- 1ICAR-Indian Institute of Pulses Research, Kanpur, India
- 2Bidhan Chandra Krishi Viswavidyalaya, Mohanpur, India
- 3International Center for Agricultural Research in the Dry Areas (ICARDA), Rabat Office, Rabat, Morocco
Food legumes are important for defeating malnutrition and sustaining agri-food systems globally. Breeding efforts in legume crops have been largely confined to the exploitation of genetic variation available within the primary genepool, resulting in narrow genetic base. Introgression as a breeding scheme has been remarkably successful for an array of inheritance and molecular studies in food legumes. Crop wild relatives (CWRs), landraces, and exotic germplasm offer great potential for introgression of novel variation not only to widen the genetic base of the elite genepool for continuous incremental gains over breeding cycles but also to discover the cryptic genetic variation hitherto unexpressed. CWRs also harbor positive quantitative trait loci (QTLs) for improving agronomic traits. However, for transferring polygenic traits, “specialized population concept” has been advocated for transferring QTLs from CWR into elite backgrounds. Recently, introgression breeding has been successful in developing improved cultivars in chickpea (Cicer arietinum), pigeonpea (Cajanus cajan), peanut (Arachis hypogaea), lentil (Lens culinaris), mungbean (Vigna radiata), urdbean (Vigna mungo), and common bean (Phaseolus vulgaris). Successful examples indicated that the usable genetic variation could be exploited by unleashing new gene recombination and hidden variability even in late filial generations. In mungbean alone, distant hybridization has been deployed to develop seven improved commercial cultivars, whereas in urdbean, three such cultivars have been reported. Similarly, in chickpea, three superior cultivars have been developed from crosses between C. arietinum and Cicer reticulatum. Pigeonpea has benefited the most where different cytoplasmic male sterility genes have been transferred from CWRs, whereas a number of disease-resistant germplasm have also been developed in Phaseolus. As vertical gene transfer has resulted in most of the useful gene introgressions of practical importance in food legumes, the horizontal gene transfer through transgenic technology, somatic hybridization, and, more recently, intragenesis also offer promise. The gains through introgression breeding are significant and underline the need of bringing it in the purview of mainstream breeding while deploying tools and techniques to increase the recombination rate in wide crosses and reduce the linkage drag. The resurgence of interest in introgression breeding needs to be capitalized for development of commercial food legume cultivars.
Introduction
Deployment of plant breeding tools has been successful for bolstering crop productivity, harmonizing crop phenology, enhancing nutritional quality, and developing resistance to multiple stresses. This became possible with identification of new combinations of genes and construction of superior populations possessing desirable novel characteristics, which have been exploited for human welfare (Anderson, 1949; Arnold, 1992). Although there are numerous examples for purposeful introgression of advantageous traits into crop varieties as a part of regular plant breeding programs, the extent and impact of either natural or farmer-aided introgression are yet to be ascertained (Jarvis and Hodgkin, 1998, 1999). With almost 20,000 species, legumes are the members of the Fabaceae/Leguminosae, the third largest family of the higher plants, which are ubiquitously present all over the temperate and tropical parts of the world (Polhill and Raven, 1981). Food legumes are important to human and animal life and occupy an important place in the global food supply chain, as well as sustainable agricultural production systems. With high protein content and 15 essential minerals, these are indispensable constituents of the cereal-based vegetarian diets and are grown traditionally with cereals, oilseeds, sugarcane, etc. Food legumes have prominent biological features and an inherent capability to fix atmospheric nitrogen owing to the presence of symbiotic association with Rhizobium bacteria in root nodules. Therefore, these crops become an indispensable part of the sustainable agricultsure strategy throughout the world (Chaturvedi et al., 2011).
A quest is on for the search of genes that can impart resistance to biotic and abiotic stresses in different food legumes, as well as to improve the physical and nutritional qualities of grains. The ever-changing climatic conditions led to emergence of new insect-pests and diseases and their biotypes and races, which are becoming a major threat limiting crop production and productivity (Chakraborty and Newton, 2011; Gautam et al., 2013). Broadening the genetic base will provide the needed armor to legume crops against these emerging challenges under climate change. The crop wild relatives (CWRs) are known to possess useful alien alleles and cryptic genetic variation, which are introgressed and expressed in cultivated genepool only when a systematic breeding scheme is put in place (Doyle, 1988; Tanksley and McCouch, 1997; Gupta and Singh, 2009; Pratap and Gupta, 2009). Recent advances in breeding and genomic tools and techniques provide an opportunity to introgress useful alleles left behind in the secondary and tertiary genepool into the elite background useful for legumes breeders. This review illustrates factors affecting wild gene introgression, population development, and success resorting wild gene introgression in cultivated food legumes.
The alien gene transfer in a crop species is paramount when the breeding value of the parental genepool no longer responds to selection, resulting in slow or no genetic gain. Conventionally, it is easier to manipulate desirable genes present within a crop species compared to the alien genes from distant relatives or exotic germplasm. This is because the gene transfer within a species is comparatively easy as there are no crossing barriers, and also it is largely free from linkage drags of unwanted traits. In food legumes, ∼3,700 improved varieties with narrow genetic base form the present varietal portfolio (Kumar et al., 2020), resulting in the genetic uniformity in farmers’ fields. These varieties have been developed by the repeated use of a handful of elite germplasm from the primary genepool and therefore resulted in narrow genetic base and limited genetic buffers (Kumar et al., 2004). Introgression of alien genes from CWR offers a viable option to diversify and widen the genetic base of legume varieties, which provide insulation against the vagaries, as well as scope for continuous genetic gains over many breeding cycles (Kumar et al., 2009).
The horizontal gene transfer from wild species and even across different genera has played a significant role in the evolution of eukaryotic genomes (Bock, 2009) as wild species have evolved through different degrees of selection pressure exerted by environmental forces and biotic agents over a long period of time. As a result, these species have acquired many useful genes/alleles imparting adaptation to environmental cues such as extreme temperature, drought, waterlogging, salinity, and mineral toxicity, as well as biotic factors such as diseases, insect-pests, parasitic weeds, etc. Thus, hybridizing wild species with elite germplasm following a proper breeding scheme offers scope for the generation of multitude of pre-bred lines with novel recombination, which can further be utilized in the mainstream breeding for continuous accelerated genetic gains.
Genetic Bottlenecks and Germplasm Redundancy
During evolution and domestication, wild progenitors have graduated to the cultivated forms passing through various genetic modifications and acquiring a combination of traits referred as “domestication syndrome.” Nevertheless, the persistence of these species in nature for a long time, largely remaining unattended, might have led to disappearance of many genes/alleles responsible for input response and higher grain yield in legume crops (Jain, 1975). Further, only limited samples of the accessions representing the narrow genetic base of the total diversity might have been brought to the center of domestication leading to the “founder effect” (Ladizinsky, 1985). The history of food legumes matches with human civilization while their evolution took place throughout many different regions of the world (Pratap and Kumar, 2011). However, keeping in view that many food legumes now have their major production base away from the actual center of diversity and also that during their domestication limited sampling might have narrowed down their genetic base, these crops might have started their domestication journey with the “founder effect.” For example, limited genetic diversity is reported in soybean outside its center of origin (Shoemaker, 1986; Pratap et al., 2012). Likewise, Phaseolus, chickpea, lentil, and pigeonpea also witnessed this bottleneck during their domestication.
While large germplasm repository of food legumes is preserved in different genebanks across the globe, mining of genetic diversity for use in mainstream breeding remains limited because of the paucity of information on economic traits and the nature of diversity itself (Kumar et al., 2007). This becomes much more alarming when we consider the use of exotic and unadapted germplasm in breeding programs. Further, the large size of germplasm collection, breeders’ preference for elite × elite crosses due to obvious advantages of their adaptability to local conditions, presence of cryptic genetic variation, and the linkage drag associated with transferring genes from wild relatives are other factors associated with restricted use of germplasm (Sharma et al., 2016).
Linkage drag is one of the major apprehensions while utilizing exotic and wild species in genetic amelioration of food legumes. In most of the cases, undesirable linkages hinder the transfer of desirable traits into cultivated backgrounds, and breaking such linkages needs dedicated efforts with a larger population and an efficient selection pressure. To overcome the problem of linkage drag, an additional generation of crossing among progenies prior to the selection or recurrent selection program over several generations is recommended. Nonetheless, it is now possible to recover or transfer into the elite germplasm the favorable alleles that were inadvertently left behind during the process of domestication. This can be done more efficiently by deploying molecular maps and integrative quantitative trait locus (QTL) analysis (for details, see Chamarthi et al., 2011) either through constructing introgression libraries that are made up of several introgression lines (ILs) or utilizing advanced-backcross QTL (AB-QTL) analysis. Introgression libraries can be constructed by crossing cultivated parent with wild donor followed by three to four times backcrossing of F1 with cultivated parent (Kumar et al., 2011). In the past, attempts have been made to develop such libraries in soybean using Glycine soja, a wild species (Concibido et al., 2003), and from synthetic tetraploids in peanut (Fonceka et al., 2012). The AB-QTL approach also deploys repeated backcrossing involving elite parent and wild accession with an aim to reduce the number and size of the donor segment transferred through alien introgression. The ultimate objective here is to minimize the effect of linkage drag in such crosses, and advanced backcrossed populations thus derived are further subjected to QTL analysis to identify desirable genes/QTL. Common bean and soybean are the best examples where this approach has been used successfully (Blair et al., 2003; Chaky et al., 2003).
Breeding Populations for Gene Introgression
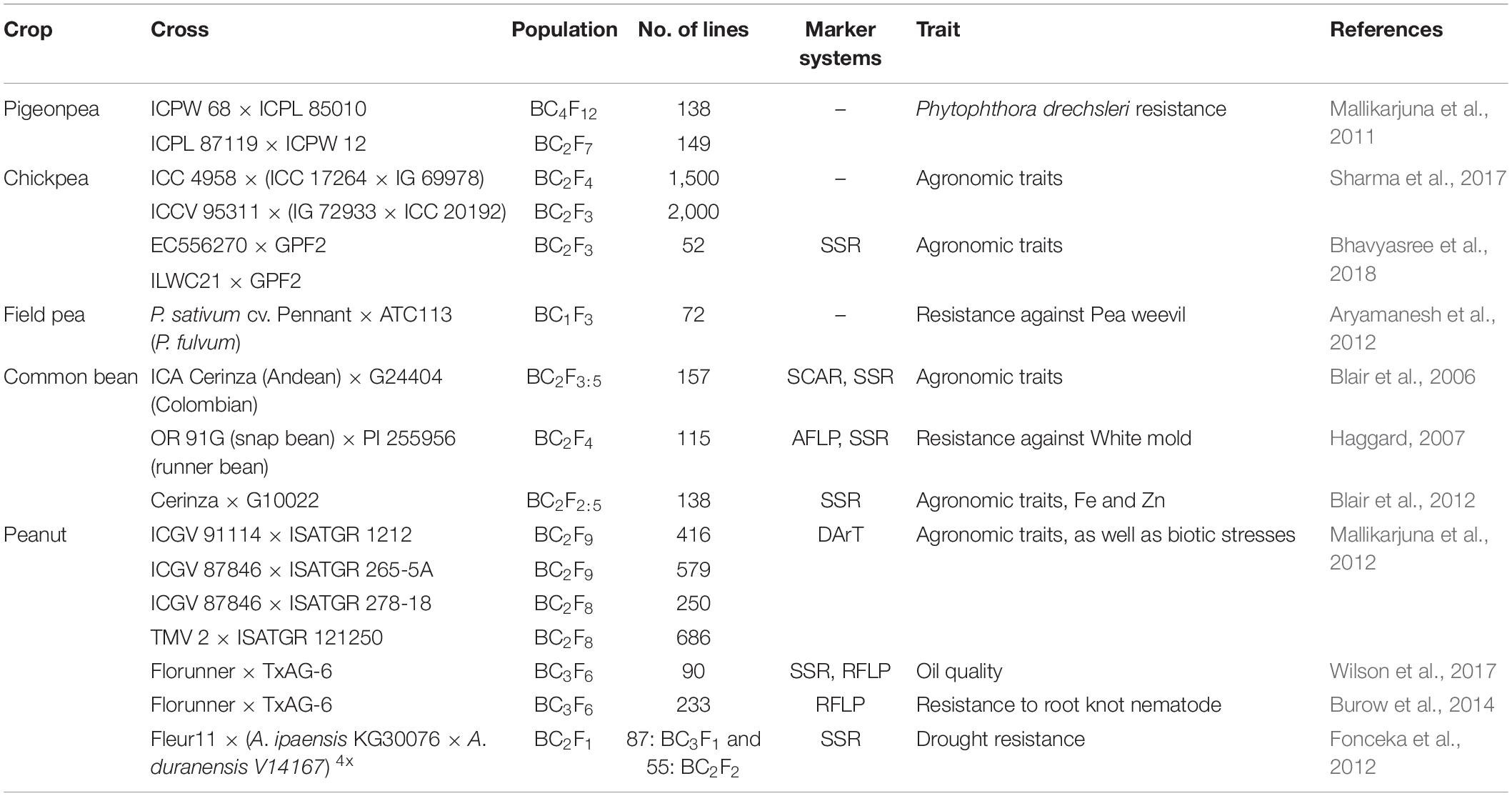
Crop wild relatives are valuable source of novel and cryptic variation for broadening the genetic base of cultivated genepool (Dwivedi et al., 2005; Pratap et al., 2014). CWRs also harbor superior QTLs for improving agronomic and yield attributing traits. However, currently available approaches for introgression are not suitable for polygenic traits because of selection bias against the alien alleles. Moreover, penetrance and expressivity of alien genes and traits when introgressed in the cultivated background are often incomplete and limited, resulting in poor genetic gains. As a result, breeding for introgression of QTLs from CWR to elite background is avoided, and emphasis has been laid upon transfer of oligogenic traits governing stress resistance mostly. Nevertheless, with the advancement in genomic tools and techniques, it has become feasible to identify and target selection for major QTLs from CWRs. For QTL analysis, mostly balanced populations (F2, BC1) have been utilized previously where alleles of both wild and elite populations are available in the same frequency, although these populations are easy to develop but are characterized by several drawbacks. Balanced populations have the most complete genetic construction and only allow for analyzing both dominant and additive effects (Wang and Chee, 2010). These populations are temporary and highly heterozygous; thus, it is difficult to use them in replicated yield trials because in every generation of either selfing or backcrossing, the genetic constitution of these populations would change. Moreover, undesirable QTLs from the unadapted wild background could lead to the linkage drag. Further, during the transfer of QTLs, epistatic component augments the complication because it is difficult to detect through statistical inference, often sensitive to environments, is difficult to manipulate, and is likely to be present in balanced populations (Bernardo, 2010). To overcome these difficulties, “specialized population concept” has been advocated for transferring QTLs from CWR into elite backgrounds. For details, please see Figure 1 The breeding population developed through different methods of gene introgression has been described in Table 1.
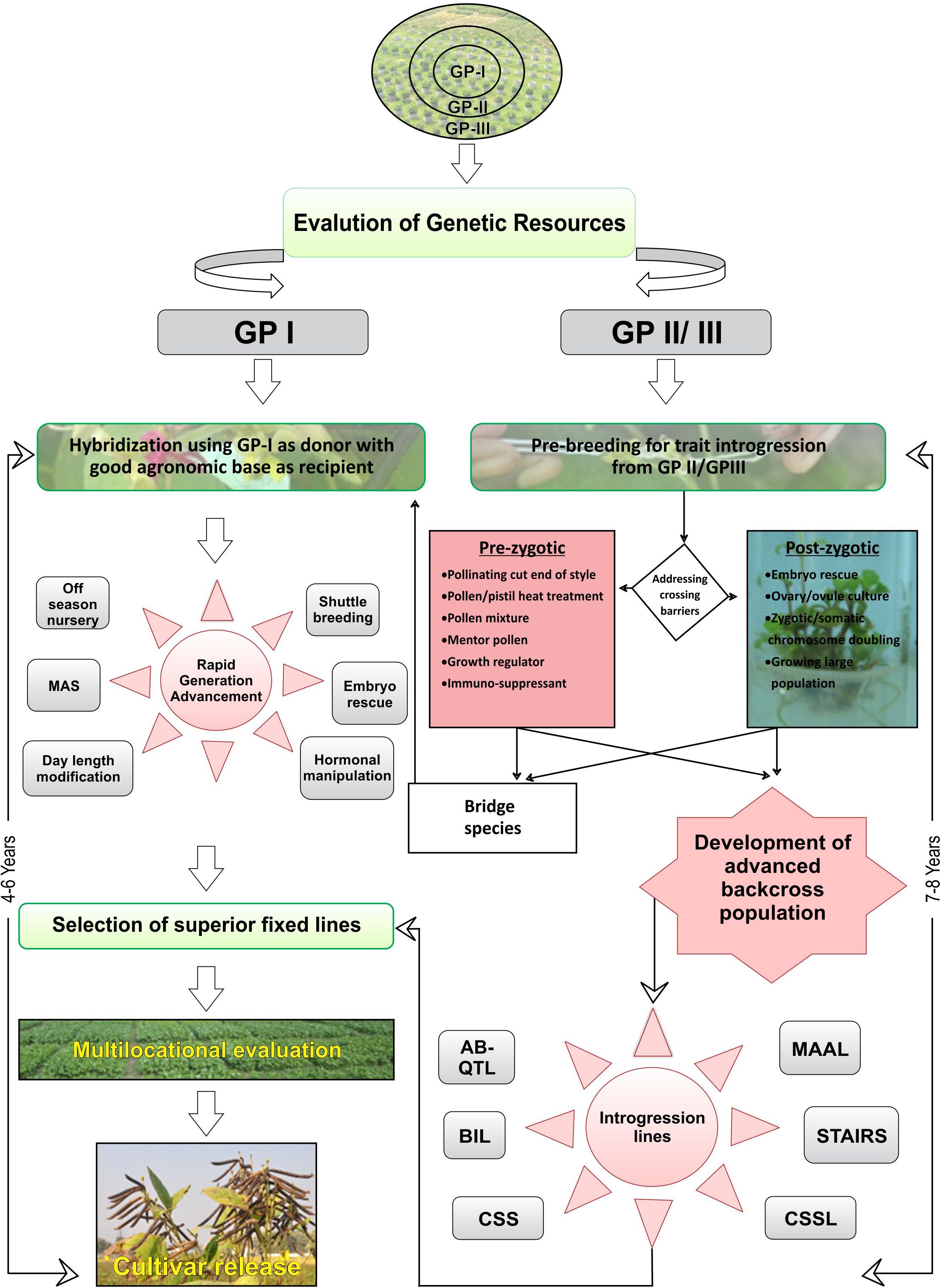
Figure 1. Scheme for gene introgression for improvement of food legumes. Left, Hybridization using GP-I as donor with good agronomic base as recipient. Right, Pre-breeding for trait introgression using GP-II and/or GP-III.
Advanced-Backcross QTL
Advanced-backcross QTL strategy was proposed by Tanksley and Nelson (1996) for concurrently mining and transferring positive QTLs from CWR into elite genepool. It is a kind of inbred backcrossing for transferring complex trait from unadapted genepool to the elite background (Sullivan and Bliss, 1983). In this methodology, QTL analysis is deferred until advanced (BC2 or BC3 and so on) generations. This is because in early generation the effects of beneficial QTLs often remain unrecognized because of the presence of epistatic interactions between favorable QTLs and other genes from the donor parent, which might be resolved in later generations, thus allowing possible silencing of the measured QTL effects (Pillen et al., 2003). The common segregating populations (F2, BC1, RIL, and DH) generally utilized for QTL analysis have some major drawbacks when involving wild species in introgression program. First, these populations represent a large segment of genes from wild parent, and the QTLs with small effects remain unseen. Second, with these populations, the discovery and further introgression of QTLs through subsequent backcrossing or intercrossing becomes a two-step process, thus becoming a time-consuming affair with mere chance of utilizing QTL information to develop superior cultivar (Tanksley and Nelson, 1996). In AB-QTL strategy, the discovery and further transfer of positive QTLs from unadapted background to elite pool are a single-step process where QTL analysis is performed in later generations to facilitate sound statistical power for detection of QTLs with small effect.
The application of AB-QTL strategy has been tested in several food legumes and recommended as a useful crop breeding venture. An attempt was made to transfer QTLs conferring yield attributing traits from two accessions of Cicer reticulatum, “EC556270” and “ILWC21” into Cicer arietinum cv. GPF2 by attempting two cross combinations. The respective BC2F3 population was tested for yield attributing traits for confirmation of introgression of productivity traits into elite chickpea cultivar (Bhavyasree et al., 2018). Field pea weevil, Bruchus pisorum, is a severe menace in cultivated field pea (Pisum sativum). An attempt was made to develop AB population (BC2F6) by involving Pisum fulvum accession “ATC113” as a resistant donor and susceptible P. sativum cv. Pennant as recipient (Aryamanesh et al., 2012). Wild beans are very diverse and useful source for enriching genetic variation of cultivated beans with low diversity. AB population (BC2F3:5) was developed in common bean involving a cross between large red-seeded commercial Columbian variety, “ICA Cerinza” as recurrent parent and wild accession “G24404” for detecting QTL toward improvement of agronomic performance. This strategy uncovered 13 QTLs for plant height, yield, and yield-attributing traits along with detection of a QTL for seed size from the wild parent (Blair et al., 2006). Another attempt has been made in common bean to transfer resistance against white mold, caused by Sclerotinia sclerotiorum. AB-QTL strategy has been undertaken to identify and transfer QTL conferring resistance to white mold into an interspecific cross of Phaseolus vulgaris cv. OR91G and Phaseolus coccineus cv. PI255956. A population of 115 BC2F4 lines were developed and genotyped using amplified fragment length polymorphism (AFLP) and simple sequence repeat (SSR) markers and screened under greenhouse for phenotypic scoring (Haggard, 2007). Wild common beans accumulate high minerals but are not commercially acceptable because of small seed size. AB population was developed for improving the mineral status of the Andean variety “Cerinza,” a large red seeded bush bean cultivar with wild genotype “G10022.” The BC2F3:5 ILs derived from this cross combination were subjected to multilocation yield trial for contemplating the role of genotype × environment interaction toward the expression of Fe and Zn content in AB population (Blair et al., 2012). The result from the study confirmed that the AB-QTL method was effective for identifying the QTL controlling Fe and Zn content, as well as their transfer into elite background and further evaluation.
Therefore, it is evident that the AB-QTL approach has been successfully applied in numerous legume species for harnessing the favorable alleles from wild into elite background, although in many other crops, viz., mungbean, urdbean, Lathyrus, and lentil, this approach is yet to be employed to explore its advantages. This strategy has paved the way for identifying more QTLs, precise measurement of the effect of individual loci, and their transfer into the cultivated background. Additionally, the formation of AB-NIL would facilitate further genetic dissection of QTLs and subsequently the map-based cloning of the underlying genes, thus opening a new vista for other legumes also.
Introgression Lines
Introgression lines are specialized populations derived through advanced backcrossing, which are nearly isogenic to recurrent parent and contain only a small fraction from donor parent (Eshed and Zamir, 1994; Tian et al., 2006). These populations are much more efficient for QTL identification and fine mapping followed by studying QTL × environment interaction due to their homozygous nature as compared to the conventional populations (Wehrhahn and Allard, 1965; Eshed and Zamir, 1995; Yin et al., 2016). The major drawback of ILs is the time taken for their development (Tanksley et al., 1989; Hospital et al., 1992; Hospital and Charcosset, 1997) and high cost for marker evaluation (Ali et al., 2010). However, with the availability of densely saturated marker systems in some legumes such as chickpea, pigeonpea, soybean, peanut, etc., the foreground as well as background selection become easier. In addition, marker-based selection facilitates detection of non-target introgression in early generation, as well as further elimination from the recipient background for speeding up IL recovery (Frisch and Melchinger, 2005).
The critical factors for reducing the problem of linkage drag in backcross population through marker-assisted selection are the tightness of the linkage between the introgressed genes and the flanking markers and the size of the population, as well as the total duration of the backcross scheme (Hospital, 2001) besides the size of the segment to be transferred. Theoretical explanation given by Hospital (2001) nicely pointed out that presence of functional markers within the genes to be introgressed or tightly linked flanking markers along with three to five generations of backcrossing would be cost-effective to minimize the length of the donor segment. In peanut, pyramiding of nematode resistance and the trait governing high oleic:linoleic acid has been introgressed successfully to develop improved Tifguard variety “Tifguard High O/L” through tightly linked cleaved amplified polymorphic sequences and SSR markers with less linkage drag problem (Chu et al., 2011). Another high oleic acid line was developed in peanut through marker-assisted introgression of two FAD2 mutant alleles conferring high oleic acid from donor parent “SunOleic95R” into the background of “ICGV 06100” (Bera et al., 2019). In chickpea, two Fusarium wilt (FW) resistance ILs, namely, “Annigeri 1” and improved “JG 74,” have been developed through marker-assisted backcrossing using “WR 315” as the donor parent (Mannur et al., 2019). Foreground selection was done with TA59, TA96, TR19, TA27, and GA16 markers, whereas background selection was done using SSR markers. Likewise, Pratap et al. (2017) developed improved “Pusa 256” using “Vijay” as the donor parent using TA 37 and TA 110 as the markers for foreground selection. Two parallel marker-assisted introgression programs have been implemented to improve both FW, as well as Ascochyta blight (AB) resistance of “C 24” cultivar by introgressing resistant locus of race 1 of FW coupled with two QTL clusters for AB resistance (Varshney et al., 2014).
Besides conventional marker-assisted introgression approaches deployed in food legumes, various IL-based strategies have been recommended like backcross inbred lines (BILs), chromosome segment substitution lines (CSSLs), stepped aligned inbred recombinant strain (STAIRS), etc., for removing background noise and measuring yield associated traits precisely. BILs are characterized by small introgression of segment from donor parent and useful for reducing background noise from donor parent, as well as for mapping interspecific variation (Eshed and Zamir, 1994). In soybean, BIL populations have been developed for overcoming abiotic stresses by mining and introgressing useful QTLs from the donor parent. Water limiting situation is one of the main restraints for soybean production (Sinclair et al., 2007). Earlier reports confirmed that root length and absorption surface area along with root architecture are prime determinants for yield performance under variable moisture regime (Price et al., 2002; Manavalan et al., 2009). As variability regarding root architecture is limited in cultivated soybean, an attempt was made to explore the potential of exotic wild species for broadening the genetic base. BIL mapping population has been developed by crossing Glycine max cv. Dunbar (PI 552538) as a recipient with a wild soybean accession “PI326582A” of G. soja. BILs have been created to minimize the magnitude of gene introgression from the wild soybean parent “PI326582A” by allowing two generations of backcrossing to produce 296 BC2F4.5 progenies (Manavalan et al., 2015). Genetic linkage map was constructed by using SSR and SNP markers, resulting in the identification of a major QTL (Satt315-I locus) on chromosome 8 that governs root traits and shoot length. It has been observed that, sometimes, the same metabolic pathway governs different stresses in plants (Xiong and Yang, 2003), and it is mostly associated with overlapping QTLs. The reason for this genetic overlapping is due to pleiotropy and linkage disequilibrium (Zhang et al., 2012). In soybean, BIL mapping population was developed by crossing a Chinese variety, “Hongfeng 11” with an American variety, “Harosoy” for mapping QTLs related to drought and low-temperature tolerance during germination. Finally, 12 QTLs were detected that were correlated with drought and low-temperature tolerance and confirmed the mechanism of partial genetic overlap between drought and low-temperature tolerance in soybean (Zhang et al., 2012). This study further validated the effectiveness of using BILs for gene introgression, trait identification, QTL mapping, and gene cloning in legume.
Chromosome Segment Substitution Line
Chromosome segment substitution lines are very robust population for QTL mapping or cloning and gene discovery, as well as for gene pyramiding (Tanksley and Nelson, 1996), and can be developed by deploying AB strategy subsequently by selfing and selection of backcross population with molecular markers. Selection of backcross population with markers leads to identification of individuals carrying the introgressed gene(s) of interest along each chromosome. CSSLs generally exclude non-targeted portion from the donor, which can create background noise due to epistatic interaction (Ali et al., 2010). In CSSLs, each line carries a single defined chromosomal section from the wild donor into the recipient background, unlike BILs, where each line carries several homozygous introgressed segments from donor parent. Unlike BIL, CSSL libraries have been developed to recover the whole genome of donor parent (Ali et al., 2010). These populations can be compared with the individual IL or recurrent parent for finding out significant differences between them. CSSLs are also useful populations for controlling allelic variation and facilitate “breeding by design” (Peleman and van der Voort, 2003; Wei et al., 2010).
This strategy has been used in many crops including wheat (Liu et al., 2006), tomato (Monforte and Tanksley, 2000), rice (Bian et al., 2010), maize (Wang et al., 2007), cotton (Wang et al., 2008), and barley (Matus et al., 2003) for gene discovery and map-based cloning and opens a new vista for exploring the potential of CSSL population in legumes for detection of genes or QTL explicitly, as well as their pyramiding into elite background. Cultivated soybean (G. max) is domesticated from wild G. soja (Broich and Palmer, 1980), which harbors useful genes governing large number of pods, richness in protein, adaptability to various biotic and abiotic factors, etc. Previous studies confirmed the versatility of G. soja as a useful donor for enriching the genetic diversity of cultivated soybean (Concibido et al., 2003; Li et al., 2008). The problem of linkage drag often circumvents the useful introgression process. Keeping these in mind, attempt has been made to construct CSSL population consisting of 151 lines by involving G. max cv. NN1138-2 as female and G. soja cv. N24852 as male. Polymorphic SSR markers between the parents were deployed for marker assisted selection (MAS) for easy recovery of CSSL. In this study, four QTLs related with plant height, as well as node numbers per plant, have been identified (Wang et al., 2013). The same CSSL population was used for mining and fine mapping of QTLs underlying seed quality traits including size and shape, as well as other agronomic traits (Wang et al., 2012, 2013; He et al., 2014).
Chromosome Substitution Strain
Another approach is the construction of chromosome substitution strain containing a large number of lines each carrying a homozygous chromosome with single crossover in such a way that the chromosome contains recurrent genotype at one end and donor genotype at the other end and known as single recombinant lines (SRLs). When the SRLs for each chromosome are sequentially stacked, they reveal a step-like progression, with each successive line having a little more donor chromosome, and constitute STAIRS libraries (Koumproglou et al., 2002). The concept was first applied in Arabidopsis thaliana for fine mapping of QTL. Although STAIRS has not yet been explored in legume crops, it is an effective strategy for comparison of genetic differences in the precise region of selected chromosome for QTL analysis, gene mining, and expression studies (Koumproglou et al., 2002). All these ILs can be maintained as an immortal representation in the form of “exotic library” for efficient detection and mapping of QTLs conferring agronomic traits (Zamir, 2001). This library is a permanent resource, which enables the researchers to explore over time and access the data generated for further use. The homozygous lines maintained in the library can be utilized as a parent for crossing with different tester lines to identify the chromosomal segments associated with heterosis. Development of an exotic library will immensely facilitate to counter the problem of linkage drag and precisely examine the phenotypic effect of QTL interaction for better insight into the epistatic effect (Eshed and Zamir, 1996). All these mapping populations along with genomic tools will be valuable for demonstrating the scope of introgression of desirable QTLs from CWR that was hitherto difficult to accomplish. The methodologies described can be extended to legume crops for harnessing the potential of CWRs for broadening the genepool through genomics-assisted genetic enhancement.
Potential Wild Species for Alien Gene Transfer of Target Traits
Most food legumes and their wild relatives (CWRs) are diploid and self-pollinated in nature. Considerable variability exists in wild species for yield contributing traits including number of pods per plant, number of seeds per pod, and seed size, as well as nutritional traits and biotic and abiotic stress resistance. The success of alien gene transfer through distant hybridization generally depends on the ploidy level of the species, pollination behavior of the plant, nature, and direction of the cross and frequency of pollination, which are further influenced by the deployment of appropriate hybridization schemes (Pratap et al., 2015a). Efforts were made to identify potential wild accessions for alien gene introgression in different food legumes by several researchers (Table 2).
Chickpea is the most important cool season grain legume and offers tremendous opportunities for its genetic improvement through introgression breeding, especially concerning biotic and abiotic stresses. Of the eight annual species, only one wild species, C. reticulatum, is readily crossable with the cultivated chickpea (Kumar et al., 2003). The success of hybridization with the remaining annual wild Cicer species requires specialized techniques such as the application of growth hormones and embryo rescue techniques (Lulsdorf et al., 2005; Mallikarjuna and Jadhav, 2008). Among the biotic stresses, FW and AB cause maximum damage to the plant and lead to severe yield reduction. FW causes up to 100% yield losses (Sharma et al., 2004; Pratap et al., 2017). AB, caused by Ascochyta rabiei usually appears at the reproductive phase, and in severe cases, the entire plant dries up suddenly. Several accessions of Cicer bijugum, Cicer echinospermum, Cicer judaicum, Cicer chorassanicum, Cicer pinnatifidum, C. reticulatum, Cicer cuneatum, Cicer yamashitae, and Cicer canariense have shown high resistance to AB (Haware et al., 1992; Kaiser et al., 1994; Collard et al., 2001; Shah et al., 2005). Simultaneously, many of them also possessed a high degree of resistance to FW (Kaiser et al., 1994; Infantino et al., 1996; Singh K. B. et al., 1998; Collard et al., 2001; Shah et al., 2005; Pande et al., 2006). Some of the accessions belonging to C. bijugum, C. echinospermum, C. judaicum, and C. reticulatum were reported to be highly resistant to Botrytis gray mold (Pande et al., 2006; Knights et al., 2008; Isenegger et al., 2011; Coyne et al., 2020). Several chickpea CWRs have shown high tolerance to abiotic stresses. For example, tolerance to drought and heat stresses was reported in six Cicer species (Toker et al., 2007a; Canci and Toker, 2009; Imtiaz et al., 2011). Likewise, tolerance to cold was reported in C. bijugum, C. echinospermum, C. pinnatifidum, and C. reticulatum (Singh et al., 1990; Robertson et al., 1995; Singh K. B. et al., 1998; Toker, 2005; Saeed et al., 2010).
Among Vigna crops, the Asiatic Vigna have tremendous scope for improvement with respect to yield and yield attributes, biotic and abiotic resistance, and nutritional quality (For review, see Pratap et al., 2020). Accessions of Vigna mungo var. silvestris have been identified as a durable source of mungbean yellow mosaic virus (MYMV) resistance (Reddy and Singh, 1993). Likewise, variation for yield contributing traits and MYMV resistance was observed in V. mungo var. silvestris, Vigna radiata var. sublobata (Singh, 1990), Vigna umbellata, Vigna trilobata, and V. mungo (Nagaraj et al., 1981; Singh and Dikshit, 2002; Pratap et al., 2018a). “PLN15,” a wild accession of V. radiata var. sublobata possessed a high number of pods per plant and seeds per pod and therefore identified as a potential donor for these traits (Reddy and Singh, 1990). V. trilobata and Vigna stipulacea was proposed as the candidates for neodomestication for drought tolerance and disease and pest resistance, respectively, by Tomooka et al. (2014), whereas resistance to diseases and pests in both the species was also reported in several other reports (Nagaraj et al., 1981; Chandel et al., 1984; Tomooka et al., 2006; Pandiyan et al., 2008; Gore et al., 2019). Accession “TC1966” of V. radiata var. sublobata was reported to possess bruchid tolerance (Tomooka et al., 1992). Likewise, resistance to legume pod borers and pod-sucking bugs was reported in Vigna vexillata and Vigna oblongifolia (Fatokun, 1991). V. mungo var. silvestris was reported as immune to bruchids (Fujii et al., 1989; Dongre et al., 1996). Many accessions of ricebean (V. umbellata) show complete resistance to bruchids and therefore have been identified as useful donors for introgressing bruchid resistance into other Vigna species. Further, as it is a cultivated species, gene introgression from ricebean into mungbean and urdbean is comparatively easier. Dense hairs on different parts of the wild cowpea, V. vexillata, are reported to impart antixenosis to pod-sucking bugs and pod borer (Oghiakhe et al., 1992; Boukar et al., 2013). Likewise, strength and hardness of the pod wall are also considered to impart pod borer resistance (Oigiangbe et al., 2002). Other wild Vigna species with resistance to Maruca vitrata and the pod-sucking bugs include Vigna unguiculata ssp. dekindtiana, Vigna luteola, Vigna oblongifolia, and Vigna reticulata. Pratap et al. (2014) reported V. umbellata (accession IC251442) and Vigna glabrescens (accession IC251372) as photo- and thermo-period insensitive as these were able to flower and set pods at temperatures as high as 43.9°C and as low as 2.7°C.
Wild Lens species have emerged as a great reservoir of useful genes for traits of breeders’ interest including resistance to strategically important diseases, insect-pests, and plant parasitic weeds. A high degree of resistance was observed for Stemphylium blight in Lens lamottei followed by Lens ervoides (Podder et al., 2013). Similarly, some accessions of Lens odemensis possessed high resistance against Sitona weevil followed by L. ervoides (El-Bouhssini et al., 2008). Some of the wild accessions with combined resistance to AB and FW or anthracnose diseases have been identified for their use in lentil breeding programs (Bayaa et al., 1995; Tullu et al., 2006). Preliminary screening of Lens CWR has indicated drought tolerance in Lens nigricans, L. odemensis, and L. ervoides (Gupta and Sharma, 2006) and cold tolerance in Lens culinaris ssp. orientalis (Hamdi et al., 1996). Simultaneously, promising donors for yield traits, viz., 100-seed weight and pods per plant were observed in L. lamottei and L. culinaris ssp. orientalis (Gupta and Sharma, 2006). Earlier reports have indicated L. ervoides as a good source of alleles for plant architectural traits such as phenology, plant growth habit, and biomass besides seed traits (Tullu et al., 2011, 2013; Kumar et al., 2014). Based on the extensive evaluation of global wild Lens taxa representing 27 countries, Kumar et al. (2014) observed wide variation for yield attributing traits, as well as resistance to multiple diseases in L. nigricans and L. ervoides. Nutritional quality traits in wild Lens showed significant diversity not only for micronutrients (Sen Gupta et al., 2016; Kumar et al., 2018) but also for prebiotics, RFO, raffinose, and verbascose (Tahir et al., 2011).
Gene Introgression in Food Legumes: Success
Wild gene introgression as a breeding strategy has been deployed successfully in food legumes for development of improved varieties, pre-bred lines, genetic stocks, mapping populations, and bridge species. Legumes such as chickpea, pigeonpea, lentil, mungbean, urdbean, and peanut have benefited from the wild gene introgression with successful examples of discovery, development, and deployment of useful traits in cultivated genepool. In chickpea, after the first report of successful interspecific crosses between C. arietinum and C. reticulatum (Ladizinsky and Adler, 1976), attempts have been made for crossing between C. arietinum and C. echinospermum (Singh and Ocampo, 1993; Pundir and Mengesha, 1995). C. reticulatum accession “ILWC119,” when involved in hybridization program, led to the development of “ILC10765” and “ILC10766,” two cyst nematode-resistant chickpea lines (Malhotra et al., 2002). Singh et al. (2005) and Ramgopal et al. (2012) utilized the diversity of C. reticulatum and C. echinospermum to transfer useful traits including tolerance to cold and resistance to diseases such as wilt, root rot, and Botrytis gray mold into cultivated chickpea. There are reports of successful interspecific crosses between C. arietinum and C. judaicum (Singh et al., 1999), C. arietinum and C. cuneatum (Singh and Singh, 1989), C. arietinum and C. pinnatifidum (Badami et al., 1997; Mallikarjuna and Jadhav, 2008), and C. arietinum and C. bijugum (Mallikarjuna et al., 2007). Successful introgression of useful genes into cultivated chickpea from these crosses has shown the transferability even from the cross-incompatible wild Cicer species.
Successful interspecific hybridization of P. vulgaris has been reported to a limited extent with the members of other wild Phaseolus species. Interspecific hybrids with Phaseolus costaricensis in the secondary genepool were reported by Singh et al. (1997), and consequently, VRW 32 was reported as the first white mold-resistant interspecific breeding line derived from P. costaricensis. Congruity backcrossing (CBC) involves recurrent backcrossing to each parent in alternate generations as opposed to the traditional recurrent backcrossing to a single recurrent parent and was first reported as a method to produce fertile intermediate hybrids between Phaseolus acutifolius and P. vulgaris (Haghighi and Ascher, 1988). This method allows substantial recombination between distant species, and new phenotypes can arise as a result of CBC (Anderson et al., 1996). Singh et al. (2009) reported the release of white mold-resistant “VCW 54” and “VCW 55” bean germplasm lines that were developed using CBC between the black bean cultivar “ICA Pijao” and the scarlet runner bean accession “G35172.” CBC has also been used to transfer traits from wild tepary species Phaseolus parvifolius into common bean (Singh S. P. et al., 1998).
Wilkinson (1983) reported a root rot-resistant line “Cornell 2114-12” derived from a cross between common bean and scarlet runner bean lines. Likewise, Miklas et al. (1999) developed the common bacterial blight-resistant bean germplasm lines “ICB-3,” “IBC-6,” “ICB-8,” and “ICB-10,” which were derived from an interspecific cross with scarlet runner bean. Beaver et al. (2012) released a bean germplasm line “PR0650-31,” which was derived from the cross BAT 93/PI 417662//VAX 6 using wild-type bean germplasm “PI 417662” collected from Jalisco, Mexico, and was resistant to web blight and common bacterial blight. Acosta-Gallegos et al. (2007) developed an inbred backcross population from a cross between G 24423, a wild bean accession from Colombia and “Negro Tacana,” a Mexican black bean cultivar. One Bc2F4:7 line from this population was later observed to produce >5,000 kg/ha seed in field trials.
Lentil CWRs have been evaluated extensively to discover and deploy traits of interest into cultivated species. These efforts have led to identification of extra early photoinsensitive (ILWL118 maturing in <90 days) and high micronutrient content (ILWL74 and ILWL80) germplasm. These CWRs have been used extensively in mainstream breeding, resulting in the development of short-duration biofortified pre-bred lines (Kumar et al., 2018). Wide crosses in lentil have also been mined for transgressive segregants for agronomically important traits (Kumar et al., 2011; 2014; Singh et al., 2013). More recently, hybridization of the cultivated lentil with L. ervoides using embryo rescue (Tullu et al., 2013) has been reported with successful transfer of resistance to Orobanche crenata and anthracnose in cultivated species (Fiala et al., 2009; Tullu et al., 2011). The International Center for Agricultural Research in the Dry Areas has successfully deployed L. orientalis and L. ervoides for introgression of resistance to key diseases, phenology, biofortification, plant habit, and other important agronomic traits toward the development of pre-bred lines. These pre-bred lines demonstrated >40% yield advantage over the best check (Bakaria) coupled with richness in micronutrients. These pre-bred lines can also fit well in short-season windows of 80–100 days (Kumar et al., 2020). These lines are currently under multilocation testing under the CWR project.
In pigeonpea, despite large visible genetic variation (Yang et al., 2006), the use of wild species in breeding programs has been rather limited to the development of cytoplasmic genic male sterility systems (Saxena et al., 2010). To date, seven cytoplasmic male sterility (CMS) systems have been reported (Saxena et al., 2010), and six of them have been developed from wild relatives belonging to the secondary genepool. The seventh system was developed utilizing Cajanus platycarpus, a member from the tertiary genepool (Saxena et al., 2010; Mallikarjuna et al., 2011). The A1 CMS system derived from Cajanus sericeus (Saxena et al., 1996) was not stable at low temperature (<10°C) as the male-sterile plants revert to male fertility (Saxena et al., 2005). However, the presence of pollen shedders in the female line and non-availability of good maintainers did not make it commercially viable for hybrid breeding. The A2 cytoplasm derived from Cajanus scarabaeoides was reported as highly stable (Tikka et al., 1997; Saxena and Kumar, 2003). Although this system is promising with respect to yield, inconsistency was observed in the fertility restoration over diverse environments, which reduced its acceptance for hybrid production. In the A3 system, the cytoplasm was derived from Cajanus volubilis (Wanjari et al., 1999). The A4 CMS system was developed from Cajanus cajanifolius, which is so far the best among different CMS systems developed. This CMS system has a good number of both maintainers and restorers. In A5 system, the cytoplasm of cultivated species of Cajanus cajan was placed along with nuclear genome of a CWR of pigeonpea, Cajanus acutifolius (Mallikarjuna and Saxena, 2005) while using C. cajan as the female parent. This system also exhibited perfect fertility restoration by cultivated accessions. The A6 cytoplasm was developed from Cajanus lineatus (A6) in 2002, from one naturally out-crossed plant with erect growth and different morphological traits. This CMS system was observed to be very stable (Saxena et al., 2010) showing perfect fertility restoration by cultivated accessions. The A7 cytoplasm derived from C. platycarpus (A7) produced good heterosis (Saxena et al., 2010). Four CMS lines, viz., “GT 288A,” “CMS 67A,” “ICRISAT CMS,” and “AKCMS 1A,” were developed from different wild sources viz., C. sericeus, C. scarabaeoides, and C. volubilis (Chauhan et al., 2008).
In Vigna species, mungbean × urdbean hybridization has been routinely practiced for mungbean and urdbean improvement programs, and the derivatives from these hybridizations exhibit many desirable features viz., resistance to vagaries, both biotic and abiotic, synchronous podding, and non-shattering pods (Pratap et al., 2019). Several traits, such as longer pods, increased seeds number (>10 seeds/pod), and erect plant type, have been transferred from mungbean to urdbean, whereas multiple clusters per peduncle and sympodial pod-bearing habit have been transferred from urdbean into mungbean (Gupta et al., 2004). Similarly, mungbean × ricebean and mungbean × V. radiata var. sublobata hybridization have also been practiced by many breeders, and progenies were derived which were resistant to MYMV (Verma and Brar, 1996). Singh et al. (2003) produced successful hybrids between V. radiata and V. umbellata with intermediate morphology and MYMV resistance. Several popular and widely adaptable cultivars have been developed as a result of wild gene introgression in both mungbean and urdbean (Table 3). These cultivars show wide adaptation, synchronous maturity, and improved plant architecture in addition to a high degree of resistance to MYMV. Recently, “IPM 312-20” and “Tripura Mung-1” have been developed as a result of mungbean × urdbean hybridization. Likewise, the resultants of mungbean × urdbean crosses were also used further to develop some of the most popular varieties of mungbean. For example, IPM 99-125 was used to develop the most popular pan India variety “IPM 205-7” (Virat) of mungbean (Pratap et al., 2013) which matures in 52–55 days and offers the farmers an excellent choice for cultivation during summer season. Earlier, “IPM 02-3” was also a highly preferred variety of mungbean by farmers (Singh et al., 2017).
In urdbean, at least five commercial varieties have been developed and deployed using wild gene introgression. The first such variety was Mash 118 developed from an urdbean × mungbean cross in 2008. This was followed by the development of four more cultivars, viz., Vamban 7, VBN 6, TU 40, and VBG04-008 in 2011. Among these, VBG04-008 showed high tolerance to heat stress, making it most popular cultivar in heat-prone environments of South India. Interspecific crosses have also been attempted successfully between V. umbellata and its wild relatives. However, the success of crosses with respect to pod set differed with the combination of parents involved in the interspecific crosses (Chen et al., 1983; Bharathi et al., 2006).
Epilogue
The narrow genetic base of the elite genepool of food legumes and resultant vulnerability of the existing varieties to climate vagaries and changing insect-pest and disease scenario warrants introgression of novel genes or alleles through hybridization and deployment of more diverse germplasm including exotic lines and CWRs in crop improvement programs. Food legumes being majorly grown by small and marginal farmers are more prone to fluctuations in the soil, water, and climate factors as compared to other crops due to limited resources at disposal of these farmers to counter these challenges. The slow process of natural evolution has been significantly replaced by human interventions of domestication, hybridization, and selection. The transformation of humans from food collector to food producer has witnessed the natural attempts of domestication to a planned and focused crop breeding, which has ultimately concluded into the modern “super domestication.” While at one side, this has ensured food and nutritional security to ever-increasing population, on the other side, it has narrowed down the genetic base of the cultivated genepool. Keeping this in view, there is a need to reorient legumes improvement programs in such a way that more diverse sources of yield contributing traits, resistance to stresses, both biotic and abiotic, and seed quality are involved in widening the genetic base of cultivated types. This requires trait discovery and deployment from CWR, exotic germplasm, and landraces in mainstream breeding programs. A huge repository of germplasm and CWR (>7 million germplasm accessions) of different crops is maintained together in more than 1,750 national and international genebanks. This includes >86,000 accessions of chickpea1, >57,000 of Phaseolus (Basavaraja et al., 2020), >43,000 of mungbean (Nair et al., 2013; Gayacharan et al., 2020), >13,500 of pigeonpea (Upadhyaya et al., 2011), >16,000 accessions of cowpea, and so on. Nonetheless, characterization and evaluation data on economically important traits are limited to a smaller set of cultivated genepool. The situation is still worse when it comes to CWRs, which needs to be addressed on priority. The genetic bottlenecks leading to narrowing down the genetic base of food legumes need to be recognized and efforts to be initiated through intensive pre-breeding programs. However, owing to pre- and post-fertilization barriers applicable to distant crosses, special tools and techniques need to be adopted. These include application of growth hormones, using mentor pollen technique, deployment of embryo rescue, and several other methods bypassing these barriers (Pratap et al., 2010, 2018b). Wild gene introgression has yielded dividends in some legume crops such as mungbean, urdbean, pigeonpea, chickpea, lentil, etc., either directly through development of commercial cultivars or indirectly through the development of breeding materials and male sterile lines helping in hybrid variety development. Nonetheless, the advantages gained are still far from the potential of gene introgression, and focused planning and implementation in this direction are needed. Development of ILs, NILs, and specialized experimental populations may help in unleashing the genetic and genomic potential of wild gene introgression in the improvement of cultivated food legumes. These populations when subjected to precise and high throughput phenotyping will provide fast and inexpensive genomic information (Pratap et al., 2015b). Wild genetic resources are enormous, opportunities are tremendous, and challenges are manifold. Thus, the need is to venture into the wild gene introgression approach as a long-term strategy with great patience and care.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work has funding support from the International Center for Agricultural Research in the Dry Areas (ICARDA), Rabat Office, Rabat-Institus for open access publication fees.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Acosta-Gallegos, J. A., Kelly, J. D., and Gepts, P. (2007). Prebreeding in common bean and use of genetic diversity from wild germplasm. Crop Sci. 47, S44-S49.
Ali, M. L., Sanchez, P. L., Yu, S., Lorieux, M., and Eizenga, G. C. (2010). Chromosome segment substitution lines: a powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa L.). Rice 3, 218–234. doi: 10.1007/s12284-010-9058-3
Amalraj, A., Taylor, J., Bithell, S., Li, Y., Moore, K., Hobson, K., et al. (2019). Mapping resistance to Phytophthora root rot identifies independent loci from cultivated (Cicer arietinum L.) and wild (Cicer echinospermum PH Davis) chickpea. Theor. Appl. Genet. 132, 1017–1033. doi: 10.1007/s00122-018-3256-6
Anderson, N.O., Ascher, P.D., and Haghighi, K. (1996). Congruity backcrossing as a means of creating genetic variability in self pollinated crops: seed morphology of Phaseolus vulgaris L. and P.acutifolius A. Gray Hybrids. Euphytica 87, 211–224 doi: 10.1007/bf00023748
Aryamanesh, N., Byrne, O., Hardie, D. C., Khan, T., Siddique, K. H. M., and Yan, G. (2012). Large-scale density-based screening for pea weevil resistance in advanced backcross lines derived from cultivated field pea (Pisum sativum) and Pisum fulvum. Crop Pasture Sci. 63(7), 612–618. doi: 10.1071/cp12225
Badami, P. S., Mallikarjuna, N., and Moss, J. P. (1997). Interspecific hybridization between Cicer arietinum and C. pinnatifidum. Plant Breed. 116, 393–395. doi: 10.1111/j.1439-0523.1997.tb01019.x
Barilli, E., Cobos, M. J., Carrillo, E., Kilian, A., Carling, J., and Rubiales, D. (2018). A high-density integrated DArTseq SNP-based genetic map of Pisum fulvum and identification of QTLs controlling rust resistance. Front. Plant Sci. 9:167. doi: 10.3389/fpls.2018.00167
Basavaraja, T., Pratap, A., Dubey, V., Gurumurthy, S., Gupta, S., Singh, N. P., et al. (2020). Molecular and Conventional Breeding Strategies for Improving Biotic Stress Resistance in Common Bean. In: S.S. Gosal- and S.H. Wani (eds.) Accelerated Plant Breeding. 3. Switzerland: Springer Nature. 389–421. doi: 10.1007/978-3-030-47306-8_13
Bayaa, B., Erskine, W., and Hamdi, A. (1994). Response of wild lentil to Ascochyta fabae f.sp. lentis from Syria. Genet. Resour. Crop Evol. 41, 61–65. doi: 10.1007/BF00053049
Beaver, J.S., Zapata, M., Alameda, M., and Porch, T.G. (2012). Registration of PR0401-259 and PR0650-31 dry bean germplasm lines. J. Plant Regist. 6, 81–84. doi: 10.3198/jpr2011.05.0283crg
Bera, S. K., Kamdar, J. H., Kasundra, S. V., Patel, S. V., Jasani, M. D., Maurya, A. K., et al. (2019). Steady expression of high oleic acid in peanut bred by marker-assisted backcrossing for fatty acid desaturase mutant alleles and its effect on seed germination along with other seedling traits. PloS One 14:e0226252. doi: 10.1371/journal.pone.0226252
Bharathi, A., Vijay Selvraj, K. S., Veerabadhiran, P., and Subba Lakshami, B. (2006). Crosability barriers in mungbean (Vigna radiata L. Wilczek): with its wild relatives. Indian J. Crop Sci. 1, 120–124.
Bhavyasree, R. K., Singh, S., and Singh, I. (2018). Advanced backcross strategy for alien introgression for productivity enhancing traits in chickpea (Cicer arietinum L.). Legume Res. 41, 379–383.
Bian, J. M., Jiang, L., Liu, L. L., Wei, X. J., Xiao, Y. H., Zhang, LJ, et al. (2010). Construction of a new set of rice chromosome segment substitution lines and identification of food weight and related traits QTLs. Breed. Sci. 60, 305–313. doi: 10.1270/jsbbs.60.305
Blair, M. W., Herrera, A. L., Sandoval, T. A., Caldas, G. V., Fileppi, M., Sparvoli, F., et al. (2012). Inheritance of seed phytate and phosphorus levels in common bean Phaseolus vulgaris L. and association with newly-mapped candidate genes for the phytic acid pathway. Mol. Breed. 30, 1265–1277. doi: 10.1007/s11032-012-9713-z
Blair, M. W., Iriarte, G., and Beebe, S. (2006). QTL analysis of yield traits in an advanced backcross population derived from a cultivated Andean× wild common bean (Phaseolus vulgaris L.) cross. Theor. Appl. Genet. 112, 1149–1163. doi: 10.1007/s00122-006-0217-2
Blair, M. W., Pedraza, F., Buendia, H. F., Gatian-Soils, E., Beebe, S. E., Gepts, P., et al. (2003). Development of a genome-wide anchors microsatellite map for common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 107, 1362–1374. doi: 10.1007/s00122-003-1398-6
Bock, R. (2009). The give and take of DNA: horizontal gene transfers in plants. Trends in Plant Sci. 15, 11–22. doi: 10.1016/j.tplants.2009.10.001
Boukar, O., Belko, N., Chamarthi, S., Togola, A., Batieno, J., Owusu, E., et al. (2019). Cowpea (Vigna unguiculata): genetics, genomics and breeding. Plant Breed. 138, 415–424.
Boukar, O., Bhattacharjee, R., Fatokun, C., Kumar, L. P., and Gueye, B. (2013). Cowpea. In Singh MUpadhyaya HDBisht IS (eds) Genetic And Genomic Resources Of Food Legume Improvement. Elsevier Inc: London. 137–157.
Broich, S. L., and Palmer, R. G. (1980). A cluster analysis of wild and domesticated soybean phenotypes. Euphytica 29, 23–32. doi: 10.1007/bf00037246
Burow, M. D., Starr, J. L., Park, C. H., Simpson, C. E., and Paterson, A. H. (2014). Introgression of homeologous quantitative trait loci (QTLs) for resistance to the root-knot nematode [Meloidogyne arenaria (Neal) Chitwood] in an advanced backcross-QTL population of peanut (Arachis hypogaea L.). Mol. Breed. 34, 393–406. doi: 10.1007/s11032-014-0042-2
Canci, H., and Toker, C. (2009). Evaluation of annual wild Cicer species for drought and heat resistance under field conditions. Genet. Resour. Crop Evol. 56, 1–6. doi: 10.1007/s10722-008-9335-9
Chakraborty, S., and Newton, A.C. (2011). Climate change, plant diseases and food security: an overview. Plant Pathol. 60, 2–14. doi: 10.1111/j.1365-3059.2010.02411.x
Chaky, J. M., Specht, J. E., and Cregan, P. B. (2003). Advanced backcross QTL analysis. In A Mating between Glycine max and Glycine soja. Plant Anim. Genome Abst. 2003:545.
Chamarthi, S. K., Kumar, A., Vuong, T. D., Blair, M. W., Gaur, P. M., Nguyen, H. T., et al. (2011). “Trait mapping and molecular breeding,” in Biology and Breeding of Food Legumes, eds A. Pratap and J. Kumar (Wallingford, UK: CABI), 296. doi: 10.1079/9781845937669.0296
Chandel, K. P. S., Lester, R. N., and Starling, R. J. (1984). The wild ancestors of urd and mungbeans (Vigna mungo (L.) Hepper and V. radiata (L.) Wilczek). Botanical J. Linnean Soc. 89, 85–96. doi: 10.1111/j.1095-8339.1984.tb01002.x
Chaturvedi, S. K., Gupta, D. S., and Jain Rashmi. (2011). “Biology of Food Legumes,” in Biology and Breeding of Food Legumes, eds A. Pratap and J. Kumar (Oxfordshire: CABI), 35–48. doi: 10.1079/9781845937669.0035
Chauhan, R. M., Panwar, L. D., Patel, P. T., and Tikka, S. B. S. (2008). Identification of heterotic combination of CMS lines and restorers of pigeonpea. J. Food Legum. 21, 25–27.
Chen, N. C., Baker, R. L., and Honma, S. (1983). Interspecific crossability among four species of Vigna food legumes. Euphytica 32, 925–937. doi: 10.1007/bf00042175
Chu, Y., Wu, C. L., Holbrook, C. C., Tillman, B. L., Person, G., Ozias-Akins, P., et al. (2011). Marker-assisted selection to pyramid nematode resistance and the high oleic trait in peanut. Plant Genome 4, 110–117. doi: 10.3835/plantgenome2011.01.0001
Clemente, A., Arques, M. C., Dalmais, M., Le Signor, C., Chinoy, C., Olias, R., et al. (2015). Eliminating anti-nutritional plant food proteins: the case of seed protease inhibitors in pea. PLoS One 10:e0134634. doi: 10.1371/journal.pone.0134634
Cobos, M. J., Satovic, Z., Rubiales, D., and Fondevilla, S. (2018). Er3 gene, conferring resistance to powdery mildew in pea, is located in pea LGIV. Euphytica 214:203.
Collard, B. C. Y., Ades, P. K., Pang, E. C. K., Brouwer, J. B., and Taylor, P. W. J. (2001). Prospecting for sources of resistance to ascochyta blight in wild Cicer species. Aus. J. Plant Pathol. 30, 271–276.
Collard, B. C. Y., Pang, E. C. K., Ades, P. K., and Taylor, P. W. J. (2003). Preliminary investigation of QTLs associated with seedling resistance to ascochyta blight from Cicer echinospermum, a wild relative of chickpea. Theor. Appl. Genet. 107, 719–729. doi: 10.1007/s00122-003-1297-x
Concibido, V. C., Vallee, B. L., Mclaird, P., Pineda, N., Meyer, J., Hummel, L., et al. (2003). Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theor. Appl. Genet. 106, 575–582 doi: 10.1007/s00122-002-1071-5
Coyne, C. J., Kumar, S., van Wettberg, E. J. B., Marques, E., Berger, J. D., Redden, R. J., Noel Ellis, T. H., Brus, J., Zoblatzka, L., and Smykal, P. (2020). Potential and limits of exploitation of crop wild relatives for pea, lentil and chickpea improvement. Legume Sci. 2:e36. doi: 10.1002/leg3.36
Devasirvatham, V., Gaur, P. M., Mallikarjuna, N., Raju, T. N., Trethowan, R. M., and Tan, D. K. (2013). Reproductive biology of chickpea response to heat stress in the field is associated with the performance in controlled environments. Field Crops Res. 142, 9–19. doi: 10.1016/j.fcr.2012.11.011
Dongre, T. K., Pawar, S. E., Thakare, R. G., and Harwalkar, M. R. (1996). Identification of resistant source to cowpea weevil [Callosobruchus maculatus (F.)] in Vigna sp. and inheritanceof their resistance in black gram (Vigna mungo var. mungo). J. Stored Prod. Res. 32, 201–204. doi: 10.1016/s0022-474x(96)00028-8
Doyle, J. J. (1988). 5S ribosomal gene variation in the soybean and its progenitor. Theor. Appl. Genet. 75, 621–624. doi: 10.1007/bf00289130
Dwivedi, S. L., Blair, M. W., Upadhyaya, H. D., Serraj, R., Buhariwalla, H. K., Ortiz, R., et al. (2005). Usinggenomics to exploit grain legume biodiversity in cropimprovement. Plant Breed. Rev. 26, 171–357. doi: 10.1002/9780470650325.ch6
Egawa, Y., Bujang, I. B., Chotechuen, S., Tomooka, N., and Tateishi, Y. (1996). Phyloegenetic differentiation of tetraploid Vigna species, V. glabrescens and V. reflexo-pilosa. Jpn. Int. Res. Center Agric. Sci. 3, 49–58.
Egawa, Y., Takeda, H., and Suzuki, K. (1999). Research plan on crop heat tolerance at the crop introduction and cultivation laboratory. Jpn. Int. Res. Center Agric. Sci. Working Rep. 14, 103–107.
El-Bouhssini, M., Sarker, A., Erskine, W., and Joubi, A. (2008). First sources of resistance to sitona weevil sitona crinitus herbst. in wild lens species. Genet. Resour. Crop Evol. 55, 1–4. doi: 10.1007/s10722-007-9297-3
Esen, A., Sari, H., Erler, F., Adak, A., Sari, D., Eker, T., et al. (2019). Screening and selection of accessions in the genus Pisum L. for resistance to pulse beetle (Callosobruchus chinensis L.). Euphytica 215:82.
Eshed, Y., and Zamir, D. (1994). A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica 79, 175–179. doi: 10.1007/BF00022516
Eshed, Y., and Zamir, D. (1995). An introgression line population of Lycopersiconpennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162.
Eshed, Y., and Zamir, D. (1996). Less than additive epistatic interactions of QTL in tomato. Genetics 143, 1807–1817.
Fernández-Aparicio, M., Sillero, J. C., and Rubiales, D. (2009). Resistance to broomrape in wild lentils (Lens spp.). Plant Breed. 128, 266–270. doi: 10.1111/j.1439-0523.2008.01559.x
Fiala, J. V., Tullu, A., Banniza, S., Séguin- Swartz, G., and Vandenberg, A. (2009). Interspecies transfer of resistance to anthracnose in lentil (Lens culinaris Medic.). Crop Sci. 49, 825–830. doi: 10.2135/cropsci2008.05.0260
Fonceka, D., Tossim, H. A., Rivallan, R., Vignes, H., Faye, I., Ndoye, O., et al. (2012). Fostered and left behind alleles in peanut: interspecific QTL mapping reveals footprints of domestication and useful natural variation for breeding. BMC Plant Biol. 12:26. doi: 10.1186/1471-2229-12-26
Frisch, M., and Melchinger, A. (2005). Selection theory for marker-assisted backcrossing. Genetics 170, 909–917. doi: 10.1534/genetics.104.035451
Fujii, K., Ishimoto, M., and Kitamura, K. (1989). Pattern of resistance to bean weevil Bruchidae. in Vigna radiata-mungo-sublobata complex inform the breeding of new resistant varieties. Appl. Entomol. Zool. 24, 126–132. doi: 10.1303/aez.24.126
Gautam, H. R., Bhardwaj, M. L., and Kumar, R. (2013). Climate change and its impact on plant diseases. Curr. Sci. 105, 1685–1691.
Gayacharan, C., Archak, S., Gupta, K., Gupta, V., Tyagi, V., Singh, K., et al. (2020). Mungbean genetic resources and utilization. In: R.M. Nair, R. Schafleitner, and S. H. Lee (eds.). The Mungbean Genome. Springer: Switzerland. 9–25. doi: 10.1007/978-3-030-20008-4_2
Gore, P. G., Tripathi, K., Pratap, A., Bhat, K. V., Umdale, S. D., Gupta, V., et al. (2019). Delineating taxonomic identity of two closely related Vigna species of section Aconitifoliae: V. trilobata (L.) Verdc. and V. stipulacea (Lam.) Kuntz in India. Genet. Resour. Crop Evol. 66, 1155–1165. doi: 10.1007/s10722-019-00767-9
Gujaria-Verma, N., Ramsay, L., Sharpe, A. G., Sanderson, L. A., Debouck, D. G., Tar’an, B., et al. (2016). Gene-based SNP discovery in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris) for diversity analysis and comparative mapping. BMC Genom. 17:239. doi: 10.1186/s12864-016-2499-3
Gupta, S., and Singh, B. B. (2009). Utilization of potential germplasm and species spectrum in improvement of pulse crops. In: M. Ali, S. Gupta, P.S. Basu, and K. Naimuddin (eds). Legumes For Ecological Sustainability. Kanpur, India: Indian Institute of Pulse Research. 332–341.
Gupta, D., and Sharma, S. K. (2006). Evaluation of wild Lens taxa for agro-morphological traits, fungal diseases and moisture stress in northwestern Indian hills. Genet. Resour. Crop Evol. 53, 1233–1241. doi: 10.1007/s10722-005-2932-y
Gupta, S., Kumar, S., and Singh, B. B. (2004). Relative Genetic Contributions of Ancestral Lines of Indian Mungbean Cultivars Based on Coefficient of Parentage Analysis. Indian J. Genet. 64, 299–302.
Haggard, J. E. (2007). Characterization of physiological resistance to white mold and search for molecular markers linked to resistance via advanced backcross QTL analysis in an interspecific cross between Phaseolus coccineus and P. vulgaris. HortSci 41:973.
Haghighi, K. R., and Ascher, P.D. (1988). Fertile, intermediate hybrids between Phaseolus vulgaris and P. acutifolius from congruity backcrossing. Sex. Plant Reprod. 1, 51–58.
Hamdi, A., and Erskine, W. (1996). Reaction of wild species of the genus Lens to drought. Euphytica 91, 173–179.
Hamdi, A., Küsmenoglu, I., and Erskine, W. (1996). Sources of winter hardiness in wild lentil. Genet. Resour. Crop Evol. 43, 63–67. doi: 10.1007/bf00126942
Harouna, D. V., Venkataramana, P. B., Matemu, A. O., and Ndakidemi, P. A. (2020). Agro-morphological exploration of some unexplored wild Vigna legumes for domestication. Agronomy 10:111. doi: 10.3390/agronomy10010111
Haware, M. P., Nene, Y. L., Pundir, R. P. S., and Narayana, R. J. (1992). Screening of world chickpea germplasm for resistance to fusarium wilt. Field Crops Res. 30, 147–154. doi: 10.1016/0378-4290(92)90063-f
He, Q., Yang, H., Xiang, S., Wang, W., Xing, G., Zhao, T., et al. (2014). QTL mapping for the number of branches and pods using wild chromosome segment substitution lines in soybean [Glycine max (L.) Merr.]. Plant Genet. Resour. 12:S172.
Hingane, A. J., Saxena, K. B., Patil, S. B., Sultana, R., Srikanth, S., Mallikarjuna, N., et al. (2015). Mechanism of water-logging tolerance in pigeonpea. Indian J. Genet. 75, 208–214. doi: 10.5958/0975-6906.2015.00032.2
Hospital, F. (2001). Size of donor chromosome segments around introgressed loci and reduction of linkage drag in marker-assisted backcross programs. Genetics 158, 1363–1379.
Hospital, F., and Charcosset, A. (1997). Marker-assisted introgression of quantitative trait loci. Genetics 147, 1469–1485.
Hospital, F., Chevalet, C., and Mulsant, P. (1992). Using markers in gene introgression breeding programs. Genetics 132, 1199–1210.
Imtiaz, M., Malhotra, R. S., and Yadav, S. S. (2011). Genetic adjustment to changing climates: chickpea. (eds). S. S. Yadav, R. Redden, J. L. Hatfield, H. Lotze-Campen, and A. Hall. In Crop Adaptation to Climate Change. Oxford: John Wiley and Sons. 251–268. doi: 10.1002/9780470960929.ch18
Infantino, A., Porta-Puglia, A., and Singh, K. B. (1996). Screening wild Cicer species for esistance to fusarium wilt. Plant Dis. 80, 42–44. doi: 10.1094/pd-80-0042
Iseki, K., Takahashi, Y., Muto, C., Naito, K., and Tomooka, N. (2016). Diversity and evolution of salt tolerance in the genus Vigna. PLoS One 11:e0164711. doi: 10.1371/journal.pone.0164711
Isenegger, D.A., Ford, R., and Taylor, P.W. (2011). Disease reaction of chickpea (Cicer spp.) genotypes to Botrytis grey mould (Botrytis cinerea). Aus. Plant Pathol. 40:583. doi: 10.1007/s13313-011-0081-7
Jain, H. K. (1975). Breeding for yield and other attributes in food legumes. Ind. J. Genet. 35, 169–187.
Jarvis, D. I., and Hodgkin, T. (1998). Wild relatives and crop cultivars: conserving the connection. (eds). N. Zencirci, Z. Kaya, Y. Anikster, and W. T. Adams. In: The Proceedings of an International Symposium on in Situ Conservation of Plant Genetic Diversity. pp. 73–80. Turkey: Central Research Institute for Field Crops.
Jarvis, D. I., and Hodgkin, T. (1999). Wild relatives and crop cultivars: detecting natural introgression and farmer selection of new genetic combinations in agroecosystems. Mol. Ecol. 8, S159–S173.
Kaiser, W. J., Alcala-Jimenez, A. R., Hervas-Vargas, A., Trapero-Casas, J. L., and Jimenez-Diaz, R. M. (1994). Screening of wild Cicer species for resistance to race 0 and 5 of Fusarium oxysporum f. sp. ciceris. Plant Dis. 78, 962–967.
Kashiwagi, J., Krishnamurthy, L., Gaur, P. M., Upadhyaya, H. D., Varshney, R. K., and Tobita, S. (2013). Traits of relevance to improve yield under terminal drought stress in chickpea (C. arietinum L.). Field Crops Res. 145, 88–95. doi: 10.1016/j.fcr.2013.02.011
Khoury, C. K., Castañeda-Alvarez, N. P., Achicanoy, H. A., Sosa, C. C., Bernau, V., Kassa, M. T., et al. (2015). Crop wild relatives of pigeonpea [Cajanus cajan (L.) Millsp.]: distributions, ex situ conservation status, and potential genetic resources for abiotic stress tolerance. Biol. Conserv. 184, 259–270. doi: 10.1016/j.biocon.2015.01.032
Knights, E. J., Southwell, R. J., Schwinghamer, M. W., and Harden, S. (2008). Resistance to Phytophthora medicaginis Hansen and Maxwell in wild Cicer species and its use in breeding root rot resistant chickpea (Cicer arietinum L.). Aus. J. Agric. Res. 59, 383–387. doi: 10.1071/ar07175
Koona, P., Osisanya, E. O., Jackai, L. E. N., Tamo, M., and Kham, R. H. (2002). Resistance in accessions of cowpea to the coreid pod bug Clavigralla tomentosicillis (Hemiptera: coreidae). J. Econ Entomol. 95, 1281–1288. doi: 10.1603/0022-0493-95.6.1281
Konarev, A. V., Tomooka, N., and Vaughan, D. A. (2002). Proteinase inhibitor polymorphism in the genus Vigna Savi subgenus Ceratotropis and its biosystematic implications. Euphytica 123, 165–177.
Konečná, E., Šafářová, D., Navratil, M., Hanáček, P., Coyne, C., Flavell, A., et al. (2014). Geographical gradient of the eIF4E alleles conferring resistance to potyviruses in pea (Pisum) germplasm. PLoS One 9:e90394. doi: 10.1371/journal.pone.0090394
Koumproglou, R., Wilkes, T. M., Townson, P., Wang, X. Y., Beynon, J., Pooni, H. S., et al. (2002). STAIRS: a new genetic resource for functional genomic studies of Arabidopsis. Plant J. 31, 355–364. doi: 10.1046/j.1365-313x.2002.01353.x
Krishnamurthy, L., Kashiwagi, J., Upadhyaya, H. D., Gowda, C. L. L., Gaur, P. M., Singh, S., et al. (2013). Partitioning coefficient—A trait that contributes to drought tolerance in chickpea. Field Crops Res. 149, 354–365. doi: 10.1016/j.fcr.2013.05.022
Kumar, D., and Kirti, P. B. (2015). Transcriptomic and proteomic analyses of resistant host responses in Arachis diogoi challenged with late leaf spot pathogen, Phaeoisariopsis personata. PLoS One 10:e0117559. doi: 10.1371/journal.pone.0117559
Kumar, N. P., Pandiyan, M., and Veerabadhiran, P. (2007). Pre-fertilization barriers in Vigna radiata × Vigna umbellata. Plant Arch. 7, 377–380.
Kumar, J., Srivastava, E., Singh, M., Mahto, D., Pratap, A., and And Kumar, S. (2014). “Lentil,” in Alien Gene Transfer in Crop Plants, Volume 2: Achievements and Impacts, eds A. Pratap and J. Kumar (New York: Springer), 191–206.
Kumar, S., Choudhary, A. K., Rana, K. S., Sarker, A., and Singh, M. (2018). Bio-fortification potential of global wild annual lentil core collection. PLoS One 13:e0191122. doi: 10.1371/journal.pone.0191122
Kumar, S., Imtiaz, M., Gupta, S., and Pratap, A. (eds.). (2011). Distant hybridization and alien gene introgression. Biology and Breeding of Food Legumes. Oxfordshire, UK: CABI. 81–110.
Kumar, S., Gupta, P., Choukri, S., and Siddique, K. H. M. (2020). Efficient breeding of pulse crops. (eds). S. S. Gosal and S. H. Wani. In: Accelerated Plant Breeding, Volume 3-Food Legumes. Switzerland: Springer Nature.
Kumar, S., Gupta, S., Chandra, S., and Singh, B. B. (2004). How wide is the genetic base of pulse crops? In: M. Ali, B. B. Singh, S. Kumar, and V. Dhar (eds), Pulses in New Perspective. Kanpur, India: Indian Institute of Pulse Research. 211–221.
Kumar, S., Rakshit, S., and Gupta, S. (2003). Genetics and cytogenetics of chickpea. In: M. Ali, S. Kumar, and N. B. Singh (eds). Chickpea Research in India. Kanpur, India: Indian Institute of Pulse Research. 31–37.
Kumar, S., Sandhu, J. S., Gupta, S. K., Dikshit, H. K., and Gupta, S. (2009). Broadening genetic base of pulse crops: present status and future strategies. In: M. Ali, S. Gupta, P.S. Basu, and K. Naimuddin (eds), Legumes for Ecological Sustainability. Kanpur, India: Indian Institute of Pulse Research. 332–341.
Kushida, A., Tazawa, A., Aoyama, S., and Tomooka, N. (2012). Novel sources of resistance to the soybean cyst nematode (Heterodera glycines) found in wild relatives of azuki bean (Vigna angularis) and their characteristics of resistance. Genet. Resour. Crop Evol. 60, 985–994. doi: 10.1007/s10722-012-9895-6
Ladizinsky, G. (1985). The genetics of hard seed coat in the genus Lens. Euphytica 34, 539–543 doi: 10.1007/bf00022952
Ladizinsky, G., and Adler, A. (1976). Genetic relationships among the annual species of Cicer L. Theor. Appl. Genet. 48, 197–203. doi: 10.1007/bf00527371
Laserna-Ruiz, I., De-Los-Mozos-Pascual, M., Santana-Meridas, O., Sanchez-Vioque, R., and Rodrıguez-Conde, M. F. (2012). Screening and selection of lentil (Lens Miller) germplasm resistant to seed bruchids (Bruchus spp.). Euphytica 188, 153–162. doi: 10.1007/s10681-012-0752-7
Li, D. D., Pfeiffer, T. W., and Cornelius, P. L. (2008). Soybean QTL for yield and yield components associated with Glycine soja alleles. Crop Sci. 48, 571–581 doi: 10.2135/cropsci2007.06.0361
Li, Y., Zheng, L., Corke, F., Smith, C., and Bevan, M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22, 1331–1336. doi: 10.1101/gad.463608
Lílian, S. T., Carmo Andressa, C. Q., Martins Cinthia, C. C., Martins Mário, A. S., Passos Luciano, P., Silva Ana, C. G., et al. (2019). Comparative proteomics and gene expression analysis in Arachis duranensis reveal stress response proteins associated to drought tolerance. J. Proteomics 192, 299–310. doi: 10.1016/j.jprot.2018.09.011
Liu, S., Zhou, R., Dong, Y., Li, P., and Jia, J. (2006). Development, utilization of introgression lines using a synthetic wheat as donor. Theor. Appl. Genet. 112, 1360–1373. doi: 10.1007/s00122-006-0238-x
Lulsdorf, M., Mallikarjuna, N., Clarke, H., and Tar’An, B. (2005). “Finding Solutions For Interspecific Hybridization Problems In Chickpea (Cicer arietinum L.),”. in 4th International Food Legumes Research Conference. (New Delhi: IFLRC).
Mallikarjuna, N., and Jadhav, D. R. (2008). Techniques to produce hybrids between Cicer arietinum L. x Cicer pinnatifidum Jaub. Ind. J. Genet. 68, 398–405.
Mallikarjuna, N., and Saxena, K. B. (2005). A new cytoplasmic nuclear male-sterility system derived from cultivated pigeonpea cytoplasm. Euphytica 142, 143–148. doi: 10.1007/s10681-005-1199-x
Mallikarjuna, N., Jadhav, D. R., Nagamani, V., Amudhavalli, C., and Hoisington, D. A. (2007). Progress in interspecific hybridization between Cicer arietinum and wild species C. bijugum. J. SAT Agric. Res. 5, 1–2. doi: 10.1007/978-3-319-23534-9_1
Mallikarjuna, N., Saxena, R. K., Gowda, M. B., and Varshney, R. K. (2017). Wide Crossing Technology for Pigeonpea Improvement the Pigeonpea Genome. Chamsford: Springer, 31–39.
Malhotra, R. S., Singh, K. B., Di Vito, M., Greco, N., and Saxena, M. C. (2002). Registration of ILC 10765 and ILC 10766 chickpea germplasm lines resistant to cyst nematode. Crop Sci. 42, 1756–1756. doi: 10.2135/cropsci2002.1756
Mallikarjuna, N., Senapathy, S., Jadhav, D. R., Saxena, K. B., Sharma, H. C., Upadhyaya, H. D., et al. (2011). Progress in the utilization of Cajanus platycarpus (Benth.) Maesen in pigeonpea improvement. Plant Breed. 130, 507–514 doi: 10.1111/j.1439-0523.2011.01870.x
Mallikarjuna, N., Senthilvel, S., and Hoisington, D. (2012). Development of new sources of tetraploid Arachis to broaden the genetic base of cultivated groundnut (Arachis hypogaea L.). Genet. Resour. Crop. Evol. 58, 889–907. doi: 10.1007/s10722-010-9627-8
Manavalan, L. P., Guttikonda, S. K., Tran, L. P., and Nguyen, H. T. (2009). Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell. Physiol. 50, 1260–1276. doi: 10.1093/pcp/pcp082
Manavalan, L.P., Prince, S. J., Musket, T. A., Chaky, J., Deshmukh, R., Vuong, T. D., et al. (2015). Identification of novel QTL governing root architectural traits in an interspecific soybean population. PLoS One 10:e0120490. doi: 10.1371/journal.pone.0120490
Mannur, D. M., Babbar, A., Thudi, M., Sabbavarapu, M. M., Roorkiwal, M., Sharanabasappa, B. Y., et al. (2019). Super Annigeri 1 and improved JG 74: two Fusarium wilt-resistant introgression lines developed using marker-assisted backcrossing approach in chickpea (Cicer arietinum L.). Mol. Breed. 39:2.
Matus, I., Corey, A., Filchkin, T., Hayes, P. M., Vales, M. I., Kling, J., et al. (2003). Development and characterization of recombinant chromosome substitution lines (RCSLs) using Hordeum vulgare subsp. vulgare background. Genome 46, 1010–1023. doi: 10.1139/g03-080
Miklas, P.N., Zapata, M., Beaver, J.S., and Grafton, K.F. (1999). Registration of four dry bean germplasms resistant to common bacterial blight: ICB-3, ICB-6, ICB-8 and ICB-10. Crop Sci. 39:594. doi: 10.2135/cropsci1999.0011183x003900020065x
Miyagi, M., Humphry, M. E., Ma, Z.Y., Lambrides, C. J., Bateson, M., Liu, C. J., et al. (2004). Construction of bacterial artificial chromosome libraries and their application in developing PCR-based markers closely linked to a major locus conditioning bruchid resistance in mungbean (Vigna radiata L. Wilczek). Theor. Appl. Genet. 110, 151–156. doi: 10.1007/s00122-004-1821-7
Monforte, A. J., and Tanksley, S. D. (2000). Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43, 803–813. doi: 10.1139/gen-43-5-803
Mwale, S. E., Shimelis, H., Mafongoya, P., and Mashilo, J. (2020). Breeding tepary bean (Phaseolus acutifolius) for drought adaptation: a review. Plant Breed. 139, 821–833. doi: 10.1111/pbr.12806
Nagaraj, N. C., Muniyappa, V., Satyan, B. A., Shanmugam, N., Jayarajan, R., and Vidhyasekaran, P. (1981). Resistance source for mungbean yellow mosaic virus. In Proceedings of the National Seminar on Disease Resistance in Crop Plants. 69–72.
Naim-Feil, E., Toren, M., Aubert, G., Rubinstein, M., Rosen, A., Eshed, R., et al. (2017). Drought response and genetic diversity in Pisum fulvum, a wild relative of domesticated pea. Crop Sci. 57, 1145–1159. doi: 10.2135/cropsci2016.10.0880
Nair, R. M., Schafleitner, R., Kenyon, L., Srinivasan, R., Easdown, W., Ebert, A.W., et al. (2013). Gnetic improvement of mungbean. SABRAO J. Breed. Genet. 44, 177–190.
Ngugi-Dawit, A., Hoang, T. M. L., Williams, B., Higgins, T. J., and Mundree, S. G. (2020). A wild Cajanus scarabaeoides (L.), Thouars, IBS 3471, for improved insect-resistance in cultivated pigeonpea. Agronomy 10:5.
Oghiakhe, S., Jackai, L. E. N., and Makanjuola, W. A. (1992). A rapid visual field screening technique for resistance of cowpea (Vigna unguiculata) to the legume pod borer Maruca testulalis (Lepidoptera: Pyralidae). Bull. Entomol. Res. 82, 507–512. doi: 10.1017/s0007485300042589
Oigiangbe, O. N., Jackai, L. E. N., Ewete, F. K., Hughes, J., and Lajide, L. (2002). Reduced consumption and use of pods of Vigna species (Leguminosae) by Maruca vitrata (Lepidoptera: Pyralidae). African Entomol. 10(2), 333–340.
Pal, S. S., Singh, J. J., and Singh, I. (2000). Transfer of YMV resistance in cultivar SML32 of Vigna radiata from other related Vigna species. Plant Dis. Res. 15, 67–69.
Pande, S., Ramgopal, D., Kishore, G. K., Mallikarjuna, N., Sharma, M., Pathak, M., et al. (2006). Evaluation of wild Cicer species for resistance to ascochyta blight and botrytis gray mold in controlled environment at icrisat, patancheru, india. J. SAT Agric. Res. 13:25-26.
Pandiyan, M., Ramamoorthi, N., Ganesh, S. K., Jebraj, S., Pagarajan, P., Balasubramanian, P., et al. (2008). Broadening the genetic base and introgression of MYMY resistance and yield improvement through unexplored genes from wild relatives in mungbean. Plant Mutation Rep. 2, 33–38.
Pillen, K., Zacharias, A., and Léon, J. (2003). Advanced backcross QTL analysis in barley (Hordeum vulgare L.). Theor. Appl. Genet. 107, 340–352. doi: 10.1007/s00122-003-1253-9
Podder, R., Banniza, S., and Vandenberg, A. (2013). Screening of wild and cultivated lentil germplasm for resistance to stemphylium blight. Plant Genet. Resour. 11:26. doi: 10.1017/s1479262112000329
Porch, T. G., Cichy, K., Wang, W., Brick, M., Beaver, J. S., Santana-Morant, D., et al. (2017). Nutritional composition and cooking characteristics of tepary bean (Phaseolus acutifolius Gray) in comparison with common bean (Phaseolus vulgaris L.). Genetic Resour. Crop Evol. 64, 935–953. doi: 10.1007/s10722-016-0413-0
Polhill, R. M., and Raven, P. H. (1981). Advances in Legume Systematics, Part 1 and 2. Kew, UK: Royal Botanical Garden.
Pratap, A., Gupta, S. K., Kumar, J., and Solanki, R. K. (2012). Soybean. In: S.K. Gupta (Ed.) Technological Innovations In Major World Oil Crops, Vol.I Breeding. Germany: Springer Science + Business Media. 293–321.
Pratap, A., and Gupta, S. K. (2009). Biotechnological Interventions in Host Plant Resistance, eds R. Peshin and A. K. Dhawan in Integrated Pest Management: Innovation, Dissemination and Impact. Dordrecht: Springer Publishers. 183–207. doi: 10.1007/978-1-4020-8992-3_8
Pratap, A., Gupta, D. S., Singh, B. B., and Kumar, S. (2013). IPM 205-7 (IC0589309-IC0589310; INGR11043-INGR11044), a mungbean (Vigna radiata (L.) Wilczek) germplasm with Super Early Maturity. Indian J. Plant Genet. Resour. 26, 89–90.
Pratap, A., Gupta, S., Basu, P. S., Tomar, R., Dubey, S., Rathore, M., et al. (2019). “Towards development of climate smart mungbean: challenges and opportunities,” in Genomic Designing of Climate-Smart Pulse Crops, ed. C. Kole (Chamsford, UK: Springer), 235–264. doi: 10.1007/978-3-319-96932-9_5
Pratap, A., and Kumar, J. (2011). History origin and evolution. (eds). A.Pratap, J.Kumar. In Biology and Breeding of Food Legumes. Oxfordshire: CABI.
Pratap, A., Basu, P. S., Gupta, S., Malviya, N., Rajan, N., Tomar, R., et al. (2014). Identification and characterization of sources for photo- and thermo-insensitivity in Vigna species. Plant Breed. 133, 756–764. doi: 10.1111/pbr.12215
Pratap, A., Chaturvedi, S.K., Tomar, R., Rajan, N., Malviya, N., Thudi, M., et al. (2017). Marker-assisted introgression of resistance to Fusarium wilt race 2 in Pusa 256, an elite cultivar of desi chickpea. Mol. Genet. Gen. 292, 1237–1245. doi: 10.1007/s00438-017-1343-z
Pratap, A., Choudhary, A. K., and Kumar, J. (2010). In vitro techniques towards genetic enhancement of food legumes-a review. J. Food Legum. 23, 169–185.
Pratap, A., Douglas, C., Prajapati, U., Kumari, G., War, A. R., Tomar, R., et al. (2020). Breeding progress and future challenges: Biotic stresses. In: R.M. Nair, R. Schafleitner, and S. H. Lee (Eds.) The Mungbean Genome. Switzerland: Springer Nature. 55–80. doi: 10.1007/978-3-030-20008-4_5
Pratap, A., Gupta, S., Malviya, N., Rajan, N., Tomar, R., Latha, M., et al. (2015a). Genome scanning of asiatic vigna species for discerning population genetic structure based on microsatellite variation. Mol. Breed. 35:178.
Pratap, A., Malviya, N., Gupta, S., Tomar, R., Pandey, V. R., Prajapati, U., et al. (2018a). Field characterization of endemic wild Vigna accessions collected from biodiversity hotspots of India to identify promising genotypes for multiple agronomic and adaptive traits. Legume Res. 41, 490–499.
Pratap, A., Prajapati, U., Singh, C. M., Gupta, S., Rathore, M., Malviya, N., et al. (2018b). Potential, constraints and applications of in vitro methods in improving food legumes. Plant Breed. 137, 235–249. doi: 10.1111/pbr.12590
Pratap, A., Tomar, R., Kumar, J., Pandey, V. R., Menhandi, S., Katiyar, P. K., et al. (2015b). High throughput plant phenotyping platforms. In: J. Kumar, A. Pratap, and S. Kumar (Eds.). Phenomics Of Crop Plants: Trends, Options And Limitations. New Delhi: Springer (India) Pvt. Ltd. 285–296 doi: 10.1007/978-81-322-2226-2_19
Price, A. H., Townend, J., Jones, M. P., Audebert, A., and Courtois, B. (2002). Mapping QTLs associated with drought avoidance in upland rice grown in the philippines and west africa. Plant Mol. Biol. 48, 683–695.
Pundir, R. P. S., and Mengesha, M. H. (1995). Cross compatibility between chickpea and its wild relative, Cicer echinospermum Davis. Euphytica 83, 241–245. doi: 10.1007/bf01678136
Ramgopal, D., Srivastava, R. K., Pande, S., Rathore, A., Jadhav, D. R., Sharma, M., et al. (2012). Introgression of Botrytis gray mold resistance gene from Cicer reticulatum (bgmr1cr) and C. echinospermum (bgmr1ce) to chickpea. Plant Genet. Resour. 11, 212–216. doi: 10.1017/s1479262112000470
Reddy, K. R., and Singh, D. P. (1990). Inheritance of resistance to mungbean yellow mosaic virus in blackgram. Madras Agric. J. 80, 199–201.
Reddy, M. V., and Singh, K. B. (1993). Rate reducing resistance to Ascochyta blight in chickpeas. Plant Dis. 77, 231–233. doi: 10.1094/pd-77-0231
Reen, R., Mumford, M., and Thompson, J. P. (2019). Novel sources of resistance to root-lesion nematode (Pratylenchus thornei) in a new collection of wild Cicer species (C. reticulatum and C. echinospermum) to improve resistance in cultivated chickpea C. arietinum. Phytopathology 109, 1270–1279. doi: 10.1094/phyto-02-19-0047-r
Robertson, L. D., Singh, K. B., and Ocampo, B. (1995). A Catalog of Annual Cicer Species. Aleppo: ICARDA.
Saeed, A., Darvishzadeh, R., Hovsepyan, H., and Asatryan, A. (2010). Tolerance to freezing stress in Cicer accessions under controlled and field conditions. African J. Biotech. 18, 2618–2626.
Saeed, A., and Darvishzadeh, R. (2017). Association analysis of biotic and abiotic stresses resistance in chickpea (Cicer s) using AFLP markers. Biotechnol. Equip. 31, 698–708. doi: 10.1080/13102818.2017.1333455
Saxena, K. B., and Kumar, R. V. (2003). Development of a cytoplasmic nuclear male-sterility system in pigeonpea using C. scarabaeoides (L.) Thouars. Ind. J. Genet. Plant Breed. 63, 225–229.
Saxena, K. B., Choudhary, A. K., Saxena, R. K., and Varshney, R. K. (2018). Breeding pigeonpea cultivars for intercropping: synthesis and strategies. Breed. Sci. 68, 159–167. doi: 10.1270/jsbbs.17105
Saxena, K. B., Kumar, R. V., Dalvi, V. A., Mallikarjuna, N., Gowda, C. L. L., Singh, B. B., et al. (2005). Hybrid breeding in food legumes– a success story of pigeonpea. (eds). M. C. Khairwal and H. K. Jain. in Proceedings Of The International Food Legumes Research Conference. (New Delhi: IFLRC-IV).
Saxena, K. B., Singh, L., Kumar, R. V., and Rao, A. N. (1996). “Development of cytoplasmic-genic male-sterility (CMS) system in pigeonpea at ICRISAT Asia Center,” in Proceedings of the Working Group on Cytoplasmic-Genic Male-Sterility (CMS) in Pigeonpea, (Andhra Pradesh: ICRISAT Center), 9–10.
Saxena, K. B., Sultana, R., Mallikarjuna, N., Saxena, R. K., Kumar, R. V., Sawargaonkar, S. L., et al. (2010). Male-sterility systems in pigeonpea and their role in enhancing yield. Plant Breed. 129, 125–134. doi: 10.1111/j.1439-0523.2009.01752.x
Schafleitner, R., Huang, S. M., Chu, S. H., Yen, J. Y., Lin, C. Y., Yan, M. R., et al. (2016). Identification of single nucleotide polymorphism markers associated with resistance to bruchids (Callosobruchus s) in wild mungbean (Vigna radiata var. sublobata) and cultivated V. radiata through genotyping by sequencing and quantitative trait locus analysis. BMC Plant Biol. 16:159. doi: 10.1186/s12870-016-0847-8
Sekhon, J., Grewal, S. K., Singh, I., and Kaur, J. (2017). Evaluation of nutritional quality and antioxidant potential of pigeonpea genotypes. J. Food Sci. Tech. 54, 3598–3611. doi: 10.1007/s13197-017-2818-y
Sen Gupta, D., Thavarajah, D., McGee, R. J., Coyne, C. J., Kumar, S., and Thavarajah, P. (2016). Genetic diversity among cultivated and wild lentils for iron, zinc, copper, calcium and magnesium concentrations. Austr. J. Crop Sci. 10:1381.
Shah, T. M., Hassan, M., Haq, M. A., Atta, B. M., Alam, S. S., Ali, H., et al. (2005). Evaluation of Cicer species for resistance to Ascochyta blight. Pakistan J. Bot. 37, 431–438.
Sharma, S., and Upadhyaya, H. D. (2015). Vernalization and photoperiod response in annual wild Cicer species and cultivated chickpea. Crop Sci. 55, 2393–2400. doi: 10.2135/cropsci2014.09.0598
Sharma, S., and Upadhyaya, H. D. (2019). Photoperiod response of annual wild Cicer species and cultivated chickpea on phenology, growth, and yield traits. Crop Sci. 59, 632–639. doi: 10.2135/cropsci2018.07.0438
Sharma, K. D., Winter, P., Kahl, G., and Muehlbauer, F. J. (2004). Molecular mapping of Fusarium oxysporum f. sp. ciceris race 3 resistance gene in chickpea. Theor. Appl. Genet. 108, 1243–1248. doi: 10.1007/s00122-003-1561-0
Sharma, R., Rawat, V., and Suresh, C. G. (2017). Genome-wide identification and tissue-specific expression analysis of nucleotide binding site-leucine rich repeat gene family in Cicer arietinum (kabuli chickpea). Genomics Data 14, 24–31. doi: 10.1016/j.gdata.2017.08.004
Sharma, S. S., Islam, M. A., Malik, A. A., Kumar, K., Negi, M. S., Tripathi, S. B., et al. (2016). Seed traits, fatty acid profile and genetic diversity assessment in Pongmia pinnata (L.) Pierre germplasm. Physiol. Mol. Biol. Plants 22, 193–205. doi: 10.1007/s12298-016-0356-0
Shoemaker, R. C. (1986). Chloroplast DNA variation in the genus Glycine subgenus soja. J. Hered. 77, 26–30. doi: 10.1093/oxfordjournals.jhered.a110161
Sinclair, T. R., Purcell, L. C., King, C. A., Sneller, C. H., Chen, P., Vadez, V., et al. (2007). Drought tolerance and yield increase of soybean resulting from improved symbiotic N fixation. Field Crops Res. 101, 68–71. doi: 10.1016/j.fcr.2006.09.010
Singh, M., Rana, M. K., Kumar, K., Bisht, I. S., Dutta, M., Gautam, N. K., et al. (2013). Broadening the genetic base of lentil cultivars through inter-sub-specific and interspecific crosses of Lens taxa. Plant Breed. 132, 667–675. doi: 10.1111/pbr.12089
Singh, B. B., and Dikshit, H. K. (2002). Possibilities and limitations of interspecific hybridization involving green gram (Phaseolus radiatus) and black gram (Phaseolus mungo). Indian J. Agril. Sci. 72, 676–678.
Singh, D. P. (1990). Distant hybridization in genus Vigna: a review. Ind. J. Genet. Plant Breed. 50, 268–276.
Singh, K. B., and Ocampo, B. (1993). Interspecific hybridization in annual Cicer species. J. Genet. Breed. 47, 199–204.
Singh, K. B., Malhotra, R. S., and Saxena, M. C. (1990). Sources for tolerance to cold in Cicer species. Crop Sci. 30, 1136–1138. doi: 10.2135/cropsci1990.0011183x003000050036x
Singh, K. B., Ocampo, B., and Robertson, L. D. (1998). Diversity for abiotic and biotic stress resistance in the wild annual Cicer species. Gene. Res. Crop Evol. 45, 9–17. doi: 10.1023/A:1008620002136
Singh, K. P., Monika, Sareen, P. K., and Kumar, A. (2003). Interspecific hybridization studies in Vigna radiata L. Wilczek and Vigna umbellata L. Nat. J. Pl. Improve. 5, 16–18.
Singh, M., Kumar, S., Basandrai, A. K., Basandrai, D., Malhotra, N., Saxena, D. R., et al. (2020). Evaluation and identification of wild lentil accessions for enhancing genetic gains of cultivated varieties. PLoS One 15:e0229554. doi: 10.1371/journal.pone.0229554
Singh, M., Mishra, J. S., and Bhatt, B. P. (2017). Effect of integrated nutrient management on production potential and quality of summer mungbean (Vigna radiata L.). J. Krishi Vigyan 5, 39–45. doi: 10.5958/2349-4433.2017.00009.5
Singh, N. P., Singh, A., Asthana, A. N., and Singh, A. (1999). Studies on inter-specific crossability barriers in chickpea. Ind. J. Pulses Res. 12, 13–19.
Singh, R. P., and Singh, B. D. (1989). Recovery of rare interspecific hybrids of gram Cicer arietinum × C. cuneatum L. through tissue culture. Curr. Sci. 58, 874–876.
Singh, S. P., Debouck, D. G., and Roca, W. W. (1998). Interspecific hybridization between Phaseolus vulgaris L. and P. parvifolius Freytag. Annu. Rep. Bean Improv. Coop. 4, 7–8.
Singh, S., Gumber, R. K., Joshi, N., and Singh, K. (2005). Introgression from wild Cicer reticulatum to cultivated chickpea for productivity and disease resistance. Plant Breed. 124, 477–480. doi: 10.1111/j.1439-0523.2005.01146.x
Singh, S.P., Debouck, D.G., and Roca, W.W. (1997). Successful interspecific hybridization between Phaseouls vulgaris L. and P. costaricensis Freytag & Debouck. Annu. Rep. Bean Improv. Coop. 40, 40–41.
Singh, S.P., Terán, H., and Beaver, J.S. (2009). Scarlet runner bean germplasm accessions G 35006 and G 35172 possess resistance to multiple diseases of common bean. Annu. Rep. Bean Improv. Coop. 52, 22–23.
Somta, C., Somta, P., Tomooka, N., Ooi, P. C., Vaughan, D. A., and Srinives, P. (2008). Characterization of new sources of mungbean (Vigna radiata (L.) Wilczek) resistance to bruchids, Callosobruchus s (Coleoptera: Bruchidae). J. Stored Products Res. 44, 316–321. doi: 10.1016/j.jspr.2008.04.002
Somta, P., Kaga, A., Tomooka, N., Kashiwaba, K., Isemura, T., Chaitieng, B., et al. (2006). Development of an interspecific vigna linkage map between Vigna umbellata (Thunb.) Ohwi & Ohashi and V. nakashimae (Ohwi) Ohwi & Ohashi and its use in analysis of bruchid resistance and comparative genomics. Plant Breed. 125, 77–84. doi: 10.1111/j.1439-0523.2006.01123.x
Souter, J. R., Gurusamy, V., Porch, T. G., and Bett, K. E. (2017). Successful introgression of abiotic stress tolerance from wild tepary bean to common bean. Crop Sci. 57, 1160–1171. doi: 10.2135/cropsci2016.10.0851
Stamigna, C., Crino, P., and Saccordo, F. (2000). Wild relatives of chickpea: multiple disease resistance and problems to introgression in the cultigen. J. Genet. Breed. 54, 213–219.
Sullivan, J. G., and Bliss, F. A. (1983). Expression of enhanced seed protein content in inbred backcross lines of common bean. J. Am. Soc. Hortic. Sci. 108, 787–791.
Tahir, M., Lindeboom, N., Båga, M., Vandenberg, A., and Chibbar, R. (2011). Composition and correlation between major seed constituents in selected lentil (Lens culinaris. Medik) genotypes. Can. J. Plant Sci. 91, 825–835. doi: 10.4141/cjps2011-010
Tanksley, S. D., and McCouch, S. R. (1997). Seed banks and molecular maps, unlocking genetic potential from the wild. Science 277, 1063–1066. doi: 10.1126/science.277.5329.1063
Tanksley, S. D., and Nelson, J. C. (1996). Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 92, 191–203. doi: 10.1007/s001220050114
Tanksley, S. D., Young, N. D., Paterson, A. H., and Bonierbale, M. W. (1989). RFLP mapping in plant breeding: new tools for an old science. Biotechnology 7, 257–264. doi: 10.1038/nbt0389-257
Tian, F., De, J. L., Fu, Q., Zuo, F. Z., Yong, C. F., Xiang, K. W., et al. (2006). Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet. 112, 570–580. doi: 10.1007/s00122-005-0165-2
Tikka, S. B. S., Parmar, L. D., and Chauhan, R. M. (1997). First record of cytoplasmic-genic male sterility system in pigeonpea (Cajanus cajan (L.) Millsp.) through wide hybridization. Gujarat Agric. Univ. Res. J. 22, 160–162.
Toker, C. (2005). Preliminary screening and selection for cold tolerance in annual wild Cicer species. Genetic. Res. Crop. Evol. 52, 1–5. doi: 10.1007/s10722-005-1743-5
Toker, C., Canci, H., and Yildirim, T. (2007a). Evaluation of perennial wild Cicer species for drought resistance. Genetic. Res. Crop. Evol. 54, 1781–1786. doi: 10.1007/s10722-006-9197-y
Toker, C., Lluch, C., Tejera, N. A., Serraj, R., and Siddique, K. H. M. (2007b). Abiotic Stresses. (eds). S.S. Yadav, R. Redden, W. Chen, and B. Sharma. In Chickpea Breeding and Management. 474–496. Wallingford: CAB International.
Tomooka, N., Kaga, A., and Vaughan, D. A. (2006). The Asian Vigna (Vigna subgenus Ceratotropis) biodiversity and evolution. In: AK Sharma and A. Sharma (eds). Plant genome: biodiversity and evolution. Part C Phanerogams (angiosperms-dicotyledons). 1. Enfield: Science Publishers. 87–126.
Tomooka, N., Kashiwaba, K., Vaughan, D. A., Ishimoto, M., and Egawa, Y. (2000). The effectiveness of evaluating wild species: searching for sources of resistance to bruchid beetle in the genus Vigna subgenus Ceratotropis. Euphytica 115, 27–41.
Tomooka, N., Lairungruang, C., Nakeeraks, P., Egawa, Y., and Thavarasook, C. (1992). Development of bruchid resistantmungbean using wild mungbean germplasm in Thailand. Plant Breed. 109, 60–66. doi: 10.1111/j.1439-0523.1992.tb00151.x
Tomooka, N., Naito, K., Kaga, A., Sakai, H., Isemura, T., OgisoTanaka, E., et al. (2014). Evolution, domestication and neo-domestication of the genus Vigna. Plant Genet. Resour. Charact. Util. 12, 168–171.
Tomooka, N., Vaughan, D. A., Xu, R. Q., Kashiwaba, K., and Kaga, A. (2001). Japanese native Vigna genetic resources. Jpn. Agric. Res. Q. 35, 1–9. doi: 10.6090/jarq.35.1
Tripathy, S. K., Nayak, P. K., Lenka, D., Swain, D., Baisakh, B., Mohanty, P., et al. (2016). Morphological diversity of local land races and wild forms of mungbean. Legume Res. 39, 485–493.
Tullu, A., Banniza, S. B., Tar’an, T., Warkentin, A., and Vandenberg (2010). Sources of resistance to ascochyta blight in wild species of lentil (Lens culinaris Medik). Genetic Resour. Crop Evol. 57, 1053–1063. doi: 10.1007/s10722-010-9547-7
Tullu, A., Bett, K., Banniza, S., Vail, S., and Vandenberg, A. (2013). Widening the genetic base of cultivated lentil through hybridization of Lens culinaris ‘Eston’ and L. ervoides accession IG 72815. Can. J. Plant Sci. 93, 1037–1047. doi: 10.4141/cjps2013-072
Tullu, A., Buchwaldt, L., Lulsdorf, M., Banniza, S., Barlow, B., Slinkard, A. E., et al. (2006). Sources of resistance to anthracnose (Colletotrichum truncatum) in wild Lens species. Genetic. Res. Crop. Evol. 53, 111–119. doi: 10.1007/s10722-004-1586-5
Tullu, A., Diederichsen, A., Suvorova, G., and Vandenberg, A. (2011). Genetic and genomic resources of lentil: status, use and prospects. Plant Genetic. Res. Crop. Evol. 9, 19–29. doi: 10.1017/s1479262110000353
Upadhyaya, H.D., Reddy, K. N., Sharma, S., Varshney, R. K., Bhattacharjee, R., Singh, S., et al. (2011). Pigeonpea composite collection for enhanced utilization of germplasm in crop improvement programs. Plant Genet. Res. 9, 97–100. doi: 10.1017/s1479262110000419
Van Zonneveld, M., Rakha, M., yee Tan, S., Chou, Y. Y., Chang, C. H., Yen, J. Y., et al. (2020). Mapping patterns of abiotic and biotic stress resilience uncovers conservation gaps and breeding potential of Vigna wild relatives. Scientific Rep. 10, 1–11. doi: 10.9755/ejfa.v27i1.17852
Varshney, R. K., Mohan, S. M., Gaur, P. M., Chamarthi, S. K., Singh, V. K., Srinivasan, S., et al. (2014). Marker-assisted backcrossing to introgress resistance to Fusarium wilt race 1 and Ascochyta blight in C 214, an elite cultivar of chickpea. Plant Gen. 7, 1–11.
Verma, M. M., and Brar, J. S. (1996). Breeding approaches for increasing yield potential of mungbean. eds A. N. Asthana and D. H. Kim. In Recent Advances in Mungbean Research. Kanpur, India: Indian Society of Pulses Research and Development. 102–123.
Wang, B., and Chee, P. W. (2010). Application of advanced backcross quantitative trait locus (QTL) analysis in crop improvement. J. Plant Breed. crop Sci. 2, 221–232.
Wang, L. Q., Zhao, Y. F., Xue, Y. D., Zhang, Z. X., Zheng, Y. L., Chen, J. T., et al. (2007). Development and evaluation of two link-up single segment introgression lines (SSILs) in Zea mays. Acta Agron. Sin. 33, 663–668.
Wang, P., Ding, Y. Z., Lu, Q. X., Guo, W. Z., and Zhang, T. Z. (2008). Development of Gossypium barbadense chromosome segment substitution lines in the genetic standard line TM-1 of Gossypium hirsutum. Chin. Sci. Bull. 53, 1512–1517 doi: 10.1007/s11434-008-0220-x
Wang, W. B., He, Q. Y., Yang, H. Y., Xiang, S. H., Xing, G. N., Zhao, T. J., et al. (2012). Detection of wild segments associated with number of branches on main stem and leafstalk angle in soybean. Sci. Agric. Sin. 45, 4749– 4758.
Wang, W. B., He, Q. Y., Yang, H. Y., Xiang, S. H., Zhao, T. J., Xing, G. N., et al. (2013). Identification of wild segments associated with stem termination, pod color, and seed coat color in soybean. Acta Agron. Sin. 39, 1155–1163 doi: 10.3724/sp.j.1006.2013.01155
Wanjari, K. B., Patil, A. N., Manapure, P., Manjayya, J. G., and Patel, M. (1999). Cytoplasmic male sterility in pigeonpea with cytoplasm from Cajanus volubilis. Annals. Plant Physiol. 13, 170–174.
Wehrhahn, C., and Allard, R. (1965). The detection and measurement of the effects of individual genes involved in the inheritance of a quantitative character in wheat. Genetics 51, 109–119.
Wei, X. J., Liu, L. L., Xu, J. F., Jiang, L., Zhang, W. W., Wang, J. K., et al. (2010). Breeding strategies for optimumheading date using genotypic information in rice. Mol. Breed. 25, 287–298. doi: 10.1007/s11032-009-9332-5
Wilkinson, R.E. (1983). Incorporation of Phaseolus coccineus germplasm may facilitate production of high yielding P. vulgaris lines. Annu. Rep. Bean Improv. Coop. 26, 28–29.
Wilson, J. N., Chopra, R., Baring, M. R., Selvaraj, M. G., Simpson, C. E., Chagoya, J., et al. (2017). Advanced backcross quantitative trait loci (QTL) analysis of oil concentration and oil quality traits in peanut (Arachis hypogaea L.). Tropical. Plant Biol. 10, 1–17. doi: 10.1007/s12042-016-9180-5
Xiong, L. Z., and Yang, Y. N. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acidinducible mitogen-activated protein kinase. Plant Cell 15, 745–759 doi: 10.1105/tpc.008714
Yang, S., Pang, W., Ash, G., Harper, J., Carling, J., Wenzel, P., et al. (2006). Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology. Thoer. Appl. Genet. 113, 585–595. doi: 10.1007/s00122-006-0317-z
Yin, X., Struik, P., Gu, J., and Wang, H. (2016). “Modelling QTL-trait-crop relationships: past experiences and future prospects,”. (eds). X. Yin and C. Stuik. in Crop Systems Biology. (Berlin: Springer).
Yoshida, J., Tomooka, N., Yee Khaing, T., Shantha, P. G. S., Naito, H., Matsuda, Y., et al. (2020). Unique responses of three highly salt-tolerant wild Vigna species against salt stress. Plant Prod. Sci. 23, 1–15. doi: 10.1080/1343943x.2019.1698968
Zamir, D. (2001). Improving plant breeding with exotic geneticlibraries. Nat. Rev. Genet. 2, 983–989. doi: 10.1038/35103590
Keywords: pre-breeding, distant hybridization, food legumes, breeding populations, introgression line
Citation: Pratap A, Das A, Kumar S and Gupta S (2021) Current Perspectives on Introgression Breeding in Food Legumes. Front. Plant Sci. 11:589189. doi: 10.3389/fpls.2020.589189
Received: 30 July 2020; Accepted: 03 December 2020;
Published: 21 January 2021.
Edited by:
Mariola Plazas, Polytechnic University of Valencia, SpainReviewed by:
Bernardo Ordas, Consejo Superior de Investigaciones Científicas (CSIC), SpainHamid Khazaei, University of Saskatchewan, Canada
Copyright © 2021 Pratap, Das, Kumar and Gupta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanjeev Gupta, c2FuaWlwckByZWRpZmZtYWlsLmNvbQ==; Shiv Kumar, c2suYWdyYXdhbEBjZ2lhci5vcmc=
 Aditya Pratap
Aditya Pratap Arpita Das
Arpita Das Shiv Kumar
Shiv Kumar Sanjeev Gupta
Sanjeev Gupta