- 1Experimental Centre of Forestry in North China, National Permanent Scientifc Research Base for Warm Temperate Zone Forestry of Jiulong Mountain, Chinese Academy of Forestry, Beijing, China
- 2State Key Laboratory of Tree Genetics and Breeding, Chinese Academy of Forestry, Beijing, China
- 3Key Laboratory of National Forestry and Grassland Administration on Ecological Landscaping of Challenging Urban Sites, Shanghai Academy of Landscape Architecture Science and Planning, Shanghai, China
- 4State Key Laboratory of Efficient Utilization of Arid and Semi-arid Arable Land in Northern China, Institute of Agricultural Resources and Regional Planning, Chinese Academy of Agricultural Sciences, Beijing, China
- 5UWA School of Agriculture and Environment, The University of Western Australia, Perth, WA, Australia
- 6Institute for Adriatic Crops and Karst Reclamation, Split, Croatia
The Acer truncatum Bunge, widely distributed in North China, shows excellent tolerance to low-P soils. However, little information is available on potential phosphate-solubilizing bacterial (PSB) strains from the A. truncatum rhizosphere. The objectives of this work were to isolate and characterize PSB from A. truncatum rhizosphere soil and to evaluate the effect of inoculation with the selected strain on A. truncatum seedlings. The strains were characterized on the basis of phenotypic characteristics, carbon source utilization pattern, fatty acid methyl esters analysis, 16S rRNA gene and the whole-genome sequence. A Gram-negative and rod-shaped bacterium, designated MQR6T, showed a high capacity to solubilize phosphate and produce indole-3-acetic acid (IAA) and siderophores. The strain can solubilize tricalcium phosphate (TCP) and rock phosphate (RP), and the solubilization of TCP was about 60% more effective than RP. Phylogenetic analyses based on the 16S rRNA gene and whole-genome sequences revealed that strain MQR6T formed a distinct phyletic lineage as a new species within the genus Pantoea. The digital DNA-DNA hybridization value between strain MQR6T and the closely related strains was 19.5-23.3%. The major cellular fatty acids were summed feature 3 (C16:1ω7c and/or C16:1ω6c), summed feature 8 (C18:1ω6c and/or C18:1ω7c), C14:0, C16:0, and C17:0 cyclo. Several genes related to IAA production, phosphonate transport, phosphate solubilization and siderophore biogenesis were found in the MQR6T genome. Furthermore, inoculation with the strain MQR6T significantly improved plant height, trunk diameter, dry weight and P accumulation in roots and shoot of A. truncatum seedlings compared to non-inoculated control. These plant parameters were improved even further in the treatment with both inoculation and P fertilization. Our results suggested that MQR6T represented a new species we named Pantoea rhizosphaerae, as a plant growth-promoting rhizobacterium that can solubilize inorganic P and improve growth of A. truncatum seedlings, emerging as a potential strategy to improve A. truncatum cultivation.
1 Introduction
Phosphorus (P) is an essential macronutrient for plant survival and reproduction, as a component of nucleic acids, membrane phospholipids and many energy-dependent metabolic processes (Vance et al., 2003). Phosphorus is frequently the most limiting element in soils because it rapidly forms insoluble complexes with cations and has low solubility and poor mobility in soils (Hinsinger, 2001; Rafique et al., 2022). The total amount of P is quite abundant in many soils, ranging from 0.02% to 0.5% (w/w), with an average of about 0.05% (w/w) (Son et al., 2006). However, plants absorb and assimilate P as inorganic (Pi) di- and monohydrogenphosphates; the average Pi concentration in the soil solution is 1 μM, which is below the Pi concentration needed for optimal plant growth (Hinsinger, 2001). Moreover, some assessments suggest world P reserves may last for only 50–200 years, which could result in a potential phosphate crisis (Herrera-Estrella and López-Arredondo, 2016). An ecologically friendly and economical approach to this problem may lie in the exploitation of the rhizosphere microbiome (De Zutter et al., 2022). Apart from P fertilization, soil P mobilization by microorganisms would be the only possible way to increase amounts of P available to plants (Etesami et al., 2021; Chouyia et al., 2022; Bouizgarne et al., 2023).
Rhizobacteria are plant-associated bacteria that colonize and persist in the proximity of roots or inside the root tissues (Backer et al., 2018). Phosphate-solubilizing bacteria (PSB) have the capacity to convert insoluble inorganic phosphates into soluble forms available to plants (Rafique et al., 2022). The principal mechanism for mineral P solubilization by PSB is associated with the production of low-molecular-weight organic acid anions, which through their hydroxyl and carboxyl groups chelate the phosphate-bound cations to liberate P in soluble forms (Hassan et al., 2019). Additionally, PSB are capable of producing physiologically active indole-3-acetic acid (IAA) and siderophores, which may have pronounced effects on plant growth (Luziatelli et al., 2020).
Strains from bacterial genera Pantoea have been reported as efficient PSB in the soil (Chen and Liu, 2019). The genus Pantoea, belonging to the family Erwiniaceae in the phylum Proteobacteria (Luziatelli et al., 2020), was first proposed by Gavini et al. (Gavini et al., 1989). The genus has been subsequently emended over the years as more species have been classified (Mergaert et al., 1993; Brady et al., 2010). The genus Pantoea showed a strong capacity of adaptation to a broad range of hosts and various environmental conditions, with strains isolated from plants, soil, fruits, seeds, the fruiting body of mushroom, humans, and animals (Castagno et al., 2011; Dutkiewicz et al., 2016; Luziatelli et al., 2020). Furthermore, many strains from Pantoea were efficient in solubilizing insoluble inorganic phosphate sources such as tricalcium phosphate (TCP) in the culturing medium (Chen and Liu, 2019; Li et al., 2020). Some isolates, including P. ananatis and P. agglomerans, were found to possess plant growth-promoting properties and reduce plant stress (Li et al., 2020; Luziatelli et al., 2020).
The genus Acer (family Aceraceae), commonly known as maple, comprises 129 species with many infraspecific taxa (Bi et al., 2016). These species are distributed in the temperate regions of Asia, Europe, northern Africa, and central and northern America (Bi et al., 2016; Wang et al., 2019). China (with 99 species reported) is considered to host the greatest diversity of the genus Acer (Bi et al., 2016). The A. truncatum Bunge is a forest tree species found in the north of China, showing excellent tolerance to P-deficiency stress (Wang et al., 2019). However, little information is available on potential PSB strains from the rhizosphere of A. truncatum growing on low-P soil. The purposes of this study were to isolate and characterize the PSB from rhizosphere soil of A. truncatum grown in the main production area in North China. In addition, the effects of inoculation of PSB with or without P fertilizer on root and shoot growth of A. truncatum seedlings and their P uptake were evaluated.
2 Materials and methods
2.1 Soil sampling and bacterial isolation
Rhizosphere soil samples of A. truncatum Bunge were taken from three sites in Jiulongshan Mountain Preserve, Beijing, People’s Republic of China (39°57′48″ N, 116°05′00″ E). At each sampling site, lateral roots of four Acer plants in the 10-30 cm soil layer were collected using a sterilized shovel and scissors. Samples of approximately 100 g of soil tightly adhering to lateral roots were collected in sterilized plastic bags, immediately placed on dry ice, and transferred to the laboratory for further work. Initial soil properties of the three sites were as follows: total N 1.3 g kg−1, total P 0.9 g kg−1, pH 8.1 (1:2.5, soil:water), Olsen-P 4.2 mg kg−1, available N (NH4+-N plus NO3–N) 17.3 mg kg−1, and exchangeable K 67 mg kg−1.
The serially diluted soil samples were plated on the TCP medium containing (per 1 liter): 5.0 g Ca3(PO4)2, 0.50 g (NH4)2SO4, 0.30 g NaCl, 0.30 g KCl, 0.30 g MgSO4, 0.03 g FeSO4, 0.03 g MgSO4, 0.50 g yeast extract, 10.0 g glucose, and 15.0 g agar. The PSB in the sampling rhizosphere were identified by clear halo zones around their colonies after 3 days of incubation at 30 °C. Experiments were performed in four replicates. The capacity of PSB to solubilize the water-insoluble phosphate was studied by the determination of solubilization index [the ratio of the total diameter (colony + halozone) and the colony diameter] (Li et al., 2020).
Single colonies with clear halos indicating P solubilization were selected and purified (Wang et al., 2020 and Bouizgarne et al., 2023). The purified strain designated MQR6T was obtained and maintained (i) on tryptone soy agar (TSA) plates at 4°C in a refrigerator for further characterization and (ii) as suspensions supplemented with 30% (w/v) glycerol at ‐80°C. Strain MQR6T was deposited at the China General Microbiological Culture Collection Center (CGMCC No. 23609T).
The reference strains Pantoea vagans DSM 23078T and Pantoea allii DSM 25133T were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ), and the strain Pantoea ananatis CICC 10283T was obtained from the China Center of Industrial Culture Collection (CICC). These strains were cultured under the same conditions as described above for comparative analyses.
2.2 Morphological characterization
Colony morphology of purified bacterial isolate was studied by streaking on TSA medium, followed by incubation of the plates at 30°C for 24 h. Cell morphology was examined by light microscopy (model 50i, Nikon), and cellular morphology was observed by a scanning electron microscope (FEI Quanta 250 FEG, USA). Gram staining was performed using a bioMérieux Gram stain kit (Hangzhou Tianhe Microorganism Reagent Co.) according to the manufacturer’s instructions.
2.3 Phenotypic characterization
The temperature range for growth was determined in tryptone soy broth (TSB) liquid medium at 4, 10, 15, 20, 25, 28, 30, 37, 40, and 45°C. Growth at different pH values (3.0–12.0 at 1.0 pH unit increments) was evaluated in the TSB medium for 24 h, using the following buffer systems: pH 3.0–5.0, 0.1 M citric acid/0.1 M sodium citrate; pH 6.0–8.0, 0.1 M KH2PO4/0.1 M NaOH; and pH 9.0–12.0, 0.1 M NaHCO3/0.1 M Na2CO3. The range of NaCl concentrations for growth was determined in the TSB medium containing 0–10% NaCl at increments of 1% (w/v). Bacterial growth was measured by an increase in turbidity at 600 nm using a spectrophotometer.
Carbon source utilization tests, enzyme activity tests, and additional physiological and biochemical tests were performed using API-20NE, API 50CH (BioMérieux), and Biolog GEN III MicroPlate systems (Reis et al., 2004). The bacterial inoculation was performed according to the manufacturer’s instructions. The type strains of P. vagans DSM 23078T, P. allii DSM 25133T and P. ananatis CICC 10283T were used as reference strains. The results for API 20NE and API 50CH were obtained after 48 h of incubation as recommended by the manufacturer. When the Biolog system was used, strains were incubated on biological universal growth medium (Biolog) at 30 °C for 24 h. GEN III microplates were inoculated according to the manufacturer’s instructions and incubated at 30 °C for 22 h. Results were captured and analyzed based on an extensive species library in the Biolog GEN III database (Woźniak et al., 2019).
For cellular fatty acid analysis, cell mass of strain MQR6T was harvested from TSA plates after incubation for 24 h at 30 °C. The fatty acid methyl esters were extracted and prepared according to the methods described by Sasser (1990). The fatty acids methyl ester mixtures were separated and analyzed on an Agilent GC-6890N gas chromatograph using the Sherlock Microbial Identification System with standard MIS Library Generation Software (version 6.0; Microbial ID Inc., Newark, DE, USA).
2.4 Phylogenetic 16S rRNA gene analysis
Genomic DNA was extracted using a Bacterial DNA Kit (Tiangen, Beijing, China) following the manufacturer’s instructions. The 16S rRNA gene was amplified by PCR using the universal primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’) (He and Wan, 2022). and the purified PCR products were sequenced by Sangon Biotech (Shanghai, PR China). The 16S rRNA gene sequences were assembled by using the SeqMan package (DNAStar). The 16S rRNA gene sequence of strain MQR6T was compared with the sequences available in the National Center for Biotechnology Information (NCBI) GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and EzBioCloud (www.ezbiocloud.net/identify) (Kim et al., 2012). Multiple alignments were carried out by using CLUSTAL_X software (Thompson et al., 1997). The phylogenetic trees were constructed using the neighbour joining (Saitou and Nei, 1987), maximum likelihood (Felsenstein, 1981) and maximum parsimony (Tamura et al., 2011) methods with the MEGA version 7.0 program (Kumar et al., 2016). The evolutionary distances were calculated using the Maximum Composite Likelihood Method (Tamura et al., 2004). Bootstrap values were calculated based on 1000 bootstrap replications in each case.
2.5 Complete genome sequencing and analysis
For genome sequencing of strain MQR6T, Illumina Hiseq TM2500 sequencing was performed at Shanghai Personal Biotechnology Co. (Beijing, PR China). The raw data were filtered and trimmed by AdapterRemoval (version 2.1.7) (Schubert et al., 2016). The trimmed reads were assembled using A5-miseq v20150522 with default parameters (Coil et al., 2015). CheckM v1.0.3 was used to estimate the completeness of the genome (Chen et al., 2020). Protein-coding open-reading frames were predicted by using Glimmer v3.02 (Delcher et al., 2007). Contigs longer than 1 kb and with read coverage of more than 10 were kept for further analysis. The G+C content of the chromosome was determined according to a draft genome sequence. The tRNA genes were predicted by tRNAscan-SE 94 (ver. 1.3.1) and the rRNA genes by Barrnap (0.9-dev) 95 (https://github.com/tseemann/barrnap) (Lowe and Eddy, 1997). Gene function prediction was performed by the Rapid Annotations using Subsystems Technology (RAST v.2.0) server (http://rast.nmpdr.org) (Overbeek et al., 2014) and eggNOG-mapper v2 (http://eggnog-mapper.embl.de) (Cantalapiedra et al., 2021). Metabolic pathways were analyzed by using the KEGG’s Blast KOALA service (Kanehisa et al., 2016).
The average nucleotide identity based on blase (ANIb) among the strain MQR6T and related species was calculated using JspeciesWS online (Richter et al., 2016). The estimated digital DNA–DNA hybridization (dDDH) values among the strains were calculated by Genome-to-Genome Distance Calculator (GGDC2.0) with the alignment method of BLAST+ (Auch et al., 2010). The partial genome files were uploaded to the GGDC 2.0 web interface (http://ggdc.dsmz.de/ggdc.php#), and Formula 2 was used as recommended for the calculation of dDDH values. The proposed and generally accepted species boundary for ANIb and dDDH values are 95~96 and 70%, respectively (Meier-Kolthoff et al., 2013).
Two methods were used to construct phylogenetic trees of strain MQR6T and the closely related Pantoea species. The first method used the classification workflows in Genome Taxonomy Database toolkit version 2.0.0 (GTDB-Tk) to identify and concatenate 120 single-copy bacterial marker genes (Chaumeil et al., 2020). The ML phylogenetic tree was established using IQ-TREE 2.2.0 program, SH-aLRT test, 1000 repeated ultrafast guidance, and ModelFinder to determine the best-fit model (Nguyen et al., 2015). The second method uploads the genome sequence data to the Type (Strain) Genome Server (https://tygs.dsmz.de/) (Meier-Kolthoff et al., 2022) for the whole-genome-based taxonomic analysis. The Bacterial Pan Genome Analysis (BPGA) pipeline was used for the pan-genome analyses of strain MQR6T and the closely related Pantoea species (Chaudhari et al., 2016).
2.6 Quantification of P-solubilizing capacity
The ability of the strain to solubilize water-insoluble phosphate was measured in liquid media containing either TCP or powdered RP. The RP medium was modified from the TCP medium by adding 5 g L-1 of rock phosphate instead of TCP. One milliliter of MQR6T culture (approximately 1×108 cfu mL-1) was transferred to a 300-mL flask containing 100 mL of medium, followed by shaking (150 rpm) at 30 °C. The non-inoculated TCP and RP media were used as controls. Quadruplicate cultivations were conducted for each medium. The suspensions were sampled up to 96 hours at 12-hour intervals. At every sampling time, 3 mL of culture liquids was sampled and centrifuged at 5000 g (Anke LXJ-IIB) for 20 min to remove biomass and insoluble matter, and the supernatants were used for determination of pH and soluble P concentration. The pH value of the medium was measured with a pH meter. Phosphorus in the supernatant was determined by the molybdenum-blue method using a spectrophotometer at 700 nm (Watanabe and Olsen, 1965).
2.7 Quantification of IAA production
The secretion of plant growth hormone IAA by strain MQR6T was measured by colorimetry. The test was performed in the presence and absence of L-tryptophan as the precursor of IAA. One milliliter of bacterial culture (approximately 1×108 cfu·mL-1) was added to 100 mL TSB medium (with 5 mM L-tryptophan or without) in 250 mL Erlenmeyer flasks, and then incubated on a shaker (30 °C, 150 rpm) for 96 h. The suspensions were sampled at 12-hour intervals. The method to collect culture supernatant was the same as that in detecting P-solubilizing capacity. The production of IAA was screened by mixing 100 µL of bacterial suspension droplets with 100 µL of Salkowski reagent (50 mL 35% v/v HClO4 + 1 mL 0.5 mol L-1 FeCl3) on a white ceramic plate based on the color change after 30 min of reaction in the dark at room temperature (Naqqash et al., 2016). Indole compounds react with Salkowski reagent to form a pink chromophore in absorbance at 530 nm using spectrophotometer (UV 3200, Shanghai, China). IAA concentrations were determined using a standard curve made from commercial IAA (Sigma), with the sterile medium as the blank.
2.8 Screening for siderophore production
Siderophore production was assayed qualitatively as described by Schwyn and Neilands (Schwyn and Neilands, 1987). Briefly, overnight culture of strain MQR6T was spot-inoculated onto a chrome azurol S (CAS) agar plate and incubated for 5 days at 30° C. The basic principle underlying the test is that when a strong ligand (for example, siderophore) is added to a highly coloured dye-Fe3+ complex, the Fe3+-ligand complex is formed, and the release of free dye is accompanied by a colour change. When a strong chelator removes the iron from the dye, its color turns from blue to orange.
2.9 Plant inoculation experiments
The experiment was carried out in a greenhouse located at the Experimental Centre of Forestry in North China, Chinese Academy of Forestry, to evaluate the effects of strain MQR6T on plant growth and nutrient uptake by A. truncatum seedlings. The soil was obtained from the Acer forest in Jiulongshan Mountain Preserve, with the properties as described above. The soil samples were air-dried, passed through a 2-mm sieve and filled into the pots at bulk density of 1.32 g cm-3. To ensure that the supply of other nutrients was adequate for plant growth, soil was supplemented with basal nutrients at the following rates (mg kg−1 soil): 200 N (NH4NO3), 50 Ca (as CaCl2), 150 K (as KCl), 28 Mg (as MgSO4), 4 Zn (as ZnSO4), and 1 Fe (as EDTA-Fe).
There were four treatments in the present study: (1) control, non-inoculated and without the application of P fertilizer (CK), (2) application of P fertilizer only, non-inoculated (P), (3) inoculation with MQR6T only, without the application of P, and (4) inoculation with MQR6T plus the application of P (MQR6+P). Monopotassium phosphate (KH2PO4) is a highly water-soluble inorganic salt, widely used as a P fertilizer in agricultural soils. The fertilizer KH2PO4 was used as P source, and the concentration was 50 mg P kg-1 soil.
The Acer truncatum seeds were surface-sterilized with sodium hypochlorite (1% w/v) for 30 min and then rinsed extensively with sterilized distilled water. The suspension of overnight bacterial culture (TSB) was diluted in sterile distilled water to a final concentration 108 cfu mL-1, and the resulting suspensions were used to treat seeds and seedlings. The surface-sterilized seeds were dipped in the inoculum (containing 108 cfu mL-1) for 15 min and then placed in pots containing 800 g of soil on 25 June 2021. Seeds dipped in medium not containing the strain were used for the non-inoculated treatments. A second inoculation was done at days 45 after seedling emergence at rates of 5 mL of bacterial suspension described above per pot. Plants were watered weekly to maintain 70–80% of field capacity.
Plants were harvested at 290 days after sowing on 10 April 2022. Plant height was recorded by measuring the length from soil surface to the tip of the main stem. Trunk diameter was measured at the base (5 cm from the ground) using a digital vernier caliper with an accuracy of 0.01 mm (Wuxi Kaibaoding Tools Co., Ltd., China). Chlorophyll content was read in the youngest fully-developed leaves using a chlorophyll meter (SPAD-502, Minolta, Osaka, Japan). The plants were separated into shoot and roots. The roots were kept in an icebox, transported to the lab, rinsed with water, and scanned by a scanner at resolution of 400 dpi. Root images were analyzed using WinRhizo Pro 2009b software (Regent Instruments Inc., Quebec, Canada) to calculate the root length and surface area. The shoots and roots were oven-dried at 105°C for 30 min and then at 70°C for 3 days to constant weight to determine dry weights and P uptake. P contents were assayed using the dry ashing digestion method.
2.10 Statistical analyses
One-way analysis of variance was performed using SAS statistical software (SAS 8.1, USA), and significant differences among means were assessed using Tukey’s test at 5% probability (P ≤ 0.05).
3 Results
3.1 Morphological and physiological characteristics of MQR6T
Colonies were circular, smooth, mucoid, convex with clear edges, and 0.8–2.0 mm in diameter after 24 h of incubation at 30°C on TSA plates (Figure 1A). Cells were rod-shaped, Gram-stain negative, single, non-spore-forming, measuring 0.5–1.4 μm×1.0–3.0 μm (Figures 1B–D). Bacterium features one or more long flagella (Figure 1D). Growth was found to occur at 10–40°C (optimum, 28–30°C) and at pH 4–11 (optimum, pH 7–9). In TSB medium, growth occurred in the presence of 0–7% (w/v) NaCl (optimum, 0–1%).
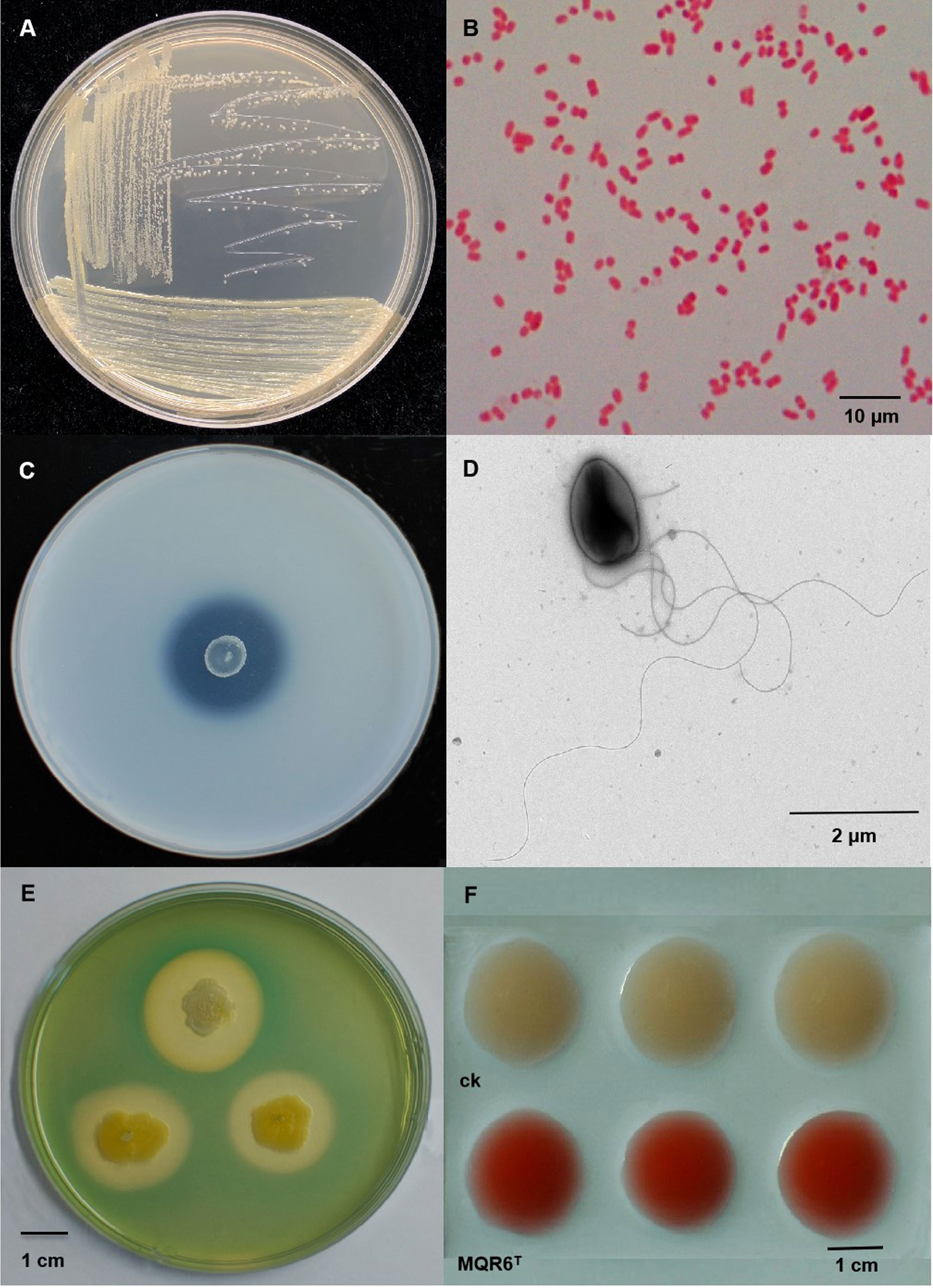
Figure 1 The morphological characteristics and plate assays of strain MQR6T. (A) The colony morphology on TSA plate; (B) The Gram-staining of the strain; (C) Halo zone on the agar medium containing TCP; (D) The cell morphology of the strain; (E) Halo zones of siderophore exudation on CAS plate; (F) Screening of IAA production with Salkowski reagent. TSA, tryptone soy agar; TCP, tricalcium phosphate; CAS, chromo azurol S; IAA, indole-3-acetic acid.
The phenotypic properties differentiating between strain MQR6T and its closest phylogenetic neighbors are shown in Table 1. According to API 50CH tests, strain MQR6T showed negative results with glycerin and sucrose, which were positive (or weakly positive) for P. vagans DSM 23078T, P. allii DSM 25133T and P. ananatis CICC 10283T. Acid was produced from the fermentation of D-glucose, ribose, D-xylose, galactose, fructose, rhamnose, mannitol, N-acetylglucosamine, maltose, and trehalose by strain MQR6T and the other three reference strains. With API 20NE, strain MQR6T was negative for sodium citrate, L-arabinose, inositol, melibiose, and rhamnose but the other three reference strains were positive (at least weakly). Strain MQR6T grew on D-maltose, but the other three reference strains did not.
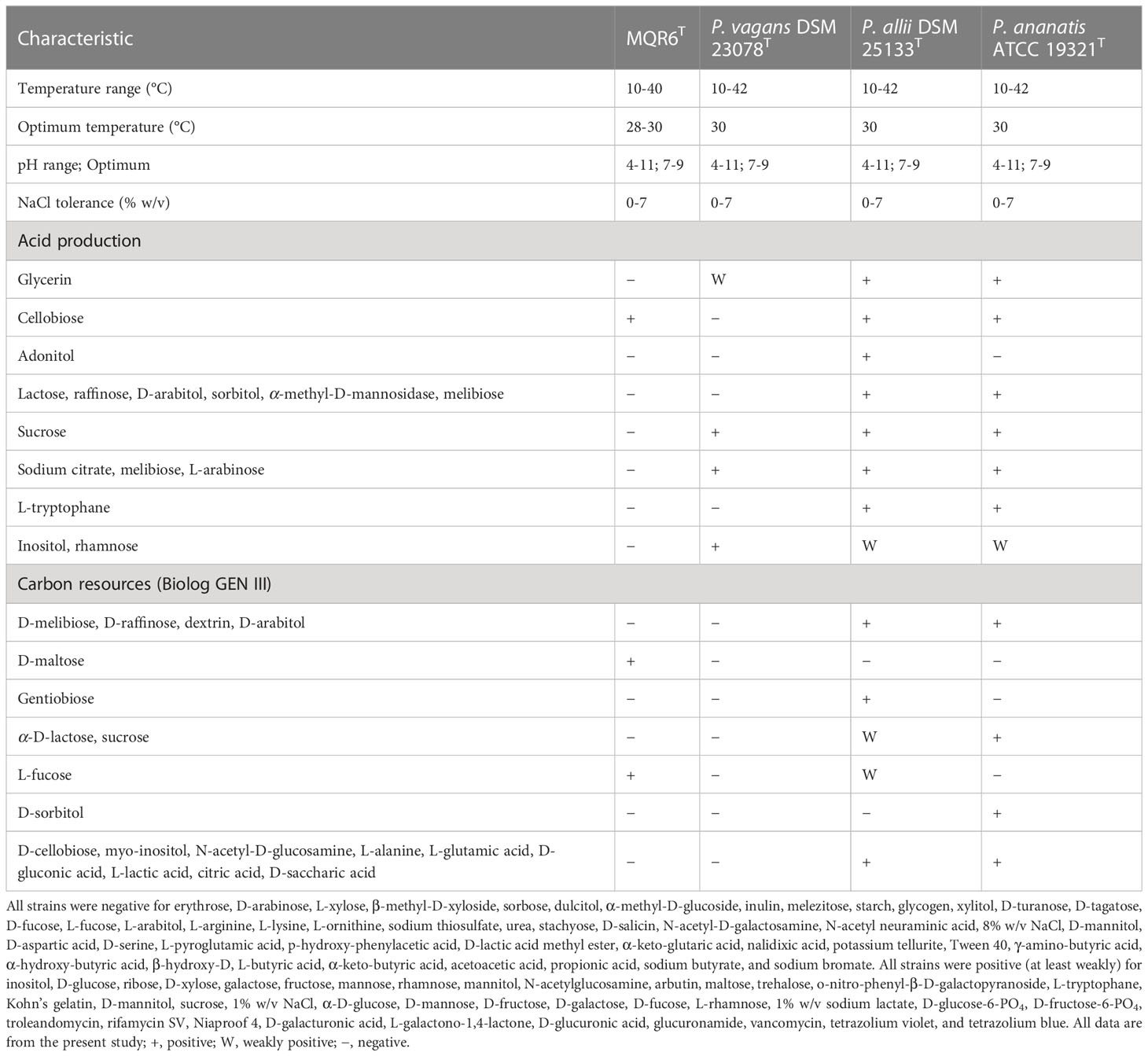
Table 1 Differential phenotypic characteristics of strain MQR6T and closely related strains in genus Pantoea.
The fatty acid analysis revealed that all strains contained C12:0, C14:0, C16:0, C17:0 cyclo, summed feature 3 (C16:1 ω7c and/or C16:1 ω6c), and summed feature 8 fatty acids (C18:1ω6c and/or C18:1ω7c) as the major components (Table 2). The major cellular fatty acid profile (>5% of total) of strain MQR6T was summed feature 3 (C16:1ω7c and/or C16:1ω6c), summed feature 8 (C18:1ω6c and/or C18:1ω7c), C14:0, C16:0, and C17:0 cyclo.
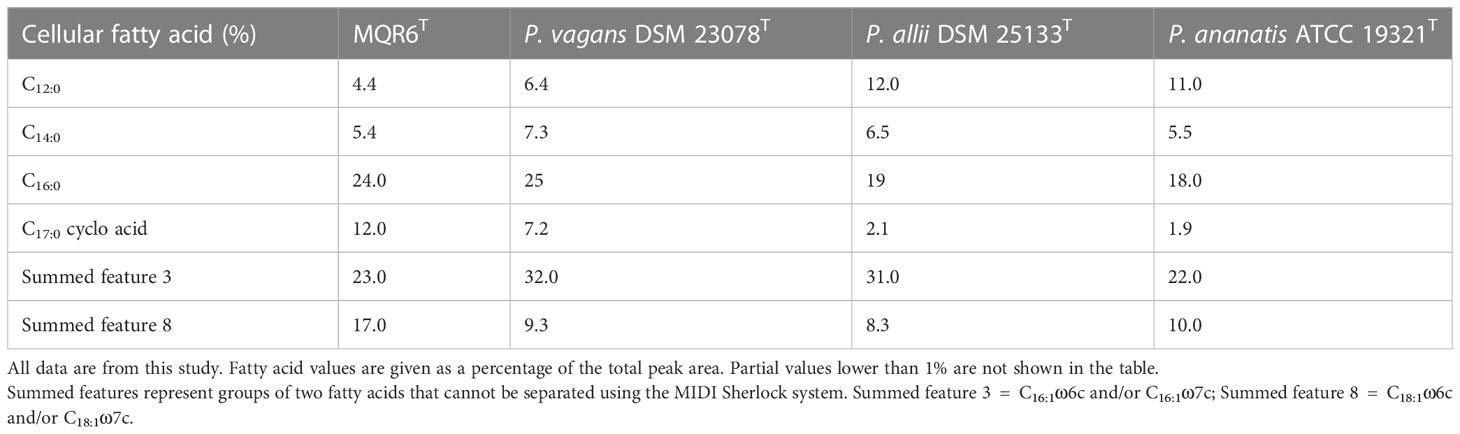
Table 2 The relative cellular fatty acid content (%) of strain MQR6T and representative strains of closely related species of genus Pantoea.
3.2 Phylogenetic analysis of 16S rRNA
The 16S rRNA gene sequence (1440 bp) of strain MQR6T was deposited in GenBank under the accession number OM826981. Based on the analysis of the EzBioCloud database, the strain MQR6T was related closely to P. vagans DSM 23078T (98.47%) and P. ananatis CICC 10283T (98.51% similarity). Phylogenetic trees were reconstructed using the neighbor joining, maximum likelihood, and maximum parsimony methods (Figure 2, Figures S1, S2). All three treeing methods yielded similar phylogeny. Strain MQR6T was located within the genus Pantoea and had a separate clade, indicating that strain MQR6T represented a member of a novel species of genus Pantoea.
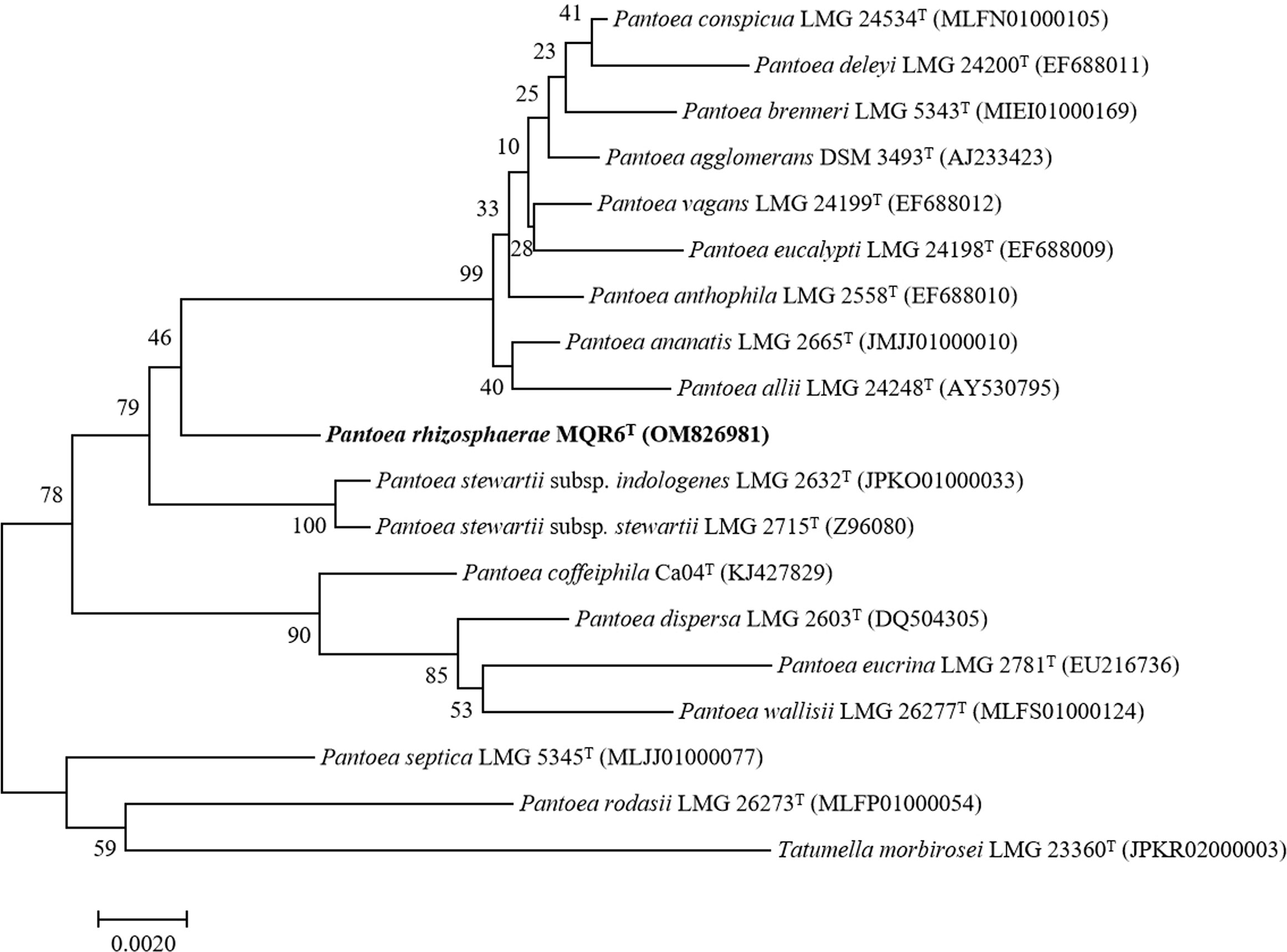
Figure 2 Neighbour-joining phylogenetic tree based on the 16S rRNA gene sequences of Pantoea rhizosphaerae strain MQR6T and other closely related species. The significance of each branch is indicated by a bootstrap value (%) calculated for 1000 subsets. Genbank accession numbers are given in parentheses. Bar = 0.0020 nucleotide substitutions per position.
3.3 Whole-genome analysis
A total of 7,989,160 reads were obtained from genome sequencing of strain MQR6T, yielding a genome of 7,674,100 reads in length. The genome was predicted to contain a total of 4548 genes, which included 4473 protein-coding genes, 4 rRNA genes and 71 tRNA genes. There were 31 contigs in strain MQR6T. The genomic DNA G+C content of strain MQR6T was 51.3%. The dDDH values between strain MQR6T and the type strains of the genus Pantoea were 19.5-23.3%, and the average nucleotide identity based on blast (ANIb) between them was lower than 79.7% (Table 3). The phylogenomic tree based on the Type (Strain) Genome Server (TYGS) web also revealed the distinct phylogeny of strain MQR6T and its close relationship with P. ananatis LMG 2665T, P. allii LMG 24248T and P. dispersa DRS002603T (Figure 3). In a phylogenetic tree based on 120 single-copy genes and the whole genome, strain MQR6T forms a separate evolutionary branch (Figure 4). We conducted a preliminary analysis of the pan-genome, which showed that 1350 shared orthologous coding sequences were clustered into the core genome of Pantoea, 65,787 were represented in the accessory genome, and 8030 were identified as strain-unique genes. The total number of genes increased in the pan-genome of Pantoea with the rise in the analyzed genome number, suggesting that the pan-genome was open (Figure S3). The previous reports showed that the gene number in the core genomes was highly conserved, while many strain-unique genomes and accessory genomes were thought to contribute to species diversity, indicating that species in the genus Pantoea were multifarious.
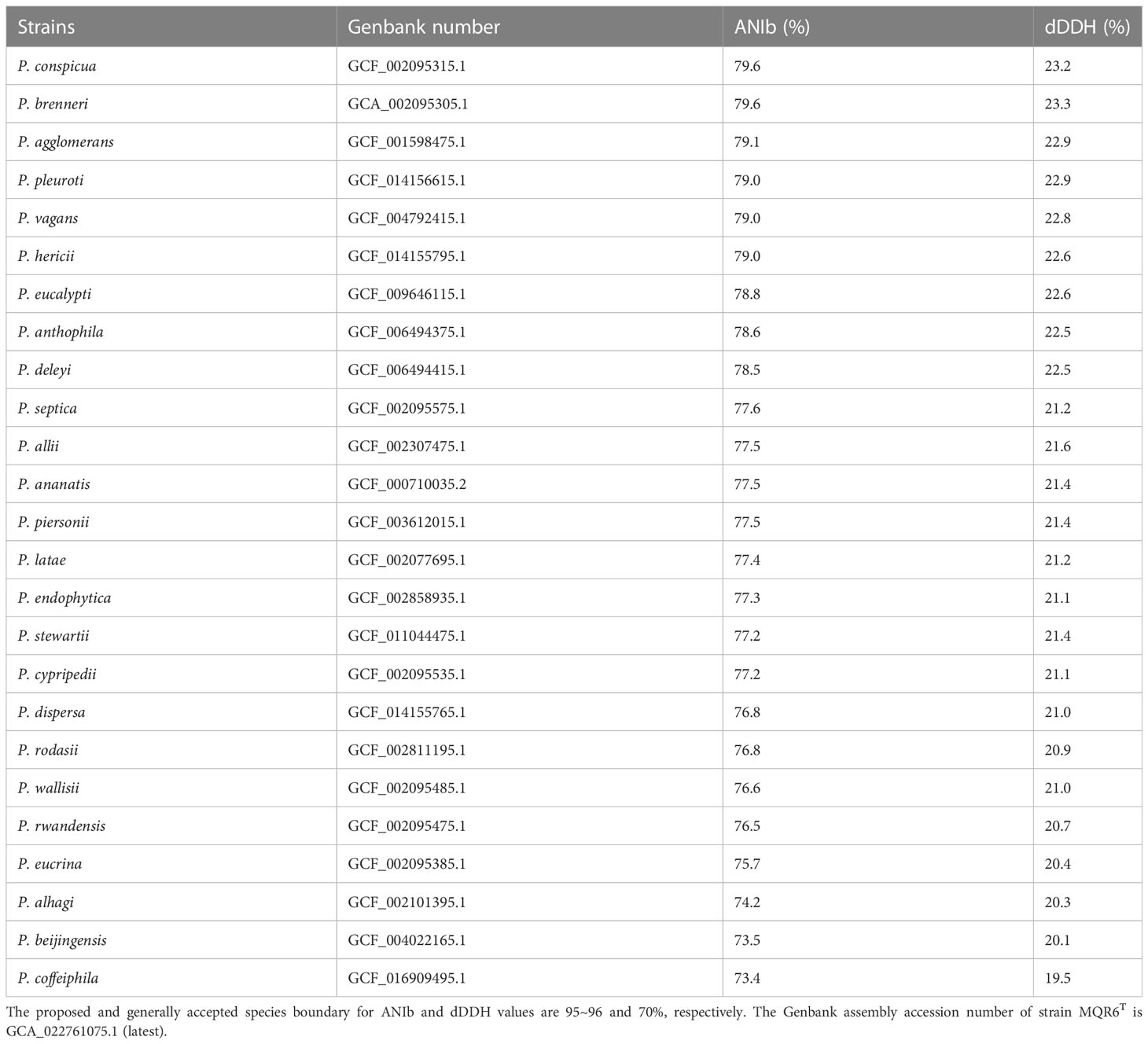
Table 3 Average Nucleotide Identity based on blast (ANIb) and digital DNA-DNA Hybridization (dDDH) values of strain MQR6T compared with all other tested Pantoea strains.
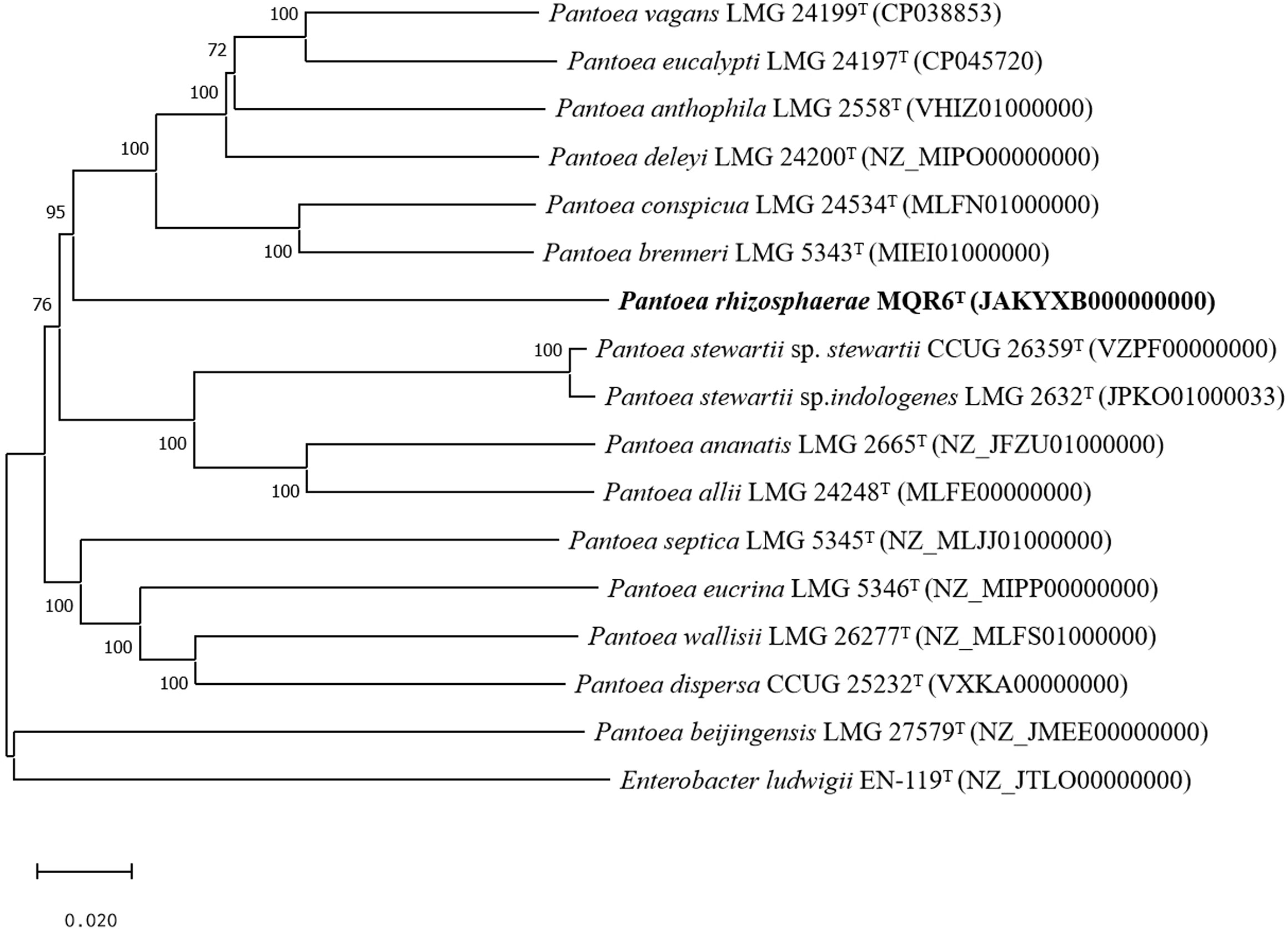
Figure 3 Tree inferred with FastME 2.1.6.1 from GBDP distances calculated from genome sequences of closely related species. The branch lengths are scaled in terms of GBDP distance formula d5. The numbers above branches are GBDP pseudo-bootstrap support values when >60% from 100 replications. GenBank genome accession numbers are given in parentheses. Bar = 0.0020 nucleotide substitutions per position.
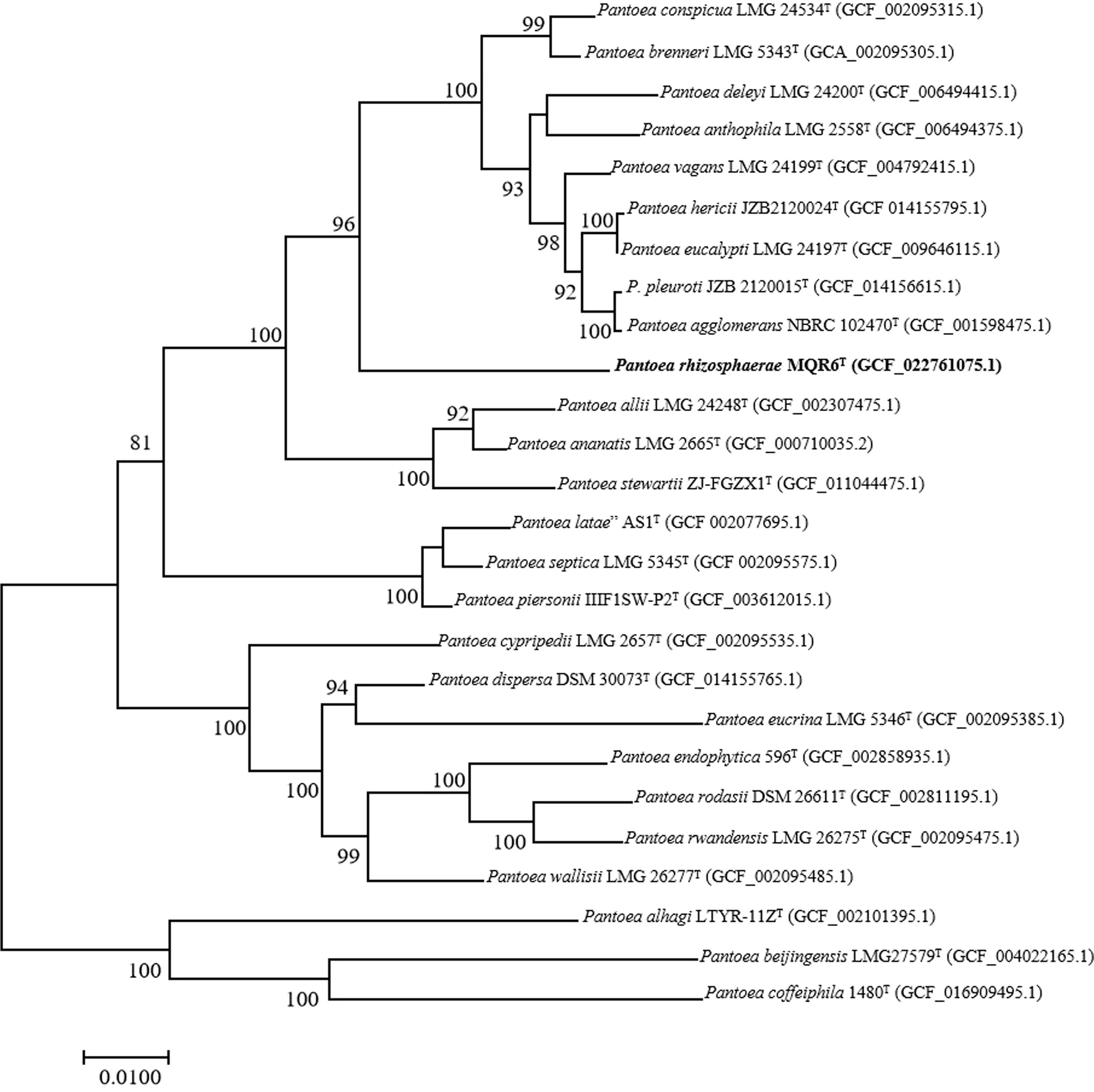
Figure 4 Phylogenomic tree inferred from the concatenation of 120 single-copy bacterial marker genes showing the phylogenetic position of strain MQR6T. A number on nodes represent bootstrap values based on the 1000 replications. Bootstrap values (≥ 70.0%) are shown at branch nodes. RefSeq assembly accession numbers are given in parentheses. Bar represents 0.01 nucleotide substitutions per position.
The whole-genome shotgun sequencing output has been deposited at DDBJ/ENA/GenBank under the accession JAKYXB000000000. The version described in this paper is JAKYXB010000000.
3.4 Identification of genes responsible for plant growth-promoting characteristics of strain MQR6T
Functional analysis of the strain MQR6T genome identified genes associated with the solubilization of phosphate and production of IAA, siderophores and phytohormones that are conducive to plant growth promotion. Eight key genes responsible for IAA production were found in MQR6T genome, including seven genes related to tryptophan operon (trpS, trpB, trpH, trpR, trpA, trpC, trpE) and the ipd C gene encoding indole pyruvate decarboxylase (Table S1). These results indicate that IPyA pathway may be the main pathway for IAA production in the strain MQR6T.
The phosphonate-related phn gene cluster is responsible for the release of biologically available phosphate through the bacterial degradation of phosphonates. Our study revealed that strain MQR6T carries several phn genes, including phnN, phnM etc., showing the capacity to hydrolyze phosphonate into phosphate and alkane (Table S2).
Gluconic acid (GA) is an organic acid that is largely responsible for the solubilization of mineral phosphates. GA biosynthesis is carried out by glucose-1-dehydrogenase along with its co-factor pyrrolo-quinolone quinine. Accordingly, MQR6T genome annotation indicated the presence of several genes related to gluconic acid biosynthesis and its co-factor genes, including pqqBDEF and gcd. Another organic acid identified in the strain MQR6T that is relevant to the phosphate-solubilizing trait is 2-ketogluconic acid produced by gluconate 2-dehydrogenase alpha/beta chain and 2-keto-D-gluconate reductase. Moreover, the strain MQR6T was found to produce other organic acids such as lactic, acetic, glycolic, and succinic (Table S3).
Genomic study showed that MQR6T may synthesize an enterobactin siderophore involving the entABCEF genes. The siderophore is then exported from the cell using entS and is responsible for recovering iron by complexing. Having several siderophore receptor genes (Table S4), strain MQR6T may take up siderophores produced by other organisms as well.
3.5 Quantification of P solubilization by strain MQR6T
Clear halozones were formed around the colonies of strain MQR6T on inorganic phosphate media, with solubilization index (SI) values from 3.20 to 3.98 (Figure 1C). The strain MQR6T was able to solubilize water-insoluble TCP and powdered rock phosphate (RP); however, amount of P solubilized was significantly higher in the TCP medium as compared to the RP medium (Figure 5A). The soluble P concentration in the TCP medium ranged between 232 and 559 mg L-1, with variations over time. By contrast, the soluble P concentration in the RP medium exhibited a range of 159–339 mg L-1. The highest concentration of soluble P in the two media was observed after 24 h incubation, then P solubilization gradually decreased over time.
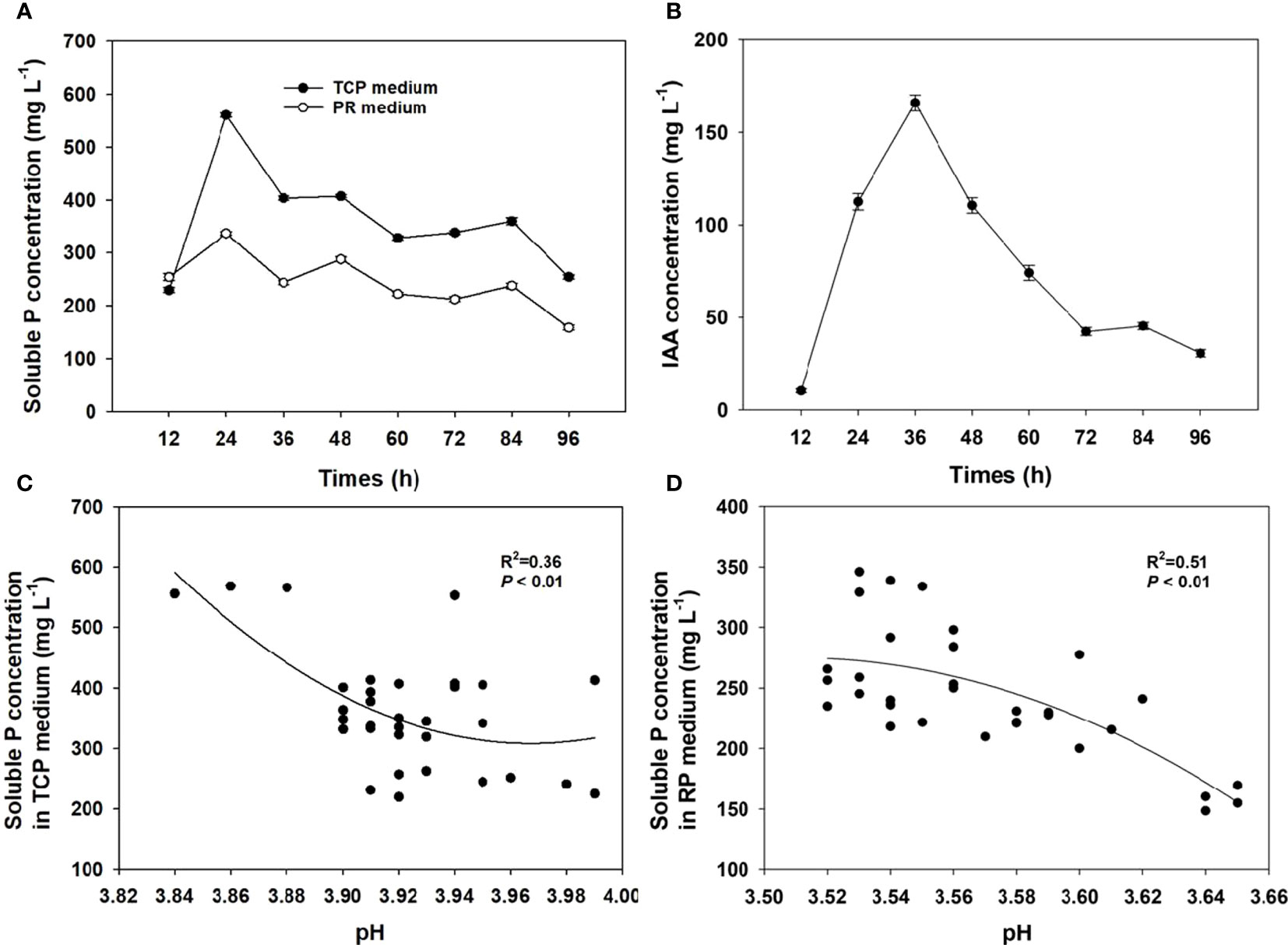
Figure 5 The solubilization of P in the TCP and RP media (A) and production of IAA (B) in the TSB medium with strain MQR6T at 12-h intervals, and the relationship between soluble P concentration and pH in the TCP and RP media (C, D). Means ± SE (where larger than the symbol), n=4. TCP, tricalcium phosphate; RP, rock phosphate.
The pH of the culture filtrates of strain MQR6T decreased from an initial level of 7.22 to 3.84 in the TCP medium and to 3.52 in the RP medium (Figures 5C, D). There was a negative correlation between soluble P concentration and pH value of the TCP medium (R2 = 0.36, P < 0.01) and RP medium (R2 = 0.51, P < 0.01).
3.6 Quantification of IAA production and screening of siderophore
Strain MQR6T showed pink color reaction with Salkowski reagent which indicated the production of IAA (Figure 1F). The highest (166 mg L-1) concentration of IAA in the medium with L-tryptophan was observed after 24 h incubation (Figure 5B). The siderophore production was detected using CAS, showing orange colonies after incubation due to siderophore-dependent removal of Fe from the dye (Figure 1E), indicating the capacity of strain MQR6T to exude siderophore.
3.7 Plant growth of and nutrient optake by A. truncatum seedlings
Inoculation with strain MQR6T significantly enhanced the shoot and root growth, dry weight accumulation and P uptake of A. truncatum seedlings compared to those grown in non-inoculated soils (Table 4 and Figure 6). Compared with the non-inoculated control treatment, plant parameters in the MQR6T inoculation treatment increased by 47% (height), 53% (trunk diameter), 15% (SPAD chlorophyll content), 100% (shoot dry weight) and 133% (root dry weight). Phosphorus accumulations in shoots and roots were, respectively, 102% and 79% greater in the MQR6T inoculated treatment than those in the non-inoculated control. Similarly, P addition alone significantly increased plant height, trunk diameter, dry weight and P uptake when compared to the non-inoculated control treatment. Moreover, combined P addition and inoculation treatment significantly increased plant height, trunk diameter, biomass accumulation and P uptake when compared to the control, the P addition only and the inoculation only treatments. No significant difference was found in root/shoot ratio among the four treatments, and there was no significant difference in shoot and root biomass, SPAD chlorophyll content and P uptake between treatments with P addition and inoculation only.
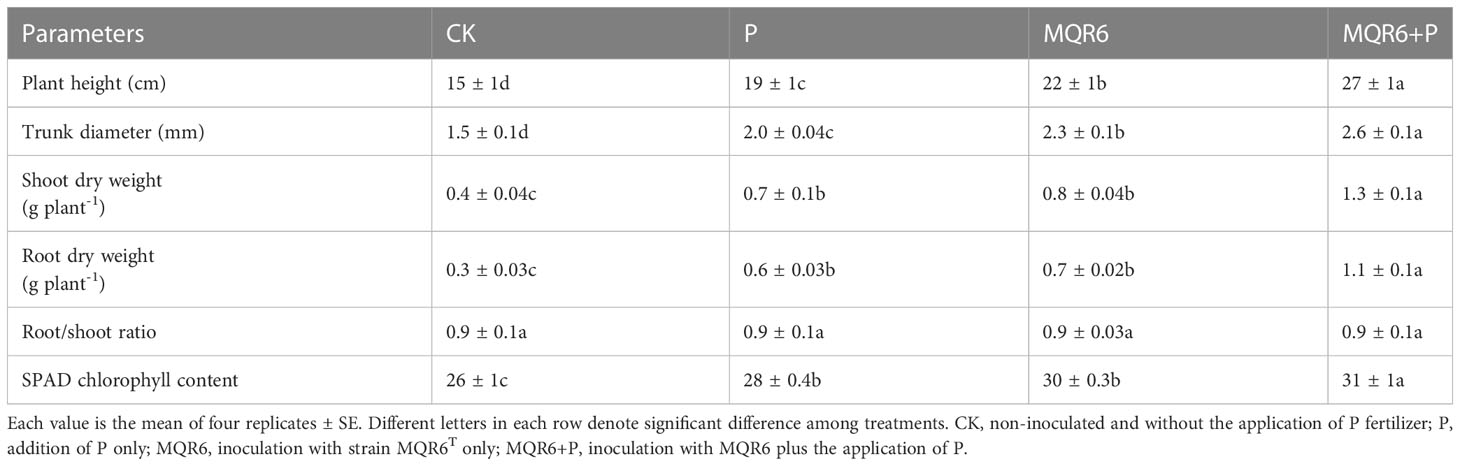
Table 4 The effect of inoculation with strain MQR6T and supply of P on the growth of Acer truncatum seedlings.
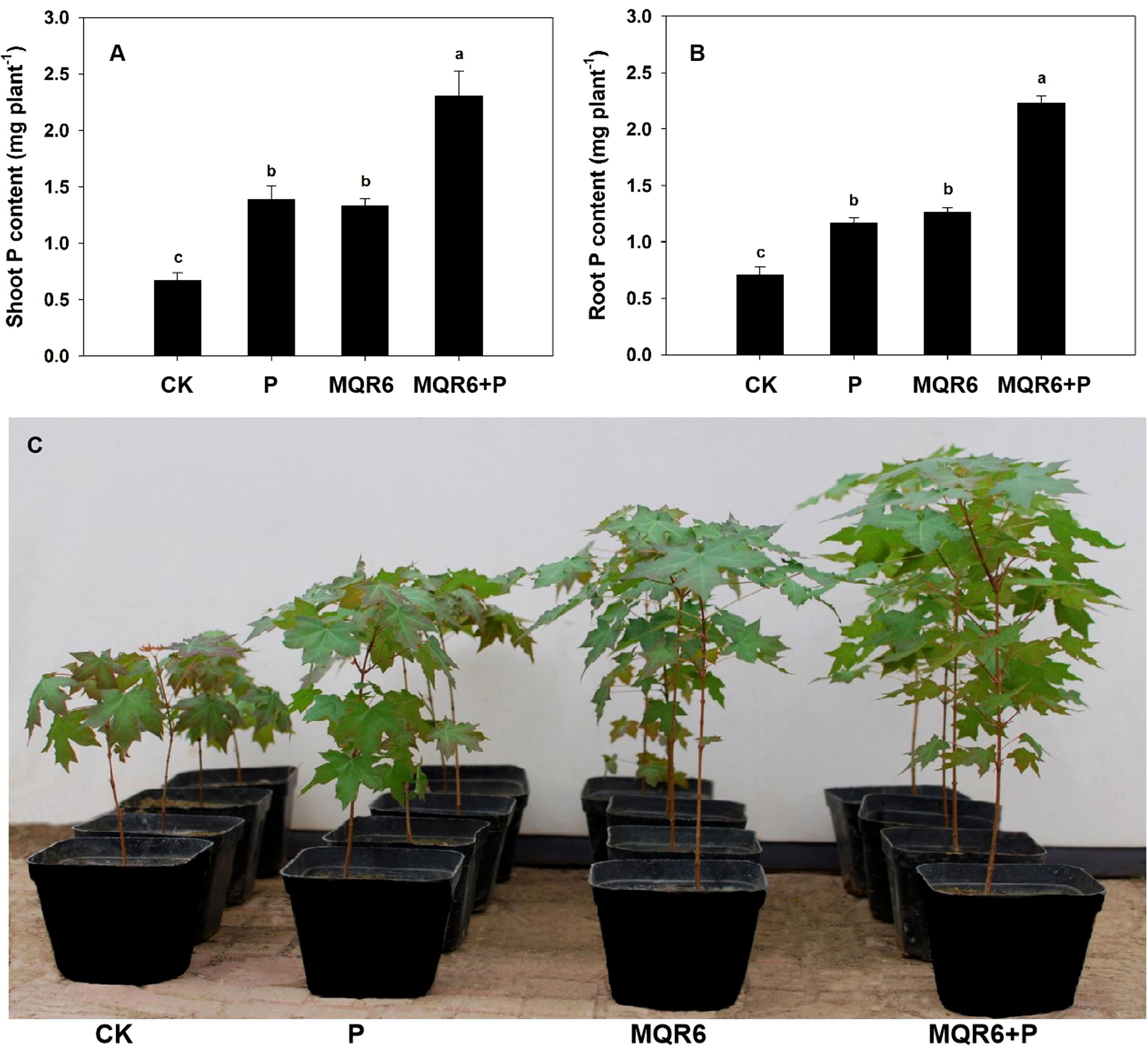
Figure 6 Effect of strain MQR6T and supply of P (as KH2PO4) on P accumulation in shoots (A) and roots (B) and shoot growth (C) of Acer truncatum 290 days after sowing. Means + SE, n=4. CK, non-inoculated and without the application of P fertilizer; P, addition of P only; MQR6, inoculation with strain MQR6T only; MQR6+P, inoculation with MQR6 plus the application of P. Different lowercase letters denote significant difference (P ≤ 0.05) among treatments.
4 Discussion
Microorganisms capable of producing clear zones (halos) around the colonies growing on solid medium were selected as potential phosphate solubilizers (Gupta et al., 1994) and were screened repeatedly by a plate assay method using either Pikovskaya agar medium or TCP medium (Yu et al., 2011). Generally, the preliminary capacity of PSM to solubilize the insoluble phosphate was determined by the SI value (Li et al., 2020). Previous study showed that among 78 isolated strains, the majority exhibited a low index (SI<2.0), but eight strains had intermediate values (2.0<SI<4.0) and none of the strains showed a high solubility index (SI>4.0) for CaHPO4 solubilization (Marra et al., 2012). Similar results were also reported with yeast strains exhibiting a P-solubilization potential with SI ranging from 1.2 to 2.8; and a significant positive correlation was found between the solubilized amounts of P and the P solubilization index (Hesham and Mohamed, 2011). By contrast, other study observed isolated Pseudomonas sp. strain PSB-2 exhibited good solubilization of TCP with a high SI (>4.0) (He and Wan, 2022). In the present study, MQR6T was isolated from the rhizosphere soil of A. truncatum, and showed clear halos of solubilizing TCP with the largest SI value of 3.98. Hence, the strain was found to be efficient phosphate solubilizer, and was selected for further evaluation.
In the present study, the general characteristics of MQR6T corresponded with the genus Pantoea as described by Brady et al. (Brady et al., 2010): Gram-negative, rod-shaped, facultatively anaerobic, non-spore-forming, and commonly motile by means of peritrichous flagella (Figure 1). Analysis of 16S rRNA and complete genome sequencing and the phylogenetic trees reconstructed by using different methods found that the isolate belonged to Pantoea spp. and had a separate clade (Figures 2–4). In addition, the main fatty acids were hexadecanoic (C16:0), cyclo-heptadecanoic (C17:0 cycle) and summed feature 3 acids (containing C16:1ω6c and/or C16:1ω7c) (Table 2), in accordance with characteristics of Pantoea spp. (Mergaert et al., 1993). The genomic DNA G+C content of strain MQR6T was 51.3 mol%, which is consistent with the DNA G+C contents ranging from 49.7 to 61.3 mol% of other members of the genus Pantoea (Brady et al., 2010). The dDDH values between strain MQR6T and the type strains of the genus Pantoea were 19.5-23.3%, well below the dDDH standard cut-off value of 70% (Wang et al., 2020). On the basis of phylogenetic, physiological and chemotaxonomic characteristics, strain MQR6T represents a novel species within the genus Pantoea, for which the name Pantoea rhizosphaerae sp. nov. is proposed.
The bacterial genus Pantoea comprises many versatile species that have been isolated from aquatic and terrestrial environments, living in association with plants, insects, humans, and animals (Castagno et al., 2011; Dutkiewicz et al., 2016; Luziatelli et al., 2020). Some isolates possess the capacities for nitrogen fixation, P solubilization, antibiotic production, and plant growth-promotion, and are being explored currently for agricultural applications (Chen and Liu, 2019; Luziatelli et al., 2020). Strain of P-dissolving Pantoea agglomerans R-42 was isolated from soybean rhizosphere had a marked insoluble phosphate-solubilizing activity (Son et al., 2006). In a different study, 50 PSB strains were isolated from the rhizosphere of Lotus tenuis grown in low-P soils (< 3 mg kg-1 of the available P) of the Salado River Basin; based on 16S rRNA gene sequencing, they belonged to Pantoea, Erwinia, Pseudomonas, Rhizobium, and Enterobacter genera; the most efficient isolate was identified as Pantoea eucalypti, a novel species of plant growth-promoting rhizobacteria (Castagno et al., 2011). Phosphate-solubilizing bacteria are known to be able to solubilize different forms of inorganic phosphates. The isolates Pantoea agglomerans ZB and Pantoea sp. S32 solubilized TCP, CaHPO4, RP, AlPO4, and FePO4 (Castagno et al., 2011; Chen and Liu, 2019; Li et al., 2020). In the present study, the tested strain had the capacity to solubilize inorganic TCP and RP, and the solubilization of TCP was about 60% more effective than RP. These results indicated that the tested strain MQR6T may be effective to release soluble P from insoluble TCP in calcareous soil, which can be a source of P for plant growth.
Some studies have showed that P solubilization by bacterial strain was significantly influenced by the sources of P used in the media (Son et al., 2006; Panhwar et al., 2011). In the work presented here, we found that the concentration of soluble P was lower in the RP medium than the TCP medium, even though the pH was slightly lower in the RP than TCP medium. Similarly in the other study, a larger drop in pH was noted in the RP that TCP medium (Panhwar et al., 2011). The lower concentration of solubilized P in RP medium (Figure 5D) may be due to hampering (or even cessation) of bacterial P-solubilization activity at low pH. The influence of initial pH on the growth of MQR6T was investigated in the pH range of 3.0 to 12.0, and pH levels below 4 resulted in a large reduction of bacterial population (data not shown). A clear relationship was established between bacterial growth and P solubilization in broth cultures (Panhwar et al., 2011). In addition, the rock phosphate has low P solubility as compared to calcium phosphate. This indicates decreased bacterial population and low pH may be associated with lower P solubilization in RP than TCP media.
Low-molecular-weight organic acids, such as acetic, oxalic and gluconic, have a high potential to solubilize water-insoluble inorganic phosphates (Hassan et al., 2019; Chen et al., 2020). Gluconic and 2-keto gluconic acids produced by bacteria play an important role in weathering and solubilization of phosphate in soil, acting as Ca2+ chelators and providing the acidification of the external environment to dissolve the sparingly soluble calcium phosphates (Yu et al., 2011). Furthermore, the glucose dehydrogenase (gcd) gene, coding for the first enzyme in the direct oxidation pathway, contributes significantly to mineral phosphate solubilization by the plant growth-promoting rhizabacteria (Shariati et al., 2017; He and Wan, 2022). Similarly, our study revealed that the genes related to gluconic and 2-keto-D-gluconic acid production were found in the strain MQR6T, including gcd, gdh, pqq, and bet (Table S3). Several genes responsible for biosynthesis of organic acids glycolic, acetic and succinic were also found in the genome of the strain MQR6T. Moreover, many PSM lower the pH of the medium by H+ extrusion (Surapat et al., 2013). A significant linear correlation was observed between culture pH and P solubilized from inorganic phosphate (Yu et al., 2011). Similar results were found between pH and solubilized P concentration in both the media tested in the present study (Figures 5C, D). Inoculation with strain MQR6T may induce excretion of organic acid anions and H+ (via separate mechanisms), increasing the concentration of organic ligands and lowering the rhizosphere pH, enhancing a capacity to mobilize P in the rhizosphere.
Inoculation with MQR6T showed a positive effect on plant height, trunk diameter, dry biomass, and P accumulation of A. truncatum Bunge seedlings in comparison with non-inoculated control (Table 4, Figure 6). Similar growth-promoting effects such as enhanced plant growth, dry weight accumulation and P uptake were also exhibited in sugar maple (A. saccharum Marsh.) seedlings inoculated with the strain MQR6T (data not shown). The possible explanations for increased growth and P accumulation under inoculation with MQR6T may include: (і) direct contribution to the mobilization of soil P by influencing rhizosphere pH and exudation of organic acid anions and phosphatases (Hinsinger, 2001; Shariati et al., 2017); (ii) the enhancement of root growth, including increased root length and root surface area, may be associated with improved spatial nutrient acquisition and chemical mobilization of P nutrients (Ma et al., 2021); (iii) several genes related to tryptophan operon (trpS, trpB, trpH, trpR, trpA, trpC, trpE) and one ipd C gene encoding indole pyruvate decarboxylase found in the strain MQR6T genome may affect the amount of IAA produced, influencing root growth and water and nutrient acquisition (Radwanski and Last, 1995); (iv) more than 10 phosphonate-related Phn genes found in the strain MQR6T genome may enable sufficient phosphate uptake to support bacterial growth (Stasi et al., 2019); and (v) several genes for siderophore receptors, such as entA, entB and entC, found in the strain MQR6T genome are responsible for iron recovery by complex formation (Saha et al., 2013). Furthermore, the transcriptomic responses of A. truncatum roots inoculated with MQR6T are currently under investigation to decipher the relevant plant-microbe interactions.
The application of P in combination with MQR6T significantly improved the plant growth, dry weight and P uptake of A. truncatum seedlings compared with the P alone and MQR6T alone treatments (Figure 6). Similarly, the most pronounced beneficial effect on growth of walnut plants was observed in the treatment with both inoculation and P addition, which was attributed partly to an increase in the population of PSB in the rhizosphere (Yu et al., 2011). Addition of inorganic P to the inoculated soil further stimulated growth of bacteria and raised the total population of PSB (Yu et al., 2011). Other study reported that competition for P did exist among PSB, arbuscular mycorrhizal (AM) fungi and plant, especially in a low-P soil, and the competition would have been alleviated by supplying an optimal amount of P to the soil (Zhang et al., 2014). In the present study, addition of 50 mg P kg-1 soil may be beneficial not only for proliferation and survival of strain MQR6T but also for growth of A. truncatum seedings. In addition, the interactions between MQR6T and P fertilizer could cause a synergistic effect that allowed improved root and shoot growth of A. truncatum. However, the molecular and physiological mechanism behind the effect of inoculation with MQR6T with or without P application on A. truncatum growth, especially in the field conditions, needs to be investigated further.
5 Conclusion
In conclusion, the new strain MQR6T, isolated from A. truncatum rhizosphere, was demonstrated to belong to a new Pantoea rhizosphaerae species on the basis of phylogenetic, physiological and chemotaxonomic characteristics. The P. rhizosphaerae strain MQR6T exhibited a high capacity to solubilize phosphate and produce IAA and siderophores, associated with the relevant genes found in the genome. The significant enhancement in shoot and root growth and P uptake of A. truncatum seedlings inoculated with MQR6T confirmed that it is a growth-promoting rhizobacterium, potentially providing a basis for a new inoculant biofertilizer for A. truncatum cultivation, especially in low-P soils.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, JAKYXB000000000.
Author contributions
QM, XX and XZ designed and performed experiments, analyzed and interpreted data, and wrote the manuscript. SH performed genomic analysis, XingW assisted with experimental design and performed the experiments, LC assisted with experimental design and supervised the study. XinghW and SP assisted with performing the experiments and analyzing the data. ZR revised the manuscript critically. XX and XZ supervised the study, designed experiments and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Science Foundation of the Chinese Academy of Forestry (CAFYBB2020ZB005) and the Agricultural Science and Technology Innovation Program (ASTIP No. CAAS-ZDRW202201).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1218445/full#supplementary-material
Abbreviations
dDDH, digital DNA–DNA hybridization; TCP, tricalcium phosphate; RP, rock phosphate; DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH; CICC, China Center of Industrial Culture Collection; NCBI, National Center for Biotechnology Information; NJ, neighbour joining; ML, maximum likelihood; MP, maximum parsimony; TSA, tryptone soy agar; TSB, tryptone soy broth; IAA, indole-3-acetic acid; CAS, chromo azurol S; SI, solubilization index [the ratio of the total diameter (colony+halozone) and the colony diameter]; ANIb, the average nucleotide identity based on blast; TYGS, Type (Strain) Genome Server; GA, Gluconic acid.
The DDBJ/ENA/GenBank accession numbers for the whole-genome sequence and the 16S rRNA gene sequence of Pantoea rhizosphaerae sp. nov. strain MQR6T are JAKYXB000000000 and OM826981, respectively.
References
Auch, A. F., von Jan, M., Klenk, H. P., Göker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. stand. Genomic Sci. 2, 117–134. doi: 10.4056/sigs.531120
Backer, R., Rokem, J. S., llangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9, 1473. doi: 10.3389/fpls.2018.01473
Bi, W., Gao, Y., Shen, J., He, C., Liu, H., Peng, Y., et al. (2016). Traditional uses, phytochemistry, and pharmacology of the genus Acer (maple): a review. J. Ethnopharmacol. 189, 31–60. doi: 10.1016/j.jep.2016.04.021
Bouizgarne, B., Bakki, M., Boutasknit, A., Banane, B., El Ouarrat, H., Ait El Maalem, S., et al. (2023). Phosphate and potash solubilizing bacteria from Moroccan phosphate mine showing antagonism to bacterial canker agent and inducing effective tomato growth promotion. Front. Plant Sci. 14, 970382. doi: 10.3389/fpls.2023.970382
Brady, C. L., Cleenwerck, I., Venter, S. N., Engelbeen, K., De Vos, P., Coutinbo, T. A. (2010). Emended description of the genus Pantoea, description of four species from human clinical samples, Pantoea septica sp. nov., Pantoea eucrina sp. nov., Pantoea brenneri sp. nov. and Pantoea conspicua sp. nov., and transfer of Pectobacterium cypripedii (Hori 1911) Brenner et al Emend. hauben et al To the genus as Pantoea cypripedii comb. nov. Int. J. Syst. Evol. Microbiol. 60, 2430–2440. doi: 10.1099/ijs.0.017301-0
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., Huerta-Cepas, J. (2021). EggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Castagno, L. N., Estrella, M. J., Sannazzaro, A. I., Grassano, A. E., Ruiz, O. A. (2011). Phosphate-solubilization mechanism and in vitro plant growth promotion activity mediated by Pantoea eucalypti isolated from Lotus tenuis rhizosphere in the salado river basin (Argentina). J. Appl. Microbiol. 110, 1151–1165. doi: 10.1111/j.1365-2672.2011.04968.x
Chaudhari, N. M., Gupta, V. K., Dutta, C. (2016). BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 6, 24373. doi: 10.1038/srep24373
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., Parks, D. H. (2020). GTDB-tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Chen, L., Gao, X., Ma, Q., Liu, H., Wang, X., Xu, Y., et al. (2020). Dyadobacter luteus sp. nov., isolated from rose rhizosphere soil. Arch. Microbiol. 202, 191–196. doi: 10.1007/s00203-019-01738-5
Chen, Q., Liu, S. (2019). Identification and characterization of the phosphate-solubilizing bacterium Pantoea sp. S32 in reclamation soil in shanxi, China. Front. Microbiol. 10, 2171. doi: 10.3389/fmicb.2019.02171
Chouyia, F. E., Ventorino, V., Pepe, O. (2022). Diversity, mechanisms and beneficial features of phosphate-solubilizing Streptomyces in sustainable agriculture: a review. Front. Plant Sci. 13, 1035358. doi: 10.3389/fpls.2022.1035358
Coil, D., Jospin, G., Darling, A. E. (2015). A5-miseq: an updated pipeline to assemble microbial genomes from illumina MiSeq data. Bioinformatics 31, 587–589. doi: 10.1093/bioinformatics/btu661
Delcher, A. L., Bratke, K. A., Powers, E. C., Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
De Zutter, N., Ameye, M., Bekaert, B., Verwaeren, J., De Gelder, L., Audenaert, K. (2022). Uncovering new insights and misconceptions on the effectiveness of phosphate solubilizing rhizobacteria in plants: a meta-analysis. Front. Plant Sci. 13, 858804. doi: 10.3389/fpls.2022.858804
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., Milanowski, J. (2016). Pantoea agglomerans: a mysterious bacterium of evil and good. part IV. beneficial effects. Ann. Agric. Environ. Med. 23, 206–222. doi: 10.5604/12321966.1203879
Etesami, H., Jeong, B. R., Glick, B. R. (2021). Contribution of arbuscular mycorrhizal fungi, phosphate–solubilizing bacteria, and silicon to p uptake by plant. Front. Plant Sci. 12, 699618. doi: 10.3389/fpls.2021.699618
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Gavini, F., Mergaert, J., Beji, A., Mielcarek, C., Izard, D., Kersters, K., et al. (1989). Transfer of Enterobacter agglomerans (Beijerinck 1988) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersa sp. nov. Int. J. Syst. Bacteriol. 39, 337–345. doi: 10.1099/00207713-39-3-337
Gupta, R., Singal, R., Shankar, A., Kuhad, R. C., Saxena, R. K. (1994). A modified plate assay for screening phosphate solubilizing microorganisms. J. Gen. Appl. Microbiol. 40, 255–260. doi: 10.2323/jgam.40.255
Hassan, M. K., Mclnroy, J. A., Kloepper, J. W. (2019). The interactions of rhizodeposits with plant growth-promoting rhizobacteria in the rhizosphere: a review. Agriculture 9, 142. doi: 10.3390/agriculture9070142
He, D., Wan, W. (2022). Distribution of culturable phosphate-solubilizing bacteria in soil aggregates and their potential for phosphorus acquisition. Microbiol. Spectr. 10, e00290–e00222. doi: 10.1128/spectrum.00290-22
Herrera-Estrella, L., López-Arredondo, D. (2016). Phosphorus: the underrated element for feeding the world. Trends Plant Sci. 21, 461–463. doi: 10.1016/j.tplants.2016.04.010
Hesham, A. E. L., Mohamed, H. (2011). Molecular genetic identification of yeast strains isolated from Egyptian soils for solubilization of inorganic phosphates and growth promotion of corn plants. J. Microbiol. Biotechnol. 21, 55–61. doi: 10.4014/jmb.1006.06045
Hinsinger, P. (2001). Bioavailability of soil inorganic p in the rhizosphere as affected by root-induced chemical changes: a review. Plant Soil 237, 173–195. doi: 10.1023/A:1013351617532
Kanehisa, M., Sato, Y., Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, L., Chen, R., Zuo, Z., Lv, Z., Yang, Z., Mao, W., et al. (2020). Evaluation and improvement of phosphate solubilization by an isolated bacterium Pantoea agglomerans ZB. World J. Microb. Biot 36, 27. doi: 10.1007/s11274-019-2744-4
Lowe, T. M., Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Luziatelli, F., Ficca, A. G., Cardarelli, M., Melini, F., Cavalieri, A., Ruzzi, M. (2020). Genome sequencing of Pantoea agglomerans C1 provides insights into molecular and genetic mechanisms of plant growth-promotion and tolerance to heavy metals. Microorganisms 8, 153. doi: 10.3390/microorganisms8020153
Ma, Q., Sun, L., Tian, H., Rengel, Z., Shen, J. (2021). Deep banding of phosphorus and nitrogen enhances Rosa multiflora growth and nutrient accumulation by improving root spatial distribution. Sci. Hortic. 277, 109800. doi: 10.1016/j.scienta.2020.109800
Marra, L. M., Soares, C. R. F. S., de Oliveira, S. M., Avelar Ferreira, P. A., Soares, B. L., de Fráguas Carvalho, R., et al. (2012). Biological nitrogen fixation and phosphate solubilization by bacteria isolated from tropical soils. Plant Soil 357, 289–307. doi: 10.1007/s11104-012-1157-z
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H., Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 14, 60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 50, 801–807. doi: 10.1093/nar/gkab902
Mergaert, J., Verdonck, L., Kersters, K. (1993). Transfer of Erwinia ananas (synonym, Erwinia uredovora) and Erwinia stewartiito the genus Pantoea emend. as Pantoea ananas (Serrano 1928) comb. nov. and Pantoea stewartii (Smith 1898) comb. nov., respectively, and description of Pantoea stewartii subsp. indologenes subsp. nov. Int. J. Syst. Bacteriol. 43, 162–173. doi: 10.1099/00207713-43-1-162
Naqqash, T., Hameed, S., Imran, A., Hanif, M., Majeed, A., van Elsas, J. (2016). Differential response of potato toward inoculation with taxonomically diverse plant growth promoting rhizobacteria. Front. Plant Sci. 7, 144. doi: 10.3389/fpls.2016.00144
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, 206–214. doi: 10.1093/nar/gkt1226
Panhwar, Q. A., Radziah, O., Zaharah, A. R., Sariah, M., Razi, I. M. (2011). Role of phosphate solubilizing bacteria on rock phosphate solubility and growth of aerobic rice. J. Environ. Biol. 32, 607–612. doi: 10.1007/s11852-010-0106-3
Radwanski, E. R., Last, R. L. (1995). Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell 7, 921–934. doi: 10.1105/tpc.7.7.921
Rafique, E., Mumtaz, M. Z., Ullah, I., Rehman, A., Qureshi, K. A., Kamran, M., et al. (2022). Potential of mineral-solubilizing bacteria for physiology and growth promotion of Chenopodium quinoa willd. Front. Plant Sci. 13, 1004833. doi: 10.3389/fpls.2022.1004833
Reis, V. M., Santos, P. E., Tenorio-Salgado, S., Vogel, J., Stoffels, M., Guyon, S., et al. (2004). Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int. J. Syst. Evol. Micr 54, 2155–2162. doi: 10.1099/ijs.0.02879-0
Richter, M., Rosselló-Móra, R., Glöckner, F. O., Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Saha, R., Saha, N., Donofrio, R. S., Bestervelt, L. L. (2013). Microbial siderophores: a mini review. J. Basic Microb. 53, 303–317. doi: 10.1002/jobm.201100552
Saitou, N., Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sasser, M. (1990). Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. Newark, DE: MIDI inc. Newark, NJ, USA: USFCC Newslett)
Schubert, M., Lindgreen, S., Orlando, L. (2016). AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes 9, 88. doi: 10.1186/s13104-016-1900-2
Schwyn, B., Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Annal Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Shariati, J. V., Malboobi, M. A., Tabrizi, Z., Tavakol, E., Owlia, P., Safari, M. (2017). Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci. Rep. 7, 15610. doi: 10.1038/s41598-017-15820-9
Son, H. J., Park, G. T., Cha, M. S., Heo, M. S. (2006). Solubilization of insoluble inorganic phosphates by a novel salt-and pH-tolerant Pantoea agglomerans r-42 isolated from soybean rhizosphere. Bioresour. Technol. 97, 204–210. doi: 10.1016/j.biortech.2005.02.021
Stasi, R., Neves, H. I., Spira, B. (2019). Phosphate uptake by the phosphonate transport system PhnCDE. BMC Microbiol. 19, 79. doi: 10.1186/s12866-019-1445-3
Surapat, W., Pukahuta, C., Rattanachaikunsopon, P., Aimi, T., Boonlue, S. (2013). Characteristics of phosphate solubilization by phosphate-solubilizing bacteria isolated from agricultural chili soil and their efficiency on the growth of chili (Capsicum frutescens l. cv. hua rua). Chiang Mai J. Sci. 40, 11–25.
Tamura, K., Nei, M., Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Vance, C. P., Uhde-Stone, C., Allan, D. L. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Wang, R., Fan, J., Chang, P., Zhu, L., Zhao, M., Li, L. (2019). Genome survey sequencing of Acer truncatum bunge to identify genomic information, simple sequence repeat (SSR) markers and complete chloroplast genome. Forests 10, 87. doi: 10.3390/f10020087
Wang, X., He, S. W., Guo, H. B., Thin, K. K., Gao, J. S., Wang, Y., et al. (2020). Pseudomonas rhizoryzae sp. nov., isolated from rice. Int. J. Syst. Evol. Microbiol. 70, 944–950. doi: 10.1099/ijsem.0.003852
Watanabe, F. S., Olsen, S. R. (1965). Test of an ascorbic acid method for determining phosphorus in water and NaHCO3 extracts from soil. Soil Sci. Soc. Am. J. 29, 677–678. doi: 10.2136/sssaj1965.03615995002900060025x
Woźniak, M., Gałązka, A., Tyśkiewicz, R., Jaroszuk-Ściseł, J. (2019). Endophytic bacteria potentially promote plant growth by synthesizing different metabolites and their Phenotypic/Physiological profiles in the biolog GEN III MicroPlate™ test. Int. J. Mol. Sci. 20, 5283. doi: 10.3390/ijms20215283
Yu, X., Liu, X., Zhu, T. H., Liu, G. H., Mao, C. (2011). Isolation and characterization of phosphate-solubilizing bacteria from walnut and their effect on growth and phosphorus mobilization. Biol. Fert. Soils 47, 437–446. doi: 10.1007/s00374-011-0548-2
Keywords: Pantoea, genome, phosphate-solubilizing bacteria, Acer truncatum, P accumulation
Citation: Ma Q, He S, Wang X, Rengel Z, Chen L, Wang X, Pei S, Xin X and Zhang X (2023) Isolation and characterization of phosphate-solubilizing bacterium Pantoea rhizosphaerae sp. nov. from Acer truncatum rhizosphere soil and its effect on Acer truncatum growth. Front. Plant Sci. 14:1218445. doi: 10.3389/fpls.2023.1218445
Received: 07 May 2023; Accepted: 29 June 2023;
Published: 14 July 2023.
Edited by:
Iftikhar Ahmad, COMSATS University Islamabad, PakistanReviewed by:
Abdelkarim Filali-Maltouf, Mohammed V University of Rabat, MoroccoAnnapurna Kannepalli, Indian Agricultural Research Institute (ICAR), India
Copyright © 2023 Ma, He, Wang, Rengel, Chen, Wang, Pei, Xin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoxia Zhang, emhhbmd4aWFveGlhQGNhYXMuY24=; Xuebing Xin, eGlueGIwMUAxNjMuY29t
 Qinghua Ma
Qinghua Ma Shanwen He3
Shanwen He3 Zed Rengel
Zed Rengel Lin Chen
Lin Chen Xinghong Wang
Xinghong Wang Xiaoxia Zhang
Xiaoxia Zhang