- 1ICAR-Central Potato Research Institute, Shimla, India
- 2School of Biotechnology, Shoolini University, Solan, India
- 3ICAR-National Research Centre for Grapes, Pune, India
- 4Department of Crop and Soil Science, Oregon State University, Corvallis, OR, United States
- 5ICAR-Central Potato Research Institute, Regional Station, Shillong, India
- 6ICAR-Central Potato Research Institute, Regional Station, Meerut, India
Potato is one of the most important food crops in the world. Late blight, viruses, soil and tuber-borne diseases, insect-pests mainly aphids, whiteflies, and potato tuber moths are the major biotic stresses affecting potato production. Potato is an irrigated and highly fertilizer-responsive crop, and therefore, heat, drought, and nutrient stresses are the key abiotic stresses. The genus Solanum is a reservoir of genetic diversity, however, a little fraction of total diversity has been utilized in potato breeding. The conventional breeding has contributed significantly to the development of potato varieties. In recent years, a tremendous progress has been achieved in the sequencing technologies from short-reads to long-reads sequence data, genomes of Solanum species (i.e., pan-genomics), bioinformatics and multi-omics platforms such as genomics, transcriptomics, proteomics, metabolomics, ionomics, and phenomics. As such, genome editing has been extensively explored as a next-generation breeding tool. With the available high-throughput genotyping facilities and tetraploid allele calling softwares, genomic selection would be a reality in potato in the near future. This mini-review covers an update on germplasm, breeding, and genomics in potato improvement for biotic and abiotic stress tolerance.
Introduction
Potato (Solanum tuberosum L.) is the third most important food crop of the world after rice and wheat. Potato suffers from various biotic and abiotic stresses, which may cause crop failure and yield loss depending on their severity. The key factors affecting potato cultivation are (a) biotic stresses including diseases like late blight, viruses, bacterial wilt, soil and tuber-borne diseases, insect-pests like aphids, whiteflies, thrips, mites, hoppers, potato tuber moths, and potato cyst nematodes (Singh et al., 2020); and (b) abiotic stresses like heat, drought, nutrient deficiency, salinity, and cold/frost stress (Handayani et al., 2019). Late blight is still the most serious disease of potato, however, in the current climate change scenario, viruses are becoming new threats especially for healthy seed production. Similarly, heat and drought stresses are major challenges in potato due to rising temperature, erratic rainfall, and drought conditions (Singh et al., 2020). Hence, management of these problems is very critical for developing climate resilient varieties through an accelerated breeding approach. Although conventional breeding has made significant progress, it is relatively slow and harnessed the limited potential of the Solanum gene pool (Hardigan et al., 2017). Now, the potato genome sequences (Potato Genome Sequencing Consortium, 2011) and resequence of wild/cultivated species are available publicly, such as de novo sequencing of two wild species namely S. commersonii (Aversano et al., 2015) and S. chacoense “M6” (Leisner et al., 2018); and resequencing of over 100 Solanum species (Hardigan et al., 2017; Kyriakidou et al., 2020; Tiwari et al., 2021). The rapid advancements in sequencing technologies, multi-omics approaches, genome editing, and genomic selection coupled with softwares/bioinformatics allow discovery of SNP markers, genes, and regulatory elements for breeding and also to enhance understanding of potato biology (Aksoy et al., 2015). This mini-review highlights the prospects of germplasm, breeding, and genomics in potato improvement for biotic and abiotic stresses tolerance.
Biotic Stresses in Potato
Late Blight and Viruses
Late blight, caused by the oomycete Phytophthora infestans (Mont.) de Bary, is the most devastating disease of potato crop worldwide. In the year 1845, this disease caused a complete loss of potato crops in the European countries mainly Ireland, and known as “Irish Famine.” More than 30 viruses are reported to infect potato crop, of which major viruses are Potato virus X (PVX), Potato virus Y (PVY), and Potato leaf roll virus (PLRV) in the world; and Tomato leaf curl New Delhi virus-potato (ToLCNDV) is a new problem in India. Potato viruses are transmitted by contact/mechanical (e.g., PVX) and insect vectors (e.g., PVY/PLRV), and cause mosaic or leaf curl and mixed symptoms (Singh et al., 2020).
Soil and Tuber-Borne Diseases
Soil and tuber-borne diseases like dry rot (Fusarium oxysporum), charcoal rot (Macrophomina phaseolina) and bacterial soft rot (Pectobacterium atrosepticum) are the main problems involved in the post-harvest, storage, and transport of potato. Black scurf (Rhizoctonia solani) and common scab (Streptomyces scabies) deteriorate tuber appearance. Bacterial wilt (Ralstonia solanacearum) is also a serious disease, while wart caused by Synchytrium endobioticum is a problem of hilly regions like Darjeeling hills in India. These diseases are managed by using healthy seeds, disinfection by boric acid treatment, cultural practices, and crop rotation (Singh et al., 2020).
Insect-Pests
Insect-pests such as aphids, whiteflies, thrips, white grubs, cutworms, leaf hopper, potato tuber moths, and mites infest potato crop. Aphids (Myzus persicae) transmit viruses in two ways i.e., persistent and circulative (PLRV), and non-persistent (PVY). Whiteflies (Bemisia tabaci) transmit ToLCNDV-potato virus. Thrips (Thrips palmi) transmit Groundnut bud necrosis virus and cause stem necrosis disease. Importantly, potato cyst nematodes (PCN) (Globodera rostochiensis and G. pallida) are key problems in temperate regions. Besides, other insect-pests are potato leaf hopper (Amrasca biguttula biguttula), white grub (Brahmina coriacea), cutworm (Agrotis segetum), potato tuber moth (Phthorimaea operculella), and mites (Polyphagotarsonemus latus) (Singh et al., 2020).
Abiotic Stress in Potato
Heat and Drought Stress
Heat stress is a great problem for potato crop, particularly in early planted crop and after the harvest of the main rabi crop under sub-tropical Indian conditions. A minimum night temperature below 20°C (day 25°C) is essential for tuber growth and development (Singh et al., 2015). Potato is mostly an irrigated crop, except in rain fed hilly regions. Therefore, all growth stages are sensitive to water availability such as germination, foliage, and root/stolon/tuber growth. Thus drought i.e., moisture deficit plays a very crucial role in determining potato yield (Dahal et al., 2019).
Nutrient Deficiency, Salinity, and Frost/Cold stress
Nutrients are very essential for plant growth, yield, and quality of potato. Potato is a heavily fertilized crop especially for nitrogen (N), and therefore reduction of N fertilizers is necessary to save the environment and reduce the production cost (Zhang et al., 2020). Nutrient deficiency drastically affects crop growth and reduces yield. Besides, salinity is another problem due to soil or irrigation water, which causes nutrient imbalance and restricts plant growth. Frost/cold is also another issue of temperate climates, where temperatures below −2°C can result in a partial or complete loss of crop (Ahmed et al., 2020).
Germplasm, Mapping, and Breeding
Potato Genetic Resources
Potato belongs to the genus Solanum (family: Solanaceae), which contains over 2,000 species, of which nearly 235 are tuber bearing potato species, where 73% are diploids (2x), 4% triploids (3x), 15% tetraploids (4x), and 8% pentaploids (5x)/hexaploids (6x) (Hawkes, 1990). The cultivated potato (S. tuberosum ssp. tuberosum) is a tetraploid (2n = 4x = 48). Potatoes are classified into four major groups (i) S. tuberosum group Andigenum of upland Andean genotypes (2x/3x/4x), and S. tuberosum group Chilotanum of lowland Chilean landraces (4x), (ii) S. ajanhuiri (2x), (iii) S. juzepczukii (3x), and (iv) S. curtilobum (5x) (Spooner et al., 2014). These species belong to different endosperm balance numbers (EBNs) like 1EBN (2x), 2EBN (2x/4x), and 4EBN (4x/6x), where hybridization within the same EBN species is successful but not with different EBN species (Hawkes, 1990). Over 98,000 potato accessions are conserved ex situ (in vitro), of which 80% are maintained in 30 key collections worldwide (FAO, 2010). To harness the potential of diverse species, a wide range of genetic variation has been recorded and deployed through breeding and ploidy manipulation techniques for potato improvement.
Linkage and Association Mapping
Gene mapping is important for molecular breeding. The complex tetrasomic inheritance, acute inbreeding depression, and high heterozygosity of potato complicate its genetic mapping. Linkage mapping is the genetic association of traits with segregating alleles of molecular markers in a defined mapping population. The first linkage map was reported in 1988 using tomato RFLP (restriction fragment length polymorphism) markers in diploid species (S. tuberosum group Phureja/Tuberosum) (Bonierbale et al., 1988). Then uncounted PCR-based molecular markers like simple sequence repeat (SSR), amplified fragment length polymorphism (AFLP), and diversity array technology (DArT) were applied for mapping. Over 10,000 AFLP markers were used to create an ultra-high-density (UHD) genetic and physical map of potato (Van Os et al., 2006), which was used in the potato genome sequencing. Sharma et al. (2013) constructed a dense genetic and physical map for a diploid backcross progeny using 2,469 markers (SSR/AFLP/DArT/SNP). Numerous genes/QTLs have been mapped in potato for various traits like late blight resistance (Hein et al., 2009) and drought stress (Anithakumari et al., 2012). On the contrary to linkage mapping, association mapping identifies genes/QTLs associated with phenotypic variation in a natural population based on the historical recombination and linkage disequilibrium (Flint-Garcia et al., 2003). In potato, diploid/tetraploid clones have been utilized in association mapping for several agronomic traits (D’hoop et al., 2014), particularly resistance to late blight (Gebhardt et al., 2004) and Verticillium wilt (Simko et al., 2004) to name a few.
Marker-Assisted Selection
Over 40 traits are considered to be important in potato breeding. Conventional breeding is a time consuming process mainly due to several years of field evaluation and clonal selection. Hence, identification of tightly linked markers with a target gene for a trait is considered to be ideal for MAS. MAS allows a significant decrease in field exposures by selection in the early stage, and thereby reduces field exposures and breeding cycles. In potato, a considerable number of linked markers have been developed and deployed mainly for simply inherited traits like late blight, viruses, and potato cyst nematode resistance (Ramakrishnan et al., 2015). However, meager information is available on MAS for complex traits like yield, nutrient use efficiency, heat, drought, and cold stress.
Progress in Genomics-Led Potato Improvement
Potato Genome Sequencing/Resequencing
In 2011, the Potato Genome Sequencing Consortium (PGSC),—formed by 26 international institutes belonging to 14 countries—successfully deciphered the potato genome (840 Mb) containing 39,031 protein-coding genes using a homozygous doubled monoploid (DM 1-3 516 R44) of S. tuberosum group Phureja (2n = 2x = 24) (Potato Genome Sequencing Consortium, 2011)1. Later Sharma et al. (2013) improved the DM potato assembly with a more accurate arrangement of scaffolds and pseudomolecules. Recently, a chromosome-scale long-read reference assembly has been constructed (Pham et al., 2020). By now over 100 potato species have been sequenced/re-sequenced mostly using Illumina platforms like wild S. commersonii (Aversano et al., 2015), tuber-bearing Solanum species (Hardigan et al., 2017), S. chacoense “M6” (Leisner et al., 2018), S. pinnatisectum-derived somatic hybrid (Tiwari et al., 2021), and cultivated potato taxa using Illumina and long-read (PacBio) technologies (Kyriakidou et al., 2020; Table 1). The rapid advancement in sequencing and bioinformatics has spurred innovation in discovery of new genes/markers/haplotypes to enable better understanding of potato biology (Zhou et al., 2020). Figure 1 illustrates different approaches used in potato germplasm, breeding, and genomics-led improvement for biotic and abiotic stresses tolerance.

Table 1. A few recent examples of application sequencing and multi-omics technologies in potato for biotic and abiotic stress resistance/tolerance.
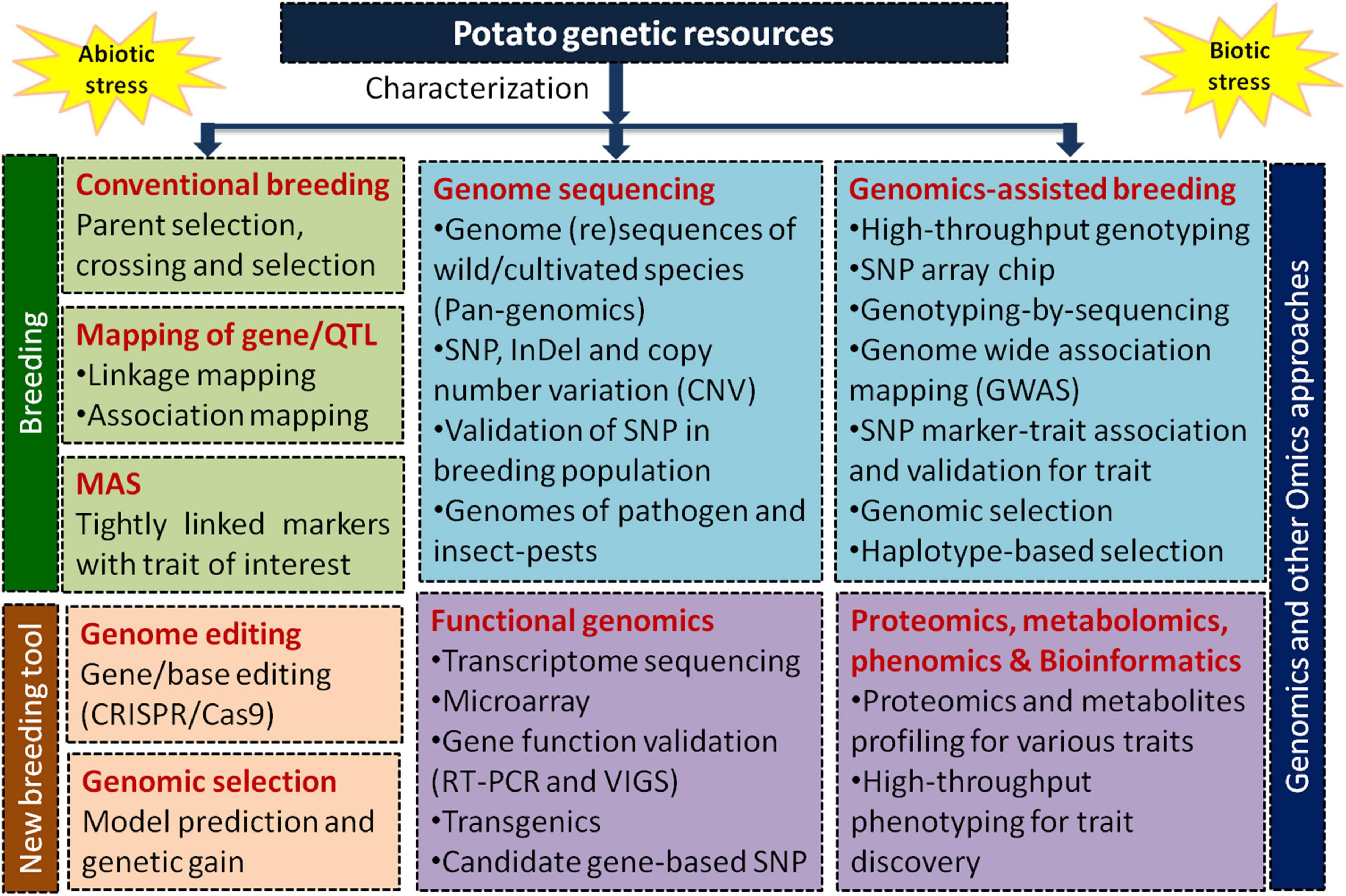
Figure 1. A schematic presentation of different approaches used for genetic enhancement and improvement of potato under various biotic and abiotic stresses applying breeding and modern genomics approaches like genome sequencing, functional genomics, genomics-assisted breeding through high-throughput genotyping by SNP markers, omics (transcriptomics, proteomics, metabolomics, and ionomics), high-throughput phenotyping, genome editing, and genomic selection.
Multi-Omics Approaches
Functional genomics allows the mining of genes for trait of interest through transcriptome analysis like RNA sequencing and microarray. Besides structural genomics, other omics approaches are transcriptomics (genes), proteomics (proteins), metabolomics (metabolites), phenomics (high-throughput phenotyping), and ionomics (mineral ions). The aims of multi-omics approaches are to acquire comprehensive and integrated understanding of biological processes (system biology) to identify various biological players/genes/regulatory elements underlying the traits like heat and drought stress (Aksoy et al., 2015). Numerous studies have been performed on transcriptomics in potato such as heat (Tang et al., 2020), drought (Moon et al., 2018; Chen et al., 2019), salinity (Li et al., 2020), and nitrogen deficiency (Tiwari et al., 2020a,b) but limited work has been carried out on proteomics, metabolomics, and ionomics (Hong et al., 2016; Boguszewska-Mańkowska et al., 2020). A few recent research works on multi-omics on biotic/abiotic stresses are mentioned in Table 1.
Genome-Wide Genetic Diversity and Association Studies Using High-Throughput Genotyping
High-Throughput Genotyping (HTG) is an essential requirement for genome-wide research on genetic diversity and association studies. First, genotyping-by-Sequencing (GBS) is a now popular method of HTG in crops including potato (Uitdewilligen et al., 2013; Bastien et al., 2018). GBS has been applied effectively in genome-wide studies in potato on genetic diversity and population structure (Pandey et al., 2021), QTL mapping (Schönhals et al., 2017), and SNP discovery (Caruana et al., 2019). Secondly, the SNP array-based HTG system has already been developed and applied in potato for population structure and SNP discovery using 20K SNP array (Vos et al., 2015, 2017), 22K SNP array for starch phosphorylation (Khlestkin et al., 2019), and 12K SNP array (Illumina) for genetic diversity in the genbank of the International Potato Centre, Peru (Ellis et al., 2018). Moreover, 8.3K SNP potato array has been demonstrated in studies on Synchytrium endobioticum resistance (Obidiegwu et al., 2015), genetic diversity (Berdugo-Cely et al., 2017), and physical mapping of yield and quality traits (Schönhals et al., 2017).
Genome-Wide Association Studies (GWAS) or linkage disequilibrium (LD) mapping is a family-based mapping approach to identify linked markers with the trait of interest in a diverse population structure. GWAS is more useful in a diverse germplasm which offers new perspectives toward the discovery of new genes and alleles especially for complex traits. The software STRUCTURE is very popular among scientific communities, and GWASpoly has been developed for tetraploid potato (Rosyara et al., 2016). GWAS has been applied in potato for QTLs/genes via LD mapping using 20K SNP array (Vos et al., 2017), wart resistance using 20K SNP array (Prodhomme et al., 2020) and common scab resistance using 12K SNP array (Yuan et al., 2020). Likewise, 8.3K SNP array has been used in LD mapping for phenotype, yield, and quality traits (Sharma et al., 2018), late blight resistance (Mosquera et al., 2016), and genetic diversity in 809 andigenum Colombian accessions (Berdugo-Cely et al., 2017). Applications of SNP array in potato for biotic and abiotic stress traits are summarized in Table 1.
Genomic Selection
Genomic selection (GS) or genome-wide selection or genomics-assisted breeding is a strategy to predict breeding model at whole-genome level for both simple and complex inherited traits. Therefore, partitioning of genetic variance and genome wide prediction with allele doses is very important in tetraploid potato (Endelman et al., 2018). GS allows the integration of phenotyping and HTG data of a training population (both genotyped and phenotyped) with a targeted breeding population (genotyped only) for the prediction of genomic models to select superior clones based on the genomic estimated breeding value (GEBV). GS accelerates the breeding cycle with an increase in genetic gain per unit time. Unlike animals and cereals, the application of GS is very limited in tetraploid potato (Caruana et al., 2019) and has been demonstrated recently for late blight and common scab resistance (Enciso-Rodriguez et al., 2018; Stich and Inghelandt, 2018). The advancement in sequencing, softwares, HTG, HTP, and marker-trait association can reduce the breeding cycle from over 10 to as few as 4 years to increase the genetic gain in potato (Slater et al., 2014; Table 1).
High-Throughput Phenotyping
Conventional phenotyping is often slow, has limited phenotyping capability, and mostly relies upon destructive sampling. Hence, modern High-Throughput Phenotyping (HTP) or phenomics is an automated precision phenotyping system allowing identification of key traits associated with phenotypic variation under different growth conditions. HTP is usually based on automation, sensors, high resolution imaging cameras (RGB, multi/hyperspectral and thermal sensors), unmanned aerial vehicle (UAV) and robotics to record real-time images and hardwares/softwares to analyze data from field or controlled growth chamber2. HTP enables measurement of phenotype, yield and its contributing traits, and physiological processes under stress such as photosynthesis, nutrient uptake and transport with precision and accuracy in a large set of genotypes with non-destructive sampling, for example the LemnaTec Scanalyzer 3D platform (LemnaTech GmbH, Germany). HTP has been applied in potato for phenology study in field (Prashar and Jones, 2014), heat and drought (Harsselaar et al., 2021), drought (Musse et al., 2021), and canopy cover using UAV (Colwell et al., 2021). Examples of HTP in heat and drought stress in potato are mentioned in Table 1.
Genome Editing
Genome editing is a powerful technology to create new variation in the genome with desirable gene combinations. Earlier sequence-specific nucleases (SSNs) methods like Zinc Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs) were used. Now, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) is the most widely used genome editing tool, which is an RNA-guided approach to target DNA/RNA sequences. CRISPR/Cas9 has revolutionized the plant research for multiple traits due to its ease in use, multiplexing capability, cost-effectiveness, and high efficiency. Although, in potato highly heterozygous and tetrasomic inheritance have complicated its deployment (Butler et al., 2015; Andersson et al., 2017) but found effective for PVY resistance using CRISPR/Cas9 (Makhotenko et al., 2019) and CRISPR/Cas13a (Zhan et al., 2019). Additionally, CRISPR/Cas9 has been demonstrated in potato for various other traits like cold-induced sweetening, glycoalkaloid content, homozygous mutants generation, acetochalactate synthase 1 and granule bound starch synthase genes (Nadakuduti et al., 2018; Dangol et al., 2019; Table 1).
Concluding Remarks
Biotic and abiotic stresses are major limiting factors of yield reduction in potato. Management of these stresses are more challenging under the climate change scenario due to emergence of new strains of pathogens and insect-pests, and erratic nature of environmental factors. Potato improvement through genomics-aided methods is essential to shorten the breeding cycle to develop new varieties. Earlier, conventional breeding, bi-parental linkage mapping, and MAS have been successfully demonstrated in potato. The potato genome sequencing and resequencing of Solanum species allow discovery of genes, markers and other regulatory elements to provide better understanding of the crop. Now, with the unprecedented advancement in sequencing technologies, genomes of Solanum species (pan-genomics), multi-omics for system biology approach (transcriptomics, proteomics, metabolomics, and ionomics), HTG by GBS and SNP array, HTP for precision phenotyping, GWAS and genomic selection would play crucial roles in genomics-led improvement of potato in the near future. There is an immense potential of genome editing for rapid breeding of climate resilient varieties resistant/tolerant to biotic and abiotic stresses. Nonetheless, the availability of an efficient CRISPR/Cas system, target gene selection, plant transformation, and off target mutants would be some challenges in tetraploid crop. Overall, designs of potato that apply genomics, particularly genome editing and genomic selection, and other omics are inevitable in the future.
Author Contributions
JT conceived idea and wrote manuscript. JT, TB, RZ, NB, DD, SN, HK, CC, SL, and VK performed research and literature collection. RS, AP, and MK edited the manuscript. All authors approved the manuscript.
Funding
This work was funded under Biotechnology Program by the ICAR-CPRI, Shimla, CABin Scheme, and ICAR-LBS Outstanding Young Scientist Project awarded to JT.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Competent Authority of ICAR-CPRI, CABin Scheme (ICAR-IASRI, New Delhi) and ICAR-LBS Outstanding Young Scientist Project for necessary support.
Footnotes
References
Ahmed, H. A. A., Şahin, N. K., Akdoğan, G., Yaman, C., Köm, D., and Uranbey, S. (2020). Variability in salinity stress tolerance of potato (Solanum tuberosum L.) varieties using in vitro screening. Ciênc. Agrotec. 44:e004220.
Aksoy, E., Demirel, U., Öztürk, Z. N., Çalişkan, S., and Çalişkan, M. E. (2015). Recent advances in potato genomics, transcriptomics, and transgenics under drought and heat stresses: a review. Turk. J. Bot. 39, 920–940.
Andersson, M., Turesson, H., Nicolia, A., Fält, A. S., Samuelsson, M., and Hofvander, P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36, 117–128. doi: 10.1007/s00299-016-2062-3
Anithakumari, A. M., Nataraja, K. N., Visser, R. G., and van der Linden, C. G. (2012). Genetic dissection of drought tolerance and recovery potential by quantitative trait locus mapping of a diploid potato population. Mol. Breed. 30, 1413–1429. doi: 10.1007/s11032-012-9728-5
Aversano, R., Contaldi, F., Ercolano, M. R., Grosso, V., Iorizzo, M., and Tatino, F. (2015). The Solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. Plant Cell 27, 954–968. doi: 10.1105/tpc.114.135954
Bastarache, P., Wajnberg, G., Dumas, P., Chacko, S., Lacroix, J., Crapoulet, N., et al. (2020). Transcriptomics-based approach identifies spinosad-associated targets in the Colorado potato beetle. Leptinotarsa decemlineata. Insects 11:820. doi: 10.3390/insects11110820
Bastien, M., Boudhrioua, C., Fortin, G., and Belzile, F. (2018). Exploring the potential and limitations of genotyping-by-sequencing for SNP discovery and genotyping in tetraploid potato. Genome 61, 449–456. doi: 10.1139/gen-2017-0236
Berdugo-Cely, J., Valbuena, R. I, Sa’nchez-Betancourt, E., Barrero, L. S., and Yockteng, R. (2017). Genetic diversity and association mapping in the Colombian central collection of Solanum tuberosum L. Andigenum group using SNPs markers. PLoS One 12:e0173039. doi: 10.1371/journal.pone.0173039
Boguszewska-Mańkowska, D., Gietler, M., and Nykiel, M. (2020). Comparative proteomic analysis of drought and high temperature response in roots of two potato cultivars. Plant Growth Regul. 92, 345–363.
Bonierbale, M. W., Plaisted, R. L., and Tanksley, S. D. (1988). RFLP Maps based on a common set of clones reveal modes of chromosomal evolution in potato and tomato. Genetics 120, 1095–1103. doi: 10.1093/genetics/120.4.1095
Butler, N. M., Atkins, P. A., Voytas, D. F., and Douches, D. S. (2015). Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PLoS One 10:e0144591. doi: 10.1371/journal.pone.0144591
Cao, W., Gan, L., Shang, K., Wang, C., Song, Y., Liu, H., et al. (2020). Global transcriptome analyses reveal the molecular signatures in the early response of potato (Solanum tuberosum L.) to Phytophthora infestans, Ralstonia solanacearum, and Potato virus Y infection. Planta 252:57. doi: 10.1007/s00425-020-03471-6
Caruana, B. M., Pembleton, L. W., Constable, F., Rodoni, B., Slater, A. T., and Cogan, N. O. I. (2019). Validation of genotyping by sequencing using transcriptomics for diversity and application of genomic selection in tetraploid potato. Front. Plant Sci. 10:670. doi: 10.3389/fpls.2019.00670
Chen, Y., Li, C., Yi, J., Yang, Y., Lei, C., and Gong, M. (2019). Transcriptome response to drought, rehydration and re-dehydration in potato. Int. J. Mol. Sci. 21:159. doi: 10.3390/ijms21010159
Colwell, F. J., Souter, J., Bryan, G. J., Compton, L. J., Boonham, N., and Prashar, A. (2021). Development and validation of methodology for estimating potato canopy structure for field crop phenotyping and improved breeding. Front. Plant Sci. 12:612843. doi: 10.3389/fpls.2021.612843
Dahal, K., Li, X.-Q., Tai, H., Creelman, A., and Bizimungu, B. (2019). Improving potato stress tolerance and tuber yield under a climate change scenario–a current overview. Front. Plant Sci. 10:563. doi: 10.3389/fpls.2019.00563
Dangol, S. D., Barakate, A., Stephens, J., Çaliskan, M. E., and Bakhsh, A. (2019). Genome editing of potato using CRISPR technologies: current development and future prospective. Plant Cell Tiss. Org. Cult. 139, 403–416. doi: 10.1007/s11240-019-01662-y
D’hoop, B., Keizer, P. C., Paulo, M. J., Visser, R. F., Eeuwijk, F., and Eck, H. (2014). Identification of agronomically important QTL in tetraploid potato cultivars using a marker–trait association analysis. Theor. Appl. Genet. 127, 731–748. doi: 10.1007/s00122-013-2254-y
Ellis, D., Chavez, O., Coombs, J., Soto, J., Gomez, R., Douches, D., et al. (2018). Genetic identity in genebanks: application of the SolCAP 12K SNP array in fingerprinting and diversity analysis in the global in trust potato collection. Genome 61, 523–537. doi: 10.1139/gen-2017-0201
Enciso-Rodriguez, F., Douches, D., Lopez-Cruz, M., Coombs, J., and de los Campos, G. (2018). Genomic selection for late blight and common scab resistance in tetraploid potato (Solanum tuberosum). G3 8, 2471–2481. doi: 10.1534/g3.118.200273
Endelman, J. B., Carley, C. A. S., Bethke, P. C., Coombs, J. J., Clough, M. E., da Silva, W. L., et al. (2018). Genetic variance partitioning and genome-wide prediction with allele dosage information in autotetraploid potato. Genetics 209, 77–87. doi: 10.1534/genetics.118.300685
Fao. (2010). The Second Report on the State of the World’s Plant Genetic Resources for Food and Agriculture. Rome: Food & Agriculture Organization of the United Nations.
Flint-Garcia, S. A., Thornsberry, J. M., and Buckler, E. S. (2003). Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54, 357–374. doi: 10.1146/annurev.arplant.54.031902.134907
Fofana, B., Somalraju, A., Fillmore, S., Zaidi, M., Main, D., and Ghose, K. (2020). Comparative transcriptome expression analysis in susceptible and resistant potato (Solanum tuberosum) cultivars to common scab (Streptomyces scabies) revealed immune priming responses in the incompatible interaction. PLoS One 15:e0235018. doi: 10.1371/journal.pone.0235018
Gebhardt, C., Ballvora, A., Walkemeier, B., Oberhagemann, P., and Schüler, K. (2004). Assessing genetic potential in germplasm collections of crop plants by marker-trait association: a case study for potatoes with quantitative variation of resistance to late blight and maturity type. Mol. Breed. 13, 93–102. doi: 10.1023/b:molb.0000012878.89855.df
Hamooh, B. T., Sattar, F. A., Wellman, G., and Mousa, M. (2021). Metabolomic and biochemical analysis of two potato (Solanum tuberosum L.) cultivars exposed to in vitro osmotic and salt stresses. Plants 10:98. doi: 10.3390/plants10010098
Handayani, T., Gilani, S. A., and Watanabe, K. N. (2019). Climatic changes and potatoes: how can we cope with the abiotic stresses? Breed. Sci. 69, 545–563. doi: 10.1270/jsbbs.19070
Hardigan, M. A., Laimbeer, F. P. E., Newton, L., Crisovan, E., Hamilton, J. P., Vaillancourt, B., et al. (2017). Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc. Natl. Acad. Sci. U.S.A. 114, E9999–E10008. doi: 10.1073/pnas.1714380114
Harsselaar, J. K. V., Claußen, J., Lübeck, J., Wörlein, N., Uhlmann, N., Sonnewald, U., et al. (2021). X-ray CT phenotyping reveals bi-phasic growth phases of potato tubers exposed to combined abiotic stress. Front. Plant Sci. 12:613108. doi: 10.3389/fpls.2021.613108
Hawkes, J. G. (1990). The Potato, Evolution, Biodiversity and Genetic Resources. London: Belhaven Press.
Hein, I., Birch, P. R. J., Danan, S., Lefebvre, V., Odeny, D. A., Gebhardt, C., et al. (2009). Progress in mapping and cloning qualitative and quantitative resistance against Phytophthora infestans in potato and its wild relatives. Potato Res. 52, 215–227.
Hong, J., Yang, L., Zhang, D., and Shi, J. (2016). Plant metabolomics: an indispensable system biology tool for plant science. Intl. J. Mol. Sci. 17:767. doi: 10.3390/ijms17060767
Jozefowicz, A. M., Hartmann, A., Matros, A., Schum, A., and Mock, H. P. (2017). Nitrogen deficiency induced alterations in the root proteome of a pair of potato (Solanum tuberosum L.) varieties contrasting for their response to low N. Proteomics 17:1700231. doi: 10.1002/pmic.201700231
Khlestkin, V. K., Rozanova, I. V., Efimov, V. M., and Khlestkina, E. K. (2019). Starch phosphorylation associated SNPs found by genome-wide association studies in the potato (Solanum tuberosum L.). BMC Genetics 20:29. doi: 10.1186/s12863-019-0729-9
Kochetov, A. V., Egorova, A. A., Glagoleva, A. Y., Strygina, K. V., Khlestkina, E. K., Gerasimova, S. V., et al. (2020). The mechanism of potato resistance to Globodera rostochiensis: comparison of root transcriptomes of resistant and susceptible Solanum phureja genotypes. BMC Plant Biol. 20:350. doi: 10.1186/s12870-020-02334-2
Kyriakidou, M., Achakkagari, S. R., Gálvez López, J. H., Zhu, X., Tang, C. Y., Tai, H. H., et al. (2020). Structural genome analysis in cultivated potato taxa. Theor. Appl. Genet. 133, 951–966. doi: 10.1007/s00122-019-03519-6
Leisner, C. P., Hamilton, J. P., Crisovan, E., Manrique-Carpintero, N. C., Marand, A. P., Newton, L., et al. (2018). Genome sequence of M6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. Plant J. 94, 562–570. doi: 10.1111/tpj.13857
Li, H., Luo, W., Ji, R., Xu, Y., Xu, G., Qiu, S., et al. (2021). A comparative proteomic study of cold responses in potato leaves. Heliyon 7:e06002. doi: 10.1016/j.heliyon.2021.e06002
Li, Q., Qin, Y., Hu, X., Li, G., Ding, H., Xiong, X., et al. (2020). Transcriptome analysis uncovers the gene expression profile of salt-stressed potato (Solanum tuberosum L.). Sci. Rep. 10:5411. doi: 10.1038/s41598-020-62057-0
Liu, B., Kong, L., Zhang, Y., and Liao, Y. (2021). Gene and metabolite integration analysis through transcriptome and metabolome brings new insight into heat stress tolerance in potato (Solanum tuberosum L.). Plants 10:103. doi: 10.3390/plants10010103
Makhotenko, A. V., Khromov, A. V., Snigir, E. A., Makarova, S. S., Makarov, V. V., Suprunova, T. P., et al. (2019). Functional analysis of coilin in virus resistance and stress tolerance of potato Solanum tuberosum using CRISPR-Cas9 editing. Dokl. Biochem. Biophys. 484, 88–91. doi: 10.1134/S1607672919010241
Moon, K. B., Ahn, D. J., Park, J. S., Jung, W. Y., Cho, H. S., Kim, H. R., et al. (2018). Transcriptome profiling and characterization of drought-tolerant potato plant (Solanum tuberosum L.). Mol. Cells 41, 979–992. doi: 10.14348/molcells.2018.0312
Mosquera, T., Alvarez, M. F., Jiménez-Gómez, J. M., Muktar, M. S., Paulo, M. J., Steinemann, S., et al. (2016). Targeted and untargeted approaches unravel novel candidate genes and diagnostic SNPs for quantitative resistance of the potato (Solanum tuberosum L.) to Phytophthora infestans causing the late blight disease. PLoS One 11:e0156254. doi: 10.1371/journal.pone.0156254
Musse, M., Hajjar, G., Ali, N., Billiot, B., Joly, G., Pépin, J., et al. (2021). A global non-invasive methodology for the phenotyping of potato under water deficit conditions using imaging, physiological and molecular tools. Plant Methods 17:81. doi: 10.1186/s13007-021-00771-0
Nadakuduti, S. S., Buell, C. R., Voytas, D. F., Starker, C. G., and Douches, D. S. (2018). Genome editing for crop improvement – applications in clonally propagated polyploids with a focus on potato (Solanum tuberosum L.). Front. Plant Sci. 9:1607. doi: 10.3389/fpls.2018.01607
Obidiegwu, J. E., Sanetomo, R., Flath, K., Tacke, E., Hofferbert, H. R., and Hofmann, A. (2015). Genomic architecture of potato resistance to Synchytrium endobioticum disentangled using SSR markers and the 8.3k SolCAP SNP genotyping array. BMC Genetics 16:38. doi: 10.1186/s12863-015-0195-y
Pandey, J., Scheuring, D. C., Koym, J. W., Coombs, J., Novy, R. G., Thompson, A. L., et al. (2021). Genetic diversity and population structure of advanced clones selected over forty years by a potato breeding program in the USA. Sci. Rep. 11:8344. doi: 10.1038/s41598-021-87284-x
Pham, G. M., Hamilton, J. P., Wood, J. C., Burke, J. T., Zhao, H., Vaillancourt, B., et al. (2020). Construction of a chromosome-scale long-read reference genome assembly for potato. Gigascience 9:giaa100. doi: 10.1093/gigascience/giaa100
Potato Genome Sequencing Consortium. (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi: 10.1038/nature10158
Prashar, A., and Jones, H. G. (2014). Infra-red thermography as a high-throughput tool for field phenotyping. Agronomy 4, 397–417.
Prodhomme, C., Vos, P. G., Paulo, M. J., Tammes, J. E., Visser, R., Vossen, J. H., et al. (2020). Distribution of P1(D1) wart disease resistance in potato germplasm and GWAS identification of haplotype-specific SNP markers. Theor. Appl. Genet. 133, 1859–1871. doi: 10.1007/s00122-020-03559-3
Rajamaki, M. L., Sikorskaite-Gudziuniene, S., Sarmah, N., Varjosalo, M., and Valkonen, J. P. T. (2020). Nuclear proteome of virus-infected and healthy potato leaves. BMC Plant Biol. 20:355. doi: 10.1186/s12870-020-02561-7
Ramakrishnan, A. P., Ritland, C. E., Sevillano, R. H. B., and Riseman, A. (2015). Review of potato molecular markers to enhance trait selection. Amer. J. Potato Res. 92, 455–472. doi: 10.1007/s12230-015-9455-7
Rosyara, U. R., De Jong, W. S., Douches, D. S., and Endelman, J. B. (2016). Software for genome-wide association studies in autopolyploids and its application to potato. Plant Genome 9, 1–10. doi: 10.3835/plantgenome2015.08.0073
Schönhals, E. M., Ding, J., Ritter, E., Paulo, M. J., Cara, N., Tacke, E., et al. (2017). Physical mapping of QTL for tuber yield, starch content and starch yield in tetraploid potato (Solanum tuberosum L.) by means of genome wide genotyping by sequencing and the 8.3 K SolCAP SNP array. BMC Genomics 18:642. doi: 10.1186/s12864-017-3979-9
Sharma, S. K., Bolser, D., de Boer, J., Sønderkær, M., Amoros, W., Carbon, M. F., et al. (2013). Construction of reference chromosome-scale pseudomolecules for potato: integrating the potato genome with genetic and physical maps. G3 3, 2031–2047. doi: 10.1534/g3.113.007153
Sharma, S. K., MacKenzie, K., McLean, K., Dale, F., Daniels, S., and Bryan, G. J. (2018). Linkage disequilibrium and evaluation of genome-wide association mapping models in tetraploid potato. G3 8, 3185–3202. doi: 10.1534/g3.118.200377
Simko, I., Costanzo, S., Haynes, K. G., Christ, B. J., and Jones, R. W. (2004). Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor. Appl. Genet. 108, 217–224. doi: 10.1007/s00122-003-1431-9
Singh, A., Siddappa, S., Bhardwaj, V., Singh, B., Kumar, D., and Singh, B. P. (2015). Expression profiling of potato cultivars with contrasting tuberization at elevated temperature using microarray analysis. Plant Physiol. Biochem. 97, 108–116. doi: 10.1016/j.plaphy.2015.09.014
Singh, A. K., Chakrabarti, S. K., Singh, B., Sharma, J., and Dua, V. K. (2020). Potato Science and Technology for Sub-Tropics. New Delhi: New India Publishing Agency.
Slater, A. T., Wilson, G. M., Cogan, N. O. I., Forster, J. W., and Hayes, B. J. (2014). Improving the analysis of low heritability complex traits for enhanced genetic gain in potato. Theor. Appl. Genet. 127, 809–820. doi: 10.1007/s00122-013-2258-7
Spooner, D. M., Ghislain, M., Simon, R., Jansky, S. H., and Gavrilenko, T. (2014). Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot. Rev. 80, 283–383.
Stich, B., and Inghelandt, D. V. (2018). Prospects and potential uses of genomic prediction of key performance traits in tetraploid potato. Front. Plant Sci. 9:159. doi: 10.3389/fpls.2018.00159
Strachan, S. M., Armstrong, M. R., Kaur, A., Wright, K. M., Lim, T. Y., Baker, K., et al. (2019). Mapping the H2 resistance effective against Globodera pallida pathotype Pa1 in tetraploid potato. Theor. Appl. Genet. 132, 1283–1294. doi: 10.1007/s00122-019-03278-4
Tang, R., Gupta, S. K., Niu, S., Li, X. Q., Yang, Q., Chen, G., et al. (2020). Transcriptome analysis of heat stress response genes in potato leaves. Mol. Biol. Rep. 47, 4311–4321. doi: 10.1007/s11033-020-05485-5
Tiwari, J. K., Buckseth, T., Singh, R. K., Kumar, M., and Kant, S. (2020a). Prospects of improving nitrogen use efficiency in potato: lessons from transgenics to genome editing strategies in plants. Front. Plant Sci. 11:597481. doi: 10.3389/fpls.2020.597481
Tiwari, J. K., Buckseth, T., Zinta, R., Saraswati, A., Singh, R. K., Rawat, S., et al. (2020b). Transcriptome analysis of potato shoots, roots and stolons under nitrogen stress. Sci. Rep. 10:1152. doi: 10.1038/s41598-020-58167-4
Tiwari, J. K., Rawat, S., Luthra, S. K., Zinta, R., Sahu, S., Varshney, S., et al. (2021). Genome sequence analysis provides insights on genomic variation and late blight resistance genes in potato somatic hybrid (parents and progeny). Mol. Biol. Rep. 48, 623–635. doi: 10.1007/s11033-020-06106-x
Uitdewilligen, J. G., Wolters, A. M., D’hoop, B. B., Borm, T. J., Visser, R. G., and van Eck, H. J. (2013). A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS One 8:e62355. doi: 10.1371/journal.pone.0062355
Van Os, H., Andrzejewski, S., Bakker, E., Barrena, I., Bryan, G., Caromel, B., et al. (2006). Construction of a 10,000-marker ultradense genetic recombination map of potato: providing a framework for accelerated gene isolation and a genomewide physical map. Genetics 73, 1075–1087. doi: 10.1534/genetics.106.055871
Vos, P. G., Paulo, M. J., Voorrips, R. E., Visser, R. G., van Eck, H. J., and van Eeuwijk, F. A. (2017). Evaluation of LD decay and various LD-decay estimators in simulated and SNP-array data of tetraploid potato. Theor. Appl. Genet. 130, 123–135. doi: 10.1007/s00122-016-2798-8
Vos, P. G., Uitdewilligen, J. G., Voorrips, R. E., Visser, R. G., and van Eck, H. J. (2015). Development and analysis of a 20K SNP array for potato (Solanum tuberosum): an insight into the breeding history. Theor. Appl. Genet. 128, 2387–2401. doi: 10.1007/s00122-015-2593-y
Wang, B., He, T., Zheng, X., Song, B., and Chen, H. (2021). Proteomic analysis of potato responding to the invasion of Ralstonia solanacearum UW551 and its type III secretion system mutant. Mol. Plant Microbe Interact. 34, 337–350. doi: 10.1094/MPMI-06-20-0144-R
Xiao, C., Huang, M., Gao, J., Wang, Z., Zhang, D., Zhang, Y., et al. (2020). Comparative proteomics of three Chinese potato cultivars to improve understanding of potato molecular response to late blight disease. BMC Genomics 21:880. doi: 10.1186/s12864-020-07286-3
Yuan, J., Bizimungu, B., Koeyer, D. D., Rosyara, U., Wen, Z., and Lagüe, M. (2020). Genome-wide association study of resistance to potato common scab. Potato Res. 63, 253–266. doi: 10.1007/s11033-019-04951-z
Zhan, X., Zhang, F., Zhong, Z., Chen, R., Wang, Y., Chang, L., et al. (2019). Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 17, 1814–1822. doi: 10.1111/pbi.13102
Zhang, J., Wang, Y., Zhao, Y., Zhang, Y., Zhang, J., Ma, H., et al. (2020). Transcriptome analysis reveals Nitrogen deficiency induced alterations in leaf and root of three cultivars of potato (Solanum tuberosum L.). PLoS One 15:e0240662. doi: 10.1371/journal.pone.0240662
Keywords: biotic, abiotic, breeding, potato, genomics, omics approaches
Citation: Tiwari JK, Buckseth T, Zinta R, Bhatia N, Dalamu D, Naik S, Poonia AK, Kardile HB, Challam C, Singh RK, Luthra SK, Kumar V and Kumar M (2022) Germplasm, Breeding, and Genomics in Potato Improvement of Biotic and Abiotic Stresses Tolerance. Front. Plant Sci. 13:805671. doi: 10.3389/fpls.2022.805671
Received: 30 October 2021; Accepted: 17 January 2022;
Published: 07 February 2022.
Edited by:
Mostafa Abdelwahed Abdelrahman, Aswan University, EgyptReviewed by:
Rupesh Kailasrao Deshmukh, National Agri-Food Biotechnology Institute, IndiaPradeep Kumar, Central Arid Zone Research Institute (ICAR), India
Copyright © 2022 Tiwari, Buckseth, Zinta, Bhatia, Dalamu, Naik, Poonia, Kardile, Challam, Singh, Luthra, Kumar and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jagesh Kumar Tiwari, amFnZXNodGl3YXJpQGdtYWlsLmNvbQ==
 Jagesh Kumar Tiwari
Jagesh Kumar Tiwari Tanuja Buckseth
Tanuja Buckseth Rasna Zinta1
Rasna Zinta1 Nisha Bhatia
Nisha Bhatia Hemant B. Kardile
Hemant B. Kardile Rajesh K. Singh
Rajesh K. Singh Manoj Kumar
Manoj Kumar