- 1Department of Botany, Hansraj College, University of Delhi, New Delhi, India
- 2EVA4.0 Unit, Faculty of Forestry and Wood Sciences, Czech University of Life Sciences Prague, Prague, Czechia
- 3Centre for Interdisciplinary Research in Basic Sciences, Jamia Millia Islamia, New Delhi, India
- 4Plant Virus and Vector Interactions Group, Crop Research Institute, Prague, Czechia
- 5Molecular Biology Research Laboratory, Department of Zoology, Deshbandhu College, University of Delhi, New Delhi, India
- 6DBC-i4 Center, Deshbandhu College, University of Delhi, New Delhi, India
The calcium (Ca2+) signaling is a crucial event during plant-herbivore interaction, which involves a transient change in cytosolic Ca2+ concentration, which is sensed by Ca2+-sensors, and the received message is transduced to downstream target proteins leading to appropriate defense response. Calmodulin-like proteins (CMLs) are calcium-sensing plant-specific proteins. Although CMLs have been identified in a few plants, they remained uncharacterized in leguminous crop plants. Therefore, a wide-range analysis of CMLs of soybean was performed, which identified 41 true CMLs with greater than 50% similarity with Arabidopsis CMLs. The phylogenetic study revealed their evolutionary relatedness with known CMLs. Further, the identification of conserved motifs, gene structure analysis, and identification of cis-acting elements strongly supported their identity as members of this family and their involvement in stress responses. Only a few Glycine max CMLs (GmCMLs) exhibited differential expression in different tissue types, and rest of them had minimal expression. Additionally, differential expression patterns of GmCMLs were observed during Spodoptera litura-feeding, wounding, and signaling compound treatments, indicating their role in plant defense. The three-dimensional structure prediction, identification of interactive domains, and docking with Ca2+ ions of S. litura-inducible GmCMLs, indicated their identity as calcium sensors. This study on the characterization of GmCMLs provided insights into their roles in calcium signaling and plant defense during herbivory.
Introduction
Plants being sessile are constantly attacked by various environmental cues that can cause abiotic or biotic stresses (Stotz et al., 2000). Among all, insect attack is crucial biotic stress, affecting the plants’ fitness, performance, and productivity (Stotz et al., 2000; Conrath et al., 2002; Dodd et al., 2010; Hancock et al., 2015; Singh et al., 2018, 2020a,b,2021c; Chen and Mao, 2020; Keshan et al., 2021; Singh and Singh, 2021). To protect themselves, plants activate their defense mechanism and produce different defensive compounds like secondary metabolites, volatile organic compounds, protease inhibitors, and other antiherbivore chemicals (Gatehouse, 2002; Halitschke and Baldwin, 2004; Kliebenstein, 2004; Schuler, 2011; Vadassery et al., 2012b; Lortzing and Steppuhn, 2016; Singh et al., 2021a,c). This activation of biosynthesis of defensive compounds is governed by certain early events that facilitate the detection of herbivory and signal transduction. However, stimulus perception and signal transduction during herbivory is not understood well. Moreover, plants must discriminate among various environmental stimuli to trigger a specific response by activating downstream cellular signaling. Therefore, it is mandatory to determine the perception molecules, crucial players of signal transduction, and molecular mechanism behind plant defense against herbivory (Kong et al., 2013; Singh et al., 2016, 2020c; Kumar et al., 2020).
Plants recognize herbivore attack by wounding patterns and interaction of herbivore/damage-associated molecular patterns (HAMPs/DAMPs) with pattern recognition receptors (PRRs) (Mithöfer and Boland, 2008). This interaction activates appropriate downstream signal transduction pathways for achieving stimulus-specific responses. The process of coupling the perception of feeding herbivores to plant adaptive response involves transient changes in the intracellular Ca2+ concentration (Maffei et al., 2004; Thor and Peiter, 2014). During the process, an increase in Ca2+ levels occurs due to the transport of Ca2+ across the membrane or from subcellular organelles, which is facilitated by plasma membrane Ca2+ -permeable channels, such as cyclic nucleotide-gated channels (CNGCs), glutamate receptor-type cation channel (GLRs), stretch-activated Ca2+ channel (OSCAs), and MID1-complementing activity (MCA) families and vacuolar channel TWO-PORE CHANNEL1 (TPC1) (Maffei et al., 2004; Dodd et al., 2010; Vadassery et al., 2012a; Kiep et al., 2015; DeFalco et al., 2016; Vincent et al., 2017; Meena et al., 2019; DeFalco and Zipfel, 2021). This transport of Ca2+ causes spatial and temporal variations in cellular distribution, frequency, amplitude, kinetics of Ca2+, and intracellular Ca2+ level leading to the generation of calcium signature, which encrypts information from primary stimuli to encode specific intracellular responses (Ranty et al., 2016). These calcium signals are decoded further by various calcium sensor proteins that consist of canonical Ca2+ binding EF-hand motif with conserved helix-loop-helix structure coordinating one Ca2+ (Lecourieux et al., 2002; Gifford et al., 2007). These proteins perceive and interpret Ca2+ signals through binding to Ca2+ ions leading to a change in their conformation and functions (Aldon et al., 2018). In plants, Ca2+ binding proteins are represented by complex families. They can be either Ca2+ responders, which convey the signal via enzymatic reactions, such as Ca2+-dependent protein kinases (CDPKs), or the non-catalytic sensor relay proteins, such as Calmodulins (CaMs), Calmodulin-like proteins (CMLs), and calcineurin B-like proteins (CBLs), which are activated upon binding to Ca2+, which promote downstream signaling (DeFalco et al., 2010).
Calmodulin like proteins are principal representative of Ca2+ sensor proteins (DeFalco et al., 2010; Kumar et al., 2020) and are characterized by two to six EF-hand motifs and are known to evolve from ancestor CaM, sharing at least 15% amino acid identity (Perochon et al., 2011; Bender and Snedden, 2013). So far, CMLs are pointed out to be involved in the developmental process, plant immunity, and stress responses by targeting many kinases, ion transporters, metabolic enzymes, phosphatases, and transcription factors (Reddy et al., 2011). The EF-hand motifs show a high affinity toward cooperative binding with Ca2+ that expose the hydrophobic surface, which allows them to interact with downstream target proteins (Vadassery et al., 2012a). CML8, CML37, CML38, and CML39 are responsive to drought, salinity, and hormonal treatment in Arabidopsis (Vanderbeld and Snedden, 2007; Park et al., 2010). CML9 is regulated by abscisic acid (ABA) and Pseudomonas syringae infection (Magnan et al., 2008; Leba et al., 2012). CML42 is known to cause aberration in trichomes (Dobney et al., 2009), and CML24 is involved in alterations in flowering time and ion stress (Delk et al., 2005; Hubbard et al., 2008). Plants possess a repertoire of CMLs, which are unique to them with 50 members in Arabidopsis (McCormack et al., 2005), 52 in Solanum lycopersicum (Munir et al., 2016), 19 in Lotus japonicas (Liao et al., 2017), and 62 in Vitis vinifera (Vandelle et al., 2018). Few CMLs of Arabidopsis thaliana (CML9, 11, 12, 16, 17, 23, and 42) are upregulated upon employing oral secretion of Spodoptera littoralis (Vadassery et al., 2012a,b). CML37 and CML42 act antagonistically to regulate stress responses in Arabidopsis by altering phytohormone signals (Heyer et al., 2021). However, the involvement of these Ca2+ sensors has not been investigated much in crop plants. To achieve more insights into the function of CMLs and recognize their involvement in plant defense against herbivory, it would be crucial to investigate them during soybean-Spodoptera litura interaction.
Glycine max, commonly known as soybean, is a dietary staple crop of Asian countries and a native legume plant of Southeastern Asia (Soyastats, 2010; Badole and Bodhankar, 2012; Badole and Mahamuni, 2013; Naresh et al., 2019). It consists of a balanced proportion of amino acids essential for the growth of the human body (Naresh et al., 2019). The genome of soybean is fully sequenced, and the processes of gene regulation, disease resistance, and nodulation are well defined (Gresshoff and Ferguson, 2017). However, insect pests can adversely affect the yield and quality of soybean (Cui et al., 1997). S. litura (common cutworm), a polyphagous insect, is one of the major destructive and widespread soybean pests throughout the Asia-Pacific region. Plants have employed various constitutive and induced defense strategies along with sophisticated signaling networks to combat predation by herbivores (Singh et al., 2021b). The induced responses are more effective for plants and lead to increased tolerance against herbivores (Stotz et al., 2000; Zhang and Yang, 2000; Fan et al., 2012). S. litura, a nocturnal moth and a serious polyphagous defoliator, infests soybean plants through the vigorous eating pattern of larvae (Singh et al., 2021b). The proteolytic activity of the gut serine protease of the larvae is mainly responsible for the significant damage to crops (Vasudev and Sohal, 2016). Therefore, it is essential to elucidate how S. litura-infestation induces defense signaling in G. max evoking the plant defense mechanism.
In the present study, a comprehensive investigation of CMLs of G. max was executed across the genome. We studied the genomic organization, evolutionary and phylogenetic relationships, gene structure analysis, and identification of EF-hand motifs of Glycine max CMLs (GmCMLs). We also evaluated the mRNA levels of GmCMLs in different tissue types. The investigation on change in mRNA levels of GmCMLs during S. litura-infestation revealed the involvement of 36 GmCMLs in plant defense against herbivory. We also screened the promoter sequences of GmCMLs to identify cis-acting elements that revealed the presence of Jasmonic acid (JA)-response elements and salicylic acid (SA)-related elements. Further, transcript profiling of S. litura-inducible CMLs upon defense-signaling compound treatments was also executed, which revealed their differential expression. We also predicted the three-dimensional structure of S. litura-inducible GmCMLs and its interaction with Ca2+, which confirmed their identity and involvement in Ca2+ signaling. This analysis has offered promising candidates that can function as an essential component of Ca2+ signaling during G. max-S. litura interaction and can be valuable for deciphering the early events involved in the upregulation of plant defense in soybean and beyond.
Materials and Methods
Sequence Retrieval, Phylogenetic Analysis, and Similarity Search of Calmodulin-Like Proteins of Glycine max
The protein and gene sequences of CMLs of G. max were identified and retrieved from the soybean database1 and phytozome database2. The DNA and peptide sequences of GmCMLs were evaluated manually utilizing BLASTN and BLASTP3. The base pair lengths and number of amino acids for each GmCMLs were retrieved from NCBI4.
To study the evolutionary relationships among model plants, monocot, and leguminous dicot, amino acid sequences of homologous GmCMLs from Medicago truncatula, Arabidopsis thaliana, and Oryza sativa were retrieved from Phytozome (see text footnote 2), TAIR5, and Oryzabase6, respectively. The amino acid sequences were then submitted to multiple sequence alignment using Clustal Omega7 (Madeira et al., 2019). The generated multiple sequence alignment was subjected to “construct” phylogenetic tree. The details of constructed phylogenetic tree were downloaded using “Download” phylogenetic data and saved in newick format. For better presentation of the phylogenetic tree, the saved phylogenetic tree was uploaded on iTOL8 (Letunic and Bork, 2021).
The identity of GmCML peptide sequences was compared with the protein sequence of CMLs of Arabidopsis thaliana using the BLAST engine of the TAIR database9.
Expression Analysis of Glycine max CMLs During Spodoptera litura-Infestation
Plant Growth
The soybean seeds (Pusa 9712) were washed, soaked for 2 h, and kept at 26−27°C in a dark chamber between 2 layers of tissue paper, with continuous moistening with distilled water for germination. After 3–4 days, the germinated seedlings were transferred to the sterilized soilrite containing pots. The seedlings were grown in a plant growth chamber under controlled conditions (photoperiod: 16 h light and 8 h dark; temperature: 26−27°C; humidity: 55–60%) and watered regularly.
Rearing and Maintenance of Spodoptera litura
The larvae of S. litura were procured from ICAR – National Bureau Of Agricultural Insect Resources, Bangalore, India, and reared in the laboratory using standard protocols at 27°C and 65–70% relative humidity on a 14/10 h light/dark cycle (Singh et al., 2021b). The freshly molted fourth instar larvae were starved for 12 h before releasing them on plants for experimentation and bioassays.
Spodoptera litura-Infestation and Mechanical Wounding on Soybean Plants and Sample Collection
For S. litura-infestation, one-month-old soybean plants were exposed to one larva per plant. Plants without any treatments were served as control. Mechanical wounding was performed according to Singh et al. (2008). The entire shoot of treated (sample) and untreated (control) plants, after 1 h of infestation/wounding from three independent experiments, was harvested and immediately snap-frozen at −80°C for further analysis.
RNA Isolation and cDNA Synthesis
Trizol (TRI) reagent (Sigma Aldrich, Spruce Street, St. Louis, MO, United States) was used to isolate RNA as per the given protocol (Singh et al., 2018, 2021b; Keshan et al., 2021). The RNA concentrations were measured on a spectrophotometer, and RNA integrity was checked by agarose gel electrophoresis. The isolated RNA was used to synthesize first-strand cDNA using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, United States).
Primer Design and Gene Expression Analysis Using qPCR
Primer 3 (v.0.4.0)10 tool was used to design real-time (RT) PCR primers. Applied Biosystems™ 7500 RT PCR System (Thermo Fisher Scientific, Waltham, MA, United States) was used to perform RT-qPCR. Each 10 μl reaction mix contained 1 μl of cDNA samples, 0.5 μl of 10 μM gene-specific primers, and 5 μl of SYBR green supermix (Bio-Rad, Hercules, CA, United States). The soybean elongation factor gene was used as an internal control. The qPCR data were analyzed utilizing the 2–ΔΔCT method. The 2–ΔΔCT values obtained by qPCR were transformed to log2 to lessen noise level. For statistical analysis, ANOVA and Tukey’s test (P < 0.05) were applied and a criterion of greater than two-fold induction/reduction level was taken into consideration to select differentially expressing genes according to a previous report (Singh et al., 2008). The heat map was generated by Graphpad Prism Software11 using qPCR data.
Expression Analysis of Glycine max CMLs Upon Treatment With Signaling Compounds
For treatment with signaling compounds, an equal volume of 100 μM of methyl-jasmonate, 50 μM ethephon, 5 μM salicylic acid, and only water/0.5% (v/v) ethanol in water (control) were sprayed on healthy soybean plants according to the previous studies (Stotz et al., 2000; Singh et al., 2008). The plants were maintained under the same conditions but in individual enclosures. The leaves of treated (sample) and untreated (control) plants were harvested after 1 h of treatment in biological triplicates and immediately snap-frozen at −80°C for further analysis. For gene expression analysis, qPCR for 36 GmCMLs (differentially expressed on S. litura-infestation) was performed, and the data were analyzed using the 2–ΔΔCT method. For statistical analysis, ANOVA and Tukey’s test (P < 0.05) were applied and a criterion of greater than two-fold induction/reduction level was taken into consideration to select differentially expressing genes. The heat map was generated by Graphpad Prism Software (see text footnote 11) using qPCR data.
In silico Analysis of True Glycine max CMLs
The subcellular localization of different GmCMLs was predicted using WoLF PSORT12. The molecular weight and isoelectric point of each GmCMLs were computed using the “Compute pI/MW tool” of ExPASy13. The presence of EF-hands was checked using the InterPro database14. The MEME software15 was used to detect and generate logo plots of the EF-hands motifs.
Exon-Intron Determination
The organization of exons and introns in GmCMLs was deciphered using Gene Structure Display server GSDS2.016 (Hu et al., 2015).
Tissue-Specific Gene Expression Profiling
The expression profile values [Reads/Kb/Million (RPKM) normalized data] for different tissues, that is, flower, young leaf, green pods, root, nodule, stem, and seed, were retrieved from SoyBase Expression Explorer17 (Waese et al., 2017). The heat map was illustrated using RPKM values for tissue-specific expression profiling of GmCMLs on Graphpad Prism Software (see text footnote 11).
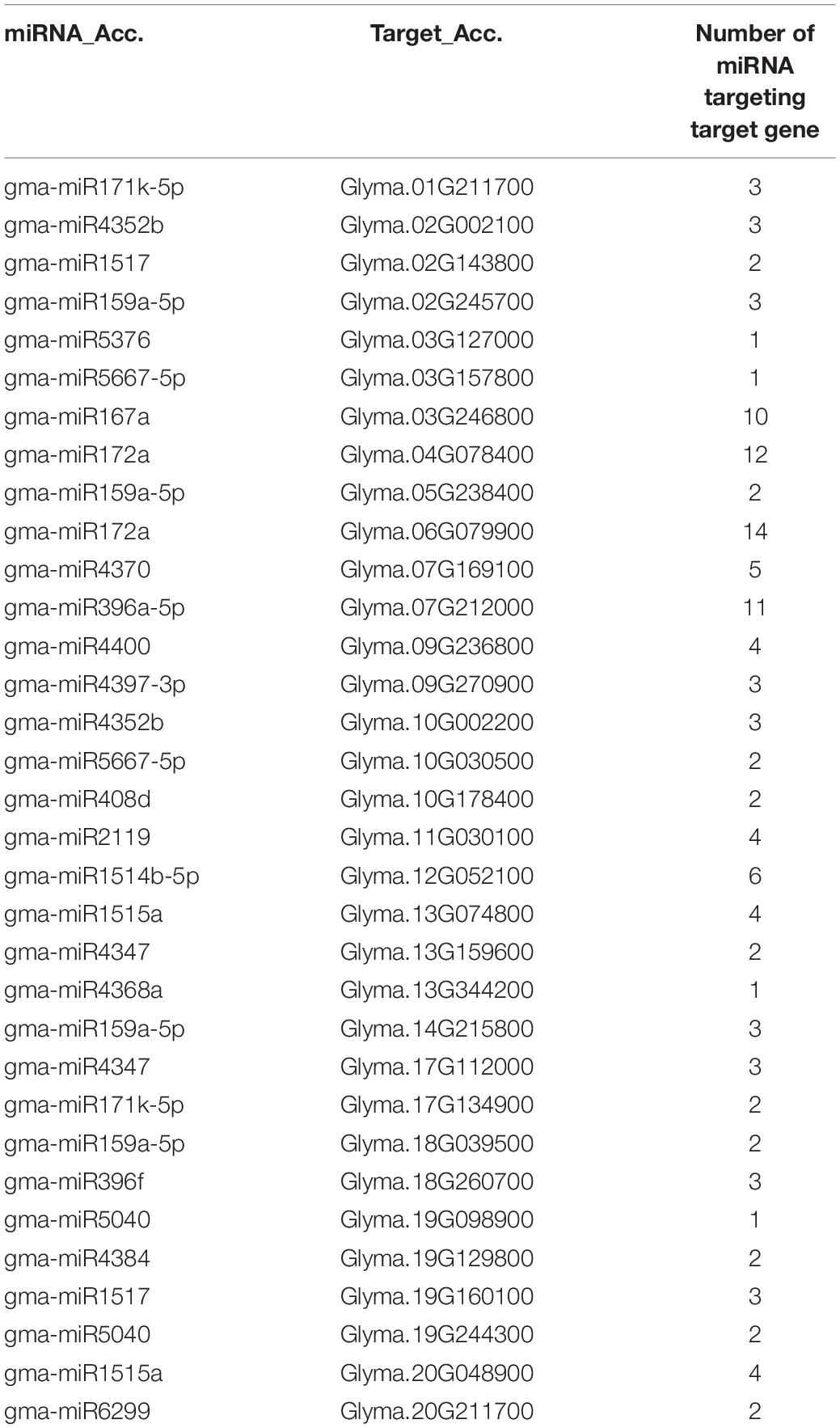
Identification of microRNA (miRNA) Targets of Glycine max CMLs
Plant small-RNA target (psRNATarget) analysis server was used to predict miRNA target sites in GmCMLs transcripts with default parameters. G. max-specific mature miRNA sequences were downloaded from the miRNA database (miRBase)18 (Bhatia et al., 2019).
Interaction of Glycine max CMLs With Other Proteins
To deduce the direct and indirect interaction of GmCMLs with other proteins, the sequences of all GmCMLs were subjected to the String database19 (Szklarczyk et al., 2021). The GmCML protein sequences were submitted as multiple proteins with organisms selected as G. max. The submission of sequences redirected to a new window, where proteins matching with input sequences along with identity, bitscore, and e-value were listed. Protein interaction list was saved by clicking on “Mapping”, and further to find interaction networks proceeded with “continue.”
Detection of Regulatory Elements in the Promoter Region of Glycine max CMLs
To detect the regulatory elements present in the 5′ upstream region of GmCMLs, 1 kb of the genomic DNA sequences before the initiation codon (ATG) of each GmCMLs were saved from NCBI and subjected to Plant Care analysis20 (Lescot et al., 2002).
Three-Dimensional Structure Prediction of Selected Glycine max CMLs by Homology Modeling
The homology modeling of S. litura-inducible GmCMLs was conducted to predict their three-dimensional structure. The selected S. litura-inducible GmCMLs were used as query sequences for homology modeling and searched against the Protein Data Bank (PDB) to identify the closely related known protein sequence. The data were submitted to the Phyre2 Protein Homology/Analogy recognition Engine http://www.sbg.bio.ic.ac.uk/∼phyre2/html/page.cgi?id=index to construct the protein 3-D structure by homology modeling under intensive modeling mode (Kelley et al., 2015). Further, to check the stereochemical quality of protein tertiary structures, the PROCHECK server was used http://saves.mbi.ucla.edu/ (Laskowski et al., 1996).
Interaction of Selected Glycine max CMLs With Ca2+
To show the interaction between Ca2+ and GmCMLs, the InChI format of Ca2+ was derived from PubChem21 (Kim et al., 2019). For docking studies, the InChI format was converted to.pdb using OpenBabel--Chemical file format converter22 (O’Boyle et al., 2011). The interaction studies were performed for true GmCMLs that were differentially regulated by S. litura-infestation using PATCHDOCK https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php (Schneidman-Duhovny et al., 2005). Different parameters used for docking studies were clustering root-mean-square deviation (RMSD) = 4, and the complex type is protein and small ligand. The top ten Patchdock docking results were further refined using FireDock23 (Mashiach et al., 2008). The GmCMLs were docked with Ca2+ based on the number of EF-hand motifs. The global energy scores of docked molecules were compared with that of the Calmodulin 2 protein of Arabidopsis thaliana with Ca2+.
Results
Genome-Wide Survey, Phylogenetic Study, and Similarity Search Identified True Glycine max CMLs in Glycine max Genome
A total of 144 CMLs were predicted in the soybean genome by Zeng et al. (2017) in a preliminary study. The protein sequences of these predicted CMLs were procured and named as GmCMLs. Phylogenetic analysis of GmCMLs with CMLs of other plants, including monocots and leguminous dicots was executed, which showed evolutionary relatedness among them. This analysis revealed that GmCML101, 118, 98, and 100 shared ancestry. GmCML113, 167, 71, and CML25 from M. truncatula and CML28 and CML29 from A. thaliana diverged recently (Figure 1). Further, the GmCMLs were compared with CMLs of A. thaliana, revealing that 41 CMLs showed 50% or more sequence similarity with CMLs of A. thaliana. Sixteen of the GmCMLs showed higher sequence similarity (85–70%), whereas 25 GmCMLs showed more than 50% sequence similarity with CMLs of A. thaliana. The rest of the CMLs (103 GmCMLs) exhibited less than 50% homology, and 3 of them did not match with any of the CMLs of A. thaliana. With an identity of 50% as a threshold, 41 GmCMLs with 50% or more identities were labeled as true GmCMLs. A list of GmCMLs and their homologous CML of A. thaliana and their percentage similarity is presented in Supplementary Table 1.
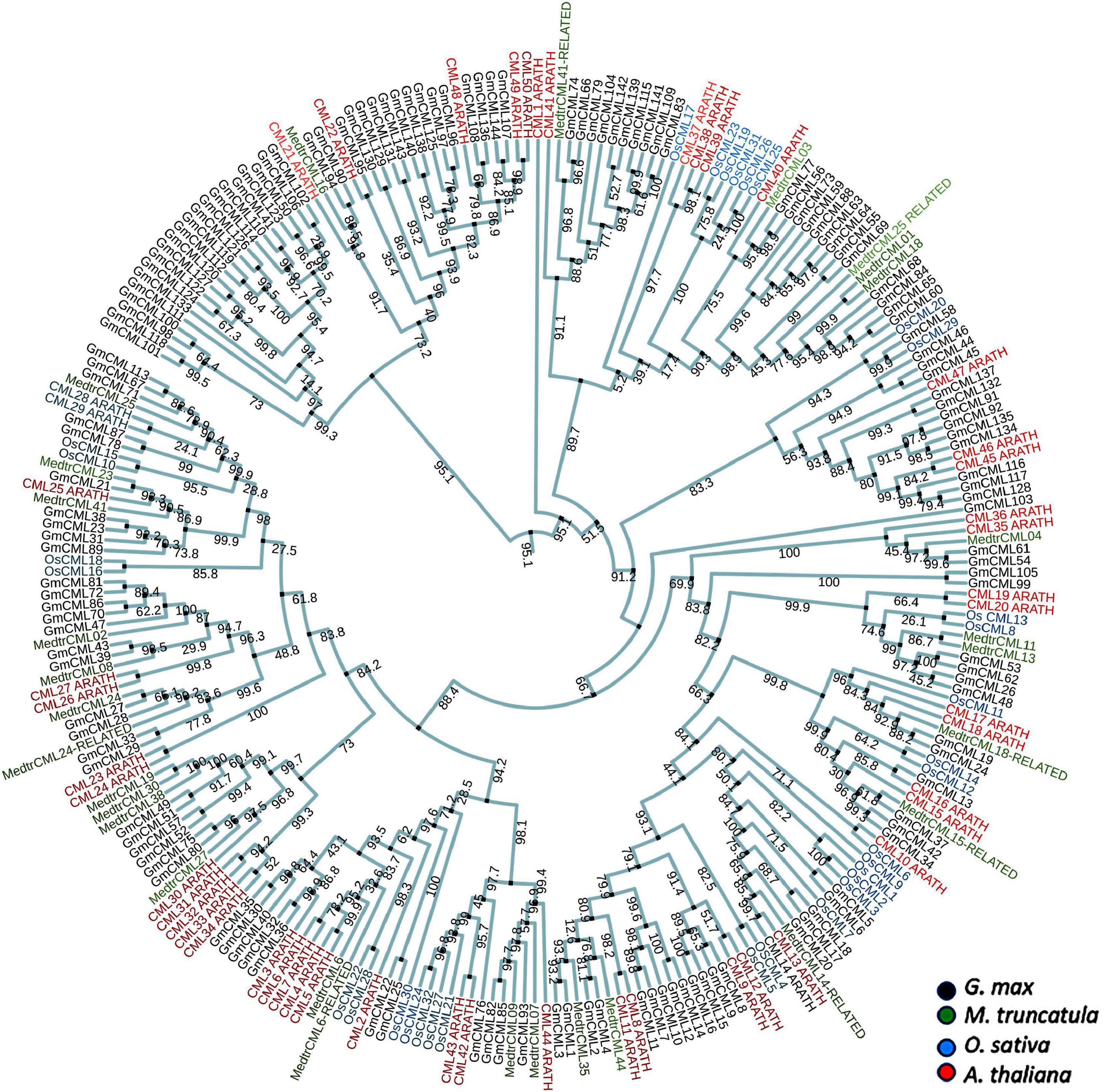
Figure 1. The phylogenetic analysis of CMLs of Glycine max, Medicago truncatula, Oryza sativa, and Arabidopsis thaliana. The alignment for phylogenetic tree was performed with Clustal Omega using full-length protein sequences. The phylogenetic tree was constructed using iTOL. All the Glycine max CMLs (GmCMLs) are presented with black, Medicago truncatula CMLs are presented with green, Oryza sativa CMLs are presented with blue, and Arabidopsis thaliana CMLs are presented with red. The bootstrap values are provided in the mid of the nodes.
Differential Expression of Glycine max CMLs During Spodoptera litura- Infestation and Wounding
To gain information on the involvement of GmCMLs in plant defense against S. litura-infestation, the expressions of true 41 GmCMLs were checked using qPCR with gene-specific primers (Supplementary Table 2), and the results indicated that 34 of the GmCMLs were upregulated while 2 of the GmCMLs were downregulated. GmCML30 showed a maximum change in expression while GmCML33, 9, 10, 1, and GmCML2, 76, 29, 3, 15, and 5 also showed a high level of expression (Figure 2; Supplementary Figure 1). The results demonstrated that these GmCMLs were responsive to S. litura-infestation. However, a different transcript pattern was observed when plants were wounded/mechanically damaged; 24 of the GmCMLs were upregulated, and 5 GmCMLs were downregulated. Seven of the GmCMLs (GmCMLs 7, 20, 21, 35, 76, 77, and 116) exhibited a significant change in mRNA expression specifically during S. litura-infestation.
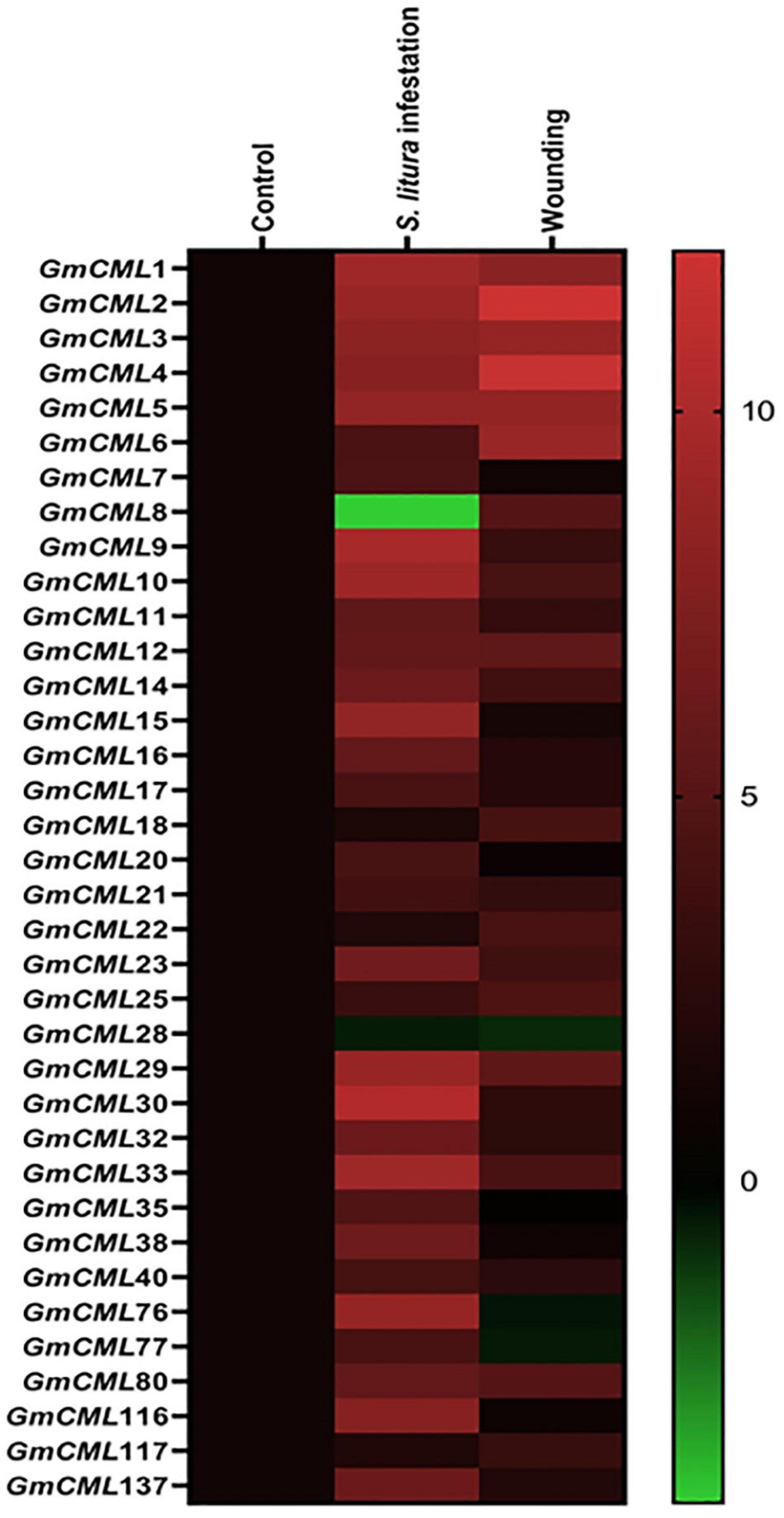
Figure 2. Heat map showing expression profile of GmCMLs upon S. litura-infestation and wounding. The expression profile of GmCMLs upon S. litura-infestation and wounding is represented as heat map, generated with the help of Graphpad Prism software using log2 values of expression data obtained from qPCR. The qPCR data were analyzed utilizing 2–ΔΔCT method and statistical analysis was performed using ANOVA and Tukey’s test (P < 0.05).
Differential Expression of Glycine max CMLs Upon Foliar Application of Signaling Compounds
To recognize the role of defense-related signaling compounds in the regulation of S. litura inducible-GmCMLs, expression analysis was performed with plants treated with JA, SA, and ethylene (ET) by qPCR. GmCMLs 1, 2, 3, 4, 5, 6, 8, 9, 10, 11, and 12 showed induced expression in all four treatments. The JA application also induced the expression of GmCMLs 14, 17, and 29. The ethylene upregulated the expression of 13 of the GmCMLs, and eight of them were commonly upregulated by JA and ethylene. In addition, eleven of the GmCMLs were upregulated by SA (Figure 3; Supplementary Figure 2).
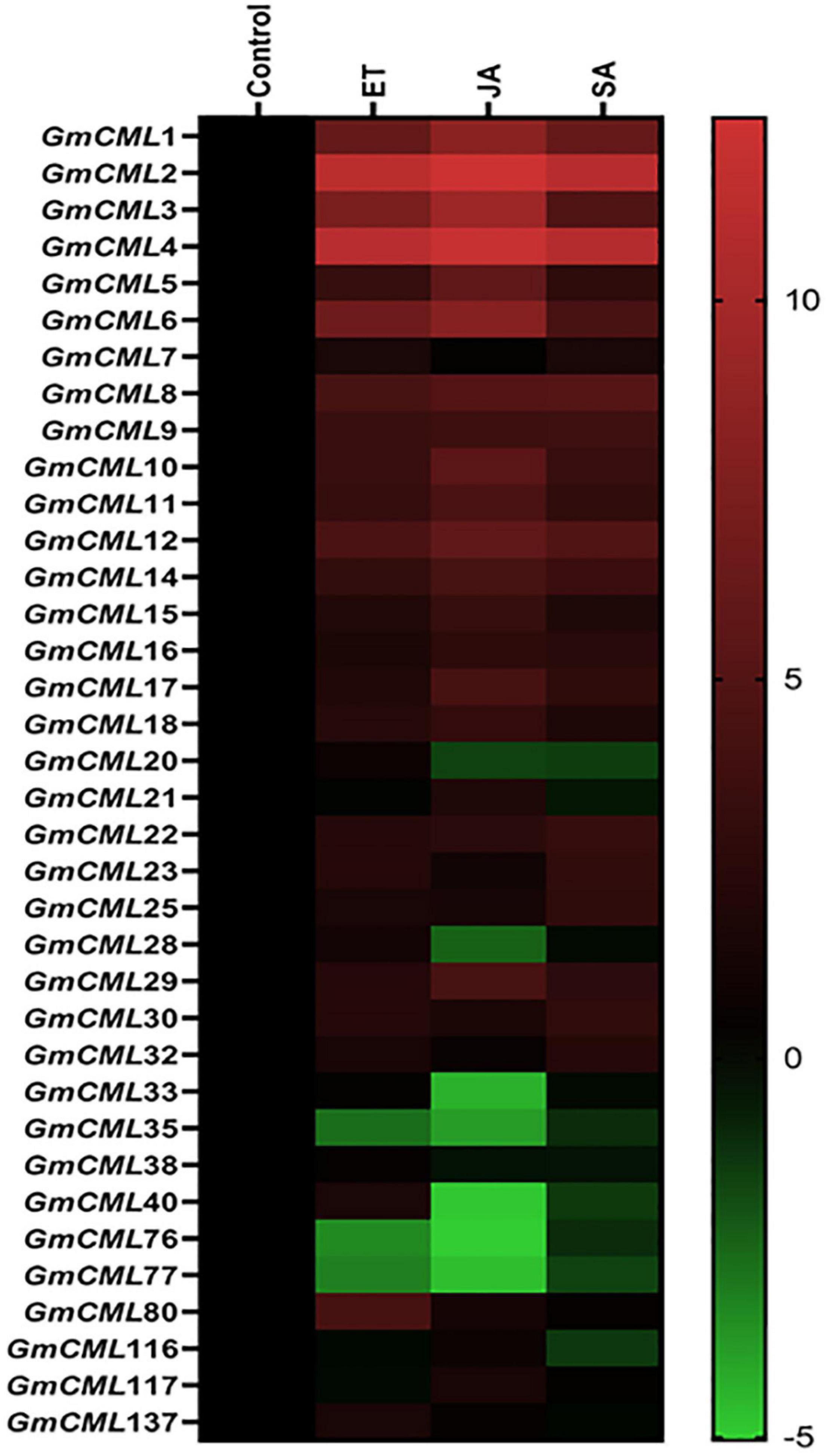
Figure 3. Heat map showing expression profile of GmCMLs on application of JA, SA, and ET. The expression profile of GmCMLs on application of JA, SA, and ET is represented as a heat map, generated with the help of Graphpad Prism software using log2 values of expression data obtained from qPCR. The qPCR data were analyzed utilizing the 2–ΔΔCT method and statistical analysis was performed using ANOVA and Tukey’s test (P < 0.05).
Subcellular Localization, Molecular Weight, Isoelectric Point, and EF-Hand Analysis of Glycine max CMLs
The subcellular localization study using in silico tools predicted GmCMLs to be located mainly in the nucleus, cytosol, or chloroplast. Only a few of them were predicted to be located on the plasma membrane. The GmCMLs ranged from 229 (GmCML22) to 102 (GmCML38) amino acids, and their molecular weight ranged between 11.26 and 25.85 KDa. GmCML38, 32, and 11 were smallest by molecular weights (11.26 KDa), whereas GmCMLs 22 and 25 were of the largest molecular weight (25.85 KDa). The isoelectric point values of the GmCMLs ranged from pI 3.94 to 6.84, where GmCML2 had a minimum isoelectric point (pI 3.94) and GmCML80 had a maximum isoelectric point (pI 6.84) (Table 1).
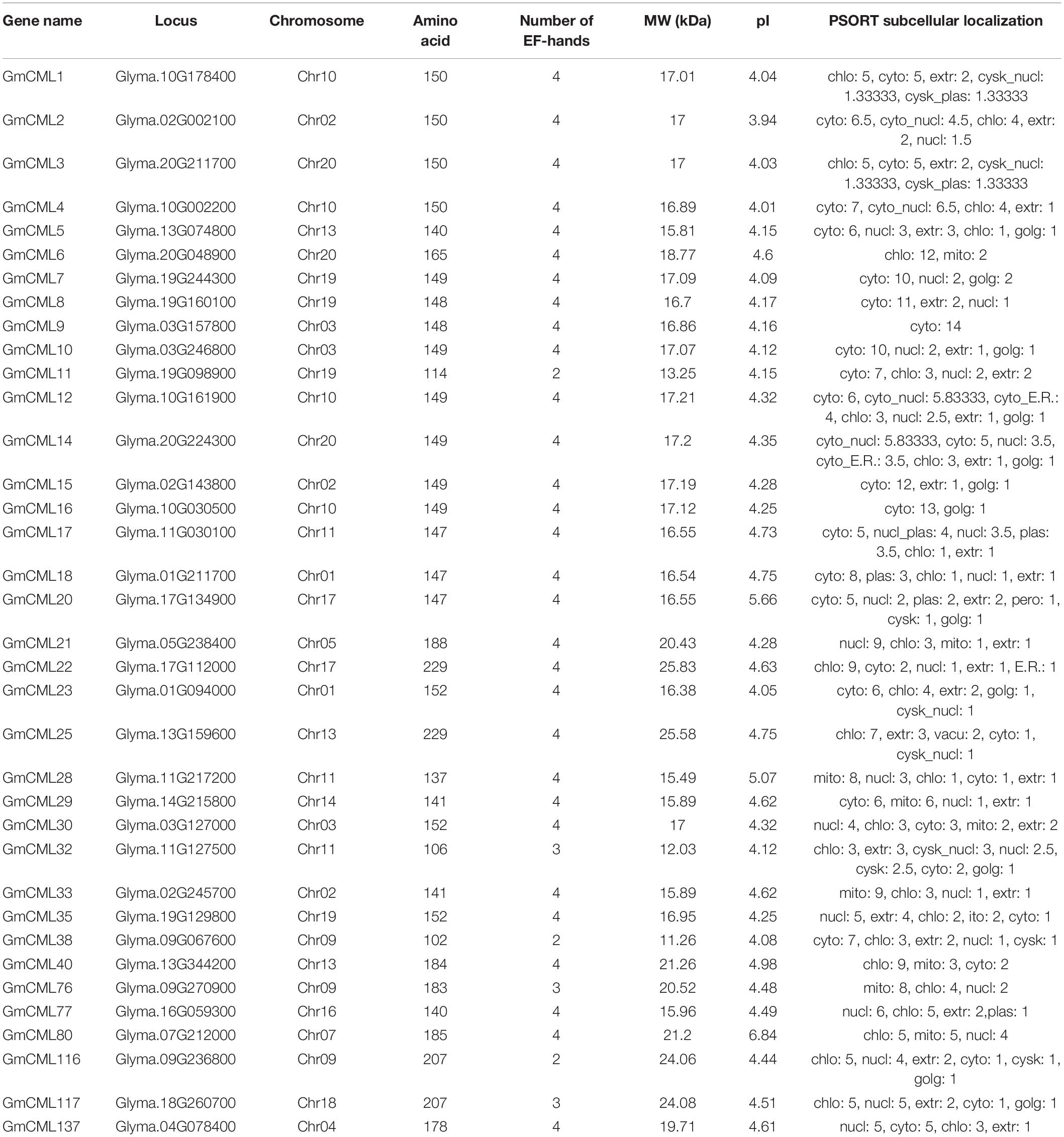
Table 1. Gene locus, chromosome number, number of amino acids, number of EF-hands, molecular weight, pI, and subcellular localization of S. litura-inducible GmCMLs.
GmCMLs Are Largely Intronless/Carry 1–3 Introns and Share EF-Hand Signature Sequence and Other Common Motifs
The intron and exon display analysis of GmCMLs revealed that a larger number of these CMLs were intronless. Among the studied 36 GmCMLs, 22 GmCMLs coding genes were intronless, whereas the rest of the GmCMLs had one to three introns. GmCMLs 1, 2, 3, 4, 7, 8, 9, 10, 14, 15, and 16 have four exons and three introns (Figure 4). Additionally, prediction of the conserved domain revealed that a couple of motifs were conserved among the members of GmCML family proteins. Many of the GmCMLs (30 GmCMLs) had four EF-hands but GmCML 32, 76, and 117 had three EF-hands and GmCML11, 38, and 137 had only two EF-hands (Figure 5). All GmCML proteins contained the EELKEAFKVFDKDGBGYISASELRHVMRSLGEKLTDEEVEQ, EAFSLFDKBGDGCITVEELATIJRSLGQNPTEEE, and MIKEA DLDGDGQVBYEEFVKM motif confirming the presence of EF-hand signature sequence.
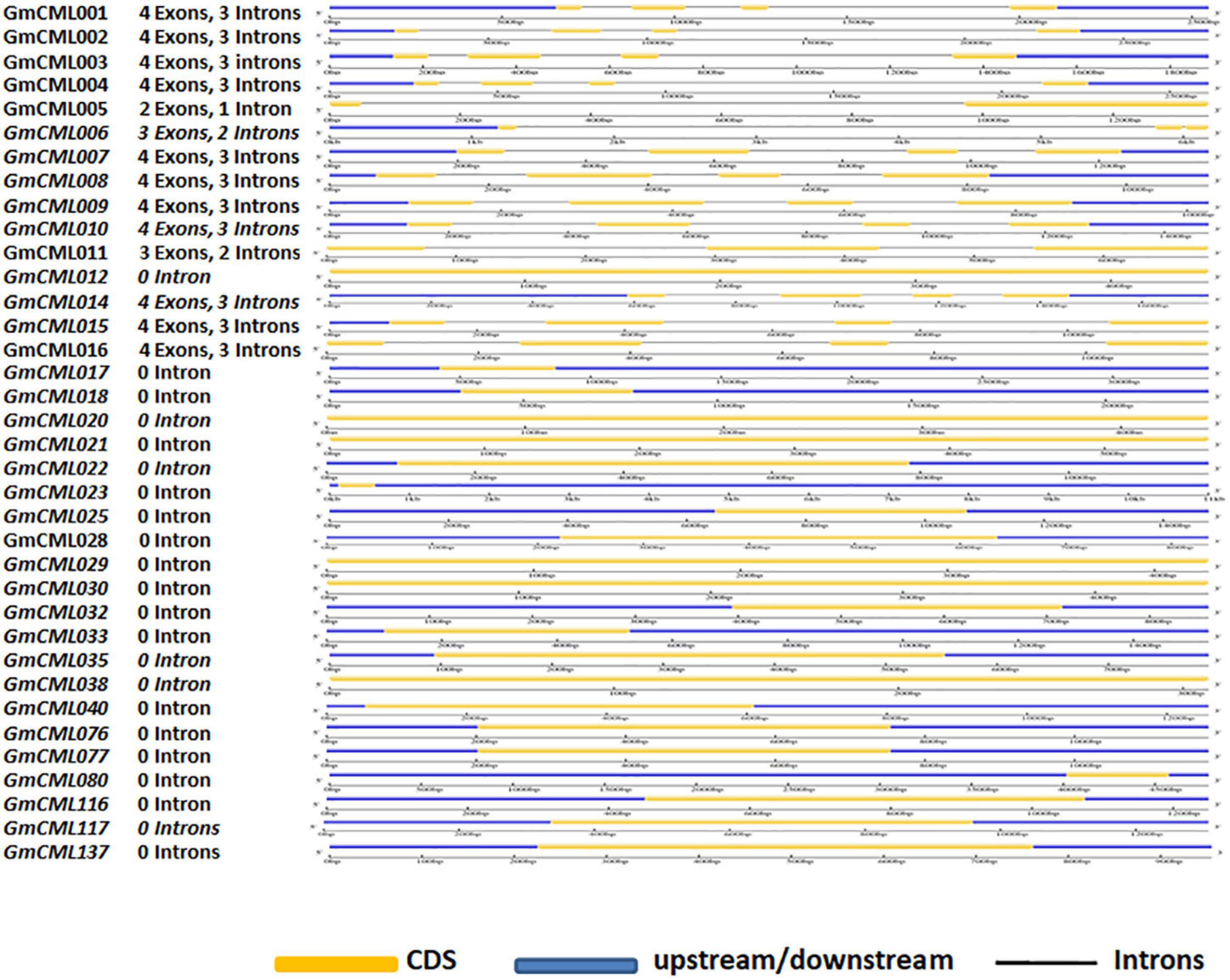
Figure 4. The gene structure analysis of GmCMLs. The gene structure analysis showing the distribution of exon-intron in GmCMLs. The exons, upstream/downstream, and introns are represented by yellow, blue, and black colors, respectively. The exon-intron distribution analysis was examined using Gene Structure Display server GSDS2.0.

Figure 5. Identification of EF-hands in GmCMLs by in silico method. The presence of EF-hands (number varied from 2 to 4) in GmCMLs are presented (A). The boxes represent the EF-hands present in each GmCMLs. The logoplot shows the presence of respective number of EF-hands in GmCMLs (generated by MEME suite) (B). The colored boxes indicate the number of EF-hand motifs at the corresponding position within GmCMLs. Different colors indicate different EF-hand motifs.
Members of Glycine max CMLs Family Differ in Their Tissue-Specific Expression Patterns
To identify the genes involved in distinct regulatory functions controlled by specific tissue, tissue-specific gene expression data were procured. The heat map was generated using RNA-seq data to examine the tissue-specific expression profile at the basal level (Figure 6; Supplementary Table 3). This analysis revealed that the relative expression level of GmCMLs 5, 6, and 40 was higher in all tissue types. GmCMLs 6, 5, 40, 28, 17, 80, 1, 4, and 29 showed their higher expression in stem. GmCMLs 5, 6, and 17 showed expression in seed and GmCMLs 6, 5, 40, 17, 80, and 137 expressed more in the nodule. GmCML17 had moderate expression in all the tissues, including flower, young leaf, green pods, root, nodule, stem, and seed. GmCMLs 6, 5, 40, 28, 17, 80, 1, 4, 29, 33, 76, and 32 exhibited maximum expression in root. GmCMLs 1, 6, 5, 40, 28, 17, and 80 exhibited higher expression in leaf. The mRNA of GmCMLs 6, 5, 40, 28, 17, and 80 were abundantly present in flower and pod. However, the rest of the genes showed minimal expression/no expression in different tissues.
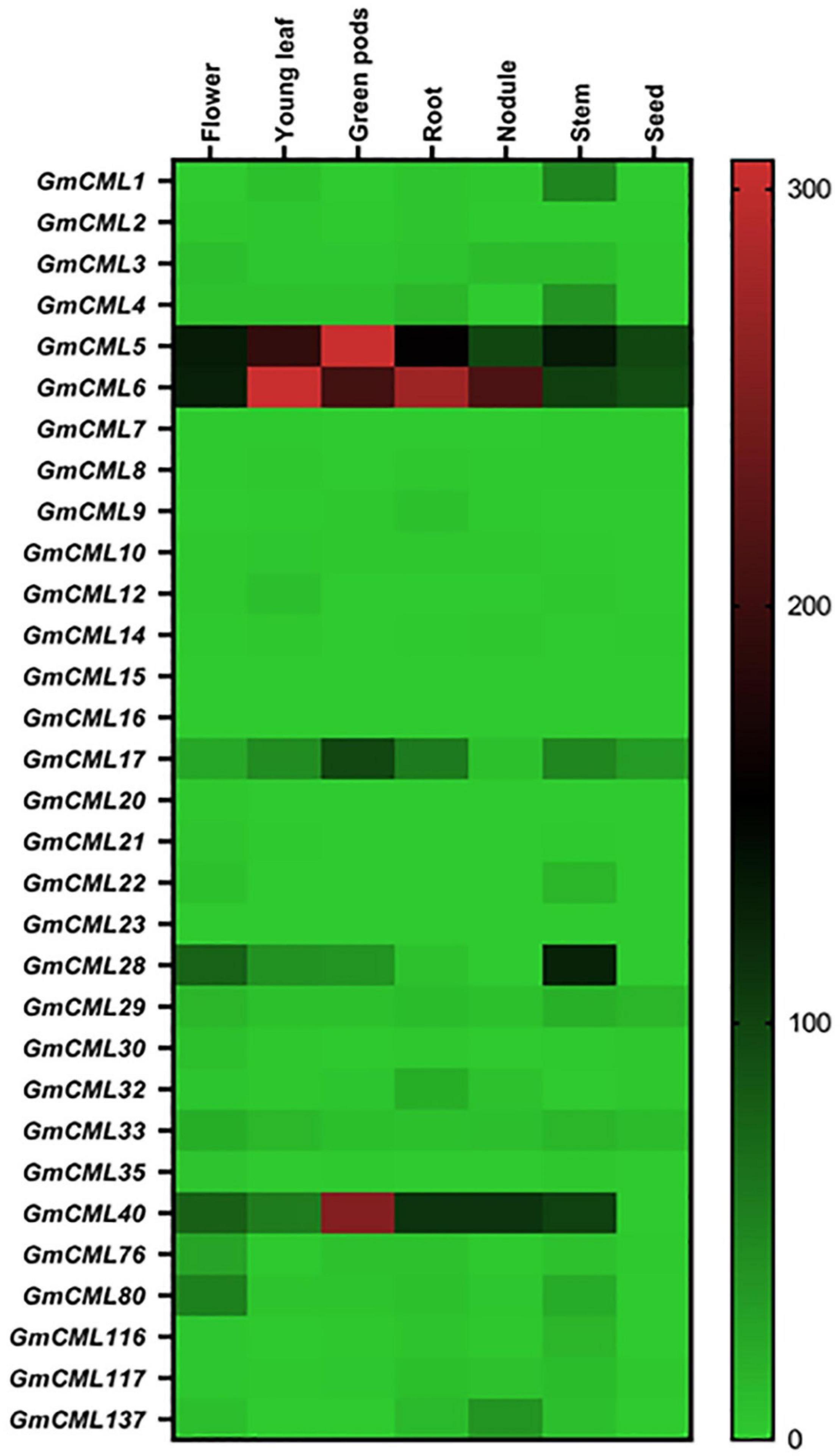
Figure 6. The expression profile of GmCMLs in different tissues of soybean. Heat map showing expression profiles of GmCMLs in different tissue types of G. max. The RPKM values were obtained from SoyBase Expression Explorer. The heat map has been generated using Graphpad Prism software. Expression data of GmCML011, GmCML018, GmCML025, GmCML038, and GmCML077 were not available.
Members of Glycine max CML Family Are Putative Targets of Known microRNAs
The miRNA target analysis (using psRNATarget) revealed 33 GmCML transcripts as putative targets of 125 miRNAs (Table 2).
Members of Glycine max CMLs Family Have an Overlapping Protein Network
To deduce the complex interplay of GmCMLs with other proteins, network and enrichment analysis of 36 selected GmCMLs was performed using the STRING database. The studies revealed that each GmCML corresponded to a particular string Id. Based on the STRING analysis, a protein protein interaction (PPI) network complex was constructed. The expected number of edges of the interaction network was 6, the average node degree of the network was 4, the average local clustering coefficient was 0.986, and the protein enrichment p-value was < 1.0e-16. The predicted results showed 40 nodes and 80 edges. Out of 36, 25 of the GmCMLs had shown their involvement as calcium-binding protein having EF-hand, where GmCML1, GmCML2, and GmCML15 had shown very high similarity with SCAM-4, SCAM-5, and Calmodulin, respectively, which were reported to be involved in plant-defense against pathogen attack (Park et al., 2004). Interestingly, the studies have shown the involvement of GmCML5 and GmCML6 in nodulation of soybean roots. Most of the GmCMLs showed to interact via GLYMA07G38930.2 (spindle-like microcephalyassociated protein isoform) and GLYMA17G01810.2 (spindle-like microcephalyassociated protein isoform). Overall, it was predicted that 24 GmCMLs have their role in plant-pathogen defense signaling and 5 of them showed their role in the phosphatidylinositol signaling and MAPK pathway (Figure 7).
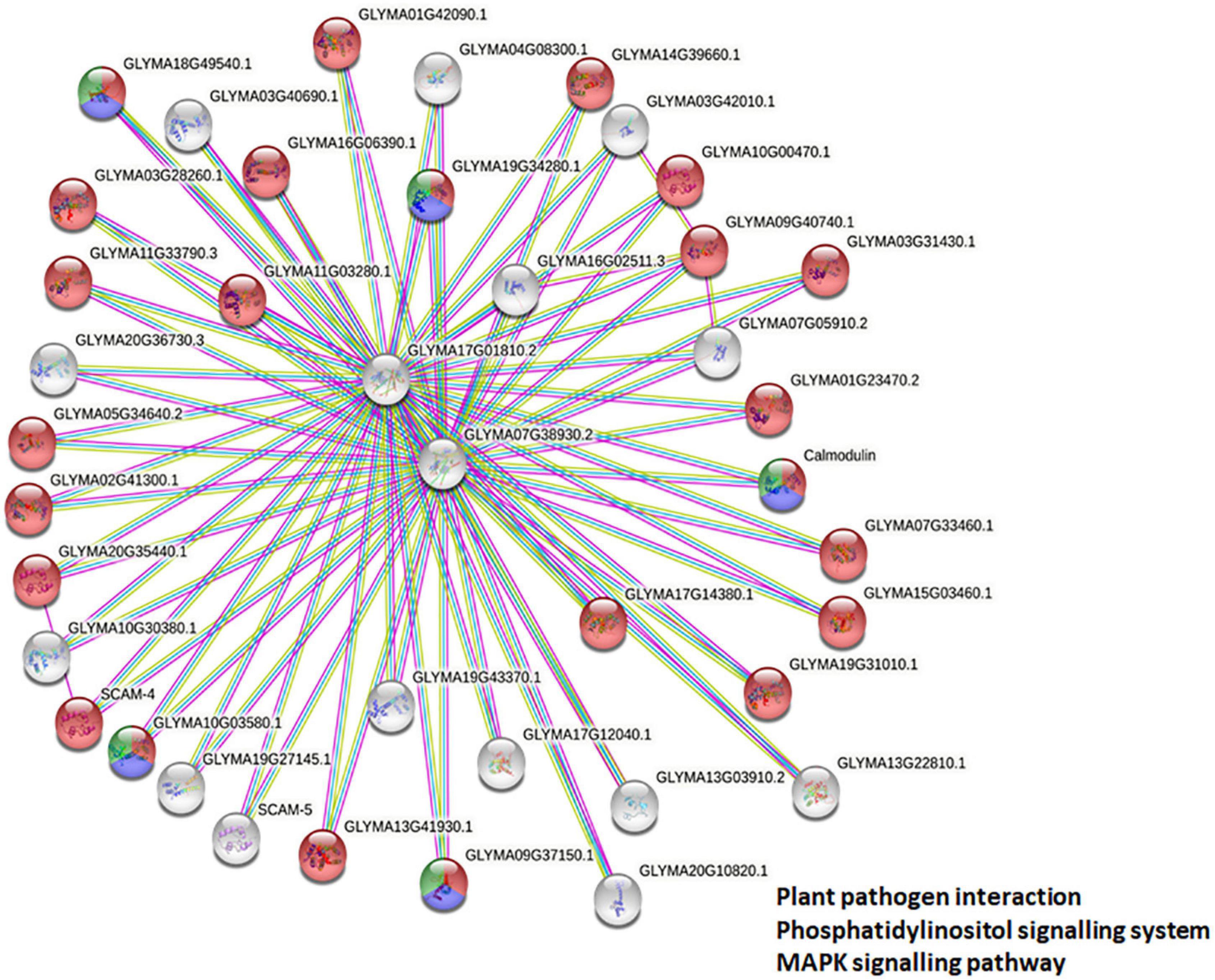
Figure 7. Protein-protein interaction network analysis. Protein-protein interaction network and enrichment analysis showing the complex interplay among GmCMLs as predicted from the STRING database. The proteins colored in red are showing their potential involvement in plant-pathogen interaction, the proteins in blue color are predicted to be involved in phosphatidylinositol signaling, and the proteins in green color are predicted to be involved in MAPK signaling pathways.
In silico Promoter Analysis Identified Multiple Hormones and Stress-Responsive Cis-Elements in Promoter Sequences of Glycine max CMLs
A broad range analysis to decipher which cis-elements were present in the promoter regions of true GmCMLs, 1 kb 5′ UTR regions of each of the GmCMLs were subjected to Plant care analysis that predicted the presence of 16 different cis-regulatory elements involved in biotic or abiotic stresses. Most of the GmCMLs promoters were predicted to harbor more than one phytohormone-related cis-regulatory elements, such as ABA (ABRE), JA (G-box, Myc binding JA response element), SA (TCA-element), Ethylene (ERE), GA (P-Box and GARE-motif – gibberellin responsiveness), and biotic and abiotic stress-related cis-elements like ARE (Anaerobic induction), wounding, Myb-binding site involved in drought-inducibility (MBS), W-box-stress inducible and LTR (a low temperature)-responsive element, TC-rich repeats –defense and stress response (Figure 8).
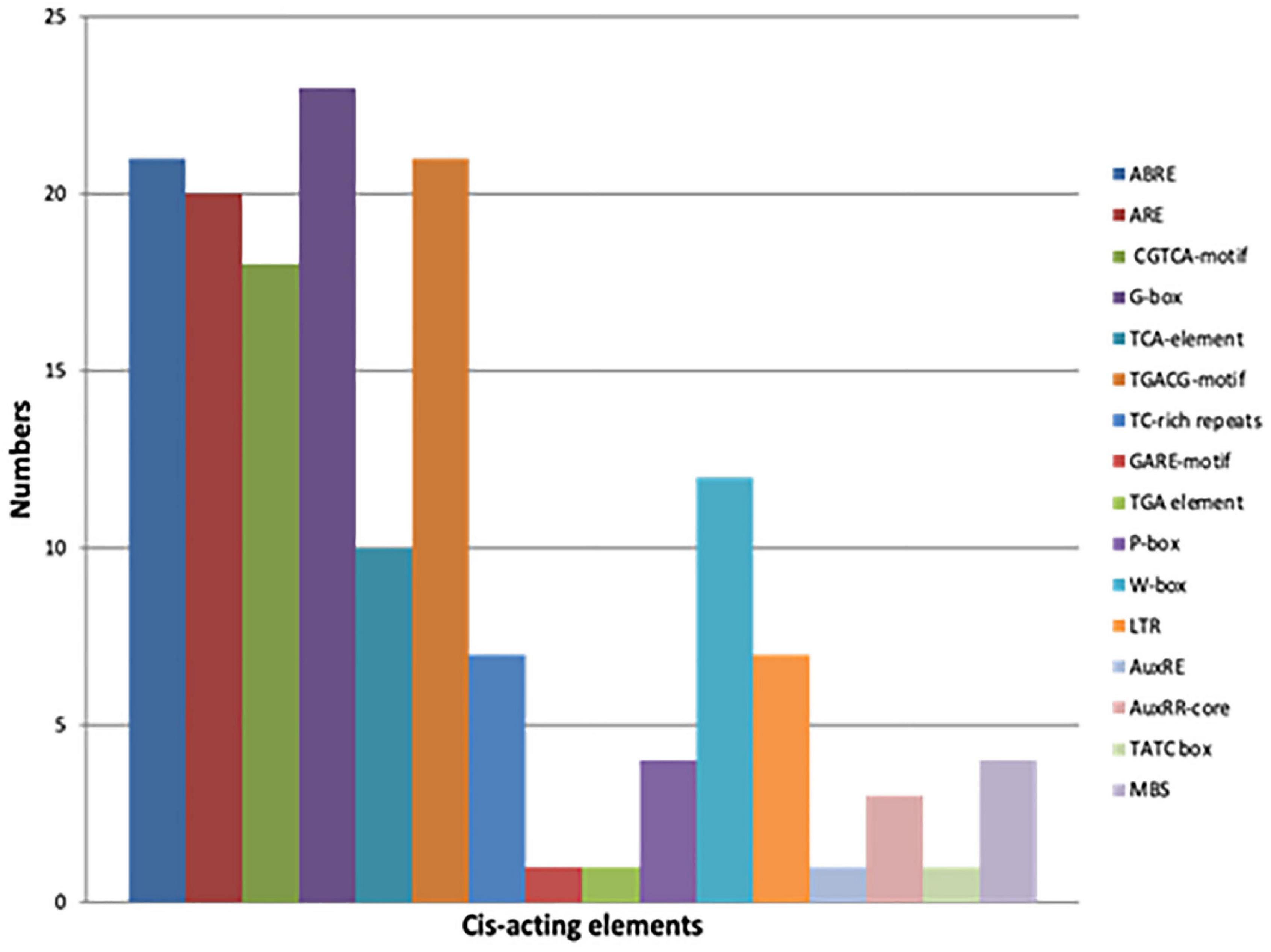
Figure 8. The graphical representation of different cis-acting elements present in the promoter sequences of GmCMLs. Different colors depict different cis-acting elements [ABRE – Abscisic acid responsiveness, ARE –Anaerobic induction (wounding), CGTCA motif – Jasmonate responsiveness (MeJA), TCA-element – Salicylic acid responsiveness, TGACG-motif - Jasmonate responsiveness (MeJA), TC-rich repeats –defense and stress response, GARE-motif – Gibberellin responsiveness, TGA element – Auxin response, P Box - Gibberellin responsiveness, W-Box - Stress inducible, LTR – Low temperature response, AuxRE- Auxin responsive element, AuxRR-core - Auxin responsive element, TATC box - Gibberellin responsiveness and MBS – Drought inducible].
Molecular Docking Indicates That Members of Glycine max CML Family Proteins Interact With Ca2+
The study of the interaction of Ca2+ with GmCMLs could be crucial for understanding their mechanism of action. The tertiary structure of GmCMLs was modeled using Phyre2, and the structures were validated by keeping a threshold value of >90% region of protein being present in the allowed region of the Ramachandran plot. The confidence level of modeled structures was more than 99%. The classic EF-hands consisting of a typical helix-loop-helix domain were detected and highlighted (Figure 9).
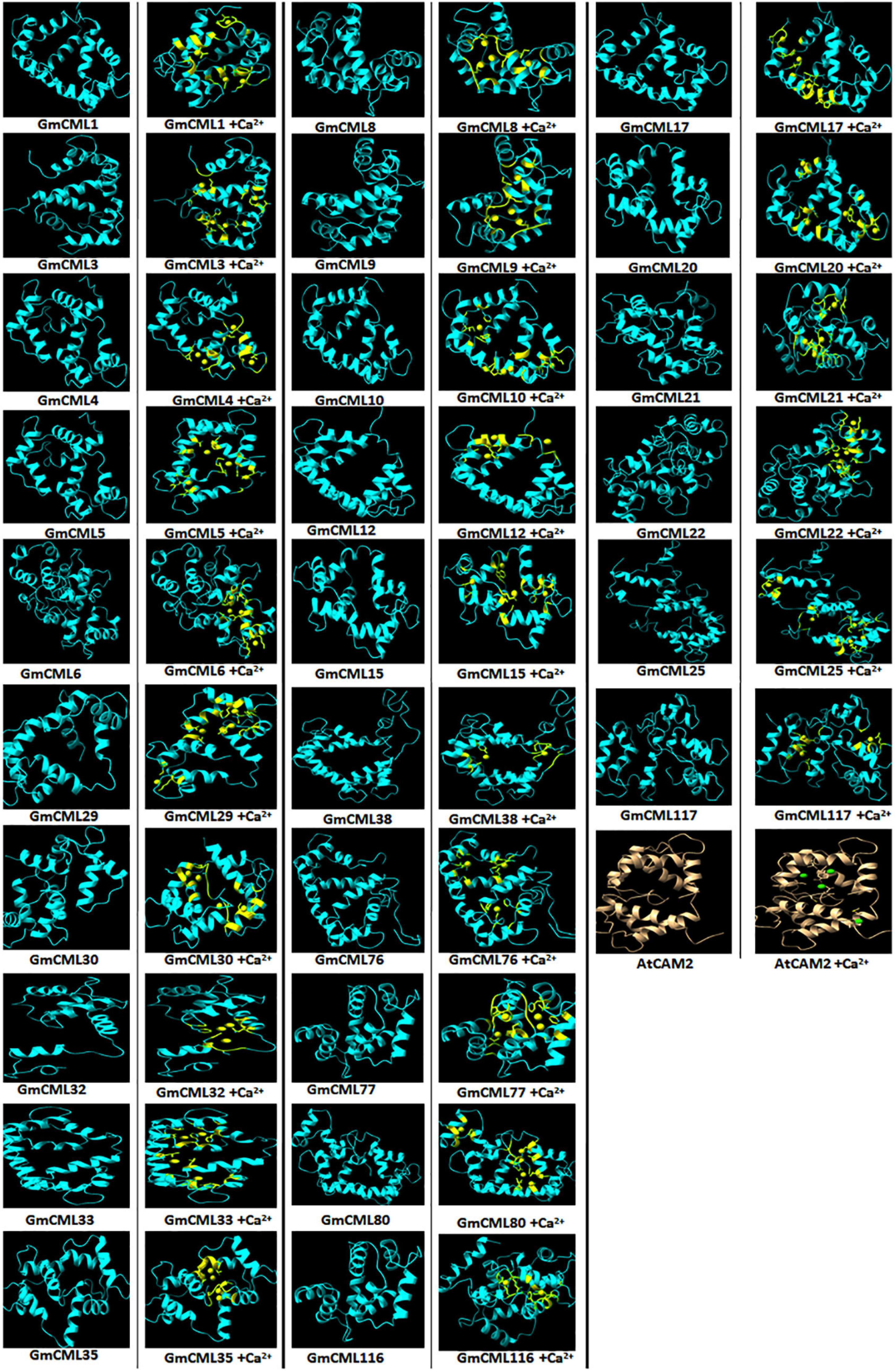
Figure 9. Interaction of GmCMLs with Ca2+ using molecular docking and their comparison with interaction of Calmodulin 2 (A. thaliana) with Ca2+. The figure shows the modeled tertiary structures of GmCMLs and their interaction with Ca2+, wherein the yellow balls indicate Ca2+ and the yellow-colored segments in the model represent EF-hands. The number of Ca2+ showing interaction with GmCMLs varied according to the number of EF-hands. The global docking energy score of interaction between 26 GmCMLs with Ca2+ is shown to have near to global docking energy score of interaction between Calmodulin 2 of A. thaliana with Ca2+. The tertiary structure of A. thaliana is shown in gray color and the green balls indicate Ca2+.
Previous studies had indicated that EF-hand motifs bind with Ca2+ for their activation. Thus, the molecular docking of 36 GmCMLs and Calmodulin 2 protein of Arabidopsis thaliana (AtCAM2) with Ca2+ were performed, and the interaction results of GmCMLs with Ca2+ were compared with the global energy score of AtCAM 2 with Ca2+. The global energy score of AtCAM2 with Ca2+ was −2.94. The global energy score for 26 of the GmCMLs with Ca2+ ranged from −2.5 to −4.5. GmCMLs 17, 15, 29, 38, 116, 117, 33, 25, 30, 8, 35, 21, 76, 32, 77, 6, 22, 5, and 80 were showing the global energy scores from −4.41 to −3.05 whereas GmCMLs 20, 9, 12, 1, 10, 3, and 4 were showing global energy scores in between −2.8 and −2.5 (Figure 9).
Discussion
Plants have to encounter numerous challenges ranging from biotic to abiotic factors during their lifespan. Therefore, they have evolved sophisticated defense mechanisms regulated by recognizing the specific type of stress and stimulation of certain early signaling events to defend themselves. One such crucial early event is the generation of Ca2+ ion pulses due to the elevation of influx and efflux, which acts as a cellular signal in response to external stimulus (Vadassery et al., 2012a,b). Calcium sensor proteins, such as CMLs, are essential components of Ca2+ signaling (DeFalco et al., 2010). We have only a little information about the involvement of CMLs in plant defense against herbivory in crop plants. Although fewer studies on the EF-hand possessing protein families, CaM, CDPK, CMLs, and CBLs (Weinl and Kudla, 2009; DeFalco et al., 2010; Poovaiah et al., 2013) have been taken into consideration in a couple of plants, such as Arabidopsis thaliana, Oryza sativa, Grapevine, Nicotiana benthamiana, and Solanum lycopersicum but significantly less information are available about CMLs in legumes (McCormack and Braam, 2003; Asano et al., 2005; Boonburapong and Buaboocha, 2007; Chen et al., 2013; Kong et al., 2013; Kleist et al., 2014).
Therefore, to characterize the CMLs of soybean and figure out their role in plant defense, the GmCMLs were subjected to phylogenetic analysis with CMLs of Medicago truncatula, Arabidopsis thaliana, and Oryza sativa, which revealed that many of the GmCMLs were grouped with CMLs of Arabidopsis and Medicago. They shared ancestry, which confirmed their evolutionary relationship. However, when GmCMLs were subjected to the BLAST search engine of TAIR against the known EF-hand protein of A. thaliana, only 41 GmCMLs had more than 50% sequence similarity. Expression analysis of true GmCMLs during S. litura-infestation was investigated, and the results demonstrated that many of these GmCMLs (36) were responsive to herbivory. Out of 41 genes, 34 of the GmCMLs were upregulated, while 2 of the GmCMLs were downregulated. Previous studies have reported that CMLs control signaling events upregulated during mechanical wounding and insect attack. It was observed that in A. thaliana, CML9, 11, 12, 16, 17, 23, and 42 genes were upregulated upon treatments with the oral secretion of larvae of the herbivorous insect S. littoralis (Vadassery et al., 2012a). In comparison to herbivory, 24 GmCMLs were upregulated 5 GmCMLs were downregulated by mechanical damage, and 8 of the GmCMLs showed induced expression exclusively during S. litura-infestation. These data suggested that wounding and herbivory had a different transcript pattern on account of the presence of insect-derived elicitors in the oral secretion during herbivory. The distinction in the transcript patterns during mechanical damage and insect attack has been detected in previous studies (Reymond et al., 2000, 2004; Singh et al., 2008). Wounding stimulated plant defense similar to herbivory; however, mechanical damage-induced responses were not the same as those activated by herbivory.
Signaling compounds, such as JA, ET, and SA, function as essential plant defense compounds against herbivory. Most of the herbivores trigger the jasmonate/ethylene pathway and the salicylate pathway (Hammerschmidt and Smith-Becker, 1999; Reymond et al., 2004). To recognize the role of defense regulators, change in the expression of GmCMLs was checked upon treatments with JA, SA, and ET. JA changed the mRNA levels of 14 of the S. litura-inducible GmCMLs, demonstrating the involvement of JA in the regulation of expression of GmCMLs. JA-responsive cis-regulatory elements, TGACG and CGTCA, were observed in the promoter regions of GmCMLs 1, 3, 9, 14, 7 and 1, 3, 9, 14, 17, respectively, which further confirmed their regulation by JA. Ethylene upregulated the expression of 13 of the GmCMLs, and eight of them were commonly upregulated by JA. ET and JA were reported to induce defense genes synergistically in Arabidopsis (Kessler and Baldwin, 2002; Singh et al., 2008). In addition, eleven of the GmCMLs were upregulated by SA, indicating the role of SA in regulation of expression of GmCMLs either directly or through a crosstalk. The presence of SA-responsive cis-regulatory element in the promoter of GmCML 8 further confirmed their regulation by SA.
The exon-intron distribution study predicted the presence of zero to three introns in GmCMLs, indicating critical evolutionary changes in the G. max genome. There were only 11 GmCMLs that carried 1–3 introns, and 22 GmCMLs were intronless. The presence of few/no introns indicated their ability to get transcribed quickly to facilitate early defense response in the host plant during stress (Keshan et al., 2021).
The subcellular localization studies predicted the presence of CMLs in the nucleus, cytosol, and chloroplast mainly. Previous studies on CMLs had reported their localization in the nucleus and cytosol in Arabidopsis (Inzé et al., 2012; Vadassery et al., 2012a). The lower values of isoelectric points revealed that GmCMLs could be acidic, making them highly hydrophilic. The motif analysis exhibited 1 to 4 EF-hand potential motifs in most of the GmCMLs, indicating their potential identity as calcium-binding proteins (Boonburapong and Buaboocha, 2007; Kong et al., 2013; Kleist et al., 2014; Munir et al., 2016). Further, identifying potential miRNA target sites in GmCMLs divulged the dynamic roles of miRNA in post-transcriptional regulation of these genes in response to normal or stressed conditions (Bhatia et al., 2019).
The promoter of every gene possesses regulatory elements that act as binding sites for transcription factors. These promoter elements govern the differential expression of a gene in a tissue-specific manner at different growth stages and during environmental stresses. Several studies have divulged a direct connection between gene expression and promoter elements present in their 5′ upstream region (Lata et al., 2014). Therefore, cis-elements present in the promoter regions of all true GmCMLs were investigated using in silico tool, which predicted the presence of 16 different cis-regulatory elements involved in biotic or abiotic stresses, indicating their role in plant responses during stress and their involvement in signal transduction via ABA, SA, and JA.
The analysis of measuring mRNA abundance of genes belonging to a family in a tissue-specific manner will allow in identifying the genes involved in the development or regulatory pathways associated with a particular tissue type. The expression data of the GmCML gene family in different organs exhibited divergence in their function. The transcript profiling of GmCMLs during several developmental stages in different organs revealed that most GmCMLs do not express vegetatively. They are inducible to exhibit a particular response, whereas some GmCMLs, such as GmCML5, GmCML6, and GmCML40, expressed highly in flower, root, pod, pod shell, young leaf, and seed, depicting their role in growth and development. Nine of the GmCMLs exhibited higher expression in the stem, illustrating their potential role in shoot development. Three of the GmCMLs showed their expression in seed, and six showed their expression in the nodule, indicating their prospective role in seed and nodule formation. Twelve GmCMLs showed higher expression in root, signifying their possible function in root development and plant-microbe interaction in soil. Seven of the GmCMLs exhibited higher expression in the leaf, specifying their role in leaf development and plant defense against pests and pathogens. Six of the GmCMLs were expressed abundantly in flower and pod, depicting their role in reproduction.
To deduce the complex interplay of GmCMLs with other proteins, network and enrichment analysis of S. litura-inducible 36 GmCMLs were performed with the STRING database. The PPI network complex indicated that out of 36 GmCMLs, 24 GmCMLs were predicted to play a role in plant-pathogen defense signaling; 5 of the GmCMLs (GmCML16, GmCML8, GmCML15, GmCML116, and GmCML117) could be involved in the phosphatidylinositol signaling system and MAPK signaling system. The plant defense signaling, phosphatidylinositol signaling system, MAPK pathway, and the components of these pathways, such as SCAM-4, SCAM-5, CaMEKK, CaZIK, CaRAF, PLA, PLC, and PLD had been reported to be involved in plant defense against biotic stresses (Park et al., 2004; Hung et al., 2014; Keshan et al., 2021). The above results are prediction-based and should be perceived with caution. Further empirical analyses and experimentation would be required to assess whether these interactions are physiologically relevant.
The interaction of CMLs with Ca2+ is crucial for their biological activity. To examine the interaction between Ca2+ and GmCMLs, the tertiary structure of S. litura-inducible GmCMLs was predicted, which demonstrated a difference in the modeled structures of GmCMLs, mainly due to the change in their amino acid sequences. This diversity in their structures could allow them to perform a specific function. However, all the modeled structures of GmCMLs had shown the EF-hand helix-loop-helix motif as described in Calmodulin of A. thaliana, which could facilitate their interaction with Ca2+. The molecular docking was performed to investigate the interaction of GmCMLs with Ca2+ and their global energy score of interaction was compared with the global energy score of interaction between AtCAM2 with Ca2+. The results revealed that the global energy score for interaction between GmCMLs with Ca2+ ranged from −2.5 to −4.5 (for 26 of the GmCMLs), which is comparable to AtCAM2 with Ca2+ interaction. Thus, in silico interaction study predicted that calcium ions interacted with GmCMLs, particularly with EF-hand motifs, indicating that GmCMLs could be Ca2+-binding proteins and could be involved in Ca2+ signaling. Further experimentation would be required to confirm whether CMLs are undergoing any conformational change to induce downstream signaling.
Conclusion
Plants encounter various environmental stresses, including insect attack during their life span. To defend themselves, plants have developed sophisticated signaling pathways that activate the biosynthesis of defensive proteins and metabolites. Among a couple of early events that instigate defense signaling, a transient change in cytosolic Ca2+ concentration is observed during herbivory. This transient change in Ca2+ concentration is identified by Ca2+ sensors, which transduce the information to downstream target proteins leading to appropriate defense response. The role of Ca2+ sensors during herbivory has been studied in model plants, but not in leguminous plants, such as G. max. This study identified 41 true GmCMLs, an important Ca2+-sensor, in soybean. The phylogenetic analysis revealed their similarity with already known CMLs from model plants. The presence of potential EF-hands further supported their identification as Ca2+-sensors. The gene structure determination of GmCMLs indicated the presence of no/few introns revealing their capability to change their expression quickly during recognition of a stimulus. The differential expression of GmCMLs in various tissues signified their involvement in growth and development. Alteration in the transcript patterns of the novel GmCMLs during S. litura-infestation suggested their significant involvement in plant defense signaling during herbivory (Figure 10). Comparative expression profiling during mechanical damage showed that wounding-regulated responses are not the same as observed during herbivory. Signaling compounds, such as JA, ET, and SA, showed their involvement in regulating the expression of S. litura-inducible GmCMLs. Network and enrichment analysis of GmCMLs with other proteins indicated their role in plant defense, phosphatidylinositol signaling system, and MAPK signaling system. The study on the interaction between calcium and GmCMLs showed their identity as Ca2+-binding proteins and their participation in signal transduction during Ca2+ signaling (Figure 10). The results of this study can be extended to confirm the role of calcium signaling in triggering the plant defense mechanism against S. litura-infestation. S. litura-inducible GmCMLs are prospective candidates for being critical components of calcium signaling during pest attacks. Thus, the structural and functional characterization of S. litura-inducible GmCMLs utilizing contemporary techniques like RNAi, overexpression, and/or gene editing is highly recommended to delineate the calcium signaling pathway and its role in plant defense during soybean-S. litura interaction.
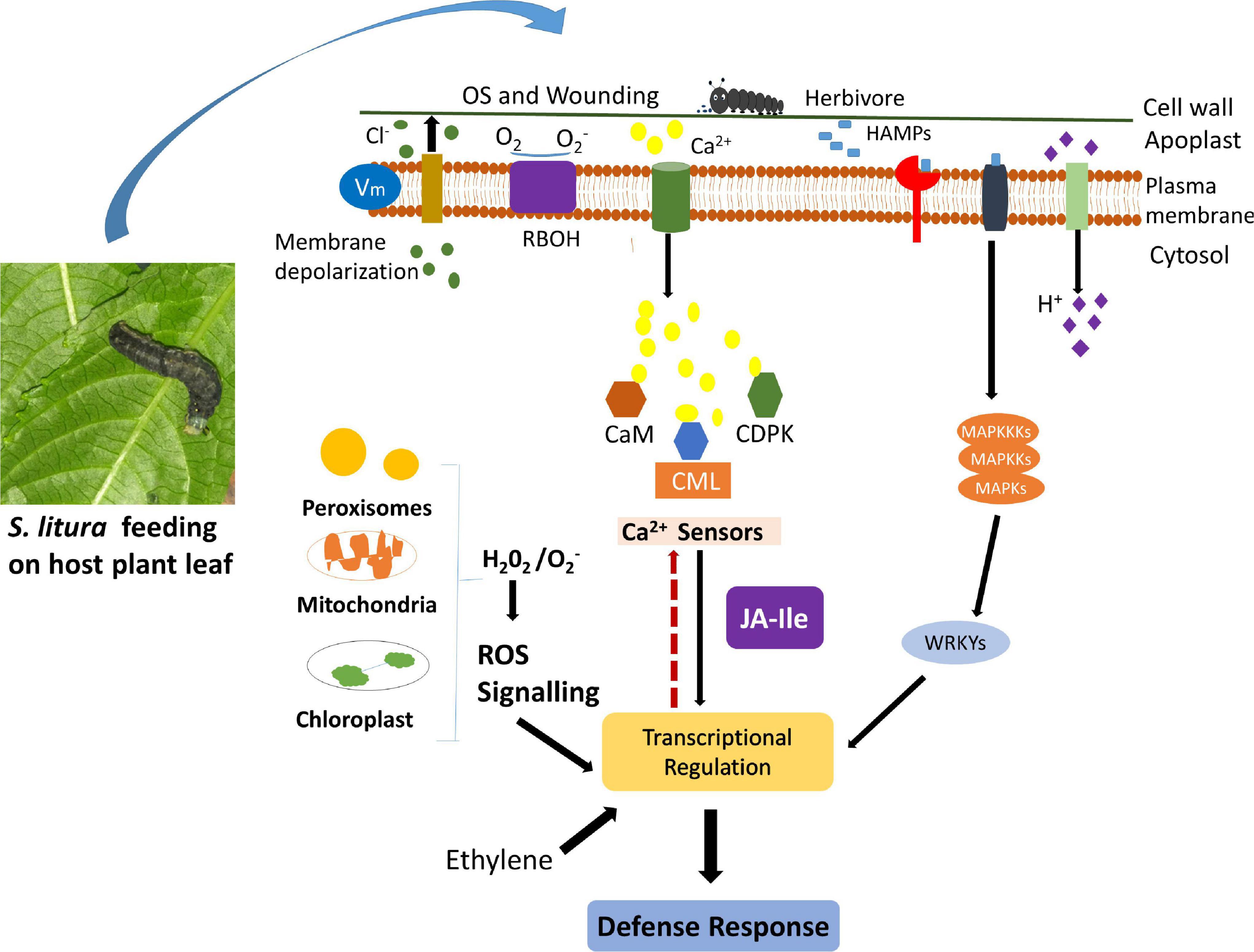
Figure 10. Schematic representation of herbivory-induced early signaling events (calcium signaling, ROS generation, activation of MAPK Pathway) and involvement of CMLs in plant defense response against insect attack. Upon herbivory, plants perceive stimuli and instigate a couple of early events, such as activation of calcium signaling, induction of mitogen-activated protein kinase pathway, and generation of reactive oxygen species, which transduce the signal and activate the biosynthesis of other signaling and defensive compounds. In soybean, the expression of few GmCMLs, one of the crucial Ca2+ sensors, is upregulated during S. litura-infestation. GmCMLs have Ca-EF hands, which allow them to sense calcium signature and bind with Ca2+, which can further regulate downstream signaling and activate plant defense response.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
AS conceptualized and supervised the study. MY, JP, AS, IKS, AC, AR, MH, and JKK contributed to the investigation. MY, JP, and AS wrote – original draft preparation. AS, IKS, AC, AR, MH, and JKK contributed to writing, reviewing, editing, and visualization. AS, IKS, and MY contributed to formal analysis. AS, AC, and AR contributed to funding acquisition. IKS and AS contributed to resources. All authors have read and agreed to the published version of the manuscript.
Funding
AS acknowledged the Science and Engineering Research Board (SERB), Department of Science and Technology, New Delhi, India, for the financial support (ECR/2017/002478). MY was thankful to CSIR, MHRD, and JP was thankful to SERB for Junior Research Fellowship. The authors thank Rama, Principal, Hansraj College, for providing all the facilities to conduct this research work and her constant support. AC and AR are financially supported by the grant “EVA 4.0,” No. CZ.02.1.01/0.0/0.0/16_019/0000803 financed by OP RDE.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.817950/full#supplementary-material
Footnotes
- ^ https://www.soybase.org/
- ^ https://phytozome.jgi.doe.gov/pz/portal.html
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://www.ncbi.nlm.nih.gov
- ^ https://www.arabidopsis.org/
- ^ https://shigen.nig.ac.jp/rice/oryzabase/
- ^ https://www.ebi.ac.uk/Tools/msa/clustalo/
- ^ https://itol.embl.de/itol.cgi
- ^ https://www.arabidopsis.org/Blast/
- ^ http://bioinfo.ut.ee/primer3-0.4.0/
- ^ https://www.graphpad.com/scientific-software/prism
- ^ https://www.genscript.com/wolf-psort.html
- ^ https://web.expasy.org/compute_pi/
- ^ https://www.ebi.ac.uk/interpro/
- ^ http://meme.sdsc.edu/meme/meme.html
- ^ http://gsds.gao-lab.org/
- ^ https://www.soybase.org/expression/
- ^ http://plantgrn.noble.org/psRNATarget/
- ^ https://string-db.org/cgi/input?sessionId=bcguyAaLcQA5&input_page_show_search=on
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare
- ^ https://pubchem.ncbi.nlm.nih.gov/
- ^ http://www.cheminfo.org/Chemistry/Cheminformatics/FormatConverter/index.html
- ^ https://bioinfo3d.cs.tau.ac.il/FireDock/
References
Aldon, D., Mbengue, M., Mazars, C., and Galaud, J.-P. (2018). Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 19:665. doi: 10.3390/ijms19030665
Asano, T., Tanaka, N., Yang, G., Hayashi, N., and Komatsu, S. (2005). Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 46, 356–366. doi: 10.1093/pcp/pci035
Badole, S. L., and Bodhankar, S. L. (2012). Glycine max (soybean) treatment for diabetes. Bioact. Food Diet. Interv. Diabetes 77, 77–82. doi: 10.1016/b978-0-12-397153-1.00008-1
Badole, S. L., and Mahamuni, S. P. (2013). “Soybean: Key Role in Skin Cancer,” in Bioactive Dietary Factors and Plant Extracts in Dermatology, eds R. Watson and S. Zibadi (Totowa, N J: Humana Press), 315–320. doi: 10.1007/978-1-62703-167-7_28
Bender, K. W., and Snedden, W. A. (2013). Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol. 163, 486–495. doi: 10.1104/pp.113.221069
Bhatia, G., Sharma, S., Upadhyay, S. K., and Singh, K. (2019). Long non-coding RNAs coordinate developmental transitions and other key biological processes in grapevine. Sci. Rep. 9:3552. doi: 10.1038/s41598-019-38989-7
Boonburapong, B., and Buaboocha, T. (2007). Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 7:4. doi: 10.1186/1471-2229-7-4
Chen, C.-Y., and Mao, Y.-B. (2020). Research advances in plant–insect molecular interaction. Faculty Rev. 9, F1000. doi: 10.12688/f1000research.21502.1
Chen, F., Fasoli, M., Tornielli, G. B., Dal Santo, S., Pezzotti, M., Zhang, L., et al. (2013). The evolutionary history and diverse physiological roles of the grapevine calcium-dependent protein kinase gene family. PLoS One 8:e80818. doi: 10.1371/journal.pone.0080818
Conrath, U., Pieterse, C. M. J., and Mauch-Mani, B. (2002). Priming in plant–pathogen interactions. Trends Plant Sci. 7, 210–216. doi: 10.1016/s1360-1385(02)02244-6
Cui, Z.-L., Gai, J.-Y., Ji, D.-F., and Ren, Z.-J. (1997). A study on leaf-feeding insect species on soybeans in Nanjing area. Soybean Sci. 16, 12–20.
DeFalco, T. A., Bender, K. W., and Snedden, W. A. (2010). Breaking the code: ca2+ sensors in plant signalling. Biochem. J. 425, 27–40. doi: 10.1042/BJ20091147
DeFalco, T. A., Moeder, W., and Yoshioka, K. (2016). Opening the gates: insights into cyclic nucleotide-gated channel-mediated signaling. Trends Plant Sci. 21, 903–906. doi: 10.1016/j.tplants.2016.08.011
DeFalco, T. A., and Zipfel, C. (2021). Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell 81, 3449–3467. doi: 10.1016/j.molcel.2021.07.029
Delk, N. A., Johnson, K. A., Chowdhury, N. I., and Braam, J. (2005). CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 139, 240–253. doi: 10.1104/pp.105.062612
Dobney, S., Chiasson, D., Lam, P., Smith, S. P., and Snedden, W. A. (2009). The calmodulin-related calcium sensor CML42 plays a role in trichome branching. J. Biol. Chem. 284, 31647–31657. doi: 10.1074/jbc.M109.056770
Dodd, A. N., Kudla, J., and Sanders, D. (2010). The language of calcium signaling. Ann. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Fan, R., Wang, H., Wang, Y., and Yu, D. (2012). Proteomic analysis of soybean defense response induced by cotton worm (Prodenia litura, fabricius) feeding. Proteome Sci. 10, 16. doi: 10.1186/1477-5956-10-16
Gatehouse, J. A. (2002). Plant resistance towards insect herbivores: a dynamic interaction. New Phytol. 156, 145–169. doi: 10.1046/j.1469-8137.2002.00519.x
Gifford, J. L., Walsh, M. P., and Vogel, H. J. (2007). Structures and metal-ion-binding properties of the Ca2+-binding helix–loop–helix EF-hand motifs. Biochem. J. 405, 199–221. doi: 10.1042/BJ20070255
Gresshoff, P. M., and Ferguson, B. J. (2017). Molecular signals in nodulation control. Int. J. Mol. Sci. 18:125. doi: 10.3390/ijms18010125
Halitschke, R., and Baldwin, I. T. (2004). Jasmonates and related compounds in plant-insect interactions. J. Plant Growth Regul. 23, 238–245. doi: 10.1007/s00344-004-0037-z
Hammerschmidt, R., and Smith-Becker, J. A. (1999). “The role of salicylic acid in disease resistance,” in Mechanisms of Resistance to Plant Diseases, eds A. Slusarenko, R. S. S. Fraser, and L. C. Van Loon (Kluwer: AcademicPublisher), 37–53.
Hancock, R. D., Hogenhout, S., and Foyer, C. H. (2015). Mechanisms of plant–insect interaction. J. Exp. Bot. 66, 421–424.
Heyer, M., Scholz, S. S., Reichelt, M., Kunert, G., Oelmüller, R., and Mithöfer, A. (2021). The Ca2+ sensor proteins CML37 and CML42 antagonistically regulate plant stress responses by altering phytohormone signals. Plant Mol. Biol. [Epub online ahead of print], doi: 10.1007/s11103-021-01184-2
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hubbard, K., Hotta, C., Gardner, M., Braam, J., and Webb, A. (2008). The Arabidopsis thaliana calmodulin-like protein CML24 is a regulator of rhythmic Ca2+ signalling and flowering time. Comp. Biochem. Physiol. Part A 150:S153.
Hung, C.-Y., Aspesi, P. Jr., Hunter, M. R., Lomax, A. W., and Perera, I. Y. (2014). Phosphoinositide-signaling is one component of a robust plant defense response. Front. Plant Sci. 5:267. doi: 10.3389/fpls.2014.00267
Inzé, A., Vanderauwera, S., Hoeberichts, F. A., Vandorpe, M., Van Gaever, T. I. M., and Van Breusegem, F. (2012). A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant. Cell Environ. 35, 308–320. doi: 10.1111/j.1365-3040.2011.02323.x
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Keshan, R., Singh, I. K., and Singh, A. (2021). Genome wide investigation of MAPKKKs from Cicer arietinum and their involvement in plant defense against Helicoverpa armigera. Physiol. Mol. Plant Pathol. 115:101685. doi: 10.1016/j.pmpp.2021.101685
Kessler, A., and Baldwin, I. T. (2002). Plant responses to insect herbivory: the emerging molecular analysis. Annu. Rev. Plant Biol. 53, 299–328. doi: 10.1146/annurev.arplant.53.100301.135207
Kiep, V., Vadassery, J., Lattke, J., Maaß, J. P., Boland, W., Peiter, E., et al. (2015). Systemic cytosolic Ca2+ elevation is activated upon wounding and herbivory in Arabidopsis. New Phytol. 207, 996–1004. doi: 10.1111/nph.13493
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., et al. (2019). PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109. doi: 10.1093/nar/gky1033
Kleist, T. J., Spencley, A. L., and Luan, S. (2014). Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front. Plant Sci. 5:187. doi: 10.3389/fpls.2014.0018
Kliebenstein, D. J. (2004). Secondary metabolites and plant/environment interactions: a view through Arabidopsis thaliana tinged glasses. Plant. Cell Environ. 27, 675–684. doi: 10.1111/j.1365-3040.2004.01180.x
Kong, X., Lv, W., Jiang, S., Zhang, D., Cai, G., Pan, J., et al. (2013). Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics 14:433. doi: 10.1186/1471-2164-14-433
Kumar, A., Panwar, R., Singh, A., and Singh, I. K. (2020). ““Role of Calcium Signalling During Plant–Herbivore Interaction”, in Plant Stress Biology, eds B. Giri and M. P. Sharma (Singapore: Springer), 491–510. doi: 10.3390/cells10092219
Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996). AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486. doi: 10.1007/BF00228148
Lata, C., Mishra, A. K., Muthamilarasan, M., Bonthala, V. S., Khan, Y., and Prasad, M. (2014). Genome-wide investigation and expression profiling of AP2/ERF transcription factor superfamily in foxtail millet (Setaria italica L.). PLoS One 9:e113092. doi: 10.1371/journal.pone.0113092
Leba, L., Cheval, C., Ortiz-Martín, I., Ranty, B., Beuzón, C. R., Galaud, J., et al. (2012). CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway. Plant J. 71, 976–989. doi: 10.1111/j.1365-313X.2012.05045.x
Lecourieux, D., Mazars, C., Pauly, N., Ranjeva, R., and Pugin, A. (2002). Analysis and effects of cytosolic free calcium increases in response to elicitors in Nicotiana plumbaginifolia cells. Plant Cell 14, 2627–2641. doi: 10.1105/tpc.005579
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Liao, J., Deng, J., Qin, Z., Tang, J., Shu, M., Ding, C., et al. (2017). Genome-wide identification and analyses of calmodulins and calmodulin-like proteins in Lotus japonicas. Front. Plant Sci. 8:482. doi: 10.3389/fpls.2017.00482
Lortzing, T., and Steppuhn, A. (2016). Jasmonate signalling in plants shapes plant–insect interaction ecology. Curr. Opin. Insect Sci. 14, 32–39. doi: 10.1016/j.cois.2016.01.002
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Maffei, M., Bossi, S., Spiteller, D., Mithofer, A., and Boland, W. (2004). Effects of feeding Spodoptera littoralis on lima bean leaves. I. Membrane potentials, intracellular calcium variations, oral secretions, and regurgitate components. Plant Physiol. 134, 1752–1762. doi: 10.1104/pp.103.034165
Magnan, F., Ranty, B., Charpenteau, M., Sotta, B., Galaud, J., and Aldon, D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56, 575–589. doi: 10.1111/j.1365-313X.2008.03622.x
Mashiach, E., Schneidman-Duhovny, D., Andrusier, N., Nussinov, R., and Wolfson, H. J. (2008). FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 36, W229–W232. doi: 10.1093/nar/gkn186
McCormack, E., and Braam, J. (2003). Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 159, 585–598. doi: 10.1046/j.1469-8137.2003.00845.x
McCormack, E., Tsai, Y.-C., and Braam, J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi: 10.1016/j.tplants.2005.07.001
Meena, M. K., Prajapati, R., Krishna, D., Divakaran, K., Pandey, Y., Reichelt, M., et al. (2019). The Ca2+ channel CNGC19 regulates Arabidopsis defense against Spodoptera herbivory. Plant Cell 31, 1539–1562. doi: 10.1105/tpc.19.00057
Mithöfer, A., and Boland, W. (2008). Recognition of herbivory-associated molecular patterns. Plant Physiol. 146, 825–831. doi: 10.1104/pp.107.113118
Munir, S., Khan, M. R. G., Song, J., Munir, S., Zhang, Y., Ye, Z., et al. (2016). Genome-wide identification, characterization and expression analysis of calmodulin-like (CML) proteins in tomato (Solanum lycopersicum). Plant Physiol. Biochem. 102, 167–179. doi: 10.1016/j.plaphy.2016.02.020
Naresh, R. K., Dhaliwal, S. S., Chaudhary, M., Chandra, M. S., and Kumar, A. (2019). Legumes: An Option to enhance Productivity and Soil Health Sustainability. Singapore: Springer, 93.
O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., and Hutchison, G. R. (2011). Open Babel: an open chemical toolbox. J. Cheminform. 3:33. doi: 10.1186/1758-2946-3-33
Park, H. C., Kim, M. L., Kang, Y. H., Jeon, J. M., Yoo, J. H., Kim, M. C., et al. (2004). Pathogen-and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 135, 2150–2161. doi: 10.1104/pp.104.041442
Park, H. C., Park, C. Y., Koo, S. C., Cheong, M. S., Kim, K. E., Kim, M. C., et al. (2010). AtCML8, a calmodulin-like protein, differentially activating CaM-dependent enzymes in Arabidopsis thaliana. Plant Cell Rep. 29, 1297–1304. doi: 10.1007/s00299-010-0916-7
Perochon, A., Aldon, D., Galaud, J.-P., and Ranty, B. (2011). Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 93, 2048–2053. doi: 10.1016/j.biochi.2011.07.012
Poovaiah, B. W., Du, L., Wang, H., and Yang, T. (2013). Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol. 163, 531–542. doi: 10.1104/pp.113.220780
Ranty, B., Aldon, D., Cotelle, V., Galaud, J.-P., Thuleau, P., and Mazars, C. (2016). Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Sci. 7:327. doi: 10.3389/fpls.2016.00327
Reddy, A. S. N., Ben-Hur, A., and Day, I. S. (2011). Experimental and computational approaches for the study of calmodulin interactions. Phytochemistry 72, 1007–1019. doi: 10.1016/j.phytochem.2010.12.022
Reymond, P., Bodenhausen, N., Van Poecke, R. M. P., Krishnamurthy, V., Dicke, M., and Farmer, E. E. (2004). A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16, 3132–3147. doi: 10.1105/tpc.104.026120
Reymond, P., Weber, H., Damond, M., and Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–719. doi: 10.1105/tpc.12.5.707
Schneidman-Duhovny, D., Inbar, Y., Nussinov, R., and Wolfson, H. J. (2005). PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 33, W363–W367. doi: 10.1093/nar/gki481
Schuler, M. A. (2011). P450s in plant–insect interactions. Biochim. Biophys. Acta 1814, 36–45. doi: 10.1016/j.bbapap.2010.09.012
Singh, A., Kumar, A., Hartley, S., and Singh, I. K. (2020a). Silicon: its ameliorative effect on plant defense against herbivory. J. Exp. Bot. 71, 6730–6743. doi: 10.1093/jxb/eraa300
Singh, S., Singh, A., Kumar, S., Mittal, P., and Singh, I. K. (2020b). Protease inhibitors: recent advancement in its usage as a potential biocontrol agent for insect pest management. Insect Sci. 27, 186–201. doi: 10.1111/1744-7917.12641
Singh, S., Tyagi, C., Rather, I. A., Sabir, J. S. M., Hassan, M., Singh, A., et al. (2020c). Molecular modeling of chemosensory protein 3 from Spodoptera litura and its binding property with plant defensive metabolites. Int. J. Mol. Sci. 21:4073. doi: 10.3390/ijms21114073
Singh, A., Panwar, R., Mittal, P., Hassan, M. I., and Singh, I. K. (2021a). Plant cytochrome P450s: role in stress tolerance and potential applications for human welfare. Int. J. Biol. Macromol. 184, 874–886. doi: 10.1016/j.ijbiomac.2021.06.125
Singh, A., Singh, S., Singh, R., Kumar, S., Singh, S. K., and Singh, I. K. (2021b). Dynamics of Zea mays transcriptome in response to a polyphagous herbivore, Spodoptera litura. Funct. Integr. Genom. 21, 571–592. doi: 10.1007/s10142-021-00796-7
Singh, S., Singh, A., and Singh, I. K. (2021c). “Transcriptomics Studies Revealing Enigma of Insect-Plant Interaction,” in Plant-Pest Interactions: From Molecular Mechanisms to Chemical Ecology, eds I. K. Singh and A. Singh (Singapore: Springer), 31–55. doi: 10.1007/978-981-15-2467-7_2
Singh, A., Singh, I. K., and Verma, P. K. (2008). Differential transcript accumulation in Cicer arietinum L. in response to a chewing insect Helicoverpa armigera and defence regulators correlate with reduced insect performance. J. Exp. Bot. 59, 2379–2392. doi: 10.1093/jxb/ern111
Singh, A., Singh, S., and Singh, I. K. (2016). Recent insights into the molecular mechanism of jasmonate signaling during insect-plant interaction. Australas. Plant Pathol. 45, 123–133. doi: 10.1007/s13313-015-0392-1
Singh, A., Tyagi, C., Nath, O., and Singh, I. K. (2018). Helicoverpa-inducible Thioredoxin h from Cicer arietinum: structural modeling and potential targets. Int. J. Biol. Macromol. 109, 231–243. doi: 10.1016/j.ijbiomac.2017.12.079
Singh, I. K., and Singh, A. (2021). Plant-Pest Interactions: From Molecular Mechanisms to Chemical Ecology. New York NY: Springer.
Soyastats (2010). A Reference Guide to Important Soybean Facts and Figure. American Soybean Association.
Stotz, H. U., Pittendrigh, B. R., Kroymann, J., Weniger, K., Fritsche, J., Bauke, A., et al. (2000). Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol. 124, 1007–1018. doi: 10.1104/pp.124.3.1007
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612.
Thor, K., and Peiter, E. (2014). Cytosolic calcium signals elicited by the pathogen-associated molecular pattern flg22 in stomatal guard cells are of an oscillatory nature. New Phytol. 204, 873–881. doi: 10.1111/nph.13064
Vadassery, J., Reichelt, M., Hause, B., Gershenzon, J., Boland, W., and Mithöfer, A. (2012a). CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 159, 1159–1175. doi: 10.1104/pp.112.198150
Vadassery, J., Scholz, S. S., and Mithöfer, A. (2012b). Multiple calmodulin-like proteins in Arabidopsis are induced by insect-derived (Spodoptera littoralis) oral secretion. Plant Signal. Behav. 7, 1277–1280. doi: 10.4161/psb.21664
Vandelle, E., Vannozzi, A., Wong, D., Danzi, D., Digby, A.-M., Dal Santo, S., et al. (2018). Identification, characterization, and expression analysis of calmodulin and calmodulin-like genes in grapevine (Vitis vinifera) reveal likely roles in stress responses. Plant Physiol. Biochem. 129, 221–237. doi: 10.1016/j.plaphy.2018.06.003
Vanderbeld, B., and Snedden, W. A. (2007). Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37 CML38 and CML39. Plant Mol. Biol. 64, 683–697. doi: 10.1007/s11103-007-9189-0
Vasudev, A., and Sohal, S. (2016). Partially purified Glycine max proteinase inhibitors: potential bioactive compounds against tobacco cutworm, Spodoptera litura (Fabricius, 1775)(Lepidoptera: Noctuidae). Turkish J. Zool. 40, 379–387. doi: 10.3906/zoo-1508-20
Vincent, T. R., Avramova, M., Canham, J., Higgins, P., Bilkey, N., Mugford, S. T., et al. (2017). Interplay of plasma membrane and vacuolar ion channels, together with BAK1, elicits rapid cytosolic calcium elevations in Arabidopsis during aphid feeding. Plant Cell 29, 1460–1479. doi: 10.1105/tpc.17.00136
Waese, J., Fan, J., Pasha, A., Yu, H., Fucile, G., Shi, R., et al. (2017). ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 29, 1806–1821. doi: 10.1105/tpc.17.00073
Weinl, S., and Kudla, J. (2009). The CBL–CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 184, 517–528. doi: 10.1111/j.1469-8137.2009.02938.x
Zeng, H., Zhang, Y., Zhang, X., Pi, E., and Zhu, Y. (2017). Analysis of EF-hand proteins in soybean genome suggests their potential roles in environmental and nutritional stress signaling. Front. Plant Sci. 8:877. doi: 10.3389/fpls.2017.00877
Keywords: Calmodulin like proteins (CMLs), soybean, Spodoptera litura, calcium signaling, signaling compounds, wounding, plant-insect interaction
Citation: Yadav M, Pandey J, Chakraborty A, Hassan MI, Kundu JK, Roy A, Singh IK and Singh A (2022) A Comprehensive Analysis of Calmodulin-Like Proteins of Glycine max Indicates Their Role in Calcium Signaling and Plant Defense Against Insect Attack. Front. Plant Sci. 13:817950. doi: 10.3389/fpls.2022.817950
Received: 18 November 2021; Accepted: 25 January 2022;
Published: 09 March 2022.
Edited by:
Raul Antonio Sperotto, Universidade do Vale do Taquari, BrazilReviewed by:
Wayne Snedden, Queen’s University, CanadaSandra Scholz, Friedrich Schiller University Jena, Germany
Copyright © 2022 Yadav, Pandey, Chakraborty, Hassan, Kundu, Roy, Singh and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amit Roy, cm95QGZsZC5jenUuY3o=; Indrakant Kumar Singh, aWtzaW5naEBkYi5kdS5hYy5pbg==; Archana Singh, YXJjaGFuYXNpbmdoQGhyYy5kdS5hYy5pbg==
 Manisha Yadav1
Manisha Yadav1 Amrita Chakraborty
Amrita Chakraborty Md. Imtaiyaz Hassan
Md. Imtaiyaz Hassan Jiban Kumar Kundu
Jiban Kumar Kundu Amit Roy
Amit Roy Indrakant Kumar Singh
Indrakant Kumar Singh Archana Singh
Archana Singh