- College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
Increasing biotic and abiotic stresses are seriously impeding the growth and yield of staple crops and threatening global food security. As one of the largest classes of regulators in vascular plants, WRKY transcription factors play critical roles governing flavonoid biosynthesis during stress responses. By binding major W-box cis-elements (TGACCA/T) in target promoters, WRKYs modulate diverse signaling pathways. In this review, we optimized existing WRKY phylogenetic trees by incorporating additional plant species with WRKY proteins implicated in stress tolerance and flavonoid regulation. Based on the improved frameworks and documented results, we aim to deduce unifying themes of distinct WRKY subfamilies governing specific stress responses and flavonoid metabolism. These analyses will generate experimentally testable hypotheses regarding the putative functions of uncharacterized WRKY homologs in tuning flavonoid accumulation to enhance stress resilience.
1 Introduction
Escalating stresses seriously impede the production of important crops, threatening global food security (Zhang et al., 2022a). Plants have evolved intricate regulatory networks. The WRKY family, as an important member of these networks, belongs to the functionally diversified transcription factor families. WRKYs are critical in governing plant diverse stress responses (Jiang and Deyholos, 2009; Li et al., 2009; Pandey and Somssich, 2009; Hu et al., 2013; Li et al., 2013; Yokotani et al., 2013; Bakshi and Oelmüller, 2014; Dai et al., 2016; Ahammed et al., 2020b; Wu et al., 2021; Ma et al., 2021c; Lin et al., 2022; Ma et al., 2023).
In sweet potato (Ipomoea batatas), after the inaugural gene was found (Ishiguro and Nakamura, 1994), WRKYs have been discovered extensively in terrestrial plants, primitive protozoans and slime molds, affirming the ancient evolutionary origin (Zhang and Wang, 2005; Pan et al., 2009). Highly conserved WRKY structural domains paralleling the zinc finger (ZF) motif is an essential feature of this protein family. The WRKY domain directly bind to the major cis-element, W-box (TGACCA/T) (Rushton et al., 2010; Chen et al., 2019). Although slight variations in some WRKYs, the conserved motif (WRKYGQK) forms the ZF to confer structural stability (Yamasaki et al., 2005; Cheng et al., 2021).
Phylogenetically, WRKYs are classified into three distinct groups depended on the number of WRKY domains together with the type of ZFs. Two WRKY domains and a C2H2 (CX4-5CX22-23HXH) ZF are present in Group I, whereas only one WRKY domain combined with C2H2 or altered C2HC (CX7CX23HXC) ZF appears in Groups II and III (Eulgem et al., 2000; Li et al., 2010; Chen et al., 2020). According to the variation of ZF, Group II can be subclassified into subgroups IIa ~ IIe (Zhang and Wang, 2005). The presence of Group I WRKYs in primitive organisms suggests that they are the ancestors of other groups (Wei et al., 2018). WRKY domains are conserved, but different WRKY groups have evolved specialized functions (He et al., 2016; Wang et al., 2022).
The importance of WRKYs in modulating plant immunological responses against pathogen invasion, including effector-triggered immunity (ETI) and pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), have been proven (Chen et al., 2013; Chi et al., 2013; Dang et al., 2013; Birkenbihl et al., 2017; Ma et al., 2021b; Wang et al., 2023b; Wang et al., 2023d; Xiao et al., 2023). Numerous WRKY genes spanning all phylogenetic groups are induced following pathogen infection or elicitor treatments across plant species, underscoring the broad defensive role (Chen et al., 2013; Dang et al., 2013; Birkenbihl et al., 2017; Wang et al., 2023b; Wang et al., 2023d; Xiao et al., 2023). Gain- and loss-of-function analyses of WRKYs have demonstrated both positive and negative regulatory functions in immune signaling (Shen et al., 2023). Additionally, WRKYs govern hormone signaling in plant (Shang et al., 2010; Chen et al., 2013; Li et al., 2013; Ding et al., 2015; Zhang et al., 2015; Chen et al., 2017; Wang et al., 2023c), regulate secondary metabolism (Suttipanta et al., 2011; Wang et al., 2023d), and dominate stress responses (Jiang and Deyholos, 2009; Li et al., 2020c; Ma et al., 2023).
Although the functional roles of plant WRKY transcription factors in stress responses are well-documented, the relationship between sequence divergences across distinct WRKY domains and their varying biological activities is still unclear. In order to fully understand the functional specificity encoded within different WRKY phylogenetic clades, further investigation is necessary Furthermore, as critical homeostatic regulators of ROS, numerous flavonoid biosynthetic genes harbor abundant W-box cis-elements within their promoters (Liu et al., 2019a; Su et al., 2022a). Growing evidence suggests that specific WRKY proteins play pivotal roles governing flavonoid metabolism to enhance plant stress adaptation (An et al., 2019; Wang et al., 2022). However, the specific contributions of individual members of the WRKY subfamily in directing flavonoid accumulation are not well understood. In this review, we will refine WRKY phylogenetic frameworks by incorporating additional plant species with WRKY genes that have been implicated in stress tolerance and flavonoid regulation. Through these analyses, we aim to elucidate the roles of specific WRKY clades in modulating flavonoid biosynthesis under defined abiotic stresses.
2 Phylogenetic analysis of WRKY proteins
The WRKYs sequences of Arabidopsis (Arabidopsis thaliana), poplar (Populus trichocarpa), maize (Zea mays), rice (Oryza sativa), and soybean (Glycine max) were downloaded from PlantTFDB (http://planttfdb.gao-lab.org/index.php). Protein sequences of WRKY with certain functions mentioned in this review were sourced from NCBI (https://www.ncbi.nlm.nih.gov). To distinguish the presence and number of structural WRKY domains, the sequences of WRKYs were examined by NCBI CD-search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) and SMART (http://smart.embl.de/). Proteins containing the WRKY domain can be used for phylogenetic analysis. By using bootstrap (1, 000 replicates), building a phylogenetic tree requires the use of NJ method by MEGA 7.0 software. Finally, the phylogenetic tree was visualized and embellished using Interactive Tree of Life (iTOL, https://itol.embl.de/) (Letunic and Bork, 2007; Nan et al., 2020).
The phylogenetic analysis showed that the 816 WRKYs were segregated into three canonical groups, designated as Roman numerals (I, II, III), with seven distinct groups and represented by colored outer circles (Figure 1), consistent with the phylogenetic system defined by Zhang and Wang (2005). Further examination reveals that Group II can be categorized into five unique subdivisions: IIa, IIb, IIc, IId, and IIe (Figure 1). Subgroup I, with 172 members, is the most numerous of the 7 subgroups. Whereas groups IIa contained only 49 members. The further division of Group II into 5 subgroups is due to specific sequence variations in the ZF motifs (Xie et al., 2005).
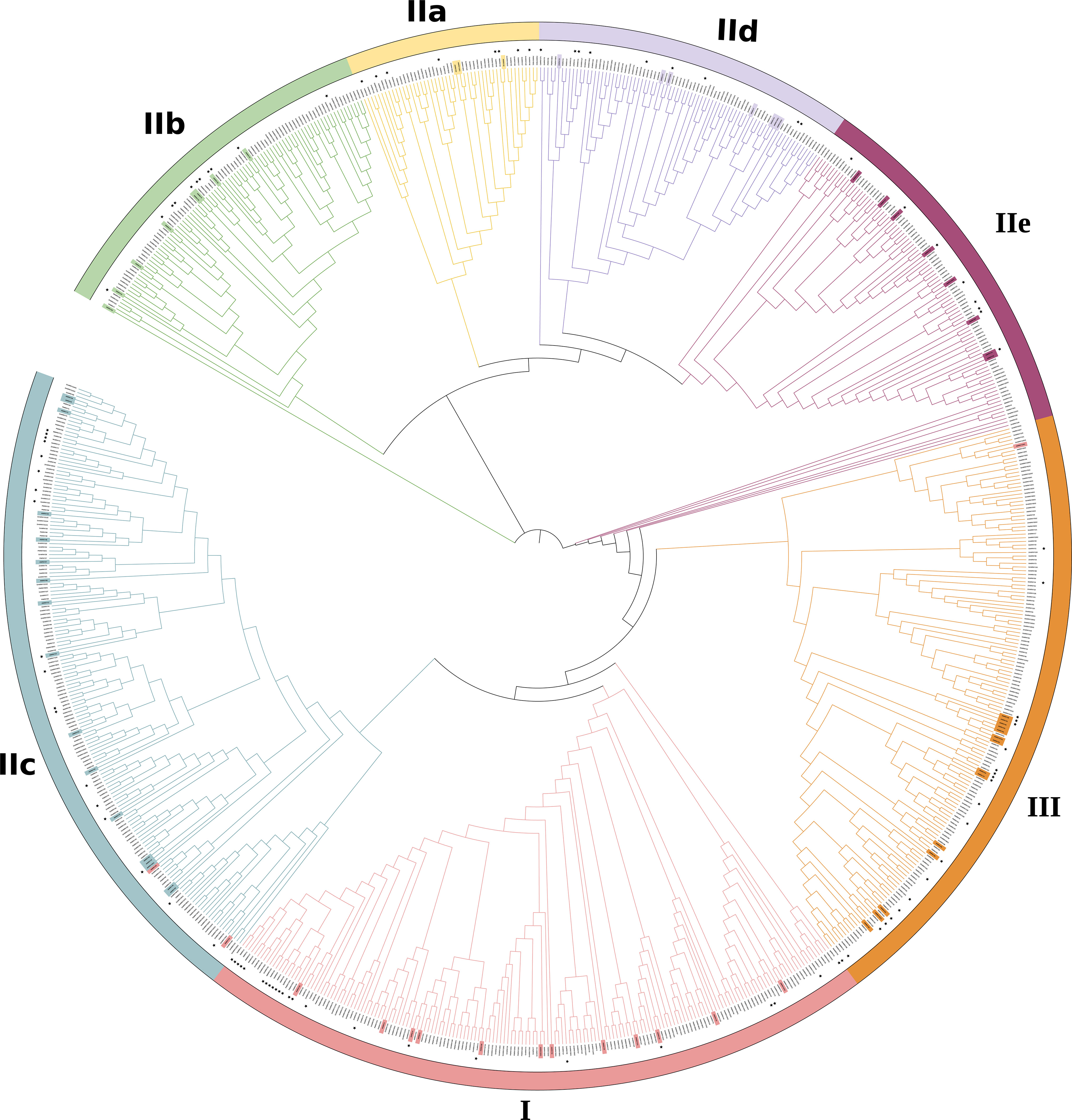
Figure 1 Phylogenetic analysis of identified WRKY proteins in Arabidopsis thaliana, Glycine max, Oryza sativa, Populus trichocarpa and Zea mays. A phylogenetic tree was established using the MEGA 7.0 program and the neighborhood joining method, drawing on WRKY domain sequences. Different colors have been utilized to distinguish between various groups or subgroups. WRKY protein sequences were obtained from NCBI (https://www.ncbi.nlm.nih.gov) database. The 72 proteins highlighted in the inner circle originate from Arabidopsis thaliana (Supplementary Table S1), while WRKY genes from Groups I to III are color-coded in Supplementary Table S2. The functional categories of WRKY, annotated in this review, are identified by black asterisks in Supplementary Table S3. Supplementary Table S4 provides a comprehensive list of the protein IDs and full sequences for all analyzed WRKYs (Jeyasri et al., 2021; He et al., 2012; Zhang et al., 2017; Yu et al., 2016).
When compared to Zhang and Wang’s system (Zhang and Wang (2005)), three WRKY proteins of the present phylogenetic analysis had distinct classes. For example, AtWRKY10, AtWRKY19, AtWRKY45 from subgroup 1 are reassigned to II c, II c, and III, respectively.
3 WRKYs bind to specific cis-acting regions of target genes
A platform for systematic investigations of WRKY family is provided by the expanding genome sequencing data from many plant species. The identification of genes crucial for specific processes has been made possible by genome sequences, which have enabled transcriptome profiling of some families (such as WRKY) during particular situations (Arndt et al., 2022; Shende et al., 2022; Zhang et al., 2022e). Furthermore, the genome-wide mapping of WRKY binding sites and target genes has been made possible by high-throughput sequencing after chromatin immunoprecipitation (ChIP-seq) (Zhou et al., 2021; Xu et al., 2022). These cutting-edge approaches have profoundly expanded the understanding of WRKY gene functions and regulatory networks.
The C-terminus contains the ZF structure C2H2 or C2HC, while the WRKYGQK motif, which forms the core region, is present at the N-terminus (Yamasaki et al., 2005; Chen et al., 2010; Song et al., 2018; Gu et al., 2019; Wang et al., 2021a). Some WRKY proteins also contain WRRY, WSKY, WKRY, WVKY, or WKKY motifs in place of the WRKY domain (Xie et al., 2005; Duan et al., 2007; Li et al., 2020b). The binding of DNA by WRKY proteins necessitates the ZF motif (Phukan et al., 2017). Replacing the zinc ion in WRKY domains with the metal chelator 1,10-phenanthroline abolishes DNA binding, indicating that WRKY proteins possess ZFs structures. The WRKY domain is composed of four β-sheets, a zinc-binding pocket, and coordination with cysteine or histidine residues make up the WRKY domain. The C-terminal of the WRKY domain stabilizes the sequence-specific interaction between the projecting N-terminal WRKYGQK sequence and the 6 bp DNA groove. Therefore, WRKY proteins may attach to the W-box clusters, primarily the TTGACC/T motif, in promoters of target genes. This enables them to regulate the dynamic network of signals and responses (Yamasaki et al., 2005; Li et al., 2020b).
The core TGAC sequence is highly conserved and principally responsible for binding by WRKY proteins (Mare et al., 2004; Ciolkowski et al., 2008; Rahman et al., 2021). In contrast, variations in the number, sequence, and nucleotide composition of the flanking bases in W-boxes of target genes contribute to the binding specificities of different WRKYs (Cheng et al., 2019).
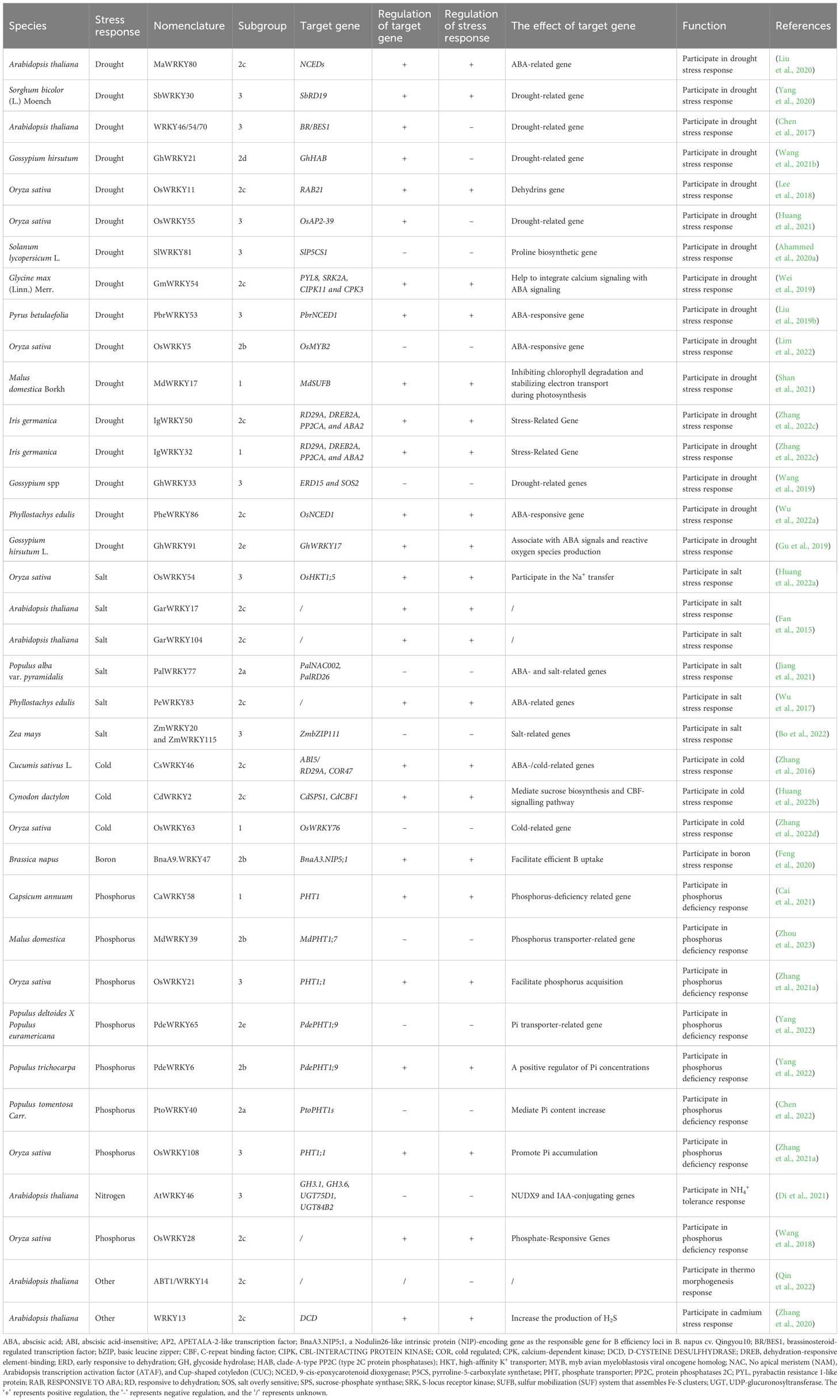
Indeed, most target genes of WRKY were discovered to contain W-box cis-elements in the promoters, as identified through various approaches. Chromatin immunoprecipitation analyses revealed that Parsley (Petroselinum crispum) WRKY1 binds to W-boxes in the promoters of PcWRKY3 and Pathogenesis-Related1-1 (PcPR1-1) (Turck et al., 2004), and pepper (Capsicum annuum) WRKY6 was able to bind to the W-boxes of the CaWRKY40, as well as defense genes Capsicum annuum defensin 1 (CaDEF1), EXTRACELLULAR PEROXIDASE 2 (CaPO2), and small heat shock protein 24 (CaHSP24) (Hussain et al., 2018). Furthermore, pull-down assays was used to identify candidate W-box containing genes (AtWRKY58, AtWRKY13, AtWRKY6) of AtWRKY53 (Miao et al., 2004). Electrophoretic mobility shift assay was applied to uncover putative W-box targets of WRKY38 in barley (Hordeum vulgare) (Mare et al., 2004), WRKY26 (Li et al., 2011), WRKY11 (Ali et al., 2018), WRKY53 (Sun and Yu, 2015) in Arabidopsis, and WRKY42 (Su et al., 2015), WRKY1, WRKY2, and WRKY4 in tobacco (Nicotiana tabacum) (Yamamoto et al., 2004), as well as WRKY71 in rice (Liu et al., 2007).
Except W-box, WRKY proteins could also recognize several other cis-elements (Figure 2). Rice OsWRKY13 binds the pathogen-responsive cis-element 4 (PRE4) (TACTGCGCTTAGT) (Xiao et al., 2013), while barley SUSIBA2/HvWRKY46 recognizes the sugar responsive element in the iso1 promoter (Sun et al. (2003). Moreover, PtoWRKY40 combines with the PHR1-binding site (P1BS) element (GNATATNC) (Zhou et al., 2008; Sun et al., 2016). The AtWRKY70 bound the WT-box sequence (YGACTTTT) of the Pep25-responsive gene expression in parsley protoplasts (Machens et al., 2014). In addition, the heat-inducible OsWRKY11 was found to attach to the promoter of heat shock elements (nGAAnnTTCnnGAAn), leading to enhanced thermotolerance of transgenic rice seedlings (Lee et al., 2018).
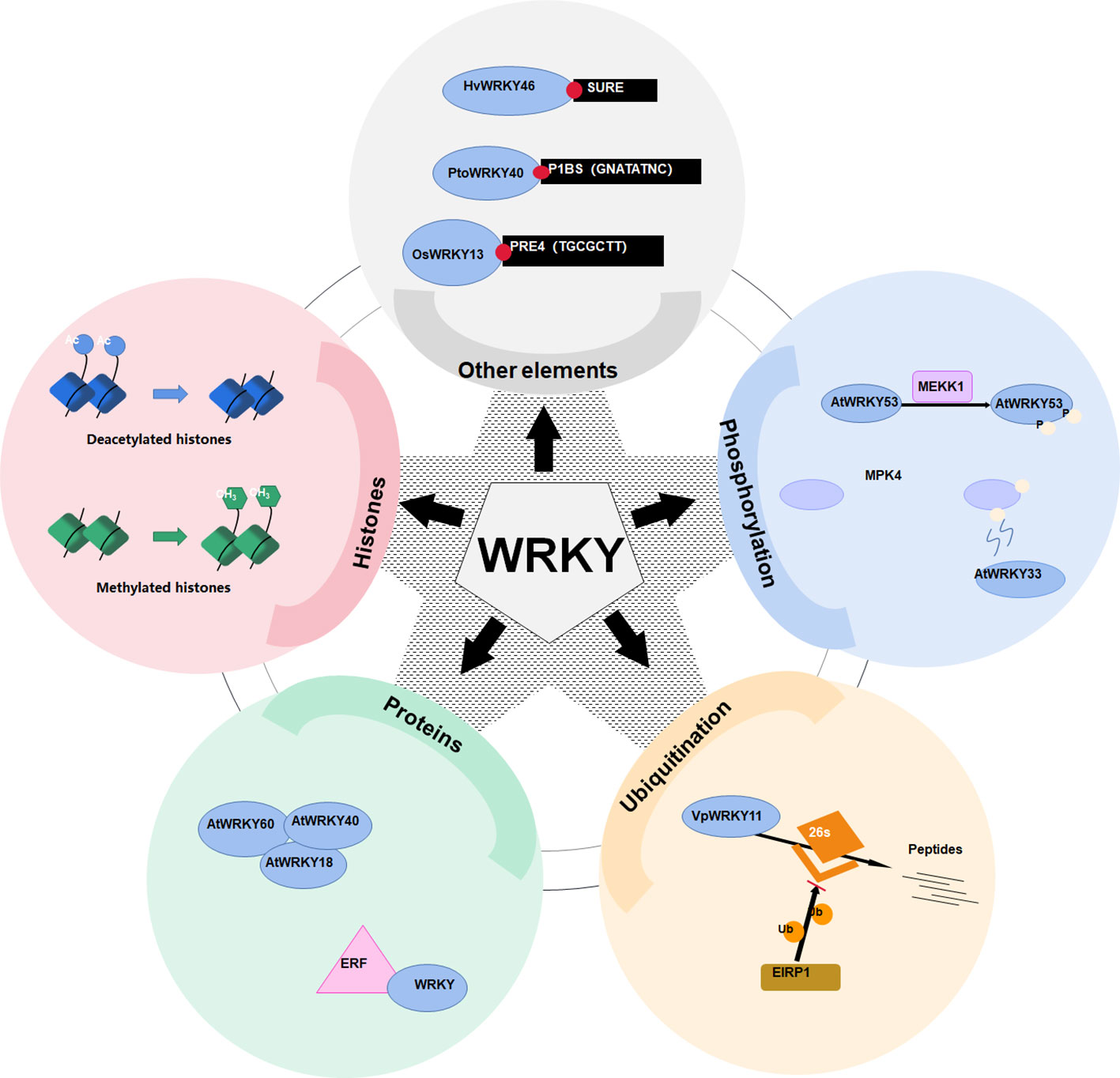
Figure 2 Various mechanisms of WRKY transcriptional activation regulation. WRKY transcription factors are regulated via a plethora of mechanisms. These range from binding to an assortment of cis-regulatory elements such as SURE, P1BS, and PRE4, to post-translational modifications including ubiquitination, methylation, and phosphorylation, which either activate them or mark them for degradation. Regulation may also involve altered chromatin conformations mediated by changes in histone acetylation or methylation, as well as modulation via interactions with other nuclear proteins like WRKY and ERF.
In essence, WRKY takes part in a variety of regulatory processes by regulating the expression of various cis-elements of target genes (Table 1). Current studies on the cis-elements of WRKY has primarily focused on six species, including Oryza sativa, Arabidopsis thaliana, Petroselinum crispum, Hordeum vulgare, Nicotiana tabacum and Capsicum annuum. It appears that members of WRKY subgroup 2e have a preference for the PRE4 element. WRKYs in subgroup 1 typically bind to the sulfur-responsive element (SURE), while the those in subgroup 3 interact with both the WT-box and the traditional W-box.
4 Regulatory mechanism of WRKYs involved in their transactivation capabilities
Extensive researches unveiled that plant WRKYs were crucial orchestrators involved in two branches of plant innate immunity (PII) (Jones and Dangl, 2006).
Upon perception of PAMPs, the mitogen-activated protein kinase (MAPK) cascade is activated, leading to upregulation of AtWRKY22 and AtWRKY29, which are two positive regulators from 2e subgroup (Asai et al., 2002; Hsu et al., 2013). Certain WRKYs also fulfill indispensable regulatory roles in ETI (Adachi et al., 2015). A plethora of WRKY genes are auto-regulated or cross-regulated through W-box elements embedded in the promoters (Robatzek and Somssich, 2002; Turck et al., 2004; Besseau et al., 2012; Yang et al., 2012; Adachi et al., 2015). For instance, CaWRKY6 activates CaWRKY40 to elevate pepper tolerance to heat and high humidity (Cai et al., 2015).
Post-translational modifications, especially phosphorylation, constitute key mechanisms orchestrating the transactivation capability of WRKYs. MAPKs phosphorylate WRKY proteins, either the DNA binding affinity is directly or indirectly altered, or the transcriptional activity is altered (Miao et al., 2007; Adachi et al., 2015). Prolonged MAPK activation increases the proportion of phosphorylated WRKYs (Figure 2), amplifying downstream signaling (Ishihama et al., 2011). For example, a MAPK kinase kinase 1 (MEKK1) phosphorylates AtWRKY53 to increase its DNA binding activity (Miao et al., 2007). In addition, MAPK 4 (MPK4) phosphorylates MKS1 (MAP kinase substrate 1) to liberate AtWRKY33 from the MKS1-WRKY33 complex, so as to targets the promoter of PHYTOALEXIN DEFICIENT3 (PAD3) for the synthesis of antimicrobial camalexin (Qiu et al., 2008).
Proteasome-mediated degradation also governs WRKY levels (Figure 2). The E3 ubiquitin protein ligase 5 (UPL5) polyubiquitinates AtWRKY53, triggering its degradation (Adachi et al., 2015). Through the 26S proteasome, the E3 ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) mediates the degradation of VpWRKY11 (Yu et al., 2013).
WRKY can fine-tune their transactivation capabilities by formation of polymers. WRKY-WRKY interactions are widespread (Xie et al., 2006; Chen et al., 2009). For example, WRKY proteins AtWRKY18, AtWRKY40 and AtWRKY60 interact both physically and functionally (Figure 2) (Xu et al., 2006; Chen et al., 2010). In addition, WRKY also interact with ethylene responsive factor (ERF) (Wang et al., 2023a) or VQ motif-containing proteins to regulate their activities (Lei et al., 2017).
In addition, histone modifications can also impact activities of WRKYs (Figure 2). For instance, the histone deacetylase 19 (HDA19) represses AtWRKY38 and AtWRKY62 (Kim et al., 2008). Additionally, the histone methyltransferase suppressor of variegation 3-9-homologous (SUVH2) epigenetically regulates AtWRKY53 (Ay et al., 2009).
In conclusion, WRKYs emerged as pivotal regulators in regulating plant responses to the environment. The diverse functions are fine-tuned through intricate interaction networks and post-translational modifications. Further elucidating these regulatory mechanisms will provide valuable insights into optimally modulating WRKY functions.
5 WRKYs regulate plant responses to abiotic stresses
Plants are routinely subjected to various stresses, which impair the normal growth and plant yield (Thomashow, 1999; Khoso et al., 2022). To surmount the stresses, sophisticated defense mechanisms have formed (Rushton et al., 2010; Schluttenhofer and Yuan, 2015; Sood et al., 2021). Recent research has shown that WRKYs have important functions in plant defenses against various stresses, including pathogens, cold, salinity, drought, and nutrition. The majority of WRKYs with known functions can be classified into three subfamilies, as shown in Figure 1; Table 2. Interestingly, WRKY members that regulate drought stress response are mostly found in subgroups 2b, 2c, 2d, and 2e, while those regulate salt resistance mainly belong to the 2a, 2c, and 3 subgroups. Additionally, WRKYs that regulate cold tolerance are mainly found in subgroups 1 and 2c, and those involved in plant responses to heavy metal poisoning are primarily found in subgroup 2c.
5.1 Involvement in drought stress
Drought stress results in cell dehydration, which threatens plant growth and yield worldwide. Long-term adaptation and evolution have led plants to have a variety of mechanisms to combat drought-induced water deficits, including closing stomata to curb water loss, a response mediated mostly by the abscisic acid (ABA) signaling (Lee et al., 1999; Finkelstein et al., 2002; Oztur et al., 2002; Shinozaki et al., 2003; Tang et al., 2016; Ahammed et al., 2020a). Under drought stress, the abscisic acid (ABA) signaling pathway often leads to stomatal closure (Ng et al., 2001; Qiu and Yu, 2009).
Actually, many WRKYs orchestrate plant drought responses by regulating ABA signaling. For instance, overexpressing of GsWRKY20, which was proven to improved response to ABA, could enhance plant tolerance to drought stress via stomatal closure (Luo et al., 2013). Besides, TaWRKY146-Overexpression also enhances drought tolerance through stomatal closure (Ma et al., 2017). In addition, soybean GmWRKY54 activates genes connected with ABA and Ca2+ pathways, alleviates water loss, and achieves drought resistance (Wei et al., 2019). PbrWRKY53 binds to and upregulates 9-cis-epoxycarotenoid dioxygenase1 (PbrNCED1), stimulating vitamin C biosynthesis and drought tolerance in Chinese white pear (Pyrus communis) (Liu et al., 2019b). In iris (Iris germanica), IgWRKY32 and IgWRKY50 together stimulate ABA signaling to upgrade drought tolerance (Zhang et al., 2022c). In contrast, the cotton GhWRKY21 and GhWRKY33, and the rice OsWRKY5, suppress this signaling, thereby undermining drought tolerance (Wang et al., 2019; Wang et al., 2021b; Lim et al., 2022).
Apart from stomatal and ABA regulation, a multitude of WRKYs can enhance plants tolerance to drought by regulating responsive genes in other pathways. Rice OsWRKY55 binds to and upregulates the APETALA-2-like transcription factor gene (OsAP2-39) to negatively modulate ethylene synthesis and drought tolerance (Huang et al., 2021). Sorghum (Sorghum bicolor) SbWRKY30 in induces the Responsive to Dehydration 19 (SbRD19), a homolog of the Arabidopsis drought-responsive RD19 (Yang et al., 2020). In apple, by modulating the iron-sulfur cluster biosynthesis protein gene (MdSUFB) expression, the MdMEK2-MdMPK6-MdWRKY17 cascade regulates chlorophyll levels during drought (Shan et al., 2021). Additionally in rice, OsWRKY11 directly upregulates RESPONSIVE TO ABA21 (RAB21), enhancing drought tolerance (Lee et al., 2018). PheWRKY86 also upregulates NCED1, encoding a rate-limiting ABA biosynthetic enzyme, conferring drought tolerance (Wu et al., 2022a).
A number of WRKYs mediate drought responses by modulating osmolyte accumulation and ROS scavenging. For instance, SlWRKY81 and WRKY46/54/70 suppress proline biosynthesis and drought responses in tomato and Arabidopsis (Ahammed et al., 2020a). Conversely, in banana (Musa acuminata), MaWRKY80 upregulates ABA biosynthesis, osmolyte accumulation, and ROS detoxification, thereby enhancing drought tolerance (Chen et al., 2017; Liu et al., 2020).
In summary, these compelling findings highlight the significant roles of WRKYs, particularly in governing ABA signaling, osmolyte metabolism and ROS homeostasis, in calibrating plant adaptation to drought stress.
5.2 Regulation of plant resistance to salinity
The growing issue of soil salinization is having a negative impact on plant growth and reducing crop yields on a global scale. Salt stress can significantly hinder plant growth, leading to harmful consequences for agricultural production worldwide (Hasegawa et al., 2000; Yan et al., 2022).
Several WRKYs were revealed as positive regulators in plant tolerance to salinity. Employing CRISPR-Cas9 to knockout OsWRKY54, Huang et al. (2022a) revealed its beneficial effect on conferring rice tolerance to salinity. Similarly, by upregulating GarWRKY17 and GarWRKY104, the salt tolerance of Arabidopsis was enhanced at different developmental stages (Fan et al., 2015). Wu et al. (2017) found that overexpressing PeWRKY83 in Arabidopsis substantiated salt tolerance, resulting in increased proline accumulation, higher germination rates, less electrolyte leakage, and lower membrane damage under salt stress.
In contrast, WRKYs could also act as negative regulators on salt resistance. Jiang et al. (2021) demonstrated that overexpressing of PalWRKY77 in poplar compromised salt tolerance through inhibition of ABA-responsive genes. Bo et al. (2022) indicated that the maize (Zea mays) ZmWRKY20-ZmWRKY115 complex in nucleus bound to promoters of basic leucine zipper (ZmbZIP111) to inhibit the expression of ZmbZIP111, which elevated the salt sensitivity of maize seedlings.
5.3 Orchestrating plant responses to cold stress
Cold stress is unfavorable to normal plant development and poses a major constraint on agricultural productivity (Andaya and Mackill, 2003). Plants have evolved various physiological, biochemical and molecular adaptation mechanisms to improve cold tolerance (Ishitani et al., 1997; Ding et al., 2019; Ding and Yang, 2022; Khoso et al., 2022). Analyzing the regulatory mechanisms and elucidating the transcriptional networks has uncovered many cold stress-related genes (Ritonga et al., 2021).
In particular, WRKYs are instrumental to cold tolerance across plant species. WRKY members in 1 and 2c, were found to regulate the chilling stress. Overexpressing CsWRKY46 from cucumber (Cucumis sativus L.) caused higher tolerance to freezing by upregulating the expression of Responsive to Desiccation gene (RD29A) and cold regulated 47 gene (COR47), and by positively regulating expression of some ABA-regulated genes under low temperature stress (Zhang et al., 2016). By contrast, rice OsWRKY63 downregulated various genes related to chilling response and ROS-scavenging, and negatively regulated chilling tolerance via the WRKY63-WRKY76-DREB1B regulatory cascade (Zhang et al., 2022d). CdWRKY2 positively regulated cold responses by binding to promoters of sucrose phosphate synthase1 (CdSPS1) and C-repeat binding factor 1 (CdCBF1) in bermudagrass (Cynodon dactylon), thereby coordinating sucrose biosynthesis and the CBF pathway (Huang et al., 2022b).
5.4 Fine-tuning plant responses to nutrient deficiency and other stresses
Insufficient or excessive levels of soil nutrients impede plant growth. Nutrient deficiency symptoms in plants vary from element to element. As one of the important plant nutrients, phosphorus deficiency affects the photosynthetic rate of plant leaves, the growth of plant stems and the formation of reproductive organs(Barry, 1988; Lauer et al., 1989). Besides, deficiencies of boron or nitrogen (NH4+) also seriously affect plant growth (Tanada, 1978; Krueger et al., 1987; Zhang et al., 2021b).
WRKY also has multiple roles in regulating these plant nutrients. For instance, suppression of OsWRKY28 resulted in decreased phosphate (Pi) accumulation in rice (Wang et al., 2018). In addition, OsWRKY21 and OsWRKY108 can positively regulate the expression level of phosphate transporter (OsPHT1;1) for Pi accumulation (Zhang et al., 2021a). In apple, overexpression of MdWRKY39 led to phosphorus deficiency through up-regulating MdPHT1;7 (Zhou et al., 2023). In poplar, a phosphate starvation response 1 (PHR1) homolog can interact with PtoWRKY40 to inhibit the transcription of PtoPHT1, thereby mediating an increase in Pi content (Chen et al., 2022). Besides, PdeWRKY6 and PdeWRKY65 modulated tissue Pi concentration by coordinating the expression of PdePHT1;9 (Yang et al., 2022). CaWRKY58 activated PHT1 and coordinated with Ca14-3-3 to improve Pi concentration under the Pi-insufficient conditions (Cai et al., 2021). In addition, BnaA9.WRKY47 enhanced the tolerance of rapeseed to boron deficiency by upregulating the boric acid channel gene (BnaA3.NIP5;1), thus increasing boron uptake (Feng et al., 2020). AtWRKY46 suppressed the expression of GDP-D-mannose pyrophosphohydrolase (NUDX9) and IAA-conjugating genes, resulting in root tolerance to NH4+ (Di et al., 2021).
WRKY also play important roles in regulating plant responses to other stresses, including heat and cadmium toxicity. Qin et al. (2022) hypothesized that Arabidopsis ABT1/WRKY14 plays a key negative regulatory role in plant thermomorphogenesis. Zhang et al. (2020) demonstrated that Cd induces WRKY13 to activate D-CYSTEINE DESULFHYDRASE (DCD) expression to elevate H2S level and enhance Cd tolerance in Arabidopsis.
6 Mechanism of WRKY affecting flavonoid synthesis
Plants are inevitably challenged by various environmental stresses including drought, high salinity, cold, ultraviolet (UV) radiation damage (Apel and Hirt, 2004; Torres and Dangl, 2005; Ferreyra et al., 2012; Choudhury et al., 2017; Song et al., 2022; Sugimoto et al., 2022). Recent evidence indicates that flavonoids, a class of antioxidants, are capable of scavenging the overproduced ROS and alleviating oxidative injury (Pi et al., 2016; Pi et al., 2018; Pi et al., 2019; Yu et al., 2020; Qian et al., 2021; Wu et al., 2022b).
6.1 Regulation of flavonoid biosynthesis in plants
Flavonoids encompass several subclasses, including anthocyanins, proanthocyanidins, flavones, flavanols, flavonols, flavanones and isoflavones (Panche et al., 2016; Durazzo et al., 2019; Luo et al., 2019). More than 10, 000 diverse variants are generated by glycosylation and other modifications (Weston and Mathesius, 2013). Anthocyanins in particular confer bright colors to plant tissues and also act as antioxidants. Flavonoid biosynthesis proceeds through a branched pathway catalyzed by sequential enzymatic reactions. The biosynthesis of flavonoid is regulated by an intricate transcriptional network. Distinct WRKYs can integrate in various ways and exert different regulatory effects on flavonoid synthesis (Table 3; Figure 3).
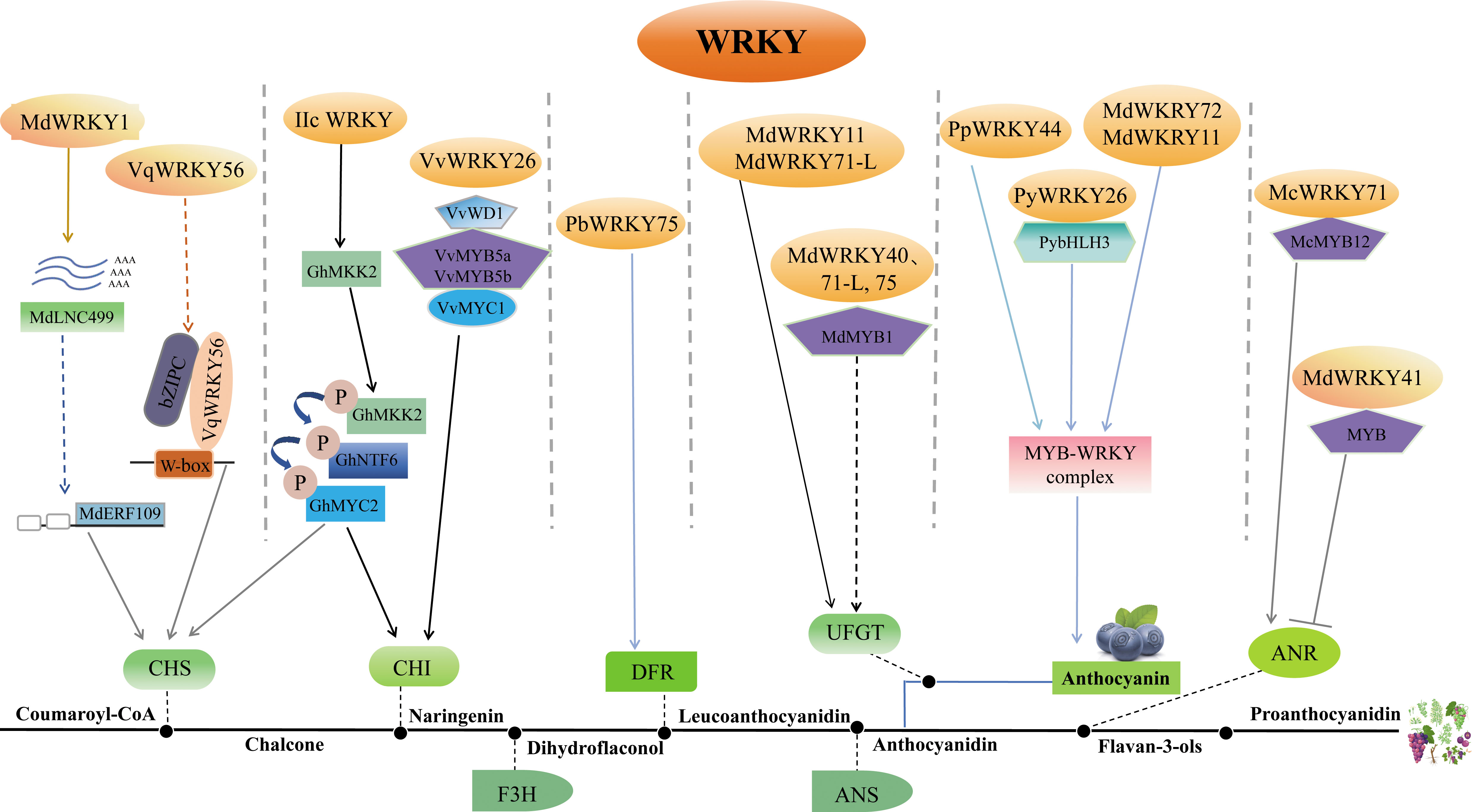
Figure 3 Depiction of the role of WRKY in the biosynthesis of flavonoid. The complex role of WRKY in regulating flavonoid biosynthesis highlights its crucial function in this pathway and subsequent stress responses in plants. The WRKY protein modulates flavonoid biosynthesis by controlling the expression of flavonoid biosynthesis genes in various ways. MdWRKY1 and VqWRKY56 can influence the transcription of CHS, while VvWRKY26 can control CHI expression by forming the MBWW complex. PbWRKY75 might enhance the expression of DFR. MdWRKY11, MdWRKY71-L, MdWRKY40, and MdWRKY75 can directly or indirectly control the expression of UFGT. MdWRKY72, MdWRKY11, PpWRKY44, and PyWRKY26 can impact anthocyanin synthesis by forming the MYB-WRKY complex. In addition to positive regulation, MdWRKY41 also has a role in negatively regulating ANR synthesis. All these pathways influence the synthesis of anthocyanin or proanthocyanidin ultimately. Arrows represent positive regulation, and T-bars represent negative regulation. CHS, chalcone synthase; CHI, chalcone isomerase; F3H: flavanone 3-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; ANR, anthocyanin reductase; UFGT, flavonoid 3-O-glycosyl-transferase.
WRKYs directly regulate flavonoid synthesis by transactivation of related enzyme genes. In apple, MdWRKY11 regulates anthocyanin synthesis through directly binding to the flavonoid 3-O-glycosyl-transferase (UFGT) promoter (Liu et al., 2019a); MdWRKY40 binds anthocyanidin synthase (MdANS) promoter (Zhang et al., 2019); and MdWRKY41 negatively regulates anthocyanin and proanthocyanidins biosynthesis by binding W-boxes of apple anthocyanin reductase (MdANR), MdUFGT, myb avian myeloblastosis viral oncogene homolog 12 (MdMYB12) (Mao et al., 2021); MdWRKY71-L targets MdUFGT (Su et al., 2022a). In crabapple (Malus crabapple), McWRKY71 controls McANR and proanthocyanidin synthesis (Zhang et al., 2022b). In grape, VqWRKY56 binds chalcone synthase 3 (VvCHS3), leucine anthocyanin reductase1 (VvLAR1) and VvANR, inducing proanthocyanidins (Wang et al., 2023d). In pear, PyMYB10 activates pear anthocyanin structural gene (Feng et al., 2010). PpWRKY44 activates PpMYB10 by binding to its promoter for light-induced anthocyanin accumulation (Alabd et al., 2022). PbWRKY75 has shown to promote anthocyanin synthesis in pear by binding the promoters of dihydroflavonol 4-reductase (PbDFR) and PbUFGT (Cong et al., 2021). FaWRKY71 stimulates anthocyanin accumulation in strawberry (Fragaria×ananassa) by upregulating genes in the synthetic pathway [flavonoid 3’-hydroxylase (FaF3’H), FaLAR, FaANR, anthocyanin transporter genes transparent testa 19 (FaTT19) and transparent testa 12 (FaTT12)] (Yue et al., 2022).
WRKYs also influence flavonoid biosynthesis indirectly through modulation of other regulators. In apple, MdWRKY1 activates a long noncoding RNA (MdLNC499) and MDERF109 expression, which in turn increases anthocyanin accumulation by inducing the expression of MdUFGT, MdCHS and basic helix-loop-helix 3 (MdbHLH3) (Ma et al., 2021a); MdWRKY71-L regulates anthocyanin synthesis via the ELONGATED HYPOCOTYL 5 (MdHY5)-MdMYB1 cascade; MdWRKY40 forms homodimers that bound two W-boxes in MdANS promoters, mitigating MdMYB111 inhibition of MdANS (Zhang et al., 2019); MdWRKY75 stimulates anthocyanins by associating with the MdMYB1 promoter (Su et al., 2022a); MdWRKY72 binds MdHY5 and MdMYB1 (Hu et al., 2020); and MdWRKY11 binds with MdHY5 (Liu et al., 2019a). In pear, PpWRKY44 upregulates PpMYB10 to stimulate anthocyanins (Alabd et al., 2022); PyWRKY26 in conjunction with PybHLH3 targets the PyMYB114 promoter, thus affecting anthocyanins accumulation (Li et al., 2020a). In Arabidopsis, the WRKY41 mutation heightens anthocyanin levels, indicating AtWRKY41 represses anthocyanin synthesis by regulating AtMYB75, AtMYB111, AtMYBD, AT1G68440 and AtGSTF12 (Duan et al., 2018). In cotton, Group IIc WRKYs induce flavonoids by controlling protein kinase kinase 2 (GhMKK2), a signaling kinase (Wang et al., 2022). In crabapple, McWRKY71 regulates proanthocyanidin synthesis by interacting with McMYB12 (Zhang et al., 2022b).
Furthermore, WRKY forms transcriptional complexes with other transcription factors. MYB, bHLH and WD40 compose the MBW complex to modulate PA synthesis and anthocyanin (Nesi et al., 2001; Ramsay and Glover, 2005; Gou et al., 2011). WRKYs participate in regulating the MBW complex. For example, VvWRKY26 is absorbed into the MBWW complex by VvMYB5a to regulate flavonoid hydroxylation in grape (Amato et al., 2017; Amato et al., 2019).
Thus, the biosynthesis of flavonoids is regulated by a complex transcriptional network. WRKYs utilize various strategies, such as directly binding to target promoters, interacting with other regulators, or forming transcriptional complexes, to regulate flavonoid biosynthesis. The combined efforts of multiple WRKYs and the interactions with other transcription factors allow for precise control over this metabolic pathway.
6.2 Elevation of plant stress tolerances through flavonoid synthesis
WRKYs play pivotal roles in modulating the synthesis of various flavonoid for regulating plant responses to various abiotic stresses caused by UV-B, O3, and wounding, etc. UV-B-induced apple MdWRKY71-L promotes in apple anthocyanin accumulation by directly activating MdUFGT and MdMYB1 (Su et al., 2022a). When exposed to UV-B radiation in the transgenic calli, MdWRKY72 directly control anthocyanin synthesis via promoting MdMYB1, or indirectly regulates by interacting with MdMYB16 (Hu et al., 2020; Mao et al., 2021). In addition, McWRKY71 directly binds to the McANR, thus regulate the PA biosynthesis in regard to O3 stress in crabapple (Zhang et al., 2022b). Moreover, MdWRKY40, interacting with MdMYB1, enhance the activation of target genes in reaction to injuries (An et al., 2019).
Apart from abiotic stresses, WRKY also partakes in biotic stresses by regulating the biosynthesis of flavonoids. For example, expression of VqWRKY56 activates PA biosynthesis genes (VvCHS3, VvLAR1 and VvANR) (Wang et al., 2023d). The upregulation of GhMYC2 by group IIc WRKYs induced GhMKK2-GhNTF6 signaling and increased cotton resistance to Fusarium oxysporum via enhancing flavonoid biosynthesis. This demonstrates a novel defense mechanism mediated by WRKY-MAPK-regulated flavonoid biosynthesis against pathogen infection in cotton (Wang et al., 2022).
In short, WRKYs participate in almost all stages of flavonoid synthesis and regulate flavonoid synthesis genes through diverse mechanisms. However, the contribution of WRKY-mediated flavonoid synthesis to plant tolerance has been poorly characterized.
7 Conclusion and perspective
In this review, we optimized existing WRKY phylogenetic trees and tried to deduce unifying themes of distinct WRKY subfamilies governing specific stress responses and flavonoid metabolism.
Analysis of documented data reveals WRKY members across all subgroups participate in flavonoid synthesis. However, WRKYs regulating salt tolerance mainly belong to subgroups 2a, 2c and 3. Given their shared protein motifs (Tables 2, 3; Figure 1), it is reasonable to hypothesize these WRKY subgroups promote flavonoid accumulation to enhance plant salinity tolerance. Similar hypotheses could be proposed for WRKY homologs tuning flavonoids to elevate plant resistance to cold, drought and nutrient deficiency. However, only a handful of studies have demonstrated direct relationships between WRKY-modulated flavonoid synthesis and stress tolerance. Perhaps the significant contribution of flavonoids has been overlooked in analyzing stress resistance, and roles of the WRKY-flavonoid interplay in plant stress tolerance deserve greater attention in future work. Nonetheless, laboratory validation remains necessary to verify these hypotheses.
On the other hand, research on WRKY-mediated flavonoid regulation has focused on few species like apple, Arabidopsis, grapevine and crabapple. Could WRKY regulate flavonoid synthesis in other species via distinct pathways? Molecular mechanisms underlying flavonoid-enhanced plant stress resilience remain largely unclear. Particularly, flavonoid regulation by WRKY transcription factors and subsequent impacts on plant stress response warrant deeper exploration.
Further research is imperative to elucidate the complex crosstalk between flavonoid metabolism and stress signaling cascades. In subsequent studies, integrating transcriptomic and metabolomic analyses could prove insightful. Transcriptomics can provide comprehensive information about WRKY transcription factors and identify flavonoid pathway target genes. Metabolomics can directly assess functional outcomes of WRKY-mediated flavonoid regulation. Together, these efforts will uncover valuable knowledge for engineering flavonoid pathways to improve multiple stress tolerance in economically important crops.
Author contributions
JZ: Writing – original draft. HZ: Writing – original draft. LC: Writing – original draft. JL: Writing – review & editing. ZW: Writing – original draft. JP: Writing – original draft. FY: Writing – original draft. XN: Writing – review & editing. YW: Writing – review & editing. YHW: Writing – review & editing. RL: Writing – review & editing. EP: Funding acquisition, Resources, Writing – original draft. SW: Funding acquisition, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Science Foundation of China (grant no. 31970286), the Natural Science Foundation of Zhejiang Province (grants no. LY22C020001 and HZSZ24C010001), A Project Supported by Scientific Research Fund of Zhejiang Provincial Education Department (grant no. Y202351641).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1303667/full#supplementary-material
References
Adachi, H., Nakano, T., Miyagawa, N., Ishihama, N., Yoshioka, M., Katou, Y., et al. (2015). WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell. 27, 2645–2663. doi: 10.1105/tpc.15.00213
Ahammed, G. J., Li, X., Wan, H. J., Zhou, G. Z., Cheng, Y. (2020a). SlWRKY81 reduces drought tolerance by attenuating proline biosynthesis in tomato. Sci. Hortic. 270, 109444. doi: 10.1016/j.scienta.2020.109444
Ahammed, G. J., Li, X., Yang, Y. X., Liu, C. C., Zhou, G. Z., Wan, H. J., et al. (2020b). Tomato WRKY81 acts as a negative regulator for drought tolerance by modulating guard cell H2O2- mediated stomatal closure. Environ. Exp. Bot. 171, 103960. doi: 10.1016/j.envexpbot.2019.103960
Alabd, A., Ahmad, M., Zhang, X., Gao, Y. H., Peng, L., Zhang, L., et al. (2022). Light-responsive transcription factor PpWRKY44 induces anthocyanin accumulation by regulating PpMYB10 expression in pear. Hortic. Res. 9, uhac199. doi: 10.1093/hr/uhac199
Ali, M. A., Azeem, F., Nawaz, M. A., Acet, T., Abbas, A., Imran, Q. M., et al. (2018). Transcription factors WRKY11 and WRKY17 are involved in abiotic stress responses in Arabidopsis. J. Plant Physiol. 226, 12–21. doi: 10.1016/j.jplph.2018.04.007
Amato, A., Cavallini, E., Walker, A. R., Pezzotti, M., Bliek, M., Quattrocchio, F., et al. (2019). The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J. 99, 1220–1241. doi: 10.1111/tpj.14419
Amato, A., Cavallini, E., Zenoni, S., Finezzo, L., Begheldo, M., Ruperti, B., et al. (2017). A grapevine TTG2-like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01979
An, J. P., Zhang, X. W., You, C. X., Bi, S. Q., Wang, X. F., Hao, Y. J. (2019). MdWRKY40 promotes wounding-induced anthocyanin biosynthesis in association with MdMYB1 and undergoes MdBT2-mediated degradation. New Phytol. 224, 380–395. doi: 10.1111/nph.16008
Andaya, V. C., Mackill, D. J. (2003). Mapping of QTLs associated with cold tolerance during the vegetative stage in rice. J. Exp. Bot. 54, 2579–2585. doi: 10.1093/jxb/erg243
Apel, K., Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Arndt, L. C., Heine, S., Wendt, L., Wegele, E., Schomerus, J. T., Schulze, J., et al. (2022). Genomic distribution and context dependent functionality of novel WRKY transcription factor binding sites. BMC Genomics 23, 673. doi: 10.1186/s12864-022-08877-y
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W.-L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 415, 977–983. doi: 10.1038/415977a
Ay, N., Irmler, K., Fischer, A., Uhlemann, R., Reuter, G., Humbeck, K. (2009). Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 58, 333–346. doi: 10.1111/j.0960-7412.2009.03782.x
Bakshi, M., Oelmüller, R. (2014). WRKY transcription factors. Plant Signal Behav. 9, 27700. doi: 10.4161/psb.27700
Barry, D. A. J. (1988). Phosphorus nutrition of maize for maximum yield: Seedling requirement and diagnosis using ear leaf and seedling samples. Agron. J. 81, 95–99. doi: 10.2134/agronj1989.00021962008100010017x
Besseau, S., Li, J., Palva, E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 63, 2667–2679. doi: 10.1093/jxb/err450
Birkenbihl, R. P., Kracher, B., Somssich, I. E. (2017). Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell. 29, 20–38. doi: 10.1105/tpc.16.00681
Bo, C., Cai, R. H., Fang, X., Wu, H., Ma, Z. X., Yuan, H. T., et al. (2022). Transcription factor ZmWRKY20 interacts with ZmWRKY115 to repress expression of ZmbZIP111 for salt tolerance in maize. Plant J. 111, 1660–1675. doi: 10.1111/tpj.15914
Cai, J. S., Cai, W. W., Huang, X. Y., Yang, S., Wen, J. Y., Xia, X. Q., et al. (2021). Ca14-3-3 interacts with CaWRKY58 to positively modulate pepper response to low-phosphorus starvation. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.607878
Cai, H. Y., Yang, S., Yan, Y., Xiao, Z. L., Cheng, J. B., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. doi: 10.1093/jxb/erv125
Chen, C. H., Chen, X. Q., Han, J., Lu, W. L., Ren, Z. H. (2020). Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 20, 443. doi: 10.1186/s12870-020-02625-8
Chen, H., Lai, Z. B., Shi, J. W., Xiao, Y., Chen, Z. X., Xu, X. P. (2010). Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10, 281. doi: 10.1186/1471-2229-10-281
Chen, X. J., Li, C., Wang, H., Guo, Z. J. (2019). WRKY transcription factors: evolution, binding, and action. Phytopathol. Res. 1, 13. doi: 10.1186/s42483-019-0022-x
Chen, Y. F., Li, L. Q., Xu, Q., Kong, Y. H., Wang, H., Wu, W. H. (2009). The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low pi stress in Arabidopsis. Plant Cell. 21, 3554–3566. doi: 10.1105/tpc.108.064980
Chen, J. N., Nolan, T. M., Ye, H. X., Zhang, M. C., Tong, H. N., Xin, P. Y., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell. 29, 1425–1439. doi: 10.1105/tpc.17.00364
Chen, N. N., Tong, S. F., Yang, J., Qin, J. J., Wang, W. W., Chen, K., et al. (2022). PtoWRKY40 interacts with PtoPHR1-LIKE3 while regulating the phosphate starvation response in poplar. Plant Physiol. 190, 2688–2705. doi: 10.1093/plphys/kiac404
Chen, L. G., Zhang, L. P., Li, D. B., Wang, F., Yu, D. Q. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, E1963–E1971. doi: 10.1073/pnas.1221347110
Cheng, Z. Y., Luan, Y. T., Meng, J. S., Sun, J., Tao, J., Zhao, D. Q. (2021). WRKY transcription factor response to high-temperature stress. Plants. 10, 2211. doi: 10.3390/plants10102211
Cheng, X. K., Zhao, Y. X., Jiang, Q. S., Yang, J., Zhao, W. S., Taylor, I. A., et al. (2019). Structural basis of dimerization and dual W-box DNA recognition by rice WRKY domain. Nucleic Acids Res. 47, 4308–4318. doi: 10.1093/nar/gkz113
Chi, Y. J., Yang, Y., Zhou, Y., Zhou, J., Fan, B. F., Yu, J. Q., et al. (2013). Protein-protein interactions in the regulation of WRKY transcription factors. Mol. Plant 6, 287–300. doi: 10.1093/mp/sst026
Choudhury, F. K., Rivero, R. M., Blumwald, E., Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. doi: 10.1111/tpj.13299
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Cong, L., Qu, Y. Y., Sha, G. Y., Zhang, S. C., Ma, Y. F., Chen, M., et al. (2021). PbWRKY75 promotes anthocyanin synthesis by activating PbDFR, PbUFGT, and PbMYB10b in pear. Physiol. Plant 173, 1841–1849. doi: 10.1111/ppl.13525
Dai, X. Y., Wang, Y. Y., Zhang, W. H. (2016). OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. doi: 10.1093/jxb/erv515
Dang, F. F., Wang, Y. N., Yu, L., Eulgem, T., Lai, Y., Liu, Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. doi: 10.1111/pce.12011
Di, D. W., Sun, L., Wang, M., Wu, J. J., Kronzucker, H. J., Fang, S., et al. (2021). WRKY46 promotes ammonium tolerance in Arabidopsis by repressing NUDX9 and indole-3-acetic acid-conjugating genes and by inhibiting ammonium efflux in the root elongation zone. New Phytol. 232, 190–207. doi: 10.1111/nph.17554
Ding, Y. L., Shi, Y. T., Yang, S. H. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Ding, Z. J., Yan, J. Y., Li, C. X., Li, G. X., Wu, Y. R., Zheng, S. J. (2015). Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 84, 56–69. doi: 10.1111/tpj.12958
Ding, Y. L., Yang, S. H. (2022). Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell. 57, 947–958. doi: 10.1016/j.devcel.2022.03.010
Duan, M. R., Nan, J., Liang, Y. H., Mao, P., Lu, L., Li, L. F., et al. (2007). DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res. 35, 1145–1154. doi: 10.1093/nar/gkm001
Duan, S. W., Wang, J. J., Gao, C. H., Jin, C. Y., Li, D., Peng, D. S., et al. (2018). Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 268, 47–53. doi: 10.1016/j.plantsci.2017.12.010
Durazzo, A., Lucarini, M., Souto, E. B., Cicala, C., Caiazzo, E., Izzo, A. A., et al. (2019). Polyphenols: A concise overview on the chemistry, occurrence, and human health. Phytother. Res. 33, 2221–2243. doi: 10.1002/ptr.6419
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/s1360-1385(00)01600-9
Fan, X. Q., Guo, Q., Xu, P., Gong, Y. Y., Shu, H. M., Yang, Y., et al. (2015). Transcriptome-wide identification of salt-responsive members of the WRKY gene family in Gossypium aridum. PloS One 10, 126148. doi: 10.1371/journal.pone.0126148
Feng, Y. N., Cui, R., Wang, S. L., He, M. L., Hua, Y. P., Shi, L., et al. (2020). Transcription factor BnaA9.WRKY47 contributes to the adaptation of Brassica napus to low boron stress by up-regulating the boric acid channel gene BnaA3.NIP5;1. Plant Biotechnol. J. 18, 1241–1254. doi: 10.1111/pbi.13288
Feng, S. Q., Wang, Y. L., Yang, S., Xu, Y. T., Chen, X. S. (2010). Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta. 232, 245–255. doi: 10.1007/s00425-010-1170-5
Ferreyra, M. L. F., Rius, S. P., Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00222
Finkelstein, R. R., Gampala, S. S. L., Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell. 14 (Suppl), S15–S45. doi: 10.1105/tpc.010441
Gou, J. Y., Felippes, F. F., Liu, C. J., Weigel, D., Wang, J. W. (2011). Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 23, 1512–1522. doi: 10.1105/tpc.111.084525
Gu, L. J., Ma, Q., Zhang, C., Wang, C. C., Wei, H. L., Wang, H. T., et al. (2019). The cotton GhWRKY91 transcription factor mediates leaf senescence and responses to drought stress in transgenic Arabidopsis thaliana. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01352
Hasegawa, P. M., Bressan, R. A., Zhu, J.-K., Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. doi: 10.1146/annurev.arplant.51.1.463
He, H. S., Dong, Q., Shao, Y. H., Jiang, H. Y., Zhu, S. W., Cheng, B. J., et al. (2012). Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 31, 1199–1217. doi: 10.1007/s00299-012-1241-0
He, G. H., Xu, J. Y., Wang, Y. X., Liu, J. M., Li, P. S., Chen, M., et al. (2016). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 16, 116. doi: 10.1186/s12870-016-0806-4
Hsu, F. C., Chou, M. Y., Chou, S. J., Li, Y. R., Peng, H. P., Shih, M. C. (2013). Submergence confers immunity mediated by the WRKY22 transcription factor in Arabidopsis. Plant Cell. 25, 2699–2713. doi: 10.1105/tpc.113.114447
Hu, Y. R., Chen, L. G., Wang, H. P., Zhang, L. P., Wang, F., Yu, D. Q. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745. doi: 10.1111/tpj.12159
Hu, J. F., Fang, H. C., Wang, J., Yue, X. X., Su, M. Y., Mao, Z. L., et al. (2020). Ultraviolet B-induced MdWRKY72 expression promotes anthocyanin synthesis in apple. Plant Sci. 292, 110377. doi: 10.1016/j.plantsci.2019.110377
Huang, X. B., Cao, L. W., Fan, J. B., Ma, G. J., Chen, L. (2022b). CdWRKY2-mediated sucrose biosynthesis and CBF-signalling pathways coordinately contribute to cold tolerance in Bermudagrass. Plant Biotechnol. J. 20, 660–675. doi: 10.1111/pbi.13745
Huang, J. J., Liu, F. H., Chao, D., Xin, B. N., Liu, K., Cao, S. L., et al. (2022a). The WRKY transcription factor OsWRKY54 is involved in salt tolerance in rice. Int. J. Mol. Sci. 23, 11999. doi: 10.3390/ijms231911999
Huang, K., Wu, T., Ma, Z. M., Li, Z., Chen, H. Y., Zhang, M. X., et al. (2021). Rice transcription factor OsWRKY55 is involved in the drought response and regulation of plant growth. Int. J. Mol. Sci. 22, 4337. doi: 10.3390/ijms22094337
Hussain, A., Li, X., Weng, Y. H., Liu, Z. Q., Ashraf, M. F., Noman, A., et al. (2018). CaWRKY22 acts as a positive regulator in pepper response to Ralstonia Solanacearum by constituting networks with CaWRKY6, CaWRKY27, CaWRKY40, and CaWRKY58. Int. J. Mol. Sci. 19, 1426. doi: 10.3390/ijms19051426
Ishiguro, S., Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5' upstream regions of genes coding for sporamin and beta-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. doi: 10.1007/BF00282746
Ishihama, N., Yamada, R., Yoshioka, M., Katou, S., Yoshioka, H. (2011). Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell. 23, 1153–1170. doi: 10.1105/tpc.110.081794
Ishitani, M., Xiong, L., Stevenson, B., Zhu, J. K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 9, 1935–1949. doi: 10.1105/tpc.9.11.1935
Jeyasri, R., Muthuramalingam, P., Satish, L., Adarshan, S., Lakshmi, M. A., Pandian, S. K., et al. (2021). The role of OsWRKY genes in rice when faced with single and multiple abiotic stresses. Agronomy. 11, 1301. doi: 10.3390/agronomy11071301
Jiang, Y. Q., Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Jiang, Y. Z., Tong, S. F., Chen, N. N., Liu, B., Bai, Q. X., Chen, Y., et al. (2021). The PalWRKY77 transcription factor negatively regulates salt tolerance and abscisic acid signaling in Populus. Plant J. 105, 1258–1273. doi: 10.1111/tpj.15109
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature. 444, 323–329. doi: 10.1038/nature05286
Khoso, M. A., Hussain, A., Ritonga, F. N., Ali, Q., Channa, M. M., Alshegaihi, R. M., et al. (2022). WRKY transcription factors (TFs): Molecular switches to regulate drought, temperature, and salinity stresses in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1039329
Kim, K. C., Lai, Z. B., Fan, B. F., Chen, Z. X. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 20, 2357–2371. doi: 10.1105/tpc.107.055566
Krueger, R. W., Lovatt, C. J., Albert, L. S. (1987). Metabolic requirement of Cucurbita pepo for boron. Plant Physiol. 83, 254–258. doi: 10.1104/pp.83.2.254
Lauer, M. J., Blevins, D. G., Sierzputowska-Gracz, H. (1989). P-nuclear magnetic resonance determination of phosphate compartmentation in leaves of reproductive soybeans (Glycine max L.) as affected by phosphate nutrition. Plant Physiol. 89, 1331–1336. doi: 10.1104/pp.89.4.1331
Lee, H., Cha, J., Choi, C., Choi, N., Ji, H. S., Park, S. R., et al. (2018). Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice. 11, 5. doi: 10.1186/s12284-018-0199-0
Lee, S., Choi, H., Suh, S., Doo, I. S., Oh, K. Y., Choi, E. J., et al. (1999). Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of tomato and Commelina communis. Plant Physiol. 121, 147–152. doi: 10.1104/pp.121.1.147
Lei, R. H., Li, X. L., Ma, Z. B., Lv, Y., Hu, Y. R., Yu, D. Q. (2017). Arabidopsis WRKY2 and WRKY34 transcription factors interact with VQ20 protein to modulate pollen development and function. Plant J. 91, 962–976. doi: 10.1111/tpj.13619
Letunic, I., Bork, P. (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23, 127–128. doi: 10.1093/bioinformatics/btl529
Li, J., Besseau, S., Törönen, P., Sipari, N., Kollist, H., Holm, L., et al. (2013). Defense-related transcription factors WRKY70 and WRKY54 modulate osmotic stress tolerance by regulating stomatal aperture in Arabidopsis. New Phytol. 200, 455–472. doi: 10.1111/nph.12378
Li, S. J., Fu, Q. T., Chen, L. G., Huang, W. D., Yu, D. Q. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Li, S. J., Fu, Q. T., Huang, W. D., Yu, D. Q. (2009). Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 28, 683–693. doi: 10.1007/s00299-008-0666-y
Li, J. J., Islam, F., Huang, Q., Wang, J., Zhou, W. J., Xu, L., et al. (2020b). Genome-wide characterization of WRKY gene family in Helianthus annuus L. and their expression profiles under biotic and abiotic stresses. PloS One 15, 241965. doi: 10.1371/journal.pone.0241965
Li, W. X., Pang, S. Y., Lu, Z. G., Jin, B. (2020c). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants. 9, 1515. doi: 10.3390/plants9111515
Li, C., Wu, J., Hu, K. D., Wei, S. W., Sun, H. Y., Hu, L. Y., et al. (2020a). PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 7, 37. doi: 10.1038/s41438-020-0254-z
Li, H. E., Xu, Y., Xiao, Y., Zhu, Z. G., Xie, X. Q., Zhao, H. Q., et al. (2010). Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta. 232, 1325–1337. doi: 10.1007/s00425-010-1258-y
Lim, C., Kang, K., Shim, Y., Yoo, S. C., Paek, N. C. (2022). Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 188, 1900–1916. doi: 10.1093/plphys/kiab492
Lin, L. K., Yuan, K. L., Huang, Y. D., Dong, H. Z., Qiao, Q. H., Xing, C. H., et al. (2022). A WRKY transcription factor PbWRKY40 from Pyrus betulaefolia functions positively in salt tolerance and modulating organic acid accumulation by regulating PbVHA-B1 expression. Environ. Exp. Bot. 196, 104782. doi: 10.1016/j.envexpbot.2022.104782
Liu, X. Q., Bai, X. Q., Wang, X. J., Chu, C. C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979. doi: 10.1016/j.jplph.2006.07.006
Liu, G. Y., Li, B., Li, X., Wei, Y. X., He, C. Z., Shi, H. T. (2020). MaWRKY80 positively regulates plant drought stress resistance through modulation of abscisic acid and redox metabolism. Plant Physiol. Biochem. 156, 155–166. doi: 10.1016/j.plaphy.2020.09.015
Liu, W. J., Wang, Y. C., Yu, L., Jiang, H. Y., Guo, Z. W., Xu, H. F., et al. (2019a). MdWRKY11 participates in anthocyanin accumulation in red-fleshed apples by affecting MYB transcription factors and the photoresponse factor MdHY5. J. Agric. Food Chem. 67, 8783–8793. doi: 10.1021/acs.jafc.9b02920
Liu, Y., Yang, T. Y., Lin, Z. K., Gu, B. J., Xing, C. H., Zhao, L. Y., et al. (2019b). A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 17, 1770–1787. doi: 10.1111/pbi.13099
Luo, X., Bai, X., Sun, X. L., Zhu, D., Liu, B. H., Ji, W., et al. (2013). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64, 2155–2169. doi: 10.1093/jxb/ert073
Luo, F., Cai, J. H., Kong, X. M., Zhou, Q., Zhou, X., Zhao, Y. B., et al. (2019). Transcriptome profiling reveals the roles of pigment mechanisms in postharvest broccoli yellowing. Hortic. Res. 6, 74. doi: 10.1038/s41438-019-0155-1
Ma, J. H., Gao, X. L., Liu, Q., Shao, Y., Zhang, D. J., Jiang, L. N., et al. (2017). Overexpression of TaWRKY146 increases drought tolerance through inducing stomatal closure in Arabidopsis thaliana. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02036
Ma, J. L., Li, C. H., Sun, L. L., Ma, X. C., Qiao, H., Zhao, W. C., et al. (2023). The SlWRKY57-SlVQ21/SlVQ16 module regulates salt stress in tomato. J. Integr. Plant Biol. 65, 2437–2455. doi: 10.1111/jipb.13562
Ma, K. W., Niu, Y. L., Jia, Y., Ordon, J., Copeland, C., Emonet, A., et al. (2021b). Coordination of microbe-host homeostasis by crosstalk with plant innate immunity. Nat. Plants. 7, 814–825. doi: 10.1038/s41477-021-00920-2
Ma, Y., Xue, H., Zhang, F., Jiang, Q., Yang, S., Yue, P. T., et al. (2021c). The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 19, 311–323. doi: 10.1111/pbi.13464
Ma, H. Y., Yang, T., Li, Y., Zhang, J., Wu, T., Song, T. T., et al. (2021a). The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. Plant Cell. 33, 3309–3330. doi: 10.1093/plcell/koab188
Machens, F., Becker, M., Umrath, F., Hehl, R. (2014). Identification of a novel type of WRKY transcription factor binding site in elicitor-responsive cis-sequences from Arabidopsis thaliana. Plant Mol. Biol. 84, 371–385. doi: 10.1007/s11103-013-0136-y
Mao, Z. L., Jiang, H. Y., Wang, S., Wang, Y. C., Yu, L., Zou, Q., et al. (2021). The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 306, 110848. doi: 10.1016/j.plantsci.2021.110848
Mare, C., Mazzucotelli, E., Crosatti, C., Francia, E., Stanca, A. M., Cattivelli, L. (2004). Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Mol. Biol. 55, 399–416. doi: 10.1007/s11103-004-0906-7
Miao, Y., Laun, T. M., Smykowski, A., Zentgraf, U. (2007). Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol. Biol. 65, 63–76. doi: 10.1007/s11103-007-9198-z
Miao, Y., Laun, T., Zimmermann, P., Zentgraf, U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55, 853–867. doi: 10.1007/s11103-005-2142-1
Nan, H., Li, W., Lin, Y. L., Gao, L. Z. (2020). Genome-wide analysis of wrky genes and their response to salt stress in the wild progenitor of Asian cultivated rice, Oryza rufipogon. Front. Genet. 11. doi: 10.3389/fgene.2020.00359
Nesi, N., Jond, C., Debeaujon, I., Caboche, M., Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 13, 2099–2114. doi: 10.1105/tpc.010098
Ng, C. K., Carr, K., McAinsh, M. R., Powell, B., Hetherington, A. M. (2001). Drought induced guard cell signal transduction involves sphingosine 1 phosphate. Nat 410, 596–599. doi: 10.1038/35069092
Oztur, Z. N., Talame, V., Deyholos, M., Michalowski, C. B., Galbraith, D. W., Gozukirmizi, N., et al. (2002). Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol. Biol. 48, 551–573. doi: 10.1023/a:1014875215580
Pan, Y. J., Cho, C. C., Kao, Y. Y., Sun, C. H. (2009). A novel WRKY-like protein involved in transcriptional activation of cyst wall protein genes in Giardia lamblia. J. Biol. Chem. 284, 17975–17988. doi: 10.1074/jbc.M109.012047
Panche, A. N., Diwan, A. D., Chandra, S. R. (2016). Flavonoids: an overview. J. Nutr. Sci. 5, e47. doi: 10.1017/jns.2016.41
Pandey, B., Grover, A., Sharma, P. (2018). Molecular dynamics simulations revealed structural differences among WRKY domain-DNA interaction in barley (Hordeum vulgare). BMC Genomics 19, 132. doi: 10.1186/s12864-018-4506-3
Pandey, S. P., Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Phukan, U. J., Jeena, G. S., Tripathi, V., Shukla, R. K. (2017). Regulation of Apetala2/Ethylene response factors in plants. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00150
Pi, E. X., Qu, L. Q., Hu, J. W., Huang, Y. Y., Qiu, L. J., Lu, H., et al. (2016). Mechanisms of soybean roots' tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol. Cell. Proteomics. 15, 266–288. doi: 10.1074/mcp.M115.051961
Pi, E. X., Xu, J., Li, H. H., Fan, W., Zhu, C. M., Zhang, T. Y., et al. (2019). Enhanced salt tolerance of rhizobia-inoculated soybean correlates with decreased phosphorylation of the transcription factor GmMYB183 and altered flavonoid biosynthesis. Mol. Cell. Proteomics. 18, 2225–2243. doi: 10.1074/mcp.RA119.001704
Pi, E. X., Zhu, C. M., Fan, W., Huang, Y. Y., Qu, L. Q., Li, Y. Y., et al. (2018). Quantitative phosphoproteomic and metabolomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt stress. Mol. Cell. Proteomics. 17, 1209–1224. doi: 10.1074/mcp.RA117.000417
Qian, Y. C., Zhang, T. Y., Yu, Y., Gou, L. P., Yang, J. T., Xu, J., et al. (2021). Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses. Front. Plant Sci. 12, 677611. doi: 10.3389/fpls.2021.677611
Qin, W. Q., Wang, N., Yin, Q., Li, H. L., Wu, A. M., Qin, G. J. (2022). Activation tagging identifies WRKY14 as a repressor of plant thermomorphogenesis in Arabidopsis. Mol. Plant 15, 1725–1743. doi: 10.1016/j.molp.2022.09.018
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221. doi: 10.1038/emboj.2008.147
Qiu, Y. P., Yu, D. Q. (2009). Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ. Exp. Bot. 65, 35–47. doi: 10.1016/j.envexpbot.2008.07.002
Rahman, M. M., Rahman, M. M., Eom, J. S., Jeon, J. S. (2021). Genome-wide identification, expression profiling and promoter analysis of trehalose-6-phosphate phosphatase gene family in rice. J. Plant Biol. 64, 55–71. doi: 10.1007/s12374-020-09279-x
Ramsay, N. A., Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Ritonga, F. N., Ngatia, J. N., Wang, Y. R., Khoso, M. A., Farooq, U., Chen, S. (2021). AP2/ERF, an important cold stress-related transcription factor family in plants: A review. Physiol. Mol. Biol. Plants. 27, 1953–1968. doi: 10.1007/s12298-021-01061-8
Robatzek, S., Somssich, I. E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16, 1139–1149. doi: 10.1101/gad.222702
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. X. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Schluttenhofer, C., Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. doi: 10.1104/pp.114.251769
Shan, D. Q., Wang, C. Y., Song, H. D., Bai, Y. X., Zhang, H. X., Hu, Z. H., et al. (2021). The MdMEK2-MdMPK6-MdWRKY17 pathway stabilizes chlorophyll levels by directly regulating MdSUFB in apple under drought stress. Plant J. 108, 814–828. doi: 10.1111/tpj.15480
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase h subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shen, Q. Q., Wang, T. J., Wang, J. G., He, L. L., Zhao, T. T., Zhao, X. T., et al. (2023). The SsWRKY1 transcription factor of Saccharum spontaneum enhances drought tolerance in transgenic Arabidopsis thaliana and interacts with 21 potential proteins to regulate drought tolerance in S. spontaneum. Plant Physiol. Biochem. 199, 107706. doi: 10.1016/j.plaphy.2023.107706
Shende, R. T., Singh, R., Kumar, A., Sengar, R. S. (2022). Genome-wide analysis of WRKY transcription factors family in chickpea (Cicer arietinum L.). Legume. Res. 45, 700–710. doi: 10.18805/Lr-4352
Shinozaki, K., Yamaguchi-Shinozaki, K., Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/s1369-5266(03)00092-x
Song, Y., Ma, B., Guo, Q. X., Zhou, L. X., Lv, C. Y., Liu, X. M., et al. (2022). UV-B induces the expression of flavonoid biosynthetic pathways in blueberry (Vaccinium corymbosum) calli. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1079087
Song, H., Sun, W. H., Yang, G. F., Sun, J. (2018). WRKY transcription factors in legumes. BMC Plant Biol. 18, 243. doi: 10.1186/s12870-018-1467-2
Sood, M., Kapoor, D., Kumar, V., Kalia, N., Bhardwaj, R., Sidhu, G. P. S., et al. (2021). Mechanisms of plant defense under pathogen stress: A review. Curr. Protein Pept. Sci. 22, 376–395. doi: 10.2174/1389203722666210125122827
Su, M. Y., Wang, S., Li, C. X., Zhang, Z. Y., Wang, N., Li, B., et al. (2022a). Ultraviolet-B-induced MdWRKY71-L expression regulates anthocyanin synthesis in apple. Environ. Exp. Bot. 201, 10500. doi: 10.1016/j.envexpbot.2022.105000
Su, T., Xu, Q., Zhang, F. C., Chen, Y., Li, L. Q., Wu, W. H., et al. (2015). WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 167, 1579–1591. doi: 10.1104/pp.114.253799
Su, M. Y., Zuo, W. F., Wang, Y. C., Liu, W. J., Zhang, Z. Y., Wang, N., et al. (2022b). The WKRY transcription factor MdWRKY75 regulates anthocyanins accumulation in apples (Malus domestica). Funct. Plant Biol. 49, 799–809. doi: 10.1071/fp21146
Sugimoto, K., Zager, J. J., St Aubin, B., Lange, B. M., Howe, G. A. (2022). Flavonoid deficiency disrupts redox homeostasis and terpenoid biosynthesis in glandular trichomes of tomato. Plant Physiol. 188, 1450–1468. doi: 10.1093/plphys/kiab488
Sun, C., Palmqvist, S., Olsson, H., Borén, M., Ahlandsberg, S., Jansson, C. (2003). A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 15, 2076–2092. doi: 10.1105/tpc.014597
Sun, L. C., Song, L., Zhang, Y., Zheng, Z., Liu, D. (2016). Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol. 170, 499–514. doi: 10.1104/pp.15.01336
Sun, Y. D., Yu, D. Q. (2015). Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 34, 1295–1306. doi: 10.1007/s00299-015-1787-8
Suttipanta, N., Pattanaik, S., Kulshrestha, M., Patra, B., Singh, S. K., Yuan, L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157, 2081–2093. doi: 10.1104/pp.111.181834
Tanada, T. (1978). Boron - Key element in the actions of phytochrome and gravity? Planta. 143, 109–111. doi: 10.1007/bf00389059
Tang, N., Ma, S. Q., Zong, W., Yang, N., Lv, Y., Yan, C., et al. (2016). MODD mediates deactivation and degradation of OsbZIP46 to negatively regulate ABA signaling and drought resistance in rice. Plant Cell. 28, 2161–2177. doi: 10.1105/tpc.16.00171
Thomashow, M. F. (1999). PLANT COLD ACCLIMATION: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Torres, M. A., Dangl, J. L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403. doi: 10.1016/j.pbi.2005.05.014
Turck, F., Zhou, A., Somssich, I. E. (2004). Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to Its native promoter and the defense-related gene PcPR1-1 in Parsley. Plant Cell. 16, 2573–2585. doi: 10.1105/tpc.104.024810
Wang, L. J., Guo, D. Z., Zhao, G. D., Wang, J. Y., Zhang, S. X., Wang, C., et al. (2022). Group IIc WRKY transcription factors regulate cotton resistance to Fusarium oxysporum by promoting GhMKK2-mediated flavonoid biosynthesis. New Phytol. 236, 249–265. doi: 10.1111/nph.18329
Wang, S., Han, S. Y., Zhou, X. G., Zhao, C. J., Guo, L. A., Zhang, J. Q., et al. (2023b). Phosphorylation and ubiquitination of OsWRKY31 are integral to OsMKK10-2-mediated defense responses in rice. Plant Cell. 35, 2391–2412. doi: 10.1093/plcell/koad064
Wang, F. Q., Li, X. R., Zuo, X., Li, M. M., Miao, C. Y., Zhi, J. Y., et al. (2021a). Transcriptome-wide identification of wrky transcription factor and functional characterization of RgWRKY37 involved in acteoside biosynthesis in Rehmannia glutinosa. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.739853
Wang, R. Y., Liu, X. Q., Zhu, H. Q., Yang, Y. M., Cui, R. F., Fan, Y. K., et al. (2023a). Transcription factors GmERF1 and GmWRKY6 synergistically regulate low phosphorus tolerance in soybean. Plant Physiol. 192, 1099–1114. doi: 10.1093/plphys/kiad170
Wang, T., Sun, Z., Wang, S. Q., Feng, S., Wang, R. M., Zhu, C. L., et al. (2023c). DcWRKY33 promotes petal senescence in carnation (Dianthus caryophyllus L.) by activating genes involved in the biosynthesis of ethylene and abscisic acid and accumulation of reactive oxygen species. Plant J. 113, 698–715. doi: 10.1111/tpj.16075
Wang, Y., Wang, X. H., Fang, J. H., Yin, W. C., Yan, X. X., Tu, M. X., et al. (2023d). VqWRKY56 interacts with VqbZIPC22 in grapevine to promote proanthocyanidin biosynthesis and increase resistance to powdery mildew. New Phytol. 237, 1856–1875. doi: 10.1111/nph.18688
Wang, J. Y., Wang, L. J., Yan, Y., Zhang, S. X., Li, H., Gao, Z., et al. (2021b). GhWRKY21 regulates ABA-mediated drought tolerance by fine-tuning the expression of GhHAB in cotton. Plant Cell Rep. 40, 2135–2150. doi: 10.1007/s00299-020-02590-4
Wang, N. N., Xu, S. W., Sun, Y. L., Liu, D., Zhou, L., Li, Y., et al. (2019). The cotton WRKY transcription factor (GhWRKY33) reduces transgenic Arabidopsis resistance to drought stress. Sci. Rep. 9, 724. doi: 10.1038/s41598-018-37035-2
Wang, P. T., Xu, X., Tang, Z., Zhang, W. W., Huang, X. Y., Zhao, F. J. (2018). OsWRKY28 regulates phosphate and arsenate accumulation, root system architecture and fertility in rice. Front. Plant Sci. 9, 1330. doi: 10.3389/fpls.2018.01330
Wei, W., Cui, M. Y., Hu, Y., Gao, K., Xie, Y. G., Jiang, Y., et al. (2018). Ectopic expression of FvWRKY42, a WRKY transcription factor from the diploid woodland strawberry (Fragaria vesca), enhances resistance to powdery mildew, improves osmotic stress resistance, and increases abscisic acid sensitivity in Arabidopsis. Plant Sci. 275, 60–74. doi: 10.1016/j.plantsci.2018.07.010
Wei, W., Liang, D. W., Bian, X. H., Shen, M., Xiao, J. H., Zhang, W. K., et al. (2019). GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 100, 384–398. doi: 10.1111/tpj.14449
Weston, L. A., Mathesius, U. (2013). Flavonoids: Their structure, biosynthesis and role in the rhizosphere, including allelopathy. J. Chem. Ecol. 39, 283–297. doi: 10.1007/s10886-013-0248-5
Wu, T. Y., Goh, H., Azodi, C. B., Krishnamoorthi, S., Liu, M. J., Urano, D. (2021). Evolutionarily conserved hierarchical gene regulatory networks for plant salt stress response. Nat. Plants. 7, 787–799. doi: 10.1038/s41477-021-00929-7
Wu, Y., Li, X., Zhang, J. N., Zhao, H. Q., Tan, S. L., Xu, W. H., et al. (2022b). ERF subfamily transcription factors and their function in plant responses to abiotic stresses. Front. Plant Sci. 13, 1042084. doi: 10.3389/fpls.2022.1042084
Wu, M., Liu, H. L., Han, G. M., Cai, R. H., Pan, F., Xiang, Y. (2017). A moso bamboo WRKY gene PeWRKY83 confers salinity tolerance in transgenic Arabidopsis plants. Sci. Rep. 7, 11721. doi: 10.1038/s41598-017-10795-z
Wu, M., Zhang, K. M., Xu, Y. Z., Wang, L. N., Liu, H. X., Qin, Z. L., et al. (2022a). The moso bamboo WRKY transcription factor, PheWRKY86, regulates drought tolerance in transgenic plants. Plant Physiol. Biochem. 170, 180–191. doi: 10.1016/j.plaphy.2021.10.024
Xiao, J., Cheng, H. T., Li, X. H., Xiao, J. H., Xu, C. G., Wang, S. P. (2013). Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol. 163, 1868–1882. doi: 10.1104/pp.113.226019
Xiao, S. H., Ming, Y. Q., Hu, Q., Ye, Z. X., Si, H., Liu, S. M., et al. (2023). GhWRKY41 forms a positive feedback regulation loop and increases cotton defence response against Verticillium dahliae by regulating phenylpropanoid metabolism. Plant Biotechnol. J. 21, 961–978. doi: 10.1111/pbi.14008
Xie, Z., Zhang, Z. L., Zou, X. L., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Xie, Z., Zhang, Z. L., Zou, X. L., Yang, G. X., Komatsu, S., Shen, Q. X. J. (2006). Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 46, 231–242. doi: 10.1111/j.1365-313X.2006.02694.x
Xu, X. P., Chen, C. H., Fan, B. F., Chen, Z. X. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell. 18, 1310–1326. doi: 10.1105/tpc.105.037523
Xu, Y., Miao, Y. X., Tian, X. J., Wang, Q. H., Hu, Y. F., Luo, Q. (2022). Transcriptomic and epigenomic assessment reveals epigenetic regulation of wrky genes in response to Magnaporthe oryzae infection in rice. Curr. Genomics 23, 182–194. doi: 10.2174/1389202923666220510195910
Yamamoto, S., Nakano, T., Suzuki, K., Shinshi, H. (2004). Elicitor-induced activation of transcription via W box-related cis-acting elements from a basic chitinase gene by WRKY transcription factors in tobacco. Biochim. Biophys. Acta Gene Structure Expression. 1679, 279–287. doi: 10.1016/j.bbaexp.2004.07.005
Yamasaki, K., Kigawa, T., Inoue, M., Tateno, M., Yamasaki, T., Yabuki, T., et al. (2005). Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell. 17, 944–956. doi: 10.1105/tpc.104.026435
Yamasaki, K., Kigawa, T., Watanabe, S., Inoue, M., Yamasaki, T., Seki, M., et al. (2012). Structural basis for sequence-specific DNA recognition by an Arabidopsis WRKY transcription factor. J. Biol. Chem. 287, 7683–7691. doi: 10.1074/jbc.M111.279844
Yan, F. Y., Zhao, H. L., Wu, L. M., Huang, Z. W., Niu, Y., Qi, B., et al. (2022). Basic cognition of melatonin regulation of plant growth under salt stress: A meta-analysis. Antioxidants. 11, 1610. doi: 10.3390/antiox11081610
Yang, Z., Chi, X. Y., Guo, F. F., Jin, X. Y., Luo, H. L., Hawar, A., et al. (2020). SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 246-247, 153142. doi: 10.1016/j.jplph.2020.153142
Yang, A., Dai, X. Y., Zhang, W. H. (2012). A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63, 2541–2556. doi: 10.1093/jxb/err431
Yang, X. Q., Zhang, K. A., Nvsvrot, T., Zhang, Y., Cai, G. H., Huang, L. Y., et al. (2022). Phosphate (Pi) stress-responsive transcription factors PdeWRKY6 and PdeWRKY65 regulate the expression of PdePHT1;9 to modulate tissue Pi concentration in poplar. Plant J. 111, 1753–1767. doi: 10.1111/tpj.15922
Yokotani, N., Sato, Y., Tanabe, S., Chujo, T., Shimizu, T., Okada, K., et al. (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64, 5085–5097. doi: 10.1093/jxb/ert298
Yu, Y., Qian, Y. C., Jiang, M. Y., Xu, J., Yang, J. T., Zhang, T. Y., et al. (2020). Regulation mechanisms of plant basic leucine zippers to various abiotic stresses. Front. Plant Sci. 11, 1258. doi: 10.3389/fpls.2020.01258
Yu, Y. C., Wang, N., Hu, R. B., Xiang, F. N. (2016). Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springerplus. 5, 920. doi: 10.1186/s40064-016-2647-x
Yu, Y. H., Xu, W. R., Wang, J., Wang, L., Yao, W. K., Yang, Y. Z., et al. (2013). The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol. 200, 834–846. doi: 10.1111/nph.12418
Yue, M. L., Jiang, L. Y., Zhang, N. T., Zhang, L. X., Liu, Y. Q., Wang, Y., et al. (2022). Importance of FaWRKY71 in strawberry (Fragaria x ananassa) fruit ripening. Int. J. Mol. Sci. 23, 12483. doi: 10.3390/ijms232012483
Zhang, Q., Cai, W., Ji, T. T., Ye, L., Lu, Y. T., Yuan, T. T. (2020). WRKY13 enhances cadmium tolerance by promoting D-CYSTEINE DESULFHYDRASE and hydrogen sulfide production. Plant Physiol. 183, 345–357. doi: 10.1104/pp.19.01504
Zhang, X. Y., Cheng, B. H., Ma, Y. Y., Liu, Y. M., Wang, N., Zhang, H., et al. (2022e). Genome-wide survey and functional analysis reveal TCF21 promotes chicken preadipocyte differentiation by directly upregulating HTR2A. Biochem. Bioph Res. Co. 587, 131–138. doi: 10.1016/j.bbrc.2021.11.103
Zhang, J., Gu, M., Liang, R. S., Shi, X. Y., Chen, L. L., Hu, X., et al. (2021a). OsWRKY21 and OsWRKY108 function redundantly to promote phosphate accumulation through maintaining the constitutive expression of OsPHT11under phosphate-replete conditions. New Phytol. 229, 1598–1614. doi: 10.1111/nph.16931
Zhang, L. Y., Gu, L. K., Ringler, P., Smith, S., Rushton, P. J., Shen, Q. J. (2015). Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci. 236, 214–222. doi: 10.1016/j.plantsci.2015.04.014
Zhang, J. W., Huang, D. Z., Zhao, X. J., Zhang, M., Wang, Q., Hou, X. Y., et al. (2022c). Drought-responsive WRKY transcription factor genes IgWRKY50 and IgWRKY32 from Iris germanica enhance drought resistance in transgenic Arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.983600
Zhang, L., Song, H. Y., Li, B. H., Wang, M., Di, D. W., Lin, X. Y., et al. (2021b). Induction of S-nitrosoglutathione reductase protects root growth from ammonium toxicity by regulating potassium homeostasis in Arabidopsis and rice. J. Exp. Bot. 72, 4548–4564. doi: 10.1093/jxb/erab140
Zhang, T., Tan, D., Zhang, L., Zhang, X., Han, Z. (2017). Phylogenetic analysis and drought-responsive expression profiles of the WRKY transcription factor family in maize. Agric. Gene. 3, 99–108. doi: 10.1016/j.aggene.2017.01.001
Zhang, Y., Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5, 1. doi: 10.1186/1471-2148-5-1
Zhang, J. K., Wang, Y. C., Mao, Z. L., Liu, W. N., Ding, L. C., Zhang, X. N., et al. (2022b). Transcription factor McWRKY71 induced by ozone stress regulates anthocyanin and proanthocyanidin biosynthesis in Malus crabapple. Ecotoxicol. Environ. Saf. 232, 113274. doi: 10.1016/j.ecoenv.2022.113274
Zhang, Z. L., Xie, Z., Zou, X. L., Casaretto, J., Ho, T. H. D., Shen, Q. X. J. (2004). A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 134, 1500–1513. doi: 10.1104/pp.103.034967
Zhang, T. L., Xu, H. F., Yang, G. X., Wang, N., Zhang, J., Wang, Y. C., et al. (2019). Molecular mechanism of MYB111 and WRKY40 involved in anthocyanin biosynthesis in red-fleshed apple callus. Plant Cell Tissue Organ Cult. 139, 467–478. doi: 10.1007/s11240-019-01637-z
Zhang, Y., Yu, H. J., Yang, X. Y., Li, Q., Ling, J., Wang, H., et al. (2016). CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Biochem. 108, 478–487. doi: 10.1016/j.plaphy.2016.08.013
Zhang, M. X., Zhao, R. R., Huang, K., Huang, S. Z., Wang, H. T., Wei, Z. Q., et al. (2022d). The OsWRKY63-OsWRKY76-OsDREB1B module regulates chilling tolerance in rice. Plant J. 112, 383–398. doi: 10.1111/tpj.15950
Zhang, H. M., Zhu, J. H., Gong, Z. Z., Zhu, J. K. (2022a). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhou, L. M., Huang, Y. Z., Wang, Q., Guo, D. J. (2021). AaHY5 ChIP-seq based on transient expression system reveals the role of AaWRKY14 in artemisinin biosynthetic gene regulation. Plant Physiol. Biochem. 168, 321–328. doi: 10.1016/j.plaphy.2021.10.010
Zhou, J., Jiao, F. C., Wu, Z. C., Li, Y. Y., Wang, X. M., He, X. W., et al. (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146, 1673–1686. doi: 10.1104/pp.107.111443
Keywords: abiotic stresses, cis-elements, flavonoids biosynthesis, transcription factor, WRKY family
Citation: Zhang J, Zhao H, Chen L, Lin J, Wang Z, Pan J, Yang F, Ni X, Wang Y, Wang Y, Li R, Pi E and Wang S (2023) Multifaceted roles of WRKY transcription factors in abiotic stress and flavonoid biosynthesis. Front. Plant Sci. 14:1303667. doi: 10.3389/fpls.2023.1303667
Received: 28 September 2023; Accepted: 04 December 2023;
Published: 15 December 2023.
Edited by:
Muthusamy Ramakrishnan, Nanjing Forestry University, ChinaReviewed by:
Charu Gupta, Amity University, IndiaMaharajan Theivanayagam, Rajagiri College of Social Sciences, India
Copyright © 2023 Zhang, Zhao, Chen, Lin, Wang, Pan, Yang, Ni, Wang, Wang, Li, Pi and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erxu Pi, ZXJ4dXBpQGh6bnUuZWR1LmNu; Shang Wang, d2FuZ3NoYW5nMDIwNUBoem51LmVkdS5jbg==
 Jinnan Zhang
Jinnan Zhang Erxu Pi
Erxu Pi Shang Wang
Shang Wang