- 1CAS Key Laboratory of Environmental and Applied Microbiology, Environmental Microbiology Key Laboratory of Sichuan Province, Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Anhui Province Key Laboratory of Horticultural Crop Quality Biology, School of Horticulture, Anhui Agricultural University, Hefei, China
- 4Pitzer College, Claremont, CA, United States
- 5Faculty of Mathematical and Physical Sciences, University College London, London, United Kingdom
- 6School of Breeding and Multiplication (Sanya Institute of Breeding and Multiplication), Hainan University, Sanya, China
Promoters are one of the most critical elements in regulating gene expression. They are considered essential biotechnological tools for heterologous protein production. The one most widely used in plants is the 35S promoter from cauliflower mosaic virus. However, our study for the first time discovered the 35S promoter reduced the expression of exogenous proteins under increased antibiotic stress. We discovered an endogenous strong promoter from duckweed named LpSUT2 that keeps higher initiation activity under antibiotic stress. Stable transformation in duckweed showed that the gene expression of eGFP in the LpSUT2:eGFP was 1.76 times that of the 35S:eGFP at 100 mg.L-1 G418 and 6.18 times at 500 mg.L-1 G418. Notably, with the increase of G418 concentration, the gene expression and the fluorescence signal of eGFP in the 35S:eGFP were weakened, while the LpSUT2:eGFP only changed slightly. This is because, under high antibiotic stress, the 35S promoter was methylated, leading to the gene silencing of the eGFP gene. Meanwhile, the LpSUT2 promoter was not methylated and maintained high activity. This is a previously unknown mechanism that provides us with new insights into screening more stable promoters that are less affected by environmental stress. These outcomes suggest that the LpSUT2 promoter has a high capacity to initiate the expression of exogenous proteins. In conclusion, our study provides a promoter tool with potential application for plant genetic engineering and also provides new insights into screening promoters.
1 Introduction
Promoters are one of the most critical regulatory elements controlling gene expression in terms of time and space. It is an invaluable and indispensable tool for investigating specific gene functions, crop yield improvement, plant stress resistance, heterologous protein production, etc (Nuccio, 2018). Promoters are widely used in synthetic biology and plant biotechnology. Many plants have been widely used as expression systems for the commercial production of heterologous proteins for their lower production cost and easier to scale-up (Sharma and Sharma, 2009; Tiwari et al., 2009).
So far, the earliest and most frequently used promoter in plant bioreactors is the 35S promoter from cauliflower mosaic virus (Nuccio, 2018; Tian et al., 2022). Examples of such applications include the envelope protein of Japanese encephalitis virus expressed in rice (Wang et al., 2009), CTB-InsB3 expressed in tobacco (Tiwari et al., 2009), human β-amyloid expressed in potato (Kim et al., 2003), etc. In addition, the 35S promoter was widely used to drive the over-expression of key genes to improve crop yield. For example, the 35S promoter driven over-expression of plasma membrane genes, such as H+ATPase, MFAP1, NAC23, FTO, and H+-pyrophosphatase IbVP1, increased rice, potato, and sweet potato yield, respectively (Fan et al., 2021; Yu et al., 2021; Zhang et al., 2021; Y. Li et al., 2022; Z. Li et al., 2022). The 35S promoter also plays an important role in the over-expression of the plant stress resistance genes. Subsequently, VvCEB1opt over-expression increases salinity tolerance in Arabidopsis (Lim et al., 2020) and IbPSS1 over-expression enhances salt tolerance in transgenic sweet potatoes at the whole-plant level (Yu et al., 2020). Nevertheless, limitations of the 35S promoter have been discovered. These include disturbing the expression of adjacent genes and their low activity in monocots (Peremarti et al., 2010; Nuccio, 2018; Somssich, 2019). Thus, researchers tended to focus on finding endogenous strong promoters in plants and discovered rice actin 1 (Actin1) and maize ubiquitin1 (Ubi1) promoters (Park et al., 2010; Nuccio, 2018). However, these promoters are only widely used in monocots (Wu et al., 2019), and their activity level decreases with the development of plants (Nuccio, 2018). Furthermore, experiments using the 35S promoter and maize Ubi1 promoter suggest that is difficult to improve their activity (Nuccio, 2018). Therefore, it is necessary and urgent to find a new endogenous strong promoter to express heterologous proteins, increase plant stress resistance, and improve crop yield, that applies to both monocots and dicots plants.
Duckweed, the smallest monocotyledonous group of flowering aquatic plants spreads worldwide, comprises 5 genera (Spirodela, Landoltia, Lemna, Wolffia, and Wolffiella) and 36 species (Bog et al., 2019). It grows fast, does not compete for land with crops, and has a high protein content (Liu et al., 2015; Tian et al., 2021). Thus, duckweed has been widely used as a bioreactor or biosynthetic chassis to produce recombinant proteins (Lam and Michael, 2022). Previous studies used duckweed as an expression system to produce protective antigen (Ko et al., 2011), M130-β-glucuronidase (Firsov et al., 2015), hemagglutinin (Bertran et al., 2015), recombinant human granulocyte colony-stimulating factor (Khvatkov et al., 2018), etc. Nevertheless, all the above studies expressed heterologous proteins with the 35S promoter. Using endogenous promoters for expressing recombinant proteins reduces potential risks associated with exogenous promoters and has demonstrated favorable outcomes. For instance, the utilization of the rice endosperm-specific Gt13a promoter drove the expression of human serum albumin and recombinant human basic fibroblast growth factor in rice seeds, showcasing the capacity of this system for large-scale production of functional human recombinant proteins (He et al., 2011; An et al., 2013). However, limited research has been dedicated to utilizing the endogenous strong promoter of duckweed for expressing heterologous or recombinant proteins. For these reasons, we aimed to screen endogenous strong promoters of duckweed for application in transgenic technology to augment the expression of exogenous proteins and explore potential molecular mechanisms. Duckweed exhibits a simple leaf–stem structure known as a frond, yet it demonstrates rapid growth, high starch accumulation capacity, and exceptional photosynthetic efficiency (Chen et al., 2022; Guo et al., 2023). In various plant species, promoters such as the sucrose transporter 2 promoter, Rubisco promoter, ubiquitin promoter, and others have demonstrated favorable outcomes in transgenic technology (Christensen and Quail, 1996; Stadler and Sauer, 2019; Boruah et al., 2023). Taking these factors into account, we considered the ADP-glucose pyrophosphorylase small subunit 1 (AGPS1) promoter, sucrose transporter 2 promoter, Rubisco promoter, and ubiquitin promoter in duckweed as candidate promoters for screening endogenous strong promoters in duckweed.
In this work, we found the duckweed sucrose transporter 2 promoter of L. punctata (LpSUT2) is an endogenous strong promoter, and expresses the exogenous proteins higher than the 35S promoter in duckweed, a monocot. It also has the same high activity as the 35S promoter in Nicotiana tabacum, a dicot. Increasing antibiotic stress can reduce the escape in transgenic events (Shin et al., 2007; Itaya et al., 2018). We discovered the LpSUT2 promoter behaved very steadily even under high antibiotic stress, while the 35S promoter did not. Together, our research offers a promoter tool with stable and effective function and application value for plant biotechnology.
2 Materials and methods
2.1 Plant materials
Landoltia punctata 0202 line and Lemna minor ZH0403 callus were stored in the duckweed resource bank at the Chengdu Institute of Biology, Chinese Academy of Science (Li et al., 2021; Tian et al., 2021). Genomic DNA was extracted from the Landoltia punctata 0202 line cultured in Hoagland medium (15 g.L-1 sucrose, pH 5.00 ± 0.05). The callus was cultured in Murashige and Skoog (MS) medium with 0.22 mg.L-1 2,4-Dichlorophenoxyacetic acid, 0.45 mg.L-1 6-Benzylaminopurine, 30 g.L-1 sucrose, and 3.5 g.L-1 gellan gum and then used for genetic transformation. These mediums were placed in a system of photon flux density cycle of 100-120/0 µmol-2s-1 (16 h light/8 h dark) and a temperature cycle of 25°C/15°C (16 h day/8 h night).
2.2 Construction of vectors
The 2000 bp promoter DNA sequences upstream from specific genes, sucrose transporter 2 gene promoter named LpSUT2 (Landoltia_punctata_GLEAN_10005267), AGPS1 gene promoter named LpA6804 (Landoltia_punctata_GLEAN_10016804), Rubisco1 gene promoter named LpR1090 (Landoltia_punctata_GLEAN_10021090), Rubisco2 gene promoter named LpR1091 (Landoltia_punctata_GLEAN_10021091), ubiquitin-like protein gene promoter named LpU4817 (Landoltia_punctata_GLEAN_10004817), and ubiquitin-1 promoter named LpU9400 (Landoltia_punctata_GLEAN_10019400) were extracted from the genome of L. punctata by TBtools software (Chen et al., 2020). The cetyltrimethylammonium bromide (CTAB) method was used to extract DNA from L. punctata (Allen et al., 2006). Next, DNA sequences of these promoters were cloned using high PCR fidelity KOD Plus enzymes (TOYOBO, Japan) and recombinant primers (Supplementary Table 1). These amplified products and the linearized vector (the binary vector pCambia2301:35S:eGFP were digested by the restriction enzymes PvuII and PstI) were connected by ClonExpress® MultiS One Step Cloning Kit (Vazyme, Nanjing, China) and then sequenced.
2.3 Western blot analysis
We referenced previous studies for the protein extraction methods of transgenic duckweed and non-transgenic duckweed (Khvatkov et al., 2018). According to the previously established method (Mahmood and Yang, 2012; Khvatkov et al., 2018), we detected the 35S: ChIL-2:His protein by the Western blot at 100 mg.L-1 and 500 mg.L-1 geneticin (G418) from the 35S: ChIL-2:His transgenic duckweed (data were not published). Anti-HIS mouse monoclonal antibody (diluted 1:2,000; Sangon Biotech, China) and Anti-HRP rabbit polyclonal antibody (diluted 1:10,000; Sangon Biotech, China) were used in this work. The blots were visualized using ECL Western blotting detection reagents (Cytiva, America).
2.4 Transient expression in dicot Nicotiana tabacum leaves
Promoter activity was often studied directly using a reporter gene (eGFP or GUS) in stable or transient transformation assays (Nuccio, 2018). As a reporter gene, β-glucuronidase (GUS) is suitable for macroscopic observation of tissue structure in promoter-initiated gene expression. However, it is not suitable for real-time observation of gene expression due to its requirement for steps such as hydrolysis reaction and destaining. On the other hand, enhanced green fluorescent protein (eGFP), as a reporter gene, enables precise mapping at the living cell level, making it suitable for observing the more subtle real-time positional structure of promoter-initiated gene expression. Therefore, we chose eGFP as the reporter gene to provide a more detailed insight into the gene expression initiated by the candidate promoters. To investigate the candidate promoters activity in dicot, we used 35S, LpSUT2, LpR1091, LpA6804, LpU9400, LpR1090, and LpU4817 promoters to respectively drive the reporter gene eGFP, and transiently expressed each in Nicotiana tabacum leaves and left for 2 days. This method was carried out as described in previous studies (Yao et al., 2015). Then we used a laser scanning confocal microscope to observe the fluorescence signal of eGFP.
2.5 Stable expression in the monocot duckweed
The vectors of the 35S:eGFP and the LpSUT2:eGFP were introduced into duckweed’s callus following Agrobacterium tumefaciens strain GV3101-mediated transformation. Then, follow the previously established method to screen and regenerate these calluses (Tan et al., 2022). Briefly, put these callus in MS medium (screening 6 weeks) and 1/2 Schenk and Hildebrandt (SH) medium (regeneration 4-5 weeks). Specially, the two-stage culture medium (MS for screening and 1/2 SH for regeneration) contained G418 concentrations of 50, 100, 200, 300, 400, and 500 mg.L-1, respectively.
2.6 Isolation of RNA and qRT−PCR analysis
Total RNAs were extracted using the Eastep Super Total RNA Extraction Kit (Promega, Shanghai, China) according to the manufacturer’s instructions. Next, we examined their concentration and quality by NanoDrop 2000 (Thermo, USA). All RNA samples were treated with RNase-free DNase to eliminate DNA contamination. The cDNA was synthesized from 1 μg of total RNAs following the manufacturer’s protocol of the GoScript™ Reverse Transcription System (Promega, Shanghai, China). These samples were harvested and immediately frozen in liquid nitrogen, subsequently stored in a -80°C freezer until being used. The L. minor 18S gene was used as an internal control. Primer sequences were listed in Supplementary Table 1. The qRT-PCR was operated by CFX Maestro Real-Time PCR (Bio-Rad, USA), using the SsoAdvanced™ Universal SYBR Green Supermix (Bio-Rad, USA). The PCR reaction system, procedures, and data analysis followed the SsoAdvanced™ Universal SYBR Green Supermix protocol.
2.7 Methylation analysis
We treated the 35S:eGFP transgenic duckweed at 500 mg.L-1 G418 with 45.64 mg.L-1 zebularine (a methylation inhibitor) for 8 days. Then, the transgenic duckweed from the other treatment groups was collected together. Transgenic duckweed genomic DNA was extracted using the CTAB method. The methylation status of promoters was determined by methylation-specific PCR. Briefly, 2 μg of genomic DNA was bisulfite-treated with the Zymo DNA Modification Kit (Zymo Research, Orange County, CA, USA). Bisulfite-treated DNA was amplified using primers specific for either methylated or unmethylated DNA (Baik et al., 2021). The sequences of the first set of methylated-specific (M) primers and unmethylated-specific (U) primers for the promoters are listed in Supplementary Table 1.
2.8 Fluorescence signal detection
In this work, the fluorescence signal of eGFP protein in all transgenic plants was observed using a laser scanning confocal microscope. To maintain the consistency of the parameters in the obtained fluorescence images, all parameters were set as follows: equipment, Leica TCS SP8; excitation and emission wavelengths, Ex488 nm/Em505-530 nm; software, LAS AF; values of intensity, 30%; the master gain, 800 V; digital gain, 1; pinhole, 1 AU.
2.9 Data analysis
We used GraphPad Prism (Version 8.0) and SPSS 26 software for data analysis.
3 Results
3.1 Screening of the endogenous strong promoter of duckweed
To find a promoter with wide application potential in plants, we selected six duckweed endogenous promoters in contrast to the widely utilized 35S promoter from cauliflower mosaic virus. In parallel, we used the 35S promoter and replaced the 35S promoter with the candidate promoters to drive the eGFP gene into the binary vector pCambia2301 (Figure 1A). To test whether the candidate promoters from monocots were suitable in dicots, we transiently expressed these plasmids were transiently expressed in dicots Nicotiana tabacum leaves for 2 days with Agrobacterium tumefaciens-mediated infiltration (Batistič et al., 2012). The images were acquired using consistent acquisition parameters. Under the same validation conditions, we observed that only the eGFP fluorescence signal initiated by the LpSUT2 promoter exhibited comparable intensity to that of the 35S promoter, indicating similar strong initiation abilities. The weaker eGFP fluorescence signals were initiated by the other endogenous promoters (LpR1091, LpA6804, LpU9400, LpR1090, and LpU4817 promoter) (Figure 1B). It is possible that they have a limited initiation ability or tissue-specific expression; further studies are necessary to distinguish these possibilities. Based on this result, only the LpSUT2 promoter fulfilled our objective of screening endogenous promoters with strong initiation ability and practicality. Subsequently, we concentrated on comparing the initiation activity and stability of the LpSUT2 promoter and the 35S promoter in duckweed.
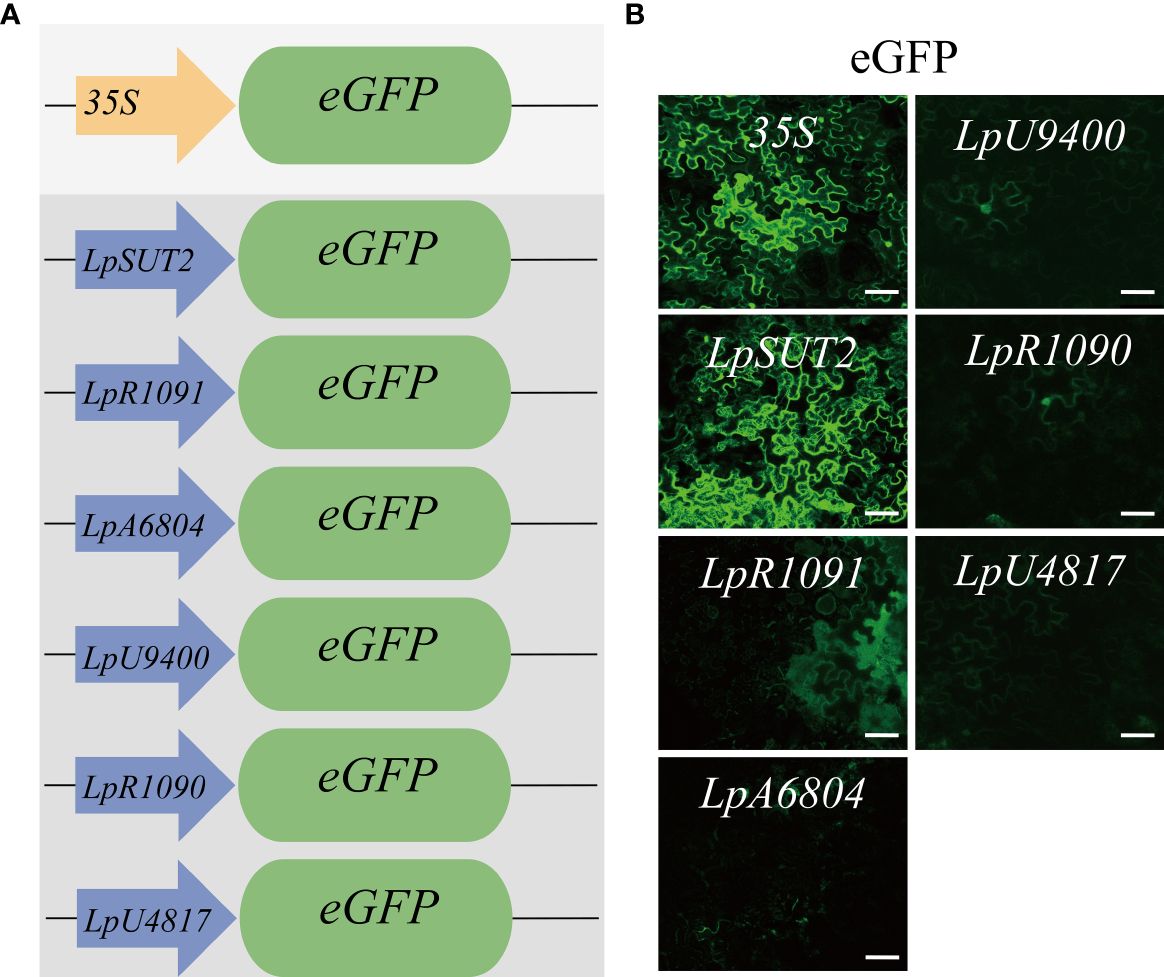
Figure 1 Initiating activity of the promoters in dicotyledonous Nicotiana tabacum leaves. (A) Schematic representation of the vector structures. The expression of the eGFP gene was driven by the 35S promoter and duckweed endogenous promoters (LpSUT2, LpR1091, LpA6804, LpU9400, LpR1090, and LpU4817 promoter). (B) The binary vectors pCambia2301:the 35S:eGFP, pCambia2301:LpSUT2:eGFP, pCambia2301: LpR1091:eGFP, pCambia2301: LpA6804:eGFP, pCambia2301: LpU9400:eGFP, pCambia2301: LpR1090:eGFP, and pCambia2301: LpU4817:eGFP were transiently expressed in Nicotiana tabacum leaves for 2 days. The green signals indicate eGFP. Scale bars: 50 μm.
3.2 Stable expression in duckweed, a monocot
Recent studies showed that duckweed has been a widely used bioreactor to express the alpha-interferon, M2e peptide of avian influenza virus H5N1 (Cox et al., 2006; Firsov et al., 2015), etc. Meanwhile, the 35S promoter has been proven to drive the expression of the target proteins in duckweed. However, the 35S promoter also raises concerns about its biosafety since it comes from a virus (Ho et al., 1999), and the low activity in the cell to recognize the sequence as foreign inactivates it (Elmayan and Vaucheret, 1996). Therefore, we expect that finding a plant endogenous promoter will become a more effective tool to produce recombinant proteins. To evaluate the activity of the LpSUT2 promoter in monocot duckweed, we stably transformed the 35S:eGFP (positive control) and the LpSUT2:eGFP vectors into duckweed by Agrobacterium tumefaciens mediated infiltration. Geneticin G418 is an aminoglycoside antibiotic that inhibits protein synthesis by binding to the 30S subunit of the ribosome of a cell. G418 is commonly employed in transgenic plant screening. In our stable transformations in duckweed, G418 was also employed as an antibiotic selection marker. We followed the G418 resistance marker carried by the pCambia2301. Consequently, plants were selected by the addition of G418 to the medium. We collected the images under the same acquisition parameters. Notably, the LpSUT2 promoter was highly active in transgenic duckweed fronds and stronger than the 35S promoter by 100 mg.L-1 G418 screening (Figure 2).
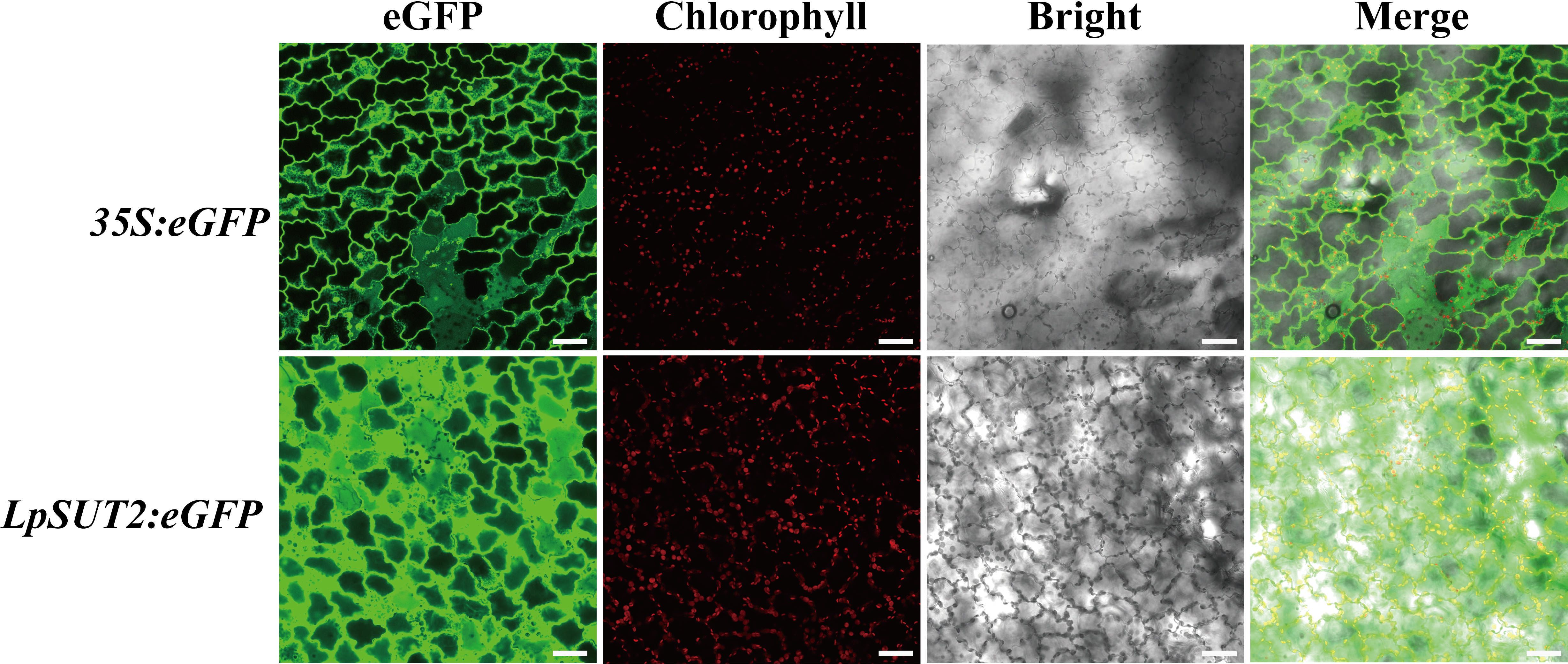
Figure 2 Comparison of the LpSUT2 and the 35S promoter activities in monocotyledonous duckweed fronds. The 35S:eGFP and the LpSUT2:eGFP in transgenic leaves of duckweed. The green signals indicate eGFP and the red signals indicate chlorophyll. Scale bars: 50 μm.
3.3 The LpSUT2 promoter drives high-level eGFP expression
To improve the efficiency of screening transgenic positive plants and test the stability of the promoter after increasing antibiotic stress, we obtained multiple stable transgenic lines using six concentrations of G418 (50, 100, 200, 300, 400, and 500 mg.L-1, respectively) during the screening and regeneration process (Figure 3A). Next, we used quantitative real-time PCR (qRT-PCR) on the stable transgenic lines from each of the six concentrations to detect the ability of promoters to initiate eGFP expression. The results showed that the expression level of eGFP driven by the 35S promoter decreased with increasing G418 concentration, while the expression level of eGFP driven by the LpSUT2 promoter did not significantly change and was all higher than that driven by the 35S promoter under each condition. Indeed, the expression level of eGFP in the LpSUT2:eGFP transgenic duckweed was 1.76 times that of the 35S:eGFP transgenic duckweed at 100 mg.L-1 G418, and 6.18 times at 500 mg.L-1 G418 (Figure 3B). Then, we observed the fluorescence signal of the eGFP protein using the laser scanning confocal microscope. The images were collected under the same parameters. Similar to the above qRT-PCR results, the fluorescence signal of the LpSUT2:eGFP transgenic lines had no significant change. However, the fluorescence intensity of the 35S:eGFP transgenic lines became weaker as the concentration of G418 increased. More importantly, the LpSUT2:eGFP transgenic lines fluorescence intensity was consistently stronger than that of the 35S:eGFP transgenic lines in each G418 concentration (Figure 3C). Therefore, these data demonstrate that the activity of the LpSUT2 promoter is higher than that of the 35S promoter in the monocotyledonous duckweed.
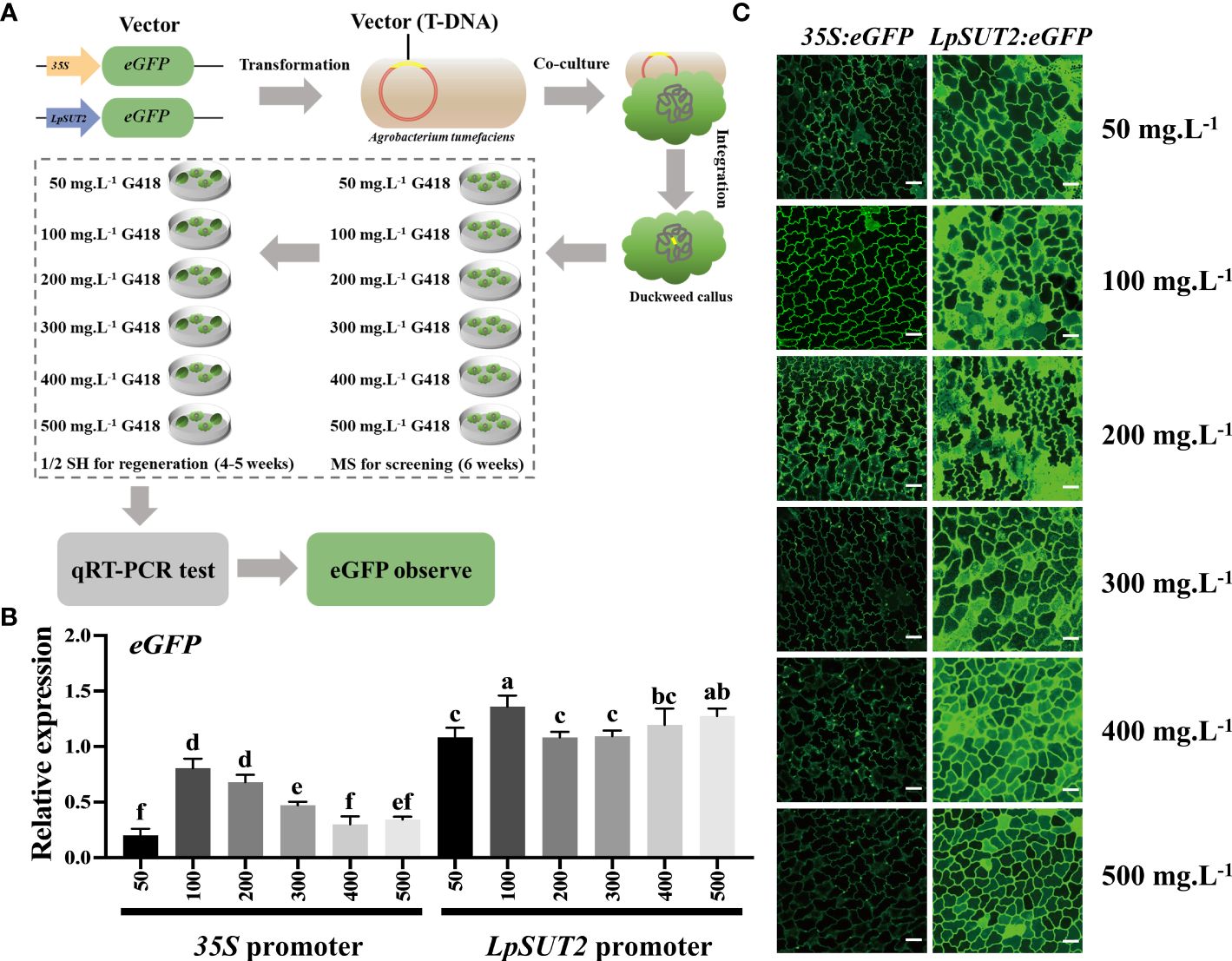
Figure 3 The ability of the LpSUT2 and the 35S promoters to initiate eGFP gene expression under different antibiotic stresses. (A) The recombinant plasmids the LpSUT2:eGFP and 35S:eGFP were transformed into duckweed callus. Screened (MS medium) and regenerated (1/2 SH medium) the transgenic duckweed in these culture medium at 50, 100, 200, 300, 400, and 500 mg.L-1 G418, respectively. The gene expression level and the fluorescence intensity of eGFP were detected by qRT-PCR and laser scanning confocal microscope, respectively. The eGFP expression levels (B) and the fluorescence signals of the eGFP (C) in the 35S:eGFP and the LpSUT2:eGFP transgenic lines grown in medium containing 50, 100, 200, 300, 400, and 500 mg.L-1 G418 were detected by qRT-PCR and laser scanning confocal microscope. Scale bars: 50 μm. For B, the 18S gene was used as an internal control. Data are the means ± SD of n=3. The letters indicate a significant difference between groups (P < 0.05 by one-way ANOVA followed by Tukey’s test).
Based on the above interesting phenomenon, we hypothesized that the ability of the 35S promoter to initiate the expression of foreign proteins would decrease after increasing the antibiotic stress. To test the hypothesis, we used seven transgenic duckweeds with the 35S promoter driving chicken interleukin 2 (ChIL-2): His fusion protein in the pCambia2301 in the laboratory and cultured them at 100 or 500 mg.L-1 concentrations of G418 (with none other treatments). Interestingly, similar to the 35S:eGFP results, we found that none of the ChIL-2:His fusion proteins in the transgenic duckweed were detectable when the G418 concentration changed from 100 mg.L-1 to 500 mg.L-1 (Supplementary Figure 1). Hence, these results indicate that the 35S promoter initiation capacity is unstable under antibiotic stress, which is not conducive to increasing antibiotic stress to rapidly screen positive transgenic plants. In contrast to the 35S promoter, the LpSUT2 promoter exhibits a very stable initiation ability (Figure 3C).
3.4 Methylation analysis in the 35S promoter and the LpSUT2 promoter
Based on the previous experimental results, the expression of the 35S:eGFP decreased with increasing concentration of G418, while the LpSUT2:eGFP remained stable. However, all transgenic duckweed lines exhibit phenotypic similarity to the wild type. There was also not much difference between 100 and 500 mg.L-1 G418 concentrations in the 35S:eGFP or LpSUT2:eGFP transgenic duckweed phenotypes. Duckweed did not exhibit abnormal growth phenotypes, such as regional albino or death (Supplementary Figure 2).
In Arabidopsis, the 35S promoter was methylated, leading to the suppression of the overexpressed exogenous proteins (Lang et al., 2015). We predicted that the 35S promoter methylation would cause the down-regulation of eGFP expression. To test this prediction, we utilized methylation-specific PCR to detect methylation in the 35S promoter region in 35S:eGFP transgenic duckweed lines grown in 100 mg.L-1 and 500 mg.L-1 G418, as well as in the LpSUT2 promoter region in LpSUT2:eGFP transgenic duckweed lines grown in 100 mg.L-1 and 500 mg.L-1 G418. The results showed that methylation of the 35S promoter region in the 35S:eGFP transgenic duckweed line grown in 500 mg.L-1 G418, and no methylation was detected in other transgenic duckweed lines (Figure 4A).
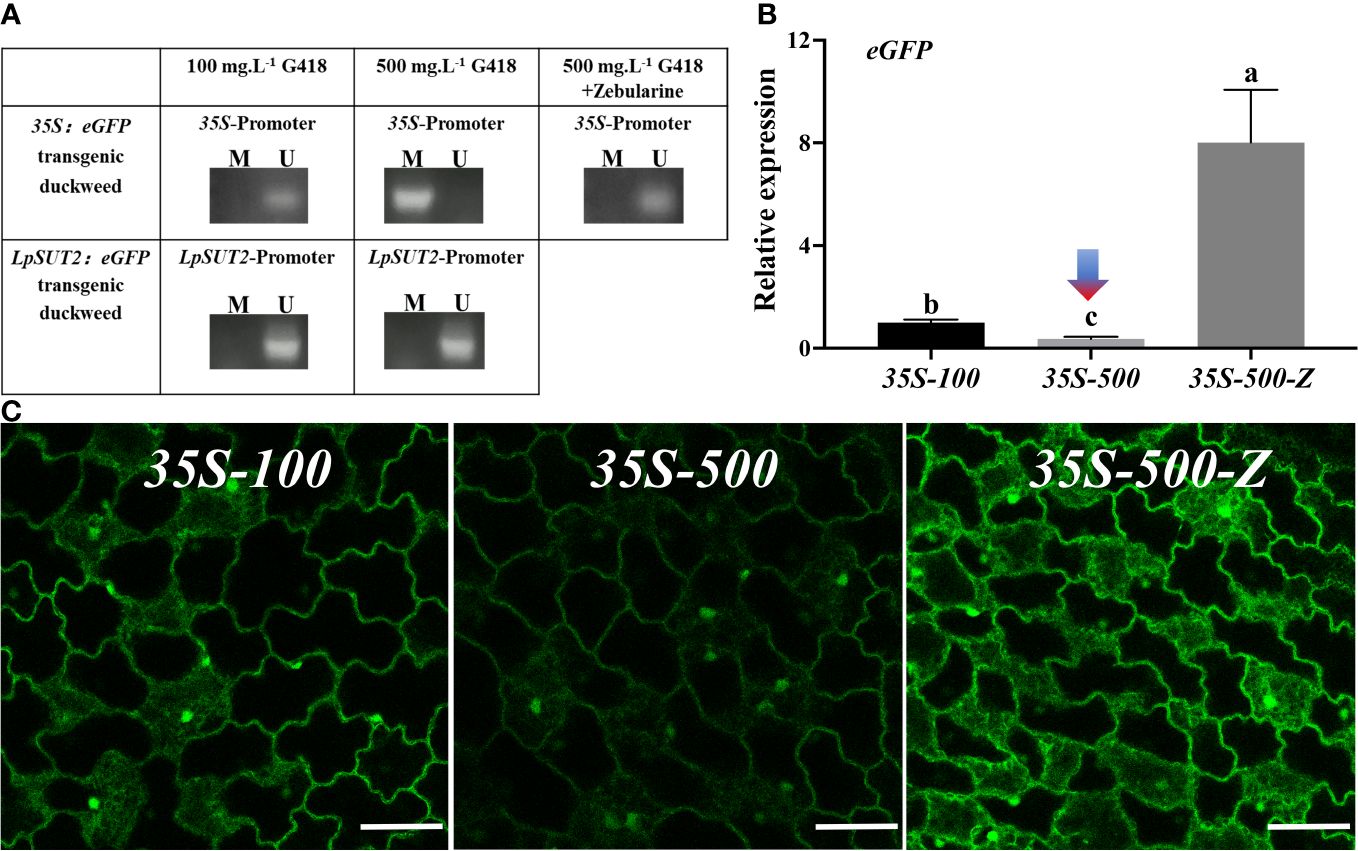
Figure 4 Methylation analysis of promoters. (A) Detection of DNA methylation in 35S and LpSUT2 promoter regions by methylation-specific PCR. M/U represents the amplified by methylation-specific primers or unmethylated-specific primers. The expression of the eGFP gene (B) and the fluorescence signal of the eGFP (C) in the 35S:eGFP transgenic duckweed under different conditions. Scale bars: 50 μm. For (B), the 18S gene was used as an internal control. Data are the means± SD of n=3. The letters indicate a significant difference between groups (P < 0.05 by one-way ANOVA followed by Tukey’s test). For (B, C), 35S-100 represents 35S:eGFP transgenic duckweed grown in 100 mg.L-1 G418. 35S-500 represents 35S:eGFP transgenic duckweed grown in 500 mg.L-1 G418. 35S-500-Z represents 35S:eGFP transgenic duckweed grown in 500 mg.L-1 G418 with 45.64 mg.L-1 zebularine.
Zebularine is a potent DNA methyltransferase inhibitor known to reduce DNA methylation in plants (Baubec et al., 2009). In our study, we supplemented 45.64 mg.L-1 Zebularine to inhibit methylation in the 35S:eGFP transgenic duckweed line continuously grown in 500 mg.L-1 G418. After adding the methylation inhibitor, we found a reversion of the 35S promoter region to a non-methylated state (Figure 4A).
Additionally, to assess the impact of DNA methylation on the initiation ability of the 35S promoter, we quantified the expression levels of eGFP in the following groups: 35S-100, representing 35S:eGFP transgenic duckweed line grown in 100 mg.L-1 G418; 35S-500, representing 35S:eGFP transgenic duckweed line grown in 500 mg.L-1 G418; 35S-500-Z, representing 35S:eGFP transgenic duckweed line grown in 500 mg.L-1 G418 with 45.64 mg.L-1 zebularine. Compared with the 35S-100, the expression of the eGFP gene was down-regulated by 2.73 times in the 35S-500 and up-regulated by 8.01 times in the 35S-500-Z (Figure 4B). Then, we also observed the eGFP fluorescence signal in these groups. All images were acquired under the same parameters. Consistent with the above qRT-PCR results, 35S-500 shows a weaker fluorescence signal, and 35S-500-Z shows a stronger fluorescence signal compared to 35S-100 (Figure 4C). These data suggest that the methylation of the 35S promoter under high levels of antibiotic stress is a significant factor contributing to the reduction in the expression of foreign proteins. This implied that the 35S promoter was methylated and reduced activity, but the LpSUT2 promoter was not (Figure 4A). Hence, the LpSUT2 promoter is more advantageous for expressing foreign proteins in plants.
4 Discussion
Transgenic technology can rapidly improve crop yield, seed nutrient content, industrial protein production, therapeutic production, disease resistance, and abiotic stress tolerance (Somssich, 2019). Particularly, a suitable and strong promoter is decisive in initiating the expression of exogenous genes in the successful application of transgenic technology. The 35S promoter is the most frequently used to drive transgene expression in plants (Somssich, 2019). Alternatively, the 35S promoter also inhibits the over-expression of foreign proteins (Lang et al., 2015). Accordingly, we screened several endogenous promoters in duckweed. The results showed that the LpSUT2 promoter had strong initiation ability and practicality, like the 35S promoter (Figure 1). Increasing antibiotic stress is an effective way to improve the screening efficiency of transgenic plants. However, when using the 35S promoter to initiate exogenous protein expression in our experiments, we found that increasing antibiotic stress reduced exogenous protein expression (Figure 3C). This impedes rapidly screening positive transgenic plants. On the contrary, we verified that the LpSUT2 promoter did not reduce when increasing antibiotic stress (Figure 3). In our work, we developed an endogenous promoter LpSUT2 to facilitate the rapid availability of positive plants.
Moreover, further experiments have been conducted to address the above interesting issue. We found that methylation was responsible for the different activities of the 35S promoter and the LpSUT2 promoter. The 35S promoter was methylated after increased antibiotic stress, thereby reducing the activity of gene expression. In contrast, the LpSUT2 promoter was not methylated and remained active under antibiotic stress (Figure 5). Exogenous promoters such as 35S, often experience the silencing effects of methylation in applications (Elmayan and Vaucheret, 1996; Shimada et al., 2017). This is consistent with the decreasing activity of the 35S promoter in increasing antibiotic stress. However, the LpSUT2 promoter exhibits stably high activity under antibiotic stress. We also found that the LpSUT2 promoter has more cis-acting elements that function like enhancers compared to the 35S promoter, meaning it has higher activity than the 35S promoter (Supplementary Table 2). In addition, compared to commonly used strong promoters (Actin1, Ubi1, and SAG12), the LpSUT2 promoter also has a certain advantage in terms of the number of cis-acting elements. For these reasons, the LpSUT2 promoter could be an alternative strong constitutive promoter and a very useful tool for transgenic plants.
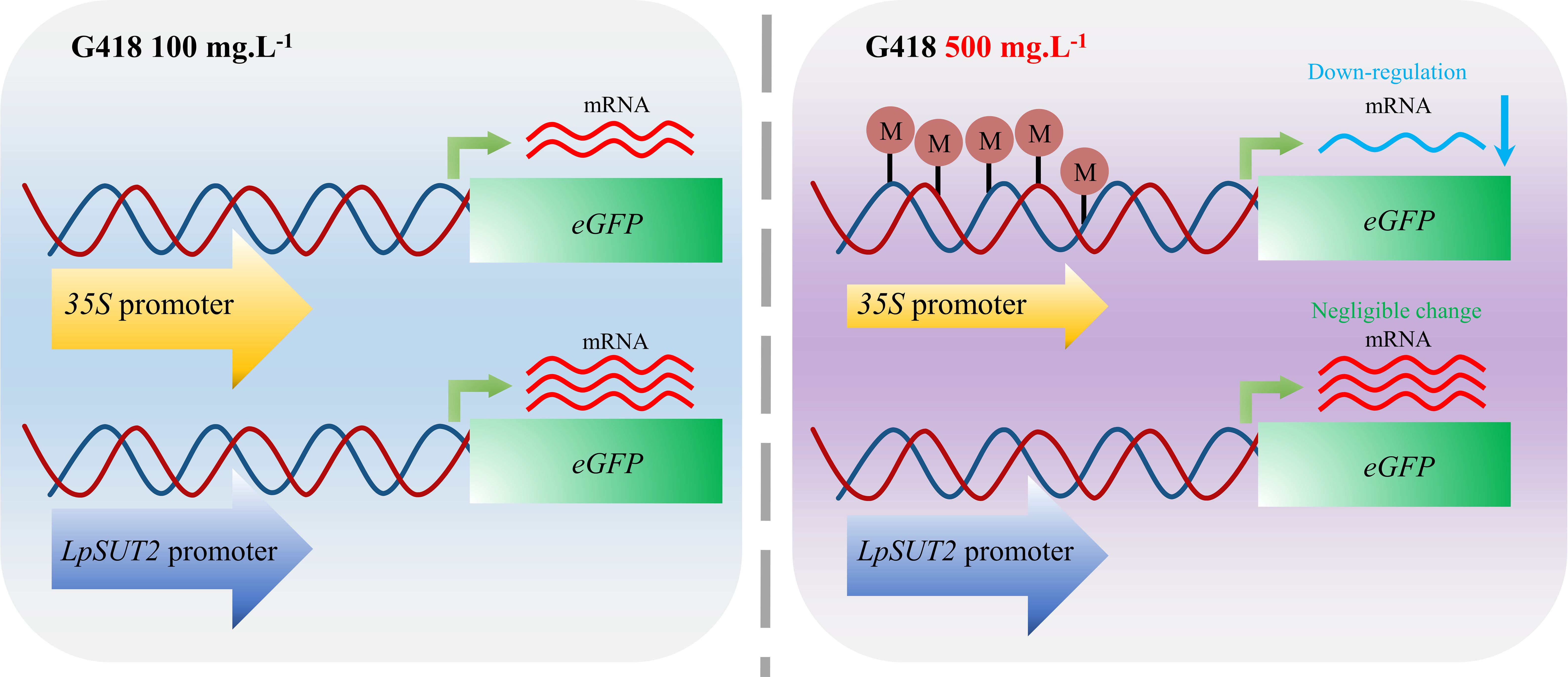
Figure 5 Schematic graphs of the 35S promoter and the LpSUT2 promoter regulating transcription under antibiotic stress. The 35S promoter is methylated and thus down-regulates gene expression when the concentration of G418 changes from 100 mg.L-1 to 500 mg.L-1. In contrast, the LpSUT2 promoter is not methylated and maintains high initiation activity under antibiotic stress. M, methylation.
Additionally, there may be clear distinctions between dicot and monocot in terms of the expression pattern and transcriptional activity of promoters. The 35S promoter is a frequently utilized constitutive promoter conferring the strong expression of transgenes in dicots, while in monocots the maize ubiquitin promoter typically drives gene expression more efficiently. However, the Ubi1 promoter is inactive in some tissues in transgenic plants, such as rice mature tissues (Cornejo et al., 1993). There are also a few promoters that displayed high promoter activity in both monocots and dicots (Jiang et al., 2018; Kummari et al., 2020), but they are not enough for the rapidly developing plant genetic engineering. In our study, we observed that the LpSUT2 promoter from duckweed demonstrated a robust initiation ability comparable to that of the 35S promoter during transient transformation in Nicotiana tabacum (dicot) (Figure 1). Furthermore, in the stable transformation of duckweed (monocot), the gene expression level initiated by the LpSUT2 promoter surpassed that of the 35S promoter (Figure 3). This may compensate for the limitation of transgenic promoters that are not applicable in both monocots and dicots. Together, few promoters are highly active in both monocots and dicots at present. In our work, we screened an endogenous strong promoter in duckweed and explored its underlying molecular mechanism by which it maintains high activity. The LpSUT2 promoter has a higher potential for application in monocots and dicots compared with the 35S promoter. Nevertheless, further experiments are required to prove whether the LpSUT2 promoter can consistently maintain high activity in more monocots and dicots.
5 Conclusions
In summary, our research has discovered a novel endogenous strong promoter in duckweed and uncovered its underlying molecular mechanism. Notably, the LpSUT2 promoter is not methylated and maintains superior activity levels compared to the commonly used 35S promoter under antibiotic stress in monocot duckweed. Specifically, in duckweed, the expression of eGFP driven by the LpSUT2 promoter was 1.76 times higher than that driven by the 35S promoter at 100 mg.L-1 G418, and 6.18 times higher at 500 mg.L-1 G418. The LpSUT2 promoter also exhibits a strong initiation ability similar to that of the 35S promoter in transient transformation in dicot Nicotiana tabacum. This work increases the diversity of promoters and facilitates the development of plant biotechnology.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
CW: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. ZH: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation. SW: Writing – review & editing, Writing – original draft, Validation, Methodology, Formal analysis, Conceptualization. XT: Writing – review & editing, Writing – original draft, Methodology, Formal analysis. YJ: Writing – review & editing, Writing – original draft, Supervision, Project administration. ZY: Writing – review & editing, Writing – original draft, Supervision, Project administration, Funding acquisition. KH: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Conceptualization. LZ: Writing – review & editing, Writing – original draft. ZC: Writing – review & editing, Writing – original draft. YF: Writing – review & editing, Writing – original draft, Supervision, Project administration, Conceptualization. SC: Writing – review & editing, Writing – original draft, Project administration, Funding acquisition, Conceptualization. PL: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. HZ: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Investigation, Data curation, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Innovation Academy for Seed Design, CAS; National Aquatic Biological Resource Center (NABRC); Biological Resources Programme, Chinese Academy of Sciences (KFJ-BRP-008) and CAS “Light of West China” Program (2020XBZG_XBQNXZ_A_001).
Acknowledgments
The authors thank the editor and the reviewers for their efforts to improve the manuscript. This study has been stored on the preprint server Research Square, and the article is linked to https://doi.org/10.21203/rs.3.rs-3703691/v2.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1368284/full#supplementary-material
References
Allen, G. C., Flores Vergara, M. A., Krasynanski, S., Kumar, S., Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
An, N., Ou, J., Jiang, D., Zhang, L., Liu, J., Fu, K., et al. (2013). Expression of a functional recombinant human basic fibroblast growth factor from transgenic rice seeds. Int. J. Mol. Sci. 14, 3556–3567. doi: 10.3390/ijms14023556
Baik, J. Y., Liu, Z., Jiao, D., Kwon, H.-J., Yan, J., Kadigamuwa, C., et al. (2021). ZBP1 not RIPK1 mediates tumor necroptosis in breast cancer. Nat. Commun. 12, 2666. doi: 10.1038/s41467-021-23004-3
Batistič, O., Rehers, M., Akerman, A., Schlücking, K., Steinhorst, L., Yalovsky, S., et al. (2012). S-acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res. 22, 1155–1168. doi: 10.1038/cr.2012.71
Baubec, T., Pecinka, A., Rozhon, W., Mittelsten Scheid, O. (2009). Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J. 57, 542–554. doi: 10.1111/j.1365-313X.2008.03699.x
Bertran, K., Thomas, C., Guo, X., Bublot, M., Pritchard, N., Regan, J. T., et al. (2015). Expression of H5 hemagglutinin vaccine antigen in common duckweed (Lemna minor) protects against H5N1 high pathogenicity avian influenza virus challenge in immunized chickens. Vaccine 33, 3456–3462. doi: 10.1016/j.vaccine.2015.05.076
Bog, M., Appenroth, K. J., Sree, K. S. (2019). Duckweed (Lemnaceae): its molecular taxonomy. Front. Sustain. Food Syst. 3. doi: 10.3389/fsufs.2019.00117
Boruah, R. R., Konwar, T., Nath, P. K., Acharjee, S., Sarmah, B. K. (2023). Activity of Arabidopsis Rubisco small subunit promoter in various tissues of chickpea. 3 Biotech. 13, 89. doi: 10.1007/s13205-023-03508-z
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G., Zhao, K., Li, W., Yan, B., Yu, Y., Li, J., et al. (2022). A review on bioenergy production from duckweed. Biomass Bioenergy 161, 106468. doi: 10.1016/j.biombioe.2022.106468
Christensen, A. H., Quail, P. H. (1996). Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218. doi: 10.1007/BF01969712
Cornejo, M. J., Luth, D., Blankenship, K. M., Anderson, O. D., Blechl, A. E. (1993). Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol. Biol. 23, 567–581. doi: 10.1007/BF00019304
Cox, K., Sterling, J., Regan, J., Gasdaska, J. R., Frantz, K. K., Peele, C. G., et al. (2006). Glycan optimization of a human monoclonal antibody in the aquatic 398 plant Lemna minor. Nat Biotechnol 24, 1591–1597. doi: 10.1038/nbt1260
Elmayan, T., Vaucheret, H. (1996). Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 9, 787–797. doi: 10.1046/j.1365-313X.1996.9060787.x
Fan, W., Zhang, Y., Wu, Y., Zhou, W., Yang, J., Yuan, L., et al. (2021). The H+-pyrophosphatase IbVP1 regulates carbon flux to influence the starch metabolism and yield of sweet potato. Hortic. Res. 8, 1–12. doi: 10.1038/s41438-020-00454-2
Firsov, A., Tarasenko, I., Mitiouchkina, T., Ismailova, N., Shaloiko, L., Vainstein, A., et al. (2015). High-yield expression of M2e peptide of avian influenza virus H5N1 in transgenic duckweed plants. Mol. Biotechnol. 57, 653–661. doi: 10.1007/s12033-015-9855-4
Guo, L., Fang, Y., Jin, Y., He, K., Zhao, H. (2023). High starch duckweed biomass production and its highly-efficient conversion to bioethanol. Environ. Technol. Innovation 32, 103296. doi: 10.1016/j.eti.2023.103296
He, Y., Ning, T., Xie, T., Qiu, Q., Zhang, L., Sun, Y., et al. (2011). Large-scale production of functional human serum albumin from transgenic rice seeds. Proc. Natl. Acad. Sci. 108, 19078–19083. doi: 10.1073/pnas.1109736108
Ho, M. W., Ryan, A., Cummins, J. (1999). Cauliflower mosaic viral promoter - a recipe for disaster? Microbial Ecol. Health Dis. 11, 194–197. doi: 10.1080/08910609908540827
Itaya, A., Zheng, S., Simmonds, D. (2018). Establishment of neomycin phosphotransferase II (nptII) selection for transformation of soybean somatic embryogenic cultures. In Vitro Cell.Dev.Biol.-Plant 54, 184–194. doi: 10.1007/s11627-017-9875-9
Jiang, P., Zhang, K., Ding, Z., He, Q., Li, W., Zhu, S., et al. (2018). Characterization of a strong and constitutive promoter from the Arabidopsis serine carboxypeptidase-like gene AtSCPL30 as a potential tool for crop transgenic breeding. BMC Biotechnol. 18, 59. doi: 10.1186/s12896-018-0470-x
Khvatkov, P., Firsov, A., Shvedova, A., Shaloiko, L., Kozlov, O., Chernobrovkina, M., et al. (2018). Development of Wolffia arrhiza as a producer for recombinant human granulocyte colony-stimulating factor. Front. Chem. 6. doi: 10.3389/fchem.2018.00304
Kim, H. S., Euym, J. W., Kim, M. S., Lee, B. C., Mook-Jung, I., Jeon, J. H., et al. (2003). Expression of human β-amyloid peptide in transgenic potato. Plant Sci. 165, 1445–1451. doi: 10.1016/j.plantsci.2003.07.007
Ko, S. M., Sun, H. J., Oh, M. J., Song, I. J., Kim, M. J., Sin, H. S., et al. (2011). Expression of the protective antigen for PEDV in transgenic duckweed, Lemna minor. Hortic. Environ. Biotechnol. 52, 511–515. doi: 10.1007/s13580-011-0007-x
Kummari, D., Palakolanu, S. R., Kishor, P. B. K., Bhatnagar-Mathur, P., Singam, P., Vadez, V., et al. (2020). An update and perspectives on the use of promoters in plant genetic engineering. J. Biosci. 45, 119. doi: 10.1007/s12038-020-00087-6
Lam, E., Michael, T. P. (2022). Wolffia, a minimalist plant and synthetic biology chassis. Trends Plant Sci. 27, 430–439. doi: 10.1016/j.tplants.2021.11.014
Lang, Z., Lei, M., Wang, X., Tang, K., Miki, D., Zhang, H., et al. (2015). The methyl-CpG-binding protein MBD7 facilitates active DNA demethylation to limit DNA hyper-methylation and transcriptional gene silencing. Mol. Cell 57, 971–983. doi: 10.1016/j.molcel.2015.01.009
Li, J., Du, A., Liu, P., Tian, X., Jin, Y., Yi, Z., et al. (2021). High starch accumulation mechanism and phosphorus utilization efficiency of duckweed (Landoltia punctata) under phosphate starvatio. Ind. Crops Products. 167, 113529. doi: 10.1016/j.indcrop.2021.113529
Li, Y., Li, T. T., He, X. R., Zhu, Y., Feng, Q., Yang, X. M., et al. (2022). Blocking Osa-miR1871 enhances rice resistance against Magnaporthe oryzae and yield. Plant Biotechnol. J. 20, 646–659. doi: 10.1111/pbi.13743
Li, Z., Wei, X., Tong, X., Zhao, J., Liu, X., Wang, H., et al. (2022). The OsNAC23-Tre6P-SnRK1a feed-forward loop regulates sugar homeostasis and grain yield in rice. Mol. Plant 15, 706–722. doi: 10.1016/j.molp.2022.01.016
Lim, S. D., Mayer, J. A., Yim, W. C., Cushman, J. C. (2020). Plant tissue succulence engineering improves water-use efficiency, water-deficit stress attenuation and salinity tolerance in Arabidopsis. Plant J. 103, 1049–1072. doi: 10.1111/tpj.14783
Liu, Y., Fang, Y., Huang, M., Jin, Y., Sun, J., Tao, X., et al. (2015). Uniconazole-induced starch accumulation in the bioenergy crop duckweed (Landoltia punctata) II: transcriptome alterations of pathways involved in carbohydrate metabolism and endogenous hormone crosstalk. Biotechnol. Biofuels 8, 64. doi: 10.1186/s13068-015-0245-8
Mahmood, T., Yang, P. C. (2012). Western blot: technique, theory, and trouble shooting. N Am. J. Med. Sci. 4, 429–434. doi: 10.4103/1947-2714.100998
Nuccio, M. L. (2018). “A brief history of promoter development for use in transgenic maize applications,” in Maize: Methods and Protocols Methods in Molecular Biology. Ed. Lagrimini, L. M. (Springer, New York, NY), 61–93. doi: 10.1007/978-1-4939-7315-6_4
Park, S. H., Yi, N., Kim, Y. S., Jeong, M. H., Bang, S. W., Choi, Y. D., et al. (2010). Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J. Exp. Bot. 61, 2459–2467. doi: 10.1093/jxb/erq076
Peremarti, A., Twyman, R. M., Gómez Galera, S., Naqvi, S., Farré, G., Sabalza, M., et al. (2010). Promoter diversity in multigene transformation. Plant Mol. Biol. 73, 363–378. doi: 10.1007/s11103-010-9628-1
Sharma, A. K., Sharma, M. K. (2009). Plants as bioreactors: Recent developments and emerging opportunities. Biotechnol. Adv. 27, 811–832. doi: 10.1016/j.bioteChadv.2009.06.004
Shimada, A., Okumura, A., Yamasaki, S., Iwata, Y., Koizumi, N., Nishihara, M., et al. (2017). A 64-bp sequence containing the GAAGA motif is essential for CaMV-35S promoter methylation in gentian. Biochim. Biophys. Acta (BBA) - Gene Regul. Mech. 1860, 861–869. doi: 10.1016/j.bbagrm.2017.06.001
Shin, Y. M., Choe, G., Shin, B., Yi, G., Yun, P-Y., Yang, K., et al. (2007). Selectionof nptII transgenic sweetpotato plants using G418 and 468 paromomycin. J. Plant Biol. 50, 206–212. doi: 10.1007/BF03030631
Somssich, M. (2019). A short history of the CaMV 35S promoter. PeerJ Preprints. doi: 10.7287/peerj.preprints.27096v3
Stadler, R., Sauer, N. (2019). “The atSUC2 Promoter: A Powerful Tool to Study Phloem Physiology and Development,” in Phloem: Methods and Protocols Methods in Molecular Biology. Ed. Liesche, J. (Springer, New York, NY), 267–287. doi: 10.1007/978-1-4939-9562-2_22
Tan, X., Chen, S., Fang, Y., Liu, P., Hu, Z., Jin, Y., et al. (2022). Rapid and highly efficient genetic transformation and application of interleukin-17B expressed in duckweed as mucosal vaccine adjuvant. Biomolecules 12, 1881. doi: 10.3390/biom12121881
Tian, C., Zhang, Y., Li, J., Wang, Y. (2022). Benchmarking intrinsic promoters and terminators for plant synthetic biology research. BioDesign Res. 2022, 9834989. doi: 10.34133/2022/9834989
Tian, X., Fang, Y., Jin, Y., Yi, Z., Li, J., Du, A., et al. (2021). Ammonium detoxification mechanism of ammonium-tolerant duckweed (Landoltia punctata) revealed by carbon and nitrogen metabolism under ammonium stress. Environ. Pollut. 277, 116834. doi: 10.1016/j.envpol.2021.116834
Tiwari, S., Verma, P. C., Singh, P. K., Tuli, R. (2009). Plants as bioreactors for the production of vaccine antigens. Biotechnol. Adv. 27, 449–467. doi: 10.1016/j.bioteChadv.2009.03.006
Wang, Y., Deng, H., Zhang, X., Xiao, H., Jiang, Y., Song, Y., et al. (2009). Generation and immunogenicity of Japanese encephalitis virus envelope protein expressed in transgenic rice. Biochem. Biophys. Res. Commun. 380, 292–297. doi: 10.1016/j.bbrc.2009.01.061
Wu, J., Lawit, S. J., Weers, B., Sun, J., Mongar, N., Van Hemert, J., et al. (2019). Overexpression of zmm28 increases maize grain yield in the field. Proc. Natl. Acad. Sci. U. S. A. 116, 23850–23858. doi: 10.1073/pnas.1902593116
Yao, J., Weng, Y., Dickey, A., Wang, K. Y. (2015). Plants as factories for human pharmaceuticals: applications and challenges. Int. J. Mol. Sci. 16, 28549–28565. doi: 10.3390/ijms161226122
Yu, Q., Liu, S., Yu, L., Xiao, Y., Zhang, S., Wang, X., et al. (2021). RNA demethylation increases the yield and biomass of rice and potato plants in field trials. Nat. Biotechnol. 39, 1581–1588. doi: 10.1038/s41587-021-00982-9
Yu, Y., Xuan, Y., Bian, X., Zhang, L., Pan, Z., Kou, M., et al. (2020). Overexpression of phosphatidylserine synthase IbPSS1 affords cellular Na+ homeostasis and salt tolerance by activating plasma membrane Na+/H+ antiport activity in sweet potato roots. Hortic. Res. 7, 1–12. doi: 10.1038/s41438-020-00358-1
Keywords: endogenous strong promoter, 35S promoter, LpSUT2 promoter, duckweed, antibiotic stress
Citation: Wei C, Hu Z, Wang S, Tan X, Jin Y, Yi Z, He K, Zhao L, Chu Z, Fang Y, Chen S, Liu P and Zhao H (2024) An endogenous promoter LpSUT2 discovered in duckweed: a promising transgenic tool for plants. Front. Plant Sci. 15:1368284. doi: 10.3389/fpls.2024.1368284
Received: 10 January 2024; Accepted: 08 March 2024;
Published: 03 April 2024.
Edited by:
Cheng Yuan, Yunnan Academy of Tobacco Agricultural Sciences, ChinaReviewed by:
Zhengyi Wei, Guangxi Academy of Agricultural Sciences, ChinaJia-Gang Wang, Shanxi Agricultural University, China
Copyright © 2024 Wei, Hu, Wang, Tan, Jin, Yi, He, Zhao, Chu, Fang, Chen, Liu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Penghui Liu, cGVuZ2h1aS5saXVAaGFpbmFudS5lZHUuY24=; Hai Zhao, emhhb2hhaUBjaWIuYWMuY24=
†These authors have contributed equally to this work and share first authorship
 Cuicui Wei
Cuicui Wei Zhubin Hu
Zhubin Hu Songhu Wang3
Songhu Wang3 Xiao Tan
Xiao Tan Yanling Jin
Yanling Jin Zhuolin Yi
Zhuolin Yi Kaize He
Kaize He Ziyue Chu
Ziyue Chu Yang Fang
Yang Fang Penghui Liu
Penghui Liu Hai Zhao
Hai Zhao