- 1College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
- 2Zhejiang Provincial Key Laboratory for Genetic Improvement and Quality Control of Medicinal Plants, Hangzhou Normal University, Hangzhou, China
- 3Kharkiv Institute, Hangzhou Normal University, Hangzhou, China
- 4College of Pharmacy, Hangzhou Normal University, Hangzhou, China
Introduction: The anti-tumor vindoline and catharanthine alkaloids are naturally existed in Catharanthus roseus (C. roseus), an ornamental plant in many tropical countries. Plant-specific TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors play important roles in various plant developmental processes. However, the roles of C. roseus TCPs (CrTCPs) in terpenoid indole alkaloid (TIA) biosynthesis are largely unknown.
Methods: Here, a total of 15 CrTCP genes were identified in the newly updated C. roseus genome and were grouped into three major classes (P-type, C-type and CYC/TB1).
Results: Gene structure and protein motif analyses showed that CrTCPs have diverse intron-exon patterns and protein motif distributions. A number of stress responsive cis-elements were identified in promoter regions of CrTCPs. Expression analysis showed that three CrTCP genes (CrTCP2, CrTCP4, and CrTCP7) were expressed specifically in leaves and four CrTCP genes (CrTCP13, CrTCP8, CrTCP6, and CrTCP10) were expressed specifically in flowers. HPLC analysis showed that the contents of three classic TIAs, vindoline, catharanthine and ajmalicine, were significantly increased by ultraviolet-B (UV-B) and methyl jasmonate (MeJA) in leaves. By analyzing the expression patterns under UV-B radiation and MeJA application with qRT-PCR, a number of CrTCP and TIA biosynthesis-related genes were identified to be responsive to UV-B and MeJA treatments. Interestingly, two TCP binding elements (GGNCCCAC and GTGGNCCC) were identified in several TIA biosynthesis-related genes, suggesting that they were potential target genes of CrTCPs.
Discussion: These results suggest that CrTCPs are involved in the regulation of the biosynthesis of TIAs, and provide a basis for further functional identification of CrTCPs.
1 Introduction
The TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) family belongs to a type of plant-specific transcription factors (TFs) that played roles in diverse aspects of growth and developmental processes (Wang et al., 2022). The TCP family was named after its firstly discovered genes in maize (TEOSINTE BRANCHED1), Antirrhinum (CYCLOIDEA), and rice (PCF) (Doebley et al., 1997; Kosugi and Ohashi, 1997; Luo et al., 1999). Based on TCP domain, TCPs were grouped into TCP-P type (mainly PCF class) and C-type type consisting of the CIN class and the CYC/TB1 class (Navaud et al., 2007). The classic TCP domain contains a noncanonical basic helix–loop–helix motif, which is defined by structural feature and is unrelated to the DNA binding bHLH domain (Xu et al., 2014).
Increasing evidences suggested that TCP genes played roles in a wide range of growth- and development-related processes (Hao et al., 2012). In Arabidopsis, several TCP genes were found to be involved in microRNA-guided controlling leaf development (Palatnik et al., 2003). AtTCP2, AtTCP3, AtTCP11, and AtTCP15 were identified to bind to different components of the core circadian clock, such as LATE ELONGATED HYPOCOTYL1 and PSEUDO RESPONSE REGULATOR5 (Giraud et al., 2010). AtTCP20 acts as an activator of nitrate assimilatory gene, NIA1, and an inhibitor of the G2/M cell-cycle-related gene, CYCB1;1, suggesting an important role of TCPs in the linking of nitrate assimilation and cell-cycle (Guan et al., 2017). AtBRANCHED1 functions as a regulator of branching signals in bud outgrowth and development by interacting with FLOWERING LOCUS T (Niwa et al., 2013). In the TCP class II, one inhibitory gene (BRANCHED-like) and two activating genes (JAW-like and TCP5-like) are involved in axillary branch outgrowth (van Es et al., 2019). Interestingly, TCP family was considered to be involved in hormone signaling pathways. A TCP class I member, AtTCP8, modulates the expression of several brassinosteroid-responsive genes, such as BRASSINAZOLE-RESISTANT1 and BRASSINAZOLE-RESISTANT2 (Spears et al., 2022). A cotton TCP, GrTCP11, inhibits root hair elongation by reducing JA signaling pathway in heterologous expressed Arabidopsis system (Hao et al., 2021).
Recent studies revealed that TCP family played essential roles in secondary metabolism. In Ginkgo biloba, three TCP family members, such as GbTCP03, GbTCP04, and GbTCP07, were reportedly involved in flavonoid biosynthesis (Yu et al., 2022b). Heterologous overexpression of GbTCP5 gene in Arabidopsis increased the lignin biosynthesis by binding to TCP II type cis-acting elements (Wang et al., 2020). Marchantia polymorpha TCP1, a TCP-P type gene, enhanced the formation of secondary metabolites, such as aminochromes (Busch et al., 2019). In tea plant, CIN-type CsTCP3/4 and PCF-type CsTCP14 modulated the transactivation activity of anthocyanin synthase and anthocyanidin reductase by forming a CsTCP3-CsTT8 heterodimer (Yu et al., 2021).
Catharanthus roseus (L.) G. Don, a diploid plant native to Madagascar, is grown as an ornamental plant in many tropical countries (Nejat et al., 2015). C. roseus extracts have historically been applied to treat a wide range of diseases like diabetes, astringent, diuretic and cough remedy for many years (Nammi et al., 2003). Researchers identified the occurrence of vinblastine and vincristine in C. roseus, belonging to a specific type of terpenoid indole alkaloids (TIAs) that were used in cancer therapy (Kellner et al., 2015). Due to their very low yield, a variety of artificial induction methods have been employed to enrich the C. roseus alkaloids, including callus suspension culture, chemical elicitation and stress inducer (Zhao et al., 2001; Rizvi et al., 2016). It is interesting that elicitors enhanced the content of key alkaloids in C. roseus by altering the expression of TIA biosynthesis-related genes (Soltani et al., 2022). Several common elicitors, such as methyl jasmonate (MeJA), salicylic acid (SA), ultraviolet-B (UV-B), and other environmental stresses, have been applied in the accumulation of C. roseus active ingredients (Ramani and Chelliah, 2007; Goklany et al., 2013; Soltani et al., 2022). However, roles of CrTCP family in the regulation of TIA biosynthesis in C. roseus are largely unknown (Hao et al., 2021).
To date, a large number of TCP genes, such as 24 TCP members in Arabidopsis, 35 TCP members in Melastoma candidum, 14 TCP members in Cymbidium goeringii, 30 TCP members in Solanum lycopersicum, 33 TCP members in Populus euphratica, and 21 TCP members in rice, were identified (Navaud et al., 2007; Hao et al., 2012; Parapunova et al., 2014; Ma et al., 2016; Liu et al., 2019; Li et al., 2022). However, TCP gene family in C. roseus genome has not been identified, and its roles in development and metabolism are largely unknown. In this study, a detailed bioinformatic analysis of TCP family genes based on the newly published C. roseus genome and their expression patterns in different tissues and in responses to elicitors were carried out. Our research will help to understand the potential roles of CrTCPs in active ingredient biosynthesis and provide valuable information for the molecular breeding of C. roseus.
2 Materials and methods
2.1 Plant materials and growth condition
Catharanthus roseus seedlings were planted in a greenhouse in Hangzhou Normal University, Hangzhou, China, at a temperature of 25 ± 2°C with a light/dark cycle of 8/16 h and 65%–75% relative humidity. Forty-five-day-old seedlings in pots were used for the abiotic stress treatments.
2.2 Abiotic stress treatments
For UV-B treatments, plants were randomly separated into different groups with three pots each. One group was kept in standard growth condition and used as control. Three other groups were treated with UV-B radiation for 3 h, 6 h, and 24 h, respectively. Artificial UV-B at 10 μmol m−2s −1 was made by a fluorescent lamp (Q-Lab narrowband UV-B type), wrapped with a 3 mm transmission filter (Schott, Mainz, Germany), and measured by an Apogee UVB intensity meter (Yang et al., 2020). For MeJA treatments, one group of seedlings sprayed with ddH2O was used as control. MeJA at a concentration of 100 μM was sprayed onto the leaves of C. roseus seedlings for 1 h, 3 h, and 6 h, respectively (Zhan et al., 2018). For each treatment, three independent repetitions were applied.
2.3 Identification of CrTCP family genes
The protein sequences of TCPs in C. roseus were searched from the published genome (Kellner et al., 2015). The local BLATX program was used to identify potential CrTCP proteins using the ‘cro_v2.proteins.fasta’ file. Using the resulting protein IDs, the full-length CrTCP family gene sequences were extracted from the ‘cro_v2.transcripts.fasta’ file. All the potential CrTCP protein sequences were analyzed using InterPro program to check their domain features.
2.4 Phylogenetic tree, gene structure and protein domain analysis
The sequences of CrTCP proteins were used for phylogenetic analysis. A multiple sequence alignment was carried out using the full length sequences of CrTCP proteins by ClustalW software. The phylogenetic tree was constructed using CrTCP protein sequences by MEGA 6.1 with neighbor-joining method. The exon-intron pattern of CrTCP genes was identified using GSDS 2.0 online tool (http://gsds.gao-lab.org/). Bootstrap values of 1000 was applied to phylogeny test and P distance of protein sequence was used to analyze substitution rates. Pfam program in InterPro database (https://www.ebi.ac.uk/interpro/entry/pfam/#table) were used to identify motifs present in the CrTCP proteins. ‘PSORT WWW Server’ (https://psort.hgc.jp/) tool was used to predict the subcellular localization of CrTCP proteins.
2.5 Cis-element analysis
The 2000 bp promoter sequences of CrTCP genes were extracted from the C. roseus genome using ‘cro_v2_asm.fasta’ file. The promoters were uploaded and scanned using the PlantCARE program.
2.6 Measurement of terpenoid indole alkaloid contents
Collected leaf samples were dried at 60°C for 24 h and grinded in a mortar. The extraction of TIAs from C. roseus leaves was performed using methanol ultrasonic extraction method. C. roseus leaves were freeze dried and screened through 40 mesh sieve. About 100 mg dry samples were measured. After added with 1.5 mL of extraction agent, samples were ultrasonically extracted at 37 °C for 30 min. The supernatants were obtained by centrifugation at 12000 rpm for 10 min. The content of TIAs is determined based on the previous published HPLC method (Zhu et al., 2015). High performance liquid chromatographic conditions: the liquid to be tested is tested by HPLC (Waters e2695) instrument, and the chromatographic column is Waters C18 (4.6 mm × 250 mm); mobile phase: A: 0.025 M ammonium acetate solution; B: acetonitrile; gradient elution procedure: 0~5 min, 70% - 60% A; 5~8 min, 60% -55% A; 8~22 min, 55% -50% A; 22~27 min, 50% -30% A; 27~30 min, 30%-70% A; flow rate: 1.0 mL/min; detection wavelength: 220 nm. The HPLC chromatograms were showed in Supplemental File 1.
2.7 RNA isolation and quantitative real-time PCR
RNAs were isolated from each sample group using a plant TRIzol reagent (Invitrogen, Shanghai, China). RNA quantity and quality were analyzed using a NanoDrop ND-430 spectrophotometer (Agilent, Santa Clara, CA, USA). About 1.0 μg RNAs of each sample were reverse transcribed to cDNA library using ReverAid First Strand cDNA Synthesis Kit (Thermo Scientific, Shanghai, China). qRT-PCR assay (three independent repeats) was applied using a SYBR Premix Ex Taq Kit (TaKaRa, Dalian, China) on an ABI PRISM 7700 DNA Sequence Detection System (Applied Biosystems, Shanghai, China). All primers were listed in Table S1.
2.8 Statistical analysis
Significant difference in TIA contents between two treatment groups was analyzed by a one-way analysis of variance method with a student’s test at a P value < 0.01. Gene expression and chemical contents were analyzed using three independent biological repeats.
3 Results
3.1 Identification of TCP family members in C. roseus
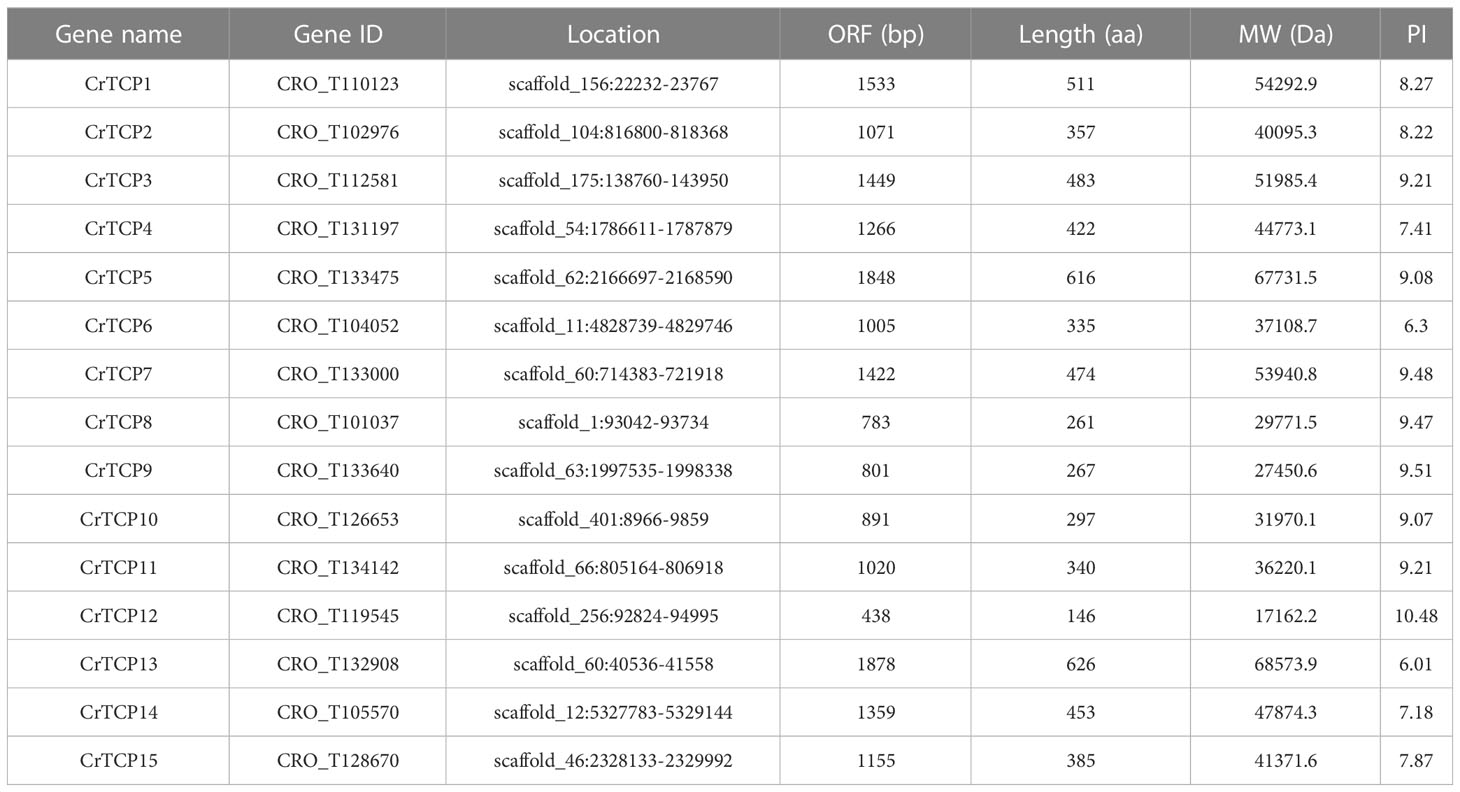
Fifteen potential CrTCP family genes were screened and identified in C. roseus based on its newly uploaded genome dataset. The open reading frame (ORF) sequence of the CrTCP genes range from 438 bp (CrTCP12) to 1878 bp (CrTCP13), the molecular weight of the proteins range from 17.2 kDa to 68.6 kDa, and the corresponding isoelectric points vary from 10.48 to 6.01. The grand average of CrTCP hydrophilic values is negative, suggesting that they are strong hydrophilic proteins. Subcellular location of all CrTCP proteins was predicted to be in the nucleus, suggesting that they function as TFs in the nucleus (Table 1).
3.2 Phylogenetic and amino acid sequence analysis
TCP proteins from C. roseus, model plant Arabidopsis thaliana and Oryza sativa, Medicago truncatula, Populus trichocarpa, and Ipomoea batatas were selected for phylogenetic tree construction. The phylogenetic analysis showed that all TCP family members could be divided into three main subclasses: TCP-P type, TCP-C type, and CYC/TB1 type. Seven CrTCPs (CrTCP3, CrTCP4, CrTCP9, CrTCP10, CrTCp11, CrTCP14, and CRTCP15) were in the TCP-P type subclass (PCF), six CrTCPs (CrTCP6, CrTCP8, CrTCP5, CrTCP1, CrTCP13, and CrTCP12) were in the TCP-C type subclass (CIN), and only two CrTCPs (CrTCP2 and CrTCP7) were in the CYC/TB1 subclass (Figure 1A).
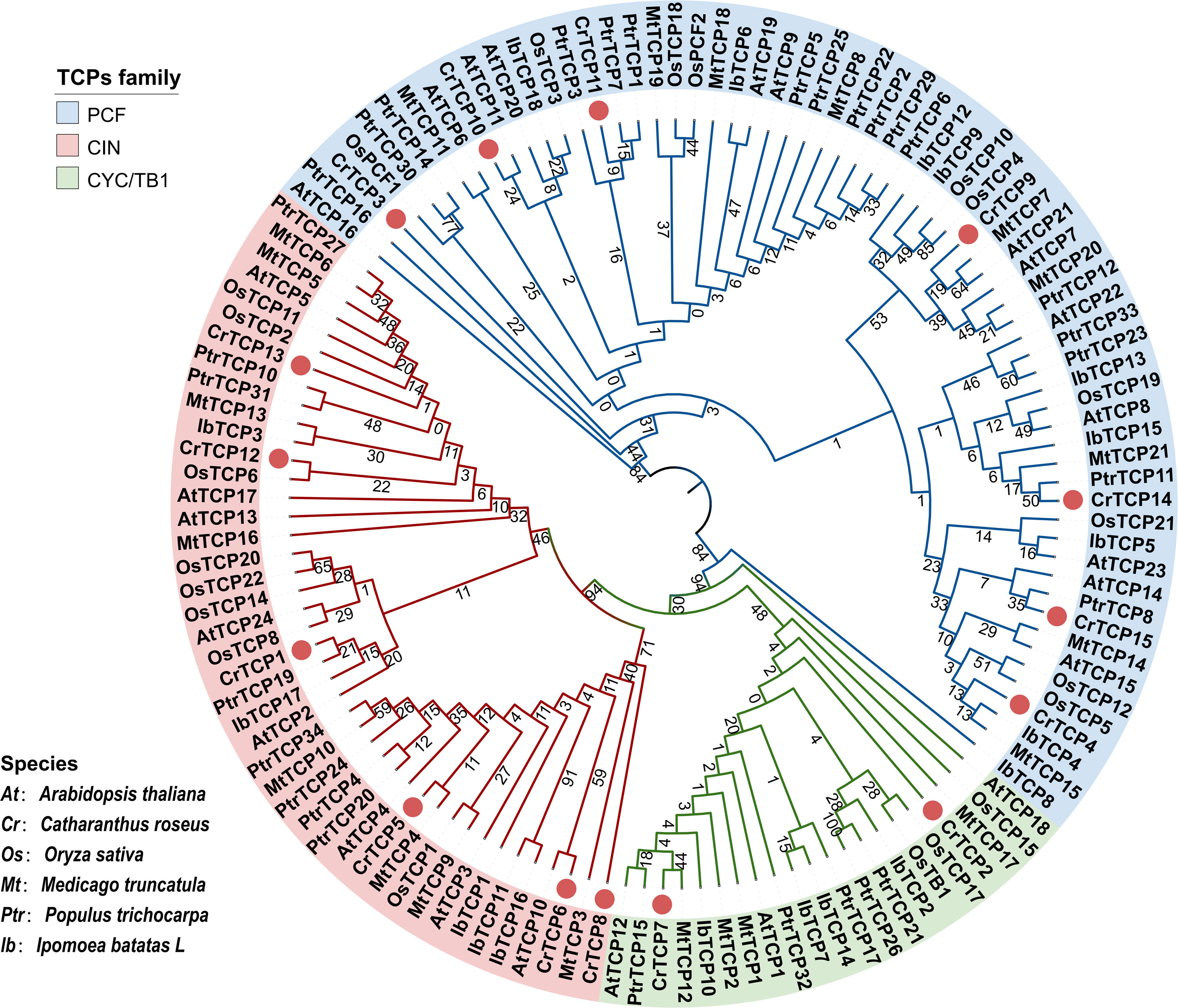
Figure 1 Phylogenetic analysis of CrTCP family members. Different colors of the outer ring denote the three main TCP groups. The blue arc represents PCF-type genes, the red arc represents CIN type, and the yellow arc represents CYC/TB1 type. ‘At’ indicated Arabidopsis thaliana, ‘Cr’ indicated Catharanthus roseus, ‘Os’ indicated Oryza sativa, ‘Mt’ indicated Medicago truncatula, ‘Ptr’ indicated Populus trichocarpa, and ‘Ib’ indicated Ipomoea batatas. Red dots indicated the CrTCPs.
The alignment analysis showed that CrTCPs possessed a series of basic amino acid residues, followed by two helix and one loop motifs, indicating that the TCP domain is highly conserved in plants (Figure S1A). The protein-protein network between all CrTCPs was predicted using STRING tool. Among the CrTCP proteins, eight CrTCPs, including CrTCP1, CrTCP2, CrTCP3, CrTCP4, CrTCP5, CrTCP10, CrTCP14, and CrTCP15, formed a protein-protein network (Figure S1B).
3.3 Gene structure and protein domain analysis of CrTCP family members
Gene structure analysis showed that only four CrTCP genes, such as CrTCP2, CrTCP3, CrTCP7, and CrTCP15, contain untranslated region annotation, indicating the weak assembling quality of C. roseus genome. Six CrTCP genes, CrTCP4, CrTCP8, CrTCP6, CrTCP10, CrTCP9, and CrTCP14, contain only one exon. CrTCP3 and CrTCP8 have the most complex gene structures with four and five introns, respectively (Figure 2A). The motifs of the CrTCP proteins were analyzed by MEME program, and five motifs were set as upper bounds. The number of motifs in CrTCPs range from one to five. Most CrTCPs contain motif 2 and 3 in the N-terminal (Figure 2B). The motif 2 and 3 corresponds to the TCP domain, and other motifs correspond to highly variable region.
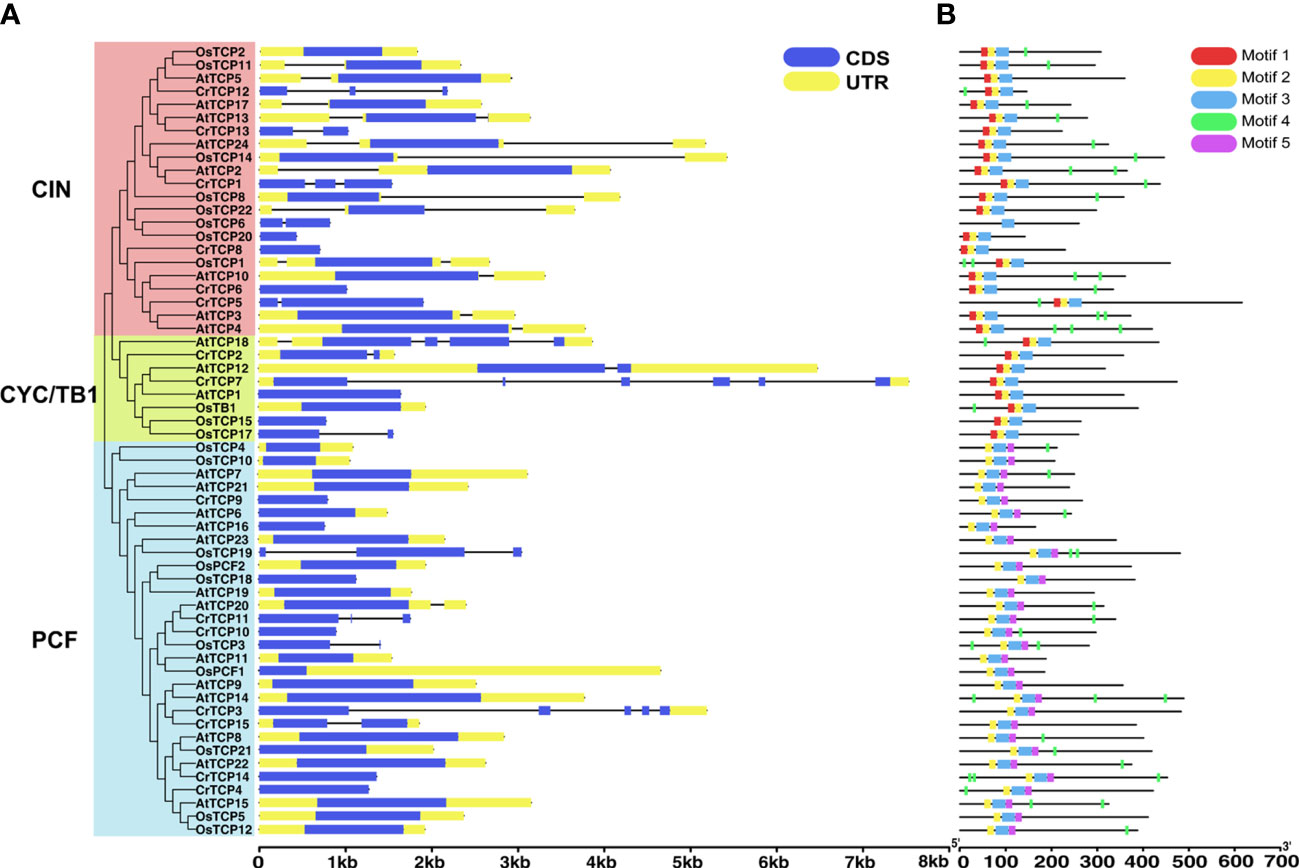
Figure 2 Gene structure and protein domain analysis of CrTCP family members. (A) Gene structure analysis of CrTCP family members. Green boxes indicate CDSs and yellow boxes indicated UTRs. All the TCPs were grouped into three classes according to their evolution relationship. (B) Rectangles with different colors represent different protein motifs.
3.4 Tissue-specific expression pattern and cis-element analysis
The tissue-specific expression of CrTCP gene family in different tissues, including flower, leaf, stem, and root, was analyzed by qRT-PCR. Generally, all CrTCP genes can be detected in at least one tissue. CrTCP13, CrTCP8, CrTCP6, and CrTCP10 specifically expressed in the flowers; CrTCP4, CrTCP2, and CrTCP7 highly expressed in the leaves; CrTCP11 mainly expressed in the stem; and CrTCP14 is a root specific expressed gene (Figure 3A).
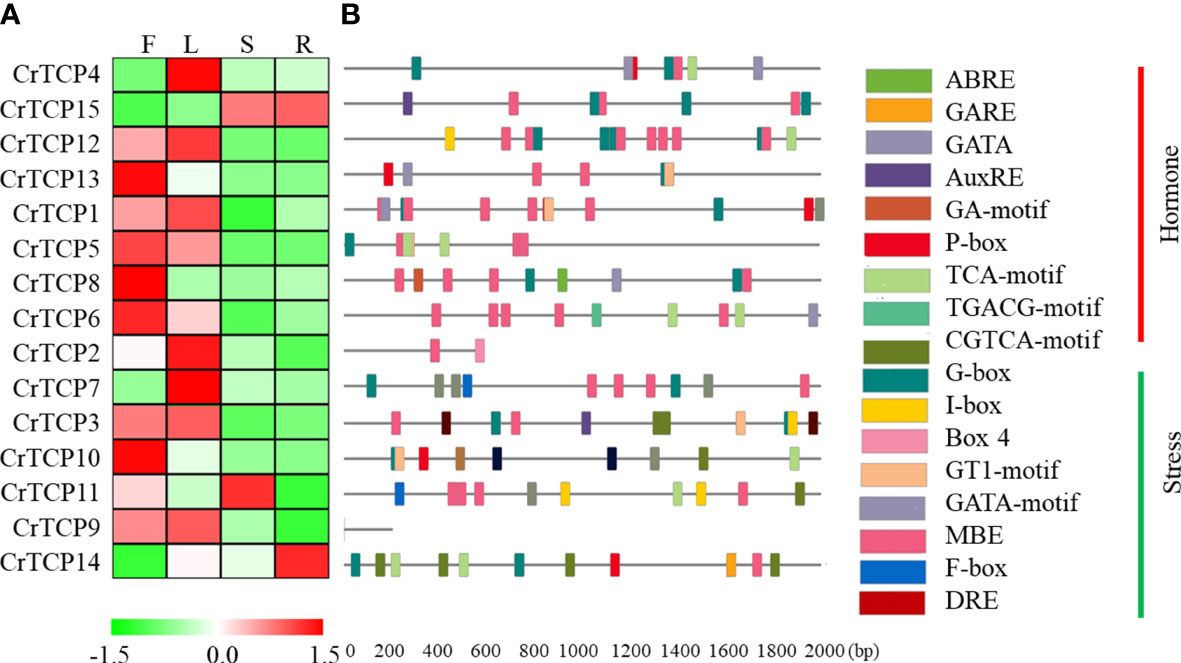
Figure 3 Tissue-specific expression pattern and cis-element analysis of CrTCP family members. (A) Expression analysis of CrTCP family genes in four different tissues by qRT-PCR assay. F: flowers, L: leaves, S: stem, and R: roots. Red indicated high expression level and green indicated low expression level. All the TCPs were grouped into different classes according to their evolution relationship. (B) Promoter analysis of CrTCP genes. Different color boxes on the black lines indicated various cis-elements. All cis-elements were grouped into two classes: hormone- and stress-related elements.
Cis-element analysis showed that the promoters of CrTCP genes had various plant specific binding elements, including nine hormone-related elements and eight stress-related elements. The MYB binding element (MBE) were significantly enriched in the promoter regions of CrTCP genes. For example, CrTCP12 (7 MBEs), CrTCP1 (5 MBEs), and CrTCP6 (5 MBEs) contained more than five MBEs in their promoter regions. The promoter sequences of several CrTCP genes contain several abiotic stress-related cis-elements. One GA-motif, one G-box, and two ABREs were found in the promoter region of CrTCP8 and three W-boxes and two G-boxes were contained in the promoter region of CrTCP7, suggesting their potential roles in abiotic stress responses (Figure 3B). Due to the low quality of C. roseus genome, the promoter sequences of CrTCP2 and CrTCP9 are temporarily unavailable. Furthermore, TGACG-motif and CGTCA-motif were cis-elements responsive to MeJA treatment. Two TGACG-motifs and one TGACG-motif were contained in the promoter sequences of CrTCP3 and CrTCP6, respectively. The CGTCA-motif was enriched in the promoter sequences of CrTCP3 and CrTCP14. Our data suggested that the expression of CrTCP3, CrTCP6 and CrTCP14 might be regulated by the MeJA treatments. I-box, GT1-motif, and GATA-motif were reported to be light response elements. The promoter of CrTCP4 contained two GATA-motifs and the promoter of CrTCP11 contained two I-boxes, suggesting that CrTCP4 and CrTCP11 might be light responsive TFs.
3.5 Expression pattern of CrTCP family genes under various elicitors
Expression pattern of CrTCP family genes under different abiotic stresses, such as the UV-B radiation and MeJA treatments, were analyzed. Under the UV-B radiation, many CrTCP genes, such as CrTCP2, CrTCP4, CrTCP5, CrTCP8, CrTCP9, CrTCP10, CrTCP12, and CrTCP13, and CrTCP15, were significantly reduced at different time points. The expression of CrTCP14 was significantly induced under 10 μmol·m-2·s-1 UV-B radiation at 3h and was significantly reduced at 24 h. The expression of CrTCP6 was significantly down-regulated at 6 h and was up-regulated at 24 h (Figure 4).
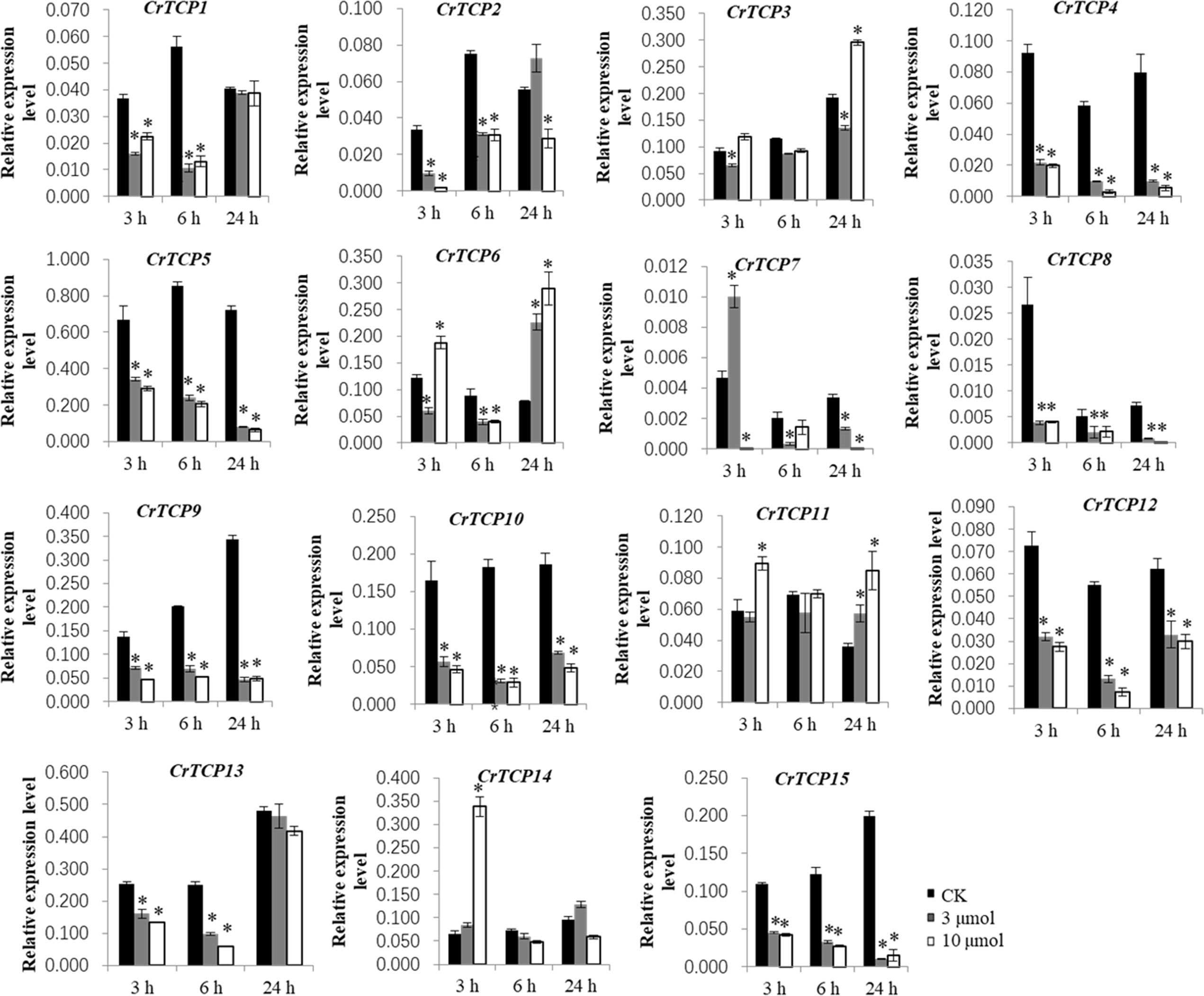
Figure 4 Expression analysis of CrTCP family genes under UV-B radiation. Black blocks indicated the control, grey blocks indicated 3 μmol UV-B treatment, and white blocks indicated 10 μmol UV-B treatment. The significant variations between treated sample and control (P < 0.05) are indicated by ‘*’. Error bars represent mean ± SD (n = 3).
Under the MeJA treatment, expression of CrTCP1, CrTCP2, CrTCP4, CrTCP7, and CrTCP14 was significantly down-regulated, while expression of CrTCP6 and CrTCP7 was significantly up-regulated under different time points. CrTCP8/11 were largely reduced at 3 h MeJA treatment and CrTCP6/13 were largely induced at 1 h MeJA treatment (Figure 5).
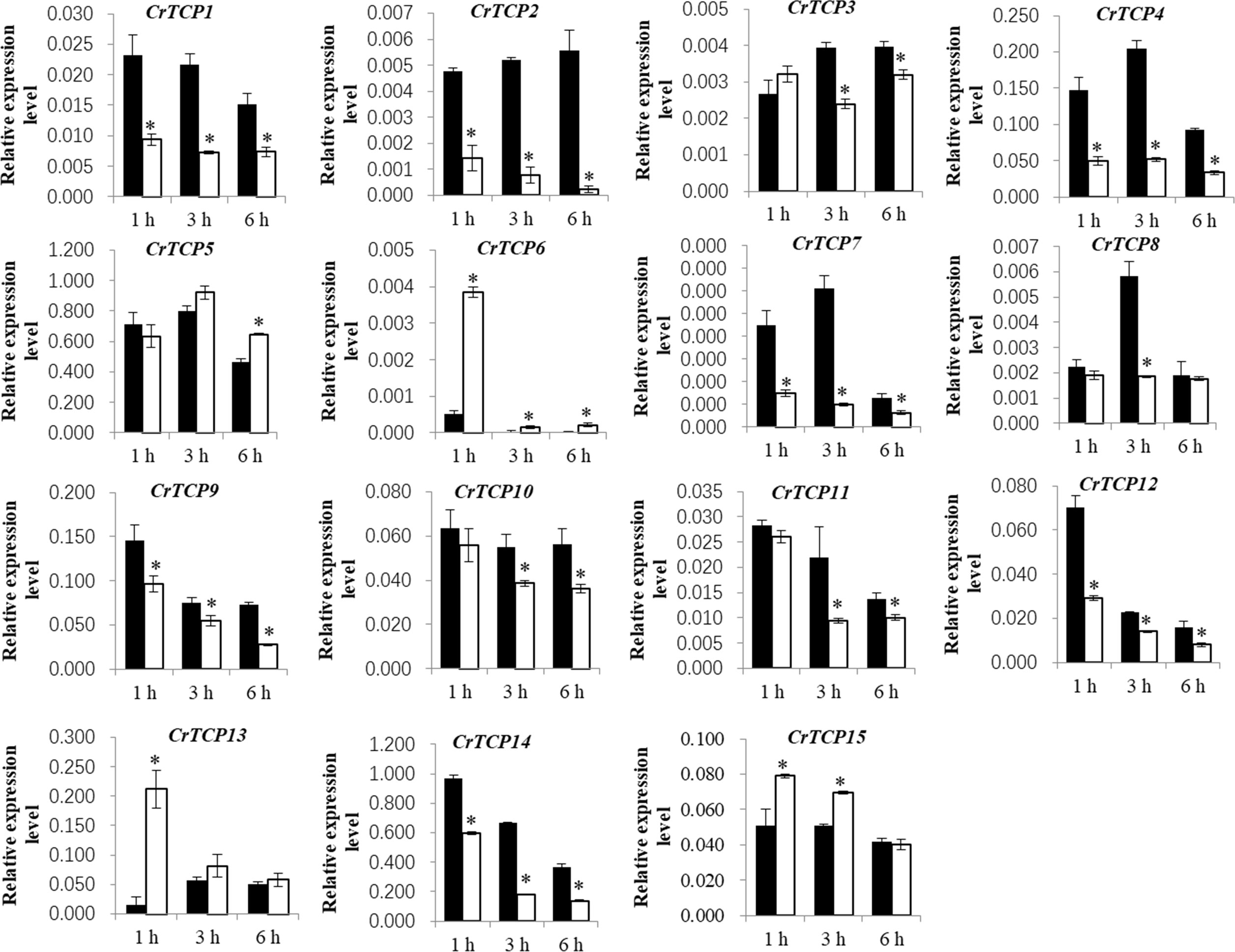
Figure 5 Expression analysis of CrTCP family genes under MeJA treatment. Black blocks indicated the control, and white blocks indicated 100 μmol MeJA treatment. The significant variations between treated sample and control (P < 0.05) are indicated by ‘*’. Error bars represent mean ± SD (n = 3).
Furthermore, the expression profile of CrTCP genes under MeJA and UV-B treatments was shown on a single heatmap. Clustering analysis showed that CrTCP6-CrTCP7, CrTCP3-CrTCP11, and CrTCP5-CrTCP10-CrTCP15 have similar expression patterns. In detail, the expression levels of CrTCP6 and CrTCP7 were significantly up-regulated by MeJA and 24 h-UV-B treatments. The expression levels of CrTCP5, CrTCP10 and CrTCP15 were highly up-regulated by the MeJA treatment and were greatly down-regulated by the UV-B treatments (Figure S2).
3.6 The TIA contents under various elicitors
To confirm the role of elicitors in TIA biosynthesis, the contents of three classic TIAs, such as vindoline, catharanthine, and ajmalicine, were determined. After 3 h, vindoline was increased from 2.39 mg.g-1 to 2.90 mg.g-1 and 2.76 mg.g-1 under 3 μmol·m-2·s-1 and 10 μmol·m-2·s-1 UV-B radiation, respectively. After 24 h UV-B radiation, vindoline was decreased from 2.75 mg·g-1 to 2.47 mg·g-1 under 10 μmol·m-2·s-1 UV-B radiation (Figure 6A). After 3 h, catharanthine was increased from 9.48 mg·g-1 to 13.11 mg·g-1 and 12.12 mg·g-1 under 3 μmol·m-2·s-1 and 10 μmol·m-2·s-1 UV-B radiation, respectively. After 24 h UV-B radiation, catharanthine was decreased from 12.24 mg.g-1 to 10.17 mg.g-1 under 10 μmol·m-2·s-1 UV-B treatment (Figure 6B). At time point 3 h, the contents of ajmalicine were induced from 0.180 mg.g-1 to 0.27 mg.g-1 and 0.27 mg.g-1 under 3 μmol·m-2·s-1 and 10 μmol·m-2·s-1 UV-B radiation, respectively. At time point 24 h, the contents of ajmalicine were up-regulated from 0.23 mg.g-1 to 0.42 mg.g-1 and 0.37 mg.g-1 under 3 μmol·m-2·s-1 and 10 μmol·m-2·s-1 UV-B treatment, respectively (Figure 6C).
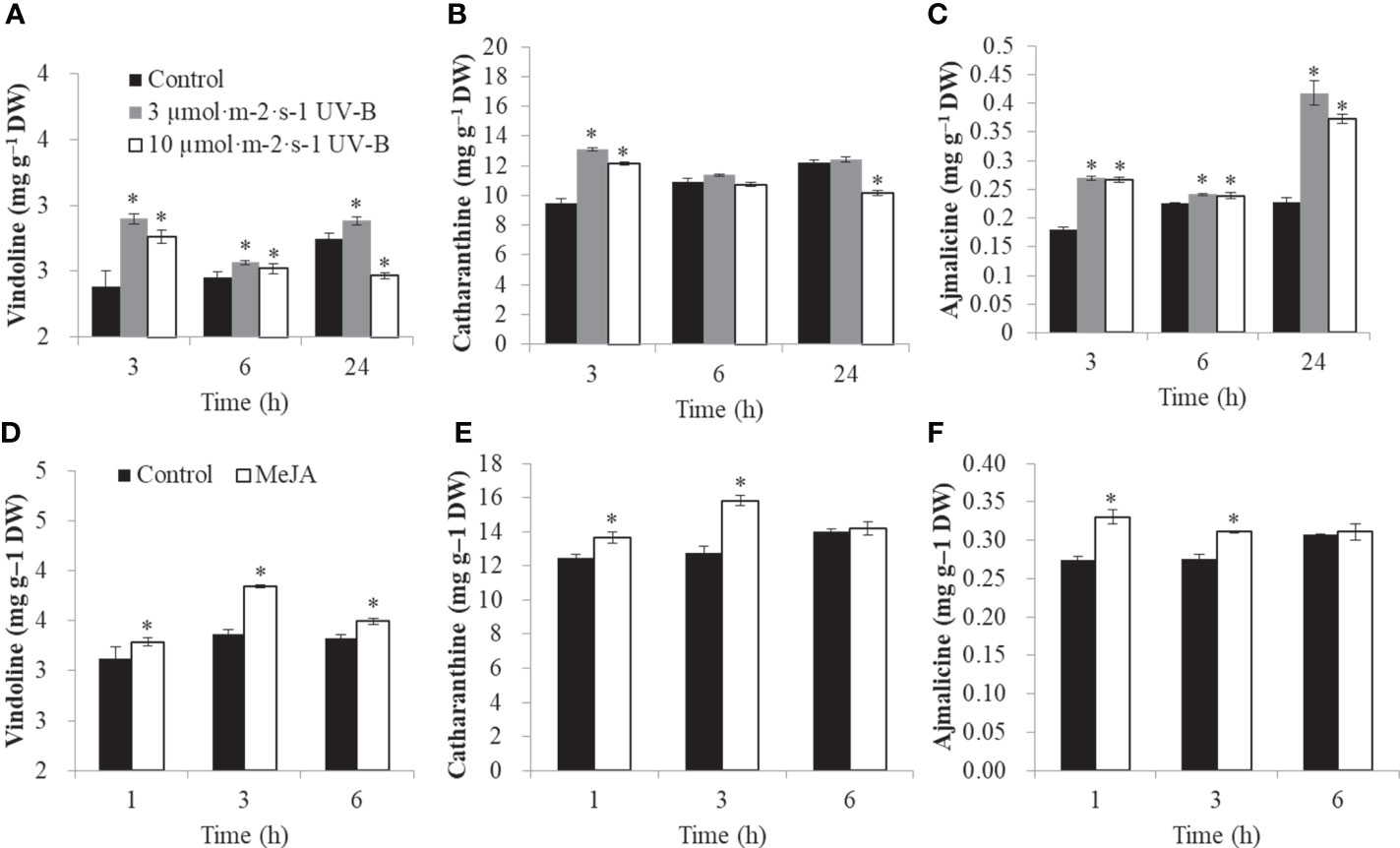
Figure 6 The TIA contents under various elicitors (UV-B and MeJA). UV-B radiation changed the contents of vindoline (A), catharanthine (B), ajmalicine (C). MeJA treatment changed the contents of vindoline (D), catharanthine (E), ajmalicine (F). The significant variations between treated sample and control (P < 0.05) are indicated by ‘*’. Error bars represent mean ± SD (n = 3).
Under MeJA treatment, vindoline was significantly up-regulated from 3.36 mg.g-1 to 3.85 mg·g-1 at 3 h (Figure 6D). Catharanthine was significantly up-regulated from 12.48 mg·g-1 to 13.67 mg.g-1 and 12.78 mg·g-1 to 15.83 mg·g-1 at 1 h and 3 h, respectively (Figure 6E). Ajmalicine was significantly induced from 0.27 mg·g-1 to 0.33 mg·g-1 and 0.28 mg·g-1 to 0.31 mg·g-1 at 1 h and 3 h, respectively (Figure 6F).
3.7 The expression levels of several TIA biosynthesis-related genes under various elicitors
Expression pattern of several TIA pathway genes under different abiotic stresses, such as the UV-B radiation and MeJA treatments, were analyzed. Our data showed that CrASα, CrTDC, and CrCPR were significantly increased and CrDXR, CrDXS, CrG10H, CrDAT, and CrPRX1 were significantly reduced under UV-B treatments (Figure S3). Under MeJA treatment, CrASa, CrSTR, CrSGD, CrDXS, CrLAMT and CrTDC were significantly up-regulated at different time points. Only CrD4H was significantly down-regulated by UV-B treatment at all three time points (Figure S4).
3.8 Prediction of potential targets of CrTCP family TFs
The consensus DNA binding sequences of TCP family TFs are two overlapping elements: GGNCCCAC and GTGGNCCC (Kosugi and Ohashi, 2002). The 2000 bp promoter regions of five JA signaling-related TF genes (CrMYC1, CrMYC2, CrORCA3, CrMPK3 and CrJAZ1), three UV-B signaling-related TF genes (CrHY5, CrUVR8 and CrCOP1), and ten TIA biosynthesis-related genes (CrASα, CrTDC, CrDXR, CrDXS, CrCPR, CrG10H, CrIS, CrIO, CrSLS, CrLAMT, CrSTR, CrSGD, CrPRX, CrD4H and CrDAT) were scanned for the TCP binding elements (TBEs) (Figure 7A). All selected TF promoters contain at least one TBS. The promoters of CrMYC2 and CrHY5 contain four TBEs. For the TIA biosynthesis-related genes, the promoters of CrASα, CrDXR, CrCPR, CrIS, CrIO, CrSTR and CrSGD contain at least one TBE (Figure 7B).
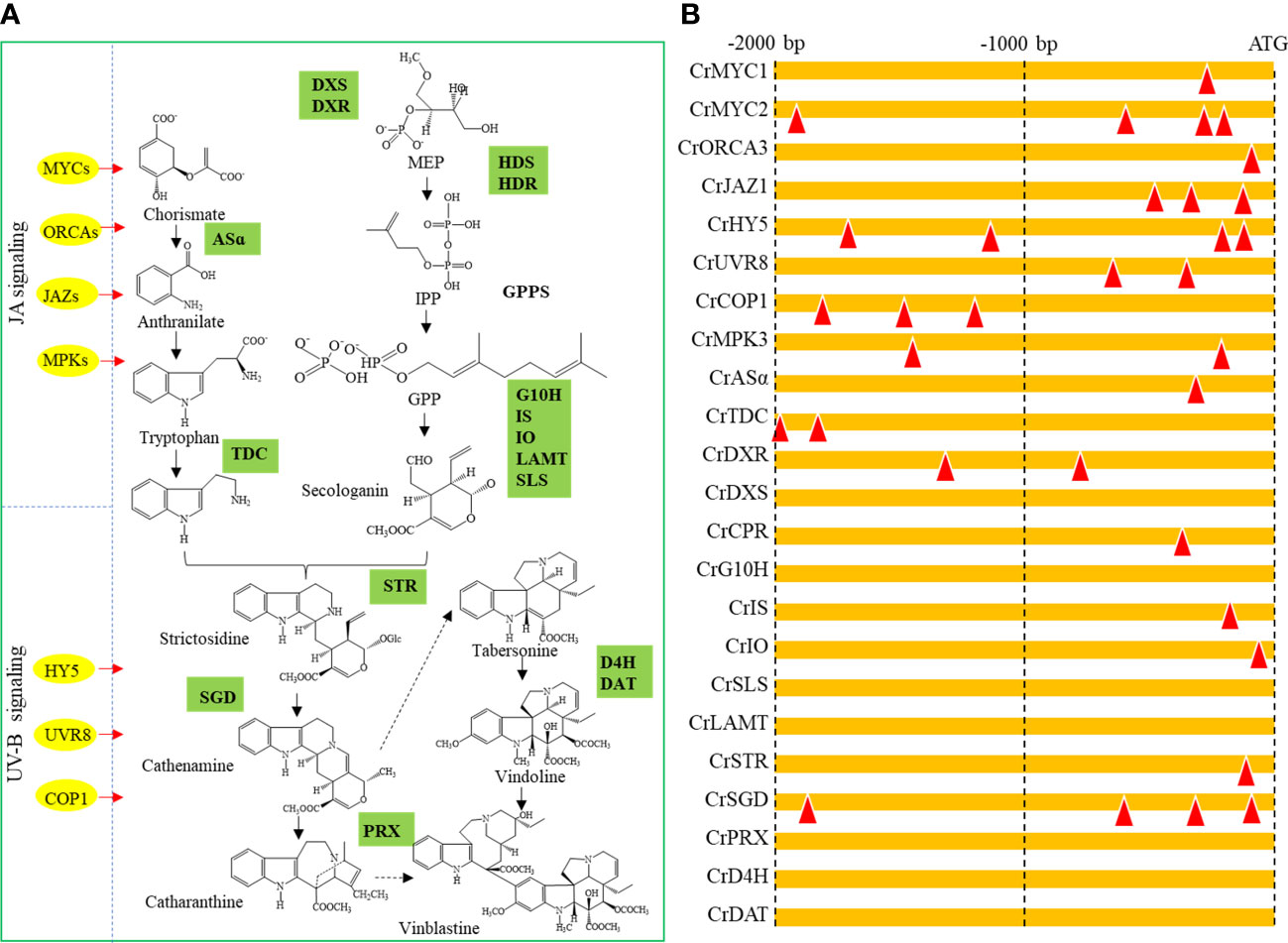
Figure 7 Prediction of potential targets of CrTCP family TFs. (A) Overview of the regulation diagram of C. roseus TIA biosynthesis. Yellow dots indicated upstream TFs, and green boxes indicated TIA biosynthesis-related genes. (B) Prediction of TCP binding element in the promoters of potential TCP downstream targets. Red triangles indicated potential TCP TF binding elements.
4 Discussion
C. roseus is a natural medicinal plant that has been used since ancient times (Kumar et al., 2022). Different fractions of C. roseus extract have been proved to have various pharmacological effects, such as anti-microbial, anti-oxidant, and anti-cancer (Das et al., 2020). Due to the extremely low contents, active ingredients from C. roseus are expressive and have great demand in pharmaceutical market (Yamamoto et al., 2016). The significance of C. roseus in a modern medical system makes the genetic breeding being an important research field.
TCP TFs are a common family with redundant functions in plants (Zhou et al., 2022). With release of the scaffolded genome of C. roseus, it is necessary to annotate the TCP genes at the whole-genome level. Recently, an improved C. roseus genome assembly composed of 2090 scaffolds with an N50 scaffold size of 2,579,454 bp was published, giving the possibility of further functional researches (Franke et al., 2019). Based on the newly released C. roseus genome, 15 TCP family members were identified, which is less than numbers of TCP family members in model plants (Navaud et al., 2007; Li, 2015). Limited to the quality of genome sequencing and assembly, not all TCP members have been identified. Once the high-quality genome at the chromosome level is available, deep analysis of TCP family can be further carried out.
Analysis of gene evolution relationship is helpful to predict gene function. AtTCP11, a class I TCP, is a developmental regulator involved in leaf, stem, petiole, and pollen development (Viola et al., 2011). CrTCP3, a homologous protein of AtTCP11, might be also a multifunctional growth regulator. In Arabidopsis, CIN-like AtTCP13 is another plant growth regulator under dehydration stresses (Urano et al., 2022). CrTCP13 is a close homologous protein of AtTCP13, indicating potential function of CrTCP13 in stress responses. Recently, AtTCP20 was reported to be involved in maintaining auxin homeostasis by modulating the expression of GH3.3 gene (Kong et al., 2022). CrTCP11 showed a close relationship to AtTCP13, indicating that CrTCP13 might be a regulator in hormone signaling pathways.
A large number of active ingredients are enriched in the leaves and flowers of C. roseus. The methanolic extracts of C. roseus leaves have incredible contributions to the field of medicinal applications. (Rajashekara et al., 2022). C. roseus flower extracts have a significant antifungal property and are used as antibacterial agents to combat different infections (Neglo et al., 2022). Tissue-specific expression analysis revealed that CrTCP2, CrTCP4, and CrTCP7 genes might be involved in leaf-specific accumulation of active ingredients and CrTCP8, CrTCP10, and CrTCP13 genes might be involved in flower-specific accumulation of active ingredients.
TFs regulated gene expression at the transcriptional level by binding to different cis-acting elements (Yu et al., 2022a; Zhan et al., 2022; Feng et al., 2023). A series of regulatory elements have been identified in the promoter region of TCP genes, including growth-related elements, stress-responsive elements, and hormone signaling-related elements (Feng et al., 2018). Due to the presence of different types of stress cis-elements in promoter regions, CrTCP12, CrTCP8, CrTCP7, CrTCP3, CrTCP10, and CrTCP11 might be associated with multiple stress responses. Several active ingredients play important roles in the adaptation of plants to the adverse environments, and some environmental stresses can be used as elicitors (Ramakrishna and Ravishankar, 2011; Hao et al., 2022). In this study, the positive roles of UV-B radiation and MeJA application in the induction of TIA contents were confirmed. However, the roles of TCP members in UV-B- and MeJA-mediated TIA biosynthesis are largely unknown. In Ginkgo biloba, the expression of GbTCP06, GbTCP11, and GbTCP13 was significantly up-regulated by exogenous of MeJA application (Yu et al., 2022b). In Chinese flowering cabbage, BrTCP7 is associated with MeJA-mediated leaf senescence by activating JA signaling pathway and chlorophyll metabolism (Xu et al., 2019). In C. roseus, several UV-B-inhibited CrTCPs, such as CrTCP4, CrTCP5, CrTCP8, CrTCP9, CrTCP10, CrTCP12, and CrTCP15, and MeJA-inhibited CrTCPs, such as CrTCP1, CrTCP2, CrTCP4, CrTCP7, and CrTCP14, were identified by qRT-PCR assay, suggesting that they might be negative regulators in the stress-induced biosynthesis of TIAs.
In C. roseus, a series of JA-responsive TF genes, such as CrMYC2 and ORCA3, were reportedly involved in the TIA biosynthesis (Paul et al., 2017). Previous studies revealed that the TCP proteins tend to form a homodimer or a heterodimer, and two variable and overlapping consensus sequences, GGNCCCAC and GTGGNCCC, were identified to be TBEs (Kosugi and Ohashi, 2002). In our study, the promoters of CrMYC2 and CrORCA3 contained four and one TBEs, respectively, suggesting that they were potential downstream targets of JA-responsive TCPs. COP1, HY5, and UVR8 are important regulators in UV-B signaling pathway (Zhang et al., 2022). The promoters of CrUVR8 and CrCOP1 contained two and three TBEs, respectively, suggesting that they were potential downstream targets of UV-B-responsive TCPs. Additionally, promoter analysis also showed that CrTCPs might be involved in TIA biosynthesis by directly binding to the promoters of CrASα, CrTDC, CrDXR, CrCPR, CrIS, CrIO, CrSTR, and CrSGD. To confirmed the roles of CrTCPs in the regulation of TIA pathway, the expression levels of several key TIA pathway genes were determined using qRT-PCR assay. As part of the iridoid pathway, CPR catalyzes the conversion of geraniol to 10-hydroxygeraniol (Miettinen et al., 2014). Tryptamine, an important precursor of TIS biosynthesis, is catalyzed from tryptophan by TDC (Goddijn et al., 1994). In our study, the expression of TDC and CPR genes was significantly up-regulated by the UV-B treatment. Considering that TDC and CPR are potential targets of TCP TFs, it showed that TCPs might plat important roles in UV-B-mediated TIA accumulation in C. roseus. The joining of secologanin and tryptamine is the key step to synthesize strictosidine, which is catalyzed by STR (Li and Gibson, 2021). Finally, SGD hydrolyses the strictosidine glucose to form a dialdehyde for Cathenamine biosynthesis (van Der Heijden et al., 2004). Under MeJA treatment, the expression of CrSTR and CrSGD were increased. Interestingly, the promoter of CrSTR contained one TBE, and the promoter of CrSGD contained four TBEs, suggesting that TCPs played a potential role in MeJA-induced TIA biosynthesis.
5 Conclusion
In the present study, a total of 15 CrTCPs were identified from in the genome of C. roseus. All CrTCP proteins were divided into three subgroups, P-type, C-type, and CYC/TB1, according to their conserved TCP domain. Analyses of gene structures, conserved motifs, and cis-elements was performed to get insights into TCP genes in C. roseus. The expression profiling of CrTCP family members was appraised in different tissues and under two elicitors. Three CrTCP genes, including CrTCP2, CrTCP4, and CrTCP7, were highly expressed in the leaves, and four CrTCP genes, including CrTCP13, CrTCP8, CrTCP6, and CrTCP10, were greatly expressed in the flowers. Several MeJA- and UV-B- responsive TCP genes were identified. Promoter analysis showed that MeJA responsive TFs (CrMYC2 and CrORCA3), UV-B radiation responsive TFs (CrUVR8 and CrCOP1), and TIA biosynthesis-related genes (CrASα, CrTDC, CrDXR, CrCPR, CrIS, CrIO, CrSTR, and CrSGD) might be potential downstream target of CrTCPs. Furthermore, the expression patterns of TIA biosynthesis-related genes under UV-B and MeJA treatments were obtained. Overall, our results provided actual data and theoretical basis for the functional identification of TCPs in C. roseus.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
JH, HQZ and CS conceptualized the initial study. CS, HQZ, JH, LZ, YH, HSZ, HK, X.L., ZTW and CC were involved in the experimental layout. JH, HK, CC, JQ, ZL and ZJW performed the lab experiments, HQZ and CS drafted the initial article. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by National Natural Science Foundation of China (31601343 and 32271905); Natural Science Foundation of Zhejiang Province under Grant No. LY18C050005, LY19C150005, LY19C160001, and LY23C160001; Opening Project of Zhejiang Provincial Key Laboratory of Forest Aromatic Plants-based Healthcare Functions (2022E10008); Open Foundation of State Key Laboratory of Subtropical Silviculture, Zhejiang A & F University (KF201708); Major Increase or Decrease Program in The Central Finance Level (Grant No. 2060302); and Zhejiang Provincial key Research and Development Project Grants (2017C02011, 2018C02030).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1161534/full#supplementary-material
References
Busch, A., Deckena, M., Almeida-Trapp, M., Kopischke, S., Kock, C., Schussler, E., et al. (2019). MpTCP1 controls cell proliferation and redox processes in marchantia polymorpha. New Phytol. 224 (4), 1627–1641. doi: 10.1111/nph.16132
Das, A., Sarkar, S., Bhattacharyya, S., Gantait, S. (2020). Biotechnological advancements in catharanthus roseus (L.) g. don. Appl. Microbiol. Biotechnol. 104 (11), 4811–4835. doi: 10.1007/s00253-020-10592-1
Doebley, J., Stec, A., Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386 (6624), 485–488. doi: 10.1038/386485a0
Feng, S., Kailin, H., Zhang, H., Chen, C., Huang, J., Wu, Q., et al. (2023). Investigation of the role of TmMYB16/123 and their targets (TmMTP1/11) in the tolerance of taxus media to cadmium. Tree Physiol. doi: 10.1093/treephys/tpad019
Feng, Z. J., Xu, S. C., Liu, N., Zhang, G. W., Hu, Q. Z., Gong, Y. M. (2018). Soybean TCP transcription factors: Evolution, classification, protein interaction and stress and hormone responsiveness. Plant Physiol. Biochem. 127, 129–142. doi: 10.1016/j.plaphy.2018.03.020
Franke, J., Kim, J., Hamilton, J. P., Zhao, D., Pham, G. M., Wiegert-Rininger, K., et al. (2019). Gene discovery in gelsemium highlights conserved gene clusters in monoterpene indole alkaloid biosynthesis. Chembiochem 20 (1), 83–87. doi: 10.1002/cbic.201800592
Giraud, E., Ng, S., Carrie, C., Duncan, O., Low, J., Lee, C. P., et al. (2010). TCP Transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in arabidopsis thaliana. Plant Cell 22 (12), 3921–3934. doi: 10.1105/tpc.110.074518
Goddijn, O. J., Lohman, F. P., de Kam, R. J., Schilperoort, R. A., Hoge, J. H. (1994). Nucleotide sequence of the tryptophan decarboxylase gene of catharanthus roseus and expression of tdc-gusA gene fusions in nicotiana tabacum. Mol. Gen. Genet. 242 (2), 217–225. doi: 10.1007/BF00391016
Goklany, S., Rizvi, N. F., Loring, R. H., Cram, E. J., Lee-Parsons, C. W. (2013). Jasmonate-dependent alkaloid biosynthesis in catharanthus roseus hairy root cultures is correlated with the relative expression of orca and zct transcription factors. Biotechnol. Prog. 29 (6), 1367–1376. doi: 10.1002/btpr.1801
Guan, P., Ripoll, J. J., Wang, R., Vuong, L., Bailey-Steinitz, L. J., Ye, D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U.S.A. 114 (9), 2419–2424. doi: 10.1073/pnas.1615676114
Hao, J., Lou, P., Han, Y., Chen, Z., Chen, J., Ni, J., et al. (2021). GrTCP11, a cotton TCP transcription factor, inhibits root hair elongation by down-regulating jasmonic acid pathway in arabidopsis thaliana. Front. Plant Sci. 12, 769675. doi: 10.3389/fpls.2021.769675
Hao, J., Lou, P., Han, Y., Zheng, L., Lu, J., Chen, Z., et al. (2022). Ultraviolet-b irradiation increases antioxidant capacity of pakchoi (Brassica rapa l.) by inducing flavonoid biosynthesis. Plants (Basel) 11 (6), 766. doi: 10.3390/plants11060766
Hao, J., Tu, L., Hu, H., Tan, J., Deng, F., Tang, W., et al. (2012). GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63 (17), 6267–6281. doi: 10.1093/jxb/ers278
Kellner, F., Kim, J., Clavijo, B. J., Hamilton, J. P., Childs, K. L., Vaillancourt, B., et al. (2015). Genome-guided investigation of plant natural product biosynthesis. Plant J. 82 (4), 680–692. doi: 10.1111/tpj.12827
Kong, Q., Low, P. M., Lim, A. R. Q., Yang, Y., Yuan, L., Ma, W. (2022). Functional antagonism of WRI1 and TCP20 modulates GH3.3 expression to maintain auxin homeostasis in roots. Plants (Basel) 11 (3), 454. doi: 10.3390/plants11030454
Kosugi, S., Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9 (9), 1607–1619. doi: 10.1105/tpc.9.9.1607
Kosugi, S., Ohashi, Y. (2002). DNA Binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30 (3), 337–348. doi: 10.1046/j.1365-313x.2002.01294.x
Kumar, S., Singh, B., Singh, R. (2022). Catharanthus roseus (L.) g. don: A review of its ethnobotany, phytochemistry, ethnopharmacology and toxicities. J. Ethnopharmacol. 284, 114647. doi: 10.1016/j.jep.2021.114647
Li, S. (2015). The arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal Behav. 10 (7), e1044192. doi: 10.1080/15592324.2015.1044192
Li, C. Y., Gibson, S. I. (2021). Repression of ZCT1, ZCT2 and ZCT3 affects expression of terpenoid indole alkaloid biosynthetic and regulatory genes. PeerJ 9, e11624. doi: 10.7717/peerj.11624
Li, H., Wen, X., Huang, X., Wei, M., Chen, H., Yu, Y., et al. (2022). Genome-wide identification and characterization of TCP gene family members in melastoma candidum. Molecules 27 (24), 9036 doi: 10.3390/molecules27249036
Liu, M. M., Wang, M. M., Yang, J., Wen, J., Guo, P. C., Wu, Y. W., et al. (2019). Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. Int. J. Mol. Sci. 20 (14), 3591. doi: 10.3390/ijms20143591
Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J., Coen, E. (1999). Control of organ asymmetry in flowers of antirrhinum. Cell 99 (4), 367–376. doi: 10.1016/s0092-8674(00)81523-8
Ma, X., Ma, J., Fan, D., Li, C., Jiang, Y., Luo, K. (2016). Genome-wide identification of TCP family transcription factors from populus euphratica and their involvement in leaf shape regulation. Sci. Rep. 6, 32795. doi: 10.1038/srep32795
Miettinen, K., Dong, L., Navrot, N., Schneider, T., Burlat, V., Pollier, J., et al. (2014). The seco-iridoid pathway from catharanthus roseus. Nat. Commun. 5, 3606. doi: 10.1038/ncomms4606
Nammi, S., Boini, M. K., Lodagala, S. D., Behara, R. B. (2003). The juice of fresh leaves of catharanthus roseus linn. reduces blood glucose in normal and alloxan diabetic rabbits. BMC Complement Altern. Med. 3, 4. doi: 10.1186/1472-6882-3-4
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., Herve, C. (2007). TCP Transcription factors predate the emergence of land plants. J. Mol. Evol. 65 (1), 23–33. doi: 10.1007/s00239-006-0174-z
Neglo, D., Adzaho, F., Agbo, I. A., Arthur, R., Sedohia, D., Tettey, C. O., et al. (2022). Antibiofilm activity of azadirachta indica and catharanthus roseus and their synergistic effects in combination with antimicrobial agents against fluconazole-resistant candida albicans strains and MRSA. Evid Based Complement Alternat Med. 2022, 9373524. doi: 10.1155/2022/9373524
Nejat, N., Valdiani, A., Cahill, D., Tan, Y. H., Maziah, M., Abiri, R. (2015). Ornamental exterior versus therapeutic interior of Madagascar periwinkle (Catharanthus roseus): the two faces of a versatile herb. ScientificWorldJournal 2015, 982412. doi: 10.1155/2015/982412
Niwa, M., Daimon, Y., Kurotani, K., Higo, A., Pruneda-Paz, J. L., Breton, G., et al. (2013). BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in arabidopsis. Plant Cell 25 (4), 1228–1242. doi: 10.1105/tpc.112.109090
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 (6955), 257–263. doi: 10.1038/nature01958
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14, 157. doi: 10.1186/1471-2229-14-157
Paul, P., Singh, S. K., Patra, B., Sui, X., Pattanaik, S., Yuan, L. (2017). A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in catharanthus roseus. New Phytol. 213 (3), 1107–1123. doi: 10.1111/nph.14252
Rajashekara, S., Reena, D., Mainavi, M. V., Sandhya, L. S., Baro, U. (2022). Biological isolation and characterization of catharanthus roseus (L.) g. don methanolic leaves extracts and their assessment for antimicrobial, cytotoxic, and apoptotic activities. BMC Complement Med. Ther. 22 (1), 328. doi: 10.1186/s12906-022-03810-y
Ramakrishna, A., Ravishankar, G. A. (2011). Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav. 6 (11), 1720–1731. doi: 10.4161/psb.6.11.17613
Ramani, S., Chelliah, J. (2007). UV-B-induced signaling events leading to enhanced-production of catharanthine in catharanthus roseus cell suspension cultures. BMC Plant Biol. 7, 61. doi: 10.1186/1471-2229-7-61
Rizvi, N. F., Weaver, J. D., Cram, E. J., Lee-Parsons, C. W. (2016). Silencing the transcriptional repressor, ZCT1, illustrates the tight regulation of terpenoid indole alkaloid biosynthesis in catharanthus roseus hairy roots. PloS One 11 (7), e0159712. doi: 10.1371/journal.pone.0159712
Soltani, N., Firouzabadi, F. N., Shafeinia, A., Shirali, M., Sadr, A. S. (2022). De novo transcriptome assembly and differential expression analysis of catharanthus roseus in response to salicylic acid. Sci. Rep. 12 (1), 17803. doi: 10.1038/s41598-022-20314-4
Spears, B. J., McInturf, S. A., Collins, C., Chlebowski, M., Cseke, L. J., Su, J., et al. (2022). Class I TCP transcription factor AtTCP8 modulates key brassinosteroid-responsive genes. Plant Physiol. 190 (2), 1457–1473. doi: 10.1093/plphys/kiac332
Urano, K., Maruyama, K., Koyama, T., Gonzalez, N., Inze, D., Yamaguchi-Shinozaki, K., et al. (2022). CIN-like TCP13 is essential for plant growth regulation under dehydration stress. Plant Mol. Biol. 108 (3), 257–275. doi: 10.1007/s11103-021-01238-5
van Der Heijden, R., Jacobs, D. I., Snoeijer, W., Hallard, D., Verpoorte, R. (2004). The catharanthus alkaloids: pharmacognosy and biotechnology. Curr. Med. Chem. 11 (5), 607–628. doi: 10.2174/0929867043455846
van Es, S. W., van der Auweraert, E. B., Silveira, S. R., Angenent, G. C., van Dijk, A. D. J., Immink, R. G. H. (2019). Comprehensive phenotyping reveals interactions and functions of arabidopsis thaliana TCP genes in yield determination. Plant J. 99 (2), 316–328. doi: 10.1111/tpj.14326
Viola, I. L., Uberti Manassero, N. G., Ripoll, R., Gonzalez, D. H. (2011). The arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 435 (1), 143–155. doi: 10.1042/BJ20101019
Wang, J. L., Wang, H. W., Cao, Y. N., Kan, S. L., Liu, Y. Y. (2022). Comprehensive evolutionary analysis of the TCP gene family: Further insights for its origin, expansion, and diversification. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.994567
Wang, Y., Yu, Y., Wang, J., Chen, Q., Ni, Z. (2020). Heterologous overexpression of the GbTCP5 gene increased root hair length, root hair and stem trichome density, and lignin content in transgenic arabidopsis. Gene 758, 144954. doi: 10.1016/j.gene.2020.144954
Xu, R., Sun, P., Jia, F., Lu, L., Li, Y., Zhang, S., et al. (2014). Genomewide analysis of TCP transcription factor gene family in malus domestica. J. Genet. 93 (3), 733–746. doi: 10.1007/s12041-014-0446-0
Xu, Y. M., Xiao, X. M., Zeng, Z. X., Tan, X. L., Liu, Z. L., Chen, J. W., et al. (2019). BrTCP7 transcription factor is associated with MeJA-promoted leaf senescence by activating the expression of BrOPR3 and BrRCCR. Int. J. Mol. Sci. 20 (16), 3963. doi: 10.3390/ijms20163963
Yamamoto, K., Takahashi, K., Mizuno, H., Anegawa, A., Ishizaki, K., Fukaki, H., et al. (2016). Cell-specific localization of alkaloids in catharanthus roseus stem tissue measured with imaging MS and single-cell MS. Proc. Natl. Acad. Sci. U.S.A. 113 (14), 3891–3896. doi: 10.1073/pnas.1521959113
Yang, Y., Guo, J., Cheng, J., Jiang, Z., Xu, M. (2020). Identification of UV-b radiation responsive microRNAs and their target genes in chrysanthemum (Chrysanthemum morifolium ramat) using high-throughput sequencing. Ind. Crops Products 151, 112484. doi: 10.1016/j.indcrop.2020.112484
Yu, L., Chen, Q., Zheng, J., Xu, F., Ye, J., Zhang, W., et al. (2022b). Genome-wide identification and expression pattern analysis of the TCP transcription factor family in ginkgo biloba. Plant Signal Behav. 17 (1), 1994248. doi: 10.1080/15592324.2021.1994248
Yu, C., Huang, J., Wu, Q., Zhang, C., Li, X. L., Xu, X., et al. (2022a). Role of female-predominant MYB39-bHLH13 complex in sexually dimorphic accumulation of taxol in taxus media. Hortic. Res. 9, uhac062. doi: 10.1093/hr/uhac062
Yu, S., Li, P., Zhao, X., Tan, M., Ahmad, M. Z., Xu, Y., et al. (2021). CsTCPs regulate shoot tip development and catechin biosynthesis in tea plant (Camellia sinensis). Hortic. Res. 8 (1), 104. doi: 10.1038/s41438-021-00538-7
Zhan, X., Chen, Z., Chen, R., Shen, C. (2022). Environmental and genetic factors involved in plant protection-associated secondary metabolite biosynthesis pathways. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.877304
Zhan, X., Liao, X., Luo, X., Zhu, Y., Feng, S., Yu, C., et al. (2018). Comparative metabolomic and proteomic analyses reveal the regulation mechanism underlying MeJA-induced bioactive compound accumulation in cutleaf groundcherry (Physalis angulata l.) hairy roots. J. Agric. Food Chem. 66 (25), 6336–6347. doi: 10.1021/acs.jafc.8b02502
Zhang, Q., Lin, L., Fang, F., Cui, B., Zhu, C., Luo, S., et al. (2022). Dissecting the functions of COP1 in the UVR8 pathway with a COP1 variant in arabidopsis. Plant J 113 (3), 478–492. doi: 10.1111/tpj.16059
Zhao, J., Zhu, W. H., Guo, Y. Q. (2001). Compact callus cluster suspension cultures of catharanthus roseus with enhanced indole alkaloid biosynthesis. IN Vitro Cell Dev. BIOL-PLANT 37, 68–72. doi: 10.1007/s11627-001-0013-2
Zhou, H., Hwarari, D., Ma, H., Xu, H., Yang, L., Luo, Y. (2022). Genomic survey of TCP transcription factors in plants: Phylogenomics, evolution and their biology. Front. Genet. 13. doi: 10.3389/fgene.2022.1060546
Keywords: cis-element, elicitor, expression pattern, stress, TCP family, qRT-PCR, terpenoid indole alkaloids
Citation: Hao J, Zheng L, Han Y, Zhang H, Hou K, Liang X, Chen C, Wang Z, Qian J, Lin Z, Wang Z, Zeng H and Shen C (2023) Genome-wide identification and expression analysis of TCP family genes in Catharanthus roseus. Front. Plant Sci. 14:1161534. doi: 10.3389/fpls.2023.1161534
Received: 08 February 2023; Accepted: 27 March 2023;
Published: 12 April 2023.
Edited by:
Sonal Mishra, Central University of Jammu, IndiaReviewed by:
Ujjal Jyoti Phukan, University of Arizona, United StatesPravin Prakash, National Botanical Research Institute (CSIR), India
Copyright © 2023 Hao, Zheng, Han, Zhang, Hou, Liang, Chen, Wang, Qian, Lin, Wang, Zeng and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chenjia Shen, c2hlbmNqQGh6bnUuZWR1LmNu; Houqing Zeng, emVuZ2hxQGh6bnUuZWR1LmNu
 Juan Hao
Juan Hao Lijun Zheng1,2
Lijun Zheng1,2 Yidie Han
Yidie Han Houqing Zeng
Houqing Zeng Chenjia Shen
Chenjia Shen