- National Institute of Agricultural Sciences, Rural Development Administration, Jeonju, Republic of Korea
Organisms regulate gene expression to produce essential proteins for numerous biological processes, from growth and development to stress responses. Transcription and translation are the major processes of gene expression. Plants evolved various transcription factors and transcriptome reprogramming mechanisms to dramatically modulate transcription in response to environmental cues. However, even the genome-wide modulation of a gene’s transcripts will not have a meaningful effect if the transcripts are not properly biosynthesized into proteins. Therefore, protein translation must also be carefully controlled. Biotic and abiotic stresses threaten global crop production, and these stresses are seriously deteriorating due to climate change. Several studies have demonstrated improved plant resistance to various stresses through modulation of protein translation regulation, which requires a deep understanding of translational control in response to environmental stresses. Here, we highlight the translation mechanisms modulated by biotic, hypoxia, heat, and drought stresses, which are becoming more serious due to climate change. This review provides a strategy to improve stress tolerance in crops by modulating translational regulation.
Introduction
Genetic information is transmitted from DNA to proteins through messenger RNA (mRNA) (with certain exceptions—i.e., reverse transcription and prions) according to the central dogma first reported by Francis Crick (Koonin, 2012; Cobb, 2017). Therefore, the strict control of gene expression determines most biological processes, including growth, development, and stress responses. Gene expression occurs in stages, including transcription and translation, that are physically and functionally connected (Orphanides and Reinberg, 2002; Buccitelli and Selbach, 2020). Transcription and translation generally occur simultaneously in prokaryotes lacking membrane-bound organelles, but are spatially separated in eukaryotes. In eukaryotic cells, mRNA is transcribed in the nucleus and then moves to the cytoplasm where translation occurs. The translation mechanism is highly conserved and comprises three major stages: initiation, elongation, and termination (Kapp and Lorsch, 2004). In eukaryotes, translation initiation relies on cap-dependent and cap-independent pathways (Merrick, 2004; Shatsky et al., 2018). Translation is initiated by the assembly of the initiation complex, which consists of eukaryotic initiation factors (eIFs), ribosomes, and the initiator methionyl-transfer RNA (Met-tRNAiMet) on mRNA (Merrick and Pavitt, 2018). The initiation complex recognizes the initiation codon AUG via the anticodon of Met-tRNAiMet in the ribosomal peptidyl (P)-site. For translation elongation, amino acids are sequentially and continuously added at the P-site with the help of eukaryotic elongation factors (eEFs) from aminoacyl-tRNAs binding to the aminoacyl (A)-site of the ribosome, and translation terminates when a stop codon (UAG, UGA, or UAA) is located in the A-site and recognized by eukaryotic release factors (Dever et al., 2018).
Climate change is raising global temperatures and shifting regional climates toward greater extremes, including increased aridity in some areas and heavier rainfall in others. In addition, the environmental stresses exacerbated by climate change can impede plant immunity and provide favorable conditions for pathogens (Velasquez et al., 2018). Crop yield and food security are threatened by these increased stresses (Chaudhry and Sidhu, 2022; Son and Park, 2022b). Because plants cannot move to avoid stresses, adaptive changes resulting from alteration of gene expression play critical roles in their survival under extreme stress conditions, and the regulation of gene expression is more crucial to the survival of individuals in plants than in animals. Indeed, not only the numbers of transcription factors but also their rates of expansion are higher in plants (Riechmann et al., 2000; Shiu et al., 2005). Plant protein translation mechanisms are also modulated in response to various stresses (Spriggs et al., 2010; Echevarría-Zomeño et al., 2013; Zlotorynski, 2022). Therefore, investigating the translation mechanisms involved in stress tolerance and engineering them in important crops are essential for sustainable agriculture. Here, we summarize and discuss the translational mechanisms that are modulated by biotic and abiotic stresses.
Translation reprogramming in response to stress stimulus
The different steps in the translation process are coordinated to ensure survival and efficient use of cellular resources under stress conditions. Stimulus-mediated translational control can be classified into global and gene-specific processes, and its mechanisms are mainly associated with translation initiation and polysome association (Gebauer and Hentze, 2004; Sonenberg and Hinnebusch, 2009). In eukaryotes, stress-induced translational regulation generally results from inhibition of canonical protein biosynthesis driven by cap-dependent translation and induction of stress-associated protein biosynthesis driven by cap-independent translation (Holcik and Sonenberg, 2005; Liu and Qian, 2014). Plant translation processes such as initiation, elongation, and termination are highly conserved, and many components involved in translation mechanisms have been well described in previous reviews (Muench et al., 2012; Roy and Von Arnim, 2013; Browning and Bailey-Serres, 2015; Merchante et al., 2017). Therefore, this review focuses on translation regulation in response to biotic and abiotic stresses.
mRNA translation in eukaryotes is initiated through two different mechanisms, cap-dependent ribosome scanning and cap-independent internal ribosome entry, which are regulated by a stress stimulus (Figure 1). In non-stress conditions, most mRNAs are translated through the cap-dependent translation pathway. Transcribed nascent pre-mRNA is modified via 5′-m7GpppN capping, RNA splicing, and 3′ poly(A) addition, and structural mRNA features are important for their translation (Von Arnim et al., 2014; Sablok et al., 2017). In cap-dependent translation, the cap-binding complex eIF4F consisting of the cap-binding protein eIF4E, the scaffolding protein eIF4G, and the ATP-dependent RNA helicase eIF4A is assembled on the 5′-cap of mRNA, while the poly(A)-binding protein (PABP) interacts with the 3′-poly(A) tail of mRNA (Browning and Bailey-Serres, 2015). In plant, there is also an eIF isoform 4F (eIFiso4F) which consists of eIFiso4E, eIFiso4G, and eIF4A. Subsequently, eIF4F interacts with eIF4B, the eIF4A cofactor, and PABP via eIF4G, resulting in mRNA unwinding and circularization. The 43S pre-initiation complex (PIC), which is composed of the 40S ribosome subunit, eIFs (i.e., eIF1, eIF1A, eIF3, and eIF5), and the ternary complex eIF2 (consisting of α, β, and γ subunits)-GTP-Met-tRNAiMet, interacts with the eIF4F complex and forms the 48S PIC, which scans mRNA in the 5′-to-3′ direction to find an initiation codon. Upon detection of an initiation codon, eIFs, including eIF1 (which is necessary for the fidelity of initiation codon selection), and hydrolyzed eIF2-GDP are released, and eIF5B-GTP and the 60S ribosome subunit are recruited to form the translation-competent 80S ribosome (Browning and Bailey-Serres, 2015; Merchante et al., 2017).
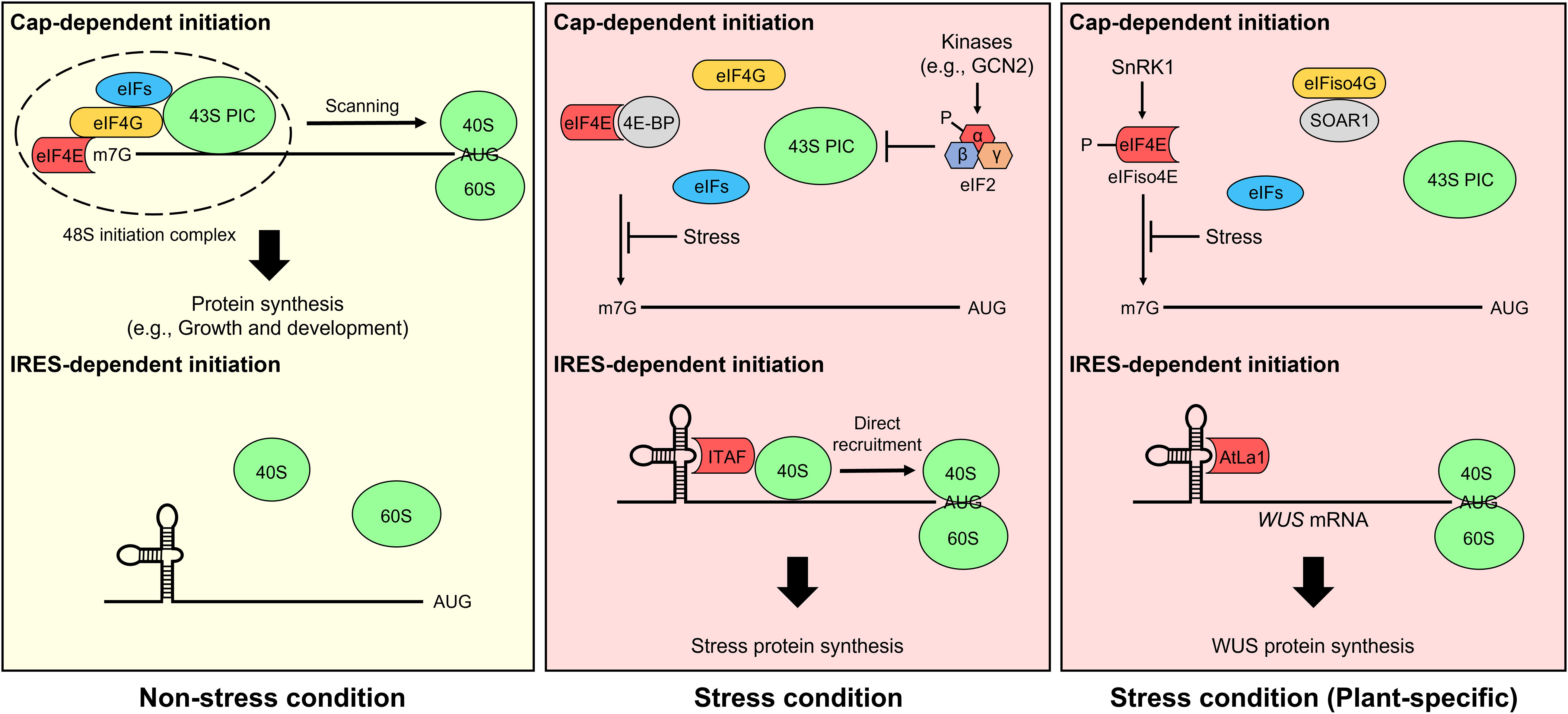
Figure 1 Cap-dependent and internal ribosomal entry site (IRES)-dependent translation initiation mechanisms. Under non-stress conditions, most mRNAs are translated through the cap-dependent translation pathway (top) in eukaryotes, including plants. The eukaryotic initiation factor 4F (eIF4F) complex, including the cap-binding protein eIF4E and the scaffolding protein eIF4G, is assembled on the 5′ cap of mRNA and recruits the 43S pre-initiation complex (PIC) to form the 48S PIC. The complex scans mRNA for an initiation codon. Finally, 60S ribosomes are recruited to form an 80S ribosome initiation complex, and mRNA translation is carried out in earnest. However, cap-dependent translation initiation is prevented under stress conditions. For example, eIF4E-binding protein (4E-BP) and phosphorylation of eIF2α inhibit the formation of eIF4F and the ternary complex, respectively, thereby suppressing cap-dependent translation (Richter and Sonenberg, 2005; Holcik, 2015). In plants lacking 4E-BP, cap-dependent translation is impaired by not only SNF1-related protein kinase 1 (SnRK1)-induced eIF4E and eIF isoform 4E (eIFiso4E) phosphorylation but also the interaction between Suppressor of the ABAR overexpressor 1 (SOAR1) and eIFiso4G (Bi et al., 2019; Bruns et al., 2019). Alternatively, the IRES-dependent translation pathway (bottom) is activated under stress conditions. In general, IRES trans-acting factors (ITAFs) mediate IRES-dependent translation in eukaryotes (Komar and Hatzoglou, 2011; Godet et al., 2019). However, this cellular mRNA translation mechanism is largely elusive in plants. Two mRNAs, maize Heat shock protein 101 (Hsp101) and Arabidopsis WUSCHEL (WUS), are reported to be translated by IRES-dependent translation under stress conditions (Dinkova et al., 2005; Cui et al., 2015b), and Cui et al. only identified Arabidopsis LA protein 1 (AtLa1) binding to the 5′ untranslated region of cellular mRNA as an IRES-dependent translation regulator.
Cap-dependent translation initiation is impaired under stress conditions. For example, in mammals, eIF4E-binding protein (4E-BP) interacts with eIF4E and prevents eIF4F formation by interfering with the binding of eIF4E and eIF4G (Richter and Sonenberg, 2005). Phosphorylation of eIF2α by multiple protein kinases (e.g., General control nondepressible 2 [GCN2]) is another mechanism that inhibits cap-dependent translation in mammals (Holcik, 2015; Hinnebusch et al., 2016). In response to a wide range of signals, stress-activated kinases phosphorylate the Ser51 of eIF2α, which inhibits ternary complex recycling by preventing the guanine nucleotide exchange factor eIF2B-mediated catalysis of GDP to GTP (Buchan and Parker, 2009). However, in plant, an orthologous genes of 4E-BP are absent (Hernandez et al., 2010). Although some proteins (e.g., Lipoxygenase 2, Basic transcription factor 3, Essential for potexvirus accumulation 1, and Conserved binding of eIF4E 1) have been identified as eIF4E- and/or eIFiso4E-binding proteins, their association with cap-dependent translation is unclear (Freire et al., 2000; Freire, 2005; Lázaro-Mixteco and Dinkova, 2012; Wu et al., 2017; Patrick et al., 2018). Even the plant eIF4E-interacting protein CERES has a positive effect on global translation, not a negative effect (Toribio et al., 2019). GCN2-eIF2α module is also controversial in plants (Wang et al., 2022). Alternatively, SNF1-related protein kinase 1 (SnRK1)-mediated eIF4E and eIFiso4E phosphorylation and the interaction of Suppressor of the ABAR overexpressor 1 (SOAR1), involved in abscisic acid signaling, with eIFiso4G inhibit cap-dependent translation initiation in plants (Bi et al., 2019; Bruns et al., 2019).
Although canonical cap-dependent translation is impeded by stress stimuli, selective mRNA translation is induced via noncanonical cap-dependent translation and cap-independent translation. eIF3d-mediated noncanonical cap-dependent translation was discovered in human cells (Lee et al., 2015; Lee et al., 2016). eIF3d, a subunit of eIF3, has cap-binding activity and induces the translation of specific mRNAs containing a stem-loop structure that inhibits the recruitment of eIF4F complex in their 5′ untranslated region (5′ UTR). Lamper et al. revealed the translation mechanism regulated by eIF3d for metabolic stress adaptation (Lamper et al., 2020). Under non-stress conditions, Casein kinase 2 (CK2) phosphorylates and inactivates eIF3d. Metabolic stresses, including glucose starvation, inhibit CK2-mediated phosphorylation of eIF3d, resulting in selective mRNA translation through cap-binding of eIF3d. In plant, Toribio et al. showed that CERES interacts with eIF4E, eIFiso4E, eIF4A, eIF3, and PABP and forms noncanonical translation initiation complex in which eIF4G or eIFiso4G is replaced by CERES (Toribio et al., 2019). They suggested the noncanonical complex supports translation initiation and regulates general translation positively when the energy and carbon supply are high. CERES is also involved in a defensive response to turnip mosaic virus regardless of its interaction with eIF4E and eIF4isoE (Toribio et al., 2021). However, the detailed mechanism of selective translation of capped mRNAs by noncanonical cap-dependent translation is unknown in plants.
Internal ribosomal entry site (IRES)-mediated cap-independent translation initiation was first discovered in viral RNA translation of picornavirus (Jackson et al., 1990) and is now recognized as an alternative translation mechanism that commonly occurs in eukaryotic cells under stress conditions (Spriggs et al., 2010; Yang and Wang, 2019). IRESs are frequently identified in cellular mRNAs of genes involved in stress responses and are subdivided into two types: Type I cellular IRESs harbor cis-regulatory elements that interact with IRES trans-acting factors (ITAFs) for ribosome recruitment (Komar and Hatzoglou, 2011; Godet et al., 2019), while Type II cellular IRESs have short cis elements that pair with 18S ribosomal RNA, a component of the 40S ribosomal subunit (Dresios et al., 2006). Since most cellular IRESs are Type I, ITAFs play a critical role in IRES-mediated translation. The mechanisms of IRES-dependent initiation are fairly well studied in animals but have been largely elusive in plants. Only a few studies have revealed the possibility of IRES-dependent translation in plants. Some plant viruses utilize IRES-dependent translation mechanisms for biosynthesis of proteins from their viral RNAs (Jaag et al., 2003; Dorokhov et al., 2006; Karetnikov and Lehto, 2007). In maize (Zea mays), the 5′ UTR of Heat shock protein 101 (HSP101) mRNA contains an IRES-like element, and its translation is increased via cap-independent translation during heat stress (Dinkova et al., 2005). In Arabidopsis (Arabidopsis thaliana), the conserved RNA-binding factor La protein 1 (La1) binds to the 5′ UTR of WUSCHEL (WUS) mRNA and improves its translation through IRES-dependent initiation under environmental hazard conditions (Cui et al., 2015b). However, further studies are needed to understand the mechanisms of IRES-dependent translation in plants.
Though translation control mechanisms in plants have been extensively studied, how this is regulated by various stresses is largely unknown. Here, we review the current knowledge of the translation mechanisms controlled by biotic, hypoxia, heat, and drought stresses in plants.
Translational control for pattern-triggered immunity
Severe crop losses caused by various pathogens (e.g., bacteria, fungi, and viruses) threaten global food and nutrition security. Therefore, it is necessary to increase plant innate immunity through effective approach such as translational regulation. Viruses use various strategies to regulate host translation mechanisms for protein synthesis (Jaafar and Kieft, 2019; Geng et al., 2021). Increasing plant antiviral immunity through manipulation of plant translation factors has been well described in previous reviews (Sanfacon, 2015; Hashimoto et al., 2016; Calil and Fontes, 2017; Zhao et al., 2020; Leonetti et al., 2021; Robertson et al., 2022). Therefore, herein, we focus on recent studies that demonstrate notable translation mechanisms associated with non-viral pathogens.
Plant immunity can be classified into pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), also called pattern-triggered immunity, and effector-triggered immunity (ETI) depending on how pathogens are recognized by the plant (Tsuda and Katagiri, 2010; Faulkner and Robatzek, 2012; Naveed et al., 2020). Sensing of conserved pathogen signatures PAMP, also referred as microbe-associated molecular pattern, by pattern recognition receptors (PRRs) in the plant plasma membrane activates downstream signaling (e.g., Receptor-like cytoplasmic kinases [RLCKs], mitogen-activated protein kinases [MAPKs or MPKs], calcium ion, reactive oxygen species, phytohormones, and transcription factors), and induces PTI which is plant defense response at basal level (Bigeard et al., 2015; DeFalco and Zipfel, 2021). Well-known PRRs are the leucine-rich repeat receptor kinases such as Flagellin sensing 2 (FLS2) and the bacterial translation Elongation factor thermo unstable (EF-Tu) receptor (EFR). Sensing of the 22-amino-acid peptide derived from bacterial flagella, flg22, by FLS2 or the 18- and 26-amino-acid peptides derived from EF-Tu, elf18 and elf26, respectively, by EFR induces elicitor-dependent complex formation through binding with somatic embryogenesis receptor-like kinases (SERKs) such as Brassinosteroid insensitive-associated kinase 1 (BAK1)/SERK3 and BAK1-like 1 (BKK1)/SERK4 (Heese et al., 2007; Schulze et al., 2010; Roux et al., 2011; Sun et al., 2013). The elicitor-dependent complex activates downstream signaling by phosphorylating the RLCK Botrytis-induced kinase 1 (BIK1) and MAPKs, resulting in PTI-related resistance responses (Wang et al., 2020).
GCN2 is a conserved serine/threonine protein kinase that participates in stress signal transduction, including nutrient starvation and immune responses (Tsalikis et al., 2013; Lokdarshi and von Arnim, 2022). Since eIF2α phosphorylation results in a stable phosphor (P)-eIF2-GDP-eIF2B complex, GCN2-mediated eIF2α phosphorylation arrests global cellular mRNA translation but induces selective translation of stress-responsive mRNAs (e.g., GCN4 in yeast and Activating transcription factor 4 in mammals) harboring upstream open reading frames (uORFs) (Dever, 2002). In plants, GCN2 regulates biotic and abiotic stress responses and also stress-induced eIF2α phosphorylation (Liu et al., 2015; Terry et al., 2015; Lokdarshi and von Arnim, 2022). Izquierdo et al. suggested that GCN1 is important for translation regulation and innate immunity in plants, while GCN2 and eIF2α phosphorylation are not (Izquierdo et al., 2018). However, Liu et al. showed that the bacteria-activated GCN2-eIF2α module induces translation of the heat-shock factor-like transcription factor TL1-binding factor 1 (TBF1) mRNA and plant immunity (Liu et al., 2019). TBF1 is a key regulator of the plant growth-defense tradeoff. TBF1 binds to the TL1 cis element (GAAGAAGAA) and controls salicylic acid- and elf18-induced transcriptional reprogramming (Pajerowska-Mukhtar et al., 2012). Interestingly, TBF1 mRNA has two uORFs conferring negative effects on its translation. Pathogen challenge results in eIF2α phosphorylation and derepresses TBF1 mRNA translation (Pajerowska-Mukhtar et al., 2012). Therefore, the translationally regulated GCN2-eIF2α-TBF1 modules may play a key role in PTI (Figure 2).
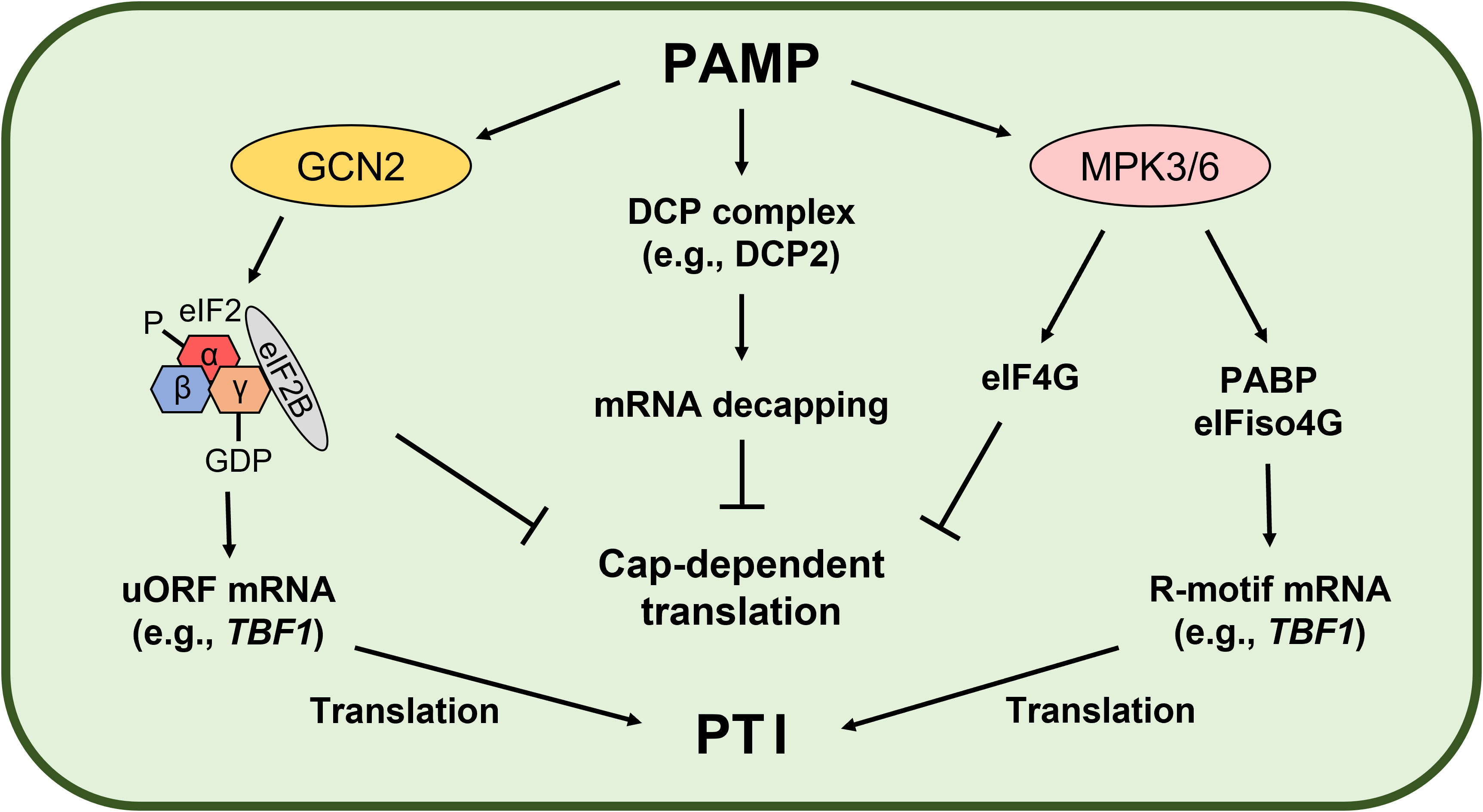
Figure 2 Pathogen-associated molecular pattern (PAMP)-induced translational reprogramming. PAMP-activated General control nondepressible 2 (GCN2) phosphorylates eukaryotic initiation factor 2α (eIF2α), leading to a stable eIF2-GDP-eIF2B complex in Arabidopsis. As a result, formation of the eIF2-GTP-Met-tRNAiMet ternary complex required for cap-dependent initiation is inhibited, while translation of defense-related mRNAs with upstream open reading frames (uORFs) conferring negative effects on translation is induced (Pajerowska-Mukhtar et al., 2012; Liu et al., 2019). Wang et al. demonstrated PAMP-mediated translational regulation mechanisms in Arabidopsis. General translation is arrested by PAMP-induced mRNA decapping and Mitogen-activated protein kinase 3 (MPK3)- and MPK6-mediated eIF4G phosphorylation (Wang et al., 2022). On the other hand, MPK3- and MPK6-mediated phosphorylation of the poly(A)-binding protein (PABP) and eIF isoform 4G (eIFiso4G) promotes PABP binding with the R-motif and eIFiso4G to drive PABP-eIFiso4G-mediated translation (Wang et al., 2022). As a result, general mRNA translation is repressed but translation of selective defense-related mRNAs is induced to promote the plant immune response (PAMP-triggered immunity, PTI).
Global translatome profiling using ribosome footprinting revealed that PTI triggers translational reprogramming to enhance plant immunity (Xu et al., 2017a). elf18 treatment increased the translational efficiency of 448 genes but decreased that of 389 genes in Arabidopsis. Interestingly, the R-motif, which consists almost exclusively of purines, is enriched on the 5′ UTR of the mRNAs, including TBF1 mRNA, whose translational efficiency is upregulated by elf18. MPK3, MPK6, and PABP is important for R-motif-mediated selective translation during PTI (Xu et al., 2017a). Processing bodies (P-bodies) also play an important role in PTI. P-bodies are cytoplasmic granules consisting of messenger ribonucleoprotein and are involved in translation arrest and mRNA decay (Decker and Parker, 2012; Maldonado-Bonilla, 2014). In eukaryotes, most mRNAs are degraded via the deadenylation-dependent mRNA decay pathway (Garneau et al., 2007). After deadenylation, mRNA is degraded immediately by 3′–5′ exonucleases in exosomes or decapped for later degradation by 5′–3′ exonucleases in P-bodies. In Arabidopsis, the decapping complex—involving mRNA-decapping enzyme Decapping protein 2 (DCP2), the co-activator DCP1, and the scaffold protein Varicose—is localized in P-bodies (Xu et al., 2006). However, PAMP-activated MPK3 and MPK6 phosphorylate DCP1, resulting in disassociation of DCP1 and DCP2 and rapid P-body disassembly within 15 to 30 minutes after flg22 treatment (Yu et al., 2019). Yu et al. suggested that phosphorylated DCP1 leads to mRNA degradation of negative regulators of plant immunity by 5′-to-3′ Exoribonuclease 4 (XRN4), an ortholog of yeast XRN1, and translation of defense-related mRNAs stored in P-body assemblies to promote the plant immune response.
Recently, the PTI-induced translational reprogramming mechanism was demonstrated in Arabidopsis (Figure 2). As global translation repression mechanism, elf18 induces DCP-complex-mediated mRNA decapping and MPK3- and MPK6-mediated eIF4G phosphorylation (Wang et al., 2022). These mechanisms compromise translation of mRNA related to growth and defense. However, for selective translation control during PTI, defense-related mRNAs containing the R-motif are translated through a PABP-eIFiso4G-mediated cap-independent pathway (Wang et al., 2022). During PTI, PAMP-activated MPK3 and MPK6 phosphorylate PABP and eIFiso4G using Receptor for activated C kinase 1 (RACK1) as a scaffold, which enhances PABP binding with the R-motif in mRNAs and with eIFiso4G, resulting in translation of defense-related mRNAs for innate immunity.
Translational control for effector-triggered immunity
Plant nucleotide-binding and leucine-rich repeat (NLR) proteins are classified into three groups (i.e., Toll/interleukin-1 receptor-like NLR, coiled coil NLR [CNL], and Resistance to powdery mildew 8-like NLR) based on their N-terminal domain and play various roles as sensors, helpers, and executors in ETI signaling (Son et al., 2021). Direct or indirect recognition of specific effectors (pathogen-secreted proteins that repress PTI and promote infection) by NLRs activates downstream signaling and triggers prolonged and robust ETI resistance responses (Cui et al., 2015a; Lolle et al., 2020).
Two CNLs, Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) and Resistance to P.syringae 2 (RPS2), are activated by post-translational modification of the plasma-membrane-localized RPM1-interacting protein (RIN4), which interacts with CNLs as well as P. syringae avirulence (Avr) effectors (e.g., AvrB, AvrRpm1, and AvrRpt2). In an incompatible interaction, AvrB and AvrRpm1 interact with RIN4, which is inactivated by the prolyl-peptidyl isomerase Rotamase cyclophilin 1, leading to RPM1-induced protein kinase-mediated RIN4 phosphorylation and RPM1 activation for ETI (Mackey et al., 2002; Liu et al., 2011; Li et al., 2014). On the other hand, the interaction of AvrRpt2 with RIN4 leads to RIN4 cleavage into AvrRpt2 cleavage products and activates RPS2 for ETI (Axtell and Staskawicz, 2003; Mackey et al., 2003; Takemoto and Jones, 2005).
Global translatome analysis revealed that RPM1- and RPS2-induced ETI are involved in translation reprogramming. Meteignier et al. showed that AvrRpm1-mediated RPM1 activation modulates the translational status of hundreds of mRNAs involved in growth-defense tradeoffs (Meteignier et al., 2017). RPM1 increases translation efficiency of defense-related mRNAs (e.g., BIG, Phosphorylcholine cytidylyltransferase 2 and Redox responsive transcription factor 1/Ethylene response factor 109) but decreases translation efficiency of mRNAs involved in growth and/or functioning as negative regulators of defense (e.g., Target of rapamycin [TOR], CBL-interacting protein kinase 5, and Homolog of BEE2 interacting with IBH 1). The TOR kinase is the conserved master regulator of the energy signaling pathway regulating growth and metabolism (Wullschleger et al., 2006). In mammals, TOR-mediated phosphorylation of 4E-BP1, Ribosomal protein S6 kinase (S6K), and La-related protein 1 has critical roles in mRNA translation (Yang et al., 2022). Although 4E-BPs remain elusive in plants, the TOR-S6K-ribosomal protein S6 (rpS6) pathway is conserved (Obomighie et al., 2021). Unlike most eukaryotes, which harbor two distinct TOR complexes, plants have only one TOR complex (TORC), consisting of TOR, Regulatory-associated protein of TOR (RAPTOR), and Lethal with sec thirteen protein 8 (LST8) (Maegawa et al., 2015). In Arabidopsis, light and auxin activate TOR-S6K-rpS6 modules to enhance translation (Schepetilnikov et al., 2013; Chen et al., 2018). In response to auxin, eIF3h phosphorylation by TOR-activated S6K1 enhances translation reinitiation of mRNAs with uORFs (Schepetilnikov et al., 2013). Furthermore, the TOR-S6K module phosphorylates MA3-domain-containing translation regulatory factor 1 (MRF1), which then interacts with eIF4A to enhance MRF1 ribosome association for mRNA translation (Lee et al., 2017). Therefore, RPM1-mediated TOR repression may be an important translational mechanism for the growth-defense tradeoff (Meteignier et al., 2017). AvrRpt2-mediated RPS2 activation also triggers translational regulation for ETI. Translational regulation by RPS2 differs from PTI-induced translational reprogramming (Yoo et al., 2020). RPS2-mediated ETI induces TBF1 translation later than PTI does, and is involved in targeted changes in active translation of specific mRNAs instead of general translation inhibition. In addition, unlike with PTI-induced translational regulation, there is a strong correlation between transcriptional and translational changes during RPS2-mediated ETI. There is overlap between RPS2-mediated and RPM1-mediated translational responses (Yoo et al., 2020). For example, 80% of upregulated and 75% of downregulated genes from the RPS2-mediated translational response overlapped with those of the RPM1-mediated translational response. Moreover, RPS2- and RPM1-mediated ETI contribute to metabolic dynamics via translational regulation (Meteignier et al., 2017; Yoo et al., 2020). Therefore, ETI-mediated translational regulation in metabolic pathways is important for plant immunity. However, the detailed mechanisms by which ETI regulates protein translation have not been elucidated.
Translation mechanisms regulated by hypoxia in plants
Hypoxia in plants is caused by flooding, submergence, and soil compaction, and its effect on modulating mRNA translation has been well studied. Hypoxia leads to energy deficiency in plant cells by inhibiting mitochondrial respiration, and plants must redistribute their energy reserves by restricting energy-consuming processes and inducing energy-conserving processes (Geigenberger, 2003; Fukao and Bailey-Serres, 2004; Bailey-Serres and Voesenek, 2008). Since mRNA translation uses an enormous amount of energy, it has to be modified under hypoxic conditions (Kafri et al., 2016; Chee et al., 2019).
In response to hypoxia, maize plants repress the translation of aerobic proteins and increase the translation of anaerobic proteins to confer flooding tolerance (Sachs et al., 1980; Sachs et al., 1996). Hypoxia-mediated selective translation induces protein biosynthesis of maize Alcohol dehydrogenase 1, and the 5′ and 3′ UTRs of its mRNA are necessary for its translation (Bailey-Serres and Dawe, 1996). In maize roots, hypoxia induces eIF4A and eIF4E phosphorylation (Webster et al., 1991; Manjunath et al., 1999), and quantitative analysis of ribosomal complexes in maize seedlings showed that translational machineries involved in initiation and elongation (i.e., eIF4E, eIF4A, eIF4B, and eEF2) are significantly more phosphorylated under oxygen deprivation (Szick-Miranda et al., 2003). In Arabidopsis, although the translation of most cellular mRNA is impaired by hypoxia due to reduced ribosome recruitment, polysome association of some stress-induced mRNAs containing a low GC nucleotide content in the 5′ UTR is increased (Branco-Price et al., 2005). Restriction of global translation in response to hypoxia is necessary for energy conservation (Branco-Price et al, 2008), and it is mainly regulated at the translation initiation level (Juntawong et al., 2014). In addition, hypoxia-induced Oligouridylate binding protein 1C binds mRNAs and forms protein-RNA complexes named stress granules, resulting in global translational arrest via mRNA sequestration (Lee et al., 2011; Sorenson and Bailey-Serres, 2014).
AMP-activated protein kinase (AMPK) is the evolutionarily conserved energy stress signaling master regulator controlling cellular energetic homeostasis in animals (Trefts and Shaw, 2021). AMPK inhibits mRNA translation in multiple steps by inhibiting TOR, eEF2, and RNA biding proteins (Liu and Chern, 2021). SnRK1 is the plant ortholog of AMPK and controls plant growth, stress tolerance, and metabolism to cope with constantly fluctuating environments (Baena-Gonzalez et al., 2007; Cho et al., 2012; Son et al., 2023). Although there is no clear evidence, some studies suggest that SnRK1-mediated TOR inhibition may occur via RAPTOR1B phosphorylation (Nietzsche et al., 2016; Nukarinen et al., 2016). Remarkably, SnRK1-mediated eIF4E and eIFiso4E phosphorylation attenuates general translation (Bruns et al., 2019). Furthermore, SnRK1 activity represses canonical protein biosynthesis without significantly changing transcript levels and protein stability (Son et al., 2022). However, under submergence conditions, SnRK1-mediated eIFiso4G1 phosphorylation confers translational enhancement of specific mRNAs, including those of core hypoxia-response genes (Cho et al., 2019). Therefore, SnRK1 plays a key role in global translational repression and specific mRNA translation under energy deficiency conditions (Figure 3). Moreover, ethylene regulates translational dynamics through both a noncanonical ethylene-signaling-activated GCN2-eIF2α module and canonical Ethylene insensitive 2-mediated ethylene signaling during submergence (Cho et al., 2022).
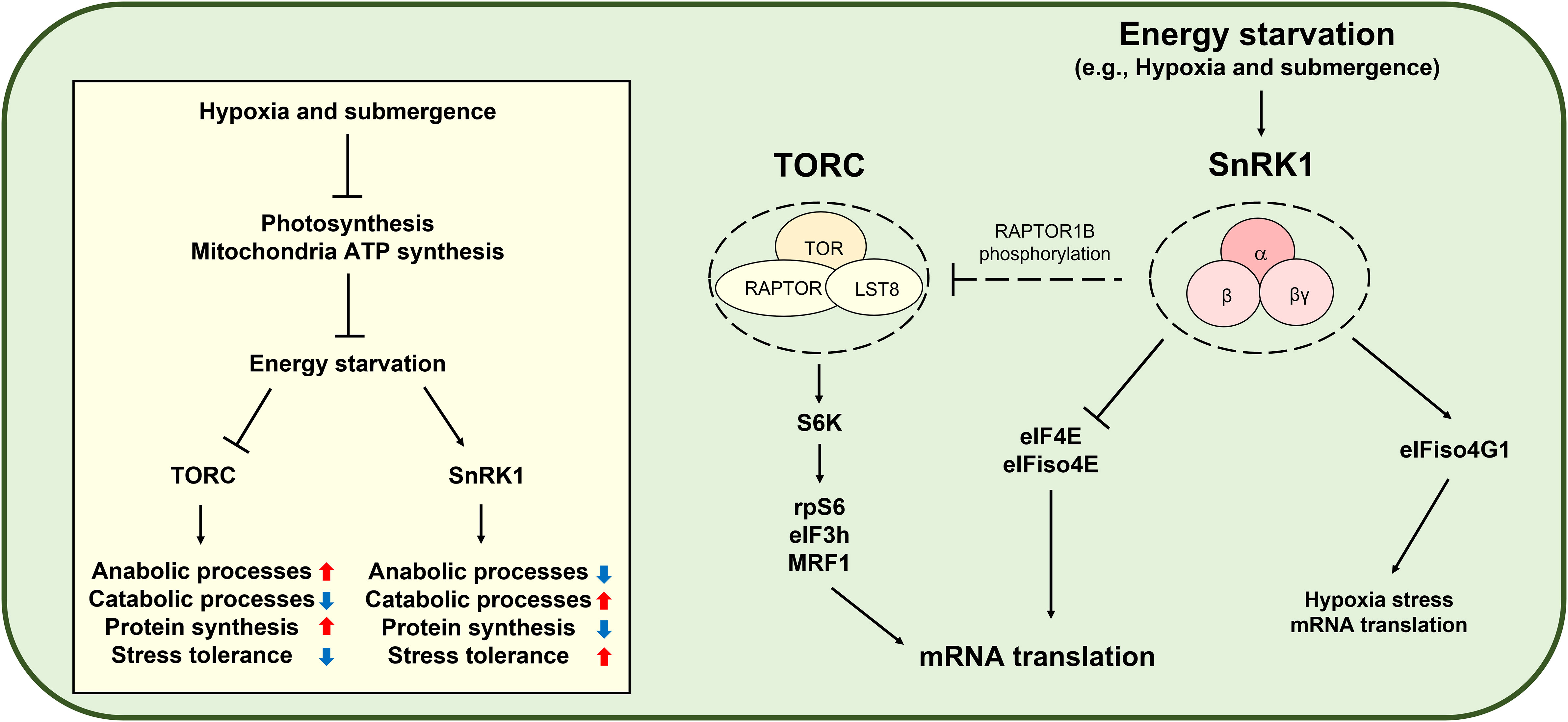
Figure 3 SNF1-related protein kinase 1 (SnRK1)-mediated translational control under hypoxia. Hypoxia and submergence in plants induce energy starvation and activate SnRK1, which consists of an α catalytic subunit and two regulatory subunits, β and βγ. SnRK1 reduces anabolic processes, including protein biosynthesis, while inducing catabolic processes and stress tolerance. Since mRNA translation is an energy-consuming process, SnRK1 modulates mRNA translation. Under submergence conditions, SnRK1 phosphorylates eukaryotic initiation factor 4E (eIF4E) and eIF isoform 4E (eIFiso4E) to repress global mRNA translation (Bruns et al., 2019), whereas SnRK1-mediated eIFiso4G1 phosphorylation induces translation of mRNAs of hypoxia-response genes (Cho et al., 2019). The Target of rapamycin complex (TORC) consists of Target of rapamycin (TOR), Regulatory-associated protein of TOR (RAPTOR), and Lethal with sec thirteen protein 8 (LST8) and is a conserved master regulator of energy signaling, including protein biosynthesis. TOR-activated ribosomal protein S6 kinase (S6K) phosphorylates ribosomal protein S6 (rpS6), eukaryotic initiation factor 3h (eIF3h), and MA3-domain-containing translation regulatory factor 1 (MRF1) to promote mRNA translation (Schepetilnikov et al., 2013; Lee et al., 2017; Chen et al., 2018). The SnRK1 ortholog in mammals is AMP-activated protein kinase (AMPK). AMPK-mediated TORC inhibition is well established in mammals. However, although studies have suggested SnRK1-mediated TOR inhibition through RAPTOR1B phosphorylation (Nietzsche et al., 2016; Nukarinen et al., 2016), it remains obscure in plants.
Translation mechanisms regulated by heat and drought stress in plants
To help plants adapt and survive under elevated temperature, canonical protein translation is significantly suppressed, while the translation of HSPs, which prevent protein denaturation and aggregation typically via a chaperone, is induced (Key et al., 1981; Nover et al., 1989; Ndimba et al., 2005; Matsuura et al., 2010; Bita and Gerats, 2013). In carrot (Daucus carota), heat stress impedes translation initiation and hampers generation of the 5′ cap and 3′ poly(A) tail conferring mRNA translation (Gallie et al., 1995). eIF4A and eIF4B are phosphorylated during heat stress in wheat (Triticum aestivum), but eIF4F, eIFiso4F, eIF2α, eIF2β, and PABP are not (Gallie et al., 1997). In Arabidopsis, mRNA sequence features, such as the GC content of the 5′ UTR and cDNA length, are important elements conferring heat-induced selective mRNA translation (Yangueez et al., 2013). Under elevated temperature, Arabidopsis XRN4 degrades mRNAs encoding HSP70 binding proteins and hydrophobic N-terminal proteins in polysomes, triggering ribosome pausing (Merret et al., 2015). eIF5B contributes to the biosynthesis of stress-protective proteins under high temperature, and eIF5B is important for heat stress tolerance in Arabidopsis (Zhang et al., 2017). Moreover, Bonnot and Nagel showed that the circadian clock and heat stress interact to prioritize the translation of the mRNA pool in Arabidopsis (Bonnot and Nagel, 2021). Translation of heat- or abiotic-stress-related mRNAs, including HSP90-3, was significantly upregulated under high temperature. They also suggested that transcription factors, including Cycling DOF factor, MYB-related, and B-box families, play important roles in heat stress-mediated plant growth dynamics dependent on the circadian clock.
Dehydration also changes mRNA translational efficiencies in plants, including Arabidopsis, maize, and soybean (Glycine max) (Hsiao, 1970; Dhindsa and Cleland, 1975; Rhodes and Matsuda, 1976; Bensen et al., 1988; Hurkman and Tanaka, 1988; Mason et al., 1988; Valluri et al., 1989; Bray, 1990; Kawaguchi et al., 2003; Kawaguchi et al., 2004). In Arabidopsis, polysome association of 71% of 2,136 genes is significantly decreased under dehydration conditions (Kawaguchi et al., 2004), and mRNA sequence features (i.e., 5′-UTR GC content, initiation codon context, and ORF length) influence the dehydration-mediated differential mRNA translation (Kawaguchi and Bailey-Serres, 2005). Furthermore, translatome profiling revealed that mRNA sequence features as well as uORFs are important for dynamic translational changes under drought conditions in maize (Lei et al., 2015). In rice (Oryza sativa), more than 50% of the genes encoding ribosomal proteins (e.g., rpS4, rpS10, rpS18a, rpL6, rpL7, rpL23A, rpL24, and rpL31) are upregulated in response to drought, and overexpressing rpL23A confers rice drought tolerance (Moin et al., 2017). In addition, silencing of rpL14B decreases drought tolerance in cotton (Gossypium hirsutum) (Shiraku et al., 2021). However, although ribosomal proteins are abundant RNA-binding proteins involved in ribosome structure and protein biosynthesis, they also have additional functions (Warner and McIntosh, 2009). Therefore, it is not clear whether the drought effects associated with ribosomal proteins are related to mRNA translation.
uORF-mediated translational control for crop improvement
The development of stress-resilient crops is needed to cope with the environmental extremes caused by climate change and the increase in food demand due to world population expansion. General protein translation is significantly decreased under stressful conditions, while translation of specific stress-associated proteins is increased (Muench et al., 2012; Merchante et al., 2017). uORFs are a conserved structure found in 30–60% of eukaryotic transcripts, including plant transcripts, and are important for mRNA translation (Von Arnim et al., 2014; Chew et al., 2016). Most mRNAs of stress-related genes upregulated by a specific stress stimulus contain uORF sequences in plants (Ebina et al., 2015; Hayashi et al., 2017). Therefore, uORF-mediated translational control represents a promising method to improve the stress resilience of crops.
Engineering plants to express specific stress-responsive genes can enhanced stress resilience (Kamthan et al., 2016). However, it can also reduce plant growth and yield due to tradeoff effects (Gurr and Rushton, 2005; Huot et al., 2014; Waadt et al., 2022). These fitness costs can be resolved by uORF-mediated translational regulation. For example, Xu et al. developed a TBF1 cassette using the TBF1 promoter and two uORFs on TBF1 mRNA that conferred pathogen-inducible translational control to overcome the fitness costs of the plant immune response (Xu et al., 2017b). They showed that pTBF1:uORFs-driven expression of Arabidopsis Nonexpressor of pathogenesis-related genes 1, encoding a master regulator of salicylic-acid-mediated defense signaling that contributes to broad-spectrum resistance, increased rice resistance to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae and the rice blast pathogen X. oryzae pv. oryzicola without fitness penalties. Therefore, developing uORF-based promoters triggering mRNA translation in response to a specific stress stimulus is an important strategy for improving crop stress resistance.
The development of the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9) technology was an innovation that allows efficient genome editing without the presence of transgenes or the regulatory issues associated with genetically modified crops (Son and Park, 2022a). Not only coding regions but also promoter regions regulating gene expression are good targets for genome editing for crop improvement (Hua et al., 2019). Since most uORFs inhibit mRNA translation via ribosome stalling, ribosome disassociation, uORF translation past the initiation codon, and mRNA decay, mutation of uORFs by CRISPR/Cas9 can change translation efficiency (Young and Wek, 2016; Um et al., 2021). Indeed, crop improvement through CRISPR/Cas9-mediated editing of uORFs regulating mRNA translation has been reported. Lettuce (Lactuca sativa) GDP-L-galactose phosphorylase 2 (LsGGP2) uORF editing enhanced ascorbic acid (vitamin C) contents and tolerance to oxidative stress in iceberg lettuce (Zhang et al., 2018). Mutation of GGP1 uORFs by CRISPR/Cas9 also increased the ascorbic acid content in wild tomato (Solanum pimpinellifolium) accession LA1589 and bacterial spot disease resistance (Li et al., 2018). Editing of the Phosphate 1 uORF induced root inorganic orthophosphate (Pi) accumulation and Pi deficiency tolerance in Arabidopsis (Reis et al., 2020). Strawberry (Fragaria vesca) S1-group basic leucine zipper protein 1.1 editing increased the sugar content (Xing et al., 2020). CRISPR/Cas9-mediated mutation of Heading date 2 delayed flowering time in rice (Liu et al., 2021). Previously, it was difficult to improve crop traits through uORF editing due to technical limitations, but more convenient methods are continuously being developed (Si et al., 2020). Therefore, uORF-mediated translational control is expected to become accessible to many scientists and contribute to stress resilience in crops.
Conclusions
Global climate change exacerbates abiotic and biotic stresses in crops, and improving crop stress resilience is essential for sustainable agriculture. Although advances in biotechnology, including CRISPR/Cas9 gene editing systems, have facilitated crop improvement, they require identification and characterization of genes and their regulatory networks. Since translational control is an efficient way to increase resistance to various stresses, it is essential to understand the mechanisms controlling protein biosynthesis. Numerous global translatome profiling technologies, such as polysome profiling, ribosome profiling, translating ribosome affinity purification, and 3′ ribosome-profiling sequencing, have been developed and utilized to study plant translation (Ingolia et al., 2009; Heiman et al., 2014; Mazzoni-Putman and Stepanova, 2018; Zhu et al., 2021) and have facilitated important advances in plant biology. Therefore, we have provided here an overview of the current understanding of protein translational control in response to pathogens, hypoxia, heat, and drought stresses in plants, and discussed a crop improvement strategy based on translational regulation through editing of uORFs. Though the translational control mechanisms involved in stress responses need further study, their exploration and application in crop breeding are important steps toward developing stress tolerant crop cultivars without fitness costs.
Author contributions
SS conceptualized and wrote the manuscript. SRP supervised. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Research Program for Agricultural Science and Technology Development (Project No. PJ01570601), and supported by the 2023 Fellowship Program (Project No. PJ01661001) of the National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Axtell, M. J., Staskawicz, B. J. (2003). Initiation of RPS2-specified disease resistance in arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 (3), 369–377. doi: 10.1016/s0092-8674(03)00036-9
Baena-Gonzalez, E., Rolland, F., Thevelein, J. M., Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448 (7156), 938–942. doi: 10.1038/nature06069
Bailey-Serres, J., Dawe, R. K. (1996). Both 5' and 3' sequences of maize adh1 mRNA are required for enhanced translation under low-oxygen conditions. Plant Physiol. 112 (2), 685–695. doi: 10.1104/pp.112.2.685
Bailey-Serres, J., Voesenek, L. A. C. J. (2008). Flooding stress: Acclimations and genetic diversity. Annu. Rev. Plant Biol. 59, 313. doi: 10.1146/annurev.arplant.59.032607.092752
Bensen, R. J., Boyer, J. S., Mullet, J. E. (1988). Water deficit-induced changes in abscisic acid, growth, polysomes, and translatable RNA in soybean hypocotyls. Plant Physiol. 88 (2), 289–294. doi: 10.1104/pp.88.2.289
Bi, C., Ma, Y., Jiang, S. C., Mei, C., Wang, X. F., Zhang, D. P. (2019). Arabidopsis translation initiation factors eIFiso4G1/2 link repression of mRNA cap-binding complex eIFiso4F assembly with RNA-binding protein SOAR1-mediated ABA signaling. New Phytol. 223 (3), 1388–1406. doi: 10.1111/nph.15880
Bigeard, J., Colcombet, J., Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8 (4), 521–539. doi: 10.1016/j.molp.2014.12.022
Bita, C. E., Gerats, T. (2013). Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00273
Bonnot, T., Nagel, D. H. (2021). Time of the day prioritizes the pool of translating mRNAs in response to heat stress. Plant Cell 33 (7), 2164–2182. doi: 10.1093/plcell/koab113
Branco-Price, C., Kaiser, K. A., Jang, C. J., Larive, C. K., Bailey-Serres, J. (2008). Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in arabidopsis thaliana. Plant J. 56 (5), 743–755. doi: 10.1111/j.1365-313X.2008.03642.x
Branco-Price, C., Kawaguchi, R., Ferreira, R. B., Bailey-Serres, J. (2005). Genome-wide analysis of transcript abundance and translation in arabidopsis seedlings subjected to oxygen deprivation. Ann. Bot. 96 (4), 647–660. doi: 10.1093/aob/mci217
Bray, E. (1990). Drought-stress-induced polypeptide accumulation in tomato leaves. Plant Cell Environ. 13 (6), 531–538. doi: 10.1111/j.1365-3040.1990.tb01069.x
Browning, K. S., Bailey-Serres, J. (2015). Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 13, e0176. doi: 10.1199/tab.0176
Bruns, A. N., Li, S., Mohannath, G., Bisaro, D. M. (2019). Phosphorylation of arabidopsis eIF4E and eIFiso4E by SnRK1 inhibits translation. FEBS J. 286 (19), 3778–3796. doi: 10.1111/febs.14935
Buccitelli, C., Selbach, M. (2020). mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 21 (10), 630–644. doi: 10.1038/s41576-020-0258-4
Buchan, J. R., Parker, R. (2009). Eukaryotic stress granules: The ins and outs of translation. Mol. Cell 36 (6), 932–941. doi: 10.1016/j.molcel.2009.11.020
Calil, I. P., Fontes, E. P. B. (2017). Plant immunity against viruses: Antiviral immune receptors in focus. Ann. Bot. 119 (5), 711–723. doi: 10.1093/aob/mcw200
Chaudhry, S., Sidhu, G. P. S. (2022). Climate change regulated abiotic stress mechanisms in plants: A comprehensive review. Plant Cell Rep. 41 (1), 1–31. doi: 10.1007/s00299-021-02759-5
Chee, N. T., Lohse, I., Brothers, S. P. (2019). mRNA-to-protein translation in hypoxia. Mol. Cancer 18 (1), 49. doi: 10.1186/s12943-019-0968-4
Chen, G. H., Liu, M. J., Xiong, Y., Sheen, J., Wu, S. H. (2018). TOR and RPS6 transmit light signals to enhance protein translation in deetiolating arabidopsis seedlings. Proc. Natl. Acad. Sci. U.S.A. 115 (50), 12823–12828. doi: 10.1073/pnas.1809526115
Chew, G. L., Pauli, A., Schier, A. F. (2016). Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat. Commun. 7, 11663. doi: 10.1038/ncomms11663
Cho, H. Y., Chou, M. Y., Ho, H. Y., Chen, W. C., Shih, M. C. (2022). Ethylene modulates translation dynamics in arabidopsis under submergence via GCN2 and EIN2. Sci. Adv. 8 (22), eabm7863. doi: 10.1126/sciadv.abm7863
Cho, Y. H., Hong, J. W., Kim, E. C., Yoo, S. D. (2012). Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 158 (4), 1955–1964. doi: 10.1104/pp.111.189829
Cho, H. Y., Lu, M. J., Shih, M. C. (2019). The SnRK1-eIFiso4G1 signaling relay regulates the translation of specific mRNAs in arabidopsis under submergence. New Phytol. 222 (1), 366–381. doi: 10.1111/nph.15589
Cobb, M. (2017). 60 years ago, Francis crick changed the logic of biology. PloS Biol. 15 (9), e2003243. doi: 10.1371/journal.pbio.2003243
Cui, Y., Rao, S., Chang, B., Wang, X., Zhang, K., Hou, X., et al. (2015b). AtLa1 protein initiates IRES-dependent translation of WUSCHEL mRNA and regulates the stem cell homeostasis of arabidopsis in response to environmental hazards. Plant Cell Environ. 38 (10), 2098–2114. doi: 10.1111/pce.12535
Cui, H., Tsuda, K., Parker, J. E. (2015a). Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Decker, C. J., Parker, R. (2012). P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4 (9), a012286. doi: 10.1101/cshperspect.a012286
DeFalco, T. A., Zipfel, C. (2021). Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell 81 (17), 3449–3467. doi: 10.1016/j.molcel.2021.07.029
Dever, T. E. (2002). Gene-specific regulation by general translation factors. Cell 108 (4), 545–556. doi: 10.1016/s0092-8674(02)00642-6
Dever, T. E., Dinman, J. D., Green, R. (2018). Translation elongation and recoding in eukaryotes. Cold Spring Harbor Perspect. Biol. 10 (8), a032649. doi: 10.1101/cshperspect.a032649
Dhindsa, R., Cleland, R. (1975). Water stress and protein synthesis: I. differential inhibition of protein synthesis. Plant Physiol. 55 (4), 778–781. doi: 10.1104/pp.55.4.778
Dinkova, T. D., Zepeda, H., Martinez-Salas, E., Martinez, L. M., Nieto-Sotelo, J., de Jimenez, E. S. (2005). Cap-independent translation of maize Hsp101. Plant J. 41 (5), 722–731. doi: 10.1111/j.1365-313X.2005.02333.x
Dorokhov, Y. L., Ivanov, P. A., Komarova, T. V., Skulachev, M. V., Atabekov, J. G. (2006). An internal ribosome entry site located upstream of the crucifer-infecting tobamovirus coat protein (CP) gene can be used for CP synthesis in vivo. J. Gen. Virol. 87 (Pt 9), 2693–2697. doi: 10.1099/vir.0.82095-0
Dresios, J., Chappell, S. A., Zhou, W., Mauro, V. P. (2006). An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 13 (1), 30–34. doi: 10.1038/nsmb1031
Ebina, I., Takemoto-Tsutsumi, M., Watanabe, S., Koyama, H., Endo, Y., Kimata, K., et al. (2015). Identification of novel arabidopsis thaliana upstream open reading frames that control expression of the main coding sequences in a peptide sequence-dependent manner. Nucleic Acids Res. 43 (3), 1562–1576. doi: 10.1093/nar/gkv018
Echevarría-Zomeño, S., Yángüez, E., Fernández-Bautista, N., Castro-Sanz, A. B., Ferrando, A., Castellano, M. M. (2013). Regulation of translation initiation under biotic and abiotic stresses. Int. J. Mol. Sci. 14 (3), 4670–4683. doi: 10.3390/ijms14034670
Faulkner, C., Robatzek, S. (2012). Plants and pathogens: Putting infection strategies and defence mechanisms on the map. Curr. Opin. Plant Biol. 15 (6), 699–707. doi: 10.1016/j.pbi.2012.08.009
Freire, M. A. (2005). Translation initiation factor (iso) 4E interacts with BTF3, the beta subunit of the nascent polypeptide-associated complex. Gene 345 (2), 271–277. doi: 10.1016/j.gene.2004.11.030
Freire, M. A., Tourneur, C., Granier, F., Camonis, J., El Amrani, A., Browning, K. S., et al. (2000). Plant lipoxygenase 2 is a translation initiation factor-4E-binding protein. Plant Mol. Biol. 44 (2), 129–140. doi: 10.1023/a:1006494628892
Fukao, T., Bailey-Serres, J. (2004). Plant responses to hypoxia–is survival a balancing act? Trends Plant Sci. 9 (9), 449–456. doi: 10.1016/j.tplants.2004.07.005
Gallie, D. R., Caldwell, C., Pitto, L. (1995). Heat shock disrupts cap and poly (A) tail function during translation and increases mRNA stability of introduced reporter mRNA. Plant Physiol. 108 (4), 1703–1713. doi: 10.1104/pp.108.4.1703
Gallie, D. R., Le, H., Caldwell, C., Tanguay, R. L., Hoang, N. X., Browning, K. S. (1997). The phosphorylation state of translation initiation factors is regulated developmentally and following heat shock in wheat. J. Biol. Chem. 272 (2), 1046–1053. doi: 10.1074/jbc.272.2.1046
Garneau, N. L., Wilusz, J., Wilusz, C. J. (2007). The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8 (2), 113–126. doi: 10.1038/nrm2104
Gebauer, F., Hentze, M. W. (2004). Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5 (10), 827–835. doi: 10.1038/nrm1488
Geigenberger, P. (2003). Response of plant metabolism to too little oxygen. Curr. Opin. Plant Biol. 6 (3), 247–256. doi: 10.1016/s1369-5266(03)00038-4
Geng, G., Wang, D., Liu, Z., Wang, Y., Zhu, M., Cao, X., et al. (2021). Translation of plant RNA viruses. Viruses 13 (12), 2499. doi: 10.3390/v13122499
Godet, A. C., David, F., Hantelys, F., Tatin, F., Lacazette, E., Garmy-Susini, B., et al. (2019). IRES trans-acting factors, key actors of the stress response. Int. J. Mol. Sci. 20 (4), 924. doi: 10.3390/ijms20040924
Gurr, S. J., Rushton, P. J. (2005). Engineering plants with increased disease resistance: how are we going to express it? Trends Biotechnol. 23 (6), 283–290. doi: 10.1016/j.tibtech.2005.04.009
Hashimoto, M., Neriya, Y., Yamaji, Y., Namba, S. (2016). Recessive resistance to plant viruses: Potential resistance genes beyond translation initiation factors. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01695
Hayashi, N., Sasaki, S., Takahashi, H., Yamashita, Y., Naito, S., Onouchi, H. (2017). Identification of arabidopsis thaliana upstream open reading frames encoding peptide sequences that cause ribosomal arrest. Nucleic Acids Res. 45 (15), 8844–8858. doi: 10.1093/nar/gkx528
Heese, A., Hann, D. R., Gimenez-Ibanez, S., Jones, A. M., He, K., Li, J., et al. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. 104 (29), 12217–12222. doi: 10.1073/pnas.0705306104
Heiman, M., Kulicke, R., Fenster, R. J., Greengard, P., Heintz, N. (2014). Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protoc. 9 (6), 1282–1291. doi: 10.1038/nprot.2014.085
Hernandez, G., Altmann, M., Lasko, P. (2010). Origins and evolution of the mechanisms regulating translation initiation in eukaryotes. Trends Biochem. Sci. 35 (2), 63–73. doi: 10.1016/j.tibs.2009.10.009
Hinnebusch, A. G., Ivanov, I. P., Sonenberg, N. (2016). Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352 (6292), 1413–1416. doi: 10.1126/science.aad9868
Holcik, M. (2015). Could the eIF2α-independent translation be the achilles heel of cancer? Front. Oncol. 5. doi: 10.3389/fonc.2015.00264
Holcik, M., Sonenberg, N. (2005). Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6 (4), 318–327. doi: 10.1038/nrm1618
Hsiao, T. C. (1970). Rapid changes in levels of polyribosomes in zea mays in response to water stress. Plant Physiol. 46 (2), 281–285. doi: 10.1104/pp.46.2.281
Hua, K., Zhang, J., Botella, J. R., Ma, C., Kong, F., Liu, B., et al. (2019). Perspectives on the application of genome-editing technologies in crop breeding. Mol. Plant 12 (8), 1047–1059. doi: 10.1016/j.molp.2019.06.009
Huot, B., Yao, J., Montgomery, B. L., He, S. Y. (2014). Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 7 (8), 1267–1287. doi: 10.1093/mp/ssu049
Hurkman, W. J., Tanaka, C. K. (1988). Polypeptide changes induced by salt stress, water deficit, and osmotic stress in barley roots: A comparison using two-dimensional gel electrophoresis. Electrophoresis 9 (11), 781–787. doi: 10.1002/elps.1150091114
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R., Weissman, J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324 (5924), 218–223. doi: 10.1126/science.1168978
Izquierdo, Y., Kulasekaran, S., Benito, P., Lopez, B., Marcos, R., Cascon, T., et al. (2018). Arabidopsis nonresponding to oxylipins locus NOXY7 encodes a yeast GCN1 homolog that mediates noncanonical translation regulation and stress adaptation. Plant Cell Environ. 41 (6), 1438–1452. doi: 10.1111/pce.13182
Jaafar, Z. A., Kieft, J. S. (2019). Viral RNA structure-based strategies to manipulate translation. Nat. Rev. Microbiol. 17 (2), 110–123. doi: 10.1038/s41579-018-0117-x
Jaag, H. M., Kawchuk, L., Rohde, W., Fischer, R., Emans, N., Prufer, D. (2003). An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc. Natl. Acad. Sci. U.S.A. 100 (15), 8939–8944. doi: 10.1073/pnas.1332697100
Jackson, R. J., Howell, M. T., Kaminski, A. (1990). The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem. Sci. 15 (12), 477–483. doi: 10.1016/0968-0004(90)90302-r
Juntawong, P., Girke, T., Bazin, J., Bailey-Serres, J. (2014). Translational dynamics revealed by genome-wide profiling of ribosome footprints in arabidopsis. Proc. Natl. Acad. Sci. 111 (1), E203–E212. doi: 10.1073/pnas.1317811111
Kafri, M., Metzl-Raz, E., Jona, G., Barkai, N. (2016). The cost of protein production. Cell Rep. 14 (1), 22–31. doi: 10.1016/j.celrep.2015.12.015
Kamthan, A., Chaudhuri, A., Kamthan, M., Datta, A. (2016). Genetically modified (GM) crops: Milestones and new advances in crop improvement. Theor. Appl. Genet. 129 (9), 1639–1655. doi: 10.1007/s00122-016-2747-6
Kapp, L. D., Lorsch, J. R. (2004). The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73, 657. doi: 10.1146/annurev.biochem.73.030403.080419
Karetnikov, A., Lehto, K. (2007). The RNA2 5′ leader of blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. J. Gen. Virol. 88 (1), 286–297. doi: 10.1099/vir.0.82307-0
Kawaguchi, R., Bailey-Serres, J. (2005). mRNA sequence features that contribute to translational regulation in arabidopsis. Nucleic Acids Res. 33 (3), 955–965. doi: 10.1093/nar/gki240
Kawaguchi, R., Girke, T., Bray, E. A., Bailey-Serres, J. (2004). Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in arabidopsis thaliana. Plant J. 38 (5), 823–839. doi: 10.1111/j.1365-313X.2004.02090.x
Kawaguchi, R., Williams, A., Bray, E., Bailey-Serres, J. (2003). Water-deficit-induced translational control in nicotiana tabacum. Plant Cell Environ. 26 (2), 221–229. doi: 10.1046/j.1365-3040.2003.00952.x
Key, J. L., Lin, C., Chen, Y. (1981). Heat shock proteins of higher plants. Proc. Natl. Acad. Sci. 78 (6), 3526–3530. doi: 10.1073/pnas.78.6.3526
Komar, A. A., Hatzoglou, M. (2011). Cellular IRES-mediated translation: The war of ITAFs in pathophysiological states. Cell Cycle 10 (2), 229–240. doi: 10.4161/cc.10.2.14472
Koonin, E. V. (2012). Does the central dogma still stand? Biol. direct 7 (1), 1–7. doi: 10.1186/1745-6150-7-27
Lamper, A. M., Fleming, R. H., Ladd, K. M., Lee, A. S. Y. (2020). A phosphorylation-regulated eIF3d translation switch mediates cellular adaptation to metabolic stress. Science 370 (6518), 853–856. doi: 10.1126/science.abb0993
Lázaro-Mixteco, P. E., Dinkova, T. D. (2012). Identification of proteins from cap-binding complexes by mass spectrometry during maize (Zea mays l.) germination. J. Mexican Chem. Soc. 56 (1), 36–50. doi: 10.29356/jmcs.v56i1.273
Lee, A. S., Kranzusch, P. J., Cate, J. H. (2015). eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522 (7554), 111–114. doi: 10.1038/nature14267
Lee, A. S., Kranzusch, P. J., Doudna, J. A., Cate, J. H. (2016). eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536 (7614), 96–99. doi: 10.1038/nature18954
Lee, S. C., Mustroph, A., Sasidharan, R., Vashisht, D., Pedersen, O., Oosumi, T., et al. (2011). Molecular characterization of the submergence response of the arabidopsis thaliana ecotype Columbia. New Phytol. 190 (2), 457–471. doi: 10.1111/j.1469-8137.2010.03590.x
Lee, D. H., Park, S. J., Ahn, C. S., Pai, H. S. (2017). MRF family genes are involved in translation control, especially under energy-deficient conditions, and their expression and functions are modulated by the TOR signaling pathway. Plant Cell 29 (11), 2895–2920. doi: 10.1105/tpc.17.00563
Lei, L., Shi, J., Chen, J., Zhang, M., Sun, S., Xie, S., et al. (2015). Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 84 (6), 1206–1218. doi: 10.1111/tpj.13073
Leonetti, P., Stuttmann, J., Pantaleo, V. (2021). Regulation of plant antiviral defense genes via host RNA-silencing mechanisms. Virol. J. 18 (1), 194. doi: 10.1186/s12985-021-01664-3
Li, M., Ma, X., Chiang, Y. H., Yadeta, K. A., Ding, P., Dong, L., et al. (2014). Proline isomerization of the immune receptor-interacting protein RIN4 by a cyclophilin inhibits effector-triggered immunity in arabidopsis. Cell Host Microbe 16 (4), 473–483. doi: 10.1016/j.chom.2014.09.007
Li, T., Yang, X., Yu, Y., Si, X., Zhai, X., Zhang, H., et al. (2018). Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36 (12), 1160–1163. doi: 10.1038/nbt.4273
Liu, X., Afrin, T., Pajerowska-Mukhtar, K. M. (2019). Arabidopsis GCN2 kinase contributes to ABA homeostasis and stomatal immunity. Commun. Biol. 2, 302. doi: 10.1038/s42003-019-0544-x
Liu, Y. J., Chern, Y. (2021). Contribution of energy dysfunction to impaired protein translation in neurodegenerative diseases. Front. Cell Neurosci. 15. doi: 10.3389/fncel.2021.668500
Liu, J., Elmore, J. M., Lin, Z. J., Coaker, G. (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9 (2), 137–146. doi: 10.1016/j.chom.2011.01.010
Liu, X., Kørner, C. J., Hajdu, D., Guo, T., Ramonell, K. M., Argueso, C. T., et al. (2015). Arabidopsis thaliana atGCN2 kinase is involved in disease resistance against pathogens with diverse life styles. Int. J. Phytopathol. 4 (2), 93–104. doi: 10.33687/phytopath.004.02.1342
Liu, X., Liu, H., Zhang, Y., He, M., Li, R., Meng, W., et al. (2021). Fine-tuning flowering time via genome editing of upstream open reading frames of heading date 2 in rice. Rice (N Y) 14 (1), 59. doi: 10.1186/s12284-021-00504-w
Liu, B., Qian, S. B. (2014). Translational reprogramming in cellular stress response. Wiley Interdiscip Rev. RNA 5 (3), 301–315. doi: 10.1002/wrna.1212
Lokdarshi, A., von Arnim, A. G. (2022). Review: Emerging roles of the signaling network of the protein kinase GCN2 in the plant stress response. Plant Sci. 320, 111280. doi: 10.1016/j.plantsci.2022.111280
Lolle, S., Stevens, D., Coaker, G. (2020). Plant NLR-triggered immunity: From receptor activation to downstream signaling. Curr. Opin. Immunol. 62, 99–105. doi: 10.1016/j.coi.2019.12.007
Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R., Dangl, J. L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 (3), 379–389. doi: 10.1016/s0092-8674(03)00040-0
Mackey, D., Holt, B. F., III, Wiig, A., Dangl, J. L. (2002). RIN4 interacts with pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in arabidopsis. Cell 108 (6), 743–754. doi: 10.1016/s0092-8674(02)00661-x
Maegawa, K., Takii, R., Ushimaru, T., Kozaki, A. (2015). Evolutionary conservation of TORC1 components, TOR, raptor, and LST8, between rice and yeast. Mol. Genet. Genomics 290 (5), 2019–2030. doi: 10.1007/s00438-015-1056-0
Maldonado-Bonilla, L. D. (2014). Composition and function of p bodies in arabidopsis thaliana. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00201
Manjunath, S., Williams, A. J., Bailey-Serres, J. (1999). Oxygen deprivation stimulates Ca2+-mediated phosphorylation of mRNA cap-binding protein eIF4E in maize roots. Plant J. 19 (1), 21–30. doi: 10.1046/j.1365-313x.1999.00489.x
Mason, H. S., Mullet, J. E., Boyer, J. S. (1988). Polysomes, messenger RNA, and growth in soybean stems during development and water deficit. Plant Physiol. 86 (3), 725–733. doi: 10.1104/pp.86.3.725
Matsuura, H., Ishibashi, Y., Shinmyo, A., Kanaya, S., Kato, K. (2010). Genome-wide analyses of early translational responses to elevated temperature and high salinity in arabidopsis thaliana. Plant Cell Physiol. 51 (3), 448–462. doi: 10.1093/pcp/pcq010
Mazzoni-Putman, S. M., Stepanova, A. N. (2018). A plant biologist's toolbox to study translation. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00873
Merchante, C., Stepanova, A. N., Alonso, J. M. (2017). Translation regulation in plants: An interesting past, an exciting present and a promising future. Plant J. 90 (4), 628–653. doi: 10.1111/tpj.13520
Merret, R., Nagarajan, V. K., Carpentier, M.-C., Park, S., Favory, J.-J., Descombin, J., et al. (2015). Heat-induced ribosome pausing triggers mRNA co-translational decay in arabidopsis thaliana. Nucleic Acids Res. 43 (8), 4121–4132. doi: 10.1093/nar/gkv234
Merrick, W. C. (2004). Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332, 1–11. doi: 10.1016/j.gene.2004.02.051
Merrick, W. C., Pavitt, G. D. (2018). Protein synthesis initiation in eukaryotic cells. Cold Spring Harbor Perspect. Biol. 10 (12), a033092. doi: 10.1101/cshperspect.a033092
Meteignier, L. V., El Oirdi, M., Cohen, M., Barff, T., Matteau, D., Lucier, J. F., et al. (2017). Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in arabidopsis. J. Exp. Bot. 68 (9), 2333–2344. doi: 10.1093/jxb/erx078
Moin, M., Bakshi, A., Madhav, M. S., Kirti, P. B. (2017). Expression profiling of ribosomal protein gene family in dehydration stress responses and characterization of transgenic rice plants overexpressing RPL23A for water-use efficiency and tolerance to drought and salt stresses. Front. Chem. 5. doi: 10.3389/fchem.2017.00097
Muench, D. G., Zhang, C., Dahodwala, M. (2012). Control of cytoplasmic translation in plants. Wiley Interdiscip Rev. RNA 3 (2), 178–194. doi: 10.1002/wrna.1104
Naveed, Z. A., Wei, X., Chen, J., Mubeen, H., Ali, G. S. (2020). The PTI to ETI continuum in phytophthora-plant interactions. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.593905
Ndimba, B. K., Chivasa, S., Simon, W. J., Slabas, A. R. (2005). Identification of arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 5 (16), 4185–4196. doi: 10.1002/pmic.200401282
Nietzsche, M., Landgraf, R., Tohge, T., Börnke, F. (2016). A protein–protein interaction network linking the energy-sensor kinase SnRK1 to multiple signaling pathways in arabidopsis thaliana. Curr. Plant Biol. 5, 36–44. doi: 10.1016/j.cpb.2015.10.004
Nover, L., Scharf, K.-D., Neumann, D. (1989). Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 9 (3), 1298–1308. doi: 10.1128/mcb.9.3.1298-1308.1989
Nukarinen, E., Nagele, T., Pedrotti, L., Wurzinger, B., Mair, A., Landgraf, R., et al. (2016). Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 6, 31697. doi: 10.1038/srep31697
Obomighie, I., Lapenas, K., Murphy, B. E., Bowles, A. M. C., Bechtold, U., Prischi, F. (2021). The role of ribosomal protein S6 kinases in plant homeostasis. Front. Mol. Biosci. 8. doi: 10.3389/fmolb.2021.636560
Orphanides, G., Reinberg, D. (2002). A unified theory of gene expression. Cell 108 (4), 439–451. doi: 10.1016/s0092-8674(02)00655-4
Pajerowska-Mukhtar, K. M., Wang, W., Tada, Y., Oka, N., Tucker, C. L., Fonseca, J. P., et al. (2012). The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr. Biol. 22 (2), 103–112. doi: 10.1016/j.cub.2011.12.015
Patrick, R. M., Lee, J. C. H., Teetsel, J. R. J., Yang, S. H., Choy, G. S., Browning, K. S. (2018). Discovery and characterization of conserved binding of eIF4E 1 (CBE1), a eukaryotic translation initiation factor 4E-binding plant protein. J. Biol. Chem. 293 (44), 17240–17247. doi: 10.1074/jbc.RA118.003945
Reis, R. S., Deforges, J., Sokoloff, T., Poirier, Y. (2020). Modulation of shoot phosphate level and growth by PHOSPHATE1 upstream open reading frame. Plant Physiol. 183 (3), 1145–1156. doi: 10.1104/pp.19.01549
Rhodes, P. R., Matsuda, K. (1976). Water stress, rapid polyribosome reductions and growth. Plant Physiol. 58 (5), 631–635. doi: 10.1104/pp.58.5.631
Richter, J. D., Sonenberg, N. (2005). Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433 (7025), 477–480. doi: 10.1038/nature03205
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290 (5499), 2105–2110. doi: 10.1126/science.290.5499.2105
Robertson, G., Burger, J., Campa, M. (2022). CRISPR/Cas-based tools for the targeted control of plant viruses. Mol. Plant Pathol. 23 (11), 1701–1718. doi: 10.1111/mpp.13252
Roux, M., Schwessinger, B., Albrecht, C., Chinchilla, D., Jones, A., Holton, N., et al. (2011). The arabidopsis leucine-rich repeat receptor–like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23 (6), 2440–2455. doi: 10.1105/tpc.111.084301
Roy, B., Von Arnim, A. G. (2013). Translational regulation of cytoplasmic mRNAs. Arabidopsis book/American Soc. Plant Biologists 11, e0165. doi: 10.1199/tab.0165
Sablok, G., Powell, J. J., Kazan, K. (2017). Emerging roles and landscape of translating mRNAs in plants. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01443
Sachs, M. M., Freeling, M., Okimoto, R. (1980). The anaerobic proteins of maize. Cell 20 (3), 761–767. doi: 10.1016/0092-8674(80)90322-0
Sachs, M. M., Subbaiah, C. C., Saab, I. N. (1996). Anaerobic gene expression and flooding tolerance in maize. J. Exp. Bot. 47 (1), 1–15. doi: 10.1093/jxb/47.1.1
Sanfacon, H. (2015). Plant translation factors and virus resistance. Viruses 7 (7), 3392–3419. doi: 10.3390/v7072778
Schepetilnikov, M., Dimitrova, M., Mancera-Martinez, E., Geldreich, A., Keller, M., Ryabova, L. A. (2013). TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 32 (8), 1087–1102. doi: 10.1038/emboj.2013.61
Schulze, B., Mentzel, T., Jehle, A. K., Mueller, K., Beeler, S., Boller, T., et al. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285 (13), 9444–9451. doi: 10.1074/jbc.M109.096842
Shatsky, I. N., Terenin, I. M., Smirnova, V. V., Andreev, D. E. (2018). Cap-independent translation: What’s in a name? Trends Biochem. Sci. 43 (11), 882–895. doi: 10.1016/j.tibs.2018.04.011
Shiraku, M. L., Magwanga, R. O., Cai, X., Kirungu, J. N., Xu, Y., Mehari, T. G., et al. (2021). Knockdown of 60S ribosomal protein L14-2 reveals their potential regulatory roles to enhance drought and salt tolerance in cotton. J. Cotton Res. 4 (1), 1–14. doi: 10.1186/s42397-021-00102-7
Shiu, S.-H., Shih, M.-C., Li, W.-H. (2005). Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol. 139 (1), 18–26. doi: 10.1104/pp.105.065110
Si, X., Zhang, H., Wang, Y., Chen, K., Gao, C. (2020). Manipulating gene translation in plants by CRISPR-Cas9-mediated genome editing of upstream open reading frames. Nat. Protoc. 15 (2), 338–363. doi: 10.1038/s41596-019-0238-3
Son, S., Im, J. H., Ko, J. H., Han, K. H. (2023). SNF1-related protein kinase 1 represses arabidopsis growth through post-translational modification of E2Fa in response to energy stress. New Phytol. 237 (3), 823–839. doi: 10.1111/nph.18597
Son, S., Im, J. H., Song, G., Park, S. R. (2022). SNF1-related protein kinase 1 activity represses the canonical translational machinery. Plants 11 (10), 1359. doi: 10.3390/plants11101359
Son, S., Kim, S., Lee, K. S., Oh, J., Choi, I., Do, J. W., et al. (2021). Identification of the capsicum baccatum NLR protein CbAR9 conferring disease resistance to anthracnose. Int. J. Mol. Sci. 22 (22), 12612. doi: 10.3390/ijms222212612
Son, S., Park, S. R. (2022a). Challenges facing CRISPR/Cas9-based genome editing in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.902413
Son, S., Park, S. R. (2022b). Climate change impedes plant immunity mechanisms. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1032820
Sonenberg, N., Hinnebusch, A. G. (2009). Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136 (4), 731–745. doi: 10.1016/j.cell.2009.01.042
Sorenson, R., Bailey-Serres, J. (2014). Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in arabidopsis. Proc. Natl. Acad. Sci. 111 (6), 2373–2378. doi: 10.1073/pnas.1314851111
Spriggs, K. A., Bushell, M., Willis, A. E. (2010). Translational regulation of gene expression during conditions of cell stress. Mol. Cell 40 (2), 228–237. doi: 10.1016/j.molcel.2010.09.028
Sun, Y., Li, L., Macho, A. P., Han, Z., Hu, Z., Zipfel, C., et al. (2013). Structural basis for flg22-induced activation of the arabidopsis FLS2-BAK1 immune complex. Science 342 (6158), 624–628. doi: 10.1126/science.1243825
Szick-Miranda, K., Jayachandran, S., Tam, A., Werner-Fraczek, J., Williams, A., Bailey-Serres, J. (2003). Evaluation of translational control mechanisms in response to oxygen deprivation in maize. Russian J. Plant Physiol. 50 (6), 774–786. doi: 10.1023/B:RUPP.0000003275.97021.2b
Takemoto, D., Jones, D. A. (2005). Membrane release and destabilization of arabidopsis RIN4 following cleavage by pseudomonas syringae AvrRpt2. Mol. Plant Microbe Interact. 18 (12), 1258–1268. doi: 10.1094/MPMI-18-1258
Terry, B. C., Liu, X., Murphy, A. M., Pajerowska-Mukhtar, K. M. (2015). Arabidopsis thaliana GCN2 is involved in responses to osmotic and heat stresses. Int. J. Plant Res. 5 (4), 87–95. doi: 10.5923/j.plant.20150504.03
Toribio, R., Muñoz, A., Castro-Sanz, A. B., Merchante, C., Castellano, M. (2019). A novel eIF4E-interacting protein that forms non-canonical translation initiation complexes. Nat. Plants 5 (12), 1283–1296. doi: 10.1038/s41477-019-0553-2
Toribio, R., Muñoz, A., Sánchez, F., Ponz, F., Castellano, M. M. (2021). High overexpression of CERES, a plant regulator of translation, induces different phenotypical defence responses during TuMV infection. Plant J. 107 (1), 256–267. doi: 10.1111/tpj.15290
Trefts, E., Shaw, R. J. (2021). AMPK: restoring metabolic homeostasis over space and time. Mol. Cell 81 (18), 3677–3690. doi: 10.1016/j.molcel.2021.08.015
Tsalikis, J., Croitoru, D. O., Philpott, D. J., Girardin, S. E. (2013). Nutrient sensing and metabolic stress pathways in innate immunity. Cell Microbiol. 15 (10), 1632–1641. doi: 10.1111/cmi.12165
Tsuda, K., Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13 (4), 459–465. doi: 10.1016/j.pbi.2010.04.006
Um, T., Park, T., Shim, J. S., Kim, Y. S., Lee, G. S., Choi, I. Y., et al. (2021). Application of upstream open reading frames (uORFs) editing for the development of stress-tolerant crops. Int. J. Mol. Sci. 22 (7), 3743. doi: 10.3390/ijms22073743
Valluri, J., Castillon, J., Newton, R., Soltes, E. (1989). Water stress-induced changes in protein synthesis of slash pine hypocotyls. J. Plant Physiol. 135 (3), 355–360. doi: 10.1016/S0176-1617(89)80132-4
Velasquez, A. C., Castroverde, C. D. M., He, S. Y. (2018). Plant-pathogen warfare under changing climate conditions. Curr. Biol. 28 (10), R619–R634. doi: 10.1016/j.cub.2018.03.054
Von Arnim, A. G., Jia, Q., Vaughn, J. N. (2014). Regulation of plant translation by upstream open reading frames. Plant Sci. 214, 1–12. doi: 10.1016/j.plantsci.2013.09.006
Waadt, R., Seller, C. A., Hsu, P. K., Takahashi, Y., Munemasa, S., Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23 (10), 680–694. doi: 10.1038/s41580-022-00479-6
Wang, W., Feng, B., Zhou, J. M., Tang, D. (2020). Plant immune signaling: Advancing on two frontiers. J. Integr. Plant Biol. 62 (1), 2–24. doi: 10.1111/jipb.12898
Wang, J., Zhang, X., Greene, G. H., Xu, G., Dong, X. (2022). PABP/purine-rich motif as an initiation module for cap-independent translation in pattern-triggered immunity. Cell 185 (17), 3186–3200. doi: 10.1016/j.cell.2022.06.037
Warner, J. R., McIntosh, K. B. (2009). How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34 (1), 3–11. doi: 10.1016/j.molcel.2009.03.006
Webster, C., Gaut, R. L., Browning, K., Ravel, J., Roberts, J. (1991). Hypoxia enhances phosphorylation of eukaryotic initiation factor 4A in maize root tips. J. Biol. Chem. 266 (34), 23341–23346. doi: 10.1016/s0021-9258(18)54502-6
Wu, Z., Huang, S., Zhang, X., Wu, D., Xia, S., Li, X. (2017). Regulation of plant immune receptor accumulation through translational repression by a glycine-tyrosine-phenylalanine (GYF) domain protein. Elife 6, e23684. doi: 10.7554/eLife.23684
Wullschleger, S., Loewith, R., Hall, M. N. (2006). TOR signaling in growth and metabolism. Cell 124 (3), 471–484. doi: 10.1016/j.cell.2006.01.016
Xing, S., Chen, K., Zhu, H., Zhang, R., Zhang, H., Li, B., et al. (2020). Fine-tuning sugar content in strawberry. Genome Biol. 21 (1), 230. doi: 10.1186/s13059-020-02146-5
Xu, G., Greene, G. H., Yoo, H., Liu, L., Marqués, J., Motley, J., et al. (2017a). Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545 (7655), 487–490. doi: 10.1038/nature22371
Xu, J., Yang, J. Y., Niu, Q. W., Chua, N. H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18 (12), 3386–3398. doi: 10.1105/tpc.106.047605
Xu, G., Yuan, M., Ai, C., Liu, L., Zhuang, E., Karapetyan, S., et al. (2017b). uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545 (7655), 491–494. doi: 10.1038/nature22372
Yang, M., Lu, Y., Piao, W., Jin, H. (2022). The translational regulation in mTOR pathway. Biomolecules 12 (6), 802. doi: 10.3390/biom12060802
Yang, Y., Wang, Z. (2019). IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 11 (10), 911–919. doi: 10.1093/jmcb/mjz091
Yangueez, E., Castro-Sanz, A. B., Fernandez-Bautista, N., Oliveros, J. C., Castellano, M. M. (2013). Analysis of genome-wide changes in the translatome of arabidopsis seedlings subjected to heat stress. PloS One 8 (8), e71425. doi: 10.1371/journal.pone.0071425
Yoo, H., Greene, G. H., Yuan, M., Xu, G., Burton, D., Liu, L., et al. (2020). Translational regulation of metabolic dynamics during effector-triggered immunity. Mol. Plant 13 (1), 88–98. doi: 10.1016/j.molp.2019.09.009
Young, S. K., Wek, R. C. (2016). Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J. Biol. Chem. 291 (33), 16927–16935. doi: 10.1074/jbc.R116.733899
Yu, X., Li, B., Jang, G. J., Jiang, S., Jiang, D., Jang, J. C., et al. (2019). Orchestration of processing body dynamics and mRNA decay in arabidopsis immunity. Cell Rep. 28 2194-2205 (8), e2196. doi: 10.1016/j.celrep.2019.07.054
Zhang, L., Liu, X., Gaikwad, K., Kou, X., Wang, F., Tian, X., et al. (2017). Mutations in eIF5B confer thermosensitive and pleiotropic phenotypes via translation defects in arabidopsis thaliana. Plant Cell 29 (8), 1952–1969. doi: 10.1105/tpc.16.00808
Zhang, H., Si, X., Ji, X., Fan, R., Liu, J., Chen, K., et al. (2018). Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36 (9), 894–898. doi: 10.1038/nbt.4202
Zhao, Y., Yang, X., Zhou, G., Zhang, T. (2020). Engineering plant virus resistance: From RNA silencing to genome editing strategies. Plant Biotechnol. J. 18 (2), 328–336. doi: 10.1111/pbi.13278
Zhu, W., Xu, J., Chen, S., Chen, J., Liang, Y., Zhang, C., et al. (2021). Large-Scale translatome profiling annotates the functional genome and reveals the key role of genic 3' untranslated regions in translatomic variation in plants. Plant Commun. 2 (4), 100181. doi: 10.1016/j.xplc.2021.100181
Keywords: abiotic stress, biotic stress, climate change, crop improvement, upstream open reading frame, translational reprogramming
Citation: Son S and Park SR (2023) Plant translational reprogramming for stress resilience. Front. Plant Sci. 14:1151587. doi: 10.3389/fpls.2023.1151587
Received: 26 January 2023; Accepted: 14 February 2023;
Published: 24 February 2023.
Edited by:
Palak Chaturvedi, University of Vienna, AustriaReviewed by:
Rémy Merret, UMR5096 Laboratoire Génome et développement des plantes, FranceRitesh Kumar, Boyce Thompson Institute (BTI), United States
Copyright © 2023 Son and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang Ryeol Park, c3JwYXJrQGtvcmVhLmty
 Seungmin Son
Seungmin Son Sang Ryeol Park
Sang Ryeol Park