- 1Department of Biology, Chungnam National University, Daejeon, South Korea
- 2Nakdonggang National Institute of Biological Resources, Sangju-si, South Korea
The genus Spumella, established by Cienkowsky in 1870, is characterized by omnivory, two (rarely three) flagella, a short stick-like structure beneath the flagella, a threadlike stalk, cell division via constriction and cyst formation. Since the first phylogenetic study of Spumella-like flagellates, their paraphyly has consistently been shown, with separation into several genera. More recently, Spumella was carefully investigated using molecular and morphological data to propose seven new species. Classification of this genus and knowledge of its species diversity remain limited because Spumella-like flagellates are extremely difficult to identify based on limited morphological characters. To understand the phylogeny and taxonomy of Spumella, we analyzed molecular and morphological data from 47 strains, including 18 strains isolated from Korean ponds or swamps. Nuclear SSU, ITS and LSU rDNA data were used for maximum likelihood and Bayesian analyses. The molecular data divided the strains into 15 clades, including seven new lineages, each with unique molecular signatures for nuclear SSU rRNA from the E23-2 to E23-5 domains, the spacer between the E23-8 and E23-9 domains of the V4 region and domain 29 of the V5 region. Our results revealed increased species diversity in Spumella. In contrast to the molecular phylogeny results, the taxa showed very similar cell morphologies, suggesting morphological convergence into simple nanoflagellates to enable heterotrophy. Three new species produced stomatocysts in culture. Aspects of stomatocyst morphology, including collar structure, surface ornamentation, and cyst shape, were very useful in differentiating the three species. The general ultrastructure of Spumella bureschii strain Baekdongje012018B8 and S. benthica strain Hwarim032418A5 showed the typical chrysophyte form for the leucoplast, a vestigial chloroplast surrounded by four envelope membranes, supporting the hypothesis that Spumella evolved from a phototroph to a heterotroph via the loss of its photosynthetic ability. Seven new species are proposed: S. benthica, S. communis, S. longicolla, S. oblata, S. rotundata, S. similis, and S. sinechrysos.
Introduction
The genus Monas was established by Müller (1773), who originally included three species: Monas termo, M. lens, and M. mica. At that time, members of the genus were delineated by being inconspicuous, mostly simple, translucent, and small in size. The first two species were transferred to Oikomonas and Heteromita (Kent, 1880) and are now known as Oikomonas termo and Bodo lens. Between creation of the genus and the middle of the nineteenth century, researchers described many Monas species (Müller, 1786; Ehrenberg, 1838; Dujardin, 1841; Pritchard, 1845, 1861; Perty, 1852; Fresenius, 1858; James-Clark, 1868). In 1838, Ehrenberg defined the genus using negative characteristics such as the lack of a tail, lip, stigma, and colony formation and added 26 new species. Dujardin (1841) redefined the genus more specifically as naked, round or oblong in shape, and possessing a flagellum and added 12 new species. James-Clark (1868) presented more details of two Monas species than provided in any previous description. Even then, the genus Monas was not well defined and was recognized as an assemblage of diverse protistan organisms.
In 1870, Cienkowsky established the new genus Spumella based on his detailed observations, characterizing the genus by its heterotrophy (feeding on algae, fungi, and starch grains), spherical or oval shape, two (rarely three) flagella, short stick-like structure beneath the flagella, one or two contractile vacuoles, short threadlike stalk, cell division via constriction at any point of the cell and cyst formation. Later, the type species, Spumella vulgaris, was considered the same as Monas guttula Ehrenberg (Stein, 1878; Kent, 1880) and finally synonymized as M. guttula (Reynold, 1934). Kent (1880) listed 26 Monas species and two Spumella species simultaneously and established a new Monas-like genus, Physomonas, described as a stalked monad possessing two unequal flagella. Three years later, Bütschli (1883) treated the genera Spumella and Physomonas as synonyms of Monas. In the early twentieth century, Pascher (1912) established another new genus, Heterochromonas, which is a counterpart of photosynthetic Ochromonas, and included two species previously known as Monas and Spumella species: Heterochromonas vivipara and H. vulgaris. Skuja (1939, 1948, 1956) followed Pascher’s classification system and added 10 new Heterochromonas species. Bourrelly (1957) restricted this genus to species that reliably produce endogenous cysts and designated H. vivipara as a lectotype. To date, more than 100 species of Monas-like flagellates (Heterochromonas, Monas, and Spumella) have been described (Supplementary Figure 1). According to Silva (1960), Monas mica is one of the originally described species and should be designated as the lectotype, but it has not observed since the time of first description. Therefore, there is no lectotype for the genus Monas. Silva (1960) suggested that the genera Monas and Heterochromonas should be abandoned due to the unknown identity of the lectotype and priority of the name Spumella, respectively. He included three species in the genus Spumella: S. vulgaris Cienkowsky, S. vivipara (Ehrenberg) Kent, and S. beauchampii (Hovasse) Silva. Most recently, studies revealed that Spumella-like flagellates are paraphyletic (Boenigk et al., 2005; Findenig et al., 2010; Grossmann et al., 2016). Therefore, considering this long taxonomic history of Spumella-like flagellates, it is thought that the genus Spumella should be defined in more detail.
Spumella-like flagellates are non-photosynthetic unicellular organisms with two unequal flagella that have a simple morphology, e.g., spherical or naked cells (Cienkowsky, 1870; Berglund et al., 2005; Boenigk et al., 2005; Lepère et al., 2006; Charvet et al., 2012). They are representative heterotrophic chrysophytes that underwent independent loss of photosynthetic ability in the chrysophyte lineage (Boenigk et al., 2005; Boenigk, 2008; Grossmann et al., 2016; Kim et al., 2020). As they are one of the major bacterivore groups in aquatic systems, their significance in carbon transfer from the microbial loop has been emphasized (Wylie and Currie, 1991; Sanders et al., 1994; Sherr and Sherr, 1994; Arndt et al., 2000; Boenigk and Arndt, 2002). Despite their important role in aquatic ecosystems and evolutionary change in nutritional mode from phototrophy to heterotrophy, relatively little attention has been given to their species diversity because of the high degree of morphological similarity. However, the genetic diversity of Spumella-like flagellates, including the genus Spumella, is high, suggesting that they are not a monophyletic lineage (Boenigk et al., 2005; Pfandl et al., 2009; Findenig et al., 2010). Recently, Spumella-like flagellates were separated into seven new genera based on nuclear SSU rDNA sequences, while the genus Spumella was finally epitypified (Findenig et al., 2010; Grossmann et al., 2016).
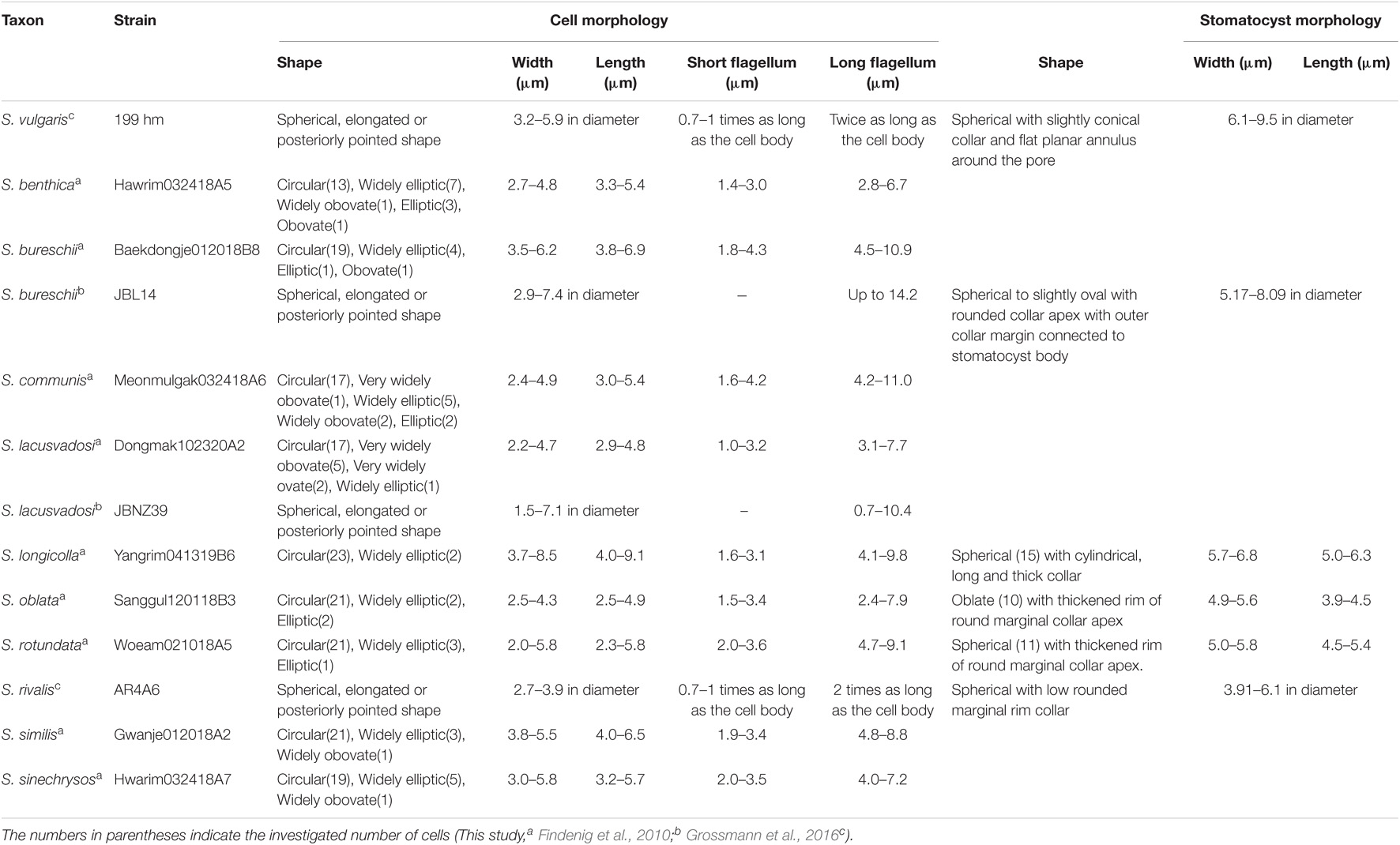
The genus Spumella is characterized by small cells (≤10 μm) with a spherical, sometimes elongated oval or oboval shape and a naked surface, two unequal flagella and colorlessness. Members of the genus lost their photosynthetic ability but still have a vestigial plastid (Preisig and Hibberd, 1983; Mylnikov et al., 2008; Grossmann et al., 2016) and retained their plastid genome (Beisser et al., 2017; Graupner et al., 2018; Dorrell et al., 2019; Kim et al., 2020). In particular, Spumella vivipara Kent, S. sphaerophora (Skuja) Mignot and S. mior (Skuja) Zhukov have strikingly large cell sizes of approximately 20 μm (Kent, 1880; Skuja, 1956; Mignot, 1977; Mylnikov and Mylnikova, 2005). Except for these three species, it is known that the vegetative cell morphologies of Spumella species are not useful in identification because of their small cell size and simple morphology. Instead, the morphology of the stomatocyst has been proposed as a useful characteristic feature for discriminating Spumella species (Findenig et al., 2010). However, only a few stomatocysts from Spumella species have been described because stomatocysts do not always develop under culture conditions (Yubuki et al., 2008; Findenig et al., 2010). The morphology of stomatocysts applies only to strains that develop stomatocysts; therefore, the identification of Spumella species based on morphological characters remains difficult.
Recently, to understand the genetic diversity of Spumella species, nuclear SSU, LSU, and 5.8S rDNA and the mitochondrial cox1 gene were used (Boenigk et al., 2005; Boenigk, 2008; Findenig et al., 2010; Grossmann et al., 2016; Bock et al., 2017). Based on molecular and morphological data, two species (Spumella rivalis Boenigk et Findenig and S. lacusvadosi Boenigk et Grossmann) were newly described, and Monas bureschii Valkanov was designated Spumella bureschii (Valkanov, 1925) Boenigk et Grossmann (Findenig et al., 2010; Grossmann et al., 2016). Compared with morphological characteristics, which are limited in their utility for species identification, molecular data are useful for identifying species on the basis of molecular signatures (Marin et al., 2003; Evans et al., 2007; Kynclova et al., 2010; Kim et al., 2013a, b; Skaloudova and Skaloud, 2013). The molecular signatures could be interpreted in the same way as generally accepted morphological criteria for classification (Marin et al., 2003), and they contain a similar phylogenetic signal and yield results largely congruent with phylogenetic relationships inferred using molecular data (Kynclova et al., 2010; Kim et al., 2013a, b; Skaloudova and Skaloud, 2013; Tragin et al., 2017).
The taxonomic definition of the genus Spumella, which has received much attention for a long time, has changed gradually over time. The currently recognized genus Spumella differs from that originally described by Cienkowsky (1870) in terms of various characters, and phylogenetic analysis of these Spumella-like flagellates using molecular data reveals that they are more diverse than previously expected. Despite the importance of Spumella as representative bacterivorous chrysophytes, their species diversity has been underestimated. To perform a taxonomic study of Spumella, we investigated 47 strains (including 18 strains isolated from Korea). Based on morphological and molecular data, we found high species diversity in Spumella, including seven new species. As key diagnostic characters for each species, we used stomatocyst morphology and molecular signatures, which were applied to each clade in the phylogenetic tree.
Materials and Methods
Cultures
Korean Spumella strains were collected from freshwater ponds or swamp sediments. We performed single-cell isolation using a Pasteur capillary pipette to establish unialgal cultures from field samples or subcultures. For unialgal clonal culture of Spumella strains, we performed two methods: single-cell isolation and dilution. For single-cell isolation, we picked up a single Spumella cell and a small aliquot of freshwater containing bacteria and then transferred them to each well of a 96-well plate. To make subcultures by the dilution method, 3 μl of field freshwater samples was mixed into 200 μl of AF-6 medium (Andersen et al., 2005) supplemented with 0.1, 0.2, or 0.5% Luria-Bertani (LB) medium (Miller, 1972). The LB broth was offered for bacterial growth. Bacteria from field freshwater samples were fed to Spumella cells as prey. When we observed the growth of Spumella cells, a single target cell was isolated and transferred to a new single-well plate by micropipette. Finally, all strains were grown in AF6 medium with a mixture of freshwater LB broth and uncharacterized bacteria from field freshwater samples. The strains were maintained at 17°C in a culture chamber in the dark. Information on the collection site and the GenBank accession number for each strain are listed in Supplementary Table 1.
DNA Extraction, Amplification, Sequencing, and Alignment
The cells were harvested by centrifugation at 11,363 g for 5 min (11,000 rpm, model 5424, Eppendorf, Hamburg, Germany). Genomic DNA was extracted by a DOKDO-PrepTM Blood Genomic DNA Purification Kit (ELPIS-Biotech Inc., Daejeon, Korea). Polymerase chain reaction (PCR) was performed for nuclear SSU, ITS and LSU rDNA using specific primers (Supplementary Table 2). PCR was performed in a total volume of 25 μl of the following: 1 μl of AccuPower® PCR premix (Bioneer Co., Daejeon, Korea), 1 μl of forward primer, 1 μl of reverse primer, 2–10 μl of template DNA, and 12–20 μl of distilled water. The genes were amplified using a T100TM Thermal Cycler (Bio-Rad Laboratories, Hercules, California, United States). The first denaturation was run at 94°C for 5 min, followed by 35 cycles of second denaturation at 94°C for 30 s, annealing at 42–52°C for 30 s-–1 min, extension at 72°C for 1–2 min, and a final extension at 72°C for 7 min, with a final hold at 12°C. All PCR products were purified using the LabopassTM PCR Purification Kit or LabopassTM Gel Purification Kit (Cosmogenetech Co., Seoul, Korea) following the protocol provided by the manufacturer. The purified PCR products were sequenced using an ABI PRISMTM (3730xL, Perkin-Elmer Applied Biosystems, Foster City, California, United States). Sequence alignments were performed visually using the Genetic Data Environment (2.6) program (Smith et al., 1994).
Molecular Data Analyses
The secondary structures of the nuclear (nr) SSU and LSU rRNA genes were aligned manually, with the alignment based on secondary structures of the nr SSU and LSU rRNA gene sequences of the dictyochophycean species Apendinella radians (Ali et al., 2001) as a guide in the Macgde (2.6) (Smith et al., 1994). As a molecular signature for Spumella-like flagellates, domain 8 from the V1 region of the nr SSU rRNA and D5 domain from the nr LSU rRNA gene were selected for identification of the genus Spumella. For discriminating species in the genus Spumella, the E23-2 to E23-5 domains, the spacer between the E23-8 and E23-9 domains of the V4 region and domain 29 of the V5 region of the nr SSU rRNA gene were selected.
Phylogenetic Analyses
The phylogenetic analysis was performed by using 1,625 nucleotides of the nr SSU rDNA from 158 chrysophycean taxa (Supplementary Figure 2). Three species (Nanochloropsis granulata, Leukarachnion sp., and Synchroma grande) were used as outgroup taxa (Grossmann et al., 2016; Pusztai and Škaloud, 2019). A combined dataset of 4,876 nucleotides (nr SSU = 1,667, nr LSU = 2,656, ITS1-5.8S-ITS2 region = 553) from 47 Spumella taxa was used for phylogenetic analysis (Figure 1 and Supplementary Table 1). For both datasets, conserved regions of genes were used only for phylogenetic analyses, and ambiguously aligned regions were excluded from phylogenetic analyses. Maximum likelihood (ML) was performed using RAxML version 8.2.10 (Stamatakis, 2014) with the general time-reversible plus gamma (GTR + I + G) model. We used 1,000 independent tree inferences, using the -# option of the program to identify the best tree. Maximum likelihood bootstrap (MLBS) values were calculated using 1,000 pseudoreplicates with the same substitution model. Bayesian analyses were performed using MrBayes version 3.7 (Ronquist et al., 2012), and the best-fitting model for the nucleotide dataset was selected by the Bayesian information criterion in jModelTest2 (Posada and Crandall, 1998), with the GTR + I + G model. Each analysis was performed using a Metropolis-coupled Markov chain Monte Carlo (MC3) approach, with 10,000,000 cycles for each chain. Trees were saved to a file every 1,000 cycles, and the burn-in point was identified graphically by tracking the likelihoods (Tracer version 1.7.1).1 The first 3,000 trees were discarded, and the remaining 7,001 trees were used to calculate the posterior probability (PP) of each clade. The trees were visualized using FigTree v.1.4.4.2
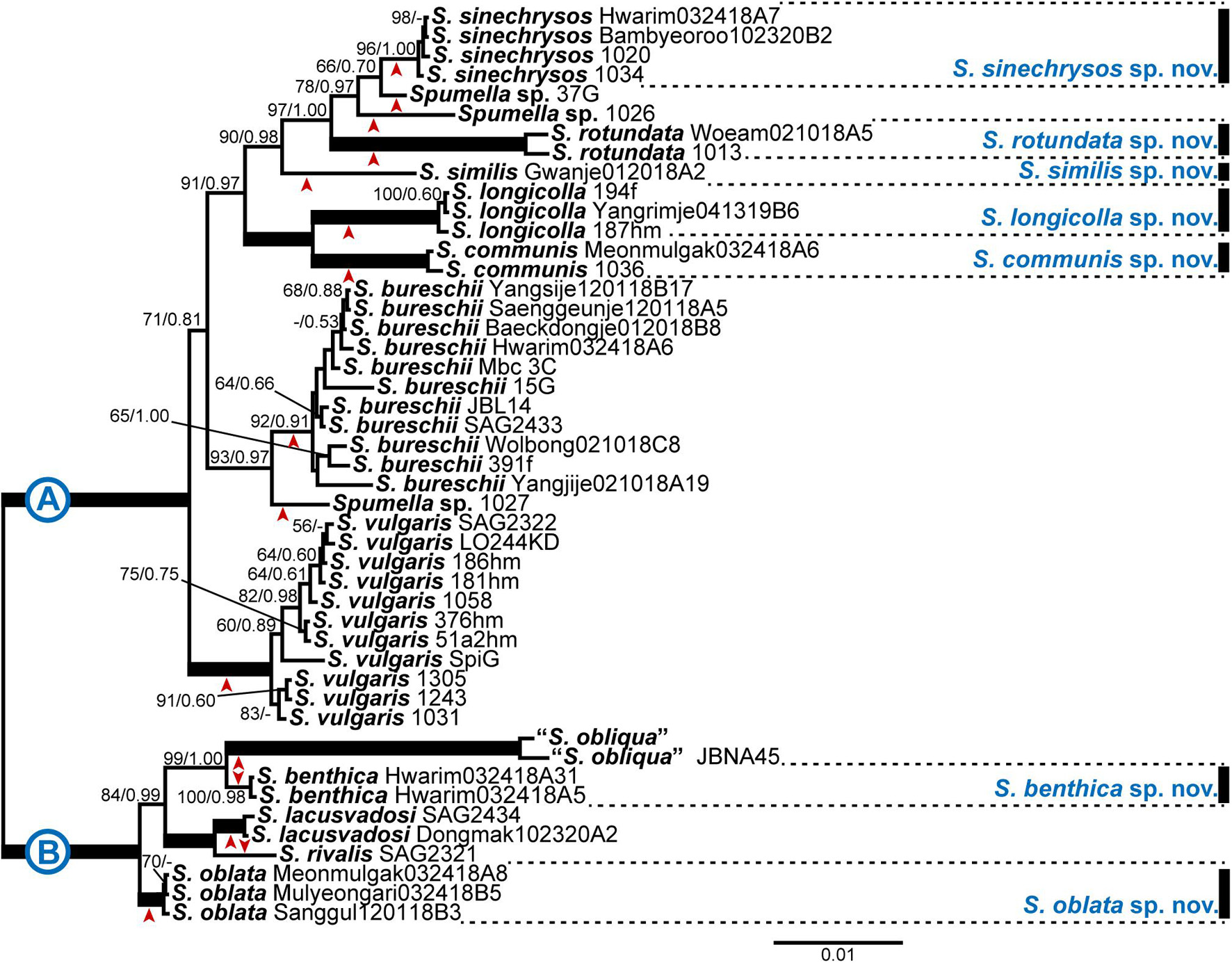
Figure 1. Bayesian tree of the genus Spumella based on combined nuclear SSU and LSU rDNA and ITS sequence data. The maximum-likelihood bootstrap values (MLBS values, left) and Bayesian posterior probabilities (PPs, right) are shown at each node. The arrow head below or above each node indicates each lineage; the scale bar indicates the number of substitutions/site; the thick line indicates full support (100% MLBS and 1.00 PP), and (–) denotes values < 50% for MLBS or 0.50 for PP.
Light Microscopy and Statistical Analysis
Culture strains were observed using an Axio Imager A2 microscope (Carl Zeiss Inc., Hallbergmoos, Germany) with differential interference contrast (DIC) optics. Images were captured with an AxioCam HRc (Carl Zeiss Inc.) and AxioCam 512 mono (Carl Zeiss Inc.) photomicrographic system connected to the microscope. Numerical values of morphological characters were obtained using photographic images of 25 cells (Table 1). The range of cell sizes was statistically analyzed using one-way ANOVA implemented in Excel 2016 to test for significantly different cell and flagellum sizes between Spumella taxa from Korea. A p-value under 0.05 was considered to indicate a statistically significant difference.
Transmission Electron Microscopy
To prepare the whole-mount transmission electron microscopy (TEM) samples, cells were fixed in 2.5% glutaraldehyde for 1 h at 4°C, and approximately 50 cells per strain were mounted on a formvar-coated copper grid. For ultrathin sectioning, the cells were collected by centrifugation for 5 min at 11,363 g (11,000 rpm, Eppendorf centrifuge 5424). After removing the supernatant, pelleted cells were fixed in 2.5% glutaraldehyde with 0.05 M cacodylate for 1 h at 4°C. After three 10 min washes with 0.05 M cacodylate buffer, the cells were fixed in 1% OsO4 for 1 h at 4°C and washed with 0.05 M cacodylate buffer. Dehydration was carried out in an ethanol series (50, 60, 70, 80, 90%) with 10 min for each step and three 10 min changes of absolute ethanol. The cells were serially transferred to three solutions of propylene oxide with ethanol (ethanol:propylene oxide = 2:1, 1:1, 1:2) for 15 min at each step and three times to absolute propylene oxide. The cells were serially transferred to three solutions of resin with propylene oxide (resin:propylene oxide = 1:2, 1:1, 2:1) for 2 h at each step and stored overnight in pure resin. The following day, the cells were transferred into new pure resin and polymerized at 70°C for 48 h. The polymerized blocks were thin-sectioned at a thickness of 70 nm. Serial sections were mounted on formvar-coated copper grids and stained with 3% (w/v) uranyl acetate and lead citrate. The grids were viewed and photographed using a JEM-1400 Plus transmission electron microscope (JEOL Ltd., Tokyo, Japan) at 120 kV.
Scanning Electron Microscopy
To record stomatocyst morphology, 100–200 stomatocysts per strain were mounted on 0.45 μm nylon membrane filters using single-cell isolation. The filters were mounted onto aluminum stubs with double-sided tape. The stubs were coated with platinum and viewed with a MIRA3 field emission scanning electron microscope (TESCAN Ltd., Brno, Czech Republic) at 10 kV.
Results
Phylogenetic Analysis
The phylogenetic analyses of Chrysophyceae based on nr SSU rDNA sequence data included nine orders (Supplementary Figure 2). The six orders of Chrysophyceae were well resolved as monophyletic, and other three orders (Chromulinales, Hydrurales, and Paraphysomonadales) got a low statistical support. The phylogenetic relationships between nine orders were not resolved. The Ochromonadales formed a monophyletic clade with high support values (MLBS = 91, PP = 1.00) and included the genus Spumella. Within the order Ochromonadales, the phylogenetic relationships among the genera were not fully resolved. The genus Spumella formed a monophyletic clade with high support values (MLBS = 100, PP = 1.00), but the phylogenetic relationships among species were not resolved, and S. bureschii was paraphyletic in the phylogeny based only on nr SSU rDNA gene sequence data.
Based on the combined dataset of nr SSU, ITS and LSU rDNA gene sequences, phylogenetic analyses of the genus Spumella revealed that the 47 strains were divided into 15 subclades, including five consisting of a single taxon, and seven subclades included newly established species with high support values (Figure 1). The phylogenetic tree was divided into two major clades (A and B) with high support values (MLBS = 100, PP = 1.00). The first clade (A) included 10 subclades with five new species (Spumella sinechrysos, S. rotundata, S. similis, S. longicolla, and S. communis). The Hwarim032418A7, Bambyeoroo102320B2, 1020 and 1034 strains were grouped into the new species S. sinechrysos (MLBS = 96, PP = 1.00). Woeam021018A5 and 1030 were grouped together as the new species S. rotundata (MLBS = 100, PP = 1.00), and S. similis formed a single-taxon lineage sister to S. sinechrysos, Spumella sp. 37G, Spumella sp. 1026 and S. rotundata (MLBS = 90, PP = 0.98). The 194f, Yangrim041319B6 and 187hm strains were grouped together as the new species S. longicolla with high support values (MLBS = 100, PP = 1.00) and showed sister relationships with the new species S. communis (MLBS = 100, PP = 1.00). S. bureschii included 11 strains (MLBS = 92, PP = 0.91) and showed sister relationships with the Spumella sp. 1027 strain (MLBS = 93, PP = 0.97). The species S. vulgaris included 11 strains and formed a monophyletic clade with high support values (MLBS = 100, PP = 1.00). The second clade (B) consisted of five taxa, including two new species. The new species S. benthica consisted of two strains (MLBS = 100, PP = 0.98) and showed sister relationships with two “S. obliqua” strains with high support values (MLBS = 99, PP = 1.00). Mulyeongari032418B5, Sanggul120118B3 and Meomulgak032418A8 were grouped together into the new species S. oblata with high support values (MLBS = 100, PP = 1.00). The two formerly described species (S. rivalis and S. lacusvadosi) showed sister relationships with “S. obliqua” and S. benthica (MLBS = 84, PP = 0.99).
Molecular Signatures
The molecular signatures of the nr SSU and LSU rRNA gene data were investigated with other representative non-photosynthetic lineages in Chrysophyceae to identify the molecular signatures unique to the genus Spumella (Figure 2). All Spumella species had unique sequences of “A:U” as the last base pair in domain 8 of the V1 region of nr SSU rRNA (Figure 2A) and “C:G” and “U:A” in domain D5 of the nr LSU rRNA gene (Figure 2B).
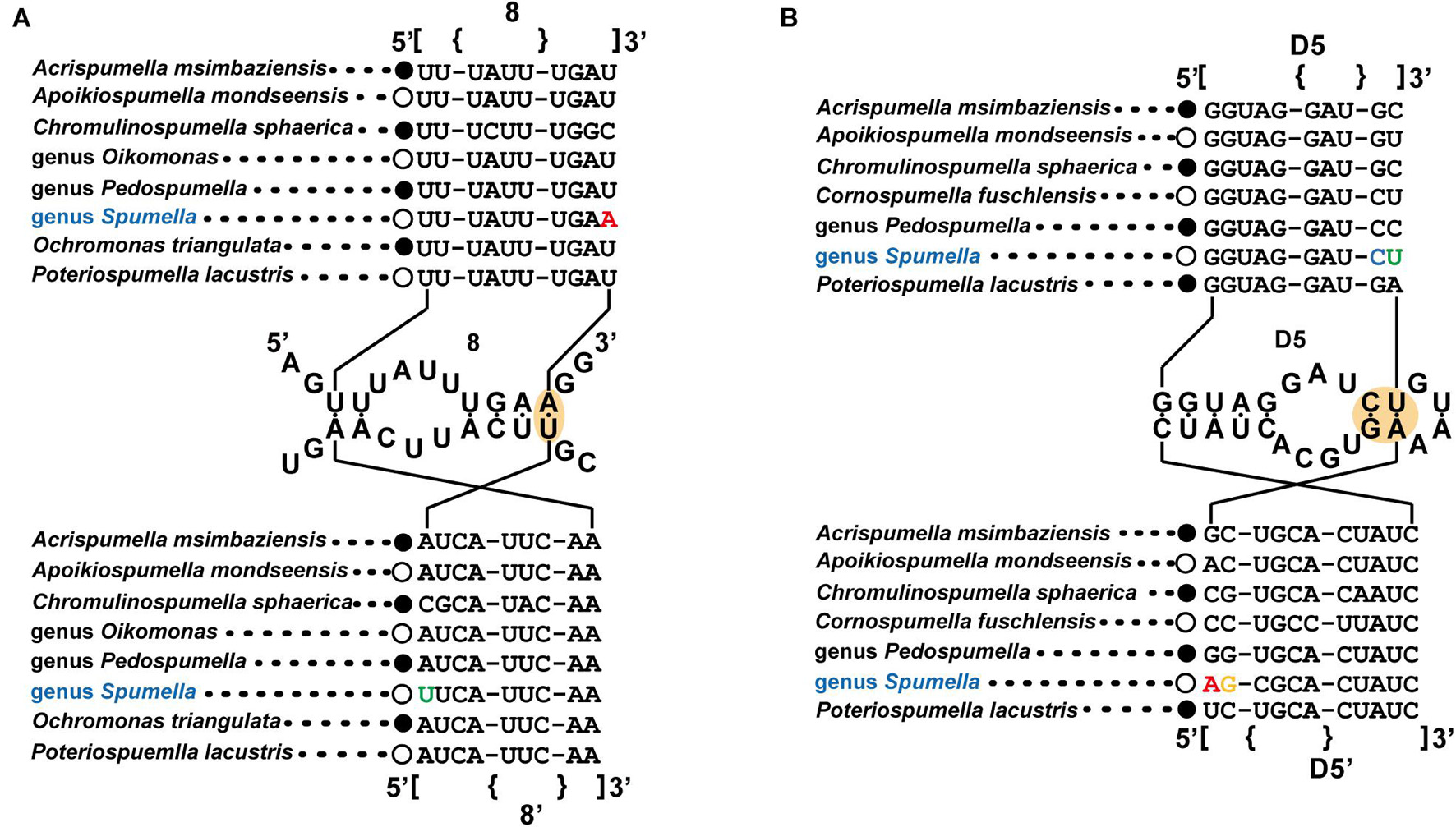
Figure 2. Molecular signatures of the nuclear SSU (A) and LSU (B) rRNA differentiating the genus Spumella from other non-photosynthetic chrysophytes. The secondary structure was constructed based on the genus Spumella. The nomenclature of nucleotides and base pairs depends on the polarity of the DNA: increasing numbers generally indicate the 5′–3′ direction. [] indicates the beginning and end of the stem, with {} indicating non-binding (loops, bulges) of the base pairs.
The nr SSU rRNA sequence combination including the E23-2 to E23-5 domain (Figure 3A), the spacer between E23-8 and E23-9 of the V4 region (Figure 3B) and domain 29 of the V5 region (Figure 3C) was selected as a molecular signature for the Spumella species. Each Spumella species had a specific molecular signature, and the signatures of new species were compared with those of closely related species. Base differences between S. sinechrysos and Spumella sp. 37G were found in E23-5 of the V4 region (G:C—A:U). Between Spumella sp. 1026 and S. rotundata, bases were different in E23-2 (A:U—G:C and G:C—A:U), E23-5 (A:U—G:C and U:G—C:G) and the spacer between E23-8 and E23-9 (G—A). S. similis was compared with S. rotundata, and bases were different in E23-2 (C:G—G:C and G:C—A:U), E23-5 (A:U—G:U and U:G—C:G) and the spacer between E23-8 and E23-9 (A—G). Two new species, S. longicolla and S. communis, were compared, and the bases were different in E23-5 (U:A—C:G). The base pair changes in the molecular signature region of S. benthica compared with that of “S. obliqua” JBNA45 were located at E23-2 (A:U—G:C). Base differences among three species, S. lacusvadosi, S. rivalis and S. oblata, were detected in E23-2 (G:C—A:U—G:C and A:U—A:U—G:C) and E23-5 (G:C—A:U—A:U). In the secondary structure of domain 29 of the V5 region, compensatory base pair changes (CBCs) occurred between the two major (A and B) clades of the phylogenetic tree of Spumella species (A:U—U:A, G:C—A:U, and A:U—U:A). In the secondary structure of the molecular signature region, several pairs of substitutions were CBCs or hemi-CBCs and conserved rRNA-base pairs of “G-C” but rarely changed to “G-U.” The base changes of each taxon are colored in Figures 2, 3.
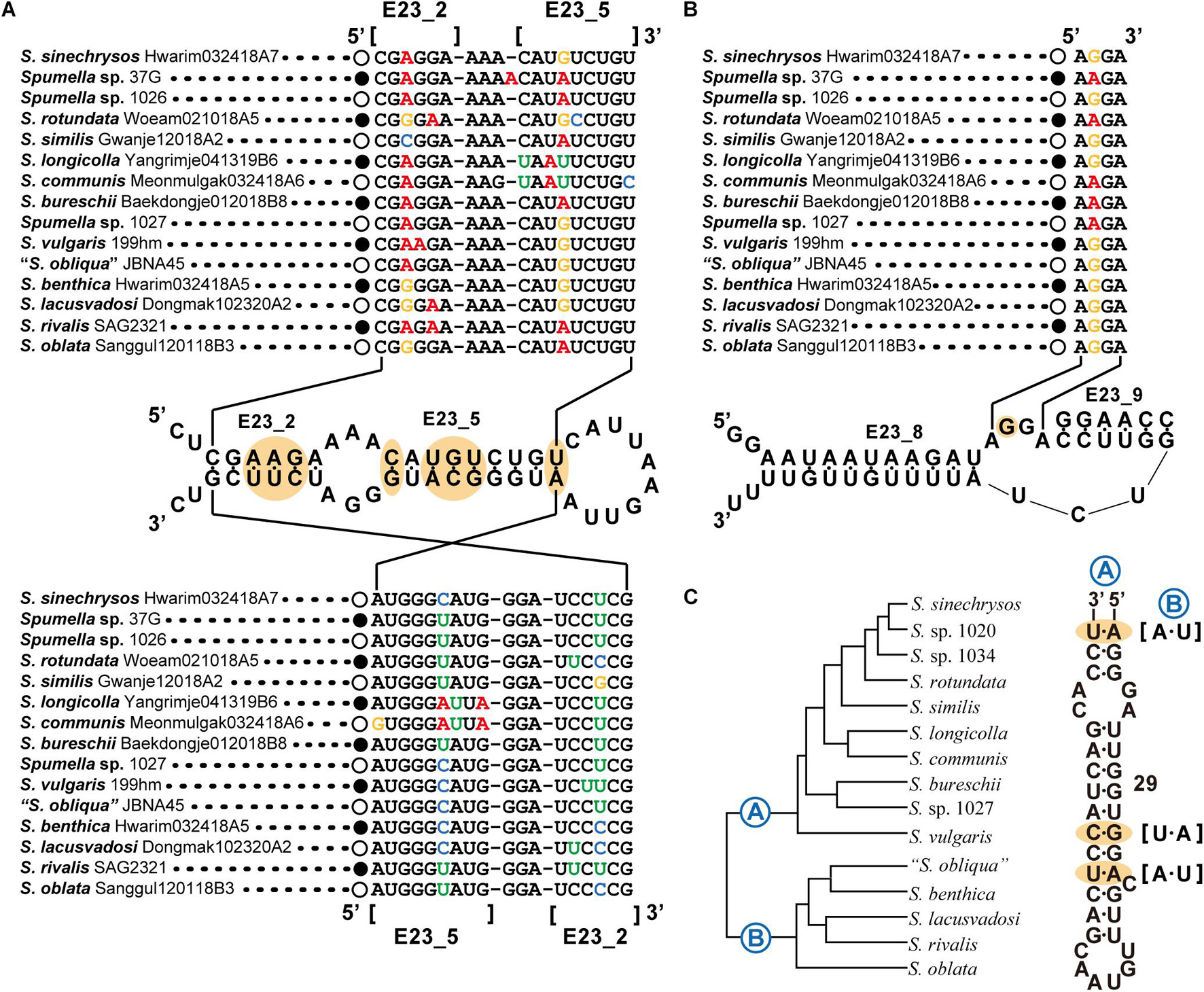
Figure 3. Molecular signatures of Spumella species using secondary structures from E23_2 to E23_5 (A), E23_8 to E23_9 of the V4 region (B) and domain 29 of the V5 region (C) in the nuclear SSU rRNA gene sequence. The secondary structure was constructed based on S. vulgaris. The nomenclature of nucleotides depends on the polarity of the DNA: increasing numbers generally indicate the 5′–3′ direction. [] indicates the beginning and end of the stem region.
Morphological Characters
The cell morphological characters of Spumella are summarized in Table 1, and the terminologies used for morphological description followed those of Radford (1986). Almost all Spumella species showed similar morphologies. The cell shapes of all Spumella species were circular to ovate, obovate or elliptic without an eyespot and cell surface ornamentation (Figure 4). The two flagella were unequal: the long flagellum had two rows of mastigonemes, but the short flagellum had no mastigonemes (Figures 4D,E,G,H,J,K,P,Q,S,T,V,W,Y,Z). The leucosin vesicle was positioned on the posterior side of the cell (Figures 4A,B,F,I,M,N,R,U,X) or on both the posterior and anterior sides of the cell when two leucosin vesicles were present in the cell (Figures 4C,O). In addition, one or two leucosin vesicles containing brilliant crystals were found only in S. benthica and S. oblata (Figures 4B,C,N,O).
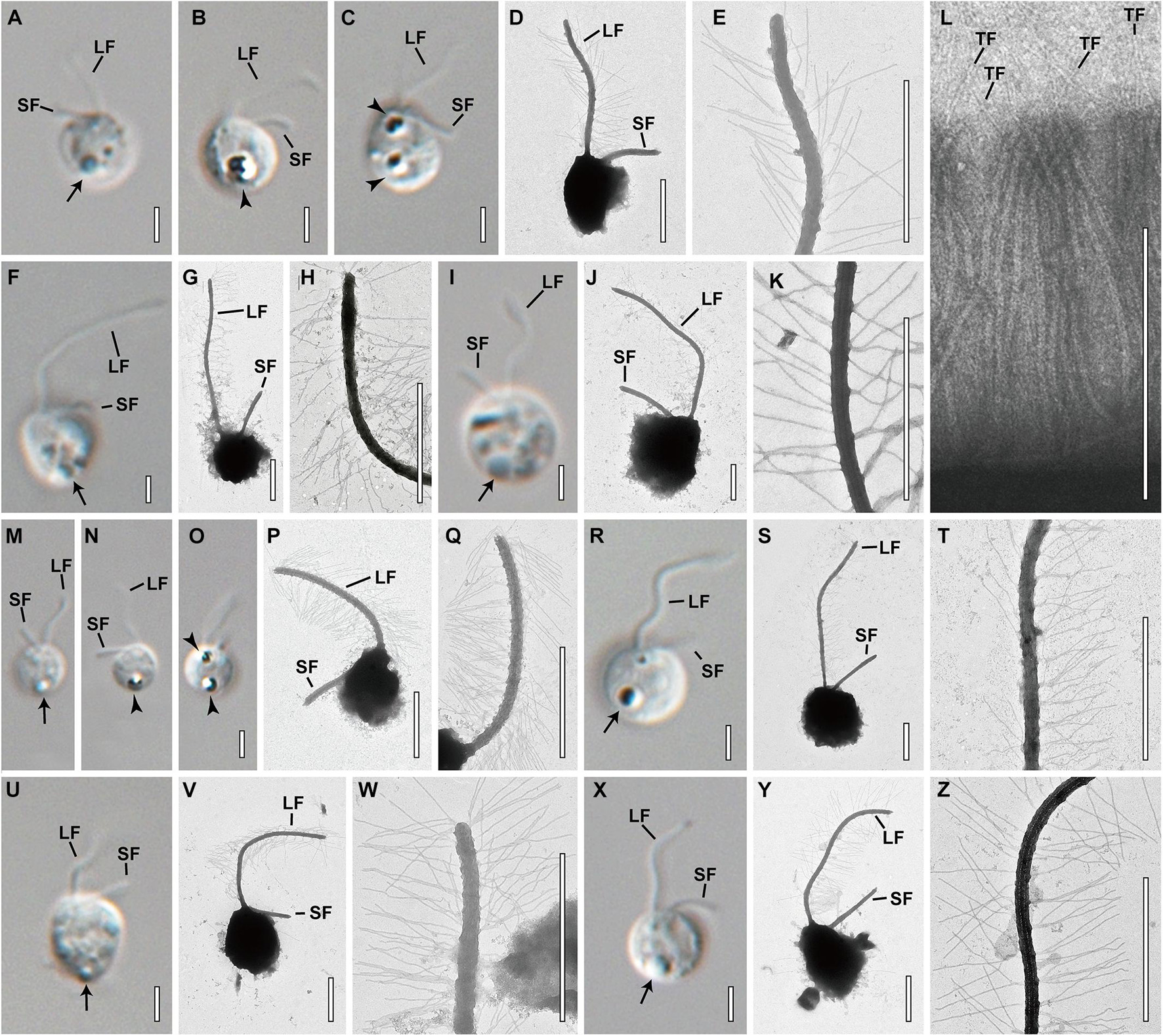
Figure 4. Light and transmission electron micrographs of Spumella species. Images showing each species having a long flagellum (LF) with two rows of mastigonemes, a terminal filament (TF) in the mastigonemes and a short flagellum (SF). Leucosin vesicle (arrow) and leucosin vesicles containing brilliant crystals (arrowhead) located on the anterior and posterior sides of the cell. (A–E) Image of S. benthica Hwarim032418A5. (F–L) Images of S. communis Meonmulgak032418A6. (I–L) Images of S. longicolla Yangrim041319B6. (M–Q) Images of S. oblata Sanggul120318B3. (R–T) Images of S. rotundata Woeam020118A5. (U–W) Images of S. similis Gwanje012018A2. (X–Z) Images of S. sinechrysos Hwarim032418A7. Scale bars = 2 μm.
Stomatocyst
Only four species, Spumella bureschii, S. longicolla, S. oblata, and S. rotundata, encysted in culture, and the stomatocyst morphologies of these species were recorded using scanning electron microscopy (SEM) (Figures 5a–f). The terminology used for stomatocyst description followed that of Duff et al. (1995) and Wilkinson et al. (2001). The stomatocyst shape of S. longicolla was spherical without surface ornamentation (Figures 5a,b). The stomatocyst of S. longicolla had a distinct, long cylindrical collar, and sometimes the collar apex had several irregular grooves along its circumference (Figure 5b). The stomatocyst shape of S. oblata was oblate without surface ornamentation (Figures 5c,d). The collar of S. oblata was low and had a thickened rim with a rounded margin (Figure 5d). The stomatocyst shape of S. rotundata was spherical without surface ornamentation (Figures 5e,f). A distinct collar was not clearly observed, but there was a shallow raised and thickened region around the pore (Figure 5f). The outer margin of this region gently sloped down and was continuous with the stomatocyst body (Figure 5f).
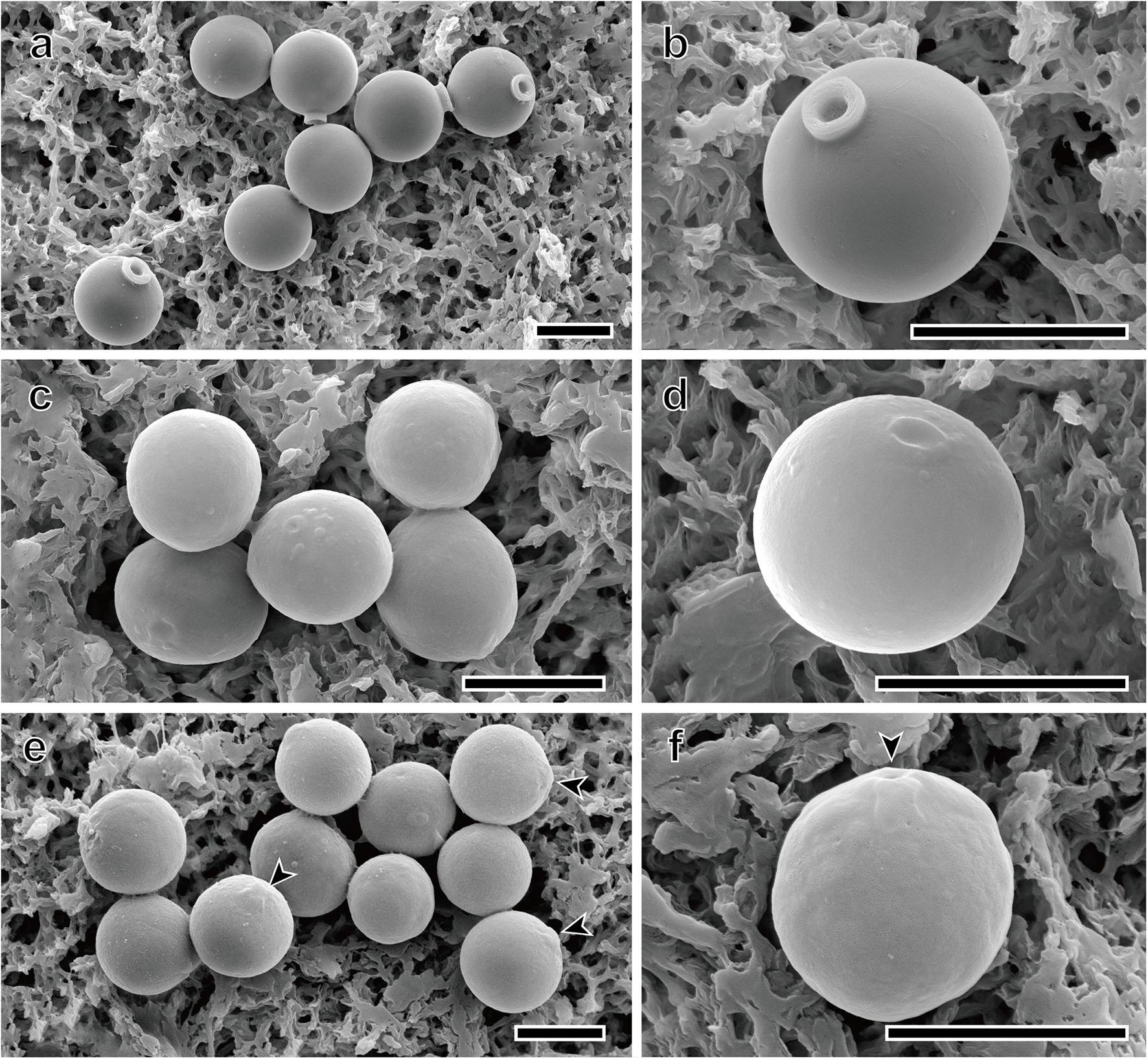
Figure 5. Scanning electron micrographs of three new Spumella species. (a,b) SEM images of the spherical stomatocyst of S. longicolla Yangrim041319B6. (c,d) SEM images of the oblate stomatocyst of S. oblata Sanggul120118B3. (e,f) SEM images of the spherical stomatocyst of S. rotundata Woeam020118A5. Arrowheads indicate a pore of the stomatocyst. Scale bars = 5 μm.
Ultrastructure
The general ultrastructure of two species, Spumella bureschii Baekdongje012018B8 and S. benthica Hwarim032418A5, as representatives of the two major clades in Figure 1, was observed with special attention to the vestigial plastid. One leucoplast was located in the perinuclear space between the inner and outer membranes of the nuclear envelope (Figures 6a–d) and was bound by four membranes. The outermost membrane was continuous with the outer membrane of the nucleus, while an amorphous mass including a membranous structure between the second and third membranes was visible. The two innermost membranes were closely appressed and formed an oval outline of the structure. The leucoplast did not show traces of a thylakoid (Figures 6a–d) and ranged from 0.43 to 0.48 μm in diameter. The two basal bodies were located perpendicular to each other, the long flagellum was directed forward, and the short flagellum was directed diagonally (Figure 6e). Swelling of the short flagellum was not found (Figure 6e). The typical structures of mitochondria, i.e., tubular cristae, a Golgi apparatus and a food vacuole, were observed (Figures 6f–h).
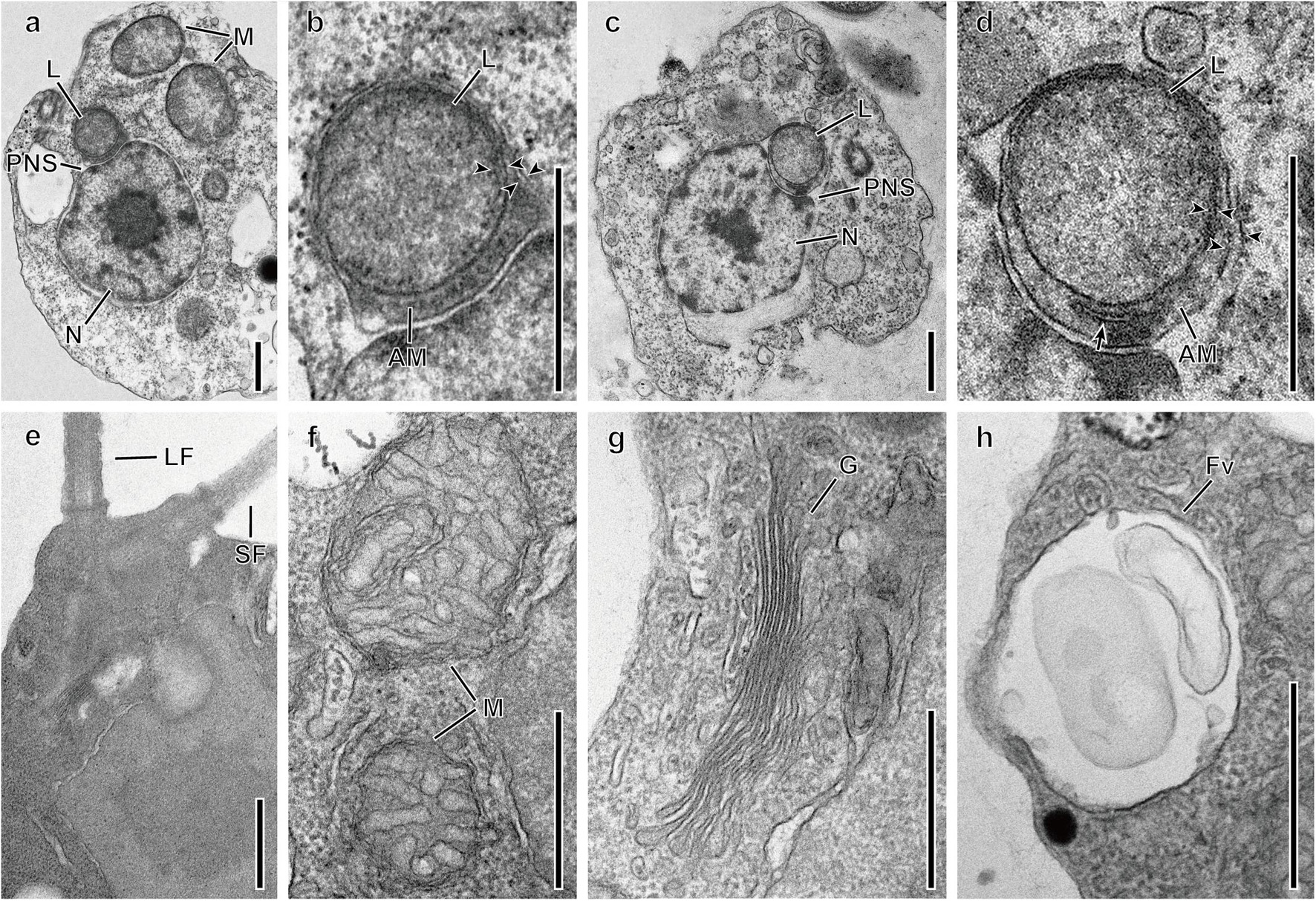
Figure 6. Transmission electron micrographs of Spumella species. (a,b) One leucoplast (L) of S. bureschii Baekdongje012018B8 located in the perinuclear space. (c,d) One leucoplast (L) of S. benthica Hwarim032418A5 located in the perinuclear space. (e) Flagellar apparatus of the long (LF) and short (SF) flagella. (f) Mitochondria with tubular cristae (M). (g) Golgi (G). (h) Food vacuole (Fv). Arrowheads indicate membrane envelops. AM, amorphous mass; PNS, perinuclear space; PR, periplastidial reticulum (arrow). Scale bars = 0.5 μm.
Feeding
Spumella species consume bacteria through phagocytosis, and the four feeding phases of S. rotundata were recorded (Figure 7 and Supplementary Figure 3). The Spumella species adhered to the substratum during the feeding phases. The long flagellum caused water currents for feeding and detected the bacterium in the contact phase. After a food particle was detected, the long flagellum pressed the bacterium closely to the cell body in the processing phase. Then, the bacterium was ingested into the cell body using pseudopodia in the ingestion phase. Finally, in the refractory phase, the bulged part of the cell body involved in phagocytosis returned, and the hunted bacterium was positioned in the food vacuole of the cell.
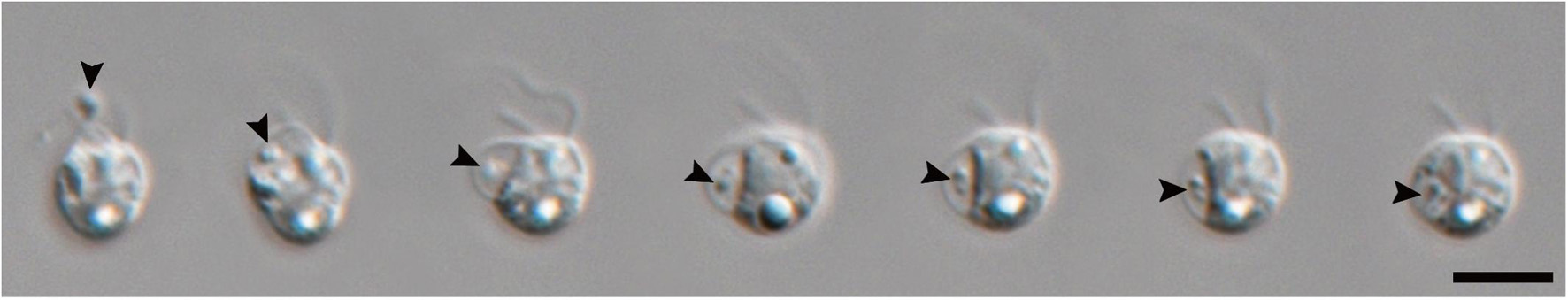
Figure 7. Serially captured LM images of four feeding phases of S. rotundata. Arrowheads indicate bacteria. Scale bar = 5 μm.
Description of New Species
Spumella benthica Jeong, M., Kim, J. I. et Shin, W. sp. nov.
DESCRIPTION: The species is colorless, non-scaled, single-celled and biflagellate without an eyespot (Figures 4A–E). Cells are circular, widely elliptic, widely obovate, elliptic or obovate, ranging in size from 2.7 to 4.8 × 3.3 to 5.4 μm. The two unequal emergent flagella are visible. The long flagellum has two rows of mastigonemes (Figures 4D,E) that are directed forward and waved or moved side to side, ranging in length from 2.8 to 6.7 μm. The short flagellum is directed diagonally or bent rearward and ranges in length from 1.4 to 3.0 μm. The leucosin vesicle is located in the posterior region of the cell (Figures 4A–C). One or two leucosin vesicles containing brilliant crystals are positioned in the cell (Figures 4B,C) and sometimes absent. The nr SSU rRNA molecular signature sequences are as follows:
E23-2 to E23-5 domain of the V4 region: CGGGGAAAACAUGUCUGU
E23-2′ to E23-5′ domain of the V4 region: AUGGGCAUGGGAUCCCCG
Spacer between the E23-8 and E23-9 domains of the V4 region: AGGA
HOLOTYPE: NNIBRPR19597, plastic TEM block Hwarim032418A5, deposited in the Nakdonggang National Institute of Biological Resources, Sangju, Korea (NNIBR).
TYPE LOCALITY: Freshwater, Hannam-ri, Namwon-eup, Seoguipo-si, Jeju-do, Korea (33°19′26.5″N, 126°39′31.9″E), 24 Mar. 2018.
ETYMOLOGY: The specific epithet “benthica” refers to the benthic habitat where this species is found.
Spumella communis Jeong, M., Kim, J. I. et Shin, W. sp. nov.
DESCRIPTION: The species is colorless, non-scaled, single-celled and biflagellate without an eyespot (Figures 4F–H). Cells are circular, very widely obovate, widely elliptic, widely obovate or elliptic, and ranging in size from 2.4 to 4.9 × 3.0 to 5.4 μm. The leucosin vesicle is located in the posterior region of the cell (Figure 4F). Two unequal emergent flagella are visible. The long flagellum has two rows of mastigonemes (Figures 4G,H) that are directed forward and waved or moved side to side, and ranging in length from 4.2 to 11.0 μm. The short flagellum is directed diagonally or bent rearward and ranges in length from 1.6 to 4.2 μm. The nr SSU rRNA molecular signature sequences are as follows:
E23-2 to E23-5 domain of the V4 region: CGAGGAAAGUAAUUCUGC
E23-2′ to E23-5′ domain of the V4 region: GUGGGAUUAGGAUCCUCG
Spacer between the E23-8 and E23-9 domains of the V4 region: AAGA
HOLOTYPE: NNIBRPR19601, plastic TEM block Meonmulgak032418A6, deposited in the Nakdonggang National Institute of Biological Resources, Sangju, Korea (NNIBR).
TYPE LOCALITY: Freshwater, Meonmulgak swamp, Seonheul-ri, Jocheon-eup, Jeju-si, Jeju-do, Korea (33°31′06.8″N 126°42′55.1″E), 24 Mar. 2018.
ETYMOLOGY: The Latin specific epithet “communis” refers to the ordinary shape of the cell.
Spumella longicolla Jeong, M., Kim, J. I. et Shin, W. sp. nov.
DESCRIPTION: The species is colorless, non-scaled, single-celled and biflagellate without an eyespot (Figures 4I–L). Cells are circular or widely elliptic, ranging in size from 3.7 to 8.5 × 4.0 to 9.1 μm. The leucosin vesicle is located in the posterior region of the cell (Figure 4I). Two unequal emergent flagella are visible. The long flagellum has two rows of mastigonemes (Figures 4J–L) that are directed forward and waved or moved side to side, ranging in length from 4.1 to 9.8 μm. The short flagellum is directed diagonally or bent rearward and ranges in length from 1.6 to 3.1 μm. The stomatocyst is spherical, ranges in size from 5.7 to 6.8 × 5.0 to 6.3 μm (n = 15), and has a smooth surface (Figures 5a,b). The collar of the stomatocyst is cylindrical and thick, ranging in height from 0.6 to 0.7 μm (n = 15) and in width from 1.0 to 1.8 μm (n = 14). The nr SSU rRNA molecular signature sequences are as follows:
E23-2 to E23-5 domain of the V4 region: CGAGGAAAAUAAUUCUGU
E23-2′ to E23-5′ domain of the V4 region: AUGGGAUUAGGAUCCUCG
Spacer between the E23-8 and E23-9 domains of the V4 region: AGGA
HOLOTYPE: NNIBRPR19599, plastic TEM block Yangrimje041319B6, deposited in the Nakdonggang National Institute of Biological Resources, Sangju, Korea (NNIBR).
TYPE LOCALITY: Freshwater, Yangrim pond, Jinyang-ri, Hampyeong-eup, Hampyeong-gun, Jeollabuk-do, Korea (35°05′16.6″N 126°29′57.6″E), 13 Apr. 2019.
ETYMOLOGY: The specific epithet “longicolla” derived from Latin “longi-” (= long) and “colla” (= collar) refers to the distinct, long collar of the stomatocyst.
Spumella oblata Jeong, M., Kim, J. I. et Shin, W. sp. nov.
DESCRIPTION: The species is colorless, non-scaled, single-celled and biflagellate without an eyespot (Figures 4M–Q). Cells are circular, widely elliptic or elliptic, ranging in size from 2.5 to 4.3 × 2.5 to 4.9 μm. The leucosin vesicle is located in the posterior region of the cell (Figures 4M–O). One or two leucosin vesicles containing brilliant crystals are positioned in the cell (Figures 4N,O) and sometimes absent (Figure 4M). Two unequal emergent flagella are visible. The long flagellum has two rows of mastigonemes (Figures 4P,Q) that are directed forward and waved or moved side to side, ranging in length from 2.4 to 7.9 μm. The short flagellum is directed diagonally or bent rearward, ranging in length from 1.5 to 3.4 μm. The stomatocyst is oblate, ranges in size from 4.9 to 5.6 × 3.9 to 4.5 μm (n = 10), and has a smooth surface (Figures 5c,d). The rim of the round marginal collar apex is slightly thickened (Figure 5d). The nr SSU rRNA molecular signature sequences are as follows:
E23-2 to E23-5 domain of the V4 region: CGGGGAAAACAUAUCUGU
E23-2′ to E23-5′ domain of the V4 region: AUGGGUAUGGGAUCCCCG
Spacer between the E23-8 and E23-9 domains of the V4 region: AGGA
HOLOTYPE: NNIBRPR19600, plastic TEM block Sanggul120118B3, deposited in the Nakdonggang National Institute of Biological Resources, Sangju, Korea (NNIBR).
TYPE LOCALITY: Freshwater, Sangdeung-ri, Booan-myeon, Gochang-gun, Jeollabuk-do, Korea (35°30′19.1″N 126°40′16.4″E), 01 December. 2018.
ETYMOLOGY: The Latin specific epithet “oblate” (= oblate) refers to the shape of the stomatocyst.
Spumella rotundata Jeong, M., Kim, J. I. et Shin, W. sp. nov.
DESCRIPTION: The species is colorless, non-scaled, single-celled and biflagellate without an eyespot (Figures 4R–T). Cells are circular, widely elliptic or elliptic, ranging in size from 2.0 to 5.8 × 2.3 to 5.8 μm. The leucosin vesicle is located in the posterior region of the cell (Figure 4F). Two unequal emergent flagella are visible. The long flagellum has two rows of mastigonemes (Figures 4S,T) that are directed forward and waved or moved side to side, ranging in length from 4.7 to 9.1 μm. The short flagellum is directed diagonally or bent rearward and ranges in length from 2.0 to 3.6 μm. The stomatocyst is spherical, ranges in size from 5.0 to 5.8 × 4.5 to 5.4 μm (n = 11), and has a smooth surface (Figures 5e,f). The rim of the round marginal collar apex is slightly thickened (Figure 5f). The collar margin gently slopes down and is continuous with the stomatocyst body (Figure 5f). The nr SSU rRNA molecular signature sequences are as follows:
E23-2 to E23-5 domain of the V4 region: CGGGAAAAACAUGCCUGU
E23-2′ to E23-5′ domain of the V4 region: AUGGGUAUGGGAUUCCCG
Spacer between the E23-8 and E23-9 domains of the V4 region: AAGA
HOLOTYPE: NNIBRPR19598, plastic TEM block Woeam021018A5, deposited in the Nakdonggang National Institute of Biological Resources, Sangju, Korea (NNIBR).
TYPE LOCALITY: Freshwater, Woeam pond, Moogo-ri, Moonnae-myeon, Haenam-gun, Jeollanam-do, Korea (34°37′45.1″N 126°18′16.4″E), 10 Feb. 2018
ETYMOLOGY: The Latin specific epithet “rotundata (= rounded)” refers to the stomatocyst shape.
Spumella similis Jeong, M., Kim, J. I. et Shin, W. sp. nov.
DESCRIPTION: The species is colorless, non-scaled, single-celled and biflagellate without an eyespot (Figures 4U–W). Cells are circular, widely elliptic or widely obovate, ranging in size from 3.8–5.5 × 4.0–6.5 μm. The leucosin vesicle is located in the posterior region of the cell (Figure 4U). Two unequal emergent flagella are visible. The long flagellum has mastigonemes (Figures 4V,W), which are directed forward and waved or moved side to side, ranging in length from 4.8 to 8.8 μm. The short flagellum is directed diagonally or bent rearward and ranges in length from 1.9 to 3.4 μm. The nr SSU rRNA molecular signature sequences are as follows:
E23-2 to E23-5 domain of the V4 region: CGCGGAAAACAUAUCUGU
E23-2′ to E23-5′ domain of the V4 region: AUGGGUAUGGGAUCCGCG
Spacer between the E23-8 and E23-9 domains of the V4 region: AGGA
HOLOTYPE: NNIBRPR19602, plastic TEM block Gwanje012018A2, deposited in the Nakdonggang National Institute of Biological Resources, Sangju, Korea (NNIBR).
TYPE LOCALITY: Freshwater, Gwanje pond, Bongeui-ri, Yongji-myeon, Kimje-si, Jeollabuk-do, Korea (35°52′18.0″N 126°57′15.2″E), 20 Jan. 2018
ETYMOLOGY: The Latin specific epithet similis (= similar) refers to the shape being extremely similar to that of other Spumella species
Spumella sinechrysos Jeong, M., Kim, J. I. et Shin, W. sp. nov.
DESCRIPTION: The species is colorless, non-scaled, single-celled and biflagellate without an eyespot (Figures 4X–Z). Cells are circular, widely elliptic or widely obovate, ranging in size from 3.0 to 5.8 × 3.2 to 5.7 μm. The leucosin vesicle is located in the posterior region of the cell (Figure 4X). Two unequal emergent flagella are visible. The long flagellum has two rows of mastigonemes (Figures 4Y,Z) that are directed forward and waved or moved side to side, ranging in length from 4.0 to 7.2 μm. The short flagellum is directed diagonally or bent rearward and ranges in length from 2.0 to 3.5 μm. The nr SSU rRNA molecular signature sequences are as follows:
E23-2 to E23-5 domain of the V4 region: CGAGGAAAACAUGUCUGU
E23-2′ to E23-5′ domain of the V4 region: AUGGGCAUGGGAUCCUCG
Spacer between the E23-8 and E23-9 domains of the V4 region: AGGA
Domain 29 of the V5 region: AGGGAUUGGUGGACGUU
Domain 29′ of the V5 region: GACUCCAUCAGCACCU
HOLOTYPE: NNIBRPR19603, plastic TEM block Hwarim032418A7, deposited in the Nakdonggang National Institute of Biological Resources, Sangju, Korea (NNIBR).
TYPE LOCALITY: Freshwater, Hannam-ri, Namwon-eup, Seoguipo-si, Jeju-do, Korea (33°19′26.5″N, 126°39′31.9″E), 24 Mar. 2018
ETYMOLOGY: The specific epithet “sinechrysos” was derived from Latin sine- (= without) and chrysos (= gold; the color of the chrysophycean plastid).
Discussion
Taxonomic Revision
In the early days after the first description of the genus Monas by Müller (1773), mostly heterotrophic and a few photosynthetic flagellates belonged to this genus (Müller, 1786; Ehrenberg, 1832, 1838; Perty, 1852). With advances in microscopy, observations of heterotrophic flagellates have become more detailed, and the limits of the genus have been changed slightly (Ehrenberg, 1838; Pritchard, 1861; Cienkowsky, 1870). Eventually, the genus Monas was divided into several Monas-related genera, and many species were transferred to other taxonomic ranks (Cienkowsky, 1870; Kent, 1880; Pascher, 1912; Grossmann et al., 2016). Furthermore, as Silva (1960) and Preisig et al. (1991) argued, the name Monas should be abandoned because the identity of its lectotype, Monas mica, designated by Diesing (1850), has not been confirmed since its original description by Müller (1773).
The genus Spumella was established by Cienkowsky (1870) and characterized by having a spherical or oval cell shape with two (or rarely three) unequal flagella, being colorless and heterotrophic (feeding on algae, fungi and starch grains), swimming or attaching to other substrata by a threadlike pedicle, and forming spherical cyst with short collar (Cienkowsky, 1870). He described Spumella vulgaris in detail and differentiated it from Monas termo, described by James-Clark (1868), according to the presence of three flagella, an anteriorly positioned nucleus, the absence of a protruding lip, and a difference in ingestion mode. When comparing the currently recognized S. vulgaris with the originally described S. vulgaris, there are also significant morphological differences. First, there is a difference in prey preference between the two species. While the current S. vulgaris is a bacterivore (Findenig et al., 2010), the original S. vulgaris appears to feed on a variety of prey, including eukaryotic cells (Cienkowsky, 1870). Second, there are large morphological differences between the two species. According to the original description of Cienkowsky (1870), S. vulgaris has two or rarely three flagella, a stick-like structure beneath the flagella, and a threadlike pedicle. However, all our strains showed two flagella and the absence of a stick-like structure and long threadlike pedicle. Lastly, cells of the originally described S. vulgaris divide into two or more parts by means of constriction (see Figures 52–54 on plate XXIV in Cienkowsky, 1870), but our strains divided into two by longitudinal cell division. A common feature of both species is the formation of cysts. Therefore, in light of these morphological differences, the currently recognized genus Spumella is clearly distinguished from the previously described genus.
Cell Size
In this study, we performed morphological and molecular analyses to understand the phylogeny and taxonomy of Spumella. The Korean Spumella strains were colorless and naked on the cell surface and had two emergent flagella: one was a long flagellum with two rows of mastigonemes, and the other was a naked short flagellum. However, these morphological characters are typical of colorless chrysophycean lineages (Grossmann et al., 2016) and cause a problem in identification not only at the species level but also at the genus level. The morphological characters of Spumella species are also highly similar to each other and do not clearly differentiate the species. Even in the one-way ANOVA based on numerical values of vegetative cell morphology, the p-values were higher than 0.05 for all of the measured cell and flagellum size dimensions (Supplementary Figure 4), suggesting that these morphological characters are not significantly different among the species.
Stomatocyst
The stomatocyst of Spumella species is a unique feature that is more reliable than vegetative cell morphology for distinguishing the species (Findenig et al., 2010). The three species among the seven new lineages also showed diagnostic characters related to stomatocyst morphology. The Spumella species with a collar include S. bureschii (rounded and thickened collar apex with the outer collar margin connected to the stomatocyst body), S. rivalis (collar with a rounded marginal rim), S. sphaerophora (thickened collar around the pore), and S. hovassei (approximately three tap-shaped collars) (Fiatte and Joyon, 1956; Skuja, 1956; Findenig et al., 2010). The slightly thickened, round marginal collar of the shallow raised area around the pore of S. rotundata is much more similar to those of S. bureschii and S. spherophora than to that of S. rivalis. However, the stomatocyst of S. rotundata is differentiated by its smaller diameter (5.0–5.8 μm) than those of S. bureschii (5.17–8.09 μm) and S. sphaerophora (9.0–11.0 μm). The cylindrical collar of S. longicolla is similar to that of S. vulgaris (Findenig et al., 2010) and S. beauchampii (Hovasse, 1943), but these two species have a flat planar annulus around the pore. In addition, the collar length of S. longicolla is approximately 2 times longer than that of S. vulgaris and is thicker than those of S. vulgaris and S. beauchampii. The low collar around the stomatocyst pore of S. oblata is similar to that of S. rivalis, but the shape in S. oblata is unique in being wider than the length. This is a striking characteristic feature that contrasts with the spherical shape of all previously reported Spumella stomatocysts, including those of S. rivalis. The newly described stomatocyst in this study also differed from the previously reported stomatocyst of S. vivipara (11.0 μm in diameter) in terms of size.
Molecular Signature
Our phylogenetic tree based on the combined dataset showed the species diversity within the genus Spumella and divided the genus into 15 lineages with seven new species. The molecular signatures of the E23-2 to E23-5 domains, the spacer between E23-8 and E23-9 in the V4 region and domain 29 of the V5 region of nr SSU rRNA were found to differentiate the Spumella species. Due to the difficult identification of microalgal taxa with simple, ambiguous morphological characters, molecular features (DNA sequences or molecular signatures) have been efficiently used to describe different ranks, including at the species level (Marin et al., 2003; Kynclova et al., 2010; Choi et al., 2013; Kim et al., 2013a, b, 2017; Skaloudova and Skaloud, 2013). In particular, the V4 region of nr SSU rRNA is known as a useful genetic marker for understanding biological diversity in various protistan groups, including chrysophytes (Zimmermann et al., 2011; Bock et al., 2017; Tragin et al., 2017). The V4 region is also known as a hypervariable region (Ali et al., 2001; Tragin et al., 2017). The sequences in the molecular signature region are well conserved within species but variable among species in the genus Spumella. Furthermore, the molecular signatures are largely congruent with the phylogenetic relationships and helpful in delimiting Spumella species. Among Spumella-like flagellates, members of the genus Spumella had two unique molecular signatures in domain 8 of the V1 region of nr SSU rRNA and the D5 domain of nr LSU rRNA. Therefore, the genus Spumella can be easily distinguished from other Spumella-like flagellates by using these specific molecular signatures.
Taxonomy of Spumella Species
Since the description of the genus Spumella by Cienkowsky (1870), many taxonomists have observed the morphological characteristics of Spumella under a light microscope or transmission electron microscope (Hovasse, 1943; Skuja, 1948; Silva, 1960; Mignot, 1977; Tanichev, 1993). Recent taxonomic studies have used both molecular and morphological data, showing that the species Spumella vulgaris,S. bureschii, S. rivalis, and S. lacusvadosi are well established as separate taxa (Findenig et al., 2010; Grossmann et al., 2016), but the identities of most traditionally recognized species remain unclear. Some previously described Spumella species, for example, S. maior, S. vivipara, S. gregaria, S. sphaerophora, and S. dinobryonis, are approximately 2–10 times larger in cell size than other Spumella species, including species newly described in this study. Mignot (1977) reported that S. sphaerophora has leucoplasts with a stigma (see Figures 4 and 8 in that article) and feeds on eukaryotic algal cells. However, none of the Spumella species investigated here provided any evidence of a stigma. In addition, the size and shape of the leucoplast in S. sphaerophora species differs from those in all Spumella species, which bear a single, small, oval-shaped leucoplast. Therefore, S. sphaerophora may not belong to Spumella. The species Spumella dinobryonis, S. gregaria, S. guttula, and S. termo have been characterized as having an anterior flagellum 2.0–3.5 times longer than their cell body length and attaching to substrates by a relatively long pedicel (Skuja, 1948; Tanichev, 1993). Another single-celled, unscaled, and stalked heterotrophic biflagellate belongs to the genus Physomonas, as established by Kent (1880). According to Kent’s classification system, the genus Physomonas is not affiliated with the Spumellidae or Monadidae but with the Dendromonadidae, suggesting that organisms bearing a long pedicel may belong to neither the genus Spumella nor other Spumella-like flagellates. The other previously described species in the genus Spumella, S. beauchampii, was originally described as a member of the genus Oikomonas (Hovasse, 1943) but later transferred to the genus Spumella (Silva, 1960) based on morphological characteristics, such as its distinct parabasal body at the bottom of the two flagella and, surprisingly, few Chlorella-like green algae in its food vacuoles. Given these characteristics, the species S. beauchampii is not considered to belong to the genus Spumella.
Feeding
The genus Spumella is a well-known genus of bacterivores in Chrysophyceae and may be adapted to high concentrations of bacteria in diverse aquatic environments (Holen and Boraas, 1991; Boenigk and Arndt, 2000, 2002). The feeding behavior of Korean Spumella was the same as that previously described and recorded in two chrysophyte genera, Ochromonas and Spumella (Boenigk and Arndt, 2000). According to previous studies, Spumella species could feed on bacteria in both benthic and pelagic environments (Boenigk and Arndt, 2000, 2002). When they are in a benthic environment, Spumella adhere to the substratum using a short, pointed plasma membrane or the posterior cell surface. The cell creates filter currents toward the cell surface, aided by the flagella, and enfolds a food particle with a pseudopodium, after which the ingested food particles move toward the posterior end of the cell. In all our Spumella species, we observed the same feeding behavior and food vacuoles in the cytoplasm (Figure 7).
Ultrastructure
The general ultrastructural features of Spumella benthica Hwarim032418A5 and S. bureschii Baekdongje012018B8, representative species of the two major clades (A and B) in the tree, were the leucoplast in the perinuclear space, mitochondria with tubular cristae, and perpendicularly arranged basal bodies but lack of photoreceptors (stigma) and swelling on the short flagellum. These morphological features resemble those of other members of the previously described Spumella (Belcher and Swale, 1976; Preisig and Hibberd, 1983; Tanichev and Karpov, 1993; Mylnikov et al., 2008). The leucoplast, known as a vestigial plastid, has been reported in the colorless chrysophycean genera Spumella, Anthophysa, and Paraphysomonas and dictyochophycean genera Pteridomonas and Ciliophrys (Belcher and Swale, 1972, 1976; Mignot, 1977; Preisig and Hibberd, 1983; Sekiguchi et al., 2002). The leucoplast in colorless chrysophycean genera is encircled by four membranes without any trace of the thylakoid membrane and is located in the perinuclear space. The outermost membrane is continuous with the outer nuclear membrane, and the periplastidial reticulum is located between the third membrane and the inner two membranes. The leucoplast observed in this study showed the same structure in terms of the number of membrane layers, presence of the periplastidial reticulum, and absence of thylakoid traces. However, the recently described genera Poteriospumella and Cornospumella do not show clear ultrastructural evidence, with the leucoplast not appearing to be encircled by the nuclear outer membrane (Grossmann et al., 2016). According to genomic and transcriptomic studies (Beisser et al., 2017; Graupner et al., 2018; Olefeld et al., 2018; Dorrell et al., 2019; Kim et al., 2020), the leucoplast of non-photosynthetic Chrysophyceae was found to have a reduced plastid genome. The plastid genome and transcriptome data showed that the non-photosynthetic lineages of chrysophytes lost most of their photosynthesis-related genes and evolved from phototrophic lineages.
Here, we studied the taxonomy of the genus Spumella based on morphological and molecular data and constructed a molecular phylogeny based on multigene data and increased taxon sampling. We offered detailed images of feeding behavior and ultrastructural images of the leucoplast and stomatocyst. Based on this evidence, we found high species diversity in the genus Spumella, including seven new species, and suggested key molecular signatures that can be used in taxonomic treatment.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MJ, JIK, and WS conceived and designed the experiments, performed LM image recording, DNA extraction, PCR amplification, and phylogenetic analysis, interpreted the data, and wrote the manuscript. MJ, SWN, and WS performed the TEM- and SEM-based image recordings. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the grants from the National Research Foundation (NRF) of Korea (2019R1I1A2A01063159) and National Institute of Biological Resources (NIBR201902207) to WS and the NRF (2018R1D1A1B07050727 and 2021R1C1C2012996) to JIK.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Robert A. Andersen for correction of the holotype and taxonomic consideration of the genera Monas and Heterochromonas.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.758067/full#supplementary-material
Footnotes
References
Ali, A. B., De Baere, R., Van der Auwera, G., De Wachter, R., and Van de Peer, Y. (2001). Phylogenetic relationships among algae based on complete large-subunit rRNA sequences. Int. J. Syst. Evol. Microbiol. 51, 737–749. doi: 10.1099/00207713-51-3-737
Andersen, R. A., Berges, J. A., Harrison, P. J., and Watanabe, M. M. (2005). “Appendix A – recipes for freshwater and seawater media,” in Algal Culturing Techniques, ed. R. A. Andersen (London: Elsevier Academic Press), 429–538.
Arndt, H., Dietrich, D., Auer, B., Cleven, E. J., Gräfenhan, T., Weitere, M., et al. (2000). “Functional diversity of heterotrophic flagellates in aquatic ecosystems,” in The Flagellates, eds B. S. C. Leadbeater and J. C. Green (London: Taylor & Francis), 240–268.
Beisser, D., Graupner, N., Bock, C., Wodniok, S., Grossmann, L., Vos, M., et al. (2017). Comprehensive transcriptome analysis provides new insights into nutritional strategies and phylogenetic relationships of chrysophytes. PeerJ 5:e2832. doi: 10.7717/peerj.2832
Belcher, J. H., and Swale, E. M. F. (1972). The morphology and fine structure of the colourless colonial flagellate Anthophysa vegetans (O. F. Müller) Stein. Br. Phycol. J. 7, 335–346. doi: 10.1080/00071617200650351
Belcher, J. H., and Swale, E. M. F. (1976). Spumella elongata (Stokes) nov. comb., a colourless flagellate from soil. Arch. Protistenk. 118, 215–220.
Berglund, J., Jürgens, K., Bruchmüller, I., Wedin, M., and Andersson, A. (2005). Use of group-specific PCR primers for identification of chrysophytes by denaturing gradient gel electrophoresis. Aquat. Microb. Ecol. 39, 171–182. doi: 10.3354/ame039171
Bock, C., Chatzinotas, A., and Boenigk, J. (2017). Genetic diversity in chrysophytes: comparison of different gene markers. Fottea 17, 209–221. doi: 10.5507/fot.2017.005
Boenigk, J. (2008). Nanoflagellates: functional groups and intraspecific variation. Denisia 23, 331–335.
Boenigk, J., and Arndt, H. (2000). Particle handling during interception feeding by four species of heterotrophic nanoflagellates. J. Eukaryot. Microbiol. 47, 350–358. doi: 10.1111/j.1550-7408.2000.tb00060.x
Boenigk, J., and Arndt, H. (2002). Bacterivory by heterotrophic flagellates: community structure and feeding strategies. Antonie Leeuwenhoek 81, 465–480. doi: 10.1023/a:1020509305868
Boenigk, J., Pfandl, K., Stadler, P., and Chatzinotas, A. (2005). High diversity of the ‘Spumella-like’ flagellates: an investigation based on the SSU rRNA gene sequences of isolates from habitats located in six different geographic regions. Environ. Microbiol. 7, 685–697. doi: 10.1111/j.1462-2920.2005.00743.x
Bourrelly, P. (1957). Recherches sur les Chrysophycées: morphologie, phylogenie et systematique. Rev. Algol. Mem. Hors. Ser. 1, 412–413.
Bütschli, O. (1883). Researches on the flagellate infusoria and allied organisms. Q. J. Microsc. Sci. 23, 53–103.
Charvet, S., Vincent, W., and Lovejoy, C. (2012). Chrysophytes and other protists in High Arctic lakes: molecular gene surveys, pigment signatures and microscopy. Polar Biol. 35, 733–748. doi: 10.1007/s00300-011-1118-7
Choi, B., Son, M., Kim, J. I., and Shin, W. (2013). Taxonomy and phylogeny of the genus Cryptomonas (Cryptophyceae, Cryptophyta) from Korea. Algae 28, 307–330. doi: 10.4490/algae.2013.28.4.307
Cienkowsky, L. (1870). Ueber palmellaceen und einige Flagellaten. Arch. Mikrosk. Anat. 6, 421–438. doi: 10.1007/BF02955988
Dorrell, R. G., Azuma, T., Nomura, M., de Kerdrel, G. A., Paoli, L., Yang, S., et al. (2019). Principles of plastid reductive evolution illuminated by nonphotosynthetic chrysophytes. Proc. Natl. Acad. Sci. U.S.A. 116, 6914–6923. doi: 10.1073/pnas.1819976116
Duff, K. E., Zeeb, B. A., and Smol, J. P. (1995). Atlas of Chrysophycean Cysts – Developments in Hydrobiology 99. Dordrecht: Kluwer Academic Publishers.
Dujardin, F. (1841). Histoire Naturelle des Zoophytes. Infusoires, Comprenant la Physiologie et la Clasification de ces Animaux et la Manière de les Étudier à L’aide du Microscope. Paris: Librarie Encyclopédique de Roret.
Ehrenberg, C. G. (1832). Über die Entwickelung und Lebensdauer der Infusionsthiere; Nebst Ferneren Beiträgen zu Einer Vergleichung Ihrer Organischen Systeme. Berlin: Abhandlungen der Königlichen Akademie Wissenschaften zu Berlin.
Ehrenberg, C. G. (1838). Die Infusionsthierchen als Vollkommene Organismen. Ein Blick in das Tiefere Organische Leben der Natur. Leipzig: Verlag von Leopold Voss.
Evans, K. M., Wortley, A. H., and Mann, D. G. (2007). An assessment of potential diatom “barcode” genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist 158, 349–364. doi: 10.1016/j.protis.2007.04.001
Fiatte, M. C., and Joyon, L. (1956). Heterochromonas hovassei (n. sp.), Chrysomonadine décolorée. Arch. Zool. Exp. Gen. 105, 273–283.
Findenig, B. M., Chatzinotas, A., and Boenigk, J. (2010). Taxonomic and ecological characterization of stomatocysts of Spumella-like flagellates (Chrysophyceae). J. Phycol. 46, 868–881. doi: 10.1111/j.1529-8817.2010.00892.x
Fresenius, G. (1858). Beiträge zur Kenntniss Mikroskopischer Organismen. Frankfurt: Druck und verlag von H. L. Brönner.
Graupner, N., Jensen, M., Bock, C., Marks, S., Rahmann, S., Beisser, D., et al. (2018). Evolution of heterotrophy in chrysophytes as reflected by comparative transcriptomics. FEMS Microbiol. Ecol. 94:fiy039. doi: 10.1093/femsec/fiy039
Grossmann, L., Bock, C., Schweikert, M., and Boenigk, J. (2016). Small but manifold - hidden diversity in “Spumella-like Flagellates”. J. Eukaryot. Microbiol. 63, 419–439. doi: 10.1111/jeu.12287
Holen, D. A., and Boraas, M. E. (1991). The feeding behavior of Spumella sp. as a function of particle size: implications for bacterial size in pelagic systems. Hydrobiologia 220, 73–88. doi: 10.1007/BF00017493
Hovasse, R. (1943). Contribution à l’étude des flagellés libres: Oicomonas beauchampi sp. nov., Protomonadine et chrysomonade. Arch. Zool. Exp. Gen. 83, 47–53.
James-Clark, H. (1868). “On the spongiae ciliatae as Infusoria flagellata; or, observations on the structure, animality, and relationship of Leucosolenia botryoides, Bowerbank,” in The Animals and Magazine of Natural History, Including Zoology, Botany and Geology, Vol. I-Fourth Series, eds C. C. Brington, J. D. Gray, W. S. Dallas, and W. Francis (London: Talyor & Francis), 133–142.
Kim, J. I., Jeong, M., Archibald, J. M., and Shin, W. (2020). Comparative plastid genomics of non-photosynthetic chrysophytes: genome reduction and compaction. Front. Plant Sci. 11:572703. doi: 10.3389/fpls.2020.572703
Kim, J. I., Nam, S., So, J. A. E., Hong, S. G., Choi, H. G., and Shin, W. (2017). Asterochloris sejongensis sp. nov. (Trebouxiophyceae, Chlorophyta) from King George Island, Antarctica. Phytotaxa 295:60. doi: 10.11646/phytotaxa.295.1.5
Kim, J. I., Shin, W., and Triemer, R. E. (2013a). Cryptic speciation in the genus Cryptoglena (Euglenaceae) revealed by nuclear and plastid SSU and LSU rRNA gene. J. Phycol. 49, 92–102. doi: 10.1111/jpy.12032
Kim, J. I., Shin, W., and Triemer, R. E. (2013b). Phylogenetic reappraisal of the genus Monomorphina (Euglenophyceae) based on molecular and morphological data. J. Phycol. 49, 82–91. doi: 10.1111/jpy.12018
Kynclova, A., Skaloud, P., and Škaloudová, M. (2010). Unveiling hidden diversity in the Synura petersenii species complex (Synurophyceae, Heterokontophyta). Nova Hedwig. 136, 283–298. doi: 10.1127/1438-9134/2010/0136-0283
Lepère, C., Boucher, D., Jardillier, L., Domaizon, I., and Debroas, D. (2006). Succession and regulation factors of small eukaryote community composition in a lacustrine ecosystem (Lake Pavin). Appl. Environ. Microbiol. 72, 2971–2981. doi: 10.1128/aem.72.4.2971-2981.2006
Marin, B., Palm, A., Klingberg, M., and Melkonian, M. (2003). Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist 154, 99–145. doi: 10.1078/143446103764928521
Mignot, J. P. (1977). E’tude ultrastructurale d’un flagelle’ du genre Spumella Cienk. (= Heterochromonas Pascher = Monas O. F. Müller), chrysomonadine leucoplastidiee. Protistologica 13, 219–231.
Müller, O. F. (1773). Vermium Terrestrium et Fluviatilium, Seu Animalium Infusoriorum, Helminthicorum et Testaceorum, Non Marinorum, Succincta Historia. Lipsiæ: Apud Heineck et Faber, typis Martini Hallager
Müller, O. F. (1786). Animalcula Infusoria Fluviatilia et Marina, Quae Detexit, Systematice Descripsit et ad Vivum Delineari Curavit. Hauniæ: Typis Nicolai Mölleri, Aulæ regiæ typographi.
Mylnikov, A. P., and Mylnikova, Z. M. (2005). The morphology of heterothrophic chrysomonads of the genus Spumella (Chrysophyta). Inland Water Biol. 3, 57–62.
Mylnikov, A. P., Mylnikova, Z. M., and Tikhonenkov, D. V. (2008). The main cell morphology of the freshwater colorless chrysomonad Spumella sp. (Ochromonadales, Chrysophyceae). Inland Water Biol. 1, 32–36. doi: 10.1007/s12212-008-1006-1
Olefeld, J. L., Majda, S., Albach, D. C., Marks, S., and Boenigk, J. (2018). Genome size of chrysophytes varies with cell size and nutritional mode. Org. Divers. Evol. 18, 163–173. doi: 10.1007/s13127-018-0365-7
Pascher, A. (1912). Über rhizopoden-und palmellastadien bei flagellaten (Chrysomonaden), nebst einer Übersicht über die braunen Flagellaten. Arch. Protistenk. 25, 153–200.
Perty, M. (1852). Zur Kenntniss Kleinster Lebensformen: Nach Bau, Funktionen, Systematik, mit Specialverzeichniss der in der Schweiz Beobachteten. Bern: Jent & Reinert.
Pfandl, K., Chatzinotas, A., Dyal, P., and Boenigk, J. (2009). SSU rRNA gene variation resolves population heterogeneity and ecophysiological differentiation within a morphospecies (Stramenopiles, Chrysophyceae). Limnol. Oceanogr. 54, 171–181. doi: 10.4319/lo.2009.54.1.0171
Posada, D., and Crandall, K. A. (1998). MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Preisig, H. R., and Hibberd, D. J. (1983). Ultrastructure and taxonomy of Paraphysomonas (Chrysophyceae) and related genera 3. Nord. J. Bot. 3, 695–723. doi: 10.1111/j.1756-1051.1983.tb01481.x
Preisig, H. R., Vørs, N., and Hallfors, G. (1991). “Diversity of heterotrophic heterokont flagellates,” in The Biology of Free-Living Heterotrophic Flagellates, eds D. J. Patterson and J. Larson (Oxford: Clarendon Press), 361–399.
Pritchard, A. (1845). A history of Infusoria, Living and Fossil: Arranged According to “Die Infusionsthierchen” of C.G. Ehrenberg. London: Whittaker and Co.
Pritchard, A. (1861). History of Infusoria, Including the Desmidiaceæ and Diatomaceæ, British and Foreign. London: Whittaker and Co.
Pusztai, M., and Škaloud, P. (2019). Elucidating the evolution and diversity of Uroglena-like colonial flagellates (Chrysophyceae): polyphyletic origin of the morphotype. Eur. J. Phycol. 54, 404–416. doi: 10.1080/09670262.2019.1574030
Reynold, B. D. (1934). Studies on monad flagellates. I. historical and taxonomic review of the genus Monas, II. Observations on Monas vestita. Arch. Protistenk. 81, 399–411.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sanders, R. W., Leeper, D. A., King, C. H., and Porter, K. G. (1994). Grazing by rotifers and crustacean zooplankton on nanoplanktonic protists. Hydrobiologia 288, 167–181. doi: 10.1007/BF00006240
Sekiguchi, H., Moriya, M., Nakayama, T., and Inouye, I. (2002). Vestigial chloroplasts in heterotrophic stramenopiles Pteridomonas danica and Ciliophrys infusionum (Dictyochophyceae). Protist 153, 157–167. doi: 10.1078/1434-4610-00094
Sherr, E. B., and Sherr, B. F. (1994). Bacterivory and herbivory: key roles of phagotrophic protists in pelagic food webs. Microb. Ecol. 28, 223–235. doi: 10.1007/BF00166812
Skaloudova, M., and Skaloud, P. (2013). A new species of Chrysosphaerella (Chrysophyceae: Chromulinales), Chrysosphaerella rotundata sp. nov., from Finland. Phytotaxa 130, 34–42. doi: 10.11646/phytotaxa.130.1.4
Skuja, H. (1948). Taxonomie des phytoplanktons einiger seen in Uppland, Schweden. Symb. Bot. Upsal. 9, 1–399.
Skuja, H. (1956). Taxonomische und biologische studien über das phytoplankton schwedischer Binnengewässer. Nova Acta Reg. Soc. Sc. Ups. Ser. IV 16, 312–321.
Smith, S., Overbeek, R., Woese, C., Gilbert, W., and Gillevet, P. (1994). The genetic data environment: an expandable GUI for multiple sequence analysis. Comput. Appl. Biosci. 10, 671–675. doi: 10.1093/bioinformatics/10.6.671
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stein, F. (1878). Der Organismus der Infusionsthiere Nach Eigenen Forschungen in Systematischere Reihenfolge Bearbeitet. Leipzig: Wlihelm Engelmann.
Tanichev, A. (1993). Morphology of the Baikal Chrysomonads, Spumella termo and S. gregaria sp. n. (Protozoa, Chrysomonadida). Zool. Zh. 72, 23–29.
Tanichev, A., and Karpov, S. (1993). Peculiar features of Baikal colorless chrysomonad Spumella termo ultrastructure. Tsitologiya 35, 3–7.
Tragin, M., Zingone, A., and Vaulot, D. (2017). Comparison of coastal phytoplankton composition estimated from the V4 and V9 regions of the 18S rRNA gene with a focus on photosynthetic groups and especially Chlorophyta. Environ. Microbiol. 20, 506–520. doi: 10.1111/1462-2920.13952
Valkanov, A. (1925). Beitrag zur kenntnis der flagellaten von bulgarien. Izv. Bulg. Bot. Drouzhestvo (Sofia) 1, 105–121.
Wilkinson, A. N., Zeeb, B. A., and Smol, J. P. (2001). Atlas of Chrysophycean Cysts Vol. II - Developments in Hydrobiology 157. Dordrecht: Kluwer Academic Publishers.
Wylie, J. L., and Currie, D. J. (1991). The relative importance of bacteria and algae as food sources for crustacean zooplankton. Limnol. Oceanogr. 36, 708–728. doi: 10.4319/lo.1991.36.4.0708
Yubuki, N., Nakayama, T., and Inouye, I. (2008). A unique life cycle and perennation in a colorless chrysophyte Spumella sp. J. Phycol. 44, 164–172. doi: 10.1111/j.1529-8817.2007.00441.x
Keywords: Chrysophyceae, heterotrophic, leucoplast, new species, phylogeny, Spumella, stomatocyst
Citation: Jeong M, Kim JI, Nam SW and Shin W (2021) Molecular Phylogeny and Taxonomy of the Genus Spumella (Chrysophyceae) Based on Morphological and Molecular Evidence. Front. Plant Sci. 12:758067. doi: 10.3389/fpls.2021.758067
Received: 13 August 2021; Accepted: 04 October 2021;
Published: 26 October 2021.
Edited by:
Denis V. Tikhonenkov, Institute of Biology of Inland Waters, Russian Academy of Sciences (RAS), RussiaReviewed by:
Pavel Skaloud, Charles University, CzechiaHaruyo Yamaguchi, National Institute for Environmental Studies (NIES), Japan
Copyright © 2021 Jeong, Kim, Nam and Shin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woongghi Shin, c2hpbndAY251LmFjLmty
 Minseok Jeong
Minseok Jeong Jong Im Kim
Jong Im Kim Seung Won Nam
Seung Won Nam Woongghi Shin
Woongghi Shin