- 1Biotechnology Research Center, Southwest University, Chongqing, China
- 2Chongqing Key Laboratory of Plant Resource Conservation and Germplasm Innovation, Southwest University, Chongqing, China
Cotton fibers are single cells that show a relatively independent developmental process of cell differentiation, elongation, and secondary wall deposition. Auxin promotes fiber cell protrusion from the surface of the ovule. However, the role of auxin at other stages of cotton fiber development remains largely unknown. To gain a deeper insight into this aspect, we measured indoleacetic acid (IAA) content in developing fibers. Results showed an increase in IAA content at the transition stage from elongation to secondary cell wall deposition. Subsequently, we investigated the differences between two transgenic cottons that show upregulated and downregulated fiber auxin levels, respectively. In planta analysis revealed that, in addition to promoting cell elongation, auxin regulated the time of initiation of reactive oxygen species (ROS) production and secondary wall deposition in cotton fibers. This was closely correlated with the upregulated expression of GhRAC13, which regulates ROS-triggered cellulose synthesis. We found multiple putative auxin-responsive elements existed within the promoter region of GhRAC13, and IAA could induce proGhRAC13 activity. The dual-luciferase reporter assay further proved the activation of proGhRAC13 by GhARF5, an auxin-signaling activator. Altogether, our results suggest a role of auxin in promoting the onset of secondary growth by directly upregulating GhRAC13 expression in cotton fibers.
Introduction
Cellulose is a linear polysaccharide polymer that provides plants with strength and flexibility. Cellulosic biomass accounts for the largest proportion of plant cell wall components, which can be used as renewable biofuels. In particular, over 90% of the cotton fiber biomass is pure cellulose (Meinert and Delmer, 1977), making it the major source of natural fiber worldwide. Further, given their single-cell structure, cotton fibers provide an excellent system to study the mechanism and regulation of cellulose synthesis (Haigler et al., 2012).
The development of cotton fiber includes four consecutive but partially overlapping stages: initiation, elongation, secondary wall deposition, and maturation. During fiber development, cellulose is massively synthesized and deposited into the secondary wall in the third stage. At the transition stage from elongation to secondary wall deposition, which normally occurs approximately 16 days post-anthesis (DPA), strong depletion of carbohydrates is coupled with a sharp increase in cellulose synthesis (Meinert and Delmer, 1977; Zhang et al., 2012) and the secondary cell wall appears highly crystalline (Basra and Saha, 1999). Ultimately, the secondary wall reaches 8–10 μm in thickness inside the primary wall, the thickness of which is normally 0.2–0.4 μm (Wilkins and Arpat, 2005). Cellulose synthases (CesAs) are the main enzyme complexes responsible for cellulose synthesis. Among them, CesA4/7/8 plays a key role in secondary wall synthesis (Kumar et al., 2017). In cotton, celA1 (a homolog to AtCesA8) and celA2 (a homolog to AtCesA4) are highly expressed in correlation to secondary wall-associated cellulose synthesis (Pear et al., 1996; Kim and Triplett, 2007). Subsequently, other members of this protein family have been analyzed. Homologs to CesA4/7/8 are shown to have a similar expression pattern in developing fibers during secondary wall deposition (Kim and Triplett, 2007; Niu et al., 2016). In addition to CesAs, actin-associated proteins GhADF (Wang et al., 2009) and GhPFN2 (Wang et al., 2010) as well as the receptor-like kinase GhRLK (Li et al., 2005) also play important roles in secondary cell wall formation. The reactive oxygen species (ROS) hydrogen peroxide (H2O2) is a signaling molecule that controls the differentiation of secondary wall during fiber development (Potikha et al., 1999). Further, the generation of H2O2 is mediated by GhRAC13, a small GTP-binding protein preferentially expressed in developing cotton fibers from 14 to 24 DPA (Delmer et al., 1995). Consistently, ectopic expression of GhRAC13 reportedly increases H2O2 levels in soybean and Arabidopsis cells (Potikha et al., 1999).
Plant hormones are pivotal regulators involved in cotton fiber development. For example, fiber cell initiation is influenced by auxin (Zhang et al., 2011), gibberellic acid (Xiao et al., 2010), cytokinins (Zeng et al., 2012), brassinosteroids (Luo et al., 2007), and jasmonic acid (Hu et al., 2016); in turn, fiber cell elongation responds to auxin (Gokani and Thaker, 2002), ethylene (Shi et al., 2006), and gibberellic acid (Xiao et al., 2010). Finally, secondary wall deposition is seemingly influenced by abscisic acid (Yang et al., 2001). Previously, we observed auxin accumulation in initiating fiber cells, of which the accumulation is possibly mediated by delocalization of auxin efflux carrier GhPIN3a (Zhang et al., 2011, 2017; Zeng et al., 2019). Increasing level of indoleacetic acid (IAA) in the ovule epidermis increases fiber number and results in fine fibers (Zhang et al., 2011). Therefore, we hypothesized that auxin plays other roles in fiber development in addition to promoting fiber initiation. Determination of plant hormone levels indicated that the level of IAA in elongating fibers will rise again as the time for secondary cell wall deposition nears (Nayyar et al., 1989; John, 1999; Gokani and Thaker, 2002), thus suggesting a possible involvement of auxin at this point. This agrees with results of a study on auxin metabolism, which reported higher IAA catabolism during secondary cell wall deposition than during cell elongation (Jasdanwala et al., 1977). Moreover, naphthalene-1-acetic acid-induced in vivo inhibition of cellulose synthesis in cotton fibers also suggests that auxin may influence secondary growth of cotton fibers (Singh et al., 2009). However, the exact role of auxin in other fiber developmental stages is still a mystery. Therefore, in this study, we used two transgenic cotton genotypes with increased or decreased auxin level in developing fibers for investigating the role of auxin in secondary cell wall deposition.
Materials and Methods
Plant Materials and Transformation
Wild-type Gossypium hirsutum ‘Jimian 14’ and transgenic cotton genotypes SCFP:iaaM and DR5:GUS have been described previously (Zhang et al., 2011, 2017). The coding sequence of iaaL was amplified with flanking sites XbaI and EcoRI to replace iaaM for generating the SCFP:iaaL construct. Cotton transformation was performed using an Agrobacterium-mediated method previously reported (Luo et al., 2007). After selection by kanamycin and β-glucuronidase (GUS) staining, positive plantlets were transferred to a glasshouse. The homogenic lines were planted in the experimental field in the summer for further observation. Mature fibers (>10 g) were sent to the Center of Cotton Fiber Quality Inspection and Testing, Chinese Ministry of Agriculture (Anyang, Henan Province, China) for quality measurement.
Histochemical Staining and Quantification of GUS Activity
Histochemical staining was performed according to a previously described method (Zhang et al., 2017). After excision, ovules with fibers were stained in the dark at 37°C for 12 h. After staining, samples were kept in 75% ethanol for observation. Fluorometric assay of GUS activity was performed using a previously reported method (Hou et al., 2008). Fresh fibers were ground in liquid nitrogen and homogenized in GUS extracting buffer [50 mM phosphate buffer solution (PBS) (pH 7.0), 100 mg/ml PVP, and 10 mM β-met]. After incubation on ice for 1 h, the extract was centrifuged at 13,000 rpm for 10 min. The supernatant was the total protein crude extract. Protein concentration was determined by the Bradford method. GUS activity was calculated as pmol 4-MU mg–1 protein min–1, and each test was performed with three biological replicates.
Extraction and Determination of IAA
IAA extraction was performed according to the method described by Zhang et al. (2011). Approximately 200 mg fresh weight of cotton fiber was homogenized in liquid nitrogen, then extracted overnight at 4°C with 7 ml of 80% methanol containing 10 ng 13C6-IAA as an internal standard. After centrifugation for 20 min at 10,000 × g at 4°C, the supernatant was collected and evaporated under vacuum at 40°C. The residue was dissolved in 0.1 M acetic acid and applied to a pre-equilibrated Sep-Pak® Plus tC18 cartridge (Waters, United States). Retained IAA was first washed with 4 ml of 17% methanol, then eluted with 6 ml of 40% methanol, and finally dried under vacuum. Each sample was redissolved in 10% methanol, filtered through a 0.22-μm filter, and analyzed on a 4000Q TRAP LC/MS/MS system (ABsciex, United States) as previously described (Zeng et al., 2019).
Quantitative PCR Analysis
Approximately 1 g cotton fiber was used for RNA extraction with the easy-spin Plant RNA Extraction Kit (Aidlab, China). cDNA synthesis and gene expression were analyzed according to a method previously described (Zhang et al., 2017). Forward and reverse primers used are listed in Supplementary Table 1. GhHIS3 (Gohir.D03G040300) and AtACT2 (AT3G18780) were used as reference genes in cotton and Arabidopsis, respectively. Each experiment was performed at least three times showing the same expression profile consistently, whereby only one result is shown here.
Ovule Culture and Measurement of Fiber Length
Ovule culture was performed as describe before (Zhang et al., 2017). Ovules at 1 DPA were floated on the BT medium (Beasley, 1973) supplemented with IAA (at concentrations ranging from 0.5 to 50 μM) and 0.5 μM GA3. After a 2-week cultivation, ovules were observed under an MVX10 stereomicroscope (Olympus). For measurement of fiber length, ovules attached with fibers were boiled in water for 10 min. Then, fibers were combed for measurement and observation. Twenty ovules were used for each treatment.
Visualization of Crystalline Cellulose
Cotton fibers were separated at different times and mounted in distilled water. Crystalline cellulose was observed by birefringence microscopy (Potikha et al., 1999) using an IX81 inverted microscope system (Olympus).
Determination of Cellulose Content
The assay was carried out using a previously described method (Updegraff, 1969), after some modifications. Cotton fibers were ground in liquid nitrogen. Homogenized samples (∼0.2 g each) were suspended in 5 ml acetic/nitric solution (acetic acid/distilled water/nitric acid = 4:1:5) and heated in boiling water for 30 min. The extract was then centrifuged at 3,000 × g for 10 min, and the supernatant was discarded. The pellet was washed by suspending and pelleting three times in 8 ml of distilled water. Then, the residue was suspended in 5 ml 80% H2SO4 (v/v) and incubated at room temperature for 1 h. A 100-μl sample was mixed with ice-cold anthrone reagent [0.2 g anthrone dissolved in 100 ml 80% H2SO4 (v/v)]. After incubation at room temperature for 10 min, optical density at 620 nm was recorded on an infinite M200PRO (TECAN) plate reader. The concentration of cellulose was determined according to a standard curve obtained using pure cellulose.
ROS Visualization
To obtain a 10-mM stock solution, 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA AM; Sigma, United States) was dissolved in DMSO. Ovules attached to fibers were washed three times in 10 mM PBS (pH 5.2). Then, they were immersed in H2DCFDA working solution (diluted in 10 mM PBS pH 5.2 at 1:1,000) for 10 min at 25°C in darkness. After staining, samples were washed again six times with 10 mM PBS (pH 5.2). The fluorescent signal of excitation at 488 nm and the emission spectrum over 500–550 nm were observed using an IX81 inverted microscope system (Olympus).
Activity Assay of the GhRAC13 Promoter
To analyze the inducible activity of the GhRAC13 promoter, the upstream sequence of the GhRAC13 (Gohir.D10G029600) coding region was amplified from upland cotton. The promoter flanked by sites HindIII and SmaI was placed in front of the β-glucuronidase (GUS) reporter gene in vector pBI121. The construct was then introduced into Arabidopsis with the mediation of Agrobacterium (strain GV3101) by the floral dip method (Clough, 2004). For dual-luciferase reporter assay, the promoter sequence of GhRAC13 was placed upstream of LUC gene in expression vector pGreen0080 using the homologous recombination method; the expression vector pGreen0080 harbored another cassette of Renilla luciferase driven by 35S promoter. GhARF5 (Gohir.A05G187900) was amplified from cDNA of elongating cotton fibers and cloned into a modified vector pLGN under the control of 35S promoter (Zeng et al., 2019). Both constructs were transformed into Agrobacterium (strain GV3101) and transiently expressed in Nicotiana benthamiana leaves. Three days later, infiltrated leaves were used for the dual-luciferase reporter assay using Dual-Glo® Luciferase Assay System (Promega, United States).
Results
Auxin Content in Developing Cotton Fibers
To investigate a possible regulatory role of auxin during cotton fiber development stages, we analyzed IAA content in developing fibers of upland cotton (Gossypium hirsutum ‘Jimian 14’) from 10 to 22 DPA (Figure 1A), which comprised fiber development stages from elongation to early secondary wall deposition. The IAA level increased remarkably (over two-fold) from 10 to 15 DPA and thereafter remained at a relatively high level until 22 DPA. GUS activity assay in DR5:GUS (β-glucuronidase) cotton, an auxin reporter system, confirmed this result: strong GUS staining was detected in fibers (Supplementary Figure 1). In addition, quantitative analysis of GUS activity showed a considerable increase from 8 to 16 DPA (Figure 1B). Both results showed an increase in IAA content at approximately 15 DPA, suggesting a possible auxin-mediated regulation of fiber development at this stage.
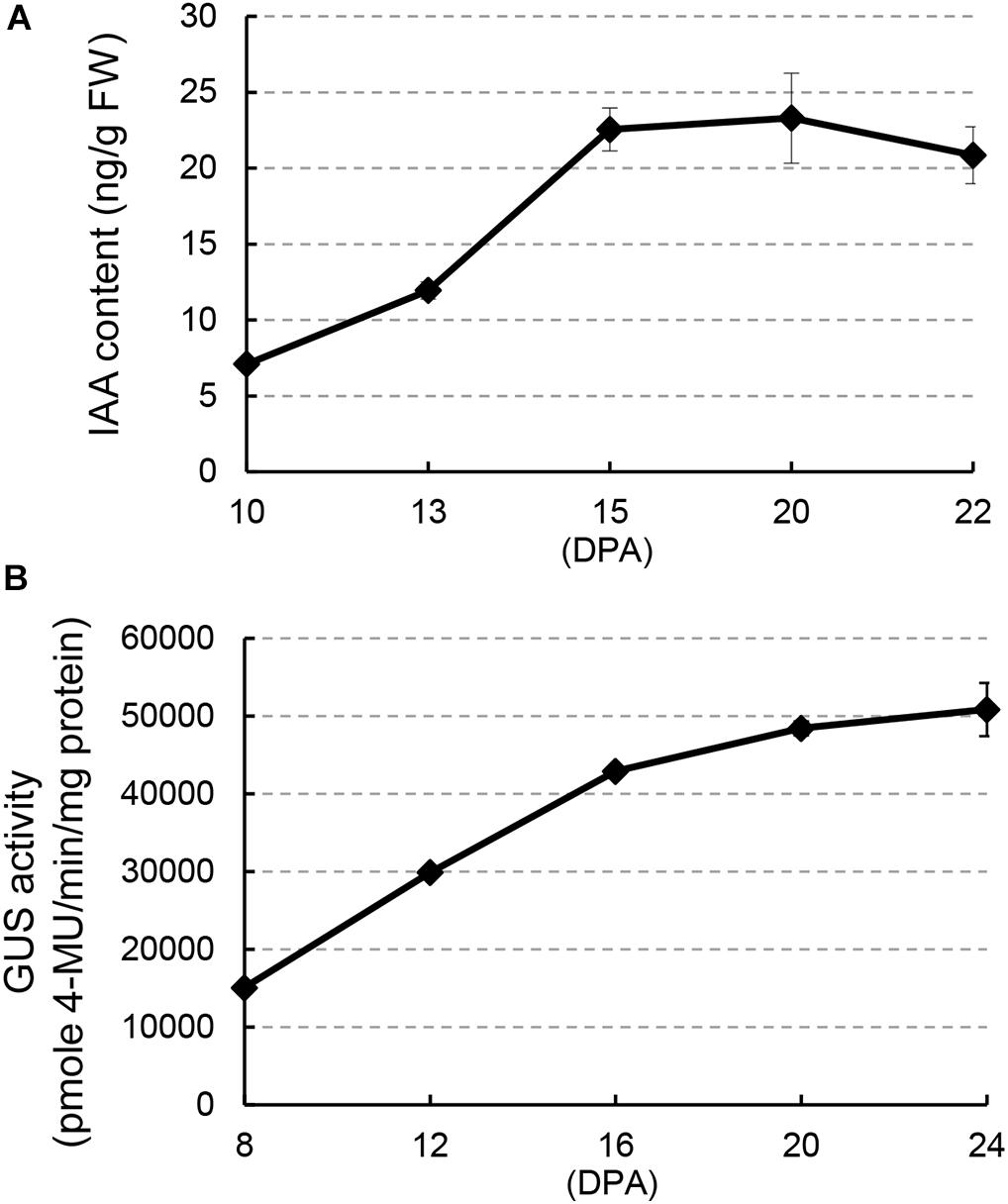
Figure 1. Auxin level increases in fiber cells during the transition between elongation and cell wall secondary growth onset. (A) Free indoleacetic acid (IAA) content in developing fibers at different days post-anthesis (DPA). (B) GUS activity in developing fibers of DR5:GUS cotton. Error bars indicate standard deviation of three biological replicates.
Changes in Endogenous IAA Content in Transgenic Cotton Fibers
We used two transgenic cottons, namely, SCFP:iaaM and SCFP:iaaL, both with modified auxin content in fibers, to test the role of auxin during early secondary wall deposition. Auxin biosynthesis-related gene iaaM and metabolism-related gene iaaL (Glass and Kosuge, 1986) were driven by a fiber-specific promoter SCFP. Two independent SCFP:iaaM lines (#26 and #27) were screened based on the transcription of iaaM in fibers at 15 DPA (Figure 2A). The transcription data showed that iaaM was highly expressed in fibers between 7 and 22 DPA (Figure 2B), resulting in a 16% and a 55% increase in IAA content in fibers of lines #26 and #27, respectively, compared with that in the wild-type (Figure 2E). Likely, we selected two lines with high expression of iaaL in fibers at 15 DPA (Figure 2C). The iaaL transcript was detected at high levels in fast elongating fibers from 4 to 15 DPA (Figure 2D). Measurements of IAA showed that fiber IAA content was 10.39 and 15.37 ng/g in lines SL8 and SL17 at 15 DPA, respectively, i.e., 72% and 58% lower than that in the wild-type (Figure 2E). As expected, all transgenic cotton lines showed modified fiber IAA content and were used for further assays.
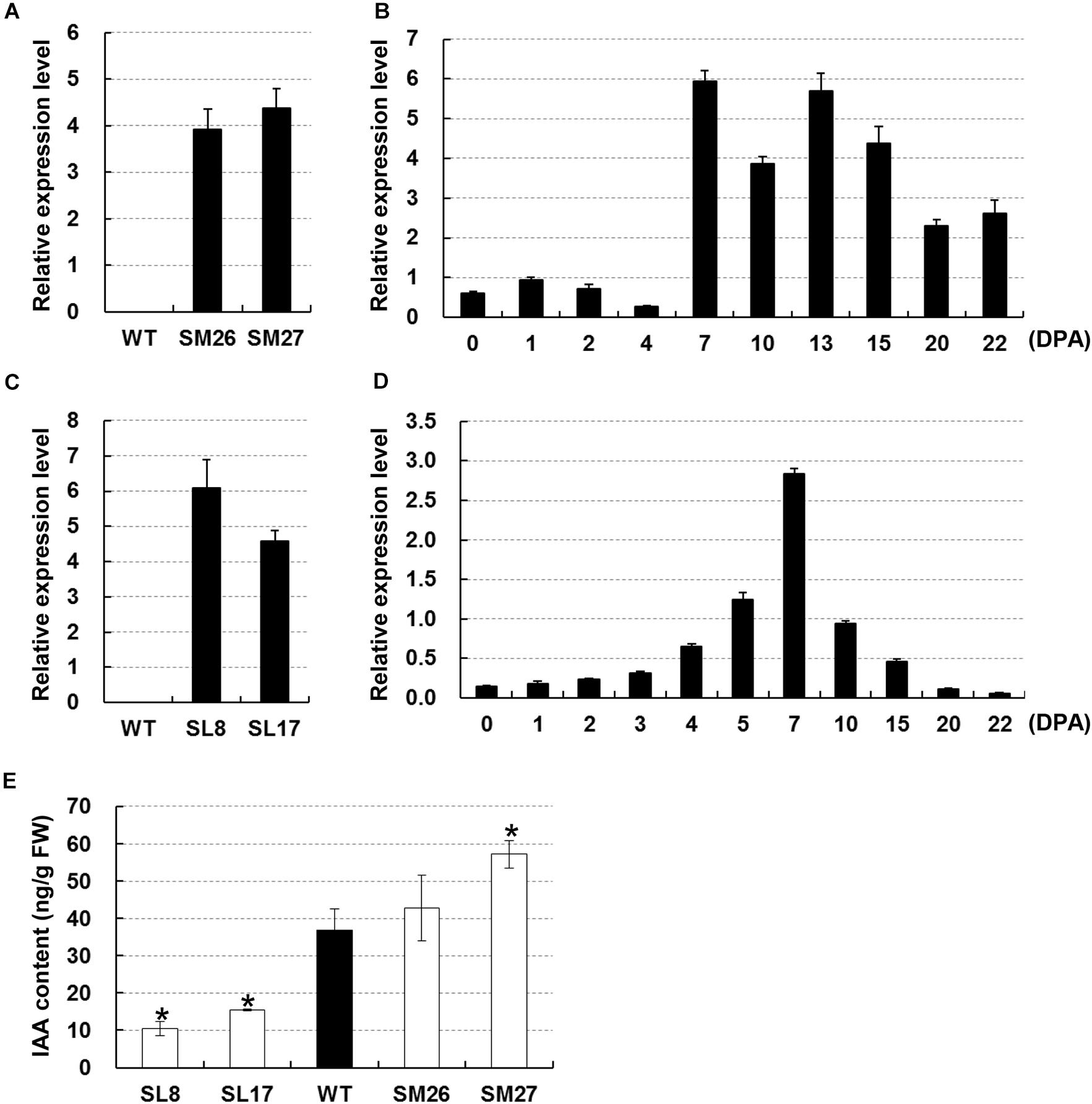
Figure 2. Manipulation of auxin level in cotton fibers. (A) Expression of iaaM in 15-DPA fibers of SCFP:iaaM cotton. (B) Expression of iaaM in developing ovules and fibers of SCFP:iaaM cotton. (C) Expression of iaaL in 15-DPA fibers of SCFP:iaaL cotton. (D) Expression of iaaL in developing ovules and fibers of SCFP:iaaL cotton. (E) Free indole acetic acid (IAA) content in wild-type and transgenic cotton fibers at 15 DPA. Error bars indicate standard deviation of three biological replicates. Wildtype in (A) and (C) was used as negative control. SM, SCFP:iaaM transgenic cotton; SL, SCFP:iaaL transgenic cotton; WT, wild-type; DPA, day post-anthesis. Asterisks indicate significant difference as determined by Student’s t-test (P < 0.05).
Expression of Development-Related Genes in Transgenic Cotton Fibers
To examine what specific event during fiber development is affected by auxin, we selected four marker genes related to elongation (GhEXP1) and secondary wall deposition (GhRAC13, GhCESA1, and GhCESA2) (Singh et al., 2009) and analyzed the corresponding level of transcription in fibers (Figure 3). Developing fibers showed a decreasing level of GhEXP1 transcript, while an increasing level of GhRAC13, GhCESA1, and GhCESA2 transcripts from 11 to 17 DPA. We thus inferred that fiber elongation slows, while secondary wall thickening begins at this time. All the four genes were remarkably upregulated in fibers expressing iaaM, particularly GhEXP1 at 11 DPA and GhRAC13, GhCESA1, and GhCESA2 after 14 DPA. This finding suggested that both fiber elongation and secondary wall deposition are promoted in iaaM-expressing fibers, in which IAA content was enhanced.
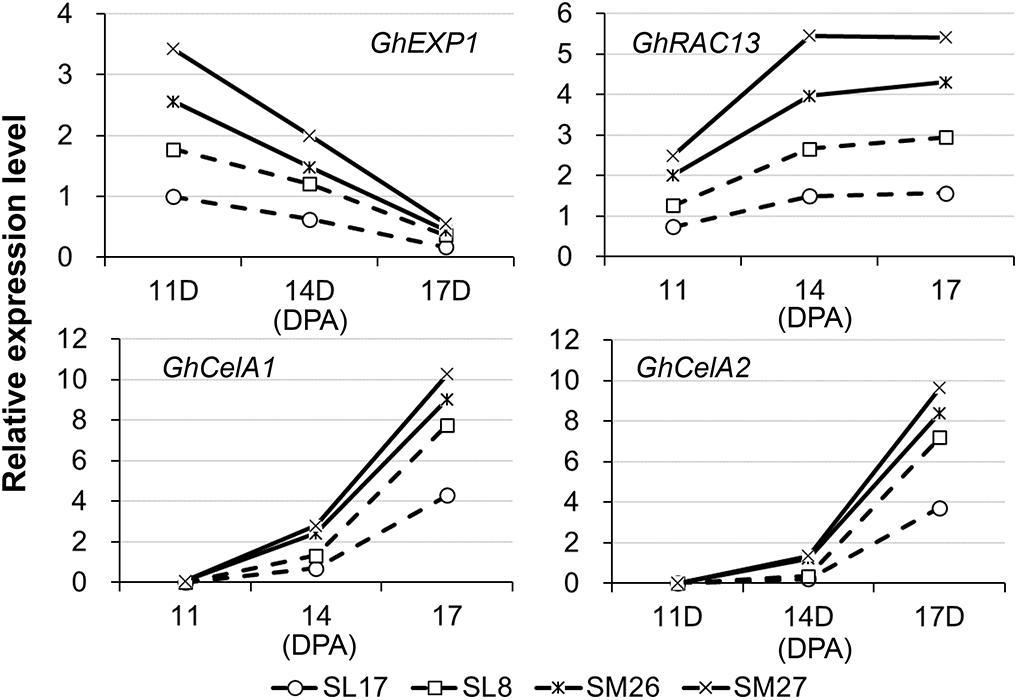
Figure 3. Expression of cell elongation- and cellulose synthesis-related genes in developing fibers. SM, SCFP:iaaM transgenic cotton; SL, SCFP:iaaL transgenic cotton; DPA, day post-anthesis.
Auxin Promotes Fiber Cell Elongation
When we measured mature fiber length to evaluate the effect of auxin on fiber elongation, we found no obvious difference in fiber length between transgenic and wild-type cotton genotypes (Supplementary Table 2). We suspected that the effect of the modified IAA level on fiber elongation might be too slight to be quantified. Therefore, we investigated the in vitro effect of IAA using cultured ovules harvested at 0 DPA, in which fiber cells already appear. Two weeks later, fiber length on cultured ovules was increased with increasing IAA supplement from 0 to 50 μM (Figures 4A,B). We then determined the length of developing fibers in transgenic cotton lines (Figure 4C). The results showed that SCFP:iaaM fibers were longer than SCFP:iaaL fibers during the elongation stage (9 to 17 DPA). However, since then, the elongation rate of fibers decreased more strikingly in the SCFP:iaaM than in the SCFP:iaaL cotton line, resulting in a similar fiber length afterward (starting at 17 DPA). These results suggested that the upregulation of IAA level could promote fiber cell elongation, but the effect might have been gradually attenuated subsequently.
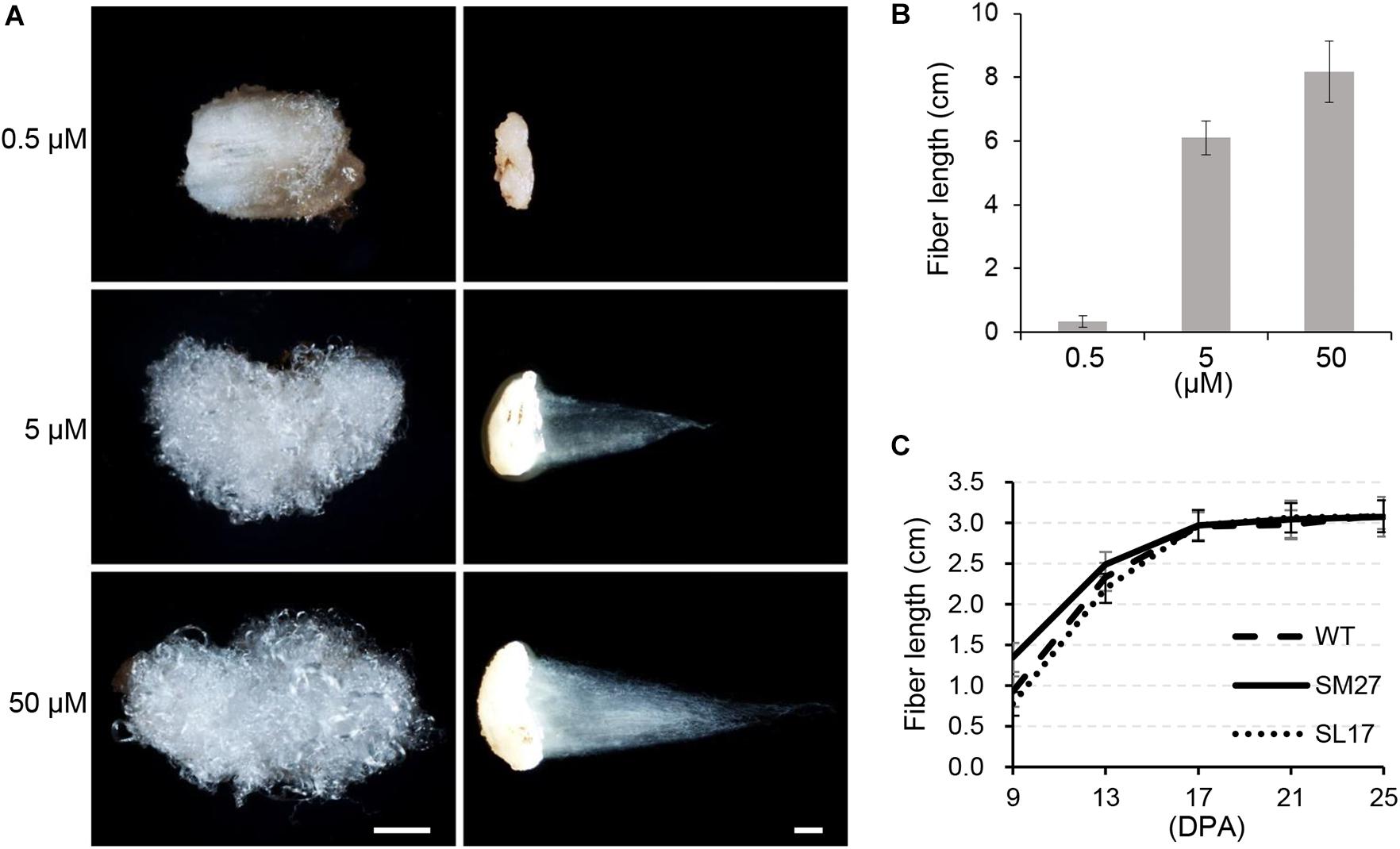
Figure 4. Auxin promotes fiber elongation. (A) Indoleacetic acid (IAA) treatment increases fiber length in vitro. Scale bar = 1 mm. (B) Fiber length under IAA treatment in vitro. (C) Fiber length of transgenic cotton. SM, SCFP:iaaM transgenic cotton; SL, SCFP:iaaL transgenic cotton. Error bars indicate standard deviation (n ≥ 10).
Auxin Activates Cellulose Synthesis for Secondary Wall Deposition
We investigated the role of auxin in secondary wall deposition through the observation of crystalline cellulose in developing fibers. Using polarization microscopy to observe birefringence of crystalline cellulose in cells, we examined cellulose synthesis in transgenic and wild-type cotton fibers (Figure 5A). The white signal of birefringence was first observed in the IAA-increased fibers (SCFP:iaaM) at 15 DPA, becoming stronger thereafter. A similar signal pattern appeared at 16 DPA in wild-type fibers, while in the IAA-decreased fibers (SCFP:iaaL), it appeared at 17 DPA. When determining fiber cellulose content at 15 DPA, we found a pronounced increase in SCFP:iaaM fibers and a decrease in SCFP:iaaL fibers relative to the wild-type (Figure 5B). In vitro culture in the presence of different IAA concentrations confirmed the stimulating effect of IAA on cellulose synthesis (Figure 5C). The change in cellulose synthesis was expected to increase fiber fineness in transgenic cotton. Surprisingly, however, micronaire values, which provide an evaluation of both fineness and maturity of cotton fibers, slightly increased in SCFP:iaaL and decreased in SCFP:iaaM fibers compared with wild-type fibers (Supplementary Table 2). Therefore, we hypothesized that auxin only activates the onset of cellulose synthesis rather than continuously enhances cellulose synthesis.
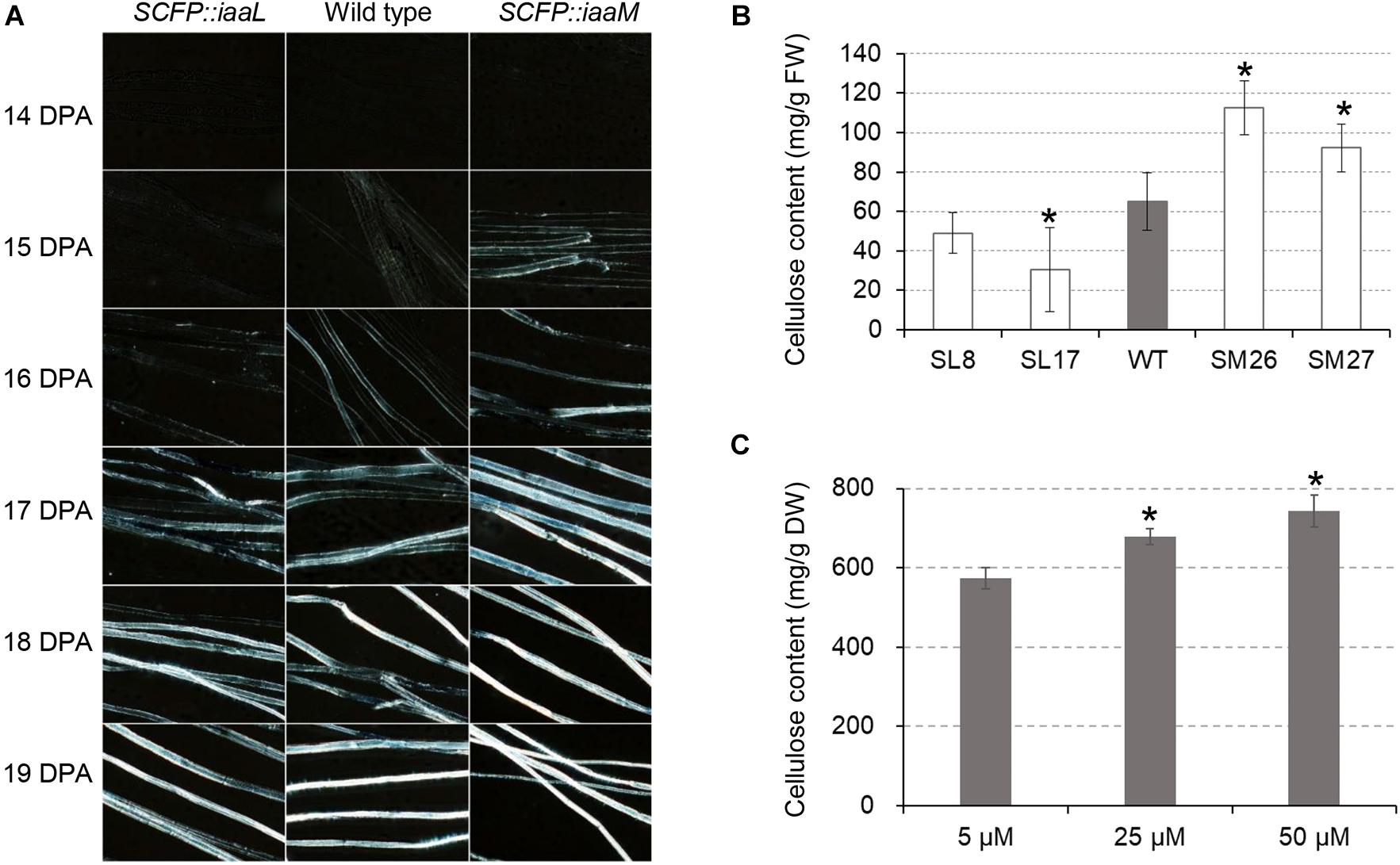
Figure 5. Auxin promotes the onset of cellulose deposition in cotton fibers. (A) Cellulose deposition in developing fibers. (B) Cellulose content in cotton fibers at 15 days post-anthesis (DPA). (C) Cellulose content in IAA-treated fibers. Ovules (0 DPA) were cultured in vitro for 30 days. WT, wild-type; SM, SCFP:iaaM transgenic cotton; SL, SCFP:iaaL transgenic cotton. Error bars indicate standard deviation based on three biological replicates. Asterisks indicate significant difference (vs. wild-type or 5 μM) as determined by Student’s t-test (P < 0.05).
Auxin Stimulates ROS Production
A previous study revealed that ROS generation is a crucial signal for triggering cellulose synthesis during secondary wall deposition of cotton fibers. Hence, we analyzed ROS production in developing fibers using fluorescent dye H2DCFDA AM (Figure 6A). A large amount of fluorescent signal indicating active ROS generation appeared first at 15 DPA in wild-type cotton fibers, remaining strong until 16 DPA and decreasing sharply after 18 DPA. This finding clearly indicated a ROS burst in developing cotton fibers from 15 to 18 DPA. In contrast, the first appearance of the ROS signal in IAA-increased fibers (SCFP:iaaM) was at 14 DPA, 1 day earlier than in the wild-type. Meanwhile in IAA-decreased fibers (SCFP:iaaL), ROS generation started at 16 DPA, 1 day later than in the wild-type. These results suggested that the ROS-signaling pathway could be activated earlier in fibers at the transition stage when intracellular IAA level was upregulated, and the pathway could be delayed when the level was downregulated.
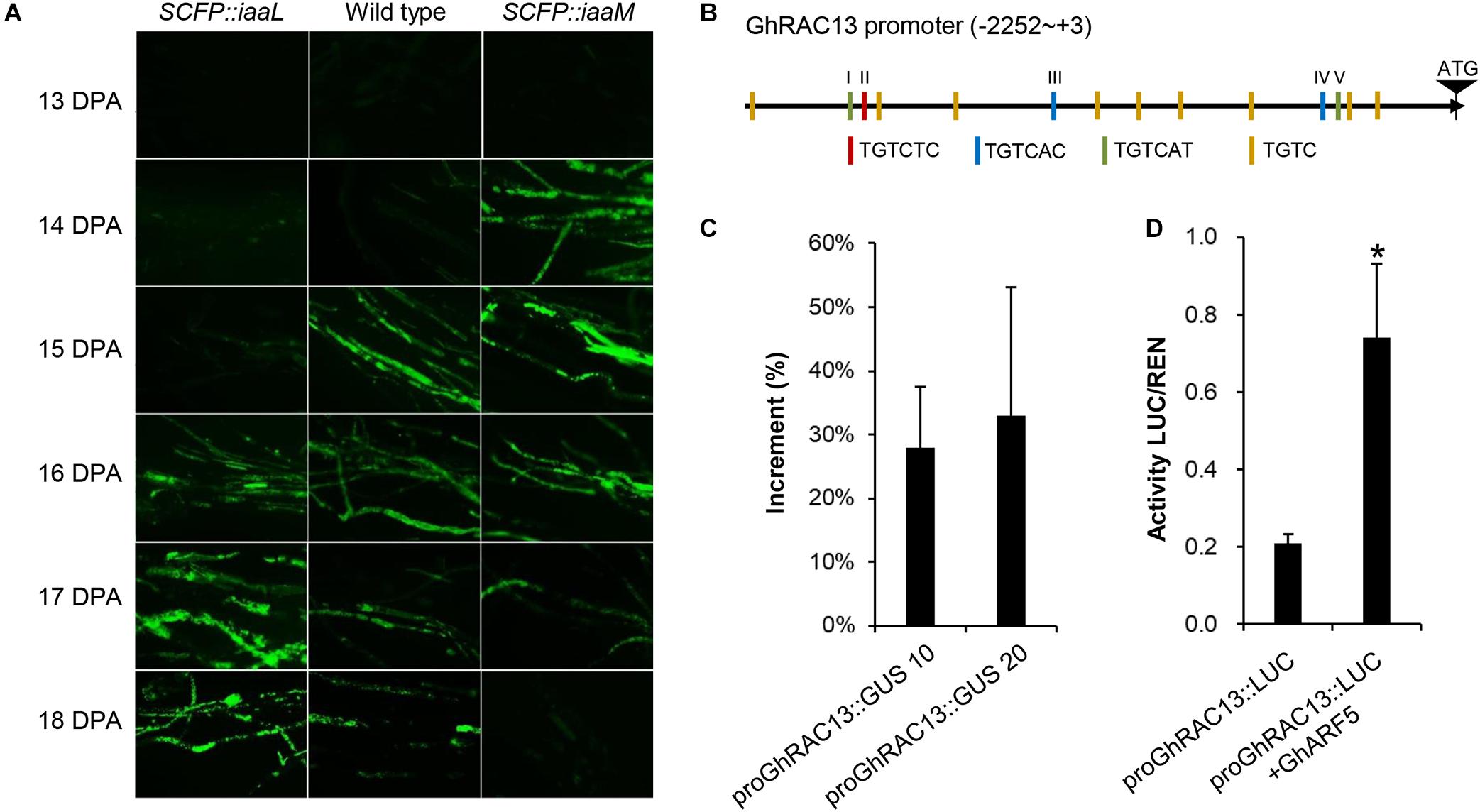
Figure 6. Auxin causes earlier occurrence of GhRAC13-mediated ROS generation in cotton fibers. (A) ROS generation occurred earlier in transgenic fibers with higher indoleacetic acid (IAA) levels. (B) Diagram of AuxREs in GhRAC13 promoter. (C) Activity of GhRAC13 promoter was upregulated by IAA treatment. (D) GhRAC13 promoter was activated under the expression of GhARF5. Error bars indicate standard deviation based on three biological replicates. Asterisks indicate significant difference as determined by Student’s t-test (P < 0.05).
GhRAC13 Expression Is Upregulated by Auxin Signaling
The earlier generation of ROS, coupled with the upregulated expression of GhRAC13 in SCFP:iaaM fibers, prompted us to verify whether IAA transcriptionally regulates GhRAC13. Thus, we amplified the ∼2.3-kb upstream regulatory region of GhRAC13 in the D subgenome. In the promoter sequence, we identified five putative auxin response elements (AuxREs) and the other nine variants containing the core motif TGTC (Figure 6B) based on the report by Sghaier et al. (2018), indicating that auxin signaling may directly regulate GhRAC13 transcription. We then fused the promoter to the GUS reporter gene and transformed it into Arabidopsis. Due to the preferential expression of GhRAC13 in fibers at the transition stage (Delmer et al., 1995), histochemical staining was not sensitive enough to detect the activity of the promoter in transgenic leaves. Alternatively, qRT-PCR was used to quantify the expression of the GUS gene. When leaves were treated with 20 μM IAA for 4 h, GUS expression was considerably upregulated by 18% to 33% (Figure 6C). We further used the dual-luciferase reporter assay to confirm this result. ARF5 is a canonical transcriptional activator in the auxin-signaling pathway (Guilfoyle and Hagen, 2007). Under co-expression with GhARF5 (Gohir.A05G187900), a homolog of ARF5 expressed in fibers at the transition stage, LUC driven by proGhRAC13 was significantly upregulated (Figure 6D). These results indicated that auxin directly regulates the expression of GhRAC13 through the auxin signaling pathway.
Discussion
Studies on the roles of auxin in cotton fiber development had been mainly limited to the stage of cell initiation (Beasley, 1973; Gialvalis and Seagull, 2001; Seagull and Giavalis, 2004; Zhang et al., 2011). In the present study, the results of in vitro experiments using transgenic cotton fibers suggest a promotive role of auxin in cell elongation (Figure 4), which is analogous to previous observations in vitro (Gokani and Thaker, 2002) and studies revealing polar growth in most cells (Cleland, 2007). Here, we clearly showed that auxin regulates the onset of secondary cell wall deposition as well. Consistent to previous studies (Nayyar et al., 1989; John, 1999; Gokani and Thaker, 2002), an increase of auxin level was found in fibers of the transition stage (Figure 1). Auxin biosynthetic pathway is not remarkably upregulated in developing fibers (Shi et al., 2006). We recently revealed that, instead of local synthesis, auxin corresponding to fiber initiation is mainly imported from the outside of ovules, and the asymmetry localization of the auxin efflux carrier GhPIN3a plays a pivotal role for the specific accumulation of IAA in fiber cells (Zhang et al., 2017; Zeng et al., 2019). The increase of auxin level was coupled with the decrease of cell expansion or elongation (Figure 4C) (Gokani and Thaker, 2002). However, this does not disagree the promotive effect of auxin on fiber elongation. Instead, the increased auxin level upregulated the expression of GhRAC13 (Figures 3, 6C), which controls ROS production in cells (Potikha et al., 1999), thereby triggering the onset of secondary growth (Potikha et al., 1999); the cell wall rigidity given by the secondary growth consequently slows down fiber elongation. Additionally, our study indicated that ROS production occurred approximately 1 day earlier than the observation of crystalline cellulose (Figures 5A, 6A). Subsequently, cellulose synthesis indicated by increased GhCelA1/2 expression was promoted (Figure 3) and much crystalline cellulose was deposited into the secondary cell wall (Figure 5A). Altogether, our data have revealed a previously unknown role of auxin in controlling the developmental transition from elongation to cellulose deposition in cotton fibers.
GhRAC13, also named GhROP7, is a homolog of small GTP-binding proteins in upland cotton. The activation of GhRAC13 reportedly increases ROS level when overexpressed in leaves (Potikha et al., 1999). In this study, we revealed that upregulation of GhRAC13 had a similar effect when induced by a high intracellular level of auxin (Figures 3, 6C). This possibly increased the amount of GhRAC13 protein to be activated. Moreover, our results further demonstrate that the expression of GhRAC13 can be activated through at least GhARF5-mediated auxin signaling pathway (Figures 6B,D). This illustrates the role of auxin–GhRAC13–ROS regulation pathway in the transition of cotton fibers from the stage of elongation to the stage of secondary wall deposition.
We found no correlation between modified cell elongation or secondary growth onset and mature fiber quality. The ultimate fiber traits agree with previous studies on transgenic cottons overexpressing auxin biosynthesis-related genes in fibers (John, 1999; Zhang et al., 2011). However, as shown herein, the decrease in elongation rate demonstrates that auxin does not determine the ultimate fiber length (Figure 4C). Moreover, earlier cellulose deposition triggered by higher auxin levels does not lead to higher, but to slightly lower, micronaire value in transgenic cotton, indicating that fibers become finer as observed for FBP7:iaaM transgenic fibers (Zhang et al., 2011). A possible explanation is that the premature deposition of secondary cell wall increases the rigidity of fiber cells and thus limits cell expansion in length and diameter (Seagull et al., 2000). This limitation is weak in developing fibers with lower IAA level, although the initial elongation rate is lower as well (Figure 4C). Consequently, no change in the length of mature fibers was observed between IAA-increased and IAA-decreased fiber. Based on these findings, it is possible to genetically modify fiber length by combination strategies; for example, by increasing auxin level in fibers at the elongation stage and simultaneously deactivating GhRAC13 in fibers at the transition stage.
Our results indicate that auxin alters the timing of ROS production in fibers at the transition stage (Figure 6A) and that ROS production will be scavenged after the activation of cellulose synthesis. It remains unclear why the relatively high level of auxin in fiber cells between 15 and 22 DPA did not result in a continuous production of ROS through the auxin–GhRAC13–ROS pathway. The comprehensive regulation via the activation of GhRAC13 by ROP guanine nucleotide exchange factors and deactivation by ROP GTPase-activating proteins should be considered in future studies to address this question. Our results suggest a direct upregulation of GhRAC13 by the GhARF5-signaling pathway (Figure 6D); however, details regarding the precise mechanisms of this regulation warrant further exploration. The effect of auxin on decreasing micronaire value of mature fibers reported here has been indicated in previous transgenic cotton expressing acsA and acsB (Li et al., 2004), two genes involved in cellulose synthesis in Acetobacter xylinum. This implies a continuous upregulation of cellulose synthesis in cotton fibers overexpressing auxin. We conclude that the cause for low micronaire value might be the same as the one that contributed to the attenuation in fiber cell elongation: the increased cell wall rigidity limits the expansion in radial direction. However, whether auxin continues to upregulate cellulose synthesis after the initial activation of cellulose via the GhRAC13–ROS pathway, as well as the underlying mechanism of auxin’s effect on fiber fineness, needs further investigation.
Altogether, our results suggest a role of auxin in the transition stage through direct upregulation of GhRAC13 expression, which in turn promotes the onset of ROS-triggered secondary growth in cotton fibers. This outlines an auxin–ROP–ROS–cellulose synthesis pathway in cell development, although details regarding this pathway need further exploration in order to improve our understanding of genetic modifications of cellulose synthesis in plants. Additionally, our study provides a basis for genetic improvement of fiber length through overexpressing auxin; however, we should develop an effective method that will prevent the inhibitory effect of auxin-induced onset of secondary wall deposition.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
YP and MZ conceived the experiments and wrote the manuscript. HC, JX, JZe, JH, and BL performed the experiments and analyzed the data. SS and JZh analyzed the agronomic trait performance. All authors reviewed and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31871676), the Ministry of Science and Technology of China (Grant No. 2013AA102601), and the Fundamental Research Funds for the Central Universities (Grant No. XDJK2020B040).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Xianlong Zhang, Huazhong Agricultural University, China, for the kind gift of the DR5:GUS cotton and Editage (www.editage.com) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.581983/full#supplementary-material
References
Basra, A. S., and Saha, S. (1999). “Growth regulation of cotton fibers,” in Cotton Fibers: Developmental Biology, Quality Improvement, and Textile Processing, ed. A. S. Basra (New York, NY: The Haworth Press), 47–63.
Beasley, C. A. (1973). Hormonal regulation of growth in unfertilized cotton ovules. Science 179, 1003–1005. doi: 10.1126/science.179.4077.1003
Cleland, R. E. (2007). “Auxin and cell elongation,” in Plant Hormones: Biosynthesis, Signal Transduction, Action!, 3rd Edn, ed. P. J. Davies (Dordrecht: Kluwer Academic), 204–220. doi: 10.1007/978-1-4020-2686-7_10
Clough, S. J. (2004). “Floral dip: Agrobacterium-mediated germ line transformation,” in Transgenic Plants: Methods and Protocols, ed. L. Peña (Totowa, NJ: Humana Press), 91–101.
Delmer, D., Pear, J., Andrawis, A., and Stalker, D. (1995). Genes encoding small GTP-binding proteins analogous to mammalian rac are preferentially expressed in developing cotton fibers. Mol. Gen. Genomics 248, 43–51. doi: 10.1007/bf02456612
Gialvalis, S., and Seagull, R. W. (2001). Plant hormones alter fiber initiation in unfertilized, cultured ovules of Gossypium hirsutum. J. Cotton Sci. 5, 252–258.
Glass, N. L., and Kosuge, T. (1986). Cloning of the gene for indoleacetic acid-lysine synthetase from Pseudomonas syringae subsp. savastanoi. J. Bacteriol. 166, 598–603. doi: 10.1128/jb.166.2.598-603.1986
Gokani, S. J., and Thaker, V. S. (2002). Physiological and biochemical changes associated with cotton fiber development: IX. role of IAA and PAA. Field Crops Res. 77, 127–136. doi: 10.1016/s0378-4290(02)00062-x
Guilfoyle, T. J., and Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460.
Haigler, C. H., Betancur, L., Stiff, M. R., and Tuttle, J. R. (2012). Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front. Plant Sci. 3:104. doi: 10.3389/fpls.2012.00104
Hou, L., Liu, H., Li, J., Yang, X., Xiao, Y., Luo, M., et al. (2008). SCFP, a novel fiber-specific promoter in cotton. Chin. Sci. Bull. 53, 2639–2645. doi: 10.1007/s11434-008-0290-9
Hu, H., He, X., Tu, L., Zhu, L., Zhu, S., Ge, Z., et al. (2016). GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J. 88, 921–935. doi: 10.1111/tpj.13273
Jasdanwala, R. T., Singh, Y. D., and Chinoy, J. J. (1977). Auxin metabolism in developing cotton hairs. J. Exp. Bot. 28, 1111–1116. doi: 10.1093/jxb/28.5.1111
John, M. E. (1999). “Genetic engineering strategies for cotton fiber modification,” in Cotton Fibers: Developmental Biology, Quality Improvement and Textile Processing, ed. A. S. Basra (New York, NY: Haworth Press), 271–292.
Kim, H. J., and Triplett, B. A. (2007). “Cellulose synthase catalytic subunit (CesA) genes associated with primary or secondary wall biosynthesis in developing cotton fibers (Gossypium hirsutum),” in Proceedings of the World Cotton Res Conference 4, Lubbock, TX.
Kumar, M., Atanassov, I., and Turner, S. (2017). Functional analysis of cellulose synthase (CESA) protein class specificity. Plant Physiol. 173, 970–983. doi: 10.1104/pp.16.01642
Li, X., Wang, X. D., Zhao, X., and Dutt, Y. (2004). Improvement of cotton fiber quality by transforming the acsA and acsB genes into Gossypium hirsutumL. by means of vacuum infiltration. Plant Cell Rep. 22, 691–697. doi: 10.1007/s00299-003-0751-1
Li, Y., Sun, J., and Xia, G. (2005). Cloning and characterization of a gene for an LRR receptor-like protein kinase associated with cotton fiber development. Mol. Gen. Genomics 273, 217–224. doi: 10.1007/s00438-005-1115-z
Luo, M., Xiao, Y., Li, X., Lu, X., Deng, W., Li, D., et al. (2007). GhDET2, a steroid 5α-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J. 51, 419–430. doi: 10.1111/j.1365-313x.2007.03144.x
Meinert, M. C., and Delmer, D. P. (1977). Changes in biochemical composition of the cell wall of the cotton fiber during development. Plant Physiol. 59, 1088–1097. doi: 10.1104/pp.59.6.1088
Nayyar, H., Kaur, K., Basra, A. S., and Malik, C. P. (1989). Hormonal regulation of cotton fibre elongation in Gossypium arboreum L. in vitro and in vivo. Biochem. Physiol. Pflanz. 185, 415–421. doi: 10.1016/s0015-3796(89)80065-4
Niu, E., Shang, X., Cheng, C., Bao, J., Zeng, Y., Cai, C., et al. (2016). Comprehensive analysis of the COBRA-Like (COBL) gene family in gossypium identifies two COBLs potentially associated with fiber quality. PLoS One 10:e0145725. doi: 10.1371/journal.pone.0145725
Pear, J. R., Kawagoe, Y., Schreckengost, W. E., Delmer, D. P., and Stalker, D. M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. U.S.A. 93, 12637–12642. doi: 10.1073/pnas.93.22.12637
Potikha, T. S., Collins, C. C., Johnson, D. I., Delmer, D. P., and Levine, A. (1999). The Involvement of hydrogen peroxide in the differentiation of secondary walls in cotton fibers. Plant Physiol. 119, 849–858. doi: 10.1104/pp.119.3.849
Seagull, R. W., and Giavalis, S. (2004). Pre- and post-anthesis application of exogenous hormones alters fiber production in Gossypium hirsutum L. cultivar Maxxa GTO. J. Cotton Sci. 8, 105–111.
Seagull, R. W., Oliveri, V., Murphy, K., Binder, A., and Kothari, S. (2000). Cotton fiber growth and development 2. Changes in cell diameter and wall birefringence. J. Cotton Sci. 4, 97–104.
Sghaier, N., Ben Ayed, R., Gorai, M., and Rebai, A. (2018). Prediction of auxin response elements based on data fusion in Arabidopsis thaliana. Mol. Biol. Rep. 45, 763–772. doi: 10.1007/s11033-018-4216-6
Shi, Y., Zhu, S., Mao, X., Feng, J., Qin, Y., Zhang, L., et al. (2006). Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18, 651–664. doi: 10.1105/tpc.105.040303
Singh, B., Cheek, H., and Haigler, C. (2009). A synthetic auxin (NAA) suppresses secondary wall cellulose synthesis and enhances elongation in cultured cotton fiber. Plant Cell Rep. 28, 1023–1032. doi: 10.1007/s00299-009-0714-2
Updegraff, D. M. (1969). Semimicro determination of cellulose inbiological materials. Anal. Biochem. 32, 420–424. doi: 10.1016/s0003-2697(69)80009-6
Wang, H. Y., Wang, J., Gao, P., Jiao, G. L., Zhao, P. M., Li, Y., et al. (2009). Down-regulation of GhADF1 gene expression affects cotton fibre properties. Plant Biotechnol. J. 7, 13–23. doi: 10.1111/j.1467-7652.2008.00367.x
Wang, J., Wang, H., Zhao, P., Han, L., Jiao, G., Zheng, Y., et al. (2010). Overexpression of a profilin (GhPFN2) promotes the progression of developmental phases in cotton fibers. Plant Cell Physiol. 51, 1276–1290. doi: 10.1093/pcp/pcq086
Wilkins, T. A., and Arpat, A. B. (2005). The cotton fiber transcriptome. Physiol. Plant. 124, 295–300. doi: 10.1111/j.1399-3054.2005.00514.x
Xiao, Y., Li, D., Yin, M., Li, X., Zhang, M., Wang, Y., et al. (2010). Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J. Plant Physiol. 167, 829–837. doi: 10.1016/j.jplph.2010.01.003
Yang, Y., Xu, C., Wang, B., and Jia, J. (2001). Effects of plant growth regulators on secondary wall thickening of cotton fibres. Plant Growth Regul. 35, 233–237.
Zeng, J., Zhang, M., Hou, L., Bai, W., Yan, X., Hou, N., et al. (2019). Cytokinin inhibits cotton fiber initiation by disrupting PIN3a-mediated asymmetric accumulation of auxin in the ovule epidermis. J. Exp. Bot. 70, 3139–3151. doi: 10.1093/jxb/erz162
Zeng, Q., Qin, S., Song, S., Zhang, M., Xiao, Y., Luo, M., et al. (2012). Molecular cloning and characterization of a cytokinin dehydrogenase gene from upland cotton (Gossypium hirsutum L.). Plant Mol. Biol. Report. 30, 1–9. doi: 10.1007/s11105-011-0308-3
Zhang, M., Song, X., Sun, X., Wang, Z., Li, Z., Ji, H., et al. (2012). The relationship between cellulose content and the contents of sugars and minerals during fiber development in colored cotton cultivars. Cellulose 19, 2003–2014. doi: 10.1007/s10570-012-9776-3
Zhang, M., Zeng, J., Long, H., Xiao, Y., Yan, X., and Pei, Y. (2017). Auxin regulates cotton fiber initiation via GhPIN-mediated auxin transport. Plant Cell Physiol. 58, 385–397.
Keywords: auxin, cellulose deposition, cotton fibers, transition, GhRAC13
Citation: Zhang M, Cao H, Xi J, Zeng J, Huang J, Li B, Song S, Zhao J and Pei Y (2020) Auxin Directly Upregulates GhRAC13 Expression to Promote the Onset of Secondary Cell Wall Deposition in Cotton Fibers. Front. Plant Sci. 11:581983. doi: 10.3389/fpls.2020.581983
Received: 10 July 2020; Accepted: 12 October 2020;
Published: 05 November 2020.
Edited by:
Laigeng Li, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Sibu Simon, Central European Institute of Technology (CEITEC), CzechiaChengbin Xiang, University of Science and Technology of China, China
Copyright © 2020 Zhang, Cao, Xi, Zeng, Huang, Li, Song, Zhao and Pei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Pei, cGVpeWFuM0Bzd3UuZWR1LmNu
 Mi Zhang
Mi Zhang Huizhen Cao1
Huizhen Cao1 Yan Pei
Yan Pei