- 1ICAR-National Bureau of Plant Genetic Resources, Regional Station, Thrissur, Kerala, India
- 2ICAR-National Bureau of Plant Genetic Resources, New Delhi, India
- 3ICAR-Indian Institute of Horticultural Research, Bengaluru, Karnataka, India
Introduction: The introgression of genetic material from one species to another through wide hybridization and repeated back-crossing, plays an important role in genetic modification and enriching the cultivated gene-pool with novel genetic variations. Okra (Abelmoschus esculentus [(L.) Moench)] is a popular vegetable crop with high dietary fibre and protein, rich in essential amino acids, lysine and tryptophan. The wild Abelmoschus genepool has many desirable traits like ornamental value, short internodal length, more number of productive branches, extended bearing, perennation tendency, reduced fruit length (more consumer preferred trait), high mucilage content (medicinal value), abiotic stress tolerances such as drought, high temperature and biotic stress resistances such as okra Yellow Vein Mosaic Virus (YVMV) and Enation Leaf Curl Virus (ELCV) diseases. The repeated use of elite breeding lines led to narrowing of the genetic base of the okra crop, one of the major factors attributed to breakdown of resistance/ tolerance to biotic stresses. YVMV and ELCV are the two major diseases, causing significant yield loss in okra. Hence, wide hybridization was attempted to transfer tolerance genes from wild species to the cultivated genepool to widen the genetic base.
Material and methods: The screening of germplasm of wild Abelmoschus species at hotspots led to the identification of tolerant species (Abelmoschus pungens var. mizoramensis, A. enbeepeegeearensis, A. caillei, A. tetraphyllus and A. angulosus var. grandiflorus), which were further used in a wide-hybridization programme to generate interspecific hybrids with the cultivated okra. Presence of pre- and post-zygotic barriers to interspecific geneflow, differences in ploidy levels and genotype specific variations in chromosome numbers led to varying degrees of sterility in F1 plants of interspecific crosses. This was overcome by doubling the chromosome number of interspecific hybrids by applying Colchicine at the seedling stage. The 113 cross derivatives generated comprising amphidiploids in the F1 generation (30), F3 (14), one each in F2 and F4 generations, back cross generation in BC1F2 (03), BC1F3 (25), and BC2F3 (02), crosses between amphidiploids (27), multi-cross combinations (07) and inter-specific cross (between A. sagittifolius × A. moschatus subsp. moschatus) selfed derivatives at F8 generation (03) were characterized in the present study. Besides they were advanced through selfing and backcrossing.
Results and Discussion: The amphidiploids were found to possess many desirable genes with a considerable magnitude of linkage drag. Majority of the wide cross derivatives had an intermediate fruit morphology and dominance of wild characters viz., hispid fruits, stem, leaves, tough fruit fibre, vigorous perennial growth habit and prolonged flowering and fruiting. The fruit morphology of three BC progenies exhibited a high morphological resemblance to the cultivated okra, confirming successful transfer of useful genes to the cultivated okra genepool. The detailed morphological characteristics of the various combinations of Abelmoschus amphidiploids and the genetic enhancement of the genepool achieved in this process is reported here.
Introduction
Introgression, the transfer of genes between species mediated primarily by backcrossing (Anderson, 1949; Twyford and Ennos, 2012) plays an important role in the evolution of plant species. It results in genetic modification and enrichment of the genepool for crop improvement programs. The extent of introgression in nature is limited though not prevented by the existence of reproductive barriers that filter natural geneflow (Mallet, 2005; Slotte et al., 2008). Several successful attempts to break interspecific barriers to gene flow have been documented in different crops, which resulted in new genomic combinations (Joseph et al., 2017). Though breeders are consistently interested in introgressing genes conferring desirable traits from wild to cultivated species, the process is elaborate due to the predominance of linkage drag exhibited by the introgressed lines.
Okra (Abelmoschus esculentus L. Moench), a popular traditional vegetable under the family Malvaceae, is grown throughout the tropical and subtropical regions of the world and in warmer parts of the temperate zone. Tender fruits are used as vegetables, in addition to their leaves, buds, and flowers being consumed especially in West Africa (Sharma, 1993). Further, the fiber extracted from the waste stems can be used to make paper pulp or fuel ( Gogoi et al., 2017), while the foliage can be used as biomass (Sharma, 1993). Okra mucilage has potential use in food and non-food products and for medicinal purposes (Ortaç et al., 2018; Peng et al., 2019). Studies also demonstrated the presence of bioactive compounds in plant parts such as leaves, flowers, and seeds (Adelakun et al., 2009; Zhang et al., 2020; Adetuyi et al., 2012; Olawuyi and Lee, 2021). Recently, the effects of okra leaves, fruits, and seed extracts on European sea bass (Dicentrarchus labrax) leukocytes and their cytotoxic, bactericidal, and antioxidant properties were reported by Guebebia et al. (2023). Okra has a vast potential for earning foreign exchange, as it has a significant share in fresh vegetable export of India (2,442.96 million US$ fresh vegetable export during 2022–2023; https://agriexchange.apeda.gov.in/IndExp/PortNew.aspx accessed on Aug 3, 2023).
Breeding efforts are in full swing to develop novel ideotypes with branching behavior, shorter internodes, multi-flory, ramiflory, and nutritional quality. However, the cultivation of okra is facing serious threats due to two major viral diseases, namely, okra Yellow Vein Mosaic Virus (YVMV) and Enation Leaf Curl Virus (ELCV) diseases. Among them, YVMV disease is the most severe constraint responsible for 90% to 100% yield loss depending upon the stage of infection. These diseases reduce the yield substantially and affect the marketability of the fruits (Sanwal et al., 2014a). The virus is readily transmitted by grafting and through the insect vector, whitefly (Bemisia tabaci Gen.) (Varma, 1952). Host-plant tolerance to viruses is one of the most practical, economical, and environment-friendly strategies for reducing yield loss in okra (Ayam et al., 2018). Intensive efforts were underway in India to develop virus-tolerant varieties, and it resulted in the release of several varieties, namely, Pusa Sawani, Arka Anamika, Arka Abhay, and Parbhani Kranti. The host-plant tolerance has been overcome by the evolution of new variants of the viruses because of the development of new strains or due to recombination in virus strains (Sanwal et al., 2014b). Another major reason for the breakdown of resistance would be due to the emergence of the polyphagous ‘B’ biotype of B. tabaci, with its increased host range resulting in the infection of the Gemini virus in previously unaffected crops (Chowda-Reddy et al., 2012). Presently, none of the cultivated okra varieties or hybrids have stable tolerance to YVMV disease (Sandeep et al., 2022).
Undomesticated crop wild relatives are rich sources of genes providing resistance to various diseases, pests, and unfavorable environmental conditions (Rattan and Kumar, 2020). In addition to disease resistance, other agronomic traits such as male sterility (Sigareva and Earle, 1997 in cabbage) and abiotic stress tolerance (Wei et al., 2015 in tobacco) have also been transferred by wide hybridization. Successful development and characterization of amphidiploids have been reported in several crops such as wheat (Zhang et al., 2016; Song et al., 2019; Klimushina et al., 2020; Zuo et al., 2020; Ren et al., 2022), rice (Wang et al., 2013; Kumar et al., 2019), peanut (de Paula et al., 2017; Li-Na et al., 2017; Favero et al., 2020; Ballén-Taborda et al., 2022), Brassica (Thomsen, 1990; Song et al., 1993), Vigna (Mjnocha et al., 1991), Dianthus (Nimura et al., 2006), Cucumis (Chen et al., 2002), and Solanum (Khan et al., 2013; Zhou et al., 2018). Even though many workers have attempted and succeeded in developing interspecific hybrids, the introgressed lines could be utilized only after eliminating the fertility barriers associated with the hybrids.
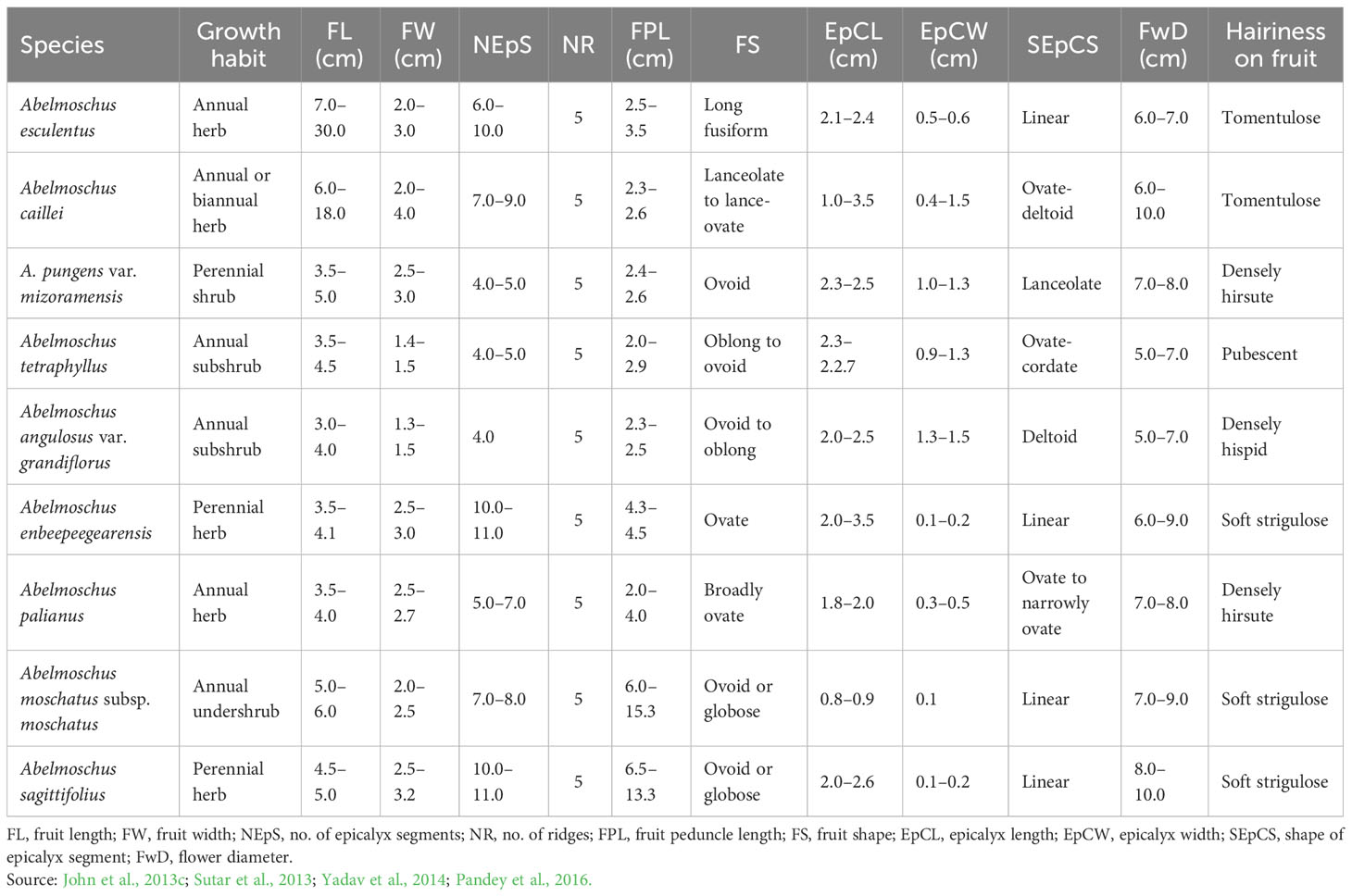
Dark green fruits, high mucilage content, extended bearing, perennation propensity, high branching, reduced fruit length, and tolerance to drought, high temperature, and okra YVMV disease are some of the favorable characteristics of the wild okra species (Sandeep et al., 2022). John et al. (2013a) reported that wild relatives, namely, Abelmoschus angulosus var. grandiflorus Thwaites, Abelmoschus crinitus Wall., Abelmoschus ficulneus (L.) Wight & Arn., Abelmoschus tetraphyllus (Roxb. ex Hornem.) Wall., Abelmoschus pungens var. mizoramensis K.J.John, Krishnaraj & K.Pradheep, and Abelmoschus enbeepeegearensis K.J.John, Scariah, Nissar, K.V.Bhat & S.R.Yadav did not express any YVMV disease symptoms under field epiphytotic conditions. Similarly, Gangopadhyay et al. (2016) reported field-level tolerance to this disease in accessions belonging to Abelmoschus caillei (A.Chev.) J.M.C.Stevels, Abelmoschus manihot (L.) Medik., and Abelmoschus moschatus Medik. species. Further, A. manihot (L.) Medik. subsp. tetraphyllus (Roxb. Ex Hornem.) Borss. Waalk. was reported as one of the most prominent species exploited worldwide as a source of resistance/tolerance to YVMV, jassids, and fruit borer (Gangopadhyay et al., 2016; Badiger and Yadav, 2019; Patel et al., 2021). Santhiya et al. (2022) also reported the promising accessions of A. moschatus (IC141055), A. tetraphyllus (IC90476-1), and A. caillei (Sikkim), which showed very low incidence of YVMV disease and no incidence of ELCV disease under natural epiphytotic screening. Accessions IC203833 and IC470751 of A. angulosus were reported by Singh et al. (2023a) to be highly resistant to YVMV disease under artificial screening conditions using viruliferous whitefly-mediated mass inoculation. However, elaborate and systematic studies on screening of available wild relatives of Abelmoschus and their utilization in breeding programs are still limited.
Hence, an intensive and systematic wide hybridization or pre-breeding program is required to identify and transfer the genes for tolerance to abiotic and biotic stresses from the wild Abelmoschus species to the cultivated species. Several earlier attempts of wide hybridization in okra have been reported, which include Pal et al. (1952) and Kuwada (1966) for A. esculentus with Abelmoschus tuberculatus; Kuwada (1974) for A. tuberculatus with A. manihot; Akhond et al. (2000) for A. esculentus with A. moschatus; and Samarajeewa (2003) for A. esculentus with A. angulosus and A. esculentus with A. caillei. Further, Manju and Gopimony (2009) succeeded in the development of the variety ‘Anjitha’ by inducing mutation through gamma irradiation of interspecific hybrids of A. esculentus cv. Kiran × A. manihot. Further, Nagaraju et al. (2019) carried out interspecific hybridization using 10 wild species and Amiteye et al. (2019) using A. esculentus and A. caillei. Rattan and Kumar (2020) developed the protocol for embryo rescue of interspecific hybrids involving A. esculentus × A. tetraphyllus. Badiger et al. (2023) elaborated the pollen germination, pollen–pistil interaction, and crossability studies in interspecific and induced colchiploid populations involving crosses between A. esculentus and wild species, viz., A. manihot var. tetraphyllus and A. moschatus. Even though several studies on wide hybridization and development of successful amphidiploids are available, this is the first-of-its-kind report on the characterization of amphidiploids developed through wide hybridizations in the Abelmoschus genus. The present report addresses the following objectives: 1) to document the morphological variability in amphidiploid derivatives among different wild Abelmoschus species and okra and 2) to identify the utility of the Abelmoschus amphidiploids as genetic stocks and sources of useful genes in a variety of development programs.
Materials and methods
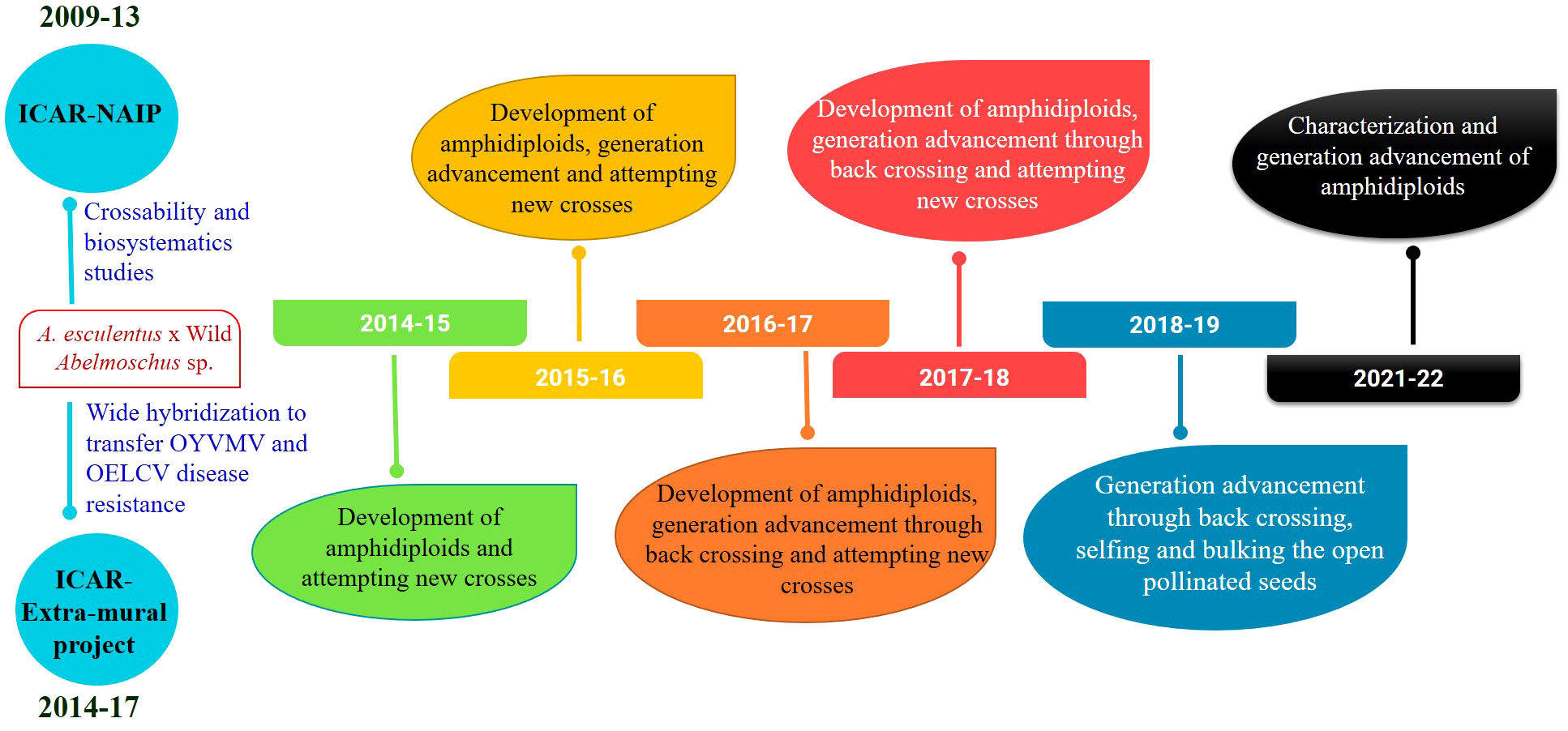
Crossability studies and wide hybridization work were started in 2009–2013 and 2015–2017 under the Indian Council of Agricultural Research (ICAR)—National Agricultural Innovation Project (NAIP) and continued under the ICAR-Extramural Project of the ICAR-Horticultural Sciences Division and later under the ICAR-Emeritus Scientist Programme (2018–2021). Initially, under the NAIP, interspecific hybridization was attempted between different Abelmoschus species, primarily for establishing crossability relationships, thereby understanding the genetic relationships between the species based on fertility and secondarily to derive new recombinants to be used in breeding programs (John et al., 2013a). Further, under the subsequent project, the available wild species (44 accns. of 11 Abelmoschus taxa) conserved in the Medium Term Storage (MTS) module of ICAR—National Bureau of Plant Genetic Resources (NBPGR), Regional Station, Thrissur, Kerala, India, were evaluated for resistance/tolerance to okra YVMV and ELCV diseases at three hot spot locations, namely, Varanasi, Guntur, and New Delhi, India. The wild Abelmoschus taxa include A. moschatus subsp. moschatus, A. caillei, A. tetraphyllus, A. pungens var. mizoramensis, A. angulosus var. grandiflorus, A. enbeepeegearensis, A. crinitus, Abelmoschus palianus, A. tuberculatus, A. ficulneus, and Abelmoschus sagittifolius. This led to the identification of tolerant accessions of A. enbeepeegearensis and A. angulosus var. grandiflorus. These tolerant accessions along with other reported sources of virus tolerance (John et al., 2013a) were selected for wide hybridization. It includes 18 accessions of A. esculentus (10 accns. and eight released varieties), one of A. caillei, and eight of wild Abelmoschus spp., the details of which are given in Table 1.
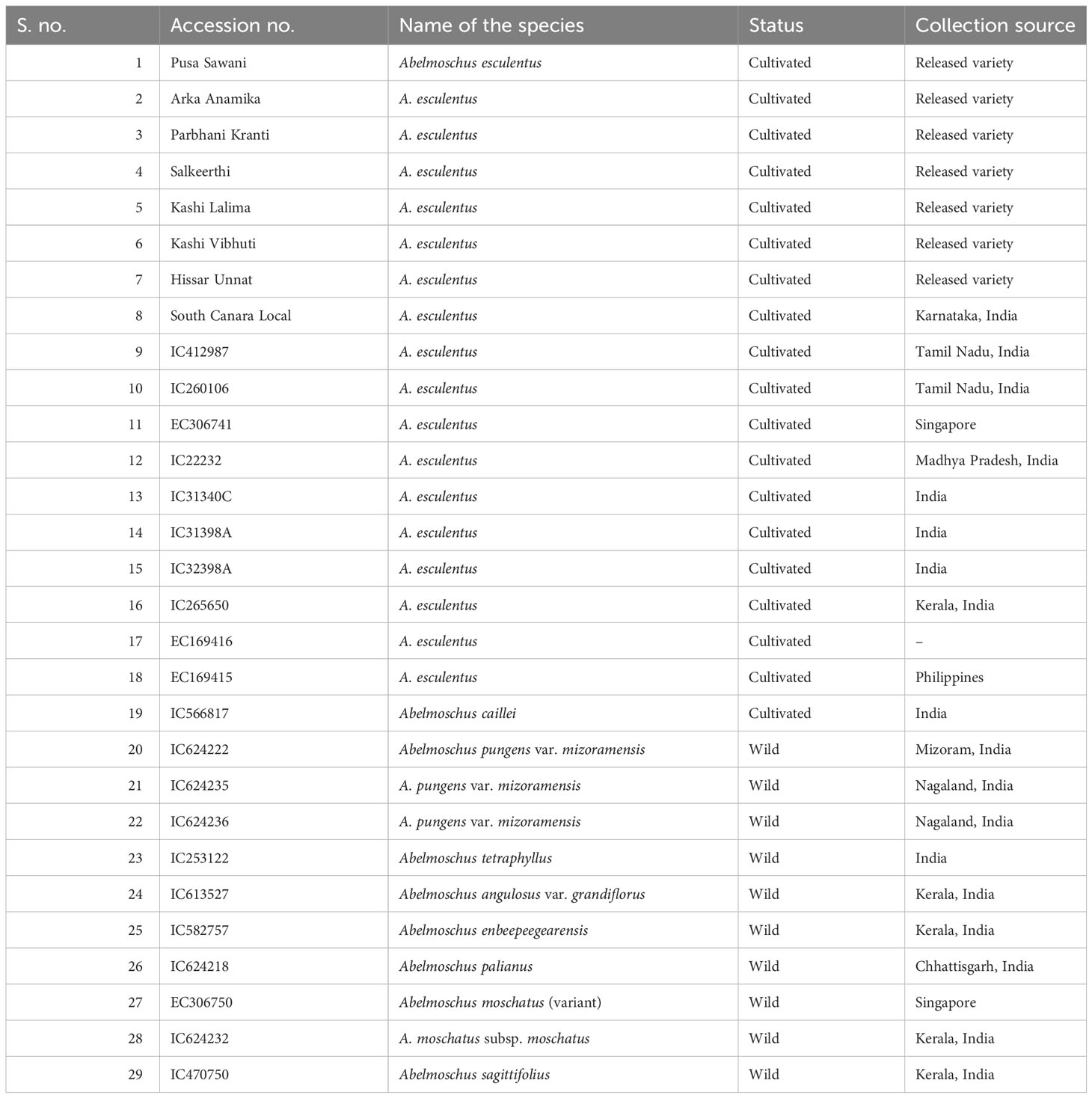
Table 1 The details of cultivated and wild Abelmoschus germplasm used in the wide hybridization program.
Hybridization
Hybridization is the critical step for obtaining successful hybrids, as anthesis time, anther dehiscence, and pollen viability of the wild species affect the success of the process. Mature flower buds of the female parent were emasculated before anthesis by shaving off the anthers in the previous evening and covered with butter paper selfing bags. Ready-to-open flower buds of the male parent were also covered at the same time to prevent pollen contamination. Pollen from the male parent was collected and dusted onto the receptive stigma of emasculated flowers between 9:00 AM and 10:00 AM (next day) under Thrissur, Kerala conditions and again covered with butter paper bags to avoid pollination with undesirable pollen and then tagged. The selfing bags were removed 3 days after pollination to allow the fruits to develop properly. The tagged fruits upon maturity were harvested, and hybrid seeds were extracted and stored for 3–6 months in a cool place.
Colchicine treatment
The interspecific F1 seeds were scarified by rubbing on sandpaper and soaked overnight in distilled water before sowing. Seedlings were raised in polybags, and half the number of seedlings in all cross combinations were subjected to colchicine treatment. The untreated seedlings were transplanted into pots and allowed to grow under optimal conditions. The untreated seedlings were grown separately, and it was found that the F1 set fruits were without seed set. The pollen grains from these hybrids were found to be pale cream colored, indicating the sterility of the pollen. In order to overcome the sterility in the interspecific hybrids, emerging seedlings at the third leaf stage (epicotyl leaves fully grown and third leaf emergence stage) were subjected to colchicine treatment, 0.10% for 3 days from 7:00 AM to 11:00 AM at 20–30-min intervals using cotton swab method (Figure 1A). As per this method, the cotton swab placed on the apical meristem was soaked with 0.10% colchicine at regular intervals with one drop of the solution. After completion of the application each day, the cotton swab was washed with distilled water, and on the fourth day, the cotton swab was removed (Joseph et al., 2017). The treated plants were regularly sprayed with artificial growth formulations of mineral supplements to boost growth. All treated seedlings were provided with support to withstand weight due to enlarged tissue on account of chromosome doubling, and plants were given an optimal growth environment by shifting them to a mist house.
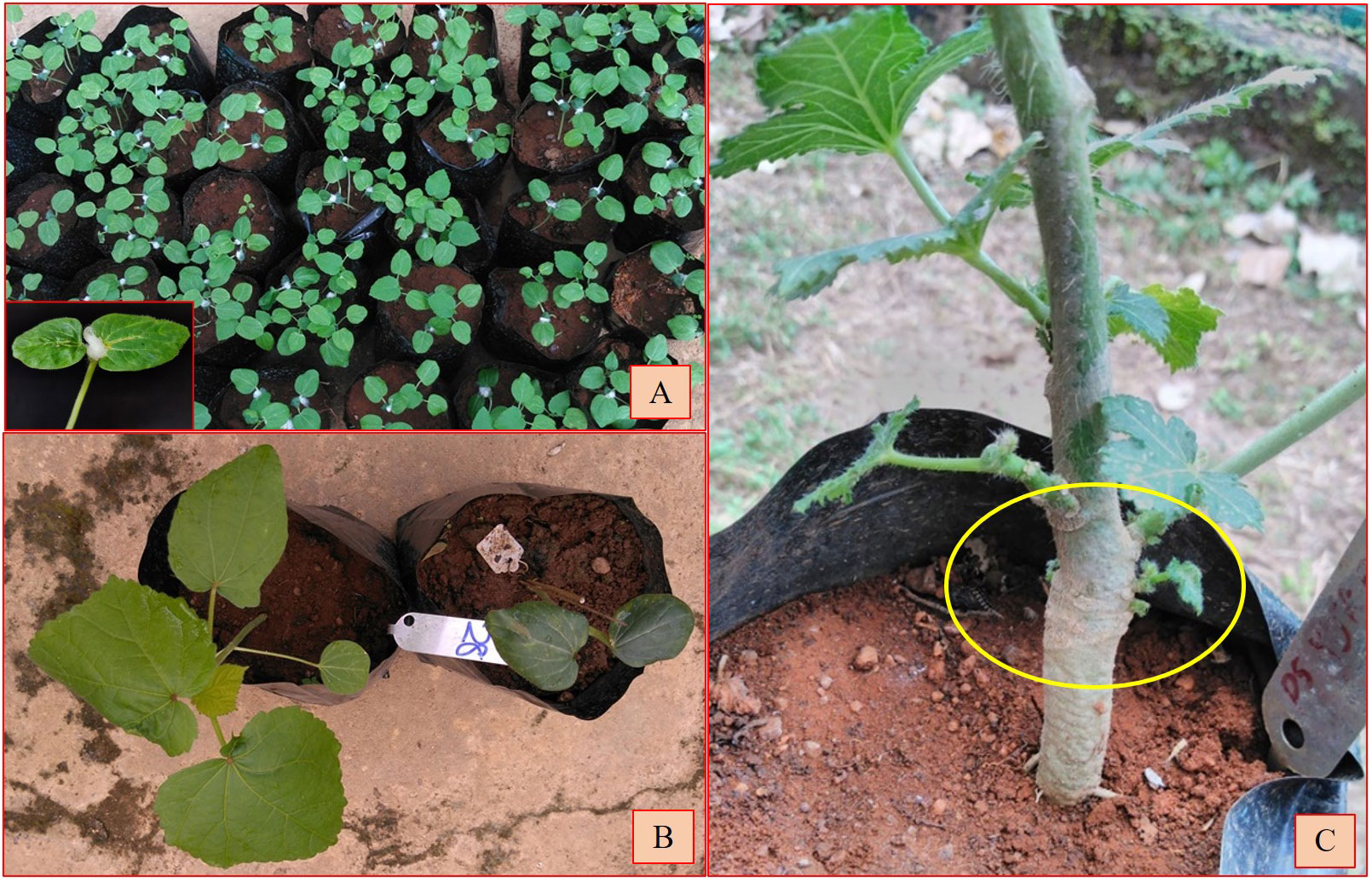
Figure 1 (A) Colchicine treatment using cotton swab method on interspecific hybrid seedlings. Inset: close-up photograph of colchicine treatment. (B) Colchicine un-treated and treated seedlings. (C) Bulging at the colchicine-treated site.
General observations on colchicine-treated seedlings
The seedlings exhibited scorching symptoms in the apical region after the colchicine treatment, but mortality depended on the genotype of the seedlings and the prevailing environmental conditions. The third leaf was crumbled or retarded in its growth and in some cases was partially dried. The epicotyl leaves became succulent and turgid and changed their color from light green to dark green (Figure 1B). The epicotyl leaf petioles became shortened compared to the normal seedlings. It took nearly 3–4 weeks for the seedling to overcome the stress induced by the colchicine treatment. Bulging near the collar region, resembling a tumor-like appearance developed at the site of colchicine treatment, was considered an indication of polyploidization (Suma et al., 2020) (Figure 1C). The surviving seedlings were transplanted to standard pots and provided optimal growth conditions following a set of practices recommended for okra. The fertile amphidiploids were further selfed for one more generation to stabilize the genetic architecture (Joseph et al., 2017). Hence, the seeds from the amphidiploids (C0 generation) were harvested and used for generation advancement for the next season. The flowchart detailing the development and advancement of amphidiploids and the timeline depicting the wide hybridization program in okra are presented in Figures 2, 3, respectively.

Figure 2 Flowchart of wide hybridization, Abelmoschus amphidiploid development, and generation advancement.
Naming of amphidiploids
The amphidiploids were named considering their generation, indigenous collection/exotic collection (IC/EC) number of the cultivated species (A. esculentus or A. caillei), and the name of the species/subspecies/variety taxon of the wild Abelmoschus used for hybridization followed by the plant number of the colchiploid selected. For instance, C3/106/mizo30-4 is an amphidiploid in which the capital letter “C” denotes the colchicine-treated offsprings (synonymous to “F”, the filial generation used in general hybridization events) and the subscript “3” the generation of the amphidiploid followed by the last two/three digits of the number of A. esculentus (e.g., 106 of IC260106 in the above-mentioned amphidiploid). It is followed by the first four letters of the lowest taxon of the wild Abelmoschus (mizo of A. pungens var. mizoramensis) species used in the hybridization, followed by the plant numbers of the amphidiploid selected in the subsequent generations. However, in C1Pusamizo and C1Arkamosc (variant) derivatives, the lowest taxon name of wild Abelmoschus species was followed by the last three digits of the IC/EC number of the wild species used in hybridization, viz., C1Pusamizo235-02 and C1Arkamosc (variant) 750-01, followed by plant number selected. Further, for convenience, the selfed derivatives in the F2 and F3 generations were coded as AM series and the multi-cross combinations as B series (Tables 2B, E, F). The surviving amphidiploids were further advanced through selfing up to the C2/C3 generation. The advanced amphidiploids were screened for tolerance/resistance to okra YVMV and ELCV diseases under natural epiphytotic conditions in New Delhi, Guntur (Andhra Pradesh) and Thrissur (Kerala), India, during 2017–2018. The amphidiploid derivatives presumed to have acquired genes from cultivated okra (with low spininess and intermediate fruit length) and found to be tolerant to YVMV disease along with a profuse and extended bearing habit were selected for generation advancement by selfing or backcrossing under the ICAR-Emeritus Scientist Programme.
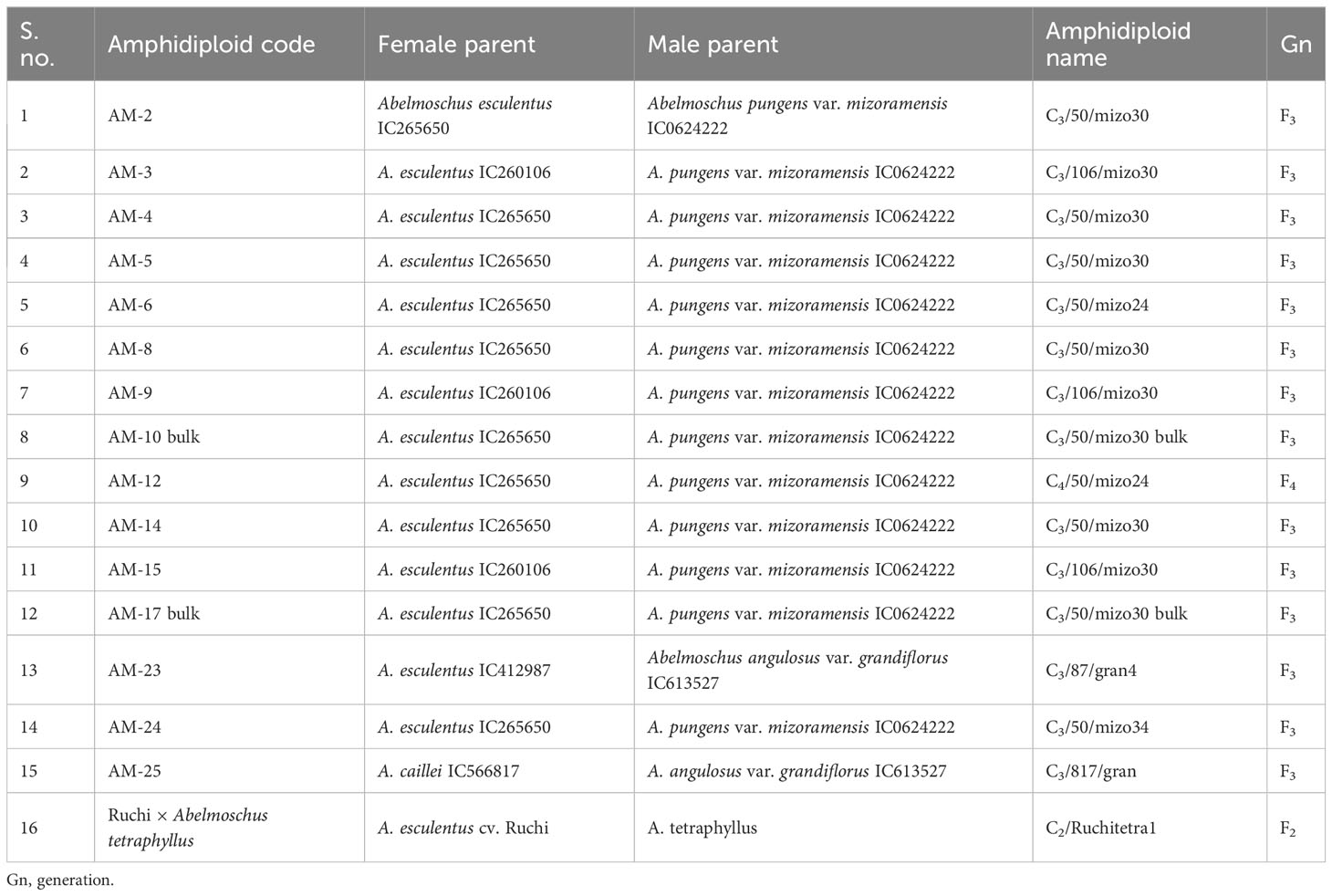
Table 2B Details of pedigree information of selfed amphidiploid derivatives in F2, F3, and F4 generations.
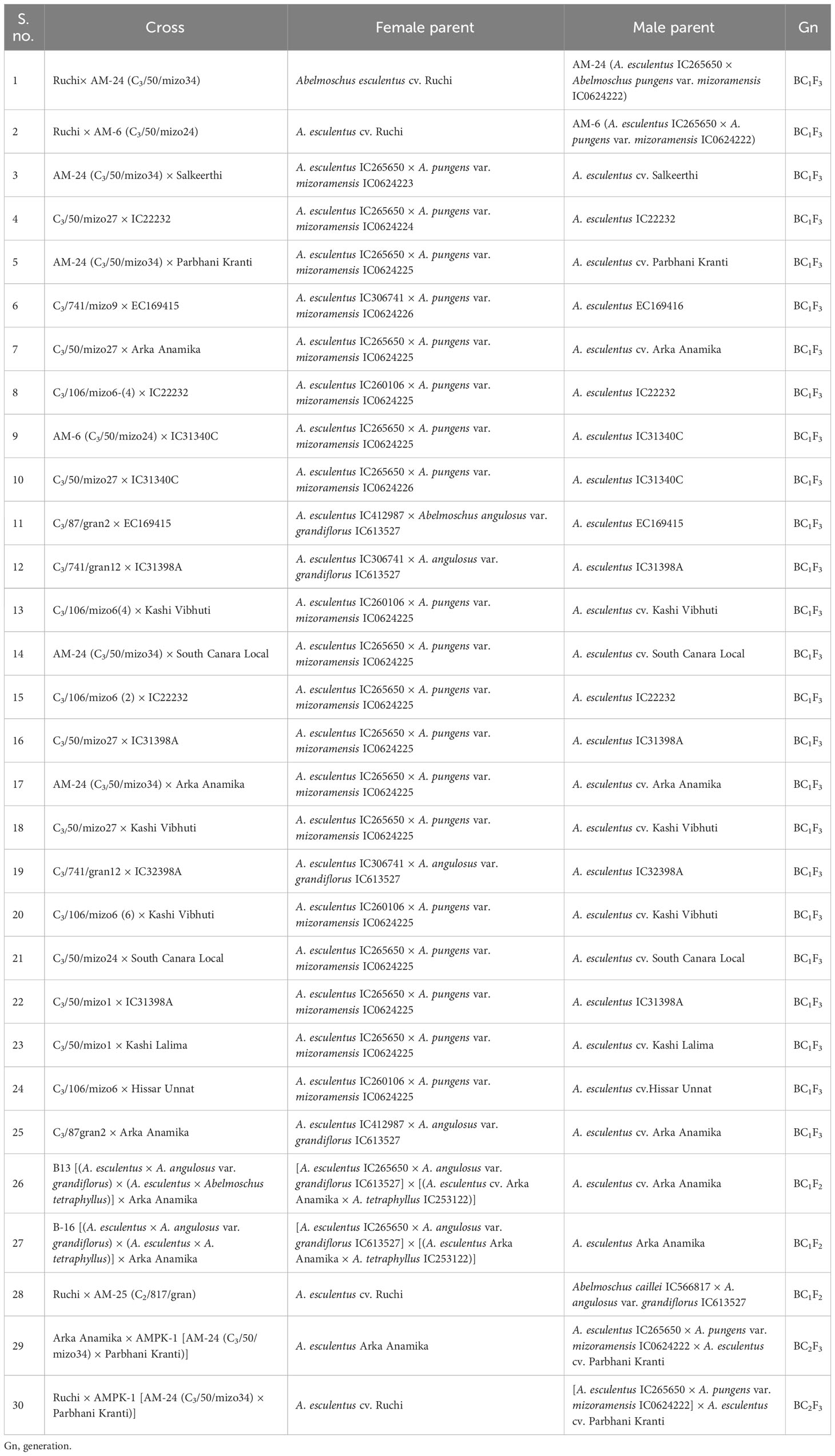
Hence, in the present study, the amphidiploids developed over the years in various generations included a total of 113 derivatives comprising amphidiploids in the F1 generation (30); F3 (14); one each in the F2 and F4 generations; backcross generation in BC1F2 (03), BC1F3 (25), and BC2F3 (02); crosses between amphidiploids (27); multi-cross combinations (07); and interspecific cross [between A. sagittifolius (Kurz) Merr. (Syn. A. moschatus subsp. tuberosus Borss.Waalk.) and A. moschatus subsp. moschatus] selfed derivatives at the F8 generation (03). Polyploidization using colchicine treatment was not undertaken in the cross between A. sagittifolius and A. moschatus subsp. moschatus, as seed setting (few) was observed in this cross. The pedigree information of the amphidiploids is given in Tables 2A–F. Data were taken for 11 quantitative (flower and fruit characteristics) and one qualitative [(presence of spiny hairs scored as absent (0), soft hairs (1), and spiny hairs (2)] characteristics following Charrier (1984) and minimal descriptors for okra (Srivastava et al., 2001). Observations on general morphology (growth habit, branching pattern, nature of lobing of leaves, and presence of red/purple color on the plant parts) were also taken. The diverse amphidiploids were raised in the field from June to November 2021 at ICAR-NBPGR, Regional Station, Vellanikkara, Thrissur, Kerala, India, located at 10.5480°N, 76.2830°E, to understand the variability pattern, to identify desirable segregants, and for generation advancement. The temperature during the crop period was between 23.4°C and 32.5°C, with a mean rainfall ranging from 19.20 mm to 626.9 mm and with an average of 18.16 rainy days per month. The data were analyzed using Microsoft Excel 2019, and summary statistics for each category of amphidiploids (mean, range, standard deviation, and coefficient of variation) were derived.
Results
Interspecific hybridization followed by polyploidization resulted in diverse amphidiploids, which in general exhibited intermediate phenotypic traits with dominant wild characteristics (Figure 4). Wild Abelmoschus species differ from the cultivated types, with respect to the growth habit, branching pattern, density of indumentum (hairiness over plant parts), length and width of the epicalyx, persistence of epicalyx at maturity, fruit length, and growth duration (perennial nature). For reference, Table 3 shows the characteristics of various Abelmoschus spp. used in this study. Basal orthotropic branching; shallow lobing of the older leaves (at bottom) and deep lobing of the top leaves; reddish coloration on the stems, petioles, and nodal region; and yellow flowers were the frequently observed characteristics in the amphidiploids. Additionally, prolific and extended bearing and densely hirsute fruits were regularly observed. The number of ridges on the fruit surface was invariably five in all the derivatives.
Selfed amphidiploid derivatives
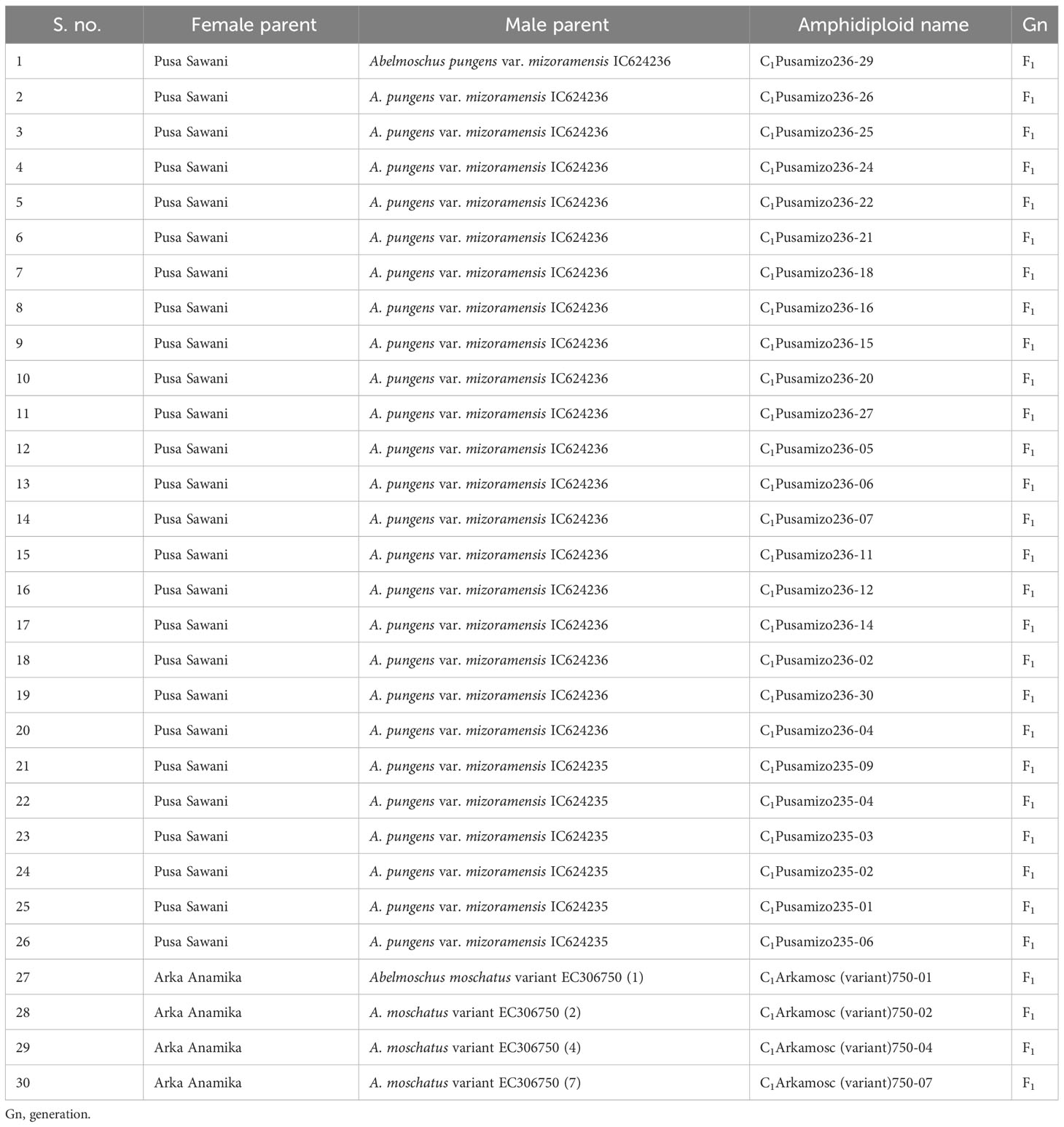
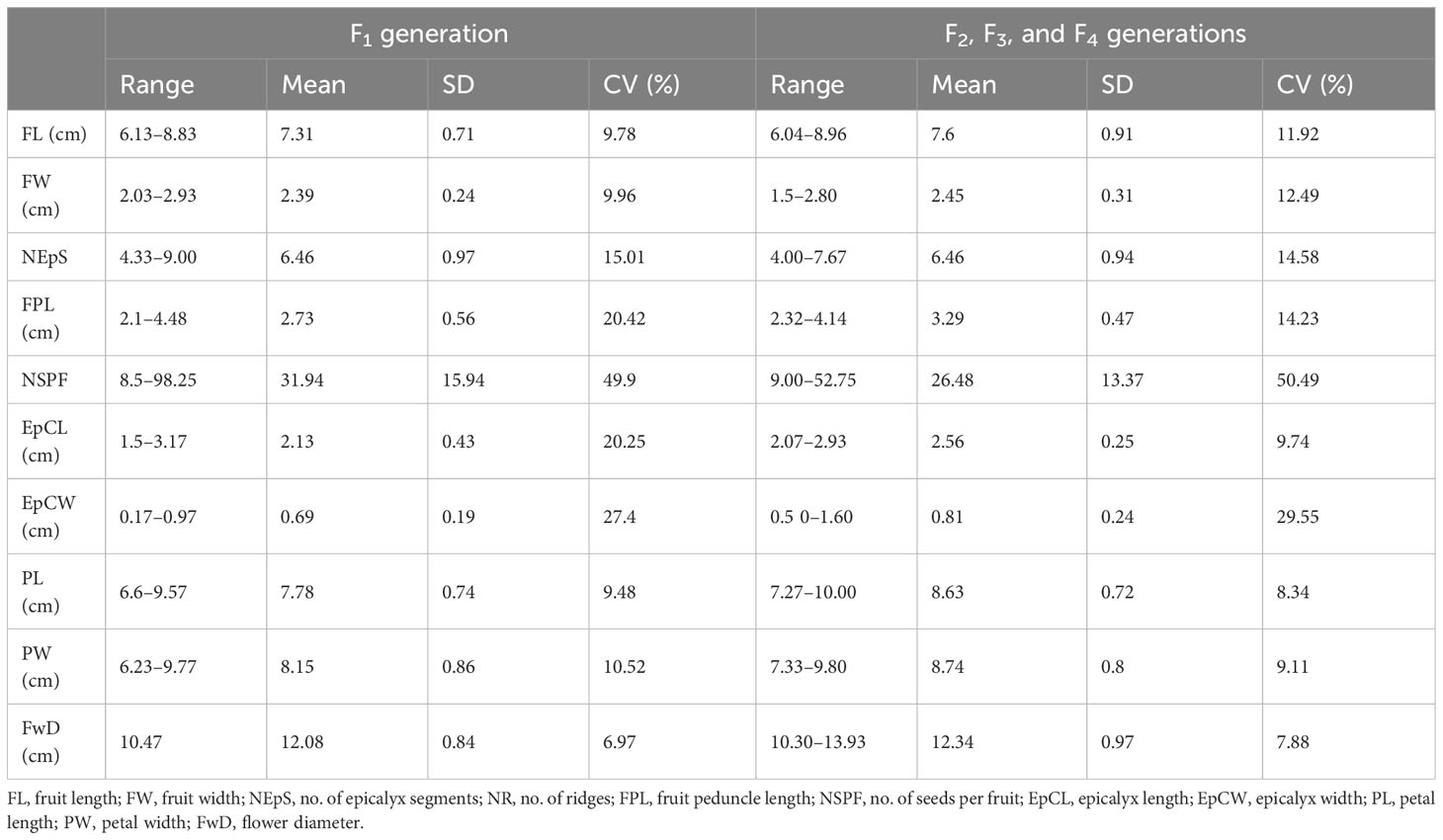
Thirty out of the 113 crosses were amphidiploids in the F1 generation. Twenty-six were crossed between Pusa Sawani and A. pungens var. mizoramensis (IC624236 and IC624235 in 20 and 6 crosses, respectively) and four between Pusa Sawani and A. moschatus variant (EC306750). In all the above crosses, Pusa Sawani was the female parent. Of them, the plants of the amphidiploid derivatives with A. pungens var. mizoramensis as male parent exhibited similar morphology. The stems, petioles, and fruits were highly hispid, and a reddish color was observed on the stems and petioles. Further, the flower buds of C1Pusamizo236 and C1Pusamizo235 derivatives were clustered at the top, resembling the wild parent A. pungens var. mizoramensis (Figures 5A–C). The fruit length and width ranged from 6.13 cm (C1Pusamizo236-26) to 8.83 cm (C1Arkamosc (variant)-750-04) and 2.03 cm (C1Pusamizo236-02) to 2.93 cm (C1Pusamizo236-18), respectively (Table 4A). The epicalyces were medium broad (0.69 cm) and became withered/detached from the fruit at maturity. The majority of them possessed spiny hairs on the fruits. However, the fruits of C1Pusamizo236-14, C1Pusamizo236-02, C1Pusamizo236-30, C1Pusamizo235-09, C1Pusamizo235-02, and C1Pusamizo23-06 were characterized by soft spiny hairs (Supplementary Table 1A). As there was a wide range in the number of seeds per fruit among the C1Pusamizo derivatives (8.50 to 98.25), the coefficient of variation (CV) was high (49.90%) compared to other characteristics.
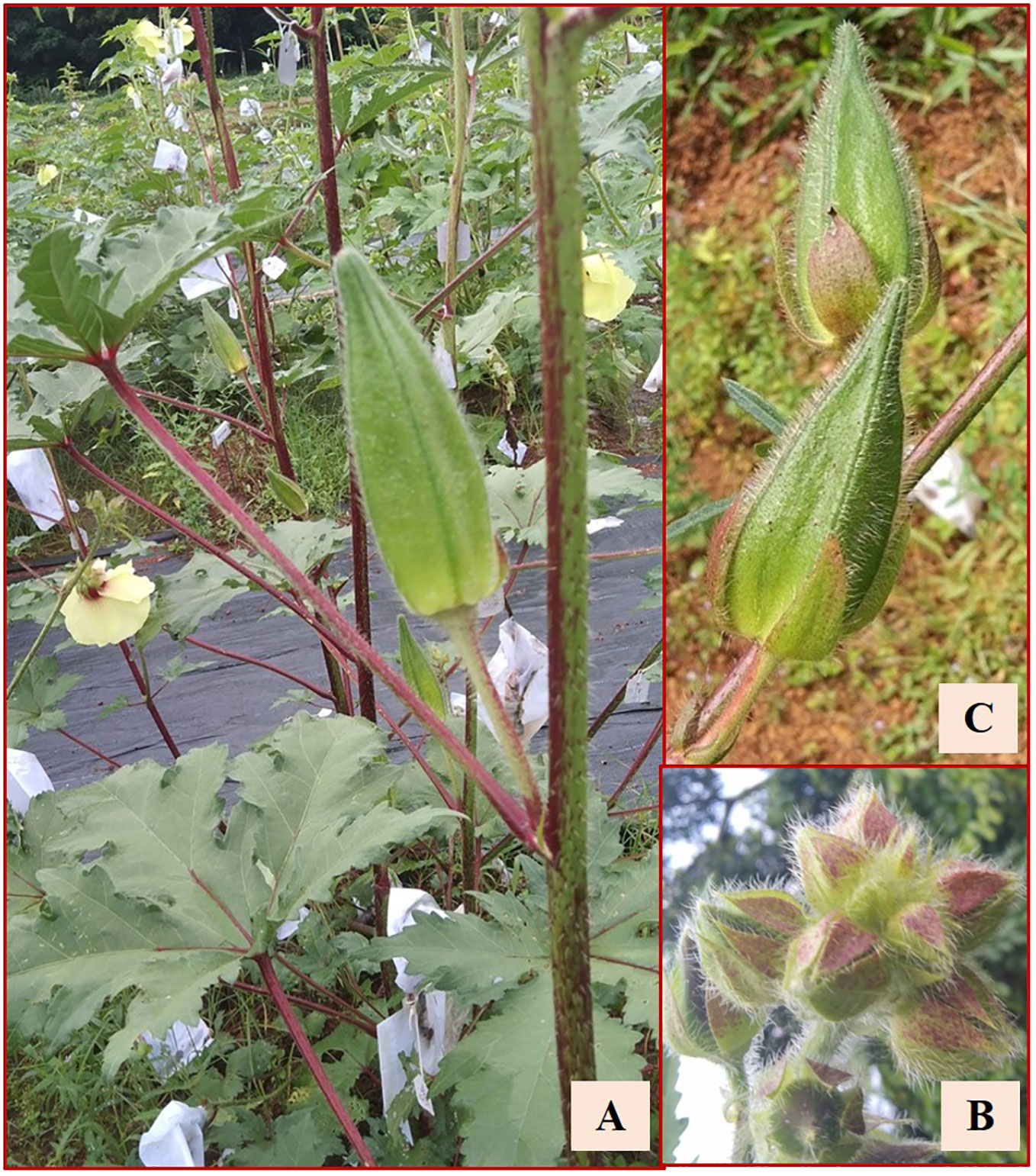
Figure 5 (A) Reddish coloration on stems and petioles. (B) Clustered flower buds in C1Pusamizo derivatives. (C) Hispid fruits.
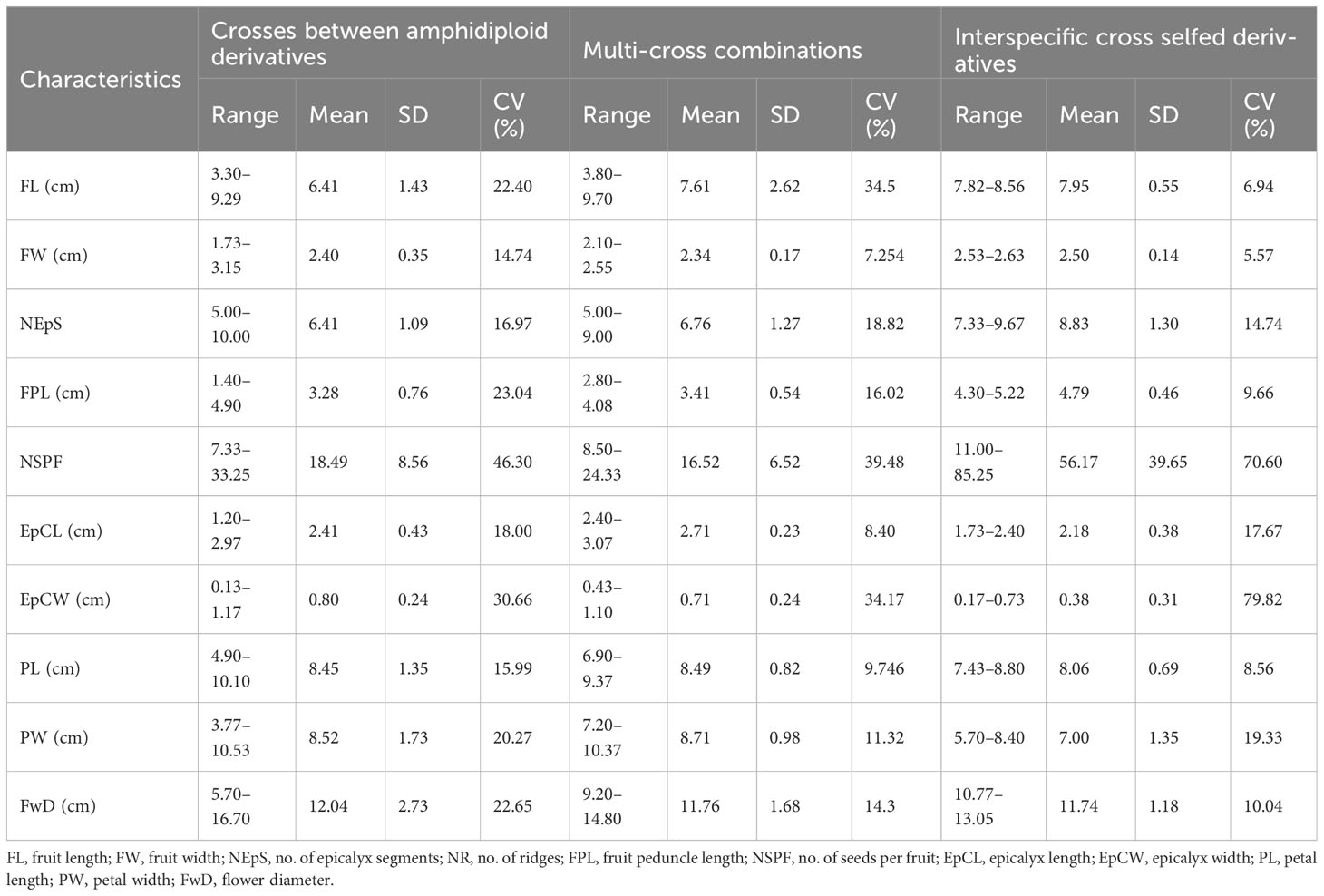
Table 4C Summary statistics of the crosses between amphidiploid derivatives, multi-cross combinations, and interspecific cross selfed derivatives.
Out of the remaining 16 amphidiploids, 14 were in the F3, and there was one each in the F2 and F4 generations. Among them, seven were progenies selected from the amphidiploid C3/50/mizo30 and three from C3/106/mizo30. Further, one derivative each from C3/50/mizo24, C4/50/mizo24, C3/50/mizo34, C3/87/gran4, C3/817/gran, and C2Ruchitetra (Table 2B) were also included. The A. esculentus genotype used in 10 amphidiploids was IC265650 (C3/50/mizo30 derivatives) and IC260106 in three (C3/106/mizo30) derivatives. IC412987 was the A. esculentus genotype in C3/87/gran4 and Ruchi in C2Ruchitetra1. An erect tall growth habit with basal branching, reddish streaks on the stems, and profuse growth were common characteristics. They also exhibited extended life span, prolific flowering and fruit set, and medium-long spiny ovoid elongate fruits. The fruits of AM-8 were reddish with spiny hairs (Figure 6A), which possessed the maximum length (8.96 cm) among the selfed derivatives. The fruit width was maximum in AM-23 (C3/87/gran4), measuring a mean value of 2.80 cm (Figure 6B) (Supplementary Table 1B). However, the young fruit surface was soft with a spongy nature in AM-3, a segregant of C3/106/mizo30 (Figure 6C). C2/Ruchi/tetra1 exhibited an intermediate fruit length (7.25 cm) with a fusiform shape (Figure 6D). AM-25 (C3/817/gran) was the only derivative with A. caillei as the female parent. The plants of this derivative exhibited green stems, red color at the nodes, broad epicalyces (0.77 cm), and fruits with a mean length of 6.23 cm. The number of seeds per fruit ranged from 9 (AM-23) to 53 (AM-3). Seed shattering was also observed in these amphidiploids. General variability in fruit characteristics among selected selfed derivatives is depicted in Figure 6E.
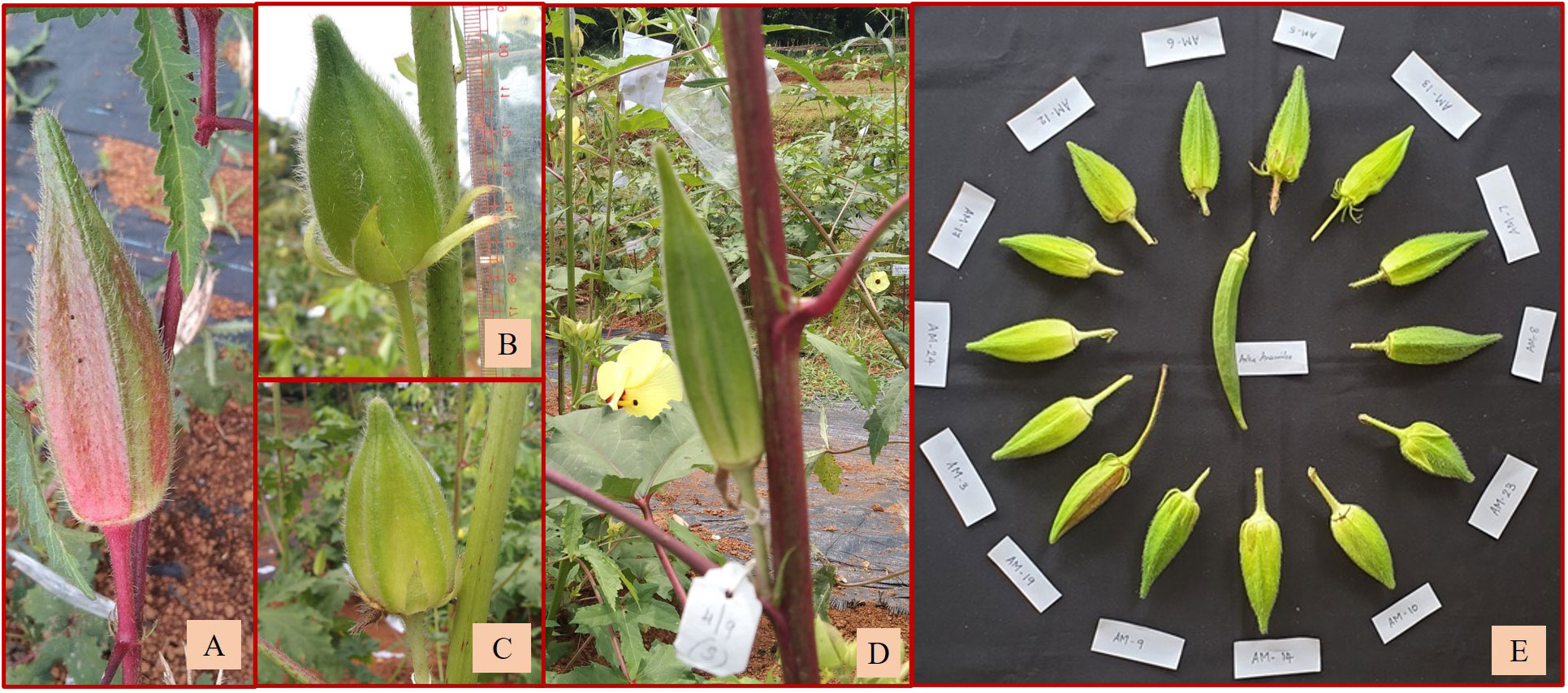
Figure 6 Fruit variability in selfed derivatives of F2, F3, and F4 generation. (A) Reddish fruits of AM-8. (B) Short fruit of AM-23. (C) Soft and spongy fruit of AM-3. (D) Fruit of C2/Ruchi/tetra1 with intermediate fruit length. (E) General variability in fruits of selfed derivatives.
Backcrossed derivatives
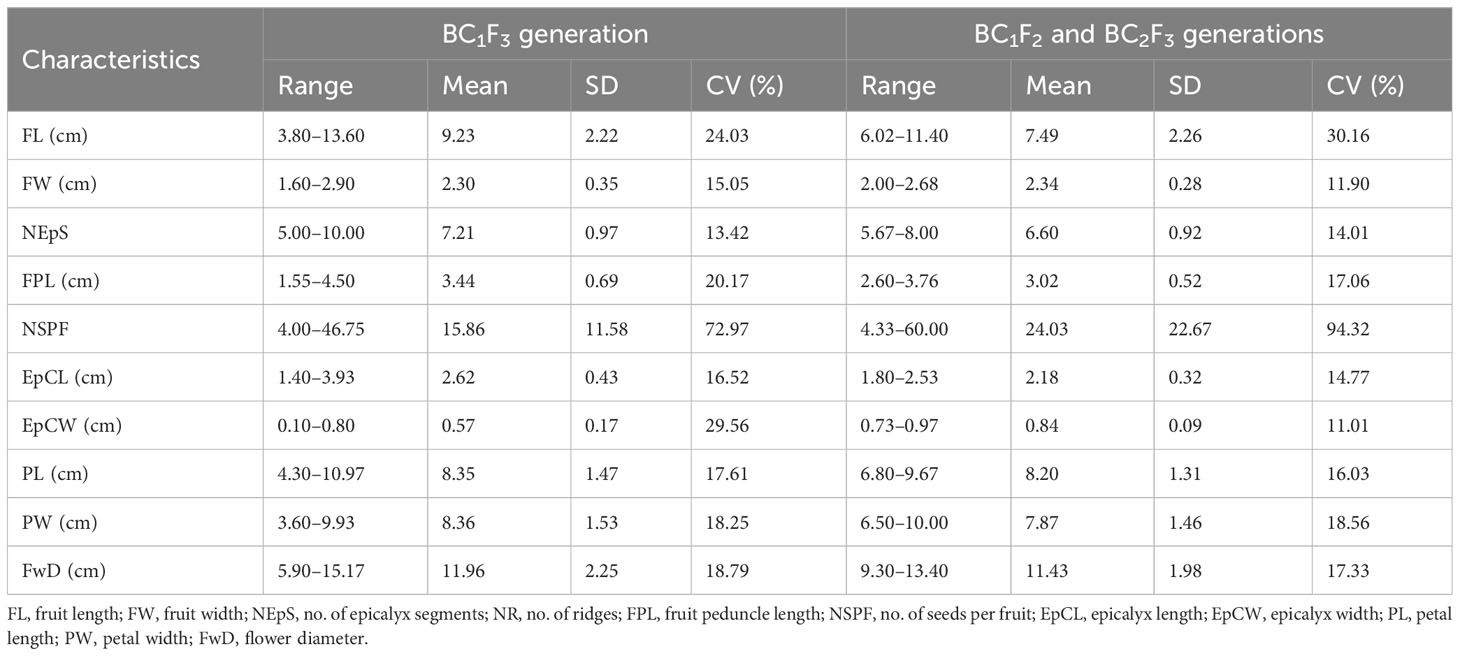
Twenty-five backcrossed derivatives that developed (BC1F3) also exhibited variability with respect to the growth habit, branching behavior, density of indumentum, and leaf and fruit characteristics. The number of seeds per fruit was the highly variable characteristic indicated by the high CV of 72.97% (Table 4B; Supplementary Table 1C). Fruit length ranged from 3.80 cm (short fruit length resembling wild types) to 13.60 cm (resembling cultivated types) with a mean of 9.23 cm (Table 4B). The plants of C3/50/mizo34 × Parbhani Kranti had purplish stems, petioles with red dorsal and green ventral sides, intermediate epicalyces (0.50 cm), and medium-long fruits (10.75 cm) (Figure 7A). Additionally, they also possessed mildly lobed leaves at the bottom and deeply lobed at the top, and hispid stems and leaves. However, in C3/50/mizo34 × Salkeerthi, the plants exhibited segregation with respect to stem color (purplish green and purple stems) (Figure 7B). They also exhibited hispid stems, petioles, and fruits; intermediate epicalyx width (0.57 cm); and prominent basal branching. Nonetheless, tall plants with green stems having light reddish patches and the absence of basal branching were the characteristic features of C3/50/mizo1 × IC31398A. It produced medium-long fruits (7.70 cm) (Figure 7C) with soft hairs. Reddish purple patches on the stems and petioles, hispid stems, petioles and leaves, and red epicalyces were observed in C3/50/mizo34 × South Canara Local and C3/50/mizo34 × South Canara Local. A similar kind of reddish coloration on the stems, petioles, and epicalyces was observed in plants of C3/50/mizo34 × Arka Anamika. C3/741/mizo9 × EC169415, C3/50/mizo27 × Arka Anamika, and C3/106 mizo6(4) × IC22232 also exhibited wild characteristics like basal branching and late flowering. In contrast to the above, the fruits of C3/50/mizo27 × IC22232 were velvety (Figure 7D), with wide epicalyces (0.80 cm), even though IC22232 and IC265650 (accessions involved in the cross) belong to the A. esculentus (with linear epicalyx) taxon.
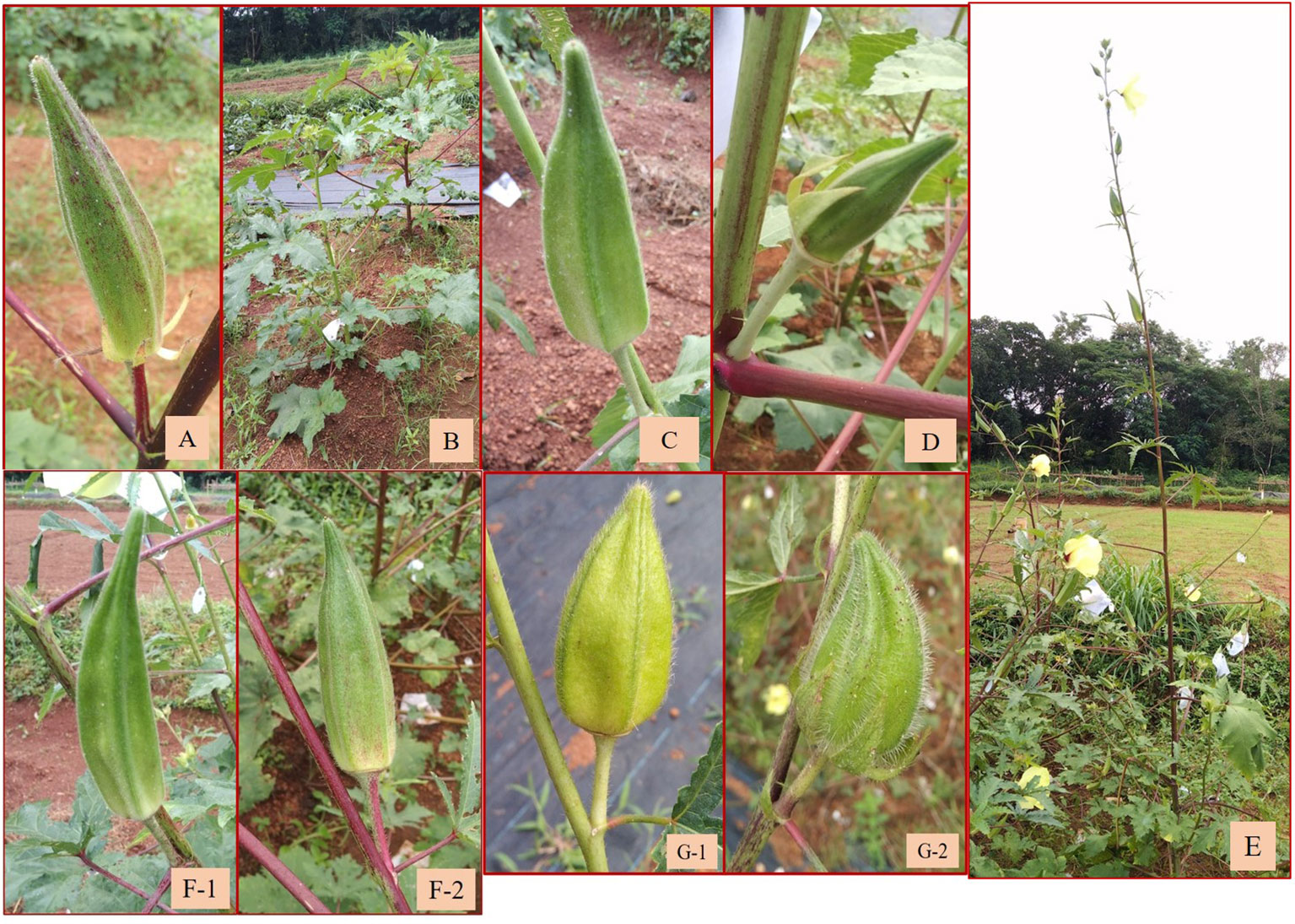
Figure 7 (A) Fruit of C3/50/mizo34 × Parbhani Kranti. (B) Segregation in stem color in C3/50/mizo34 × Salkeerthi. (C) Medium-long fruits of C3/50/mizo1 × IC31398A. (D) Velvety fruits of C3/50/mizo34 × IC22232. (E) Tall growth habit of C3/106/mizo6 × Hissar Unnat. (F-1, F-2) Fruits of C3/106/mizo6 × Kashi Vibhuti and C3/106/mizo6 × Hissar Unnat with low trichome density. (G-1, G-2) Short fruits of B16 × Arka Anamika and B13 × Arka Anamika.
Similarly, a robust tall growth habit, green stem, reddish spots and streaks along the stems, and reddish color at the nodes were observed among the plants of the derivatives C3/87/gran2 × EC169415. Epicalyx was broader (0.60 cm) and non-persistent; fruits were longer than those of the other amphidiploids (12.33 cm). It also exhibited mildly lobed leaves at the base and deeply lobed toward the top. C3/741/gran12 × IC31398A also had stems, petioles, fruits, and leaves covered with thick spiny hairs. Leaf lobing was like the former one, with red coloration at the node, with an average of 19 seeds per fruit; however, epicalyx persisted up to a certain maturity of the fruit. Branching extended up to half the height of the plants. The plants of C3/106/mizo6 × Hissar Unnat were also very tall with basal branching (Figure 7E), reddish stems and petioles, and prominent spiny hairs on them. The tall nature, robust growth, and low trichome density in fruits (Figures 7F-1, F-2) were common in all derivatives involving C3/106/mizo6. The fruit hairs were soft in B-16 × Arka Anamika and B-13 × Arka Anamika (BC1F2) (Figures 7G-1, G-2). C3/50/mizo1 × Kashi Lalima exhibited reddish coloration on the stems, epicalyces, flower buds, and petioles but with green hispid fruits and medium broad epicalyces (Figures 8A–C). The plants of derivative C3/87/gran2 × Arka Anamika were an intermediate type with a semi-dwarf habit but with highly spiny and reddish stems, basal branching, medium-long fruits (8.53 cm), and medium broad epicalyces (0.43 cm). However, the fruit surface was soft and hairy. Non-spiny fruit surface was observed in four amphidiploids (Ruchi × AM-24, Ruchi × AM-6, C3/50/mizo27 × IC22232, and C3/50/mizo27 × IC31398A). The fruits of the first two resembled the morphology of cultivated okra having mean fruit length of 12.55 and 13.60 cm and with slender linear epicalyces. The number of seeds per fruit was 40 in Ruchi × AM-24 and 24 in Ruchi × AM-6.
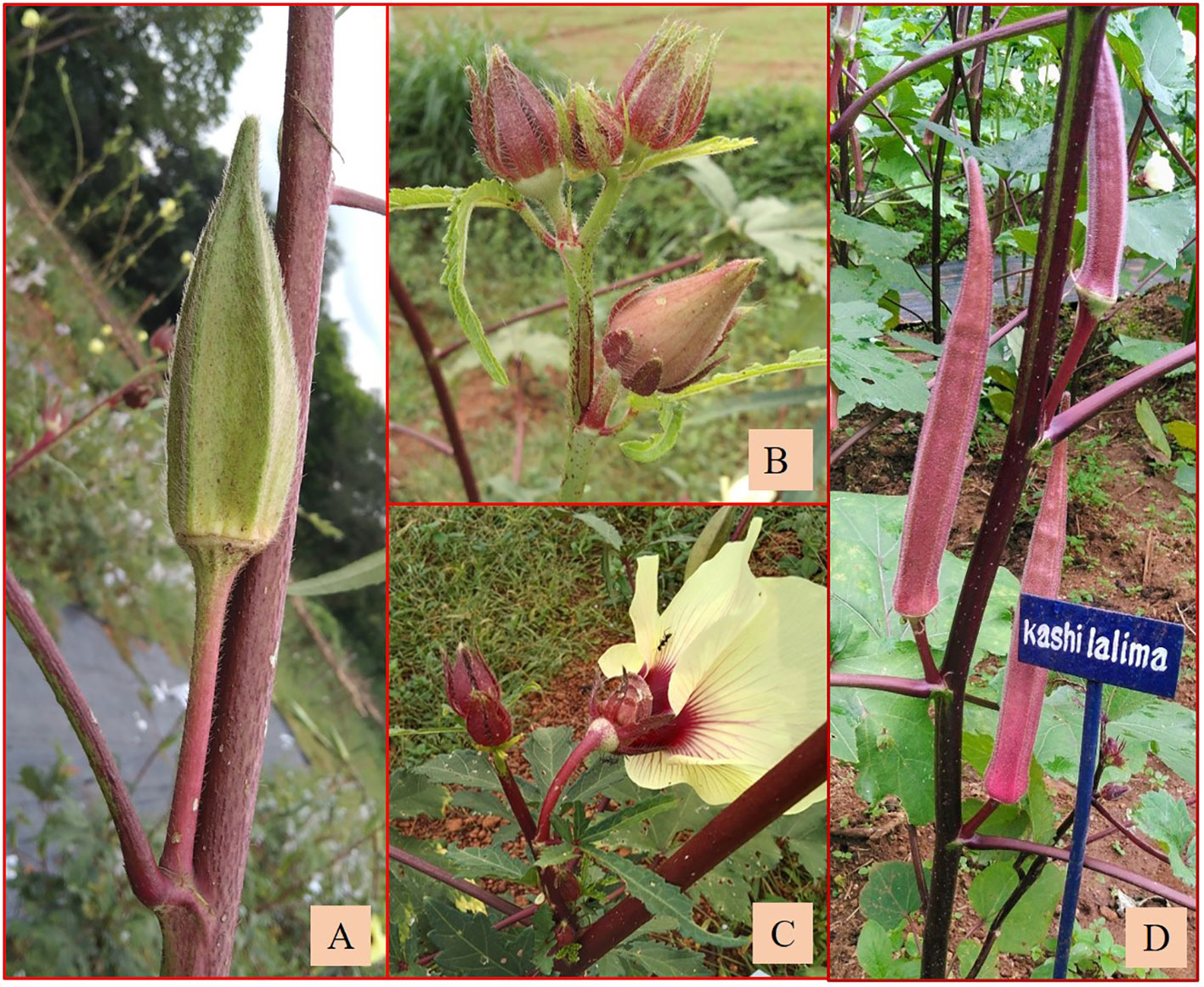
Figure 8 (A) Fruit. (B) Flower bud with epicalyx. (C) Flower base of C3/50/mizo1 × A.esculentus cv. Kashi Lalima. (D) Kashi Lalima.
Among the derivatives in the BC1F2 and BC2F3 generations, Ruchi × AMPK-1, Ruchi × AM25, B-13 × Arka Anamika, and B-16 × Arka Anamika were wild types with short to medium fruits (6.02 cm–7.46 cm) (Supplementary Table 1D) with the presence of soft to nil hairs. However, in B-16 × Arka Anamika, fewer branching was a distinct characteristic. Arka Anamika × AMPK-1 (BC2F3) exhibited morphology of cultivated A. esculentus with long fruits (11.40 cm) devoid of spiny hairs on the fruit, with the number of seeds as high as 60 per fruit. Narrow variability was exhibited among the derivatives for epicalyx width (0.73 cm–0.97 cm) and fruit width (2.00–2.68 cm) as demonstrated by the narrow range and low CV of 11.01% and 11.90%, respectively (Table 4B). However, the range of seed setting in fruits was wide (4.33–60.00). Figures 9A–C depict the backcross derivatives with cultivated okra-type fruits.
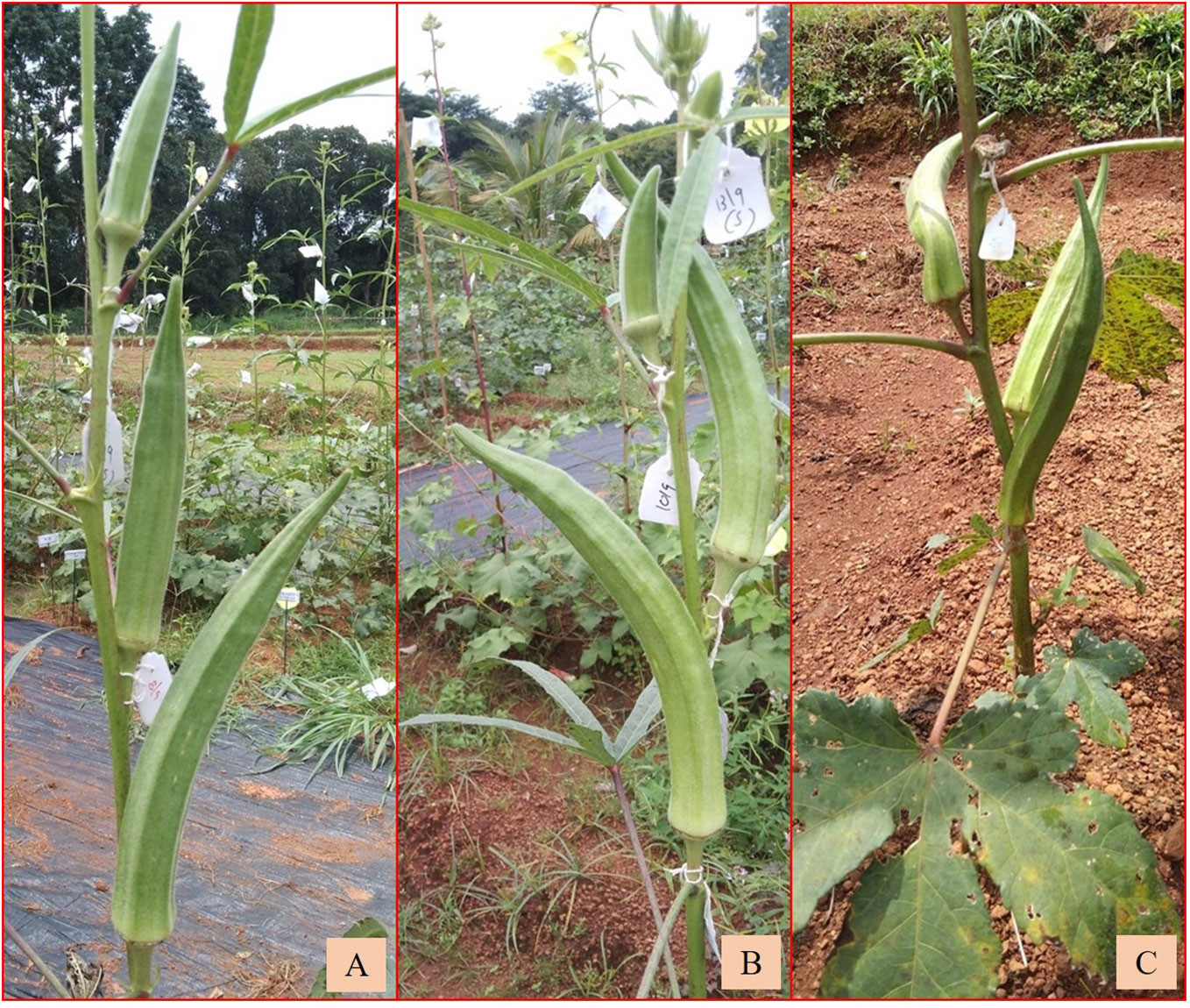
Figure 9 Backcrossed derivatives with cultivated okra morphology (A) Ruchi × AM-6. (B) Ruchi × AM-24. (C) Arka Anamika × AMPK-1.
Cross between amphidiploids
Twenty-seven cross derivatives developed by crossing between amphidiploids had also exhibited variation, as revealed by the wide range in characteristics like fruit length (3.30–9.29 cm), number of epicalyx segments (5.00–10.00), number of seeds per fruit (7.33–33.25), petal length (4.90–10.10 cm), petal width (3.77–10.53 cm), and flower diameter (5.70–16.70 cm) (Table 4C). B-13 × AM-24, a derivative of cross involving A. esculentus, A. angulosus var. grandiflorus, A. tetraphyllus, and A. pungens var. mizoramensis (Table 2D), exhibited maximum values for fruit length (9.29 cm) and number of epicalyx segments (10.0). The minimum values for fruit width (1.73 cm), epicalyx length (1.20 cm), epicalyx width (0.13 cm), petal length (3.77 cm), petal width (3.77 cm), flower diameter (5.70 cm), and the absence of spiny hairs on the fruit surface were also shown by this derivative (Supplementary Table 1E). The plants of B-4 × AM-24 and B9 × AM-25 were vigorous, with profuse branching and fruiting. However, B-9 × AM-23 plants were distinct, with comparatively small flowers (diameter 7.50 cm), broad epicalyces, and broadly lobed leaves and without spiny hairs on the plant parts. Thirteen derivatives possessed either red color throughout or red spots or red striations on the stems. Red coloration at the nodal region was also a frequently observed characteristic in many derivatives.
AM-24 × B-4 was a cross between two amphidiploids in which B4 was a progeny of a three-way cross involving Arka Anamika, A. tetraphyllus (IC253122), and A. palianus (IC0624218). The fruits had an average length of 5.60 cm, a width of 2.70 cm, and six epicalyces measuring 2.76 cm × 0.83 cm, and soft trichomes. The increase in the length of fruits was evident in the derivatives, namely, B-16 × AM-24, B-15 × AM-25, AM-25 × CR-1, and B-13 × AM-24 (Figures 10A–D). AM-25 × CR-1 (A. esculentus IC260106 × A. sagittifolius IC470750)]) was the only cross involving A. sagittifolius in this category; however, none of the distinct characteristics of A. sagittifolius (pink flowers, slender epicalyces, soft fruits, and long peduncles) were observed.

Figure 10 Increase in fruit length in derivatives of cross between amphidiploids (A) B-15 × AM-25, (B) AM-25 × CR-1, (C) B-13 × AM-24, and (D) B-16 × AM-24.
Multi-cross combinations
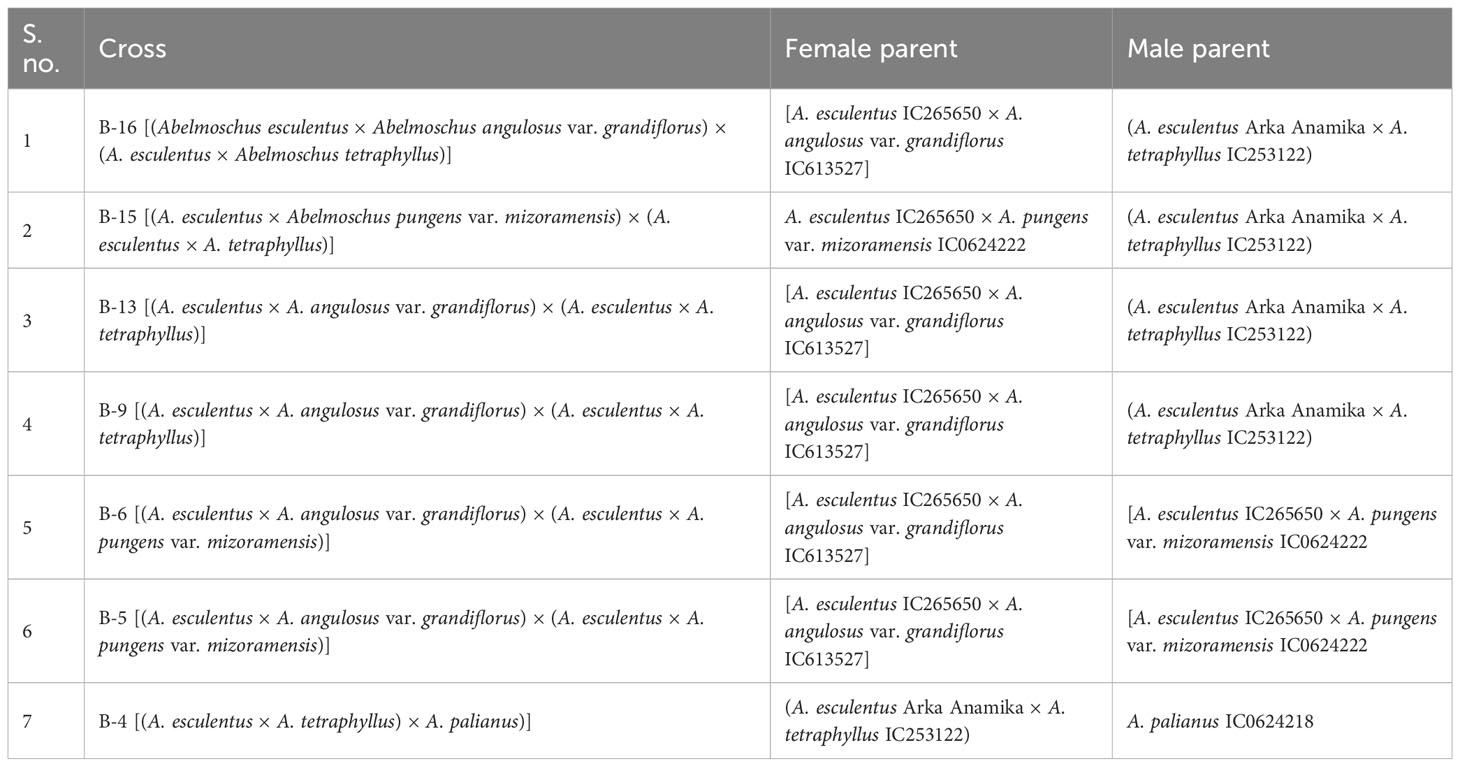
All the derivatives of multi-cross combinations had medium-wide epicalyces to wide epicalyces (range 0.43–1.10 cm) with medium spiny hairs on the fruit surface. A wide range was observed in fruit length (3.80–9.70 cm), in contrast to a narrow range in fruit width (2.10–2.55 cm). The number of seeds per fruit exhibited maximum CV (39.48%), and the values ranged from 8.50 (B-9) to 24.33 (B-4). The fruits of B-5 had on average nine epicalyx segments per fruit and fruit length of 9.04 cm. The number of epicalyx segments in the derivatives under this category ranged from 5.0 (B-15) to 9.0 (B-5). However, the fruits of B-15 were short (3.80 cm), with only five epicalyx segments with length and width ranging from 2.90 cm and 0.90 cm, respectively. Fruit peduncle length was maximum (4.08 cm) in B-5 (Table 4C; Supplementary Table 1F). B-9 exhibited maximum values for flower diameter (14.80 cm); however, B-5 and B-13 exhibited maximum values for petal length and width, respectively.
Interspecific cross selfed derivatives
Three interspecific cross selfed derivatives of the cross, i.e., A. sagittifolius × A. moschatus subsp. moschatus, advanced to the F8 generation (Table 4C; Supplementary Table 1G) were also included in the study. Two of them (AM-13 and AM-19) exhibited the characteristics of A. moschatus having fruits with slender linear epicalyces, soft to no hairs, and oblong capsules with acute apex and bright yellow flowers (Figures 11A–C). The fruit length and width ranged from 7.82 to 8.56 cm and 2.53 to 2.63 cm, respectively. Similar to A. moschatus, the number of epicalyx segments varied from 7.33 to 9.67 with a mean of 8.83 per fruit. The length and width of epicalyx segments were 2.18 cm and 0.38 cm, respectively. In contrast to basal branching, branching throughout the nodes was a peculiar characteristic observed in these derivatives. The flower diameter ranged from 10.77 to 13.05 cm with a mean of 11.74 cm (Table 4C). AM-13 did not exhibit spiny hairs on the fruits, and the number of seeds also varied significantly from the other two derivatives (Supplementary Table 1G). The fruit peduncle length ranged from 4.30 cm to 5.22 cm with an average of 4.79 cm in the derivatives.
Discussion
A promising breeding method for the creation of new genetic variability in a crop species is wide hybridization, which became a routine practice in crops such as wheat after the advancement of hybridization techniques (Mujeeb-Kazi and Rajaram, 2002). The dynamics of geneflow existing between the cultivated and wild species of Abelmoschus is less studied as compared to other vegetable crops. Earlier studies revealed that “it is more difficult to cross cultivated species with wild species of Abelmoschus” as evident from several attempts made for interspecific hybridization by various authors (Joshi and Hardas, 1956; Kuwada, 1957; Hamon and Yapo, 1985). Wild relatives of okra represent a good source of variation for breeding programs, particularly for traits related to biotic and abiotic stresses, and fruit quality (Sandeep et al., 2022). Hence, wild species that remain unexploited may be used via pre-breeding approaches for developing desirable introgressed lines.
In the present investigation, interspecific crosses involving A. esculentus and wild relatives, followed by polyploidization, generated considerable variability in the amphidiploid derivatives. These results reaffirmed the earlier reports on the successful interspecific hybridization to generate considerable variability (Reddy, 2010; Arunkumar, 2015; Patel et al., 2021; Badiger et al., 2023; Singh et al., 2023b) and the existence of barriers to the transmission of gene flow from wild species to the cultivated ones (Rajamony et al., 2006; Jatkar et al., 2007; Badiger et al., 2023). Widely variable amphidiploids have also been generated from the hybrids of wheat and related species of the genera Aegilops, Secale, and Thinopyrum by Nemeth et al. (2015). Rapid phenotypic changes in synthetic allopolyploids in Brassica napus have also been reported by several workers, which were attributed to mechanisms such as recombination between homoeologous chromosomes and alterations in the expression pattern and DNA methylation status of genes (Song et al., 1995; Schranz and Osborn, 2000; Lukens et al., 2006; Gaeta et al., 2007; Xu et al., 2009; Szadkowski et al., 2010). However, in contrast to this, Sheela (1994) reported a strong genetic mechanism preventing free recombination in the interspecific hybrid population of Abelmoschus.
In the present study, the majority of the okra amphidiploids exhibited intermediate morphology, especially with respect to the fruit characteristics, which is in conjunction with the observations in amphidiploids developed between Cucumis sativus L. and Cucumis hystrix Chakrav. (Chen et al., 2002) and Solanum melongena L. and Solanum aculeatissimum Jacq. (Zhou et al., 2018), where the synthetic amphidiploid plants possessed some characteristics common either to cultivated or to the wild parents. Further, the greater vigor of the amphidiploids may be attributed to the heterozygosity of different genomes (Nasrallah et al., 2000; Chen, 2010; Fujimoto et al., 2011). Badiger et al. (2023) observed that the interspecific hybrids developed between A. esculentus and wild species (A. manihot var. tetraphyllus and A. moschatus) showed intermediate morphological traits with greater alliance toward their wild parents. In the present study, distinct variation was also apparent in the indumentum density among the amphidiploid derivatives. The purple coloration on the stems, petioles, and nodal region in the majority of the amphidiploids was inherited from the wild parent A. pungens var. mizoramensis. This taxon as described originally by John et al. (2020) exhibits dark-purple blotches at nodes, and these blotches extend to the internodes. It was interesting to note that the recovery and survival rate of the amphidiploids with A. pungens var. mizoramensis were high compared to those of other combinations (A. esculentus with other wild species) after colchiploidization, reaffirming the findings of Rana and Arora (1991) that there is a genetic affinity between A. esculentus and A. pungens.
Selfed amphidiploid derivatives
Wild characteristics, namely, shorter fruit length, hispid nature, and seed dehiscence from the apex of the fruit, were prominent in the amphidiploids in the F1 generation; however, profuse and extended bearing were observed. As linkage drag was prominent, they required repeated backcrossing to accumulate the desirable genome of the recurrent parent. The similarity in morphology among the C1Pusamizo derivatives may be attributed to the heterozygosity in all the F1 individuals leading to a uniform homogenous population. The increase in fruit length was evident (range of fruit length 6.13–8.83 cm) (Table 4A) against the range of fruit length in A. pungens var. mizoramensis (3.5–5.0 cm) (Table 3). IC624235 and IC624236 (both collections from Nagaland, India) were the A. pungens var. mizoramensis genotypes used. Among them, a higher seed setting (more than 45 seeds) was obtained in amphidiploids involving IC624235, except in C1Pusamizo235-03. This reveals that there is genotype-specific cross compatibility between A. esculentus and A. pungens var. mizoramensis. The persistence of epicalyces at fruit maturity was not observed in the derivatives in contrast to the persistent epicalyces in the wild parent (John et al., 2020). Regarding the derivatives involving A. esculentus and A. moschatus variant, the seed setting ranged from 22.75 to 98.25, indicating that the seed setting was possible by using A. esculentus as a female parent. Badiger et al. (2023) also confirmed that viable seeds were obtained in the direct cross. They reported that A. moschatus × A. esculentus crosses (reciprocal cross) suffered from severe pre-zygotic barriers that resulted in the formation of aborted embryos with partially filled seeds. C1Arkamosc (variant)750-02 produced fruits like A. moschatus, confirming the dominance of genes from the wild parent. As A. moschatus, a perennial species, is reported to be tolerant to other multiple biotic stresses such as powdery mildew, jassids, whitefly, fruit, and shoot borer (Badiger and Yadav, 2019; Yadav et al., 2019), it can be utilized in multiple pest and disease resistance breeding (Badiger et al., 2023). The perennation tendency and profuse-bearing habit of amphidiploids can also be exploited for selecting segregants to be used as a home garden vegetable, as a single plant in the kitchen garden can suffice the okra requirement of a family throughout the year. For commercial cultivation, genotypes bearing during the off-season will also fetch higher prices for the farmers.
The amphidiploids in the F2, F3, and F4 generations also exhibited wide variations with respect to the number of seeds per fruit. The seed set was even as low as nine seeds per fruit in AM-23, a cross between A. esculentus and A. angulosus var. grandiflorus. This low seed set might be owing to genetic imbalances due to improper bivalent formations during meiosis because of genomic differences in parents (A. angulosus var. grandiflorus as 2n = 66; A. esculentus as 2n = 130) (Merita et al., 2012). The epicalyx width, one of the important taxonomic characteristics, was maximum in the amphidiploid AM-23 (C3/87/gran4) (1.60 cm) (Supplementary Table 1B), indicating that the trait might have been transferred from the male parent IC613527 (A. angulosus var. grandiflorus), which has comparable epicalyx width ranging from 1.30 to 1.50 cm in general (Table 3) (Yadav et al., 2014). C2/Ruchi/tetra1 had plant morphology similar to that of A. tetraphyllus, with purplish stems, very vigorous growth, and orthotropic basal branching. It developed fruits with an average length of 7.25 cm and without prickly spines (the only amphidiploid without spiny hairs among the advanced selfed derivatives). This contrasted with the fruit length in A. tetraphyllus, which ranges from 3.50 to 5.50 cm (Pandey et al., 2016), indicating the transfer of useful genes from cultivar Ruchi. Similar kinds of naturally introgressed lines (with enhanced fruit length and fusiform shape) can be observed when A. tetraphyllus and A. esculentus are grown in the adjacent or same fields in the previous season. AM-4, a derivative selected from the cross between A. esculentus IC265650 and A. pungens var. mizoramensis IC0624222, was observed to have a maximum value for flower diameter (13.93 cm), which exceeded the mean flower diameter of both the parents.
All the derivatives produced showed yellow flowers with extended flowering periods, giving scope for identifying segregants with ornamental qualities with perennial nature. We also identified novel types with profuse flowering, suitable for deriving ornamental types in the selections among C1Pusamizo14, C1Pusamizo02, C1Pusamizo30, C1Pusamizo09, and C1Pusamizo06, which were less hispid too. However, horticultural practices like pruning need to be standardized to maintain them as a bushy plant. Cui et al. (2018) reported the prospects of the development of perennial wheat through hybridization between wheat and wheatgrasses (Thinopyrum spp.). Similar valuable trait-specific amphidiploids of Triticum turgidum subsp. durum–Aegilops longissima with high iron and zinc contents (Tiwari et al., 2008) and T. turgidum–Aegilops umbellulata with strong tillering ability, stripe rust resistance, and seed size-related traits (Song et al., 2019) have also been reported. Similarly in vegetables, the hull-less seed trait was transferred from Cucurbita pepo to Cucurbita moschata through interspecific hybridization (Kaur et al., 2023).
Backcrossed derivatives
The traits from wild species exhibit dominance; accordingly, the amphidiploids developed possessed a high hispid nature of the plant parts. To recover the traits of a cultivated parent, backcrossing was initiated in the F2/F3 stage. Irrespective of the recurrent parent used in backcrossing, our focus was on the selection of segregants with intermediate to long fruits with less spiny hairs on the fruit surface, extended and profuse bearing, and a greater number of seeds per fruit. Wide variability was observed among the BC derivatives with respect to the fruit morphology varying from true wild types to the cultivated types, in addition to the intermediate types. Similar results of morphological divergence among backcrossed progenies developed from the interspecific hybrids of Brassica oleracea var. alboglabra × Brassica rapa L. var. purpurea were reported by Zhang et al. (2016). The increase in fruit length indicates the gene flow from the cultivated types. Basal branching observed among the derivatives may be a dominant characteristic, as the majority of the derivatives exhibited this characteristic. The segregants without basal branching may be selected in the advanced generations, as this characteristic impedes easy harvesting and reduces the number of plants that can be accommodated in unit areas during commercial cultivation. The characteristic reddish spots and patches along the stems and petioles and tall nature in derivatives with A. pungens var. mizoramensis as one of the parents indicated that these characteristics might have transferred from this wild parent. A. pungens var. mizoramensis exhibit purplish spots on the younger stems and attain height up to c. 3 m as reported by Yadav et al. (2014) and John et al. (2020). Similarly, the characteristics, namely, the persistence of epicalyces at fruit maturity and red coloration at the nodal region in C3/741/gran12 × IC31398A, might have been inherited from the wild parent, A. angulosus var. grandiflorus, which is reported to have these characteristics (Yadav et al., 2014). C3/50/mizo34 × IC31340C and C3/50/mizo27 × IC31340C, the derivatives from the same male parent, had similar morphology. The plants shared characteristics like the presence of reddish spots on the stems, medium-wide epicalyces, and soft to spiny hairs on the stems, leaves, petioles, and fruits. It was observed that seed setting was less than 10 per fruit in 12 backcrossed derivatives (Supplementary Table 1C), which may be attributed to the genotype differentiation in crossability behavior.
Among the BC1F3s, Ruchi × AM-24 and Ruchi × AM-6 were the desirable segregants with long fusiform fruits with as high as 60 seeds per fruit. Arka Anamika × AMPK-1, a derivative in the BC2F3 generation, also exhibited the morphology of cultivated A. esculentus (Figures 9A–C). However, more backcrossing will be needed to re-constitute the genome of cultivated okra. The flow of genes from the cultivated to the amphidiploids was apparent in the BC derivative involving Kashi Lalima also. Kashi Lalima is a variety of okra released from ICAR-Indian Institute of Vegetable Research, Varanasi, Uttar Pradesh, India, bearing reddish-purple fruits rich in anthocyanin and phenolics and red-colored stems, petioles, and leaves (Figure 8D) (https://iivr.icar.gov.in/kashi-lalima-vror-157 accessed on July 12, 2023). C3/50/mizo1 × Kashi Lalima exhibited reddish-colored stems, epicalyces, and petioles, which might have been inherited from the cultivated variety. However, it produced green hispid fruits (8.10 × 1.80 cm) with medium broad epicalyces (0.35 cm) inherited from the wild parent (Figures 8A–D). The desirable backcrossed derivatives may be screened at hot spot locations of YVMV and ELCV diseases, and the tolerant/resistant lines may be included in the breeding programs. Arunkumar (2015) and Singh et al. (2023b) identified resistant backcross derivatives of the cross between A. esculentus and A. caillei and A. esculentus and A. manihot, respectively.
Crosses between amphidiploids and multi-cross combinations
As a penultimate objective, the amphidiploids were crossed among themselves to induce maximum recombination between the different genomes so that there is a greater probability of obtaining desirable segregants. This was supported by the views of Favero et al. (2020) in Arachis sp. that there is a possibility to obtain complex hybrid derivatives generated by crossing between the amphidiploids with high crossability rates. In the present investigation, all the derivatives developed by crossing between amphidiploids yielded plants with prominent basal branching with broader mildly lobed leaves at the bottom and deeply lobed ones at the top. This is in concurrence with the report of Sutar et al. (2013) regarding the presence of angular or palmatifid lower leaves and palmisect upper leaves in A. palianus, palmatifid lower leaves in A. angulosus Wall. ex Wight et Arn. var. mahendragiriensis R.C.Misra var. nov. (Misra et al., 2018), and broadly orbicular lower leaves in A. pungens var. mizoramensis (John et al., 2020).
B-16 × AM-24 was one of the desirable segregants, with medium-long fruits (8.88 cm), closer to the cultivated type, though with some wild-type morphology. The average fruit length of the wild species used in the above cross, viz., A. tetraphyllus, A. angulosus var. grandiflorus, and A. pungens var. mizoramensis, was 3.50 to 4.50 cm, 3.00 to 4.00 cm, and 3.50 to 5.00 cm, respectively (Table 3). Hence, the increase in fruit length can be attributed to the transfer of genes conferring fruit length from the A. esculentus genome. This trend also manifested in B-15 × AM-25 (8.53 cm), AM-25 × CR-1 (8.37 cm), and B-13 × AM-24 (9.29 cm). However, this increase was not exhibited in the characteristic number of seeds per fruit, which ranged from 7.33 to 33.25 with an average of 18.50 seeds (Table 4C). B-16 × AM-23, a derivative of cross involving A. esculentus, A. angulosus var. grandiflorus, and A. tetraphyllus, possessed the largest flower among all the derivatives with an average flower diameter of 16.70 cm. Nonetheless, the flower diameter was minimum (5.70 cm) in B-13 × AM-24, a smaller value than any of its parents. The flower diameter of the parental species ranged from 6.00 to 7.00 cm in A. esculentus and A. angulosus var. grandiflorus, 7.00 to 8.00 cm in A. pungens var. mizoramensis, and 5.00 to 7.00 cm in A. tetraphyllus (Table 3). Except for B-9 × AM-25 and B-15 × AM-25, all the derivatives had A. pungens var. mizoramensis as one of their parents. B-9 × AM-25 recorded the maximum value for the width of epicalyx segments among the crosses between amphidiploids. Seven derivatives, namely, B-16 × AM-24, B-15 × AM-25, B-13 × AM-24, B-16 × AM-23, B-2 × AM-25, B-9 × AM-23, and B-4 × Was-15, were devoid of spiny trichomes on their fruit.
All the multi-cross combinations had medium spiny fruits (Supplementary Table 1F). Three derivatives, namely, B-5, B-6, and B-9, had intermediate fruit characteristics, indicating the probability of increasing fruit length through backcrossing. The results are encouraging considering that simultaneous introgression of genes from four wild Arachis species into peanuts was possible through the development of complex hybrids by crossing between amphidiploids of Arachis spp. (Favero et al., 2020). Further, the derivatives from these complex multi-crosses can be used for conducting basic studies to understand the genetic mechanisms of virus tolerance in Abelmoschus. Such derivatives can help in the identification of alternate genes for tolerance in okra.
Interspecific cross selfed derivatives
In addition to searching germplasm for virus tolerance, interspecific crosses were also made to investigate the prospects of obtaining other desirable traits from the wild germplasm. A. sagittifolius (a wild taxon bearing tuberous roots, slender epicalyces, and white to pale yellow to dark pinkish flowers) was crossed with A. moschatus subsp. moschatus (bearing non-tuberous tap roots, slender epicalyces, and yellow flowers with dark purple-centered corolla). The F1s developed yielded plants with deep red color with ornamental potential, both as a candidate for flowerbeds and as potted plants. The ornamental F1 hybrids had tuberous tap roots and perennation ability, which can be easily propagated through stem cuttings. They hardly needed any care and could easily grow throughout the year in tropical climates (John et al., 2013a; John et al., 2013b). The F2s of the cross produced flowers of various shades of pink, yellow, light red, and dark red and combinations of these colors. However, the selfed derivatives of the present study produced bright yellow flowers only, as yellow-flowered segregants were selected from the early segregating generation. Further, they have attained homozygosity for the characteristics at the F8 generation. Both direct and reciprocal crosses can be attempted further to derive more diverse segregants. The derivatives have the potential to act as a source of powdery mildew tolerance in okra breeding.
Fertility restoration in amphidiploids
The number of seeds set per fruit increased significantly in the amphidiploids, and it ranged from 4.00 to 98.00 per fruit. Seven derivatives produced more than 50 seeds per fruit, which include four selfed amphidiploid derivatives, one backcrossed, and two interspecific cross selfed derivatives. However, the number of seeds was less than 10 in 22 derivatives, which comprised 13 BCs, two selfed, and seven derivatives of the crosses between amphidiploids. The wide range in value for this trait also indicates that there is genotype specificity in crossability between different species, as crosses of wild Abelmoschus with different genotypes of A. esculentus had different degrees of fertility restoration leading to differed numbers of seeds per fruit (Merita et al., 2012). Profuse fruit bearing was observed in the majority of the amphidiploids even though seed setting propensity was different. Bhat et al. (2014) reported high irregularities in meiotic chromosome pairing such as the formation of multivalent associations, lower frequency of bivalents with more rod over ring bivalents, and higher number of univalents in the interspecific hybrids involving various Abelmoschus species. Similar reports of low frequency of multivalent formation and lagging chromosomes were observed during meiosis in synthetic amphidiploids developed from interspecific hybrids between S. melongena and S. aculeatissimum (Zhou et al., 2018). These results also highlight the need to conduct detailed cytogenetic analyses of interspecific hybrids and their derivatives in Abelmoschus. Reports on the chromosome counts of all the available taxa are also limited. Table 5 depicts the chromosome counts of wild Abelmoschus species used in the present study.
In general, shorter fruit stalk observed in amphidiploids revealed that the gene controlling this trait has been incorporated from cultivated types. A. esculentus is reported to have fruit stalk length ranging from 1.99 to 3.00 cm (Reddy et al., 2022) and that of A. caillei ranging from 1.55 to 6.11 cm (AdeOluwa and Kehinde, 2011). However, the fruit stalk length of wild species is longer, viz., A. palianus with 4 cm (Sutar et al., 2013), A. moschatus with 6.0–15.3 cm (Yadav et al., 2014; Gangopadhyay et al., 2016), and A. enbeepeegearensis with 4.3–4.5 cm (John et al., 2013c).
Conclusion
The interspecific hybridization and subsequent polyploidization created tremendous variability among the amphidiploids. A considerable number of amphidiploids exhibited wild characteristics. However, amphidiploids with cultivated species morphology and intermediate types with longer fruits and less trichome density were also recorded. As the wild relatives used in the wide hybridization are reported to be the source of field resistance/tolerance to okra YVMV and ELCV diseases, the new synthetic amphidiploids would be a valuable source for okra improvement. Out of the 113 amphidiploids characterized, three derivatives, viz., Ruchi × AM-24, Ruchi × AM-6, and Arka Anamika × AMPK-1, expressed the phenotype of the cultivated okra with long fruits, less trichome density and basal branching, and low spininess, which can be directly used as breeding lines. The ornamental potential of interspecific crosses such as AM-13 and AM-19 may also be tapped. Profusely flowering C1Pusamizo derivatives could be advanced further for deriving suitable ornamental types. The presence of more seeds per fruit in certain derivatives indicated that fertility restoration and advancing of generation through backcrossing can yield desirable segregants with okra morphology. Other desirable traits like profuse tillering, perennation nature thereby extended flowering and fruiting, off-season bearing, and ornamental qualities can also be exploited. All the derivatives were advanced by backcrossing (72 lines), selfing (48 lines), and bulking the open-pollinated seeds (all 113 lines). Multi-location testing for YVMV disease tolerance of the advanced lines is in progress, which will help to study the reaction of resistance gene(s) in hosts to various strains of YVMV, which in turn will help the breeders to identify major genes controlling known physiological basis of resistance. Hence, the genetic stocks in the form of interspecific derivatives developed here involve seven different wild species and are a reservoir of sources of genes with tolerance to YVMV and ELCV as well as several other characteristics of importance to okra improvement. However, more systematic analyses are required so that genes for tolerance and other traits can be identified and tagged with molecular markers to facilitate their marker-assisted transfer in okra improvement programs. The variations among derivatives for strain level tolerances are to be investigated using this material to enhance their utilization.
The amphidiploid derivatives can also be analyzed through a genome-wide association study (GWAS) so that trait-linked associations can be identified. As reported by Daunay et al. (1993) in eggplant, there is the possibility of bringing back the amphidiploid status to the diploid level, followed by backcrossing with the cultivated okra to easily introgress the valuable traits. Further, there are still chances of evolving diverse derivatives from the bulked population, as segregation was observed in some of the derivatives. As stated by Stalker (2017), the incorporation of genes from the wild species will also contribute to the reduction of production costs, since the introduction of these genes contributes to a decrease in the incidence of diseases, thereby reducing pesticide use and, thus, generating great savings for the producer/farmer.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
AS: Data curation, Formal Analysis, Investigation, Writing – original draft, Funding acquisition, Software. JJK: Conceptualization, Data curation, Investigation, Supervision, Writing – review & editing, Resources. KVB: Conceptualization, Methodology, Resources, Validation, Writing – review & editing. ML: Formal Analysis, Investigation, Writing – review & editing. CJL: Data curation, Investigation, Writing – review & editing. MP: Investigation, Resources, Validation, Writing – review & editing. VAMN: Data curation, Formal Analysis, Investigation, Resources, Writing – review & editing. PPT: Data curation, Formal Analysis, Investigation, Writing – review & editing. CP: Resources, Writing – review & editing. SP: Resources, Supervision, Writing – review & editing. AK: Writing – review & editing. RKG: Supervision, Validation, Writing – review & editing. GPS: Project administration, Supervision, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This characterization study was undertaken under the institutional project.
Acknowledgments
The funding received from the ICAR-National Agricultural Innovation Project, Extramural Project of the ICAR-Horticultural Sciences Division, and ICAR-Education Division Emeritus Scientist to KVB, JJK, MP and VAMN for developing the amphidiploids is gratefully acknowledged. The authors are also thankful to ICAR-NBPGR, New Delhi, for providing the facilities for undertaking the present work on the morphological characterization of okra amphidiploids. Thanks are also due to ICAR (Indian Council of Agricultural Research) for carrying forward this work under the project Consortium Research Platform on Agro-Biodiversity—Component III (Okra).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1284070/full#supplementary-material
References
Adelakun, O. E., Oyelade, O. J., Ade-Omowaye, B. I. O., Adeyemi, I. A., Venter, M. V. (2009). Chemical composition and the antioxidative properties of Nigerian Okra Seed (Abelmoschus esculentus Moench) Flour. Food Chem. Toxicol. 47, 1123–1126. doi: 10.1016/j.fct.2009.01.036
AdeOluwa, O. O., Kehinde, O. B. (2011). Genetic variability studies in West African okra (Abelmoschus caillei). Agric. Biol. J. N. Am. 2, 1326–1335. doi: 10.5251/abjna.2011.2.10.1326-1335
Adetuyi, F., Ajala, L., Ibrahim, T. (2012). Effect of the addition of defatted okra seed (Abelmoschus esculentus) flour on the chemical composition, functional properties and Zn bioavailability of plantain (Musa paradisiacal Linn) flour. J. Microbiol. Biotechnol. Food Sci. 2, 69–82. Available at: https://office2.jmbfs.org/index.php/JMBFS/article/view/7188.
Akhond, Y. A. M., Molla, M. A. H., Islam, M. O., Ali, M. (2000). Cross compatibility between Abelmoschus esculentus and A. moschatus. Euphytica 114 (3), 175 –180. doi: 10.1023/A:1003985728630
Amiteye, S., Amitaaba, T., Akama, C., Amoatey, H. M. (2019). Hybridization studies of okra (Abelmoschus spp. (L.) Moench) accessions. IJBTT 9, 42–47. doi: 10.14445/22490183/IJBTT-V9I4P606
Arunkumar, B. (2015). Evaluation and molecular characterization of advanced genetaion distant hybridization selections of okra [Abelmoschus esculentus (L.) Moench]. MSc Thesis. Kerala Agric. Univ. Thrissur Kerala., 177. Available at: http://krishikosh.egranth.ac.in/handle/1/5810029729
Ayam, P. D., Swadesh, B., Praveen, K. M., Tridip, B., Arup, C., Imtinungsang, J. (2018). Assessment of breeding potential of cultivated okra (Abelmoschus esculentus L. Moench) for selecting donor parent aiming at Enation Leaf Curl Virus disease tolerance in Eastern India. Agri Res. Tech: Open Access J. 18, 556072. doi: 10.19080/ARTOAJ.2018.18.556072
Badiger, M., Yadav, R. K. (2019). Screening of germplasm of Abelmoschus against biotic stresses. Indian J. Agric. Sci. 89 (12), 2085–2090. doi: 10.56093/ijas.v89i12.96279
Badiger, M., Yadav, R. K., Sharma, B. B., Bhat, K. V., Tomar, B. S., Lata, S., et al. (2023). Pollen germination, pollen–pistil interaction and crossability studies in interspecific and induced colchiploid population of Abelmoschus species. Genet. Resour. Crop Evol. doi: 10.1007/s10722-023-01610-y
Ballén-Taborda, C., Chu, Y., Ozias-Akins, P., Holbrook, C. C., Timper, P., Jackson, S. A., et al. (2022). Development and genetic characterization of peanut advanced backcross lines that incorporate Root-Knot Nematode resistance from Arachis stenosperma. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.785358
Bhat, K. V., Yadav, S. R., Rao, S. R., John, K. J., Latha, M., Malik, S. K., et al. (2014). NAIP project report on Biosystematics of the genera Vigna, Cucumis and Abelmoschus. (New Delhi-110012: National Bureau of Plant Genetic Resources).
Breslavetz, L., Medvedeva, G., Magitt, M. (1934). Zytologische untersuchungen der bastpflanzen. Z. Züchtung 19, 229–234.
Charrier, A. (1984). Genetic resources of abelmoschus medik. (Okra) (Rome, Italy: International Board of Plant Genetic Resources), 61.
Chen, Z. J. (2010). Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 15, 57–71. doi: 10.1016/j.tplants.2009.12.003
Chen, J. F., Staub, J., Adelberg, J., Lewis, S., Kunkle, B. (2002). Synthesis and preliminary characterization of a new species (amphidiploid) in Cucumis. Euphytica 123, 315–322. doi: 10.1023/A:1015095430624
Chowda-Reddy, R. V., Kirankumar, M., Seal, S. E., Muniyappa, V., Girish, B., Valand Govindappa, M. R., et al. (2012). Bemisia tabaci phylogenetic groups in India and the relative transmission efficacy of tomato leaf curl Bangalore virus by an indigenous and an exotic population. J. Integr. Agric. 11, 235–248. doi: 10.1016/S2095-3119(12)60008-2
Cui, L., Ren, Y., Murray, T., Yan, W., Guo, Q., Niu, Y., et al. (2018). Development of perennial wheat through hybridization between wheat and wheatgrasses: A review. Engineering 4, 507–513. doi: 10.1016/j.eng.2018.07.003
Daunay, M. C., Chaput, M. H., Sihachakr, D., Allot, M., Vedel, F., Ducreux, G. (1993). Production and characterization of fertile somatic hybrids of eggplant (Solanum melongena L.) with Solanum aethiopicum. Theor. Appl. Genet. 85, 841–850. doi: 10.1007/BF00225027
de Paula, A. F., Dinato, N. B., Vigna, B. B. Z., Fávero, A. P. (2017). Recombinants from the crosses between amphidiploid and cultivated peanut (Arachis hypogaea) for pest-resistance breeding programs. PloS One 12, e0175940. doi: 10.1371/journal.pone.0175940
Favero, A. P., Custodio, A. R., Dinato, N. B., de Godoy, I. J., Seijo, J. G., Michelotto, M. D. (2020). Transference of multiple resistance to peanut through the development of cross-compatible complex hybrids of wild Arachis. Genet. Mol. Biol. 43 (2), e20190099. doi: 10.1590/1678-4685-GMB-2019-0099
Ford, C. E. (1938). A contribution to a cytogenetical survey of the Malvaceae. Genetica 20, 431 –452. doi: 10.1007/BF01531777
Fujimoto, R., Taylor, J. M., Sasaki, T., Kawanabe, T., Dennis, E. S. (2011). Genome wide gene expression in artificially synthesized amphidiploids of Arabidopsis. Plant Mol. Biol. 77, 419–431. doi: 10.1007/s11103-011-9820-y
Gadwal, V. R., Joshi, A. B., Iyer, R. D. (1968). Interspecific hybrids in Abelmoschus through ovule and embryo culture. Indian J. Genet. Plant Breed. 28, 269–274.
Gaeta, R. T., Pires, J. C., Iniguez-Luy, F., Leon, E., Osborn, T. C. (2007). Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19, 3403–3417. doi: 10.1105/tpc.107.054346
Gangopadhyay, K. K., Singh, A., Bag, M. K., Ranjan, P., Prasad, T. V., Roy, A., et al. (2016). Diversity analysis and evaluation of wild Abelmoschus species for agro-morphological traits and major biotic stresses under the north western agro-climatic condition of India. Genet. Resour. Crop Evol. 64, 775–790. doi: 10.1007/s10722-016-0400-5
Gogoi, N., Gogoi, M., Choudhury., S. (2017). Utilization of agro waste-okra and its potentiality. Asian J. Home Sci. 12250–256. doi: 10.15740/HAS/AJHS/12.1/250256
Guebebia, S., Espinosa-Ruiz, C., Zourgui, L., Cuesta, A., Romdhane, M., Esteban, MÁ. (2023). Effects of okra (Abelmoschus esculentus L.) leaves, fruits and seeds extracts on European sea bass (Dicentrarchus labrax) leukocytes, and their cytotoxic, bactericidal and antioxidant properties. Fish Shellfish Immunol. 138, 108799. doi: 10.1016/j.fsi.2023.108799
Hamon, S., Yapo, A. (1985). Perturbations induced within the genus Abelmoschus by the discovery of a second edible okra species in West Africa. Acta Hortic. 182, 133–144. doi: 10.17660/ActaHortic.1986.182.16
Jatkar, M. A., Prabu, T., Warade, S. D. (2007). Induction of colchiploidy in sterile interspecific okra F1 hybrids. Crop Res. 34, 133–136.
John, J. K., Krishnaraj, M. V., Pradheep, K., Patil, P., Harish, G. D., Bhat, K. V. (2020). A new variety of Abelmoschus pungens (Malvaceae) from Indo-Burma Biodiversity Hotspot. Rheedea 30, 456–462. doi: 10.22244/rheedea.2020.30.04.05
John, J. K., Latha, M., Sheen, S., Nissar, V. A. M., Dwivedi, N. K., Bhat, K. V. (2013b). Okra for aesthetic beauty. Indian Horticulture. 58(2).
John, J. K., Nissar, V. A. M., Latha, M., Sutar, S., Patil, P., Malik, S. K., et al. (2013a). Genetic resources and crossability relationship among various species of Abelmoschus. Curr. Horticulture 1, 35–46.
John, J. K., Scariah, S., Nissar, V. A. M., Bhat, K. V., Yadav, S. R. (2013c). Abelmoschus enbeepeegearense sp. nov. (Malvaceae), an endemic species of okra from Western Ghats, India. Nord. J. Bot. 30, 1–6. doi: 10.1111/j.1756-1051.2012.01624.x
Joseph, J. K., Roy, Y. C., Latha, M., Bhat, K. V. (2017). “Wide hybridization in the genus abelmoschus medikus des,” in Distant hybridization in horticultural crops. Eds. Dinesh, M. R., Sankaran, M. (New Delhi: Astral International Pvt. Ltd), 189–199.
Joshi, A. B., Gadwal, V. R., Hardas, M. W. (1974). “Okra,” in Evolutionary studies in world crops: diversity and change in the Indian sub-continent. Ed. Hutchinson, J. B. (London, UK: Cambridge University Press), 99–105.
Joshi, A. B., Hardas, M. W. (1953). Chromosome number in Abelmoschus tuberculatus Pal and Singh - a species related to cultivated bhindi. Curr. Sci. 22384–385. Available at: chrome-extension://efaidnbmnnnibpcajpcglclefindmkaj/https://www.nature.com/articles/1781190a0.pdf.
Joshi, A. B., Hardas, M. W. (1956). Allopolyploid nature of okra (Abelmoschus esculentus (L.) Moench). Nature 178, 1190. doi: 10.1038/1781190a0
Joshi, A. B., Hardas, M. W. (1976). “Okra,” in Evolution of crop plants. Ed. Simmonds, N. W. (London: Longman), 194 –195.
Kamalova, G. V. (1977). Cytological studies of some species of the Malvaceae. Uzbekistan Biologija Zurnali 266–69.
Kaur, B., Garcha, K. S., Sandhu, J. S., Sharma, M., Dhatt, A. S. (2023). Interspecific hybridization for transfer of hull-less seed trait from Cucurbita pepo to C. moschata. Sci. Rep. 13, 4627. doi: 10.1038/s41598-023-29935-9
Khan, M. M. R., Hasnunnahar, M., Isshiki., S. (2013). Production of amphidiploids of the hybrids between Solanum macrocarpon and Eggplant. HortScience 48, 422–424. doi: 10.21273/HORTSCI.48.4.422
Klimushina, M. V., Kroupin, P. Y., Bazhenov, M. S., Karlov, G. I., Divashuk, M. G. (2020). Waxy gene-orthologs in Wheat × Thinopyrum amphidiploids. Agronomy 10, 963. doi: 10.3390/agronomy10070963
Kumar, K., Neelam, K., Singh, G., Mathan, J., Ranjan, A., Brar, D. S., et al. (2019). Production and cytological characterization of a synthetic amphiploid derived from a cross between Oryza sativa and Oryza punctata. Genome 62, 705–714. doi: 10.1139/gen-2019-0062
Kuwada, H. (1957). Cross compatibility in the reciprocal crosses between Abelmoschus esculentus and A. manihot, and the characters and meiosis in F1 hybrids. Jpn. J. BREED 7, 93–102. doi: 10.1270/jsbbs1951.7.93
Kuwada, H. (1966). The new amphidiploid plant named Abelmoschus tubercularesculentus obtained from the progeny of the reciprocal crossing between A. tuberculatus and A. esculentus. Jpn. J. BREED 16, 21–30. doi: 10.1270/jsbbs1951.16.21
Kuwada, H. (1974). F1 hybrids of Abelmoschus tuberculatus x A. manihot with reference to the genome relationship. Jpn. J. BREED 24, 207–210. doi: 10.1270/jsbbs1951.24.207
Li-Na, L., Du, P., Fu, L. Y., Liu, H., Xu, J., Qin, L., et al. (2017). Development and characterization of amphidiploid derived from interspecific cross between cultivated peanut and its wild relative Arachis oteroi. Acta Agron. Sin. 43, 133–140. doi: 10.3724/SP.J.1006.2017.00133
Lukens, L. N., Pires, J. C., Leon, E., Vogelzang, R., Oslach, L., Osborn, T. (2006). Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140, 336–348. doi: 10.1104/pp.105.066308
Mallet, J. (2005). Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237. doi: 10.1016/j.tree.2005.02.010
Manju, S., Gopimony, R. (2009). “Anjitha - A new okra variety through induced mutation in interspecific hybrids of Abelmoschus spp,” in Induced plant mutations in the genomics era. Ed. Shu, Q. Y. (Rome: Food and Agriculture Organization of the United Nations), 87–90.
Medwedewa, G. B. (1936). Karyological review of 15 species of the genus Hibiscus. J. Botanique l’URSS 21, 533–550.
Merita, K., Joseph, J. K., Yadav, S. R., Bhat, K. V., Rao, S. R. (2012). Chromosome counts in wild and cultivated species of Abelmoschus Medikus. from the Indian sub-continent. J. Hortic. Sci. Biotechnol. 87 (6), 593 –599. doi: 10.1080/14620316.2012.11512917
Mjnocha, J. L., Ravi, R., Kumar, R., Metha, A. (1991). Morphological, cytological and peroxidase isozyme studies in diploids and amphidiploids of V. radiata and V. mungo. Indian J. Genet. 51, 429–437.
Misra, R. C., Pani, D. R., Bharathi, L. K., Ahlawat, S. P. (2018). Abelmoschus angulosus var. mahendragiriensis (Malvaceae): a new taxonomic variety of wild okra from Eastern Ghats of India. Genet. Resour Crop Evol. 65, 993–1002. doi: 10.1007/s10722-017-0590-5
Mujeeb-Kazi, A., Rajaram (2002). Transferring alien genes from related species and genera for wheat improvement. in Bread wheat improvement and production, vol. 30. (Rome: FAO Plant Production and Protection Series), 199–215.
Nagaraju, K., Pitchaimuthu, M., Sadashiva, A. T., Rao, E. S., Rekha, A., Venugopalan, R. (2019). Interspecific hybridization in Genus abelmoschus medikus. Int. J. Curr. Microbiol. App. Sci. 8, 425–438. doi: 10.20546/ijcmas.2019.808.048
Nasrallah, M. E., Yogeeswaran, K., Snyder, S., Nasrallah, J. B. (2000). Arabidopsis species hybrids in the study of species differences and evolution of amphiploidy in plants. Plant Physiol. 124, 1605–1614. doi: 10.1104/pp.124.4.1615
Nemeth, C., Yang, C. Y., Kasprzak, P., Hubbart, S., Scholefield, D., Mehra, S., et al. (2015). Generation of amphidiploids from hybrids of wheat and related species from the genera Aegilops, Secale, Thinopyrum, and Triticum as a source of genetic variation for wheat improvement. Genome 58, 71–79. doi: 10.1139/gen-2015-0002
Nimura, M., Kato, J., Horaguchi, H., Mii, M., Sakai, K., Katoh, T. (2006). Induction of fertile amphidiploids by artificial chromosome - doubling in interspecific hybrid between Dianthus caryophyllus L. and D. japonicus Thunb. Breed. Sci. 56, 303–310. doi: 10.1270/jsbbs.56.303
Olawuyi, I. F., Lee, W. Y. (2021). Structural characterization, functional properties and antioxidant activities of polysaccharide extract obtained from okra leaves (Abelmoschus esculentus). Food Chem. 354, 129437. doi: 10.1016/j.foodchem.2021.129437
Ortaç, D., Cemek, M., Karaca, T., Büyükokuroglu, M., Özdemir, ZÖ., Kocaman, A. T., et al. (2018). In vivo anti-ulcerogenic effect of okra (Abelmoschus esculentus) on ethanol-induced acute gastric mucosal lesions. Pharm. Bio. 56, 165–175. doi: 10.1080/13880209.2018.1442481
Pal, B. P., Singh, H. B., Swarup, V. (1952). Taxonomic relationships and breeding possibilities of species of Abelmoschus related to okra (A. esculentus). Bot. Gaz. 113, 455–464. doi: 10.1086/335734
Pandey, A., Semwal, D. P., Bhatt, K. C., Gupta, R., Ahlawat, S. P. (2016). A new report on cultivation of ‘‘Sukhlai’’ (Abelmoschus manihot (L.) Medik. subsp. tetraphyllus (Roxb. ex Hornem.) Borss. Waalk.): a species used as organic clearant in jaggery industry in India. Genet. Resour. Crop Evol. doi: 10.1007/s10722-016-0451-7
Patel, B., Hegde, G., Manu, T. (2021). Interspecific hybridization as a way of resistance transfer against viruses in okra: Hindrances and way forward. Plant Genet. Resour. 19 (4), 357–362. doi: 10.1017/S1479262121000423
Peng, C., Lin, H., Lin, C., Wang, C., Huang, C. (2019). Abelmoschus esculentus subfractions improved nephropathy with regulating dipeptidyl peptidase-4 and type1glucagon-like peptide receptor in type 2 diabetic rats. J. Food Drug Anal. 27, 135–144. doi: 10.1016/j.jfda.2018.07.004
Purewal, S. S., Randhawa, G. S. (1947). Studies in Hibiscus esculentus (Lady’s finger): Chromosome and pollination studies. Indian J. Agric. Sci. 17129 –136.
Rajamony, L., Chandran, M., Rajmohan, K. (2006). In vitro embryo rescue of interspecific crosses for transferring virus resistance in Okra (Abemoschus esculentus (L.) Moench). Acta Hortic. 725, 235–240. doi: 10.17660/ActaHortic.2006.725.30
Rana, R. S., Arora, R. K. (1991). Plant genetic resources activities in okra - an Indian perspective. (Rome: International Board for Plant Genetic Resources).
Rattan, P., Kumar, S. (2020). Development of interspecific hybrids (Abelmoschus esculentus × A. tetraphyllus) in okra using embryo rescue approach. Biosci. Biotechnol. Res. Asia 17, 517–523. doi: 10.13005/bbra/2855
Reddy, M. T. (2010). Genetic diversity, heterosis combining ability and stability in okra (Abelmoschus esculentus (L.) Moench) (Rajendranagar, Hyderabad, Andhra Pradesh, India: Acharya Ranga Agricultural University).
Reddy, J. P., Anpanandan, V., Kumar, S. S. (2022). Genotypic, phenotypic variability and evaluation of okra [Abelmoschus esculentus (L.) Moench] genotypes for yield components. J. Appl. Nat. Sci. 14, 180 –187. doi: 10.31018/jans.v14i1.3322
Ren, T., Jiang, Q., Sun, Z., Ren, Z., Tan, F., Yang, W., et al. (2022). Development and characterization of novel wheat-rye 1RS·1BL translocation lines with high resistance to Puccinia striiformis f. sp. tritici. Phytopathology 112, 1310–1315. doi: 10.1094/PHYTO-07-21-0313-R
Roy, R. P., Jha, R. P. (1958). A semi-asynaptic plant of Abelmoschus esculentus (L.) Moench (= Hibiscus esculentus L.). Cytologia 23, 356–361. doi: 10.1508/cytologia.23.356
Samarajeewa, P. K. (2003). “Wild Abelmoschus species in the improvement of okra,” in Conservation and use of crop wild relatives. Eds. Jayasuriya, A. H. M., Vaughan, D. A. (Japan: Proceedings of the Joint Department of Agriculture, Sri Lanka and National Institute of Agrobiological Sciences), 97–108.
Sandeep, N., Dushyanthakumar, B. M., Sridhara, S., Dasaiah, L., Satish, K. M., El-Shehawi, A. M., et al. (2022). Characterization of okra Species, their hybrids and crossability relationships among Abelmoschus species of the Western Ghats Region. Horticulturae 8, 587. doi: 10.3390/horticulturae8070587
Santhiya, S., Yadav, R. K., Lata, S., Sharma, B. B., Talukdar, A., Ghosh, A., et al. (2022). Screening of wild okra (Abelmoschus esculentus) against yellow vein mosaic and enation leaf curl diseases. Indian J. Agric. Sci. 92 (10), 1268–1273. doi: 10.56093/ijas.v92i10.125710
Sanwal, K., Singh, M., Singh, B., Naik, P. S. (2014a). Resistance to Yellow Vein Mosaic Virus and Okra Enation Leaf Curl Virus: Challenges and future strategies. Curr. Sci. 106, 1470–1471.
Sanwal, S. K., Venkataravanappa, V., Chauhan, N. S. (2014b). “Screening of wild and cultivated okra germplasm against yellow vein mosaic disease,” in Paper presented on International conference on horticulture for nutritional livelihood and environmental security in hills: opportunity and challenges at Kalimpongm Darjeeling(India).
Schranz, M. E., Osborn, T. C. (2000). Novel flowering time variation in the resynthesized polyploid Brassica napus. J. Hered. 91, 242–246. doi: 10.1093/jhered/91.3.242
Sharma, B. R. (1993). “Okra: abelmoschus spp,” in Genetic Improvement of Vegetable Crops. Eds. Kalloo, G., Bergh, B. O. (Oxford and New York: Pergamon press), 751–769.
Sheela, M. N. (1994). Induction of genetic recombination in interspecific crosses of Abelmoschus (Thrissur: Kerala Agric. Univ.).
Siemonsma, J. S. (1982a). West-African okra – Morphological and cytogenetical indications for the existence of a natural amphidiploid of Abelmoschus esculentus (L.) Moench and A. manihot (L.) Medikus. Euphytica 31, 241–252. doi: 10.1007/BF00028327
Siemonsma, J. S. (1982b). La Culture du Gombo (Abelmoschus spp.), Legume-Fruit Tropical (avec Reference Speciale à la Cote d’Ivoire). (Wageningen, The Netherlands: Wageningen Agricultural University), 279.
Sigareva, M. A., Earle, E. D. (1997). Direct transfer of a cold-tolerant Ogura male sterile cytoplasm into cabbage (Brassica oleracea ssp. capitata) via protoplast fusion. Theor. Appl. Genet. 94, 213–220. doi: 10.1007/s001220050402
Singh, H. B., Bhatnagar, A. (1975). Chromosome number in okra from Ghana. Indian J. Genet. Plant Breed. 3626–27.
Singh, G., Pathak, M., Pathak, D., Sharma, A., Chawla, N. (2023b). Performance of okra backcross derivatives for agronomic traits and yellow vein mosaic disease resistance. Indian Phytopathol. doi: 10.1007/s42360-023-00671-4
Singh, G., Pathak, M., Sharma, A. (2023a). dentification of novel sources of yellow vein mosaic disease resistance in okra (Abelmoschus esculentus L. Moench). Euphytica 219, 111. doi: 10.1007/s10681-023-03242-w
Skovsted, A. (1935). Chromosome numbers in the family Malvceae 1. J. Genet. 31, 263–296. doi: 10.1007/BF02982344
Slotte, T., Huang, H., Lascoux, M., Ceplitis, A. (2008). Polyploid speciation did not confer instant reproductive isolation in Capsella (Brassicaceae). Mol. Biol. Evol. 25, 1472–1481. doi: 10.1093/molbev/msn092
Song, Z., Dai, S., Jia, Y., Zhao, L., Kang, L., Liu, D., et al. (2019). Development and characterization of Triticum turgidum - Aegilops umbellulata amphidiploids. Plant Genet. Resour 17 (1), 24–32. doi: 10.1017/S1479262118000254
Song, K., Lu, P., Tang, K., Osborn, T. C. (1995). Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. U.S.A. 92, 7719–7723. doi: 10.1073/pnas.92.17.7719
Song, K., Tang, K., Osborn, T. C. (1993). Development of synthetic Brassica amphidiploids by reciprocal hybridization and comparison to natural amphidiploids. Theor. Appl. Genet. 86, 811–821. doi: 10.1007/BF00212606
Srivastava, U., Mahajan, R. K., Gangopadhyay, K. K., Singh, M., Dhillon, B. S. (2001). Minimal descriptors of agri-horticultural crops, part II: vegetable crops (New Delhi: NBPGR), 247–251.
Stalker, T. H. (2017). Utilizing wild species for peanut improvement. Crop Sci. 57, 1102–1120. doi: 10.2135/cropsci2016.09.0824
Suma, A., Joseph, J. K., Latha, M., Bhat, K. V. (2020). “Wide hybridization and development of amphidiploids for incorporation of Yellow Vein Mosaic Virus tolerance,” in Prospects in Plant Biology. Eds. Okra Viswanath, S., Mohana, K. V., Hrideek, T. K., Raghu, A. V., Radhakrishnan, V. V., Amruth, M. (Kerala, India: Kerala State Council for Science, Technology and Environment (KSCSTE), Department of Science and Technology, Government of Kerala), 108–116.
Sutar, S., Patil, P., Aitawade, M., John, J., Malik, S., Rao, S., et al. (2013). A new species of Abelmoschus Medik. (Malvaceae) from Chhattisgarh, India. Genet. Resour Crop Evol. 60, 1953–1958. doi: 10.1007/s10722-013-0023-z
Szadkowski, E., Eber, F., Huteau, V., Lode, M., Huneau, C., Belcram, H., et al. (2010). The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186, 102–112. doi: 10.1111/j.1469-8137.2010.03182.x
Teshima, T. (1933). Genetical and cytological studies in an interspecific hybrid of Hibiscus esculentus and H. manihot. J. Fac. Agric. Hokkaido Univ. 4, 1 –155.
Thomsen, T. (1990). Synthesis of an amphidiploid Brassica species. HortScience 25 (9), 1116. doi: 10.21273/HORTSCI.25.9.1116c
Tiwari, V. K., Rawat, N., Neelam, K., Randhawa, G. S., Singh, K., Chhuneja, P., et al. (2008). Development of Triticum turgidum ssp. durum - Aegilops longissima amphiploids with high iron and zinc content through unreduced gamete formation in F1 hybrids. Genome 51, 757–766. doi: 10.1139/G08-057
Twyford, A., Ennos, R. (2012). Next-generation hybridization and introgression. Heredity 108, 179–189. doi: 10.1038/hdy.2011.68
Ugale, S. D., Patil, R. C., Khupse, S. S. (1976). Cytogenetic studies in the cross between Abelmoschus esculentus and A. tetraphyllus. J. Maharashtra Agric. Univ. 1, 106 –110.
Varma, P. M. (1952). Studies on the relationship of bhindi yellow vein mosaic virus and its vector, the white fly (Bemisia tabaci). Indian J. Agric. Sci. 25, 75 –91.
Wang, A., Zhang, X., Yang, C., Song, Z., Du, C., Chen, D., et al. (2013). Development and characterization of synthetic amphiploid (AABB) between Oryza sativa and Oryza punctata. Euphytica 189, 1–8. doi: 10.1007/s10681-012-0821-y
Wei, K., Pang, S., Yang, J., Wei, Z. (2015). Enhancement of cadmium tolerance and accumulation by introducing Perilla frutescens (L.) Britt var. frutescens genes in Nicotiana tabacum L. plants. Environ. Sci. pollut. Res. Int. 22, 5405–5416. doi: 10.1007/s11356-014-4048-4
Xu, Y., Zhong, L., Wu, X., Fang, X., Wang, J. (2009). Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 229, 471–483. doi: 10.1007/s00425-008-0844-8
Yadav, S. R., Bhat, K. V., Latha, M., John, J. K., Malik, S. K., Aitwase, M., et al. (2014). An illustrated guide for the identification of Vigna Savi, Cucumis L. and Abelmoschus Medik. species in India (Shillong: National Bureau of Plant Genetic Resources, New Delhi, Shivaji University, Kolhapur and North Eastern Hill University), 145.
Yadav, R. K., Mahesh, B., Suman, L. (2019). Studies of genetic variability and association of yield traits and YVMV disease in Abelmoschus species. Indian J. Agric. Sci. 89 (4), 688–694. doi: 10.56093/ijas.v89i4.88868
Zhang, X., Liu, T., Li, X., Duan, M., Wang, J., Qiu, Y., et al. (2016). Interspecific hybridization, polyploidization, and backcross of Brassica oleracea var. alboglabra with B. rapa var. purpurea morphologically recapitulate the evolution of Brassica vegetables. Sci. Rep. 6, 18618. doi: 10.1038/srep18618
Zhang, W., Xiang, Q., Zhao, J., Mao, G., Feng, W., Chen, Y., et al. (2020). Purification, structural elucidation and physicochemical properties of a polysaccharide from Abelmoschus esculentus L. (okra) flowers. Int. J. Biol. Macromol. 155, 740–750. doi: 10.1016/j.ijbiomac.2020.03.235
Zhou, X., Bao, S., Liu, J., Yang, Y., Zhuang, Y. (2018). Production and characterization of an amphidiploid derived from interspecific hybridization between Solanum melongena L. and Solanum aculeatissimum Jacq. Sci. Hortic. 230, 102–106. doi: 10.1016/j.scienta.2017.11.024
Keywords: pre-breeding, crop wild relatives, amphidiploids, polyploidization, morphological characterization, colchicine
Citation: Suma A, Joseph John K, Bhat KV, Latha M, Lakshmi CJ, Pitchaimuthu M, Nissar VAM, Thirumalaisamy PP, Pandey CD, Pandey S, Kumar A, Gautam RK and Singh GP (2023) Genetic enhancement of okra [Abelmoschus esculentus (L.) Moench] germplasm through wide hybridization. Front. Plant Sci. 14:1284070. doi: 10.3389/fpls.2023.1284070
Received: 27 August 2023; Accepted: 16 October 2023;
Published: 03 November 2023.
Edited by:
Rodomiro Ortiz, Swedish University of Agricultural Sciences, SwedenReviewed by:
Pritam Kalia, Indian Agricultural Research Institute (ICAR), IndiaAjay Kumar Thakur, Central Potato Research Institute (ICAR), India
Ajoy Kumar Roy, Indian Council of Agricultural Research (ICAR), India
Copyright © 2023 Suma, Joseph John, Bhat, Latha, Lakshmi, Pitchaimuthu, Nissar, Thirumalaisamy, Pandey, Pandey, Kumar, Gautam and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. Suma, U3VtYS5BQGljYXIuZ292Lmlu
†Present address: V. A. M. Nissar ICAR-Indian Institute of Spices Research, Kozhikode, Kerala, India
 A. Suma
A. Suma K. Joseph John1
K. Joseph John1 Kangila Venkataramana Bhat
Kangila Venkataramana Bhat Sushil Pandey
Sushil Pandey Ashok Kumar
Ashok Kumar Raj Kumar Gautam
Raj Kumar Gautam Gyanendra Pratap Singh
Gyanendra Pratap Singh