- 1College of Horticulture, Shenyang Agricultural University, Shenyang, China
- 2College of Horticulture Science and Technology, Hebei Normal University of Science and Technology, Qinhuangdao, China
- 3Hebei Key Laboratory of Horticultural Germplasm Excavation and Innovative Utilization, Qinhuangdao, China
- 4National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design and Application Technology (Liaoning), Shenyang, China
Grape gray mold disease (Botrytis cinerea) is widespread during grape production especially in Vitis vinifera and causes enormous losses to the grape industry. In nature, the grapevine cultivar ‘Beta ‘ (Vitis riparia × Vitis labrusca) showed high resistance to grape gray mold. Until now, the candidate genes and their mechanism of gray mold resistance were poorly understood. In this study, we firstly conducted quantitative trait locus (QTL) mapping for grape gray mold resistance based on two hybrid offspring populations that showed wide separation in gray mold resistance. Notably, two stable QTL related to gray mold resistance were detected and located on linkage groups LG2 and LG7. The phenotypic variance ranged from 6.86% to 13.70% on LG2 and 4.40% to 11.40% on LG7. Combined with RNA sequencing (RNA-seq), one structural gene VlEDR2 (Vitvi02g00982) and three transcription factors VlERF039 (Vitvi00g00859), VlNAC047 (Vitvi08g01843), and VlWRKY51 (Vitvi07g01847) that may be involved in VlEDR2 expression and grape gray mold resistance were selected. This discovery of candidate gray mold resistance genes will provide an important theoretical reference for grape gray mold resistance mechanisms, research, and gray mold-resistant grape cultivar breeding in the future.
1 Introduction
Vitis vinifera L. belongs to genus Vitis of the family Vitaceae. As a major table grape resource, it possesses important social and economic values in the world. While in China, due to the temperate continental climate, it is easily infected by many fungal diseases among which the grape gray mold that is caused by Botrytis cinerea Pers. was one of the major pathogens (Choquer et al., 2007). In most grape production regions, in case of infection by grape gray mold, the yield would reduce by 20%–60%, and the berry quality would also face huge damages (Martínez-Romero et al., 2007; Dean et al., 2012; Saito et al., 2019). During the grape production process, antifungal agents could inhibit the occurrence of diseases to a certain extent, but this is not recommended due to environmental pollution and food safety. At present, the breeding of high gray mold resistance grapevine cultivar became a hot point. In nature, many grapevine resources possess higher gray mold resistance than V. vinifera L., including Vitis amurensis Rupr., Vitis quinquangularis Rehd., Vitis piasezkii Maxim., Vitis riparia Michx, Vitis rupestris Scheele, and Vitis labrusca L. (Gabler et al., 2003; Wan et al., 2015).
Marker-assisted selection based on genetic linkage map construction and quantitative trait locus (QTL) mapping has been widely used to screen high disease resistance grapevine cultivars through traditional crossbreeding strategies such as ripe rot, downy mildew, powdery mildew, and white rot (Barba et al., 2014; Teh et al., 2017; Fu et al., 2019; Sapkota et al., 2019; Tello et al., 2019; Su et al., 2021) for its high breeding efficiency. Until now, there were no QTL mapping reports related to grape gray mold resistance, and research on gray mold resistance transcriptional regulation mechanism was majorly focused on the transcription factor ERF and MYB families in Arabidopsis and tomato (Lorenzo et al., 2003; Pre et al., 2008; Zhao et al., 2012; Liu et al., 2021). In grapevine, there have been some reports related to gray mold resistance including structure genes VvSWEE4, VvSWEE15, VvSWEET7, and VvAMP2 and some transcription factors including VvWRKY52, VqERF072, VqERF112, VqERF114, VaERF20, VaERF16, and VaMYB306 (Nanni et al., 2014; Jiao et al., 2015; Wang et al., 2018a; Wang et al., 2018b; Breia et al., 2019; Zhu et al., 2019; Wang et al., 2020; Zhu et al., 2022). While the quantitative trait was controlled by many genes, candidate genes related to gray mold resistance in grapevine still need to be explored.
In this study, we selected three gray mold resistance grapevine cultivars, ‘Zhuosexiang’ (‘ZSX’) (V. vinifera × V. labrusca), ‘Venus seedless’ (‘VS’) (V. vinifera × V. labrusca), and ‘Beta’ (“BT”) (V. riparia × V. labrusca), and two susceptible cultivars, ‘Red Globe’ (‘RG’) and ‘Victoria’ (‘VT’), which belong to V. vinifera. Among these grapevine cultivars, ‘RG’ was identified as one of the highly susceptible grape cultivars to B. cinerea (Wan et al., 2015), and ‘BT’ was usually used as rootstock for its high cold and disease resistance character. Based on the hybrid population and high-density genetic linkage map (Zhu et al., 2018; Su et al., 2021), which was created through interspecific crossing of ‘ZSX’ × ‘VT’ and ‘RG’ × ‘VS,’ we firstly conducted QTL mapping for gray mold resistance, and then transcriptome analysis was conducted for ‘RG’ and ‘BT’ at different infection stages on account of their most distinct resistance level of grapevine gray mold. Finally, candidate genes related to grapevine gray mold resistance were screened by QTL mapping and RNA sequencing (RNA-seq).
2 Materials and methods
2.1 Plant material and gray mold resistance identification
Grape cultivars ‘RG’ (V. vinifera L.), ‘VT’ (V. vinifera L.), ‘ZSX’ (V. vinifera × V. labrusca), ‘VS’ (V. vinifera × V. labrusca), and ‘Beta’ (‘BT’) (V. riparia × V. labrusca) and two hybrid populations were cultivated in the Grape Experimental Garden of Shenyang Agricultural University (23°24’N, 41°50’E), China. Interspecific hybridization of ‘RG’ × ‘VS’ was conducted in May 2009; ‘RG’ was used as the female parent, and ‘VS’ was used as the male parent. ‘ZSX’ × ‘VT’ was conducted in May 2014; ‘ZSX’ was used as the female parent, and ‘VT’ was used as the male parent. A total of 177 and 176 individuals from ‘RG’ × ‘VS’ and ‘ZSX’ × ‘VT’ were used for the gray mold resistance identification in 2019 and 2020. The third-to-fourth leaf from the tip of an annual branch was selected (three leaves per individual). The collected leaves were rinsed with 70% ethanol for 1 min, followed by 10% sodium hypochlorite for 1 min, and rinsed three times with ultrapure water. Next, the leaves were placed in plastic culture dishes and punctured in the left, middle, and right regions. Ten microliters of 107/ml gray mold spore suspension was then dripped on the wound points to induce gray mold infection. Leaves with gray mold spores were incubated in a moist chamber at 28°C with 95% relative humidity. The lesion area of the infected region of each leaf was measured with a YMJ-C smart leaf area meter (Tuopu Instrument, Guangdong, China) (Su et al., 2021). Leaf samples of ‘RG’ and ‘BT’ that showed distinct resistance to gray mold at 0, 72, and 120 h after infection were collected for RNA-seq. Three biological replicates were collected at different infection periods of each cultivar with at least three leaves per replicate.
2.2 Gray mold resistance quantitative trait locus mapping
The lesion area (mean value of three replicates) of each genotype collected in 2019 and 2020 was used for QTL mapping. The integrated genetic linkage maps of ‘RG’ × ‘VS’ and ‘ZSX’ × ‘VT’ used in this research were constructed by using Restriction-site Associated DNA (RAD)-Sequencing, including 6,249 and 70,061 single nucleotide polymorphism (SNP) markers (Zhu et al., 2018; Su et al., 2021). A multiple QTL mapping (MQM) method was used to find significant QTL after a 1,000-permutation test (α = 0.05) based on the R/qtl package (Broman et al., 2003), and finally, the Logarithm of odds (LOD) threshold was set to 3. The max.qtl was set to 10 for forward selection. A 1-LOD confidence interval corresponding to the 95% confidence interval was calculated by using the “lodint” function. The explained phenotypic variation of each QTL phenotypic variation explained (PVE) was estimated using the “fitqtl” function. Candidate genes within the confidence interval of each QTL on the integrated map were selected according to 12X.v2 version of the Grape Genome database (https://urgi.versailles.inra.fr/Species/Vitis/Data-Sequences/Genome-sequences).
2.3 Gray mold resistance transcriptome analysis
RNA integrity was assessed using the RNA Nano 6000 Assay Kit and the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). The input material for the RNA sample preparation was 1-μg RNA per sample. Sequencing libraries were generated using the NEBNext® Ultra™ RNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA) and then sequenced on an Illumina Novaseq platform. Finally, 150-bp paired-end reads were generated. Clean reads were obtained by removing reads containing adapter, ploy-N, and low-quality reads from the raw data. The high-quality and paired-end clean reads were aligned to the reference genome (https://urgi.versailles.inra.fr/Species/Vitis/Data-Sequences/Genome-sequences) using HISAT 2v2.0.5 software, and the mapped reads of each sample were assembled by StringTie. The fragments per kilobase per million (FPKM) value of each gene was calculated based on the length of the gene and the number of reads mapped to this gene. Differential expression analysis was performed using the DESeq2 R package (1.20.0), and genes with an adjusted P-value <0.05 found by DESeq2 were assigned as differentially expressed. Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) was implemented by the clusterProfiler R package.
2.4 qRT-PCR validation of candidate genes
Infected leaves of grape cultivars ‘RG’ and ‘BT’ at 0, 72, and 120 h after gray mold infection were collected, and then these samples were used for total RNA extraction according to the manufacturer’s instructions of Plant Total RNA Isolation Kit (SK8631; Sangon Biotech, Shanghai, China). The PrimeScript™ RT-PCR Kit (RR047A; TaKaRa Bio, Kusatsu, Japan) was used to conduct cDNA synthesis, and the cDNA was diluted five times. Quantitative real-time PCR (qRT-PCR) was conducted in ABI QuantStudio™ 6 Flex System (Applied Biosystems). The relative expression level of selected genes was normalized to grapevine β-actin (Fujimori et al., 2016) and calculated using the 2-ΔΔCT method. All reactions were performed using three biological replicates. The primers used in this study are listed in Table S1.
3 Results
3.1 Identification of grapevine gray mold resistance
Gray mold resistance identification of five grape cultivars, ‘RG,’ ‘VT,’ ‘ZSX,’ ‘VS,’ and ‘BT,’ at different infection stages was evaluated based on the lesion area (Figure 1A). Among these five cultivars, ‘BT’ showed the highest resistance to gray mold infection, and ‘ZSX’ also showed higher resistance compared with the other three cultivars. Furthermore, 176 hybrid progenies of ‘RG’ × ‘VS’ and 177 hybrid progenies of ‘ZSX’ × ‘VT’ were identified for gray mold resistance in 2019 and 2020; the results of these two hybrid progenies showed continuous variation (Figure 1B; Table S2). These results indicated that gray mold resistance in grapevine was a typical quantitative trait controlled by multiple genes.
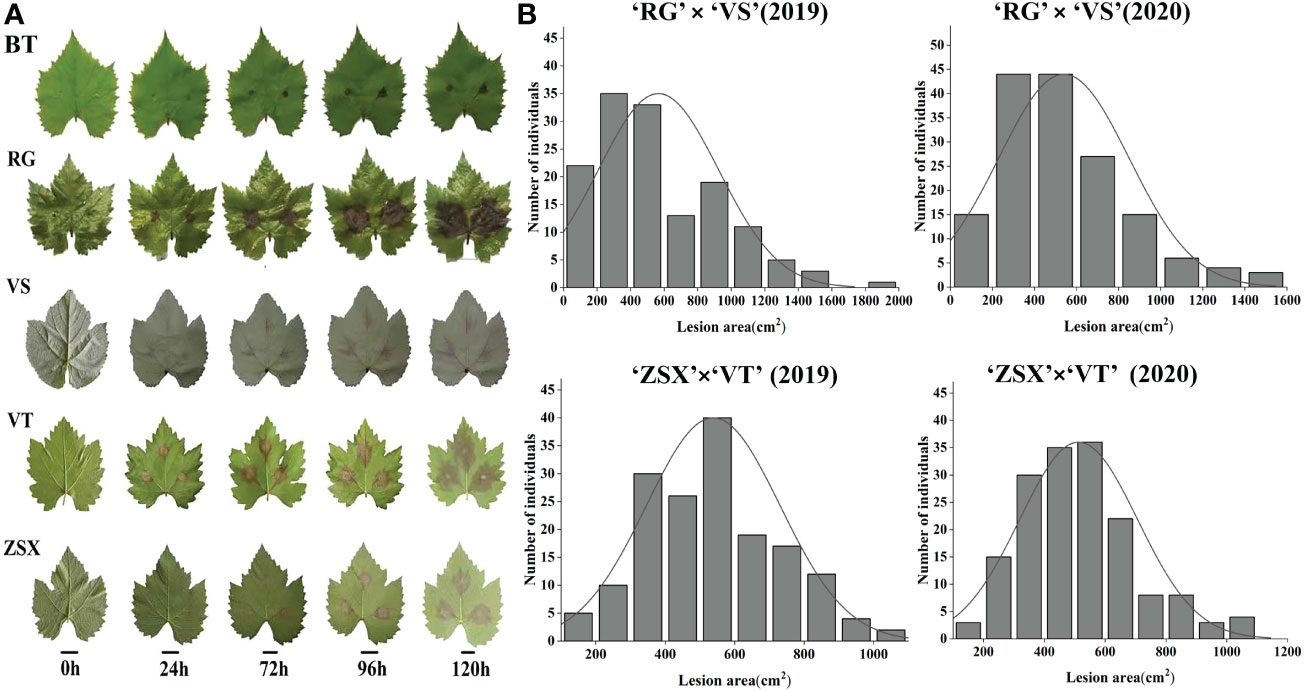
Figure 1 Grapevine gray mold resistance identification of five grape cultivars and two hybrid populations. (A) Gray mold lesion area identification of five grape cultivars. ‘BT,’ ‘RG,’ ‘VS,’ ‘ZSX,’ and ‘VT’ represent grape cultivars ‘Beta,’ ‘Red Globe,’ ‘Venus seedless,’ ‘Zhuosexiang,’ and’Victoria,’ respectively. (B) Gray mold lesion area distribution of two hybrid populations in 2019 and 2020.
3.2 Gene function annotation and differential expression analysis
To identify candidate genes involved in grape gray mold resistance, we conducted RNA-seq for grapevine cultivars ‘RG’ and ‘BT’ at 0, 72, and 120 h after infection. After removing low-quality reads and adapters, a total of 124.20 Gb Clean Data were harvested and retained for further analysis. The average clean data of each sample were 6.17 Gb and have been uploaded to NCBI Sequence Read Archive (SRA) with the Accession Number PRJNA788159. The clean data were assembled using StringTie software. In total, 50,817 annotated transcripts from 42,416 gene loci were obtained through aligning with Swiss-Prot, GO, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Pfam databases by using BLAST and HMMER software (Tables S3, S4). The FPKM value that was calculated by the comparison of sequenced reads with obtained RNA-seq database represents the expression of each transcript (Table S4). To confirm the reliability and rationality of the experiment, we calculated the Pearson’s correlation coefficients for all gene expression levels between each sample and reflected these coefficients in the form of a correlation matrix map (Figure 2A). A total of 5,407 genes were differentially expressed in RG0 vs. BT0 {|[log2 (fold change)]| >1 and adjusted P < 0.05} after differential expression analysis, among which 2,838 were upregulated and 2,569 were downregulated; 7,642 genes were differentially expressed in RG72 vs. BT72, among which 3,693 were upregulated and 3,949 were downregulated; 6,529 genes were differentially expressed in RG120 vs. BT120, among which 2,887 were upregulated and 3,642 were downregulated (Figure 2B).
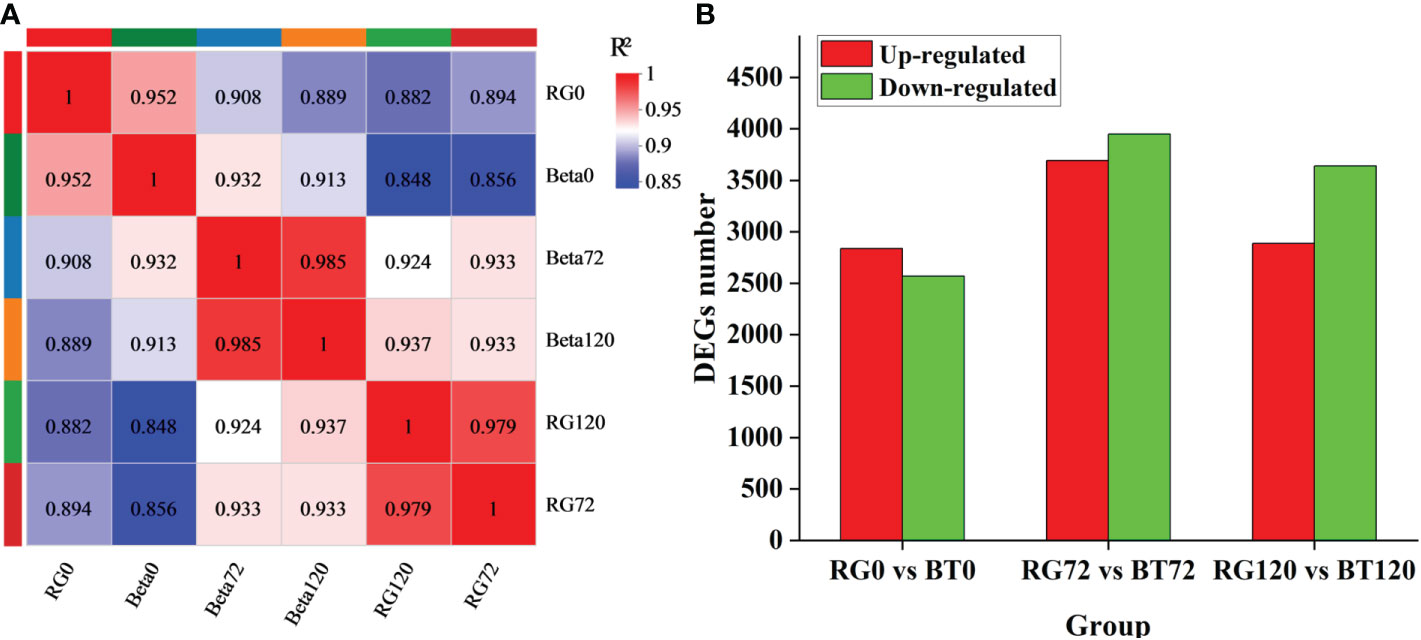
Figure 2 Transcriptome and differentially expressed gene analysis. (A) Pearson’s correlation coefficient analysis for gene expression levels between each sample. (B) Differentially expressed gene statistics in RG0 vs. BT0d, RG72 vs. BT72, and RG120 vs. BT120.
3.3 Gray mold resistance gene discovery based on QTL mapping
Based on the gray mold identification of hybrid offspring in 2019 and 2020 and our constructed genetic linkage maps, we conducted QTL mapping to further discover the candidate genes related to grape gray mold resistance (Figure 3; Table 1). Eight potential QTL related to grape gray mold resistance were identified on LG2, LG7, LG9, LG12, and LG14 in the integrated map of ‘ZSX’ × ‘VT’ (Figure 3A), and the phenotypic variation they explained ranged from 6.70% to 16.50%. Seven potential QTL were identified on LG2, LG7, LG8, LG13, and LG16 in the integrated map of ‘RG’ × ‘VS’ (Figure 3B), and the phenotypic variation they explained ranged from 4.40% to 15.10%. Interestingly, four potential QTL on LG2 were detected stable in the two integrated maps in 2019 and 2020, and these stable QTL accounted for 6.86%–13.70% of the phenotypic variation in the gray mold resistance. Two potential QTL on LG7 were detected stable in the integrated map of ‘RG’ × ‘VS’ in 2019 and 2020. These stable QTL accounted for 4.40%–11.40% of the phenotypic variation in the gray mold resistance.
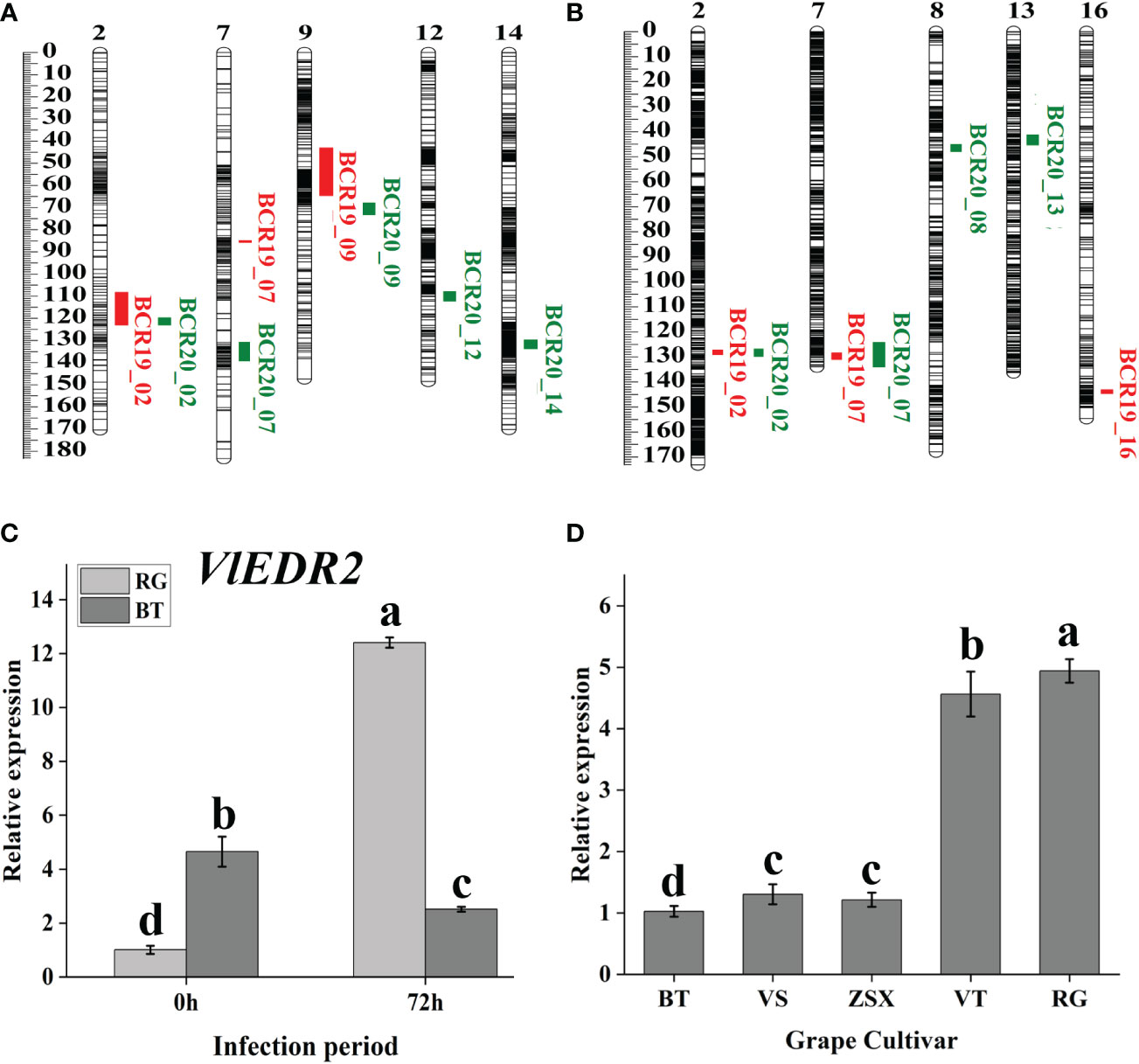
Figure 3 Candidate gray mold resistance gene discovery based on QTL mapping. (A, B) Gray mold resistance QTL mapping based on the hybrid population ‘ZSX’ × ‘VT’ and ‘RG’ × ‘VS’. (C) Cluster heat map of gene expression involved in the common interval of stable QTL. (D) qRT-PCR analysis of candidate gray mold resistance gene VlEDR2 at different infection periods. Light-gray bars represent cultivar ‘RG,’ and dark-gray bars represent cultivar ‘BT.’ Error bars represent the standard deviation of three biological replicates. Lowercase letters on the bar chart represent significant differences between the two cultivars and different developmental stages according to Duncan’s multiple range test at P < 0.05.
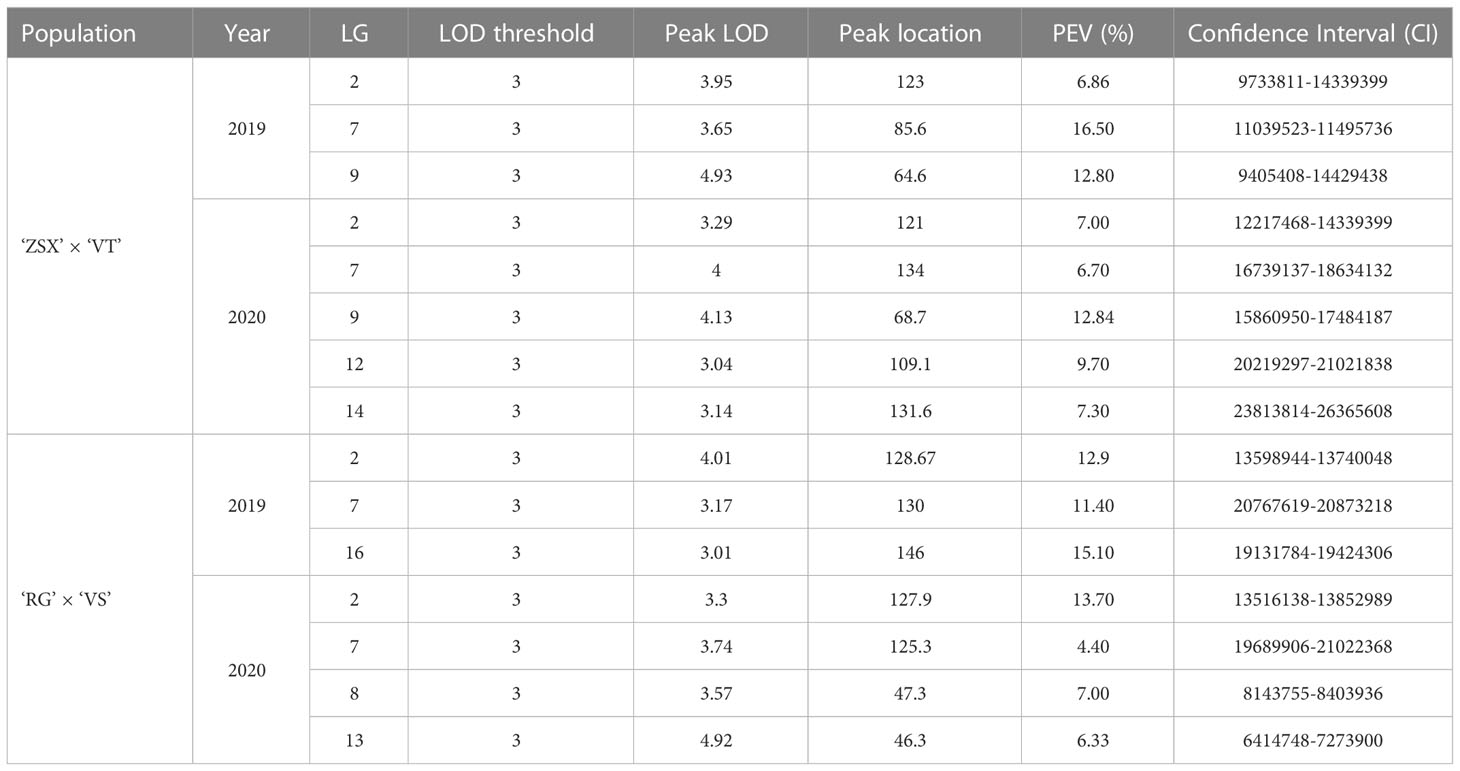
Table 1 Gray mold resistance QTL mapping based on the hybrid offspring of ‘ZSX’ × ‘VT’ and ‘RG’ × ‘VS’.
According to the QTL mapping, the common physical intervals of stable QTL were 13598944-13740048 in chromosome 2 and 20767619-20873218 in chromosome 7. In this study, we majorly focused on the candidate genes that were involved in the common intervals, and finally, 17 genes were discovered (Table S5). After analyzing the differential expression of the selected genes in different comparison groups (RG0 vs. BT0, RG120 vs. BT120, RG0 vs. RG120, and BT0 vs. BT120) with |[log2FC]| >1 and adjusted P < 0.05 (Figure 4A; Table S3), we finally screened the candidate gene Vitvi02g00982 that annotated as enhanced disease resistance 2 (VlEDR2) for further analysis (Figure 3C). The results showed that the expression of VlEDR2 in ‘RG’ was significantly upregulated after gray mold infection, and the expression level in ‘BT’ was significantly downregulated; the expression level of VlEDR2 in ‘RG’ was significantly higher than that in ‘BT’ at 72 h (P < 0.05). After that, the expression level of VlEDR2 in grapevine cultivars ‘VT,’ ‘ZSX,’ and ‘VS’ was also identified (Figure 3D). The result showed that the expression of VlEDR2 in sensitive cultivars was significantly higher than that in resistant cultivars (Figure 3D). The Kruskal–Wallis test was employed to analyze the relationships between the phenotypic values and genotypes of the markers on LG2 and LG7, which showed a significant correlation at P < 0.05. Markers chr2_12269488 and chr2_13516138 were most significantly linked to gray mold resistance in the population of ‘ZSX’ × ‘VT’ and ‘RG’ × ‘VS’ according to the Kruskal–Wallis test (Figure 5). These two markers were located at 12,269,488 bp and 13,516,138 bp on chromosome 2. Raw sequencing data related to these markers were analyzed, and the nucleotides were A/A in ‘VT,’ G/A in ‘ZSX,’ A/A in ‘VS,’ and G/A in ‘RG.’ Progeny carrying A/A in the population of ‘ZSX’ × ‘VT’ generally showed susceptible phenotypes, and the average lesion area of A/A individuals in 2019 and 2020 was 554.7 mm2 and 530.2 mm2, respectively; whereas G/A individuals generally showed resistance, and the average lesion area of G/A individuals in 2019 and 2020 was 517.6 mm2 and 467.7 mm2, respectively. Progeny carrying G/G in the population of ‘RG’ × ‘VS’ generally showed susceptible phenotypes, and the average lesion area of G/G individuals in 2019 and 2020 was 624.1 mm2 and 525.3 mm2, respectively; whereas A/A individuals generally showed resistance, and the average lesion area of A/A individuals in 2019 and 2020 was 470.9 mm2 and 484.9 mm2, respectively.
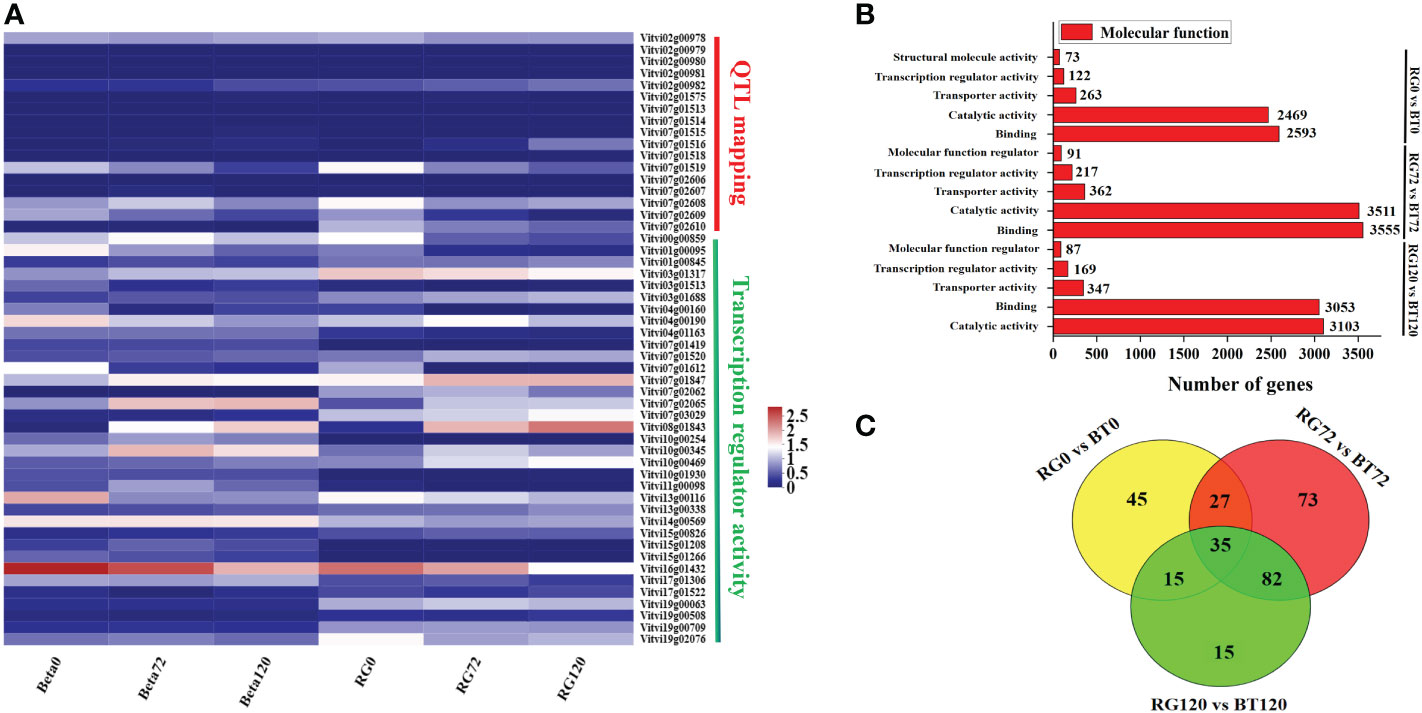
Figure 4 Differentially expressed structural gene and transcription factor analyses based on QTL mapping and GO enrichment. Analysis for grape cultivars ‘RG’ and ‘BT’ at different infection periods. (A) Cluster heat map of structural gene and transcription factor expression. (B) Number of differentially expressed genes in “Molecular function” catalog at different infection periods. (C) Common differentially expressed transcription factor identification in RG0 vs. BT0d, RG72 vs. BT72, and RG120 vs. BT120.
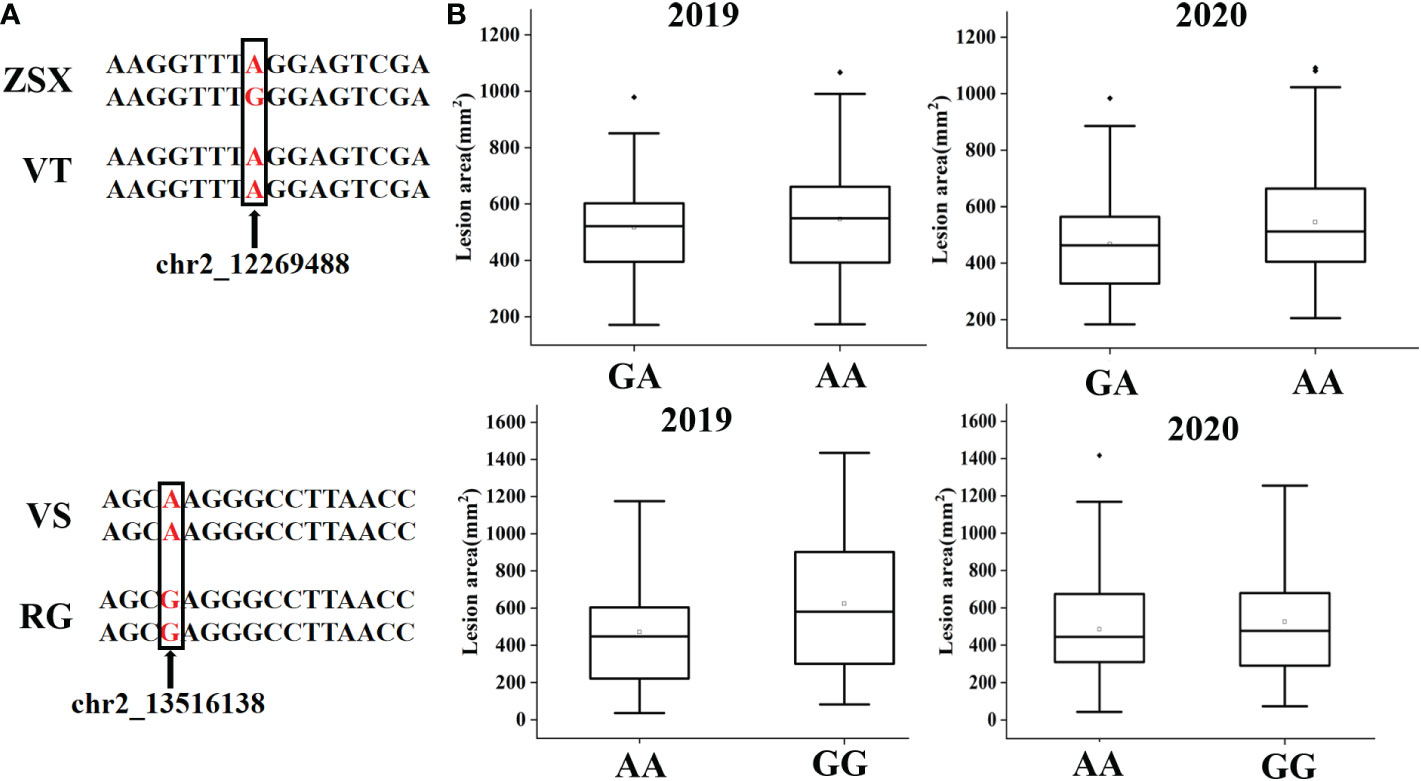
Figure 5 Distributions of grape hybrid gray mold lesion area according to the markers chr2_12269488 and chr2_13516138 in the population of ‘ZSX’ × ‘VT’ and ‘RG’ × ‘VS.’ (A) Base information of markers chr2_12269488 and chr2_13516138 in different cultivars and the flanking sequence. (B) Gray mold lesion area distribution of F1 progeny from the population of ‘ZSX’ × ‘VT’ and ‘RG’ × ‘VS’ in different years.
3.4 Transcription factor discovery related to VlEDR2 regulation
In our study, we selected a candidate grape gray mold-sensitive gene VlEDR2 based on QTL mapping (Figure 3). To further identify transcription factors involved in VlEDR2 regulation, we conducted GO enrichment analysis for these DEGs in the group of RG0 vs. BT0, RG72 vs. BT72, and RG120 vs. BT120. A total of 122, 217, and 169 genes in “Transcription regulator activity” cataloged under “Molecular function” were discovered, respectively (Figure 4B). Finally, 35 DEGs were selected for their significantly different expression in RG0 vs. BT0, RG72 vs. BT72, and RG120 vs. BT120 (Figures 4A–C; Table S6), among which 21 annotated genes were from ERF, MYB, MAD-box, NAC, and WRKY families, and we majorly focused on these 21 transcription factors.
To further select relevant transcription factors related to VlEDR2 expression, the FPKM values of these 21 transcription factors and VlEDR2 at 0 and 72 h were used to conduct the correlation analysis (Figure 6A). Finally, three candidate transcription factors, VlERF039 (Vitvi00g00859), VlNAC047 (Vitvi08g01843), and VlWRKY51 (Vitvi07g01847), from ERF, NAC, and WRKY families that showed a significant correlation (P < 0.05) with the expression of VvEDR2 were selected. The qRT-PCR verification showed that VlERF039 was repressed in ‘RG’ and ‘BT’ after gray mold infection, and the expression level in ‘BT’ was significantly higher than that in ‘RG.’ VlNAC047 and VlWRKY51 that showed a positive correlation with VlEDR2 were also identified. The expression of VlNAC047 and VlWRKY51 was induced in ‘BT’ and ‘RG,’ and the expression level of these two candidate genes in ‘RG’ was significantly higher than that in ‘BT’ (Figure 6B).
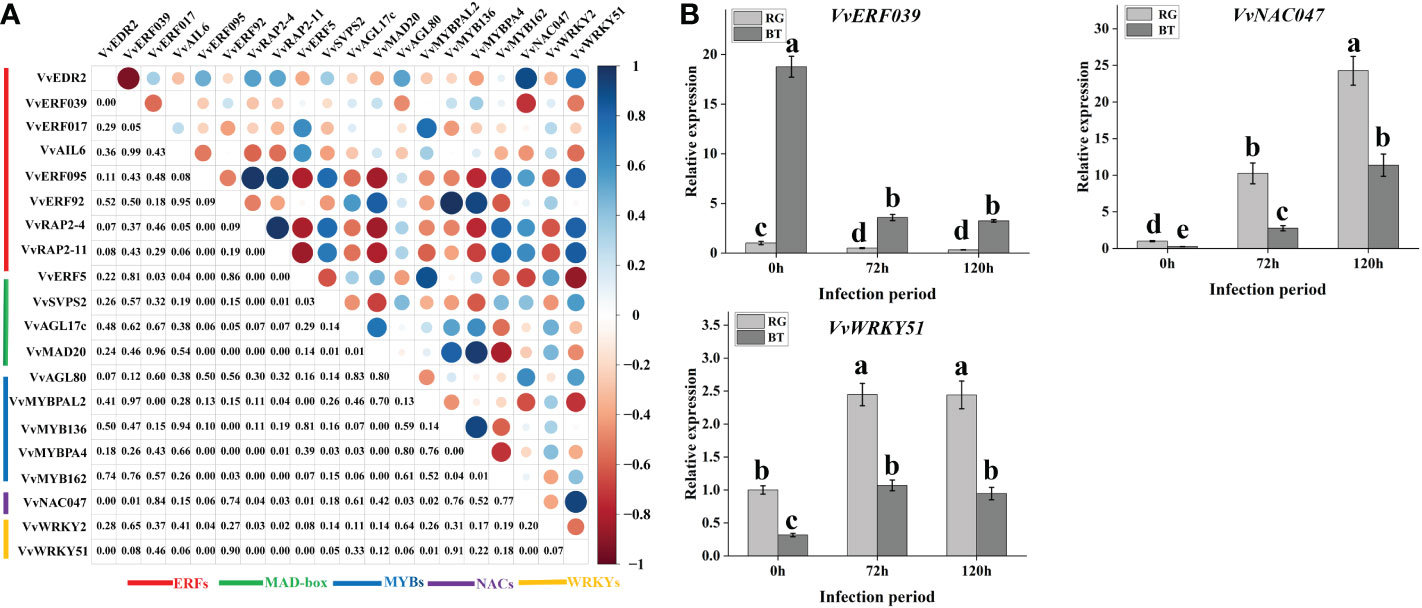
Figure 6 Candidate transcription factor filter related to VlEDR2 expression and grapevine gray mold resistance. (A) Correlation analysis of transcription factors from different families with candidate gray mold resistance gene VlEDR2 at different infection periods. (B) qRT-PCR analysis of candidate gray mold resistance transcription factors at different infection periods. Light-gray bars represent cultivar ‘RG,’ and dark-gray bars represent cultivar ‘BT.’ Error bars represent the standard deviation of three biological replicates. Lowercase letters on the bar chart represent significant differences between the two cultivars and different developmental stages according to Duncan’s multiple range test at P < 0.05.
4 Discussion
4.1 The formation of heterobeltiosis and lower QTL effect
In this study, some individuals from our two constructed hybrid offspring showed higher gray mold resistance than their parent cultivar ‘ZSX’ and ‘VS.’ The additive effects of several desired dominant alleles or the combined effect of different alleles at the same gene locus, or a combination of both, may have formed heterobeltiosis, and the genetic differences between parents are the primary cause of it. According to heterobeltiosis, we can screen for superior parents and predict the heterosis of parental combinations. In our study, a total of 12 individuals that showed higher gray mold resistance than their resistant parents from the hybrid progenies of ‘RG’ × ‘VS’ and ‘ZSX’ × ‘VT’ were identified, and transgressive offspring in our study provided important grape gray mold resistance resources, and they can also be used as material for underlying genetic and molecular mechanisms of grape gray mold resistance.
QTL mapping and candidate gene discovery of grapevine gray mold resistance are important for grape breeding. In our study, we discovered two stable QTL related to gray mold resistance that were located on linkage groups LG2 and LG7. While the phenotypic variance of these QTL ranged from 6.86% to 13.70% on LG2 and 4.40% to 11.40% on LG7, the smaller QTL effect may be due to the quantitative nature of the host resistance, and according to the Beavis effect, when the sample size was small, the QTL effect would be greatly inflated, and the larger the sample size, the smaller the QTL effect and the closer to the true value (Beavis, 1994; Göring et al., 2001; Slate, 2013).
4.2 Discovery of structural genes related to gray mold resistance
Structural genes related to gray mold resistance were majorly involved in the pattern recognition receptor (PRR)-triggered immunity (PTI) that could mediate gray mold resistance through recognizing pathogen-associated molecular patterns (PAMPs) and host damage-associated molecular patterns (DAMPs), such as chitin elicitor receptor kinase 1 (CERK1), LysMdomain-containing glycosylphosphate ethylinositol-anchored protein 2 (LYM2), and wall-associated kinase 1 (WAK1), and polygalacturonidase-inhibiting proteins (PGIPs) Botrytis-induced kinase 1 (BIK1), MPK2/3/6, PAD3, and Arabidopsis histidine kinase 5 (AHK5) (Miya et al., 2007; Qiu et al., 2008; Ren et al., 2008; De Lorenzo et al., 2011; Eckardt, 2011; Galletti et al., 2011; Birkenbihl et al., 2012; Pham et al., 2012; Faulkner et al., 2013; Zhang et al., 2014; Guan et al., 2015; Liu et al., 2015). In grapevine, some structural genes related to gray mold resistance have also been reported (Agüero et al., 2005; Agudelo-Romero et al., 2015; Jiao et al., 2015; Rubio et al., 2015; Wang Y. et al., 2017; Wan et al., 2021), but most of these genes were selected through either previous research or transcriptome analysis based on two different gray mold resistance cultivars. In our study, we firstly conducted grape gray mold resistance QTL mapping supplemented by transcriptomic analysis, and finally, a new candidate resistance gene VvEDR2 was selected. Based on previous research, EDR played a negative role and the edr mutants display high resistance (HR)-like lesions in response to a pathogen attack stimulus such as powdery mildew in plant that is involved in the salicylic acid (SA) defense pathway (Frye and Innes, 1998; Tang et al., 2005a; Tang et al., 2005b; Tang et al., 2006). Moreover, some studies have also shown the SA-independent phenotype of EDR2 that is involved in hypersensitivity to ethylene-induced senescence, implicating EDR2 in the regulation of senescence and defense signaling (Frye et al., 2001; Tang et al., 2005b). In our study, we preliminarily identified the potential role of VlEDR2 in negatively regulated grapevine gray mold resistance, and this discovered resistance gene will provide new reference for the research on grapevine gray mold resistance.
4.3 Candidate transcription factors involved in the regulation mechanism of gray mold resistance
Many reports have shown the role of ERFs in plant gray mold resistance, such as RAP2.2, ORA59, ERF1, ERF5, and ERF6 in Arabidopsis thaliana; overexpression of these genes could enhance the resistance to gray mold through binding to GCC-box elements of defense marker gene PDF1.2 and promoting its expression in jasmonic acid (JA) and ethylene (ET) signaling pathways (Berrocal-Lobo et al., 2002; Pre et al., 2008; Zarei et al., 2011; Moffat et al., 2012; Zhao et al., 2012). In tomato, silencing of SlERF.A1, SlERF.A3, SlERF.B4, or SlERF.C3 resulted in increased susceptibility to B. cinerea (Ouyang et al., 2016). In grapevine, overexpression of VqERF072, VqERF112, VqERF114, and VaERF20 in A. thaliana could also enhance the resistance to B. cinerea in JA and ET signaling pathways (Wang et al., 2018a; Wang et al., 2020). WRKY TFs could also regulate gray mold resistance through activating the expression of structural genes involved in SA and JA signaling, such as LrWRKY4, LrWRKY12, and LrWRKY39 in Lilium (Cui et al., 2018; Fu et al., 2022), SlDRW1 and SlWRKY46 in tomato (Liu et al., 2014; Shu et al., 2021), RcWRKY41 in rose (Liu et al., 2019), and VqWRKY52 in grapevine (Wang X. et al., 2017). Moreover, TFs from the MYB family could also play positive and negative regulatory roles in gray mold resistance, such as RcMYB84, RcMYB123, and MYB108 in JA signaling pathway (Mengiste et al., 2003; Ren et al., 2020; Cui et al., 2022) and MYB72 in induced systemic resistance signaling pathway (Van der Ent et al., 2008). MYB46 negatively mediated gray mold resistance through repressing the synthesis of cellulose synthases (Ramirez et al., 2011), and BjMYB1 positively regulated gray mold resistance through activating the expression of BjCHI1 (Gao and Zhao, 2017). In grapevine, the interaction of VaERF16 and VaMYB306 could increase the expression level of VaPDF1.2 and then enhance gray mold resistance (Zhu et al., 2022). In our study, based on the expression pattern of VlEDR2, we screened out a new candidate ERF gene VlERF039 and WRKY gene VlWRKY51, while their potential possibility in regulating the expression of VlEDR2 and grape gray mold resistance still needs a deep exploration. Moreover, NAC gene VlNAC047 was also discovered, and until now, there was no report focused on its function in gray mold resistance; this discovery can provide a new insight on transcriptional regulation mechanisms of grape gray mold resistance.
5 Conclusion
Based on QTL mapping and transcriptome analysis, we discovered one structural gene, VlEDR2 (Vitvi02g00982), which may play a negative role in grapevine resistance to gray mold. Moreover, three potential transcription factors including VlERF039 (Vitvi00g00859), VlNAC047 (Vitvi08g01843), and VlWRKY51 (Vitvi07g01847) that may influence the expression of VlEDR2 and grapevine gray mold resistance in positive and negative ways were also discovered. The candidate genes identified in our study will provide an important reference for research into grapevine gray mold resistance mechanisms and breeding in grape species.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA788159.
Author contributions
YG, KS, and YZ contributed to experimental design, KS and WZ performed the experiments and KS wrote the article. KS and WZ performed grapevine gray mold identification and KS, HL, and CJ contributed to data analysis. All authors contributed to the article and approved the submitted version.
Funding
The research was supported by the National Natural Science Foundation of China (Grant No.31972368), the China Agriculture Research System (Grant No. CARS-29-yc-6), the Department of Science and Technology of Liaoning Province (Grant No. 2022030723-JH5/104), the Shenyang Science and Technology Bureau Funds (Grant No. 21-116-3-27) and the Liaoning key R&D Program (Grant No. 2020JH2/10200032).Funding bodies were not involved in the design of the study and collection, analysis, interpretation of data and in writing the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1127206/full#supplementary-material
References
Agudelo-Romero, P., Erban, A., Rego, C., Carbonell-Bejerano, P., Nascimento, T., Sousa, L., et al. (2015). Transcriptome and metabolome reprogramming in Vitis vinifera cv. trincadeira berries upon infection with Botrytis cinerea. J. Exp. Bot. 66 (7), 1769–1785. doi: 10.1093/jxb/eru517
Agüero, C. B., Uratsu, S. L., Greve, C., Powell, A. L., Labavitch, J. M., Meredith, C. P., et al. (2005). Evaluation of tolerance to pierce's disease and Botrytis in transgenic plants of Vitis vinifera l. expressing the pear PGIP gene. Mol. Plant Pathol. 6 (1), 43–51. doi: 10.1111/j.1364-3703.2004.00262.x
Barba, P., Cadle-Davidson, L., Harriman, J., Glaubitz, J. C., Brooks, S., Hyma, K., et al. (2014). Grapevine powdery mildew resistance and susceptibility loci identified on a high-resolution SNP map. Theor. Appl. Genet. 127 (1), 73–84. doi: 10.1007/s00122-013-2202-x
Beavis, W. D. (1994). “The power and deceit of QTL experiments: Lessons from comparative QTL studies,” in Proceedings of the Forty-Ninth Annual Corn and Sorghum Industry Research Conference. (Washington, DC, USA: American SeedTrade Association), 250–266.
Berrocal-Lobo, M., Molina, A., Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29 (1), 23–32. doi: 10.1046/j.1365-313x.2002.01191.x
Birkenbihl, R. P., Diezel, C., Somssich, I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159 (1), 266–285. doi: 10.1104/pp.111.192641
Breia, R., Conde, A., Pimentel, D., Conde, C., Fortes, A. M., Granell, A., et al. (2019). VvSWEET7 is a mono- and disaccharide transporter up-regulated in response to Botrytis cinerea infection in grape berries. Front. Plant Sci. 10, 1753. doi: 10.3389/fpls.2019.01753
Broman, K. W., Wu, H., Sen, S., Churchill, G. A. (2003). R/qtl: QTL mapping in experimental crosses. Bioinformatics 19 (7), 889–890. doi: 10.1093/bioinformatics/btg112
Choquer, M., Fournier, E., Kunz, C., Levis, C., Pradier, J. M., Simon, A., et al. (2007). Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 277 (1), 1–10. doi: 10.1111/j.1574-6968.2007.00930.x
Cui, F., Li, X., Wu, W., Luo, W., Wu, Y., Brosché, M., et al. (2022). Ectopic expression of BOTRYTIS SUSCEPTIBLE1 reveals its function as a positive regulator of wound-induced cell death and plant susceptibility to Botrytis. Plant Cell 34 (10), 4105–4116. doi: 10.1093/plcell/koac206
Cui, Q., Yan, X., Gao, X., Zhang, D. M., He, H. B., Jia, G. X. (2018). Analysis of WRKY transcription factors and characterization of two Botrytis cinerea-responsive LrWRKY genes from Lilium regale. Plant Physiol. Biochem. 127, 525–536. doi: 10.1016/j.plaphy.2018.04.027
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13 (4), 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
De Lorenzo, G., Brutus, A., Savatin, D. V., Sicilia, F., Cervone, F. (2011). Engineering plant resistance by constructing chimeric receptors that recognize damage-associated molecular patterns (DAMPs). FEBS Lett. 585 (11), 1521–1528. doi: 10.1016/j.febslet.2011.04.043
Eckardt, N. A. (2011). BIK1 function in plant growth and defense signaling. Plant Cell 23 (8), 2806–2086. doi: 10.1105/tpc.111.230811
Faulkner, C., Petutschnig, E., Benitez-Alfonso, Y., Beck, M., Robatzek, S., Lipka, V., et al. (2013). LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc. Natl. Acad. Sci. U.S.A. 110 (22), 9166–9170. doi: 10.1073/pnas.1203458110
Frye, C. A., Innes, R. W. (1998). An arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10 (6), 947–956. doi: 10.1105/tpc.10.6.947
Frye, C. A., Tang, D., Innes, R. W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. U.S.A. 98 (1), 373–378. doi: 10.1073/pnas.98.1.373
Fujimori, N., Enoki, S., Suzuki, A., Naznin, H. A., Shimizu, M., Suzuki, S. (2016). Grape apoplasmic β-1,3-glucanase confers fungal disease resistance in Arabidopsis. Sci. Hortic- Amsterdam 200, 105–110. doi: 10.1016/j.scienta.2016.01.008
Fu, P., Tian, Q., Lai, G., Li, R., Song, S., Lu, J. (2019). Cgr1, a ripe rot resistance QTL in Vitis amurensis 'Shuang hong' grapevine. Hortic. Res. 6, 67. doi: 10.1038/s41438-019-0148-0
Fu, Y., Li, J., Wu, H., Jiang, S., Zhu, Y., Liu, C., et al. (2022). Analyses of Botrytis cinerea-responsive LrWRKY genes from lilium regale reveal distinct roles of two LrWRKY transcription factors in mediating responses to B. cinerea. Plant Cell Rep. 41 (4), 995–1012. doi: 10.1007/s00299-022-02833-6
Gabler, F. M., Smilanick, J. L., Mansour, M., Ramming, D. W., Mackey, B. E. (2003). Correlations of morphological, anatomical, and chemical features of grape berries with resistance to Botrytis cinerea. Phytopathology 93 (10), 1263–1273. doi: 10.1094/phyto.2003.93.10.1263
Galletti, R., Ferrari, S., De Lorenzo, G. (2011). Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157 (2), 804–814. doi: 10.1104/pp.111.174003
Gao, Y., Zhao, K. (2017). Molecular mechanism of BjCHI1-mediated plant defense against Botrytis cinerea infection. Plant Signal Behav. 12 (1), e1271859. doi: 10.1080/15592324.2016.1271859
Göring, H. H., Terwilliger, J. D., Blangero, J. (2001). Large Upward bias in estimation of locus-specific effects from genomewide scans. Am. J. Hum. Genet. 69 (6), 1357–1369. doi: 10.1086/324471
Guan, R., Su, J., Meng, X., Li, S., Liu, Y., Xu, J., et al. (2015). Multilayered regulation of ethylene induction plays a positive role in Arabidopsis resistance against Pseudomonas syringae. Plant Physiol. 169 (1), 299–312. doi: 10.1104/pp.15.00659
Jiao, C., Gao, M., Wang, X., Fei, Z. (2015). Transcriptome characterization of three wild Chinese Vitis uncovers a large number of distinct disease related genes. BMC Genomics 16 (1), 223. doi: 10.1186/s12864-015-1442-3
Liu, B., Hong, Y. B., Zhang, Y. F., Li, X. H., Huang, L., Zhang, H. J., et al. (2014). Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. 227, 145–156. doi: 10.1016/j.plantsci.2014.08.001
Liu, M., Zhang, Z., Xu, Z., Wang, L., Chen, C., Ren, Z. (2021). Overexpression of SlMYB75 enhances resistance to Botrytis cinerea and prolongs fruit storage life in tomato. Plant Cell Rep. 40 (1), 43–58. doi: 10.1007/s00299-020-02609-w
Liu, S., Kracher, B., Ziegler, J., Birkenbihl, R. P., Somssich, I. E. (2015). Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife 4, e07295. doi: 10.7554/eLife.07295
Liu, X., Li, D., Zhang, S., Xu, Y., Zhang, Z. (2019). Genome-wide characterization of the rose (Rosa chinensis) WRKY family and role of RcWRKY41 in gray mold resistance. BMC Plant Biol. 19 (1), 522. doi: 10.1186/s12870-019-2139-6
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15 (1), 165–178. doi: 10.1105/tpc.007468
Martínez-Romero, D., Guillén, F., Valverde, J. M., Bailén, G., Zapata, P., Serrano, M., et al. (2007). Influence of carvacrol on survival of Botrytis cinerea inoculated in table grapes. Int. J. Food Microbiol. 115 (2), 144–148. doi: 10.1016/j.ijfoodmicro.2006.10.015
Mengiste, T., Chen, X., Salmeron, J., Dietrich, R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15 (11), 2551–2565. doi: 10.1105/tpc.014167
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104 (49), 19613–19618. doi: 10.1073/pnas.0705147104
Moffat, C. S., Ingle, R. A., Wathugala, D. L., Saunders, N. J., Knight, H., Knight, M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in arabidopsis. PloS One 7 (4), e35995. doi: 10.1371/journal.pone.0035995
Nanni, V., Schumacher, J., Giacomelli, L., Brazzale, D., Sbolci, L., Moser, C., et al. (2014). VvAMP2, a grapevine flower-specific defensin capable of inhibiting Botrytis cinerea growth: insights into its mode of action. Plant Pathol. 63 (4), 899–910. doi: 10.1111/ppa.12170
Ouyang, Z., Liu, S., Huang, L., Hong, Y., Li, X., Huang, L., et al. (2016). Tomato SlERF.A1, SlERF.B4, SlERF.C3 and SlERF.A3, members of B3 group of ERF family, are required for resistance to Botrytis cinerea. Front. Plant Sci. 7, 1964. doi: 10.3389/fpls.2016.01964
Pham, J., Liu, J., Bennett, M. H., Mansfield, J. W., Desikan, R. (2012). Arabidopsis histidine kinase 5 regulates salt sensitivity and resistance against bacterial and fungal infection. New Phytol. 194 (1), 168–180. doi: 10.1111/j.1469-8137.2011.04033.x
Pre, M., Atallah, M., Champion, A., De Vos, M., Pieterse, C. M., Memelink, J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147 (3), 1347–1357. doi: 10.1104/pp.108.117523
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27 (16), 2214–2221. doi: 10.1038/emboj.2008.147
Ramirez, V., Garcia-Andrade, J., Vera, P. (2011). Enhanced disease resistance to Botrytis cinerea in myb46 arabidopsis plants is associated to an early down-regulation of CesA genes. Plant Signal Behav. 6 (6), 911–913. doi: 10.4161/psb.6.6.15354
Ren, D., Liu, Y., Yang, K. Y., Han, L., Mao, G., Glazebrook, J., et al. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105 (14), 5638–5643. doi: 10.1073/pnas.0711301105
Ren, H., Bai, M., Sun, J., Liu, J., Ren, M., Dong, Y., et al. (2020). RcMYB84 and RcMYB123 mediate jasmonate-induced defense responses against Botrytis cinerea in rose (Rosa chinensis). Plant J. 103 (5), 1839–1849. doi: 10.1111/tpj.14871
Rubio, J., Montes, C., Castro, Á., Álvarez, C., Olmedo, B., Muñoz, M., et al. (2015). Genetically engineered Thompson seedless grapevine plants designed for fungal tolerance: selection and characterization of the best performing individuals in a field trial. Transgenic Res. 24 (1), 43–60. doi: 10.1007/s11248-014-9811-2
Saito, S., Michailides, T. J., Xiao, C. L. (2019). Fungicide-resistant phenotypes in Botrytis cinerea populations and their impact on control of gray mold on stored table grapes in California. Eur. J. Plant Pathol. 154 (2), 203–213. doi: 10.1007/s10658-018-01649-z
Sapkota, S., Chen, L. L., Yang, S., Hyma, K. E., Cadle-Davidson, L., Hwang, C. F. (2019). Construction of a high-density linkage map and QTL detection of downy mildew resistance in Vitis aestivalis-derived 'Norton'. Theor. Appl. Genet. 132 (1), 137–147. doi: 10.1007/s00122-018-3203-6
Shu, P., Zhang, S., Li, Y., Wang, X., Yao, L., Sheng, J., et al. (2021). Over-expression of SlWRKY46 in tomato plants increases susceptibility to Botrytis cinerea by modulating ROS homeostasis and SA and JA signaling pathways. Plant Physiol. Biochem. 166, 1–9. doi: 10.1016/j.plaphy.2021.05.021
Slate, J. (2013). From beavis to beak color: a simulation study to examine how much qtl mapping can reveal about the genetic architecture of quantitative traits. Evolution 67 (5), 1251–1262. doi: 10.1111/evo.12060
Su, K., Guo, Y., Zhong, W., Lin, H., Liu, Z., Li, K., et al. (2021). High-density genetic linkage map construction and white rot resistance quantitative trait loci mapping for genus vitis based on restriction site-associated DNA sequencing. Phytopathology 111 (4), 659–670. doi: 10.1094/phyto-12-19-0480-r
Tang, D., Ade, J., Frye, C. A., Innes, R. W. (2005a). Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 44 (2), 245–257. doi: 10.1111/j.1365-313X.2005.02523.x
Tang, D., Ade, J., Frye, C. A., Innes, R. W. (2006). A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J. 47 (1), 75–84. doi: 10.1111/j.1365-313X.2006.02769.x
Tang, D., Christiansen, K. M., Innes, R. W. (2005b). Regulation of plant disease resistance, stress responses, cell death, and ethylene signaling in arabidopsis by the EDR1 protein kinase. Plant Physiol. 138 (2), 1018–1026. doi: 10.1104/pp.105.060400
Teh, S. L., Fresnedo-Ramírez, J., Clark, M. D., Gadoury, D. M., Sun, Q., Cadle-Davidson, L., et al. (2017). Genetic dissection of powdery mildew resistance in interspecific half-sib grapevine families using SNP-based maps. Mol. Breed 37 (1), 1. doi: 10.1007/s11032-016-0586-4
Tello, J., Roux, C., Chouiki, H., Laucou, V., Sarah, G., Weber, A., et al. (2019). A novel high-density grapevine (Vitis vinifera l.) integrated linkage map using GBS in a half-diallel population. Theor. Appl. Genet. 132 (8), 2237–2252. doi: 10.1007/s00122-019-03351-y
Van der Ent, S., Verhagen, B. W., Van Doorn, R., Bakker, D., Verlaan, M. G., Pel, M. J., et al. (2008). MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 146 (3), 1293–1304. doi: 10.1104/pp.107.113829
Wang, X., Guo, R., Tu, M., Wang, D., Guo, C., Wan, R., et al. (2017). Ectopic expression of the wild grape WRKY transcription factor VqWRKY52 in Arabidopsis thaliana enhances resistance to the biotrophic pathogen powdery mildew but not to the necrotrophic pathogen Botrytis cinerea. Front. Plant Sci. 8, 97. doi: 10.3389/fpls.2017.00097
Wang, L., Liu, W., Wang, Y. (2020). Heterologous expression of Chinese wild grapevine VqERFs in Arabidopsis thaliana enhance resistance to Pseudomonas syringae pv. tomato DC3000 and to botrytis cinerea. Plant Sci. 293, 110421. doi: 10.1016/j.plantsci.2020.110421
Wang, X., Tu, M., Wang, D., Liu, J., Li, Y., Li, Z., et al. (2018b). CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16 (4), 844–855. doi: 10.1111/pbi.12832
Wan, R., Guo, C., Hou, X., Zhu, Y., Gao, M., Hu, X., et al. (2021). Comparative transcriptomic analysis highlights contrasting levels of resistance of vitis vinifera and vitis amurensis to Botrytis cinerea. Hortic. Res. 8 (1), 103. doi: 10.1038/s41438-021-00537-8
Wang, Y., Wang, D., Wang, F., Huang, L., Tian, X., van Nocker, S., et al. (2017). Expression of the grape VaSTS19 gene in Arabidopsis improves resistance to powdery mildew and Botrytis cinerea but increases susceptibility to Pseudomonas syringe pv tomato DC3000. Int. J. Mol. Sci. 18 (9). doi: 10.3390/ijms18092000
Wang, M., Zhu, Y., Han, R., Yin, W., Guo, C., Li, Z., et al. (2018a). Expression of vitis amurensis VaERF20 in Arabidopsis thaliana improves resistance to Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000. Int. J. Mol. Sci. 19 (3), 696. doi: 10.3390/ijms19030696
Wan, R., Hou, X., Wang, X., Qu, J., Singer, S. D., Wang, Y., et al. (2015). Resistance evaluation of Chinese wild vitis genotypes against Botrytis cinerea and different responses of resistant and susceptible hosts to the infection. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00854
Zarei, A., Korbes, A. P., Younessi, P., Montiel, G., Champion, A., Memelink, J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75 (4-5), 321–331. doi: 10.1007/s11103-010-9728-y
Zhang, L., Kars, I., Essenstam, B., Liebrand, T. W., Wagemakers, L., Elberse, J., et al. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164 (1), 352–364. doi: 10.1104/pp.113.230698
Zhao, Y., Wei, T., Yin, K. Q., Chen, Z., Gu, H., Qu, L. J., et al. (2012). Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol. 195 (2), 450–460. doi: 10.1111/j.1469-8137.2012.04160.x
Zhu, J., Guo, Y., Su, K., Liu, Z., Ren, Z., Li, K., et al. (2018). Construction of a highly saturated genetic map for vitis by next-generation restriction site-associated DNA sequencing. BMC Plant Biol. 18 (1), 347. doi: 10.1186/s12870-018-1575-z
Zhu, Y., Li, Y., Zhang, S., Zhang, X., Yao, J., Luo, Q., et al. (2019). Genome-wide identification and expression analysis reveal the potential function of ethylene responsive factor gene family in response to botrytis cinerea infection and ovule development in grapes (Vitis vinifera l.). Plant Biol. (Stuttg) 21 (4), 571–584. doi: 10.1111/plb.12943
Keywords: Vitis vinifera, gray mold, resistance breeding, transcriptome analysis, QTL mapping
Citation: Su K, Zhao W, Lin H, Jiang C, Zhao Y and Guo Y (2023) Candidate gene discovery of Botrytis cinerea resistance in grapevine based on QTL mapping and RNA-seq. Front. Plant Sci. 14:1127206. doi: 10.3389/fpls.2023.1127206
Received: 19 December 2022; Accepted: 17 January 2023;
Published: 07 February 2023.
Edited by:
Jianfu Jiang, Zhengzhou Fruit Research Institute (CAAS), ChinaReviewed by:
Lei Sun, Zhengzhou Fruit Research Institute (CAAS), ChinaPeining Fu, Shanghai Jiao Tong University, China
Shen Fengying, Hebei North University, China
Copyright © 2023 Su, Zhao, Lin, Jiang, Zhao and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuhui Zhao, emhhb3l1aHVpNzZAc3lhdS5lZHUuY24=; Yinshan Guo, Z3VveWluc2hhbjc3QHN5YXUuZWR1LmNu
 Kai Su1,2,3
Kai Su1,2,3 Yinshan Guo
Yinshan Guo