- 1Dipartimento di Scienze Agrarie e Forestali (DAFNE), Università della Tuscia, Viterbo, Italy
- 2Dipartimento di Scienze Agrarie, Forestali e Alimentari (DISAFA), Università degli Studi di Torino, Grugliasco, Italy
Among the New Plant Breeding Techniques (NPBTs), the CRISPR/Cas9 system represents a useful tool for target gene editing, improving the traits of the plants rapidly. This technology allows targeting one or more sequences simultaneously, as well as introducing new genetic variations by homology-directed recombination. However, the technology of CRISPR/Cas9 remains a challenge for some polyploid woody species, since all the different alleles for which the mutation is required must be simultaneously targeted. In this work we describe improved protocols adapting the CRISPR/Cas9 system to highbush blueberry (Vaccinium corymbosum L.), using Agrobacterium-mediated transformation. As a proof of concept, we targeted the gene encoding for phytoene desaturase, whose mutation disrupts chlorophyll biosynthesis allowing for the visual assessment of knockout efficiency. Leaf explants of in vitro-cultured blueberry cv. Berkeley has been transformed with a CRISPR/Cas9 construct containing two guide RNAs (gRNA1 and gRNA2) targeting two conserved gene regions of pds and subsequently maintained on a selection medium enriched with kanamycin. After 4 weeks in culture on the selection medium, the kanamycin-resistant lines were isolated, and the genotyping of these lines through Sanger sequencing revealed successful gene editing. Some of mutant shoot lines included albino phenotypes, even if the editing efficiencies were quite low for both gRNAs, ranging between 2.1 and 9.6% for gRNA1 and 3.0 and 23.8 for gRNA2. Here we showed a very effective adventitious shoot regeneration protocol for the commercial cultivar of highbush blueberry “Berkeley”, and a further improvement in the use of CRISPR/Cas9 system in Vaccinium corymbosum L., opening the way to the breeding mediated by biotechnological approaches.
Introduction
Blueberry plants are species with expanding production worldwide, and a commercially valuable minor fruit crop in Italy (especially highbush blueberry) since it is a much sought-after species to differentiate crops, utilizes marginal areas and meets the growing consumer demand (Marangelli et al., 2022)., Moreover it is interesting for both fresh market or to be processed as a food ingredients but also for pharmaceutical and cosmetics industries, since it represents an important source of antioxidants.
Blueberry fruits are used for fresh consumption to process for juices, preserves and jams, and even for the pharmaceutical and cosmetics industries, since they are one of the richest sources of antioxidant metabolites which possess a high potential to reduce the incidence of several degenerative diseases (Ferrao et al., 2021). The genetic engineering in blueberry has also been studied in depth, and many successful works have been reported describing novel transgenic traits such as freezing-tolerance obtained by the overexpression of Vcddf1 gene (Walworth and Song, 2018), trans-grafting by an induced constitutive expression of flowering locus T gene (Song et al., 2019), increase of blueberry yield obtained by manipulating the expression of Soc1-MADS-Box (Song and Chen, 2018), and an interesting transgenic line which overexpresses the gene Vcrr2, useful to study the dormancy in highbush blueberry (Lin et al., 2019; Surridge, 2019).
Among the New Plant Breeding Techniques (NPBTs), the CRISPR/Cas9 has become part of the commonly used methodologies for obtaining new varieties (Bortesi and Fischer, 2015; Kim et al., 2021), has the advantage of accelerating and enhancing classical breeding, and it could be very interesting also for genetic improvement of highbush blueberry. Very little information about CRISPR/Cas9 and V. corymbosum L. has been reported. The first work reported the knockout of the Centroradialis (Cen) gene (Omori et al., 2021), and the second one studied the editing efficiency of CRISPR/Cas9 and CRISPR/Cas12a, both driven by a 35S promoter or AtUbi promoter, on a marker gene (gusA), previously introduced in transgenic lines as a single copy (Han et al., 2022). The last work demonstrated that the Cas-12a was not able to edit and that the factors to be considered during the process are numerous (pH, tissue culture conditions, temperature, etc.). Furthermore, highbush blueberry is a polyploid crop; traditional breeding often takes advantage of polyploidy. However, some methods such as random mutation breeding and genome editing are more difficult to be applied due to the presence of multiple homoeologous copies (or alleles) of each gene (Schaart et al., 2021). Hence, in polyploid species such as Vaccinium corymbosum L., guide RNAs must be carefully designed, since they must target all the different alleles simultaneously for which the mutation is required (Schaart et al., 2021). Therefore the application of editing technologies has also already been applied to many polyploid crops, including herbaceous and woody species (Zaman et al., 2019; Schaart et al., 2021). The number of new manuscripts published increased rapidly (Corte et al., 2019; Zhou et al., 2020; Fizikova et al., 2021; Karmakar et al., 2022; Liu et al., 2022; Nerkar et al., 2022; Parsaeimehr et al., 2022; Rao et al., 2022).
Phytoene desaturase (pds), a gene involved in carotenoid biosynthesis, has been targeted in most gene editing studies. Pds may be used as a visible marker to demonstrate specific genome editing effects (Savadi et al., 2021) since its knockout in plants results in albino phenotypes due to the absence of chlorophylls. CRISPR/Cas9-mediated pds gene knock-out has been found to confer an albino phenotype in many fruit crops such as grape (Nakajima et al., 2017), chestnut (2022; Pavese et al., 2021), strawberry (Wilson et al., 2019), cavendish banana (Naim et al., 2018; Ntui et al., 2020), apple (Nishitani et al., 2016) and pear (Charrier et al., 2019; Malabarba et al., 2021).
In this study, we reported the establishment of a CRISPR/Cas9-based transformation protocol through de novo shoot organogenesis in highbush blueberry by targeting the Vcpds endogenous reporter gene as a proof-of-concept of gene editing. The methodology here reported represents a suitable platform for the application of CRISPR/Cas9 technology in highbush blueberry.
Materials and methods
Plant material
The plant material used in our study were shoots of the cv Berkeley, cultivated on proliferation medium (PM) consisting of WPM basal medium including vitamins (Lloyd and McCown, 1980), 0.2 mgL-1 of L-glutamine, 3% (w/v) sucrose 3%, and 2.28 µM of zeatin trans-isomers (Duchefa, NL) (pH 5.6, gelled with 0.7% w/v agar - B&V, Italy) and grown under a 16 h photoperiod, provided by Fluora L58W/77 lamps (Osram, Germany), at photosynthetic photon lux (PPF) of 40 μmol m−2 s−1 and at 24 ± 1°C.
Mining of pds in the Vaccinium genome and phylogenetic analysis
Vaccinium myrtillus pds (Vmpds) coding sequence was detected from the Genome Database for Vaccinium (GDV; https://www.vaccinium.org/ accessed on 1st december 2021). The PROSITE database (https://prosite.expasy.org/ accessed on 20th january 2022) was then used to analyze and annotate Vmpds functional domains. Subsequently, using NGPhilogeny online tool (https://ngphylogeny.fr, accessed on 31st july 2022), the Vmpds coding sequence was aligned using MAFFT Alignment with 44 other plant pds coding sequence (Supplementary File S1), available on NCBI database, to produce the phylogenetic tree. The statistical significance of each branch was achieved by bootstrap analysis with 1,000 iterations. The phylogenetic tree was then visualized using PRESTO (Phylogenetic tReE viSualisaTiOn https://ngphylogeny.fr/data/displaytree, accessed on 31st January 2022) tool.
Vector design
To knock-out the Vcpds gene sequence, two specific guide RNA (gRNAs) (Table 1) were designed on the CDS of Vaccinium myrtillus pds, available on the Genome Database for Vaccinium, using CRISPR-P 2.0 online tool (http://crispr.hzau.edu.cn/CRISPR2, accessed on 31st january 2022).
Two pairs of primers (Table 2) designed on V. myrtillus pds gene were used to amplify V. corymbosum pds cv Berkeley to confirm the gRNAs sequences conservation among species.

Table 2 Primer sequences used for PCR of pds of Vaccinium corymbosum L. and for gRNAs flanking regions.
The gRNAs were then assembled into a CRISPR/Cas9 vector using the GoldenBraid 4.0 assembly system (Vázquez-Vilar et al., 2021). The Cas9 coding sequence was placed under the control of 35S promoter, the nptII gene for kanamycin resistance was under pNos promoter, while gRNAs were under pU6-26 promoter.
The final vector, named alpha_nptII_Cas9_gRNA1_gRNA2 (Supplementary File S2), was transferred into the Agrobacterium tumefaciens EHA105 strain (Hood et al., 1993) by using the freeze/thaw method (Xu and Li, 2008).
De novo shoot organogenesis and genetic transformation
Approximately 1144 leaf explants were excised from 3-week-old in vitro cultured shoots of cv. Berkeley and placed on 44 Petri dishes (26 leaf each one), following the protocol described by Marangelli et al. (2022). Briefly, the leaf explants were placed abaxial side up on the adventitious shoots organogenesis medium (SOM), consisting of WPM including vitamins, sucrose 3%, and 11.4 µM of zeatin riboside (ZR), (pH 5.6, gelled with 0.7% w/v agar - B&V, Italy), supplemented with 100 µM Acetosyringone (AS).
For genetic transformation experiments, A. tumefaciens EHA105-alpha1_NptII_Cas9_gRNA1_gRNA2 was grown overnight in Luria-Bertani (LB) selection medium supplemented with 50 mg L-1 kanamycin and 50 mg L-1 rifampicin at 28°C with shaking (100 rpm). Then, an aliquot (0.2-0.5 ml) of actively growing bacterial suspension was resuspended into 10 ml of LB (with kanamycin and rifampicin), under the same conditions, until an OD600 = 0.6 was reached. In a next step, centrifugation was used to recover the bacteria (4,000 rpm for 15 min) and the pellet was then resuspended in 10 ml of hormone-free SOM supplemented with 100 µM AS.
After 24 h in dark conditions at 28°C, a few drops of the Agrobacterium suspension were poured over the leaves and, after 10-15 minutes, the excess fluid was removed by absorption with sterile paper discs. The explants were then co-cultured with the engineered Agrobacterium EHA105 on the same medium (SOM + 100 µM AS) for 48, 72 and 96 hours, in dark conditions at 24°C.
After co-cultivation, half of the explants were placed in the same medium above described supplemented with 150 mg L-1 Cefotaxime (Cfx) alone, or in combination with 50 mg L-1 kanamycin (Kan). The same media have been used to induce adventitious shoot organogenesis on untreated leaf explants (as control). Since for the explants cultured on the medium enriched with kanamycin, no organogenesis occurs, all the explants have been transferred to the SOM medium containing 200 mg L-1 cefotaxime alone. The plates with the organogenetic tissues were checked daily for possible contaminations due to Agrobacterium strain used or other contaminants, and to check for the appearance of any mutant phenotypes. Throughout the experiment, regenerated shoots showing mutant phenotypes were isolated and after six weeks from the start of the experiments, all the regenerated shoots have been transferred to a PM supplemented with 200 mg L-1 Cfx and 50 mg L-1 Kan. Some of the mutants have been selected for molecular analysis.
Scanning of electron microscopy
To better understand when the shoot organogenesis occurs, early observations have been done by SEM analyses. Samples were fixed overnight at 4°C with 2.5% (v/v) glutaraldehyde + 2% (v/v) paraformaldehyde in 0.1 M cacodylate buffer, pH 7.2. After 3x20 min washings at 4°C in the same buffer, samples were post-fixed with 2% (v/v) osmium tetroxide in 0.1M cacodylate buffer, pH 7.2 for 2 h at 4°C. Specimens were washed in the same buffer (3 changes for 15 min each at 4°C), and then dehydrated in a graded ethanol series. Samples were dried by the critical point method using CO2 in a Balzers Union CPD 020. Then samples were attached to aluminum stubs using carbon tape and sputter-coated with gold in a Balzers MED 010 unit. The observations were made by a JEOL JSM 6010LA electron microscope.
Molecular analysis of kanamycin-resistant mutants
Genomic DNAs were extracted from 0.1 g of plant tissue obtained through transformation with nptII_Cas9_pds_gRNA1_gRNA2 vector using EZNA® Plant DNA kit (Omega Bio-tek, Norcross, GA, USA). The kanamycin-resistant shoots include not only the albino phenotype but, in general, all the shoots showing an atypical phenotype and/or resistance to the presence of kanamycin in the growing medium.
Mutation frequencies at the target sites were evaluated through PCR amplification using primers designed on gRNAs flanking regions (Table 2). DNA was amplified using KAPA HIFI Taq (KapaBiosystems, Roche, Basel, Switzerland) and the following PCR program: 95°C/3 min, followed by 30 cycles of 98°C/20 sec, 60°C/20 sec, 72°C/45 sec and 72°C/3 min. The PCR products were purified using DNA/RNA Clean Up E.Z.N.A.® kit (Omega Bio-tek, Norcross, GA, USA). Samples were sequenced using the Sanger method and the chromatograms obtained were analyzed using the TIDE online software (https://tide.deskgen.com, accessed on 31st July 2022).
Statistical analysis
The data was subjected to analysis of variance (ANOVA) performed using the software SPSS version 20.0. Percentage data were transformed as the arcsin of the square root of p/100 before analysis of variance. Data were presented as means of three replications at least. Significance was determined using Duncan’s test (p < 0.05).
Results and discussion
Gene structure and phylogenetic analysis
The Vmpds coding sequence is 1,737 bp long, and the protein size is 578 amino acids. Within the protein sequence a single amino_oxidase domain is present (Figure 1A). The amino_oxidase domain starts at the amino acid position 49 and ends at position 274 and has the molecular function of catalyzing oxidation-reduction (redox) reactions. Available full-length NCBI pds coding sequence orthologues (Supplementary File 1) were used for phylogenetic tree construction, and the resulting unrooted maximum-likelihood tree shown in Figure 1B suggests the phylogenetic relationship of gene sequence between V. myrtillus L. and Rhododendron kiusianum L. (94% bootstrap value). Both are species belonging to the Ericacee family, and they represent species-rich groups within the Ericacee (Rose et al., 2018). Affinity also emerged with Camellia sinensis L. and Diospyros kaki L. which belong to different families but from Ericales order.
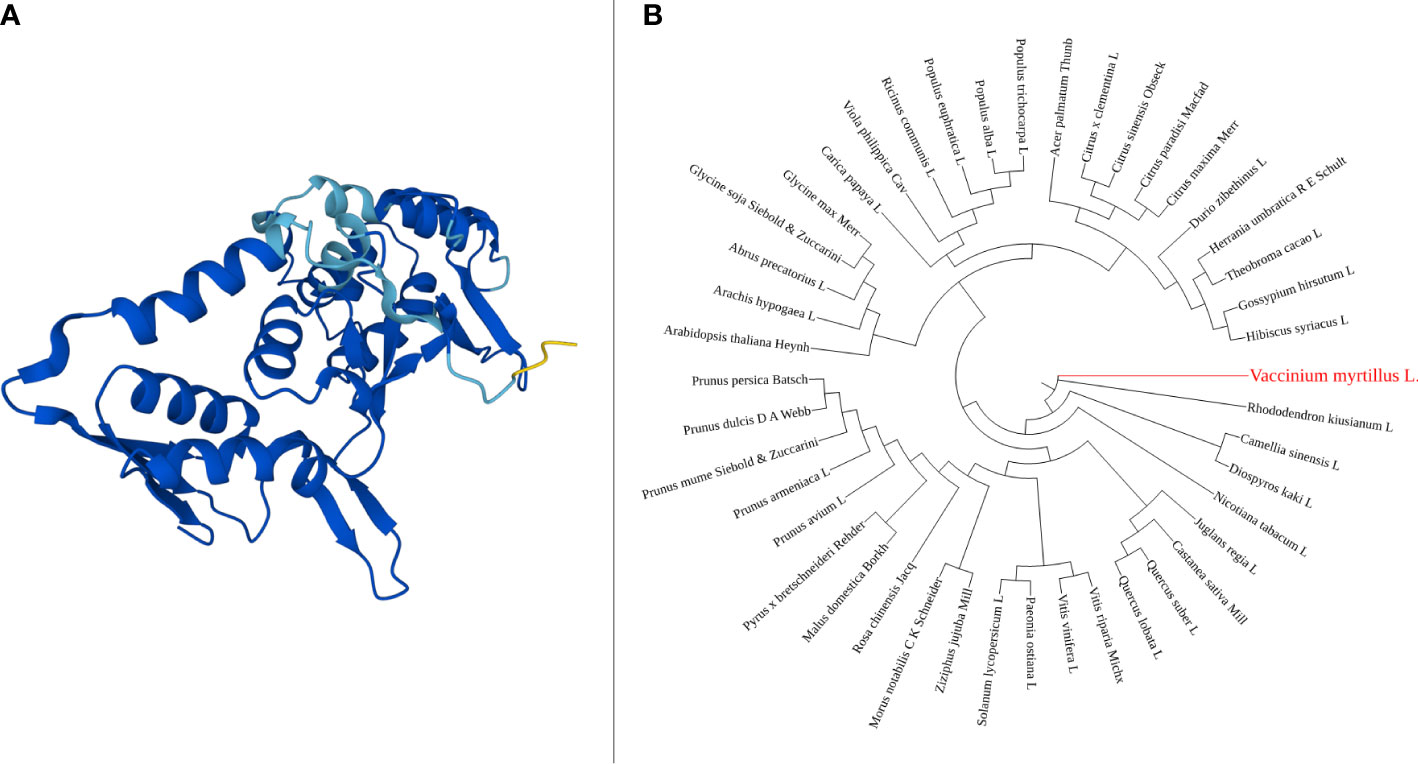
Figure 1 PDS amino_oxidase domain 3D structure, generated using AlphaFold online tool (A). Phylogenetic tree of pds gene, constructed using NGPhylogeny (B).
CRISPR/Cas9-mediated transformation of highbush blueberry
The presence of de novo shoot organogenesis was simultaneously observed just 5–6 days after the begin of the experiments on the control, in which single (Figure 2A) or grouped bud primordia (Figure 2B) were observed, confirming that the induction and differentiation phases in this species take place early as recently highlighted (Marangelli et al., 2022).
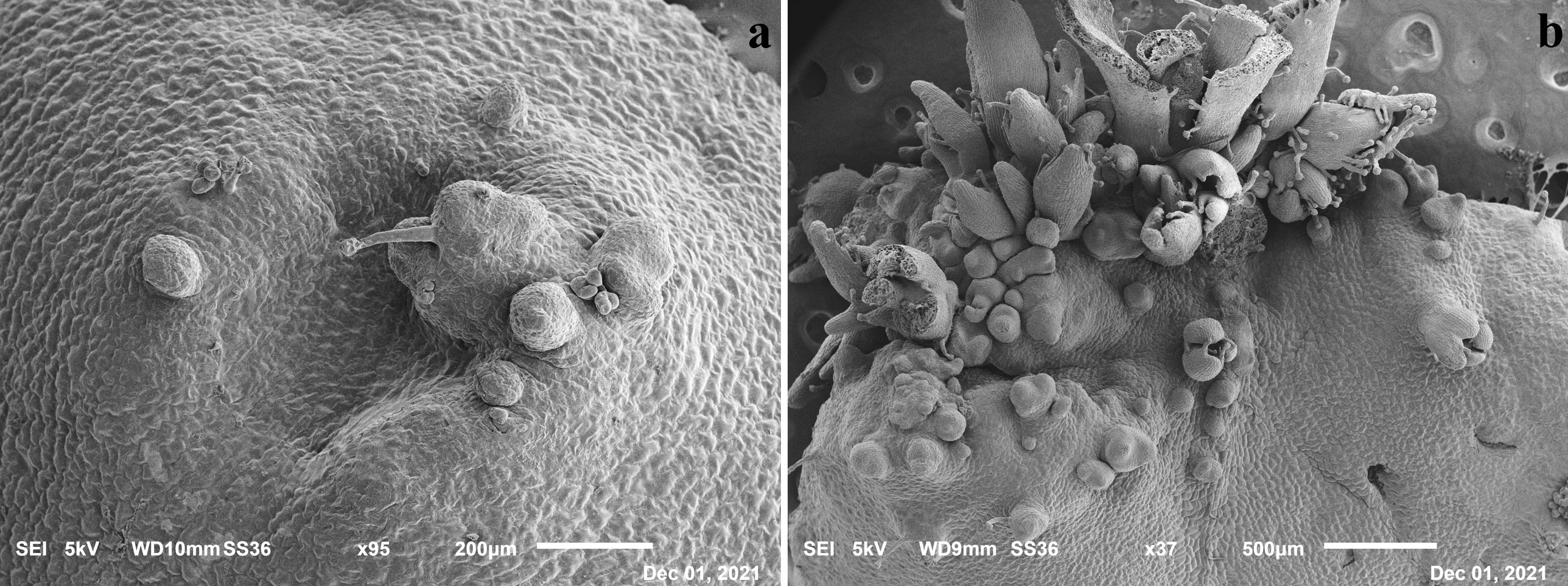
Figure 2 Scanning Electron Microscope (SEM) images of leaf tissues after 5 days in SOM medium with evidence of single (A) and grouped primordia (B).
Different from other species, shoot organogenesis did not occur frequently from the wounds but on the whole leaf surface, as also reported in Marangelli et al. (2022). These results were also observed by Liu et al. (2010), who described two different patterns during the de novo organogenesis processes of various cultivars of southern highbush blueberry: pattern I, where regenerants emerged mainly from the portion closer to the cutting area, and pattern II, where regeneration occurred directly from the entire leaf blade.
As shown in Table 3, a total of 936 explants has been used for genetic transformation, and 208 explants used as control (no co-culture with A. tumefaciens strain). Furthermore, in a preliminary experiment we also tested the effect of cefotaxime on the regeneration potentiality of highbush blueberry leaves cv. Berkeley (data not shown). The results of the experiment display that the presence of Cfx at 150 mg L-1 does not affect the regeneration process. As previously observed some antibiotics can affect morphogenesis, while kanamycin exerts a negative effect in some species, including in highbush blueberry, a positive effect has been highlighted when β-lactams such as cefotaxime are added to the regeneration media (Bosela, 2009; Silvestri et al., 2016).
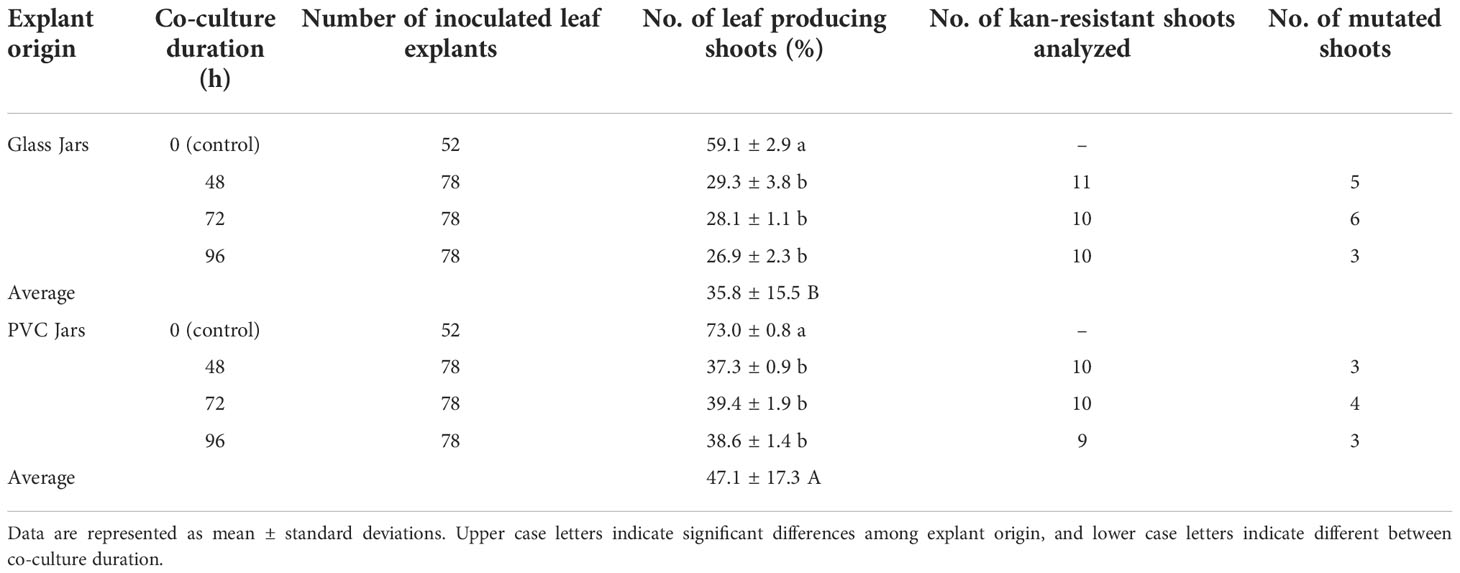
Table 3 Summary of Berkeley transformation (data considered only the explants regenerated on the medium containing only cefotaxime, because in the regeneration medium containing kanamycin, no regenerations occur).
After about ten days, when the first regenerations were observed in explants cultured on media enriched with only Cfx, no regeneration events were detected on the explants cultured on media also enriched with kanamycin. In general, a significant difference was observed in the regeneration ability of the explants derived from proliferation in glass jars or in PVC-jars, both in the controls and in the Agrobacterium-inoculated leaves. In fact, the average number of leaf-forming shoots was higher for the leaves harvested in PVC-Jars than the ones derived from glass jars (47.1 ± 17.3 vs 35.8 ± 15.5 respectively) (Table 3).
As confirmed by statistical analysis, co-culture with the bacteria resulted in a significant reduction in the regeneration efficiency of leaf explants, irrespective of both the origin (glass- or PVC jars) and the duration of co-culture (48, 72 and 96 hours). On the regeneration media containing only Cfx, the regenerating tissues produce numerous adventitious buds just after a couple of weeks (Figure 3A) which develop into tiny shoots (up to 30 shoots per explant) (Figure 3B). When they were transferred to the PM supplemented with 200 mg L-1 Cfx and 50 mg L-1 Kan, most of the explants continue to grow but, after two weeks, only the explants showing altered phenotypes were able to grow in kanamycin-enriched medium, while most of the shoots stop the growth and, in some cases, they rapidly turn brown. A broad phenotypic variation was observed among the regenerated shoots. In few cases, regenerated shoots showed the expected albino phenotype (Figures 3C, D), some shoots were characterized by a uniform red pigmentation (Figure 3E), and in many cases, instead, shoots with chimeric phenotype were observed, presenting green, white, and red colorations simultaneously (Figures 3F-I), emphasizing the difficulty to obtain fully edited shoots. It appears that explants exhibiting an albino phenotype showed stunted growth also if cultivated in in vitro conditions on an artificial medium. This characteristic could be ascribed to the fact that PDS represents not only an essential plant carotenoid biosynthetic enzyme and a prominent target of certain inhibitors, such as norflurazon, acting as bleaching herbicides (Koschmieder et al., 2017), but also an endogenous plant growth regulator, due to its role in gibberellin biosynthesis (Qin et al., 2007). Indeed, it has been demonstrated how PDS disruption could lead to a GA deficiency. Since the processes of adventitious organogenesis are particularly sensitive to hormonal balances, this aspect should be further investigated.
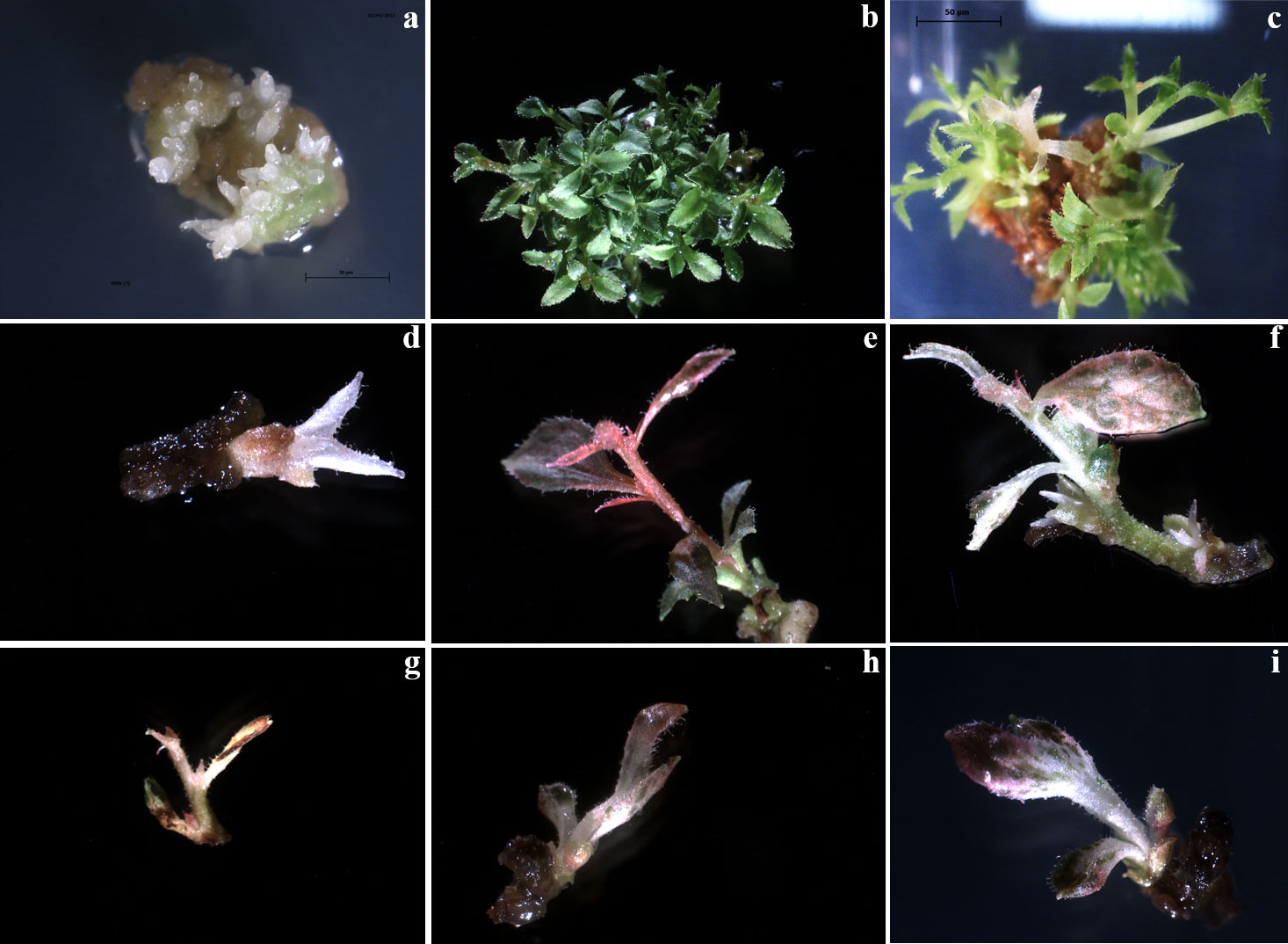
Figure 3 Editing of highbush blueberry cv Berkeley by knockout of pds gene with CRISPR/Cas9. (A) regenerating tissues produces numerous adventitious buds, (B) that rapidly evolve in tiny shoots (up to 30 shoots per explant). (C-D) regenerated shoots showed the expected albino phenotype. (E) shoot characterized by a red pigmentation and, (F, G, H, I) regenerated shoots showing chimeric phenotypes.
Mutant screening
To confirm the editing event, kanamycin resistant selected samples were sequenced through the Sanger sequencing and the resulting chromatograms were analyzed using the TIDE online software. The editing efficiencies were different among the two gRNAs (Table 4) ranging between 2.1 and 9.6% for gRNA1 and 3.0 and 23.8 for gRNA2. It reinforces the necessity to use different guides simultaneously to ensure an efficient gene knockout (Pavese et al., 2021).
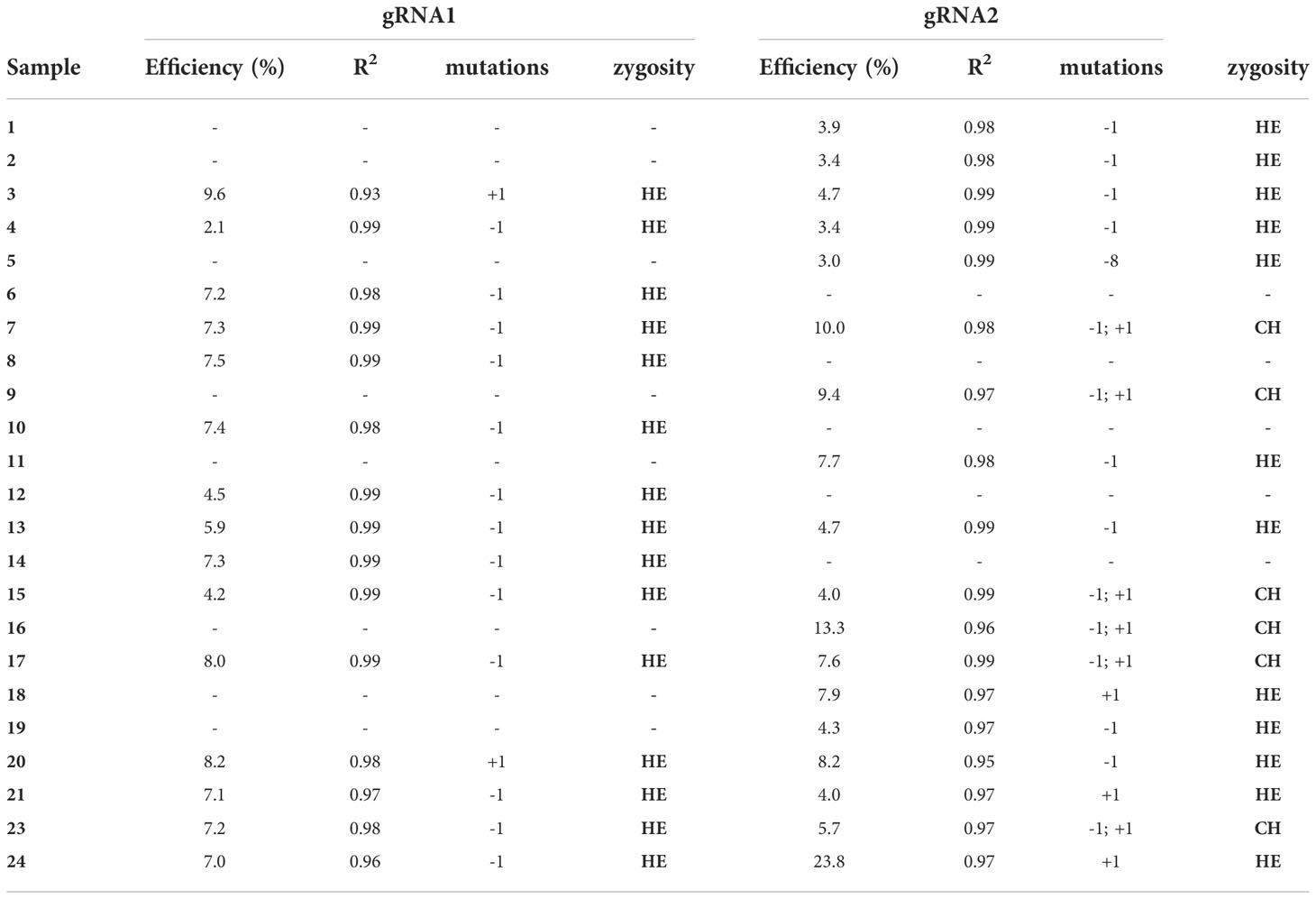
Table 4 Genotyping of targeted gene mutations induced by CRISPR/Cas9 in the transformed samples: editing efficiency, goodness-of-fit (R2), mutations and zygosity (HE – heterozygosity or CH – chimeric).
As reported in Table 4, of the 24 lines analyzed, 16 lines were edited for gRNA1 (66.6%) while 19 for gRNA2 (79%). While for gRNA1 only heterozygous mutants were observed, for gRNA2 6 out of 19 edited lines (31%) resulted chimeric. Our results are in agreement with the results reported by Pavese et al. (2021) for European chestnut, where they found almost all heterozygous lines, contrarily to other woody crops, in which most of the mutants were homologous and biallelic (Nishitani et al., 2016; Charrier et al., 2019; Dai et al., 2021). Chimeric genotypes represent one of the major concerns in genome editing, especially for heterozygous species such as perennial fruit crops characterized by high degree of heterozygosity, the length of their juvenile phase, the long generation times and auto-incompatibility systems (Ding et al., 2020). Anyway, this problem could be minimized through the second round of adventitious shoot organogenesis, as recently reported for highbush blueberry by Han et al. (2022).
The most common mutations observed in the edited lines were single insertions or deletions (+/-1 bp) and, only in one case (line 5) a -8 bp deletion was observed. Previous observations (Bortesi et al., 2016) showed that, in most cases, one bp insertions and deletions are the predominant ones, especially interesting T and A (Bortesi et al., 2016). Anyway, the type and size of mutations could vary considerably, probably due to different factors such as target sequences, vectors, and plant species (Jacobs et al., 2015; Xu et al., 2015; Pavese et al., 2021).
The editing efficiency we reported (up to 23.8%) is in line with the only two manuscripts so far published by other authors on highbush blueberry. The first work described the knockout of the Centroradialis (Cen) gene, selected because it is present in a single-copy gene in the polyploid genome of the highbush blueberry plant. The authors demonstrated for the first time the applicability of this technique to highbush blueberry, obtaining different edited lines with an average mutated allele ratio of 19-22%, on the basis of the selected gRNA. Nevertheless, the editing efficiency of fruit crops is very variable: up to 90% in apple (Osakabe et al., 2018; Charrier et al., 2019), less than 10% in callus of kiwifruit (Wang et al., 2018), 51% in Populus spp. (Fan et al., 2017), 31% in grape (Wang et al., 2018), 35% in orange (Peng et al., 2017) and higher than 90% in Castanea sativa Mill (Pavese et al., 2021).
In our experiments, a complete albino phenotype was observed even with low editing efficiency (23.8%) (Figure 4A), as well chimeric mutants showing green islands (Figure 4B) or anthocyanins (Figure 4C), that often appears with the albinism, as also observed in other species (Henderson et al., 2020). Anyway, the CRISPR/Cas9 in polyploids is more challenging, due to the presence of multiple homoeologous copies (or alleles) of each gene (Schaart et al., 2021), and low editing efficiency could be ascribed to different reasons. In fact, Wilson et al. (2019) demonstrated that by targeting pds gene in octoploid strawberry, some albino shoots were obtained also when wild-type pds alleles were still present and, in the same way, also they observed pale-green shoots containing the 11% of the wild-type alleles, assuming that probably just one allele is responsible for enzyme production and the others are non-functional alleles. Martin-Pizarro et al. (2019), by targeting FaTM6 gene to study the function of the MADS box transcription factor during flower development of Fragaria x ananassa obtained the expected phenotype with modified anthers even though none of the edited lines showed mutations in all present alleles. Similarly, Gao et al. (2020) targeted the Reduced Anthocyanins in Petioles (RAP) gene in octoploid strawberry, obtaining changes in fruit colour without the occurrence of the editing in all the alleles, confirming that the loss of functional alleles is a common phenomenon in allopolyploids (Wendel et al., 2018).

Figure 4 Edited phenotypes of highbush blueberry cv Berkeley. A developed albino phenotype (A), albino/chimeric phenotype showing island of green tissues (B) and an edited chimeric shoot showing red color due to the presence of anthocyanins (C).
Here we found a very effective adventitious shoot regeneration protocol for highbush blueberry, leading to the regeneration of up to 30 adventitious shoots from each leaf tissue. These results confirmed that zeatin and zeatin riboside are very useful to obtain high-efficient protocols for adventitious shoot organogenesis in some blueberry varieties (Rowland and Ogden, 1992; Cao and Hammerschlag, 2000; Marangelli et al., 2022).
Regarding the experiments of genetic transformation and genome editing, our results indicate that the knockout of pds gene by using CRISPR/Cas9 technology is efficient in highbush blueberry, opening the opportunity to the functional knockout of interesting candidate genes.
Conclusions
The CRISPR/Cas technology is representing the most powerful tool in the era of New Plant Breeding Techniques (NPBTs), able to speed up the breeding and genetic improvement of plants, especially nowadays, as the agriculture is facing the climate change accompanied by an increase of potential climate impacts, with the urgency to address and guide agricultural adaptation.
Our results represent an update of the genome editing technology applied to highbush blueberry, and the first example of the pds knockout gene in this species. The protocol is efficient and reliable, but further improvements are needed to make this technique a ready-to-use tool in the genetic improvement of this high-value species, by studying other important factors such as pH of the medium, culture conditions, light, temperature and considering the different response of the genotypes.
Moreover, chimerism is not necessarily a disadvantage, but it can become particularly attractive in species for which efficient protocols of adventitious organogenesis exist, where a second round of regeneration could eliminate the chimerism rate and increase the editing efficiency, as described by Han et al. (2022).
This protocol can be used for the genetic improvement of some important traits for the highbush blueberry, such as low chilling requirements, superior fruit quality (Ferrao et al., 2021), increased shelf-life and tolerance/resistance to the most important pests and diseases.
However, the breakthrough in the application of genome editing technology lies precisely in the possibility of producing transgene-free mutants. To avoid the stable integration of recombinant DNA, the CRISPR/Cas9 RNP approach combined with protoplasts can represent an innovative way to obtain transgene-free plants leading to potential better acceptance by consumers and opening at legislative level.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
CS, VP and AM: conceptualization. GV, VP and CS: data curation and writing—original draft preparation. GV, VP and CS: investigation. AM, CS and VC: supervision and writing—review and editing preparation. All authors contributed to the article and approved the submitted version.
Funding
The research was supported by funding of MIUR (Ministry for Education, University and Research), Law 232/2016, “Department of Excellence”.
Acknowledgments
The authors thank Antonella Minandri for her technical assistance and Anna Rita Taddei for assistance in using the scanning electron microscope. We do not endorse any of the companies or products mentioned; names of manufacturers and products are mentioned for information only.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1074541/full#supplementary-material
Supplementary File 1 | #pds coding sequences used for phylogenetic tree construction using NGPhylogeny (https://ngphylogeny.fr/, accessed on 31st July 2022).
Supplementary File 2 | Final vector containing gRNAs, hCas9 and nptII, assembled using GoldenBraid technology.
References
Bortesi, L., Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotech. Adv. 33 (1), 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Bortesi, L., Zhu, C., Zischewski, J., Perez, L., Bassié, L., Nadi, R., et al. (2016). Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol. J. 14, 2203–2216. doi: 10.1111/pbi.12634
Bosela, M. J. (2009). Effect of b-lactam antibiotics, auxins, and cytokinins on shoot regeneration from callus cultures of two hybrid aspens, Populus tremuloides x p. tremula and P. xcanescens x P. gradidentata. Plant Cell Tissue Organ 98, 249–261. doi: 10.1007/s11240-009-9555-5
Cao, X., Hammerschlag, F. A. (2000). Improved shoot organogenesis from leaf explants of highbush blueberry. HortSci 35, 945–947. doi: 10.21273/HORTSCI.35.5.945
Charrier, A., Vergne, E., Dousset, N., Richer, A., Petiteau, A., Chevreau, E. (2019). Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00040
Corte, L. E., Mahmoud, L. M., Moraes, T. S., Mou, Z., Grosser, J. W., Dutt, M. (2019). Development of improved fruit, vegetable, and ornamental crops using the CRISPR/cas9 genome editing technique. Plants 8 (12). doi: 10.3390/plants8120601
Dai, X., Yang, X., Wang, C., Fan, Y., Xin, S., Hua, Y., et al. (2021). CRISPR/Cas9-mediated genome editing in hevea brasiliensis. Ind. Crop Prod. 164, 113418. doi: 10.1016/j.indcrop.2021.113418
Ding, L., Chen, Y., Ma, Y., Wang, H., Wei, J. (2020). Effective reduction in chimeric mutants of poplar trees produced by CRISPR/Cas9 through a second round of shoot regeneration. Plant Biotechnol. Rep. 14, 549–558. doi: 10.1007/s11816-020-00629-2
Fan, S., Jian, D., Wei, X., Chen, J., Beeson, R. C., Zhou, Z., et al. (2017). Micropropagation of blueberry ‘Bluejay’ and ‘Pink lemonade’ through in vitro shoot culture. Sci. Hortic. 226, 277–284. doi: 10.1016/j.scienta.2017.08.052
Ferrao, L. F. V., Amadeu, R. R., Benvenuto, J., De Bem Oliveira, I., Munoz, P. R. (2021). Genomic selection in an outcrossing autotetraploid fruit crop: Lesson from blueberry breeding. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.676326
Fizikova, A., Tikhonova, N., Ukhatova, Y., Ivanov, R., Khlestkina, E. (2021). Applications of CRISPR/Cas9 system in vegetatively propagated fruit and berry crops. Agronomy 11 (9). doi: 10.3390/agronomy11091849
Gao, Q., Luo, H., Li, Y., Liu, Z., Kang, C. (2020). Genetic modulation of RAP alters fruit coloration in both wild and cultivated strawberry. Plant Biotechnol. J. 18, 1550–1561. doi: 10.1111/pbi.13317
Han, X., Yang, Y., Han, X., Ryner, J. T., Ahmed, E. A. H., Qi, Y., et al. (2022). CRISPR Cas9- and Cas12a- mediated gusA editing in transgenic blueberry. Plant Cell Tissue Organ Cult. 148, 217–229. doi: 10.1007/s11240-021-02177-1
Henderson, S. W., Henderson, S. T., Goetz, M., Koltunow, A. M. G. (2020). Efficient CRISPR/Cas9-mediated knockout of an endogenous PHYTOENE DESATURASE gene in T1 progeny of apomictic hieracium enables new strategies for apomixis gene identification. Genes 11, 1064. doi: 10.3390/genes11091064
Hood, E. E., Gelvin, S. B., Melchers, L. S., Hoekema, A. (1993). New agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2, 208–218. doi: 10.1007/BF01977351
Jacobs, T. B., Lafayette, P. R., Schmitz, R. J., Parrott, W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15, 16. doi: 10.1186/s12896-015-0131-2
Karmakar, S., Das, P., Panda, D., Xie, K., Baig, M. J., Molla, K. A. (2022). A detailed landscape of CRISPR-cas-mediated plant disease and pest management. Plant Sci. 323. doi: 10.1016/j.plantsci.2022.111376
Kim, Y. C., Kang, Y., Yang, E. Y., Cho, M. C., Schafleitner, R., Lee, J. H., et al. (2021). Applications and major achievements of genome editing in vegetable crops: A review. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.688980
Koschmieder, J., Fehling-Kaschek, M., Schaub, P., Ghisla, S., Brausemann, A., Timmer, J., et al. (2017). Plant-type phytoene desaturase: Functional evaluation of structural implications. PloS One 12 (11). doi: 10.1371/journal.pone.0187628
Lin, T. Y., Walworth, A., Zong, X. J., Danial, G. H., Tomaszewski, E. M., Callow, P., et al. (2019). VcRR2 regulates chilling-mediated flowering through expression of hormone genes in a transgenic blueberry mutant. Hortic-Res-England 96, 6. doi: 10.1038/s41438-019-0180-0
Liu, C., Callow, P., Rowland, L. J., Hancock, J. F., Song, G. (2010). Adventitious shoot regeneration from leaf explants of southern highbush blueberry cultivars. Plant Cell Tissue Organ Cult. 103, 137–144. doi: 10.1007/s11240-010-9755-z
Liu, Y., Zhang, C., Wang, X., Li, X., You, C. (2022). CRISPR/Cas9 technology and its application in horticultural crops. Hortic. Plant J. 8 (4), 395–407. doi: 10.1016/j.hpj.2022.04.007
Lloyd, G., McCown, B. (1980). Commercially-feasible micropropagation of mountain laurel, kalmia latifolia, by use of shoot-tip culture combined proceedings. Int. Plant Propagators’ Soc 30, 421–427.
Malabarba, J., Chevreau, E., Dousset, N., Veillet, F., Moizan, J., Vergne, E. (2021). New strategies to overcome present CRISPR/CAS9 limitations in apple and pear: Efficient dechimerization and base editing. Int. J. Mol. Sci. 22 (1), 1–20. doi: 10.3390/ijms22010319
Marangelli, F., Pavese, V., Vaia, G., Lupo, M., Bashir, M. A., Cristofori, V., et al. (2022). In vitro polyploid induction of highbush blueberry through De novo shoot organogenesis. Plants 11, 2349. doi: 10.3390/plants11182349
Martin-Pizarro, C., Trivino, J. C., Pose, D. (2019). Functional analysis of the TM6MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 70, 885–895. doi: 10.1093/jxb/ery400
Naim, F., Dugdale, B., Kleidon, J., Brinin, A., Shand, K., Waterhouse, P., et al. (2018). Gene editing the phytoene desaturase alleles of cavendish banana using CRISPR/Cas9. Transgenic Res. 27 (5), 451–460. doi: 10.1007/s11248-018-0083-0
Nakajima, I., Ban, Y., Azuma, A., Onoue, N., Moriguchi, T., Yamamoto, T., et al. (2017). CRISPR/Cas9-mediated targeted mutagenesis in grape. PloS One 12 (5). doi: 10.1371/journal.pone.0177966
Nerkar, G., Devarumath, S., Purankar, M., Kumar, A., Valarmathi, R., Devarumath, R., et al. (2022). Advances in crop breeding through precision genome editing. Front. Genet. 13. doi: 10.3389/fgene.2022.880195
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6, 31481. doi: 10.1038/srep31481
Ntui, V. O., Tripathi, J. N., Tripathi, L. (2020). Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa spp.). Curr. Plant Biol. 21. doi: 10.1016/j.cpb.2019.100128
Omori, M., Yamane, H., Osakabe, K., Osakabe, Y., Tao, R. (2021). Targeted mutagenesis of CENTRORADIALIS using CRISPR/Cas9 system through the improvement of genetic transformation efficiency of tetraploid highbush blueberry. J. Hortic. Sci. Biotechnol. 96, 153–161. doi: 10.1080/14620316.2020.1822760
Osakabe, Y., Liang, Z., Ren, C., Nishitani, C., Osakabe, K., Wada, M., et al. (2018). CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 13 (12), 2844–2863. doi: 10.1038/s41596-018-0067-9
Parsaeimehr, A., Ebirim, R. I., Ozbay, G. (2022). CRISPR-cas technology a new era in genomic engineering. Biotechnol. Rep. 34, e00731. doi: 10.1016/j.btre.2022.e00731
Pavese, V., Moglia, A., Abbà, S., Milani, A. M., Marinoni, D. T., Corredoira, E., et al. (2022). First report on genome editing via ribonucleoprotein (RNP) in Castanea sativa mill. Int. J. Mol. Sci. 23 (10). doi: 10.3390/ijms23105762
Pavese, V., Moglia, A., Corredoira, E., Martínez, M. T., Torello Marinoni, D., Botta, R. (2021). First report of CRISPR/Cas9 gene editing in castanea sativa mill. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.728516
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Qin, G., Gu, H., Ma, L., Peng, Y., Deng, X. W., Chen, Z., et al. (2007). Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17 (5), 471–482. doi: 10.1038/cr.2007.40
Rao, Y., Yang, X., Pan, C., Wang, C., Wang, K. (2022). Advance of clustered regularly interspaced short palindromic repeats-Cas9 system and its application in crop improvement. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.839001
Rose, J. P., Kleist, T. J., Löfstrand, S. D., Drew, B. T., Schönenberger, J., Sytsma, K. J. (2018). Phylogeny, historical biogeography, and diversification of angiosperm order ericales suggest ancient neotropical and east asian connections. Mol. Phylogenet. Evol. 122, 59–79. doi: 10.1016/j.ympev.2018.01.014
Rowland, L. J., Ogden, E. L. (1992). Use of a cytokinin conjugate for efficient shoot regeneration from leaf sections of highbush blueberry. HortSci 27, 1127–1129. doi: 10.21273/HORTSCI.27.10.1127
Savadi, S., Mangalassery, S., Sandesh, M. S. (2021). Advances in genomics and genome editing for breeding next generation of fruit and nut crops. Genomics 113 (6), 3718–3734. doi: 10.1016/j.ygeno.2021.09.001
Schaart, J. G., Van De Wiel, C. C. M., Smulders, M. J. M. (2021). Genome editing of polyploid crops: Prospects, achievements and bottlenecks. Transgenic Res. 30 (4), 337–351. doi: 10.1007/s11248-021-00251-0
Silvestri, C., Cristofori, V., Ceccarelli, M., Caceres, M. E., Escribà-Lacuesta, J., Rugini, E. (2016). Adventitious shoot organogenesis from leaf and petiole explants of European hazelnut. Plant Cell Tissue Organ Cult. 126 (1), 59–65. doi: 10.1007/s11240-016-0976-7
Song, G. Q., Chen, Q. (2018). Overexpression of the MADS-box gene K-domain increases the yield potential of blueberry. Plant Sci. 276, 10. doi: 10.1016/j.plantsci.2018.07.018
Song, G. Q., Walworth, G. Q., Walworth, A., Lin, T., Chen, Q., Han, X., et al. (2019). VcFT-induced mobile florigenic signals in transgenic and transgrafted blueberries. Hortic. Res. 6, 105. doi: 10.1038/s41438-019-0188-5
Surridge, C. (2019). Blueberry fooled into flowering. Nat. Plants 5, 910–910. doi: 10.1038/s41477-019-0520-y
Vázquez-Vilar, M., García-Carpintero, V., Selma, S., Bernabé-Orts, J. M., Sánchez-Vicente, J., Salazar-Sarasua, B., et al. (2021). The GB4.0 platform, an all-In-One tool for CRISPR/Cas-based multiplex genome engineering in plants. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.689937
Walworth, A., Song, G. Q. (2018). The cold-regulated genes of blueberry and their response to overexpression of VcDDF1 in several tissues. Int. J. Mol. Sci. 19. doi: 10.3390/ijms19061553
Wang, X., Tu, M., Wang, D., Liu, J., Li, Y., Li, Z., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16, 844–855. doi: 10.1111/pbi.12832
Wendel, J. F., Lisch, D., Hu, G. J., Mason, A. S. (2018). The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr. Opin. Genet. Dev. 49, 1–7. doi: 10.1016/j.gde.2018.01.004
Wilson, F. M., Harrison, K., Armitage, A. D., Simkin, A. J., Harrison, R. J. (2019). CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 15, 45. doi: 10.1186/s13007-019-0428-6
Xu, R., Li, Q. Q. (2008). Protocol: streamline cloning of genes into binary vectors in agrobacterium via the Gateway® TOPO vector system. Plant Methods 4, 4. doi: 10.1186/1746-4811-4-4
Xu, R. F., Li, H., Qin, R. Y., Li, J., Qiu, C.-H., Yang, Y.-C., et al. (2015). Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5, 11491. doi: 10.1038/srep11491
Zaman, Q. U., Li, C., Cheng, H., Hu, Q. (2019). Genome editing opens a new era of genetic improvement in polyploid crops. Crop J. 7 (2), 141–150. doi: 10.1016/j.cj.2018.07.004
Keywords: Vaccinium corymbosum L., gene editing, phytoene desaturase, gene loss-of-function, de novo shoot organogenesis
Citation: Vaia G, Pavese V, Moglia A, Cristofori V and Silvestri C (2022) Knockout of phytoene desaturase gene using CRISPR/Cas9 in highbush blueberry. Front. Plant Sci. 13:1074541. doi: 10.3389/fpls.2022.1074541
Received: 19 October 2022; Accepted: 28 November 2022;
Published: 15 December 2022.
Edited by:
Kaijun Zhao, Institute of Crop Sciences (CAAS), ChinaReviewed by:
Rukmini Mishra, Centurion University of Technology and Management, IndiaJunhua Peng, Huazhi Bio-Tech Co., Ltd., China
Copyright © 2022 Vaia, Pavese, Moglia, Cristofori and Silvestri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vera Pavese, dmVyYS5wYXZlc2VAdW5pdG8uaXQ=; Cristian Silvestri, c2lsdmVzdHJpLmNAdW5pdHVzLml0
 Giuseppe Vaia
Giuseppe Vaia Vera Pavese
Vera Pavese Andrea Moglia
Andrea Moglia Valerio Cristofori
Valerio Cristofori Cristian Silvestri
Cristian Silvestri