- 1UMR Qualisud, Avignon Université, Avignon, France
- 2UR GAFL, INRAE PACA, Avignon, France
The transcriptomic and metabolomic responses of peach to Myzus persicae infestation were studied in Rubira, an accession carrying the major resistance gene Rm2 causing antixenosis, and GF305, a susceptible accession. Transcriptome and metabolome showed both a massive reconfiguration in Rubira 48 hours after infestation while GF305 displayed very limited changes. The Rubira immune system was massively stimulated, with simultaneous activation of genes encoding cell surface receptors involved in pattern-triggered immunity and cytoplasmic NLRs (nucleotide-binding domain, leucine-rich repeat containing proteins) involved in effector-triggered immunity. Hypersensitive reaction featured by necrotic lesions surrounding stylet punctures was supported by the induction of cell death stimulating NLRs/helpers couples, as well as the activation of H2O2-generating metabolic pathways: photorespiratory glyoxylate synthesis and activation of the futile P5C/proline cycle. The triggering of systemic acquired resistance was suggested by the activation of pipecolate pathway and accumulation of this defense hormone together with salicylate. Important reduction in carbon, nitrogen and sulphur metabolic pools and the repression of many genes related to cell division and growth, consistent with reduced apices elongation, suggested a decline in the nutritional value of apices. Finally, the accumulation of caffeic acid conjugates pointed toward their contribution as deterrent and/or toxic compounds in the mechanisms of resistance.
Introduction
The green peach aphid (GPA), Myzus persicae, is a polyphagous sap-sucking pest found throughout the world, attacking many crop species and whose harmful nature is largely due to its ability to transmit plant viruses. The primary host of this aphid is peach (Prunus persica L. Batsch), on which it overwinters in the egg stage. Among the peach genetic resources only a few accessions showing strong resistance to GPA have been characterized, all carrying a locus located at the bottom of chromosome 1: “Weeping Flower Peach”, an ornamental genotype carrying Rm1 (Resistance to Myzus 1) gene (Monet and Massonié, 1994; Pascal et al., 2017), Rubira, a red-leaf rootstock carrying Rm2 (Lambert and Pascal, 2011) and “Fen Shouxing”, a semi-wild selection carrying Rm3 (Niu et al., 2018). Major aphid resistance loci have also been described and cloned in other species: the root-knot nematode resistance gene Mi-1.2 (Milligan et al., 1998) confers resistance to Macrosiphum euphorbiae in tomato (Rossi et al., 1998; Vos et al., 1998) and Vat against Aphis gossypii in melon (Dogimont et al., 2014). Mi-1.2 was the first insect-specific resistance gene to be cloned and the first example of a resistance gene active against distant organisms since it confers resistance to root-knot nematode, M. euphorbiae, psyllids (Casteel et al., 2006) and whiteflies (Nombela et al., 2003) in tomato. Furthermore, Pallipparambil et al. (2015) demonstrated an extended spectrum of resistance controlled by this gene, as Mi-1.2 negatively impacts non-phloem sap-feeding organisms like larvae of Oirus insidiosus, which are beneficial zoophytophagous predators that prey on aphids. The Vat gene was found to confer dual resistance to Aphis gossypii and to viruses it transmits (Dogimont et al., 2014). This resistance involved a localised hypersensitive response (HR) (Villada et al., 2009), negatively influenced aphid nutrition and has been shown effective against most Aphis gossypii clones (Boissot et al., 2016a; Boissot et al., 2016b).
Mi-1.2 and Vat have been found to encode effector-triggered Immunity (ETI) receptors, i.e. resistance proteins (R) containing remarkable Nucleotide-Binding and Leucine-Rich Repeat domains (NLR or NBS-LRR) (see Ngou et al., 2022a and Ngou et al., 2022b for review on NLRs). These immunity proteins are divided into three classes according to their N-terminal domain: Toll/interleukin-1 receptor (TNLs), coiled coil domain (CNLs) or RPW8-like coiled coil domain (RNLs) (Jones et al., 2016). Both Mi-1.2 and Vat were characterised as CNLs (Dogimont et al., 2014) and a strong TNL candidate for peach Rm3 has been recently identified (Pan et al., 2022). ETI is based on intracellular NLR sensors acting alone or in pairs (Feehan et al., 2020) and is one of the two major layers of the plant immune system. The other layer is the pattern-triggered Immunity (PTI), made up of pattern recognition receptors (PRRs), i.e. cell surface sensors activated by pathogenesis-associated molecular patterns (PAMPs), herbivore-associated molecular patterns (HAMPS) or damage-associated molecular patterns (DAMPS). PRRs include receptor-like kinases (RLKs) and receptor-like proteins (RLPs), which comprise a variable extracellular domain allowing the recognition of various ligands, a transmembrane domain and a cytoplasmic kinase in the case of RLKs (Tang et al., 2017) which transmits a signal to intracellular proteins once activated. The selection pressure exerted by PTI on pathogen populations has led to the emergence of populations carrying effector proteins that neutralize resistance mechanisms and restore virulence through Effector Triggered Susceptibility (ETS, Ngou et al., 2022a). This, in turn, resulted in the evolution of plant populations that carry cytosolic receptors capable of specifically recognize pathogen effectors and initiating the strong defenses that constitute ETI (Jones and Dangl, 2006).
Aphids are piercing-sucking insects that feed exclusively on phloem sap, which they collect with their specialized mouthpiece, the stylet. Their feeding behavior comprises several phases generating signals capable of triggering plant defense: the stylet and the secreted gel-like saliva can produce PAMPs and DAMPs during intercellular insertion and mesophyll cells probing phases, while the watery saliva secreted when sucking phloem sap from sieve elements contains protein effectors that suppress plant defense, (Will et al., 2007; Bos et al., 2010), contributing to the stealthy nature of aphids, but that may potentially trigger ETI. Indeed, defense against aphids proved to involve both PTI and ETI. The role of PTI in defense against aphid infestation was demonstrated by application of eliciting extracts of GPA (Prince et al., 2014) and by the study of the Arabidopsis bak1 mutant, deficient in the BAK1 RLK coreceptor (Prince et al., 2014; Chaudhary et al., 2014; Tungadi et al., 2021).
Omic studies have uncovered other molecular constituents of plant-sap-sucking insects including aphids interplay (reviewed by Zogli et al., 2020) and many of them concern the model Arabidopsis thaliana-GPA interaction (Louis and Shah, 2013). Kerchev et al. (2013), for example, showed the importance of redox control, SA and abscisic acid (ABA) signalling pathways in Arabidopsis after infestation by GPA. Analysis of transcriptional responses to Macrosiphum euphorbiae in tomato also highlighted the involvement of antioxidant mechanisms and hormonal regulations: SA, jasmonic acid (JA), ethylene and brassinosteroids (BRs) and the activation of callose synthesis genes, dedicated to blocking symplastic connections (Kuśnierczyk et al., 2008; Coppola et al., 2013). In peach, a transcriptomic study of “Fen Shouxing” after GPA infestation revealed the activation of thousands of genes involved in signalling cascades or resistance mechanisms, such as redox modifications, calcium fluxes, mitogen-activated protein kinases (MAPKs), phytohormones, transcription factors, pathogenesis-related proteins (PRs) and enzymes of secondary metabolic pathways (Niu et al., 2018). Metabolomic studies on plant-aphid interactions are less common but crucial to uncovering metabolites involved in stress signalling and defense (Zogli et al., 2020) as well as reconfigurations of carbon and nitrogen fluxes controlling the growth and nutritional value of infested plants. Gao et al. (2021) reported alterations in central and secondary metabolism of rose in response to Macrosiphum rosivorum and Kuśnierczyk et al. (2008) confirmed the antibiotic role of secondary metabolites such as glucosinolates and the alkaloid camalexin in Arabidopsis.
Molecular responses of Arabidopsis to GPA have been extensively studied, but ecotypes of this secondary host exhibit only partial antibiosis and antixenosis (Cabrera y Poch et al., 1998) and major R genes capable of triggering ETI have never been described in this species. In this respect, peach, as primary host of GPA with lines owning Rm genes conferring total antixenosis, is a relevant species to decipher all layers of the plant immune mechanisms involved in strong resistance to these phloem-feeders. In the present study, we establish a detailed picture of signal transduction and metabolic mechanisms triggered by activation of Rm2 upon GPA infestation. We analyzed the responses of Rubira, carrying Rm2, and compared it to the susceptible accession GF305, after 48 hours of infestation, a duration required and sufficient to trigger induced resistance in Rubira (Sauge et al., 2011). We evaluated the global metabolic and transcriptomic reprogramming and looked for differentially expressed genes involved in PTI and ETI, hypersensitive response (HR) and systemic acquired resistance (SAR). We also analyzed metabolic reconfigurations underlying the production of hormonal signals and defense compounds and examined pathways reflecting a reorientation of growth fluxes and activation of oxidative burst.
Materials and methods
Plant material
Two highly homozygous seedling rootstocks selected by INRAE and contrasting for their resistance to GPA were used: the susceptible rootstock GF305 and the red-leaf rootstock Rubira, carrying the dominant resistance gene Rm2 (Lambert & Pascal, 2011). Seeds were produced by natural inbreeding in isolated orchards of a commercial nursery (Pépinières Lafond, Valréas, France). Disinfected seeds were placed in petri dishes containing humidified perlite and were stratified during 3 months in dark cold room at 4°C. After radicle emergence, seeds were sown, grown in a greenhouse and kept free of pests and diseases without spraying or biological control. Axillary shoots were removed to keep only the main stem. 8 weeks after sowing, 10 plants of each genotype were acclimated during one week in the air-conditioned room where experiments took place, at 22°C with a 16 h day/8 h night photoperiod. The pots were placed in large trays filled with water to prevent the movement of aphids from one plant to another during the experiment.
Aphids
The Myzus persicae clonal line used, Mp06, was obtained in 2013 from a single egg collected in a peach orchard (Avignon, France). Since then, parthenogenetic apterous females were continuously reared on GF305 seedlings in an air-conditioned room at 22°C with 16h day/8h night photoperiod.
Infestation and plant sampling
One week before infestation, parthenogenetic females were installed on GF305 seedlings for laying and removed after 48 hours to obtain a nymph population of the same age. The day of infestation, the synchronized adult females obtained were placed for 4 h at 22°C to generate a fasting period. Ten plants of GF305 and Rubira were infested by placing carefully with a brush 10 aphids on their apex. After 48h, aphids and nymphs were pulled out with a brush, control plants (10 for each genotype) were equally stimulated with the brush, and the apices of control and infested plants were cut below the second elongated internode (approximately 100 mg of fresh material per plant), immediately frozen in liquid nitrogen and then stored at -80 C until analysis. Each apex was analysed individually and a set of 4 replicates were dedicated to transcriptomic analysis and another 5 to metabolomics.
A second experiment was conducted in the same conditions for a kinetic study of phytohormones with harvest of apex 12, 24 and 48 hours post-infestation (hpi) and for measure of plant elongation 7 days post-infestation (dpi).
RNA extraction and sequencing
Each sample was manually ground in liquid nitrogen with disposable pestles and total RNA was extracted with the RNeasy Plant Mini Kit (QUIAGEN, France) according to the manufacturer’s instructions, which comprises a guanidine isothiocyanate lysis and purification with silica membrane. RNA concentration and purity were checked with a Nanodrop (Thermo Fisher Scientific, Wilmington, USA), a QuBit 3.0 Fluorometer (Thermo Fisher Scientific, Wilmington, USA) and RNA integrity with a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, USA). Next generation sequencing of RNA (RNAseq) was realized by the GeT-PlaGe platform (INRAE, Toulouse, France). Sixteen RNAseq libraries were prepared with the mRNA TruSeq Stranded kit (Illumina, San Diego, USA), according to Illumina’s protocols. Briefly, mRNAs were isolated from total RNA using poly-T beads, fragmented and converted to cDNA. Specifics adapters and multiplexing indexes were ligated before PCR amplification. Libraries quality was checked using a Fragment Analyser (AATI, Ankeny, USA) and they were quantified by qPCR using Quant Studio 6 Real-Time PCR system (Thermo Fisher Scientific, Wilmington, USA). Finally, libraries were pooled on a single flowcell line (Illumina HiSeq3000 sequencer) for paired-end sequencing (2 x 150 bp).
RNAseq analysis
Sequencer output raw data files (binary base call format) were converted to Fastq format using the CASAVA (Consensus Assessment of Sequence And VAriation) Illumina software. The global quality of sequences was assessed using FastQ Illumina filter and FastQC software (Babraham Institute, Cambridge, UK). Sequencing adapters were removed using Cutadapt version 1.14 (Martin, 2011). Then, the reads were splice-aligned on the peach reference genome version 2.0.a1 (Verde et al., 2017) using STAR version 2.5.1b software (Dobin et al., 2013). Finally, transcript expression was quantified using RSEM version 1.3.0 (Li and Dewey, 2011).
The raw sequencing data were filtered by excluding transcripts with zero or less than 8 counts (corresponding to at least one count for half of the samples), thus reducing the number of transcripts detected to 20606, out of the 26873 protein-coding genes predicted in version 2.0.a1 of the peach reference genome. To evaluate the statistically significant changes in gene expression, an “Independent hypothesis weighting” test was applied according to the DESeq2 R package protocol, with a max fold change of 2, a p.value threshold set to 0.05 and an adjusted false discovery rate (FDR). To overcome the poor annotation of the peach transcriptome, we performed a blastp of the full peach proteome version 2.0.a1 against Arabidopsis thaliana Araport11 protein sequences. The set of best Arabidopsis homologs obtained for each peach protein was used to conduct overrepresentation tests and enrichment of expression data via panther.org (Thomas et al., 2003) and gProfiler (Raudvere et al., 2019) through the Gene Ontology (GO) and KEGG (Kyoto Encyclopaedia of Genes and Genomes) databases. A P. persica protein was considered as the “true” ortholog of an Arabidopsis protein when it was also the best match of a reverse blastp (from Arabidopsis to P. persica proteomes); a total of 13097 Arabidopsis orthologs were found. Detailed gene function was also retrieved from The Universal Protein Resource (UniProt) and The Arabidopsis Information Resource (TAIR) online databases (Berardini et al., 2015; The UniProt Consortium, 2021). Enrichment analysis was conducted using fold enrichment cut-off of 1.5 and p-value cut-off of 0.05. Fold enrichment is the ratio between the frequency of term genes in a given list (here the lists of induced or repressed genes) and the frequency of term genes expected in this list, based on the frequency in the reference list (complete list of annotated genes). The p-value is the probability that the number of term genes observed occurred by chance (randomly), as determined by the reference list. The full dataset of transcripts can be found in Supplementary Table S1 and the raw (BioProject ID PRJNA877419) is available in the NCBI Sequence Read Archive.
Metabolites extraction
Solvents were purchased from Honeywell (Charlotte, USA) and Thermo Fisher Scientific (Wilmington, USA) and ultra-pure water was obtained from a Milli-Q system (Merck, Darmstadt, Germany). Frozen fresh samples were ground in 2 mL microtubes with ball mills for 1 min at 30 Hz (Mixer mill MM200, Retsch, Eragny, France), then 10 mg of fresh powder was extracted for 15 min at 70°C under stirring (940 rpm) with 1.5 mL of a methanol/water (80:20, v/v) solution containing 50 µM of ribitol as internal standard. Volume was adjusted to keep the same ratio mass to volume in every sample. Samples were centrifuged 5 min at 26 400 g and the supernatants containing polar metabolites were filtered before analysis (0.22 µm filters Millex-Lg, PTFE hydrophile, 4 mm, Sigma Aldrich, Merck, Darmstadt, Germany).
Untargeted GC-EI-TOFMS profiling
The method was adapted from Roessner et al. (2000). Samples were derivatized online before injection with a MultiPurpose Sampler (Gerstel MPS, CTC Analytics AG, Mülheim an der Ruhr, Switzerland): dry extracts were incubated in 50 µL of a pyridine solution containing 20 mg/mL of methoxyamine hydrochloride under constant shaking at 900 rpm and 80°C for 90 min. Then, 80 µL of BSTFA containing a mixture of 9 n-alkanes were added before heating for 30 min at 80°C under constant shaking at 900 rpm. Data acquisition was performed with a gas chromatograph system (7890B GC, Agilent Technologies, Santa Clara, CA, USA) equipped with a capillary column (ZB-SemiVolatiles, 34.59 m, internal diameter 250 µm, film thickness 250 µm, Phenomenex, Torrance, USA) hyphenated to a time-of-flight (TOF) mass spectrometer (Pegasus BT, Leco, Saint Joseph, Benton Harbor, MI, USA). One microliter of sample was injected in split mode (1:50) at 230°C. Helium was used as carrier gas at 0.6 mL/min. The initial oven temperature was kept at 70°C for 1 min and then increased to 320°C (9°C/min) and maintained for 10 min. The m/z scan range was 70–600 with a cycle time of 20 scans/s. Source temperature and transfer line were set at 250°C. The MultiPurpose Sampler was controlled by Maestro Version 1.4.40.1. Gerstel and gas chromatography system with mass spectrometer were controlled by ChromaTOF Version 5.20.38.0.54864 (LECO, Saint Joseph, MI, USA). GC–EI–TOFMS data were deconvoluted with the LECO NTD software (LECO, St.Joseph, MI, USA), then the peak list was curated manually to remove incorrectly deconvoluted peaks and contaminants. Peak annotation was based on spectral and retention index (RI) similarity using mass spectral libraries (Golm database, NIST 2014, Leco-fiehn rtx5). Identification level of each metabolites was determined according to criteria inspired by Schymanski et al. (2014): level 1, confirmed structure by comparison with authentic standard, reverse match > 850, difference between retention index (RI) < 1%; level 2, probable structure, reverse match > 800 and difference between RI < 1%; level 3, tentative candidate, 600 < reverse match < 1000 and difference between RI > 1%. A specific extracted ion chromatogram (XIC) was chosen for each molecule for integration; then peak areas were normalized against the internal standard so the final dataset consisted of semi-quantitative information. The list of metabolites detected and their analytical features can be found in Supplementary Table S2; raw dataset (accession number MSV000084377) can be downloaded from the publicly available MassIVE repository at the UCSD Center for Computational Mass Spectrometry website.
Untargeted UPLC-ESI-QTOF-MS/MS profiling
Analyses were performed with an Acquity I-Class UPLC system (Waters, Mildorf, MA) hyphenated to a Synapt G2-Si quadrupole time-of-flight (QTOF) mass spectrometer (Waters, Mildorf, MA) equipped with an electrospray ionization source (ESI). Chromatographic separation was achieved using a Kinetex 1.7 µm F5 Core-shell LC columns (150 x 2.1 mm, Phenomenex, California, USA). The mobile phase consisted of water (A) and acetronitrile (B), both containing 0.1% formic acid. One microliter of sample was injected before running the solvent gradient: 2% B for 1 min, then up to 100% B in 18 min followed by 2 min at 100% B and then back to initial conditions in 1 min (total run time 23 min). The column was maintained at 35°C with a flow rate of 0.3 mL/min. The source temperature was set to 120°C and the desolvation temperature to 600°C. The capillary voltage was set to 0.8 kV and the cone voltage to 40 V. Nitrogen was used as the drying and nebulizing gas, with 50 L/h gas flow and 800 L/h desolvation gas flow. Analysis was performed twice, in negative and positive ionization modes, with a resolution of 40 000. Data independent acquisitions were performed simultaneously in MS and MS/MS modes (MSe, continuum), with a collision energy ramp from 20 V to 70 V. Mass spectra were recorded at 0.2 second per scan from 50 to 1200 m/z. Chromatograms from negative mode acquisition were scanned to extract the major compounds. Metabolites were identified at the level 3 (Schymanski et al., 2014) by a manual examination of MS and MS/MS spectra in positive and negative modes and spectral comparison with in-house database and literature (Supplementary Table S3).
Targeted UPLC-DAD-ESI-TQMS profiling
Analyses were performed with an Acquity I-Class UPLC system (Waters, Mildorf, MA) equipped with a diode array detector (DAD) and hyphenated to a Xevo TQ-XS (Waters, Mildorf, MA) equipped with an ESI source. Pure standards (Merck KGaA, Darmstadt, Germany) were injected and MRM (multiple reactions monitoring) methods were optimized by testing ESI polarity, cone voltage and collision energy. Data acquisition and analysis were performed in using MassLynx software (Waters, Mildorf, USA). Identification level of each metabolites was determined according to Schymanski et al. (2014) as follows: level 1, confirmed structure by comparison with authentic standard; level 2, probable structure by comparison with data bases and/or literature; level 3, MS2 spectrum interpretation and light absorbance spectrum matched a tentative candidate. List of metabolites with detailed analytical parameters are provided in Supplementary Table S4.
Phenolic compounds
Chromatographic separation was achieved using a Kinetex 1.7 µm F5 Core-shell LC columns (150 x 2.1 mm, Phenomenex, California, USA). The mobile phase consisted of water (A) and acetronitrile (B), both containing 0.1% formic acid. One microliter of sample was injected before running the solvent gradient: 2% B for 1 min, then up to 55% B in 18 min, 1 min to reach 100% B followed 2 min at 100% B and then back to initial conditions in 3 min (total run time 25 min). The column was maintained at 35°C with a flow rate of 0.25 mL/min. The source temperature was set to 120°C and the desolvation temperature was set to 600°C. The capillary voltage was set to 1.2 kV. Nitrogen was used as the drying and nebulizing gas, with 50 L/h gas flow and 800 L/h desolvation gas flow.
Amino acids and polyamines
Polyamines and amino acids were analyzed after derivatization with 6-Aminoquinolyl-N-hydroxysuccinimidyl carbamate (AccQ-Tag Ultra Derivitization Kit, Waters, Mildorf, USA) according to the manufacturer’s instructions. Chromatographic separation was achieved using an Acquity UPLC BEH C18 1.7 µm column (2.1 x 50 mm, Waters, Mildorf, USA) with a pre-column. One microliter of sample was injected before running the solvent gradient: 0.01% B for 0.54 min, then up to 9.1% B in 6.5 min, 2 min at 21.2% B followed by 0.4 min at 59.6% B and then back to initial conditions in 0.6 min (total run time 10.1 min). The column was maintained at 55°C with a flow rate of 0.7 mL/min. The source temperature was set to 150°C and the desolvation temperature was set to 650°C. The capillary voltage was set to 3.0 kV. Nitrogen was used as the drying and nebulizing gas, with 600 L/h gas flow and 1200 L/h desolvation gas flow.
Kinetic analysis of salicylic acid and salicylic acid glucoside
One milliliter of polar extract was evaporated under vacuum overnight. The dry residue was concentrated in 100 µL of a methanol/water (80:20, v/v) solution. Chromatographic separation was achieved using an Acquity 1.7 µm C18 CSH LC column (100 x 2.1 mm, Waters, Mildorf, USA). The mobile phase consisted of water (A) and methanol (B), both containing 0.01% formic acid. Five microliters of sample were injected before running the solvent gradient: 25% B for 0,5 min, then up to 100% B in 6 min followed 2 min at 100% B and then back to initial conditions in 1,5 min (total run time 9,5 min). The column was maintained at 45°C with a flow rate of 0.4 mL/min. The source temperature was set to 120°C and the desolvation temperature was set to 550°C. The capillary voltage was set to 2.8 kV. Nitrogen was used as the drying and nebulizing gas, with 150 L/h gas flow and 1000 L/h desolvation gas flow.
Data analysis
Boxplots were generated with the R packages “PMCMRplus” and “ggoplot” and statistically significant differences were found with a Kruskal-Wallis test and Conover post-hoc test with Bonferroni adjustment. Principal component analysis (PCA) and Heatmap were generated with the R package “FactoMineR” and “Pheatmap” respectively, after performing log transformation and Pareto scaling. Metabolites were mapped to metabolic pathways using the Plant Metabolic Network database (Hawkins et al., 2021).
Results
Response to infestation: Aphid behavior and plant symptoms
After infestation, the aphids deposited on the susceptible GF305 remained on the plant, with an average of nine aphids per plant, while they decreased rapidly on the resistant Rubira over 7 days, down to five aphids per plant 48 hpi, until all of them escaped 96 hpi (Figure 1A). The number of nymphs increased constantly on GF305 up to more than 150 (120 hpi), while after a moderate increase during 24 h on Rubira, it fell down to less than five 120 hpi (Figure 1B). The evolution of honeydew abundance on the leaves corroborates these trends: it was high on GF305 and low on Rubira, indicating an aphid feeding failure on the resistant plants (Figure 1C). GF305 plants also displayed typical symptoms of susceptibility 48 hpi, with young twisted leaves, whereas Rubira developed necrotic lesions on shoots and leaves and showed wilting leaves, (Figure 1D and Supplementary Figure S1). Seven dpi, infested Rubira plants had a 39% lower increase in stem height compared to control plants, while stem height increase of GF305 plants was not affected by infestation (Figures 1E, F).
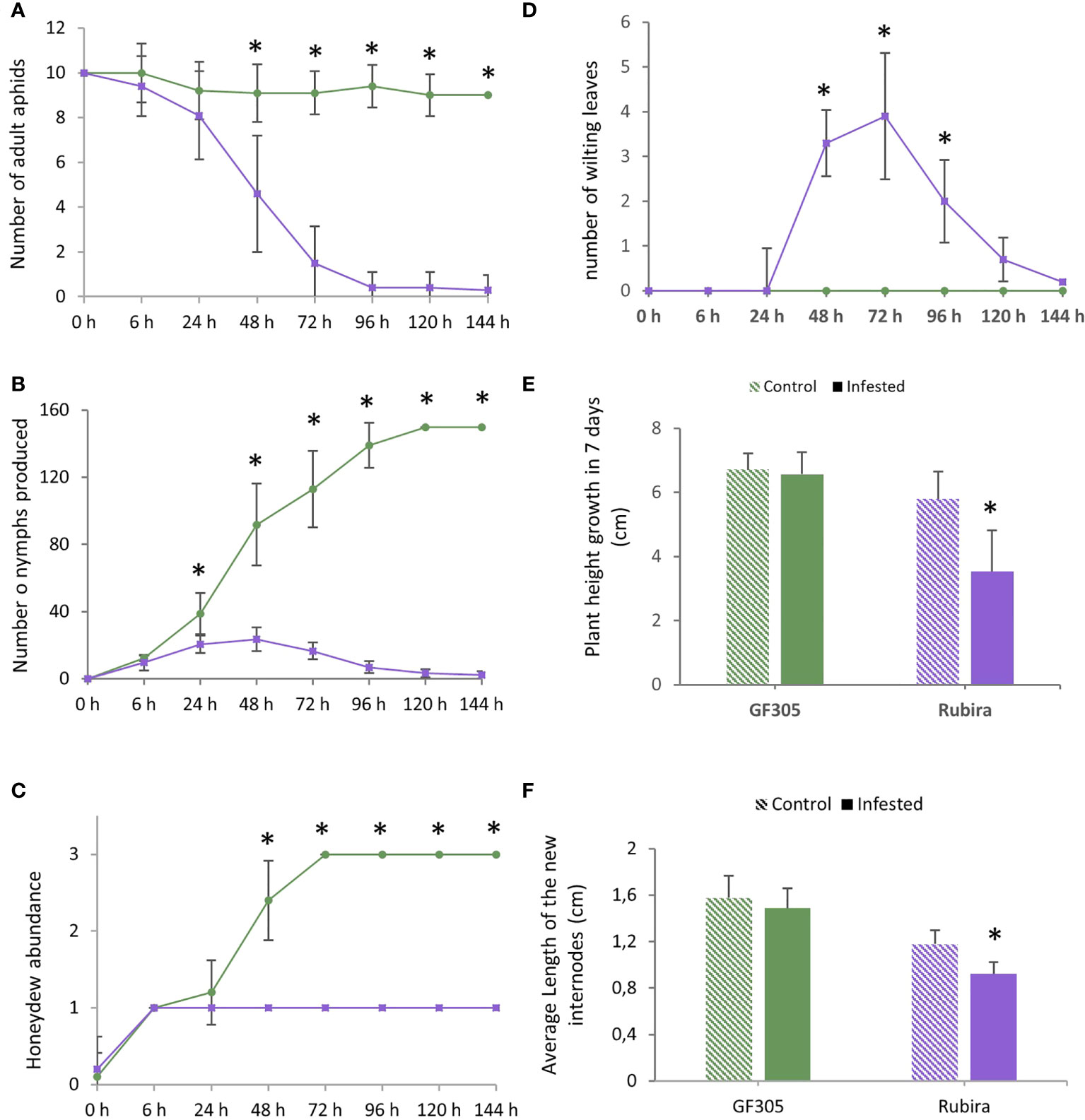
Figure 1 Aphid behavior and plant symptoms after infestation by GPA. Significant differences according to a Mann Whitney test are indicated (p.value<0.01 *). (A) Number of remaining aphids (out of ten adult deposited) on the susceptible GF305 (green circles) and the resistant Rubira (purple squares) seedlings over 6 days. (B) Number of nymphs produced. (C) Honeydew abundance expressed per classes (0: absence; 1: low; 2: medium; 3: high). (D) Number of wilting leaves on the growing shoot. (E) Height growth of the plants in seven days. (F) Average length of new internodes formed in 7 days (cm).
Overall trends in the transcriptomic and metabolomic responses to GPA 48 hpi
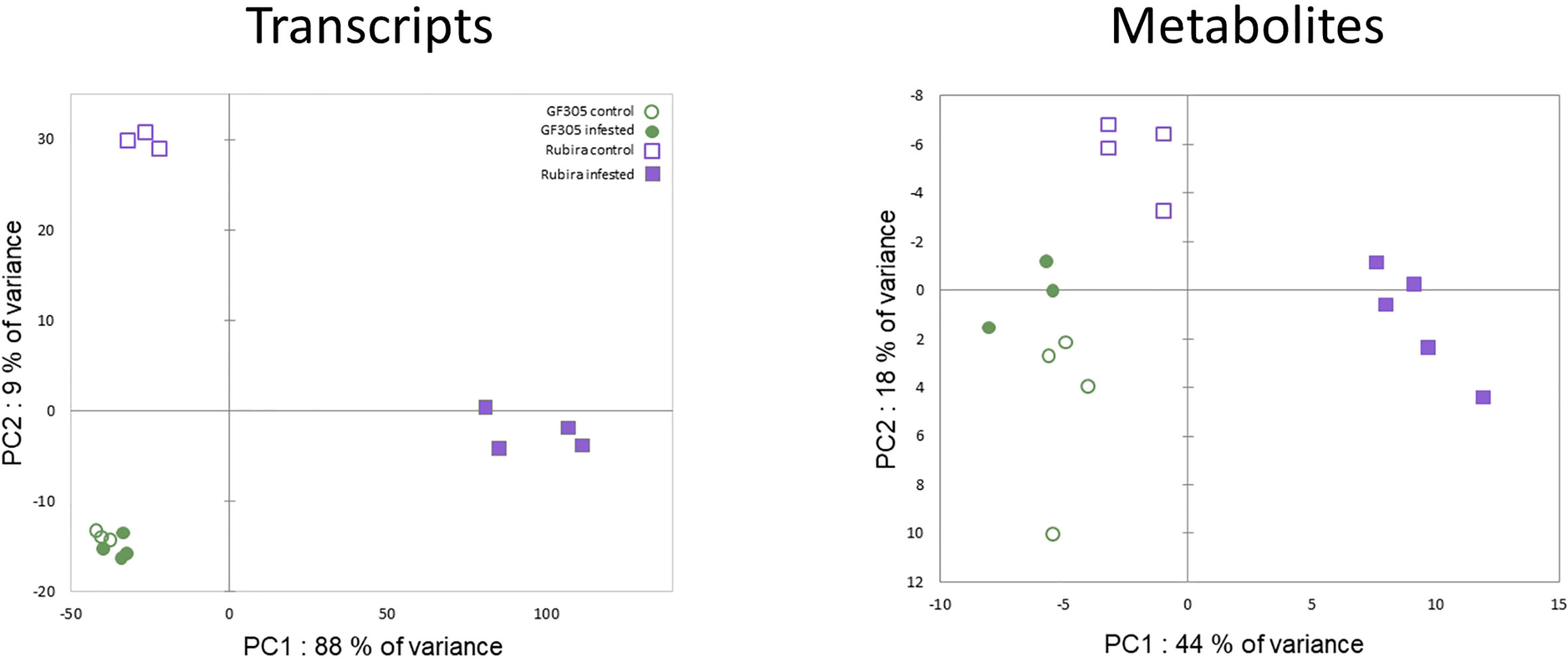
A PCA performed on the transcriptomic raw dataset (Figure 2) revealed that 88% of the total variance associated to PC1 was driven by aphid-induced differentially expressed genes (DEGs) in the resistant genotype Rubira. The second PC only expressed 9% of the total variance and was driven by the genotype differential gene expression. Among the 20 606 expressed genes, we found a total of 5743 DEGs: 284 genes were found differentially expressed between the 2 genotypes under control condition (175 DEGs upregulated in Rubira and 109 downregulated, Supplementary Tables S1, S5), only 35 DEGs were found between control and infested apices of the susceptible GF305 (34 upregulated and 1 downregulated, Supplementary Table S6) and 5424 DEGs between control and infested apices of the resistant Rubira (2990 upregulated and 2434 downregulated, Supplementary Tables S1, S7 and Table 1).
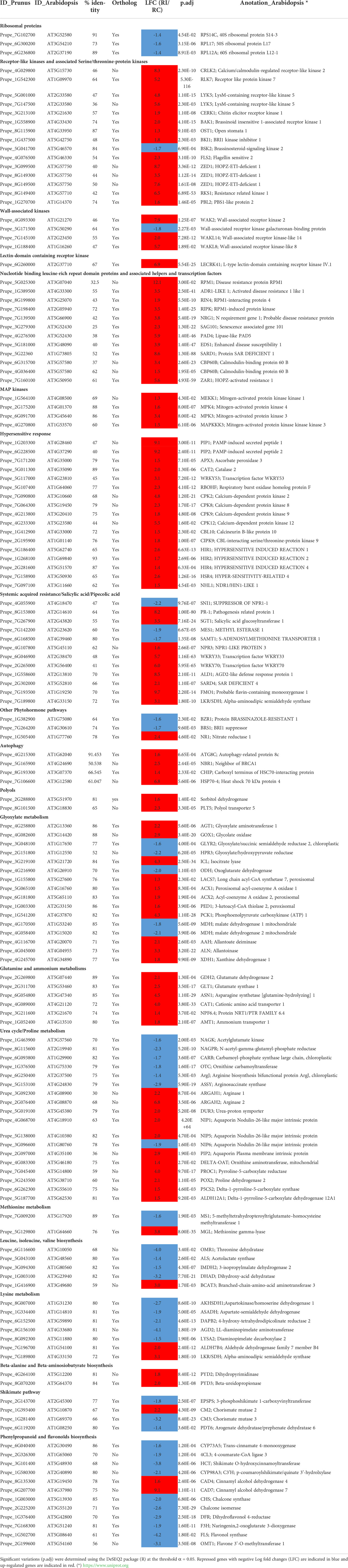
Table 1 Selection of DEGs in the resistant genotype Rubira after GPA infestation, cited in the text and figures.
A combination of targeted and non-targeted metabolomic approaches provided the relative contents of 138 metabolites, including 3 defense phytohormones, 69 primary and 66 secondary compounds. The reliability of identification was determined as recommended by Schymanski et al. (2014): 69 compounds were identified at level 1, 22 at level 2 and 47 at level 3. The distribution of these compounds in structural families is shown in the Supplementary Figure S2. A total of 94 compounds showed significant variation in at least one condition: 73 discriminated the uninfested genotypes (Supplementary Table S8), 12 changed after infestation in GF305 (Supplementary Table S9) and 54 in Rubira (Supplementary Table S10). Interestingly, the trends of metabolomics data revealed by a PCA was similar to that observed for gene expression (Figure 2): PC1 explained 44% of the variance and discriminated aphid-infested Rubira samples from other samples, while PC2 explained only 18% of the variance and was associated to the discrimination of the uninfested genotypes, confirming that the molecular response of Rubira to GPA was several orders of magnitude greater than that of GF305 and also exceeded the constitutive difference between the genotypes. A hierarchical clustering analysis conducted on the 94 discriminant compounds revealed clusters of metabolites discriminating the genotypes and/or responding to infestation (Figure 3). The clusters 2.2.2 and 1.2 gather mostly secondary metabolites remaining stable after infestation but with higher constitutive content in GF305 and Rubira respectively. The cluster 2.2.2 is a mixture of flavonoids and coumaric acid derivatives while the cluster 1.2 contains mainly flavonols and anthocyanins. The cluster 2.1 gathers mainly caffeic acid esters that were constitutively more abundant in GF305 but decreased in this genotype after infestation while they increased in Rubira. Finally, the clusters 1.1 and 2.2.1 show respectively the specific aphid-induced accumulation and depletion of primary metabolites in Rubira.
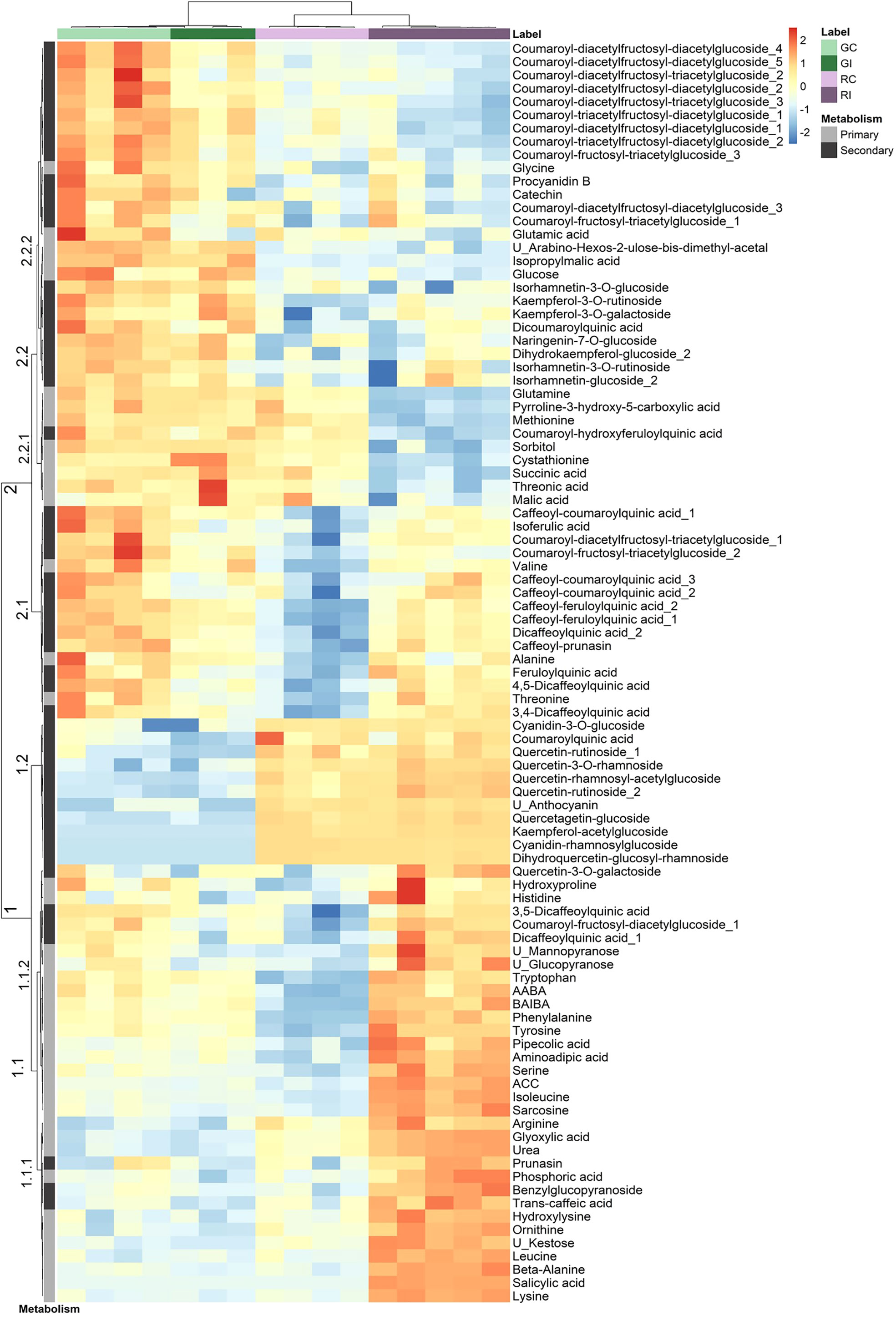
Figure 3 Hierarchical clustering analysis of a subset of metabolites displaying a significant difference (Kruskall-Wallis, p<0.05) between uninfested GF305 and Rubira control plants (GC vs RC) or between control and GPA-infested plants (Gc vs GI and RC vs RI) 48 hpi. HCA was performed using the Euclidian distance and the Ward algorithm on data after log transformation and Pareto normalization (mean-centered and divided by the square root of the standard deviation). Primary and secondary metabolites are indicated by a gray scale on the left. GC, GF305 control; GI, GF305 infested; RC, Rubira control; RI, Rubira infested.
Constitutive differences between the susceptible and resistant genotypes
The Gene Ontology (GO) enrichment analysis of the 284 DEGs highlighted five functions, most upregulated genes being found in Rubira (Figure 4; Supplementary Table S5). The greater differences relate to flavonoid metabolism (GO:0019748, GO:0009812), with most genes globally induced in Rubira. This is consistent with the accumulation of quercetin-3-O-rhamnoside and especially cyanidin-3-O-glucoside (Supplementary Figure S3), responsible for the red colour of Rubira’s foliage. This trait is controlled by the Gr locus which is independent of aphid resistance (Pascal et al., 2002) and located on chromosome 6 (Lambert and Pascal, 2011). Indeed, PpMYB10.4 (Prupe_6G175900), a Gr gene candidate encoding a MYB transcription factor (Zhou et al., 2014b) was highly induced in Rubira. The high anthocyanin content is further explained by the strong expression of the orthologs of leucoanthocyanidin dioxygenase (Prupe_5G086700) and anthocyanidin-3-O-glucosyltransferase (Prupe_2G324700), involved in the anthocyanin biosynthesis pathway, and PpGST1 (Prupe_3G013600), a glutathione S-transferase, orthologous to GSTF12 and involved in the transport of anthocyanins essential for peach coloration (Zhao et al., 2020). In contrast, GF305 had higher levels of isorhamnetin-3-O-glucoside, isorhamnetin-3-O-rutinoside and kaempferol-3-O-rutinoside, as well as caffeoylquinic acids, phenylpropanoid-acetyl-sucrose esters and the three aromatic amino acids, phenylalanine, tyrosine and tryptophan.

Figure 4 Gene Ontology (GO) enrichment analysis for the comparison of constitutive gene expression in the uninfested resistant Rubira compared to the susceptible GF305. Bars indicate the percentage of upregulated genes associated to GO terms. A total of 217 markers were mapped in this enrichment analysis. GO terms with at least a 1.5-fold over-representation and an enrichment adjusted p.value lower than 0.05 are presented. For each GO term, details are given in parentheses: (i) number of P. persica GO annotated genes, (ii) adjusted p.value of enrichment, (iii) GO groups (BP: Biological process).
Responses to infestation in the susceptible genotype
Only 35 genes were differentially expressed in GF305 after infestation, all but one induced (Supplementary Table S6). Of the 34 genes induced, 27 were also induced in Rubira after infestation. The small number of genes involved did not allow for an enrichment study but many genes could be a priori assigned to a few functional groups. The most remarkable DEG, Prupe_5G025300, is a NLR gene that shares homology (32.5% identity) with its Blastp best hit in Arabidopsis, the resistance protein RPM1 (Gao et al., 2011). It was the only gene repressed in GF305 after infestation while it was induced in Rubira. Among the genes that were induced, several are involved in brassinosteroid (BR) regulation, like Prupe_1G520800, a close homolog of EXORDIUM, which is a central coordinator of BR-dependent growth control (Coll-Garcia et al., 2004), also involved in response to herbivory (Mohanta et al., 2012), and Prupe_5G222200, ortholog of the basic helix-loop-helix (bHLH) transcription factor BEE3, a positive and early regulator of BR signalling (Friedrichsen et al., 2002). Several DEGs are homologs of Arabidopsis genes regulated by BRs and also regulated by auxin, like KRP1 (Gupta et al., 2015), coding for a Calcium binding EF-hand family protein linked to calcium homeostasis during the photoperiod and possibly controlling the diurnal sucrose synthase activity (Solomon et al., 2010). Finally, only twelve metabolites showed statistically significant variations after infestation (Supplementary Table S9). All of them decreased: glutamate, threonine and nine secondary metabolites related to defense, including caffeoyl-prunasin, caffeoylquinic and dicaffeoylquinic acids and coumaroyl-acetyl-sucrose esters (Supplementary Figure S4).
Responses to infestation in the resistant genotype
24 GO terms including stress and metabolism were mostly upregulated and 31 GO terms including cell division were mostly downregulated in Rubira after infestation (Figure 5). DEGs commented below are exposed in the Table 1, the full annotation is presented in the Supplementary Table S7.
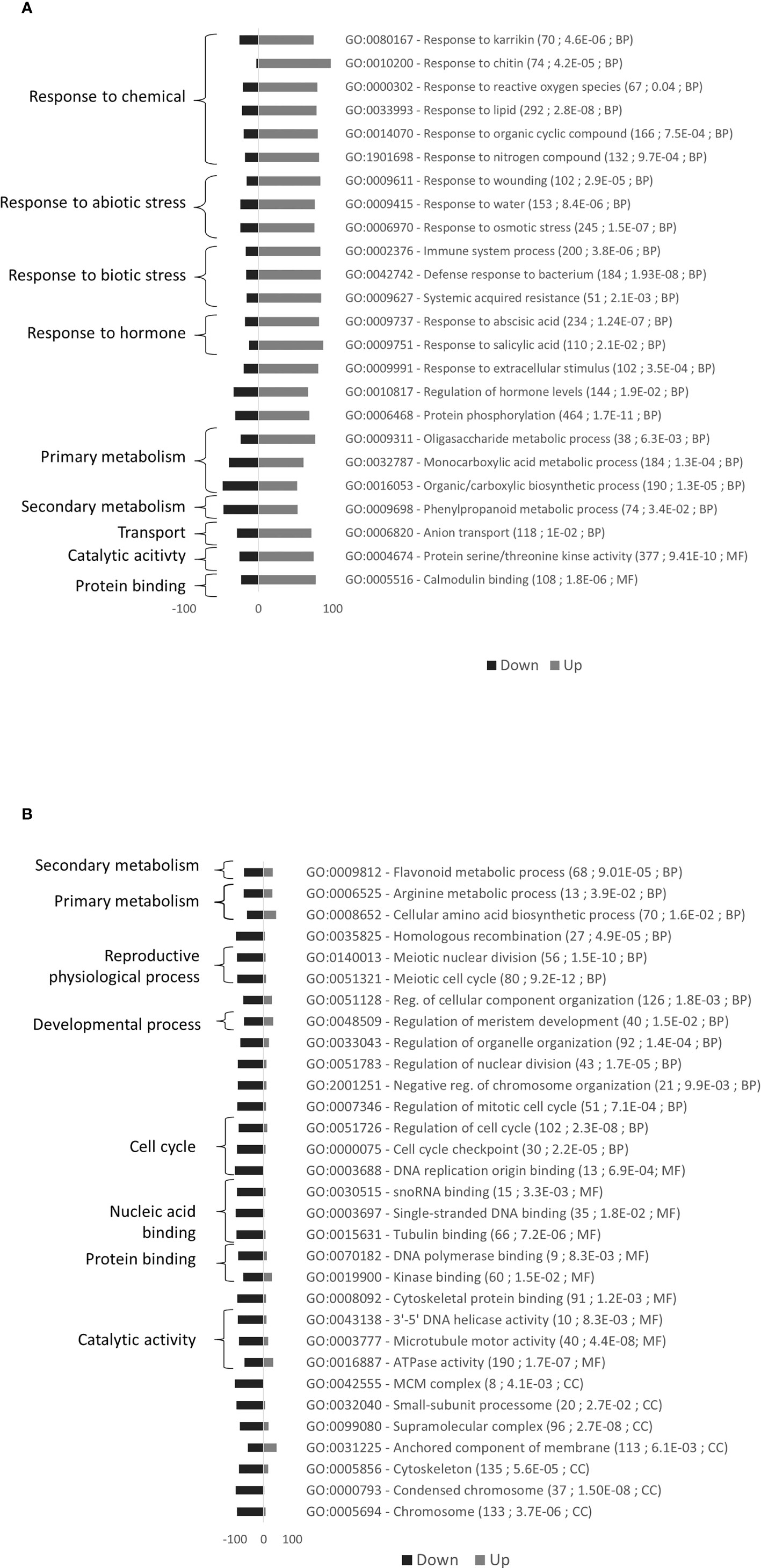
Figure 5 Gene Ontology (GO) enrichment analysis for the comparison of gene expression variations induced by GPA infestation of Rubira. (A) Upregulated genes. (B) Downregulated genes. Bars indicate the percentage of genes up- or down-regulated associated to GO terms. A total of 3376 markers were mapped in this enrichment analysis. GO terms with at least a 1.5-fold over-representation and an enrichment adjusted p.value lower than 0.05 are presented. For each GO term, details are given in parentheses: (i) number of P. persica GO annotated genes, (ii) adjusted p.value of enrichment, (iii) GO groups (BP, Biological process; CC, Cellular compartment; MF, Molecular Function).
Repression of cell division and growth
Most downregulated genes are operating in the many processes of cell division and growth (Figure 5), including supramolecular complex (GO:0099080), chromosomes (GO:0005694) and protein complexes such as the mini-chromosome maintenance (MCM) complex (GO:0042555), a protein complex necessary for the initiation and regulation of DNA replication (Tuteja et al., 2011), or the small-subunit processome (GO:0032040), a preribosomal complex required for the 18S ribosomal RNA biogenesis (Bernstein et al., 2004). The molecular functions involved concern interaction with nucleic acids for DNA replication, RNA transcription, and cell division, such as DNA polymerase binding (GO:0070182) and single-stranded DNA binding (GO:0003697). The downregulated genes are also associated to the regulation of cell cycle (GO:0051726) and meristem development (GO:0048507). Accordingly, nearly sixty genes coding for ribosomal proteins were downregulated, such as Prupe_7G102700, Prupe_6G300200, and Prupe_6G236800, the respective orthologs of RPS14C, RPL17 and RPL12A (Table 1).
Induced expression of immune receptor genes
The genes associated with protein serine/threonine kinase activity (GO:0004674) and calmodulin binding (GO:0005516) were mostly upregulated (Figure 5). Biotic stress is first perceived by transmembrane protein kinases, such as RLKs, comprising an extracellular domain that perceives stimuli and an intracellular domain transmitting information to several cytoplasmic protein kinases subfamilies: mitogen-activated protein kinases (MAPKs), calcium-dependent protein kinases (CPKs) and calcineurin B-like proteins (CBLs) (Kurusu et al., 2010; Tang et al., 2017). Many genes coding for these proteins were upregulated in Rubira after infestation, probably involving reactive oxygen species (ROS)-mediated signal transduction (response to reactive oxygen species, GO:0000302).
Homologs of typical PRR genes involved in PTI were activated (Table 1), like Prupe_4G029800, an homolog of CLRK1/CLRK2 in Arabidopsis and homolog of Csa5M642150 in cucumber, identified as a candidate for resistance to Aphis gossypii (Liang et al., 2016). Prupe_1G542300 is the ortholog of RLK7, a cell surface receptor which triggers immune response upon detection of the PAMP-induced secreted peptide 1 (PIP1) and controls the accumulation of ROS detoxifying enzymes such as the superoxide dismutase (Pitorre et al., 2010; Hou et al., 2014). Prupe_3G213100 and Prupe_5G001000, orthologs of chitin elicitor receptor kinase 1 (CERK1) and Lysin motif-containing receptor-like kinase 5 (LYK5) respectively, both involved in chitin detection (Erwig et al., 2017), were up-regulated as well. Prupe_1G558900 encoding a homolog of the co-receptor BAK1, involved in brassinosteroid signalling and PTI, through its association with BRI1 or FLS2 respectively (Li et al., 2002; Chinchilla et al., 2007) was upregulated, as well as BKI1 (Wang and Chory, 2006) and homologs of FLS2 (Gómez-Gómez and Boller, 2000). BAK1 also associates to OST1 (Prupe_8G115900, ortholog), a ROS/ABA dependent activator of RBOHF (Mittler and Blumwald, 2015). On the contrary Prupe_5G041700, ortholog of BSK2 involved in BR signalling downstream of BRI1, was downregulated (Tang et al., 2008). It is also worth mentioning the impact of infestation of Rubira on the expression of genes encoding WAKs (Wall-associated kinases, like Prupe_4G093300, the ortholog of WAK2) and WAKLs (WAK-like, such as Prupe_7G145100, the ortholog of WAKL14), with 14 out of 15 homologs being induced and only one repressed.
Many NLR genes were also up-regulated, like thirteen homologs of RPM1 and the orthologs of functionally associated defense genes Prupe_8G19980 (RIN4) and Prupe_7G198400 (RIPK). Prupe_1G389500, the ortholog of ADR1-like1, and NRG1 (Table 1) are CNLs, working downstream in parallel branches as helpers of TNLs necessary for TNL-mediated immunity (Wu et al., 2019). These proteins associate with nucleocytoplasmic lipase-like proteins, like EDS1 and PAD4 or SAG101, that enhance TNL responses (Lapin et al., 2019; Wu et al., 2019). The orthologs of EDS1 (Prupe_5G181000), PAD4 (Prupe_4G276500) and SAG101 (Prupe_3G279300) were all upregulated in infested Rubira (Table 1). In addition, Prupe_7G160100, the ortholog of the CNL ZAR1, was also induced, as were homologs of the associated RLCKs: ZED1, RKS1 (Prupe_8G149400, orthologs) and PBL2 (Prupe_1G270700, ortholog) (Duxbury et al., 2021).
Upregulation of ROS signalling, phosphorylation cascades and HR
Downstream detection systems, defense signal transduction mechanisms were activated in Rubira. Upregulated MAPK phosphorylation cascades included Prupe_1G564100 an homolog of the H2O2 inducible gene MEKK1, which in turn induced MPK4 (Prupe_2G175200, ortholog) expression (Pitzschke et al., 2009) (Table 1). These two central regulators of redox homeostasis in plants control detoxification enzymes, such as the catalase CAT2 (Prupe_5G011300, ortholog) and can induce expression of WRKY53 (Prupe_5G117000, ortholog) (Pitzschke et al., 2009), a transcription factor responding to infection or abiotic stress (Hu et al., 2012). Prupe_6G091700, encoding the ortholog of the important pathogen-responsive MPK3 (Lang et al., 2022) was also upregulated. Other peach genes encoding protein kinases subfamilies were strongly represented and generally induced: CPK (e.g. Prupe_4G213800, ortholog of CPK9), CBL (e.g. Prupe_1G412900, ortholog of CBL10) and CBL-interacting protein kinases (CIPKs, (e.g. Prupe_2G195900, ortholog of CIPK9). These proteins share a Ca2+ activated EF-hand motif and a kinase domain triggering phosphorylation events (Marcec et al., 2019). CPKs can activate respiratory burst oxidases (RBOH), such as RBOHF (Prupe_5G107400, ortholog), a family of NADPH-oxidase producing in the apoplasm, which turns into H2O2 (Kimura et al., 2017).
The presence of necrotic spots on Rubira near the secondary veins and on the stems after GPA infestation suggests the establishment of a hypersensitive response (HR). This was associated to the transcriptional upregulation of genes involved in response to reactive oxygen species (GO:0000302) and protein phosphorylation (GO:0006468), which are rapid events generally preceding HR. Orthologs of important HR genes were upregulated, like the orthologs of HIR1, HIR2 and HIR4 encoding hypersensitive-induced reaction (HIR) proteins, members of the Proliferation, Ion and Death superfamily and involved in the development of spontaneous lesions (Choi et al., 2011; Qi et al., 2011). Other induced HR markers were Prupe_7G158900, the ortholog of HSR4 (Zhang et al., 2014), Prupe_7G097100 the ortholog of NHL1, a member of NDR1/HIN1-like (NHL) gene family including NDR1 in Arabidopsis (Knepper et al., 2011), required for HR and for resistance conferred by R genes, and HIN1, initially identified in tobacco as an HR marker based on its induction by harpins (Pontier et al., 1999).
Changes in phytohormone pathways
In aphid-infested Rubira, 84% of the DEGs classified in SAR (GO:0009627) were upregulated (Figure 5), including the two main homologs of the transcription factor WRKY33 (the ortholog Prupe_6G046900 and Prupe_6G286000), a global regulator of SAR (Wang et al., 2018a; Barco and Clay, 2020) and the two main homologs of EDS1 (the ortholog Prupe_5G181000 and Prupe_5G180900), a positive regulator of HR and SAR through SA signalling (Gao et al., 2015; Dongus and Parker, 2021). Moreover, one of the nine repressed genes in this GO category was SNI1, which encodes a core protein capable of suppressing the expression of PRs and more generally of SAR (Li et al., 1999). SAR is controlled by two immune pathways, one involves SA and PR proteins, the other pipecolic acid. The activation of the SA pathway was demonstrated by accumulation of SA and SAG in Rubira 48 hpi (Figure 3, cluster 1.1., Supplementary Table S10, Supplementary Figure S5) as well as the induction of 87% of SA response genes (GO:0009751) and induction of the ortholog (Prupe_8G153800) and three homologs of PR-1. The transcription of many genes implicated in SA regulation was impacted (Table 1), like Prupe_7G267900, the ortholog of SGT1 coding for a glycosyltransferase catalyzing the formation of SAG and SA glucose ester (SGE), Prupe_7G142200 and Prupe_6G168500, the respective orthologs of MES1 and SAMT1 involved in the production of methyl-salicylate (MeSA). Upstream, the ortholog of a gene encoding a transcription factor recruited in the promoter of many SA and PR biosynthesis genes, SARD1 (Prupe_5G22360), was upregulated (Sun et al., 2015), as well as homologs of CBP60b, that positively regulates immunity genes (Li et al., 2021), including SARD1 (Huang et al., 2021). The ortholog of WRKY70 (Prupe_2G265000) was up-regulated as well. This transcription factor regulating the balance between SA and JA signalling defense pathways (Li et al., 2006) is required for the resistance to aphids and nematodes mediated by Mi-1 in tomato and was found to be upregulated by SA and downregulated by JA (Atamian et al., 2012; Chen et al., 2021). Interestingly the ortholog of the MYB44 transcription factor Prupe_1G430000 was also induced. MYB44 is a transcriptional activator of WRKY70 expression that activates SA-mediated defenses and represses JA-mediated defenses (Shim et al., 2013). MYB44 plays a critical role in resistance to GPA in Arabidopsis (Lü et al., 2013) and is highly expressed in response to Sitobion avenae and associated to phloem-based defenses in wheat (Zhai et al., 2017).
The second SAR pathway depends on the accumulation of N-hydroxypipecolic acid (NHP) (Chen et al., 2018; Hartmann and Zeier, 2018) and its precursor pipecolic acid (Návarová et al., 2012). NHP could not be detected in this study but its direct precursor, pipecolic acid, was found to accumulate 48 hpi in Rubira (Supplementary Figure S5). Orthologs of NPH biosynthetic pathway genes were all upregulated: ALD1 (Prupe_1G558600), coding for an aminotransferase that catalyzes the first step of lysine catabolism to ϵ-amino-α-keto caproic acid (Návarová et al., 2012; Ding et al., 2016; Hartmann et al., 2017), SARD4 (Prupe_2G302000), which allows the formation of pipecolate from Δ1-piperidine-2-carboxylate (Ding et al., 2016) and FMO1 (Prupe_7G193500) allowing the hydroxylation of pipecolate to NHP (Hartmann et al., 2018).
Other phytohormone pathways were regulated as well upon infestation: orthologs of BZR1 (Prupe_1G382900), a positive regulator of BR signalling pathways and BRS1 (Prupe_7G264200), a carboxypeptidase involved in BR signalling, were repressed after infestation (Table 1). On the contrary, NO signalling was probably activated, with the upregulation of the Nitrate reductase 1 ortholog (NR1, Prupe_1G505400).
Upregulation of autophagy
Several genes involved in autophagy were upregulated upon infestation (Table 1). Autophagy-related 8 genes (e.g. Prupe_4G215300, an ATG8c isoform) produce proteins that covalently attach to autophagic membranes and can bind autophagy receptors such as NBR1 (Prupe_5G165900, homolog) to deliver cargo in vacuole for degradation (Kirkin et al., 2009; Yoshimoto and Ohsumi, 2018). Other markers of autophagy include Prupe_8G193300, the ortholog of CHIP, coding for a ubiquitin ligase and Prupe_7G106600, the homolog of a heat shock protein HSC70-4 gene, which both mediate cytosolic protein aggregate degradation (Lee et al., 2009; Zhou et al., 2014a).
Reconfiguration of central metabolism
A handful of metabolites and genes indicate that the central carbon metabolism was affected by infestation in Rubira (Table 1; Supplementary Table S10). Sorbitol level decreased massively 48 hpi (Figure 3, cluster 2.2.1. and Supplementary Figure S6A), while the upregulation of homologs of polyol transporters and Prupe_2G288800, the ortholog of sorbitol dehydrogenase converting sorbitol to fructose (Nosarzewski et al., 2012), suggest an active depletion of the sorbitol pool in infested apices. On the contrary, glyoxylate accumulated 48 hpi (Figure 3, cluster 1.1., Supplementary Figure S7A) and several DEGs indicate that the three pathways contributing to glyoxylate biosynthesis, i.e. photorespiration, the glyoxylate shunt, and purine catabolism, were all activated. The photorespiratory production of glyoxylate (Dellero et al., 2016) is revealed by several cues: (i) the accumulation of serine, (ii) the induction of anabolic reactions genes: the ortholog of serine-glyoxylate aminotransferase 1 (AGT1, Prupe_4G258800) and homolog of glyoxylate oxidase (GOX1) and (iii) the repression of catabolic reactions: Prupe_3G048100, the ortholog of the Glyoxylate/succinic semialdehyde reductase 2 (GLYR2) and a homolog of Glyoxylate/hydroxypyruvate reductase (HPR3) (Table 1). A key enzyme of the glyoxylate shunt produces glyoxylate and succinate from isocitrate: the isocitrate lyase (ICL), whose ortholog gene, Prupe_3G219100, was also induced in Rubira 48 hpi. This anaplerotic pathway shunts the tricarboxylic acid cycle and indeed, orthologs of the TCA cycle ODH (Prupe_4G216900) and MDH (Prupe_4G170500) were downregulated after infestation of Rubira. Consistently, citrate and glyoxylate increased and succinate and malate pools were reduced, supporting the idea of a downregulation of the TCA cycle in favor of the glyoxylate shunt. The shunt is fed by fatty acid-derived acetyl-CoA, condensed onto oxaloacetate to produce citrate (Smith, 2002). In our data, the induction of orthologs of the long chain acyl-CoA synthase 7 (LACS7, Prupe_1G155800), acyl-CoA oxidases (ACX1 Prupe_5G065100, ACX2, Prupe_6G181800) and 3-ketoacyl-CoA thiolase 2 (PED1, Prupe_1G003300), traduced the activation of lipid beta-oxidation. Moreover, the strong upregulation of Prupe_1G541200, the ortholog of PCK1, revealed an activation of gluconeogenesis, a primary function of the glyoxylate shunt (Smith, 2002). The third pathway of glyoxylate production involves the degradation of purine (Werner and Witte, 2011), attested here by the induction of orthologs of allantoate deaminase (AAH, Prupe_4G116700), allantoinase (ALN, Prupe_4G045000) and xanthine dehydrogenase 1 (XDH1, Prupe_4G245700) (Table 1), the latter producing glyoxylate and urea, which both accumulated 48 hpi (Supplementary Figure S7A, C).
The reconfiguration of central N metabolism involved genes in the categories Arginine metabolic process (GO: 0006525) and Cellular amino acid biosynthesis process (GO: 0008652), mostly downregulated (Figure 5). Indeed, in infested Rubira, the major amino acids involved in nitrogen assimilation and distribution were either stable, for aspartate and asparagine, or reduced for glutamate and glutamine (Supplementary Figure S6B, S7B). Decrease in glutamate could be related to the upregulation of GDH2 ortholog (Prupe_2G269800), and the decrease in glutamine could be linked to the upregulation of GLT1 ortholog (Prupe_2G311700) and glutamine hydrolyzing asparagine synthetase ortholog (ASN1, Prupe_6G054800) (Table 1). The strong induction of ASN1 after infestation, while the asparagine pool remained constant, suggests the activation of a mechanism exporting nitrogen out of the infested apices. This hypothesis is supported by the upregulation of homologs of cationic amino acid transporter genes (CATs) and nitrate or ammonium transporter genes (NPF, AMT1). Nitrogen metabolism was also impacted at the level of the urea cycle, a pathway producing urea from arginine and recycling ornithine in the process (Winter et al., 2015). Citrulline decreased 48 hpi but ornithine and arginine accumulated (Supplementary Figure S6C, S7C). Paradoxically, the upstream pathways participating in arginine biosynthesis were transcriptionally downregulated: NAGK, CARB, OCT, ArgJ and ASSY orthologs, whereas ARGAH homologs, involved in the last step turning arginine into ornithine and urea, were upregulated. Urea content actually increased, concomitantly with the induction of the DUR3 ortholog Prupe_5G019100, encoding an active urea transporter (Bohner et al., 2015) as well as some aquaporin orthologs (NIPs) allowing the passive transport of this molecule (Matiz et al., 2019). The catabolism of ornithine was also activated toward the biosynthesis of proline, since the ortholog of the ornithine aminotransferase (OAT, Prupe_4G083300) and a homolog of the pyrroline-5-carboxylate reductase (PROC1) were upregulated (Table 1). Proline level however remained stable, perhaps because of the simultaneous transcriptional activation of its degradation via Δ-1-pyrroline-5-carboxylate dehydrogenase (Prupe_5G187700, ortholog of ALDH12A1). The sulfur amino acid methionine and its precursor cystathionine were notably reduced after infestation (Figure 3, cluster 2.2.1. and Supplementary Figure S6D) and consistently, genes involved in methionine biosynthesis and degradation, i.e. the orthologs of MS1 and MGL, were respectively down- and upregulated.
The branched-chain amino acids (BCAAs) isoleucine, leucine and valine were accumulated in Rubira tissues after infestation, as well as the isoleucine precursor threonine (Figure 3, cluster 1.1. and Supplementary Figure S7D). The genes coding for enzymes involved in early steps of biosynthesis were all downregulated by infestation: a homolog of OMR1 and the ortholog of ALS (Prupe_5G043100) for isoleucine, orthologs of IMDH2 and DHAD for leucine and valine respectively. On the contrary, homologs of BCAT3, encoding the enzyme catalyzing the last step of BCAAs biosynthesis, were upregulated (Table 1). Another discrepancy concerns lysine metabolism, since DEGs involved in its biosynthesis were downregulated while lysine and its catabolites, hydroxylysine and pipecolate, accumulated Supplementary Figure S7E. Beta-alanine and beta-aminoisobutyrate accumulation (BAIBA) (Supplementary Figure S7F) could be explained by the activation of the uracil degradation pathway (Parthasarathy et al., 2019), with the upregulation of the dihydropyrimidase homolog (PYD2) and Beta-ureidopropionase ortholog (PYD3, Prupe_8G070200).
Finally, it is worth mentioning the trace detection of aphid-induced alpha-aminobutyric acid (AABA) and sarcosine (N-methyl-glycine) (Supplementary Figure S7F). No DEG could be associated to these compounds.
Reconfigurations of secondary metabolism
Enrichment in the categories Phenylpropanoid metabolic process (GO: 0009698) and Flavonoid metabolic process (GO:0009812) underlines that significant modifications occurred in secondary metabolism in Rubira after infestation. The transcription of most DEGs implicated in the phenylpropanoid pathway was downregulated (Table 1). Upstream, the shikimic acid pathway genes involved in chorismic acid and aromatic amino acids were downregulated with the exception of the chorismate mutase 2 (CM2) ortholog (Prupe_1G393400). The two peach phenylalanine ammonia lyase (PAL) genes were not differentially expressed, but the CYP73A5 ortholog, Prupe_6G040400, encoding a trans-cinnamate 4-monooxygenase (C4H) producing coumaric acid from cinnamic acid and an homolog of 4CL3, encoding a coumarate-coA ligase specifically involved in flavonoid biosynthesis (Li et al., 2015), were both repressed. Homologs of genes involved in the biosynthesis of hydroxycinnamic derivatives, HCT (Prupe_3G101400), C3’H (Prupe_1G580300), and putative caffeoyl-CoA O-methyltransferases (Prupe_2G199600, Prupe_2G199800) were also downregulated, as well as downstream genes, operating at the entrance of the flavonoid pathway (ortholog of CHS, Prupe_1G003000) or for the subsequent methylation of flavonoids (Prupe_2G199600 and Prupe_2G199800, homologs of OMT1, a multifunctional flavone 3’-O-methyltransferase also involved in formation of lignins and sinapoyl esters (Muzac et al., 2000). The only exception was the upregulation of the ortholog of CAD4 (Prupe_8G135300) and a poorly annotated homolog of CAD7, coding respectively for cinnamyl alcohol dehydrogenases catalyzing the last step of monolignols biosynthesis (Tronchet et al., 2009) and for a NADPH-dependent aldehyde reductase converting (Z)-3-hexenal to (Z)-3-hexen-1-ol in Arabidopsis (Tanaka et al., 2018). The metabolites however present a different pattern: while most flavonoids did not change in response to infestation, many hydroxycinnamoylquinate derivatives were accumulated (Figure 3, cluster 1.1., Supplementary Figure S8; Supplementary Table S10). Aromatic amino acids increased, as well as the simple phenylpropanoids caffeic and isoferulic acids. Quinic acid esters increased while their direct precursor, free quinic acid, decreased. Many of the phenylpropanoids accumulated 48 hpi were caffeic conjugates, like caffeoyl-feruloyl-quinic acid and especially 5 isomers of dicaffeoylquinic acid. Finally, a few coumaroyl-acetyl-glucose esters increased, as well as the defense cyanogenic glycosides prunasin and caffeoyl-prunasin derived from phenylalanine (Shimomura et al., 1987; Yamaguchi et al., 2014). Interestingly, some of these compounds increased in Rubira whereas they decreased in the susceptible genotype GF305 after infestation (Figure 3, cluster 2.1. and Supplementary Figure S4).
Discussion
In this work, we assessed the transcriptomic and metabolomic responses of peach to infestation by the green peach aphid GPA, a major global threat to horticultural and field crops. The study of peach is of particular interest since it is almost the only primary host of this polyphagous aphid species and peach accessions carry major resistance genes in contrast to secondary hosts of GPA. The responses of two rootstocks cultivars, GF305, susceptible to GPA and Rubira, carrying the major resistance gene Rm2, were studied 48 hpi, once the induced resistance of Rubira is fully established.
GF305 and Rubira have highly contrasting responses to GPA infestation
Large differences in the number of DEGs induced by aphid infestation between susceptible and resistant genotypes are found in transcriptomic studies, but generally not to the same extent as in our investigation, which involved 5424 DEGs in Rubira, i.e. 20% of the predicted protein-coding sequences of peach genome, and only 35 DEGs in GF305. In peach, a previous study compared the transcriptomic response to GPA of the resistant peach cultivar “Fen Shouxing”, bearing the single dominant gene Rm3, to the response of a susceptible genotype (Niu et al., 2018). Infestation resulted in 1177 DEGs in the resistant line 48 hpi, compared to the aphid-free control plants, i.e. almost 5 times less than in the present study, and 282 in the susceptible line, that is 8 times more than in our work. Comparisons between studies should be made with caution since environmental conditions, plant age, genetic characteristics, initial number of aphids and RNAseq data filtering parameters, may not be identical across experiments. However, the much lower number of DEGs induced by infestation in the susceptible strain in our experiment could be explained by the very high susceptibility of GF305 to GPA (Sauge et al., 1998a), and/or by the high pre-adaptation of the aphids, since they were reared on this genotype before the experiment. Indeed, aphids continuously adapt their behavior to the plant characteristics, like their resistance level (Pompon and Pelletier, 2012; Jhou et al., 2021). Their experience on the plants they are reared on modulates aphids physiology temporarily, like duration of phloem intake, probing frequency, salivation duration (ten Broeke et al., 2014) and the gene expression level of particular effectors related to host utilization (Eyres et al., 2016; Thorpe et al., 2020). The second main difference concerns the number of repressed DEGs in the infested resistant line: Niu et al. (2018) found only a small proportion of repressed genes (a minimum of 6% 12 hpi and a maximum of 38% 72 hpi), whereas in our experiment half of the DEGs of the resistant genotype Rubira were repressed by infestation. This difference is due to the repression of hundreds of genes involved in metabolism and cell division (Figure 5) and is probably related to the strong inhibition of plant elongation measured post infestation in our experiment.
GF305 weak response to infestation may reflect manipulation by GPA, repressing defense and promoting growth
The very limited transcriptional response of the susceptible genotype 48 hpi included only one underexpressed gene: Prupe_5G025300, a homolog of the CNL RPM1, a R-protein triggering HR-mediated defense against P. syringae (Kim et al., 2009). In Arabidopsis, RPM1 was found to interact functionally with the important defense protein RIN4, which is activated by an RPM1-induced protein kinase (RIPK) (Liu et al., 2009; Chung et al., 2011). Prupe_5G025300 and 12 other RPM1 homologs, as well as the RIN4 and RIPK orthologs (Prupe_8G19980 and Prupe_7G198400 respectively), were upregulated in Rubira after infestation, which might lead to the detection of a particular aphid effector, thereby activating a set of defense responses controlled by RPM1. By contrast, RPM1 downregulation and RIN4 and RIPK not being differentially expressed in GF305 could traduce a neutralization of defense mechanisms by the aphid.
The induction of several growth-related hormones genes, specially brassinosteroid-related (e.g. EXORDIUM, BEE3, KRP1, (Table 1 and Supplementary Table S6) in both susceptible and resistant genotypes are manifestations of the complex interplay between defense activation, resources allocation and growth that could either indicate a role in defense or the activation of a local physiological sink beneficial to the aphid (Züst and Agrawal, 2016). The metabolic profiles of GF305 did not show any significant change in the levels of primary metabolites that could improve the diet of the aphids, excepted maybe a decrease in glutamate (Supplementary Table S9), reported to lower the nutritional quality of phloem sap at high concentration (Karley et al., 2002). It is conversely possible that glutamate reduction impaired defense, since this compound is involved in long distance wounding signalling (Toyota et al., 2018). The observed decrease in the pool of defense compounds such as caffeoyl derivatives also supports the hypothesis of efficient manipulation of metabolism orchestrated by GPA.
Overall, the susceptibility was manifested by a quasi-absence of transcriptional response and a controlled inhibition of plant defenses by GPA in GF305, in contrast with the massive activation of defense responses observed in the resistant genotype.
The Rm2 gene triggered PTI, ETI, SAR and HR markers upon infestation
Three genes conferring high-level resistance to GPA, Rm1, Rm2 and Rm3, have been detected in peach so far, located in the same genomic region at the bottom of chromosome 1 (Lambert and Pascal, 2011; Pascal et al., 2017; Niu et al., 2018). These three overlapping regions contain TNL candidate genes (Pan et al., 2022) which are likely receptors activated by recognition of a GPA saliva effector that triggers ETI. Our study suggests that Rm2 activation by GPA effectors triggered coordinated processes in a dynamic network of PTI, ETI, pipecolate- and SA-mediated SAR contributing to aphid resistance through HR and defense metabolites accumulation (Figure 6).
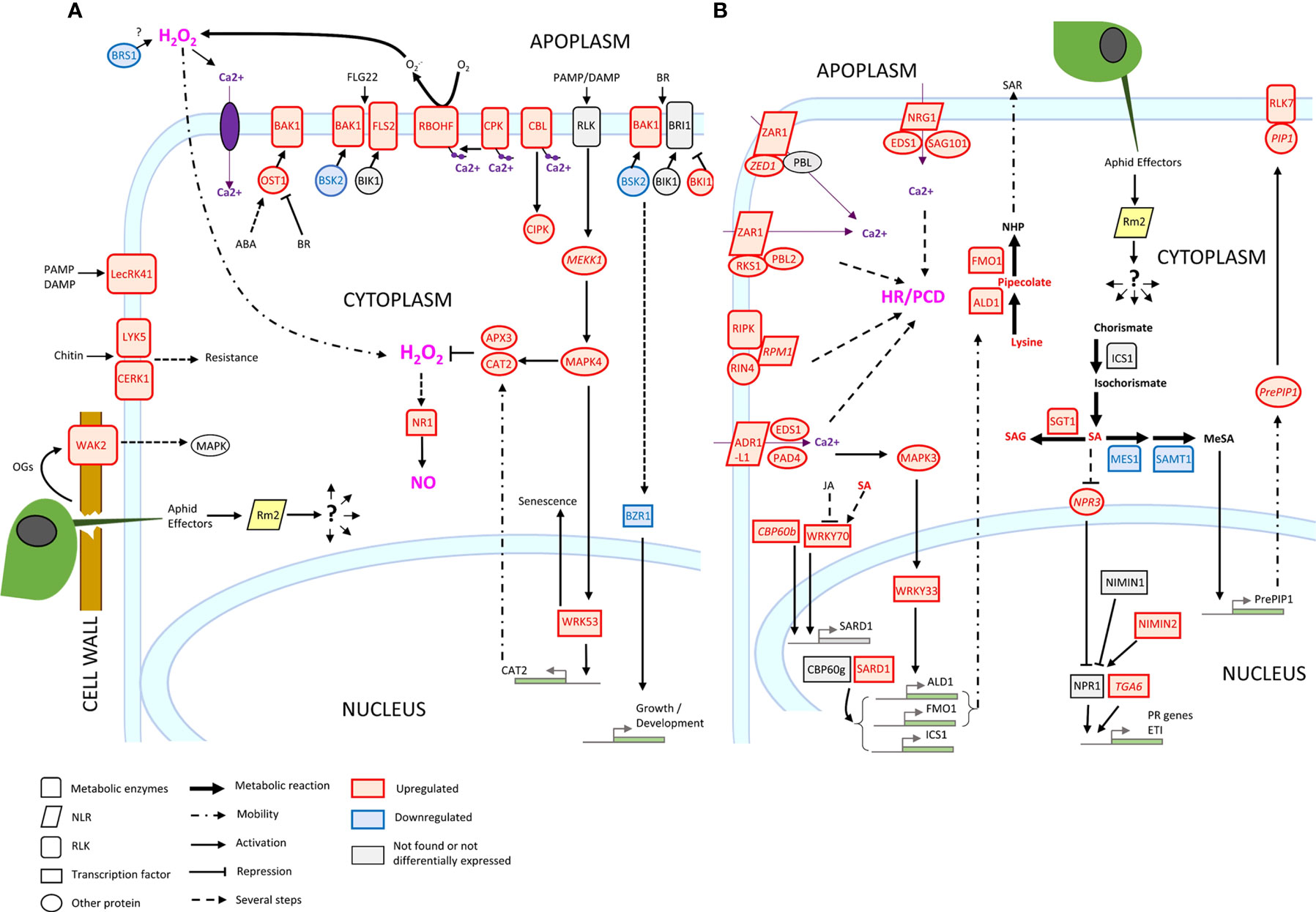
Figure 6 Transcriptional regulation of ETI, PTI, HR and SAR in Rubira 48 hours after infestation with GPA. (A) PTI genes. (B) ETI genes. Genes are annotated according to their Arabidopsis orthologs, non-orthologs (homologs) are noted in italics. Upregulated genes/metabolites are figured in red, downregulated genes/metabolites in blue and not found or not differentially expressed genes/metabolites in black. BIK1, Botrytis induced kinase 1; BRI1, Brassinosteroid insensitive 1; ICS1, Isochorismate synthase 1; NPR1, Nonexpresser of PR genes 1; OST1, Open stomata 1; RIN4, RPM1-interacting protein 4; RPM1-induced protein kinase; Disease resistance protein RPM1; PCD, programmed cell death; ADR1-LIKE 1, Activated disease resistance 1 like 1; ALD1, AGD2-like defense response protein 1; APX3, Ascorbate peroxidase 3; BAK1, Brassinoid insensitive 1-associated receptor kinase 1; BKI1, BRI1 kinase inhibitor 1; BRS1, BRI1 suppressor; BSK2, Brassinosteroid-signalling kinase 2; BZR1, Protein BRASSINAZOLE-RESISTANT 1; CAT2, Catalase 2; CBL10, Calcineurin B-like protein 10; CBP60B, Calmodulin-binding protein 60 B; CERK1, Chitin elicitor receptor kinase 1; CIPK25, CBL-interacting serine/threonine-protein kinase 25; CPK12, Calcium-dependent protein kinase 12; EDS1, Enhanced disease susceptibility 1; FLS2, Flagellin sensitive 2; FMO1, Probable flavin-containing monooxygenase 1; LECRK41, L-type lectin-domain containing receptor kinase IV.1; LYK5, LysM-containing receptor-like kinase 5; MEKK1, Mitogen-activated protein kinase kinase 1; MES1, Methyl esterase 1; MPK3, Mitogen-activated protein kinase 3; MPK4, Mitogen-activated protein kinase 4; NIMIN1, NIM1-interacting 1; NIMIN2, NIM1-interacting 2; NPR3, NPR1-like protein 3; NRG1, Probable disease resistance protein; PAD4, Lipase-like PAD4; PIP1, PAMP-induced secreted peptide 1; RBOHF, Respiratory burst oxidase homolog protein F; RLK7, Receptor like protein kinase 7; SAG101, Senescence associated gene 101; SAMT1, S-adenosylmethionine transporter 1; SARD1, Protein SAR deficient 1; SGT1, Salicylic acid glucosyltransferase 1; TGA6, TGACG motif-binding factor 6; WKRY33, Transcription factor WKRY33; WRKY53, Transcription factor WKRY53; ABA, Abscisic acid; SA, Salicylic acid; SAG, Salicylic acid glucoside; OGs, oligogalacturonides; PAMP, Pathogenesis-associated molecular pattern; DAMP, Damage-associated molecular pattern.
Induction of peach homologs of cell surface receptors, potentiating elicitors detection such as flagellin (FLS2) chitin (CERK1, LYK5) and secreted peptides PIP1/PIP2 (RLK7), indicates that PTI was strongly activated. Among these induced RLKs, BAK1 is a key regulatory co-receptor required downstream of numerous immune responses through complex phosphorylation cascades of cofactors, whose implication in aphid detection has already been extensively demonstrated (Prince et al., 2014; Chaudhary et al., 2014; Vincent et al., 2017; Tungadi et al., 2021). BAK1 co-receptor interacts with other RLKs, like FLS2 and BRI1 and other co-receptors BKI1, BIK1 and BSKs, essential for BR-signalling transduction. We found an increase in the expression of BKI1, a negative regulator of BRI1, and a decrease of the positive regulator BSK2 (Tang et al., 2011), suggesting a reduction of BZR1 dephosphorylation and thus of its activity as a positive regulator of BR-dependent gene expression (Ortiz-Morea et al., 2020). The downregulation of BZR1 and BSR1 indicates a general shutdown of BR pathways that could contribute to explain the slowdown of apices elongation measured 7 dpi. Furthermore, BRS1 was described as an apoplastic regulator of redox status and stress signalling (Zhang et al., 2021), while BAK1 also associates to OST1 to stimulate RBOHF and trigger ABA/NO regulated stomatal closure (Sierla et al., 2016). Several genes encoding wall-associated receptors, WAKs, were also induced. These proteins possess an extracellular domain that binds pectin (Kohorn et al., 2009) and are activated by oligogalacturonides (Brutus et al., 2010). Foyer et al. (2015), reported in a meta-analysis the induction of these receptors, in particular WAK1 and WAK2, in tissues attacked by phloem-feeding insects. It has been proposed that aphid salivary effectors such as pectin-methylesterases and polygalacturonases released during stylet penetration between cells would produce oligogalacturonides, acting as DAMPS and detected by WAKs to trigger MAPK signalling cascades (Silva-Sanzana et al., 2020).
Downstream consequences of Rm2 activation involved the helper RNLs ADR1-L1 and NRG1, whose peach orthologs were induced upon infestation of Rubira. ADR1 interacts with the lipase-like EDS1 and PAD4 to activate SAR and was recently found to localize at the plasma membrane by interacting with phospholipids, whereas NRG1 interacts with EDS1 and SAG101 to activate programmed cell death (Collier et al., 2011; Lapin et al., 2019; Saile et al., 2021). The peach orthologs of EDS1, PAD4 and SAG101 induced in infested Rubira could thus play a major role in the activation of the resistance to GPA. This is supported by the induction of the ortholog of SARD1 and of a close homolog of its regulator, CBP60b. The transcription factor SARD1 is thought to act downstream of EDS1 and PAD4 (Wang et al., 2011), by binding to the promoters of defense genes, notably ALD1, FMO1 and ICS1, directly involved in the biosynthesis of pipecolate and SA (Sun et al., 2015). Consistently, Rubira showed a significant accumulation of SA and pipecolate 48 hpi, demonstrating the establishment of SAR: the biosynthesis of SA upregulates NHP biosynthesis genes, then the mobile NHP triggers de novo SA biosynthesis in systemic tissues and amplifies ETI and PTI (Schnake et al., 2020; Yildiz et al., 2021). A response similar to the one we report here has been already observed by Donze-Reiner et al. (2017) in switchgrass (Panicum virgatum) leaves infested by the greenbug Schizaphis graminum, suggesting that pipecolate could be a major defense player against aphids in very distant species. Moreover, the transcriptional activation of MPK3 and WRKY33 indicates that the SAR positive regulatory loop revealed by Wang et al. (2018a) and involving MPK3, WRKY33, ALD1 and pipecolate was also triggered in Rubira 48 hpi. The upregulation of several DEGs involved in SA catabolism (SGT1, MES1, SAMT1) or in the repression of its biosynthesis (NPR3), suggest a downregulation of SA, possibly as a feedback loop after a strong stimulation of its biosynthesis. Infestation also induced the expression of the different elements of the resistosome, including another NLR, coding for ZAR1, which does not require a helper (Adachi et al., 2019), but whose molecular association with kinases was recently discovered (Bi and Zhou, 2021): ZED1, a decoy pseudokinase that captures a Pseudomonas effector (Lewis et al., 2013) and ZAR1/RKS1, which has been shown to recognize PBL2 (Wang et al., 2015) and form a pentameric calcium-permeable complex associated to the membrane, the resistosome, that promotes cell death when activated by an effector (Wang et al., 2019).
Overall, our observations fit well with the emerging model of a strong interplay between ETI and PTI. Ngou et al. (2021) showed that a strong immune response requires both ETI and PTI, via upregulation of PTI signalling components at transcriptional and post transcriptional levels, ETI being insufficient to activate ROS production alone, but enhancing it upon elicitation by PAMPs and elevated protein levels of PTI signalling components. The same finding was made by Yuan et al. (2021), who showed that ETI could not be established without the activation of PTI, in particular the establishment of an efficient oxidative burst, with the PRRs and NLRs receptors working in synergy. Consistently, downstream signalling networks were activated in Rubira upon infestation, involving calcium-dependent proteins CBLs, CPKs, CIPK and NADPH oxidase, which can be inferred to have led to a strong production of apoplastic ROS, mitigated by the accumulation of detoxification enzymes. CPK2 is involved in ROS signalling through its direct interaction with RBOHD (Wang et al., 2018b) and CPK8, CPK9 and CPK12 participate, directly or indirectly, in H2O2 homeostasis (Zhao et al., 2011; Zou et al., 2015; Chen et al., 2019). The role of ROS in plant-aphid interaction is well documented (see Goggin and Fischer, 2022, for review). Interestingly, we found the specific activation of a RBOHF ortholog consistent with the work on Arabidopsis/GPA reported by Jaouannet et al. (2015), but different from Kuśnierczyk et al. (2007), who reported a transcriptional upregulation of RBOHD in Arabidopsis Cvi ecotype but not in Ws after infestation by GPA or Brevicoryne brassicae. There thus seems to be a specialization of the RBOHs in the defense against aphids, which would be dependent on plant genotypes. More implication of ROS in response to GPA was revealed by the upregulation of Prupe_5G117000, the ortholog of WRKY53. This transcription factor, inducible by H2O2, is a key regulator of senescence and controls the expression of catalase genes (Miao et al., 2004). Although direct essays of ROS in tissues are lacking here to formally establish the onset of HR, it is attested by the upregulation of the NO pathway, with upregulation of NR1, specifically responsible for NO generation under the control of ABA and H2O2 (Bright et al., 2006). Early NO induction was previously reported in wheat responding to the Russian wheat aphid (Diuraphis noxia) and nitrate reductase transcriptional upregulation was observed in Arabidopsis upon infestation by GPA and B. brassicae (Kuśnierczyk et al., 2007).
Aphid-induced mobilization of glyoxylate and P5C/proline metabolisms likely promotes ROS burst in Rubira
An important metabolic consequence of aphid infestation in Rubira 48 hpi was the accumulation of glyoxylate and the upregulation of several afferent metabolic pathways (Figure 7). An activation of peroxisomal photorespiration was observed both at transcriptomic and metabolomic levels. Rojas et al. (2012) has shown the role of GOX2, which is a major source of H2O2, in the control of SA, JA and ethylene pathways, while Ahammed et al. (2018) demonstrated its role in resistance to P. syringae and its involvement in the regulation of SA-mediated signalling. Peroxisomal H2O2 production thus appears to be a key component of redox-mediated defense (Taler et al., 2004; Sandalio et al., 2021), and our data confirm the recent finding of its implication in Arabidopsis response to GPA (Xu et al., 2021). Our work suggests that the glyoxylate pool was also supplied by the glyoxylate shunt (Figure 7). Very little information is available about the involvement of this pathway in defense, but the upregulation of genes involved in lipid beta-oxidation feeding this pathway might be related to defense as well, since ACXs produce H2O2 (Kong et al., 2017) and ACX1 and PED1 are required for JA biosynthesis in the peroxisome (Cruz Castillo et al., 2004). Moreover, induction of ICL and MS have been reported in switchgrass infested by greenbugs (Donze-Reiner et al., 2017) and in soybean under attack by B. cinerea (Cots et al., 2002). It is therefore possible that the glyoxylate shunt operates in defense, beyond the fatty acid catabolism. Finally, the induction of purine and pyrimidine catabolism genes (homolog of PYD2; ortholog of PYD3 and ortholog of XDH1) and the accumulation of uracil and thymine degradation products, i.e. beta-alanine and BAIBA (Zrenner et al., 2009), suggests the possibility that glyoxylate was also synthesized from urate and was a marker of an accelerated senescence (Figure 7 and Table 1). Interestingly, the implication of XDH1 in defense-related ROS generation has been reported in epidermal cells of Arabidopsis leaves under powdery mildew attack (Ma et al., 2016).
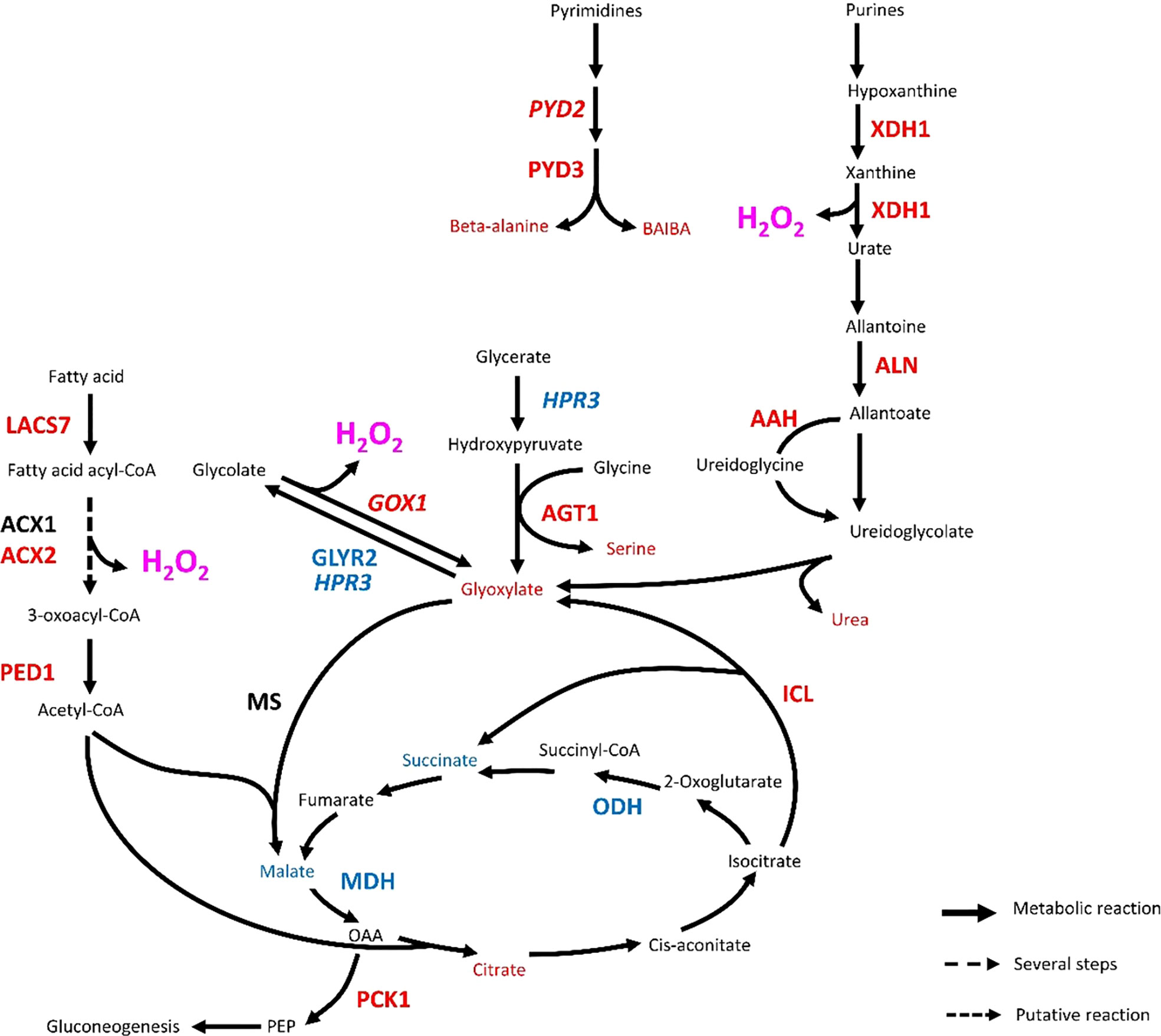
Figure 7 Transcriptional regulation of glyoxylate metabolic pathways in Rubira 48 hours after infestation with GPA. Genes are annotated according to their Arabidopsis orthologs, non-orthologs (homologs) are noted in italics. Upregulated genes/metabolites are figured in red, downregulated genes/metabolites in blue and not found or not differentially expressed genes/metabolites in black. AAH, Allantoate deiminase; ACX1, Peroxisomal acyl-coenzyme A oxidase 1; ACX2, Acyl-coenzyme A oxidase 2; AGT1, Glyoxylate aminotransferase 1; ALN, Allantoinase; GLYR2, Glyoxylate/succinic semialdehyde reductase 2; GOX1, Glycolate oxidase; HPR3, Glyoxylate/hydroxypyruvate reductase; ICL, Isocitrate lyase; LACS7, Long chain acyl-CoA synthetase 7; MDH, malate dehydrogenase; MS, Malate synthase; ODH, Oxoglutarate dehydrogenase; PCK1, Phosphoenolpyruvate carboxykinase 1; PED1, 3-ketoacyl-CoA thiolase 2, peroxisomal; PYD2, Dihydropyrimidinase; PYD3, Beta-ureidopropionase; XDH1, Xanthine dehydrogenase 1; BAIBA, Beta-aminoisobutyrate; PEP, Phosphoenol pyruvate.
Other metabolic reconfigurations induced in Rubira by GPA infestation concern the arginine, proline and lysine pathways, which revolve around the Δ1-Pyrroline-5-Carboxylate (P5C). This compound is produced by the catabolism of proline, under the action of POXs, or by the conversion of glutamate into glutamic-γ-semialdehyde (GSA), the latter being in spontaneous equilibrium with P5C. A third biosynthetic pathway of P5C is the catabolism of ornithine through the action of OAT (Miller et al., 2009). All P5C biosynthetic pathways were transcriptionally activated 48 hpi, whereas ALDH12A1 (P5CDH), which catalyzes its degradation to glutamate, was repressed (Figure 8). Furthermore, a net conversion of P5C to proline can be excluded as the proline pool remained stable: the simultaneous induction of POX2 and PROC1 (P5CR) rather indicate a high flux in the futile proline/P5C cycle. This cycle is thought to be ROS-generating because POX2 activity increases electron transfer from its cofactor FAD to the electron transfer chain and ultimately O2, resulting in mitochondrial ROS production (Miller et al., 2009). In animal cells, the proline/P5C cycle is central to the control of redox potential and cell death (Chalecka et al., 2021) and some studies have shown that it is involved in HR in plants (Monteoliva et al., 2014), in particular because the transcription of POXs in Arabidopsis was stimulated by SA and their activity was found increased in cells undergoing programmed cell death while the pox mutants, displaying reduced ROS levels and cell death, were more susceptible to P. syringae (Cecchini et al., 2011). These authors reported the same pattern as in our study: POX activation was accompanied by an increase in P5CR transcription, i.e. the proline/P5C cycle, but not P5CDH, while proline levels remained constant. The importance of OAT activity to PTI and ETI as well as to resistance to P. syringae has been demonstrated in Nicotiana benthamiana (Senthil-Kumar and Mysore, 2012). In our case, the urea cycle seemed to be oriented towards P5C formation, with induction of the ornithine-producing arginases ARGAH1 and ARGAH2, and induction of OAT, generating P5C, while citrulline formation collapsed due to the repression of OTC and CARB. The urea cycle therefore appeared to operate in an open mode to convert arginine to P5C, which is consistent with increased susceptibility to clubroot disease (Plasmodiophora brassicae) and B. cinerea reported in the Arabidopsis argah mutants (Brauc et al., 2012; Gravot et al., 2012), and with the JA-dependant induction of arginase shown by Gravot et al. (2012). Another hypothetical role for PROC1 could be the biosynthesis of pipecolate. The ALD1 pathway is thought to be the only effective pipecolate pathway in plants but an alternative route exists, with 3 intermediates: saccharopine, α-aminoadipate-δ-semialdehyde (AASA), and Δ1 –piperidine-6-carboxylate (P6C) (Hartmann and Zeier, 2018). None of these compounds were detected in our study but a strong accumulation of α-aminoadipate was observed, which is a direct catabolite of AASA, and a gene coding for an enzyme of the saccharopine pathway was also upregulated: the lysine-ketoglutarate reductase/saccharopine dehydrogenase (LKR/SDH), that produces AASA. In human, P6C was found to be in spontaneous equilibrium with AASA and P5CR (PROC1/PYCR1) was proved to be able to turn P6C into pipecolate (Struys et al., 2014). Though not very likely, an involvement of this pathway in pipecolate accumulation after aphid infestation cannot be entirely ruled out. Finally, the accumulation of hydroxyproline (Supplementary Figure S9), which may be derived from turnover of parietal proteins, as well as pyrroline-3-hydroxy-5-carboxylate, suggests the existence of an infestation-stimulated hydroxyproline oxidase activity. Such an enzyme is known in animals (Cooper et al., 2008) but has no known equivalent in plants yet.
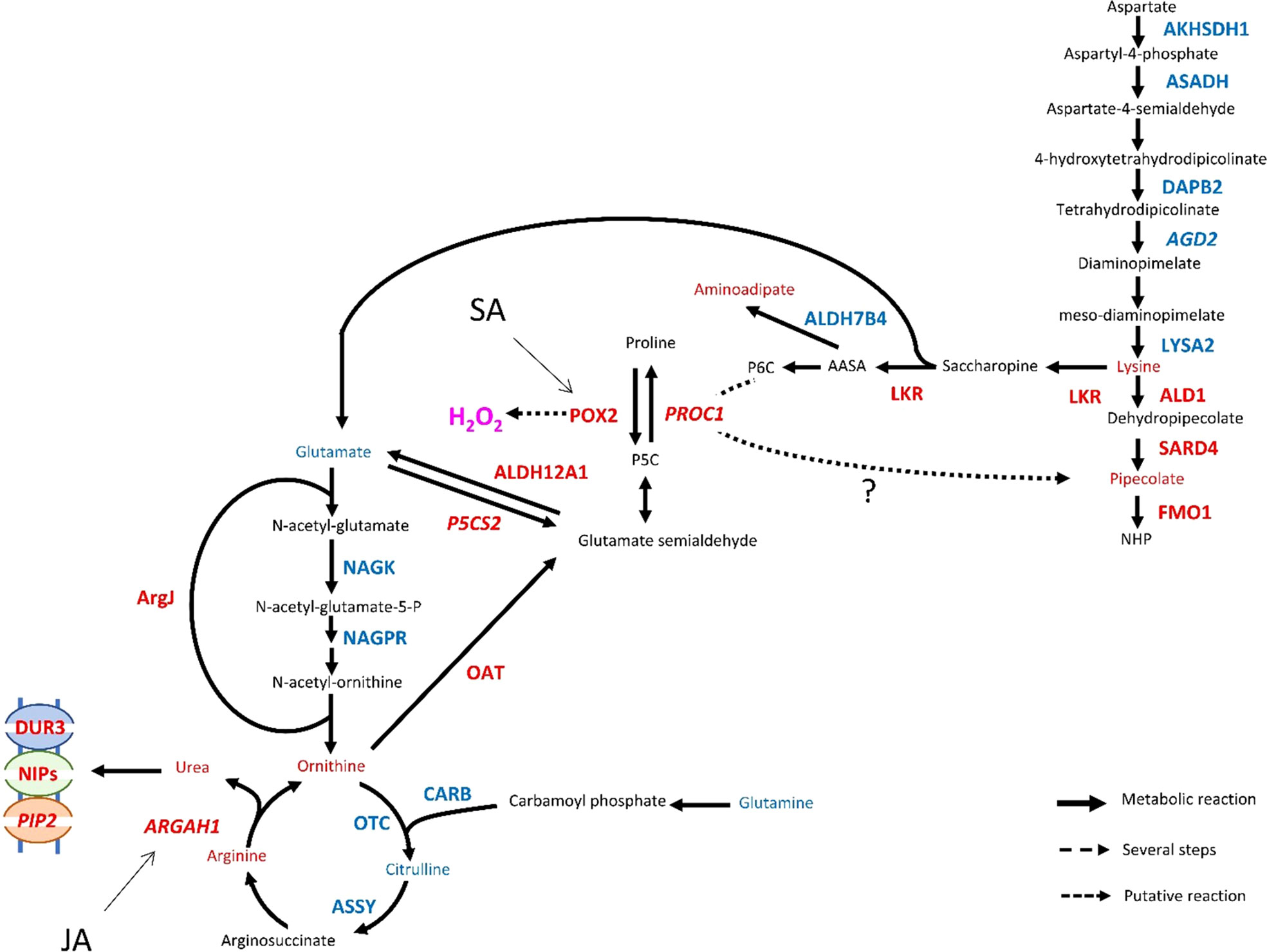
Figure 8 Transcriptional regulation of Arginine, proline and lysine metabolic pathways in Rubira 48 hours after infestation with GPA. Genes are annotated according to their Arabidopsis orthologs, non-orthologs (homologs) are noted in italics. Upregulated genes/metabolites are figured in red, downregulated genes/metabolites in blue and not found or not differentially expressed genes/metabolites in black. AGD2, LL-diaminopimelate aminotransferase; AKHSDH1, Aspartokinase/homoserine dehydrogenase 1; ALD1, AGD2-like defense response protein 1; ALDH12A1, Delta-1-pyrroline-5-carboxylate dehydrogenase 12A1; ALDH7B4, Aldehyde dehydrogenase family 7 member B4; ARGAH1, Arginase 1; ArgJ, Arginine biosynthesis bifunctional protein; Aspartate-semialdehyde dehydrogenase; ASSY, Arginosuccinate synthase; CARB, Carbamoyl-phosphate synthase large chain; DAPB2, 4-hydroxy-tetrahydrodipicolinate reductase 2; DUR3, Urea-proton symporter; FMO1, Probable flavin-containing monooxygenase 1; LKR/SDH, Alpha-aminoadipic semialdehyde synthase; LYSA2, Diaminopimelate decarboxylase 2; NAGK, Acetylglutamate kinase; NAGPR, N-acetyl-gamma-glutamyl-phosphate reductase; NIP1, Aquaporin Nodulin-26-like major intrinsic protein; OCT, Ornithine carbamoyltransferase; P5CS2, Delta-1-pyrroline-5-carboxylate synthase; PIP2, Aquaporin Plasma membrane intrinsic protein; POX2, Proline dehydrogenase 2; PROC1, Pyrroline-5-carboxylate reductase; SARD4, SAR deficient 4; AASA, alpha-amino adipate; NHP, N-hydroxypipecolate; P5C, Pyrroline-5-carboxylate; P6c, Pyrroline-6-carboxylate.
Response to GPA activated protein recycling through autophagy in Rubira
The observed accumulation of lysine, arginine and ornithine contrasted singularly with the repression of genes involved in their biosynthesis (Figure 8). This was also true to some extent for BCAAs and histidine. The work of Niu et al. (2018) showed the similar repression of these biosynthetic pathways 48 hpi in the “Fen Shouxing” accession carrying the major GPA resistance gene Rm3. One might think that this repression followed a strong activation earlier, but the genes of the lysine biosynthetic pathway for example were all repressed between 6 and 72 hpi (Niu et al., 2018). The origin of these amino acids could therefore lie in the recycling of proteins from the infestation-induced reconfiguration of the proteome, or even from a state of senescence, as suggested by the accumulation of urea, which is frequently accumulated in this situation (Bohner et al., 2015). The upregulation of autophagy, revealed by the induced expression of ATGs, NBR1 and genes involved in protein aggregate degradation (CHIP and HSP70-4) support the hypothesis of protein recycling. Interestingly, NBR1 was also found to target viral proteins, implying a possible coactivation of defenses against the aphid and the viruses it could transmit (Hafrén and Hofius, 2017).
Metabolic depletion of key metabolites might have reduced the nutritional value of Rubira for GPA
Some amino acids pools diminished 48 hpi. This was the case for glutamine and methionine, in a manner consistent with the transcriptional regulation of their biosynthetic pathway. When the decrease in sorbitol is also considered, it appears that the aphid’s main sources of carbon, nitrogen and sulfur nutrition have been reduced in Rubira. Jordan et al. (2020) reported peach infestation levels by GPA proportional to their amino acid and carbohydrate content, stressing that their reduction has the potential to impair aphid growth, a similar conclusion to that of Karley et al. (2002), who compared aphid performance on young developing and old mature potato plants with contrasted nutritional value. The importance of plant nutritional quality was demonstrated in feeding choice tests of the specialists aphids Uroleucon tanaceti and Macrosiphoniella tanacetaria fed on tansy (Tanacetum vulgare), as both species showed their preference for plants highly fertilized with nitrogen, while infestation with Uroleucon tanaceti even increased phloem essential amino acids, especially methionine (Nowak and Komor, 2010; Jakobs et al., 2019). Infestation by sucking insects is well known to modify amino acid levels: GPA for example increases the amino acid/carbohydrate ratio in Brassica pekinensis sap, and the absolute amino acid content in whole leaves (Cao et al., 2016). This study even indicated that nutritional value was at least as important a criterion for food choice as the level of plant defense. It is therefore probable that the reduction in key nutrients and the coincidental slowing of plant elongation negatively impacted aphid performance on Rubira and contributed to limit colony settlement.
Secondary metabolites accumulation may have contributed to antixenosis in Rubira
In compatible interactions, the very short exploration phase (only a few punctures before reaching the phloem), precedes a long ingestion phase as observed by Sauge et al. (1998b) on GF305 and a massive colonization associated with leaf curling. However, on Rubira, Weeping Flower Peach and “Fen Shouxing”, carrying Rm2, Rm1, Rm3 resistance genes respectively, aphids perform numerous punctures before reaching the phloem in which they fail to feed for long periods (Sauge et al., 1998a; Niu et al., 2018). This leads eventually to the rejection of the plant as a host, a phenomenon characteristic of phloem-based antixenosis (Sauge et al., 1998a), described as well by Le Roux et al. (2010) in the interaction between GPA and various wild Solanum species. Antixenosis may result from the presence of soluble secondary metabolites and indeed we noted in Rubira a particular accumulation of caffeoyl derivatives of quinic acid. Many of them increased in Rubira while they decreased in GF305, albeit reaching equal levels due to their higher constitutive content of GF305. Other compounds were more abundant in Rubira, such as 3,5 dicaffeoylquinic acid, whose accumulation has already been demonstrated in infested Rubira (Poëssel et al., 2002). The deterrent effects of phenolic compounds on aphid feeding have been demonstrated in vitro (Dreyer and Jones, 1981). Antixenosis, measured by choice tests and manifested by the escape of aphids, or even antibiosis, manifested by a direct toxic effect of molecules on the insect, has also been demonstrated in vivo: in pepper (Capsicum annuum) the leaf contents of direct hydroxycinnamic acid derivatives, notably caffeic acid, increased after infestation by GPA (Florencio-Ortiz et al., 2021), while in peach a negative correlation was found between the content of phenolic compounds in leaves and the level of infestation by GPA (Jordan et al., 2020). Similarly, in apple, resistant cultivars showed higher levels of hydroxycinnamic acids, especially 4-caffeoylquinic acid (Berrueta et al., 2018). The mode of action of these compounds remains elusive. The presence of an orthodiphenol group on the caffeoyl residue makes them potential substrates for polyphenol oxidases and peroxidases in H2O2 detoxification reactions, as well as chelating agents for metals such as iron that can modulate the pro-oxidative Fenton reaction. Such metabolites could also feed the deposition of oxidized polyphenols along the stylet track, as shown by in the interaction between GPA and potato (Kerchev et al., 2012).
Concerning the phenolic pathways, there was an apparent contradiction between the transcriptomic and metabolomic data: we observed an overall repression of the phenolic pathway genes in Rubira 48 hpi, while aromatic amino acids clearly accumulated as well as some phenylpropanoids. This may result from the activation kinetics of the shikimate pathway, as Niu et al. (2018) showed a slight induction of this pathway genes in the first hours after infestation of “ Fen shouxing”, but then, 3 hpi, repression was gradually set up, until it reached a gene expression profile close to the one observed in Rubira 48 hpi. An alternative or complementary explanation is that the pathway was fed by aromatic amino acid originating from the activation of proteolysis/autophagy in infested tissues mentioned above.
Increased prunasin and its caffeoyl derivative may also have played a role in Rubira’s resistance, as suggested by the in vitro suppressive effect of prunasin on the duration of ingestion phase of bird cherry-oat aphid (Rhopalosiphum padi), whose primary host is the cyanogenic glycosides rich bird cherry (Prunus padus) (Halarewicz and Gabryś, 2012). On top of a direct toxic effect of cyanogenic glycosides, their turnover has also been shown to contribute to SA biosynthesis in peach (Diaz-Vivancos et al., 2017), thereby possibly reinforcing defense signalling.
Conclusion
In conclusion, our results show that peach-GPA system provides a relevant model to study plant-aphid interactions and to decipher by omics approaches the molecular functions involved in susceptibility and in R-gene-mediated resistance.
These findings highlight the stealthy action of GPA on peach susceptible plants, likely resulting from repression of peach basal resistance by a very efficient effectors panel. They also illustrated the major transcriptome and metabolome reprogramming occurring during expression of the Rm2-mediated resistance, involving jointly ETI and PTI, SAR establishment by salicylic and pipecolic acids and primary and secondary metabolism changes resulting in enhanced defense (Figure 9). Further studies are needed to clarify which processes or defense compounds are key elements in triggering the antixenosis conferred by Rm2 gene in peach.
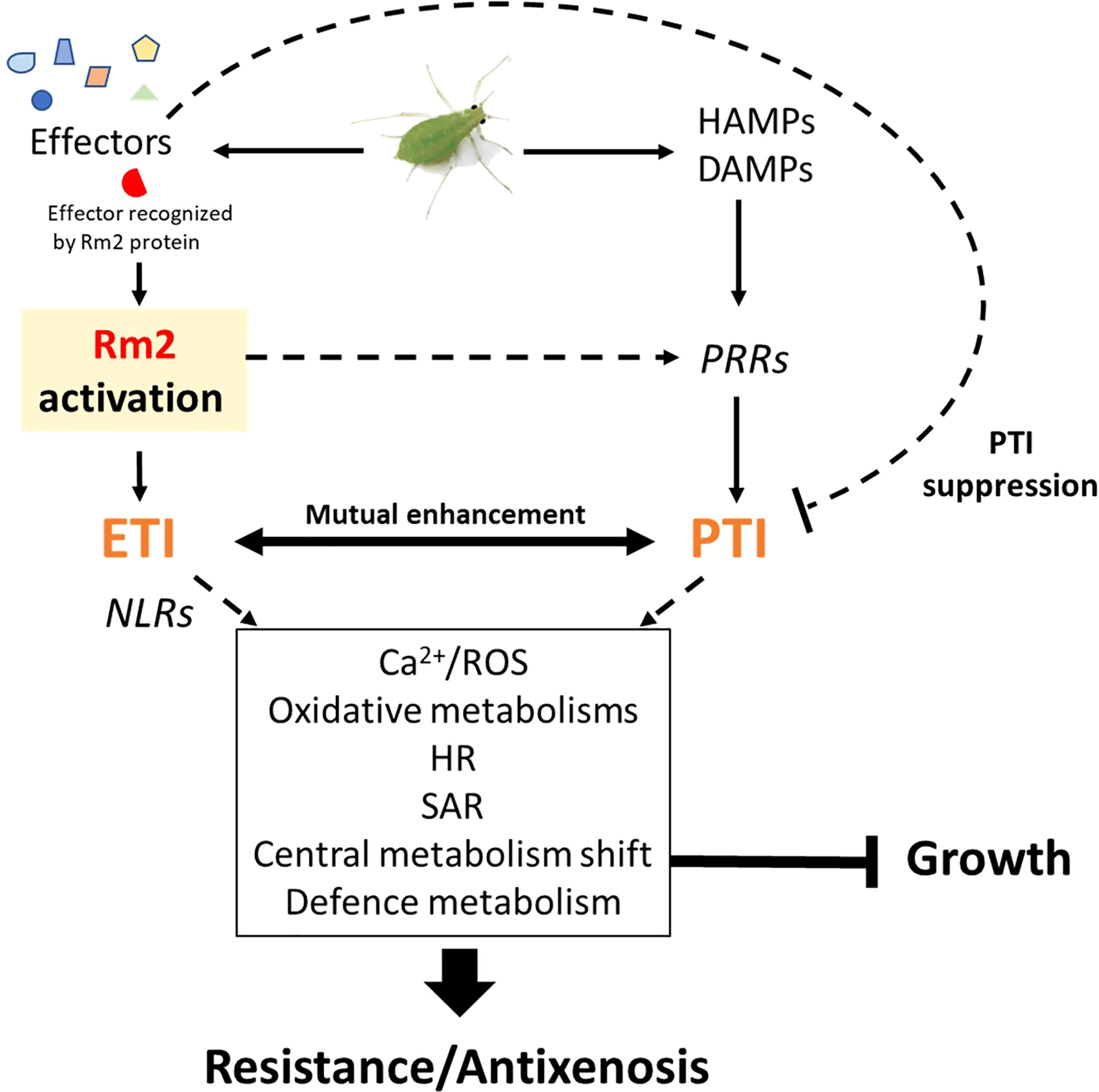
Figure 9 Conceptual model of the immune response to aphids in Rubira apices. Activation of the NLR receptor protein Rm2 by one of the M. persicae effectors triggers the transcription of numerous ETI and PTI genes as well as SAR and HR markers, resulting in metabolic reconfigurations favoring defense at the expense of growth: activation of H2O2-producing metabolic pathways, depletion of central metabolite pools and accumulation of caffeic acid conjugates.
Data availability statement
The full dataset of transcripts is available in the NCBI Sequence Read Archive. (BioProject ID PRJNA877419): https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA877419 Raw metabolomics dataset (accession number MSV000084377) can be downloaded from the publicly available MassIVE repository at the UCSD Center for Computational Mass Spectrometry website: https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=56e8ba4bcb814e3d92863c88702f235c.
Author contributions
RL, DR, J-LP designed the research and participated in setting up the experiments. PB performed analytical chemistry, datamining and contributed to write the paper. DR generated and analyzed the transcriptomic data. J-LP contributed to write the paper. RL, performed non-targeted metabolomics and wrote the paper. All authors contributed to the article and approved the submitted version.
Acknowledgments
We warmly thanks Virginie Ledoux who took care of the Myzus persicae rearing and who contributed to experiments. This work took place in the experimental facilities of AHM INRAE Unit and we thank the staff for growing the plants.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.992544/full#supplementary-material
Abbreviations
PAMPS, Pathogen-Associated Molecular Patterns; HAMPS, Herbivore-Associated Molecular Patterns; DAMPS, Damage-Associated Molecular Patterns; ETI, Effector Triggered Immunity, PTI, Pattern-triggered immunity; TNL, TIR Nucleotide-Binding and Leucine-rich repeat domains receptor; CNL Coiled-coil Nucleotide-Binding and Leucine-rich repeat domains receptor; DEG, Differentially expressed gene; hpi, hours post infestation; dpi, days post infestation; ROS, Reactive oxygene species; JA, Jasmonic acid; SA, Salicylic acid; ABA, Abscissic acid; BR, Brassinostéroïdes; NHP, N-hydroxypipecolate; GPA, Green peach aphid; HR, Hypersensible response; SAR, Systemic acquired resistance.
References
Adachi, H., Derevnina, L., Kamoun, S. (2019). NLR singletons, pairs, and networks: Evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 50, 121–131. doi: 10.1016/j.pbi.2019.04.007
Ahammed, G. J., Li, X., Zhang, G., Zhang, H., Shi, J., Pan, C., et al. (2018). Tomato photorespiratory glycolate-oxidase-derived H2O2 production contributes to basal defence against Pseudomonas syringae. Plant Cell Environ. 41, 1126–1138. doi: 10.1111/pce.12932
Atamian, H. S., Eulgem, T., Kaloshian, I. (2012). SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta 235, 299–309. doi: 10.1007/s00425-011-1509-6
Barco, B., Clay, N. K. (2020). Hierarchical and dynamic regulation of defense-responsive specialized metabolism by WRKY and MYB transcription factors. Front. Plant Sci. 10, 1775. doi: 10.3389/fpls.2019.01775
Berardini, T. Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The arabidopsis information resource: Making and mining the ‘gold standard’ annotated reference plant genome. Genesis 53, 474–485. doi: 10.1002/dvg.22877
Bernstein, K. A., Gallagher, J. E. G., Mitchell, B. M., Granneman, S., Baserga, S. J. (2004). The small-subunit processome is a ribosome assembly intermediate. Eukaryotic Cell 3, 1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004
Berrueta, L. A., Sasía-Arriba, A., Miñarro, M., Antón, M. J., Alonso-Salces, R. M., Micheletti, D., et al. (2018). Relationship between hydroxycinnamic acids and the resistance of apple cultivars to rosy apple aphid. Talanta 187, 330–336. doi: 10.1016/j.talanta.2018.05.040
Bi, G., Zhou, J.-M. (2021). Regulation of cell death and signaling by pore-forming resistosomes. Annu. Rev. Phytopathol. 59, 239–263. doi: 10.1146/annurev-phyto-020620-095952
Bohner, A., Kojima, S., Hajirezaei, M., Melzer, M., von Wirén, N. (2015). Urea retranslocation from senescing arabidopsis leaves is promoted by DUR3-mediated urea retrieval from leaf apoplast. Plant J. 81, 377–387. doi: 10.1111/tpj.12740
Boissot, N., Schoeny, A., Vanlerberghe-Masutti, F. (2016a). Vat, an amazing gene conferring resistance to aphids and viruses they carry: From molecular structure to field effects. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01420
Boissot, N., Thomas, S., Chovelon, V., Lecoq, H. (2016b). NBS-LRR-mediated resistance triggered by aphids: viruses do not adapt; aphids adapt via different mechanisms. BMC Plant Biol. 16, 25. doi: 10.1186/s12870-016-0708-5
Bos, J. I. B., Prince, D., Pitino, M., Maffei, M. E., Win, J., Hogenhout, S. A. (2010). A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (Green peach aphid). PloS Genet. 6, e1001216. doi: 10.1371/journal.pgen.1001216
Brauc, S., De Vooght, E., Claeys, M., Geuns, J. M. C., Höfte, M., Angenon, G. (2012). Overexpression of arginase in Arabidopsis thaliana influences defence responses against Botrytis cinerea. Plant Biol. 14 Suppl 1, 39–45. doi: 10.1111/j.1438-8677.2011.00520.x
Bright, J., Desikan, R., Hancock, J. T., Weir, I. S., Neill, S. J. (2006). ABA-induced NO generation and stomatal closure in arabidopsis are dependent on H2O2 synthesis. Plant J. 45, 113–122. doi: 10.1111/j.1365-313X.2005.02615.x
Brutus, A., Sicilia, F., Macone, A., Cervone, F., De Lorenzo, G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. 107, 9452–9457. doi: 10.1073/pnas.1000675107
Cabrera y Poch, H. L., Ponz, F., Fereres, A. (1998). Searching for resistance in Arabidopsis thaliana to the green peach aphid Myzus persicae. Plant Sci. 138, 209–216. doi: 10.1016/S0168-9452(98)00144-7
Cao, H.-H., Liu, H.-R., Zhang, Z.-F., Liu, T.-X. (2016). The green peach aphid Myzus persicae perform better on pre-infested Chinese cabbage Brassica pekinensis by enhancing host plant nutritional quality. Sci. Rep. 6, 21954. doi: 10.1038/srep21954
Casteel, C. L., Walling, L. L., Paine, T. D. (2006). Behavior and biology of the tomato psyllid, Bactericerca cockerelli, in response to the Mi-1.2 gene. Entomol Experimentalis Applicata 121, 67–72. doi: 10.1111/j.1570-8703.2006.00458.x
Cecchini, N. M., Monteoliva, M. I., Alvarez, M. E. (2011). Proline dehydrogenase contributes to pathogen defense in arabidopsis. Plant Physiol. 155, 1947–1959. doi: 10.1104/pp.110.167163
Chalecka, M., Kazberuk, A., Palka, J., Surazynski, A. (2021). P5C as an interface of proline interconvertible amino acids and its role in regulation of cell survival and apoptosis. Int. J. Mol. Sci. 22, 11763. doi: 10.3390/ijms222111763
Chaudhary, R., Atamian, H. S., Shen, Z., Briggs, S. P., Kaloshian, I. (2014). GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc. Natl. Acad. Sci. 111, 8919–8924. doi: 10.1073/pnas.1407687111
Chen, S., Ding, Y., Tian, H., Wang, S., Zhang, Y. (2021). WRKY54 and WRKY70 positively regulate SARD1 and CBP60g expression in plant immunity. Plant Signaling Behav. 16, 1932142. doi: 10.1080/15592324.2021.1932142
Chen, Y.-C., Holmes, E. C., Rajniak, J., Kim, J.-G., Tang, S., Fischer, C. R., et al. (2018). N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in arabidopsis. Proc. Natl. Acad. Sci. 115, E4920–E4929. doi: 10.1073/pnas.1805291115
Chen, D.-H., Liu, H.-P., Li, C.-L. (2019). Calcium-dependent protein kinase CPK9 negatively functions in stomatal abscisic acid signaling by regulating ion channel activity in arabidopsis. Plant Mol. Biol. 99, 113–122. doi: 10.1007/s11103-018-0805-y
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., Jones, J. D. G., et al. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Choi, H. W., Kim, Y. J., Hwang, B. K. (2011). The hypersensitive induced reaction and leucine-rich repeat proteins regulate plant cell death associated with disease and plant immunity. Mol. Plant-Microbe Interact. 24, 68–78. doi: 10.1094/MPMI-02-10-0030
Chung, E.-H., da Cunha, L., Wu, A.-J., Gao, Z., Cherkis, K., Afzal, A. J., et al. (2011). Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9, 125–136. doi: 10.1016/j.chom.2011.01.009
Coll-Garcia, D., Mazuch, J., Altmann, T., Müssig, C. (2004). EXORDIUM regulates brassinosteroid-responsive genes. FEBS Lett. 563, 82–86. doi: 10.1016/S0014-5793(04)00255-8
Collier, S. M., Hamel, L.-P., Moffett, P. (2011). Cell death mediated by the n-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant-Microbe Interact. 24, 918–931. doi: 10.1094/MPMI-03-11-0050
Cooper, S. K., Pandhare, J., Donald, S. P., Phang, J. M. (2008). A novel function for hydroxyproline oxidase in apoptosis through generation of reactive oxygen species. J. Biol. Chem. 283, 10485–10492. doi: 10.1074/jbc.M702181200
Coppola, V., Coppola, M., Rocco, M., Digilio, M. C., D'Ambrosio, C., Renzone, G., et al. (2013). Transcriptomic and proteomic analysis of a compatible tomato-aphid interaction reveals a predominant salicylic acid-dependent plant response. BMC Genomics 14, 515. doi: 10.1186/1471-2164-14-515
Cots, J., Fargeix, C., Gindro, K., Widmer, F. (2002). Pathogenic attack and carbon reallocation in soybean leaves (Glycine max. l.): Reinitiation of the glyoxylate cycle as a defence reaction. J. Plant Physiol. 159, 91–96. doi: 10.1078/0176-1617-00620
Cruz Castillo, M., Martínez, C., Buchala, A., Métraux, J.-P., León, J. (2004). Gene-specific involvement of beta-oxidation in wound-activated responses in arabidopsis. Plant Physiol. 135, 85–94. doi: 10.1104/pp.104.039925
Dellero, Y., Jossier, M., Schmitz, J., Maurino, V. G., Hodges, M. (2016). Photorespiratory glycolate-glyoxylate metabolism. J. Exp. Bot. 67, 3041–3052. doi: 10.1093/jxb/erw090
Diaz-Vivancos, P., Bernal-Vicente, A., Cantabella, D., Petri, C., Hernández, J. A. (2017). Metabolomics and biochemical approaches link salicylic acid biosynthesis to cyanogenesis in peach plants. Plant Cell Physiol. 58, 2057–2066. doi: 10.1093/pcp/pcx135
Ding, P., Rekhter, D., Ding, Y., Feussner, K., Busta, L., Haroth, S., et al. (2016). Characterization of a pipecolic acid biosynthesis pathway required for systemic acquired resistance. Plant Cell 28, 2603–2615. doi: 10.1105/tpc.16.00486
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
Dogimont, C., Chovelon, V., Pauquet, J., Boualem, A., Bendahmane, A. (2014). The Vat locus encodes for a CC-NBS-LRR protein that confers resistance to Aphis gossypii infestation and A. gossypii-mediated virus resistance. Plant J. 80, 993–1004. doi: 10.1111/tpj.12690
Dongus, J. A., Parker, J. E. (2021). EDS1 signalling: At the nexus of intracellular and surface receptor immunity. Curr. Opin. Plant Biol. 62, 102039. doi: 10.1016/j.pbi.2021.102039
Donze-Reiner, T., Palmer, N. A., Scully, E. D., Prochaska, T. J., Koch, K. G., Heng-Moss, T., et al. (2017). Transcriptional analysis of defense mechanisms in upland tetraploid switchgrass to greenbugs. BMC Plant Biol. 17, 46. doi: 10.1186/s12870-017-0998-2
Dreyer, D. L., Jones, K. C. (1981). Feeding deterrency of flavonoids and related phenolics towards Schizaphis graminum and Myzus persicae: Aphid feeding deterrents in wheat. Phytochemistry 20, 2489–2493. doi: 10.1016/0031-9422(81)83078-6
Duxbury, Z., Wu, C., Ding, P. (2021). A comparative overview of the intracellular guardians of plants and animals: NLRs in innate immunity and beyond. Annu. Rev. Plant Biol. 72, 155–184. doi: 10.1146/annurev-arplant-080620-104948
Erwig, J., Ghareeb, H., Kopischke, M., Hacke, R., Matei, A., Petutschnig, E., et al. (2017). Chitin-induced and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) phosphorylation-dependent endocytosis of Arabidopsis thaliana LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE5 (LYK5). New Phytol. 215, 382–396. doi: 10.1111/nph.14592
Eyres, I., Jaquiéry, J., Sugio, A., Duvaux, L., Gharbi, K., Zhou, J. J., et al. (2016). Differential gene expression according to race and host plant in the pea aphid. Mol. Ecol. 25, 4197–4215. doi: 10.1111/mec.13771
Feehan, J. M., Castel, B., Bentham, A. R., Jones, J. D. (2020). Plant NLRs get by with a little help from their friends. Curr. Opin. Plant Biol. 56, 99–108. doi: 10.1016/j.pbi.2020.04.006
Florencio-Ortiz, V., Gruz, J., Casas, J. L. (2021). Changes in the free phenolic acid composition of pepper (Capsicum annuum l.) leaves in response to green peach aphid (Myzus persicae sulzer) infestation. Arthropod-Plant Interact. 15, 329–336. doi: 10.1007/s11829-021-09820-w
Foyer, C. H., Verrall, S. R., Hancock, R. D. (2015). Systematic analysis of phloem-feeding insect-induced transcriptional reprogramming in arabidopsis highlights common features and reveals distinct responses to specialist and generalist insects. J. Exp. Bot. 66, 495–512. doi: 10.1093/jxb/eru491
Friedrichsen, D. M., Nemhauser, J., Muramitsu, T., Maloof, J. N., Alonso, J., Ecker, J. R., et al. (2002). Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162, 1445–1456. doi: 10.1093/genetics/162.3.1445
Gao, Z., Chung, E.-H., Eitas, T. K., Dangl, J. L. (2011). Plant intracellular innate immune receptor resistance to pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc. Natl. Acad. Sci. 108, 7619–7624. doi: 10.1073/pnas.1104410108
Gao, P., Zhang, H., Yan, H., Zhou, N., Yan, B., Fan, Y., et al. (2021). Transcriptomic and metabolomic changes triggered by Macrosiphum rosivorum in rose (Rosa longicuspis). BMC Genomics 22, 885. doi: 10.1186/s12864-021-08198-6
Gao, Q.-M., Zhu, S., Kachroo, P., Kachroo, A. (2015). Signal regulators of systemic acquired resistance. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00228
Goggin, F. L., Fischer, H. D. (2022). Reactive oxygen species in plant interactions with aphids. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.811105
Gómez-Gómez, L., Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/s1097-2765(00)80265-8
Gravot, A., Deleu, C., Wagner, G., Lariagon, C., Lugan, R., Todd, C., et al. (2012). Arginase induction represses gall development during clubroot infection in arabidopsis. Plant Cell Physiol. 53, 901–911. doi: 10.1093/pcp/pcs037
Gupta, A., Singh, M., Laxmi, A. (2015). Multiple interactions between glucose and brassinosteroid signal transduction pathways in arabidopsis are uncovered by whole-genome transcriptional profiling. Plant Physiol. 168, 1091–1105. doi: 10.1104/pp.15.00495
Hafrén, A., Hofius, D. (2017). NBR1-mediated antiviral xenophagy in plant immunity. Autophagy 13, 2000–2001. doi: 10.1080/15548627.2017.1339005
Halarewicz, A., Gabryś, B. (2012). Probing behavior of bird cherry-oat aphid Rhopalosiphum padi (L.) on native bird cherry Prunus padus l. and alien invasive black cherry Prunus serotina erhr. in Europe and the role of cyanogenic glycosides. Arthropod-Plant Interact. 6, 497–505. doi: 10.1007/s11829-012-9228-x
Hartmann, M., Kim, D., Bernsdorff, F., Ajami-Rashidi, Z., Scholten, N., Schreiber, S., et al. (2017). Biochemical principles and functional aspects of pipecolic acid biosynthesis in plant immunity. Plant Physiol. 174, 124–153. doi: 10.1104/pp.17.00222
Hartmann, M., Zeier, J. (2018). L-lysine metabolism to n-hydroxypipecolic acid: an integral immune-activating pathway in plants. Plant J. 96, 5–21. doi: 10.1111/tpj.14037
Hartmann, M., Zeier, T., Bernsdorff, F., Reichel-Deland, V., Kim, D., Hohmann, M., et al. (2018). Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 173, 456–469. doi: 10.1016/j.cell.2018.02.049
Hawkins, C., Ginzburg, D., Zhao, K., Dwyer, W., Xue, B., Xu, A., et al. (2021). Plant metabolic network 15: A resource of genome-wide metabolism databases for 126 plants and algae. J. Integr. Plant Biol. 63, 1888–1905. doi: 10.1111/jipb.13163
Hou, S., Wang, X., Chen, D., Yang, X., Wang, M., Turrà, D., et al. (2014). The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PloS Pathog. 10, e1004331. doi: 10.1371/journal.ppat.1004331
Huang, W., Wu, Z., Tian, H., Li, X., Zhang, Y. (2021). Arabidopsis CALMODULIN-BINDING PROTEIN 60b plays dual roles in plant immunity. Plant Commun. 2, 100213. doi: 10.1016/j.xplc.2021.100213
Hu, Y. R., Dong, Q. Y., Yu, D. Q. (2012). Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185185, 288–297. doi: 10.1016/j.plantsci.2011.12.003
Jakobs, R., Schweiger, R., Müller, C. (2019). Aphid infestation leads to plant part-specific changes in phloem sap chemistry, which may indicate niche construction. New Phytol. 221, 503–514. doi: 10.1111/nph.15335
Jaouannet, M., Morris, J. A., Hedley, P. E., Bos, J. I. B. (2015). Characterization of arabidopsis transcriptional responses to different aphid species reveals genes that contribute to host susceptibility and non-host resistance. PloS Pathog. 11, e1004918. doi: 10.1371/journal.ppat.1004918
Jhou, Y.-S., Poovendhan, S., Huang, L.-H., Tsai, C.-W. (2021). Host acceptance and plant resistance: A comparative behavioral study of Myzus persicae and. Acyrthosiphon Pisum Insects 12, 975. doi: 10.3390/insects12110975
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jones, J. D. G., Vance, R. E., Dangl, J. L. (2016). Intracellular innate immune surveillance devices in plants and animals. Science 354, (6316), aaf6395-1- aaf6395-8. doi: 10.1126/science.aaf6395
Jordan, M.-O., Sauge, M.-H., Vercambre, G. (2020). Chemical and growth traits of the peach tree may induce higher infestation rates of the green peach aphid, Myzus persicae (Sulzer). Pest Manage. Sci. 76, 797–806. doi: 10.1002/ps.5583
Karley, A. J., Douglas, A. E., Parker, W. E. (2002). Amino acid composition and nutritional quality of potato leaf phloem sap for aphids. J. Exp. Biol. 205, 3009–3018. doi: 10.1242/jeb.205.19.3009
Kerchev, P. I., Fenton, B., Foyer, C. H., Hancock, R. D. (2012). Infestation of potato (Solanum tuberosum l.) by the peach-potato aphid (Myzus persicae sulzer) alters cellular redox status and is influenced by ascorbate. Plant Cell Environ. 35, 430–440. doi: 10.1111/j.1365-3040.2011.02395.x
Kerchev, P. I., Karpińska, B., Morris, J. A., Hussain, A., Verrall, S. R., Hedley, P. E., et al. (2013). Vitamin c and the abscisic acid-insensitive 4 transcription factor are important determinants of aphid resistance in arabidopsis. Antioxid Redox Signaling 18, 2091–2105. doi: 10.1089/ars.2012.5097
Kim, M. G., Geng, X., Lee, S. Y., Mackey, D. (2009). The Pseudomonas syringae type III effector AvrRpm1 induces significant defenses by activating the arabidopsis nucleotide-binding leucine-rich repeat protein RPS2. Plant J. 57, 645–653. doi: 10.1111/j.1365-313X.2008.03716.x
Kimura, S., Waszczak, C., Hunter, K., Wrzaczek, M. (2017). Bound by fate: The role of reactive oxygen species in receptor-like kinase signaling. Plant Cell 29, 638–654. doi: 10.1105/tpc.16.00947
Kirkin, V., Lamark, T., Sou, Y.-S., Bjorkoy, G., Nunn, J. L., Bruun, J. A., et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516. doi: 10.1016/j.molcel.2009.01.020
Knepper, C., Savory, E. A., Day, B. (2011). The role of NDR1 in pathogen perception and plant defense signaling. Plant Signaling Behav. 6, 1114–1116. doi: 10.4161/psb.6.8.15843
Kohorn, B. D., Johansen, S., Shishido, A., Todorova, T., Martinez, R., Defeo, E., et al. (2009). Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 60, 974–982. doi: 10.1111/j.1365-313X.2009.04016.x
Kong, F., Liang, Y., Légeret, B., Beyly-Adriano, A., Blangy, S., Haslam, R. P., et al. (2017). Chlamydomonas carries out fatty acid β-oxidation in ancestral peroxisomes using a bona fide acyl-CoA oxidase. Plant J. 90, 358–371. doi: 10.1111/tpj.13498
Kuśnierczyk, A., Winge, P., Jørstad, T. S., Troczyńska, J., Rossiter, J. T., Bones, A. M. (2008). Towards global understanding of plant defence against aphids-timing and dynamics of early arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ. 31, 1097–1115. doi: 10.1111/j.1365-3040.2008.01823.x
Kuśnierczyk, A., Winge, P., Midelfart, H., Armbruster, W. S., Rossiter, J. T., Bones, A. M. (2007). Transcriptional responses of Arabidopsis thaliana ecotypes with different glucosinolate profiles after attack by polyphagous Myzus persicae and oligophagous Brevicoryne brassicae. J. Exp. Bot. 58, 2537–2552. doi: 10.1093/jxb/erm043
Kurusu, T., Hamada, J., Hamada, H., Hanamata, S., Kuchitsu, K. (2010). Roles of calcineurin b-like protein-interacting protein kinases in innate immunity in rice. Plant Signaling Behav. 5, 1045–1047. doi: 10.4161/psb.5.8.12407
Lambert, P., Pascal, T. (2011). Mapping Rm2 gene conferring resistance to the green peach aphid (Myzus persicae sulzer) in the peach cultivar “Rubira®”. Tree Genet. Genomes 7, 1057–1068. doi: 10.1007/s11295-011-0394-2
Lang, J., Genot, B., Bigeard, J., Colcombet, J. (2022). MPK3 and MPK6 control salicylic acid signaling by up-regulating NLR receptors during pattern- and effector-triggered immunity. J. Exp. Bot. 73, 2190–2205. doi: 10.1093/jxb/erab544
Lapin, D., Kovacova, V., Sun, X., Dongus, J. A., Bhandari, D., von Born, P., et al. (2019). A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell 31, 2430–2455. doi: 10.1105/tpc.19.00118
Lee, S., Lee, D. W., Lee, Y., Mayer, U., Stierhof, Y.-D., Lee, S., et al. (2009). Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in arabidopsis. Plant Cell 21, 3984–4001. doi: 10.1105/tpc.109.071548
Le Roux, V., Dugravot, S., Brunissen, L., Vincent, C., Pelletier, Y., Giordanengo, P. (2010). Antixenosis phloem-based resistance to aphids: is it the rule? Ecol. Entomol 35, 407–416. doi: 10.1111/j.1365-2311.2010.01192.x
Lewis, J. D., Lee, A. H.-Y., Hassan, J. A., Wan, J., Hurley, B., Jhingree, J. R., et al. (2013). The arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc. Natl. Acad. Sci. 110, 18722–18727. doi: 10.1073/pnas.131552011
Liang, D., Chen, M., Qi, X., Xu, Q., Zhou, F., Chen, X. (2016). QTL mapping by SLAF-seq and expression analysis of candidate genes for aphid resistance in cucumber. Front. Plant Sci. 7, 1000. doi: 10.3389/fpls.2016.01000
Li, J., Brader, G., Kariola, T., Palva, E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491. doi: 10.1111/j.1365-313X.2006.02712.x
Li, B., Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li, Y., Kim, J. I., Pysh, L., Chapple, C. (2015). Four isoforms of arabidopsis 4-Coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 169, 2409–2421. doi: 10.1104/pp.15.00838
Liu, J., Elmore, J. M., Fuglsang, A. T., Palmgren, M. G., Staskawicz, B. J., Coaker, G. (2009). RIN4 functions with plasma membrane h+-ATPases to regulate stomatal apertures during pathogen attack. PloS Biol. 7, e1000139. doi: 10.1371/journal.pbio.1000139
Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E., Walker, J. C. (2002). BAK1, an arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. doi: 10.1016/s0092-8674(02)00812-7
Li, L.-S., Ying, J., Li, E., Ma, T., Li, M., Gong, L.-M., et al. (2021). Arabidopsis CBP60b is a central transcriptional activator of immunity. Plant Physiol. 186, 1645–1659. doi: 10.1093/plphys/kiab164
Li, X., Zhang, Y., Clarke, J. D., Li, Y., Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98, 329–339. doi: 10.1016/s0092-8674(00)81962-5
Louis, J., Shah, J. (2013). Arabidopsis thaliana-myzus persicae interaction: shaping the understanding of plant defense against phloem-feeding aphids. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00213
Lü, B.-B., Li, X.-J., Sun, W.-W., Li, L., Gao, R., Zhu, Q., et al. (2013). AtMYB44 regulates resistance to the green peach aphid and diamondback moth by activating EIN2-affected defences in arabidopsis. Plant Biol. 15, 841–850. doi: 10.1111/j.1438-8677.2012.00675.x
Marcec, M. J., Gilroy, S., Poovaiah, B. W., Tanaka, K. (2019). Mutual interplay of Ca2+ and ROS signaling in plant immune response. Plant Sci. 283, 343–354. doi: 10.1016/j.plantsci.2019.03.004
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Matiz, A., Cambuí, C. A., Richet, N., Mioto, P. T., Gomes, F., Pikart, F. C., et al. (2019). Involvement of aquaporins on nitrogen-acquisition strategies of juvenile and adult plants of an epiphytic tank-forming bromeliad. Planta 250, 319–332. doi: 10.1007/s00425-019-03174-7
Ma, X., Wang, W., Bittner, F., Schmidt, N., Berkey, R., Zhang, L. L., et al. (2016). Dual and opposing roles of xanthine dehydrogenase in defense-associated reactive oxygen species metabolism in arabidopsis. Plant Cell 28, 1108–1126. doi: 10.1105/tpc.15.00880
Miao, Y., Laun, T., Zimmermann, P., Zentgraf, U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in arabidopsis. Plant Mol. Biol. 55, 853–867. doi: 10.1007/s11103-004-2142-6
Miller, G., Honig, A., Stein, H., Suzuki, N., Mittler, R., Zilberstein, A. (2009). Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J. Biol. Chem. 284, 26482–26492. doi: 10.1074/jbc.M109.009340
Milligan, S. B., Bodeau, J., Yaghoobi, J., Kaloshian, I., Zabel, P., Williamson, V. M. (1998). The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10, 1307–1319. doi: 10.1105/tpc.10.8.1307
Mittler, R., Blumwald, E. (2015). The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27, 64–70. doi: 10.1105/tpc.114.133090
Mohanta, T. K., Occhipinti, A., Atsbaha Zebelo, S., Foti, M., Fliegmann, J., Bossi, S., et al. (2012). Ginkgo biloba responds to herbivory by activating early signaling and direct defenses. PloS One 7, e32822. doi: 10.1371/journal.pone.0032822
Monet, R., Massonié, G. (1994). Déterminisme génétique de la résistance au puceron vert (Myzus persicae) chez le pêcher. Résultats Complémentaires Agronom. 14, 177–182. doi: 10.1051/agro:19940304
Monteoliva, M. I., Rizzi, Y. S., Cecchini, N. M., Hajirezaei, M.-R., Alvarez, M. E. (2014). Context of action of proline dehydrogenase (ProDH) in the hypersensitive response of arabidopsis. BMC Plant Biol. 14, 21. doi: 10.1186/1471-2229-14-21
Muzac, I., Wang, J., Anzellotti, D., Zhang, H., Ibrahim, R. K. (2000). Functional expression of an arabidopsis cDNA clone encoding a flavonol 3’-o-methyltransferase and characterization of the gene product. Arch. Biochem. Biophysics 375, 385–388. doi: 10.1006/abbi.1999.1681
Návarová, H., Bernsdorff, F., Döring, A.-C., Zeier, J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24, 5123–5141. doi: 10.1105/tpc.112.103564
Ngou, B. P. M., Ahn, H.-K., Ding, P., Jones, J. D. G. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Ngou, B. P. M., Ding, P., Jones, J. D. G. (2022a). Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/plcell/koac041
Ngou, B. P. M., Jones, J. D. G., Ding, P. (2022b). Plant immune networks. Trends Plant Sci. 27, 255–273. doi: 10.1016/j.tplants.2021.08.012
Niu, L., Pan, L., Zeng, W., Lu, Z., Cui, G., Fan, M., et al. (2018). Dynamic transcriptomes of resistant and susceptible peach lines after infestation by green peach aphids (Myzus persicae sülzer) reveal defence responses controlled by the Rm3 locus. BMC Genomics 19, 846. doi: 10.1186/s12864-018-5215-7
Nombela, G., Williamson, V. M., Muñiz, M. (2003). The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol. Plant-Microbe Interact. 16, 645–649. doi: 10.1094/MPMI.2003.16.7.645
Nosarzewski, M., Downie, A. B., Wu, B., Archbold, D. D. (2012). The role of SORBITOL DEHYDROGENASE in Arabidopsis thaliana. Funct. Plant Biol. 39, 462–470. doi: 10.1071/FP12008
Nowak, H., Komor, E. (2010). How aphids decide what is good for them: experiments to test aphid feeding behaviour on Tanacetum vulgare (L.) using different nitrogen regimes. Oecologia 163, 973–984. doi: 10.1007/s00442-010-1652-y
Ortiz-Morea, F. A., He, P., Shan, L., Russinova, E. (2020). It takes two to tango – molecular links between plant immunity and brassinosteroid signalling. J. Cell Sci. 133, jcs246728. doi: 10.1242/jcs.246728
Pallipparambil, G. R., Sayler, R. J., Shapiro, J. P., Thomas, J. M. G., Kring, T. J., Goggin, F. L. (2015). Mi-1.2, an R gene for aphid resistance in tomato, has direct negative effects on a zoophytophagous biocontrol agent, Orius insidiosus. J. Exp. Bot. 66, 549–557. doi: 10.1093/jxb/eru361
Pan, L., Lu, Z., Yan, L., Zeng, W., Shen, Z., Yu, M., et al. (2022). NLR1 is a strong candidate for the Rm3 dominant green peach aphid (Myzus persicae) resistance trait in peach. J. Exp. Bot. 73, 1357–1369. doi: 10.1093/jxb/erab506
Parthasarathy, A., Savka, M. A., Hudson, A. O. (2019). The synthesis and role of β-alanine in plants. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00921
Pascal, T., Aberlenc, R., Confolent, C., Hoerter, M., Lecerf, E., Tuéro, C., et al. (2017). Mapping of new resistance (Vr2, Rm1) and ornamental (Di2, pl) mendelian trait loci in peach. Euphytica 213, 132. doi: 10.1007/s10681-017-1921-5
Pascal, T., Pfeiffer, F., Kervella, J., Lacroze, J. P., Sauge, M. H., Weber, W. E. (2002). Inheritance of green peach aphid resistance in the peach cultivar ‘Rubira’. Plant Breed. 121, 459–461. doi: 10.1046/j.1439-0523.2002.734333.x
Pitorre, D., Llauro, C., Jobet, E., Guilleminot, J., Brizard, J.-P., Delseny, M., et al. (2010). RLK7, a leucine-rich repeat receptor-like kinase, is required for proper germination speed and tolerance to oxidative stress in Arabidopsis thaliana. Planta 232, 1339–1353. doi: 10.1007/s00425-010-1260-4
Pitzschke, A., Djamei, A., Bitton, F., Hirt, H. (2009). A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant 2, 120–137. doi: 10.1093/mp/ssn079
Poëssel, J. L., Corre, M. N., Kervella, J., Lacroze, J. P., Sauge, M. H. (2002). “Increase in phenolic content in the resistant peach cultivar 'Rubira' infested by the green peach aphid, myzus persicae,” in XXI international conference on polyphenols, vol. 1 . Polyphenols Communications 2002. Ed. Hadrami, I. E.(Groupe Polyphénols Publisher: Marrakech, Morocco), 131–132.
Pompon, J., Pelletier, Y. (2012). Changes in aphid probing behaviour as a function of insect age and plant resistance level. Bull. Entomol Res. 102, 550–557. doi: 10.1017/S0007485312000120
Pontier, D., Gan, S., Amasino, R. M., Roby, D., Lam, E. (1999). Markers for hypersensitive response and senescence show distinct patterns of expression. Plant Mol. Biol. 39, 1243–1255. doi: 10.1023/a1006133311402
Prince, D. C., Drurey, C., Zipfel, C., Hogenhout, S. A. (2014). The leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in arabidopsis. Plant Physiol. 164, 2207–2219. doi: 10.1104/pp.114.235598
Qi, Y., Tsuda, K., Nguyen, L. V., Wang, X., Lin, J., Murphy, A. S., et al. (2011). Physical association of arabidopsis hypersensitive induced reaction proteins (HIRs) with the immune receptor RPS2. J. Biol. Chem. 286, 31297–31307. doi: 10.1074/jbc.M110.211615
Raudvere, U., Kolberg, L., Kuzmin, I., Arak, T., Adler, P., Peterson, H., et al. (2019). gProfiler: a web server for functional enrichment analysis and conversions of gene list, (2019 Update). Nucleic Acids Res. 47, 191–198. doi: 10.1093/nar/gkz369
Roessner, U., Wagner, C., Kopka, J., Trethewey, R. N., Willmitzer, L. (2000). Simultaneous analysis of metabolites in potato tuber by gas chromatography–mass spectrometry. Plant J. 23, 131–142. doi: 10.1046/j.1365-313x.2000.00774.x
Rojas, C. M., Senthil-Kumar, M., Wang, K., Ryu, C.-M., Kaundal, A., Mysore, K. S. (2012). Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in Nicotiana benthamiana and arabidopsis. Plant Cell 24, 336–352. doi: 10.1105/tpc.111.093245
Rossi, M., Goggin, F. L., Milligan, S. B., Kaloshian, I., Ullman, D. E., Williamson, V. M. (1998). The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. 95, 9750–9754. doi: 10.1073/pnas.95.17.9750
Saile, S. C., Ackermann, F. M., Sunil, S., Keicher, J., Bayless, A., Bonardi, V., et al. (2021). Arabidopsis ADR1 helper NLR immune receptors localize and function at the plasma membrane in a phospholipid dependent manner. New Phytol. 232, 2440–2456. doi: 10.1111/nph.17788
Sandalio, L. M., Peláez-Vico, M. A., Molina-Moya, E., Romero-Puertas, M. C. (2021). Peroxisomes as redox-signaling nodes in intracellular communication and stress responses. Plant Physiol. 186, 22–35. doi: 10.1093/plphys/kiab060
Sauge, M.-H., Kervella, J., Pascal, T. (1998a). Settling behaviour and reproductive potential of the green peach aphid myzus persicae on peach varieties and a related wild prunus. Entomol Experimentalis Applicata 89, 233–242. doi: 10.1046/j.1570-7458.1998.00404.x
Sauge, M.-H., Kervella, J., Rahbé, Y. (1998b). Probing behaviour of the green peach aphid myzus persicae on resistant prunus genotypes. Entomol Experimentalis Applicata 89, 223–232. doi: 10.1046/j.1570-7458.1998.00403.x
Sauge, M.-H., Poëssel, J.-L., Guillemaud, T., Lapchin, L. (2011). Resistance induction and herbivore virulence in the interaction between Myzus persicae (Sulzer) and a major aphid resistance gene (Rm2) from peach. Arthropod-Plant Interact. 5, 369–377. doi: 10.1007/s11829-011-9141-8
Schnake, A., Hartmann, M., Schreiber, S., Malik, J., Brahmann, L., Yildiz, I., et al. (2020). Inducible biosynthesis and immune function of the systemic acquired resistance inducer N-hydroxypipecolic acid in monocotyledonous and dicotyledonous plants. J. Exp. Bot. 71, 6444–6459. doi: 10.1093/jxb/eraa317
Schymanski, E. L., Jeon, J., Gulde, R., Fenner, K., Ruff, M., Singer, H. P., et al. (2014). Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ. Sci. Technol. 48, 2097–2098. doi: 10.1021/es5002105
Senthil-Kumar, M., Mysore, K. S. (2012). Ornithine-delta-aminotransferase and proline dehydrogenase genes play a role in non-host disease resistance by regulating pyrroline-5-carboxylate metabolism-induced hypersensitive response. Plant Cell Environ. 35, 1329–1343. doi: 10.1111/j.1365-3040.2012.02492.x
Shim, J. S., Jung, C., Lee, S., Min, K., Lee, Y.-W., Choi, Y., et al. (2013). AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 73, 483–495. doi: 10.1111/tpj.12051
Shimomura, H., Sashida, Y., Adachi, T. (1987). Cyanogenic and phenylpropanoid glucosides from Prunus grayana. Phytochemistry 26, 2363–2366. doi: 10.1016/S0031-9422(00)84720-2
Sierla, M., Waszczak, C., Vahisalu, T., Kangasjärvi, J. (2016). Reactive oxygen species in the regulation of stomatal movements. Plant Physiol. 171, 1569–1580. doi: 10.1104/pp.16.00328
Silva-Sanzana, C., Estevez, J. M., Blanco-Herrera, F. (2020). Influence of cell wall polymers and their modifying enzymes during plant-aphid interactions. J. Exp. Bot. 71, 3854–3864. doi: 10.1093/jxb/erz550
Smith, S. M. (2002). Does the glyoxylate cycle have an anaplerotic function in plants? Trends Plant Sci. 7, 12–13. doi: 10.1016/S1360-1385(01)02189-6
Solomon, O. L., Berger, D. K., Myburg, A. A. (2010). Diurnal and circadian patterns of gene expression in the developing xylem of eucalyptus trees. South Afr. J. Bot. 76, 425–439. doi: 10.1016/j.sajb.2010.02.087
Struys, E. A., Jansen, E. E. W., Salomons, G. S. (2014). Human pyrroline-5-carboxylate reductase (PYCR1) acts on Δ(1)-piperideine-6-carboxylate generating l-pipecolic acid. J. Inherited Metab. Dis. 37, 327–332. doi: 10.1007/s10545-013-9673-4
Sun, T., Zhang, Y., Li, Y., Zhang, Q., Ding, Y., Zhang, Y. (2015). ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 6, 10159. doi: 10.1038/ncomms10159
Taler, D., Galperin, M., Benjamin, I., Cohen, Y., Kenigsbuch, D. (2004). Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell 16, 172–184. doi: 10.1105/tpc.016352
Tanaka, T., Ikeda, A., Shiojiri, K., Ozawa, R., Shiki, K., Nagai-Kunihiro, N., et al. (2018). Identification of a hexenal reductase that modulates the composition of green leaf volatiles. Plant Physiol. 178, 552–564. doi: 10.1104/pp.18.00632
Tang, W., Kim, T.-W., Oses-Prieto, J. A., Sun, Y., Deng, Z., Zhu, S., et al. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in arabidopsis. Science 321, 557–560. doi: 10.1126/science.1156973
Tang, D., Wang, G., Zhou, J.-M. (2017). Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 29, 618–637. doi: 10.1105/tpc.16.00891
Tang, W., Yuan, M., Wang, R., Yang, Y. H., Wang, C. M., Oses-Prieto, J. A., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13, 124–131. doi: 10.1038/ncb2151
ten Broeke, C. J. M., Dicke, M., van Loon, J. J. A. (2014). Rearing history affects behaviour and performance of two virulent Nasonovia ribisnigri populations on two lettuce cultivars. Entomol Experimentalis Applicata 151, 97–105. doi: 10.1111/eea.12181
The UniProt Consortium (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, 480–489. doi: 10.1093/nar/gkaa1100
Thomas, P. D., Campbell, M. J., Kejariwal, A., Mi, H., Karlak, B., Daverman, R., et al. (2003). PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141. doi: 10.1101/gr.772403
Thorpe, P., Escudero-Martinez, C. M., Eves-van den Akker, S., Bos, J. I. B. (2020). Transcriptional changes in the aphid species Myzus cerasi under different host and environmental conditions. Insect Mol. Biol. 29, 271–282. doi: 10.1111/imb.12631
Toyota, M., Spencer, D., Sawai-Toyota, S., Jiaqi, W., Zhang, T., Koo, A. J., et al. (2018). Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361, 1112–1115. doi: 10.1126/science.aat774
Tronchet, M., Balagué, C., Kroj, T., Jouanin, L., Roby, D. (2009). Cinnamyl alcohol dehydrogenases-c and d, key enzymes in lignin biosynthesis, play an essential role in disease resistance in arabidopsis. Mol. Plant Pathol. 11, 83–92. doi: 10.1111/j.1364-3703.2009.00578.x
Tungadi, T., Watt, L. G., Groen, S. C., Murphy, A. M., Du, Z., Pate, A. E., et al. (2021). Infection of arabidopsis by cucumber mosaic virus triggers jasmonate-dependent resistance to aphids that relies partly on the pattern-triggered immunity factor BAK1. Mol. Plant Pathol. 22, 1082–1091. doi: 10.1111/mpp.13098
Tuteja, N., Tran, N. Q., Dang, H. Q., Tuteja, R. (2011). Plant MCM proteins: role in DNA replication and beyond. Plant Mol. Biol. 77, 537–545. doi: 10.1007/s11103-011-9836-3
Verde, I., Jenkins, J., Dondini, L., Micali, S., Pagliarani, G., Vendramin, E., et al. (2017). The peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics 18, 225. doi: 10.1186/s12864-017-3606-9
Villada, E. S., González, E. G., López-Sesé, A. I., Castiel, A. F., Gómez-Guillamón, M. L. (2009). Hypersensitive response to Aphis gossypii glover in melon genotypes carrying the Vat gene. J. Exp. Bot. 60, 3269–3277. doi: 10.1093/jxb/erp163
Vincent, T. R., Avramova, M., Canham, J., Higgins, P., Bilkey, N., Mugford, S. T., et al. (2017). Interplay of plasma membrane and vacuolar ion channels, together with BAK1, elicits rapid cytosolic calcium elevations in arabidopsis during aphid feeding. Plant Cell 29, 1460–1479. doi: 10.1105/tpc.17.00136
Vos, P., Simons, G., Jesse, T., Wijbrandi, J., Heinen, L., Hogers, R., et al. (1998). The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotechnol. 16, 1365–1369. doi: 10.1038/4350
Wang, X., Chory, J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313, 1118–1122. doi: 10.1126/science.1127593
Wang, G., Roux, B., Feng, F., Guy, E., Li, L., Li, N. N., et al. (2015). The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18, 285–295. doi: 10.1016/j.chom.2015.08.004
Wang, Y., Schuck, S., Wu, J., Yang, P., Döring, A.-C., Zeier, J., et al. (2018a). A MPK3/6-WRKY33-ALD1-Pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell 30, 2480–2494. doi: 10.1105/tpc.18.00547
Wang, L., Tsuda, K., Truman, W., Sato, M., Nguyen, L. V., Katagiri, F., et al. (2011). CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 67, 1029–1041. doi: 10.1111/j.1365-313X.2011.04655.x
Wang, J., Wang, J., Hu, M., Wu, S., Qi, J. F., Wang, G. X., et al. (2019). Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364, 5868–5870. doi: 10.1126/science.aav5868
Wang, W., Zhang, H., Wei, X., Yang, L., Yang, B., Zhang, L., et al. (2018b). Functional characterization of calcium-dependent protein kinase (CPK) 2 gene from oilseed rape (Brassica napus l.) in regulating reactive oxygen species signaling and cell death control. Gene 651, 49–56. doi: 10.1016/j.gene.2018.02.006
Werner, A. K., Witte, C.-P. (2011). The biochemistry of nitrogen mobilization: purine ring catabolism. Trends Plant Sci. 16, 381–387. doi: 10.1016/j.tplants.2011.03.012
Will, T., Tjallingii, W. F., Thönnessen, A., van Bel, A. J. E. (2007). Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. 104, 10536–10541. doi: 10.1073/pnas.0703535104
Winter, G., Todd, C. D., Trovato, M., Forlani, G., Funck, D. (2015). Physiological implications of arginine metabolism in plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00534
Wu, Z., Li, M., Dong, O. X., Xia, S., Liang, W., Bao, Y., et al. (2019). Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol. 222, 938–953. doi: 10.1111/nph.15665
Xu, J., Padilla, C. S., Li, J., Wickramanayake, J., Fischer, H. D., Goggin, F. L. (2021). Redox responses of Arabidopsis thaliana to the green peach aphid, Myzus persicae. Mol. Plant Pathol. 22, 727–736. doi: 10.1111/mpp.13054
Yamaguchi, T., Yamamoto, K., Asano, Y. (2014). Identification and characterization of CYP79D16 and CYP71AN24 catalyzing the first and second steps in l-phenylalanine-derived cyanogenic glycoside biosynthesis in the Japanese apricot, Prunus mume sieb. et zucc. Plant Mol. Biol. 86, 215–223. doi: 10.1007/s11103-014-0225-6
Yildiz, I., Mantz, M., Hartmann, M., Zeier, T., Kessel, J., Thurow, C., et al. (2021). The mobile SAR signal N-hydroxypipecolic acid induces NPR1-dependent transcriptional reprogramming and immune priming. Plant Physiol. 186, 1679–1705. doi: 10.1093/plphys/kiab166
Yoshimoto, K., Ohsumi, Y. (2018). Unveiling the molecular mechanisms of plant autophagy–from autophagosomes to vacuoles in plants. Plant Cell Physiol. 59, 1337–1344. doi: 10.1093/pcp/pcy112
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Zhai, Y., Li, P., Mei, Y., Chen, M., Chen, X., Xu, H., et al. (2017). Three MYB genes co-regulate the phloem-based defence against English grain aphid in wheat. J. Exp. Bot. 68, 4153–4169. doi: 10.1093/jxb/erx204
Zhang, B., Van Aken, O., Thatcher, L., De Clercq, I., Duncan, O., Law, S. R., et al. (2014). The mitochondrial outer membrane AAA ATPase AtOM66 affects cell death and pathogen resistance in Arabidopsis thaliana. Plant J. 80, 709–727. doi: 10.1111/tpj.12665
Zhang, D., Zhao, Y., Wang, J., Zhao, P., Xu, S. (2021). BRS1 mediates plant redox regulation and cold responses. BMC Plant Biol. 21, 268. doi: 10.1186/s12870-021-03045-y
Zhao, Y., Dong, W., Zhu, Y., Allan, A. C., Lin-Wang, K., Xu, C. (2020). PpGST1, an anthocyanin-related glutathione s-transferase gene, is essential for fruit coloration in peach. Plant Biotechnol. J. 18, 1284–1295. doi: 10.1111/pbi.13291
Zhao, R., Wang, X., Zhang, D.-P. (2011). CPK12: A Ca2+-dependent protein kinase balancer in abscisic acid signaling. Plant Signaling Behav. 6, 1687–1690. doi: 10.4161/psb.6.11.17954
Zhou, J., Zhang, Y., Qi, J., Chi, Y., Fan, B., Yu, J.-Q., et al. (2014a). E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PloS Genet. 10, e1004116. doi: 10.1371/journal.pgen.1004116
Zhou, Y., Zhou, H., Lin-Wang, K., Vimolmangkang, S., Espley, R. V., Wang, L., et al. (2014b). Transcriptome analysis and transient transformation suggest an ancient duplicated MYB transcription factor as a candidate gene for leaf red coloration in peach. BMC Plant Biol. 14, 388. doi: 10.1186/s12870-014-0388-y
Zogli, P., Pingault, L., Grover, S., Louis, J. (2020). Ento(o)mics: the intersection of ‘omic’ approaches to decipher plant defense against sap-sucking insect pests. Curr. Opin. Plant Biol. 56, 153–161. doi: 10.1016/j.pbi.2020.06.002
Zou, J.-J., Li, X.-D., Ratnasekera, D., Wang, C., Liu, W.-X., Song, L.-F., et al. (2015). Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27, 1445–1460. doi: 10.1105/tpc.15.00144
Zrenner, R., Riegler, H., Marquard, C. R., Lange, P. R., Geserick, C., Bartosz, C. E., et al. (2009). A functional analysis of the pyrimidine catabolic pathway in arabidopsis. New Phytol. 183, 117–132. doi: 10.1111/j.1469-8137.2009.02843.x
Keywords: Green peach aphid (Myzus persicae), Effector-triggered immunity (ETI), Prunus persica, Metabolomics, Pattern-triggered immunity (PTI), Transcriptomics (RNA-Seq), Systemic acquired resistance (SAR), Hypersensitive response (HR)
Citation: Le Boulch P, Poëssel J-L, Roux D and Lugan R (2022) Molecular mechanisms of resistance to Myzus persicae conferred by the peach Rm2 gene: A multi-omics view. Front. Plant Sci. 13:992544. doi: 10.3389/fpls.2022.992544
Received: 12 July 2022; Accepted: 08 September 2022;
Published: 05 October 2022.
Edited by:
Zuhua He, Center for Excellence in Molecular Plant Sciences (CAS), ChinaReviewed by:
Jiancai Li, Key Laboratory of Insect Developmental and Evolutionary Biology (CAS), Shanghai, ChinaXue-xin Chen, Zhejiang University, China
Copyright © 2022 Le Boulch, Poëssel, Roux and Lugan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raphaël Lugan, cmFwaGFlbC5sdWdhbkB1bml2LWF2aWdub24uZnI=
 Pauline Le Boulch
Pauline Le Boulch Jean-Luc Poëssel
Jean-Luc Poëssel David Roux
David Roux Raphaël Lugan
Raphaël Lugan