- 1Department of Agronomy, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University, Nakhon Pathom, Thailand
- 2Center for Advanced Studies for Agriculture and Food, Kasetsart University Institute for Advanced Studies, Kasetsart University, Bangkok, Thailand
- 3Institute of Industrial Crops, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 4Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China
- 5Center for Agricultural Biotechnology, Kasetsart University, Nakhon Pathom, Thailand
- 6Center of Excellence on Agricultural Biotechnology: (AG-BIO/MHESI), Bangkok, Thailand
Seed dormancy in wild mungbean (Vigna radiata var. sublobata) may be useful for the breeding of cultivated mungbean (var. radiata) with pre-harvest sprouting resistance. Previous studies have identified two major quantitative trait loci (QTLs) for seed dormancy, HsA and Sdwa5.1.1+, in wild mungbean that are possibly having the same locus or linked. However, these QTLs have not been confirmed/verified and a molecular basis of seed dormancy in mungbean is not yet known. In this study, we aimed to finely map the Sdwa5.1.1+ and identify candidate gene(s) for this locus. Microscopic observations revealed that wild mungbean “ACC41” seeds had a palisade cuticle layer, while cultivated mungbean “Kamphaeng Saen 2” (KPS2) seeds lacked this layer. Fine mapping using an F2 population developed from a cross between ACC41 and KPS2 revealed two linked QTLs, Sdwa5.1.1+ and Sdwa5.1.2+, controlling seed dormancy. The Sdwa5.1.1+ was confirmed in an F2:3 population derived from the same cross and mapped to a 3.298-Kb region containing only one gene LOC106767068, designated as VrKNAT7-1, which encodes the transcription factor KNOTTED ARABIDOPSIS THALIANA7 (KNAT7), a class II KNOTTED1-LIKE HOMEOBOX (KNOX II) protein. VrKNAX7 sequence alignment between ACC41 and KPS2 revealed several polymorphisms in the coding, untranslated, and promoter regions. Quantitative real-time PCR (qRT-PCR) analysis revealed that the expression of VrKNAT7-1 and VrCYP86A, a putative downstream regulation of VrKNAT7-1, in the seed coat of ACC41 is statistically much higher than that of KPS2. Altogether, these results indicate that VrKNAT7-1 controls physical seed dormancy in the wild mungbean ACC41.
Introduction
Plant domestication, the earliest form of plant breeding, is an evolutionary change of wild plants to domesticated plants that serve human needs, including foods, fibers, medicines, feeds, energies, cosmetics, and ornamentals. Plant domestication is one of the most important events in human history as it transformed human ways of life from the hunting-gathering society to agricultural society approximately 12,000 years ago, which eventually led to present modern and civilized society (Diamond, 1999, 2002; Larson et al., 2014). Processes of plant domestication are associated with forces stemmed from human-mediated selection, both conscious and unconscious selections, under human-manipulated environments. The changes of wild plants include morphological, developmental, and physiological traits. For example, in general, compared with wild plants, domesticated plants possess non-dormant seeds, indehiscent seeds or fruits, larger organs (fruit, seed, leaf, and stem), reduced or no branching, earlier flowering and maturity, tastier, and lower or no toxic and more colorful edible part(s). Those domestication-related traits are called “domestication syndrome” (Hammer, 1984). In the plant domestication, seed dormancy and seed/fruit indehiscence are believed to be the first trait selection as the traits are for advantageous cultivation.
Mungbean [Vigna radiata (L.) Wilczek], one of several legume crops of the genus Vigna, is an important crop of Asia. The crop is grown into various cropping systems due to its fast growth, early maturity (60–75 days), relatively tolerance to drought, ability to improve soil fertility through atmospheric nitrogen (N2) fixation in symbiosis with Rhizobium species in the soil (Somta and Srinives, 2007). Mungbean seeds are the sources of protein, amino acids, carbohydrates, vitamins, and minerals. Dry seeds of mungbean contain about 20–25% proteins and 60–70% carbohydrates. Whole and split seeds of mungbean are cooked and consumed in a variety of ways. The seeds are an important protein source for people in the cereal-based society, especially in South Asia. The seeds are also used to produce bean sprouts, paste, starches, noodles, protein isolates, and protein concentrates (Nair and Schreinemachers, 2020). Due to its high protein content, mungbean has become an important and a popular source of plant-based proteins. The world production area of mungbean is about 7.2 million ha, of which about 90% is in Asia (Nair and Schreinemachers, 2020). The crop is now gaining increasing popularity in Australia, America, and Africa.
Seed dormancy is a crucial trait for the domestication of cereal and legume crops. Non-dormant seeds are advantageous for uniform and timely cultivation. An understanding of the genetic basis underlying seed dormancy may be useful for exploiting wild genetic resources for the improvement of crops (Imrie et al., 1988; Lawn et al., 1988), especially in the face of climate change. For example, the dormancy may provide the protection of pre-harvest sprouting. In the genus, Vigna that comprises 10 domesticated legume crops, there are not many studies reported on quantitative trait locus (QTL) mapping of domestication syndrome, including adzuki bean (Isemura et al., 2007; Kaga et al., 2008), black gram (Somta et al., 2020), cowpea (Kongjaimun et al., 2012; Lo et al., 2018), mungbean (Isemura et al., 2012), moth bean (Yundaeng et al., 2019), and zombi pea (Dachapak et al., 2018; Amkul et al., 2020). One to six QTLs controlled seed dormancy in these legume species. Candidate genes for seed dormancy have only been reported for zombi pea (Amkul et al., 2020). In mungbean, Mendelian genetic analysis showed that seed dormancy in wild mungbean (V. radiata var. sublobata) accession “Pantnagar” from India, having about 95% dormant seeds is controlled by a single dominant gene, Hd1 (Singh et al., 1983). Lawn et al. (1988) demonstrated that wild mungbean accession “ACC41” from Australia, with > 95% dormant seeds is likely to be controlled by a single major gene and some modifying factors. QTL mapping in a recombinant inbred line population derived from a cross between commercial mungbean cultivar and ACC41 that was grown under field and glasshouse conditions using restriction fragment length polymorphism (RFLP) markers revealed that the dormancy in ACC41 is controlled by one major and three minor QTLs located on different linkage groups (Humphry et al., 2005). However, only the major QTL, HsA, was found in both environments. The HsA was located between RFLP markers cgO103 and VrCS364 and explained 23.2% of the dormancy variation. Broad-sense heritability (H2) was estimated for the dormancy in this population is 90% for both field and glasshouse conditions (Humphry et al., 2005). Similarly, a study using an F2 population of a cross between landrace mungbean and wild mungbean accession “JP211874” (99% dormant seeds and from Myanmar) grown in a single environment demonstrated that H2 for seed dormancy is 99% and the trait is conditioned by two major and two minor QTLs located on different linkage groups (Isemura et al., 2012). Among those QTLs, Sdwa5.1.1+ showed the largest effect, explaining 33.7% of the seed dormancy variation. The Sdwa5.1.1+ was located on linkage group 1 between simple sequence repeat (SSR) markers cp05137 and CEDG074b. The distance between these markers was large, being 14.2 cM. However, the Sdwa5.1.1+ has not been confirmed/validated and a genetic basis of this locus is not yet known. Nonetheless, based on the mungbean linkage map reported by Wang et al. (2016) and BLASTN search revealed that cgO103, VrCS364, cp05137, and CEDG074b are linked (P. Somta, unpublished data), suggesting that HsA and Sdwa5.1.1+ may be the same locus or different locus but linked. In addition, markers associating with Sdwa5.1.1+ also showed a linkage with Sdg3.1.2, which is a major QTL conferring seed dormancy in wild adzuki bean (Vigna angularis var. nipponensis) (Kaga et al., 2008).
In this paper, we report fine mapping of the Sdwa5.1.1+ and the identification of candidate genes(s) at this QTL and a novel QTL linked with Sdwa5.1.1+. The objectives of this study were to (i) finely map the Sdwa5.1.1+ in wild mungbean ACC41 and (ii) identify candidate(s) gene for the Sdwa5.1.1+. We demonstrated that a class II KNOTTED1-LIKE HOMEOBOX (KNOX II) gene, KNAX7-1, is associated with physical dormancy in ACC41.
Materials and Methods
Plant Materials and Population Development
Two mungbean populations, F2 and F2:3 generations, were used in this study. The F2 population comprised 575 individuals developed from the hybridization between ACC41 (female parent) and Kamphaeng Saen 2 (male parent; hereafter called KPS2). ACC41 is a wild mungbean (var. sublobata) from Australia and possesses dormant seeds, while KPS2 is a cultivated mungbean (var. radiata) from Thailand and possesses non-dormant seeds. This F2 population has been previously used for gene mapping of bruchid resistance (Kaewwongwal et al., 2020). The F2 plants and parents (10 plants each) were grown under field conditions at Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom, Thailand from February to May 2018. Mature pods were harvested from each plant and the dry seeds were used for dormancy evaluation. Details of the F2:3 generation is described in the section “Confirmation of Sdwa5.1.1+ for seed dormancy.”
Characterization of Seed Dormancy and Morphology
In total, 50 intact seeds of ACC41 and KPS1, and scarified seeds of ACC41 were used for the observation of imbibition and germination test. The seeds were soaked in deionized water, and then imbibition and germination of the seeds were observed and photographed at 0, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, and 24 h.
Seed coat structure of ACC41 and KPS1 was investigated using microscope following the procedures described by Chai et al. (2016). In brief, mature seeds of both accessions were soaked in sterile water and then fixed with 2.5% glutaraldehyde and 4% paraformaldehyde in a PBS buffer. Then, the seeds were washed with PBS and postfixed in 1% osmium tetroxide, dehydrated in a series of ethanol dilutions, embedded in LR White resin, and polymerized. Cross-sections were cut in the middle of the seed using Leica EM UC7 Ultramicrotome (Leica Microsystems, Germany). Subsequently, semithin sections were stained with 1% toluidine blue O and observed under Nikon Microphot-2 (Nikon Corporation, Japan).
Evaluation of Seed Dormancy in the F2 Population
In total, 50 intact seeds of each F2 plant that had been stored at room temperature for 60–80 days after harvest were placed into a hole of a germination tray. Each tray contained 35–40 holes. Deionized water was added into each hole until the seeds were submerged. The tray was then incubated at 25°C with 12-h light and 12-h darkness for 7 days. Deionized water was added to the holes daily, if necessary, to maintain the level of water submergence of the seeds. The number of seeds that not imbibed water was recorded. Percentage of seed dormancy (PSD) of each plant was calculated.
Development of New DNA Markers for Fine Mapping the Sdwa5.1.1+ and Identification of New Quantitative Trait Loci Conferring Seed Dormancy
Sdwa5.1.1+ conferring seed dormancy in wild mungbean was previously detected between SSR markers cp05137 and CEDG074b (Isemura et al., 2012). To finely map the Sdwa5.1.1+ locus, we determined physical locations of these markers by performing BLASTN search of the primer sequences of cp05137 and CEDG074b against mungbean reference genome (Kang et al., 2014). After the physical genome locations of these two markers were identified, DNA sequence between and around the two locations was searched for SSRs using the software SSRIT (Temnykh et al., 2001). Primers for the SSRs were designed using the software Primer3 (Untergasser et al., 2012).
It has been reported that Sdg3.1.2 is a major QTL conferring seed dormancy in wild adzuki bean (Vigna angularis var. nipponensis) (Kaga et al., 2008). SSR markers CEDG214 and CEDG256 associating with the Sdg3.1.2 (Kaga et al., 2008) have been shown to be linked with the marker cp05137 in the mungbean (Isemura et al., 2012). Existence of QTL homologous to the Sdg3.1.2 in the wild mungbean ACC41 was investigated. To do so, locations of the CEDG214 and CEDG256 on the mungbean reference genome (Kang et al., 2014) were determined by BLASTN (Altschul et al., 1990). Subsequently, SSRs lying between and around these markers were searched and used to develop SSR markers using the same software described above.
In total, 405 SSR markers were screened for polymorphism between ACC41 and KPS2 (Supplementary Table 1). A marker analysis was carried out as per Yundaeng et al. (2021). In brief, PCR was carried out in a total volume of 10 μl containing 5 ng of DNA template, 1 × Taq buffer, 2 mM MgCl2, 0.2 mM dNTPs, 1 U Taq DNA polymerase, and 2.5 μM each of forward and reverse primers. Amplification was performed at 94°C for 3 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 30 s, and 72°C for 10 min. PCR products were electrophoresed on 5% polyacrylamide gel electrophoresis and visualized by silver staining. In total, 24 markers showing polymorphic and unambiguous DNA bands were used to genotype the F2 population.
Linkage Map Construction and Quantitative Trait Loci Analysis in the F2 Population
A genetic linkage map was constructed for the F2 population using the software QTL IciMapping 4.2 (Meng et al., 2015). The markers were grouped with a logarithm of the odds (LODs) value of 5.0. Orders of the markers on the linkage group were determined by the REcombination Counting and ORDering (RECORD) algorithm (Van Os et al., 2005) and rippled by the Sum of Adjacent Recombination Frequencies (SARF) function (Falk, 1989). Genetic distance in centimorgan unit (cM) between the markers was calculated using the Kosambi mapping function. Location of the QTLs was determined by an inclusive composite interval mapping (ICIM) (Li et al., 2007) using the same software for a linkage analysis. ICIM was performed at every 0.1 cM. Significant LOD score threshold for the QTL was determined by running a 5,000-permutation test at p = 0.001.
Confirmation of Sdwa5.1.1+ for Seed Dormancy
Based on the results from the QTL analysis in the F2 population that two linked QTLs, Sdwa5.1.1+ and Sdwa5.1.2+, were identified for seed dormancy, we selected 15 F2 plants that were heterozygous at flanking markers of the QTLs Sdwa5.1.1+. In total, 10–15 F3 seeds from the selected F2 plants were grown under field conditions at Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom, Thailand from February to May 2020. Finally, 112 F3 plants were used to confirm the Sdwa5.1.1+. Five markers were selected and used for confirmation. DNA extraction was carried out as per Lodhi et al. (1994). A marker analysis, seed dormancy evaluation analysis, and QTL analysis were the same as described above.
Sequencing of Candidate Gene, VrKNAT7-1
Once the QTLs for seed dormancy were identified, mungbean reference genome (Kang et al., 2014) was inspected to identify candidate gene(s). Annotated genes locating between markers flanking each QTL were considered as candidate gene(s) for the dormancy. Only candidate gene, LOC106767068 (VrKNAT7-1), at the Sdwa5.1.1+ was sequenced because its location was confirmed. Coding sequence (CDS), 5′-untranslated region (5′UTR), 3′-untranslated region (3′UTR), and 884-bp upstream sequence of the gene were amplified from the genomic DNA of ACC41 and KPS2 using the primers listed in Supplementary Table 1. PCR was carried out in a total volume of 10 μl containing 5 ng of DNA template, 1 × Taq buffer, 2 mM MgCl2, 0.2 mM dNTPs, 1 U KOD-Plus-Neo DNA polymerase (TOYOBO, China), and 0.5 μM each of forward and reward primers. PCR was performed in SimpliAmp thermal cycler (Applied Biosystems, United States) programmed as follows: 94°C for 2 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, and 72°C for 10 min. PCR products were run on 1.5% agarose gel electrophoresis to confirm the single DNA fragment was amplified. The fragments were Sanger sequenced using ABI 3730xl DNA Analyzer (Applied Biosystems, United States) by Tsingke (Beijing, China). The sequences of KPS2, ACC41, and reference sequence (VC1973A; Kang et al., 2014) were aligned to identify polymorphism(s) using Clustal Omega (Sievers et al., 2011). The CDSs of ACC41 and KPS2 were translated into protein sequences and aligned to find amino acid polymorphism.
Expression Analysis of the Candidate Gene
ACC41 and KPS2 were grown in a crossing block. Total RNA was extracted from flowers, pods, the seed coat, and seeds (cotyledons and embryos) of both accessions following the protocol described by Laksana and Chanprame (2015). The RNA was converted into complementary DNA (cDNA) using the RevertAid H Minus First Strand cDNA Synthesis kit (Thermo Scientific, United States). cDNA concentration was quantified by ND-1000 Spectrophotometer (NanoDrop Technologies, Inc., United States). The cDNA was subjected to a gene expression analysis by a quantitative real-time PCR (qRT-PCR). Primers for qRT-PCR of the candidate gene VrKNAT7-1 (LOC106767068) and reference gene VrACTIN (LOC106770112) were designed using the Primer 3 (Supplementary Table 1). qRT-PCR was performed using ViiA 7 Real-Time PCR System (Applied Biosystems, United States). Three biological and technical replicates were conducted for ACC41 and KPS2. Reaction mixtures contained water, 1 × Master mix of Fast SYBR™ Green Master Mix (Thermo Fisher Scientific, United States), 5 μM of forward primer, 5 μM of reverse primer, and 50 ng cDNA. Thermocycle conditions included initial denaturation at 95°C for 20 s, followed by 40 cycles at 95°C for 3 s and 60°C for 30 s. After 40 cycles, a melting curve was generated by slowly increasing (0.5°C per 1 s) the temperature from 60 to 95°C, while the fluorescence was measured. Fluorescent data were acquired during each extension phase. Expression levels of the VrKNAT7-1 were calculated based on the ΔCT method by using ACTIN as the reference (Livak and Schmittgen, 2001). Statistical differences in the gene expression level between ACC41 and KPS2 were tested by a t-test at 1% probability using R program version 2.10.0.
Because VrKNAT7-1 is an ortholog of Medicago truncatula KNOX4 (MtKNOX4; Medtr5g011070) in which that latter is a transcription factor and has been reported to regulate the expression of CYP86A (MtCYP86A; Medtr8g030590) that is involved in cuticle biosynthesis (Chai et al., 2016), we determined whether the expression of VrCYP86A (LOC106774045) which is an ortholog of MtCYP86A in ACC41 and KPS2 is different. Expression analysis of VrCYP86A was the same as that of the VrKNAT7-1.
Phylogenetic Analysis of KNOXs
VrKNAT7-1 (XP_014507374.1) and VrKNAT7-2 (XP_014521195.1) together with KNOX protein sequences of Arabidopsis thaliana [AtKNAT1 (AT4G08150.1), AtKNAT2 (AT1G70510.1), AtKNAT3 (AT5G25220.1), AtKNAT4 (AT5G11060.1), AtKNAT5 (AT4G32040.1), AtKNAT6 (AT1G23380.1), AtKNAT7 (AT1G62990.1), and AtKNATM (AT1G14760.1)] and of M. truncatula (MtKNOX1 (Medtr2g024390.1), MtKNOX2 (Medtr1g017080.1), MtKNOX3 (Medtr1g012960.1), MtKNOX4 (Medtr5g011070.1), MtKNOX5 (Medtr3g106400.1), MtKNOX6 (Medtr5g085860.1), MtKNOX7 (Medtr5g033720.1), MtKNOX8 (Medtr1g084060.1), MtKNOX9 (Medtr4g116545.1), MtKNOX10 (Medtr2g461240.1), MtFCL1 (Medtr6g071190.1), and MtFCL2 (Medtr1g032750.1) were used for a phylogenetic analysis. The KNOX sequences of mungbean were from the GenBank database, whereas those of A. thaliana and M. truncatula were from TAIR1 and Phytozome132 databases, respectively. The phylogenetic analysis was carried out by the software using Phylogeny.fr (Dereeper et al., 2008) in which the sequences were aligned by MUSCLE 3.7 and constructed into a tree by maximum likelihood with 1,000 bootstraps.
Results
Seed Morphology and Dormancy of ACC41 and KPS2
The wild mungbean ACC41 and the cultivated mungbean KPS2 showed a clear difference in seed dormancy (Figure 1A). In the germination test (Figure 1B), nearly all the intact seeds of ACC41 were static throughout the course of the germination test, 7 days. However, when the seeds of ACC41 were scarified and subjected to germination test, all the seeds imbibed water rapidly and germinated at 12 h (Figure 1B). In contrast, all intact seeds of KPS2 started imbibing water at 4 h as shown by swelling of the seeds (Figure 1B). The seed coat of KPS2 became wrinkle at 8 h and cracked at 14 h. The seeds eventually germinated at 16 h as shown by a protrusion of the radicle. The percentage of dormant seeds (PDSs) in ACC41 was 96.0%, while that in KPS2 was 0%.
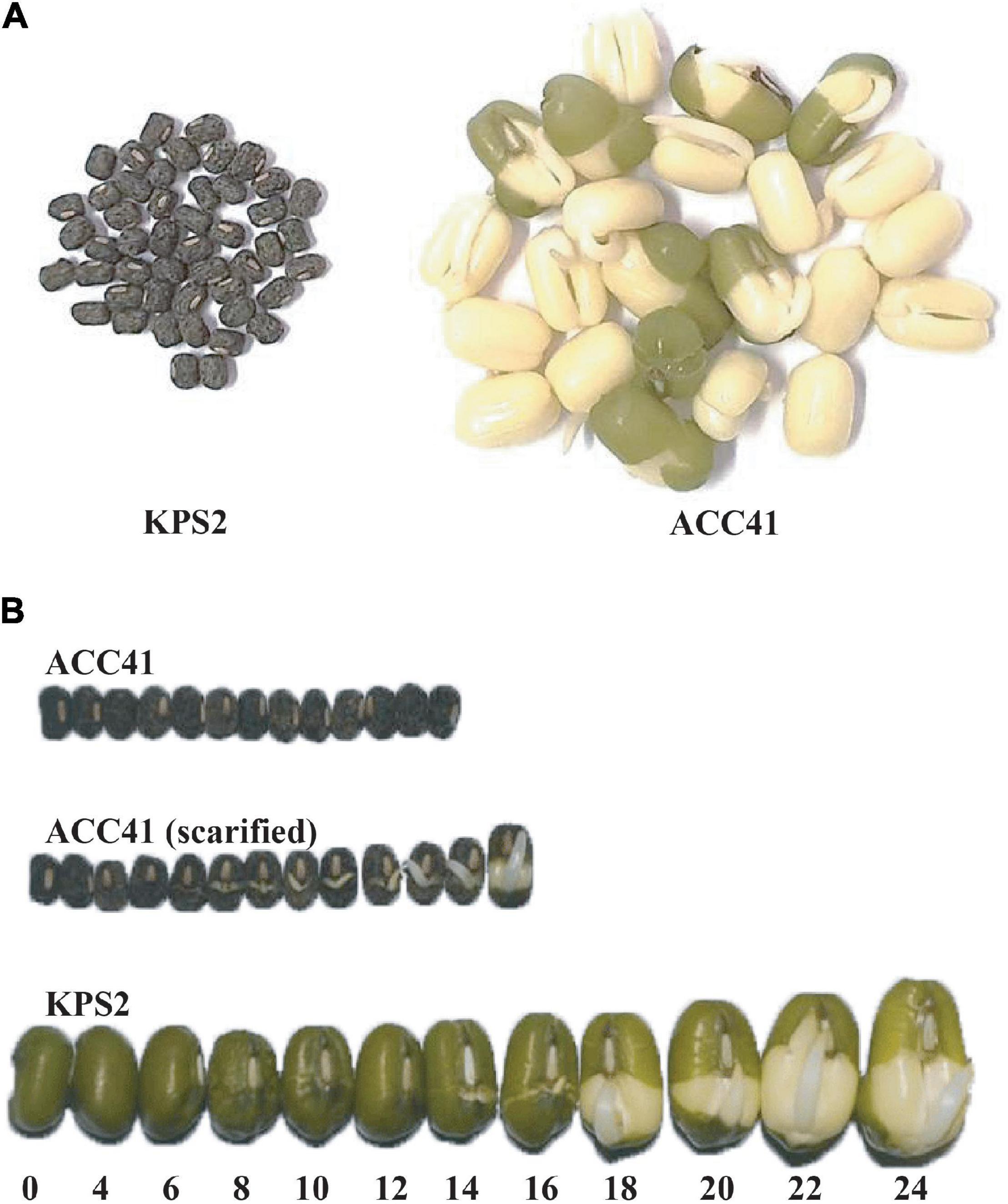
Figure 1. Seed germination test of the wild mungbean ACC41 and the cultivated mungbean Kamphaeng Saen 2 (KPS2). The germination of intact seeds of ACC41 and KPS2 at 7 days after the test (A). The germination of intact seeds of KPS2 and ACC41 and scarified seeds of ACC41 at different time series (0–24 h) (B).
Microscopic observations of cross-sections of the intact seeds of ACC41 and KPS2 revealed that the seed coat of the two accessions possessed similar parenchyma, hourglass, and palisade cells (Figure 2). Nonetheless, the seed coat of ACC41 had a palisade cuticle layer, whereas that of KPS2 lacked the palisade cuticle layer (Figure 2). In ACC41, the thickness of a cuticle layer was almost the same as that of a palisade cell.
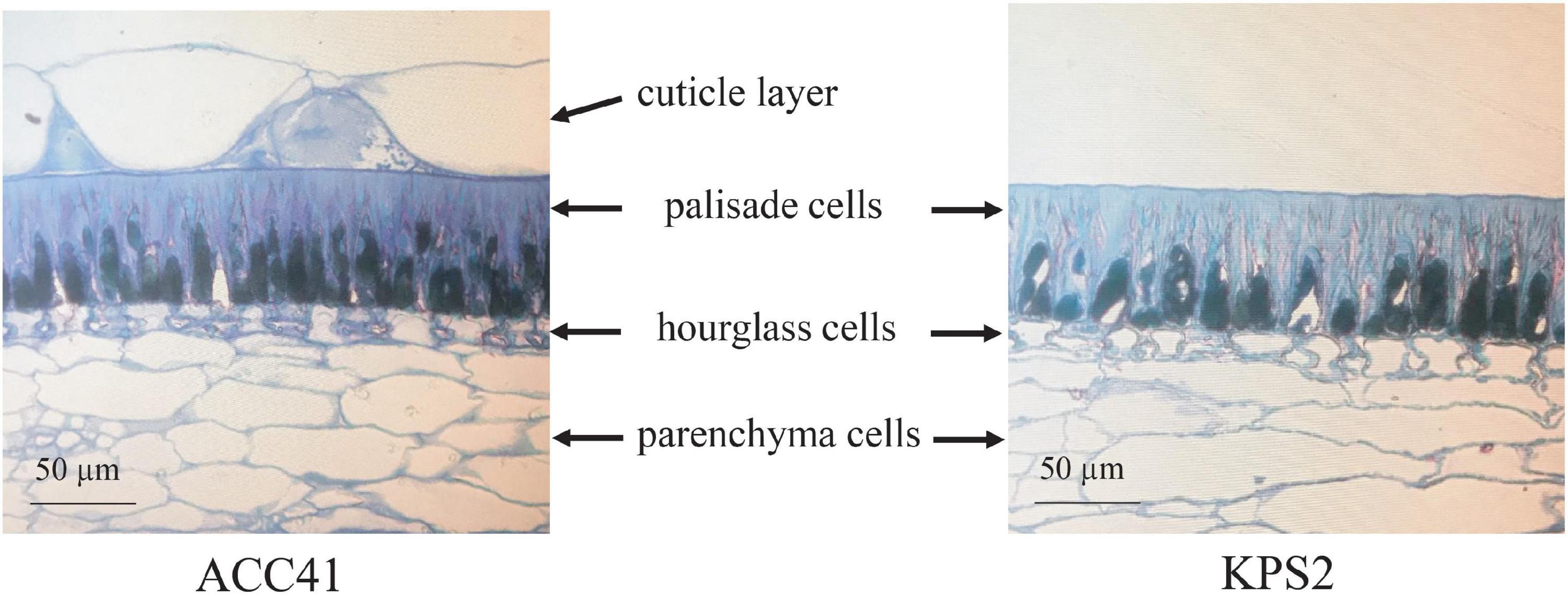
Figure 2. Transverse sections of the seed coat at the maturation stage of the cultivated mungbean of wild mungbean ACC41 and cultivated mungbean KPS2, stained with 0.05% toluidine blue O.
Fine Mapping for the Sdwa5.1.1+ and Identification of New QTLs for Seed Dormancy
In the F2 population of KPS2 × ACC41, PDSs varied between 0% and 100% with a mean of 62.68%. ACC41 had a PDS of 95.34%, while KPS2 had a PDS of 0%. The PDS of the F2 population showed continuous distribution, but skewed into the direction of ACC41 (Figure 3A). Broad-sense H2 estimated for this population was high, being 97.01%.
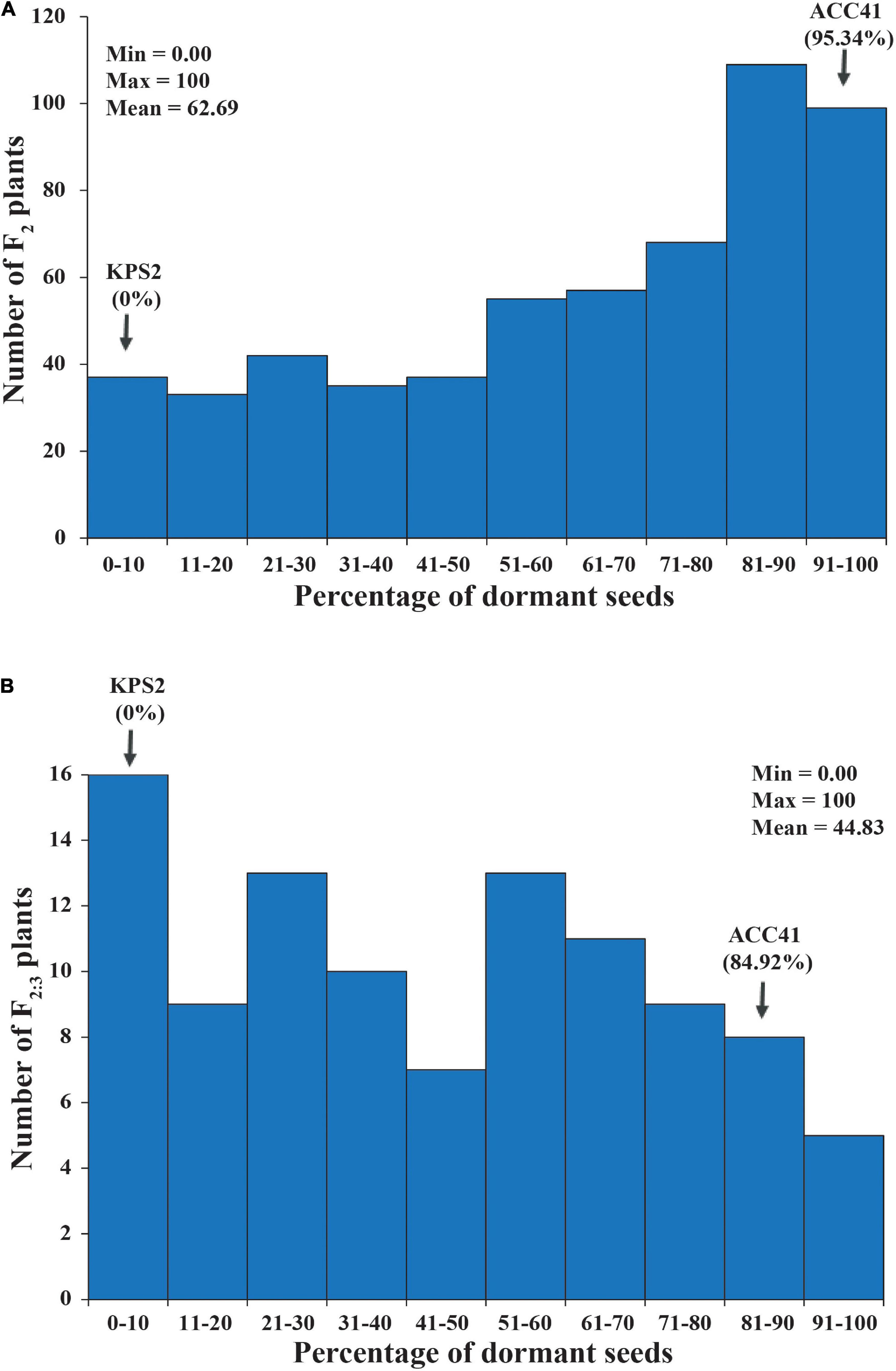
Figure 3. Frequency distribution of the percentage of dormant seeds (PDSs) in F2 population of 573 individuals (A) and F2:3 population of 100 individuals (B) derived from the cross between KPS2 and ACC41.
The BLASTN search revealed that physical locations of the markers cp05137 and CEDG074b that delimited the QTL Sdwa5.1.1+ in the study of Isemura et al. (2012) on the mungbean reference genome were at the positions 31,187,230 and 31,822,876 Mbp, respectively, on chromosome 7. Thus, the two markers were 635.65 Kb apart. In addition, to identify a new QTL for seed dormancy that may linked with the Sdwa5.1.1+, locations of the CEDG214 and CEDG256 associated with the Sdg3.1.2 controlling seed dormancy in adzuki bean on the mungbean reference genome were determined. The results showed that they were on the chromosome 7 at the positions 26,548,328 and 27,596,788 in that order. In total, 361 newly developed SSR and Indel markers together with 10 SSR markers reported for seed dormancy QTLs in mungbean and adzuki bean were screened for polymorphism between KPS2 and ACC41. In total, 23 markers showing unambiguous and polymorphic DNA bands were used to analyze the F2 population. A linkage group constructed from these markers spanned 21.69 cM in length.
An inclusive composite interval mapping analysis in the F2 population revealed two linked QTLs controlling seed dormancy, designated as Sdwa5.1.1+ and Sdwa5.1.2+ (Figure 4A and Table 1). Sdwa5.1.1+ was located between the markers VrSdp-SSR5 and VrKNAT7-SSR4, while Sdwa5.1.2+ was located between the markers VrSdp-SSR102 and VrSdp-SSR104. These QTLs were 21.1 cM apart. Sdwa5.1.1+ accounted for 17.53% of the dormancy variation in the population and possessed additive effect of −9.12 and dominant effect of 11.55. Sdwa5.1.2+ explained 12.27% of the dormancy variation in the population and had additive and dominant effects of −10.91 and 2.27, respectively. At both QTLs, allele(s) from the wild mungbean ACC41 increased the PDS.
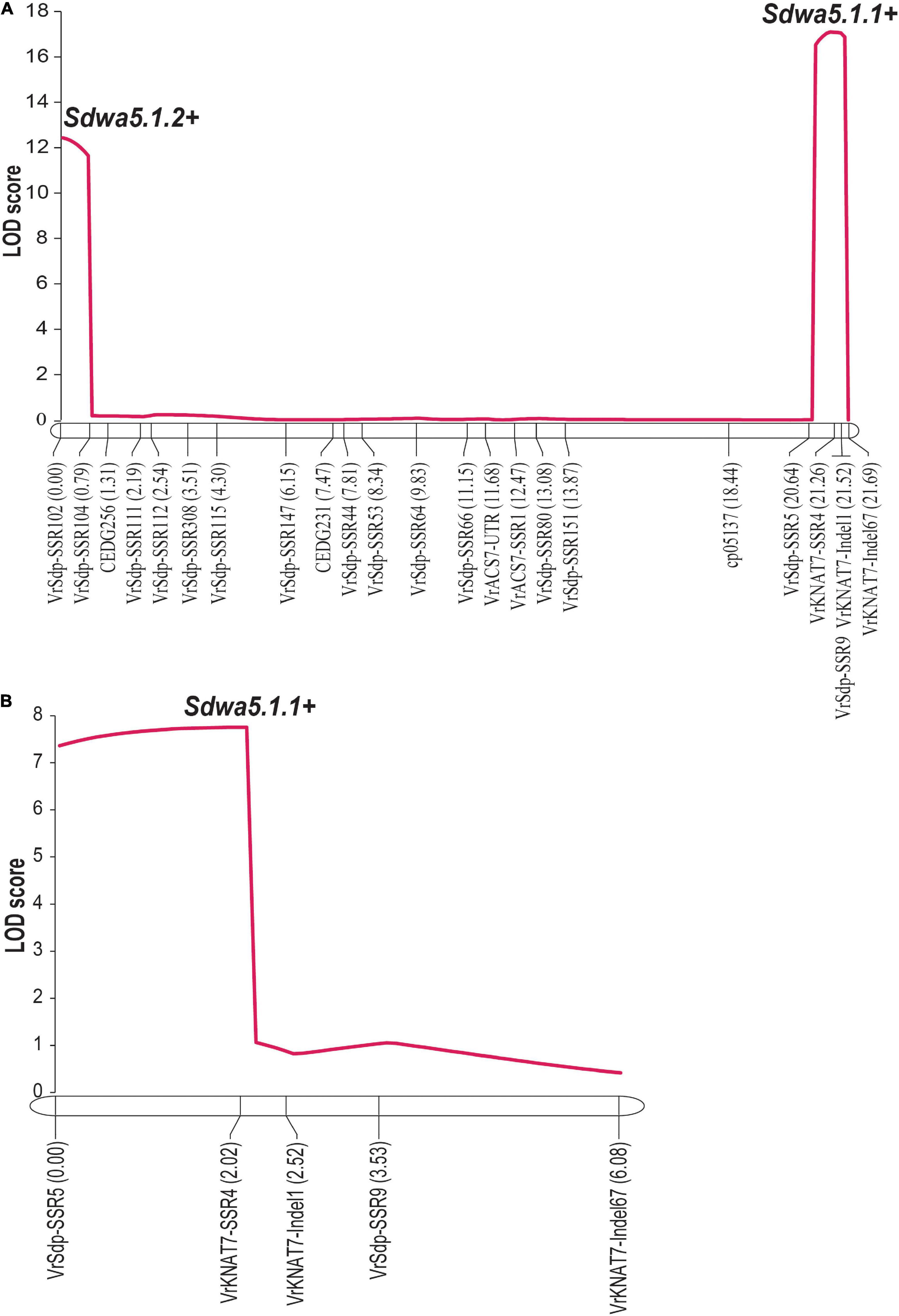
Figure 4. Logarithm of the odd (LOD) graphs of quantitative trait loci (QTLs) for seed dormancy detected on linkage group 1 of F2 population of 573 individuals (A) and F2:3 population of 100 individuals (B) derived from the cross between KPS2 and ACC41. Dotted line parallel to x-axis represents LOD threshold for the QTLs.

Table 1. Locations and effects of quantitative trait loci on linkage group 2 controlling seed dormancy detected in the F2 population of 575 individuals derived from hybridization between cultivated mungbean Kamphaeng Saen 2 (KPS2) and wild mungbean ACC41.
The QTL Sdwa5.1.1+ identified in the F2 population was confirmed using an F3 population of 112 individuals. PDS in the F2:3 population ranged from 0 to 100% with a mean of 62.68%. ACC41 had a PDS of 95.34%, while KPS2 had a PDS of 0%. The PDS showed continuous distribution and skewed toward KPS2 (Figure 3B). The H2 value estimated for the F2:3 population was nearly the same with that of the F2 population, 97.08%. The ICIM analysis in the F2:3 population confirmed that the Sdwa5.1.1+ located between the markers VrSdp-SSR5 and VrKNAT7-SSR4 (Figure 4B and Supplementary Table 2). The Sdwa5.1.1+ accounted for 31.75% of the dormancy variation in the F2:3A population and possessed an additive effect of −18.42 and a dominant effect of 11.85. Again, the allele(s) from ACC41 increased the dormancy.
Identification of Candidate Genes for the Sdwa5.1.1+ and Sdwa5.1.2+
The Sdwa5.1.1+ was identified and confirmed in two populations. Based on the mungbean reference genome, the physical region of the marker interval VrSdp-SSR5 and VrKNAT7-SSR4 covering the Sdwa5.1.1+ was only 3,298 bp (Figure 5). There was only one annotated gene between the two markers, LOC106767068. LOC106767068 encodes a transcription factor KNOTTED ARABIDOPSIS THALIANA 7 (KNAT7). We designated LOC106767068 as VrKNAT7-1 and considered it as the candidate gene for the Sdwa5.1.1+. It is noteworthy that the marker VrKNAT7-SSR4 was developed from VrKNAT7-1.
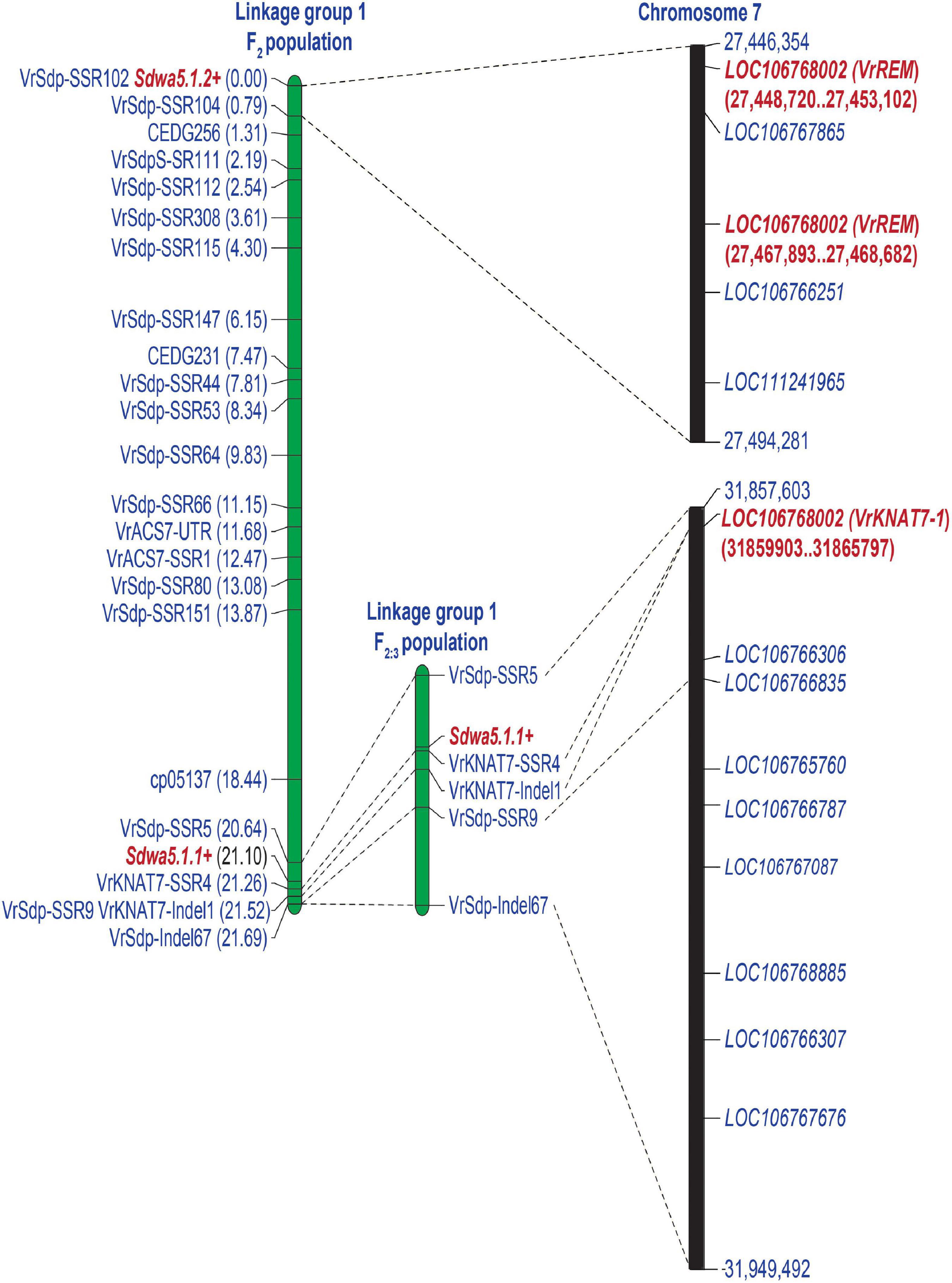
Figure 5. A comparative map illustrating the position of the QTLs Sda5.1.1+ and Sdwa5.1.2+ controlling seed dormancy on the reference genome of mungbean (VC1973A). Candidate genes at the Sdwa5.1.1+ and Sdwa5.1.2+ are highlighted in red and bold.
The Sdwa5.1.2+ was identified in only one population. Physical location of the marker interval VrSdp-SSR102 and VrSdp-SSR104 delimited the Sdwa5.1.2+ was 47,927 bp (Figure 5). There were 5 annotated genes in this region, including LOC106768002, LOC106767865, LOC106767275, LOC106766251, and LOC111241965 (Figure 5 and Supplementary Table 3). Among these genes, LOC106768002 encoding B3 domain-containing protein, with homology to Os01g0234100 from rice, and LOC106767275 encoding calmodulin-like (CML) protein 1 were considered as the candidate gene for the Sdwa5.1.2+. BLASTN search against the TAIR database revealed that LOC106768002 showed the best hit with Reproductive Meristem 1 (At3g19184), so we designated LOC106768002 as VrREM1. In case of LOC106767275, we designated it as VrCML1.
Nucleotide Polymorphisms in VrKNAT7-1
VrKNAT7-1 of ACC41 and KPS2 were sequenced and compared with the reference sequence (VC1973A). VrKNAT7-1 sequence alignment revealed no polymorphism between the cultivated mungbeans KPS2 and VC1973A, but showed several polymorphisms between ACC41 and KPS2 (Figure 6); one single-nucleotide polymorphism (SNP) in the CDS, one SNP and a 2-bp insertion/deletion (Indel) in the 5′UTR, and four SNPs in the 3′UTR. In addition, an alignment of the VrKNAT7-1 upstream sequences showed several SNPs and InDels between ACC41 and KPS2 (Supplementary Figure 1). Nonetheless, the SNP in the CDS was a synonymous mutation (Supplementary Figures 2, 3).
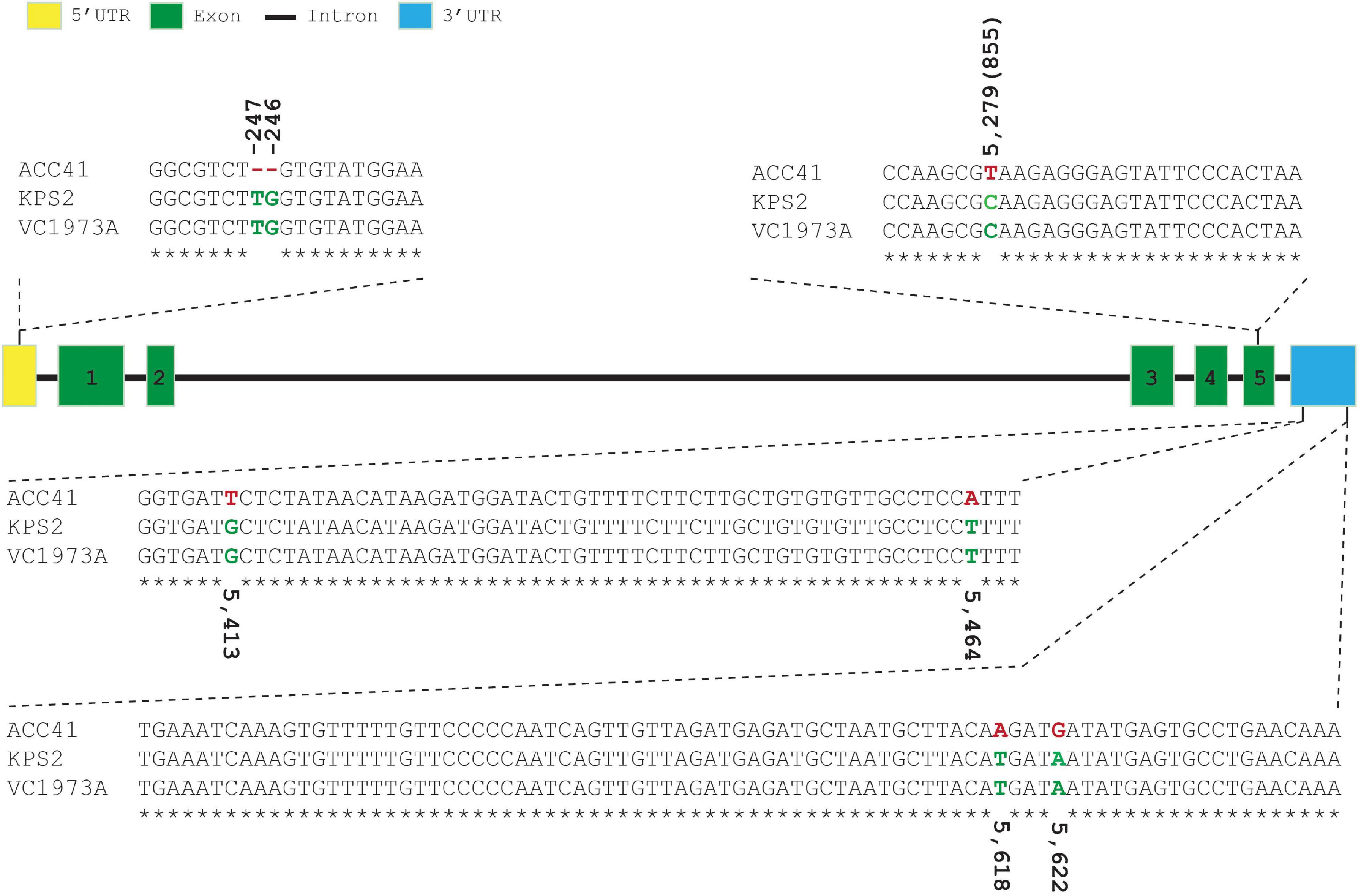
Figure 6. Sequence alignment of 5′-untranslated region, coding sequencing, and 3′-untranslated region of the LOC106767068 (VrKNAT7-1) among ACC41, KPS2 and mungbean reference genome sequence (VC1973A). The number in parenthesis upper/under nucleotide(s) indicates its position from the first nucleotide of the start codon.
Gene Expression Analysis
The expression of the VrKNAT7-1 and VrCYP86A in a seed without the seed coat and with the seed coat of ACC41 and KPS2 was determined by qRT-PCR. In case of VrKNAT7-1, the analysis revealed no significant difference in the seed without seed coat between ACC41 and KPS2, but showed a statistical difference in the seed coat between the two mungbeans (Figure 7A). The expression in ACC41 was about 10-fold higher than KPS2. For VrCYP86A, the analysis revealed that the expression in ACC41 was significantly higher than that in KPS2 in both seeds without the seed coat and with the seed coat (Figure 7B).
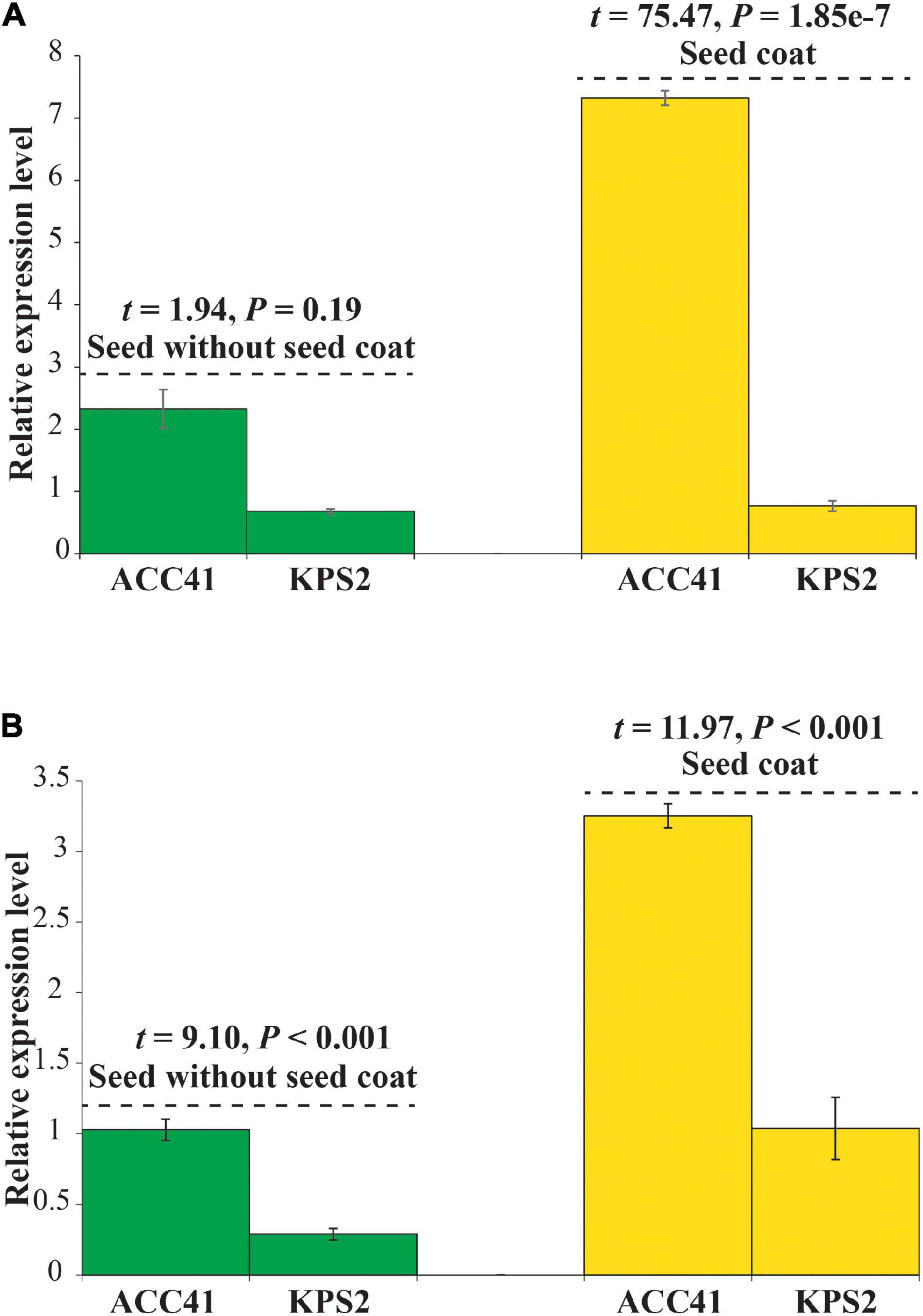
Figure 7. Relative gene expression level of VrKNAT7-1 (LOC106767068) (A) and VrCYP86A (LOC106774045) (B) in a seed without seed coat and with the seed coat of ACC41 and KPS2.
Phylogenetic Analysis of VrKNAT7 Proteins
Mungbean contained two KNAT7 proteins, VrKNAT7-1 and VrKNAT7-2. A phylogenetic analysis revealed three major clusters (classes) of KNOX proteins, namely class M KNOX, class I KNOX, and class II KNOX (Figure 8). The analysis also confirmed the close relationship between the VrKNAT7-1 and VrKNAT7-2 and demonstrated that both of them were sub-clustered with other KNAT7-like proteins, MtKNOX4 and AtKNAT7 (Figure 8). These KNAT7-like proteins were clustered with the KNOX sub-cluster KNAT3/4/5-like proteins, forming the class II KNOX proteins.
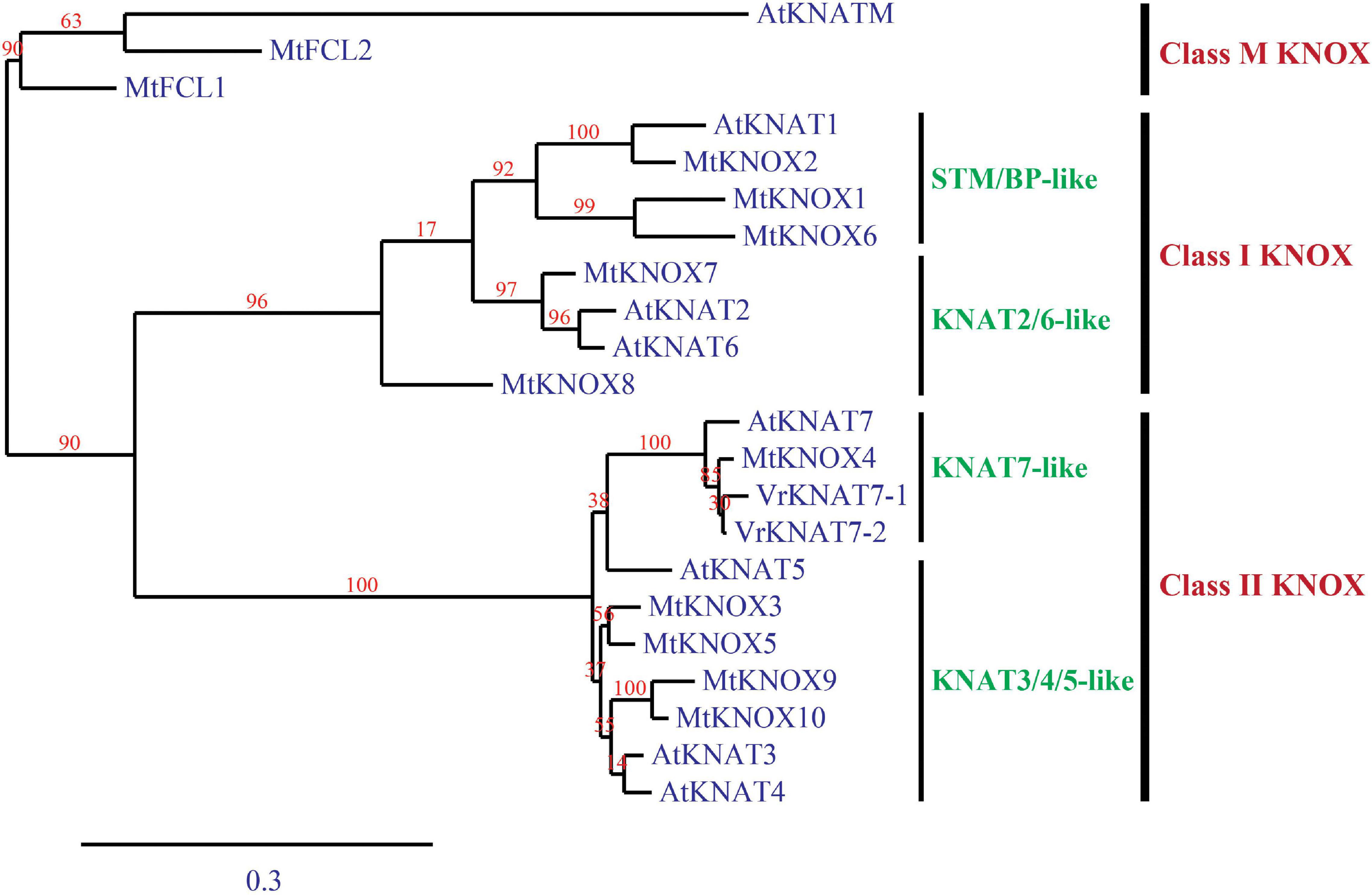
Figure 8. A phylogenic tree of Vigna radiata KNAT7-1 and KNAT7-2 proteins, and Arabidopsis thaliana and Medicago truncatula KNOX proteins. The tree is constructed by a maximum-likelihood method.
Discussion
A decrease in or a loss of seed dormancy is an important biological mechanism of crop domestication that enables seeds to germinate uniformly and timely. Although seed dormancy is problematic for crop cultivation, it may be useful for preventing pre-harvest sprouting of seed yield. In general, cultivated mungbeans are suppressed for seed dormancy although a few mungbean germplasms exhibits fresh-seed dormancy for a short period of time (4 days) (Lamichaney et al., 2018). In this study, the H2 estimated for the seed dormancy in the F2 and F3 populations was H2 (90–91%). This result agrees with those from the previous studies that seed dormancy in mungbean is a highly heritable trait with a H2 value of 95–99% (Lawn et al., 1988; Humphry et al., 2005; Isemura et al., 2012).
Seed dormancy in wild mungbean is controlled by one or two major QTLs together with 2–3 minor QTLs (Humphry et al., 2005; Isemura et al., 2012). Isemura et al. (2012) reported that Sdwa5.1.1+ is the major QTL controlling seed dormancy in wild mungbean. However, this QTL was identified in only one environment and has not been confirmed/validated. In this study, we validated the major QTL Sdwa5.1.1+ using wild mungbean accession ACC41 as the source of seed dormancy. The Sdwa5.1.1+ was previously mapped between the markers cp05137 and CEDG074b (Isemura et al., 2012). Based on the mungbean reference genome sequence (Kang et al., 2014), these flanking markers were located on chromosome 7 at the positions 31,187,230 and 31,822,876, respectively. However, our high-resolution mapping results in both F2 and F3 populations consistently demonstrated that the Sdwa5.1.1+ resides between the markers VrSpd-SSR5 and VrKNAT7-SSR4 (Figure 4), which corresponded to the positions 31,857,603 and 31,860,901 of chromosome 7, respectively (Figure 5). Thus, the region of the Sdwa5.1.1+ mapped in our study is different from that reported by Isemura et al. (2012), albeit the two QTL regions are only about 34.7 Kb apart. The contrasting results are possibly due to different sizes of mapping populations used in two studies. The population used in our study comprised 575 individuals, which is about 2.3-fold larger than the one used by Isemura et al. (2012). Larger mapping population size provides a better accuracy of the QTL location (Tanksley, 1993; Charmet, 2000).
Physical seed dormancy, also called hard-seededness, is an adaptive trait for the survival of wild progenitors of seed crops. This type of dormancy is caused by the existence of a water-impermeable layer in the seed coat (Finch-Savage and Leubner-Metzger, 2006). In the genus Vigna subgenus Ceratotrapis, to which mungbean belongs, seeds of species in this taxon imbibe water through the lens (strophiole) near the hilum (Gopinathan and Babu, 1985; Kikuchi et al., 2006). In this study, intact seeds of ACC41 and KPS2 showed a contrasting degree of dormancy (Figure 1A), while scarified seeds of ACC41 germinated rapidly (Figure 1B). Microscopic observation of seeds clearly showed that the seed cuticle layer was present in ACC41, but absent in KPS2 (Figure 2). These results indicated that the presence of cuticle/cutin causes the physical dormancy in the wild mungbean ACC41. VrKNAT7-1 was identified as the only candidate gene for seed dormancy at the locus Sdwa5.1.1+ (Figure 5). VrKNAT7-1 encodes a transcription factor homeobox protein HD1 (KNAT7), a KNOX II protein. In M. truncatula L., a model legume species, genetic, and molecular analyses revealed that MtKNOX4, a class II KNOX gene, controls seed physical dormancy (Chai et al., 2016). NCBI BLASTP search of MtKNOX4 protein against the reference protein database of mungbean showed that the MtKNOX4 was best matched with XP_014521195.1 (95% query coverage, E-value = 0.0, and 89.04% identity), followed by VrKNAT7-1 (96% query coverage, E-value = 9e–169, and 80.95% identity). XP_014521195.1 protein is encoded by LOC106777892, designated VrKNAT7-2, localizing on mungbean chromosome 11. A phylogenetic analysis further supports a close relationship among VrKNAT7-1, VrKNAT7-2, MtKNOX4, and MtKNAT7 (Figure 8). These results demonstrated that there are two VrKNAT7 genes in mungbean and both VrKNAT7-1 and VrKNAT7-2 are orthologs of MtKNOX4. A mutation causing the loss of function of the MtKNOX4 resulted in the reduction of hydroxylated fatty acids in the seed coat (especially18:2 ω-hydroxy fatty acid), a group of lipid polyester monomer composition of seed cutin, that alters cuticle layer permeability, and thus the physical dormancy of M. truncatula (Chai et al., 2016). MtKNOX4 controls seed dormancy by regulating the expression of MtCYP86A gene (Chai et al., 2016) that plays an important role in the cuticle biosynthesis. In Arabidopsis, CYP86A2, a cytochrome P450 monooxygenase catalyzing fatty acid oxidation, is required for the biosynthesis of cutin and cuticle development (Xiao et al., 2004). In M. truncatula, MtKNOX4 protein directly binds to the promoter of MtCYP86A, and thus MtKNOX4 is believed to regulate cuticle biosynthesis pathway in the seed coat (Chai et al., 2016). In our study, the expressions of VrKNAT7-1 and VrCYP86A in the seed coat of ACC41 and KPS2 collected at a yellow-pod stage revealed that expressions of these genes in ACC41 were statistically much higher than those in KPS2 (Figure 7). The different expression of VrKNAT7-1 between ACC41 and KPS2 is very likely to be caused by nucleotide polymorphisms in the upstream sequence, 5′ UTR and 3′ UTR of this gene (Figure 6 and Supplementary Figure 1). Nonetheless, these results demonstrated that the VrKNAT7-1 regulate cutin biosynthesis during seed coat development of the wild mungbean by controlling the expression level of the VrCYP86A, and that a common mechanism of physical seed dormancy exists between the different wild species of the genus Vigna and Medicago.
Apart from physical dormancy, physiological dormancy widely exists in seed-plant species. In fact, the majority of seeds exhibit physiological dormancy in which germination is prevented by using germination-inhibiting hormones, including abscisic acid (ABA) and gibberellins (GAs) (Penfield, 2017). In the previous studies, Sdwa5.1.1+ (HsA) was identified as a single QTL on the LG1 controlling seed dormancy (Humphry et al., 2005; Isemura et al., 2012). Nonetheless, our QTL analysis revealed that apart from Sdwa5.1.1+, Sdwa5.1.2+ was also detected on LG1 for the trait (Figure 4). The genetic effects of Sdwa5.1.1+ and Sdwa5.1.2+ are comparable. The environmental factor(s) may explain contrasting results between the present study and the previous ones regarding the Sdwa5.1.2+. Although we did not confirm the Sdwa5.1.2+, based on physical location of Sdwa5.1.2+ and function of the genes in this QTL region, we considered VrREM1 and VrCML1 as candidate genes at this QTL (Figure 6 and Supplementary Table 2). As VrREM1 encodes a B3 domain-containing protein, with a homolog to Os01g0234100 from rice, it is likely that VrREM1 is a transcription factor. B3 transcription factors regulate expressions of seed genes during embryogenesis, maturation, dormancy, and germination [reviewed in Carbonero et al. (2017)]. VrREM1 is a homolog to A. thaliana REM1 (AtREM1). Although REM1 has not been reported to be associated with seed dormancy, a recent study showed that REM1 is among 7 TFs possibly regulated the expression of 17 hub genes involved in dormancy transition of Polygonatum kingianum corm (Wang et al., 2020). VrCML1 encodes a CML protein 1. Calmodulin (CaM) and CaM-like proteins are the major Ca2+-binding proteins, and their signaling has been shown to be involved in the ABA-induced inhibition of seed germination and seedling growth. In A. thaliana, transgenic lines with under-expressing AtCML24 shows resistance to ABA-induced inhibition of seed germination and seedling growth (Delk et al., 2005). The expression of AtCML9 is affected by ABA and abiotic stress, and the cml9 null mutant shows a hypersensitive response to ABA during seed germination and seedling growth (Magnan et al., 2008). A. thaliana transgenic lines possessing a novel CML gene, OsMSR2, from rice (Oryza sativa L.) exhibited hypersensitivity to ABA during seed germination and post-germination growth (Xu et al., 2011). Recently, the genes that are homologous to calcium signaling pathway-related genes, including CALMODULIN-BINDING RECEPTOR-LIKE CYTOPLASMIC KINASE 3, CALCIUM-BINDING PROTEIN, CALMODULIN-RELATED PROTEIN, CBL-INTERACTING PROTEIN KINASE 31, CALMODULIN-BINDING PROTEIN 60D-LIKE, and CALCIUM-DEPENDENT PROTEIN KINASE were found to be related with seed dormancy in a wheat (Triticum aestivum L.) mutant (Rikiishi et al., 2021). In case, VrREM1 and/or VrCML1 are truly involved in the dormancy, it would indicate that both physical and physiological dormancy controls the dormancy in ACC41.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
PS and KL conceived the idea, designed the studies, and wrote and revised the manuscript. KL, KA, and TY carried out field experiments and phenotyping. KL, KA, JC, YL, XY, and LW conducted a DNA marker analysis, a gene expression analysis, and DNA sequencing. KL and KA conducted a microscopic observation. PS and XC secured research funding and coordinated the study. KL analyzed data. All authors approved the final version of the manuscript.
Funding
This research was supported by the Office of National Higher Education Science Research and Innovation Policy Council via the Program Management Unit for Human Resources and Institutional Development, Research and Innovation (Grant No. B16F630088), Center for Advanced Studies for Agriculture and Food, Kasetsart University Institute for Advanced Studies, Kasetsart University, and the National Key Research and Development Program of China (Grant No. 2019YFD1001300).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Apinan Sonong for technical support in microscopic observation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.852373/full#supplementary-material
Footnotes
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410.
Amkul, K., Somta, P., Laosatit, K., and Wang, L. (2020). Identification of QTLs for domestication-related traits in zombi pea [Vigna vexillata (L.) A. Rich], a lost crop of Africa. Front. Genet. 11:803. doi: 10.3389/fgene.2020.00803
Carbonero, P., Iglesias-Fernández, R., and Vicente-Carbajosa, J. (2017). The AFL subfamily of B3 transcription factors: evolution and function in angiosperm seeds. J. Exp. Bot. 68, 871–880. doi: 10.1093/jxb/erw458
Chai, M., Zhou, C., Molina, I., Fu, C., Nakashima, J., Li, G., et al. (2016). A class II KNOX gene, KNOX4, controls seed physical dormancy. Proc. Natl. Acad. Sci. U S A. 113, 6997–7002. doi: 10.1073/pnas.1601256113
Charmet, G. (2000). Power and accuracy of QTL detection: simulation studies of one-QTL models. Agronomie 20, 309–323.
Dachapak, S., Tomooka, N., Somta, P., Naito, K., Kaga, A., and Srinives, P. (2018). QTL analysis of domestication syndrome in zombi pea (Vigna vexillata), an underutilized legume crop. PLoS One 13:e0200116. doi: 10.1371/journal.pone.0200116
Delk, N. A., Johnson, K. A., Chowdhury, N. I., and Braam, J. (2005). CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 139, 240–253. doi: 10.1104/pp.105.062612
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Diamond, J. (1999). Guns, germs and steel: The fates of human societies. New York, NY: W.W. Norton and Company.
Diamond, J. (2002). Evolution, consequences and future of plant and animal domestication. Nature 418, 700–707. doi: 10.1038/nature01019
Falk, C. T. (1989). “A simple scheme for preliminary ordering of multiple loci: application to 45 CF families,” in Multipoint mapping and linkage based upon affected pedigree members, eds R. C. Elston, M. A. Spence, S. E. Hodge, and J. W. MacCluer (New York, NY: Liss), 17–22.
Finch-Savage, W. E., and Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171, 501–523. doi: 10.1111/j.1469-8137.2006.01787.x
Gopinathan, M. C., and Babu, C. R. (1985). Structural diversity and its adaptive significance in seeds of Vigna minima (Roxb.) Ohwi & Ohashi and allies (Leguminosae-Papilionoideae). Ann. Bot. 56, 723–732.
Humphry, M. E., Lambrides, C. J., Chapman, S. C., Aitken, E. A. B., Imrie, B. C., Lawn, R. J., et al. (2005). Relationships between hard-seededness and seed weight in mungbean (Vigna radiata) assessed by QTL analysis. Plant Breed. 124, 292–298.
Imrie, B. C., Williams, R. W., and Lawn, R. J. (1988). “Breeding for resistance to weather damage in mungbean,” in ‘Mungbean: Proceedings of the Second International Symposium’, eds S. Shanmugasundaram and B. T. McLean (Shanhua: Asian Vegetable Research and Development Centre), 130–135. doi: 10.1002/(SICI)1097-0029(19960601)34:2<177::AID-JEMT12>3.0.CO;2-N
Isemura, T., Kaga, A., Konishi, S., Ando, T., Tomooka, N., et al. (2007). Genome dissection of traits related to domestication in azuki bean (Vigna angularis) and their comparison with other warm season legumes. Ann. Bot. 100, 1053–1071. doi: 10.1093/aob/mcm155
Isemura, T., Kaga, A., Tabata, S., Somta, P., Srinives, P., Shimizu, T., et al. (2012). Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS One 7:e41304. doi: 10.1371/journal.pone.0041304
Kaewwongwal, A., Liu, C., Somta, P., Chen, J., Tian, J., Yuan, X., et al. (2020). A second VrPGIP1 allele is associated with bruchid resistance (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) accession ACC41. Mol. Genet. Genomics 295, 275–286. doi: 10.1007/s00438-019-01619-y
Kaga, A., Isemura, T., Tomooka, N., and Vaughan, D. A. (2008). The genetics of domestication of azuki bean (Vigna angularis). Genetics 178, 1013–1036. doi: 10.1534/genetics.107.078451
Kang, Y. J., Kim, S. K., Kim, M. Y., Lestari, P., Kim, K. H., Ha, B. K., et al. (2014). Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 5:5443. doi: 10.1038/ncomms6443
Kikuchi, K., Koizumi, M., Ishida, N., and Kano, H. (2006). Water uptake by dry beans observed by micro-magnetic resonance imaging. Ann. Bot. 98, 545–553. doi: 10.1093/aob/mcl145
Kongjaimun, A., Kaga, A., Tomooka, N., Somta, P., Vaughan, D. A., and Srinives, P. (2012). The genetics of domestication of yardlong bean, Vigna unguiculata (L.) Walp. ssp. unguiculata cv.-gr. sesquipedalis. Ann. Bot. 109, 1185–1200. doi: 10.1093/aob/mcs048
Laksana, C., and Chanprame, S. (2015). A simple and rapid method for RNA extraction from young and mature leaves of oil palm (Elaeis guineensis Jacq.). J. ISSAAS 21, 96–106.
Lamichaney, A., Katiyar, P., Laxmi, V., and Pratap, A. (2018). Variation in pre-harvest sprouting tolerance and fresh seed germination in mungbean (Vigna radiata L.) genotypes. Plant Genet. Resour. 16, 437–445.
Larson, G., Piperno, D. R., Allaby, R. G., Purugganan, M. D., Andersson, L., Arroyo-Kalin, M., et al. (2014). Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. U S A. 111, 6139–6146. doi: 10.1073/pnas.1323964111
Lawn, R. J., Williams, R. W., and Imrie, B. C. (1988). “Potential of wild germplasm as a source of tolerance to environmental stresses in mungbean,” in Mungbean: Proceedings of the Second International Symposium, eds S. Shanmugasundaram and B. T. McLean (Shanhua: Asian Vegetable Research and Development Centre), 136–145.
Li, H., Ye, G., and Wang, J. (2007). A modified algorithm for the improvement of composite interval mapping. Genetics 175, 361–374. doi: 10.1534/genetics.106.066811
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lo, S., Muñoz-Amatriaín, M., Boukar, O., et al. (2018). Identification of QTL controlling domestication-related traits in cowpea (Vigna unguiculata L. Walp). Sci. Rep. 8:6261. doi: 10.1038/s41598-018-24349-4
Lodhi, M. A., Ye, G. N., Weeden, N. F., and Reisch, B. I. (1994). A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 12, 6–13. doi: 10.1007/bf02668658
Magnan, F., Ranty, B., Charpenteau, M., Sotta, B., Galaud, J. P., and Aldon, D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56, 575–589. doi: 10.1111/j.1365-313X.2008.03622.x
Meng, L., Li, H., Zhang, L., and Wang, J. (2015). QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Nair, R., and Schreinemachers, P. (2020). “Global status and economic importance of mungbean,” in The Mungbean Genome, eds R. M. Nair, R. Schafleitner, and S. H. Lee (Amsterdam: Springer International Publishing), 1–8. doi: 10.1007/978-3-030-20008-4_12
Rikiishi, K., Sugimoto, M., and Maekawa, M. (2021). Transcriptomic analysis of developing seeds in a wheat (Triticum aestivum L.) mutant RSD32 with reduced seed dormancy. Breed Sci. 71, 155–166. doi: 10.1270/jsbbs.20016
Sievers, F., Wilm, A., Dineen, D. G., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Singh, D. P., Sharma, B. L., and Dwivedi, S. (1983). Inheritance of hard seeds in interspecific crosses of Mungbean. Indian J. Genet. 43, 378–379.
Somta, P., and Srinives, P. (2007). Genome research in mungbean [Vigna radiata (L.) Wilczek] and blackgram [V. mungo (L.) Hepper]. ScienceAsia 33, 69–74. doi: 10.2306/scienceasia1513-1874.2007.33(s1).069
Somta, P., Chen, J., Yimram, T., Yundaeng, C., Yuan, X., Tomooka, N., et al. (2020). QTL mapping for agronomic and adaptive traits confirmed pleiotropic effect of mog gene in black gram [Vigna mungo (L.) Hepper]. Front. Genet. 11:635. doi: 10.3389/fgene.2020.00635
Temnykh, S., DeClerck, G., Lukashova, A., Lipovich, L., Cartinhour, S., and McCouch, S. (2001). Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11, 1441–1452. doi: 10.1101/gr.184001
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Van Os, H., Stam, P., Visser, R. G., and Van Eck, H. J. (2005). RECORD: a novel method for ordering loci on a genetic linkage map. Theor. Appl. Genet. 112, 30–40. doi: 10.1007/s00122-005-0097-x
Wang, L., Wu, C., Zhong, M., Zhao, D., Mei, L., Chen, H., et al. (2016). Construction of an integrated map and location of a bruchid resistance gene in mung bean. Crop J. 4, 360–366.
Wang, Y., Bailey, D. C., Yin, S., and Dong, X. (2020). Characterizing rhizome bud dormancy in Polygonatum kingianum: Development of novel chill models and determination of dormancy release mechanisms by weighted correlation network analysis. PLoS One 15:e0231867. doi: 10.1371/journal.pone.0231867
Xu, G. Y., Rocha, P. S., Wang, M. L., Xu, M. L., Cui, Y. C., Li, L. Y., et al. (2011). A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta 234, 47–59. doi: 10.1007/s00425-011-1386-z
Xiao, F., Goodwin, S. M., Xiao, Y., Sun, Z., Baker, D., Tang, X., et al. (2004). Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 23, 2903–2913. doi: 10.1038/sj.emboj.7600290
Yundaeng, C., Somta, P., Amkul, K., Kongjaimun, A., Kaga, A., and Tomooka, N. (2019). Construction of genetic linkage map and genome dissection of domestication-related traits of moth bean (Vigna aconitifolia), a legume crop of arid areas. Mol. Genet. Genomics 294, 621–635. doi: 10.1007/s00438-019-01536-0
Keywords: mungbean, seed dormancy, hardseededness, KNOX II, KNAT7
Citation: Laosatit K, Amkul K, Yimram T, Chen J, Lin Y, Yuan X, Wang L, Chen X and Somta P (2022) A Class II KNOX Gene, KNAT7-1, Regulates Physical Seed Dormancy in Mungbean [Vigna radiata (L.) Wilczek]. Front. Plant Sci. 13:852373. doi: 10.3389/fpls.2022.852373
Received: 11 January 2022; Accepted: 31 January 2022;
Published: 15 March 2022.
Edited by:
Jose Maria Barrero, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaReviewed by:
Hifzur Rahman, International Center for Biosaline Agriculture (ICBA), United Arab EmiratesSteven B. Cannon, Agricultural Research Service (USDA), United States
Copyright © 2022 Laosatit, Amkul, Yimram, Chen, Lin, Yuan, Wang, Chen and Somta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Chen, Y3hAamFhcy5hYy5jbg==; Prakit Somta, YWdycGtzQGt1LmFjLnRo
 Kularb Laosatit
Kularb Laosatit Kitiya Amkul1
Kitiya Amkul1 Jingbin Chen
Jingbin Chen Yun Lin
Yun Lin Xingxing Yuan
Xingxing Yuan Lixia Wang
Lixia Wang Xin Chen
Xin Chen Prakit Somta
Prakit Somta