- 1Leibniz Institute of Vegetable and Ornamental Crops, Großbeeren, Germany
- 2Federal Research Center the N.I. Vavilov All-Russian Institute of Plant Genetic Resources (VIR), St. Petersburg, Russia
Germplasm collections reflect the genetic variability in crops and their wild relatives. Hence, those genetic resources are tremendously valuable for breeders and researchers, especially in light of climatic change and stagnant crop production rates. In order to achieve improvements in crop production and end-use quality, favorable traits and donor alleles present in germplasm collections need to be identified and utilized. This review covers recent reports on the utilization of germplasm material to isolate genotypes of Brassica oleracea and B. rapa vegetables, focusing on high nutrient use efficiency, accumulation of biologically active metabolites, pest resistance, and favorable phenotypic appearance. We discuss the current state of Brassica vegetable collections in genebanks and summarize studies directed to the molecular characterization of those collections.
Introduction
National and international botanical genebanks collect, conserve, investigate, and provide genetic resources of plant species. It is crucial to preserve plant materials, such as landraces, old and advanced cultivars, inbred and double haploid lines, and hybrid populations, and to identify their characteristics. Ex situ conservation is most efficient in terms of ease for users to access the plant material and because of its cost efficiency. Genebanks have the task of providing precisely classified living plant material for basic research and various applied research areas, such as plant breeding (Halewood et al., 2020). By integrating morphological and anatomical analysis with molecular fingerprinting approaches, important questions about the evolution of plant diversity, species boundaries and intra-species variability are addressed and form an essential basis for reliable classification and identification of plants. More recently, high-throughput genotyping, coupled with phenotype characterization, has been introduced as a next step for efficiently mining germplasm material (Nguyen and Norton, 2020). Ideally, quality traits should also be provided for each accession, such as productivity, earliness, stress resistance, and biochemical composition (Davies and Allender, 2017), in addition to epiphenotype, the characterization of the plant-environment relationship. Hammer et al. (2018) highlight the importance of both ex situ and in situ local biodiversity conservation for a range of vegetable species, and describe the evolution of broccoli (Brassica oleracea var. italica) cultivars from a landrace. Genebanks are a valuable resource for providing plant accessions with a high level of resistance to biotic and abiotic stresses, valuable biochemical profiles or preferable sensory traits in an era of large climate changes (Pignone and Hammer, 2013; Anglin et al., 2018).
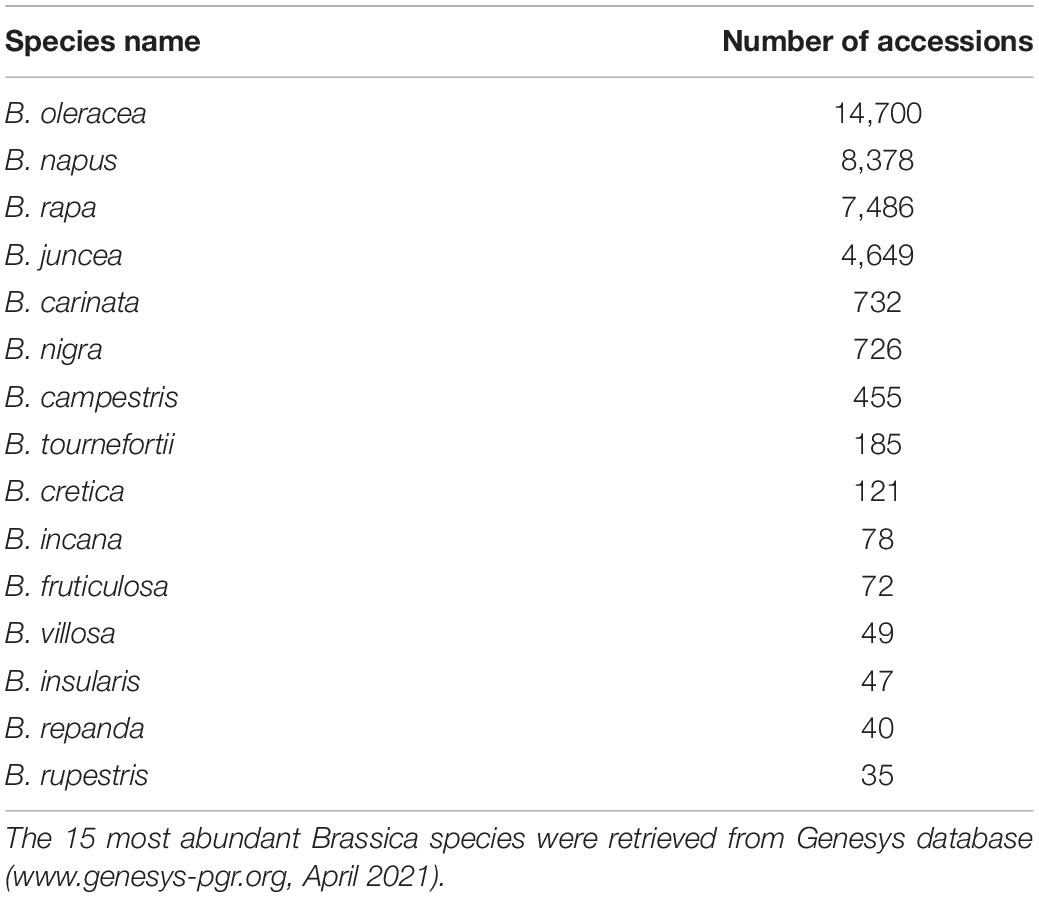
Cultivars of Brassica species are horticulture and field crops worldwide. They are vegetable, oil, fodder, green manure or spice plants. Brassicaceous vegetables belong to the taxa B. oleracea (cabbage, broccoli, cauliflower, kale, Brussels sprouts, collard greens, savoy, kohlrabi, and Chinese kale), B. rapa (turnip, mizuna, napa cabbage, cime di rapa, turnip rape), B. napus (swede), and B. juncea (Indian mustard), etc. These plants are grown for their root (turnip, rutabaga/swede), their swollen stem base (kohlrabi), their leaves (cabbage, kale, pak choi) or their inflorescence (cauliflower, broccoli). The importance of this class of vegetables to human health lies not only in their contribution to the vitamin and mineral components of the human diet, but also in the beneficial effects of their numerous classes of plant secondary metabolites. Just as for crops generally, maintaining their productivity in the light of increasing climate change and the dynamically evolving pest and pathogen communities represents major challenges requiring the genetic diversity in genebank accessions (Table 1).
After briefly outlining the importance of Brassica vegetables in human nutrition, the current state of international efforts in Brassica vegetable collections will be discussed. Subsequently, we will summarize different quality traits that represent possible breeding targets for crop improvement and were the focus of recent research efforts, and pinpoint strategies to overcome current shortcomings in applying genebank material for crop improvement. This review focuses on developments and achievements between 2009 and 2021 in four areas where genebank accessions have been employed: to analyze nutrient use efficiency, to gain insight into the accumulation of special metabolites, to isolate sources of pest resistance and for phenotype studies.
Nutritional and Biologically Active Compounds in Brassica Vegetable Crops
In developed societies, the dietary and medicinal properties of food has become the most important factors when choosing food. Brassica crops have a high content of water and low fat, and consequently a low caloric content. They are characterized by a relatively high level of carbohydrates, proteins that include essential amino acids, and many mineral elements. The high value of Brassica crops for human nutrition is primarily determined by their wide variety of biologically active compounds – enzymes, pigments, vitamins, and secondary metabolites (Manchali et al., 2012). Breeding for biologically active compounds has been considered a priority (Prange and Hewett, 2003). Fresh Brassica vegetables contain antioxidants, such as vitamins C and E, carotenoids and antioxidant enzymes such as catalase, superoxide dismutase and peroxidase (Singh et al., 2010). Furthermore, they are also rich in beneficial plant metabolites, including glucosinolates, anthocyanins, flavonoids, terpenes, S-methylcysteine sulfoxide, coumarins, and other minor compounds (Manchali et al., 2012; Neugart et al., 2018).
The beneficial effects of Brassica vegetables on human health have been partially linked to these phytochemicals, which may act at different and complementary levels. They prevent oxidative stress, induce detoxification enzymes, stimulate the immune system, inhibit malignant transformation and carcinogenic mutations, and reduce proliferation of cancer cells (Boivin et al., 2009; Herr and Buchler, 2010; Kestwal et al., 2011). Consumption of cruciferous vegetables appears to lower the risk for some kinds of cancers, including renal, prostate and possibly colorectal cancers. Consumption of vegetables, including Brassica species, has been strongly associated with a reduced risk of chronic conditions such as cardiovascular disease, diabetes, Alzheimer’s disease, cataracts and age-related functional decline (Cohen et al., 2000; Knekt et al., 2002; Higdon et al., 2007; Sergentanis et al., 2018).
Greater fundamental knowledge of the accumulation of nutrients and biologically active substances by plants is required to permit the development of a strategy for creating new varieties of Brassicaceae family vegetables with improved consumer properties. Genebank germplasm represents a valuable natural genetic resource that can be exploited for the improvement of traits related to plant growth, pest resistance, and nutrient use efficiency.
Current Status of B. Oleracea and B. Rapa Vegetable Collections
Wheat, rice, barley and maize account for a total of about 1,656,000 genebank accessions (retrieved from http://www.fao.org/wiews/en/on October 15th 2020). In Europe, around 31% of agricultural production value is represented by less than 20 crops. These include the four main European staple crops (wheat, potato, maize, and barley), several vegetable crops (tomato, sugar beet, cucumbers, melons/watermelons, Brassicas, and onion) and fruits (apples, citrus). Around one million accessions of these crops, held in worldwide genebanks, are listed in the Genesys online catalog, among them 31,653 accessions of Brassica vegetables1. For example, germplasm of broccoli (B. oleracea var. italica) is hold in genebank in the United Kingdom (448 accessions), Taiwan (316 accessions), Russia (172 accessions), United States (165 accessions), Germany (136 accessions), Italy (105 accessions), India (94 accessions), Australia (60 accessions), and Spain (56 accession), among others (see text footnote 1, April 2021). Most biological diversity of Brassica crops is now conserved ex situ in genebanks and breeders’ materials. The largest national collections of B. oleracea are located at the Russian N.I.Vavilov Institute of Plant Genetic Resources (VIR) and the German Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), while the largest B. rapa collections are maintained at the Australian Grains Genebank (AGG) and VIR. The history of the 97-years old Russian Brassica collection, and results and perspectives of its study were summarized recently (Artemyeva et al., 2019). Botanical and morphological descriptions of four Brassica species from the German national collection were presented by Gladis and Hammer (1992).
The formation of Russian state worldwide Brassicaceae collection kept at VIR had begun in 1923 after N. I. Vavilov‘s visit of West-European countries, United States, and Canada (1921-1922). After that, collecting missions had continued in Russia and all centers of plant origin and diversity worldwide. 1,500 accessions of vegetable Brassicas and 450 oilseed accessions were collected by N. I. Vavilov himself and his colleagues until 1940. As a result of the complex evaluation in the different ecologo-geographical zones of Russia and the phylogenetic studies of subspecies, varieties, forms, cultivar groups using many biological traits, descriptors were developed for B. oleracea, B. rapa, B. napus, Raphanus sativus, B. juncea, Sinapis alba, S. nigra, S. arvensis, among others (Sinskaya, 1928; Lizgunova, 1984). On this basis, Lizgunova divided the white cabbages into 7 eco-geographical groups and 33 cultivar types, cauliflower and broccoli into 7 groups and 21 cultivar types, savoy into 5 groups and 16 types, etc. Later, Artemyeva divided the Chinese cabbages (B. rapa ssp. pekinensis) into 14 cultivar types (Artemyeva, 2004). Nowadays the VIR Brassicaceae collection consists of 10,997 accessions from 11 genera and 32 species: vegetable, fodder, oilseed, spicy, ornamental crops from 98 countries. The collection includes the accessions of different status: wild species, landraces (30%), old and advanced breeding cultivars (58%), inbred and double haploid lines, hybrid populations, and mapping populations (12%).
The genetic diversity of local and traditional Brassica cultivars can be used to genetically improve plants, expand their base and create new cultivars adapted to growing in less favorable environmental conditions. Currently, the international movement of genetic resources takes place for a variety of purposes and in different forms: exchange of ex situ germplasm accessions, sales of varietal seeds, exchange through seed companies, and management of in situ storage. Within the framework of the international exchange of genebank accessions, several tens of thousands of transactions are carried out annually; such exchange plays an important role in the conservation, research and development of genetic resources. Continuous availability for research and development is a prerequisite for improving crops, including Brassica crops. The genetic resources of Brassica plants have the potential to create a variety of traits that can help address potential future challenges, such as the need for crops to adapt to changing climatic conditions or disease and insects resistance. Therefore, continued access to them is necessary to meet the growing demand for food, as well as access to neglected and underutilized crops, given their important role in nutrition. Until now, several domesticated and wild Brassica species have been subjected to genome analysis for the identification of beneficial alleles (Saad et al., 2021). Merging available genome information, reverse genetics resources and genome editing tools with the diversity in germplasm pools is the most promising strategy for crop improvement.
Most conserved accessions are labeled with basic descriptors of morphological traits and information on the origin of collected material. For example, the VIR passport database is based on 28 characteristics and includes all accessions from the collections; the characterization and evaluation databases include 60–90% accessions depending of crops, while also a separate trait database is operated for characteristics of genetic and breeding interest. Hence, a proportion of material consists of unwanted duplications that need to be identified and minimized within and between collections (Solberg et al., 2018; Palme et al., 2020). A further issue is the taxonomic misclassification of accessions. Molecular characterization of plant material has become a valuable tool to increase the efficiency of genebank management and to identify duplicated or misclassified accessions (Mason et al., 2015; Pelc et al., 2015; Koh et al., 2017; Stansell et al., 2018; Yousef et al., 2018). The genetic molecular resources represented by Brassicaceae seed collections have been reviewed (Knee et al., 2011). In order to assess the genetic diversity present in Brassica collections, amplified fragment length polymorphisms have been studied in 50 accessions of B. oleracea (van Hintum et al., 2007), 17 accessions of B. oleracea convar. acephala (Christensen et al., 2011) and 20 accessions of B. oleracea var. capitata (Faltusova et al., 2011). Sequence-specific amplification polymorphism has been shown to be useful for phylogenetic analysis of the B. rapa core collection of VIR, encompassing 96 accessions (Budahn et al., 2013). Others have used single-sequence repeat markers to investigate genebank germplasms, such as 25 accessions of different B. oleracea subspecies (El-Esawi et al., 2016) and 239 accessions of B. rapa divided into the main morphotypes (Zhao et al., 2010), as well as several core collections from the VIR B. oleracea and B. rapa accessions: 18 Russian local white cabbage accessions by SSRs were divided into Northern and Central Russian, Dutch and Central European, Southern European cultivars, when the level of single-sequence repeat DNA polymorphism within the Russian gene pool was higher than within the European gene pool (Artemyeva et al., 2006), 96 accessions of B. rapa (Artemyeva et al., 2008), 50 accessions from different B. oleracea varieties – cabbages, cauliflower, kales (Artemyeva et al., 2009) and 40 broccoli accessions were divided into four clusters according to vegetation period length, curd size and sprout development (Fateev and Artemyeva, 2020).
In order to determine genotype-phenotype correlations in germplasm collections, association mapping using mostly SSR markers have been performed in B. rapa, B. oleracea and R. sativus accessions with respect to agronomic traits and morphological characteristics: bolting time, growth related traits, morphological characters connected with quality and productivity (Artemyeva et al., 2013) as well as biochemical traits: content of dry matter, sugars, protein, ascorbic acid, carotenoids, chlorophylls (Artemyeva et al., 2016, 2017). For instance, in B. rapa the most important loci are located on the top of chromosome A02, on the bottom of A03, in the higher region of A10 that correspond to BrFLC2, BrFLC5, BrFLC1 positions, supporting the importance of flowering time for the development of many morphological and biochemical traits in Brassica crops.
Research Focus of the Last Years in Utilizing Germplasm Material
The large potential of germplasm material, sometimes referred to as “diamond in the rough,” needs extensive work to make use of accessions more feasible. To establish the true value of genetic resources, broad evaluation is required by curators with respect to desired plant characteristics, e.g., agronomic, nutritional or resistance traits. For example, more than 500 vegetable Brassica accessions per year have been trialed at five VIR experimental stations.
Most genebanks have applied trait analyses. For example, systematic study of its worldwide collection of cultivated plants has been conducted at the VIR Department of Biochemistry since 1922, as summarized by Ivanov et al. (1938), Ermakov et al. (1972), Ermakov and Arasimovich (1961), and many others. Study of the biochemical composition of the Brassicaceae collection at VIR began in 1933. Accessions of cole crops were assessed by dry matter, sugars, protein, ascorbic acid, carotenoids, and chlorophylls. In 1951, studies on varieties of cabbage were performed to assess ascorbic acid, dry matter, sugars, fiber, mineral elements, carotene, and proteins, and the dynamics of accumulation and consumption of nutrients. Later, the genetic and geographical variabilities of the accumulation of biochemical compounds were established including vitamins, pigments, mustard oils, in cabbage (Lukovnikova, 1959; Lukovnikova and Lizgunova, 1965), turnips and rutabagas (Solovyeva et al., 2013), and radishes (Shebalina and Sazonova, 1985). In the 1970s, studies of the biochemical composition and varietal diversity of the Brassica collection were deepened (Lizgunova et al., 1978). In recent years, the focus of attention has been on accumulation of the major biochemical components and biologically active substances (Artemyeva et al., 2006; Solovyova et al., 2014).
Nutrient Use Efficiency
The optimal use of essential nutrients is very important for plant production in terms of productivity and sustainability. Increasing nutrient use efficiency is critical to improve crop yield, reduce fertilizer demand and alleviate environmental pollution. In order to determine the natural variation for shoot zinc content, 376 accessions of B. oleracea and 74 commercial cultivars were investigated, revealing high variation and also an interaction with soil phosphorus availability (Broadley et al., 2010). The same set of lines was tested for potassium use efficiency in the field and in the greenhouse, and a twofold variation in shoot potassium content was found, encouraging further breeding approaches (White et al., 2010).
Another aspect of plant nutrition is the use of organic farming production systems. Here, cultivars with high nutrient use efficiency are in demand due to the reduced input of fertilizers. Yousef et al. (2015) monitored the growth of 178 B. oleracea var. botrytis accessions under organic and conventional farming conditions, and identified genotypes that are suitable as parental lines for breeding under organic farming conditions.
Accumulation of Metabolites
As outlined above, Brassica vegetables are rich in biologically active metabolites that account for their health-promoting effects. Hence, there is a special interest in increasing the accumulation of these value-adding compounds in vegetables. Glucosinolates have been a focus of research activity for many years and numerous studies have evaluated germplasm material genotypes of diverse geographical origin.
In a field trail, a total of 146 cabbage (B. oleracea var. capitata) genotypes were analyzed and accessions identified with increased concentrations of glucosinolates (Bhandari et al., 2020). A collection of 70 Turkish white head cabbages (B. oleracea var. capitata sub. var. alba) grown in the field in two harvesting seasons revealed a large variation in glucosinolate composition and concentration, leading to the identification of promising genotypes for further breeding activities (Sarikamis et al., 2009).
Lee et al. (2013) profiled 48 B. rapa turnips for glucosinolate accumulation and found a grouping of genotypes. Similarly, the glucosinolate patterns and antioxidant activities of 62 varieties of Chinese cabbage (B. rapa ssp. pekinensis) were determined and several genotypes for further breeding were identified (Lee et al., 2014). Leaves and tubers of 16 B. rapa turnip accessions were assayed in parallel for their accumulation of glucosinolates and glucosinolate hydrolysis products (Klopsch et al., 2017). Besides a tissue-specific accumulation of glucosinolates and their breakdown products, the study revealed that clustering patterns of accessions based on their glucosinolate pattern differed from those based on the hydrolysis products, indicating the importance of profiling the latter bioactive compounds. The same observation was made when profiling 91 leafy B. rapa accessions from different subspecies (Klopsch et al., 2018). Correlations in abundance of specific glucosinolates and of their breakdown products were identified, with implications for future breeding activities.
Fifty-one accessions from 5 genera of family Brassicaceae from the VIR collection (Sinapis alba, Lepidium sativum, Eruca sativa, Diplotaxis muralis, B. juncea, B. rapa) were profiled for 21 glucosinolates. The highest content (more than 40 μM/g) was found in all accessions of white mustard (sinalbin) and in six accessions of Indian mustard (sinigrin), hence, these species should be useful for biofumigation approaches. Seventeen glucosinolates components have been determined in B. rapa, among them were indol glucosinolates, which are more useful for human, and three local accessions of Chinese cabbage had threefold higher levels of indole glucosinolates as compared to the remaining tested cultivars (Solovyeva et al., 2013).
A set of 19 Italian kale (B. oleracea var. acephala) was profiled for the accumulation of phenols, flavonoids, and anthocyanins, representing the basis for further nutritional use of locally adapted accessions (Lotti et al., 2018). A set of 70 white cabbage and 30 cauliflower accessions were characterized for variability of biochemical compounds at VIR; the accumulation by different genotypes of protein, sugars, ascorbic acid, carotenoids, chlorophylls, amino acids, organic acids, fatty acids, and phenolic compounds was determined. The sources of high levels of nutritive and biologically active substances were found, mostly within central Russian and Dutch white cabbage accessions, and within German and French cauliflower accessions (Solovyova et al., 2014). The connection between primary and secondary metabolism in different colored cauliflower curds was investigated and revealed closely related metabolic networks (Park et al., 2013). The complex of morphological and biochemical traits of an Italian broccoli and cauliflower landraces collection compared with F1 was studied by Branca et al. (2018) who detected a large diversity of glucosinolate, anthocyanin, carotenoids, polyphenols and ascorbic acid contents. Single sequence repeat analysis divided the collection into five clusters; all Sicilian accessions were distinct. A core collection of 168 B. rapa accessions of different morphotypes and origins was explored to find genetic association between markers and tocopherols, carotenoids, chlorophylls and folate (Del Carpio et al., 2011).
Pest Resistance
Crop production is severely threatened by herbivores feeding on Brassica vegetables and the distribution of pests has increased in the recent years due to the expansion of rapeseed. Hence, the role of genetic sources of resistance, including landraces with high adaptability, is increasing in breeding programs. At the same time, the specificity of the human’s consumption of these crops as food requires a maximum degree of ecological safety of products, and modern ecological thinking in plant protection requires minimizing the anthropogenic impact on the components of agrobiocenosis, including useful entomofauna.
The most common pests of Brassicaceae family crops are thrips (Thrips tabaci), cabbage root fly (Delia radicum), cabbage moth (Mamestra brassicae), cabbage aphid (Brevicoryne brassicae) and diamond back moth (Plutella xylostella). A screen of 27 cabbage (B. oleracea var. capitata) cultivars revealed a range of preference of cabbage aphid toward the tested accessions (de Melo et al., 2013). Plant resistance tests of eight genotypes of collard greens (B. oleraceae var. acephala) against cabbage aphid revealed an interactive effect of biochemical and morphological resistance mechanisms (Canassa et al., 2020). Sources of resistance against cabbage moth were detected in a screen of 21 local and commercial cabbage varieties and interactive effects of leaf traits, head compactness, and leaf glucosinolate content were identified (Cartea et al., 2010). A set of 56 turnip (B. rapa ssp. rapa) accessions were screened for resistance against cabbage root fly under controlled conditions and in the field, leading to the isolation of resistant cultivars and resistance plant traits (Santolamazza-Carbone et al., 2017). Resistance against cabbage whitefly (Aleyrodes proletella) was tested in a collection of 432 accessions of B. oleracea and its wild relatives. It was shown that the wild relatives exhibited an earlier resistance than the breeding cultivars, which is probably due to the earlier formation of trichomes (Pelgrom et al., 2015). The effect of epicuticular waxes on plant resistance against the green peach aphid (Myzus persicae) was minor in twelve kale genotypes (Costa et al., 2014).
Phenotypic Appearance
Besides health and welfare benefits, an important marketing goal of Brassica producers is the attractive appearance, size and shape of crops. Hence, genebank accessions are screened for phenotypic traits. Thirty kale genotypes were investigated for 44 morphological traits, revealing a limited extent of phenotypical variation albeit from a wide extent of genetical divergence (Azevedo et al., 2014). Thorwarth et al. (2018) selected 174 cauliflower accessions to gain insight into six curd-related traits. Further, they identified associations between these traits and genetic markers by genome-wide association mapping.
At VIR, a complete phenotyping of all accessions in the different eco-geographical zones of Russia is performed according to the same methodic during three years. The VIR evaluation databases include 47–55 morphological and phenological characters for vegetable crops, 18–23 characters for oilseeds, as well as separate immunological, physiological, and biochemical databases. Since pathogenic organisms spoil the phenotypic appearance of crops, trait collections have been established based on resistance/susceptibility to black rot, clubroot, downy mildew, and leaf spot disease. The range of responses was dependent on the crop, the pathogen race and geographical origin of the accessions. For instance, maximum frequency of B. rapa resistance to black rot originates from accessions from Central-Asian subpopulation, which is considered as the most genetically variable part of the species genepool, and to a subpopulation from Japan (Ignatov et al., 2011).
The variability of ten accessions of Chinese broccoli was investigated for morphological, biochemical, genetic properties and a large diversity was noted in this set, generally on plant habit, stem diameter and chlorophyll content (Fotev et al., 2018). Morphological and biochemical traits linked to consumer value of 96 B. rapa accessions from different morphotypes from the VIR core collection were screened in the field and under greenhouse conditions in the North-West Russia and in the field in South China. The study revealed correlations between characteristics, and sources for breeding and genetic markers for traits investigated (Artemyeva et al., 2017). A study of the morphological traits of the plant, leaf lamina and root of 16 local Scandinavian turnip accessions from the VIR collection allowed determination of typical (cvs. Petrovskaya, Karelskaya, Tankard yellow) and rare types (cv. Kostenevskaya), and mixed populations, as well as valuable stock for breeding projects (Kornyukhin and Artemyeva, 2017).
Conclusion
Genebanks have many tasks and challenges (Fu, 2017; Diez et al., 2018). At present, the high-throughput use of omic tools for extensive germplasm collections is challenging due to the laborious sample preparation (nucleic acids, proteins, and metabolites), as well as suboptimal handling and analysis approaches of large-scale datasets. Further, unharmonized laboratory-specific analytical techniques hinder the exchange of results between labs. However, assimilating large amounts of genome, expression and metabolite data, in conjunction with phenotype information, will truly accelerate breeding processes and increase our understanding of the molecular background of trait formation. These enormous efforts need to be supported by increased public funding. Although the public awareness on safeguarding the agricultural and horticultural heritage has increased in recent years, funding of germplasm banks did not catch up with the demands, currently covering basically permanent activities (collect, inventory, and propagate, etc.), but insufficiently financing genome and phenome investigations. The utilization of Brassica vegetable germplasm collections to isolate promising genotypes for breeding activities has been achieved successfully. However, there is still underutilization of germplasms, and the link between conservation and use of collections must be strengthened further. Recent activities aimed at developing user-friendly databases to aid the access to germplasm collections, ultimately centralizing database management. Currently, besides international platforms, such as Genesys, Crop Wild Relatives Global Portal, EURISCO, New World Fruits Database, and ECPGR, also numerous national databases, such as GRIN, GBIS/I, and SeedStor are available. Centralized and detailed information about genotype, phenotype, stress resistance and metabolite accumulation of accessions would increase use of collections. Applying high-throughput molecular and phenotypic screening methods will allow the deep characterization of collections and yield valuable trait information for genebank curators as well as for researchers and breeders.
Author Contributions
KW, AK, and AA wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was prepared in accordance with the state assignment of Russian Federation No. 0662-2019-0003 “Genetic resources of vegetable and cucurbit crops of the VIR worldwide collection: effective ways to expand diversity, disclose the patterns of hereditary variability, use the adaptive potential,” state registration number of R&D (RK), according to the plan of scientific research work of VIR AAAA-A19-11-9013090157-1.
Footnotes
References
Anglin, N. L., Amri, A., Kehel, Z., and Ellis, D. (2018). A case of need: linking traits to genebank accessions. Biopreserv. Biobank. 16, 337–349. doi: 10.1089/bio.2018.0033
Artemyeva, A. (2004). “Donors and sources for a breeding of Brassica rapa L. leafy vegetables,” in Catalogue of VIR Worldwide Collection (St. Petersburg: VIR Press).
Artemyeva, A. M., Chesnokov, Y. V., and Klocke, E. (2008). Genetic diversity and intraspecific phylogenetic relationships of Brassica rapa L. species crops based on microsatellite analysis. Newsletter Vavilov Soc. Genet. Breed. 12, 608–619.
Artemyeva, A. M., Chesnokov, Y. V., and Klocke, E. (2009). Analysis of phylogenetic relationships of Brassica oleracea L. Newsletter Vavilov Soc. Genet. Breed. 13, 759–771.
Artemyeva, A. M., Chesnokov, Y. V., Budahn, H., and Bonnema, G. (2013). “Association mapping of agronomically important traits in Brassica rapa L,” in: Branca F, Tribulato A, eds Acta Horticulturae. Belgium: International Society for Horticultural Science doi: 10.17660/ActaHortic.2013.1005.17
Artemyeva, A. M., Solovjova, A. E., Kocherina, N. V., Berensen, F. A., Rudneva, E. N., and Chesnokov, Y. V. (2016). Mapping of chromosome loci determined manifestation of morphological and biochemical traits of quality in Brassica rapa L. crops. Russ. J. Plant Physiol. 63, 259–272. doi: 10.1134/S1021443716020047
Artemyeva, A. M., Solovyeva, A. E., Berensen, F. A., Kocherina, N. V., and Chesnokov, Y. V. (2017). Ecologo-genetic evaluation of morphological and biochemical traits in VIR Brassica rapa L. collection. Agric. Biol. 52, 129–142.
Artemyeva, A., Dubovskaya, A., and Kon’kova, N. (2019). The Russian brassicaceae collection - from NI Vavilov and EN sinskaya till nowadays. Newsletter Vavilov Soc. Genet. Breed. 23, 157–164. doi: 10.18699/VJ19.553
Artemyeva, A., Solovyeva, A. E., and Chesnokov, Y. V. (2006). Genetic properties of Russian white cabbage cultivars. Agric. Biol. 5, 53–61.
Azevedo, A. M., Junior, V. C. A., and Fernandes, J. S. C. (2014). Genetic divergence and importance of morphological characters in genotypes of kale. Hortic. Bras. 32, 48–54. doi: 10.1590/S0102-05362014000100008
Bhandari, S. R., Rhee, J., Choi, C. S., Jo, J. S., Shin, Y. K., and Lee, J. G. (2020). Profiling of individual desulfo-glucosinolate content in cabbage head (Brassica oleracea var. capitata) germplasm. Molecules 25:1860 doi: 10.3390/molecules25081860
Boivin, D., Lamy, S., Lord-Dufour, S., Jackson, J., Beaulieu, E., and Cote, M. (2009). Antiproliferative and antioxidant activities of common vegetables: a comparative study. Food Chem. 112, 374–380. doi: 10.1016/j.foodchem.2008.05.084
Branca, F., Chiarenza, G. L., Cavallaro, C., Gu, H., Zhao, Z., and Tribulato, A. (2018). Diversity of Sicilian broccoli (Brassica oleracea var. italica) and cauliflower (Brassica oleracea var. botrytis) landraces and their distinctive bio-morphological, antioxidant, and genetic traits. Genet. Resour. Crop Evol. 65, 485–502. doi: 10.1007/s10722-017-0547-8
Broadley, M. R., Lochlainn, S. O., Hammond, J. P., Cakmak, I., Eker, S., and Erdem, H. (2010). Shoot zinc (Zn) concentration varies widely within Brassica oleracea L. and is affected by soil Zn and phosphorus (P) levels. J. Horticult. Sci. Biotechnol. 85, 375–380. doi: 10.1080/14620316.2010.11512683
Budahn, H., Artemyeva, A. M., and Chesnokov, Y. V. (2013). S-SAP markers - a valuable tool for genetic distance analysis in Brassica rapa L. Acta Hortic. 1005, 133–139. doi: 10.17660/ActaHortic.2013.1005.12
Canassa, V. F., Baldin, E. L. L., Lourencao, A. L., Barros, D. R. P., Lopes, N. P., and Sartori, M. M. P. (2020). Feeding behavior of Brevicoryne brassicae in resistant and susceptible collard greens genotypes: interactions among morphological and chemical factors. Entomol. Exp. Appl. 168, 228–239. doi: 10.1111/eea.12897
Cartea, M. E., Francisco, M., Lema, M., Soengas, P., and Velasco, P. (2010). Resistance of cabbage (Brassica oleracea capitata group) crops to Mamestra brassicae. J. Econ. Entomol. 103, 1866–1874. doi: 10.1603/EC09375
Christensen, S., Von Bothmer, R., Poulsen, G., Maggioni, L., Phillip, M., and Andersen, R. A. (2011). AFLP analysis of genetic diversity in leafy kale (Brassica oleracea L. convar. acephala (DC.) Alef.) landraces, cultivars and wild populations in Europe. Genet. Resour. Crop Evol. 58, 657–666. doi: 10.1007/s10722-010-9607-z
Cohen, J. H., Kristal, A. R., and Stanford, J. L. (2000). Fruit and vegetable intakes and prostate cancer risk. J. Natl. Cancer Inst. 92, 61–68. doi: 10.1093/jnci/92.1.61
Costa, E. M. R., Marchese, A., Maluf, W. R., and Silva, A. A. (2014). Resistance of kale genotypes to the green peach aphid and its relation to leaf wax. Rev. Cienc. Agron. 45, 146–154. doi: 10.1590/S1806-66902014000100018
Davies, L. R., and Allender, C. J. (2017). Who is sowing our seeds? A systematic review of the use of plant genetic resources in research. Genet. Res. Crop Evol. 64, 1999–2008. doi: 10.1007/s10722-017-0491-7
de Melo, B. D. C., Bleicher, E., Bertini, C., and Da Silva, J. F. (2013). Genetic divergence between cabbage commercial cultivars as to cabbage aphid preference. Pesqui. Agropecu. Bras. 48, 459–465. doi: 10.1590/S0100-204X2013000500001
Del Carpio, D. P., Basnet, R. K., De Vos, R. C. H., Maliepaard, C., Paulo, M. J., and Bonnema, G. (2011). Comparative methods for association studies: a case study on metabolite variation in a Brassica rapa core collection. PLoS One 6:e19624 doi: 10.1371/journal.pone.0019624
Diez, M. J., De La Rosa, L., Martin, I., Guasch, L., Cartea, M. E., and Mallor, C. (2018). Plant genebanks: present situation and proposals for their improvement. the case of the Spanish network. Front. Plant Sci. 9:1794. doi: 10.3389/fpls.2018.01794
El-Esawi, M. A., Germaine, K., Bourke, P., and Malone, R. (2016). Genetic diversity and population structure of Brassica oleracea germplasm in Ireland using SSR markers. C. R. Biol. 339, 133–140. doi: 10.1016/j.crvi.2016.02.002
Ermakov, A. I., and Arasimovich, V. V. (1961). Biochemistry of Vegetable Crops. Leningrad: John Wiley & Sons Ltd
Ermakov, A. I., Arasimovich, V. V., Smirnova-Ikonnikova, M. I., Yarosh, N. P., and Lukovnikova, G. A. (1972). Methods for the Biochemical Analysis of Plants. Springer: Leningrad.
Faltusova, Z., Kucera, L., and Ovesna, J. (2011). Genetic diversity of Brassica oleracea var. capitata gene bank accessions assessed by AFLP. Electron. J. Biotechnol. 14:10. doi: 10.2225/vol14-issue3-fulltext-4
Fateev, D. A., and Artemyeva, A. M. (2020). Molecular genetic characterisatics of broccoli (Brassica oleracea L. var. italica Plenck) from the VIR collection. Proc. Appl. Bot. Genet. Breed. 181, 91–99. doi: 10.30901/2227-8834-2020-3-91-99
Fotev, Y. V., Artemyeva, A. M., Fateev, D. A., Bugrovskaya, G. A., Belousova, V. P., and Kukushkina, T. A. (2018). Results of SSR analysis, properties of plant morphology and biochemical composition of Chinese broccoli – a new vegetable crop for Russia. Veg. Crops Russia 1, 12–19. doi: 10.18619/2072-9146-2018-1-12-19
Fu, Y. B. (2017). The vulnerability of plant genetic resources conserved ex situ. Crop Sci. 57, 2314–2328. doi: 10.2135/cropsci2017.01.0014
Gladis, T., and Hammer, K. (1992). Die gaterslebener brassica-kollektion: Brassica juncea, B. napus, B. nigra und B. rapa. Feddes Repert. 103, 469–507. doi: 10.1002/fedr.19921030704
Halewood, M., Jamora, N., Noriega, I. L., Anglin, N. L., Wenzl, P., and Payne, T. (2020). Germplasm acquisition and distribution by CGIAR genebanks. Plants Basel 9:1296 doi: 10.3390/plants9101296
Hammer, K., Montesano, V., Direnzo, P., and Laghetti, G. (2018). Conservation of crop genetic resources in Italy with a focus on vegetables and a case study of a neglected race of Brassica oleracea. Agric. Basel 8:105. doi: 10.3390/agriculture8070105
Herr, I., and Buchler, M. W. (2010). Dietary constituents of broccoli and other cruciferous vegetables: implications for prevention and therapy of cancer. Cancer Treat. Rev. 36, 377–383. doi: 10.1016/j.ctrv.2010.01.002
Higdon, J. V., Delage, B., Williams, D. E., and Dashwood, R. H. (2007). Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol. Res. 55, 224–236. doi: 10.1016/j.phrs.2007.01.009
Ignatov, A., Artemyeva, A., Chesnokov, Y. V., Polityko, V. A., Matveeva, E. V., and Oraevskiy, A. A. (2011). Resistance of Brassica rapa L. and B. napus L. to black rot and leaf spot pathogens. Agric. Biol. 1, 85–92.
Ivanov, N., Knjaginicev, M., and Smirnova, M. (1938). Biochemistry of cultivated plants. Leningrad: Institute of Plant Industry.
Kestwal, R. M., Lin, J. C., Bagal-Kestwal, D., and Chiang, B. H. (2011). Glucosinolates fortification of cruciferous sprouts by sulphur supplementation during cultivation to enhance anti-cancer activity. Food Chem. 126, 1164–1171. doi: 10.1016/j.foodchem.2010.11.152
Klopsch, R., Witzel, K., Artemyeva, A. M., Ruppel, S., and Hanschen, F. S. (2018). Genotypic variation of glucosinolates and their breakdown products in leaves of Brassica rapa. J. Agric. Food Chem. 66, 5481–5490. doi: 10.1021/acs.jafc.8b01038
Klopsch, R., Witzel, K., Börner, A., Schreiner, M., and Hanschen, F. S. (2017). Metabolic profiling of glucosinolates and their hydrolysis products in a germplasm collection of Brassica rapa turnips. Food Res. Int. 100, 392–403. doi: 10.1016/j.foodres.2017.04.016
Knee, E. M., Rivero, L., Crist, D., Grotewold, E., and Scholl, R. (2011). “Germplasm and molecular resources,” in. Genetics and Genomics of the Brassicaceae, eds R. Schmidt. and I. Bancroft. Berlin: Springer doi: 10.1007/978-1-4419-7118-0_16
Knekt, P., Kumpulainen, J., Jarvinen, R., Rissanen, H., Heliövaara, M., and Reunanen, A. (2002). Flavonoid intake and risk of chronic diseases. Am. J. Clin. Nutr. 76, 560–568. doi: 10.1093/ajcn/76.3.560
Koh, J. C. O., Barbulescu, D. M., Norton, S., Redden, B., Salisbury, P. A., and Kaur, S. (2017). A multiplex PCR for rapid identification of Brassica species in the triangle of U. Plant Methods 13:49. doi: 10.1186/s13007-017-0200-8
Kornyukhin, D. L., and Artemyeva, A. M. (2017). Scandinavian table and stubble turnips: variability and value for breeding. Veg. Crops Russia 4, 58–61. doi: 10.18619/2072-9146-2017-4-58-61
Lee, J. G., Bonnema, G., Zhang, N. W., Kwak, J. H., De Vos, R. C. H., and Beekwilder, J. (2013). Evaluation of glucosinolate variation in a collection of turnip (Brassica rapa) germplasm by the analysis of intact and desulfo glucosinolates. J. Agric. Food Chem. 61, 3984–3993. doi: 10.1021/jf400890p
Lee, M. K., Chun, J. H., Byeon, D. H., Chung, S. O., Park, S. U., and Park, S. (2014). Variation of glucosinolates in 62 varieties of Chinese cabbage (Brassica rapa L. ssp pekinensis) and their antioxidant activity. LWT-Food Sci. Technol. 58, 93–101. doi: 10.1016/j.lwt.2014.03.001
Lizgunova, T. V. (1984). Cultural Flora of the USSR. XI: Cabbage (Brassica oleracea L.). Leningrad: Kolos Press.
Lizgunova, T. V., Boos, G. V., and Dzhokhadze, T. I. (1978). Formation, results of study and use of VIR cabbage collection. VIR Bulletin 85, 33–56.
Lotti, C., Iovieno, P., Centomani, I., Marcotrigiano, A. R., Fanelli, V., and Mimiola, G. (2018). Genetic, bio-agronomic, and nutritional characterization of kale (Brassica oleracea L. var. acephala) diversity in Apulia, Southern Italy. Divers. Basel 10:11. doi: 10.3390/d10020025
Lukovnikova, G. A. (1959). Variability of the amount and quality of the nitrogenous substances in cabbage species and varieties. Proc. Appl. Bot. Genet. Breed. 32, 149–158.
Lukovnikova, G. A., and Lizgunova, T. V. (1965). Comparative biochemical characteristic of cabbage organs. Proc. Appl. Bot. Genet. Breed. 37, 17–25.
Manchali, S., Murthy, K. N. C., and Patil, B. S. (2012). Crucial facts about health benefits of popular cruciferous vegetables. J. Funct. Foods 4, 94–106. doi: 10.1016/j.jff.2011.08.004
Mason, A. S., Zhang, J., Tollenaere, R., Vasquez Teuber, P., Dalton-Morgan, J., and Hu, L. (2015). High-throughput genotyping for species identification and diversity assessment in germplasm collections. Mol. Ecol. Resour. 15, 1091–1101. doi: 10.1111/1755-0998.12379
Neugart, S., Baldermann, S., Hanschen, F. S., Klopsch, R., Wiesner-Reinhold, M., and Schreiner, M. (2018). The intrinsic quality of brassicaceous vegetables: how secondary plant metabolites are affected by genetic, environmental, and agronomic factors. Sci. Hortic. 233, 460–478. doi: 10.1016/j.scienta.2017.12.038
Nguyen, G. N., and Norton, S. L. (2020). Genebank phenomics: a strategic approach to enhance value and utilization of crop germplasm. Plants Basel 9:817. doi: 10.3390/plants9070817
Palme, A. E., Hagenblad, J., Solberg, S. O., Aloisi, K., and Artemyeva, A. (2020). SNP markers and evaluation of duplicate holdings of Brassica oleracea in two European genebanks. Plants Basel 9:925. doi: 10.3390/plants9080925
Park, S. Y., Lim, S. H., Ha, S. H., Yeo, Y., Park, W. T., and Kwon, D. Y. (2013). Metabolite profiling approach reveals the interface of primary and secondary metabolism in colored cauliflowers (Brassica oleracea L. ssp botrytis). J. Agric. Food Chem. 61, 6999–7007. doi: 10.1021/jf401330e
Pelc, S. E., Couillard, D. M., Stansell, Z. J., and Farnham, M. W. (2015). Genetic diversity and population structure of collard landraces and their relationship to other Brassica oleracea crops. Plant Genome 8:eplantgenome2015.04.0023 doi: 10.3835/plantgenome2015.04.0023
Pelgrom, K. T. B., Broekgaarden, C., Voorrips, R. E., Bas, N., Visser, R. G. F., and Vosman, B. (2015). Host plant resistance towards the cabbage whitefly in Brassica oleracea and its wild relatives. Euphytica 202, 297–306. doi: 10.1007/s10681-014-1306-y
Pignone, D., and Hammer, K. (2013). “Conservation, evaluation, and utilization of biodiversity,” in: Kole C, ed. Genomics and Breeding for Climate-Resilient Crops: Vol. 1 Concepts and Strategies. Berlin: Springer doi: 10.1007/978-3-642-37045-8_2
Prange, P. K., and Hewett, E. W. (2003). A proceedings of the XXVI International Horticultural Congress, Toronto, Canada, 11-17 August, 2002: Issues and Advances in Postharvest Horticulture. Leuven: International Society for Horticultural Science Press.
Saad, N. S. M., Severn-Ellis, A. A., Pradhan, A., Edwards, D., and Batley, J. (2021). Genomics armed with diversity leads the way in Brassica improvement in a changing global environment. Front. Genet. 12:28. doi: 10.3389/fgene.2021.600789
Santolamazza-Carbone, S., Velasco, P., and Cartea, M. E. (2017). Resistance to the cabbage root fly, Delia radicum (Diptera, Anthomyiidae), of turnip varieties (Brassica rapa subsp rapa). Euphytica 213:274 doi: 10.1007/s10681-017-2069-z
Sarikamis, G., Balkaya, A., and Yanmaz, R. (2009). Glucosinolates within a collection of white head cabbages (Brassica oleracea var. capitata sub.var. alba) from Turkey. Afr. J. Biotechnol. 8, 5046–5052.
Sergentanis, T. N., Psaltopoulou, T., Ntanasis-Stathopoulos, I., Liaskas, A., Tzanninis, I. G., and Dimopoulos, M. A. (2018). Consumption of fruits, vegetables, and risk of hematological malignancies: a systematic review and meta-analysis of prospective studies. Leuk. Lymphoma 59, 434–447. doi: 10.1080/10428194.2017.1339873
Shebalina, M., and Sazonova, L. (1985). Cultural Flora of the USSR. XVIII: Root crops (Brassica – Turnip, Rutabaga, Radish, Small Radish). Leningrad: Ahropromyzdat Press. 324.
Singh, B. K., Sharma, S. R., and Singh, B. (2010). Antioxidant enzymes in cabbage: variability and inheritance of superoxide dismutase, peroxidase and catalase. Sci. Hortic. 124, 9–13. doi: 10.1016/j.scienta.2009.12.011
Sinskaya, E. N. (1928). The oleiferous plants and root crops of the family Cruciferae. Proc. Appl. Bot. Genet. Breed. 19:656.
Solberg, S. O., Artemyeva, A., Yndgaard, F., Dorre, M., Niss, J., and Burleigh, S. (2018). Duplication assessments in Brassica vegetable accessions. Plant Genet. Resour. 16, 201–208. doi: 10.1017/S1479262117000156
Solovyeva, A. E., Artemyeva, A. M., and Schütze, W. (2013). Peculiar properties of glucosinolate accumulation in the Brassicaceae family. Russian Agric. Sci. 4, 12–14. doi: 10.3103/S1068367413050169
Solovyova, A. E., Sokolova, D. V., Piskunova, T. M., and Artemyeva, A. M. (2014). Nutrients and biologically active substances in vegetable crops and their role in improving nutrition. Proc. Appl. Bot. Genet. Breed. 175, 5–19.
Stansell, Z., Hyma, K., Fresnedo-Ramirez, J., Sun, Q., Mitchell, S., and Bjorkman, T. (2018). Genotyping-by-sequencing of Brassica oleracea vegetables reveals unique phylogenetic patterns, population structure and domestication footprints. Hortic. Res. 5:38. doi: 10.1038/s41438-018-0040-3
Thorwarth, P., Yousef, E. A. A., and Schmid, K. J. (2018). Genomic prediction and association mapping of curd-related traits in gene bank accessions of cauliflower. G3 8, 707–718. doi: 10.1534/g3.117.300199
van Hintum, T. J. L., De Wiel, C., Visser, D. L., Van Treuren, R., and Vosman, B. (2007). The distribution of genetic diversity in a Brassica oleracea gene bank collection related to the effects on diversity of regeneration, as measured with AFLPs. Theor. Appl. Genet. 114, 777–786. doi: 10.1007/s00122-006-0456-2
White, P. J., Hammond, J. P., King, G. J., Bowen, H. C., Hayden, R. M., and Meacham, M. C. (2010). Genetic analysis of potassium use efficiency in Brassica oleracea. Ann. Bot. 105, 1199–1210. doi: 10.1093/aob/mcp253
Yousef, E. A. A., Muller, T., Borner, A., and Schmid, K. J. (2018). Comparative analysis of genetic diversity and differentiation of cauliflower (Brassica oleracea var. botrytis) accessions from two ex situ genebanks. PLoS One 13:e0192062. doi: 10.1371/journal.pone.0192062
Yousef, E., Lampei, C., and Schmid, K. (2015). Evaluation of cauliflower genebank accessions under organic and conventional cultivation in Southern Germany. Euphytica 201, 389–400. doi: 10.1007/s10681-014-1225-y
Keywords: Brassica, genebank, glucosinolate, phenotyping, vegetables
Citation: Witzel K, Kurina AB and Artemyeva AM (2021) Opening the Treasure Chest: The Current Status of Research on Brassica oleracea and B. rapa Vegetables From ex situ Germplasm Collections. Front. Plant Sci. 12:643047. doi: 10.3389/fpls.2021.643047
Received: 17 December 2020; Accepted: 26 April 2021;
Published: 20 May 2021.
Edited by:
Carla S. Santos, Catholic University of Portugal, PortugalReviewed by:
Karl Schmid, University of Hohenheim, GermanyNian Wang, Huazhong Agricultural University, China
Zhansheng Li, Insititute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, China
Copyright © 2021 Witzel, Kurina and Artemyeva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katja Witzel, d2l0emVsQGlnemV2LmRl; Anna M. Artemyeva, YWttZTExQHlhbmRleC5ydQ==
 Katja Witzel
Katja Witzel Anastasia B. Kurina
Anastasia B. Kurina Anna M. Artemyeva
Anna M. Artemyeva