- 1Ministry of Agriculture and Rural Affairs (MARA) Key Laboratory of Sustainable Crop Production in the Middle Reaches of the Yangtze River (Co-Construction by Ministry and Province)/Engineering Research Center of Ecology and Agricultural Use of Wetland, Ministry of Education, Hubei Collaborative Innovation Center for Grain Industry, College of Agriculture, Yangtze University, Jingzhou, China
- 2Nanchong Academy of Agriculture Sciences, Nanchong, Sichuan, China
- 3Hubei Key Laboratory of Quality Control of Characteristic Fruits and Vegetables, Hubei Engineering University, Xiaogan, Hubei, China
- 4College of Plant Protection, Northwest A&F University, Yangling, Shaanxi, China
L-aspartate oxidase (AO) is the first enzyme in NAD+ biosynthesis and is widely distributed in plants, animals, and microorganisms. Recently, AO family members have been reported in several plants, including Arabidopsis thaliana and Zea mays. Research on AO in these plants has revealed that AO plays important roles in plant growth, development, and biotic stresses; however, the nature and functions of AO proteins in wheat are still unclear. In this study, nine AO genes were identified in the wheat genome via sequence alignment and conserved protein domain analysis. These nine wheat AO genes (TaAOs) were distributed on chromosomes 2, 5, and 6 of sub-genomes A, B, and D. Analysis of the phylogenetic relationships, conserved motifs, and gene structure showed that the nine TaAOs were clustered into three groups, and the TaAOs in each group had similar conserved motifs and gene structure. Meanwhile, the subcellular localization analysis of transient expression mediated by Agrobacterium tumetioniens indicated that TaAO3-6D was localized to chloroplasts. Prediction of cis-elements indicated that a large number of cis-elements involved in responses to ABA, SA, and antioxidants/electrophiles, as well as photoregulatory responses, were found in TaAO promoters, which suggests that the expression of TaAOs may be regulated by these factors. Finally, transcriptome and real-time PCR analysis showed that the expression of TaAOs belonging to Group III was strongly induced in wheat infected by F. graminearum during anthesis, while the expression of TaAOs belonging to Group I was heavily suppressed. Additionally, the inducible expression of TaAOs belonging to Group III during anthesis in wheat spikelets infected by F. graminearum was repressed by ABA. Finally, expression of almost all TaAOs was induced by exposure to cold treatment. These results indicate that TaAOs may participate in the response of wheat to F. graminearum infection and cold stress, and ABA may play a negative role in this process. This study lays a foundation for further investigation of TaAO genes and provides novel insights into their biological functions.
1 Introduction
L-aspartate oxidase (AO), a kind of flavin oxidase, converts aspartate to iminoaspartic acid using either molecular oxygen or fumarate as electron acceptors. It plays an indispensable role in the biosynthesis of nicotinamide adenine dinucleotide (NAD+) (Mattevi et al., 1999). NAD+ biosynthesis consists of five steps, of which the first is oxidation of L-aspartate into iminoaspartate catalyzed by L-aspartate oxidase (Katoh et al., 2006). NAD+ is an important component of the respiratory chain, so it plays an important role in biological energy metabolism. It additionally participates in reduction–oxidation reactions, DNA repair, ADP-ribosylation, and a series of metabolic processes (Gakiere et al., 2018). L-aspartate oxidase, as the enzyme of the first reaction in de novo synthesis of NAD+, is also thought to play an important role in the energy metabolism system and other metabolic pathways in organisms. Therefore, AO has been researched extensively over the years.
AO was initially reported in Escherichia coli, where the B protein of quinolinic synthase was identified as a L-aspartate oxidase (Nasu et al., 1982). Later, two conserved domains, the FAD binding domain and Succ_DH_flav_C, were found in AO proteins (Mattevi et al., 1999). Subsequently, AO was identified in Pyrococcu shorikishii OT-3 (Sakuraba et al., 2002), Sulfolobus tokodaii (Sakuraba et al., 2008), Bacillus subtilis (Marinoni et al., 2008), and Pseudomonas putida (Leese et al., 2013). Since then, the physiological and biochemical properties of AO have been extensively investigated in bacteria (Mortarino et al., 1996; Bifulco et al., 2013; Armenia et al., 2017; Chow et al., 2017). AO has the following two features: (a) in vitro, it is able to use different electron acceptors, such as oxygen, fumarate, cytochrome c, and quinones, suggesting that it is involved in NAD biosynthesis in anaerobic as well as aerobic conditions (Tedeschi et al., 1996); and (b) the primary and tertiary structures are not similar to those of other flavo-oxidases, but rather are similar to those of the flavoprotein subunit of the succinate dehydrogenase/fumarate reductase class of enzymes. AO can reduce fumarate and oxidize L-aspartate, but cannot oxidize succinate (Bacchella et al., 1999; Tedeschi et al., 2010). It has been reported that Shigella, a nicotinic acid auxotroph, is unable to synthesize NAD via the de novo pathway due to AO gene mutations. When AO function is restored in Shigella, sustained loss of virulence and inability to invade host cells are observed, which points toward AO as a locus of antivirulence (Prunier et al., 2007).
Compared with microorganisms, little is still known about AO in plants. Up to this point, AO proteins have been reported only in maize and A. thaliana. In maize, a gene GRMZM2G139689 has been reported to encode AO protein. At the mononuclear stage of microspore development, the expression level of this gene was found to be greatly downregulated in male sterile line C48-2 compared with maintainer line 48-2 (Dong, 2019). This suggests that AO protein may be involved in pollen abortion in maize. In A. thaliana, At5g14760 has been identified as an AO (Macho et al., 2012). Overexpression of AtAO increases NAD+ content, and loss of AtAO activity results in a decrease in NAD+ levels (Hao et al., 2018). It is worth noting that the expression of AOs is upregulated in A. thaliana leaves infected by avirulent Pseudomonas syringae pv. tomato strain (Petriacq et al., 2012). Furthermore, research with an AO A. thaliana mutant has shown that AO is required for reactive oxygen species (ROS) bursts triggered by pathogen-associated molecular patterns and for stomatal immunity (Macho et al., 2012). These studies indicate that AO genes play important roles in regulating plant development and response to biotic stresses.
Wheat is one of the top three crops worldwide (Guo et al., 2018). Almost 60% of the wheat produced globally is consumed as food (Sobolewska et al., 2020), and global demand for wheat is expected to grow by approximately 70% over the next 30 years with growing populations, rising income levels, and increasing household consumption (Abedi and Mojiri, 2020). Wheat often suffers from exposure to biotic and abiotic stresses during growth; it is unclear whether wheat AO participates in the plant’s response to biotic/abiotic stresses, and the molecular characteristics of wheat AO are also unclear. In this study, TaAO genes were identified via sequence alignment and protein domain analysis, and the gene structure, phylogenetic relationships, and chromosome distribution of TaAO family genes were subsequently analyzed systematically using bioinformatics methods. Finally, the expression patterns of TaAO family genes were quantified via transcriptome analysis and qRT-PCR. This study lays a foundation for further analysis of AO genes in wheat.
2 Materials and methods
2.1 Genome-wide identification of AO genes in T. aestivum, Ae. tauschii, T. urartu, and T. dicoccoides
Genome data for T. aestivum (IWGSCv2.1), Ae. tauschii (v4.0.43), T. urartu (v1.43), and T. dicoccum (v1.0.43) were downloaded from Ensembl Plants database (http://plants.ensembl:index.html). Hidden Markov models (HMMs) for the FAD binding domain (PF00890.27) and the Succ_DH_flav_C domain (PF02910.23), obtained from the Pfam database (http://pfam.xfam), were used as queries to identify wheat AOs using HMMER3.0 (http://hmmer:download.html) with hit sequences specified as those with an e-value below 1e-5. Three Arabidopsis AOs (AtAOs), three maize AOs (ZmAOs), and two rice AOs (OsAOs), where the identification method was similar to that of TaAOs, were retrieved from genome databases for Arabidopsis (http://www.arabidopsis:index.jsp), maize (https://www.maizegdb.org), and rice (http://rice.plantbiology.msu.edu), respectively (Table S1). These AO proteins were used as queries to search for AO proteins in the genomes of T. aestivum, Ae. tauschii, T. urartu, and T. dicoccoides via BLASTp. Hit sequences with an e-value below 1e-5 were retained. The wheat AO candidates obtained using the above two methods were combined, and the non-redundant proteins were further analyzed using Pfam (v31.05) (http://pfam.sanger.ac.uk/search) and SMART (http://smart.embl-heidelberg.de/) (Letunic and Bork, 2018). Only these proteins that contained both the FAD binding domain and the Succ_DH_flav_C domain were considered to be wheat AOs. The AO genes were named according to their distribution on the chromosomes.
2.2 Characteristics of TaAO proteins
The protein sequence length, isoelectric point (pI), molecular weight (MW), instability index, and grand average of hydropathicity (GRAVY) of TaAOs were predicted using the ExPASy online tool (https://www.expasy.org/) (Gasteiger et al., 2003). Subcellular localization of TaAOs was predicted using the WoLF PSORT online tool (https://wolfpsort.hgc.jp/) (Chou and Shen, 2010).
2.3 Phylogenetic analysis of TaAOs
The sequences of AO proteins from A. thaliana, rice, maize, T. aestivum, Ae. tauschii, T. urartu, and T. dicoccoides were aligned using the ClustalW2 software package (Thompson et al., 1994). A neighbor-joining (NJ) phylogenetic tree was constructed using the MEGA X software package (Mega Limited, Auckland, New Zealand) (Kumar et al., 2018) with 1000 bootstrap repetitions. Finally, the tree was modified using the Interactive Tree of Life tool (iTOL, v6, http://itol.embl.de) (Letunic and Bork, 2021).
2.4 Genomic organization of TaAOs in wheat
Information on the position of TaAOs in wheat chromosomes was extracted from annotated information on the wheat genome. The physical map was drawn using the MapInspect software package. Information on the exon–intron structure of TaAO genes was visualized using the TBtools software package (Chen et al., 2020). Conserved motifs of TaAOs were identified using the MEME suite, with the following parameter settings: number of motifs, up to 15; width range, from 6 to 50 amino acids. The outputs on the motif structures of TaAO proteins were displayed using TBtools. Gene duplication events were analyzed according to the method described by Panchy et al. (2016) and illustrated using the Circos package in TBtools. For further examination of the footprints of selection during the processes of domestication (wild emmer and Ae. tauschii versus landraces) and improvement (landraces versus varieties), we overlapped the identified AO genes with the sweep region identified by Cheng et al. (2019) to check whether they were selected. The Ka and Ks values and the Ka/Ks ratio were calculated using TBtools. A Ka/Ks value of 1 indicates a neutral selection effect; a Ka/Ks value >1 indicates positive selection for evolutionary acceleration; and Ka/Ks <1 indicates purifying selection under function constraints.
2.5 RNA isolation and cDNA first-strand synthesis
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Subsequently, 1 μg RNA was used for cDNA first-strand synthesis using a PrimeScript RT reagent kit with gDNA Eraser (Takara, China) according to the manufacturer’s instruction.
2.6 Subcellular localization analysis of TaAO3-6D protein
Subcellular localization analysis of a TaAO protein was performed using an Agrobacterium tumefaciens mediated transient expression system in leaves of Nicotiana benthamiana. First, the coding sequence of a TaAO, TaAO3-6D, was amplified (the primers are listed in Table S2) and cloned into a GFP fusion protein expression vector pCAMBIA1300-GFP. Next, the recombinant vector was transformed into an Agrobacterium tumefaciens GV3101 strain. Positive clones were cultured and injected into the leaves of 5- to 6-week-old Nicotiana benthamiana. These were observed using a fluorescence microscope (Olympus FV3000, Tokyo, Japan) 48 h after injection.
2.7 Cis-acting element and protein interaction network analyses of TaAOs
The 1.5 kb sequence upstream of the start codon of each of the TaAO genes was obtained, and these sequences were used for the prediction of cis-acting elements via the PlantCARE website (http://bioinformatics.psb.ugent.be/webto-ols/plantcare/html/). The cis-acting elements were arranged and displayed using the R software package “pheatmap” (Rombauts et al., 1999). To study the protein–protein interactions (PPIs) between TaAOs and other proteins, a protein network was generated using the STRING v11.5 webserver (https://cn.string-db.org/).
2.8 Expression profiling of TaAO genes via transcriptome analysis
Transcriptome data on sequences involved in wheat growth and wheat responses to biotic stresses (Fusarium graminearum, stripe rust, and wheat powdery mildew) and abiotic stresses (phosphorous starvation, cold, heat, and drought) were downloaded from the NCBI database (Table S3) and mapped to the wheat reference genome via Hisat2 (Fang et al., 2020). The expression levels of TaAOs were calculated using Cufflinks (Trapnell et al., 2012). All transcript values were standardized by log2 (TPM + 1) transformation, and the expression profiles of TaAOs were generated using the R package “pheatmap”.
2.9 Growth and stress treatments of wheat seedlings
Jingshuang 16, a wheat cultivar moderately susceptible to powdery mildew and rust (Ren et al., 2015), was used in this study. The wheat seeds were disinfected with 1% hydrogen peroxide; subsequently, after washing with distilled water, the seeds were kept at 25°C for 2 days for germination. The seedlings were cultured in quarter-strength Hoagland nutrient solution for 3 days, and then transferred to one-half Hoagland nutrient solution (pH=6.0) (Bishop and Bugbee, 1998). To examine the differential expression of TaAOs in response to drought and abscisic acid (ABA) treatment, seedlings were cultured in a greenhouse at 25/20°C under a 16 h light/8 h dark cycle. The seedlings were treated with 20% PEG6000, once at the heart stage and once at the leaf stage. Wheat leaves were sampled at 0, 2, 12, 24, 36, and 48 h after treatment. For the ABA treatment, ABA was added to one-half strength Hoagland nutrient solution at a final concentration of 100 μM. After 0, 2, 6, 12, 24, and 48 h of treatment, leaves were harvested for further research. Finally, for the stripe rust infection treatment, seedlings were cultured in a plant growth chamber under a 16 h light/8 h dark cycle at 16°C. Wheat leaves were inoculated with fresh uredospores of stripe rust CYR32 using the smearing method and kept in dark, moist conditions for 24 hours to promote infection (Santra et al., 2008). Leaves were collected at 0, 6, 12, 24, and 48 h. All samples were immediately frozen in liquid nitrogen and stored at -80°C for future use.
2.10 qRT-PCR analysis
Real-time PCR reaction systems were used to carry out reaction schemes following the manufacturer’s instruction (Vazyme, China). Gene-specific primers (Table S4) were designed using the Primer 5.0 software package. The ADP-ribosylation factor Ta2291 was used as the internal reference gene for qRT-PCR analysis. Each experiment was carried out with three biological replicates, and three technical repeats were performed for each replicate.
3 Results
3.1 Identification and classification of AO genes in wheat
Ten AO candidates were obtained from the wheat genome via HMM search. Meanwhile, the same ten AO candidates were retrieved from the wheat genome via BLASTp. Of these, one gene without the Succ_DH_flav_C domain was excluded, and the remaining nine candidate genes containing both of the FAD binding domain and the Succ_DH_flav_C domain were identified as TaAOs (Table 1; Table S5). Using the same procedure, four, three, and six AOs were identified for T. urartu, Ae. Tauschii, and T. dicoccoides, respectively (Table S4). The locations of AO genes on the wheat chromosomes were determined and analyzed for genomic homology; this analysis indicated that nine TaAO genes were distributed evenly on chromosomes 2, 5, and 6 of sub-genomes A, B, and D, while no tandem duplication or segmental duplication events were found (Figure 1).
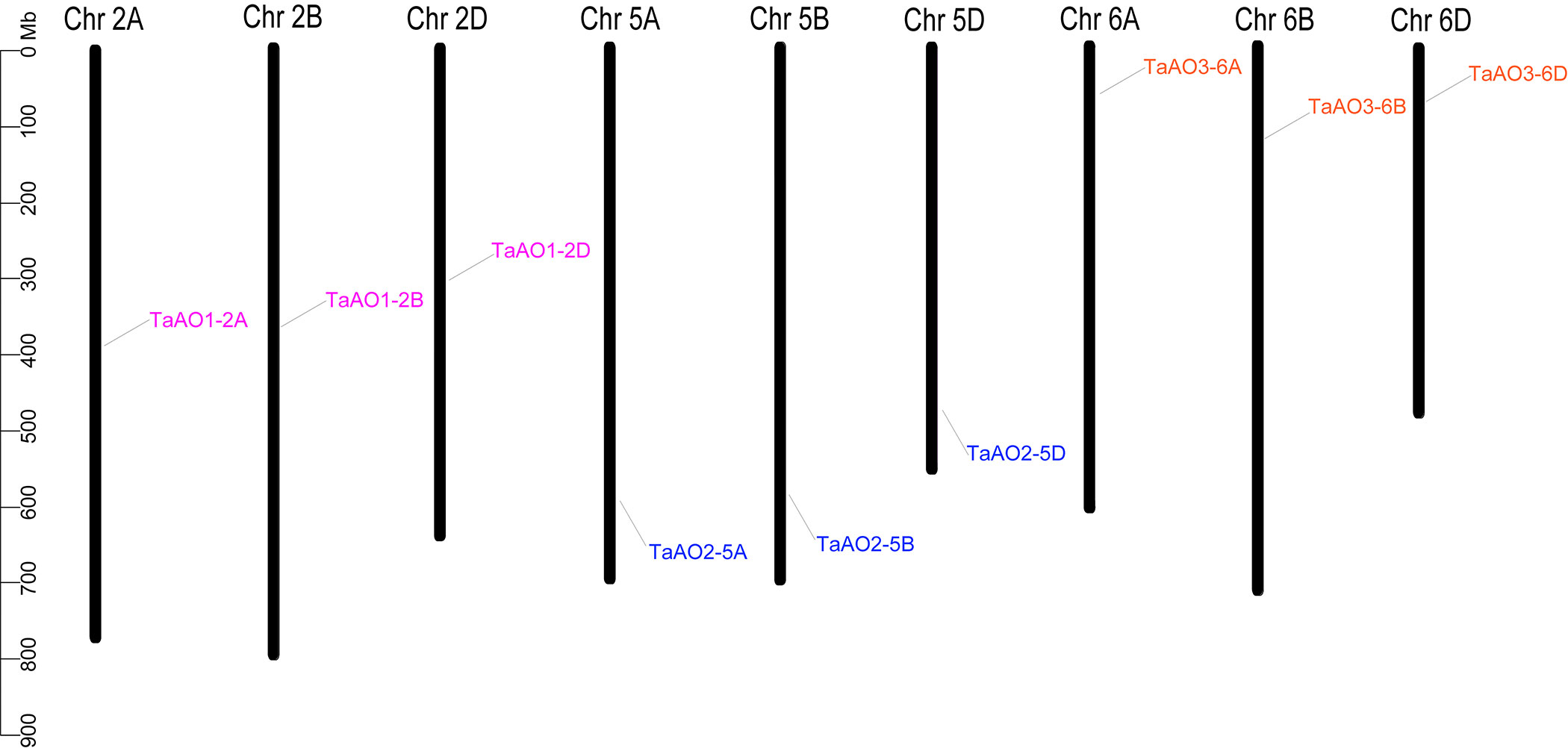
Figure 1 Chromosome locations of the nine TaAOs in wheat. The ruler on the left represents the physical distance between genes (Mbp).
3.2 Analysis of TaAO protein characteristics
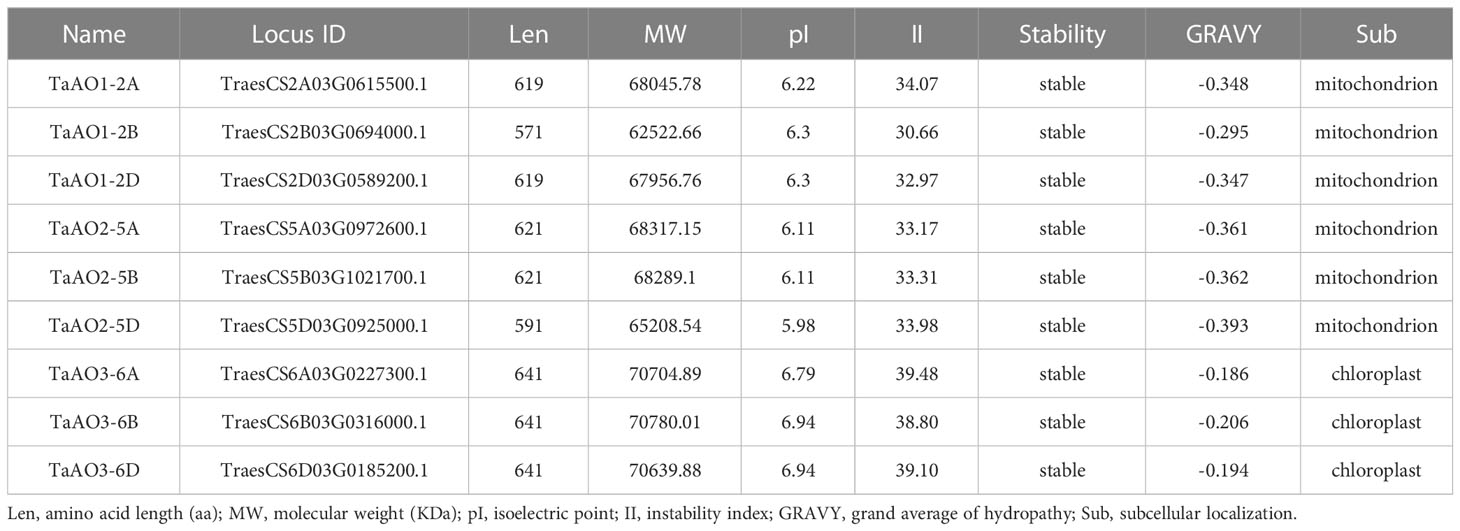
To further understand the characteristics of the TaAO proteins, protein length, molecular weight, instability index, isoelectric points, average hydrophilicity coefficients, and predicted subcellular localization were analyzed. As shown in Table 1, the protein length of the TaAOs ranged from 571 to 641 aa, and molecular weight ranged from 62.5 to 70.7 kDa. Instability index ranged from 30.66 to 39.48, indicating that these TaAOs were all stable proteins (instability index < 40). The isoelectric points of these TaAOs fell between 5.98 and 6.94, which showed that they were acidic proteins. Their average hydrophilicity coefficients ranged from 0.186 to 0.393, indicating that they were hydrophilic proteins. Subcellular localization prediction via the WoLF PSORT software package indicated that TaAO3-6A, TaAO3-6B, and TaAO3-6D were localized to chloroplasts, while TaAO1-2A, TaAO1-2B, TaAO3-3D, TaAO2-5A, TaAO2-5B, and TaAO2-5D were localized to mitochondria. The results of TaAO3-6D-GFP fusion protein expression assays showed that TaAO3-6D was localized to chloroplasts (Figure 2), which was consistent with the predicted results.
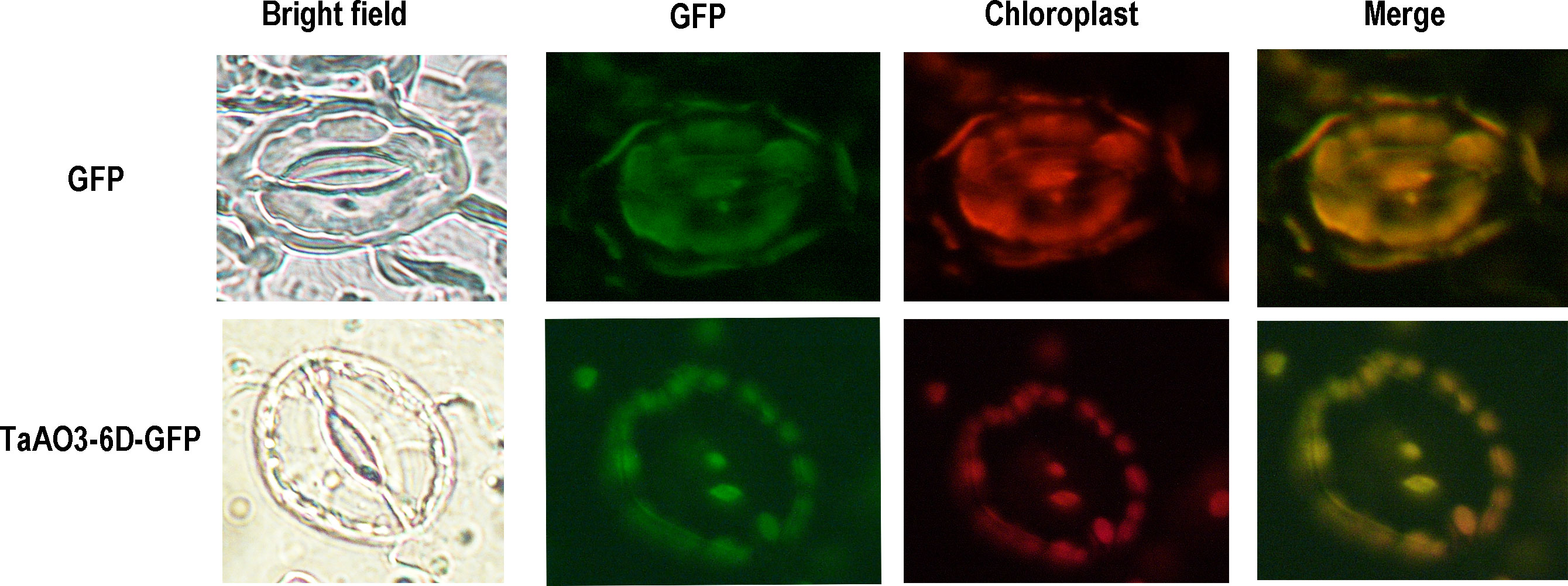
Figure 2 GFP-fused proteins of TaAO3-6D were localized to the chloroplasts of Arabidopsis guard cells. Chloroplast autofluorescence (red) and GFP fluorescence (green) in guard cells were simultaneously observed. Bright-field images (on the left) show the shapes of guard cells. Unfused GFP was found in the cytoplasm and nucleus, whereas TaAO3-6D-GFP was found in the chloroplasts. Scale bar = 10 μm.
3.3 Conserved motifs and gene structures of TaAOs
To understand the evolutionary relationships between TaAOs, a phylogenetic tree was constructed. As shown in Figure 3A, three TaAOs located on chromosome 2 were grouped into Group I, three TaAOs located on chromosome 5 were grouped into Group II, and the three genes located on chromosome 6 were clustered into Group III. The results of a conserved motifs analysis of the TaAOs (Figure 3B; Table S6) showed that all TaAO proteins contained Motifs 1, 2, 3, 5, 7, 8, 10, 11, and 12. Motifs 7, 8, and 10 were contained in the Succ_DH_flav_C domain and Motifs 1, 2, 3, 5, 11, and 12 were contained in the FAD binding domain. The number and order of motifs in TaAOs belonging to Groups I and II were essentially consistent, with the exceptions of TaAO1-2B lacking Motif 4 and TaAO2-5D lacking Motif 13. The number of motifs of Group III members differed from that of members of Groups I and II. TaAOs in Group III only had 13 motifs, from which Motifs 13 and 15 were absent. Furthermore, the order of motifs in Group III also differed from that of the other two groups. Additionally, the results on intron/exon distribution patterns of TaAO genes appeared to indicate that the number of exons was significantly greater in Groups I and II than in Group III. Generally, TaAOs belonging to Groups I and II contained 16 exons, with the exception of TaAO1-2D, which only contained 15 exons. In contrast, the TaAOs belonging to Group III only contained seven exons (Figure 3C).
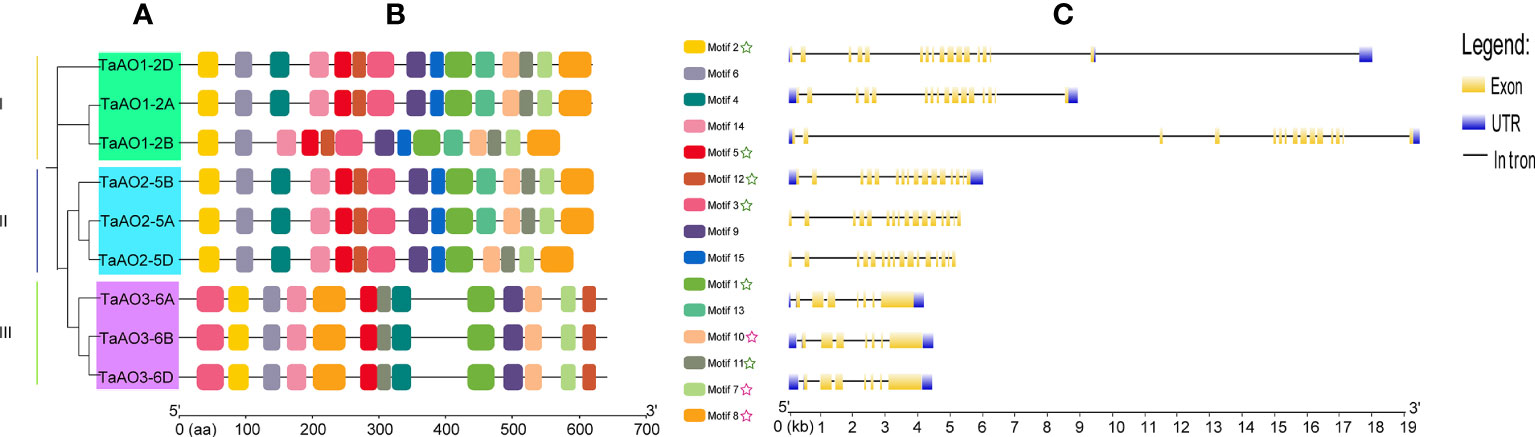
Figure 3 Phylogenetic analysis, conserved motifs, and gene structure of TaAOs. (A) Phylogenetic tree of TaAOs. The tree was built using the neighbor-joining (NJ) method with 1000 bootstrap repetitions in MEGA7. (B) Motifs of TaAO identified using MEME; MAST was used to visualize the patterns. Each motif is indicated with a specific color. The green star symbol represents the FAD binding domain; the purple star symbol represents the Succ_DH_flav_C domain. (C) Exon–intron structure of TaAOs, analyzed using GSDS. Untranslated regions (UTRs) are represented by blue frames; exons are represented by yellow frames; and introns are represented by black lines.
3.4 Phylogenetic and Ka/Ks analysis of AO genes of T. aestivum and its ancestor species
To further evaluate the phylogenetic relationships of TaAOs with other plant AOs, nine AOs from T. aestivum, four from T. urartu, three from Ae. Tauschii, six from T. dicoccoides, two from Oryza sativa, three from Zea mays, and three from A. thaliana were used to construct a phylogenetic tree. As shown in Figure 4, the set of all AOs could be divided into three groups, which was consistent with the above-described results on the phylogenetic tree of wheat AOs. TaAOs located on chromosome 2 in different sub-genomes were classified into Group I, TaAOs located on chromosome 5 were in Group II, and TaAOs on chromosome 6 were in Group III (Table S7). It is interesting that all three AtAOs were clustered into Group III, which was found to have the closest relationship with TaAOs on chromosome 6. The AOs of rice and Zea mays were classified into Groups I and III, with none falling into Group II. With regard to sub-genome donor species, members were distributed across the three groups. These results indicate that AOs in these species may have evolved under different evolutionary directions, and furthermore, the functions of AOs in different groups may be at variance with one another.
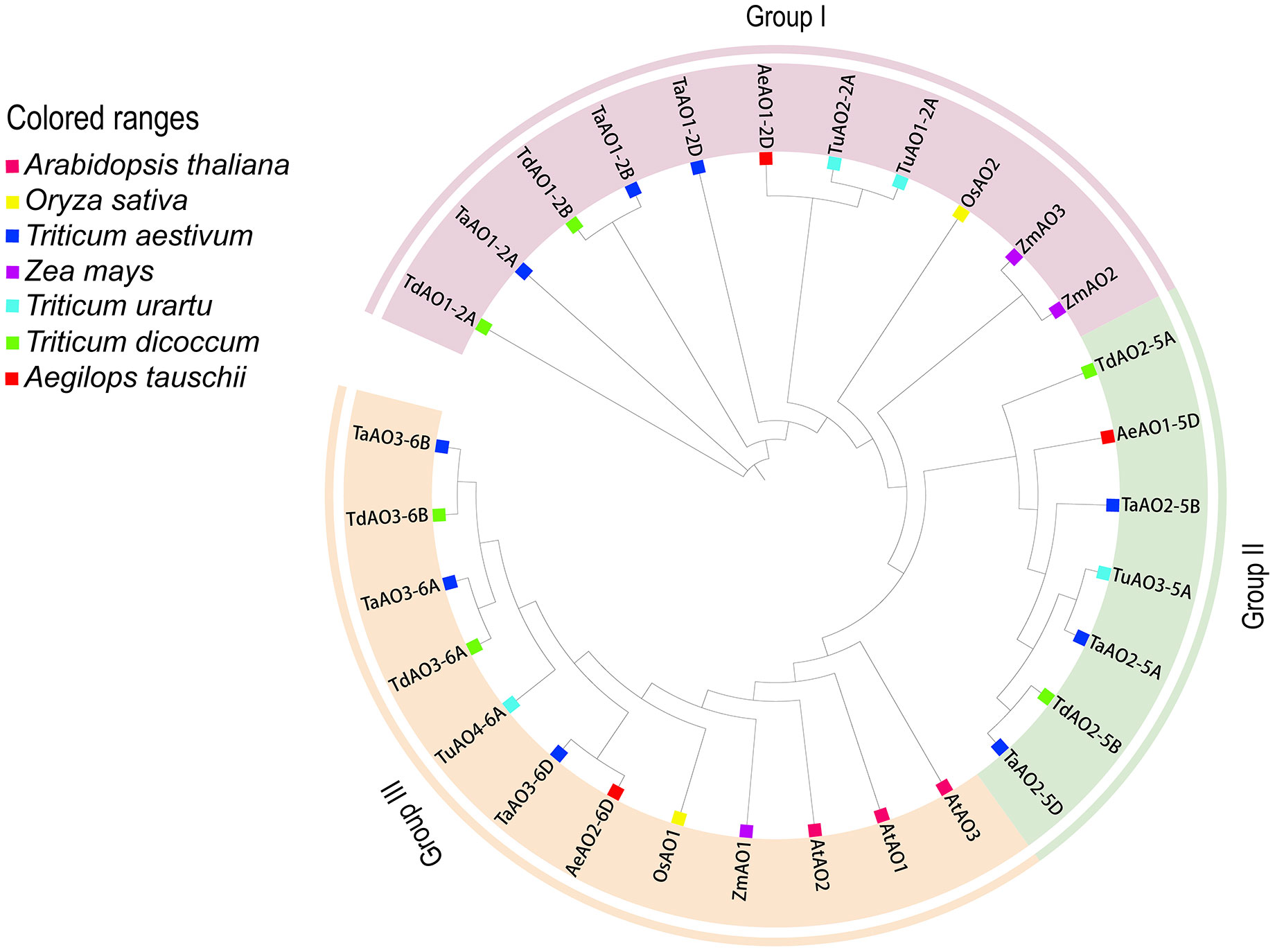
Figure 4 Phylogenetic tree of AOs from wheat, maize, rice, Arabidopsis, T. urartu, T. dicoccoides, and Ae. tauschii. The protein sequences were aligned using ClustalW2, and the phylogenetic tree was constructed using neighbor-joining (NJ) (1000 repeats) in the MEGA 7.0 software package. The AOs of wheat, maize, rice, Arabidopsis, T. urartu, T. dicoccoides, and Ae. tauschii can be distinguished by the use of different colors and shapes.
A homology analysis of wheat and three sub-genome donor species was conducted, and the orthologs and paralogs were clustered. Orthologs are defined as genes in different species that are derived from a single gene in the last common ancestor, and paralogs are homologous genes within a single species, resulting from gene duplication (Remm et al., 2001). A total of 53 homologous gene pairs were associated with T. aestivum (Figure 5A), of which nine were paralog gene pairs and 44 were ortholog gene pairs were found. Among these 44 ortholog gene pairs, there were 7, 22, and 15 ortholog genes between T. aestivum and each of its ancestor species (Ae. Tauschii, T. dicoccoides, and T. Urartu, respectively). TaAO genes the AOs of Ae. tauschii, T. dicoccoides, T. urartu and T. aestivum can be divided into three groups (Figure 5B) were not found in either the domestication-related or the improvement-related sweep regions, and all the Ka/Ks values for AO replication gene pairs were <1 (Figure 5C; Table S8), indicating that TaAO genes were purified and selected, and their functions may be conserved.
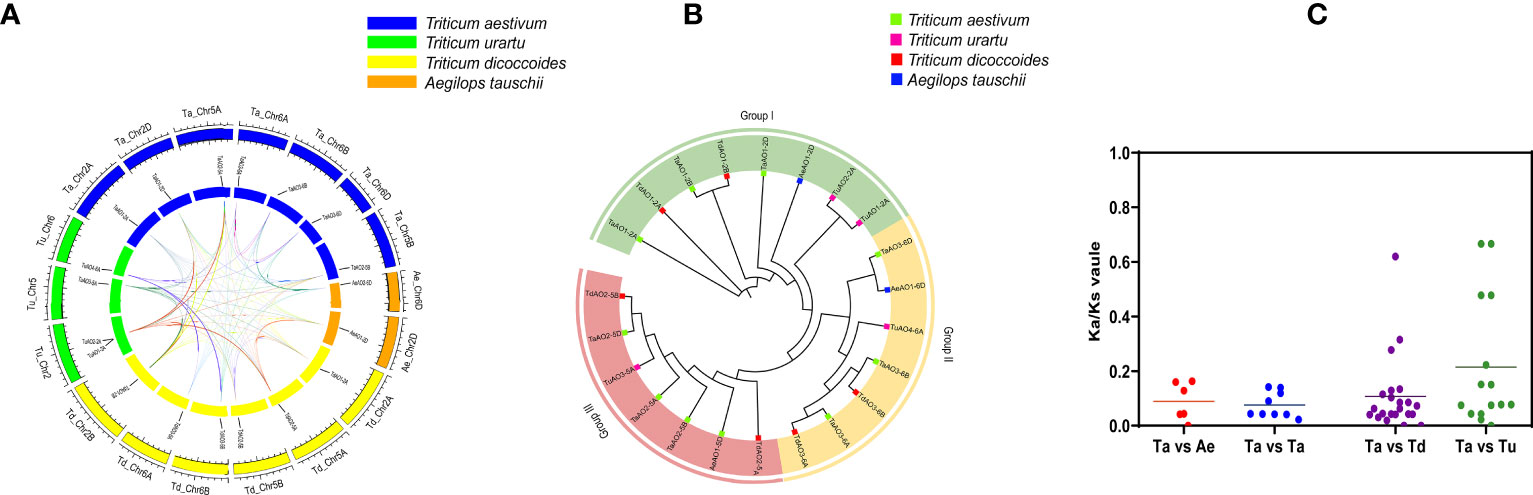
Figure 5 Analysis of (A) synteny and (B) phylogeny for AO genes in T. aestivum and its sub-genomic progenitors T. urartu, T. dicoccoides, and Ae. Tauschii. (A) Orange rectangles represent Ae. tauschii chromosomes, green rectangles represent T. urartu chromosomes, blue rectangles represent T. aestivum chromosomes, and yellow rectangles represent T. dicoccoides chromosomes. (B) This phylogenetic tree was constructed using 1000 bootstrap repetitions under the neighbor-joining (NJ) method in MEGA7. Blue, red, purple, and green squares represent Ae. Tauschii, T. dicoccoides, T. urartu, and T. aestivum, respectively. (C) Ka/Ks values for AO orthologous gene pairs between T. aestivum, T. urartu, T. dicoccoides, and Ae. Tauschii.
3.5 Cis-element analysis of TaAO genes
In the process of plant growth and development, not only can cis-regulatory elements regulate the spatio-temporal expression of genes, but they also are involved in responses to phytohormone exposure and abiotic stresses (Davis, 2009). In the present study, 39 kinds of cis-elements were identified in the promoter regions of AO genes in wheat (Figure 6; Table S9). Cis-elements involved in growth and development, including the TATA box and CAAT box, were found in all TaAO promoters. For cis-elements associated with plant hormone responses, the largest set of cis-elements was that of abscisic acid response elements (ABRE), of which 13 were identified. The second largest sets of cis-elements were salicylic acid-associated elements (CGTCA motif and TCA elements); the number of elements identified for each of these was 10. This suggests that the expression of TaAOs may be regulated by ABA and SA. The TGACG motif (methyl jasmonate), TGA element (auxin), CGTCA motif, and TCA element (salicylic acid) were identified in most of the TaAO gene promoters. In terms of biotic and abiotic stresses, the most abundant cis-element was the G-box, with 14 elements identified, followed by ARE, with 11. This suggests that the expression of TaAOs may be regulated by antioxidant/electrophile and photoregulatory factors. In summary, in addition to biotic stress regulation of AO, AO may also be regulated by ABA and SA hormones, as well as antioxidant/electrophile and photoregulatory abiotic stresses.
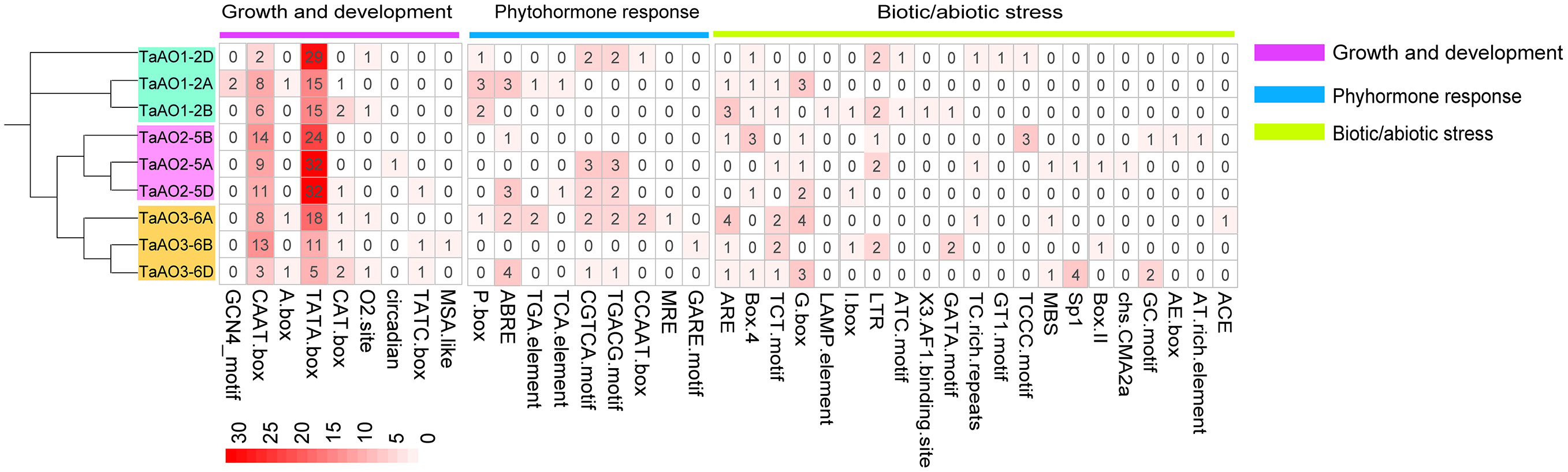
Figure 6 Identification of cis-acting elements of TaAO genes. The color gradient in each grid represents the number of promoter elements of these AO genes.
3.6 Molecular interaction networks
A network of the interactions between TaAOs and other wheat proteins was built using STRING v11.0. The results showed that all nine TaAO proteins interacted with 17 wheat proteins. Among these 17 wheat proteins, seven (Traes_5BL_402FB4A3F.1, Traes_7AL_C0609756B.1, Traes_7BL_8F49CE9D6.2, Traes_2AS_4AB51ACE2.1, Traes_2AS_BA55E613D.2, Traes_7AL_6405AB56F.2, Traes_4AS_2DCA42965.1) were unknown proteins, and the remaining ten were Succinate-CoA ligases (Traes_2DS_3AC11B9D8.1, Traes_2AS_3BA916807.1, Traes_2BS_8863D42E7.1), 4Fe-4S ferredoxin-type protein (Traes_5DL_885A58CBA.3), lactate/malate dehydrogenase (Traes_1BL_A93F9F079.2, Traes_1DL_5A31A68D3.2, Traes_1AL_2EC98608D.2), malate dehydrogenase (Traes_1BL_BD3E22844.1), succinate dehydrogenase (Traes_7DL_91E866851), and transket_pyr protein (Traes_2AL_1E2B26B7F.1) (Figure 7; Table S10). Succinate-CoA ligase (SUCL) can promote the production of ATP during the conversion of malate to succinate in the TCA cycle (Ostergaard, 2008). 4Fe-4S ferredoxin-type protein (4Fe-4S Fed) is involved in various redox processes in organisms, such as DNA repair, RNA and protein modification, and cofactor synthesis (Feng et al., 2021). Lactate/malate dehydrogenases (LDH/MDH) are involved in energy metabolism. Lactate dehydrogenase (LDH) operates at the final stage of aerobic glycolysis. Malate dehydrogenase (MDH) is a key enzyme in the regulation of malate metabolism; it catalyzes the reversible oxidative decarboxylation of malate to produce pyruvate and CO2, as well as the reduction of NAD(P)+ (Zhao et al., 2022). Succinate dehydrogenase (SDH) is the only enzyme that participates in both the tricarboxylic acid or citric acid cycle and the electron transport chain (Gill, 2012). Transket_pyr protein (TK) plays an important role in carbon metabolism (Liu et al., 2023). These AO-interacting proteins all play important functions in energy metabolism, which explains why AO also plays such an important role in energy metabolism. These results provide clues for further study of the function of TaAO genes.
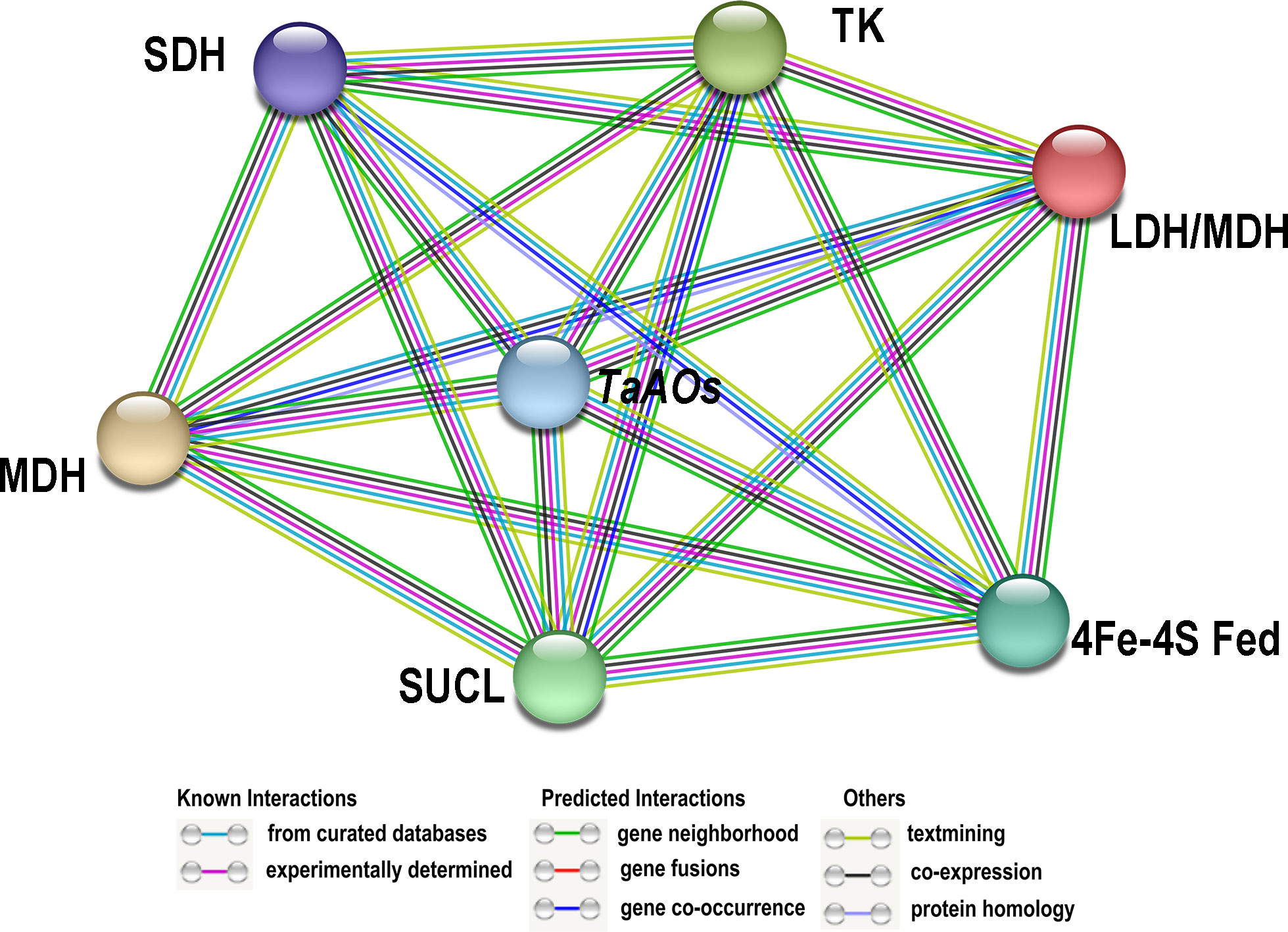
Figure 7 Protein–protein interaction (PPI) analysis of TaAO proteins. PPI network generated using STRINGV 11.5. Each node denotes a protein, and each edge represents an interaction.
3.7 Transcriptome analysis of TaAOs
In order to understand the expression patterns of TaAOs, wheat transcriptome data taken from different tissues and under exposure to different abiotic and biotic stresses were analyzed. From Figure 8, it is clearly evident that the expression levels of TaAOs belonging to Group II were the highest in all selected transcriptomes, which indicates that these three TaAOs may play essential roles in plant growth and response to various treatments. In terms of the different tissues, TaAOs belonging to Group I were relatively highly expressed in the spike, grain, stem, and root, and TaAOs belonging to Group III were relatively highly expressed in the root and seedling, while TaAOs belonging to Group II were highly expressed in all tissues, with no clear tissue specificity observed (Figure 8A).
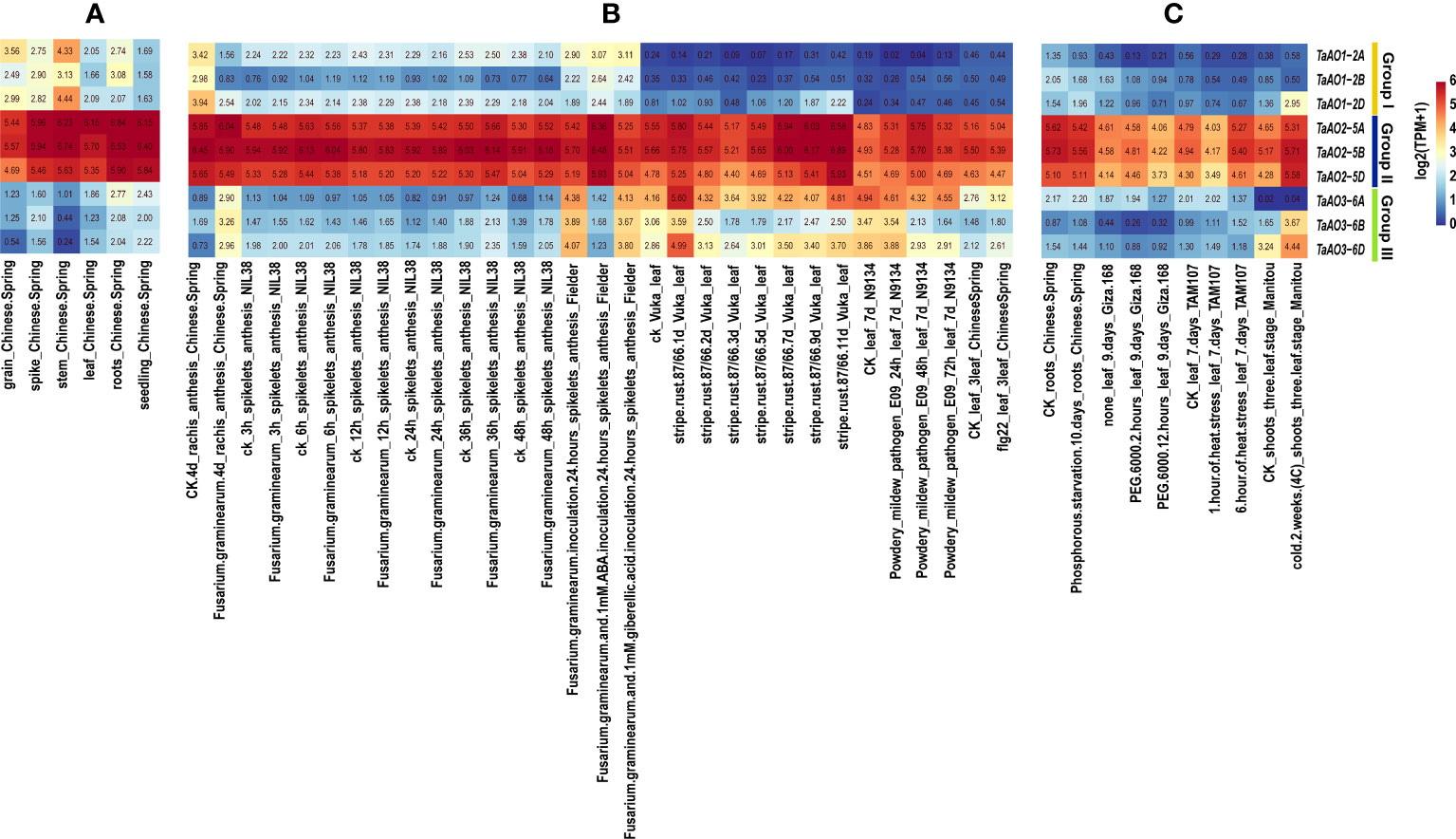
Figure 8 Transcriptome analysis of TaAOs. (A) Expression levels of nine TaAO genes in different tissues of Chinese Spring wheat. (B) Gene expression profiles of TaAOs under exposure to biological stress, plant hormone treatment, and pathogen-associated molecular patterns (PAMP). (C) Expression patterns of TaAO genes in wheat plants under exposure to different abiotic stresses. Different colors indicate the gene expression level: red indicates higher levels of expression, and blue lower.
As shown in Figure 8B, the transcriptomes of TaAOs under biotic stress were analyzed. In general, the expression of TaAOs changed slightly after infection by stripe rust. With the exceptions of TaAO1-2A and TaAO3-6B, the expression of TaAOs was induced by this form of stress. Interestingly, expression of TaAO1-2D was induced to a large extent at the late stage of infection (9 d and 11 d), while expression of TaAO3-6D was significantly induced at the early stage of infection (1d). These findings suggest that TaAOs might play a role in the response to stripe rust infection, but the patterns of expression were different for different TaAOs. In wheat N9134 from leaf tissue infected by powdery mildew, the expression levels of TaAOs also changed slightly. The expression of TaAO1-2A and of TaAOs belonging to Group III was suppressed, while the expression of other TaAOs was induced. The expression of TaAOs was also slightly altered in wheat NIL38 from spikelet tissue infected by F. graminearum, but it was greatly altered in F. graminearum-infected rachis tissue of Chinese Spring. In rachis tissue infected by F. graminearum, the expression of TaAOs belonging to Group I was repressed. The TaAOs with the greatest decrease in expression were TaAO1-2B, the expression levels of which decreased to 11.2% of the levels observed in CK, while the expression of TaAOs belonging to Group III was induced. The TaAOs with the greatest increase in expression were TaAO3-6D, the expression level of which was 10.33 times that observed in CK. These findings indicate that TaAOs may be involved in the pathogenesis of Fusarium head blight, and that TaAOs belonging to Groups I and III play different roles. Additionally, we found that the expression levels of TaAOs belonging to Group III were much lower in F. graminearum-infected wheat spikelet tissue treated with 1 mM ABA than in non-ABA-treated wheat spikelet tissue. This indicates that ABA negatively regulates the expression of TaAOs belonging to Group III in F. graminearum-infected wheat spikelets.
As shown in Figure 8C, the transcriptomes of TaAOs under abiotic stress were also analyzed. The results showed that the most significant changes in expression levels of TaAOs were found in wheat in response to cold treatment. The expression of all TaAOs was induced, with the exception of TaAO1-2B, the expression level of which was decreased by half. In contrast, the expression of most TaAOs was repressed during exposure to heat stress and drought stress. These results indicate that most TaAOs may participate in the response to cold and that they play different roles in the responses to heat and drought.
3.8 Quantitative real-time PCR analysis
To further understand the potential role of TaAO genes in biotic and abiotic stresses, the patterns of expression of TaAO1-2D, TaAO2-5A, and TaAO3-6D in response to stripe rust infection, ABA, and PEG stress were quantified via qRT-PCR.
After PEG treatment (Figure 9), the expression levels of TaAO1-2D and TaAO2-5A were decreased for 36 hours, but they were subsequently increased at 48 hours after treatment. In contrast, the expression levels of TaAO3-6D were decreased for 48 hours following treatment. In the case of ABA treatment, similar expression patterns were detected in TaAO2-5A and TaAO3-6D, expression of which was suppressed for 48 hours following ABA treatment. The pattern of expression of TaAO1-2D differed slightly from that of the two aforementioned genes: an increase in the expression level of TaAO1-2D was observed 2 hours after ABA treatment.
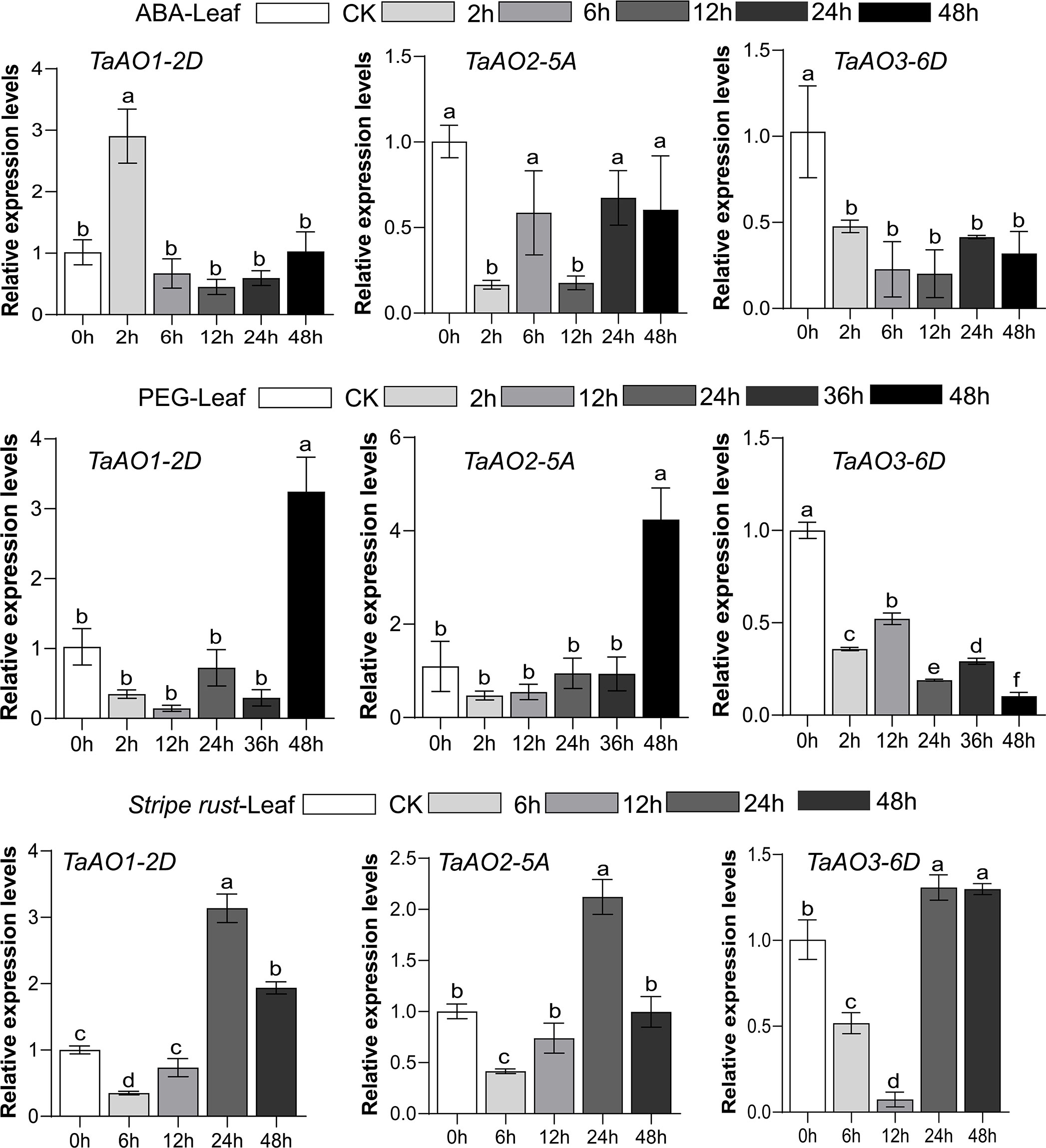
Figure 9 Expression levels of three TaAO genes in wheat leaves under exposure to abiotic stress (PEG), plant hormone treatment (ABA treatment), and biological stress (stripe rust infection), as indicated by qRT-PCR. The x axis represents time point, and the y axis represents expression level. Data from three independent replicates were analyzed; error bars represent the standard deviation. Lowercase letters (a-f) on the bars indicate significant differences determined via one-way ANOVA (P <.05). Plots created using GraphPad Prism 5.
Finally, in wheat inoculated with stripe rust, the general tendency of expression levels of TaAO1-2D, TaAO2-5A,and TaAO3-6D was to increase after an initial decrease. The lowest level of expression of TaAO3-6D was observed at 12 hpi, but the lowest levels for TaAO1-2D and TaAO2-5A were observed at 6 hpi. Compared with the expression level at 24 hpi, the expression levels of TaAO1-2D and TaAO2-5A were decreased at 48 hpi, while the expression level of TaAO3-6D at 48 hpi was roughly equal to that at 24 hpi.
4 Discussion
NAD+ is an essential cofactor in energy metabolism and electron transfer. Additionally, several reports suggest that NAD+ may be involved in plant defense responses (Dutilleul et al., 2003; Zhang and Mou, 2009; Djebbar et al., 2012; Petriacq et al., 2012). Therefore, as the first enzyme of NAD+ biosynthesis, AO may influence energy metabolism and plant defense by regulating NAD+ content. However, this enzyme has previously only been reported in Arabidopsis and maize (Macho et al., 2012; Dong, 2019). In this study, we first conducted a systematic analysis of AO-family genes in wheat. The gene architectures, gene duplication events, chromosomal distributions, cis-elements in promoter regions, and expression patterns of AO genes in wheat were then further analyzed.
In this study, nine AOs were identified in wheat. Compared with other plants, the wheat genome encodes many more AOs, which may be due to the heterologous hexaploidy of the wheat genome. In this paper, homology analyses of wheat and three sub-genome donor species were carried out, as a result of which 44 homologous gene pairs were identified, accounting for 90.56% of the wheat orthologous gene pairs. These findings suggest that TaAOs are derived from three sub-genome donor species of wheat. Phylogenetic tree analysis of maize, Arabidopsis, rice, T. aestivum, and its ancestor species (Ae. tauschii, T. dicoccoides, and T. urartu) revealed that Group II only contained T. aestivum and its ancestor species. The expression of AOs in Group II was high during growth and development, and under biotic and abiotic stress (Figure 8), indicating that AOs of Group II play an important role in wheat, and that this group has been conserved in the evolution of wheat.
Although AOs are thought to be widely distributed in plants, there has been little investigation of the function of AOs in plants. Up to this point, only two reports on the function of plant AOs have been published (Macho et al., 2012; Dong, 2019). In one of these studies, a differentially expressed ZmAO gene involved in energy metabolism was screened from maize CMS-C sterile line C48-2. Compared with the control maintenance line 48-2, ZmAO was found to be significantly downregulated in C48-2 during the mononuclear stage of anther development. The ZmAO gene may be a positive energy regulator involved in plant growth and development through the NAD+ synthesis pathway (Dong, 2019). Furthermore, AtAO2 of Arabidopsis is thought to participate in PTI and in resistance to Pst DC3000. The expression of AO has been found to be increased in non-virulent DC3000-inoculated Arabidopsis (Petriacq et al., 2012). Compared with the wild-type, flg22-triggered ROS bursts are significantly suppressed in AtAO2 mutants, and AtAO2 mutants are more susceptible to Pst DC3000 (Macho et al., 2012). These two AOs were both found to be members of Group III in the present study and are localized to chloroplasts (Katoh et al., 2006; Dong, 2019). Among the TaAOs identified in this study, TaAO3-6A, TaAO3-6B, and TaAO3-6D were also found to be clustered into Group III, and also located in chloroplasts, which suggests that TaAO3-6A, TaAO3-6B, and TaAO3-6D may have similar functions to those of the AOs reported in Arabidopsis and maize. Additionally, transcriptome analysis showed that the expression levels of TaAO3-6A, TaAO3-6B, and TaAO3-6D in flg22-treated wheat were increased by 33% to 53% compared with CK. This suggests that TaAO3-6A, TaAO3-6B, and TaAO3-6D may also play a role in PTI in wheat. Moreover, in our study, we found that the expression levels of TaAOs belonging to Group III were decreased in wheat treated with powdery mildew and increased in wheat treated with F. graminearum (Figure 8). As we know, the pathogen of wheat powdery mildew is a biotrophic parasite (Spanu et al., 2010), while F. graminearum is a hemi-biotrophic pathogen (Ma et al., 2020). These results suggest that TaAO may play opposing roles in the pathogenesis of hemi-biotrophic and biotrophic pathogens.
Previous studies have revealed that AO plays important roles in biotic stresses and plant development. In this study, we also found that AO may work in response to abiotic stress. The expression of most TaAOs was significantly upregulated under exposure to cold stress and downregulated under combined drought and heat stress. This indicates that AOs play an important role in the adaptation of plants to cold, heat, and drought stress. In summary, this research lays a foundation for further investigation of the function of TaAOs.
5 Conclusions
In this study, we systematically identified AO genes in wheat genomes. A total of nine TaAO genes were identified, which were distributed on three chromosomes of three sub-genomes. TaAOs were clustered into three groups. Gene structure and conserved motifs were similar within each group, but differed among the groups. Transcriptome analysis and real-time PCR assay indicated that TaAOs belonging to Group II were highly expressed in all tissues. TaAOs of Group III were found to be involved in PTI response and in the response to ABA treatment, and were found to play a positive role in wheat resistance to F. graminearum infection. Furthermore, TaAOs might positively regulate the response to cold treatment. These results provide systematic information on AO in wheat and lay a foundation for further research on the functions of TaAOs.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
WC, ZF, and WW designed the experiments and directed the writing of the manuscript. YF, MT, and JX performed the experiments and wrote the first draft. YF, PL, LW, and WC revised the manuscript. YF and MT contributed to the data analysis. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Open Program of Engineering Research Center of Ecology and Agricultural Use of Wetland Ministry of Education (KFT202107), the open project program of the State Key Laboratory for Biology of Plant Diseases and Insect Pests (NO. SKLOF202008), and the National Natural Science Foundation of China (31672088).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer XN declared a shared affiliation with the author WW to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1210632/full#supplementary-material
References
Abedi, T., Mojiri, A. (2020). Cadmium uptake by wheat (Triticum aestivum l.): an overview. Plants (Basel) 9, 500. doi: 10.3390/plants9040500
Armenia, I., Balzaretti, R., Pirrone, C., Allegretti, C., D'arrigo, P., Valentino, M., et al. (2017). L-aspartate oxidase magnetic nanoparticles: synthesis, characterization and l-aspartate bioconversion. Rsc Adv. 7, 21136–21143. doi: 10.1039/c7ra00384f
Bacchella, L., Lina, C., Todone, F., Negri, A., Tedeschi, G., Ronchi, S., et al. (1999). Crystallization of l-aspartate oxidase, the first enzyme in the bacterial de novo biosynthesis of NAD. Acta Crystallogr. D Biol. Crystallogr. 55, 549–551. doi: 10.1107/s0907444998011913
Bifulco, D., Pollegioni, L., Tessaro, D., Servi, S., Molla, G. (2013). A thermostable l-aspartate oxidase: a new tool for biotechnological applications. Appl. Microbiol. Biotechnol. 97, 7285–7295. doi: 10.1007/s00253-013-4688-1
Bishop, D. L., Bugbee, B. G. (1998). Photosynthetic capacity and dry mass partitioning in dwarf and semi-dwarf wheat (Triticum aestivum l.). J. Plant Physiol. 153, 558–565. doi: 10.1016/s0176-1617(98)80204-6
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, H., Liu, J., Wen, J., Nie, X., Xu, L., Chen, N., et al. (2019). Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol. 20, 136. doi: 10.1186/s13059-019-1744-x
Chou, K. C., Shen, H. B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5, e11335. doi: 10.1371/journal.pone.0011335
Chow, C., Hegde, S., Blanchard, J. S. (2017). Mechanistic characterization of escherichia coli l-aspartate oxidase from kinetic isotope effects. Biochemistry 56, 4044–4052. doi: 10.1021/acs.biochem.7b00307
Davis, S. J. (2009). Integrating hormones into the floral-transition pathway of arabidopsis thaliana. Plant Cell Environ. 32, 1201–1210. doi: 10.1111/j.1365-3040.2009.01968.x
Djebbar, R., Rzigui, T., Petriacq, P., Mauve, C., Priault, P., Fresneau, C., et al. (2012). Respiratory complex I deficiency induces drought tolerance by impacting leaf stomatal and hydraulic conductances. Planta 235, 603–614. doi: 10.1007/s00425-011-1524-7
Dong, B. (2019). The functional analysis of differentially expressed genes ZmAI and ZmAO in maize CMS-c CNKI. CNKI 7, 7–10. doi: 10.27345/d.cnki.gsnyu.2019.001308
Dutilleul, C., Garmier, M., Noctor, G., Mathieu, C., Chetrit, P., Foyer, C. H., et al. (2003). Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15, 1212–1226. doi: 10.1105/tpc.009464
Fang, Z. W., He, Y. Q., Liu, Y. K., Jiang, W. Q., Song, J. H., Wang, S. P., Yin, J. L., et al. (2020). Bioinformatic identification and analyses of the non-specific lipid transfer proteins in wheat. J. Integr. Agric. 19, 1170–1185. doi: 10.1016/S2095-3119(19)62776-0
Feng, J. Q., Shaik, S., Wang, B. J. (2021). Spin-regulated electron transfer and exchange-enhanced reactivity in Fe4S4-mediated redox reaction of the Dph2 enzyme during the biosynthesis of diphthamide. Angewandte Chemie-International Edition 60, 20430–20436. doi: 10.1002/anie.202107008
Gakiere, B., Hao, J. F., De Bont, L., Petriacq, P., Nunes-Nesi, A., Fernie, A. R. (2018). NAD(+) biosynthesis and signaling in plants. Crit. Rev. Plant Sci. 37, 259–307. doi: 10.1080/07352689.2018.1505591
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gill, A. J. (2012). Succinate dehydrogenase (SDH) and mitochondrial driven neoplasia. Pathology 44, 285–292. doi: 10.1097/PAT.0b013e3283539932
Guo, G., Lei, M., Wang, Y., Song, B., Yang, J. (2018). Accumulation of as, cd, and Pb in sixteen wheat cultivars grown in contaminated soils and associated health risk assessment. Int. J. Environ. Res. Public Health 15, 2601. doi: 10.3390/ijerph15112601
Hao, J., Petriacq, P., De Bont, L., Hodges, M., Gakiere, B. (2018). Characterization of l-aspartate oxidase from arabidopsis thaliana. Plant Sci. 271, 133–142. doi: 10.1016/j.plantsci.2018.03.016
Katoh, A., Uenohara, K., Akita, M., Hashimoto, T. (2006). Early steps in the biosynthesis of NAD in arabidopsis start with aspartate and occur in the plastid. Plant Physiol. 141, 851–857. doi: 10.1104/pp.106.081091
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Leese, C., Fotheringham, I., Escalettes, F., Speight, R., Grogan, G. (2013). Cloning, expression, characterisation and mutational analysis of l-aspartate oxidase from pseudomonas putida. J. Mol. Catalysis B-Enzymatic 85-86, 17–22. doi: 10.1016/j.molcatb.2012.07.008
Letunic, I., Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Liu, G., Liu, Q., Han, Z., Wang, P., Li, Y. (2023). Comparative proteomics analysis of adult haemonchus contortus isolates from ovis ammon. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1087210
Ma, Z., Xie, Q., Li, G., Jia, H., Zhou, J., Kong, Z., et al. (2020). Germplasms, genetics and genomics for better control of disastrous wheat fusarium head blight. Theor. Appl. Genet. 133, 1541–1568. doi: 10.1007/s00122-019-03525-8
Macho, A. P., Boutrot, F., Rathjen, J. P., Zipfel, C. (2012). Aspartate oxidase plays an important role in arabidopsis stomatal immunity. Plant Physiol. 159, 1845–1856. doi: 10.1104/pp.112.199810
Marinoni, I., Nonnis, S., Monteferrante, C., Heathcote, P., Hartig, E., Bottger, L. H., et al. (2008). Characterization of l-aspartate oxidase and quinolinate synthase from bacillus subtilis. FEBS J. 275, 5090–5107. doi: 10.1111/j.1742-4658.2008.06641.x
Mattevi, A., Tedeschi, G., Bacchella, L., Coda, A., Negri, A., Ronchi, S. (1999). Structure of l-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family. Structure 7, 745–756. doi: 10.1016/s0969-2126(99)80099-9
Mortarino, M., Negri, A., Tedeschi, G., Simonic, T., Duga, S., Gassen, H. G., et al. (1996). L-aspartate oxidase from escherichia coli. i. characterization of coenzyme binding and product inhibition. Eur. J. Biochem. 239, 418–426. doi: 10.1111/j.1432-1033.1996.0418u.x
Nasu, S., Wicks, F. D., Gholson, R. K. (1982). L-aspartate oxidase, a newly discovered enzyme of escherichia coli, is the b protein of quinolinate synthetase. J. Biol. Chem. 257, 626–632. doi: 10.1016/S0021-9258(19)68239-6
Ostergaard, E. (2008). Disorders caused by deficiency of succinate-CoA ligase. J. Inherit Metab. Dis. 31, 226–229. doi: 10.1007/s10545-008-0828-7
Panchy, N., Lehti-Shiu, M., Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. doi: 10.1104/pp.16.00523
Petriacq, P., De Bont, L., Hager, J., Didierlaurent, L., Mauve, C., Guerard, F., et al. (2012). Inducible NAD overproduction in arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to pst-AvrRpm1. Plant J. 70, 650–665. doi: 10.1111/j.1365-313X.2012.04920.x
Prunier, A. L., Schuch, R., Fernandez, R. E., Mumy, K. L., Kohler, H., Mccormick, B. A., et al. (2007). nadA and nadB of shigella flexneri 5a are antivirulence loci responsible for the synthesis of quinolinate, a small molecule inhibitor of shigella pathogenicity. Microbiol. (Reading) 153, 2363–2372. doi: 10.1099/mic.0.2007/006916-0
Remm, M., Storm, C. E., Sonnhammer, E. L. (2001). Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314, 1041–1052. doi: 10.1006/jmbi.2000.5197
Ren, Y., Liu, L. S., He, Z. H., Wu, L., Bai, B., Xia, X. C. (2015). QTL mapping of adult-plant resistance to stripe rust in a "Lumai 21xJingshuang 16' wheat population. Plant Breed. 134, 501–507. doi: 10.1111/pbr.12290
Rombauts, S., Dehais, P., Van Montagu, M., Rouze, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Sakuraba, H., Satomura, T., Kawakami, R., Yamamoto, S., Kawarabayasi, Y., Kikuchi, H., et al. (2002). L-aspartate oxidase is present in the anaerobic hyperthermophilic archaeon pyrococcus horikoshii OT-3: characteristics and role in the de novo biosynthesis of nicotinamide adenine dinucleotide proposed by genome sequencing. Extremophiles 6, 275–281. doi: 10.1007/s00792-001-0254-3
Sakuraba, H., Yoneda, K., Asai, I., Tsuge, H., Katunuma, N., Ohshima, T. (2008). Structure of l-aspartate oxidase from the hyperthermophilic archaeon sulfolobus tokodaii. Biochim. Biophys. Acta 1784, 563–571. doi: 10.1016/j.bbapap.2007.12.012
Santra, D. K., Chen, X. M., Santra, M., Campbell, K. G., Kidwell, K. K. (2008). Identification and mapping QTL for high-temperature adult-plant resistance to stripe rust in winter wheat (Triticum aestivum l.) cultivar 'Stephens'. Theor. Appl. Genet. 117, 793–802. doi: 10.1007/s00122-008-0820-5
Sobolewska, M., Wenda-Piesik, A., Jaroszewska, A., Stankowski, S. (2020). Effect of habitat and foliar fertilization with K, zn and Mn on winter wheat grain and baking qualities. Agronomy-Basel 10, 276. doi: 10.3390/agronomy10020276
Spanu, P. D., Abbott, J. C., Amselem, J., Burgis, T. A., Soanes, D. M., Stuber, K., et al. (2010). Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330, 1543–1546. doi: 10.1126/science.1194573
Tedeschi, G., Negri, A., Mortarino, M., Ceciliani, F., Simonic, T., Faotto, L., et al. (1996). L-aspartate oxidase from escherichia coli. II. interaction with C4 dicarboxylic acids and identification of a novel l-aspartate: fumarate oxidoreductase activity. Eur. J. Biochem. 239, 427–433. doi: 10.1111/j.1432-1033.1996.0427u.x
Tedeschi, G., Nonnis, S., Strumbo, B., Cruciani, G., Carosati, E., Negri, A. (2010). On the catalytic role of the active site residue E121 of e. coli l-aspartate oxidase. Biochimie 92, 1335–1342. doi: 10.1016/j.biochi.2010.06.015
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Zhang, X., Mou, Z. (2009). Extracellular pyridine nucleotides induce PR gene expression and disease resistance in arabidopsis. Plant J. 57, 302–312. doi: 10.1111/j.1365-313X.2008.03687.x
Keywords: TaAO, gene structure, abiotic stresses, gene expression, quantitative PCR, biological functions
Citation: Feng Y, Tang M, Xiang J, Liu P, Wang Y, Chen W, Fang Z and Wang W (2023) Genome-wide characterization of L-aspartate oxidase genes in wheat and their potential roles in the responses to wheat disease and abiotic stresses. Front. Plant Sci. 14:1210632. doi: 10.3389/fpls.2023.1210632
Received: 23 April 2023; Accepted: 09 June 2023;
Published: 05 July 2023.
Edited by:
Xinli Zhou, Southwest University of Science and Technology, ChinaReviewed by:
Haifeng Liu, Chonnam National University, Republic of KoreaXiaojun Nie, Northwest A&F University, China
Xiaodong Wang, Hebei Agricultural University, China
Huan Luo, Chungnam National University, Republic of Korea
Copyright © 2023 Feng, Tang, Xiang, Liu, Wang, Chen, Fang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wang Chen, Y2hlbndhbmdjaHdAMTYzLmNvbQ==; Zhengwu Fang, ZmFuZ3poZW5nd3U4OEAxNjMuY29t; Wenli Wang, d2xpd0AxNjMuY29t
†These authors have contributed equally to this work
 Yanqun Feng1†
Yanqun Feng1† Wang Chen
Wang Chen