- 1CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 2Center of Conservation Biology, Core Botanical Gardens, Chinese Academy of Sciences, Wuhan, China
- 3University of Chinese Academy of Sciences, Beijing, China
- 4Section of Plant Biology and the L.H. Bailey Hortorium, School of Integrative Plant Science, Cornell University, Ithaca, NY, United States
- 5BTI Computational Biology Center, Boyce Thompson Institute, Ithaca, NY, United States
- 6CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 7Yunnan International Joint Laboratory for Biodiversity of Central Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
Polygonaceae has a complex taxonomic history, although a few studies using plastid or nuclear DNA fragments have explored relationships within this family, intrafamilial relationships remain controversial. Here, we newly sequenced and annotated 17 plastomes representing 12 genera within Polygonaceae. Combined with previously published data, a total of 49 plastomes representing 22/46 Polygonaceae genera and 16/20 Polygonoideae genera were collected to infer the phylogeny of Polygonaceae, with an emphasis on Polygonoideae. Plastome comparisons revealed high conservation within Polygonoideae in structure and gene order. Phylogenetic analyses using both Maximum Likelihood and Bayesian methods revealed two major clades and seven tribes within Polygonoideae. BEAST and S-DIVA analyses suggested a Paleocene origin of Polygonoideae in Asia. While most genera of Polygonoideae originated and further diversified in Asia, a few genera experienced multiple long-distance dispersal events from Eurasia to North America after the Miocene, with a few dispersal events to the Southern Hemisphere also being detected. Both ancient vicariance and long-distance events have played important roles in shaping the current distribution pattern of Polygonoideae.
Introduction
Polygonaceae Juss. (Caryophyllales) contains approximately 46 genera and 1200 species with morphological disparity, with life forms varying from herbs, lianas, woody vines, shrubs to trees (Kubitzki et al., 1990; Sanchez et al., 2009; Burke and Sanchez, 2011; Schuster et al., 2015). Polygonaceae is widely distributed around the world, mainly in temperate North America, Europe and Southeast Asia, but species can also be found in South America, the Caribbean, Africa and Australasia (Frye and Kron, 2003; Burke and Sanchez, 2011; Sanchez et al., 2011). Classification within Polygonaceae, specifically the definitions of different subfamilies or genera, has long been controversial (Burke and Sanchez, 2011; Sanchez et al., 2011; Schuster et al., 2013). In previous studies, subfamilies were primarily differentiated based on morphological characters, e.g., the presence or absence of ocreas, tepal arrangement and habitat, with two to four subfamilies having been proposed (Meisner, 1856; Jaretzky, 1925). Meisner (1856) suggested four subfamilies including Polygonoideae Eaton, Eriogonoideae Arn, Brunnichioideae Meisn. and Symmerioideae Meisn. Later, a proposal of three subfamilies including Polygonoideae, Eriogonoideae and Coccoloboideae Luerss. was suggested (Perdrigeat, 1900). Afterwards Coccoloboideae was merged with Polygonoideae by Jaretzky (1925). Generally, the recognition of Polygonoideae and Eriogonoideae as subfamilies of Polygonaceae has been widely accepted. Typically, Polygonoideae is characterized by possessing ocreae, while Eriogonoideae is characterized by lacking ocreae (Jaretzky, 1925).
In general, Polygonoideae is recognized to comprise 20 genera (Schuster et al., 2015). The subfamily is further divided into different tribes step by step (Sanchez and Kron, 2008; Sanchez et al., 2011). Sanchez and Kron (2008), using sequences of three chloroplast DNA segments and LEAFY, revealed three tribes: Persicarieae Dumort., Rumiceae Dumort., and Polygoneae Rchb. Galasso et al. (2009) divided the subfamily into four tribes including Persicarieae, Rumiceae, Polygoneae and Fagopyreae Yonke. based only on rbcL sequences. Afterwards, Sanchez et al. (2011) proposed five tribes Persicarieae, Rumiceae, Polygoneae, Fagopyreae and Calligoneae based on matK, ndhF and rbcL sequences with a more comprehensive sampling. More recently, an updated phylogenetic analysis suggested seven tribes with two tribes, Oxygoneae T.M.Schust. & Reveal and Pteroxygoneae T.M.Schust. & Reveal, added using matK, trnL-F and ITS sequences (Schuster et al., 2015). Polygonoideae has a worldwide distribution, covering North America, South America, Europe, Asia, Africa and Oceania. Previous biogeographical studies revealed that taxa displaying intercontinental disjunction usually have an Asian origin, followed by long-distance dispersal and vicariance (e.g., Givnish and Renner, 2004; Yoder and Nowak, 2006; Nie et al., 2012; Fritsch et al., 2015; Yang et al., 2018). Notably, 17 Polygonoideae genera can be found in Asia; however, up till now, studies investigating the area of origin and dispersal of the subfamily are absent and thus limiting a comprehensive understanding of the evolutionary history of Polygonoideae.
Whole plastome sequencing is a cost-effective method that has been widely applied to resolve phylogenetic relationships at different taxonomic levels (Dong et al., 2020; Jung et al., 2021; Wen et al., 2021). Recently, such approaches have been applied to investigate the phylogenetic relationships of genera within Polygonaceae (Song et al., 2020; Zhou et al., 2020; Yang et al., 2021; Zhang et al., 2021). However, a comprehensive analysis, including plastome structure, phylogenomics and biogeography of Polygonoideae is lacking. In this study, we newly assembled 17 Polygonaceae plastomes to explore phylogenetic relationships and trace the origins of Polygonoideae along with previously published plastome sequences. The main objectives of this study are to (1) analyze and compare plastomes to track plastome evolution within Polygonoideae, and (2) elucidate the phylogeny and the biogeographic history of Polygonoideae to gain insights into the origin and the drivers of diversification of the subfamily.
Materials and methods
Taxon sampling, DNA extraction and sequencing
A total of 49 plastomes representing 22 genera of Polygonaceae were sampled, covering most genera of Polygonoideae and six genera from other subfamilies of Polygonaceae as of the most recent treatment of Schuster et al. (2015). Five individuals from three genera in Plumbaginaceae were selected as outgroups. Seventeen samples of Polygonaceae were newly sequenced and collected from around the world (Table S1). Fresh leaves were collected in silica-gel for DNA extraction. Voucher specimens were deposited at the Wuhan Botanical Garden (Table S1). All plastome sequences used in this study (including downloaded and newly sequenced) are listed in Table 1. Genomic DNA was extracted with a modified CTAB method (Li et al., 2013). DNA concentration was measured using the Qubit® DNA Assay Kit with a Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). For library preparation, 1.5 µg of DNA per sample was used and fragmented by sonication to a size of 350 bp. Generated DNA fragments were end polished, A-tailed, and ligated with full-length adapters for Illumina sequencing with further PCR amplification. Finally, purified PCR products were analyzed for size distribution with an Agilent2100 Bioanalyzer and quantified using real-time PCR. Constructed libraries were sequenced on an Illumina HiSeq using 150 bp paired-end reads with an insert size around 350 bp. A minimum of 2 GB of raw sequencing data was generated for each accession.
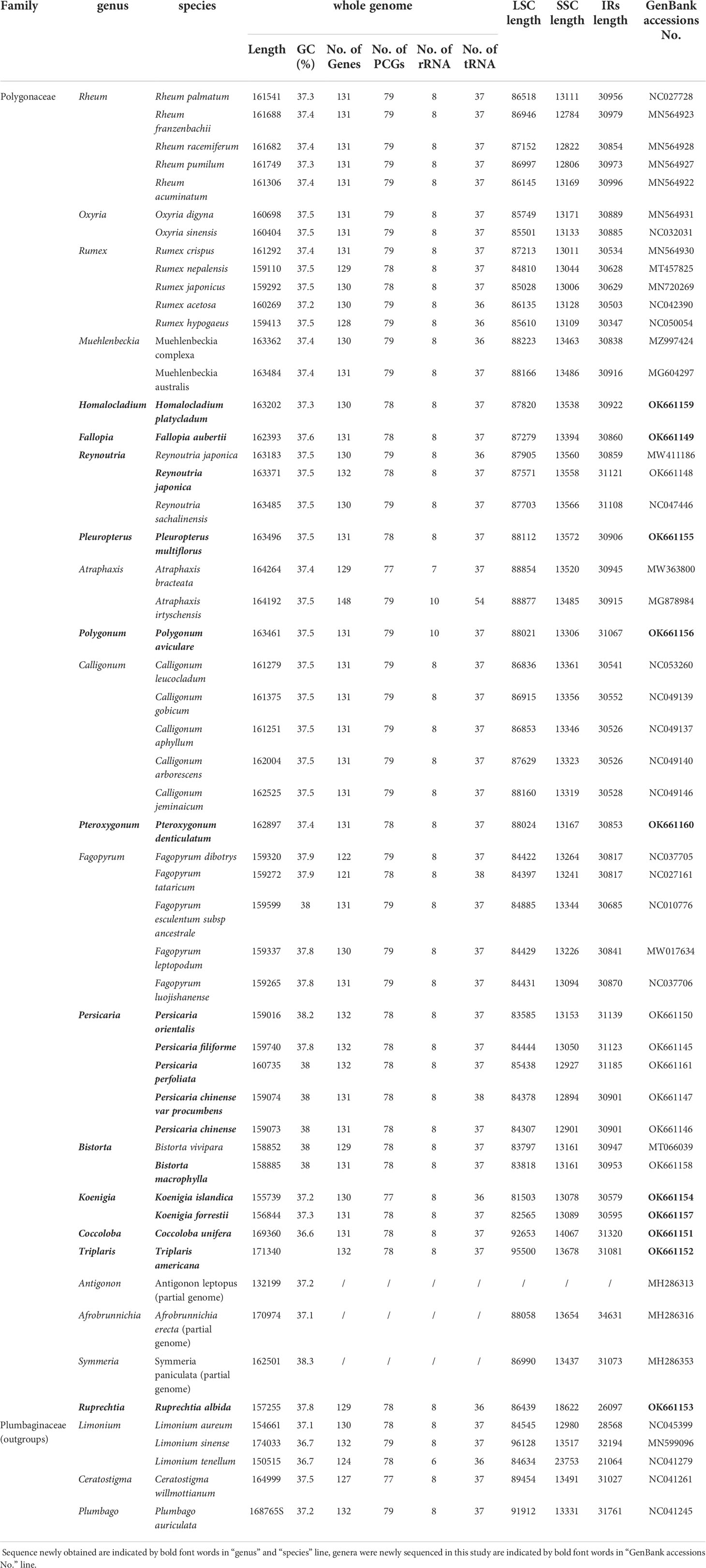
Table 1 Summary of major characteristics of all 54 chloroplast genome sequences, including sequence length (bp), numbers of genes, GC content (%), and GenBank accession number.
Plastome assembly, annotation and comparison
Raw sequencing reads were filtered using Trimmomatic v0.39 (Bolger et al., 2014) to remove adapters, low quality and unidentified nucleotides reads with the following parameters: LEADING=5, TRAILING=5, SLIDINGWINDOW=4:5, MINLEN=25. Clean reads were then de novo assembled using NOVOPlasty v4.3.1 (Dierckxsens et al., 2017) with the RUBP sequences as the seed for chloroplast assembly. The plastome sequence of Rheum palmatum (NCBI accession NC027728) was set as the reference. Assembled plastomes were annotated with PGA (Qu et al., 2019), again using R. palmatum (NCBI accession NC027728) as the reference. The preliminary annotated sequences were imported into Geneious v.9.0.2 to check start/stop codons and intron/exon boundaries (Kearse et al., 2012). Newly determined plastome sequences were submitted to NCBI (National Center for Biotechnology information) GeneBank (Table 1).
To detect inverted repeat (IR) expansion or contraction of plastomes and compare the boundary genes of the SC and IR among Polygonoideae, we chose R. palmatum (NCBI accession NC027728) as the reference and visualized the borders of the LSC, SSC, and IR in 17 genera in Polygonoideae using IRscope (Amiryousefi et al., 2018). Among the 17 plastomes, eight were downloaded from NCBI and nine accessions were newly sequenced in this study.
Phylogenetic analyses
To explore the phylogenetic relationships of Polygonoideae, a total of 54 plastomes representing 23 Polygonaceae genera were included in the phylogenomic analysis. Among these, 37 plastomes were download from Genebank and 17 plastomes were newly sequenced. Five species of Plumbaginaceae were selected as outgroups, including three species of Limonium, one species of Plumbago and one species of Ceratostigma (Table 1). For each plastome, we extracted 74 shared PCGs and aligned them with the codon-aware program MACSE v2.03 (Ranwez et al., 2018) followed by manual examination and adjustment in Mega X (Kumar et al., 2018). Alignments of PCGs were concatenated into a super-matrix with PhyloSuite v1.2.2 (Zhang et al., 2019a). The complete plastome nucleotide sequences, including only one copy of the IR regions were also used for the phylogenetic analyses. Both maximum-likelihood (ML) and Bayesian inference (BI) analyses were conducted for phylogenetic inference. For the ML analysis, RAxML v8.2.12 (Stamatakis, 2014) was used with the general time reversible model for nucleotide substitution, the gamma model of rate heterogeneity (GTR+G), and 500 rapid bootstrap replicates. Bayesian analyses were conducted with MrBayes v3.2.7 (Huelsenbeck and Ronquist, 2001). The best-fit model was calculated with ModelTest-NG (Darriba et al., 2020) under the Bayesian information criterion (BIC). Two runs with four Markov chains were applied with 2,000,000 generations from a random starting tree with sampling every 500 generations. The initial 25% of sampled trees were discarded as burn-in and the remaining trees were used to construct a majority-rule consensus tree and calculate the posterior probability. Bootstrap support (BS) and posterior probability (PP) were used to measure the support of the generated phylogenetic trees. Additionally, since we were not able to generate plastomes of Oxygonum and Knorringia in Polygonoideae, three chloroplast fragments (matK, trnL and rbcL) were extracted from the plastomes used here and combined with available data online to explore their phylogenetic positions (Table S3). A tree based on our results combined with previous studies were generated to exhibit a comprehensive phylogeny of Polygonoideae. The final phylogenetic topologies were viewed in FigTree v1.3.1 (Rambaut, 2009).
Estimation of divergence time
BEAST v1.10.4 (Drummond and Rambaut, 2007) was used to estimate the divergence time between lineages of Polygonaceae using the concatenated three chloroplast loci data matrix for inclusion of the largest number of genera. We chose the GTR+G substitution model, with a relaxed molecular clock model and Yule process as the tree prior. According to previous studies (Manchester and O’leary, 2010; Schuster et al., 2013; Yao et al., 2019), two fossil calibrations and two secondary calibrations were used. A detailed discussion of the fossil calibrations is provided in Supplementary Methods S1. The crown age of Polygonaceae was set to an age range of 72.1-66.0 Ma with a lognormal calibration prior (Manchester and O’leary, 2010). The crown age of Muehlenbeckia was set to an age range of 22.0–19.0 Ma with a lognormal calibration prior (Pole, 1992; Schuster et al., 2013). The crown age of Plumbaginaceae was set to a mean age of 60.0 Ma with a SD of 3.0 Ma and the crown age of all included species was set to a mean age of 91.8 Ma with a SD of 0.5 Ma (Yao et al., 2019). Both secondary calibration priors were set as a normal distribution. The MCMC was run for 1 x 109 generations, sampling every 10,000 generations. The convergence of the two runs and stationarity of the chains were checked in Tracer v1.7 (Rambaut et al., 2018), with a sufficient effective sample size (ESS) > 200 for all relevant parameters. The first 25% trees were discarded as burn-in, and a maximum clade credibility tree with mean heights reported for node heights and 95% highest posterior density intervals (95% HPDs) was generated with TreeAnnotator v1.10.4 (Suchard et al., 2018). FigTree v1.3.1 was used for visualizing the resulting phylogenetic tree (Rambaut, 2009).
Biogeographic analysis
We collected species distribution data of Polygonoideae from monographs, regional floras and online databases such as GBIF (https://www.gbif.org) and JSTOR Global Plants (https://plants.jstor.org/). Based on the distribution data and related geological history, we defined seven biogeographic areas: (A) North America, (B) South America, (C) Europe, (D) Asia, (E) Africa, and (F) Oceania. We used BioGeoBEARS as implemented in RASP 4.0 (Matzke, 2014; Yu et al., 2015) to explore the ancestral area of Polygonoideae with a Statistical Dispersal-Vicariance Analysis (S-DIVA) (Yu et al., 2010) and a condensed tree derived from the BEAST analysis (outgroups were excluded).
Results
Genome assembly and plastomes features
Illumina sequencing generated a total of 12,997,916- 20,520,168 paired-end clean reads for each species (Table S1). The mean sequencing coverage of the observed plastomes ranged from 209× to 3,956× (Table S1). The 17 newly sequenced plastomes, ranging in size from 155,739 to 171,340 bp, displayed a typical quadripartite structure and similar gene order consisting of a LSC (81,503-95,500 bp), SSC (12,806- 18,622 bp), and two IRs (IRa and IRb; 26,097-31,320 bp) (Table 1). The plastomes contained 77-79 protein coding genes, seven to 10 rRNA genes and 36-38 tRNA genes arranged in the same order. Plastome comparisons revealed no clear expansions or contractions in the IR regions (Figure 1). The LSC/IR boundaries of 17 Polygonoideae plastomes were all located at the rps19 and trnH-GUG, while the SSC/IR boundaries were all located at ndhF and rps15 (Figure 1).
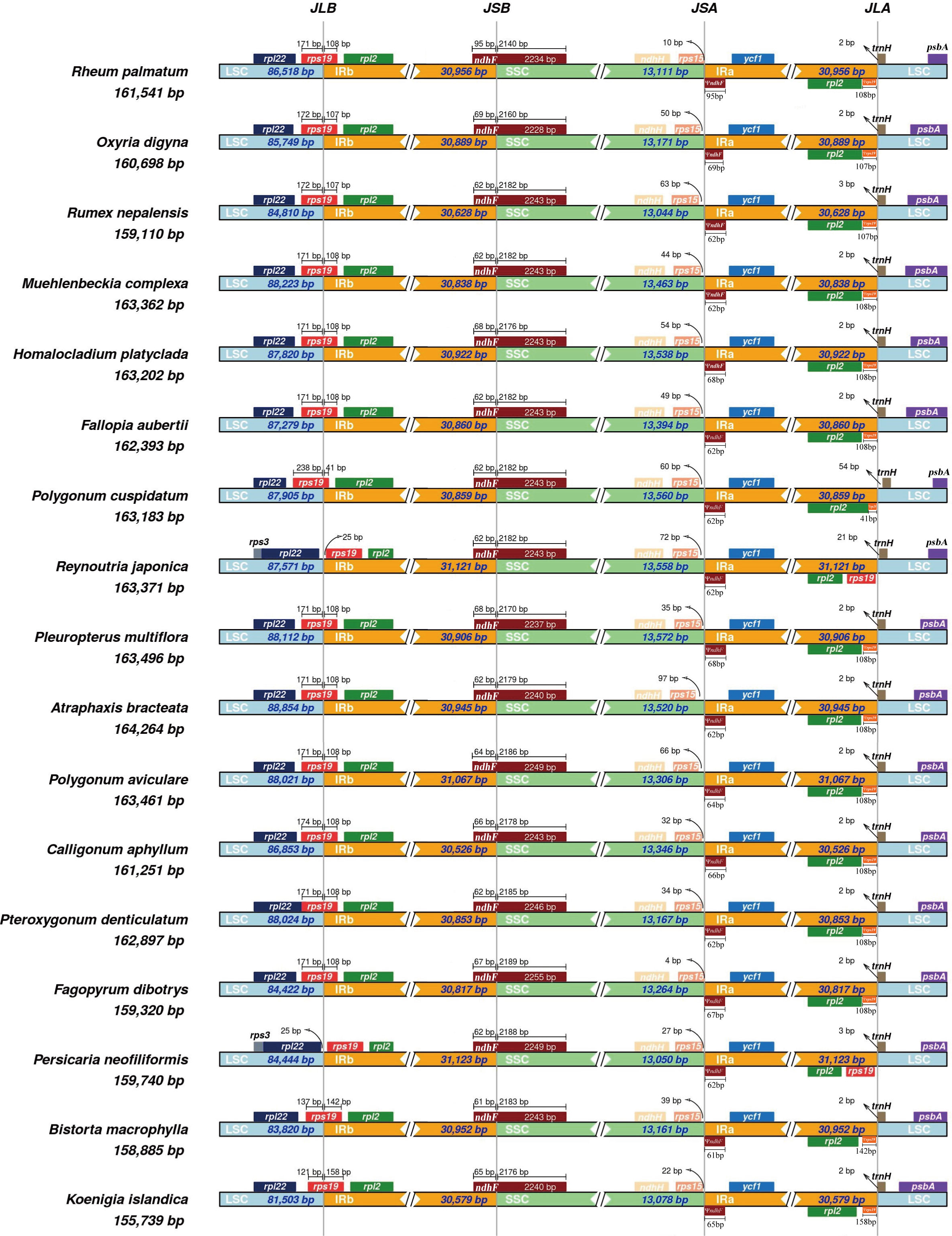
Figure 1 The IR/SSC borders in the plastomes of 17 genera of Polygonoideae. JLB, JSB, JSA and JLA denote the junction sites of LSC/IRb, SSC/IRb, SSC/IRa and LSC/IRa respectively.
Evolutionary rates of plastomes
Nucleotide diversity (pi) and Watterson’s theta estimator (θ) of the 74 shared PCGs were calculated in DNAsp v6 to assess the sequence divergence level of Polygonoideae (Rozas et al., 2017). Among the 74 PCGs, pi values ranged from 0.0066 (ndhB) to 0.0991 (ndhF) (Figure S1A, Table S2), with highly divergent genes identified as having pi > 0.08. Four genes were detected as highly divergent including ccsA, matK, ndhF and rps15 (Figure S1A, Table S2), which can be potential molecular markers for phylogenetic analyses.
Phylogenetic relationships
The alignment matrix of the 74 PCGs was 73,025 bp in length, with 14,430 Parsimony-informative sites and 22,084 variable sites. The analyses of the concatenated matrix and the complete plastome nucleotide sequences matrix generated identical topologies at every node (Figures 2, Figure S2). The phylogenetic analysis with three chloroplast DNA fragments also generated identical relationships although some clades were weakly supported (Figure S3, Table S3). Hence an updated phylogeny was generated by combing all of the above phylogenetic trees (Figure 3). Generally, the phylogenetic analyses revealed Polygonoideae as monophyletic and composed of two subclades, clade A and clade B (Figures 2, 3). Clade A consists of Rumiceae Dumort., Polygoneae Rchb., Calligoneae C. A. Mey., Pteroxygonea T.M.Schust. & Reveal and Fagopyreae Yonek. While Clade B contains only the tribe Persicarieae Dumort. Rumiceae consists of three genera: Rheum, Oxyria and Rumex. Polygoneae consists of seven genera. Calligoneae, Pteroxygonea and Fagopyreae each consist of one genus. Persicarieae consists of three genera. Both the 74-PCGs tree and the tree based on three chloroplast loci revealed that Eriogonoideae is not monophyletic (Figures 2, S2).
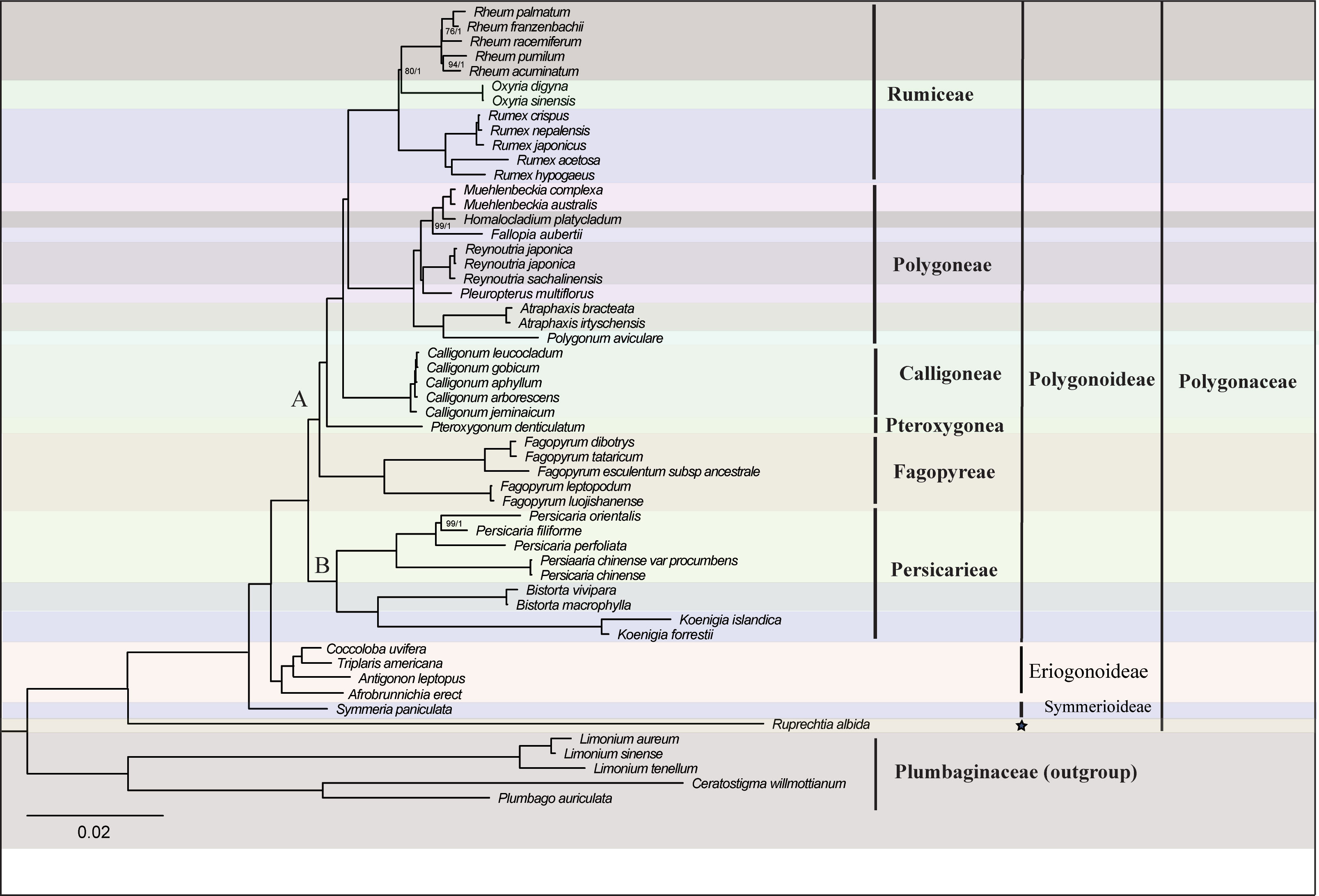
Figure 2 Phylogenetic tree of 54 taxa using maximum likelihood (ML) and Bayesian inference (BI) based on 74 shared genes. Maximum likelihood bootstrap values (BS) and posterior probabilities (PP) are shown at nodes. Branches with no values listed have 100% BS and PP of 1.0. Pentagram represents uncertain classification treatment.
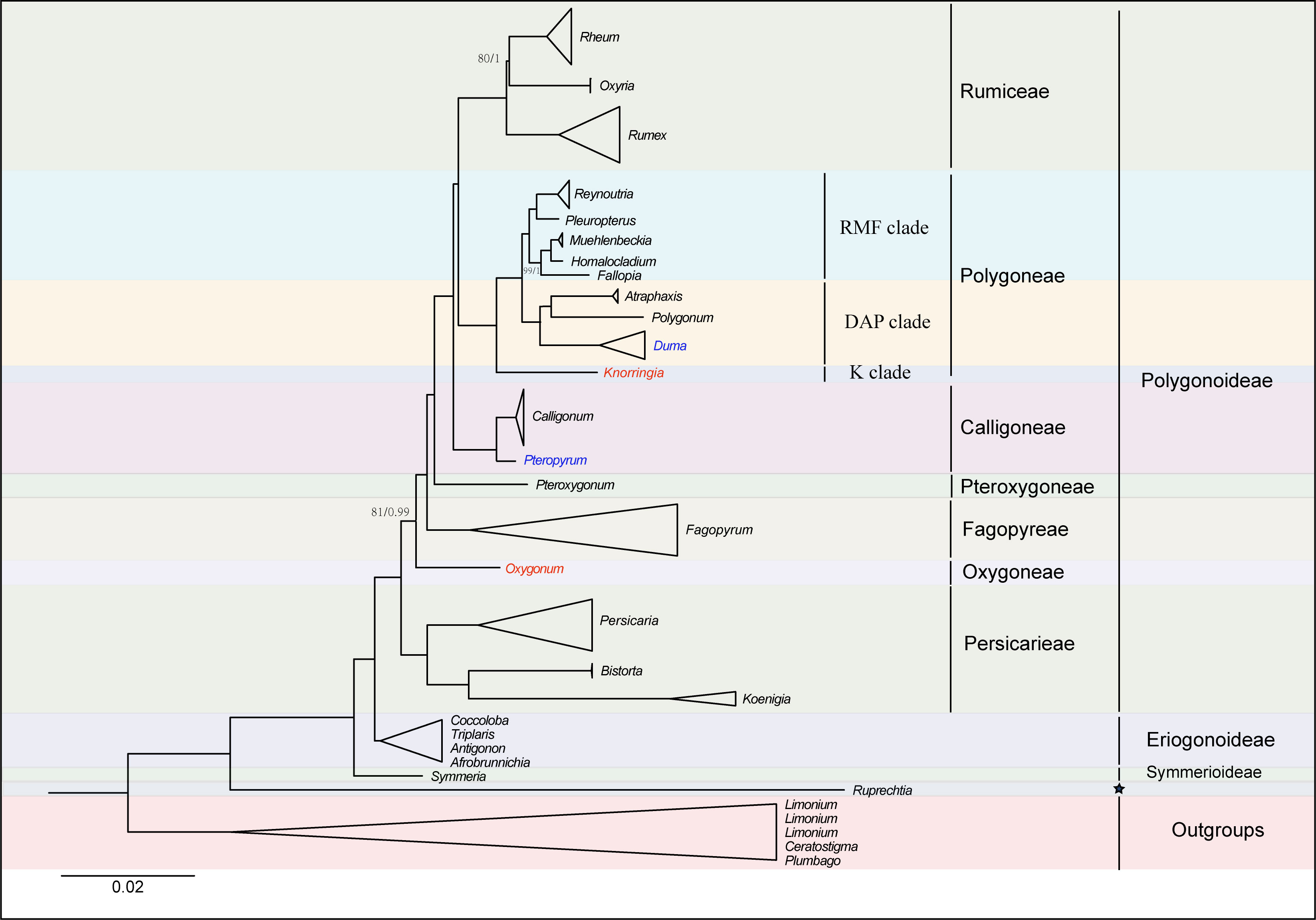
Figure 3 An update phylogenetic tree based on 74 shared PCGs and chloroplast fragments, with positions of species in red being generated from chloroplast fragments. Maximum likelihood bootstrap values (BS) and posterior probabilities (PP) are shown at nodes. Branches with no values listed have 100% BS and PP of 1.0 except for species with blue. Positions of species with blue were generated from previous studies (Schuster et al., 2011, 2015). Pentagram represents uncertain classification treatment.
Time estimation and biogeographic analysis
Results of the dating analyses suggested that Polygonoideae diverged from other members of Polygonaceae during the late Paleocene (54.9 Ma, 95% HPD: 45.2-65.6 Ma). The split of the two major clades in Polygonoideae occurred in the early Eocene (50.45 Ma, 95% HPD: 41.6-60.1 Ma). In Clade B (Persicarieae), Persicaria and the other two genera (Bistora and Koenigia) diverged during the Eocene (42.5 Ma, 95% HPD: 32.3-52.4 Ma) (Figure 4). In Clade A, Fagopyreae and the remaining tribes also diverged during the Eocene (46.8 Ma, 95% HPD: 37.6-55.6 Ma); Pteroxygonea diverged during the Eocene (43.5 Ma, 95% HPD: 34.9-51.6 Ma) and Calligoneae diverged during the late Eocene (39.5 Ma, 95% HPD: 32.1-47.2 Ma). The divergence of Rumiceae and Polygoneae was estimated to have occurred during the late Eocene (37.1 Ma, 95% HPD: 29.9-44.3 Ma). In Polygoneae, the included genera diverged during the Oligocene (30.7 Ma, 95% HPD: 25.2-36.9 Ma) (Figure 4). In Rumiceae, genera diverged during the late Oligocene (25.9 Ma, 95% HPD: 16.8-34.4 Ma); Rheum and Oxyria diverged during the early Miocene (22.4 Ma, 95% HPD: 13.3-31.7 Ma) (Figure 4).
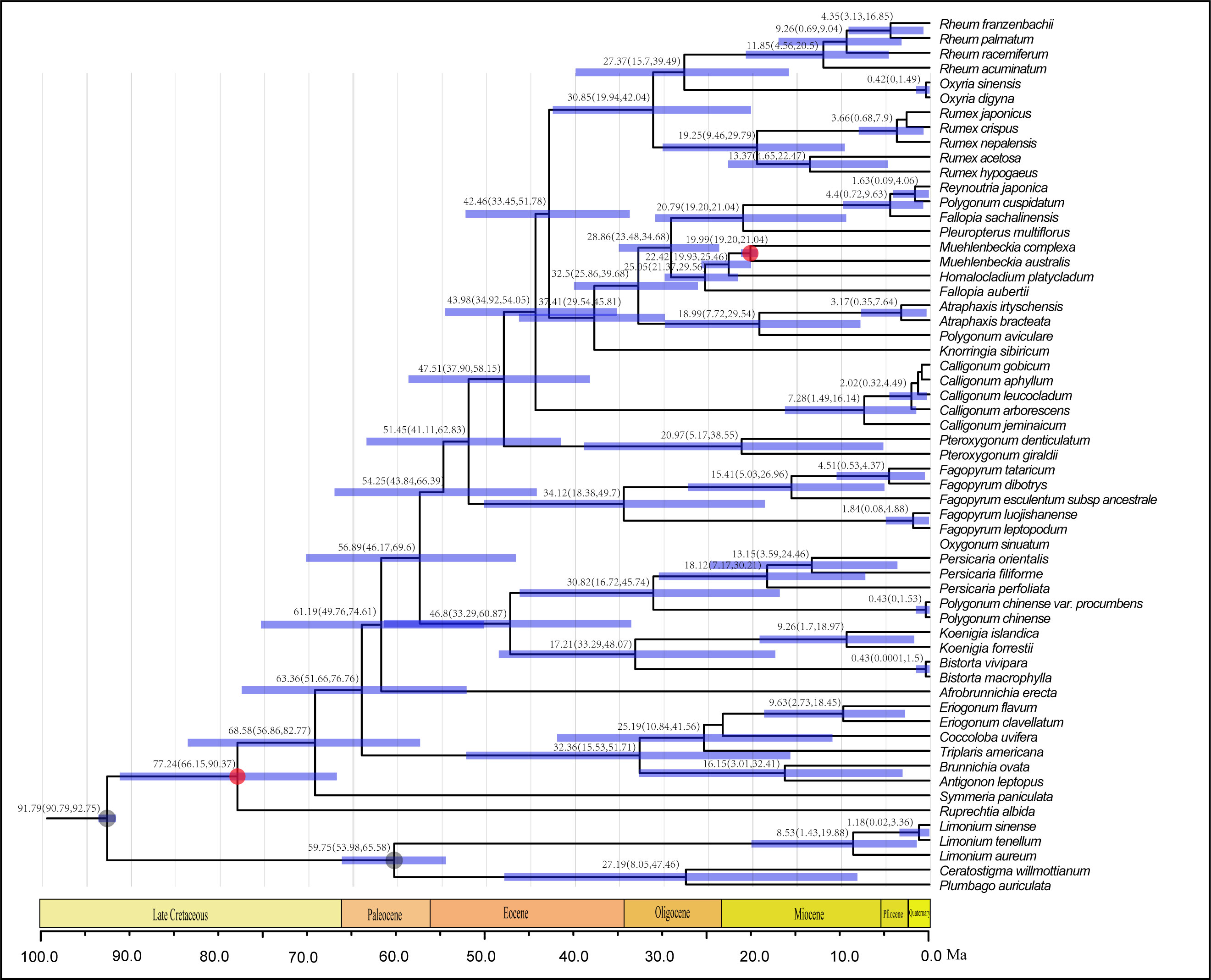
Figure 4 BEAST-derived chronogram of Polygonaceae based on three chloroplast fragments with two fossil calibration (red circles) and two second calibration point (gray circles), fossils’ information were provided in Supplementary Methods S1. Numbers above the tree branches represented mean divergent ages and 95% confidence interval of each node. Blue bars indicate the 95% highest posterior density (HPD) confidence intervals for node ages (Ma).
Results from the biogeographic analysis suggest that the ancestor of Polygonoideae is likely from Asia. Much of the lineage of Polygonoideae was reconstructed with an Asian origin (Figure 5). A total of two vicariance and 26 dispersal events were detected (Figure 5). Migrations to Europe and North America were common and most occurred after the Miocene. Dispersals to South America and Oceania were also detected, with one vicariance event occurring in Asia and Africa during the Eocene, while another vicariance event in this area occurred during the Oligocene.
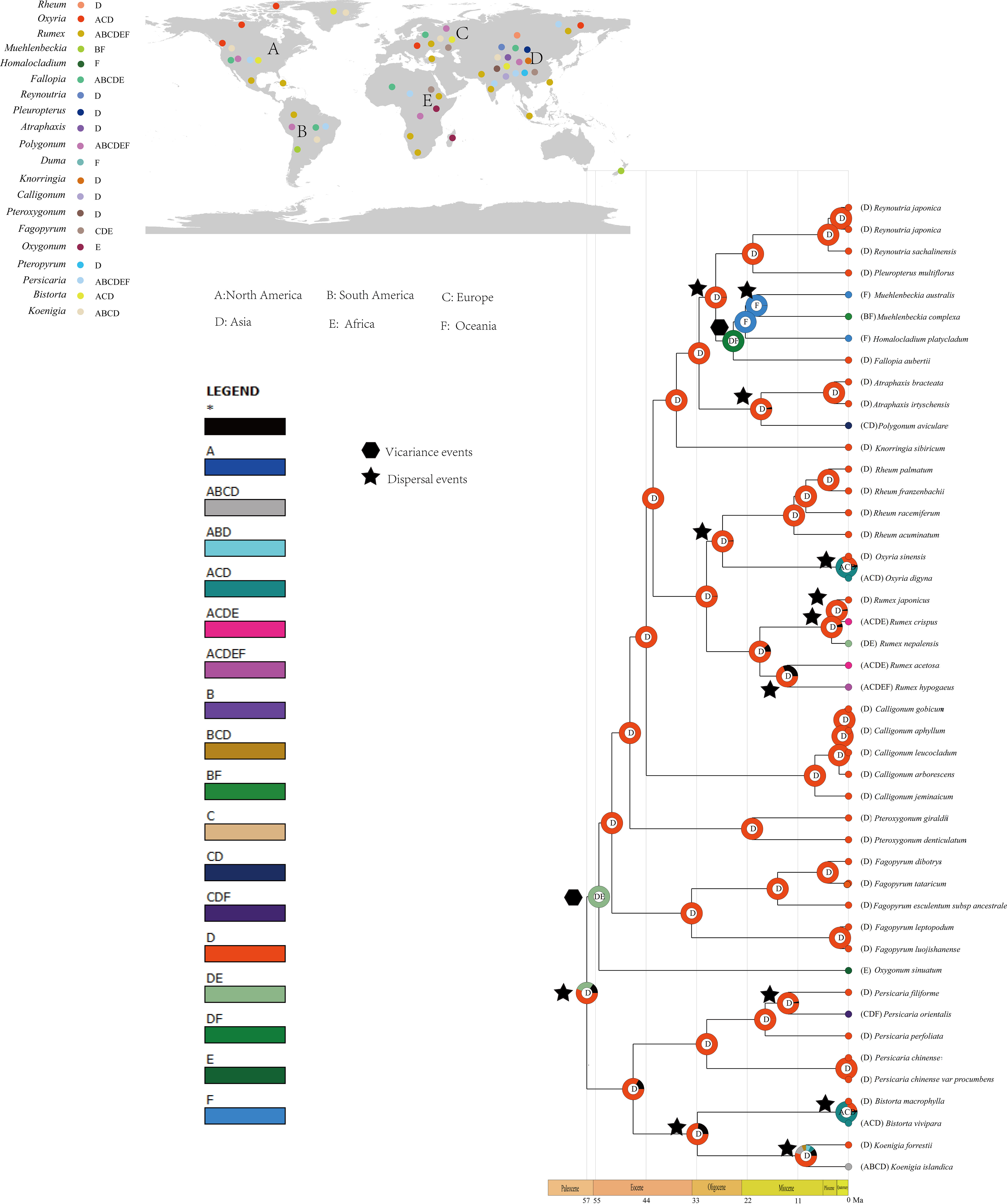
Figure 5 Ancestral area reconstruction of Polygonaceae. Figure of upper left shows the distribution of each genus.
Discussion
Phylogenetic analyses
The monophyly of Polygonaceae is supported by the analyses in the current study and those in previous studies (Sanchez and Kron, 2008; Sanchez et al., 2009; Schuster et al., 2015). The definition of different subfamilies within Polygonaceae based on morphological characters has been controversial over the past 200 years (e.g., Meisner, 1856; Perdrigeat, 1900; Jaretzky, 1925). However, the subfamily Polygonoideae and Eriogonoideae have been widely supported by subsequent molecular analyses (Frye and Kron, 2003; Galasso et al., 2009; Schuster et al., 2015). The monophyly of Polygonoideae is confirmed in the current study (Figures 2, S2) and our results show Polygonoideae consists of seven clades, corresponding to the seven tribes defined by Schuster et al. (2015). Oxygoneae was previously shown as the earliest-diverging clade in Schuster et al. (2015), while our analyses show Persicarieae is the earliest-diverging clade of Polygonoideae, followed by Oxygoneae (Figures 2, S2, S3). The relationships among Oxygoneae, Fagopyreae, Pteroxygoneae, Calligoneae, Polygoneae and Rumiceae in the current analyses are identical with those revealed by Schuster et al. (2015). Our analyses also identified similar clades within Polygoneae as suggested previously (Schuster et al., 2015). Notably, Pleuropterus is included here for the first time in a phylogenetic analysis of Polygonoideae. Unlike previous studies (Sanchez et al., 2009; Sanchez et al., 2011; Schuster et al., 2013; Schuster et al., 2015), the current analyses resolved relationships within Rumiceae; Rheum and Oxyria form a sister relationship with 80/1 (bootstrap and posterior probability) support value. Although several genera including Pteropyrum and Parogonum are absent from the phylogenetic analyses of Polygonoideae here, the relationships among different Polygonoideae tribes are well resolved with high support. Despite our sampling of Eriogonoideae being inadequate for phylogenetic analyses of the subfamily, a nonmonophyletic signal for Eriogonoideae is revealed (Figures 2, S2). The nonmonophyly of Eriogonoideae has also been reported in Sanchez et al. (2009). Both Symmeria and Ruprechtia fall outside of the large clade comprised by Polygonoideae and some Eriogonoideae genera (Figures 2, S2), suggesting dividing Polygonaceae into three or more subfamilies is reasonable, which needs to be confirmed by more adequate sampling and sequencing in the future.
Divergence times and biogeography
Divergence time estimation and S-DIVA analysis revealed an Asia origin of Polygonoideae during the Eocene, with subsequent migrations primarily to Europe and North America (Figure 5). Asia was inferred as the ancestral distribution area of all Polygonoideae tribes except for Oxygoneae (Figure 5). Previous studies of taxa displaying intercontinental disjunction also found these taxa more often originated in Asia, especially in the Qinghai-Tibet Plateau (QTP) region (Morley, 2003; Nie et al., 2013; Sun et al., 2017; Zhang et al., 2019b). Some taxa, e.g., Carex (Cyperaceae), Urtica (Urticaceae) and Balsaminaceae, have similar distributions as Polygonoideae (Figure 5), following dispersal and vicariance events after origins in Asia, which have played key roles in shaping current distribution patterns (Yuan et al., 2004; Huang et al., 2019; Martín‐Bravo et al., 2019). Although the breakup of the Gondwanan supercontinent may have resulted in a disjunct distribution pattern in lineages (Raven and Axelrod, 1974; Conti et al., 2002), many lineages are found to originate more recently (Figure 5), indicating the possibility of long-distance dispersal occurring during their evolutionary histories. All dispersal events detected in Polygonoideae occurred after the Paleocene, suggesting the Beringia and North Atlantic Land Bridge may have been important routes facilitating the dispersal of Polygonoideae. Overall, we propose seven dispersal routes for Polygonoideae: Asia→North America (Oxyria, Bistortia), Asia→North America→South America (Koenigia), Asia→Europe (Oxyria, Bistorta, Polygonum, Rumex, Persicaria, Koenigia), Asia→Europe→North America (Oxyria, Koenigia, Bistorta), Asia→Africa/Asia→Europe→Africa (Rumex), Asia→Oceania (Rumex, Persicaria), Oceania→South America (Muehlenbeckia).
The Asia to North America route has been widely employed by both gymnosperm and angiosperm lineages (Wang and Ran, 2014). From the Eocene to the late Miocene, the Bering Land Bridge connected East Asia and western North America, making migration between the two continents possible (Hopkins, 1967; Tiffney and Manchester, 2001). All Asia to North America dispersal events detected in Polygonoideae happened after the Eocene (Figures 4, 5), suggesting an important role of the Beringia Land Bridge as a corridor for the dispersals. The Asia to North America then to South America route could have been easily established once the spread from Asia to North America was successful, as previously reported in Chrysosplenium (Saxifragaceae), Munroa (Poaceae), Ephedra (Ephedraceae) and Gunnera (Gunneraceae) (Soltis et al., 2001; Wanntorp and Wanntorp, 2003; Ickert‐Bond et al., 2009; Amarilla et al., 2015). Similarly, long distance dispersal form Asia to Europe is achievable via the Himalayas to the West Pamir Mountains (Tajikistan), passing through the northern Iranian Plateau then to the Caucasus (Figure 6), this route was also detected in Oryza (Poaceae) and Triticum (Poaceae) (Liu et al., 2017; Spengler et al., 2021).
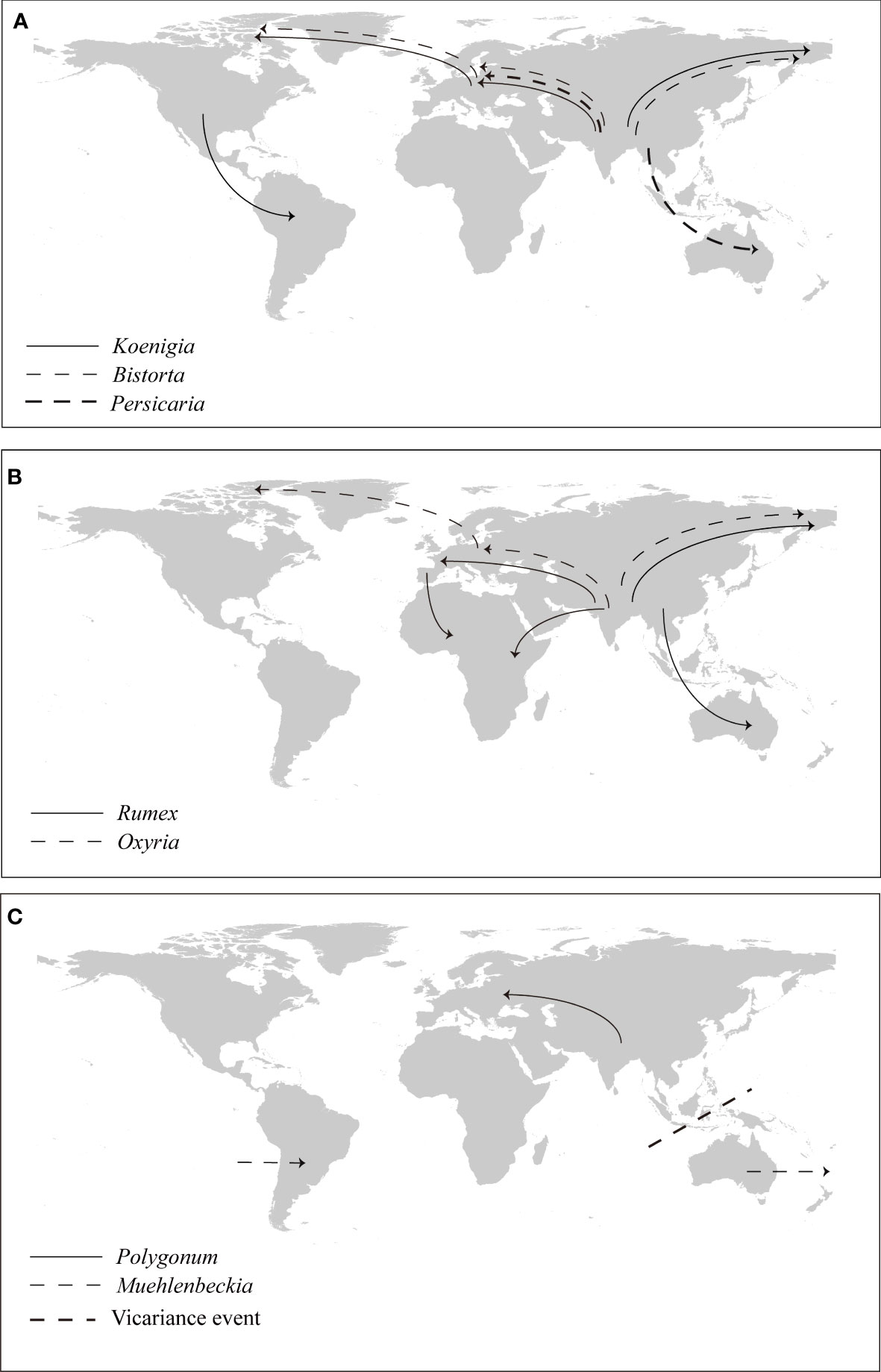
Figure 6 Vicariance and dispersal events in Polygonoideae. (A) Dispersal events in Persicarieae, (B) dispersal events in Rumiceae, (C) Vicariance and dispersal events in Polygoneae.
The Asia to Europe then to North America route was detected for species with a circumarctic distribution in Polygonoideae, after spreading from Asia to the Caucasus of Europe, species continued to spread westward to the Balkan and Carpathian Mountains, along the Alpine Mountains to the Scandinavian Mountains, finally arriving to Greenland, and then to eastern North America to attain a circumarctic distribution (Figure 6). A similar route to a circumarctic distribution has been detected in Sibbaldia (Rosaceae) (Zhang et al., 2019b). As revealed in a previous study, migration from Asia to Africa is likely to have occurred via the rifting of the Indian subcontinent during the Late Cretaceous/Early Paleocene (Conti et al., 2002). However, the Asia to Africa dispersals in Polygonoideae happened during or after the Miocene, ruling out ancient rifting scenarios and indicating a more recent dispersal via steppingstones to the Seychelles, the Comoros, and the Chagos archipelago, about halfway between Africa and Indonesia. The Miocene collision of the Afro-Arabian plate with Asia opened another channel of migration between the continents. For the Asia to Europe then to Africa route, after species spread to Europe, they migrated mainly through Mediterranean regions including the Balkans, Apennines and Iberia to Africa (Figure 6).
Biogeographic connections between Asia and the Southern Hemisphere are rare (Wen et al., 2014), but a series of islands including the Malay island chain and the new Guinea island chain resulting from the collision between Australia and Southeast Asia could have promoted the Asia to Oceania dispersal (2009; Hall, 2002). Generally the breakup of Gondwana can explain the disjunctive distribution pattern among New Zealand, Australia and South America (Givnish and Renner, 2004). A dispersal via seed dispersal related to ocean currents is more reasonable for the America dispersal in Polygonoideae.
Conclusion
This study explored both plastome phylogenomic and biogeographic analyses of the Polygonoideae. The phylogenomic analyses revealed seven lineages corresponding to seven tribes in Polygonoideae, with Persicarieae representing the earliest-diverging lineage. The biogeographic analyses indicated Polygonoideae originated in Asia during the Paleocene, following diversification via long-distance dispersal and vicariance mainly after the Eocene. Generally, the results from the current study provide insights into our comprehensive understanding of the evolution, including origin, dispersal and diversification of Polygonoideae. This study also provides a good example for further study to investigate the evolution pattern of intercontinental disjunctions in a broader phylogenetic framework on a global scale.
Perspective
Considering the extensive distribution of Polygonoideae and some still controversial relationships, e.g., the positions of Persicarieae and Oxygoneae, further sampling covering all genera and distribution ranges of Polygonoideae is necessary. Comprehensive sampling is also a basis for more accurate estimations in the timing of origin and dispersal. Additional studies focused on comparative morphology and transcriptome sequencing are needed to better understand the evolutionary relationships and history of Polygonoideae.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HZ, TD, HS, and HW designed the study. HZ, XZ, YS, and BT analyzed the data and wrote the manuscript. JL, LL, GH, JS, and TK assisted with the sampling and grammatical modifications. All authors read and approved the final manuscript.
Funding
This study was supported by grants from the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502), the Key Projects of the Joint Fund of the National Natural Science Foundation of China (U1802232), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA20050203), the Youth Innovation Promotion Association of Chinese Academy of Sciences (2019382), the Ten Thousand Talents Program of Yunnan Province (202005AB160005) and Project funded by China Postdoctoral Science Foundation (2022M713333).
Acknowledgments
We thank Peter Brownless from Royal Botanic Garden Edinburgh, Xianhui Shen from Xishuangbanna Tropical Botanical Garden and Xiaodong Li from Wuhan Botanical Garden for helping with samples collecting.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.893201/full#supplementary-material
Supplementary Figure 1 | (A) Nucleotide diversity (pi) of 74 protein-coding genes among Polygonaceae and outgroups, genes with high nucleotide diversity (pi > 0.08) are colored in red. (B) Watterson’s theta (θ) of 74 protein-coding genes among Polygonaceae and outgroups.
Supplementary Figure 2 | Phylogenetic tree of 54 taxa using maximum likelihood (ML) and Bayesian inference (BI) based on complete plastome sequences and remove one copy of the IR regions. Maximum likelihood bootstrap values (BS) and posterior probabilities (PP) are shown at nodes. Branches with no values listed have 100% BS and PP of 1.0.
Supplementary Figure 3 | Phylogenetic tree of 60 taxa using maximum likelihood (ML) and Bayesian inference (BI) based on three chloroplast fragments (matK, trnL and rbcL).
References
Amarilla, L. D., Chiapella, J. O., Sosa, V., Moreno, N. C., Anton, A. M. (2015). A tale of north and south America: Time and mode of dispersal of the amphitropical genus Munroa (Poaceae, chloridoideae). Bot. J. Linn. Soc 179, 110–125. doi: 10.1111/boj.12304
Amiryousefi, A., Hyvönen, J., Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Burke, J. M., Sanchez, A. (2011). Revised subfamily classification for polygonaceae, with a tribal classification for eriogonoideae. Brittonia 63, 510–520. doi: 10.1007/s12228-011-9197-x
Conti, E., Eriksson, T., Schönenberger, J., Sytsma, K. J., Baum, D. A. (2002). Early tertiary out-of-India dispersal of crypteroniaceae: evidence from phylogeny and molecular dating. Evolution 56, 1931–1942. doi: 10.2307/3094636
Darriba, D., Posada, D., Kozlov, A. M., Stamatakis, A., Morel, B., Flouri, T. (2020). ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37, 291–294. doi: 10.1093/molbev/msz189
Dierckxsens, N., Mardulyn, P., Smits, G. (2017). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18–e18. doi: 10.1093/nar/gkw955
Dong, W., Xu, C., Wen, J., Zhou, S. (2020). Evolutionary directions of single nucleotide substitutions and structural mutations in the chloroplast genomes of the family calycanthaceae. BMC Evol. Biol. 20, 1–12. doi: 10.1186/s12862-020-01661-0
Drummond, A. J., Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 1–8. doi: 10.1186/1471-2148-7-214
Fritsch, P. W., Manchester, S. R., Stone, R. D., Cruz, B. C., Almeda, F. (2015). Northern hemisphere origins of the amphi-pacific tropical plant family symplocaceae. J. Biogeogr. 42, 891–901. doi: 10.1111/jbi.12442
Frye, A. S. L., Kron, K. A. (2003). rbcL phylogeny and character evolution in polygonaceae. Syst. Bot. 28, 326–332. doi: 10.2307/3094001
Galasso, G., Banfi, E., De Mattia, F., Grassi, F., Sgorbati, S., Labra, M. (2009). Molecular phylogeny of Polygonum l. sl (Polygonoideae, polygonaceae), focusing on European taxa: preliminary results and systematic considerations based on rbcL plastidial sequence data. Atti della Società italiana di Sci. naturali e del Museo civico di storia naturale di Milano 150, 113–148.
Givnish, T. J., Renner, S. S. (2004). Tropical intercontinental disjunctions: Gondwana breakup, immigration from the boreotropics, and transoceanic dispersal. Int. J. Plant Sci. 165, S1–S6. doi: 10.1086/424022
Hall, R. (2002). Cenozoic Geological and plate tectonic evolution of SE Asia and the SW pacific: computer-based reconstructions, model and animations. J. Asian Earth Sci. 20, 353–431. doi: 10.1016/s1367-9120(01)00069-4
Hall, R. (2009). Southeast asia's changing palaeogeography. blumea-biodiversity. Evol. Biogeogr. Plants 54, 148–161. doi: 10.3767/000651909x475941
Huang, X., Deng, T., Moore, M. J., Wang, H., Li, Z., Lin, N., et al. (2019). Tropical Asian origin, boreotropical migration and long-distance dispersal in nettles (Urticeae, urticaceae). Mol. Phylogen. Evol. 137, 190–199. doi: 10.1016/j.ympev.2019.05.007
Huelsenbeck, J. P., Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Ickert-Bond, S. M., Rydin, C., Renner, S. S. (2009). A fossil-calibrated relaxed clock for ephedra indicates an oligocene age for the divergence of Asian and new world clades and Miocene dispersal into south America. J. Syst. Evol. 47, 444–456. doi: 10.1111/j.1759-6831.2009.00053.x
Jaretzky, R. (1925). Beiträge zur systematik der polygonaceae unter berücksichtigung des oxymethylanthrachinon-vorkommens. Repert. novarum specierum regni vegetabilis 22, 49–83. doi: 10.1002/fedr.19250220402
Jung, J., Kim, C., Kim, J.-H. (2021). Insights into phylogenetic relationships and genome evolution of subfamily commelinoideae (Commelinaceae mirb.) inferred from complete chloroplast genomes. BMC Genomics 22, 1–12. doi: 10.1186/s12864-021-07541-1
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kubitzki, K., Rohwer, J., Bittrich, V. (1990). The families and genera of vascular plants. Taxon 54, 574. doi: 10.2307/25065407
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547. doi: 10.1093/molbev/msy096
Liu, X., Lister, D. L., Zhao, Z., Petrie, C. A., Zeng, X., Jones, P. J., et al. (2017). Journey to the east: Diverse routes and variable flowering times for wheat and barley en route to prehistoric China. PloS One 12, e0187405. doi: 10.1371/journal.pone.0187405
Li, J., Wang, S., Yu, J., Wang, L., Zhou, S. (2013). A modified CTAB protocol for plant DNA extraction. Chin. Bull. Bot. 48, 72. doi: 10.1101/pdb.prot5177
Manchester, S. R., O’leary, E. L. (2010). Phylogenetic distribution and identification of fin-winged fruits. Bot. Rev. 76, 1–82. doi: 10.1007/s12229-010-9041-0
Martín-Bravo, S., Jiménez-Mejías, P., Villaverde, T., Escudero, M., Hahn, M., Spalink, D., et al. (2019). A tale of worldwide success: Behind the scenes of Carex (Cyperaceae) biogeography and diversification. J. Syst. Evol. 57, 695–718. doi: 10.1111/jse.12549
Matzke, N. J. (2014). Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 63, 951–970. doi: 10.1093/sysbio/syu056
Meisner, C. F. (1856). “Polygonaceae”, in de Candolle, A, ed. Prodomus systematis naturalis regni vegetabilis. (Paris: V Masson) 14, 1–186
Morley, R. J. (2003). Interplate dispersal paths for megathermal angiosperms. Perspect. Plant Ecol. Evol. Syst. 6, 5–20. doi: 10.1078/1433-8319-00039
Nie, Z. L., Deng, T., Meng, Y., Sun, H., Wen, J. (2013). Post-boreotropical dispersals explain the pantropical disjunction in Paederia (Rubiaceae). Ann. Bot. 111, 873–886. doi: 10.1093/aob/mct053
Nie, Z. L., Sun, H., Manchester, S. R., Meng, Y., Luke, Q., Wen, J. (2012). Evolution of the intercontinental disjunctions in six continents in the ampelopsis clade of the grape family (Vitaceae). BMC Evol. Biol. 12, 1–13. doi: 10.1186/1471-2148-12-17
Perdrigeat, C. A. (1900). Anatomie comparée des polygonées et ses rapports avec la morphologie et la classification. Actes Soc Linn Bordeaux. 55, 1–93.
Pole, M. (1992). Early Miocene flora of the manuherikia group, new zealand. 2. conifers. J. R. Soc N. Z. 22, 287–302. doi: 10.1080/03036758.1992.10420822
Qu, X. J., Moore, M. J., Li, D. Z., Yi, T. S. (2019). PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15, 1–12. doi: 10.1186/s13007-019-0435-7
Rambaut, A. (2009) FigTree v1. 3.1. Available at: http://tree.bio.ed.ac.uk/software/figtree/.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901. doi: 10.1093/sysbio/syy032
Ranwez, V., Douzery, E. J., Cambon, C., Chantret, N., Delsuc, F. (2018). MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 35, 2582–2584. doi: 10.1093/molbev/msy159
Raven, P. H., Axelrod, D. I. (1974). Angiosperm biogeography and past continental movements. Ann. Missouri Bot. Garden 61, 539–673. doi: 10.2307/2395021
Rozas, J., Ferrer-Mata, A., Sánchez-Delbarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Sanchez, A., Kron, K. A. (2008). Phylogenetics of polygonaceae with an emphasis on the evolution of eriogonoideae. Syst. Bot. 33, 87–96. doi: 10.1600/036364408783887456
Sanchez, A., Schuster, T. M., Burke, J. M., Kron, K. A. (2011). Taxonomy of polygonoideae (Polygonaceae): a new tribal classification. Taxon 60, 151–160. doi: 10.1002/tax.601013
Sanchez, A., Schuster, T. M., Kron, K. A. (2009). A large-scale phylogeny of polygonaceae based on molecular data. Int. J. Plant Sci. 170, 1044–1055. doi: 10.1086/605121
Schuster, T. M., Reveal, J. L., Bayly, M. J., Kron, K. A. (2015). An updated molecular phylogeny of polygonoideae (Polygonaceae): Relationships of Oxygonum, Pteroxygonum, and Rumex, and a new circumscription of Koenigia. Taxon 64, 1188–1208. doi: 10.12705/646.5
Schuster, T. M., Setaro, S. D., Kron, K. A. (2013). Age estimates for the buckwheat family polygonaceae based on sequence data calibrated by fossils and with a focus on the amphi-pacific Muehlenbeckia. PloS One 8, e61261. doi: 10.1371/journal.pone.0061261
Schuster, T. M., Reveal, J. L., Kron, K. A. (2011). Phylogeny of Polygoneae (Polygonaceae: Polygonoideae). Taxon 60, 1653–1666. doi: 10.1002/tax.606010
Soltis, D. E., Tago-Nakazawa, M., Xiang, Q. Y., Kawano, S., Murata, J., Wakabayashi, M., et al. (2001). Phylogenetic relationships and evolution in Chrysosplenium (Saxifragaceae) based on matK sequence data. Am. J. Bot. 88, 883–893. doi: 10.2307/2657040
Song, F., Li, T., Burgess, K. S., Feng, Y., Ge, X.-J. (2020). Complete plastome sequencing resolves taxonomic relationships among species of Calligonum L.(Polygonaceae) in China. BMC Plant Biol. 20, 1–15. doi: 10.1186/s12870-020-02466-5
Spengler, R. N., Stark, S., Zhou, X., Fuks, D., Tang, L., Mir-Makhamad, B., et al. (2021). A journey to the West: The ancient dispersal of rice out of East Asia. Rice 14, 1–18. doi: 10.1186/s12284-021-00518-4
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Suchard, M. A., Lemey, P., Baele, G., Ayres, D. L., Drummond, A. J., Rambaut, A. (2018). Bayesian Phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016. doi: 10.1093/ve/vey016
Sun, H., Zhang, J. W., Deng, T., Boufford, D. E. (2017). Origins and evolution of plant diversity in the hengduan mountains, China. Plant Diversity 39, 161. doi: 10.1016/j.pld.2017.09.004
Tiffney, B. H., Manchester, S. R. (2001). The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the northern hemisphere tertiary. Int. J. Plant Sci. 162, S3–S17. doi: 10.1086/323880
Wang, X. Q., Ran, J. H. (2014). Evolution and biogeography of gymnosperms. Mol. Phylogen. Evol. 75, 24–40. doi: 10.1016/j.ympev.2014.02.005
Wanntorp, L., Wanntorp, H. E. (2003). The biogeography of Gunnera l.: vicariance and dispersal. J. Biogeogr. 30, 979–987. doi: 10.1046/j.1365-2699.2003.00895.x
Wen, J., Xie, D. F., Price, M., Ren, T., Deng, Y. Q., Gui, L. J., et al. (2021). Backbone phylogeny and evolution of apioideae (Apiaceae): New insights from phylogenomic analyses of plastome data. Mol. Phylogen. Evol. 161, 107183. doi: 10.1016/j.ympev.2021.107183
Wen, J., Zhang, J. Q., Nie, Z. L., Zhong, Y., Sun, H. (2014). Evolutionary diversifications of plants on the qinghai-Tibetan plateau. Front. Genet. 5. doi: 10.3389/fgene.2014.00004
Yang, B. B., Li, L. D., Liu, J. Q., Zhang, L. S. (2021). Plastome and phylogenetic relationship of the woody buckwheat Fagopyrum tibeticum in the qinghai-Tibet plateau. Plant Diversity 43, 198–205. doi: 10.1016/j.pld.2020.10.001
Yang, T., Lu, L. M., Wang, W., Li, J. H., Manchester, S. R., Wen, J., et al. (2018). Boreotropical range expansion and long-distance dispersal explain two amphi-pacific tropical disjunctions in sabiaceae. Mol. Phylogen. Evol. 124, 181–191. doi: 10.1016/j.ympev.2018.03.005
Yao, G., Jin, J. J., Li, H. T., Yang, J. B., Mandala, V. S., Croley, M., et al. (2019). Plastid phylogenomic insights into the evolution of caryophyllales. Mol. Phylogen. Evol. 134, 74–86. doi: 10.1016/j.ympev.2018.12.023
Yoder, A. D., Nowak, M. D. (2006). Has vicariance or dispersal been the predominant biogeographic force in Madagascar? only time will tell. Annu. Rev. Ecol. Evol. Syst. 37, 405–431. doi: 10.2307/30033838
Yuan, Y. M., Song, Y., Geuten, K., Rahelivololona, E., Wohlhauser, S., Fischer, E., et al. (2004). Phylogeny and biogeography of balsaminaceae inferred from ITS sequences. Taxon 53, 391–404. doi: 10.2307/4135617
Yu, Y., Harris, A. J., Blair, C., He, X. (2015). RASP (Reconstruct ancestral state in phylogenies): a tool for historical biogeography. Mol. Phylogen. Evol. 87, 46–49. doi: 10.1016/j.ympev.2015.03.008
Yu, Y., Harris, A., He, X. (2010). S-DIVA (Statistical dispersal-vicariance analysis): a tool for inferring biogeographic histories. Mol. Phylogen. Evol. 56, 848–850. doi: 10.1016/j.ympev.2010.04.011
Zhang, H. J., Feng, T., Landis, J. B., Deng, T., Zhang, X., Meng, A. P., et al. (2019b). Molecular phylogeography and ecological niche modeling of Sibbaldia procumbens sl (Rosaceae). Front. Genet. 10. doi: 10.3389/fgene.2019.00201
Zhang, D., Gao, F. L., Jakovlić, I., Zou, H., Zhang, J., Li, W. X., et al. (2019a). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, H. J., Zhang, X., Landis, J. B., Sun, Y. X., Sun, J., Kuang, T. H., et al. (2021). Phylogenomic and comparative analyses of Rheum (Polygonaceae, polygonoideae). J. Syst. Evol. doi: 10.1111/jse.12814
Keywords: dispersal routes, phylogenomic, plastomes, Polygonoideae, biogeography
Citation: Zhang H, Zhang X, Sun Y, Landis JB, Li L, Hu G, Sun J, Tiamiyu BB, Kuang T, Deng T, Sun H and Wang H (2022) Plastome phylogenomics and biogeography of the subfam. Polygonoideae (Polygonaceae). Front. Plant Sci. 13:893201. doi: 10.3389/fpls.2022.893201
Received: 10 March 2022; Accepted: 12 September 2022;
Published: 05 October 2022.
Edited by:
Manjusha Verma, Division of Genomic Resources, (ICAR), IndiaReviewed by:
Wen-Bin Yu, Xishuangbanna Tropical Botanical Garden, (CAS), ChinaAndrés J. Cortés, Colombian Corporation for Agricultural Research (AGROSAVIA), Colombia
Copyright © 2022 Zhang, Zhang, Sun, Landis, Li, Hu, Sun, Tiamiyu, Kuang, Deng, Sun and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Deng, ZGVuZ3Rhb0BtYWlsLmtpYi5hYy5jbg==; Hang Sun, c3VuaGFuZ0BtYWlsLmtpYi5hYy5jbg==; Hengchang Wang, aGN3YW5nQHdiZ2Nhcy5jbg==
†These authors contributed equally to this study
 Huajie Zhang
Huajie Zhang Xu Zhang
Xu Zhang Yanxia Sun
Yanxia Sun Jacob B. Landis
Jacob B. Landis Lijuan Li1,2,3
Lijuan Li1,2,3 Guangwan Hu
Guangwan Hu Jiao Sun
Jiao Sun Bashir B. Tiamiyu
Bashir B. Tiamiyu Tao Deng
Tao Deng Hang Sun
Hang Sun Hengchang Wang
Hengchang Wang