- 1Key Laboratory of Cultivation and Protection for Non-Wood Forest Trees, Ministry of Education, Central South University of Forestry and Technology, Changsha, China
- 2Academy of Camellia Oil Tree, Central South University of Forestry and Technology, Changsha, China
- 3The Belt and Road International Union Research Center for Tropical Arid Non-wood Forest in Hunan Province, Changsha, China
Introduction: Self-incompatibility (SI) is an important strategy for plants to maintain abundant variation to enhance their adaptability to the environment. Camellia oleifera is one of the most important woody oil plants and is widely cultivated in China. Late acting self-incompatibility (LSI) in C. oleifera results in a relatively poor fruit yield in the natural state, and understanding of the LSI mechanism remains limited.
Methods: To better understand the molecular expression and gene coexpression network in the LSI reaction in C. oleifera, we conducted self- and cross-pollination experiments at two different flower bud developmental stages (3–4 d before flowering and 1 d before flowering), and cytological observation, fruit setting rate (FSR) investigation and RNA-Seq analysis were performed to investigate the mechanism of the male −female interaction and identify hub genes responsible for the LSI in C. oleifera.
Results: Based on the 21 ovary transcriptomes, a total of 7669 DEGs were identified after filtering out low-expression genes. Weighted gene coexpression network analysis (WGCNA) divided the DEGs into 15 modules. Genes in the blue module (1163 genes) were positively correlated with FSR, and genes in the pink module (339 genes) were negatively correlated with FSR. KEGG analysis indicated that flavonoid biosynthesis, plant MAPK signaling pathways, ubiquitin-mediated proteolysis, and plant-pathogen interaction were the crucial pathways for the LSI reaction. Fifty four transcription factors (TFs) were obtained in the two key modules, and WRKY and MYB were potentially involved in the LSI reaction in C. oleifera. Network establishment indicated that genes encoding G-type lectin S-receptor-like serine (lecRLK), isoflavone 3’-hydroxylase-like (CYP81Q32), cytochrome P450 87A3-like (CYP87A3), and probable calcium-binding protein (CML41) were the hub genes that positively responded to the LSI reaction. The other DEGs inside the two modules, including protein RALF-like 10 (RALF), F-box and pectin acetylesterase (MTERF5), might also play vital roles in the LSI reaction in C. oleifera.
Discussion: Overall, our study provides a meaningful resource for gene network studies of the LSI reaction process and subsequent analyses of pollen−pistil interactions and TF roles in the LSI reaction, and it also provides new insights for exploring the mechanisms of the LSI response.
1 Introduction
The Camellia oil tree (Camellia oleifera Abel.), one of the four major woody oil tree species in the world, is widely cultivated as an oil crop in many countries, such as China, India, Brazil, Japan, the Philippines and South Korea (Chen et al., 2017; Lin et al., 2022b). Camellia oil is a high-grade edible oil that scavenges free radicals and prevents cardiovascular disease and has been used as an edible oil for more than 2300 years in China (Quan et al., 2022). The unsaturated fatty acid content in Camellia oil reaches 90%, and Camellia oil also contains sterols, squalene, polyphenols, sterols, vitamin E and other compounds that are good for human health (Lee and Yen, 2006; Hosokawa et al., 2013; Zhang et al., 2021). It is widely used in products such as ink, lubricants, and cosmetics (Lin et al., 2018). Although C. oleifera has been cultivated on approximately 3 million hectares, 70% of its planting area is low-yield, and the annual production cannot meet the demand for this healthy oil (Hajjar and Hodgkin, 2007). The main reason for the low yield of C. oleifera is its self-incompatibility (SI) property and the improper configuration of pollination trees. Thus, it is meaningful to study the SI mechanism of C. oleifera to improve its seed yield.
SI is a complex reproductive barrier mechanism between organisms that is critical to speciation (Broz and Bedinger, 2021), and it has been widely adopted by flowering plants to promote cross-pollination and prevent self-pollination (Wu et al., 2020b). The SI mechanism has been extensively studied and can be divided into three types: sporophytic self-incompatibility (SSI), gametophytic SI (GSI) and late-acting self-incompatibility (LSI) (Zhou et al., 2020). In SSI, such as in Asteraceae and Convolvulaceae, the SI reaction was determined by two polymorphic S loci that were genetically linked: pollen-coat-specific cysteine-rich protein (SCR) and stigma-specific serine/threonine receptor kinase (SRK), which could inhibit the germination and growth of self-pollen (Wang et al., 2014; Nasrallah, 2019). In GSI, such as in Solanaceae and Rosaceae, S-specificity was determined by haploid pollen (Duan et al., 2020) and controlled by a single S-locus composed of two linked polymorphic genes: pollen-specific F-box protein (SFB/SLF) and style-specific T2 ribonuclease (S-RNase) (Gordillo-Romero et al., 2020). Another GSI mechanism observed in Papaver rhoeas showed that incompatible pollen could trigger the interaction between the male S-loci PrpS and the female S-loci PrsS, which leads to Ca2+ signaling cascades and initiates programmed cell death (PCD) (Wheeler et al., 2009; Eaves et al., 2014; Bedinger et al., 2017). LSI has been found in many plants, such as C. sinensis, Nymphaea spp. and Theobroma cacao, in which the pollen tube can grow successfully in the styles, but the inhibition occurs either prezygotically or postzygotically (Hao et al., 2012). Many studies have been performed to identify the key factors involved in the LSI response, but the mechanism remains unclear (Chen et al., 2012; Sun et al., 2019). The genetic basis of LSI was hypothesized to be gametophytic under genetic control (Zhou and Zheng, 2015; Zhang et al., 2016). However, no effort has been made to identify the genes of SI loci.
The SI reaction is a relatively complex signal transduction process. In addition to the S factor, non-S factors are also required during the recognition between pollen and pistils (McClure et al., 2011). In Petunia hybrida, the S-RNase binding protein (SBP1) from pollen could bind to S-RNase, and it was proposed that SBP1 can form a complex with SLF and allow nonself S-RNase degradation. SBP1 can also interact with other non-S factors, such as arabinogalactan proteins (AGPs) and 120-kDa glycoprotein (120K) (O’Brien et al., 2004; Ben-Naim et al, 2006). The participation of thioredoxin h proteins (THL) was involved in pollen rejection, where the downregulation of THL1 could lead to the breakdown of SI (Haffani et al., 2004). In Nicotiana, suppressing HT-B expression in the pistil could prevent S-specific pollen rejection (Covey et al., 2010). Moreover, there were many other identified non-S factors, such as calcium-binding protein (CML) (Miao et al., 2013), actin-binding proteins (ABPs) (Poulter et al., 2010), RALF (Bedinger et al., 2010) and actin-depolymerizing factor (ADF) (Dong et al., 2021).
In C. oleifera, recent studies showed that pollen tubes in self-pollinated pistils could grow normally through the style but failed to complete double fertilization (He et al., 2020; Zhou et al., 2020). Several candidate genes and pathways have been found that might participate in the LSI reaction by comparing the transcriptomes of self- and cross-pollination (He et al., 2020). However, the molecular mechanism of the LSI in C. oleifera remains unclear. In this study, we investigated the fruit setting rates under self- and cross-pollination in two different developmental stages of flower buds, observed the differences in pollen tube growth between self- and cross-pollination, and performed transcriptome sequencing. Furthermore, weighted gene coexpression network analysis (WGCNA) was used to identify the hub genes and transcription factors (TFs) related to the LSI response of C. oleifera. This study provides new insights into the mechanisms of the LSI response and is valuable for improving the yield and haploid breeding of C. oleifera.
2 Materials and methods
2.1 Plant materials
The C. oleifera cultivars ‘Hua Jin’ (HJ) and ‘Hua Xin’ (HX) were used in this study. They were selected from among 84 C. oleifera clones by a comparative regional assessment and were identified by Professors Tan Xiaofeng and Yuan Deyi at Central South University of Forestry and Technology (Zhang et al., 2021). The cultivar was planted in Huju Forest Farm in the Chaling district, Zhuzhou city, Hunan Province, China (26°C47’24’ N, 113°C32’24’ E). During the peak flowering period of HJ in October, flower buds at different developmental stages were selected for pollination, including 3-4 d before flowering (stage 3), 1 d before flowering (stage 1) and 1 d after flowering (stage 0).
2.2 Pollination treatment and sample collection
We designed six treatments for pollination, including self-pollination at stage 3 (SPT3), self-pollination at stage 1 (SPT1), cross-pollination at stage 3 (CPT3), self-pollination at stage 1 (CPT1), natural pollination (NPT0), no pollination at stage 1 (CKT1), and no pollination at stage 3 (CKT3). Among them, self-pollination was based on HJ as the parent, cross-pollination was based on HJ as the female parent, and HX as the male parent. Healthy and strong plants at the same growth stage were selected as female parents for pollination, and 500 flowers in different ten trees were pollinated in each treatment. Before artificial pollination, mature pollen of HJ and HX was collected from anthers in advance during the bud stage, placed in sulfate paper bags and kept at 25°C for 8 h. The experiment was conducted at midmorning on sunny days. The flower buds at the correct developmental stage were selected, pollinated before removing the corolla and anthers, and then bagged in sulfate paper bags. At 48 h after pollination, seven groups of samples for RNA-seq were collected from the same ten trees, and each group contained three biological replicates. Each replicate contained 30 ovaries. The collected samples were immediately placed into liquid nitrogen for preservation. In addition, thirty pistils for pollen tube observations were collected at different intervals (0, 12, 24, 36, 48, 60, 72, 84 h) after pollination from the same ten trees and then stored in Carnoy’s fixative.
2.3 Cytological observation, stigma acceptability test and fruit set survey
The fixed pistils were macerated with sodium hypochlorite for 2 h to soften and were then washed with deionized water and macerated in 8 M NaOH for 2 h. Then, the pistils were stained with 0.1% aniline blue for 6 h. After staining, the styles were split along the vertical axis into 3-5 sections with a surgical blade. Finally, we tracked pollen tube growth using fluorescence microscopy (Nikon Instech Co., Ltd., Kawasaki, Japan).
Pollen viability was determined by benzidine staining. Fresh style sections were immersed in a reaction solution containing benzidine-hydrogen peroxide (1% benzidine: 3% hydrogen peroxide: water = 4:11:22, v/v) for observation. If the styles were receptive, the reaction solution and style gradually turned brown or black as the active pollen. The fruit setting rate was investigated 60 d after pollination.
2.4 Transcriptome sequencing and alternative splicing analysis
The total RNA of samples was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol, and rRNA was subsequently removed using a Ribo-Zero™ Magnetic Kit (Epicenter, Madison, WI, USA). Then, the cDNA fragments were purified with a QIAquick PCR extraction kit (Qiagen, Venlo, The Netherlands), end-repaired, polyadenylated, and ligated to Illumina sequencing adapters. The ligation products were subjected to size selection via agarose gel electrophoresis, amplified via PCR, and sequenced using the Illumina NovaSeq 6000 platform.
Reads obtained from the sequencing machines included raw reads, which affected the subsequent assembly and analysis. To obtain high-quality clean reads, the reads were further filtered by fastp (Chen et al., 2018), and rRNA sequences were removed by alignment. The genome data of C. oleifera ‘Huashuo’ have been obtained (unpublished data). An index of the reference genome was built, and the paired-end clean reads were mapped to the reference genome using HISAT 2.2.4 (Kim et al., 2015) with “-rna-strandness RF” and other parameters set as a default. The mapped reads of each sample were assembled by using StringTie v1.3.1 (Pertea et al., 2015) in a reference-based approach. Based on the comparison results of all the reads (Total_Mapped reads) that can be located on the genome, we calculate the distribution of reads in the reference genome. We divided the regions mapped to the genome into exon, intron and intergenic regions. The unannotated transcripts were blasted against databases including, including the NR, Swiss-Prot, GO and KEGG databases, the whole pineline of the read assembly, alignment and annonation were present as a flowchart (Supplementary Figure S1A). RNA differential expression analysis was performed by DESeq2 (Love et al., 2014) software between two different groups (and by edgeR (Robinson et al., 2010) between two samples), and the genes with the parameters of false discovery rate (FDR) ≤ 0.05 and absolute fold change ≥ 2 were considered differentially expressed genes.
The software rMATS (Shen et al., 2014) (version 4.0.1) (http://rnaseq-mats.sourceforge.net/index.html ) was used to identify alternative splicing events and analyze differential alternative splicing events between samples. We identified AS events with a false discovery rate (FDR) <0.05 in a comparison as significant AS events.
2.5 Weighted gene coexpression network analysis
To identify the hub genes related to the LSI reaction in C. oleifera, weight gene coexpression network analysis (WGCNA) was performed on the OmicShare platform (https://www.omicsmart.com/WGCNA/home.html#/taskPandect) based on the differentially expressed genes (DEGs) and fruit setting rates. The DEGs with FPKM values < 1 in all samples were filtered out before WGCNA analysis (Lin et al., 2022a), so as to improve the accuracy of network construction. A total of 7669 DEGs were obtained to construct a Gene hierarchical cluster diagram based on the correlation of gene expression, coexpression modules were constructed with a soft threshold of 8.0, mini-num module size of 50, and Similarity of module eigenvalue of 0.9 for merging similar transcripts. The key modules were selected based on the correlation coefficients between traits and modules, and the gene coexpression network was constructed based on the top 10% of DEGs ordered by the edge weight coefficient within the module. Pearson correlation coefficients between the hub genes and TFs of each module, include blue module and pink module, were computed by OmicShare tools (https://www.omicshare.com/tools). Regulatory networks were visualized by Cytoscape 3.7.2 (Shannon et al., 2003). The whole process of the WGCNA was present as a flowchart (Supplementary Figure S1B).
2.6 Quantitative real-time reverse transcription PCR analysis
To verify the accuracy of the RNA-seq data, a total of 12 DEGs related to SI were selected and evaluated by qRT−PCR. The total RNA of the 21 samples in seven comparison groups (NPT0, CKT3, CKT1, SPT1, SPT3, CPT1, CPT3) was extracted by the Goldenstar™ RT6 cDNA Synthesis Kit Ver.2 (Beijing TsingKe Biotech Co., Ltd, China), and the remaining elimination of potential gDNA and reverse transcription were performed with HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing, China). Specific primers for the 12 DEGs were designed by Primer Premier 5.0 (Supplementary Table S1). The qRT−PCR was conducted in conjunction with ChamQ Universal SYBR qPCR Master Mix #Q711 (Vazyme, Nanjing, China). The experiment was performed on the CFX96 Real Time PCR System (Bio-Rad, Hercules, CA, USA), and CoGAPDH (KC337052.1) was chosen as the reference gene for Camellia oil tree. All analyses were repeated with three biological replicates, and the relative expression level of 12 DEGs were calculated with the 2-ΔΔCt method. The results were analyzed using Microsoft Office Excel 2013 (Microsoft, Redmond, WA, USA) and SPSS (ver. 20.0; SPSS, Inc., Chicago, IL, United States) software.
3 Results
3.1 The stigma of immature flower buds (3 DBFs) exhibiting high acceptability in C.oleifera
The external morphological characteristics of flower buds of C. oleifera were observed and recorded starting at 6 d before flowering (DBF) to determine the exact flower bud stage for bud pollination (Figures 1A, B). The C. oleifera flower buds began to swell and entered the middle bud stage three days before flowering. Self-pollination at the early bud stage could reduce the reaction of self-incompatibility, promote the fruit setting rate and help to create inbred offspring in many plants with SI properties (Zhang et al., 2019). Since then, immature flower buds (3 DBFs) and mature flower buds (1 DBF) were both selected for self- and cross- pollination in this study.
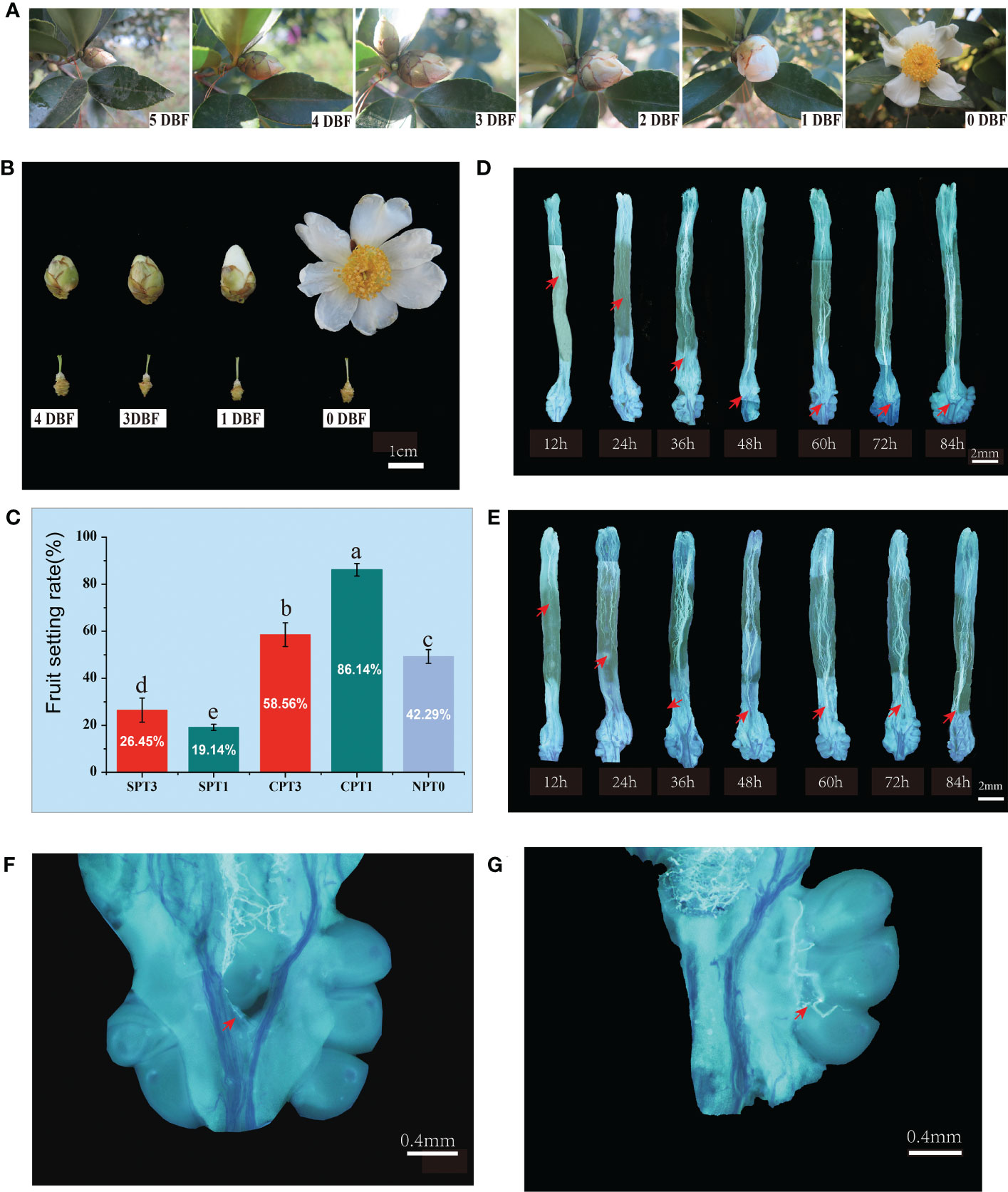
Figure 1 Investigation of fruit setting rates and observation of pollen tube growth in pistils in Camellia oleifera HJ. (A) Observation of flower bud development in nature; 5 DBF: 5 days before flowering, 4 DBF: 4 days before flowering, 3 DBF: 3 days before flowering, 2 DBF: 2 days before flowering, 1 DBF: 1 day before flowering. (B) Flower buds at different developmental stages. (C) Fruit setting rates of SPT1, SPT3, CPT1, CPT3 and NPT0. (D) Pollen tube growth from 12 h to 84 h in the pistil after cross-pollination. (E) Pollen tube growth from 12 h to 84 h in the pistil after self-pollination. (F, G) Successful pollen tube growth to the ovary 60 h after self-pollination.
The acceptability of stigmas greatly affects the efficiency of hybrid pollination. To test the stigma receptivity of the four main flower development stages of C. oleifera, the most reliable method, called the benzidine staining method, was used in this study. The results showed that all stigmas from the four flower development stages were receptive, as the stigmas were bubbly and brown, and the style was the most receptive on the day of flowering. The color of the stigma became dark brown, and there were more bubbles (Supplementary Figure S2A).
3.2 Transcriptome profiles of 21 RNA libraries from camellia oil tree ovaries pollinated with different treatments
Self- and cross-pollination were performed on pistils at 3 DBF and 1 DBF. The fruiting rates were investigated 60 d after pollination. The results showed that there was a significant difference in the fruit setting rate between self-pollination and cross-pollination, regardless of pollination at earlier or later developmental stages. The fruiting rates of CPT1 and CPT3 reached 86.14% and 58.56%, respectively, which were significantly higher than those of SPT1 (19.14%) and SPT3 (26.45%). Moreover, the early natural fruit setting rates of C. oleifera were relatively moderate, with a fruit setting rate of 49.29% (Figure 1C). The results indicated that the pollen from different varieties was more easily accepted by pistils than by self-pollen. Furthermore, the fruit setting rate of SPT3 was higher than that of SPT1, and the fruit setting rate of CPT1 was significantly higher than that of CPT3. This suggested that the transcription level and translation level of SI-related genes in 3 DBF buds were lower than those in 1 DBF bud, and mature pistils were more beneficial to the successful germination, growth and effective fertilization of cross-pollinated pollen tubes.
To evaluate the pollen germination rates and pollen tube growth between self- and cross-pollination and to determine the actual time when the self-incompatibility reaction occurred after self-pollination, fluorescence microscopic observation of pollen tube growth was performed (Figures 1D, E). The results showed that there were no differences in pollen tube growth in pistils at different developmental stages. The pollen grains germinated normally at 12 h after self- and cross-pollination. At 48 h after pollination, all of the pollen tubes grew smoothly to the upper part of the ovary. Thereafter, the self-pollinated pollen tubes stopped growing with the features of tissue distortion and folding, which are characteristic of programmed cell death (PCD). The cross-pollinated pollen tubes continued to grow downward until the double fertilization reaction was completed at approximately 72 h after cross-pollination (Figures 1F, G). The cytological observation showed that the SI of Camellia oil tree belonged to the LSI. Moreover, these results supported the SI reaction occurred at 48h of self-pollination, which was the best sampling duration for transcriptome sequencing.
3.3 Transcriptome sequencing revealed differentially expressed genes related to LSI in C. oleifera
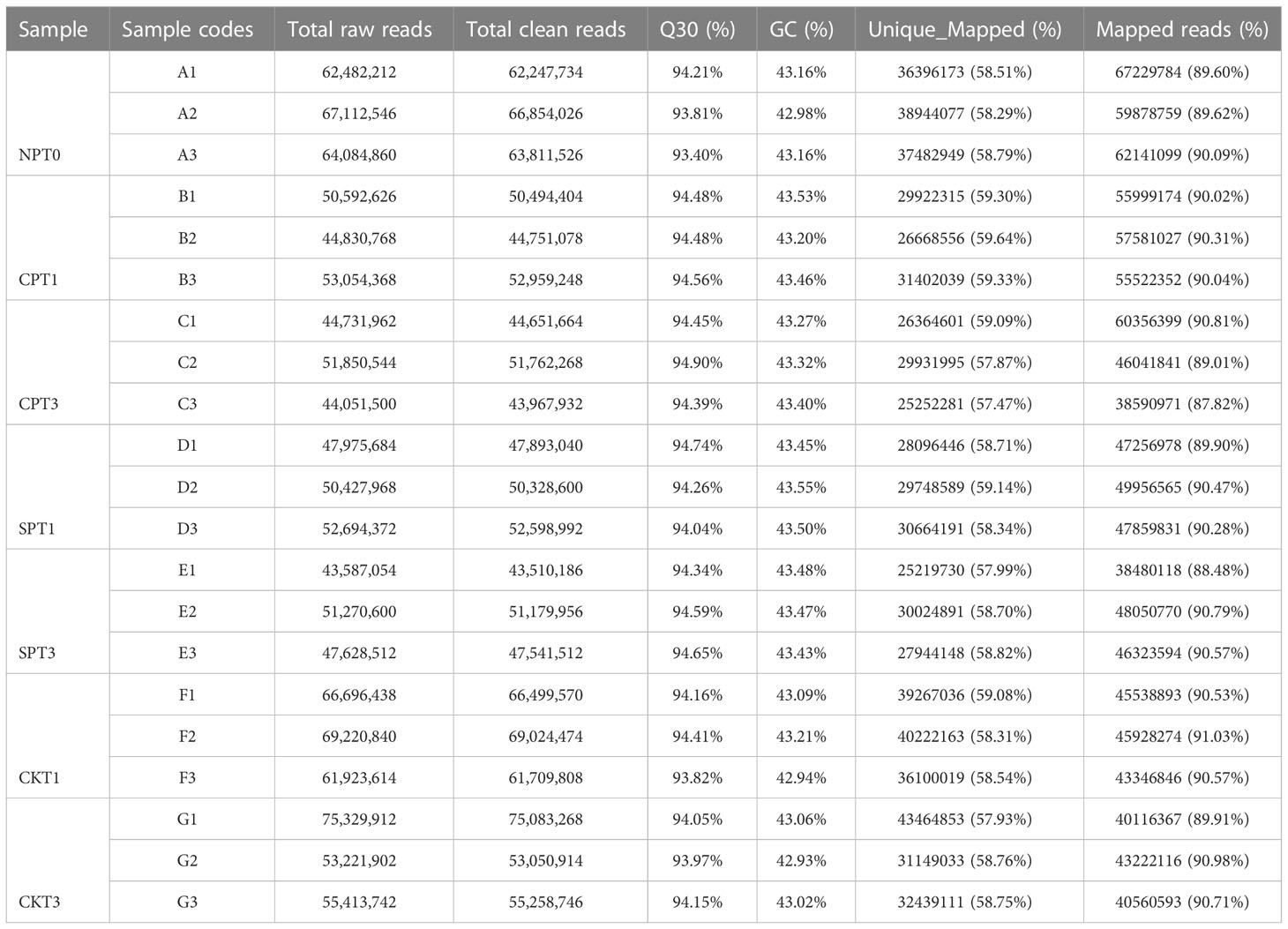
To further explore the molecular mechanism of LSI of C. oleifera, comparative transcriptomic sequencing analyses of the ovaries after different pollination treatments (SPT3, SPT1, CPT3, CPT1, NPT0, CKT1, CKT3) were performed. A total of 1.15 GB of high-quality data were obtained from the 21 cDNA libraries, with an average of 54.97 MB of data per sample. The Q30 percentages were greater than 93.19%, while the GC percentage of each sample was approximately 43% (Table 1). Approximately 87.82–91.03% of reads were mapped to the genome of C. oleifera. A total of 125,475 genes were identified including 1,5091 novel genes. The DEGs related to the LSI of Camellia oil tree were identified by the FPKM values from each sample library, and the expression levels of genes in the ovaries after compatible and self-incompatible pollination were assessed. The high reproducibility of the replicates was shown in a PCA plot (Figure 2A). Among these samples, there was a significant difference in FSR under each pollination treatment, and the FSR under cross-pollination was significantly higher than that under self-pollination. Next, 21 samples were divided into six groups for analysis (CKT1 vs. SPT1, CKT3 vs. SPT3, SPT3 vs. SPT1, CPT1 vs. CPT3, CPT3 vs. SPT3, CPT1 vs. SPT1), and the number of DEGs for the six groups is shown in Figure 2B. Among these comparisons, the CKT3 vs. SPT3 group had the most DEGs, reaching 5213 (3300 upregulated, 1913 downregulated). Next, there were 5082 DEGs (3730 upregulated, 1352 downregulated) in the CKT1 vs. SPT1 group. However, only 371, 225, 343, and 615 DEGs were identified in the SPT3 vs. SPT1, CPT1 vs. CPT3, CPT3 vs. SPT3, and CPT1 vs. SPT1 comparisons, respectively (Figures 2C–E). The comparison analysis showed that 121 DEGs overlapped among CPT1 vs. SPT1 and CPT3 vs. SPT3 (Figure 2F). The results suggested that these DEGs in six comparison groups should be involved in LSI reaction of C. oleifera.
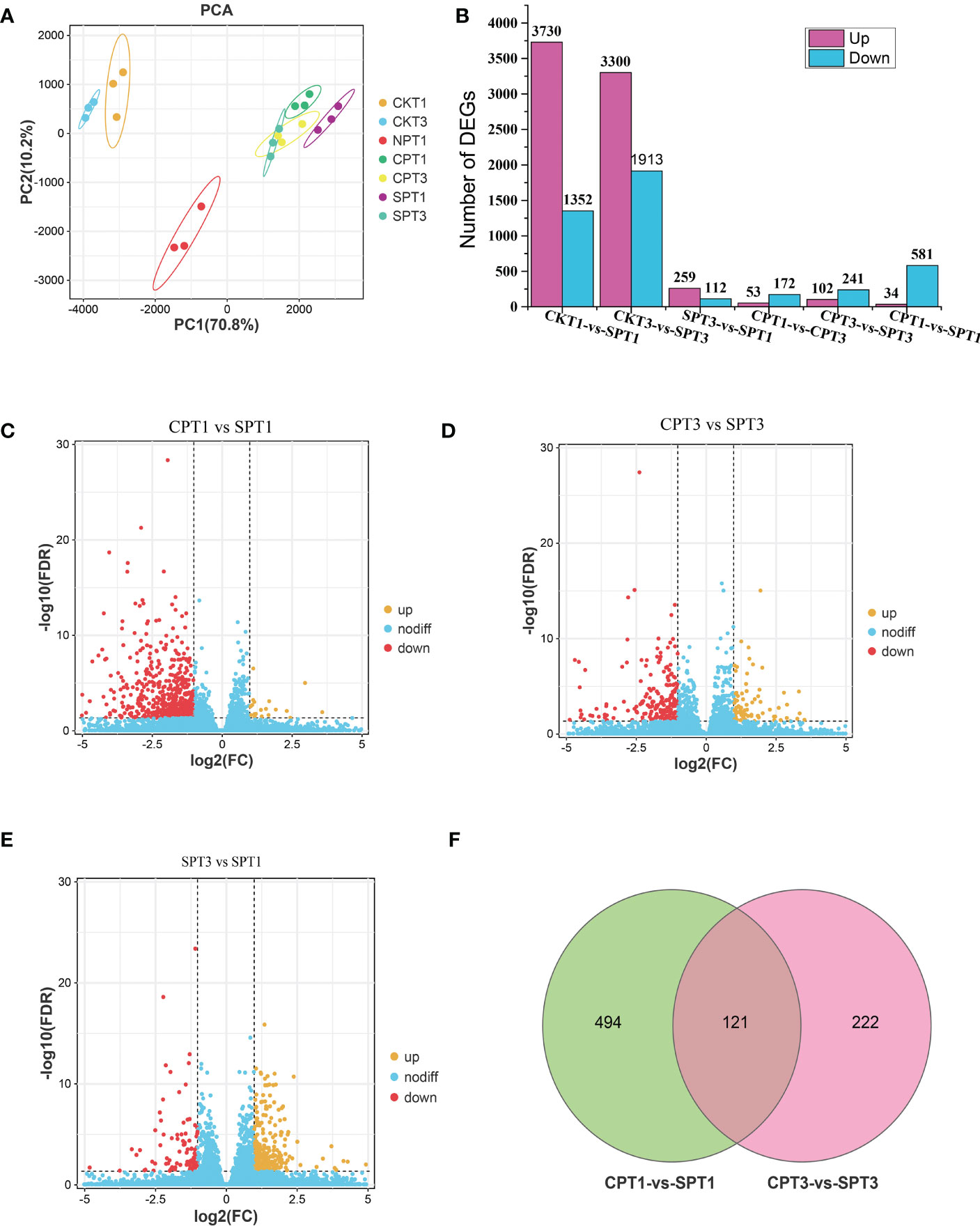
Figure 2 Graphical representation of DEGs related to late-acting self-incompatibility in different treatments of Camellia oleifera ‘Huajin’. (A) PCA score plot. (B) Number of up- and downregulated DEGs among the six gene sets. (C) Volcano plot of all DEGs in CPT1 vs. SPT1. (D) Volcano plot of all DEGs in CPT3 vs. SPT3. (E) Volcano plot of all DEGs in SPT3 vs. SPT1. (F) The Venn diagram represents the number of overlapping DEGs between self- and cross-pollination.
3.4 GO and KEGG enrichment analysis of DEGs
Gene Ontology (GO) enrichment analyses can help to determine which biological functions the DEGs are significantly associated with. For GO functional analysis, all the DEGs in the six comparison groups were divided into 53 functional groups in the molecular function (MF, 12 subcategories), cellular component (CC, 17 subcategories), and biological process (BP, 24 subcategories) categories. In the CC category, the terms that were significantly enriched were “cell”, “cell part”, “organelle” and “membrane”. In the MF category, the DEGs were enriched in “catalytic activity”, “binding” and “transporter activity”. In the BP category, “metabolic process”, “cellular process”, “single-organism process” and “response to stimulus” were more abundant than other terms (Figure 3A and Supplementary Table S2). For the DEGs in the six comparison groups, most GO terms were enriched in the BP and MF categories. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was conducted to gain insight into the actual biological function of DEGs. All the DEGs in the six comparison groups were significantly mapped to 37 KEGG pathways in five categories: metabolism, environmental information, genetic information processing, organismal systems, and cellular processing. The results showed that most DEGs identified were enriched in “Plant hormone signal transduction”, “Flavonoid biosynthesis”, “Brassinosteroid biosynthesis”, “ABC transporters”, “Ubiquitin mediated proteolysis” and “Stilbenoid, diarylheptanoid and gingerol biosynthesis”. Of note, the DEGs were significantly enriched in “plant hormone signal transduction” in all group comparisons except for CPT1 vs. CPT3 (Figure 3B; Supplementary Figure S2B and Table S3), especially in ethylene, brassinosteroid and jasmonic acid signal transduction pathways, indicating that DEGs enriched in plant hormone signal transduction was involved in the LSI reaction of Camellia oil tree. In addition, DEGs were significantly enriched in brassinosteroid biosynthesis in three groups (CPT1 vs. SPT1, CPT3 vs. SPT3 and CKT1 vs. SPT1), and DEGs were also significantly enriched in flavonoid biosynthesis in the other three comparison groups (CKT1 vs. SPT1, CKT3 vs. SPT3 and CPT1 vs. CPT3). Therefore, the results indicated that molecular and cellular events, including plant hormone signal transduction, brassinosteroid biosynthesis, flavonoid biosynthesis, etc., occurred in the LSI reaction in Camellia oil tree.
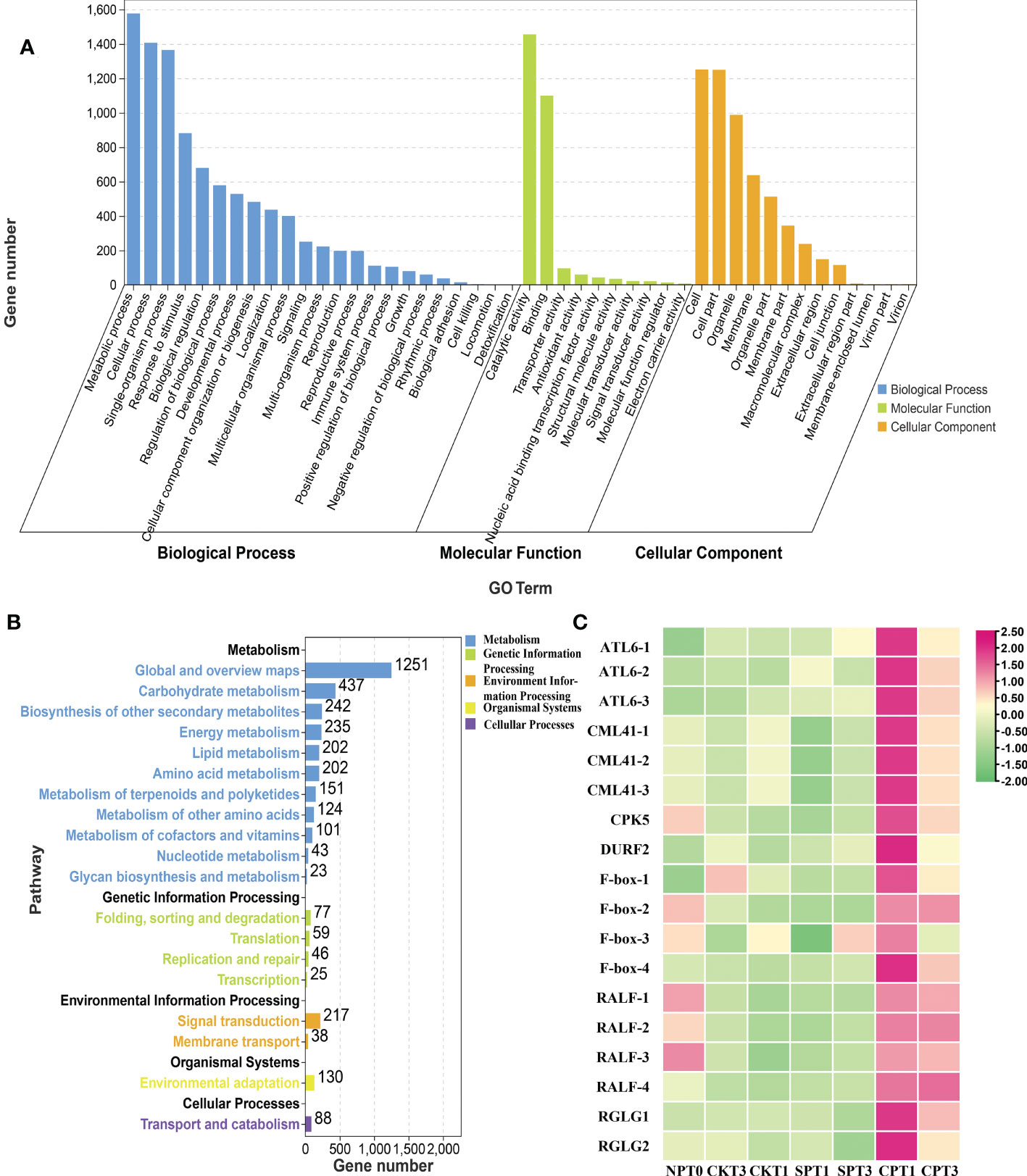
Figure 3 Analysis of DEGs in Camellia oleifera HJ. (A) GO enrichment analysis of the DEGs in C. oleifera HJ; (B) KEGG enrichment analysis of the DEGs in (C) oleifera HJ; (C) Expression profiles of crucial DEGs related to LSI in (C) oleifera HJ.
3.5 Co-expression network analysis identified key modules correlated with LSI reaction in C. oleifera
To explore the specific genes and the genetic regulatory network in the LSI reaction in C. oleifera ovaries, WGCNA was performed based on 7669 DEGs, which with FPKM values < 1 in all samples were filtered out, among the six comparison groups and the fruit setting rate phenotypes (Supplementary Table S4). A cluster dendrogram was generated to cluster the genes with similar expression patterns into the same module (Figure 4A). A total of 15 coexpressed modules (M1–M15) were obtained after dynamic tree cutting and merging, and each module was marked with different colors, including module 1 (turquoise, 1,479 genes), module 2 (blue, 1,163 genes), module 3 (black, 1,102 genes), module 4 (salmon, 885 genes), module 5 (brown, 733 genes), module 6 (red, 480 genes), module 7 (magenta, 444 genes), module 8 (pink, 339 genes), module 9 (purple, 284 genes), module 10 (greenyellow, 207 genes), module 11 (tan, 165 genes), module 12 (cyan, 147 genes), module 13 (lightcyan, 117 genes), and module 14 (gray60, 81 genes) (Figure 4C). Forty-three genes did not belong to any of the modules and were assigned to the gray module. Moreover, the correlation between the fruit setting rate and the modules was analyzed, and the correlation coefficients between them ranged from -0.71 to 0.74. Among the 15 modules, three modules (M2, M13, and M8) had high correlation coefficients and showed significant associations (p < 0.05) with the fruit setting rates (Figure 4B). We also constructed a module-sample correlation heatmap to intuitively visualize the expression pattern of each module in each sample (Figure 4D).
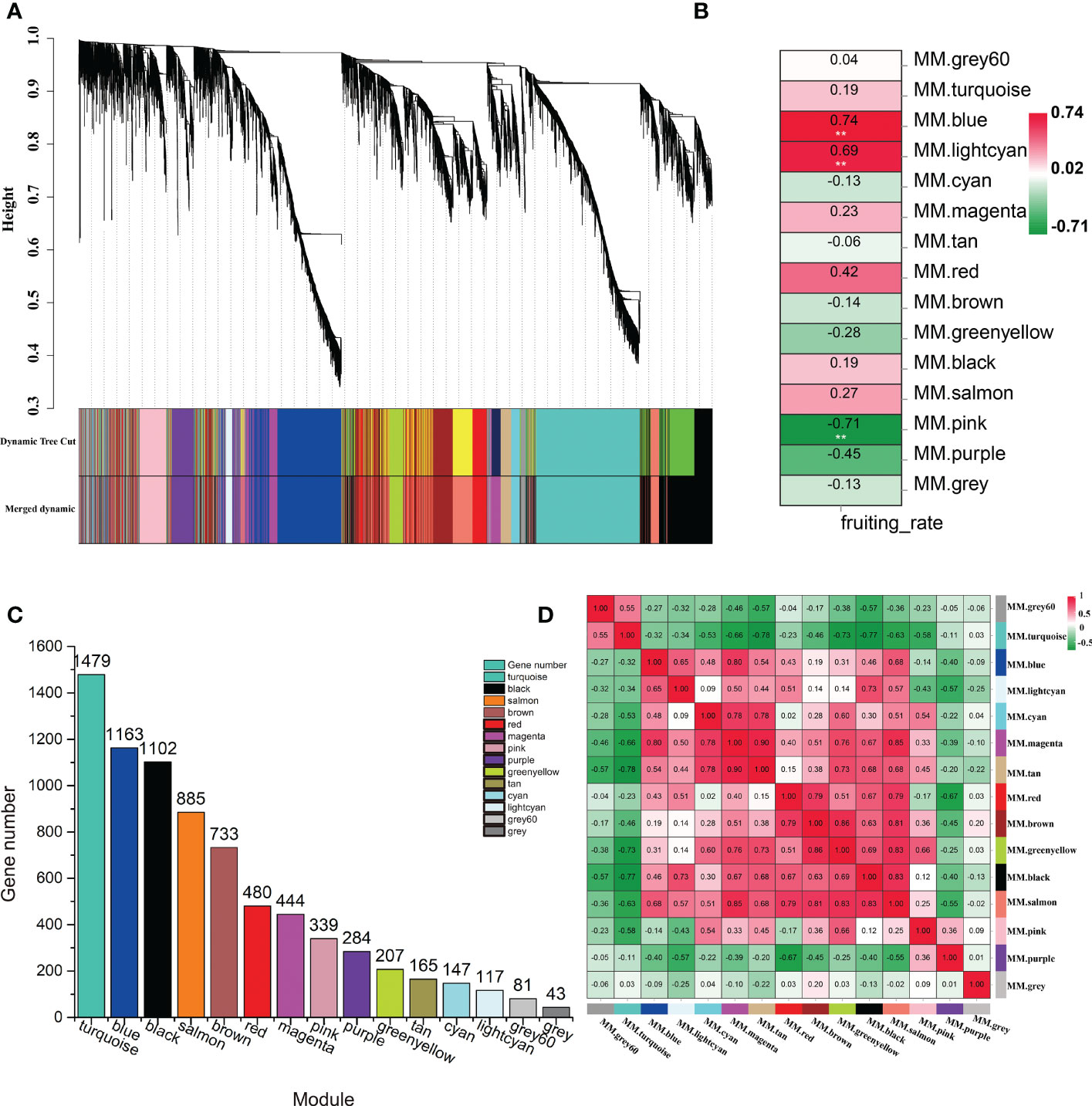
Figure 4 Identification of coexpression gene modules using differentially expressed genes. (A) Dendrogram of all filtered genes enriched according to a dissimilarity measure (1-TOM) and the cluster module colors. (B) Heatmap of the correlation between the fruit setting rates and module eigengenes (MEs) of bladder cancer. The redder the module color, the more significant the relationship. (C) Number of members in the 15 modules. (D) Module-sample correlation heatmap to intuitively visualize the expression pattern of each module in each sample.
3.6 Functional specific enrichment analysis of the two key modules
To further understand the specific functions of the key module, GO and KEGG enrichment analyses of the DEGs from the two key modules (blue and pink) were performed. GO analysis revealed that “response to stress”, “response to endogenous stimulus”, “hormone-mediated signaling pathway”, “regulation of programmed cell death”, “oxidoreductase activity”, and “protein kinase activity” were the main enriched terms in the blue module (Supplementary Figure S3A and Supplementary Table S5). KEGG enrichment analysis indicated that “Phenylpropanoid biosynthesis”, “MAPK signaling pathway”, “Biosynthesis of secondary metabolites”, “Plant-pathogen interaction”, “Brassinosteroid biosynthesis”, and “Flavonoid biosynthesis” were the main significantly enriched metabolic pathways, indicating that these genes in blue module involved in the LSI reaction in C. oleifera by regulating “Phenylpropanoid biosynthesis”, “MAPK signaling pathway”, “Biosynthesis of secondary metabolites”, “Plant-pathogen interaction”, “Brassinosteroid biosynthesis”, and “Flavonoid biosynthesis” (Supplementary Figure S3B and Supplementary Table S6). In pink, statistics on GO terms showed that “tubulin binding”, “cytoskeletal protein binding”, “protein kinase regulator activity”, “cytoskeletal part”, and “microtubule cytoskeleton” were the main significantly enriched terms (Supplementary Figure S3C and Supplementary Table S7). KEGG enrichment analysis revealed that most DEGs were enriched in “Phenylpropanoid biosynthesis”, “Flavonoid biosynthesis”, “Biosynthesis of secondary metabolites”, “Ubiquitin-mediated proteolysis”, which show that genes in Pink module contribute to reject self-pollen of C. oleifera by regulating Flavonoid biosynthesis and Ubiquitin-mediated proteolysis (Supplementary Figure S3D and Supplementary Table S8).
3.7 Hub genes and TFs involved in the LSI of C. oleifera via WGCNA
To fur identify the specific gene s and the regulatory relationship within the modules related to LSI in the blue and pink modules, the top 10% of edges, which are ordered by the edge weight coefficient, were selected to create the gene coexpression network by CYTOSCAPE software (Figures 5A–D; Supplementary Tables S9, S10). After deleting the unknown genes, the top 50 genes ordered by degree with high connectivity are shown in Figure 5 (Supplementary Tables S11, S12). From our results, we indeed discovered some important genes that might be related to LSI in Camellia oil trees. The blue modules were significantly positively correlated with FSR under self-pollination. Five genes encoding CML41, LECRK3, CRP, P450 (CYP87A3), WRKY, and ERF1B were identified as candidate hub genes based on the degree values (Figures 5A, C). It has been reported that the apical Ca2+ gradient has an important influence on the growth of pollen tubes, and the calcium-binding protein (CML) protein plays a key role in the Ca2+ signaling system (Zhao et al., 2020). In heteromorphic SI, cytochrome P450 (CYP) is one of five important S-locus genes that can determine the length of the style (Zhao et al., 2022). ERF1B is an ethylene-responsive transcription factor that plays a key role in the ethylene signaling pathway, and it has been reported that ETH is related to PCD of pollen tube. LECRK3 encodes a G-type lectin S-receptor-like serine/threonine-protein kinase and is considered to be involved in the SI signal transduction mechanism in tea plant (Ma et al., 2018). In the pink modules, KIN12F and GDSL were identified as candidate hub genes (Figures 5B, D), which were significantly negatively correlated with the fruit setting rates of Camellia oil tree. KIN12F encodes a kinesin-like protein that can regulate on several important stress-responsive genes, such as ethylene-responsive transcription factors and RNA polymerases. (Islam et al., 2020). The GDSL esterase/lipase protein is vital for anther and pollen development (Yuan et al., 2020). The result indicated that these hub genes play a positively role in the LSI reaction according to the gene correlation networks.
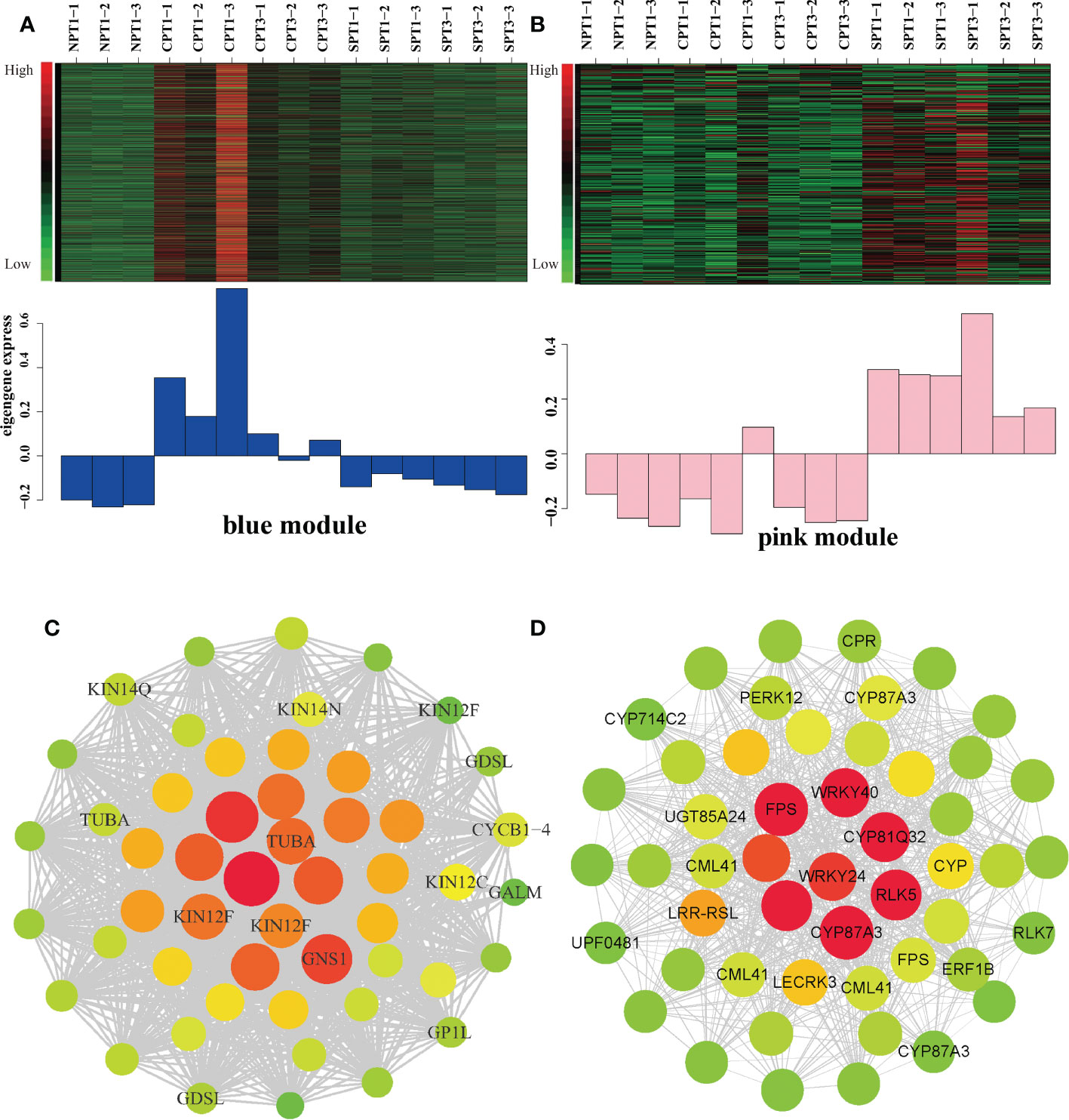
Figure 5 Analysis of the blue and pink modules. (A) Expression pattern in blue modules under different pollination treatments. (B) Expression pattern in pink modules under different pollination treatments. (C) Correlation network of the blue module. Each node represents a gene, the edge between genes represents the coexpression correlation, and the top 50 genes ordered by degree with high connectivity were visualized by Cytoscape. The color and size of each circle represent the number of edges. (D) Correlation network of the pink module. Each node represents a gene, the edge between genes represents the coexpression correlation, and the top 50 genes ordered by degree with high connectivity were visualized by Cytoscape. The color and size of each circle represent the number of edges.
Moreover, we also found that several genes were related to the LSI of C. oleifera in the pink and blue modules, although the connectivity degree was not in the top 50. Four DEGs encoding F-box proteins were identified and had relatively high expression levels in CPT1 and CPT3 (Figure 3C). The F-box is the pollen S-determinant in GSI, and SLF from different S-haplotypes in pollen can protect pollen tube growth from its own S-RNase, resulting in the breakdown of incompatibility (Zhao et al., 2022). Four DEGs encoding RALF were identified in these two modules, and their expression in cross-pollination was significantly higher than that in self-pollination. RALF and AGP are involved in normal pollen tube (PT) elongation in the style and PT-ovular guidance (Seth et al., 2019). The expression levels of ATL6, DURF2, ICR4, SIZ1 and ERD4, which are three types of E3 ubiquitin ligases related to ubiquitin-mediated proteolysis, were significantly higher in cross-pollination than in self-pollination. Moreover, 1 calcium-dependent protein kinase gene, CPK5, and 3 probable calcium-binding protein CML41 genes were identified to be involved in plant-pathogen interactions. Overall, these genes were identified as candidate genes related to the inhibition of pollen tube growth in pistils, revealing the molecular mechanism of the LSI of C. oleifera.
Various TFs and their regulated gene expression networks play vital roles in plant growth, biosynthesis of secondary metabolites and development (Wu et al., 2020a). In this study, 597 TFs were identified among all of the DEGs (Supplementary Table S13). The MYB, ERF, bHLH, WRKY and C2H2 families were the top 5 TF families, and the MYB family was one of the largest families, containing 83 members. However, members of the SRS, WOX, RAV, LFY, FAR1, YABBY and GeBP families had fewer than 4 genes. To systematically study the TFs and their regulatory relationship with hub genes related to the LSI of C. oleifera, a total of 54 TFs were obtained in the pink and blue modules, and a heatmap of their expression patterns was presented in Figure 6A. By calculating the correlation coefficient between TFs in modules and hub genes, and then screening the related pairs as edge information based on the correlation coefficient (blue module:|correlation coefficient| ≥ 0.9, p < 0.05; pink module: |correlation coefficient| ≥ 0.7, p < 0.05; Supplementary Tables S14, S15), we constructed an inferred regulatory network to explore the regulatory relationship between the TFs, hub genes and genes related to LSI in pink or blue modules. In the blue module, 12 hub TFs mainly in the WRKY, NAC, MYB, ERF, bHLH, and FAR1 families showed regulatory relationships with hub genes (Figure 6B). It has been reported that the WRKY TF family played an important role in pollen germination, pollen viability and pollen tube growth in several plants (Zou et al., 2010; Guan et al., 2014), such as cotton (Wang, 2019) and Arabidopsis (Guan et al., 2014). In addition, a regulatory relationship between hub TFs and hub genes was also observed in the pink module (Figure 6C), including the TF with the highest connective degree, MYB3R4, which was involved in the SI process (He et al., 2020). More importantly, other TFs (ATHB-40, BHLH62 and ERF060) associated with the stimulus response were also detected in the regulatory relationship network. The results suggested that TFs in two modules might be pivotal regulators in the LSI response of C. oleifera, but this finding needs further verification.
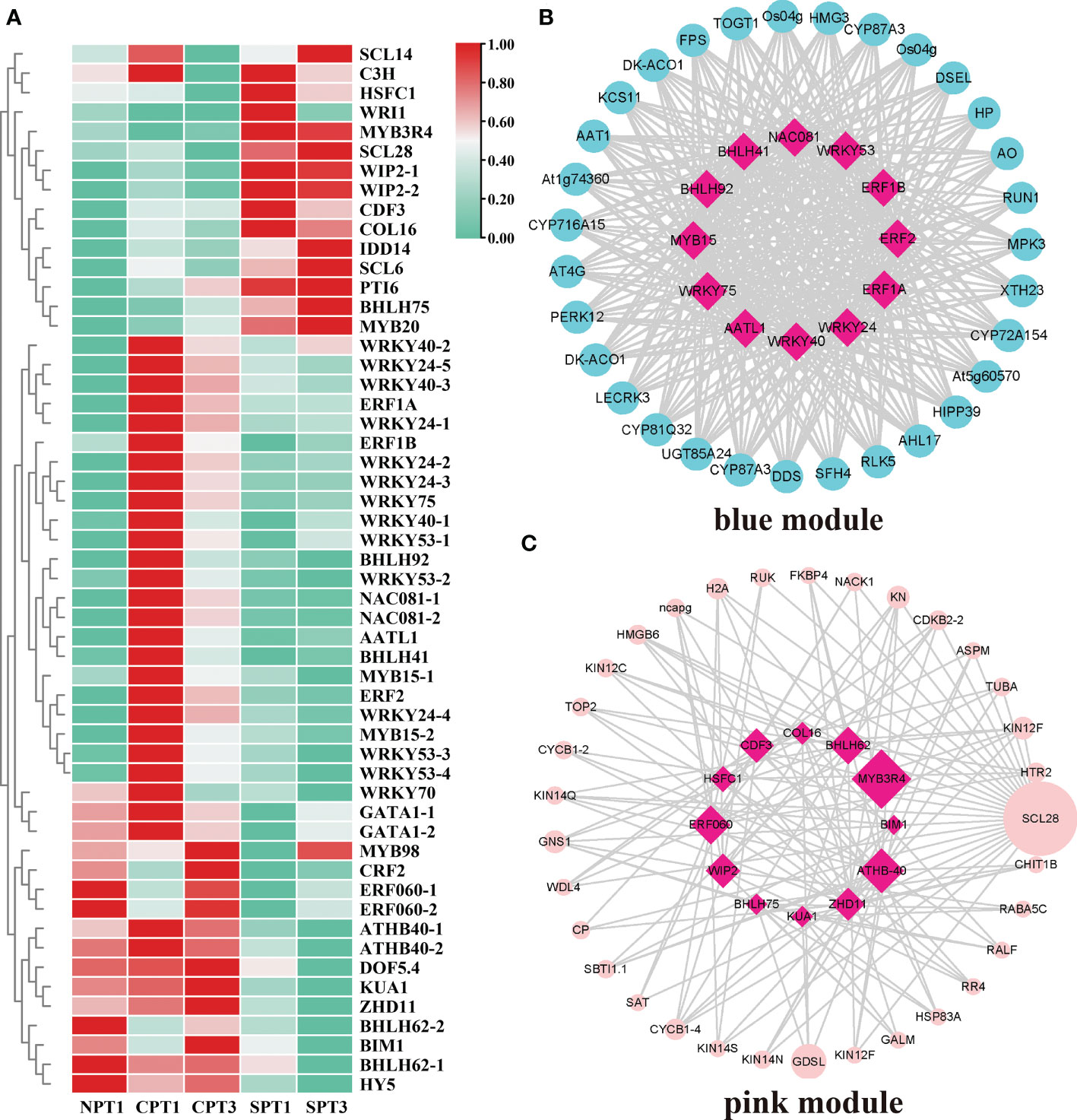
Figure 6 Analysis of TFs in the blue and pink modules. (A) Heatmap showing the expression profiles of the TFs in the self- and cross-pollination treatments. (B) Genetic regulatory networks (GRNs) between hub genes in blue modules. (C) Genetic regulatory networks (GRNs) between hub genes in the pink module.
3.8 Alternative splicing events analysis during the LSI reaction in C. oleifera
Alternative splicing (AS) is an important gene regulation mechanism in eukaryotic plants. In this study, five AS events were detected, including: skipping exon (SE), retained intron (RI), mutually exlusion exon (MXE), alternative 5′ splice site (A5SS), and alternative 3′ splice site (A3SS) in the ovary of C. oleifera (Figure 7A). As shown in the Figure 7B, 72,299 AS events were identified, and among which SE was the most abundant, accounting for 82.43% of AS events. RI (3.22%), together with A3SS (3.09%), A5SS (2.02%) or MXE (9.24%), these four types of AS events only accounted for (17.58%) (Figure 7B). To further investigate whether the AS types with different functional annotations exhibited bias in self- and cross- pollination, the GO and KEGG pathway analyses were performed. The SE isoforms were mainly enriched in the GO terms “salicylic acid metabolic process”, “gene expression”, “response to chemical” and “signal transduction”. KEGG revealed that “Plant-pathogen interaction”, “Ubiquitin mediated proteolysis”, “Endocytosis” and “Plant hormone signal transduction” were the main pathways for the AE-type transcripts. For A3SS-type transcripts, “developmental process involved in reproduction” was the most enriched GO term, and significant pathways of “SNARE interactions in vesicular transport”, “Ubiquitin mediated proteolysis”, and “RNA transport” were identified in A3SS type transcripts. The Metabolic pathways was significantly enriched in three AS types including A3SS-, MEX- and RI-type transcripts.
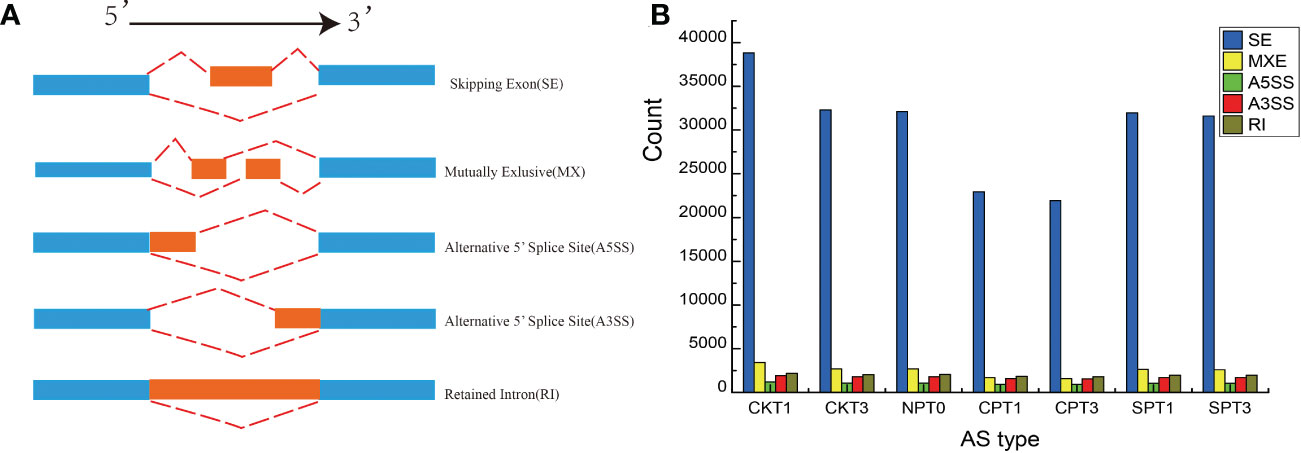
Figure 7 AS analysis of C.oleifera ovaries. (A) Visualization of seven AS types. (B) Numbers of different AS events detected in transcriptomes from ovaries.
Among the DEGs identified that might be involved in the LSI reaction of C. oleifera in this study, approximately 25 DEGs exhibited SE-type alternative splicing events, including F-Box, CYP87A3, E3 ubiquitin-protein ligase RGLG2-like, G-type lectin S-receptor-like serine CES101, WRKY40, and BHLH75. Two types of AS both had one DEG each, the A3SS-type gene CSLD5 and the A5SS-type gene AK2. Two DEGs, BHLH75 and callose synthase 2-like isoform X1, exhibited MXE-type alternative splicing events. Three DEGs, including CDC20-1 and 2 WRKY transcription factors, exhibited RI-type alternative splicing events. Overall, the results suggested that the DEGs identified in this study, which potential involved in the LSI reaction, undergo variable splicing events, it enhanced the plasticity of transcriptome and also might play a role in regulating the level of different transcripts.
3.9 Quantitative real-time PCR validation of RNA-Seq data
To validate the reliability of our RNA-seq data, a total of 12 DEGs mentioned in this study were selected and subjected to qRT–PCR. Among the 12 DEGs, F-Box (ID: oil_tea_GLEAN_10305490) was related to ubiquitin-mediated proteolysis. RALF-1 (ID: MSTRG.114451), RALF-3 (ID: MSTRG.55710) and CAML41 (ID: oil_tea_GLEAN_10201557) were related to the growth of pollen tube. The other 8 genes were transcription factors involved in LSI reaction of C. oleifera, including DOF5 (ID: oil_tea_GLEAN_10115754), CPK29 (ID: oil_tea_GLEAN_10394599), WRKY24 (ID: oil_tea_GLEAN_10391421), ATHB-1 (ID: oil_tea_GLEAN_10231301), MYB20 (ID: oil_tea_GLEAN_10287798), BHLH62 (ID: oil_tea_GLEAN_10050733), ERF1A (ID: oil_tea_GLEAN_10347617), ATHB-3 (ID: oil_tea_GLEAN_10174212). We compared the expression data of the 12 DEGs obtained by qRT-PCR and RNA-seq, and the expression patterns of these 12 selected DEGs obtained by qRT-PCR were corresponded well with RPKM values obtained by RNA-seq (Figure 8). The experiment demonstrated that the RNA-seq data used in this study were highly reliable.
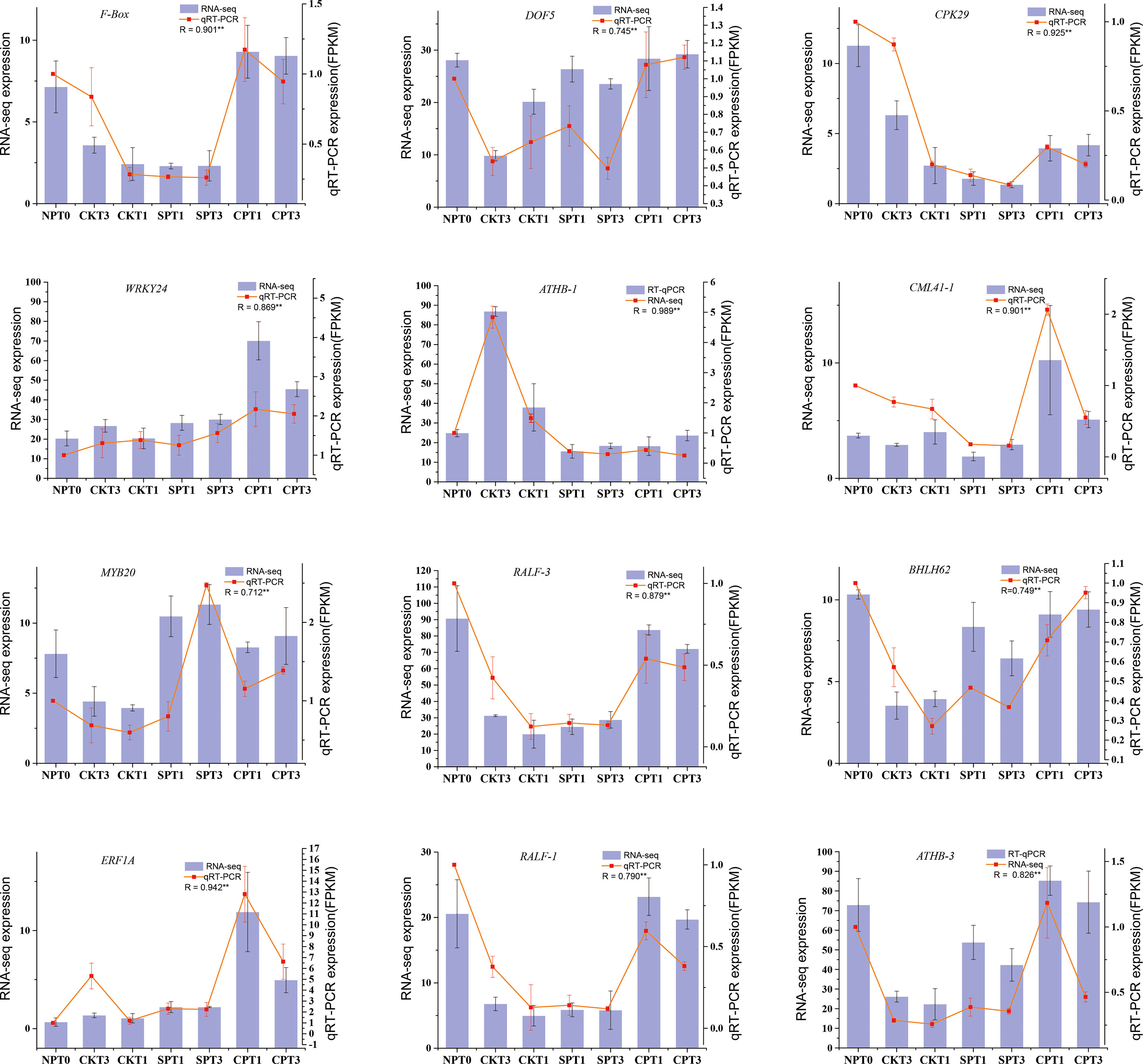
Figure 8 qRT-PCR validation of 12 DEGs identified as related to self-incompatibility. *p < 0.05; **p < 0.01.
4 Discussion
The oil of Camellia oil tree seed has been used as a high-quality edible oil and a traditional medicine in China (Cheng et al., 2015). However, the yield of C. oleifera seeds is affected by the climate and by its LSI biological characteristics (He et al., 2020). The molecular mechanism of SI has been widely studied in plants (Zhao et al., 2015; Ma et al., 2018; Shin et al., 2022), but little is known about the SI of C. oleifera. In this study, we found that self-pollination at earlier bud developmental stages increased the fruit setting rate in C. oleifera to a certain extent. The present study reported the fruit setting rates of different treatments with self-and cross-pollination at early bud developmental stages, and the molecular regulatory mechanism of Camellia oil tree was preliminarily revealed by transcriptomic analysis. The results led to a focus on some hub genes and vital pathways, including plant MAPK signaling pathways, ubiquitin-mediated proteolysis, plant-pathogen interaction, vital signal molecules and TFs, which are discussed below.
4.1 Pollination at different bud developmental stages affects the fruit setting rate
It has been reported that the incongruity in SI or interspecific crossing could be overcome by bud pollination in breeding programs (Goldman, 2015). In brassica, pollination using mature pollen was performed on immature flowers (3–4 DBF) to bypass the influence of SI (Hiscock and Dickinson, 1993). In S-RNase-based GSI plants, it was reported that the degree of SI was positively correlated with the content of S protein in the style, and the content of S protein in the style was lower in the bud stage and higher in mature flowers (Anderson et al., 1986; McClure et al., 1999; Roldán et al., 2010). Therefore, many self-incompatible plants were self-pollinated in the immature bud stage to create homozygous offspring. In this study, the fruit setting rate of SPT3 (3–4 DBF) was higher than that of SPT1 (1 DBF), but it did not reach a significant level, suggesting that the fruit setting rate might be higher in the case of self-pollination in the earlier developmental stage of flower buds.
Effective pollination with a high fruit setting rate is determined by multiple factors, including pollen tube kinetics, stigma receptivity, and ovule development (Sanzol and Herrero, 2001). In the cross-pollination comparison, the fruit setting rate of CPT3 was lower than that of CPT1. Studies have shown that the developmental stage of the style could influence the number of growing pollen tubes, and mature stigmas have more energy and nutrients to maintain more pollen growth (Goldman, 2015), which is also an important reason for the lower fruit setting rate in cross-pollination with immature stigmas.
4.2 F-box and ubiquitin-mediated proteolysis are involved in the LSI of C. oleifera
Protein hydrolysis mediated by ubiquitination plays an important role in the GSI reaction, and this mechanism was first proposed in the study of SI in petunia (Zhang et al., 2016; Chen et al., 2018; Ren et al., 2020; Zhao et al., 2021). In self-incompatible Petunia and Antirrhinum species, S-RNase, which is the S-determinant of pistils, could be recognized by pollen S-determinant SLFs and ubiquitinated by forming functional SCF complexes in a nonself-recognition manner and then degraded by the 26S proteasome pathway, restricting the cytotoxicity of S-RNase and allowing pollen tubes to grow normally (Qiao et al., 2004; Hua and Kao, 2006; Hua et al., 2008; Kubo et al., 2010; Liu et al., 2014). F-box, Cullin-1 (CUL1), S-phase kinase-associated protein 1 (Skp-1), and RING box protein-1 (Rbx1) are the functional components of the SCF complex (Hua and Kao, 2006). In this study, we found that the two key modules (pink and blue), which had a high correlation with fruit setting rates, were both significantly enriched in the ubiquitin-mediated proteolysis pathway, including three genes, RZPF34, CDC20-1, and UBC20. In addition, we also found 4 DEGs encoding F-boxes and three types of E3 ubiquitin ligases related to ubiquitin-mediated proteolysis in self- and cross-pollination. Research has shown that ubiquitin ligase can reduce the production of reactive oxygen species (ROS) and cell death, positively regulating plant salt tolerance (Potocký et al., 2007). Furthermore, we identified nine RNase T2 family genes in the self-pollination transcriptome data of C. oleifera Huashuo in our previous study. The discovery of these DEGs encoding F-box and ubiquitin-mediated proteolysis proteins implied that the LSI in Camellia oil tree might be under gametophytic control, and the enhanced understanding of the mechanism needs further verification.
4.3 TFs and hub genes related to LSI selected by WGCNA
The LSI reaction in C. oleifera is not controlled by a single gene. It is a rather a complex genetic network containing multiple genes. It has been reported that TFs play an important role in plant biological processes by regulating relative gene expression, including plant growth, synthesis of secondary metabolites (Song et al., 2022), stress/stimulus responses (Luo et al., 2022), flower senescence (Ji et al., 2022), SI reaction processes (Hou et al., 2022), pollen tube growth (Zou et al., 2010) and other important biological processes. In tomato, progressive mutation of TFs led to a transition from outcrossing to self-pollination (Benoit, 2021). In this study, 54 TFs were identified by WGCNA that were involved in the LSI process in the two key modules, and they were significantly differentially expressed. Among these TFs, WRKY, MYB and bHLH were the major TFs with high connectivity degrees to other hub genes. WRKY TFs have been demonstrated to regulate the plant response to biotic stress, such as salicylic acid (SA)-mediated defense and disease responses (Dellagi et al., 2000; Asai et al., 2002; Lai et al., 2008). In Arabidopsis, WRKY34 has been demonstrated to be a pollen-specific TF and is also a substrate of MPK/MPK6 (Guan et al., 2014). Moreover, MAPK signaling pathways were significantly enriched in both the SPT1 vs. CPT1 and SPT3 vs. SPT3 comparisons. MYB is a large TF family in plants, and it has been reported that MYB TFs are associated with the response to external or internal stimuli and plant development (Ito, 2005). MYB TFs are also involved in regulating several important activities associated with female reproduction in Arabidopsis (Punwani et al., 2008). In addition, the bHLH TFs JAZs and MYB could form a complex that may play an important role in seed production and stamen development in Arabidopsis (Qi et al., 2015). The results suggested that the three major transcription factors (WRKY, MYB and bHLH) might play an important role in the transcriptional activation of genes involved in the LSI reaction in C. oleifera.
The pollination and fertilization of plants can be divided into four stages, namely, pollen germination, tube growth, tube guidance and double fertilization (Dresselhaus and Franklin-Tong, 2013). We observed that the pollen tubes of C. oleifera stopped growing at the bottom of the style, where cell–cell communication events should take place at the late stage of pollen tube growth, just before double fertilization. The rapid alkalinization factor (RALF) is a ligand−receptor pair with FERONIA (FER). The RALF-FER pathway can mediate cell elongation (Zhu et al., 2021), and it also plays a key role in communication between the pollen tube and the synergid cell during pollen tube reception (Li et al., 2016; Liang and Zhou, 2018; Ge et al., 2019). In this study, we identified two RALF genes in the pink module that were expressed at significantly higher levels in cross-pollination than in self-pollination. Moreover, two hub genes were identified in the blue module: UTG and G-type lecRLK. A study in Arabidopsis suggested that the mutation of EVAN and TURAN led to overgrowth of pollen tubes inside the female gametophyte without rupture of pollen tubes. UTG superfamily proteins are encoded by TURNA, which is very important for pollen tube growth and integrity by affecting the stability of pollen-specific FERONIA RLKs (Escobar-Restrepo et al., 2007; Lindner et al., 2015). Our results suggested that the low expression of RALF, UTG and G-type lecRLK in self-pollination treatments was one of the main reasons why pollen tubes stopped growing and failed to fertilize.
Cytochrome P450 has wide-range catalytic activity and is involved in the synthesis and metabolism of sterols, terpenoids, fatty acids, alkaloids, isoflavones, flavonoids and plant hormones (Sun et al., 2019). In our study, three cytochrome P450 87A3-like genes (CYPs) were identified as candidate hub genes selected by WGCNA, and the expression of CYPs in cross-pollination was significantly higher than that in self-pollination. Furthermore, the flavonoid biosynthesis pathway was significantly enriched in both the blue module and pink module. Since it has been reported that flavonoids play an important role in flower development, especially in the growth of pollen tubes (Lan et al., 2017; Paupière et al., 2017), studies in maize and petunia suggested that pollen deficient in flavonoids could not produce functional pollen tubes, while the defect can be overcome by applying specific flavonols to the stigma during pollination (Hosokawa et al., 2013; Han et al., 2016). In addition, DEGs in the brassinosteroid biosynthesis pathway were significantly enriched in both the SPT1 vs. CPT1 and SPT3 vs. CPT3 comparisons. In Primula, brassinosteroids have been demonstrated to regulate pistil development and SI reactions (Manghwar et al., 2022). CYP734A50 was reported to be a key S-locus gene, and its products can degrade brassinosteroids (Zhao et al., 2019). In this work, how CYP, flavonoids and brassinosteroids work together during fertilization in Camellia oil trees remained unknown, but we suggested that these genes might be involved in the LSI reaction in C. oleifera. In addition, the qRT–PCR results demonstrated the accuracy of our transcriptomic data analysis.
In this study, we identified a set of candidate genes and major enrichment metabolic pathways related to self-incompatibility, we developed a hypothetical model based on the results. When the self- and nonself-pollens fall on the stigma of Camellia oil tree, both can germinate and grow normally. When the pollen stretches to the base of the style and the upper part of the ovary, the self-incompatible factor enters the pollen tube to exercise the function of cytotoxicity, degrades the pollen tube RNA by the way of ubiquitinated protein hydrolysis, and starts a series of downstream molecular signal transmissions, and at the same time, plants themselves can also resist damage from self-pollination by regulating flavonoid biosynthesis, plant MAPK signaling pathways, ubiquitin-mediated proteolysis, and plant-pathogen interaction pathways. While finally the pollen tube stopped growth and programmed cell death was occurred (Figure 9).
5 Conclusion
First, transcription analysis of early-development self- and cross-pollinated ovaries was performed in C. oleifera. The MAPK signaling pathway, ubiquitin-mediated proteolysis, flavonoid biosynthesis, brassinosteroid biosynthesis, and plant–pathogen interactions were significantly enriched during the pollination process. Through WGCNA, we confirmed hub genes and TFs involved in the LSI reaction in C. oleifera. Thus, our study provided a useful transcriptomic resource for revealing the molecular mechanism of LSI in C. oleifera and also provided a solid theoretical basis for comprehensively increasing the output of C. oleifera.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
Methodology, CL, SW, JZ and XT. Data analysis, CL, YL. Data curation, YL. Resources, YL and YX. Validation, CL and YL. Writing-review and editing, CL, ML and JZ. Writing-original manuscript, CL. Supervision, JZ, SW and XT All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Natural Science Foundation of China (Grant number 31730016).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1065872/full#supplementary-material
Abbreviations
ATL6, E3 ubiquitin-protein ligase ATL31-like; DURF2, E3 ubiquitin-protein ligase RDUF2-like; RGLG1, E3 ubiquitin-protein ligase RGLG2-like; RHC1A, probable E3 ubiquitin-protein ligase RHC1A; CML41, probable calcium-binding protein CML41; CRP, receptor-like protein kinase HSL1; CYP87A3, NADPH-cytochrome P450 reductase-like; LECRK3, probable receptor-like serine/threonine-protein kinase; ERF1B, ethylene-responsive transcription factor 1B-like; KIN12F, tubulin alpha chain; ASPM, serine/threonine-protein kinase RUNKEL-like isoform X1; TUBA, GDSL esterase/lipase At5g14450-like; F-box, F-box/kelch-repeat protein At1g74510-like; OXI1, serine/threonine-protein kinase OXI1-like; CDC20-1, cofactor of APC complex-like; UBC20, ubiquitin-conjugating enzyme E2 20 isoform X1; RZPF34, E3 ubiquitin-protein ligase RZFP34-like isoform X3; PTI6, pathogenesis-related genes transcriptional activator PTI6-like; HSP83A, heat shock protein 83; KINUC, endocytosis kinesin-like protein; RABA5C, ras-related protein RABA5c-like; WRKY, WRKY transcription factor; MAPK, mitogen-activated protein kinase.
References
Anderson, M. A., Cornish, E. C., Mau, S.-L., Williams, E. G., Hoggart, R., Atkinson, A., et al. (1986). Cloning of cDNA for a stylar glycoprotein associated with expression of self-incompatibility in nicotiana alata. Nature 321, 38–44. doi: 10.1038/321038a0
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W.-L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Bedinger, P. A., Broz, A. K., Tovar-Mendez, A., McClure, B. (2017). Pollen-pistil interactions and their role in mate selection. Plant Physiol. 173, 79–90. doi: 10.1104/pp.16.01286
Bedinger, P. A., Pearce, G., Covey, P. A. (2010). RALFs: Peptide regulators of plant growth. Plant Signaling Behav. 5, 1342–1346. doi: 10.4161/psb.5.11.12954
Ben-Naim, O., Eshed, R., Parnis, A., Teper-Bamnolker, P., Shalit, A., Coupland, G., et al. (2006). The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 46, 462–476. doi: 10.1111/j.1365-313X.2006.02706.x
Benoit, M. (2021). From non-self to self: stepwise mutations in transcription factors promote the transition to self-pollination in tomato. Plant Cell 33, 3183–3184. doi: 10.1093/plcell/koab200
Broz, A. K., Bedinger, P. A. (2021). Pollen-pistil interactions as reproductive barriers. Annu. Rev. Plant Biol. 72, 615–639. doi: 10.1146/annurev-arplant-080620-102159
Cheng, Y.-T., Lu, C.-C., Yen, G.-C. (2015). Beneficial effects of camellia oil (Camellia oleifera abel.) on hepatoprotective and gastroprotective activities. J. Nutr. Sci. Vitaminology J. Nutr. Sci. Vitaminol 61, S100–S102. doi: 10.3177/jnsv.61.S100
Chen, X., Hao, S., Wang, L., Fang, W., Wang, Y., Li, X. (2012). Late-acting self-incompatibility in tea plant (Camellia sinensis). Biologia 67, 347–351. doi: 10.2478/s11756-012-0018-9
Chen, Q., Meng, D., Gu, Z., Li, W., Yuan, H., Duan, X., et al. (2018). SLFL genes participate in the ubiquitination and degradation reaction of s-RNase in self-compatible peach. Front. Plant Sci. 9, 227. doi: 10.3389/fpls.2018.00227
Chen, J., Yang, X., Huang, X., Duan, S., Long, C., Chen, J., et al. (2017). Leaf transcriptome analysis of a subtropical evergreen broadleaf plant, wild oil-tea camellia (Camellia oleifera), revealing candidate genes for cold acclimation. BMC Genomics 18, 211. doi: 10.1186/s12864-017-3570-4
Chen, S., Zhou, Y., Chen, Y., Gu, J. h. fastp: An ultra-fast all-in-one FASTQ preprocessor. Biotechnology 34, 884-890. doi: 10.1093/bioinformatics/bty560
Covey, P. A., Kondo, K., Welch, L., Frank, E., Sianta, S., Kumar, A., et al. (2010). Multiple features that distinguish unilateral incongruity and self-incompatibility in the tomato clade: Unilateral pollen rejection in tomato. Plant J. 64, 367–378. doi: 10.1111/j.1365-313X.2010.04340.x
Dellagi, A., Heilbronn, J., Avrova, A. O., Montesano, M., Palva, E. T., Stewart, H. E. (2000). A potato gene encoding a WRKY-like transcription factor is induced in interactions with Erwinia carotovora subsp. atroseptica and Phytophthora infestans and is coregulated with class I endochitinase expression. MPMI 13, 1092–1101. doi: 10.1094/MPMI.2000.13.10.1092
Dong, B., Yang, Q., Song, Z., Niu, L., Cao, H., Liu, T., et al. (2021). Hyperoside promotes pollen tube growth by regulating the depolymerization effect of actin-depolymerizing factor 1 on microfilaments in okra. Hortic. Res. 8, 145. doi: 10.1038/s41438-021-00578-z
Dresselhaus, T., Franklin-Tong, N. (2013). Male–Female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol. Plant 6, 1018–1036. doi: 10.1093/mp/sst061
Duan, Z., Dou, S., Liu, Z., Li, B., Yi, B., Shen, J., et al. (2020). Comparative phosphoproteomic analysis of compatible and incompatible pollination in brassica napus l. Acta Biochim. Biophys. Sin. 52, 446–456. doi: 10.1093/abbs/gmaa011
Eaves, D. J., Flores-Ortiz, C., Haque, T., Lin, Z., Teng, N., Franklin-Tong, V. E. (2014). Self-incompatibility in Papaver : advances in integrating the signalling network. Biochem. Soc. Trans. 42, 370–376. doi: 10.1042/BST20130248
Escobar-Restrepo, J.-M., Huck, N., Kessler, S., Gagliardini, V., Gheyselinck, J., Yang, W.-C., et al. (2007). The FERONIA receptor-like kinase mediates Male-female interactions during pollen tube reception. Science 317, 656-660. doi: 10.1126/science.1143562
Ge, Z., Cheung, A. Y., Qu, L. (2019). Pollen tube integrity regulation in flowering plants: Insights from molecular assemblies on the pollen tube surface. New Phytol. 222, 687–693. doi: 10.1111/nph.15645
Goldman, M. H. S. (2015). Pollination triggers female gametophyte development in immature nicotiana tabacum flowers. Front. Plant Sci. 6, 561. doi: 10.3389/fpls.2015.00561
Gordillo-Romero, M., Correa-Baus, L., Baquero-Méndez, V., Torres, M., de, L., Vintimilla, C., et al. (2020). Gametophytic self-incompatibility in Andean capuli (Prunus serotina subsp. capuli): Allelic diversity at the s-RNase locus influences normal pollen-tube formation during fertilization. PeerJ 8, e9597. doi: 10.7717/peerj.9597
Guan, Y., Meng, X., Khanna, R., LaMontagne, E., Liu, Y., Zhang, S. (2014). Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in arabidopsis. PloS Genet. 10, e1004384. doi: 10.1371/journal.pgen.1004384
Haffani, Y., Gaude, T., Cock, J., Goring, D. (2004). Antisense suppression of thioredoxinhmRNA in brassica napus cv. Plant Mol. Biol. 55, 619–630. doi: 10.1007/s11103-004-1126-x
Hajjar, R., Hodgkin, T. (2007). The use of wild relatives in crop improvement: A survey of developments over the last 20 years. Euphytica 156, 1–13. doi: 10.1007/s10681-007-9363-0
Han, Y., Ding, T., Su, B., Jiang, H. (2016). Genome-wide identification, characterization and expression analysis of the chalcone synthase family in maize. IJMS 17, 161. doi: 10.3390/ijms17020161
Hao, Y.-Q., Zhao, X.-F., She, D.-Y., Xu, B., Zhang, D.-Y., Liao, W.-J. (2012). The role of late-acting self-incompatibility and early-acting inbreeding depression in governing female fertility in monkshood, aconitum kusnezoffii. PloS One 7, e47034. doi: 10.1371/journal.pone.0047034
He, Y., Song, Q., Chen, S., Wu, Y., Zheng, G., Feng, J., et al. (2020). Transcriptome analysis of self- and cross-pollinated pistils revealing candidate unigenes of self-incompatibility in Camellia oleifera. J. Hortic. Sci. Biotechnol. 95, 19–31. doi: 10.1080/14620316.2019.1632749
He, Y., Song, Q., Wu, Y., Ye, S., Chen, S., Chen, H. TMT-based quantitative proteomic analysis reveals the crucial biological pathways involved in self-incompatibility responses in camellia oleifera. IJMS 21, 1987. doi: 10.3390/ijms21061987
Hiscock, S. J., Dickinson, H. G. (1993). Unilateral incompatibility within the brassicaceae: further evidence for the involvement of the self-incompatibility (S)-locus. Theoret. Appl. Genet. 86, 744–753. doi: 10.1007/BF00222665
Hosokawa, M., Yamauchi, T., Takahama, M., Goto, M., Mikano, S., Yamaguchi, Y., et al. (2013). Phosphorus starvation induces post-transcriptional CHS gene silencing in petunia corolla. Plant Cell Rep. 32, 601–609. doi: 10.1007/s00299-013-1391-8
Hou, S., Zhao, T., Yang, Z., Liang, L., Ma, W., Wang, G., et al. (2022). Stigmatic transcriptome analysis of self-incompatible and compatible pollination in corylus heterophylla fisch. × Corylus avellana L. Front. Plant Sci. 13, 800768. doi: 10.3389/fpls.2022.800768
Hua, Z.-H., Fields, A., Kao, T. (2008). Biochemical models for s-RNase-Based self-incompatibility. Mol. Plant 1, 575–585. doi: 10.1093/mp/ssn032
Hua, Z., Kao, T. (2006). Identification and characterization of components of a putative Petunia s -locus F-Box–containing E3 ligase complex involved in S-RNase–based self-incompatibility. Plant Cell 18, 2531–2553. doi: 10.1105/tpc.106.041061
Islam, M. M., Sandhu, J., Walia, H., Saha, R. (2020). Transcriptomic data-driven discovery of global regulatory features of rice seeds developing under heat stress. Comput. Struct. Biotechnol. J. 18, 2556–2567. doi: 10.1016/j.csbj.2020.09.022
Ito, M. (2005) Conservation and diversification of three-repeat Myb transcription factors in plants. J Plant Res. 118, 61-69. doi: 10.1007/s10265-005-0192-8
Ji, X., Wang, M., Xu, Z., Wang, , Sun, D., Niu, L., et al (2022). PlMYB308 regulates flower senescence by modulating ethylene biosynthesis in herbaceous peony. Front. Plant Sci. 13, 872442. doi: 10.3389/fpls.2022.872442
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kubo, K., Entani, T., Takara, A., Wang, N., Fields, A. M., Hua, Z., et al. (2010). Collaborative non-self recognition system in S-RNase–based self-incompatibility. Science 330, 796–799. doi: 10.1126/science.1195243
Lai, Z., Vinod, K., Zheng, Z., Fan, B., Chen, Z. (2008). Roles of arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol. 8, 68. doi: 10.1186/1471-2229-8-68
Lan, X., Yang, J., Abhinandan, K., Nie, Y., Li, X., Li, Y., et al. (2017). Flavonoids and ROS play opposing roles in mediating pollination in ornamental kale (Brassica oleracea var. acephala). Mol. Plant 10, 1361–1364. doi: 10.1016/j.molp.2017.08.002
Lee, C.-P., Yen, G.-C. (2006) Antioxidant activity and bioactive compounds of tea seed (Camellia oleifera abel.). J Agric Food Chem. 54, 779-784.
Liang, X., Zhou, J.-M. (2018). The secret of fertilization in flowering plants unveiled. Sci. Bull. 63, 408–410. doi: 10.1016/j.scib.2018.02.010
Lindner, H., Kessler, S. A., Müller, L. M., Shimosato-Asano, H., Boisson-Dernier, A., Grossniklaus, U. (2015). TURAN and EVAN mediate pollen tube reception in arabidopsis synergids through protein glycosylation. PloS Biol. 13, e1002139. doi: 10.1371/journal.pbio.1002139
Lin, M., Wang, S., Liu, Y., Li, J., Zhong, H., Zou, F., et al. (2022a). 11-hydrogen cyanamide enhances flowering time in tea oil camellia (Camellia oleifera abel.). Ind. Crops Products 176, 114313. doi: 10.1016/j.indcrop.2021.114313
Lin, P., Wang, K., Wang, Y., Hu, Z., Yan, C., Huang, H., et al. (2022b). 2 the genome of oil-camellia and population genomics analysis provide insights into seed oil domestication. Genome Biol. 23, 14. doi: 10.1186/s13059-021-02599-2
Lin, P., Wang, K., Zhou, C., Xie, Y., Yao, X., Yin, H. (2018). Seed transcriptomics analysis in camellia oleifera uncovers genes associated with oil content and fatty acid composition. Int. J. Mol. Sci. 19, 118. doi: 10.3390/ijms19010118
Liu, W., Fan, J., Li, J., Song, Y., Li, Q., Zhang, Y., et al. (2014). SCFSLF-mediated cytosolic degradation of s-RNase is required for cross-pollen compatibility in s-RNase-based self-incompatibility in petunia hybrida. Front. Genet. 5, 228. doi: 10.3389/fgene.2014.00228
Li, C., Wu, H.-M., Cheung, A. Y. (2016). FERONIA and her pals: Functions and mechanisms. Plant Physiol. 171, 2379–2392. doi: 10.1104/pp.16.00667
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Luo, Z., Zhou, Z., Li, Y., Tao, S., Hu, Z.-R., Yang, J.-S., et al. (2022). Transcriptome-based gene regulatory network analyses of differential cold tolerance of two tobacco cultivars. BMC Plant Biol. 22, 369. doi: 10.1186/s12870-022-03767-7
Ma, Q., Chen, C., Zeng, Z., Zou, Z., Li, H., Zhou, Q., et al. (2018). Transcriptomic analysis between self- and cross-pollinated pistils of tea plants (Camellia sinensis). BMC Genomics 19, 289. doi: 10.1186/s12864-018-4674-1
Manghwar, H., Hussain, A., Ali, Q., Liu, F. (2022). Brassinosteroids (BRs) role in plant development and coping with different stresses. IJMS 23, 1012. doi: 10.3390/ijms23031012
McClure, B., Cruz-García, F., Romero, C. (2011). Compatibility and incompatibility in s-RNase-based systems. Ann. Bot. 108, 647–658. doi: 10.1093/aob/mcr179
McClure, B., Mou, B., Canevascini, S., Bernatzky, R. (1999). A small asparagine-rich protein required for s-Allele-Specific pollen rejection in nicotiana. Proc. Natl. Acad. Sci. United States America 96, 13548–13553.
Miao, H., Qin, Y., da Silva, J. A. T., Ye, Z., Hu, G. (2013). Identification of differentially expressed genes in pistils from self-incompatible citrus reticulata by suppression subtractive hybridization. Mol. Biol. Rep. 40, 159–169. doi: 10.1007/s11033-012-2045-6
Nasrallah, J. B. (2019). “Self-incompatibility in the brassicaceae: Regulation and mechanism of self-recognition,” Curr Top Dev Biol. 131, 435-451. doi: 10.1016/bs.ctdb.2018.10.002
O’Brien, M., Major, G., Chantha, S.-C., Matton, D. P. (2004). Isolation of s-RNase binding proteins from solanum chacoense: identification of an SBP1 (RING finger protein) orthologue. Sex Plant Reprod. 17, 81–87. doi: 10.1007/s00497-004-0218-8
Paupière, M. J., Müller, F., Li, H., Rieu, I., Tikunov, Y. M., Visser, R. G. F., et al. (2017). Untargeted metabolomic analysis of tomato pollen development and heat stress response. Plant Reprod. 30, 81–94. doi: 10.1007/s00497-017-0301-6
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Potocký, M., Jones, M. A., Bezvoda, R., Smirnoff, N., Žárský, V. (2007). Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phytol. 174, 742–751. doi: 10.1111/j.1469-8137.2007.02042.x
Poulter, N. S., Staiger, C. J., Rappoport, J. Z., Franklin-Tong, V. E. (2010). Actin-binding proteins implicated in the formation of the punctate actin foci stimulated by the self-incompatibility response in Papaver. Plant Physiol. 152, 1274–1283. doi: 10.1104/pp.109.152066
Punwani, J. A., Rabiger, D. S., Lloyd, A., Drews, G. N. (2008). The MYB98 subcircuit of the synergid gene regulatory network includes genes directly and indirectly regulated by MYB98. Plant J. 55, 406–414. doi: 10.1111/j.1365-313X.2008.03514.x
Qiao, H., Wang, H., Zhao, L., Zhou, J., Huang, J., Zhang, Y., et al. (2004). The f-box protein AhSLF-s 2 physically interacts with s-RNases that may be inhibited by the Ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in antirrhinum. Plant Cell 16, 582–595. doi: 10.1105/tpc.017673
Qi, T., Huang, H., Song, S., Xie, D. (2015) Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in arabidopsis. Plant Cell. 27, 1620-1633. doi: 10.1105/tpc.15.00116
Quan, W., Wang, A., Gao, C., Li, C. (2022). Applications of Chinese camellia oleifera and its by-products: A review. Front. Chem. 10, 921246. doi: 10.3389/fchem.2022.921246
Ren, Y., Hua, Q., Pan, J., Zhang, Z., Zhao, J., He, X., et al. (2020). SKP1-like protein, CrSKP1-e, interacts with pollen-specific f-box proteins and assembles into SCF-type E3 complex in ‘Wuzishatangju’ (Citrus reticulata blanco) pollen. PeerJ 8, e10578. doi: 10.7717/peerj.10578
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Roldán, J. A., Quiroga, R., Goldraij, A. (2010). Molecular and genetic characterization of novel s-RNases from a natural population of nicotiana alata. Plant Cell Rep. 29, 735–746. doi: 10.1007/s00299-010-0860-6
Sanzol, J., Herrero, M. (2001). The ‘‘effective pollination period’’ in fruit trees. Scientia Hortic. 90, 7-12.
Seth, R., Bhandawat, A., Parmar, R., Singh, P., Kumar, S., Sharma, R. (2019). Global transcriptional insights of pollen-pistil interactions commencing self-incompatibility and fertilization in tea [Camellia sinensis (L.) o. kuntze]. IJMS 20, 539. doi: 10.3390/ijms20030539
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shen, S., Park, J. W., Lu, Z., Lin, L., Henry, M. D., Wu, Y. N., et al. (2014). rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl. Acad. Sci. U.S.A. 111, E5593-E5601. doi: 10.1073/pnas.1419161111
Shin, N.-R., Shin, Y.-H., Kim, H.-S., Park, Y.-D. (2022). Function analysis of the PR55/B gene related to self-incompatibility in Chinese cabbage using CRISPR/Cas9. IJMS 23, 5062. doi: 10.3390/ijms23095062
Song, Q., Gong, W., Yu, X., Ji, K., Chang, Y., Wang, L., et al. (2022). Integrative analysis of the metabolome and transcriptome provides novel insights into the mechanisms of flavonoid biosynthesis in camellia lanceoleosa. Scientia Hortic. 304, 111357. doi: 10.1016/j.scienta.2022.111357
Sun, C.-Q., Chen, F.-D., Teng, N.-J., Yao, Y.-M., Shan, X., Dai, Z.-L. (2019). Transcriptomic and proteomic analysis reveals mechanisms of low pollen-pistil compatibility during water lily cross breeding. BMC Plant Biol. 19, 542. doi: 10.1186/s12870-019-2166-3
Wang, Y. (2019). A cotton (Gossypium hirsutum) WRKY transcription factor (GhWRKY22) participates in regulating anther/pollen development. Plant Physiol. Biochem. 9 141, 231–239. doi: 10.1016/j.plaphy.2019.06.005
Wang, L., Peng, H., Ge, T., Liu, T., Hou, X., Li, Y. (2014). Identification of differentially accumulating pistil proteins associated with self-incompatibility of non-heading Chinese cabbage. Plant Biol. J. 16, 49–57. doi: 10.1111/plb.12016
Wheeler, M. J., de Graaf, B. H. J., Hadjiosif, N., Perry, R. M., Poulter, N. S., Osman, K., et al. (2009). Identification of the pollen self-incompatibility determinant in papaver rhoeas. Nature 459, 992–995. doi: 10.1038/nature08027
Wu, L., Li, J., Li, Z., Zhang, F., Tan, X. (2020a). Transcriptomic analyses of camellia oleifera ‘Huaxin’ leaf reveal candidate genes related to long-term cold stress. IJMS 21, 846. doi: 10.3390/ijms21030846
Wu, L., Williams, J. S., Sun, L., Kao, T. (2020b). Sequence analysis of the Petunia inflata s -locus region containing 17 S-locus f-box genes and the S-RNase gene involved in self-incompatibility. Plant J. 104, 1348–1368. doi: 10.1111/tpj.15005
Yuan, G., Zou, T., Zhang, X., Liu, M., Luo, T., He, Z., et al. (2020). A rice GDSL esterase/lipase protein (GELP) is required for anther and pollen development. Mol. Breed. 40, 90. doi: 10.1007/s11032-020-01170-4
Zhang, F., Li, Z., Zhou, J., Gu, Y., Tan, X. (2021). Comparative study on fruit development and oil synthesis in two cultivars of camellia oleifera. BMC Plant Biol. 21, 348. doi: 10.1186/s12870-021-03114-2
Zhang, C., Wang, P., Tang, D., Yang, Z., Lu, F., Qi, J., et al. (2019). The genetic basis of inbreeding depression in potato. Nat. Genet. 51, 374–378. doi: 10.1038/s41588-018-0319-1
Zhang, C.-C., Wang, L.-Y., Wei, K., Wu, L.-Y., Li, H.-L., Zhang, F., et al. (2016). Transcriptome analysis reveals self-incompatibility in the tea plant (Camellia sinensis) might be under gametophytic control. BMC Genomics 17, 359. doi: 10.1186/s12864-016-2703-5
Zhao, Z., Lou, Z., Yuan, S., Mei, L., Zhang, D. (2019) Global transcriptome and gene co-expression network analyses on the development of distyly in primula oreodoxa. HEREDITY 123, 784-794. doi: 10.1038/s41437-019-0250-y
Zhao, W., Qu, X., Zhuang, Y., Wang, L., Bosch, M., Franklin-Tong, V. E., et al. (2020). Villin controls the formation and enlargement of punctate actin foci in pollen tubes. J. Cell Science jcs 133, 6. doi: 10.1242/jcs.237404
Zhao, H., Song, Y., Li, J., Zhang, Y., Huang, H., Li, Q., et al. (2021). Primary restriction of s-RNase cytotoxicity by a stepwise ubiquitination and degradation pathway in Petunia hybrida. New Phytol. 231, 1249–1264. doi: 10.1111/nph.17438
Zhao, H., Zhang, Y., Zhang, H., Song, Y., Zhao, F., Zhang, Y., et al. (2022). Origin, loss, and regain of self-incompatibility in angiosperms. Plant Cell 34, 579–596. doi: 10.1093/plcell/koab266
Zhao, P., Zhang, L., Zhao, L. (2015). Dissection of the style’s response to pollination using transcriptome profiling in self-compatible (Solanum pimpinellifolium) and self-incompatible (Solanum chilense) tomato species. BMC Plant Biol. 15, 119. doi: 10.1186/s12870-015-0492-7
Zhou, J., Lu, M., Yu, S., Liu, Y., Yang, J., Tan, X. (2020). In-depth understanding of camellia oleifera self-incompatibility by comparative transcriptome, proteome and metabolome. IJMS 21, 1600. doi: 10.3390/ijms21051600
Zhou, Q., Zheng, Y. (2015). Comparative De novo transcriptome analysis of fertilized ovules in xanthoceras sorbifolium uncovered a pool of genes expressed specifically or preferentially in the selfed ovule that are potentially involved in late-acting self-incompatibility. PloS One 10, e0140507. doi: 10.1371/journal.pone.0140507
Zhu, S., Fu, Q., Xu, F., Zheng, H., Yu, F. (2021). New paradigms in cell adaptation: decades of discoveries on the Cr RLK1L receptor kinase signalling network. New Phytol. 232, 1168–1183. doi: 10.1111/nph.17683
Keywords: camellia oleifera, late-acting self-incompatibility, transcriptome, pollination, hub genes
Citation: Li C, Long Y, Lu M, Zhou J, Wang S, Xu Y and Tan X (2023) Gene coexpression analysis reveals key pathways and hub genes related to late-acting self-incompatibility in Camellia oleifera. Front. Plant Sci. 13:1065872. doi: 10.3389/fpls.2022.1065872
Received: 10 October 2022; Accepted: 29 December 2022;
Published: 24 January 2023.
Edited by:
Guang Sheng Zhou, Huazhong Agricultural University, ChinaReviewed by:
Vinay Randhawa, Brigham and Women’s Hospital and Harvard Medical School, United StatesHai Du, Southwest University, China
Copyright © 2023 Li, Long, Lu, Zhou, Wang, Xu and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junqin Zhou, emhvdWp1bnFpbkBjc3VmdC5lZHUuY24=; Xiaofeng Tan, dGFueGlhb2ZlbmdjbkAxMjYuY29t
†These authors have contributed equally to this work
 Chang Li
Chang Li Yi Long1,2†
Yi Long1,2†