- Department of Biological Sciences, University of Alberta, Edmonton, AB, Canada
Nectaries are a promising frontier for plant evo-devo research, and are particularly fascinating given their diversity in form, position, and secretion methods across angiosperms. Emerging model systems permit investigations of the molecular basis for nectary development and nectar secretion across a range of taxa, which addresses fundamental questions about underlying parallelisms and convergence. Herein, we explore nectary development and nectar secretion in the emerging model taxa, Cleome violacea (Cleomaceae), which exhibits a prominent adaxial nectary. First, we characterized nectary anatomy and quantified nectar secretion to establish a foundation for quantitative and functional gene experiments. Next, we leveraged RNA-seq to establish gene expression profiles of nectaries across three key stages of development: pre-anthesis, anthesis, and post-fertilization. We then performed functional studies on five genes that were putatively involved in nectary and nectar formation: CvCRABSCLAW (CvCRC), CvAGAMOUS (CvAG), CvSHATTERPROOF (CvSHP), CvSWEET9, and a highly expressed but uncharacterized transcript. These experiments revealed a high degree of functional convergence to homologues from other core Eudicots, especially Arabidopsis. CvCRC, redundantly with CvAG and CvSHP, are required for nectary initiation. Concordantly, CvSWEET9 is essential for nectar formation and secretion, which indicates that the process is eccrine based in C. violacea. While demonstration of conservation is informative to our understanding of nectary evolution, questions remain. For example, it is unknown which genes are downstream of the developmental initiators CvCRC, CvAG, and CvSHP, or what role the TCP gene family plays in nectary initiation in this family. Further to this, we have initiated a characterization of associations between nectaries, yeast, and bacteria, but more research is required beyond establishing their presence. Cleome violacea is an excellent model for continued research into nectary development because of its conspicuous nectaries, short generation time, and close taxonomic distance to Arabidopsis.
1 Introduction
Flowers exhibit tremendous diversity of form, much of which is driven by plant-pollinator interactions. Responses to similar pollinator environments have resulted in repeated evolution of floral forms across angiosperms (reviewed in: (Endress, 2011; Sauquet et al., 2017; Wessinger and Hileman, 2020)). Such traits include, but are not limited to, monosymmetry (zygomorphy), organ fusion, spurs, and heterostyly (Specht and Howarth, 2015; Kramer, 2019; Phillips et al., 2020; Wessinger and Hileman, 2020). This repeated evolution raises fundamental questions about the developmental and genetic bases of their evolutionary shifts (Sobel and Streisfeld, 2013; Specht and Howarth, 2015; Kramer, 2019; Wessinger and Hileman, 2020). Among these questions is whether the same genetic pathways have been recruited in independent origins of these traits (Specht and Howarth, 2015; Wessinger and Hileman, 2020). Remarkable and repeated recruitment of the same genetic pathway is clear with certain traits, notably monosymmetry [reviewed in: (Preston et al., 2009; Preston et al., 2011; Hileman, 2014a; Hileman, 2014b; Wessinger and Hileman, 2020)], but whether the genetic basis of other features is conserved remains unclear.
Nectaries, and the nectar they secrete, are integral to plant-animal interactions and, as such, warrant detailed investigation across taxa (Liao et al., 2021). They have evolved multiple times across angiosperms and are remarkably variable in position, structure, and morphology (Bernardello, 2007; Nepi, 2007; Nepi et al., 2018; Liao et al., 2021; Slavkovic et al., 2021). Despite this variation, nectaries are united by the simple function of producing nectar, a complex sugar-rich solution that contains a wide range of metabolites and microbes (Nepi, 2007; Heil, 2011; Nepi et al., 2018; Slavkovic et al., 2021; Liao et al., 2021). As a critical reward to insects, and potential attractor, nectaries and their nectar drive many macroevolutionary patterns via relationships with pollinators and other animals (Parachnowitsch et al., 2019; Liao et al., 2021). Nectaries are associated with all plant organs except for roots, and floral nectaries can be associated with any floral organ (Nepi, 2007; Liao et al., 2021). Nectary morphology can be structured (i.e., distinct morphology with identifiable cell types) or unstructured (i.e., no specialized morphology) (Nepi, 2007; Slavkovic et al., 2021). Nectar secretion ranges from modified stomata (nectarostomata), to specialized trichomes, and even cell rupture (reviewed in: (Nepi, 2007; Slavkovic et al., 2021)). This diversity in morphology and secretion mechanisms differs across families and within genera (Bernardello, 2007). Also, variable nectar composition impacts pollinator interactions (Nepi et al., 2018; Parachnowitsch et al., 2019). This extensive diversity calls into question whether nectary development is underpinned by similar or different developmental programs in taxa with variable nectaries.
A genetic breakthrough in nectary research was the establishment of CRABS CLAW (CRC), a YABBY family transcription factor, as essential for nectary initiation (Alvarez and Smyth, 1998; Bowman and Smyth, 1999; Lee et al., 2005a). In Arabidopsis, CRC knockouts do not develop nectaries (Bowman and Smyth, 1999). CRC has since been shown as essential for nectary formation across the core eudicots (Lee et al., 2005b; Fourquin et al., 2014), and is expressed in extrafloral nectaries (Lee et al., 2005b). CRC protein dimerizes with other YABBY transcription factors and also has an important role in Arabidopsis carpel development (Alvarez and Smyth, 1998; Alvarez and Smyth, 1999; Alvarez and Smyth, 2002; Lee et al., 2005a) that is widely conserved (Orashakova et al., 2009; Fourquin et al., 2014; Pfannebecker et al., 2017). The expression of CRC across core eudicot nectaries, regardless of morphology or position, suggests that CRC regulation of nectary development is consistent across the clade (Lee et al., 2005b). To the best of our knowledge, functional studies of nectaries have only been conducted in four core Eudicot taxa: Petunia (Solonales) (Lee et al., 2005b; Morel et al., 2018), Gossypium (Malvales) (Pei et al., 2021), Pisum (Fabales) (Fourquin et al., 2014), and Arabidopsis (Brassicales) (Bowman and Smyth, 1999). CRC is shown as essential for nectary development in the aforementioned taxa, except for Gossypium where the gene GoNe is required for both floral and extra floral nectaries (Pei et al., 2021). Thus, investigations of additional taxa are needed to uncover the extent of this potential conserved role of CRC.
The role of CRC as essential for nectary development does not extend beyond the core eudicots. For example, all petals of Aquilegia have elongated spurs, which bear nectaries in their distal tips. In this taxa, three STYLISH (STY) homologs, a member of the SHORT INTERNODES (SHI) gene family, are redundantly necessary for the formation of nectaries in the spurs as well as style development (Min et al., 2019). Thus, both CRC and STY are involved in nectary and gynoecial development (Pfannebecker et al., 2017), which raises questions about developmental pathways shared between nectaries and carpels.
Upstream regulators of CRC are also shared between Petunia and Arabidopsis (Morel et al., 2018). CRC is insufficient for ectopic nectary formation (Baum et al., 2001), which reveals a necessity for upstream regulators. In Arabidopsis, these regulators include ABC(E) class genes APETALA2/3 (AP2/3), PISTILLATA (PI), AGAMOUS (AG), and SEPALLATA1/2/3 (SEP1/2/3), as well as MADS-box gene SHATTERPROOF 1/2 (SHP1/2) (Reviewed in: (Slavkovic et al., 2021)). In sum, SHP1/2 and AG act redundantly to promote CRC, such that knockouts of each one alone does not prevent nectary formation, although combined they do (Lee et al., 2005a). The floral meristem identity genes LEAFY (LFY) and UNUSUAL FLORAL ORGANS (UFO) are also upstream of CRC and function to restrict CRC expression to nectaries and carpels (Bowman and Smyth, 1999; Slavkovic et al., 2021). Loss of SEP1/2/3 also prevents nectaries from developing (Lee et al., 2005a). Individual knockouts of any aforementioned gene do not prevent nectary formation, although they can impact shape and size (e.g., lfy, ufo, pi, ag) (Baum et al., 2001). Double and triple knockouts however cause a loss of nectaries (e.g., lfy & ufo, sep1/2/3) (reviewed in: (Slavkovic et al., 2021)). Nectary inhibition may be indirect because meristem identity genes act upstream of ABC class genes, i.e., nectary formation may be halted because their associated organs fail to form. In Petunia, nectary formation is also dependent on C class genes, i.e., euAG and PLEN are essential for nectary formation (Morel et al., 2018). This redundancy of MADS-box genes implies that the entire regulatory pathway was established prior to the Rosid/Asterid split (Morel et al., 2018; Slavkovic et al., 2021).
Beyond nectary formation, genes have been identified that are important for nectary size and growth. In Petunia, two euAP2 genes, BLIND ENHANCER (BEN) and REPRESSOR OF B FUNCTION (ROB), impact floral nectary size such that rob1 rob2 rob3 triple mutants have flowers with larger nectaries than wildtype (Morel et al., 2018). This phenotype is enhanced when BEN is also knocked out, such that much of the carpel is converted to nectary tissue (Morel et al., 2018). Whereas in Arabidopsis, BLADE ON PETIOLE 1/2 (BOP1/2) are essential for nectary growth independent of CRC (Mckim et al., 2008). Knockouts of BOP1/2 result in nectaries that are small and not fully differentiated into parenchyma and secretory tissue (Mckim et al., 2008).
Phytohormones also play an important role in nectary development, composition, and secretion. AUXIN RESPONSE FACTOR 6/8 (ARF6/8) promote and coordinate nectary formation in Arabidopsis (Reeves et al., 2012) and Aquilegia (Zhang et al., 2020). Thus, while these taxa differ in which key regulator promotes nectary formation, they have a shared response to hormone signalling, which reflects the central role of plant hormones in floral evolution (Wessinger and Hileman, 2020). Auxin plays an additional role in nectar secretion via PIN FORMED 6 (PIN6) expression, which is positively correlated with nectar production (Bender et al., 2013). Also correlated with an increase in nectar production is jasmonic acid (JA), which peaks in concentration just prior to nectar secretion in Brassica napus (Radhika et al., 2010). Further, both auxin and JA are regulated by gibberellic acid (GA) (Reeves et al., 2012), which speaks to the complex interplay between auxin, JA and GA.
Investigations of additional taxa are critical for assessing not only the extent of the conserved role of CRC, but also how it is regulated, and the potential pathway deviations across taxa with different nectary shapes and positions. Towards addressing these outstanding questions, Cleomaceae is an excellent model for investigating floral development. Cleomaceae is a small, cosmopolitan family of circa 270 species placed in 25 genera (Bayat et al., 2018). This family houses floral variation in traits likely associated with pollinator interactions, including petal color, petal size, and gynophores/androgynophores (Iltis et al., 2011; Higuera-Diaz et al., 2015; Bayat et al., 2018). Importantly, members of the family exhibit a wide range of nectary size, shape, and position. Across the family, nectaries may be absent, adaxially positioned, or annular (Tucker and Vanderpool, 2010). Cleomaceae is sister to Brassicaceae and the phylogenetic framework within the family is established (Patchell et al., 2014; Barrett et al., 2017; Bayat et al., 2018). While some floral developmental patterns are described (Erbar and Leins, 1997b; Erbar and Leins, 1997a; Patchell et al., 2011), most information regarding nectaries is based on floristic work (e.g., (Tucker and Vanderpool, 2010)). There is also limited empirical information on pollinators, which has revealed generalist and specialist systems across the family (Cane, 2008; Fleming et al., 2009; Higuera-Diaz et al., 2015; Raju and Rani, 2016). Of note, functional approaches have been established for Cleome violacea (Carey et al., 2021). This species is amenable to investigations of nectaries as their flowers have prominent, 3-lobed nectaries adaxially positioned between petals and stamen (Figure 1).

Figure 1 Cleome violacea flowers at various stages of development. (A) Large undissected floral bud. (B) Large dissected floral bud showing nectary. (C) Newly anthetic flower. (D) Post-anthetic flower with developing fruit. (E) Magnified view of anthetic nectary. Scale bars = 1 mm.
The overarching goal of this study was to determine the genetic basis of floral nectaries of Cleome violacea. Towards this end, we first characterized nectary anatomy and nectar volume. Second, we conducted a detailed transcriptomic analysis of nectaries from pre-anthetic, anthetic, and post-anthetic (post-fertilization) flowers to document gene expression patterns during nectary development and assess possible convergences in underlying genetic pathways. Finally, we conducted functional studies on key genes to test their direct role and putative interactions in nectary development and nectar production.
2 Materials and methods
2.1 Plant growth conditions
Inbred lines of C. violacea were grown from lab seed stock. A voucher was deposited in the vascular plant herbarium at the University of Alberta (ALTA; Hall & Bolton s.n., 20 February 2008; #813 from Hortus Botanicus, Amsterdam). Seeds were sown individually in 7.5 cm diameter pots containing sterilized (20 min, liquid, 121.1°C) Sun Gro Sunshine Mix (Agawam, Massachusetts, USA). All plants were grown in a growth chamber at the University of Alberta, Department of Biological Sciences with 16 h of full spectrum LED light at 22°C and 8 h of darkness at 18°C.
2.2 Histology and scanning electron microscopy (SEM)
Inflorescence tips, small buds (<1 mm wide), medium buds (1-1.5 mm wide), large buds (2.5-3 mm wide), flowers, and post-anthesis flowers were collected and fixed in FAA solution (50% EtOH, 5% glacial acetic acid, 10% formalin, 35% MilliQ water) and vacuum infiltrated as outlined previously (Hall et al., 2006; Patchell et al., 2011). Plant samples were then dehydrated in an ethanol series (50% to 100%). All ethanol solutions were kept at 4°C and samples were incubated for 2 hours. The 100% ethanol solutions were left overnight. Samples were infiltrated with CitriSolv (Decon Labs, USA) by changing to a 1:1 ethanol:CitriSolv solution, then changing to 100% CitriSolv. Each CitriSolv solution was incubated for two hours at room temperature with shaking. Samples were infiltrated with Tissue-Prep paraplast (Leica Biosystems, Canada) with 2-3 changes daily for five days then embedded in paraplast. Samples were sectioned to 8 μm using a Microm HM 325 (GMI, Inc., Ramsey, MN, USA) microtome prior to mounting. Slides were cleared with CitriSolv and dehydrated in isopropanol before staining with 0.025% Alcian blue and 0.01% Safranin O in 0.1M acetate buffer for two hours. Slides were examined using a Nikon (Tokyo, Japan) Eclipse 801 microscope with a Nikon DS-Ri1 photo system.
Samples used for scanning electron microscopy (SEM) were fixed and dehydrated as indicated above. After dehydration, samples were critical point dried with carbon dioxide using a CPD 030 critical point dryer (Bal-Tec AG, Liechtenstein, Germany). Specimens were then dissected and mounted on scanning electron microscopy stubs with conductive carbon tabs and sputter coated with gold using a Hummer 6.2 sputter coater (Anatech USA, Sparks, Nevada, USA). Finally, specimens were imaged using a ZEISS EVO 10 scanning electron microscope (Carl Zeiss AG, Oberkochen, Germany). Contrast and brightness of micrographs were adjusted using GIMP version 2.10.18 (https://gimp.org).
2.3 Nectar volume
Nectar volume of C. violacea was measured by pooling nectar from all the open flowers on each plant (2-7 flowers) in a capillary tube (Morrant et al., 2009). This measurement was taken for 20 plants and repeated at the same time each day for four consecutive days. Only flowers with visible nectar were measured. Individual flowers typically senesce three days after anthesis and stop producing nectar. Average nectar volume was calculated for each day and then graphed. A student’s t-test was run for binary comparisons between day 1-4.
2.4 RNA isolation and cDNA library preparation
Nectaries were collected from C. violacea flowers at three stages of development: pre-anthetic (buds 2.5-3 mm wide), anthetic (first day of anthesis) and post-anthetic (fertilized flowers with fruits at approximately 10 mm in length). RNA from these three developmental stages of four biological replicates were extracted to provide 1) an overview of gene expression at the end of nectary development and 2) insight into how gene expression changes before, at, and after anthesis. Nectary tissue was excised, flash frozen in liquid nitrogen, and stored at -80°C. Following: Carey et al., (2019), RNA was extracted from manually-ground frozen tissue using a Qiagen RNeasy micro kit (Hilden, Germany) and cDNA was generated using the Illumina TruSeq stranded mRNA LT sample prep kit RS-122-2101 (California, U.S.). In this case, mRNA for each sample was isolated using nucleomag beads (Macherey-Nagel, Düren, Germany). Samples were sent to The Center for Applied Genetics (TCAG) at the Toronto Sick Kids Hospital in Ontario, Canada where they were normalized, pooled and sequenced on a HiSeq 2500.
2.5 De novo transcript assembly, differential expression, and annotation
Raw reads were downloaded from the TCAG webserver and processed as in (Carey et al., 2019) using updated software (Table S1). The raw reads are available at the Sequence Read Archive (SRA) database (BioProject: PRJNA912718). After differential expression analysis with edgeR (Robinson et al., 2009), transcripts were classified as significantly differentially expressed if they had a log2 (fold-change) greater than four and a False Discovery Rate (FDR)-corrected p-value (α) less than 0.001. The ‘analyze_diff_expr.pl’ script, provided with Trinity (Haas et al., 2013), was used to generate a matrix of all significantly differentially expressed contig clustered transcripts, which was then used to generate a z-score heatmap in R (R Core Team, 2013).
We performed an additional z-score analysis with trinity transcripts filtered using TransDecoder.LongOrfs and TransDecoder.Predict to remove potential misassemblies. In total, 81,151 of 143,919 transcripts remained. A list of the 81,151 transcripts was used to extract significant transcripts from the original matrix file produced by ‘analyze_diff_expr.pl’, as well as from a list of all transcripts with expression greater than 100 TPM. Additionally, any transcripts with one or more biological groupings below 10 TPM, or with a coefficient of variation greater than or equal to 50, were removed.
All transcripts from the larger (original) trinity fasta file were annotated using BLASTx (Altschul et al., 1990) with default parameters and a local copy of the Araport11 protein database. Transcripts with the highest bit-score from the TAIR database were used as representative transcripts. Gene specific heatmaps were generated using ggplot2 and ggplot in R (R Core Team, 2013), respectively. Assembly completeness was determined using Benchmarking Universal Single Copy Orthologs (BUSCO) (Simao et al., 2015), and an ExN90 profile.
Transcripts unique to each stage were uploaded to the KEGG automatic annotation server (KAAS) using the bi-directional best hit against the following organism databases: Arabidopsis thaliana (Brassicaceae), Brassica napa (Brassicaceae), and Tarenaya hassleriana (Cleomaceae). Transcripts were considered unique if their expression was ≥ 10 TPM with a coefficient of variation < 50. A list of all KEGG entries was compiled, excluding most human diseases and other mammalian-exclusive categories. Of note, some categories were kept because they are convergent with pathways in plants and/or yeast.
2.6 Virus-induced gene silencing (VIGS)
Viral vector constructs were designed following (Carey et al., 2021). Tobacco rattle virus vectors pTRV1 (donor stock no. YL192) and pTRV2 (donor stock no. YL156) were obtained from The Arabidopsis Information Resource (TAIR; https://www.arabidopsis.org) using their stock center (Arabidopsis Biological Resource Center [ABRC], Ohio State University, Columbus, Ohio, USA; https://abrc.osu.edu). The pTRV2 vector is used for downregulating genes of interest (Ratcliff et al., 2001) and the pTRV1 vector assists with viral movement (Ziegler-Graff et al., 1991). Six new endogenous constructs were generated for this study using C. violacea mRNA: pTRV2-CvANS, pTRV2-CvAG, pTRV2-CvAG-CvSHP, pTRV2-CvCRC-CvANS, pTRV2-CvSWEET9-CvANS, and pTRV2-DN802_c0_g1_i4-CvANS. The CvANS construct was used as a marker gene and positive control. Treatment with pTRV2-CvSHP was explored in a preliminary round of VIGS but produced no remarkable floral phenotype and, as such, was abandoned in future trials.
All constructs were generated as follows. All cDNA was synthesized following manufacturer instructions using SuperScript III Reverse Transcriptase (Invitrogen), poly(T) primers, and random hexamer primers. All primers were designed using the transcriptomic data from this study. All amplification was done using Invitrogen recombinant Taq DNA Polymerase (Waltham, Massachusetts, USA) using the manufacturer protocol, 50 μL reaction volumes, and 35 cycles. All amplicons were verified using agarose gel electrophoresis, and colonies were screened via PCR with primers, 156F and 156R that span the TRV2 multiple cloning site (Gould and Kramer, 2007). Manufacturer protocols were used for each step unless otherwise noted. First, a 533 bp insert of CvANS was amplified using forward and reverse primers with added BAMHI [G^GATCC] and XHOI [C^TCGAG] restriction sites, respectively. Amplicons were purified using a QIAquick PCR purification kit and digested alongside empty TRV2 vector with NEB BAMHI and XHOI restriction enzymes (Ipswich, Massachusetts, USA). Digests were purified using the Quantum Prep Freeze ‘N Squeeze DNA Gel Extraction protocol with 200 μL pipette tips and 2 mL tubes in lieu of spin columns. Eluate was further purified using ethanol precipitation (https://projects.iq.harvard.edu/hlalab/resources-0). Digests were ligated together using NEB T4 DNA ligase and immediately transformed using One Shot™ TOP10 Chemically Competent E. coli. Escherichia coli was incubated for 24 h at 37°C in Miller LB broth (Sigma-Aldrich, Burlington, Massachusetts, USA) containing 50 μg/mL kanamycin. The pTRV2-CvANS construct was verified using colony PCR, and colonies containing the appropriately sized plasmids were extracted using a GeneJET Plasmid Miniprep Kit (Thermo Fisher, Waltham, Massachusetts, USA), verified using agarose gel electrophoresis, and transformed into chemically competent Agrobacterium GV3101; cells were prepared and transformed according to protocol (Luo et al., 2008). All media used to grow Agrobacterium contained 50 μg/mL kanamycin, 50 μg/mL gentamycin, and 25 μg/mL rifampicin. Plasmids from transformed Agrobacterium were verified via restriction digestion and agarose gel electrophoresis, and finally sanger sequencing. Agrobacterium containing the appropriate pTRV2-CvANS vector were grown for 48 h at 28°C and mixed 1:1 with sterile 50% glycerol prior to storage at -80°C.
All other vectors were made following the same protocol. Amplicons from CvCRC, CvSWEET9, and DN802_c0_g1_i4 were ligated to pTRV2-CvANS vectors using XBAI [T^CTAGA] and BAMHI restriction sites; ANS acting both as a positive control and marker gene for facilitated phenotyping. The pTRV2-CvAG construct was generated using BAMHI and XHOI restriction sites, as with pTRV2-CvANS. The CvSHP amplicon was then ligated to the pTRV2-CvAG vector using XBAI and BAMHI restriction sites. No CvANS marker was used for the CvAG or CvAG-CvSHP constructs because downregulation of CvAG is distinct. Viral constructs were verified for off‐target silencing using siFi21 (Lück et al., 2019).
Agrobacterium tumefaciens was prepared for DNA transformation as previously described (Carey et al., 2021). All vectors were transformed into A. tumefaciens using calcium chloride heat‐shock transformation. For each, 100 ng of purified construct was combined with 250 µL of competent A. tumefaciens. Transformants were plated on LB media containing the aforementioned antibiotics. Transformants were then screened as before using 156F and 156R primers, and glycerol stocks were made and stored at −80°C (1:1 ratio of 50% glycerol and overnight A. tumefaciens culture).
The vacuum infiltration protocol, which has been shown to be an effective infiltration method with C. violacea, was modified from Carey et al. (2021). For each vector, A. tumefaciens cultures were serially inoculated up to 1000 mL cultures containing antibiotics, 1mM MES buffer and 0.02 mM acetosynringone. A 1:1 ratio of pTRV1 cultures were also serially inoculated up to 1000 mL. The final cultures were grown until they reached an OD600 between 0.8-1, and then immediately centrifuged and resuspended in infiltration buffer (10 mM MES, 10 mM MgCL2 and 0.2 mM acetosyringone) to an OD600 of 4.0 ± 0.1 and left for four hours to acclimatize. Agrobacterium containing pTRV1 should be inoculated 1-2 hours prior to pTRV2 cultures because they have a slower growth rate. An OD600 of four was chosen because it has been reported to achieve greater yields, and when pTRV2 and pTRV1 are combined their OD600 values half to an optimal OD600 of 2.0 (Wang et al., 2006). The serial inoculation was halted at OD600 0.8 to capture log-phase growth. The pTRV2 and pTRV1 suspensions were combined prior to infiltration at a 1:1 ratio. Silwet L-77 surfactant was added to each mixture at 100 μL/L. Groups of seedlings were extracted from the soil, rinsed in reverse osmosis water, briefly air-dried, submerged in Agrobacterium, and placed in a vacuum chamber. The chamber was evacuated to -20 inHg and held for 2 minutes. Vacuum pressure was then quickly released, and plants were rinsed and planted in fresh soil. Finally, plants were grown at 22°C for 16 h and 18°C for 8 h because it was found that lower temperatures consistently resulted in better VIGS efficacy in Petunia (Broderick and Jones, 2014).
All treated plants began showing phenotypes five weeks post-inoculation, and phenotypes lasted until plant senescence. Phenotypes for construct pTRV2-CvANS were scored based on reduction in maroon pigmentation in petals, which is hereafter referred to as yellowing, i.e., a reduction in anthocyanins resulted in petals with increased yellow pigmentation. A flower was scored as having a moderate phenotype when at least two petals displayed obvious yellowing. Flowers were scored as having a strong phenotype if all four petals displayed obvious yellowing. Flowers with less yellowing than moderate flowers, but which were distinct from untreated flowers, were scored as having a mild phenotype. There were no observed instances of only a single petal yellowing.
The yellowing phenotype assisted with scoring of CvCRC, CvSWEET9, and DN802_c0_g1_i4 constructs. Phenotypes for pTRV2-CvCRC-CvANS were scored based on complete or partial absence of nectary. Yellowed flowers with complete nectaries, and non-yellow flowers without nectaries were also recorded because it is possible for only a single gene to be silenced even with multiple gene constructs. Phenotypes for pTRV2-CvSWEET9-CvANS were scored based on visual inspection of nectary gland for presence of nectary droplets using a dissection microscope. Phenotypes for pTRV2- DN802_c0_g1_i4-CvANS were indistinguishable from pTRV2-CvANS.
Phenotypes for pTRV2-CvAG and pTRV2-CvAG-CvSHP were scored based on AG silenced phenotypes in Arabidopsis because of conservation of ABC gene function (Mizukami and Ma 1997). Silencing efficacy was based on the extent of repetition of perianth whorls and the absence of reproductive whorls. For both constructs, presence/absence of nectaries, absence of reproductive whorls, and repetition of perianth whorls were noted. Plant tissue from treated and control plants was excised, flash-frozen in liquid nitrogen, and stored at -80°C. Phenotypes were imaged using a Nikon SMZ 1500 dissecting microscope (Nikon, Tokyo, Japan) and a handheld digital Canon DS126181 camera (Canon, Tokyo, Japan). Images were standardized, scaled, color balanced, and assembled into figures using Inkscape version 0.92.5 (https://inkscape.org) and GIMP.
3 Results
3.1 Morphology and nectar production in Cleome violacea
Anthetic nectaries of C. violacea are adaxially positioned between petals and stamen. These nectaries are prominent due to their relatively large size (i.e., roughly half the size of an abaxial petal). Nectaries are tri-lobulate with two larger lateral lobes and a smaller central lobe at anthesis (Figures 1B–E). Following the terminology of Nepi (2007), these structured nectaries have prominent epidermis, and mostly consist of specialized parenchyma with vascular tissue interspersed throughout (Figure 2). Nectar is secreted via nectarostomata, which are present prior to anthesis (Figure 3). Floral nectaries are first visible late in development when developing stamens reach the same length as petals. At this stage, sepals and petals are growing, stamens have differentiated into filaments and anthers, and the gynoecium is formed with papillate stigma. Nectaries mature concordantly with stamens and reach maturity just prior to anthesis (Figure 1B). Nectary primordia are visible in small buds (Figure 2) when sepals are maturing.
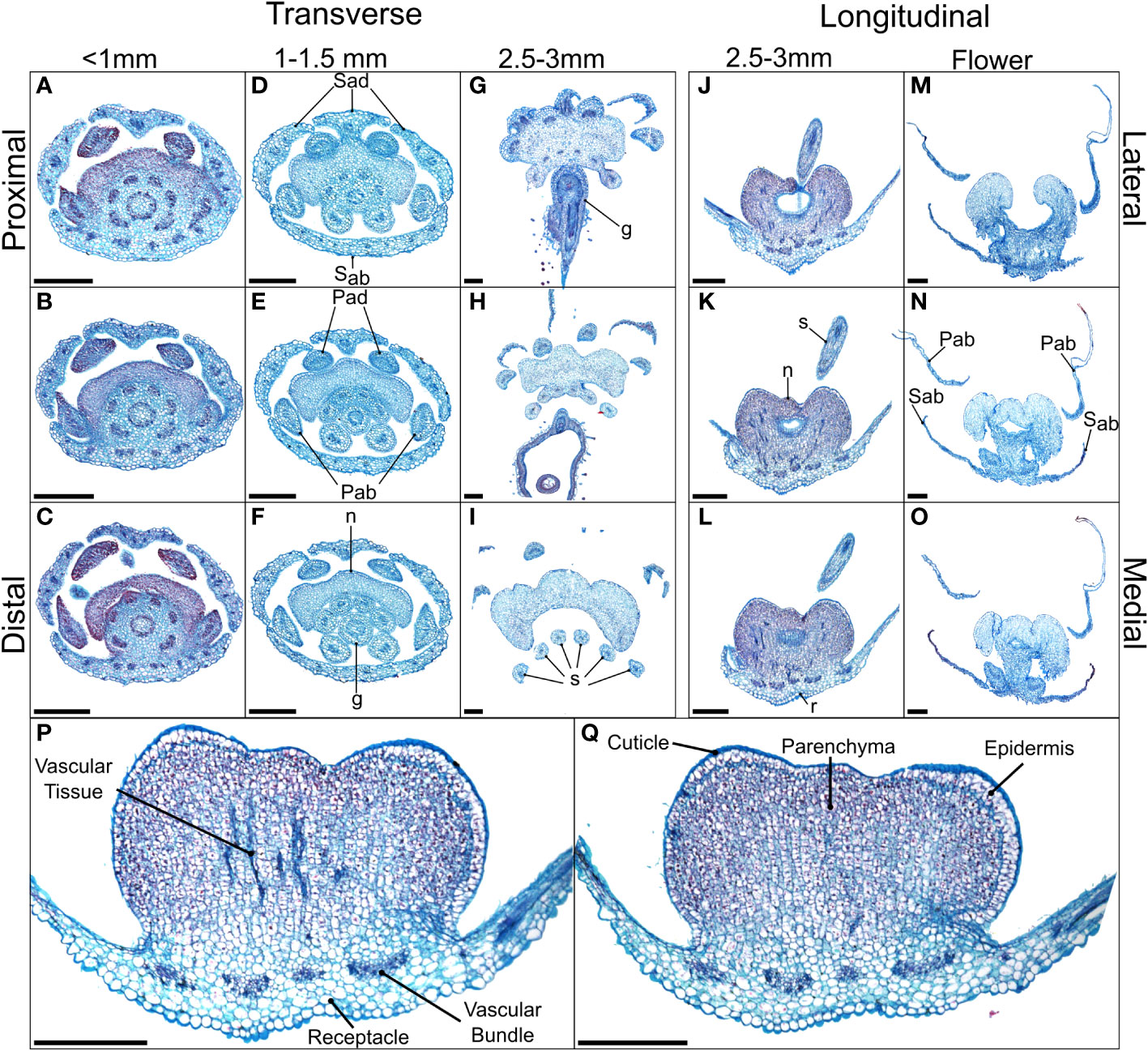
Figure 2 Alcian blue/safranin O-stained sections of Cleome violacea nectaries at pre-anthetic, anthetic and post-anthetic stages. From left to right: (A–C) small, (D–F) medium, and (G–I) large buds in transverse view with proximal-distal indicating relative distance to receptacle. (J–L) Large bud and (M–O) flowers in longitudinal view with lateral-medial indicating relative distance from center. (P, Q) Longitudinal view of 20 μm slices of the same large floral bud with and without vascular tissue, respectively. Scale bars = 250 μm. Sad = adaxial sepal; Sab = abaxial sepal; Pad = adaxial petal; Pab = abaxial petal; s = stamen; g = gynoecium, r = receptacle.
In small buds (<1.0 mm wide), nectaries are oblong and marginally lobed. At this stage, cells appear parenchymal with no differentiation of epidermis or vascular tissue, although the cuticle is apparent (Figures 2A–C). Medium buds (1-1.5 mm wide) have more pronounced lobes with differentiated epidermis (Figures 2D–F). Large buds (2.5-3.0 mm wide) have larger lobes comprised of parenchymal cells which make up the bulk of the nectary (Figures 2G–I). Epidermal cells are 1-2 layers thick on medial and lateral nectary lobes (Figure 2Q). In large buds, vascular tissue is distributed throughout the specialized parenchyma and likely connects with other vasculature near the receptacle base (Figure 2P) and with nectarostomata on the nectary surface (Figure 3). These nectarostomata are present on large buds (Figure 3A) and anthetic flowers (Figure 3B). Nectaries produce a low volume of nectar that decreases in volume after anthesis (Figure 4). Anthetic nectaries produce an average of 0.17 ± 0.07 μL) of nectar (Figure 4). Nectar volume decreases after day 1 but remains stable over three consecutive days of sampling at ~ 0.11 ± 0.04 μL) (Figure 4).
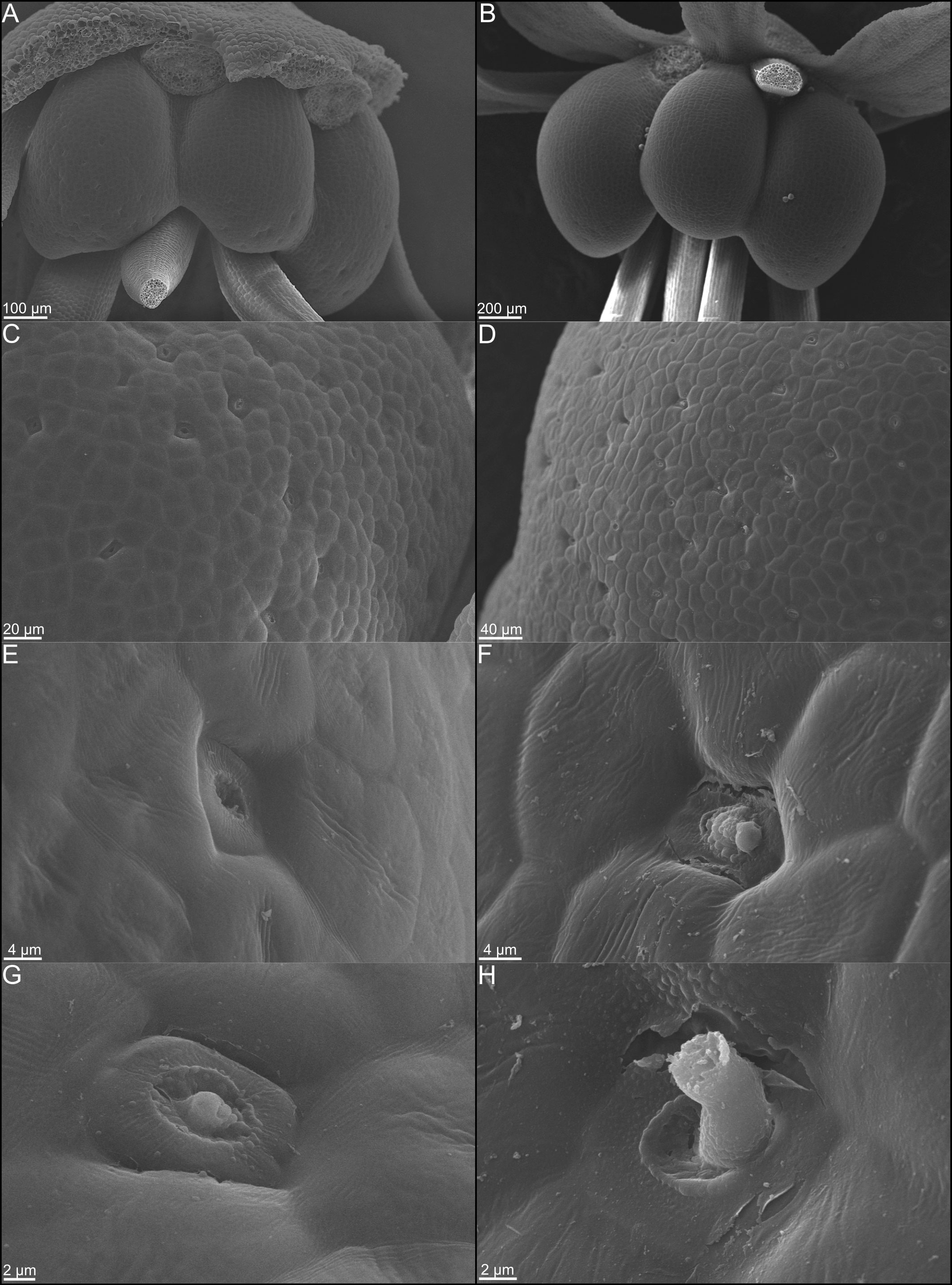
Figure 3 Scanning electron micrographs of whole nectaries from Cleome violacea at (A) pre-anthetic and (B) anthetic stages. (C) Distribution of nectarostomata on pre-anthetic nectary lobe and (D) anthetic nectary lobe. Examples of nectarostomata from (E–G) bud and (F–H) anthetic flowers.
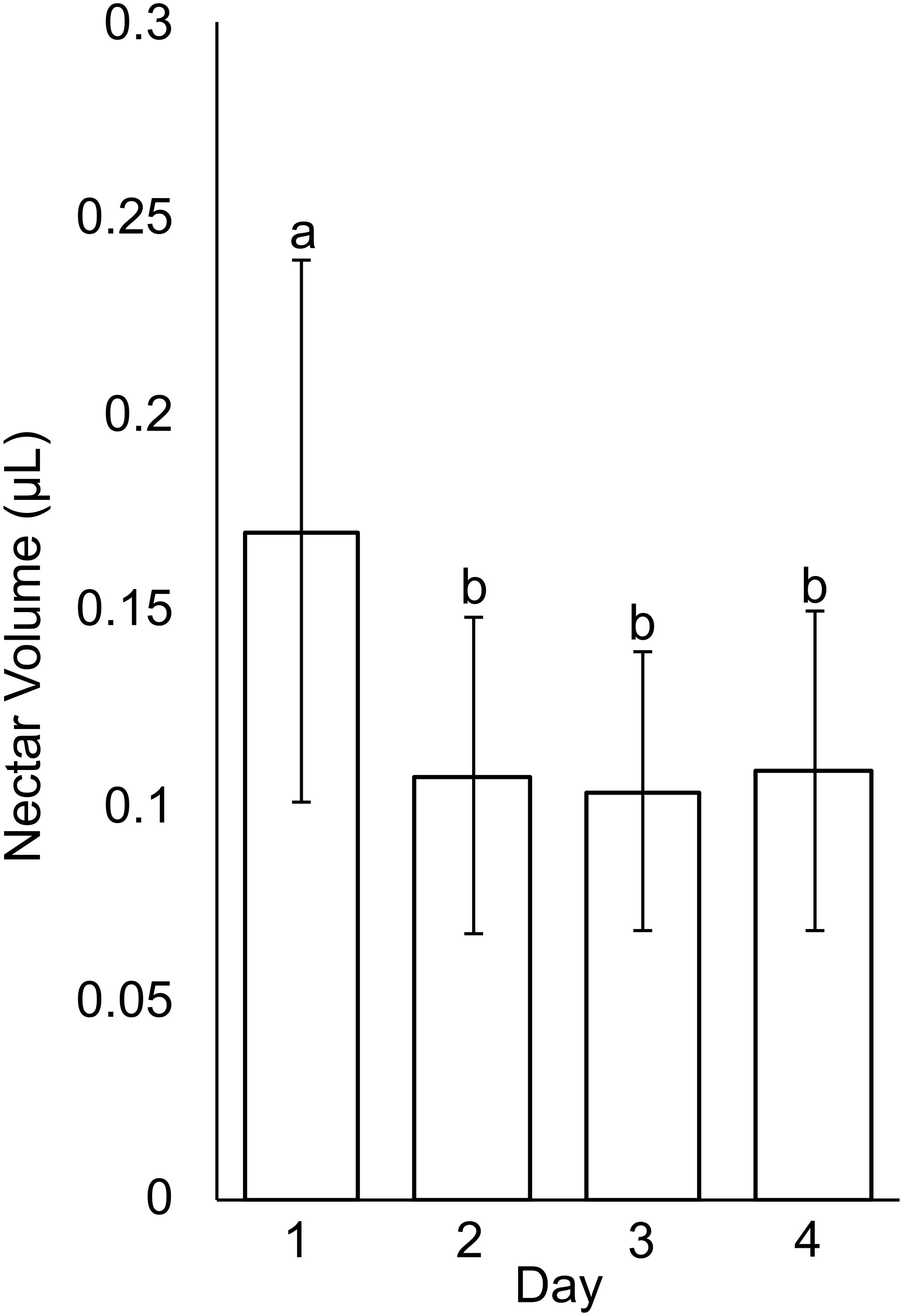
Figure 4 Nectar volume from Cleome violacea flowers taken on first day of anthesis (Day 1) and three days post-anthesis (Days 2, 3, and 4). Averaged value of flowers from 20 plants. Significance measured using paired, one-tailed, student’s t-tests (α < 0.01).
3.2 Expression profiles show distinct gene expression patterns pre to post anthesis
The transcriptome is of suitable quality and completeness for downstream analyses. The transcriptomic read depth averaged 19.9 million reads across 12 biological replicates, totaling just over 239M paired end trimmed reads. Median Phred scores are between 34 and 39 for each base pair of all 143,919 Trinity transcripts (Table S2), which indicates a base call accuracy between 99.7% and 99.99% (data not shown). The E90N50 value of the assembled transcriptome is 2227, and peaks at 2259 for Ex93 and Ex94 (Figure S1). A peak around Ex90 generally indicates a high level of transcriptome completeness. Further, the Benchmarking Universal Single Copy Orthologues (BUSCO) analysis of Viridiplantae orthologues (Simao et al., 2015) revealed that the transcriptome was 99.6% complete with 2 fragmented BUSCOs (Table S3).
Heatmap patterns of gene expression of pre-anthetic, anthetic and post-anthetic nectaries are consistent across two distinct thresholds. We compared all 4521 significantly differentially expressed transcripts, as well as the 1214 transcripts above 100 TPM (with a coefficient of variation < 50 in one or more biological groupings) (Figure 5). Pre and post-anthetic nectaries have opposing expression profiles, such that transcripts upregulated in pre-anthetic nectaries are generally downregulated in post-anthetic nectaries. Anthetic nectaries have no large clusters of up or downregulated transcripts and appear to be partially transitional, although they have a few unique clusters of differential expression (Figure 5). Expression patterns of transcripts filtered using TransDecoder were similar to the unfiltered list for significantly differentially expressed transcripts, as well as those above 100 TPM (Figure S5). In sum, each of the three developmental stages is genetically distinct.
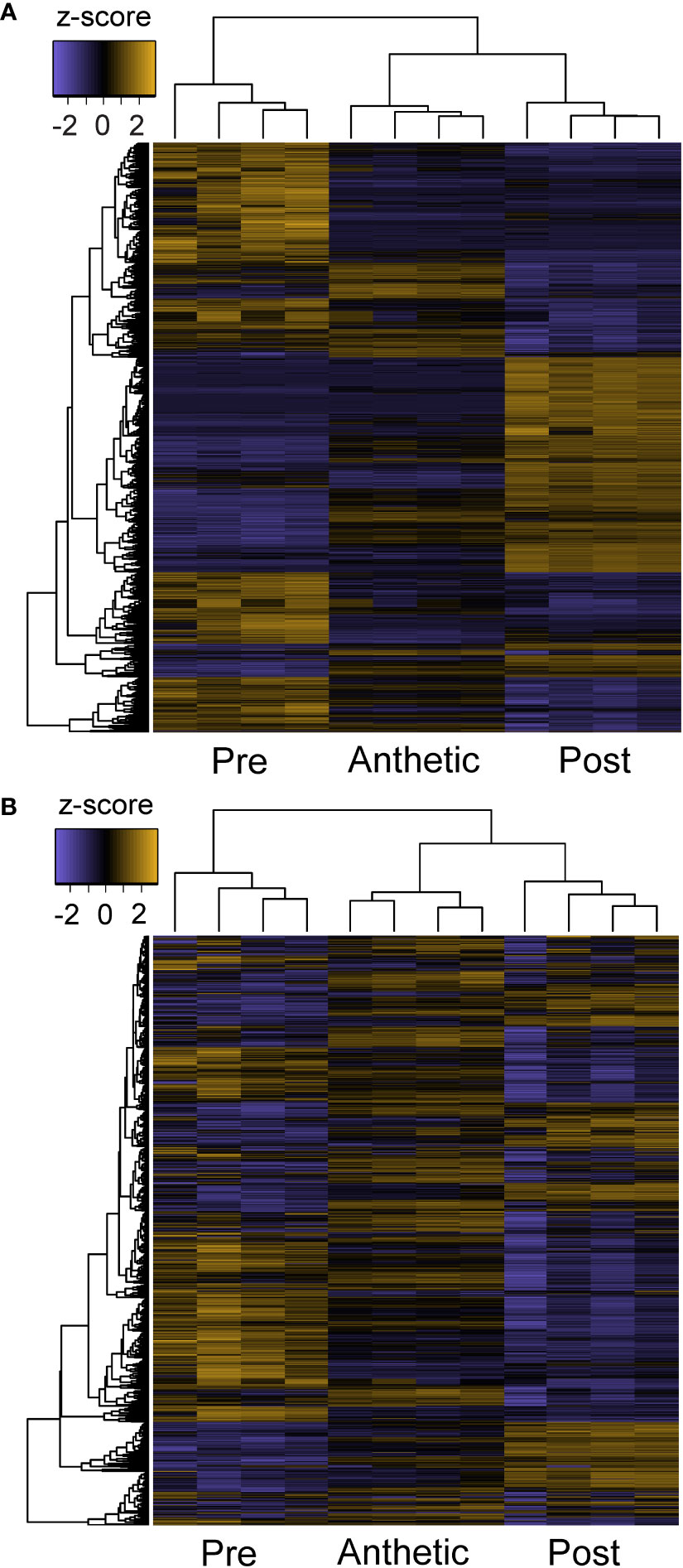
Figure 5 Z-score heatmaps of (A) all differentially expressed transcripts and (B) transcripts with TPM > 100 from Cleome violacea pre-anthetic, anthetic, and post-anthetic nectaries.
To provide additional insight into gene regulatory networks governing nectar secretion and nectary development, we also assembled the highest expressed transcripts across all stages from the TransDecoder-filtered dataset. This list included 20 of the highest expressed transcripts from pre-anthetic, anthetic and post-anthetic stages that were significantly differentially expressed in pairwise comparisons, and the top 20 highest overall expressed transcripts that were not differentially expressed. Due to overlap, there were 56 transcripts in total (40 among the differentially expressed and 16 non-differentially highly expressed transcripts). A few transcripts matched to the same genes leaving 51 unique accessions. Out of the 51, eight had no obvious role specific to nectary function (e.g., ubiquitous cellular process; Table S4). Five were related to photosynthesis, 14 to water transport and sugar production, 16 to stress response and six to cell growth. YABBY5, which can dimerize with CRC (Gross et al., 2018) was also among the highest expressed (Table S4). Putative gene function was estimated using gene description information from TAIR (www.arabidopsis.org), and a brief review of the literature.
3.3 Energy metabolism and hormonal regulation across nectary development
Genetic networks were assessed to determine which categories were active at each sampled stage of nectary development. KEGG analyses revealed several categories that had different relative counts (i.e., putative orthologs) in one or more stages: energy metabolism, biosynthesis of secondary metabolites, translation, replication and repair, environmental adaptation, and cell growth and death. We interpret greater counts as greater biological activity. A difference of three or less was disregarded to account for any potential noise in the data, e.g., invalid isoforms created during the assembly process. In pre-anthetic nectaries, oxidative phosphorylation (35 relative to 25 in the other two stages), and thermogenesis (40 relative to 32 in the other two stages) are the only subcategories with a greater number of putative orthologs (Table S5). Mitochondrial oxidative phosphorylation in plants is known to provide ATP for cellular functions (e.g., sucrose metabolism) and is tightly linked to photosynthesis (Braun, 2020). However, photosynthetic processes are similar between all stages (Table S5). Anthetic nectaries have no categories with greater hits than the other two stages (Table S5). Post-anthetic nectaries have increased biological activity in two categories, replication and repair, and cell growth and death; three of the five subcategories for cell growth and death are directly related to yeast (Table S5), i.e., nectary yeast are likely contributing to ortholog abundance in this category. Overall, most categories have a similar number of putative orthologues across all stages.
Although plant hormone signalling is important in nectary development and nectar secretion, there was no indication of differences between hormone signalling related orthologs between stages, based on the KEGG analysis (Table S5). However, expression analyses indicated significant differential expression in genes related to these pathways (Figure 6). Auxin, JA, and GA are known to regulate transcriptional expression related to nectar secretion (Slavkovic et al., 2021), and ethylene interacts synergistically with auxin (Muday et al., 2012), although to our knowledge has no direct link to nectaries. Examples of highly expressed transcripts were AUXIN RESPONSE FACTOR 6 (ARF6) and JASMONATE ZIM-DOMAIN PROTEIN 1 (JAZ1) (Figures 6A, C). Three GIBBERELLIN 2-OXIDASE genes were expressed in pre-anthetic, anthetic and post-anthetic nectaries, respectively (Figure 6B). We also found significant upregulation of multiple ethylene related transcripts in pre-anthetic nectaries (e.g., ETHYLENE RESPONSE FACTOR 1 (ERF1) and ETHYLENE FORMING ENZYME (EFE1). (Figure 6D). These data suggest that a combination of auxin, JA, and GA influence nectary development and nectar secretion in C. violacea.
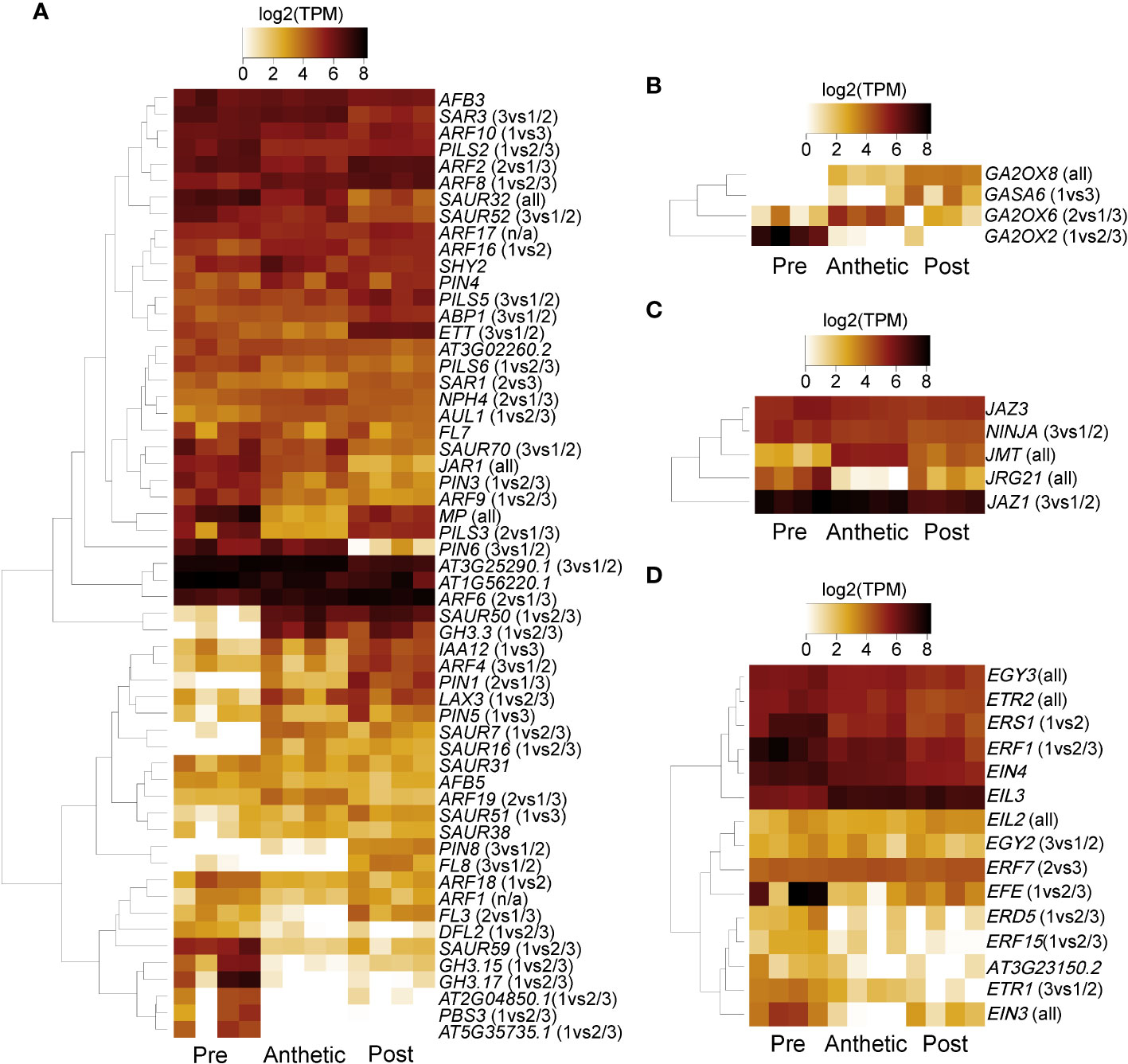
Figure 6 A heatmap of phytohormone-related transcripts expressed in pre-anthetic, anthetic, and post-anthetic nectaries of Cleome violacea displayed in log2(TPM). Representative transcripts of genes related to (A) auxin, (B) gibberellic acid (C) jasmonic acid, and (D) ethylene. Significance displayed in brackets. 1 = pre-anthetic; 2 = anthetic; 3 = post-anthetic.
3.4 Yeast and bacteria are present on Cleome violacea nectaries
Transcriptomic and SEM data provide evidence that yeast and bacteria colonize C. violacea nectaries. There are a total of 46 and 44 hits (e-value < 1e-50) to bacterial and yeast-related rRNA in the C. violacea transcriptome, respectively (Figure 7). Ribosomal rRNA can still be present in poly(A)-enriched libraries in appreciable percentages (Kim et al., 2019), which is valuable for finding non-plant related expression. Generally, expression of fungal and bacterial rRNA was inconsistent across biological replicates and stages; suggesting that colonization may be replicate specific. However, there are a few instances where expression is consistent across replicates and stages, which may indicate an established biological interaction (Figure 7). These data are further supported by the obstructions surrounding and within the nectarostomata (e.g., what appears to be budding yeast cells) (Figures 3F–H) and are consistent with the abundance of yeast related KEGG terms (Table S5). For example, there are nearly twofold more KEGG terms related to the yeast cell cycle in post-anthetic nectaries (43) than pre-anthetic (22) or anthetic (20) nectaries (Table S5). Carotenoid related genes FLAVONOL SYNTHASE 1 (FLS1), PHYTOENE SYNTHASE (PSY), and CHALCONE SYNTHASE (CHS) are also highly expressed at various developmental stages (Table 1 and Figure S2). All three genes have purported roles in combating biotic stress (Dao et al., 2011; Havaux, 2014; Naparlo et al., 2019).
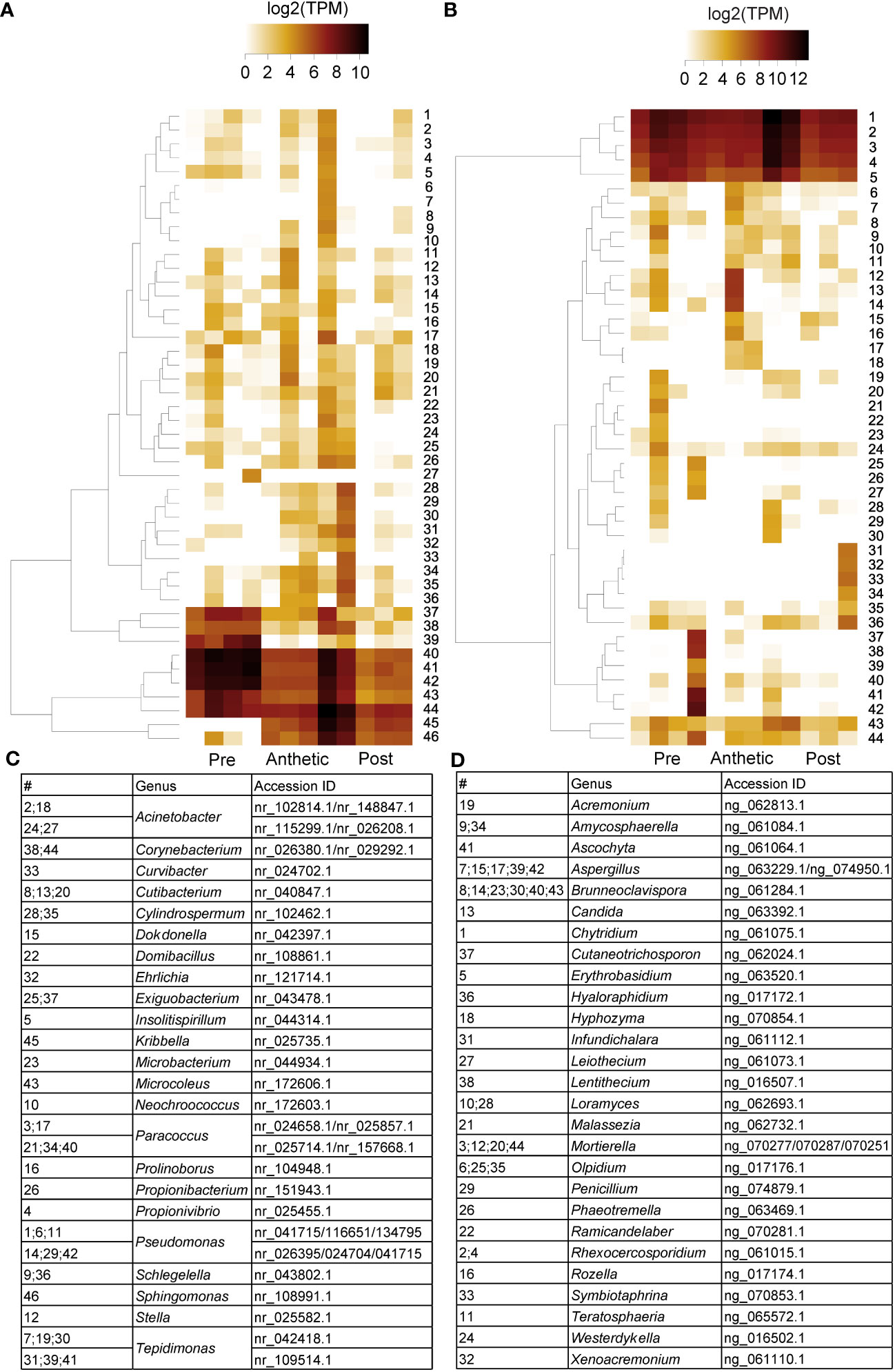
Figure 7 Heatmap of (A) 16s bacterial rRNA and (B) 18s fungal rRNA related transcripts in pre-anthetic, anthetic, and post-anthetic nectaries of Cleome violacea displayed in log2(TPM). Genera and NCBI accessions of respective transcripts for bacteria and fungi outlined in (C, D), respectively.
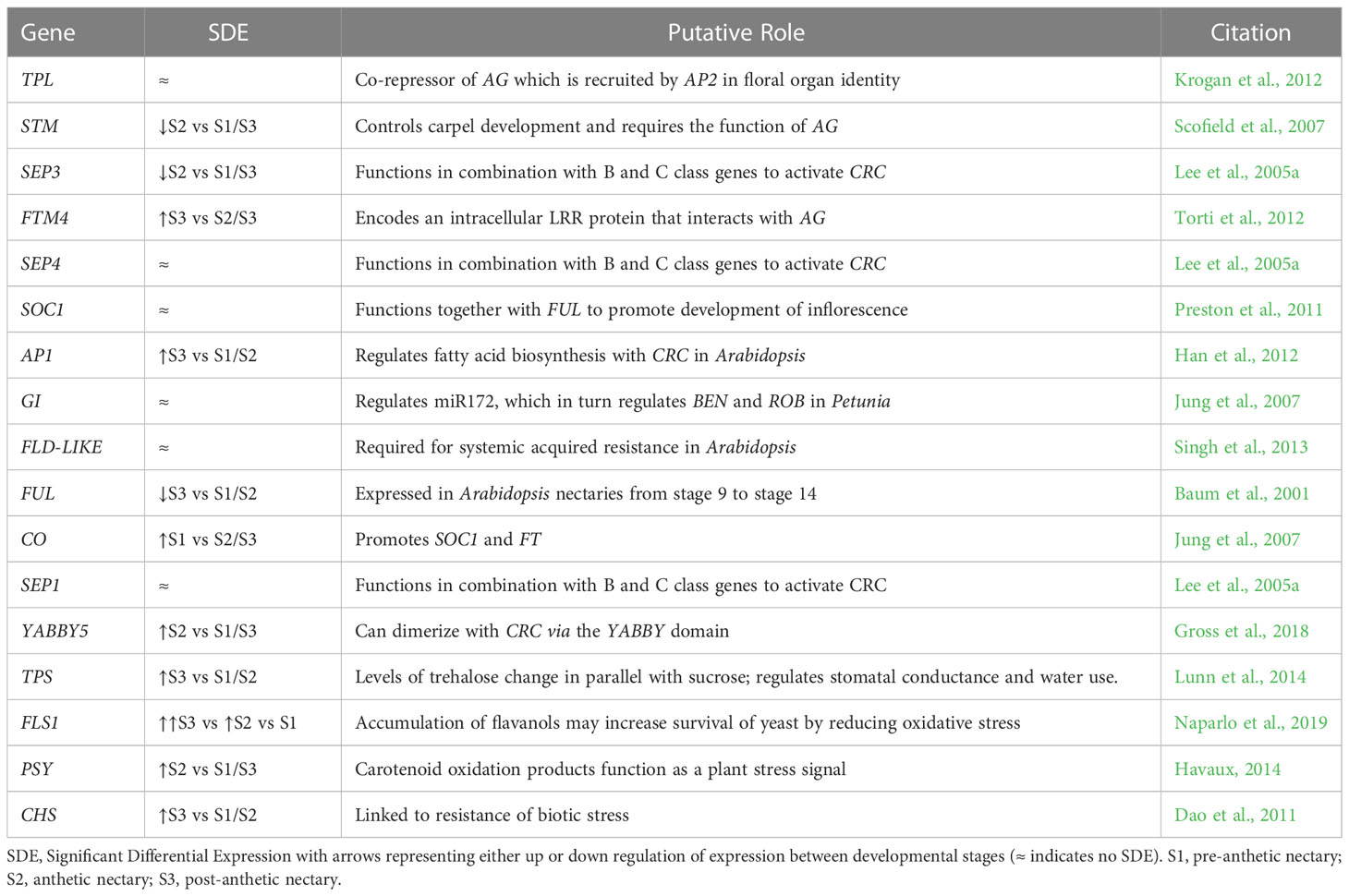
Table 1 Genes of interest not directly implicated in nectary development with relative expression values from our transcriptomic dataset, putative roles, and relevant citations.
3.5 Dynamic expression patterns of genes involved in nectar and nectary formation
After establishing global patterns and active biological networks, we examined expression patterns of 17 genes of interest with uncertain roles, nine genes known to be involved in nectar production, and ten genes with direct roles in nectary formation (e.g., expression in Arabidopsis nectaries) (Tables 1–3; Figure S2). These analyses reveal dynamic expression patterns from pre to post-anthetic nectaries. Seven genes linked to nectar production are significantly upregulated in either pre-anthetic and/or anthetic nectaries: BAM1, PIN6, MYB21, SWEET9, JAZ, G20X and JMT (Table 2). This pattern mirrors the onset of nectar production. Interestingly, expression profiles for nectary development genes (Table 3) are generally opposite to nectar production (Table 2). That is, of the genes examined with established roles in nectary development, most transcripts are downregulated in pre-anthetic nectaries, and three of ten genes explored are evenly expressed across all stages (Table 3). Downregulated genes include PI, AG, ARF6, ARF8, and STY (Table 3). Of the 17 genes with uncertain roles, six are evenly expressed across all three developmental stages: SEP1/4, TOPLESS (TPL), SUPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), GIGANTEA (GI), and FLOWERING LOCUS D LIKE (FLD-like). Of the remaining, no clear pattern emerges (Table 1).
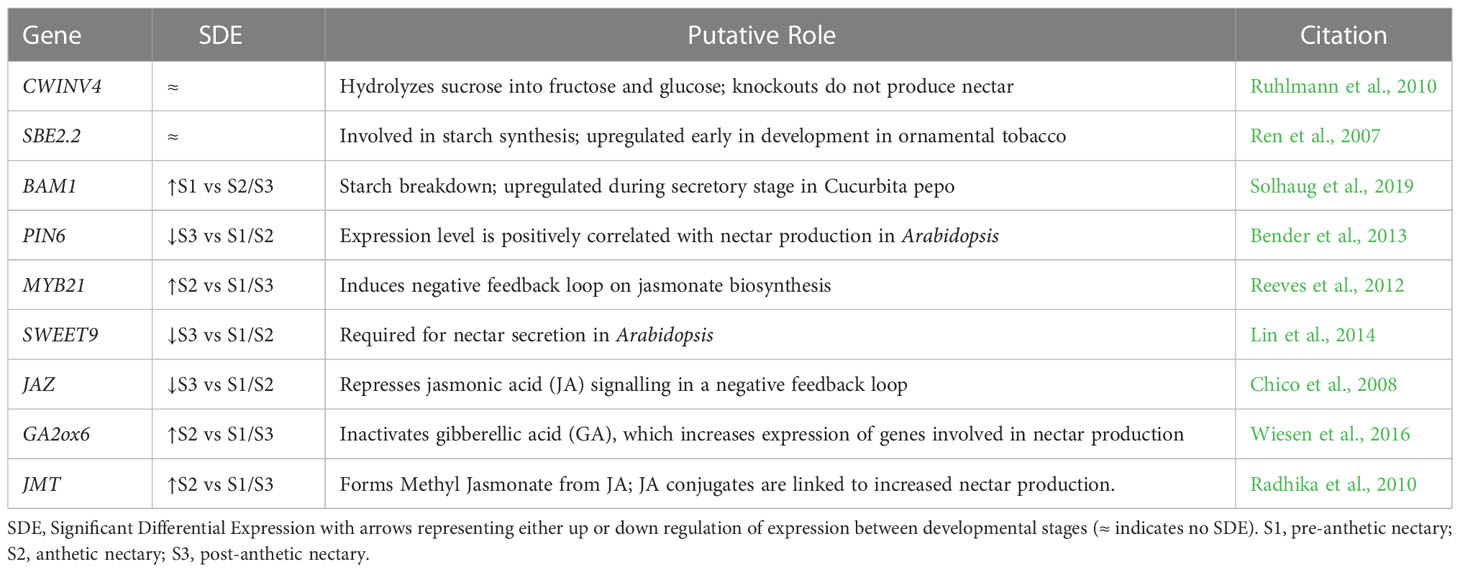
Table 2 Genes of interest with direct roles in nectar production with relative expression values from our transcriptomic dataset, putative roles, and relevant citations.
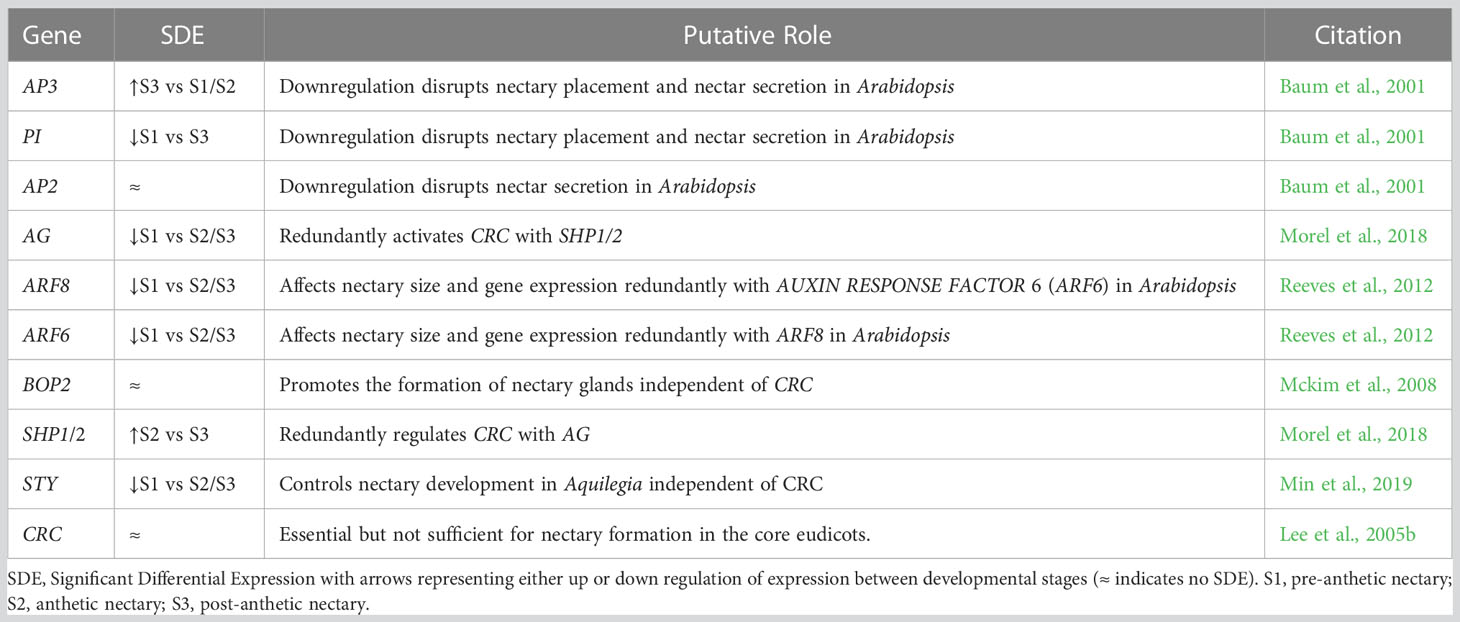
Table 3 Genes of interest with direct roles in nectary formation with relative expression values from our transcriptomic dataset, putative roles and relevant citations.
3.6 Functional studies demonstrate key roles of CvAG, CvSHP, CvCRC, and CvSWEET9 in nectary development and nectar secretion
VIGS experiments tested the putative function of five genes. Four of the five genes targeted for downregulation were highly expressed and have established roles in nectary development and nectar production: CvAG, CvSHP, CvCRC, and CvSWEET9 (Tables 2, 3 and Figure S2). We also downregulated an uncharacterized transcript (DN802_c0_g1_i4) because it was among the highest expressed in the transcriptome, has a similar profile to CvSWEET9, and has no significant match to either the TAIR11 database or the nr database, despite an ORF of 375 bp (Figure S3). It partially matches AT412520.1 (e-value = 8.19e-4) from the TAIR11 database and MW419336 (e-value = 6.33e-7) from the nr database. These hits were not considered further because of their large e-values.
ANS was used both as a positive control vector (pTRV2-CvANS) and as a marker gene to facilitate scoring of phenotypes for CvCRC (pTRV2-CvCRC-CvANS), CvSWEET9 (pTRV2-CvSWEET9-CvANS), and DN802_c0_g1_i4 (pTRV2-DN802-CvANS). Untreated C. violacea and plants treated with pTRV2-MCS constructs were also used as controls (Figures 8A–D). Treatment with pTRV2-CvANS produced flowers with primarily yellow pigmentation on adaxial and abaxial petals (Figures 8E, F). Yellowing is purported to be from a disruption of the anthocyanin production pathway and was the visual marker used for other constructs because it is not expected to alter the form or function of nectaries. CvANS is only moderately expressed in post-anthetic nectaries (Figure S2).

Figure 8 Cleome violacea flowers from untreated and treatment control groups. (A) Untreated newly anthetic flower and (B) maturing flower. pTRV2-MCS treated flower displaying (C) moderate and (D) mild viral phenotype. (E, F) pTRV2-CvANS treated flowers displaying moderate yellowing petal phenotypes. Scale bars = 1 mm.
With exception of pTRV2-DN802_c0_g1_i4 (Figure S4), all other treatment groups had marked phenotypes related to nectary and nectar formation. DN802_c0_g1_i4 was highly expressed in pre-anthetic and anthetic nectaries, and relatively downregulated in post-anthetic nectaries (Figure S2). Treatment with pTRV2-DN802_c0_g1_i4-CvANS resulted in flowers that were phenotypically indistinguishable from the CvANS control (Figure S4; Figures 8E, F), despite a high efficacy and mortality among treated flowers relative to control (Table S6). It is plausible that this transcript is related to water and/or nutrient transport because of its unusually high mortality during silencing (Table S6), absence of discernible silencing phenotype (Figure S4), and similar expression profile to CvSWEET9 (Figure S2), but further research is required.
Functional studies suggest the role of CvCRC and CvSWEET9 in nectary and nectar formation, respectively. CvCRC is expressed across all three developmental stages investigated, but with no significant difference between stages (Table 3). In contrast, CvSWEET9 was downregulated in post-anthetic nectaries as compared to pre-anthetic and anthetic nectaries (Table 2). Treatment with pTRV2-CvCRC-CvANS resulted in either partial or total loss of nectaries in all flowers with yellowing phenotype (Figure 9; Table S6). Treatment with pTRV2-CvSWEET9-CvANS produced flowers with a visible reduction in nectar on their nectaries (Figure 10), and no detectable sugar using a refractometer (n = 10; data not shown).

Figure 9 Flowers of Cleome violacea treated with pTRV2-CvCRC-CvANS constructs. (A) Flower with strong yellowing phenotype and no nectary. (B) Flower with moderate yellowing phenotype and no nectary. (C) Flower with moderate yellowing phenotype, no nectary, and enlarged gynoecium. (D) Flower with moderate yellowing phenotype and no nectary. (E) Flower with half normal and half yellowing petals with partially absent nectary. (F) Flower with strong yellowing phenotype and reduced lateral nectary lobes. Scale bars = 1 mm.

Figure 10 Flowers of Cleome violacea treated with pTRV2-CvSWEET9-CvANS constructs. (A) Flower with moderate yellowing and nectary with reduced nectar accumulation. (B) Magnified view of nectary in A. (C) Flower with partial yellowing and partial normal phenotype. (D) Flower with near-normal pigmentation and reduced nectar production. (E) Magnified nectary from C displaying decreased nectar accumulation correlating with yellowing phenotype. Scale bars = 1 mm.
Individual and combined constructs of CvAG and CvSHP demonstrate these genes are functionally redundant in regulation of nectary formation. Treatment with pTRV2-CvAG and pTRV2-CvAG-CvSHP produced flowers without reproductive whorls and repeating perianth (Figures 11, 12). Flowers treated with pTRV2-CvAG still produced nectaries, but their position and structure were altered relative to untreated flowers (Figure 11). This misplacement is likely due to a loss of reproductive whorls and repeated morphology. Flowers treated with pTRV2-CvAG-CvSHP generally produced no nectaries, although occasionally they were present and reduced in size (Figures 12A, F).

Figure 11 Flowers of Cleome violacea treated with pTRV2-CvAG constructs. (A) Flower with repeating perianth whorls. (B) Nectary from flower similar to A with petals removed. (C) Flower with normal adaxial petals, repeating perianth whorls and adaxial nectary. (D) Flower with repeating perianth whorls and distally positioned nectary. (E) Flower with petaloid stamens and adaxial nectary. (F) Flower with repeating perianth whorls and adaxial nectary. White arrowheads indicate nectary position. Scale bars = 1 mm.

Figure 12 Flowers of Cleome violacea treated with pTRV2-CvAG-CvSHP constructs. (A) Flower with partial nectary. (B–E) Flowers with repeating perianth whorls and no nectary. (F) Flower with repeating perianth whorls and partial nectary. Black and white arrowheads represent reduced and absent nectary, respectively. Scale bars = 1 mm.
4 Discussion
4.1 Cleome violacea have structured nectaries that produce nectar secreted via nectarostomata
The nectary of C. violacea is striking in that it is a prominent feature of the flower, due to its large size and location (Figure 1). The nectary is adaxially positioned between stamens and adaxial petals, contributing to monosymmetry of the flower in addition to petal color patterning and reproductive organ curvature. Nectaries appear late in development, well after initiation of stamens and gynoecium. Once formed, mature nectaries are a large 3-lobulate structure (Figures 1–3). Cleome violacea nectaries are characteristic of many other structured nectaries (Nepi, 2007): (1) the nectary epidermis has thick cuticle, (2) the nectary parenchyma is made up of small, dense cells, and (3) the vasculature is interspersed throughout the nectary and likely connects with vascular bundles in the receptacle (Figure 2P). Presence of nectarostomata on nectaries of C. violacea has been described previously (Erbar and Leins, 1997b). In the annular nectary of Cleomella sparsifolia (= Cleome sparsifolia), nectarostomata appear on abaxial tips (Lee et al., 2005b), suggesting nectarostomata may be common in Cleomaceae. Nectarostomata are modified stomata that secrete carbohydrate rich solutions for pollinator reward; a similar genetic pathway regulates both nectarostomata and unmodified stomata (Pillitteri et al., 2008; Baylis et al., 2013).
Nectar is known to be secreted via a few different methods, and most commonly via nectarostomata in a granulocrine or eccrine based manner (Nepi, 2007). Outlined by Roy et al. (2017) eccrine based secretion begins with the breakdown of starch and subsequent synthesis of sucrose, which is then transported out of the cell and hydrolyzed before secretion out of nectarostomata in droplets of nectar. Key to the export of sucrose is SWEET9, which is essential for sugar transport in nectaries of Arabidopsis, Brassica rapa, and Nicotiana (Lin et al., 2014). Another relevant gene, CWINV4 is important for nectar formation in Arabidopsis, specifically cleaving sucrose in the extracellular space which has the effect of moving water towards sugar, forming nectary droplets (Ruhlmann et al., 2010). In C. violacea, multiple lines of evidence support eccrine-based nectar secretion. First, there are nectarostomata on the nectary surface (Figures 3C–H) which likely connect with the vasculature present throughout parenchymal tissue (Figure 2P). Second, both CvSWEET9 and CvCWINV4 are highly expressed in pre-anthetic and anthetic nectaries (Table 2 and Figure S2). Additionally, we identified 14 highly expressed transcripts that are related to sugar production or water transport, e.g., five of which are related to aquaporins found in Aquilegia (Singh et al., 2020) (Figure S5 and Table S4). Finally, nectar secretion is lessened when CvSWEET9 is completely downregulated (Figure 10). In sum, nectary secretion in C. violacea is dependent on CvSWEET9, as demonstrated for Arabidopsis, Brassica and Nicotiana, which supports its key role in sucrose export across the core eudicots (Lin et al., 2014).
Nectar is secreted at anthesis, accumulates on C. violacea nectary lobes (Figure 1E) and has a low average secretion volume (0.17 ± 0.07 μL) (Figure 4). This volume is lower than averages of wild populations of other species of Cleomaceae: Cleomella serrulata (0.85 ± 0.96 μL) and Polanisia dodecandra (0.63 ± 0.32 μL) (Higuera-Diaz et al., 2015). However, it is similar to the average volume produced by one species of Brassicaceae: Erysimum mediohispanicum (0.136 ± 0.010 μL). The differences in nectar volume may be in part explained by flower size as C. violacea has much smaller flowers than C. serrulata and P. dodecandra. It may also reflect different pollinator environments; flowers of C. serrulata and P. dodecandra have a wide range of visitors and somewhat overlap in geography in some areas of North America (Higuera-Diaz et al., 2015). It is also unclear if there is a reduction of nectar in lab-grown inbred lines of C. violacea relative to wild populations. No empirical pollination study has been conducted on C. violacea to date, which is native to Spain (GBIF.org), so there is no information on which pollinators would be attracted to and rewarded by its nectar.
4.2 CvCRC, CvSHP, and CvAG, exhibit conserved roles with other core eudicots in nectary formation
CRC is essential for nectary formation in Arabidopsis, Petunia, Pisum, and Medicago in addition to having an important role in carpel formation (Baum et al., 2001; Lee et al., 2005b; Fourquin et al., 2014). As with these taxa and other core eudicots (Lee et al., 2005b; Slavkovic et al., 2021), CvCRC is expressed in developing nectaries without any significant difference in gene expression patterns from late-stage buds to post-anthetic flowers (Table 3 and Figure S2). Like with other species, loss of CvCRC resulted in an absence of nectaries (Figure 9), which demonstrates that CvCRC is essential for nectary formation in C. violacea. While strong CsCRC expression in nectaries of C. sparsifolia implied the conserved role of CRC (Lee et al., 2005b), this study provides the first functional evidence of the direct contribution of CRC to nectary formation in Cleomaceae. Since the nectaries of C. sparsifolia are annular, forming a ring around the stamen base, as compared to the adaxial position of C. violacea nectaries (Figure 1), these data indicate that upstream regulators of CRC are likely important for nectary position and morphology within Cleomaceae flowers. In Medicago and Pisum, inconspicuous nectaries form at the base of the staminal tube. Like with Fabaceae, nectaries of Arabidopsis are found at the base of stamens, although in this instance forming six glands on the abaxial side. Nectaries in Petunia form a ring at the base of the gynoecium (Morel et al., 2018). Interestingly, unlike knockout or knockdowns of CRC in Arabidopsis (Alvarez and Smyth, 1999; Alvarez and Smyth, 2002), Fabaceae (Ferrandiz and Fourquin, 2014), and poppy (Orashakova et al., 2009), we did not observe many notable changes to gynoecium or fruit formation in CvCRC knockdowns (but see Figure 9C). Additional studies are necessary to explore the extent of CRC’s role in gynoecium development and its conservation in C. violacea. The highly expressed YABBY5 (Table 1, Figure S2) should also be explored due to its ability to dimerize with CRC (Gross et al., 2018), i.e., it may share a role with CRC in C. violacea. CRC homologs have variable importance in carpel formation across core eudicots (Morel et al., 2018), which also warrants further examination in C. violacea. As shown in Arabidopsis (Pinyopich et al., 2003), we predict a high redundancy of gene function for gynoecial formation in C. violacea given its importance to plant fitness.
MADS-box genes AG and SHP act redundantly upstream of CRC in both Arabidopsis and Petunia to initiate nectary development (Morel et al., 2018). The regulatory roles of these genes appear to be conserved in nectary formation of C. violacea. Both CvAG and CvSHP are strongly expressed across all stages of development (Table 3 and Figure S2). Treatment with pTRV2-CvSHP or pTRV2-CvAG alone is insufficient to prevent nectaries from forming (Figure 11 & Table S6). Treatment with pTRV2-CvSHP alone has no effect on floral phenotype (Table S6) but treatment with pTRV2-CvAG disrupts the formation of whorls 3 and 4 (Figure 11). These phenotypes in C. violacea are like Arabidopsis ag-1 mutants (Baum et al., 2001). Only doubly silenced flowers do not produce nectaries, although they are otherwise like flowers treated with pTRV2-CvAG (Figure 12). Our data is consistent with the model from Wollmann et al. (2010) which shows a balance between AP2 and AG activities, i.e., in pTRV2-CvAG treated flowers, stamens occasionally appear petaloid (Figure 11F). Thus, there is the possibility that the overlapping of whorls may be the condition which contributes to nectary formation because all the ABC genes are expressed in nectaries to some degree (Table 3 and Figure S2). When flowers are treated with pTRV2-CvAG, and petals form haphazardly, nectary tissue surrounds each petal at the base of the flower and the lobe-like structure is lost. Perhaps this is because nectary tissue here has no boundary due to the absence of reproductive whorl (Figure 11B). These results are consistent with those observed in Arabidopsis and Petunia in that CRC expression is dependent on both AG and SHP lineages (Morel et al., 2018). It is striking that the upstream regulators are likely shared between these three taxa. However, like Morel et al. (2018) our data cannot distinguish whether this shared regulation is due to a single evolutionary origin of nectaries or due to the conservation of CRC in carpel development.
Intriguingly, when CvTCP1 is downregulated in C. violacea, nectaries are altered with phenotypes ranging from reduced lobes to complete absence (Carey et al., in prep). Like CRC, the regulatory pathway upstream of TCP1 is unclear, although the key contribution of TCP homologs towards many types of floral monosymmetry has been demonstrated across angiosperms (Preston and Hileman, 2009; Hileman, 2014b; Wessinger and Hileman, 2020). Given that expression domains of AG and SHP are much broader across the flowers, other genetic factors are required for restriction of nectaries to a single whorl (Morel et al., 2018). In C. violacea, CvTCP1 may be involved, at least indirectly. As noted above, functional data for floral nectaries to date has been conducted on flowers whose nectaries are distributed evenly around floral organs (e.g., circular around Petunia gynoecium and at the base of all stamens in Arabidopsis), unlike the adaxial positioning of the nectary in C. violacea. Thus, adaxial floral identity may be required for nectary formation in C. violacea, although it is unclear if TCP1 has a direct role in nectary initiation. Functional studies of Cleomaceae with annular nectaries, such as Tarenaya hassleriana, would inform on decoupling nectary position and identity in the family.
Less is known about nectary size than initiation. In Petunia, BEN and ROB are important for nectary size (Morel et al., 2018), whereas BOP1/2 impacts nectary size in Arabidopsis (Mckim et al., 2008). It is perhaps unsurprising that no BEN or ROB homologs were expressed in nectaries of C. violacea, but CvBOP2 is expressed throughout all stages examined (Table 3 and Figure S2). Unlike BEN and ROB, the interactions between BOP1/2 and other floral homeotic genes, with regards to nectary formation and size, are not as well understood (Slavkovic et al., 2021). Further experiments are needed to determine whether CvBOP2 contributes to nectary size in C. violacea. In our analysis of highly expressed transcripts (Figure S5 and Table S4), six transcripts are potentially linked to cell growth in nectaries, although they have only been characterized in leaves (e.g., EXL2) and roots (e.g., PRX44) (Schröder et al., 2009; Marzol et al., 2022). Future studies should explore genes similar to those identified in this study, as well as earlier stages of nectary development. Altogether, these expression patterns suggest that pathways determining nectary size are not conserved across the core eudicots.
Gene expression data suggests additional conservation as well as deviation in the genetic pathway of nectary development between Arabidopsis and C. violacea. Transcriptomic data shows many genes important for nectary formation are conserved across Arabidopsis and Cleome, including ABC genes AG, AP2, AP3, PI, and MADS-box gene SHP (Table 3 and Figure S4.2). Notably, AqSTY has been shown as essential for nectary formation in Aquilegia (Min et al., 2019) and CvSTY is expressed in nectaries of C. violacea. While expression is low, it is significantly differentially expressed and down regulated in pre-anthetic flowers (Table 3 and Figure S2). This co-expression presents a tantalizing hypothesis that CvSTY and CvCRC are not mutually exclusive pathways in C. violacea nectary development. STY likely interacts with CRC in developing carpels of Arabidopsis (Kuusk et al., 2002) such that interactions in other floral structures are feasible. In addition, STY is also linked to auxin biosynthesis (Baylis et al., 2013), which is important to nectary development.
4.3 The nectar of Cleome violacea is complex, as is its secretion method
Nectar is a multifaceted sugar solution that changes in composition over time and includes microorganisms as well as secondary metabolites made by both plant and microbes (Alvarez-Perez et al., 2012; Chappell and Fukami, 2018; Parachnowitsch et al., 2019; Liao et al., 2021; Jacquemyn et al., 2021). Our data are consistent with bacteria and fungi colonization of C. violacea nectaries (Figure 7) and reflect complexities in these interactions. Unsurprisingly, many of the identified microorganisms from this study are commonplace in soil and/or have been previously isolated from nectar (e.g., Sphingomonas, Pseudomonas, and Erythrobasidium) (Figure 7) (Alvarez-Perez et al., 2012; Jacquemyn et al., 2013). However, the exact nature of the relationship (i.e., mutualism, commensalism, or parasitism) cannot be determined with gene expression data alone, especially since there was variation across replicates. Nonetheless, we found six transcripts that potentially play a role in combating biotic stress from our analysis of highly expressed transcripts (Figure S5 and Table S4), e.g., LIPID TRANSFER PROTEIN 2 (LTP2) and β-GLUCOSIDASE 19 (BGLU19) (Molina and García-Olmedo, 1997; Li et al., 2019) KEGG counts also showed enriched plant-pathogen interactions (26, 25, and 26 in pre-anthetic, anthetic, and post-anthetic nectaries, respectively.) (Table S5). Further, compounds typically produced by nectar-associated microbial communities (e.g., alcohols, isoprenoids, and ketones) (Rering et al., 2018) are difficult to distinguish with transcriptomics because many of these metabolites are also produced by the plant. Additionally, yeasts are known to chemically alter metabolites already present in nectar (Vannette and Fukami, 2016).
In all stages of developing nectaries of C. violacea, we found roughly even KEGG counts of carotenoid and flavonoid biosynthesis (~14 and ~8 across stages, respectively) (Table S5). Flavonoids have antioxidant activity, which reduces reactive oxygen species (ROS) in Arabidopsis (Vannette and Fukami, 2016), and they have also been linked to the reduction of E.coli fimbria, which may reduce biofilm formation (Lee et al., 2011). FLS1 is significantly upregulated in anthetic and post-anthetic nectaries (Table 1 and Figure S2). Additionally, accumulation of flavanols have also been shown to increase survival of yeast by reducing oxidative stress (Naparlo et al., 2019). Thus, flavonoid accumulation may be a way to inhibit bacterial biofilms while simultaneously supporting symbiotic yeast. CHS, which is highly expressed in our transcriptome (Table 1, S4 and Figure S2), is also linked to resistance against biotic and abiotic stress such as UV, temperature, wounding, and bacteria (Dao et al., 2011). However, even though their role in the reduction of ROS can potentially impact biotic stress, carotenoids are more commonly linked to abiotic stress (Havaux, 2014), pollinator attraction (Cazzonelli, 2011), and photoprotection (Demmig-Adams, 1990), so further research is required.
Phytohormone expression in C. violacea is complex with evidence supporting convergence to other eudicots. In Brassicales, phytohormones play an important role in gland development and nectar secretion (Slavkovic et al., 2021). In both Aquilegia and Arabidopsis, auxin is linked to nectary initiation via ARF6 and ARF8 (Nagpal et al., 2005; Reeves et al., 2012) to nectar production via PIN6 (Bender et al., 2013). Cleome violacea nectaries express multiple ARFs and PINs across development (Figure 6A), although not all expression is identical to that in Arabidopsis (Tables 2, 3). All PINs serve to promote the flow of auxin between cells (Křeček et al., 2009), so there is likely conservation of function between Cleomaceae and Brassicaceae. Auxin however does not function alone, and gland development is complicated by phytohormone interactions. For example, JA is positively and negatively regulated by auxin and GA, respectively. Both GA (Wiesen et al., 2016) and JA (Radhika et al., 2010) are linked to nectar secretion (Slavkovic et al., 2021), e.g., JA is positively correlated with nectar production. For other biological processes (e.g., seedling development) there is substantial crosstalk between auxin and ethylene (Muday et al., 2012). To our knowledge there have been no studies to date that characterize ethylene function in nectaries, although few do show ethylene-related genes present in nectary tissues (Tang et al., 1994) or ethylene production with CRC promoters (Switzenberg et al., 2015). Our transcriptome has multiple ethylene-related genes that are expressed across all developmental stages, e.g., CvEIN4 (ETHYLENE INSENSITIVE 4) and CvEIL3 (ETHYLENE INSENSITIVE-LIKE 3) (Figure 6C). Further, CHITINASE LIKE 1 (CLK1), which modulates ethylene biosynthesis in root development, is among the highest expressed transcripts (Table S4). However, it is yet unclear what role these genes play, and whether they have a function unique to nectaries.
5 Conclusions
As no explicit ancestral reconstruction states of nectaries have been performed in Brassicales or core eudicots, it remains unknown whether nectaries in Cleomaceae and Brassicaceae represent a single or independent origin of nectaries. The data presented in this study demonstrate a high degree of conservation between Cleomaceae and Brassicaceae, which would be consistent with a single origin of nectaries in these sister lineages. CvCRC functions as it does in Arabidopsis and is regulated redundantly by MADS-box genes AG and SHP. Cleome violacea nectaries are eccrine-based and appear to regulate their own energy production. Given multiple origins of other traits (e.g., monosymmetry), we cannot exclude the possibility of independent recruitment in the roles of CRC, AG, SHP and SWEET9 for nectary development and nectar secretion, respectively. Research on the evolution and development of nectaries and on nectar biology is ripe for interdisciplinary research (Liao et al., 2021). Here we show that Cleome violacea is a promising model for nectary development in the Cleomaceae that will pave the way forward for future nectary research on other key factors such as morphology and pollination.
Data availability statement
The raw reads used in this study are available at the sequence read archive (SRA) database under BioProject ID: PRJNA912718.
Author contributions
SC and JH conceived the study. SC collected and analysed the VIGS and transcriptomic data. AD collected nectar volume data. AD and SC prepared histological slides. BZ produced SEM micrographs. SC wrote the initial manuscript draft. SC, BZ, and JH edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (funding reference number 5014131).
Acknowledgments
The authors thank Nathan Gerein (Scanning Electron Microscopy Laboratory, Department of Earth and Atmospheric Sciences, University of Alberta) and Kacie Norton (Advanced Microscopy Facility, Department of Biological Sciences, University of Alberta) for assistance with scanning electron microscopy. We also thank Ida John for their help with generating SEM photos. We thank all Hall lab members for help with plant care and tissue collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1085900/full#supplementary-material
Supplementary Figure 1 | ExN50 graph generated by the ‘contig_ExN50_statistic.pl’ and ‘plot_ExN50_statistic.Rscript’ scripts provided with Trinity.
Supplementary Figure 2 | A heatmap of nectary-related genes displayed as log2(TPM).
Supplementary Figure 3 | Sequence and ORF of uncharacterized Trinity transcript DN802_c0_g1_i4.. The ORF is highlighted and begins at bp 182.
Supplementary Figure 4 | Flowers of Cleome violacea treated with pTRV2-DN802_c0_g1_i4-CvANS constructs. Phenotypes were indiscernible from pTRV2-CvANS control. (A) Flower with mild yellowing and underdeveloped stamens. (B) Flower with strong yellowing and no nectar production. (C) Flower with mild yellowing and underdeveloped stamens. (D) Flower with moderate yellowing. (E) Flower with moderate yellowing and underdeveloped stamen. (F) Flower with moderate yellowing. Scale bars = 1 mm.
Supplementary Figure 5 | Z-score heatmaps of TransDecoder filtered transcripts; (A) All differentially expressed transcripts and (B) transcripts with TPM > 100 from Cleome violacea pre-anthetic, anthetic, and post-anthetic nectaries.
References
Altschul, S., Gish, W., Miller, W., Myers, E., Lipman, D. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alvarez-Perez, S., Herrera, C. M., De Vega, C. (2012). Zooming-in on floral nectar: a first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol. Ecol. 80, 591–602. doi: 10.1111/j.1574-6941.2012.01329.x
Alvarez, J., Smyth, D. R. (1998). Genetic pathways controlling carpel development in Arabidopsis thaliana. J. Plant Res. 111, 295–298. doi: 10.1007/BF02512187
Alvarez, J., Smyth, D. R. (1999). CRABS CLAW and SPATULA, two arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386. doi: 10.1242/dev.126.11.2377
Alvarez, J., Smyth, D. R. (2002). CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int. J. Plant Sci. 163, 17–41. doi: 10.1086/324178
Andrews, S. (2010) FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
Barrett, R. L., Roalson, E. H., Ottewell, K., Byrne, M., Govindwar, S. P., Yadav, S. R., et al. (2017). Resolving generic boundaries in Indian-Australasian cleomaceae: Circumscription of Areocleome, Arivela, and Corynandra as distinct genera. Systemat. Bot. 42, 15. doi: 10.1600/036364417X696401
Baum, S. F., Eshed, Y., Bowman, J. L. (2001). The arabidopsis nectary is an ABC-independent floral structure. Development 128, 4657–4667. doi: 10.1242/dev.128.22.4657
Bayat, S., Schranz, M. E., Roalson, E. H., Hall, J. C. (2018). Lessons from cleomaceae, the sister of crucifers. Trends Plant Sci. 23, 808–821. doi: 10.1016/j.tplants.2018.06.010
Baylis, T., Cierlik, I., Sundberg, E., Mattsson, J. (2013). SHORT INTERNODES/STYLISH genes, regulators of auxin biosynthesis, are involved in leaf vein development in Arabidopsis thaliana. New Phytol. 197, 737–750. doi: 10.1111/nph.12084
Bender, R. L., Fekete, M. L., Klinkenberg, P. M., Hampton, M., Bauer, B., Malecha, M., et al. (2013). PIN6 is required for nectary auxin response and short stamen development. Plant J. 74, 893–904. doi: 10.1111/tpj.12184
Bernardello, G. (2007). A systematic survey of floral nectaries (Netherlands: Springer, Po Box 17, 3300 Aa Dordrecht).
Bowman, J. L., Smyth, D. R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. doi: 10.1242/dev.126.11.2387
Braun, H. P. (2020). The oxidative phosphorylation system of the mitochondria in plants. Mitochondrion 53, 66–75. doi: 10.1016/j.mito.2020.04.007
Broderick, S. R., Jones, M. L. (2014). An optimized protocol to increase virus-induced gene silencing efficiency and minimize viral symptoms in petunia. Plant Mol. Biol. Rep. 32, 219–233. doi: 10.1007/s11105-013-0647-3
Cane, J. H. (2008). Breeding biologies, seed production and species-rich bee guilds of Cleome lutea and Cleome serrulata (Cleomaceae). Plant Species Biol. 23, 152–158. doi: 10.1111/j.1442-1984.2008.00224.x
Carey, S., Higuera-Díaz, M., Mankowski, P., Rocca, A., Hall, J. C. (2021). Virus-induced gene silencing as a tool for functional studies in Cleome violacea. Appl. Plant Sci. 9. doi: 10.1002/aps3.11435
Carey, S., Mendler, K., Hall, J. C. (2019). How to build a fruit: Transcriptomics of a novel fruit type in the brassiceae. PloS One 14, 18. doi: 10.1371/journal.pone.0209535
Cazzonelli, C. I. (2011). Carotenoids in nature: insights from plants and beyond. Funct. Plant Biol. 38, 833–847. doi: 10.1071/FP11192
Chappell, C. R., Fukami, T. (2018). Nectar yeasts: a natural microcosm for ecology. Yeast 35, 417–423. doi: 10.1002/yea.3311
Chico, J. M., Chini, A., Fonseca, S., And Solano, R. (2008). JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 11, 486–494. doi: 10.1016/j.pbi.2008.06.003
Dao, T. T. H., Linthorst, H. J. M., And Verpoorte, R. (2011). Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10, 397–412. doi: 10.1007/s11101-011-9211-7
Demmig-Adams, B. (1990). Carotenoids and photoprotection in plants: a role for the xanthophyll zeaxanthin. Biochim. Biophys. Acta (BBA)-Bioenerget. 1020, 1–24. doi: 10.1016/0005-2728(90)90088-L
Endress, P. K. (2011). Evolutionary diversification of the flowers in angiosperms. Am. J. Bot. 98, 370–396. doi: 10.3732/ajb.1000299
Erbar, C., Leins, P. (1997a). Different patterns of floral development in whorled flowers, exemplified by apiaceae and brassicaceae. Int. J. Plant Sci. 158, 49–64. doi: 10.1086/297506
Erbar, C., Leins, P. (1997b). Studies on the early floral development in cleomoideae (Capparaceae) with emphasis on the androecial development. Plant Systematics Evol. 206, 119–132. doi: 10.1007/BF00987944
Ferrandiz, C., Fourquin, C. (2014). Role of the FUL-SHP network in the evolution of fruit morphology and function. J. Exp. Bot. 65, 4505–4513. doi: 10.1093/jxb/ert479
Fleming, T. H., Geiselman, C., Kress, W. J. (2009). The evolution of bat pollination: a phylogenetic perspective. Ann. Bot. 104, 1017–1043. doi: 10.1093/aob/mcp197
Fourquin, C., Primo, A., Martinez-Fernandez, I., Huet-Trujillo, E., Ferrandiz, C. (2014). The CRC orthologue from Pisum sativum shows conserved functions in carpel morphogenesis and vascular development. Ann. Bot. 114, 1535–1544. doi: 10.1093/aob/mcu129
Gould, B., Kramer, E. M. (2007). Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, ranunculaceae). BMC Plant Methods 3, 6. doi: 10.1186/1746-4811-3-6
Gross, T., Broholm, S., And Becker, A. (2018). CRABS CLAW acts as a bifunctional transcription factor in flower development. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00835
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494. doi: 10.1038/nprot.2013.084
Hall, J. C., Tisdale, T. E., Donohue, K., Kramer, E. M. (2006). Developmental basis of an anatomical novelty: heteroarthrocarpy in Cakile lanceolata and Erucaria erucarioides (Brassicaceae). Int. J. Plant Sci. 167, 771–789. doi: 10.1086/504928
Han, X., Yin, L., And Xue, H. (2012). Co-Expression analysis identifies CRC and AP1 the regulator of arabidopsis fatty acid biosynthesis. J. Integr. Plant Biol. 54, 486–499. doi: 10.1111/j.1744-7909.2012.01132.x
Havaux, M. (2014). Carotenoid oxidation products as stress signals in plants. Plant J. 79, 597–606. doi: 10.1111/tpj.12386
Heil, M. (2011). Nectar: generation, regulation, and ecological functions. Trends Plant Sci. 16, 191–200. doi: 10.1016/j.tplants.2011.01.003
Higuera-Diaz, M., Manson, J. S., Hall, J. C. (2015). Pollination biology of Cleomella serrulata and Polanisia dodecandra in a protected natural prairie in southern Alberta, Canada. Botany 93, 745–757. doi: 10.1139/cjb-2015-0084
Hileman, L. C. (2014a). Bilateral flower symmetry - how, when and why? Curr. Opin. Plant Biol. 17, 146–152. doi: 10.1016/j.pbi.2013.12.002
Hileman, L. C. (2014b). Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos. Trans. R Soc. Lond B Biol. Sci. 369. doi: 10.1098/rstb.2013.0348
Iltis, H. H., Hall, J. C., Cochrane, T. S., Sytsma, K. J. (2011). Studies in the cleomaceae I: On the separate recognition of capparaceae, cleomaceae, and brassicaceae. Ann. Missouri Botan. Garden 98, 28–36. doi: 10.3417/2007017
Jacquemyn, H., Lenaerts, M., Tyteca, D., Lievens, B. (2013). Microbial diversity in the floral nectar of seven epipactis (Orchidaceae) species. Microbiologyopen 2, 644–658. doi: 10.1002/mbo3.103
Jacquemyn, H., Pozo, M. I., Alvarez-Perez, S., Lievens, B., Fukami, T. (2021). Yeast-nectar interactions: metacommunities and effects on pollinators. Curr. Opin. Insect Sci. 44, 35–40. doi: 10.1016/j.cois.2020.09.014
Jung, J. H., Seo, Y. H., Pil, J. S., Reyes, J. L., Yun, J., Chua, N. H., et al. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in arabidopsis. Plant Cell 19, 2736–2748. doi: 10.1105/tpc.107.054528
Kim, I. V., Ross, E. J., Dietrich, S., Doring, K., Alvarado, A. S., Kuhn, C. D. (2019). Efficient depletion of ribosomal RNA for RNA sequencing in planarians. BMC Genomics 20, 12. doi: 10.1186/s12864-019-6292-y
Kramer, E. M. (2019). Plus ca change, plus c’est la meme chose: The developmental evolution of flowers. Curr. Top. Dev. Biol. 131, 211–238. doi: 10.1016/bs.ctdb.2018.11.015
Křeček, P., Skůpa, P., Libus, J., Naramoto, S., Tejos, R., Friml, J., et al. (2009). Protein family review the PIN-FORMED (PIN) protein family of auxin transporters. Genome Biology 10, 1–11. doi: 10.1186/gb-2009-10-12-249
Krogan, N. T., Hogan, K., And Long, J. A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139, 4180–4190. doi: 10.1242/dev.085407
Krueger, F. (2012) Trim galore!. Available at: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
Kuusk, S., Sohlberg, J. J., Long, J. A., Fridborg, I., Sundberg, E. (2002). STY1 and STY2 promote the formation of apical tissues during Arabidopsis gynoecium development. Development 129, 4707–4717. doi: 10.1242/dev.129.20.4707
Lee, J. Y., Baum, S. F., Alvarez, J., Patel, A., Chitwood, D. H., Bowman, J. L. (2005a). Activation of CRABS CLAW in the nectaries and carpels of arabidopsis. Plant Cell 17, 25–36. doi: 10.1105/tpc.104.026666
Lee, J. Y., Baum, S. F., Oh, S. H., Jiang, C. Z., Chen, J. C., Bowman, J. L. (2005b). Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development 132, 5021–5032. doi: 10.1242/dev.02067
Lee, J. H., Regmi, S. C., Kim, J. A., Cho, M. H., Yun, H., Lee, C. S., et al (2011). Apple flavonoid phloretin inhibits Escherichia coli O157: H7 biofilm formation and ameliorates colon inflammation in rats. Infection and immunity, 79(12), 4819–4827.
Liao, I. T., Hileman, L. C., Roy, R. (2021). On the horizon for nectar-related research. Am. J. Bot. 108, 2326–2330. doi: 10.1002/ajb2.1767
Li, B. B., Liu, Y. G., Tao, W. U., Wang, J. P., Xie, G. R., Chu, Z. H., et al. (2019). OsBGLU19 and OsBGLU23 regulate disease resistance to bacterial leaf streak in rice. J. Integr. Agric. 18, 1199–1210. doi: 10.1016/S2095-3119(18)62117-3
Lin, I. W., Sosso, D., Chen, L. Q., Gase, K., Kim, S. G., Kessler, D., et al. (2014). Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 508, 546–549. doi: 10.1038/nature13082
Lück, S., Kreszies, T., Strickert, M., Schweizer, P., Kuhlmann, M., Douchkov, D. (2019). siRNA-finder (si-Fi) software for RNAi-target design and off-target prediction. Front. Plant Sci. 10, 12. doi: 10.3389/fpls.2019.01023
Lunn, J. E., Delorge, I., Figueroa, C. M., Van Dijck, P., And Stitt, M. (2014). Trehalose metabolism in plants. Plant J. 79, 544–567. doi: 10.1111/tpj.12509
Luo, K. M., Harding, S. A., Tsai, C. J. (2008). A modified T-vector for simplified assembly of hairpin RNAi constructs. Biotechnol. Lett. 30, 1271–1274. doi: 10.1007/s10529-008-9673-x
Marzol, E., Borassi, C., Sardoy, M. C., Ranocha, P., Aptekmann, A. A., Bringas, M., et al. (2022). Class III peroxidases PRX01, PRX44, and PRX73 control root hair growth in arabidopsis thaliana. Int. J. Mol. Sci. 23, 5375. doi: 10.3390/ijms23105375
Mckim, S. M., Stenvik, G. E., Butenko, M. A., Kristiansen, W., Cho, S. K., Hepworth, S. R., et al. (2008). The BLADE-ON-PETIOLE genes are essential for abscission zone formation in arabidopsis. Development 135, 1537–1546. doi: 10.1242/dev.012807
Min, Y., Bunn, J. I., Kramer, E. M. (2019). Homologs of the STYLISH gene family control nectary development in aquilegia. New Phytol. 221, 1090–1100. doi: 10.1111/nph.15406
Mizukami, Y., Ma, H. (1997). Determination of Arabidopsis floral meristem identity by AGAMOUS. The Plant Cell 9(3), 393–408. doi: 10.1105/tpc.9.3.393
Molina, A., García-Olmedo, F. (1997). Enhanced tolerance to bacterial pathogens caused by the transgenic expression of barley lipid transfer protein LTP2. Plant J. 12, 669–675. doi: 10.1046/j.1365-313X.1997.00605.x
Morel, P., Heijmans, K., Ament, K., Chopy, M., Trehin, C., Chambrier, P., et al. (2018). The floral c-lineage genes trigger nectary development in petunia and arabidopsis. Plant Cell 30, 2020–2037. doi: 10.1105/tpc.18.00425
Morrant, D. S., Schumann, R., And Petit, S. (2009). Field methods for sampling and storing nectar from flowers with low nectar volumes. Ann. Bot. 103, 533–542. doi: 10.1093/aob/mcn241
Muday, G. K., Rahman, A., Binder, B. M. (2012). Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 17, 181–195. doi: 10.1016/j.tplants.2012.02.001
Nagpal, P., Ellis, C. M., Weber, H., Ploense, S. E., Barkawi, L. S., Guilfoyle, T. J., et al. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107–4118. doi: 10.1242/dev.01955
Naparlo, K., Zyracka, E., Bartosz, G., And Sadowska-Bartosz, I. (2019). Flavanols protect the yeast Saccharomyces cerevisiae against heating and freezing/thawing injury. J. Appl. Microbiol. 126, 872–880. doi: 10.1111/jam.14170
Nepi, M. (2007). Nectary structure and ultrastructure (Netherlands: Springer, Po Box 17, 3300 Aa Dordrecht).
Nepi, M., Grasso, D. A., Mancuso, S. (2018). Nectar in plant–insect mutualistic relationships: From food reward to partner manipulation. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01063
Orashakova, S., Lange, M., Lange, S., Wege, S., Becker, A. (2009). The CRABS CLAW ortholog from California poppy (Eschscholzia californica, papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J. 58, 682–693. doi: 10.1111/j.1365-313X.2009.03807.x
Parachnowitsch, A. L., Manson, J. S., Sletvold, N. (2019). Evolutionary ecology of nectar. Ann. Bot. 123, 247–261. doi: 10.1093/aob/mcy132
Patchell, M. J., Bolton, M. C., Mankowski, P., Hall, J. C. (2011). Comparative floral development in cleomaceae reveals two distinct pathways leading to monosymmetry. Int. J. Plant Sci. 172, 352–365. doi: 10.1086/658158
Patchell, M. J., Roalson, E. H., Hall, J. C. (2014). Resolved phylogeny of cleomaceae based on all three genomes. Taxon 63, 315–328. doi: 10.12705/632.17
Pei, Y. F., Zhang, J., Wu, P., Ye, L., Yang, D. F., Chen, J. D., et al. (2021). GoNe encoding a class VIIIb AP2/ERF is required for both extrafloral and floral nectary development in gossypium. Plant J. 106, 1116–1127. doi: 10.1111/tpj.15223
Pfannebecker, K. C., Lange, M., Rupp, O., Becker, A. (2017). Seed plant-specific gene lineages involved in carpel development. Mol. Biol. Evol. 34, 925–942. doi: 10.1093/molbev/msw297
Phillips, H. R., Landis, J. B., Specht, C. D. (2020). Revisiting floral fusion: the evolution and molecular basis of a developmental innovation. J. Exp. Bot. 71, 3390–3404. doi: 10.1093/jxb/eraa125
Pillitteri, L. J., Bogenschutz, N. L., Torii, K. U. (2008). The bHLH protein, MUTE, controls differentiation of stomata and the hydathode pore in arabidopsis. Plant Cell Physiol. 49, 934–943. doi: 10.1093/pcp/pcn067
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Preston, J. C., Hileman, L. C. (2009). Developmental genetics of floral symmetry evolution. Trends Plant Sci. 14, 147–154. doi: 10.1016/j.tplants.2008.12.005
Preston, J. C., Hileman, L. C., Cubas, P. (2011). Reduce, resuse and recycle: developmental evolution of trait diversification. Am. J. Bot. 98, 397–403. doi: 10.3732/ajb.1000279
Preston, J. C., Kost, M. A., Hileman, L. C. (2009). Conservation and diversification of the symmetry developmental program among close relatives of snapdragon with divergent floral morphologies. New Phytol. 182, 751–762. doi: 10.1111/j.1469-8137.2009.02794.x
Radhika, V., Kost, C., Boland, W., Heil, M. (2010). The role of jasmonates in floral nectar secretion. PloS One 5, 6. doi: 10.1371/journal.pone.0009265
Raju, A., Rani, D. (2016). Reproductive ecology of Cleome gynandra and Cleome viscosa (Capparaceae). Phytol. Balcanica 22, 15–28.
Ratcliff, F., Martin-Hernandez, A. M., Baulcombe, D. C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. doi: 10.1046/j.0960-7412.2000.00942.x
R Core Team (2013). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: http://www.r-project.org/.
Reeves, P. H., Ellis, C. M., Ploense, S. E., Wu, M. F., Yadav, V., Tholl, D., et al. (2012). A regulatory network for coordinated flower maturation. PloS Genet. 8, 17. doi: 10.1371/journal.pgen.1002506
Ren, G., Healy, R. A., Horner, H. T., James, M. G., And Thornburg, R. W. (2007). Expression of starch metabolic genes in the developing nectaries of ornamental tobacco plants. Plant Sci. 173, 621–637. doi: 10.1016/j.plantsci.2007.08.012
Rering, C. C., Beck, J. J., Hall, G. W., Mccartney, M. M., Vannette, R. L. (2018). Nectar-inhabiting microorganisms influence nectar volatile composition and attractiveness to a generalist pollinator. New Phytol. 220, 750–759. doi: 10.1111/nph.14809
Robinson, M., Mccarthy, D., Smyth, G. (2009). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Roy, R., Schmitt, A. J., Thomas, J. B., Carter, C. J. (2017). Nectar biology: from molecules to ecosystems. Plant Science 262, 148–164.
Ruhlmann, J. M., Kram, B. W., Carter, C. J. (2010). CELL WALL INVERTASE 4 is required for nectar production in arabidopsis. J. Exp. Bot. 61, 395–404. doi: 10.1093/jxb/erp309
Sauquet, H., Von Balthazar, M., Magallón, S., Doyle, J. A., Endress, P. K., Bailes, E. J., et al. (2017). The ancestral flower of angiosperms and its early diversification. Nat. Commun. 8, 16047. doi: 10.1038/ncomms16047
Schröder, F., Lisso, J., Lange, P., Müssig, C. (2009). The extracellular EXO protein mediates cell expansion in arabidopsis leaves. BMC Plant Biol. 9, 1–12. doi: 10.1186/1471-2229-9-20
Scofield, S., Dewitte, W., And Murray, J. A. H. (2007). The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in arabidopsis. Plant J. 50, 767–781. doi: 10.1111/j.1365-313X.2007.03095.x
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Singh, S., Bhatt, V., Kumar, V., Kumawat, S., Khatri, P., Singla, P., et al. (2020). Evolutionary understanding of aquaporin transport system in the basal eudicot model species aquilegia coerulea. Plants 9, 799. doi: 10.3390/plants9060799
Singh, V., Roy, S., Giri, M. K., Chaturvedi, R., Chowdhury, Z., Shah, J., et al. (2013). Arabidopsis thaliana FLOWERING LOCUS d is required for systemic acquired resistance. Mol. Plant-Microbe Interact. 26, 1079–1088. doi: 10.1094/MPMI-04-13-0096-R
Slavkovic, F., Dogimont, C., Morin, H., Boualem, A., Bendahmane, A. (2021). The genetic control of nectary development. Trends Plant Sci. 26, 260–271. doi: 10.1016/j.tplants.2020.11.002
Sobel, J. M., Streisfeld, M. A. (2013). Flower color as a model system for studies of plant evo-devo. Front. Plant Sci. 4, 321. doi: 10.3389/fpls.2013.00321
Solhaug, E. M., Roy, R., Chatt, E. C., Klinkenberg, P. M., Mohd-Fadzil, N., Hampton, M., et al. (2019). An integrated transcriptomics and metabolomics analysis of the Cucurbita pepo nectary implicates key modules of primary metabolism involved in nectar synthesis and secretion. Plant Direct 3, e00120. doi: 10.1002/pld3.120
Specht, C. D., Howarth, D. G. (2015). Adaptation in flower form: a comparative evodevo approach. New Phytol. 206, 74–90. doi: 10.1111/nph.13198
Switzenberg, J. A., Beaudry, R. M., And Grumet, R. (2015). Effect of CRC::etr1-1 transgene expression on ethylene production, sex expression, fruit set and fruit ripening in transgenic melon (Cucumis melo l.). Transgenic Res. 24, 497–507. doi: 10.1007/s11248-014-9853-5
Tang, X., Gomes, R., Bhatia, A., And Woodson, W. R. (1994). Pistil-specific and ethylene-regulated expression of l-Aminocyclopropane-l-Caaboxylate oxidase genes in petunia flowers. Plant Cell 6, 1227–1239. doi: 10.2307/3869821
Torti, S., Fornara, F., Vincent, C., Andrés, F., Nordström, K., Göbel, U., et al. (2012). Analysis of the arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24, 444–462. doi: 10.1105/tpc.111.092791
Tucker, G. C., Vanderpool, S. S. (2010). “Cleomaceae,” in Flora of north America north of Mexico: Magnoliophyta: Salicaceae to brassicaceae (New York: Oxford University Press).
Vannette, R. L., Fukami, T. (2016). Nectar microbes can reduce secondary metabolites in nectar and alter effects on nectar consumption by pollinators. Ecology 97, 1410–1419. doi: 10.1890/15-0858.1
Wang, C. C., Cai, X. Z., Wang, X. M., Zheng, Z. (2006). Optimisation of tobacco rattle virus-induced gene silencing in arabidopsis. Funct. Plant Biol. 33, 347–355. doi: 10.1071/FP05096
Wessinger, C. A., Hileman, L. C. (2020). Parallelism in flower evolution and development. Annu. Rev. Ecol. Evol. Syst. 51, 387–408. doi: 10.1146/annurev-ecolsys-011720-124511
Wiesen, L. B., Bender, R. L., Paradis, T., Larson, A., Perera, M. A. D. N., Nikolau, B. J., et al. (2016). A role for GIBBERELLIN 2-OXIDASE6 and gibberellins in regulating nectar production. Mol. Plant 9, 753–756. doi: 10.1016/j.molp.2015.12.019
Wollmann, H., Mica, E., Todesco, M., Long, J. A., Weigel, D. (2010). On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137(21), 3633–3642.
Zhang, R., Min, Y., Holappa, L. D., Walcher-Chevillet, C. L., Duan, X. S., Donaldson, E., et al. (2020). A role for the auxin response factors ARF6 andARF8 homologs in petal spur elongation and nectary maturation in Aquilegia. New Phytol. 227, 1392–1405. doi: 10.1111/nph.16633
Keywords: Cleomaceae, nectaries, nectar, transcriptomics, VIGS, parallel evolution, RNA-seq
Citation: Carey S, Zenchyzen B, Deneka AJ and Hall JC (2023) Nectary development in Cleome violacea. Front. Plant Sci. 13:1085900. doi: 10.3389/fpls.2022.1085900
Received: 31 October 2022; Accepted: 22 December 2022;
Published: 09 February 2023.
Edited by:
Verónica S. Di Stilio, University of Washington, United StatesReviewed by:
Yaowu Yuan, University of Connecticut, United StatesAna Maria Rocha De Almeida, California State University, East Bay, United States
Copyright © 2023 Carey, Zenchyzen, Deneka and Hall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shane Carey, c2NhcmV5QHVhbGJlcnRhLmNh
 Shane Carey
Shane Carey Brandi Zenchyzen
Brandi Zenchyzen A. J. Deneka
A. J. Deneka Jocelyn C. Hall
Jocelyn C. Hall