- 1Key Laboratory of Plant-Soil Interactions, College of Resources and Environmental Sciences, Ministry of Education, National Academy of Agriculture Green Development, China Agricultural University, Beijing, China
- 2Department of Vegetable Sciences, China Agricultural University, Beijing, China
Magnesium (Mg) deficiency is becoming a widespread limiting factor for crop production. How crops adapt to Mg limitation remains largely unclear at the molecular level. Using hydroponic-cultured tomato seedlings, we found that total Mg2+ content significantly decreased by ∼80% under Mg limitation while K+ and Ca2+ concentrations increased. Phylogenetic analysis suggested that Mg transporters (MRS2/MGTs) constitute a previously uncharacterized 3-clade tree in planta with two rounds of asymmetric duplications, providing evolutionary evidence for further molecular investigation. In adaptation to internal Mg deficiency, the expression of six representative MGTs (two in the shoot and four in the root) was up-regulated in Mg-deficient plants. Contradictory to the transcriptional elevation of most of MGTs, Mg limitation resulted in the ∼50% smaller root system. Auxin concentrations particularly decreased by ∼23% in the Mg-deficient root, despite the enhanced accumulation of gibberellin, cytokinin, and ABA. In accordance with such auxin reduction was overall transcriptional down-regulation of thirteen genes controlling auxin biosynthesis (TAR/YUCs), transport (LAXs, PINs), and signaling (IAAs, ARFs). Together, systemic down-tuning of gene expression in the auxin signaling pathway under Mg limitation preconditions a smaller tomato root system, expectedly stimulating MGT transcription for Mg uptake or translocation.
Introduction
Magnesium (Mg), a non-substitutive component, performs a vast array of physiological and biochemical functions in plants. The well-addressed function of Mg is its participation in photosynthetic CO2 assimilation (Gerendás and Führs, 2013; Li J. et al., 2020). A large proportion (15–35%) of total available Mg within a plant is confined to the light-capturing complex of chloroplasts, where it not only functions as a structural element of chlorophyll (Chl) but also underpins photosynthetic performance (Farhat et al., 2016; Chen et al., 2018). Mg also acts as a cofactor of various enzymes (>300) involved in essential biological processes such as Chl biosynthesis, sugar transportation, and energy metabolism (Ma et al., 2016; Chen et al., 2018; Koch et al., 2019). Recent meta-analysis-based studies highlighted that Mg deficiency is becoming a rising concern in most production systems due to the large removal of Mg in intensive crop production systems and injudicious fertilization (Hauer-Jákli and Tränkner, 2019; Wang et al., 2020).
Plant roots employ adaptive mechanisms in response to environmental stimuli or nutrient variations. For instance, mild nitrogen or phosphate deficiency tends to enhance root growth for a greater nutrient absorption or translocation system (Ahmad et al., 2018; Meier et al., 2020; Nadeem et al., 2020). However, how roots respond to the Mg deficiency stress is inconsistent: root growth is clearly reduced by Mg deficiency in bean plants (Vicia faba L.) (Neuhaus et al., 2014), Arabidopsis thaliana (Gruber et al., 2013; Li D. et al., 2020), and potato (Solanum tuberosum L.) (Koch et al., 2019, 2020). However, in other studies, Mg deficiency does not considerably affect root growth in Arabidopsis (Hermans and Verbruggen, 2005) and Chinese cabbage (Verbruggen and Hermans, 2013). Notably, low Mg supply leads to more root hair development in Arabidopsis as an adapted strategy (Niu et al., 2014; Liu M. et al., 2018). Hence, further study is required to unravel root adaptation to Mg deficiency stress at morphological and molecular levels in different plant species, especially in model crop plants.
Mg transporter genes, homologues of CorA Mg transporters, have been identified across crop plants and classified into the CorA/MRS2/MGT family (Chen et al., 2018; Yan et al., 2018; Zhang et al., 2019a). MRS2/MGT genes are supposed to be primary transporters for Mg2+ uptake, distribution, and homeostasis in plants (Li et al., 2001, 2016; Marschner, 2012; Saito et al., 2013). In Arabidopsis, the MRS2/MGT gene family is composed of ten members, and several genes are important for Mg2+ uptake and transportation under normal or Mg limitation (Oda et al., 2016; Yan et al., 2018). However, the activity of these MGTs relies on the genetic makeup of plant species (Cui et al., 2016). Mg deficiency alters MRS2/MGT expression in the rice root (Chen et al., 2017; Zhang et al., 2019b); however, moderate Mg deficiency had no impact on MRS2/MGT transcription in the Arabidopsis root (Hermans et al., 2010; Ogura et al., 2018). Therefore, further molecular investigation is needed to understand the evolutionary dynamics, and transcription patterns of these MRS2/MGT family transporters in plant roots under Mg limitation.
Auxin is a key player regulating root growth and development (Porco et al., 2016; Stoeckle et al., 2018; Meier et al., 2020; Moret et al., 2020; Hu et al., 2021). During auxin biosynthesis, TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS/TRYPTOPHAN AMINOTRANSFERASE-RELATED (TAA/TAR) controls indole-3-pyruvic acid (IPA) generation and YUCCA (YUC) acts to convert IPA into Indole-3-acetic acid (IAA) (Mashiguchi et al., 2011; Song et al., 2017). Auxin then moves across membranes by AUXIN1/LIKE-AUX1 (AUX1/LAX) influx carriers, PIN-FORMED (PIN) efflux carrier, PIN-LIKES (PILS), or ATP-binding cassette subfamily B (ABCB)-type transporters (Sauer and Kleine-Vehn, 2019; Vosolsobe et al., 2020). PIN transporters largely control intercellular and intracellular auxin transport (Zhang Y. et al., 2020); LAX3 acts as an auxin influx carrier during root development (Maghiaoui et al., 2020). Finally, AUXIN RESPONSE FACTORS (ARFs) together with AUXIN/INDOLE ACETIC ACID (Aux/IAA) regulate downstream gene expression in the auxin signaling pathway (Roosjen et al., 2018). Under low availability of IAA, Aux/IAA proteins bind to AUXIN RESPONSE FACTORS (ARFs) to prevent their expression (Lavenus et al., 2013; Mironova et al., 2017); while higher levels of IAA promotes Aux/IAA degradation to release the suppression of auxin-responsive genes (Wang and Estelle, 2014; Weijers and Wagner, 2016). Such Aux/IAA degradation is strongly linked with lateral root development (Guseman et al., 2015).
Auxin biosynthesis, transport, and signaling are essential components related to root growth and root system architecture. Nutrient deficiency stresses frequently enhance auxin accumulation in roots. For instance, low levels of nitrate stimulate auxin accumulation in roots of Arabidopsis and maize (Ma et al., 2014; Sun et al., 2020), and low concentrations of ammonium also increase auxin content in the Arabidopsis root (Meier et al., 2020). Similarly, phosphorus deficiency improves auxin levels in roots of Arabidopsis and Foxtail millet (Ahmad et al., 2018; Bhosale et al., 2018), and potassium limitation modulates the expression of auxin-responsive genes in the root of rice (Ma et al., 2012). Zinc supply alters auxin homeostasis in the roots of Arabidopsis (Zhang et al., 2018; Wang et al., 2021). Few studies show the distribution and transcriptional variations of auxin in response to Mg supply. For instance, P and Mg interactively affect the redistribution and accumulation of auxin in Arabidopsis roots through altered signaling functions of AUX1, PIN2, and PIN3, which modulate primary root elongation and growth direction (Niu et al., 2015). In another study, optimum Mg supply promotes PIN2-based polar auxin transport and distribution under aluminum toxicity, which restores root growth by regulating root surface pH (Zhang Z. et al., 2020). Transcriptomic data show that Mg deficiency differentially regulates expression of auxin-responsive genes in Arabidopsis and Citrus sinensis (Hermans et al., 2010; Yang et al., 2019). For instance, Mg deficiency altered expression of auxin efflux carrier proteins and AUX/IAA (IAA1, IAA5, IAA6, IAA14, IAA17, IAA19) in the leaves of Arabidopsis (Hermans et al., 2010). Similarly, under Mg deficiency, IAA11, IAA13, IAA29, and ARF4 are upregulated by 2-, 1. 5-, 2. 8-, and 2.5-fold, respectively, in the leaves of Citrus sinensis (Yang et al., 2019). However, how Mg limitation affects auxin homeostasis and related gene expression in plant roots remains elusive.
Tomato (Solanum lycopersicum L.) is one of the most important vegetable plants worldwide and it requires more Mg to form the same biomass than grass or grain crops (Broadley and White, 2010; Gerendás and Führs, 2013). In this study, we used tomato as a model plant to investigate the response of seedlings, especially the root system, to low external Mg supply at the molecular level. We performed the evolutionary analysis of the MRS2/MGT-type Mg transporter family for further molecular investigation. We found overall up-regulation of Mg transporters in the smaller root and consistent down-regulation of auxin accumulation and related gene expression in the root under Mg limitation.
Materials and Methods
Plant Growth Conditions and Experimental Set-Up
This study was conducted in a standard greenhouse at China Agricultural University, Beijing, China. The greenhouse environment was as follows: 28/22°C temperature, 60% relative humidity, 14/10 light/dark photoperiod, and natural daylight. Tomato (Solanum lycopersicum L.) cv. Xianliang, procured from Dalian Lida Seed Company (China), was used as the plant material in this study. Seeds were first surface-sterilized by heating in a water bath at 55°C for 15 min, and soaking in 10% Na3PO2 for 20 min. Seeds were then nurtured by imbibing in distilled water (DW) for 6 h, and germinated on moist filter paper covered with black plastic wrap in a growth chamber. The sprouted seeds with a 2-cm primary root were sown in 50-cell seedling plug trays filled with commercially available potting soil containing low indigenous Mg (46 mg kg–1), and were irrigated regularly with DW.
At the four fully unfolded compound leaf stage (∼23-days after sowing), consistent and uniformly sized seedlings were transferred into continuously aerated 10-L (in 28 × 25 × 17 cm pots) nutrient solution. Nutrient solution composition was slightly modified from a published protocol for tomato culture (Cámara-Zapata et al., 2019). The final nutrient solution contained macro- and micronutrients as follows: 4 mM Ca (NO3)2.4H2O, 0.6 mM KNO3, 1 mM KH2PO4, 1 mM MgSO4.7H2O, 90 μM Na2Fe-EDTA, 25 μM H3BO3, 2 μM MnSO4.H2O, 2 μM ZnSO4.7H2O, 0.5 μM CuSO4.5H2O, and 0.5 μM (NH4)6Mo7O24.4H2O. The experiment contained two levels of Mg, one control (Ctrl) having optimum Mg as 1 mM MgSO4.7H2O, and another low Mg (LMG) consisting of 0.02 mM MgSO4.7H2O. The LMG level was determined according to our preliminary phenotypic screen by growing tomato seedlings at varying Mg concentrations (0.02, 0.06, 0.25, 1, 4 mM). The onset of the treatment was established at the time of transplanting with the intention to avoid Mg accumulation in the vacuole at the very initial growth stages (Hauer-Jákli and Tränkner, 2019). In the base solution, MgSO4 was the only source of SO42–, meanwhile, equivalent moles of Na2SO4 was applied to Mg-depleted plants to maintain the relatively constant cation/anion balance and compensate the SO42– deficiency.
To avoid osmotic shock, seedlings were first grown in the 50%-strength nutrient solution for 3 days, and then full-strength nutrient solution was supplied till harvest. This study was performed with six biological replicates and each biological replicate had six plants (technical replicates). The nutrient solution was replaced at an interval of 3 days to avoid ion depletion. The pH was maintained at 6.0 with 1M TRIS (C4H11NO3). After a 3-week treatment, seedlings were dissected at the root-shoot junction, flash-frozen in liquid-N2, and stored at –80°C for physiological and molecular analysis. For root growth analysis, 2-week samples were also harvested. The experiment was reproduced at least two times.
Elemental Analysis and Root-to-Shoot Translocation of Mg2+
For elemental analysis (Mg2+, K+, Ca2+, and Na+) in the plant material, oven-dried root and shoot samples were grounded separately by an electric grinder. The 100 mg of tissue powder was weighed using a high-accuracy balance, and transferred in a Teflon digestion tube. Followed by, 6 ml concentrated HNO3 was added and stood overnight until the vigorous reaction phase was over. Then, 2 ml H2O2 was added and digested in the microwave digestion system (MARS 6, CEM Microwave Technology LTD, United States). The digested solution was transferred into a 25−ml volumetric flask and filled up to the calibration mark with ultrapure water. The concentrations of minerals in the solution were determined by an inductively coupled plasma-optical emission spectrometer (OPTIMA 3300 DV, Perkin-Elmer, MA, United States). After every 20 samples, the blank sample was included to ensure the accuracy of measurements. At least two sets of samples were run with six replicates, and concentrations of each element were quantified as described in our earlier study (Ishfaq et al., 2021). The total Mg2+ content and root-to-shoot translocation of Mg2+ were successively quantified as follow: Total Mg2+ content = [(root dry biomass × root Mg2+ concentration) + (shoot dry biomass × shoot Mg2+ concentration)], and root-to-shoot translocation of Mg2+ = [(shoot dry biomass × shoot Mg2+ concentration)/Total Mg2+ content] × 100%.
Sequence Blast, Alignment, and Phylogenetic Construction
To retrieve the protein sequences in angiosperms, lycophytes, mosses, liverworts, algae and fungi, MRS2/MGT proteins in Arabidopsis thaliana were used as queries to blast in Phytozome1 and National Center for Biotechnology Information.2 The sequences were scored and aligned sequentially using the MUSCLE 3.6 program,3 GeneDoc 3.24 (Nicholas, 1997), and CLUSTALX.5 The protein matrix was used to plot the MRS2/MGT family tree. The phylogenetic tree was mapped by the MEGA 6.0 program6 following the neighbor-joining method, and the bootstrap was analyzed in 1,000 replications. The gene structure was plotted by using GSDS.7 Gene accession numbers were listed in Supplementary Table 1.
Plant Phenotyping and Root System Analysis
The root and shoot dry weight was determined after separately drying in the oven at 65°C till constant weight. The root-to-shoot ratio was calculated on a dry matter basis. Each sample had six independent biological replicates, and each replicate was averaged by two samples harvested from the same pot.
At harvest, the entire root system dissected from the root-shoot junction was rinsed three times by DW and stored at –4°C until scan analysis. Each root was placed on a transparent plastic slide and thoroughly combed apart in DW. The desktop scanner (Epson Perfection V850 Pro) was used to scan the roots at a resolution of 400 dpi. The scanned images were analyzed using the WinRHIZO software (version 5.0) (Regent Instruments Inc., Quebec City, QC, Canada) to quantify root indexes as described (Freschet et al., 2020). The relative root growth rate (RRGR) was computed as a given equation (Samal et al., 2010).
Where, ln is natural log, TRL is total root length in cm and t is time of harvest in days; and the subscripts 1 and 2 refer to the 14 and 21 days of harvesting after transfer, respectively.
Extraction and Quantification of Endogenous Hormones
To analyze dynamic hormonal distribution under Mg limitation, enzyme-linked immunosorbent assay (ELISA) was performed having 96-well microtitration plates filled with extract of plant tissues or hormone standards and corresponding antibodies, according to the standard protocol (EL310, Bio-TEK, Winooski, VT) and the published method (Dai et al., 2009). Briefly, ∼0.5 g frozen plant tissue was fine grounded in a pre-cooled mortar, and homogenized in 10 ml 80% (v/v) methanol extraction medium containing 1 mM butylated hydroxytoluene (BHT) as an antioxidant. Subsequently, the collected extract was incubated at 4°C for 4 h, and then centrifuged at 4,000 rpm for 20 min at 2–4°C. The supernatant was passed through Chromosep C18 columns (C18 SepPak Cartridges, Waters, Millford, MA), prewashed with absolute methanol. The hormonal fraction was eluted with 10 ml of 100% (v/v) methanol and then 10 ml ether. The elute was then N2-dried, and dissolved in 2.0 ml phosphate-buffered saline (PBS) containing 0.1% (w/v) gelatin and 0.1% (v/v) Tween-20 (pH 7.5). The concentrations of IAA (indole-3-acetic acid), ABA (abscisic acid), GA3 (gibberellic acid 3), and ZR (zeatin riboside) were quantified at an optical density of A490 (Weiler et al., 1981).
RNA Extraction and Quantification by RT-qPCR
Total RNA was extracted from ∼100 mg powdered (in liquid N2) tomato samples using the Trizol reagent, following the manufacturer’s instructions (Invitrogen). The quality of extracted RNA was checked by a NanoDrop Spectrophotometer. Next, 4–5 g RNA was digested by DNase 1 (Takara Biomedicals, Kyoto, Japan) to eliminate potential DNA contamination. Reverse transcription of RNA samples into cDNA was carried out using M-MLV reverse transcriptase (Thermo Fisher Scientific, Waltham, MA United States). Quantitative PCR (RT-qPCR) in a Bio-Rad iCycler iQ5 system (Bio-Rad, Hercules, CA, United States) was operated to quantify relative gene expression by using cDNA, SYBR Premix Ex Taq™ (Takara), and designed primers of genes of interest (Supplementary Table 2). The qPCR was programmed for 10 min of pre-incubation at 95°C, 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. Four biological and two technical replicates were analyzed for each gene. The housekeeping gene Slubi3 was used as an internal control (Liu S. et al., 2018). The equation 2^-(ΔΔCt) was used to determine relative gene expression (Livak and Schmittgen, 2001).
Statistical Analysis
Data processing with simple calculations (e.g., mean, standard deviation [SD], maximum and minimum) was performed by Microsoft Excel-2019. One-way analysis of variance (ANOVA) along with the Tukey honest significant difference (HSD) test was performed by Statistix 8.1 (Analytical Software, Tallahassee, FL, United States) to analyze the statistical difference (*P < 0.05, **P < 0.01, ***P < 0.001) across treatments, as specified in figure legends. Bar graphs were plotted by GraphPad Prism 9.
Results
Elemental Homeostasis Under Magnesium Limitation
The data of elemental homeostasis showed an obvious variation among treatments. The Mg2+ concentration significantly reduced from 2.6 to 1.29 mg g–1 (∼50%) in the tomato root, and 4.27–mg g–1 (∼61%) in the shoot under Mg limitation, compared to control (Figure 1A). The total Mg2+ content in the Mg-sufficient seedling was significantly higher by ∼80%, compared to LMG plants (Figure 1B). Out of whole plant Mg2+ uptake, we did not find significant variations in percent root-to-shoot translocation of Mg2+ among treatments (Figure 1C). Importantly, during Mg limitation, the concentration of other competing cations such as K+, Ca2+, and Na+ significantly increased by 34, 33, and 85% in the root, respectively. In the shoot, K+, Ca2+, and Na+ concentrations significantly increased by 9, 22%, and ∼2-fold, respectively, compared to control (Figures 1D–F).
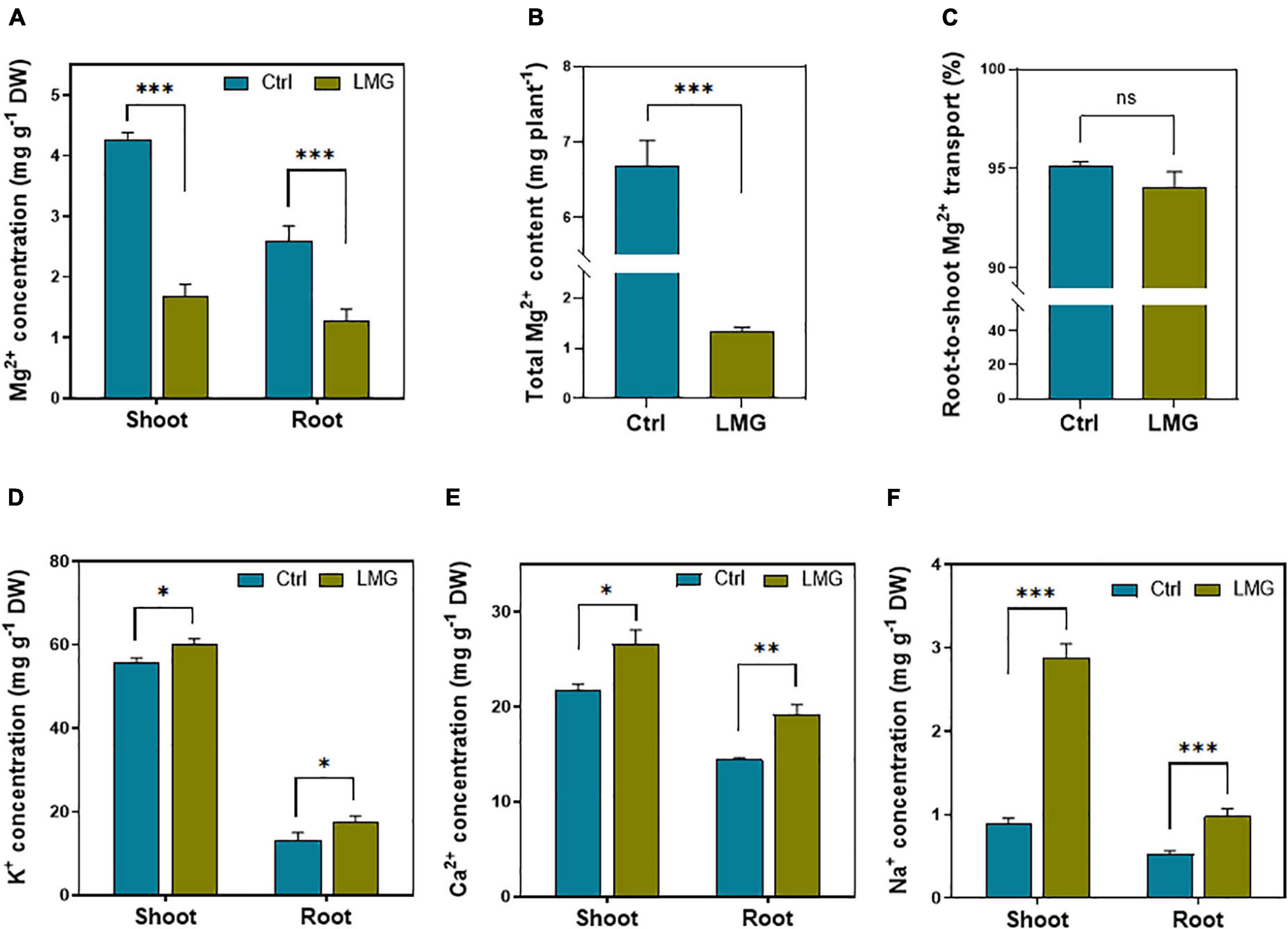
Figure 1. Mg limitation altered Mg2+ uptake and elemental homeostasis in tomato seedlings. (A) Mg2+ concentration in shoot and root (mg g–1 DM), (B) total Mg2+ content (mg pot–1), (C) root-to-shoot translocation of Mg2+ (%), (D) K+ concentration in shoot and root (mg g–1 DM), (E) Ca2+ concentration in shoot and root (mg g–1 DM), and (F) Na+ concentration in shoot and root (mg g–1 DM). The bar graph showed the mean value while whiskers represented the maximum/minimum values of six independent biological replicates. Asterisks indicated significant differences at *P < 0.05, **P < 0.01, and ***P < 0.001, according to Tukey’s HSD test. Where, Ctrl, control; LMG, low Mg; ns, non-significant.
Evolutionary Analysis of the MRS2/MGT-Type Magnesium Transporter Family and MGTs Expression in Response to Magnesium Limitation
To better select and analyze expression levels of representative Mg transporters, we constructed the phylogenetic tree of the MRS2/MGT-type Mg transporter family by retrieving corresponding protein sequences of Solanum lycopersicum, Arabidopsis thaliana, Oryza sativa, Zea mays, Amborella trichopoda, Selaginella moellendorffii, Physcomitrella patens, Marchantia polymorpha, and Chlamydomonas reinhardtii, with fungal MRSs as an out-group. Phylogenetic analysis showed that MRS2/MGT constitutes a previously uncharacterized 3-basal clade in planta with two rounds of asymmetric duplications. Briefly, Clade I split into four subclades (MGT1/2/3, MGT4, MGT5/6, MGT7/8/9), and Clade II(MGT1/2/3, MGT4the MGT10 subfamily. Clade I and Clade II contained members across angiosperms, lycophytes, mosses, liverworts, and algae, while Clade III had no members from angiosperms (Figure 2). The gene structure of the MRS2/MGT family also supported such a three-clade pattern, as shown in Figure 3A. The average exon number showed obvious changes across clades I, II, and III and average exon 1–12, 11–14, 6–18 exons, respectively. Less exons of MGT1/2/3, MGT4, and MGT5/6 are probably due to intron losses in these subfamilies during evolution.
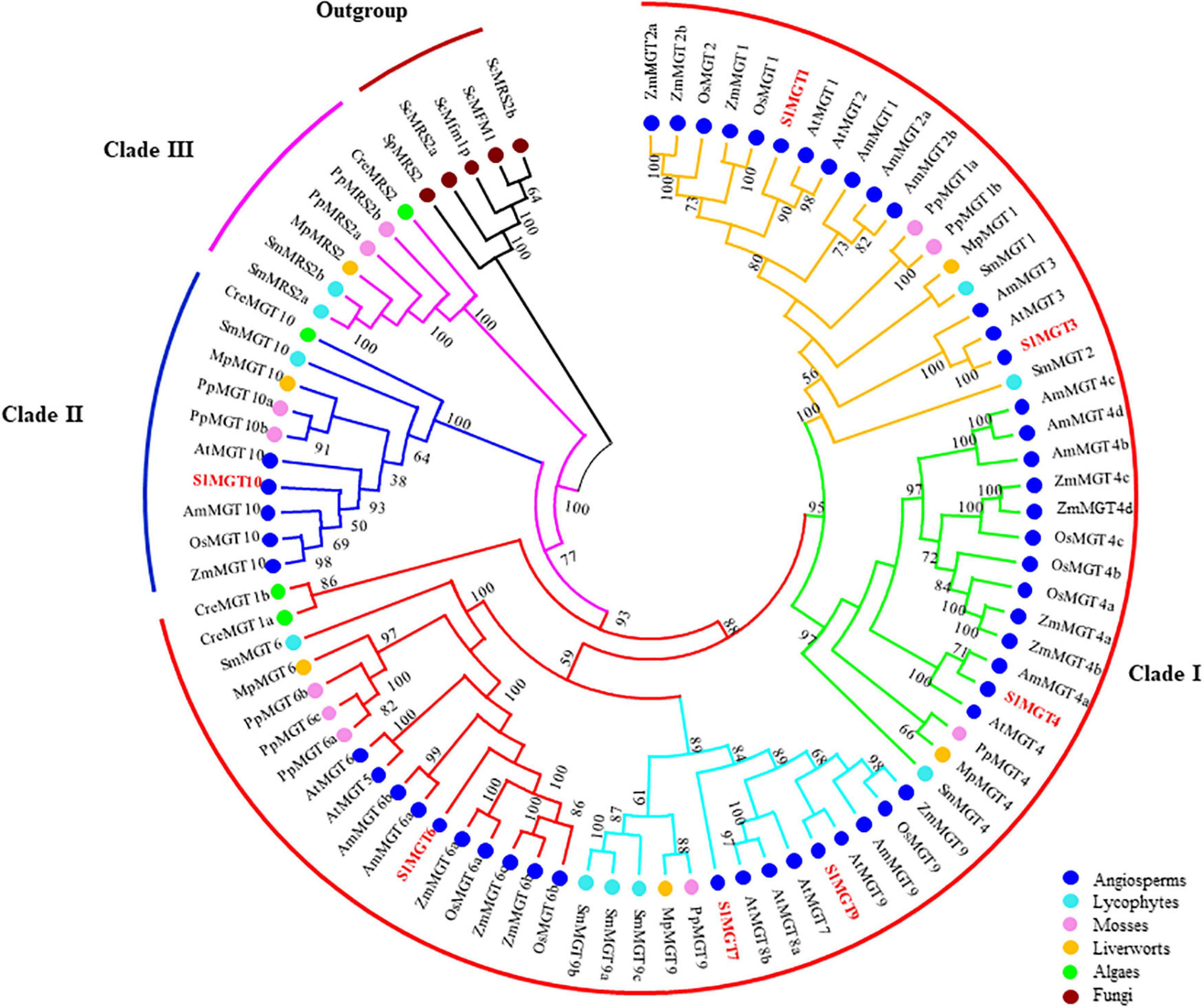
Figure 2. Phylogenetic analysis of the MRS2/MGT gene family in angiosperms, lycophytes, mosses, liverworts, algae and fungi. Three basal clades were indicated by red (Clade 1), blue (Clade II), and purple (Clade III) curves. In Clade 1, MGT1/2/3, MGT4/5, MGT7/8/9, and MGT5/6 branches were in yellow, green, light blue, and red, respectively. Numbers on branches indicated bootstrap values above 50.
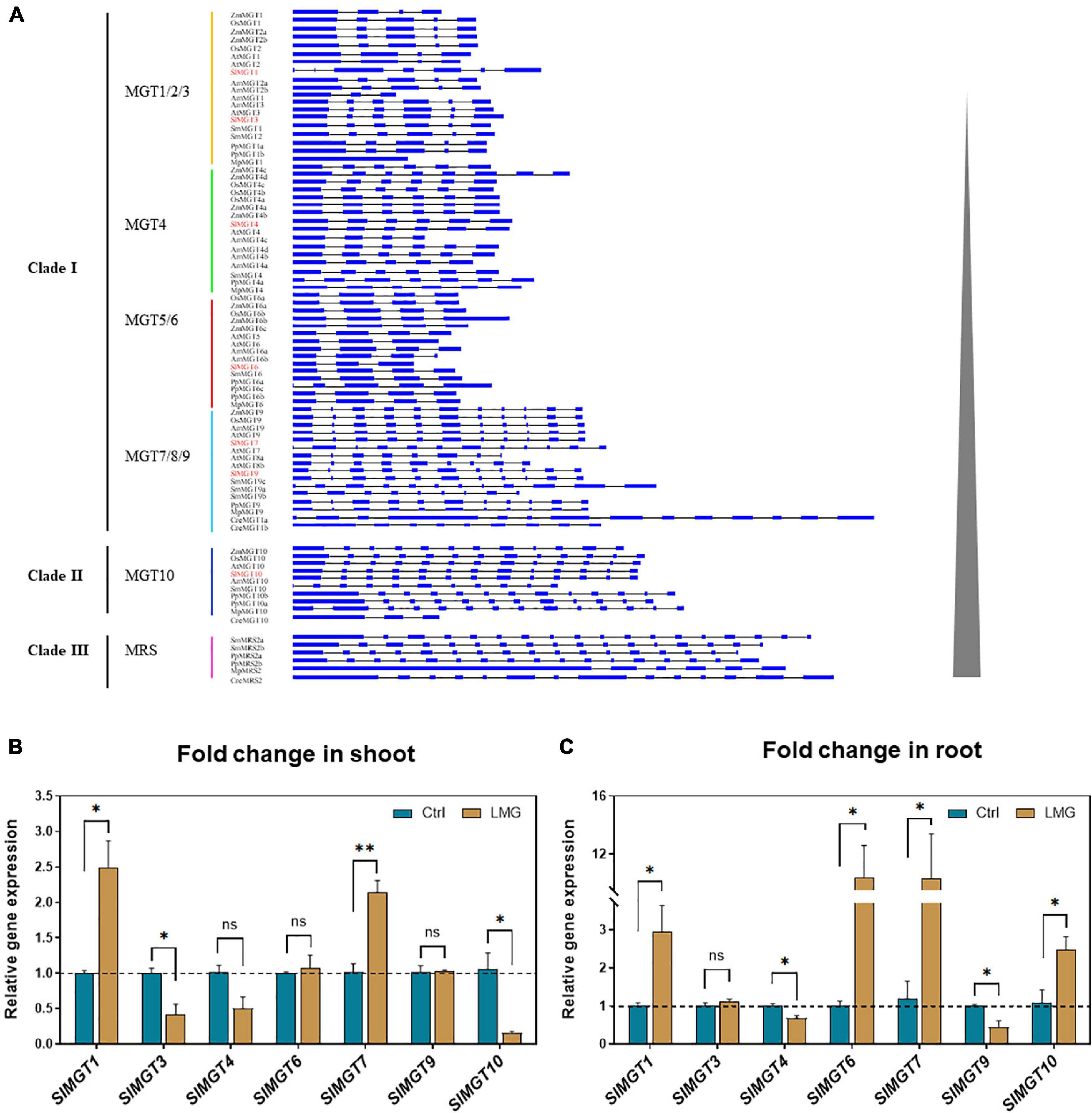
Figure 3. The gene structure of MGTs, and transcription patterns of MGTs in LMG seedlings. (A) Gene structure of MRS2/MGT-type Mg transporters, (B) relative gene expression of MGTs in the shoot, and (C) relative gene expression of MGTs in the root. The values of MGTs were reported as relative fold change from control, which was normalized to 1 as shown by dashed horizontal lines. The bar graph showed the mean value while whiskers represented the maximum/minimum values of four independent biological replicates. Asterisks indicated significant differences at *P < 0.05 and **P < 0.01, according to Tukey’s HSD test. Where, Ctrl, control; LMG, low Mg; MGT, Mg transporter; ns, non-significant.
We suspected that reduced Mg accumulation in LMG seedlings was primarily attributed to differential expression (and further functioning) of Mg transporters. RT-qPCR results showed that Mg limitation stimulated expression MGT1 and MGT7 by 2.5- and 2.1-fold, respectively, in the shoot. Meanwhile, MGT3 and MGT10 expression was significantly down-regulated to 0.42- and 0.16-fold, respectively, in the shoot of LMG seedlings (Figure 3B). In the root, MGT1, MGT6, MGT7, and MGT10 had 2. 9-, 10. 4-, 10. 3-, and 2.5-fold higher levels of transcription under Mg limitation, respectively. Meanwhile, the expression of MGT4 and MGT9 was down-regulated to 0.68- and 0.44-fold, respectively, in the LMG root, as shown in Figure 3C. Further, the alignment of SIMGT and ATMGT proteins showed that all differentially expressed MRS2/MGTs in tomato had two typical transmembrane domains and a GMN (Gly-Met-Asn) tripeptide motif (Figure 4).
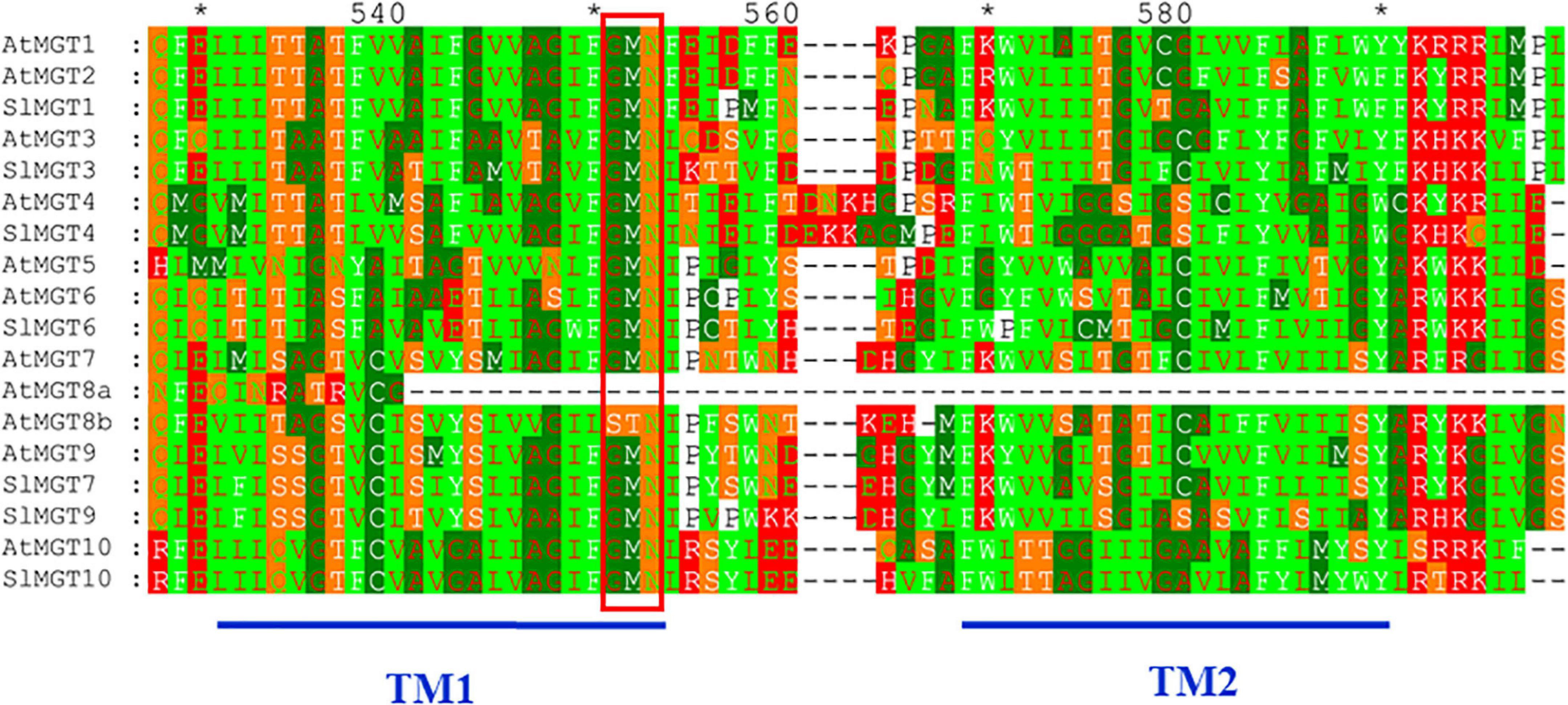
Figure 4. The alignment of SIMGT and ATMGT proteins. Conserved residues of each group of MRS2/MGTs were shaded in different colors. GMN (Gly-Met-Asn) tripeptide motif was indicated by red bar. Dashes represented gaps in the sequence and conserved residues that constituted the active sites were marked by asterisk. Numbers above the alignment gave the position of an amino acid within the alignment. Where, TM, transmembrane localization.
Plant Biomass and Root Architectural Indexes of Low Magnesium Seedlings
Mg limitation significantly reduced root and shoot dry biomass compared to control, with ∼50 and 36% decreases, respectively (Figures 5A–C). Interestingly, the percentage reduction in root biomass was higher compared to that in shoot biomass, which contributed to a ∼47% decrease of the root/shoot ratio of Mg-depleted plants, compared to control plants (Figure 5D).
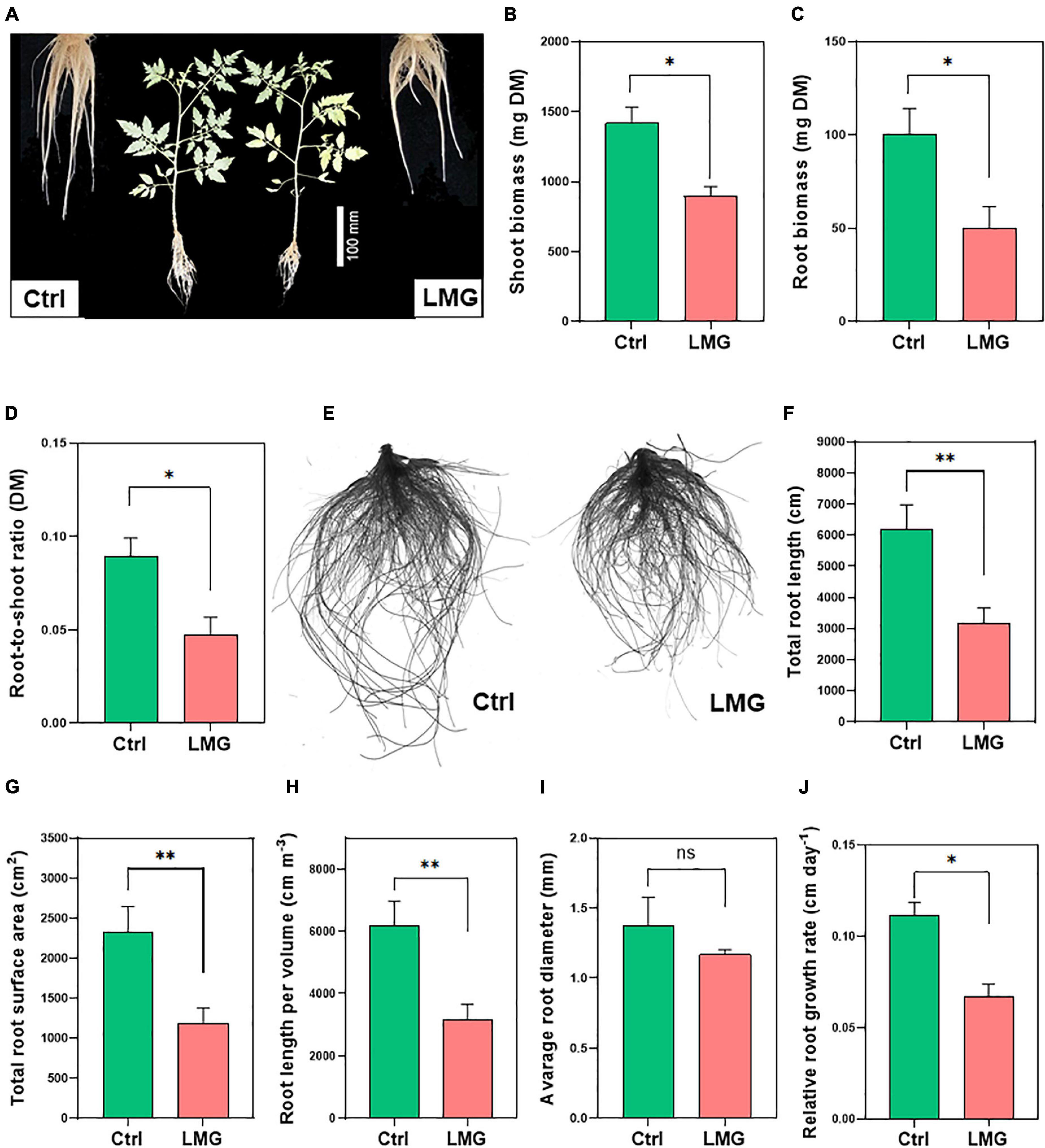
Figure 5. Mg limitation inhibited tomato growth and root architectural indexes. (A) Plant phenotypic responses after 3 weeks of treatments, (B) Root biomass (mg plant–1 DM), (C) Shoot biomass (mg plant–1 DM), (D) Root-to-shoot ratio (DM), (E) Scanned image of control and LMG treated roots (F) Total root length (cm), (G) Total root surface area (cm2), (H) Root length per volume (cm m–3), (I) Average root diameter (mm), and (J) Relative root growth rate (cm day–1). The bar graph showed the mean value while whiskers represented the maximum/minimum values of six independent biological replicates. Asterisks indicated a significant difference at *P < 0.05 and **P < 0.01, according to Tukey’s HSD test. Where, Ctrl, control; LMG, low Mg; ns, non-significant.
Entire roots were scanned to examine treatment effects on root system architecture (Figure 5E). Root morphological indexes revealed a significant (P < 0.01) reduction in total root length (∼49%), root surface area (∼50%), and length per volume (∼49%) in Mg-depleted plants, compared to control (Figures 5F–H). Whereas, average root diameter (mm) did not show significant differences as shown in Figure 5I. We also observed a sharp decline in the overall root system after the 14-day treatment (Supplementary Figure 1). To better understand the dynamic change of root growth, we computed relative root growth rate (RRGR) by harvesting tomato seedlings on day 14 and 21 after treatments. Results showed 0.07 cm day–1 root growth under Mg limitation in contrast to 0.11 cm day–1 growth of control roots (Figure 5J).
Magnesium Deficiency-Induced Hormonal Distribution and Expression Patterns of Auxin Biosynthesis
Below- and above-ground hormonal accumulation differed apparently under LMG, as shown in Figures 6A–D. The concentration of endogenous indole-3-acetic acid (IAA) was 40% higher (increased from 25 to 35 ng g–1) in the shoot of Mg-deficient plants. However, the IAA concentration surprisingly decreased by 23% (to 8.69 ng g–1) in the root of Mg-deficient plants, compared to control plants (Figure 6A), possibly due to diminished shoot-to-root IAA translocation under Mg limitation. Contradictory to the IAA accumulation pattern, other hormones displayed significantly higher accumulation in the root as compared to that in the shoot under Mg limitation: concentrations of ABA, GA3 and ZR reduced by 18, 27, and 20%, respectively, in LMG shoot. Meanwhile, approximately 6, 14, and 36% more ABA, GA3, and ZR, respectively, were detected in the LMG root, compared to control (Figures 6B–D).
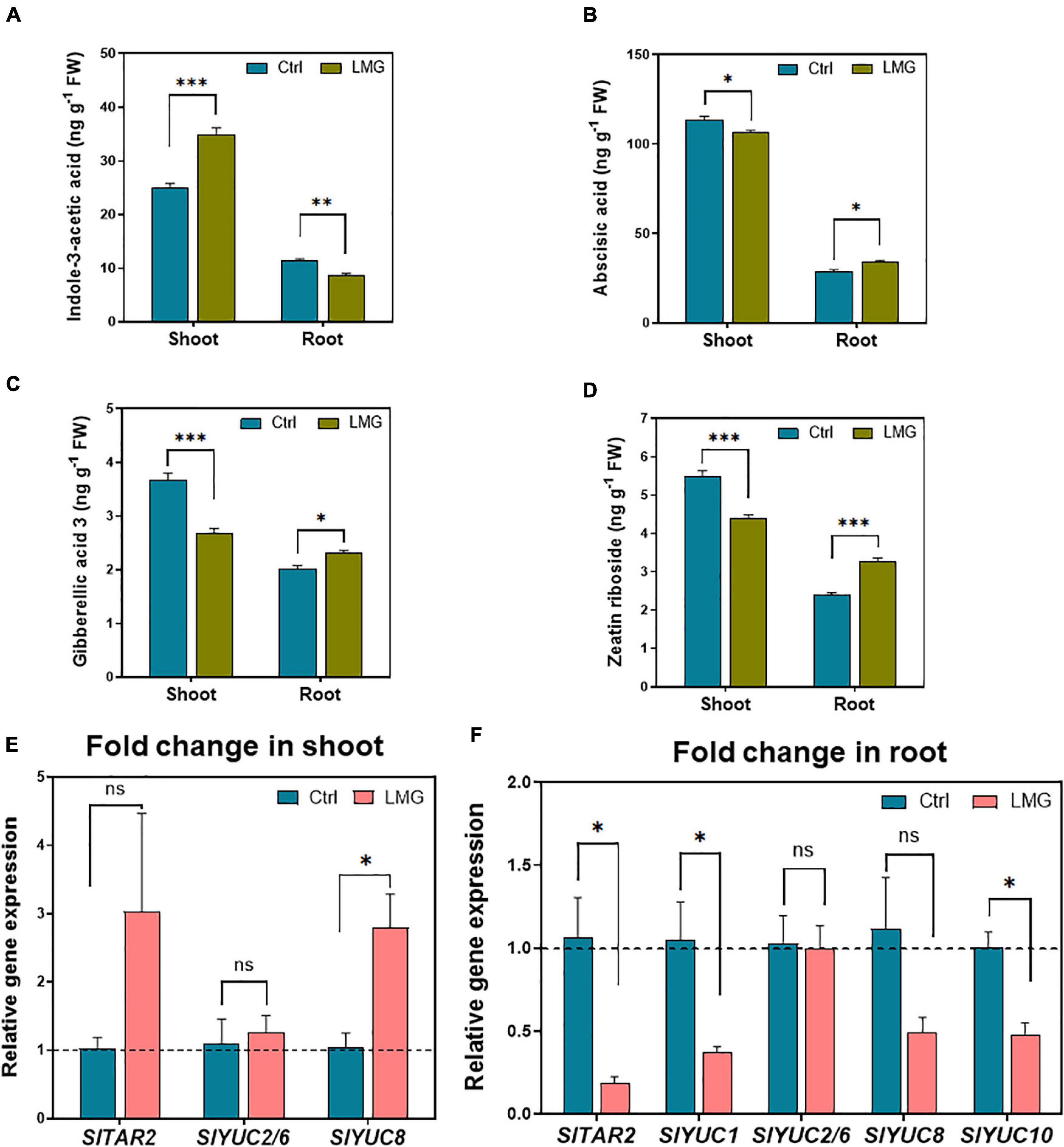
Figure 6. LMG disturbed the hormonal distribution and expression patterns of auxin biosynthesis in the root. (A) Indole-3-acetic acid concentration in shoot and root (ng g–1 FW), (B) Abscisic acid concentration in shoot and root (ng g–1 FW), (C) Gibberellic acid 3 concentration in shoot and root (ng g–1 FW), (D) Zeatin riboside concentration in shoot and root (ng g–1 FW), (E) Relative gene expression of TAR/YUCs in the shoot, (F) Relative gene expression of TAR/YUCs in the root. The bar graph showed the mean value while whiskers represented the maximum/minimum values of four independent biological replicates. Asterisks indicated significant differences at *P < 0.05, **P < 0.01, and ***P < 0.001, according to Tukey’s HSD test. Where, Ctrl, control; LMG, low Mg; ns, non-significant; YUC, YUCCA.
In LMG shoot, the YUC8 was upregulated by 2.8-fold and the expression of other auxin synthesis related genes, i.e., TAR2, YUC2/6, and YUC8 remained unchanged (Figure 6E). Consistent with lower IAA concentrations in LMG roots, TAR2, YUC1, YUC8, and YUC10 showed 0. 18-, 0. 37-, 0. 49-, and 0.48-fold down-regulation in the root under Mg limitation. However, YUC2/6 and YUC8 showed no significant change in expression (Figure 6F).
Transcriptional Alterations of Auxin Transport and Signaling Genes Under Magnesium Limitation
To further investigate the potential implications of auxin signaling in modulating root growth under Mg limitation, we quantified the expression of genes regulating auxin transport and signaling by RT-qPCR. With regard to auxin transport, PIN2 and PIN7 showed higher expression levels in the LMG shoot compared with control. No significant variation in expression of LAX1, PIN1, PIN3, and PIN4 was observed in the Mg-depleted shoot (Figure 7A). In the LMG root, transcription of LAX1, PIN1, PIN2, and PIN7 was significantly repressed to 0. 54-, 0. 45-, 0. 51-, and 0.64-fold, respectively, compared with control. However, PIN3 and PIN4 showed no significant change in expression (Figure 7B).
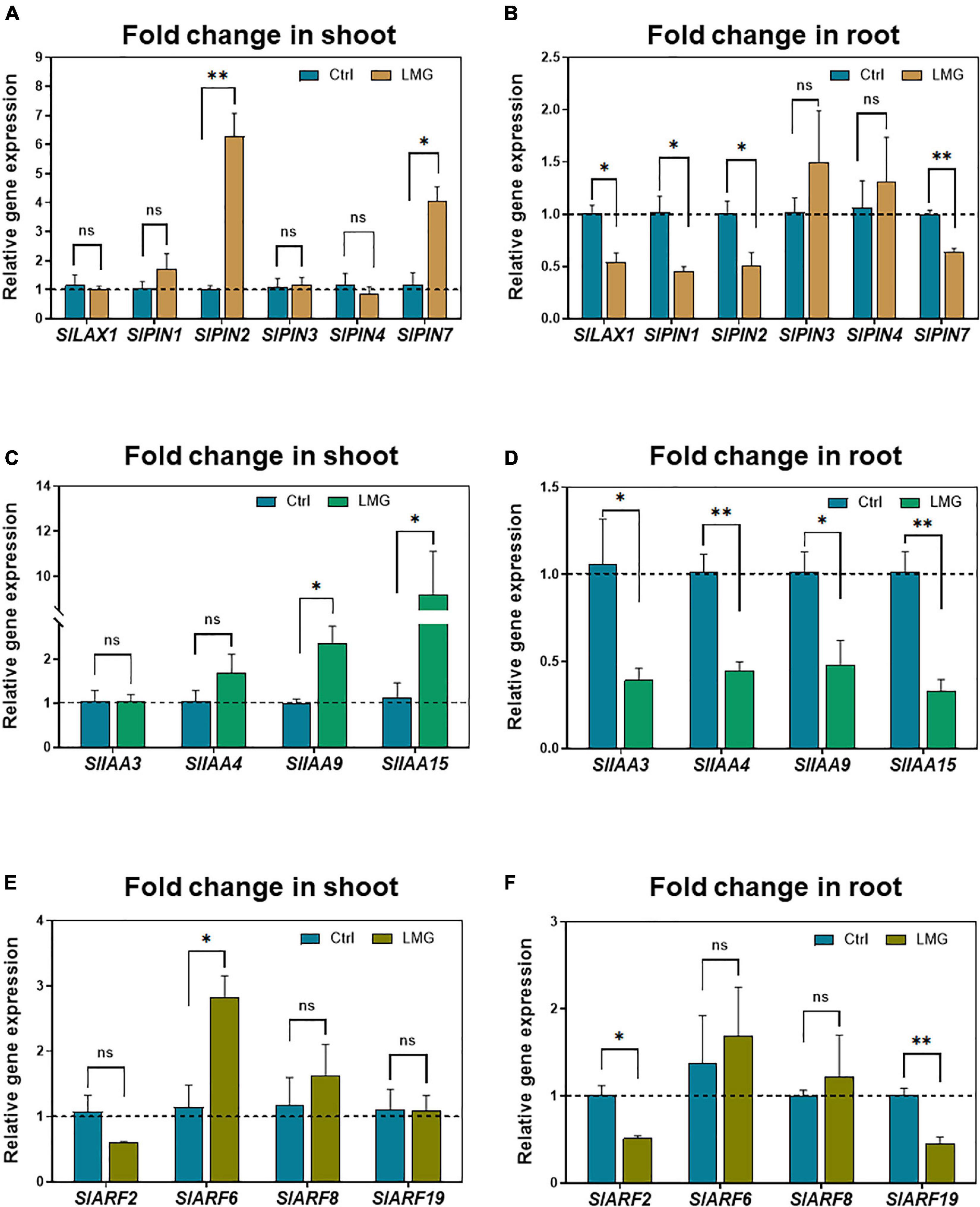
Figure 7. Transcriptional alterations of auxin transport and signaling genes under Mg limitation. (A) Relative gene expression of LAX/PINs in the shoot, (B) Relative gene expression of LAX/PINs in the root, (C) Relative gene expression of IAAs in the shoot, (D) Relative gene expression of IAAs in the root, (E) Relative gene expression of ARFs in the shoot, (F) Relative gene expression of ARFs in the root. The values were reported as relative fold change from control, which was normalized to 1 as shown by dashed horizontal lines. The bar graph showed the mean value while whiskers represented the maximum/minimum values of four independent biological replicates. Asterisks indicated a significant difference at *P < 0.05, **P < 0.01, according to Tukey’s HSD test. Where, Ctrl, control; LMG, low Mg; ns, non-significant; LAX, LIKE-AUX1; PIN, PIN-FORMED; Aux/IAA, AUXIN/INDOLE ACETIC ACID; ARFs, AUXIN RESPONSE FACTORS.
Beyond auxin transport is its signaling that orchestrates complicated physiological outcomes (Lv et al., 2020; Meier et al., 2020; Hu et al., 2021), therefore we analyzed expression of related IAA and ARF genes. Mg limitation led to significant up-regulation of IAA9 and IAA15 by 2.35- and 9.2-fold, respectively, in the LMG shoot (Figure 7C) in contrast to down-regulation of IAA3, IAA4, IAA9, and IAA15 by 0. 39-, 0. 45-, 0. 48-, and 0.33-fold, respectively, in the LMG root (Figure 7D). Lastly, ARF genes had differential expression in the shoot and root under Mg limitation, too. ARF6 had 2.8-fold higher expression in the LMG shoot, with no change in expression of ARF2, ARF8, and ARF19, compared with control (Figure 7E). Different from expression patterns in the shoot, no ARF expression was stimulated by Mg limitation in the root, while ARF2 and ARF19 expression was down-regulated by 0.51- and 0.45-fold, respectively, compared to control (Figure 7F).
Discussion
Mg plays fundamental roles in regulating crop production and produce quality, and Mg deficiency is emerging as an increasing agricultural and nutritional issue required to be tackled worldwide (Hauer-Jákli and Tränkner, 2019; Wang et al., 2020; Fiorentini et al., 2021). However, how plants adapt to Mg deficiency stress remains largely unclear or inconsistent. Here, we presented a previously uncharacterized 3-clade tree for Mg transporters in planta, revealed up-regulation of six representative MGTs in Mg-deficient tomato seedlings, and characterized a smaller root system harboring coherent down-regulation of auxin accumulation and expression of related genes in auxin signaling in an important horticultural model crop, providing new valuable insights for future studies.
Differential Expression of MGTs in Tomato Seedlings Favors Mg2+ Uptake and Translocation in Response to Low Magnesium
In our study, tomato seedlings suffered from Mg deficiency from the first day of transplanting into the nutrient solution which may avoid Mg2+ accumulation in the vacuole during initial growth stages (Hauer-Jákli and Tränkner, 2019). Mg limitation reduced Mg2+ uptake by tomato roots (Figures 1A,B). Low eternal Mg conditions also disturbed elemental homeostasis by increasing uptake of other competing cations, i.e., K+, Ca2+, and Na+. To analyze expression of representative MGTs, we generated a unified three-clade phylogenetic tree of the MRS2/MGT gene family (Figure 2), which provided the evolutionary foundation for future functional characterization of MGTs. In Arabidopsis, MGT6 regulates cellular homeostasis of Mg2+ under Mg limitation (Mao et al., 2014; Oda et al., 2016; Yan et al., 2018). Mg limitation results in higher MGT1 expression in Arabidopsis (Lenz et al., 2013) and rice (Zhang et al., 2019b). In tomato, all differentially expressed MGTs fell into clade I except MGT10 in clade II (Figures 2, 3A). In total, six MGTs showed differential expression between control and LMG seedlings, and four up-regulated (MGT1, MGT6, MGT7, and MGT10) transporters in root probably favored Mg2+ uptake and translocation (Figure 3C). Differential expression of MGT1, MGT3, MGT7, and MGT10 in the shoot possibly manipulate Mg2+ translocation within plant tissues under Mg limitation (Figure 3B). The relative functional importance of these MGTs in tomato adaptation to Mg limitation calls for further studies to unravel underlying molecular mechanisms.
Transcriptional Down-Tuning of Genes Regulating Auxin Biosynthesis, Transport, and Signaling Conditioned a Smaller Seedling Root Under Magnesium Limitation
Root growth is largely programmed by internal genetic, developmental cues and various environmental stimuli primarily via hierarchical hormone signaling cascades (Lakehal and Bellini, 2019; Alaguero-Cordovilla et al., 2021; Hu et al., 2021). Auxin plays a central role in root initiation, elongation, and architectural configuration (Olatunji et al., 2017; Meier et al., 2020; Sun et al., 2020). YUCs overexpression promotes IAA production in Arabidopsis (Mashiguchi et al., 2011; Novak et al., 2012). Here, TAR2, YUC1, YUC8, and YUC10 downregulated by 0. 18-, 0. 37-, 0. 49-, and 0.48-fold in Mg-depleted roots (Figure 6F), suggesting that Mg limitation impairs IAA synthesis. Consistently, IAA accumulation decreased by ∼23% in the root in contrast to the ∼40% increase in the shoot under Mg limitation (Figure 6A), clearly suggesting that Mg limitation reshaped auxin synthesis and distribution pattern between above- and below-ground. Such significant lower levels of auxin ultimately converted into developmental cues to slow down root growth.
Auxin transport and signaling components play essential roles in root system development (Porco et al., 2016; Du and Scheres, 2018; Hu et al., 2021). AUX/LAX, PIN2, PIN3, and PIN7 all participate in root development (Paponov et al., 2005; Marhavy et al., 2013). Our results showed significant transcriptional repression of LAX1, PIN1, PIN2, and PIN7 in the LMG root (Figure 7A), in agreement with lower levels of auxin. Aux/IAA is a key regulator of auxin-modulated signal transduction (Chandler, 2016; Israeli et al., 2020; Lv et al., 2020). In our study, expression of IAA3, IAA4, IAA9, and IAA15 was depressed in the LMG root (Figure 7D). Further, ARF2 and ARF19 expression was highly down-regulated in the root under Mg limitation (Figure 7F). The transcriptional activators ARF7 and ARF19 play an important role in root branching (Lee et al., 2019). Based on the actual expression levels, it is speculated that the relative strong expressed genes such as MGT7, TAR2, YUC1, PIN1, PIN2, PIN7, IAA4, IAA15, and ARF2 may altered the auxin-related process in Mg-deficient roots. Hence, on the basis of consistent decreases in auxin accumulation and attenuation of related gene expression along auxin signaling in the LMG root (Figures 6, 7), we propose that LMG led to weakened auxin functioning, i.e., synthesis, transport, accumulation, and signal transduction in the root system (Figure 8).
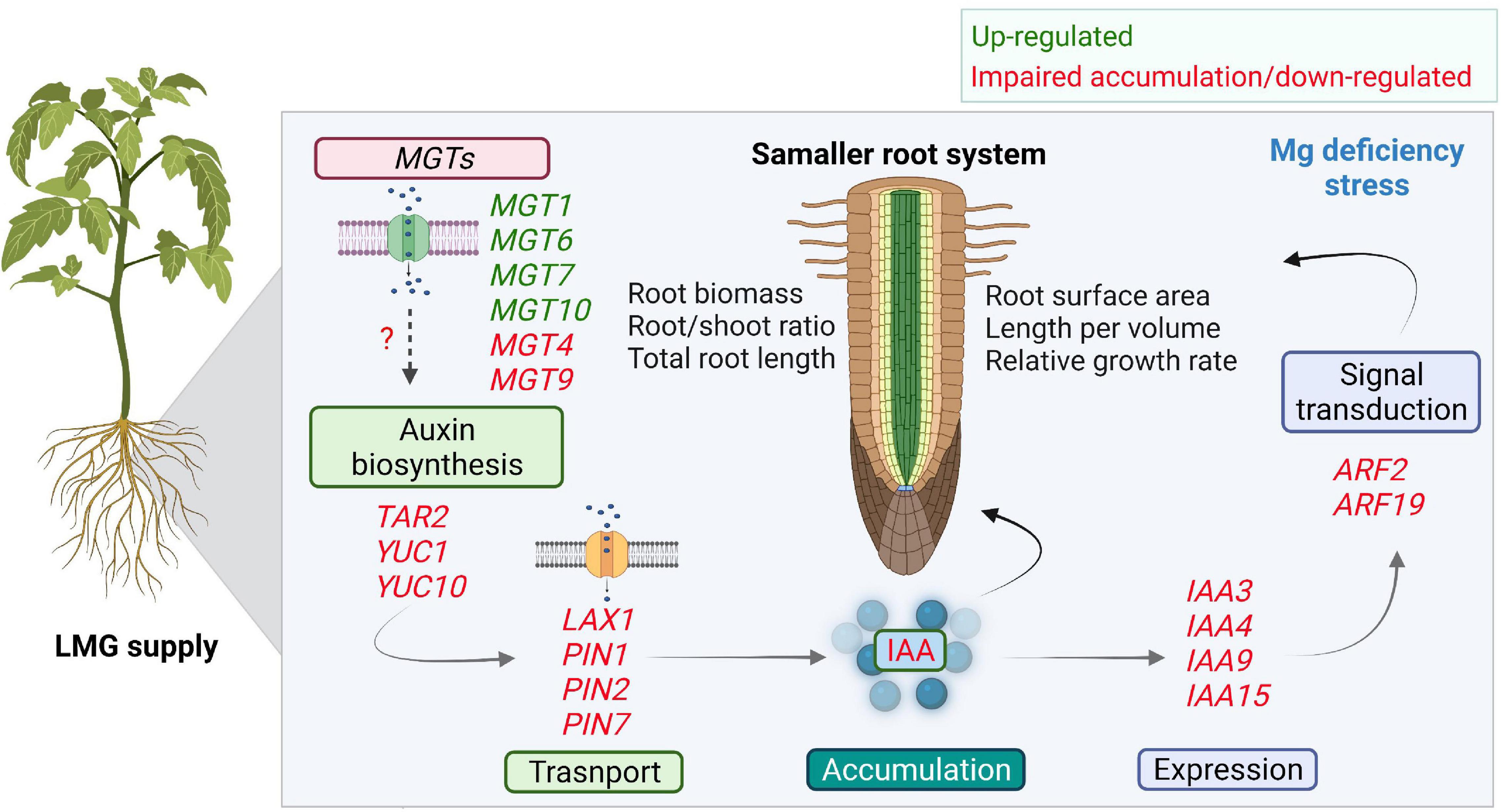
Figure 8. Scheme depicting the impaired auxin accumulation and signaling in Mg-deficient root. Differential expression of MGTs in the root favors Mg uptake and translocation under Mg limitation. Low Mg leads to smaller root related indexes. In the root, Mg limitation down-regulates gene expression along auxin synthesis, transport, and signal transduction. Where, MGTs, Mg transporters; YUC, YUCCA; LAX, LIKE-AUX1; PIN, PIN-FORMED; Aux/IAA, AUXIN/INDOLE ACETIC ACID; ARFs, AUXIN RESPONSE FACTORS.
The work presented in this study provides a new valuable insight that LMG supply disturbs transcription of auxin signaling genes, which well fit in the phenotype of smaller roots. However, in future detailed molecular investigation with mutant lines and protein functions is required to directly link such phenotypes to auxin signaling. In summary, given the crucial roles of auxin in modulating root growth, it is speculated that down-tuning of auxin accumulation and signaling in the root preconditions smaller root related indexes, while expression of MGTs are mostly up-regulated for Mg uptake and translocation.
Conclusion
Phylogenetic analysis suggested that Mg transporters (MRS2/MGTs) constitute a previously uncharacterized 3-clade tree in planta. In adaptation to internal Mg deficiency, tomato seedlings altered MGTs expression under Mg limitation. Notably, lower auxin (IAA) accumulation in Mg-deficient roots was consistent with systemic down-tuning of gene expression in auxin synthesis (TAR/YUCs), transport (LAXs, PINs), and signaling (IAAs, ARFs). Given the crucial roles of auxin in modulating root growth, it is speculated that weakened auxin functioning under Mg limitation may precondition a smaller root system. Further efforts are required to better understand the molecular functioning of MRS2/MGTs, auxin signaling and its involvement in modulating root growth under Mg limitation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MI and XL conceived, designed the study, wrote, and revised the manuscript. MI and YW performed the experiments. MI and YZ analyzed the data. All authors have reviewed and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32172663).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.802399/full#supplementary-material
Footnotes
- ^ https://phytozome.jgi.doe.gov
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://www.drive5.com/muscle/
- ^ https://www.softpedia.com/get/Science-CAD/GeneDoc.shtml
- ^ http://www.clustal.org/clustal2/
- ^ https://www.megasoftware.net/history.php
- ^ http://gsds.cbi.pku.edu.cn/
References
Ahmad, Z., Nadeem, F., Wang, R., Diao, X., Han, Y., Wang, X., et al. (2018). A larger root system is coupled with contrasting expression patterns of phosphate and nitrate transporters in foxtail millet [Setaria italica (L.) Beauv.] under phosphate limitation. Front. Plant Sci. 9:1367. doi: 10.3389/fpls.2018.01367
Alaguero-Cordovilla, A., Belen Sanchez-Garcia, A., Ibanez, S., Albacete, A., Cano, A., Acosta, M., et al. (2021). An auxin-mediated regulatory framework for wound-induced adventitious root formation in tomato shoot explants. Plant Cell Environ. 44, 1642–1662. doi: 10.1111/pce.14001
Bhosale, R., Giri, J., Pandey, B. K., Giehl, R. F. H., Hartmann, A., Traini, R., et al. (2018). A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat. Commun. 9:1818. doi: 10.1038/s41467-018-04281-x
Broadley, M. R., and White, P. J. (2010). Eats roots and leaves. Can edible horticultural crops address dietary calcium, magnesium and potassium deficiencies? Proc. Nutr. Soc. 69, 601–612. doi: 10.1017/S0029665110001588
Cámara-Zapata, J. M., Brotons-Martínez, J. M., Simón-Grao, S., Martinez-Nicolás, J. J., and García-Sánchez, F. (2019). Cost–benefit analysis of tomato in soilless culture systems with saline water under greenhouse conditions. J. Sci. Food Agric. 99, 5842–5851. doi: 10.1002/jsfa.9857
Chandler, J. W. (2016). Auxin response factors. Plant Cell Environ. 39, 1014–1028. doi: 10.1111/pce.12662
Chen, Z. C., Peng, W. T., Li, J., and Liao, H. (2018). Functional dissection and transport mechanism of magnesium in plants. Semin. Cell Dev. Biol. 74, 142–152. doi: 10.1016/j.semcdb.2017.08.005
Chen, Z. C., Yamaji, N., Horie, T., Che, J., Li, J., An, G., et al. (2017). A magnesium transporter OsMGT1 plays a critical role in salt tolerance in rice. Plant Physiol. 174, 1837–1849. doi: 10.1104/pp.17.00532
Cui, Y., Zhao, S., Wang, X., and Zhou, B. (2016). A novel Drosophila mitochondrial carrier protein acts as a Mg2+ exporter in fine-tuning mitochondrial Mg2+ homeostasis. Biochim. Biophys. Acta 1863, 30–39. doi: 10.1016/j.bbamcr.2015.10.004
Dai, X. Y., Su, Y. R., Wei, W. X., Wu, J. S., and Fan, Y. K. (2009). Effects of top excision on the potassium accumulation and expression of potassium channel genes in tobacco. J. Exp. Bot. 60, 279–289. doi: 10.1093/jxb/ern285
Du, Y., and Scheres, B. (2018). Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 69, 155–167. doi: 10.1093/jxb/erx223
Farhat, N., Elkhouni, A., Zorrig, W., Smaoui, A., Abdelly, C., and Rabhi, M. (2016). Effects of magnesium deficiency on photosynthesis and carbohydrate partitioning. Acta Physiol. Plant. 38:145. doi: 10.1007/s11738-016-2165-z
Fiorentini, D., Cappadone, C., Farruggia, G., and Prata, C. (2021). Magnesium: biochemistry, nutrition, detection, and social impact of diseases linked to its deficiency. Nutrients 13:1136. doi: 10.3390/nu13041136
Freschet, G., Pagès, L., Iversen, C., Comas, L., Rewald, B., Roumet, C., et al. (2020). A starting guide to root ecology: strengthening ecological concepts and standardizing root classification, sampling, processing and trait measurements. New Phytol. 232, 973–1122. doi: 10.1111/nph.17572
Gerendás, J., and Führs, H. (2013). The significance of magnesium for crop quality. Plant Soil 368, 101–128. doi: 10.1007/s11104-012-1555-2
Gruber, B. D., Giehl, R. F. H., Friedel, S., and von Wirén, N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179. doi: 10.1104/pp.113.218453
Guseman, J. M., Hellmuth, A., Lanctot, A., Feldman, T. P., Moss, B. L., Klavins, E., et al. (2015). Auxin-induced degradation dynamics set the pace for lateral root development. Development 142, 905–909. doi: 10.1242/dev.117234
Hauer-Jákli, M., and Tränkner, M. (2019). Critical leaf magnesium thresholds and the impact of magnesium on plant growth and photo-oxidative defense: a systematic review and meta-analysis from 70 years of research. Front. Plant Sci. 10:766. doi: 10.3389/fpls.2019.00766
Hermans, C., and Verbruggen, N. (2005). Physiological characterization of Mg deficiency in Arabidopsis thaliana. J. Exp. Bot. 56, 2153–2161. doi: 10.1093/jxb/eri215
Hermans, C., Vuylsteke, M., Coppens, F., Cristescu, S. M., Harren, F. J. M., Inzé, D., et al. (2010). Systems analysis of the responses to long-term magnesium deficiency and restoration in Arabidopsis thaliana. New Phytol. 187, 132–144. doi: 10.1111/j.1469-8137.2010.03257.x
Hu, Y., Omary, M., Hu, Y., Doron, O., Hoermayer, L., Chen, Q., et al. (2021). Cell kinetics of auxin transport and activity in Arabidopsis root growth and skewing. Nat. Commun. 12:1657. doi: 10.1038/s41467-021-21802-3
Ishfaq, M., Wakeel, A., Shahzad, M. N., Kiran, A., and Li, X. (2021). Severity of zinc and iron malnutrition linked to low intake through a staple crop: a case study in east-central Pakistan. Environ. Geochem. Health 43, 4219–4233. doi: 10.1007/s10653-021-00912-3
Israeli, A., Reed, J. W., and Ori, N. (2020). Genetic dissection of the auxin response network. Nat. Plants 6, 1082–1090. doi: 10.1038/s41477-020-0739-7
Koch, M., Busse, M., Naumann, M., Jákli, B., Smit, I., Cakmak, I., et al. (2019). Differential effects of varied potassium and magnesium nutrition on production and partitioning of photoassimilates in potato plants. Physiol. Plant. 166, 921–935. doi: 10.1111/ppl.12846
Koch, M., Winkelmann, M. K., Hasler, M., Pawelzik, E., and Naumann, M. (2020). Root growth in light of changing magnesium distribution and transport between source and sink tissues in potato (Solanum tuberosum L.). Sci. Rep. 10:15192. doi: 10.1038/s41598-020-72313-y
Lakehal, A., and Bellini, C. (2019). Control of adventitious root formation: insights into synergistic and antagonistic hormonal interactions. Physiol. Plant. 165, 90–100. doi: 10.1111/ppl.12823
Lavenus, J., Goh, T., Roberts, I., Guyomarc’h, S., Lucas, M., De Smet, I., et al. (2013). Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18, 455–463. doi: 10.1016/j.tplants.2013.04.006
Lee, H. W., Cho, C., Pandey, S. K., Park, Y., Kim, M. J., and Kim, J. (2019). LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 19:46. doi: 10.1186/s12870-019-1659-4
Lenz, H., Dombinov, V., Dreistein, J., Reinhard, M. R., Gebert, M., and Knoop, V. (2013). Magnesium deficiency phenotypes upon multiple knockout of Arabidopsis thaliana MRS2 clade B genes can be ameliorated by concomitantly reduced calcium supply. Plant Cell Physiol. 54, 1118–1131. doi: 10.1093/pcp/pct062
Li, D., Ma, W., Wei, J., Mao, Y., Peng, Z., Zhang, J., et al. (2020). Magnesium promotes root growth and increases aluminum tolerance via modulation of nitric oxide production in Arabidopsis. Plant Soil 457, 83–95. doi: 10.1007/s11104-019-04274-9
Li, H., Du, H., Huang, K., Chen, X., Liu, T., Gao, S., et al. (2016). Identification, and functional and expression analyses of the CorA/MRS2/MGT-type magnesium transporter family in maize. Plant Cell Physiol. 57, 1153–1168. doi: 10.1093/pcp/pcw064
Li, J., Yokosho, K., Liu, S., Cao, H. R., Yamaji, N., Zhu, X. G., et al. (2020). Diel magnesium fluctuations in chloroplasts contribute to photosynthesis in rice. Nat. Plants 6, 848–859. doi: 10.1038/s41477-020-0686-3
Li, L., Tutone, A. F., Drummond, R. S. M., Gardner, R. C., and Luan, S. (2001). A novel family of magnesium transport genes in Arabidopsis. Plant Cell 13, 2761–2775. doi: 10.1105/tpc.13.12.2761
Liu, M., Zhang, H., Fang, X., Zhang, Y., and Jin, C. (2018). Auxin acts downstream of ethylene and nitric oxide to regulate magnesium deficiency-induced root hair development in Arabidopsis thaliana. Plant Cell Physiol. 59, 1452–1465. doi: 10.1093/pcp/pcy078
Liu, S., Zhang, Y., Feng, Q., Qin, L., Pan, C., Lamin-Samu, A. T., et al. (2018). Tomato AUXIN RESPONSE FACTOR 5 regulates fruit set and development via the mediation of auxin and gibberellin signaling. Sci. Rep. 8:2971. doi: 10.1038/s41598-018-21315-y
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, B., Yu, Q., Liu, J., Wen, X., Yan, Z., Hu, K., et al. (2020). Non-canonical AUX/IAA protein IAA33 competes with canonical AUX/IAA repressor IAA5 to negatively regulate auxin signaling. EMBO J. 39, e101515. doi: 10.15252/embj.2019101515
Ma, C. L., Qi, Y. P., Liang, W. W., Yang, L. T., Lu, Y. B., Guo, P., et al. (2016). MicroRNA regulatory mechanisms on citrus sinensis leaves to magnesium-deficiency. Front. Plant Sci. 7:201. doi: 10.3389/fpls.2016.00201
Ma, T. L., Wu, W. H., and Wang, Y. (2012). Transcriptome analysis of rice root responses to potassium deficiency. BMC Plant Biol. 12:13. doi: 10.1186/1471-2229-12-161
Ma, W., Li, J., Qu, B., He, X., Zhao, X., Li, B., et al. (2014). Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 78, 70–79. doi: 10.1111/tpj.12448
Maghiaoui, A., Bouguyon, E., Cuesta, C., Section, F. P. W., Alcon, C., Krouk, G., et al. (2020). The Arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J. Exp. Bot. 71, 4480–4494. doi: 10.1093/jxb/eraa242
Mao, D., Chen, J., Tian, L., Liu, Z., Yang, L., Tang, R., et al. (2014). Arabidopsis transporter MGT6 mediates magnesium uptake and is required for growth under magnesium limitation. Plant Cell 26, 2234–2248. doi: 10.1105/tpc.114.124628
Marhavy, P., Vanstraelen, M., De Rybel, B., Ding, Z., Bennett, M. J., Beeckman, T., et al. (2013). Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO J. 32, 149–158. doi: 10.1038/emboj.2012.303
Marschner, H. (2012). Marschner’s Mineral Nutrition of Higher Plants, Vol. 89. Cambridge, MA: Academic press.
Mashiguchi, K., Tanaka, K., Sakai, T., Sugawara, S., Kawaide, H., Natsume, M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Nat. Acad. Sci. U.S.A. 108, 18512–18517. doi: 10.1073/pnas.1108434108
Meier, M., Liu, Y., Lay-Pruitt, K. S., Takahashi, H., and von Wiren, N. (2020). Auxin-mediated root branching is determined by the form of available nitrogen. Nat. Plants 6, 1136–1145. doi: 10.1038/s41477-020-00756-2
Mironova, V., Teale, W., Shahriari, M., Dawson, J., and Palme, K. (2017). The systems biology of auxin in development emoryos. Trends Plant Sci. 22, 225–235. doi: 10.1016/j.tplants.2016.11.010
Moret, B., Marhava, P., Fandino, A. C. A., Hardtke, C. S., and ten Tusscher, K. H. W. (2020). Local auxin competition explains fragmented differentiation patterns. Nat. Commun. 11:2965. doi: 10.1038/s41467-020-16803-7
Nadeem, F., Ahmad, Z., Ul Hassan, M., Ruifeng, W., Diao, X., and Li, X. (2020). Adaptation of foxtail millet (Setaria italica L.) to abiotic stresses: a special perspective of responses to nitrogen and phosphate limitations. Front. Plant Sci. 11:187. doi: 10.3389/fpls.2020.00187
Neuhaus, C., Geilfus, C. M., and Muehling, K. H. (2014). Increasing root and leaf growth and yield in Mg-deficient faba beans (Vicia faba) by MgSO4 foliar fertilization. J. Plant Nutr. Soil Sci. 177, 741–747. doi: 10.1002/jpln.201300127
Niu, Y., Chai, R., Liu, L., Jin, G., Liu, M., Tang, C., et al. (2014). Magnesium availability regulates the development of root hairs in Arabidopsis thaliana (L.) Heynh. Plant Cell Environ. 37, 2795–2813. doi: 10.1111/pce.12362
Niu, Y., Jin, G., Li, X., Tang, C., Zhang, Y., Liang, Y., et al. (2015). Phosphorus and magnesium interactively modulate the elongation and directional growth of primary roots in Arabidopsis thaliana (L.) Heynh. J. Exp. Bot. 66, 3841–3854. doi: 10.1093/jxb/erv181
Novak, O., Henykova, E., Sairanen, I., Kowalczyk, M., Pospisil, T., and Ljung, K. (2012). Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 72, 523–536. doi: 10.1111/j.1365-313X.2012.05085.x
Oda, K., Kamiya, T., Shikanai, Y., Shigenobu, S., Yamaguchi, K., and Fujiwara, T. (2016). The Arabidopsis mg transporter, MRS2-4, is essential for mg homeostasis under both low and high mg conditions. Plant Cell Physiol. 57, 754–763. doi: 10.1093/pcp/pcv196
Ogura, T., Kobayashi, N. I., Suzuki, H., Iwata, R., Nakanishi, T. M., and Tanoi, K. (2018). Magnesium uptake characteristics in Arabidopsis revealed by Mg-28 tracer studies. Planta 248, 745–750. doi: 10.1007/s00425-018-2936-4
Olatunji, D., Geelen, D., and Verstraeten, I. (2017). Control of endogenous auxin levels in plant root development. Int. J. Mol. Sci. 18:2587. doi: 10.3390/ijms18122587
Paponov, I. A., Teale, W. D., Trebar, M., Blilou, K., and Palme, K. (2005). The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends Plant Sci. 10, 170–177. doi: 10.1016/j.tplants.2005.02.009
Porco, S., Larrieu, A., Du, Y., Gaudinier, A., Goh, T., Swarup, K., et al. (2016). Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development 143, 3340–3349. doi: 10.1242/dev.136283
Roosjen, M., Paque, S., and Weijers, D. (2018). Auxin response factors: output control in auxin biology. J. Exp. Bot. 69, 179–188. doi: 10.1093/jxb/erx237
Saito, T., Kobayashi, N. I., Tanoi, K., Iwata, N., Suzuki, H., Iwata, R., et al. (2013). Expression and functional analysis of the CorA-MRS2-ALR-type magnesium transporter family in rice. Plant Cell Physiol. 54, 1673–1683. doi: 10.1093/pcp/pct112
Samal, D., Kovar, J. L., Steingrobe, B., Sadana, U. S., Bhadoria, P. S., and Claassen, N. (2010). Potassium uptake efficiency and dynamics in the rhizosphere of maize (Zea mays L.), wheat (Triticum aestivum L.), and sugar beet (Beta vulgaris L.) evaluated with a mechanistic model. Plant Soil 332, 105–121. doi: 10.1007/s11104-009-0277-6
Sauer, M., and Kleine-Vehn, J. (2019). PIN-FORMED and PIN-LIKES auxin transport facilitators. Development 146:dev168088. doi: 10.1242/dev.168088
Song, Y., Zhou, L., Yang, S., Wang, C., Zhang, T., and Wang, J. (2017). Dose-dependent sensitivity of Arabidopsis thaliana seedling root to copper is regulated by auxin homeostasis. Environ. Exp. Bot. 139, 23–30. doi: 10.1016/j.envexpbot.2017.04.003
Stoeckle, D., Thellmann, M., and Vermeer, J. E. M. (2018). Breakout–lateral root emergence in Arabidopsis thaliana. Curr. Opin. Plant Biol. 41, 67–72. doi: 10.1016/j.pbi.2017.09.005
Sun, X., Chen, H., Wang, P., Chen, F., Yuan, L., and Mi, G. (2020). Low nitrogen induces root elongation via auxin-induced acid growth and auxin-regulated target of rapamycin (TOR) pathway in maize. J. Plant Physiol. 254:153281. doi: 10.1016/j.jplph.2020.153281
Verbruggen, N., and Hermans, C. (2013). Physiological and molecular responses to magnesium nutritional imbalance in plants. Plant Soil 368, 87–99. doi: 10.1007/s11104-013-1589-0
Vosolsobe, S., Skokan, R., and Petrasek, J. (2020). The evolutionary origins of auxin transport: what we know and what we need to know. J. Exp. Bot. 71, 3287–3295. doi: 10.1093/jxb/eraa169
Wang, J., Moeen-ud-din, M., and Yang, S. (2021). Dose-dependent responses of Arabidopsis thaliana to zinc are mediated by auxin homeostasis and transport. Environ. Exp. Bot. 189:104554. doi: 10.1016/j.envexpbot.2021.104554
Wang, R., and Estelle, M. (2014). Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 21, 51–58. doi: 10.1016/j.pbi.2014.06.006
Wang, Z., Ul Hassan, M., Nadeem, F., Wu, L., Zhang, F., and Li, X. (2020). Magnesium fertilization improves crop yield in most production systems: a meta-analysis. Front. Plant Sci. 10:1727. doi: 10.3389/fpls.2019.01727
Weijers, D., and Wagner, D. (2016). Transcriptional responses to the auxin hormone, in: Merchant, S.S. (Ed.). Annu. Rev. Plant Biol. 67, 539–574. doi: 10.1146/annurev-arplant-043015-112122
Weiler, E. W., Jourdan, P. S., and Conrad, W. (1981). Use of immunoassay in plant-science. Levels of indole-3-acetic-acid in intact and decapitated coleoptiles as determined by a specific and highly sensitive solid-phase enzyme-immunoassay. Planta 153, 561–571. doi: 10.1007/bf00385542
Yan, Y. W., Mao, D. D., Yang, L., Qi, J. L., Zhang, X. X., Tang, Q. L., et al. (2018). Magnesium transporter MGT6 plays an essential role in maintaining magnesium homeostasis and regulating high magnesium tolerance in Arabidopsis. Front. Plant Sci. 9:274. doi: 10.3389/fpls.2018.00274
Yang, L.-T., Zhou, Y.-F., Wang, Y.-Y., Wu, Y.-M., Ye, X., Guo, J.-X., et al. (2019). Magnesium deficiency induced global transcriptome change in Citrus sinensis leaves revealed by RNA-Seq. Int. J. Mol. Sci. 20, 3129. doi: 10.3390/ijms20133129
Zhang, L., Wen, A., Wu, X., Pan, X., Wu, N., Chen, X., et al. (2019a). Molecular identification of the magnesium transport gene family in Brassica napus. Plant Physiol. Biochem. 136, 204–214. doi: 10.1016/j.plaphy.2019.01.017
Zhang, L., Peng, Y., Li, J., Tian, X., and Chen, Z. (2019b). OsMGT1 confers resistance to magnesium deficiency by enhancing the import of mg in rice. Int. J. Mol. Sci. 20:207. doi: 10.3390/ijms20010207
Zhang, P., Sun, L., Qin, J., Wan, J., Wang, R., and Li, S. (2018). cGMP is involved in Zn tolerance through the modulation of auxin redistribution in root tips. Environ. Exp. Bot. 147, 22–30. doi: 10.1016/j.envexpbot.2017.10.025
Zhang, Y., Hartinger, C., Wang, X., and Friml, J. (2020). Directional auxin fluxes in plants by intramolecular domain-domain coevolution of PIN auxin transporters. New Phytol. 227, 1406–1416. doi: 10.1111/nph.16629
Keywords: magnesium limitation, MRS2/MGT gene family, auxin, auxin signaling, PIN family, root system
Citation: Ishfaq M, Zhong Y, Wang Y and Li X (2021) Magnesium Limitation Leads to Transcriptional Down-Tuning of Auxin Synthesis, Transport, and Signaling in the Tomato Root. Front. Plant Sci. 12:802399. doi: 10.3389/fpls.2021.802399
Received: 26 October 2021; Accepted: 06 December 2021;
Published: 23 December 2021.
Edited by:
Antonio Lupini, Mediterranea University of Reggio Calabria, ItalyReviewed by:
Sheliang Wang, Huazhong Agricultural University, ChinaBipin Kumar Pandey, University of Nottingham, United Kingdom
Copyright © 2021 Ishfaq, Zhong, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuexian Li, c3RldmVAY2F1LmVkdS5jbg==
 Muhammad Ishfaq
Muhammad Ishfaq Yanting Zhong
Yanting Zhong Yongqi Wang
Yongqi Wang Xuexian Li
Xuexian Li