- 1State Key Laboratory of Aridland Crop Science, College of Agronomy, Gansu Agricultural University, Lanzhou, China
- 2Gansu Key Laboratory of Crop Genetic Improvement and Germplasm Innovation, Gansu Agricultural University, Lanzhou, China
- 3Gansu Yasheng Agricultural Research Institute Co. Ltd, Crop Office, Lanzhou, China
The CUP-SHAPED COTYLEDON2 (CUC2) gene plays an important role in the formation of apical meristem and organ edges in plants. The apical meristematic tissue of Brassica rapa (B. rapa) is associated with cold resistance, however, the role of the CUC2 gene in cold resistance of B.rapa is unclear. In this study, we used bioinformatics software to analyze the structure of BrCUC2 gene, real-time fluorescence quantitative PCR to detect the expression level of BrCUC2, constructed transgenic Arabidopsis thaliana by the flower dipping method and subcellular localization for functional validation. The results showed that, we isolated a 1104 bp open reading frame of BrCUC2 from the winter B. rapa cultivar ‘Longyou 7’. The BrCUC2 contains a highly conserved domain belonging to the NAM superfamily. Its homologus CUC genes contain similar conserved motifs and are closely related to Brassica oleracea (B.oleracea), and the N-terminal of amino acid sequence contains NAC domain. BrCUC2 protein was localized in the nucleus and self-activation tests showed that pGBKT7-BrCUC2 had self-activation. Tissue-specific expression analysis and promoter β-Glucuronidase (GUS) activity showed that BrCUC2 had high expression levels in B. rapa growth points and A. thaliana leaf edges, stems and growth points. After low-temperature stress, BrCUC2 showed greater expression in ‘Longyou 7,’ which presents strong cold resistance and concave growth points, than in ‘Longyou 99,’ which presents weak cold resistance and protruding growth points. BrCUC2 promoter contains multiple elements related to stress responses. BrCUC2 overexpression revealed that the phenotype did not differ from that of the wild type during the seedling stage but showed weak growth and a dwarf phenotype during the flowering and mature stages. After low-temperature treatment, the physiological indexes and survival rate of BrCUC2-overexpression lines of Arabidopsis thaliana (A. thaliana) were better than those of the wild type within 12 h, although differences were not observed after 24 h. These results showed that BrCUC2 improved the low-temperature tolerance of transgenic A. thaliana within a short time. It can provide a foundation for the study of cold resistance in winter B. rapa.
Introduction
Rapeseed production is important for the sustainable supply of edible oil in China (Li et al., 2022). In northern China, the temperature can reach -32°C in winter, drought occurs in spring, and ecological conditions are harsh (Zhou et al., 2014). Wind erosion and farmland degradation are severe, and desertified areas are expanding. The successful northward migration of winter rapeseed, which represents the only winter oil seed crop, has provided economic benefits in the north (Ma et al., 2019b). Moreover, the plant has been utilized as a winter cover crop, which can effectively increase vegetation coverage in winter and spring, improve the multiple cropping index and land utilization rate, reduce soil wind erosion, increase organic matter content, protect farmlands, promote balanced development between local economic production, and achieve ecological and environmental protection(Guo et al., 2023).
As the main production region for winter rapeseed, the planting area of northern China accounts for approximately 90% of the total rapeseed area. Brassica rapa (B. rapa) is mainly distributed on the western Loess Plateau (Wu et al., 2022). Before overwintering, the aboveground parts of strong cold-resistant varieties grow slowly, and the leaves grow prostrate and enter the withered leaf stage earlier (Ma et al., 2019a). During the overwintering period, the growth point of winter rapeseed varieties with strong cold resistance sinks, and the growth occurs belowground. The central leaves cover the surface, such that the growth point can maintain a specific moisture content and remain in the soil layer at a relatively stable temperature. This can effectively prevent damage to rapeseed when the temperature changes dramatically and allow the plant to safely overwinter. Sun et al (Sun et al., 2011a) found that winter rapeseed varieties with different cold resistance have great differences in the growth points, and the strong cold resistance B. rapa varieties that can overwinter in northern regions such as Hexi Corridor in Gansu Province have the characteristics of concave growth points and creeping growth. (Niu et al., 2021). Therefore, an unknown regulatory mechanism may occur between the morphological characteristics of winter rapeseed with concave and raised growth points and their cold resistance.
The NAC family transcription factor CUP-SHAPED COTYLEDON2 (CUC2) regulates the formation of various tissues and organs during plant growth and development (Hu et al., 2015), and plays a particularly important role in the formation of the apical meristem and maintenance of organ primordium edges (Aida et al., 1997; Olsen et al., 2005). In 1997, the CUC2 gene was first isolated and identified in Arabidopsis thaliana (A. thaliana) (Niu et al., 2021), and its mutation caused developmental defects in the apical meristem (Aida et al., 1999; Maugarny et al., 2016). The CUC2 gene can promote the aggregation of auxins in the organ primordium by regulating PIN-FORMED 1 (PIN1), thus participating in the regulation of early ovule primordium formation (Kwon et al., 2006). In addition, CUC2 can indirectly regulate the synthesis of LATERAL SUPPRESSOR (LAS) genes for lateral branch development (Reinhardt et al., 2003).
CUC2 is a negative regulator of plant growth that antagonizes cytokinins and auxins (Motte et al., 2011). It slows plant growth by inhibiting the division of plant cells and promoting the initiation of apical meristem development under the regulation of auxin. CUC2 is gradually expressed and leads to changes in the plant growth stage, beginning in the spherical embryo stage and proceeding to the tip of the embryo during seed germination and the edge of the cotyledon after cotyledon formation (Nikovics et al., 2006). As the upstream gene of the apical meristem-forming factor Shoot meristemless (STM) (Balkunde et al., 2017), the CUC2 gene is regulated by the activity of the STM gene (Spinelli et al., 2011) during the growth and development of the apical meristem of A. thaliana plants and restricted to the margins of the cotyledon and apical meristem. Apical meristem formation is regulated by regulating the expression of STM (Balkunde et al., 2017). Studies on CUC2 promoters of Betula platyphylla (B. platyphylla) found that they contain many cis-acting elements associated with tissue-specific expression, hormone synthesis, environmental stress response, and transcription factors and are highly expressed in the apical meristem, leaf margins, and flower tissue organs (Aida et al., 1999).
The regulatory role of the CUC2 gene in A. thaliana and B. platyphylla meristems has been studied. CUC2 is a negative regulator of plant growth that plays a role in the apical meristem and organ edge formation, leaf edge morphogenesis, and growth and development (Aida et al., 1997). However, its role in the growth point of winter B. rapa remains unclear.
In this study, we cloned the BrCUC2 gene from the growing point of winter B. rapa and analyzed its bioinformatics and tissue expression. The promoter sequence 2000 bp upstream of BrCUC2 was cloned using Gateway technology. Online software was used to predict the cis-acting regulatory element of the promoter, and a promoter expression vector was constructed and transformed into A. thaliana. Transgenic plants were then identified by β-Glucuronidase (GUS) histochemical staining. This study reveals the role of BrCUC2 at the growth point of winter B. rapa and lays a foundation for further studies on the transcriptional regulation of BrCUC2.
Materials and methods
Plant materials and growth conditions
The experimental B. rapa materials included the ‘Longyou 7’ (strong cold resistance) and ‘Longyou 99’ (weak cold resistance) varieties. Rapeseed seeds with excellent vitality were selected, treated with 10% H2O2 for 30 min, rinsed with sterile water 2–3 times, and germinated in Petri dishes containing wet filter paper under light conditions for 14 h at 30°C and dark conditions for 10 h at 28°C. After germination, the seeds were sown in pots and cultured in an artificial incubator (14 h light at 25°C, 10 h dark at 20°C) until the seven–leaves stage. Rapeseed seedlings were placed in incubators at 4, 0, and -4°C for the low-temperature treatment and 25°C (room temperature) as the control. After 72 h of treatment, the roots, stems, leaves, and growth cones of rapeseed were collected, immediately frozen in liquid nitrogen, and stored at -80°C for real-time fluorescence quantitative expression analysis of extracted RNA. Three biological replicates were performed for each treatment.
Nicotiana benthamiana (N.benthamiana) seeds were vernalized at 4°C for three days and seeded in a sterilized soil matrix. A subcellular localization test was performed during the 4th week of growth. The growth chamber conditions were as follows: 16 h light, 8 h darkness, 25°C, and 65% relative humidity.
Cloning of the BrCUC2 gene
The basic local allignment search tool (BLAST) alignment method of the national center for biotechnology information (NCBI) was applied to the BrCUC2 gene coding region sequence reported in the GenBank database, and a coding sequence with 100% homology to B. rapa was obtained. Primer Premier 5.0 software (Premier Biosoft International, Palo Alto, CA, USA) was used to design the gene cloning primers CUC2-A: 5′-ATGGACATTCCGTACTACCACTAT-3′ and CUC2-S: 5′-TAGTAATTCCATACGCAATCAAGT-3′. The cDNA at the ‘Longyou 7’ growing point at room temperature was used as a template for in vitro amplification by polymerase chain reaction (PCR). The amplification process was as follows: pre-denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 58.4°C for 30 s, and extension at 72.0°C for 60 s; terminal extension at 72°C for 10 min; and preservation at 4°C. The size of the PCR products was determined by 1% agarose gel electrophoresis. Target bands were recovered using an agar DNA gel recovery kit (TaKaRa, Dalian, China). The purified PCR products were connected with the pMD-19T vector, incubated at 25°C overnight for 18 h, and transferred to E. coli DH5α receptor cells. Positive recombinants were screened and confirmed using sequencing.
Sequence alignment and evolutionary analyses of BrCUC2 proteins from different plant species
Bioinformatics was used to predict the molecular properties of the protein encoded by the BrCUC2 gene. We used ProtParam’s Expasy software (https://web.expasy.org/protparam/) to predict protein physicochemical properties, ProtScale (Expasy - ProtScale) (Wilkins et al., 1999) to analyze hydrophobicity, SignaIP (https://services.healthtech.dtu.dk/services/SignalP-5.0/) (Teufel et al., 2022) to predict signal peptides, TMHMM (https://services.healthtech.dtu.dk/services/TMHMM-2.0/) (Krogh et al., 2001) to predict transmembrane domains, SMART tool to predict conserved structural domains, SOPMA(https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa%20_sopma.html) (Geourjon and Deléage, 1995) and SWISS-MODEL (Waterhouse et al., 2018) (https://swissmodel.expasy.org/) to predict protein secondary and tertiary structures.
Download the homologous gene sequence of BrCUC2 from NCBI database. Determine and visualize the distribution position of BrCUC2 homologous gene on chromosome with TBtools software. Utilizing online software Gene Structure Display Server (GSDS)(Guo et al., 2007) (http://gsds.gao-lab.org/index.php) to analyse the gene structure of BrCUC2 homologous gene in B. rapa. Based on the location information of the BrCUC2 homologous gene on chromosomes, the chromosome location map of the CUC genes were drawn using by MapChart software (Voorrips, 2002). The conservative motifs of homologous gene were predicted by MEME (https://meme-suite.org/meme/tools/meme) (Bailey et al., 2009). and the protein sequences of different species of Brassica napus, Arabidopsis lyrate, Asparagus officinalis, Arabidopsis thaliana, Brassica oleracea, Camelina sativa, Glycine max, Medicago truncatula, Nicotiana attenuata, Raphanus sativus, Ricinus communis, Sesamum indicum and ‘Longyou 7’ were obtained by NCBI database BLAST comparison, The amino acid sequences were compared by DNAMAN software. Neighbor–joining method was used to construct the phylogenetic tree by MEGA7.0, and the value of Bootstrap method was 1000 (Hall, 2013; Kumar et al., 2016).
Real-time fluorescence quantitative PCR
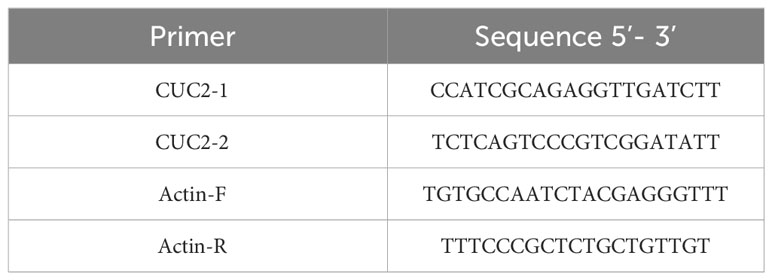
RNA was extracted from the leaves, roots, stems, and growth cones of winter B. rapa ‘Longyou 7’ and ‘Longyou 99’ after low-temperature treatment and reverse-transcribed into cDNA using a real-time fluorescence quantification biotechnology kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The specifically designed primers are listed in Table 1. RT-qPCR was performed by adding a 20 μL system containing the target gene and reference gene to a 96–well plate using a fluorescence quantitative PCR apparatus. The reaction was repeated three times, and the reaction procedure was determined according to the manufacturer’s instructions for the specific reagent. The relative gene expression was calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Subcellular localization of BrCUC2
Using the coding sequence (CDS) amplification product of BrCUC2 as a template, Gateway technology (Liang et al., 2013) was used to amplify the product using aatB joint-specific primers, and the amplified product was cloned into the pDNOR entry vector via the BP reaction. After the LR reaction, the gene was introduced into the pEarlygate101-GFP vector, and the constructed vector plasmid was transferred into Agrobacterium GV3101 using the electrical conversion method. Bacterial fluid was collected and resuspended in 1 mL of infiltration buffer, and set the optical density (OD) at 600 nm of the solution to 0.6. The lower epidermis of N. Benthamiana leaves was labeled with the bacterial solution using a sterile needle tube and then incubated in a 22°C culture room for 48 h. Labeled N. Benthamiana leaves were observed and photographed under a laser confocal microscope to determine the location of the genes.
Self-activation validation of BrCUC2
The constructed positive control plasmids pGBKT7-53+pGADT7-T, negative control plasmids pGBKT7-Lam+pGADT7-T, and pGBKT7-CUC2+pGADT7 plasmids were transformed into yeast strain Y2HGold, respectively. The transformed strains were cultured on DDO/X(SD/-Leu/-Trp/-X-α-gal) medium and incubate in a 30°C incubator for 3–5 days. Select single colonies on QDO/X/A(SD/Leu/-Trp/-His/-Ade/-X-α-gal/AbA) medium. Observe the self-activation phenomenon of pGBKT7-CUC2 based on the growth status of yeast.
Promoter cloning and cis-acting regulatory element analysis
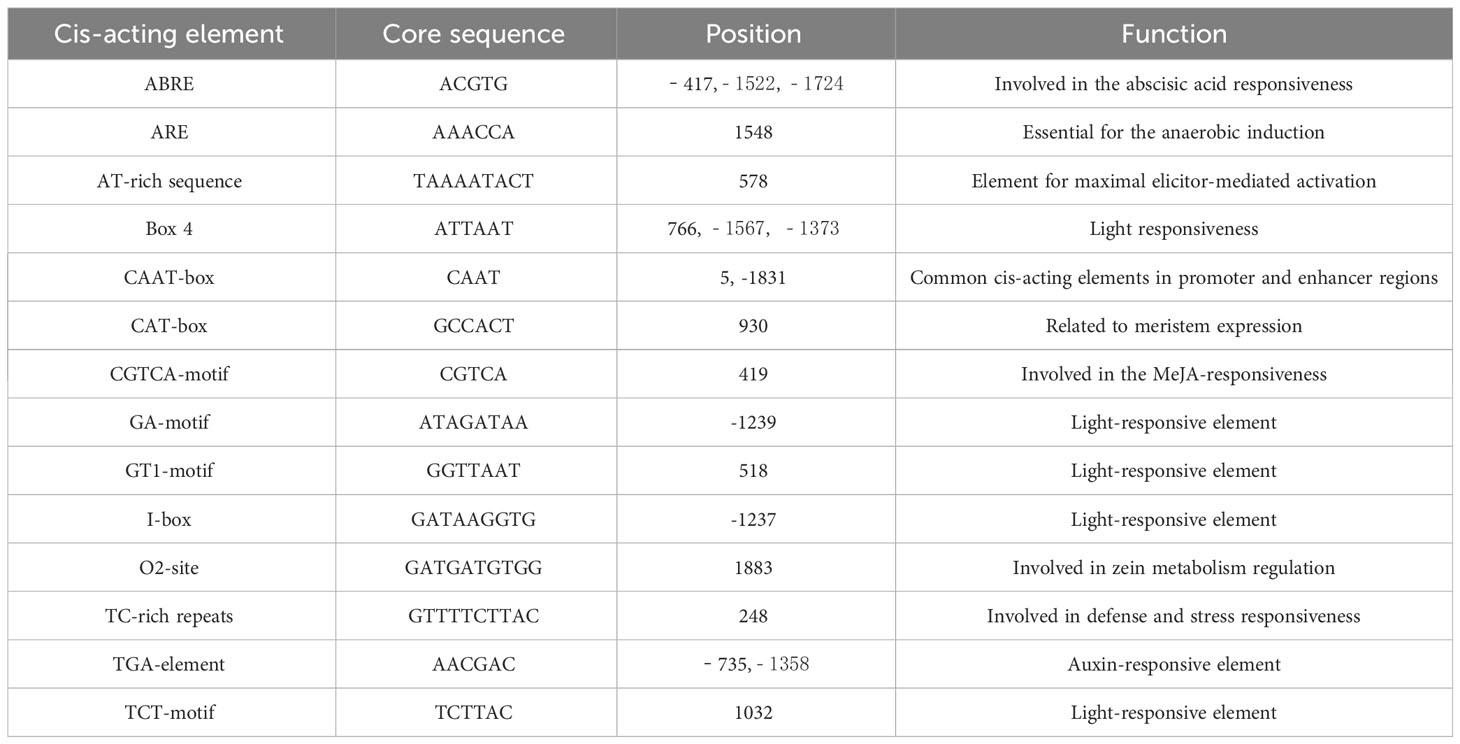
We searched the region approximately 2000 bp upstream of the BrCUC2 sequence in the genome of B. rapa and designed specific primers using Primer Premier 5.0 software. The following primers were added to the Gateway vector connector sequence: CUC2 F(5’-3’): AAAAAAGCAGGCTTCCAATATGACACTAATTATGC; CUC2 R(5’-3’): AGAAAGCTGGGTCGAAGAACTGATGTTAAAAC. Amplification based on the Gateway technique was performed in the following PCR reaction system (total 25 μL): Prime STAR DNA polymerase, 0.25 μL; 5×PrimeSTAR buffer, 5 μL; 2.5 mM dNTP, 1 μL, forward and reverse primers, each 1 μL; template DNA, 2 μL; and ddH2O, 14.75 μL. The PCR reaction procedure was as follows: pre-denaturation at 98°C for 10 s and a total of 30 cycles of denaturation at 98°C for 10 s and extension at 68°C for 1 min. The PCR reactions were performed in two rounds, and after two rounds of PCR, the target bands were cut and recovered, connected to the PMD19-T vector, and transformed into E. coli DH5α. The promoter sequences were then sequenced after PCR detection of the bacterial solution. BrCUC2 promoter sequences were analyzed using the PlantCARE database (Lescot et al., 2002) to investigate cis-acting regulatory elements.
Genetic transformation and β-glucuronidase staining in A. thaliana
The constructed plant overexpression vector pEarlyGate101 was transformed into Agrobacterium GV3101 using the freeze–thaw method (Weigel and Glazebrook, 2006). When A. thaliana grew to the flowering stage, the Agrobacterium solution was activated and resuspended in a 5% sucrose solution. The absorbance at 600 nm was adjusted to 0.8, and 3/10000 surfactant (Silwet L-77) was added for later use. Culturing was performed in the dark at 25°C for 24 h and then in the light. The herbicide Glufosinate Ammonium (Basta) (1/10000) (Sangon Biotecch, shanghai, China) was sprayed for screening. After maturation, A. thaliana seeds were harvested from each plant.
Resistant A. thaliana plants were placed in a GUS staining solution, and wild-type A. thaliana was used as the control. The plants were kept at 37°C overnight, rinsed and soaked with 70% ethanol, and photographed under a microscope to observe GUS staining (Liu et al., 2017).
Screening, identification, and functional analysis of transgenic A. thaliana
After vernalization, the first generation T0 transgenic A. thaliana seeds were planted in sterilized nutrient soil, sprayed with Basta (10%) two weeks later for screening, and sprayed again three times a week. The surviving A. thaliana plants were harvested from the T1 generation seeds. The T1 generation seeds were seeded in 1/2 MS medium containing Basta and screened according to the 3:1 principle. T2 generation seeds were harvested after transplantation. The T2 generation seeds were planted in 1/2 MS medium containing Basta, and all surviving plants were transgenic homozygous plants. Subsequent experiments were conducted after transplantation. Genomic DNA of transgenic A. thaliana was amplified using the primers CUC2 F(5’-3’): CTATCCTTCGCAAGACCTTC and CUC2 R(5’-3’): TAATTCCATACGCAATCAAGT and identified via PCR and RT-qPCR analyses.
Phenotypic changes were observed in the third-generation transgenic lines and wild-type A. thaliana during the seedling, flowering, and mature stages. After four weeks of growth, they were exposed to -4°C for 3, 6, 12, and 24 h and then transferred to room temperature (25°C) to recover growth for seven days, and their survival rate was counted. A. thaliana leaves were collected at different treatment times, and their physiological indicators and gene expression levels were measured. Growth at room temperature (25°C) was used as a control. Superoxide dismutase (SOD) activity was determined using the nitroblue tetrazolium (NBT) photoreduction method (Durak et al., 1993). peroxidase (POD) activity was determined using the guaiacol method (Senthilkumar et al., 2021). Proline (PRO) content was measured using the acid ninhydrin method (Abrahám et al., 2010).
Results
Identification and analysis of CUC2 gene family in winter B. rapa
The NAC (PF02365) domain of CUC2 was determined using the Pfam database. Ten CUC genes of B.rapa were selected, and the homologous BrCUC2 protein sequences of different species were screened by BLAST-Protein (BlASTP) tool. DNAMAN software was used to compare the protein sequences of BrCUC2 and its homologous genes CUC1、CUC3, and BrCUC2 (B.rapa, B.napus, B.oleracea, R.sativus, A. thaliana) in different species (Figure 1). The results showed that all CUC protein sequences contained a highly conserved NAC domain at the N-terminus, including A, B, C, D and E subdomains. the subdomains A, C and D were highly conserved, while the subdomains B and E were structurally variable and might be involved in different biological functions. The integrity of the NAC domain is essential for CUC2 to promote the formation of adventitious buds in callus tissue (Taoka et al., 2004). The absence of any of the five motifs affects the formation of adventitious buds. The C-terminal domain of CUC2 protein contains three conserved motifs, namely the V motif (TEHVSCFS), L motif (SLPP), W motif (WNY), and two serine rich regions (S). Among them, the W motif is essential for the transactivation of the CUC2 protein, and the V motif contains miRNA binding sites, which is responsible for regulating CUC2 gene activity. The intact CUC2 protein plays an important role in Shoot apical meristem (SAM) formation or plant organ separation. The BrCUC2 (Bra022685) protein sequence of B.rapa was 99.46% similar to the CUC2 protein of ‘Longyou 7’, and there were two amino acid mutation sites, alanine (A) of ‘Longyou 7’ was changed to valine (V), glycine (G) was changed to glutamic acid (E).The homology was 89.01% with Raphanus sativus (R. sativus), 97.28% with Brassica oleracea (B. oleracea) and 99.46% with Brassica napus (B. napus). Suggesting that the BrCUC2 (Bra022685) protein sequence is highly conserved during evolution process.
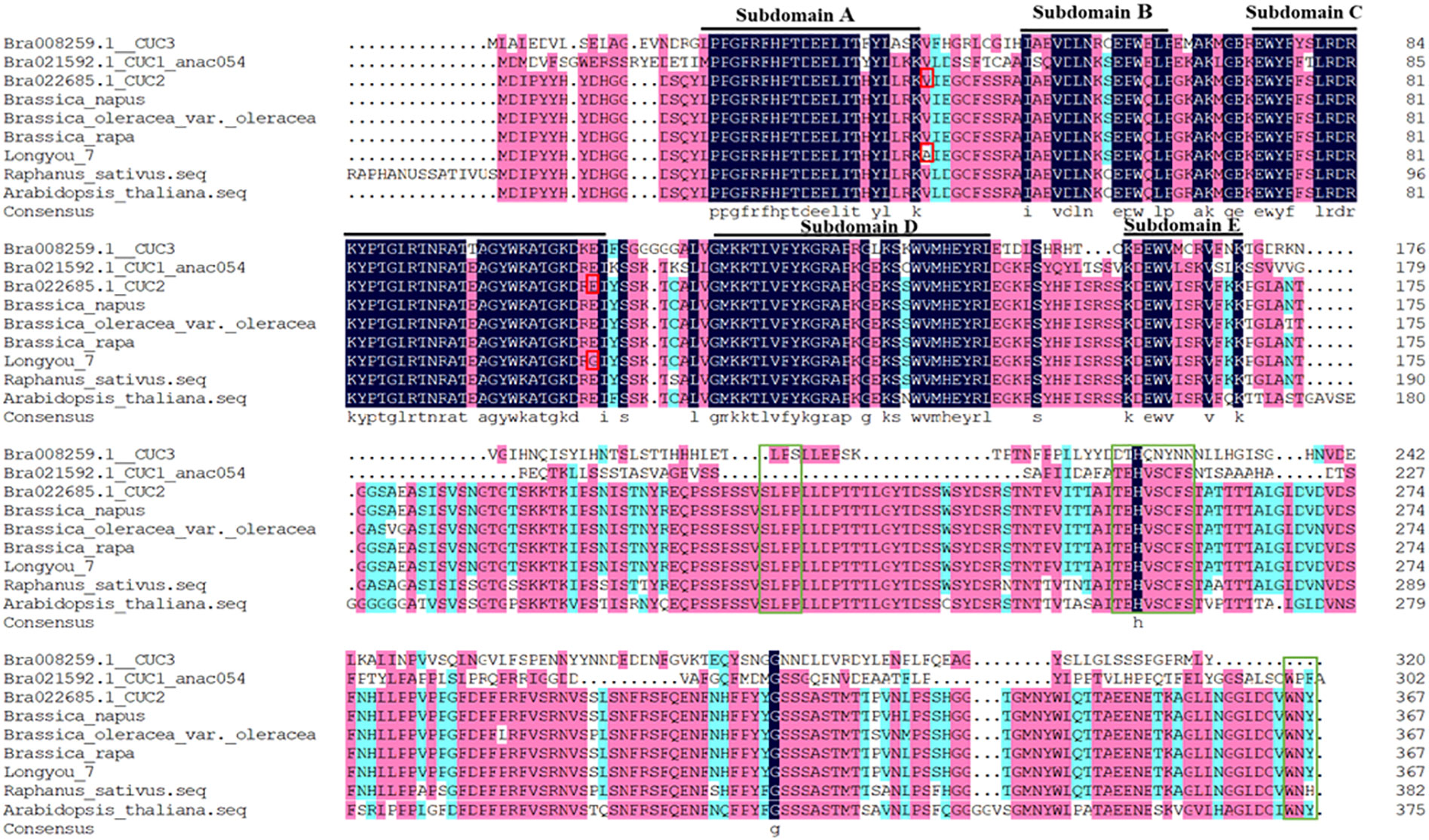
Figure 1 Amino acid sequence comparison of CUC2 protein in different species and homologous protein CUC. Highly conserved subdomains (A–E) are indicated by black lines. Amino acid mutation sites are indicated by red boxes. The L motif, V motif and W motif of C-terminal domain are represented by green boxes.
In order to further clarify the functional and evolutionary relationships of the BrCUC2 gene in the growth cone of winter B.rapa, MEGA11 was used to construct a phylogenetic tree, and the evolutionary relationships were compared between CUCs and BrCUC2 proteins in 13 different species (including B. napus, B. rapa, C. sativa, B. oleracea, R. sativus, etc.) (Figure 2). The results showed that they were mainly divided into nine subfamilies, among which BrCUC2 (Bra002685.1) was more closely related to B. oleracea, B. napus, R. sativus and Bra003023. CUC1 (Bra001586) and CUC3 (Bra008259) were closely related to Asparagus officinalis (A. officinalis). It is speculated that the BrCUC2 gene in winter B. rapa ‘Longyou 7’ is functionally similar to that in B. napus, B. oleracea and R. sativus.
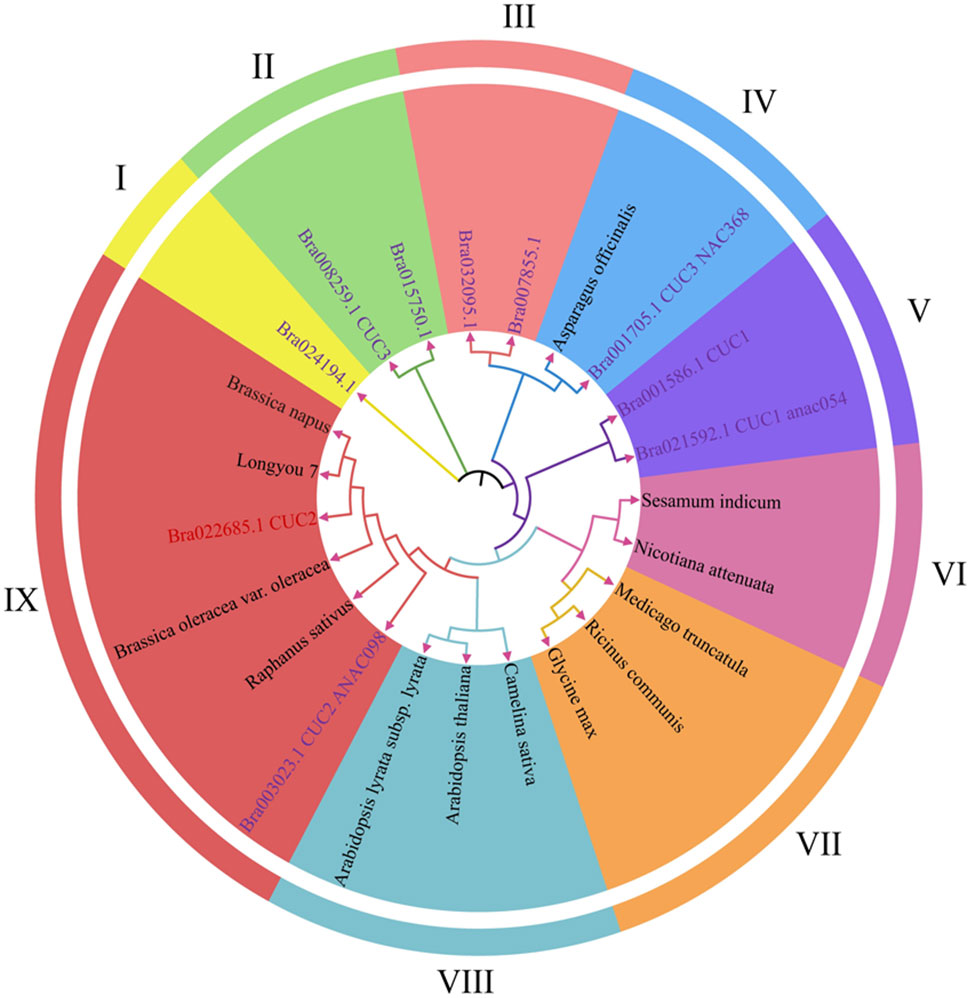
Figure 2 Evolutionary analysis of CUC2 in different species. The 9 subfamilies have different colors. Purple font represents members of the BrCUC2 homologous genes, The others are BrCUC2 in different species.
Chromosome mapping and structure analysis of CUCs gene in winter B. rapa
Ten CUC genes in B. rapa were distributed on seven chromosomes (Figure 3A), two of which were distributed on chromosome A02, three on chromosome A03, and the rest on chromosomes A01, A04, A07, A09 and A10, respectively. The positions of introns and exons were determined by comparing the genomic DNA with the full-length cDNA of CUCs (Figure 3D). the results showed that Bra003023 contained three introns and the rest nine genes contained two introns. Ten CUC genes contained a total of 18 motifs, and all contained motif 1, motif 3 and motif 4, of which Bra022685 and Bra003023, Bra007855 and Bra032095, Bra021592 and Bra001586 contained the same motif, respectively. and the same subfamily of CUC proteins in the phylogenetic tree had the same motif, indicating that they were highly conserved (Figures 3B, C, S1; Table S1).
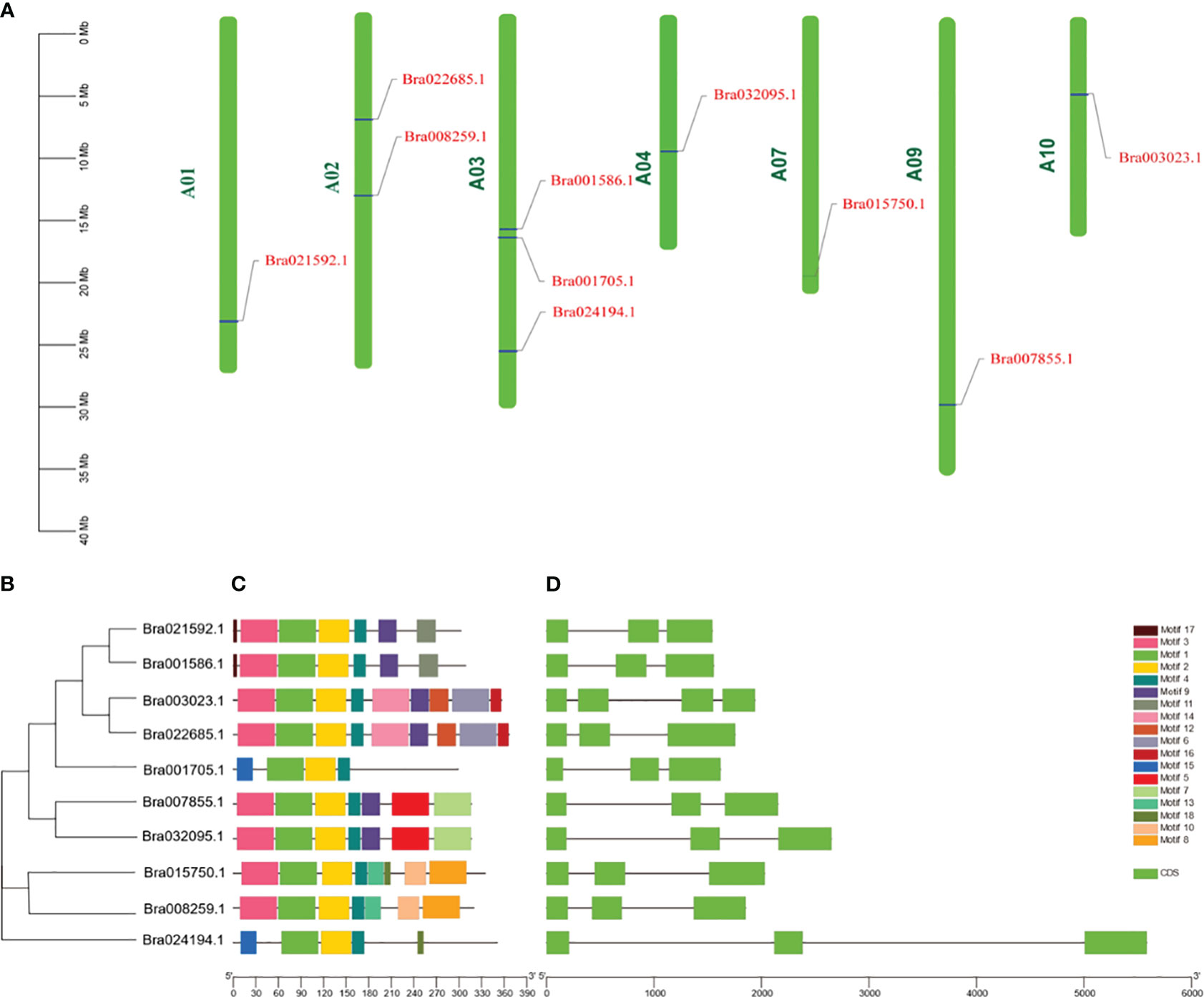
Figure 3 Chromosomal localization and structural analysis of the CUCs gene. (A) Chromosomal location distribution of CUCs genes. (B) Phylogenetic relationships of CUCs genes. (C) Schematic representation of conserved motifs in CUCs proteins. Different colored boxes represent different patterns. The black lines represent non-conserved sequences. (D) CUCs exon/intron structure. The green boxes represent exons; The space between boxes is the intron. The scale bars at the bottom of (C, D) represent the lengths of genes and motifs, respectively.Cloning and Sequence Analysis of the BrCUC2 (Bra022685) Gene.
The BrCUC2 gene (1104 bp) was amplified using the cDNA template from the growing site of ‘Longyou 7,’ and the result was consistent with the predicted fragment size. The BrCUC2 gene sequence encodes 367 amino acids. The theoretical isoelectric point (PI) was 8.66, fat index was 58.71, instability index was 44.07, and total average hydrophilic value was -0.569. The BrCUC2 protein was predicted as an unstable hydrophilic protein (Figure S2A). No transmembrane regions and signaling peptides were detected (Figures S2B, C). The conserved domain was distributed among 18–144 amino acids and composed of 127 amino acids, and the conserved sequences belonged to the NAM superfamily (Figure S1). The secondary and tertiary structures of the BrCUC2 protein mainly consisted of irregular curling (65.12%), extended chain (16.25%), α helix (14.17%), and β angle (4.36%) (Figures S2D, E).
Prediction and functional analysis of cis-acting regulatory elements in the BrCUC2 promoter region
Promoters are important gene elements that regulate gene expression. By analyzing the cis-acting elements, the pathway genes that may participate in abiotic stress-tolerance mechanisms can be better understood (Hernandez-Garcia and Finer, 2014). Using the genomic DNA of the ‘Longyou 7’ growth cone of winter B. rapa as a template, the BrCUC2 promoter sequence with a length of approximately 2000 bp was amplified and sequenced after connecting to the vector.The sequencing was correctly used to prepare Agrobacterium liquid transformed Arabidopsis. PlantCARE and GSDS2.0 online software were used to analyze the BrCUC2 promoter sequence and predict the cis-acting regulatory elements (Table 2; Figure 4). The results showed that the promoter region of this gene contains ABRE-acting elements involved in abscisic acid and stress responses, TGA elements involved in the auxin response, CAT-box elements involved in meristem expression. Tc-rich repeats are involved in stress response; ARE active elements are involved in anaerobic responses; Box4, GA-motif, GT1-motif, I-box, and TCT-motif active elements are involved in light response; and several CAAT-box and CAT-box elements are core promoters.
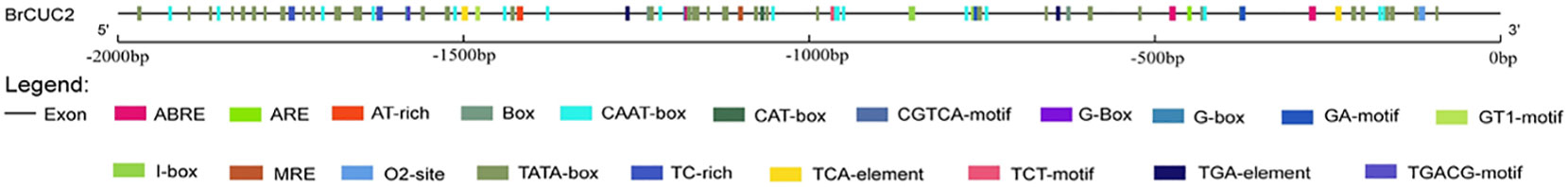
Figure 4 Promoter cis-acting regulatory element distribution. Different colored rectangular boxes represent individual cis-acting components.
Subcellular localization and self-activation validation of BrCUC2
The fusion expression vector PEarlyGate101 BrCUC2 GFP was constructed using Gateway technology, and tobacco leaves were transformed instantaneously with empty GFP as a control. After 48 h of growth in the dark, scanning and photography were performed using a confocal laser microscope. The results showed that the BrCUC2 GFP fusion protein was distributed in the nucleus (Figure 5A).
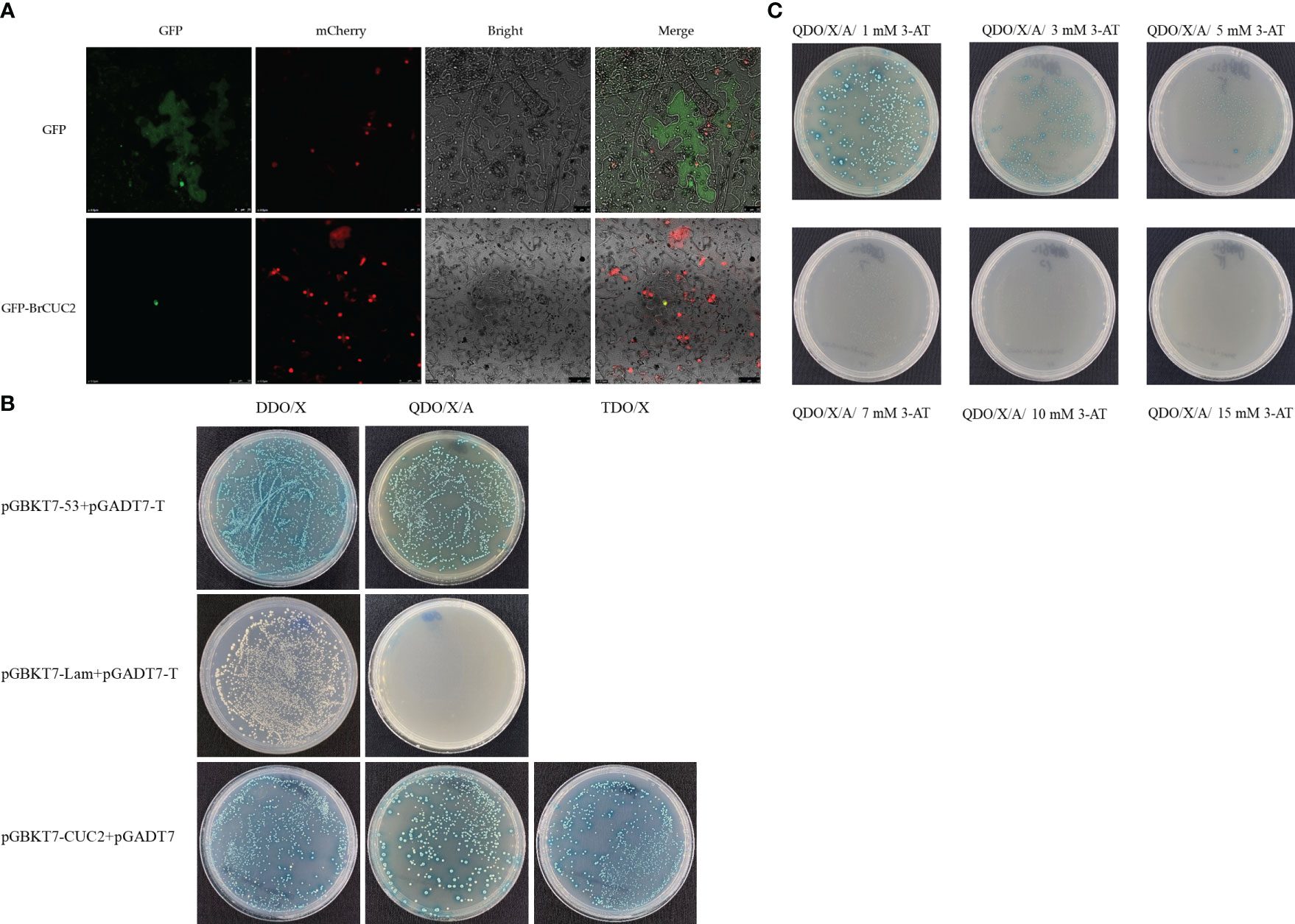
Figure 5 (A) Subcellular localization of PEarlyGate101-BrCUC2-GFP protein, empty GFP as a control, mCherry, a nuclear localization protein. (B) Self-activation verification of BrCUC2. The positive control pGBKT7-53+pGADT7-T, the negative control pGBKT7-Lam+pGADT7-T, and the experimental group pGBKT7-CUC2+pGADT7 were transferred to yeast strain Y2HGold, respectively. Cultured on DDO/X (SD/-Leu/-Trp/- X-α-Gal) and QDO/X/A (SD/Leu/-Trp/-His/-Ade/-X-α-Gal) medium, respectively. TDO/X (SD/-Leu/-Trp/-His/-X-α-Gal). (C) Screening of 3-AT inhibition at different concentrations in QDO/X/A medium. 3-amino-1,2,4-triazole(3-AT), 3-AT is a competitive inhibitor of histidine and can inhibit mild self-activation.
To further characterize the function of BrCUC2, we investigated whether BrCUC2 has self-activating activity in yeast cells. The positive control pGBKT7-53+pGADT7-T, negative control pGBKT7-Lam+pGADT7-T, and pGBKT7-CUC2+pGADT7 were transformed into Y2HGold yeast cells and cultured on DDO/X (SD/-Leu/-Trp/-X-α-gal) and QDO/X/A (SD/-Leu/-Trp/-His/-Ade/-X-α-gal/AbA) medium, respectively. The results showed that the positive control combination grew and turned blue on DDO/X and QDO/X/A medium, while the negative control group grew but remained blue on DDO/X media, and did not grow on QDO/X/A media, indicating the success of the positive control and negative control experiments. The experimental group pGBKT7-CUC2+pGADT7 grew on DDO/X medium, grew and turned blue on both TDO/X (SD/-Leu/-Trp/-His/-X-α-gal) and QDO/X/A media (Figure 5B), indicating that the pGBKT7-CUC2 plasmid was successfully transferred into the yeast strain and showed self-activation in the yeast strain. Further inhibition experiments were conducted using (3-amino-1,2,4-triazole)3-AT, and the results showed that the growth turned blue on QDO/X/A/(1-5) mM 3-AT and weakened with the increase of 3-AT concentration. However, it did not grow on QDO/X/A/(7-15) mM 3-AT medium (Figure 5C), indicating that the QDO/X/A/7 mM 3-AT screening conditions can inhibit the self-activation phenomenon of pGBKT7-CUC2.
Analysis of BrCUC2 gene expression
To clarify the differences in the expression of the BrCUC2 gene in various winter rapeseed tissues, RT-qPCR was performed to determine the relative expression levels of BrCUC2 in the leaves, stems, growth cones, and roots (Figure 6A). The results showed that the expression of BrCUC2 in different tissues of ‘Longyou 7’ and ‘Longyou 99’ was significantly different. The expression level in ‘Longyou 99’ was 10.9-times higher in the growth points than in the leaves, whereas that in ‘Longyou 7’ was 16.2-times higher in the growth cone than in the leaves, indicating that the BrCUC2 gene mainly acts on the growth cone of winter B. rapa.
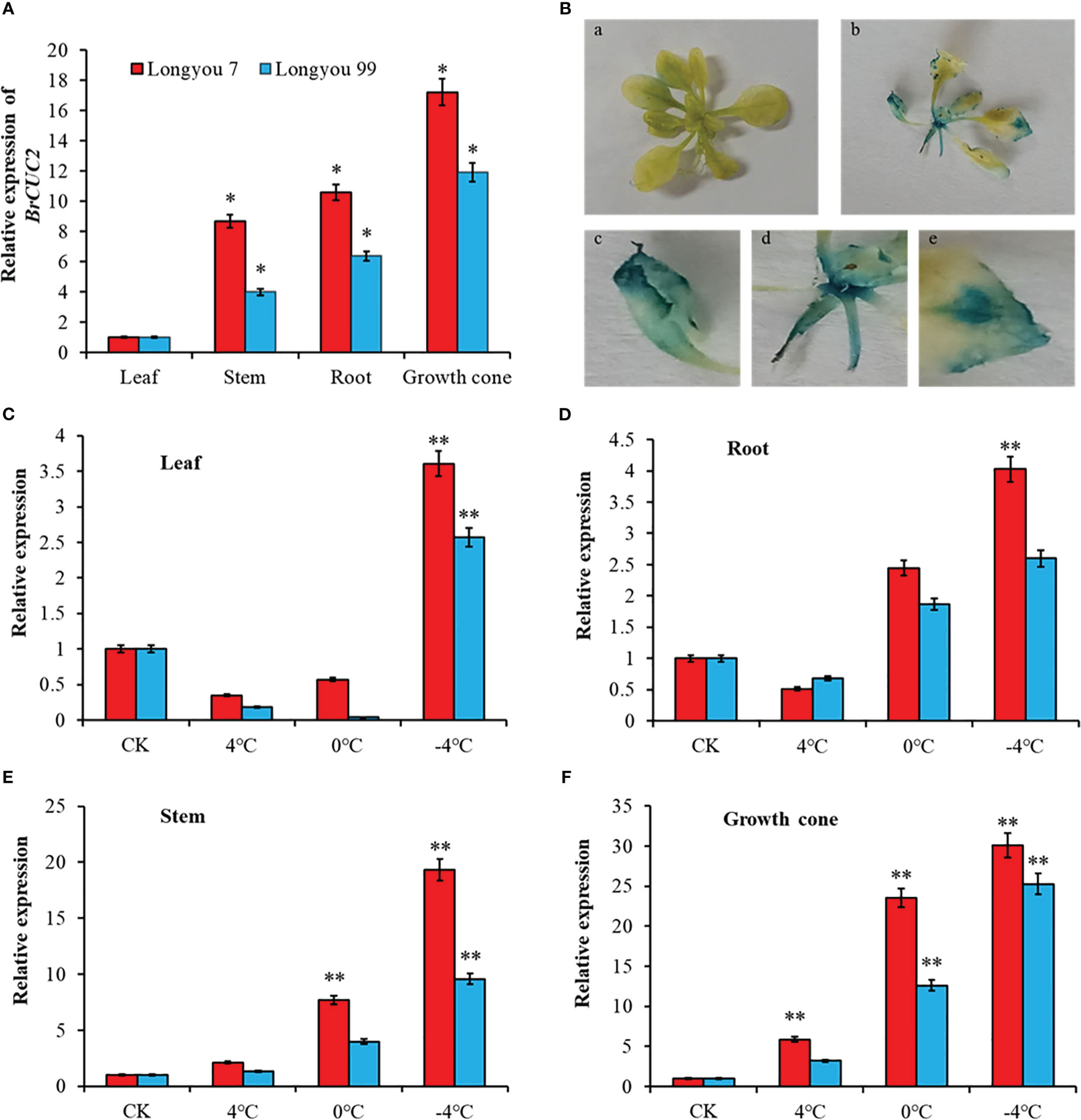
Figure 6 BrCUC2 tissue expression specificity. (A) Expression level of BrCUC2 in different tissues. (B) GUS mapping of BrCUC2 transgenic Arabidopsis thaliana. a, GUS staining of whole wild - type A. thaliana; b, GUS staining of whole transgenic A. thaliana; c-e, Partial magnification of b (C) Expression of BrCUC2 gene in leaf under low temperature stress. (D) Expression of BrCUC2 gene in Root under low temperature stress. (E) Expression of BrCUC2 gene in Stem under low temperature stress. (F) Expression of BrCUC2 gene in growth cone under low temperature stress. Symbols “*”and “**” indicate significant differences at P values of 0.05 and 0.01, respectively.
To verify the expression pattern of BrCUC2, GUS tissue staining was performed on wild-type and transgenic A. thaliana seedlings (Figure 6B). The results revealed that the GUS signal was absent in the wild-type A. thaliana but present on the leaf edges, stems, and growth points of transgenic A. thaliana seedlings, indicating that the BrCUC2 promoter cloned from winter B. rapa could drive downstream GUS reporter gene expression with tissue specificity.
To investigate the effect of cold stress on BrCUC2 expression, ‘Longyou 7’ and ‘Longyou 99’ were treated at 0, 4, and -4°C, RT-qPCR was performed to measure the expression. The results showed differences in the relative expression of the BrCUC2 gene among different tissues after low-temperature stress. As the temperature decreased, the expression of the BrCUC2 gene was upregulated and reached the highest value at -4°C. The difference was most significant in stem and growth cone and higher in ‘Longyou 7,’ which presents strong cold resistance and a concave growth cone, than in ‘Longyou 99,’ which presents weak cold resistance and a convex growth cone. In the leaves, under normal temperature control, the expression of BrCUC2 in ‘Longyou 7’ and ‘Longyou 99’ was downregulated at 4°C and 0°C, and after treatment at -4°C, the expression was 2.6- and 1.5- times higher than those in the control, respectively (Figure 6C). In the roots, the expression of BrCUC2 in ‘Longyou 7’ and ‘Longyou 99’ was downregulated at 4°C and upregulated at -4°C and 0°C, respectively. After treatment at -4°C, the expression was 3.0- and 1.6- times higher than those in the control, respectively (Figure 6D). With a decrease in temperature, BrCUC2 was upregulated in the stem and growth cone compared with that in the control (Figures 6E, F). After treatment at -4°C, the expression levels of BrCUC2 in ‘Longyou 7’ and ‘Longyou 99’ were 18.3- and 8.5-times higher in the stem and 29.1 and 24.3-times higher in the growth cone than those in the control, respectively.
Screening and identification of the transgenic BrCUC2 gene in A.thaliana
The first generation T0 BrCUC2-overexpressing plants were vernalized and planted in nutrient-rich soil. Two weeks later, they were sprayed with a 10% Basta solution. After exposure to the spray three times, seedlings that did not exhibit resistance to the herbicides turned yellow and died. Normal surviving seedlings were transplanted, and seeds were harvested from individual plants to obtain the T1 generation. The T1 generation of A. thaliana was selected on 1/2MS medium containing Basta according to the 3:1 principle. Seedlings with normal leaf and root growth were transplanted, and individual plants were harvested to obtain the T2 generation. The T2 generation of A. thaliana was screened for all surviving seedlings on 1/2MS medium containing Basta to obtain homozygous transgenic lines, which were then transplanted into pots for subsequent tests (Figure S3A).
After extracting BrCUC2 transgenic A. thaliana DNA, PCR amplification results showed no amplification bands in the wild-type A. thaliana single plants; however, 10 bands of approximately 1100 bp were amplified from 11 single transgenic plants. Among them, C2 had no bands, indicating that the transformation of the BrCUC2 gene in C2 single plants failed, while the 10 remaining single plants were successfully transformed, indicating that they were transgenic plants, with C6 and C10 showing lighter bands (Figure S3B).
RT-qPCR identification was performed on 10 overexpression plants and wild-type transgenic plants, and the results showed that the BrCUC2 gene was detected in all overexpression plants. The expression levels varied among different individual plants, with the highest expression levels of the CUC2 gene in C3 and C9, which were 7.53 and 7.79 times higher than those in the WT (Figure S3C).
Phenotypic differences associated with the BrCUC2 gene in A. thaliana treated at low temperature
Wild-type and BrCUC2-overexpressed A. thaliana phenotypes were observed at the seedling (4 weeks old), flowering, and mature stages under the same growth conditions (16h light/8 h dark cycle at 25°C). Comparisons between the wild-type and BrCUC2-overexpression A. thaliana plants showed that significant phenotype differences did not occur at the seedling stage but did occur at the flowering stage, with weaker growth and development and a dwarf phenotype (Figures 7A, B). The overexpression A. thaliana lines developed slowly and were set later than the wild-type at the mature stage. When wild-type A. thaliana plants were clamped and matured, the lateral branches of overexpression lines were still in the flowering stage, and the seed-setting rate was lower than that of the wild type (Figure 7C).
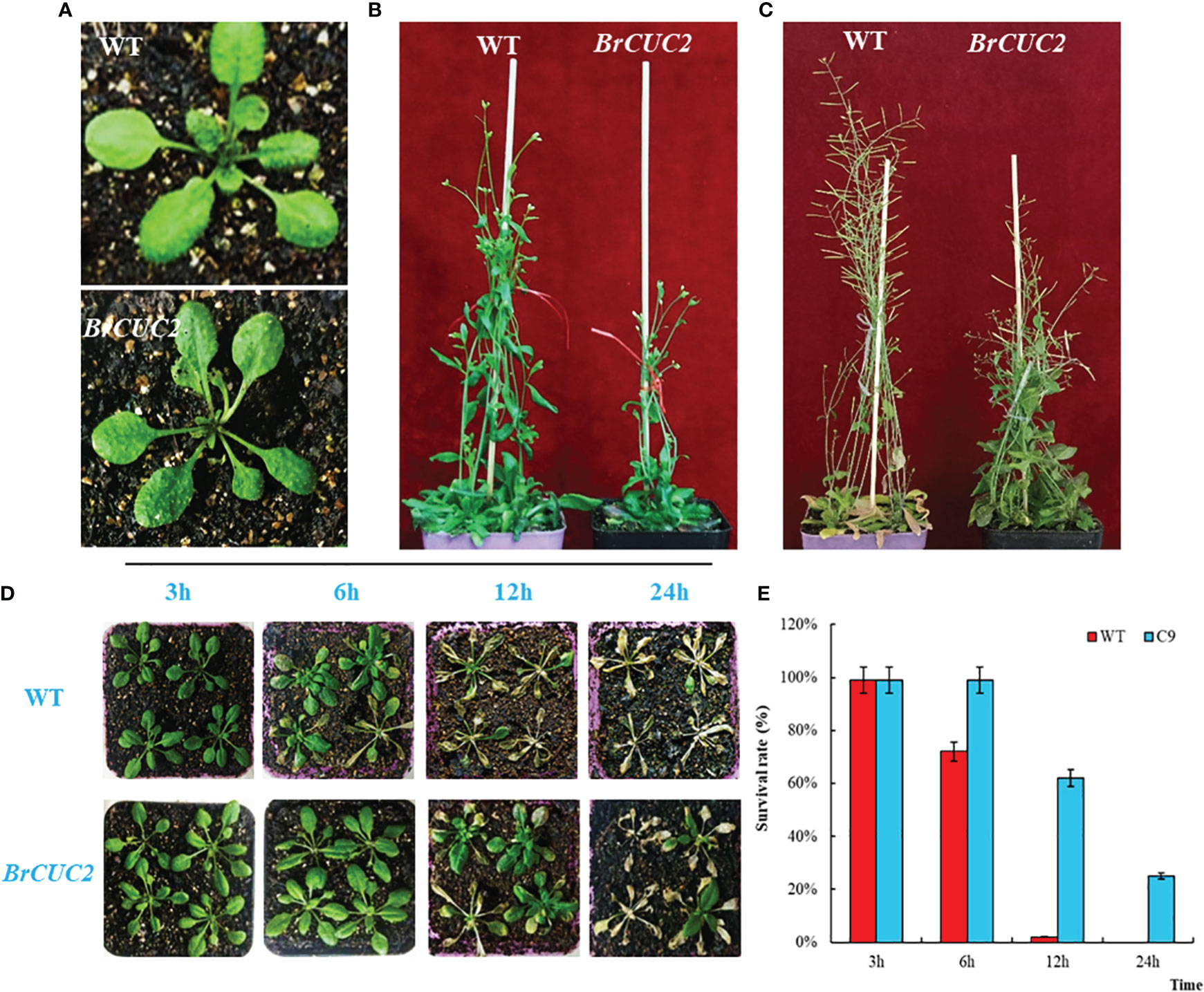
Figure 7 BrCUC2 transgenic A. thaliana phenotype. (A) Seedling stage, (B) Flowering stage, and (C) Maturity stage. (D) Phenotypes of transgenic and wild-type A. thaliana after low-temperature treatment at -4°C for 3, 6, 12, and 24 h. (E) Survival rate of plants after low-temperature treatment. C9: BrCUC2 overexpressed plant.
To verify the response of the BrCUC2 gene to low-temperature stress in plants, 4-week-old wild-type and BrCUC2-overexpression plants were treated at -4°C for 3, 6, 12, and 24 h and then returned to room temperature (25°C) for 7 d to observe the phenotypes (Figures 7D, E). All plants grew normally after low-temperature treatment for 3 h; however, the survival rate of the wild-type plants was 72% after low-temperature treatment for 6 h and decreased to 8% after 12 h, with most of the leaves dying. After low-temperature treatment for 6 h, the survival rate of BrCUC2-overexpression plants was 99%; after 12 h, some leaves yellowed, and the survival rate was 62%, and after 24 h, the survival rate was 25%. These results indicate that BrCUC2 overexpression enhances plant low-temperature tolerance over a short period.
Physiological parameters of BrCUC2 gene transfer in A. thaliana
The physiological indicators of wild-type and overexpressed A. thaliana after low-temperature treatment were analyzed after treatment at -4°C. The enzyme content in both plants gradually increased over time. The superoxide dismutase(SOD)activity、peroxidase (POD) activity and proline content in the BrCUC2-overexpressing plants were higher than those in the wild-type plants, reaching the highest values after 24 h (16.9%, 41.9%, and 22.9% higher than those in the wild-type, respectively) (Figure 8). This result indicates that the BrCUC2 gene is involved in the response of winter B. rapa to low-temperature stress.
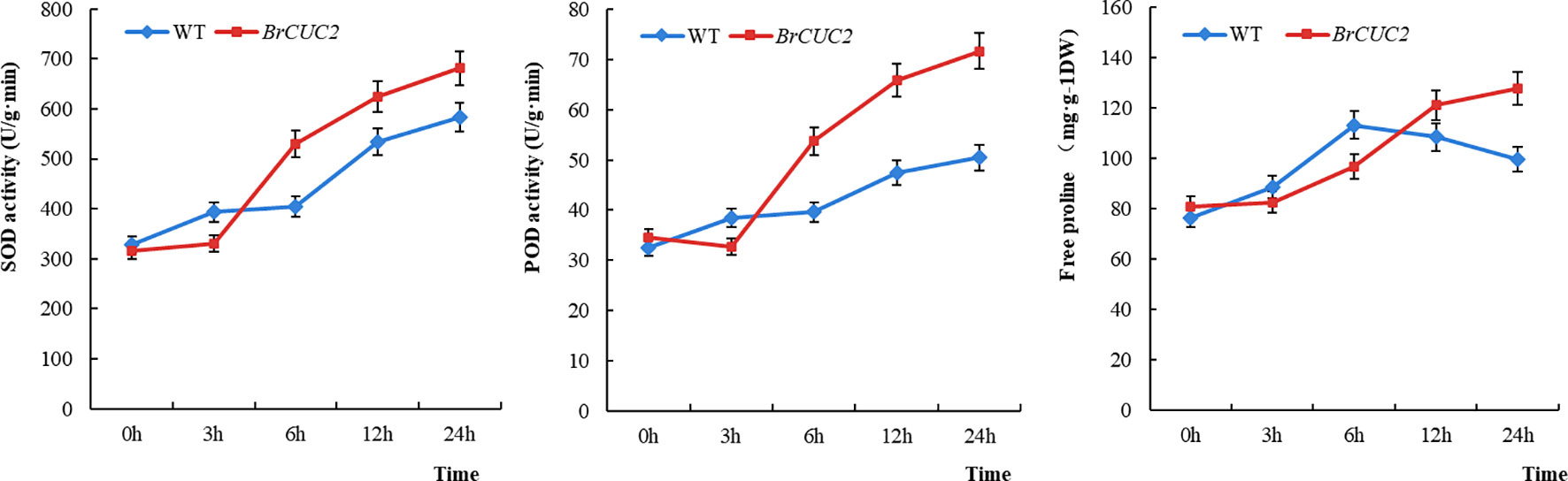
Figure 8 Physiological indices SOD、POD、Proline of Arabidopsis thaliana overexpressing BrCUC2 gene at low temperature. WT: wild type, BrCUC2: overexpressed plant.
Discussion
The A. thaliana NAC family transcription factor CUC2, a negative regulator of plant growth (Takada et al., 2001), plays an important role in the formation of apical meristems and organ margins, leaf edge morphological formation, growth, and development. The formation of meristem edges in A. thaliana is mediated by the NAC family transcription factors CUC1, CUC2, and CUC3 (Bertrand et al., 2003; Kamiuchi et al., 2014). The CUC1 and CUC2 are negatively regulated by miR164 (Bhatt et al., 2004). Kwon et al (Kwon et al., 2006) found that the CUC2 gene could fuse cotyledons and flower organogenesis in A. thaliana. Souer et al (Souer et al., 1996) showed that CUC2 is highly homologous to petunia genes necessary for flower organ development, cotyledon separation, and apical meristem embryogenesis. In A. thaliana, the CUC2 gene is necessary for the activation of the STM gene during embryonic development (Lenhard et al., 2002). Studies have shown that STM can directly activate CUC2 (Scofield and Murray, 2006), and mutations in the CUC2 gene can change the cotyledon traits of A. thaliana and cause developmental defects in its apical meristem (Vroemen et al., 2003). STM or CUC2 gene mutations affect the number of plant stem cells and lead to cotyledon fusion (Mallory et al., 2004). However, the regulatory mechanism of the CUC2 gene in winter rapeseed growth sites has not yet been reported.
In the present study, the BrCUC2 gene was cloned from the growth point of winter B. rapa, which controls the growth point differentiation of A. thaliana. The BrCUC2 gene was expressed in the leaves, stems, roots, and growing points of winter B. rapa, with the highest expression level observed at the growing points. The relative expression level was as follows: the expression level of BrCUC2 in the growing point of ‘Longyou 99’ was 10.9 times that in the leaves, and the expression level of BrCUC2 at the growing point of ‘Longyou 7’ was 16.2 times that in the leaves. After 72 h of low-temperature treatment, the expression in the leaves was initially downregulated and then upregulated, while the expression in roots, stems, and growth points was upregulated and reached the highest value at -4 °C. The differences were most significant in the stems and growth points. The expression in ‘Longyou 99,’ which presents weak cold resistance and a raised growth point, was lower than that in ‘Longyou 7,’ which presents strong cold resistance and a depressed growth point. These results indicate that the BrCUC2 gene might be induced by low-temperature stress and inhibit the growth of growing sites. Moreover, the high BrCUC2 content caused the ‘Longyou 7’ growth point to sag and grow below the ground.
Bioinformatics analysis and functional verification showed that the BrCUC2 gene contains a conserved NAM superfamily domain, CUC2 is a transcription factor in the NAC family, and NAC domain proteins usually contain two relatively independent domains, one is a highly conserved N-terminal domain, divided into five (A-E) sub domains, responsible for DNA binding. The C-terminal region is highly divergent and serves as a transcriptional activation region, which endows NAC protein with functional variations (Bian et al., 2020). By comparing the protein sequences of BrCUC2 in different species and its homeotic genes, we found that the N-terminal and C-terminal of BrCUC2 contain NAC domains. Systematic evolution analysis shows that BrCUC2 is closely related to B. oleracea, and the CUCs gene contains similar conserved motifs, indicating that functional similarity was maintained during evolution.
Cloning the BrCUC2 promoter sequence at the growth site of winter B. rapa showed that BrCUC2 contained meristem-forming elements, indicating that the expression of BrCUC2 would affect the formation of the meristem. The BrCUC2 gene mainly present in the nucleus, and yeast self-activation verifies its self-activation activity, which may acts as a transcription factor. The BrCUC2 promoter contains various photoresponsive elements, hormone- and stress-responsive elements. When plants are subjected to low temperature stress, transcription factors activate the expression of low-temperature -responsive genes by binding cis-acting elements on the gene promoter, thus regulating the signaling pathways in the plant to improve low temperature tolerance. The bZIP transcription factor binds to the ABRE element (ACGT) on the target gene promoter to regulate the expression of downstream low temperature response genes (Hwang et al., 2016). CsbZIP6 is a negative regulatory factor for low temperature stress response in tea plants, leading to a downregulation of downstream low temperature response gene expression and affecting the sensitivity of tea plants to ABA (Wang et al., 2017). The Arabidopsis gene AtbZIP1 binding ABRE action element and regulates plant response to low temperature stress through ABA-dependent signal transduction pathway (Sun et al., 2011b). BrCUC2 promoter contains ABRE, ARE, CGTCA-motif, TC-rich repeats and TGA-element elements, which may be involved in the regulation of low temperature stress response. GUS is mainly observed in the leaf margins, stems, and meristems. Research on the expression pattern of the BrCUC2 gene in winter rapeseed under low-temperature stress and GUS tissue staining showed that the light response elements, meristem formation, and stress-related elements contained in BrCUC2 played an important role, which is consistent with previous research results on the CUC2 promoter of B. platyphylla.
The natural environment is an important factor influencing plant growth and development. To ensure normal growth, plants must change their molecular and physiological mechanisms in response to external stress. SOD and POD, as scavengers of reactive oxygen species (ROS), can effectively protect plants from low temperatures and other abiotic stresses (Nadarajah, 2020).
Zhang et al (Zhang et al., 2020) found that overexpression of the SikRbcs2 gene significantly increased SOD, POD, and CAT enzyme activity in tomato SikRbcs2 transgenic tobacco under 4°C stress. The transgenic tomato ZPR1 gene significantly enhanced Arabidopsis physiological activity and cold resistance (Li et al., 2012). The cuc1-2 double mutant of A. thaliana has an abnormal stem end meristem tissue formation (Aida et al., 1997; Uauy et al., 2006). The plant transformed from B. platyphylla to 35S::BPCUC2 was shorter than that of the normal plant (Liu et al., 2018). This phenomenon may be due to CUC2 overexpression, which affects plant growth by affecting the growth of plant meristems. In this study, we found no difference in the phenotypes of the wild-type and CUC2-overexpression strains at the seedling stage, whereas the growth characteristics of the overexpression plants were weaker and shorter during the flowering and maturation stages. This finding indicates that the BrCUC2 gene, as a negative regulatory factor for plant growth, has an inhibitory effect on the growth cones of winter B. rapa. After low-temperature treatment, the physiological activity of the overexpression plants was significantly higher than that of wild-type A. thaliana only at 6 and 12 h, and there was no difference after 24 h, indicating that the BrCUC2 gene can improve the low-temperature tolerance of transgenic A. thaliana over a short period.
Conclusion
The BrCUC2 gene was cloned from the growth point of winter B. rapa. It encodes 367 amino acids, and the conserved sequence belongs to the NAM superfamily. Multiple sequence alignment revealed that the N-terminal amino acid sequence of BrCUC2 homologous CUC proteins contained a highly conserved NAC domain. Systematic evolution analysis showed that BrCUC2 in winter B. rapa had the highest affinity for B. oleracea, B. napus, and R. sativus. BrCUC2 is expressed in the nucleus and has self-activition. After low-temperature stress, the expression level of BrCUC2 was the highest at the growth site. As the temperature decreased, BrCUC2 was upregulated and presented higher expression in ‘Longyou 7,’ which presents strong cold resistance and a concave growth site, than in ‘Longyou 99,’ which presents weak cold resistance and a protruding growth site. The BrCUC2 promoter sequence contains a variety of action elements related to light, hormone, and stress responses and meristem expression. GUS chemical tissue staining mainly revealed expression at the edges of the leaves, stems, and growth points in transgenic A. thaliana. The overexpression of the BrCUC2 gene in A. thaliana is a characteristic of stunted plant development. After 12 h of low-temperature treatment, some leaves of the BrCUC2-overexpression A. thaliana withered and yellowed, and the survival rate was 62%; however, the survival rate of wild-type A. thaliana decreased to 8%, and most of the leaves died. After 24 h of low-temperature treatment, the physiological indicators SOD, POD, and PRO reached their highest values (16.9%, 41.9%, and 22.9% higher than that of the wild type, respectively). This result indicates that the BrCUC2 gene can improve the low-temperature tolerance of transgenic A. thaliana. This study aimed to explore the regulatory mechanism of BrCUC2 at the growth site of winter B. rapa to provide a foundation for studying cold resistance in winter B. rapa.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XT: Data curation, Writing – original draft. YZ: Data curation, Writing – review & editing. JW: Data curation, Funding acquisition, Methodology, Writing – review & editing. RZ: Data curation, Writing – review & editing. JJ: Data curation, Writing – review & editing. RL: Data curation, Writing – review & editing. WM: Data curation, Writing – review & editing. YL: Data curation, Writing – review & editing. WW: Methodology, Writing – review & editing. YP: Methodology, Writing – review & editing. GY: Methodology, Writing – review & editing. LL: Methodology, Writing – review & editing. XL: Methodology, Writing – review & editing. LM: Funding acquisition, Methodology, Writing – review & editing. WS: Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (32260519), Gansu Provincial Technology Innovation Guidance Program (23CXNA0041), Science and Technology Program of Gansu Province (22ZD6NA009), Gansu Province Modern Cold and Arid Agriculture Science and Technology Support (KJZC-2023-12), and the China Agriculture Research System of MOF and MARA (CARS-12).
Acknowledgments
We wish to thank Master Jia Xu, Master Na Zhang of Gansu Agricultural University for help with the experiments. We thank Editage (www.editage.cn) for English language editing during the preparation of this manuscript.
Conflict of interest
Author YZ is employed by Gansu Yasheng Agricultural Research Institute Co. LTD Lanzhou, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1274567/full#supplementary-material
Supplementary Figure 1 | Conserved motifs logo in CUCs proteins.
Supplementary Figure 2 | Structural characteristics of the BrCUC2 gene.
Supplementary Figure 3 | Screening of BrCUC2 transgenic Arabidopsis thaliana.
Supplementary Table 1 | Motif prediction of BrCUC2 protein by MEME server.
References
Abrahám, E., Hourton-Cabassa, C., Erdei, L., Szabados, L. (2010). Methods for determination of proline in plants. Methods Mol. Biol. 639, 317–331. doi: 10.1007/978-1-60761-702-0_20
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 9, 841–857. doi: 10.1105/tpc.9.6.841
Aida, M., Ishida, T., Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Dev. (Cambridge England). 126, 1563–1570. doi: 10.1242/dev.126.8.1563
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Balkunde, R., Kitagawa, M., Xu, X. M., Wang, J., Jackson, D. (2017). SHOOT MERISTEMLESS trafficking controls axillary meristem formation, meristem size and organ boundaries in Arabidopsis. Plant J. 90, 435–446. doi: 10.1111/tpj.13504
Bertrand, C., Bergounioux, C., Domenichini, S., Delarue, M., Zhou, D. X. (2003). Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278, 28246–28251. doi: 10.1074/jbc.M302787200
Bhatt, A. M., Etchells, J. P., Canales, C., Lagodienko, A., Dickinson, H. (2004). VAAMANA—a BEL1-like homeodomain protein, interacts with KNOX proteins BP and STM and regulates inflorescence stem growth in Arabidopsis. Gene 328, 103–111. doi: 10.1016/j.gene.2003.12.033
Bian, Z., Gao, H., Wang, C. (2020). NAC transcription factors as positive or negative regulators during ongoing battle between pathogens and our food crops. Int. J. Mol. Sci. 22, 81. doi: 10.3390/ijms22010081
Durak, I., Yurtarslanl, Z., Canbolat, O., Akyol, O. (1993). A methodological approach to superoxide dismutase (SOD) activity assay based on inhibition of nitroblue tetrazolium (NBT) reduction. Clin. Chim. Acta 214, 103–104. doi: 10.1016/0009-8981(93)90307-p
Geourjon, C., Deléage, G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosciences: CABIOS. 11, 681–684. doi: 10.1093/bioinformatics/11.6.681
Guo, S., Zhang, Y., Zhai, L., Liu, J., Wang, H., Liu, H. (2023). The environmental benefit and farmer adoption of winter cover crops in the North China Plain. Pedosphere doi: 10.1016/j.pedsph.2023.03.011
Guo, A. Y., Zhu, Q. H., Chen, X., Luo, J. C. (2007). [GSDS: a gene structure display server]. Yi Chuan. 29, 1023–1026. doi: 10.1360/yc-007-1023
Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evolution. 30, 1229–1235. doi: 10.1093/molbev/mst012
Hernandez-Garcia, C. M., Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217-218, 109–119. doi: 10.1016/j.plantsci.2013.12.007
Hu, W., Wei, Y., Xia, Z., Yan, Y., Hou, X., Zou, M., et al. (2015). Genome-wide identification and expression analysis of the NAC transcription factor family in cassava. PloS One 10, e0136993. doi: 10.1371/journal.pone.0136993
Hwang, I., Manoharan, R. K., Kang, J. G., Chung, M. Y., Kim, Y. W., Nou, I. S. (2016). Genome-Wide Identification and Characterization of bZIP Transcription Factors in Brassica oleracea under Cold Stress. BioMed. Res. Int. 2016, 4376598. doi: 10.1155/2016/4376598
Kamiuchi, Y., Yamamoto, K., Furutani, M., Tasaka, M., Aida, M. (2014). The CUC1 and CUC2 genes promote carpel margin meristem formation during Arabidopsis gynoecium development. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00165
Krogh, A., Larsson, B., Von Heijne, G., Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kwon, C. S., Pfluger, J., Bezhani, S., Metha, H., Aida, M., Tasaka, M., et al. (2006). A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Dev. (Cambridge England). 133, 3223–3230. doi: 10.1242/dev.02508
Lenhard, M., Jürgens, G., Laux, T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129, 3195–3206. doi: 10.1242/dev.129.13.3195
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, J., Han, Z., Xian, M. (2022). Exploration and application of agriculture-tourism technologies based on rape flowers in rural revitalization of China. Oil Crop Sci. 7, 122–126. doi: 10.1016/j.ocsci.2022.08.002
Li, J., Sima, W., Ouyang, B., Luo, Z., Yang, C., Ye, Z., et al. (2012). Identification and expression pattern of a ZPR1 gene in wild tomato (Solanum pennellii). Plant Mol. Biol. Rep. 31, 409–417. doi: 10.1007/s11105-012-0509-4
Liang, X., Peng, L., Baek, C. H., Katzen, F. (2013). Single step BP/LR combined Gateway reactions. Biotechniques 55, 265–268. doi: 10.2144/000114101
Liu, Z., Wang, W., Zhang, C. G., Zhao, J. F., Chen, Y. L. (2017). GUS staining of guard cells to identify localised guard cell gene expression. Bio Protoc. 7, e2446. doi: 10.21769/BioProtoc.2446
Liu, C., Xu, H., Jiang, J., Wang, S., Liu, G. (2018). Analysis of the promoter features of BpCUC2 in Betula platyphylla × Betula pendula. Plant Cell Tissue Organ Cult. 132, 191–199. doi: 10.1007/s11240-017-1324-2
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, L., Coulter, J. A., Liu, L., Zhao, Y., Chang, Y., Pu, Y., et al. (2019a). Transcriptome analysis reveals key cold-stress-responsive genes in winter rapeseed (Brassica rapa L.). Int. J. Mol. Sci. 20, 1071. doi: 10.3390/ijms20051071
Ma, L., Wang, X., Pu, Y., Wu, J., Coulter, J. A., Li, X., et al. (2019b). Ecological and economic benefits of planting winter rapeseed (Brassica rapa L.) in the wind erosion area of northern China. Sci. Rep. 9, 20272. doi: 10.1038/s41598-019-56678-3
Mallory, A. C., Reinhart, B. J., Jones-Rhoades, M. W., Tang, G., Zamore, P. D., Barton, M. K., et al. (2004). MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' region. EMBO J. 23, 3356–3364. doi: 10.1038/sj.emboj.7600340
Maugarny, A., Gonçalves, B., Arnaud, N., Laufs, P. (2016). Chapter 15 - CUC transcription factors: to the meristem and beyond//Gonzalez, D.H. Plant transcription factors. Acad. Press. 15, 229–247. doi: 10.1016/B978-0-12-800854-6.00015-4
Motte, H., Verstraeten, I., Werbrouck, S., Geelen, D. (2011). CUC2 as an early marker for regeneration competence in Arabidopsis root explants. J. Plant Physiol. 168, 1598–1601. doi: 10.1016/j.jplph.2011.02.014
Nadarajah, K. K. (2020). ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 21, 5208. doi: 10.3390/ijms21155208
Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in arabidopsis. Plant Cell. 18, 2929–2945. doi: 10.1105/tpc.106.045617
Niu, Z., Liu, L., Pu, Y., Ma, L., Wu, J., Hu, F., et al. (2021). iTRAQ-based quantitative proteome analysis insights into cold stress of Winter Rapeseed (Brassica rapa L.) grown in the field. Sci. Rep. 11, 23434. doi: 10.1038/s41598-021-02707-z
Olsen, A. N., Ernst, H. A., Leggio, L. L., Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Reinhardt, D., Pesce, E. R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., et al. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260. doi: 10.1038/nature02081
Scofield, S., Murray, J. A. (2006). KNOX gene function in plant stem cell niches. Plant Mol. Biol. 60, 929–946. doi: 10.1007/s11103-005-4478-y
Senthilkumar, M., Amaresan, N., Sankaranarayanan, A. (2021). “Estimation of peroxidase (POD),” in Plant-Microbe Interactions: Laboratory Techniques. Eds. Senthilkumar, M., Amaresan, N., Sankaranarayanan, A. (US: Springer), 123–125. doi: 10.1007/978-1-0716-1080-0_31
Souer, E., Van Houwelingen, A., Kloos, D., Mol, J., Koes, R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. doi: 10.1016/s0092-8674(00)81093-4
Spinelli, S., Martin, A., Viola, I., Gonzalez, D., Palatnik, J. (2011). A mechanistic link between STM and CUC1 during arabidopsis development. Plant Physiol. 156, 1894–1904. doi: 10.1104/pp.111.177709
Sun, X.-L., Li, Y., Cai, H., Bai, X., Ji, W., Ji, Z.-J., et al. (2011b). Arabidopsis bZIP1 Transcription Factor Binding to ABRE cis-Element Regulates Abscisic Acid Signal Transduction. Acta Agron. Sini. 37, 612–619. doi: 10.1016/S1875-2780(11)60016-3
Sun, W., Wu, J., Fang, Y., Liu, Q., Yang, R., Mao, W., et al. (2011a). Growth and development characteristics of winter rapeseed northern-extended from the cold and arid regions in China. Acta Agrono Sini. 36, 2124–2134. doi: 10.3724/SP.J.1006.2010.02124
Takada, S., Hibara, K., Ishida, T., Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128, 1127–1135. doi: 10.1242/dev.128.7.1127
Taoka, K., Yanagimoto, Y., Daimon, Y., Hibara, K., Aida, M., Tasaka, M. (2004). The NAC domain mediates functional specificity of CUP-SHAPED COTYLEDON proteins. Plant J. 40, 462–473. doi: 10.1111/j.1365-313X.2004.02238.x
Teufel, F., Almagro Armenteros, J. J., Johansen, A. R., Gíslason, M. H., Pihl, S. I., Tsirigos, K. D., et al. (2022). SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025. doi: 10.1038/s41587-021-01156-3
Uauy, C., Distelfeld, A., Fahima, T., Blechl, A., Dubcovsky, J. (2006). A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301. doi: 10.1126/science.1133649
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Vroemen, C. W., Mordhorst, A. P., Albrecht, C., Kwaaitaal, M. A. C. J., De Vries, S. C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell. 15, 1563–1577. doi: 10.1105/tpc.012203
Wang, L., Cao, H., Qian, W., Yao, L., Hao, X., Li, N., et al. (2017). Identification of a novel bZIP transcription factor in Camellia sinensis as a negative regulator of freezing tolerance in transgenic Arabidopsis. Ann. Bot. 119, 1195–1209. doi: 10.1093/aob/mcx011
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–w303. doi: 10.1093/nar/gky427
Weigel, D., Glazebrook, J. (2006). Transformation of agrobacterium using the freeze-thaw method. CSH Protoc. 2006, 4666. doi: 10.1101/pdb.prot4666
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Wu, J., Xu, X. D., Liu, L., Ma, L., Pu, Y., Wang, W., et al. (2022). A chromosome level genome assembly of a winter turnip rape (Brassica rapa L.) to explore the genetic basis of cold tolerance. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.936958
Zhang, L., Yang, J., Guo, X., Wang, A., Zhu, J. (2020). Overexpression of SikRbcs2 gene promotes chilling tolerance of tomato by improving photosynthetic enzyme activity, reducing oxidative damage, and stabilizing cell membrane structure. Food Sci. Nutr. 8, 3479–3491. doi: 10.1002/fsn3.1631
Keywords: Brassica rapa, BrCUC2, functional analysis, low-temperature stress, transgenic Arabidopsis thaliana
Citation: Tao X, Zhao Y, Ma L, Wu J, Zeng R, Jiao J, Li R, Ma W, Lian Y, Wang W, Pu Y, Yang G, Liu L, Li X and Sun W (2023) Cloning and functional analysis of the BrCUC2 gene in Brassica rapa L. Front. Plant Sci. 14:1274567. doi: 10.3389/fpls.2023.1274567
Received: 08 August 2023; Accepted: 16 October 2023;
Published: 30 October 2023.
Edited by:
Douglas S. Domingues, University of São Paulo, BrazilReviewed by:
Sachin Teotia, Sharda University, IndiaSwarup Roy Choudhury, Indian Institute of Science Education and Research, Tirupati, India
Copyright © 2023 Tao, Zhao, Ma, Wu, Zeng, Jiao, Li, Ma, Lian, Wang, Pu, Yang, Liu, Li and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Ma, bWFsQGdzYXUuZWR1LmNu; Wancang Sun, MTgyOTMxMjE4NTFAMTYzLmNvbQ==
 Xiaolei Tao
Xiaolei Tao Yuhong Zhao3
Yuhong Zhao3 Li Ma
Li Ma Lijun Liu
Lijun Liu Wancang Sun
Wancang Sun