- 1Guangdong Provincial Key Laboratory of Plant Resources, School of Life Sciences, Sun Yat-sen University, Guangzhou, China
- 2Institute of Medical Plant Physiology and Ecology, School of Pharmaceutical Sciences, Guangzhou University of Chinese Medicine, Guangzhou, China
A phosphorylation/dephosphorylation cycle at tyrosine 428 of CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) plays an essential role in chitin triggered immunity in Arabidopsis thaliana. In this study, we used a differential peptide pull-down (PPD) assay to identify factors that could participate downstream of this cycle. We identified ZYGOTIC ARREST 1 (ZAR1) and showed that it interacts with CERK1 specifically when the tyrosine 428 (Y428) residue of CERK1 is dephosphorylated. ZAR1 was originally characterized as an integrator for calmodulin and G-protein signals to regulate zygotic division in Arabidopsis. Our current results established that ZAR1 also negatively contributed to defense against the fungus Botrytis cinerea and played a redundant role with its homolog ZAR2 in this process. The zar1-3 zar2-1 double mutant exhibited stronger resistance to B. cinerea compared with zar1-3 single mutant, zar2-1 single mutant, and wild-type plants. Moreover, the inducible expression of numerous defense response genes upon B. cinerea infection was increased in the zar1-3zar2-1 double mutant, consistent with a repressive role for ZAR proteins in the defense response. Therefore, our findings provided insight into the function of ZAR1 in multiple defenses and developmental regulation pathways.
Introduction
Plants confront attacks from various pathogens and have developed immunity mechanisms to defend against pathogen infection. Cell-surface localized receptors termed pattern recognition receptors (PRRs) detect microbe-associated molecular patterns (MAMPs) and mediate signal transduction (Couto and Zipfel, 2016; Zipfel and Oldroyd, 2017). In Arabidopsis thaliana, the bacterial flagellin peptide (flg22), elongation factor Tu peptide (elf18), and fungal chitin are the best-characterized MAMPs (Boller and Felix, 2009). The cognate receptors for flg22, elf18, and chitin have been identified as flagellin-sensitive 2 (FLS2), EF-TU receptor (EFR), and LYSM-containing receptor-like kinase 5 (LYK5), respectively (Gomez-Gomez and Boller, 2000; Zipfel et al., 2006; Cao et al., 2014). The perception of MAMPs by PRRs activates downstream signal transduction networks, resulting in various immune responses including mitogen-activated protein kinase (MAPK) activation, the burst of reactive oxygen species (ROS), and inducible expression of defense genes (Couto and Zipfel, 2016; Gong et al., 2020).
In Arabidopsis, the chitin elicitor receptor kinase 1 (CERK1)/LysM-RLK1 is an indispensable receptor-like kinase (RLK) for chitin-triggered immunity signaling (Miya et al., 2007; Wan et al., 2008; Liu et al., 2012). Both heterodimerization between CERK1 and LYK5 and chitin-triggered homodimerization of CERK1 play essential roles in chitin signaling in Arabidopsis (Liu et al., 2012; Cao et al., 2014). Chitin-activated CERK1 phosphorylates downstream receptor-like cytoplasmic kinases (RLCKs) to regulate the chitin triggered MAPK activation and ROS burst (Zhang et al., 2010; Yamada et al., 2016; Liu et al., 2018). CERK1 activation is regulated by a phosphorylation/dephosphorylation cycle at the tyrosine 428 (Y428) residue to dynamically control immunity signaling (Liu et al., 2018). After chitin elicitation, CERK1 recruits the CERK1-interacting protein phosphatase 1 (CIPP1) to dephosphorylate Y428, which suppresses CERK1 signaling. CIPP1 subsequently dissociates from CERK1 harboring the dephosphorylated Y428 residue, allowing CERK1 to return to a standby state (Liu et al., 2018).
Candidate chitin-signaling regulators have been identified in Arabidopsis based on their ability to interact with CERK1 (Gong et al., 2020). For example, the RLK IOS1 can physically associate with CERK1 to facilitate chitin signaling (Yeh et al., 2016). Similarly, the RLK FERONIA can promote responses to chitin signal, whereas the RALF23 ligand-bound FERONIA plays the opposite role (Stegmann et al., 2017). In addition, Arabidopsis LIK1 is an interacting partner of CERK1 that plays a negative regulatory role in chitin-induced responses (Le et al., 2014). However, the factors involved in detecting and binding to the CERK1 motif containing the phosphorylated/dephosphorylated-Y428 residue remain largely unknown. In this study, we reported that the leucine-rich repeat RLK (LRR-RLK) ZYGOTIC ARREST 1 (ZAR1) could interact with CERK1 when it harbored the dephosphorylated Y428 residue and that ZAR1 negatively contributed to the defense against Botrytis cinerea in Arabidopsis.
Materials and Methods
Plant Materials and Growth Conditions
The seeds of A. thaliana used in this study, including the wild-type Columbia-0 (Col-0) ecotype, zar1-4 mutant (SALK_143663), and zar2-1 mutant (SALK-110111c) were obtained from the Nottingham Arabidopsis Stock Centre.1 The T-DNA insertion mutant zar1-3 (SALK_021338)was kindly provided by Wei-Cai Yang from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing China. The cerk1 mutant was kindly provided by Gary Stacey. All plants were grown under 22°C, a 12-h/12-h light/dark cycle, and relative air humidity of 60–70%.
Isolation of Protoplasts
Protoplast isolation from Arabidopsis seedlings was performed as previously described (Zhai et al., 2009) with modifications. Briefly, 14-day-old Arabidopsis seedlings were cut into strips and then incubated in the digestion solution containing.5 M mannitol, 10 mM MES-KOH (pH 5.7), 20 mM CaCl2, 40 mM KCl,1% Cellulase R-10 (Yakult Honsha Co., Ltd., Japan), and.3% Macerozyme R-10 (Yakult Honsha Co., Ltd., Japan) with gentle shaking for 3 h in darkness. The protoplasts were collected by centrifugation at 100 g for 7 min and washed twice with the W5 solution containing.1% glucose, 0.08% KCl, 0.9% NaCl, 1.84% CaCl2, and 2 mM MES-KOH (pH 5.7).
Peptide Pull-Down Assay
For differential peptide pull-down (PPD) screening, total proteins derived from.5 g 10-day-old Arabidopsis seedlings were extracted with protein extraction buffer (10 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5%Triton X-100, 1 × piece protease inhibitor EDTA-free (Thermo, Fisher Scientific Inc., MA, United States), and 1 × piece phosphatase inhibitor Mini tablets (Thermo Fisher Scientific Inc., MA, United States), and incubated with 30 μl peptidepY428-beads or peptideY428-beads for 4 h at 4°C with gentle shaking. Proteins not binding with beads were removed with extensive washing, and then the beads-binding proteins in each sample were then analyzed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE), followed by silver staining (Fast Silver Stain Kit, Beyotime, China). The differential protein bands between two samples were identified through mass spectrometry.
For pull-down verification of ZAR1-peptideY428 interaction, 2 ml Arabidopsis protoplasts (∼106 cells) were transfected with 200 μg plasmids DNA (35S:ZAR1-HA) and cultured in darkness. After 12 h of expression, the total proteins of protoplasts were extracted with 300 μl protein extraction buffer [10 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, 1 × piece protease inhibitor EDTA-free, and 1 × piece phosphatase inhibitor Mini tablets]. The total cell extracts were incubated with 30 μl peptidepY428-beads or peptideY428-beads for 4 h at 4°C with gentle shaking. The beads-binding protein was detected by immunoblotting with HRP-conjugated anti-HA antibody.
Co-immunoprecipitation Assay
For co-immunoprecipitation (co-IP) assay in Figure 1D, 2 ml of protoplasts were transfected with 200 μg plasmids DNA, to co-express the ZAR1-FLAG protein with CERK1-HA, CERK1 Y428F-HA, or CERK1 △ RD-HA proteins, respectively. The protoplasts transfected with CERK1-HA plasmids only were used as the negative control. After protein expression for 12 h, the protoplasts were harvested and total proteins were extracted with 300 μl protein extraction buffer [10 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, 1 × piece protease inhibitor/EDTA-free, and 1 × piece phosphatase inhibitor Mini tablets]. The co-IP assay in Arabidopsis protoplasts was conducted according to a detailed protocol described previously (Cheng et al., 2015). Afterward, 10 μl of protein extracts of each sample was used as the input fraction, the remaining crude extracts of each sample were further incubated with 20 μl anti-FLAG M2 affinity gel (Sigma-Aldrich, United States) for 4 h at 4°C with gentle shaking. After washing three times with ice-cold extraction buffer, the gel-bound proteins were eluted by boiling with 30 μl 2 × SDS-PAGE loading buffer. The presence of ZAR1-FLAG and CERK1-HA in the eluate (IP fraction) and input fraction was analyzed by SDS-PAGE and immunoblotting with HRP-conjugated anti-HA or HRP-conjugated anti-FLAG antibodies.
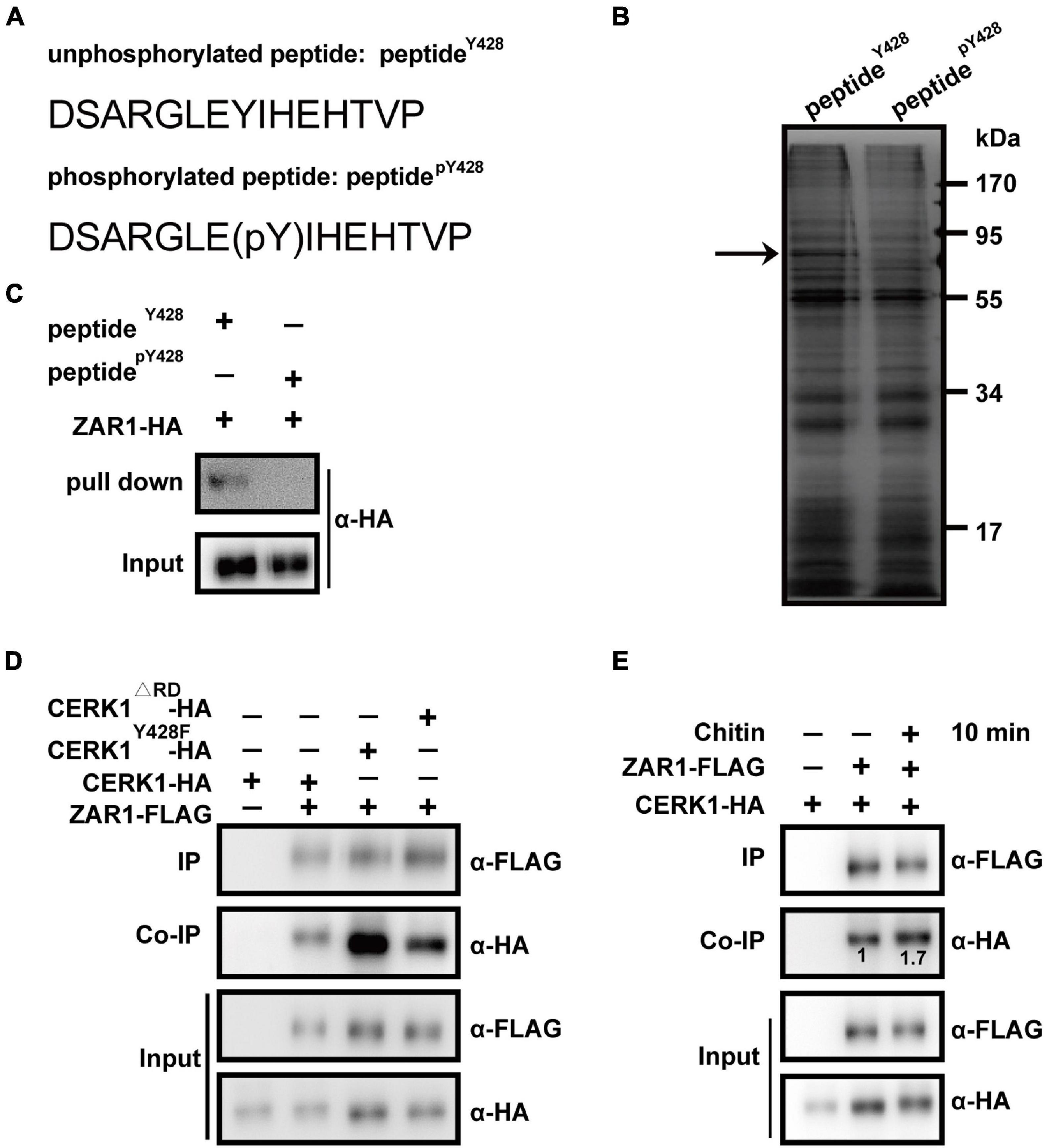
Figure 1. Zygotic arrest 1 (ZAR1) interacts with chitin elicitor receptor KINASE 1 (CERK1) depending on the dephosphorylation of Y428 residue and functions in defense against Botrytis cinerea. (A) Two “bait” peptides, peptidepY428 fragment “DSARGLE(pY)IHEHTVP” and peptideY428 fragment “DSARGLEYIHEHTVP” was designed via the web-based motif-x program (http://motif-x.med.harvard.edu/). (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of differential peptide-pulling down (PPD) proteins. After the PPD assay, the peptideY428 beads-binding proteins and peptidepY428 beads-binding proteins were analyzed with SDS-PAGE and followed by silver staining. The differential protein bands were observed. The differential band was illustrated with an arrow. (C) Pulling-down of ZAR1 protein with peptideY428 beads. The HA-tagged ZAR1 protein expressed in Arabidopsis protoplasts was incubated with peptideY428 beads, and the pulling-down proteins were detected by immunoblotting using an anti-HA antibody. Total protoplast proteins were used as input. The experiments were repeated three times with similar results. (D) Co-immunoprecipitation (co-IP) of ZAR1 and entire CERK1 protein. FLAG-tagged ZAR1 with HA-tagged CERK1Y428F, FLAG-tagged ZAR1 with CERK1Δ RD, or FLAG-tagged ZAR1 with wild type CERK1 was co-expressed in Arabidopsis protoplasts for 12 h, respectively. CERK1Y428F and CERK1Δ RD protein showed enhanced interaction with ZAR1 protein. The experiments were repeated three times with similar results. (E) Interaction between ZAR1 and CERK1 was influenced by chitin elicitation. HA-tagged CERK1 and FLAG-tagged ZAR1 were co-expressed in protoplasts for 12 h, then the co-IP was performed before chitin treatment or after 10 min of chitin treatment. Numbers indicate relative amounts of CERK1-HA co-immunoprecipitated with ZAR1-FLAG, measured using the ImageJ program. The experiments were repeated three times with similar results.
For co-IP assay in Figure 1E, HA-tagged CERK1 and FLAG-tagged ZAR1 were co-expressed in protoplasts for 12 h. After being treated with 50 μg/ml chitin or not, the protoplasts were harvested, and total proteins were extracted according to the previous description. The protoplasts transfected with CERK1-HA plasmids were only used as the negative control.
Botrytis cinerea Infection
Botrytis cinerea (CECT2100, Spanish Type Culture Collection, University de Valencia, Valencia, Spain) was cultured on PDA plates at 22°C for 10 days. The spores were washed, collected with potato dextrose broth (PDB), and diluted to 107 spores/ml. For each infection, 5 μl droplet of spores was inoculated onto the surface of one leaf of the 4-week-old Arabidopsis plants. The leaves were detached from plants and cultivated in a high-humidity environment. After 48–72 h of inoculation, the lesion size was recorded and measured with Image J software.
For measurement of photosystem II (PSII) activity and cell death phenotype, the 4-week-old Arabidopsis plants were sprayed with a B. cinerea spore suspension and incubated under high humidity in darkness (Zhang et al., 2018).
Analysis of Photosystem II Activity
The PSII activity was analyzed by measuring chlorophyll fluorescence. Chlorophyll fluorescence of Arabidopsis plants was measured using the MAXI version of the IMAGING-PAM M-Series chlorophyll fluorescence system (Heinz-Walz Instruments) according to the previous description (Lu et al., 2011; Zhang et al., 2018). The quantum efficiency of light-adapted leaves (ΦPSII) was calculated as (Fm′–F)/Fm′ according to the previous description (Lu et al., 2011).
Trypan Blue Staining
Trypan blue solution contained 10 g phenol, 10 ml glycerol, 10 ml lactic acid, 10 ml water, and.02 g of trypan blue together (stock solution). The working solution was prepared by diluting the stock solution with ethanol (1:3 V/V). The leaves of 4-week-old Arabidopsis plants which were sprayed with B. cinerea spore suspension were boiled in staining solution at 95°C for 10 min. After cooling to room temperature, the leaves were de-stained in chloral hydrate solution (chloral hydrate:H2O:glycerol; 4:2:1 W/V/V) overnight. Images were captured under a Zeiss stereomicroscope (SteREO Lumar version 12, Germany).
Mitogen-Activated Protein Kinase Assay
Ten-day-old Arabidopsis seedlings grown in the.5 × MS liquid medium were treated with 200 μg/ml chitin at different times before the seedlings were frozen in liquid nitrogen. Total proteins were extracted with the protein extraction buffer [10 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, 1 × piece protease inhibitor EDTA-free, and 1 × piece phosphatase inhibitor Mini tablets]. The protein lysates were resolved in a 10% SDS-PAGE gel and activated MAPKs were visualized by immunoblotting with anti-pERK antibodies (Cell Signaling Technology, United States).
Oxidative Burst Assay
Reactive oxygen species measurement was performed as previously described (Zhang et al., 2007). In brief, rosette leaves from 6-week-old Arabidopsis plants were sliced into approximately 1 mm strip and incubated overnight in water in a 96-well culture plate. The next day, the water was replaced by a reaction solution containing.5 mM L-012 (Wako, Japan), 1 mg/ml horseradish peroxidase, and 200 μg/ml chitin (or water as the mock), and the chemiluminescence was measured in a time-course manner using a microplate reader (Varioskan LUX, Thermo Fisher Scientific Inc., United States or Tecan-Spark, Tecan, Switzerland). Each data point represents the average of six replicates.
Transcriptome Sequencing and Data Analysis
Previous studies revealed a major transcriptional shift occurred around 20–28 h post-infection coinciding with the lag phase in B. cinerea growth (Windram et al., 2012). Therefore, 28 HPI was chosen as the time point of transcriptome sequencing in this study. Moreover, the zar1-3 zar2-1 double mutant has shown stronger B. cinerea resistance compared with Col-0 in the early growth stage, which is suitable for the isolation of high-quality RNA. Then RNA-Seq analysis was performed with 12-day-old Col-0 and zar1-3 zar2-1 Arabidopsis seedlings sprayed with B. cinerea or PDB. After 28 h of treatment, the total RNA derived from leaves of each sample was purified for transcriptome sequencing by Beijing Genomics Institute (BGI, Shenzhen, China). Three independent biological replicates were being performed in each experiment group. Gene Ontology (GO) enrichment analysis was performed on a website2 based on a hypergeometric test on the proportion of GO categories between the differentially expressed genes (DEGs) and the whole genome; while the FDR of the test was <0.05. GO categories were considered enriched. The sum of all transcript per million (TPM) values is the same in all samples, such that a TPM value represents a relative expression level that is comparable between Col-0 and zar1-3zar2-1 double mutant, or different treatment conditions.
Raw Illumina sequence reads were available in the Sequence Read Archive (SRA) of the NCBI under BioProject ID: PRJNA757130.3
Quantitative RT-PCR
The 12-day-old Arabidopsis seedlings grown in the 1/2 MS medium were sprayed with B. cinerea or PDB for 28 h. Total RNA derived from leaves of each sample was extracted using a Plant RNA Kit (Magen, China) and 5 mg total RNA was used for cDNA synthesis using the PrimeScript RT reagent kit (Takara, Japan). qPCR was performed using SYBR Premix Ex Taq (Takara, Japan), and amplification was monitored in real-time on the LightCycler 480 (Roche). Reactions were performed in triplicate for each sample, and relative expression levels were normalized using the UBQ5 gene. The primers used for qRT-qPCR are listed in Supplementary Table 2.
Results
ZAR1 Interacts With the CERK1 Motif Containing the Dephosphorylated Y428 Residue
To screen for candidate proteins interacting with the CERK1 motif, specifically with or without phosphorylation of the Y428 residue, we performed a differential PPD assay. A ‘‘bait’’ peptide fragment of CERK1 containing the Y428 residue was designed via the web-based motif-x program4 (Schwartz and Gygi, 2005; Chou and Schwartz, 2011). The peptidepY428 fragment “DSARGLE(pY)IHEHTVP” and the peptideY428 fragment “DSARGLEYIHEHTVP” were synthesized and individually conjugated to agarose beads (Figure 1A). For PPD screening, total proteins derived from 10-day-old Arabidopsis seedlings were incubated with peptidepY428- or peptideY428-conjugated beads. The peptidepY428- or peptideY428-bound proteins were analyzed with SDS-PAGE and the differential protein bands between the samples were further analyzed using mass spectrometry (Figure 1B). Notably, the results indicated that ZAR1 (AT2G01210) could interact with peptideY428, but not peptidepY428 (Figure 1C).
ZAR1 Interacts With CERK1 Protein-Containing Dephosphorylated Y428
We investigated the interaction between ZAR1 and the entire CERK1 protein containing the dephosphorylated Y428 site in vivo. The Y428 phosphorylation is abolished in CERK1Y428F, in which the 428th tyrosine residue of CERK1 is substituted with phenylalanine (F), and in a catalytically inactive CERK1 mutant (CERK1Δ RD; Liu et al., 2018). We evaluated the co-IP of FLAG-tagged ZAR1 protein with HA-tagged wild-type CERK1, CERK1Y428F, or CERK1Δ RD in the Arabidopsis protoplasts. While ZAR1 showed weak interaction with the wild-type CERK1-HA protein, the ZAR1-CERK1 interaction was enhanced in the presence of the Y428F or ΔRD mutation in CERK1 protein (Figure 1D).
Notably, Y428 phosphorylation is a consequence of CERK1 autophosphorylation, and the Y428 phosphorylation level was dynamic during chitin signaling (Liu et al., 2018). We hypothesized that the interaction between ZAR1 and CEKR1 could be influenced by chitin elicitation. Indeed, ZAR1-CERK1 interaction was enhanced after 10 min of chitin treatment (Figure 1E), which was consistent with our earlier observation that phosphorylation of the Y428 residue gradually declined within 0–10 min upon chitin treatment (Liu et al., 2018).
The minor ZAR1-CERK1 interaction was also observed in the absence of chitin (Figures 1D,E). Most of the CERK1 proteins might be autophosphorylated at the Y428 residue in the standby state without chitin signal, while minor dephosphorylation of Y428 residue occurred on occasion, possibilities that need further investigation. Taken together, our differential PPD assay and co-IP data indicated that ZAR1 interacted with CERK1 protein harboring dephosphorylated Y428 residue.
ZAR1 Negatively Contributes to Plant Defense Against Botrytis cinerea Independently of Chitin-Triggered Responses
ZAR1 encodes a receptor-like kinase and regulate the division of zygote in Arabidopsis (Yu et al., 2016). The homozygous zar1-1–/– mutation (in ecotype Landsberg erecta) is embryo lethal, and the strong phenotype is most likely caused by the expression of the truncated ZAR1 protein, which functions in a dominant-negative manner. By contrast, plants harboring the null allele zar1-2 (in ecotype Landsberg erecta) and zar1-3 (in ecotype Col-0) are fertile, which may be due to the functional redundancy of ZAR1 homologs (Yu et al., 2016). To date, whether ZAR1 is involved in biological processes other than zygotic division regulation is still uncovered.
To explore whether ZAR1 is also involved in plant defense, we evaluated the sensitivity to fungal pathogen Botrytis cinerea of two zar1 mutant lines in the Col-0 background, zar1-3 (SALK_021338) and zar1-4 (SALK_143663). After 48 h of B. cinerea infection, both zar1-3 and zar1-4 mutants showed smaller lesion sizes on leaves compared with those of Col-0, indicating the stronger resistance against B. cinerea in both zar1-3 and zar1-4 mutants (Figure 2A). We further characterized this phenotype by measuring photosystem II (PSII) activity and cell death in zar1-3, cerk1, and Col-0 plants that had been sprayed with B. cinerea spore suspension, as previously described (Zhang et al., 2018). The zar1-3 mutant showed stronger resistance against B. cinerea, including smaller lesion sizes, higher PSII system activity, and less cell death compared with Col-0 plants (Figures 2B,C). By contrast, the cerk1 mutant was more sensitive to B. cinerea compared with Col-0 plants (Figures 2B,C). These results imply that ZAR1 negatively contributes to defense against B. cinerea in Arabidopsis.
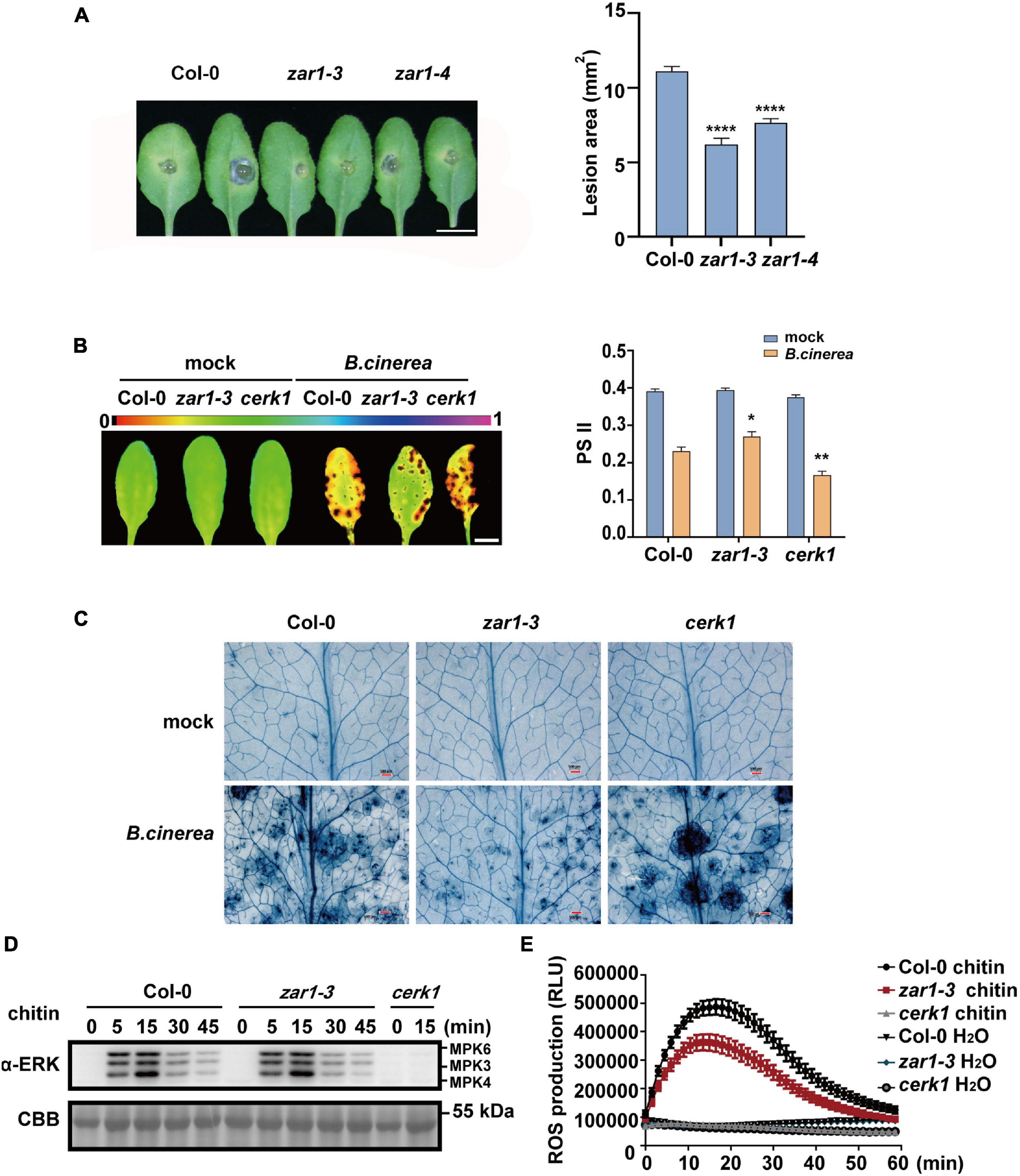
Figure 2. ZAR1 functions in the resistance to the fungal pathogen Botrytis cinerea. (A) ZAR1 functions in the resistance to the fungal pathogen Botrytis cinerea. Lesions on the leaves of zar1-3, zar1-4 mutant, and Col-0 plant were observed at 48 h after inoculation with B. cinerea. Lesion sizes were analyzed. Error bars represent SEM from means of 16 leaves from each line. Asterisks indicate significant differences compared with Col-0 plant (****P < 0.0001, Student’s t-test). Scale bar = 1 cm. (B) Chlorophyll fluorescence imaging and quantification of ΦPSII was measured in zar1-3 mutant, cerk1 mutant, and Col-0 plant, at 36 h after B. cinerea spray. Error bars represent SEM of 16 leaves from each line. Asterisks indicate significant differences between each mutant and Col-0 plant (*P < 0.05, **P < 0.01, Student’s t-test). (C) Visualization of cell death in infected leaves at 36 h post-infection. Representative images of trypan blue (TPB) stained leaves are shown. Scale bar = 500 μm. (D) Chitin-triggered MAPK activation in zar1-3 mutant, cerk1 mutant, and wild type Col-0. 10-day-old seedlings were treated with 200 μg/mL chitin at the indicated time points. MAPK activation was detected with an anti-pERK antibody. cerk1 mutant was included as a control. Equal loading is demonstrated by CBB (Coomassie brilliant blue) staining of Rubisco (below). The experiments were repeated three times, with similar results. (E) Leaves of the 4-week-old zar1-3 mutant, cerk1 mutant, and Col-0 plants were treated with 200 μg/ml chitin and incubated with luminol and horseradish peroxidase to detect ROS. Luminescence was recorded at different time points as indicated. Error bars represent SEM of data derived from replicate samples (n = 12). The chemiluminescence was measured in a time-course manner using a microplate reader (Varioskan LUX, Thermo Fisher Scientific Inc., MA, United States).
As chitin is the best-characterized MAMP of fungal pathogens, we evaluated chitin-triggered immunity responses, focusing on the early stage. We assessed mitogen-activated protein kinases (MAPK) activation and reactive oxygen species (ROS) burst in the zar1-3 mutant upon chitin treatment. As the negative control, the cerk1 mutant completely lost the chitin-induced MAPK activation and ROS burst, as previously observed (Figures 2D,E; Liu et al., 2018). However, there was no difference in MAPK activation between zar1-3 and Col-0 plants (Figure 2D). In addition, the zar1-3 mutant exhibited compromised ROS burst compared with Col-0 upon chitin treatment (Figure 2E). Thus, ZAR1 may play a minor role in the positive regulation of chitin signaling, although ZAR1 contributes negatively to resistance against B. cinerea. Taking these data together, we conclude that the enhanced resistance to B. cinerea in zar1-3 mutant seems to be independent of early chitin-triggered responses such as MAPK activation and ROS burst.
ZAR1 and ZAR2 Function Redundantly in Defense Against Botrytis cinerea
Zygotic arrest 1 functions redundantly in zygotic division regulation (Yu et al., 2016). Although the zar1-3 single mutant showed stronger B. cinerea resistance compared with Col-0, we hypothesized that there could be some functional redundancy of ZAR1 in plant defense. We performed phylogenetic analysis of ZAR1 homologs based on the full-length amino acid sequences obtained from the NCBI non-redundant protein database and identified a ZAR1-homologous kinase named ZAR2 (AT1G25320) (Figure 3A). We obtained the homozygous zar1-3 zar2-1 double mutant by crossing zar1-3 with zar2-1 (Figures 3B,C). The zar1-3 zar2-1 double mutant plants exhibited almost normal growth and development compared with single mutants, despite a minor reduction in the leaf size. After 72 h of B. cinerea infection, the average lesion size on Col-0 leaves was about 40 mm2, whereas the average lesion size on zar1-3 zar2-1 double mutant leaves was about 20 mm2, much smaller than that of zar1-3 single mutant, zar2-1 single mutant, cerk1, and Col-0 plants (Figure 3D). This result indicated that ZAR1 and ZAR2 might function redundantly in defense against B. cinerea in Arabidopsis.
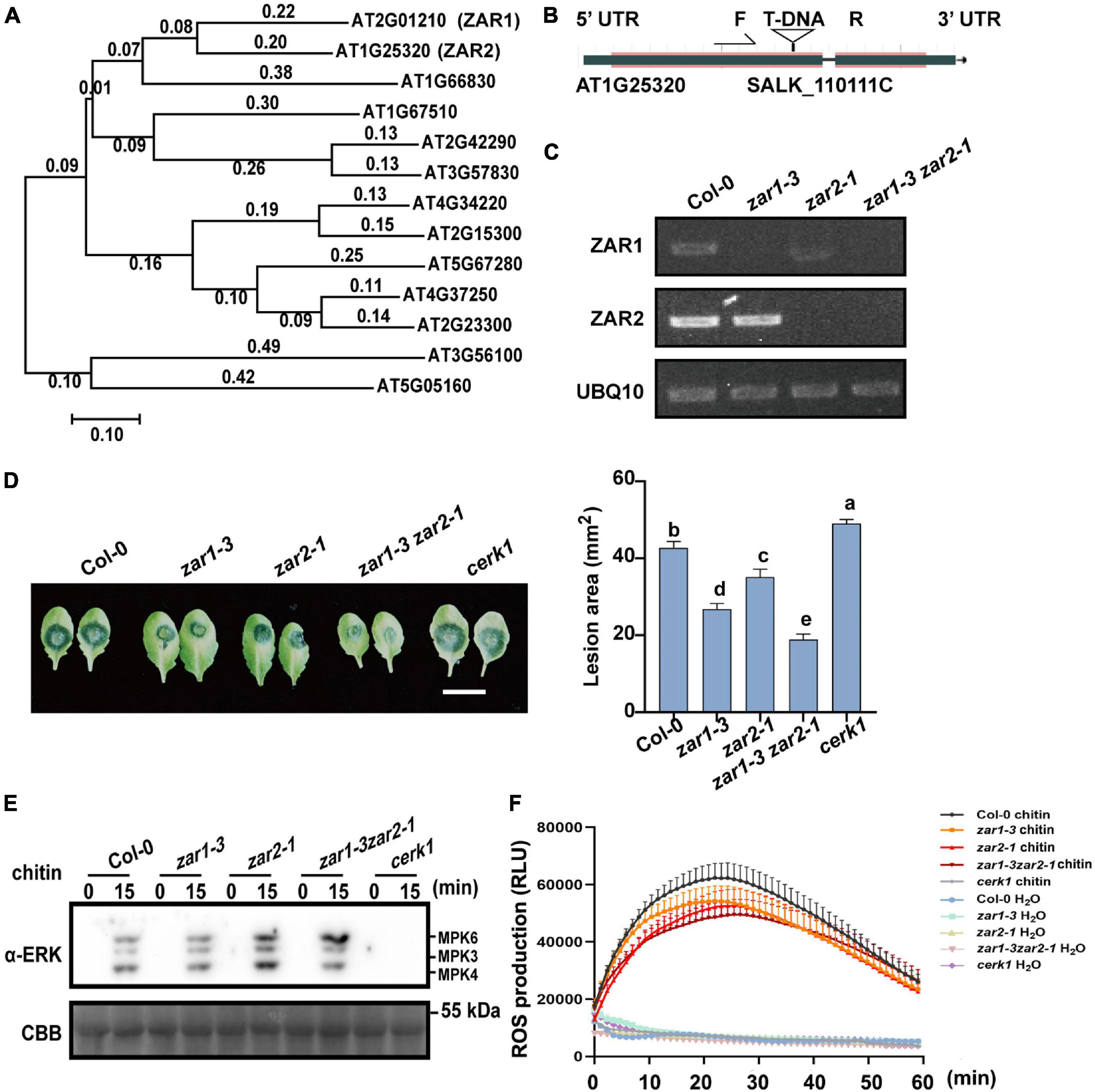
Figure 3. ZAR1 and ZAR2 function redundantly in defense against B. cinerea. (A) The unrooted phylogenetic tree was constructed based on the full-length amino acid sequences of Arabidopsis proteins using MEGA 7 software by the Neighbor-Joining method. AT1G25320 (ZAR2) is most homologous to ZAR1. Scale bar is indicated. (B,C) Identification of zar1-3zar2-1 double mutant. zar2-1 (SALK-110111c) is a T-DNA insertion mutant in the Col-0 ecotype. zar1-3zar2-1 double mutant was obtained by crossing zar1-3 mutant with zar2-1 mutant. The transcription of ZAR1 and ZAR2 genes in this double mutant was examined by RT-PCR. UBQ10 was used as an internal control. (D) Both ZAR1 and ZAR2 negatively contribute to resistance against B. cinerea. Lesions on the leaves of zar1-3 mutant, zar2-1 mutant, cerk1 mutant, zar1-3zar2-1 double mutant, and Col-0 plants were observed at 72 h after inoculation with B. cinerea. Lesion sizes were analyzed. Error bars represent SEM from means of 16 leaves from each line. Different letters indicate significant difference at P < 0.05, as determined by one-way ANOVA with Tukey’s post-test. Scale bar = 1 cm. (E) Chitin-triggered MAPK activation in zar1-3 mutant, zar2-1 mutant, zar1-3zar2-1 double mutant, cerk1 mutant, and wild type Col-0. 10-day-old seedlings were treated with 200 μg/mL chitin at the indicated time points. MAPK activation was detected with an anti-pERK antibody. cerk1 mutant was included as a control. Equal loading is demonstrated by CBB (Coomassie brilliant blue) staining of Rubisco (below). The experiments were repeated three times, with similar results. (F) Leaves of the 4-week-old zar1-3 mutant, zar2-1 mutant, zar1-3zar2-1 double mutant, cerk1 mutant, and Col-0 plants were treated with 200 μg/mL chitin and incubated with luminol and horseradish peroxidase to detect ROS. Luminescence was recorded at different time points as indicated. Error bars represent SEM of data derived from replicate samples (n = 12). The chemiluminescence was measured in a time-course manner using a microplate reader (Tecan-Spark, Tecan).
Since the negative role of ZAR1 in defense against B. cinerea seems to be independent of the chitin-triggered early PTI responses (Figures 2D,E), we further explored the chitin-triggered early responses in zar1-3 zar2-1 double mutant plants. Notably, the chitin-triggered MAPK activation was slightly enhanced in zar2-1 single mutant and zar1-3 zar2-1 double mutant (Figure 3E). In addition, both zar2-1 single mutant and zar1-3 zar2-1 double mutant exhibited compromised ROS burst upon chitin treatment, similar to that of zar1-3 plants (Figure 3F). These results indicated that ZAR1 and ZAR2 may play variable roles in chitin-triggered MAPK activation, as well as minor positive roles in ROS burst. Thus, the remarkably enhanced resistance against B. cinerea in zar1-3 zar2-1 double mutant might be due to the potential regulatory pathways other than pattern triggered immunity (PTI) responses.
We further explored the downstream mechanism of ZAR1/ZAR2-related defense against B. cinerea using transcriptome analysis. We performed transcriptome sequencing in Col-0 and zar1-3 zar2-1 double mutant plants 28h after treatment with B. cinerea. Total RNAs derived from Col-0 or zar1-3 zar2-1 double mutant plants, treated with B. cinerea spore or PDB (as a mock treatment) spray, were compared between different genotypes and treatments. The numbers of up- and down-regulated genes (P-value < 0.05, fold change > 2) in the samples of 1) PDB-treated Col-0 compared with B. cinerea-treated Col-0 plants, 2) PDB-treated zar1-3 zar2-1 compared with B. cinerea-treated zar1-3 zar2-1 plants, 3) PDB-treated Col-0 compared with PDB-treated zar1-3 zar2-1 plants, and 4) B. cinerea-treated Col-0 compared with B. cinerea-treated zar1-3 zar2-1 plants, are illustrated in Figures 4A,B (Supplementary Table 1). Interestingly, there were many genes encoding transcription factors (TFs) among the DEGs between Col-0 and zar1-3 zar2-1 after B. cinerea treatment. The top four enriched clusters analyzed through GO were defense-related transcription factors, including ethylene response factors (ERFs), MYB domain proteins (MYBs), NAC domain containing proteins (NACs), and WRKY DNA-binding proteins (WRKYs) (Figures 5A,B).
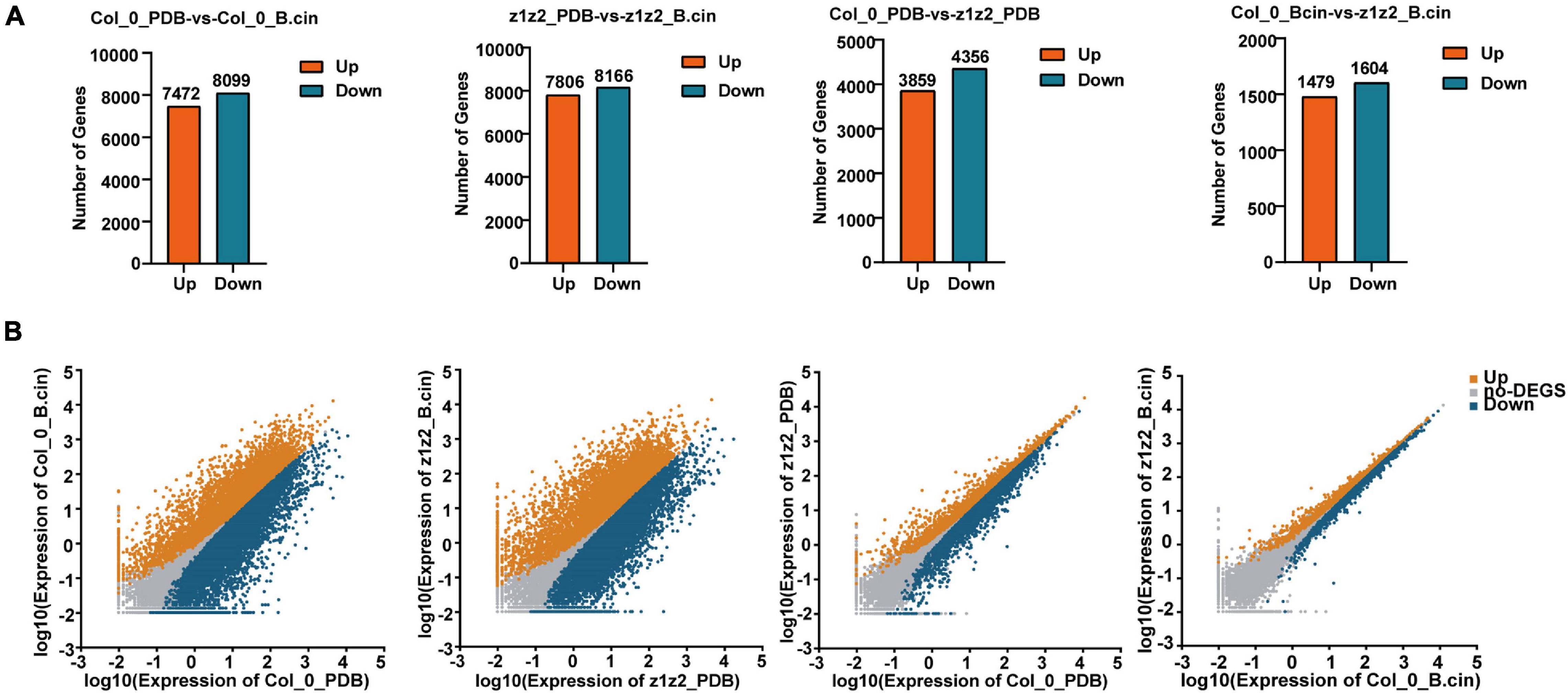
Figure 4. Data analysis of RNA-sequencing. (A) RNA-sequencing was performed in Col-0 and zar1-3zar2-1 double mutant after 28 h of treatment with B. cinerea spray or PDB. Each treatment contained three biological replicates. Total RNAs derived from Col-0 or zar1-3zar2-1 double mutant plants upon B. cinerea infection or PDB were compared between different genotypes or treatment conditions. Numbers of up- and down-regulated genes were illustrated (P-value < 0.05, fold change > 2). z1z2_PDB: zar1-3zar2-1 double mutant plants treated with PDB; z1z2_B.cin: zar1-3zar2-1 double mutant plants treated with B. cinerea; Col_0_PDB: Col-0 plants treated with PDB; Col_0_B.cin: Col-0 plants treated with B. cinerea. (B) The scatter plots illustrate differentially expressed genes between Col-0 and zar1-3 zar2 double mutant. Genes up-regulated, down-regulated, or unchanged were shown in orange, blue, or gray, respectively. | log2FC| > 1 and FDR < 0.05 were set as a cut off.
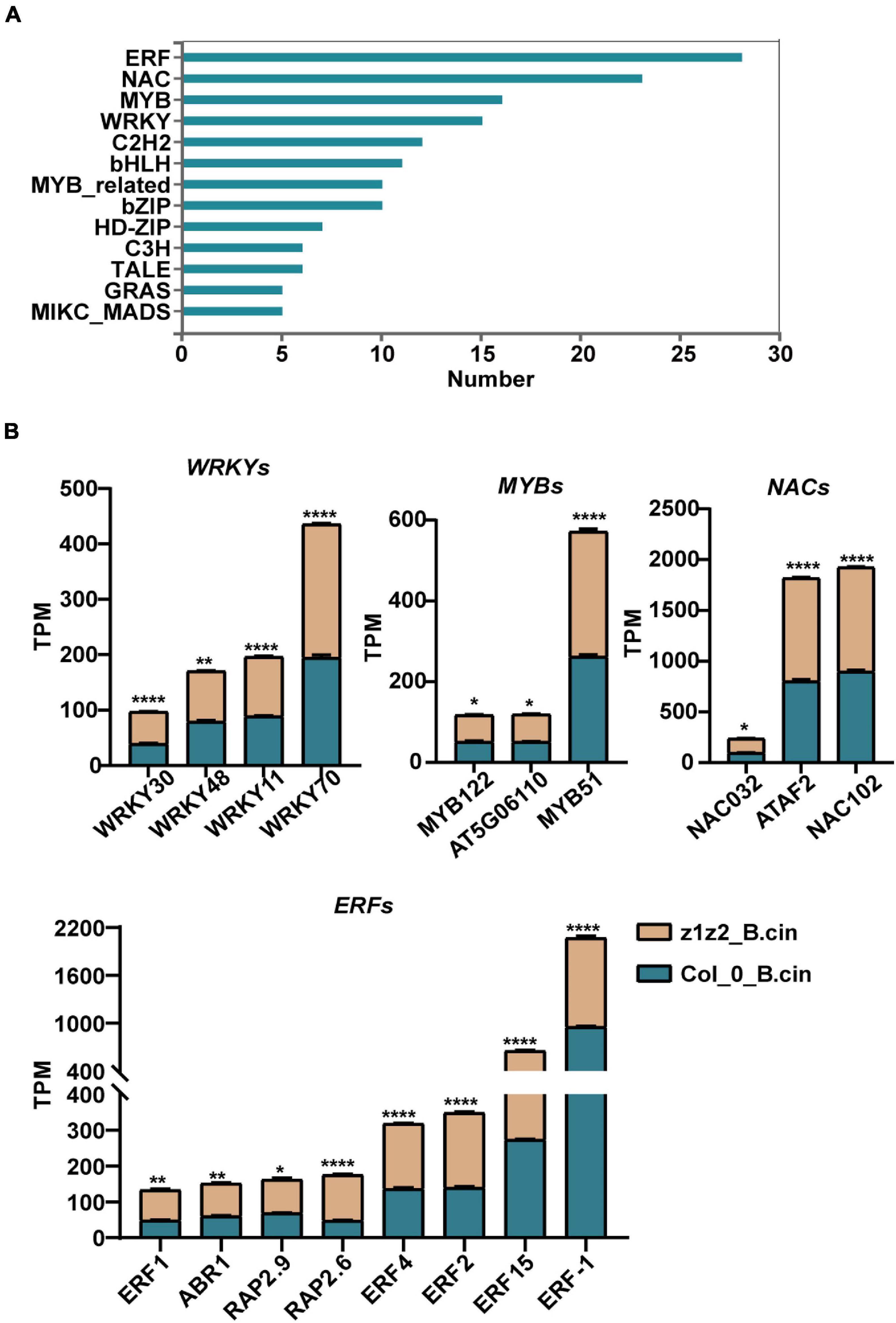
Figure 5. Gene Ontology (GO) analyses of differentially expressed transcription factor genes between zar1-3zar2-1 and Col-0 plants upon B. cinerea infection. (A) Top 10 enriched clusters (GO analysis) of transcription factors genes based on data of the differentially expressed genes between Col-0 (B. cinerea treatment) and zar1-3 zar2 (B. cinerea treatment) detected in the RNA-sequencing. (B) TPM (transcripts per Million) value of four defense-related transcription factor gene subgroups (WRKYs, MYBs, NACs, and ERFs) derived from the differentially expressed genes between Col-0 and zar1-3zar2-1 mutant after B. cinerea infection. Asterisks indicate significant differences between zar1-3zar2-1 mutant and Col-0 plant (*P < 0.05, **P < 0.01, ****P < 0.0001, Student’s t-test).
The expression profiles of these TF genes were further validated with quantitative RT-PCR. In the absence of B. cinerea (PDB treatment), the basal transcriptional levels of these investigated genes were much lower than that of the UBQ5 gene (Figure 6). After B. cinerea infection, the expression of WRKY30, WRKY48, WRKY11, and WRKY70 was remarkably increased in zar1-3 zar2-1, compared with single mutants and Col-0 plants (Figure 6). In addition, the expression levels of MYB122, MYB51, NAC032, NAC102, as well as several ERF genes, RAP2.9, RAP2.6, ERF4, and FER15, were strongly up-regulated upon B. cinerea infection in the zar1-3 zar2-1 double mutant. The relative expression level of these genes in zar1-3 zar2-1 was much higher than those of Col-0 plants (Figure 6). Thus, upon B. cinerea infection, the transcript levels of multiple defense-related TF genes in the zar1-3 zar2-1 double mutant were increased to much higher levels compared with those in single mutants and Col-0 plants, which was consistent with the remarkably enhanced resistance observed in the zar1-3 zar2-1 double mutant.
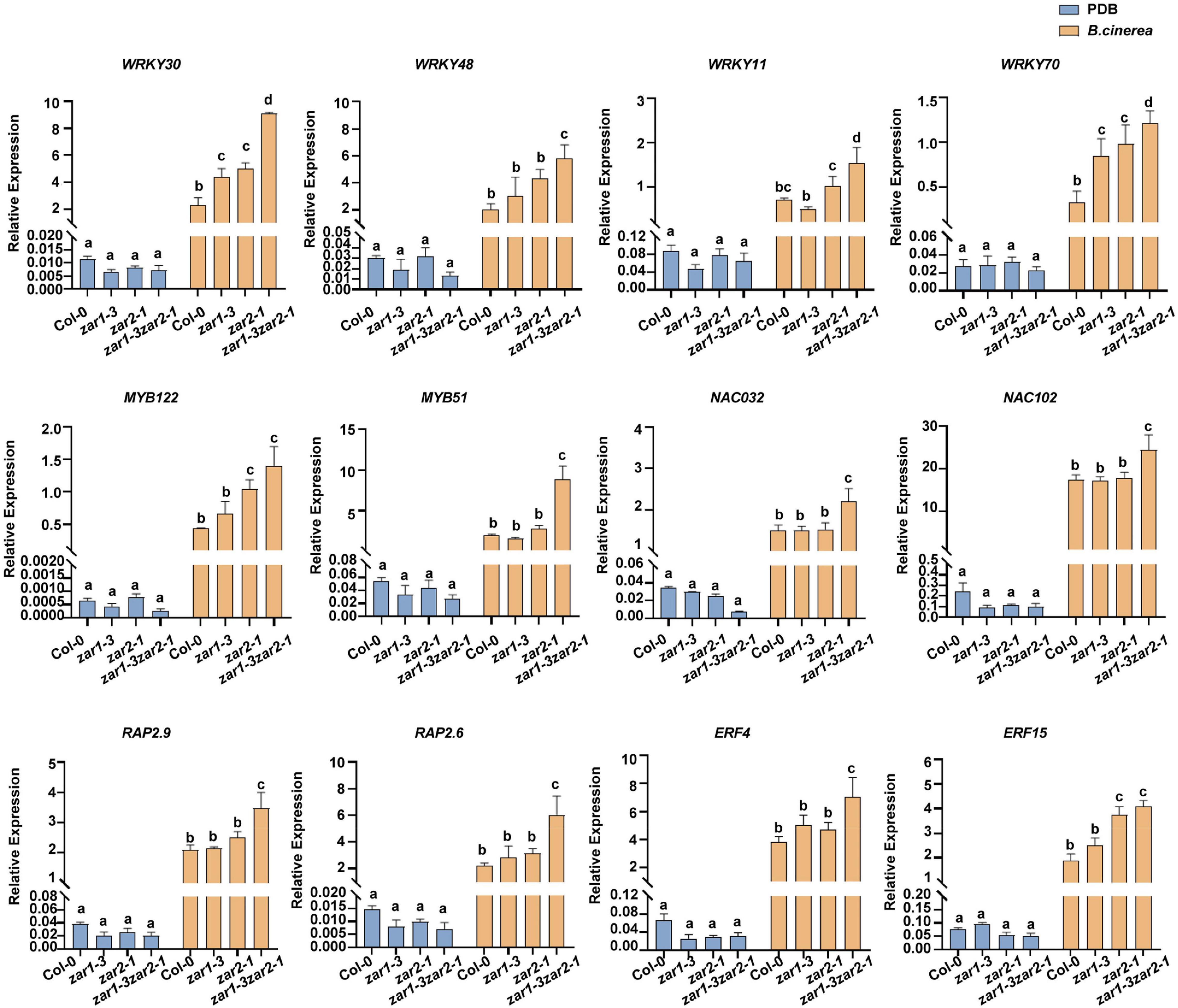
Figure 6. The relative expression levels of defense-related transcription factor genes induced by B. cinerea were enhanced in zar1-3zar2-1 double mutant. The relative expression levels of defense-related transcription factor genes upon B. cinerea infection or PDB treatment were evaluated in zar1-3 mutant, zar2-1 mutant, zar1-3zar2-1 double mutant, and Col-0 plant. Twelve-day-old seedlings were inoculated with B. cinerea spores or PDB for 28 h. The relative expression levels of defense-related TF genes were analyzed by qPCR, UBQ5 was used as an internal control. The relative expression was calculated by the transcription level of each gene relative to UBQ5 upon B. cinerea or PDB treatment. The different letter indicates significant difference (p < 0.05), as determined by one-way ANOVA with Tukey’s post-test.
Discussion
Plants have evolved various cell-surface RLKs that perceive diverse signals, such as phytohormones and peptides derived from plants, and pattern molecules derived from microbes. The coordination of growth and defense that depends on such receptors improves the ability of plants to adapt to environmental changes. In our current study, we demonstrated that the zygotic division regulator ZAR1 could directly bind to the defense protein CERK1. Furthermore, we showed that this interaction depended on dephosphorylation at the Y428 residue of CERK1 (Figure 1). Moreover, ZAR1 functioned redundantly with its homolog ZAR2 in plant defense against B. cinerea (Figures 2–6). Our results suggested a possible functional connection between ZAR1 and CERK1, each of which was originally identified as being involved in separate processes. Notably, ZAR1 is an LRR-RLK (Yu et al., 2016). LRR-RLKs are critical proteins involved in processes of plant development and stress responses in Arabidopsis. For example, the bacterial MAMP cognate receptors FLS2 and EFR are LRR-RLKs that modulate plant immune signaling (Gomez-Gomez and Boller, 2000; Zipfel et al., 2006). Moreover, LRR-RLKs play multiple roles during plant growth and development. For example, the BRI1-associated receptor kinase 1 (BAK1) functions in both brassinosteroid (BR)-dependent growth regulation and plant innate immunity in Arabidopsis (Chinchilla et al., 2007; Heese et al., 2007; Kim et al., 2013). The LRR-RLK ERECTA recognizes epidermal patterning factors (EPFs)/EPF-like proteins (EPFLs) and is essential in plant growth and development (Torii et al., 1996; Shpak et al., 2003; Meng et al., 2012). ERECTA can also regulate immune responses in Arabidopsis (Godiard et al., 2003; Jorda et al., 2016). In the current study, our findings identified crosstalk between the LRR-RLK ZAR1 and the LysM-RLK CERK1, as well as revealed that ZAR1 has multiple roles, functioning in both zygotic division and pathogen defense.
ZYGOTIC ARREST 1 can interact physically with the heterotrimeric G-protein Gβ (AGB1) and calmodulin to regulate the zygote development (Yu et al., 2016). ZAR1 and AGB1 form a complex, and the kinase activity of ZAR1 is activated by the interaction of AGB1 (Yu et al., 2016). Interestingly, AGB1 has been reported to be a minor positive regulator of chitin-induced ROS burst (Liu et al., 2013). We similarly observed that loss of ZAR1 function impaired the chitin-induced ROS burst in the zar1-3 mutant (Figure 2E). Thus, both ZAR1 and AGB1 may play minor roles in the positive regulation of chitin-triggered early responses. However, it was paradoxical that loss of function of ZAR1 homologs enhanced the plant defense against B. cinerea. We speculated that ZAR1 was involved in the regulation of defense against B. cinerea independent of the chitin- trigged PTI responses, although ZAR1 could be recruited by the chitin receptor complex depending on the dephosphorylation of the residue 428 tyrosine of CERK1. Moreover, the ambivalent effects of ZAR2 deficiency on chitin signaling in the zar1-3 zar2-1 double mutant further implied that ZAR1 homologs contributed to B. cinerea resistance beyond chitin signaling.
In host plants upon pathogen infection, defense signals driven by the plant immune responses are ultimately transmitted into the nucleus and the massive transcriptional reprogramming was governed by a complex gene regulatory network. TFs are key components in this regulatory cascade and function in plant resistance to pathogens by activating and repressing the expression of multiple genes. Upon B. cinerea infection in the zar1-3 zar2-1 double mutant, the levels of induced transcription for various defense-related transcription factors were much higher than those of Col-0 plants, suggesting global activation of defense-related gene expression (Figures 5, 6). Furthermore, genes in pathways associated with defense-related hormones, such as the jasmonate (JA), salicylic acid (SA), ethylene (ET), and abscisic acid (ABA) pathway, were much more highly activated in the zar1-3 zar2-1 double mutant compared with Col-0 plants upon B. cinerea infection (Figure 7), which still needs further validation.
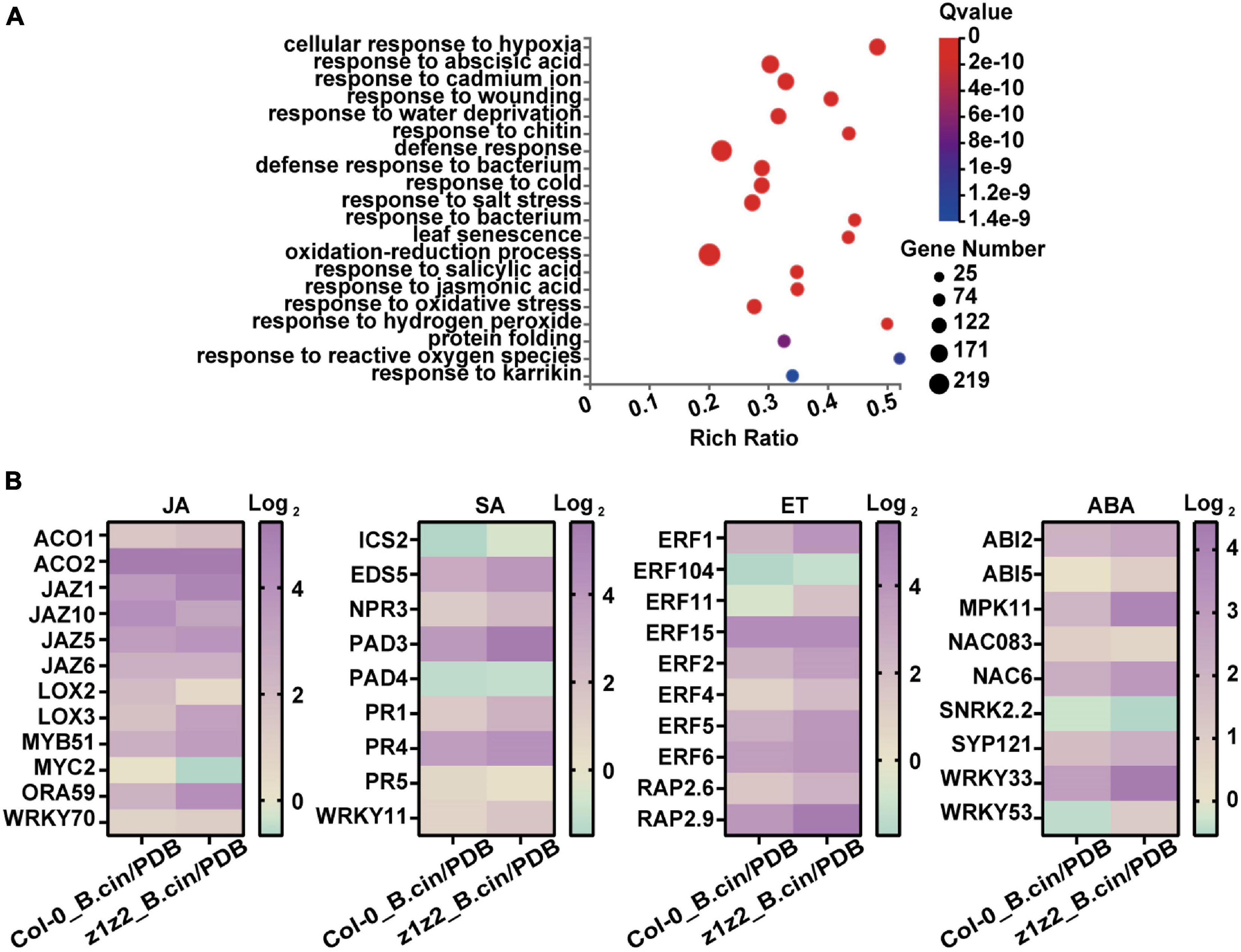
Figure 7. Analysis of differentially expressed genes involved in defense-related hormone pathways in zar1-3 zar2-1 and Col-0 plants upon B. cinerea infection. (A) Top 20 GO enrichment of the genes detected in the transcriptomes that have significant differences in Col-0 plants (B. cinerea treatment) and zar1-3zar2 plants (B. cinerea treatment). (B) Expression profiles of various JA, SA, ET, and ABA-related genes in Col-0 and zar1-3zar2-1 double mutant compared between different treatment conditions (Col-0_ B.cin/PDB: Col-0 samples treated with B. cinerea compared with Col-0 samples treated with PDB, z1z2_B.cin/PDB: zar1-3zar2-1 double mutant samples treated with B. cinerea compared with zar1-3zar2-1 double mutant samples treated with PDB). Heatmap showed TPM levels of DEG genes from RNA-sequencing.
In conclusion, our study established that ZAR1 played multifaceted functions in addition to zygotic division regulation in Arabidopsis. It was curious that loss of function of ZAR1 enhanced resistance against B. cinerea, independent of the intact chitin-induced MAPK activation and compromised chitin-elicited ROS burst in the zar1-3 mutant. Our results highlighted the possibility that ZAR1 homologs could contribute to fungal resistance beyond chitin signaling. Recently, the important role of crosstalk between RLKs in regulating plant immunity has been recognized (Tang et al., 2017; Gong et al., 2019). Receptors can integrate their signaling to regulate defense responses (Macho and Zipfel, 2014; Tang et al., 2017; Miao et al., 2020). As LRR-RLKs, ZAR1 homologs could perceive ligand signals besides chitin and would have the potential to integrate differential signaling pathways to precisely regulate defense responses in plants upon pathogen infection, a concept that needs further investigation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: http://www.ncbi.nlm.nih.gov/bioproject/, PRJNA757130.
Author Contributions
LC and BL designed the study and wrote the manuscript. LC, JX, YS, YL, JL, and HC performed the research. H-BW analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (Nos. 31970521 and 31571249).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Wei-Cai Yang for the kind gift of the zar1-3 seeds, Gary Stacey for the kind gift of the cerk1 mutant seeds, Shouan Liu for the kind gift of Botrytis cinerea.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.736560/full#supplementary-material
Footnotes
- ^ www.Arabidopsis.org.uk
- ^ http://geneontology.org/
- ^ http://www.ncbi.nlm.nih.gov/bioproject/757130
- ^ http://motif-x.med.harvard.edu/
References
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Cao, Y. R., Liang, Y., Tanaka, K., Nguyen, C. T., Jedrzejczak, R. P., Joachimiak, A., et al. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3:e03766. doi: 10.7554/eLife.03766
Cheng, Z., Li, J. F., Niu, Y., Zhang, X. C., Woody, O. Z., Xiong, Y., et al. (2015). Pathogen-secreted proteases activate a novel plant immune pathway. Nature 521, 213–216. doi: 10.1038/nature14243
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nurnberger, T., Jones, J. D., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Chou, M. F., and Schwartz, D. (2011). Biological sequence motif discovery using motif-x. Curr. Protocols Bioinform. 13, 15–24. doi: 10.1002/0471250953.bi1315s35
Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor signaling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Godiard, L., Sauviac, L., Torii, K. U., Grenon, O., Mangin, B., Grimsley, N. H., et al. (2003). ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 36, 353–365. doi: 10.1046/j.1365-313X.2003.01877.x
Gomez-Gomez, L., and Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Gong, B. Q., Guo, J., Zhang, N., Yao, X., Wang, H. B., and Li, J. F. (2019). Cross-microbial protection via priming a conserved immune co-receptor through juxtamembrane phosphorylation in plants. Cell Host Microbe 26, 810–822. doi: 10.1016/j.chom.2019.10.010
Gong, B. Q., Wang, F. Z., and Li, J. F. (2020). Hide-and-Seek: Chitin-Triggered Plant Immunity and Fungal Counterstrategies. Trends Plant Sci. 25, 805–816. doi: 10.1016/j.tplants.2020.03.006
Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M., He, K., Li, J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. P. Natl. Acad. Sci. USA 104, 12217–12222. doi: 10.1073/pnas.0705306104
Jorda, L., Sopena-Torres, S., Escudero, V., Nunez-Corcuera, B., Delgado-Cerezo, M., Torii, K. U., et al. (2016). ERECTA and BAK1 Receptor Like Kinases Interact to regulate immune responses in Arabidopsis. Front. Plant Sci. 28:897. doi: 10.3389/fpls.2016.00897
Kim, B. H., Kim, S. Y., and Nam, K. H. (2013). Assessing the diverse functions of BAK1 and its homologs in Arabidopsis, beyond BR signaling and PTI responses. Mol. Cell 35, 7–16. doi: 10.1007/s10059-013-2255-3
Le, M. H., Cao, Y., Zhang, X. C., and Stacey, G. (2014). LIK1, a CERK1-interacting kinase, regulates plant immune responses in Arabidopsis. PLoS One 9:e102245. doi: 10.1371/journal.pone.0102245
Liu, J., Ding, P., Sun, T., Nitta, Y., Dong, O., Huang, X., et al. (2013). Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol. 161, 2146–2158. doi: 10.1104/pp.112.212431
Liu, J., Liu, B., Chen, S., Gong, B. Q., Chen, L., Zhou, Q., et al. (2018). A tyrosine phosphorylation cycle regulates fungal activation of a plant receptor Ser/Thr kinase. Cell Host Microbe 23, 241–253. doi: 10.1016/j.chom.2017.12.005
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. doi: 10.1126/science.1218867
Lu, Y., Hall, D. A., and Last, R. L. (2011). A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23, 1861–1875. doi: 10.1105/tpc.111.085456
Macho, A. P., and Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Meng, X., Wang, H., He, Y., Liu, Y., Walker, J. C., Torii, K. U., et al. (2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24, 4948–4960. doi: 10.1105/tpc.112.104695
Miao, S., Liu, J., Guo, J., and Li, J. F. (2020). Engineering plants to secrete affinity-tagged pathogen elicitors for deciphering immune receptor complex or inducing enhanced immunity. J. Integr. Plant Biol. 62, 761–776. doi: 10.1111/jipb.12859
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104, 19613–19618. doi: 10.1073/pnas.0705147104
Schwartz, D., and Gygi, S. P. (2005). An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398. doi: 10.1038/nbt1146
Shpak, E. D., Lakeman, M. B., and Torii, K. U. (2003). Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA Leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15, 1095–1110. doi: 10.1105/tpc.010413
Stegmann, M., Monaghan, J., Smakowska-Luzan, E., Rovenich, H., Lehner, A., Holton, N., et al. (2017). The receptor kinase FER is a RALFregulated scaffold controlling plant immune signaling. Science 355, 287–289. doi: 10.1126/science.aal2541
Tang, D., Wang, G., and Zhou, J. M. (2017). Receptor Kinases in Plant-Pathogen Interactions: More Than Pattern Recognition. Plant Cell 29, 618–637.
Torii, K. U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R. F., et al. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. doi: 10.1105/tpc.8.4.735
Wan, J., Zhang, X. C., Neece, D., Ramonell, K. M., Clough, S., Kim, S. Y., et al. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20, 471–481. doi: 10.1105/tpc.107.056754
Windram, O., Madhou, P., McHattie, S., Hill, C., Hickman, R., Cooke, E., et al. (2012). Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. Plant Cell 24, 3530–3557. doi: 10.1105/tpc.112.102046
Yamada, K., Yamaguchi, K., Shirakawa, T., Nakagami, H., Mine, A., Ishikawa, K., et al. (2016). The Arabidopsis CERK1-associated kinase PBL27 connects chitin perception to MAPK activation. EMBO J. 35, 2468–2483. doi: 10.15252/embj.201694248
Yeh, Y. H., Panzeri, D., Kadota, Y., Huang, Y. C., Huang, P. Y., Tao, C. N., et al. (2016). The Arabidopsis malectinlike/LRR-RLK IOS1 is critical for BAK1-dependent and BAK1-independent pattern-triggered immunity. Plant Cell 28, 1701–1721. doi: 10.1105/tpc.16.00313
Yu, T. Y., Shi, D. Q., Jia, P. F., Tang, J., Li, H. J., Liu, J., et al. (2016). The Arabidopsis Receptor Kinase ZAR1 Is Required for Zygote Asymmetric Division and Its Daughter Cell Fate. PLoS Genet. 12:e1005933. doi: 10.1371/journal.pgen.1005933
Zhai, Z., Jung, H. I., and Vatamaniuk, O. K. (2009). Isolation of protoplasts from tissues of 14-day-old seedlings of Arabidopsis thaliana. J. Vis. Exp. 30:1149. doi: 10.3791/1149
Zhang, H., Hu, Z., Lei, C., Zheng, C., Wang, J., Shao, S., et al. (2018). A Plant Phytosulfokine Peptide Initiates Auxin-Dependent Immunity through Cytosolic Ca(2+) Signaling in Tomato. Plant Cell 30, 652–667. doi: 10.1105/tpc.17.00537
Zhang, J., Li, W., Xiang, T., Liu, Z., Laluk, K., Ding, X., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7, 290–301. doi: 10.1016/j.chom.2010.03.007
Zhang, J., Shao, F., Li, Y., Cui, H., Chen, L., Li, H., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1, 175–185. doi: 10.1016/j.chom.2007.03.006
Zipfel, C., Kunze, G., Chinchilla, D., Caniard, A., Jones, J. D., Boller, T., et al. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760.
Keywords: ZYGOTIC ARREST 1, CERK1, plant resistance, Botrytis cinerea, Arabidopsis
Citation: Chen L, Xiao J, Song Y, Li Y, Liu J, Cai H, Wang H-B and Liu B (2021) The Zygotic Division Regulator ZAR1 Plays a Negative Role in Defense Against Botrytis cinerea in Arabidopsis. Front. Plant Sci. 12:736560. doi: 10.3389/fpls.2021.736560
Received: 14 July 2021; Accepted: 29 September 2021;
Published: 26 October 2021.
Edited by:
Simone Ferrari, Sapienza University of Rome, ItalyReviewed by:
Mario Serrano, National Autonomous University of Mexico, MexicoDaniel V. Savatin, University of Tuscia, Italy
Copyright © 2021 Chen, Xiao, Song, Li, Liu, Cai, Wang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Liu, bGl1YmluZ0BtYWlsLnN5c3UuZWR1LmNu
 Lijuan Chen
Lijuan Chen Jiahui Xiao1
Jiahui Xiao1 Yuxiao Song
Yuxiao Song You Li
You Li Huiren Cai
Huiren Cai Hong-Bin Wang
Hong-Bin Wang Bing Liu
Bing Liu