson (Cham: Springer).
Imamura, J., and Harada, H. (1980). Effects of abscisic acid and water stress on the embryo and plantlet production in anther culture of Nicotiana tabacum cv. Samsun. Z. Pflanzenphysiol. 100, 285–289. doi: 10.1016/S0044-328X(80)80232-7
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jefferson, R. A., Klass, M., Wolf, N., and Hirsh, D. (1987). Expression of chimeric genes in Caenorhabditis elegans. J. Mol. Biol. 193, 41–46. doi: 10.1016/0022-2836(87)90624-3
Joosen, R., Cordewener, J., Supena, E. D. J., Vorst, O., Lammers, M., Maliepaard, C., et al. (2007). Combined transcriptome and proteome analysis identifies pathways and markers associated with the establishment of rapeseed microspore-derived embryo development. Plant Physiol. 144, 155–172. doi: 10.1104/pp.107.098723
Karlin-Neumann, G. A., and Tobin, E. M. (1986). Transit peptides of nuclear-encoded chloroplast proteins shares a common amino acid framework. EMBO J. 5, 9–13.
Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11, 557–570. doi: 10.1105/tpc.11.4.557
Kelley, L. A., and Sternberg, M. J. E. (2009). Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4, 363–371. doi: 10.1038/nprot.2009.2
Kiran, K., Ansari, S. A., Srivastava, R., Lodhi, N., Chaturvedi, C. P., Sawant, S. V., et al. (2006). The TATA-box sequence in the basal promoter contributes to determining light-dependent gene expression in plants. Plant Physiol. 142, 364–376. doi: 10.1104/pp.106.084319
Klingler, J. P., Batelli, G., and Zhu, J. (2010). ABA receptors: the START of a new paradigm in phytohormone signaling. J. Exp. Bot. 61, 3199–3210. doi: 10.1093/jxb/erq151
Kodama, Y., and Sano, H. (2006). Evolution of a basic helix-loop-helix protein from a transcriptional repressor to a plastid-resident regulatory factor: involvement in hypersensitive cell death in tobacco plants. J. Biol. Chem. 281, 35369–35380. doi: 10.1074/jbc.M604140200
Kodama, Y., and Sano, H. (2007). A comparative analysis of basic helix-loop-helix proteins, AtPTF1 and NtWIN4, with reference to plastid localization. Plant Biotech. 24, 335–338. doi: 10.5511/plantbiotechnology.24.335
Komarnytsky, S., and Borisjuk, N. (2003). Functional analysis of promoter elements in plants. Genet. Eng. 25, 113–141. doi: 10.1007/978-1-4615-0073-5_6
Koo, A. J. K., and Ohlrogge, J. B. (2002). The predicted candidates of Arabidopsis plastid inner envelope membrane proteins and their expression profiles. Plant Physiol. 130, 823–836. doi: 10.1104/pp.008052
Kozak, M. (1981). Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 9, 5233–5252. doi: 10.1093/nar/9.20.5233
Kuno, N., and Furuya, M. (2000). Phytochrome regulation of nuclear gene expression in plants. Stem Cell Dev. Biol. 11, 485–493. doi: 10.1006/scdb.2000.0205
Kyo, M., and Harada, H. (1986). Control of the developmental pathway of tobacco pollen in vitro. Planta 168, 427–432. doi: 10.1007/BF00392260
Lagacé, M., and Matton, D. P. (2004). Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta 219, 185–189. doi: 10.1007/s00425-004-1253-2
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). Plant CARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Goodstadt, L., Dickens, N. J., Doerks, T., Schultz, J., Mott, R., et al. (2002). Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30, 242–244. doi: 10.1093/nar/30.1.242
Leymarie, J., Robayo-Romero, M., Gendreau, E., Benech-Arnold, R., and Corbineau, F. (2008). Involvement of ABA in induction of secondary dormancy in barley (Hordeum vulgare L.) seeds. Plant Cell Physiol. 49, 1830–1838. doi: 10.1093/pcp/pcn164
Lichter, R. (1982). Induction of haploid plants from isolated pollen of Brassica napus L. Z. Pflanzenphysiol. 105, 427–434. doi: 10.1016/S0044-328X(82)80040-8
Lopez-Molina, L., Mongrand, S., and Chua, N. (2001). A post-germination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 98, 4782–4787. doi: 10.1073/pnas.081594298
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Malik, M. R., Wang, F., Dirpaul, J. M., Zhou, N., Hammerlindl, J., Keller, W., et al. (2008). Isolation of an embryogenic line from non-embryogenic Brassica napus cv. Westar through microspore embryogenesis. J. Exp. Bot. 59, 2857–2873. doi: 10.1093/jxb/ern149
Malik, M. R., Wang, F., Dirpaul, J. M., Zhou, N., Polowick, P. L., Ferrie, A. M. R., et al. (2007). Transcript profiling and identification of molecular markers for early microspore embryogenesis in Brassica napus. Plant Physiol. 144, 134–154. doi: 10.1104/pp.106.092932
McCourt, P. (1999). Genetic analysis of hormone signaling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 219–243. doi: 10.1146/annurev.arplant.50.1.219
Meggio, F., and Pinna, L. A. (2003). One-thousand-and-one substrates of protein kinase CK2. FASEB J. 17, 349–368. doi: 10.1096/fj.02-0473rev
Miller, M. R. (2009). The importance of being flexible: the case of basic region leucine zipper transcriptional regulators. Curr. Protein Pept. Sci. 10, 244–269. doi: 10.2174/138920309788452164
Nag, R., Maity, M. K., and DasGupta, M. (2005). Dual DNA binding property of ABA insensitive 3 like factors targeted to promoters responsive to ABA and auxin. Plant Mol. Biol. 59, 821–838. doi: 10.1007/s11103-005-1387-z
Nagahara, U. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jap. J. Bot. 7, 389–452.
Nakano, T., Nagata, N., Kimura, T., Sekimoto, M., Kawaide, H., Murakami, S., et al. (2003). CND41, a chloroplast nucleoid protein that regulates plastid development, causes reduced gibberellin content and dwarfism in tobacco. Physiol. Plant. 117, 130–136. doi: 10.1034/j.1399-3054.2003.1170116.x
Nakashima, K., Fujita, Y., Kanamori, N., Katagiri, T., Umezawa, T., Kidokoro, S., et al. (2009). Three Arabidopsis SNRK2 protein kinases, srk2d/snrk2.2, srk2e/snrk2.6/ost1 and srk2i/snrk2.3, involved in aba signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363. doi: 10.1093/pcp/pcp083
Nambara, E., and Marion-Poll, A. (2003). ABA action and interactions in seeds. Trends Plant Sci. 8, 213–217. doi: 10.1016/S1360-1385(03)00060-8
Nitsch, C., and Nitsch, J. (1967). The induction of flowering in vitro in stem segments of Plumbago indica L. Planta 72, 355–370. doi: 10.1007/BF00390146
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of start proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Pauls, K. P. (1996). “The utility of doubled haploid populations for studying the genetic control of traits determinated by recessive alleles” in In vitro haploid production in higher plants. eds. S. M. Jain, S. K. Sopory, and R. E. Veilleux (The Netherlands: Kluwer Academic Publishers), 125–144.
Pauls, K. P., Chan, J., Woronuk, G., Schulze, D., and Brazolot, J. (2006). When microspores decide to become embryos-cellular and molecular changes. Can. J. Bot. 84, 668–678. doi: 10.1139/b06-064
Perry, S. E., Lehti, M. D., and Fernandez, D. E. (1999). The MADS-domain protein AGAMOUS-like 15 accumulates in embryonic tissues with diverse origins. Plant Physiol. 120, 121–130. doi: 10.1104/pp.120.1.121
Possingham, J., and Saurer, W. (1969). Changes in chloroplast number per cell during leaf development in spinach. Planta 86, 186–194. doi: 10.1007/BF00379826
Prem, D., Solís, M., Bárány, I., Rodríguez-Sanz, H., Risueño, M. C., and Testillano, P. S. (2012). A new microspore embryogenesis system under low temperature which mimics zygotic embryogenesis initials, expresses auxin and efficiently regenerates doubled-haploid plants in Brassica napus. BMC Plant Biol. 12:127. doi: 10.1186/1471-2229-12-127
Pyke, K. A. (1999). Plastid division and development. Plant Cell 11, 549–556. doi: 10.1105/tpc.11.4.549
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., and Grill, E. (2010). ABA perception and signaling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Reeves, W., Lynch, T., Mobin, R., and Finkelstein, R. (2011). Direct targets of the transcription factors ABA-insensitive (ABI4) and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol. Biol. 75, 347–363. doi: 10.1007/s11103-011-9733-9
Rodríguez-Gacio, M. D. C., Matilla-Vázquez, M. A., and Matilla, A. J. (2009). Seed dormancy and ABA signaling: the breakthrough goes on. Plant Signal. Behav. 4, 1035–1049. doi: 10.4161/psb.4.11.9902
Rose, R. J., Nolan, K. E., and Lakshmanan, P. (2006). Genetic regulation of somatic embryogenesis with particular reference to Arabidopsis thaliana and Medicago truncatula. In Vitro Cell. Dev. Biol. Plant 42, 473–481. doi: 10.1079/IVP2006806
Rost, B., Casadio, R., Fariselli, P., and Sander, C. (1995). Prediction of helical transmembrane segments at 95% accuracy. Protein Sci. 4, 521–533. doi: 10.1002/pro.5560040318
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. doi: 10.3389/fpls.2016.00571
Sato, N. (2001). Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci. 6, 151–155. doi: 10.1016/S1360-1385(01)01888-X
Sato, N., Albrieux, C., Joyard, J., Douce, R., and Kuroiwa, T. (1993). Detection and characterization of a plastid envelope DNA binding protein which may anchor plastid nucleoids. EMBO J. 12, 555–581.
Sato, N., Misumi, O., Joyard, J., and Douce, R. (1995). “DNA-binding proteins mediate interaction of nucleoids with envelope membrane in developing plastids” in Photosynthesis: From light to biosphere. ed. P. Mathis (Dordrecht, The Netherlands: Kluwer Academic Publishers), 635–638.
Sato, N., Ohshima, K., Watanabe, A., Ohta, N., Nishiyama, Y., Joyard, J., et al. (1998). Molecular characterization of the PEND protein, a novel bZIP protein present in the envelope membrane that is the site of nucleoid replication in developing plastids. Plant Cell 10, 859–872. doi: 10.1105/tpc.10.5.859
Schulze, D., and Pauls, K. P. (1997). Flow cytometric characterization of embryogenic and gametophytic development in Brassica napus microspore cultures. Plant Cell Physiol. 39, 226–234. doi: 10.1093/oxfordjournals.pcp.a029361
Seguí-Simarro, J. M., and Nuez, F. (2008). How microspores transform into haploid embryos: changes associated with embryogenesis induction and microspore-derived embryogenesis. Physiol. Plant. 134, 1–12. doi: 10.1111/j.1399-3054.2008.01113.x
Siemering, K. R., Golbick, R., Sever, R., and Haseloff, J. (1996). Mutations that suppress the thermos-sensitivity of green fluorescent protein. Curr. Biol. 27, 1653–1663. doi: 10.1016/s0960-9822(02)70789-6
Shariatpanahi, M. E., and Ahmadi, B. (2016). “Isolated microspore culture and its applications in plant breeding and genetics” in plant Tissue culture: Propagation, conservation and crop improvement. 2nd Edn. Vol. 22. eds. M. Anis and N. Ahmad (Singapore: Springer, 487–507.
Soriano, M., Li, H., and Boutilier, K. (2013). Microspore embryogenesis: establishment of embryo identity and pattern in culture. Plant Reprod. 26, 181–196. doi: 10.1007/s00497-013-0226-7
Soriano, M., Li, H., Jacquard, C., Angenent, G. C., Krochko, J., Offringa, R., et al. (2014). Plasticity in cell division patterns and auxin transport dependency during in vitro embryogenesis in Brassica napus. Plant Cell 26, 2568–2581. doi: 10.1105/tpc.114.126300
Takabe, T., Nishimtjra, M., and Akazawa, T. (1979). Isolation of intact chloroplasts from spinach leaf by centrifugation in gradients of the modified silica “Percoll.” Agric. Biol. Chem. 43, 2137–2142. doi: 10.1271/bbb1961.43.2137
Tejos, R. I., Mercado, A. V., and Meisel, L. A. (2010). Analysis of chlorophyll fluorescence reveals stage specific patterns of chloroplast-containing cells during Arabidopsis embryogenesis. Biol. Res. 43, 99–111. doi: 10.4067/S0716-97602010000100012
Terasawa, K., and Sato, N. (2005). Occurrence and characterization of PEND proteins in angiosperms. J. Plant Res. 118, 111–119. doi: 10.1007/s10265-005-0200-z
Terasawa, K., and Sato, N. (2009). Plastid localization of the PEND protein is mediated by a noncanonical transit peptide. FEBS J. 276, 1709–1719. doi: 10.1111/j.1742-4658.2009.06901.x
Thurling, N., and Chay, P. M. (1984). The influence of donor plant genotype and environment on production of multicellular microspores in cultured anthers of Brassica napus ssp. oleifera. Ann. Bot. 54, 681–693. doi: 10.1093/oxfordjournals.aob.a086838
Tsuwamoto, R., Fukuoka, H., and Takahata, Y. (2007). Identification and characterization of genes expressed in early embryogenesis from microspores of Brassica napus. Planta 225, 641–652. doi: 10.1007/s00425-006-0388-8
Tzafrir, I., Pena-Muralla, R., Dickerman, A., Berg, M., Rogers, R., Hutchens, S., et al. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135, 1206–1220. doi: 10.1104/pp.104.045179
Umezawa, T., Nakashima, K., Miyakawa, T., Kuromori, T., Tanokura, M., Shinozaki, K., et al. (2010). Molecular basis of the core regulatory network in aba responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839. doi: 10.1093/pcp/pcq156
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., and Leunissen, J. A. M. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, 71–74. doi: 10.1093/nar/gkm306
van Bergen, S., Kottenhagen, M. J., van der Meulen, R. M., and Wang, M. (1999). The role of abscisic acid in induction of androgenesis: a comparative study between Hordeum vulgare L-cvs. Igri and digger. J. Plant Growth Regul. 18, 135–143. doi: 10.1007/pl00007061
van der Krol, A. R., and Chua, N. H. (1991). The basic domain of plant bZIP proteins facilitates import of a reporter protein into plant nuclei. Plant Cell 7, 667–675. doi: 10.1105/tpc.3.7.667
Vettore, A. L., Yunes, J. A., Cord Neto, G., da Silva, M. J., Arruda, P., and Leite, A. (1998). The molecular and functional characterization of an Opaque2 homologue gene from Coix and a new classification of plant bZIP proteins. Plant Mol. Biol. 36, 249–263. doi: 10.1023/A:1005995806897
Vlad, F., Rubio, S., Rodrigues, A., Sirichandra, C., Belin, C., Robert, N., et al. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis (W). Plant Cell 21, 3170–3184. doi: 10.1105/tpc.109.069179
Wang, G., and Fiers, M. (2010). CLE peptide signaling during plant development. Protoplasma 240, 33–43. doi: 10.1007/s00709-009-0095-y
Wang, Y., Li, L., Ye, T., Lu, Y., Chen, X., and Wu, Y. (2013). The inhibitory effect of ABA on floral transition is mediated by ABI5 in Arabidopsis. J. Exp. Bot. 64, 675–684. doi: 10.1093/jxb/ers361
Wasternack, C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697. doi: 10.1093/aob/mcm079
Wędzony, M., Forster, B. P., Žur, I., Golemiec, E., Szechyńska-Hebda, M., Dubas, E., et al. (2009). “Progress in doubled haploid technology in higher plants” in Advances in haploid production in higher plants. eds. A. Touraev, B. P. Forster, and S. M. Jain (Dorchester: Springer), 1–34.
Whittle, C., Malik, M., Li, R., and Krochko, J. (2010). Comparative transcript analyses of the ovule, microspore, and mature pollen in Brassica napus. Plant Mol. Biol. 72, 279–299. doi: 10.1007/s11103-009-9567-x
Williams, M. E., Foster, R., and Chua, N. H. (1992). Sequences flanking the hexameric G-box core CACGTG affect the specificity of protein binding. Plant Cell 4, 485–496. doi: 10.1105/tpc.4.4.485
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718. doi: 10.1371/journal.pone.0000718
Xu, D., Duan, X., Wang, B., Hong, B., Ho, T., and Wu, R. (1996). Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic Rice. Plant Physiol. 110, 249–257. doi: 10.1104/pp.110.1.249
Xu, T. T., Ren, S. C., Song, X. F., and Liu, C. M. (2015). CLE19 expressed in the embryo regulates both cotyledon establishment and endosperm development in Arabidopsis. J. Exp. Bot. 66, 5217–5227. doi: 10.1093/jxb/erv293
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yokoyama, R., Yamamoto, H., Kondo, M., Takeda, S., Ifuku, K., Fukao, Y., et al. (2016). Grana-localized proteins, RIQ1 and RIQ2, affect the organization of light-harvesting complex II and grana stacking in Arabidopsis. Plant Cell 28, 2261–2275. doi: 10.1105/tpc.16.00296
Zhang, X., and Glaser, E. (2002). Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends Plant Sci. 7, 14–21. doi: 10.1016/S1360-1385(01)02180-X
Keywords: microspore embryogenesis, Brassica napus, Arabidopsis, transcription factor, bZIP, gene expression, chloroplast localization
Citation: Shahmir F and Pauls KP (2021) Identification, Gene Structure, and Expression of BnMicEmUP: A Gene Upregulated in Embryogenic Brassica napus Microspores. Front. Plant Sci. 11:576008. doi: 10.3389/fpls.2020.576008
Edited by:
Goetz Hensel, Heinrich Heine University Düsseldorf, GermanyReviewed by:
Palak Chaturvedi, University of Vienna, AustriaAtanas Ivanov Atanassov, Joint Genomic Center, Bulgaria
Copyright © 2021 Shahmir and Pauls. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: K. Peter Pauls, cHBhdWxzQHVvZ3VlbHBoLmNh
 Fariba Shahmir
Fariba Shahmir K. Peter Pauls
K. Peter Pauls