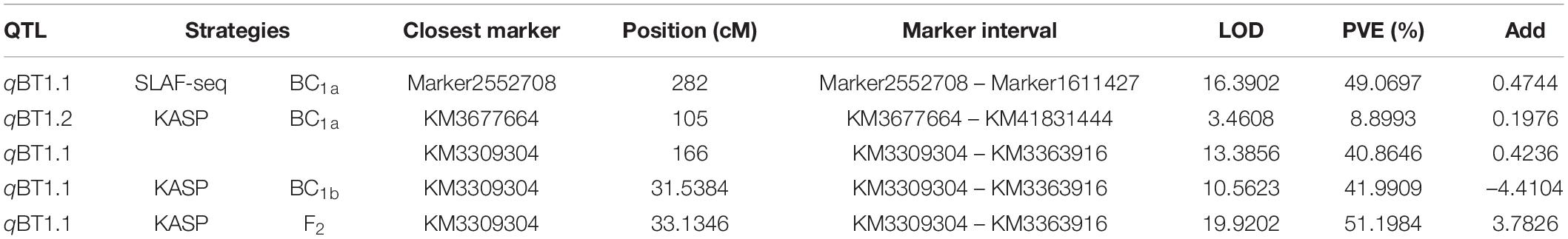
- 1Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Ilera Healthcare LLC, Waterfall, PA, United States
Spinach is a typical light-sensitive plant. Long days can induce early bolting, thereby influencing the regional adaptation, quality, and vegetative yield of spinach. However, the genes and genetic mechanisms underlying this trait in spinach remain unclear. In this study, a major quantitative trait locus (QTL) qBT1.1, was mapped on chromosome 1 using a BC1 population (BC1a) derived from 12S3 (late-bolting recurrent lines) and 12S4 (early bolting lines) with specific-locus amplified fragment (SLAF) markers and Kompetitive Allele Specific PCR (KASP) markers. The qBT1.1 locus was further confirmed and narrowed down to 0.56 Mb by using a large BC1 (BC1b) population and an F2 population using the above KASP markers and the other 20 KASP markers. Within this region, two putative genes, namely, SpFLC and SpCOL14, were of interest due to their relationship with flower regulatory pathways. For SpCOL14, we found multiple variations in the promoter, and the expression pattern was consistent with bolting stages. SpCOL14 was therefore assumed to the best candidate gene for bolting. Overall, our results provide a basis for understanding the molecular mechanisms of bolting in spinach and contribute to the breeding of diverse spinach germplasms for adaptation to different regions.
Introduction
Spinach (Spinacia oleracea L.) is a diploid plant (2n = 2x = 12) of the Amaranthaceae family (Morelock and Correll, 2008). It was domesticated in Iran around 2,000 years ago (Rubatzky and Yamaguchi, 1997) and was first mentioned as the “herb of Persia” in China approximately 600 A.D. (Kuwahara et al., 2014). Spinach is an important and nutritious green leafy vegetable that is rich in carotenoids, folate, vitamin C, calcium, and iron (Lester et al., 2013). Spinach is also a good source of antioxidants and has one of the highest ORAC (oxygen radical absorbance capacity) values of any vegetable (Koh et al., 2012). It is typically consumed as a fresh, cooked or canned vegetable (Morelock and Correll, 2008; Ma et al., 2016). The reproductive process usually begins with bolting (the elongation of the stem) (Mutasa-Göttgens et al., 2010), which leads to decreased yields and low quality (Abe et al., 2014). Spinach is easily influenced by the photoperiod (Chun et al., 2000b), and bolts in spring (Tang et al., 2018). New slow-bolting spinach cultivars available to that can adapt to a wide range of photoperiods and climatic conditions (Bhattarai and Shi, 2021). Selecting the appropriate cultivars will improve the efficiency of breeding and production in spinach (Goreta and Leskovar, 2006).
Bolting refers to the rapid lengthening of the plant stem and is due to the coordinated effects of developmental and environmental factors (Chen et al., 2019). As a complex quantitative trait, bolting shows continuous phenotypic variation in many crops (Melchinger, 1998). Bolting is a transitional stage between vegetative growth and reproductive growth, and thus evaluating the genetics of bolting is essential for elucidating this phenomenon. Many key bolting and flowering genes have been identified and functionally characterized in Arabidopsis. Arabidopsis is characterized by inflorescence axis elongation-type bolting (Chen et al., 2019), which provides a reference for the study of bolting and flowering genes in spinach. Genetic studies in Arabidopsis have revealed that the genes controlling bolting and flowering are involved in, and can be assigned to, distinct regulatory pathways, including photoperiod, vernalization, gibberellin, autonomous, ambient temperature, and age (Fornara et al., 2010). These pathways are associated with plant developmental and environmental cues, such as photoperiod and temperature (Cho et al., 2017). One of the key genes affecting bolting and flowering is FLOWERING LOCUS C (FLC), which represses bolting and flowering by encoding the MADS-box protein in the vernalization pathway and FLC is expressed widely in the shoot apical meristem and leaves (Sheldon et al., 1999, 2000). The other key genes affecting bolting and flowering include CONSTANS (CO), which is involved in the photoperiod pathway. CO is the key gene accelerating bolting and flowering during long days (Suarez-Lopez et al., 2001), which acts upstream of FLOWERING LOCUS T (FT) in the photoperiod pathway (Dally et al., 2014). CO belongs to CONSTANS-LIKE (COL) proteins, called B-box (BBX) proteins (Griffiths et al., 2003). COLs are a class of zinc finger transcription factors that consist of a CO, COL, and TIMING OF CAB1 (CCT) domain (Abe et al., 2014). One COL (SoCOL1) and two FLOWERING LOCUS T (FT) homologs were isolated and characterized in the photoperiodic regulation of spinach (Abe et al., 2014).
It has been reported that flowering and bolting traits in spinach are greatly affected by long-day photoperiods and gibberellin (Zeevaart, 1971; Wu et al., 1996; Kim et al., 2000). Thus far, a few molecular markers and genes related to bolting and flowering in spinach have been reported. Chitwood et al. (2016) used 288 United States Department of Agriculture (USDA) spinach accessions as the association panel in this research and found three single nucleotide polymorphism (SNP) markers associated with bolting through genotyping-by-sequencing (GBS) technology and genome wide association study (GWAS). A draft genome sequence of spinach has been reported, and two quantitative trait loci (QTLs) associated with bolting have been obtained in the region from 44.7 to 50.5 Mb of chromosome 2 (Xu et al., 2017). Bhattarai et al. (2020) identified SNP sites associated with bolting and flowering on chromosomes 2, 3, and 5 by GWAS techniques with 300 USDA spinach accessions. Recently, a new spinach genome SOL_r1.1 have revealed three QTLs connected with bolting by double-digest restriction-site-associated DNA sequencing (ddRAD-seq) (Hideki et al., 2021). GWAS analyses of bolting and flowering traits yielded several associated regions across the six chromosomes and detected a region harboring genes encoding MADS-box transcription factors (SOV6g023690 and SOV4g008150) by the Monoe-Viroflay spinach genome (Cai et al., 2021). With the transcriptome sequencing of spinach bolting (Abolghasemi et al., 2021), more genes will be detected in the future research. These results suggest that spinach bolting is controlled by multiple QTLs or genes. However, details on the genetic mechanisms of bolting and flowering remained unclear in spinach, and no reliable molecular markers have been developed for the molecular marker-assisted selection (MAS) of slow bolting traits in spinach breeding.
Quantitative trait locus mapping is a powerful approach to dissect the genetic architecture of complex traits (Mauricio, 2001), and has been used to identify potential genes by revealing the relationship between the genotype (based on molecular markers) and phenotype (Salvi and Tuberosa, 2005). In spinach, QTL mapping has largely been used to investigate in: sex-determining locus (Khattak et al., 2006), nitrogen use efficiency (Chan-Navarrete et al., 2016), leaf color (Cai et al., 2018), fruit spines (Liu et al., 2021a), and leaf-related traits (Liu et al., 2021b). Moreover, the bolting trait has been reported in many crops by QTL mapping, such as Brassica napus L. (Fu et al., 2020; Xu et al., 2021), Beta vulgaris (Tränkner et al., 2017), wheat (Buerstmayr et al., 2009) and so forth. In our previous study, the early bolting inbred line 12S4 and the late-bolting line 12S3 were used as parents to develop segregated populations, and a high-density spinach genetic linkage map with 4080 specific-locus amplified fragment (SLAF) markers (Qian et al., 2017) was constructed using a derived BC1a population (N = 148). The objectives of the current study were to map the bolting gene through SLAF-based and KASP-based QTL mapping approaches and identify the candidate genes controlling the bolting trait using the BC1b and F2 populations. This study will be help elucidate the genetic mechanisms of bolting, which may lay the foundation in MAS bolting behavior in spinach breeding.
Materials and Methods
Plant Material and Phenotyping Evaluation of Bolting Time
Two inbred lines, 12S3 and 12S4, which exhibit significant differences in bolting, were selected as the parents. Line 12S3, with extreme resistance to bolting, was used as the female and recurrent parent, while the early bolting line 12S4 was used as the male parent to develop a BC1 and an F2 population (Qian et al., 2017). The two parental lines, the derived F1 line, and the 148 BC1 individuals (BC1a) were planted in a field in spring 2015 for primary mapping. In addition, 200 BC1 progenies (BC1b) and 150 F2 progenies were planted in the same location in spring 2020 in natural conditions for validation of QTLs and narrowing down of the QTL regions. All of these materials were developed and tested by the Spinach Research Group, Institute of Vegetables and Flowers (IVF), and Chinese Academy of Agricultural Sciences (CAAS).
Each individual plant was visually inspected daily, and the bolting date was determined as the date that a stem of a plant was seen to be at least 5 cm in length (Fu et al., 2020). The bolting time (BOT) was then determined as the period from the sowing date to the bolting date. The phenotypic data of all plant materials in these experiments were analyzed with Excel 2013 (Microsoft Corp., Redmond, United States) for calculating the mean, standard error (SE), and coefficient of variation (CV) in each line or population.
DNA Extraction
At the four true-leaf stage, fresh young leaves were collected from each plant of the F1, BC1a, BC1b, F2 populations, and parents, immediately frozen in liquid nitrogen, and stored in a –80°C freezer. Genomic DNA was extracted from each plant using the cetyltrimethyl ammonium bromide (CTAB) method (Murray and Thompson, 1980). The DNA concentration and quality were assessed using a ND-2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States) and 1.0% agarose gel electrophoresis, respectively.
Specific-Locus Amplified Fragment Library Construction for High-Throughput Sequencing
Specific length amplified fragment sequencing (SLAF-seq) is an efficient method of large-scale genotyping developed on the basis of high-throughput sequencing technology and reduced representation library (RRL). In brief, an SLAF pilot-experiment was first designed to improve the efficiency of SLAF-seq, which considered the uniform distribution and avoided the duplication of SLAFs. Next, according to the pre-experiment, the SLAF library was conducted as follows The genomic DNA from each sample was completely digested by the two restriction enzymes - RsaI and HaeIII (New England Biolabs, NEB). After digestion, the DNA fragments were repaired with adenine and duplex tag-labeled sequencing adapters. Twenty polymerase chain reaction (PCR) cycles were used to enrich the concentration of fragments and the PCR products were then purified and pooled. The sample was performed by 2% agarose gel electrophresis (120 V, 60 min). After gel purifcation, DNA fragments of 364–414 bp were excised and diluted for paired-end sequencing. Finally, the selected SLAFs were sequenced on an Illumina High-seq 2500 sequencing platform (Illumina, Inc.; San Diego, CA, United States).
The analysis of SLAF-markers followed the procedures described by Sun et al. (2013). All SLAF paired-end reads were clustered on the basis of sequence similarity, which was detected by BLAST (-tileSize = 10, -stepSize = 5). Sequences with over 95% identity were grouped in one SLAF locus. SLAFs with two to four tags were deemed as polymorphic SLAFs.
Single Nucleotide Polymorphism Molecular Marker Analysis and Genotyping
The SNP molecular markers were obtained from 4080 SLAF markers from the spinach high-density genetic map constructed by Qian et al. (2017), following which a total of 300 KASP primers was designed by the LGC company (Shanghai, China), and the slow bolting parent 12S3 and early bolting parent 12S4 were tested (Liu et al., 2021b). A subset of KASP primers were selected and used to genotype the BC1a.
For the KASP assays, each sample contained 2.5 μL 2 × KASP Master mix, 0.07 μL KASP Assay mix, and 2.5 μL genomic DNA diluted to 20–30 ng/μL. The reaction system was as follows: 94°C for 15 min, 10 cycles of 94°C for 20 s and 61°C (0.6°C drop per cycle) for 60 s and a further 26 cycles of 94°C for 20 s and 55°C for 60 s. An additional three cycles of 20 s at 94°C and 60 s at 55°C were executed if the results of the initial KASP thermal cycles did not acquire sufficiently defined genotype clusters. In addition to DNA samples, two no-template controls (NTCs) were included on each 384-well PCR plate. All plates were read below 40°C in a 7900 HT Fast Real-Time PCR System (Applied Biosystems), and the data were analyzed using SDS2.3 software (supplied by Applied Biosystems) (Semagn et al., 2014).
Linkage Map Construction and Quantitative Trait Locus Mapping
The SNP markers were selected with no segregation distortion, and markers with more than 25% missing data were also excluded. The valid markers were then used to construct the linkage map from the BC1a population using JoinMap 4.0 software (Van Ooijen, 2006). All markers were firstly grouped based on a threshold of LOD = 3.0, while all other settings were left at their default values.
The BC1b and F2 populations were used to confirm and narrow down the predicted region, and other KASP markers were developed based on the SNP variation between the two parents around the initial QTL area. The QTLs for bolting were also detected using QTL IciMapping 4.2 software (Meng et al., 2015) based on the phenotype of 148 BC1a individuals. The Composite Interval Mapping of ADDitive QTL (ICIM-ADD) method was used for QTLs. The parameters were as follows: a step in 1 cM, probability in stepwise regression of 0.001, and LOD = 3.0. The final QTLs were named based on the method of McCouch et al. (1997): “ q” + the English abbreviation of the trait + the chromosome number + “. “ the QTL number.
Candidate Gene Analysis and Real-Time Polymerase Chain Reaction of Bolting Time
Based on the of spinach genome annotations (version Sp75) in SpinachBase,1 the genes related to bolting and flowering within the identified interval were selected for further analysis. The full-length RNA was extracted at the 12-leaf-stage and the promoters of the candidate genes were sequenced between the two parents. The specific primers were designed by Primer3 plus2 (Table 2). The candidate genes were cloned and the sequences were aligned by MUSCLE software.3 Finally, the gene structure was elucidated based on the re-sequenced result.4
Quantitative real-time PCR was employed to evaluate the expression of the candidate genes from the seedling to bolting stages. Leaf tissue of the 12S3 and 12S4 lines was collected at 6, 9, 12, 15, and 18 weeks until both parents began bolting in spring of 2021 (12S3 bolted 18 weeks after planting; 12S4 bolted 15 weeks after planting) (Figure 7B), and the total RNA was extracted using a Plant Total RNA Mini Kit (GeneBetter Biotech, Beijing, China5). The cDNA was synthesized from 500 ng total RNA with a TranScript One-Step gDNA Removal and cDNA Synthesis Kit (TransGen Biotech, Beijing, China6). Three independent biological and three technical replicates of each period were performed and analyzed. The synthesized cDNA was subjected to quantitative real-time (qRT)-PCR analysis using a QuantStudio™ 12 K Flex Real-Time PCR System (Applied Biosystems) with SYBR Fast qPCR Mix (TaKaRa7). The reaction mixture contained 70 ng template cDNA, 0.2 μM of gene-specific primer (Table 2), 0.2 μM ROX Reference Dye II, 3.4 μL ddH2O, and 5 μL 2 × SYBR Fast qPCR Mix in a 20 μL volume. The qRT-PCR was performed at 95°C for 30 s, followed by 35 cycles of 95°C for 15 s and 60°C for 1 min. The relative expression was calculated using the 2–ΔΔCT method. SpActin was used as the reference gene (Lee and Zeevaart, 2002).
Results
Bolting Time Analysis and Mapping of Quantitative Trait Loci Controlling Spinach Bolting
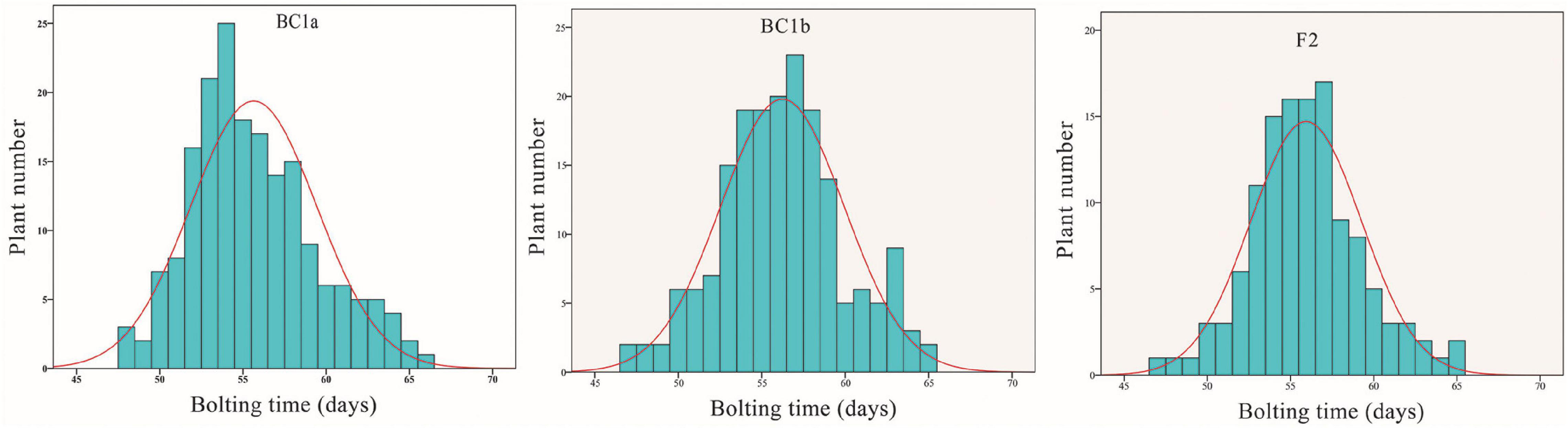
In 2015, the bolting time of line 12S3 and 12S4 was on average 62 (60–65) and 46.5 (45–48) days, respectively, indicating differences in the bolting time of the parents. From the BC1a line, the bolting time ranged from 48 to 66 days, with an average of 54.5 days. The bolting time of 155 BC1b and the 123 F2 individuals was from 47 to 65 days, with a mean value of 57.6 days in BC1b and 55.8 days in the F2 populations. The phenotypic traits are summarized in Table 1 and Supplementary Table 1. Moreover, these segregating populations showed continuous variation in bolting time, suggesting that the bolting trait has a quantitatively inherited character in spinach (Figure 1 and Supplementary Table 2).
In our previous study (Qian et al., 2017), a total of 4080 SLAF markers for 148 BC1a individuals were acquired by SLAF-seq, and the linkage groups were coded with six linkage groups (P01–P06) in a total length of 1125.97 cM, which matches the spinach chromosome numbers (Supplementary Table 3). After the exclusion of missing and disqualified data, 130 BC1a individuals were finally used to map the QTLs (Supplementary Table 3). Combining the SLAF high-density genetic map with bolting time in 130 BC1a progenies, a major QTL (named qBT1.1), which contributed 49.07% of the phenotypic variance (PVE) (Table 1), was identified at the interval 15.82–18.97 cM on LG3 between two adjacent SLAF markers (Marker 2552708 and Marker 1611427), with an LOD score of 16.39. Based on the spinach genome Sp75 (Xu et al., 2017), this QTL was mapped on chromosome 1 in the region of 47.72–50.61 Mb (Figure 2).
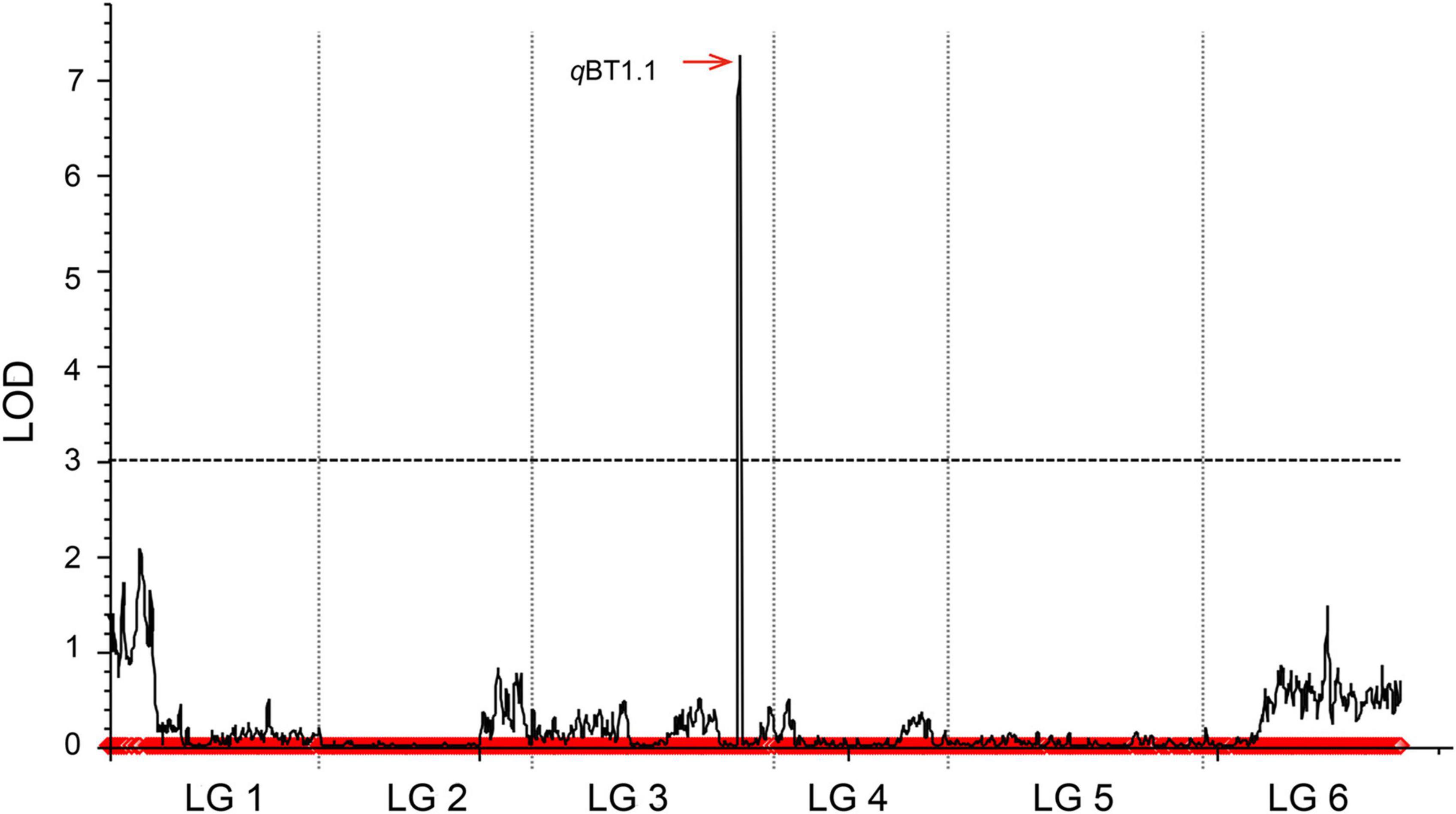
Figure 2. Mapping QTLs controlling spinach bolting trait using a high-density genetic linkage map constructed with SLAF markers.
A total of 181 informative SNP markers (Liu et al., 2021b) and 147 BC1a individuals were selected for KASP-based linkage analysis (Supplementary Table 4). Based on the 181 KASP markers and screened 127 BC1a plants, two QTLs (qBT1.2 and qBT1.1) were mapped to 103.5–105.5 cM and 163.5–166.0 cM on LG3 (Figure 3) and were located at 41.44–42.02 Mb and 46.76–49.12 Mb on chromosome 1, respectively. The LOD scores were 3.46 and 13.39, explaining 8.90 and 40.86% PVE, separately (Table 1). In 2020, the same 181 KASP markers were used in 185 BC1b and 112 F2 populations and they were co-located in the same area between KM706861 and KM3309304 (Figure 3). The LOD score was 10.5623 and explained 41.99% PVE in BC1b individuals while the figures were 19.9202 and 51.20% in F2 plants, respectively (Table 1).
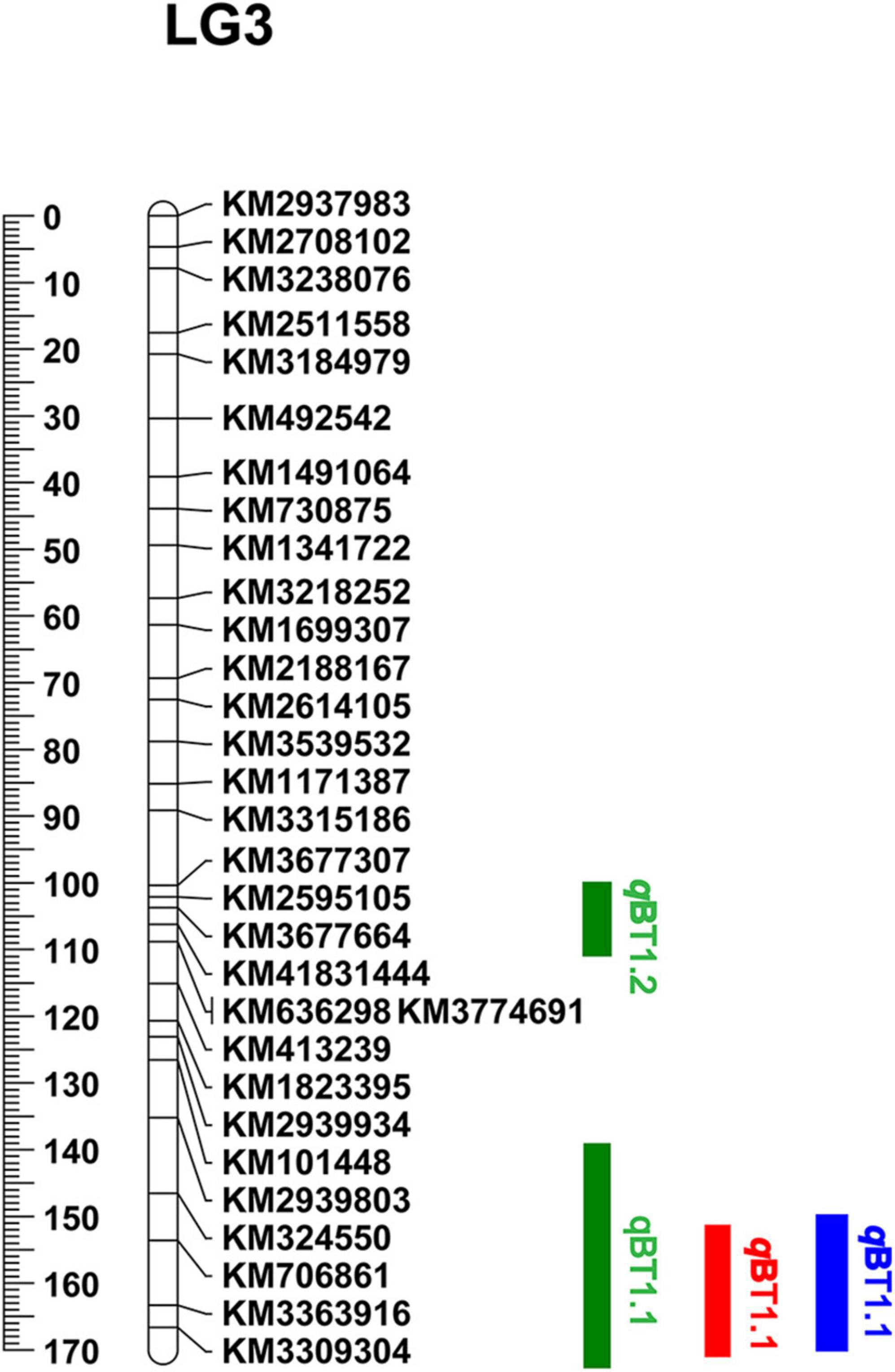
Figure 3. QTL results of bolting using KASP genetic linkage map. The color Green, Red, and Blue respect BC1a, BC1b, and F2 populations, respectively.
Fine-Mapping of Spinach Bolting
The SLAF-based and KASP-based QTLs in the two strategies indicated that qBT1.1 was a stable locus that could be used for fine-mapping and cloning. The SNP variations were explored in the sequences at this region of 12S3 and 12S4. The raw reads were first-filtered by fastp 0.12.0 (Chen et al., 2018) and the alignment data were obtained on the spinach genome Sp75 (Xu et al., 2017) by BWA 0.7.17-r1188 (Li and Durbin, 2009). The vcf files were finally generated by Samtools/Bcftools 0.1.19 – 44428 cd (Li et al., 2009). To further refine the mapping region, the KASP markers were developed from 40 to 51 Mb on chromosome 1 by the file.
After eliminating the invalid segregation data, 20 efficient KASP markers were designed for fine mapping (Supplementary Table 5). In the expanded BC1 population (BC1b), 185 individuals were obtained in 2020, and then 16 recombinant plants were ultimately acquired by the KASP genotyping. A major QTL (qBT1.1) for bolting time was verified between KMBL53 (31.0 cM) and KM3309304 (31.5 cM) (Figure 4). Furthermore, in the F2 population in 2020, the 112 plants were used to map the QTLs for bolting time, and nine recombinant individuals were obtained. A major QTL in the F2 populations was also fine-mapped in the interval of KMBL53 (32.4 cM) and KM3309304 (33.1 cM). In conclusion, a QTL named qBT1.1 was detected with a 0.56-Mb region between KMBL53 (47.56 Mb) and KM3309304 (48.12 Mb) on chromosome 1 (Figure 4).
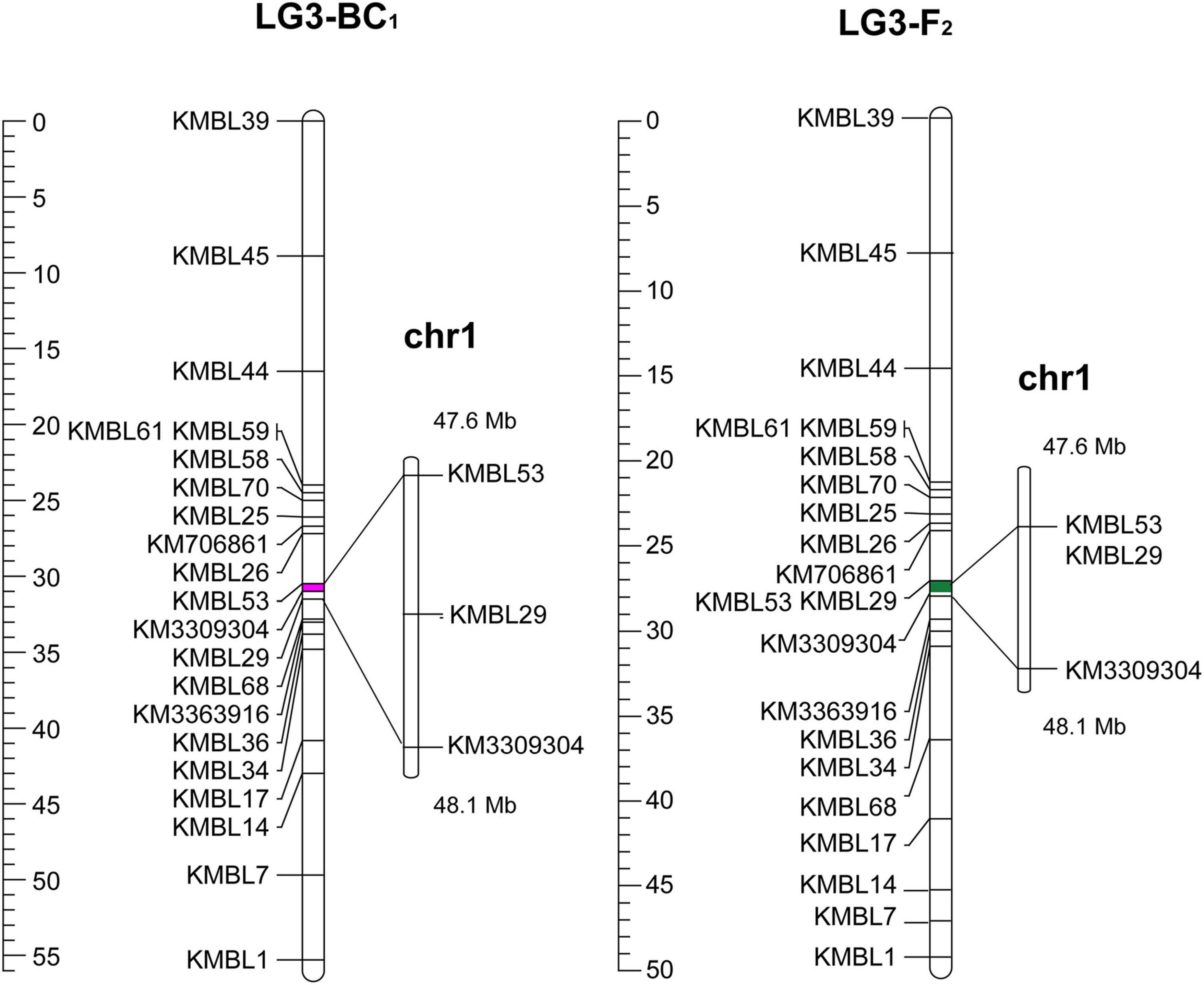
Figure 4. QTL mapping of BC1b and F2 population: the left was the fine mapping of BC1b population; the right was the fine mapping of F2 population.
Screening for Candidate Genes Controlling Spinach Bolting Trait
A total of 68 genes were located in a 560 kb region based on the spinach genome (version Sp75)8 (Supplementary Figure 1 and Supplementary Table 6). Among these genes, two genes were unannotated, 23 genes encoded various enzymes, and four genes had transmembrane structure. Two genes Spo04942 and Spo04967 were found to be homologs of bolting and flowering genes in Arabidopsis (Table 2, Figure 5, and Supplementary Table 6), and thus could potentially be the candidate genes controlling the spinach bolting trait.
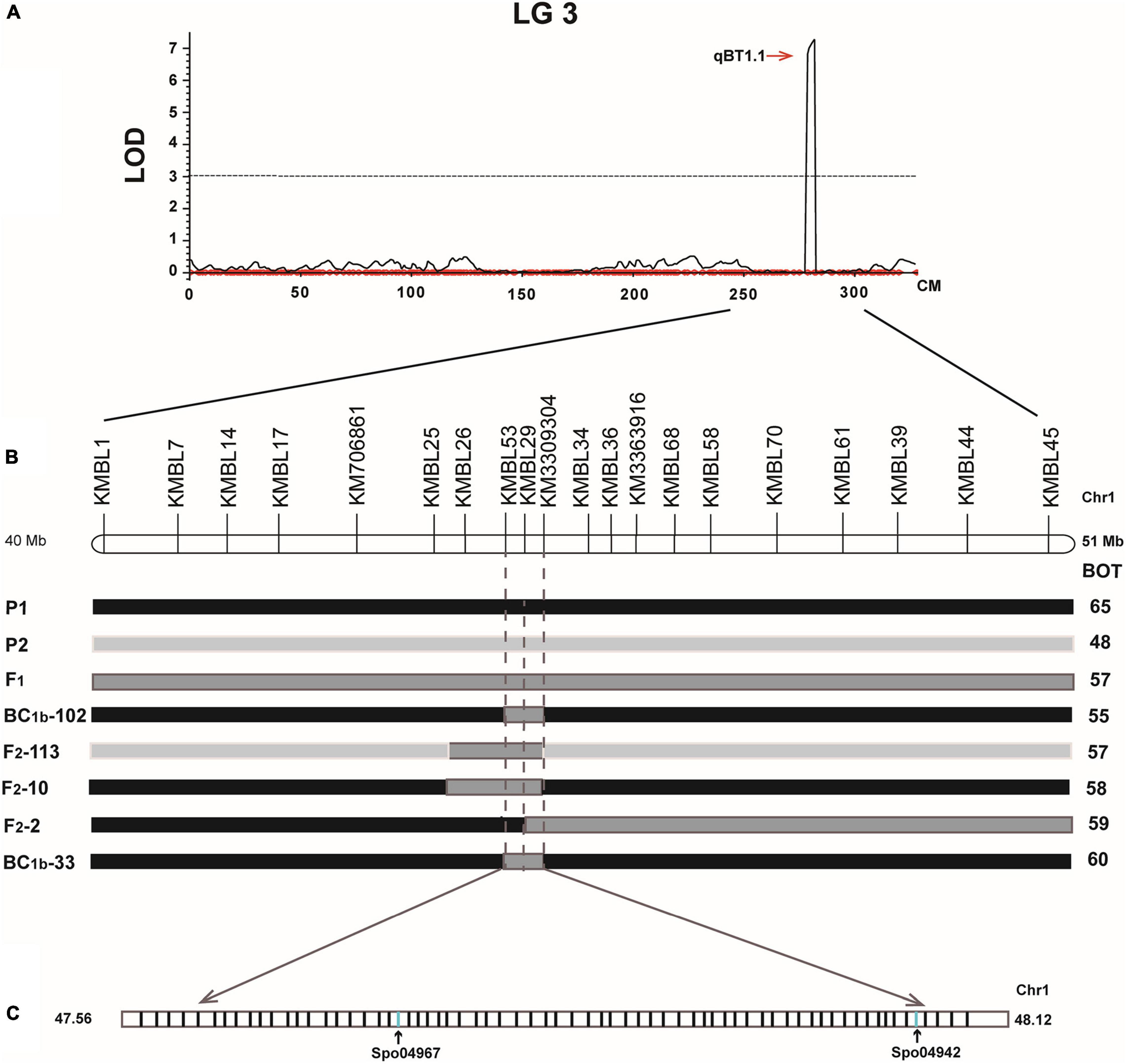
Figure 5. Fine mapping QTLs controlling spinach bolting trait. (A) One QTL was mapped in LG3 using the BC1a population; (B) fine mapping of the QTL controlling spinach bolting trait using the BC1b and F2 populations; (C) two candidate gene were identified in the interval from 47.56 Mb and 48.12 Mb of spinach chromosome 1.
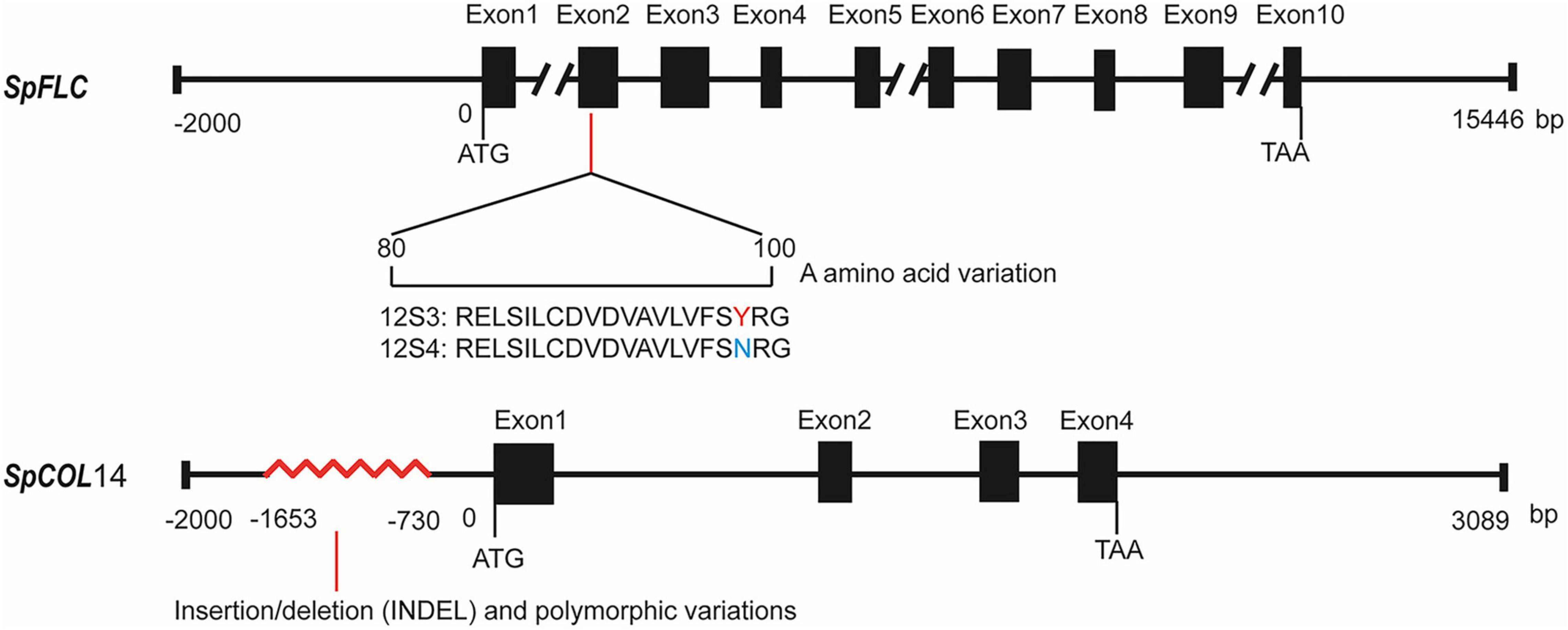
Spo04942 is MADS-box transcription factor that is homologous to Arabidopsis FLC and the sugar beet FLC homolog FLC-LIKE 1, thus was renamed as SpFLC. In the coding area, there were two synonymous SNP variations and one non-synonymous SNP variation that led to the change from tyrosine (12S3) to asparagine (12S4) at position 98 in the domain area (Figure 6). In the 2-kb upstream non-coding sequences, no difference was found between the two parents. The gene Spo04967 may encode a zinc finger protein similar to CONSTANS-LIKE 14, which belonged to the COL family, and thus was named as SpCOL14. With Sanger sequencing, no variation was found in the coding region of SpCOL14 between 12S3 and 12S4. However, from the 2-kb upstream non-coding region, variations of about 900 bp were found between 12S3 and 12S4 (–730 bp to –1653 bp) (Figure 6) that may affect the expression of SpCOL14.
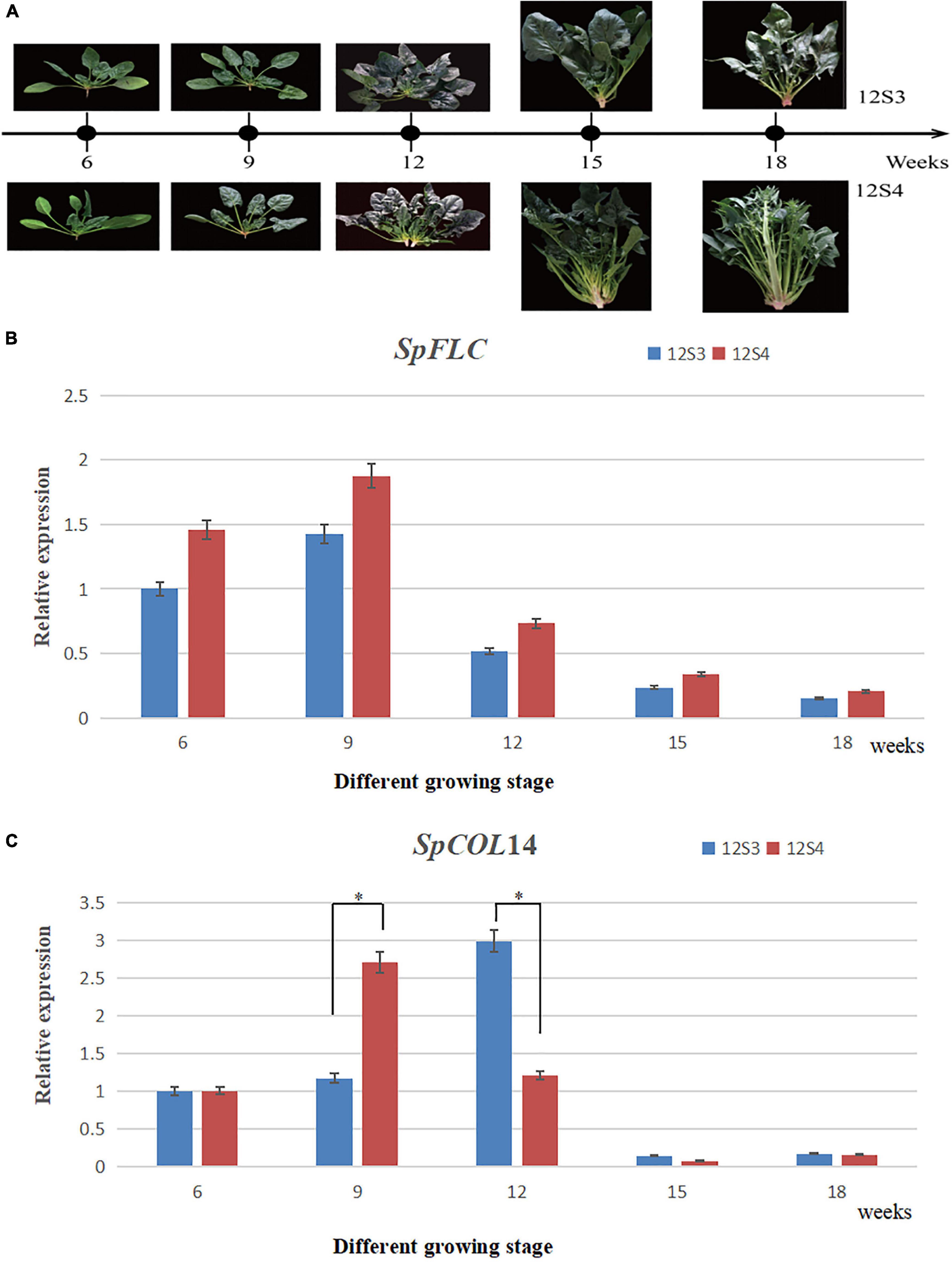
Figure 7. (A) Plant materials used in qRT-PCR planted in 2021 spring; (B) the expression of SpFLC; (C) the expression of SpCOL14. * Represented significant difference (P < 0.05).
Except for SpFLC and SpCOL14, no other genes were found to be related to bolting and flowering genes, such as FT, SOC1, and FLOWERING LOCUS D (FD) that were reported in other plants (Hideki et al., 2021). Interestingly, in this 0.56 Mb region, we found some transcription factors that regulate the various stages of plant growth and development, including Spo04911, which is an NAC domain-containing protein that plays a role in regulating plant growth and stress resistance, and Spo04943, which is a MADS-box factor with an AGAMOUS-LIKE 9 homolog to floral organ development. The factors may play some minor roles in affecting bolting regulation, but the gene regulatory network will be identified in the future with technological progress.
Expression Analysis of the Candidate Genes Controlling the Spinach Bolting Trait
We assessed the expression patterns using qRT-PCR analysis between 12S3 and 12S4 at different growth stages to further assess the two candidate genes (Table 2). 12S4 bolted after 15 weeks, while the bolting time of 12S3 was after 18 weeks (Figure 7B). Although the expression of these two candidate genes at the five stages were significantly different, a similar change trend of expression levels of SpFLC was found both in 12S3 and 12S4, with a high level observed in both at week 9. However, the expression level of SpCOL14 between the 12S3 and 12S4 plants was associated with differences in the phenotype; in 12S4 (Figure 7A), the expression of SpCOL14 showed a high level at week 9, while 12S3 showed high expression at week 12 (Figure 7C). These results suggested that SpCOL14 could potentially be the key candidate gene controlling bolting in spinach.
Discussion
As a green leafy vegetable, spinach can lose its flavor and thus commodity value in the reproductive stage (Abe et al., 2014). Bolting signifies the first transition between the vegetative and reproductive periods, thus rendering it a criterion of the reproductive stage. This study identified a novel QTL strongly associated with bolting time in BC1 and F2 populations by KASP and SLAF technology within 2 years.
In an earlier study, the QTLs for bolting in spinach were verified in many groups. Based on a SNP linkage genetic map, three QTLs associated with bolting and flowering (two at P01 and one at P02) were found in Chan-Navarrete et al. (2016), and three SNP markers (AYZV02001321_398, AYZV02041012_1060, and AYZV02118171_95) were screened by Chitwood et al. (2016). With the genome Sp75 sequences, QTLs for bolting were mapped to 44.7 to 50.5 Mb of chromosome 2 (Xu et al., 2017), and SNPs were also discovered on chromosome 2, chromosome 3, and chromosome 5 (Bhattarai et al., 2020). Recently, Hideki et al. (2021) identified three QTLs for bolting time (qBt2.1 on LG2; qBt3.1, and qBt3.2 on LG3) based on the new spinach genome SOL_r1.1. In this study, we fine-mapped a novel QTL qBT1.1 for spinach bolting located at 47.56 – 48.12 Mb on chromosome 1, which revealed 45.5% PVE in the two-year average results. The physical location of qBT1.1 was close to KMBL29, which had the highest LOD score. The new stable QTL facilitated the confirmation of the major genes responsible for bolting time and allowed for reliable molecular markers for the breeding of bolting resistance in spinach to be explored.
Genes that affect bolting and flowering time have been identified by flower regulatory pathways in Arabidopsis (Fornara et al., 2010), which provides a reference for detecting the bolting gene in spinach. Bolting and flowering in spinach are mainly related to the photoperiod pathway. Two FT and one COL homolog have been isolated in spinach (Abe et al., 2014). Xu et al. (2017) discovered one gene (Spo00403) showing high homology to the bolting and flowering gene of Arabidopsis AGAMOUS-LIKE 20, and three QTLs (qBt2.1, qBt3.1, and qBt3.2) reported by Hideki et al. (2021) contained FT, FLC, AGAMOUS-LIKE 24 homologs, and AGAMOUS-LIKE 22/SVP genes. In the present study, 68 genes were in the major QTL area qBT1.1. We detected the target genes using the Flowering Interactive Database9 and only found one gene similar to the Arabidopsis gene FLC, namely the sugar beet FLC-LIKE 1 (BvFL1). FLC is a MADS-box transcription factor that acts as a repressor of floral transition in both the autonomous and vernalization pathways (Sheldon et al., 1999). In sugar beet, which is in the same family as spinach (Amaranthaceae), a notable gene BvFL1, which is responsible for bolting in many studies, was shown to act as a repressor of flowering when transformed into an Arabidopsis FLC null mutant (Reeves et al., 2007; Mutasa-Göttgens et al., 2010). FLC is the key gene in the vernalization requirement as a flowering repressor (Sheldon et al., 2000), and the bolting and flowering of spinach mainly depend on the photoperiod-dependent flowering pathway (Chun et al., 2000a). In our study, the re-sequencing results suggested one SNP variation on SpFLC, while the expression of SpFLC did not show the same expression levels between 12S3 and 12S4. Given this, SpFLC may not be the candidate gene controlling the bolting trait in this study.
Bolting and flowering in spinach are closely related to the photoperiod pathway, The photoperiod pathway gene identified in Arabidopsis were not detected in these 68 genes. SpCOL14 (Spo04967, CONSTANS LIKE 14), belongs to the COL family and has similar functional domains to COL14. The CO transcription factor is critical in the photoperiod response and shows characteristic patterns of transcription required for day-length sensing. There are 17 COL gene members in Arabidopsis, which can be divided into four groups (Group I to Group IV) that have a CO, COL, and TOC1 (CCT) domain respectively, mediating the interactions with DNA (Robson et al., 2001). The CO family genes have different functions; for example, the expression of COL1 and COL2 in Arabidopsis has no role in bolting and flowering, but delays bolting and flowering in sugar beet (Chia et al., 2008; Ledger et al., 2010), and in soybean, COL2 has no significant effect on flowering rhythm, while COL5 can promote flowering. However, the functions of individual COL genes in Arabidopsis have not been fully determined. COL14 belongs to Group III of the COL family, and comprise one B-box and one CCT domain (Griffiths et al., 2003). With Sanger sequencing, no variations were detected in the coding region of SpCOL14 between the early and late flowering parents, while about 900-bp variations were found in the promoters (–730 bp to –1653 bp). According to our qRT-PCR results, the expression of SpCOL14 exhibited significant differences between the two parents in different phases, and the expression peak of this gene in the early bolting line appeared several weeks before that of the slow bolting line. In conclusion, SpCOL14 is very likely the candidate gene controlling bolting trait in spinach. Further functional analysis of these candidate genes will help elucidate the regulatory mechanism of bolting in spinach. In the further study, we can focus on the different varieties of spinach to take full advantage of the bolting genetic information for breeding.
Conclusion
In the present study, a major QTL, qBT1.1, controlling the bolting trait in spinach, was detected in the BC1 and F2 populations in two years using KASP and SLAF-seq methods. This QTL was mapped to the same region between 47.56 Mb and 48.12 Mb on spinach chromosome 1 in different segregation populations. This qBT1.1 is a novel QTL. In this interval, one gene Spo04967 (renamed SpCOL14) is very likely the candidate gene controlling bolting in spinach. These findings lay a foundation for analysis of the genetic mechanisms underlying spinach bolting and flowering time and can be applied for MAS in spinach breeding.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
WQ designed the study. QM and ZL conducted the experiments and analyzed the data. QM wrote the manuscript. WQ, CF, XW, and JW made the revision of the manuscript. ZL, HZ, HS, and ZX prepared and collected the samples. All authors contributed to the article and approved the submitted version.
Funding
This work was performed at the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, Beijing, China, and was supported by Central Public-interest Scientific Institution. Basal Research Fund (IVF-BRF2021004), the Chinese Academy of Agricultural Sciences Innovation Project (CAAS-ASTIP-IVFCAAS), and China Agricultural Research System (CARS-23-A-17).
Conflict of Interest
CF is employed by Ilera Healthcare LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.850810/full#supplementary-material
Footnotes
- ^ http://www.spinachbase.org/
- ^ http://www.primer3plus.com
- ^ https://www.ebi.ac.uk/Tools/msa/muscle
- ^ https://www.ncbi.nlm.nih.gov/Structure.cgi
- ^ http://www.gene-better.cn
- ^ https://www.transgen.com.cn
- ^ http://www.takara-bio.com
- ^ http://spinachbase.org/
- ^ http://www.phytosystems.ulg.ac.be/florid/
References
Abe, E., Fujino, K., Masuda, K., and Yamaguchi, Y. (2014). Isolation and expression profiling of a Constans-Like gene and two Flowering Locus T-Like genes from Spinacia oleracea L. Am. J. Plant Sci. 5, 4018–4028. doi: 10.4236/ajps.2014.526420
Abolghasemi, R., Haghighi, M., Etemadi, N., Wang, S., and Soorni, A. (2021). Transcriptome architecture reveals genetic networks of bolting regulation in spinach. BMC Plant Biol. 21:179. doi: 10.1186/s12870-021-02956-0
Bhattarai, G., and Shi, A. (2021). Research advances and prospects of spinach breeding, genetics, and genomics. Veg. Res. 1:9. doi: 10.48130/VR-2021-0009
Bhattarai, G., Shi, A., Correll, J. C., and Poude, B. (2020). “Identification of genomic regions associated with bolting and flowering time in spinach,” in Proceedings of the 2020 ASHS Annual Conference (Alexandria, VA: ASHS).
Buerstmayr, H., Ban, T., and Anderson, J. A. (2009). QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed. 128, 1–26. doi: 10.1111/j.1439-0523.2008.01550.x
Cai, X., Sun, X., Xu, C., Sun, H., Wang, X., Ge, C., et al. (2021). Genomic analyses provide insights into spinach domestication and the genetic basis of agronomic traits. Nat. Commun. 12:7246. doi: 10.1038/s41467-021-27432
Cai, X., Xu, C., Wang, X., Wang, S., Zhang, Z., Fei, Z., et al. (2018). Construction of genetic linkage map using genotyping-by-sequencing and identification of QTLs associated with leaf color in spinach. Euphytica 214:229. doi: 10.1007/s10681-018-2312-2
Chan-Navarrete, R., Dolstra, O., van Kaauwen, M., van Bueren, E. T. L., and van der Linden, C. G. (2016). Genetic map construction and QTL analysis of nitrogen use efficiency in spinach (Spinacia oleracea L.). Euphytica 208, 621–636. doi: 10.1007/s10681-015-1618-6
Chen, C., Huang, W., Hou, K., and Wu, W. (2019). Bolting, an important process in plant development, two types in plants. J. Plant Biol. 62, 161–169. doi: 10.1007/s12374-018-0408-9
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chia, T. Y. P., Müller, A., Jung, C., and Mutasa-Göttgens, E. S. (2008). Sugar beet contains a large CONSTANS-LIKE gene family including a CO homologue that is independent of the early-bolting (B) gene locus. J. Exp. Bot. 59, 2735–2748. doi: 10.1093/jxb/ern129
Chitwood, J., Shi, A., Mou, B., Evans, M., Clark, J., Motes, D., et al. (2016). Population structure and association analysis of bolting, plant height, and leaf erectness in spinach. HortScience 51, 481–486. doi: 10.21273/HORTSCI.51.5.481
Cho, L. H., Yoon, J., and An, G. (2017). The control of flowering time by environmental factors. Plant J. 90, 708–719. doi: 10.1111/tpj.13461
Chun, C., Watanabe, A., Koza, T., Kim, H. H., and Fuse, J. (2000b). Bolting and growth of Spinacia oleracea L. can be altered by modifying the photoperiod during transplant production. HortScience 35, 624–626. doi: 10.21273/HORTSCI.35.4.624
Chun, C., Kozai, T., Kubota, C., and Okabe, K. (2000a). Manipulation of bolting and flowering in a spinach (Spinacia oleracea L.) transplant production system using artificial light. Acta Hortic. 515, 201–206. doi: 10.17660/ActaHortic.2000.515.25
Dally, N., Xiao, K., Holtgräwe, D., and Jung, C. (2014). The B2 flowering time locus of beet encodes a zinc finger transcription factor. Proc. Natl. Acad. Sci. U.S.A. 111, 10365–10370. doi: 10.1073/pnas.1404829111
Fornara, F., Montaigu, A. D., and Coupland, G. (2010). Snapshot: control of flowering in Arabidopsis. Cell 141, 550–550. doi: 10.1016/j.cell.2010.04.024
Fu, W., Huang, S., Gao, Y., Zhang, M., Qu, G., Wang, N., et al. (2020). Role of BrSDG8 on bolting in Chinese cabbage (Brassica rapa). Theor. Appl. Genet. 133, 2937–2948. doi: 10.1007/s00122-020-03647-4
Goreta, S., and Leskovar, D. I. (2006). Screening spinach cultivars for white rust resistance and bolting. HortTechnology 16, 162–166. doi: 10.21273/HORTTECH.16.1.0162
Griffiths, S., Dunford, R. P., and Laurie, C. D. A. (2003). The evolution of Constans-Like gene families in barley, rice, and Arabidopsis. Plant Physiol. 131, 1855–1867. doi: 10.1104/pp.102.016188
Hideki, H., Atsushi, T., Takehiko, I., Yutaka, S., Nagano, A. J., Suguru, S., et al. (2021). A spinach genome assembly with remarkable completeness, and its use for rapid identification of candidate genes for agronomic traits. DNA Res. 28:dsab004. doi: 10.1093/dnares/dsab004
Khattak, J. Z., Torp, A. M., and Andersen, S. B. (2006). A genetic linkage map of Spinacia oleracea and localization of a sex determination locus. Euphytica 148, 311–318. doi: 10.1007/s10681-005-9031-1
Kim, H. H., Chun, C., Kozai, T., and Fuse, J. (2000). The potential use of photoperiod during transplant production under artificial lighting conditions on floral development and bolting, using spinach as a model. HortScience 35, 43–45. doi: 10.1016/j.cplett.2010.04.027
Koh, E., Charoenprasert, S., and Mitchell, A. E. (2012). Effect of organic and conventional cropping systems on ascorbic acid, vitamin C, flavonoids, nitrate, and oxalate in 27 varieties of spinach (Spinacia oleracea L.). J. Agric. Food Chem. 60, 3144–3150. doi: 10.1021/jf300051f
Kuwahara, K., Suzuki, R., Ito, Y., Mikami, T., and Onodera, Y. (2014). An analysis of genetic differentiation and geographical variation of spinach germplasm using SSR markers. Plant Genet. Resour. 12, 185–190. doi: 10.1017/S1479262113000464
Ledger, S., Strayer, C., Ashton, F., Kay, S. A., and Putterill, J. (2010). Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 26, 15–22. doi: 10.1046/j.1365-313x.2001.01003.x
Lee, D. J., and Zeevaart, J. A. D. (2002). Differential regulation of RNA levels of gibberellin dioxygenases by photoperiod in spinach. Plant Physiol. 130, 2085–2094. doi: 10.1104/pp.008581
Lester, G. E., Makus, D. J., Hodges, D. M., and Jifon, J. L. (2013). Summer (subarctic) versus winter (subtropic) production affects spinach (Spinacia oleracea L.) leaf bionutrients: vitamins (C, E, Folate, K1, provitamin A), lutein, phenolics, and antioxidants. J. Agric. Food Chem. 61, 7019–7027. doi: 10.1021/jf401461z
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu, Z., Lu, T., Feng, C., Zhang, H., Xu, Z., Correll, J. C., et al. (2021a). Fine mapping and molecular marker development of the Fs gene controlling fruit spines in spinach (Spinacia oleracea L.). Theor. Appl. Genet. 134, 1319–1328. doi: 10.1007/s00122-021-03772-8
Liu, Z., She, H., Xu, Z., Zhang, H., Li, G., Zhang, S., et al. (2021b). Quantitative trait loci (QTL) analysis of leaf related traits in spinach (Spinacia oleracea L.). BMC Plant Biol. 21:290. doi: 10.1186/s12870-021-03092-5
Ma, J., Shi, A., Mou, B., Evans, M., Clark, J. R., Motes, D., et al. (2016). Association mapping of leaf traits in spinach (Spinacia oleracea L.). Plant Breed. 135, 399–404. doi: 10.1111/pbr.12369
Mauricio, R. (2001). Mapping quantitative trait loci in plants: uses and caveats for evolutionary biology. Nat. Rev. Genet. 2, 370–381. doi: 10.1038/35072085
McCouch, S., Cho, Y., Yano, M., Paul, E., Blinstrub, M., Morishima, H., et al. (1997). Report on QTL nomenclature. Rice Genet. Newsl. 14, 11–13.
Melchinger, A. E. (1998). “Advances in the analysis of data on quantitative trait loci,” in Proceedings of 2nd International Crop Science Congress, New Delhi.
Meng, L., Li, H., Zhang, L., and Wang, J. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Morelock, T., and Correll, J. (2008). “Spinach,” in Vegetables I: Asteraceae, Brassicaceae, Chenopodiaceae, and Cucurbitaceae, eds J. Prohens and F. Nuez (New York, NY: Springer), 189–218.
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Mutasa-Göttgens, E. S., Qi, A., Zhang, W., Schulze-Buxloh, G., Jennings, A., Hohmann, U., et al. (2010). Bolting and flowering control in sugar beet: relationships and effects of gibberellin, the bolting gene b and vernalization. Aob Plants 2010:plq012. doi: 10.1093/aobpla/plq012
Qian, W., Fan, G., Liu, D., Zhang, H., Wang, X., Wu, J., et al. (2017). Construction of a high-density genetic map and the X/Y sex-determining gene mapping in spinach based on large-scale markers developed by specific-locus amplified fragment sequencing (SLAF-seq). BMC Genomics 18:276. doi: 10.1186/s12864-017-3659-9
Reeves, P. A., He, Y., Schmitz, R. J., Amasino, R. M., Panella, L. W., and Richards, C. M. (2007). Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: evidence from the sugar beet (Beta vulgaris). Genetics 176, 295–307. doi: 10.1534/genetics.106.069336
Robson, F., Costa, M. M. R., Hepworth, S. R., Vizir, I., Pineiro, M., Reeves, P. H., et al. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28, 619–631. doi: 10.1046/j.1365-313x.2001.01163.x
Rubatzky, V. E., and Yamaguchi, M. (1997). “Spinach, table beets, and other vegetable chenopods,” in World Vegetables, eds V. E. Rubatzky and M. Yamaguchi (Boston, MA: Springer US), 457–473. doi: 10.1007/978-1-4615-6015-9.21
Salvi, S., and Tuberosa, R. (2005). To clone or not to clone plant QTLs: present and future challenges. Trends Plant Sci. 10, 297–304. doi: 10.1016/j.tplants.2005.04.008
Semagn, K., Babu, R., Hearne, S., and Olsen, M. (2014). Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol. Breed. 33, 1–14. doi: 10.1007/s11032-013-9917-x
Sheldon, C. C., Burn, J. E., Perez, P. P., Metzger, J., Edwards, J. A., Peacock, W. J., et al. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. doi: 10.2307/3870872
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., and Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U.S.A. 97, 3753–3758. doi: 10.1073/pnas.060023597
Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. doi: 10.1038/35074138
Sun, X., Liu, D., Zhang, X., Li, W., Liu, H., Hong, W., et al. (2013). SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS One 8:e58700. doi: 10.1371/journal.pone.0058700
Tang, X., Gong, R., Sun, W., Zhang, C., and Yu, S. (2018). Genetic dissection and validation of candidate genes for flag leaf size in rice (Oryza sativa L.). Theor. Appl. Genet. 131, 801–815. doi: 10.1007/s00122-017-3036-8
Tränkner, C., Pfeiffer, N., Kirchhoff, M., Kopisch-Obuch, F. J., van Dijk, H., Schilhabel, M., et al. (2017). Deciphering the complex nature of bolting time regulation in Beta vulgaris. Theor. Appl. Genet. 130, 1649–1667. doi: 10.1007/s00122-017-2916-2
Van Ooijen, J. W. (2006). JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wagening, The Netherlands: Kyazma BV. 59.
Wu, K., Li, L., Gage, D. A., and Zeevaart, J. A. (1996). Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from the long-day plant spinach. Plant Physiol. 110, 547–554. doi: 10.1104/pp.110.2.547
Xu, C., Jiao, C., Sun, H., Cai, X., Wang, X., and Ge, C. (2017). Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat. Commun. 8:1527. doi: 10.1038/ncomms15275
Xu, Y., Zhang, B., Ma, N., Liu, X., Qin, M., Zhang, Y., et al. (2021). Quantitative trait locus mapping and identification of candidate genes controlling flowering time in Brassica napus L. Front. Plant Sci. 11:626205. doi: 10.3389/fpls.2020.626205
Keywords: spinach, bolting time, quantitative trait locus (QTL), QTL mapping, candidate gene
Citation: Meng Q, Liu Z, Feng C, Zhang H, Xu Z, Wang X, Wu J, She H and Qian W (2022) Quantitative Trait Locus Mapping and Identification of Candidate Genes Controlling Bolting in Spinach (Spinacia oleracea L.). Front. Plant Sci. 13:850810. doi: 10.3389/fpls.2022.850810
Received: 10 January 2022; Accepted: 28 February 2022;
Published: 30 March 2022.
Edited by:
Ainong Shi, University of Arkansas, United StatesReviewed by:
Gehendra Bhattarai, University of Arkansas, United StatesQuanhua Wang, Shanghai Normal University, China
Copyright © 2022 Meng, Liu, Feng, Zhang, Xu, Wang, Wu, She and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Qian, cWlhbndlaUBjYWFzLmNu
†These authors have contributed equally to this work and share first authorship
 Qing Meng1†
Qing Meng1† Zhiyuan Liu
Zhiyuan Liu Xiaowu Wang
Xiaowu Wang Jian Wu
Jian Wu Wei Qian
Wei Qian