- 1School of Health and Nursing, Wuchang University of Technology, Wuhan, China
- 2Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 3College of Forestry, Central South University of Forestry and Technology, Changsha, Hunan, China
- 4College of Basic Medical Sciences, Hubei University of Chinese Medicine, Wuhan, China
Woody oil plants are the most productive oil-bearing species that produce seeds with high levels of valuable triacylglycerols (TAGs). TAGs and their derivatives are the raw materials for many macromolecular bio-based products, such as nylon precursors, and biomass-based diesel. Here, we identified 280 genes encoding seven distinct classes of enzymes (i.e., G3PAT, LPAAT, PAP, DGAT, PDCT, PDAT, and CPT) involved in TAGs-biosynthesis. Several multigene families are expanded by large-scale duplication events, such as G3PATs, and PAPs. RNA-seq was used to survey the expression profiles of these TAG pathway-related genes in different tissues or development, indicating functional redundancy for some duplicated genes originated from the large-scale duplication events, and neo-functionalization or sub-functionalization for some of them. Sixty-two genes showed strong, preferential expression during the period of rapid seed lipid synthesis, suggesting that their might represented the core TAG-toolbox. We also revealed for the first time that there is no PDCT pathway in Vernicia fordii and Xanthoceras sorbifolium. The identification of key genes involved in lipid biosynthesis will be the foundation to plan strategies to develop woody oil plant varieties with enhanced processing properties and high oil content.
1 Introduction
Triacylglycerols (TAGs) are the primary form of energy storage in most living organisms (Gibbons et al., 2000; Durrett et al., 2008). They accumulate in plant seeds and provide nutrients and energy for subsequent germination and seedling development (Yang and Benning, 2018). These storage lipids are mainly composed of polysaturated fatty acids (Mu and Porsgaard, 2005; Lieb et al., 2019), which have great nutraceutical and nutritional value (Lung and Weselake, 2006; Zhang et al., 2019). Therefore, they are important precursors for chemical industry products, also providing sources for human nutrition (Lung and Weselake, 2006; Liu et al., 2011). At present, TAGs are also increasingly becoming important raw materials for the production of nylon precursors, biofuels, lubricants, detergents, and paints, as well as biomass diesel (Durrett et al., 2008; Barreira et al., 2009; Nabizadeh et al., 2011; Xu and Shanklin, 2016).
TAGs exist in all eukaryotes, such as protists, fungi, animals, and plants (Coleman and Lee, 2004). As a common metabolic pathway, the biosynthesis of TAGs seems to be conserved from bacteria to plants and humans (Yang et al., 2012). The biosynthesis of TAG mainly occurs in two different cell compartments, so it can be divided into two independent sets of reactions (Buchanan et al., 2015). In the first step, the fatty acids, or acyl, chains of TAG are primarily produced from precursor carbon molecules in the plastid. These compounds are then transported to the endoplasmic reticulum (ER), where they act as precursors for membrane and storage lipids production. In general, the TAG assembly sequentially consumes acyl-CoA using substrate glycerol-3-phosphate (G3P) with diverse enzymes. The phosphatidic acids and lysophosphatidic acids catalyzed by lysophosphatidic acid acyltransferase (LPAAT) and Glycerol-3-phosphoglycerol acyltransferase (G3PAT) were produced, respectively (Takeuchi and Reue, 2009; Athenstaedt, 2021). After the removal of phosphate, phosphatidic acids are converted to diacylglycerols, the precursor of triacylglycerols. The 3-sn-phosphatidate phosphohydrolase or phosphatidic acid phosphohydrolase (PAP) catalyzes the dephosphorylation of phosphatidic acid to produce diacylglycerols, which is the precursor of TAGs (Neumann and Römheld, 1999; Nakamura et al., 2007). Both phospholipid: diacylglycerol acyltransferase (PDAT) and acyl-CoA: diacylglycerol acyltransferase (DGAT) as acyl donors, convert diacylglycerols (DAGs) to TAGs (Stobart et al., 1997; Dahlqvist et al., 2000). Finally, the oil bodies are produced from oleosins which are assembled by TAGs in plant seeds (Kocsis and Weselake, 1996).
It is known that the global oil reserves in oilfield reservoirs will be exhausted in the next 40 to 50 years (Wenrui et al., 2013), so there is an urgent need to find renewable alternative fuels. Biodiesel is considered to be a way to solve this problem and enters the field of scientific research because it can be made from a variety of raw materials, such as microalgae, animal fat, and plant oil (Demirbas and Demirbas, 2011; Hsieh et al., 2012; Rodionova et al., 2017; Chen et al., 2018). Among the various lipids synthesized by plant cells, TAGs have the highest biofuel value. Woody oil plants, including purpleblow maple (Acer truncatum), walnut (Juglans regia), physic nut (Jatropha curcas), tung tree (Vernicia fordii), oil palm (Elaeis guineensis), yellowhorn (Xanthoceras sorbifolium), and olive tree (Olea europaea), are most popular in a variety of applications, such as bioremediation, cosmetics, and biofuel. In the present study, we report the identification of the genes putatively involved in TAGs biosynthesis in woody oil plants. Subsequently, we carried out the expression patterns of these genes in different tissues and/or seed development, and then discussed the functional roles of these different gene members in various enzyme classes. Our data provide the basis for further understanding the TAGs biosynthesis and will ultimately allow the validation of biotechnological strategies to design of future studies involving manipulation of oil production in woody oil plants.
2 Materials and methods
2.1 In silico identification of genes encoding enzymes from the TAGs biosynthesis pathway
We analyzed and collected the A. thaliana enzymes for each of seven gene families of the TAG-pathway from Plant Metabolic Network (http://www.plantcyc.org) and downloaded the TAIR database (https://www.arabidopsis.org/). The protein sequences of purpleblow maple (A. truncatum), physic nut (J. curcas), walnut (J. regia), yellowhorn (X. sorbifolium), tung tree (V. fordii), oil palm (E. guineensis), and olive tree (O. europaea) were obtained from https://doi.org/10.6084/m9.figshare.12986237.v2, https://plantgenomics.snu.ac.kr, http://dendrome.ucdavis.edu/ftp/Genome_Data/genome/Reju/, http://www.gigadb.org, https://bigd.big.ac.cn/gsa/, http://genomsawit.mpob.gov.my, and http://www.gigadb.org, respectively. Subsequently, these A. thaliana enzymes were examined by InterProScan (http://www.ebi.ac.uk/interpro/) (Jones et al., 2014) and PFAM (http://pfam.xfam.org/) (Mistry et al., 2021). Finally, we used the BLAST and HMM model searches to identify the TAG-pathway enzymes by scanning these seven woody oil plants genome database, including purpleblow maple (A. truncatum) (Ma et al., 2020), walnut (J. regia) (Martínez-García et al., 2016), yellowhorn (X. sorbifolium) (Bi et al., 2019; Liang et al., 2019), physic nut (J. curcas) (Ha et al., 2019), tung tree (V. fordii) (Zhang et al., 2019), oil palm (E. guineensis) (Singh et al., 2013), and olive tree (O. europaea) (Unver et al., 2017). The inclusion criteria of putative TAG-pathway enzymes required that they have more than 50% amino acid identity with the Arabidopsis query and contain corresponding domains using PFAM (Mistry et al., 2021) and InterProScan (Mistry et al., 2021).
2.2 Phylogenetic and sequence analyses
MAFFT software was used to align the deduced amino acid sequences from identified enzymes (Katoh et al., 2019). Available literature data were used to perform the characteristic signatures of enzyme classes and in silico characterization of enzyme domains (Schläpfer et al., 2017). TmPred server was used to predict the transmembrane domain. Dendrograms were drawn using IQ-tree with Maximum likelihood (ML) method according to the best model as implemented in IQ-tree (Minh et al., 2020).
2.3 RNA‐seq expression analysis
Transcriptome or RNA-seq data for different tissues from woody oil plants were obtained and downloaded from the NCBI SRA database with accession numbers: PRJNA318350, PRJNA483508, PRJNA643637, PRJNA399212, PRJNA557096, PRJNA590386, and PRJNA445068. The fastp (https://github.com/OpenGene/fastp) was used to perform the quality-based trimming. The obtained clean reads were mapped to the corresponding genomes using the HISAT2 with default parameters. The absolute transcription abundance values of all identified TAG biosynthesis genes were estimated from fragments per kilobase million (FPKM) values obtained by StringTiec (Pertea et al., 2016; Jiang et al., 2022). The expression level values used in our study are the log2-transformed FPKM values, as described in previously published papers (Carocha et al., 2015; Cao et al., 2019; Cao et al., 2022).
3 Result and discussion
3.1 In silico identification of TAG genes encoding enzymes
In the present study, both HMM model and BLAST searches were used to identify genes of seven different metabolic lipid families involved in TAG biosynthesis using known lipid enzymes as queries. The steps for gene identification contained: (1) A. thaliana enzymes were examined by PFAM to identify HMM model, which can scan the putative TAG-pathway enzymes; (2) BLASTp was used to further identify the putative TAG-pathway enzymes, only enzymes that have more than 50% amino acid identity with these enzymes scanned in A. thaliana were used as queries; (3) amino acid sequence alignment was used to identify regions of homology between putative TAG-pathway enzymes, so as to finally determine the characteristics and motifs of each type of enzyme. Finally, 280 TAG genes encoding enzymes were identified and used for further analyses (Figure 1, Tables S1, S2).
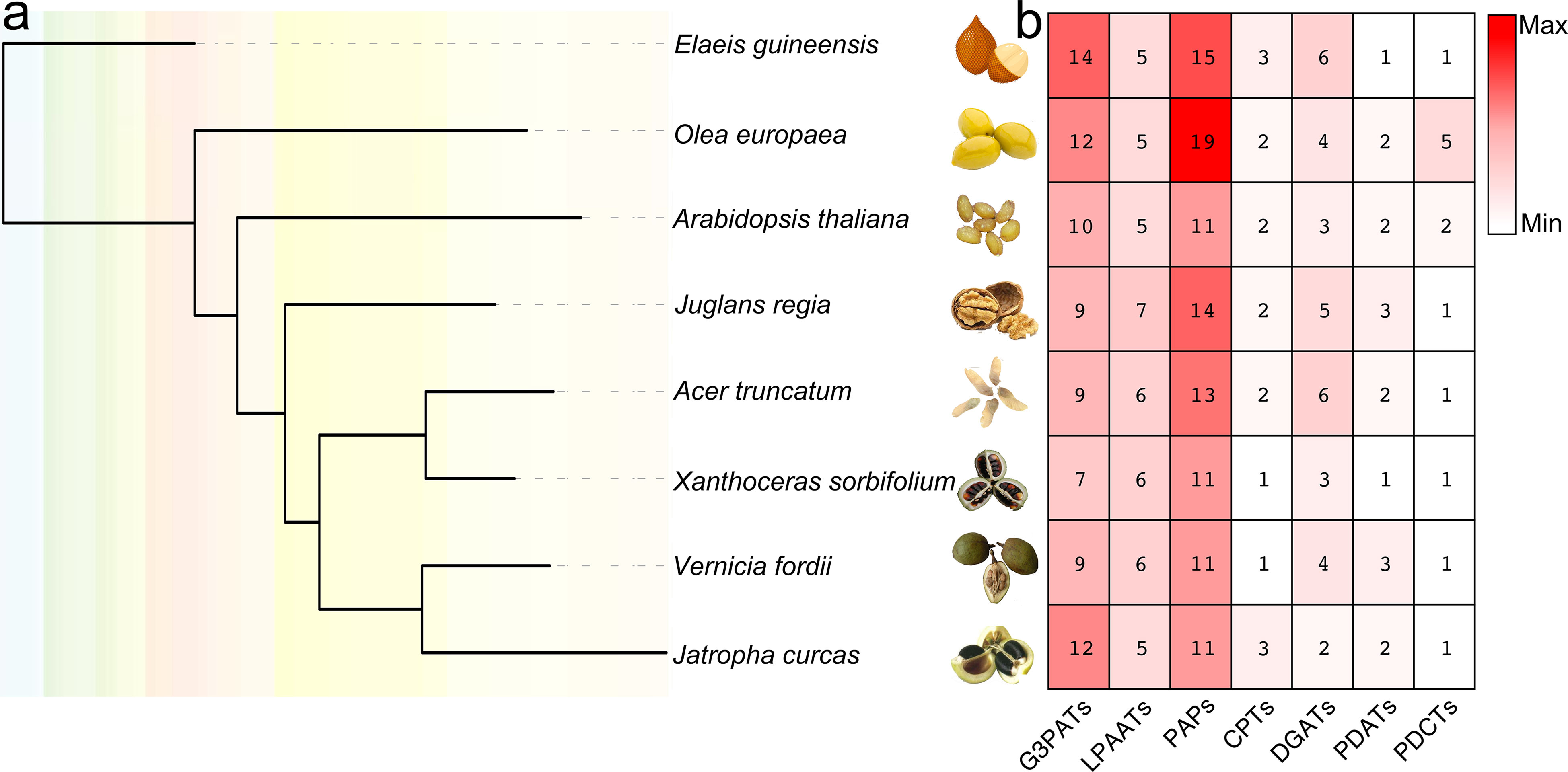
Figure 1 Species tree of eight plant genomes and TAG-related biosynthesis gene numbers of each species. (A) one-to-one orthologs were detected between genomes by Orthofinder (Emms and Kelly, 2015), and species tree was constructed by IQ-tree (Nguyen et al., 2015). (B) the numbers of TAG-related biosynthesis genes in each species were listed accordingly.
3.2 Glycerol-3-phosphate-1-acyltransferase
G3PAT is the first enzyme in the Kennedy Pathway that catalyzes acyl-CoAs onto the C1 hydroxyl group of glycerol-3-phosphate or the attachment of an acyl group from either acyl-carrier proteins (acyl-ACPs) to give 1-acylglycerol-3-phosphate (Zheng et al., 2003). As early as 60 years ago, researchers have characterized the biochemical functions of G3PATs from plant and animal tissues (Kornberg and Pricer, 1953; Weiss et al., 1960). In plants, the activity of G3PATs was observed in three different plant subcellular compartments, including plastid, endoplasmic reticulum (ER), and mitochondria (Gidda et al., 2009). Seventy-two G3PATs were detected by BLASTP and HMM model with reported A. thaliana AtG3PATs as queries in seven woody oil plant reference genomes. The ML tree was divided these G3PATs into three clades, which was consistent with previously published manuscripts (Waschburger et al., 2018; Wang et al., 2020). The large-scale duplication events were found in the EguG3PATs, OeuG3PATs, and JcuG3PATs, resulting in more G3PAT family members than the other four woody oil plants (Figure 2).
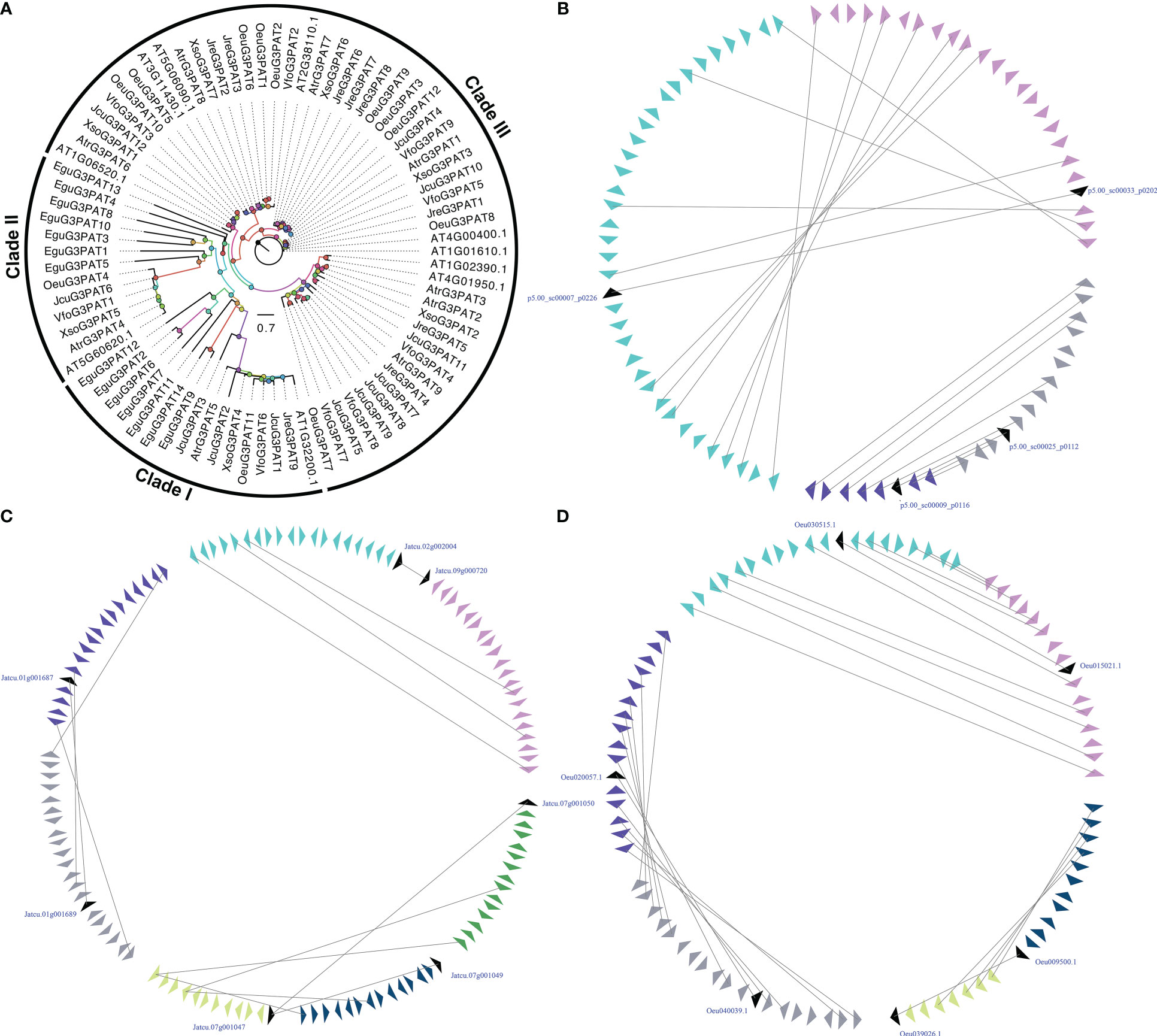
Figure 2 The phylogenetic tree (A) and microsynteny related to G3PAT genes in (B) E guineensis; (C) O. europaea; and (D) J curcas. The triangles indicated a gene in a family and its flanking genes, as well as the gene’s orientation. The homologous genes were connected by gray lines between two fragments.
The functions of most A. thaliana AtG3PAT genes have been characterized and identified in detail. However, the functional mechanism of woody oil plants G3PAT genes has not been uncovered so far, especially their role in seed oil biosynthesis. In the present study, expression profiling of G3PATs in different tissues or development exhibited that the transcripts accumulated in tissue-specific patterns (Figure S1). For example, some G3PATs demonstrated specific transcript accumulations in leaf, root, flower, stem, and seeds, such as JcuG3PAT8 in stem, JreG3PAT1 in root, and OeuG3PAT10 in flower. The expression patterns of duplicated G3PAT genes exhibited distinct tissue-specific, implying that these G3PATs were derived from the large-scale duplication events followed by functional divergence. To further determine G3PAT genes that might contribute to the seed oil biosynthesis, we carried out the absolute expression level of G3PATs in seeds. Compared to other G3PATs, AtrG3PAT1 from A. truncatum (2-fold higher), XsoG3PAT2, XsoG3PAT3 and XsoG3PAT4 from X. sorbifolium (100-fold higher), JcuG3PAT1, JcuG3PAT3 and JcuG3PAT6 from J. curcas (5-fold higher), JreG3PAT1 and JreG3PAT9 from J. regia (100-fold higher), OeuG3PAT3, OeuG3PAT4 and OeuG3PAT8 from O. europaea (20-fold higher), and VfoG3PAT3, VfoG3PAT4 and VfoG3PAT6 from V. fordii (3-fold higher) were relatively highly expressed in seeds, indicating these G3PATs might be involved in seed oil biosynthesis (Figure S1).
3.3 Lysophosphatidic acid acyltransferase
LPAATs perform essential cellular functions by controlling the conversion of 1-acyl-sn-G3P (LPA) to phosphatidic acid (PA) (Hills and Roscoe, 2006). LPAAT was first characterized biochemically about 20 years ago, and then many homologous of LPAATs were identified in plants and animals (Yu et al., 2004; Körbes et al., 2016; Wang et al., 2017). In plants, there are several different isoforms of LPAAT, so their activity is related to a variety of membrane systems, such as ER, chloroplasts, and outer membrane of mitochondria (Bourgis et al., 1999; Yu et al., 2004). The LPAAT is an ancient and very larger gene family. The first LPAAT gene was cloned with containing activities about 20 years ago, then homologous of LPAATs have been found and characterized in many plants (Ichihara et al., 1987; Körbes et al., 2016; Bradley and Duncan, 2018; Meesapyodsuk et al., 2021). To identify the gene encoding LPAAT, we used both HMM model and BLAST to search the local genome database using proteins of A. thaliana At4g30580, At3g57650, At1g51260, At3g18850, and At1g75020 as the query sequences. Finally, we identified forty LPAATs, including 5 EguLPAATs in E. guineensis, 5 AtrLPAATs in A. truncatum, 7 JreLPAATs in J. regia, 5 OeuLPAATs in O. europaea, 6 VfoLPAATs in V. fordii, 6 XsoLPAATs in X. sorbifolium and 5 JcuLPAATs in J. curcas, respectively. Among them, JreLPAAT3 and JreLPAAT5 were located in chromosome 13 and 15 and evolved from a same ancestor after undergoing large-scale duplication events (Figure 3).
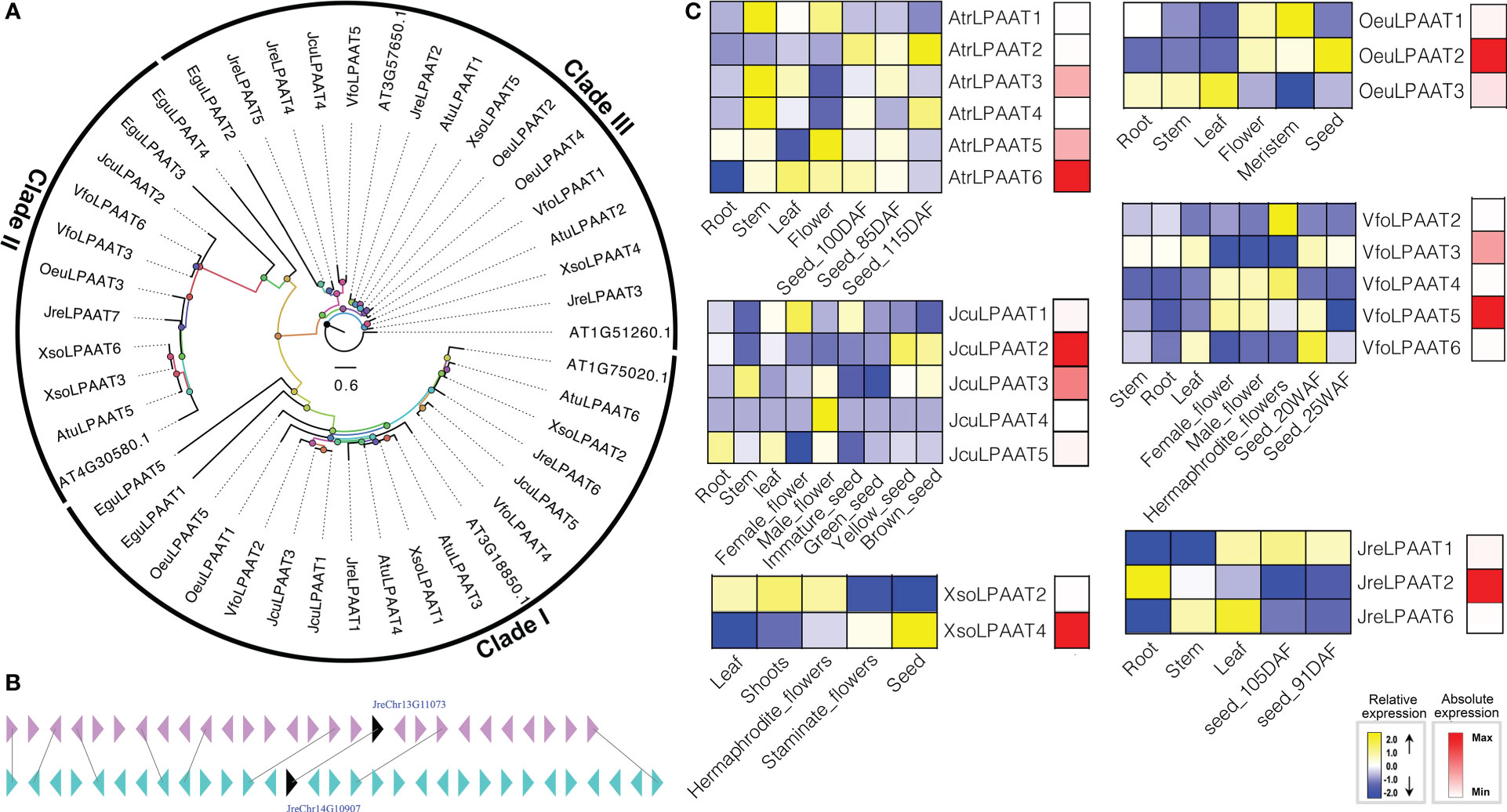
Figure 3 The LPAAT subfamily: phylogenetic tree (A), microsynteny (B), and expression profiles (C). The triangles indicated a gene in a family and its flanking genes, as well as the gene’s orientation. The homologous genes were connected by gray lines between two fragments. The expression profiles were obtained by RNA-Seq. Blue and yellow colors indicated low-expression and high-expression, respectively. Red color suggested absolute high-expression.
The biological functions of LPAATs have been proven to be diverse (Bradley and Duncan, 2018). For example, the role of the same enzyme in affecting the downstream lipid biosynthesis pathways was significantly diverse in different tissues, highlighting the importance of tissue-specific research (Bradley and Duncan, 2018; Fahs et al., 2019). In the present study, the expression of JreLPAAT3, JreLPAAT4, JreLPAAT5 and JreLPAAT7, OeuLPAAT5, VfoLPAAT1, XsoLPAAT1, XsoLPAAT3, XsoLPAAT5, and XsoLPAAT6 were not detected in any tissues or seed development. However, the expression patterns of other LPAAT genes exhibited different tissue-specific, and some of them showed specific expression in seeds, which indicated their potential functions in seed oil biosynthesis. The absolute expression levels of LPAATs were performed to investigate which LPAAT might contribute to seed oil synthesis. Finally, seven LPAATs, including AtrLPAAT6, JcuLPAAT2, JcuLPAAT3, JreLPAAT2, OeuLPAAT2, VfoLPAAT5, and XsoLPAAT4, were determined to be involved in the biosynthesis of seed oil (Figure 3).
3.4 Phosphatidic acid phosphatase
PAP is the three enzyme in the Kennedy Pathway that performs essential cellular function by controlling the conversion of PA to inorganic phosphate and sn-1,2-diacylglycerol (DAG) (Neumann and Römheld, 1999; Nakamura et al., 2007). PAP is related to the inner membrane of ER, cytoplasm, and chloroplast, its intracellular translocation may play an important role in both intracellular signaling and lipid metabolism mechanisms (Kocsis and Weselake, 1996; Perry et al., 1999; Carman and Henry, 2007). PAP activity located in the inner membrane of the plastid envelopes of Pisum sativum and Spinacia oleracea is believed to be associated with the prokaryotic glycolipid synthesis pathway, while the activity related to the microsome-associated fractions of Persea Americana and Carthamus tinctorius is believed to be related to the so-called eukaryotic lipid biosynthesis pathway (França et al., 2008). Using eleven A. thaliana AtPAPs (At1g15080, At3g02600, At2g01180, At3g09560, At5g42870, At3g18220, At3g50920, At3g58490, At4g22550, At5g03080 and At5g66450) as BLAST queries, we identified 94 PAPs in seven woody oil plants. The phylogenetic analysis of PAPs from seven woody oil plants and A. thaliana resulted in the formation of an ML tree with three different clades for Clade I, Clade II, and Clade III. Each of the seven woody oil plants and A. thaliana contributed at least one PAP to these three clades, we can infer that woody oil plants PAP genes shared a common ancestor. To determine whether the PAPs have undergone the large-scale duplication events, we analyzed the relationship of the PAPs in these seven woody oil plants and identified two, three, four, nine, and four collinear gene pairs in A. truncatum, E. guineensis, J. curcas, J. regia, and O. europaea, respectively, while only one and one collinear gene pair in X. sorbifolium and V. fordii, respectively (Figure 4). These results might have resulted from ancient processes during the long evolutionary period (Figure 4).
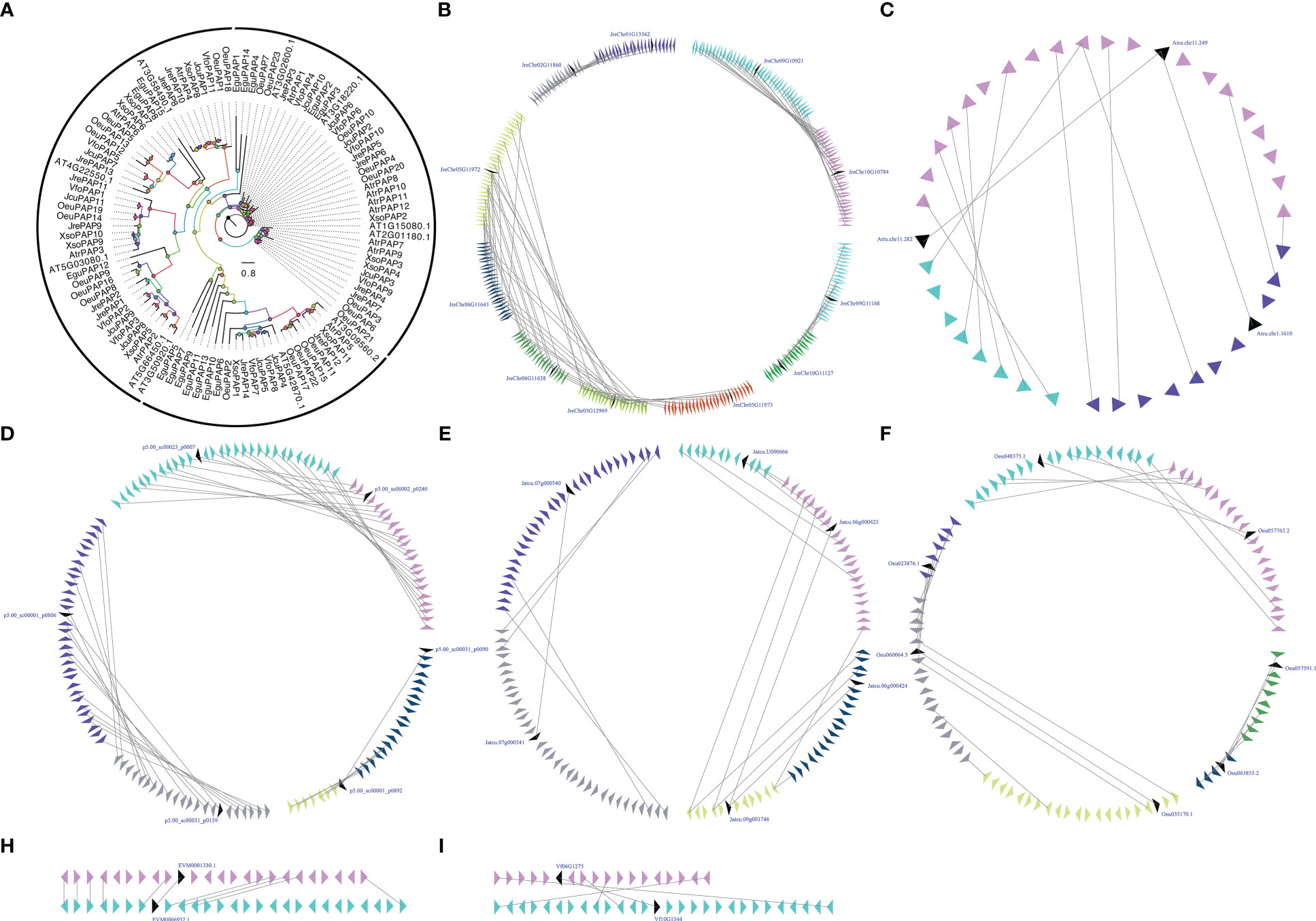
Figure 4 The phylogenetic tree (A) and microsynteny related to PAP genes in (B) J regia; (C) A truncatum; (D) E guineensis; (E) J curcas; (F) O. europaea; (H) X. sorbifolium; and (I) V. fordii. The triangles indicated a gene in a family and its flanking genes, as well as the gene’s orientation. The homologous genes were connected by gray lines between two fragments.
Gene expression patterns can provide clues to gene function (Cao et al., 2020; Cao et al., 2021). To further understand the potential functions of PAPs, the RNA-seq sets were used to determine the expression patterns of these genes in different tissues. With respect to JrePAP11, VfoPAP2, VfoPAP3, VfoPAP6, OeuPAP10, OeuPAP13, OeuPAP16, OeuPAP17, OeuPAP18, OeuPAP19, OeuPAP20, OeuPAP21, OeuPAP22, OeuPAP23, XsoPAP2, XsoPAP3, XsoPAP4, XsoPAP10, and XsoPAP11, we were unable to detect any expression in tested different tissues. The involvement of PAPs in other cellular processes or the induction of other biotic and abiotic stresses might explain their undetectable expression in these tested tissues. For instance, PAPs are not only involved in glycerolipid biosynthesis, but also play important roles in the turnover and degradation of phospholipids (Cagliari et al., 2010). The expression patterns of duplicated PAP pairs showed different tissue-specific, suggesting that neo-functionalization or sub-functionalization occurred after the large-scale duplication events (Figure S2). The absolute expression levels of PAPs were carried out to survey which PAP might be involved in seed oil biosynthesis. Finally, twelve PAPs, including VfoPAP5, VfoPAP9, VfoPAP10, XsoPAP1, AtrPAP4, AtrPAP10, JcuPAP2, JcuPAP3, OeuPAP1, OeuPAP3, JrePAP3, and JrePAP10, were determined to be involved in the biosynthesis of seed oil (Figure S2).
3.5 Diacylglycerol acyltransferase
DGAT is a transmembrane enzyme that plays an essential role in the final rate-limiting step of TAG biosynthesis. As a key enzyme for biotechnology purposes, DGAT has great prospects for being used to increase the oil content of oil crops (Slabas et al., 2001; Lung and Weselake, 2006; Xu et al., 2008; Liu et al., 2020; Gao et al., 2021). Overexpression of DGAT in A. thaliana can significantly increase seed oil content (Jako et al., 2001). The substrate preference of DGAT mainly depends on the temperature, acyl-CoA concentration, and the acyl composition of the diacylglycerol (DAG) pool (Lung and Weselake, 2006). A. thaliana contains three genes (At3g51520, At2g19450, and At1g48300) that encode enzymes with DGAT activity. Through HMM model and BLAST searches, six, six, five, four, four, three and two DGATs were identified in E. guineensis, A. truncatum, J. regia, O. europaea, V. fordii, X. sorbifolium and J. curcas, respectively (Figure 1).
Different types of DGAT enzymes with different membrane-bound polypeptides have been identified in several species (Cases et al., 1998; Cases et al., 2001; Bhatt-Wessel et al., 2018; Chitraju et al., 2019), which is consistent with our results that DGAT genes were divided into three clades by ML tree (Figure 5). The roles of different types of DGAT enzymes can be species-dependent in oil production (Shockey et al., 2006). In our study, we investigated the expression patterns of DGATs in different tissues or development. In O. europaea, OeuDGAT1 and OeuDGAT3 were preferentially expressed seeds. Additionally, OeuDGAT3 exhibited eightfold higher expression than OeuDGAT1 in seeds tissues, which may consistent with its contributed to seed oil biosynthesis. Similarly, AtrDGAT1, AtrDGAT3, AtrDGAT5, JcuDGAT2, JreDGAT1, VfoDGAT3, and XsoDGAT2 were identified that probably involved in seed oil biosynthesis (Figure 5).
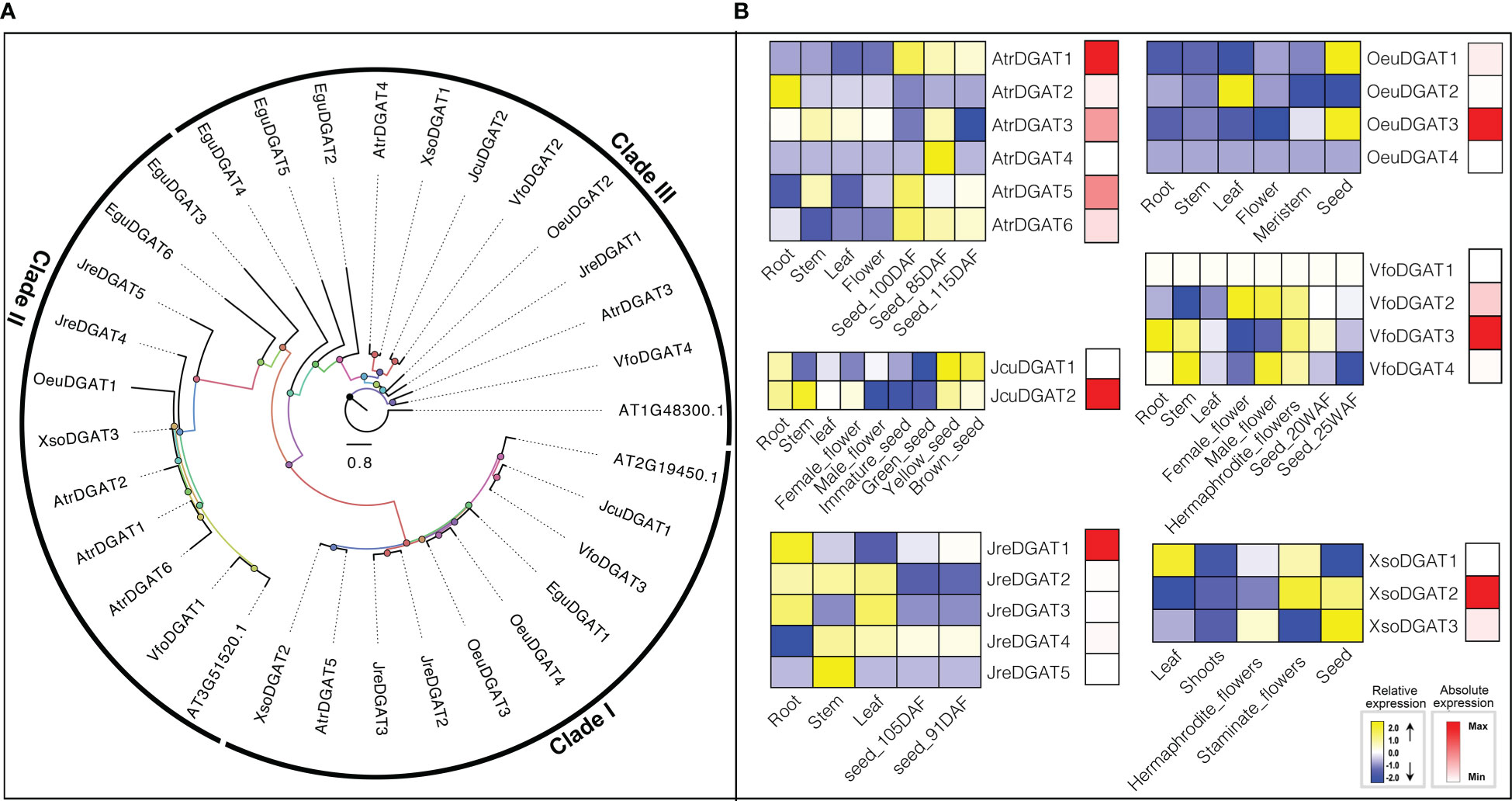
Figure 5 The DGAT subfamily: phylogenetic tree (A), and expression profiles (B). The expression profiles were obtained by RNA-Seq. Blue and yellow colors indicated low-expression and high-expression, respectively. Red color suggested absolute high-expression.
3.6 Phospholipid diacylglycerol acyltransferase
Phospholipid diacylglycerol acyltransferase (PDAT) utilizes an acyl-CoA independent mechanism to transfer a fatty acid from the sn-2 position of phosphatidylcholine (PC) to -diacylglyerol (DAG) forming TAG and a lysophospholipid (Dahlqvist et al., 2000). Therefore, PDAT can contribute to the high levels of polyunsaturated fatty acid (PUFA) in seed oils of A. thaliana (Fan et al., 2013), flax (Linum usitatissimum) (Wickramarathna et al., 2015), and Camelina sativa (Marmon et al., 2017). A. thaliana has two PDATs (At3g44830 and At5g13640). To detect the gene encoding PDAT in seven woody oil plants, both HMM model and BLAST were used to search the local genome databases. Finally, fourteen PDATs were identified, including 1 in E. guineensis, 2 in J. curcas, 2 in O. europaea, 3 in J. regia, 2 in A. truncatum, 1 in X. sorbifolium, and 3 in V. fordii, respectively (Figure 6).
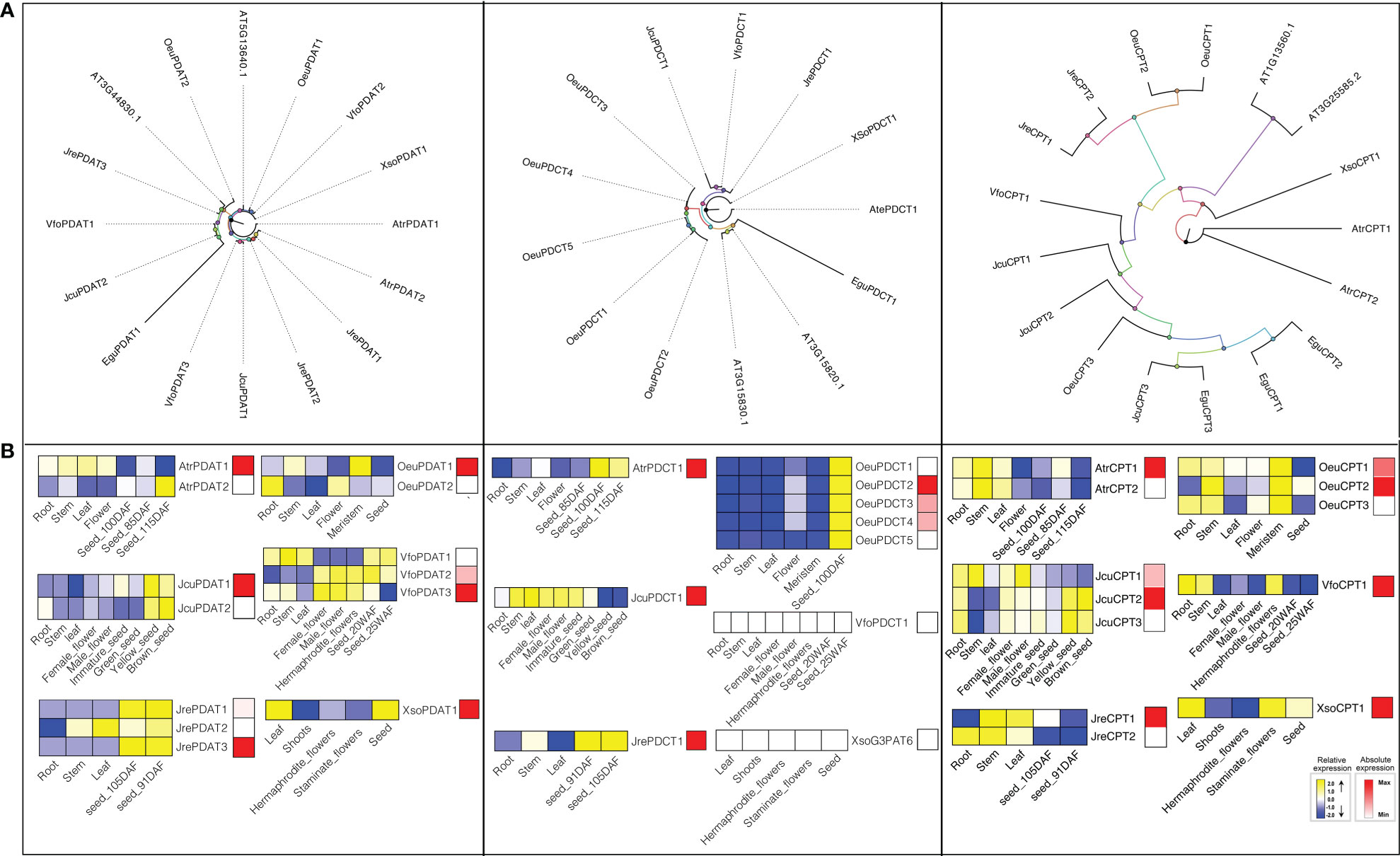
Figure 6 The PDAT, PDCT, and CPT subfamilies: phylogenetic trees (A), and expression profiles (B). The expression profiles were obtained by RNA-Seq. Blue and yellow colors indicated low-expression and high-expression, respectively. Red color suggested absolute high-expression.
To further understand the potential function of PDAT genes, we analyzed the expression profile of this gene family. The expression patterns suggested that VfoPDAT3 was highly expressed in seed at 20 WAF (Seed oil was synthesized in large quantities during this period) compared to VfoPDAT2 (2-fold higher) and VfoPDAT1 (50-fold higher), indicating VfoPDAT3 might involve the oil biosynthesis of V. fordii seed. Similarly, AtrPDAT1, JcuPDAT1, JrePDAT3, OeuPDAT1, VfoPDAT3, and XsoPDAT1 probably involved in seed oil biosynthesis (Figure 6).
3.7 Phosphatidylcholine diacylglycerol cholinephosphotransferase
Phosphatidylcholine diacylglycerol cholinephosphotransferase (PDCT) is an enzyme that can catalyze the reversible transfer of the phosphocholine headgroup from PUFA-enriched PC to PUFA-enriched DAG (Bates and Browse, 2011). The acyl groups on DAG entering PC are driven by the action of PDCT, which are then returned to DAG after modification (Cahoon et al., 1999), suggesting that PDCT might contribute to the biosynthesis of TAG and determine the seed oil fatty acids composition (Wickramarathna et al., 2015). To determine the gene encoding PDCT, we used both HMM model and BLAST to search the local genome database using proteins of A. thaliana At3g15830 and At3g15820 as the query sequences. Consequently, eleven candidate PDCTs were identified from the oil woody plant genome databases, namely AtrPDCT1, EguPDCT1, JcuPDCT1, JrePDCT1, VfoPDCT1, XSoPDCT1, and OeuPDCT1-5 (Table S1, Figure 6).
E. guineensis, J. curcas, J. regia, A. truncatum, X. sorbifolium, and V. fordii contained single copy PDCT, while the O. europaea genome had five members. OeuPDCT1/OeuPDCT2 and OeuPDCT3/OeuPDCT4/OeuPDCT5 were located in chromosome 10 and 21, respectively, which were derived from the lineage-specific tandem duplication followed by functional redundancy (Table S1, Figure 6). For example, the expression profiles of these five OeuPDCTs were very similar and were weakly expressed root, stem, leaf and meristem, and highly expressed seeds (Figure 6). Compared to the other four PDCTs, OeuPDCT2 was highly expressed in seed (2.2-fold higher), indicating that this gene was probably involved in seed oil biosynthesis of O. europaea. Remarkably, the expression of VfoPDCT1 and XsoPDCT1 were not detected in different tissues, suggesting the PDCT pathway might not be involved in the seed oil biosynthesis of X. sorbifolium and V. fordii. Taken together, our results suggested that AtrPDCT1, EguPDCT1, JcuPDCT1, JrePDCT1, and OeuPDCT2 might contribute to seed oil biosynthesis (Figure 6).
3.8 Choline phosphotransferase
CPT or aminoalcohol phosphotransferase (AAPT) is a key enzyme in oilseed metabolism, which can promote the TAG biosynthesis through the reversible conversion of phosphatidylcholine (PC) to diacylglycerol (DAG) (Voelker and Kinney, 2001). A. thaliana contains two genes (At1g13560 and At3g25585) that encode enzymes with both CDP-ethanolamine: diacylglycerol choline phosphotransferase (EPT) activity and CPT activity (Liu et al., 2015b). By using A. thaliana CPT enzymes as queries, we have identified three, three, three, two, two, one, and one CPTs in E. guineensis, J. curcas, O. europaea, J. regia, A. truncatum, X. sorbifolium, and V. fordii, respectively. Previous studies have shown that CPT proteins from Brassica rapa, yeast, soybean, A. thaliana, and Ricinus communis are highly conserved, that is, they generally contain a highly conserved CDP-alcohol phosphotransferase group sequences: DGxxARxxxxxxxxGxxxDxxxD (Mcmaster and Bell, 1997; Williams and Mcmaster, 1998; Goode and Dewey, 1999; Cagliari et al., 2010). This motif was also highly conserved in CPTs among these seven woody oil plants (Table S2).
To further understand how CPTs evolved, the gene duplication events of this gene family were investigated in woody oil plants. JcuCPT2/JcuCPT3 were located in chromosome 11, which were derived from the lineage-specific tandem duplication followed by functional redundancy (Table S1). For example, the expression profile of JcuCPT2/JcuCPT3 exhibited a strong and preferential expression in seed development (Figure 6). The absolute expression levels suggested that JcuCPT2 and to a lesser extent JcuCPT1 are probably involved in seed oil biosynthesis in J. curcas. Similarly, AtrCPT1, JreCPT1, VfoCPT1, XsoCPT1, OeuCPT1, and OeuCPT2 were also probably involved in seed oil biosynthesis (Figure 6).
3.9 Hypothetical pathways involved in the biosynthesis of lipid in woody oil plants
The improvement and modification of seed oil composition can help to produce the production of industrially and nutritionally desirable oils (Kocsis and Weselake, 1996). To achieve this goal, it is necessary to manipulate the biosynthesis of fatty acids and TAGs in such a way that the specific fatty acids are synthesized at high rates and then effectively inserted into each position of TAGs (Yu et al., 2004). Although it is now possible to obtain genes encoding suitable fatty acid modifying enzymes from the model species A. thaliana and other species, the expression of these genes in transgenic plants does not produce high yields of the required oil components, making progress limited (Yu et al., 2004; Burgal et al., 2008). The biosynthesis of TAG requires the participation of multiple enzymes and the synergy of regulatory factors (Cagliari et al., 2010). Overexpression of one gene from the TAG synthesis pathway has a limited effect on increasing the oil content of seeds (Elhiti et al., 2012; Shi et al., 2012). Recently, some researchers have found that co-expression of multiple genes in transgenic lines can significantly increase the oil content. For example, Liu et al., (2015a) overexpressed ScLPAAT, BnDGAT, BnGPAT, and BnGPDH genes in Brassica napus, and found that the seed oil content of the overexpression lines was significantly higher than that of the control lines (Liu et al., 2015a). Here, our data highlighted 62 genes encompassing seven enzyme families involved in the core TAG biosynthesis of seeds (Figure 7), such as XsoCPT1, AtrPDCT1, and VfoPDAT3. To gain understand the expression profiles of strongly expressed and/or preferentially expressed genes, a unique expression pattern was also observed by using RNA-seq. Additionally, some other genes, such as OeuG3PAT10 and AtrPAP13, were expressed in these tested tissues, indicating that these genes might be involved in other biosynthesis other than the TAG pathway.
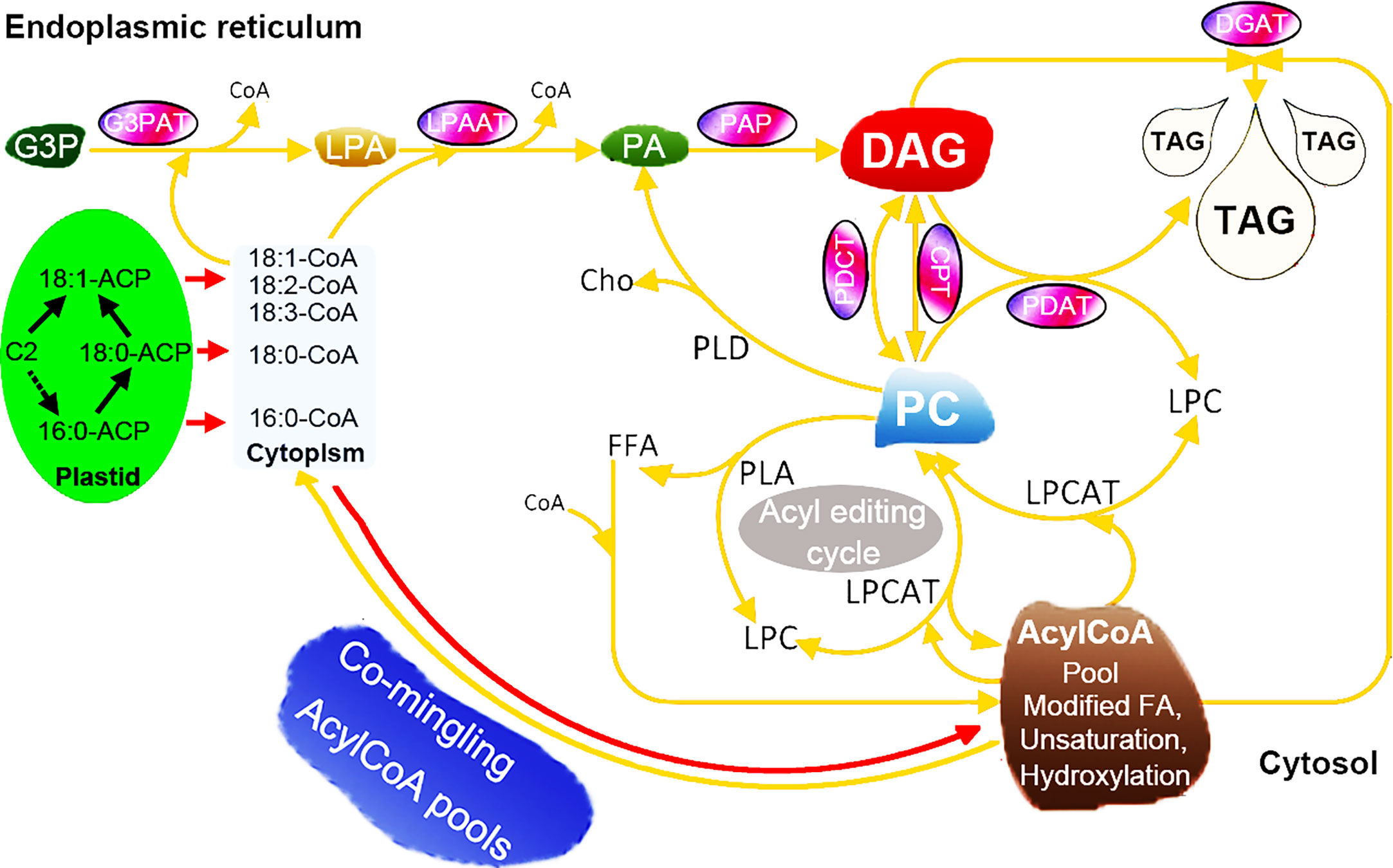
Figure 7 Hypothetical pathways involved in the TAG biosynthesis in seeds of woody oil plants. The TAG biosynthesis pathway was adapted from previous papers (Coleman and Lee, 2004; Cagliari et al., 2011; Bates, 2012; Parajuli et al., 2020).
4 Conclusion
Seeds of woody oil plants have been used for producing biofuels, biomass diesel, paints, and edible oil. In this study, we provide key information regarding core TAG biosynthesis, which has been limited to date. Although further studies encompassing enzymatic assays and posttranscriptional analyses are needed to determine the activity of the enzymes encoded by these genes from the TAG biosynthesis. However, our data exhibited here may help identify potential targets for future biotechnological approaches that may play crucial roles in the development of genetic strategies to control lipid biosynthesis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YC designed this research and then wrote the manuscript. YC, QL, and LZ participated in the evaluation of the manuscript revision. YC contributed to the provided guidance of the whole study. All authors contributed to the article and approved the submitted version.
Acknowledgments
We extend our thanks to the editors and reviewers for their careful reading and helpful comments on this article. This work was supported by the National Natural Science Foundation of China (Grant No. 32201602), the Natural Science Fund Youth Project of Hunan Province (Grant No. 2021JJ41067), and the Outstanding Youth of the Education Department of Hunan Province (Grant No. 20B624).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1170723/full#supplementary-material
Supplementary Table 1 | Identification of TAG-related biosynthesis genes in woody oil plants
Supplementary Table 2 | Amino acid sequence of TAG-related biosynthesis genes in woody oil plants
Supplementary Figure 1 | The expression profiles of G3PAT genes in woody oil plants. The expression profiles were obtained by RNA-Seq. Blue and yellow colors indicated low-expression and high-expression, respectively. Red color suggested absolute high-expression.
Supplementary Figure 2 | The expression profiles of PAP genes in woody oil plants. The expression profiles were obtained by RNA-Seq. Blue and yellow colors indicated low-expression and high-expression, respectively. Red color suggested absolute high-expression.
References
Athenstaedt, K. (2021). Phosphatidic acid biosynthesis in the model organism yeast saccharomyces cerevisiae-a survey. Biochim. Biophys. Acta (BBA)-Molecular Cell Biol. Lipids 1866, 158907. doi: 10.1016/j.bbalip.2021.158907
Barreira, J. O. C. M., Casal, S., Ferreira, I. C. F. R., Oliveira, M. B. P. P., Pereira, J. A. (2009). Nutritional, fatty acid and triacylglycerol profiles of castanea sativa mill. cultivars: a compositional and chemometric approach. J. Agric. Food Chem. 57, 2836–2842. doi: 10.1021/jf803754u
Bates, P. D. (2012). The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front. Plant Sci. 3, 147. doi: 10.3389/fpls.2012.00147
Bates, P. D., Browse, J. (2011). The pathway of triacylglycerol synthesis through phosphatidylcholine in arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J. 68, 387–399. doi: 10.1111/j.1365-313X.2011.04693.x
Bhatt-Wessel, B., Jordan, T. W., Miller, J. H., Peng, L. (2018). Role of DGAT enzymes in triacylglycerol metabolism. Arch. Biochem. biophysics 655, 1–11. doi: 10.1016/j.abb.2018.08.001
Bi, Q., Zhao, Y., Du, W., Lu, Y., Gui, L., Zheng, Z., et al. (2019). Pseudomolecule-level assembly of the Chinese oil tree yellowhorn (Xanthoceras sorbifolium) genome. GigaScience 8, giz070. doi: 10.1093/gigascience/giz070
Bourgis, F., Kader, J.-C., Barret, P., Renard, M., Robinson, D., Robinson, C., et al. (1999). A plastidial lysophosphatidic acid acyltransferase from oilseed rape. Plant Physiol. 120, 913–922. doi: 10.1104/pp.120.3.913
Bradley, R. M., Duncan, R. E. (2018). The lysophosphatidic acid acyltransferases (acylglycerophosphate acyltransferases) family: one reaction, five enzymes, many roles. Curr. Opin. lipidology 29, 110–115. doi: 10.1097/MOL.0000000000000492
Buchanan, B. B., Gruissem, W., Jones, R. L. (2015). Biochemistry and molecular biology of plants (John wiley & sons).
Burgal, J., Shockey, J., Lu, C., Dyer, J., Larson, T., Graham, I., et al. (2008). Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol. J. 6, 819–831. doi: 10.1111/j.1467-7652.2008.00361.x
Cagliari, A., Margis, R., Dos Santos Maraschin, F., Turchetto-Zolet, A. C., Loss, G., Margis-Pinheiro, M. (2011). Biosynthesis of triacylglycerols (TAGs) in plants and algae. Int. J. Plant Biol. 2, e10–e10. doi: 10.4081/pb.2011.e10
Cagliari, A., Margis-Pinheiro, M., Loss, G., Mastroberti, A. A., De Araujo Mariath, J. E., Margis, R. (2010). Identification and expression analysis of castor bean (Ricinus communis) genes encoding enzymes from the triacylglycerol biosynthesis pathway. Plant Sci. 179, 499–509. doi: 10.1016/j.plantsci.2010.07.015
Cahoon, E. B., Carlson, T. J., Ripp, K. G., Schweiger, B. J., Cook, G. A., Hall, S. E., et al. (1999). Biosynthetic origin of conjugated double bonds: production of fatty acid components of high-value drying oils in transgenic soybean embryos. Proc. Natl. Acad. Sci. 96, 12935–12940. doi: 10.1073/pnas.96.22.12935
Cao, Y., Li, Y., Wang, L., Zhang, L., Jiang, L. (2022). Evolution and function of ubiquitin-specific proteases (UBPs): Insight into seed development roles in tung tree (Vernicia fordii). Int. J. Biol. Macromolecules. 221, 796–805. doi: 10.1016/j.ijbiomac.2022.08.163
Cao, Y., Liu, M., Long, H., Zhao, Q., Jiang, L., Zhang, L. (2020). Hidden in plain sight: Systematic investigation of leucine-rich repeat containing genes unveil the their regulatory network in response to fusarium wilt in tung tree. Int. J. Biol. Macromolecules 163, 1759–1767. doi: 10.1016/j.ijbiomac.2020.09.106
Cao, Y., Liu, W., Zhao, Q., Long, H., Li, Z., Liu, M., et al. (2019). Integrative analysis reveals evolutionary patterns and potential functions of SWEET transporters in euphorbiaceae. Int. J. Biol. macromolecules 139, 1–11. doi: 10.1016/j.ijbiomac.2019.07.102
Cao, Y., Mo, W., Li, Y., Li, W., Dong, X., Liu, M., et al. (2021). Deciphering the roles of leucine-rich repeat receptor-like protein kinases (LRR-RLKs) in response to fusarium wilt in the vernicia fordii (Tung tree). Phytochemistry 185, 112686. doi: 10.1016/j.phytochem.2021.112686
Carman, G. M., Henry, S. A. (2007). Regulation of lipid metabolism in yeast. Biochim. Biophys. Acta 1771, 239.
Carocha, V., Soler, M., Hefer, C., Cassan-Wang, H., Fevereiro, P., Myburg, A. A., et al. (2015). Genome-wide analysis of the lignin toolbox of eucalyptus grandis. New Phytol. 206, 1297–1313. doi: 10.1111/nph.13313
Cases, S., Smith, S. J., Zheng, Y.-W., Myers, H. M., Lear, S. R., Sande, E., et al. (1998). Identification of a gene encoding an acyl CoA: diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. 95, 13018–13023. doi: 10.1073/pnas.95.22.13018
Cases, S., Stone, S. J., Zhou, P., Yen, E., Tow, B., Lardizabal, K. D., et al. (2001). Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J. Biol. Chem. 276, 38870–38876. doi: 10.1074/jbc.M106219200
Chen, J., Li, J., Dong, W., Zhang, X., Tyagi, R. D., Drogui, P., et al. (2018). The potential of microalgae in biodiesel production. Renewable Sustain. Energy Rev. 90, 336–346. doi: 10.1016/j.rser.2018.03.073
Chitraju, C., Walther, T. C., Farese, J. R. V. (2019). The triglyceride synthesis enzymes DGAT1 and DGAT2 have distinct and overlapping functions in adipocytes. J. Lipid Res. 60, 1112–1120. doi: 10.1194/jlr.M093112
Coleman, R. A., Lee, D. P. (2004). Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43, 134–176. doi: 10.1016/S0163-7827(03)00051-1
Dahlqvist, A., Ståhl, U., Lenman, M., Banas, A., Lee, M., Sandager, L., et al. (2000). Phospholipid: diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. 97, 6487–6492. doi: 10.1073/pnas.120067297
Demirbas, A., Demirbas, M. F. (2011). Importance of algae oil as a source of biodiesel. Energy conversion Manage. 52, 163–170. doi: 10.1016/j.enconman.2010.06.055
Durrett, T. P., Benning, C., Ohlrogge, J. (2008). Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. doi: 10.1111/j.1365-313X.2008.03442.x
Elhiti, M., Yang, C., Chan, A., Durnin, D. C., Belmonte, M. F., Ayele, B. T., et al. (2012). Altered seed oil and glucosinolate levels in transgenic plants overexpressing the brassica napus SHOOTMERISTEMLESS gene. J. Exp. Bot. 63, 4447–4461. doi: 10.1093/jxb/ers125
Emms, D. M., Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16, 157. doi: 10.1186/s13059-015-0721-2
Fahs, Z., Rossez, Y., Guénin, S., Gutierrez, L., Thomasset, B., Perrin, Y. (2019). Cloning and molecular characterization of three lysophosphatidic acid acyltransferases expressed in flax seeds. Plant Sci. 280, 41–50. doi: 10.1016/j.plantsci.2018.10.026
Fan, J., Yan, C., Xu, C. (2013). Phospholipid: diacylglycerol acyltransferase-mediated triacylglycerol biosynthesis is crucial for protection against fatty acid-induced cell death in growing tissues of a rabidopsis. Plant J. 76, 930–942. doi: 10.1111/tpj.12343
França, M. G. C., Matos, A. R., D'arcy-Lameta, A., Passaquet, C., Lichtlé, C., Zuily-Fodil, Y., et al. (2008). Cloning and characterization of drought-stimulated phosphatidic acid phosphatase genes from vigna unguiculata. Plant Physiol. Biochem. 46, 1093–1100. doi: 10.1016/j.plaphy.2008.07.004
Gao, Y., Sun, Y., Gao, H., Chen, Y., Wang, X., Xue, J., et al. (2021). Ectopic overexpression of a type-II DGAT (CeDGAT2-2) derived from oil-rich tuber of cyperus esculentus enhances accumulation of oil and oleic acid in tobacco leaves. Biotechnol. Biofuels 14, 1–16. doi: 10.1186/s13068-021-01928-8
Gibbons, G. F., Islam, K., Pease, R. J. (2000). Mobilisation of triacylglycerol stores. Biochim. Biophys. Acta (BBA)-Molecular Cell Biol. Lipids 1483, 37–57. doi: 10.1016/S1388-1981(99)00182-1
Gidda, S. K., Shockey, J. M., Rothstein, S. J., Dyer, J. M., Mullen, R. T. (2009). Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol. Biochem. 47, 867–879. doi: 10.1016/j.plaphy.2009.05.008
Goode, J. H., Dewey, R. E. (1999). Characterization of aminoalcoholphosphotransferases from arabidopsis thaliana and soybean. Plant Physiol. Biochem. 37, 445–457. doi: 10.1016/S0981-9428(99)80049-7
Ha, J., Shim, S., Lee, T., Kang, Y. J., Hwang, W. J., Jeong, H., et al. (2019). Genome sequence of a non-edible biodiesel plant, provides a resource to improve seed-related traits. Plant Biotechnol. J. 17, 517–530. doi: 10.1111/pbi.12995
Hills, M. J., Roscoe, T. J. (2006). “"Synthesis of structural and storage lipids by the ER,",” in The plant endoplasmic reticulum (Springer).
Hsieh, H.-J., Su, C.-H., Chien, L.-J. (2012). Accumulation of lipid production in chlorella minutissima by triacylglycerol biosynthesis-related genes cloned from saccharomyces cerevisiae and yarrowia lipolytica. J. Microbiol. 50, 526–534. doi: 10.1007/s12275-012-2041-5
Ichihara, K. I., Asahi, T., Fujii, S. (1987). 1-Acyl-sn-glycerol-3-phosphate acyltransferase in maturing safflower seeds and its contribution to the non-random fatty acid distribution of triacylglycerol. Eur. J. Biochem. 167, 339–347. doi: 10.1111/j.1432-1033.1987.tb13342.x
Jako, C., Kumar, A., Wei, Y., Zou, J., Barton, D. L., Giblin, E. M., et al. (2001). Seed-specific over-expression of an arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 126, 861–874. doi: 10.1104/pp.126.2.861
Jiang, L., Lin, M., Wang, H., Song, H., Zhang, L., Huang, Q., et al. (2022). Haplotype-resolved genome assembly of bletilla striata (Thunb.) reichb.f. to elucidate medicinal value. Plant J. 115, 1340–1353. doi: 10.1111/tpj.15892
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., Mcanulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kocsis, M. G., Weselake, R. J. (1996). Phosphatidate phosphatases of mammals, yeast, and higher plants. Lipids 31, 785–802. doi: 10.1007/BF02522974
Körbes, A. P., Kulcheski, F. R., Margis, R., Margis-Pinheiro, M., Turchetto-Zolet, A. C. (2016). Molecular evolution of the lysophosphatidic acid acyltransferase (LPAAT) gene family. Mol. Phylogenet. Evol. 96, 55–69. doi: 10.1016/j.ympev.2015.12.001
Kornberg, A., Pricer, J. W. E. (1953). Enzymatic synthesis of the coenzyme a derivatives of long chain fatty acids. J. Biol. Chem. 204, 329–343. doi: 10.1016/S0021-9258(18)66142-3
Liang, Q., Li, H., Li, S., Yuan, F., Sun, J., Duan, Q., et al. (2019). The genome assembly and annotation of yellowhorn (Xanthoceras sorbifolium bunge). GigaScience 8, giz071. doi: 10.1093/gigascience/giz071
Lieb, V. M., Schuster, L. K., Kronmüller, A., Schmarr, H.-G., Carle, R., Steingass, C. B. (2019). Fatty acids, triacylglycerols, and thermal behaviour of various mango (Mangifera indica l.) kernel fats. Food Res. Int. 116, 527–537. doi: 10.1016/j.foodres.2018.08.070
Liu, T. H., Chyan, C. L., Li, F. Y., Chen, Y. J., Tzen, J. T. C. (2011). Engineering lysine-rich caleosins as carrier proteins to render biotin as a hapten on artificial oil bodies for antibody production. Biotechnol. Prog. 27, 1760–1767. doi: 10.1002/btpr.684
Liu, D., Ji, H., Yang, Z. (2020). Functional characterization of three novel genes encoding diacylglycerol acyltransferase (DGAT) from oil-rich tubers of cyperus esculentus. Plant Cell Physiol. 61, 118–129. doi: 10.1093/pcp/pcz184
Liu, Y., Wang, G., Wang, X. (2015b). Role of aminoalcoholphosphotransferases 1 and 2 in phospholipid homeostasis in arabidopsis. Plant Cell 27, 1512–1528. doi: 10.1105/tpc.15.00180
Liu, F., Xia, Y., Wu, L., Fu, D., Hayward, A., Luo, J., et al. (2015a). Enhanced seed oil content by overexpressing genes related to triacylglyceride synthesis. Gene 557, 163–171. doi: 10.1016/j.gene.2014.12.029
Lung, S.-C., Weselake, R. J. (2006). Diacylglycerol acyltransferase: a key mediator of plant triacylglycerol synthesis. Lipids 41, 1073–1088. doi: 10.1007/s11745-006-5057-y
Ma, Q., Sun, T., Li, S., Wen, J., Zhu, L., Yin, T., et al. (2020). The acer truncatum genome provides insights into nervonic acid biosynthesis. Plant J. 104, 662–678. doi: 10.1111/tpj.14954
Marmon, S., Sturtevant, D., Herrfurth, C., Chapman, K., Stymne, S., Feussner, I. (2017). Two acyltransferases contribute differently to linolenic acid levels in seed oil. Plant Physiol. 173, 2081–2095. doi: 10.1104/pp.16.01865
Martínez-García, P. J., Crepeau, M. W., Puiu, D., Gonzalez-Ibeas, D., Whalen, J., Stevens, K. A., et al. (2016). The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 87, 507–532. doi: 10.1111/tpj.13207
Mcmaster, C. R., Bell, R. M. (1997). CDP-choline: 1, 2-diacylglycerol cholinephosphotransferase. Biochim. Biophys. Acta (BBA)-Lipids Lipid Metab. 1348, 100–110. doi: 10.1016/S0005-2760(97)00097-0
Meesapyodsuk, D., Chen, Y., Ye, S., Chapman, R. G., Qiu, X. (2021). Co-Expressing eranthis hyemalis lysophosphatidic acid acyltranferase 2 and elongase improves two very long chain polyunsaturated fatty acid production in brassica carinata. Metab. Eng. Commun., 16, e00171. doi: 10.1016/j.mec.2021.e00171
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Mu, H., Porsgaard, T. (2005). The metabolism of structured triacylglycerols. Prog. Lipid Res. 44, 430–448. doi: 10.1016/j.plipres.2005.09.002
Nabizadeh, E., Taherifard, E., Gerami, F. (2011). Effect of pruning lateral branches on four varieties of medicinal castor bean plant (Ricinuscommunis l.) yield, growth and development. J. Medicinal Plants Res. 5, 5828–5834.
Nakamura, Y., Tsuchiya, M., Ohta, H. (2007). Plastidic phosphatidic acid phosphatases identified in a distinct subfamily of lipid phosphate phosphatases with prokaryotic origin. J. Biol. Chem. 282, 29013–29021. doi: 10.1074/jbc.M704385200
Neumann, G., Römheld, V. (1999). Root excretion of carboxylic acids and protons in phosphorus-deficient plants. Plant Soil 211, 121–130. doi: 10.1023/A:1004380832118
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Parajuli, S., Kannan, B., Karan, R., Sanahuja, G., Liu, H., Garcia-Ruiz, E., et al. (2020). Towards oilcane: Engineering hyperaccumulation of triacylglycerol into sugarcane stems. GCB Bioenergy 12, 476–490. doi: 10.1111/gcbb.12684
Perry, H. J., Bligny, R., Gout, E., Harwood, J. L. (1999). Changes in Kennedy pathway intermediates associated with increased triacylglycerol synthesis in oil-seed rape. Phytochemistry 52, 799–804. doi: 10.1016/S0031-9422(99)00294-0
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Rodionova, M. V., Poudyal, R. S., Tiwari, I., Voloshin, R. A., Zharmukhamedov, S. K., Nam, H. G., et al. (2017). Biofuel production: challenges and opportunities. Int. J. Hydrogen Energy 42, 8450–8461. doi: 10.1016/j.ijhydene.2016.11.125
Schläpfer, P., Zhang, P., Wang, C., Kim, T., Banf, M., Chae, L., et al. (2017). Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 173, 2041–2059. doi: 10.1104/pp.16.01942
Shi, L., Katavic, V., Yu, Y., Kunst, L., Haughn, G. (2012). Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J. 69, 37–46. doi: 10.1111/j.1365-313X.2011.04768.x
Shockey, J. M., Gidda, S. K., Chapital, D. C., Kuan, J.-C., Dhanoa, P. K., Bland, J. M., et al. (2006). Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18, 2294–2313. doi: 10.1105/tpc.106.043695
Singh, R., Ong-Abdullah, M., Low, E.-T. L., Manaf, M., Rosli, R., Nookiah, R., et al. (2013). Oil palm genome sequence reveals divergence of interfertile species in old and new worlds. Nature 500, 335–339. doi: 10.1038/nature12309
Slabas, A. R., Simon, J. W., Brown, A. P. (2001). Biosynthesis and regulation of fatty acids and triglycerides in oil seed rape. current status and future trends. Eur. J. Lipid Sci. Technol. 103, 455–466. doi: 10.1002/1438-9312(200107)103:7<455::AID-EJLT455>3.0.CO;2-U
Stobart, K., Mancha, M., Lenman, M., Dahlqvist, A., Stymne, S. (1997). Triacylglycerols are synthesised and utilized by transacylation reactions in microsomal preparations of developing safflower (Carthamus tinctorius L.) seeds. Planta 203, 58–66. doi: 10.1007/s00050165
Takeuchi, K., Reue, K. (2009). Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am. J. Physiol-Endocrinol And Metab. 296, E1195–E1209. doi: 10.1152/ajpendo.90958.2008
Unver, T., Wu, Z., Sterck, L., Turktas, M., Lohaus, R., Li, Z., et al. (2017). Genome of wild olive and the evolution of oil biosynthesis. Proc. Natl. Acad. Sci. 114, E9413–E9422. doi: 10.1073/pnas.1708621114
Voelker, T., Kinney, A. J. (2001). Variations in the biosynthesis of seed-storage lipids. Annu. Rev. Plant Biol. 52, 335–361. doi: 10.1146/annurev.arplant.52.1.335
Wang, N., Ma, J., Pei, W., Wu, M., Li, H., Li, X., et al. (2017). A genome-wide analysis of the lysophosphatidate acyltransferase (LPAAT) gene family in cotton: organization, expression, sequence variation, and association with seed oil content and fiber quality. BMC Genomics 18, 1–18. doi: 10.1186/s12864-017-3594-9
Wang, J., Singh, S. K., Geng, S., Zhang, S., Yuan, L. (2020). Genome-wide analysis of glycerol-3-phosphate O-acyltransferase gene family and functional characterization of two cutin group GPATs in brassica napus. Planta 251, 1–16. doi: 10.1007/s00425-020-03384-4
Waschburger, E., Kulcheski, F. R., Veto, N. M., Margis, R., Margis-Pinheiro, M., Turchetto-Zolet, A. C. (2018). Genome-wide analysis of the glycerol-3-Phosphate acyltransferase (GPAT) gene family reveals the evolution and diversification of plant GPATs. Genet. Mol. Biol. 41, 355–370. doi: 10.1590/1678-4685-gmb-2017-0076
Weiss, S. B., Kennedy, E. P., Kiyasu, J. Y. (1960). The enzymatic synthesis of triglycerides. J. Biol. Chem. 235, 40–44. doi: 10.1016/S0021-9258(18)69581-X
Wenrui, H., Jingwei, B., Bin, H. (2013). Trend and progress in global oil and gas exploration. Petroleum Explor. Dev. 40, 439–443. doi: 10.1016/S1876-3804(13)60055-5
Wickramarathna, A. D., Siloto, R. M. P., Mietkiewska, E., Singer, S. D., Pan, X., Weselake, R. J. (2015). Heterologous expression of flax PHOSPHOLIPID: DIACYLGLYCEROL CHOLINEPHOSPHOTRANSFERASE (PDCT) increases polyunsaturated fatty acid content in yeast and arabidopsis seeds. BMC Biotechnol. 15, 1–15. doi: 10.1186/s12896-015-0156-6
Williams, J. G., Mcmaster, C. R. (1998). Scanning alanine mutagenesis of the CDP-alcohol phosphotransferase motif of saccharomyces cerevisiaeCholinephosphotransferase. J. Biol. Chem. 273, 13482–13487. doi: 10.1074/jbc.273.22.13482
Xu, J., Francis, T., Mietkiewska, E., Giblin, E. M., Barton, D. L., Zhang, Y., et al. (2008). Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol. J. 6, 799–818. doi: 10.1111/j.1467-7652.2008.00358.x
Xu, C., Shanklin, J. (2016). Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu. Rev. Plant Biol. 67, 179–206. doi: 10.1146/annurev-arplant-043015-111641
Yang, Y., Benning, C. (2018). Functions of triacylglycerols during plant development and stress. Curr. Opin. Biotechnol. 49, 191–198. doi: 10.1016/j.copbio.2017.09.003
Yang, L., Ding, Y., Chen, Y., Zhang, S., Huo, C., Wang, Y., et al. (2012). The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J. Lipid Res. 53, 1245–1253. doi: 10.1194/jlr.R024117
Yu, B., Wakao, S., Fan, J., Benning, C. (2004). Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in arabidopsis. Plant Cell Physiol. 45, 503–510. doi: 10.1093/pcp/pch064
Zhang, L., Liu, M., Long, H., Dong, W., Pasha, A., Esteban, E., et al. (2019). Tung tree (Vernicia fordii) genome provides a resource for understanding genome evolution and improved oil production. Genomics Proteomics Bioinf. 17, 558–575. doi: 10.1016/j.gpb.2019.03.006
Keywords: woody oil plants, lipid biosynthesis, triacylglycerol pathway, seeds, expression
Citation: Cao Y, Li Q and Zhang L (2023) The core triacylglycerol toolbox in woody oil plants reveals targets for oil production bioengineering. Front. Plant Sci. 14:1170723. doi: 10.3389/fpls.2023.1170723
Received: 21 February 2023; Accepted: 21 March 2023;
Published: 03 April 2023.
Edited by:
Lixin Wang, Hebei Agricultural University, ChinaReviewed by:
Zishan Ahmad, Nanjing Forestry University, ChinaSafina Naz, Bahauddin Zakariya University, Pakistan
Norihiro Sato, Tokyo University of Pharmacy and Life Sciences, Japan
Copyright © 2023 Cao, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunpeng Cao, eGZjeXBlbmdAMTI2LmNvbQ==; Lin Zhang, bHpoYW5nc3NAbXNuLmNvbQ==
 Yunpeng Cao
Yunpeng Cao Qiang Li1
Qiang Li1 Lin Zhang
Lin Zhang