- 1Department of Agricultural, Forest and Food Sciences (DISAFA), Plant Genetics, University of Torino, Grugliasco, Italy
- 2Department of Agricultural, Food, Environmental and Animal Sciences (DI4A), University of Udine, Udine, Italy
- 3Department of Agronomy, Food, Natural Resources, Animals and Environment (DAFNAE), Laboratory of Plant Genetics and Breeding, University of Padua, Legnaro, Italy
This review highlights -omics research in Solanaceae family, with a particular focus on resilient traits. Extensive research has enriched our understanding of Solanaceae genomics and genetics, with historical varietal development mainly focusing on disease resistance and cultivar improvement but shifting the emphasis towards unveiling resilience mechanisms in genebank-preserved germplasm is nowadays crucial. Collecting such information, might help researchers and breeders developing new experimental design, providing an overview of the state of the art of the most advanced approaches for the identification of the genetic elements laying behind resilience. Building this starting point, we aim at providing a useful tool for tackling the global agricultural resilience goals in these crops.
1 Introduction
The Solanaceae family comprises significant crops, including, within the most economically and culturally important, tomato (Solanum lycopersicum), potato (Solanum tuberosum) pepper (Capsicum annuum), and eggplant (Solanum melongena), which are cultivated worldwide (FAOSTAT, 2021). Climate changes, such as rising temperatures, altered precipitation patterns, and increased occurrence of extreme weather events, have already demonstrated a significant impact on natural ecosystems and agricultural productivity (Abbass et al., 2022). Therefore, they pose a threat to the distinctive qualities and geographical indications associated with food products.
Over the past few decades, climate change has started impacting Solanaceae crops, and extreme emergent weather patterns are going to significantly affect yield and quality of tomato, pepper, and eggplant (Lee et al., 2018; Bhandari et al., 2021; Suman, 2022; Toppino et al., 2022). While certain agricultural practices and cultivation techniques may provide temporary coping mechanisms, long-term strategies need to be implemented to address the challenges of climate change in vulnerable regions. Breeding strategies play a vital role in developing climate-resilient varieties, and both conventional breeding techniques (CBTs) and new breeding techniques (NBTs) offer powerful tools for enhancing crop resilience within low-input production systems (Razzaq et al., 2021; Xiong et al., 2022). Historically, breeding programs have been focused on developing disease-resistant varieties to ensure sustainable production (Poczai et al., 2022). By selectively breeding for natural resistance or incorporating resistance genes from wild relatives, breeders can enhance the crops’ resilience to common diseases such as late blight, bacterial wilt, and viral infections. Breeding efforts also target agronomic traits that can mitigate the impact of climate change on Solanaceae crops, including drought tolerance, heat tolerance, water-use efficiency (WUE), and nutrient uptake efficiency (NUE). At the same time, enhancing fruit quality attributes is a crucial breeding objective for tomato, pepper, and eggplant (Bebeli and Mazzucato, 2009). For this reason, the main breeding focus has been to improve traits such as flavor, nutritional content, texture, and shelf life, incorporating them into new varieties, ensuring that these crops maintain their appeal to consumers and adapt to changing market demands.
In this paper, the state of the art on next-generation genotyping and -omics technologies for the molecular prediction of multiple resilient traits in the Solanaceae family will be reviewed, with the aim of building a starting point for the research activities within the Recovery and Resilience Facility (RRF) NextGenerationEU founding plan.
2 Genetic resources
The Solanaceae Genomics Network (SGN – www.solgenomics.net; Mueller et al., 2005), as well as the G2P-SOL gateway (www.g2p-sol.eu) and Genesys platform (https://www.genesys-pgr.org) are the main centers for genetic and genomic resources for Solanaceae plants, mainly for cultivated species and their crop wild relatives (CWRs). Through these databases, genetic resources can be retrieved, and useful information on their botanical origin and main agronomic characteristics can be collected.
2.1 Tomato
Tomato germplasm research has made significant contributions to understanding the genetic and phenotypic diversity at variety level, providing valuable insights for crop improvement of the species through breeding. In one study conducted by Roohanitaziani et al. (2020), a tomato core collection of 122 genebank accessions was extensively explored. The study focused on variation in phenotypic traits, comparing cultivated varieties with wild accessions. The researchers identified key genes and mutations responsible for traits such as fruit size, shape, color, and plant architecture, shedding light on the role of these genes in tomato domestication and breeding, and offering valuable information for precision breeding strategies. Mata-Nicolás et al. (2021) conducted research on a collection of 163 accessions, representing the genetic and morphological variability of tomato from its centers of origin and domestication. This study aimed to explore the genomic landscape of these accessions using sequencing technologies. A substantial number of single nucleotide polymorphisms (SNPs) was observed, and their impact on tomato traits was investigated. The availability of these genomic data, coupled with the creation of F2 populations, provides a valuable resource for further genetic studies and the testing of identified SNPs in the context of tomato breeding and selection. In a separate study, Jin et al. (2019) investigated the morphological and molecular diversity of 324 cultivated inbred tomato lines. By analyzing them for 17 agronomic traits, the significant phenotypic variation was assessed, including traits related to disease resistance, fruit characteristics, and plant growth. Genotyping using insertion-deletion (indels) markers further revealed a rich genetic background and the existence of distinct groups within the cultivated tomato lines. This study highlights the potential of employing cluster analysis and molecular markers in guiding parental selection for hybrid breeding and improving tomato cultivars.
2.2 Pepper
Pepper germplasm have been extensively investigated by several authors, with each study offering valuable insights into different aspects of genetic diversity and population structure. Lee et al. (2016) focused on exploring the genetic diversity within a large pepper germplasm collection comprising 3,821 accessions. Their investigation revealed that C. annuum holds the highest genetic diversity among the 11 species studied. Additionally, they successfully selected a core collection (named CC240), consisting of 240 accessions that represented a wide range of phenotypic variation and displayed higher genetic diversity compared to other core collections. In a study by Zhang et al. (2016), the genetic structure of Chinese pepper germplasm, including both local cultivars and landraces, was examined. By genotyping 372 genebank accessions and analyzing their relationships, they found that the genetic structure corresponded not only to geographic origin, but also to distinct cultivar types. This suggested that migration and human selection played key roles in shaping the genetic diversity of Chinese pepper landraces. Mongkolporn et al. (2015) directed their attention to chili germplasm and aimed to establish a representative core collection. Through the evaluation of 230 chili accessions using microsatellite markers, they highlighted two major groups within the collection. Group I mainly consisted of C. annuum, while Group II included various Capsicum species, with C. frutescens, C. chinense, and C. baccatum being the most prevalent. This core collection, comprising 28 representative chili accessions, harbored a similar level of diversity to that of the entire collection. Gu et al. (2019) delved into the genetic diversity of pepper accessions in China and investigated the relationship between genetic structure and fruit type, as well as geographic origin. Their findings unveiled the presence of two distinct genetic groups, which aligned with differences in fruit type and geographical distribution, highlighting the impact of both natural and human-driven factors on the genetic structure of pepper germplasm. In a study by Christov et al. (2021), attention was shifted to Balkan pepper accessions. By genotyping 179 C. annuum accessions from different locations in the Balkan Peninsula, the authors identified three main clusters based on fruit traits such as shape, size, and pungency. Moreover, a further sub-clustering observed within the main clusters, emphasized a more intricate genetic structure of Balkan pepper germplasm. Nankar et al. (2020) conducted an extensive evaluation of 180 pepper accessions collected from diverse locations across six Balkan countries. Through the examination of agro-morphological traits, fruit quality, and virus resistance, they observed considerable variation and identified six distinct clusters. Their findings indicated the potential of the Balkan pepper collection as a valuable gene source for pre-breeding and cultivar development, particularly those adapted to local conditions. In 2021, Tripodi et al. (2021) conducted a comprehensive genomic analysis of more than 10,000 pepper accessions sourced from genebanks worldwide. Through this investigation, they successfully identified instances of duplicate accessions and taxonomic misassignments, illustrating the intricate challenges associated with classifying interspecific hybrids. Notably, their study unveiled a substantial convergence of pepper types collected across diverse geographical regions, underscoring the global significance of pepper as a cultural commodity. To complement traditional population genetic analyses, the ReMIXTURE method was developed, which effectively quantifies the resemblance between pepper populations originating from different regions. Furthermore, they observed marker-trait associations and selective sweeps affecting pivotal characteristics, such as pungency, which exhibited a nonuniform distribution on a global scale, suggesting a predominant influence of human preferences in shaping the genetic architecture of domesticated pepper varieties. This investigation significantly enriches our understanding of pepper’s genetic history, consequently facilitating advancements in breeding and conservation endeavors. Lastly, Guo et al. (2023) compared the diversity of local landraces and current breeding lines of pepper in China. Through an analysis of qualitative and quantitative traits, traits, as well as molecular markers, they found that current breeding lines exhibited higher diversity in some quantitative traits, especially those related to fruit organs. However, molecular marker analysis revealed that the genetic diversity in breeding lines was lower compared to local landraces. The authors suggested the need for a balanced approach in future breeding processes, focusing not only on target traits but also on molecular marker-based background selection and the incorporation of genetic information from wild and domesticated species.
2.3 Eggplant
Research on eggplant germplasm has been conducted by several authors, each shedding light on different aspects of genetic diversity and population structure. Oladosu et al. (2021), in their comprehensive review, emphasized the global occurrences and genebank holdings of cultivated eggplants and their wild relatives. Among such genebanks, the World Vegetable Center (WorldVeg) is the largest public collection of the three cultivated eggplant species (S. melongena, S. aethiopicum and S. macrocarpon), including accessions from related species. As reported by Taher et al. (2017), such material is essential for the development of breeding programs aiming at the enhancement of fruit quality, yield performance, and pest and disease resistance, since wild relatives can harbor undomesticated alleles that can confer novel traits to the commercial varieties. A core collection is a limited set of accessions representing, with a minimum of repetitiveness, the genetic diversity of a crop species and its wild relatives (Frankel, 1984). To this purpose, Naegele et al. (2014) evaluated 99 eggplant genotypes from different landraces, including heirloom cultivars and wild relative species, identifying four different clusters. Successively, by analyzing a dataset of 893 accessions from NARO Genebank, Miyatake et al. (2019) highlighted that the genetic diversity of the Asian accessions, particularly those from South Asian and Southeast Asian countries, was higher compared to other regions, also identifying four distinct clusters (S1–S4), each corresponding to a specific geographical group. Through SPET technology, the collection of 422 accessions, gathered by the G2P-SOL consortium, was genotyped (Barchi et al., 2019a), allowing the identification of duplicates and mislabeled accessions in genebanks. Lastly, Fallahi et al. (2022) assessed the genetic and morphological diversity of eggplant cultivars comprising four Iranian lines and 13 non-Iranian genotypes. They employed morphological analysis of 16 traits and molecular analysis using five SSR markers, revealing significant variations in morphological traits, clear differentiation between eggplants from different origins, and identifying three distinct clusters.
2.4 Potato
Cultivated potatoes can be categorized into two primary groups: i) landraces, which are indigenous varieties still cultivated in South America, and ii) improved varieties, grown globally for market production. Potato landraces exhibit significant diversity in tuber shapes, as well as skin and flesh colors. They are cultivated in the upland Andes, spanning from western Venezuela to northern Argentina, and in the lowlands of south-central Chile, with an adaptation to middle to high elevations ranging from 3,000 to 4,000 meters above sea level (Contreras et al., 1993). While genebanks stand as a cost-effective method for conservation, their operations encounter challenges such as potential loss of unique materials and the risk of genetic erosion under suboptimal maintenance or regeneration conditions, and in cases where reorganization is necessitated due to political or environmental factors (Fu, 2017). The management of potato collections varies according to the type of accession, whether it be a variety, breeding line, landrace, or wild species, and the preservation is linked to the corresponding plant organ, including seeds, tubers, in vitro plantlets, or shoot tips. The systematic collection of potato genetic resources in South America, encompassing Colombia, Ecuador, Peru, Bolivia, and Chile, was initiated by Russian scientists between 1925 and 1933 (Loskutov and Institute I. P. G. R, 1999). This effort laid the foundation for the first potato germplasm collection at the N. I. Vavilov Institute of Plant Genetic Resources (VIR) in St. Petersburg (Ovchinnikova et al., 2011). Similarly, German scientists undertook collection missions in Chile and Bolivia in 1930, giving rise to the Groß Lüsewitz Potato Collections (GLKS), now integrated into the IPK Federal ex situ Gene Bank. This collection comprises 6,300 accessions, including over 2,800 named cultivars, 510 short-day adapted materials, and more than 2,950 accessions of over 130 wild and cultivated Solanum species gathered from South and Central America. From 1938 onwards, British expeditions were dispatched to Mexico and South America (Argentina, Bolivia, Peru, Ecuador, Colombia), resulting in the acquisition of 1,164 accessions of wild and cultivated potato species (Hawkes, 1941). This collection formed the basis for the Commonwealth Potato Collection (CPC) currently housed at the James Hutton Institute in Scotland. In the United States, the U.S. Potato Genebank was established in the late 1940s to preclude the importation of varieties that might pose threats to the potato industry or endemic wild species. Since the 1990s, wild potato germplasm has been collected in nearly all Latin American countries (Spooner and Hijmans, 2001). The CGN (Centre for Genetic Resources) potato collection in the Netherlands, originating from Dutch expeditions in 1955 and 1974, was further enriched with germplasm from German missions in 1959, Argentine genebank of INTA-Balcarce, and a collecting mission in Bolivia in 1980 (Van Soest et al., 1983). Collaborative missions between the USA and the Netherlands led to the collection of 2,700 potato accessions from 12 American countries. Presently, approximately 55% of the collection complies with EU plant health requirements for germplasm distribution. Lastly, the International Germplasm Bank for potato was established at the International Potato Center (CIP) in Lima, Peru, during the early 1970s. In collaboration with the Peruvian National Institute for Agricultural Research (INIA), over 300 systematic exploration missions and more than 100 collecting missions were conducted across 12 countries(Huaman et al., 2000; Spooner et al., 2014). In total, 82,293 potato accessions are maintained globally in 89 institutions and four international/regional centers across 59 countries. Among these, 47 institutes in 36 countries hold more than 100 accessions, with just five countries (France, Germany, India, Russia, USA), along with CIP, collectively possessing over 50% of all potato accessions (Nagel et al., 2022).
These findings have significant implications for crop improvement, evolutionary studies, and understanding plant-pathogen interactions. The availability of large collections of genetic resources and advanced genomic technologies has paved the way for further research and the development of improved varieties. The collective efforts of researchers and genebanks worldwide are crucial for the conservation, utilization, and sustainable management of Solanaceae genetic resources to address the challenges of agricultural productivity and food security in the future.
3 Genomic resources
Genome sequencing projects have been carried out for several Solanaceae species, serving as fundamental tools for comparative genomics, enabling researchers to identify and analyze genes, regulatory elements, and other functional elements across different species. Moreover, gene expression data and transcriptome analyses have shed light on the dynamic regulation of genes during various developmental stages and in response to environmental stimuli. These resources collectively contribute to a deeper understanding of the genetic diversity, adaptive traits, and agronomically important characteristics of Solanaceae plants, ultimately aiding in the development of improved crop varieties with desirable traits and increased resilience. Moreover, the recent advances in the development of pangenomes allow the overcoming of the limitations imposed using a single genotype as reference, making it possible to discover novel genetic elements.
3.1 Tomato
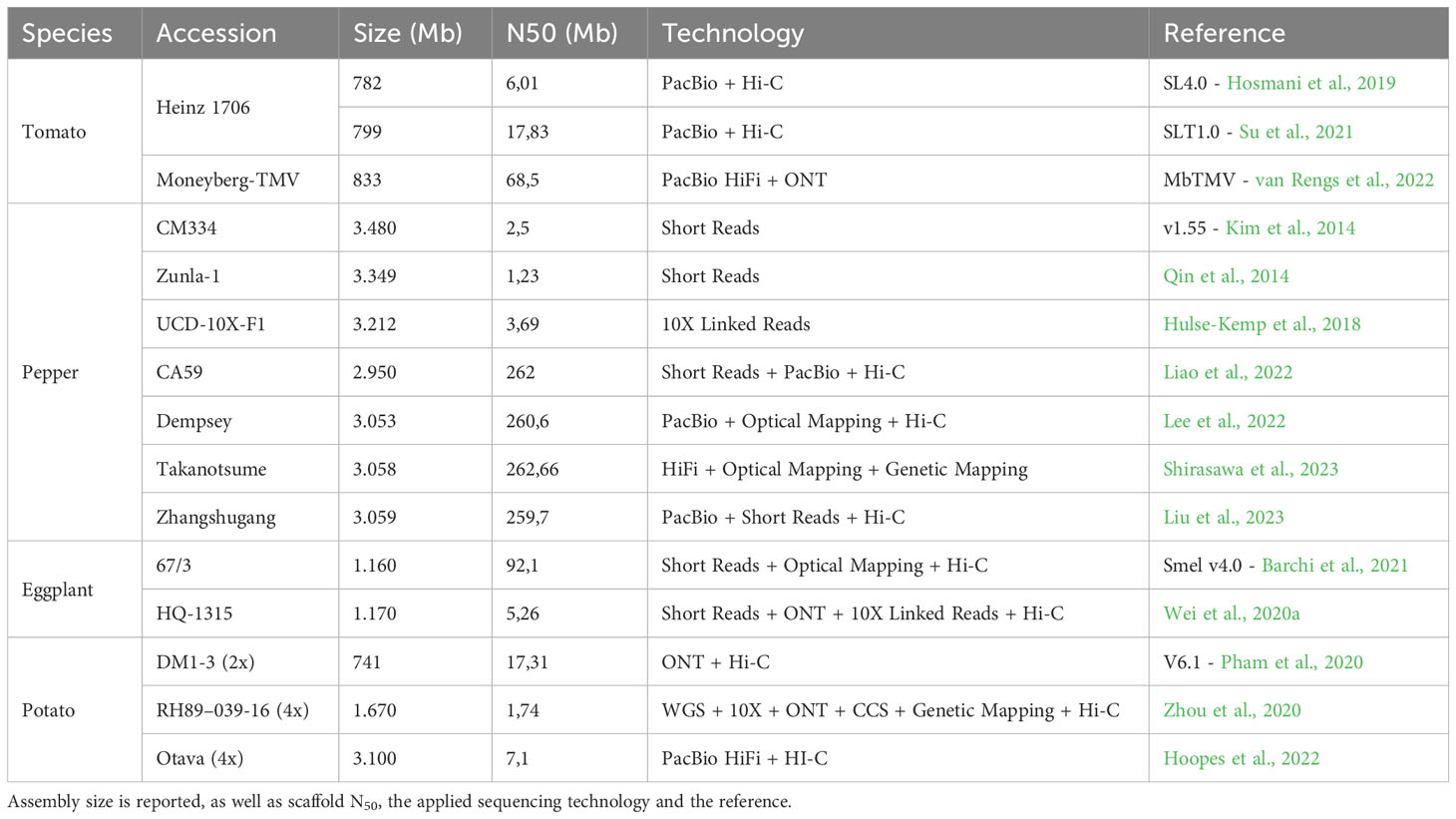
The Tomato Genome Consortium played a pivotal role in presenting a high-quality genome sequence of domesticated tomato (S. lycopersicum, cultivar ‘Heinz 1706’; Tomato Genome Consortium, 2012). Later, several upgrades led to the reference version SL4.0 (Hosmani et al., 2019), based on single-molecule sequencing, Hi-C proximity ligation and optical maps. Later, Su et al. (2021) focused on the assembly of an updated reference genome of the tomato cultivar ‘Heinz 1706’ and its comparison with S. pimpinellifolium ‘LA2093’. The updated genome assembly (SLT1.0) was 799.09 Mb in length and encompassed 34,384 predicted protein-coding genes, along with 65.66% repetitive sequences. Through comparative analysis, numerous genomic fragments in S. lycopersicum that were likely associated with human selection, indicating their potential role in the domestication process of tomato, were identified (Frary et al., 2000; van der Knaap et al., 2014; Pereira et al., 2021). The study conducted by (van Rengs et al., 2022) utilized advanced sequencing technologies, specifically PacBio HiFi and Oxford Nanopore Technology (ONT) long read sequencing, to assemble the genome of an inbred TMV-resistant tomato variety. By combining data from both platforms, highly contiguous and complementary genome assemblies were achieved. The merged assembly, consisting of 12 chromosomes represented as 12 contiguous sequences (N50 = 68.5 Mbp), did not require scaffolding using an orthogonal data type. The merged assembly was validated using chromosome conformation capture data and found it to be consistent with previous tomato genome assemblies that utilized genetic maps and Hi-C technology for scaffolding. This long-read-only assembly provided valuable insights into the structural variations associated with TMV resistance and their impact on the tomato breeding process. While the previously reported study by Roohanitaziani et al. (2020) investigated cultivated tomato accession, Yu et al. (2022) explored the genomes of various tomato species, including S. habrochaites, S. galapagense, S. lycopersicum, S. pimpinellifolium, and S. pennellii. They focused on genes related to isoprenoid biosynthesis and resistance gene analogs (RGAs). They discovered that the genomes of S. habrochaites and S. galapagense exhibited an enrichment of genes involved in the terpenoid biosynthetic process. Specifically, they identified a remarkable expansion of the TPS-a subfamily, which encodes sesquiterpene synthases, in S. habrochaites, suggesting the potential for diverse or unique sesquiterpene synthesis in this species. Furthermore, they also identified many RGAs in the genomes of all five tomato species, with certain species showing additional RGAs in regions associated with insertion or expansion. These findings highlight the reservoir of resistance genes present in wild tomato relatives acting in networks with varying degrees of complexity, akin to observations in other species and supporting their potential significance in tomato breeding programs. Table 1 presents a comprehensive list of the reference genomes released for tomato, together with their main characteristics.
The pangenome is a recent powerful tool for identifying causal SVs and improving the accuracy of heritability estimates for complex traits (Sherman and Salzberg, 2020; Usadel, 2022). Gao et al. (2019) constructed a pangenome using 725 diverse accessions, identifying 4,873 genes that were not present in the SL3.0 reference genome and revealing PAVs useful for breeding purposes and a rare allele in the TomLoxC promoter whose selection apparently modified tomato aroma during domestication. Alonge et al. (2020) described the discovery of thousands of SVs in the tomato genome through the analysis of 100 lines, including 14 new reference-level genome assemblies, showing how SVs can be used to improve tomato crops through genome editing. Furthermore, Zhou et al. (2022) described the construction of a graph pangenome of tomato, by including the genetic information from 838 different tomato lines, and the identification of SVs that are not present in the single linear reference genome. Such pangenome can be used to improve the accuracy of heritability estimates for complex traits, such as fruit flavor and soluble solid content.
3.2 Pepper
Pepper reference genome development has been a subject of extensive research, providing valuable insights into the genetic makeup and evolution of this important crop. The first whole-genome sequences of C. annuum (‘CM334’) and C. chinense (PI159236) were published by Kim et al. (2014). In the same year, Qin et al. (2014) released the genome sequences of the C. annuum ‘Zunla-1’ variety and of ‘Chiltepin’ pepper (C. annuum var. glabriusculum). These studies revealed that the pepper genome is ~3–3.5 Gb in size, contains over 80% repetitive elements, and encodes 35k genes. A resequencing effort (Han et al., 2016) made available the raw sequences of two C. annuum lines: ‘Dempsey’, a large bell-type non-pungent genotype, and ‘Perennial’, a pungent genotype with small elongated fruits. In 2017, an improved version of the reference genome of both ‘CM334’ and C. chinense ‘PI159236’ was published (Kim et al., 2017), together with the genome sequence of the domesticated C. baccatum. This study contributed to deciphering the evolutionary relationships among the three species as well as to estimating the lineage-divergence times occurring in Capsicum. In 2018, Hulse-Kemp et al. (2018) obtained the genome sequence of an F1 hybrid from a cross between ‘CM334’ with a non-pungent blocky accession of C. annuum, by adopting the linked-read sequencing technology. In 2019, Du et al. (2019b) produced low-pass resequencing data from 35 different C. annuum lines, to identify a panel of 92 SNPs for population structure analysis. In 2020, Acquadro et al. (2020) conducted whole-genome resequencing of four Italian sweet pepper landraces to gain insights into sequence variation in genes of agronomic value. Some current works (Ou et al., 2018; Lee et al., 2022) developed a pan-genome and a graph-based pangenome approach for pepper, assembling genomes of multiple lines of different Capsicum species; these study unveiled informative variants, including presence–absence variants (PAVs), copy-number variants (CNVs), and inversions, and provided insights into the role of transposable elements in shaping the genomic landscape of peppers and insights into Capsicum evolution. In 2022, Liao et al. (2022) integrated PacBio long-reads with Hi-C maps with epigenomics, transcriptomic, and genetic variation data to develop a 3D structure of the cultivar “Ca59”. More recently, Shirasawa et al. (2023) presented a study on a Japanese chili pepper landrace (‘Takanotsume’) using long-read sequencing, optical mapping, and genetic mapping techniques to achieve a chromosome-scale genome assembly. Additionally, they conducted a comparative genomics analysis involving two C. chinense lines, and 13 genotypes among Capsicum species, highlighting nucleotide and structural variations providing valuable information for pan-genomics, breeding, and the understanding of genetic mechanisms underlying important traits in Capsicum. Table 1 contains a list of the reference genomes developed in pepper.
3.3 Eggplant
The development of eggplant genomics has been the focus of several research endeavors. In 2014, Hirakawa et al. (2014) presented a study in which they dissected the eggplant genome and constructed a draft genome dataset termed SME_r2.5.1. This dataset covered a significant portion of the eggplant genome, providing valuable insights into its genomic nature. The study estimated that approximately 90% of the gene space was covered, resulting in the prediction of 85,446 genes in the eggplant genome. Comparative analysis of eggplant genes with those of other solanaceous plants and Arabidopsis thaliana revealed the presence of eggplant-specific gene clusters, potentially responsible for conferring unique traits to the crop. Later, Barchi et al. (2019b) made significant contributions to eggplant genomics by obtaining a high-quality genome assembly of eggplant, providing a chromosome-anchored reference. This assembly contained 34,916 genes, confirming the approximate diploid gene number in the Solanaceae family. The study conducted comparative genomic analyses between eggplant and other solanaceous crops such as tomato, potato, and pepper. These analyses highlighted the rapid evolution of miRNA:mRNA regulatory pairs and defense genes within the Solanaceae family. The research also offered insights into the lack of steroidal glycoalkaloid compounds in the Capsicum genus. By reconstructing the chromosomal complements of key ancestors, the study shed light on the rearrangements that led to the karyotypes of present-day species and their ancestors. This assembly was enhanced in 2021 by Barchi et al. (2021) through Hi-C retrofitting of the previously released assembly. In 2020, Wei et al. (2020a) released a high-quality reference genome for the eggplant inbred line HQ-1315. This assembly was based on a combination of Illumina, Nanopore, and 10X genomics sequencing technologies, as well as Hi-C technology for genome assembly. The assembled genome exhibited a total size of approximately 1.17 Gb and comprised 12 chromosomes. The study identified various genomic variations, including SNPs, indels, and structural variants (SVs). Furthermore, they developed the first eggplant pangenome, based on 23 cultivated accessions and one accession of S. insanum and S. incanum, assembling 51.5 additional megabases, and annotating 816 additional genes compared with the reference genome (Table 1).
3.4 Potato
Sequencing tetraploid plant material, particularly those exhibiting heterozygosity, such as cultivated potatoes, presents a formidable challenge. In addressing this challenge, the haploid potato germplasm DM BARD 1–3 516 R44 (referred to as DM), was employed to construct a reference genome (Veilleux, 2017). The establishment of these monoploids originated from originally heterozygous adapted S. tuberosum group Phureja clones, which underwent chromosome doubling procedures (Paz and Veilleux, 1997). After sequencing, the final assembly encompassed approximately 86% of the potato genome, amounting to 844 Mb, and yielded predictions for approximately 39,000 genes. A comparative analysis was then conducted between the sequence of DM and that of the heterozygous diploid potato genome (RH89–039-16, abbreviated as RH), characterized by a pronounced degree of heterozygosity. The alignment between the RH genome and the DM genome revealed a coverage of only 55%, with the occurrence of potentially deleterious mutations being prevalent and identified as a plausible contributor to inbreeding depression (The Potato Genome Consortium, 2011). Trying to further enhance to the genome assembly’s quality and its relevance to potato breeding, a comprehensive marker analysis of a backcross segregation population of DD, a heterozygous clone belonging to the S. tuberosum ‘Andigenum Group’, x (DM x DD) was conducted, applying in silico anchoring approaches, and integrating data from both physical and genetic maps derived from RH and tomato (Sharma et al., 2013). In 2015, the draft genome of the wild species S. commersonii, which diverged from the cultivated potato approximately 2.3 million years ago, was reported. Such assembly, comparable in size to the potato genome at 830 Mb, exhibited markedly reduced heterozygosity, thereby affording valuable insights into evolutionary dynamics and environmental adaptation (Aversano et al., 2015). Further contributions to the understanding of cultivated potato and its wild counterparts were made by Leisner et al. (2018), who sequenced the genome of the diploid inbred clone (M6) of S. chacoense. This effort yielded an assembly of 882 Mb and the annotation of 37,740 functional genes. Recent advancements in sequencing technologies have culminated in multiple publications presenting genome assemblies for various diploid and polyploid potato varieties (Table 1; Kyriakidou et al., 2020; Pham et al., 2020; Yang et al., 2020; Zhou et al., 2020; Freire et al., 2021; Tiwari et al., 2021; Sun et al., 2022). Finally, two pangenome assembly were obtained through the implementation of long-read sequencing technology: Hoopes et al. (2022) encompassed six potato varieties tailored for distinct markets - fresh (‘Colomba’, ‘Spunta’), chip processing (‘Atlantic’), frozen processing (‘Castle Russet’), and starch markets (‘Altus’, ‘Avenger’), developing the first pan-genome of polyploid potato varieties. In 2023, Bozan et al. (2023) constructed a super-pangenome of 296 diverse potato accessions belonging to the Solanum section Petota, with a ploidy level ranging from 2x to 5x, constructing a phylogenetic tree using PAVs, and identifying clades for abiotic stress response and flowering and tuberization.
The genome sequencing projects for Solanaceae species, coupled with genetic maps, gene expression data, and transcriptome analyses, have significantly advanced our understanding of their genetic diversity, adaptive traits, and agronomically important characteristics. These resources, along with comprehensive online platforms like the SGN, contribute to the development of improved crop varieties with desirable traits and increased resilience, offering promising solutions for agricultural challenges.
4 Genotyping tools and their evolution
The integration of genotyping tools in crop breeding is instrumental in accelerating crop improvement endeavors. By employing tailored techniques and protocols for each crop, breeders can effectively analyze genetic markers, identify desirable traits, and selectively breed plants with superior characteristics. Advances in genotyping make breeding crops faster and more precise, leading to the development of crops with higher yields, better disease resistance, and greater tolerance to environmental stressors. Furthermore, the amalgamation of genotyping data with advanced bioinformatics tools enriches our comprehension of intricate genetic interactions. Consequently, genotyping tools and customized approaches assume a pivotal role in ensuring global food security and addressing pressing agricultural challenges.
4.1 Tomato
Genotyping technologies in tomato have advanced significantly over the years, providing valuable insights into the genetic diversity and traits of this important crop. In 2012, Sim et al. (2012) used a high-density SNP array (SolCAP SNP Array) to genotype a collection of 426 tomato accessions, revealing seven sub-populations with distinct genetic characteristics. The study demonstrated the impact of selection on genome variation and identified candidate loci under positive selection. In 2013, Víquez-Zamora et al. (2013) employed next-generation sequencing and high-throughput genotyping to identify a set of 5,528 SNPs, allowing for the evaluation of tomato germplasm at various levels. They discovered markers distinguishing commercial varieties and wild relatives, providing valuable tools for genotyping and phylogenetic analysis. Celik et al. (2017) focused on S. pimpinellifolium, a wild species with high breeding potential; using a genotyping by sequencing (GBS) approach, they identified 3,125 unique SNP loci and demonstrated the efficiency of GBS for QTL mapping and introgression of favorable alleles. In 2019, Barchi et al. (2019a) introduced single primer enrichment technology (SPET) for targeted genotyping in tomato and eggplant. Their panel of 5k target SNPs enabled high-throughput genotyping, generating many SNPs, including novel ones, with cross-transferability between related species. Finally, Ziarsolo et al. (2021) presented the K-seq, a cost-effective genotyping methodology based on Klenow amplification and Illumina sequencing. K-seq demonstrated reproducibility and reliability, providing valuable genetic data for diverse species, including tomato.
4.2 Pepper
Research on pepper genotyping technologies has made significant strides over the years, contributing to our understanding of genetic diversity and population structure in Capsicum spp., also investigating the genetic segregation exploded within several mapping populations (Mimura et al., 2012; Borovsky et al., 2019; Zhu et al., 2019; Lozada et al., 2022; Yi et al., 2022). In 2016, Taranto et al. (2016) employed genotyping by sequencing (GBS) to identify SNPs in a collection of Capsicum spp. accessions, with a focus on cultivated pepper (C. annuum) genotypes. The GBS analysis resulted in the detection of 108,591 SNP markers, of which 105,184 were specific to C. annuum accessions. By further analyzing a subset of 222 C. annuum accessions using 32,950 high-quality SNPs, the genetic diversity and population structure of the collection was investigated, and the clustering analysis revealed three distinct clusters, with cluster I encompassing varieties and landraces primarily from Southern and Northern Italy, and Eastern Europe. Clusters II and III comprised accessions from various geographical origins, demonstrating the genetic differentiation of pepper based on both geographical origin and fruit-related features. Moving ahead, Cheng et al. (2016) focused on the development of a high-density interspecific genetic map and the characterization of genetic diversity in pepper. They selected 15,000 SNPs from resequencing data and synthesized an array with 12,720 loci, covering approximately 81.18% of the pepper genome. Using this array, they constructed a high-density genetic map with 5,569 SNPs, utilizing 297 F2 individuals. Lastly (Siddique et al. (2022) aimed to understand the genetics of resistance to Pepper yellow leaf curl virus (PepYLCV) in C. annuum. By utilizing GBS and constructing a high-density linkage map, they identified three QTLs associated with PepYLCV resistance on chromosomes P1, P7, and P12, respectively. Candidate genes within these QTL regions were inferred, and single markers derived from the QTLs were developed and validated in additional populations and commercial varieties.
4.3 Eggplant
Different approaches have been applied also in eggplant, not only investigating natural populations but also analyzing segregation patterns within mapping crosses, as reviewed by Gaccione et al. (2023). In 2012, Barchi et al. (2012) utilized the restriction-site associated DNA strategy combined with high throughput sequencing (RADseq) to develop a large number of functional SNP markers. This approach enabled the construction of a linkage map and the discovery of the genetic basis of anthocyanin-related traits in eggplant. In 2017, Acquadro et al. (2017) used a genotyping by sequencing (GBS) approach to detect SNPs polymorphisms in eggplant, working with a set of 76 accessions of species belonging to the brinjal (S. melongena), scarlet (S. aethiopicum) and gboma (S. macrocarpon) eggplant complexes, to detect the genetic relationships within and between the genepools. Moving to 2019, Liu et al. (2019b) employed the SNP genotyping technique (SNP-seq) to validate genome-wide perfect SNPs in eggplant. This high-throughput SNP genotyping technology allowed a genome-wide association study based on the 219 SNPs identified five associated SNPs located near the SUN and OVATE homologs, which had conserved function in controlling the fruit shape. In the same year, Barchi et al. (2019a) introduced SPET as a targeted genotyping solution, relying. This assay relies on sequencing a region flanking a single primer and simplifies panel design, allowing for higher levels of multiplexing. SPET was used to genotype a panel of target SNPs, both on coding regions and introns/UTRs, in tomato and eggplant, resulting in the discovery of thousands of closely linked, novel SNPs. Lastly, Wei et al. (2020b) utilized specific length amplified fragment (SLAF) sequencing for high-throughput SNP discovery in the eggplant genome. They constructed a high-density genetic map using these SNP markers for QTL analysis of multiple traits in eggplant.
4.4 Potato
Analyzing survey data derived from 32 genebanks, it is noteworthy that only two repositories, namely the International Potato Center in Peru and the Raphoe Potato Laboratory in Ireland, have conducted comprehensive genotyping on their respective potato germplasm. In contrast, 20 collections have undergone partial genotyping, and data dissemination has been limited, with only 10 genebanks having made their information publicly accessible thus far (Nagel et al., 2022). Historically, simple sequence repeat (SSR) markers, also identified as microsatellites, have been the predominant choice. together with amplified fragment length polymorphism (AFLP). Nevertheless, a shift toward the adoption of SNPs has been observed, facilitated by genotyping-by-sequencing (GBS) or the deployment of the SolCAP 8K, 12K, or 20K Infinium arrays (Nagel et al., 2022). Distinct regions exhibit unique foci in their genotyping endeavors. For instance, in Asia, specific screening of Japanese germplasm for the potato cyst nematode resistance has taken precedence, with over 1,000 accessions evaluated and potential resistant varieties identified after the initial detection of the nematode in Japan in 2015 (Asano et al., 2021). Conversely, European genebank collections are driven by diverse objectives such as conservation, breeding, and the establishment of reference collections. Consequently, the emphasis in genotyping efforts resides in the assessment of genetic diversity within the established genebanks. A case in point is the Nordic potato collection (NordGen), where 133 potato accessions, varieties, and breeding clones, predominantly cultivated under long daylength conditions in Europe, were genotyped using the Infinium Illumina 20K SNP array; it revealed striking genetic homogeneity, suggesting the need for the introduction of new genetic material for Nordic variety breeding (Selga et al., 2022). Similarly. the Estonian Crop Research Institute has undertaken the fingerprinting of over 450 potato varieties and landraces for the purpose of validating varieties and ascertaining the origin of the material. Employing 8 SSR markers, this endeavor unveiled unique accessions and identified instances of duplicated varieties (Ivanova-Pozdejeva et al., 2022), and the INRAE collection in France sought to develop core collections and facilitate marker-assisted selection, resulting in the fingerprinting of approximately 2,000 accessions using Cleaved Amplified Polymorphic Sequence (CAPS) markers and the SolCAP 8K array. Notably, also the Commonwealth Potato Collection, while only partially genotyped, has been extensively employed in diverse studies encompassing taxonomic and breeding aspects (Hawkes et al., 1994; Bradshaw and Ramsay, 2005; Spooner et al., 2005), as well as evaluations for resistances and tolerances against both biotic and abiotic stress, exemplified by its assessment for resistance to Potato Virus Y (Torrance et al., 2020). The International Potato Center’s germplasm collection has undergone thorough genotyping efforts, with specific findings detailed in (Ellis et al. (2020). Both cultivated and wild potato collections have been subjected to partial genotyping, utilizing techniques such as AFLP and SSR markers. For instance, 1,000 landrace accessions were genotyped (Ghislain et al., 2004), and 742 landraces along with select wild progenitors were also included in this genotyping initiative (Spooner et al., 2007). Moreover, the entire cultivated collection underwent genotyping using the SolCAP 12K array. Intriguingly, a comparative analysis of genotypic information from 250 accessions revealed mismatches between field-maintained plants and in vitro clones in slow-growth storage, with a comparable rate of genetic mismatches to other stock centers (Anastasio et al., 2011); a quarter of the cultivated collection was further genotyped using Diversity Arrays Technology sequencing (DarTseq), and approximately 2,000 accessions, spanning both cultivated and wild species, were subjected to genotyping through GBS. Of particular interest is the discernment that the exclusive utilization of data from the SolCAP 12K array can reliably predict the species, aligning with Hawkes taxonomy with a commendable degree of accuracy in North America. A comprehensively reviewed by Nagel et al. (2022), genotyping initiatives have been initiated across diverse collections, albeit with data accessibility remaining a partial facet. In anticipation of future breeding programs, the integration of contemporary technologies such as SolCAP, DArTs, or GBS approaches across entire collections is imperative. Furthermore, the strategic storage of emerging genotypic information in easily searchable databases is crucial for the informed progression of scientific endeavors in the field.
5 Classical mapping approaches for biotic and abiotic resistance
Developing mapping populations in horticultural crops holds significant importance for the advancement of plant breeding and genetic research. These populations, created through controlled crosses between selected parental lines, help to identify and locate specific genes or genomic regions associated with traits such as disease resistance, yield, quality attributes, and environmental adaptability. This information not only enhances our understanding of the underlying genetics but also enables breeders to make informed decisions in selecting parental combinations as well as for marker-assisted selection (MAS) for breeding programs to enhance overall crop productivity and sustainability. As resistance traits have been generally eroded by domestication, introgression breeding has emerged as a powerful tool in the field of plant breeding, facilitating the development of crop varieties with improved resistance to abiotic and biotic stresses (Rajpal et al., 2023). Using wild relatives, or related species that have naturally evolved tolerance to both stresses, introgression breeding enables the transfer of favorable traits into crops, broadens the genetic diversity of crop plants and enhances their adaptability and resilience to adverse environmental conditions when interspecific barriers are not present.
5.1 Tomato
While hundreds of research articles on the identification of QTLs for quality and productivity traits in tomato have been published in the last decades, starting from Weide et al. (1993), (Eshed and Zamir, 1994), and (Alpert et al., 1995), the focus on biotic and abiotic resistance for QTLs identification has been relatively limited; this could be potentially attributed to the challenges associated with designing robust experimental schemes for evaluating these traits. It is worth noting how, since the 1970s, the genetic diversity of tomato has witnessed a remarkable expansion, primarily due to the implementation of introgression breeding programs involving wild tomato relatives. Notably, the initial surge in genetic diversity occurred through the introgression of resistance traits such tomato mosaic virus (ToMV), southern root-knot nematode (Meloidogyne incognita), and leaf mold disease (Cladosporium fulvum) from S. pennelli and S. pimpinellifolium. These introgressions ranged in size, with some spanning approximately 5% of a chromosome (such as the introgressions of Cf-4 and Cf-9 on chromosome 1, conferring resistance to C. fulvum), while others covered half of chromosome 9 (ToMV). Collectively, these introgressions significantly enhanced the overall genetic diversity. A subsequent wave of diversity, starting in late ‘80s, had a dual impact on fruit size and quality traits (Schouten et al., 2019). One of the earliest studies identifying resistance QTLs to biotic stress (Bai et al., 2003) investigated the resistance of tomato to powdery mildew caused by Oidium lycopersici, reporting three QTLs on chromosomes 6 and 12. Panthee et al. (2017) focused on late blight resistance caused by Phytophthora infestans; in this work, the major genes (Ph-1, Ph-2, and Ph-3), along with additional minor QTLs associated with resistance, were successfully mapped in an inter-specific cross (S. lycorpersicum x S. pimpinellifolium). Qi et al. (2022) focused on Tomato spotted wilt virus (TSWV) resistance and fine-mapped the NLR gene Sl5R-1, which regulates resistance to TSWV, by identifying a gene cluster consisting of three genes (Sl5R-1, Sl5R-2, and Sl5R-3) that code for NBS-LRR proteins, along with the transcription factors regulating the resistance mechanism. Moving to abiotic stress resistance, Arms et al. (2015) explored the tolerance of wild tomato (S. habrochaites) root chilling, identifying a major QTL (stm9), on chromosome 9, which controlled shoot turgor maintenance under root chilling. Successively, Wen et al. (2019) identified five major QTLs related to heat tolerance and identified candidate genes within these QTLs using RNA-seq analysis. Table 2 shows a list of loci that have been associated with candidate genes for resistance to abiotic and biotic stress in tomato.
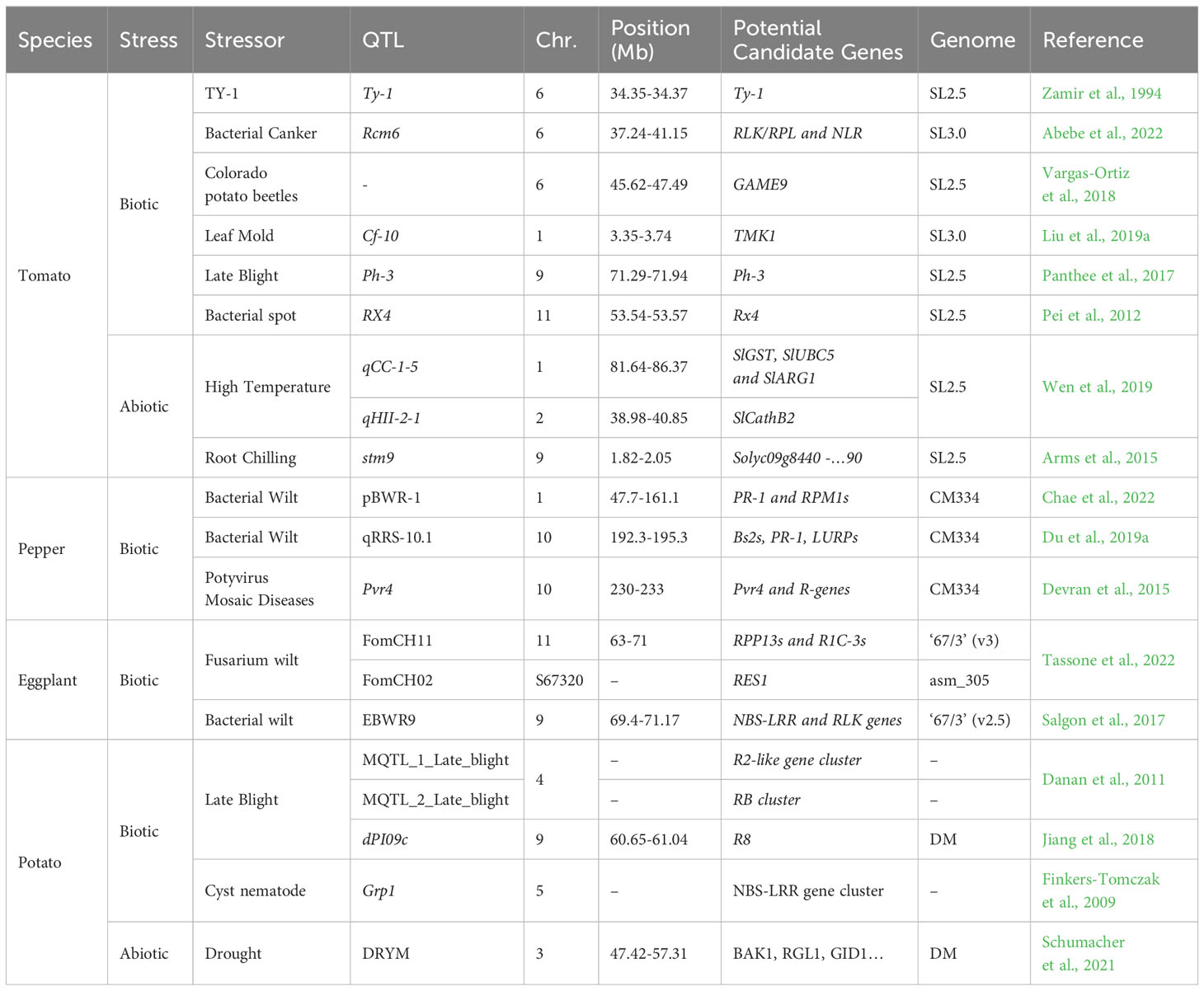
Table 2 List of QTLs and potential candidate genes for resistance to abiotic and biotic stress in the four Solanaceae.
5.2 Pepper
As explained, wild relatives are the best reservoir of resistance alleles from which a breeder can draw, and have been used since mid ‘90s to introgress resistances in the cultivated pepper genetic background (Lefebvre et al., 1995). However, interspecific crosses could be hard to obtain, according to species, and in pepper hybridization is usually complicated by interspecific barriers, which can preclude crossing within the Capsicum genus (Parry et al., 2021). Thus, searching for alleles already available in the C. annuum genetic background has represented a valuable option since early 2000 (Ben-Chaim and Paran, 2000). Intraspecific mapping populations have been applied also for resistance breeding: Eun et al. (2016) searched for cucumber mosaic virus (CMV) resistance in C. annuum, identifying two QTLs accounting for a notable proportion of phenotypic variation within the mapping population, and providing valuable information for the development of CMV-resistant pepper cultivars. More recently, Du et al. (2019a) aimed to uncover the genetic basis of pepper’s resistance to bacterial wilt. Their findings demonstrated that resistant rootstocks effectively suppressed the spread of Ralstonia solanacearum into the scion. Through specific-locus amplified fragment sequencing and bulked segregant analysis, two adjacent resistance-associated regions on chromosome 10 were identified. Anthracnose, a destructive disease affecting pepper crops, was the focus of investigation by Zhao et al. (2020). The study aimed to identify the gene responsible for anthracnose resistance in pepper through re-sequencing genotyping. The anti-anthracnose locus AnRGO5 was mapped to a specific region on the P5 chromosome, in a region containing five genes. These findings provide valuable information for future studies on the genetic basis of anthracnose resistance and marker-assisted selection in pepper breeding. Sun et al. (2020) focused on aphid resistance in Capsicum baccatum. The study conducted a QTL analysis for Myzus persicae resistance and identified two QTLs on chromosome 2, affecting aphid survival and reproduction. Fine mapping narrowed down the major QTL, Rmprp-1, to a genomic region encoding analogues of resistance genes belonging to the leucine-rich repeat domain (LRR-RLKs) family. The research conducted by van Haperen et al. (2021) focused on thrips resistance in Capsicum, fine-mapping a major QTL for thrips resistance on chromosome 6 and identifying 15 candidate genes within the QTL region. Lastly, in Chae et al. (2022), the genetic regions associated with bacterial wilt (BW) resistance in pepper crops were explored. Through QTL analysis, genomic loci and alleles critical for BW resistance were identified. Genotyping-by-sequencing was employed, resulting in the construction of a pepper genetic linkage map revealing the presence of a significant QTL, pBWR-1, on chromosome 1, explaining a substantial proportion of phenotypic variance. As the main findings on biotic resistance in pepper have been extensively reviewed by Parisi et al., 2020, including several lists of alleles donors and identified QTLs, Table 2 presents a brief overview of the latest findings in pepper genetic resistances.
5.3 Eggplant
Recently, Toppino et al. (2022) comprehensively reviewed the state of the art of the eggplant research on introgression breeding for resistances trait, reporting that, while strides have been made in mapping traits related to fruit features and biotic stress resistance, the full potential of marker-assisted breeding for a wider range of abiotic and biotic stresses remains largely unexplored. The availability of introgression lines carrying stress-resistance genes from wild relatives like S. linneanum (Doganlar et al., 2002; Frary et al., 2003) and S. incanum presents a promising avenue for uncovering chromosomal regions responsible for conferring tolerance, but further research and population screening hold the potential to unveil these mechanisms and contribute to the development of more robust and resilient eggplant varieties. Table 2 reports the identified QTLs for biotic and abiotic resistance for which candidate genes have been identified.
5.4 Potato
Several wild potato species has been reported to be suitable as introgression material, and have been used for mapping since early ‘90s (Barone et al., 1990; Kreike et al., 1993), both for biotic and abiotic resistances. Among them, seven species have been classified as primary gene pool (i.e. closely related species that can be generally crossed to cultivated varieties), 62 as secondary gene pool, and 24 as tertiary gene pool (Castañeda-Álvarez et al., 2015). Moreover, the species belonging to the primary and secondary gene pool are the mostly used for breeding and introgression of favorable traits (Maxted et al., 2012). The species belonging to the primary gene pool have been reported to be suitable for introgressing a number of desirable alleles, including frost tolerance (S. acaule), blackleg and soft rot resistance (S. berthaultii and S. candolleanum), Verticillium wilt (S. berthaultii and S. vernei), Globodera pallida, and cyst nematode (S. brevicaule and S. vernei) resistance, while the ones belonging to the secondary pool have been used as introgression material for late blight resistance (S. stoloniferum, S. raphanifolium, S. demissum and S. verrucosum), Potato Virus Y resistance (S. palustre), Potato Leaf Roll virus resistance (S. demissum), drought and heat tolerance (S. chacoense), frost tolerance (S. boliviense), blackleg and soft rot resistance (and S. hjertingii), and; Verticillium wilt resistance (S. chacoense; Nagel et al., 2022). For example, Dong et al. (2023) delved into the abiotic challenge of frost tolerance in potatoes. By constructing a BC1 population from cold-resistant (S. commersonii) and cold-sensitive (S. verrucosum) wild species, the study identified four QTLs on chromosomes 2 and 11 associated with frost tolerance. The inclusion of bulked segregant analysis and identification of candidate genes adds depth to the understanding of the complex genetic architecture governing frost resistance in potatoes under varying low-temperature conditions. Not only wild species, but also commercial material can be a reserve for resistance and tolerance alleles. Santa et al. (2018)focused on addressing the phytosanitary challenges posed by late blight and Guatemalan potato tuber moth in Colombia and Ecuador. By utilizing a F1 tetraploid segregant population, they successfully identified six QTLs linked to resistance against Phytophthora infestans and four QTLs related to Tecia solanivora. The genetic linkage map constructed demonstrated good distribution across the genome, providing valuable insights for breeding programs in regions where these pests limit potato production. Odilbekov et al. (2020) concentrated on early blight caused by Alternaria solani, causing an economic significant foliar disease in potato-growing regions. Through the characterization of a bi-parental tetraploid population, the study identified several QTLs associated with resistance to early blight in both leaves and tubers. Notably, the absence of a statistically significant correlation between resistance scores in leaves and tubers highlights the complexity of the genetic basis for this trait. In 2021, da Silva Pereira et al. (2021) addressed the common scab caused by Streptomyces spp., an understudied bacterial disease affecting the potato industry, identifying a QTL on chromosome 3 contributing to disease resistance, providing valuable information for genomics-assisted breeding approaches. Collectively, these studies contribute significantly to the development of resistant cultivars, addressing both biotic and abiotic challenges faced by potato crops globally.
6 Modern approaches for the identification and selection of resilience alleles
In recent years, several innovative approaches have been developed for the identification and selection of resilience alleles (Magon et al., 2023; Villalobos-López et al., 2022), enabling a deeper understanding of the genetic basis of resilience in various organisms. Transcriptomics have played a crucial role. By comparing the transcriptomes of resilient and susceptible individuals, candidate genes involved in such traits can be identified. Additionally, modern mapping approaches such as genome wide association (GWA) studies and bulked segregant analysis (BSA) combined with sequencing technologies have proven effective in identifying genetic markers associated with resilience traits. These approaches provide insights into the specific genomic regions and alleles contributing to plant adaptability. Furthermore, Genomic Prediction leverages genomic data to predict the resilience potential of individuals, facilitating targeted breeding programs. Finally, the integration of phenomics, which involves high-throughput phenotypic data collection, enables the comprehensive characterization of resilience traits and their associations with genomic information. The combination of all these approaches offers promising avenues for accelerating the identification, selection, and breeding of resilient individuals across various organisms, ultimately contributing to the development of more resilient and sustainable agricultural and ecological systems.
6.1 Transcriptomics for abiotic/biotic resilience-related genes identification
Transcriptomics is a powerful tool for identifying candidate genes associated with abiotic and biotic resilience. By analyzing gene expression patterns, researchers can uncover genes involved in tolerance to drought, heat, salinity, pathogen resistance, and pest resistance. Comparative transcriptomic analyses between resilient and susceptible individuals enable the identification of differentially expressed genes as potential candidates, without the need for developing and testing complex mapping populations. These findings have significant implications for enhancing crops’ resilience, but further research is usually needed to characterize the precise roles of these candidate genes and develop targeted strategies for resilience enhancement. As set of examples, here we collected a sample of the latest research applying RNAseq for the identification of abiotic and biotic resilience-related genes in the four crop Solanaceae.
6.1.1 Tomato
- Ashrafi-Dehkordi et al. (2018) conducted a meta-analysis of gene expression integrating data from 391 microarray samples derived from 23 different experiments, identifying 2,336 differentially expressed genes (DEGs) involved in multiple stresses. Overall, 361 DEGs exhibiting conserved expression patterns between biotic and abiotic stresses, indicated some level of shared mechanisms in response to different stressors, but, in general, the number of genes differentially regulated in response to biotic stresses (1,862) far exceeded those regulated by abiotic stresses (835) gene were categorized and some enrichments on transcription factor (TF) families were spotted, emphasizing the crucial role of TFs in orchestrating stress-responsive gene expression. In a more recent study, Zhou et al. (2019) focused specifically on the combined effects of cold and drought stress in tomatoes, investigating the physiological and genetic tomato responses. The study found that the combined stress condition altered physiological parameters such as water loss rate, H2O2 content, and hormone levels (e.g., ABA, IAA, ZR, melatonin), highlighting the close interaction between physiological and genetic responses, and the related complex regulatory mechanisms. More recently, Amoroso et al. (2023), resembling the work by Ashrafi-Dehkordi et al. (2018) but going further the limits of microarray technologies, applied RNA-seq for the investigation of tomato plants exposed to seven biotic stresses; the authors included Cladosporium fulvum, Phytophthora infestans, Pseudomonas syringae, Ralstonia solanacearum, Sclerotinia sclerotiorum, Tomato spotted wilt virus (TSWV), Tuta absoluta, and five abiotic stresses (namely drought, salinity, low temperatures, and oxidative stress). Following the analysis, they discovered a total of 1,474 DEGs that were commonly shared between biotic and abiotic stresses. Among these, 67 DEGs exhibited response patterns in at least four different stress conditions, signifying their pivotal roles in stress adaptation. The identified DEGs encompassed a diverse set of genes associated with stress response pathways. Notable examples included Receptor-Like Kinases (RLKs), Mitogen-Activated Protein Kinases (MAPKs), Fasciclin-like arabinogalactans (FLAs), glycosyltransferases, genes involved in auxin, ethylene (ET), and jasmonic acid (JA) pathways, as well as MYBs, basic leucine zippers (bZIPs), WRKYs, and Ethylene Responsive Factor (ERF) genes. All the latter gene families play crucial roles in mediating signal transduction, hormone regulation, and transcriptional reprogramming during stress response. The identified DEGs hold immense potential for further exploration using biotechnological approaches to enhance plant tolerance in real-world agricultural settings.
6.1.2 Pepper
Li et al. (2016) explored through RNAseq the role of brassinosteroids (BRs) and in particular of 24-epibrassinolide (EBR), during chilling stress. By supplementing EBR under low temperatures, the study identified some DEGs, likely regulated by EBR, associated with photosynthesis, hormone metabolism, redox homeostasis, signaling pathways, transcription, and defense. The results highlighted the synergistic crosstalk between BRs and other signaling pathways, such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ETH), occurring after chilling. More recently, Zhang et al. (2022b) focused on deciphering the molecular mechanisms underlying pepper’s response to cold stress. They conducted transcriptomic and metabolomic analyses on two pepper cultivars, XS (cold-sensitive) and GZ (cold-resistant), identifying key players for the response to cold stress. López-Serrano et al. (2021) compared the transcriptomic responses of a salt-tolerant accession (A25) and a salt-sensitive accession (A6) to NaCl stress. They identified DEGs and pathways associated with salt stress tolerance, highlighting how the up-regulated in the tolerant genotypes were involved in osmotic potential regulation, ion homeostasis, growth regulation, photosynthetic protection, antioxidant activity, and stress signaling. Notably, the ABA signaling pathway played a prominent role in salt tolerance, with key genes involved in ABA biosynthesis and signaling being upregulated in the salt-tolerant accession. Ma et al. (2023) investigated the alleviation of salt stress in pepper through the application of SA through aa transcriptomic analysis The study revealed that SA treatment regulated the expression of genes involved ion absorption and homeostasis, redox regulation, stress signaling, and transcriptional control. The authors suggested that SA acts as a key modulator in enhancing pepper’s tolerance to salt stress by orchestrating multiple physiological and molecular responses. Borràs et al. (2021) examined drought stress effects on bell pepper, identifying two NAC transcription factors (CaNAC072 and CaNAC104) with contrasting responses. A validation approach through gene silencing was attempted, confirming the role of CaNAC072 in enhancing drought tolerance, while not for CaNAC104. Lastly, Padilla et al. (2023) focused on understanding the tolerance mechanisms of NIBER®, a water stress and salt-tolerant pepper hybrid rootstock compared to a sensitive pepper accession (A10) to short-term water stress. Under water stress, NIBER® exhibited up-regulation of transcription factors like DREBs and MYC, as well as increased levels of auxins, abscisic acid (ABA), and jasmonic acid (JA). The rootstock also showed an increase in osmoprotectant sugars and antioxidants, along with enhanced expression of aquaporins and chaperones. These findings indicate the strategies employed by NIBER® to overcome water stress, involving osmotic regulation, hormone signaling, and stress response pathways, and offer potential targets for enhancing water stress tolerance in pepper.
6.1.3 Eggplant
Recently, Gaccione et al. (2023) comprehensively reviewed the state of the art of transcriptomic research for biotic and abiotic stress in eggplant, combining such information with QTLs data, when possible, with the aim of identifying potential candidates for eggplant resilience. For instance, QTLs and candidate genes associated with bacterial wilt resistance were organized in quantitative genomic regions (QGRs) associated with resistance traits on chromosome 4 and 1. For resistance to R. solanacearum, SmMYB44 (SMEL4.1_04g019540.1), on chromosome4, was reported to lead to spermidine accumulation and resistance genotypes. For fungal wilt resistance, QGRs were identified on chromosome 2 and 11, were Dirigent protein 23 have been associated with the FomE02.01 resistance locus.
6.1.4 Potato
Chandrasekar et al. (2022) investigated the impact of potato cyst nematodes (PCNs), a significant pest, on the resistant cultivar ‘Kufri Swarna’ and the susceptible cultivar ‘Kufri Jyoti’. Employing high-throughput RNA sequencing, the study revealed 791 differentially expressed genes (DEGs) in the resistant cultivar, including upregulated disease resistance genes (KIN) and transcription factors (WRKY, HMG, and MYB), as well as enriched pathways like mitogen-activated protein kinase (MAPK) signaling. In 2023, Agho et al. (2023) assessed the field resistance of the Estonian potato cultivar ‘Ando’ to Phytophthora infestans, compared with the cultivar ‘Arielle’, identifying 5927 DEGs in the first on variety, and 1161 DEGs in the second one. Compared with ‘Arielle’, signal transduction, endocytosis, MAPK, autophagy, serine/threonine kinase activity, and plant-pathogen interaction pathways were reported to be significantly upregulated in ‘Ando’. Similarly, investigating the transcriptomic response to abiotic stress, Li et al. (2020) focused on the response of the salt-tolerant genotype ‘Longshu No. 5’ to salt stress. The study identified 5,508 DEGs and highlighted the significant enrichment of categories related to nucleic acid binding, transporter activity, ion/molecule transport, ion binding, kinase activity, and oxidative phosphorylation. More recently, Jian et al. (2022) and Tang et al. (2023) delved into the molecular responses of potatoes to drought stress and ubiquitination, respectively. Jian et al. identified 12,798 DEGs in response to simulated drought stress, elucidating pathways such as galactose metabolism, fatty acid metabolism, and plant-pathogen interaction. Tang et al. comprehensively analyzed deubiquitinase (DUB) genes under PEG-induced drought stress, revealing 6,067 down-regulated and 4,950 up-regulated DEGs, with DUBs implicated in the regulation of osmotic stress.
Collectively, these studies have significantly advanced our understanding of the molecular mechanisms underlying Solanaceae’s resistance to biotic and abiotic stresses. By employing transcriptomic approaches, researchers have identified differentially expressed genes, enriched gene ontologies, and key molecular pathways associated with stress responses. Continued research in this field will definitively contribute to the development of increasingly sustainable agriculture.
6.2 Association studies
Effectively harnessing the potential of crop germplasm collections necessitates a full comprehension of the genetic diversity contained within them. Such understanding allows the identification of essential “core collections,” which consist of a limited number of accessions that represent as much diversity as possible. These core collections hold immense significance in evolutionary biology and breeding, playing a crucial role in planning phenotypic characterization, seeking genetic resources adapted to varying environmental conditions, selecting materials for breeding programs and hybridization experiments, as well as guiding the selection of accessions for genome sequencing and comparative studies. The advent of NGS technologies allowed the exploitation of GWAS (genome-wide association study) as a routine practice in uncovering associations between genotype and phenotype across many species. GWAS serves as an alternative to traditional bi-parental linkage mapping, enabling the dissection of quantitative traits in crops. Within the framework of GWAS, the prevalence of polymorphisms is influenced by the overall genetic variation present within the germplasm panel. Similarly, the non-random associations between genetic markers, known as linkage disequilibrium, are influenced by factors beyond recombination frequency, including genetic drift, the plant’s mating system, and historical selection events. It is worth noting that population structure can introduce misleading connections between potential markers and phenotypes, especially when there are variations in trait means or frequencies across subpopulations. However, understanding the population structure within germplasm collections can help mitigate this risk.
6.2.1 Tomato
Since the first GWAS in tomato (Sauvage et al., 2014),several investigations have been conducted to investigate various aspects of tomato genetics and resistance to diseases. For example, Nguyen et al. (2021) investigated the genetic factors associated with resistance to bacterial wilt (BW) in tomato. Their research focused on a core collection of 191 tomato varieties and employed the 51 K Axiom® tomato array for genotyping. The team identified eight marker-trait associations (MTAs) linked to BW resistance, with two consistently observed on chromosomes 4 and 12. These QTLs explained a significant proportion of the observed variations. Notably, the study successfully demonstrated the effectiveness of the identified SNP markers in distinguishing resistant tomatoes within commercial cultivars. Additionally, the researchers identified four MTAs specific to disease assays, implying the existence of environment-specific QTLs on chromosomes 1 and 8–10. More recently, Park et al. (2022) identified novel QTLs for Powdery Mildew Resistance (PMR) through a GWA approach. They discovered five novel QTL regions associated with PMR, clearly distinct from previously reported ones, and identified three promising candidate genes (two nucleotide binding site-leucine rich repeat class genes and a receptor-like kinase gene).
6.2.2 Pepper
GWA studies have been performed also in pepper, identifying mainly agronomical and quality traits like fruit shape, fruit size, and male sterility (Nimmakayala et al., 2016; Colonna et al., 2019; Wu et al., 2019; Wu et al., 2022). While Potnis et al. (2019) identified loci associated with defoliation in response to infection with Xanthomonas gardneri in the USDA core collection, (Ro et al., 2022a) focused on Phytophthora blight resistance in pepper and performed a GWAS to identify SNPs associated with resistance to Phytophthora capsici and identifying by high-resolution melting (HRM) two markers: Chr02-1126 marker predicted P. capsici resistance with 78.5% accuracy, while the QTL5-1 marker predicted resistance with 80.2% accuracy.
6.2.3 Eggplant
In the same way, few GWAS analyses have been conducted in eggplant, mainly on agronomical and quality traits (Cericola et al., 2014; Portis et al., 2015; Arrones et al., 2022; Ro et al., 2022b), but studies focusing on resistance traits have not been published yet. On the other hand,
The results from these studies, especially when sustained by QTL analysis data, provide useful information for the understanding of the genomic regions mainly involved in resistance traits, and might be targeted in future experiments to depict candidate genes and understand the genetic mechanisms regulating tolerance in high-value genotypes within core-collections.
6.2.4 Potato
As for other species, several studies have identified SNPs associated with agronomical and quality traits, such as tuber shape, eye depth, and fry color retention during low-temperature storage (Sharma et al., 2018; Byrne et al., 2020; Zhang et al., 2022a; Pandey et al., 2023; Zhao et al., 2023). Moreover, multiple genetic markers and candidate genes aimed at enhancing disease resistance to late blight have been identified by Wang et al. (2021) and Sood et al. (2023), while Díaz et al. (2021) and Alvarez-Morezuelas et al. (2023) investigated plant’s response to abiotic stressors, revealing genes and QTLs that may aid in developing varieties better suited to changing environmental conditions and drought seasons. Additionally, Vos et al. (2022) identified two haplotypes (namely SGT1 and SGT2) as strictly associated with the α-solanine and α-chaconine, two steroidal glycoalkaloids— plant defense molecules with anti-nutritional properties for humans— that interact play a key role in plant’s defense against pathogens.
6.3 Innovative mapping approaches
Novel mapping strategies have emerged as alternatives to the conventional bi-parental populations for genetic mapping in crops, aiming to overcome the limitations associated with mapping precision and genetic diversity (Arrones et al., 2020; Scott et al., 2020; Yang et al., 2021). One such innovative approach is the advanced inter-cross (AIC; Rockman and Kruglyak, 2008) design, which entails the introduction of additional crossing generations before conducting genetic mapping. Despite its potential benefits, AIC has not been extensively adopted in crop studies, potentially due to the increased complexity it entails, and the labor-intensive manual crossing required for self-fertilizing species. Indeed, two main strategies have gained attention by the research community: nested association mapping (NAM; Yu et al., 2008) and multi-parent advanced generation inter-cross (MAGIC; Cavanagh et al., 2008) populations. As depicted in Figure 1, NAM populations involve crosses between a recurrent founder line and multiple alternative founders, akin to sets of bi-parental populations interlinked through a common parent, thereby making it conceptually familiar to researchers accustomed to working with bi-parental populations. While NAM populations offer increased genetic diversity, their approach to capturing greater genetic recombination largely relies on expanding the number of lines screened, much like bi-parental populations. In contrast, the MAGIC design represents a more intricate approach, building upon the principles of AIC. It involves the inter-crossing of several founders over multiple generations, followed by silting to generate inbred lines. Typically, MAGIC populations descend from a variable number of parents, commonly 4, 8, or 16, adhering to a straightforward funnel breeding design, and each line inherits alleles from all parents, resulting in the formation of random mosaics of founder haplotypes. By harnessing increased genetic recombination and genetic variation, MAGIC populations strive to overcome the limitations inherent to bi-parental populations in QTL mapping endeavors. The advantages offered by these novel mapping population strategies encompass heightened mapping power and resolution, expanded genetic diversity vis-à-vis bi-parental populations, and the minimal presence of population structure. Consequently, these strategies have witnessed an upsurge in popularity, leading to the development of multiple populations catering to various major crops. Their availability presents opportunities for QTL replication across diverse populations and enables the amalgamation of different populations to augment mapping power. However, it is worth noting that such populations are conceived with a long-term perspective, serving as enduring platforms for mapping non-target traits and exploring the genetic architecture of complex traits and the interrelationships between traits in the source population.
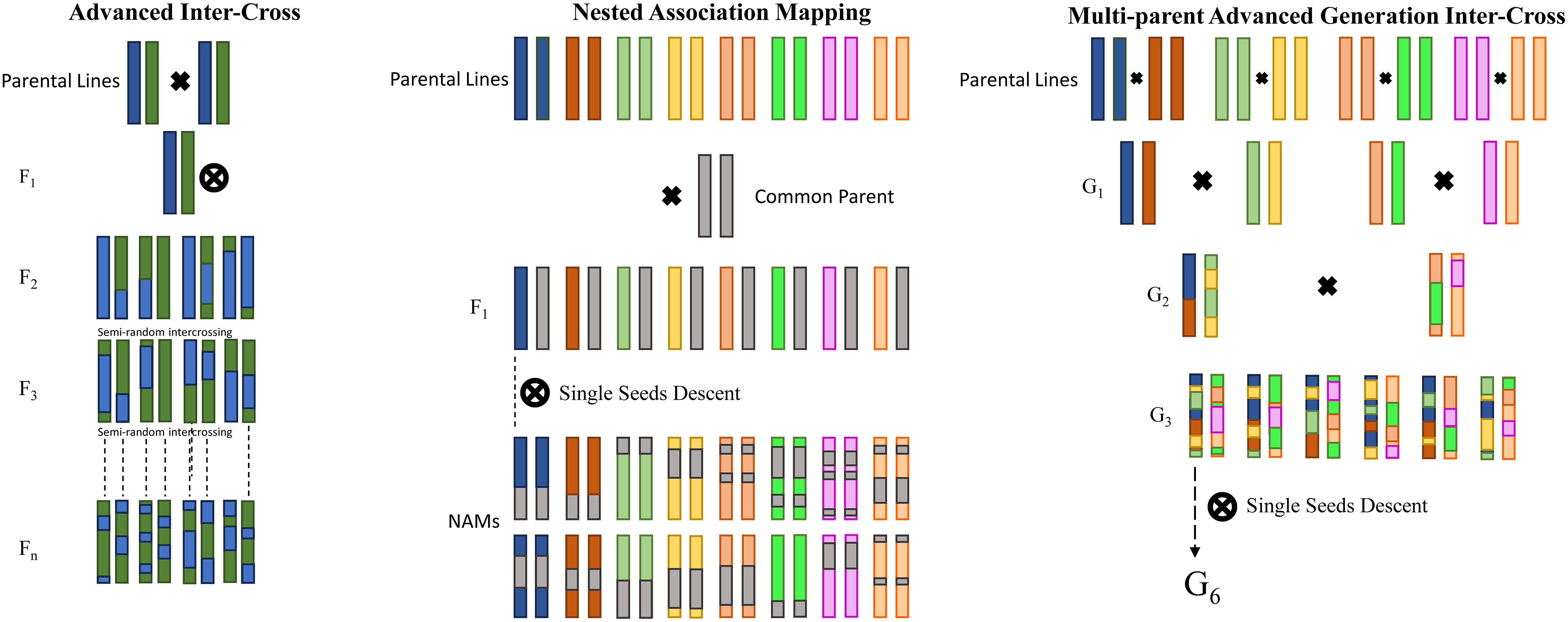
Figure 1 Graphical presentation of three advanced mapping approaches. From the left: advanced inter-cross (AIC), nested association mapping (NAM), and multi-parent advanced generation inter-cross (MAGIC).
6.3.1 Tomato
In 2015, Pascual et al. (2015) introduced the first tomato MAGIC population. Through resequencing of the founder genomes and their selection based on the obtained informative SNPs, they achieved a significant increase in recombination frequencies compared to biparental populations, facilitating haplotype prediction, and allowing QTL detection for fruit weight. Moreover, the combination of the MAGIC population with whole genome sequencing of the founders enabled the identification of candidate polymorphisms underlying QTLs, reducing the number of candidates from thousands to a handful. Using the same population, Diouf et al. (2018) assessed the impact of water deficit and salinity on fruit quality in contrasting environmental conditions. By evaluating 250 individual lines over two years, they identified QTLs for fruit quality and yield components, phenology traits, and a vegetative trait. The study mapped a total of 54 QTLs, with a considerable proportion (65%) being treatment specific. The MAGIC population facilitated high-resolution mapping and the projection of QTL intervals on the genome, enabling the identification of co-localized QTL regions. Candidate gene detection was performed based on the allelic effects of parental lines and sequence information, demonstrating the potential of the MAGIC population for dissecting the genetic basis of complex traits like fruit quality and elucidating genotype × environment interactions. Lastly, in a study by Bineau et al. (2021), the impact of high-temperature stress in tomato was investigated using both the MAGIC population developed by Pascual et al. (2015) and a core collection of small-fruited tomato accessions. The populations were evaluated for various yield components, phenology, and fruit quality traits under optimal and high-temperature conditions. The research demonstrated that high temperature significantly affected all traits, but a few genotypes with stable yield under high-temperature stress were identified. Plasticity indices were calculated to measure the extent of the heat impact on each trait. QTL analysis in the MAGIC and core collection populations led to the identification of a total of 69 plasticity QTLs associated with tomato heat response. Transcriptome analysis of the ovary in genotypes with contrasting heat responses revealed differentially expressed genes, providing insights into the molecular mechanisms underlying heat stress tolerance.
In 2019, Campanelli et al. (2019) further expanded the application of the MAGIC tomato population by leveraging it for the development of genotypes with desirable agronomic traits through Participatory Plant Breeding (PPB). Their MAGIC population was grown in an organic field experiment and, through phenotypic selection and molecular analysis, individuals with resistance genes and desired fruit shape were identified. Selected plants exhibiting desirable traits were used to generate stable lines in subsequent generations, providing an important genetic resource for the tomato scientific community. The MAGIC population was also cultivated in different organic farms, revealing significant phenotypic variations in development, productivity, and fruit color. This variability was harnessed to select tomato families adapted to specific environmental conditions, agricultural practices, and market requirements.
6.3.2 Pepper
While only two eight-way pepper population obtained using landraces as parental lines has been reported as “in progress” state at the Universitat Politècnica de València (Arrones et al., 2020) and Sativa (www.sativa.it), in collaboration with University of Torino (Martina et al., 2023), without further reports, in tomato two MAGIC populations, originated by crossing eight founder genotypes, have been reported.
6.3.3 Eggplant
Mangino et al. (2022) reported on the development of the first eggplant MAGIC population (S3MEGGIC), whose parental lines were previously genomically investigated by Gramazio et al. (2019). It consists of 420 individuals obtained by crossing seven cultivated eggplant varieties with one wild relative, S. incanum. After genotyping with eggplant SPET 5k probes GWA analysis revealed strong associations between specific genes involved in anthocyanin biosynthesis and the target traits, and two MYBs, similar to MYB113, a key transcription factor in the pathway, showed significant associations with anthocyanin presence in vegetative plant tissues (PA) and fruit epidermis (FA). Additionally, a COP1 gene encoding a photo-regulatory protein exhibited significant associations with the light-insensitive anthocyanin pigmentation under the calyx (PUC) trait. Comparing the eggplant genome with the tomato genome, duplication events of an ancestral MYB113 gene were detected and its translocation from chromosome 10 to chromosome 1 was identified. These findings provide insights into the evolutionary history of anthocyanin-related genes in eggplant. The S3MEGGIC population represents the largest recombinant population available for eggplant research. Its extensive genetic diversity and trait associations make it a powerful tool for future eggplant genetics and breeding studies.
6.3.4 Potato
The polyploid nature of the potato genome, together with its tendency to inbreeding depression (Zhang et al., 2019), has reduced the possibilities of developing novel mapping approaches, and the only population that have been reported so far is a NAM population that uses M6, a diploid self-compatible inbred line S. chachoense harboring several resistances, including early blight (Alternaria solani), common scab (Streptomyces scabies), late blight (Phytophthora infestans), and cold-induced sweetening, as common progenitor (Jansky et al., 2014). However, recent developments in diploid self-compatible clones offer a promising solution to overcome inbreeding depression and replace the current out-breeding method with a true seed-based F1 hybrid system (de Vries et al., 2023). While this idea has been previously regarded as unrealistic, diploid breeding program has successfully produced self-compatible offspring by employing a homozygous progenitor with the Sli gene (Clot et al., 2020; Eggers et al., 2021). F1 hybrid potato breeding opens doors for the development of new potato products with desirable trait combinations, benefiting all stakeholders in the potato chain (Lindhout et al., 2011).
6.4 Phenomics and genomic prediction
Phenomics and genomic prediction (GP), followed by genomic selection (GS), are two groundbreaking approaches in the development of plant varieties (Zhao et al., 2019; Pasala and Pandey, 2020; Crossa et al., 2021, R2D2 Consortium et al., 2021; Budhlakoti et al., 2022; Parveen et al., 2023). Even if the vast amount of data provided by genotyping technologies has made phenotyping the current bottleneck in plant breeding, the deployment of remote sensing technologies has revolutionized phenotyping by enabling the collection of large volumes of precise and efficient data. By combining phenomic data with genomic prediction, which utilizes genome-wide marker datasets, researchers can significantly enhance the accuracy of yield predictions.
6.4.1 Tomato
In 2020, Cappetta et al. (2020) reviewed the potentiality of genomic prediction in tomato, highlighting how implementing GP requires optimizing field trial management, agricultural practices, seed production, phenotyping, and sequencing, but also that a careful evaluation of parameters such as inbreeding levels, marker metrics, and the number of individuals to assess is essential. According to the authors, the integration of GP in the Single Seed Descendent scheme and backcrossing programs can reduce the number of generations and shorten the selection process in tomato, and the identification of candidate genotypes and undesirable segments through genotyping platforms can enhance trait introgression. Successively, Cappetta et al. (2021) applied GP on a F4 population for the prediction of yield and soluble solid content (SCC). The developed models exhibited a high prediction accuracy for both yield production (0.729) and SCC (0.715). Implementing these predictive models improved the efficiency of selection in subsequent breeding cycles, showcasing the potential of GS in expediting the breeding of heat-tolerant tomato varieties. Furthermore, by annotating SNPs located in gene body regions and conducting QTL analysis, the study identified five potential candidate genes involved in the response to high temperatures. This information was integrated into a GS model, utilizing a calibrated panel of markers.
By applying phenomic tools, Johansen et al. (2020) conducted field and UAV-based phenotyping of a wild tomato species to predict biomass and yield based on UAV imagery. Shape features derived from UAV, such as plant area and border length, were found to be important predictors. Multispectral UAV imagery collected 2 weeks prior to harvest produced the highest explained variances for biomass and yield. The study demonstrated the feasibility of predicting biomass and yield up to 8 weeks prior to harvest.
6.4.2 Pepper
While no works have been previously reported in eggplant for these two novel applications, their potentialities were investigated in pepper by few researchers. Hong et al. (2020) evaluated the potential of genomic selection in predicting fruit traits such as length, shape, width, weight, and pericarp thickness. By using a core collection consisting of 302 C. annuum complex accessions and testing various genomic prediction models, the researchers identified the Reproducing Kernel Hilbert Space (RKHS) model as producing the highest prediction accuracies across the fruit traits. Such model was further validated using a separate population of recombinant inbred lines, achieving moderate prediction accuracies. Successively, Kim et al. (2022) focused on capsaicinoid contents in chili peppers and employed genomic selection to predict these traits. Capsaicinoids are responsible for the pungency of chili peppers, and their regulation has not been completely deciphered. The researchers utilized a core collection of 351 Capsicum accessions as a training population and 96 breeding lines as a testing population. They tested different numbers of genome-wide SNP markers and identified the optimal marker set for genomic selection, achieving the highest mean prediction accuracy. Through GWAS, they also identified markers associated with capsaicinoid biosynthesis genes and quantitative trait loci. Integrating these markers as fixed-effect markers improved the prediction models, leading to higher accuracies. This study laid the groundwork for developing pepper cultivars with different capsaicinoid levels using genomic selection for capsaicinoid contents.
On the phenomics side, Pereira-Dias et al. (2020) focused on the characterization of a diverse collection of C. annuum accessions from various regions in Spain, particularly the bell types known as Pimiento Morrón. By employing both conventional descriptors and high-throughput digital phenotyping using Tomato Analyzer software, the researchers evaluated 32 conventional traits and 35 digital traits. They found substantial variation in both types of traits, highlighting the diversity of Spanish peppers in terms of plant and fruit morphology. Digital phenotyping proved to be more accurate in discriminating varietal groups, with digital parameters outperforming conventional traits in distinguishing categories and revealing significant differences among closely related groups. Furthermore, a subset of four conventional descriptors and 13 Tomato Analyzer traits were identified as the most discriminant in distinguishing closely related C. annuum accessions. More recently, Fumia et al. (2023) investigated different methods of genomic and phenomic selection in the Capsicum core collection within the G2P-sol project, with the aim of identifying genotypes with heat tolerance traits for pepper breeding. By combining classical and multispectral phenotyping methods and integrating genomic data, they identified 33 promising genotypes for heat tolerance breeding, with 13 genotypes being selected by multiple methods. The study revealed that the combination of phenomic and genomic approaches yielded better selection results compared to individual methods. Interestingly, when each method was used independently, similar results were obtained, suggesting the robustness of the selection methods. This study exemplified the potential of utilizing high-throughput data, including phenomic and genomic information, to compensate for limited germplasm knowledge and overcome logistical or financial constraints in pepper breeding programs.
6.4.3 Potato
The progression of genomic prediction techniques in tetraploid potato breeding is evident through a series of innovative studies. Initial research conducted by Wilson et al. (2021) signified a shift towards models that integrate dominance for certain traits, with an inclination towards non-parametric models to better capture complex genetic interactions. Selga et al. (2022) then scrutinized the predictive power of Bayesian Ridge Regression for genomic estimated breeding values, shedding light on the intricacies involved in early selection phases. Subsequent investigations by Ortiz et al. (2022) offered fresh insights into various genome-based prediction strategies, emphasizing the role of genotype-environment dynamics, and supporting both linear and kernel-based models for traits assessment. Advancing these concepts, Adams et al. (2023) assessed prediction models within diploid hybrid potato breeding, aiming to enhance genetic improvements across compound traits. The most recent contribution from Cuevas et al. (2022) involved examining both single-trait and multi-trait multi-environment models, underscoring the efficacy of multi-trait approaches in complex trait forecasting. This succession of research underscores a clear trend towards more nuanced and robust predictive methodologies that accommodate the inherent complexities within potato breeding programs.
Similarly, potato phenomics research has made significant strides in overcoming traditional limitations associated with labor-intensive and time-consuming phenotyping techniques. de Jesus Colwell et al. (2021) addressed these challenges by employing point cloud data obtained from low-cost UAV imaging to create 3D surface models of potato plant canopies. This approach facilitated detailed and accurate measurements of plant height distribution, canopy ground cover, and canopy volume throughout the growing season. The study identified distinct patterns of canopy development, including growth, lodging, maturity, and senescence, exemplified by varieties ‘Nadine’, ‘Bonnie’, and ‘Bounty’. This innovative method holds promise for alleviating current bottlenecks in plant development studies, enabling larger-scale population-based field studies in the future. Neilson et al. (2021) focused on the importance of potato tuber shape as a quality trait for breeding and variety development. They introduced an image analysis pipeline using commercially available cameras to extract tuber shape statistics, offering a more efficient alternative to manual measurements. The study successfully classified mean tuber shape from field-grown plants within ideal parameters for processing markets with high accuracy. The identification of parents with higher breeding value for tuber shape based on progeny performance demonstrates the potential for improving breeding selection processes. Johnson et al. (2021) addressed the urgent need for rapid and automated identification of blight disease in potatoes. They introduced an automated system utilizing Mask Region-based Convolutional Neural Network (Mask R-CNN) architecture for detecting blight disease patches on potato leaves in field conditions. The model, trained on a dataset of 1,423 images, demonstrated high precision in differentiating between diseased and healthy portions of the leaf, offering a practical solution for timely disease management in potato farming. Liu et al. (2021) highlighted the importance of accurate phenotyping for external quality attributes of potato tubers in breeding. They developed a 3D image analysis method for counting potato eyes and estimating eye depth, overcoming the limitations of subjective naked-eye visual evaluation. The study identified shape uniformity traits with better discriminatory power between varieties, demonstrating the potential of 3D image analysis in improving the accuracy of phenotyping complex geometrical traits in potato breeding.
7 Concluding remarks and future perspectives
Altogether, research and breeding efforts have produced solid knowledge of Solanaceae genomics and genetics. Decades of varietal development have only tackled the surface of breeding for resilience traits, mainly focusing on diseases resistance and on the development of superior varieties, qualitatively fulfilling consumer liking and agronomically satisfying producers. Nowadays, it is pivotal to switch such tendencies, investing research efforts in the understanding of the resilience mechanisms to both biotic and abiotic stressors already available in the genebank-preserved germplasm. By reviewing the state of the art of the -omics research on Solanaceae, as summarized in Figure 2, with a special focus on research for resilient traits, our work provides a starting point for the development of the future experimental designs.
Author contributions
MM: Writing – review & editing, Writing – original draft. VDR: Writing – original draft, Writing – review & editing. GM: Writing – original draft, Writing – review & editing. AA: Writing – review & editing. LB: Writing – review & editing. GB: Writing – review & editing, Conceptualization. EDP: Writing – review & editing, Conceptualization. AV: Conceptualization, Writing – review & editing, Project administration. EP: Conceptualization, Writing – review & editing, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The overall work fulfils some goals of the Agritech National Research Center and received funding from the European Union Next-Generation EU (PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR)–MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4—D.D. 1032 17/06/2022, CN00000022). In particular, this study represents a review paper within Spoke 4 (Task4.1.1.) ‘Next-generation genotyping and -omics technologies for the molecular prediction of multiple resilient traits in crop plants’.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbass, K., Qasim, M. Z., Song, H., Murshed, M., Mahmood, H., Younis, I. (2022). A review of the global climate change impacts, adaptation, and sustainable mitigation measures. Environ. Sci. pollut. Res. 29, 42539–42559. doi: 10.1007/s11356-022-19718-6
Abebe, A. M., Oh, C.-S., Kim, H. T., Choi, G., Seo, E., Yeam, I., et al. (2022). QTL-seq analysis for identification of resistance loci to bacterial canker in tomato. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.809959
Acquadro, A., Barchi, L., Gramazio, P., Portis, E., Vilanova, S., Comino, C., et al. (2017). Coding SNPs analysis highlights genetic relationships and evolution pattern in eggplant complexes. PloS One 12, e0180774. doi: 10.1371/journal.pone.0180774
Acquadro, A., Barchi, L., Portis, E., Nourdine, M., Carli, C., Monge, S., et al. (2020). Whole genome resequencing of four Italian sweet pepper landraces provides insights on sequence variation in genes of agronomic value. Sci. Rep. 10, 9189. doi: 10.1038/s41598-020-66053-2
Adams, J., de Vries, M., van Eeuwijk, F. (2023). Efficient genomic prediction of yield and dry matter in hybrid potato. Plants 12, 2617. doi: 10.3390/plants12142617
Agho, C. A., Kaurilind, E., Tähtjärv, T., Runno-Paurson, E., Niinemets, Ü. (2023). Comparative transcriptome profiling of potato cultivars infected by late blight pathogen Phytophthora infestans: Diversity of quantitative and qualitative responses. Genomics 115, 110678. doi: 10.1016/j.ygeno.2023.110678
Alonge, M., Wang, X., Benoit, M., Soyk, S., Pereira, L., Zhang, L., et al. (2020). Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182, 145–161.e23. doi: 10.1016/j.cell.2020.05.021
Alpert, K. B., Grandillo, S., Tanksley, S. D. (1995). fw 2.2:a major QTL controlling fruit weight is common to both red- and green-fruited tomato species. Theoret. Appl. Genet. 91, 994–1000. doi: 10.1007/BF00223911
Alvarez-Morezuelas, A., Barandalla, L., Ritter, E., Ruiz de Galarreta, J. I. (2023). Genome-wide association study of agronomic and physiological traits related to drought tolerance in potato. Plants (Basel) 12, 734. doi: 10.3390/plants12040734
Amoroso, C. G., D’Esposito, D., Aiese Cigliano, R., Ercolano, M. R. (2023). Comparison of tomato transcriptomic profiles reveals overlapping patterns in abiotic and biotic stress responses. Int. J. Mol. Sci. 24, 4061. doi: 10.3390/ijms24044061
Anastasio, A. E., Platt, A., Horton, M., Grotewold, E., Scholl, R., Borevitz, J. O., et al. (2011). Source verification of mis-identified Arabidopsis thaliana accessions. Plant J. 67, 554–566. doi: 10.1111/j.1365-313X.2011.04606.x
Arms, E. M., Bloom, A. J., St. Clair, D. A. (2015). High-resolution mapping of a major effect QTL from wild tomato Solanum habrochaites that influences water relations under root chilling. Theor. Appl. Genet. 128, 1713–1724. doi: 10.1007/s00122-015-2540-y
Arrones, A., Mangino, G., Alonso, D., Plazas, M., Prohens, J., Portis, E., et al. (2022). Mutations in the SmAPRR2 transcription factor suppressing chlorophyll pigmentation in the eggplant fruit peel are key drivers of a diversified colour palette. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1025951
Arrones, A., Vilanova, S., Plazas, M., Mangino, G., Pascual, L., Díez, M. J., et al. (2020). The dawn of the age of multi-parent MAGIC populations in plant breeding: novel powerful next-generation resources for genetic analysis and selection of recombinant elite material. Biol. (Basel) 9, 229. doi: 10.3390/biology9080229
Asano, K., Shimosaka, E., Yamashita, Y., Narabu, T., Aiba, S., Sakata, I., et al. (2021). Improvement of diagnostic markers for resistance to Globodera pallida and application for selection of resistant germplasms in potato breeding. Breed. Sci. 71, 354–364. doi: 10.1270/jsbbs.20134
Ashrafi-Dehkordi, E., Alemzadeh, A., Tanaka, N., Razi, H. (2018). Meta-analysis of transcriptomic responses to biotic and abiotic stress in tomato. PeerJ 6, e4631. doi: 10.7717/peerj.4631
Aversano, R., Contaldi, F., Ercolano, M. R., Grosso, V., Iorizzo, M., Tatino, F., et al. (2015). The Solanum commersonii Genome Sequence Provides Insights into Adaptation to Stress Conditions and Genome Evolution of Wild Potato Relatives. Plant Cell 27, 954. doi: 10.1105/tpc.114.135954
Bai, Y., Huang, C.-C., van der Hulst, R., Meijer-Dekens, F., Bonnema, G., Lindhout, P. (2003). QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorum G1.1601 co-localize with two qualitative powdery mildew resistance genes. Mol. Plant Microbe Interact. 16, 169–176. doi: 10.1094/MPMI.2003.16.2.169
Barchi, L., Acquadro, A., Alonso, D., Aprea, G., Bassolino, L., Demurtas, O., et al. (2019a). Single primer enrichment technology (SPET) for high-throughput genotyping in tomato and eggplant germplasm. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01005
Barchi, L., Lanteri, S., Portis, E., Valè, G., Volante, A., Pulcini, L., et al. (2012). A RAD tag derived marker based eggplant linkage map and the location of QTLs determining anthocyanin pigmentation. PloS One 7, e43740. doi: 10.1371/journal.pone.0043740
Barchi, L., Pietrella, M., Venturini, L., Minio, A., Toppino, L., Acquadro, A., et al. (2019b). A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 9, 11769. doi: 10.1038/s41598-019-47985-w
Barchi, L., Rabanus-Wallace, M. T., Prohens, J., Toppino, L., Padmarasu, S., Portis, E., et al. (2021). Improved genome assembly and pan-genome provide key insights into eggplant domestication and breeding. Plant J. 107, 579–596. doi: 10.1111/tpj.15313
Barone, A., Ritter, E., Schachtschabel, U., Debener, T., Salamini, F., Gebhardt, C. (1990). Localization by restriction fragment length polymorphism mapping in potato of a major dominant gene conferring resistance to the potato cyst nematode Globodera rostochiensis. Mol. Gen. Genet. 224, 177–182. doi: 10.1007/BF00271550
Bebeli, P., Mazzucato, A. (2009) The Solanaceae – A Review of Recent Research on Genetic Resources and Advances in the Breeding of Tomato, Pepper and Eggplant. Available at: https://www.semanticscholar.org/paper/The-Solanaceae-%E2%80%93-A-Review-of-Recent-Research-on-and-Bebeli-Mazzucato/e8f13859977eaf6d4934aa06456e2f133db19dd6 (Accessed August 4, 2023).
Ben-Chaim, A., Paran, I. (2000). Genetic analysis of quantitative traits in pepper (Capsicum annuum). J. Am. Soc. Hortic. Sci. 125, 66–70. doi: 10.21273/JASHS.125.1.66
Bhandari, R., Neupane, N., Adhikari, D. P. (2021). Climatic change and its impact on tomato (lycopersicum esculentum l.) production in plain area of Nepal. Environ. Challenges 4, 100129. doi: 10.1016/j.envc.2021.100129
Bineau, E., Diouf, I., Carretero, Y., Duboscq, R., Bitton, F., Djari, A., et al. (2021). Genetic diversity of tomato response to heat stress at the QTL and transcriptome levels. Plant J. 107, 1213–1227. doi: 10.1111/tpj.15379
Borovsky, Y., Monsonego, N., Mohan, V., Shabtai, S., Kamara, I., Faigenboim, A., et al. (2019). The zinc-finger transcription factor CcLOL1 controls chloroplast development and immature pepper fruit color in Capsicum chinense and its function is conserved in tomato. Plant J. 99, 41–55. doi: 10.1111/tpj.14305
Borràs, D., Barchi, L., Schulz, K., Moglia, A., Acquadro, A., Kamranfar, I., et al. (2021). Transcriptome-based identification and functional characterization of NAC transcription factors responsive to drought stress in capsicum annuum L. Front. Genet. 12. doi: 10.3389/fgene.2021.743902
Bozan, I., Achakkagari, S. R., Anglin, N. L., Ellis, D., Tai, H. H., Strömvik, M. V. (2023). Pangenome analyses reveal impact of transposable elements and ploidy on the evolution of potato species. Proc. Natl. Acad. Sci. 120, e2211117120. doi: 10.1073/pnas.2211117120
Bradshaw, J. E., Ramsay, G. (2005). Utilisation of the Commonwealth Potato Collection in potato breeding. Euphytica 146, 9–19. doi: 10.1007/s10681-005-3881-4
Budhlakoti, N., Kushwaha, A. K., Rai, A., Chaturvedi, K. K., Kumar, A., Pradhan, A. K., et al. (2022). Genomic selection: A tool for accelerating the efficiency of molecular breeding for development of climate-resilient crops. Front. Genet. 13. doi: 10.3389/fgene.2022.832153
Byrne, S., Meade, F., Mesiti, F., Griffin, D., Kennedy, C., Milbourne, D. (2020). Genome-wide association and genomic prediction for fry color in potato. Agronomy 10, 90. doi: 10.3390/agronomy10010090
Campanelli, G., Sestili, S., Acciarri, N., Montemurro, F., Palma, D., Leteo, F., et al. (2019). Multi-parental advances generation inter-cross population, to develop organic tomato genotypes by participatory plant breeding. Agronomy 9, 119. doi: 10.3390/agronomy9030119
Cappetta, E., Andolfo, G., Di Matteo, A., Barone, A., Frusciante, L., Ercolano, M. R. (2020). Accelerating tomato breeding by exploiting genomic selection approaches. Plants 9, 1236. doi: 10.3390/plants9091236
Cappetta, E., Andolfo, G., Guadagno, A., Di Matteo, A., Barone, A., Frusciante, L., et al. (2021). Tomato genomic prediction for good performance under high-temperature and identification of loci involved in thermotolerance response. Hortic. Res. 8, 1–16. doi: 10.1038/s41438-021-00647-3
Castañeda-Álvarez, N. P., Haan, S., Juárez, H., Khoury, C. K., Achicanoy, H. A., Sosa, C. C., et al. (2015). Ex situ conservation priorities for the wild relatives of potato (Solanum L. Section petota). PloS One 10, e0122599. doi: 10.1371/journal.pone.0122599
Cavanagh, C., Morell, M., Mackay, I., Powell, W. (2008). From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11, 215–221. doi: 10.1016/j.pbi.2008.01.002
Celik, I., Gurbuz, N., Uncu, A. T., Frary, A., Doganlar, S. (2017). Genome-wide SNP discovery and QTL mapping for fruit quality traits in inbred backcross lines (IBLs) of solanum pimpinellifolium using genotyping by sequencing. BMC Genomics 18, 1. doi: 10.1186/s12864-016-3406-7
Cericola, F., Portis, E., Lanteri, S., Toppino, L., Barchi, L., Acciarri, N., et al. (2014). Linkage disequilibrium and genome-wide association analysis for anthocyanin pigmentation and fruit color in eggplant. BMC Genomics 15, 896. doi: 10.1186/1471-2164-15-896
Chae, S.-Y., Lee, K., Do, J.-W., Hong, S.-C., Lee, K.-H., Cho, M.-C., et al. (2022). QTL mapping of resistance to bacterial wilt in pepper plants (Capsicum annuum) using genotyping-by-sequencing (GBS). Horticulturae 8, 115. doi: 10.3390/horticulturae8020115
Chandrasekar, S., Natarajan, P., Mhatre, P. H., Mahajan, M., Nivitha, S., Palanisamy, V. E., et al. (2022). RNA-seq of cyst nematode infestation of potato (Solanum tuberosum L.): A comparative transcriptome analysis of resistant and susceptible cultivars. Plants 11(8), 1008. doi: 10.3390/plants11081008
Cheng, J., Qin, C., Tang, X., Zhou, H., Hu, Y., Zhao, Z., et al. (2016). Development of a SNP array and its application to genetic mapping and diversity assessment in pepper (Capsicum spp.). Sci. Rep. 6, 33293. doi: 10.1038/srep33293
Christov, N. K., Tsonev, S., Todorova, V., Todorovska, E. G. (2021). Genetic diversity and population structure analysis – a prerequisite for constructing a mini core collection of Balkan Capsicum annuum germplasm. Biotechnol. Biotechnol. Equip. 35, 1010–1023. doi: 10.1080/13102818.2021.1946428
Clot, C. R., Polzer, C., Prodhomme, C., Schuit, C., Engelen, C. J. M., Hutten, R. C. B., et al. (2020). The origin and widespread occurrence of Sli-based self-compatibility in potato. Theor. Appl. Genet. 133, 2713–2728. doi: 10.1007/s00122-020-03627-8
Colonna, V., D’Agostino, N., Garrison, E., Albrechtsen, A., Meisner, J., Facchiano, A., et al. (2019). Genomic diversity and novel genome-wide association with fruit morphology in Capsicum, from 746k polymorphic sites. Sci. Rep. 9, 10067. doi: 10.1038/s41598-019-46136-5
Contreras, A., Ciampi, L., Padulosi, S., Spooner, D. M. (1993). Potato germplasm collecting expedition to the Guaitecas and chonos Archipelagos, Chil. Potato Res. 36, 309–316. doi: 10.1007/BF02361797
Crossa, J., Fritsche-Neto, R., Montesinos-Lopez, O. A., Costa-Neto, G., Dreisigacker, S., Montesinos-Lopez, A., et al. (2021). The modern plant breeding triangle: optimizing the use of genomics, phenomics, and enviromics data. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.651480
Cuevas, J., Reslow, F., Crossa, J., Ortiz, R. (2022). Modeling genotype × environment interaction for single and multitrait genomic prediction in potato (Solanum tuberosum L.). G3 (Bethesda) 13, jkac322. doi: 10.1093/g3journal/jkac322
Danan, S., Veyrieras, J.-B., Lefebvre, V. (2011). Construction of a potato consensus map and QTL meta-analysis offer new insights into the genetic architecture of late blight resistance and plant maturity traits. BMC Plant Biol. 11, 16. doi: 10.1186/1471-2229-11-16
da Silva Pereira, G., Mollinari, M., Schumann, M. J., Clough, M. E., Zeng, Z.-B., Yencho, G. C. (2021). The recombination landscape and multiple QTL mapping in a Solanum tuberosum cv. ‘Atlantic’-derived F1 population. Heredity 126, 817–830. doi: 10.1038/s41437-021-00416-x
de Jesus Colwell, F., Souter, J., Bryan, G. J., Compton, L. J., Boonham, N., Prashar, A. (2021). Development and validation of methodology for estimating potato canopy structure for field crop phenotyping and improved breeding. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.612843
Devran, Z., Kahveci, E., Özkaynak, E., Studholme, D. J., Tör, M. (2015). Development of molecular markers tightly linked to Pvr4 gene in pepper using next-generation sequencing. Mol. Breed 35, 101. doi: 10.1007/s11032-015-0294-5
de Vries, M. E., Adams, J. R., Eggers, E., Ying, S., Stockem, J. E., Kacheyo, O. C., et al. (2023). Converting hybrid potato breeding science into practice. Plants 12, 230. doi: 10.3390/plants12020230
Díaz, P., Sarmiento, F., Mathew, B., Ballvora, A., Vásquez, T. M. (2021). Genomic regions associated with physiological, biochemical and yield-related responses under water deficit in diploid potato at the tuber initiation stage revealed by GWAS. PloS One 16, e0259690. doi: 10.1371/journal.pone.0259690
Diouf, I. A., Derivot, L., Bitton, F., Pascual, L., Causse, M. (2018). Water deficit and salinity stress reveal many specific QTL for plant growth and fruit quality traits in tomato. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00279
Doganlar, S., Frary, A., Daunay, M.-C., Lester, R. N., Tanksley, S. D. (2002). A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the solanaceae. Genetics 161, 1697–1711. doi: 10.1093/genetics/161.4.1697
Dong, J., Li, J., Deng, G., Chen, C., Jing, S., Song, B., et al. (2023). QTL analysis for low temperature tolerance of wild potato species Solanum commersonii in natural field trials. Scientia Hortic. 310, 111689. doi: 10.1016/j.scienta.2022.111689
Du, H., Wen, C., Zhang, X., Xu, X., Yang, J., Chen, B., et al. (2019a). Identification of a Major QTL (qRRs-10.1) That Confers Resistance to Ralstonia solanacearum in Pepper (Capsicum annuum) Using SLAF-BSA and QTL Mapping. Int. J. Mol. Sci. 20, 5887. doi: 10.3390/ijms20235887
Du, H., Yang, J., Chen, B., Zhang, X., Zhang, J., Yang, K., et al. (2019b). Target sequencing reveals genetic diversity, population structure, core-SNP markers, and fruit shape-associated loci in pepper varieties. BMC Plant Biol. 19, 578. doi: 10.1186/s12870-019-2122-2
Eggers, E.-J., van der Burgt, A., van Heusden, S. A. W., de Vries, M. E., Visser, R. G. F., Bachem, C. W. B., et al. (2021). Neofunctionalisation of the Sli gene leads to self-compatibility and facilitates precision breeding in potato. Nat. Commun. 12 (1), 4141. doi: 10.1038/s41467-021-24267-6
Ellis, D., Salas, A., Chavez, O., Gomez, R., Anglin, N. (2020). “Ex situ conservation of potato [Solanum section petota (Solanaceae)] genetic resources in genebanks,” in The Potato Crop: Its Agricultural, Nutritional and Social Contribution to Humankind. Eds. Campos, H., Ortiz, O. (Cham: Springer International Publishing), 109–138. doi: 10.1007/978-3-030-28683-5_4
Eshed, Y., Zamir, D. (1994). A genomic library of Lycopersicon pennellii in L. esculentum: A tool for fine mapping of genes. Euphytica 79, 175–179. doi: 10.1007/BF00022516
Eun, M. H., Han, J.-H., Yoon, J. B., Lee, J. (2016). QTL mapping of resistance to the Cucumber mosaic virus P1 strain in pepper using a genotyping-by-sequencing analysis. Hortic. Environ. Biotechnol. 57, 589–597. doi: 10.1007/s13580-016-0128-3
Fallahi, F., Abdossi, V., Bagheri, M., Ghanbari Jahromi, M., Mozafari, H. (2022). Genetic diversity analysis of Eggplant Germplasm from Iran: assessments by morphological and SSR markers. Mol. Biol. Rep. 49, 11705–11714. doi: 10.1007/s11033-022-07768-5
FAOSTAT (2021). Food and Agriculture Organization Corporate Statistical Database. Available at: https://www.fao.org/faostat/en/#data/QC
Finkers-Tomczak, A., Danan, S., van Dijk, T., Beyene, A., Bouwman, L., Overmars, H., et al. (2009). A high-resolution map of the Grp1 locus on chromosome V of potato harbouring broad-spectrum resistance to the cyst nematode species Globodera pallida and Globodera rostochiensis. Theor. Appl. Genet. 119, 165–173. doi: 10.1007/s00122-009-1026-1
Frankel, O. H. (1984). “Genetic perspectives of germplasm conservation,” in Genetic Manipulation: Impact on Man and Society (Cambridge: Cambridge University Press), 161–170.
Frary, A., Doganlar, S., Daunay, M. C., Tanksley, S. D. (2003). QTL analysis of morphological traits in eggplant and implications for conservation of gene function during evolution of solanaceous species. Theor. Appl. Genet. 107, 359–370. doi: 10.1007/s00122-003-1257-5
Frary, A., Nesbitt, T. C., Grandillo, S., Knaap, E., Cong, B., Liu, J., et al. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. doi: 10.1126/science.289.5476.85
Freire, R., Weisweiler, M., Guerreiro, R., Baig, N., Hüttel, B., Obeng-Hinneh, E., et al. (2021). Chromosome-scale reference genome assembly of a diploid potato clone derived from an elite variety. G3 Genes|Genomes|Genetics 11, jkab330. doi: 10.1093/g3journal/jkab330
Fu, Y.-B. (2017). The vulnerability of plant genetic resources conserved ex situ. Crop Sci. 57, 2314–2328. doi: 10.2135/cropsci2017.01.0014
Fumia, N., Kantar, M., Lin, Y., Schafleitner, R., Lefebvre, V., Paran, I., et al. (2023). Exploration of high-throughput data for heat tolerance selection in Capsicum annuum. Plant Phenome J. 6, e20071. doi: 10.1002/ppj2.20071
Gaccione, L., Martina, M., Barchi, L., Portis, E. (2023). A compendium for novel marker-based breeding strategies in eggplant. Plants 12, 1016. doi: 10.3390/plants12051016
Gao, L., Gonda, I., Sun, H., Ma, Q., Bao, K., Tieman, D. M., et al. (2019). The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 51, 1044–1051. doi: 10.1038/s41588-019-0410-2
Ghislain, M., Spooner, D. M., Rodríguez, F., Villamón, F., Núñez, J., Vásquez, C., et al. (2004). Selection of highly informative and user-friendly microsatellites (SSRs) for genotyping of cultivated potato. Theor. Appl. Genet. 108, 881–890. doi: 10.1007/s00122-003-1494-7
Gramazio, P., Yan, H., Hasing, T., Vilanova, S., Prohens, J., Bombarely, A. (2019). Whole-Genome Resequencing of Seven Eggplant (Solanum melongena) and One Wild Relative (S. incanum) Accessions Provides New Insights and Breeding Tools for Eggplant Enhancement. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01220
Gu, X., Cao, Y., Zhang, Z., Zhang, B., Zhao, H., Zhang, X., et al. (2019). Genetic diversity and population structure analysis of Capsicum germplasm accessions. J. Integr. Agric. 18, 1312–1320. doi: 10.1016/S2095-3119(18)62132-X
Guo, G., Pan, B., Yi, X., Khan, A., Zhu, X., Ge, W., et al. (2023). Genetic diversity between local landraces and current breeding lines of pepper in China. Sci. Rep. 13(1), 4058. doi: 10.1038/s41598-023-29716-4
Han, K., Jeong, H.-J., Yang, H.-B., Kang, S.-M., Kwon, J.-K., Kim, S., et al. (2016). An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 23, 81–91. doi: 10.1093/dnares/dsv038
Hawkes, J. G. (1941) Potato collecting expeditions in Mexico and South America. Available at: https://www.cabdirect.org/cabdirect/abstract/19411601242 (Accessed November 24, 2023).
Hawkes, J., Bradshaw, J., Mackay, G. (1994) Origins of cultivated potatoes and species relationships. Available at: https://www.semanticscholar.org/paper/Origins-of-cultivated-potatoes-and-species-Hawkes-Bradshaw/63b40b47de6d494fe3054c521aa6aad02f70d4d2 (Accessed November 24, 2023).
Hirakawa, H., Shirasawa, K., Miyatake, K., Nunome, T., Negoro, S., Ohyama, A., et al. (2014). Draft genome sequence of eggplant (Solanum melongena L.): the representative solanum species indigenous to the old world. DNA Res. 21, 649–660. doi: 10.1093/dnares/dsu027
Hong, J.-P., Ro, N., Lee, H.-Y., Kim, G. W., Kwon, J.-K., Yamamoto, E., et al. (2020). Genomic selection for prediction of fruit-related traits in pepper (Capsicum spp.). Front. Plant Sci. 11. doi: 10.3389/fpls.2020.570871
Hoopes, G., Meng, X., Hamilton, J. P., Achakkagari, S. R., de Alves Freitas Guesdes, F., Bolger, M. E., et al. (2022). Phased, chromosome-scale genome assemblies of tetraploid potato reveal a complex genome, transcriptome, and predicted proteome landscape underpinning genetic diversity. Mol. Plant 15, 520–536. doi: 10.1016/j.molp.2022.01.003
Hosmani, P. S., Flores-Gonzalez, M., Geest, H., Maumus, F., Bakker, L. V., Schijlen, E., et al. (2019). An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. bioRxiv 767764. doi: 10.1101/767764
Hulse-Kemp, A. M., Maheshwari, S., Stoffel, K., Hill, T. A., Jaffe, D., Williams, S. R., et al. (2018). Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic. Res. 5, 4. doi: 10.1038/s41438-017-0011-0
Ivanova-Pozdejeva, A., Kivistik, A., Kübarsepp, L., Tähtjärv, T., Tsahkna, A., Droz, E., et al. (2022). Fingerprinting of potato genotypes from Estonian genebank collection using SSR markers. Potato Res. 65, 153–170. doi: 10.1007/s11540-021-09514-z
Jansky, S., Chung, Y. S., Kittipadakul, P. (2014). M6: A diploid potato inbred line for use in breeding and genetics research. J. Plant Registrations 8, 195–199. doi: 10.3198/jpr2013.05.0024crg
Jian, H., Sun, H., Liu, R., Zhang, W., Shang, L., Wang, J., et al. (2022). Construction of drought stress regulation networks in potato based on SMRT and RNA sequencing data. BMC Plant Biol. 22, 381. doi: 10.1186/s12870-022-03758-8
Jiang, R., Li, J., Tian, Z., Du, J., Armstrong, M., Baker, K., et al. (2018). Potato late blight field resistance from QTL dPI09c is conferred by the NB-LRR gene R8. J. Exp. Bot. 69, 1545–1555. doi: 10.1093/jxb/ery021
Jin, L., Zhao, L., Wang, Y., Zhou, R., Song, L., Xu, L., et al. (2019). Genetic diversity of 324 cultivated tomato germplasm resources using agronomic traits and InDel markers. Euphytica 215, 69. doi: 10.1007/s10681-019-2391-8
Johansen, K., Morton, M. J. L., Malbeteau, Y., Aragon, B., Al-Mashharawi, S., Ziliani, M. G., et al. (2020). Predicting biomass and yield in a tomato phenotyping experiment using UAV imagery and random forest. Front. Artif. Intell. 3. doi: 10.3389/frai.2020.00028
Johnson, J., Sharma, G., Srinivasan, S., Masakapalli, S. K., Sharma, S., Sharma, J., et al. (2021). Enhanced field-based detection of potato blight in complex backgrounds using deep learning. Plant Phenomics 2021. doi: 10.34133/2021/9835724
Kim, G. W., Hong, J.-P., Lee, H.-Y., Kwon, J.-K., Kim, D.-A., Kang, B.-C. (2022). Genomic selection with fixed-effect markers improves the prediction accuracy for Capsaicinoid contents in Capsicum annuum. Horticulture Res. 9, uhac204. doi: 10.1093/hr/uhac204
Kim, S., Park, M., Yeom, S.-I., Kim, Y.-M., Lee, J. M., Lee, H.-A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Kim, S., Park, J., Yeom, S.-I., Kim, Y.-M., Seo, E., Kim, K.-T., et al. (2017). New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 18, 210. doi: 10.1186/s13059-017-1341-9
Kreike, C. M., de Koning, J. R., Vinke, J. H., van Ooijen, J. W., Gebhardt, C., Stiekema, W. J. (1993). Mapping of loci involved in quantitatively inherited resistance to the potato cyst-nematode Globodera rostochiensis pathotype Ro1. Theor. Appl. Genet. 87, 464–470. doi: 10.1007/BF00215092
Kyriakidou, M., Achakkagari, S. R., Gálvez López, J. H., Zhu, X., Tang, C. Y., Tai, H. H., et al. (2020). Structural genome analysis in cultivated potato taxa. Theor. Appl. Genet. 133, 951–966. doi: 10.1007/s00122-019-03519-6
Lee, S. G., Kim, S. K., Lee, H. J., Lee, H. S., Lee, J. H. (2018). Impact of moderate and extreme climate change scenarios on growth, morphological features, photosynthesis, and fruit production of hot pepper. Ecol. Evol. 8, 197–206. doi: 10.1002/ece3.3647
Lee, H.-Y., Ro, N.-Y., Jeong, H.-J., Kwon, J.-K., Jo, J., Ha, Y., et al. (2016). Genetic diversity and population structure analysis to construct a core collection from a large Capsicum germplasm. BMC Genet. 17, 142. doi: 10.1186/s12863-016-0452-8
Lee, J.-H., Venkatesh, J., Jo, J., Jang, S., Kim, G. W., Kim, J.-M., et al. (2022). High-quality chromosome-scale genomes facilitate effective identification of large structural variations in hot and sweet peppers. Horticulture Res. 9, uhac210. doi: 10.1093/hr/uhac210
Lefebvre, V., Palloix, A., Caranta, C., Pochard, E. (1995). Construction of an intraspecific integrated linkage map of pepper using molecular markers and doubled-haploid progenies. Genome 38, 112–121. doi: 10.1139/g95-014
Leisner, C. P., Hamilton, J. P., Crisovan, E., Manrique-Carpintero, N. C., Marand, A. P., Newton, L., et al. (2018). Genome sequence of M6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. Plant J. 94, 562–570. doi: 10.1111/tpj.13857
Li, Q., Qin, Y., Hu, X., Li, G., Ding, H., Xiong, X., et al. (2020). Transcriptome analysis uncovers the gene expression profile of salt-stressed potato (Solanum tuberosum L.). Sci. Rep. 10, 5411. doi: 10.1038/s41598-020-62057-0
Li, J., Yang, P., Kang, J., Gan, Y., Yu, J., Calderón-Urrea, A., et al. (2016). Transcriptome analysis of pepper (Capsicum annuum) revealed a role of 24-epibrassinolide in response to chilling. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01281
Liao, Y., Wang, J., Zhu, Z., Liu, Y., Chen, J., Zhou, Y., et al. (2022). The 3D architecture of the pepper genome and its relationship to function and evolution. Nat. Commun. 13, 3479. doi: 10.1038/s41467-022-31112-x
Lindhout, P., Meijer, D., Schotte, T., Hutten, R. C. B., Visser, R. G. F., van Eck, H. J. (2011). Towards F1 hybrid seed potato breeding. Potato Res. 54, 301–312. doi: 10.1007/s11540-011-9196-z
Liu, W., Qian, Z., Zhang, J., Yang, J., Wu, M., Barchi, L., et al. (2019b). Impact of fruit shape selection on genetic structure and diversity uncovered from genome-wide perfect SNPs genotyping in eggplant. Mol. Breed. 39, 140. doi: 10.1007/s11032-019-1051-y
Liu, J., Xu, X., Liu, Y., Rao, Z., Smith, M. L., Jin, L., et al. (2021). Quantitative potato tuber phenotyping by 3D imaging. Biosyst. Eng. 210, 48–59. doi: 10.1016/j.biosystemseng.2021.08.001
Liu, F., Zhao, J., Sun, H., Xiong, C., Sun, X., Wang, X., et al. (2023). Genomes of cultivated and wild Capsicum species provide insights into pepper domestication and population differentiation. Nat. Commun. 14, 5487. doi: 10.1038/s41467-023-41251-4
Liu, G., Zhao, T., You, X., Jiang, J., Li, J., Xu, X. (2019a). Molecular mapping of the Cf-10 gene by combining SNP/InDel-index and linkage analysis in tomato (Solanum lycopersicum). BMC Plant Biol. 19, 15. doi: 10.1186/s12870-018-1616-7
López-Serrano, L., Calatayud, Á., López-Galarza, S., Serrano, R., Bueso, E. (2021). Uncovering salt tolerance mechanisms in pepper plants: a physiological and transcriptomic approach. BMC Plant Biol. 21, 169. doi: 10.1186/s12870-021-02938-2
Loskutov, I. G., Institute I. P. G. R (1999) Vavilov and his institute: a history of the world collection of plant genetic resources in Russia. Available at: https://cgspace.cgiar.org/handle/10568/104272 (Accessed November 24, 2023).
Lozada, D. N., Bosland, P. W., Barchenger, D. W., Haghshenas-Jaryani, M., Sanogo, S., Walker, S. (2022). Chile pepper (Capsicum) breeding and improvement in the “Multi-omics” Era. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.879182
Ma, J., Wang, Y., Wang, L., Lin, D., Yang, Y. (2023). Transcriptomic analysis reveals the mechanism of the alleviation of salt stress by salicylic acid in pepper (Capsicum annuum L.). Mol. Biol. Rep. 50, 3593–3606. doi: 10.1007/s11033-022-08064-y
Mangino, G., Arrones, A., Plazas, M., Pook, T., Prohens, J., Gramazio, P., et al. (2022). Newly developed MAGIC population allows identification of strong associations and candidate genes for anthocyanin pigmentation in eggplant. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.847789
Martina, M., Gianoglio, N., Barchi, L., Milanesi, C., Acquadro, A., Portis, E. (2023). Establishing and characterizing a Capsicum annuum MAGIC population: phenotypic and genomic insights. LXVI SIGA Annual Congress, “Climate-smart plants to feed the future”. (Bari, Italy: Poster Presentation).
Mata-Nicolás, E., Montero-Pau, J., Gimeno-Paez, E., García-Pérez, A., Ziarsolo, P., Blanca, J., et al. (2021). Discovery of a major QTL controlling trichome IV density in tomato using K-seq genotyping. Genes 12, 243. doi: 10.3390/genes12020243
Maxted, N., Kell, S., Ford-Lloyd, B., Dulloo, E., Toledo, Á. (2012). Toward the systematic conservation of global crop wild relative diversity. Crop Sci. 52, 774–785. doi: 10.2135/cropsci2011.08.0415
Mimura, Y., Inoue, T., Minamiyama, Y., Kubo, N. (2012). An SSR-based genetic map of pepper (Capsicum annuum L.) serves as an anchor for the alignment of major pepper maps. Breed Sci. 62, 93–98. doi: 10.1270/jsbbs.62.93
Miyatake, K., Shinmura, Y., Matsunaga, H., Fukuoka, H., Saito, T. (2019). Construction of a core collection of eggplant (Solanum melongena L.) based on genome-wide SNP and SSR genotypes. Breed Sci. 69, 498–502. doi: 10.1270/jsbbs.18202
Magon, G., De Rosa, V., Martina, M., Falchi, R., Acquadro, A., Barcaccia, G., et al. (2023). Boosting grapevine breeding for climate-smart viticulture: from genetic resources to predictive genomics. Front. Plant Sci. 14:1293186. doi: 10.3389/fpls.2023.1293186
Mongkolporn, O., Hanyong, S., Chunwongse, J., Wasee, S. (2015). Establishment of a core collection of chilli germplasm using microsatellite analysis. Plant Genet. Resour. 13, 104–110. doi: 10.1017/S1479262114000768
Mueller, L. A., Solow, T. H., Taylor, N., Skwarecki, B., Buels, R., Binns, J., et al. (2005). The SOL genomics network. A comparative resource for solanaceae biology and beyond. Plant Physiol. 138, 1310–1317. doi: 10.1104/pp.105.060707
Naegele, R. P., Boyle, S., Quesada-Ocampo, L. M., Hausbeck, M. K. (2014). Genetic diversity, population structure, and resistance to phytophthora capsici of a worldwide collection of eggplant germplasm. PloS One 9, e95930. doi: 10.1371/journal.pone.0095930
Nagel, M., Dulloo, M. E., Bissessur, P., Gavrilenko, T., Bamberg, J., Ellis, D., et al. (2022). Global Strategy for the Conservation of Potato (Bonn, Germany: Global Crop Diversity Trust). doi: 10.5447/ipk/2022/29
Nankar, A. N., Todorova, V., Tringovska, I., Pasev, G., Radeva-Ivanova, V., Ivanova, V., et al. (2020). A step towards Balkan Capsicum annuum L. core collection: Phenotypic and biochemical characterization of 180 accessions for agronomic, fruit quality, and virus resistance traits. PloS One 15, e0237741. doi: 10.1371/journal.pone.0237741
Neilson, J. A. D., Smith, A. M., Mesina, L., Vivian, R., Smienk, S., De Koyer, D. (2021). Potato tuber shape phenotyping using RGB imaging. Agronomy 11(9), 1781. doi: 10.3390/agronomy11091781
Nguyen, T. T., Le, N. T., Sim, S.-C. (2021). Genome-wide association study and marker development for bacterial wilt resistance in tomato (Solanum lycopersicum L.). Scientia Hortic. 289, 110418. doi: 10.1016/j.scienta.2021.110418
Nimmakayala, P., Abburi, V. L., Saminathan, T., Alaparthi, S. B., Almeida, A., Davenport, B., et al. (2016). Genome-wide diversity and association mapping for capsaicinoids and fruit weight in capsicum annuum L. Sci. Rep. 6, 38081. doi: 10.1038/srep38081
Odilbekov, F., Selga, C., Ortiz, R., Chawade, A., Liljeroth, E. (2020). QTL mapping for resistance to early blight in a tetraploid potato population. Agronomy 10, 728. doi: 10.3390/agronomy10050728
Oladosu, Y., Rafii, M. Y., Arolu, F., Chukwu, S. C., Salisu, M. A., Olaniyan, B. A., et al. (2021). Genetic diversity and utilization of cultivated eggplant germplasm in varietal improvement. Plants 10, 1714. doi: 10.3390/plants10081714
Ortiz, R., Crossa, J., Reslow, F., Perez-Rodriguez, P., Cuevas, J. (2022). Genome-based genotype × Environment prediction enhances potato (Solanum tuberosum L.) improvement using pseudo-diploid and polysomic tetraploid modeling. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.785196
Ou, L., Li, D., Lv, J., Chen, W., Zhang, Z., Li, X., et al. (2018). Pan-genome of cultivated pepper (Capsicum) and its use in gene presence–absence variation analyses. New Phytol. 220, 360–363. doi: 10.1111/nph.15413
Ovchinnikova, A., Krylova, E., Gavrilenko, T., Smekalova, T., Zhuk, M., Knapp, S., et al. (2011). Taxonomy of cultivated potatoes (Solanum section Petota: Solanaceae). Botanical J. Linn. Soc. 165, 107–155. doi: 10.1111/j.1095-8339.2010.01107.x
Padilla, Y. G., Gisbert-Mullor, R., Bueso, E., Zhang, L., Forment, J., Lucini, L., et al. (2023). New insights into short-term water stress tolerance through transcriptomic and metabolomic analyses on pepper roots. Plant Sci. 333, 111731. doi: 10.1016/j.plantsci.2023.111731
Pandey, J., Thompson, D., Joshi, M., Scheuring, D. C., Koym, J. W., Joshi, V., et al. (2023). Genetic architecture of tuber-bound free amino acids in potato and effect of growing environment on the amino acid content. Sci. Rep. 13, 13940. doi: 10.1038/s41598-023-40880-5
Panthee, D. R., Piotrowski, A., Ibrahem, R. (2017). Mapping quantitative trait loci (QTL) for resistance to late blight in tomato. Int. J. Mol. Sci. 18, 1589. doi: 10.3390/ijms18071589
Park, J., Lee, S., Choi, Y., Park, G., Park, S., Je, B., et al. (2022). Germplasm screening using DNA markers and genome-wide association study for the identification of powdery mildew resistance loci in tomato. Int. J. Mol. Sci. 23, 13610. doi: 10.3390/ijms232113610
Parry, C., Wang, Y.-W., Lin, S., Barchenger, D. W. (2021). Reproductive compatibility in Capsicum is not necessarily reflected in genetic or phenotypic similarity between species complexes. PloS One 16, e0243689. doi: 10.1371/journal.pone.0243689
Parveen, R., Kumar, M., Swapnil, n., Singh, D., Shahani, M., Imam, Z., et al. (2023). Understanding the genomic selection for crop improvement: current progress and future prospects. Mol. Genet. Genomics 298, 813–821. doi: 10.1007/s00438-023-02026-0
Pasala, R., Pandey, B. B. (2020). Plant phenomics: High-throughput technology for accelerating genomics. J. Biosci. 45, 111. doi: 10.1007/s12038-020-00083-w
Pascual, L., Desplat, N., Huang, B. E., Desgroux, A., Bruguier, L., Bouchet, J.-P., et al. (2015). Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol. J. 13, 565–577. doi: 10.1111/pbi.12282
Paz, M. M., Veilleux, R. E. (1997). Genetic diversity based on randomly amplified polymorphic DNA (RAPD) and its relationship with the performance of diploid potato hybrids. J. Am. Soc. Hortic. Sci. 122, 740–747. doi: 10.21273/JASHS.122.6.740
Pei, C., Wang, H., Zhang, J., Wang, Y., Francis, D. M., Yang, W. (2012). Fine mapping and analysis of a candidate gene in tomato accession PI128216 conferring hypersensitive resistance to bacterial spot race T3. Theor. Appl. Genet. 124, 533–542. doi: 10.1007/s00122-011-1726-1
Pereira, L., Sapkota, M., Alonge, M., Zheng, Y., Zhang, Y., Razifard, H., et al. (2021). Natural genetic diversity in tomato flavor genes. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.642828
Pereira-Dias, L., Fita, A., Vilanova, S., Sánchez-López, E., Rodríguez-Burruezo, A. (2020). Phenomics of elite heirlooms of peppers (Capsicum annuum L.) from the Spanish centre of diversity: Conventional and high-throughput digital tools towards varietal typification. Scientia Hortic. 265, 109245. doi: 10.1016/j.scienta.2020.109245
Pham, G. M., Hamilton, J. P., Wood, J. C., Burke, J. T., Zhao, H., Vaillancourt, B., et al. (2020). Construction of a chromosome-scale long-read reference genome assembly for potato. GigaScience 9, giaa100. doi: 10.1093/gigascience/giaa100
Poczai, P., D’Agostino, N., Deanna, R., Portis, E. (2022). Editorial: solanaceae VII: biology, genetics, and evolution. Front. Genet. 13. doi: 10.3389/fgene.2022.932421
Portis, E., Cericola, F., Barchi, L., Toppino, L., Acciarri, N., Pulcini, L., et al. (2015). Association mapping for fruit, plant and leaf morphology traits in eggplant. PloS One 10, e0135200. doi: 10.1371/journal.pone.0135200
Potnis, N., Branham, S. E., Jones, J. B., Wechter, W. P. (2019). Genome-wide association study of resistance to xanthomonas gardneri in the USDA pepper (Capsicum) collection. Phytopathology® 109, 1217–1225. doi: 10.1094/PHYTO-06-18-0211-R
Qi, S., Shen, Y., Wang, X., Zhang, S., Li, Y., Islam, M. M., et al. (2022). A new NLR gene for resistance to Tomato spotted wilt virus in tomato (Solanum lycopersicum). Theor. Appl. Genet. 135, 1493–1509. doi: 10.1007/s00122-022-04049-4
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111, 5135–5140. doi: 10.1073/pnas.1400975111
R2D2 Consortium, Fugeray-Scarbel, A., Bastien, C., Dupont-Nivet, M., Lemarié, S. (2021). Why and how to switch to genomic selection: lessons from plant and animal breeding experience. Front. Genet. 12. doi: 10.3389/fgene.2021.629737
Rajpal, V. R., Singh, A., Kathpalia, R., Thakur, R. K., Khan, M. K., Pandey, A., et al. (2023). The Prospects of gene introgression from crop wild relatives into cultivated lentil for climate change mitigation. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1127239
Razzaq, A., Kaur, P., Akhter, N., Wani, S. H., Saleem, F. (2021). Next-generation breeding strategies for climate-ready crops. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.620420
Ro, N., Haile, M., Hur, O., Geum, B., Rhee, J., Hwang, A., et al. (2022a). Genome-Wide Association Study of Resistance to Phytophthora capsici in the Pepper (Capsicum spp.) Collection. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.902464
Ro, N., Haile, M., Kim, B., Cho, G.-T., Lee, J., Lee, Y.-J., et al. (2022b). Genome-wide association study for agro-morphological traits in eggplant core collection. Plants 11, 2627. doi: 10.3390/plants11192627
Rockman, M. V., Kruglyak, L. (2008). Breeding designs for recombinant inbred advanced intercross lines. Genetics 179, 1069–1078. doi: 10.1534/genetics.107.083873
Roohanitaziani, R., de Maagd, R. A., Lammers, M., Molthoff, J., Meijer-Dekens, F., van Kaauwen, M. P. W., et al. (2020). Exploration of a resequenced tomato core collection for phenotypic and genotypic variation in plant growth and fruit quality traits. Genes 11, 1278. doi: 10.3390/genes11111278
Salgon, S., Jourda, C., Sauvage, C., Daunay, M.-C., Reynaud, B., Wicker, E., et al. (2017). Eggplant resistance to the ralstonia solanacearum species complex involves both broad-spectrum and strain-specific quantitative trait loci. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00828
Santa, J. D., Berdugo-Cely, J., Cely-Pardo, L., Soto-Suárez, M., Mosquera, T., M, C. H. G. (2018). QTL analysis reveals quantitative resistant loci for Phytophthora infestans and Tecia solanivora in tetraploid potato (Solanum tuberosum L.). PloS One 13, e0199716. doi: 10.1371/journal.pone.0199716
Sauvage, C., Segura, V., Bauchet, G., Stevens, R., Do, P. T., Nikoloski, Z., et al. (2014). Genome-wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol. 165, 1120–1132. doi: 10.1104/pp.114.241521
Schouten, H. J., Tikunov, Y., Verkerke, W., Finkers, R., Bovy, A., Bai, Y., et al. (2019). Breeding has increased the diversity of cultivated tomato in the Netherlands. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01606
Schumacher, C., Krannich, C. T., Maletzki, L., Köhl, K., Kopka, J., Sprenger, H., et al. (2021). Unravelling differences in candidate genes for drought tolerance in potato (Solanum tuberosum L.) by use of new functional microsatellite markers. Genes (Basel) 12, 494. doi: 10.3390/genes12040494
Scott, M. F., Ladejobi, O., Amer, S., Bentley, A. R., Biernaskie, J., Boden, S. A., et al. (2020). Multi-parent populations in crops: a toolbox integrating genomics and genetic mapping with breeding. Heredity 125, 396–416. doi: 10.1038/s41437-020-0336-6
Selga, C., Chrominski, P., Carlson-Nilsson, U., Andersson, M., Chawade, A., Ortiz, R. (2022). Diversity and population structure of Nordic potato cultivars and breeding clones. BMC Plant Biol. 22, 350. doi: 10.1186/s12870-022-03726-2
Sharma, S. K., Bolser, D., Boer, J., Sonderkaer, M., Amoros, W., Carboni, M. F., et al. (2013). Construction of reference chromosome-scale pseudomolecules for potato: Integrating the potato genome with genetic and physical maps. G3: Genes Genomes Genet. 3(11), 2031–2047. doi: 10.1534/g3.113.007153
Sharma, S. K., MacKenzie, K., McLean, K., Dale, F., Daniels, S., Bryan, G. J. (2018). Linkage disequilibrium and evaluation of genome-wide association mapping models in tetraploid potato. G3 (Bethesda) 8, 3185–3202. doi: 10.1534/g3.118.200377
Sherman, R. M., Salzberg, S. L. (2020). Pan-genomics in the human genome era. Nat. Rev. Genet. 21, 243–254. doi: 10.1038/s41576-020-0210-7
Shirasawa, K., Hosokawa, M., Yasui, Y., Toyoda, A., Isobe, S. (2023). Chromosome-scale genome assembly of a Japanese chili pepper landrace, Capsicum annuum ‘Takanotsume.’. DNA Res. 30, dsac052. doi: 10.1093/dnares/dsac052
Siddique, M. I., Lee, J.-H., Ahn, J.-H., Kusumawardhani, M. K., Safitri, R., Harpenas, A., et al. (2022). Genotyping-by-sequencing-based QTL mapping reveals novel loci for Pepper yellow leaf curl virus (PepYLCV) resistance in Capsicum annuum. PloS One 17, e0264026. doi: 10.1371/journal.pone.0264026
Sim, S.-C., Van Deynze, A., Stoffel, K., Douches, D. S., Zarka, D., Ganal, M. W., et al. (2012). High-density SNP genotyping of tomato (Solanum lycopersicum L.) reveals patterns of genetic variation due to breeding. PloS One 7, e45520. doi: 10.1371/journal.pone.0045520
Sood, S., Bhardwaj, V., Bairwa, A., Sharma, S., Sharma, A. K., Kumar, A., et al. (2023). Genome-wide association mapping and genomic prediction for late blight and potato cyst nematode resistance in potato (Solanum tuberosum L.). Front. Plant Sci. 14:1211472. doi: 10.3389/fpls.2023.1211472
Spooner, D. M., Ghislain, M., Simon, R., Jansky, S. H., Gavrilenko, T. (2014). Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot. Rev. 80, 283–383. doi: 10.1007/s12229-014-9146-y
Spooner, D. M., Hijmans, R. J. (2001). Potato systematics and germplasm collecting 1989–2000. Amer J. Potato Res. 78, 237–268. doi: 10.1007/BF02875691
Spooner, D. M., McLean, K., Ramsay, G., Waugh, R., Bryan, G. J. (2005). A single domestication for potato based on multilocus amplified fragment length polymorphism genotyping. Proc. Natl. Acad. Sci. 102, 14694–14699. doi: 10.1073/pnas.0507400102
Spooner, D. M., Núñez, J., Trujillo, G., del Rosario Herrera, M., Guzmán, F., Ghislain, M. (2007). Extensive simple sequence repeat genotyping of potato landraces supports a major reevaluation of their gene pool structure and classification. Proc. Natl. Acad. Sci. 104, 19398–19403. doi: 10.1073/pnas.0709796104
Su, X., Wang, B., Geng, X., Du, Y., Yang, Q., Liang, B., et al. (2021). A high-continuity and annotated tomato reference genome. BMC Genomics 22, 898. doi: 10.1186/s12864-021-08212-x
Suman, F. (2022). Climate change to hit Italian tomato production. Nat. Italy. doi: 10.1038/d43978-022-00079-0
Sun, H., Jiao, W.-B., Krause, K., Campoy, J. A., Goel, M., Folz-Donahue, K., et al. (2022). Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. Nat. Genet. 54, 342. doi: 10.1038/s41588-022-01015-0
Sun, M., Voorrips, R. E., van’t Westende, W., van Kaauwen, M., Visser, R. G. F., Vosman, B. (2020). Aphid resistance in Capsicum maps to a locus containing LRR-RLK gene analogues. Theor. Appl. Genet. 133, 227–237. doi: 10.1007/s00122-019-03453-7
Taher, D., Solberg, S.Ø., Prohens, J., Chou, Y., Rakha, M., Wu, T. (2017). World vegetable center eggplant collection: origin, composition, seed dissemination and utilization in breeding. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01484
Tang, X., Sun, F., Zhang, N., Rana, B. B., Kharel, R., Luo, P., et al. (2023). RNA-seq provides insights into potato deubiquitinase responses to drought stress in seedling stage. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1268448
Taranto, F., D’Agostino, N., Greco, B., Cardi, T., Tripodi, P. (2016). Genome-wide SNP discovery and population structure analysis in pepper (Capsicum annuum) using genotyping by sequencing. BMC Genomics 17, 943. doi: 10.1186/s12864-016-3297-7
Tassone, M. R., Bagnaresi, P., Desiderio, F., Bassolino, L., Barchi, L., Florio, F. E., et al. (2022). A genomic BSAseq approach for the characterization of QTLs underlying resistance to fusarium oxysporum in eggplant. Cells 11(16), 2548. doi: 10.3390/cells11162548
Tiwari, J. K., Rawat, S., Luthra, S. K., Zinta, R., Sahu, S., Varshney, S., et al. (2021). Genome sequence analysis provides insights on genomic variation and late blight resistance genes in potato somatic hybrid (parents and progeny). Mol. Biol. Rep. 48, 623–635. doi: 10.1007/s11033-020-06106-x
Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 485(7400):635–41. doi: 10.1038/nature11119
Toppino, L., Barchi, L., Rotino, G. L. (2022). “Next generation breeding for abiotic stress resistance in eggplant,” in Genomic Designing for Abiotic Stress Resistant Vegetable Crops. Ed. Kole, C. (Cham: Springer International Publishing), 115–151. doi: 10.1007/978-3-031-03964-5_4
Torrance, L., Cowan, G. H., McLean, K., MacFarlane, S., Al-Abedy, A. N., Armstrong, M., et al. (2020). Natural resistance to Potato virus Y in Solanum tuberosum Group Phureja. Theor. Appl. Genet. 133, 967–980. doi: 10.1007/s00122-019-03521-y
Tripodi, P., Rabanus-Wallace, M. T., Barchi, L., Kale, S., Esposito, S., Acquadro, A., et al. (2021). Global range expansion history of pepper (Capsicum spp.) revealed by over 10,000 genebank accessions. Proc. Natl. Acad. Sci. 118, e2104315118. doi: 10.1073/pnas.2104315118
Usadel, B. (2022). Solanaceae pangenomes are coming of graphical age to bring heritability back. aBIOTECH 3, 233. doi: 10.1007/s42994-022-00087-0
van der Knaap, E., Chakrabarti, M., Chu, Y. H., Clevenger, J. P., Illa-Berenguer, E., Huang, Z., et al. (2014). What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00227
van Haperen, P., Voorrips, R. E., van Kaauwen, M., van Eekelen, H. D. L. M., de Vos, R. C. H., van Loon, J. J. A., et al. (2021). Fine mapping of a thrips resistance QTL in Capsicum and the role of diterpene glycosides in the underlying mechanism. Theor. Appl. Genet. 134, 1557–1573. doi: 10.1007/s00122-021-03790-6
van Rengs, W. M. J., Schmidt, M. H.-W., Effgen, S., Le, D. B., Wang, Y., Zaidan, M. W. A. M., et al. (2022). A chromosome scale tomato genome built from complementary PacBio and Nanopore sequences alone reveals extensive linkage drag during breeding. Plant J. 110, 572–588. doi: 10.1111/tpj.15690
Van Soest, L. J. M., Hawkes, J. G., Hondelmann, W. (1983). Potato collecting expedition to Bolivia and the importance of Bolivian germplasm for plant breeding. Z. Pflanzenzücht 91, 154–168. Available at: https://pascal-francis.inist.fr/vibad/index.php?action=getRecordDetail&idt=9315567.
Vargas-Ortiz, E., Gonda, I., Smeda, J. R., Mutschler, M. A., Giovannoni, J. J., Jander, G. (2018). Genetic mapping identifies loci that influence tomato resistance against Colorado potato beetles. Sci. Rep. 8, 7429. doi: 10.1038/s41598-018-24998-5
Veilleux, R. E. (2017). “Genetic stocks used for potato genome sequencing,” in The Potato Genome Compendium of Plant Genomes. Eds. Kumar Chakrabarti, S., Xie, C., Kumar Tiwari, J. (Cham: Springer International Publishing), 73–79. doi: 10.1007/978-3-319-66135-3_4
Villalobos-López, M. A., Arroyo-Becerra, A., Quintero-Jiménez, A., Iturriaga, G. (2022). Biotechnological advances to improve abiotic stress tolerance in crops. Int. J. Mol. Sci. 23, 12053. doi: 10.3390/ijms231912053
Víquez-Zamora, M., Vosman, B., van de Geest, H., Bovy, A., Visser, R. G., Finkers, R., et al. (2013). Tomato breeding in the genomics era: insights from a SNP array. BMC Genomics 14, 354. doi: 10.1186/1471-2164-14-354
Vos, P. G., Paulo, M. J., Bourke, P. M., Maliepaard, C. A., van Eeuwijk, F. A., Visser, R. G. F., et al. (2022). GWAS in tetraploid potato: identification and validation of SNP markers associated with glycoalkaloid content. Mol. Breed 42, 76. doi: 10.1007/s11032-022-01344-2
Wang, F., Zou, M., Zhao, L., Xia, Z., Wang, J. (2021). Genome-wide association mapping of late blight tolerance trait in potato (Solanum tuberosum L.). Front. Genet. 12. doi: 10.3389/fgene.2021.714575
Wei, Q., Wang, W., Hu, T., Hu, H., Wang, J., Bao, C. (2020b). Construction of a SNP-based genetic map using SLAF-seq and QTL analysis of morphological traits in eggplant. Front. Genet. 11. doi: 10.3389/fgene.2020.00178
Wei, Q., Wang, J., Wang, W., Hu, T., Hu, H., Bao, C. (2020a). A high-quality chromosome-level genome assembly reveals genetics for important traits in eggplant. Hortic. Res. 7, 1–15. doi: 10.1038/s41438-020-00391-0
Weide, R., van Wordragen, M. F., Lankhorst, R. K., Verkerk, R., Hanhart, C., Liharska, T., et al. (1993). Integration of the classical and molecular linkage maps of tomato chromosome 6. Genetics 135, 1175–1186. doi: 10.1093/genetics/135.4.1175
Wen, J., Jiang, F., Weng, Y., Sun, M., Shi, X., Zhou, Y., et al. (2019). Identification of heat-tolerance QTLs and high-temperature stress-responsive genes through conventional QTL mapping, QTL-seq and RNA-seq in tomato. BMC Plant Biol. 19, 398. doi: 10.1186/s12870-019-2008-3
Wilson, S., Zheng, C., Maliepaard, C., Mulder, H. A., Visser, R. G. F., van der Burgt, A., et al. (2021). Understanding the effectiveness of genomic prediction in tetraploid potato. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.672417
Wu, L., Wang, H., Liu, S., Liu, M., Liu, J., Wang, Y., et al. (2022). Mapping of CaPP2C35 involved in the formation of light-green immature pepper (Capsicum annuum L.) fruits via GWAS and BSA. Theor. Appl. Genet. 135, 591–604. doi: 10.1007/s00122-021-03987-9
Wu, L., Wang, P., Wang, Y., Cheng, Q., Lu, Q., Liu, J., et al. (2019). Genome-wide correlation of 36 agronomic traits in the 287 pepper (Capsicum) accessions obtained from the SLAF-seq-based GWAS. Int. J. Mol. Sci. 20, 5675. doi: 10.3390/ijms20225675
Xiong, W., Reynolds, M., Xu, Y. (2022). Climate change challenges plant breeding. Curr. Opin. Plant Biol. 70, 102308. doi: 10.1016/j.pbi.2022.102308
The Potato Genome Sequencing Consortium. (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi: 10.1038/nature10158
Yang, C. J., Edmondson, R. N., Piepho, H.-P., Powell, W., Mackay, I. (2021). Crafting for a better MAGIC: systematic design and test for Multiparental Advanced Generation Inter-Cross population. G3 Genes|Genomes|Genetics 11, jkab295. doi: 10.1093/g3journal/jkab295
Yang, X., Yang, Y., Ling, J., Guan, J., Guo, X., Dong, D., et al. (2020). A high-throughput BAC end analysis protocol (BAC-anchor) for profiling genome assembly and physical mapping. Plant Biotechnol. J. 18, 364. doi: 10.1111/pbi.13203
Yi, S., Lee, D.-G., Back, S., Hong, J.-P., Jang, S., Han, K., et al. (2022). Genetic mapping revealed that the Pun2 gene in Capsicum chacoense encodes a putative aminotransferase. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1039393
Yu, J., Holland, J. B., McMullen, M. D., Buckler, E. S. (2008). Genetic design and statistical power of nested association mapping in maize. Genetics 178, 539–551. doi: 10.1534/genetics.107.074245
Yu, X., Qu, M., Shi, Y., Hao, C., Guo, S., Fei, Z., et al. (2022). Chromosome-scale genome assemblies of wild tomato relatives Solanum habrochaites and Solanum galapagense reveal structural variants associated with stress tolerance and terpene biosynthesis. Horticulture Res. 9, uhac139. doi: 10.1093/hr/uhac139
Zamir, D., Ekstein-Michelson, I., Zakay, Y., Navot, N., Zeidan, M., Sarfatti, M., et al. (1994). Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, TY-1. Theor. Appl. Genet. 88, 141–146. doi: 10.1007/BF00225889
Zhang, J., Liang, L., Xie, Y., Zhao, Z., Su, L., Tang, Y., et al. (2022b). Transcriptome and metabolome analyses reveal molecular responses of two pepper (Capsicum annuum L.) cultivars to cold stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.819630
Zhang, F., Qu, L., Gu, Y., Xu, Z.-H., Xue, H.-W. (2022a). Resequencing and genome-wide association studies of autotetraploid potato. Mol. Horticulture 2, 6. doi: 10.1186/s43897-022-00027-y
Zhang, C., Wang, P., Tang, D., Yang, Z., Lu, F., Qi, J., et al. (2019). The genetic basis of inbreeding depression in potato. Nat. Genet. 51, 374–378. doi: 10.1038/s41588-018-0319-1
Zhang, X., Zhang, Z., Gu, X., Mao, S., Li, X., Chadœuf, J., et al. (2016). Genetic diversity of pepper (Capsicum spp.) germplasm resources in China reflects selection for cultivar types and spatial distribution. J. Integr. Agric. 15, 1991–2001. doi: 10.1016/S2095-3119(16)61364-3
Zhao, Y., Liu, Y., Zhang, Z., Cao, Y., Yu, H., Ma, W., et al. (2020). Fine mapping of the major anthracnose resistance QTL AnRGO5 in Capsicum chinense ‘PBC932.’. BMC Plant Biol. 20, 189. doi: 10.1186/s12870-019-2115-1
Zhao, C., Zhang, Y., Du, J., Guo, X., Wen, W., Gu, S., et al. (2019). Crop phenomics: current status and perspectives. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00714
Zhao, L., Zou, M., Deng, K., Xia, C., Jiang, S., Zhang, C., et al. (2023). Insights into the genetic determination of tuber shape and eye depth in potato natural population based on autotetraploid potato genome. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1080666
Zhou, Q., Tang, D., Huang, W., Yang, Z., Zhang, Y., Hamilton, J. P., et al. (2020). Haplotype-resolved genome analyses of a heterozygous diploid potato. Nat. Genet. 52, 1018–1023. doi: 10.1038/s41588-020-0699-x
Zhou, R., Yu, X., Zhao, T., Ottosen, C.-O., Rosenqvist, E., Wu, Z. (2019). Physiological analysis and transcriptome sequencing reveal the effects of combined cold and drought on tomato leaf. BMC Plant Biol. 19, 377. doi: 10.1186/s12870-019-1982-9
Zhou, Y., Zhang, Z., Bao, Z., Li, H., Lyu, Y., Zan, Y., et al. (2022). Graph pangenome captures missing heritability and empowers tomato breeding. Nature 606, 527–534. doi: 10.1038/s41586-022-04808-9
Zhu, Z., Sun, B., Wei, J., Cai, W., Huang, Z., Chen, C., et al. (2019). Construction of a high density genetic map of an interspecific cross of Capsicum chinense and Capsicum annuum and QTL analysis of floral traits. Sci. Rep. 9, 1054. doi: 10.1038/s41598-018-38370-0
Keywords: biotic and abiotic resistance, eggplant, genetic resources, potato, genomic resources, pepper, resilient traits, tomato
Citation: Martina M, De Rosa V, Magon G, Acquadro A, Barchi L, Barcaccia G, De Paoli E, Vannozzi A and Portis E (2024) Revitalizing agriculture: next-generation genotyping and -omics technologies enabling molecular prediction of resilient traits in the Solanaceae family. Front. Plant Sci. 15:1278760. doi: 10.3389/fpls.2024.1278760
Received: 17 August 2023; Accepted: 22 January 2024;
Published: 05 February 2024.
Edited by:
Byoung-Cheorl Kang, Seoul National University, Republic of KoreaReviewed by:
Lei Gao, Chinese Academy of Sciences (CAS), ChinaYongfu Tao, Chinese Academy of Agricultural Sciences, China
Uday Chand Jha, Indian Institute of Pulses Research (ICAR), India
Copyright © 2024 Martina, De Rosa, Magon, Acquadro, Barchi, Barcaccia, De Paoli, Vannozzi and Portis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ezio Portis, ZXppby5wb3J0aXNAdW5pdG8uaXQ=; Alessandro Vannozzi, YWxlc3NhbmRyby52YW5ub3p6aUB1bmlwZC5pdA==
†These authors have contributed equally to this work
 Matteo Martina
Matteo Martina Valeria De Rosa
Valeria De Rosa Gabriele Magon
Gabriele Magon Alberto Acquadro
Alberto Acquadro Lorenzo Barchi
Lorenzo Barchi Gianni Barcaccia
Gianni Barcaccia Emanuele De Paoli
Emanuele De Paoli Alessandro Vannozzi
Alessandro Vannozzi Ezio Portis
Ezio Portis